hexsha
stringlengths 40
40
| size
int64 6
14.9M
| ext
stringclasses 1
value | lang
stringclasses 1
value | max_stars_repo_path
stringlengths 6
260
| max_stars_repo_name
stringlengths 6
119
| max_stars_repo_head_hexsha
stringlengths 40
41
| max_stars_repo_licenses
list | max_stars_count
int64 1
191k
⌀ | max_stars_repo_stars_event_min_datetime
stringlengths 24
24
⌀ | max_stars_repo_stars_event_max_datetime
stringlengths 24
24
⌀ | max_issues_repo_path
stringlengths 6
260
| max_issues_repo_name
stringlengths 6
119
| max_issues_repo_head_hexsha
stringlengths 40
41
| max_issues_repo_licenses
list | max_issues_count
int64 1
67k
⌀ | max_issues_repo_issues_event_min_datetime
stringlengths 24
24
⌀ | max_issues_repo_issues_event_max_datetime
stringlengths 24
24
⌀ | max_forks_repo_path
stringlengths 6
260
| max_forks_repo_name
stringlengths 6
119
| max_forks_repo_head_hexsha
stringlengths 40
41
| max_forks_repo_licenses
list | max_forks_count
int64 1
105k
⌀ | max_forks_repo_forks_event_min_datetime
stringlengths 24
24
⌀ | max_forks_repo_forks_event_max_datetime
stringlengths 24
24
⌀ | avg_line_length
float64 2
1.04M
| max_line_length
int64 2
11.2M
| alphanum_fraction
float64 0
1
| cells
list | cell_types
list | cell_type_groups
list |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
cbea331d4dac18ceb158c38ea1f74b4b06c281ae
| 190,718 |
ipynb
|
Jupyter Notebook
|
Notebooks/Slides_vs_Transcribes_Frequency.ipynb
|
Eoli-an/Exam-topic-prediction
|
57f64a8c89109352a9ca8cebd165bd6a65a18130
|
[
"MIT"
] | 1 |
2022-01-23T11:05:08.000Z
|
2022-01-23T11:05:08.000Z
|
Notebooks/Slides_vs_Transcribes_Frequency.ipynb
|
Eoli-an/Exam-topic-prediction
|
57f64a8c89109352a9ca8cebd165bd6a65a18130
|
[
"MIT"
] | 14 |
2022-02-05T17:06:24.000Z
|
2022-02-06T16:25:56.000Z
|
Notebooks/Slides_vs_Transcribes_Frequency.ipynb
|
Eoli-an/Exam-topic-prediction
|
57f64a8c89109352a9ca8cebd165bd6a65a18130
|
[
"MIT"
] | null | null | null | 225.434988 | 153,482 | 0.859615 |
[
[
[
"<a href=\"https://colab.research.google.com/github/Eoli-an/Exam-topic-prediction/blob/main/Slides_vs_Transcribes_Frequency.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# Plot for Dense Ranks of Word Usage in Slides and Transcribes of Relevant Words\n\nFor this plot we analyse the relationship between the word frequency of the slides versus the word frequency of the transcribes of the lecture. We only analyse hand picked words that are relevant for predicting exam topics or their difficulties.",
"_____no_output_____"
]
],
[
[
"!pip install scattertext\n!pip install tika\n!pip install textblob",
"Collecting scattertext\n Downloading scattertext-0.1.5-py3-none-any.whl (7.3 MB)\n\u001b[K |████████████████████████████████| 7.3 MB 3.7 MB/s \n\u001b[?25hRequirement already satisfied: scikit-learn in /usr/local/lib/python3.7/dist-packages (from scattertext) (1.0.2)\nRequirement already satisfied: six in /usr/local/lib/python3.7/dist-packages (from scattertext) (1.15.0)\nRequirement already satisfied: pandas in /usr/local/lib/python3.7/dist-packages (from scattertext) (1.3.5)\nRequirement already satisfied: numpy in /usr/local/lib/python3.7/dist-packages (from scattertext) (1.19.5)\nCollecting gensim>=4.0.0\n Downloading gensim-4.1.2-cp37-cp37m-manylinux_2_12_x86_64.manylinux2010_x86_64.whl (24.1 MB)\n\u001b[K |████████████████████████████████| 24.1 MB 6.0 MB/s \n\u001b[?25hCollecting mock\n Downloading mock-4.0.3-py3-none-any.whl (28 kB)\nRequirement already satisfied: statsmodels in /usr/local/lib/python3.7/dist-packages (from scattertext) (0.10.2)\nCollecting flashtext\n Downloading flashtext-2.7.tar.gz (14 kB)\nRequirement already satisfied: scipy in /usr/local/lib/python3.7/dist-packages (from scattertext) (1.4.1)\nRequirement already satisfied: smart-open>=1.8.1 in /usr/local/lib/python3.7/dist-packages (from gensim>=4.0.0->scattertext) (5.2.1)\nRequirement already satisfied: python-dateutil>=2.7.3 in /usr/local/lib/python3.7/dist-packages (from pandas->scattertext) (2.8.2)\nRequirement already satisfied: pytz>=2017.3 in /usr/local/lib/python3.7/dist-packages (from pandas->scattertext) (2018.9)\nRequirement already satisfied: threadpoolctl>=2.0.0 in /usr/local/lib/python3.7/dist-packages (from scikit-learn->scattertext) (3.1.0)\nRequirement already satisfied: joblib>=0.11 in /usr/local/lib/python3.7/dist-packages (from scikit-learn->scattertext) (1.1.0)\nRequirement already satisfied: patsy>=0.4.0 in /usr/local/lib/python3.7/dist-packages (from statsmodels->scattertext) (0.5.2)\nBuilding wheels for collected packages: flashtext\n Building wheel for flashtext (setup.py) ... \u001b[?25l\u001b[?25hdone\n Created wheel for flashtext: filename=flashtext-2.7-py2.py3-none-any.whl size=9309 sha256=d7ddfda4838a50a0e01255e7eb289e13c2e5ff760f9b942a589faf9bd070e251\n Stored in directory: /root/.cache/pip/wheels/cb/19/58/4e8fdd0009a7f89dbce3c18fff2e0d0fa201d5cdfd16f113b7\nSuccessfully built flashtext\nInstalling collected packages: mock, gensim, flashtext, scattertext\n Attempting uninstall: gensim\n Found existing installation: gensim 3.6.0\n Uninstalling gensim-3.6.0:\n Successfully uninstalled gensim-3.6.0\nSuccessfully installed flashtext-2.7 gensim-4.1.2 mock-4.0.3 scattertext-0.1.5\nCollecting tika\n Downloading tika-1.24.tar.gz (28 kB)\nRequirement already satisfied: setuptools in /usr/local/lib/python3.7/dist-packages (from tika) (57.4.0)\nRequirement already satisfied: requests in /usr/local/lib/python3.7/dist-packages (from tika) (2.23.0)\nRequirement already satisfied: idna<3,>=2.5 in /usr/local/lib/python3.7/dist-packages (from requests->tika) (2.10)\nRequirement already satisfied: certifi>=2017.4.17 in /usr/local/lib/python3.7/dist-packages (from requests->tika) (2021.10.8)\nRequirement already satisfied: chardet<4,>=3.0.2 in /usr/local/lib/python3.7/dist-packages (from requests->tika) (3.0.4)\nRequirement already satisfied: urllib3!=1.25.0,!=1.25.1,<1.26,>=1.21.1 in /usr/local/lib/python3.7/dist-packages (from requests->tika) (1.24.3)\nBuilding wheels for collected packages: tika\n Building wheel for tika (setup.py) ... \u001b[?25l\u001b[?25hdone\n Created wheel for tika: filename=tika-1.24-py3-none-any.whl size=32893 sha256=510c46fdd39035d4d9a50aedbd8895289a6de5921988025067f7ca544e9c18a5\n Stored in directory: /root/.cache/pip/wheels/ec/2b/38/58ff05467a742e32f67f5d0de048fa046e764e2fbb25ac93f3\nSuccessfully built tika\nInstalling collected packages: tika\nSuccessfully installed tika-1.24\nRequirement already satisfied: textblob in /usr/local/lib/python3.7/dist-packages (0.15.3)\nRequirement already satisfied: nltk>=3.1 in /usr/local/lib/python3.7/dist-packages (from textblob) (3.2.5)\nRequirement already satisfied: six in /usr/local/lib/python3.7/dist-packages (from nltk>=3.1->textblob) (1.15.0)\n"
],
[
"import pandas as pd\nimport glob\nimport seaborn as sns\nimport matplotlib.pyplot as plt\n%matplotlib inline\nimport scattertext as st\nfrom tika import parser\nfrom textblob import TextBlob\nimport nltk\nnltk.download('punkt')\nnltk.download('averaged_perceptron_tagger')\nnltk.download('brown')",
"[nltk_data] Downloading package punkt to /root/nltk_data...\n[nltk_data] Unzipping tokenizers/punkt.zip.\n[nltk_data] Downloading package averaged_perceptron_tagger to\n[nltk_data] /root/nltk_data...\n[nltk_data] Unzipping taggers/averaged_perceptron_tagger.zip.\n[nltk_data] Downloading package brown to /root/nltk_data...\n[nltk_data] Unzipping corpora/brown.zip.\n"
]
],
[
[
"The Slides are expected to be in a folder called Slides. The Transcibes are expected to be in a folder called Transcribes",
"_____no_output_____"
]
],
[
[
"lectures_spoken = []\npath = 'Transcribes/*.txt'\nfiles=glob.glob(path)\nfor file in sorted(files):\n with open(file, 'r') as f:\n lectures_spoken.append(f.read())\nlectures_spoken = \" \".join(lectures_spoken)",
"_____no_output_____"
],
[
"lectures_pdf = []\npath = 'Slides/*.pdf'\nfiles=glob.glob(path)\nfor file in sorted(files):\n lectures_pdf.append(parser.from_file(file)[\"content\"])\nlectures_pdf = \" \".join(lectures_pdf)",
"_____no_output_____"
]
],
[
[
"Create a texblob of the text. This is used to extract the noun phrases.",
"_____no_output_____"
]
],
[
[
"blob_spoken = TextBlob(lectures_spoken)\nfreq_spoken = nltk.FreqDist(blob_spoken.noun_phrases)",
"_____no_output_____"
],
[
"blob_pdf = TextBlob(lectures_pdf)\nfreq_pdf = nltk.FreqDist(blob_pdf.noun_phrases)",
"_____no_output_____"
]
],
[
[
"This function checks if a noun phrase is sufficiently similar to a relevant word(templates). Sufficiently similar is defined as that the template is a substring of the noun phrase.",
"_____no_output_____"
]
],
[
[
"def convert_to_template(df_element, template):\n for template_element in template:\n if template_element in df_element:\n return template_element\n return \"None\"\n",
"_____no_output_____"
]
],
[
[
"We first create a pandas dataframe of all the noun phrases and their frequencies in both slides and transcribes. After that, we extract all words that are similar to a relevant word (as of the convert_to_template function). Then we group by the relevant words",
"_____no_output_____"
]
],
[
[
"relevant_words = ['bayes', 'frequentist', 'fairness', 'divergence', 'reproduc', 'regulariz', 'pca', 'principal c' 'bootstrap', 'nonlinear function', 'linear function', 'entropy', 'maximum likelihood estimat', 'significa', 'iid', 'bayes theorem', 'visualization', 'score function', 'dimensionality reduction', 'estimat', 'bayes', 'consumption', 'fisher', 'independence', 'logistic regression', 'bias', 'standard deviation', 'linear discriminant analysis', 'information matrix', 'null hypothesis', 'log likelihood', 'linear regression', 'hypothesis test', 'confidence', 'variance', 'sustainability', 'gaussian', 'linear model', 'climate', 'laplace', ]",
"_____no_output_____"
],
[
"df_spoken = pd.DataFrame.from_dict({\"word\": list(freq_spoken.keys()), \"freq_spoken\" : list(freq_spoken.values())})\ndf_pdf = pd.DataFrame.from_dict({\"word\": list(freq_pdf.keys()), \"freq_pdf\" : list(freq_pdf.values())})\ndf = df_spoken.merge(df_pdf,how=\"outer\",on=\"word\")\ndf[\"word\"] = df[\"word\"].apply(lambda x: convert_to_template(x,relevant_words))\ndf = df.groupby([\"word\"]).sum().reset_index()\ndf = df[df[\"word\"] != \"None\"].reset_index()",
"_____no_output_____"
]
],
[
[
"We use the dense_rank functionality of the scattertext library to convert the absolute number of occurances of a word to a dense rank. This means that we only consider the relative order of the frequencies of the word and discard all information that tells us how far apart two word frequencies are. ",
"_____no_output_____"
]
],
[
[
"df[\"freq_spoken\"] = st.Scalers.dense_rank(df[\"freq_spoken\"])\ndf[\"freq_pdf\"] = st.Scalers.dense_rank(df[\"freq_pdf\"])",
"_____no_output_____"
],
[
"df",
"_____no_output_____"
],
[
"\n\nplt.figure(figsize=(20,12))\nsns.set_theme(style=\"dark\")\np1 = sns.scatterplot(x='freq_spoken', # Horizontal axis\n y='freq_pdf', # Vertical axis\n data=df, # Data source\n s = 80,\n legend=False,\n color=\"orange\",\n #marker = \"s\"\n ) \n\nfor line in range(0,df.shape[0]):\n if line == 6:#divergence\n p1.text(df.freq_spoken[line]-0.12, df.freq_pdf[line]-0.007, \n df.word[line], horizontalalignment='left', \n size='xx-large', color='black', weight='normal')\n elif line == 21:#linear regression\n p1.text(df.freq_spoken[line]-0.18, df.freq_pdf[line]-0.007, \n df.word[line], horizontalalignment='left', \n size='xx-large', color='black', weight='normal')\n elif line == 18:#linear discriminant analysis\n p1.text(df.freq_spoken[line]-0.05, df.freq_pdf[line]-0.05, \n df.word[line], horizontalalignment='left', \n size='xx-large', color='black', weight='normal')\n elif line == 19:#linear function\n p1.text(df.freq_spoken[line]-0.02, df.freq_pdf[line]-0.04, \n df.word[line], horizontalalignment='left', \n size='xx-large', color='black', weight='normal')\n elif line == 29:#reproduce\n p1.text(df.freq_spoken[line]-0.03, df.freq_pdf[line]+0.03, \n df.word[line], horizontalalignment='left', \n size='xx-large', color='black', weight='normal')\n elif line == 12:#gaussian:\n p1.text(df.freq_spoken[line]-0.1, df.freq_pdf[line]-0.007, \n df.word[line], horizontalalignment='left', \n size='xx-large', color='black', weight='normal')\n elif line == 16:#information matrix:\n p1.text(df.freq_spoken[line]+0.01, df.freq_pdf[line]-0.025, \n df.word[line], horizontalalignment='left', \n size='xx-large', color='black', weight='normal')\n elif line == 25:#nonlinear function:\n p1.text(df.freq_spoken[line]+0.01, df.freq_pdf[line]-0.025, \n df.word[line], horizontalalignment='left', \n size='xx-large', color='black', weight='normal')\n elif line == 24:#maximum likelihood estimat:\n p1.text(df.freq_spoken[line]-0.07, df.freq_pdf[line]+0.02, \n df.word[line], horizontalalignment='left', \n size='xx-large', color='black', weight='normal')\n elif line == 17:#laplace:\n p1.text(df.freq_spoken[line]-0.08, df.freq_pdf[line]-0.007, \n df.word[line], horizontalalignment='left', \n size='xx-large', color='black', weight='normal')\n else:\n p1.text(df.freq_spoken[line]+0.01, df.freq_pdf[line]-0.007, \n df.word[line], horizontalalignment='left', \n size='xx-large', color='black', weight='normal')\n\n\n#plt.title('Dense Ranks of Word Usage in Slides and Transcribes of Relevant Words',size = \"xx-large\")\n# Set x-axis label\nplt.xlabel('Transcribes Frequency',size = \"xx-large\")\n# Set y-axis label\nplt.ylabel('Slides Frequency',size = \"xx-large\")\n\np1.set_xticks([0,0.5,1]) # <--- set the ticks first\np1.set_xticklabels([\"Infrequent\", \"Average\", \"Frequent\"],size = \"x-large\")\n\np1.set_yticks([0,0.5,1]) # <--- set the ticks first\np1.set_yticklabels([\"Infrequent\", \"Average\", \"Frequent\"],size = \"x-large\")\nplt.show()",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
cbea4bb3da2790a0018e23f960887ae026f06d6e
| 90,477 |
ipynb
|
Jupyter Notebook
|
lecture_01.ipynb
|
nicholascar/comp7230-training
|
ad37c7203233fb14ea373fb08a362ee367672288
|
[
"CC-BY-4.0"
] | 2 |
2021-09-07T05:07:38.000Z
|
2022-01-09T09:48:25.000Z
|
lecture_01.ipynb
|
nicholascar/comp7230-training
|
ad37c7203233fb14ea373fb08a362ee367672288
|
[
"CC-BY-4.0"
] | null | null | null |
lecture_01.ipynb
|
nicholascar/comp7230-training
|
ad37c7203233fb14ea373fb08a362ee367672288
|
[
"CC-BY-4.0"
] | 1 |
2021-09-07T05:06:18.000Z
|
2021-09-07T05:06:18.000Z
| 52.480858 | 25,388 | 0.622247 |
[
[
[
"## Guest Lecture COMP7230\n# Using Python packages for Linked Data & spatial data\n#### by Dr Nicholas Car\n\nThis Notebook is the resource used to deliver a guest lecture for the [Australian National University](https://www.anu.edu.au)'s course [COMP7230](https://programsandcourses.anu.edu.au/2020/course/COMP7230): *Introduction to Programming for Data Scientists*\n\nClick here to run this lecture in your web browser:\n[](https://mybinder.org/v2/gh/nicholascar/comp7230-training/HEAD?filepath=lecture_01.ipynb)\n\n## About the lecturer\n**Nicholas Car**:\n* PhD in informatics for irrigation\n* A former CSIRO informatics researcher\n * worked on integrating environmental data across government / industry\n * developed data standards\n* Has worked in operation IT in government\n* Now in a private IT consulting company, [SURROUND Australia Pty Ltd](https://surroundaustralia.com) supplying Data Science solutions\n\nRelevant current work:\n\n* building data processing systems for government & industry\n* mainly using Python\n * due to its large number of web and data science packages\n* maintains the [RDFlib](https://rdflib.net) Python toolkit\n * for processing [RDF](https://en.wikipedia.org/wiki/Resource_Description_Framework)\n* co-chairs the [Australian Government Linked Data Working Group](https://www.linked.data.gov.au) with Armin Haller\n * plans for multi-agency data integration\n* still developing data standards\n * in particular GeoSPARQL 1.1 (https://opengeospatial.github.io/ogc-geosparql/geosparql11/spec.html) \n * for graph representations of spatial information\n\n\n## 0. Lecture Outline\n1. Notes about this training material\n2. Accessing RDF data\n3. Parsing RDF data\n4. Data 'mash up'\n5. Data Conversions & Display\n\n\n## 1. Notes about this training material\n\n#### This tool\n* This is a Jupyter Notebook - interactive Python scripting\n* You will cover Jupyter Notebooks more, later in this course\n* Access this material online at:\n * GitHub: <https://github.com/nicholascar/comp7230-training>\n\n[](https://mybinder.org/v2/gh/nicholascar/comp7230-training/?filepath=lecture_01.ipynb)\n\n#### Background data concepts - RDF\n\n_Nick will talk RDF using these web pages:_\n\n* [Semantic Web](https://www.w3.org/standards/semanticweb/) - the concept\n* [RDF](https://en.wikipedia.org/wiki/Resource_Description_Framework) - the data model\n * refer to the RDF image below\n* [RDFlib](https://rdflib.net) - the (Python) toolkit\n* [RDFlib training Notebooks are available](https://github.com/surroundaustralia/rdflib-training)\n\nThe LocI project:\n* The Location Index project: <http://loci.cat>\n\nRDF image, from [the RDF Primer](https://www.w3.org/TR/rdf11-primer/), for discussion:\n\n\n\nNote that:\n* _everything_ is \"strongly\" identified\n * including all relationships\n * unlike lots of related data\n* many of the identifiers resolve\n * to more info (on the web)\n\n## 2. Accessing RDF data\n\n* Here we use an online structured dataset, the Geocoded National Address File for Australia\n * Dataset Persistent Identifier: <https://linked.data.gov.au/dataset/gnaf>\n * The above link redirects to the API at <https://gnafld.net>\n* GNAF-LD Data is presented according to *Linked Data* principles\n * online\n * in HTML & machine-readable form, RDF\n * RDF is a Knowledge Graph: a graph containing data + model\n * each resource is available via a URI\n * e.g. <https://linked.data.gov.au/dataset/gnaf/address/GAACT714845933>\n\n\n\n\n2.1. Get the Address GAACT714845933 using the *requests* package",
"_____no_output_____"
]
],
[
[
"import requests # NOTE: you must have installed requests first, it's not a standard package\nr = requests.get(\n \"https://linked.data.gov.au/dataset/gnaf/address/GAACT714845933\"\n)\nprint(r.text)",
"<!DOCTYPE html>\n<html>\n<head lang=\"en\">\n <meta charset=\"UTF-8\">\n <title>Address API</title>\n <link rel=\"stylesheet\" href=\"/static/css/psma_theme.css\" />\n</head>\n<body>\n <div id=\"widther\">\n <div id=\"header\">\n <div style=\"float:left;\">\n <a href=\"https://www.psma.com.au/\">PSMA Australia Ltd.</a>\n </div>\n <div style=\"float:right;\">\n <a href=\"/\">Home</a>\n <a href=\"/?_view=reg\">Registers</a>\n <a href=\"/sparql\">SPARQL endpoint</a>\n <a href=\"http://linked.data.gov.au/def/gnaf\">GNAF ontology</a>\n <a href=\"http://linked.data.gov.au/def/gnaf/code/\">GNAF codes</a>\n <a href=\"/about\">About</a>\n </div>\n <div style=\"clear:both;\"></div>\n </div>\n <div id=\"container-content\">\n \n <h1>Address GAACT714845933</h1>\n <script type=\"application/ld+json\">\n {\"@type\": \"Place\", \"name\": \"Geocoded Address GAACT714845933\", \"geo\": {\"latitude\": -35.20113263, \"@type\": \"GeoCoordinates\", \"longitude\": 149.03865604}, \"@context\": \"http://schema.org\", \"address\": {\"addressRegion\": \"Australian Capital Territory\", \"postalCode\": \"2615\", \"streetAddress\": \"6 Packham Place\", \"addressCountry\": \"AU\", \"@type\": \"PostalAddress\", \"addressLocality\": \"Charnwood\"}}\n </script>\n <h2>G-NAF View</h2>\n <table class=\"content\">\n <tr><th>Property</th><th>Value</th></tr>\n <tr><td>Address Line</td><td><code>6 Packham Place, Charnwood, ACT 2615</code></td></tr>\n \n\n \n\n \n\n \n\n \n\n \n\n \n <tr>\n <td><a href=\"http://linked.data.gov.au/def/gnaf#FirstStreetNumber\">First Street Number</a></td>\n <td><code>6</code></td>\n </tr>\n \n\n \n\n \n <tr><td><a href=\"http://linked.data.gov.au/def/gnaf#hasStreetLocality\">Street Locality</a></td><td><code><a href=\"/streetLocality/ACT3857\">Packham Place</a></code></td></tr>\n \n \n <tr><td><a href=\"http://linked.data.gov.au/def/gnaf#hasLocality\">Locality</a></td><td><code><a href=\"/locality/ACT570\">Charnwood</a></code></td></tr>\n \n \n <tr><td><a href=\"http://linked.data.gov.au/def/gnaf#hasState\">State/Territory</a></td><td><code>ACT</code></td></tr>\n \n \n <tr><td><a href=\"http://linked.data.gov.au/def/gnaf#hasPostcode\">Postcode</a></td><td><code>2615</code></td></tr>\n \n \n <tr><td><a href=\"http://linked.data.gov.au/def/gnaf#hasLegalParcelId\">Legal Parcel ID</a></td><td><code>BELC/CHAR/15/16/</code></td></tr>\n \n \n <tr><td><a href=\"http://linked.data.gov.au/def/gnaf#hasAddressSite\">Address Site PID</a></td><td><code><a href=\"/addressSite/710446419\">710446419</a></code></td></tr>\n \n \n <tr><td><a href=\"http://linked.data.gov.au/def/gnaf#\">Level Geocoded Code</a></td><td><code>7</code></td></tr>\n \n \n \n <tr><td><a href=\"http://linked.data.gov.au/def/gnaf#hasGnafConfidence\">GNAF Confidence</a></td><td><code><a href=\"http://gnafld.net/def/gnaf/GnafConfidence_2\">Confidence level 2</a></code></td></tr>\n \n \n <tr><td><a href=\"http://linked.data.gov.au/def/gnaf#hasCreatedDate\">Date Created</a></td><td><code>2004-04-29</code></td></tr>\n \n \n <tr><td><a href=\"http://linked.data.gov.au/def/gnaf#hasLastModifiedDate\">Date Last Modified</a></td><td><code>2018-02-01</code></td></tr>\n \n \n \n <tr>\n <td><a href=\"http://www.opengis.net/ont/geosparql#hasGeometry\">Geometry</a></td>\n <td><code><a href=\"http://gnafld.net/def/gnaf/code/GeocodeTypes#FrontageCentreSetback\">Frontage Centre Setback</a> →<br /><http://www.opengis.net/def/crs/EPSG/0/4283> POINT(149.03865604 -35.20113263)</code></td>\n </tr>\n \n \n \n \n \n \n <tr>\n <td><a href=\"http://linked.data.gov.au/def/gnaf#hasMeshBlockMatch\">Mesh Blocks 2011</a></td><td>\n \n <code><a href=\"http://gnafld.net/def/gnaf/code/MeshBlockMatchTypes#ParcelLevel\">Parcel Level Match</a> → <a href=\"http://linked.data.gov.au/dataset/asgs/MB2011/80006300000\">80006300000</a></code><br />\n \n </td>\n </tr>\n \n \n <tr>\n <td><a href=\"http://linked.data.gov.au/def/gnaf#hasMeshBlockMatch\">Mesh Blocks 2016</a></td><td>\n \n <code><a href=\"http://gnafld.net/def/gnaf/code/MeshBlockMatchTypes#ParcelLevel\">Parcel Level Match</a> → <a href=\"http://linked.data.gov.au/dataset/asgs/MB2016/80006300000\">80006300000</a></code><br />\n \n </td>\n </tr>\n \n </table>\n\n <h2>Other views</h2>\n <p>Other model views of a Address are listed in the <a href=\"/address/GAACT714845933?_view=alternates\">Alternates View</a>.</p>\n\n <h2>Citation</h2>\n <p>If you wish to cite this Address as you would a publication, please use the following format:</p>\n <code style=\"display:block; margin: 0 5em 0 5em;\">\n PSMA Australia Limited (2017). Address GAACT714845933. Address object from the Geocoded National Address File (G-NAF). http://linked.data.gov.au/dataset/gnaf/address/GAACT714845933\n </code>\n\n\n </div>\n <div id=\"footer\"></div>\n </div>\n</body>\n</html>\n"
]
],
[
[
"2.2 Get machine-readable data, RDF triples\nUse HTTP Content Negotiation\nSame URI, different *format* of data",
"_____no_output_____"
]
],
[
[
"r = requests.get(\n \"https://linked.data.gov.au/dataset/gnaf/address/GAACT714845933\",\n headers={\"Accept\": \"application/n-triples\"}\n)\nprint(r.text)",
"<http://linked.data.gov.au/dataset/gnaf/address/GAACT714845933> <http://purl.org/dc/terms/identifier> \"GAACT714845933\"^^<http://www.w3.org/2001/XMLSchema#string> .\n<http://linked.data.gov.au/dataset/gnaf/address/GAACT714845933> <http://www.w3.org/2000/01/rdf-schema#comment> \"6 Packham Place, Charnwood, ACT 2615\"^^<http://www.w3.org/2001/XMLSchema#string> .\n_:Nfbdb238ffe9d4fa4bd6dd6f8cced7318 <http://www.w3.org/1999/02/22-rdf-syntax-ns#type> <http://www.opengis.net/ont/sf#Point> .\n_:N3677002343da47bbb35c569fc67f349f <http://www.w3.org/ns/prov#value> \"6\"^^<http://www.w3.org/2001/XMLSchema#integer> .\n<http://linked.data.gov.au/dataset/gnaf/address/GAACT714845933> <http://linked.data.gov.au/def/gnaf#hasStreetLocality> <http://linked.data.gov.au/dataset/gnaf/streetLocality/ACT3857> .\n<http://linked.data.gov.au/dataset/gnaf/address/GAACT714845933> <http://purl.org/dc/terms/modified> \"2018-02-01\"^^<http://www.w3.org/2001/XMLSchema#date> .\n<http://linked.data.gov.au/dataset/gnaf/address/GAACT714845933> <http://www.w3.org/2000/01/rdf-schema#label> \"Address GAACT714845933 of Unknown type\"^^<http://www.w3.org/2001/XMLSchema#string> .\n_:Nfbdb238ffe9d4fa4bd6dd6f8cced7318 <http://linked.data.gov.au/def/gnaf#gnafType> <http://gnafld.net/def/gnaf/code/GeocodeTypes#FrontageCentreSetback> .\n<http://linked.data.gov.au/dataset/gnaf/address/GAACT714845933> <http://linked.data.gov.au/def/gnaf#hasDateCreated> \"2004-04-29\"^^<http://www.w3.org/2001/XMLSchema#date> .\n<http://linked.data.gov.au/dataset/gnaf/address/GAACT714845933> <http://www.opengis.net/ont/geosparql#hasGeometry> _:Nfbdb238ffe9d4fa4bd6dd6f8cced7318 .\n<http://linked.data.gov.au/dataset/gnaf/address/GAACT714845933> <http://linked.data.gov.au/def/gnaf#hasNumber> _:N3677002343da47bbb35c569fc67f349f .\n<http://linked.data.gov.au/dataset/gnaf/address/GAACT714845933> <http://purl.org/dc/terms/type> <http://gnafld.net/def/gnaf/code/AddressTypes#Unknown> .\n<http://linked.data.gov.au/dataset/gnaf/address/GAACT714845933> <http://linked.data.gov.au/def/loci#isMemberOf> <http://linked.data.gov.au/dataset/gnaf/address/> .\n<http://linked.data.gov.au/dataset/gnaf/address/GAACT714845933> <http://linked.data.gov.au/def/gnaf#hasDateLastModified> \"2018-02-01\"^^<http://www.w3.org/2001/XMLSchema#date> .\n<http://linked.data.gov.au/dataset/gnaf/address/GAACT714845933> <http://linked.data.gov.au/def/gnaf#hasGnafConfidence> <http://gnafld.net/def/gnaf/GnafConfidence_2> .\n<http://linked.data.gov.au/dataset/gnaf/address/GAACT714845933> <http://www.w3.org/1999/02/22-rdf-syntax-ns#type> <http://linked.data.gov.au/def/gnaf#Address> .\n<http://linked.data.gov.au/dataset/gnaf/address/GAACT714845933> <http://purl.org/dc/terms/created> \"2004-04-29\"^^<http://www.w3.org/2001/XMLSchema#date> .\n<http://gnafld.net/def/gnaf/GnafConfidence_2> <http://www.w3.org/2000/01/rdf-schema#label> \"Confidence level 2\"^^<http://www.w3.org/2001/XMLSchema#string> .\n_:N3677002343da47bbb35c569fc67f349f <http://linked.data.gov.au/def/gnaf#gnafType> <http://linked.data.gov.au/def/gnaf/code/NumberTypes#FirstStreet> .\n_:Nfbdb238ffe9d4fa4bd6dd6f8cced7318 <http://www.w3.org/2000/01/rdf-schema#label> \"Frontage Centre Setback\"^^<http://www.w3.org/2001/XMLSchema#string> .\n<http://linked.data.gov.au/dataset/gnaf/address/GAACT714845933> <http://linked.data.gov.au/def/gnaf#hasAddressSite> <http://linked.data.gov.au/dataset/gnaf/addressSite/710446419> .\n<http://linked.data.gov.au/dataset/gnaf/address/GAACT714845933> <http://linked.data.gov.au/def/gnaf#hasState> <http://www.geonames.org/2177478> .\n_:N3677002343da47bbb35c569fc67f349f <http://www.w3.org/1999/02/22-rdf-syntax-ns#type> <http://linked.data.gov.au/def/gnaf#Number> .\n<http://www.geonames.org/2177478> <http://www.w3.org/2000/01/rdf-schema#label> \"Australian Capital Territory\"^^<http://www.w3.org/2001/XMLSchema#string> .\n_:Nfbdb238ffe9d4fa4bd6dd6f8cced7318 <http://purl.org/dc/terms/type> <http://gnafld.net/def/gnaf/code/GeocodeTypes#FrontageCentreSetback> .\n_:Nfbdb238ffe9d4fa4bd6dd6f8cced7318 <http://www.opengis.net/ont/geosparql#asWKT> \"<http://www.opengis.net/def/crs/EPSG/0/4283> POINT(149.03865604 -35.20113263)\"^^<http://www.opengis.net/ont/geosparql#wktLiteral> .\n<http://linked.data.gov.au/dataset/gnaf/address/GAACT714845933> <http://linked.data.gov.au/def/gnaf#hasLocality> <http://linked.data.gov.au/dataset/gnaf/locality/ACT570> .\n<http://linked.data.gov.au/dataset/gnaf/address/GAACT714845933> <http://linked.data.gov.au/def/gnaf#hasPostcode> \"2615\"^^<http://www.w3.org/2001/XMLSchema#integer> .\n\n\n"
]
],
[
[
"2.3 Get machine-readable data, Turtle\nEasier to read",
"_____no_output_____"
]
],
[
[
"r = requests.get(\n \"https://linked.data.gov.au/dataset/gnaf/address/GAACT714845933\",\n headers={\"Accept\": \"text/turtle\"}\n)\nprint(r.text)",
"@prefix dct: <http://purl.org/dc/terms/> .\n@prefix geo: <http://www.opengis.net/ont/geosparql#> .\n@prefix gnaf: <http://linked.data.gov.au/def/gnaf#> .\n@prefix loci: <http://linked.data.gov.au/def/loci#> .\n@prefix prov: <http://www.w3.org/ns/prov#> .\n@prefix rdf: <http://www.w3.org/1999/02/22-rdf-syntax-ns#> .\n@prefix rdfs: <http://www.w3.org/2000/01/rdf-schema#> .\n@prefix sf: <http://www.opengis.net/ont/sf#> .\n@prefix xml: <http://www.w3.org/XML/1998/namespace> .\n@prefix xsd: <http://www.w3.org/2001/XMLSchema#> .\n\n<http://linked.data.gov.au/dataset/gnaf/address/GAACT714845933> a gnaf:Address ;\n rdfs:label \"Address GAACT714845933 of Unknown type\"^^xsd:string ;\n gnaf:hasAddressSite <http://linked.data.gov.au/dataset/gnaf/addressSite/710446419> ;\n gnaf:hasDateCreated \"2004-04-29\"^^xsd:date ;\n gnaf:hasDateLastModified \"2018-02-01\"^^xsd:date ;\n gnaf:hasGnafConfidence <http://gnafld.net/def/gnaf/GnafConfidence_2> ;\n gnaf:hasLocality <http://linked.data.gov.au/dataset/gnaf/locality/ACT570> ;\n gnaf:hasNumber [ a gnaf:Number ;\n gnaf:gnafType <http://linked.data.gov.au/def/gnaf/code/NumberTypes#FirstStreet> ;\n prov:value 6 ] ;\n gnaf:hasPostcode 2615 ;\n gnaf:hasState <http://www.geonames.org/2177478> ;\n gnaf:hasStreetLocality <http://linked.data.gov.au/dataset/gnaf/streetLocality/ACT3857> ;\n loci:isMemberOf <http://linked.data.gov.au/dataset/gnaf/address/> ;\n dct:created \"2004-04-29\"^^xsd:date ;\n dct:identifier \"GAACT714845933\"^^xsd:string ;\n dct:modified \"2018-02-01\"^^xsd:date ;\n dct:type <http://gnafld.net/def/gnaf/code/AddressTypes#Unknown> ;\n geo:hasGeometry [ a sf:Point ;\n rdfs:label \"Frontage Centre Setback\"^^xsd:string ;\n gnaf:gnafType <http://gnafld.net/def/gnaf/code/GeocodeTypes#FrontageCentreSetback> ;\n dct:type <http://gnafld.net/def/gnaf/code/GeocodeTypes#FrontageCentreSetback> ;\n geo:asWKT \"<http://www.opengis.net/def/crs/EPSG/0/4283> POINT(149.03865604 -35.20113263)\"^^geo:wktLiteral ] ;\n rdfs:comment \"6 Packham Place, Charnwood, ACT 2615\"^^xsd:string .\n\n<http://gnafld.net/def/gnaf/GnafConfidence_2> rdfs:label \"Confidence level 2\"^^xsd:string .\n\n<http://www.geonames.org/2177478> rdfs:label \"Australian Capital Territory\"^^xsd:string .\n\n\n"
]
],
[
[
"## 3. Parsing RDF data\n\nImport the RDFlib library for manipulating RDF data\nAdd some namespaces to shorten URIs",
"_____no_output_____"
]
],
[
[
"import rdflib\nfrom rdflib.namespace import RDF, RDFS\nGNAF = rdflib.Namespace(\"http://linked.data.gov.au/def/gnaf#\")\nADDR = rdflib.Namespace(\"http://linked.data.gov.au/dataset/gnaf/address/\")\nGEO = rdflib.Namespace(\"http://www.opengis.net/ont/geosparql#\")\nprint(GEO)",
"http://www.opengis.net/ont/geosparql#\n"
]
],
[
[
"Create a graph and add the namespaces to it",
"_____no_output_____"
]
],
[
[
"g = rdflib.Graph()\ng.bind(\"gnaf\", GNAF)\ng.bind(\"addr\", ADDR)\ng.bind(\"geo\", GEO)",
"_____no_output_____"
]
],
[
[
"Parse in the machine-readable data from the GNAF-LD",
"_____no_output_____"
]
],
[
[
"r = requests.get(\n \"https://linked.data.gov.au/dataset/gnaf/address/GAACT714845933\",\n headers={\"Accept\": \"text/turtle\"}\n)\ng.parse(data=r.text, format=\"text/turtle\")",
"_____no_output_____"
]
],
[
[
"Print graph length (no. of triples) to check",
"_____no_output_____"
]
],
[
[
"print(len(g))",
"28\n"
]
],
[
[
"Print graph content, in Turtle",
"_____no_output_____"
]
],
[
[
"print(g.serialize(format=\"text/turtle\").decode())",
"@prefix addr: <http://linked.data.gov.au/dataset/gnaf/address/> .\n@prefix dct: <http://purl.org/dc/terms/> .\n@prefix geo: <http://www.opengis.net/ont/geosparql#> .\n@prefix gnaf: <http://linked.data.gov.au/def/gnaf#> .\n@prefix loci: <http://linked.data.gov.au/def/loci#> .\n@prefix prov: <http://www.w3.org/ns/prov#> .\n@prefix rdfs: <http://www.w3.org/2000/01/rdf-schema#> .\n@prefix sf: <http://www.opengis.net/ont/sf#> .\n@prefix xsd: <http://www.w3.org/2001/XMLSchema#> .\n\naddr:GAACT714845933 a gnaf:Address ;\n rdfs:label \"Address GAACT714845933 of Unknown type\"^^xsd:string ;\n gnaf:hasAddressSite <http://linked.data.gov.au/dataset/gnaf/addressSite/710446419> ;\n gnaf:hasDateCreated \"2004-04-29\"^^xsd:date ;\n gnaf:hasDateLastModified \"2018-02-01\"^^xsd:date ;\n gnaf:hasGnafConfidence <http://gnafld.net/def/gnaf/GnafConfidence_2> ;\n gnaf:hasLocality <http://linked.data.gov.au/dataset/gnaf/locality/ACT570> ;\n gnaf:hasNumber [ a gnaf:Number ;\n gnaf:gnafType <http://linked.data.gov.au/def/gnaf/code/NumberTypes#FirstStreet> ;\n prov:value 6 ] ;\n gnaf:hasPostcode 2615 ;\n gnaf:hasState <http://www.geonames.org/2177478> ;\n gnaf:hasStreetLocality <http://linked.data.gov.au/dataset/gnaf/streetLocality/ACT3857> ;\n loci:isMemberOf addr: ;\n dct:created \"2004-04-29\"^^xsd:date ;\n dct:identifier \"GAACT714845933\"^^xsd:string ;\n dct:modified \"2018-02-01\"^^xsd:date ;\n dct:type <http://gnafld.net/def/gnaf/code/AddressTypes#Unknown> ;\n geo:hasGeometry [ a sf:Point ;\n rdfs:label \"Frontage Centre Setback\"^^xsd:string ;\n gnaf:gnafType <http://gnafld.net/def/gnaf/code/GeocodeTypes#FrontageCentreSetback> ;\n dct:type <http://gnafld.net/def/gnaf/code/GeocodeTypes#FrontageCentreSetback> ;\n geo:asWKT \"<http://www.opengis.net/def/crs/EPSG/0/4283> POINT(149.03865604 -35.20113263)\"^^geo:wktLiteral ] ;\n rdfs:comment \"6 Packham Place, Charnwood, ACT 2615\"^^xsd:string .\n\n<http://gnafld.net/def/gnaf/GnafConfidence_2> rdfs:label \"Confidence level 2\"^^xsd:string .\n\n<http://www.geonames.org/2177478> rdfs:label \"Australian Capital Territory\"^^xsd:string .\n\n\n"
]
],
[
[
"### 3.1 Getting multi-address data:\n3.1.1. Retrieve an index of 10 addresses, in RDF\n3.1.2. For each address in the index, get each Address' data\n* use paging URI: <https://linked.data.gov.au/dataset/gnaf/address/?page=1>\n3.1.3. Get only the street address and map coordinates\n\n#### 3.1.1. Retrieve index",
"_____no_output_____"
]
],
[
[
"# clear the graph\ng = rdflib.Graph()\n\nr = requests.get(\n \"https://linked.data.gov.au/dataset/gnaf/address/?page=1\",\n headers={\"Accept\": \"text/turtle\"}\n)\ng.parse(data=r.text, format=\"text/turtle\")\nprint(len(g))",
"70\n"
]
],
[
[
"#### 3.1.2. Parse in each address' data",
"_____no_output_____"
]
],
[
[
"for s, p, o in g.triples((None, RDF.type, GNAF.Address)):\n print(s.split(\"/\")[-1])\n r = requests.get(\n str(s),\n headers={\"Accept\": \"text/turtle\"}\n )\n g.parse(data=r.text, format=\"turtle\")\n print(len(g))",
"GAACT714845953\n97\nGAACT714845955\n122\nGAACT714845945\n147\nGAACT714845941\n172\nGAACT714845935\n197\nGAACT714845951\n222\nGAACT714845949\n247\nGAACT714845954\n272\nGAACT714845950\n297\nGAACT714845942\n322\nGAACT714845943\n347\nGAACT714845946\n372\nGAACT714845947\n397\nGAACT714845938\n422\nGAACT714845944\n447\nGAACT714845933\n472\nGAACT714845936\n497\nGAACT714845934\n522\nGAACT714845952\n547\nGAACT714845939\n572\n"
]
],
[
[
"The graph model used by the GNAF-LD is based on [GeoSPARQL 1.1](https://opengeospatial.github.io/ogc-geosparql/geosparql11/spec.html) and looks like this:\n\n\n\n#### 3.1.3. Extract (& print) street address text & coordinates\n(CSV)",
"_____no_output_____"
]
],
[
[
"addresses_tsv = \"GNAF ID\\tAddress\\tCoordinates\\n\"\nfor s, p, o in g.triples((None, RDF.type, GNAF.Address)):\n for s2, p2, o2 in g.triples((s, RDFS.comment, None)):\n txt = str(o2)\n for s2, p2, o2 in g.triples((s, GEO.hasGeometry, None)):\n for s3, p3, o3 in g.triples((o2, GEO.asWKT, None)):\n coords = str(o3).replace(\"<http://www.opengis.net/def/crs/EPSG/0/4283> \", \"\")\n\n addresses_tsv += \"{}\\t{}\\t{}\\n\".format(str(s).split(\"/\")[-1], txt, coords)\n\nprint(addresses_tsv)",
"GNAF ID\tAddress\tCoordinates\nGAACT714845953\t5 Jamieson Crescent, Kambah, ACT 2902\tPOINT(149.06864966 -35.37733591)\nGAACT714845955\t3 Baylis Place, Charnwood, ACT 2615\tPOINT(149.03046282 -35.20202762)\nGAACT714845945\t9 Baylis Place, Charnwood, ACT 2615\tPOINT(149.03047333 -35.20156767)\nGAACT714845941\t7 Mcdowall Place, Kambah, ACT 2902\tPOINT(149.06860919 -35.37833726)\nGAACT714845935\t26 Jauncey Court, Charnwood, ACT 2615\tPOINT(149.03640841 -35.19777173)\nGAACT714845951\t15 Mcdowall Place, Kambah, ACT 2902\tPOINT(149.06946494 -35.37908886)\nGAACT714845949\t13 Mcdowall Place, Kambah, ACT 2902\tPOINT(149.06908395 -35.37882495)\nGAACT714845954\t5 Baylis Place, Charnwood, ACT 2615\tPOINT(149.03048051 -35.20185603)\nGAACT714845950\t7 Baylis Place, Charnwood, ACT 2615\tPOINT(149.03049843 -35.20169346)\nGAACT714845942\t5 Bunker Place, Charnwood, ACT 2615\tPOINT(149.04029706 -35.19999611)\nGAACT714845943\t22 Jauncey Court, Charnwood, ACT 2615\tPOINT(149.03688520 -35.19795303)\nGAACT714845946\t11 Mcdowall Place, Kambah, ACT 2902\tPOINT(149.06895786 -35.37862878)\nGAACT714845947\t20 Jauncey Court, Charnwood, ACT 2615\tPOINT(149.03705032 -35.19796828)\nGAACT714845938\t5 Mcdowall Place, Kambah, ACT 2902\tPOINT(149.06851657 -35.37815855)\nGAACT714845944\t9 Mcdowall Place, Kambah, ACT 2902\tPOINT(149.06872290 -35.37847955)\nGAACT714845933\t6 Packham Place, Charnwood, ACT 2615\tPOINT(149.03865604 -35.20113263)\nGAACT714845936\t17 Geeves Court, Charnwood, ACT 2615\tPOINT(149.03687042 -35.20395740)\nGAACT714845934\t3 Bunker Place, Charnwood, ACT 2615\tPOINT(149.04011870 -35.19989093)\nGAACT714845952\t18 Jauncey Court, Charnwood, ACT 2615\tPOINT(149.03721725 -35.19805563)\nGAACT714845939\t24 Jauncey Court, Charnwood, ACT 2615\tPOINT(149.03661902 -35.19784933)\n\n"
]
],
[
[
"#### 3.1.4. Convert CSV data to PANDAS DataFrame\n(CSV)",
"_____no_output_____"
]
],
[
[
"import pandas\nfrom io import StringIO\ns = StringIO(addresses_tsv)\ndf1 = pandas.read_csv(s, sep=\"\\t\")\nprint(df1)\n",
" GNAF ID Address \\\n0 GAACT714845953 5 Jamieson Crescent, Kambah, ACT 2902 \n1 GAACT714845955 3 Baylis Place, Charnwood, ACT 2615 \n2 GAACT714845945 9 Baylis Place, Charnwood, ACT 2615 \n3 GAACT714845941 7 Mcdowall Place, Kambah, ACT 2902 \n4 GAACT714845935 26 Jauncey Court, Charnwood, ACT 2615 \n5 GAACT714845951 15 Mcdowall Place, Kambah, ACT 2902 \n6 GAACT714845949 13 Mcdowall Place, Kambah, ACT 2902 \n7 GAACT714845954 5 Baylis Place, Charnwood, ACT 2615 \n8 GAACT714845950 7 Baylis Place, Charnwood, ACT 2615 \n9 GAACT714845942 5 Bunker Place, Charnwood, ACT 2615 \n10 GAACT714845943 22 Jauncey Court, Charnwood, ACT 2615 \n11 GAACT714845946 11 Mcdowall Place, Kambah, ACT 2902 \n12 GAACT714845947 20 Jauncey Court, Charnwood, ACT 2615 \n13 GAACT714845938 5 Mcdowall Place, Kambah, ACT 2902 \n14 GAACT714845944 9 Mcdowall Place, Kambah, ACT 2902 \n15 GAACT714845933 6 Packham Place, Charnwood, ACT 2615 \n16 GAACT714845936 17 Geeves Court, Charnwood, ACT 2615 \n17 GAACT714845934 3 Bunker Place, Charnwood, ACT 2615 \n18 GAACT714845952 18 Jauncey Court, Charnwood, ACT 2615 \n19 GAACT714845939 24 Jauncey Court, Charnwood, ACT 2615 \n\n Coordinates \n0 POINT(149.06864966 -35.37733591) \n1 POINT(149.03046282 -35.20202762) \n2 POINT(149.03047333 -35.20156767) \n3 POINT(149.06860919 -35.37833726) \n4 POINT(149.03640841 -35.19777173) \n5 POINT(149.06946494 -35.37908886) \n6 POINT(149.06908395 -35.37882495) \n7 POINT(149.03048051 -35.20185603) \n8 POINT(149.03049843 -35.20169346) \n9 POINT(149.04029706 -35.19999611) \n10 POINT(149.03688520 -35.19795303) \n11 POINT(149.06895786 -35.37862878) \n12 POINT(149.03705032 -35.19796828) \n13 POINT(149.06851657 -35.37815855) \n14 POINT(149.06872290 -35.37847955) \n15 POINT(149.03865604 -35.20113263) \n16 POINT(149.03687042 -35.20395740) \n17 POINT(149.04011870 -35.19989093) \n18 POINT(149.03721725 -35.19805563) \n19 POINT(149.03661902 -35.19784933) \n"
]
],
[
[
"#### 3.1.5. SPARQL querying RDF data\nA graph query, similar to a database SQL query, can traverse the graph and retrieve the same details as the multiple\nloops and Python code above in 3.1.3.",
"_____no_output_____"
]
],
[
[
"q = \"\"\"\nSELECT ?id ?addr ?coords\nWHERE {\n ?uri a gnaf:Address ;\n rdfs:comment ?addr .\n\n ?uri geo:hasGeometry/geo:asWKT ?coords_dirty .\n\n BIND (STRAFTER(STR(?uri), \"address/\") AS ?id)\n BIND (STRAFTER(STR(?coords_dirty), \"4283> \") AS ?coords)\n}\nORDER BY ?id\n\"\"\"\nfor r in g.query(q):\n print(\"{}, {}, {}\".format(r[\"id\"], r[\"addr\"], r[\"coords\"]))",
"GAACT714845933, 6 Packham Place, Charnwood, ACT 2615, POINT(149.03865604 -35.20113263)\nGAACT714845934, 3 Bunker Place, Charnwood, ACT 2615, POINT(149.04011870 -35.19989093)\nGAACT714845935, 26 Jauncey Court, Charnwood, ACT 2615, POINT(149.03640841 -35.19777173)\nGAACT714845936, 17 Geeves Court, Charnwood, ACT 2615, POINT(149.03687042 -35.20395740)\nGAACT714845938, 5 Mcdowall Place, Kambah, ACT 2902, POINT(149.06851657 -35.37815855)\nGAACT714845939, 24 Jauncey Court, Charnwood, ACT 2615, POINT(149.03661902 -35.19784933)\nGAACT714845941, 7 Mcdowall Place, Kambah, ACT 2902, POINT(149.06860919 -35.37833726)\nGAACT714845942, 5 Bunker Place, Charnwood, ACT 2615, POINT(149.04029706 -35.19999611)\nGAACT714845943, 22 Jauncey Court, Charnwood, ACT 2615, POINT(149.03688520 -35.19795303)\nGAACT714845944, 9 Mcdowall Place, Kambah, ACT 2902, POINT(149.06872290 -35.37847955)\nGAACT714845945, 9 Baylis Place, Charnwood, ACT 2615, POINT(149.03047333 -35.20156767)\nGAACT714845946, 11 Mcdowall Place, Kambah, ACT 2902, POINT(149.06895786 -35.37862878)\nGAACT714845947, 20 Jauncey Court, Charnwood, ACT 2615, POINT(149.03705032 -35.19796828)\nGAACT714845949, 13 Mcdowall Place, Kambah, ACT 2902, POINT(149.06908395 -35.37882495)\nGAACT714845950, 7 Baylis Place, Charnwood, ACT 2615, POINT(149.03049843 -35.20169346)\nGAACT714845951, 15 Mcdowall Place, Kambah, ACT 2902, POINT(149.06946494 -35.37908886)\nGAACT714845952, 18 Jauncey Court, Charnwood, ACT 2615, POINT(149.03721725 -35.19805563)\nGAACT714845953, 5 Jamieson Crescent, Kambah, ACT 2902, POINT(149.06864966 -35.37733591)\nGAACT714845954, 5 Baylis Place, Charnwood, ACT 2615, POINT(149.03048051 -35.20185603)\nGAACT714845955, 3 Baylis Place, Charnwood, ACT 2615, POINT(149.03046282 -35.20202762)\n"
]
],
[
[
"## 4. Data 'mash up'\nAdd some fake data to the GNAF data - people count per address.\n\nThe GeoSPARQL model extension used is:\n\n\n\nNote that for real Semantic Web work, the `xxx:` properties and classes would be \"properly defined\", removing any ambiguity of use.",
"_____no_output_____"
]
],
[
[
"import pandas\ndf2 = pandas.read_csv('fake_data.csv')\nprint(df2)",
" GNAF ID Persons\n0 GAACT714845944 3\n1 GAACT714845934 5\n2 GAACT714845943 10\n3 GAACT714845949 1\n4 GAACT714845955 2\n5 GAACT714845935 1\n6 GAACT714845947 4\n7 GAACT714845950 3\n8 GAACT714845933 4\n9 GAACT714845953 2\n10 GAACT714845945 3\n11 GAACT714845946 3\n12 GAACT714845939 4\n13 GAACT714845941 2\n14 GAACT714845942 1\n15 GAACT714845954 0\n16 GAACT714845952 5\n17 GAACT714845938 3\n18 GAACT714845936 4\n19 GAACT714845951 3\n"
]
],
[
[
"Merge DataFrames",
"_____no_output_____"
]
],
[
[
"df3 = pandas.merge(df1, df2)\nprint(df3.head())",
" GNAF ID Address \\\n0 GAACT714845953 5 Jamieson Crescent, Kambah, ACT 2902 \n1 GAACT714845955 3 Baylis Place, Charnwood, ACT 2615 \n2 GAACT714845945 9 Baylis Place, Charnwood, ACT 2615 \n3 GAACT714845941 7 Mcdowall Place, Kambah, ACT 2902 \n4 GAACT714845935 26 Jauncey Court, Charnwood, ACT 2615 \n\n Coordinates Persons \n0 POINT(149.06864966 -35.37733591) 2 \n1 POINT(149.03046282 -35.20202762) 2 \n2 POINT(149.03047333 -35.20156767) 3 \n3 POINT(149.06860919 -35.37833726) 2 \n4 POINT(149.03640841 -35.19777173) 1 \n"
]
],
[
[
"## 5. Spatial Data Conversions & Display\n\nOften you will want to display or export data.\n\n#### 5.1 Display directly in Jupyter\nUsing standard Python plotting (matplotlib).\n\nFirst, extract addresses, longitudes & latitudes into a dataframe using a SPARQL query to build a CSV string.",
"_____no_output_____"
]
],
[
[
"import re\naddresses_csv = \"Address,Longitude,Latitude\\n\"\n\nq = \"\"\"\n SELECT ?addr ?coords\n WHERE {\n ?uri a gnaf:Address ;\n rdfs:comment ?addr .\n\n ?uri geo:hasGeometry/geo:asWKT ?coords .\n\n BIND (STRAFTER(STR(?uri), \"address/\") AS ?id)\n BIND (STRAFTER(STR(?coords_dirty), \"4283> \") AS ?coords)\n }\n ORDER BY ?id\n \"\"\"\nfor r in g.query(q):\n match = re.search(\"POINT\\((\\d+\\.\\d+)\\s(\\-\\d+\\.\\d+)\\)\", r[\"coords\"])\n long = float(match.group(1))\n lat = float(match.group(2))\n addresses_csv += f'\\\"{r[\"addr\"]}\\\",{long},{lat}\\n'\n\nprint(addresses_csv)",
"Address,Longitude,Latitude\n\"6 Packham Place, Charnwood, ACT 2615\",149.03865604,-35.20113263\n\"3 Bunker Place, Charnwood, ACT 2615\",149.0401187,-35.19989093\n\"26 Jauncey Court, Charnwood, ACT 2615\",149.03640841,-35.19777173\n\"17 Geeves Court, Charnwood, ACT 2615\",149.03687042,-35.2039574\n\"5 Mcdowall Place, Kambah, ACT 2902\",149.06851657,-35.37815855\n\"24 Jauncey Court, Charnwood, ACT 2615\",149.03661902,-35.19784933\n\"7 Mcdowall Place, Kambah, ACT 2902\",149.06860919,-35.37833726\n\"5 Bunker Place, Charnwood, ACT 2615\",149.04029706,-35.19999611\n\"22 Jauncey Court, Charnwood, ACT 2615\",149.0368852,-35.19795303\n\"9 Mcdowall Place, Kambah, ACT 2902\",149.0687229,-35.37847955\n\"9 Baylis Place, Charnwood, ACT 2615\",149.03047333,-35.20156767\n\"11 Mcdowall Place, Kambah, ACT 2902\",149.06895786,-35.37862878\n\"20 Jauncey Court, Charnwood, ACT 2615\",149.03705032,-35.19796828\n\"13 Mcdowall Place, Kambah, ACT 2902\",149.06908395,-35.37882495\n\"7 Baylis Place, Charnwood, ACT 2615\",149.03049843,-35.20169346\n\"15 Mcdowall Place, Kambah, ACT 2902\",149.06946494,-35.37908886\n\"18 Jauncey Court, Charnwood, ACT 2615\",149.03721725,-35.19805563\n\"5 Jamieson Crescent, Kambah, ACT 2902\",149.06864966,-35.37733591\n\"5 Baylis Place, Charnwood, ACT 2615\",149.03048051,-35.20185603\n\"3 Baylis Place, Charnwood, ACT 2615\",149.03046282,-35.20202762\n\n"
]
],
[
[
"Read the CSV into a DataFrame.",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nfrom io import StringIO\naddresses_df = pd.read_csv(StringIO(addresses_csv))\n\nprint(addresses_df[\"Longitude\"])",
"0 149.038656\n1 149.040119\n2 149.036408\n3 149.036870\n4 149.068517\n5 149.036619\n6 149.068609\n7 149.040297\n8 149.036885\n9 149.068723\n10 149.030473\n11 149.068958\n12 149.037050\n13 149.069084\n14 149.030498\n15 149.069465\n16 149.037217\n17 149.068650\n18 149.030481\n19 149.030463\nName: Longitude, dtype: float64\n"
]
],
[
[
"Display the first 5 rows of the DataFrame directly using matplotlib.",
"_____no_output_____"
]
],
[
[
"from matplotlib import pyplot as plt\naddresses_df[:5].plot(kind=\"scatter\", x=\"Longitude\", y=\"Latitude\", s=50, figsize=(10,10))\n\nfor i, label in enumerate(addresses_df[:5]):\n plt.annotate(addresses_df[\"Address\"][i], (addresses_df[\"Longitude\"][i], addresses_df[\"Latitude\"][i]))\n \nplt.show()",
"_____no_output_____"
]
],
[
[
"#### 5.2 Convert to common format - GeoJSON\n\nImport Python conversion tools (shapely).",
"_____no_output_____"
]
],
[
[
"import shapely.wkt\nfrom shapely.geometry import MultiPoint\nimport json",
"_____no_output_____"
]
],
[
[
"Loop through the graph using ordinary Python loops, not a query.",
"_____no_output_____"
]
],
[
[
"points_list = []\n\nfor s, p, o in g.triples((None, RDF.type, GNAF.Address)):\n for s2, p2, o2 in g.triples((s, GEO.hasGeometry, None)):\n for s3, p3, o3 in g.triples((o2, GEO.asWKT, None)):\n points_list.append(\n shapely.wkt.loads(str(o3).replace(\"<http://www.opengis.net/def/crs/EPSG/0/4283> \", \"\"))\n )\n\nmp = MultiPoint(points=points_list)\n\ngeojson = shapely.geometry.mapping(mp)\nprint(json.dumps(geojson, indent=4))",
"{\n \"type\": \"MultiPoint\",\n \"coordinates\": [\n [\n 149.06864966,\n -35.37733591\n ],\n [\n 149.03046282,\n -35.20202762\n ],\n [\n 149.03047333,\n -35.20156767\n ],\n [\n 149.06860919,\n -35.37833726\n ],\n [\n 149.03640841,\n -35.19777173\n ],\n [\n 149.06946494,\n -35.37908886\n ],\n [\n 149.06908395,\n -35.37882495\n ],\n [\n 149.03048051,\n -35.20185603\n ],\n [\n 149.03049843,\n -35.20169346\n ],\n [\n 149.04029706,\n -35.19999611\n ],\n [\n 149.0368852,\n -35.19795303\n ],\n [\n 149.06895786,\n -35.37862878\n ],\n [\n 149.03705032,\n -35.19796828\n ],\n [\n 149.06851657,\n -35.37815855\n ],\n [\n 149.0687229,\n -35.37847955\n ],\n [\n 149.03865604,\n -35.20113263\n ],\n [\n 149.03687042,\n -35.2039574\n ],\n [\n 149.0401187,\n -35.19989093\n ],\n [\n 149.03721725,\n -35.19805563\n ],\n [\n 149.03661902,\n -35.19784933\n ]\n ]\n}\n"
]
],
[
[
"Another, better, GeoJSON export - including Feature information.\n\nFirst, build a Python dictionary matching the GeoJSON specification, then export it to JSON.",
"_____no_output_____"
]
],
[
[
"geo_json_features = []\n\n# same query as above\nfor r in g.query(q):\n match = re.search(\"POINT\\((\\d+\\.\\d+)\\s(\\-\\d+\\.\\d+)\\)\", r[\"coords\"])\n long = float(match.group(1))\n lat = float(match.group(2))\n geo_json_features.append({\n \"type\": \"Feature\", \n \"properties\": { \"name\": r[\"addr\"] },\n \"geometry\": { \n \"type\": \"Point\", \n \"coordinates\": [ long, lat ] \n } \n })\n \ngeo_json_data = {\n \"type\": \"FeatureCollection\",\n \"name\": \"test-points-short-named\",\n \"crs\": { \"type\": \"name\", \"properties\": { \"name\": \"urn:ogc:def:crs:OGC:1.3:CRS84\" } },\n \"features\": geo_json_features\n}\n\nimport json\ngeo_json = json.dumps(geo_json_data, indent=4)\nprint(geo_json)",
"{\n \"type\": \"FeatureCollection\",\n \"name\": \"test-points-short-named\",\n \"crs\": {\n \"type\": \"name\",\n \"properties\": {\n \"name\": \"urn:ogc:def:crs:OGC:1.3:CRS84\"\n }\n },\n \"features\": [\n {\n \"type\": \"Feature\",\n \"properties\": {\n \"name\": \"6 Packham Place, Charnwood, ACT 2615\"\n },\n \"geometry\": {\n \"type\": \"Point\",\n \"coordinates\": [\n 149.03865604,\n -35.20113263\n ]\n }\n },\n {\n \"type\": \"Feature\",\n \"properties\": {\n \"name\": \"3 Bunker Place, Charnwood, ACT 2615\"\n },\n \"geometry\": {\n \"type\": \"Point\",\n \"coordinates\": [\n 149.0401187,\n -35.19989093\n ]\n }\n },\n {\n \"type\": \"Feature\",\n \"properties\": {\n \"name\": \"26 Jauncey Court, Charnwood, ACT 2615\"\n },\n \"geometry\": {\n \"type\": \"Point\",\n \"coordinates\": [\n 149.03640841,\n -35.19777173\n ]\n }\n },\n {\n \"type\": \"Feature\",\n \"properties\": {\n \"name\": \"17 Geeves Court, Charnwood, ACT 2615\"\n },\n \"geometry\": {\n \"type\": \"Point\",\n \"coordinates\": [\n 149.03687042,\n -35.2039574\n ]\n }\n },\n {\n \"type\": \"Feature\",\n \"properties\": {\n \"name\": \"5 Mcdowall Place, Kambah, ACT 2902\"\n },\n \"geometry\": {\n \"type\": \"Point\",\n \"coordinates\": [\n 149.06851657,\n -35.37815855\n ]\n }\n },\n {\n \"type\": \"Feature\",\n \"properties\": {\n \"name\": \"24 Jauncey Court, Charnwood, ACT 2615\"\n },\n \"geometry\": {\n \"type\": \"Point\",\n \"coordinates\": [\n 149.03661902,\n -35.19784933\n ]\n }\n },\n {\n \"type\": \"Feature\",\n \"properties\": {\n \"name\": \"7 Mcdowall Place, Kambah, ACT 2902\"\n },\n \"geometry\": {\n \"type\": \"Point\",\n \"coordinates\": [\n 149.06860919,\n -35.37833726\n ]\n }\n },\n {\n \"type\": \"Feature\",\n \"properties\": {\n \"name\": \"5 Bunker Place, Charnwood, ACT 2615\"\n },\n \"geometry\": {\n \"type\": \"Point\",\n \"coordinates\": [\n 149.04029706,\n -35.19999611\n ]\n }\n },\n {\n \"type\": \"Feature\",\n \"properties\": {\n \"name\": \"22 Jauncey Court, Charnwood, ACT 2615\"\n },\n \"geometry\": {\n \"type\": \"Point\",\n \"coordinates\": [\n 149.0368852,\n -35.19795303\n ]\n }\n },\n {\n \"type\": \"Feature\",\n \"properties\": {\n \"name\": \"9 Mcdowall Place, Kambah, ACT 2902\"\n },\n \"geometry\": {\n \"type\": \"Point\",\n \"coordinates\": [\n 149.0687229,\n -35.37847955\n ]\n }\n },\n {\n \"type\": \"Feature\",\n \"properties\": {\n \"name\": \"9 Baylis Place, Charnwood, ACT 2615\"\n },\n \"geometry\": {\n \"type\": \"Point\",\n \"coordinates\": [\n 149.03047333,\n -35.20156767\n ]\n }\n },\n {\n \"type\": \"Feature\",\n \"properties\": {\n \"name\": \"11 Mcdowall Place, Kambah, ACT 2902\"\n },\n \"geometry\": {\n \"type\": \"Point\",\n \"coordinates\": [\n 149.06895786,\n -35.37862878\n ]\n }\n },\n {\n \"type\": \"Feature\",\n \"properties\": {\n \"name\": \"20 Jauncey Court, Charnwood, ACT 2615\"\n },\n \"geometry\": {\n \"type\": \"Point\",\n \"coordinates\": [\n 149.03705032,\n -35.19796828\n ]\n }\n },\n {\n \"type\": \"Feature\",\n \"properties\": {\n \"name\": \"13 Mcdowall Place, Kambah, ACT 2902\"\n },\n \"geometry\": {\n \"type\": \"Point\",\n \"coordinates\": [\n 149.06908395,\n -35.37882495\n ]\n }\n },\n {\n \"type\": \"Feature\",\n \"properties\": {\n \"name\": \"7 Baylis Place, Charnwood, ACT 2615\"\n },\n \"geometry\": {\n \"type\": \"Point\",\n \"coordinates\": [\n 149.03049843,\n -35.20169346\n ]\n }\n },\n {\n \"type\": \"Feature\",\n \"properties\": {\n \"name\": \"15 Mcdowall Place, Kambah, ACT 2902\"\n },\n \"geometry\": {\n \"type\": \"Point\",\n \"coordinates\": [\n 149.06946494,\n -35.37908886\n ]\n }\n },\n {\n \"type\": \"Feature\",\n \"properties\": {\n \"name\": \"18 Jauncey Court, Charnwood, ACT 2615\"\n },\n \"geometry\": {\n \"type\": \"Point\",\n \"coordinates\": [\n 149.03721725,\n -35.19805563\n ]\n }\n },\n {\n \"type\": \"Feature\",\n \"properties\": {\n \"name\": \"5 Jamieson Crescent, Kambah, ACT 2902\"\n },\n \"geometry\": {\n \"type\": \"Point\",\n \"coordinates\": [\n 149.06864966,\n -35.37733591\n ]\n }\n },\n {\n \"type\": \"Feature\",\n \"properties\": {\n \"name\": \"5 Baylis Place, Charnwood, ACT 2615\"\n },\n \"geometry\": {\n \"type\": \"Point\",\n \"coordinates\": [\n 149.03048051,\n -35.20185603\n ]\n }\n },\n {\n \"type\": \"Feature\",\n \"properties\": {\n \"name\": \"3 Baylis Place, Charnwood, ACT 2615\"\n },\n \"geometry\": {\n \"type\": \"Point\",\n \"coordinates\": [\n 149.03046282,\n -35.20202762\n ]\n }\n }\n ]\n}\n"
]
],
[
[
"Export the data and view it in a GeoJSON map viewer, such as http://geojsonviewer.nsspot.net/ or QGIS (desktop_.\n\n## Concluding remarks\n\n* Semantic Web, realised through Linked Data, builds a global machine-readable data system\n* the RDF data structure is used\n * to link things\n * to define things, and the links\n* specialised parts of the Sem Web can represent a/any domain\n * e.g. spatial\n * e.g. Addresses\n* powerful graph pattern matching queries, SPARQL, can be used to subset (federated) Sem Web data\n* RDF manipulation libraries exist\n * can convert to other, common forms, e.g. CSV GeoJSON\n* _do as much data science work as you can with well-defined models!_\n\n## License\nAll the content in this repository is licensed under the [CC BY 4.0 license](https://creativecommons.org/licenses/by/4.0/). Basically, you can:\n\n* copy and redistribute the material in any medium or format\n* remix, transform, and build upon the material for any purpose, even commercially\n\nYou just need to:\n\n* give appropriate credit, provide a link to the license, and indicate if changes were made\n* not apply legal terms or technological measures that legally restrict others from doing anything the license permits\n\n## Contact Information\n**Dr Nicholas J. Car**<br />\n*Data Systems Architect*<br />\n[SURROUND Australia Pty Ltd](https://surroundaustralia.com)<br />\n<[email protected]><br />\nGitHub: [nicholascar](https://github.com/nicholascar)<br />\nORCID: <https://orcid.org/0000-0002-8742-7730><br />",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
cbea68ae71bfd5af237d42bf984ad7d718c5a140
| 221,227 |
ipynb
|
Jupyter Notebook
|
2-Working-With-Data/07-python/notebook.ipynb
|
qxdn/Data-Science-For-Beginners
|
aa431ef23a71a2cb8b33cede2d1d731d1e19ae08
|
[
"MIT"
] | null | null | null |
2-Working-With-Data/07-python/notebook.ipynb
|
qxdn/Data-Science-For-Beginners
|
aa431ef23a71a2cb8b33cede2d1d731d1e19ae08
|
[
"MIT"
] | null | null | null |
2-Working-With-Data/07-python/notebook.ipynb
|
qxdn/Data-Science-For-Beginners
|
aa431ef23a71a2cb8b33cede2d1d731d1e19ae08
|
[
"MIT"
] | null | null | null | 146.994684 | 79,734 | 0.863005 |
[
[
[
"## Basic Pandas Examples\n\nThis notebook will walk you through some very basic Pandas concepts. We will start with importing typical data science libraries:",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt",
"_____no_output_____"
]
],
[
[
"### Series\n\nSeries is like a list or 1D-array, but with index. All operations are index-aligned.",
"_____no_output_____"
]
],
[
[
"a = pd.Series(range(1,10))\nb = pd.Series([\"I\",\"like\",\"to\",\"use\",\"Python\",\"and\",\"Pandas\",\"very\",\"much\"],index=range(0,9))\nprint(a,b)",
"0 1\n1 2\n2 3\n3 4\n4 5\n5 6\n6 7\n7 8\n8 9\ndtype: int64 0 I\n1 like\n2 to\n3 use\n4 Python\n5 and\n6 Pandas\n7 very\n8 much\ndtype: object\n"
]
],
[
[
"One of the frequent usage of series is **time series**. In time series, index has a special structure - typically a range of dates or datetimes. We can create such an index with `pd.date_range`.\n\nSuppose we have a series that shows the amount of product bought every day, and we know that every sunday we also need to take one item for ourselves. Here is how to model that using series:",
"_____no_output_____"
]
],
[
[
"start_date = \"Jan 1, 2020\"\nend_date = \"Dec 31, 2020\"\nidx = pd.date_range(start_date,end_date)\nprint(f\"Length of index is {len(idx)}\")\nitems_sold = pd.Series(np.random.randint(25,50,size=len(idx)),index=idx)\nitems_sold.plot(figsize=(10,3))\nplt.show()",
"Length of index is 366\n"
],
[
"additional_items = pd.Series(10,index=pd.date_range(start_date,end_date,freq=\"W\"))\nprint(f\"Additional items (10 item each week):\\n{additional_items}\")\ntotal_items = items_sold+additional_items\nprint(f\"Total items (sum of two series):\\n{total_items}\")",
"Additional items (10 item each week):\n2020-01-05 10\n2020-01-12 10\n2020-01-19 10\n2020-01-26 10\n2020-02-02 10\n2020-02-09 10\n2020-02-16 10\n2020-02-23 10\n2020-03-01 10\n2020-03-08 10\n2020-03-15 10\n2020-03-22 10\n2020-03-29 10\n2020-04-05 10\n2020-04-12 10\n2020-04-19 10\n2020-04-26 10\n2020-05-03 10\n2020-05-10 10\n2020-05-17 10\n2020-05-24 10\n2020-05-31 10\n2020-06-07 10\n2020-06-14 10\n2020-06-21 10\n2020-06-28 10\n2020-07-05 10\n2020-07-12 10\n2020-07-19 10\n2020-07-26 10\n2020-08-02 10\n2020-08-09 10\n2020-08-16 10\n2020-08-23 10\n2020-08-30 10\n2020-09-06 10\n2020-09-13 10\n2020-09-20 10\n2020-09-27 10\n2020-10-04 10\n2020-10-11 10\n2020-10-18 10\n2020-10-25 10\n2020-11-01 10\n2020-11-08 10\n2020-11-15 10\n2020-11-22 10\n2020-11-29 10\n2020-12-06 10\n2020-12-13 10\n2020-12-20 10\n2020-12-27 10\nFreq: W-SUN, dtype: int64\nTotal items (sum of two series):\n2020-01-01 NaN\n2020-01-02 NaN\n2020-01-03 NaN\n2020-01-04 NaN\n2020-01-05 45.0\n ... \n2020-12-27 49.0\n2020-12-28 NaN\n2020-12-29 NaN\n2020-12-30 NaN\n2020-12-31 NaN\nLength: 366, dtype: float64\n"
]
],
[
[
"As you can see, we are having problems here, because in the weekly series non-mentioned days are considered to be missing (`NaN`), and adding `NaN` to a number gives us `NaN`. In order to get correct result, we need to specify `fill_value` when adding series:",
"_____no_output_____"
]
],
[
[
"total_items = items_sold.add(additional_items,fill_value=0)\nprint(total_items)\ntotal_items.plot(figsize=(10,3))\nplt.show()",
"2020-01-01 27.0\n2020-01-02 49.0\n2020-01-03 34.0\n2020-01-04 29.0\n2020-01-05 45.0\n ... \n2020-12-27 49.0\n2020-12-28 26.0\n2020-12-29 49.0\n2020-12-30 48.0\n2020-12-31 35.0\nLength: 366, dtype: float64\n"
],
[
"monthly = total_items.resample(\"1M\").mean()\nax = monthly.plot(kind='bar',figsize=(10,3))\nax.set_xticklabels([x.strftime(\"%b-%Y\") for x in monthly.index], rotation=45)\nplt.show()",
"_____no_output_____"
]
],
[
[
"## DataFrame\n\nA dataframe is essentially a collection of series with the same index. We can combine several series together into a dataframe. Given `a` and `b` series defined above:",
"_____no_output_____"
]
],
[
[
"df = pd.DataFrame([a,b])\ndf",
"_____no_output_____"
]
],
[
[
"We can also use Series as columns, and specify column names using dictionary:",
"_____no_output_____"
]
],
[
[
"df = pd.DataFrame({ 'A' : a, 'B' : b })\ndf",
"_____no_output_____"
]
],
[
[
"The same result can be achieved by transposing (and then renaming columns, to match the previous example):",
"_____no_output_____"
]
],
[
[
"pd.DataFrame([a,b]).T.rename(columns={ 0 : 'A', 1 : 'B' })",
"_____no_output_____"
]
],
[
[
"**Selecting columns** from DataFrame can be done like this:",
"_____no_output_____"
]
],
[
[
"print(f\"Column A (series):\\n{df['A']}\")\nprint(f\"Columns B and A (DataFrame):\\n{df[['B','A']]}\")",
"Column A (series):\n0 1\n1 2\n2 3\n3 4\n4 5\n5 6\n6 7\n7 8\n8 9\nName: A, dtype: int64\nColumns B and A (DataFrame):\n B A\n0 I 1\n1 like 2\n2 to 3\n3 use 4\n4 Python 5\n5 and 6\n6 Pandas 7\n7 very 8\n8 much 9\n"
]
],
[
[
"**Selecting rows** based on filter expression:",
"_____no_output_____"
]
],
[
[
"df[df['A']<5]",
"_____no_output_____"
]
],
[
[
"The way it works is that expression `df['A']<5` returns a boolean series, which indicates whether expression is `True` or `False` for each elemens of the series. When series is used as an index, it returns subset of rows in the DataFrame. Thus it is not possible to use arbitrary Python boolean expression, for example, writing `df[df['A']>5 and df['A']<7]` would be wrong. Instead, you should use special `&` operation on boolean series:",
"_____no_output_____"
]
],
[
[
"df[(df['A']>5) & (df['A']<7)]",
"_____no_output_____"
]
],
[
[
"**Creating new computable columns**. We can easily create new computable columns for our DataFrame by using intuitive expressions. The code below calculates divergence of A from its mean value.",
"_____no_output_____"
]
],
[
[
"df['DivA'] = df['A']-df['A'].mean()\ndf",
"_____no_output_____"
]
],
[
[
"What actually happens is we are computing a series, and then assigning this series to the left-hand-side, creating another column.",
"_____no_output_____"
]
],
[
[
"# WRONG: df['ADescr'] = \"Low\" if df['A'] < 5 else \"Hi\"\ndf['LenB'] = len(df['B']) # Wrong result",
"_____no_output_____"
],
[
"df['LenB'] = df['B'].apply(lambda x: len(x))\n# or\ndf['LenB'] = df['B'].apply(len)\ndf",
"_____no_output_____"
]
],
[
[
"**Selecting rows based on numbers** can be done using `iloc` construct. For example, to select first 5 rows from the DataFrame:",
"_____no_output_____"
]
],
[
[
"df.iloc[:5]",
"_____no_output_____"
]
],
[
[
"**Grouping** is often used to get a result similar to *pivot tables* in Excel. Suppose that we want to compute mean value of column `A` for each given number of `LenB`. Then we can group our DataFrame by `LenB`, and call `mean`:",
"_____no_output_____"
]
],
[
[
"df.groupby(by='LenB').mean()",
"_____no_output_____"
]
],
[
[
"If we need to compute mean and the number of elements in the group, then we can use more complex `aggregate` function:",
"_____no_output_____"
]
],
[
[
"df.groupby(by='LenB') \\\n .aggregate({ 'DivA' : len, 'A' : lambda x: x.mean() }) \\\n .rename(columns={ 'DivA' : 'Count', 'A' : 'Mean'})",
"_____no_output_____"
]
],
[
[
"## Printing and Plotting\n\nData Scientist often has to explore the data, thus it is important to be able to visualize it. When DataFrame is big, manytimes we want just to make sure we are doing everything correctly by printing out the first few rows. This can be done by calling `df.head()`. If you are running it from Jupyter Notebook, it will print out the DataFrame in a nice tabular form.",
"_____no_output_____"
]
],
[
[
"df.head()",
"_____no_output_____"
]
],
[
[
"\nWe have also seen the usage of `plot` function to visualize some columns. While `plot` is very useful for many tasks, and supports many different graph types via `kind=` parameter, you can always use raw `matplotlib` library to plot something more complex. We will cover data visualization in detail in separate course lessons.\n",
"_____no_output_____"
]
],
[
[
"df['A'].plot()\nplt.show()",
"_____no_output_____"
],
[
"df['A'].plot(kind='bar')\nplt.show()",
"_____no_output_____"
]
],
[
[
"\nThis overview covers most important concepts of Pandas, however, the library is very rich, and there is no limit to what you can do with it! Let's now apply this knowledge for solving specific problem.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
cbea6ebc74967f2950b69042d320418e7d2653dd
| 363,857 |
ipynb
|
Jupyter Notebook
|
module4/assignment_regression_classification_4e.ipynb
|
JimKing100/DS-Unit-2-Regression-Classification
|
8343e6b78be2e129cde6b5a0be01f5ed6eaae851
|
[
"MIT"
] | null | null | null |
module4/assignment_regression_classification_4e.ipynb
|
JimKing100/DS-Unit-2-Regression-Classification
|
8343e6b78be2e129cde6b5a0be01f5ed6eaae851
|
[
"MIT"
] | null | null | null |
module4/assignment_regression_classification_4e.ipynb
|
JimKing100/DS-Unit-2-Regression-Classification
|
8343e6b78be2e129cde6b5a0be01f5ed6eaae851
|
[
"MIT"
] | null | null | null | 158.820166 | 19,040 | 0.854734 |
[
[
[
"<a href=\"https://colab.research.google.com/github/JimKing100/DS-Unit-2-Regression-Classification/blob/master/module4/assignment_regression_classification_4e.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"# Installs\n%%capture\n!pip install --upgrade category_encoders plotly",
"_____no_output_____"
],
[
"# Imports\nimport os, sys\n\nos.chdir('/content')\n!git init .\n!git remote add origin https://github.com/LambdaSchool/DS-Unit-2-Regression-Classification.git\n!git pull origin master\n \nos.chdir('module4')",
"Reinitialized existing Git repository in /content/.git/\nfatal: remote origin already exists.\nFrom https://github.com/LambdaSchool/DS-Unit-2-Regression-Classification\n * branch master -> FETCH_HEAD\nAlready up to date.\n"
],
[
"# Disable warning\nimport warnings\nwarnings.filterwarnings(action='ignore', category=FutureWarning, module='numpy')",
"_____no_output_____"
]
],
[
[
"### Load Data",
"_____no_output_____"
]
],
[
[
"import pandas as pd\n\ntrain_features = pd.read_csv('../data/tanzania/train_features.csv')\ntrain_labels = pd.read_csv('../data/tanzania/train_labels.csv')\ntest_features = pd.read_csv('../data/tanzania/test_features.csv')\nsample_submission = pd.read_csv('../data/tanzania/sample_submission.csv')\n\nassert train_features.shape == (59400, 40)\nassert train_labels.shape == (59400, 2)\nassert test_features.shape == (14358, 40)\nassert sample_submission.shape == (14358, 2)",
"_____no_output_____"
]
],
[
[
"### Train/Validate/Test Split",
"_____no_output_____"
]
],
[
[
"# Load initial train features and labels\nfrom sklearn.model_selection import train_test_split\nX_train = train_features\ny_train = train_labels['status_group']\n\nX_train.shape, y_train.shape",
"_____no_output_____"
],
[
"# Split the initial train features and labels 80% into new train and new validation\nX_train, X_val, y_train, y_val = train_test_split(\n X_train, y_train, train_size = 0.80, test_size = 0.20,\n stratify = y_train, random_state=42\n)\n\nX_train.shape, X_val.shape, y_train.shape, y_val.shape",
"_____no_output_____"
],
[
"# Check values of new train labels\ny_train.value_counts(normalize=True)",
"_____no_output_____"
],
[
"# Check values of new validation labels\ny_val.value_counts(normalize=True)",
"_____no_output_____"
]
],
[
[
"### One-Hot Encoding - Quantity",
"_____no_output_____"
]
],
[
[
"# Check values of quantity feature\nX_train['quantity'].value_counts(normalize=True)",
"_____no_output_____"
],
[
"# Recombine X_train and y_train, for exploratory data analysis\ntrain = X_train.copy()\ntrain['status_group'] = y_train\n\ntrain.groupby('quantity')['status_group'].value_counts(normalize=True)",
"_____no_output_____"
],
[
"# Plot the values, dry shows a strong relationship to functional\nimport matplotlib.pyplot as plt\nimport seaborn as sns\n\ntrain['functional']= (train['status_group'] == 'functional').astype(int)\ntrain[['status_group', 'functional']]\n\nsns.catplot(x='quantity', y='functional', data=train, kind='bar', color='grey')\nplt.title('% of Waterpumps Functional by Water Quantity')",
"_____no_output_____"
]
],
[
[
"### One-Hot Encoding - Waterpoint Type",
"_____no_output_____"
]
],
[
[
"X_train['waterpoint_type'].value_counts(normalize=True)",
"_____no_output_____"
],
[
"# Recombine X_train and y_train, for exploratory data analysis\ntrain = X_train.copy()\ntrain['status_group'] = y_train\ntrain.groupby('waterpoint_type')['status_group'].value_counts(normalize=True)\ntrain['functional']= (train['status_group'] == 'functional').astype(int)\ntrain[['status_group', 'functional']]\n\nsns.catplot(x='waterpoint_type', y='functional', data=train, kind='bar', color='grey')\nplt.title('% of Waterpumps Functional by Waterpoint Type')",
"_____no_output_____"
]
],
[
[
"### One-Hot Encoding - Extraction Type",
"_____no_output_____"
]
],
[
[
"X_train['extraction_type'].value_counts(normalize=True)\ntrain = X_train.copy()\ntrain['status_group'] = y_train\ntrain.groupby('extraction_type')['status_group'].value_counts(normalize=True)\ntrain['functional']= (train['status_group'] == 'functional').astype(int)\ntrain[['status_group', 'functional']]\n\nsns.catplot(x='extraction_type', y='functional', data=train, kind='bar', color='grey')\nplt.title('% of Waterpumps Functional by Extraction Type')",
"_____no_output_____"
]
],
[
[
"### Bin and One-Hot Encoding - Installer",
"_____no_output_____"
]
],
[
[
"X_train['installer'] = X_train['installer'].str.lower()\nX_val['installer'] = X_val['installer'].str.lower()\n\nX_train['installer'] = X_train['installer'].str.lower()\nX_val['installer'] = X_val['installer'].str.lower()\n\nX_train['installer'] = X_train['installer'].str.replace('danid', 'danida')\nX_val['installer'] = X_val['installer'].str.replace('danid', 'danida')\n\nX_train['installer'] = X_train['installer'].str.replace('disti', 'district council')\nX_val['installer'] = X_val['installer'].str.replace('disti', 'district council')\n\nX_train['installer'] = X_train['installer'].str.replace('commu', 'community')\nX_val['installer'] = X_val['installer'].str.replace('commu', 'community')\n\nX_train['installer'] = X_train['installer'].str.replace('central government', 'government')\nX_val['installer'] = X_val['installer'].str.replace('central government', 'government')\n\nX_train['installer'] = X_train['installer'].str.replace('kkkt _ konde and dwe', 'kkkt')\nX_val['installer'] = X_val['installer'].str.replace('kkkt _ konde and dwe', 'kkkt')\n\nX_train['installer'].value_counts(normalize=True)\ntop10 = X_train['installer'].value_counts()[:5].index\nX_train.loc[~X_train['installer'].isin(top10), 'installer'] = 'Other'\nX_val.loc[~X_val['installer'].isin(top10), 'installer'] = 'Other'\n\ntrain = X_train.copy()\ntrain['status_group'] = y_train\n\ntrain.groupby('installer')['status_group'].value_counts(normalize=True)",
"/usr/local/lib/python3.6/dist-packages/ipykernel_launcher.py:1: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n \"\"\"Entry point for launching an IPython kernel.\n/usr/local/lib/python3.6/dist-packages/ipykernel_launcher.py:2: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n \n/usr/local/lib/python3.6/dist-packages/ipykernel_launcher.py:4: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n after removing the cwd from sys.path.\n/usr/local/lib/python3.6/dist-packages/ipykernel_launcher.py:5: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n \"\"\"\n/usr/local/lib/python3.6/dist-packages/ipykernel_launcher.py:7: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n import sys\n/usr/local/lib/python3.6/dist-packages/ipykernel_launcher.py:8: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n \n/usr/local/lib/python3.6/dist-packages/ipykernel_launcher.py:10: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n # Remove the CWD from sys.path while we load stuff.\n/usr/local/lib/python3.6/dist-packages/ipykernel_launcher.py:11: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n # This is added back by InteractiveShellApp.init_path()\n/usr/local/lib/python3.6/dist-packages/ipykernel_launcher.py:13: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n del sys.path[0]\n/usr/local/lib/python3.6/dist-packages/ipykernel_launcher.py:14: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n \n/usr/local/lib/python3.6/dist-packages/ipykernel_launcher.py:16: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n app.launch_new_instance()\n/usr/local/lib/python3.6/dist-packages/ipykernel_launcher.py:17: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n/usr/local/lib/python3.6/dist-packages/ipykernel_launcher.py:19: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n/usr/local/lib/python3.6/dist-packages/ipykernel_launcher.py:20: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n/usr/local/lib/python3.6/dist-packages/pandas/core/indexing.py:543: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n self.obj[item] = s\n"
],
[
"train['functional']= (train['status_group'] == 'functional').astype(int)\ntrain[['status_group', 'functional']]\n\nsns.catplot(x='installer', y='functional', data=train, kind='bar', color='grey')\nplt.title('% of Waterpumps Functional by Installer')",
"_____no_output_____"
]
],
[
[
"### New Feature - Pump Age",
"_____no_output_____"
]
],
[
[
"X_train['pump_age'] = 2013 - X_train['construction_year']\nX_train.loc[X_train['pump_age'] == 2013, 'pump_age'] = 0\nX_val['pump_age'] = 2013 - X_val['construction_year']\nX_val.loc[X_val['pump_age'] == 2013, 'pump_age'] = 0\n\nX_train.loc[X_train['pump_age'] == 0, 'pump_age'] = 10\nX_val.loc[X_val['pump_age'] == 0, 'pump_age'] = 10\n\ntrain = X_train.copy()\ntrain['status_group'] = y_train\n\ntrain.groupby('pump_age')['status_group'].value_counts(normalize=True)\ntrain['functional']= (train['status_group'] == 'functional').astype(int)\ntrain[['status_group', 'functional']]\n\nsns.catplot(x='pump_age', y='functional', data=train, kind='bar', color='grey')\nplt.title('% of Waterpumps Functional by Pump Age')",
"/usr/local/lib/python3.6/dist-packages/ipykernel_launcher.py:1: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n \"\"\"Entry point for launching an IPython kernel.\n/usr/local/lib/python3.6/dist-packages/pandas/core/indexing.py:543: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n self.obj[item] = s\n/usr/local/lib/python3.6/dist-packages/ipykernel_launcher.py:3: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n This is separate from the ipykernel package so we can avoid doing imports until\n"
]
],
[
[
"### Bin and One-Hot Encoding - Funder",
"_____no_output_____"
]
],
[
[
"X_train['funder'] = X_train['funder'].str.lower()\nX_val['funder'] = X_val['funder'].str.lower()\n\nX_train['funder'] = X_train['funder'].str[:3]\nX_val['funder'] = X_val['funder'].str[:3]\n\nX_train['funder'].value_counts(normalize=True)\ntop10 = X_train['funder'].value_counts()[:20].index\nX_train.loc[~X_train['funder'].isin(top10), 'funder'] = 'Other'\nX_val.loc[~X_val['funder'].isin(top10), 'funder'] = 'Other'\n\ntrain = X_train.copy()\ntrain['status_group'] = y_train\n\ntrain.groupby('funder')['status_group'].value_counts(normalize=True)\n\ntrain['functional']= (train['status_group'] == 'functional').astype(int)\ntrain[['status_group', 'functional']]\n\nsns.catplot(x='funder', y='functional', data=train, kind='bar', color='grey')\nplt.title('% of Waterpumps Functional by Funder')",
"/usr/local/lib/python3.6/dist-packages/ipykernel_launcher.py:1: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n \"\"\"Entry point for launching an IPython kernel.\n/usr/local/lib/python3.6/dist-packages/ipykernel_launcher.py:2: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n \n/usr/local/lib/python3.6/dist-packages/ipykernel_launcher.py:4: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n after removing the cwd from sys.path.\n/usr/local/lib/python3.6/dist-packages/ipykernel_launcher.py:5: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n \"\"\"\n/usr/local/lib/python3.6/dist-packages/pandas/core/indexing.py:543: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n self.obj[item] = s\n"
]
],
[
[
"### One-Hot Encoding - Water Quality",
"_____no_output_____"
]
],
[
[
"X_train['water_quality'].value_counts(normalize=True)\ntrain = X_train.copy()\ntrain['status_group'] = y_train\ntrain.groupby('water_quality')['status_group'].value_counts(normalize=True)\ntrain['functional']= (train['status_group'] == 'functional').astype(int)\ntrain[['status_group', 'functional']]\n\nsns.catplot(x='water_quality', y='functional', data=train, kind='bar', color='grey')\nplt.title('% of Waterpumps Functional by Water Quality')",
"_____no_output_____"
]
],
[
[
"### One-Hot Encoding - Basin",
"_____no_output_____"
]
],
[
[
"X_train['basin'].value_counts(normalize=True)\ntrain = X_train.copy()\ntrain['status_group'] = y_train\ntrain.groupby('basin')['status_group'].value_counts(normalize=True)\ntrain['functional']= (train['status_group'] == 'functional').astype(int)\ntrain[['status_group', 'functional']]\n\nsns.catplot(x='basin', y='functional', data=train, kind='bar', color='grey')\nplt.title('% of Waterpumps Functional by Basin')",
"_____no_output_____"
]
],
[
[
"### One-Hot Encoding - Region",
"_____no_output_____"
]
],
[
[
"X_train['region'].value_counts(normalize=True)\ntrain = X_train.copy()\ntrain['status_group'] = y_train\ntrain.groupby('region')['status_group'].value_counts(normalize=True)\ntrain['functional']= (train['status_group'] == 'functional').astype(int)\ntrain[['status_group', 'functional']]\n\nsns.catplot(x='region', y='functional', data=train, kind='bar', color='grey')\nplt.title('% of Waterpumps Functional by Region')",
"_____no_output_____"
]
],
[
[
"### Use Mean for GPS Height Missing Values",
"_____no_output_____"
]
],
[
[
"X_train.loc[X_train['gps_height'] == 0, 'gps_height'] = X_train['gps_height'].mean()\nX_val.loc[X_val['gps_height'] == 0, 'gps_height'] = X_val['gps_height'].mean()\n\ntrain = X_train.copy()\ntrain['status_group'] = y_train\n\ntrain.groupby('gps_height')['status_group'].value_counts(normalize=True)\ntrain['functional']= (train['status_group'] == 'functional').astype(int)\ntrain[['status_group', 'functional']];\n\n#sns.catplot(x='amount_tsh', y='functional', data=train, kind='bar', color='grey')\n#plt.title('% of Waterpumps Functional by Pump Age')",
"/usr/local/lib/python3.6/dist-packages/pandas/core/indexing.py:543: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n self.obj[item] = s\n"
]
],
[
[
"### One-Hot Encoding - Payment",
"_____no_output_____"
]
],
[
[
"X_train['payment'].value_counts(normalize=True)\ntrain = X_train.copy()\ntrain['status_group'] = y_train\ntrain.groupby('payment')['status_group'].value_counts(normalize=True)\ntrain['functional']= (train['status_group'] == 'functional').astype(int)\ntrain[['status_group', 'functional']]\n\nsns.catplot(x='payment', y='functional', data=train, kind='bar', color='grey')\nplt.title('% of Waterpumps Functional by Payment')",
"_____no_output_____"
]
],
[
[
"### One-Hot Encoded - Source",
"_____no_output_____"
]
],
[
[
"X_train['source'].value_counts(normalize=True)\ntrain = X_train.copy()\ntrain['status_group'] = y_train\ntrain.groupby('source')['status_group'].value_counts(normalize=True)\ntrain['functional']= (train['status_group'] == 'functional').astype(int)\ntrain[['status_group', 'functional']]\n\nsns.catplot(x='source', y='functional', data=train, kind='bar', color='grey')\nplt.title('% of Waterpumps Functional by Source')",
"_____no_output_____"
]
],
[
[
"### Bin and One-Hot Encoded - LGA",
"_____no_output_____"
]
],
[
[
"X_train['lga'].value_counts(normalize=True)\ntop10 = X_train['lga'].value_counts()[:10].index\nX_train.loc[~X_train['lga'].isin(top10), 'lga'] = 'Other'\nX_val.loc[~X_val['lga'].isin(top10), 'lga'] = 'Other'\n\ntrain = X_train.copy()\ntrain['status_group'] = y_train\n\ntrain.groupby('lga')['status_group'].value_counts(normalize=True)\n\ntrain['functional']= (train['status_group'] == 'functional').astype(int)\ntrain[['status_group', 'functional']]\n\nsns.catplot(x='lga', y='functional', data=train, kind='bar', color='grey')\nplt.title('% of Waterpumps Functional by LGA')",
"/usr/local/lib/python3.6/dist-packages/pandas/core/indexing.py:543: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n self.obj[item] = s\n"
]
],
[
[
"### Bin and One-Hot Encoded - Ward",
"_____no_output_____"
]
],
[
[
"X_train['ward'].value_counts(normalize=True)\ntop10 = X_train['ward'].value_counts()[:20].index\nX_train.loc[~X_train['ward'].isin(top10), 'ward'] = 'Other'\nX_val.loc[~X_val['ward'].isin(top10), 'ward'] = 'Other'\n\ntrain = X_train.copy()\ntrain['status_group'] = y_train\n\ntrain.groupby('ward')['status_group'].value_counts(normalize=True)\n\ntrain['functional']= (train['status_group'] == 'functional').astype(int)\ntrain[['status_group', 'functional']]\n\nsns.catplot(x='ward', y='functional', data=train, kind='bar', color='grey')\nplt.title('% of Waterpumps Functional by Ward')",
"/usr/local/lib/python3.6/dist-packages/pandas/core/indexing.py:543: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n self.obj[item] = s\n"
]
],
[
[
"### One-Hot Encode - Scheme Management",
"_____no_output_____"
]
],
[
[
"X_train['scheme_management'].value_counts(normalize=True)\ntrain = X_train.copy()\ntrain['status_group'] = y_train\ntrain.groupby('scheme_management')['status_group'].value_counts(normalize=True)\ntrain['functional']= (train['status_group'] == 'functional').astype(int)\ntrain[['status_group', 'functional']]\n\nsns.catplot(x='scheme_management', y='functional', data=train, kind='bar', color='grey')\nplt.title('% of Waterpumps Functional by Scheme Management')",
"_____no_output_____"
]
],
[
[
"### One-Hot Encode - Management",
"_____no_output_____"
]
],
[
[
"X_train['management'].value_counts(normalize=True)\ntrain = X_train.copy()\ntrain['status_group'] = y_train\ntrain.groupby('management')['status_group'].value_counts(normalize=True)\ntrain['functional']= (train['status_group'] == 'functional').astype(int)\ntrain[['status_group', 'functional']]\n\nsns.catplot(x='management', y='functional', data=train, kind='bar', color='grey')\nplt.title('% of Waterpumps Functional by Management')",
"_____no_output_____"
]
],
[
[
"### Create a Region/District Feature",
"_____no_output_____"
]
],
[
[
"X_train['region_code'].value_counts(normalize=True)\ntrain = X_train.copy()\ntrain['status_group'] = y_train\ntrain.groupby('region_code')['status_group'].value_counts(normalize=True)\ntrain['functional']= (train['status_group'] == 'functional').astype(int)\ntrain[['status_group', 'functional']]\n\nsns.catplot(x='region_code', y='functional', data=train, kind='bar', color='grey')\nplt.title('% of Waterpumps Functional by Region Code')",
"_____no_output_____"
],
[
"X_train['region_district'] = X_train['region_code'].astype(str) + X_train['district_code'].astype(str)\nX_val['region_district'] = X_val['region_code'].astype(str) + X_val['district_code'].astype(str)\n\ntrain = X_train.copy()\ntrain['status_group'] = y_train\n\ntrain.groupby('region_district')['status_group'].value_counts(normalize=True)\ntrain['functional']= (train['status_group'] == 'functional').astype(int)\ntrain[['status_group', 'functional']]\n\nsns.catplot(x='region_district', y='functional', data=train, kind='bar', color='grey')\nplt.title('% of Waterpumps Functional by Region/District')",
"/usr/local/lib/python3.6/dist-packages/ipykernel_launcher.py:1: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n \"\"\"Entry point for launching an IPython kernel.\n/usr/local/lib/python3.6/dist-packages/ipykernel_launcher.py:2: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n \n"
]
],
[
[
"### One-Hot Encode - Subvillage",
"_____no_output_____"
]
],
[
[
"X_train['subvillage'].value_counts(normalize=True)\ntop10 = X_train['subvillage'].value_counts()[:10].index\nX_train.loc[~X_train['subvillage'].isin(top10), 'subvillage'] = 'Other'\nX_val.loc[~X_val['subvillage'].isin(top10), 'subvillage'] = 'Other'\n\ntrain = X_train.copy()\ntrain['status_group'] = y_train\n\ntrain.groupby('subvillage')['status_group'].value_counts(normalize=True)\n\ntrain['functional']= (train['status_group'] == 'functional').astype(int)\ntrain[['status_group', 'functional']]\n\nsns.catplot(x='subvillage', y='functional', data=train, kind='bar', color='grey')\nplt.title('% of Waterpumps Functional by Subvillage')",
"/usr/local/lib/python3.6/dist-packages/pandas/core/indexing.py:543: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n self.obj[item] = s\n"
]
],
[
[
"### One-Hot Encoding - Water Quality",
"_____no_output_____"
]
],
[
[
"X_train['water_quality'].value_counts(normalize=True)\ntrain = X_train.copy()\ntrain['status_group'] = y_train\ntrain.groupby('water_quality')['status_group'].value_counts(normalize=True)\ntrain['functional']= (train['status_group'] == 'functional').astype(int)\ntrain[['status_group', 'functional']]\n\nsns.catplot(x='water_quality', y='functional', data=train, kind='bar', color='grey')\nplt.title('% of Waterpumps Functional by Quality')",
"_____no_output_____"
]
],
[
[
"### Lat/Long Cleanup",
"_____no_output_____"
]
],
[
[
"#test['region'].value_counts()\naverage_lat = X_train.groupby('region').latitude.mean().reset_index()\naverage_long = X_train.groupby('region').longitude.mean().reset_index()\n\nshinyanga_lat = average_lat.loc[average_lat['region'] == 'Shinyanga', 'latitude']\nshinyanga_long = average_long.loc[average_lat['region'] == 'Shinyanga', 'longitude']\n\nX_train.loc[(X_train['region'] == 'Shinyanga') & (X_train['latitude'] > -1), ['latitude']] = shinyanga_lat[17]\nX_train.loc[(X_train['region'] == 'Shinyanga') & (X_train['longitude'] == 0), ['longitude']] = shinyanga_long[17]\n\nmwanza_lat = average_lat.loc[average_lat['region'] == 'Mwanza', 'latitude']\nmwanza_long = average_long.loc[average_lat['region'] == 'Mwanza', 'longitude']\n\nX_train.loc[(X_train['region'] == 'Mwanza') & (X_train['latitude'] > -1), ['latitude']] = mwanza_lat[13]\nX_train.loc[(X_train['region'] == 'Mwanza') & (X_train['longitude'] == 0) , ['longitude']] = mwanza_long[13]\n",
"/usr/local/lib/python3.6/dist-packages/pandas/core/indexing.py:543: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n self.obj[item] = s\n"
]
],
[
[
"### Impute Amount TSH",
"_____no_output_____"
]
],
[
[
"def tsh_calc(tsh, source, base, waterpoint):\n if tsh == 0:\n if (source, base, waterpoint) in tsh_dict:\n new_tsh = tsh_dict[source, base, waterpoint]\n return new_tsh\n else:\n return tsh\n \n return tsh",
"_____no_output_____"
],
[
"temp = X_train[X_train['amount_tsh'] != 0].groupby(['source_class',\n 'basin',\n 'waterpoint_type_group'])['amount_tsh'].mean()\n\ntsh_dict = dict(temp)\nX_train['amount_tsh'] = X_train.apply(lambda x: tsh_calc(x['amount_tsh'], x['source_class'], x['basin'], x['waterpoint_type_group']), axis=1)\n#X_train",
"/usr/local/lib/python3.6/dist-packages/ipykernel_launcher.py:6: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n \n"
],
[
"#X_train.loc[X_train['amount_tsh'] == 0, 'amount_tsh'] = X_train['amount_tsh'].median()\n#X_val.loc[X_val['amount_tsh'] == 0, 'amount_tsh'] = X_val['amount_tsh'].median()\n\ntrain = X_train.copy()\ntrain['status_group'] = y_train\n\ntrain.groupby('amount_tsh')['status_group'].value_counts(normalize=True)\ntrain['functional']= (train['status_group'] == 'functional').astype(int)\ntrain[['status_group', 'functional']];",
"_____no_output_____"
],
[
"#X_train.loc[X_train['public_meeting'].isnull(), 'public_meeting'] = False\n#X_val.loc[X_val['public_meeting'].isnull(), 'public_meeting'] = False\n\n#train = X_train.copy()\n#train['status_group'] = y_train\n\n#train.groupby('public_meeting')['status_group'].value_counts(normalize=True)\n#train['functional']= (train['status_group'] == 'functional').astype(int)\n#train[['status_group', 'functional']];\n\n#sns.catplot(x='public_meeting', y='functional', data=train, kind='bar', color='grey')\n#plt.title('% of Waterpumps Functional by Region/District')",
"_____no_output_____"
]
],
[
[
"### Run the Logistic Regression",
"_____no_output_____"
]
],
[
[
"import sklearn\nsklearn.__version__\n\n# Import the class\nfrom sklearn.linear_model import LogisticRegressionCV\n\n# Import package and scaler\nimport category_encoders as ce\nfrom sklearn.preprocessing import StandardScaler",
"_____no_output_____"
],
[
"# use quantity feature and the numerical features but drop id\ncategorical_features = ['quantity', 'waterpoint_type', 'extraction_type', 'installer',\n 'basin', 'region', 'payment', 'source', 'lga', 'public_meeting',\n 'scheme_management', 'permit', 'management', 'region_district',\n 'subvillage', 'funder', 'water_quality', 'ward']\n \n# \nnumeric_features = X_train.select_dtypes('number').columns.drop('id').drop('num_private').tolist()\nfeatures = categorical_features + numeric_features\n\n# make subsets using the quantity feature all numeric features except id\nX_train_subset = X_train[features]\nX_val_subset = X_val[features]\n\n# Do the encoding\nencoder = ce.OneHotEncoder(use_cat_names=True)\nX_train_encoded = encoder.fit_transform(X_train_subset)\nX_val_encoded = encoder.transform(X_val_subset)\n\n# Use the scaler\nscaler = StandardScaler()\nX_train_scaled = scaler.fit_transform(X_train_encoded)\nX_val_scaled = scaler.transform(X_val_encoded)\n\n# Fit the model and check the accuracy\nmodel = LogisticRegressionCV(n_jobs = -1)\nmodel.fit(X_train_scaled, y_train)\n\nprint('Validation Accuracy', model.score(X_val_scaled, y_val));\n\n",
"/usr/local/lib/python3.6/dist-packages/sklearn/linear_model/logistic.py:469: FutureWarning: Default multi_class will be changed to 'auto' in 0.22. Specify the multi_class option to silence this warning.\n \"this warning.\", FutureWarning)\n/usr/local/lib/python3.6/dist-packages/sklearn/model_selection/_split.py:1978: FutureWarning: The default value of cv will change from 3 to 5 in version 0.22. Specify it explicitly to silence this warning.\n warnings.warn(CV_WARNING, FutureWarning)\n"
]
],
[
[
"### Run RandomForestClassifier",
"_____no_output_____"
]
],
[
[
"from sklearn.ensemble import RandomForestClassifier\n\nmodel = RandomForestClassifier(n_estimators=1000, \n random_state=42,\n max_features = 'auto',\n n_jobs=-1,\n verbose = 1)\n\nmodel.fit(X_train_scaled, y_train)\n\nprint('Validation Accuracy', model.score(X_val_scaled, y_val));\n\n",
"[Parallel(n_jobs=-1)]: Using backend ThreadingBackend with 2 concurrent workers.\n[Parallel(n_jobs=-1)]: Done 46 tasks | elapsed: 8.2s\n[Parallel(n_jobs=-1)]: Done 196 tasks | elapsed: 34.9s\n[Parallel(n_jobs=-1)]: Done 446 tasks | elapsed: 1.3min\n[Parallel(n_jobs=-1)]: Done 796 tasks | elapsed: 2.4min\n[Parallel(n_jobs=-1)]: Done 1000 out of 1000 | elapsed: 3.0min finished\n[Parallel(n_jobs=2)]: Using backend ThreadingBackend with 2 concurrent workers.\n[Parallel(n_jobs=2)]: Done 46 tasks | elapsed: 0.2s\n[Parallel(n_jobs=2)]: Done 196 tasks | elapsed: 1.0s\n[Parallel(n_jobs=2)]: Done 446 tasks | elapsed: 2.2s\n[Parallel(n_jobs=2)]: Done 796 tasks | elapsed: 3.8s\n"
],
[
"test_features['pump_age'] = 2013 - test_features['construction_year']\ntest_features.loc[test_features['pump_age'] == 2013, 'pump_age'] = 0\n\ntest_features['region_district'] = test_features['region_code'].astype(str) + test_features['district_code'].astype(str)\n\ntest_features.drop(columns=['num_private'])\n\nX_test_subset = test_features[features]\nX_test_encoded = encoder.transform(X_test_subset)\nX_test_scaled = scaler.transform(X_test_encoded)\nassert all(X_test_encoded.columns == X_train_encoded.columns)\n\ny_pred = model.predict(X_test_scaled)",
"[Parallel(n_jobs=2)]: Using backend ThreadingBackend with 2 concurrent workers.\n[Parallel(n_jobs=2)]: Done 46 tasks | elapsed: 0.2s\n[Parallel(n_jobs=2)]: Done 196 tasks | elapsed: 0.8s\n[Parallel(n_jobs=2)]: Done 446 tasks | elapsed: 1.9s\n[Parallel(n_jobs=2)]: Done 796 tasks | elapsed: 3.4s\n[Parallel(n_jobs=2)]: Done 1000 out of 1000 | elapsed: 4.4s finished\n"
],
[
"submission = sample_submission.copy()\nsubmission['status_group'] = y_pred\nsubmission.to_csv('/content/submission-01.csv', index=False)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
cbea71afb4442c49fd738050562bd15390913191
| 113,422 |
ipynb
|
Jupyter Notebook
|
mivia.ipynb
|
squillero/foie-gras
|
c166d92361a838aa0ee50136767174ebc7bc8bb0
|
[
"Unlicense"
] | 1 |
2021-05-29T20:43:43.000Z
|
2021-05-29T20:43:43.000Z
|
mivia.ipynb
|
squillero/foie-gras
|
c166d92361a838aa0ee50136767174ebc7bc8bb0
|
[
"Unlicense"
] | null | null | null |
mivia.ipynb
|
squillero/foie-gras
|
c166d92361a838aa0ee50136767174ebc7bc8bb0
|
[
"Unlicense"
] | 1 |
2021-06-23T14:27:53.000Z
|
2021-06-23T14:27:53.000Z
| 872.476923 | 110,174 | 0.952813 |
[
[
[
"> **`(!)`** 2021 Written by Giovanni Squillero `<[email protected]>` \r\n> This is free and unencumbered software released into the public domain. ",
"_____no_output_____"
],
[
"ARG Database: [https://mivia.unisa.it/datasets/graph-database/arg-database/documentation/](https://mivia.unisa.it/datasets/graph-database/arg-database/documentation/)",
"_____no_output_____"
]
],
[
[
"import struct\r\nimport logging\r\n\r\nlogging.basicConfig(format='[%(asctime)s] %(levelname)s: %(message)s', datefmt='%H:%M:%S')\r\nlogging.getLogger().setLevel(level=logging.INFO)\r\n\r\nimport networkx as nx\r\nfrom matplotlib import pyplot as plt\r\nplt.rcParams[\"figure.figsize\"] = [21, 9]",
"_____no_output_____"
],
[
"with open('graphs/si2_r001_m200.A42', 'rb') as input:\r\n raw_data = input.read()\r\ncooked_data = [struct.unpack('<h', raw_data[i:i+2])[0] for i in range(0, len(raw_data), 2)]",
"_____no_output_____"
],
[
"graph = nx.DiGraph()\r\n\r\nnum_nodes = cooked_data.pop(0)\r\nlogging.info(f\"Graph has {num_nodes:,} nodes\")\r\n\r\nfor node in range(num_nodes):\r\n graph.add_node(node)\r\n\r\nfor node in range(num_nodes):\r\n num_edges = cooked_data.pop(0)\r\n logging.debug(f\"Node {node} has {num_edges} edges\")\r\n for edge in range(num_edges):\r\n target = cooked_data.pop(0)\r\n label = None # or label = cooked_data.pop(0)\r\n graph.add_edge(node, target, label=label)\r\n logging.debug(f\" {node} =[{label}]=> {target}\")\r\nassert not cooked_data",
"[19:12:34] INFO: Graph has 40 nodes\n"
],
[
"nx.draw(graph, node_size=20)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
cbea733b1fac400793f8cc4fe19cf10a3285c477
| 27,665 |
ipynb
|
Jupyter Notebook
|
site/pt-br/tutorials/keras/text_classification.ipynb
|
gmb-ftcont/docs-l10n
|
8b24263ca37dbf5cb4b0c15070a3d32c7284729d
|
[
"Apache-2.0"
] | 1 |
2020-08-05T05:52:57.000Z
|
2020-08-05T05:52:57.000Z
|
site/pt-br/tutorials/keras/text_classification.ipynb
|
gmb-ftcont/docs-l10n
|
8b24263ca37dbf5cb4b0c15070a3d32c7284729d
|
[
"Apache-2.0"
] | null | null | null |
site/pt-br/tutorials/keras/text_classification.ipynb
|
gmb-ftcont/docs-l10n
|
8b24263ca37dbf5cb4b0c15070a3d32c7284729d
|
[
"Apache-2.0"
] | null | null | null | 38.211326 | 604 | 0.559913 |
[
[
[
"##### Copyright 2018 The TensorFlow Authors.",
"_____no_output_____"
]
],
[
[
"#@title Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# https://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.",
"_____no_output_____"
],
[
"#@title MIT License\n#\n# Copyright (c) 2017 François Chollet\n#\n# Permission is hereby granted, free of charge, to any person obtaining a\n# copy of this software and associated documentation files (the \"Software\"),\n# to deal in the Software without restriction, including without limitation\n# the rights to use, copy, modify, merge, publish, distribute, sublicense,\n# and/or sell copies of the Software, and to permit persons to whom the\n# Software is furnished to do so, subject to the following conditions:\n#\n# The above copyright notice and this permission notice shall be included in\n# all copies or substantial portions of the Software.\n#\n# THE SOFTWARE IS PROVIDED \"AS IS\", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR\n# IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,\n# FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL\n# THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER\n# LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING\n# FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER\n# DEALINGS IN THE SOFTWARE.",
"_____no_output_____"
]
],
[
[
"# Classificação de texto com avaliações de filmes",
"_____no_output_____"
],
[
"<table class=\"tfo-notebook-buttons\" align=\"left\">\n <td>\n <a target=\"_blank\" href=\"https://www.tensorflow.org/tutorials/keras/text_classification\"><img src=\"https://www.tensorflow.org/images/tf_logo_32px.png\" />Veja em TensorFlow.org</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://colab.research.google.com/github/tensorflow/docs-l10n/blob/master/site/pt-br/tutorials/keras/text_classification.ipynb\"><img src=\"https://www.tensorflow.org/images/colab_logo_32px.png\" />Execute em Google Colab</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://github.com/tensorflow/docs-l10n/blob/master/site/pt-br/tutorials/keras/text_classification.ipynb\"><img src=\"https://www.tensorflow.org/images/GitHub-Mark-32px.png\" />Veja fonte em GitHub</a>\n </td>\n <td>\n <a href=\"https://storage.googleapis.com/tensorflow_docs/docs-l10n/site/pt-br/tutorials/keras/text_classification.ipynb\"><img src=\"https://www.tensorflow.org/images/download_logo_32px.png\" />Baixe o notebook</a>\n </td>\n</table>",
"_____no_output_____"
],
[
"Note: A nossa comunidade TensorFlow traduziu estes documentos. Como as traduções da comunidade são *o melhor esforço*, não há garantias de que sejam uma reflexão exata e atualizada da [documentação oficial em Inglês](https://www.tensorflow.org/?hl=en). Se tem alguma sugestão para melhorar esta tradução, por favor envie um pull request para o repositório do GitHub [tensorflow/docs](https://github.com/tensorflow/docs). Para se voluntariar para escrever ou rever as traduções da comunidade, contacte a [lista [email protected]](https://groups.google.com/a/tensorflow.org/forum/#!forum/docs).",
"_____no_output_____"
],
[
"Este *notebook* classifica avaliações de filmes como **positiva** ou **negativa** usando o texto da avaliação. Isto é um exemplo de classificação *binária* —ou duas-classes—, um importante e bastante aplicado tipo de problema de aprendizado de máquina.\n\nUsaremos a base de dados [IMDB](https://www.tensorflow.org/api_docs/python/tf/keras/datasets/imdb) que contém avaliaçòes de mais de 50000 filmes do bando de dados [Internet Movie Database](https://www.imdb.com/). A base é dividida em 25000 avaliações para treinamento e 25000 para teste. Os conjuntos de treinamentos e testes são *balanceados*, ou seja, eles possuem a mesma quantidade de avaliações positivas e negativas.\n\nO notebook utiliza [tf.keras](https://www.tensorflow.org/guide/keras), uma API alto-nível para construir e treinar modelos com TensorFlow. Para mais tutoriais avançados de classificação de textos usando `tf.keras`, veja em [MLCC Text Classification Guide](https://developers.google.com/machine-learning/guides/text-classification/).",
"_____no_output_____"
]
],
[
[
"import tensorflow as tf\nfrom tensorflow import keras\n\nimport numpy as np\n\nprint(tf.__version__)",
"_____no_output_____"
]
],
[
[
"## Baixe a base de dados IMDB\n\nA base de dados vem empacotada com TensorFlow. Ela já vem pré-processada de forma que as avaliações (sequências de palavras) foram convertidas em sequências de inteiros, onde cada inteiro representa uma palavra específica no dicionário.\n\nO código abaixo baixa a base de dados IMDB para a sua máquina (ou usa a cópia em *cache*, caso já tenha baixado):\"",
"_____no_output_____"
]
],
[
[
"imdb = keras.datasets.imdb\n\n(train_data, train_labels), (test_data, test_labels) = imdb.load_data(num_words=10000)",
"_____no_output_____"
]
],
[
[
"O argumento `num_words=10000` mantém as 10000 palavras mais frequentes no conjunto de treinamento. As palavras mais raras são descartadas para preservar o tamanho dos dados de forma maleável.",
"_____no_output_____"
],
[
"## Explore os dados\n\nVamos parar um momento para entender o formato dos dados. O conjunto de dados vem pré-processado: cada exemplo é um *array* de inteiros representando as palavras da avaliação do filme. Cada *label* é um inteiro com valor ou de 0 ou 1, onde 0 é uma avaliação negativa e 1 é uma avaliação positiva.",
"_____no_output_____"
]
],
[
[
"print(\"Training entries: {}, labels: {}\".format(len(train_data), len(train_labels)))",
"_____no_output_____"
]
],
[
[
"O texto das avaliações foi convertido para inteiros, onde cada inteiro representa uma palavra específica no dicionário. Isso é como se parece a primeira revisão:",
"_____no_output_____"
]
],
[
[
"print(train_data[0])",
"_____no_output_____"
]
],
[
[
"As avaliações dos filmes têm diferentes tamanhos. O código abaixo mostra o número de palavras da primeira e segunda avaliação. Sabendo que o número de entradas da rede neural tem que ser de mesmo também, temos que resolver isto mais tarde.",
"_____no_output_____"
]
],
[
[
"len(train_data[0]), len(train_data[1])",
"_____no_output_____"
]
],
[
[
"### Converta os inteiros de volta a palavras\n\nÉ util saber como converter inteiros de volta a texto. Aqui, criaremos uma função de ajuda para consultar um objeto *dictionary* que contenha inteiros mapeados em strings:",
"_____no_output_____"
]
],
[
[
"# Um dicionário mapeando palavras em índices inteiros\nword_index = imdb.get_word_index()\n\n# Os primeiros índices são reservados\nword_index = {k:(v+3) for k,v in word_index.items()}\nword_index[\"<PAD>\"] = 0\nword_index[\"<START>\"] = 1\nword_index[\"<UNK>\"] = 2 # unknown\nword_index[\"<UNUSED>\"] = 3\n\nreverse_word_index = dict([(value, key) for (key, value) in word_index.items()])\n\ndef decode_review(text):\n return ' '.join([reverse_word_index.get(i, '?') for i in text])",
"_____no_output_____"
]
],
[
[
"Agora, podemos usar a função `decode_review` para mostrar o texto da primeira avaliação:",
"_____no_output_____"
]
],
[
[
"decode_review(train_data[0])",
"_____no_output_____"
]
],
[
[
"## Prepare os dados\n\nAs avaliações —os *arrays* de inteiros— devem ser convertidas em tensores (*tensors*) antes de alimentar a rede neural. Essa conversão pode ser feita de duas formas:\n\n* Converter os arrays em vetores de 0s e 1s indicando a ocorrência da palavra, similar com one-hot encoding. Por exemplo, a sequência [3, 5] se tornaria um vetor de 10000 dimensões, onde todos seriam 0s, tirando 3 e 5, que são 1s. Depois, faça disso a primeira camada da nossa rede neural — a Dense layer — que pode trabalhar com dados em ponto flutuante. Essa abordagem é intensa em relação a memória, logo requer uma matriz de tamanho `num_words * num_reviews`.\n\n* Alternativamente, podemos preencher o array para que todos tenho o mesmo comprimento, e depois criar um tensor inteiro de formato `max_length * num_reviews`. Podemos usar uma camada *embedding* capaz de lidar com o formato como a primeira camada da nossa rede.\n\nNesse tutorial, usaremos a segunda abordagem.\n\nJá que as avaliações dos filmes devem ter o mesmo tamanho, usaremos a função [pad_sequences](https://www.tensorflow.org/api_docs/python/tf/keras/preprocessing/sequence/pad_sequences) para padronizar os tamanhos:",
"_____no_output_____"
]
],
[
[
"train_data = keras.preprocessing.sequence.pad_sequences(train_data,\n value=word_index[\"<PAD>\"],\n padding='post',\n maxlen=256)\n\ntest_data = keras.preprocessing.sequence.pad_sequences(test_data,\n value=word_index[\"<PAD>\"],\n padding='post',\n maxlen=256)",
"_____no_output_____"
]
],
[
[
"Agora, vamos olhar o tamanho dos exemplos:",
"_____no_output_____"
]
],
[
[
"len(train_data[0]), len(train_data[1])",
"_____no_output_____"
]
],
[
[
"E inspecionar as primeiras avaliações (agora preenchidos):",
"_____no_output_____"
]
],
[
[
"print(train_data[0])",
"_____no_output_____"
]
],
[
[
"## Construindo o modelo\n\nA rede neural é criada por camadas empilhadas —isso necessita duas decisões arquiteturais principais:\n\n* Quantas camadas serão usadas no modelo?\n* Quantas *hidden units* são usadas em cada camada?\n\nNeste exemplo, os dados de entrada são um *array* de palavras-índices. As *labels* para predizer são ou 0 ou 1. Vamos construir um modelo para este problema:",
"_____no_output_____"
]
],
[
[
"# O formato de entrada é a contagem vocabulário usados pelas avaliações dos filmes (10000 palavras)\nvocab_size = 10000\n\nmodel = keras.Sequential()\nmodel.add(keras.layers.Embedding(vocab_size, 16))\nmodel.add(keras.layers.GlobalAveragePooling1D())\nmodel.add(keras.layers.Dense(16, activation='relu'))\nmodel.add(keras.layers.Dense(1, activation='sigmoid'))\n\nmodel.summary()",
"_____no_output_____"
]
],
[
[
"As camadas são empilhadas sequencialmente para construir o classificador:\n\n1. A primeira camada é uma camada `Embedding` (*`Embedding` layer*). Essa camada pega o vocabulário em inteiros e olha o vetor *embedding* em cada palavra-index. Esses vetores são aprendidos pelo modelo, ao longo do treinamento. Os vetores adicionam a dimensão ao *array* de saída. As dimensões resultantes são: `(batch, sequence, embedding)`.\n2. Depois, uma camada `GlobalAveragePooling1D` retorna um vetor de saída com comprimento fixo para cada exemplo fazendo a média da sequência da dimensão. Isso permite o modelo de lidar com entradas de tamanhos diferentes da maneira mais simples possível.\n3. Esse vetor de saída com tamanho fixo passa por uma camada *fully-connected* (`Dense`) layer com 16 *hidden units*.\n4. A última camada é uma *densely connected* com um único nó de saída. Usando uma função de ativação `sigmoid`, esse valor é um float que varia entre 0 e 1, representando a probabilidade, ou nível de confiança.",
"_____no_output_____"
],
[
"### Hidden units\n\nO modelo abaixo tem duas camadas intermediárias ou _\\\"hidden\\\"_ (hidden layers), entre a entrada e saída. O número de saídas (unidades— *units*—, nós ou neurônios) é a dimensão do espaço representacional para a camada. Em outras palavras, a quantidade de liberdade que a rede é permitida enquanto aprende uma representação interna.\n\nSe o modelo tem mais *hidden units* (um espaço representacional de maior dimensão), e/ou mais camadas, então a rede pode aprender representações mais complexas. Entretanto, isso faz com que a rede seja computacionalmente mais custosa e pode levar ao aprendizado de padrões não desejados— padrões que melhoram a performance com os dados de treinamento, mas não com os de teste. Isso se chama *overfitting*, e exploraremos mais tarde.",
"_____no_output_____"
],
[
"### Função Loss e otimizadores (optimizer)\n\nO modelo precisa de uma função *loss* e um otimizador (*optimizer*) para treinamento. Já que é um problema de classificação binário e o modelo tem como saída uma probabilidade (uma única camada com ativação sigmoide), usaremos a função loss `binary_crossentropy`.\n\nEssa não é a única escolha de função loss, você poderia escolher, no lugar, a `mean_squared_error`. Mas, geralmente, `binary_crossentropy` é melhor para tratar probabilidades— ela mede a \\\"distância\\\" entre as distribuições de probabilidade, ou, no nosso caso, sobre a distribuição real e as previsões.\n\nMais tarde, quando explorarmos problemas de regressão (como, predizer preço de uma casa), veremos como usar outra função loss chamada *mean squared error*.\n\nAgora, configure o modelo para usar o *optimizer* a função loss:",
"_____no_output_____"
]
],
[
[
"model.compile(optimizer='adam',\n loss='binary_crossentropy',\n metrics=['accuracy'])",
"_____no_output_____"
]
],
[
[
"## Crie um conjunto de validação\n\nQuando treinando. queremos checar a acurácia do modelo com os dados que ele nunca viu. Crie uma conjunto de *validação* tirando 10000 exemplos do conjunto de treinamento original. (Por que não usar o de teste agora? Nosso objetivo é desenvolver e melhorar (tunar) nosso modelo usando somente os dados de treinamento, depois usar o de teste uma única vez para avaliar a acurácia).",
"_____no_output_____"
]
],
[
[
"x_val = train_data[:10000]\npartial_x_train = train_data[10000:]\n\ny_val = train_labels[:10000]\npartial_y_train = train_labels[10000:]",
"_____no_output_____"
]
],
[
[
"## Treine o modelo\n\nTreine o modelo em 40 *epochs* com *mini-batches* de 512 exemplos. Essas 40 iterações sobre todos os exemplos nos tensores `x_train` e `y_train`. Enquanto treina, monitore os valores do loss e da acurácia do modelo nos 10000 exemplos do conjunto de validação:",
"_____no_output_____"
]
],
[
[
"history = model.fit(partial_x_train,\n partial_y_train,\n epochs=40,\n batch_size=512,\n validation_data=(x_val, y_val),\n verbose=1)",
"_____no_output_____"
]
],
[
[
"## Avalie o modelo\n\nE vamos ver como o modelo se saiu. Dois valores serão retornados. Loss (um número que representa o nosso erro, valores mais baixos são melhores), e acurácia.",
"_____no_output_____"
]
],
[
[
"results = model.evaluate(test_data, test_labels, verbose=2)\n\nprint(results)",
"_____no_output_____"
]
],
[
[
"Esta é uma abordagem ingênua que conseguiu uma acurácia de 87%. Com abordagens mais avançadas, o modelo deve chegar em 95%.",
"_____no_output_____"
],
[
"## Crie um gráfico de acurácia e loss por tempo\n\n`model.fit()` retorna um objeto `History` que contém um dicionário de tudo o que aconteceu durante o treinamento:",
"_____no_output_____"
]
],
[
[
"history_dict = history.history\nhistory_dict.keys()",
"_____no_output_____"
]
],
[
[
"Tem 4 entradas: uma para cada métrica monitorada durante a validação e treinamento. Podemos usá-las para plotar a comparação do loss de treinamento e validação, assim como a acurácia de treinamento e validação:",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\n\nacc = history_dict['accuracy']\nval_acc = history_dict['val_accuracy']\nloss = history_dict['loss']\nval_loss = history_dict['val_loss']\n\nepochs = range(1, len(acc) + 1)\n\n# \"bo\" is for \"blue dot\"\nplt.plot(epochs, loss, 'bo', label='Training loss')\n# b is for \"solid blue line\"\nplt.plot(epochs, val_loss, 'b', label='Validation loss')\nplt.title('Training and validation loss')\nplt.xlabel('Epochs')\nplt.ylabel('Loss')\nplt.legend()\n\nplt.show()",
"_____no_output_____"
],
[
"plt.clf() # clear figure\n\nplt.plot(epochs, acc, 'bo', label='Training acc')\nplt.plot(epochs, val_acc, 'b', label='Validation acc')\nplt.title('Training and validation accuracy')\nplt.xlabel('Epochs')\nplt.ylabel('Accuracy')\nplt.legend()\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"No gráfico, os pontos representam o loss e acurácia de treinamento, e as linhas são o loss e a acurácia de validação.\n\nNote: que o loss de treinamento *diminui* a cada *epoch* e a acurácia *aumenta*. Isso é esperado quando usado um gradient descent optimization—ele deve minimizar a quantidade desejada a cada iteração.\n\nEsse não é o caso do loss e da acurácia de validação— eles parecem ter um pico depois de 20 epochs. Isso é um exemplo de *overfitting*: o modelo desempenha melhor nos dados de treinamento do que quando usado com dados nunca vistos. Depois desse ponto, o modelo otimiza além da conta e aprende uma representação *especifica* para os dados de treinamento e não *generaliza* para os dados de teste.\n\nPara esse caso particular, podemos prevenir o *overfitting* simplesmente parando o treinamento após mais ou menos 20 epochs. Depois, você verá como fazer isso automaticamente com um *callback*.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
cbea7b1029d7552d3100c704cfa64e353908ecdb
| 1,969 |
ipynb
|
Jupyter Notebook
|
sample.ipynb
|
sourav-roni/binder_test
|
f635e683d2d14ec733b30fccf325f35ff723f0cc
|
[
"MIT"
] | null | null | null |
sample.ipynb
|
sourav-roni/binder_test
|
f635e683d2d14ec733b30fccf325f35ff723f0cc
|
[
"MIT"
] | null | null | null |
sample.ipynb
|
sourav-roni/binder_test
|
f635e683d2d14ec733b30fccf325f35ff723f0cc
|
[
"MIT"
] | null | null | null | 19.49505 | 52 | 0.512951 |
[
[
[
"!pip install plotly==4.14.1\n!pip install python-igraph",
"_____no_output_____"
],
[
"import csv\nimport ast\nimport plotly.graph_objects as go\nfrom platform import python_version\nprint(python_version())\nimport random\nimport igraph\nfrom igraph import Graph, EdgeSeq\n\nimport plotly",
"_____no_output_____"
],
[
"plotly.__version__",
"_____no_output_____"
],
[
"igraph.__version__",
"_____no_output_____"
],
[
"# plotly==4.14.1\n# igraph==0.8.3",
"_____no_output_____"
],
[
"with open(\"./data/Books.csv\",\"r\") as f:\n file = csv.reader(f)\n file_content = []\n for row in file:\n file_content.append(row)\nf.close()\nprint(len(file_content))\nheader = file_content[0]\ndata = file_content[1:]\nprint(\"all data: \", len(data))\nprint(header)",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbea7f47bd41203660c204d22f74d2e957e193f7
| 17,287 |
ipynb
|
Jupyter Notebook
|
intro-to-pytorch/Part 1 - Tensors in PyTorch (Exercises).ipynb
|
Shahid1993/udacity-deep-learning-v2-pytorch
|
ec627d008a7fda5f758aff55bb8b289327e0d5ae
|
[
"MIT"
] | null | null | null |
intro-to-pytorch/Part 1 - Tensors in PyTorch (Exercises).ipynb
|
Shahid1993/udacity-deep-learning-v2-pytorch
|
ec627d008a7fda5f758aff55bb8b289327e0d5ae
|
[
"MIT"
] | 6 |
2020-01-28T23:14:41.000Z
|
2022-02-10T01:10:39.000Z
|
intro-to-pytorch/Part 1 - Tensors in PyTorch (Exercises).ipynb
|
Shahid1993/udacity-deep-learning-v2-pytorch
|
ec627d008a7fda5f758aff55bb8b289327e0d5ae
|
[
"MIT"
] | null | null | null | 38.415556 | 674 | 0.598427 |
[
[
[
"# Introduction to Deep Learning with PyTorch\n\nIn this notebook, you'll get introduced to [PyTorch](http://pytorch.org/), a framework for building and training neural networks. PyTorch in a lot of ways behaves like the arrays you love from Numpy. These Numpy arrays, after all, are just tensors. PyTorch takes these tensors and makes it simple to move them to GPUs for the faster processing needed when training neural networks. It also provides a module that automatically calculates gradients (for backpropagation!) and another module specifically for building neural networks. All together, PyTorch ends up being more coherent with Python and the Numpy/Scipy stack compared to TensorFlow and other frameworks.\n\n",
"_____no_output_____"
],
[
"## Neural Networks\n\nDeep Learning is based on artificial neural networks which have been around in some form since the late 1950s. The networks are built from individual parts approximating neurons, typically called units or simply \"neurons.\" Each unit has some number of weighted inputs. These weighted inputs are summed together (a linear combination) then passed through an activation function to get the unit's output.\n\n<img src=\"assets/simple_neuron.png\" width=400px>\n\nMathematically this looks like: \n\n$$\n\\begin{align}\ny &= f(w_1 x_1 + w_2 x_2 + b) \\\\\ny &= f\\left(\\sum_i w_i x_i +b \\right)\n\\end{align}\n$$\n\nWith vectors this is the dot/inner product of two vectors:\n\n$$\nh = \\begin{bmatrix}\nx_1 \\, x_2 \\cdots x_n\n\\end{bmatrix}\n\\cdot \n\\begin{bmatrix}\n w_1 \\\\\n w_2 \\\\\n \\vdots \\\\\n w_n\n\\end{bmatrix}\n$$",
"_____no_output_____"
],
[
"## Tensors\n\nIt turns out neural network computations are just a bunch of linear algebra operations on *tensors*, a generalization of matrices. A vector is a 1-dimensional tensor, a matrix is a 2-dimensional tensor, an array with three indices is a 3-dimensional tensor (RGB color images for example). The fundamental data structure for neural networks are tensors and PyTorch (as well as pretty much every other deep learning framework) is built around tensors.\n\n<img src=\"assets/tensor_examples.svg\" width=600px>\n\nWith the basics covered, it's time to explore how we can use PyTorch to build a simple neural network.",
"_____no_output_____"
]
],
[
[
"# First, import PyTorch\nimport torch",
"_____no_output_____"
],
[
"def activation(x):\n \"\"\" Sigmoid activation function \n \n Arguments\n ---------\n x: torch.Tensor\n \"\"\"\n return 1/(1+torch.exp(-x))",
"_____no_output_____"
],
[
"### Generate some data\ntorch.manual_seed(7) # Set the random seed so things are predictable\n\n# Features are 5 random normal variables\nfeatures = torch.randn((1, 5))\n# True weights for our data, random normal variables again\nweights = torch.randn_like(features)\n# and a true bias term\nbias = torch.randn((1, 1))",
"_____no_output_____"
]
],
[
[
"Above I generated data we can use to get the output of our simple network. This is all just random for now, going forward we'll start using normal data. Going through each relevant line:\n\n`features = torch.randn((1, 5))` creates a tensor with shape `(1, 5)`, one row and five columns, that contains values randomly distributed according to the normal distribution with a mean of zero and standard deviation of one. \n\n`weights = torch.randn_like(features)` creates another tensor with the same shape as `features`, again containing values from a normal distribution.\n\nFinally, `bias = torch.randn((1, 1))` creates a single value from a normal distribution.\n\nPyTorch tensors can be added, multiplied, subtracted, etc, just like Numpy arrays. In general, you'll use PyTorch tensors pretty much the same way you'd use Numpy arrays. They come with some nice benefits though such as GPU acceleration which we'll get to later. For now, use the generated data to calculate the output of this simple single layer network. \n> **Exercise**: Calculate the output of the network with input features `features`, weights `weights`, and bias `bias`. Similar to Numpy, PyTorch has a [`torch.sum()`](https://pytorch.org/docs/stable/torch.html#torch.sum) function, as well as a `.sum()` method on tensors, for taking sums. Use the function `activation` defined above as the activation function.",
"_____no_output_____"
]
],
[
[
"## Calculate the output of this network using the weights and bias tensors\n\ny = activation(torch.sum(features * weights) + bias)\ny = activation((features * weights).sum() + bias)",
"_____no_output_____"
]
],
[
[
"You can do the multiplication and sum in the same operation using a matrix multiplication. In general, you'll want to use matrix multiplications since they are more efficient and accelerated using modern libraries and high-performance computing on GPUs.\n\nHere, we want to do a matrix multiplication of the features and the weights. For this we can use [`torch.mm()`](https://pytorch.org/docs/stable/torch.html#torch.mm) or [`torch.matmul()`](https://pytorch.org/docs/stable/torch.html#torch.matmul) which is somewhat more complicated and supports broadcasting. If we try to do it with `features` and `weights` as they are, we'll get an error\n\n```python\n>> torch.mm(features, weights)\n\n---------------------------------------------------------------------------\nRuntimeError Traceback (most recent call last)\n<ipython-input-13-15d592eb5279> in <module>()\n----> 1 torch.mm(features, weights)\n\nRuntimeError: size mismatch, m1: [1 x 5], m2: [1 x 5] at /Users/soumith/minicondabuild3/conda-bld/pytorch_1524590658547/work/aten/src/TH/generic/THTensorMath.c:2033\n```\n\nAs you're building neural networks in any framework, you'll see this often. Really often. What's happening here is our tensors aren't the correct shapes to perform a matrix multiplication. Remember that for matrix multiplications, the number of columns in the first tensor must equal to the number of rows in the second column. Both `features` and `weights` have the same shape, `(1, 5)`. This means we need to change the shape of `weights` to get the matrix multiplication to work.\n\n**Note:** To see the shape of a tensor called `tensor`, use `tensor.shape`. If you're building neural networks, you'll be using this method often.\n\nThere are a few options here: [`weights.reshape()`](https://pytorch.org/docs/stable/tensors.html#torch.Tensor.reshape), [`weights.resize_()`](https://pytorch.org/docs/stable/tensors.html#torch.Tensor.resize_), and [`weights.view()`](https://pytorch.org/docs/stable/tensors.html#torch.Tensor.view).\n\n* `weights.reshape(a, b)` will return a new tensor with the same data as `weights` with size `(a, b)` sometimes, and sometimes a clone, as in it copies the data to another part of memory.\n* `weights.resize_(a, b)` returns the same tensor with a different shape. However, if the new shape results in fewer elements than the original tensor, some elements will be removed from the tensor (but not from memory). If the new shape results in more elements than the original tensor, new elements will be uninitialized in memory. Here I should note that the underscore at the end of the method denotes that this method is performed **in-place**. Here is a great forum thread to [read more about in-place operations](https://discuss.pytorch.org/t/what-is-in-place-operation/16244) in PyTorch.\n* `weights.view(a, b)` will return a new tensor with the same data as `weights` with size `(a, b)`.\n\nI usually use `.view()`, but any of the three methods will work for this. So, now we can reshape `weights` to have five rows and one column with something like `weights.view(5, 1)`.\n\n> **Exercise**: Calculate the output of our little network using matrix multiplication.",
"_____no_output_____"
]
],
[
[
"## Calculate the output of this network using matrix multiplication\nprint(features.shape, weights.shape)\n\ny = activation(torch.mm(features, weights.view(5,1)) + bias)",
"torch.Size([1, 5]) torch.Size([1, 5])\n"
]
],
[
[
"### Stack them up!\n\nThat's how you can calculate the output for a single neuron. The real power of this algorithm happens when you start stacking these individual units into layers and stacks of layers, into a network of neurons. The output of one layer of neurons becomes the input for the next layer. With multiple input units and output units, we now need to express the weights as a matrix.\n\n<img src='assets/multilayer_diagram_weights.png' width=450px>\n\nThe first layer shown on the bottom here are the inputs, understandably called the **input layer**. The middle layer is called the **hidden layer**, and the final layer (on the right) is the **output layer**. We can express this network mathematically with matrices again and use matrix multiplication to get linear combinations for each unit in one operation. For example, the hidden layer ($h_1$ and $h_2$ here) can be calculated \n\n$$\n\\vec{h} = [h_1 \\, h_2] = \n\\begin{bmatrix}\nx_1 \\, x_2 \\cdots \\, x_n\n\\end{bmatrix}\n\\cdot \n\\begin{bmatrix}\n w_{11} & w_{12} \\\\\n w_{21} &w_{22} \\\\\n \\vdots &\\vdots \\\\\n w_{n1} &w_{n2}\n\\end{bmatrix}\n$$\n\nThe output for this small network is found by treating the hidden layer as inputs for the output unit. The network output is expressed simply\n\n$$\ny = f_2 \\! \\left(\\, f_1 \\! \\left(\\vec{x} \\, \\mathbf{W_1}\\right) \\mathbf{W_2} \\right)\n$$",
"_____no_output_____"
]
],
[
[
"### Generate some data\ntorch.manual_seed(7) # Set the random seed so things are predictable\n\n# Features are 3 random normal variables\nfeatures = torch.randn((1, 3))\n\n# Define the size of each layer in our network\nn_input = features.shape[1] # Number of input units, must match number of input features\nn_hidden = 2 # Number of hidden units \nn_output = 1 # Number of output units\n\n# Weights for inputs to hidden layer\nW1 = torch.randn(n_input, n_hidden)\n# Weights for hidden layer to output layer\nW2 = torch.randn(n_hidden, n_output)\n\n# and bias terms for hidden and output layers\nB1 = torch.randn((1, n_hidden))\nB2 = torch.randn((1, n_output))",
"_____no_output_____"
]
],
[
[
"> **Exercise:** Calculate the output for this multi-layer network using the weights `W1` & `W2`, and the biases, `B1` & `B2`. ",
"_____no_output_____"
]
],
[
[
"## Your solution here\n\nh = activation(torch.mm(features, W1) + B1)\noutput = activation(torch.mm(h, W2) + B2)\n\nprint(output)",
"tensor([[0.3171]])\n"
]
],
[
[
"If you did this correctly, you should see the output `tensor([[ 0.3171]])`.\n\nThe number of hidden units a parameter of the network, often called a **hyperparameter** to differentiate it from the weights and biases parameters. As you'll see later when we discuss training a neural network, the more hidden units a network has, and the more layers, the better able it is to learn from data and make accurate predictions.",
"_____no_output_____"
],
[
"## Numpy to Torch and back\n\nSpecial bonus section! PyTorch has a great feature for converting between Numpy arrays and Torch tensors. To create a tensor from a Numpy array, use `torch.from_numpy()`. To convert a tensor to a Numpy array, use the `.numpy()` method.",
"_____no_output_____"
]
],
[
[
"import numpy as np\na = np.random.rand(4,3)\na",
"_____no_output_____"
],
[
"b = torch.from_numpy(a)\nb",
"_____no_output_____"
],
[
"b.numpy()",
"_____no_output_____"
]
],
[
[
"The memory is shared between the Numpy array and Torch tensor, so if you change the values in-place of one object, the other will change as well.",
"_____no_output_____"
]
],
[
[
"# Multiply PyTorch Tensor by 2, in place\nb.mul_(2)",
"_____no_output_____"
],
[
"# Numpy array matches new values from Tensor\na",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
cbea7ffda03a462f08cb168d323170410a6134f4
| 47,103 |
ipynb
|
Jupyter Notebook
|
src/ml_basic/parameter_server_training.ipynb
|
lisy09/research-to-applied-ml
|
c6f1b660757dd8178e5c3b4027136e996c6c96de
|
[
"Apache-2.0"
] | null | null | null |
src/ml_basic/parameter_server_training.ipynb
|
lisy09/research-to-applied-ml
|
c6f1b660757dd8178e5c3b4027136e996c6c96de
|
[
"Apache-2.0"
] | null | null | null |
src/ml_basic/parameter_server_training.ipynb
|
lisy09/research-to-applied-ml
|
c6f1b660757dd8178e5c3b4027136e996c6c96de
|
[
"Apache-2.0"
] | null | null | null | 59.548673 | 740 | 0.674034 |
[
[
[
"import tensorflow as tf\nimport tensorflow.keras as keras\nimport portpicker\nimport multiprocessing",
"_____no_output_____"
],
[
"def create_in_process_cluster(num_workers: int, num_ps: int):\n \"\"\"\n Create and start local servers and return the cluster_resolver.\n \"\"\"\n worker_ports = [portpicker.pick_unused_port() for _ in range(num_workers)]\n ps_ports = [portpicker.pick_unused_port() for _ in range(num_ps)]\n\n cluster_dict = {}\n cluster_dict[\"worker\"] = [f\"localhost:{port}\" for port in worker_ports ]\n if num_ps > 0:\n cluster_dict[\"ps\"] = [f\"localhost:{port}\" for port in ps_ports]\n \n cluster_spec = tf.train.ClusterSpec(cluster_dict)\n\n # workers need some inter_ops threads to work properly\n worker_config = tf.compat.v1.ConfigProto()\n if multiprocessing.cpu_count() < num_workers + 1:\n worker_config.inter_op_parallelism_threads = num_workers + 1\n \n for i in range(num_workers):\n tf.distribute.Server(\n cluster_spec,\n job_name=\"worker\",\n task_index=i,\n protocol='grpc'\n )\n\n for i in range(num_ps):\n tf.distribute.Server(\n cluster_spec,\n job_name=\"ps\",\n task_index=i,\n protocol='grpc'\n )\n \n cluster_resolver = tf.distribute.cluster_resolver.SimpleClusterResolver(\n cluster_spec=cluster_spec,\n rpc_layer='grpc'\n )\n return cluster_resolver\n\n\n# Set the environment variable to allow reporting worker and ps failure to the\n# coordinator. This is a workaround and won't be necessary in the future.\nos.environ[\"GRPC_FAIL_FAST\"] = \"use_caller\"\n\nNUM_WORKERS = 3\nNUM_PS = 2\ncluster_resolver = create_in_process_cluster(NUM_WORKERS, NUM_PS)\n",
"2022-03-26 15:47:36.021073: I tensorflow/core/platform/cpu_feature_guard.cc:151] This TensorFlow binary is optimized with oneAPI Deep Neural Network Library (oneDNN) to use the following CPU instructions in performance-critical operations: AVX2 FMA\nTo enable them in other operations, rebuild TensorFlow with the appropriate compiler flags.\n2022-03-26 15:47:36.079179: I tensorflow/stream_executor/cuda/cuda_gpu_executor.cc:922] could not open file to read NUMA node: /sys/bus/pci/devices/0000:01:00.0/numa_node\nYour kernel may have been built without NUMA support.\n2022-03-26 15:47:36.121087: I tensorflow/stream_executor/cuda/cuda_gpu_executor.cc:922] could not open file to read NUMA node: /sys/bus/pci/devices/0000:01:00.0/numa_node\nYour kernel may have been built without NUMA support.\n2022-03-26 15:47:36.121451: I tensorflow/stream_executor/cuda/cuda_gpu_executor.cc:922] could not open file to read NUMA node: /sys/bus/pci/devices/0000:01:00.0/numa_node\nYour kernel may have been built without NUMA support.\n2022-03-26 15:47:36.500713: I tensorflow/stream_executor/cuda/cuda_gpu_executor.cc:922] could not open file to read NUMA node: /sys/bus/pci/devices/0000:01:00.0/numa_node\nYour kernel may have been built without NUMA support.\n2022-03-26 15:47:36.500998: I tensorflow/stream_executor/cuda/cuda_gpu_executor.cc:922] could not open file to read NUMA node: /sys/bus/pci/devices/0000:01:00.0/numa_node\nYour kernel may have been built without NUMA support.\n2022-03-26 15:47:36.501009: I tensorflow/core/common_runtime/gpu/gpu_device.cc:1609] Could not identify NUMA node of platform GPU id 0, defaulting to 0. Your kernel may not have been built with NUMA support.\n2022-03-26 15:47:36.501226: I tensorflow/stream_executor/cuda/cuda_gpu_executor.cc:922] could not open file to read NUMA node: /sys/bus/pci/devices/0000:01:00.0/numa_node\nYour kernel may have been built without NUMA support.\n2022-03-26 15:47:36.501270: I tensorflow/core/common_runtime/gpu/gpu_device.cc:1525] Created device /job:worker/replica:0/task:0/device:GPU:0 with 7403 MB memory: -> device: 0, name: NVIDIA GeForce RTX 3080, pci bus id: 0000:01:00.0, compute capability: 8.6\n2022-03-26 15:47:36.508681: I tensorflow/core/distributed_runtime/rpc/grpc_channel.cc:272] Initialize GrpcChannelCache for job ps -> {0 -> localhost:22929, 1 -> localhost:18536}\n2022-03-26 15:47:36.508707: I tensorflow/core/distributed_runtime/rpc/grpc_channel.cc:272] Initialize GrpcChannelCache for job worker -> {0 -> localhost:20447, 1 -> localhost:21839, 2 -> localhost:16457}\n2022-03-26 15:47:36.509739: I tensorflow/core/distributed_runtime/rpc/grpc_server_lib.cc:437] Started server with target: grpc://localhost:20447\n2022-03-26 15:47:36.513299: I tensorflow/stream_executor/cuda/cuda_gpu_executor.cc:922] could not open file to read NUMA node: /sys/bus/pci/devices/0000:01:00.0/numa_node\nYour kernel may have been built without NUMA support.\n2022-03-26 15:47:36.513608: I tensorflow/stream_executor/cuda/cuda_gpu_executor.cc:922] could not open file to read NUMA node: /sys/bus/pci/devices/0000:01:00.0/numa_node\nYour kernel may have been built without NUMA support.\n2022-03-26 15:47:36.513778: I tensorflow/stream_executor/cuda/cuda_gpu_executor.cc:922] could not open file to read NUMA node: /sys/bus/pci/devices/0000:01:00.0/numa_node\nYour kernel may have been built without NUMA support.\n2022-03-26 15:47:36.514123: I tensorflow/stream_executor/cuda/cuda_gpu_executor.cc:922] could not open file to read NUMA node: /sys/bus/pci/devices/0000:01:00.0/numa_node\nYour kernel may have been built without NUMA support.\n2022-03-26 15:47:36.514138: I tensorflow/core/common_runtime/gpu/gpu_device.cc:1609] Could not identify NUMA node of platform GPU id 0, defaulting to 0. Your kernel may not have been built with NUMA support.\n2022-03-26 15:47:36.514332: I tensorflow/stream_executor/cuda/cuda_gpu_executor.cc:922] could not open file to read NUMA node: /sys/bus/pci/devices/0000:01:00.0/numa_node\nYour kernel may have been built without NUMA support.\n2022-03-26 15:47:36.514356: I tensorflow/core/common_runtime/gpu/gpu_device.cc:1525] Created device /job:worker/replica:0/task:1/device:GPU:0 with 7403 MB memory: -> device: 0, name: NVIDIA GeForce RTX 3080, pci bus id: 0000:01:00.0, compute capability: 8.6\n2022-03-26 15:47:36.532338: I tensorflow/core/distributed_runtime/rpc/grpc_channel.cc:272] Initialize GrpcChannelCache for job ps -> {0 -> localhost:22929, 1 -> localhost:18536}\n2022-03-26 15:47:36.532365: I tensorflow/core/distributed_runtime/rpc/grpc_channel.cc:272] Initialize GrpcChannelCache for job worker -> {0 -> localhost:20447, 1 -> localhost:21839, 2 -> localhost:16457}\n2022-03-26 15:47:36.532732: I tensorflow/core/distributed_runtime/rpc/grpc_server_lib.cc:437] Started server with target: grpc://localhost:21839\n2022-03-26 15:47:36.533848: I tensorflow/stream_executor/cuda/cuda_gpu_executor.cc:922] could not open file to read NUMA node: /sys/bus/pci/devices/0000:01:00.0/numa_node\nYour kernel may have been built without NUMA support.\n2022-03-26 15:47:36.534148: I tensorflow/stream_executor/cuda/cuda_gpu_executor.cc:922] could not open file to read NUMA node: /sys/bus/pci/devices/0000:01:00.0/numa_node\nYour kernel may have been built without NUMA support.\n2022-03-26 15:47:36.534326: I tensorflow/stream_executor/cuda/cuda_gpu_executor.cc:922] could not open file to read NUMA node: /sys/bus/pci/devices/0000:01:00.0/numa_node\nYour kernel may have been built without NUMA support.\n2022-03-26 15:47:36.534670: I tensorflow/stream_executor/cuda/cuda_gpu_executor.cc:922] could not open file to read NUMA node: /sys/bus/pci/devices/0000:01:00.0/numa_node\nYour kernel may have been built without NUMA support.\n2022-03-26 15:47:36.534684: I tensorflow/core/common_runtime/gpu/gpu_device.cc:1609] Could not identify NUMA node of platform GPU id 0, defaulting to 0. Your kernel may not have been built with NUMA support.\n2022-03-26 15:47:36.534908: I tensorflow/stream_executor/cuda/cuda_gpu_executor.cc:922] could not open file to read NUMA node: /sys/bus/pci/devices/0000:01:00.0/numa_node\nYour kernel may have been built without NUMA support.\n2022-03-26 15:47:36.534936: I tensorflow/core/common_runtime/gpu/gpu_device.cc:1525] Created device /job:worker/replica:0/task:2/device:GPU:0 with 7403 MB memory: -> device: 0, name: NVIDIA GeForce RTX 3080, pci bus id: 0000:01:00.0, compute capability: 8.6\n2022-03-26 15:47:36.559450: I tensorflow/core/distributed_runtime/rpc/grpc_channel.cc:272] Initialize GrpcChannelCache for job ps -> {0 -> localhost:22929, 1 -> localhost:18536}\n2022-03-26 15:47:36.559470: I tensorflow/core/distributed_runtime/rpc/grpc_channel.cc:272] Initialize GrpcChannelCache for job worker -> {0 -> localhost:20447, 1 -> localhost:21839, 2 -> localhost:16457}\n2022-03-26 15:47:36.559757: I tensorflow/core/distributed_runtime/rpc/grpc_server_lib.cc:437] Started server with target: grpc://localhost:16457\n2022-03-26 15:47:36.560782: I tensorflow/stream_executor/cuda/cuda_gpu_executor.cc:922] could not open file to read NUMA node: /sys/bus/pci/devices/0000:01:00.0/numa_node\nYour kernel may have been built without NUMA support.\n2022-03-26 15:47:36.561040: I tensorflow/stream_executor/cuda/cuda_gpu_executor.cc:922] could not open file to read NUMA node: /sys/bus/pci/devices/0000:01:00.0/numa_node\nYour kernel may have been built without NUMA support.\n2022-03-26 15:47:36.561208: I tensorflow/stream_executor/cuda/cuda_gpu_executor.cc:922] could not open file to read NUMA node: /sys/bus/pci/devices/0000:01:00.0/numa_node\nYour kernel may have been built without NUMA support.\n2022-03-26 15:47:36.561502: I tensorflow/stream_executor/cuda/cuda_gpu_executor.cc:922] could not open file to read NUMA node: /sys/bus/pci/devices/0000:01:00.0/numa_node\nYour kernel may have been built without NUMA support.\n2022-03-26 15:47:36.561513: I tensorflow/core/common_runtime/gpu/gpu_device.cc:1609] Could not identify NUMA node of platform GPU id 0, defaulting to 0. Your kernel may not have been built with NUMA support.\n2022-03-26 15:47:36.561674: I tensorflow/stream_executor/cuda/cuda_gpu_executor.cc:922] could not open file to read NUMA node: /sys/bus/pci/devices/0000:01:00.0/numa_node\nYour kernel may have been built without NUMA support.\n2022-03-26 15:47:36.561711: I tensorflow/core/common_runtime/gpu/gpu_device.cc:1525] Created device /job:ps/replica:0/task:0/device:GPU:0 with 7403 MB memory: -> device: 0, name: NVIDIA GeForce RTX 3080, pci bus id: 0000:01:00.0, compute capability: 8.6\n2022-03-26 15:47:36.571176: I tensorflow/core/distributed_runtime/rpc/grpc_channel.cc:272] Initialize GrpcChannelCache for job ps -> {0 -> localhost:22929, 1 -> localhost:18536}\n2022-03-26 15:47:36.571201: I tensorflow/core/distributed_runtime/rpc/grpc_channel.cc:272] Initialize GrpcChannelCache for job worker -> {0 -> localhost:20447, 1 -> localhost:21839, 2 -> localhost:16457}\n2022-03-26 15:47:36.572598: I tensorflow/core/distributed_runtime/rpc/grpc_server_lib.cc:437] Started server with target: grpc://localhost:22929\n2022-03-26 15:47:36.573343: I tensorflow/stream_executor/cuda/cuda_gpu_executor.cc:922] could not open file to read NUMA node: /sys/bus/pci/devices/0000:01:00.0/numa_node\nYour kernel may have been built without NUMA support.\n2022-03-26 15:47:36.573601: I tensorflow/stream_executor/cuda/cuda_gpu_executor.cc:922] could not open file to read NUMA node: /sys/bus/pci/devices/0000:01:00.0/numa_node\nYour kernel may have been built without NUMA support.\n2022-03-26 15:47:36.573782: I tensorflow/stream_executor/cuda/cuda_gpu_executor.cc:922] could not open file to read NUMA node: /sys/bus/pci/devices/0000:01:00.0/numa_node\nYour kernel may have been built without NUMA support.\n2022-03-26 15:47:36.574120: I tensorflow/stream_executor/cuda/cuda_gpu_executor.cc:922] could not open file to read NUMA node: /sys/bus/pci/devices/0000:01:00.0/numa_node\nYour kernel may have been built without NUMA support.\n2022-03-26 15:47:36.574136: I tensorflow/core/common_runtime/gpu/gpu_device.cc:1609] Could not identify NUMA node of platform GPU id 0, defaulting to 0. Your kernel may not have been built with NUMA support.\n2022-03-26 15:47:36.574320: I tensorflow/stream_executor/cuda/cuda_gpu_executor.cc:922] could not open file to read NUMA node: /sys/bus/pci/devices/0000:01:00.0/numa_node\nYour kernel may have been built without NUMA support.\n2022-03-26 15:47:36.574352: I tensorflow/core/common_runtime/gpu/gpu_device.cc:1525] Created device /job:ps/replica:0/task:1/device:GPU:0 with 7403 MB memory: -> device: 0, name: NVIDIA GeForce RTX 3080, pci bus id: 0000:01:00.0, compute capability: 8.6\n2022-03-26 15:47:36.598395: I tensorflow/core/distributed_runtime/rpc/grpc_channel.cc:272] Initialize GrpcChannelCache for job ps -> {0 -> localhost:22929, 1 -> localhost:18536}\n2022-03-26 15:47:36.598418: I tensorflow/core/distributed_runtime/rpc/grpc_channel.cc:272] Initialize GrpcChannelCache for job worker -> {0 -> localhost:20447, 1 -> localhost:21839, 2 -> localhost:16457}\n2022-03-26 15:47:36.598850: I tensorflow/core/distributed_runtime/rpc/grpc_server_lib.cc:437] Started server with target: grpc://localhost:18536\n"
],
[
"variable_partitioner = (\n tf.distribute.experimental.partitioners.MinSizePartitioner(\n min_shard_bytes=(256<<10),\n max_shards=NUM_PS\n )\n)\nstrategy = tf.distribute.experimental.ParameterServerStrategy(\n cluster_resolver,\n variable_partitioner=variable_partitioner)\n",
"INFO:tensorflow:`tf.distribute.experimental.ParameterServerStrategy` is initialized with cluster_spec: ClusterSpec({'ps': ['localhost:22929', 'localhost:18536'], 'worker': ['localhost:20447', 'localhost:21839', 'localhost:16457']})\nINFO:tensorflow:ParameterServerStrategyV2 is now connecting to cluster with cluster_spec: ClusterSpec({'ps': ['localhost:22929', 'localhost:18536'], 'worker': ['localhost:20447', 'localhost:21839', 'localhost:16457']})\n"
],
[
"def dataset_fn(input_context):\n global_batch_size = 64\n batch_size = input_context.get_per_replica_batch_size(global_batch_size)\n\n x = tf.random.uniform((10, 10))\n y = tf.random.uniform((10,))\n\n dataset = tf.data.Dataset.from_tensor_slices((x, y)).shuffle(10).repeat()\n dataset = dataset.shard(\n input_context.num_input_pipelines,\n input_context.input_pipeline_id)\n dataset = dataset.batch(batch_size)\n dataset = dataset.prefetch(2)\n\n return dataset\n\n\ndc = tf.keras.utils.experimental.DatasetCreator(dataset_fn)\n",
"_____no_output_____"
],
[
"with strategy.scope():\n model = tf.keras.models.Sequential([tf.keras.layers.Dense(10)])\n\n model.compile(tf.keras.optimizers.SGD(), loss='mse', steps_per_execution=10)\n",
"_____no_output_____"
],
[
"working_dir = '/tmp/my_working_dir'\nlog_dir = os.path.join(working_dir, 'log')\nckpt_filepath = os.path.join(working_dir, 'ckpt')\nbackup_dir = os.path.join(working_dir, 'backup')\n\ncallbacks = [\n tf.keras.callbacks.TensorBoard(log_dir=log_dir),\n tf.keras.callbacks.ModelCheckpoint(filepath=ckpt_filepath),\n tf.keras.callbacks.BackupAndRestore(backup_dir=backup_dir),\n]\n\nmodel.fit(dc, epochs=5, steps_per_epoch=20, callbacks=callbacks)\n",
"Epoch 1/5\nINFO:tensorflow:Reduce to /device:CPU:0 then broadcast to ('/replica:0/device:CPU:0',).\nINFO:tensorflow:Reduce to /device:CPU:0 then broadcast to ('/replica:0/device:CPU:0',).\nINFO:tensorflow:Reduce to /device:CPU:0 then broadcast to ('/replica:0/device:CPU:0',).\nINFO:tensorflow:Reduce to /device:CPU:0 then broadcast to ('/replica:0/device:CPU:0',).\nINFO:tensorflow:Reduce to /device:CPU:0 then broadcast to ('/replica:0/device:CPU:0',).\nINFO:tensorflow:Reduce to /device:CPU:0 then broadcast to ('/replica:0/device:CPU:0',).\nINFO:tensorflow:Reduce to /device:CPU:0 then broadcast to ('/replica:0/device:CPU:0',).\nINFO:tensorflow:Reduce to /device:CPU:0 then broadcast to ('/replica:0/device:CPU:0',).\n"
],
[
"feature_vocab = [\n \"avenger\", \"ironman\", \"batman\", \"hulk\", \"spiderman\", \"kingkong\", \"wonder_woman\"\n]\nlabel_vocab = [\"yes\", \"no\"]\n\nwith strategy.scope():\n feature_lookup_layer = tf.keras.layers.StringLookup(\n vocabulary=feature_vocab,\n mask_token=None)\n label_lookup_layer = tf.keras.layers.StringLookup(\n vocabulary=label_vocab,\n num_oov_indices=0,\n mask_token=None)\n\n raw_feature_input = tf.keras.layers.Input(\n shape=(3,),\n dtype=tf.string,\n name=\"feature\")\n feature_id_input = feature_lookup_layer(raw_feature_input)\n feature_preprocess_stage = tf.keras.Model(\n {\"features\": raw_feature_input},\n feature_id_input)\n\n raw_label_input = tf.keras.layers.Input(\n shape=(1,),\n dtype=tf.string,\n name=\"label\")\n label_id_input = label_lookup_layer(raw_label_input)\n\n label_preprocess_stage = tf.keras.Model(\n {\"label\": raw_label_input},\n label_id_input)\n",
"/home/lisy09/develop/repo/research-to-applied-ml/venv/lib/python3.8/site-packages/numpy/core/numeric.py:2449: FutureWarning: elementwise comparison failed; returning scalar instead, but in the future will perform elementwise comparison\n return bool(asarray(a1 == a2).all())\n"
],
[
"import random\ndef feature_and_label_gen(num_examples=200):\n examples = {\"features\": [], \"label\": []}\n for _ in range(num_examples):\n features = random.sample(feature_vocab, 3)\n label = [\"yes\"] if \"avenger\" in features else [\"no\"]\n examples[\"features\"].append(features)\n examples[\"label\"].append(label)\n return examples\n\n\nexamples = feature_and_label_gen()\n",
"_____no_output_____"
],
[
"def dataset_fn(_):\n raw_dataset = tf.data.Dataset.from_tensor_slices(examples)\n\n train_dataset = raw_dataset.map(\n lambda x: (\n {\"features\": feature_preprocess_stage(x[\"features\"])},\n label_preprocess_stage(x[\"label\"])\n )).shuffle(200).batch(32).repeat()\n return train_dataset\n",
"_____no_output_____"
],
[
"# These variables created under the `Strategy.scope` will be placed on parameter\n# servers in a round-robin fashion.\nwith strategy.scope():\n # Create the model. The input needs to be compatible with Keras processing layers.\n model_input = tf.keras.layers.Input(\n shape=(3,), dtype=tf.int64, name=\"model_input\")\n\n emb_layer = tf.keras.layers.Embedding(\n input_dim=len(feature_lookup_layer.get_vocabulary()), output_dim=16384)\n emb_output = tf.reduce_mean(emb_layer(model_input), axis=1)\n dense_output = tf.keras.layers.Dense(\n units=1, activation=\"sigmoid\")(emb_output)\n model = tf.keras.Model({\"features\": model_input}, dense_output)\n\n optimizer = tf.keras.optimizers.RMSprop(learning_rate=0.1)\n accuracy = tf.keras.metrics.Accuracy()\n",
"_____no_output_____"
],
[
"assert len(emb_layer.weights) == 2\nassert emb_layer.weights[0].shape == (4, 16384)\nassert emb_layer.weights[1].shape == (4, 16384)\nassert emb_layer.weights[0].device == \"/job:ps/replica:0/task:0/device:CPU:0\"\nassert emb_layer.weights[1].device == \"/job:ps/replica:0/task:1/device:CPU:0\"\n",
"_____no_output_____"
],
[
"@tf.function\ndef step_fn(iterator):\n\n def replica_fn(batch_data, labels):\n with tf.GradientTape() as tape:\n pred = model(batch_data, training=True)\n per_example_loss = tf.keras.losses.BinaryCrossentropy(\n reduction=tf.keras.losses.Reduction.NONE)(labels, pred)\n loss = tf.nn.compute_average_loss(per_example_loss)\n gradients = tape.gradient(loss, model.trainable_variables)\n\n optimizer.apply_gradients(zip(gradients, model.trainable_variables))\n\n actual_pred = tf.cast(tf.greater(pred, 0.5), tf.int64)\n accuracy.update_state(labels, actual_pred)\n return loss\n\n batch_data, labels = next(iterator)\n losses = strategy.run(replica_fn, args=(batch_data, labels))\n return strategy.reduce(tf.distribute.ReduceOp.SUM, losses, axis=None)\n",
"_____no_output_____"
],
[
"coordinator = tf.distribute.experimental.coordinator.ClusterCoordinator(\n strategy)\n",
"_____no_output_____"
],
[
"@tf.function\ndef per_worker_dataset_fn():\n return strategy.distribute_datasets_from_function(dataset_fn)\n\n\nper_worker_dataset = coordinator.create_per_worker_dataset(\n per_worker_dataset_fn)\nper_worker_iterator = iter(per_worker_dataset)\n",
"WARNING:tensorflow:Model was constructed with shape (None, 3) for input KerasTensor(type_spec=TensorSpec(shape=(None, 3), dtype=tf.string, name='feature'), name='feature', description=\"created by layer 'feature'\"), but it was called on an input with incompatible shape (3,).\n"
],
[
"num_epoches = 4\nsteps_per_epoch = 5\nfor i in range(num_epoches):\n accuracy.reset_states()\n for _ in range(steps_per_epoch):\n coordinator.schedule(step_fn, args=(per_worker_iterator,))\n # Wait at epoch boundaries.\n coordinator.join()\n print(\"Finished epoch %d, accuracy is %f.\" % (i, accuracy.result().numpy()))\n",
"INFO:tensorflow:Reduce to /device:CPU:0 then broadcast to ('/replica:0/device:CPU:0',).\nINFO:tensorflow:Reduce to /device:CPU:0 then broadcast to ('/replica:0/device:CPU:0',).\nFinished epoch 0, accuracy is 0.675000.\nFinished epoch 1, accuracy is 0.500000.\nFinished epoch 2, accuracy is 0.781250.\nFinished epoch 3, accuracy is 1.000000.\n"
],
[
"loss = coordinator.schedule(step_fn, args=(per_worker_iterator,))\nprint(\"Final loss is %f\" % loss.fetch())\n",
"Final loss is 0.000000\n"
],
[
"eval_dataset = tf.data.Dataset.from_tensor_slices(\n feature_and_label_gen(num_examples=16)).map(\n lambda x: (\n {\"features\": feature_preprocess_stage(x[\"features\"])},\n label_preprocess_stage(x[\"label\"])\n )).batch(8)\n\neval_accuracy = tf.keras.metrics.Accuracy()\n\nfor batch_data, labels in eval_dataset:\n pred = model(batch_data, training=False)\n actual_pred = tf.cast(tf.greater(pred, 0.5), tf.int64)\n eval_accuracy.update_state(labels, actual_pred)\n\nprint(\"Evaluation accuracy: %f\" % eval_accuracy.result())\n",
"WARNING:tensorflow:Model was constructed with shape (None, 3) for input KerasTensor(type_spec=TensorSpec(shape=(None, 3), dtype=tf.string, name='feature'), name='feature', description=\"created by layer 'feature'\"), but it was called on an input with incompatible shape (3,).\nEvaluation accuracy: 1.000000\n"
],
[
"with strategy.scope():\n # Define the eval metric on parameter servers.\n eval_accuracy = tf.keras.metrics.Accuracy()\n\n\[email protected]\ndef eval_step(iterator):\n def replica_fn(batch_data, labels):\n pred = model(batch_data, training=False)\n actual_pred = tf.cast(tf.greater(pred, 0.5), tf.int64)\n eval_accuracy.update_state(labels, actual_pred)\n batch_data, labels = next(iterator)\n strategy.run(replica_fn, args=(batch_data, labels))\n\n\ndef eval_dataset_fn():\n return tf.data.Dataset.from_tensor_slices(\n feature_and_label_gen(num_examples=16)).map(\n lambda x: (\n {\"features\": feature_preprocess_stage(x[\"features\"])},\n label_preprocess_stage(x[\"label\"])\n )).shuffle(16).repeat().batch(8)\n\n\nper_worker_eval_dataset = coordinator.create_per_worker_dataset(\n eval_dataset_fn)\nper_worker_eval_iterator = iter(per_worker_eval_dataset)\n\neval_steps_per_epoch = 2\nfor _ in range(eval_steps_per_epoch):\n coordinator.schedule(eval_step, args=(per_worker_eval_iterator,))\ncoordinator.join()\nprint(\"Evaluation accuracy: %f\" % eval_accuracy.result())\n",
"WARNING:tensorflow:Model was constructed with shape (None, 3) for input KerasTensor(type_spec=TensorSpec(shape=(None, 3), dtype=tf.string, name='feature'), name='feature', description=\"created by layer 'feature'\"), but it was called on an input with incompatible shape (3,).\nWARNING:tensorflow:1 GPUs are allocated per worker. Please use DistributedDataset by calling strategy.experimental_distribute_dataset or strategy.distribute_datasets_from_function to make best use of GPU resources\nWARNING:tensorflow:1 GPUs are allocated per worker. Please use DistributedDataset by calling strategy.experimental_distribute_dataset or strategy.distribute_datasets_from_function to make best use of GPU resources\nWARNING:tensorflow:1 GPUs are allocated per worker. Please use DistributedDataset by calling strategy.experimental_distribute_dataset or strategy.distribute_datasets_from_function to make best use of GPU resources\nEvaluation accuracy: 1.000000\n"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbeaa83d7c1d7b664c6172ac7e965a2a956c300f
| 209,362 |
ipynb
|
Jupyter Notebook
|
examples/misc.ipynb
|
mforets/polyhedron_tools
|
d95a75c76af84bc75737fcdaa076e579a69bef69
|
[
"MIT"
] | null | null | null |
examples/misc.ipynb
|
mforets/polyhedron_tools
|
d95a75c76af84bc75737fcdaa076e579a69bef69
|
[
"MIT"
] | 2 |
2017-05-10T12:05:55.000Z
|
2017-05-19T10:58:14.000Z
|
examples/misc.ipynb
|
mforets/polyhedron_tools
|
d95a75c76af84bc75737fcdaa076e579a69bef69
|
[
"MIT"
] | null | null | null | 157.415038 | 43,592 | 0.864937 |
[
[
[
"**Objective**. This notebook contains illustrating examples for the utilities in the [polyhedron_tools](https://github.com/mforets/polyhedron_tools) module. ",
"_____no_output_____"
]
],
[
[
"%display typeset",
"_____no_output_____"
]
],
[
[
"## 1. Modeling with Polyhedra: back and forth with half-space representation \n\nWe present examples for creating Polyhedra from matrices and conversely to obtain matrices from Polyhedra.",
"_____no_output_____"
]
],
[
[
"from polyhedron_tools.misc import polyhedron_from_Hrep, polyhedron_to_Hrep\n\nA = matrix([[-1.0, 0.0, 0.0, 0.0, 0.0, 0.0],\n[ 1.0, 0.0, 0.0, 0.0, 0.0, 0.0],\n[ 0.0, 1.0, 0.0, 0.0, 0.0, 0.0],\n[ 0.0, -1.0, 0.0, 0.0, 0.0, 0.0],\n[ 0.0, 0.0, -1.0, 0.0, 0.0, 0.0],\n[ 0.0, 0.0, 1.0, 0.0, 0.0, 0.0],\n[ 0.0, 0.0, 0.0, -1.0, 0.0, 0.0],\n[ 0.0, 0.0, 0.0, 1.0, 0.0, 0.0],\n[ 0.0, 0.0, 0.0, 0.0, 1.0, 0.0],\n[ 0.0, 0.0, 0.0, 0.0, -1.0, 0.0],\n[ 0.0, 0.0, 0.0, 0.0, 0.0, 1.0],\n[ 0.0, 0.0, 0.0, 0.0, 0.0, -1.0]])\n\nb = vector([0.0, 10.0, 0.0, 0.0, 0.2, 0.2, 0.1, 0.1, 0.0, 0.0, 0.0, 0.0])\n\nP = polyhedron_from_Hrep(A, b); \nP",
"_____no_output_____"
],
[
"P.inequalities()",
"_____no_output_____"
],
[
"P.equations()",
"_____no_output_____"
]
],
[
[
"It is possible to obtain the matrices that represent the inequality and the equality constraints separately, using the keyword argument `separate_equality_constraints`. This type of information is somtimes useful for optimization solvers.",
"_____no_output_____"
]
],
[
[
"[A, b, Aeq, beq] = polyhedron_to_Hrep(P, separate_equality_constraints = True)",
"_____no_output_____"
],
[
"A, b",
"_____no_output_____"
],
[
"Aeq, beq",
"_____no_output_____"
]
],
[
[
"## 2. Generating polyhedra",
"_____no_output_____"
],
[
"### Constructing hyperrectangles",
"_____no_output_____"
],
[
"Let's construct a ball in the infinity norm, specifying the center and radius. \n\nWe remark that the case of a hyperbox can be done in Sage's library `polytopes.hypercube(n)` where `n` is the dimension. However, as of Sage v7.6. there is no such hyperrectangle function (or n-orthotope, see the [wikipedia page](https://en.wikipedia.org/wiki/Hyperrectangle)), so we use `BoxInfty`.",
"_____no_output_____"
]
],
[
[
"from polyhedron_tools.misc import BoxInfty",
"_____no_output_____"
],
[
"P = BoxInfty(center=[1,2,3], radius=0.1); P.plot(aspect_ratio=1)",
"_____no_output_____"
]
],
[
[
"As a side note, the function also works when the arguments are not named, as in",
"_____no_output_____"
]
],
[
[
"P = BoxInfty([1,2,3], 0.1); P",
"_____no_output_____"
]
],
[
[
"Another use of `BoxInfty` is to specify the lengths of the sides. For example:",
"_____no_output_____"
]
],
[
[
"P = BoxInfty([[0,1], [2,3]]); P",
"_____no_output_____"
]
],
[
[
"### Random polyhedra",
"_____no_output_____"
],
[
"The `random_polygon_2d` receives the number of arguments as input and produces a polygon whose vertices are randomly sampled from the unit circle. See the docstring for more another options.",
"_____no_output_____"
]
],
[
[
"from polyhedron_tools.polygons import random_polygon_2d\n\nrandom_polygon_2d(5)",
"_____no_output_____"
]
],
[
[
"### Opposite polyhedron",
"_____no_output_____"
]
],
[
[
"from polyhedron_tools.misc import BoxInfty, opposite_polyhedron\n\nP = BoxInfty([1,1], 0.5);\nmp = opposite_polyhedron(P);",
"_____no_output_____"
],
[
"P.plot(aspect_ratio=1) + mp.plot(color='red')",
"_____no_output_____"
]
],
[
[
"## 3. Miscelaneous functions",
"_____no_output_____"
],
[
"### Support function of a polytope",
"_____no_output_____"
]
],
[
[
"from polyhedron_tools.misc import support_function\n\nP = BoxInfty([1,2,3,4,5], 1); P\nsupport_function(P, [1,-1,1,-1,1], verbose=1)",
"**** Solve LP ****\nMaximization:\n x_0 - x_1 + x_2 - x_3 + x_4 \n\nConstraints:\n x_0 <= 2.0\n x_1 <= 3.0\n x_2 <= 4.0\n x_3 <= 5.0\n x_4 <= 6.0\n - x_0 <= 0.0\n - x_4 <= -4.0\n - x_3 <= -3.0\n - x_2 <= -2.0\n - x_1 <= -1.0\nVariables:\n x_0 is a continuous variable (min=-oo, max=+oo)\n x_1 is a continuous variable (min=-oo, max=+oo)\n x_2 is a continuous variable (min=-oo, max=+oo)\n x_3 is a continuous variable (min=-oo, max=+oo)\n x_4 is a continuous variable (min=-oo, max=+oo)\nObjective Value: 8.0\nx_0 = 2.000000\nx_1 = 1.000000\nx_2 = 4.000000\nx_3 = 3.000000\nx_4 = 6.000000\n\n\n"
]
],
[
[
"It is also possible to input the polyhedron in matrix form, $[A, b]$. If this is possible, it is preferable, since it is often faster. Below is an example with $12$ variables. We get beteen 3x and 4x improvement in the second case. ",
"_____no_output_____"
]
],
[
[
"reset('P, A, b')\nP = BoxInfty([1,2,3,4,5,6,7,8,9,10,11,12], 1); P\n[A, b] = polyhedron_to_Hrep(P)",
"_____no_output_____"
],
[
"timeit('support_function(P, [1,-1,1,-1,1,-1,1,-1,1,-1,1,-1])')",
"125 loops, best of 3: 1.77 ms per loop\n"
],
[
"support_function([A, b], [1,-1,1,-1,1,-1,1,-1,1,-1,1,-1])",
"_____no_output_____"
],
[
"timeit('support_function([A, b], [1,-1,1,-1,1,-1,1,-1,1,-1,1,-1])')",
"625 loops, best of 3: 421 µs per loop\n"
]
],
[
[
"### Support function of an ellipsoid",
"_____no_output_____"
]
],
[
[
"from polyhedron_tools.misc import support_function, support_function_ellipsoid \nimport random\n\n# Generate a random ellipsoid and check support function outer approximation.\n# Define an ellipse as: x^T*Q*x <= 1\nM = random_matrix(RR, 2, distribution=\"uniform\")\nQ = M.T*M\nf = lambda x, y : Q[0,0]*x^2 + Q[1,1]*y^2 + (Q[0,1]+Q[1,0])*x*y-1\nE = implicit_plot(f,(-5,5),(-3,3),fill=True,alpha=0.5,plot_points=600)\n\n# generate at random k directions, and compute the overapproximation of E using support functions\n# It works 'in average': we might get unbounded domains (random choice did not enclose the ellipsoid).\n# It is recommended to use QQ as base_ring to avoid 'frozen set' issues.\nk=15\nA = matrix(RR,k,2); b = vector(RR,k)\nfor i in range(k):\n theta = random.uniform(0, 2*pi.n(digits=5))\n d = vector(RR,[cos(theta), sin(theta)])\n s_fun = support_function_ellipsoid(Q, d)\n A.set_row(i,d); b[i] = s_fun\n\nOmegaApprox = polyhedron_from_Hrep(A, b, base_ring = QQ)\nE + OmegaApprox.plot(fill=False, color='red')",
"_____no_output_____"
]
],
[
[
"### Supremum norm of a polyhedron",
"_____no_output_____"
]
],
[
[
"from polyhedron_tools.misc import BoxInfty, radius",
"_____no_output_____"
],
[
"P = BoxInfty([-13,24,-51,18.54,309],27.04);\nradius(P)",
"_____no_output_____"
],
[
"got_lengths, got_center_and_radius = False, False",
"_____no_output_____"
],
[
"got_lengths is not False",
"_____no_output_____"
],
[
"radius(polyhedron_to_Hrep(P))",
"_____no_output_____"
],
[
"8401/25.n()",
"_____no_output_____"
]
],
[
[
"Consider a higher-dimensional system. We obtain almost a 200x improvement for a 15-dimensional set. This is because in the case we call ```poly_sup_norm``` with a polytope, the ```bounding_box()``` function consumes time. ",
"_____no_output_____"
]
],
[
[
"%%time \nP = BoxInfty([1,1,1,1,1,1,1,1,1,1,1,1,1,1,1], 14.28);\nradius(P)",
"CPU times: user 1.4 s, sys: 45.5 ms, total: 1.44 s\nWall time: 1.44 s\n"
],
[
"%%time \n[A, b] = BoxInfty([1,1,1,1,1,1,1,1,1,1,1,1,1,1,1], 14.28, base_ring = RDF, return_HSpaceRep = True)\nradius([A, b])",
"CPU times: user 20.4 ms, sys: 1.19 ms, total: 21.6 ms\nWall time: 20.9 ms\n"
]
],
[
[
"The improvement in speed is quite interesting! Moreover, for a 20-dimensional set, the polyhedron construct does not finish.",
"_____no_output_____"
],
[
"### Linear map of a polyhedron",
"_____no_output_____"
],
[
"The operation `matrix x polyhedron` can be performed in Sage with the usual command, `*`.",
"_____no_output_____"
]
],
[
[
"U = BoxInfty([[0,1],[0,1]])\nB = matrix([[1,1],[1,-1]])\nP = B * U\nP.plot(color='red', alpha=0.5) + U.plot()",
"_____no_output_____"
]
],
[
[
"### Chebyshev center",
"_____no_output_____"
]
],
[
[
"from polyhedron_tools.misc import chebyshev_center, BoxInfty\nfrom polyhedron_tools.polygons import random_polygon_2d\n\nP = random_polygon_2d(10, base_ring = QQ)\nc = chebyshev_center(P); \n\nB = BoxInfty([[1,2],[0,1]])\nb = chebyshev_center(B)\n\nfig = point(c, color='blue') + P.plot(color='blue', alpha=0.2)\nfig += point(b, color='red') + B.plot(color='red', alpha=0.2)\n\nfig += point(P.center().n(), color='green',marker='x')\nfig += point(B.center().n(), color='green',marker='x')\n\nfig",
"_____no_output_____"
]
],
[
[
"The method ```center()``` existent in the Polyhedra class, computes the average of the vertices. In contrast, the Chebyshev center is the center of the largest box enclosed by the polytope.",
"_____no_output_____"
]
],
[
[
"B.bounding_box()",
"_____no_output_____"
],
[
"P.bounding_box()",
"_____no_output_____"
],
[
"e = [ (P.bounding_box()[0][0] + P.bounding_box()[1][0])/2, (P.bounding_box()[0][1] + P.bounding_box()[1][1])/2]\nl = [[P.bounding_box()[0][0], P.bounding_box()[1][0]], [P.bounding_box()[0][1], P.bounding_box()[1][1]] ]\n\nfig += point(e,color='black') + BoxInfty(lengths=l).plot(alpha=0.1,color='grey')\n\nfig",
"_____no_output_____"
]
],
[
[
"Here we have added in grey the bounding box that is obtained from the method ```bounding_box()```. To make the picture complete, we should also add the box of center Cheby center, and of maximal radius which is included in it.",
"_____no_output_____"
],
[
"## 4. Approximate projections",
"_____no_output_____"
]
],
[
[
"from polyhedron_tools.projections import lotov_algo\nfrom polyhedron_tools.misc import polyhedron_to_Hrep, polyhedron_from_Hrep \n\nA = matrix([[-1.0, 0.0, 0.0, 0.0, 0.0, 0.0],\n[ 1.0, 0.0, 0.0, 0.0, 0.0, 0.0],\n[ 0.0, 1.0, 0.0, 0.0, 0.0, 0.0],\n[ 0.0, -1.0, 0.0, 0.0, 0.0, 0.0],\n[ 0.0, 0.0, -1.0, 0.0, 0.0, 0.0],\n[ 0.0, 0.0, 1.0, 0.0, 0.0, 0.0],\n[ 0.0, 0.0, 0.0, -1.0, 0.0, 0.0],\n[ 0.0, 0.0, 0.0, 1.0, 0.0, 0.0],\n[ 0.0, 0.0, 0.0, 0.0, 1.0, 0.0],\n[ 0.0, 0.0, 0.0, 0.0, -1.0, 0.0],\n[ 0.0, 0.0, 0.0, 0.0, 0.0, 1.0],\n[ 0.0, 0.0, 0.0, 0.0, 0.0, -1.0]])\n\nb = vector([0.0, 10.0, 0.0, 0.0, 0.2, 0.2, 0.1, 0.1, 0.0, 0.0, 0.0, 0.0])\n\nP = polyhedron_from_Hrep(A, b); P",
"_____no_output_____"
],
[
"lotov_algo(A, b, [1,0,0], [0,1,0], 0.5)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
]
] |
cbeaac8909610f8ae71ef6734d1c1cdd53ca8995
| 17,106 |
ipynb
|
Jupyter Notebook
|
Content Based.ipynb
|
PhonnPyaeKyaw/Recommendation_system-Movie-Lens-dataset-
|
1635528e5d4674a1479c0fcec987f80abd9f4697
|
[
"MIT"
] | null | null | null |
Content Based.ipynb
|
PhonnPyaeKyaw/Recommendation_system-Movie-Lens-dataset-
|
1635528e5d4674a1479c0fcec987f80abd9f4697
|
[
"MIT"
] | null | null | null |
Content Based.ipynb
|
PhonnPyaeKyaw/Recommendation_system-Movie-Lens-dataset-
|
1635528e5d4674a1479c0fcec987f80abd9f4697
|
[
"MIT"
] | null | null | null | 36.62955 | 138 | 0.371624 |
[
[
[
"import numpy as np\nimport pandas as pd",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"ratings = pd.read_csv('ratings.csv', sep='\\t', encoding='latin-1', usecols=['user_id', 'movie_id', 'rating'])\n\nusers = pd.read_csv('users.csv', sep='\\t', encoding='latin-1', usecols=['user_id', 'gender', 'zipcode', 'age_desc', 'occ_desc'])\n\nmovies = pd.read_csv('movies.csv', sep='\\t', encoding='latin-1', usecols=['movie_id', 'title', 'genres'])",
"_____no_output_____"
],
[
"dataset = pd.merge(pd.merge(movies, ratings),users)\n\ndataset[['title','genres','rating']].sort_values('rating', ascending=False).head(20)",
"_____no_output_____"
],
[
"movies['genres'] = movies['genres'].str.split('|')\n\nmovies['genres'] = movies['genres'].fillna(\"\").astype('str')",
"_____no_output_____"
],
[
"from sklearn.feature_extraction.text import TfidfVectorizer #read_more about TfidVectorizer\ntf = TfidfVectorizer(analyzer='word',ngram_range=(1, 2),min_df=0, stop_words='english')\ntfidf_matrix = tf.fit_transform(movies['genres'])\ntfidf_matrix.shape",
"_____no_output_____"
],
[
"from sklearn.metrics.pairwise import linear_kernel\ncosine_sim = linear_kernel(tfidf_matrix, tfidf_matrix)\ncosine_sim[:4, :4]",
"_____no_output_____"
],
[
"titles = movies['title']\nindices = pd.Series(movies.index, index=movies['title'])\n\ndef genre_recommendations(title):\n idx = indices[title]\n sim_scores = list(enumerate(cosine_sim[idx]))\n sim_scores = sorted(sim_scores, key=lambda x: x[1], reverse=True)\n sim_scores = sim_scores[1:21]\n movie_indices = [i[0] for i in sim_scores]\n return titles.iloc[movie_indices]",
"_____no_output_____"
],
[
"indices",
"_____no_output_____"
],
[
"genre_recommendations('Jumanji (1995)').head(10)",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbeac4c6d7107e92b92a9012948b29c3f2076c9c
| 43,011 |
ipynb
|
Jupyter Notebook
|
matrix_one/day3.ipynb
|
jmical-mc/dw_matrix
|
e79226ec2a344d3a1175d2ed2fba5b84fccf303a
|
[
"MIT"
] | null | null | null |
matrix_one/day3.ipynb
|
jmical-mc/dw_matrix
|
e79226ec2a344d3a1175d2ed2fba5b84fccf303a
|
[
"MIT"
] | null | null | null |
matrix_one/day3.ipynb
|
jmical-mc/dw_matrix
|
e79226ec2a344d3a1175d2ed2fba5b84fccf303a
|
[
"MIT"
] | null | null | null | 43,011 | 43,011 | 0.720862 |
[
[
[
"#!pip install datadotworld\n#!pip install datadotworld[pandas]",
"_____no_output_____"
],
[
"#!dw configure",
"_____no_output_____"
],
[
"from google.colab import drive\nimport pandas as pd\nimport numpy as np\n\nimport datadotworld as dw",
"_____no_output_____"
],
[
"#drive.mount(\"/content/drive\")",
"_____no_output_____"
],
[
"ls",
"\u001b[0m\u001b[01;34mdrive\u001b[0m/ \u001b[01;34msample_data\u001b[0m/\n"
],
[
"cd \"drive/My Drive/Colab Notebooks/dw_matrix\"",
"/content/drive/My Drive/Colab Notebooks/dw_matrix\n"
],
[
"!mkdir data",
"mkdir: cannot create directory ‘data’: File exists\n"
],
[
"!echo 'data' > .gitignore",
"_____no_output_____"
],
[
"!git add .gitignore",
"_____no_output_____"
],
[
"data = dw.load_dataset('datafiniti/mens-shoe-prices')",
"_____no_output_____"
],
[
"df = data.dataframes['7004_1']\ndf.shape",
"/usr/local/lib/python3.6/dist-packages/datadotworld/models/dataset.py:209: UserWarning: Unable to set data frame dtypes automatically using 7004_1 schema. Data types may need to be adjusted manually. Error: Integer column has NA values in column 10\n 'Error: {}'.format(resource_name, e))\n/usr/local/lib/python3.6/dist-packages/datadotworld/util.py:121: DtypeWarning: Columns (39,45) have mixed types. Specify dtype option on import or set low_memory=False.\n return self._loader_func()\n"
],
[
"df.sample(5)",
"_____no_output_____"
],
[
"df.columns",
"_____no_output_____"
],
[
"df.prices_currency.unique()",
"_____no_output_____"
],
[
"df.prices_currency.value_counts()",
"_____no_output_____"
],
[
"df.prices_currency.value_counts(normalize=True)",
"_____no_output_____"
],
[
"df_usd = df[df.prices_currency == 'USD'].copy()\ndf_usd.shape",
"_____no_output_____"
],
[
"df_usd['prices_amountmin'] = df_usd.prices_amountmin.astype(np.float)\ndf_usd['prices_amountmin'].hist();",
"_____no_output_____"
],
[
"filter_max = np.percentile(df_usd['prices_amountmin'], 99)\nfilter_max",
"_____no_output_____"
],
[
"df_usd_filter = df_usd[df_usd['prices_amountmin'] < filter_max]",
"_____no_output_____"
],
[
"df_usd_filter.prices_amountmin.hist(bins=100);",
"_____no_output_____"
],
[
"df.to_csv('data/shoes_prices.csv', index=False)",
"_____no_output_____"
],
[
"!git add matrix_one/day3.ipynb",
"_____no_output_____"
],
[
"!git commit -m \"Read Men's Shoe Prices dataset from data.world part 2\"",
"\n*** Please tell me who you are.\n\nRun\n\n git config --global user.email \"[email protected]\"\n git config --global user.name \"Your Name\"\n\nto set your account's default identity.\nOmit --global to set the identity only in this repository.\n\nfatal: unable to auto-detect email address (got 'root@e3a20f28dd7a.(none)')\n"
],
[
"!git config --global user.email \"[email protected]\"\n!git config --global user.name \"Your Name\"",
"_____no_output_____"
],
[
"!git push -u origin master",
"Counting objects: 9, done.\nDelta compression using up to 2 threads.\nCompressing objects: 16% (1/6) \rCompressing objects: 33% (2/6) \rCompressing objects: 50% (3/6) \rCompressing objects: 66% (4/6) \rCompressing objects: 83% (5/6) \rCompressing objects: 100% (6/6) \rCompressing objects: 100% (6/6), done.\nWriting objects: 11% (1/9) \rWriting objects: 22% (2/9) \rWriting objects: 33% (3/9) \rWriting objects: 44% (4/9) \rWriting objects: 55% (5/9) \rWriting objects: 77% (7/9) \rWriting objects: 88% (8/9) \rWriting objects: 100% (9/9) \rWriting objects: 100% (9/9), 21.67 KiB | 1.81 MiB/s, done.\nTotal 9 (delta 2), reused 0 (delta 0)\nremote: Resolving deltas: 100% (2/2), done.\u001b[K\nTo https://github.com/jmical-mc/dw_matrix.git\n dc33f33..12504a7 master -> master\nBranch 'master' set up to track remote branch 'master' from 'origin'.\n"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbeac6ca44e84d02bf937e2623a1173212637ef1
| 2,505 |
ipynb
|
Jupyter Notebook
|
tactile/example.ipynb
|
lab-api/tactile
|
d71c1d13cba79bda1cd9a15753583141f18ebde2
|
[
"MIT"
] | null | null | null |
tactile/example.ipynb
|
lab-api/tactile
|
d71c1d13cba79bda1cd9a15753583141f18ebde2
|
[
"MIT"
] | null | null | null |
tactile/example.ipynb
|
lab-api/tactile
|
d71c1d13cba79bda1cd9a15753583141f18ebde2
|
[
"MIT"
] | null | null | null | 25.30303 | 138 | 0.577246 |
[
[
[
"from parametric import Parameter\nfrom tactile import AbsoluteStream, RelativeStream\n \n# stream = AbsoluteStream('VI61 Out', display_events=True).start()\n# stream = RelativeStream('Midi Fighter Twister', plus_value=65, minus_value=63, display_events=True, resolution=256).start()\nstream = RelativeStream(plus_value=65, minus_value=63, display_events=True, resolution=256).start() # auto-detect first device\n \n## attach a few parameters to the stream\nx = Parameter('x', 0, bounds = (-1, 1))\ny = Parameter('y', 0, bounds = (-1, 1))\n\n# stream.assign(20, x)\n# stream.assign(21, y)",
"_____no_output_____"
],
[
"stream.bind(x)",
"_____no_output_____"
]
],
[
[
"In the following example, we'll map a pair of knobs to the X and Y axes of a Mirrorcle PicoAmp MEMS mirror driver:",
"_____no_output_____"
]
],
[
[
"from ybdrivers.mirrorcle import PicoAmp\nfrom tactile import RelativeStream\n\n# mirror = PicoAmp('470016934')\n\nstream = RelativeStream('Midi Fighter Twister', plus_value=65, minus_value=63, display_events=True, resolution=256).start()\n# stream.assign(0, mirror.X)\n# stream.assign(1, mirror.Y)",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code"
] |
[
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
cbeaca682c7fa95a47beb33962b9f4863d46f768
| 278,381 |
ipynb
|
Jupyter Notebook
|
Classification_Cat_VS_Dogs_Transfer_Learning.ipynb
|
R-aryan/Image_Classification_VGG16
|
c22f6e43c4c75cd881c57c92c42c3d8a7e293321
|
[
"MIT"
] | 1 |
2021-12-23T10:58:43.000Z
|
2021-12-23T10:58:43.000Z
|
Classification_Cat_VS_Dogs_Transfer_Learning.ipynb
|
R-aryan/Image_Classification_VGG16
|
c22f6e43c4c75cd881c57c92c42c3d8a7e293321
|
[
"MIT"
] | null | null | null |
Classification_Cat_VS_Dogs_Transfer_Learning.ipynb
|
R-aryan/Image_Classification_VGG16
|
c22f6e43c4c75cd881c57c92c42c3d8a7e293321
|
[
"MIT"
] | 1 |
2021-07-07T20:03:49.000Z
|
2021-07-07T20:03:49.000Z
| 285.226434 | 131,332 | 0.886454 |
[
[
[
"<a href=\"https://colab.research.google.com/github/R-aryan/Image_Classification_VGG16/blob/master/Classification_Cat_VS_Dogs_Transfer_Learning.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"import keras,os\nfrom keras.models import Sequential\nfrom keras.layers import Dense, Conv2D, MaxPool2D , Flatten\nfrom keras.preprocessing.image import ImageDataGenerator\nfrom keras.applications.vgg16 import VGG16\nfrom keras.models import Model\nfrom keras import optimizers\nfrom keras.models import load_model\nimport numpy as np\n\nimport shutil\nfrom os import listdir\nfrom os.path import splitext\n\n\n\nfrom keras.preprocessing import image\n\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"train_directory= \"/content/drive/My Drive/classification_Dataset/cat_VS_dogs/train\"\ntest_directory=\"/content/drive/My Drive/classification_Dataset/cat_VS_dogs/test1\"",
"_____no_output_____"
],
[
"src= '/content/drive/My Drive/classification_Dataset/cat_VS_dogs/train'\ndest_d='/content/drive/My Drive/classification_Dataset/cat_VS_dogs/train/Dogs'\ndest_c='/content/drive/My Drive/classification_Dataset/cat_VS_dogs/train/Cats'\nvalidation_set='/content/drive/My Drive/classification_Dataset/cat_VS_dogs/validation_data'",
"_____no_output_____"
],
[
"trdata = ImageDataGenerator()\ntraindata = trdata.flow_from_directory(directory=src,target_size=(224,224),batch_size=32)",
"Found 20438 images belonging to 2 classes.\n"
],
[
"tsdata = ImageDataGenerator()\ntestdata = tsdata.flow_from_directory(directory=validation_set, target_size=(224,224),batch_size=32)",
"Found 98 images belonging to 2 classes.\n"
],
[
"",
"_____no_output_____"
]
],
[
[
"Here using the ImageDataGenerator method in keras I will import all the images of cat and dog in the model. ImageDataGenerator will automatically label the data and map all the labels to its specific data.",
"_____no_output_____"
]
],
[
[
"\nvggmodel = VGG16(weights='imagenet', include_top=True)",
"WARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/keras/backend/tensorflow_backend.py:66: The name tf.get_default_graph is deprecated. Please use tf.compat.v1.get_default_graph instead.\n\nWARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/keras/backend/tensorflow_backend.py:541: The name tf.placeholder is deprecated. Please use tf.compat.v1.placeholder instead.\n\nWARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/keras/backend/tensorflow_backend.py:4432: The name tf.random_uniform is deprecated. Please use tf.random.uniform instead.\n\nWARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/keras/backend/tensorflow_backend.py:4267: The name tf.nn.max_pool is deprecated. Please use tf.nn.max_pool2d instead.\n\nDownloading data from https://github.com/fchollet/deep-learning-models/releases/download/v0.1/vgg16_weights_tf_dim_ordering_tf_kernels.h5\n553467904/553467096 [==============================] - 16s 0us/step\nWARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/keras/backend/tensorflow_backend.py:190: The name tf.get_default_session is deprecated. Please use tf.compat.v1.get_default_session instead.\n\nWARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/keras/backend/tensorflow_backend.py:197: The name tf.ConfigProto is deprecated. Please use tf.compat.v1.ConfigProto instead.\n\nWARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/keras/backend/tensorflow_backend.py:203: The name tf.Session is deprecated. Please use tf.compat.v1.Session instead.\n\nWARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/keras/backend/tensorflow_backend.py:207: The name tf.global_variables is deprecated. Please use tf.compat.v1.global_variables instead.\n\nWARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/keras/backend/tensorflow_backend.py:216: The name tf.is_variable_initialized is deprecated. Please use tf.compat.v1.is_variable_initialized instead.\n\nWARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/keras/backend/tensorflow_backend.py:223: The name tf.variables_initializer is deprecated. Please use tf.compat.v1.variables_initializer instead.\n\n"
],
[
"",
"_____no_output_____"
]
],
[
[
"Here in this part I will import VGG16 from keras with pre-trained weights which was trained on imagenet. Here as you can see that include top parameter is set to true. This means that weights for our whole model will be downloaded. If this is set to false then the pre-trained weights will only be downloaded for convolution layers and no weights will be downloaded for dense layers.",
"_____no_output_____"
]
],
[
[
"vggmodel.summary()",
"Model: \"vgg16\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\ninput_1 (InputLayer) (None, 224, 224, 3) 0 \n_________________________________________________________________\nblock1_conv1 (Conv2D) (None, 224, 224, 64) 1792 \n_________________________________________________________________\nblock1_conv2 (Conv2D) (None, 224, 224, 64) 36928 \n_________________________________________________________________\nblock1_pool (MaxPooling2D) (None, 112, 112, 64) 0 \n_________________________________________________________________\nblock2_conv1 (Conv2D) (None, 112, 112, 128) 73856 \n_________________________________________________________________\nblock2_conv2 (Conv2D) (None, 112, 112, 128) 147584 \n_________________________________________________________________\nblock2_pool (MaxPooling2D) (None, 56, 56, 128) 0 \n_________________________________________________________________\nblock3_conv1 (Conv2D) (None, 56, 56, 256) 295168 \n_________________________________________________________________\nblock3_conv2 (Conv2D) (None, 56, 56, 256) 590080 \n_________________________________________________________________\nblock3_conv3 (Conv2D) (None, 56, 56, 256) 590080 \n_________________________________________________________________\nblock3_pool (MaxPooling2D) (None, 28, 28, 256) 0 \n_________________________________________________________________\nblock4_conv1 (Conv2D) (None, 28, 28, 512) 1180160 \n_________________________________________________________________\nblock4_conv2 (Conv2D) (None, 28, 28, 512) 2359808 \n_________________________________________________________________\nblock4_conv3 (Conv2D) (None, 28, 28, 512) 2359808 \n_________________________________________________________________\nblock4_pool (MaxPooling2D) (None, 14, 14, 512) 0 \n_________________________________________________________________\nblock5_conv1 (Conv2D) (None, 14, 14, 512) 2359808 \n_________________________________________________________________\nblock5_conv2 (Conv2D) (None, 14, 14, 512) 2359808 \n_________________________________________________________________\nblock5_conv3 (Conv2D) (None, 14, 14, 512) 2359808 \n_________________________________________________________________\nblock5_pool (MaxPooling2D) (None, 7, 7, 512) 0 \n_________________________________________________________________\nflatten (Flatten) (None, 25088) 0 \n_________________________________________________________________\nfc1 (Dense) (None, 4096) 102764544 \n_________________________________________________________________\nfc2 (Dense) (None, 4096) 16781312 \n_________________________________________________________________\npredictions (Dense) (None, 1000) 4097000 \n=================================================================\nTotal params: 138,357,544\nTrainable params: 138,357,544\nNon-trainable params: 0\n_________________________________________________________________\n"
]
],
[
[
"Now as I run vggmodel.summary() then the summary of the whole VGG model which was downloaded will be printed. Its output is attached below.",
"_____no_output_____"
]
],
[
[
"",
"_____no_output_____"
]
],
[
[
"After the model has been downloaded then I need to use this model for my problem statement which is to detect cats and dogs. So here I will set that I will not be training the weights of the first 19 layers and use it as it is. Therefore i am setting the trainable parameter to False for first 19 layers.",
"_____no_output_____"
]
],
[
[
"vggmodel.layers\n",
"_____no_output_____"
],
[
"for layers in (vggmodel.layers)[:19]:\n print(layers)\n layers.trainable = False",
"<keras.engine.input_layer.InputLayer object at 0x7fb35fb36128>\n<keras.layers.convolutional.Conv2D object at 0x7fb35fb36198>\n<keras.layers.convolutional.Conv2D object at 0x7fb324f381d0>\n<keras.layers.pooling.MaxPooling2D object at 0x7fb3244b5518>\n<keras.layers.convolutional.Conv2D object at 0x7fb324f46978>\n<keras.layers.convolutional.Conv2D object at 0x7fb324f4eb38>\n<keras.layers.pooling.MaxPooling2D object at 0x7fb324f52b00>\n<keras.layers.convolutional.Conv2D object at 0x7fb324f5a2b0>\n<keras.layers.convolutional.Conv2D object at 0x7fb324f5ae80>\n<keras.layers.convolutional.Conv2D object at 0x7fb3244c00b8>\n<keras.layers.pooling.MaxPooling2D object at 0x7fb3244c6a20>\n<keras.layers.convolutional.Conv2D object at 0x7fb3244d21d0>\n<keras.layers.convolutional.Conv2D object at 0x7fb3244d2eb8>\n<keras.layers.convolutional.Conv2D object at 0x7fb3244d8e80>\n<keras.layers.pooling.MaxPooling2D object at 0x7fb32264c978>\n<keras.layers.convolutional.Conv2D object at 0x7fb322659160>\n<keras.layers.convolutional.Conv2D object at 0x7fb322659f60>\n<keras.layers.convolutional.Conv2D object at 0x7fb32261fdd8>\n<keras.layers.pooling.MaxPooling2D object at 0x7fb32262c940>\n"
]
],
[
[
"Since my problem is to detect cats and dogs and it has two classes so the last dense layer of my model should be a 2 unit softmax dense layer. Here I am taking the second last layer of the model which is dense layer with 4096 units and adding a dense softmax layer of 2 units in the end. In this way I will remove the last layer of the VGG16 model which is made to predict 1000 classes.",
"_____no_output_____"
]
],
[
[
"\nX= vggmodel.layers[-2].output\npredictions = Dense(2, activation=\"softmax\")(X)\nmodel_final = Model(input = vggmodel.input, output = predictions)",
"/usr/local/lib/python3.6/dist-packages/ipykernel_launcher.py:4: UserWarning: Update your `Model` call to the Keras 2 API: `Model(inputs=Tensor(\"in..., outputs=Tensor(\"de...)`\n after removing the cwd from sys.path.\n"
]
],
[
[
"Now I will compile my new model. I will set the learning rate of SGD (Stochastic Gradient Descent) optimiser using lr parameter and since i have a 2 unit dense layer in the end so i will be using categorical_crossentropy as loss since the output of the model is categorical.",
"_____no_output_____"
]
],
[
[
"model_final.compile(loss = \"categorical_crossentropy\", optimizer = optimizers.SGD(lr=0.0001, momentum=0.9), metrics=[\"accuracy\"])",
"WARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/keras/optimizers.py:793: The name tf.train.Optimizer is deprecated. Please use tf.compat.v1.train.Optimizer instead.\n\nWARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/keras/backend/tensorflow_backend.py:3576: The name tf.log is deprecated. Please use tf.math.log instead.\n\n"
],
[
"model_final.summary()",
"Model: \"model_1\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\ninput_1 (InputLayer) (None, 224, 224, 3) 0 \n_________________________________________________________________\nblock1_conv1 (Conv2D) (None, 224, 224, 64) 1792 \n_________________________________________________________________\nblock1_conv2 (Conv2D) (None, 224, 224, 64) 36928 \n_________________________________________________________________\nblock1_pool (MaxPooling2D) (None, 112, 112, 64) 0 \n_________________________________________________________________\nblock2_conv1 (Conv2D) (None, 112, 112, 128) 73856 \n_________________________________________________________________\nblock2_conv2 (Conv2D) (None, 112, 112, 128) 147584 \n_________________________________________________________________\nblock2_pool (MaxPooling2D) (None, 56, 56, 128) 0 \n_________________________________________________________________\nblock3_conv1 (Conv2D) (None, 56, 56, 256) 295168 \n_________________________________________________________________\nblock3_conv2 (Conv2D) (None, 56, 56, 256) 590080 \n_________________________________________________________________\nblock3_conv3 (Conv2D) (None, 56, 56, 256) 590080 \n_________________________________________________________________\nblock3_pool (MaxPooling2D) (None, 28, 28, 256) 0 \n_________________________________________________________________\nblock4_conv1 (Conv2D) (None, 28, 28, 512) 1180160 \n_________________________________________________________________\nblock4_conv2 (Conv2D) (None, 28, 28, 512) 2359808 \n_________________________________________________________________\nblock4_conv3 (Conv2D) (None, 28, 28, 512) 2359808 \n_________________________________________________________________\nblock4_pool (MaxPooling2D) (None, 14, 14, 512) 0 \n_________________________________________________________________\nblock5_conv1 (Conv2D) (None, 14, 14, 512) 2359808 \n_________________________________________________________________\nblock5_conv2 (Conv2D) (None, 14, 14, 512) 2359808 \n_________________________________________________________________\nblock5_conv3 (Conv2D) (None, 14, 14, 512) 2359808 \n_________________________________________________________________\nblock5_pool (MaxPooling2D) (None, 7, 7, 512) 0 \n_________________________________________________________________\nflatten (Flatten) (None, 25088) 0 \n_________________________________________________________________\nfc1 (Dense) (None, 4096) 102764544 \n_________________________________________________________________\nfc2 (Dense) (None, 4096) 16781312 \n_________________________________________________________________\ndense_7 (Dense) (None, 2) 8194 \n=================================================================\nTotal params: 134,268,738\nTrainable params: 119,554,050\nNon-trainable params: 14,714,688\n_________________________________________________________________\n"
],
[
"from keras.callbacks import ModelCheckpoint, EarlyStopping\n\ncheckpoint = ModelCheckpoint(\"/content/drive/My Drive/classification_Dataset/vgg16_tl.h5\", monitor='val_acc', verbose=1, save_best_only=True, save_weights_only=False, mode='auto', period=1)\n\nearly = EarlyStopping(monitor='val_acc', min_delta=0, patience=40, verbose=1, mode='auto')\n\nmodel_final.fit_generator(generator= traindata, steps_per_epoch= 2, epochs= 100, validation_data= testdata, validation_steps=1, callbacks=[checkpoint,early])\n\nmodel_final.save_weights(\"/content/drive/My Drive/classification_Dataset/vgg16_tl.h5\")",
"Epoch 1/100\n2/2 [==============================] - 33s 17s/step - loss: 0.0410 - acc: 0.9844 - val_loss: 0.1768 - val_acc: 0.9688\n\nEpoch 00001: val_acc improved from -inf to 0.96875, saving model to /content/drive/My Drive/classification_Dataset/vgg16_tl.h5\nEpoch 2/100\n2/2 [==============================] - 33s 17s/step - loss: 0.0613 - acc: 0.9688 - val_loss: 0.0639 - val_acc: 0.9375\n\nEpoch 00002: val_acc did not improve from 0.96875\nEpoch 3/100\n2/2 [==============================] - 16s 8s/step - loss: 0.0630 - acc: 0.9688 - val_loss: 0.0261 - val_acc: 1.0000\n\nEpoch 00003: val_acc improved from 0.96875 to 1.00000, saving model to /content/drive/My Drive/classification_Dataset/vgg16_tl.h5\nEpoch 4/100\n2/2 [==============================] - 34s 17s/step - loss: 0.1175 - acc: 0.9688 - val_loss: 3.4574e-04 - val_acc: 1.0000\n\nEpoch 00004: val_acc did not improve from 1.00000\nEpoch 5/100\n2/2 [==============================] - 18s 9s/step - loss: 0.0174 - acc: 1.0000 - val_loss: 0.1632 - val_acc: 0.9688\n\nEpoch 00005: val_acc did not improve from 1.00000\nEpoch 6/100\n2/2 [==============================] - 17s 9s/step - loss: 0.0370 - acc: 0.9688 - val_loss: 0.0383 - val_acc: 1.0000\n\nEpoch 00006: val_acc did not improve from 1.00000\nEpoch 7/100\n2/2 [==============================] - 33s 17s/step - loss: 0.1946 - acc: 0.9688 - val_loss: 0.0509 - val_acc: 0.9688\n\nEpoch 00007: val_acc did not improve from 1.00000\nEpoch 8/100\n2/2 [==============================] - 37s 19s/step - loss: 0.1751 - acc: 0.9219 - val_loss: 5.1495e-04 - val_acc: 1.0000\n\nEpoch 00008: val_acc did not improve from 1.00000\nEpoch 9/100\n2/2 [==============================] - 40s 20s/step - loss: 0.1401 - acc: 0.9375 - val_loss: 0.2020 - val_acc: 0.9688\n\nEpoch 00009: val_acc did not improve from 1.00000\nEpoch 10/100\n2/2 [==============================] - 36s 18s/step - loss: 0.0923 - acc: 0.9531 - val_loss: 0.0186 - val_acc: 1.0000\n\nEpoch 00010: val_acc did not improve from 1.00000\nEpoch 11/100\n2/2 [==============================] - 36s 18s/step - loss: 0.1257 - acc: 0.9219 - val_loss: 0.0120 - val_acc: 1.0000\n\nEpoch 00011: val_acc did not improve from 1.00000\nEpoch 12/100\n2/2 [==============================] - 33s 16s/step - loss: 0.1794 - acc: 0.9688 - val_loss: 3.3763e-04 - val_acc: 1.0000\n\nEpoch 00012: val_acc did not improve from 1.00000\nEpoch 13/100\n2/2 [==============================] - 36s 18s/step - loss: 0.1376 - acc: 0.9688 - val_loss: 0.0325 - val_acc: 0.9688\n\nEpoch 00013: val_acc did not improve from 1.00000\nEpoch 14/100\n2/2 [==============================] - 18s 9s/step - loss: 0.0391 - acc: 0.9844 - val_loss: 0.0073 - val_acc: 1.0000\n\nEpoch 00014: val_acc did not improve from 1.00000\nEpoch 15/100\n2/2 [==============================] - 39s 20s/step - loss: 0.0782 - acc: 0.9688 - val_loss: 0.2249 - val_acc: 0.9688\n\nEpoch 00015: val_acc did not improve from 1.00000\nEpoch 16/100\n2/2 [==============================] - 37s 18s/step - loss: 0.0741 - acc: 0.9531 - val_loss: 0.0313 - val_acc: 1.0000\n\nEpoch 00016: val_acc did not improve from 1.00000\nEpoch 17/100\n2/2 [==============================] - 34s 17s/step - loss: 0.0833 - acc: 0.9375 - val_loss: 0.0394 - val_acc: 0.9688\n\nEpoch 00017: val_acc did not improve from 1.00000\nEpoch 18/100\n2/2 [==============================] - 20s 10s/step - loss: 0.0023 - acc: 1.0000 - val_loss: 0.0065 - val_acc: 1.0000\n\nEpoch 00018: val_acc did not improve from 1.00000\nEpoch 19/100\n2/2 [==============================] - 34s 17s/step - loss: 0.0213 - acc: 1.0000 - val_loss: 0.1982 - val_acc: 0.9688\n\nEpoch 00019: val_acc did not improve from 1.00000\nEpoch 20/100\n2/2 [==============================] - 36s 18s/step - loss: 0.0353 - acc: 0.9844 - val_loss: 2.0862e-07 - val_acc: 1.0000\n\nEpoch 00020: val_acc did not improve from 1.00000\nEpoch 21/100\n2/2 [==============================] - 34s 17s/step - loss: 0.0312 - acc: 0.9844 - val_loss: 0.2128 - val_acc: 0.9688\n\nEpoch 00021: val_acc did not improve from 1.00000\nEpoch 22/100\n2/2 [==============================] - 17s 9s/step - loss: 0.0367 - acc: 0.9844 - val_loss: 0.0090 - val_acc: 1.0000\n\nEpoch 00022: val_acc did not improve from 1.00000\nEpoch 23/100\n2/2 [==============================] - 35s 17s/step - loss: 0.1912 - acc: 0.9531 - val_loss: 0.0102 - val_acc: 1.0000\n\nEpoch 00023: val_acc did not improve from 1.00000\nEpoch 24/100\n2/2 [==============================] - 36s 18s/step - loss: 0.1545 - acc: 0.9531 - val_loss: 7.0010e-05 - val_acc: 1.0000\n\nEpoch 00024: val_acc did not improve from 1.00000\nEpoch 25/100\n2/2 [==============================] - 35s 17s/step - loss: 0.0566 - acc: 0.9688 - val_loss: 0.1862 - val_acc: 0.9688\n\nEpoch 00025: val_acc did not improve from 1.00000\nEpoch 26/100\n2/2 [==============================] - 38s 19s/step - loss: 0.0442 - acc: 0.9844 - val_loss: 0.0300 - val_acc: 1.0000\n\nEpoch 00026: val_acc did not improve from 1.00000\nEpoch 27/100\n2/2 [==============================] - 36s 18s/step - loss: 0.0669 - acc: 0.9844 - val_loss: 0.0146 - val_acc: 1.0000\n\nEpoch 00027: val_acc did not improve from 1.00000\nEpoch 28/100\n2/2 [==============================] - 37s 18s/step - loss: 0.0870 - acc: 0.9688 - val_loss: 7.1526e-07 - val_acc: 1.0000\n\nEpoch 00028: val_acc did not improve from 1.00000\nEpoch 29/100\n2/2 [==============================] - 36s 18s/step - loss: 0.0189 - acc: 1.0000 - val_loss: 0.0169 - val_acc: 1.0000\n\nEpoch 00029: val_acc did not improve from 1.00000\nEpoch 30/100\n2/2 [==============================] - 18s 9s/step - loss: 0.0881 - acc: 0.9531 - val_loss: 0.0467 - val_acc: 0.9688\n\nEpoch 00030: val_acc did not improve from 1.00000\nEpoch 31/100\n2/2 [==============================] - 33s 17s/step - loss: 0.1250 - acc: 0.9531 - val_loss: 0.2150 - val_acc: 0.9688\n\nEpoch 00031: val_acc did not improve from 1.00000\nEpoch 32/100\n2/2 [==============================] - 33s 17s/step - loss: 0.0451 - acc: 0.9844 - val_loss: 0.0012 - val_acc: 1.0000\n\nEpoch 00032: val_acc did not improve from 1.00000\nEpoch 33/100\n2/2 [==============================] - 35s 17s/step - loss: 0.0774 - acc: 0.9688 - val_loss: 0.2604 - val_acc: 0.9375\n\nEpoch 00033: val_acc did not improve from 1.00000\nEpoch 34/100\n2/2 [==============================] - 18s 9s/step - loss: 0.0099 - acc: 1.0000 - val_loss: 4.7287e-04 - val_acc: 1.0000\n\nEpoch 00034: val_acc did not improve from 1.00000\nEpoch 35/100\n2/2 [==============================] - 16s 8s/step - loss: 0.0589 - acc: 0.9688 - val_loss: 0.0199 - val_acc: 1.0000\n\nEpoch 00035: val_acc did not improve from 1.00000\nEpoch 36/100\n2/2 [==============================] - 34s 17s/step - loss: 0.0390 - acc: 0.9688 - val_loss: 9.8381e-04 - val_acc: 1.0000\n\nEpoch 00036: val_acc did not improve from 1.00000\nEpoch 37/100\n2/2 [==============================] - 36s 18s/step - loss: 0.0920 - acc: 0.9531 - val_loss: 0.0473 - val_acc: 0.9688\n\nEpoch 00037: val_acc did not improve from 1.00000\nEpoch 38/100\n2/2 [==============================] - 36s 18s/step - loss: 0.1967 - acc: 0.9531 - val_loss: 0.0027 - val_acc: 1.0000\n\nEpoch 00038: val_acc did not improve from 1.00000\nEpoch 39/100\n2/2 [==============================] - 18s 9s/step - loss: 0.0478 - acc: 0.9688 - val_loss: 0.1658 - val_acc: 0.9688\n\nEpoch 00039: val_acc did not improve from 1.00000\nEpoch 40/100\n2/2 [==============================] - 34s 17s/step - loss: 0.1162 - acc: 0.9531 - val_loss: 0.0020 - val_acc: 1.0000\n\nEpoch 00040: val_acc did not improve from 1.00000\nEpoch 41/100\n2/2 [==============================] - 17s 9s/step - loss: 0.0205 - acc: 1.0000 - val_loss: 0.1426 - val_acc: 0.9688\n\nEpoch 00041: val_acc did not improve from 1.00000\nEpoch 42/100\n2/2 [==============================] - 19s 9s/step - loss: 0.0108 - acc: 1.0000 - val_loss: 0.0129 - val_acc: 1.0000\n\nEpoch 00042: val_acc did not improve from 1.00000\nEpoch 43/100\n2/2 [==============================] - 34s 17s/step - loss: 0.0352 - acc: 0.9844 - val_loss: 0.0553 - val_acc: 0.9688\n\nEpoch 00043: val_acc did not improve from 1.00000\nEpoch 00043: early stopping\n"
],
[
"",
"_____no_output_____"
]
],
[
[
"Predicting the output",
"_____no_output_____"
]
],
[
[
"# from keras.preprocessing import image\n\n# import matplotlib.pyplot as plt\n\nimg = image.load_img(\"/content/drive/My Drive/classification_Dataset/cat_VS_dogs/test1/12500.jpg\",target_size=(224,224))\nimg = np.asarray(img)\n\nplt.imshow(img)\nimg = np.expand_dims(img, axis=0)\nfrom keras.models import load_model\nmodel_final.load_weights(\"/content/drive/My Drive/classification_Dataset/vgg16_tl.h5\")\n#saved_model.compile()\n\noutput = model_final.predict(img)\nif output[0][0] > output[0][1]:\n print(\"cat\")\nelse:\n print('dog')",
"_____no_output_____"
],
[
"def prediction(path_image):\n img = image.load_img(path_image,target_size=(224,224))\n img = np.asarray(img)\n\n\n plt.imshow(img)\n img = np.expand_dims(img, axis=0)\n\n model_final.load_weights(\"/content/drive/My Drive/classification_Dataset/vgg16_tl.h5\")\n\n output = model_final.predict(img)\n if output[0][0] > output[0][1]:\n print(\"cat\")\n else:\n \n print('dog')\n\n\n",
"_____no_output_____"
],
[
"prediction(\"/content/drive/My Drive/classification_Dataset/cat_VS_dogs/test1/12500.jpg\")\n",
"cat\n"
],
[
"prediction(\"/content/drive/My Drive/classification_Dataset/cat_VS_dogs/test1/12499.jpg\")",
"dog\n"
],
[
"",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
cbead6a0b36bbcc9c495cd8cf37709477fa25cfc
| 9,753 |
ipynb
|
Jupyter Notebook
|
primer.ipynb
|
PervykhDarya/TRO4
|
f9d7ab2656a6ad40ade2eef109b95a4483240c1e
|
[
"MIT"
] | null | null | null |
primer.ipynb
|
PervykhDarya/TRO4
|
f9d7ab2656a6ad40ade2eef109b95a4483240c1e
|
[
"MIT"
] | null | null | null |
primer.ipynb
|
PervykhDarya/TRO4
|
f9d7ab2656a6ad40ade2eef109b95a4483240c1e
|
[
"MIT"
] | null | null | null | 23.166271 | 82 | 0.490721 |
[
[
[
"import matplotlib.pyplot as plt\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"### Построение графика",
"_____no_output_____"
]
],
[
[
"import numpy as np\n# Независимая (x) и зависимая (y) переменные\nx = np.linspace(0, 10, 50)\ny = x\n# Построение графика\nplt.title(\"Линейная зависимость y = x\") # заголовок\nplt.xlabel(\"x\") # ось абсцисс\nplt.ylabel(\"y\") # ось ординат\nplt.grid() # включение отображение сетки\nplt.plot(x, y) # построение графика",
"_____no_output_____"
],
[
"# Построение графика\nplt.title(\"Линейная зависимость y = x\") # заголовок\nplt.xlabel(\"x\") # ось абсцисс\nplt.ylabel(\"y\") # ось ординат\nplt.grid() # включение отображение сетки\nplt.plot(x, y, \"r--\") # построение графика",
"_____no_output_____"
]
],
[
[
"### Несколько графиков на одном поле",
"_____no_output_____"
]
],
[
[
"# Линейная зависимость\nx = np.linspace(0, 10, 50)\ny1 = x\n# Квадратичная зависимость\ny2 = [i**2 for i in x]\n# Построение графика\nplt.title(\"Зависимости: y1 = x, y2 = x^2\") # заголовок\nplt.xlabel(\"x\") # ось абсцисс\nplt.ylabel(\"y1, y2\") # ось ординат\nplt.grid() # включение отображение сетки\nplt.plot(x, y1, x, y2) # построение графика",
"_____no_output_____"
]
],
[
[
"### Несколько разделенных полей с графиками",
"_____no_output_____"
]
],
[
[
"# Линейная зависимость\nx = np.linspace(0, 10, 50)\ny1 = x\n# Квадратичная зависимость\ny2 = [i**2 for i in x]\n# Построение графиков\nplt.figure(figsize=(9, 9))\nplt.subplot(2, 1, 1)\nplt.plot(x, y1) # построение графика\nplt.title(\"Зависимости: y1 = x, y2 = x^2\") # заголовок\nplt.ylabel(\"y1\", fontsize=14) # ось ординат\nplt.grid(True) # включение отображение сетки\nplt.subplot(2, 1, 2)\nplt.plot(x, y2) # построение графика\nplt.xlabel(\"x\", fontsize=14) # ось абсцисс\nplt.ylabel(\"y2\", fontsize=14) # ось ординат\nplt.grid(True) # включение отображения сетки",
"_____no_output_____"
]
],
[
[
"### Построение диаграммы для категориальных данных",
"_____no_output_____"
]
],
[
[
"fruits = [\"apple\", \"peach\", \"orange\", \"bannana\", \"melon\"]\ncounts = [34, 25, 43, 31, 17]\nplt.bar(fruits, counts)\nplt.title(\"Fruits!\")\nplt.xlabel(\"Fruit\")\nplt.ylabel(\"Count\")",
"_____no_output_____"
]
],
[
[
"### Основные элементы графика",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nfrom matplotlib.ticker import (MultipleLocator, FormatStrFormatter,\n\nAutoMinorLocator)\n\nimport numpy as np\nx = np.linspace(0, 10, 10)\ny1 = 4*x\ny2 = [i**2 for i in x]\nfig, ax = plt.subplots(figsize=(8, 6))\nax.set_title(\"Графики зависимостей: y1=4*x, y2=x^2\", fontsize=16)\nax.set_xlabel(\"x\", fontsize=14)\nax.set_ylabel(\"y1, y2\", fontsize=14)\nax.grid(which=\"major\", linewidth=1.2)\nax.grid(which=\"minor\", linestyle=\"--\", color=\"gray\", linewidth=0.5)\nax.scatter(x, y1, c=\"red\", label=\"y1 = 4*x\")\nax.plot(x, y2, label=\"y2 = x^2\")\nax.legend()\nax.xaxis.set_minor_locator(AutoMinorLocator())\nax.yaxis.set_minor_locator(AutoMinorLocator())\nax.tick_params(which='major', length=10, width=2)\nax.tick_params(which='minor', length=5, width=1)\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Текстовые надписи на графике",
"_____no_output_____"
]
],
[
[
"x = [1, 5, 10, 15, 20]\ny = [1, 7, 3, 5, 11]\nplt.plot(x, y, label='steel price')\nplt.title('Chart price', fontsize=15)\nplt.xlabel('Day', fontsize=12, color='blue')\nplt.ylabel('Price', fontsize=12, color='blue')\nplt.legend()\nplt.grid(True)\nplt.text(15, 4, 'grow up!')",
"_____no_output_____"
]
],
[
[
"### Работа с линейным графиком",
"_____no_output_____"
]
],
[
[
"x = [1, 5, 10, 15, 20]\ny = [1, 7, 3, 5, 11]\nplt.plot(x, y, '--')",
"_____no_output_____"
],
[
"x = [1, 5, 10, 15, 20]\ny = [1, 7, 3, 5, 11]\nline = plt.plot(x, y)\nplt.setp(line, linestyle='--')",
"_____no_output_____"
],
[
"x = [1, 5, 10, 15, 20]\ny1 = [1, 7, 3, 5, 11]\ny2 = [i*1.2 + 1 for i in y1]\ny3 = [i*1.2 + 1 for i in y2]\ny4 = [i*1.2 + 1 for i in y3]\nplt.plot(x, y1, '-', x, y2, '--', x, y3, '-.', x, y4, ':')",
"_____no_output_____"
],
[
"plt.plot(x, y1, '-')\nplt.plot(x, y2, '--')\nplt.plot(x, y3, '-.')\nplt.plot(x, y4, ':')",
"_____no_output_____"
]
],
[
[
"### Цвет линии",
"_____no_output_____"
]
],
[
[
"x = [1, 5, 10, 15, 20]\ny = [1, 7, 3, 5, 11]\nplt.plot(x, y, '--r')",
"_____no_output_____"
]
],
[
[
"### Тип графика",
"_____no_output_____"
]
],
[
[
"plt.plot(x, y, 'ro')",
"_____no_output_____"
],
[
"plt.plot(x, y, 'bx')",
"_____no_output_____"
]
],
[
[
"### Работа с функцией subplot()",
"_____no_output_____"
]
],
[
[
"# Исходный набор данных\nx = [1, 5, 10, 15, 20]\ny1 = [1, 7, 3, 5, 11]\ny2 = [i*1.2 + 1 for i in y1]\ny3 = [i*1.2 + 1 for i in y2]\ny4 = [i*1.2 + 1 for i in y3]\n# Настройка размеров подложки\nplt.figure(figsize=(12, 7))\n# Вывод графиков\nplt.subplot(2, 2, 1)\nplt.plot(x, y1, '-')\nplt.subplot(2, 2, 2)\nplt.plot(x, y2, '--')\nplt.subplot(2, 2, 3)\nplt.plot(x, y3, '-.')\nplt.subplot(2, 2, 4)\nplt.plot(x, y4, ':')",
"_____no_output_____"
]
],
[
[
"### Второй вариант использования subplot()",
"_____no_output_____"
]
],
[
[
"# Вывод графиков\nplt.subplot(221)\nplt.plot(x, y1, '-')\nplt.subplot(222)\nplt.plot(x, y2, '--')\nplt.subplot(223)\nplt.plot(x, y3, '-.')\nplt.subplot(224)\nplt.plot(x, y4, ':')",
"_____no_output_____"
]
],
[
[
"### Работа с функцией subplots()",
"_____no_output_____"
]
],
[
[
"fig, axs = plt.subplots(2, 2, figsize=(12, 7))\naxs[0, 0].plot(x, y1, '-')\naxs[0, 1].plot(x, y2, '--')\naxs[1, 0].plot(x, y3, '-.')\naxs[1, 1].plot(x, y4, ':')",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cbeaec3f70d80cebd575a37f3cca2faeb391bbd9
| 18,701 |
ipynb
|
Jupyter Notebook
|
Interactions.ipynb
|
Mayakshanesht/Kalman-and-bayesian-filters
|
967091f1c9640b4b3f96bbf5f422fee1b3e69778
|
[
"CC-BY-4.0"
] | null | null | null |
Interactions.ipynb
|
Mayakshanesht/Kalman-and-bayesian-filters
|
967091f1c9640b4b3f96bbf5f422fee1b3e69778
|
[
"CC-BY-4.0"
] | null | null | null |
Interactions.ipynb
|
Mayakshanesht/Kalman-and-bayesian-filters
|
967091f1c9640b4b3f96bbf5f422fee1b3e69778
|
[
"CC-BY-4.0"
] | null | null | null | 130.776224 | 13,356 | 0.844607 |
[
[
[
"# Interactions\n\nThis is a collection of interactions, mostly from the book. If you have are reading a print version of the book, or are reading it online via Github or nbviewer you will be unable to run the interactions.\n\nSo I have created this notebook. Here is how you run an interaction if you do not have IPython installed on your computer.\n\n1. Go to try.juptyer.org in your browser. It will launch a temporary notebook server for you.\n\n2. Click the **New** button and select `Python 3`. This will create a new notebook that will run Python 3 for you in your browser.\n\n3. Copy the entire contents of a cell from this notebook and paste it into a 'code' cell in the notebook on your browser. \n\n4. Press CTRL+ENTER to execute the cell.\n\n5. Have fun! Change code. Play. Experiment. Hack.\n\nYour server and notebook is not permanently saved. Once you close the session your data is lost. Yes, it says it is saving your file if you press save, and you can see it in the directory. But that is just happening in a Docker container that will be deleted as soon as you close the window. Copy and paste any changes you want to keep to an external file.\n\nOf course if you have IPython installed you can download this notebook and run it on your own computer. Type\n\n ipython notebook\n \nin a command prompt from the directory where you downloaded this file. Click on the name of this file to open it.",
"_____no_output_____"
],
[
"# Experimenting with FPF'\n\n\nThe Kalman filter uses the equation $P^- = FPF^\\mathsf{T}$ to compute the prior of the covariance matrix during the prediction step, where P is the covariance matrix and F is the system transistion function. For a Newtonian system $x = \\dot{x}\\Delta t + x_0$ F might look like\n\n$$F = \\begin{bmatrix}1 & \\Delta t\\\\0 & 1\\end{bmatrix}$$\n\n$FPF^\\mathsf{T}$ alters P by taking the correlation between the position ($x$) and velocity ($\\dot{x}$). This interactive plot lets you see the effect of different designs of F has on this value. For example,\n\n* what if $x$ is not correlated to $\\dot{x}$? (set F01 to 0)\n\n* what if $x = 2\\dot{x}\\Delta t + x_0$? (set F01 to 2)\n\n* what if $x = \\dot{x}\\Delta t + 2*x_0$? (set F00 to 2)\n\n* what if $x = \\dot{x}\\Delta t$? (set F00 to 0)",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\nfrom IPython.html.widgets import interact, interactive, fixed\nimport IPython.html.widgets as widgets\nimport numpy as np\nimport numpy.linalg as linalg\nimport math\nimport matplotlib.pyplot as plt\nfrom matplotlib.patches import Ellipse\n\ndef plot_covariance_ellipse(x, P, edgecolor='k', ls='solid'):\n U,s,v = linalg.svd(P)\n angle = math.atan2(U[1,0],U[0,0])\n width = math.sqrt(s[0]) * 2\n height = math.sqrt(s[1]) * 2\n\n ax = plt.gca()\n e = Ellipse(xy=(0, 0), width=width, height=height, angle=angle,\n edgecolor=edgecolor, facecolor='none',\n lw=2, ls=ls)\n ax.add_patch(e)\n ax.set_aspect('equal')\n \n \ndef plot_FPFT(F00, F01, F10, F11, covar):\n \n dt = 1.\n x = np.array((0, 0.))\n P = np.array(((1, covar), (covar, 2)))\n F = np.array(((F00, F01), (F10, F11)))\n\n plot_covariance_ellipse(x, P)\n plot_covariance_ellipse(x, np.dot(F, P).dot(F.T), edgecolor='r')\n #plt.axis('equal')\n plt.xlim(-4, 4)\n plt.ylim(-4, 4)\n plt.title(str(F))\n plt.xlabel('position')\n plt.ylabel('velocity')\n \ninteract(plot_FPFT, \n F00=widgets.IntSliderWidget(value=1, min=0, max=2.), \n F01=widgets.FloatSliderWidget(value=1, min=0., max=2., description='F01(dt)'),\n F10=widgets.FloatSliderWidget(value=0, min=0., max=2.),\n F11=widgets.FloatSliderWidget(value=1, min=0., max=2.),\n covar=widgets.FloatSliderWidget(value=0, min=0, max=1.));",
"_____no_output_____"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
]
] |
cbeaff5707d104e7612c6048bd41000ec59694fe
| 3,341 |
ipynb
|
Jupyter Notebook
|
if-else/blocos-identacao.ipynb
|
amarelopiupiu/python
|
bbd07a4b5e52d011c77f20622e17126f78fa3051
|
[
"MIT"
] | null | null | null |
if-else/blocos-identacao.ipynb
|
amarelopiupiu/python
|
bbd07a4b5e52d011c77f20622e17126f78fa3051
|
[
"MIT"
] | null | null | null |
if-else/blocos-identacao.ipynb
|
amarelopiupiu/python
|
bbd07a4b5e52d011c77f20622e17126f78fa3051
|
[
"MIT"
] | 1 |
2021-06-09T01:05:59.000Z
|
2021-06-09T01:05:59.000Z
| 25.899225 | 173 | 0.55223 |
[
[
[
"# Blocos e Identação\n\n## Estrutura\nSempre que usamos o if ou qualquer outra estrutura no Python, devemos usar a identação para dizer para o Programa onde a estrutura começa e onde ela termina.\n\nIsso vai ajudar muito quando tivermos mais de 1 condição ao mesmo tempo e quando tivermos várias ações para fazer dentro de um if.\n\n### Várias ações em 1 if:",
"_____no_output_____"
]
],
[
[
"if condicao:\n alguma coisa\n outra coisa\n outra coisa mais\n outra coisa ainda mais\nelse:\n uma coisa\n uma coisa mais\n coisa final",
"_____no_output_____"
]
],
[
[
"## Exemplo\nVamos fazer um novo exemplo abaixo:\n\nDigamos que você precisa criar um programa para um fundo de investimentos conseguir avaliar o resultado de uma carteira de ações e o quanto de taxa deverá ser pago.\n\nA regra desse fundo de investimentos é:\n\n- O fundo se compromete a entregar no mínimo 5% de retorno ao ano.\n- Caso o fundo não consiga entregar os 5% de retorno, ele não pode cobrar taxa dos seus investidores.\n- Caso o fundo consiga entregar mais de 5% de retorno, ele irá cobrar 2% de taxa dos seus investidores.\n- Caso o fundo consiga mais de 20% de retorno, ele irá cobrar 4% de taxa dos seus investidores.",
"_____no_output_____"
]
],
[
[
"retorno = 0.05\nfundo = 0.25\nif fundo > retorno:\n if fundo > 0.20:\n taxa = 0.04\n else:\n taxa = 0.50\nelse:\n taxa = 0\nprint(f'A taxa é de {taxa}')",
"A taxa é de 0.04\n"
]
],
[
[
"### Mais de uma condição ao mesmo tempo:",
"_____no_output_____"
]
],
[
[
"if condicao_1:\n o que fazer se a condição 1 for verdadeira\n if condicao_2:\n o que fazer se a condição 1 e 2 for verdadeira\n else:\n o que fazer se a condição 2 for falsa (mas a condição 1 é verdadeira)\nelse:\n o que fazer se a condição 1 for falsa",
"_____no_output_____"
]
],
[
[
"## Cuidado importante: repetição de código\n\nSempre que possível, evite repetir código. De forma geral:<br>\n\"Se você está repetindo um código, existe uma forma melhor de fazer\"",
"_____no_output_____"
]
]
] |
[
"markdown",
"raw",
"markdown",
"code",
"markdown",
"raw",
"markdown"
] |
[
[
"markdown"
],
[
"raw"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"raw"
],
[
"markdown"
]
] |
cbeb04533c5ba08ab9c28aa9268b5a532af9017d
| 10,812 |
ipynb
|
Jupyter Notebook
|
homwork2.ipynb
|
Fairy45/jxh
|
5781410e3b8314dc60435ffbc54269bedc474cd4
|
[
"Apache-2.0"
] | null | null | null |
homwork2.ipynb
|
Fairy45/jxh
|
5781410e3b8314dc60435ffbc54269bedc474cd4
|
[
"Apache-2.0"
] | null | null | null |
homwork2.ipynb
|
Fairy45/jxh
|
5781410e3b8314dc60435ffbc54269bedc474cd4
|
[
"Apache-2.0"
] | null | null | null | 25.027778 | 89 | 0.424898 |
[
[
[
"1",
"_____no_output_____"
]
],
[
[
"class Rectangle(object):\n def __init__(self,width,height):\n self.width=width\n self.height=height\n def getArea(self):\n q=self.height*self.width\n print(\"这个矩形的面积为:\",q)\n def getPerimeter(self):\n w=2*self.height+2*self.width\n print(\"这个矩形的周长为:\",w)\nqwe = Rectangle(10,11)\nqwe.getArea()\nqwe.getPerimeter()",
"这个矩形的面积为: 110\n这个矩形的周长为: 42\n"
]
],
[
[
"2",
"_____no_output_____"
]
],
[
[
"class Account(object):\n def __init__(self):\n self.lilv=0\n self.ann=100\n self.lixi=0\n def shuju(self,id,ann):\n self.id=id\n self.ann=ann\n def getMonthlyInterestRate(self,lilv):\n self.lilv=lilv\n def getMonthlyInterest(self):\n q=self.ann*self.lilv\n self.lixi=q\n def withdraw(self):\n print(\"请输入取钱金额\")\n res = input(\"输入\")\n self.ann = self.ann - int(res)\n print(\"您成功取出\",res,\"元\")\n def deposit(self):\n print(\"请输入存钱金额\")\n res1=input(\"输入\")\n self.ann=self.ann+int(res1)\n print(\"您成功存入\",res1,\"元\")\n def dayin(self):\n print(self.id,\"您账户余额为:\",self.ann,\"利率为:\",self.lilv,\"利息为\",self.lixi)\n \nqwe = Account()\nqwe.shuju(1122,20000)\nqwe.getMonthlyInterestRate(0.045)\nqwe.getMonthlyInterest()\nqwe.withdraw()\nqwe.deposit()\nqwe.dayin()\n",
"请输入取钱金额\n输入10000\n您成功取出 10000 元\n请输入存钱金额\n输入8000\n您成功存入 8000 元\n1122 您账户余额为: 18000 利率为: 0.045 利息为 900.0\n"
]
],
[
[
"3",
"_____no_output_____"
]
],
[
[
"class Fan(object):\n def __init__(self,speed=1,on=False,r=5,color='blue'):\n self.SLOW=1\n self.MEDIUM=2\n self.FAST=3\n self.__speed=int(speed)\n self.__on=bool(on)\n self.__r=float(r)\n self.__color=str(color) \n def prints(self):\n if self.__speed==1:\n print('SLOW')\n elif self.__speed==2:\n print('MEDIUM')\n elif self.__speed==3:\n print('FAST')\n if self.__on=='on':\n print('打开')\n else:\n print('关闭') \n print(self.__r)\n print(self.__color)\nif __name__ == \"__main__\":\n speed=int(input('选择速度:'))\n on=bool(input('on or off'))\n r=float(input('半径是:'))\n color=str(input('颜色是:'))\n fan=Fan(speed,on,r,color)\n fan.prints()",
"选择速度:2\non or off5\n半径是:2\n颜色是:1\nMEDIUM\n关闭\n2.0\n1\n"
]
],
[
[
"4",
"_____no_output_____"
]
],
[
[
"import math\nclass RegularPolygon(object):\n def __init__(self,n,side,x,y):\n self.n=n\n self.side=side\n self.x=x\n self.y=y\n def getPerimenter(self):\n print(self.n*self.side)\n def getArea(self):\n Area = self.n*self.side/(4*math.tan(math.pi/self.n))\n print(Area)\nqwe = RegularPolygon(20,5,10.6,7.8)\nqwe.getPerimenter()\nqwe.getArea()",
"100\n157.84378786687608\n"
]
],
[
[
"5",
"_____no_output_____"
]
],
[
[
"class LinearEquation(object): \n def __init__(self,a,b,c,d,e,f):\n self.__a=a\n self.__b=b\n self.__c=c\n self.__d=d\n self.__e=e\n self.__f=f \n def set_a(self,a):\n self.__a=a\n def get_a(self):\n return self.__a\n def set_b(self,b):\n self.__b=b\n def get_b(self):\n return self.___b\n def set_c(self,c):\n self.__c=c\n def get_c(self):\n return self.__c\n def set_d(self,d):\n self.__d=d\n def get_d(self):\n return self.__d\n def set_e(self,e):\n self.__e=e\n def get_e(self):\n return self.__e\n def set_f(self,f):\n self.__f=f\n def get_f(self):\n return self.__f\n def isSolvable(self):\n if (self.__a*self.__d)-(self.__c*self.__b)!=0:\n return True\n else:\n return print('此方程无解')\n def getX(self):\n s=(self.__a*self.__d)-(self.__b*self.__c)\n x=(self.__e*self.__d)-(self.__b*self.__f)/s\n print('X的值为:%.2f'% x)\n def getY(self):\n s=(self.__a*self.__d)-(self.__b*self.__c)\n y=(self.__a*self.__f)-(self.__e*self.__c)/s\n print('Y的值为:%.2f'% y)\nif __name__==\"__main__\":\n a=int(input('a的值是:'))\n b=int(input('b的值是:'))\n c=int(input('c的值是:'))\n d=int(input('d的值是:'))\n e=int(input('e的值是:'))\n f=int(input('f的值是:'))\n l=LinearEquation(a,b,c,d,e,f)\n l.isSolvable()\n l.getX()\n l.getY()",
"a的值是:4\nb的值是:5\nc的值是:4\nd的值是:4\ne的值是:7\nf的值是:8\nX的值为:38.00\nY的值为:39.00\n"
]
],
[
[
"6",
"_____no_output_____"
]
],
[
[
"class zuobao:\n def shur(self):\n import math \n x1,y1,x2,y2=map(float,input().split())\n x3,y3,x4,y4=map(float,input().split())\n u1=(x4-x3)*(y1-y3)-(x1-x3)*(y4-y3)\n v1=(x4-x3)*(y2-y3)-(x2-x3)*(y4-y3)\n u=math.fabs(u1)\n v=math.fabs(v1)\n\n x5=(x1*v+x2*u)/(u+v)\n y5=(y1*v+y2*u)/(u+v)\n print(x5,y5) \nre=zuobao()\nre.shur()",
"_____no_output_____"
]
],
[
[
"7",
"_____no_output_____"
]
],
[
[
"class LinearEquation(object): \n def __init__(self,a,b,c,d,e,f):\n self.__a=a\n self.__b=b\n self.__c=c\n self.__d=d\n self.__e=e\n self.__f=f \n def set_a(self,a):\n self.__a=a\n def get_a(self):\n return self.__a\n def set_b(self,b):\n self.__b=b\n def get_b(self):\n return self.___b\n def set_c(self,c):\n self.__c=c\n def get_c(self):\n return self.__c\n def set_d(self,d):\n self.__d=d\n def get_d(self):\n return self.__d\n def set_e(self,e):\n self.__e=e\n def get_e(self):\n return self.__e\n def set_f(self,f):\n self.__f=f\n def get_f(self):\n return self.__f\n def isSolvable(self):\n if (self.__a*self.__d)-(self.__c*self.__b)!=0:\n return True\n else:\n return print('此方程无解')\n def getX(self):\n s=(self.__a*self.__d)-(self.__b*self.__c)\n x=(self.__e*self.__d)-(self.__b*self.__f)/s\n print('X的值为:%.2f'% x)\n def getY(self):\n s=(self.__a*self.__d)-(self.__b*self.__c)\n y=(self.__a*self.__f)-(self.__e*self.__c)/s\n print('Y的值为:%.2f'% y)\nif __name__==\"__main__\":\n a=int(input('a的值是:'))\n b=int(input('b的值是:'))\n c=int(input('c的值是:'))\n d=int(input('d的值是:'))\n e=int(input('e的值是:'))\n f=int(input('f的值是:'))\n l=LinearEquation(a,b,c,d,e,f)\n l.isSolvable()\n l.getX()\n l.getY()",
"a的值是:7\nb的值是:8\nc的值是:9\nd的值是:10\ne的值是:77\nf的值是:88\nX的值为:1122.00\nY的值为:962.50\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cbeb05ca4c9f9c50d84cfc74f3836643564aa249
| 10,213 |
ipynb
|
Jupyter Notebook
|
notebooks/caip-training-hypertune.ipynb
|
jarokaz/caip-demo
|
60738b00f6024988c0702b1a0efe2322d8ebd5b1
|
[
"Apache-2.0"
] | null | null | null |
notebooks/caip-training-hypertune.ipynb
|
jarokaz/caip-demo
|
60738b00f6024988c0702b1a0efe2322d8ebd5b1
|
[
"Apache-2.0"
] | null | null | null |
notebooks/caip-training-hypertune.ipynb
|
jarokaz/caip-demo
|
60738b00f6024988c0702b1a0efe2322d8ebd5b1
|
[
"Apache-2.0"
] | null | null | null | 26.805774 | 166 | 0.542544 |
[
[
[
"# Hyperparameter tuning with AI Platform ",
"_____no_output_____"
]
],
[
[
"import os\nimport time",
"_____no_output_____"
]
],
[
[
"## Configure environment\n*You need to walk through the `local-experimentation.ipynb` notebook to create training and validation datasets.*\n",
"_____no_output_____"
]
],
[
[
"PROJECT_ID = !(gcloud config get-value core/project)\nARTIFACT_STORE = 'gs://{}-artifact-store'.format(PROJECT_ID[0])",
"_____no_output_____"
],
[
"TRAINING_DATA_PATH = '{}/datasets/training.csv'.format(ARTIFACT_STORE)\nTESTING_DATA_PATH = '{}/datasets/testing.csv'.format(ARTIFACT_STORE)\nREGION = \"us-central1\"\nJOBDIR_BUCKET = '{}/jobs'.format(ARTIFACT_STORE)",
"_____no_output_____"
]
],
[
[
"## Create a training application package\n### Create a training module",
"_____no_output_____"
]
],
[
[
"TRAINING_APP_FOLDER = '../hypertune_app/trainer'\n\nos.makedirs(TRAINING_APP_FOLDER, exist_ok=True)\n!touch $TRAINING_APP_FOLDER/__init__.py",
"_____no_output_____"
],
[
"%%writefile $TRAINING_APP_FOLDER/train.py\n\nimport logging\nimport os\nimport subprocess\nimport sys\n\nimport fire\nimport numpy as np\nimport pandas as pd\n\nimport hypertune\n\nfrom sklearn.model_selection import cross_val_score\nfrom sklearn.externals import joblib\nfrom sklearn.decomposition import PCA\nfrom sklearn.linear_model import Ridge\nfrom sklearn.pipeline import Pipeline\nfrom sklearn.preprocessing import StandardScaler\n\n\ndef train(job_dir, data_path, n_components, alpha):\n \n \n # Load data from GCS\n df_train = pd.read_csv(data_path)\n\n y = df_train.octane\n X = df_train.drop('octane', axis=1)\n \n # Configure a training pipeline\n pipeline = Pipeline([\n ('scale', StandardScaler()),\n ('reduce_dim', PCA(n_components=n_components)),\n ('regress', Ridge(alpha=alpha))\n ])\n\n # Calculate the performance metric\n scores = cross_val_score(pipeline, X, y, cv=10, scoring='neg_mean_squared_error')\n \n # Log it with hypertune\n hpt = hypertune.HyperTune()\n hpt.report_hyperparameter_tuning_metric(\n hyperparameter_metric_tag='neg_mean_squared_error',\n metric_value=scores.mean()\n )\n\n # Fit the model on a full dataset\n pipeline.fit(X, y)\n\n # Save the model\n model_filename = 'model.joblib'\n joblib.dump(value=pipeline, filename=model_filename)\n gcs_model_path = \"{}/{}\".format(job_dir, model_filename)\n subprocess.check_call(['gsutil', 'cp', model_filename, gcs_model_path], stderr=sys.stdout)\n logging.info(\"Saved model in: {}\".format(gcs_model_path)) \n \nif __name__ == \"__main__\":\n logging.basicConfig(level=logging.INFO)\n fire.Fire(train)",
"Overwriting ../hypertune_app/trainer/train.py\n"
]
],
[
[
"### Create hyperparameter configuration file",
"_____no_output_____"
]
],
[
[
"%%writefile $TRAINING_APP_FOLDER/hptuning_config.yaml \n\ntrainingInput:\n hyperparameters:\n goal: MAXIMIZE\n maxTrials: 12\n maxParallelTrials: 3\n hyperparameterMetricTag: neg_mean_squared_error\n enableTrialEarlyStopping: TRUE \n params:\n - parameterName: n_components\n type: DISCRETE\n discreteValues: [\n 2,\n 3,\n 4,\n 5,\n 6,\n 7,\n 8\n ]\n - parameterName: alpha\n type: DOUBLE\n minValue: 0.0001\n maxValue: 0.1\n scaleType: UNIT_LINEAR_SCALE\n ",
"Overwriting ../hypertune_app/trainer/hptuning_config.yaml\n"
]
],
[
[
"### Configure dependencies",
"_____no_output_____"
]
],
[
[
"%%writefile $TRAINING_APP_FOLDER/../setup.py\n\nfrom setuptools import find_packages\nfrom setuptools import setup\n\nREQUIRED_PACKAGES = ['fire', 'gcsfs', 'cloudml-hypertune']\n\nsetup(\n name='trainer',\n version='0.1',\n install_requires=REQUIRED_PACKAGES,\n packages=find_packages(),\n include_package_data=True,\n description='My training application package.'\n)",
"Overwriting ../hypertune_app/trainer/../setup.py\n"
]
],
[
[
"## Submit a training job",
"_____no_output_____"
]
],
[
[
"JOB_NAME = \"JOB_{}\".format(time.strftime(\"%Y%m%d_%H%M%S\"))\nSCALE_TIER = \"BASIC\"\nMODULE_NAME = \"trainer.train\"\nRUNTIME_VERSION = \"2.1\"\nPYTHON_VERSION = \"3.7\"",
"_____no_output_____"
],
[
"!gcloud ai-platform jobs submit training $JOB_NAME \\\n--region $REGION \\\n--job-dir $JOBDIR_BUCKET/$JOB_NAME \\\n--package-path $TRAINING_APP_FOLDER \\\n--module-name $MODULE_NAME \\\n--scale-tier $SCALE_TIER \\\n--python-version $PYTHON_VERSION \\\n--runtime-version $RUNTIME_VERSION \\\n--config $TRAINING_APP_FOLDER/hptuning_config.yaml \\\n-- \\\n--data_path $TRAINING_DATA_PATH ",
"Job [JOB_20200312_221336] submitted successfully.\nYour job is still active. You may view the status of your job with the command\n\n $ gcloud ai-platform jobs describe JOB_20200312_221336\n\nor continue streaming the logs with the command\n\n $ gcloud ai-platform jobs stream-logs JOB_20200312_221336\njobId: JOB_20200312_221336\nstate: QUEUED\n"
],
[
"!gcloud ai-platform jobs describe $JOB_NAME",
"createTime: '2020-03-12T22:13:38Z'\netag: 0aS-cH9IZ0U=\njobId: JOB_20200312_221336\nstate: PREPARING\ntrainingInput:\n args:\n - --data_path\n - gs://mlops-dev-100-artifact-store/datasets/training.csv\n hyperparameters:\n enableTrialEarlyStopping: true\n goal: MAXIMIZE\n hyperparameterMetricTag: neg_mean_squared_error\n maxParallelTrials: 3\n maxTrials: 12\n params:\n - discreteValues:\n - 2.0\n - 3.0\n - 4.0\n - 5.0\n - 6.0\n - 7.0\n - 8.0\n parameterName: n_components\n type: DISCRETE\n - maxValue: 0.1\n minValue: 0.0001\n parameterName: alpha\n scaleType: UNIT_LINEAR_SCALE\n type: DOUBLE\n jobDir: gs://mlops-dev-100-artifact-store/jobs/JOB_20200312_221336\n packageUris:\n - gs://mlops-dev-100-artifact-store/jobs/JOB_20200312_221336/packages/34eae760f2f652ef2b982f0a29cdc3cf2b5df0cddd31be408e1f03c0b9cde456/trainer-0.1.tar.gz\n pythonModule: trainer.train\n pythonVersion: '3.7'\n region: us-central1\n runtimeVersion: '2.1'\ntrainingOutput:\n isHyperparameterTuningJob: true\n\nView job in the Cloud Console at:\nhttps://console.cloud.google.com/mlengine/jobs/JOB_20200312_221336?project=mlops-dev-100\n\nView logs at:\nhttps://console.cloud.google.com/logs?resource=ml.googleapis.com%2Fjob_id%2FJOB_20200312_221336&project=mlops-dev-100\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
cbeb091263c00cc5c16a0a8ea4f9b192d66af41c
| 43,556 |
ipynb
|
Jupyter Notebook
|
lab1/Part1_TensorFlow.ipynb
|
IkhlasJihad/introtodeeplearning
|
e34cef9e832472af538b3be2e5b9bb789680566e
|
[
"MIT"
] | null | null | null |
lab1/Part1_TensorFlow.ipynb
|
IkhlasJihad/introtodeeplearning
|
e34cef9e832472af538b3be2e5b9bb789680566e
|
[
"MIT"
] | null | null | null |
lab1/Part1_TensorFlow.ipynb
|
IkhlasJihad/introtodeeplearning
|
e34cef9e832472af538b3be2e5b9bb789680566e
|
[
"MIT"
] | null | null | null | 54.377029 | 12,098 | 0.664776 |
[
[
[
"<table align=\"center\">\n <td align=\"center\"><a target=\"_blank\" href=\"http://introtodeeplearning.com\">\n <img src=\"https://i.ibb.co/Jr88sn2/mit.png\" style=\"padding-bottom:5px;\" />\n Visit MIT Deep Learning</a></td>\n <td align=\"center\"><a target=\"_blank\" href=\"https://colab.research.google.com/github/aamini/introtodeeplearning/blob/master/lab1/Part1_TensorFlow.ipynb\">\n <img src=\"https://i.ibb.co/2P3SLwK/colab.png\" style=\"padding-bottom:5px;\" />Run in Google Colab</a></td>\n <td align=\"center\"><a target=\"_blank\" href=\"https://github.com/aamini/introtodeeplearning/blob/master/lab1/Part1_TensorFlow.ipynb\">\n <img src=\"https://i.ibb.co/xfJbPmL/github.png\" height=\"70px\" style=\"padding-bottom:5px;\" />View Source on GitHub</a></td>\n</table>\n\n\n# Copyright Information\n",
"_____no_output_____"
]
],
[
[
"# Copyright 2022 MIT 6.S191 Introduction to Deep Learning. All Rights Reserved.\n# \n# Licensed under the MIT License. You may not use this file except in compliance\n# with the License. Use and/or modification of this code outside of 6.S191 must\n# reference:\n#\n# © MIT 6.S191: Introduction to Deep Learning\n# http://introtodeeplearning.com\n#",
"_____no_output_____"
]
],
[
[
"# Lab 1: Intro to TensorFlow and Music Generation with RNNs\n\nIn this lab, you'll get exposure to using TensorFlow and learn how it can be used for solving deep learning tasks. Go through the code and run each cell. Along the way, you'll encounter several ***TODO*** blocks -- follow the instructions to fill them out before running those cells and continuing.\n\n\n# Part 1: Intro to TensorFlow\n\n## 0.1 Install TensorFlow\n\nTensorFlow is a software library extensively used in machine learning. Here we'll learn how computations are represented and how to define a simple neural network in TensorFlow. For all the labs in 6.S191 2022, we'll be using the latest version of TensorFlow, TensorFlow 2, which affords great flexibility and the ability to imperatively execute operations, just like in Python. You'll notice that TensorFlow 2 is quite similar to Python in its syntax and imperative execution. Let's install TensorFlow and a couple of dependencies.\n",
"_____no_output_____"
]
],
[
[
"%tensorflow_version 2.x\nimport tensorflow as tf\n\n# Download and import the MIT 6.S191 package\n!pip install mitdeeplearning\nimport mitdeeplearning as mdl\n\nimport numpy as np\nimport matplotlib.pyplot as plt",
"Collecting mitdeeplearning\n Downloading mitdeeplearning-0.2.0.tar.gz (2.1 MB)\n\u001b[K |████████████████████████████████| 2.1 MB 12.5 MB/s \n\u001b[?25hRequirement already satisfied: numpy in /usr/local/lib/python3.7/dist-packages (from mitdeeplearning) (1.21.5)\nRequirement already satisfied: regex in /usr/local/lib/python3.7/dist-packages (from mitdeeplearning) (2019.12.20)\nRequirement already satisfied: tqdm in /usr/local/lib/python3.7/dist-packages (from mitdeeplearning) (4.62.3)\nRequirement already satisfied: gym in /usr/local/lib/python3.7/dist-packages (from mitdeeplearning) (0.17.3)\nRequirement already satisfied: cloudpickle<1.7.0,>=1.2.0 in /usr/local/lib/python3.7/dist-packages (from gym->mitdeeplearning) (1.3.0)\nRequirement already satisfied: scipy in /usr/local/lib/python3.7/dist-packages (from gym->mitdeeplearning) (1.4.1)\nRequirement already satisfied: pyglet<=1.5.0,>=1.4.0 in /usr/local/lib/python3.7/dist-packages (from gym->mitdeeplearning) (1.5.0)\nRequirement already satisfied: future in /usr/local/lib/python3.7/dist-packages (from pyglet<=1.5.0,>=1.4.0->gym->mitdeeplearning) (0.16.0)\nBuilding wheels for collected packages: mitdeeplearning\n Building wheel for mitdeeplearning (setup.py) ... \u001b[?25l\u001b[?25hdone\n Created wheel for mitdeeplearning: filename=mitdeeplearning-0.2.0-py3-none-any.whl size=2115442 sha256=7eaa984f4e7575f36f943ebbcb5aa71b356826b6c40f6396c8db4f2155dc4e6d\n Stored in directory: /root/.cache/pip/wheels/9a/b9/4f/99b7c8c5c75355550b83e1fcfc02956fb40c35eb01e2262877\nSuccessfully built mitdeeplearning\nInstalling collected packages: mitdeeplearning\nSuccessfully installed mitdeeplearning-0.2.0\n"
]
],
[
[
"## 1.1 Why is TensorFlow called TensorFlow?\n\nTensorFlow is called 'TensorFlow' because it handles the flow (node/mathematical operation) of Tensors, which are data structures that you can think of as multi-dimensional arrays. Tensors are represented as n-dimensional arrays of base dataypes such as a string or integer -- they provide a way to generalize vectors and matrices to higher dimensions.\n\nThe ```shape``` of a Tensor defines its number of dimensions and the size of each dimension. The ```rank``` of a Tensor provides the number of dimensions (n-dimensions) -- you can also think of this as the Tensor's order or degree.\n\nLet's first look at 0-d Tensors, of which a scalar is an example:",
"_____no_output_____"
]
],
[
[
"sport = tf.constant(\"Tennis\", tf.string)\nnumber = tf.constant(1.41421356237, tf.float64)\n\nprint(\"`sport` is a {}-d Tensor\".format(tf.rank(sport).numpy()))\nprint(\"`number` is a {}-d Tensor\".format(tf.rank(number).numpy()))",
"`sport` is a 0-d Tensor\n`number` is a 0-d Tensor\n"
]
],
[
[
"Vectors and lists can be used to create 1-d Tensors:",
"_____no_output_____"
]
],
[
[
"sports = tf.constant([\"Tennis\", \"Basketball\"], tf.string)\nnumbers = tf.constant([3.141592, 1.414213, 2.71821], tf.float64)\n\nprint(\"`sports` is a {}-d Tensor with shape: {}\".format(tf.rank(sports).numpy(), tf.shape(sports)))\nprint(\"`numbers` is a {}-d Tensor with shape: {}\".format(tf.rank(numbers).numpy(), tf.shape(numbers)))",
"`sports` is a 1-d Tensor with shape: [2]\n`numbers` is a 1-d Tensor with shape: [3]\n"
]
],
[
[
"Next we consider creating 2-d (i.e., matrices) and higher-rank Tensors. For examples, in future labs involving image processing and computer vision, we will use 4-d Tensors. Here the dimensions correspond to the number of example images in our batch, image height, image width, and the number of color channels.",
"_____no_output_____"
]
],
[
[
"### Defining higher-order Tensors ###\n\n'''TODO: Define a 2-d Tensor'''\nmatrix = tf.constant([[1,2,3],[4,5,6]], tf.int32)\n\nassert isinstance(matrix, tf.Tensor), \"matrix must be a tf Tensor object\"\nassert tf.rank(matrix).numpy() == 2\nprint(tf.shape(matrix))",
"tf.Tensor([2 3], shape=(2,), dtype=int32)\n"
],
[
"'''TODO: Define a 4-d Tensor.'''\n# Use tf.zeros to initialize a 4-d Tensor of zeros with size 10 x 256 x 256 x 3. \n# You can think of this as 10 images where each image is RGB 256 x 256.\nimages = tf.zeros((10, 256, 256, 3))\n\nassert isinstance(images, tf.Tensor), \"matrix must be a tf Tensor object\"\nassert tf.rank(images).numpy() == 4, \"matrix must be of rank 4\"\nassert tf.shape(images).numpy().tolist() == [10, 256, 256, 3], \"matrix is incorrect shape\"",
"_____no_output_____"
]
],
[
[
"As you have seen, the ```shape``` of a Tensor provides the number of elements in each Tensor dimension. The ```shape``` is quite useful, and we'll use it often. You can also use slicing to access subtensors within a higher-rank Tensor:",
"_____no_output_____"
]
],
[
[
"row_vector = matrix[1]\ncolumn_vector = matrix[:,2]\nscalar = matrix[1, 2]\n\nprint(\"`row_vector`: {}\".format(row_vector.numpy()))\nprint(\"`column_vector`: {}\".format(column_vector.numpy()))\nprint(\"`scalar`: {}\".format(scalar.numpy()))",
"`row_vector`: [4 5 6]\n`column_vector`: [3 6]\n`scalar`: 6\n"
]
],
[
[
"## 1.2 Computations on Tensors\n\nA convenient way to think about and visualize computations in TensorFlow is in terms of graphs. We can define this graph in terms of Tensors, which hold data, and the mathematical operations that act on these Tensors in some order. Let's look at a simple example, and define this computation using TensorFlow:\n\n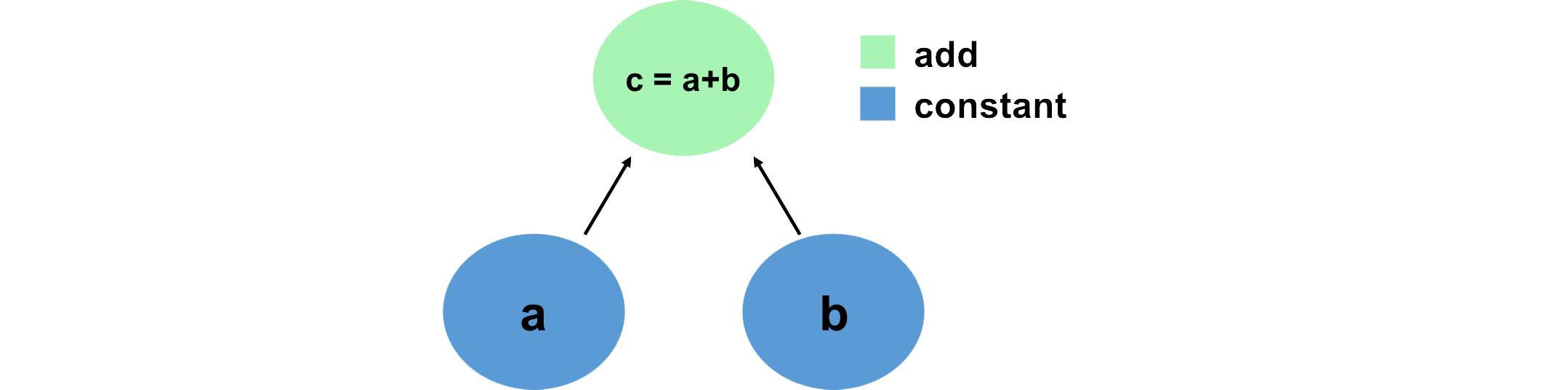",
"_____no_output_____"
]
],
[
[
"# Create the nodes in the graph, and initialize values\na = tf.constant(15)\nb = tf.constant(61)\n\n# Add them!\nc1 = tf.add(a,b)\nc2 = a + b # TensorFlow overrides the \"+\" operation so that it is able to act on Tensors\nprint(c1)\nprint(c2)",
"tf.Tensor(76, shape=(), dtype=int32)\ntf.Tensor(76, shape=(), dtype=int32)\n"
]
],
[
[
"Notice how we've created a computation graph consisting of TensorFlow operations, and how the output is a Tensor with value 76 -- we've just created a computation graph consisting of operations, and it's executed them and given us back the result.\n\nNow let's consider a slightly more complicated example:\n\n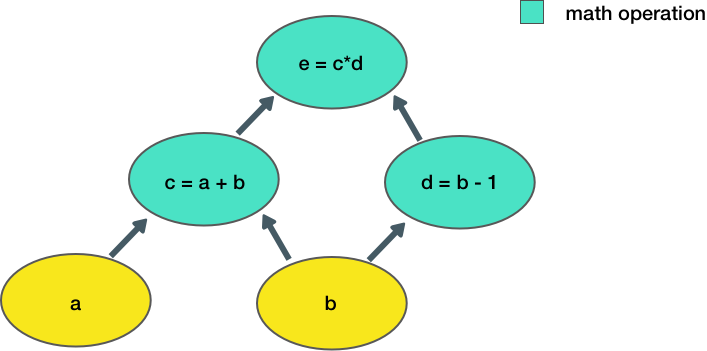\n\nHere, we take two inputs, `a, b`, and compute an output `e`. Each node in the graph represents an operation that takes some input, does some computation, and passes its output to another node.\n\nLet's define a simple function in TensorFlow to construct this computation function:",
"_____no_output_____"
]
],
[
[
"### Defining Tensor computations ###\n\n# Construct a simple computation function\ndef func(a,b):\n '''TODO: Define the operation for c, d, e (use tf.add, tf.subtract, tf.multiply).'''\n c = a+b\n d = b-1\n e = c*d\n return e",
"_____no_output_____"
]
],
[
[
"Now, we can call this function to execute the computation graph given some inputs `a,b`:",
"_____no_output_____"
]
],
[
[
"# Consider example values for a,b\na, b = 1.5, 2.5\n# Execute the computation\ne_out = func(a,b)\nprint(e_out)",
"6.0\n"
]
],
[
[
"Notice how our output is a Tensor with value defined by the output of the computation, and that the output has no shape as it is a single scalar value.",
"_____no_output_____"
],
[
"## 1.3 Neural networks in TensorFlow\nWe can also define neural networks in TensorFlow. TensorFlow uses a high-level API called [Keras](https://www.tensorflow.org/guide/keras) that provides a powerful, intuitive framework for building and training deep learning models.\n\nLet's first consider the example of a simple perceptron defined by just one dense layer: $ y = \\sigma(Wx + b)$, where $W$ represents a matrix of weights, $b$ is a bias, $x$ is the input, $\\sigma$ is the sigmoid activation function, and $y$ is the output. We can also visualize this operation using a graph: \n\n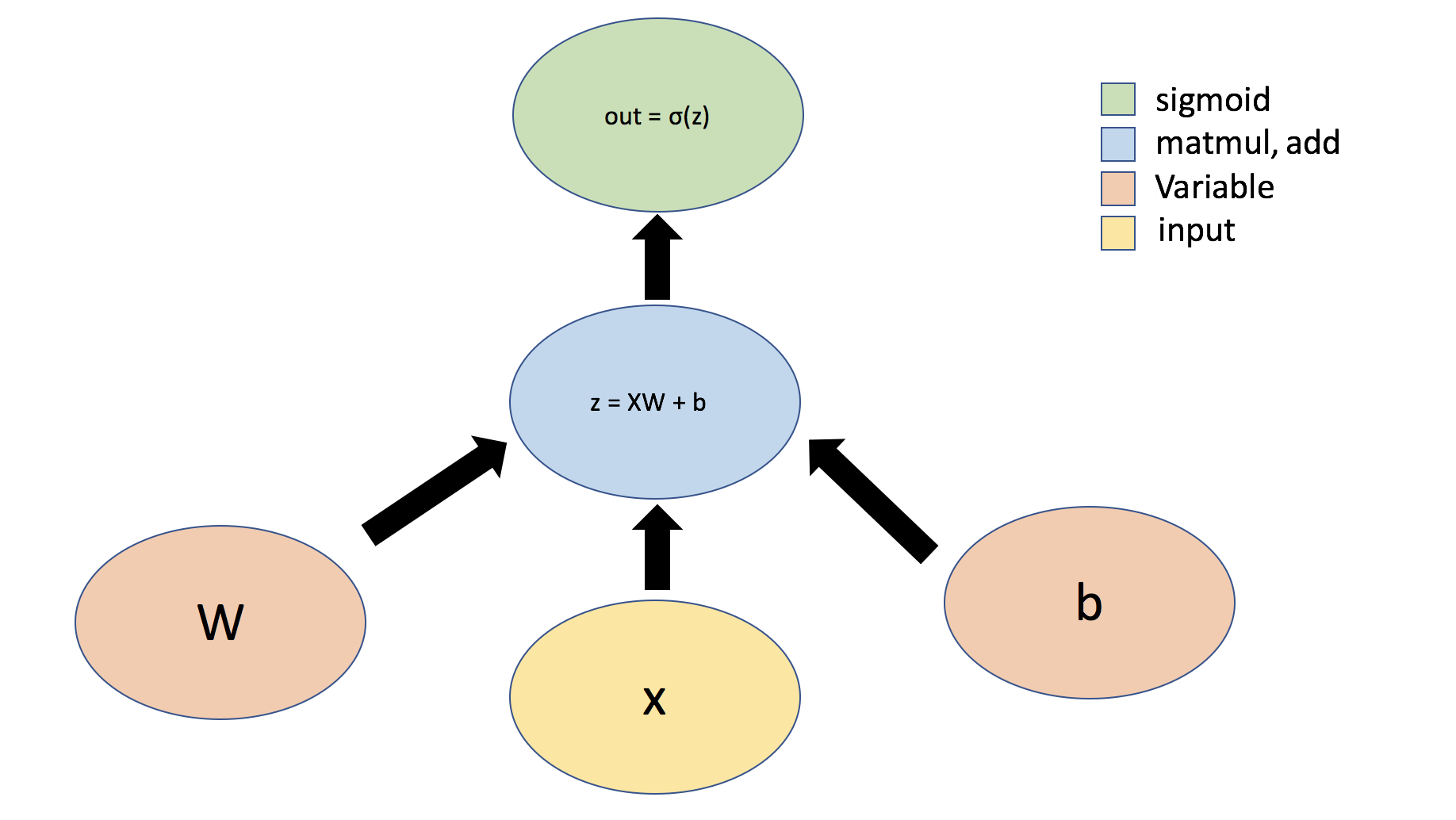\n\nTensors can flow through abstract types called [```Layers```](https://www.tensorflow.org/api_docs/python/tf/keras/layers/Layer) -- the building blocks of neural networks. ```Layers``` implement common neural networks operations, and are used to update weights, compute losses, and define inter-layer connectivity. We will first define a ```Layer``` to implement the simple perceptron defined above.",
"_____no_output_____"
]
],
[
[
"### Defining a network Layer ###\n\n# n_output_nodes: number of output nodes\n# input_shape: shape of the input\n# x: input to the layer\n\nclass OurDenseLayer(tf.keras.layers.Layer):\n def __init__(self, n_output_nodes):\n super(OurDenseLayer, self).__init__()\n self.n_output_nodes = n_output_nodes\n\n def build(self, input_shape):\n d = int(input_shape[-1])\n # Define and initialize parameters: a weight matrix W and bias b\n # Note that parameter initialization is random!\n self.W = self.add_weight(\"weight\", shape=[d, self.n_output_nodes]) # note the dimensionality\n self.b = self.add_weight(\"bias\", shape=[1, self.n_output_nodes]) # note the dimensionality\n\n def call(self, x):\n '''TODO: define the operation for z (hint: use tf.matmul)'''\n z = tf.matmul(x, self.W) + self.b\n\n '''TODO: define the operation for out (hint: use tf.sigmoid)'''\n y = tf.sigmoid(z)\n return y\n\n# Since layer parameters are initialized randomly, we will set a random seed for reproducibility\ntf.random.set_seed(1)\nlayer = OurDenseLayer(3)\nlayer.build((1,2))\nx_input = tf.constant([[1,2.]], shape=(1,2))\ny = layer.call(x_input)\n\n# test the output!\nprint(y.numpy())\nmdl.lab1.test_custom_dense_layer_output(y)",
"[[0.2697859 0.45750412 0.66536945]]\n[PASS] test_custom_dense_layer_output\n"
]
],
[
[
"Conveniently, TensorFlow has defined a number of ```Layers``` that are commonly used in neural networks, for example a [```Dense```](https://www.tensorflow.org/api_docs/python/tf/keras/layers/Dense?version=stable). Now, instead of using a single ```Layer``` to define our simple neural network, we'll use the [`Sequential`](https://www.tensorflow.org/versions/r2.0/api_docs/python/tf/keras/Sequential) model from Keras and a single [`Dense` ](https://www.tensorflow.org/versions/r2.0/api_docs/python/tf/keras/layers/Dense) layer to define our network. With the `Sequential` API, you can readily create neural networks by stacking together layers like building blocks. ",
"_____no_output_____"
]
],
[
[
"### Defining a neural network using the Sequential API ###\n\n# Import relevant packages\nfrom tensorflow.keras import Sequential\nfrom tensorflow.keras.layers import Dense\n\n# Define the number of outputs\nn_output_nodes = 3\n\n# First define the model \nmodel = Sequential()\n\n'''TODO: Define a dense (fully connected) layer to compute z'''\n# https://www.tensorflow.org/api_docs/python/tf/keras/layers/Dense?version=stable\ndense_layer = Dense(n_output_nodes, activation='sigmoid', input_shape=(2, ))\n\n# Add the dense layer to the model\nmodel.add(dense_layer)\n",
"_____no_output_____"
]
],
[
[
"That's it! We've defined our model using the Sequential API. Now, we can test it out using an example input:",
"_____no_output_____"
]
],
[
[
"# Test model with example input\nx_input = tf.constant([[1,2.]], shape=(1,2))\n\n'''TODO: feed input into the model and predict the output!'''\nprint(model(x_input))",
"tf.Tensor([[0.6504887 0.47828162 0.8373661 ]], shape=(1, 3), dtype=float32)\n"
]
],
[
[
"In addition to defining models using the `Sequential` API, we can also define neural networks by directly subclassing the [`Model`](https://www.tensorflow.org/api_docs/python/tf/keras/Model?version=stable) class, which groups layers together to enable model training and inference. The `Model` class captures what we refer to as a \"model\" or as a \"network\". Using Subclassing, we can create a class for our model, and then define the forward pass through the network using the `call` function. Subclassing affords the flexibility to define custom layers, custom training loops, custom activation functions, and custom models. Let's define the same neural network as above now using Subclassing rather than the `Sequential` model.",
"_____no_output_____"
]
],
[
[
"### Defining a model using subclassing ###\n\nfrom tensorflow.keras import Model\nfrom tensorflow.keras.layers import Dense\n\nclass SubclassModel(tf.keras.Model):\n\n # In __init__, we define the Model's layers\n def __init__(self, n_output_nodes):\n super(SubclassModel, self).__init__()\n '''Our model consists of a single Dense layer''' \n self.dense_layer = Dense(n_output_nodes, activation='sigmoid', input_shape=(2, ))\n\n # In the call function, we define the Model's forward pass.\n def call(self, inputs):\n return self.dense_layer(inputs)",
"_____no_output_____"
]
],
[
[
"Just like the model we built using the `Sequential` API, let's test out our `SubclassModel` using an example input.\n\n",
"_____no_output_____"
]
],
[
[
"n_output_nodes = 3\nmodel = SubclassModel(n_output_nodes)\n\nx_input = tf.constant([[1,2.]], shape=(1,2))\n\nprint(model.call(x_input))",
"tf.Tensor([[0.29996255 0.62776643 0.48460066]], shape=(1, 3), dtype=float32)\n"
]
],
[
[
"## 1.4 Automatic differentiation in TensorFlow\n\n[Automatic differentiation](https://en.wikipedia.org/wiki/Automatic_differentiation)\nis one of the most important parts of TensorFlow and is the backbone of training with \n[backpropagation](https://en.wikipedia.org/wiki/Backpropagation). We will use the TensorFlow GradientTape [`tf.GradientTape`](https://www.tensorflow.org/api_docs/python/tf/GradientTape?version=stable) to trace operations for computing gradients later. \n\nWhen a forward pass is made through the network, all forward-pass operations get recorded to a \"tape\"; then, to compute the gradient, the tape is played backwards. By default, the tape is discarded after it is played backwards; this means that a particular `tf.GradientTape` can only\ncompute one gradient, and subsequent calls throw a runtime error. However, we can compute multiple gradients over the same computation by creating a ```persistent``` gradient tape. \n\nFirst, we will look at how we can compute gradients using GradientTape and access them for computation. We define the simple function $ y = x^2$ and compute the gradient:",
"_____no_output_____"
]
],
[
[
"### Gradient computation with GradientTape ###\n\n# y = x^2\n# Example: x = 3.0\nx = tf.Variable(3.0)\n\n# Initiate the gradient tape\nwith tf.GradientTape() as tape:\n # Define the function\n y = x * x\n# Access the gradient -- derivative of y with respect to x\ndy_dx = tape.gradient(y, x)\n\nassert dy_dx.numpy() == 6.0",
"_____no_output_____"
]
],
[
[
"In training neural networks, we use differentiation and stochastic gradient descent (SGD) to optimize a loss function. Now that we have a sense of how `GradientTape` can be used to compute and access derivatives, we will look at an example where we use automatic differentiation and SGD to find the minimum of $L=(x-x_f)^2$. Here $x_f$ is a variable for a desired value we are trying to optimize for; $L$ represents a loss that we are trying to minimize. While we can clearly solve this problem analytically ($x_{min}=x_f$), considering how we can compute this using `GradientTape` sets us up nicely for future labs where we use gradient descent to optimize entire neural network losses.",
"_____no_output_____"
]
],
[
[
"### Function minimization with automatic differentiation and SGD ###\n\n# Initialize a random value for our initial x\nx = tf.Variable([tf.random.normal([1])])\nprint(\"Initializing x={}\".format(x.numpy()))\n\nlearning_rate = 1e-2 # learning rate for SGD\nhistory = []\n# Define the target value\nx_f = 4\n\n# We will run SGD for a number of iterations. At each iteration, we compute the loss, \n# compute the derivative of the loss with respect to x, and perform the SGD update.\nfor i in range(500):\n with tf.GradientTape() as tape:\n '''TODO: define the loss as described above'''\n loss = (x - x_f)*(x - x_f)\n\n # loss minimization using gradient tape\n grad = tape.gradient(loss, x) # compute the derivative of the loss with respect to x\n new_x = x - learning_rate*grad # sgd update\n x.assign(new_x) # update the value of x\n history.append(x.numpy()[0])\n\n# Plot the evolution of x as we optimize towards x_f!\nplt.plot(history)\nplt.plot([0, 500],[x_f,x_f])\nplt.legend(('Predicted', 'True'))\nplt.xlabel('Iteration')\nplt.ylabel('x value')",
"Initializing x=[[-1.1771784]]\n"
]
],
[
[
"`GradientTape` provides an extremely flexible framework for automatic differentiation. In order to back propagate errors through a neural network, we track forward passes on the Tape, use this information to determine the gradients, and then use these gradients for optimization using SGD.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
cbeb111440357753a193e7cd5949b1354859d546
| 188,201 |
ipynb
|
Jupyter Notebook
|
Aula 28 - interconexao e props no dom da freq/.ipynb_checkpoints/Interconexao de SLITS - dom da freq-checkpoint.ipynb
|
RicardoGMSilveira/codes_proc_de_sinais
|
e6a44d6322f95be3ac288c6f1bc4f7cfeb481ac0
|
[
"CC0-1.0"
] | 8 |
2020-10-01T20:59:33.000Z
|
2021-07-27T22:46:58.000Z
|
Aula 28 - interconexao e props no dom da freq/.ipynb_checkpoints/Interconexao de SLITS - dom da freq-checkpoint.ipynb
|
RicardoGMSilveira/codes_proc_de_sinais
|
e6a44d6322f95be3ac288c6f1bc4f7cfeb481ac0
|
[
"CC0-1.0"
] | null | null | null |
Aula 28 - interconexao e props no dom da freq/.ipynb_checkpoints/Interconexao de SLITS - dom da freq-checkpoint.ipynb
|
RicardoGMSilveira/codes_proc_de_sinais
|
e6a44d6322f95be3ac288c6f1bc4f7cfeb481ac0
|
[
"CC0-1.0"
] | 9 |
2020-10-15T12:08:22.000Z
|
2021-04-12T12:26:53.000Z
| 818.265217 | 68,528 | 0.950069 |
[
[
[
"# Neste notebook vamos simular a interconexão entre SLITs",
"_____no_output_____"
]
],
[
[
"# importar as bibliotecas necessárias\nimport numpy as np # arrays\nimport matplotlib.pyplot as plt # plots\nplt.rcParams.update({'font.size': 14})\nimport IPython.display as ipd # to play signals\nimport sounddevice as sd\nimport soundfile as sf\n# Os próximos módulos são usados pra criar nosso SLIT\nfrom scipy.signal import butter, lfilter, freqz, chirp, impulse",
"_____no_output_____"
]
],
[
[
"# Vamos criar 2 SLITs\n\nPrimeiro vamos criar dois SLITs. Um filtro passa alta e um passa-baixa. Você pode depois mudar a ordem de um dos filtros e sua frequência de corte e, então, observar o que acontece na FRF do SLIT concatenado.",
"_____no_output_____"
]
],
[
[
"# Variáveis do filtro\norder1 = 6\nfs = 44100 # sample rate, Hz\ncutoff1 = 1000 # desired cutoff frequency of the filter, Hz\n\n# Passa baixa\nb, a = butter(order1, 2*cutoff1/fs, btype='low', analog=False)\nw, H1 = freqz(b, a)\n\n# Passa alta\ncutoff2 = 1000\norder2 = 6\nb, a = butter(order2, 2*cutoff2/fs, btype='high', analog=False)\nw, H2 = freqz(b, a)\n\nplt.figure(figsize=(15,5))\nplt.subplot(1,2,1)\nplt.semilogx(fs*w/(2*np.pi), 20 * np.log10(abs(H1)), 'b', linewidth = 2, label = 'Passa-baixa')\nplt.semilogx(fs*w/(2*np.pi), 20 * np.log10(abs(H2)), 'r', linewidth = 2, label = 'Passa-alta')\nplt.title('Magnitude')\nplt.xlabel('Frequency [Hz]')\nplt.ylabel('Amplitude [dB]')\nplt.margins(0, 0.1)\nplt.grid(which='both', axis='both')\nplt.ylim((-100, 20))\n\nplt.subplot(1,2,2)\nplt.semilogx(fs*w/(2*np.pi), np.angle(H1), 'b', linewidth = 2, label = 'Passa-baixa')\nplt.semilogx(fs*w/(2*np.pi), np.angle(H2), 'r', linewidth = 2, label = 'Passa-alta')\nplt.legend(loc = 'upper right')\nplt.title('Fase')\nplt.xlabel('Frequency [Hz]')\nplt.ylabel('Amplitude [dB]')\nplt.margins(0, 0.1)\nplt.grid(which='both', axis='both')\nplt.show()",
"_____no_output_____"
]
],
[
[
"# Interconexão em série\n\n\\begin{equation}\nH(\\mathrm{j}\\omega) = H_1(\\mathrm{j}\\omega)H_2(\\mathrm{j}\\omega)\n\\end{equation}",
"_____no_output_____"
]
],
[
[
"Hs = H1*H2\n\nplt.figure(figsize=(15,5))\nplt.subplot(1,2,1)\nplt.semilogx(fs*w/(2*np.pi), 20 * np.log10(abs(H1)), '--k', linewidth = 2)\nplt.semilogx(fs*w/(2*np.pi), 20 * np.log10(abs(H2)), '--k', linewidth = 2)\nplt.semilogx(fs*w/(2*np.pi), 20 * np.log10(abs(Hs)), 'b', linewidth = 2, label = 'R: Band pass')\nplt.title('Magnitude')\nplt.xlabel('Frequency [Hz]')\nplt.ylabel('Amplitude [dB]')\nplt.margins(0, 0.1)\nplt.grid(which='both', axis='both')\nplt.ylim((-100, 20))\n\nplt.subplot(1,2,2)\nplt.semilogx(fs*w/(2*np.pi), np.angle(H1), '--k', linewidth = 2)\nplt.semilogx(fs*w/(2*np.pi), np.angle(H2), '--k', linewidth = 2)\nplt.semilogx(fs*w/(2*np.pi), np.angle(Hs), 'b', linewidth = 2, label = 'R: Band pass')\nplt.legend(loc = 'upper right')\nplt.title('Fase')\nplt.xlabel('Frequency [Hz]')\nplt.ylabel('Amplitude [dB]')\nplt.margins(0, 0.1)\nplt.grid(which='both', axis='both')\nplt.show()\n",
"_____no_output_____"
]
],
[
[
"# Interconexão em paralelo\n\n\\begin{equation}\nH(\\mathrm{j}\\omega) = H_1(\\mathrm{j}\\omega)+H_2(\\mathrm{j}\\omega)\n\\end{equation}",
"_____no_output_____"
]
],
[
[
"Hs = H1+H2\n\nplt.figure(figsize=(15,5))\nplt.subplot(1,2,1)\nplt.semilogx(fs*w/(2*np.pi), 20 * np.log10(abs(H1)), '--k', linewidth = 2)\nplt.semilogx(fs*w/(2*np.pi), 20 * np.log10(abs(H2)), '--k', linewidth = 2)\nplt.semilogx(fs*w/(2*np.pi), 20 * np.log10(abs(Hs)), 'b', linewidth = 2, label = 'R: All pass')\nplt.title('Magnitude')\nplt.xlabel('Frequency [Hz]')\nplt.ylabel('Amplitude [dB]')\nplt.margins(0, 0.1)\nplt.grid(which='both', axis='both')\nplt.ylim((-100, 20))\n\nplt.subplot(1,2,2)\nplt.semilogx(fs*w/(2*np.pi), np.angle(H1), '--k', linewidth = 2)\nplt.semilogx(fs*w/(2*np.pi), np.angle(H2), '--k', linewidth = 2)\nplt.semilogx(fs*w/(2*np.pi), np.angle(Hs), 'b', linewidth = 2, label = 'R: All pass')\nplt.legend(loc = 'upper right')\nplt.title('Fase')\nplt.xlabel('Frequency [Hz]')\nplt.ylabel('Amplitude [dB]')\nplt.margins(0, 0.1)\nplt.grid(which='both', axis='both')\nplt.show()",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cbeb1732baef352375fe08763756e2918c880d8e
| 4,779 |
ipynb
|
Jupyter Notebook
|
tutormagic.ipynb
|
pfsq/notebooks
|
3ca04dcaeb3ce7dd5d63ed58f76c1004d1ab3fe7
|
[
"BSD-2-Clause"
] | 8 |
2015-05-15T16:09:15.000Z
|
2021-08-21T14:46:15.000Z
|
tutormagic.ipynb
|
pfsq/notebooks
|
3ca04dcaeb3ce7dd5d63ed58f76c1004d1ab3fe7
|
[
"BSD-2-Clause"
] | 2 |
2015-04-04T19:33:58.000Z
|
2015-11-10T14:22:07.000Z
|
tutormagic.ipynb
|
pfsq/notebooks
|
3ca04dcaeb3ce7dd5d63ed58f76c1004d1ab3fe7
|
[
"BSD-2-Clause"
] | 22 |
2015-04-25T00:40:37.000Z
|
2022-03-12T11:00:00.000Z
| 26.114754 | 364 | 0.54321 |
[
[
[
"Esta será una microentrada para presentar una extensión para el notebook que estoy usando en un curso interno que estoy dando en mi empresa.\n\nSi a alguno más os puede valer para mostrar cosas básicas de Python (2 y 3, además de Java y Javascript) para muy principiantes me alegro.",
"_____no_output_____"
],
[
"# Nombre en clave: tutormagic",
"_____no_output_____"
],
[
"Esta extensión lo único que hace es embeber dentro de un IFrame la página de [pythontutor](http://www.pythontutor.com) usando el código que hayamos definido en una celda de código precedida de la *cell magic* `%%tutor`.\n\nComo he comentado anteriormente, se puede escribir código Python2, Python3, Java y Javascript, que son los lenguajes soportados por pythontutor.",
"_____no_output_____"
],
[
"# Ejemplo",
"_____no_output_____"
],
[
"Primero deberemos instalar la extensión. Está disponible en pypi por lo que la podéis instalar usando `pip install tutormagic`. Una vez instalada, dentro de un notebook de IPython la deberías cargar usando:",
"_____no_output_____"
]
],
[
[
"%load_ext tutormagic",
"_____no_output_____"
]
],
[
[
"Una vez hecho esto ya deberiamos tener disponible la *cell magic* para ser usada:",
"_____no_output_____"
]
],
[
[
"%%tutor --lang python3\na = 1\nb = 2\n\ndef add(x, y):\n return x + y\n\nc = add(a, b)",
"_____no_output_____"
]
],
[
[
"Ahora un ejemplo con javascript:",
"_____no_output_____"
]
],
[
[
"%%tutor --lang javascript\nvar a = 1;\nvar b = 1;\nconsole.log(a + b);",
"_____no_output_____"
]
],
[
[
"# Y eso es todo",
"_____no_output_____"
],
[
"Lo dicho, espero que sea útil para alguien.\n\n* [tutormagic en pypi](https://pypi.python.org/pypi/tutormagic/0.1.0).\n* [tutormagic en github](https://github.com/kikocorreoso/tutormagic)\n\nSaludos.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
cbeb2829a7e156915c515cba2584e2fad6727b51
| 2,737 |
ipynb
|
Jupyter Notebook
|
examples/noaa/noaa_compare.ipynb
|
LLNL/Sina
|
f3e9bb3a122cfae2a9fd82c3c5613cff939d3aa1
|
[
"MIT"
] | 5 |
2019-06-28T22:52:19.000Z
|
2021-09-03T04:28:24.000Z
|
examples/noaa/noaa_compare.ipynb
|
LLNL/Sina
|
f3e9bb3a122cfae2a9fd82c3c5613cff939d3aa1
|
[
"MIT"
] | 2 |
2019-07-03T11:40:38.000Z
|
2020-10-28T17:26:50.000Z
|
examples/noaa/noaa_compare.ipynb
|
LLNL/Sina
|
f3e9bb3a122cfae2a9fd82c3c5613cff939d3aa1
|
[
"MIT"
] | 1 |
2019-06-28T22:52:25.000Z
|
2019-06-28T22:52:25.000Z
| 24.008772 | 332 | 0.546218 |
[
[
[
"Comparing Records\n================\nA comparison function in `sina.model` allows you to do a simple comparison between two records. It will compare the differences between two records and print them out in an easy to read table. The only requirement is to have two similar structured records. In this example we will be comparing records of the NOAA data set.\n\nAccessing the Data\n-----------------------\nWe'll first create a Sina DAOFactory that's aware of our database.",
"_____no_output_____"
]
],
[
[
"import sina\nimport sina.utils\nimport sina.cli.diff\n\n# Load the database\ndatabase = sina.utils.get_example_path('noaa/data.sqlite')\nprint(\"Using database {}\".format(database))\nds = sina.connect(database)\nrecords = ds.records\n\nprint(\"The data access object factory has been created. Proceed to the next cell.\")",
"_____no_output_____"
]
],
[
[
"Get the records\n------------------",
"_____no_output_____"
]
],
[
[
"record_one_id = \"WCOA2011-13-95-1-7\"\nrecord_two_id = \"WCOA2011-13-95-1-8\"\nrecord_one = records.get(record_one_id)\nrecord_two = records.get(record_two_id)\nprint('Retrieved records: {} and {}.'.format(record_one, record_two))",
"_____no_output_____"
]
],
[
[
"Compare the records\n-------------------------",
"_____no_output_____"
]
],
[
[
"sina.cli.diff.print_diff_records(record_one, record_two)",
"_____no_output_____"
]
],
[
[
"Releasing Resources\n-------------------------\nDon't forget to release resources when you're all done!",
"_____no_output_____"
]
],
[
[
"ds.close()",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cbeb3400347d7aa45767f2676da892b37e0ef7b5
| 15,888 |
ipynb
|
Jupyter Notebook
|
machine-learning/coursera-machine-learning/week2.ipynb
|
cutamar/machine-learning-curriculum
|
082f61cdbe24a4abe027f3ac68ceba4b150bff37
|
[
"MIT"
] | null | null | null |
machine-learning/coursera-machine-learning/week2.ipynb
|
cutamar/machine-learning-curriculum
|
082f61cdbe24a4abe027f3ac68ceba4b150bff37
|
[
"MIT"
] | null | null | null |
machine-learning/coursera-machine-learning/week2.ipynb
|
cutamar/machine-learning-curriculum
|
082f61cdbe24a4abe027f3ac68ceba4b150bff37
|
[
"MIT"
] | null | null | null | 32.557377 | 212 | 0.529645 |
[
[
[
"# Week 2\n## Matlab Resources\n### Onramp\n- Go to: https://matlabacademy.mathworks.com/ and click on the MATLAB Onramp button to start learning MATLAB\n\n### Tutorials\n#### Get Started with MATLAB and MATLAB Online\n- [What is MATLAB?](https://youtu.be/WYG2ZZjgp5M)\\*\n- [MATLAB Variables](https://youtu.be/0w9NKt6Fixk)\\*\n- [MATLAB as a Calculator](https://youtu.be/aRSkNpCSgWY)\\*\n- [MATLAB Functions](https://youtu.be/RJp46UVQBic)\\*\n- [Getting Started with MATLAB Online](https://youtu.be/XjzxCVWKz58)\n- [Managing Files in MATLAB Online](https://youtu.be/B3lWLIrYjC0)\n\n#### Vectors\n- [Creating Vectors](https://youtu.be/R5Mnkrk9Mos)\\*\n- [Creating Uniformly Spaced Vectors](https://youtu.be/_zqTOV5yl8Y)\\*\n- [Accessing Elements of a Vector Using Conditions](https://youtu.be/8D04GW_foQ0)\\*\n- [Calculations with Vectors](https://youtu.be/VQaZ0TvjF0c)\\*\n- [Vector Transpose](https://youtu.be/vgRLwjHBmsg)\n\n#### Visualization\n- [Line Plots](https://youtu.be/-hhJoveE4sY)\\*\n- [Annotating Graphs](https://youtu.be/JyovEGPSdoI)\\*\n- [Multiple Plots](https://youtu.be/fBx8EFuXFLM)\\*\n\n#### Matrices\n- [Creating Matrices](https://youtu.be/qdTdwTh6jMo)\\*\n- [Calculations with Matrices](https://youtu.be/mzzJ9gnMrYE)\\*\n- [Accessing Elements of a Matrix](https://youtu.be/uWPHxpTuZRA)\\*\n- [Matrix Creation Functions](https://youtu.be/VPcbpVd_mPA)\\*\n- [Combining Matrices](https://youtu.be/ejTr3ekTTyA)\n- [Determining Array Size and Length](https://youtu.be/IF9-ffmxuy8)\n- [Matrix Multiplication](https://youtu.be/4hsx3bdNjGk)\n- [Reshaping Arrays](https://youtu.be/UQpDIHlFo8A)\n- [Statistical Functions with Matrices](https://youtu.be/Y97W3_u7cM4)\n\n#### MATLAB Programming\n- [Logical Variables](https://youtu.be/bRMg4GsFDQ8)\\*\n- [If-Else Statements](https://youtu.be/JZSuU-Laigo)\\*\n- [Writing a FOR loop](https://youtu.be/lg65bzgvI5c)\\*\n- [Writing a WHILE Loop](https://youtu.be/PKH5lCMJXbk)\n- [Writing Functions](https://youtu.be/GrcNN04eqXU)\n- [Passing Functions as Inputs](https://youtu.be/aNCwR9dRjHs)\n\n#### Troubleshooting\n- [Using Online Documentation](https://youtu.be/54n5zJwR8aM)\\*\n- [Which File or Variable Am I Using?](https://youtu.be/Z09BvGeYNdE)\n- [Troubleshooting Code with the Debugger](https://youtu.be/DB4aJMnZtNQ)\n\n***Indicates content covered in Onramp**\n\n## Multivariate Linear Regression\n### Notation\n- $n$ = nubmer of features\n- $x^{(i)}$ = input (features) of $i^{th}$ training example\n- $x_j^{(i)}$ = value of feature $j$ in $i^{th}$ training example\n\n### Hypothesis\n- For convenience of notation, we define $(x_0^{(i)}=1)$, so all $x_0$'s are equal to 1\n- $h_{\\theta}(x) = \\theta_0 x_0 + \\theta_1 x_1 + \\theta_2 x_2 + \\cdots + \\theta_n x_n$\n- $x, \\theta \\in \\mathbb{R}^{n+1}$\n- This way it can be also written in vector form:\n - $h_{\\theta}(x)=\\theta^T x=\\begin{bmatrix}\\theta_0 & \\theta_1 & \\cdots & \\theta_n\\end{bmatrix} \\cdot \\begin{bmatrix}x_0 \\\\ x_1 \\\\ \\vdots \\\\ x_n \\end{bmatrix}$\n \n### Cost Function\n$$J(\\theta)=\\frac{1}{2m}\\sum_{i=1}^{m}(h_{\\theta}(x^{(i)})-y^{(i)})^2$$\n \n### Gradient Descent $(n\\geq 1)$\n- Repeat the following for $j=0,\\dots, n$:\n$$\\theta_j := \\theta_j - \\alpha\\frac{1}{m}\\sum_{i=1}^m(h_{\\theta}(x^{(i)})-y^{(i)})x_j^{(i)}$$\n\n### Feature Scaling\n- Speeds gradient descent up, because $\\theta$ will descend quickly on small ranges and slowly on large ranges, and so will oscillate inefficiently down to the optimum when the variables are very uneven\n- To prevent this we modify the ranges to roughly the same\n - $-1 \\leq x_{(i)} \\leq 1$ or\n - $-0.5 \\leq x_{(i)} \\leq 0.5$\n- We use **feature scaling** (division part) and **mean normalization** (subtraction part):\n$$x_i := \\frac{x_i - \\mu_i}{s_i}$$\n- $\\mu_i$ is the **average** of all the values for feature (i)\n- $s_i$ is the **range** of values (max - min) or the **standard deviation**\n\n### Debugging Gradient Descent\n- Make a plot with *number of iterations* on the x-axis and plot the cost function $J(\\theta)$ over the number of iterations of gradient descent\n- If $J(\\theta)$ ever increases, then you probably need to decrease the learning rate $\\alpha$\n- **Automatic convergence tests** are also possible:\n - You declare convergence if $J(\\theta)$ decreases by less than E in one iteration, where E is some small value such as $10^{-3}$\n - However in practice it's difficult to choose E\n \n### Features and Polynomial Regression\n- We can combine multiple features into one feature, e.g. width and height into one feature, area (= width x height)\n- Also our hypothesis function need not be linear, if that doesn't fit the data well\n- We could use a quadratic, cubic, square function etc.\n - Square function example: $h_{\\theta}(x) = \\theta_0 + \\theta_1 x_1 + \\theta_2 \\sqrt{x_1}$\n- Choosing features this way, don't forget that feature scaling becomes even more important\n\n### Normal Equation\n#### How it Works\n- Another way of minimizing $J$\n- Explicit, non-iterative, analytical way\n- We minimize $J$ by explicitly taking its derivatives with respect to the $\\theta_j$'s, and set them to zero\n$$\\theta = (X^T X)^{-1}X^Ty$$\n- Don't forget to add $x_0^{(i)}$ to the $X$ matrix (which equals 1)\n- There is **no need** to do feature scaling with the normal equation\n- Because it needs to calculate the inverse of $X^T X$, it's slow if $n$ is very large\n- As a broad rule, you should switch to gradient descend for $n \\geq 10000$\n- Normal equation has a runtime of $O(n^3)$, as compared to gradient descend, which has a runtime of $O(kn^2)$\n\n#### Noninvertibility\n- Using `pinv` in octave/matlab will give us a value of $\\theta$ even if $X^T X$ is not invertible (singular/degenerate)\n- If $X^T X$ is **noninvertible**, possible reasons are:\n - Redundant features, where two features are very closely related (i.e. linearly dependent)\n - Too many features (e.g. $m \\leq n$)\n - In this case delete some features or\n - Use **regularization**\n \n## Octave/Matlab Commands\n### Basic Operations\n```\n% equal\n1 == 2\n\n% not equal\n1 ~= 2\n\n% and\n1 && 0\n\n% or\n1 || 0\n\n% xor\nxor(1,0)\n\n% change prompt\nPS1('>> ')\n\n% semicolon supresses output\na = 3;\n\n% display (print)\ndisp('Hello World')\n\n% string format\nsprintf('2 decimals: %0.2f', pi)\n\n% change how many digits should be shown\nformat short\nformat long\n\n% generate row vector start:step:end\n1:0.25:2\n\n% you can also leave out the step param i.e. start:end, this will by default increment by 1\n1:5\n\n% generate matrix consisting of ones (row count, column count)\nones(2,3)\n\n% generate matrix consisting of zeros (row count, column count)\nzeros(1,3)\n\n% generate matrix consisting of random values between 0 and 1 (row count, column count)\nrand(1,3)\n\n% generate matrix consisting of normally distributed random values (row count, column count)\nrandn(1,3)\n\n% plot histogram (data, optional: bin/bucket count) NOTE: in matlab histogram should be used instead of hist\nhist(randn(1, 10000))\n\n% generate identity matrix for the given dimension\neye(6)\n\n% help for given function\nhelp eye\n```\n\n### Moving Data Around\n```\n% number of rows\nsize(A, 1)\n\n% number of columns\nsize(A, 2)\n\n% gives the size of the longest dimension, but usually only used on vectors\nlength(A)\n\n% current working directory\npwd\n\n% change directory\ncd\n\n% list files and folders\nls\n\n% load data\nload featuresX.dat\n\n% or the same calling\nload('featuresX.dat')\n\n% shows variables in current scope\nwho\n\n% or for the detailed view\nwhos\n\n% remove variable from scope\nclear featuresX\n\n% get first 10 elements\npriceY(1:10)\n\n% saves variable v into file hello.mat\nsave hello.mat v\n\n% clear all variables\nclear\n\n% saves in a human readable format (no metadata like variable name)\nsave hello.txt v -ascii\n\n% fetch everything in the second row (\":\" means every element along that row/column)\nA(2,:)\n\n% fetch everything from first and third row\nA([1 3], :)\n\n% can also be used for assignments\nA(:,2) = [10; 11; 12]\n\n% append another column vector to right\nA = [A, [100; 101; 102]]\n\n% put all elements of A into a single vector\nA(:)\n\n% concat two matrices\nC = [A B]\n\n% or the same as\nC = [A, B]\n\n% or put it on the bottom\nC = [A; B]\n```\n\n### Computing on Data\n```\n% multiple A11 with B11, A12 with B12 etc. (element-wise)\nA .* B\n\n% element-wise squaring\nA .^ 2\n\n% element-wise inverse\n1 ./ A\n\n% element-wise log\nlog(v)\n\n% element-wise exp\nexp(v)\n\n% element-wise abs\nabs(v)\n\n% same as -1*v\n-v\n\n% element-wise incremental by e.g. 1\nv + ones(length(v), 1)\n\n% or use this (+ and - are element-wise)\nv + 1\n\n% returns max value and index\n[val, ind] = max(a)\n\n% element-wise comparison\na < 3\n\n% tells the indexes of the variables for which the condition is true\nfind(a < 3)\n\n% generates a matrix of n x n dimension, where all rows, columns and diagonals sum up to the same value\nmagic(3)\n\n% find used on matrices, returns rows and columns\n[r,c] = find(A >= 7)\n\n% adds up all elements\nsum(a)\n\n% product of all elements\nprod(a)\n\n% round down\nfloor(a)\n\n% round up\nceil(a)\n\n% element-wise max\nmax(A, B)\n\n% column-wise max\nmax(A,[],1)\n\n% or use\nmax(A)\n\n% row-wise max\nmax(A,[],2)\n\n% max element\nmax(max(A))\n\n% or turn A into a vector\nmax(A(:))\n\n% column-wise sum\nsum(A,1)\n\n% row-wise sum\nsum(A,2)\n\n% diagonal sum\nsum(sum(A .* eye(length(A))))\n\n% other diagonal sum\nsum(sum(A .* flipud(eye(length(A)))))\n```\n\n### Plotting Data\n```\nt=[0:0.01:0.98]\n\n% plot given x and y data\nplot(t, sin(t))\n\n% plots next figures on top of the open one (old one)\nhold on\n\n% sets x-axis label\nxlabel('time')\n\n% sets y-axis label\nylabel('value')\n\n% show legend\nlegend('sin', 'cos')\n\n% show title\ntitle('my plot')\n\n% saves open plot as png\nprint -dpng 'myPlot.png'\n\n% close open plot\nclose\n\n% multiple plots\nfigure(1); plot(t, sin(t));\nfigure(2); plot(t, cos(t));\n\n% divides plot into a 1x2 grid, access first element\nsubplot(1,2,1)\n\n% set x-axis range to 0.5 -> 1 and y-axis range to -1 -> 1\naxis([0.5 1 -1 1])\n\n% clear plot\nclf\n\n% plot matrix\nimagesc(A)\n\n% show colorbar with values\ncolorbar\n\n% change to gray colormap\ncolormap gray\n\n% comma chaining of commands, useful e.g. if you want to change colormap etc. (output doesn't get surpressed like when using \";\")\na=1, b=2, c=3\n```\n\n### Control Statements\n```\n% initialize zero vector of dimension 10\nv = zeros(10,1)\n\n% for loop\nfor i=1:10, v(i) = 2^i; end;\n\n% same for loop\nindices = 1:10;\nfor i=indices, v(i) = 2^i; end;\n\n% break and continue also work in octave/matlab\n\n% while loop\ni = 1;\nwhile i <= 5,\n v(i) = 100;\n i = i + 1;\nend;\n\n% if and break\ni = 1;\nwhile true,\n v(i) = 999;\n i = i + 1;\n if i == 6,\n break;\n end;\nend;\n\n% if, elseif, else\nv(1) = 2;\nif v(1)==1,\n disp('The value is one');\nelseif v(1)==2,\n disp('The value is two');\nelse\n disp('The value is not one or two')\nend;\n\n% functions need to be defined in a .m file\n% also note you need to cd into this directory\n% to call the function, or add the folder to\n% your search path using addpath\n\n% the file name only matters as function identification\n\n% squareThisNumber.m\nfunction y = squareThisNumber(x)\ny = x^2;\n\n% return multiple values\n% squareAndCubeThisNumber.m\nfunction [y1,y2] = squareAndCubeThisNumber(x)\ny1 = x^2;\ny2 = x^3;\n```\n\n### Vectorization\n```\n% it's simpler and more efficient \n% instead of writing a for loop, you write\nprediction = theta' * x\n```",
"_____no_output_____"
]
]
] |
[
"markdown"
] |
[
[
"markdown"
]
] |
cbeb35f26dfed281e342da0b885062db1a61ec64
| 10,519 |
ipynb
|
Jupyter Notebook
|
downloaded_kernels/house_sales/kernel_1.ipynb
|
josepablocam/common-code-extraction
|
a6978fae73eee8ece6f1db09f2f38cf92f03b3ad
|
[
"MIT"
] | null | null | null |
downloaded_kernels/house_sales/kernel_1.ipynb
|
josepablocam/common-code-extraction
|
a6978fae73eee8ece6f1db09f2f38cf92f03b3ad
|
[
"MIT"
] | null | null | null |
downloaded_kernels/house_sales/kernel_1.ipynb
|
josepablocam/common-code-extraction
|
a6978fae73eee8ece6f1db09f2f38cf92f03b3ad
|
[
"MIT"
] | 2 |
2021-07-12T00:48:08.000Z
|
2021-08-11T12:53:05.000Z
| 27.608924 | 169 | 0.513832 |
[
[
[
"import numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\nimport xgboost\nimport math\nfrom __future__ import division\nfrom scipy.stats import pearsonr\nfrom sklearn.linear_model import LinearRegression\nfrom sklearn import cross_validation, tree, linear_model\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.metrics import explained_variance_score",
"_____no_output_____"
]
],
[
[
"# 1. Exploratory Data Analysis",
"_____no_output_____"
]
],
[
[
"# Read the data into a data frame\ndata = pd.read_csv('../input/kc_house_data.csv')",
"_____no_output_____"
],
[
"# Check the number of data points in the data set\nprint(len(data))\n# Check the number of features in the data set\nprint(len(data.columns))\n# Check the data types\nprint(data.dtypes.unique())",
"_____no_output_____"
]
],
[
[
"- Since there are Python objects in the data set, we may have some categorical features. Let's check them. ",
"_____no_output_____"
]
],
[
[
"data.select_dtypes(include=['O']).columns.tolist()",
"_____no_output_____"
]
],
[
[
"- We only have the date column which is a timestamp that we will ignore.",
"_____no_output_____"
]
],
[
[
"# Check any number of columns with NaN\nprint(data.isnull().any().sum(), ' / ', len(data.columns))\n# Check any number of data points with NaN\nprint(data.isnull().any(axis=1).sum(), ' / ', len(data))",
"_____no_output_____"
]
],
[
[
"- The data set is pretty much structured and doesn't have any NaN values. So we can jump into finding correlations between the features and the target variable",
"_____no_output_____"
],
[
"# 2. Correlations between features and target",
"_____no_output_____"
]
],
[
[
"features = data.iloc[:,3:].columns.tolist()\ntarget = data.iloc[:,2].name",
"_____no_output_____"
],
[
"correlations = {}\nfor f in features:\n data_temp = data[[f,target]]\n x1 = data_temp[f].values\n x2 = data_temp[target].values\n key = f + ' vs ' + target\n correlations[key] = pearsonr(x1,x2)[0]",
"_____no_output_____"
],
[
"data_correlations = pd.DataFrame(correlations, index=['Value']).T\ndata_correlations.loc[data_correlations['Value'].abs().sort_values(ascending=False).index]",
"_____no_output_____"
]
],
[
[
"- We can see that the top 5 features are the most correlated features with the target \"price\"\n- Let's plot the best 2 regressors jointly",
"_____no_output_____"
]
],
[
[
"y = data.loc[:,['sqft_living','grade',target]].sort_values(target, ascending=True).values\nx = np.arange(y.shape[0])",
"_____no_output_____"
],
[
"%matplotlib inline\nplt.subplot(3,1,1)\nplt.plot(x,y[:,0])\nplt.title('Sqft and Grade vs Price')\nplt.ylabel('Sqft')\n\nplt.subplot(3,1,2)\nplt.plot(x,y[:,1])\nplt.ylabel('Grade')\n\nplt.subplot(3,1,3)\nplt.plot(x,y[:,2],'r')\nplt.ylabel(\"Price\")\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"# 3. Predicting House Sales Prices",
"_____no_output_____"
]
],
[
[
"# Train a simple linear regression model\nregr = linear_model.LinearRegression()\nnew_data = data[['sqft_living','grade', 'sqft_above', 'sqft_living15','bathrooms','view','sqft_basement','lat','waterfront','yr_built','bedrooms']]",
"_____no_output_____"
],
[
"X = new_data.values\ny = data.price.values",
"_____no_output_____"
],
[
"X_train, X_test, y_train, y_test = cross_validation.train_test_split(X, y ,test_size=0.2)",
"_____no_output_____"
],
[
"regr.fit(X_train, y_train)\nprint(regr.predict(X_test))",
"_____no_output_____"
],
[
"regr.score(X_test,y_test)",
"_____no_output_____"
]
],
[
[
"- Prediction score is about 70 which is not really optimal",
"_____no_output_____"
]
],
[
[
"# Calculate the Root Mean Squared Error\nprint(\"RMSE: %.2f\"\n % math.sqrt(np.mean((regr.predict(X_test) - y_test) ** 2)))",
"_____no_output_____"
],
[
"# Let's try XGboost algorithm to see if we can get better results\nxgb = xgboost.XGBRegressor(n_estimators=100, learning_rate=0.08, gamma=0, subsample=0.75,\n colsample_bytree=1, max_depth=7)",
"_____no_output_____"
],
[
"traindf, testdf = train_test_split(X_train, test_size = 0.3)\nxgb.fit(X_train,y_train)",
"_____no_output_____"
],
[
"predictions = xgb.predict(X_test)\nprint(explained_variance_score(predictions,y_test))",
"_____no_output_____"
]
],
[
[
"- Our accuracy is changing between 79%-84%. I think it is close to an optimal solution.",
"_____no_output_____"
]
],
[
[
"",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
cbeb3c85c3871a9a880659880ed8408291e26122
| 9,627 |
ipynb
|
Jupyter Notebook
|
work/JRC_excel_data.ipynb
|
Vizzuality/helix-scope-data
|
567f77b1a8e3a3066450ab4f939d7af281005168
|
[
"MIT"
] | null | null | null |
work/JRC_excel_data.ipynb
|
Vizzuality/helix-scope-data
|
567f77b1a8e3a3066450ab4f939d7af281005168
|
[
"MIT"
] | null | null | null |
work/JRC_excel_data.ipynb
|
Vizzuality/helix-scope-data
|
567f77b1a8e3a3066450ab4f939d7af281005168
|
[
"MIT"
] | null | null | null | 35.655556 | 217 | 0.505038 |
[
[
[
"# Process the JRC Excel files\n\n### JRC Data\n\nExpDam is direct expected damage per year from river flooding in Euro (2010 values). Data includes baseline values (average 1976-2005) and impact at SWLs.\n\nAll figures are multi-model averages based on EC-EARTH r1 to r7 (7 models)\n\nPopAff is population affected per year from river flooding. Data includes baseline values (average 1976-2005) and impact at SWLs.\n\nAll figures are multi-model averages based on EC-EARTH r1 to r7 (7 models)\n\nReference\nAlfieri, L., Bisselink, B., Dottori, F., Naumann, G., de Roo, A., Salamon, P., Wyser, K. and Feyen, L.: Global projections of river flood risk in a warmer world, Earths Future, doi:10.1002/2016EF000485, 2017.\n\n### Note:\n\nWe need to calculate anomalies against the historical base period.",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport geopandas as gpd\nfrom iso3166 import countries\nimport os\nimport warnings\nwarnings.filterwarnings('ignore')",
"_____no_output_____"
],
[
"def identify_netcdf_and_csv_files(path='data/'):\n \"\"\"Crawl through a specified folder and return a dict of the netcdf d['nc']\n and csv d['csv'] files contained within.\n Returns something like\n {'nc':'data/CNRS_data/cSoil/orchidee-giss-ecearth.SWL_15.eco.cSoil.nc'}\n \"\"\"\n netcdf_files = []\n csv_files = []\n for root, dirs, files in os.walk(path):\n if isinstance([], type(files)):\n for f in files:\n if f.split('.')[-1] in ['nc']:\n netcdf_files.append(''.join([root,'/',f]))\n elif f.split('.')[-1] in ['csv']:\n csv_files.append(''.join([root,'/',f]))\n return {'nc':netcdf_files,'csv':csv_files}\n",
"_____no_output_____"
],
[
"\ndef extract_value(df, swl, verbose =False):\n \"\"\"Extract the historical and absolute SWL values and calculate\n an anomaly.\n \"\"\"\n if verbose: print(df[swl].values)\n if 'PopAff_1976-2005' in df:\n historical_key = 'PopAff_1976-2005'\n #print(\"In pop aff\")\n elif 'ExpDam_1976-2005' in df:\n historical_key = 'ExpDam_1976-2005'\n else:\n raise ValueError('Found no historical data in the file')\n # Get the SWL mean\n try:\n tmp_abs = float(''.join(df[swl].values[0].split(\",\")))\n except:\n tmp_abs = None\n # Get the historical mean\n try:\n tmp_historical = float(''.join(df[historical_key].values[0].split(\",\")))\n if tmp_historical == 0: tmp_historical = None\n except:\n tmp_historical = None\n #print(tmp_historical, tmp_abs)\n if all([tmp_historical, tmp_abs]):\n anomaly = int(tmp_abs - tmp_historical)\n else:\n anomaly = None\n return anomaly\n\n\ndef gen_output_fname(fnm, swl_label):\n path = '/'.join(fnm.split('/')[1:3])\n file_name = swl_label+'_'+fnm.split('/')[-1]\n tmp_out = '/'.join(['./processed/admin0/', path, file_name])\n return tmp_out\n\n\ndef process_JRC_excel(fnm, verbose=False):\n # I should loop over the set of shapes in gadams8 shapefile and look for the country in the data...\n # SIMPLIFIED SHAPES FOR ADMIN 0 LEVEL\n s = gpd.read_file(\"./data/gadm28_countries/gadm28_countries.shp\")\n raw_data = pd.read_csv(fnm)\n # Note 184 are how many valid admin 0 areas we got with the netcdf data.\n keys =['name_0','iso','variable','swl_info',\n 'count', 'max','min','mean','std','impact_tag','institution',\n 'model_long_name','model_short_name','model_taxonomy',\n 'is_multi_model_summary','is_seasonal','season','is_monthly',\n 'month']\n swl_dic = {'SWL1.5':1.5, 'SWL2':2.0, 'SWL4':4.0}\n\n possible_vars = {'data/JRC_data/river_floods/PopAff_SWLs_Country.csv':'river_floods_PopAff',\n 'data/JRC_data/river_floods/ExpDam_SWLs_Country.csv':'river_floods_ExpDam'}\n num_swls = 0\n for swl in ['SWL1.5','SWL2', 'SWL4']:\n num_swls += 1\n tot = 0\n valid = 0\n extracted_values = []\n meta_level1 = {'variable':possible_vars[fnm],\n 'swl_info':swl_dic[swl],\n 'is_multi_model_summary':True,\n 'model_short_name':'EC-EARTH',\n 'model_long_name': \"Projections of average changes in river flood risk per country at SWLs, obtained with the JRC impact model based on EC-EARTH r1-r7 climate projections.\",\n 'model_taxonomy': 'EC-EARTH',\n 'is_seasonal': False,\n 'season': None,\n 'is_monthly':False,\n 'month': None,\n 'impact_tag': 'w',\n 'institution': \"European Commission - Joint Research Centre\",\n }\n for i in s.index:\n tot += 1\n meta_level2 = {'name_0': s['name_engli'][i],\n 'iso': s['iso'][i],}\n tmp_mask = raw_data['ISO3_countryname'] == meta_level2['iso']\n data_slice = raw_data[tmp_mask]\n if len(data_slice) == 1:\n #print(meta_level2['iso'])\n #return data_slice\n extracted = extract_value(data_slice, swl)\n if verbose: print(meta_level2['iso'], meta_level1['swl_info'], extracted)\n dic_level3 = {'min':None,\n 'mean': extracted,\n 'max': None,\n 'count':None,\n 'std':None}\n valid += 1\n # FIND ALL VALUES NEEDED BY KEY\n # WRITE TO EXTRACTED_VALUES\n d = {**meta_level1, **meta_level2, **dic_level3}\n extracted_values.append([d[key] for key in keys])\n tmp_df = pd.DataFrame(extracted_values, columns=keys)\n output_filename = gen_output_fname(fnm, swl)\n path_check ='/'.join(output_filename.split('/')[:-1])\n # WRITE EXTRACTED VALUES TO A SPECIFIC SWL CSV FILE IN PROCESSED\n if not os.path.exists(path_check):\n os.makedirs(path_check)\n #return tmp_df\n tmp_df.to_csv(output_filename, index=False)\n if verbose: print('Created ', output_filename)\n print('TOTAL in loop:', tot)\n print('valid:', valid)\n print(\"Looped for\", num_swls, 'swls')",
"_____no_output_____"
],
[
"fs = identify_netcdf_and_csv_files(path='data/JRC_data')\nfs['csv']\nfor fnm in fs['csv']:\n print(fnm)\n process_JRC_excel(fnm)",
"data/JRC_data/river_floods/PopAff_SWLs_Country.csv\nTOTAL in loop: 256\nvalid: 186\nLooped for 3 swls\ndata/JRC_data/river_floods/ExpDam_SWLs_Country.csv\nTOTAL in loop: 256\nvalid: 186\nLooped for 3 swls\n"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
cbeb4532d24643bab0c9d313a41dfe6e035cf9d0
| 8,133 |
ipynb
|
Jupyter Notebook
|
M220P: MongoDB for Python Developers/mflix-python/notebooks/change_streams.ipynb
|
helpthx/MongoDB_University
|
aec6e1d215cb6191470e6ee0a693f45c0e8764c5
|
[
"MIT"
] | null | null | null |
M220P: MongoDB for Python Developers/mflix-python/notebooks/change_streams.ipynb
|
helpthx/MongoDB_University
|
aec6e1d215cb6191470e6ee0a693f45c0e8764c5
|
[
"MIT"
] | null | null | null |
M220P: MongoDB for Python Developers/mflix-python/notebooks/change_streams.ipynb
|
helpthx/MongoDB_University
|
aec6e1d215cb6191470e6ee0a693f45c0e8764c5
|
[
"MIT"
] | null | null | null | 29.900735 | 274 | 0.594246 |
[
[
[
"<img src=\"https://s3.amazonaws.com/edu-static.mongodb.com/lessons/M220/notebook_assets/screen_align.png\" style=\"margin: 0 auto;\">\n",
"_____no_output_____"
],
[
"<h1 style=\"text-align: center; font-size=58px;\">Change Streams</h1>",
"_____no_output_____"
],
[
"In this lesson, we're going to use change streams to track real-time changes to the data that our application's using.",
"_____no_output_____"
],
[
"### Change Streams\n\n- Report changes at the collection level\n- Accept pipelines to transform change events",
"_____no_output_____"
],
[
"As of MongoDB 3.6, change streams report changes at the collection level, so we open a change stream against a specific collection.\n\nBut by default it will return any change to the data in that collection regardless of what it is, so we can also pass a pipeline to transform the change events we get back from the stream.",
"_____no_output_____"
]
],
[
[
"from pymongo import MongoClient, errors\nuri = \"mongodb+srv://m220-user:[email protected]/test\"\nclient = MongoClient(uri)",
"_____no_output_____"
]
],
[
[
"So here I'm just initializing my MongoClient object,",
"_____no_output_____"
]
],
[
[
"lessons = client.lessons\ninventory = lessons.inventory\ninventory.drop()\n\nfruits = [ \"strawberries\", \"bananas\", \"apples\" ]\nfor fruit in fruits:\n inventory.insert_one( { \"type\": fruit, \"quantity\": 100 } )\n \nlist(inventory.find())",
"_____no_output_____"
]
],
[
[
"And I'm using a new collection for this lesson, `inventory`. If you imagine we have a store that sells fruits, this collection will store the total quanities of every fruit that we have in stock.\n\nIn this case, we have a very small store that only sells three types of fruits, and I've just updated the inventory to reflect that we just got a shipment for 100 of each fruit.\n\nNow I'm just going to verify that our collection looks the way we expect.\n\n(run cell)\n\nAnd it looks like we have 100 of each fruit in the collection.\n\nBut people will start buying them, cause you know, people like fruit. They'll go pretty quickly, and we want to make sure we don't run out. So I'm going to open a change stream against this collection, and track data changes to the `inventory` collection in real time.",
"_____no_output_____"
]
],
[
[
"try:\n with inventory.watch(full_document='updateLookup') as change_stream_cursor:\n for data_change in change_stream_cursor:\n print(data_change)\nexcept pymongo.errors.PyMongoError:\n print('Change stream closed because of an error.')",
"_____no_output_____"
]
],
[
[
"So here I'm opening a change stream against the `inventory` (point) collection, using the `watch()` method. `watch()` (point) returns a cursor object, so we can iterate through it in Python to return whatever document is next in the cursor.\n\nWe've wrapped this in a try-catch block so if something happens to the connection used for the change stream, we'll know immediately.\n\n(start the while loop)\n\n(go to `updates_every_one_second` notebook and start up process)\n\n(come back here)\n\nSo the change stream cursor is just gonna spit out anything it gets, with no filter. Any change to the data in the `inventory` collection will appear in this output.\n\nBut really, this is noise. We don't care when the quantity drops to 71 (point) or 60 (point), we only want to know when it's close to zero.",
"_____no_output_____"
]
],
[
[
"low_quantity_pipeline = [ { \"$match\": { \"fullDocument.quantity\": { \"$lt\": 20 } } } ]\n\ntry:\n with inventory.watch(pipeline=low_quantity_pipeline, full_document='updateLookup') as change_stream_cursor:\n for data_change in change_stream_cursor:\n current_quantity = data_change[\"fullDocument\"].get(\"quantity\")\n fruit = data_change[\"fullDocument\"].get(\"type\")\n msg = \"There are only {0} units left of {1}!\".format(current_quantity, fruit)\n print(msg)\nexcept pymongo.errors.PyMongoError:\n logging.error('Change stream closed because of an error.')",
"_____no_output_____"
]
],
[
[
"Let's say we want to know if any of our quantities (point to quantity values) dip below 20 units, so we know when to buy more.\n\nHere I've defined a pipeline for the change event documents returned by the cursor. In this case, if the cursor returns a change event to me, it's because that event caused one of our quantities to fall below 10 units.\n\n(open the change stream)\n\n(go to `updates_every_one_second` and start the third cell)\n\n(come back here)\n\nSo if we just wait for the customers to go about their business...\n\n(wait for a print statement)\n\nAnd now we know that we need to buy more strawberries!",
"_____no_output_____"
],
[
"## Summary\n\n- Change streams can be opened against a collection\n - Tracks data changes in real time\n- Aggregation pipelines can be used to transform change event documents",
"_____no_output_____"
],
[
"So change streams are a great way to track changes to the data in a collection. And if you're using Mongo 4.0, you can open a change stream against a whole database, and even a whole cluster.\n\nWe also have the flexibility to pass an aggregation pipeline to the change stream, to transform or filter out some of the change event documents.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
cbeb456ccd2689322dfeecc9aed3a99a93dcb322
| 766,500 |
ipynb
|
Jupyter Notebook
|
pyflux/Dynamic Linear Regression Models in Python.ipynb
|
amirudin07/pythondata
|
f8978bf33368ce1ecb1838cb35c6f8a54f3ce59d
|
[
"MIT"
] | 122 |
2017-08-27T16:32:57.000Z
|
2022-01-09T04:19:19.000Z
|
pyflux/Dynamic Linear Regression Models in Python.ipynb
|
hossein20s/Random-Forest-forecasting
|
48934a19575d3c3652631d3043c55a7b1554d459
|
[
"MIT"
] | 1 |
2021-02-02T23:00:44.000Z
|
2021-02-02T23:00:44.000Z
|
pyflux/Dynamic Linear Regression Models in Python.ipynb
|
hossein20s/Random-Forest-forecasting
|
48934a19575d3c3652631d3043c55a7b1554d459
|
[
"MIT"
] | 215 |
2017-08-28T14:18:39.000Z
|
2022-03-26T15:13:16.000Z
| 859.304933 | 129,984 | 0.936228 |
[
[
[
"import pandas as pd\nimport numpy as np\nimport pyflux as pf\nimport matplotlib.pyplot as plt\nfrom fbprophet import Prophet\n \n%matplotlib inline\nplt.rcParams['figure.figsize']=(20,10)\nplt.style.use('ggplot')",
"_____no_output_____"
]
],
[
[
"### Load the data\n\nFor this work, we're going to use the same retail sales data that we've used before. It can be found in the examples directory of this repository.",
"_____no_output_____"
]
],
[
[
"sales_df = pd.read_csv('../examples/retail_sales.csv', index_col='date', parse_dates=True)",
"_____no_output_____"
],
[
"sales_df.head()",
"_____no_output_____"
]
],
[
[
"Like all good modeling projects, we need to take a look at the data to get an idea of what it looks like.",
"_____no_output_____"
]
],
[
[
"sales_df.plot()",
"_____no_output_____"
]
],
[
[
"It's pretty clear from this data that we are looking at a trending dataset with some seasonality. This is actually a pretty good datset for prophet since the additive model and prophet's implemention does well with this type of data. \n\nWith that in mind, let's take look at what prophet does from a modeling standpoint to compare with the dynamic linear regression model. For more details on this, you can take a look at my blog post titled **Forecasting Time Series data with Prophet – Part 4** (http://pythondata.com/forecasting-time-series-data-prophet-part-4/)",
"_____no_output_____"
]
],
[
[
"# Prep data for prophet and run prophet\ndf = sales_df.reset_index()\ndf=df.rename(columns={'date':'ds', 'sales':'y'})\n\nmodel = Prophet(weekly_seasonality=True)\nmodel.fit(df);\nfuture = model.make_future_dataframe(periods=24, freq = 'm')\n\nforecast = model.predict(future)\nmodel.plot(forecast);",
"INFO:fbprophet.forecaster:Disabling daily seasonality. Run prophet with daily_seasonality=True to override this.\n"
]
],
[
[
"With our prophet model ready for comparison, let's build a model with pyflux's dynamic linear regresion model.\n",
"_____no_output_____"
],
[
"### More Data Viz \n\nNow that we've run our prophet model and can see what it has done, its time to walk through what I call the 'long form' of model building. This is more involved than throwing data at a library and accepting the results.\n\nFor this data, let's first look at the differenced log values of our sales data (to try to make it more stationary).",
"_____no_output_____"
]
],
[
[
"diff_log = pd.DataFrame(np.diff(np.log(sales_df['sales'].values)))\ndiff_log.index = sales_df.index.values[1:sales_df.index.values.shape[0]]\ndiff_log.columns = [\"Sales DiffLog\"]\n",
"_____no_output_____"
],
[
"sales_df['logged']=np.log(sales_df['sales'])",
"_____no_output_____"
],
[
"sales_df.tail()",
"_____no_output_____"
],
[
"sales_df.plot(subplots=True)",
"_____no_output_____"
]
],
[
[
"With our original data (top pane in orange), we can see a very pronounced trend. With the differenced log values (bottom pane in blue), we've removed that trend and made the data staionary (or hopefully we have).\n\nNow, lets take a look at an autocorrelation plot, which will tell us whether the future sales is correlated with the past data. I won't go into detail on autocorrelation, but if you don't understand whether you have autocorrelation (and to what degree), you might be in for a hard time :)\n\nLet's take a look at the autocorrelation plot (acf) if the differenced log values as well as the ACF of the square of the differenced log values.\n",
"_____no_output_____"
]
],
[
[
"pf.acf_plot(diff_log.values.T[0])\npf.acf_plot(np.square(diff_log.values.T[0]))",
"_____no_output_____"
]
],
[
[
"We can see that at a lag of 1 and 2 months, there are positive correlations for sales but as time goes on, that correlation drops quickly to a negative correlation that stays in place over time, which hints at the fact that there are some autoregressive effects within this data. \n\nBecause of this fact, we can start our modeling by using an ARMA model of some sort.",
"_____no_output_____"
]
],
[
[
"Logged = pd.DataFrame(np.log(sales_df['sales']))\nLogged.index = pd.to_datetime(sales_df.index)\nLogged.columns = ['Sales - Logged']",
"_____no_output_____"
],
[
"Logged.head()",
"_____no_output_____"
],
[
"modelLLT = pf.LLT(data=Logged)",
"_____no_output_____"
],
[
"x = modelLLT.fit()\nx.summary()",
"LLT \n======================================================= ==================================================\nDependent Variable: Sales - Logged Method: MLE \nStart Date: 2009-10-01 00:00:00 Log Likelihood: 79.4161 \nEnd Date: 2015-09-01 00:00:00 AIC: -152.8322 \nNumber of observations: 72 BIC: -146.0022 \n==========================================================================================================\nLatent Variable Estimate Std Error z P>|z| 95% C.I. \n======================================== ========== ========== ======== ======== =========================\nSigma^2 irregular 0.00404690 \nSigma^2 level 2.2047e-05 \nSigma^2 trend 0.0 \n==========================================================================================================\n"
],
[
"model.plot_fit(figsize=(20,10))",
"_____no_output_____"
],
[
"modelLLT.plot_predict_is(h=len(Logged)-1, figsize=(20,10))",
"_____no_output_____"
],
[
"predicted = modelLLT.predict_is(h=len(Logged)-1)\npredicted.columns = ['Predicted']",
"_____no_output_____"
],
[
"predicted.tail()",
"_____no_output_____"
],
[
"np.exp(predicted).plot()",
"_____no_output_____"
],
[
"sales_df_future=sales_df\nsales_df",
"_____no_output_____"
],
[
"final_sales=sales_df.merge(np.exp(predicted),right_on=predicted.index)",
"_____no_output_____"
],
[
"final_sales = sales_df.merge()",
"_____no_output_____"
],
[
"final_sales.tail()",
"_____no_output_____"
],
[
"final_sales.plot()",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbeb50723d69a3bf06bd5e851a770d1343668610
| 34,765 |
ipynb
|
Jupyter Notebook
|
PassagensDecolar/FindPassagens.ipynb
|
joaocamargo/estudos-python
|
c5fbf59a1f06131d9789dca7dbdfdcf2200d0227
|
[
"MIT"
] | 1 |
2019-10-09T12:56:13.000Z
|
2019-10-09T12:56:13.000Z
|
PassagensDecolar/FindPassagens.ipynb
|
joaocamargo/estudos-python
|
c5fbf59a1f06131d9789dca7dbdfdcf2200d0227
|
[
"MIT"
] | null | null | null |
PassagensDecolar/FindPassagens.ipynb
|
joaocamargo/estudos-python
|
c5fbf59a1f06131d9789dca7dbdfdcf2200d0227
|
[
"MIT"
] | null | null | null | 46.79004 | 4,195 | 0.587689 |
[
[
[
"import urllib.request\nimport json\nimport pandas as pd\nfrom datetime import datetime\nimport seaborn as sns\ncm = sns.light_palette(\"red\", as_cmap=True)\n#https://www.trilhaseaventuras.com.br/siglas-dos-principais-aeroportos-do-mundo-iata/\n\n#urlOneWay\n#https://www.decolar.com/shop/flights-busquets/api/v1/web/search?adults=1&children=0&infants=0&limit=4&site=BR&channel=site&from=POA&to=MIA&departureDate=2020-03-04&groupBy=default&orderBy=total_price_ascending&viewMode=CLUSTER&language=pt_BR&airlineSummary=false&chargesDespegar=false&user=e1861e3a-3357-4a76-861e-3a3357ea76c0&h=38dc1f66dbf4f5c8df105321c3286b5c&flow=SEARCH&di=1-0&clientType=WEB&disambiguationApplied=true&newDisambiguationService=true&initialOrigins=POA&initialDestinations=MIA&pageViewId=62ef8aab-ab53-406c-8429-885702acecbd",
"_____no_output_____"
],
[
"import requests\n\nurl = \"https://www.pontosmultiplus.com.br/service/facilities/handle-points\"\n\npayload = \"logado=&select-name=1000&points=1000&action=calculate\"\nheaders = {\n 'authority': 'www.pontosmultiplus.com.br',\n 'accept': 'application/json, text/javascript, */*; q=0.01',\n 'origin': 'https://www.pontosmultiplus.com.br',\n 'x-requested-with': 'XMLHttpRequest',\n 'user-agent': 'Mozilla/5.0 (Windows NT 10.0; Win64; x64) AppleWebKit/537.36 (KHTML, like Gecko) Chrome/79.0.3945.88 Safari/537.36',\n 'dnt': '1',\n 'content-type': 'application/x-www-form-urlencoded; charset=UTF-8',\n 'sec-fetch-site': 'same-origin',\n 'sec-fetch-mode': 'cors',\n 'referer': 'https://www.pontosmultiplus.com.br/facilidades/compradepontos',\n 'accept-encoding': 'gzip, deflate, br',\n 'accept-language': 'pt-BR,pt;q=0.9,en-US;q=0.8,en;q=0.7',\n 'cookie': 'userPrefLanguage=pt_BR; sback_client=573a40fecdbbbb66963e544d; sback_partner=false; sb_days=1549545557254; sback_browser=0-14236400-1548261174b4075e5fdbd390aa38772d39e7c7a352593b045121165093285c48973622c2c1-45877488-170246360, 20525122160-1549545560; sback_customer=$2QUxcVTzd0dOVENUtmd6dlTmp3RjlHVF90bxETQ1oWOad0dWF0QUN3T0hnYBlFVx0UO5BVWTRFVNZTblh2YqRkW2$12; chaordic_browserId=09b01e60-2300-11e9-8ced-6fbc9e419cda; chaordic_anonymousUserId=anon-09b01e60-2300-11e9-8ced-6fbc9e419cda; sback_total_sessions=2; _ducp=eyJfZHVjcCI6ImE4MzY0NWM2LTI3ZWYtNGUzZS1iMzNjLTI3YmY4ZTY4MDMwOCIsIl9kdWNwcHQiOiIifQ==; _fbp=fb.2.1550499169207.1066950068; cto_h2h=B; s_fid=2E4956A0C0C14E48-2CB286BB7EF81637; cto_lwid=01abc4e4-21f3-450f-9f35-57dee229928a; __utmz=196304045.1569964079.10.2.utmcsr=multiplus-emkt|utmccn=20190930_EMAIL_INSTITUCIONAL_BOAS_VINDAS_NOVA_MARCA_BRASIL-20191001|utmcmd=emkt|utmctr=14602|utmcct=cabecalho-ver_extrato_deslogado; s_vnum=1596641437499%26vn%3D2; s_lv=1569964112519; optionExchange=0; origin=[{%22city%22:{%22iataCode%22:%22POA%22%2C%22name%22:%22Porto%20Alegre%22}%2C%22type%22:%22airport%22%2C%22iataCode%22:%22POA%22%2C%22name%22:%22Salgado%20Filho%22%2C%22value%22:%22POA_airport%22%2C%22orderCodeNumber%22:%222%22%2C%22orderCode%22:%22Porto%20Alegre2%22%2C%22label%22:%22Porto%20Alegre%20(POA)%2C%20Salgado%20Filho%20(POA)%2C%20Brasil%22%2C%22position%22:%2200002Porto%20Alegre%20(POA)%2C%20Salgado%20Filho%20(POA)%2C%20Brasil%22}]; destiny=[{%22city%22:{%22iataCode%22:%22FRA%22%2C%22name%22:%22Frankfurt%22}%2C%22type%22:%22airport%22%2C%22iataCode%22:%22FRA%22%2C%22name%22:%22Frankfurt%20Intl.%22%2C%22value%22:%22FRA_airport%22%2C%22orderCodeNumber%22:%222%22%2C%22orderCode%22:%22Frankfurt2%22%2C%22label%22:%22Frankfurt%20(FRA)%2C%20Frankfurt%20Intl.%20(FRA)%2C%20Alemanha%22%2C%22position%22:%2200002Frankfurt%20(FRA)%2C%20Frankfurt%20Intl.%20(FRA)%2C%20Alemanha%22}]; cabinClass=Y; classesSuggestions=[{%22idCabin%22:1%2C%22cabinClass%22:%22Y%22%2C%22cabinName%22:%22Economy%22}%2C{%22idCabin%22:2%2C%22cabinClass%22:%22W%22%2C%22cabinName%22:%22Premium%20Economy%22}%2C{%22idCabin%22:3%2C%22cabinClass%22:%22J%22%2C%22cabinName%22:%22Premium%20Business%22}]; _gcl_au=1.1.278670892.1578924604; _hjid=59ae5b53-f6c8-48b1-bc67-fb8182856ead; chaordic_testGroup=%7B%22experiment%22%3Anull%2C%22group%22%3Anull%2C%22testCode%22%3Anull%2C%22code%22%3Anull%2C%22session%22%3Anull%7D; country_code=br; language_code=pt; __utmc=196304045; _esvan_ref.50060.=; language_country=pt_br; _ga=GA1.3.1171237216.1579530427; _gid=GA1.3.911523691.1579530427; _gaZ=GA1.3.1171237216.1579530427; _gaZ_gid=GA1.3.911523691.1579530427; return=Sat%20Apr%2011%202020%2012:00:00%20GMT-0300%20(Hor%C3%A1rio%20Padr%C3%A3o%20de%20Bras%C3%ADlia); trip=ida_vuelta; departure=Sat%20Apr%2004%202020%2012:00:00%20GMT-0300%20(Hor%C3%A1rio%20Padr%C3%A3o%20de%20Bras%C3%ADlia); SMSESSION=LOGGEDOFF; userIdZ=; __utma=196304045.1744687836.1549545551.1579547569.1579636257.15; analyticsHelper.cd38=ef144e288de8d22700e20cda9fce9ee5ee61b5d25b61bd0dab35f4ddc72e95ce; ATGSESSIONID=yiPNORqQ9P7PZ74G-Syy7CLAjB8uk3Tw0Wc4dHWdUyC7KjCIe4s0u0021-680739279; __zjc7749=4962761565; userTags=%7B%22id%22%3A%22Anonimo%22%2C%22age%22%3A0%2C%22gender%22%3Anull%2C%22email%22%3Anull%2C%22emailHash%22%3Anull%2C%22country%22%3Anull%2C%22city%22%3Anull%2C%22state%22%3Anull%2C%22zipCode%22%3Anull%2C%22typeOfParticipation%22%3Anull%2C%22balance%22%3Anull%2C%22status%22%3A%22deslogado%22%7D; _gac_UA-83192457-1=1.1579696070.CjwKCAiAgqDxBRBTEiwA59eEN-j8nGbsIpfJMIrCCHTfzUi4saF5CmN227pOPsXIuXAOZmOQs_DMSRoCBtMQAvD_BwE; _gcl_aw=GCL.1579696070.CjwKCAiAgqDxBRBTEiwA59eEN-j8nGbsIpfJMIrCCHTfzUi4saF5CmN227pOPsXIuXAOZmOQs_DMSRoCBtMQAvD_BwE; _dc_gtm_UA-83192457-1=1; _gac_UA-83192457-13=1.1579696070.CjwKCAiAgqDxBRBTEiwA59eEN-j8nGbsIpfJMIrCCHTfzUi4saF5CmN227pOPsXIuXAOZmOQs_DMSRoCBtMQAvD_BwE; _dc_gtm_UA-83192457-13=1; __z_a=3200530082274793935727479; JSESSIONID=_hHNOSuko30OZo1X7XyjT4_6rnAXanFcwA7M9PShrPBBjztzhMrIu0021-1010243761; SS_X_JSESSIONID=KoLNOSzOIq0SooUobVecEo7ju0GL-8Y2O_kOVlqjZsm5rKnmkG33u0021-183582721; akavpau_multiplusgeral=1579696676~id=48e1b4d4309a5f9f09664afd46406b0e; __zjc872=4962761577; _gat=1'\n}\n\nresponse = requests.request(\"POST\", url, headers=headers, data = payload)\nresultPontos = response.text.encode('utf8')\nresPontos = json.loads(resultPontos.decode('utf-8'))\nprint(resPontos['data']['total'])\nPONTOSMULTIPLUS = resPontos['data']['total']",
"_____no_output_____"
],
[
"dataInicial = '2020-07-03'\ndataFinal = '2020-07-19'\n\nidaEvolta=True\n#tripType=''\n\n#dataInicial = '2020-04-08'\n#dataFinal = '2020-04-22'\n\n#if idaEvolta:\n# tripType = 'roundtrip'\n#else:\n# tripType = 'oneway'\n\nspecificDate = False",
"_____no_output_____"
],
[
"origens = ['POA','GRU','GIG']\ndestinos = ['ATL','MIA','MDZ','BRC','LIM','CTG','ADZ','FRA']\n\n#dfDict.append({'de':origem,'para':destino,'Ida': p['departureDate'],'Volta':arr['arrivalDate'],'preco':arr['price'][\"amount\"]})",
"_____no_output_____"
],
[
"resumo = []\ndfDict =[]\n\nfor origem in origens:\n for destino in destinos: \n minValue = 999999999\n fraseFinal= ''\n print(origem + ' -> '+ destino)\n urlDecolar = '''https://www.decolar.com/shop/flights-busquets/api/v1/web/calendar-prices/matrix?adults=1&children=0&infants=0&limit=4&site=BR&channel=site&from={origem}&to={destino}&departureDate={dataInicial}&returnDate={dataFinal}&orderBy=total_price_ascending&viewMode=CLUSTER&language=pt_BR&clientType=WEB&initialOrigins={origem}&initialDestinations={destino}&pageViewId=b35e67df-abc9-4308-875f-c3810b3729e4&mustIncludeDates=NA_NA¤cy=BRL&breakdownType=TOTAL_FARE_ONLY'''.format(dataInicial=dataInicial,dataFinal=dataFinal,origem=origem,destino=destino)\n #print(urlDecolar)\n with urllib.request.urlopen(urlDecolar) as url:\n s = url.read()\n data = json.loads(s.decode('utf-8'))\n #print(data)\n for p in data['departures']:\n for arr in p['arrivals']:\n if 'price' in arr:\n dfDict.append({'DataPesquisa':datetime.now().strftime(\"%d/%m/%Y %H:%M:%S\"),'de':origem,'para':destino,'Ida': p['departureDate'],'Volta':arr['arrivalDate'],'preco':arr['price'][\"amount\"]})\n if specificDate:\n if p['departureDate'] == dataInicial and arr['arrivalDate'] == dataFinal: \n if minValue > arr['price'][\"amount\"]:\n minValue = arr['price'][\"amount\"]\n fraseFinal = 'Voo mais barato '+origem + ' -> '+ destino+' de:' + p['departureDate'], ' ate ',arr['arrivalDate'],'- valor: ' + str(arr['price'][\"amount\"]) \n resumo.append(fraseFinal)\n print('de:' + p['departureDate'], ' ate ',arr['arrivalDate'],'- valor: ' + str(arr['price'][\"amount\"])) \n else: \n if minValue > arr['price'][\"amount\"]:\n minValue = arr['price'][\"amount\"]\n fraseFinal = 'Voo mais barato '+origem + ' -> '+ destino+' de:' + p['departureDate'], ' ate ',arr['arrivalDate'],'- valor: ' + str(arr['price'][\"amount\"]) \n resumo.append(fraseFinal)\n print('de:' + p['departureDate'], ' ate ',arr['arrivalDate'],'- valor: ' + str(arr['price'][\"amount\"])) \n print('')\n print(fraseFinal)\n print(minValue)\n print('')\n \nfor r in resumo:\n print(r)\n ",
"_____no_output_____"
],
[
"df = pd.DataFrame.from_dict(dfDict)\n\nif specificDate:\n df = df[df['Ida']==dataInicial]\n df = df[df['Volta']==dataFinal]\ndisplay(df.describe())\ndf.sort_values(by='preco',ascending=True).head(5).style.background_gradient(cmap='OrRd')",
"_____no_output_____"
],
[
"with open('historicoPesquisaPrecos.csv', 'a') as f:\n df.to_csv(f, mode='a',header=f.tell()==0)\n ",
"_____no_output_____"
],
[
"dfGrafico = pd.read_csv(\"historicoPesquisaPrecos.csv\") \n",
"_____no_output_____"
],
[
"dfGrafico = dfGrafico[dfGrafico['Ida']>='2020-07-03']\ndfGrafico = dfGrafico[dfGrafico['Ida']<='2020-07-07']\ndfGrafico = dfGrafico[dfGrafico['Volta']>='2020-07-17']\ndfGrafico = dfGrafico[dfGrafico['Volta']<='2020-07-20']",
"_____no_output_____"
],
[
"dfGrafico['DataPesquisa'] = dfGrafico['DataPesquisa'].apply(lambda x:x[0:13])\ndfGrafico['DataPesquisaDATA']=dfGrafico['DataPesquisa'].apply(lambda x:pd.to_datetime(x[0:10]))\ndfGrafico['Dias'] = dfGrafico.apply(lambda x: int(str(pd.to_datetime(x['Volta'])- pd.to_datetime(x['Ida']))[0:2]),axis=1)",
"_____no_output_____"
],
[
"dfGrafico['OrigemDestino'] = dfGrafico.apply(lambda x: x['de'] + x['para'],axis=1)\ndfGrafico['EspecificoIda'] = dfGrafico.apply(lambda x: x['de'] + x['para']+'-'+x['Ida'],axis=1)\ndfGrafico['EspecificoVolta'] = dfGrafico.apply(lambda x: x['de'] + x['para']+'-'+x['Volta'],axis=1)\ndfGrafico['EspecificoTodos'] = dfGrafico.apply(lambda x: x['de'] + x['para']+'-'+x['Ida']+'-'+x['Volta'],axis=1)\ndisplay(dfGrafico)",
"_____no_output_____"
],
[
"#dfGraficoPOA_ATL = dfGrafico.query('de == \"POA\" & para == \"ATL\"')\n#dfGraficoPOA_MIA = dfGrafico.query('de == \"POA\" & para == \"MIA\"')\n#dfGraficoGRU_MIA = dfGrafico.query('de == \"GRU\" & para == \"MIA\"')\n#dfGraficoGRU_ATL = dfGrafico.query('de == \"GRU\" & para == \"ATL\"')\n#dfGraficoGRU_MDZ = dfGrafico.query('de == \"GRU\" & para == \"MDZ\"')\n#dfGraficoPOA_MDZ = dfGrafico.query('de == \"POA\" & para == \"MDZ\"')\n\n#datasets = [dfGrafico,dfGraficoPOA_ATL,dfGraficoPOA_MIA,dfGraficoGRU_MIA,dfGraficoGRU_ATL,dfGraficoGRU_MDZ,dfGraficoPOA_MDZ]\n\n#print(dfGraficoPOA_ATL['Ida'].count())\n#print(dfGraficoPOA_MIA['Ida'].count())\n#print(dfGraficoGRU_MIA['Ida'].count())\n#print(dfGraficoGRU_ATL['Ida'].count())\n#print(dfGraficoGRU_MDZ['Ida'].count())\n#print(dfGraficoPOA_MDZ['Ida'].count())",
"_____no_output_____"
],
[
"#import plotly.express as px\n\n#for graph in datasets: \n# #graph = graph.query('Ida ==\"2020-07-05\" & Volta ==\"2020-07-20\"')\n# graph = graph.query('de ==\"POA\" & Dias >=14 & Dias <=17')# | de ==\"GRU\"')\n# fig = px.line(graph.drop_duplicates(), x=\"DataPesquisa\", y=\"preco\", color=\"EspecificoTodos\",hover_data=['de','para','Ida', 'Volta','preco'])\n# fig.show()",
"_____no_output_____"
],
[
"import pandas_profiling\nprint(dfGraficoPOA_MIA.columns)",
"_____no_output_____"
],
[
"#pandasDf=dfGraficoPOA_MIA[['Ida', 'Volta', 'de', 'para', 'preco','DataPesquisaDATA', 'Dias']]\n#display(pandasDf.head(3))\n#pandas_profiling.ProfileReport(pandasDf)\n",
"_____no_output_____"
],
[
"dfPivot = dfGrafico.query('de == \"POA\" or de==\"GRU\"')\n#display(dfPivot.head(3))",
"_____no_output_____"
],
[
"dfPivot = pd.pivot_table(dfPivot,values='preco',index=['de','para','Dias','Ida'],columns='DataPesquisa')",
"_____no_output_____"
]
],
[
[
"## Maiores valores da serie historica",
"_____no_output_____"
]
],
[
[
"#display(dfPivot)\n#dfPivot.style.apply(highlight_max)",
"_____no_output_____"
]
],
[
[
"## Menores valores da serie historica",
"_____no_output_____"
]
],
[
[
"#dfPivot.style.apply(highlight_min)",
"_____no_output_____"
],
[
"#dfLastSearch = dfGrafico.query('de == \"POA\" or de==\"GRU\"')\n#print(dfLastSearch.groupby(['de','para']).count())\n#dfLastSearch = dfLastSearch[dfLastSearch['DataPesquisaDATA']>='21/01/2020']\n#dfLastSearchPivot = pd.pivot_table(dfLastSearch,values='preco',index=['de','para','Dias','Ida','Volta'],columns='DataPesquisa')",
"_____no_output_____"
],
[
"#dfLastSearchPivot.style.apply(highlight_min)",
"_____no_output_____"
],
[
"import pandas as pd\nimport matplotlib.pyplot as plt\nfrom matplotlib import colors\n\ndef background_gradient(s, m, M, cmap='PuBu', low=0, high=0):\n rng = M - m\n norm = colors.Normalize(m - (rng * low),\n M + (rng * high))\n normed = norm(s.values)\n c = [colors.rgb2hex(x) for x in plt.cm.get_cmap(cmap)(normed)]\n return ['background-color: %s' % color for color in c]\n\n#df = pd.DataFrame([[3,2,10,4],[20,1,3,2],[5,4,6,1]])\n",
"_____no_output_____"
],
[
"#dfLastSearchPivot.fillna(0,inplace=True)\n#dfLastSearchPivot.query('para == \"MIA\"').style.background_gradient(cmap='OrRd')\n#display(dfLastSearchPivot.style.background_gradient(cmap='OrRd'))\n#print(dfLastSearchPivot.groupby(['de','para']).count())\n#dfLastSearchPivot.style.apply(background_gradient,cmap='OrRd',m=dfLastSearchPivot.min().min(),M=dfLastSearchPivot.max().max(),low=0,high=7000)",
"_____no_output_____"
],
[
"urlPontoLatam = 'https://bff.latam.com/ws/proxy/booking-webapp-bff/v1/public/redemption/recommendations/outbound?departure={dataInicial}&origin={origem}&destination={destino}&cabin=Y&country=BR&language=PT&home=pt_br&return={dataFinal}&adult=1&tierCode=LTAM&tierType=low'\norigensPontos = ['POA','GRU','GIG']\ndestinosPontos = ['ATL','MIA','MDZ','BRC','LIM','CTG','ADZ','FRA']\ndataPesquisa = datetime.now().strftime(\"%d/%m/%Y %H:%M:%S\")\ndfPontosListIda =[]\ndfPontosListVolta =[]\nmeuSaldoAtual = 22000\n\nfor origem in origensPontos:\n for destino in destinosPontos: \n minValue = 999999999\n fraseFinal= ''\n print(origem + ' -> '+ destino)\n urlPontos = urlPontoLatam.format(dataInicial=dataInicial,dataFinal=dataFinal,origem=origem,destino=destino)\n #print(urlDecolar)\n with urllib.request.urlopen(urlPontos) as url:\n s = url.read()\n data = json.loads(s.decode('utf-8'))\n try:\n \n for flight in data['data']: \n for cabins in flight['flights']: \n paradas = cabins['stops']\n dataChegada=cabins['arrival']['date']\n horaChegada = cabins['arrival']['time']['hours']\n minutoChegada = cabins['arrival']['time']['minutes']\n overnight = cabins['arrival']['overnights']\n #partida\n dataPartida=cabins['departure']['date']\n horaPartida = cabins['departure']['time']['hours']\n minutoPartida = cabins['departure']['time']['minutes'] \n for price in cabins['cabins']:\n dfPontosListIda.append({'DataPesquisa':dataPesquisa,'De':origem,'Para':destino,'PartidaData':dataPartida,'PartidaHora':horaPartida,'PartidaMinuto':minutoPartida,'ChegadaData':dataChegada,'ChegadaHora':horaChegada,'ChegadaMinuto':minutoChegada,'overnight':overnight,'Paradas':paradas,'pontos':price['displayPrice'],'preco':(PONTOSMULTIPLUS *price['displayPrice'])/1000,'precoMenosSaldo':(PONTOSMULTIPLUS *(price['displayPrice']-meuSaldoAtual))/1000}) \n dfPontosIda = pd.DataFrame.from_dict(dfPontosListIda)\n except:\n print('erro')\n \n print(destino + ' -> '+ origem)\n urlPontos = urlPontoLatam.format(dataInicial=dataFinal,dataFinal=dataFinal,origem=destino,destino=origem)\n with urllib.request.urlopen(urlPontos) as url:\n s = url.read()\n data = json.loads(s.decode('utf-8'))\n try:\n \n for flight in data['data']: \n for cabins in flight['flights']: \n paradas = cabins['stops']\n dataChegada=cabins['arrival']['date']\n horaChegada = cabins['arrival']['time']['hours']\n minutoChegada = cabins['arrival']['time']['minutes']\n overnight = cabins['arrival']['overnights']\n #partida\n dataPartida=cabins['departure']['date']\n horaPartida = cabins['departure']['time']['hours']\n minutoPartida = cabins['departure']['time']['minutes'] \n for price in cabins['cabins']:\n dfPontosListVolta.append({'DataPesquisa':dataPesquisa,'De':destino,'Para':origem,'PartidaData':dataPartida,'PartidaHora':horaPartida,'PartidaMinuto':minutoPartida,'ChegadaData':dataChegada,'ChegadaHora':horaChegada,'ChegadaMinuto':minutoChegada,'overnight':overnight,'Paradas':paradas,'pontos':price['displayPrice'],'valorPontos':PONTOSMULTIPLUS,'preco':(PONTOSMULTIPLUS *price['displayPrice'])/1000,'precoMenosSaldo':(PONTOSMULTIPLUS *(price['displayPrice']-meuSaldoAtual))/1000}) \n dfPontosVolta = pd.DataFrame.from_dict(dfPontosListVolta) \n except:\n print('erro')",
"_____no_output_____"
],
[
"with open('historicoPesquisaPontosIda.csv', 'a') as f:\n dfPontosIda.to_csv(f, mode='a',header=f.tell()==0)\n\nwith open('historicoPesquisaPontosVolta.csv', 'a') as f:\n dfPontosVolta.to_csv(f, mode='a',header=f.tell()==0)",
"_____no_output_____"
],
[
"#dfLoadPontosIda = pd.read_csv(\"historicoPesquisaPontosIda.csv\") \n#dfLoadPontosVolta = pd.read_csv(\"historicoPesquisaPontosVolta.csv\") ",
"_____no_output_____"
],
[
"#dfPontosC = dfLoadPontosVolta[['DataPesquisa','De','Para','PartidaData','PartidaHora', 'PartidaMinuto','ChegadaData', 'ChegadaHora', 'ChegadaMinuto','Paradas','overnight', 'pontos', 'preco','precoMenosSaldo']]\n#display(dfPontosC.sort_values(by='preco',ascending=True).style.background_gradient(cmap='OrRd'))\n",
"_____no_output_____"
],
[
"uriPontos = 'https://www.pontosmultiplus.com.br/service/facilities/handle-points'",
"_____no_output_____"
],
[
"#dfT = dfLastSearch\n#dfTeste = dfT[dfT['DataPesquisaDATA']=='24/01/2020']\n#dfTeste = pd.pivot_table(dfLastSearch,values='preco',index=['de','para','Ida'],columns='Volta')\n#dfTeste.fillna(0,inplace=True)",
"_____no_output_____"
],
[
"#display(dfTeste.style.background_gradient(cmap='OrRd'))",
"_____no_output_____"
],
[
"aa",
"_____no_output_____"
],
[
"#POSTMAN ONE WAY ",
"_____no_output_____"
],
[
"import requests\n\ndataInicial = '2020-04-08'\ndataFinal = '2020-04-22'\n\norigens = ['POA','GRU','GIG','BSB','FOR']\ndestinos = ['ATL','MIA']\n\nurl = \"https://www.decolar.com/shop/flights-busquets/api/v1/web/search\"\n\nfor origem in origens:\n for destino in destinos: \n querystring = {\"adults\":\"1\",\"limit\":\"4\",\"site\":\"BR\",\"channel\":\"site\",\"from\":\"{origem}\".format(origem=origem),\"to\":\"{destino}\".format(destino=destino),\"departureDate\":\"2020-03-04\",\"orderBy\":\"total_price_ascending\",\"viewMode\":\"CLUSTER\",\"language\":\"pt_BR\",\"h\":\"38dc1f66dbf4f5c8df105321c3286b5c\",\"flow\":\"SEARCH\",\"clientType\":\"WEB\",\"initialOrigins\":\"{origem}\".format(origem=origem),\"initialDestinations\":\"{destino}\".format(destino=destino)}\n \n headers = {\n 'Connection': \"keep-alive\",\n 'DNT': \"1\",\n 'X-UOW': \"results-13-1579106681089\",\n 'X-RequestId': \"xzTTJ6fDfw\",\n 'User-Agent': \"Mozilla/5.0 (Windows NT 10.0; Win64; x64) AppleWebKit/537.36 (KHTML, like Gecko) Chrome/79.0.3945.88 Safari/537.36\",\n 'Accept': \"application/json, text/javascript, */*; q=0.01\",\n 'X-Requested-With': \"XMLHttpRequest\",\n 'XDESP-REFERRER': \"https://www.decolar.com/shop/flights/search/oneway/{origem}/{destino}/2020-03-04/2/0/0/NA/NA/NA/NA/?from=SB&di=2-0\".format(origem=origem,destino=destino),\n 'Sec-Fetch-Site': \"same-origin\",\n 'Sec-Fetch-Mode': \"cors\",\n 'Referer': \"https://www.decolar.com/shop/flights/search/oneway/{origem}/{destino}/2020-03-04/1/0/0/NA/NA/NA/NA/?from=SB&di=1-0\".format(origem=origem,destino=destino),\n 'Accept-Encoding': \"gzip, deflate, br\",\n 'Accept-Language': \"pt-BR,pt;q=0.9,en-US;q=0.8,en;q=0.7\",\n 'Cookie': 'trackerid=e1861e3a-3357-4a76-861e-3a3357ea76c0; xdesp-rand-usr=292; xdsid=C632CEAAF251AE2A72F165ECA9A4A2CA; xduid=1727A02D2FAA249C654A094113369154; _ga=GA1.2.772144563.1579011917; _gid=GA1.2.317154519.1579011917; trackeame_cookie=%7B%22id%22%3A%22UPA_e1861e3a-3357-4a76-861e-3a3357ea76c0%22%2C%22version%22%3A%225.0%22%2C%22upa_id%22%3A%22e1861e3a-3357-4a76-861e-3a3357ea76c0%22%2C%22creation_date%22%3A%222020-01-14T14%3A25%3A17Z%22%7D; __ssid=41de76d348be0e334af8e657f6801b8; _gcl_au=1.1.1367791908.1579011932; _fbp=fb.1.1579011933564.1470255143; __gads=ID=9139db3a836078f5:T=1579011933:S=ALNI_MawboBo55i9nPvoDvzaF396HudEKg; abzTestingId=\"{\\\"flightsFisherAB\\\":90,\\\"pkgImbatibleBrand_ctrl\\\":76,\\\"s_flights_s_violet_sbox_v1\\\":21,\\\"upsellingConfig\\\":58,\\\"twoOneWayForceMX\\\":0,\\\"filterLandingFlights\\\":41,\\\"s_loyalty_v2_ctrl\\\":5,\\\"s_flights_l_violet_sbox_v1\\\":0,\\\"s_flights_l_loyalty_v2\\\":58,\\\"mostProfitablePromotion\\\":0,\\\"despechecks\\\":72,\\\"s_loyalty_v2_review\\\":33,\\\"platform\\\":55,\\\"selected_radio_button\\\":0,\\\"fisher_2ow\\\":0,\\\"loyalty_non_adherents\\\":63,\\\"paymentMethod\\\":55,\\\"shifuMobileProductLabels\\\":0,\\\"obFee\\\":40,\\\"twoOneWay\\\":0,\\\"s_violet_sbox_v1\\\":17,\\\"s_flights_s_loyalty_v2\\\":14,\\\"flights_loyalty_non_adherents\\\":63,\\\"pkgImbatibleBrand-ctrl\\\":60,\\\"crossBorderTicketing\\\":0}; chktkn=ask3r5kj6ed0ksqrs7eio4cebk; searchId=243920d8-49cc-4271-972a-60d05221ef20; _gat_UA-36944350-2=1,trackerid=e1861e3a-3357-4a76-861e-3a3357ea76c0; xdesp-rand-usr=292; xdsid=C632CEAAF251AE2A72F165ECA9A4A2CA; xduid=1727A02D2FAA249C654A094113369154; _ga=GA1.2.772144563.1579011917; _gid=GA1.2.317154519.1579011917; trackeame_cookie=%7B%22id%22%3A%22UPA_e1861e3a-3357-4a76-861e-3a3357ea76c0%22%2C%22version%22%3A%225.0%22%2C%22upa_id%22%3A%22e1861e3a-3357-4a76-861e-3a3357ea76c0%22%2C%22creation_date%22%3A%222020-01-14T14%3A25%3A17Z%22%7D; __ssid=41de76d348be0e334af8e657f6801b8; _gcl_au=1.1.1367791908.1579011932; _fbp=fb.1.1579011933564.1470255143; __gads=ID=9139db3a836078f5:T=1579011933:S=ALNI_MawboBo55i9nPvoDvzaF396HudEKg; abzTestingId=\"{\\\"flightsFisherAB\\\":90,\\\"pkgImbatibleBrand_ctrl\\\":76,\\\"s_flights_s_violet_sbox_v1\\\":21,\\\"upsellingConfig\\\":58,\\\"twoOneWayForceMX\\\":0,\\\"filterLandingFlights\\\":41,\\\"s_loyalty_v2_ctrl\\\":5,\\\"s_flights_l_violet_sbox_v1\\\":0,\\\"s_flights_l_loyalty_v2\\\":58,\\\"mostProfitablePromotion\\\":0,\\\"despechecks\\\":72,\\\"s_loyalty_v2_review\\\":33,\\\"platform\\\":55,\\\"selected_radio_button\\\":0,\\\"fisher_2ow\\\":0,\\\"loyalty_non_adherents\\\":63,\\\"paymentMethod\\\":55,\\\"shifuMobileProductLabels\\\":0,\\\"obFee\\\":40,\\\"twoOneWay\\\":0,\\\"s_violet_sbox_v1\\\":17,\\\"s_flights_s_loyalty_v2\\\":14,\\\"flights_loyalty_non_adherents\\\":63,\\\"pkgImbatibleBrand-ctrl\\\":60,\\\"crossBorderTicketing\\\":0}\"; chktkn=ask3r5kj6ed0ksqrs7eio4cebk; searchId=243920d8-49cc-4271-972a-60d05221ef20; _gat_UA-36944350-2=1; xdsid=DCF9EDC0035E07BEDBFEE30E55F725C5; xduid=55D857BEFC5E27A8B84A7407D4A86B38; xdesp-rand-usr=292; abzTestingId=\"{\\\"flightsFisherAB\\\":90,\\\"pkgImbatibleBrand_ctrl\\\":76,\\\"s_flights_s_violet_sbox_v1\\\":21,\\\"upsellingConfig\\\":58,\\\"twoOneWayForceMX\\\":0,\\\"filterLandingFlights\\\":41,\\\"s_loyalty_v2_ctrl\\\":5,\\\"s_flights_l_violet_sbox_v1\\\":0,\\\"s_flights_l_loyalty_v2\\\":58,\\\"mostProfitablePromotion\\\":0,\\\"despechecks\\\":72,\\\"s_loyalty_v2_review\\\":33,\\\"platform\\\":55,\\\"selected_radio_button\\\":0,\\\"fisher_2ow\\\":0,\\\"loyalty_non_adherents\\\":63,\\\"paymentMethod\\\":55,\\\"shifuMobileProductLabels\\\":0,\\\"obFee\\\":40,\\\"twoOneWay\\\":0,\\\"s_violet_sbox_v1\\\":17,\\\"s_flights_s_loyalty_v2\\\":14,\\\"flights_loyalty_non_adherents\\\":63,\\\"pkgImbatibleBrand-ctrl\\\":60,\\\"crossBorderTicketing\\\":0}',\n 'Cache-Control': \"no-cache\",\n 'Postman-Token': \"4c6c6b9f-ed0a-477f-a787-c8cde039475b,4e35a9da-93ed-4602-825a-283f619d543b\",\n 'Host': \"www.decolar.com\",\n 'cache-control': \"no-cache\"\n }\n\n response = requests.request(\"GET\", url, headers=headers, params=querystring)\n dataOneWay = json.loads(response.text)\n print(origem, '->' , destino)\n print(querystring)\n print(dataOneWay)\n if 'clusters' in dataOneWay:\n for i in dataOneWay['clusters']: \n print(i['priceDetail']['mainFare']['amount'])\n\n\n",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbeb539235420579cea8431cabe89fa7d46aa436
| 408,645 |
ipynb
|
Jupyter Notebook
|
Experiment.ipynb
|
cmougan/FairEncoder
|
9814faa78583dbdbd287ee911abff22354184df8
|
[
"MIT"
] | null | null | null |
Experiment.ipynb
|
cmougan/FairEncoder
|
9814faa78583dbdbd287ee911abff22354184df8
|
[
"MIT"
] | 1 |
2021-12-23T13:10:53.000Z
|
2021-12-23T13:10:53.000Z
|
Experiment.ipynb
|
cc-jalvarez/FairEncoder
|
c1b3a1b72f0d0fb3ddae9e7d05f567e4a394224f
|
[
"MIT"
] | 2 |
2021-12-07T15:09:20.000Z
|
2021-12-16T16:06:35.000Z
| 443.215835 | 33,706 | 0.657297 |
[
[
[
"import pandas as pd\npd.set_option('display.max_columns', None)\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nplt.style.use('seaborn')\n\n#from pandas_profiling import ProfileReportofileReport\n\nimport warnings\nwarnings.filterwarnings('ignore')",
"_____no_output_____"
],
[
"from fairlearn.metrics import MetricFrame\nfrom fairlearn.metrics import selection_rate, false_positive_rate,true_positive_rate,count\n\n\nfrom sklearn.metrics import accuracy_score\nfrom sklearn.pipeline import Pipeline\nfrom sklearn.linear_model import LogisticRegression,Lasso\nfrom sklearn.preprocessing import LabelEncoder\nfrom sklearn.metrics import accuracy_score,precision_score,recall_score,roc_auc_score\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.svm import SVC\nfrom sklearn.ensemble import RandomForestClassifier,GradientBoostingClassifier\n\nfrom xgboost import XGBRegressor,XGBClassifier\nfrom catboost import CatBoostClassifier\nimport shap\n\nfrom category_encoders.target_encoder import TargetEncoder\nfrom category_encoders.m_estimate import MEstimateEncoder\nfrom category_encoders.cat_boost import CatBoostEncoder\nfrom category_encoders.leave_one_out import LeaveOneOutEncoder\n\nfrom tqdm.notebook import tqdm\nfrom collections import defaultdict ",
"_____no_output_____"
],
[
"#pd.read_csv('propublica_data_for_fairml.csv').head()",
"_____no_output_____"
],
[
"df = pd.read_csv(\"data/compas-scores-raw.csv\")\n\ndf[\"Score\"] = df[\"DecileScore\"]\n\n# df.loc[df[\"DecileScore\"] > 7, \"Score\"] = 2\n# df.loc[(df[\"DecileScore\"] > 4) & (df[\"DecileScore\"] < 8), \"Score\"] = 1\n# df.loc[df[\"DecileScore\"] < 5, \"Score\"] = 0\n\ndf.loc[df[\"DecileScore\"] > 4, \"Score\"] = 1\ndf.loc[df[\"DecileScore\"] <= 4, \"Score\"] = 0\n\n\ncols = [\n \"Person_ID\",\n \"AssessmentID\",\n \"Case_ID\",\n \"LastName\",\n \"FirstName\",\n \"MiddleName\",\n \"DateOfBirth\",\n \"ScaleSet_ID\",\n \"Screening_Date\",\n \"RecSupervisionLevel\",\n \"Agency_Text\",\n \"AssessmentReason\",\n \"Language\",\n \"Scale_ID\",\n \"IsCompleted\",\n \"IsDeleted\",\n \"AssessmentType\",\n \"DecileScore\",\n]\n\n\ndf = df.drop(columns=cols)\n\npossible_targets = [\"RawScore\", \"ScoreText\", \"Score\"]\n\nX = df.drop(columns=possible_targets)\ny = df[[\"Score\"]]",
"_____no_output_____"
],
[
"X_tr, X_te, y_tr, y_te = train_test_split(X, y, test_size=0.33, random_state=42)",
"_____no_output_____"
],
[
"te = CatBoostEncoder(sigma=3)\nmodel = XGBClassifier()\npipe = Pipeline([('encoder', te), ('model', model)])\n\npipe.fit(X_tr,y_tr)\n\npreds = pipe.predict(X_te)\n\n",
"[22:05:58] WARNING: /Users/travis/build/dmlc/xgboost/src/learner.cc:1115: Starting in XGBoost 1.3.0, the default evaluation metric used with the objective 'binary:logistic' was changed from 'error' to 'logloss'. Explicitly set eval_metric if you'd like to restore the old behavior.\n"
],
[
"explainer = shap.TreeExplainer(pipe.named_steps[\"model\"])\nshap_values = explainer.shap_values(pipe[:-1].transform(X_tr))\n",
"_____no_output_____"
],
[
"shap_values",
"_____no_output_____"
],
[
"shap.initjs()\nshap.force_plot(explainer.expected_value, shap_values[0,:], X.iloc[0,:])",
"_____no_output_____"
],
[
"shap_values.squeeze()",
"_____no_output_____"
],
[
"shap.plots.bar(shap_values.values)",
"_____no_output_____"
],
[
"gm = MetricFrame(\n metrics=accuracy_score,\n y_true=y_te,\n y_pred=preds,\n sensitive_features=X_te[\"Sex_Code_Text\"],\n)\nprint(gm.overall)\nprint(gm.by_group)\n\ngm = MetricFrame(\n metrics=accuracy_score,\n y_true=y_te,\n y_pred=preds,\n sensitive_features=X_te[\"Ethnic_Code_Text\"],\n)\n\nprint(gm.by_group)\n\ngm = MetricFrame(\n metrics=accuracy_score,\n y_true=y_te,\n y_pred=preds,\n sensitive_features=X_te[\"RecSupervisionLevelText\"],\n)\n\nprint(gm.by_group)",
"0.6805119776881319\nSex_Code_Text\nFemale 0.735368\nMale 0.66531\nName: accuracy_score, dtype: object\nEthnic_Code_Text\nAfrican-Am 0.416667\nAfrican-American 0.572278\nArabic 0.833333\nAsian 0.846847\nCaucasian 0.748179\nHispanic 0.793374\nNative American 0.671053\nOriental 0.818182\nOther 0.841608\nName: accuracy_score, dtype: object\nRecSupervisionLevelText\nHigh 0.167894\nLow 0.892282\nMedium 0.413852\nMedium with Override Consideration 0.267319\nName: accuracy_score, dtype: object\n"
],
[
"def fit_predict(modelo, enc, data, target, test):\n pipe = Pipeline([(\"encoder\", enc), (\"model\", modelo)])\n pipe.fit(data, target)\n return pipe.predict(test)\n\n\ndef auc_group(model, data, y_true, dicc, group: str = \"\"):\n aux = data.copy()\n aux[\"target\"] = y_true\n cats = aux[group].unique().tolist()\n cats = cats + [\"all\"]\n\n if len(dicc) == 0:\n dicc = defaultdict(list, {k: [] for k in cats})\n\n for cat in cats:\n if cat != \"all\":\n aux2 = aux[aux[group] == cat]\n preds = model.predict_proba(aux2.drop(columns=\"target\"))[:, 1]\n truth = aux2[\"target\"]\n dicc[cat].append(roc_auc_score(truth, preds))\n else:\n dicc[cat].append(roc_auc_score(y_true, model.predict_proba(data)[:, 1]))\n\n return dicc",
"_____no_output_____"
],
[
"for metrics in [selection_rate, false_positive_rate, true_positive_rate]:\n gms = []\n gms_rec = []\n ms = []\n auc = {}\n\n #param = [0, 1, 2, 5, 10, 20, 50, 100, 200, 500, 1000, 2000, 5000, 10_000, 20_000]\n param = np.linspace(0,1,20)\n for m in tqdm(param):\n # encoder = MEstimateEncoder(m=m)\n # encoder = TargetEncoder(smoothing=m)\n encoder = LeaveOneOutEncoder(sigma=m)\n model = LogisticRegression()\n #model = GradientBoostingClassifier()\n\n pipe = Pipeline([(\"encoder\", encoder), (\"model\", model)])\n pipe.fit(X_tr, y_tr)\n preds = pipe.predict(X_te)\n\n gm = MetricFrame(\n metrics=metrics,\n y_true=y_te,\n y_pred=preds,\n sensitive_features=X_te[\"Ethnic_Code_Text\"],\n )\n auc = auc_group(\n model=pipe, data=X_te, y_true=y_te, dicc=auc, group=\"Ethnic_Code_Text\"\n )\n gm_rec = MetricFrame(\n metrics=metrics,\n y_true=y_te,\n y_pred=preds,\n sensitive_features=X_te[\"RecSupervisionLevelText\"],\n )\n\n gms.append(gm)\n gms_rec.append(gm_rec)\n ms.append(m)\n\n # Impact Score\n plt.figure()\n title = \"Impact of encoding regularization in category fairness \" + str(\n metrics.__name__\n )\n plt.title(title)\n plt.xlabel(\"M parameter\")\n plt.plot(ms, [gm.overall for gm in gms_rec], label=\"Overall\")\n plt.plot(ms, [gm.by_group[\"Low\"] for gm in gms_rec], label=\"Low\")\n plt.plot(ms, [gm.by_group[\"High\"] for gm in gms_rec], label=\"High\")\n plt.plot(ms, [gm.by_group[\"Medium\"] for gm in gms_rec], label=\"Medium\")\n plt.plot(\n ms,\n [gm.by_group[\"Medium with Override Consideration\"] for gm in gms_rec],\n label=\"Medium with Override Consideration\",\n )\n\n plt.legend(bbox_to_anchor=(1.1, 1))\n plt.show()\n\n # Ethnic\n plt.figure()\n title = \"Impact of encoding regularization in category fairness \" + str(\n metrics.__name__\n )\n plt.title(title)\n plt.xlabel(\"M parameter\")\n plt.plot(ms, [gm.overall for gm in gms], label=\"Overall\")\n plt.plot(ms, [gm.by_group[\"Caucasian\"] for gm in gms], label=\"Caucasian\")\n plt.plot(\n ms, [gm.by_group[\"African-American\"] for gm in gms], label=\"AfricanAmerican\"\n )\n plt.plot(ms, [gm.by_group[\"Arabic\"] for gm in gms], label=\"Arabic\")\n plt.plot(ms, [gm.by_group[\"Hispanic\"] for gm in gms], label=\"Hispanic\")\n # plt.plot(ms,[gm.by_group['Oriental'] for gm in gms],label='Oriental')\n plt.legend(bbox_to_anchor=(1.1, 1))\n plt.show()\n\n # AUC ROC\n plt.title(\"AUC ROC\")\n plt.xlabel(\"M parameter\")\n plt.plot(ms, auc[\"all\"], label=\"Overall\")\n plt.plot(ms, auc[\"Caucasian\"], label=\"Caucasian\")\n plt.plot(ms, auc[\"African-American\"], label=\"AfricanAmerican\")\n plt.plot(ms, auc[\"Arabic\"], label=\"Arabic\")\n plt.plot(ms, auc[\"Hispanic\"], label=\"Hispanic\")\n plt.legend(bbox_to_anchor=(1.1, 1))\n plt.show()",
"_____no_output_____"
],
[
"kk",
"_____no_output_____"
],
[
"X_tr.head()",
"_____no_output_____"
],
[
"d = {}\n\nfor i in range(0,2):\n d = auc_group(model=pipe, data=X_tr, y_true=y_tr, dicc=d,group='Sex_Code_Text')",
"_____no_output_____"
],
[
"d",
"_____no_output_____"
],
[
"aa",
"_____no_output_____"
]
],
[
[
"Métricas reales, no-normalizadas.\nUsar una métrica tipo AUC.\nAñadir alguna métrica más intuitiva.",
"_____no_output_____"
]
],
[
[
"$\\frac{group\\_mean * n\\_samples + global\\_mean * m}{n\\_samples + m}$",
"_____no_output_____"
]
],
[
[
"X,y\ngroupby(X['feat_cat'])[y].mean()",
"_____no_output_____"
],
[
"y.global = 0.5\n\ny.spana = 0.2\ny.francia = 0.8",
"_____no_output_____"
],
[
"n = 10000\nm-regularizador",
"_____no_output_____"
]
],
[
[
"# Other Data",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\nfrom sklearn.datasets import fetch_openml\ndata = fetch_openml(data_id=1590, as_frame=True)\nX = pd.get_dummies(data.data)\ny_true = (data.target == '>50K') * 1\nsex = data.data['sex']\nsex.value_counts()",
"_____no_output_____"
],
[
"from fairlearn.metrics import MetricFrame\nfrom sklearn.metrics import accuracy_score\nfrom sklearn.tree import DecisionTreeClassifier",
"_____no_output_____"
],
[
"classifier = DecisionTreeClassifier(min_samples_leaf=10, max_depth=4)\nclassifier.fit(X, y_true)\nDecisionTreeClassifier()\ny_pred = classifier.predict(X)\ngm = MetricFrame(metrics=accuracy_score, y_true=y_true, y_pred=y_pred, sensitive_features=sex)",
"_____no_output_____"
],
[
"data.data['sex']",
"_____no_output_____"
]
],
[
[
"gms = []\ngms_rec = []\nms = []\n\nparam = [0, 1, 2, 5, 10, 20, 50, 100, 200, 500, 1000, 2000, 5000, 10_000, 20_000]\n# np.linspace(0,X_tr.shape[0],20)\nfor m in tqdm(param):\n encoder = MEstimateEncoder(m=m)\n model = LogisticRegression()\n # model = GradientBoostingClassifier()\n\n preds = fit_predict(modelo=model, enc=encoder, data=X_tr, target=y_tr, test=X_te)\n gm = MetricFrame(\n metrics=selection_rate,\n y_true=y_te,\n y_pred=preds,\n sensitive_features=X_te[\"Ethnic_Code_Text\"],\n )\n gm_rec = MetricFrame(\n metrics=selection_rate,\n y_true=y_te,\n y_pred=preds,\n sensitive_features=X_te[\"RecSupervisionLevelText\"],\n )\n\n gms.append(gm)\n gms_rec.append(gm_rec)\n ms.append(m)\n\nplt.figure()\nplt.title('Impact of encoding regularization in category fairness ')\nplt.xlabel('M parameter')\nplt.plot(ms,[gm.overall for gm in gms_rec],label='Overall')\nplt.plot(ms,[gm.by_group['Low'] for gm in gms_rec],label='Low')\nplt.plot(ms,[gm.by_group['High'] for gm in gms_rec],label='High')\nplt.plot(ms,[gm.by_group['Medium'] for gm in gms_rec],label='Medium')\nplt.plot(ms,[gm.by_group['Medium with Override Consideration'] for gm in gms_rec],label='Medium with Override Consideration')\n\nplt.legend(bbox_to_anchor=(1.1, 1))\nplt.show()",
"_____no_output_____"
]
]
] |
[
"code",
"raw",
"markdown",
"code",
"markdown",
"code",
"raw"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"raw"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"raw"
]
] |
cbeb722d4a38a75a9218b4a196bac20135323fcd
| 61,914 |
ipynb
|
Jupyter Notebook
|
cold_posterior_bnn/plot_results.ipynb
|
ojInc/google-research
|
9929c88b664800a25b8716c22068dd77d80bd5ee
|
[
"Apache-2.0"
] | 23,901 |
2018-10-04T19:48:53.000Z
|
2022-03-31T21:27:42.000Z
|
cold_posterior_bnn/plot_results.ipynb
|
ojInc/google-research
|
9929c88b664800a25b8716c22068dd77d80bd5ee
|
[
"Apache-2.0"
] | 891 |
2018-11-10T06:16:13.000Z
|
2022-03-31T10:42:34.000Z
|
cold_posterior_bnn/plot_results.ipynb
|
ojInc/google-research
|
9929c88b664800a25b8716c22068dd77d80bd5ee
|
[
"Apache-2.0"
] | 6,047 |
2018-10-12T06:31:02.000Z
|
2022-03-31T13:59:28.000Z
| 315.887755 | 28,626 | 0.907533 |
[
[
[
"Copyright 2020 The Google Research Authors.\n\nLicensed under the Apache License, Version 2.0 (the \"License\"); You may not use this file except in compliance with the License. You may obtain a copy of the License at\n\nhttp://www.apache.org/licenses/LICENSE-2.0\n\nUnless required by applicable law or agreed to in writing, software\ndistributed under the License is distributed on an \"AS IS\" BASIS,\nWITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\nSee the License for the specific language governing permissions and\nlimitations under the License.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nimport pandas as pd\nfrom pathlib import Path",
"_____no_output_____"
],
[
"# dictionary of metrics to plot (each metric is shown in an individual plot)\n# dictionary key is name of metric in logs.csv, dict value is label in the final plot\nplot_metrics = {'ens_acc': 'Test accuracy', # Ensemble accuracy\n 'ens_ce': 'Test cross entropy'} # Ensemble cross entropy\n\n# directory of results\n# should include 'run_sweeps.csv', generated by run_resnet_experiments.sh/run_resnet_experiments.sh\nresults_dir = '/tmp/google_research/cold_posterior_bnn/results_resnet/'",
"_____no_output_____"
],
[
"# load csv with results of all runs\nsweeps_df = pd.read_csv(results_dir+'run_sweeps.csv').set_index('id')\n\n# add final performance of run as columns to sweep_df\nfor metric in plot_metrics.keys():\n sweeps_df[metric] = [0.] * len(sweeps_df)\n\nfor i in range(len(sweeps_df)):\n # get logs of run\n log_dir = sweeps_df.loc[i, 'dir']\n logs_df = pd.read_csv('{}{}/logs.csv'.format(results_dir, log_dir))\n for metric in plot_metrics:\n # get final performace of run and add to df\n idx = 0\n final_metric = float('nan')\n while np.isnan(final_metric):\n idx += 1\n final_metric = logs_df.tail(idx)[metric].values[0] # indexing starts with 1\n sweeps_df.at[i, metric] = final_metric\n\n# save/update csv file\nsweeps_df.to_csv(results_dir+'run_sweeps.csv')",
"_____no_output_____"
],
[
"# plot\nfont_scale = 1.1\nline_width = 3\nmarker_size = 7\ncm_lines = sns.color_palette('deep')\ncm_points = sns.color_palette('bright')\n\n# style settings\nsns.reset_defaults()\nsns.set_context(\"notebook\", font_scale=font_scale,\n rc={\"lines.linewidth\": line_width,\n \"lines.markersize\" :marker_size}\n )\nsns.set_style(\"whitegrid\")\n\nfor metric, metric_label in plot_metrics.items():\n # plot SG-MCMC\n fig, ax = plt.subplots(figsize=(7.0, 2.85))\n g = sns.lineplot(x='temperature', y=metric, data=sweeps_df, marker='o', label='SG-MCMC', color=cm_lines[0], zorder=2, ci='sd')\n\n # finalize plot\n plt.legend(loc=3, fontsize=14)\n g.set_xscale('log')\n #g.set_ylim(bottom=0.88, top=0.94)\n g.set_xlim(left=1e-4, right=1)\n fig.tight_layout()\n ax.set_frame_on(False)\n ax.set_xlabel('Temperature $T$')\n ax.set_ylabel(metric_label)\n\n ax.margins(0,0)\n plt.savefig('{}resnet_{}.pdf'.format(results_dir, metric_label), format=\"pdf\", dpi=300, bbox_inches=\"tight\", pad_inches=0)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
cbeb7de7fe0cc1cae1c89edcfe8a5d8204a7a5d6
| 84,064 |
ipynb
|
Jupyter Notebook
|
src/tmp.ipynb
|
yifan-fanyi/DCST
|
d068ebd9281a21fcb9ed263500994723fb289fcf
|
[
"MIT"
] | null | null | null |
src/tmp.ipynb
|
yifan-fanyi/DCST
|
d068ebd9281a21fcb9ed263500994723fb289fcf
|
[
"MIT"
] | null | null | null |
src/tmp.ipynb
|
yifan-fanyi/DCST
|
d068ebd9281a21fcb9ed263500994723fb289fcf
|
[
"MIT"
] | null | null | null | 53.441831 | 167 | 0.570494 |
[
[
[
"\nfrom skimage.measure import block_reduce\n\nfrom framework.dependency import *\nfrom framework.utli import *",
"_____no_output_____"
],
[
"def pipeline(X, Qstep=16, Q_mode=1, ML_inv=True, write=False, name='tmp.txt', tPCA=None, isDCT=True):\n H, W = X.shape[0], X.shape[1]\n X = X.reshape(1, H, W, -1)\n X_p, X_q, X_r = X[:,:,:,0:1], block_reduce(X[:,:,:,1:2], (1, 2, 2, 1), np.mean), block_reduce(X[:,:,:,2:], (1, 2, 2, 1), np.mean)\n # P\n def proP(X_p, Qstep, Q_mode, isDCT):\n X_block = Shrink(X_p, win=8)\n if isDCT == False:\n trans_pca = myPCA(is2D=True, H=8, W=8)\n trans_pca.fit(X_block)\n else:\n trans_pca = DCT(8,8)\n tX = trans_pca.transform(X_block)\n tX = Q(trans_pca, tX, Qstep, mode=Q_mode)\n return trans_pca, tX\n # Quant\n def inv_proP(trans_pca, tX, Qstep, Xraw, ML_inv, Q_mode):\n Xraw = Shrink(Xraw, win=8)\n tX = dQ(trans_pca, tX, Qstep, mode=Q_mode)\n if ML_inv == True:\n iX = trans_pca.ML_inverse_transform(Xraw, tX)\n else:\n iX = trans_pca.inverse_transform(tX)\n iX_p = invShrink(iX, win=8)\n return iX_p\n\n # QR\n def proQR(trans_pca, X_q, Qstep, Q_mode):\n X_block = Shrink(X_q, win=8)\n tX = trans_pca.transform(X_block)\n tX = Q(trans_pca, tX, Qstep, mode=Q_mode)\n return tX\n \n if tPCA == None:\n tPCA, tX_p = proP(X_p, Qstep, Q_mode, isDCT)\n else:\n tX_p = proQR(tPCA, X_p, Qstep, Q_mode)\n if write == True:\n write_to_txt(tX_p, name)\n iX_p = inv_proP(tPCA, tX_p, Qstep, X_p, ML_inv, Q_mode)\n\n tX_q = proQR(tPCA, X_q, Qstep, Q_mode)\n if write == True:\n write_to_txt(tX_q, name)\n iX_q = inv_proP(tPCA, tX_q, Qstep, X_q, ML_inv, Q_mode)\n iX_q = cv2.resize(iX_q[0,:,:,0], (W, H)).reshape(1, H, W, 1)\n\n tX_r = proQR(tPCA, X_r, Qstep, Q_mode)\n if write == True:\n write_to_txt(tX_r, name)\n with open(name, 'a') as f:\n f.write('-1111')\n iX_r = inv_proP(tPCA, tX_r, Qstep, X_r, ML_inv, Q_mode)\n iX_r = cv2.resize(iX_r[0,:,:,0], (W, H)).reshape(1, H, W, 1)\n return np.concatenate((iX_p, iX_q, iX_r), axis=-1)\n \n\ndef run(tPCA=None, ML_inv=True, ML_color_inv=True, img=0, write=False, isYUV=True, name=None, Q_mode=0, isDCT=False):\n psnr = []\n if isDCT == True:\n q = np.arange(5, 99, 5)\n else:\n q = [200, 160, 140, 120, 100, 90, 80, 70, 60, 50, 40.0,32.0,26.6,22.8,20.0,17.7,16.0,14.4,12.8,11.2,9.6,8.0,6.4,4.8,3.2]\n q = np.arange(5, 99, 5)\n for i in range(len(q)): \n X_bgr = cv2.imread('/Users/alex/Desktop/proj/compression/data/Kodak/Kodak/'+str(img)+'.bmp')\n if isYUV == False:\n color_pca, X = BGR2PQR(X_bgr)\n else:\n X = BGR2YUV(X_bgr)\n iX = pipeline(X, Qstep=q[i], Q_mode=Q_mode, ML_inv=ML_inv, write=write, name='../result/'+name+'/'+str(img)+'_'+str(i)+'.txt', tPCA=tPCA, isDCT=isDCT)\n if ML_color_inv == True:\n iX = ML_inv_color(X_bgr, iX)\n #cv2.imwrite(str(img)+'_'+str(i)+'.png', copy.deepcopy(iX))\n psnr.append(PSNR(iX, X_bgr))\n else:\n if isYUV == True:\n iX = YUV2BGR(iX)\n else:\n iX = PQR2BGR(iX, color_pca)\n psnr.append(PSNR(iX[0], X_bgr))\n #break\n return psnr\n \n \npsnr = []\nname = 'tmp'\nfor i in range(24):\n psnr.append(run(None, 1, 1, img=i, write=1, isYUV=False, name=name, Q_mode=3, isDCT=False))\n #break\npsnr = np.array(psnr)\nwith open('../result/psnr_'+name+'.pkl', 'wb') as f:\n pickle.dump(psnr,f)",
"write 393216 to ../result/tmp/0_0.txt 3 -9\nwrite 98304 to ../result/tmp/0_0.txt 1 -3\nwrite 98304 to ../result/tmp/0_0.txt 0 0\nwrite 393216 to ../result/tmp/0_1.txt 5 -18\nwrite 98304 to ../result/tmp/0_1.txt 2 -7\nwrite 98304 to ../result/tmp/0_1.txt 1 -1\nwrite 393216 to ../result/tmp/0_2.txt 8 -26\nwrite 98304 to ../result/tmp/0_2.txt 3 -10\nwrite 98304 to ../result/tmp/0_2.txt 1 -1\nwrite 393216 to ../result/tmp/0_3.txt 10 -35\nwrite 98304 to ../result/tmp/0_3.txt 4 -13\nwrite 98304 to ../result/tmp/0_3.txt 2 -2\nwrite 393216 to ../result/tmp/0_4.txt 13 -44\nwrite 98304 to ../result/tmp/0_4.txt 4 -17\nwrite 98304 to ../result/tmp/0_4.txt 2 -2\nwrite 393216 to ../result/tmp/0_5.txt 15 -53\nwrite 98304 to ../result/tmp/0_5.txt 5 -20\nwrite 98304 to ../result/tmp/0_5.txt 3 -2\nwrite 393216 to ../result/tmp/0_6.txt 18 -62\nwrite 98304 to ../result/tmp/0_6.txt 6 -23\nwrite 98304 to ../result/tmp/0_6.txt 3 -3\nwrite 393216 to ../result/tmp/0_7.txt 20 -70\nwrite 98304 to ../result/tmp/0_7.txt 7 -27\nwrite 98304 to ../result/tmp/0_7.txt 4 -3\nwrite 393216 to ../result/tmp/0_8.txt 23 -79\nwrite 98304 to ../result/tmp/0_8.txt 8 -30\nwrite 98304 to ../result/tmp/0_8.txt 4 -3\nwrite 393216 to ../result/tmp/0_9.txt 26 -88\nwrite 98304 to ../result/tmp/0_9.txt 9 -33\nwrite 98304 to ../result/tmp/0_9.txt 4 -4\nwrite 393216 to ../result/tmp/0_10.txt 28 -98\nwrite 98304 to ../result/tmp/0_10.txt 10 -37\nwrite 98304 to ../result/tmp/0_10.txt 5 -4\nwrite 393216 to ../result/tmp/0_11.txt 32 -110\nwrite 98304 to ../result/tmp/0_11.txt 11 -42\nwrite 98304 to ../result/tmp/0_11.txt 6 -5\nwrite 393216 to ../result/tmp/0_12.txt 37 -126\nwrite 98304 to ../result/tmp/0_12.txt 13 -48\nwrite 98304 to ../result/tmp/0_12.txt 6 -6\nwrite 393216 to ../result/tmp/0_13.txt 43 -147\nwrite 98304 to ../result/tmp/0_13.txt 15 -56\nwrite 98304 to ../result/tmp/0_13.txt 7 -6\nwrite 393216 to ../result/tmp/0_14.txt 51 -176\nwrite 98304 to ../result/tmp/0_14.txt 18 -67\nwrite 98304 to ../result/tmp/0_14.txt 9 -8\nwrite 393216 to ../result/tmp/0_15.txt 64 -220\nwrite 98304 to ../result/tmp/0_15.txt 22 -84\nwrite 98304 to ../result/tmp/0_15.txt 11 -10\nwrite 393216 to ../result/tmp/0_16.txt 85 -293\nwrite 98304 to ../result/tmp/0_16.txt 30 -111\nwrite 98304 to ../result/tmp/0_16.txt 15 -13\nwrite 393216 to ../result/tmp/0_17.txt 128 -440\nwrite 98304 to ../result/tmp/0_17.txt 44 -167\nwrite 98304 to ../result/tmp/0_17.txt 22 -19\nwrite 393216 to ../result/tmp/0_18.txt 256 -880\nwrite 98304 to ../result/tmp/0_18.txt 89 -334\nwrite 98304 to ../result/tmp/0_18.txt 44 -39\nwrite 393216 to ../result/tmp/1_0.txt 3 -10\nwrite 98304 to ../result/tmp/1_0.txt 1 -3\nwrite 98304 to ../result/tmp/1_0.txt 1 -1\nwrite 393216 to ../result/tmp/1_1.txt 7 -20\nwrite 98304 to ../result/tmp/1_1.txt 2 -6\nwrite 98304 to ../result/tmp/1_1.txt 2 -3\nwrite 393216 to ../result/tmp/1_2.txt 10 -30\nwrite 98304 to ../result/tmp/1_2.txt 4 -9\nwrite 98304 to ../result/tmp/1_2.txt 4 -4\nwrite 393216 to ../result/tmp/1_3.txt 14 -41\nwrite 98304 to ../result/tmp/1_3.txt 5 -12\nwrite 98304 to ../result/tmp/1_3.txt 5 -6\nwrite 393216 to ../result/tmp/1_4.txt 17 -51\nwrite 98304 to ../result/tmp/1_4.txt 6 -16\nwrite 98304 to ../result/tmp/1_4.txt 6 -7\nwrite 393216 to ../result/tmp/1_5.txt 21 -61\nwrite 98304 to ../result/tmp/1_5.txt 7 -19\nwrite 98304 to ../result/tmp/1_5.txt 7 -8\nwrite 393216 to ../result/tmp/1_6.txt 24 -71\nwrite 98304 to ../result/tmp/1_6.txt 9 -22\nwrite 98304 to ../result/tmp/1_6.txt 8 -10\nwrite 393216 to ../result/tmp/1_7.txt 28 -81\nwrite 98304 to ../result/tmp/1_7.txt 10 -25\nwrite 98304 to ../result/tmp/1_7.txt 10 -11\nwrite 393216 to ../result/tmp/1_8.txt 31 -91\nwrite 98304 to ../result/tmp/1_8.txt 11 -28\nwrite 98304 to ../result/tmp/1_8.txt 11 -13\nwrite 393216 to ../result/tmp/1_9.txt 35 -101\nwrite 98304 to ../result/tmp/1_9.txt 12 -31\nwrite 98304 to ../result/tmp/1_9.txt 12 -14\nwrite 393216 to ../result/tmp/1_10.txt 39 -113\nwrite 98304 to ../result/tmp/1_10.txt 14 -35\nwrite 98304 to ../result/tmp/1_10.txt 13 -15\nwrite 393216 to ../result/tmp/1_11.txt 44 -127\nwrite 98304 to ../result/tmp/1_11.txt 15 -39\nwrite 98304 to ../result/tmp/1_11.txt 15 -17\nwrite 393216 to ../result/tmp/1_12.txt 50 -145\nwrite 98304 to ../result/tmp/1_12.txt 18 -45\nwrite 98304 to ../result/tmp/1_12.txt 17 -20\nwrite 393216 to ../result/tmp/1_13.txt 58 -169\nwrite 98304 to ../result/tmp/1_13.txt 21 -52\nwrite 98304 to ../result/tmp/1_13.txt 20 -23\nwrite 393216 to ../result/tmp/1_14.txt 70 -203\nwrite 98304 to ../result/tmp/1_14.txt 25 -62\nwrite 98304 to ../result/tmp/1_14.txt 24 -28\nwrite 393216 to ../result/tmp/1_15.txt 87 -254\nwrite 98304 to ../result/tmp/1_15.txt 31 -78\nwrite 98304 to ../result/tmp/1_15.txt 30 -35\nwrite 393216 to ../result/tmp/1_16.txt 116 -338\nwrite 98304 to ../result/tmp/1_16.txt 41 -104\nwrite 98304 to ../result/tmp/1_16.txt 40 -46\nwrite 393216 to ../result/tmp/1_17.txt 175 -507\nwrite 98304 to ../result/tmp/1_17.txt 62 -156\nwrite 98304 to ../result/tmp/1_17.txt 60 -70\nwrite 393216 to ../result/tmp/1_18.txt 349 -1014\nwrite 98304 to ../result/tmp/1_18.txt 124 -312\nwrite 98304 to ../result/tmp/1_18.txt 119 -139\nwrite 393216 to ../result/tmp/2_0.txt 10 -3\nwrite 98304 to ../result/tmp/2_0.txt 5 -1\nwrite 98304 to ../result/tmp/2_0.txt 0 -1\nwrite 393216 to ../result/tmp/2_1.txt 20 -6\nwrite 98304 to ../result/tmp/2_1.txt 9 -2\nwrite 98304 to ../result/tmp/2_1.txt 1 -2\nwrite 393216 to ../result/tmp/2_2.txt 30 -9\nwrite 98304 to ../result/tmp/2_2.txt 14 -4\nwrite 98304 to ../result/tmp/2_2.txt 1 -3\nwrite 393216 to ../result/tmp/2_3.txt 40 -13\nwrite 98304 to ../result/tmp/2_3.txt 19 -5\nwrite 98304 to ../result/tmp/2_3.txt 1 -4\nwrite 393216 to ../result/tmp/2_4.txt 50 -16\nwrite 98304 to ../result/tmp/2_4.txt 24 -6\nwrite 98304 to ../result/tmp/2_4.txt 2 -5\nwrite 393216 to ../result/tmp/2_5.txt 60 -19\nwrite 98304 to ../result/tmp/2_5.txt 28 -7\nwrite 98304 to ../result/tmp/2_5.txt 2 -6\nwrite 393216 to ../result/tmp/2_6.txt 70 -22\nwrite 98304 to ../result/tmp/2_6.txt 33 -9\nwrite 98304 to ../result/tmp/2_6.txt 2 -7\nwrite 393216 to ../result/tmp/2_7.txt 80 -25\nwrite 98304 to ../result/tmp/2_7.txt 38 -10\nwrite 98304 to ../result/tmp/2_7.txt 2 -8\nwrite 393216 to ../result/tmp/2_8.txt 90 -28\nwrite 98304 to ../result/tmp/2_8.txt 42 -11\nwrite 98304 to ../result/tmp/2_8.txt 3 -9\nwrite 393216 to ../result/tmp/2_9.txt 100 -32\nwrite 98304 to ../result/tmp/2_9.txt 47 -12\nwrite 98304 to ../result/tmp/2_9.txt 3 -10\nwrite 393216 to ../result/tmp/2_10.txt 111 -35\nwrite 98304 to ../result/tmp/2_10.txt 52 -14\nwrite 98304 to ../result/tmp/2_10.txt 3 -11\nwrite 393216 to ../result/tmp/2_11.txt 124 -39\nwrite 98304 to ../result/tmp/2_11.txt 59 -15\nwrite 98304 to ../result/tmp/2_11.txt 4 -13\nwrite 393216 to ../result/tmp/2_12.txt 142 -45\nwrite 98304 to ../result/tmp/2_12.txt 67 -17\nwrite 98304 to ../result/tmp/2_12.txt 4 -15\nwrite 393216 to ../result/tmp/2_13.txt 166 -53\nwrite 98304 to ../result/tmp/2_13.txt 79 -20\nwrite 98304 to ../result/tmp/2_13.txt 5 -17\nwrite 393216 to ../result/tmp/2_14.txt 199 -63\nwrite 98304 to ../result/tmp/2_14.txt 94 -24\nwrite 98304 to ../result/tmp/2_14.txt 6 -20\nwrite 393216 to ../result/tmp/2_15.txt 249 -79\nwrite 98304 to ../result/tmp/2_15.txt 118 -30\nwrite 98304 to ../result/tmp/2_15.txt 8 -26\nwrite 393216 to ../result/tmp/2_16.txt 332 -105\nwrite 98304 to ../result/tmp/2_16.txt 157 -41\nwrite 98304 to ../result/tmp/2_16.txt 10 -34\nwrite 393216 to ../result/tmp/2_17.txt 498 -158\nwrite 98304 to ../result/tmp/2_17.txt 236 -61\nwrite 98304 to ../result/tmp/2_17.txt 15 -51\nwrite 393216 to ../result/tmp/2_18.txt 996 -315\nwrite 98304 to ../result/tmp/2_18.txt 472 -122\nwrite 98304 to ../result/tmp/2_18.txt 31 -102\nwrite 393216 to ../result/tmp/3_0.txt 11 -4\nwrite 98304 to ../result/tmp/3_0.txt 3 -1\nwrite 98304 to ../result/tmp/3_0.txt 2 0\nwrite 393216 to ../result/tmp/3_1.txt 22 -7\nwrite 98304 to ../result/tmp/3_1.txt 6 -2\nwrite 98304 to ../result/tmp/3_1.txt 3 -1\nwrite 393216 to ../result/tmp/3_2.txt 33 -11\nwrite 98304 to ../result/tmp/3_2.txt 8 -4\nwrite 98304 to ../result/tmp/3_2.txt 5 -1\nwrite 393216 to ../result/tmp/3_3.txt 44 -15\n"
]
]
] |
[
"code"
] |
[
[
"code",
"code"
]
] |
cbeb878e8154826717368d49bc628db99feba309
| 456,994 |
ipynb
|
Jupyter Notebook
|
KD1/.ipynb_checkpoints/KD1-pp-checkpoint.ipynb
|
Hansxsourse/Cross-Modality-Knowledge-Distillation-for-Multi-modal-Aerial-View-Object-Classification
|
78caf3aec50291f839e9f5fc123c972e4c30a65c
|
[
"MIT"
] | 2 |
2021-07-27T03:08:13.000Z
|
2022-02-06T05:14:09.000Z
|
KD1/KD1-pp.ipynb
|
Hansxsourse/Cross-Modality-Knowledge-Distillation-for-Multi-modal-Aerial-View-Object-Classification
|
78caf3aec50291f839e9f5fc123c972e4c30a65c
|
[
"MIT"
] | null | null | null |
KD1/KD1-pp.ipynb
|
Hansxsourse/Cross-Modality-Knowledge-Distillation-for-Multi-modal-Aerial-View-Object-Classification
|
78caf3aec50291f839e9f5fc123c972e4c30a65c
|
[
"MIT"
] | 2 |
2022-01-07T20:40:11.000Z
|
2022-01-11T12:01:05.000Z
| 41.609214 | 26,968 | 0.600971 |
[
[
[
"# from numba import jit\n# from tqdm import trange\n# import pandas as pd\n# eo_df = pd.read_csv(\"/mnt/sda1/cvpr21/Classification/Aerial-View-Object-Classification/data/train_EO.csv\")\n# eo_df = eo_df.sort_values(by='img_name')\n\n# sar_df = pd.read_csv(\"/mnt/sda1/cvpr21/Classification/Aerial-View-Object-Classification/data/train_SAR.csv\") \n# sar_df = sar_df.sort_values(by='img_name')\n\n# @jit()\n# def equal():\n# notsame_image = 0\n# notsame_label = 0\n# t = trange(len(sar_df))\n \n\n \n# for i in t:\n# t.set_postfix({'nums of not same label:': notsame_label})\n# eo_label = next(eo_df.iterrows())[1].class_id\n# sar_label = next(sar_df.iterrows())[1].class_id\n# # if not eo_image == sar_image:\n# # notsame_image += 1\n# if not eo_label == sar_label:\n# notsame_label += 1\n# # notsame_label += 1\n# # print(\"nums of not same imageid:\", notsame_image)\n# #print(\"nums of not same label:\", notsame_label)\n\n# equal()\n\n ",
"_____no_output_____"
],
[
"from __future__ import print_function, division\n\nimport torch\nimport math\nimport torch.nn as nn\nimport torch.optim as optim\nfrom torch.optim import lr_scheduler\nimport numpy as np\nimport torchvision\nfrom torchvision import datasets, models, transforms\nimport matplotlib.pyplot as plt\nimport time\nimport os\nimport copy\nfrom torch.autograd import Variable\nimport random\nimport torch.nn.functional as F\n\nexp_num = \"45_kd_sar-teacher_eo-student_pretrain-on-sar\"\n",
"_____no_output_____"
],
[
"def seed_everything(seed):\n random.seed(seed)\n os.environ['PYTHONHASHSEED'] = str(seed)\n np.random.seed(seed)\n torch.manual_seed(seed)\n torch.cuda.manual_seed(seed)\n torch.backends.cudnn.deterministic = True\n# tf.set_random_seed(seed)\n\nseed = 2019\nseed_everything(seed)",
"_____no_output_____"
],
[
"#https://github.com/4uiiurz1/pytorch-auto-augment\n\nimport random\nimport numpy as np\nimport scipy\nfrom scipy import ndimage\nfrom PIL import Image, ImageEnhance, ImageOps\n\n\nclass AutoAugment(object):\n def __init__(self):\n self.policies = [\n ['Invert', 0.1, 7, 'Contrast', 0.2, 6],\n ['Rotate', 0.7, 2, 'TranslateX', 0.3, 9],\n ['Sharpness', 0.8, 1, 'Sharpness', 0.9, 3],\n ['ShearY', 0.5, 8, 'TranslateY', 0.7, 9],\n ['AutoContrast', 0.5, 8, 'Equalize', 0.9, 2],\n ['ShearY', 0.2, 7, 'Posterize', 0.3, 7],\n ['Color', 0.4, 3, 'Brightness', 0.6, 7],\n ['Sharpness', 0.3, 9, 'Brightness', 0.7, 9],\n ['Equalize', 0.6, 5, 'Equalize', 0.5, 1],\n ['Contrast', 0.6, 7, 'Sharpness', 0.6, 5],\n ['Color', 0.7, 7, 'TranslateX', 0.5, 8],\n ['Equalize', 0.3, 7, 'AutoContrast', 0.4, 8],\n ['TranslateY', 0.4, 3, 'Sharpness', 0.2, 6],\n ['Brightness', 0.9, 6, 'Color', 0.2, 8],\n ['Solarize', 0.5, 2, 'Invert', 0.0, 3],\n ['Equalize', 0.2, 0, 'AutoContrast', 0.6, 0],\n ['Equalize', 0.2, 8, 'Equalize', 0.6, 4],\n ['Color', 0.9, 9, 'Equalize', 0.6, 6],\n ['AutoContrast', 0.8, 4, 'Solarize', 0.2, 8],\n ['Brightness', 0.1, 3, 'Color', 0.7, 0],\n ['Solarize', 0.4, 5, 'AutoContrast', 0.9, 3],\n ['TranslateY', 0.9, 9, 'TranslateY', 0.7, 9],\n ['AutoContrast', 0.9, 2, 'Solarize', 0.8, 3],\n ['Equalize', 0.8, 8, 'Invert', 0.1, 3],\n ['TranslateY', 0.7, 9, 'AutoContrast', 0.9, 1],\n ]\n\n def __call__(self, img):\n img = apply_policy(img, self.policies[random.randrange(len(self.policies))])\n return img\n\n\noperations = {\n 'ShearX': lambda img, magnitude: shear_x(img, magnitude),\n 'ShearY': lambda img, magnitude: shear_y(img, magnitude),\n 'TranslateX': lambda img, magnitude: translate_x(img, magnitude),\n 'TranslateY': lambda img, magnitude: translate_y(img, magnitude),\n 'Rotate': lambda img, magnitude: rotate(img, magnitude),\n 'AutoContrast': lambda img, magnitude: auto_contrast(img, magnitude),\n 'Invert': lambda img, magnitude: invert(img, magnitude),\n 'Equalize': lambda img, magnitude: equalize(img, magnitude),\n 'Solarize': lambda img, magnitude: solarize(img, magnitude),\n 'Posterize': lambda img, magnitude: posterize(img, magnitude),\n 'Contrast': lambda img, magnitude: contrast(img, magnitude),\n 'Color': lambda img, magnitude: color(img, magnitude),\n 'Brightness': lambda img, magnitude: brightness(img, magnitude),\n 'Sharpness': lambda img, magnitude: sharpness(img, magnitude),\n 'Cutout': lambda img, magnitude: cutout(img, magnitude),\n}\n\n\ndef apply_policy(img, policy):\n if random.random() < policy[1]:\n img = operations[policy[0]](img, policy[2])\n if random.random() < policy[4]:\n img = operations[policy[3]](img, policy[5])\n\n return img\n\n\ndef transform_matrix_offset_center(matrix, x, y):\n o_x = float(x) / 2 + 0.5\n o_y = float(y) / 2 + 0.5\n offset_matrix = np.array([[1, 0, o_x], [0, 1, o_y], [0, 0, 1]])\n reset_matrix = np.array([[1, 0, -o_x], [0, 1, -o_y], [0, 0, 1]])\n transform_matrix = offset_matrix @ matrix @ reset_matrix\n return transform_matrix\n\n\ndef shear_x(img, magnitude):\n img = np.array(img)\n magnitudes = np.linspace(-0.3, 0.3, 11)\n\n transform_matrix = np.array([[1, random.uniform(magnitudes[magnitude], magnitudes[magnitude+1]), 0],\n [0, 1, 0],\n [0, 0, 1]])\n transform_matrix = transform_matrix_offset_center(transform_matrix, img.shape[0], img.shape[1])\n affine_matrix = transform_matrix[:2, :2]\n offset = transform_matrix[:2, 2]\n img = np.stack([ndimage.interpolation.affine_transform(\n img[:, :, c],\n affine_matrix,\n offset) for c in range(img.shape[2])], axis=2)\n img = Image.fromarray(img)\n return img\n\n\ndef shear_y(img, magnitude):\n img = np.array(img)\n magnitudes = np.linspace(-0.3, 0.3, 11)\n\n transform_matrix = np.array([[1, 0, 0],\n [random.uniform(magnitudes[magnitude], magnitudes[magnitude+1]), 1, 0],\n [0, 0, 1]])\n transform_matrix = transform_matrix_offset_center(transform_matrix, img.shape[0], img.shape[1])\n affine_matrix = transform_matrix[:2, :2]\n offset = transform_matrix[:2, 2]\n img = np.stack([ndimage.interpolation.affine_transform(\n img[:, :, c],\n affine_matrix,\n offset) for c in range(img.shape[2])], axis=2)\n img = Image.fromarray(img)\n return img\n\n\ndef translate_x(img, magnitude):\n img = np.array(img)\n magnitudes = np.linspace(-150/331, 150/331, 11)\n\n transform_matrix = np.array([[1, 0, 0],\n [0, 1, img.shape[1]*random.uniform(magnitudes[magnitude], magnitudes[magnitude+1])],\n [0, 0, 1]])\n transform_matrix = transform_matrix_offset_center(transform_matrix, img.shape[0], img.shape[1])\n affine_matrix = transform_matrix[:2, :2]\n offset = transform_matrix[:2, 2]\n img = np.stack([ndimage.interpolation.affine_transform(\n img[:, :, c],\n affine_matrix,\n offset) for c in range(img.shape[2])], axis=2)\n img = Image.fromarray(img)\n return img\n\n\ndef translate_y(img, magnitude):\n img = np.array(img)\n magnitudes = np.linspace(-150/331, 150/331, 11)\n\n transform_matrix = np.array([[1, 0, img.shape[0]*random.uniform(magnitudes[magnitude], magnitudes[magnitude+1])],\n [0, 1, 0],\n [0, 0, 1]])\n transform_matrix = transform_matrix_offset_center(transform_matrix, img.shape[0], img.shape[1])\n affine_matrix = transform_matrix[:2, :2]\n offset = transform_matrix[:2, 2]\n img = np.stack([ndimage.interpolation.affine_transform(\n img[:, :, c],\n affine_matrix,\n offset) for c in range(img.shape[2])], axis=2)\n img = Image.fromarray(img)\n return img\n\n\ndef rotate(img, magnitude):\n img = np.array(img)\n magnitudes = np.linspace(-30, 30, 11)\n theta = np.deg2rad(random.uniform(magnitudes[magnitude], magnitudes[magnitude+1]))\n transform_matrix = np.array([[np.cos(theta), -np.sin(theta), 0],\n [np.sin(theta), np.cos(theta), 0],\n [0, 0, 1]])\n transform_matrix = transform_matrix_offset_center(transform_matrix, img.shape[0], img.shape[1])\n affine_matrix = transform_matrix[:2, :2]\n offset = transform_matrix[:2, 2]\n img = np.stack([ndimage.interpolation.affine_transform(\n img[:, :, c],\n affine_matrix,\n offset) for c in range(img.shape[2])], axis=2)\n img = Image.fromarray(img)\n return img\n\n\ndef auto_contrast(img, magnitude):\n img = ImageOps.autocontrast(img)\n return img\n\n\ndef invert(img, magnitude):\n img = ImageOps.invert(img)\n return img\n\n\ndef equalize(img, magnitude):\n img = ImageOps.equalize(img)\n return img\n\n\ndef solarize(img, magnitude):\n magnitudes = np.linspace(0, 256, 11)\n img = ImageOps.solarize(img, random.uniform(magnitudes[magnitude], magnitudes[magnitude+1]))\n return img\n\n\ndef posterize(img, magnitude):\n magnitudes = np.linspace(4, 8, 11)\n img = ImageOps.posterize(img, int(round(random.uniform(magnitudes[magnitude], magnitudes[magnitude+1]))))\n return img\n\n\ndef contrast(img, magnitude):\n magnitudes = np.linspace(0.1, 1.9, 11)\n img = ImageEnhance.Contrast(img).enhance(random.uniform(magnitudes[magnitude], magnitudes[magnitude+1]))\n return img\n\n\ndef color(img, magnitude):\n magnitudes = np.linspace(0.1, 1.9, 11)\n img = ImageEnhance.Color(img).enhance(random.uniform(magnitudes[magnitude], magnitudes[magnitude+1]))\n return img\n\n\ndef brightness(img, magnitude):\n magnitudes = np.linspace(0.1, 1.9, 11)\n img = ImageEnhance.Brightness(img).enhance(random.uniform(magnitudes[magnitude], magnitudes[magnitude+1]))\n return img\n\n\ndef sharpness(img, magnitude):\n magnitudes = np.linspace(0.1, 1.9, 11)\n img = ImageEnhance.Sharpness(img).enhance(random.uniform(magnitudes[magnitude], magnitudes[magnitude+1]))\n return img\n\n\ndef cutout(org_img, magnitude=None):\n img = np.array(img)\n\n magnitudes = np.linspace(0, 60/331, 11)\n\n img = np.copy(org_img)\n mask_val = img.mean()\n\n if magnitude is None:\n mask_size = 16\n else:\n mask_size = int(round(img.shape[0]*random.uniform(magnitudes[magnitude], magnitudes[magnitude+1])))\n top = np.random.randint(0 - mask_size//2, img.shape[0] - mask_size)\n left = np.random.randint(0 - mask_size//2, img.shape[1] - mask_size)\n bottom = top + mask_size\n right = left + mask_size\n\n if top < 0:\n top = 0\n if left < 0:\n left = 0\n\n img[top:bottom, left:right, :].fill(mask_val)\n\n img = Image.fromarray(img)\n\n return img\n\n\n\nclass Cutout(object):\n def __init__(self, length=16):\n self.length = length\n\n def __call__(self, img):\n img = np.array(img)\n\n mask_val = img.mean()\n\n top = np.random.randint(0 - self.length//2, img.shape[0] - self.length)\n left = np.random.randint(0 - self.length//2, img.shape[1] - self.length)\n bottom = top + self.length\n right = left + self.length\n\n top = 0 if top < 0 else top\n left = 0 if left < 0 else top\n\n img[top:bottom, left:right, :] = mask_val\n\n img = Image.fromarray(img)\n\n return img",
"_____no_output_____"
]
],
[
[
"### MIXUP",
"_____no_output_____"
]
],
[
[
"alpha_ = 0.4\n\n\n# def mixup_data(x, y, alpha=alpha_, use_cuda=True):\n# if alpha > 0:\n# lam = np.random.beta(alpha, alpha)\n# else:\n# lam = 1\n\n# batch_size = x.size()[0]\n# if use_cuda:\n# index = torch.randperm(batch_size).cuda()\n# else:\n# index = torch.randperm(batch_size)\n\n# mixed_x = lam * x + (1 - lam) * x[index, :]\n# y_a, y_b = y, y[index]\n# return mixed_x, y_a, y_b, lam\n\n# def mixup_criterion(criterion, pred, y_a, y_b, lam):\n# return lam * criterion(pred.float().cuda(), y_a.float().cuda()) + (1 - lam) * criterion(pred.float().cuda(), y_b.float().cuda())\n\n\ndef mixup_data(x, y, alpha=1.0, use_cuda=True):\n '''Returns mixed inputs, pairs of targets, and lambda'''\n if alpha > 0:\n lam = np.random.beta(alpha, alpha)\n else:\n lam = 1\n\n batch_size = x.size()[0]\n if use_cuda:\n index = torch.randperm(batch_size).cuda()\n else:\n index = torch.randperm(batch_size)\n\n mixed_x = lam * x + (1 - lam) * x[index, :]\n# print(y)\n y_a, y_b = y, y[index]\n \n return mixed_x, y_a, y_b, lam\n\n\ndef mixup_criterion(criterion, pred, y_a, y_b, lam):\n return lam * criterion(pred, y_a) + (1 - lam) * criterion(pred, y_b)",
"_____no_output_____"
],
[
"class ConcatDataset(torch.utils.data.Dataset):\n def __init__(self, *datasets):\n self.datasets = datasets\n\n def __getitem__(self, i):\n return tuple(d[i] for d in self.datasets)\n\n def __len__(self):\n return min(len(d) for d in self.datasets)",
"_____no_output_____"
],
[
"\n\nplt.ion() # interactive mode\n\nEO_data_transforms = {\n 'Training': transforms.Compose([\n transforms.Grayscale(num_output_channels=3),\n transforms.Resize((30,30)),\n AutoAugment(),\n Cutout(),\n# transforms.RandomRotation(15,),\n# transforms.RandomResizedCrop(30),\n# transforms.RandomHorizontalFlip(),\n# transforms.RandomVerticalFlip(),\n transforms.Grayscale(num_output_channels=1),\n transforms.ToTensor(),\n transforms.Normalize([0.2913437], [0.12694514])\n \n \n #transforms.Normalize(mean=[0.485, 0.456, 0.406], std=[0.229, 0.224, 0.225]),\n# transforms.Lambda(lambda x: x.repeat(3, 1, 1)),\n #transforms.Normalize(mean=[0.507, 0.487, 0.441], std=[0.267, 0.256, 0.276])\n ]),\n 'Test': transforms.Compose([\n transforms.Grayscale(num_output_channels=1),\n transforms.Resize(30),\n transforms.ToTensor(),\n transforms.Normalize([0.2913437], [0.12694514])\n #transforms.Normalize(mean=[0.485, 0.456, 0.406], std=[0.229, 0.224, 0.225]),\n# transforms.Lambda(lambda x: x.repeat(3, 1, 1)),\n # transforms.Normalize(mean=[0.507, 0.487, 0.441], std=[0.267, 0.256, 0.276])\n ]),\n 'valid_EO': transforms.Compose([\n transforms.Grayscale(num_output_channels=1),\n transforms.Resize((30,30)),\n# AutoAugment(),\n \n# transforms.RandomRotation(15,),\n# transforms.RandomResizedCrop(48),\n# transforms.RandomHorizontalFlip(),\n# transforms.RandomVerticalFlip(),\n transforms.ToTensor(),\n transforms.Normalize([0.2913437], [0.12694514])\n \n# transforms.Grayscale(num_output_channels=1),\n# transforms.Resize(48),\n# transforms.ToTensor(),\n# transforms.Normalize([0.5], [0.5])\n# transforms.Lambda(lambda x: x.repeat(3, 1, 1)),\n # transforms.Normalize(mean=[0.507, 0.487, 0.441], std=[0.267, 0.256, 0.276])\n ]),\n}\n\n\n\n# Data augmentation and normalization for training\n# Just normalization for validation\ndata_transforms = {\n 'Training': transforms.Compose([\n transforms.Grayscale(num_output_channels=1),\n transforms.Resize((52,52)),\n transforms.RandomRotation(15,),\n transforms.RandomResizedCrop(48),\n transforms.RandomHorizontalFlip(),\n transforms.RandomVerticalFlip(),\n transforms.ToTensor(),\n transforms.Normalize([0.4062625], [0.12694514])\n #transforms.Normalize(mean=[0.485, 0.456, 0.406], std=[0.229, 0.224, 0.225]),\n# transforms.Lambda(lambda x: x.repeat(3, 1, 1)),\n #transforms.Normalize(mean=[0.507, 0.487, 0.441], std=[0.267, 0.256, 0.276])\n ]),\n 'Test': transforms.Compose([\n transforms.Grayscale(num_output_channels=1),\n transforms.Resize(48),\n transforms.ToTensor(),\n transforms.Normalize([0.4062625], [0.12694514])\n #transforms.Normalize(mean=[0.485, 0.456, 0.406], std=[0.229, 0.224, 0.225]),\n# transforms.Lambda(lambda x: x.repeat(3, 1, 1)),\n # transforms.Normalize(mean=[0.507, 0.487, 0.441], std=[0.267, 0.256, 0.276])\n ]),\n 'valid': transforms.Compose([\n transforms.Grayscale(num_output_channels=1),\n transforms.Resize((52,52)),\n transforms.RandomRotation(15,),\n transforms.RandomResizedCrop(48),\n transforms.RandomHorizontalFlip(),\n transforms.RandomVerticalFlip(),\n transforms.ToTensor(),\n transforms.Normalize([0.4062625], [0.12694514])\n \n# transforms.Grayscale(num_output_channels=1),\n# transforms.Resize(48),\n# transforms.ToTensor(),\n# transforms.Normalize([0.5], [0.5])\n# transforms.Lambda(lambda x: x.repeat(3, 1, 1)),\n # transforms.Normalize(mean=[0.507, 0.487, 0.441], std=[0.267, 0.256, 0.276])\n ]),\n}\n\n\n# data_dir = '/mnt/sda1/cvpr21/Classification/ram'\n# EO_image_datasets = {x: datasets.ImageFolder(os.path.join(data_dir, x),\n# EO_data_transforms[x])\n# for x in ['Training', 'Test']}\n# EO_dataloaders = {x: torch.utils.data.DataLoader(EO_image_datasets[x], batch_size=256,\n# shuffle=True, num_workers=64, pin_memory=True)\n# for x in ['Training', 'Test']}\n# EO_dataset_sizes = {x: len(EO_image_datasets[x]) for x in ['Training', 'Test']}\n# EO_class_names = EO_image_datasets['Training'].classes\n\n\n\n# image_datasets = {x: datasets.ImageFolder(os.path.join(data_dir, x),\n# data_transforms[x])\n# for x in ['Training', 'Test']}\n\n# combine_dataset = ConcatDataset(EO_image_datasets, image_datasets)\n# dataloaders = {x: torch.utils.data.DataLoader(combine_dataset[x], batch_size=256,\n# shuffle=True, num_workers=64, pin_memory=True)\n# for x in ['Training', 'Test']}\n# dataset_sizes = {x: len(image_datasets[x]) for x in ['Training', 'Test']}\n# class_names = image_datasets['Training'].classes\n\ndevice = torch.device(\"cuda:0\" if torch.cuda.is_available() else \"cpu\")\n\n# def imshow(inp, title=None):\n# \"\"\"Imshow for Tensor.\"\"\"\n# inp = inp.numpy().transpose((1, 2, 0))\n# # mean = np.array([0.1786, 0.4739, 0.5329])\n# # std = np.array([[0.0632, 0.1361, 0.0606]])\n# # inp = std * inp + mean\n# inp = np.clip(inp, 0, 1)\n# plt.imshow(inp)\n# if title is not None:\n# plt.title(title)\n# plt.pause(0.001) # pause a bit so that plots are updated\n\n\n \n# # Get a batch of training data\n# EO_inputs, EO_classes = next(iter(EO_dataloaders['Training']))\n\n\n# inputs, classes, k ,_= next(iter(dataloaders))\n\n\n\n\n\n\n# # Make a grid from batch\n# EO_out = torchvision.utils.make_grid(EO_inputs)\n\n# out = torchvision.utils.make_grid(inputs)\n# imshow(EO_out, title=[EO_class_names[x] for x in classes])\n\n# imshow(out, title=[class_names[x] for x in classes])",
"_____no_output_____"
],
[
"from torch.utils import data\nfrom tqdm import tqdm\nfrom PIL import Image\noutput_dim = 10\n\nclass SAR_EO_Combine_Dataset(data.Dataset):\n def __init__(self,df_sar,dirpath_sar,transform_sar,df_eo=None,dirpath_eo=None,transform_eo=None,test = False):\n self.df_sar = df_sar\n self.test = test\n self.dirpath_sar = dirpath_sar\n self.transform_sar = transform_sar\n \n self.df_eo = df_eo\n# self.test = test\n self.dirpath_eo = dirpath_eo\n self.transform_eo = transform_eo\n #image data \n# if not self.test:\n# self.image_arr = np.asarray(str(self.dirpath)+'/'+self.df.iloc[:, 0]+'.png')\n# else:\n# self.image_arr = np.asarray(str(self.dirpath)+'/'+self.df.iloc[:, 0])\n \n# #labels data\n# if not self.test:\n# self.label_df = self.df.iloc[:,1]\n \n # Calculate length of df\n self.data_len = len(self.df_sar.index)\n\n def __len__(self):\n return self.data_len\n \n \n def __getitem__(self, idx):\n image_name_sar = self.df_sar.img_name[idx]\n image_name_sar = os.path.join(self.dirpath_sar, image_name_sar)\n img_sar = Image.open(image_name_sar)#.convert('RGB')\n img_tensor_sar = self.transform_sar(img_sar)\n \n image_name_eo = self.df_eo.img_name[idx]\n image_name_eo = os.path.join(self.dirpath_eo, image_name_eo)\n img_eo = Image.open(image_name_eo)#.convert('RGB')\n img_tensor_eo = self.transform_eo(img_eo)\n \n# image_name = self.df.img_name[idx]\n# img = Image.open(image_name)#.convert('RGB')\n# img_tensor = self.transform(img)\n \n \n if not self.test:\n image_labels = int(self.df_sar.class_id[idx])\n# label_tensor = torch.zeros((1, output_dim))\n# for label in image_labels.split():\n# label_tensor[0, int(label)] = 1\n image_label = torch.tensor(image_labels,dtype= torch.long)\n image_label = image_label.squeeze()\n \n image_labels_eo = int(self.df_eo.class_id[idx])\n# label_tensor_eo = torch.zeros((1, output_dim))\n# for label_eo in image_labels_eo.split():\n# label_tensor_eo[0, int(label_eo)] = 1\n image_label_eo = torch.tensor(image_labels_eo,dtype= torch.long)\n image_label_eo = image_label_eo.squeeze()\n# print(image_label_eo)\n \n \n \n \n return (img_tensor_sar,image_label), (img_tensor_eo, image_label_eo)\n \n return (img_tensor_sar)\n",
"_____no_output_____"
],
[
"class SAR_EO_Combine_Dataset2(data.Dataset):\n def __init__(self,df_sar,dirpath_sar,transform_sar,df_eo=None,dirpath_eo=None,transform_eo=None,test = False):\n self.df_sar = df_sar\n self.test = test\n self.dirpath_sar = dirpath_sar\n self.transform_sar = transform_sar\n \n self.df_eo = df_eo\n# self.test = test\n self.dirpath_eo = dirpath_eo\n self.transform_eo = transform_eo\n #image data \n# if not self.test:\n# self.image_arr = np.asarray(str(self.dirpath)+'/'+self.df.iloc[:, 0]+'.png')\n# else:\n# self.image_arr = np.asarray(str(self.dirpath)+'/'+self.df.iloc[:, 0])\n \n# #labels data\n# if not self.test:\n# self.label_df = self.df.iloc[:,1]\n \n # Calculate length of df\n self.data_len = len(self.df_sar.index)\n\n def __len__(self):\n return self.data_len\n \n \n def __getitem__(self, idx):\n image_name_sar = self.df_sar.img_name[idx]\n image_name_sar = os.path.join(self.dirpath_sar, image_name_sar)\n img_sar = Image.open(image_name_sar)#.convert('RGB')\n img_tensor_sar = self.transform_sar(img_sar)\n \n image_name_eo = self.df_eo.img_name[idx]\n image_name_eo = os.path.join(self.dirpath_eo, image_name_eo)\n img_eo = Image.open(image_name_eo)#.convert('RGB')\n img_tensor_eo = self.transform_eo(img_eo)\n \n# image_name = self.df.img_name[idx]\n# img = Image.open(image_name)#.convert('RGB')\n# img_tensor = self.transform(img)\n \n \n if not self.test:\n image_labels = int(self.df_sar.class_id[idx])\n# label_tensor = torch.zeros((1, output_dim))\n# for label in image_labels.split():\n# label_tensor[0, int(label)] = 1\n image_label = torch.tensor(image_labels,dtype= torch.long)\n image_label = image_label.squeeze()\n \n image_labels_eo = int(self.df_eo.class_id[idx])\n# label_tensor_eo = torch.zeros((1, output_dim))\n# for label_eo in image_labels_eo.split():\n# label_tensor_eo[0, int(label_eo)] = 1\n image_label_eo = torch.tensor(image_labels_eo,dtype= torch.long)\n image_label_eo = image_label_eo.squeeze()\n# print(image_label_eo)\n \n \n \n \n return (img_tensor_sar,image_label), (img_tensor_eo, image_label_eo)\n \n return (img_tensor_sar)",
"_____no_output_____"
],
[
"import pandas as pd\neo_df = pd.read_csv(\"/home/hans/sandisk/dataset_mover/kd_train_EO.csv\")\neo_df = eo_df.sort_values(by='img_name')\n\nsar_df = pd.read_csv(\"/home/hans/sandisk/dataset_mover/kd_train_SAR.csv\") \nsar_df = sar_df.sort_values(by='img_name')",
"_____no_output_____"
],
[
"\n\n\neo_test_df = pd.read_csv(\"/home/hans/sandisk/dataset_mover/kd_test_EO.csv\")\neo_test_df = eo_test_df.sort_values(by='img_name')\n\nsar_test_df = pd.read_csv(\"/home/hans/sandisk/dataset_mover/kd_test_SAR.csv\") \nsar_test_df = sar_test_df.sort_values(by='img_name')",
"_____no_output_____"
],
[
"BATCH_SIZE = 512\n\ndirpath_sar = \"/home/hans/sandisk/dataset_mover/kd_train_SAR\"\ndirpath_eo = \"/home/hans/sandisk/dataset_mover/kd_train_EO\"\nSAR_EO_Combine = SAR_EO_Combine_Dataset(sar_df,dirpath_sar,data_transforms[\"Test\"],eo_df,dirpath_eo,EO_data_transforms[\"Training\"],test = False)\n",
"_____no_output_____"
],
[
"\n\n\ntestpath_sar = \"/home/hans/sandisk/dataset_mover/kd_val_SAR\"\ntestpath_eo = \"/home/hans/sandisk/dataset_mover/kd_val_EO\"\n\ntest_set = SAR_EO_Combine_Dataset(sar_test_df,testpath_sar,data_transforms[\"Test\"],eo_test_df,testpath_eo,EO_data_transforms[\"Test\"],test = False)\n# test_loader = data.DataLoader(dataset=test_dataset,batch_size=BATCH_SIZE,shuffle=False)",
"_____no_output_____"
],
[
"train_size = len(SAR_EO_Combine)",
"_____no_output_____"
],
[
"\ntest_size = len(test_set)\n\n# from sklearn.model_selection import train_test_split\n\n# train_dataset, test_dataset = train_test_split(SAR_EO_Combine[0], SAR_EO_Combine[2], test_size=0.2, random_state=2017, stratify = SAR_EO_Combine[2])\n# train_dataset, test_dataset = torch.utils.data.random_split(SAR_EO_Combine, [train_size, test_size])",
"_____no_output_____"
],
[
"data_loader = data.DataLoader(dataset=SAR_EO_Combine,batch_size=BATCH_SIZE,shuffle=True,pin_memory = True)",
"_____no_output_____"
],
[
"\ntest_loader = data.DataLoader(dataset=test_set,batch_size=BATCH_SIZE,shuffle=True,pin_memory = True)",
"_____no_output_____"
],
[
"def imshow(inp, title=None):\n \"\"\"Imshow for Tensor.\"\"\"\n inp = inp.numpy().transpose((1, 2, 0))\n# mean = np.array([0.1786, 0.4739, 0.5329])\n# std = np.array([[0.0632, 0.1361, 0.0606]])\n# inp = std * inp + mean\n inp = np.clip(inp, 0, 1)\n plt.imshow(inp)\n if title is not None:\n plt.title(title)\n plt.pause(0.001) # pause a bit so that plots are updated\n\n\nSAR, EO = next(iter(data_loader))\n# Get a batch of training data\nEO_inputs, EO_classes = EO[0],EO[1]\n\n\ninputs, classes = SAR[0],SAR[1]\n\n\n# EO_class_names = SAR_EO_Combine.image_label\n\n\n\n# Make a grid from batch\nEO_out = torchvision.utils.make_grid(EO_inputs)\n\nout = torchvision.utils.make_grid(inputs)\nimshow(EO_out)#, title=[EO_class_names[x] for x in classes])\n\nimshow(out)#, title=[class_names[x] for x in classes])\nprint(len(EO_classes))\nprint(classes)",
"_____no_output_____"
],
[
"# a = 0\n# for i in range(500):\n# SAR, EO = next(iter(data_loader))\n# # Get a batch of training data\n# EO_inputs, EO_classes = EO[0],EO[1]\n\n\n# inputs, classes = SAR[0],SAR[1]\n# if 9 in classes:\n# a+=1\n# print(a)",
"_____no_output_____"
]
],
[
[
"### check if paired succeed",
"_____no_output_____"
]
],
[
[
"# from tqdm import trange\n# def equal():\n# notsame_image = 0\n# notsame_label = 0\n# t = trange(len(sar_df))\n \n\n \n# for i in t:\n# t.set_postfix({'nums of not same label:': notsame_label})\n# sar, eo = next(iter(data_loader))\n# eo_label = eo[1][0].tolist()\n# sar_label = sar[1][0].tolist()\n# # print(eo_label)\n# # print(sar_label)\n# # if not eo_image == sar_image:\n# # notsame_image += 1\n \n# # eoval = next(eo_label) \n# # sarval = next(sar_label) \n# if not eo_label==sar_label:\n# notsame_label += 1\n \n# # notsame_label += 1\n# # print(\"nums of not same imageid:\", notsame_image)\n# #print(\"nums of not same label:\", notsame_label)\n\n# equal()\n# #next(iter(data_loader))",
"_____no_output_____"
],
[
"len(sar_df) == len(eo_df)",
"_____no_output_____"
],
[
"next(eo_df.iterrows())[1]",
"_____no_output_____"
],
[
"Num_class=10\nnum_classes = Num_class\nnum_channel = 1\n\n# model_ft = models.resnet34(pretrained=False)\nmodel_ft = torch.load(\"10/pre_resnet34_model_epoch99.pt\") ## Attention: you need to change to the path of pre_EO.pt file, which located in the repo folder pre-train\n# model_ft.conv1 = nn.Conv2d(num_channel, 64, kernel_size=7, stride=2, padding=3,bias=False)\n# # model_ft.avgpool = SpatialPyramidPooling((3,3))\n# model_ft.fc = nn.Linear(512, Num_class)\n# model_ft.conv0 = nn.Conv2d(\n# model_ft.features[0] = nn.Conv2d(num_channel, 16, kernel_size=3, stride=2, padding=1,bias=False)\n# model_ft.classifier[3] = nn.Linear(1024, Num_class, bias=True)\nmodel_ft.eval()",
"_____no_output_____"
],
[
"data_dir = '/mnt/sda1/cvpr21/Classification/ram'\nweights = []\nfor i in range(len(os.listdir(os.path.join(data_dir, \"Training\")))):\n img_num = len([lists for lists in os.listdir(os.path.join(data_dir, \"Training\",str(i)))])\n print('filenum:',len([lists for lists in os.listdir(os.path.join(data_dir, \"Training\",str(i)))]))# if os.path.isfile(os.path.join(data_dir, lists))]))\n weights.append(img_num)\nprint(weights)\nweights = torch.tensor(weights, dtype=torch.float32).cuda()\nweights = weights / weights.sum()\nprint(weights)\nweights = 1.0 / weights\nweights = weights / weights.sum()\nprint(weights)",
"filenum: 234135\nfilenum: 28030\nfilenum: 15234\nfilenum: 10587\nfilenum: 1668\nfilenum: 782\nfilenum: 768\nfilenum: 562\nfilenum: 783\nfilenum: 573\n[234135, 28030, 15234, 10587, 1668, 782, 768, 562, 783, 573]\ntensor([0.7988, 0.0956, 0.0520, 0.0361, 0.0057, 0.0027, 0.0026, 0.0019, 0.0027,\n 0.0020], device='cuda:0')\ntensor([0.0005, 0.0044, 0.0080, 0.0115, 0.0733, 0.1563, 0.1591, 0.2175, 0.1561,\n 0.2133], device='cuda:0')\n"
]
],
[
[
"### Teacher model (SAR)",
"_____no_output_____"
]
],
[
[
"netT = torch.load(\"10/resnet34_model_epoch119.pt\") ## Attention: you need to change to the path of pre_SAR.pt file, which located in the repo folder pre-train\n\n# netT = torch.load('29_auto_aug_eo_sar_noimagenet/pre_resnet34_eo_epoch99.pt')\ncriterion2 = nn.KLDivLoss()\nnetT.eval()",
"_____no_output_____"
],
[
"from tqdm.notebook import trange\nfrom tqdm import tqdm_notebook as tqdm \nimport warnings\nwarnings.filterwarnings('ignore')\ndef train_model(model, criterion, optimizer, scheduler, num_epochs=25):\n since = time.time()\n print(\"---------------Start KD FIT( TEACHER AND STUDENT )-----------------\")\n best_model_wts = copy.deepcopy(model.state_dict())\n best_acc = 0.0\n best_train_acc = 0.0\n \n kd_alpha = 0.2\n \n Loss_list = []\n Accuracy_list = []\n T_Loss_list = []\n T_Accuracy_list = []\n\n for epoch in trange(num_epochs):\n print('Epoch {}/{}'.format(epoch, num_epochs - 1))\n \n # Each epoch has a training and validation phase\n for phase in ['Training', 'Test']:#['Test','Training']: #:\n if phase == 'Training':\n model.train() # Set model to training mode\n else:\n model.eval() # Set model to evaluate mode\n\n running_loss = 0.0\n running_corrects = 0\n \n \n\n\n # Iterate over data.\n \n \n if phase == 'Training':\n \n for SAR, EO in tqdm(data_loader):\n inputs, labels = EO[0], EO[1]\n inputs = inputs.to(device)\n\n labels = labels.to(device)\n\n T_input, T_labels = SAR[0], SAR[1]\n T_input = T_input.to(device)\n T_labels = T_labels.to(device)\n # print(T_labels, labels)\n # labels = torch.argmax(labels, 0)\n # T_labels = torch.argmax(T_labels, 0)\n\n# confusion_matrix = torch.zeros(10, 10)\n \n # zero the parameter gradients\n optimizer.zero_grad()\n\n # forward\n # track history if only in train\n with torch.no_grad():\n soft_target = netT(T_input)\n with torch.set_grad_enabled(phase == 'Training'):\n outputs = model(inputs) # _, \n #print(outputs.dim())\n\n _, preds = torch.max(outputs, 1)\n\n# for t, p in zip(labels.view(-1), preds.view(-1)):\n# confusion_matrix[t.long(), p.long()] += 1\n# print(confusion_matrix.diag()/confusion_matrix.sum(1))\n \n # _, T_preds = torch.max(soft_target, 1)\n T = 2\n outputs_S = F.log_softmax(outputs/T, dim=1)\n outputs_T = F.softmax(soft_target/T, dim=1)\n # print(outputs_S.size())\n # print(outputs_T.size())\n\n loss2 = criterion2(outputs_S, outputs_T) * T * T\n #print(preds)\n\n if phase == 'Training':\n inputs, y_a, y_b, lam = mixup_data(inputs, labels)\n inputs, y_a, y_b = map(Variable, (inputs, y_a, y_b))\n # print(y_a)\n # print(y_b)\n\n loss = mixup_criterion(criterion, outputs, y_a, y_b, lam)\n loss = loss*(1-kd_alpha) + loss2*kd_alpha\n else:\n\n loss = criterion(outputs, labels)\n loss = loss*(1-kd_alpha) + loss2*kd_alpha\n\n # backward + optimize only if in training phase\n if phase == 'Training':\n loss.backward()\n optimizer.step()\n\n\n # statistics\n running_loss += loss.item() * inputs.size(0)\n running_corrects += torch.sum(preds == labels.data)\n if phase == 'Training':\n scheduler.step()\n\n epoch_loss = running_loss / train_size\n epoch_acc = running_corrects.double() / train_size\n\n print('{} Loss: {:.4f} Acc: {:.4f}'.format(\n phase, epoch_loss, epoch_acc))\n\n if phase == \"Training\":\n Loss_list.append(epoch_loss)\n Accuracy_list.append(100 * epoch_acc)\n else:\n T_Loss_list.append(epoch_loss)\n T_Accuracy_list.append(100 * epoch_acc)\n\n\n\n # deep copy the model\n if phase == 'Test' and epoch_acc > best_acc:\n best_acc = epoch_acc\n best_model_wts = copy.deepcopy(model.state_dict())\n PATH = os.path.join(str(exp_num), \"resnet34_kd_best.pt\")#\"resnet18_model_epoch{}.pt\".format(epoch)\n if not os.path.exists(str(exp_num)):\n os.makedirs(str(exp_num))\n torch.save(model, PATH)\n time_elapsed = time.time() - since\n print('Time from Start {:.0f}m {:.0f}s'.format(\n time_elapsed // 60, time_elapsed % 60))\n\n if phase == 'Training' and epoch_acc > best_train_acc:\n best_train_acc = epoch_acc\n # PATH = os.path.join(str(exp_num), \"resnet34_kd_best.pt\")#\"resnet18_model_epoch{}.pt\".format(epoch)\n # if not os.path.exists(str(exp_num)):\n # os.makedirs(str(exp_num))\n # torch.save(model, PATH)\n \n \n #############################################################################\n elif phase == 'Test':\n acc_matrix_sum = torch.zeros(10)\n \n for SAR, EO in tqdm(test_loader):\n inputs, labels = EO[0], EO[1]\n inputs = inputs.to(device)\n# print(inputs)\n\n labels = labels.to(device)\n\n T_input, T_labels = SAR[0], SAR[1]\n T_input = T_input.to(device)\n T_labels = T_labels.to(device)\n # print(T_labels, labels)\n # labels = torch.argmax(labels, 0)\n # T_labels = torch.argmax(T_labels, 0)\n\n\n # zero the parameter gradients\n optimizer.zero_grad()\n\n # forward\n # track history if only in train\n with torch.no_grad():\n soft_target = netT(T_input)\n with torch.set_grad_enabled(phase == 'Training'):\n outputs = model(inputs) # _, \n #print(outputs.dim())\n\n _, preds = torch.max(outputs, 1)\n \n confusion_matrix = torch.zeros(10, 10)\n for t, p in zip(labels.view(-1), preds.view(-1)):\n confusion_matrix[t.long(), p.long()] += 1\n acc_matrix_batch = (confusion_matrix.diag()/confusion_matrix.sum(1))\n \n\n\n # _, T_preds = torch.max(soft_target, 1)\n T = 2\n outputs_S = F.log_softmax(outputs/T, dim=1)\n outputs_T = F.softmax(soft_target/T, dim=1)\n # print(outputs_S.size())\n # print(outputs_T.size())\n\n loss2 = criterion2(outputs_S, outputs_T) * T * T\n #print(preds)\n\n if phase == 'Training':\n inputs, y_a, y_b, lam = mixup_data(inputs, labels)\n inputs, y_a, y_b = map(Variable, (inputs, y_a, y_b))\n # print(y_a)\n # print(y_b)\n\n loss = mixup_criterion(criterion, outputs, y_a, y_b, lam)\n loss = loss*(1-kd_alpha) + loss2*kd_alpha\n else:\n\n loss = criterion(outputs, labels)\n loss = loss*(1-kd_alpha) + loss2*kd_alpha\n\n # backward + optimize only if in training phase\n if phase == 'Training':\n loss.backward()\n optimizer.step()\n\n\n # statistics\n running_loss += loss.item() * inputs.size(0)\n running_corrects += torch.sum(preds == labels.data)\n acc_matrix_sum += acc_matrix_batch\n \n acc_matrix = acc_matrix_sum / test_size\n print(\"acc for each class: {}\".format(acc_matrix)) \n #################\n \n if phase == 'Training':\n scheduler.step()\n\n epoch_loss = running_loss / test_size\n epoch_acc = running_corrects.double() / test_size\n\n print('{} Loss: {:.4f} Acc: {:.4f}'.format(\n phase, epoch_loss, epoch_acc))\n\n if phase == \"Training\":\n Loss_list.append(epoch_loss)\n Accuracy_list.append(100 * epoch_acc)\n else:\n T_Loss_list.append(epoch_loss)\n T_Accuracy_list.append(100 * epoch_acc)\n\n\n\n # deep copy the model\n if phase == 'Test' and epoch_acc > best_acc:\n best_acc = epoch_acc\n best_model_wts = copy.deepcopy(model.state_dict())\n PATH = os.path.join(str(exp_num), \"resnet34_kd_best.pt\")#\"resnet18_model_epoch{}.pt\".format(epoch)\n if not os.path.exists(str(exp_num)):\n os.makedirs(str(exp_num))\n torch.save(model, PATH)\n time_elapsed = time.time() - since\n print('Time from Start {:.0f}m {:.0f}s'.format(\n time_elapsed // 60, time_elapsed % 60))\n\n if phase == 'Training' and epoch_acc > best_train_acc:\n best_train_acc = epoch_acc\n # PATH = os.path.join(str(exp_num), \"resnet34_kd_best.pt\")#\"resnet18_model_epoch{}.pt\".format(epoch)\n # if not os.path.exists(str(exp_num)):\n # os.makedirs(str(exp_num))\n # torch.save(model, PATH)\n\n\n print()\n PATH = os.path.join(str(exp_num), \"resnet34_kd{}.pt\".format(epoch))#\"resnet18_model_epoch{}.pt\".format(epoch)\n if not os.path.exists(str(exp_num)):\n os.makedirs(str(exp_num))\n torch.save(model, PATH)\n# torch.save(best_model_wts, \"best.pt\")\n\n time_elapsed = time.time() - since\n print('Training complete in {:.0f}m {:.0f}s'.format(\n time_elapsed // 60, time_elapsed % 60))\n print('Best train Acc: {:4f}'.format(best_train_acc))\n print('Best val Acc: {:4f}'.format(best_acc))\n \n\n # load best model weights\n model.load_state_dict(best_model_wts)\n \n \n ##### PLOT\n x1 = range(0, num_epochs)\n x2 = range(0, num_epochs)\n y1 = Accuracy_list\n y2 = Loss_list\n plt.subplot(2, 1, 1)\n plt.plot(x1, y1, 'o-')\n plt.title('Train accuracy vs. epoches')\n plt.ylabel('Train accuracy')\n plt.subplot(2, 1, 2)\n plt.plot(x2, y2, '.-')\n plt.xlabel('Train loss vs. epoches')\n plt.ylabel('Train loss')\n plt.show()\n plt.savefig(\"Train_accuracy_loss.jpg\")\n \n x1 = range(0, num_epochs)\n x2 = range(0, num_epochs)\n y1 = T_Accuracy_list\n y2 = T_Loss_list\n plt.subplot(2, 1, 1)\n plt.plot(x1, y1, 'o-')\n plt.title('Test accuracy vs. epoches')\n plt.ylabel('Test accuracy')\n plt.subplot(2, 1, 2)\n plt.plot(x2, y2, '.-')\n plt.xlabel('Test loss vs. epoches')\n plt.ylabel('Test loss')\n plt.show()\n plt.savefig(\"Test_accuracy_loss.jpg\")\n \n return model\n\n\nmodel_ft = model_ft.to(device)\n\n# #criterion = nn.CrossEntropyLoss()\ncriterion = nn.CrossEntropyLoss(weight=weights) #weight=weights, ",
"_____no_output_____"
],
[
"# def train_model(model, criterion, optimizer, scheduler, num_epochs=25):\n# since = time.time()\n# print(\"---------------Start KD FIT( TEACHER AND STUDENT )-----------------\")\n# best_model_wts = copy.deepcopy(model.state_dict())\n# best_acc = 0.0\n# best_train_acc = 0.0\n \n# kd_alpha = 0.8\n \n# Loss_list = []\n# Accuracy_list = []\n# T_Loss_list = []\n# T_Accuracy_list = []\n\n# for epoch in trange(num_epochs):\n# print('Epoch {}/{}'.format(epoch, num_epochs - 1))\n \n# # Each epoch has a training and validation phase\n# for phase in ['Training', 'Test']:\n# ##################################### train#############################\n# if phase == 'Training':\n# model.train() # Set model to training mode\n# else:\n# model.eval() # Set model to evaluate mode\n\n# running_loss = 0.0\n# running_corrects = 0\n\n# # Iterate over data.\n# if phase == \"Training\":\n# for SAR, EO in tqdm(data_loader):\n# inputs, labels = SAR[0], SAR[1]\n# inputs = inputs.to(device)\n\n# labels = labels.to(device)\n\n# T_input, T_labels = EO[0], EO[1]\n# T_input = T_input.to(device)\n# T_labels = T_labels.to(device)\n# # print(T_labels, labels)\n# # labels = torch.argmax(labels, 0)\n# # T_labels = torch.argmax(T_labels, 0)\n\n\n# # zero the parameter gradients\n# optimizer.zero_grad()\n\n# # forward\n# # track history if only in train\n# with torch.no_grad():\n# soft_target = netT(T_input)\n# with torch.set_grad_enabled():\n# outputs = model(inputs) # _, \n# #print(outputs.dim())\n\n# _, preds = torch.max(outputs, 1)\n\n\n# # _, T_preds = torch.max(soft_target, 1)\n# T = 2\n# outputs_S = F.log_softmax(outputs/T, dim=1)\n# outputs_T = F.softmax(soft_target/T, dim=1)\n# # print(outputs_S.size())\n# # print(outputs_T.size())\n\n# loss2 = criterion2(outputs_S, outputs_T) * T * T\n# #print(preds)\n\n\n# inputs, y_a, y_b, lam = mixup_data(inputs, labels)\n# inputs, y_a, y_b = map(Variable, (inputs, y_a, y_b))\n# # print(y_a)\n# # print(y_b)\n\n# loss = mixup_criterion(criterion, outputs, y_a, y_b, lam)\n# loss = loss*(1-kd_alpha) + loss2*kd_alpha\n# running_loss += loss.item() * inputs.size(0)\n# running_corrects += torch.sum(preds == labels.data) \n\n# loss.backward()\n# optimizer.step()\n# scheduler.step()\n\n \n \n# ##############################test#############################\n# else:\n# for SAR, EO in tqdm(test_data_loader):\n# inputs, labels = SAR[0], SAR[1]\n# inputs = inputs.to(device)\n\n# labels = labels.to(device)\n\n# T_input, T_labels = EO[0], EO[1]\n# T_input = T_input.to(device)\n# T_labels = T_labels.to(device)\n\n\n\n\n# optimizer.zero_grad()\n\n# # forward\n# # track history if only in train\n# with torch.no_grad():\n# soft_target = netT(T_input)\n\n# outputs = model(inputs) # _, \n# #print(outputs.dim())\n\n# _, preds = torch.max(outputs, 1)\n\n\n# # _, T_preds = torch.max(soft_target, 1)\n# T = 2\n# outputs_S = F.log_softmax(outputs/T, dim=1)\n# outputs_T = F.softmax(soft_target/T, dim=1)\n# # print(outputs_S.size())\n# # print(outputs_T.size())\n\n# loss2 = criterion2(outputs_S, outputs_T) * T * T\n\n# loss = criterion(outputs, labels)\n# loss = loss*(1-kd_alpha) + loss2*kd_alpha\n# ################################\n\n# running_loss += loss.item() * inputs.size(0)\n# running_corrects += torch.sum(preds == labels.data) \n \n# epoch_loss = running_loss / dataset_sizes[phase]\n# epoch_acc = running_corrects.double() / dataset_sizes[phase]\n\n# print('{} Loss: {:.4f} Acc: {:.4f}'.format(\n# phase, epoch_loss, epoch_acc))\n\n# if phase == \"Training\":\n# Loss_list.append(epoch_loss)\n# Accuracy_list.append(100 * epoch_acc)\n# else:\n# T_Loss_list.append(epoch_loss)\n# T_Accuracy_list.append(100 * epoch_acc)\n\n\n\n# # deep copy the model\n# if phase == 'Test' and epoch_acc > best_acc:\n# best_acc = epoch_acc\n# best_model_wts = copy.deepcopy(model.state_dict())\n# PATH = os.path.join(str(exp_num), \"resnet34_kd_best.pt\")#\"resnet18_model_epoch{}.pt\".format(epoch)\n# if not os.path.exists(str(exp_num)):\n# os.makedirs(str(exp_num))\n# torch.save(model, PATH)\n# time_elapsed = time.time() - since\n# print('Time from Start {:.0f}m {:.0f}s'.format(\n# time_elapsed // 60, time_elapsed % 60))\n\n# if phase == 'Training' and epoch_acc > best_train_acc:\n# best_train_acc = epoch_acc\n# # PATH = os.path.join(str(exp_num), \"resnet34_kd_best.pt\")#\"resnet18_model_epoch{}.pt\".format(epoch)\n# # if not os.path.exists(str(exp_num)):\n# # os.makedirs(str(exp_num))\n# # torch.save(model, PATH)\n# print()\n# PATH = os.path.join(str(exp_num), \"resnet34_kd{}.pt\".format(epoch))#\"resnet18_model_epoch{}.pt\".format(epoch)\n# if not os.path.exists(str(exp_num)):\n# os.makedirs(str(exp_num))\n# torch.save(model, PATH)\n \n# time_elapsed = time.time() - since\n# print('Training complete in {:.0f}m {:.0f}s'.format(\n# time_elapsed // 60, time_elapsed % 60))\n# print('Best train Acc: {:4f}'.format(best_train_acc))\n# print('Best val Acc: {:4f}'.format(best_acc))\n \n\n# # load best model weights\n# model.load_state_dict(best_model_wts)\n \n \n# ##### PLOT\n# x1 = range(0, num_epochs)\n# x2 = range(0, num_epochs)\n# y1 = Accuracy_list\n# y2 = Loss_list\n# plt.subplot(2, 1, 1)\n# plt.plot(x1, y1, 'o-')\n# plt.title('Train accuracy vs. epoches')\n# plt.ylabel('Train accuracy')\n# plt.subplot(2, 1, 2)\n# plt.plot(x2, y2, '.-')\n# plt.xlabel('Train loss vs. epoches')\n# plt.ylabel('Train loss')\n# plt.show()\n# plt.savefig(\"Train_accuracy_loss.jpg\")\n \n# x1 = range(0, num_epochs)\n# x2 = range(0, num_epochs)\n# y1 = T_Accuracy_list\n# y2 = T_Loss_list\n# plt.subplot(2, 1, 1)\n# plt.plot(x1, y1, 'o-')\n# plt.title('Test accuracy vs. epoches')\n# plt.ylabel('Test accuracy')\n# plt.subplot(2, 1, 2)\n# plt.plot(x2, y2, '.-')\n# plt.xlabel('Test loss vs. epoches')\n# plt.ylabel('Test loss')\n# plt.show()\n# plt.savefig(\"Test_accuracy_loss.jpg\")\n \n# return model\n\n\n# model_ft = model_ft.to(device)\n\n# # #criterion = nn.CrossEntropyLoss()\n# criterion = nn.CrossEntropyLoss(weight=weights) #weight=weights, \n \n \n ",
"_____no_output_____"
],
[
"# os.environ[\"CUDA_LAUNCH_BLOCKING\"] = \"1\"\n",
"_____no_output_____"
],
[
"optimizer_ft = optim.SGD(model_ft.parameters(), lr=0.01, momentum=0.9)\n\n# Decay LR by a factor of 0.1 every 7 epochs\nexp_lr_scheduler = lr_scheduler.StepLR(optimizer_ft, step_size=20, gamma=0.5)\n\nmodel_ft = train_model(model_ft, criterion, optimizer_ft, exp_lr_scheduler,\n num_epochs=120)",
"---------------Start KD FIT( TEACHER AND STUDENT )-----------------\n"
],
[
"!mkdir test",
"_____no_output_____"
],
[
"!unzip NTIRE2021_Class_test_images_EO.zip -d ./test",
"Archive: NTIRE2021_Class_test_images_EO.zip\r\n extracting: ./test/EO_279235.png \r\n extracting: ./test/EO_326948.png \r\n extracting: ./test/EO_318186.png \r\n extracting: ./test/EO_395537.png \r\n extracting: ./test/EO_26324.png \r\n extracting: ./test/EO_182959.png \r\n extracting: ./test/EO_293762.png \r\n extracting: ./test/EO_363012.png \r\n extracting: ./test/EO_290993.png \r\n extracting: ./test/EO_263157.png \r\n extracting: ./test/EO_19944.png \r\n extracting: ./test/EO_361455.png \r\n extracting: ./test/EO_87145.png \r\n extracting: ./test/EO_389934.png \r\n extracting: ./test/EO_383950.png \r\n extracting: ./test/EO_165871.png \r\n extracting: ./test/EO_161349.png \r\n extracting: ./test/EO_249232.png \r\n extracting: ./test/EO_315505.png \r\n extracting: ./test/EO_428962.png \r\n extracting: ./test/EO_142365.png \r\n extracting: ./test/EO_353153.png \r\n extracting: ./test/EO_158148.png \r\n extracting: ./test/EO_349381.png \r\n extracting: ./test/EO_79831.png \r\n extracting: ./test/EO_183515.png \r\n extracting: ./test/EO_472701.png \r\n extracting: ./test/EO_274208.png \r\n extracting: ./test/EO_414667.png \r\n extracting: ./test/EO_186948.png \r\n extracting: ./test/EO_37165.png \r\n extracting: ./test/EO_407399.png \r\n extracting: ./test/EO_203111.png \r\n extracting: ./test/EO_380358.png \r\n extracting: ./test/EO_13136.png \r\n extracting: ./test/EO_394032.png \r\n extracting: ./test/EO_109488.png \r\n extracting: ./test/EO_21764.png \r\n extracting: ./test/EO_251712.png \r\n extracting: ./test/EO_180427.png \r\n extracting: ./test/EO_290297.png \r\n extracting: ./test/EO_433955.png \r\n extracting: ./test/EO_432190.png \r\n extracting: ./test/EO_88142.png \r\n extracting: ./test/EO_394589.png \r\n extracting: ./test/EO_246449.png \r\n extracting: ./test/EO_119208.png \r\n extracting: ./test/EO_353374.png \r\n extracting: ./test/EO_159537.png \r\n extracting: ./test/EO_273399.png \r\n extracting: ./test/EO_114485.png \r\n extracting: ./test/EO_357927.png \r\n extracting: ./test/EO_73713.png \r\n extracting: ./test/EO_381522.png \r\n extracting: ./test/EO_78978.png \r\n extracting: ./test/EO_198776.png \r\n extracting: ./test/EO_93885.png \r\n extracting: ./test/EO_131401.png \r\n extracting: ./test/EO_37886.png \r\n extracting: ./test/EO_416604.png \r\n extracting: ./test/EO_276064.png \r\n extracting: ./test/EO_382285.png \r\n extracting: ./test/EO_58977.png \r\n extracting: ./test/EO_439483.png \r\n extracting: ./test/EO_293047.png \r\n extracting: ./test/EO_347624.png \r\n extracting: ./test/EO_195873.png \r\n extracting: ./test/EO_260223.png \r\n extracting: ./test/EO_316062.png \r\n extracting: ./test/EO_366652.png \r\n extracting: ./test/EO_386517.png \r\n extracting: ./test/EO_152307.png \r\n extracting: ./test/EO_372571.png \r\n extracting: ./test/EO_350758.png \r\n extracting: ./test/EO_201211.png \r\n extracting: ./test/EO_330231.png \r\n extracting: ./test/EO_255691.png \r\n extracting: ./test/EO_376760.png \r\n extracting: ./test/EO_292656.png \r\n extracting: ./test/EO_96662.png \r\n extracting: ./test/EO_381384.png \r\n extracting: ./test/EO_305103.png \r\n extracting: ./test/EO_467279.png \r\n extracting: ./test/EO_412703.png \r\n extracting: ./test/EO_132951.png \r\n extracting: ./test/EO_52052.png \r\n extracting: ./test/EO_317214.png \r\n extracting: ./test/EO_53878.png \r\n extracting: ./test/EO_72942.png \r\n extracting: ./test/EO_436042.png \r\n extracting: ./test/EO_280467.png \r\n extracting: ./test/EO_461669.png \r\n extracting: ./test/EO_267008.png \r\n extracting: ./test/EO_52630.png \r\n extracting: ./test/EO_186000.png \r\n extracting: ./test/EO_329205.png \r\n extracting: ./test/EO_391184.png \r\n extracting: ./test/EO_205228.png \r\n extracting: ./test/EO_470584.png \r\n extracting: ./test/EO_122923.png \r\n extracting: ./test/EO_469540.png \r\n extracting: ./test/EO_434333.png \r\n extracting: ./test/EO_166743.png \r\n extracting: ./test/EO_371498.png \r\n extracting: ./test/EO_363674.png \r\n extracting: ./test/EO_53615.png \r\n extracting: ./test/EO_321397.png \r\n extracting: ./test/EO_78463.png \r\n extracting: ./test/EO_384735.png \r\n extracting: ./test/EO_174612.png \r\n extracting: ./test/EO_170137.png \r\n extracting: ./test/EO_252744.png \r\n extracting: ./test/EO_322684.png \r\n extracting: ./test/EO_293068.png \r\n extracting: ./test/EO_279418.png \r\n extracting: ./test/EO_188964.png \r\n extracting: ./test/EO_388025.png \r\n extracting: ./test/EO_53288.png \r\n extracting: ./test/EO_28070.png \r\n extracting: ./test/EO_18425.png \r\n extracting: ./test/EO_345263.png \r\n extracting: ./test/EO_91090.png \r\n extracting: ./test/EO_115888.png \r\n extracting: ./test/EO_425926.png \r\n extracting: ./test/EO_386067.png \r\n extracting: ./test/EO_249881.png \r\n extracting: ./test/EO_163572.png \r\n extracting: ./test/EO_253001.png \r\n extracting: ./test/EO_365223.png \r\n extracting: ./test/EO_422907.png \r\n extracting: ./test/EO_348839.png \r\n extracting: ./test/EO_309051.png \r\n extracting: ./test/EO_265529.png \r\n extracting: ./test/EO_193122.png \r\n extracting: ./test/EO_434280.png \r\n extracting: ./test/EO_108636.png \r\n extracting: ./test/EO_181463.png \r\n extracting: ./test/EO_378432.png \r\n extracting: ./test/EO_115380.png \r\n extracting: ./test/EO_398050.png \r\n extracting: ./test/EO_424993.png \r\n extracting: ./test/EO_462422.png \r\n extracting: ./test/EO_113225.png \r\n extracting: ./test/EO_443523.png \r\n extracting: ./test/EO_449936.png \r\n extracting: ./test/EO_17344.png \r\n extracting: ./test/EO_404719.png \r\n extracting: ./test/EO_356919.png \r\n extracting: ./test/EO_333035.png \r\n extracting: ./test/EO_454061.png \r\n extracting: ./test/EO_288647.png \r\n extracting: ./test/EO_361929.png \r\n extracting: ./test/EO_307021.png \r\n extracting: ./test/EO_346164.png \r\n extracting: ./test/EO_340329.png \r\n extracting: ./test/EO_10079.png \r\n extracting: ./test/EO_100578.png \r\n extracting: ./test/EO_285173.png \r\n extracting: ./test/EO_53579.png \r\n extracting: ./test/EO_342729.png \r\n extracting: ./test/EO_124731.png \r\n extracting: ./test/EO_434360.png \r\n extracting: ./test/EO_303784.png \r\n extracting: ./test/EO_31990.png \r\n extracting: ./test/EO_457479.png \r\n extracting: ./test/EO_117610.png \r\n extracting: ./test/EO_328620.png \r\n extracting: ./test/EO_171921.png \r\n extracting: ./test/EO_335125.png \r\n extracting: ./test/EO_310532.png \r\n extracting: ./test/EO_371754.png \r\n extracting: ./test/EO_353300.png \r\n extracting: ./test/EO_270116.png \r\n extracting: ./test/EO_452191.png \r\n extracting: ./test/EO_454920.png \r\n extracting: ./test/EO_121325.png \r\n extracting: ./test/EO_421944.png \r\n extracting: ./test/EO_280377.png \r\n extracting: ./test/EO_187308.png \r\n extracting: ./test/EO_162593.png \r\n extracting: ./test/EO_382613.png \r\n extracting: ./test/EO_380468.png \r\n extracting: ./test/EO_146077.png \r\n extracting: ./test/EO_410316.png \r\n extracting: ./test/EO_88725.png \r\n extracting: ./test/EO_109961.png \r\n extracting: ./test/EO_199199.png \r\n extracting: ./test/EO_363042.png \r\n extracting: ./test/EO_288036.png \r\n extracting: ./test/EO_445251.png \r\n extracting: ./test/EO_97302.png \r\n extracting: ./test/EO_178655.png \r\n extracting: ./test/EO_180634.png \r\n extracting: ./test/EO_298141.png \r\n extracting: ./test/EO_361613.png \r\n extracting: ./test/EO_125897.png \r\n extracting: ./test/EO_81468.png \r\n extracting: ./test/EO_359617.png \r\n extracting: ./test/EO_382667.png \r\n extracting: ./test/EO_387752.png \r\n extracting: ./test/EO_405028.png \r\n extracting: ./test/EO_274419.png \r\n extracting: ./test/EO_285686.png \r\n extracting: ./test/EO_49508.png \r\n extracting: ./test/EO_419729.png \r\n extracting: ./test/EO_353547.png \r\n extracting: ./test/EO_442720.png \r\n extracting: ./test/EO_398577.png \r\n extracting: ./test/EO_163079.png \r\n extracting: ./test/EO_448467.png \r\n extracting: ./test/EO_363403.png \r\n extracting: ./test/EO_198331.png \r\n extracting: ./test/EO_206590.png \r\n extracting: ./test/EO_64125.png \r\n extracting: ./test/EO_16771.png \r\n extracting: ./test/EO_197839.png \r\n extracting: ./test/EO_6483.png \r\n extracting: ./test/EO_32940.png \r\n extracting: ./test/EO_156671.png \r\n extracting: ./test/EO_198379.png \r\n extracting: ./test/EO_81420.png \r\n extracting: ./test/EO_173171.png \r\n extracting: ./test/EO_120249.png \r\n extracting: ./test/EO_300240.png \r\n extracting: ./test/EO_354953.png \r\n extracting: ./test/EO_464740.png \r\n extracting: ./test/EO_348090.png \r\n extracting: ./test/EO_78450.png \r\n extracting: ./test/EO_429594.png \r\n extracting: ./test/EO_251691.png \r\n extracting: ./test/EO_348565.png \r\n extracting: ./test/EO_357504.png \r\n extracting: ./test/EO_138478.png \r\n extracting: ./test/EO_366261.png \r\n extracting: ./test/EO_185248.png \r\n extracting: ./test/EO_83336.png \r\n extracting: ./test/EO_352935.png \r\n extracting: ./test/EO_301736.png \r\n extracting: ./test/EO_285023.png \r\n extracting: ./test/EO_206314.png \r\n extracting: ./test/EO_153531.png \r\n extracting: ./test/EO_409792.png \r\n extracting: ./test/EO_167585.png \r\n extracting: ./test/EO_157564.png \r\n extracting: ./test/EO_57405.png \r\n extracting: ./test/EO_270478.png \r\n extracting: ./test/EO_297168.png \r\n extracting: ./test/EO_80972.png \r\n extracting: ./test/EO_157777.png \r\n extracting: ./test/EO_324183.png \r\n extracting: ./test/EO_115892.png \r\n extracting: ./test/EO_283768.png \r\n extracting: ./test/EO_88495.png \r\n extracting: ./test/EO_195362.png \r\n extracting: ./test/EO_355855.png \r\n extracting: ./test/EO_374616.png \r\n extracting: ./test/EO_8083.png \r\n extracting: ./test/EO_330396.png \r\n extracting: ./test/EO_443030.png \r\n extracting: ./test/EO_93128.png \r\n extracting: ./test/EO_433654.png \r\n extracting: ./test/EO_32945.png \r\n extracting: ./test/EO_267871.png \r\n extracting: ./test/EO_459278.png \r\n extracting: ./test/EO_369987.png \r\n extracting: ./test/EO_299146.png \r\n extracting: ./test/EO_352419.png \r\n extracting: ./test/EO_2137.png \r\n extracting: ./test/EO_16441.png \r\n extracting: ./test/EO_245976.png \r\n extracting: ./test/EO_123487.png \r\n extracting: ./test/EO_415258.png \r\n extracting: ./test/EO_302146.png \r\n extracting: ./test/EO_400560.png \r\n extracting: ./test/EO_202558.png \r\n extracting: ./test/EO_352580.png \r\n extracting: ./test/EO_390587.png \r\n extracting: ./test/EO_295551.png \r\n extracting: ./test/EO_444996.png \r\n extracting: ./test/EO_63547.png \r\n extracting: ./test/EO_429265.png \r\n extracting: ./test/EO_437849.png \r\n extracting: ./test/EO_200905.png \r\n extracting: ./test/EO_209287.png \r\n extracting: ./test/EO_337114.png \r\n extracting: ./test/EO_121047.png \r\n extracting: ./test/EO_437349.png \r\n extracting: ./test/EO_47677.png \r\n extracting: ./test/EO_460917.png \r\n extracting: ./test/EO_1066.png \r\n extracting: ./test/EO_82184.png \r\n extracting: ./test/EO_375663.png \r\n extracting: ./test/EO_270369.png \r\n extracting: ./test/EO_196460.png \r\n extracting: ./test/EO_349886.png \r\n extracting: ./test/EO_363965.png \r\n extracting: ./test/EO_281722.png \r\n extracting: ./test/EO_117621.png \r\n extracting: ./test/EO_208475.png \r\n extracting: ./test/EO_301471.png \r\n extracting: ./test/EO_376267.png \r\n extracting: ./test/EO_241928.png \r\n extracting: ./test/EO_244271.png \r\n extracting: ./test/EO_354247.png \r\n extracting: ./test/EO_361467.png \r\n extracting: ./test/EO_11532.png \r\n extracting: ./test/EO_132612.png \r\n extracting: ./test/EO_343038.png \r\n extracting: ./test/EO_4368.png \r\n extracting: ./test/EO_319704.png \r\n extracting: ./test/EO_458247.png \r\n extracting: ./test/EO_130067.png \r\n extracting: ./test/EO_38865.png \r\n extracting: ./test/EO_426958.png \r\n extracting: ./test/EO_372107.png \r\n extracting: ./test/EO_122648.png \r\n extracting: ./test/EO_430232.png \r\n extracting: ./test/EO_473089.png \r\n extracting: ./test/EO_347921.png \r\n extracting: ./test/EO_468944.png \r\n extracting: ./test/EO_288116.png \r\n extracting: ./test/EO_330287.png \r\n extracting: ./test/EO_72580.png \r\n extracting: ./test/EO_358284.png \r\n extracting: ./test/EO_361623.png \r\n extracting: ./test/EO_283457.png \r\n extracting: ./test/EO_255877.png \r\n extracting: ./test/EO_243582.png \r\n extracting: ./test/EO_207446.png \r\n extracting: ./test/EO_15811.png \r\n extracting: ./test/EO_410428.png \r\n extracting: ./test/EO_294830.png \r\n extracting: ./test/EO_133128.png \r\n extracting: ./test/EO_64071.png \r\n extracting: ./test/EO_53479.png \r\n extracting: ./test/EO_412565.png \r\n extracting: ./test/EO_467933.png \r\n extracting: ./test/EO_444417.png \r\n extracting: ./test/EO_110955.png \r\n extracting: ./test/EO_145010.png \r\n extracting: ./test/EO_468939.png \r\n extracting: ./test/EO_360833.png \r\n extracting: ./test/EO_471572.png \r\n extracting: ./test/EO_359988.png \r\n extracting: ./test/EO_203419.png \r\n extracting: ./test/EO_123190.png \r\n extracting: ./test/EO_163044.png \r\n extracting: ./test/EO_210705.png \r\n extracting: ./test/EO_450979.png \r\n extracting: ./test/EO_453172.png \r\n extracting: ./test/EO_364425.png \r\n extracting: ./test/EO_429095.png \r\n extracting: ./test/EO_103494.png \r\n extracting: ./test/EO_308685.png \r\n extracting: ./test/EO_121236.png \r\n extracting: ./test/EO_16333.png \r\n extracting: ./test/EO_288057.png \r\n extracting: ./test/EO_313730.png \r\n extracting: ./test/EO_326825.png \r\n extracting: ./test/EO_326065.png \r\n extracting: ./test/EO_246515.png \r\n extracting: ./test/EO_298619.png \r\n extracting: ./test/EO_309230.png \r\n extracting: ./test/EO_164160.png \r\n extracting: ./test/EO_54542.png \r\n extracting: ./test/EO_431878.png \r\n extracting: ./test/EO_206661.png \r\n extracting: ./test/EO_432901.png \r\n extracting: ./test/EO_338309.png \r\n extracting: ./test/EO_294748.png \r\n extracting: ./test/EO_38611.png \r\n extracting: ./test/EO_404361.png \r\n extracting: ./test/EO_409893.png \r\n extracting: ./test/EO_444089.png \r\n extracting: ./test/EO_272178.png \r\n extracting: ./test/EO_26602.png \r\n extracting: ./test/EO_286231.png \r\n extracting: ./test/EO_429972.png \r\n extracting: ./test/EO_69985.png \r\n extracting: ./test/EO_394778.png \r\n extracting: ./test/EO_280013.png \r\n extracting: ./test/EO_284926.png \r\n extracting: ./test/EO_197586.png \r\n extracting: ./test/EO_428296.png \r\n extracting: ./test/EO_314881.png \r\n extracting: ./test/EO_173635.png \r\n extracting: ./test/EO_90285.png \r\n extracting: ./test/EO_411273.png \r\n extracting: ./test/EO_360499.png \r\n extracting: ./test/EO_59554.png \r\n extracting: ./test/EO_278549.png \r\n extracting: ./test/EO_293062.png \r\n extracting: ./test/EO_61276.png \r\n extracting: ./test/EO_274604.png \r\n extracting: ./test/EO_280469.png \r\n extracting: ./test/EO_165501.png \r\n extracting: ./test/EO_256266.png \r\n extracting: ./test/EO_339288.png \r\n extracting: ./test/EO_331412.png \r\n extracting: ./test/EO_334455.png \r\n extracting: ./test/EO_321787.png \r\n extracting: ./test/EO_95890.png \r\n extracting: ./test/EO_365850.png \r\n extracting: ./test/EO_428732.png \r\n extracting: ./test/EO_175772.png \r\n extracting: ./test/EO_349658.png \r\n extracting: ./test/EO_472149.png \r\n extracting: ./test/EO_170240.png \r\n extracting: ./test/EO_290132.png \r\n extracting: ./test/EO_48545.png \r\n extracting: ./test/EO_36266.png \r\n extracting: ./test/EO_432126.png \r\n extracting: ./test/EO_187149.png \r\n extracting: ./test/EO_75850.png \r\n extracting: ./test/EO_164491.png \r\n extracting: ./test/EO_248151.png \r\n extracting: ./test/EO_140401.png \r\n extracting: ./test/EO_370784.png \r\n extracting: ./test/EO_199233.png \r\n extracting: ./test/EO_256322.png \r\n extracting: ./test/EO_189622.png \r\n extracting: ./test/EO_360350.png \r\n extracting: ./test/EO_252301.png \r\n extracting: ./test/EO_6349.png \r\n extracting: ./test/EO_23326.png \r\n extracting: ./test/EO_462031.png \r\n extracting: ./test/EO_387078.png \r\n extracting: ./test/EO_74658.png \r\n extracting: ./test/EO_241967.png \r\n extracting: ./test/EO_358975.png \r\n extracting: ./test/EO_431011.png \r\n extracting: ./test/EO_376231.png \r\n extracting: ./test/EO_432798.png \r\n extracting: ./test/EO_196456.png \r\n extracting: ./test/EO_340395.png \r\n extracting: ./test/EO_141992.png \r\n extracting: ./test/EO_327515.png \r\n extracting: ./test/EO_66584.png \r\n extracting: ./test/EO_373646.png \r\n extracting: ./test/EO_60143.png \r\n extracting: ./test/EO_71587.png \r\n extracting: ./test/EO_13333.png \r\n extracting: ./test/EO_349052.png \r\n extracting: ./test/EO_110148.png \r\n extracting: ./test/EO_21954.png \r\n extracting: ./test/EO_70877.png \r\n extracting: ./test/EO_269133.png \r\n extracting: ./test/EO_293290.png \r\n extracting: ./test/EO_83998.png \r\n extracting: ./test/EO_99431.png \r\n extracting: ./test/EO_465512.png \r\n extracting: ./test/EO_380715.png \r\n extracting: ./test/EO_285193.png \r\n extracting: ./test/EO_386820.png \r\n extracting: ./test/EO_342193.png \r\n extracting: ./test/EO_115104.png \r\n extracting: ./test/EO_432137.png \r\n extracting: ./test/EO_365020.png \r\n extracting: ./test/EO_83591.png \r\n extracting: ./test/EO_466183.png \r\n extracting: ./test/EO_183287.png \r\n extracting: ./test/EO_167214.png \r\n extracting: ./test/EO_463276.png \r\n extracting: ./test/EO_268873.png \r\n extracting: ./test/EO_1102.png \r\n extracting: ./test/EO_14972.png \r\n extracting: ./test/EO_11660.png \r\n extracting: ./test/EO_275396.png \r\n extracting: ./test/EO_405067.png \r\n extracting: ./test/EO_67456.png \r\n extracting: ./test/EO_34678.png \r\n extracting: ./test/EO_335428.png \r\n extracting: ./test/EO_394572.png \r\n extracting: ./test/EO_117598.png \r\n extracting: ./test/EO_361029.png \r\n extracting: ./test/EO_115102.png \r\n extracting: ./test/EO_372567.png \r\n extracting: ./test/EO_25045.png \r\n extracting: ./test/EO_409350.png \r\n extracting: ./test/EO_403105.png \r\n extracting: ./test/EO_437675.png \r\n extracting: ./test/EO_429527.png \r\n extracting: ./test/EO_192295.png \r\n extracting: ./test/EO_444735.png \r\n extracting: ./test/EO_107465.png \r\n extracting: ./test/EO_372790.png \r\n extracting: ./test/EO_450460.png \r\n extracting: ./test/EO_140754.png \r\n extracting: ./test/EO_352460.png \r\n extracting: ./test/EO_12814.png \r\n extracting: ./test/EO_36239.png \r\n extracting: ./test/EO_421410.png \r\n extracting: ./test/EO_392000.png \r\n extracting: ./test/EO_428206.png \r\n extracting: ./test/EO_30207.png \r\n extracting: ./test/EO_64819.png \r\n extracting: ./test/EO_337442.png \r\n extracting: ./test/EO_304778.png \r\n extracting: ./test/EO_282331.png \r\n extracting: ./test/EO_188333.png \r\n extracting: ./test/EO_92948.png \r\n extracting: ./test/EO_356548.png \r\n extracting: ./test/EO_256964.png \r\n extracting: ./test/EO_288239.png \r\n extracting: ./test/EO_338235.png \r\n extracting: ./test/EO_467964.png \r\n extracting: ./test/EO_127548.png \r\n extracting: ./test/EO_301233.png \r\n extracting: ./test/EO_41757.png \r\n extracting: ./test/EO_356152.png \r\n extracting: ./test/EO_332510.png \r\n extracting: ./test/EO_146975.png \r\n extracting: ./test/EO_387372.png \r\n extracting: ./test/EO_353768.png \r\n extracting: ./test/EO_246446.png \r\n extracting: ./test/EO_390994.png \r\n extracting: ./test/EO_255212.png \r\n extracting: ./test/EO_71529.png \r\n extracting: ./test/EO_200627.png \r\n extracting: ./test/EO_470267.png \r\n extracting: ./test/EO_356827.png \r\n extracting: ./test/EO_73289.png \r\n extracting: ./test/EO_64049.png \r\n extracting: ./test/EO_444435.png \r\n extracting: ./test/EO_362544.png \r\n extracting: ./test/EO_199095.png \r\n extracting: ./test/EO_367604.png \r\n extracting: ./test/EO_134241.png \r\n extracting: ./test/EO_343548.png \r\n extracting: ./test/EO_439053.png \r\n extracting: ./test/EO_83784.png \r\n extracting: ./test/EO_332518.png \r\n extracting: ./test/EO_431328.png \r\n extracting: ./test/EO_463195.png \r\n extracting: ./test/EO_71736.png \r\n extracting: ./test/EO_147837.png \r\n extracting: ./test/EO_155605.png \r\n extracting: ./test/EO_409842.png \r\n extracting: ./test/EO_158458.png \r\n extracting: ./test/EO_322029.png \r\n extracting: ./test/EO_115826.png \r\n extracting: ./test/EO_37992.png \r\n extracting: ./test/EO_356360.png \r\n extracting: ./test/EO_145549.png \r\n extracting: ./test/EO_363422.png \r\n extracting: ./test/EO_372528.png \r\n extracting: ./test/EO_423094.png \r\n extracting: ./test/EO_401751.png \r\n extracting: ./test/EO_389486.png \r\n extracting: ./test/EO_200129.png \r\n extracting: ./test/EO_127004.png \r\n extracting: ./test/EO_416897.png \r\n extracting: ./test/EO_157368.png \r\n extracting: ./test/EO_54152.png \r\n extracting: ./test/EO_437811.png \r\n extracting: ./test/EO_450618.png \r\n extracting: ./test/EO_331939.png \r\n extracting: ./test/EO_370910.png \r\n extracting: ./test/EO_382277.png \r\n extracting: ./test/EO_91193.png \r\n extracting: ./test/EO_83566.png \r\n extracting: ./test/EO_430242.png \r\n extracting: ./test/EO_80853.png \r\n extracting: ./test/EO_284063.png \r\n extracting: ./test/EO_335380.png \r\n extracting: ./test/EO_321014.png \r\n extracting: ./test/EO_427998.png \r\n extracting: ./test/EO_161914.png \r\n extracting: ./test/EO_464695.png \r\n extracting: ./test/EO_278589.png \r\n extracting: ./test/EO_324683.png \r\n extracting: ./test/EO_124012.png \r\n extracting: ./test/EO_297607.png \r\n extracting: ./test/EO_252500.png \r\n extracting: ./test/EO_265188.png \r\n extracting: ./test/EO_44968.png \r\n extracting: ./test/EO_244315.png \r\n extracting: ./test/EO_160084.png \r\n extracting: ./test/EO_242595.png \r\n extracting: ./test/EO_356225.png \r\n extracting: ./test/EO_201814.png \r\n extracting: ./test/EO_326134.png \r\n extracting: ./test/EO_359311.png \r\n extracting: ./test/EO_65306.png \r\n extracting: ./test/EO_6117.png \r\n extracting: ./test/EO_247524.png \r\n extracting: ./test/EO_274820.png \r\n extracting: ./test/EO_385097.png \r\n extracting: ./test/EO_357550.png \r\n extracting: ./test/EO_365772.png \r\n extracting: ./test/EO_274614.png \r\n extracting: ./test/EO_17858.png \r\n extracting: ./test/EO_470449.png \r\n extracting: ./test/EO_273656.png \r\n extracting: ./test/EO_64525.png \r\n extracting: ./test/EO_40569.png \r\n extracting: ./test/EO_308353.png \r\n extracting: ./test/EO_246776.png \r\n extracting: ./test/EO_297922.png \r\n extracting: ./test/EO_243285.png \r\n extracting: ./test/EO_150480.png \r\n extracting: ./test/EO_270863.png \r\n extracting: ./test/EO_442219.png \r\n extracting: ./test/EO_355712.png \r\n extracting: ./test/EO_197127.png \r\n extracting: ./test/EO_287531.png \r\n extracting: ./test/EO_315813.png \r\n extracting: ./test/EO_329484.png \r\n extracting: ./test/EO_63414.png \r\n extracting: ./test/EO_334510.png \r\n extracting: ./test/EO_118390.png \r\n extracting: ./test/EO_423097.png \r\n extracting: ./test/EO_286629.png \r\n extracting: ./test/EO_248765.png \r\n extracting: ./test/EO_272413.png \r\n extracting: ./test/EO_286210.png \r\n extracting: ./test/EO_118149.png \r\n extracting: ./test/EO_164184.png \r\n extracting: ./test/EO_382873.png \r\n extracting: ./test/EO_431195.png \r\n extracting: ./test/EO_47970.png \r\n extracting: ./test/EO_431422.png \r\n extracting: ./test/EO_289479.png \r\n extracting: ./test/EO_126843.png \r\n extracting: ./test/EO_258496.png \r\n extracting: ./test/EO_287521.png \r\n extracting: ./test/EO_302833.png \r\n extracting: ./test/EO_278577.png \r\n extracting: ./test/EO_259747.png \r\n extracting: ./test/EO_192908.png \r\n extracting: ./test/EO_61116.png \r\n extracting: ./test/EO_84212.png \r\n extracting: ./test/EO_93027.png \r\n extracting: ./test/EO_390588.png \r\n extracting: ./test/EO_436126.png \r\n extracting: ./test/EO_118109.png \r\n extracting: ./test/EO_5287.png \r\n extracting: ./test/EO_387380.png \r\n extracting: ./test/EO_379011.png \r\n extracting: ./test/EO_356061.png \r\n extracting: ./test/EO_312548.png \r\n extracting: ./test/EO_386247.png \r\n extracting: ./test/EO_366916.png \r\n extracting: ./test/EO_256848.png \r\n extracting: ./test/EO_167951.png \r\n extracting: ./test/EO_302792.png \r\n extracting: ./test/EO_112660.png \r\n extracting: ./test/EO_375717.png \r\n extracting: ./test/EO_42691.png \r\n extracting: ./test/EO_40558.png \r\n extracting: ./test/EO_107809.png \r\n extracting: ./test/EO_75916.png \r\n extracting: ./test/EO_433095.png \r\n extracting: ./test/EO_396208.png \r\n extracting: ./test/EO_362376.png \r\n extracting: ./test/EO_210722.png \r\n extracting: ./test/EO_324371.png \r\n extracting: ./test/EO_431684.png \r\n extracting: ./test/EO_336631.png \r\n extracting: ./test/EO_307685.png \r\n extracting: ./test/EO_390999.png \r\n extracting: ./test/EO_392866.png \r\n extracting: ./test/EO_327946.png \r\n extracting: ./test/EO_362191.png \r\n extracting: ./test/EO_242369.png \r\n extracting: ./test/EO_182991.png \r\n extracting: ./test/EO_395689.png \r\n extracting: ./test/EO_2004.png \r\n extracting: ./test/EO_324875.png \r\n extracting: ./test/EO_127686.png \r\n extracting: ./test/EO_467368.png \r\n extracting: ./test/EO_430940.png \r\n extracting: ./test/EO_65839.png \r\n extracting: ./test/EO_344032.png \r\n extracting: ./test/EO_112879.png \r\n extracting: ./test/EO_123484.png \r\n extracting: ./test/EO_33936.png \r\n extracting: ./test/EO_454647.png \r\n extracting: ./test/EO_274787.png \r\n extracting: ./test/EO_279733.png \r\n extracting: ./test/EO_340508.png \r\n extracting: ./test/EO_357458.png \r\n extracting: ./test/EO_370267.png \r\n extracting: ./test/EO_250746.png \r\n extracting: ./test/EO_372085.png \r\n extracting: ./test/EO_251198.png \r\n extracting: ./test/EO_425918.png \r\n extracting: ./test/EO_267483.png \r\n extracting: ./test/EO_268369.png \r\n extracting: ./test/EO_360902.png \r\n extracting: ./test/EO_159085.png \r\n extracting: ./test/EO_417159.png \r\n extracting: ./test/EO_49871.png \r\n extracting: ./test/EO_283307.png \r\n extracting: ./test/EO_289138.png \r\n extracting: ./test/EO_317867.png \r\n extracting: ./test/EO_115167.png \r\n extracting: ./test/EO_14459.png \r\n extracting: ./test/EO_117352.png \r\n extracting: ./test/EO_444737.png \r\n extracting: ./test/EO_91423.png \r\n extracting: ./test/EO_113618.png \r\n extracting: ./test/EO_301876.png \r\n extracting: ./test/EO_39094.png \r\n extracting: ./test/EO_430659.png \r\n extracting: ./test/EO_466519.png \r\n extracting: ./test/EO_328261.png \r\n extracting: ./test/EO_41845.png \r\n extracting: ./test/EO_200579.png \r\n extracting: ./test/EO_430495.png \r\n extracting: ./test/EO_423944.png \r\n extracting: ./test/EO_73275.png \r\n extracting: ./test/EO_145832.png \r\n extracting: ./test/EO_466617.png \r\n extracting: ./test/EO_259014.png \r\n extracting: ./test/EO_88018.png \r\n extracting: ./test/EO_300807.png \r\n extracting: ./test/EO_462317.png \r\n extracting: ./test/EO_300220.png \r\n extracting: ./test/EO_202168.png \r\n extracting: ./test/EO_86541.png \r\n extracting: ./test/EO_125542.png \r\n extracting: ./test/EO_427323.png \r\n extracting: ./test/EO_89821.png \r\n extracting: ./test/EO_468395.png \r\n extracting: ./test/EO_246170.png \r\n extracting: ./test/EO_142888.png \r\n extracting: ./test/EO_420214.png \r\n extracting: ./test/EO_409915.png \r\n extracting: ./test/EO_375554.png \r\n extracting: ./test/EO_280304.png \r\n extracting: ./test/EO_83253.png \r\n extracting: ./test/EO_466231.png \r\n extracting: ./test/EO_119974.png \r\n extracting: ./test/EO_184255.png \r\n extracting: ./test/EO_52090.png \r\n extracting: ./test/EO_90308.png \r\n extracting: ./test/EO_117855.png \r\n extracting: ./test/EO_424265.png \r\n extracting: ./test/EO_76872.png \r\n extracting: ./test/EO_448742.png \r\n extracting: ./test/EO_107094.png \r\n extracting: ./test/EO_331353.png \r\n extracting: ./test/EO_248669.png \r\n extracting: ./test/EO_328236.png \r\n extracting: ./test/EO_193312.png \r\n extracting: ./test/EO_435495.png \r\n extracting: ./test/EO_189175.png \r\n extracting: ./test/EO_277219.png \r\n extracting: ./test/EO_209615.png \r\n extracting: ./test/EO_439942.png \r\n extracting: ./test/EO_396045.png \r\n extracting: ./test/EO_92256.png \r\n extracting: ./test/EO_115129.png \r\n extracting: ./test/EO_270620.png \r\n extracting: ./test/EO_38135.png \r\n extracting: ./test/EO_450818.png \r\n extracting: ./test/EO_73071.png \r\n extracting: ./test/EO_406977.png \r\n extracting: ./test/EO_114176.png \r\n extracting: ./test/EO_247554.png \r\n extracting: ./test/EO_273646.png \r\n extracting: ./test/EO_118660.png \r\n extracting: ./test/EO_378672.png \r\n extracting: ./test/EO_424144.png \r\n extracting: ./test/EO_422582.png \r\n extracting: ./test/EO_60411.png \r\n extracting: ./test/EO_424166.png \r\n extracting: ./test/EO_290492.png \r\n extracting: ./test/EO_102385.png \r\n extracting: ./test/EO_10264.png \r\n extracting: ./test/EO_111447.png \r\n extracting: ./test/EO_274846.png \r\n extracting: ./test/EO_18472.png \r\n extracting: ./test/EO_368255.png \r\n extracting: ./test/EO_201446.png \r\n extracting: ./test/EO_377362.png \r\n extracting: ./test/EO_285704.png \r\n extracting: ./test/EO_293294.png \r\n extracting: ./test/EO_269519.png \r\n extracting: ./test/EO_61050.png \r\n extracting: ./test/EO_432513.png \r\n extracting: ./test/EO_341239.png \r\n extracting: ./test/EO_123485.png \r\n extracting: ./test/EO_376878.png \r\n extracting: ./test/EO_283464.png \r\n extracting: ./test/EO_315242.png \r\n extracting: ./test/EO_42523.png \r\n extracting: ./test/EO_465056.png \r\n extracting: ./test/EO_277177.png \r\n extracting: ./test/EO_128173.png \r\n extracting: ./test/EO_125807.png \r\n extracting: ./test/EO_404447.png \r\n extracting: ./test/EO_180976.png \r\n extracting: ./test/EO_449683.png \r\n extracting: ./test/EO_249341.png \r\n extracting: ./test/EO_440377.png \r\n extracting: ./test/EO_385740.png \r\n extracting: ./test/EO_433569.png \r\n extracting: ./test/EO_427588.png \r\n extracting: ./test/EO_438715.png \r\n extracting: ./test/EO_74064.png \r\n extracting: ./test/EO_69199.png \r\n extracting: ./test/EO_319437.png \r\n extracting: ./test/EO_415756.png \r\n extracting: ./test/EO_40111.png \r\n extracting: ./test/EO_71731.png \r\n extracting: ./test/EO_434508.png \r\n extracting: ./test/EO_55922.png \r\n extracting: ./test/EO_251947.png \r\n extracting: ./test/EO_368496.png \r\n extracting: ./test/EO_456546.png \r\n extracting: ./test/EO_167170.png \r\n extracting: ./test/EO_360715.png \r\n extracting: ./test/EO_334254.png \r\n extracting: ./test/EO_385432.png \r\n extracting: ./test/EO_371529.png \r\n extracting: ./test/EO_443313.png \r\n extracting: ./test/EO_296733.png \r\n extracting: ./test/EO_364731.png \r\n extracting: ./test/EO_416047.png \r\n extracting: ./test/EO_267392.png \r\n extracting: ./test/EO_396780.png \r\n extracting: ./test/EO_308583.png \r\n extracting: ./test/EO_284407.png \r\n extracting: ./test/EO_192974.png \r\n"
],
[
"model = torch.load(\"45_kd_sar-teacher_eo-student_pretrain-on-sar/resnet34_kd114.pt\")\n\nimport pandas as pd\nfrom torch.utils import data\nfrom tqdm import tqdm\nfrom PIL import Image\nclass ImageData(data.Dataset):\n def __init__(self,df,dirpath,transform,test = False):\n self.df = df\n self.test = test\n self.dirpath = dirpath\n self.conv_to_tensor = transform\n #image data \n if not self.test:\n self.image_arr = np.asarray(str(self.dirpath)+'/'+self.df.iloc[:, 0]+'.png')\n else:\n self.image_arr = np.asarray(str(self.dirpath)+'/'+self.df.iloc[:, 0])\n \n #labels data\n if not self.test:\n self.label_df = self.df.iloc[:,1]\n \n # Calculate length of df\n self.data_len = len(self.df.index)\n\n def __len__(self):\n return self.data_len\n \n def __getitem__(self, idx):\n image_name = self.image_arr[idx]\n img = Image.open(image_name)#.convert('RGB')\n img_tensor = self.conv_to_tensor(img)\n if not self.test:\n image_labels = self.label_df[idx]\n label_tensor = torch.zeros((1, output_dim))\n for label in image_labels.split():\n label_tensor[0, int(label)] = 1\n image_label = torch.tensor(label_tensor,dtype= torch.float32)\n return (img_tensor,image_label.squeeze())\n return (img_tensor)\n\n \nBATCH_SIZE = 1\ntest_dir = \"./test\"\ntest_dir_ls = os.listdir(test_dir)\ntest_dir_ls.sort()\ntest_df = pd.DataFrame(test_dir_ls)\n\ntest_dataset = ImageData(test_df,test_dir,EO_data_transforms[\"valid_EO\"],test = True)\ntest_loader = data.DataLoader(dataset=test_dataset,batch_size=BATCH_SIZE,shuffle=False)\n\noutput_dim = 10\n\nDISABLE_TQDM = False\npredictions = np.zeros((len(test_dataset), output_dim))\ni = 0\nfor test_batch in tqdm(test_loader,disable = DISABLE_TQDM):\n test_batch = test_batch.to(device)\n batch_prediction = model(test_batch).detach().cpu().numpy()\n predictions[i * BATCH_SIZE:(i+1) * BATCH_SIZE, :] = batch_prediction\n i+=1\n \n",
"100%|██████████| 826/826 [00:13<00:00, 60.65it/s]\n"
],
[
"predictions[170]",
"_____no_output_____"
]
],
[
[
"### submission balance for class 0",
"_____no_output_____"
]
],
[
[
"m = nn.Softmax(dim=1)\npredictions_tensor = torch.from_numpy(predictions)\noutput_softmax = m(predictions_tensor)\n# output_softmax = output_softmax/output_softmax.sum()\n\npred = np.argmax(predictions,axis = 1)\n\nplot_ls = []\nidx = 0\nfor each_pred in pred:\n if each_pred == 0:\n plot_ls.append(output_softmax[idx][0].item())\n idx+=1\n# plot_ls\n \n\n# idx = 0\n# # print(output_softmax)\n# for i in pred:\n# # print(predictions_tensor[idx])\n# each_output_softmax = output_softmax[idx]/output_softmax[idx].sum()\n# print(each_output_softmax)\n# if i == 0:\n# new_list = set(predictions[idx])\n# new_list.remove(max(new_list))\n# index = predictions[idx].tolist().index(max(new_list))\n# # index = predictions[idx].index()\n# # print(index)\n\n \n# idx+=1\n\nimport matplotlib.pyplot as plt\n \nplt.hist(plot_ls, bins=80, histtype=\"stepfilled\", alpha=.8)\nplot_ls.sort()\nval = plot_ls[-85]\nprint(val)\nplt.vlines(val, ymin = 0, ymax = 22, colors = 'r')\n\n\n \n \n ",
"0.2121564553048015\n"
],
[
"# print(output_softmax)\nidx = 0\ncounter = 0\nfor i in pred:\n# print(predictions_tensor[idx])\n# each_output_softmax = output_softmax[idx]/output_softmax[idx].sum()\n# print(each_output_softmax)\n if i == 0 and output_softmax[idx][0] < val:\n \n new_list = set(predictions[idx])\n new_list.remove(max(new_list))\n index = predictions[idx].tolist().index(max(new_list))\n# index = predictions[idx].index()\n# print(index)\n pred[idx] = index\n output_softmax[idx][0] = -100.0\n counter += 1\n \n\n \n idx+=1\nprint(counter)",
"374\n"
]
],
[
[
"### submission balance for class 1",
"_____no_output_____"
]
],
[
[
"\n\n\n\nplot_ls = []\nidx = 0\nfor each_pred in pred:\n if each_pred == 1:\n plot_ls.append(output_softmax[idx][1].item())\n idx+=1\n# plot_ls\n \n\n# idx = 0\n# # print(output_softmax)\n# for i in pred:\n# # print(predictions_tensor[idx])\n# each_output_softmax = output_softmax[idx]/output_softmax[idx].sum()\n# print(each_output_softmax)\n# if i == 0:\n# new_list = set(predictions[idx])\n# new_list.remove(max(new_list))\n# index = predictions[idx].tolist().index(max(new_list))\n# # index = predictions[idx].index()\n# # print(index)\n\n \n# idx+=1\n\nimport matplotlib.pyplot as plt\n \nplt.hist(plot_ls, bins=80, histtype=\"stepfilled\", alpha=.8)\nplot_ls.sort()\nval = plot_ls[-85]\nprint(val)\nplt.vlines(val, ymin = 0, ymax = 22, colors = 'r')",
"0.21454049970403302\n"
],
[
"# print(output_softmax)\nidx = 0\ncounter = 0\nfor i in pred:\n# print(predictions_tensor[idx])\n# each_output_softmax = output_softmax[idx]/output_softmax[idx].sum()\n# print(each_output_softmax)\n if i == 1 and output_softmax[idx][1] < val:\n new_list = set(output_softmax[idx])\n new_list.remove(max(new_list))\n index = output_softmax[idx].tolist().index(max(new_list))\n# index = predictions[idx].index()\n# print(index)\n pred[idx] = index\n output_softmax[idx][1] = -100.0\n counter += 1\n \n\n \n idx+=1\nprint(counter)",
"154\n"
]
],
[
[
"### submission balance for class 2",
"_____no_output_____"
]
],
[
[
"\n\nplot_ls = []\nidx = 0\nfor each_pred in pred:\n if each_pred == 2:\n plot_ls.append(output_softmax[idx][2].item())\n idx+=1\n# plot_ls\n \n\n# idx = 0\n# # print(output_softmax)\n# for i in pred:\n# # print(predictions_tensor[idx])\n# each_output_softmax = output_softmax[idx]/output_softmax[idx].sum()\n# print(each_output_softmax)\n# if i == 0:\n# new_list = set(predictions[idx])\n# new_list.remove(max(new_list))\n# index = predictions[idx].tolist().index(max(new_list))\n# # index = predictions[idx].index()\n# # print(index)\n\n \n# idx+=1\n\nimport matplotlib.pyplot as plt\n \nplt.hist(plot_ls, bins=80, histtype=\"stepfilled\", alpha=.8)\nplot_ls.sort()\nval = plot_ls[-85]\nprint(val)\nplt.vlines(val, ymin = 0, ymax = 22, colors = 'r')",
"0.11162966531637959\n"
],
[
"# print(output_softmax)\nidx = 0\ncounter = 0\nfor i in pred:\n# print(predictions_tensor[idx])\n# each_output_softmax = output_softmax[idx]/output_softmax[idx].sum()\n# print(each_output_softmax)\n if i == 2 and output_softmax[idx][2] < val:\n new_list = set(output_softmax[idx])\n new_list.remove(max(new_list))\n index = output_softmax[idx].tolist().index(max(new_list))\n# index = predictions[idx].index()\n# print(index)\n pred[idx] = index\n output_softmax[idx][2] = -100.0\n counter += 1\n \n\n \n idx+=1\nprint(counter)",
"63\n"
]
],
[
[
"### submission balance for class 3",
"_____no_output_____"
]
],
[
[
"\nplot_ls = []\nidx = 0\nfor each_pred in pred:\n if each_pred == 3:\n plot_ls.append(output_softmax[idx][3].item())\n idx+=1\n# plot_ls\n \n\n# idx = 0\n# # print(output_softmax)\n# for i in pred:\n# # print(predictions_tensor[idx])\n# each_output_softmax = output_softmax[idx]/output_softmax[idx].sum()\n# print(each_output_softmax)\n# if i == 0:\n# new_list = set(predictions[idx])\n# new_list.remove(max(new_list))\n# index = predictions[idx].tolist().index(max(new_list))\n# # index = predictions[idx].index()\n# # print(index)\n\n \n# idx+=1\n\nimport matplotlib.pyplot as plt\n \nplt.hist(plot_ls, bins=80, histtype=\"stepfilled\", alpha=.8)\nplot_ls.sort()\nval = plot_ls[-85]\nprint(val)\nplt.vlines(val, ymin = 0, ymax = 22, colors = 'r')",
"0.09716976256894183\n"
],
[
"# print(output_softmax)\nidx = 0\ncounter = 0\nfor i in pred:\n# print(predictions_tensor[idx])\n# each_output_softmax = output_softmax[idx]/output_softmax[idx].sum()\n# print(each_output_softmax)\n if i == 3 and output_softmax[idx][3] < val:\n new_list = set(output_softmax[idx])\n new_list.remove(max(new_list))\n index = output_softmax[idx].tolist().index(max(new_list))\n# index = predictions[idx].index()\n# print(index)\n pred[idx] = index\n output_softmax[idx][3] = -100.0\n counter += 1\n \n\n \n idx+=1\nprint(counter)",
"47\n"
]
],
[
[
"### submission balance for class 4\n",
"_____no_output_____"
]
],
[
[
"\n# plot_ls = []\n# idx = 0\n# for each_pred in pred:\n# if each_pred == 4:\n# plot_ls.append(output_softmax[idx][4].item())\n# idx+=1\n# # plot_ls\n \n\n# # idx = 0\n# # # print(output_softmax)\n# # for i in pred:\n# # # print(predictions_tensor[idx])\n# # each_output_softmax = output_softmax[idx]/output_softmax[idx].sum()\n# # print(each_output_softmax)\n# # if i == 0:\n# # new_list = set(predictions[idx])\n# # new_list.remove(max(new_list))\n# # index = predictions[idx].tolist().index(max(new_list))\n# # # index = predictions[idx].index()\n# # # print(index)\n\n \n# # idx+=1\n\n# import matplotlib.pyplot as plt\n \n# plt.hist(plot_ls, bins=80, histtype=\"stepfilled\", alpha=.8)\n# plot_ls.sort()\n# val = plot_ls[-85]\n# print(val)\n# plt.vlines(val, ymin = 0, ymax = 22, colors = 'r')",
"_____no_output_____"
],
[
"# # print(output_softmax)\n# idx = 0\n# counter = 0\n# for i in pred:\n# # print(predictions_tensor[idx])\n# # each_output_softmax = output_softmax[idx]/output_softmax[idx].sum()\n# # print(each_output_softmax)\n# if i == 4 and output_softmax[idx][4] < val:\n# new_list = set(output_softmax[idx])\n# new_list.remove(max(new_list))\n# index = output_softmax[idx].tolist().index(max(new_list))\n# # index = predictions[idx].index()\n# # print(index)\n# pred[idx] = index\n# output_softmax[idx][4] = -100.0\n# counter += 1\n \n\n \n# idx+=1\n# print(counter)",
"_____no_output_____"
]
],
[
[
"### submission balance for class 5\n",
"_____no_output_____"
]
],
[
[
"# plot_ls = []\n# idx = 0\n# for each_pred in pred:\n# if each_pred == 5:\n# plot_ls.append(output_softmax[idx][5].item())\n# idx+=1\n# # plot_ls\n \n\n# # idx = 0\n# # # print(output_softmax)\n# # for i in pred:\n# # # print(predictions_tensor[idx])\n# # each_output_softmax = output_softmax[idx]/output_softmax[idx].sum()\n# # print(each_output_softmax)\n# # if i == 0:\n# # new_list = set(predictions[idx])\n# # new_list.remove(max(new_list))\n# # index = predictions[idx].tolist().index(max(new_list))\n# # # index = predictions[idx].index()\n# # # print(index)\n\n \n# # idx+=1\n\n# import matplotlib.pyplot as plt\n \n# plt.hist(plot_ls, bins=80, histtype=\"stepfilled\", alpha=.8)\n# plot_ls.sort()\n# val = plot_ls[-85]\n# print(val)\n# plt.vlines(val, ymin = 0, ymax = 22, colors = 'r')",
"_____no_output_____"
],
[
"# # print(output_softmax)\n# idx = 0\n# counter = 0\n# for i in pred:\n# # print(predictions_tensor[idx])\n# # each_output_softmax = output_softmax[idx]/output_softmax[idx].sum()\n# # print(each_output_softmax)\n# if i == 5 and output_softmax[idx][5] < val:\n# new_list = set(output_softmax[idx])\n# new_list.remove(max(new_list))\n# index = output_softmax[idx].tolist().index(max(new_list))\n# # index = predictions[idx].index()\n# # print(index)\n# pred[idx] = index\n# output_softmax[idx][5] = -100.0\n# counter += 1\n \n\n \n# idx+=1\n# print(counter)",
"_____no_output_____"
]
],
[
[
"### submission balance for class 6 not arrange\n",
"_____no_output_____"
]
],
[
[
"# plot_ls = []\n# idx = 0\n# for each_pred in pred:\n# if each_pred == 6:\n# plot_ls.append(output_softmax[idx][6].item())\n# idx+=1\n# # plot_ls\n \n\n# # idx = 0\n# # # print(output_softmax)\n# # for i in pred:\n# # # print(predictions_tensor[idx])\n# # each_output_softmax = output_softmax[idx]/output_softmax[idx].sum()\n# # print(each_output_softmax)\n# # if i == 0:\n# # new_list = set(predictions[idx])\n# # new_list.remove(max(new_list))\n# # index = predictions[idx].tolist().index(max(new_list))\n# # # index = predictions[idx].index()\n# # # print(index)\n\n \n# # idx+=1\n\n# import matplotlib.pyplot as plt\n \n# plt.hist(plot_ls, bins=80, histtype=\"stepfilled\", alpha=.8)\n# plot_ls.sort()\n# val = plot_ls[-85]\n# print(val)\n# plt.vlines(val, ymin = 0, ymax = 22, colors = 'r')\n\n# # print(output_softmax)\n# idx = 0\n# counter = 0\n# for i in pred:\n# # print(predictions_tensor[idx])\n# # each_output_softmax = output_softmax[idx]/output_softmax[idx].sum()\n# # print(each_output_softmax)\n# if i == 6 and output_softmax[idx][6] < val:\n# new_list = set(output_softmax[idx])\n# new_list.remove(max(new_list))\n# index = output_softmax[idx].tolist().index(max(new_list))\n# # index = predictions[idx].index()\n# # print(index)\n# pred[idx] = index\n# output_softmax[idx][6] = -100.0\n# counter += 1\n \n\n \n# idx+=1\n# print(counter)",
"_____no_output_____"
],
[
"len(plot_ls)",
"_____no_output_____"
]
],
[
[
"### submission balance for class 7\n",
"_____no_output_____"
]
],
[
[
"# plot_ls = []\n# idx = 0\n# for each_pred in pred:\n# if each_pred == 7:\n# plot_ls.append(output_softmax[idx][7].item())\n# idx+=1\n# # plot_ls\n \n\n# # idx = 0\n# # # print(output_softmax)\n# # for i in pred:\n# # # print(predictions_tensor[idx])\n# # each_output_softmax = output_softmax[idx]/output_softmax[idx].sum()\n# # print(each_output_softmax)\n# # if i == 0:\n# # new_list = set(predictions[idx])\n# # new_list.remove(max(new_list))\n# # index = predictions[idx].tolist().index(max(new_list))\n# # # index = predictions[idx].index()\n# # # print(index)\n\n \n# # idx+=1\n\n# import matplotlib.pyplot as plt\n \n# plt.hist(plot_ls, bins=80, histtype=\"stepfilled\", alpha=.8)\n# plot_ls.sort()\n# val = plot_ls[-85]\n# print(val)\n# plt.vlines(val, ymin = 0, ymax = 22, colors = 'r')\n\n# # print(output_softmax)\n# idx = 0\n# counter = 0\n# for i in pred:\n# # print(predictions_tensor[idx])\n# # each_output_softmax = output_softmax[idx]/output_softmax[idx].sum()\n# # print(each_output_softmax)\n# if i == 7 and output_softmax[idx][7] < val:\n# new_list = set(output_softmax[idx])\n# new_list.remove(max(new_list))\n# index = output_softmax[idx].tolist().index(max(new_list))\n# # index = predictions[idx].index()\n# # print(index)\n# pred[idx] = index\n# output_softmax[idx][7] = -100.0\n# counter += 1\n \n\n \n# idx+=1\n# print(counter)",
"_____no_output_____"
]
],
[
[
"### submission balance for class 8\n",
"_____no_output_____"
],
[
"### submission balance for class 9\n",
"_____no_output_____"
]
],
[
[
"\n# plot_ls = []\n# idx = 0\n# for each_pred in pred:\n# if each_pred == 9:\n# plot_ls.append(output_softmax[idx][9].item())\n# idx+=1\n# # plot_ls\n \n\n# # idx = 0\n# # # print(output_softmax)\n# # for i in pred:\n# # # print(predictions_tensor[idx])\n# # each_output_softmax = output_softmax[idx]/output_softmax[idx].sum()\n# # print(each_output_softmax)\n# # if i == 0:\n# # new_list = set(predictions[idx])\n# # new_list.remove(max(new_list))\n# # index = predictions[idx].tolist().index(max(new_list))\n# # # index = predictions[idx].index()\n# # # print(index)\n\n \n# # idx+=1\n\n# import matplotlib.pyplot as plt\n \n# plt.hist(plot_ls, bins=80, histtype=\"stepfilled\", alpha=.8)\n# plot_ls.sort()\n# val = plot_ls[-85]\n# print(val)\n# plt.vlines(val, ymin = 0, ymax = 22, colors = 'r')\n",
"_____no_output_____"
],
[
"\npred",
"_____no_output_____"
],
[
"# pred = np.argmax(predictions,axis = 1)\npred_list = []\nfor i in range(len(pred)):\n result = [pred[i]]\n pred_list.append(result)\npred_list\n\n",
"_____no_output_____"
],
[
"predicted_class_idx = pred_list\n\ntest_df['class_id'] = predicted_class_idx\ntest_df['class_id'] = test_df['class_id'].apply(lambda x : ' '.join(map(str,list(x))))\ntest_df = test_df.rename(columns={0: 'image_id'})\ntest_df['image_id'] = test_df['image_id'].apply(lambda x : x.split('.')[0])\ntest_df\n",
"_____no_output_____"
],
[
"for (idx, row) in test_df.iterrows():\n row.image_id = row.image_id.split(\"_\")[1]\n",
"_____no_output_____"
],
[
"for k in range(10):\n i = 0\n for (idx, row) in test_df.iterrows():\n if row.class_id == str(k):\n i+=1\n print(i)",
"113\n103\n95\n85\n132\n135\n21\n119\n18\n5\n"
],
[
"test_df",
"_____no_output_____"
],
[
"test_df.to_csv('results.csv',index = False)",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbeb8dfd2e06a2cd3c42bc4fb7bc7c76b6e2422e
| 18,969 |
ipynb
|
Jupyter Notebook
|
DataAnalysis/.ipynb_checkpoints/Analysis-checkpoint.ipynb
|
includeavaneesh/Song-Genre-Identifier
|
4cae465f18e9e88426b211eb297c70dd24499e77
|
[
"MIT"
] | null | null | null |
DataAnalysis/.ipynb_checkpoints/Analysis-checkpoint.ipynb
|
includeavaneesh/Song-Genre-Identifier
|
4cae465f18e9e88426b211eb297c70dd24499e77
|
[
"MIT"
] | null | null | null |
DataAnalysis/.ipynb_checkpoints/Analysis-checkpoint.ipynb
|
includeavaneesh/Song-Genre-Identifier
|
4cae465f18e9e88426b211eb297c70dd24499e77
|
[
"MIT"
] | null | null | null | 32.150847 | 124 | 0.391903 |
[
[
[
"import pandas as pd\nimport numpy as np\nimport json\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"f = open(\"C:/Navneet/College/Fall21/NLP/Song-Genre-Identifier/DataProcessor/dataset/processed/songList.json\")\ndata = json.load(f)\nf.close()\ndf = pd.json_normalize(data['songs'])\ndf.head()",
"_____no_output_____"
],
[
"df['Lyrics'][0][0][1]",
"_____no_output_____"
],
[
"def totalWords(arr):\n tot = 0\n freq = {}\n print(arr)\n for x in arr:\n tot = tot + len(x)\n for y in x:\n if(y in freq):\n freq[y]=freq[y]+1\n else:\n freq[y]=1\n WordMax = max(zip(freq.values(), freq.keys()))[1]\n WordFreq = max(zip(freq.values(), freq.keys()))[0]\n return tot, len(freq), WordMax, WordFreq",
"_____no_output_____"
],
[
"lengthArray = []\nuniqueArray = []\nmostFrequentWord = []\nfreqOfMFW = []\nfor index,row in df.iterrows():\n n,freq,mfw,fmfw = totalWords(row.Lyrics)\n lengthArray.append(n)\n uniqueArray.append(freq)\n mostFrequentWord.append(mfw)\n freqOfMFW.append(fmfw)\ndf['TotalWords'] = pd.Series(lengthArray, index=df.index)\ndf['UniqueWords'] = pd.Series(uniqueArray, index = df.index)\ndf['MostFrequentWords'] = pd.Series(mostFrequentWord, index=df.index)\ndf['FrequencyOfMFW'] = pd.Series(freqOfMFW, index=df.index)",
"_____no_output_____"
],
[
"df['MostFrequentWords']",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
],
[
"def sentenceProcessing(arr):\n freq = {}\n total = 0\n for x in arr:\n sent = ' '.join(x)\n if sent in freq:\n freq[sent] = freq[sent]+1\n else:\n freq[sent] = 1\n total = total+1\n sentMax = max(zip(freq.values(), freq.keys()))[1]\n sentCount = max(zip(freq.values(), freq.keys()))[0]\n return total, sentMax, sentCount",
"_____no_output_____"
],
[
"sentM = []\nsentC = []\ntotalS = []\nfor index,row in df.iterrows():\n total, sentMax, sentCount = sentenceProcessing(row['Lyrics'])\n sentM.append(sentMax)\n sentC.append(sentCount)\n totalS.append(total)\ndf['TotalSentences'] = pd.Series(totalS, index = df.index)\ndf['MostFrequentSentence'] = pd.Series(sentM, index = df.index)\ndf['CountMostSentence'] = pd.Series(sentC, index = df.index)",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
],
[
"df.to_json(r'C:\\Navneet\\College\\Fall21\\NLP\\Song-Genre-Identifier\\DataAnalysis\\LyricsAnalysis\\dataset.json')\ndf.to_csv(r'C:\\Navneet\\College\\Fall21\\NLP\\Song-Genre-Identifier\\DataAnalysis\\LyricsAnalysis\\dataset.csv')",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbeba1bbcc01a68a9312da06fb78a7b0daafd1b1
| 39,145 |
ipynb
|
Jupyter Notebook
|
Linear Regression With Pyspark On Databricks.ipynb
|
Hacker-Davinci/Pyspark_Basics_and_Projects
|
61b00b6619d56ecc298fb30ccbfe0ac364e59bf4
|
[
"MIT"
] | null | null | null |
Linear Regression With Pyspark On Databricks.ipynb
|
Hacker-Davinci/Pyspark_Basics_and_Projects
|
61b00b6619d56ecc298fb30ccbfe0ac364e59bf4
|
[
"MIT"
] | null | null | null |
Linear Regression With Pyspark On Databricks.ipynb
|
Hacker-Davinci/Pyspark_Basics_and_Projects
|
61b00b6619d56ecc298fb30ccbfe0ac364e59bf4
|
[
"MIT"
] | null | null | null | 19,572.5 | 39,144 | 0.516235 |
[
[
[
"## Overview\n\nThis notebook will show you how to create and query a table or DataFrame that you uploaded to DBFS. [DBFS](https://docs.databricks.com/user-guide/dbfs-databricks-file-system.html) is a Databricks File System that allows you to store data for querying inside of Databricks. This notebook assumes that you have a file already inside of DBFS that you would like to read from.\n\nThis notebook is written in **Python** so the default cell type is Python. However, you can use different languages by using the `%LANGUAGE` syntax. Python, Scala, SQL, and R are all supported.",
"_____no_output_____"
]
],
[
[
"# File location and type\nfile_location = \"/FileStore/tables/tips.csv\"\nfile_type = \"csv\"\n\n# The applied options are for CSV files. For other file types, these will be ignored.\ndf =spark.read.csv(file_location,header=True,inferSchema=True)\ndf.show()",
"_____no_output_____"
],
[
"df.printSchema()",
"_____no_output_____"
],
[
"df.columns",
"_____no_output_____"
],
[
"### Handling Categorical Features\nfrom pyspark.ml.feature import StringIndexer",
"_____no_output_____"
],
[
"indexer=StringIndexer(inputCol=\"sex\",outputCol=\"sex_indexed\")\ndf_r=indexer.fit(df).transform(df)\ndf_r.show()",
"_____no_output_____"
],
[
"indexer=StringIndexer(inputCols=[\"smoker\",\"day\",\"time\"],outputCols=[\"smoker_indexed\",\"day_indexed\",\n \"time_index\"])\ndf_r=indexer.fit(df_r).transform(df_r)\ndf_r.show()",
"_____no_output_____"
],
[
"df_r.columns",
"_____no_output_____"
],
[
"from pyspark.ml.feature import VectorAssembler\nfeatureassembler=VectorAssembler(inputCols=['tip','size','sex_indexed','smoker_indexed','day_indexed',\n 'time_index'],outputCol=\"Independent Features\")\noutput=featureassembler.transform(df_r)",
"_____no_output_____"
],
[
"output.select('Independent Features').show()",
"_____no_output_____"
],
[
"output.show()",
"_____no_output_____"
],
[
"finalized_data=output.select(\"Independent Features\",\"total_bill\")",
"_____no_output_____"
],
[
"type(finalized_data)",
"_____no_output_____"
],
[
"finalized_data.show()",
"_____no_output_____"
],
[
"from pyspark.ml.regression import LinearRegression\n##train test split\ntrain_data,test_data=finalized_data.randomSplit([0.75,0.25])\nregressor=LinearRegression(featuresCol='Independent Features', labelCol='total_bill')\nregressor=regressor.fit(train_data)",
"_____no_output_____"
],
[
"regressor.coefficients",
"_____no_output_____"
],
[
"regressor.intercept # the initialized value",
"_____no_output_____"
],
[
"### Predictions\npred_results=regressor.evaluate(test_data)",
"_____no_output_____"
],
[
"print(type(regressor))\nprint(type(pred_results))\nprint(type(pred_results.predictions))",
"_____no_output_____"
],
[
"## Final comparison\npred_results.predictions.show()",
"_____no_output_____"
],
[
"### PErformance Metrics\npred_results.r2,pred_results.meanAbsoluteError,pred_results.meanSquaredError",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbebb313595bfd7ad5127049203a51c4b21a04d8
| 99,763 |
ipynb
|
Jupyter Notebook
|
II Machine Learning & Deep Learning/01_Why Neural Networks Deeply Learn a Mathematical Formula/04session.ipynb
|
algoriv92/machine-learning-program
|
47969691c4e1dcd54c91f47838aa7a4421a0c69c
|
[
"MIT"
] | null | null | null |
II Machine Learning & Deep Learning/01_Why Neural Networks Deeply Learn a Mathematical Formula/04session.ipynb
|
algoriv92/machine-learning-program
|
47969691c4e1dcd54c91f47838aa7a4421a0c69c
|
[
"MIT"
] | null | null | null |
II Machine Learning & Deep Learning/01_Why Neural Networks Deeply Learn a Mathematical Formula/04session.ipynb
|
algoriv92/machine-learning-program
|
47969691c4e1dcd54c91f47838aa7a4421a0c69c
|
[
"MIT"
] | null | null | null | 36.356778 | 570 | 0.395868 |
[
[
[
"<font size=\"+5\">#04. Why Neural Networks Deeply Learn a Mathematical Formula?</font>",
"_____no_output_____"
],
[
"- Book + Private Lessons [Here ↗](https://sotastica.com/reservar)\n- Subscribe to my [Blog ↗](https://blog.pythonassembly.com/)\n- Let's keep in touch on [LinkedIn ↗](www.linkedin.com/in/jsulopz) 😄",
"_____no_output_____"
],
[
"# Machine Learning, what does it mean?",
"_____no_output_____"
],
[
"> - The Machine Learns...\n>\n> But, **what does it learn?**",
"_____no_output_____"
]
],
[
[
"%%HTML\n<blockquote class=\"twitter-tweet\" data-lang=\"en\"><p lang=\"en\" dir=\"ltr\">Machine Learning, what does it mean? ⏯<br><br>· The machine learns...<br><br>Ha ha, not funny! 🤨 What does it learn?<br><br>· A mathematical equation. For example: <a href=\"https://t.co/sjtq9F2pq7\">pic.twitter.com/sjtq9F2pq7</a></p>— Jesús López (@sotastica) <a href=\"https://twitter.com/sotastica/status/1449735653328031745?ref_src=twsrc%5Etfw\">October 17, 2021</a></blockquote> <script async src=\"https://platform.twitter.com/widgets.js\" charset=\"utf-8\"></script>",
"_____no_output_____"
]
],
[
[
"# How does the Machine Learn?",
"_____no_output_____"
],
[
"## In a Linear Regression",
"_____no_output_____"
]
],
[
[
"%%HTML\n<iframe width=\"560\" height=\"315\" src=\"https://www.youtube.com/embed/Ht3rYS-JilE\" title=\"YouTube video player\" frameborder=\"0\" allow=\"accelerometer; autoplay; clipboard-write; encrypted-media; gyroscope; picture-in-picture\" allowfullscreen></iframe>",
"_____no_output_____"
]
],
[
[
"## In a Neural Network",
"_____no_output_____"
]
],
[
[
"%%HTML\n<iframe width=\"560\" height=\"315\" src=\"https://www.youtube.com/embed/IHZwWFHWa-w?start=329\" title=\"YouTube video player\" frameborder=\"0\" allow=\"accelerometer; autoplay; clipboard-write; encrypted-media; gyroscope; picture-in-picture\" allowfullscreen></iframe>",
"_____no_output_____"
]
],
[
[
"A Practical Example → [Tesla Autopilot](https://www.tesla.com/AI)",
"_____no_output_____"
],
[
"An Example where It Fails → [Tesla Confuses Moon with Semaphore](https://twitter.com/Carnage4Life/status/1418920100086784000?s=20)",
"_____no_output_____"
],
[
"# Load the Data",
"_____no_output_____"
],
[
"> - Simply execute the following lines of code to load the data.\n> - This dataset contains **statistics about Car Accidents** (columns)\n> - In each one of **USA States** (rows)",
"_____no_output_____"
],
[
"https://www.kaggle.com/fivethirtyeight/fivethirtyeight-bad-drivers-dataset/",
"_____no_output_____"
]
],
[
[
"import seaborn as sns\n\ndf = sns.load_dataset(name='car_crashes', index_col='abbrev')\ndf.sample(5)",
"_____no_output_____"
]
],
[
[
"# Neural Network Concepts in Python",
"_____no_output_____"
],
[
"## Initializing the `Weights`",
"_____no_output_____"
],
[
"> - https://keras.io/api/layers/initializers/",
"_____no_output_____"
],
[
"### How to `kernel_initializer` the weights?",
"_____no_output_____"
],
[
"$$\naccidents = speeding \\cdot w_1 + alcohol \\cdot w_2 \\ + ... + \\ ins\\_losses \\cdot w_7\n$$",
"_____no_output_____"
]
],
[
[
"from tensorflow.keras import Sequential, Input\nfrom tensorflow.keras.layers import Dense",
"_____no_output_____"
],
[
"df.shape",
"_____no_output_____"
],
[
"model = Sequential()\nmodel.add(layer=Input(shape=(6,)))\nmodel.add(layer=Dense(units=3, kernel_initializer='zeros'))\nmodel.add(layer=Dense(units=1))",
"Metal device set to: Apple M1\n"
]
],
[
[
"#### Make a Prediction with the Neural Network",
"_____no_output_____"
],
[
"> - Can we make a prediction for for `Washington DC` accidents\n> - With the already initialized Mathematical Equation?",
"_____no_output_____"
]
],
[
[
"X = df.drop(columns='total')\ny = df.total",
"_____no_output_____"
],
[
"AL = X[:1]",
"_____no_output_____"
],
[
"AL",
"_____no_output_____"
]
],
[
[
"#### Observe the numbers for the `weights`",
"_____no_output_____"
]
],
[
[
"model.get_weights()",
"_____no_output_____"
]
],
[
[
"#### Predictions vs Reality",
"_____no_output_____"
],
[
"> 1. Calculate the Predicted Accidents and\n> 2. Compare it with the Real Total Accidents",
"_____no_output_____"
]
],
[
[
"model.predict(x=AL)",
"2022-01-05 09:59:02.342564: I tensorflow/compiler/mlir/mlir_graph_optimization_pass.cc:185] None of the MLIR Optimization Passes are enabled (registered 2)\n2022-01-05 09:59:02.343180: W tensorflow/core/platform/profile_utils/cpu_utils.cc:128] Failed to get CPU frequency: 0 Hz\n2022-01-05 09:59:02.375183: I tensorflow/core/grappler/optimizers/custom_graph_optimizer_registry.cc:112] Plugin optimizer for device_type GPU is enabled.\n"
]
],
[
[
"#### `fit()` the `model` and compare again",
"_____no_output_____"
]
],
[
[
"model.compile(loss='mse', metrics=['mse'])",
"_____no_output_____"
],
[
"model.fit(X, y, epochs=500, verbose=1)",
"Epoch 1/500\n"
]
],
[
[
"##### Observe the numbers for the `weights`",
"_____no_output_____"
]
],
[
[
"model.get_weights()",
"_____no_output_____"
]
],
[
[
"##### Predictions vs Reality",
"_____no_output_____"
],
[
"> 1. Calculate the Predicted Accidents and\n> 2. Compare it with the Real Total Accidents",
"_____no_output_____"
]
],
[
[
"y_pred = model.predict(X)",
"2022-01-05 09:59:29.374264: I tensorflow/core/grappler/optimizers/custom_graph_optimizer_registry.cc:112] Plugin optimizer for device_type GPU is enabled.\n"
],
[
"dfsel = df[['total']].copy()\ndfsel['pred_zeros_after_fit'] = y_pred\ndfsel.head()",
"_____no_output_____"
],
[
"mse = ((dfsel.total - dfsel.pred_zeros_after_fit)**2).mean()\nmse",
"_____no_output_____"
]
],
[
[
"### How to `kernel_initializer` the weights to 1?",
"_____no_output_____"
]
],
[
[
"dfsel['pred_ones_after_fit'] = y_pred\ndfsel.head()",
"_____no_output_____"
],
[
"mse = ((dfsel.total - dfsel.pred_ones_after_fit)**2).mean()\nmse",
"_____no_output_____"
]
],
[
[
"### How to `kernel_initializer` the weights to `glorot_uniform` (default)?",
"_____no_output_____"
],
[
"## Play with the Activation Function",
"_____no_output_____"
],
[
"> - https://keras.io/api/layers/activations/",
"_____no_output_____"
]
],
[
[
"%%HTML\n<iframe width=\"560\" height=\"315\" src=\"https://www.youtube.com/embed/IHZwWFHWa-w?start=558\" title=\"YouTube video player\" frameborder=\"0\" allow=\"accelerometer; autoplay; clipboard-write; encrypted-media; gyroscope; picture-in-picture\" allowfullscreen></iframe>",
"_____no_output_____"
]
],
[
[
"### Use `sigmoid` activation in last layer",
"_____no_output_____"
]
],
[
[
"model = Sequential()\nmodel.add(layer=Input(shape=(6,)))\nmodel.add(layer=Dense(units=3, kernel_initializer='glorot_uniform'))\nmodel.add(layer=Dense(units=1, activation='sigmoid'))",
"_____no_output_____"
],
[
"model.compile(loss='mse', metrics=['mse'])",
"_____no_output_____"
]
],
[
[
"#### `fit()` the Model",
"_____no_output_____"
]
],
[
[
"model.fit(X, y, epochs=500, verbose=0)",
"_____no_output_____"
]
],
[
[
"#### Predictions vs Reality",
"_____no_output_____"
],
[
"> 1. Calculate the Predicted Accidents and\n> 2. Compare it with the Real Total Accidents",
"_____no_output_____"
]
],
[
[
"y_pred = model.predict(X)",
"_____no_output_____"
],
[
"dfsel['pred_sigmoid'] = y_pred\ndfsel.head()",
"_____no_output_____"
],
[
"mse = ((dfsel.total - dfsel.pred_sigmoid)**2).mean()\nmse",
"_____no_output_____"
]
],
[
[
"#### Observe the numbers for the `weights`\n\n> - Have they changed?",
"_____no_output_____"
]
],
[
[
"model.get_weights()",
"_____no_output_____"
]
],
[
[
"### Use `linear` activation in last layer",
"_____no_output_____"
],
[
"### Use `tanh` activation in last layer",
"_____no_output_____"
],
[
"### Use `relu` activation in last layer",
"_____no_output_____"
],
[
"### How are the predictions changing? Why?",
"_____no_output_____"
],
[
"## Optimizer",
"_____no_output_____"
],
[
"> - https://keras.io/api/optimizers/#available-optimizers",
"_____no_output_____"
],
[
"Optimizers comparison in GIF → https://mlfromscratch.com/optimizers-explained/#adam",
"_____no_output_____"
],
[
"Tesla's Neural Network Models is composed of 48 models trainned in 70.000 hours of GPU → https://tesla.com/ai",
"_____no_output_____"
],
[
"1 Year with a 8 GPU Computer → https://twitter.com/thirdrowtesla/status/1252723358342377472",
"_____no_output_____"
],
[
"### Use Gradient Descent `SGD`",
"_____no_output_____"
]
],
[
[
"model = Sequential()\nmodel.add(layer=Input(shape=(6,)))\nmodel.add(layer=Dense(units=3, kernel_initializer='glorot_uniform'))\nmodel.add(layer=Dense(units=1, activation='sigmoid'))",
"_____no_output_____"
]
],
[
[
"#### `compile()` the model",
"_____no_output_____"
]
],
[
[
"model.compile(optimizer='sgd', loss='mse', metrics=['mse'])",
"_____no_output_____"
]
],
[
[
"#### `fit()` the Model",
"_____no_output_____"
]
],
[
[
"history = model.fit(X, y, epochs=500, verbose=0)",
"_____no_output_____"
]
],
[
[
"#### Predictions vs Reality",
"_____no_output_____"
],
[
"> 1. Calculate the Predicted Accidents and\n> 2. Compare it with the Real Total Accidents",
"_____no_output_____"
]
],
[
[
"y_pred = model.predict(X)",
"_____no_output_____"
],
[
"dfsel['pred_gsd'] = y_pred\ndfsel.head()",
"_____no_output_____"
],
[
"mse = ((dfsel.total - dfsel.pred_sgd)**2).mean()\nmse",
"_____no_output_____"
]
],
[
[
"#### Observe the numbers for the `weights`\n\n> - Have they changed?",
"_____no_output_____"
]
],
[
[
"model.get_weights()",
"_____no_output_____"
]
],
[
[
"\n#### View History",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"plt.plot(history.history['loss'])\nplt.plot(history.history['val_loss'])\nplt.title('model loss')\nplt.ylabel('loss')\nplt.xlabel('epoch')\nplt.legend(['train', 'val'], loc='upper left')\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Use `ADAM`",
"_____no_output_____"
],
[
"### Use `RMSPROP`",
"_____no_output_____"
],
[
"### Does it take different times to get the best accuracy? Why?",
"_____no_output_____"
],
[
"## Loss Functions",
"_____no_output_____"
],
[
"> - https://keras.io/api/losses/",
"_____no_output_____"
],
[
"### `binary_crossentropy`",
"_____no_output_____"
],
[
"### `sparse_categorical_crossentropy`",
"_____no_output_____"
],
[
"### `mean_absolute_error`",
"_____no_output_____"
],
[
"### `mean_squared_error`",
"_____no_output_____"
],
[
"## In the end, what should be a feasible configuration of the Neural Network for this data?",
"_____no_output_____"
],
[
"# Common Errors",
"_____no_output_____"
],
[
"## The `kernel_initializer` Matters",
"_____no_output_____"
],
[
"## The `activation` Function Matters",
"_____no_output_____"
],
[
"## The `optimizer` Matters",
"_____no_output_____"
],
[
"## The Number of `epochs` Matters",
"_____no_output_____"
],
[
"## The `loss` Function Matters",
"_____no_output_____"
],
[
"## The Number of `epochs` Matters",
"_____no_output_____"
],
[
"# Neural Network's importance to find **Non-Linear Patterns** in the Data\n\n> - The number of Neurons & Hidden Layers",
"_____no_output_____"
],
[
"https://towardsdatascience.com/beginners-ask-how-many-hidden-layers-neurons-to-use-in-artificial-neural-networks-51466afa0d3e",
"_____no_output_____"
],
[
"https://playground.tensorflow.org/#activation=tanh&batchSize=10&dataset=circle®Dataset=reg-plane&learningRate=0.03®ularizationRate=0&noise=0&networkShape=4,2&seed=0.87287&showTestData=false&discretize=false&percTrainData=50&x=true&y=true&xTimesY=false&xSquared=false&ySquared=false&cosX=false&sinX=false&cosY=false&sinY=false&collectStats=false&problem=classification&initZero=false&hideText=false",
"_____no_output_____"
],
[
"## Summary\n\n- Mathematical Formula\n- Weights / Kernel Initializer\n- Loss Function\n- Activation Function\n- Optimizers",
"_____no_output_____"
],
[
"## What cannot you change arbitrarily of a Neural Network?\n\n- Input Neurons\n- Output Neurons\n- Loss Functions\n- Activation Functions",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
cbebb4e8e2eeb4dd9235269ac796fd8963749683
| 21,975 |
ipynb
|
Jupyter Notebook
|
DataBase/Neural_Networks/VGG/vgg.ipynb
|
J0AZZ/chord-detection-challenge
|
e0648d235ee0fbbf48d692911032aba7e4fedb31
|
[
"MIT"
] | 2 |
2021-04-02T16:36:09.000Z
|
2021-04-14T14:30:45.000Z
|
DataBase/Neural_Networks/VGG/vgg.ipynb
|
J0AZZ/chord-detection-challenge
|
e0648d235ee0fbbf48d692911032aba7e4fedb31
|
[
"MIT"
] | null | null | null |
DataBase/Neural_Networks/VGG/vgg.ipynb
|
J0AZZ/chord-detection-challenge
|
e0648d235ee0fbbf48d692911032aba7e4fedb31
|
[
"MIT"
] | null | null | null | 38.552632 | 635 | 0.580068 |
[
[
[
"<h1>Table of Contents<span class=\"tocSkip\"></span></h1>\n<div class=\"toc\"><ul class=\"toc-item\"><li><span><a href=\"#Particionando-bases-de-treino-e-teste-com-split-70-30%\" data-toc-modified-id=\"Particionando-bases-de-treino-e-teste-com-split-70-30%-1\"><span class=\"toc-item-num\">1 </span>Particionando bases de treino e teste com split 70-30%</a></span></li><li><span><a href=\"#Particionando-bases-de-treino-e-teste-com-diferentes-músicas\" data-toc-modified-id=\"Particionando-bases-de-treino-e-teste-com-diferentes-músicas-2\"><span class=\"toc-item-num\">2 </span>Particionando bases de treino e teste com diferentes músicas</a></span></li></ul></div>",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport tensorflow as tf\nfrom tensorflow.keras.preprocessing.image import ImageDataGenerator\nimport glob\nimport numpy as np\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.metrics import classification_report\nfrom sklearn.metrics import accuracy_score\nfrom sklearn.utils import class_weight\nfrom imblearn.under_sampling import RandomUnderSampler",
"_____no_output_____"
],
[
"tf.__version__",
"_____no_output_____"
],
[
"from tensorflow.compat.v1 import ConfigProto\nfrom tensorflow.compat.v1 import InteractiveSession\n\nconfig = ConfigProto()\nconfig.gpu_options.allow_growth = True\nsession = InteractiveSession(config=config)",
"_____no_output_____"
],
[
"df = pd.read_csv('E:\\chord-detection-challenge\\DataBase\\CSV/status_A1.csv')\ndf.sort_values(by='title', inplace=True)\nimages_path = sorted(glob.glob('E:\\chord-detection-challenge\\DataBase\\clean_windows/Train/*'))\ndf['Unnamed: 0'] = np.array(images_path)\ndf.reset_index(inplace=True, drop=True)\ndf['status'] = df['status'].astype(str)",
"_____no_output_____"
]
],
[
[
"### Particionando bases de treino e teste com split 70-30%",
"_____no_output_____"
]
],
[
[
"### particiona considerando 70-30% e mantendo a frequência de amostras para treino, validação e teste de acordo com as colunas título e status (rótulo da rede)\n\nX_train, X_test, y_train, y_test = train_test_split(df[['Unnamed: 0', 'title']], df['status'], test_size=0.30, random_state=42, stratify=df[['status', 'title']])\ndf_train = pd.concat([X_train, y_train], axis=1)\nX_train, X_val, y_train, y_val = train_test_split(df_train[['Unnamed: 0', 'title']], df_train['status'], test_size=0.30, random_state=42, stratify=df_train[['status', 'title']])\n\n### contatena atributos de entrada e rótulo em um único dataframe para utilizar o flow_from_dataframe do tensorflow\ndf_test = pd.concat([X_test, y_test], axis=1)\ndf_train = pd.concat([X_train, y_train], axis=1)\ndf_val = pd.concat([X_val, y_val], axis=1)\n\nprint('Total de imagens de treinamento', len(df_train))\nprint('Total de imagens de validação', len(df_val))\nprint('Total de imagens de teste', len(df_test))",
"Total de imagens de treinamento 7648\nTotal de imagens de validação 3278\nTotal de imagens de teste 4683\n"
],
[
"undersample_train = RandomUnderSampler(sampling_strategy='majority')\nundersample_validation = RandomUnderSampler(sampling_strategy='majority')\n\nX_undertrain, y_undertrain = undersample_train.fit_resample(df_train[['Unnamed: 0', 'title']], df_train['status'])\nX_undervalidation, y_undervalidation = undersample_validation.fit_resample(df_val[['Unnamed: 0', 'title']], df_val['status'])",
"_____no_output_____"
],
[
"df_train = pd.concat([X_undertrain, y_undertrain], axis=1)\ndf_val = pd.concat([X_undervalidation, y_undervalidation], axis=1)",
"_____no_output_____"
]
],
[
[
"### Particionando bases de treino e teste com diferentes músicas",
"_____no_output_____"
]
],
[
[
"songs, n = df['title'].unique(), 5\nindex = np.random.choice(len(songs), 5, replace=False) \nselected_songs = songs[index] ## seleciona n músicas disponíveis para teste\ndf_test = df[df['title'].isin(selected_songs)] ## banco de teste contém todos os espectrogramas das n músicas selecionadas anteriormemente\ndf_train = df[~(df['title'].isin(selected_songs))] ## banco de treino contém os espectrogramas de todas as músicas EXCETO as selecionadas anteriormente para teste\n\nX_train, X_val, y_train, y_val = train_test_split(df_train[['Unnamed: 0', 'title']], df_train['status'], test_size=0.30, random_state=42, stratify=df_train[['status', 'title']]) ## divide em validação considerando 30% e balanceamento de acordo com título e status\n\n### contatena atributos de entrada e rótulo em um único dataframe para utilizar o flow_from_dataframe do tensorflow\ndf_train = pd.concat([X_train, y_train], axis=1)\ndf_val = pd.concat([X_val, y_val], axis=1)\n\nprint('Total de imagens de treinamento', len(df_train))\nprint('Total de imagens de validação', len(df_val))\nprint('Total de imagens de teste', len(df_test))\n",
"Total de imagens de treinamento 9591\nTotal de imagens de validação 4111\nTotal de imagens de teste 1907\n"
],
[
"datagen=ImageDataGenerator(rescale=1./255)\ntrain_generator=datagen.flow_from_dataframe(dataframe=df_train, directory='E:\\chord-detection-challenge\\DataBase\\clean_windows/Train/', x_col='Unnamed: 0', y_col=\"status\", class_mode=\"binary\", target_size=(224,224), batch_size=32)\nvalid_generator=datagen.flow_from_dataframe(dataframe=df_val, directory='E:\\chord-detection-challenge\\DataBase\\clean_windows/Train/', x_col='Unnamed: 0', y_col=\"status\", class_mode=\"binary\", target_size=(224,224), batch_size=32)\ntest_generator=datagen.flow_from_dataframe(dataframe=df_test, directory='E:\\chord-detection-challenge\\DataBase\\clean_windows/Train/', x_col='Unnamed: 0', y_col=\"status\", class_mode=\"binary\", target_size=(224,224), batch_size=1,shuffle=False)",
"Found 4494 validated image filenames belonging to 2 classes.\nFound 1926 validated image filenames belonging to 2 classes.\nFound 4683 validated image filenames belonging to 2 classes.\n"
],
[
"#from tensorflow.keras.models import Model\nrestnet = tf.keras.applications.VGG16(\n include_top=False, # não vai aproveitar a camada de saída \n weights=None, #não pega os pesso da imagenet\n input_shape=(224,224,3)\n)\noutput = restnet.layers[-1].output\noutput = tf.keras.layers.Flatten()(output)\nrestnet = tf.keras.Model(inputs=restnet.input, outputs=output)\nfor layer in restnet.layers: #treina tudo do zero\n layer.trainable = True\nrestnet.summary()",
"Model: \"model\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\ninput_1 (InputLayer) [(None, 224, 224, 3)] 0 \n_________________________________________________________________\nblock1_conv1 (Conv2D) (None, 224, 224, 64) 1792 \n_________________________________________________________________\nblock1_conv2 (Conv2D) (None, 224, 224, 64) 36928 \n_________________________________________________________________\nblock1_pool (MaxPooling2D) (None, 112, 112, 64) 0 \n_________________________________________________________________\nblock2_conv1 (Conv2D) (None, 112, 112, 128) 73856 \n_________________________________________________________________\nblock2_conv2 (Conv2D) (None, 112, 112, 128) 147584 \n_________________________________________________________________\nblock2_pool (MaxPooling2D) (None, 56, 56, 128) 0 \n_________________________________________________________________\nblock3_conv1 (Conv2D) (None, 56, 56, 256) 295168 \n_________________________________________________________________\nblock3_conv2 (Conv2D) (None, 56, 56, 256) 590080 \n_________________________________________________________________\nblock3_conv3 (Conv2D) (None, 56, 56, 256) 590080 \n_________________________________________________________________\nblock3_pool (MaxPooling2D) (None, 28, 28, 256) 0 \n_________________________________________________________________\nblock4_conv1 (Conv2D) (None, 28, 28, 512) 1180160 \n_________________________________________________________________\nblock4_conv2 (Conv2D) (None, 28, 28, 512) 2359808 \n_________________________________________________________________\nblock4_conv3 (Conv2D) (None, 28, 28, 512) 2359808 \n_________________________________________________________________\nblock4_pool (MaxPooling2D) (None, 14, 14, 512) 0 \n_________________________________________________________________\nblock5_conv1 (Conv2D) (None, 14, 14, 512) 2359808 \n_________________________________________________________________\nblock5_conv2 (Conv2D) (None, 14, 14, 512) 2359808 \n_________________________________________________________________\nblock5_conv3 (Conv2D) (None, 14, 14, 512) 2359808 \n_________________________________________________________________\nblock5_pool (MaxPooling2D) (None, 7, 7, 512) 0 \n_________________________________________________________________\nflatten (Flatten) (None, 25088) 0 \n=================================================================\nTotal params: 14,714,688\nTrainable params: 14,714,688\nNon-trainable params: 0\n_________________________________________________________________\n"
],
[
"mc = tf.keras.callbacks.ModelCheckpoint('resnet_model.h5', monitor='val_binary_accuracy', mode='max', save_best_only=True)\n\nmodel = tf.keras.models.Sequential()\nmodel.add(restnet)\nmodel.add(tf.keras.layers.Dense(128, activation='relu', input_dim=(224,224,3)))\nmodel.add(tf.keras.layers.Dropout(0.5))\nmodel.add(tf.keras.layers.Dense(1, activation='sigmoid'))\n# tf.keras.layers.Conv2D(32, (3, 3), padding='same',\n# input_shape=(32,32,3)),\n# tf.keras.layers.MaxPool2D(),\n# tf.keras.layers.Conv2D(64, (3, 3)),\n# tf.keras.layers.Conv2D(128, (3, 3)),\n# tf.keras.layers.Flatten(),\n# tf.keras.layers.Dense(128,activation='relu'),\n# tf.keras.layers.Dense(2)\n#)\n\nmodel.compile(\n optimizer=tf.keras.optimizers.Adamax(),\n loss=tf.keras.losses.BinaryCrossentropy(),\n metrics=[tf.keras.metrics.BinaryAccuracy()]\n #weighted_metrics=[tf.keras.metrics.BinaryAccuracy()]\n)\nclass_weights = class_weight.compute_class_weight('balanced',\n np.unique(df_train['status']),\n df_train['status'])\nclass_weights = dict(enumerate(class_weights))\n\nSTEP_SIZE_TRAIN=train_generator.n//train_generator.batch_size\nSTEP_SIZE_VALID=valid_generator.n//valid_generator.batch_size\nmodel.fit(train_generator,\n steps_per_epoch=STEP_SIZE_TRAIN,\n validation_data=valid_generator,\n validation_steps=STEP_SIZE_VALID,\n #class_weight=class_weights,\n epochs=10,\n callbacks = [mc])",
"C:\\Users\\cflav\\anaconda3\\lib\\site-packages\\sklearn\\utils\\validation.py:70: FutureWarning: Pass classes=['0' '1'], y=0 0\n1 0\n2 0\n3 0\n4 0\n ..\n4489 1\n4490 1\n4491 1\n4492 1\n4493 1\nName: status, Length: 4494, dtype: object as keyword args. From version 1.0 (renaming of 0.25) passing these as positional arguments will result in an error\n warnings.warn(f\"Pass {args_msg} as keyword args. From version \"\n"
],
[
"STEP_SIZE_TEST=test_generator.n//test_generator.batch_size\n\nprint('---------------Teste-------------')\ntest_generator.reset()\npredictions = model.predict(test_generator,\n steps=STEP_SIZE_TEST,\n verbose=1)",
"---------------Teste-------------\n4683/4683 [==============================] - 122s 26ms/step\n"
],
[
"predictions",
"_____no_output_____"
],
[
"y_pred = predictions > 0.5",
"_____no_output_____"
],
[
"predicted_class_indices=np.argmax(predictions,axis=1)\n",
"_____no_output_____"
],
[
"predicted_class_indices",
"_____no_output_____"
],
[
"print(accuracy_score(test_generator.labels, y_pred))",
"0.29404228058936577\n"
],
[
"print(classification_report(test_generator.labels, y_pred))",
" precision recall f1-score support\n\n 0 0.00 0.00 0.00 3306\n 1 0.29 1.00 0.45 1377\n\n accuracy 0.29 4683\n macro avg 0.15 0.50 0.23 4683\nweighted avg 0.09 0.29 0.13 4683\n\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbebb4fdf34729d930b1feec0bb6967466c221bb
| 95,516 |
ipynb
|
Jupyter Notebook
|
examples/Example notebook.ipynb
|
peendebak/AutoDepGraph
|
93efb54a2b46ec03d84c3dc17f71e567ae152ab4
|
[
"MIT"
] | null | null | null |
examples/Example notebook.ipynb
|
peendebak/AutoDepGraph
|
93efb54a2b46ec03d84c3dc17f71e567ae152ab4
|
[
"MIT"
] | null | null | null |
examples/Example notebook.ipynb
|
peendebak/AutoDepGraph
|
93efb54a2b46ec03d84c3dc17f71e567ae152ab4
|
[
"MIT"
] | null | null | null | 60.95469 | 11,780 | 0.590644 |
[
[
[
"\n# Example notebook\n\nThis example will contain the following examples\n- Creating and saving a graph \n- Plotting the graph \n- Executing a node\n- Loading a graph from disk",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\nimport matplotlib.pyplot as plt\nimport networkx as nx\nfrom importlib import reload\nimport os\nimport autodepgraph as adg\nfrom autodepgraph import AutoDepGraph_DAG",
"_____no_output_____"
]
],
[
[
"## Creatinga custom graph\nA graph can be instantiated and nodes can be added to the graph as with any `networkx` graph object. \nIt is important to specify a `calibrat",
"_____no_output_____"
]
],
[
[
"cal_True_delayed= 'autodepgraph.node_functions.calibration_functions.test_calibration_True_delayed'\ntest_graph = AutoDepGraph_DAG('test graph')\nfor node in ['A', 'B', 'C', 'D', 'E']:\n test_graph.add_node(node, \n calibrate_function=cal_True_delayed)",
"_____no_output_____"
],
[
"test_graph.add_node?",
"_____no_output_____"
]
],
[
[
"Some nodes require other nodes to be in a `good` or calibrated state. Such dependencies are defined by setting edges in the graph. ",
"_____no_output_____"
]
],
[
[
"test_graph.add_edge('C', 'A')\ntest_graph.add_edge('C', 'B')\ntest_graph.add_edge('B', 'A')\ntest_graph.add_edge('D', 'A')\ntest_graph.add_edge('E', 'D')",
"_____no_output_____"
]
],
[
[
"## Visualizing the graph\nWe support two ways of visualizing graphs: \n- matplotlib in the notebook \n- an svg in an html page that updates in real-time",
"_____no_output_____"
],
[
"### Realtime svg/html visualization ",
"_____no_output_____"
]
],
[
[
"# The default plotting mode is SVG \ntest_graph.cfg_plot_mode = 'svg'\n# Updates the monitor, in this case the svg/html page\ntest_graph.update_monitor()\n\n# Updating the monitor overwrites an svg file whose location is determined by the attribute:\ntest_graph.cfg_svg_filename",
"_____no_output_____"
],
[
"from IPython.display import display, SVG\ndisplay(SVG(test_graph.cfg_svg_filename))",
"_____no_output_____"
],
[
"# The html page is located at the location specified by the url. \n# The page generated based on a template when the open_html_viewer command is called. \nurl = test_graph.open_html_viewer()\nprint(url)",
"/Users/adriaanrol/GitHubRepos/Personal/AutoDepGraph/autodepgraph/svg_viewer/svgviewer-etruszyd.html\n"
]
],
[
[
"### Matplotlib drawing of the graph\n",
"_____no_output_____"
]
],
[
[
"\n\n# Alternatively a render in matplotlib can be drawn \ntest_graph.draw_mpl()",
"_____no_output_____"
]
],
[
[
"# Maintaining the graph ",
"_____no_output_____"
]
],
[
[
"test_graph.set_all_node_states('needs calibration')",
"_____no_output_____"
],
[
"test_graph.maintain_B()",
"Maintaining node \"B\".\nMaintaining node \"A\".\n\tCalibrating node A.\n\tCalibration of node A successful.\n\tCalibrating node B.\n\tCalibration of node B successful.\n"
],
[
"display(SVG(test_graph.cfg_svg_filename))",
"_____no_output_____"
],
[
"# Update the plotting monitor (default matplotlib) to show your graph\ntest_graph.update_monitor()",
"_____no_output_____"
],
[
"test_graph.set_all_node_states('needs calibration')",
"_____no_output_____"
],
[
"test_graph.maintain_node('E')",
"Maintaining node \"E\".\nMaintaining node \"D\".\nMaintaining node \"A\".\n\tCalibrating node A.\n\tCalibration of node A successful.\n\tCalibrating node D.\n\tCalibration of node D successful.\n\tCalibrating node E.\n\tCalibration of node E successful.\n"
],
[
"display(SVG(test_graph.cfg_svg_filename))",
"_____no_output_____"
]
],
[
[
"### Three qubit example\n\nThis example shows a more realistic graph. \nThe examples below show ways of exploring the graph",
"_____no_output_____"
]
],
[
[
"test_dir = os.path.join(adg.__path__[0], 'tests', 'test_data')\nfn = os.path.join(test_dir, 'three_qubit_graph.yaml')\nDAG = nx.readwrite.read_yaml(fn)",
"_____no_output_____"
],
[
"test_graph.cfg_plot_mode = 'svg'\nDAG.update_monitor()\n# This graph is so big, the html visualization is more suitable. \ndisplay(SVG(DAG.cfg_svg_filename))",
"_____no_output_____"
],
[
"url = test_graph.open_html_viewer()\nurl",
"_____no_output_____"
]
],
[
[
"\n### Reset the state of all nodes",
"_____no_output_____"
]
],
[
[
"DAG.nodes['CZ q0-q1']",
"_____no_output_____"
],
[
"DAG.set_all_node_states('needs calibration')\n# DAG.set_all_node_states('unknown')\nDAG.update_monitor()",
"_____no_output_____"
],
[
"DAG._construct_maintenance_methods(DAG.nodes.keys())",
"_____no_output_____"
],
[
"DAG.maintain_CZ_q0_q1()",
"Maintaining node \"CZ q0-q1\".\nMaintaining node \"Chevron q0-q1\".\nMaintaining node \"3 qubit device multiplexed readout\".\nMaintaining node \"q0 optimal weights\".\nMaintaining node \"q0 pulse amplitude med\".\nMaintaining node \"q0 frequency fine\".\nMaintaining node \"q0 pulse amplitude coarse\".\nMaintaining node \"AWG8 MW-staircase\".\n\tCalibrating node AWG8 MW-staircase.\n\tCalibration of node AWG8 MW-staircase successful.\nMaintaining node \"q0 frequency coarse\".\nMaintaining node \"q0 resonator frequency\".\nMaintaining node \"3 qubit device resonator frequencies coarse\".\n\tCalibrating node 3 qubit device resonator frequencies coarse.\n\tCalibration of node 3 qubit device resonator frequencies coarse successful.\n\tCalibrating node q0 resonator frequency.\n\tCalibration of node q0 resonator frequency successful.\n\tCalibrating node q0 frequency coarse.\n\tCalibration of node q0 frequency coarse successful.\nMaintaining node \"q0 mixer offsets drive\".\n\tCalibrating node q0 mixer offsets drive.\n\tCalibration of node q0 mixer offsets drive successful.\nMaintaining node \"q0 mixer offsets readout\".\n\tCalibrating node q0 mixer offsets readout.\n\tCalibration of node q0 mixer offsets readout successful.\nMaintaining node \"q0 mixer skewness drive\".\n\tCalibrating node q0 mixer skewness drive.\n\tCalibration of node q0 mixer skewness drive successful.\n\tCalibrating node q0 pulse amplitude coarse.\n\tCalibration of node q0 pulse amplitude coarse successful.\n\tCalibrating node q0 frequency fine.\n\tCalibration of node q0 frequency fine successful.\n\tCalibrating node q0 pulse amplitude med.\n\tCalibration of node q0 pulse amplitude med successful.\n\tCalibrating node q0 optimal weights.\n\tCalibration of node q0 optimal weights successful.\nMaintaining node \"q1 optimal weights\".\nMaintaining node \"q1 pulse amplitude med\".\nMaintaining node \"q1 frequency fine\".\nMaintaining node \"q1 pulse amplitude coarse\".\nMaintaining node \"q1 frequency coarse\".\nMaintaining node \"q1 resonator frequency\".\n\tCalibrating node q1 resonator frequency.\n\tCalibration of node q1 resonator frequency successful.\n\tCalibrating node q1 frequency coarse.\n\tCalibration of node q1 frequency coarse successful.\nMaintaining node \"q1 mixer offsets drive\".\n\tCalibrating node q1 mixer offsets drive.\n\tCalibration of node q1 mixer offsets drive successful.\nMaintaining node \"q1 mixer offsets readout\".\n\tCalibrating node q1 mixer offsets readout.\n\tCalibration of node q1 mixer offsets readout successful.\nMaintaining node \"q1 mixer skewness drive\".\n\tCalibrating node q1 mixer skewness drive.\n\tCalibration of node q1 mixer skewness drive successful.\n\tCalibrating node q1 pulse amplitude coarse.\n\tCalibration of node q1 pulse amplitude coarse successful.\n\tCalibrating node q1 frequency fine.\n\tCalibration of node q1 frequency fine successful.\n\tCalibrating node q1 pulse amplitude med.\n\tCalibration of node q1 pulse amplitude med successful.\n\tCalibrating node q1 optimal weights.\n\tCalibration of node q1 optimal weights successful.\nMaintaining node \"q2 optimal weights\".\nMaintaining node \"q2 pulse amplitude med\".\nMaintaining node \"q2 frequency fine\".\nMaintaining node \"q2 pulse amplitude coarse\".\nMaintaining node \"q2 frequency coarse\".\nMaintaining node \"q2 resonator frequency\".\n\tCalibrating node q2 resonator frequency.\n\tCalibration of node q2 resonator frequency successful.\n\tCalibrating node q2 frequency coarse.\n\tCalibration of node q2 frequency coarse successful.\nMaintaining node \"q2 mixer offsets drive\".\n\tCalibrating node q2 mixer offsets drive.\n\tCalibration of node q2 mixer offsets drive successful.\nMaintaining node \"q2 mixer offsets readout\".\n\tCalibrating node q2 mixer offsets readout.\n\tCalibration of node q2 mixer offsets readout successful.\nMaintaining node \"q2 mixer skewness drive\".\n\tCalibrating node q2 mixer skewness drive.\n\tCalibration of node q2 mixer skewness drive successful.\n\tCalibrating node q2 pulse amplitude coarse.\n\tCalibration of node q2 pulse amplitude coarse successful.\n\tCalibrating node q2 frequency fine.\n\tCalibration of node q2 frequency fine successful.\n\tCalibrating node q2 pulse amplitude med.\n\tCalibration of node q2 pulse amplitude med successful.\n\tCalibrating node q2 optimal weights.\n\tCalibration of node q2 optimal weights successful.\n\tCalibrating node 3 qubit device multiplexed readout.\n\tCalibration of node 3 qubit device multiplexed readout successful.\nMaintaining node \"AWG8 Flux-staircase\".\n\tCalibrating node AWG8 Flux-staircase.\n\tCalibration of node AWG8 Flux-staircase successful.\nMaintaining node \"q0 gates restless\".\n\tCalibrating node q0 gates restless.\n\tCalibration of node q0 gates restless successful.\nMaintaining node \"q1 gates restless\".\n\tCalibrating node q1 gates restless.\n\tCalibration of node q1 gates restless successful.\n\tCalibrating node Chevron q0-q1.\n\tCalibration of node Chevron q0-q1 successful.\nMaintaining node \"q0 cryo dist. corr.\".\nMaintaining node \"q0 room temp. dist. corr.\".\n\tCalibrating node q0 room temp. dist. corr..\n\tCalibration of node q0 room temp. dist. corr. successful.\n\tCalibrating node q0 cryo dist. corr..\n\tCalibration of node q0 cryo dist. corr. successful.\nMaintaining node \"q1 cryo dist. corr.\".\nMaintaining node \"q1 room temp. dist. corr.\".\n\tCalibrating node q1 room temp. dist. corr..\n\tCalibration of node q1 room temp. dist. corr. successful.\n\tCalibrating node q1 cryo dist. corr..\n\tCalibration of node q1 cryo dist. corr. successful.\n\tCalibrating node CZ q0-q1.\n\tCalibration of node CZ q0-q1 successful.\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
cbebb74ca3cc40b6248bf04fc024229668272736
| 30,251 |
ipynb
|
Jupyter Notebook
|
nbs/post-process.ipynb
|
KeremTurgutlu/tse
|
c08bd33dc40a61db3a57355e8580bf38ad8304be
|
[
"Apache-2.0"
] | 1 |
2020-05-19T15:35:45.000Z
|
2020-05-19T15:35:45.000Z
|
nbs/post-process.ipynb
|
KeremTurgutlu/tse
|
c08bd33dc40a61db3a57355e8580bf38ad8304be
|
[
"Apache-2.0"
] | 2 |
2021-09-28T01:31:04.000Z
|
2022-02-26T07:05:37.000Z
|
nbs/post-process.ipynb
|
KeremTurgutlu/tse
|
c08bd33dc40a61db3a57355e8580bf38ad8304be
|
[
"Apache-2.0"
] | null | null | null | 49.429739 | 8,140 | 0.698027 |
[
[
[
"from fastai.core import *",
"_____no_output_____"
],
[
"test_df = pd.read_csv(\"../data/test.csv\")\nsub_df = pd.read_csv(\"../data/submission.csv\")",
"_____no_output_____"
],
[
"test_df['selected_text'] = sub_df['selected_text']",
"_____no_output_____"
],
[
"test_df",
"_____no_output_____"
],
[
"for text, selected_text, sent in zip(test_df['text'], test_df['selected_text'], test_df['sentiment']):\n if sent != 'neutral':\n\n i = text.find(selected_text)\n selected_text_set = set(selected_text.split())\n text_set = set(text[i:].split())\n \n for w in selected_text_set:\n if w not in text_set:\n break",
"_____no_output_____"
],
[
"selected_text_set",
"_____no_output_____"
],
[
"text_set",
"_____no_output_____"
],
[
"(0.698436 + 0.692732 + 0.691364 + 0.695767 + 0.706681) /5",
"_____no_output_____"
]
],
[
[
"### backward",
"_____no_output_____"
]
],
[
[
"from fastai.vision import *",
"_____no_output_____"
],
[
"y = 4\nx = 2",
"_____no_output_____"
],
[
"# y = x**2\nx1 = torch.randn(100)\ny1 = x1**2 ",
"_____no_output_____"
],
[
"# y = -(x-5)**2 \nx2 = torch.randn(100)+2\ny2 = (-(x2-2)**2 ) + 5 ",
"_____no_output_____"
],
[
"axes = subplots(1,1)\naxes[0][0].scatter(x1,y1)\naxes[0][0].scatter(x2,y2)",
"_____no_output_____"
],
[
"def loss_fn(y_hat, y): return torch.sqrt(((y_hat - y)**2).mean())",
"_____no_output_____"
],
[
"class SimpleNet(Module):\n def __init__(self):\n self.lin0 = nn.Linear(1,1)\n self.lin1 = nn.Linear(1,1)\n self.lin2 = nn.Linear(1,1)\n \n def forward(self,x): \n return self.lin2(F.relu(self.lin1(F.relu(self.lin0(x)))))",
"_____no_output_____"
],
[
"model = SimpleNet()",
"_____no_output_____"
],
[
"xs = torch.cat([x1,x2]).view(-1,1)\nys = torch.cat([y1,y2]).view(-1,1)",
"_____no_output_____"
],
[
"lr = 0.001\nfor i in range(100):\n loss = loss_fn(model(xs), ys)\n print(loss.item())\n loss.backward()\n for p in model.parameters(): p.data.add_(-lr*p.grad)\n for p in model.parameters(): p.grad.zero_()",
"2.026779890060425\n2.026779890060425\n2.026779890060425\n2.026779890060425\n2.026779890060425\n2.0267796516418457\n2.026779890060425\n2.026779890060425\n2.026779890060425\n2.026779890060425\n2.026779890060425\n2.026779890060425\n2.0267796516418457\n2.0267796516418457\n2.0267796516418457\n2.026779890060425\n2.026779890060425\n2.026779890060425\n2.026779890060425\n2.0267796516418457\n2.026779890060425\n2.0267796516418457\n2.026779890060425\n2.026779890060425\n2.0267796516418457\n2.0267796516418457\n2.026779890060425\n2.0267796516418457\n2.026779890060425\n2.026779890060425\n2.026779890060425\n2.0267796516418457\n2.0267796516418457\n2.0267796516418457\n2.0267796516418457\n2.0267796516418457\n2.0267796516418457\n2.0267796516418457\n2.0267796516418457\n2.0267796516418457\n2.026779890060425\n2.0267796516418457\n2.026779890060425\n2.026779890060425\n2.026779890060425\n2.0267796516418457\n2.0267796516418457\n2.0267796516418457\n2.026779890060425\n2.0267796516418457\n2.026779890060425\n2.0267796516418457\n2.0267796516418457\n2.0267796516418457\n2.0267796516418457\n2.0267796516418457\n2.0267796516418457\n2.0267796516418457\n2.0267796516418457\n2.0267796516418457\n2.026779890060425\n2.0267796516418457\n2.0267796516418457\n2.0267796516418457\n2.0267796516418457\n2.0267796516418457\n2.0267796516418457\n2.0267796516418457\n2.0267796516418457\n2.0267796516418457\n2.0267796516418457\n2.0267794132232666\n2.0267796516418457\n2.0267796516418457\n2.0267796516418457\n2.0267796516418457\n2.0267796516418457\n2.0267796516418457\n2.0267796516418457\n2.0267796516418457\n2.0267796516418457\n2.0267796516418457\n2.0267796516418457\n2.0267796516418457\n2.0267796516418457\n2.0267796516418457\n2.0267796516418457\n2.0267796516418457\n2.0267796516418457\n2.0267796516418457\n2.0267796516418457\n2.0267796516418457\n2.0267796516418457\n2.0267796516418457\n2.0267796516418457\n2.0267796516418457\n2.0267796516418457\n2.0267796516418457\n2.0267796516418457\n2.0267796516418457\n"
],
[
"yhats = model(xs).view(-1).detach()",
"_____no_output_____"
],
[
"plt.scatter(xs, yhats)",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbebb95a260be37d70a33040ffbb1bd4b481d95f
| 354,049 |
ipynb
|
Jupyter Notebook
|
python/Session5_dimensionality_reduction_Notebook2.ipynb
|
alimadani/ML_FromTheoy_ToModeling
|
376c4b864411432f988996899e7249ffeb491738
|
[
"MIT"
] | 2 |
2020-11-25T08:57:57.000Z
|
2020-11-25T23:01:25.000Z
|
python/Session5_dimensionality_reduction_Notebook2.ipynb
|
alimadani/ML_FromTheoy_ToModeling
|
376c4b864411432f988996899e7249ffeb491738
|
[
"MIT"
] | null | null | null |
python/Session5_dimensionality_reduction_Notebook2.ipynb
|
alimadani/ML_FromTheoy_ToModeling
|
376c4b864411432f988996899e7249ffeb491738
|
[
"MIT"
] | 2 |
2020-11-25T23:23:03.000Z
|
2022-03-22T01:47:11.000Z
| 318.389388 | 61,370 | 0.920624 |
[
[
[
" # Dimensionality Reduction \n\n\nReducing number of dimensions whcih means that the number of new features is lower than the number of original features.\nFirst, we need to import numpy, matplotlib, and scikit-learn and get the UCI ML digit image data. Scikit-learn already comes with this data (or will automatically download it for you) so we don’t have to deal with uncompressing it ourselves! Additionally, I’ve provided a function that will produce a nice visualization of our data.\n\nWe are going to use the following libraries and packages:\n\n* **numpy**: \"NumPy is the fundamental package for scientific computing with Python.\" (http://www.numpy.org/)\n* **matplotlib**: \"Matplotlib is a Python 2D plotting library which produces publication quality figures in a variety of hardcopy formats and interactive environments across platforms.\" (https://matplotlib.org/)\n* **sklearn**: Scikit-learn is a machine learning library for Python programming language. (https://scikit-learn.org/stable/)\n* **pandas**: \"Pandas provides easy-to-use data structures and data analysis tools for Python.\" (https://pandas.pydata.org/)",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\nimport matplotlib.cm as cm\nfrom matplotlib import offsetbox \nimport pandas as pd\n",
"_____no_output_____"
]
],
[
[
"# t-distributed Stochastic Neighbor Embedding (t-SNE)\n\nt-SNE is an algorithm to optimally map the higher dimensional space to lower dimensions paying attention to short distances. The trasformation is different for different regions. SNE is the general concept behind this type of mapping and \"t\" shows usage of t-distribution in t-SNE. ",
"_____no_output_____"
],
[
"## Synthetic data \n\nLet's generate synthetic data as follows:\n1) Points are scattered in 2 dimensional space as follows. There are N-2 other dimensions that all the points have same values in each dimension\n2) We will reduce the dimensionality of the data to 2D\n",
"_____no_output_____"
]
],
[
[
"group_1_X = np.repeat(2,90)+np.random.normal(loc=0, scale=1,size=90)\ngroup_1_Y = np.repeat(2,90)+np.random.normal(loc=0, scale=1,size=90)\n\ngroup_2_X = np.repeat(10,90)+np.random.normal(loc=0, scale=1,size=90)\ngroup_2_Y = np.repeat(10,90)+np.random.normal(loc=0, scale=1,size=90)\n\nplt.scatter(group_1_X,group_1_Y, c='blue')\nplt.scatter(group_2_X,group_2_Y,c='green')\nplt.xlabel('1st dimension')\nplt.ylabel('2nd dimension')",
"_____no_output_____"
]
],
[
[
"### Implementing t-SNE on the synthetic data",
"_____no_output_____"
]
],
[
[
"####\ncombined = np.column_stack((np.concatenate([group_1_X,group_2_X]),np.concatenate([group_1_Y,group_2_Y])))\nprint(combined.shape)\n####\nfrom sklearn import manifold\ncombined_tSNE = manifold.TSNE(n_components=2, init='pca',perplexity=30,learning_rate=200,n_iter=500,random_state=2).fit_transform(combined)\n####\nimport umap\ncombined_UMAP = umap.UMAP(n_neighbors=10, min_dist=0.3, n_components=2,random_state=2).fit_transform(combined)\n\nfig, (ax1, ax2) = plt.subplots(1, 2, sharey=True)\n\nax1.scatter(combined_tSNE[0:90,0], combined_tSNE[0:90,1], c='blue')\nax1.scatter(combined_tSNE[90:180,0], combined_tSNE[90:180,1], c='green')\nax1.set_title('t-SNE')\n\nax2.scatter(combined_UMAP[0:90,0], combined_UMAP[0:90,1], c='blue')\nax2.scatter(combined_UMAP[90:180,0], combined_UMAP[90:180,1], c='green')\nax2.set_title('UMAP')",
"(180, 2)\n"
]
],
[
[
"**Parameters of t-SNE:**\n\n* ***Perplexity (perplexity)***: somehow shows the number of close neighbors each point has. Hence, perplexity should be smaller than the number of points. There is a suggested range for perplexity in the original paper: \"The performance of SNE is fairly robust to changes in the perplexity, and typical values are between 5 and 50.\". Although perplexity=5 is usually not optimal, values higher than 50 also may result in weird grouping of the data points and shapes in 2 dimensional space. \n\n* ***Number of iterations (n_iter)*** required for converagence of the approach is another important parameter that depened on the input dataset. There are no fixed number to make sure of the convergence but there are some rule of thumb to check that. As an example, if there are pinched shapes in the t-SNE plot, it is better to run the approach for higher iteration number to makes sure that the resulted shapes and clusters are not artifacts of an unconverged t-SNE.\n\n**Parameters of UMAP:**\n\n* ***Number of neighbors (n_neighbors)***: Number of neighboring data points used in the process of local manifold approximation. This parameters is suggested to be between 5 and 50.\n* ***Minimum distance (min_dist)***: It is a measure of allowed compression of points together in low dimensional space. This parameters is suggested to be between 0.001 and 0.5.\n",
"_____no_output_____"
],
[
"### Let's change the structure of synthetic data\n\nLet's generate synthetic data as follows:",
"_____no_output_____"
]
],
[
[
"group_1_X = np.arange(10,100)\ngroup_1_Y = np.arange(10,100)+np.random.normal(loc=0, scale=0.3,size=90)-np.repeat(4,90)\n\ngroup_2_X = np.arange(10,100)\ngroup_2_Y = np.arange(10,100)+np.random.normal(loc=0, scale=0.3,size=90)+np.repeat(4,90)\n\nplt.scatter(group_1_X,group_1_Y, c='blue')\nplt.scatter(group_2_X,group_2_Y,c='green')\nplt.xlabel('1st dimension')\nplt.ylabel('2nd dimension')",
"_____no_output_____"
]
],
[
[
"### Implementing t-SNE and UMAP on the synthetic data\n\n\n",
"_____no_output_____"
]
],
[
[
"####\ncombined = np.column_stack((np.concatenate([group_1_X,group_2_X]),np.concatenate([group_1_Y,group_2_Y])))\nprint(combined.shape)\n####\nfrom sklearn import manifold\ncombined_tSNE = manifold.TSNE(n_components=2, init='pca',perplexity=30,learning_rate=200,n_iter=500,random_state=2).fit_transform(combined)\n####\nimport umap\ncombined_UMAP = umap.UMAP(n_neighbors=5, min_dist=0.01, n_components=2,random_state=2).fit_transform(combined)\n\nfig, (ax1, ax2) = plt.subplots(1, 2, sharey=True)\n\nax1.scatter(combined_tSNE[0:90,0], combined_tSNE[0:90,1], c='blue')\nax1.scatter(combined_tSNE[90:180,0], combined_tSNE[90:180,1], c='green')\nax1.set_title('t-SNE')\n\nax2.scatter(combined_UMAP[0:90,0], combined_UMAP[0:90,1], c='blue')\nax2.scatter(combined_UMAP[90:180,0], combined_UMAP[90:180,1], c='green')\nax2.set_title('UMAP')",
"(180, 2)\n"
]
],
[
[
"### Another synthetic data\n\nLet's generate synthetic data as follows:\n",
"_____no_output_____"
]
],
[
[
"group_1_X = np.arange(start=0,stop=1**2,step=0.001)\ngroup_1_Y = np.sqrt(np.repeat(1**2,1000)-group_1_X**2)\n\ngroup_2_X = np.arange(start=0,stop=1.5,step=0.001)\ngroup_2_Y = np.sqrt(np.repeat(1.5**2,1500)-group_2_X**2)\n\nplt.scatter(group_1_X,group_1_Y, c='blue', )\nplt.scatter(group_2_X,group_2_Y,c='green')\nplt.xlabel('1st dimension')\nplt.ylabel('2nd dimension')\nplt.xlim(0,2.5)\nplt.ylim(0,2.5)",
"_____no_output_____"
]
],
[
[
"### Implementing t-SNE on the synthetic data",
"_____no_output_____"
]
],
[
[
"####\ncombined = np.column_stack((np.concatenate([group_1_X,group_2_X]),np.concatenate([group_1_Y,group_2_Y])))\nprint(combined.shape)\n####\nfrom sklearn import manifold\ncombined_tSNE = manifold.TSNE(n_components=2, init='pca',perplexity=30,learning_rate=200,n_iter=500,random_state=2).fit_transform(combined)\n####\nimport umap\ncombined_UMAP = umap.UMAP(n_neighbors=10, min_dist=0.9, n_components=2,random_state=2).fit_transform(combined)\n\nfig, (ax1, ax2) = plt.subplots(1, 2, sharey=True)\n\nax1.scatter(combined_tSNE[0:1000,0], combined_tSNE[0:1000,1], c='blue')\nax1.scatter(combined_tSNE[1000:2500,0], combined_tSNE[1000:2500,1], c='green')\nax1.set_title('t-SNE')\n\nax2.scatter(combined_UMAP[0:1000,0], combined_UMAP[0:1000,1], c='blue')\nax2.scatter(combined_UMAP[1000:2500,0], combined_UMAP[1000:2500,1], c='green')\nax2.set_title('UMAP')",
"(2500, 2)\n"
]
],
[
[
"### UCI ML digit image data\n\n* load and return digit data set",
"_____no_output_____"
]
],
[
[
"from sklearn import datasets\n\n# Loading digit images\ndigits = datasets.load_digits()\n\nX = digits.data\ny = digits.target\nn_samples, n_features = X.shape\nprint(\"number of samples (data points):\", n_samples)\nprint(\"number of features:\", n_features)",
"number of samples (data points): 1797\nnumber of features: 64\n"
]
],
[
[
"Pixels of images have values between 0 and 16:",
"_____no_output_____"
]
],
[
[
"np.max(X)",
"_____no_output_____"
]
],
[
[
"Let's write a function to use it for visualization of the results of all the dimension reduction methods.",
"_____no_output_____"
],
[
"#### Let's visualize some of the images",
"_____no_output_____"
]
],
[
[
"fig, ax_array = plt.subplots(1,10)\naxes = ax_array.flatten()\nfor i, ax in enumerate(axes):\n ax.imshow(digits.images[i])\nplt.setp(axes, xticks=[], yticks=[])\nplt.tight_layout(h_pad=0.5, w_pad=0.01)",
"_____no_output_____"
]
],
[
[
"Now that we understood how t-SNE works, let's implement it on the UCI ML digit image data:",
"_____no_output_____"
]
],
[
[
"from sklearn import manifold\n\nX_tsne = manifold.TSNE(n_components=2, init='pca',perplexity=30,learning_rate=200,n_iter=500,random_state=2).fit_transform(X)",
"_____no_output_____"
]
],
[
[
"Now, we use the plotting function to show the first 2 principle component scores of all teh data points.",
"_____no_output_____"
]
],
[
[
"def embedding_plot(X,labels,title):\n plt.scatter(X[:, 0], X[:, 1], c=labels, cmap='Spectral', s=5)\n plt.gca().set_facecolor((1, 1, 1))\n plt.xlabel('1st dimension', fontsize=24)\n plt.ylabel('2nd dimension', fontsize=24)\n plt.colorbar(boundaries=np.arange(11)-0.5).set_ticks(np.arange(10))\n plt.grid(False)\n plt.title(title, fontsize=24);",
"_____no_output_____"
],
[
"embedding_plot(X_tsne, y,\"t-SNE\")",
"_____no_output_____"
]
],
[
[
"**t-SNE is an unsupervised approach similar to PCA and ICA. We add color for the sample labels afterward.**",
"_____no_output_____"
],
[
"## Normalizing data before dimensionality reduction\n\nIt is a good idea usually to normalize the data so that the scale of values for different features would become similar. ",
"_____no_output_____"
]
],
[
[
"from sklearn import preprocessing\nX_norm = pd.DataFrame(preprocessing.scale(X)) \n\nXnorm_tsne = manifold.TSNE(n_components=2, init='pca',perplexity=30,learning_rate=200,n_iter=500,random_state=2).fit_transform(X_norm)\n\nembedding_plot(Xnorm_tsne, y,\"t-SNE\")",
"_____no_output_____"
]
],
[
[
"# Uniform Manifold Approximation and Projection (UMAP)\n\nUMAP is a manifold learning method that is comptetitive to t-SNE for visualization quality while preserving the global structure of data, unlike t-SNE. UMAP has no computational restriction and is scalable to extremely large dataset, like GoogleNews, unlike t-SNE.\n\nUMAP uses k-nearest neighbor and uses Stochastic Gradient Descent to minimize the difference between the distances in the high dimensional and low dimensional spaces.\n\n\n**Definitions**\n\n* A n-dimensional manifold (n-manifold) M is a topological space that is locally homeomorphic to the Euclidean space of dimension n.\n* Locally homeomorphic means that every point in the space M is contained in an open set U such that there is a one-to-one onto map f:U -> M.\n* One-to-one onto map f:U -> M means that each element of M is mapped by exactly one element of U.\n* A topological space is a collection of open sets (with some mathematical properties).\n* A Riemannian (smooth) manifold M is a real smooth manifold with an inner product that varies smoothly from point to point in the tangent space of M.\n* Riemannian metric is collection of all the inner products of the points in the manifold M on the tangent space of M.\n\n* A simplicial complex K in n-dimensional real space is a collection of simplices in the space such that 1) Every face of a simplex of K is in K, and 2) The intersection of any two simplices of K is a face of each of them (Munkres 1993, p. 7; http://mathworld.wolfram.com/).\n* A simplex is the generalization of a tetrahedral region of space to n dimensions(http://mathworld.wolfram.com/).",
"_____no_output_____"
]
],
[
[
"import umap\n\nX_umap = umap.UMAP(n_neighbors=10, min_dist=0.3, n_components=2, random_state=2).fit_transform(X)\nembedding_plot(X_umap, y,\"umap\")",
"_____no_output_____"
]
],
[
[
"## Boston housing dataset",
"_____no_output_____"
]
],
[
[
"from sklearn import datasets\n\n# Loading digit images\nhousing = datasets.load_boston()\n\nXhouse = pd.DataFrame(housing.data)\nXhouse.columns = housing.feature_names\n\nyhouse = housing.target\nn_samples, n_features = Xhouse.shape\n\nprint(\"number of samples (data points):\", n_samples)\nprint(\"number of features:\", n_features)",
"number of samples (data points): 506\nnumber of features: 13\n"
]
],
[
[
"### Normalizing the data",
"_____no_output_____"
]
],
[
[
"from sklearn import preprocessing\nXhouse_norm = pd.DataFrame(preprocessing.scale(Xhouse), columns=Xhouse.columns) ",
"_____no_output_____"
]
],
[
[
"## Implementing t-SNE on the California housing data",
"_____no_output_____"
]
],
[
[
"Xhousenorm_tSNE = manifold.TSNE(n_components=2, init='pca',perplexity=30,learning_rate=200,n_iter=500,random_state=2).fit_transform(Xhouse_norm)\nXhousenorm_tSNE.shape",
"_____no_output_____"
]
],
[
[
"### Visualizing the results of t-SNE implemented on the California housing dataset",
"_____no_output_____"
]
],
[
[
"import seaborn as sns\n\ncmap = sns.cubehelix_palette(as_cmap=True)\n\n\nfig, ax = plt.subplots()\npoints = ax.scatter(x=Xhousenorm_tSNE[:,0], y=Xhousenorm_tSNE[:,1], c=yhouse, s=10, cmap=cmap)\nfig.colorbar(points)",
"_____no_output_____"
]
],
[
[
"## Implementing UMAP on the Boston housing data",
"_____no_output_____"
]
],
[
[
"Xhousenorm_umap = umap.UMAP(n_neighbors=10, min_dist=0.4, n_components=2, random_state=2).fit_transform(Xhouse_norm)\n\nfig, ax = plt.subplots()\npoints = ax.scatter(x=Xhousenorm_umap[:,0], y=Xhousenorm_umap[:,1], c=yhouse, s=10, cmap=cmap)\nfig.colorbar(points)",
"_____no_output_____"
]
],
[
[
"### Removing outliers and repeating the analysis",
"_____no_output_____"
]
],
[
[
"Xhouse_norm_noout = Xhouse_norm.iloc[np.where((Xhouse_norm.max(axis=1) < 3)==True)[0],:]\n\nXhousenorm_noout_umap = umap.UMAP(n_neighbors=5, min_dist=0.4, n_components=2, random_state=2).fit_transform(Xhouse_norm_noout)\n\nfig, ax = plt.subplots()\npoints = ax.scatter(x=Xhousenorm_noout_umap[:,0], y=Xhousenorm_noout_umap[:,1], c=yhouse[np.where((Xhouse_norm.max(axis=1) < 3)==True)[0]], s=10, cmap=cmap)\nfig.colorbar(points)",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
cbebbb8b32aaff3221f3c3899f8a0100f3b5c454
| 654,380 |
ipynb
|
Jupyter Notebook
|
3. Facial Keypoint Detection, Complete Pipeline.ipynb
|
thanhthtran/Facial_keypoint_cnv
|
0328bd096246db28e2349f54aa93f0fff6bac3b1
|
[
"MIT"
] | null | null | null |
3. Facial Keypoint Detection, Complete Pipeline.ipynb
|
thanhthtran/Facial_keypoint_cnv
|
0328bd096246db28e2349f54aa93f0fff6bac3b1
|
[
"MIT"
] | 1 |
2018-12-04T13:38:48.000Z
|
2019-02-26T14:47:17.000Z
|
3. Facial Keypoint Detection, Complete Pipeline.ipynb
|
thanhthtran/Facial_keypoint_cnv
|
0328bd096246db28e2349f54aa93f0fff6bac3b1
|
[
"MIT"
] | null | null | null | 1,936.035503 | 323,020 | 0.959016 |
[
[
[
"## Face and Facial Keypoint detection\n\nAfter you've trained a neural network to detect facial keypoints, you can then apply this network to *any* image that includes faces. The neural network expects a Tensor of a certain size as input and, so, to detect any face, you'll first have to do some pre-processing.\n\n1. Detect all the faces in an image using a face detector (we'll be using a Haar Cascade detector in this notebook).\n2. Pre-process those face images so that they are grayscale, and transformed to a Tensor of the input size that your net expects. This step will be similar to the `data_transform` you created and applied in Notebook 2, whose job was tp rescale, normalize, and turn any iimage into a Tensor to be accepted as input to your CNN.\n3. Use your trained model to detect facial keypoints on the image.\n\n---",
"_____no_output_____"
],
[
"In the next python cell we load in required libraries for this section of the project.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\nimport matplotlib.image as mpimg\n%matplotlib inline\n",
"_____no_output_____"
]
],
[
[
"#### Select an image \n\nSelect an image to perform facial keypoint detection on; you can select any image of faces in the `images/` directory.",
"_____no_output_____"
]
],
[
[
"import cv2\n# load in color image for face detection\nimage = cv2.imread('images/obamas.jpg')\n\n# switch red and blue color channels \n# --> by default OpenCV assumes BLUE comes first, not RED as in many images\nimage = cv2.cvtColor(image, cv2.COLOR_BGR2RGB)\n\n# plot the image\nfig = plt.figure(figsize=(9,9))\nplt.imshow(image)",
"_____no_output_____"
]
],
[
[
"## Detect all faces in an image\n\nNext, you'll use one of OpenCV's pre-trained Haar Cascade classifiers, all of which can be found in the `detector_architectures/` directory, to find any faces in your selected image.\n\nIn the code below, we loop over each face in the original image and draw a red square on each face (in a copy of the original image, so as not to modify the original). You can even [add eye detections](https://docs.opencv.org/3.4.1/d7/d8b/tutorial_py_face_detection.html) as an *optional* exercise in using Haar detectors.\n\nAn example of face detection on a variety of images is shown below.\n\n<img src='images/haar_cascade_ex.png' width=80% height=80%/>\n",
"_____no_output_____"
]
],
[
[
"# load in a haar cascade classifier for detecting frontal faces\nface_cascade = cv2.CascadeClassifier('detector_architectures/haarcascade_frontalface_default.xml')\n\n# run the detector\n# the output here is an array of detections; the corners of each detection box\n# if necessary, modify these parameters until you successfully identify every face in a given image\nfaces = face_cascade.detectMultiScale(image, 1.2, 2)\n\n# make a copy of the original image to plot detections on\nimage_with_detections = image.copy()\n\n# loop over the detected faces, mark the image where each face is found\nfor (x,y,w,h) in faces:\n # draw a rectangle around each detected face\n # you may also need to change the width of the rectangle drawn depending on image resolution\n cv2.rectangle(image_with_detections,(x,y),(x+w,y+h),(255,0,0),3) \n\nfig = plt.figure(figsize=(9,9))\n\nplt.imshow(image_with_detections)",
"_____no_output_____"
]
],
[
[
"## Loading in a trained model\n\nOnce you have an image to work with (and, again, you can select any image of faces in the `images/` directory), the next step is to pre-process that image and feed it into your CNN facial keypoint detector.\n\nFirst, load your best model by its filename.",
"_____no_output_____"
]
],
[
[
"import torch\nfrom models import Net\n\nnet = Net()\n\n## TODO: load the best saved model parameters (by your path name)\n## You'll need to un-comment the line below and add the correct name for *your* saved model\nnet.load_state_dict(torch.load('saved_models/model_3200_1600_smoothL1.pt'))\n\n## print out your net and prepare it for testing (uncomment the line below)\n",
"_____no_output_____"
]
],
[
[
"## Keypoint detection\n\nNow, we'll loop over each detected face in an image (again!) only this time, you'll transform those faces in Tensors that your CNN can accept as input images.\n\n### TODO: Transform each detected face into an input Tensor\n\nYou'll need to perform the following steps for each detected face:\n1. Convert the face from RGB to grayscale\n2. Normalize the grayscale image so that its color range falls in [0,1] instead of [0,255]\n3. Rescale the detected face to be the expected square size for your CNN (224x224, suggested)\n4. Reshape the numpy image into a torch image.\n\nYou may find it useful to consult to transformation code in `data_load.py` to help you perform these processing steps.\n\n\n### TODO: Detect and display the predicted keypoints\n\nAfter each face has been appropriately converted into an input Tensor for your network to see as input, you'll wrap that Tensor in a Variable() and can apply your `net` to each face. The ouput should be the predicted the facial keypoints. These keypoints will need to be \"un-normalized\" for display, and you may find it helpful to write a helper function like `show_keypoints`. You should end up with an image like the following with facial keypoints that closely match the facial features on each individual face:\n\n<img src='images/michelle_detected.png' width=30% height=30%/>\n\n\n",
"_____no_output_____"
]
],
[
[
"net.eval()",
"_____no_output_____"
],
[
"image_copy = np.copy(image)\nfrom torchvision import transforms, utils\nfrom data_load import Rescale, RandomCrop, Normalize, ToTensor\n\nimage_transform = transforms.Compose([Normalize(),Rescale((224,224)), ToTensor()])\n\n# loop over the detected faces from your haar cascade\nfor (x,y,w,h) in faces:\n # Select the region of interest that is the face in the image \n roi = image_copy[y:y+h, x:x+w]\n \n image_copy = np.copy(roi)\n \n ## TODO: Convert the face region from RGB to grayscale\n image_copy = cv2.cvtColor(roi, cv2.COLOR_RGB2GRAY)\n \n ## TODO: Normalize the grayscale image so that its color range falls in [0,1] instead of [0,255]\n image_copy= image_copy/255.0\n \n ## TODO: Rescale the detected face to be the expected square size for your CNN (224x224, suggested)\n image_copy = cv2.resize(image_copy, (224, 224))\n \n ## TODO: Reshape the numpy image shape (H x W x C) into a torch image shape (C x H x W)\n if(len(image_copy.shape) == 2):\n image_copy = image_copy.reshape(image_copy.shape[0], image_copy.shape[1], 1)\n \n image_copy = image_copy.transpose((2, 0, 1))\n image_copy = torch.from_numpy(image_copy)\n image_copy = image_copy.type(torch.FloatTensor)\n image_copy.unsqueeze_(0)\n print(image_copy.size())\n \n ## TODO: Make facial keypoint predictions using your loaded, trained network \n ## perform a forward pass to get the predicted facial keypoints\n output_pts = net(image_copy)\n\n\n ## TODO: Display each detected face and the corresponding keypoints \n \n",
"torch.Size([1, 1, 224, 224])\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
cbebcf73d67f7b3b5e856fc90c83db777f0ed2e2
| 237,377 |
ipynb
|
Jupyter Notebook
|
notebooks/demo.ipynb
|
Ezibenroc/pycewise
|
7f7608eec7082deef4c12c52761f4aed4f408d6c
|
[
"MIT"
] | 1 |
2020-05-16T05:10:35.000Z
|
2020-05-16T05:10:35.000Z
|
notebooks/demo.ipynb
|
Ezibenroc/pycewise
|
7f7608eec7082deef4c12c52761f4aed4f408d6c
|
[
"MIT"
] | null | null | null |
notebooks/demo.ipynb
|
Ezibenroc/pycewise
|
7f7608eec7082deef4c12c52761f4aed4f408d6c
|
[
"MIT"
] | 1 |
2019-02-14T07:32:43.000Z
|
2019-02-14T07:32:43.000Z
| 428.478339 | 54,884 | 0.929066 |
[
[
[
"# Simple test",
"_____no_output_____"
]
],
[
[
"%load_ext autoreload\n%autoreload 2",
"_____no_output_____"
],
[
"from pycewise import *\nfrom test import *\nimport random\nimport matplotlib.pyplot as plt\nprint(__version__)\nprint(__git_version__)",
"0.0.5\n55040ff6f76244888ccf905d7f6fef0321253750\n"
],
[
"random.seed(42)\ndataset = generate_dataset(0, 1, 30, 0, 10) +\\\n generate_dataset(0, 2, 30, 10, 20) +\\\n generate_dataset(20, 0.5, 30, 20, 30)\n\ndef add_normal_noise(dataset, sigma):\n return [(d[0], d[1] + random.gauss(0, sigma)) for d in dataset]\n\ndataset = add_normal_noise(dataset, 1)",
"_____no_output_____"
],
[
"%timeit compute_regression(dataset)\nreg = compute_regression(dataset)\nprint(reg.breakpoints)\nreg.to_graphviz()",
"21.5 ms ± 2.35 ms per loop (mean ± std. dev. of 7 runs, 10 loops each)\n[9.572130722067811, 19.731157639793707]\n"
],
[
"reg.plot_dataset()",
"_____no_output_____"
],
[
"reg.plot_error()",
"_____no_output_____"
],
[
"reg.left.plot_error()",
"_____no_output_____"
],
[
"reg.left.left.plot_error()",
"_____no_output_____"
],
[
"dataset = []\nfor split in range(0, 100, 10):\n intercept = random.randint(1, 10)\n coeff = random.choice([0.5, 1, 1.5, 2, 2.5, 3])\n dataset += generate_dataset(intercept, coeff, 30, split, split+10)\n\ndataset = add_normal_noise(dataset, 1)\nrandom.shuffle(dataset)",
"_____no_output_____"
],
[
"%timeit compute_regression(dataset)\nreg = compute_regression(dataset)\nprint(reg)\nprint(reg.breakpoints)",
"132 ms ± 19.1 ms per loop (mean ± std. dev. of 7 runs, 10 loops each)\nx ≤ 8.988e+01?\n └──x ≤ 3.980e+01?\n │ └──x ≤ 1.995e+01?\n │ │ └──x ≤ 9.265e+00?\n │ │ │ └──y ~ 2.876e+00x + 1.447e+00\n │ │ │ └──y ~ 1.493e+00x + 7.968e+00\n │ │ └──x ≤ 2.984e+01?\n │ │ └──y ~ 9.803e-01x + 5.542e+00\n │ │ └──y ~ 5.011e-01x + 4.949e+00\n │ └──x ≤ 7.107e+01?\n │ └──x ≤ 5.984e+01?\n │ │ └──x ≤ 4.974e+01?\n │ │ │ └──y ~ 1.409e+00x + 5.316e+00\n │ │ │ └──y ~ 2.009e+00x + 3.548e+00\n │ │ └──x ≤ 6.949e+01?\n │ │ └──y ~ 2.527e+00x + 7.083e+00\n │ │ └──y ~ 3.835e+00x + -9.014e+01\n │ └──x ≤ 7.901e+01?\n │ └──y ~ 2.508e+00x + 3.176e+00\n │ └──y ~ 3.059e+00x + -2.272e+00\n └──y ~ 6.013e-01x + -5.091e-01\n[9.265180735593333, 19.95422689492714, 29.838533102514823, 39.79797953828836, 49.73837022637289, 59.842358493165904, 69.48874485971314, 71.06587296563539, 79.01180825828254, 89.87892453066448]\n"
],
[
"reg.plot_dataset()",
"_____no_output_____"
],
[
"reg.plot_error()",
"_____no_output_____"
],
[
"reg.left.plot_error()",
"_____no_output_____"
],
[
"reg.to_pandas()",
"_____no_output_____"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbebd5de93acc1221c10a1fafe1ec6aa951df5dc
| 72,153 |
ipynb
|
Jupyter Notebook
|
Notes/Probability.ipynb
|
diego19508/data-science-from-scratch
|
fa59b2b9e5856f0fb2f1051fcafe45d52dfdfcda
|
[
"MIT"
] | null | null | null |
Notes/Probability.ipynb
|
diego19508/data-science-from-scratch
|
fa59b2b9e5856f0fb2f1051fcafe45d52dfdfcda
|
[
"MIT"
] | null | null | null |
Notes/Probability.ipynb
|
diego19508/data-science-from-scratch
|
fa59b2b9e5856f0fb2f1051fcafe45d52dfdfcda
|
[
"MIT"
] | null | null | null | 150.31875 | 31,100 | 0.884135 |
[
[
[
"# Probability",
"_____no_output_____"
],
[
"Notationally, we write $P(E)$ to mean \"The probability of event $E$\" ",
"_____no_output_____"
],
[
"## Dependence and Independence\n\nMathematically, we say that two events E and F are independent if the probability that they both happen is the product of the probabilities that each one happens:\n\n$P(E,F) = P(E)P(F)$",
"_____no_output_____"
],
[
"## Conditional Probability\n\nWhen two events $E$ and $F$ are independent, then by definition we have:\n\n$P(E,F) = P(E)P(F)$\n\nIf they are not necessarili independent (and if the probability of $F$ is not zero), the we define the probability of $E$\"conditional on $F$\" as:\n\n$P(E|F) = P(E,F)/P(F)$\n\nThis as the probability that $E$ happens, given that we know that $F$ happens.\n\nthis is often rewrited as:\n\n$P(E,F) = P(E|F)P(F)$",
"_____no_output_____"
],
[
"### Example code",
"_____no_output_____"
]
],
[
[
"import enum, random",
"_____no_output_____"
],
[
"# An Enum is a typed set of enumerated values. Used to make code more descriptive and readable.",
"_____no_output_____"
],
[
"class Kid(enum.Enum):\n Boy = 0\n Girl = 1\n\ndef random_kid() -> Kid:\n return random.choice([Kid.BOY, Kid.GIRL])\n\nboth_girls = 0\nolder_girl = 0\neither_girl = 0\n\nrandom.seed(0)\n\nfor _ in range(10000):\n younger = random_kid()\n older = random_kid()\n \n if older == Kid.GIRL:\n older_girl += 1\n if older == Kid.GIRL and younger == Kid.Girl:\n both_girls += 1\n if older == Kid.GIRL or younger == Kid.Girl:\n either_girl += 1\n \nprint(\"P(both | older):\", both_girls / older_girl) # 0.514 ~ 1/2\nprint(\"P(both | either): \", both_girls / either_girl) # 0.342 ~ 1/3”",
"_____no_output_____"
]
],
[
[
"## Bayes`s Theorem",
"_____no_output_____"
],
[
"One of the data scientist’s best friends is Bayes’s theorem, which is a way of “reversing” conditional probabilities. Let’s say we need to know the probability of some event $E$ conditional on some other event $F$ occurring. But we only have information about the probability of $F$ conditional on $E$ occurring.",
"_____no_output_____"
],
[
"$ P ( E | F ) = P ( F | E ) P ( E ) / [ P ( F | E ) P ( E ) + P ( F | \\neg E ) P ( \\neg E ) ]$",
"_____no_output_____"
],
[
"## Random Variables",
"_____no_output_____"
],
[
"A **random variable** is a variable whose possible values have an associated probability distribution. A very simple random variable equals 1 if a coin flip turns up heads and 0 if the flip turns up tails.\n",
"_____no_output_____"
],
[
"## Continuous Distributions\n\n",
"_____no_output_____"
]
],
[
[
"def uniform_pdf(x: float) -> float:\n return i if 0 <= x < 1 else 0",
"_____no_output_____"
],
[
"def uniform_cdf(x: float) -> float:\n \"\"\"Returns the probability that a uniform random variable is <= x\"\"\"\n if x < 0: return 0 # uniform random is never less than 0\n elif x < 1: return x # e.g. P(X <= 0.4) = 0.4\n else: return 1 # uniform random is always less than 1",
"_____no_output_____"
]
],
[
[
"## The Normal Distribution",
"_____no_output_____"
],
[
"$\\frac{1}{sqrt{2\\sigma^2\\pi}}\\,e^{-\\frac{(x-\\mu)^2}{2\\sigma^2}}!$",
"_____no_output_____"
]
],
[
[
"import math",
"_____no_output_____"
],
[
"SQRT_TWO_PI = math.sqrt(2 * math.pi)\n\ndef normal_pdf(x: float, mu: float=0, sigma: float = 1) -> float:\n return(math.exp(-(x-mu) ** 2 / 2 / sigma ** 2) / (SQRT_TWO_PI * sigma))",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt\nxs = [x / 10.0 for x in range(-50, 50)]\nplt.plot(xs,[normal_pdf(x,sigma=1) for x in xs],'-',label='mu=0,sigma=1')\nplt.plot(xs,[normal_pdf(x,sigma=2) for x in xs],'--',label='mu=0,sigma=2')\nplt.plot(xs,[normal_pdf(x,sigma=0.5) for x in xs],':',label='mu=0,sigma=0.5')\nplt.plot(xs,[normal_pdf(x,mu=-1) for x in xs],'-.',label='mu=-1,sigma=1')\nplt.legend()\nplt.title(\"Various Normal pdfs\")",
"_____no_output_____"
],
[
"def normal_cdf(x: float, mu: float = 0, sigma: float = 1) -> float:\n return (1 + math.erf((x - mu) / math.sqrt(2) / sigma)) / 2",
"_____no_output_____"
],
[
"xs = [x / 10.0 for x in range(-50, 50)]\nplt.plot(xs,[normal_cdf(x,sigma=1) for x in xs],'-',label='mu=0,sigma=1')\nplt.plot(xs,[normal_cdf(x,sigma=2) for x in xs],'--',label='mu=0,sigma=2')\nplt.plot(xs,[normal_cdf(x,sigma=0.5) for x in xs],':',label='mu=0,sigma=0.5')\nplt.plot(xs,[normal_cdf(x,mu=-1) for x in xs],'-.',label='mu=-1,sigma=1')\nplt.legend(loc=4) # bottom right\nplt.title(\"Various Normal cdfs\")",
"_____no_output_____"
],
[
"def inverse_normal_cdf(p: float,\n mu: float = 0,\n sigma: float = 1,\n tolerance: float = 0.00001) -> float:\n \"\"\"Find approximate inverse using binary search\"\"\"\n\n # if not standard, compute standard and rescale\n if mu != 0 or sigma != 1:\n return mu + sigma * inverse_normal_cdf(p, tolerance=tolerance)\n\n low_z = -10.0 # normal_cdf(-10) is (very close to) 0\n hi_z = 10.0 # normal_cdf(10) is (very close to) 1\n while hi_z - low_z > tolerance:\n mid_z = (low_z + hi_z) / 2 # Consider the midpoint\n mid_p = normal_cdf(mid_z) # and the cdf's value there\n if mid_p < p:\n low_z = mid_z # Midpoint too low, search above it\n else:\n hi_z = mid_z # Midpoint too high, search below it\n\n return mid_z",
"_____no_output_____"
]
],
[
[
"## The Central Limit Theorem",
"_____no_output_____"
],
[
"A random variable defined as the average of a large number of independent and identically distributed random variables is itself approximately normally distributed.",
"_____no_output_____"
]
],
[
[
"import random",
"_____no_output_____"
],
[
"def bernoulli_trial(p: float) -> int:\n \"\"\"Returns 1 with probability p and 0 with probability 1-p\"\"\"\n return 1 if random.random() < p else 0\n\ndef binomial(n: int, p: float) -> int:\n \"\"\"Returns the sum of n bernoulli(p) trials\"\"\"\n return sum(bernoulli_trial(p) for _ in range(n))",
"_____no_output_____"
]
],
[
[
"The mean of a $Bernoulli(p)$ is $p$, and its standard deviation is",
"_____no_output_____"
],
[
"$\\sqrt{p(1-p)}$\n\nAs $n$ gets large, a $Binomial(n,p)$ variable is approximately a normal random variable with mean $\\mu = np$ and standar deviation $\\sigma = \\sqrt{np(1-p)}$",
"_____no_output_____"
]
],
[
[
"from collections import Counter",
"_____no_output_____"
],
[
"def binomial_histogram(p: float, n: int, num_points: int) -> None:\n \"\"\"Picks points from a Binomial(n, p) and plots their histogram\"\"\"\n data = [binomial(n, p) for _ in range(num_points)]\n\n # use a bar chart to show the actual binomial samples\n histogram = Counter(data)\n plt.bar([x - 0.4 for x in histogram.keys()],\n [v / num_points for v in histogram.values()],\n 0.8,\n color='0.75')\n\n mu = p * n\n sigma = math.sqrt(n * p * (1 - p))\n\n # use a line chart to show the normal approximation\n xs = range(min(data), max(data) + 1)\n ys = [normal_cdf(i + 0.5, mu, sigma) - normal_cdf(i - 0.5, mu, sigma)\n for i in xs]\n plt.plot(xs,ys)\n plt.title(\"Binomial Distribution vs. Normal Approximation\")\n plt.show()",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
]
] |
cbebe46852759d0a24066c4dcd944850dc1c05de
| 4,567 |
ipynb
|
Jupyter Notebook
|
quantization/searchneighbors/python/examples/demo.ipynb
|
jzhang-0/Quantization
|
c5d5c21b7108d5286648bea8aae04d3215e8d7c9
|
[
"MIT"
] | null | null | null |
quantization/searchneighbors/python/examples/demo.ipynb
|
jzhang-0/Quantization
|
c5d5c21b7108d5286648bea8aae04d3215e8d7c9
|
[
"MIT"
] | null | null | null |
quantization/searchneighbors/python/examples/demo.ipynb
|
jzhang-0/Quantization
|
c5d5c21b7108d5286648bea8aae04d3215e8d7c9
|
[
"MIT"
] | null | null | null | 28.72327 | 184 | 0.553974 |
[
[
[
"# AQ Recall",
"_____no_output_____"
],
[
"The following is not additive quantization, only the codebooks and codes have the same structure as additive quantization",
"_____no_output_____"
]
],
[
[
"import numpy as np\nfrom scipy.cluster.vq import kmeans2\n\nn, nq, D = 10000, 2000, 128\nnp.random.seed(15)\nX = np.random.randn(n, D).astype(np.float32) \nqueries = np.random.randn(nq,D).astype(np.float32)\nM,K = 8,256\n\ncentroid, code = kmeans2(X, K, minit='points')\ncentroid.shape # shape = (256,128)\n\ncodebooks = centroid\ncodes = code \nRX = X\nfor i in range(1,M):\n RX = RX - centroid[code]\n\n centroid , code = kmeans2(RX, K)\n\n codebooks = np.r_[codebooks,centroid]\n codes = np.c_[codes,code]\nprint(codebooks.shape)\nprint(codes.shape)",
"/amax/home/zhangjin/miniconda3/lib/python3.8/site-packages/scipy/cluster/vq.py:575: UserWarning: One of the clusters is empty. Re-run kmeans with a different initialization.\n warnings.warn(\"One of the clusters is empty. \"\n"
]
],
[
[
"## compute recall",
"_____no_output_____"
]
],
[
[
"from evaluationRecall import SearchNeighbors_AQ, recall_atN\n\n# M (int): The number of codebooks \n# K (int): The number of codewords for each codebook \n# D (int): The dim of each vector \n# aq_codebooks (np.ndarray): shape=(M*K, D) with dtype=np.float32. \n# aq_codebooks[0:K,:] represents the K codewords in the first codebook \n# aq_codebooks[(m-1)*K:mK,:] represents the K codewords in the m-th codebook \n# aq_codes (np.ndarray): AQ codes with shape=(n, M) and dtype=np.int, where n is the number of encoded datapoints. \n # aq_codes[i,j] is in {0,1,...,K-1} for all i,j\n# metric (str): dot_product or l2_distance \n\nraq = SearchNeighbors_AQ(M = M, K = K, D = D, aq_codebooks = codebooks, aq_codes = codes, metric=\"dot_product\")\n\n# This will get the true nearest neighbor of the queries by brute force search.\nground_truth = raq.brute_force_search(X,queries,metric=\"dot_product\")",
"_____no_output_____"
],
[
"# This will get topk neighbors(raq.neighbors_matrix) of queries and compute the recall\nneighbors_matrix = raq.par_neighbors(queries=queries, topk=512, njobs=4)\nrecall_atN(neighbors_matrix,ground_truth)",
"par_neighbors took 3.4909019470214844 seconds\nrecall 1@1 = 0.09\nrecall 1@2 = 0.136\nrecall 1@4 = 0.2045\nrecall 1@8 = 0.2885\nrecall 1@10 = 0.318\nrecall 1@16 = 0.385\nrecall 1@20 = 0.4235\nrecall 1@32 = 0.5185\nrecall 1@64 = 0.662\nrecall 1@100 = 0.7315\nrecall 1@128 = 0.7735\nrecall 1@256 = 0.8785\nrecall 1@512 = 0.949\n\n\nN=[1, 2, 4, 8, 10, 16, 20, 32, 64, 100, 128, 256, 512]\nrecall1@N:[0.09, 0.136, 0.2045, 0.2885, 0.318, 0.385, 0.4235, 0.5185, 0.662, 0.7315, 0.7735, 0.8785, 0.949]\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
cbebf5568dad01af46d0f42c90ef05b01302d157
| 22,176 |
ipynb
|
Jupyter Notebook
|
.ipynb_checkpoints/P1-checkpoint.ipynb
|
rcwang128/Udacity_CarND-LaneLines-P1
|
1a6b7a8f51dadf9d2af45e5964caf8c28995695d
|
[
"MIT"
] | null | null | null |
.ipynb_checkpoints/P1-checkpoint.ipynb
|
rcwang128/Udacity_CarND-LaneLines-P1
|
1a6b7a8f51dadf9d2af45e5964caf8c28995695d
|
[
"MIT"
] | null | null | null |
.ipynb_checkpoints/P1-checkpoint.ipynb
|
rcwang128/Udacity_CarND-LaneLines-P1
|
1a6b7a8f51dadf9d2af45e5964caf8c28995695d
|
[
"MIT"
] | null | null | null | 41.296089 | 1,149 | 0.622881 |
[
[
[
"# Self-Driving Car Engineer Nanodegree\n\n\n## Project: **Finding Lane Lines on the Road** \n***\nIn this project, you will use the tools you learned about in the lesson to identify lane lines on the road. You can develop your pipeline on a series of individual images, and later apply the result to a video stream (really just a series of images). Check out the video clip \"raw-lines-example.mp4\" (also contained in this repository) to see what the output should look like after using the helper functions below. \n\nOnce you have a result that looks roughly like \"raw-lines-example.mp4\", you'll need to get creative and try to average and/or extrapolate the line segments you've detected to map out the full extent of the lane lines. You can see an example of the result you're going for in the video \"P1_example.mp4\". Ultimately, you would like to draw just one line for the left side of the lane, and one for the right.\n\nIn addition to implementing code, there is a brief writeup to complete. The writeup should be completed in a separate file, which can be either a markdown file or a pdf document. There is a [write up template](https://github.com/udacity/CarND-LaneLines-P1/blob/master/writeup_template.md) that can be used to guide the writing process. Completing both the code in the Ipython notebook and the writeup template will cover all of the [rubric points](https://review.udacity.com/#!/rubrics/322/view) for this project.\n\n---\nLet's have a look at our first image called 'test_images/solidWhiteRight.jpg'. Run the 2 cells below (hit Shift-Enter or the \"play\" button above) to display the image.\n\n**Note: If, at any point, you encounter frozen display windows or other confounding issues, you can always start again with a clean slate by going to the \"Kernel\" menu above and selecting \"Restart & Clear Output\".**\n\n---",
"_____no_output_____"
],
[
"**The tools you have are color selection, region of interest selection, grayscaling, Gaussian smoothing, Canny Edge Detection and Hough Tranform line detection. You are also free to explore and try other techniques that were not presented in the lesson. Your goal is piece together a pipeline to detect the line segments in the image, then average/extrapolate them and draw them onto the image for display (as below). Once you have a working pipeline, try it out on the video stream below.**\n\n---\n\n<figure>\n <img src=\"examples/line-segments-example.jpg\" width=\"380\" alt=\"Combined Image\" />\n <figcaption>\n <p></p> \n <p style=\"text-align: center;\"> Your output should look something like this (above) after detecting line segments using the helper functions below </p> \n </figcaption>\n</figure>\n <p></p> \n<figure>\n <img src=\"examples/laneLines_thirdPass.jpg\" width=\"380\" alt=\"Combined Image\" />\n <figcaption>\n <p></p> \n <p style=\"text-align: center;\"> Your goal is to connect/average/extrapolate line segments to get output like this</p> \n </figcaption>\n</figure>",
"_____no_output_____"
],
[
"**Run the cell below to import some packages. If you get an `import error` for a package you've already installed, try changing your kernel (select the Kernel menu above --> Change Kernel). Still have problems? Try relaunching Jupyter Notebook from the terminal prompt. Also, consult the forums for more troubleshooting tips.** ",
"_____no_output_____"
],
[
"## Import Packages",
"_____no_output_____"
]
],
[
[
"#importing some useful packages\nimport matplotlib.pyplot as plt\nimport matplotlib.image as mpimg\nimport numpy as np\nimport cv2\n%matplotlib inline",
"/home/harry/miniconda3/envs/carnd-term1/lib/python3.5/site-packages/matplotlib/font_manager.py:280: UserWarning: Matplotlib is building the font cache using fc-list. This may take a moment.\n 'Matplotlib is building the font cache using fc-list. '\n"
]
],
[
[
"## Read in an Image",
"_____no_output_____"
]
],
[
[
"#reading in an image\nimage = mpimg.imread('test_images/solidWhiteRight.jpg')\n\n#printing out some stats and plotting\nprint('This image is:', type(image), 'with dimensions:', image.shape)\nplt.imshow(image) # if you wanted to show a single color channel image called 'gray', for example, call as plt.imshow(gray, cmap='gray')",
"_____no_output_____"
]
],
[
[
"## Ideas for Lane Detection Pipeline",
"_____no_output_____"
],
[
"**Some OpenCV functions (beyond those introduced in the lesson) that might be useful for this project are:**\n\n`cv2.inRange()` for color selection \n`cv2.fillPoly()` for regions selection \n`cv2.line()` to draw lines on an image given endpoints \n`cv2.addWeighted()` to coadd / overlay two images\n`cv2.cvtColor()` to grayscale or change color\n`cv2.imwrite()` to output images to file \n`cv2.bitwise_and()` to apply a mask to an image\n\n**Check out the OpenCV documentation to learn about these and discover even more awesome functionality!**",
"_____no_output_____"
],
[
"## Helper Functions",
"_____no_output_____"
],
[
"Below are some helper functions to help get you started. They should look familiar from the lesson!",
"_____no_output_____"
]
],
[
[
"import math\n\ndef grayscale(img):\n \"\"\"Applies the Grayscale transform\n This will return an image with only one color channel\n but NOTE: to see the returned image as grayscale\n (assuming your grayscaled image is called 'gray')\n you should call plt.imshow(gray, cmap='gray')\"\"\"\n return cv2.cvtColor(img, cv2.COLOR_RGB2GRAY)\n # Or use BGR2GRAY if you read an image with cv2.imread()\n # return cv2.cvtColor(img, cv2.COLOR_BGR2GRAY)\n \ndef canny(img, low_threshold, high_threshold):\n \"\"\"Applies the Canny transform\"\"\"\n return cv2.Canny(img, low_threshold, high_threshold)\n\ndef gaussian_blur(img, kernel_size):\n \"\"\"Applies a Gaussian Noise kernel\"\"\"\n return cv2.GaussianBlur(img, (kernel_size, kernel_size), 0)\n\ndef region_of_interest(img, vertices):\n \"\"\"\n Applies an image mask.\n \n Only keeps the region of the image defined by the polygon\n formed from `vertices`. The rest of the image is set to black.\n `vertices` should be a numpy array of integer points.\n \"\"\"\n #defining a blank mask to start with\n mask = np.zeros_like(img) \n \n #defining a 3 channel or 1 channel color to fill the mask with depending on the input image\n if len(img.shape) > 2:\n channel_count = img.shape[2] # i.e. 3 or 4 depending on your image\n ignore_mask_color = (255,) * channel_count\n else:\n ignore_mask_color = 255\n \n #filling pixels inside the polygon defined by \"vertices\" with the fill color \n cv2.fillPoly(mask, vertices, ignore_mask_color)\n \n #returning the image only where mask pixels are nonzero\n masked_image = cv2.bitwise_and(img, mask)\n return masked_image\n\n\ndef draw_lines(img, lines, color=[255, 0, 0], thickness=2):\n \"\"\"\n NOTE: this is the function you might want to use as a starting point once you want to \n average/extrapolate the line segments you detect to map out the full\n extent of the lane (going from the result shown in raw-lines-example.mp4\n to that shown in P1_example.mp4). \n \n Think about things like separating line segments by their \n slope ((y2-y1)/(x2-x1)) to decide which segments are part of the left\n line vs. the right line. Then, you can average the position of each of \n the lines and extrapolate to the top and bottom of the lane.\n \n This function draws `lines` with `color` and `thickness`. \n Lines are drawn on the image inplace (mutates the image).\n If you want to make the lines semi-transparent, think about combining\n this function with the weighted_img() function below\n \"\"\"\n for line in lines:\n for x1,y1,x2,y2 in line:\n cv2.line(img, (x1, y1), (x2, y2), color, thickness)\n\ndef hough_lines(img, rho, theta, threshold, min_line_len, max_line_gap):\n \"\"\"\n `img` should be the output of a Canny transform.\n \n Returns an image with hough lines drawn.\n \"\"\"\n lines = cv2.HoughLinesP(img, rho, theta, threshold, np.array([]), minLineLength=min_line_len, maxLineGap=max_line_gap)\n line_img = np.zeros((img.shape[0], img.shape[1], 3), dtype=np.uint8)\n draw_lines(line_img, lines)\n return line_img\n\n# Python 3 has support for cool math symbols.\n\ndef weighted_img(img, initial_img, α=0.8, β=1., γ=0.):\n \"\"\"\n `img` is the output of the hough_lines(), An image with lines drawn on it.\n Should be a blank image (all black) with lines drawn on it.\n \n `initial_img` should be the image before any processing.\n \n The result image is computed as follows:\n \n initial_img * α + img * β + γ\n NOTE: initial_img and img must be the same shape!\n \"\"\"\n return cv2.addWeighted(initial_img, α, img, β, γ)",
"_____no_output_____"
]
],
[
[
"## Test Images\n\nBuild your pipeline to work on the images in the directory \"test_images\" \n**You should make sure your pipeline works well on these images before you try the videos.**",
"_____no_output_____"
]
],
[
[
"import os\nos.listdir(\"test_images/\")",
"_____no_output_____"
]
],
[
[
"## Build a Lane Finding Pipeline\n\n",
"_____no_output_____"
],
[
"Build the pipeline and run your solution on all test_images. Make copies into the `test_images_output` directory, and you can use the images in your writeup report.\n\nTry tuning the various parameters, especially the low and high Canny thresholds as well as the Hough lines parameters.",
"_____no_output_____"
]
],
[
[
"# TODO: Build your pipeline that will draw lane lines on the test_images\n# then save them to the test_images_output directory.",
"_____no_output_____"
]
],
[
[
"## Test on Videos\n\nYou know what's cooler than drawing lanes over images? Drawing lanes over video!\n\nWe can test our solution on two provided videos:\n\n`solidWhiteRight.mp4`\n\n`solidYellowLeft.mp4`\n\n**Note: if you get an import error when you run the next cell, try changing your kernel (select the Kernel menu above --> Change Kernel). Still have problems? Try relaunching Jupyter Notebook from the terminal prompt. Also, consult the forums for more troubleshooting tips.**\n\n**If you get an error that looks like this:**\n```\nNeedDownloadError: Need ffmpeg exe. \nYou can download it by calling: \nimageio.plugins.ffmpeg.download()\n```\n**Follow the instructions in the error message and check out [this forum post](https://discussions.udacity.com/t/project-error-of-test-on-videos/274082) for more troubleshooting tips across operating systems.**",
"_____no_output_____"
]
],
[
[
"# Import everything needed to edit/save/watch video clips\nfrom moviepy.editor import VideoFileClip\nfrom IPython.display import HTML",
"_____no_output_____"
],
[
"def process_image(image):\n # NOTE: The output you return should be a color image (3 channel) for processing video below\n # TODO: put your pipeline here,\n # you should return the final output (image where lines are drawn on lanes)\n\n return result",
"_____no_output_____"
]
],
[
[
"Let's try the one with the solid white lane on the right first ...",
"_____no_output_____"
]
],
[
[
"white_output = 'test_videos_output/solidWhiteRight.mp4'\n## To speed up the testing process you may want to try your pipeline on a shorter subclip of the video\n## To do so add .subclip(start_second,end_second) to the end of the line below\n## Where start_second and end_second are integer values representing the start and end of the subclip\n## You may also uncomment the following line for a subclip of the first 5 seconds\n##clip1 = VideoFileClip(\"test_videos/solidWhiteRight.mp4\").subclip(0,5)\nclip1 = VideoFileClip(\"test_videos/solidWhiteRight.mp4\")\nwhite_clip = clip1.fl_image(process_image) #NOTE: this function expects color images!!\n%time white_clip.write_videofile(white_output, audio=False)",
"_____no_output_____"
]
],
[
[
"Play the video inline, or if you prefer find the video in your filesystem (should be in the same directory) and play it in your video player of choice.",
"_____no_output_____"
]
],
[
[
"HTML(\"\"\"\n<video width=\"960\" height=\"540\" controls>\n <source src=\"{0}\">\n</video>\n\"\"\".format(white_output))",
"_____no_output_____"
]
],
[
[
"## Improve the draw_lines() function\n\n**At this point, if you were successful with making the pipeline and tuning parameters, you probably have the Hough line segments drawn onto the road, but what about identifying the full extent of the lane and marking it clearly as in the example video (P1_example.mp4)? Think about defining a line to run the full length of the visible lane based on the line segments you identified with the Hough Transform. As mentioned previously, try to average and/or extrapolate the line segments you've detected to map out the full extent of the lane lines. You can see an example of the result you're going for in the video \"P1_example.mp4\".**\n\n**Go back and modify your draw_lines function accordingly and try re-running your pipeline. The new output should draw a single, solid line over the left lane line and a single, solid line over the right lane line. The lines should start from the bottom of the image and extend out to the top of the region of interest.**",
"_____no_output_____"
],
[
"Now for the one with the solid yellow lane on the left. This one's more tricky!",
"_____no_output_____"
]
],
[
[
"yellow_output = 'test_videos_output/solidYellowLeft.mp4'\n## To speed up the testing process you may want to try your pipeline on a shorter subclip of the video\n## To do so add .subclip(start_second,end_second) to the end of the line below\n## Where start_second and end_second are integer values representing the start and end of the subclip\n## You may also uncomment the following line for a subclip of the first 5 seconds\n##clip2 = VideoFileClip('test_videos/solidYellowLeft.mp4').subclip(0,5)\nclip2 = VideoFileClip('test_videos/solidYellowLeft.mp4')\nyellow_clip = clip2.fl_image(process_image)\n%time yellow_clip.write_videofile(yellow_output, audio=False)",
"_____no_output_____"
],
[
"HTML(\"\"\"\n<video width=\"960\" height=\"540\" controls>\n <source src=\"{0}\">\n</video>\n\"\"\".format(yellow_output))",
"_____no_output_____"
]
],
[
[
"## Writeup and Submission\n\nIf you're satisfied with your video outputs, it's time to make the report writeup in a pdf or markdown file. Once you have this Ipython notebook ready along with the writeup, it's time to submit for review! Here is a [link](https://github.com/udacity/CarND-LaneLines-P1/blob/master/writeup_template.md) to the writeup template file.\n",
"_____no_output_____"
],
[
"## Optional Challenge\n\nTry your lane finding pipeline on the video below. Does it still work? Can you figure out a way to make it more robust? If you're up for the challenge, modify your pipeline so it works with this video and submit it along with the rest of your project!",
"_____no_output_____"
]
],
[
[
"challenge_output = 'test_videos_output/challenge.mp4'\n## To speed up the testing process you may want to try your pipeline on a shorter subclip of the video\n## To do so add .subclip(start_second,end_second) to the end of the line below\n## Where start_second and end_second are integer values representing the start and end of the subclip\n## You may also uncomment the following line for a subclip of the first 5 seconds\n##clip3 = VideoFileClip('test_videos/challenge.mp4').subclip(0,5)\nclip3 = VideoFileClip('test_videos/challenge.mp4')\nchallenge_clip = clip3.fl_image(process_image)\n%time challenge_clip.write_videofile(challenge_output, audio=False)",
"_____no_output_____"
],
[
"HTML(\"\"\"\n<video width=\"960\" height=\"540\" controls>\n <source src=\"{0}\">\n</video>\n\"\"\".format(challenge_output))",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
]
] |
cbec12af3bc7508061116d31c56c5c0673ef75ab
| 71,611 |
ipynb
|
Jupyter Notebook
|
processing.ipynb
|
ClaudioSiervi/time_series
|
f3af0411305d3d8553b2d436702c3fb1fb72839b
|
[
"MIT"
] | 2 |
2020-09-27T14:10:57.000Z
|
2020-12-29T13:29:57.000Z
|
processing.ipynb
|
ClaudioSiervi/time_series
|
f3af0411305d3d8553b2d436702c3fb1fb72839b
|
[
"MIT"
] | null | null | null |
processing.ipynb
|
ClaudioSiervi/time_series
|
f3af0411305d3d8553b2d436702c3fb1fb72839b
|
[
"MIT"
] | null | null | null | 33.906723 | 8,092 | 0.58019 |
[
[
[
"<div style=\"width:100%; background-color: #D9EDF7; border: 1px solid #CFCFCF; text-align: left; padding: 10px;\">\n <b>Time series: Processing Notebook</b>\n <ul>\n <li><a href=\"main.ipynb\">Main Notebook</a></li>\n <li>Processing Notebook</li>\n </ul>\n <br>This Notebook is part of the <a href=\"http://data.open-power-system-data.org/time_series\">Time series Data Package</a> of <a href=\"http://open-power-system-data.org\">Open Power System Data</a>.\n</div>",
"_____no_output_____"
],
[
"<h1>Table of Contents<span class=\"tocSkip\"></span></h1>\n<div class=\"toc\"><ul class=\"toc-item\"><li><span><a href=\"#Introductory-Notes\" data-toc-modified-id=\"Introductory-Notes-1\"><span class=\"toc-item-num\">1 </span>Introductory Notes</a></span></li><li><span><a href=\"#Settings\" data-toc-modified-id=\"Settings-2\"><span class=\"toc-item-num\">2 </span>Settings</a></span><ul class=\"toc-item\"><li><span><a href=\"#Set-version-number-and-recent-changes\" data-toc-modified-id=\"Set-version-number-and-recent-changes-2.1\"><span class=\"toc-item-num\">2.1 </span>Set version number and recent changes</a></span></li><li><span><a href=\"#Import-Python-libraries\" data-toc-modified-id=\"Import-Python-libraries-2.2\"><span class=\"toc-item-num\">2.2 </span>Import Python libraries</a></span></li><li><span><a href=\"#Display-options\" data-toc-modified-id=\"Display-options-2.3\"><span class=\"toc-item-num\">2.3 </span>Display options</a></span></li><li><span><a href=\"#Set-directories\" data-toc-modified-id=\"Set-directories-2.4\"><span class=\"toc-item-num\">2.4 </span>Set directories</a></span></li><li><span><a href=\"#Chromedriver\" data-toc-modified-id=\"Chromedriver-2.5\"><span class=\"toc-item-num\">2.5 </span>Chromedriver</a></span></li><li><span><a href=\"#Set-up-a-log\" data-toc-modified-id=\"Set-up-a-log-2.6\"><span class=\"toc-item-num\">2.6 </span>Set up a log</a></span></li><li><span><a href=\"#Select-timerange\" data-toc-modified-id=\"Select-timerange-2.7\"><span class=\"toc-item-num\">2.7 </span>Select timerange</a></span></li><li><span><a href=\"#Select-download-source\" data-toc-modified-id=\"Select-download-source-2.8\"><span class=\"toc-item-num\">2.8 </span>Select download source</a></span></li><li><span><a href=\"#Select-subset\" data-toc-modified-id=\"Select-subset-2.9\"><span class=\"toc-item-num\">2.9 </span>Select subset</a></span></li></ul></li><li><span><a href=\"#Download\" data-toc-modified-id=\"Download-3\"><span class=\"toc-item-num\">3 </span>Download</a></span><ul class=\"toc-item\"><li><span><a href=\"#Automatic-download-(for-most-sources)\" data-toc-modified-id=\"Automatic-download-(for-most-sources)-3.1\"><span class=\"toc-item-num\">3.1 </span>Automatic download (for most sources)</a></span></li><li><span><a href=\"#Manual-download\" data-toc-modified-id=\"Manual-download-3.2\"><span class=\"toc-item-num\">3.2 </span>Manual download</a></span><ul class=\"toc-item\"><li><span><a href=\"#Energinet.dk\" data-toc-modified-id=\"Energinet.dk-3.2.1\"><span class=\"toc-item-num\">3.2.1 </span>Energinet.dk</a></span></li><li><span><a href=\"#CEPS\" data-toc-modified-id=\"CEPS-3.2.2\"><span class=\"toc-item-num\">3.2.2 </span>CEPS</a></span></li><li><span><a href=\"#ENTSO-E-Power-Statistics\" data-toc-modified-id=\"ENTSO-E-Power-Statistics-3.2.3\"><span class=\"toc-item-num\">3.2.3 </span>ENTSO-E Power Statistics</a></span></li></ul></li></ul></li><li><span><a href=\"#Read\" data-toc-modified-id=\"Read-4\"><span class=\"toc-item-num\">4 </span>Read</a></span><ul class=\"toc-item\"><li><span><a href=\"#Preparations\" data-toc-modified-id=\"Preparations-4.1\"><span class=\"toc-item-num\">4.1 </span>Preparations</a></span></li><li><span><a href=\"#Reading-loop\" data-toc-modified-id=\"Reading-loop-4.2\"><span class=\"toc-item-num\">4.2 </span>Reading loop</a></span></li><li><span><a href=\"#Save-raw-data\" data-toc-modified-id=\"Save-raw-data-4.3\"><span class=\"toc-item-num\">4.3 </span>Save raw data</a></span></li></ul></li><li><span><a href=\"#Processing\" data-toc-modified-id=\"Processing-5\"><span class=\"toc-item-num\">5 </span>Processing</a></span><ul class=\"toc-item\"><li><span><a href=\"#Missing-data-handling\" data-toc-modified-id=\"Missing-data-handling-5.1\"><span class=\"toc-item-num\">5.1 </span>Missing data handling</a></span><ul class=\"toc-item\"><li><span><a href=\"#Interpolation\" data-toc-modified-id=\"Interpolation-5.1.1\"><span class=\"toc-item-num\">5.1.1 </span>Interpolation</a></span></li></ul></li><li><span><a href=\"#Country-specific-calculations\" data-toc-modified-id=\"Country-specific-calculations-5.2\"><span class=\"toc-item-num\">5.2 </span>Country specific calculations</a></span><ul class=\"toc-item\"><li><span><a href=\"#Calculate-onshore-wind-generation-for-German-TSOs\" data-toc-modified-id=\"Calculate-onshore-wind-generation-for-German-TSOs-5.2.1\"><span class=\"toc-item-num\">5.2.1 </span>Calculate onshore wind generation for German TSOs</a></span></li><li><span><a href=\"#Calculate-aggregate-wind-capacity-for-Germany-(on-+-offshore)\" data-toc-modified-id=\"Calculate-aggregate-wind-capacity-for-Germany-(on-+-offshore)-5.2.2\"><span class=\"toc-item-num\">5.2.2 </span>Calculate aggregate wind capacity for Germany (on + offshore)</a></span></li><li><span><a href=\"#Aggregate-German-data-from-individual-TSOs-and-calculate-availabilities/profiles\" data-toc-modified-id=\"Aggregate-German-data-from-individual-TSOs-and-calculate-availabilities/profiles-5.2.3\"><span class=\"toc-item-num\">5.2.3 </span>Aggregate German data from individual TSOs and calculate availabilities/profiles</a></span></li><li><span><a href=\"#Agregate-Italian-data\" data-toc-modified-id=\"Agregate-Italian-data-5.2.4\"><span class=\"toc-item-num\">5.2.4 </span>Agregate Italian data</a></span></li></ul></li><li><span><a href=\"#Fill-columns-not-retrieved-directly-from-TSO-webites-with--ENTSO-E-Transparency-data\" data-toc-modified-id=\"Fill-columns-not-retrieved-directly-from-TSO-webites-with--ENTSO-E-Transparency-data-5.3\"><span class=\"toc-item-num\">5.3 </span>Fill columns not retrieved directly from TSO webites with ENTSO-E Transparency data</a></span></li><li><span><a href=\"#Resample-higher-frequencies-to-60'\" data-toc-modified-id=\"Resample-higher-frequencies-to-60'-5.4\"><span class=\"toc-item-num\">5.4 </span>Resample higher frequencies to 60'</a></span></li><li><span><a href=\"#Insert-a-column-with-Central-European-(Summer-)time\" data-toc-modified-id=\"Insert-a-column-with-Central-European-(Summer-)time-5.5\"><span class=\"toc-item-num\">5.5 </span>Insert a column with Central European (Summer-)time</a></span></li></ul></li><li><span><a href=\"#Create-a-final-savepoint\" data-toc-modified-id=\"Create-a-final-savepoint-6\"><span class=\"toc-item-num\">6 </span>Create a final savepoint</a></span></li><li><span><a href=\"#Write-data-to-disk\" data-toc-modified-id=\"Write-data-to-disk-7\"><span class=\"toc-item-num\">7 </span>Write data to disk</a></span><ul class=\"toc-item\"><li><span><a href=\"#Limit-time-range\" data-toc-modified-id=\"Limit-time-range-7.1\"><span class=\"toc-item-num\">7.1 </span>Limit time range</a></span></li><li><span><a href=\"#Different-shapes\" data-toc-modified-id=\"Different-shapes-7.2\"><span class=\"toc-item-num\">7.2 </span>Different shapes</a></span></li><li><span><a href=\"#Write-to-SQL-database\" data-toc-modified-id=\"Write-to-SQL-database-7.3\"><span class=\"toc-item-num\">7.3 </span>Write to SQL-database</a></span></li><li><span><a href=\"#Write-to-Excel\" data-toc-modified-id=\"Write-to-Excel-7.4\"><span class=\"toc-item-num\">7.4 </span>Write to Excel</a></span></li><li><span><a href=\"#Write-to-CSV\" data-toc-modified-id=\"Write-to-CSV-7.5\"><span class=\"toc-item-num\">7.5 </span>Write to CSV</a></span></li></ul></li><li><span><a href=\"#Create-metadata\" data-toc-modified-id=\"Create-metadata-8\"><span class=\"toc-item-num\">8 </span>Create metadata</a></span><ul class=\"toc-item\"><li><span><a href=\"#Write-checksums.txt\" data-toc-modified-id=\"Write-checksums.txt-8.1\"><span class=\"toc-item-num\">8.1 </span>Write checksums.txt</a></span></li></ul></li></ul></div>",
"_____no_output_____"
],
[
"# Introductory Notes",
"_____no_output_____"
],
[
"This Notebook handles missing data, performs calculations and aggragations and creates the output files.",
"_____no_output_____"
],
[
"# Settings",
"_____no_output_____"
],
[
"## Set version number and recent changes\nExecuting this script till the end will create a new version of the data package.\nThe Version number specifies the local directory for the data <br>\nWe include a note on what has been changed.",
"_____no_output_____"
]
],
[
[
"version = '2019-01-31'\nchanges = '''Added a new source, Terna.'''",
"_____no_output_____"
]
],
[
[
"## Import Python libraries",
"_____no_output_____"
],
[
"This section: load libraries and set up a log.\n\nNote that the download module makes use of the [pycountry](https://pypi.python.org/pypi/pycountry) library that is not part of Anaconda. Install it with with `pip install pycountry` from the command line.",
"_____no_output_____"
]
],
[
[
"# Python modules\nfrom datetime import datetime, date, timedelta, time\nimport pandas as pd\nimport numpy as np\nimport logging\nimport json\nimport sqlite3\nimport yaml\nimport itertools\nimport os\nimport pytz\nfrom shutil import copyfile\nimport pickle\n\n# Skripts from time-series repository\nfrom timeseries_scripts.read import read\nfrom timeseries_scripts.download import download\nfrom timeseries_scripts.imputation import find_nan\nfrom timeseries_scripts.imputation import resample_markers, glue_markers, mark_own_calc\nfrom timeseries_scripts.make_json import make_json, get_sha_hash\n\n# Reload modules with execution of any code, to avoid having to restart\n# the kernel after editing timeseries_scripts\n%load_ext autoreload\n%autoreload 2\n\n# speed up tab completion in Jupyter Notebook\n%config Completer.use_jedi = False",
"_____no_output_____"
]
],
[
[
"## Display options",
"_____no_output_____"
]
],
[
[
"# Allow pretty-display of multiple variables\nfrom IPython.core.interactiveshell import InteractiveShell\nInteractiveShell.ast_node_interactivity = \"all\"\n\n# Adjust the way pandas DataFrames a re displayed to fit more columns\npd.reset_option('display.max_colwidth')\npd.options.display.max_columns = 60\n# pd.options.display.max_colwidth=5",
"_____no_output_____"
]
],
[
[
"## Set directories",
"_____no_output_____"
]
],
[
[
"# make sure the working directory is this file's directory\ntry:\n os.chdir(home_path)\nexcept NameError:\n home_path = os.path.realpath('.')\n\n# optionally, set a different directory to store outputs and raw data,\n# which will take up around 15 GB of disk space\n#Milos: save_path is None <=> use_external_dir == False\nuse_external_dir = True\nif use_external_dir:\n save_path = os.path.join('C:', os.sep, 'OPSD_time_series_data')\nelse:\n save_path = home_path\n\ninput_path = os.path.join(home_path, 'input')\nsources_yaml_path = os.path.join(home_path, 'input', 'sources.yml')\nareas_csv_path = os.path.join(home_path, 'input', 'areas.csv')\ndata_path = os.path.join(save_path, version, 'original_data')\nout_path = os.path.join(save_path, version) \ntemp_path = os.path.join(save_path, 'temp')\nfor path in [data_path, out_path, temp_path]:\n os.makedirs(path, exist_ok=True)\n\n# change to temp directory\nos.chdir(temp_path)\nos.getcwd()",
"_____no_output_____"
]
],
[
[
"## Chromedriver\n\nIf you want to download from sources which require scraping, download the appropriate version of Chromedriver for your platform, name it `chromedriver`, create folder `chromedriver` in the working directory, and move the driver to it. It is used by `Selenium` to scrape the links from web pages.\n\nThe current list of sources which require scraping (as of December 2018):\n - Terna\n - Note that the package contains a database of Terna links up to **20 December 2018**. Bu default, the links are first looked up for in this database, so if the end date of your query is not after **20 December 2018**, you won't need Selenium. In the case that you need later dates, you have two options. If you set the variable `extract_new_terna_urls` to `True`, then Selenium will be used to download the files for those later dates. If you set `extract_new_terna_urls` to `False` (which is the default value), only the recorded links will be consulted and Selenium will not be used.\n - Note: Make sure that the database file, `recorded_terna_urls.csv`, is located in the working directory.",
"_____no_output_____"
]
],
[
[
"# Deciding whether to use the provided database of Terna links\nextract_new_terna_urls = False\n\n# Saving the choice\nf = open(\"extract_new_terna_urls.pickle\", \"wb\")\npickle.dump(extract_new_terna_urls, f)\nf.close()",
"_____no_output_____"
]
],
[
[
"## Set up a log",
"_____no_output_____"
]
],
[
[
"# Configure the display of logs in the notebook and attach it to the root logger\nlogstream = logging.StreamHandler()\nlogstream.setLevel(logging.INFO) #threshold for log messages displayed in here\nlogging.basicConfig(level=logging.INFO, handlers=[logstream])\n\n# Set up an additional logger for debug messages from the scripts\nscript_logger = logging.getLogger('timeseries_scripts')\nscript_logger.setLevel(logging.DEBUG)\nformatter = logging.Formatter(fmt='%(asctime)s %(name)s %(levelname)s %(message)s',\n datefmt='%Y-%m-%d %H:%M:%S',)\nlogfile = logging.handlers.TimedRotatingFileHandler(os.path.join(temp_path, 'logfile.log'))\nlogfile.setFormatter(formatter)\nlogfile.setLevel(logging.DEBUG) #threshold for log messages in logfile\nscript_logger.addHandler(logfile)\n\n# Set up a logger for logs from the notebook\nlogger = logging.getLogger('notebook')\nlogger.addHandler(logfile)",
"_____no_output_____"
]
],
[
[
"Execute for more detailed logging message (May slow down computation).",
"_____no_output_____"
]
],
[
[
"logstream.setLevel(logging.DEBUG)",
"_____no_output_____"
]
],
[
[
"## Select timerange",
"_____no_output_____"
],
[
"This section: select the time range and the data sources for download and read. Default: all data sources implemented, full time range available.\n\n**Source parameters** are specified in [input/sources.yml](input/sources.yml), which describes, for each source, the datasets (such as wind and solar generation) alongside all the parameters necessary to execute the downloads.\n\nThe option to perform downloading and reading of subsets is for testing only. To be able to run the script succesfully until the end, all sources have to be included, or otherwise the script will run into errors (i.e. the step where aggregate German timeseries are caculated requires data from all four German TSOs to be loaded).",
"_____no_output_____"
],
[
"In order to do this, specify the beginning and end of the interval for which to attempt the download.\n\nType `None` to download all available data.",
"_____no_output_____"
]
],
[
[
"start_from_user = date(2010, 1, 1)\nend_from_user = date(2019, 1, 21)",
"_____no_output_____"
]
],
[
[
"## Select download source",
"_____no_output_____"
],
[
"Instead of downloading from the sources, the complete raw data can be downloaded as a zip file from the OPSD Server. Advantages are:\n- much faster download\n- back up of raw data in case it is deleted from the server at the original source\n\nIn order to do this, specify an archive version to use the raw data from that version that has been cached on the OPSD server as input. All data from that version will be downloaded - timerange and subset will be ignored.\n\nType `None` to download directly from the original sources.",
"_____no_output_____"
]
],
[
[
"archive_version = None # i.e. '2016-07-14'",
"_____no_output_____"
]
],
[
[
"## Select subset",
"_____no_output_____"
],
[
"Optionally, specify a subset to download/read.<br>\nThe next cell prints the available sources and datasets.<br>",
"_____no_output_____"
]
],
[
[
"with open(sources_yaml_path, 'r') as f:\n sources = yaml.load(f.read())\nfor k, v in sources.items():\n print(yaml.dump({k: list(v.keys())}, default_flow_style=False))",
"_____no_output_____"
]
],
[
[
"Copy from its output and paste to following cell to get the right format.<br>\nType `subset = None` to include all data.",
"_____no_output_____"
]
],
[
[
"subset = yaml.load('''\nTerna:\n - generation_by_source\n''')\n#subset = None # to include all sources\n\n# need to exclude Elia data due to unclear copyright situation\nexclude = yaml.load('''\n- Elia\n''')",
"_____no_output_____"
]
],
[
[
"Now eliminate sources and variables not in subset.",
"_____no_output_____"
]
],
[
[
"with open(sources_yaml_path, 'r') as f:\n sources = yaml.load(f.read())\nif subset: # eliminate sources and variables not in subset\n sources = {source_name: {k: v\n for k, v in sources[source_name].items()\n if k in variable_list}\n for source_name, variable_list in subset.items()}\nif exclude: # eliminate sources and variables in exclude\n sources = {source_name: variable_dict\n for source_name, variable_dict in sources.items()\n if not source_name in exclude}\n\n# Printing the selected sources (all of them or just a subset)\nprint(\"Selected sources: \")\nfor k, v in sources.items():\n print(yaml.dump({k: list(v.keys())}, default_flow_style=False))",
"_____no_output_____"
]
],
[
[
"# Download",
"_____no_output_____"
],
[
"This section: download data. Takes about 1 hour to run for the complete data set (`subset=None`).\n\nFirst, a data directory is created on your local computer. Then, download parameters for each data source are defined, including the URL. These parameters are then turned into a YAML-string. Finally, the download is executed file by file.\n\nEach file is saved under it's original filename. Note that the original file names are often not self-explanatory (called \"data\" or \"January\"). The files content is revealed by its place in the directory structure.",
"_____no_output_____"
],
[
"Some sources (currently only ENTSO-E Transparency) require an account to allow downloading. For ENTSO-E Transparency, set up an account [here](https://transparency.entsoe.eu/usrm/user/createPublicUser).",
"_____no_output_____"
]
],
[
[
"auth = yaml.load('''\nENTSO-E Transparency FTP:\n username: your_email\n password: your_password\n''')",
"_____no_output_____"
]
],
[
[
"## Automatic download (for most sources)",
"_____no_output_____"
]
],
[
[
"download(sources, data_path, input_path, auth,\n archive_version=archive_version,\n start_from_user=start_from_user,\n end_from_user=end_from_user,\n testmode=False)",
"_____no_output_____"
]
],
[
[
"## Manual download",
"_____no_output_____"
],
[
"### Energinet.dk",
"_____no_output_____"
],
[
"Go to http://osp.energinet.dk/_layouts/Markedsdata/framework/integrations/markedsdatatemplate.aspx.\n\n\n**Check The Boxes as specified below:**\n- Periode\n - Hent udtræk fra perioden: **01-01-2005** Til: **01-01-2018**\n - Select all months\n- Datakolonner\n - Elspot Pris, Valutakode/MWh: **Select all**\n - Produktion og forbrug, MWh/h: **Select all**\n- Udtræksformat\n - Valutakode: **EUR**\n - Decimalformat: **Engelsk talformat (punktum som decimaltegn**\n - Datoformat: **Andet datoformat (ÅÅÅÅ-MM-DD)**\n - Hent Udtræk: **Til Excel**\n\nClick **Hent Udtræk**\n\nYou will receive a file `Markedsata.xls` of about 50 MB. Open the file in Excel. There will be a warning from Excel saying that file extension and content are in conflict. Select \"open anyways\" and and save the file as `.xlsx`.\n\nIn order to be found by the read-function, place the downloaded file in the following subdirectory: \n**`{{data_path}}{{os.sep}}Energinet.dk{{os.sep}}prices_wind_solar{{os.sep}}2005-01-01_2017-12-31`**",
"_____no_output_____"
],
[
"### CEPS\n\nGo to http://www.ceps.cz/en/all-data#GenerationRES\n\n**check boxes as specified below:**\n\nDISPLAY DATA FOR: **Generation RES** \nTURN ON FILTER **checked** \nFILTER SETTINGS: \n- Set the date range\n - interval\n - from: **2012** to: **2018**\n- Agregation and data version\n - Aggregation: **Hour**\n - Agregation function: **average (AVG)**\n - Data version: **real data**\n- Filter\n - Type of power plant: **ALL**\n- Click **USE FILTER**\n- DOWNLOAD DATA: **DATA V TXT**\n\nYou will receive a file `data.txt` of about 1.5 MB.\n\nIn order to be found by the read-function, place the downloaded file in the following subdirectory: \n**`{{data_path}}{{os.sep}}CEPS{{os.sep}}wind_pv{{os.sep}}2012-01-01_2018-01-31`**",
"_____no_output_____"
],
[
"### ENTSO-E Power Statistics\n\nGo to https://www.entsoe.eu/data/statistics/Pages/monthly_hourly_load.aspx\n\n**check boxes as specified below:**\n\n- Date From: **01-01-2016** Date To: **30-04-2016**\n- Country: **(Select All)**\n- Scale values to 100% using coverage ratio: **NO**\n- **View Report**\n- Click the Save symbol and select **Excel**\n\nYou will receive a file `1.01 Monthly%5FHourly Load%5FValues%5FStatistical.xlsx` of about 1 MB.\n\nIn order to be found by the read-function, place the downloaded file in the following subdirectory: \n**`{{os.sep}}original_data{{os.sep}}ENTSO-E Power Statistics{{os.sep}}load{{os.sep}}2016-01-01_2016-04-30`**\n\nThe data covers the period from 01-01-2016 up to the present, but 4 months of data seems to be the maximum that interface supports for a single download request, so you have to repeat the download procedure for 4-Month periods to cover the whole period until the present.",
"_____no_output_____"
],
[
"# Read",
"_____no_output_____"
],
[
"This section: Read each downloaded file into a pandas-DataFrame and merge data from different sources if it has the same time resolution. Takes ~15 minutes to run.",
"_____no_output_____"
],
[
"## Preparations",
"_____no_output_____"
],
[
"Set the title of the rows at the top of the data used to store metadata internally. The order of this list determines the order of the levels in the resulting output.",
"_____no_output_____"
]
],
[
[
"headers = ['region', 'variable', 'attribute', 'source', 'web', 'unit']",
"_____no_output_____"
]
],
[
[
"Read a prepared table containing meta data on the geographical areas",
"_____no_output_____"
]
],
[
[
"areas = pd.read_csv(areas_csv_path)",
"_____no_output_____"
]
],
[
[
"View the areas table",
"_____no_output_____"
]
],
[
[
"areas.loc[areas['area ID'].notnull(), :'EIC'].fillna('')",
"_____no_output_____"
]
],
[
[
"## Reading loop",
"_____no_output_____"
],
[
"Loop through sources and variables to do the reading.\nFirst read the original CSV, Excel etc. files into pandas DataFrames.",
"_____no_output_____"
]
],
[
[
"areas = pd.read_csv(areas_csv_path)\n\n# For each source in the source dictionary\nfor source_name, source_dict in sources.items():\n # For each variable from source_name\n for variable_name, param_dict in source_dict.items():\n # variable_dir = os.path.join(data_path, source_name, variable_name)\n res_list = param_dict['resolution']\n for res_key in res_list:\n df = read(data_path, areas, source_name, variable_name,\n res_key, headers, param_dict,\n start_from_user=start_from_user,\n end_from_user=end_from_user)\n\n os.makedirs(res_key, exist_ok=True)\n filename = '_'.join([source_name, variable_name]) + '.pickle'\n df.to_pickle(os.path.join(res_key, filename))",
"_____no_output_____"
]
],
[
[
"Then combine the DataFrames that have the same temporal resolution",
"_____no_output_____"
]
],
[
[
"# Create a dictionary of empty DataFrames to be populated with data\ndata_sets = {'15min': pd.DataFrame(),\n '30min': pd.DataFrame(),\n '60min': pd.DataFrame()}\nentso_e = {'15min': pd.DataFrame(),\n '30min': pd.DataFrame(),\n '60min': pd.DataFrame()}\nfor res_key in data_sets.keys():\n if not os.path.isdir(res_key):\n continue\n for filename in os.listdir(res_key):\n source_name = filename.split('_')[0]\n if subset and not source_name in subset.keys():\n continue\n logger.info('include %s', filename)\n df_portion = pd.read_pickle(os.path.join(res_key, filename))\n\n if source_name == 'ENTSO-E Transparency FTP':\n dfs = entso_e\n else:\n dfs = data_sets\n\n if dfs[res_key].empty:\n dfs[res_key] = df_portion\n elif not df_portion.empty:\n dfs[res_key] = dfs[res_key].combine_first(df_portion)\n else:\n logger.warning(filename + ' WAS EMPTY')",
"_____no_output_____"
],
[
"for res_key, df in data_sets.items():\n logger.info(res_key + ': %s', df.shape)\nfor res_key, df in entso_e.items():\n logger.info('ENTSO-E ' + res_key + ': %s', df.shape)",
"_____no_output_____"
]
],
[
[
"Display some rows of the dataframes to get a first impression of the data.",
"_____no_output_____"
]
],
[
[
"data_sets['60min'].head()",
"_____no_output_____"
]
],
[
[
"## Save raw data",
"_____no_output_____"
],
[
"Save the DataFrames created by the read function to disk. This way you have the raw data to fall back to if something goes wrong in the ramainder of this notebook without having to repeat the previos steps.",
"_____no_output_____"
]
],
[
[
"data_sets['15min'].to_pickle('raw_data_15.pickle')\ndata_sets['30min'].to_pickle('raw_data_30.pickle')\ndata_sets['60min'].to_pickle('raw_data_60.pickle')\nentso_e['15min'].to_pickle('raw_entso_e_15.pickle')\nentso_e['30min'].to_pickle('raw_entso_e_30.pickle')\nentso_e['60min'].to_pickle('raw_entso_e_60.pickle')",
"_____no_output_____"
]
],
[
[
"Load the DataFrames saved above",
"_____no_output_____"
]
],
[
[
"data_sets = {}\ndata_sets['15min'] = pd.read_pickle('raw_data_15.pickle')\ndata_sets['30min'] = pd.read_pickle('raw_data_30.pickle')\ndata_sets['60min'] = pd.read_pickle('raw_data_60.pickle')\nentso_e = {}\nentso_e['15min'] = pd.read_pickle('raw_entso_e_15.pickle')\nentso_e['30min'] = pd.read_pickle('raw_entso_e_30.pickle')\nentso_e['60min'] = pd.read_pickle('raw_entso_e_60.pickle')",
"_____no_output_____"
]
],
[
[
"# Processing",
"_____no_output_____"
],
[
"This section: missing data handling, aggregation of sub-national to national data, aggregate 15'-data to 60'-resolution. Takes 30 minutes to run.",
"_____no_output_____"
],
[
"## Missing data handling",
"_____no_output_____"
],
[
"### Interpolation",
"_____no_output_____"
],
[
"Patch missing data. At this stage, only small gaps (up to 2 hours) are filled by linear interpolation. This catched most of the missing data due to daylight savings time transitions, while leaving bigger gaps untouched\n\nThe exact locations of missing data are stored in the `nan_table` DataFrames.\n\nWhere data has been interpolated, it is marked in a new column `comment`. For eaxample the comment `solar_DE-transnetbw_generation;` means that in the original data, there is a gap in the solar generation timeseries from TransnetBW in the time period where the marker appears.",
"_____no_output_____"
],
[
"Patch the datasets and display the location of missing Data in the original data. Takes ~5 minutes to run.",
"_____no_output_____"
]
],
[
[
"nan_tables = {}\noverviews = {}\nfor res_key, df in data_sets.items():\n data_sets[res_key], nan_tables[res_key], overviews[res_key] = find_nan(\n df, res_key, headers, patch=True)",
"_____no_output_____"
],
[
"for res_key, df in entso_e.items():\n entso_e[res_key], nan_tables[res_key + ' ENTSO-E'], overviews[res_key + ' ENTSO-E'] = find_nan(\n df, res_key, headers, patch=True)",
"_____no_output_____"
]
],
[
[
"Execute this to see an example of where the data has been patched.",
"_____no_output_____"
]
],
[
[
"data_sets['60min'][data_sets['60min']['interpolated_values'].notnull()].tail()",
"_____no_output_____"
]
],
[
[
"Display the table of regions of missing values",
"_____no_output_____"
]
],
[
[
"nan_tables['60min']",
"_____no_output_____"
]
],
[
[
"You can export the NaN-tables to Excel in order to inspect where there are NaNs",
"_____no_output_____"
]
],
[
[
"writer = pd.ExcelWriter('NaN_table.xlsx')\nfor res_key, df in nan_tables.items():\n df.to_excel(writer, res_key)\nwriter.save()\n\nwriter = pd.ExcelWriter('Overview.xlsx')\nfor res_key, df in overviews.items():\n df.to_excel(writer, res_key)\nwriter.save()",
"_____no_output_____"
]
],
[
[
"Save/Load the patched data sets",
"_____no_output_____"
]
],
[
[
"data_sets['15min'].to_pickle('patched_15.pickle')\ndata_sets['30min'].to_pickle('patched_30.pickle')\ndata_sets['60min'].to_pickle('patched_60.pickle')\nentso_e['15min'].to_pickle('patched_entso_e_15.pickle')\nentso_e['30min'].to_pickle('patched_entso_e_30.pickle')\nentso_e['60min'].to_pickle('patched_entso_e_60.pickle')",
"_____no_output_____"
],
[
"data_sets = {}\ndata_sets['15min'] = pd.read_pickle('patched_15.pickle')\ndata_sets['30min'] = pd.read_pickle('patched_30.pickle')\ndata_sets['60min'] = pd.read_pickle('patched_60.pickle')\nentso_e = {}\nentso_e['15min'] = pd.read_pickle('patched_entso_e_15.pickle')\nentso_e['30min'] = pd.read_pickle('patched_entso_e_30.pickle')\nentso_e['60min'] = pd.read_pickle('patched_entso_e_60.pickle')",
"_____no_output_____"
]
],
[
[
"## Country specific calculations",
"_____no_output_____"
],
[
"### Calculate onshore wind generation for German TSOs",
"_____no_output_____"
],
[
"For 50 Hertz, it is already in the data.\nFor TenneT, it is calculated by substracting offshore from total generation.\nFor Amprion and TransnetBW, onshore wind generation is just total wind generation.\nTakes <1 second to run.",
"_____no_output_____"
]
],
[
[
"# Some of the following operations require the Dataframes to be lexsorted in\n# the columns\nfor res_key, df in data_sets.items():\n df.sort_index(axis=1, inplace=True)",
"_____no_output_____"
],
[
"for area, source, url in zip(\n ['DE_amprion', 'DE_tennet', 'DE_transnetbw'],\n ['Amprion', 'TenneT', 'TransnetBW'],\n ['http://www.amprion.net/en/wind-feed-in',\n 'http://www.tennettso.de/site/en/Transparency/publications/network-figures/actual-and-forecast-wind-energy-feed-in',\n 'https://www.transnetbw.com/en/transparency/market-data/key-figures']):\n\n new_col_header = {\n 'variable': 'wind_onshore',\n 'region': '{area}',\n 'attribute': 'generation_actual',\n 'source': '{source}',\n 'web': '{url}',\n 'unit': 'MW'\n }\n\n if area == 'DE_tennet':\n colname = ('DE_tennet', 'wind_offshore', 'generation_actual', 'TenneT')\n offshore = data_sets['15min'].loc[:, colname]\n else:\n offshore = 0\n\n data_sets['15min'][\n tuple(new_col_header[level].format(area=area, source=source, url=url)\n for level in headers)\n ] = (data_sets['15min'][(area, 'wind', 'generation_actual', source)] - offshore)\n\n # Sort again\n data_sets['15min'].sort_index(axis=1, inplace=True)",
"_____no_output_____"
]
],
[
[
"### Calculate aggregate wind capacity for Germany (on + offshore)",
"_____no_output_____"
],
[
"Apart from being interesting on it's own, this is also required to calculate an aggregated wind-profile for Germany",
"_____no_output_____"
]
],
[
[
"new_col_header = {\n 'variable': 'wind',\n 'region': 'DE',\n 'attribute': 'capacity',\n 'source': 'own calculation based on BNetzA and netztransparenz.de',\n 'web': 'http://data.open-power-system-data.org/renewable_power_plants',\n 'unit': 'MW'\n}\nnew_col_header = tuple(new_col_header[level] for level in headers)\n\ndata_sets['15min'][new_col_header] = (\n data_sets['15min']\n .loc[:, ('DE', ['wind_onshore', 'wind_offshore'], 'capacity')]\n .sum(axis=1, skipna=False))\n\n# Sort again\ndata_sets['15min'].sort_index(axis=1, inplace=True)",
"_____no_output_____"
]
],
[
[
"### Aggregate German data from individual TSOs and calculate availabilities/profiles",
"_____no_output_____"
],
[
"The wind and solar in-feed data for the 4 German balancing areas is summed up and stored in in new columns, which are then used to calculate profiles, that is, the share of wind/solar capacity producing at a given time. The column headers are created in the fashion introduced in the read script. Takes 5 seconds to run.",
"_____no_output_____"
]
],
[
[
"control_areas_DE = ['DE_50hertz', 'DE_amprion', 'DE_tennet', 'DE_transnetbw']\n\nfor variable in ['solar', 'wind', 'wind_onshore', 'wind_offshore']:\n # we could also include 'generation_forecast'\n for attribute in ['generation_actual']:\n # Calculate aggregate German generation\n sum_col = data_sets['15min'].loc(axis=1)[(control_areas_DE,\n variable, attribute)].sum(axis=1, skipna=False).to_frame()\n\n # Create a new MultiIndex\n new_col_header = {\n 'variable': '{variable}',\n 'region': 'DE',\n 'attribute': '{attribute}',\n 'source': 'own calculation based on German TSOs',\n 'web': '',\n 'unit': 'MW'\n }\n tuples = [tuple(new_col_header[level].format(\n variable=variable, attribute=attribute) for level in headers)]\n sum_col.columns = pd.MultiIndex.from_tuples(tuples, names=headers)\n\n # append aggregate German generation to the dataset after rounding\n data_sets['15min'] = data_sets['15min'].combine_first(sum_col.round(0))\n\n if attribute == 'generation_actual':\n # Calculate the profile column\n profile_col = (sum_col.values /\n data_sets['15min']['DE', variable, 'capacity']).round(4)\n\n # Create a new MultiIndex and append profile to the dataset\n new_col_header = {\n 'variable': '{variable}',\n 'region': 'DE',\n 'attribute': 'profile',\n 'source': 'own calculation based on German TSOs, BNetzA and netztranzparenz.de',\n 'web': '',\n 'unit': 'fraction'\n }\n tuples = [tuple(new_col_header[level].format(variable=variable)\n for level in headers)]\n profile_col.columns = pd.MultiIndex.from_tuples(\n tuples, names=headers)\n data_sets['15min'] = data_sets['15min'].combine_first(profile_col)",
"_____no_output_____"
]
],
[
[
"### Agregate Italian data",
"_____no_output_____"
],
[
"The data for Italy come by regions (North, Central North, Sicily, etc.) so they need to be agregated in order to get the data for Italy as a whole. In the next cell, we sum up the data by region and for each variable-attribute pair present in the Terna dataset header.",
"_____no_output_____"
]
],
[
[
"bidding_zones_IT = [\"IT_CNOR\", \"IT_CSUD\", \"IT_NORD\", \"IT_SARD\", \"IT_SICI\", \"IT_SUD\"]\n\nfor variable in [\"solar\", \"wind_onshore\"]:\n sum_col = data_sets['60min'].loc(axis=1)[(bidding_zones_IT,\n variable)].sum(axis=1, skipna=False)#.to_frame()\n \n # Create a new MultiIndex\n new_col_header = {\n \"region\" : \"IT\",\n \"variable\" : variable,\n \"attribute\" : \"generation_actual\",\n \"source\": \"own calculation based on Terna\",\n \"web\" : \"\",\n \"unit\" : \"MW\"\n }\n tuples = tuple(new_col_header[level] for level in headers)\n\n data_sets['60min'][tuples] = sum_col#\\\n# data_sets['60min'].loc[:, (italian_regions, variable, attribute)].sum(axis=1)\n\n# Sort again\ndata_sets['60min'].sort_index(axis=1, inplace=True)",
"_____no_output_____"
]
],
[
[
"Another savepoint",
"_____no_output_____"
]
],
[
[
"data_sets['15min'].to_pickle('calc_15.pickle')\ndata_sets['30min'].to_pickle('calc_30.pickle')\ndata_sets['60min'].to_pickle('calc_60.pickle')",
"_____no_output_____"
],
[
"os.chdir(temp_path)\ndata_sets = {}\ndata_sets['15min'] = pd.read_pickle('calc_15.pickle')\ndata_sets['30min'] = pd.read_pickle('calc_30.pickle')\ndata_sets['60min'] = pd.read_pickle('calc_60.pickle')\nentso_e = {}\nentso_e['15min'] = pd.read_pickle('patched_entso_e_15.pickle')\nentso_e['30min'] = pd.read_pickle('patched_entso_e_30.pickle')\nentso_e['60min'] = pd.read_pickle('patched_entso_e_60.pickle')",
"_____no_output_____"
]
],
[
[
"## Fill columns not retrieved directly from TSO webites with ENTSO-E Transparency data",
"_____no_output_____"
]
],
[
[
"for res_key, df in entso_e.items():\n # Combine with TSO data\n\n # Copy entire 30min data from ENTSO-E\n if data_sets[res_key].empty:\n data_sets[res_key] = df\n\n else:\n # Keep only region, variable, attribute in MultiIndex for comparison\n data_cols = data_sets[res_key].columns.droplevel(\n ['source', 'web', 'unit'])\n # Compare columns from ENTSO-E against ours, keep which we don't have yet\n tuples = [col for col in df.columns if not col[:3] in data_cols]\n add_cols = pd.MultiIndex.from_tuples(tuples, names=headers)\n data_sets[res_key] = data_sets[res_key].combine_first(df[add_cols])\n\n # Add the ENTSO-E markers (but only for the columns actually copied)\n add_cols = ['_'.join(col[:3]) for col in tuples]\n # Spread marker column out over a DataFrame for easiser comparison\n # Filter out everey second column, which contains the delimiter \" | \"\n # from the marker\n marker_table = (df['interpolated_values'].str.split(' | ', expand=True)\n .filter(regex='^\\d*[02468]$', axis='columns'))\n # Replace cells with markers marking columns not copied with NaNs\n marker_table[~marker_table.isin(add_cols)] = np.nan\n\n for col_name, col in marker_table.iteritems():\n if col_name == 0:\n marker_entso_e = col\n else:\n marker_entso_e = glue_markers(marker_entso_e, col)\n\n # Glue ENTSO-E marker onto our old marker\n marker = data_sets[res_key]['interpolated_values']\n data_sets[res_key].loc[:, 'interpolated_values'] = glue_markers(\n marker, df['interpolated_values'].reindex(marker.index))",
"_____no_output_____"
]
],
[
[
"## Resample higher frequencies to 60'",
"_____no_output_____"
],
[
"Some data comes in 15 or 30-minute intervals (i.e. German or British renewable generation), other in 60-minutes (i.e. load data from ENTSO-E and Prices). We resample the 15 and 30-minute data to hourly resolution and append it to the 60-minutes dataset.\n\nThe marker column is resampled separately in such a way that all information on where data has been interpolated is preserved.\n\nThe `.resample('H').mean()` methods calculates the means from the values for 4 quarter hours [:00, :15, :30, :45] of an hour values, inserts that for :00 and drops the other 3 entries. Takes 15 seconds to run.",
"_____no_output_____"
]
],
[
[
"#marker_60 = data_sets['60min']['interpolated_values']\nfor res_key, df in data_sets.items():\n if res_key == '60min':\n break\n # Resample first the marker column\n marker_resampled = df['interpolated_values'].groupby(\n pd.Grouper(freq='60Min', closed='left', label='left')\n ).agg(resample_markers, drop_region='DE_AT_LU')\n marker_resampled = marker_resampled.reindex(data_sets['60min'].index)\n\n # Glue condensed 15 min marker onto 60 min marker\n data_sets['60min'].loc[:, 'interpolated_values'] = glue_markers(\n data_sets['60min']['interpolated_values'],\n marker_resampled.reindex(data_sets['60min'].index))\n\n # Drop DE_AT_LU bidding zone data from the 15 minute resolution data to\n # be resampled since it is already provided in 60 min resolution by\n # ENTSO-E Transparency\n df = df.drop('DE_AT_LU', axis=1, errors='ignore')\n\n # Do the resampling\n resampled = df.resample('H').mean()\n resampled.columns = resampled.columns.map(mark_own_calc)\n resampled.columns.names = headers\n\n # Round the resampled columns\n for col in resampled.columns:\n if col[2] == 'profile':\n resampled.loc[:, col] = resampled.loc[:, col].round(4)\n else:\n resampled.loc[:, col] = resampled.loc[:, col].round(0)\n\n data_sets['60min'] = data_sets['60min'].combine_first(resampled)",
"_____no_output_____"
]
],
[
[
"## Insert a column with Central European (Summer-)time",
"_____no_output_____"
],
[
"The index column of th data sets defines the start of the timeperiod represented by each row of that data set in **UTC** time. We include an additional column for the **CE(S)T** Central European (Summer-) Time, as this might help aligning the output data with other data sources.",
"_____no_output_____"
]
],
[
[
"info_cols = {'utc': 'utc_timestamp',\n 'cet': 'cet_cest_timestamp',\n 'marker': 'interpolated_values'}",
"_____no_output_____"
],
[
"for res_key, df in data_sets.items():\n if df.empty:\n continue\n df.index.rename(info_cols['utc'], inplace=True)\n df.insert(0, info_cols['cet'],\n df.index.tz_localize('UTC').tz_convert('Europe/Brussels'))",
"_____no_output_____"
]
],
[
[
"# Create a final savepoint",
"_____no_output_____"
]
],
[
[
"data_sets['15min'].to_pickle('final_15.pickle')\ndata_sets['30min'].to_pickle('final_30.pickle')\ndata_sets['60min'].to_pickle('final_60.pickle')",
"_____no_output_____"
],
[
"os.chdir(temp_path)\ndata_sets = {}\ndata_sets['15min'] = pd.read_pickle('final_15.pickle')\ndata_sets['30min'] = pd.read_pickle('final_30.pickle')\ndata_sets['60min'] = pd.read_pickle('final_60.pickle')",
"_____no_output_____"
]
],
[
[
"Show the column names contained in the final DataFrame in a table",
"_____no_output_____"
]
],
[
[
"col_info = pd.DataFrame()\ndf = data_sets['60min']\nfor level in df.columns.names:\n col_info[level] = df.columns.get_level_values(level)\n\ncol_info",
"_____no_output_____"
]
],
[
[
"# Write data to disk",
"_____no_output_____"
],
[
"This section: Save as [Data Package](http://data.okfn.org/doc/tabular-data-package) (data in CSV, metadata in JSON file). All files are saved in the directory of this notebook. Alternative file formats (SQL, XLSX) are also exported. Takes about 1 hour to run.",
"_____no_output_____"
],
[
"## Limit time range\nCut off the data outside of `[start_from_user:end_from_user]`",
"_____no_output_____"
]
],
[
[
"for res_key, df in data_sets.items():\n # In order to make sure that the respective time period is covered in both\n # UTC and CE(S)T, we set the start in CE(S)T, but the end in UTC\n if start_from_user:\n start_from_user = (\n pytz.timezone('Europe/Brussels')\n .localize(datetime.combine(start_from_user, time()))\n .astimezone(pytz.timezone('UTC')))\n if end_from_user:\n end_from_user = (\n pytz.timezone('UTC')\n .localize(datetime.combine(end_from_user, time()))\n # Appropriate offset to inlude the end of period\n + timedelta(days=1, minutes=-int(res_key[:2])))\n # Then cut off the data_set\n data_sets[res_key] = df.loc[start_from_user:end_from_user, :]",
"_____no_output_____"
]
],
[
[
"## Different shapes",
"_____no_output_____"
],
[
"Data are provided in three different \"shapes\": \n- SingleIndex (easy to read for humans, compatible with datapackage standard, small file size)\n - Fileformat: CSV, SQLite\n- MultiIndex (easy to read into GAMS, not compatible with datapackage standard, small file size)\n - Fileformat: CSV, Excel\n- Stacked (compatible with data package standard, large file size, many rows, too many for Excel) \n - Fileformat: CSV\n\nThe different shapes need to be created internally befor they can be saved to files. Takes about 1 minute to run.",
"_____no_output_____"
]
],
[
[
"data_sets_singleindex = {}\ndata_sets_multiindex = {}\ndata_sets_stacked = {}\nfor res_key, df in data_sets.items():\n if df.empty:\n continue\n\n# # Round floating point numbers to 2 digits\n# for col_name, col in df.iteritems():\n# if col_name[0] in info_cols.values():\n# pass\n# elif col_name[2] == 'profile':\n# df[col_name] = col.round(4)\n# else:\n# df[col_name] = col.round(3)\n\n # MultIndex\n data_sets_multiindex[res_key + '_multiindex'] = df\n\n # SingleIndex\n df_singleindex = df.copy()\n # use first 3 levels of multiindex to create singleindex\n df_singleindex.columns = [\n col_name[0] if col_name[0] in info_cols.values()\n else '_'.join([level for level in col_name[0:3] if not level == ''])\n for col_name in df.columns.values]\n\n data_sets_singleindex[res_key + '_singleindex'] = df_singleindex\n\n # Stacked\n stacked = df.copy().drop(columns=info_cols['cet'], level=0)\n stacked.columns = stacked.columns.droplevel(['source', 'web', 'unit'])\n # Concatrenate all columns below each other (=\"stack\").\n # df.transpose().stack() is faster than stacking all column levels\n # seperately\n stacked = stacked.transpose().stack(dropna=True).to_frame(name='data')\n data_sets_stacked[res_key + '_stacked'] = stacked",
"_____no_output_____"
]
],
[
[
"## Write to SQL-database",
"_____no_output_____"
],
[
"This file format is required for the filtering function on the OPSD website. This takes ~3 minutes to complete.",
"_____no_output_____"
]
],
[
[
"os.chdir(out_path)\nfor res_key, df in data_sets_singleindex.items():\n table = 'time_series_' + res_key\n df = df.copy()\n df.index = df.index.strftime('%Y-%m-%dT%H:%M:%SZ')\n cet_col_name = info_cols['cet']\n df[cet_col_name] = (df[cet_col_name].dt.strftime('%Y-%m-%dT%H:%M:%S%z'))\n df.to_sql(table, sqlite3.connect('time_series.sqlite'),\n if_exists='replace', index_label=info_cols['utc'])",
"_____no_output_____"
]
],
[
[
"## Write to Excel",
"_____no_output_____"
],
[
"Writing the full tables to Excel takes extremely long. As a workaround, only the timestamp-columns are exported. The rest of the data can than be inserted manually from the `_multindex.csv` files.",
"_____no_output_____"
]
],
[
[
"os.chdir(out_path)\nwriter = pd.ExcelWriter('time_series1.xlsx')\nfor res_key, df in data_sets_multiindex.items():\n # Need to convert CE(S)T-timestamps to tz-naive, otherwise Excel converts\n # them back to UTC\n excel_timestamps = df.loc[:,(info_cols['cet'], '', '', '', '', '')]\n excel_timestamps = excel_timestamps.dt.tz_localize(None)\n excel_timestamps.to_excel(writer, res_key.split('_')[0],\n float_format='%.2f', merge_cells=True)\n # merge_cells=False doesn't work properly with multiindex\nwriter.save()",
"_____no_output_____"
]
],
[
[
"## Write to CSV",
"_____no_output_____"
],
[
"This takes about 10 minutes to complete.",
"_____no_output_____"
]
],
[
[
"os.chdir(out_path)\n# itertoools.chain() allows iterating over multiple dicts at once\nfor res_stacking_key, df in itertools.chain(\n data_sets_singleindex.items(),\n data_sets_multiindex.items(),\n data_sets_stacked.items()\n):\n\n df = df.copy()\n\n # convert the format of the cet_cest-timestamp to ISO-8601\n if not res_stacking_key.split('_')[1] == 'stacked':\n df.iloc[:, 0] = df.iloc[:, 0].dt.strftime('%Y-%m-%dT%H:%M:%S%z') # https://frictionlessdata.io/specs/table-schema/#date\n filename = 'time_series_' + res_stacking_key + '.csv'\n df.to_csv(filename, float_format='%.4f',\n date_format='%Y-%m-%dT%H:%M:%SZ')",
"_____no_output_____"
]
],
[
[
"# Create metadata",
"_____no_output_____"
],
[
"This section: create the metadata, both general and column-specific. All metadata we be stored as a JSON file. Takes 10s to run.",
"_____no_output_____"
]
],
[
[
"os.chdir(out_path)\nmake_json(data_sets, info_cols, version, changes, headers, areas,\n start_from_user, end_from_user)",
"_____no_output_____"
]
],
[
[
"## Write checksums.txt",
"_____no_output_____"
],
[
"We publish SHA-checksums for the outputfiles on GitHub to allow verifying the integrity of outputfiles on the OPSD server.",
"_____no_output_____"
]
],
[
[
"os.chdir(out_path)\nfiles = os.listdir(out_path)\n\n# Create checksums.txt in the output directory\nwith open('checksums.txt', 'w') as f:\n for file_name in files:\n if file_name.split('.')[-1] in ['csv', 'sqlite', 'xlsx']:\n file_hash = get_sha_hash(file_name)\n f.write('{},{}\\n'.format(file_name, file_hash))\n\n# Copy the file to root directory from where it will be pushed to GitHub,\n# leaving a copy in the version directory for reference\ncopyfile('checksums.txt', os.path.join(home_path, 'checksums.txt'))",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
]
] |
cbec1ad11685b67a6afa36b2b52417c06368f37e
| 13,898 |
ipynb
|
Jupyter Notebook
|
assets/tutorials/tut11_policy_gradient_cartpole.ipynb
|
uoft-csc413/2022
|
769a215505e4fdb706e85bcae37f7bbf662389eb
|
[
"MIT"
] | 2 |
2022-01-11T23:51:03.000Z
|
2022-01-31T14:41:01.000Z
|
assets/tutorials/tut11_policy_gradient_cartpole.ipynb
|
uoft-csc413/2022
|
769a215505e4fdb706e85bcae37f7bbf662389eb
|
[
"MIT"
] | 4 |
2022-02-05T00:25:13.000Z
|
2022-02-26T21:38:45.000Z
|
assets/tutorials/tut11_policy_gradient_cartpole.ipynb
|
uoft-csc413/2022
|
769a215505e4fdb706e85bcae37f7bbf662389eb
|
[
"MIT"
] | 3 |
2022-02-04T23:29:49.000Z
|
2022-02-26T19:33:45.000Z
| 51.665428 | 1,628 | 0.556843 |
[
[
[
"import gym\n\nimport torch\nfrom torch import nn\nimport torch.nn.functional as F\nfrom torch.autograd import Variable\nfrom torch.distributions import Bernoulli\n\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"class PolicyNet(nn.Module):\n def __init__(self, input_dim, output_dim):\n super(PolicyNet, self).__init__()\n\n self.input_dim = input_dim\n self.output_dim = output_dim\n\n self.fc1 = nn.Linear(self.input_dim, 32)\n self.fc2 = nn.Linear(32, 32)\n self.output = nn.Linear(32, self.output_dim)\n\n def forward(self, x):\n output = F.relu(self.fc1(x))\n output = F.relu(self.fc2(output))\n output = torch.sigmoid(self.output(output))\n\n return output",
"_____no_output_____"
],
[
"def convert_to_torch_variable(arr):\n \"\"\"Converts a numpy array to torch variable\"\"\"\n return Variable(torch.from_numpy(arr).float())\n\n",
"_____no_output_____"
],
[
"# Define environment\nenv = gym.make(\"CartPole-v0\")\nenv.seed(0)\n\n# Create environment monitor for video recording\nvideo_monitor_callable = lambda _: True\n# monitored_env = gym.wrappers.Monitor(env, './cartpole_videos', force=True, video_callable=video_monitor_callable)\n\nstate_dim = env.observation_space.shape[0]\naction_dim = env.action_space.n\nbernoulli_action_dim = 1\n\n# Initialize policy network\npolicy_net = PolicyNet(input_dim=state_dim, output_dim=bernoulli_action_dim)\n\n# Hyperparameters\nNUM_EPISODES = 500\nGAMMA = 0.99\nBATCH_SIZE = 5\nLEARNING_RATE = 0.01\n\n# Let baseline be 0 for now\nbaseline = 0.0\n\n# Define optimizer\noptimizer = torch.optim.RMSprop(policy_net.parameters(), lr=LEARNING_RATE)",
"_____no_output_____"
],
[
"# Collect trajectory rewards for plotting purpose\ntraj_reward_history = []\n\n# training loop\nfor ep_i in range(NUM_EPISODES):\n loss = 0.0\n\n # Record states, actions and discounted rewards of this episode\n states = []\n actions = []\n rewards = []\n cumulative_undiscounted_reward = 0.0\n\n for traj_i in range(BATCH_SIZE):\n time_step = 0\n done = False\n\n # initialize environment\n cur_state = env.reset()\n cur_state = convert_to_torch_variable(cur_state)\n\n discount_factor = 1.0\n discounted_rewards = []\n\n grad_log_params = []\n\n while not done:\n # Compute action probability using the current policy\n action_prob = policy_net(cur_state)\n\n # Sample action according to action probability\n action_sampler = Bernoulli(probs=action_prob)\n action = action_sampler.sample()\n action = action.numpy().astype(int)[0]\n\n # Record the states and actions -- will be used for policy gradient later\n states.append(cur_state)\n actions.append(action)\n\n # take a step in the environment, and collect data\n next_state, reward, done, _ = env.step(action)\n\n # Discount the reward, and append to reward list\n discounted_reward = reward * discount_factor\n discounted_rewards.append(discounted_reward)\n cumulative_undiscounted_reward += reward\n\n # Prepare for taking the next step\n cur_state = convert_to_torch_variable(next_state)\n\n time_step += 1\n discount_factor *= GAMMA\n\n # Finished collecting data for the current trajectory. \n # Recall temporal structure in policy gradient.\n # Donstruct the \"cumulative future discounted reward\" at each time step.\n for time_i in range(time_step):\n # relevant reward is the sum of rewards from time t to the end of trajectory\n relevant_reward = sum(discounted_rewards[time_i:])\n rewards.append(relevant_reward)\n\n # Finished collecting data for this batch. Update policy using policy gradient.\n avg_traj_reward = cumulative_undiscounted_reward / BATCH_SIZE\n traj_reward_history.append(avg_traj_reward)\n\n if (ep_i + 1) % 10 == 0:\n print(\"Episode {}: Average reward per trajectory = {}\".format(ep_i + 1, avg_traj_reward))\n\n #if (ep_i + 1) % 100 == 0:\n # record_video()\n\n optimizer.zero_grad()\n data_len = len(states)\n loss = 0.0\n\n # Compute the policy gradient\n for data_i in range(data_len):\n action_prob = policy_net(states[data_i])\n action_sampler = Bernoulli(probs=action_prob)\n\n loss -= action_sampler.log_prob(torch.Tensor([actions[data_i]])) * (rewards[data_i] - baseline)\n loss /= float(data_len)\n loss.backward()\n optimizer.step()",
"Episode 10: Average reward per trajectory = 123.6\nEpisode 20: Average reward per trajectory = 200.0\nEpisode 30: Average reward per trajectory = 200.0\nEpisode 40: Average reward per trajectory = 200.0\nEpisode 50: Average reward per trajectory = 200.0\nEpisode 60: Average reward per trajectory = 200.0\nEpisode 70: Average reward per trajectory = 200.0\nEpisode 80: Average reward per trajectory = 200.0\nEpisode 90: Average reward per trajectory = 200.0\nEpisode 100: Average reward per trajectory = 190.6\n"
],
[
"# Don't forget to close the environments.\n#monitored_env.close()\nenv.close()\n\n# Plot learning curve\nplt.figure()\nplt.plot(traj_reward_history)\nplt.title(\"Learning to Solve CartPole-v1 with Policy Gradient\")\nplt.xlabel(\"Episode\")\nplt.ylabel(\"Average Reward per Trajectory\")\nplt.savefig(\"CartPole-pg.png\")\nplt.show()\nplt.close()",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbec1ee8fb4d89069ccbf4f282b6a11f8698e75d
| 33,645 |
ipynb
|
Jupyter Notebook
|
examples/notebooks/statespace_news.ipynb
|
KishManani/statsmodels
|
300b6fba90c65c8e94b4f83e04f7ae1b0ceeac2e
|
[
"BSD-3-Clause"
] | 6,931 |
2015-01-01T11:41:55.000Z
|
2022-03-31T17:03:24.000Z
|
examples/notebooks/statespace_news.ipynb
|
Ajisusanto136/statsmodels
|
e741f3b22302199121090822353f20d794a02148
|
[
"BSD-3-Clause"
] | 6,137 |
2015-01-01T00:33:45.000Z
|
2022-03-31T22:53:17.000Z
|
examples/notebooks/statespace_news.ipynb
|
Ajisusanto136/statsmodels
|
e741f3b22302199121090822353f20d794a02148
|
[
"BSD-3-Clause"
] | 2,608 |
2015-01-02T21:32:31.000Z
|
2022-03-31T07:38:30.000Z
| 46.089041 | 572 | 0.648952 |
[
[
[
"## Forecasting, updating datasets, and the \"news\"\n\nIn this notebook, we describe how to use Statsmodels to compute the impacts of updated or revised datasets on out-of-sample forecasts or in-sample estimates of missing data. We follow the approach of the \"Nowcasting\" literature (see references at the end), by using a state space model to compute the \"news\" and impacts of incoming data.\n\n**Note**: this notebook applies to Statsmodels v0.12+. In addition, it only applies to the state space models or related classes, which are: `sm.tsa.statespace.ExponentialSmoothing`, `sm.tsa.arima.ARIMA`, `sm.tsa.SARIMAX`, `sm.tsa.UnobservedComponents`, `sm.tsa.VARMAX`, and `sm.tsa.DynamicFactor`.",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\n\nimport numpy as np\nimport pandas as pd\nimport statsmodels.api as sm\nimport matplotlib.pyplot as plt\n\nmacrodata = sm.datasets.macrodata.load_pandas().data\nmacrodata.index = pd.period_range('1959Q1', '2009Q3', freq='Q')",
"_____no_output_____"
]
],
[
[
"Forecasting exercises often start with a fixed set of historical data that is used for model selection and parameter estimation. Then, the fitted selected model (or models) can be used to create out-of-sample forecasts. Most of the time, this is not the end of the story. As new data comes in, you may need to evaluate your forecast errors, possibly update your models, and create updated out-of-sample forecasts. This is sometimes called a \"real-time\" forecasting exercise (by contrast, a pseudo real-time exercise is one in which you simulate this procedure).\n\nIf all that matters is minimizing some loss function based on forecast errors (like MSE), then when new data comes in you may just want to completely redo model selection, parameter estimation and out-of-sample forecasting, using the updated datapoints. If you do this, your new forecasts will have changed for two reasons:\n\n1. You have received new data that gives you new information\n2. Your forecasting model or the estimated parameters are different\n\nIn this notebook, we focus on methods for isolating the first effect. The way we do this comes from the so-called \"nowcasting\" literature, and in particular Bańbura, Giannone, and Reichlin (2011), Bańbura and Modugno (2014), and Bańbura et al. (2014). They describe this exercise as computing the \"**news**\", and we follow them in using this language in Statsmodels.\n\nThese methods are perhaps most useful with multivariate models, since there multiple variables may update at the same time, and it is not immediately obvious what forecast change was created by what updated variable. However, they can still be useful for thinking about forecast revisions in univariate models. We will therefore start with the simpler univariate case to explain how things work, and then move to the multivariate case afterwards.",
"_____no_output_____"
],
[
"**Note on revisions**: the framework that we are using is designed to decompose changes to forecasts from newly observed datapoints. It can also take into account *revisions* to previously published datapoints, but it does not decompose them separately. Instead, it only shows the aggregate effect of \"revisions\".",
"_____no_output_____"
],
[
"**Note on `exog` data**: the framework that we are using only decomposes changes to forecasts from newly observed datapoints for *modeled* variables. These are the \"left-hand-side\" variables that in Statsmodels are given in the `endog` arguments. This framework does not decompose or account for changes to unmodeled \"right-hand-side\" variables, like those included in the `exog` argument.",
"_____no_output_____"
],
[
"### Simple univariate example: AR(1)\n\nWe will begin with a simple autoregressive model, an AR(1):\n\n$$y_t = \\phi y_{t-1} + \\varepsilon_t$$\n\n- The parameter $\\phi$ captures the persistence of the series\n\nWe will use this model to forecast inflation.\n\nTo make it simpler to describe the forecast updates in this notebook, we will work with inflation data that has been de-meaned, but it is straightforward in practice to augment the model with a mean term.\n",
"_____no_output_____"
]
],
[
[
"# De-mean the inflation series\ny = macrodata['infl'] - macrodata['infl'].mean()",
"_____no_output_____"
]
],
[
[
"#### Step 1: fitting the model on the available dataset",
"_____no_output_____"
],
[
"Here, we'll simulate an out-of-sample exercise, by constructing and fitting our model using all of the data except the last five observations. We'll assume that we haven't observed these values yet, and then in subsequent steps we'll add them back into the analysis.",
"_____no_output_____"
]
],
[
[
"y_pre = y.iloc[:-5]\ny_pre.plot(figsize=(15, 3), title='Inflation');",
"_____no_output_____"
]
],
[
[
"To construct forecasts, we first estimate the parameters of the model. This returns a results object that we will be able to use produce forecasts.",
"_____no_output_____"
]
],
[
[
"mod_pre = sm.tsa.arima.ARIMA(y_pre, order=(1, 0, 0), trend='n')\nres_pre = mod_pre.fit()\nprint(res_pre.summary())",
"_____no_output_____"
]
],
[
[
"Creating the forecasts from the results object `res` is easy - you can just call the `forecast` method with the number of forecasts you want to construct. In this case, we'll construct four out-of-sample forecasts.",
"_____no_output_____"
]
],
[
[
"# Compute the forecasts\nforecasts_pre = res_pre.forecast(4)\n\n# Plot the last 3 years of data and the four out-of-sample forecasts\ny_pre.iloc[-12:].plot(figsize=(15, 3), label='Data', legend=True)\nforecasts_pre.plot(label='Forecast', legend=True);",
"_____no_output_____"
]
],
[
[
"For the AR(1) model, it is also easy to manually construct the forecasts. Denoting the last observed variable as $y_T$ and the $h$-step-ahead forecast as $y_{T+h|T}$, we have:\n\n$$y_{T+h|T} = \\hat \\phi^h y_T$$\n\nWhere $\\hat \\phi$ is our estimated value for the AR(1) coefficient. From the summary output above, we can see that this is the first parameter of the model, which we can access from the `params` attribute of the results object.",
"_____no_output_____"
]
],
[
[
"# Get the estimated AR(1) coefficient\nphi_hat = res_pre.params[0]\n\n# Get the last observed value of the variable\ny_T = y_pre.iloc[-1]\n\n# Directly compute the forecasts at the horizons h=1,2,3,4\nmanual_forecasts = pd.Series([phi_hat * y_T, phi_hat**2 * y_T,\n phi_hat**3 * y_T, phi_hat**4 * y_T],\n index=forecasts_pre.index)\n\n# We'll print the two to double-check that they're the same\nprint(pd.concat([forecasts_pre, manual_forecasts], axis=1))",
"_____no_output_____"
]
],
[
[
"#### Step 2: computing the \"news\" from a new observation\n\nSuppose that time has passed, and we have now received another observation. Our dataset is now larger, and we can evaluate our forecast error and produce updated forecasts for the subsequent quarters.",
"_____no_output_____"
]
],
[
[
"# Get the next observation after the \"pre\" dataset\ny_update = y.iloc[-5:-4]\n\n# Print the forecast error\nprint('Forecast error: %.2f' % (y_update.iloc[0] - forecasts_pre.iloc[0]))",
"_____no_output_____"
]
],
[
[
"To compute forecasts based on our updated dataset, we will create an updated results object `res_post` using the `append` method, to append on our new observation to the previous dataset.\n\nNote that by default, the `append` method does not re-estimate the parameters of the model. This is exactly what we want here, since we want to isolate the effect on the forecasts of the new information only.",
"_____no_output_____"
]
],
[
[
"# Create a new results object by passing the new observations to the `append` method\nres_post = res_pre.append(y_update)\n\n# Since we now know the value for 2008Q3, we will only use `res_post` to\n# produce forecasts for 2008Q4 through 2009Q2\nforecasts_post = pd.concat([y_update, res_post.forecast('2009Q2')])\nprint(forecasts_post)",
"_____no_output_____"
]
],
[
[
"In this case, the forecast error is quite large - inflation was more than 10 percentage points below the AR(1) models' forecast. (This was largely because of large swings in oil prices around the global financial crisis).",
"_____no_output_____"
],
[
"To analyse this in more depth, we can use Statsmodels to isolate the effect of the new information - or the \"**news**\" - on our forecasts. This means that we do not yet want to change our model or re-estimate the parameters. Instead, we will use the `news` method that is available in the results objects of state space models.\n\nComputing the news in Statsmodels always requires a *previous* results object or dataset, and an *updated* results object or dataset. Here we will use the original results object `res_pre` as the previous results and the `res_post` results object that we just created as the updated results.",
"_____no_output_____"
],
[
"Once we have previous and updated results objects or datasets, we can compute the news by calling the `news` method. Here, we will call `res_pre.news`, and the first argument will be the updated results, `res_post` (however, if you have two results objects, the `news` method could can be called on either one).\n\nIn addition to specifying the comparison object or dataset as the first argument, there are a variety of other arguments that are accepted. The most important specify the \"impact periods\" that you want to consider. These \"impact periods\" correspond to the forecasted periods of interest; i.e. these dates specify with periods will have forecast revisions decomposed.\n\nTo specify the impact periods, you must pass two of `start`, `end`, and `periods` (similar to the Pandas `date_range` method). If your time series was a Pandas object with an associated date or period index, then you can pass dates as values for `start` and `end`, as we do below.",
"_____no_output_____"
]
],
[
[
"# Compute the impact of the news on the four periods that we previously\n# forecasted: 2008Q3 through 2009Q2\nnews = res_pre.news(res_post, start='2008Q3', end='2009Q2')\n# Note: one alternative way to specify these impact dates is\n# `start='2008Q3', periods=4`",
"_____no_output_____"
]
],
[
[
"The variable `news` is an object of the class `NewsResults`, and it contains details about the updates to the data in `res_post` compared to `res_pre`, the new information in the updated dataset, and the impact that the new information had on the forecasts in the period between `start` and `end`.\n\nOne easy way to summarize the results are with the `summary` method.",
"_____no_output_____"
]
],
[
[
"print(news.summary())",
"_____no_output_____"
]
],
[
[
"**Summary output**: the default summary for this news results object printed four tables:\n\n1. Summary of the model and datasets\n2. Details of the news from updated data\n3. Summary of the impacts of the new information on the forecasts between `start='2008Q3'` and `end='2009Q2'`\n4. Details of how the updated data led to the impacts on the forecasts between `start='2008Q3'` and `end='2009Q2'`\n\nThese are described in more detail below.\n\n*Notes*:\n\n- There are a number of arguments that can be passed to the `summary` method to control this output. Check the documentation / docstring for details.\n- Table (4), showing details of the updates and impacts, can become quite large if the model is multivariate, there are multiple updates, or a large number of impact dates are selected. It is only shown by default for univariate models.",
"_____no_output_____"
],
[
"**First table: summary of the model and datasets**\n\nThe first table, above, shows:\n\n- The type of model from which the forecasts were made. Here this is an ARIMA model, since an AR(1) is a special case of an ARIMA(p,d,q) model.\n- The date and time at which the analysis was computed.\n- The original sample period, which here corresponds to `y_pre`\n- The endpoint of the updated sample period, which here is the last date in `y_post`",
"_____no_output_____"
],
[
"**Second table: the news from updated data**\n\nThis table simply shows the forecasts from the previous results for observations that were updated in the updated sample.\n\n*Notes*:\n\n- Our updated dataset `y_post` did not contain any *revisions* to previously observed datapoints. If it had, there would be an additional table showing the previous and updated values of each such revision.",
"_____no_output_____"
],
[
"**Third table: summary of the impacts of the new information**\n\n*Columns*:\n\nThe third table, above, shows:\n\n- The previous forecast for each of the impact dates, in the \"estimate (prev)\" column\n- The impact that the new information (the \"news\") had on the forecasts for each of the impact dates, in the \"impact of news\" column\n- The updated forecast for each of the impact dates, in the \"estimate (new)\" column\n\n*Notes*:\n\n- In multivariate models, this table contains additional columns describing the relevant impacted variable for each row.\n- Our updated dataset `y_post` did not contain any *revisions* to previously observed datapoints. If it had, there would be additional columns in this table showing the impact of those revisions on the forecasts for the impact dates.\n- Note that `estimate (new) = estimate (prev) + impact of news`\n- This table can be accessed independently using the `summary_impacts` method.\n\n*In our example*:\n\nNotice that in our example, the table shows the values that we computed earlier:\n\n- The \"estimate (prev)\" column is identical to the forecasts from our previous model, contained in the `forecasts_pre` variable.\n- The \"estimate (new)\" column is identical to our `forecasts_post` variable, which contains the observed value for 2008Q3 and the forecasts from the updated model for 2008Q4 - 2009Q2.",
"_____no_output_____"
],
[
"**Fourth table: details of updates and their impacts**\n\nThe fourth table, above, shows how each new observation translated into specific impacts at each impact date.\n\n*Columns*:\n\nThe first three columns table described the relevant **update** (an \"updated\" is a new observation):\n\n- The first column (\"update date\") shows the date of the variable that was updated.\n- The second column (\"forecast (prev)\") shows the value that would have been forecasted for the update variable at the update date based on the previous results / dataset.\n- The third column (\"observed\") shows the actual observed value of that updated variable / update date in the updated results / dataset.\n\nThe last four columns described the **impact** of a given update (an impact is a changed forecast within the \"impact periods\").\n\n- The fourth column (\"impact date\") gives the date at which the given update made an impact.\n- The fifth column (\"news\") shows the \"news\" associated with the given update (this is the same for each impact of a given update, but is just not sparsified by default)\n- The sixth column (\"weight\") describes the weight that the \"news\" from the given update has on the impacted variable at the impact date. In general, weights will be different between each \"updated variable\" / \"update date\" / \"impacted variable\" / \"impact date\" combination.\n- The seventh column (\"impact\") shows the impact that the given update had on the given \"impacted variable\" / \"impact date\".\n\n*Notes*:\n\n- In multivariate models, this table contains additional columns to show the relevant variable that was updated and variable that was impacted for each row. Here, there is only one variable (\"infl\"), so those columns are suppressed to save space.\n- By default, the updates in this table are \"sparsified\" with blanks, to avoid repeating the same values for \"update date\", \"forecast (prev)\", and \"observed\" for each row of the table. This behavior can be overridden using the `sparsify` argument.\n- Note that `impact = news * weight`.\n- This table can be accessed independently using the `summary_details` method.\n\n*In our example*:\n\n- For the update to 2008Q3 and impact date 2008Q3, the weight is equal to 1. This is because we only have one variable, and once we have incorporated the data for 2008Q3, there is no no remaining ambiguity about the \"forecast\" for this date. Thus all of the \"news\" about this variable at 2008Q3 passes through to the \"forecast\" directly.",
"_____no_output_____"
],
[
"#### Addendum: manually computing the news, weights, and impacts\n\nFor this simple example with a univariate model, it is straightforward to compute all of the values shown above by hand. First, recall the formula for forecasting $y_{T+h|T} = \\phi^h y_T$, and note that it follows that we also have $y_{T+h|T+1} = \\phi^h y_{T+1}$. Finally, note that $y_{T|T+1} = y_T$, because if we know the value of the observations through $T+1$, we know the value of $y_T$.\n\n**News**: The \"news\" is nothing more than the forecast error associated with one of the new observations. So the news associated with observation $T+1$ is:\n\n$$n_{T+1} = y_{T+1} - y_{T+1|T} = Y_{T+1} - \\phi Y_T$$\n\n**Impacts**: The impact of the news is the difference between the updated and previous forecasts, $i_h \\equiv y_{T+h|T+1} - y_{T+h|T}$.\n\n- The previous forecasts for $h=1, \\dots, 4$ are: $\\begin{pmatrix} \\phi y_T & \\phi^2 y_T & \\phi^3 y_T & \\phi^4 y_T \\end{pmatrix}'$. \n- The updated forecasts for $h=1, \\dots, 4$ are: $\\begin{pmatrix} y_{T+1} & \\phi y_{T+1} & \\phi^2 y_{T+1} & \\phi^3 y_{T+1} \\end{pmatrix}'$.\n\nThe impacts are therefore:\n\n$$\\{ i_h \\}_{h=1}^4 = \\begin{pmatrix} y_{T+1} - \\phi y_T \\\\ \\phi (Y_{T+1} - \\phi y_T) \\\\ \\phi^2 (Y_{T+1} - \\phi y_T) \\\\ \\phi^3 (Y_{T+1} - \\phi y_T) \\end{pmatrix}$$\n\n**Weights**: To compute the weights, we just need to note that it is immediate that we can rewrite the impacts in terms of the forecast errors, $n_{T+1}$.\n\n$$\\{ i_h \\}_{h=1}^4 = \\begin{pmatrix} 1 \\\\ \\phi \\\\ \\phi^2 \\\\ \\phi^3 \\end{pmatrix} n_{T+1}$$\n\nThe weights are then simply $w = \\begin{pmatrix} 1 \\\\ \\phi \\\\ \\phi^2 \\\\ \\phi^3 \\end{pmatrix}$",
"_____no_output_____"
],
[
"We can check that this is what the `news` method has computed.",
"_____no_output_____"
]
],
[
[
"# Print the news, computed by the `news` method\nprint(news.news)\n\n# Manually compute the news\nprint()\nprint((y_update.iloc[0] - phi_hat * y_pre.iloc[-1]).round(6))",
"_____no_output_____"
],
[
"# Print the total impacts, computed by the `news` method\n# (Note: news.total_impacts = news.revision_impacts + news.update_impacts, but\n# here there are no data revisions, so total and update impacts are the same)\nprint(news.total_impacts)\n\n# Manually compute the impacts\nprint()\nprint(forecasts_post - forecasts_pre)",
"_____no_output_____"
],
[
"# Print the weights, computed by the `news` method\nprint(news.weights)\n\n# Manually compute the weights\nprint()\nprint(np.array([1, phi_hat, phi_hat**2, phi_hat**3]).round(6))",
"_____no_output_____"
]
],
[
[
"### Multivariate example: dynamic factor\n\nIn this example, we'll consider forecasting monthly core price inflation based on the Personal Consumption Expenditures (PCE) price index and the Consumer Price Index (CPI), using a Dynamic Factor model. Both of these measures track prices in the US economy and are based on similar source data, but they have a number of definitional differences. Nonetheless, they track each other relatively well, so modeling them jointly using a single dynamic factor seems reasonable.\n\nOne reason that this kind of approach can be useful is that the CPI is released earlier in the month than the PCE. One the CPI is released, therefore, we can update our dynamic factor model with that additional datapoint, and obtain an improved forecast for that month's PCE release. A more involved version of this kind of analysis is available in Knotek and Zaman (2017).",
"_____no_output_____"
],
[
"We start by downloading the core CPI and PCE price index data from [FRED](https://fred.stlouisfed.org/), converting them to annualized monthly inflation rates, removing two outliers, and de-meaning each series (the dynamic factor model does not ",
"_____no_output_____"
]
],
[
[
"import pandas_datareader as pdr\nlevels = pdr.get_data_fred(['PCEPILFE', 'CPILFESL'], start='1999', end='2019').to_period('M')\ninfl = np.log(levels).diff().iloc[1:] * 1200\ninfl.columns = ['PCE', 'CPI']\n\n# Remove two outliers and de-mean the series\ninfl['PCE'].loc['2001-09':'2001-10'] = np.nan",
"_____no_output_____"
]
],
[
[
"To show how this works, we'll imagine that it is April 14, 2017, which is the data of the March 2017 CPI release. So that we can show the effect of multiple updates at once, we'll assume that we haven't updated our data since the end of January, so that:\n\n- Our **previous dataset** will consist of all values for the PCE and CPI through January 2017\n- Our **updated dataset** will additionally incorporate the CPI for February and March 2017 and the PCE data for February 2017. But it will not yet the PCE (the March 2017 PCE price index was not released until May 1, 2017).",
"_____no_output_____"
]
],
[
[
"# Previous dataset runs through 2017-02\ny_pre = infl.loc[:'2017-01'].copy()\nconst_pre = np.ones(len(y_pre))\nprint(y_pre.tail())",
"_____no_output_____"
],
[
"# For the updated dataset, we'll just add in the\n# CPI value for 2017-03\ny_post = infl.loc[:'2017-03'].copy()\ny_post.loc['2017-03', 'PCE'] = np.nan\nconst_post = np.ones(len(y_post))\n\n# Notice the missing value for PCE in 2017-03\nprint(y_post.tail())",
"_____no_output_____"
]
],
[
[
"We chose this particular example because in March 2017, core CPI prices fell for the first time since 2010, and this information may be useful in forecast core PCE prices for that month. The graph below shows the CPI and PCE price data as it would have been observed on April 14th$^\\dagger$.\n\n-----\n\n$\\dagger$ This statement is not entirely true, because both the CPI and PCE price indexes can be revised to a certain extent after the fact. As a result, the series that we're pulling are not exactly like those observed on April 14, 2017. This could be fixed by pulling the archived data from [ALFRED](https://alfred.stlouisfed.org/) instead of [FRED](https://fred.stlouisfed.org/), but the data we have is good enough for this tutorial.",
"_____no_output_____"
]
],
[
[
"# Plot the updated dataset\nfig, ax = plt.subplots(figsize=(15, 3))\ny_post.plot(ax=ax)\nax.hlines(0, '2009', '2017-06', linewidth=1.0)\nax.set_xlim('2009', '2017-06');",
"_____no_output_____"
]
],
[
[
"To perform the exercise, we first construct and fit a `DynamicFactor` model. Specifically:\n\n- We are using a single dynamic factor (`k_factors=1`)\n- We are modeling the factor's dynamics with an AR(6) model (`factor_order=6`)\n- We have included a vector of ones as an exogenous variable (`exog=const_pre`), because the inflation series we are working with are not mean-zero.",
"_____no_output_____"
]
],
[
[
"mod_pre = sm.tsa.DynamicFactor(y_pre, exog=const_pre, k_factors=1, factor_order=6)\nres_pre = mod_pre.fit()\nprint(res_pre.summary())",
"_____no_output_____"
]
],
[
[
"With the fitted model in hand, we now construct the news and impacts associated with observing the CPI for March 2017. The updated data is for February 2017 and part of March 2017, and we'll examining the impacts on both March and April.\n\nIn the univariate example, we first created an updated results object, and then passed that to the `news` method. Here, we're creating the news by directly passing the updated dataset.\n\nNotice that:\n\n1. `y_post` contains the entire updated dataset (not just the new datapoints)\n2. We also had to pass an updated `exog` array. This array must cover **both**:\n - The entire period associated with `y_post`\n - Any additional datapoints after the end of `y_post` through the last impact date, specified by `end`\n\n Here, `y_post` ends in March 2017, so we needed our `exog` to extend one more period, to April 2017.",
"_____no_output_____"
]
],
[
[
"# Create the news results\n# Note\nconst_post_plus1 = np.ones(len(y_post) + 1)\nnews = res_pre.news(y_post, exog=const_post_plus1, start='2017-03', end='2017-04')",
"_____no_output_____"
]
],
[
[
"> **Note**:\n>\n> In the univariate example, above, we first constructed a new results object, and then passed that to the `news` method. We could have done that here too, although there is an extra step required. Since we are requesting an impact for a period beyond the end of `y_post`, we would still need to pass the additional value for the `exog` variable during that period to `news`:\n> \n> ```python\nres_post = res_pre.apply(y_post, exog=const_post)\nnews = res_pre.news(res_post, exog=[1.], start='2017-03', end='2017-04')\n```",
"_____no_output_____"
],
[
"Now that we have computed the `news`, printing `summary` is a convenient way to see the results.",
"_____no_output_____"
]
],
[
[
"# Show the summary of the news results\nprint(news.summary())",
"_____no_output_____"
]
],
[
[
"Because we have multiple variables, by default the summary only shows the news from updated data along and the total impacts.\n\nFrom the first table, we can see that our updated dataset contains three new data points, with most of the \"news\" from these data coming from the very low reading in March 2017.\n\nThe second table shows that these three datapoints substantially impacted the estimate for PCE in March 2017 (which was not yet observed). This estimate revised down by nearly 1.5 percentage points.\n\nThe updated data also impacted the forecasts in the first out-of-sample month, April 2017. After incorporating the new data, the model's forecasts for CPI and PCE inflation in that month revised down 0.29 and 0.17 percentage point, respectively.",
"_____no_output_____"
],
[
"While these tables show the \"news\" and the total impacts, they do not show how much of each impact was caused by each updated datapoint. To see that information, we need to look at the details tables.\n\nOne way to see the details tables is to pass `include_details=True` to the `summary` method. To avoid repeating the tables above, however, we'll just call the `summary_details` method directly.",
"_____no_output_____"
]
],
[
[
"print(news.summary_details())",
"_____no_output_____"
]
],
[
[
"This table shows that most of the revisions to the estimate of PCE in April 2017, described above, came from the news associated with the CPI release in March 2017. By contrast, the CPI release in February had only a little effect on the April forecast, and the PCE release in February had essentially no effect.",
"_____no_output_____"
],
[
"### Bibliography\n\nBańbura, Marta, Domenico Giannone, and Lucrezia Reichlin. \"Nowcasting.\" The Oxford Handbook of Economic Forecasting. July 8, 2011.\n\nBańbura, Marta, Domenico Giannone, Michele Modugno, and Lucrezia Reichlin. \"Now-casting and the real-time data flow.\" In Handbook of economic forecasting, vol. 2, pp. 195-237. Elsevier, 2013.\n\nBańbura, Marta, and Michele Modugno. \"Maximum likelihood estimation of factor models on datasets with arbitrary pattern of missing data.\" Journal of Applied Econometrics 29, no. 1 (2014): 133-160.\n\nKnotek, Edward S., and Saeed Zaman. \"Nowcasting US headline and core inflation.\" Journal of Money, Credit and Banking 49, no. 5 (2017): 931-968.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
cbec268b79c7d6f0f56a6e81df59b3d43e3456a7
| 1,183 |
ipynb
|
Jupyter Notebook
|
10 Days of Statistics/Day_2. Basic Probability (Q & A).ipynb
|
Nam-SH/HackerRank
|
d1ced5cdad3eae7661f39af4d12aa33f460821cb
|
[
"MIT"
] | null | null | null |
10 Days of Statistics/Day_2. Basic Probability (Q & A).ipynb
|
Nam-SH/HackerRank
|
d1ced5cdad3eae7661f39af4d12aa33f460821cb
|
[
"MIT"
] | null | null | null |
10 Days of Statistics/Day_2. Basic Probability (Q & A).ipynb
|
Nam-SH/HackerRank
|
d1ced5cdad3eae7661f39af4d12aa33f460821cb
|
[
"MIT"
] | null | null | null | 19.716667 | 128 | 0.508876 |
[
[
[
"# Day 2: Basic Probability (Q & A)\n\n\n`Task `\nIn a single toss of `2` fair (evenly-weighted) six-sided dice, find the probability that their sum will be at most `9`.\n\n```\n 1: 2 / 3\n\n* 2: 5 / 6 \n\n 3: 1 / 4\n\n 4: 1 / 6\n```",
"_____no_output_____"
]
]
] |
[
"markdown"
] |
[
[
"markdown"
]
] |
cbec36f7f6bfce9c5729e1380bfc1b8cabf4d08c
| 300,677 |
ipynb
|
Jupyter Notebook
|
udacity-dand-p2-investigate-a-dataset.ipynb
|
Zhenmao/udacity-dand-p2-investigate-a-dataset
|
bf25f28c46bbbc034f0c3d5aa71ef423b69daf5a
|
[
"MIT"
] | null | null | null |
udacity-dand-p2-investigate-a-dataset.ipynb
|
Zhenmao/udacity-dand-p2-investigate-a-dataset
|
bf25f28c46bbbc034f0c3d5aa71ef423b69daf5a
|
[
"MIT"
] | null | null | null |
udacity-dand-p2-investigate-a-dataset.ipynb
|
Zhenmao/udacity-dand-p2-investigate-a-dataset
|
bf25f28c46bbbc034f0c3d5aa71ef423b69daf5a
|
[
"MIT"
] | null | null | null | 125.438882 | 40,440 | 0.843583 |
[
[
[
"# Investigate a Dataset: Titanic Data",
"_____no_output_____"
],
[
"## Table of Contents\n1. [Introduction](#1-Introduction) \n2. [Data Wrangling](#2-Data-Wrangling) \n 2.1 [Handling Data Types](#2.1-Handling-Data-Types) \n 2.2 [Handling Missing Values](#2.2-Handling-Missing-Values) \n 2.2.1[Age](#2.2.1-Age) \n 2.2.2[Cabin](#2.2.2-Cabin) \n 2.2.3[Embarked (Port)](#2.2.3-Embarked-(Port)) \n3. [Data Exploration 1: Initial Examination of Single Variables](#3-Data-Exploration-1:-Initial-Examination-of-Single-Variables) \n 3.1 [Survival](#3.1-Survival) \n 3.2 [Class](#3.2-Class) \n 3.3 [Sex](#3.3-Sex) \n 3.4 [Age](#3.4-Age) \n 3.5 [SibSp](#3.5-SibSp) \n 3.6 [ParCh](#3.6-ParCh) \n 3.7 [Fare](#3.7-Fare) \n 3.8 [Cabin](#3.8-Cabin) \n 3.9 [Port](#3.9-Port) \n4. [Data Exploration 2: What factors make people more likely to survive?](#4-Data-Exploration-2:-What-factors-make-people-more-likely-to-survive?)\n 4.1 [Survived vs Class](#4.1-Survived-vs-Class) \n 4.2 [Survived vs Sex](#4.2-Survived-vs-Sex) \n 4.3 [Survived vs Age](#4.3-Survived-vs-Age) \n 4.4 [Survived vs SibSp](#4.4-Survived-vs-SibSp) \n 4.5 [Survived vs ParCh](#4.5-Survived-vs-ParCh) \n 4.6 [Survived vs Fare](#4.6-Survived-vs-Fare) \n5. [Data Exploration 3: What money can buy? Explore relations among passenger class, cabin, and fare](#5-Data-Exploration-3:-What-money-can-buy?-Explore-relations-among-passenger-class,-cabin,-and-fare) \n 5.1 [Class vs Fare](#5.1-Class-vs-Fare) \n 5.2 [Carbin vs Fare vs Class](#5.2-Carbin-vs-Fare-vs-Class) \n6. [Conclusion](#6-Conclusion) ",
"_____no_output_____"
],
[
"## 1 Introduction\n\nIn this report, I will investigate the Titanic survivor data using exploratory data analysis.\n\nIn the data wrangling phase, I will determine the appropriate datatypes for our dataset, and I will also show how to handle missing values. \n\nIn the data exploration phase, I will first look at each variable and its distribution. After that, I will answer two question:\n\n1. What factors make people more likely to survive?\n\n2. What money can buy? -- Explore relations among passenger class, cabin, and fare. \n\nLast, I will conclude this report by summarizing the findings and stating the limitations of my analysis.",
"_____no_output_____"
],
[
"## 2 Data Wrangling",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nimport seaborn as sns\nimport matplotlib.pyplot as plt\n%pylab inline\n# Change figure size into 8 by 6 inches\nmatplotlib.rcParams['figure.figsize'] = (8, 6)",
"Populating the interactive namespace from numpy and matplotlib\n"
]
],
[
[
"### 2.1 Handling Data Types\n\nAfter reviewing the original description of the dataset from [the Kaggle website](https://www.kaggle.com/c/titanic/data), the data type of each variable is chosen as following, and categorical variables will be converted to more descriptive labels:\n\n| Variable | Definition | Key | Type |\n|----------|--------------------------------------------|------------------------------------------------|-----------------|\n| Survived | Survival | 0 = No, 1 = Yes | int (Survived)* |\n| Pclass | Ticket class | 1 = 1st, 2 = 2nd, 3 = 3rd | int (Class) |\n| Sex | Sex | | str |\n| Age | Age in years | | float |\n| SibSp | # of siblings / spouses aboard the Titanic | | int |\n| ParCh | # of parents / children aboard the Titanic | | int |\n| Ticket | Ticket number | | int |\n| Fare | Passenger fare | | float |\n| Cabin | Cabin number | | str |\n| Embarked | Port of embarkation | C = Cherbourg, Q = Queenstown, S = Southampton | str (Port) |\n\n\\* indicate the name of converted categorical variable",
"_____no_output_____"
]
],
[
[
"data_file = 'titanic-data.csv'\ntitanic_df = pd.read_csv(\n data_file, \n dtype = {'PassengerId': str}\n)",
"_____no_output_____"
],
[
"# Convert categorical variables to more descriptive labels.\n\n# Create descriptive Survival column from Survived column\ntitanic_df['Survival'] = titanic_df['Survived'].map({0: 'Died', \n 1: 'Survived'})\n\n# Create descriptive Class column from Pclass column \ntitanic_df['Class'] = titanic_df['Pclass'].map({1: 'First Class', \n 2: 'Second Class', \n 3: 'Third Class'})\n\n# Create descriptive Port column from Embarked column\ntitanic_df['Port'] = titanic_df['Embarked'].map({'C': 'Cherbourg', \n 'Q': 'Queenstown', \n 'S': 'Southampton'})",
"_____no_output_____"
]
],
[
[
"### 2.2 Handling Missing Values",
"_____no_output_____"
]
],
[
[
"total_passengers = len(titanic_df)\nprint(\"There are {} passengers on board.\".format(total_passengers))",
"There are 891 passengers on board.\n"
],
[
"titanic_df.isnull().sum()",
"_____no_output_____"
]
],
[
[
"There are three columns with missing values: Age, Cabin, and Embarked(Port).",
"_____no_output_____"
],
[
"#### 2.2.1 Age\n\nThere are 117 out of 891 passengers missing Age values. Because filling in the missing Age values will affect the distribution of Age variable, and this analysis is mainly exploratory, I will **drop rows with missing Age values**.\n\nBut as an exercise, I will show here a way of filling in the missing Age values based on the assumption that Age values can be infered from SibSp, ParCh, Class, and Sex.\n\nWe'll store the filled Age values in a new column called AgeFilled.",
"_____no_output_____"
]
],
[
[
"titanic_df['AgeFilled'] = titanic_df['Age']",
"_____no_output_____"
]
],
[
[
"##### SibSp and ParCh\n\nSibSp(number of siblings/spouses): The number of spouses can only be 0 or 1, therefore any SibSp value greater than 1 can be implied that this person is traveling with sibling(s).\n\nParCh(number of parents/children): The number of parents can only be 2 maximum, any ParCh value greater than 2 can be implied that this person is traveling with child(ren).\n\nFrom these two observations, we can infer two rules:\n\n1. If SibSp >= 2, this passenger has at least 1 sibling on board, and if also his/her ParCh >= 1, then he/she is most likly a **child** traveling with sibling(s) and parent(s).\n\n2. If Parch >= 3, this passenger has at least 1 child, then he/she has to be an **adult** parent traveling with child(ren).\n\nFrom these two rules, we can divide all rows into three categories: **Child**, **Adult**, **Unknown**, and store this categorical value in a new column ChAd. And we can fill in the Age value of the row categoried with child or parent with its category's median age value.",
"_____no_output_____"
]
],
[
[
"def child_or_adult(sibsp, parch):\n '''Categorize a person as Child, Adult or Unknown\n \n Arg:\n sibsp: the number of siblings/spouses\n parch: the number of parents/children\n \n Return:\n A string denotes Child, Adult or Unknown\n '''\n if sibsp >= 2 and parch >= 1:\n return 'Child'\n if parch >= 3:\n return 'Adult'\n return 'Unknown'",
"_____no_output_____"
],
[
"titanic_df['ChAd'] = titanic_df[['SibSp', 'ParCh']].apply(lambda x: child_or_adult(*x), axis=1)",
"_____no_output_____"
],
[
"sns.boxplot(x='ChAd', y='AgeFilled', data=titanic_df)",
"_____no_output_____"
]
],
[
[
"Fill Adult, Child groups' missing Age values with groups' median values. Leave Unknown group' missing Age values as NaN.",
"_____no_output_____"
]
],
[
[
"for group_name, group in titanic_df.groupby('ChAd'):\n if group_name == 'Adult' or group_name == 'Child':\n group['AgeFilled'].fillna(group['AgeFilled'].median())",
"_____no_output_____"
]
],
[
[
"##### Class and Sex\n\nFor the passengers categorized as Unknown in the ChAd column, I will fill the missing Age values with the median value of the same Class and Sex group.",
"_____no_output_____"
]
],
[
[
"sns.boxplot(x='Class', y='AgeFilled', hue='Sex', data=titanic_df)",
"_____no_output_____"
],
[
"fillna_with_median = lambda x: x.fillna(x.median(), inplace=False)\ntitanic_df['AgeFilled'] = titanic_df.groupby(['Sex', 'Pclass'])['AgeFilled'].transform(fillna_with_median)",
"_____no_output_____"
]
],
[
[
"I will look at how the distribution of Age has changed after filling the missing values in the next section.",
"_____no_output_____"
],
[
"#### 2.2.2 Cabin\n\nThere are 689 out of 891 passengers missing Cabin values. Because the majority of rows are missing cabin values, I decide to exclude these rows during analysis when cabin value is considered.",
"_____no_output_____"
],
[
"#### 2.2.3 Embarked (Port)\n\nThere are 2 out of 891 passengers missing Embarked values. Here I choose to fill these two missing values with the most frequent value (mode).",
"_____no_output_____"
]
],
[
[
"titanic_df['Embarked'] = titanic_df['Embarked'].fillna(titanic_df['Embarked'].mode().iloc[0]);\ntitanic_df['Port'] = titanic_df['Port'].fillna(titanic_df['Port'].mode().iloc[0]);",
"_____no_output_____"
]
],
[
[
"## 3 Data Exploration 1: Initial Examination of Single Variables",
"_____no_output_____"
],
[
"For the first exploration phase, I will look at some of the variables individually.",
"_____no_output_____"
]
],
[
[
"def categorical_count_and_frequency(series):\n '''Calculate count and frequency table\n \n Given an categorical variable pandas Series, return a DataFrame containing\n counts and frequencies of each possible value.\n \n Arg:\n series: A pandas Series from a categorical variable\n \n Returns:\n A DataFrame containing counts and frequencies\n '''\n \n counts = series.value_counts()\n frequencies = series.value_counts(normalize=True)\n return counts.to_frame(name='Counts').join(frequencies.to_frame(name='Frequencies')).sort_index()",
"_____no_output_____"
]
],
[
[
"### 3.1 Survival",
"_____no_output_____"
]
],
[
[
"categorical_count_and_frequency(titanic_df['Survival'])",
"_____no_output_____"
]
],
[
[
"Out of 891 passengers, there are 342 of them survived, and 549 died. The overall survival rate is about 38.3%",
"_____no_output_____"
],
[
"### 3.2 Class",
"_____no_output_____"
]
],
[
[
"categorical_count_and_frequency(titanic_df['Class'])",
"_____no_output_____"
]
],
[
[
"There are 216 first class ticket passengers (24%), 184 second class ticket passengers (21%), and 491 third class ticket passengers (55%).",
"_____no_output_____"
],
[
"### 3.3 Sex",
"_____no_output_____"
]
],
[
[
"categorical_count_and_frequency(titanic_df['Sex'])",
"_____no_output_____"
]
],
[
[
"There are 577 male passengers (65%) and 314 female passengers (35%).",
"_____no_output_____"
],
[
"### 3.4 Age",
"_____no_output_____"
]
],
[
[
"titanic_df['Age'].describe()",
"_____no_output_____"
],
[
"ax = sns.distplot(titanic_df['Age'].dropna())\nax.set_title('Histogram of Age')\nax.set_ylabel('Frequency')\nplt.xticks(linspace(0, 80, 9))",
"_____no_output_____"
]
],
[
[
"The age distribution is bimodal. One mode is centered around 5 years old representing children. The other mode is positively skewed with a peak between 20 and 30 representing teenagers and adults.\n\nI will also take look at the age distribtion with missing values filled from the previous section.",
"_____no_output_____"
]
],
[
[
"sns.distplot(titanic_df['Age'].dropna())\nax = sns.distplot(titanic_df['AgeFilled'].dropna())\nax.set_title('Histogram of Age vs AgeFilled')\nax.set_ylabel('Frequency')\nplt.xticks(np.linspace(0, 80, 9))\nplt.yticks(np.linspace(0, 0.065, 14))",
"_____no_output_____"
]
],
[
[
"Comparing Age (blue) and AgeFilled (orange) histograms, one can clearly see that filling the Age missing values changed the distribution. Most of the Age values filled are concentrated around the largest peak of the original age distribution.",
"_____no_output_____"
],
[
"### 3.5 SibSp",
"_____no_output_____"
]
],
[
[
"categorical_count_and_frequency(titanic_df['SibSp'])",
"_____no_output_____"
]
],
[
[
"608 passengers (68%) travel without any sibling or spouse.\n\n209 passengers (23%) passengers travel with only 1 sibling or spouse, and in this case, it is likely to be a spouse.\n\nThe remaining 74 passengers (9%) travel more than ! sibling or spouse, and in this case, it is like to be siblings.",
"_____no_output_____"
],
[
"### 3.6 ParCh",
"_____no_output_____"
]
],
[
[
"categorical_count_and_frequency(titanic_df['ParCh'])",
"_____no_output_____"
]
],
[
[
"678 passengers (76%) travel without any parent or child.\n\nThe remaining 213 passengers travel with 1 to 6 parent(s) or child(ren).",
"_____no_output_____"
],
[
"### 3.7 Fare",
"_____no_output_____"
]
],
[
[
"titanic_df['Fare'].describe()",
"_____no_output_____"
],
[
"ax = sns.distplot(titanic_df['Fare'])\nax.set_title('Histogram of Fare')\nax.set_ylabel('Frequency')",
"_____no_output_____"
]
],
[
[
"From the historgram we can see the fare distribution is positively skewed.",
"_____no_output_____"
],
[
"### 3.8 Cabin",
"_____no_output_____"
]
],
[
[
"titanic_df['Cabin'].describe()",
"_____no_output_____"
]
],
[
[
"Out of the 891 passengers on board, there are only 204 of them with their cabin information.\n\nThe cabin number can be interpreted as the following:\n- The first letter indicating the cabin level.\n- The following digits indicating the room number. \n\nFor our analysis purpose, we are going to only look at the cabin level here for passengers with known cabin number, and keep NaN as NaN for passenger without any cabin number.\n\nAdd a column 'CabinLevel' that contains the cabin level (the first letter of 'Cabin' column).",
"_____no_output_____"
]
],
[
[
"titanic_df['CabinLevel'] = titanic_df['Cabin'].str[0]",
"_____no_output_____"
],
[
"categorical_count_and_frequency(titanic_df['CabinLevel'])",
"_____no_output_____"
]
],
[
[
"### 3.9 Port",
"_____no_output_____"
]
],
[
[
"categorical_count_and_frequency(titanic_df['Port'])",
"_____no_output_____"
]
],
[
[
"After filling the 2 passengers who are missing their port of embarkation information, 168 (19%) boarded at Cherbourg, 77 (9%) boarded at Queenstown, and 646 (72%) boarded at Southampton.",
"_____no_output_____"
],
[
"## 4 Data Exploration 2: What factors make people more likely to survive?",
"_____no_output_____"
]
],
[
[
"def survival_by(variable_name):\n '''Calculate survival rate for a given variable\n \n For the titanic pandas DataFrame titanic_df, calculate the survival count and \n survival rate grouped by a given categorical variable.\n \n Arg:\n variable_name: a categorical variable name to group by\n \n Return:\n survival_by_df: a pandas DataFrame that contains the total number of passengers,\n survivied number of passengers, and survival rate for each category of\n the given categorical variable\n '''\n \n grouped = titanic_df.groupby(variable_name)\n total = grouped.size() # number of total passengers by variable\n survived = grouped['Survived'].sum() # number of survived passengers by variable\n survival_rate = survived / total # survival rate by variable\n survival_by_df = pd.DataFrame(\n data = [total, survived, survival_rate], \n index = ['Total', 'Survived', 'Survival Rate']\n )\n return survival_by_df",
"_____no_output_____"
],
[
"def survival_by_plot(survival_by_df):\n '''Plot a bar graph showing Survival Rate vs a categorical variable\n \n For the survial_by_df, a pandas DataFrame showing the survival count and survival rate\n for the titanic_df pandas DataFrame, this funcions plots a bar graph showing the\n survival rate across a given categorical variable.\n \n Arg:\n survival_by_df: a pandas DataFrame generated by the survival_by(variable_name) function.\n '''\n \n ax = survival_by_df.T['Survival Rate'].plot(kind = 'bar')\n ax.set_ylabel('Survival Rate')\n ax.set_title('Barplot of Survival Rate vs {}'.format(survival_by_df.columns.name))",
"_____no_output_____"
]
],
[
[
"### 4.1 Survived vs Class",
"_____no_output_____"
]
],
[
[
"survival_by_class_df = survival_by('Class')\nsurvival_by_class_df",
"_____no_output_____"
],
[
"survival_by_plot(survival_by_class_df)",
"_____no_output_____"
]
],
[
[
"Class (passenger class) is an important factor that correlates with survival rate. \n\nFirst class passenger survival rate (63%) > second class passenger survival rate (47%) > third class passenger survival rate. \n\nSince Class is a proxy for socio-economic status (1st ~ Upper; 2nd ~ Middle; 3rd ~ Lower), the result suggests that the higher your socio-economic status is, the higher your survival rate is.",
"_____no_output_____"
],
[
"### 4.2 Survived vs Sex",
"_____no_output_____"
]
],
[
[
"survival_by_sex_df = survival_by('Sex')\nsurvival_by_sex_df",
"_____no_output_____"
],
[
"survival_by_plot(survival_by_sex_df)",
"_____no_output_____"
]
],
[
[
"Sex is one important factor that correlates with survival rate. Female survival rate (74%) is much higher than male survival rate (19%)",
"_____no_output_____"
],
[
"### 4.3 Survived vs Age",
"_____no_output_____"
],
[
"Because Age is a continuous variable, I need to convert it into descrete groups first. Here I choose Age groups in 10 year interval ranging from 0 to 80 in order to include all passgeners with Age values. These Age groups are stored in a new column called 'AgeGrp'",
"_____no_output_____"
]
],
[
[
"titanic_df['AgeGrp'] = pd.cut(\n titanic_df['Age'], \n bins = np.linspace(0, 80, 9), \n include_lowest = True\n)",
"_____no_output_____"
],
[
"survival_by_age_df = survival_by('AgeGrp')\nsurvival_by_age_df",
"_____no_output_____"
],
[
"survival_by_plot(survival_by_age_df)",
"_____no_output_____"
]
],
[
[
"Young kids aged below 10 has a high survival rate close to 60%, old people aged above 60 has a low survival rate around 20%, and everyone else has a similar survival rate around 40%.",
"_____no_output_____"
],
[
"### 4.4 Survived vs SibSp",
"_____no_output_____"
]
],
[
[
"survival_by_sibsp_df = survival_by('SibSp')\nsurvival_by_sibsp_df",
"_____no_output_____"
],
[
"survival_by_plot(survival_by_sibsp_df)",
"_____no_output_____"
]
],
[
[
"Passengers who travel without any sibling/spouse has a survival rate around 35%, which is lower than the survival rate of passengers who travel with 1 (54%) or 2 (46%) siblings/spouses. The number of passengers who travel with 3 or more siblings/spouses is too small to draw any definitive conclusion.",
"_____no_output_____"
],
[
"### 4.5 Survived vs ParCh",
"_____no_output_____"
]
],
[
[
"survival_by_parch_df = survival_by('ParCh')\nsurvival_by_parch_df",
"_____no_output_____"
],
[
"survival_by_plot(survival_by_parch_df)",
"_____no_output_____"
]
],
[
[
"Passengers who travel without any parent/child has a survival rate of 34%, which is lower than the survival rate of passengers who travel with 1 (55%) or 2 (50%) parents/children. The number of passengers who travel with 3 or more parents/children is too small to draw any definitive conclusion.",
"_____no_output_____"
],
[
"### 4.6 Survived vs Fare",
"_____no_output_____"
],
[
"As the histogram from previous section shows that the fare distribution is highly positively skewed. If we categorize fare into fare groups with fixed width bins, the low fare bin will end up with too many passengers while the high fare bin will only have very few passengers.\n\nHere I decide to choose bins corresponding to quartiles, so that every bin has similar number of passengers. The quartile categorization is stored in a new column called 'FareGrp'.",
"_____no_output_____"
]
],
[
[
"titanic_df['FareGrp'] = pd.cut(\n titanic_df['Fare'], \n bins = titanic_df['Fare'].quantile([0, 0.25, 0.5, 0.75, 1]), \n labels = ['1Q', '2Q', '3Q', '4Q'], \n include_lowest = True\n)",
"_____no_output_____"
],
[
"survival_by_fare_df = survival_by('FareGrp')\nsurvival_by_fare_df",
"_____no_output_____"
],
[
"survival_by_plot(survival_by_fare_df)",
"_____no_output_____"
]
],
[
[
"From the bar plot one can clearly see that a higher fare correlates to a higher survival rate.",
"_____no_output_____"
],
[
"## 5 Data Exploration 3: What money can buy? Explore relations among passenger class, cabin, and fare",
"_____no_output_____"
],
[
"### 5.1 Class vs Fare",
"_____no_output_____"
],
[
"Because there're only 204 out of 891 passengers with Cabin information, we'll look at only 'Class' and 'Fare' first.",
"_____no_output_____"
]
],
[
[
"ax = sns.boxplot(x = 'Class', y = 'Fare', data = titanic_df, \n order=['First Class', 'Second Class', 'Third Class']);\nax.set_title('Boxplot of Fare vs Class')",
"_____no_output_____"
],
[
"titanic_df.groupby('Class')['Fare'].median()",
"_____no_output_____"
]
],
[
[
"The median First Class fare is about 4 times of the median Second Class fare, and the median Second Class fare is about 2 times of the median Third Class fare.",
"_____no_output_____"
],
[
"### 5.2 Carbin vs Fare vs Class",
"_____no_output_____"
],
[
"Now I will include Cabin into the analysis, and only include the 204 passengers with givin Cabin values.\n\nBecause the number of data points is not too large, I will use a swarmplot to show all data points and to look at all 3 variables at once.",
"_____no_output_____"
]
],
[
[
"ax = sns.swarmplot(\n x = 'CabinLevel', \n y = 'Fare', \n hue = 'Class', \n order = list('TABCDEFG'),\n hue_order = ['First Class', 'Second Class', 'Third Class'],\n data = titanic_df\n)\nax.set_title('Swarmplot of Fare vs CabinLevel vs Class')",
"_____no_output_____"
]
],
[
[
"There are some interesting findings from this plot:\n\n1. Most of the cabin numbers are recorded for First Class passengers, and very few Second and Third Class passengers have their cabin numbers on the record.\n2. From the records, cabin level T, A, B and C are exclusively for First Class passengers, and cabin level F and G only accommodate Second and Third Class passengers.",
"_____no_output_____"
],
[
"## 6 Conclusion",
"_____no_output_____"
],
[
"By ananlyzing the Titanic dataset, I tried to answer two intersting questions:\n\nQ1. What factors make people more likely to survive?\n\nA1. The survival rates of the Titanic disaster:\n\n- First Class > Second Class > Third Class\n- Female > Male\n- Age 0-10 (Children) > Age 10-60 > age 60-80 (Senior Citizens) \n- Traveling with 1 or 2 sibling(s)/spouse(s) > traveling without sibling/spouse\n- Traveling with 1 or 2 parent(s)/child(ren) > traveling without parent/child\n- Expensive fare > Cheap fare\n\nQ2. What money can buy?\n\nA2. A typical First Class passenger pays a higher fare than Second and Third Class passengers, but he/she gets to stay on a higher cabin level where none or very few Second and Third class passengers stay.\n\nAlthough the findings of the anaylsis are clear, there are a few limitations that can be improved in the future. \n\n- When dealing with missing values, I used simple direct methods (filling with group median (Age), droping rows (Cabin), filling with mode (Embarked). To imporve, I can use some machine learning method to build a model to fill the missing values more rigorously. \n\n- I did not include any statistical testing in the report since I am only exploring the dataset to find some interesting observations. \n\n- This report does not include any machine learning algorithm to mind the dataset due to the limited scope of this report.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
cbec3e70ff1b2a6f4a8c558f779a3492b21bb35d
| 172,953 |
ipynb
|
Jupyter Notebook
|
notebooks/pytorch/introspection.ipynb
|
Petr-By/qtpyvis
|
0b9a151ee6b9a56b486c2bece9c1f03414629efc
|
[
"MIT"
] | 3 |
2017-10-04T14:51:26.000Z
|
2017-10-22T09:35:50.000Z
|
notebooks/pytorch/introspection.ipynb
|
CogSciUOS/DeepLearningToolbox
|
bf07578b9486d8c48e25df357bc4b9963b513b46
|
[
"MIT"
] | 13 |
2017-11-26T10:05:00.000Z
|
2018-03-11T14:08:40.000Z
|
notebooks/pytorch/introspection.ipynb
|
CogSciUOS/DeepLearningToolbox
|
bf07578b9486d8c48e25df357bc4b9963b513b46
|
[
"MIT"
] | 2 |
2017-09-24T21:39:42.000Z
|
2017-10-04T15:29:54.000Z
| 201.10814 | 4,892 | 0.909097 |
[
[
[
"import torch\nimport torch.nn as nn\nimport torch.nn.functional as F\nfrom torchvision import datasets, transforms\nfrom torch.autograd import Variable\nfrom collections import OrderedDict\nimport numpy as np\nimport matplotlib.pyplot as plt\nplt.rcParams['image.cmap'] = 'gray'\n%matplotlib inline",
"_____no_output_____"
],
[
"# input batch size for training (default: 64)\nbatch_size = 64\n\n# input batch size for testing (default: 1000)\ntest_batch_size = 1000\n\n# number of epochs to train (default: 10)\nepochs = 10\n\n# learning rate (default: 0.01)\nlr = 0.01\n\n# SGD momentum (default: 0.5)\nmomentum = 0.5\n\n# disables CUDA training\nno_cuda = True\n\n# random seed (default: 1)\nseed = 1\n\n# how many batches to wait before logging training status\nlog_interval = 10\n\n# Setting seed for reproducibility.\ntorch.manual_seed(seed)\n\ncuda = not no_cuda and torch.cuda.is_available()\nprint(\"CUDA: {}\".format(cuda))",
"CUDA: False\n"
],
[
"if cuda:\n torch.cuda.manual_seed(seed)\ncudakwargs = {'num_workers': 1, 'pin_memory': True} if cuda else {}\n\nmnist_transform = transforms.Compose([\n transforms.ToTensor(),\n transforms.Normalize((0.1307,), (0.3081,)) # Precalcualted values.\n])\n\ntrain_set = datasets.MNIST(\n root='data',\n train=True,\n transform=mnist_transform,\n download=True,\n)\n\ntest_set = datasets.MNIST(\n root='data',\n train=False,\n transform=mnist_transform,\n download=True,\n)\n\ntrain_loader = torch.utils.data.DataLoader(\n dataset=train_set,\n batch_size=batch_size,\n shuffle=True,\n **cudakwargs\n)\n\ntest_loader = torch.utils.data.DataLoader(\n dataset=test_set,\n batch_size=test_batch_size,\n shuffle=True,\n **cudakwargs\n)\n",
"_____no_output_____"
]
],
[
[
"## Loading the model.\nHere we will focus only on `nn.Sequential` model types as they are easier to deal with. Generalizing the methods described here to `nn.Module` will require more work.",
"_____no_output_____"
]
],
[
[
"class Flatten(nn.Module):\n def forward(self, x):\n return x.view(x.size(0), -1)\n \n def __str__(self):\n return 'Flatten()'\n\nmodel = nn.Sequential(OrderedDict([\n ('conv2d_1', nn.Conv2d(in_channels=1, out_channels=32, kernel_size=3)),\n ('relu_1', nn.ReLU()),\n ('max_pooling2d_1', nn.MaxPool2d(kernel_size=2)),\n ('conv2d_2', nn.Conv2d(in_channels=32, out_channels=32, kernel_size=3)),\n ('relu_2', nn.ReLU()),\n ('dropout_1', nn.Dropout(p=0.25)),\n ('flatten_1', Flatten()),\n ('dense_1', nn.Linear(3872, 64)),\n ('relu_3', nn.ReLU()),\n ('dropout_2', nn.Dropout(p=0.5)),\n ('dense_2', nn.Linear(64, 10)),\n ('readout', nn.LogSoftmax())\n]))\n\nmodel.load_state_dict(torch.load('example_torch_mnist_model.pth'))",
"_____no_output_____"
]
],
[
[
"## Accessing the layers\nA `torch.nn.Sequential` module serves itself as an iterable and subscriptable container for all its children modules.\n",
"_____no_output_____"
]
],
[
[
"for i, layer in enumerate(model):\n print('{}\\t{}'.format(i, layer))",
"0\tConv2d(1, 32, kernel_size=(3, 3), stride=(1, 1))\n1\tReLU ()\n2\tMaxPool2d (size=(2, 2), stride=(2, 2), dilation=(1, 1))\n3\tConv2d(32, 32, kernel_size=(3, 3), stride=(1, 1))\n4\tReLU ()\n5\tDropout (p = 0.25)\n6\tFlatten()\n7\tLinear (3872 -> 64)\n8\tReLU ()\n9\tDropout (p = 0.5)\n10\tLinear (64 -> 10)\n11\tLogSoftmax ()\n"
]
],
[
[
"Moreover `.modules` and `.children` provide generators for accessing layers.",
"_____no_output_____"
]
],
[
[
"for m in model.modules():\n print(m)",
"Sequential (\n (conv2d_1): Conv2d(1, 32, kernel_size=(3, 3), stride=(1, 1))\n (relu_1): ReLU ()\n (max_pooling2d_1): MaxPool2d (size=(2, 2), stride=(2, 2), dilation=(1, 1))\n (conv2d_2): Conv2d(32, 32, kernel_size=(3, 3), stride=(1, 1))\n (relu_2): ReLU ()\n (dropout_1): Dropout (p = 0.25)\n (flatten_1): Flatten (\n )\n (dense_1): Linear (3872 -> 64)\n (relu_3): ReLU ()\n (dropout_2): Dropout (p = 0.5)\n (dense_2): Linear (64 -> 10)\n (readout): LogSoftmax ()\n)\nConv2d(1, 32, kernel_size=(3, 3), stride=(1, 1))\nReLU ()\nMaxPool2d (size=(2, 2), stride=(2, 2), dilation=(1, 1))\nConv2d(32, 32, kernel_size=(3, 3), stride=(1, 1))\nReLU ()\nDropout (p = 0.25)\nFlatten (\n)\nLinear (3872 -> 64)\nReLU ()\nDropout (p = 0.5)\nLinear (64 -> 10)\nLogSoftmax ()\n"
],
[
"for c in model.children():\n print(c)",
"Conv2d(1, 32, kernel_size=(3, 3), stride=(1, 1))\nReLU ()\nMaxPool2d (size=(2, 2), stride=(2, 2), dilation=(1, 1))\nConv2d(32, 32, kernel_size=(3, 3), stride=(1, 1))\nReLU ()\nDropout (p = 0.25)\nFlatten (\n)\nLinear (3872 -> 64)\nReLU ()\nDropout (p = 0.5)\nLinear (64 -> 10)\nLogSoftmax ()\n"
]
],
[
[
"## Getting the weigths.",
"_____no_output_____"
]
],
[
[
"conv2d_1_weight = model[0].weight.data.numpy()\nconv2d_1_weight.shape",
"_____no_output_____"
],
[
"for i in range(32):\n plt.imshow(conv2d_1_weight[i, 0])\n plt.show()",
"_____no_output_____"
]
],
[
[
"### Getting layer properties",
"_____no_output_____"
],
[
"The layer objects themselfs expose most properties as attributes.",
"_____no_output_____"
]
],
[
[
"conv2d_1 = model[0]",
"_____no_output_____"
],
[
"conv2d_1.kernel_size",
"_____no_output_____"
],
[
"conv2d_1.stride",
"_____no_output_____"
],
[
"conv2d_1.dilation",
"_____no_output_____"
],
[
"conv2d_1.in_channels, conv2d_1.out_channels",
"_____no_output_____"
],
[
"conv2d_1.padding",
"_____no_output_____"
],
[
"conv2d_1.output_padding",
"_____no_output_____"
],
[
"dropout_1 = model[5]",
"_____no_output_____"
],
[
"dropout_1.p",
"_____no_output_____"
],
[
"dense_1 = model[7]",
"_____no_output_____"
],
[
"dense_1.in_features, dense_1.out_features",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbec4a662d110c50d69cd179f4ea5bdd7f628b55
| 126,327 |
ipynb
|
Jupyter Notebook
|
notebook/lm_model.ipynb
|
wckdouglas/tgirt_smRNA_correction
|
4820055cccd12cf286d4c154a4d05cdfb5da2538
|
[
"MIT"
] | null | null | null |
notebook/lm_model.ipynb
|
wckdouglas/tgirt_smRNA_correction
|
4820055cccd12cf286d4c154a4d05cdfb5da2538
|
[
"MIT"
] | null | null | null |
notebook/lm_model.ipynb
|
wckdouglas/tgirt_smRNA_correction
|
4820055cccd12cf286d4c154a4d05cdfb5da2538
|
[
"MIT"
] | null | null | null | 242.936538 | 107,044 | 0.883327 |
[
[
[
"%matplotlib inline\n\nimport pandas as pd\nimport numpy as np\nfrom sklearn.linear_model import Ridge\nfrom sklearn.preprocessing import LabelEncoder, StandardScaler\nfrom sklearn.model_selection import KFold\nfrom sklearn.ensemble import RandomForestRegressor\nimport matplotlib.pyplot as plt\nfrom sklearn.metrics import explained_variance_score\nfrom scipy.stats import pearsonr, ranksums\nimport seaborn as sns\nimport os\nfrom scipy.cluster import hierarchy as sch\n\nsns.set_style('white')\n\ndef positioning(x):\n return x[-1]\n\ndef count_to_cpm(count_array):\n count_array = np.true_divide(count_array,count_array.sum()) * 1e6 \n return count_array\n\ndef get_end(x):\n if 'head' in x:\n return \"5'\"\n elif 'tail' in x:\n return \"3'\"\n\ndef make_column_name(colnames):\n col_d = pd.DataFrame({'nucleotide':colnames.str.slice(-1),\n 'position':colnames.str.slice(4,5),\n 'end':colnames.map(get_end)}) \\\n .assign(offset = lambda d: np.where(d.end==\"5'\",-1, 3)) \\\n .assign(adjusted_position = lambda d: np.abs(d.position.astype(int) - d.offset))\\\n .assign(colnames = colnames)\n #print col_d\n return col_d.end + '-position:'+col_d.adjusted_position.astype(str) +':'+ col_d.nucleotide\n \ndef preprocess_dataframe(df):\n nucleotides = df.columns[df.columns.str.contains('head|tail')]\n dummies = pd.get_dummies(df[nucleotides])\n dummies.columns = make_column_name(dummies.columns)\n df = pd.concat([df,dummies],axis=1) \\\n .drop(nucleotides, axis=1) \n return df",
"_____no_output_____"
],
[
"df = pd.read_table('../test/test_train_set.tsv')\\\n .assign(expected_cpm = lambda d: count_to_cpm(d['expected_count']))\\\n .assign(cpm = lambda d: count_to_cpm(d['experimental_count']))\\\n .assign(log_cpm = lambda d: np.log2(d.cpm+1) - np.log2(d.expected_cpm+1))\\\n .pipe(preprocess_dataframe) \\\n .drop(['experimental_count','cpm'], axis=1)\ndf.head()",
"_____no_output_____"
],
[
"X = df.filter(regex='^5|^3')\nX.columns",
"_____no_output_____"
],
[
"def train_lm(d, ax):\n X = d.drop(['seq_id','log_cpm','expected_count','expected_cpm'], axis=1)\n Y = d['log_cpm'].values\n lm = Ridge()\n lm.fit( X, Y)\n pred_Y = lm.predict(X)\n rsqrd = explained_variance_score(Y, pred_Y)\n rho, pval = pearsonr(pred_Y, Y)\n ax.scatter(Y, pred_Y)\n ax.text(0,-5, '$R^2$ = %.3f' %(rsqrd), fontsize=15)\n ax.text(0,-6, r'$\\rho$ = %.3f' %rho, fontsize=15)\n ax.spines['top'].set_visible(False)\n ax.spines['right'].set_visible(False)\n ax.set_xlabel(r'True $\\Delta$CPM from TGIRT-seq (log2)')\n ax.set_ylabel(r'Predicted $\\Delta$CPM from end nucleotides (log2)')\n sns.despine()\n return lm, X, Y\n\n#.assign(side = lambda d: d.X.str.replace('-position:[0-9]-[ACTG]$','')) \\\n# .assign(X = lambda d: d.X.str.replace('-[ACTG]$','')) \\\ndef coefficient_plot(d, lm, ax):\n X_factor = d.drop(['log_cpm','seq_id','expected_count','expected_cpm'], axis=1).columns\n coefficient = lm.coef_\n colors = sns.color_palette('Dark2',10)\n d = pd.DataFrame({'X': X_factor, 'coef':coefficient}) \\\n .assign(side = lambda d: d.X.str.replace('','')) \\\n .assign(color = lambda d: list(map(lambda x: colors[0] if \"5'\" in x else colors[1], d.side))) \\\n .sort_values('coef') \\\n .assign(bar_color = lambda d: list(map(lambda x: 'green' if x < 0 else 'purple', d.coef)))\n ax.bar(np.arange(len(d.coef)),\n d.coef,\n color = d.bar_color)\n ax.xaxis.set_ticks(np.arange(len(d.coef)))\n x = ax.set_xticklabels(d.X,rotation=90)\n for xt, col in zip(ax.get_xticklabels(), d.color):\n xt.set_color(col)\n ax.legend(title =' ')\n ax.set_ylabel('Coefficients')\n #ax.set_title('Coefficients for positional bases')\n ax.set_xlabel(' ')\n ax.spines['top'].set_visible(False)\n ax.spines['right'].set_visible(False)\n ax.hlines(xmin=-4, xmax=len(d.coef), y = 0)\n ax.set_xlim(-1, len(d.coef))\n\ndef get_sim(df):\n sim = poisson.rvs(10000, range(df.shape[0]))\n sim = np.log2(count_to_cpm(sim)+1) - df['expected_count']\n return sim\n\ndef cross_validation(X_train, X_test, Y_train, Y_test, i):\n lm = Ridge()\n lm.fit(X_train, Y_train)\n\n err = X_test * lm.coef_\n err = err.sum(axis=1)\n error_df = pd.DataFrame({'Corrected':err, \n 'Uncorrected': Y_test})\\\n .pipe(pd.melt, \n var_name = 'correction', \n value_name = 'error') \\\n .groupby('correction', as_index=False) \\\n .agg({'error': lambda x: np.sqrt(x.pow(2).mean())}) \n return error_df\n\ndef plot_error(ax, X, Y, df):\n X = df.drop(['seq_id','log_cpm','expected_count','expected_cpm'], axis=1).values\n Y = df['log_cpm'].values\n kf = KFold(n_splits=20, random_state=0)\n dfs = []\n for i, (train_index, test_index) in enumerate(kf.split(X)):\n X_train, X_test = X[train_index], X[test_index] \n Y_train, Y_test = Y[train_index], Y[test_index]\n df = cross_validation(X_train, X_test, Y_train, Y_test, i)\n dfs.append(df)\n error_df = pd.concat(dfs)\n sns.swarmplot(data=error_df, x = 'correction', \n y = 'error', order=['Uncorrected','Corrected'])\n ax.set_xlabel(' ')\n ax.set_ylabel('Root-mean-square error (log2 CPM)')\n ax.spines['top'].set_visible(False) \n ax.spines['right'].set_visible(False) \n return error_df\n\ndef remove_grid(ax):\n for o in ['top','bottom','left','right']:\n ax.spines[o].set_visible(False) \n\ndef make_cluster(d, ax):\n linkage_color = ['black','darkcyan','darkseagreen','darkgoldenrod']\n sch.set_link_color_palette(linkage_color)\n z = sch.linkage(compare_df)\n l = sch.dendrogram(z, orientation='left',\n color_threshold=100,\n no_labels=True)\n ax.xaxis.set_visible(False)\n ax.yaxis.set_visible(False)\n remove_grid(ax)\n return l\n\ndef plot_heat(compare_df, cl, ax, cax):\n plot_df = compare_df.iloc[cl['leaves'],[1,0]] + np.log2(1040.5)\n hm = ax.imshow(plot_df, cmap='inferno',aspect='auto')\n ax.xaxis.set_ticks(range(0,2))\n ax.set_xticklabels(plot_df.columns)\n ax.yaxis.set_visible(False)\n cb = plt.colorbar(hm, cax=cax)\n cb.set_label('log2(CPM)',rotation=270,labelpad=18)\n remove_grid(ax)\n remove_grid(cax)\n\nplt.rc('xtick',labelsize=14)\nplt.rc('ytick',labelsize=14)\nplt.rc('axes',labelsize=14)\nfig = plt.figure(figsize=(9,12))\nax = fig.add_axes([0,0.55,0.45,0.5])\nax2 = fig.add_axes([0.55,0.6,0.48,0.4])\nlm, X, Y = train_lm(df,ax)\ncoefficient_plot(df, lm, ax2)\n\n\n#plot heatmap and error\ncompare_df = pd.DataFrame({'Corrected':Y - np.sum(lm.coef_ * X, axis=1),\n 'Uncorrected':df.log_cpm})\nd_ax = fig.add_axes([-0.05,0,0.1,0.45])\ncl = make_cluster(compare_df, d_ax)\nmap_ax = fig.add_axes([0.05,0,0.3,0.45])\ncbar_ax = fig.add_axes([0.36,0,0.02,0.45])\nplot_heat(compare_df, cl, map_ax, cbar_ax)\n\nax3 = fig.add_axes([0.58,0,0.45,0.45])\nerror_df = plot_error(ax3, X, Y, df)\n\n#fig.tight_layout()\nfig.text(-0.05,1.03,'A',size=15)\nfig.text(0.52,1.03,'B',size=15)\nfig.text(-0.05, 0.47,'C',size=15)\nfig.text(0.5, 0.47,'D',size=15)\nfigurename = os.getcwd() + '/expression_prediction.pdf'\nfig.savefig(figurename, bbox_inches='tight', transparent=True)\nprint('plotted ', figurename )",
"No handles with labels found to put in legend.\n"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code"
]
] |
cbec4e7087599442ff6662aa5d28e42749883231
| 24,856 |
ipynb
|
Jupyter Notebook
|
fonte_original_tunning/Tunning_Version2.ipynb
|
marciodelima/case1_santander_engml_geral
|
2f76838b362d5d4c9d4e2f3cf464b5d78e0270d1
|
[
"Apache-2.0"
] | null | null | null |
fonte_original_tunning/Tunning_Version2.ipynb
|
marciodelima/case1_santander_engml_geral
|
2f76838b362d5d4c9d4e2f3cf464b5d78e0270d1
|
[
"Apache-2.0"
] | null | null | null |
fonte_original_tunning/Tunning_Version2.ipynb
|
marciodelima/case1_santander_engml_geral
|
2f76838b362d5d4c9d4e2f3cf464b5d78e0270d1
|
[
"Apache-2.0"
] | null | null | null | 32.791557 | 209 | 0.526593 |
[
[
[
"# Case 1 - Santander - Tunning Hiper-Parametros do Modelo Original\n## Marcio de Lima",
"_____no_output_____"
],
[
"<img style=\"float: left;\" src=\"https://guardian.ng/wp-content/uploads/2016/08/Heart-diseases.jpg\" width=\"350px\"/>",
"_____no_output_____"
]
],
[
[
"import warnings\nwarnings.filterwarnings('ignore')",
"_____no_output_____"
],
[
"#%pip install -U scikit-learn",
"_____no_output_____"
],
[
"import numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\nimport seaborn as sns #for plotting\nfrom sklearn.ensemble import RandomForestClassifier #for the model\nfrom sklearn.tree import DecisionTreeClassifier\nfrom sklearn.tree import export_graphviz #plot tree\nfrom sklearn.metrics import roc_curve, auc #for model evaluation\nfrom sklearn.metrics import classification_report #for model evaluation\nfrom sklearn.metrics import confusion_matrix #for model evaluation\nfrom sklearn.model_selection import train_test_split #for data splitting\nimport eli5 #for purmutation importance\nfrom eli5.sklearn import PermutationImportance\nimport shap #for SHAP values\nfrom pdpbox import pdp, info_plots #for partial plots\nnp.random.seed(123) #ensure reproducibility\n\npd.options.mode.chained_assignment = None #hide any pandas warnings\n\n#Marcio de Lima\nfrom sklearn.model_selection import GridSearchCV\nfrom sklearn.model_selection import RandomizedSearchCV\n",
"Using TensorFlow backend.\n"
]
],
[
[
"<a id='section2'></a>",
"_____no_output_____"
],
[
"# The Data",
"_____no_output_____"
]
],
[
[
"dt = pd.read_csv(\"../dados/heart.csv\")",
"_____no_output_____"
],
[
"dt.columns = ['age', 'sex', 'chest_pain_type', 'resting_blood_pressure', 'cholesterol', 'fasting_blood_sugar', 'rest_ecg', 'max_heart_rate_achieved',\n 'exercise_induced_angina', 'st_depression', 'st_slope', 'num_major_vessels', 'thalassemia', 'target']",
"_____no_output_____"
],
[
"dt['sex'][dt['sex'] == 0] = 'female'\ndt['sex'][dt['sex'] == 1] = 'male'\n\ndt['chest_pain_type'][dt['chest_pain_type'] == 1] = 'typical angina'\ndt['chest_pain_type'][dt['chest_pain_type'] == 2] = 'atypical angina'\ndt['chest_pain_type'][dt['chest_pain_type'] == 3] = 'non-anginal pain'\ndt['chest_pain_type'][dt['chest_pain_type'] == 4] = 'asymptomatic'\n\ndt['fasting_blood_sugar'][dt['fasting_blood_sugar'] == 0] = 'lower than 120mg/ml'\ndt['fasting_blood_sugar'][dt['fasting_blood_sugar'] == 1] = 'greater than 120mg/ml'\n\ndt['rest_ecg'][dt['rest_ecg'] == 0] = 'normal'\ndt['rest_ecg'][dt['rest_ecg'] == 1] = 'ST-T wave abnormality'\ndt['rest_ecg'][dt['rest_ecg'] == 2] = 'left ventricular hypertrophy'\n\ndt['exercise_induced_angina'][dt['exercise_induced_angina'] == 0] = 'no'\ndt['exercise_induced_angina'][dt['exercise_induced_angina'] == 1] = 'yes'\n\ndt['st_slope'][dt['st_slope'] == 1] = 'upsloping'\ndt['st_slope'][dt['st_slope'] == 2] = 'flat'\ndt['st_slope'][dt['st_slope'] == 3] = 'downsloping'\n\ndt['thalassemia'][dt['thalassemia'] == 1] = 'normal'\ndt['thalassemia'][dt['thalassemia'] == 2] = 'fixed defect'\ndt['thalassemia'][dt['thalassemia'] == 3] = 'reversable defect'",
"_____no_output_____"
],
[
"dt['sex'] = dt['sex'].astype('object')\ndt['chest_pain_type'] = dt['chest_pain_type'].astype('object')\ndt['fasting_blood_sugar'] = dt['fasting_blood_sugar'].astype('object')\ndt['rest_ecg'] = dt['rest_ecg'].astype('object')\ndt['exercise_induced_angina'] = dt['exercise_induced_angina'].astype('object')\ndt['st_slope'] = dt['st_slope'].astype('object')\ndt['thalassemia'] = dt['thalassemia'].astype('object')",
"_____no_output_____"
],
[
"dt = pd.get_dummies(dt, drop_first=True)",
"_____no_output_____"
]
],
[
[
"# The Model\n\nThe next part fits a random forest model to the data,",
"_____no_output_____"
]
],
[
[
"X_train, X_test, y_train, y_test = train_test_split(dt.drop('target', 1), dt['target'], test_size = .2, random_state=10) #split the data",
"_____no_output_____"
],
[
"model = RandomForestClassifier(max_depth=5)\nmodel.fit(X_train, y_train)",
"_____no_output_____"
],
[
"y_predict = model.predict(X_test)\ny_pred_quant = model.predict_proba(X_test)\ny_pred_bin = model.predict(X_test)",
"_____no_output_____"
],
[
"confusion_matrix = confusion_matrix(y_test, y_pred_bin)\nconfusion_matrix",
"_____no_output_____"
],
[
"total=sum(sum(confusion_matrix))\n\nsensitivity = confusion_matrix[0,0]/(confusion_matrix[0,0]+confusion_matrix[1,0])\nprint('Sensitivity : ', sensitivity )\n\nspecificity = confusion_matrix[1,1]/(confusion_matrix[1,1]+confusion_matrix[0,1])\nprint('Specificity : ', specificity)",
"Sensitivity : 0.8787878787878788\nSpecificity : 0.7857142857142857\n"
],
[
"print('Accuracy of RandomForest Regression Classifier on train set: {:.2f}'.format(model.score(X_train, y_train)*100))\nprint('Accuracy of RandomForest Regression Classifier on test set: {:.2f}'.format(model.score(X_test, y_test)*100))",
"Accuracy of RandomForest Regression Classifier on train set: 92.98\nAccuracy of RandomForest Regression Classifier on test set: 83.61\n"
],
[
"print(classification_report(y_test, model.predict(X_test)))",
" precision recall f1-score support\n\n 0 0.88 0.83 0.85 35\n 1 0.79 0.85 0.81 26\n\n accuracy 0.84 61\n macro avg 0.83 0.84 0.83 61\nweighted avg 0.84 0.84 0.84 61\n\n"
]
],
[
[
"<a id='section4'></a>",
"_____no_output_____"
],
[
"# Tunning Model - Version 1",
"_____no_output_____"
]
],
[
[
"def rodarTunning(X_train, y_train, X_test, y_test, rf_classifier):\n \n param_grid = {'n_estimators': [50, 75, 100, 125, 150, 175],\n 'min_samples_split':[2,4,6,8,10],\n 'min_samples_leaf': [1, 2, 3, 4],\n 'max_depth': [5, 10, 15, 20, 25]}\n\n grid_obj = GridSearchCV(rf_classifier,\n return_train_score=True,\n param_grid=param_grid,\n scoring='roc_auc',\n cv=10)\n\n grid_fit = grid_obj.fit(X_train, y_train)\n rf_opt = grid_fit.best_estimator_\n\n print('='*20)\n print(\"best params: \" + str(grid_obj.best_estimator_))\n print(\"best params: \" + str(grid_obj.best_params_))\n print('best score:', grid_obj.best_score_)\n print('='*20)\n \n print(classification_report(y_test, rf_opt.predict(X_test)))\n\n print('New Accuracy of Model on train set: {:.2f}'.format(rf_opt.score(X_train, y_train)*100))\n print('New Accuracy of Model on test set: {:.2f}'.format(rf_opt.score(X_test, y_test)*100))\n\n return rf_opt",
"_____no_output_____"
],
[
"rf_classifier = RandomForestClassifier(class_weight = \"balanced\", random_state=7)\nrf_opt = rodarTunning(X_train, y_train, X_test, y_test, rf_classifier)",
"====================\nbest params: RandomForestClassifier(class_weight='balanced', max_depth=15,\n min_samples_leaf=2, n_estimators=50, random_state=7)\nbest params: {'max_depth': 15, 'min_samples_leaf': 2, 'min_samples_split': 2, 'n_estimators': 50}\nbest score: 0.925094905094905\n====================\n precision recall f1-score support\n\n 0 0.85 0.83 0.84 35\n 1 0.78 0.81 0.79 26\n\n accuracy 0.82 61\n macro avg 0.82 0.82 0.82 61\nweighted avg 0.82 0.82 0.82 61\n\nNew Accuracy of Model on train set: 97.93\nNew Accuracy of Model on test set: 81.97\n"
]
],
[
[
"# Tunning Model - Version 2",
"_____no_output_____"
],
[
"### Dados com escalas diferentes - Aplicando MinMaxScaler",
"_____no_output_____"
]
],
[
[
"from sklearn.preprocessing import MinMaxScaler\n\nscaler = MinMaxScaler(feature_range=(0, 5))\ndf_HR = dt\nHR_col = list(df_HR.columns)\nHR_col.remove('target')\nfor col in HR_col:\n df_HR[col] = df_HR[col].astype(float)\n df_HR[[col]] = scaler.fit_transform(df_HR[[col]])\ndf_HR['target'] = pd.to_numeric(df_HR['target'], downcast='float')",
"_____no_output_____"
],
[
"X_train_hr, X_test_hr, y_train_hr, y_test_hr = train_test_split(df_HR.drop('target', 1), df_HR['target'], test_size = .2, random_state=10) #split the data\nrf_classifier = RandomForestClassifier(class_weight = \"balanced\", random_state=7)\nrf_opt2 = rodarTunning(X_train_hr, y_train_hr, X_test_hr, y_test_hr, rf_classifier)",
"====================\nbest params: RandomForestClassifier(class_weight='balanced', max_depth=15,\n min_samples_leaf=2, n_estimators=50, random_state=7)\nbest params: {'max_depth': 15, 'min_samples_leaf': 2, 'min_samples_split': 2, 'n_estimators': 50}\nbest score: 0.9256143856143856\n====================\n precision recall f1-score support\n\n 0.0 0.85 0.83 0.84 35\n 1.0 0.78 0.81 0.79 26\n\n accuracy 0.82 61\n macro avg 0.82 0.82 0.82 61\nweighted avg 0.82 0.82 0.82 61\n\nNew Accuracy of Model on train set: 97.93\nNew Accuracy of Model on test set: 81.97\n"
]
],
[
[
"# Tunning Model - Version 3",
"_____no_output_____"
],
[
"## Avaliando outros modelos",
"_____no_output_____"
]
],
[
[
"from sklearn import svm, tree, linear_model, neighbors\nfrom sklearn import naive_bayes, ensemble, discriminant_analysis, gaussian_process\nfrom sklearn.linear_model import LogisticRegression\nfrom sklearn.tree import DecisionTreeClassifier\nfrom sklearn.neighbors import KNeighborsClassifier\nfrom sklearn.discriminant_analysis import LinearDiscriminantAnalysis\nfrom sklearn.naive_bayes import GaussianNB\nfrom sklearn.svm import SVC\nfrom sklearn.neighbors import KNeighborsClassifier\nfrom sklearn.ensemble import RandomForestClassifier\nfrom xgboost import XGBClassifier\nfrom sklearn import feature_selection\nfrom sklearn import model_selection\nfrom sklearn import metrics\n\ndef testarModelos(X_train, X_test, y_train, y_test):\n\n models = []\n models.append(('Logistic Regression', LogisticRegression(solver='liblinear', class_weight='balanced')))\n models.append(('SVM', SVC(gamma='auto')))\n models.append(('KNN', KNeighborsClassifier()))\n models.append(('Decision Tree Classifier', DecisionTreeClassifier()))\n models.append(('Gaussian NB', GaussianNB()))\n models.append(('Xgboost', XGBClassifier(learning_rate=0.02, n_estimators=600, objective='binary:logistic',\n silent=True, nthread=1)))\n models.append(('RandomForestClassifier', RandomForestClassifier(max_depth=5)))\n\n acc_results = []\n auc_results = []\n names = []\n\n col = ['Algorithm', 'ROC AUC Mean', 'ROC AUC STD', \n 'Accuracy Mean', 'Accuracy STD']\n df_results = pd.DataFrame(columns=col)\n i = 0\n\n for name, model in models:\n kfold = model_selection.KFold(\n n_splits=10, shuffle=True) # 10-fold cross-validation\n\n cv_acc_results = model_selection.cross_val_score( # accuracy scoring\n model, X_train, y_train, cv=kfold, scoring='accuracy')\n\n cv_auc_results = model_selection.cross_val_score( # roc_auc scoring\n model, X_train, y_train, cv=kfold, scoring='roc_auc')\n\n acc_results.append(cv_acc_results)\n auc_results.append(cv_auc_results)\n names.append(name)\n df_results.loc[i] = [name,\n round(cv_auc_results.mean()*100, 2),\n round(cv_auc_results.std()*100, 2),\n round(cv_acc_results.mean()*100, 2),\n round(cv_acc_results.std()*100, 2)\n ]\n i += 1\n return df_results.sort_values(by=['ROC AUC Mean'], ascending=False)\n",
"_____no_output_____"
],
[
"#Sem MinMaxScaler\nrf_classifier = RandomForestClassifier(class_weight = \"balanced\", random_state=7)\nrf_opt2 = rodarTunning(X_train, y_train, X_test, y_test, rf_classifier)",
"====================\nbest params: RandomForestClassifier(class_weight='balanced', max_depth=15,\n min_samples_leaf=2, n_estimators=50, random_state=7)\nbest params: {'max_depth': 15, 'min_samples_leaf': 2, 'min_samples_split': 2, 'n_estimators': 50}\nbest score: 0.925094905094905\n====================\n precision recall f1-score support\n\n 0 0.85 0.83 0.84 35\n 1 0.78 0.81 0.79 26\n\n accuracy 0.82 61\n macro avg 0.82 0.82 0.82 61\nweighted avg 0.82 0.82 0.82 61\n\nNew Accuracy of Model on train set: 97.93\nNew Accuracy of Model on test set: 81.97\n"
],
[
"df_results = testarModelos(X_train, X_test, y_train, y_test)\nprint(df_results)",
" Algorithm ROC AUC Mean ROC AUC STD Accuracy Mean \\\n0 Logistic Regression 92.33 3.94 83.95 \n6 RandomForestClassifier 89.03 7.45 84.27 \n5 Xgboost 88.34 5.54 80.60 \n4 Gaussian NB 86.95 6.56 78.50 \n3 Decision Tree Classifier 76.38 8.31 76.03 \n2 KNN 67.81 10.18 62.35 \n1 SVM 57.19 15.65 57.45 \n\n Accuracy STD \n0 7.16 \n6 4.90 \n5 8.48 \n4 10.18 \n3 8.26 \n2 5.94 \n1 9.73 \n"
],
[
"#Com MinMaxScaler\ndf_results = testarModelos(X_train_hr, X_test_hr, y_train_hr, y_test_hr)\nprint(df_results)",
" Algorithm ROC AUC Mean ROC AUC STD Accuracy Mean \\\n6 RandomForestClassifier 91.42 6.03 81.80 \n0 Logistic Regression 90.56 8.54 82.68 \n5 Xgboost 88.64 5.18 82.63 \n4 Gaussian NB 88.32 7.47 81.33 \n1 SVM 86.69 8.03 78.47 \n2 KNN 86.55 5.04 80.55 \n3 Decision Tree Classifier 70.44 7.66 74.35 \n\n Accuracy STD \n6 6.53 \n0 8.35 \n5 7.17 \n4 9.22 \n1 7.00 \n2 7.21 \n3 10.20 \n"
],
[
"X_train, X_test, y_train, y_test = train_test_split(dt.drop('target', 1), dt['target'], test_size = .2, random_state=10) \nrf_classifier = XGBClassifier(learning_rate=0.02, objective='binary:logistic')\nrf_opt3 = rodarTunning(X_train, y_train, X_test, y_test, rf_classifier)",
"====================\nbest params: XGBClassifier(learning_rate=0.02, max_depth=5, min_samples_leaf=1,\n min_samples_split=2, n_estimators=150)\nbest params: {'max_depth': 5, 'min_samples_leaf': 1, 'min_samples_split': 2, 'n_estimators': 150}\nbest score: 0.9123876123876122\n====================\n precision recall f1-score support\n\n 0.0 0.85 0.83 0.84 35\n 1.0 0.78 0.81 0.79 26\n\n accuracy 0.82 61\n macro avg 0.82 0.82 0.82 61\nweighted avg 0.82 0.82 0.82 61\n\nNew Accuracy of Model on train set: 97.93\nNew Accuracy of Model on test set: 81.97\n"
]
],
[
[
"## Tunning 1 demonstrou melhor acurária e maior acertos nas 2 target (0 e 1)\n#### Tivemos um aumento de 3% no Treinamento e o mesmo resultado no Teste, mas pela métrica de matriz de confusão e relatório de classificação o acerto entre as classes foi equalizado, mais genérico. \n#### Não foi muita diferença na aplicação de escala no dataset, desta forma, foi ignorada. \n#### O modelo XGBClassifier aparece como promissor, mas para o case, vamos seguir com a decisão do Data Science (Autor) com o RandomForestClassifier",
"_____no_output_____"
]
],
[
[
"#Save Modelo Tunning Version 1 - Marcio de Lima\nimport pickle\n\nfilename = 'modelo/tunning_model_v2.pkl'\npickle.dump(rf_opt, open(filename, 'wb'))",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
cbec68a27a05d82fe8d7b4ab87e4d9c2d357b527
| 249,144 |
ipynb
|
Jupyter Notebook
|
radiomicsfeatureextractionpipeline/tools/test_mapping.ipynb
|
Maastro-CDS-Imaging-Group/SQLite4Radiomics
|
e3a7afc181eec0fe04c18da00edc3772064e6758
|
[
"Apache-2.0"
] | null | null | null |
radiomicsfeatureextractionpipeline/tools/test_mapping.ipynb
|
Maastro-CDS-Imaging-Group/SQLite4Radiomics
|
e3a7afc181eec0fe04c18da00edc3772064e6758
|
[
"Apache-2.0"
] | 6 |
2021-06-09T19:39:27.000Z
|
2021-09-30T16:41:40.000Z
|
radiomicsfeatureextractionpipeline/tools/test_mapping.ipynb
|
Maastro-CDS-Imaging-Group/SQLite4Radiomics
|
e3a7afc181eec0fe04c18da00edc3772064e6758
|
[
"Apache-2.0"
] | null | null | null | 25.863594 | 182 | 0.412773 |
[
[
[
"# %load IBSI_benchmark_evaluator.py\nimport pandas as pd\nimport argparse\n\n\ndef main(args):\n try:\n pipeline_df = pd.read_csv(args.pipeline_csv_file)\n benchmark_df = pd.read_csv(args.benchmark_csv_file)\n mapping_df = pd.read_csv(args.mapping_csv_file)\n\n except:\n print(\"Error in reading csv files.\")\n exit()\n\n tags_of_interest = []\n\n benchmark_df[\"pyradiomics_tag\"] = benchmark_df[\"tag\"]\n\n\n for f_ibsi, f_pyradiomics in zip(mapping_df[\"IBSIName\"], mapping_df[\"PyradiomicsFeature\"]):\n f_ibsi = f_ibsi.lstrip(\"F\").replace(\".\", \"_\")\n match_condition = benchmark_df['tag'].str.contains(f_ibsi)\n\n benchmark_df['your_result'][match_condition] = pipeline_df[f_pyradiomics].values[0]\n\n benchmark_df['pyradiomics_tag'][match_condition] = f_pyradiomics\n\n tags_of_interest.append(benchmark_df[match_condition & benchmark_df['benchmark_value'].notnull()])\n\n\n matched_df = pd.concat(tags_of_interest)\n\n matched_df[\"difference\"] = (matched_df[\"your_result\"] - matched_df[\"benchmark_value\"]).abs()\n matched_df[\"check\"] = matched_df[\"difference\"] <= matched_df[\"tolerance\"] \n \n matched_df.to_csv(args.save_csv)\n\n\nif __name__ == \"__main__\":\n parser = argparse.ArgumentParser()\n parser.add_argument(\"--pipeline_csv_file\", help=\"Path to the pipeline generated CSV file\")\n parser.add_argument(\"--benchmark_csv_file\", help=\"Path to CSV file provided by IBSI for benchmarking\", default='IBSI-1-configA.csv')\n parser.add_argument(\"--mapping_csv_file\", help=\"Mapping file with correspondences between tags of IBSI and pyradiomics\", default='Pyradiomics2IBSIFeatures.csv')\n parser.add_argument(\"--save_csv\", help=\"Output csv file path\", default=\"out.csv\")\n\n args = parser.parse_args()\n main(args)",
"_____no_output_____"
],
[
"import pandas as pd\n\n# input files\npipeline_csv_file = r\"C:\\Users\\ivan.zhovannik\\OneDrive - Maastro - Clinic\\_PhD_stuff\\Papers\\2021\\SQLite4Radiomics\\_SQLite4Radiomics_benchmarking\\1 CT_configE.csv\"\nbenchmark_csv_file = \"IBSI-1-configE.csv\"\nmapping_csv_file = \"Pyradiomics2IBSIFeatures.csv\"\nsave_csv = \"_test_E.csv\"\n\n\npipeline_df = pd.read_csv(pipeline_csv_file)\nbenchmark_df = pd.read_csv(benchmark_csv_file)\nmapping_df = pd.read_csv(mapping_csv_file)\n\ntags_of_interest = []\n\nbenchmark_df[\"pyradiomics_tag\"] = benchmark_df[\"tag\"]\n\n\nfor f_ibsi, f_pyradiomics in zip(mapping_df[\"IBSIName\"], mapping_df[\"PyradiomicsFeature\"]):\n \n print(f_ibsi)\n f_ibsi = f_ibsi.lstrip(\"F\").replace(\".\", \"_\")\n print(f_ibsi)\n \n match_condition = benchmark_df['tag'].str.contains(f_ibsi)\n print(match_condition.loc[match_condition])\n \n display(benchmark_df['your_result'][match_condition])\n benchmark_df.loc[match_condition, 'your_result'] = pipeline_df[f_pyradiomics].values[0]\n display(benchmark_df['your_result'][match_condition])\n \n print(f_pyradiomics)\n benchmark_df.loc[match_condition, 'pyradiomics_tag'] = f_pyradiomics\n\n tags_of_interest.append(benchmark_df[match_condition & benchmark_df['benchmark_value'].notnull()])\n display(benchmark_df[match_condition & benchmark_df['benchmark_value'].notnull()])\n \n\nmatched_df = pd.concat(tags_of_interest)\n\nmatched_df[\"difference\"] = (matched_df[\"your_result\"] - matched_df[\"benchmark_value\"]).abs()\nmatched_df[\"check\"] = matched_df[\"difference\"] <= matched_df[\"tolerance\"] \n\nmatched_df.to_csv(save_csv)",
"Fmorph.pca.elongation\nmorph_pca_elongation\n74 True\nName: tag, dtype: bool\n"
],
[
"matched_df.loc[matched_df.check ]",
"_____no_output_____"
],
[
"matched_df.check.sum()",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code"
]
] |
cbec6d5653b65a218213b8afba655e494c4dabd7
| 293,679 |
ipynb
|
Jupyter Notebook
|
ez_code/sim_paths.ipynb
|
jngod2011/asset_pricing_code
|
406510d72a26e296444bdc2d728883afe308eb41
|
[
"MIT"
] | 1 |
2020-03-14T15:51:33.000Z
|
2020-03-14T15:51:33.000Z
|
ez_code/sim_paths.ipynb
|
jngod2011/asset_pricing_code
|
406510d72a26e296444bdc2d728883afe308eb41
|
[
"MIT"
] | null | null | null |
ez_code/sim_paths.ipynb
|
jngod2011/asset_pricing_code
|
406510d72a26e296444bdc2d728883afe308eb41
|
[
"MIT"
] | null | null | null | 1,203.602459 | 33,370 | 0.948348 |
[
[
[
"import numpy as np\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"\nfrom by_model import BY",
"_____no_output_____"
],
[
"by = BY()",
"_____no_output_____"
],
[
"z, σ = by.simulate_state(ts_length=1000)\ng_c = by.simulate_consumption_given_state(z, σ)\ng_d = by.simulate_dividends_given_state(z, σ)\nout = g_c, g_d, z, σ\nlabels = \"$g_c$\", \"$g_d$\", \"$z$\", \"$\\sigma$\"",
"_____no_output_____"
],
[
"for x, la in zip(out, labels):\n fig, ax = plt.subplots()\n ax.plot(x, label=la)\n ax.legend()\n plt.show()",
"_____no_output_____"
],
[
"from ssy_model import SSY",
"_____no_output_____"
],
[
"ssy = SSY()",
"_____no_output_____"
],
[
"z, h_z, h_c, h_d = ssy.simulate_state(ts_length=1000)\ng_c = ssy.simulate_consumption_given_state(z, h_z, h_c, h_d)\ng_d = ssy.simulate_dividends_given_state(z, h_z, h_c, h_d)\nout = g_c, g_d, z, h_z, h_c, h_d\nlabels = \"$g_c$\", \"$g_d$\", \"$z$\", \"$h_z$\", \"$h_c$\", \"$h_d$\"",
"_____no_output_____"
],
[
"for x, la in zip(out, labels):\n fig, ax = plt.subplots()\n ax.plot(x, label=la)\n ax.legend()\n plt.show()",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbec87c9d7ecdad97779e6f1cb67f30e35b10151
| 54,801 |
ipynb
|
Jupyter Notebook
|
ADS_FINAL_Project.ipynb
|
hirenpatel27/ADS
|
0a0c42f9045cf48fd6994f1cded8fb3ec11a9478
|
[
"MIT"
] | null | null | null |
ADS_FINAL_Project.ipynb
|
hirenpatel27/ADS
|
0a0c42f9045cf48fd6994f1cded8fb3ec11a9478
|
[
"MIT"
] | null | null | null |
ADS_FINAL_Project.ipynb
|
hirenpatel27/ADS
|
0a0c42f9045cf48fd6994f1cded8fb3ec11a9478
|
[
"MIT"
] | null | null | null | 41.642097 | 393 | 0.593931 |
[
[
[
"# Image Classification using GoogLeNet Architecture from Scratch\n\n#### In this notebook we are trying to make Object Classification using CNN like [GoogleNet](https://arxiv.org/pdf/1409.4842.pdf).\n- We have used **GOOGLE CLOUD Platform** to test and train our model\n- We have also used image augmentation to boost the performance of deep networks.\n- Due to overfitting of the model and getting less accuracy in the [Dataset](http://vision.stanford.edu/aditya86/ImageNetDogs/) of Dog (Subset of Imagenet Dataset) in our CNN model so we also try to other CNN and run on CIFAR-10 and got 68% accuracy in 50 Epochs",
"_____no_output_____"
],
[
"#### Steps to setup GCP(Google Cloud Platform) for Keras and Tensorflow\n\n1. Setup Virtual Machine Instance in GCP using [link](https://cloud.google.com/compute/docs/instances/). Make sure your instance have GPU.\n- Follow all the steps in [link](https://medium.com/google-cloud/running-jupyter-notebooks-on-gpu-on-google-cloud-d44f57d22dbd) to setup Anaconda, Tensorflow and Keras with GPU driver.\n- To import dataset to VM use ssh to your VM run:\n - `gcloud compute scp ~/localdirectory/ example-instance:~/destinationdirectory`\n- Navigate to you jupyter on local browser and make a new notebook.\n",
"_____no_output_____"
],
[
"**Some more library to install to run the notebook:**\n- Install PIL : pip install pillow --> Python Imaging Library which adds image processing capabilities to your python interpreter.\n\n- Install tqdm : pip install tqdm --> tqdm is used to show progress bar\n- Install h5py : pip install h5py --> used to store weights in local",
"_____no_output_____"
]
],
[
[
"#Import library\nimport keras\nfrom keras.datasets import cifar10\nfrom keras.layers import Input\nfrom keras.models import Model\nfrom keras.layers import Dense, Dropout, Flatten, Input, AveragePooling2D, merge\nfrom keras.layers import Conv2D, MaxPooling2D, BatchNormalization,GlobalAveragePooling2D\nfrom keras.layers import Concatenate\nfrom keras.optimizers import SGD\nfrom keras.models import model_from_json\n\n#pre-processing Images\nfrom sklearn.datasets import load_files \nfrom keras.utils import np_utils\nimport numpy as np\nfrom glob import glob\n\n\nfrom keras.preprocessing.image import ImageDataGenerator\n#\nfrom PIL import ImageFile \nImageFile.LOAD_TRUNCATED_IMAGES = True \n\nfrom keras.preprocessing import image \nfrom tqdm import tqdm",
"Using TensorFlow backend.\n"
]
],
[
[
"### Allow the GPU as memory is needed rather than pre-allocate memory\n- You can find more details of tensorflow GPU [here](https://www.tensorflow.org/programmers_guide/tensors)",
"_____no_output_____"
]
],
[
[
"# backend\nimport tensorflow as tf\nfrom keras import backend as k\n\n# Don't pre-allocate memory; allocate as-needed\nconfig = tf.ConfigProto()\nconfig.gpu_options.allow_growth = True\n\n# Create a session with the above options specified.\nk.tensorflow_backend.set_session(tf.Session(config=config))",
"_____no_output_____"
],
[
"# function to load dataset\ndef load_dataset(path):\n #load files from path\n data = load_files(path)\n #takes the filename and put in array\n dog_files = np.array(data['filenames'])\n #one hot encoding\n dog_targets = np_utils.to_categorical(np.array(data['target']), 133)\n return dog_files, dog_targets\n",
"_____no_output_____"
],
[
"train_files, train_targets = load_dataset('Dog/train')\nvalid_files, valid_targets = load_dataset('Dog/valid')\ntest_files, test_targets = load_dataset('Dog/test')",
"_____no_output_____"
],
[
"# Just getting first 5 dogs breads\ndog_names = [item.split('.')[1].rstrip('\\/') for item in sorted(glob(\"Dog/train/*/\"))]\ndog_names[:5]",
"_____no_output_____"
],
[
"\nprint('There are %d total dog categories.' % len(dog_names))\nprint('There are %s total dog images.\\n' % len(np.hstack([train_files, valid_files, test_files])))\nprint('There are %d training dog images.' % len(train_files))\nprint('There are %d validation dog images.' % len(valid_files))\nprint('There are %d test dog images.'% len(test_files))",
"There are 133 total dog categories.\nThere are 8341 total dog images.\n\nThere are 6670 training dog images.\nThere are 835 validation dog images.\nThere are 836 test dog images.\n"
]
],
[
[
"##### This dataset is already split into train, validation and test parts. As the traning set consits of 6670 images, there are only 50 dogs per breed on average.",
"_____no_output_____"
],
[
"#### PreProcess the Data\n- Path_to_tensor is the function that takes the image path, convert into array and return the 4D tensor with shape (1,224,224,3) (batch,height, width, color)\n- paths_to_tensor array of image path return the tensor of image in array ",
"_____no_output_____"
]
],
[
[
"\ndef path_to_tensor(img_path):\n # loads RGB image as PIL.Image.Image type\n img = image.load_img(img_path, target_size=(224, 224))\n # convert PIL.Image.Image type to 3D tensor with shape (224, 224, 3)\n x = image.img_to_array(img)\n # convert 3D tensor to 4D tensor with shape (1, 224, 224, 3) and return 4D tensor\n return np.expand_dims(x, axis=0)\n\ndef paths_to_tensor(img_paths):\n list_of_tensors = [path_to_tensor(img_path) for img_path in tqdm(img_paths)]\n return np.vstack(list_of_tensors)\n",
"_____no_output_____"
]
],
[
[
"Rescale the images by dividing every pixel in every image by 255.",
"_____no_output_____"
]
],
[
[
"from PIL import ImageFile \nImageFile.LOAD_TRUNCATED_IMAGES = True \n\ntrain_tensors = paths_to_tensor(train_files).astype('float32')/255\nvalid_tensors = paths_to_tensor(valid_files).astype('float32')/255\ntest_tensors = paths_to_tensor(test_files).astype('float32')/255",
"100%|██████████| 6670/6670 [01:03<00:00, 104.63it/s]\n100%|██████████| 835/835 [00:07<00:00, 119.19it/s]\n100%|██████████| 836/836 [00:07<00:00, 116.94it/s]\n"
]
],
[
[
"## Image Augmentation\n#### While we have train the CNN found that it was overfitting by huge number where train accuracy was 50% and validation accuracy was only 18% in the 70 epochs. So to reduce the overfitting we try to do image augmentation.\n#### This helps prevent overfitting and helps the model generalize better.",
"_____no_output_____"
]
],
[
[
" from keras.preprocessing.image import ImageDataGenerator",
"_____no_output_____"
],
[
"#this is the augmentation configuration we will use for training\ntrain_datagen = ImageDataGenerator(\n rescale=1./255, # Rescaling factor\n shear_range=0.2, # Shear angle in counter-clockwise direction in degrees\n zoom_range=0.2, # Range for random zoom\n horizontal_flip=True,# Randomly flip inputs horizontally.\n fill_mode='nearest' #fill_mode is the strategy used for filling in newly created pixels,\n ) \nbatch_size=16",
"_____no_output_____"
],
[
"# this is the augmentation configuration we will use for testing\n# only rescaling\ntest_datagen = ImageDataGenerator(rescale=1./255)",
"_____no_output_____"
]
],
[
[
"##### This is a generator that will read pictures found in subfolers of 'dogs/train', and indefinitely generate batches of augmented image data",
"_____no_output_____"
]
],
[
[
"train_generator = train_datagen.flow_from_directory(\n 'Dog/train', # this is the target directory\n target_size=(224, 224), # all images will be resized to 224 x 224\n batch_size=batch_size,\n class_mode='categorical') # since we use categorical value",
"Found 6670 images belonging to 133 classes.\n"
]
],
[
[
"##### This is a similar generator, for validation data",
"_____no_output_____"
]
],
[
[
"validation_generator = test_datagen.flow_from_directory(\n 'Dog/valid',\n target_size=(224, 224), # all images will be resized to 224 x 224\n batch_size=batch_size,\n class_mode='categorical') ",
"Found 835 images belonging to 133 classes.\n"
]
],
[
[
"<center> **GoogLeNet Inception Architecture** \n\n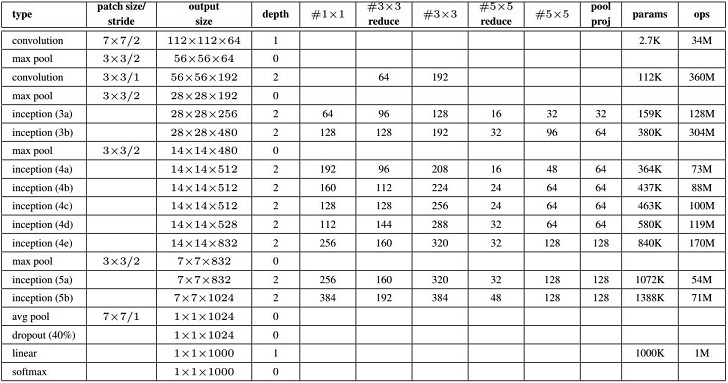",
"_____no_output_____"
],
[
"#### It is generally difficult to decide which architecture will be good for the particular dataset it's most of the time trail and error if you are making CNN from scratch. Pre-train CNN with it's will give more accuracy in less iteration compare to training from scratch because network has to learn from beginning.",
"_____no_output_____"
],
[
"**This notebook will only work on tensorflow not with theano** **Theano use channels as first whereas tensorflow uses channels as last**\n- Lets start with input tensor which will be: ",
"_____no_output_____"
]
],
[
[
"input = Input(shape = (224, 224, 3))",
"_____no_output_____"
],
[
"## So let's start to make CNN for Our dataset which is Dog's dataset which contains 133 classes with total 8341",
"_____no_output_____"
]
],
[
[
"- Starting with CNN first layer from the diagram it would be `convolution` with `7 x 7` patch size and `stride` of `(2,2)` with input image of `224 x 244` followed by `BatchNormalization` for faster learning and higher overall accuracy. If want to know about [this](https://medium.com/deeper-learning/glossary-of-deep-learning-batch-normalisation-8266dcd2fa82) blog has good explanation.",
"_____no_output_____"
]
],
[
[
"x = Conv2D(64,( 7, 7), strides=(2, 2), padding='same',activation='relu')(input)\nx = BatchNormalization()(x) #default axis is 3 is you are using theano it would be 1.",
"_____no_output_____"
]
],
[
[
"- `MaxPooling` with `3 x 3` with strides as `2`",
"_____no_output_____"
]
],
[
[
"x = MaxPooling2D((3, 3), strides=(2, 2), padding='same')(x) ",
"_____no_output_____"
]
],
[
[
"- Next is `convolution` with `3 x 3` with stride 1 it has two convolution 3 x 3 reduce with 64 layers and 3 x 3 192 layers but as our dataset is way small compare to ImageNet we try to reduce the layer from 64 layers to 48 and 192 to 128.",
"_____no_output_____"
]
],
[
[
"x = Conv2D(48,(1,1),strides=(1,1),padding='same',activation='relu')(x)\nx = BatchNormalization()(x)\nx = Conv2D(64,(1,1),strides=(1,1),padding='same',activation='relu')(x)\nx = BatchNormalization()(x)",
"_____no_output_____"
]
],
[
[
"### Inception 3a type in the GoogLeNet Architecture\n- It is couple steps to complete this layers so making function so we can able to reuse it.\n- `1 x 1` 64 convolution layers followed by BatchNormalization\n- `3 x 3` 80 convolution layers where input is as out of `1 x 1` convolution layer followed by BatchNormalization\n- `5 x 5` 16 convolution layers where input is as out of `1 x 1` convolution layer followed by BatchNormalization\n- Last convolution layers is pooling which is 32 convolution layers with `1 x 1`\n- Merge ouptut of `1 x 1` , `3 x 3` and `5 x 5` with respect to last axis \n\n\n So while calling function I would call as add_module(input, 64, 80, 16, 32, 32) ",
"_____no_output_____"
]
],
[
[
"def add_module(input,reduce_1, onex1, threex3, fivex5, pool):\n #print(input.shape)\n \n Conv2D_reduce = Conv2D(reduce_1, (1,1), strides=(2,2), activation='relu', padding='same')(input)\n Conv2D_reduce = BatchNormalization()(Conv2D_reduce)\n #print(Conv2D_reduce.shape)\n \n Conv2D_1_1 = Conv2D(onex1, (1,1), activation='relu', padding='same')(input)\n Conv2D_1_1 = BatchNormalization()(Conv2D_1_1)\n #print(Conv2D_1_1.shape)\n Conv2D_3_3 = Conv2D(threex3, (3,3),strides=(2,2), activation='relu', padding='same')(Conv2D_1_1)\n Conv2D_3_3 = BatchNormalization()(Conv2D_3_3)\n #print(Conv2D_3_3.shape)\n Conv2D_5_5 = Conv2D(fivex5, (5,5),strides=(2,2), activation='relu', padding='same')(Conv2D_1_1)\n Conv2D_5_5 = BatchNormalization()(Conv2D_5_5)\n #print(Conv2D_5_5.shape)\n \n MaxPool2D_3_3 = MaxPooling2D(pool_size=(2,2), strides=(2,2))(input)\n #print(MaxPool2D_3_3.shape)\n Cov2D_Pool = Conv2D(pool, (1,1), activation='relu', padding='same')(MaxPool2D_3_3)\n Cov2D_Pool = BatchNormalization()(Cov2D_Pool)\n #print(Cov2D_Pool.shape)\n \n concat = Concatenate(axis=-1)([Conv2D_reduce,Conv2D_3_3,Conv2D_5_5,Cov2D_Pool])\n #print(concat.shape)\n \n return concat",
"_____no_output_____"
]
],
[
[
"### Inception 3b \n- It is couple steps to complete this layers.\n- `1 x 1` 80 convolution layers followed by BatchNormalization\n- `3 x 3` 16 convolution layers where input is as out of `1 x 1` convolution layer followed by BatchNormalization\n- `5 x 5` 48 convolution layers where input is as out of `1 x 1` convolution layer followed by BatchNormalization\n- Last convolution layers is pooling which is 64 convolution layers with `1 x 1`\n- Merge ouptut of `1 x 1` , `3 x 3` and `5 x 5` with respect to last axis\n\n So while calling function I would call as add_module(input, 48, 80, 16, 48, 64) \n \nAnd than adding maxpooling with `3 x 3` with strides of 3\n### So putting all together\nI am not using more complex architecture because that might overfitting model as per my dataset as shown in diagram because imagenet hase 1000 categorical images with each images have more than 1000 images of each category whereas in our case we have small dataset and for that the whole architecture implementation would overfit the model.\n#### Final layer I am using activation funtion as softmax with dense of 133 (num_classes).",
"_____no_output_____"
]
],
[
[
"input = Input(shape=(224,224,3))\nx = Conv2D(64,( 7, 7), strides=(2, 2), padding='same',activation='relu')(input)\nx = BatchNormalization()(x)\nx = MaxPooling2D((3, 3), strides=(2, 2), padding='same')(x) \nx = Conv2D(48,(1,1),strides=(1,1),padding='same',activation='relu')(x)\nx = BatchNormalization()(x)\nx = Conv2D(64,(1,1),strides=(1,1),padding='same',activation='relu')(x)\nx = BatchNormalization()(x)\nx = MaxPooling2D((3, 3), strides=(2, 2), padding='same')(x)\nx = add_module(x, 64, 80, 16, 32, 32) \n# x = MaxPooling2D((3, 3), strides=(2, 2), padding='same')(x)\nx = add_module(x, 48, 80, 16, 48, 64)\n# x = MaxPooling2D((3, 3), strides=(2, 2), padding='same')(x)\n# x = add_module(x)\n# --- Last Layer --- \n\n\n# Now commes 3 level inception\n\nx = AveragePooling2D((7, 7), strides=(1, 1), padding='valid')(x) \nx = Dropout(0.5)(x)\nx = GlobalAveragePooling2D()(x)\nx = Dense(1024, activation='linear')(x)\nOutput = Dense(133, activation='softmax')(x)",
"_____no_output_____"
]
],
[
[
"# Lets make the model ",
"_____no_output_____"
]
],
[
[
"model = Model(inputs= input, outputs = Output)",
"_____no_output_____"
],
[
"model.summary()",
"__________________________________________________________________________________________________\nLayer (type) Output Shape Param # Connected to \n==================================================================================================\ninput_2 (InputLayer) (None, 224, 224, 3) 0 \n__________________________________________________________________________________________________\nconv2d_2 (Conv2D) (None, 112, 112, 64) 9472 input_2[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_2 (BatchNor (None, 112, 112, 64) 256 conv2d_2[0][0] \n__________________________________________________________________________________________________\nmax_pooling2d_2 (MaxPooling2D) (None, 56, 56, 64) 0 batch_normalization_2[0][0] \n__________________________________________________________________________________________________\nconv2d_3 (Conv2D) (None, 56, 56, 48) 3120 max_pooling2d_2[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_3 (BatchNor (None, 56, 56, 48) 192 conv2d_3[0][0] \n__________________________________________________________________________________________________\nconv2d_4 (Conv2D) (None, 56, 56, 64) 3136 batch_normalization_3[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_4 (BatchNor (None, 56, 56, 64) 256 conv2d_4[0][0] \n__________________________________________________________________________________________________\nmax_pooling2d_3 (MaxPooling2D) (None, 28, 28, 64) 0 batch_normalization_4[0][0] \n__________________________________________________________________________________________________\nconv2d_6 (Conv2D) (None, 28, 28, 80) 5200 max_pooling2d_3[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_6 (BatchNor (None, 28, 28, 80) 320 conv2d_6[0][0] \n__________________________________________________________________________________________________\nmax_pooling2d_4 (MaxPooling2D) (None, 14, 14, 64) 0 max_pooling2d_3[0][0] \n__________________________________________________________________________________________________\nconv2d_5 (Conv2D) (None, 14, 14, 64) 4160 max_pooling2d_3[0][0] \n__________________________________________________________________________________________________\nconv2d_7 (Conv2D) (None, 14, 14, 16) 11536 batch_normalization_6[0][0] \n__________________________________________________________________________________________________\nconv2d_8 (Conv2D) (None, 14, 14, 32) 64032 batch_normalization_6[0][0] \n__________________________________________________________________________________________________\nconv2d_9 (Conv2D) (None, 14, 14, 32) 2080 max_pooling2d_4[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_5 (BatchNor (None, 14, 14, 64) 256 conv2d_5[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_7 (BatchNor (None, 14, 14, 16) 64 conv2d_7[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_8 (BatchNor (None, 14, 14, 32) 128 conv2d_8[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_9 (BatchNor (None, 14, 14, 32) 128 conv2d_9[0][0] \n__________________________________________________________________________________________________\nconcatenate_1 (Concatenate) (None, 14, 14, 144) 0 batch_normalization_5[0][0] \n batch_normalization_7[0][0] \n batch_normalization_8[0][0] \n batch_normalization_9[0][0] \n__________________________________________________________________________________________________\nconv2d_11 (Conv2D) (None, 14, 14, 80) 11600 concatenate_1[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_11 (BatchNo (None, 14, 14, 80) 320 conv2d_11[0][0] \n__________________________________________________________________________________________________\nmax_pooling2d_5 (MaxPooling2D) (None, 7, 7, 144) 0 concatenate_1[0][0] \n__________________________________________________________________________________________________\nconv2d_10 (Conv2D) (None, 7, 7, 48) 6960 concatenate_1[0][0] \n__________________________________________________________________________________________________\nconv2d_12 (Conv2D) (None, 7, 7, 16) 11536 batch_normalization_11[0][0] \n__________________________________________________________________________________________________\nconv2d_13 (Conv2D) (None, 7, 7, 48) 96048 batch_normalization_11[0][0] \n__________________________________________________________________________________________________\nconv2d_14 (Conv2D) (None, 7, 7, 64) 9280 max_pooling2d_5[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_10 (BatchNo (None, 7, 7, 48) 192 conv2d_10[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_12 (BatchNo (None, 7, 7, 16) 64 conv2d_12[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_13 (BatchNo (None, 7, 7, 48) 192 conv2d_13[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_14 (BatchNo (None, 7, 7, 64) 256 conv2d_14[0][0] \n__________________________________________________________________________________________________\nconcatenate_2 (Concatenate) (None, 7, 7, 176) 0 batch_normalization_10[0][0] \n batch_normalization_12[0][0] \n batch_normalization_13[0][0] \n batch_normalization_14[0][0] \n__________________________________________________________________________________________________\naverage_pooling2d_1 (AveragePoo (None, 1, 1, 176) 0 concatenate_2[0][0] \n__________________________________________________________________________________________________\ndropout_1 (Dropout) (None, 1, 1, 176) 0 average_pooling2d_1[0][0] \n__________________________________________________________________________________________________\nglobal_average_pooling2d_1 (Glo (None, 176) 0 dropout_1[0][0] \n__________________________________________________________________________________________________\ndense_1 (Dense) (None, 1024) 181248 global_average_pooling2d_1[0][0] \n__________________________________________________________________________________________________\ndense_2 (Dense) (None, 133) 136325 dense_1[0][0] \n==================================================================================================\nTotal params: 558,357\nTrainable params: 557,045\nNon-trainable params: 1,312\n__________________________________________________________________________________________________\n"
]
],
[
[
"#### Traning Starts\n We have setup VM with two GPU attach to instance so we are going to use parallel model of gpu",
"_____no_output_____"
]
],
[
[
"from keras.utils import multi_gpu_model\n\nparallel_model = multi_gpu_model(model, gpus=2)",
"_____no_output_____"
]
],
[
[
"### Optimizers selecting is very important to get the model good accuracy as per the paper I am using SGD optimizers as it gives the best result incase of the imagenet.",
"_____no_output_____"
]
],
[
[
"parallel_model.compile(optimizer=SGD(lr=0.0001, momentum=0.9), loss='categorical_crossentropy',metrics=[\"accuracy\"])",
"_____no_output_____"
]
],
[
[
"#### First try to run for 20 epochs it takes ~ 145s for 2 GPUs K-80 tesla with 16 memory and where enresult loss_function for training is 3.8295 and for validation is 4.2274.\n#### Run only 20 epochs to check the model is working.\n#### Still running on 40 more epochs but got validation_accuracy of 12% and training_accuracy as 20%\n#### Reasons for running less epochs to check if model is overfitting or not. ",
"_____no_output_____"
],
[
"#### Commented out because I don't want to run my training by mistake in this file because it takes forever and will not able to do anything else in this file.\n",
"_____no_output_____"
]
],
[
[
"parallel_model.fit_generator(\n train_generator,\n steps_per_epoch=6670 // batch_size,\n epochs=20,\n validation_data=validation_generator,\n validation_steps=835 // batch_size)\nmodel.save_weights('testing_fina1.h5')",
"_____no_output_____"
]
],
[
[
"#### Save model in Json Format",
"_____no_output_____"
]
],
[
[
"model_json = model.to_json()\nwith open(\"final_ model.json\", \"w\") as json_file:\n json_file.write(model_json)",
"_____no_output_____"
]
],
[
[
"#### Load model from json with its weight.",
"_____no_output_____"
]
],
[
[
"from keras.models import model_from_json\njson_file = open('final_ model.json', 'r')\nloaded_model_json = json_file.read()\njson_file.close()\nloaded_model = model_from_json(loaded_model_json)\nloaded_model.load_weights(\"testing_fina17.h5\")",
"_____no_output_____"
]
],
[
[
"#### Don't forget to compile the model before using it or else it will give error",
"_____no_output_____"
]
],
[
[
"parallel_model = multi_gpu_model(loaded_model, gpus=2)\nloaded_model.compile(optimizer=SGD(lr=0.0001, momentum=0.9), loss='categorical_crossentropy',metrics=[\"accuracy\"])",
"_____no_output_____"
]
],
[
[
"### Create a generator to get the accuracy and from the model you trained",
"_____no_output_____"
]
],
[
[
"generator = train_datagen.flow_from_directory(\n 'Dog/train',\n target_size=(224, 224),\n batch_size=batch_size,\n class_mode=None, # this means our generator will only yield batches of data, no labels\n shuffle=False) ",
"Found 6670 images belonging to 133 classes.\n"
],
[
"score = loaded_model.evaluate_generator(validation_generator, 800/16, workers=12)\n\nscores = loaded_model.predict_generator(validation_generator, 800/16, workers=12)\n",
"_____no_output_____"
],
[
"correct=0\nfor i, n in enumerate(validation_generator.filenames):\n if \"Affenpinscher\" in n and scores[i][0] <= 0.5:\n correct += 1\n\nprint(\"Correct:\", correct, \" Total: \", len(validation_generator.filenames))\nprint(\"Loss: \", score[0], \"Accuracy: \", score[1]*100,\"%\")",
"Correct: 8 Total: 835\nLoss: 3.401902123070753 Accuracy: 16.95965176890109 %\n"
]
],
[
[
"#### This will give you the accuracy of the dog bread we search as `_Affenpinscher_`",
"_____no_output_____"
],
[
"# Trying CIFAR-10 for GoogleNet\n- As the image size is small in CIFAR-10 i.e. 32x32 we are using one layer as mention in figure.\n\n",
"_____no_output_____"
]
],
[
[
"# Hyperparameters\nbatch_size = 128\nnum_classes = 10\nepochs = 50",
"_____no_output_____"
],
[
"from keras.datasets import cifar10\n# Load CIFAR10 Data\n(x_train, y_train), (x_test, y_test) = cifar10.load_data()\nimg_height, img_width, channel = x_train.shape[1],x_train.shape[2],x_train.shape[3]\n\n# convert to one hot encoing \ny_train = keras.utils.to_categorical(y_train, num_classes)\ny_test = keras.utils.to_categorical(y_test, num_classes)",
"_____no_output_____"
],
[
"input = Input(shape=(img_height, img_width, channel,))\n\nConv2D_1 = Conv2D(64, (3,3), activation='relu', padding='same')(input)\nMaxPool2D_1 = MaxPooling2D(pool_size=(2, 2), strides=(2,2))(Conv2D_1)\nBatchNorm_1 = BatchNormalization()(MaxPool2D_1)\n\nModule_1 = add_module(BatchNorm_1, 16, 16, 16, 16,16)\nModule_1 = add_module(Module_1,16, 16, 16, 16,16)\n\nOutput = Flatten()(Module_1)\nOutput = Dense(num_classes, activation='softmax')(Output)",
"_____no_output_____"
],
[
"model = Model(inputs=[input], outputs=[Output])\nmodel.summary()",
"__________________________________________________________________________________________________\nLayer (type) Output Shape Param # Connected to \n==================================================================================================\ninput_11 (InputLayer) (None, 32, 32, 3) 0 \n__________________________________________________________________________________________________\nconv2d_79 (Conv2D) (None, 32, 32, 64) 1792 input_11[0][0] \n__________________________________________________________________________________________________\nmax_pooling2d_28 (MaxPooling2D) (None, 16, 16, 64) 0 conv2d_79[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_71 (BatchNo (None, 16, 16, 64) 256 max_pooling2d_28[0][0] \n__________________________________________________________________________________________________\nconv2d_81 (Conv2D) (None, 16, 16, 16) 1040 batch_normalization_71[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_73 (BatchNo (None, 16, 16, 16) 64 conv2d_81[0][0] \n__________________________________________________________________________________________________\nconv2d_80 (Conv2D) (None, 8, 8, 16) 1040 batch_normalization_71[0][0] \n__________________________________________________________________________________________________\nconv2d_82 (Conv2D) (None, 8, 8, 16) 2320 batch_normalization_73[0][0] \n__________________________________________________________________________________________________\nconv2d_83 (Conv2D) (None, 8, 8, 16) 6416 batch_normalization_73[0][0] \n__________________________________________________________________________________________________\nmax_pooling2d_29 (MaxPooling2D) (None, 8, 8, 64) 0 batch_normalization_71[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_72 (BatchNo (None, 8, 8, 16) 64 conv2d_80[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_74 (BatchNo (None, 8, 8, 16) 64 conv2d_82[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_75 (BatchNo (None, 8, 8, 16) 64 conv2d_83[0][0] \n__________________________________________________________________________________________________\nconv2d_84 (Conv2D) (None, 8, 8, 16) 1040 max_pooling2d_29[0][0] \n__________________________________________________________________________________________________\nconcatenate_11 (Concatenate) (None, 8, 8, 64) 0 batch_normalization_72[0][0] \n batch_normalization_74[0][0] \n batch_normalization_75[0][0] \n conv2d_84[0][0] \n__________________________________________________________________________________________________\nconv2d_86 (Conv2D) (None, 8, 8, 16) 1040 concatenate_11[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_77 (BatchNo (None, 8, 8, 16) 64 conv2d_86[0][0] \n__________________________________________________________________________________________________\nconv2d_85 (Conv2D) (None, 4, 4, 16) 1040 concatenate_11[0][0] \n__________________________________________________________________________________________________\nconv2d_87 (Conv2D) (None, 4, 4, 16) 2320 batch_normalization_77[0][0] \n__________________________________________________________________________________________________\nconv2d_88 (Conv2D) (None, 4, 4, 16) 6416 batch_normalization_77[0][0] \n__________________________________________________________________________________________________\nmax_pooling2d_30 (MaxPooling2D) (None, 4, 4, 64) 0 concatenate_11[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_76 (BatchNo (None, 4, 4, 16) 64 conv2d_85[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_78 (BatchNo (None, 4, 4, 16) 64 conv2d_87[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_79 (BatchNo (None, 4, 4, 16) 64 conv2d_88[0][0] \n__________________________________________________________________________________________________\nconv2d_89 (Conv2D) (None, 4, 4, 16) 1040 max_pooling2d_30[0][0] \n__________________________________________________________________________________________________\nconcatenate_12 (Concatenate) (None, 4, 4, 64) 0 batch_normalization_76[0][0] \n batch_normalization_78[0][0] \n batch_normalization_79[0][0] \n conv2d_89[0][0] \n__________________________________________________________________________________________________\nflatten_2 (Flatten) (None, 1024) 0 concatenate_12[0][0] \n__________________________________________________________________________________________________\ndense_4 (Dense) (None, 10) 10250 flatten_2[0][0] \n==================================================================================================\nTotal params: 36,522\nTrainable params: 36,138\nNon-trainable params: 384\n__________________________________________________________________________________________________\n"
],
[
"parallel_model = multi_gpu_model(model, gpus=2)\nRMsprop=RMSprop(lr=0.0001, rho=0.9, epsilon=None, decay=0.0)\nparallel_model.compile(loss='categorical_crossentropy', optimizer=RMsprop, metrics=['accuracy'])",
"_____no_output_____"
],
[
"parallel_model.fit(x_train, y_train, epochs=epochs, validation_data=(x_test, y_test))",
"Train on 50000 samples, validate on 10000 samples\nEpoch 1/50\n50000/50000 [==============================] - 52s 1ms/step - loss: 1.7030 - acc: 0.4012 - val_loss: 1.5079 - val_acc: 0.4636\nEpoch 2/50\n50000/50000 [==============================] - 49s 988us/step - loss: 1.4144 - acc: 0.4974 - val_loss: 1.3259 - val_acc: 0.5272\nEpoch 3/50\n50000/50000 [==============================] - 50s 992us/step - loss: 1.2600 - acc: 0.5505 - val_loss: 1.2323 - val_acc: 0.5544\nEpoch 4/50\n50000/50000 [==============================] - 50s 991us/step - loss: 1.1661 - acc: 0.5866 - val_loss: 1.2349 - val_acc: 0.5603\nEpoch 5/50\n50000/50000 [==============================] - 50s 991us/step - loss: 1.0984 - acc: 0.6126 - val_loss: 1.1461 - val_acc: 0.5873\nEpoch 6/50\n50000/50000 [==============================] - 50s 991us/step - loss: 1.0439 - acc: 0.6332 - val_loss: 1.1066 - val_acc: 0.6004\nEpoch 7/50\n50000/50000 [==============================] - 49s 989us/step - loss: 0.9957 - acc: 0.6478 - val_loss: 1.0820 - val_acc: 0.6116\nEpoch 8/50\n50000/50000 [==============================] - 49s 988us/step - loss: 0.9565 - acc: 0.6640 - val_loss: 1.0496 - val_acc: 0.6222\nEpoch 9/50\n50000/50000 [==============================] - 50s 990us/step - loss: 0.9220 - acc: 0.6759 - val_loss: 1.0571 - val_acc: 0.6264\nEpoch 10/50\n50000/50000 [==============================] - 49s 989us/step - loss: 0.8907 - acc: 0.6888 - val_loss: 1.0301 - val_acc: 0.6337\nEpoch 11/50\n50000/50000 [==============================] - 49s 980us/step - loss: 0.8649 - acc: 0.6984 - val_loss: 1.0468 - val_acc: 0.6350\nEpoch 12/50\n50000/50000 [==============================] - 48s 963us/step - loss: 0.8382 - acc: 0.7060 - val_loss: 1.0742 - val_acc: 0.6246\nEpoch 13/50\n50000/50000 [==============================] - 49s 973us/step - loss: 0.8163 - acc: 0.7177 - val_loss: 0.9941 - val_acc: 0.6518\nEpoch 14/50\n50000/50000 [==============================] - 49s 983us/step - loss: 0.7951 - acc: 0.7236 - val_loss: 0.9804 - val_acc: 0.6591\nEpoch 15/50\n50000/50000 [==============================] - 49s 987us/step - loss: 0.7775 - acc: 0.7300 - val_loss: 1.0012 - val_acc: 0.6540\nEpoch 16/50\n50000/50000 [==============================] - 48s 967us/step - loss: 0.7636 - acc: 0.7355 - val_loss: 0.9677 - val_acc: 0.6620\nEpoch 17/50\n50000/50000 [==============================] - 48s 960us/step - loss: 0.7456 - acc: 0.7421 - val_loss: 1.0059 - val_acc: 0.6613\nEpoch 18/50\n 9952/50000 [====>.........................] - ETA: 35s - loss: 0.7262 - acc: 0.7468"
],
[
"# This I think stop because I keep running my VM and sleep my laptop so it didnot autosave.",
"_____no_output_____"
],
[
"scores = model.evaluate(x_test, y_test, verbose=0)\nprint(\"Accuracy: %.2f%%\" % (scores[1]*100))",
"Accuracy: 69.57%\n"
]
],
[
[
"###### Challenges we face doing this project so might help to others who want to do similar type of Project :\n\n- Make sure you have good computation power with atleast two GPU.\n\n- Trying to run model in parallel so it takes less time to train and test.\n\n- Start with simple architecture try to run 30-40 echos first and check the model is overfitting or not If it is overfitting than you don't need to break the trainning. \n\n- Try to take best weight in every epochs I know it kind of tricky in parallel model but you can do this by following the links \n\n- Use jupyter notebook in background using nohup the documentation for that in this [link](https://hackernoon.com/aws-ec2-part-4-starting-a-jupyter-ipython-notebook-server-on-aws-549d87a55ba9)\n\n- If you to try notebook which contain code of THEANO you can do it in 3 easy steps:\n 1. First [install](http://deeplearning.net/software/theano/install.html) theano with GPU.\n \n 2. `nano ~/.keras/keras.json ` Change following:\n \n {\n \"image_dim_ordering\": \"th\",\n \"backend\": \"theano\",\n \"image_data_format\": \"channels_first\"\n } \n \n - Restart the jupyter notebook\n- My model was overfitting and I was not able to figureout what it was overfitting so first thing I tried is Image Augmentation , than change the learning rate than change the layers and than so on. It's I guess **trail and error** methods and In my case each epochs takes around 160-180s with two K-80 tesla GPU it was time cosuming but good way to learn. Shoud have patience.\n \n ",
"_____no_output_____"
],
[
"The content of this project itself is licensed under the, and the underlying source code used to format and display that content is licensed under the [MIT LICENSE](https://github.com/hirenpatel27/ADS/blob/master/LICENSE)",
"_____no_output_____"
],
[
"## Citations\n\n<a id='google-net'>\n[1] Alex Krizhevsky, Ilya Sutskever, and Geoffrey E. Hinton. \"ImageNet Classification with Deep Convolutional Neural Networks.\" NIPS 2012\n<br>\n\n<a id='inception-v1-paper'>\n[2] Christian Szegedy, Wei Liu, Yangqing Jia, Pierre Sermanet, Scott Reed,\nDragomir Anguelov, Dumitru Erhan, Andrew Rabinovich.\n\"Going Deeper with Convolutions.\" CVPR 2015.\n<br>\n\n<a id='vgg-paper'>\n[3] Karen Simonyan and Andrew Zisserman. \"Very Deep Convolutional Networks for Large-Scale Image Recognition.\" ICLR 2015\n<br>\n\n<a id='resnet-cvpr'>\n[4] Kaiming He, Xiangyu Zhang, Shaoqing Ren, and Jian Sun. \"Deep Residual Learning for Image Recognition.\" CVPR 2016.\n<br>\n\n<a id='resnet-eccv'>\n[5] Kaiming He, Xiangyu Zhang, Shaoqing Ren, and Jian Sun. \"Identity Mappings in Deep Residual Networks.\" ECCV 2016.",
"_____no_output_____"
],
[
"## References\n\n[1] [GoogleNet in Keras](http://joelouismarino.github.io/blog_posts/blog_googlenet_keras.html) for understanding of GoogleNet Architecture.\n\n[2] [Keras Documentation](https://keras.io/) for how to use Keras\n\n[3] [Convolution Neural Networks for Visual Recognition](http://cs231n.github.io/convolutional-networks/) for understanding how CNN works\n\n[4] [How convolution neural network Works](https://www.youtube.com/watch?v=FmpDIaiMIeA&t=634s)\n\n[5] [Dog breed classification with Keras](http://machinememos.com/python/keras/artificial%20intelligence/machine%20learning/transfer%20learning/dog%20breed/neural%20networks/convolutional%20neural%20network/tensorflow/image%20classification/imagenet/2017/07/11/dog-breed-image-classification.html)\n\n[6] Keras blog [Image Augmentation](https://blog.keras.io/building-powerful-image-classification-models-using-very-little-data.html)\n\n[7] [Image Datagenertor-Methods](https://keras.io/preprocessing/image/#imagedatagenerator-methods)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
cbec99757a0ba705c30e139f2bfb338b270d3362
| 23,860 |
ipynb
|
Jupyter Notebook
|
notebook.ipynb
|
MLVPRASAD/Analyze-Your-Runkeeper-Fitness-Data
|
7baaa4fcdc0f03bebcda7c645a05e87777083d02
|
[
"MIT"
] | null | null | null |
notebook.ipynb
|
MLVPRASAD/Analyze-Your-Runkeeper-Fitness-Data
|
7baaa4fcdc0f03bebcda7c645a05e87777083d02
|
[
"MIT"
] | null | null | null |
notebook.ipynb
|
MLVPRASAD/Analyze-Your-Runkeeper-Fitness-Data
|
7baaa4fcdc0f03bebcda7c645a05e87777083d02
|
[
"MIT"
] | null | null | null | 23,860 | 23,860 | 0.666806 |
[
[
[
"## 1. Obtain and review raw data\n<p>One day, my old running friend and I were chatting about our running styles, training habits, and achievements, when I suddenly realized that I could take an in-depth analytical look at my training. I have been using a popular GPS fitness tracker called <a href=\"https://runkeeper.com/\">Runkeeper</a> for years and decided it was time to analyze my running data to see how I was doing.</p>\n<p>Since 2012, I've been using the Runkeeper app, and it's great. One key feature: its excellent data export. Anyone who has a smartphone can download the app and analyze their data like we will in this notebook.</p>\n<p><img src=\"https://assets.datacamp.com/production/project_727/img/runner_in_blue.jpg\" alt=\"Runner in blue\" title=\"Explore world, explore your data!\"></p>\n<p>After logging your run, the first step is to export the data from Runkeeper (which I've done already). Then import the data and start exploring to find potential problems. After that, create data cleaning strategies to fix the issues. Finally, analyze and visualize the clean time-series data.</p>\n<p>I exported seven years worth of my training data, from 2012 through 2018. The data is a CSV file where each row is a single training activity. Let's load and inspect it.</p>",
"_____no_output_____"
]
],
[
[
"# Import pandas\nimport pandas as pd\n\n# Define file containing dataset\nrunkeeper_file = 'datasets/cardioActivities.csv'\n\n# Create DataFrame with parse_dates and index_col parameters \ndf_activities = pd.read_csv(runkeeper_file, parse_dates=True, index_col='Date')\n\n# First look at exported data: select sample of 3 random rows \ndisplay(df_activities.sample(3))\n\n# Print DataFrame summary\ndf_activities.info()",
"_____no_output_____"
]
],
[
[
"## 2. Data preprocessing\n<p>Lucky for us, the column names Runkeeper provides are informative, and we don't need to rename any columns.</p>\n<p>But, we do notice missing values using the <code>info()</code> method. What are the reasons for these missing values? It depends. Some heart rate information is missing because I didn't always use a cardio sensor. In the case of the <code>Notes</code> column, it is an optional field that I sometimes left blank. Also, I only used the <code>Route Name</code> column once, and never used the <code>Friend's Tagged</code> column.</p>\n<p>We'll fill in missing values in the heart rate column to avoid misleading results later, but right now, our first data preprocessing steps will be to:</p>\n<ul>\n<li>Remove columns not useful for our analysis.</li>\n<li>Replace the \"Other\" activity type to \"Unicycling\" because that was always the \"Other\" activity.</li>\n<li>Count missing values.</li>\n</ul>",
"_____no_output_____"
]
],
[
[
"# Define list of columns to be deleted\ncols_to_drop = ['Friend\\'s Tagged','Route Name','GPX File','Activity Id','Calories Burned', 'Notes']\n\n# Delete unnecessary columns\ndf_activities.drop(cols_to_drop, axis=1, inplace=True)\n\n# Count types of training activities\ndisplay(df_activities['Type'].value_counts())\n\n# Rename 'Other' type to 'Unicycling'\ndf_activities['Type'] = df_activities['Type'].str.replace('Other', 'Unicycling')\n\n# Count missing values for each column\ndf_activities.isnull().sum()",
"_____no_output_____"
]
],
[
[
"## 3. Dealing with missing values\n<p>As we can see from the last output, there are 214 missing entries for my average heart rate.</p>\n<p>We can't go back in time to get those data, but we can fill in the missing values with an average value. This process is called <em>mean imputation</em>. When imputing the mean to fill in missing data, we need to consider that the average heart rate varies for different activities (e.g., walking vs. running). We'll filter the DataFrames by activity type (<code>Type</code>) and calculate each activity's mean heart rate, then fill in the missing values with those means.</p>",
"_____no_output_____"
]
],
[
[
"# Calculate sample means for heart rate for each training activity type \navg_hr_run = df_activities[df_activities['Type'] == 'Running']['Average Heart Rate (bpm)'].mean()\navg_hr_cycle = df_activities[df_activities['Type'] == 'Cycling']['Average Heart Rate (bpm)'].mean()\n\n# Split whole DataFrame into several, specific for different activities\ndf_run = df_activities[df_activities['Type'] == 'Running'].copy()\ndf_walk = df_activities[df_activities['Type'] == 'Walking'].copy()\ndf_cycle = df_activities[df_activities['Type'] == 'Cycling'].copy()\n\n# Filling missing values with counted means \ndf_walk['Average Heart Rate (bpm)'].fillna(110, inplace=True)\ndf_run['Average Heart Rate (bpm)'].fillna(int(avg_hr_run), inplace=True)\ndf_cycle['Average Heart Rate (bpm)'].fillna(int(avg_hr_cycle), inplace=True)\n\n# Count missing values for each column in running data\ndf_run.isnull().sum()",
"_____no_output_____"
]
],
[
[
"## 4. Plot running data\n<p>Now we can create our first plot! As we found earlier, most of the activities in my data were running (459 of them to be exact). There are only 29, 18, and two instances for cycling, walking, and unicycling, respectively. So for now, let's focus on plotting the different running metrics.</p>\n<p>An excellent first visualization is a figure with four subplots, one for each running metric (each numerical column). Each subplot will have a different y-axis, which is explained in each legend. The x-axis, <code>Date</code>, is shared among all subplots.</p>",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\n\n# Import matplotlib, set style and ignore warning\nimport matplotlib.pyplot as plt\n%matplotlib inline\nimport warnings\nplt.style.use('ggplot')\nwarnings.filterwarnings(\n action='ignore', module='matplotlib.figure', category=UserWarning,\n message=('This figure includes Axes that are not compatible with tight_layout, so results might be incorrect.')\n)\n\n# Prepare data subsetting period from 2013 till 2018\nruns_subset_2013_2018 = df_run.loc['20190101':'20130101']\n\n# Create, plot and customize in one step\nruns_subset_2013_2018.plot(subplots=True,\n sharex=False,\n figsize=(12,16),\n linestyle='none',\n marker='o',\n markersize=3,\n )\n\n# Show plot\nplt.show()",
"_____no_output_____"
]
],
[
[
"## 5. Running statistics\n<p>No doubt, running helps people stay mentally and physically healthy and productive at any age. And it is great fun! When runners talk to each other about their hobby, we not only discuss our results, but we also discuss different training strategies. </p>\n<p>You'll know you're with a group of runners if you commonly hear questions like:</p>\n<ul>\n<li>What is your average distance?</li>\n<li>How fast do you run?</li>\n<li>Do you measure your heart rate?</li>\n<li>How often do you train?</li>\n</ul>\n<p>Let's find the answers to these questions in my data. If you look back at plots in Task 4, you can see the answer to, <em>Do you measure your heart rate?</em> Before 2015: no. To look at the averages, let's only use the data from 2015 through 2018.</p>\n<p>In pandas, the <code>resample()</code> method is similar to the <code>groupby()</code> method - with <code>resample()</code> you group by a specific time span. We'll use <code>resample()</code> to group the time series data by a sampling period and apply several methods to each sampling period. In our case, we'll resample annually and weekly.</p>",
"_____no_output_____"
]
],
[
[
"# Prepare running data for the last 4 years\nruns_subset_2015_2018 = df_run.loc['20190101':'20150101']\n\n# Calculate annual statistics\nprint('How my average run looks in last 4 years:')\ndisplay(runs_subset_2015_2018.resample('A').mean())\n\n# Calculate weekly statistics\nprint('Weekly averages of last 4 years:')\ndisplay(runs_subset_2015_2018.resample('W').mean().mean())\n\n# Mean weekly counts\nweekly_counts_average = runs_subset_2015_2018['Distance (km)'].resample('W').count().mean()\nprint('How many trainings per week I had on average:', weekly_counts_average)",
"_____no_output_____"
]
],
[
[
"## 6. Visualization with averages\n<p>Let's plot the long term averages of my distance run and my heart rate with their raw data to visually compare the averages to each training session. Again, we'll use the data from 2015 through 2018.</p>\n<p>In this task, we will use <code>matplotlib</code> functionality for plot creation and customization.</p>",
"_____no_output_____"
]
],
[
[
"# Prepare data\nruns_distance = runs_subset_2015_2018['Distance (km)']\nruns_hr = runs_subset_2015_2018['Average Heart Rate (bpm)']\n\n# Create plot\nfig, (ax1, ax2) = plt.subplots(2, sharex=True, figsize=(12, 8))\n\n# Plot and customize first subplot\nruns_distance.plot(ax=ax1)\nax1.set(ylabel='Distance (km)', title='Historical data with averages')\nax1.axhline(runs_distance.mean(), color='blue', linewidth=1, linestyle='-.')\n\n# Plot and customize second subplot\nruns_hr.plot(ax=ax2, color='gray')\nax2.set(xlabel='Date', ylabel='Average Heart Rate (bpm)')\nax2.axhline(runs_hr.mean(), color='blue', linewidth=1, linestyle='-.')\n\n# Show plot\nplt.show()",
"_____no_output_____"
]
],
[
[
"## 7. Did I reach my goals?\n<p>To motivate myself to run regularly, I set a target goal of running 1000 km per year. Let's visualize my annual running distance (km) from 2013 through 2018 to see if I reached my goal each year. Only stars in the green region indicate success.</p>",
"_____no_output_____"
]
],
[
[
"# Prepare data\ndf_run_dist_annual = df_run.sort_index()['20130101':'20181231']['Distance (km)'] \\\n .resample('A').sum()\n\n# Create plot\nfig = plt.figure(figsize=(8, 5))\n\n# Plot and customize\nax = df_run_dist_annual.plot(marker='*', markersize=14, linewidth=0, color='blue')\nax.set(ylim=[0, 1210], \n xlim=['2012','2019'],\n ylabel='Distance (km)',\n xlabel='Years',\n title='Annual totals for distance')\n\nax.axhspan(1000, 1210, color='green', alpha=0.4)\nax.axhspan(800, 1000, color='yellow', alpha=0.3)\nax.axhspan(0, 800, color='red', alpha=0.2)\n\n# Show plot\nplt.show()",
"_____no_output_____"
]
],
[
[
"## 8. Am I progressing?\n<p>Let's dive a little deeper into the data to answer a tricky question: am I progressing in terms of my running skills? </p>\n<p>To answer this question, we'll decompose my weekly distance run and visually compare it to the raw data. A red trend line will represent the weekly distance run.</p>\n<p>We are going to use <code>statsmodels</code> library to decompose the weekly trend.</p>",
"_____no_output_____"
]
],
[
[
"# Import required library\nimport statsmodels.api as sm\n\n# Prepare data\ndf_run_dist_wkly = df_run.loc['20190101':'20130101']['Distance (km)'] \\\n .resample('W').bfill()\ndecomposed = sm.tsa.seasonal_decompose(df_run_dist_wkly, extrapolate_trend=1, freq=52)\n\n# Create plot\nfig = plt.figure(figsize=(12, 5))\n\n# Plot and customize\nax = decomposed.trend.plot(label='Trend', linewidth=2)\nax = decomposed.observed.plot(label='Observed', linewidth=0.5)\n\nax.legend()\nax.set_title('Running distance trend')\n\n# Show plot\nplt.show()",
"_____no_output_____"
]
],
[
[
"## 9. Training intensity\n<p>Heart rate is a popular metric used to measure training intensity. Depending on age and fitness level, heart rates are grouped into different zones that people can target depending on training goals. A target heart rate during moderate-intensity activities is about 50-70% of maximum heart rate, while during vigorous physical activity it’s about 70-85% of maximum.</p>\n<p>We'll create a distribution plot of my heart rate data by training intensity. It will be a visual presentation for the number of activities from predefined training zones. </p>",
"_____no_output_____"
]
],
[
[
"# Prepare data\nhr_zones = [100, 125, 133, 142, 151, 173]\nzone_names = ['Easy', 'Moderate', 'Hard', 'Very hard', 'Maximal']\nzone_colors = ['green', 'yellow', 'orange', 'tomato', 'red']\ndf_run_hr_all = ...\n\n# Create plot\nfig, ax = ...\n\n# Plot and customize\nn, bins, patches = ax.hist(df_run_hr_all, bins=hr_zones, alpha=0.5)\nfor i in range(0, len(patches)):\n patches[i].set_facecolor(zone_colors[i])\n\nax.set(title='Distribution of HR', ylabel='Number of runs')\nax.xaxis.set(ticks=hr_zones)\n# ... YOUR CODE FOR TASK 9 ...\n\n# Show plot\n# ... YOUR CODE FOR TASK 8 ...",
"_____no_output_____"
]
],
[
[
"## 10. Detailed summary report\n<p>With all this data cleaning, analysis, and visualization, let's create detailed summary tables of my training. </p>\n<p>To do this, we'll create two tables. The first table will be a summary of the distance (km) and climb (m) variables for each training activity. The second table will list the summary statistics for the average speed (km/hr), climb (m), and distance (km) variables for each training activity.</p>",
"_____no_output_____"
]
],
[
[
"# Concatenating three DataFrames\ndf_run_walk_cycle = ...\n\ndist_climb_cols, speed_col = ['Distance (km)', 'Climb (m)'], ['Average Speed (km/h)']\n\n# Calculating total distance and climb in each type of activities\ndf_totals = ...\n\nprint('Totals for different training types:')\ndisplay(df_totals)\n\n# Calculating summary statistics for each type of activities \ndf_summary = df_run_walk_cycle.groupby('Type')[dist_climb_cols + speed_col].describe()\n\n# Combine totals with summary\nfor i in dist_climb_cols:\n df_summary[i, 'total'] = df_totals[i]\n\nprint('Summary statistics for different training types:')\n# ... YOUR CODE FOR TASK 10 ...",
"_____no_output_____"
]
],
[
[
"## 11. Fun facts\n<p>To wrap up, let’s pick some fun facts out of the summary tables and solve the last exercise.</p>\n<p>These data (my running history) represent 6 years, 2 months and 21 days. And I remember how many running shoes I went through–7.</p>\n<pre><code>FUN FACTS\n- Average distance: 11.38 km\n- Longest distance: 38.32 km\n- Highest climb: 982 m\n- Total climb: 57,278 m\n- Total number of km run: 5,224 km\n- Total runs: 459\n- Number of running shoes gone through: 7 pairs\n</code></pre>\n<p>The story of Forrest Gump is well known–the man, who for no particular reason decided to go for a \"little run.\" His epic run duration was 3 years, 2 months and 14 days (1169 days). In the picture you can see Forrest’s route of 24,700 km. </p>\n<pre><code>FORREST RUN FACTS\n- Average distance: 21.13 km\n- Total number of km run: 24,700 km\n- Total runs: 1169\n- Number of running shoes gone through: ...\n</code></pre>\n<p>Assuming Forest and I go through running shoes at the same rate, figure out how many pairs of shoes Forrest needed for his run.</p>\n<p><img src=\"https://assets.datacamp.com/production/project_727/img/Forrest_Gump_running_route.png\" alt=\"Forrest's route\" title=\"Little run of Forrest Gump\"></p>",
"_____no_output_____"
]
],
[
[
"# Count average shoes per lifetime (as km per pair) using our fun facts\naverage_shoes_lifetime = ...\n\n# Count number of shoes for Forrest's run distance\nshoes_for_forrest_run = ...\n\nprint('Forrest Gump would need {} pairs of shoes!'.format(shoes_for_forrest_run))",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cbecae68b27beae038a869adf1e7ce20e75ae6b8
| 9,117 |
ipynb
|
Jupyter Notebook
|
old_dsfs/clustering.ipynb
|
giordafrancis/DSfS
|
e854db2da376e1c3efe7740073b55f8692cb0863
|
[
"MIT"
] | null | null | null |
old_dsfs/clustering.ipynb
|
giordafrancis/DSfS
|
e854db2da376e1c3efe7740073b55f8692cb0863
|
[
"MIT"
] | null | null | null |
old_dsfs/clustering.ipynb
|
giordafrancis/DSfS
|
e854db2da376e1c3efe7740073b55f8692cb0863
|
[
"MIT"
] | null | null | null | 27.460843 | 295 | 0.519688 |
[
[
[
"One of the simplest clustering methods is k-means, in which the number of clusters is chosen in advance, after shich the goal is to partition the inputs into sets S1, ... Sk in a way that minimizes the total sum of squared distances from each point to the mean of its assigned cluster. \n\nWe will set for an iterative algorithm, that usually finds a good clustering:\n\n- Start with a set of k-means randomly assigned, which are points in d-dimensional space\n- assign each point to the mean to which is closest centerpoint\n- if no point assigment has changed, stop and keep the clusters.\n- if some point assignment has changed, recompute the means and return to step2",
"_____no_output_____"
]
],
[
[
"from linear_algebra import Vector",
"_____no_output_____"
],
[
"# helper function that dectects if any centerpoint assigmmnet has changed\ndef num_differences(v1: Vector, v2: Vector) -> int:\n assert len(v1) == len(v2)\n return len([x1 for x1, x2 in zip(v1, v2) if x1 != x2])\n\nassert num_differences([1, 2, 3], [2, 1, 3]) == 2\nassert num_differences([1, 2], [1, 2]) == 0",
"_____no_output_____"
],
[
"from typing import List\nfrom linear_algebra import vector_mean\nimport random",
"_____no_output_____"
],
[
"def cluster_means(k: int, inputs: List[Vector], assignments: List[int]) -> List[Vector]:\n # clusters[i] contains the inputs whose assignment is i\n clusters = [[] for i in range(k)]\n for input, assignment in zip(inputs, assignments):\n #print(input, assignment)\n clusters[assignment].append(input)\n #print(clusters)\n #break\n # if a cluster is empty, just use a ramdom point\n return [vector_mean(cluster) if cluster else random.choice(inputs)\n for cluster in clusters]",
"_____no_output_____"
],
[
"inputs = [[-1, 1], [-2,3], [-3, 4], [4, 5], [-2, 6], [0, 3]]\nassignments = [0, 0, 2, 2, 2, 1]\ncluster_means(6, inputs, assignments)",
"_____no_output_____"
],
[
"import itertools\n\nfrom linear_algebra import squared_distance",
"_____no_output_____"
],
[
"# I undertand the intuition and the code main points\n#\n\nclass KMeans:\n def __init__(self, k: int) -> None:\n self.k = k #number of clusters\n self.means = None\n \n def classify(self, input: Vector) -> int:\n \"\"\"return the index of the cluster closest to the input\"\"\"\n # means method woudl be already computed as claissfy method is called\n # means len == k\n return min(range(self.k),\n key= lambda i: squared_distance(input, self.means[i]))\n def train(self, inputs: List[Vector]) -> None:\n # start with a random assignments\n assignments = [random.randrange(self.k) for _ in inputs]\n for _ in itertools.count():\n # print(assignments)\n # compute means\n self.means = cluster_means(self.k, inputs, assignments)\n # and find new assignments\n new_assignments = [self.classify(input) for input in inputs]\n # check how many assignments have changed and if we're done\n num_changed = num_differences(assignments, new_assignments)\n if num_changed == 0: \n return\n # otherwise keep the new assignments, and compute new means\n assignments = new_assignments\n self.means = cluster_means(self.k, inputs, assignments)\n print(f\"changed: {num_changed} / {len(inputs)}\")",
"_____no_output_____"
],
[
"inputs: List[List[float]] = [[-14,-5],[13,13],[20,23],[-19,-11],[-9,-16],[21,27],[-49,15],[26,13],[-46,5],[-34,-1],[11,15],\n [-49,0],[-22,-16],[19,28],[-12,-8],[-13,-19],[-41,8],[-11,-6],[-25,-9],[-18,-3]]",
"_____no_output_____"
],
[
"k = 2\nrandom.seed(0)\nclusterer = KMeans(k)\nclusterer.train(inputs)\nmeans = sorted(clusterer.means)\n\nassert len(means) == k\nmeans",
"changed: 8 / 20\nchanged: 4 / 20\n"
],
[
"# check that the measures are close to what we expect\nsquared_distance(means[0], [-44, 5]) ",
"_____no_output_____"
]
],
[
[
"#### Chosing K\n\nThere are various ways to choose a k. One that is reasonably easy to develop intuition involves plotting the sum of squared errors (between each \n",
"_____no_output_____"
]
],
[
[
"from matplotlib import pyplot as plt\n\ndef squared_clustering_errors(inputs: List[Vector], k: int) -> float:\n \"\"\"finds the total squared error from k-means clustering the inputs\n \"\"\"\n clusterer = KMeans(k)\n clusterer.train(inputs)\n means = clusterer.means\n # there isnt an assignment attribute\n assignments = [clusterer.classify(input) for input in inputs]\n return sum(squared_distance(input, means[cluster])\n for input, cluster in zip(inputs, assignments))",
"_____no_output_____"
],
[
"clusterer = KMeans(3)\nclusterer.train(inputs)",
"changed: 12 / 20\n"
],
[
"means = clusterer.means\nmeans",
"_____no_output_____"
],
[
"assignments = [clusterer.classify(input) for input in inputs]\nassignments",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
cbecdc088ec879805dcbbba2947580d49c565302
| 268,833 |
ipynb
|
Jupyter Notebook
|
notebooks/gan_training.ipynb
|
AlexanderKlemps/simple-gan
|
8915baf8d50875fe51586540333887654db11764
|
[
"Apache-2.0"
] | null | null | null |
notebooks/gan_training.ipynb
|
AlexanderKlemps/simple-gan
|
8915baf8d50875fe51586540333887654db11764
|
[
"Apache-2.0"
] | null | null | null |
notebooks/gan_training.ipynb
|
AlexanderKlemps/simple-gan
|
8915baf8d50875fe51586540333887654db11764
|
[
"Apache-2.0"
] | null | null | null | 858.891374 | 76,496 | 0.954563 |
[
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\nfrom simple_GAN.gan import GAN\nfrom simple_GAN.utils import sample_data, relu, sigmoid, tanh, DataChunk",
"_____no_output_____"
],
[
"data, labels = sample_data(1000, 2)\nplt.scatter(data[:, 0], data[:, 1], c=labels, s=3)\nplt.show()",
"_____no_output_____"
],
[
"max_value = np.max(np.abs(data))\nmax_value",
"_____no_output_____"
],
[
"normed_data = data / max_value",
"_____no_output_____"
],
[
"plt.scatter(normed_data[:, 0], normed_data[:, 1], c=labels, s=3)\nplt.show()",
"_____no_output_____"
],
[
"chunk = DataChunk(normed_data, labels)\nnet = GAN(batch_size=128)",
"_____no_output_____"
],
[
"net.train(chunk, epochs=1000, 1e-4)",
"_____no_output_____"
],
[
"plt.scatter(data[:, 0], data[:, 1], c=labels, s=3)\nfor i in range(4):\n noise = np.random.uniform(-1, 1, [net.batch_size, 100]).astype(np.float32)\n gen_output = net.generate(noise) * max_value\n plt.scatter(gen_output[:, 0], gen_output[:, 1], c='r', s=3)\nplt.show()",
"_____no_output_____"
],
[
"fake_probs = net.discriminator.infer(gen_output[:16]/max_value)\nfake_probs",
"_____no_output_____"
],
[
"np.mean(fake_probs)",
"_____no_output_____"
],
[
"net.generator_loss(fake_probs)",
"_____no_output_____"
],
[
"batch, _ = next(chunk.iter_batches(net.batch_size))\nreal_probs = net.discriminator.infer(batch[:16])\nreal_probs",
"_____no_output_____"
],
[
"np.mean(real_probs)",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbece00f4ab1ca443ba9b0b0e1dd4c12c446ca48
| 360,312 |
ipynb
|
Jupyter Notebook
|
WeatherPy/WeatherPy.ipynb
|
mariaalzaga/python_api_challenge
|
69ba64234619902e4bc42dd9202d556ab1d9e3cc
|
[
"ADSL"
] | null | null | null |
WeatherPy/WeatherPy.ipynb
|
mariaalzaga/python_api_challenge
|
69ba64234619902e4bc42dd9202d556ab1d9e3cc
|
[
"ADSL"
] | null | null | null |
WeatherPy/WeatherPy.ipynb
|
mariaalzaga/python_api_challenge
|
69ba64234619902e4bc42dd9202d556ab1d9e3cc
|
[
"ADSL"
] | null | null | null | 284.831621 | 43,880 | 0.916023 |
[
[
[
"# Dependencies and Setup\nimport matplotlib.pyplot as plt\nimport pandas as pd\nimport numpy as np\nimport requests\nimport time\nimport json\nimport scipy.stats as st\nfrom scipy.stats import linregress\n",
"_____no_output_____"
],
[
"# Import API key\nfrom api_keys import g_key",
"_____no_output_____"
],
[
"from citipy import citipy\n# https://pypi.org/project/citipy/ (pip install citipy)\n# Range of latitudes and longitudes\nlat_range = (-90, 90)\nlng_range = (-180, 180)",
"_____no_output_____"
],
[
"\n# List for holding latitudes-longitudes and cities\nlat_lngs = []\ncities = []\n\n# Create a set of random lat and lng combinations\nlatitudes = np.random.uniform(low=-90.000, high=90.000, size=1500)\nlongitudes = np.random.uniform(low=-180.000, high=180.000, size=1500)\nlatitudes_longitudes = zip(latitudes, longitudes)\n\n# Identify nearest city for each latitudes-longitudes combination\nfor everylat_lng in latitudes_longitudes:\n city = citipy.nearest_city(everylat_lng[0], everylat_lng[1]).city_name\n \n # If the city is not already present, then add it to a our cities list\n if city not in cities:\n cities.append(city)\n\n# Print the city count to confirm sufficient count\nlen(cities)",
"_____no_output_____"
],
[
"#Perform API Calls\ncloudiness = []\ncountry = []\ndate = []\nhumidity = []\nlatitude_list = []\nlongitude_list = []\nmaximum_temp = []\nwind_speed = []\n",
"_____no_output_____"
],
[
"from api_keys import weather_api_key\nindex_counter = 0\nset_counter = 1",
"_____no_output_____"
],
[
"# Save config information.\nurl = \"http://api.openweathermap.org/data/2.5/weather?\"\nunits = \"imperial\"\n\n# Build partial query URL\nquery_url = f\"{url}appid={weather_api_key}&units={units}&q=\"\n",
"_____no_output_____"
],
[
"# For each city name in cities list, do below things...used exception handling\nfor index, city in enumerate(cities, start = 1):\n try:\n response = requests.get(query_url + city).json()\n cloudiness.append(response[\"clouds\"][\"all\"])\n country.append(response[\"sys\"][\"country\"])\n date.append(response[\"dt\"])\n humidity.append(response[\"main\"][\"humidity\"])\n latitude_list.append(response[\"coord\"][\"lat\"])\n longitude_list.append(response[\"coord\"][\"lon\"])\n maximum_temp.append(response['main']['temp_max'])\n wind_speed.append(response[\"wind\"][\"speed\"])\n if index_counter > 49:\n index_counter = 0\n set_counter = set_counter + 1\n \n else:\n index_counter = index_counter + 1\n \n print(f\"Processing Record {index_counter} of Set {set_counter} : {city}\") \n \n except(KeyError, IndexError):\n print(\"City not found. Skipping...\")",
"Processing Record 1 of Set 1 : hilo\nProcessing Record 2 of Set 1 : new norfolk\nProcessing Record 3 of Set 1 : rikitea\nProcessing Record 4 of Set 1 : punta arenas\nProcessing Record 5 of Set 1 : east london\nProcessing Record 6 of Set 1 : puerto ayora\nProcessing Record 7 of Set 1 : longyearbyen\nProcessing Record 8 of Set 1 : dikson\nProcessing Record 9 of Set 1 : mishan\nProcessing Record 10 of Set 1 : narsaq\nProcessing Record 11 of Set 1 : tiksi\nProcessing Record 12 of Set 1 : hermanus\nProcessing Record 13 of Set 1 : ribeira grande\nProcessing Record 14 of Set 1 : talnakh\nProcessing Record 15 of Set 1 : huarmey\nProcessing Record 16 of Set 1 : evensk\nProcessing Record 17 of Set 1 : vaini\nProcessing Record 18 of Set 1 : provideniya\nProcessing Record 19 of Set 1 : buin\nProcessing Record 20 of Set 1 : porto novo\nProcessing Record 21 of Set 1 : meulaboh\nProcessing Record 22 of Set 1 : torbay\nProcessing Record 23 of Set 1 : hobart\nProcessing Record 24 of Set 1 : dhidhdhoo\nProcessing Record 25 of Set 1 : copiapo\nProcessing Record 26 of Set 1 : victoria\nProcessing Record 27 of Set 1 : mataura\nCity not found. Skipping...\nProcessing Record 28 of Set 1 : coria\nProcessing Record 29 of Set 1 : albany\nProcessing Record 30 of Set 1 : barrow\nProcessing Record 31 of Set 1 : lavrentiya\nProcessing Record 32 of Set 1 : upernavik\nProcessing Record 33 of Set 1 : katobu\nProcessing Record 34 of Set 1 : villachuato\nProcessing Record 35 of Set 1 : bukama\nCity not found. Skipping...\nProcessing Record 36 of Set 1 : myitkyina\nCity not found. Skipping...\nProcessing Record 37 of Set 1 : tuktoyaktuk\nProcessing Record 38 of Set 1 : oranjemund\nProcessing Record 39 of Set 1 : waingapu\nProcessing Record 40 of Set 1 : puerto leguizamo\nProcessing Record 41 of Set 1 : tabou\nCity not found. Skipping...\nProcessing Record 42 of Set 1 : cockburn town\nProcessing Record 43 of Set 1 : pringsewu\nProcessing Record 44 of Set 1 : conceicao do araguaia\nProcessing Record 45 of Set 1 : fairhope\nProcessing Record 46 of Set 1 : coihaique\nProcessing Record 47 of Set 1 : elat\nProcessing Record 48 of Set 1 : norman wells\nProcessing Record 49 of Set 1 : calabar\nProcessing Record 50 of Set 1 : nyrob\nProcessing Record 0 of Set 2 : morondava\nProcessing Record 1 of Set 2 : port alfred\nProcessing Record 2 of Set 2 : khandbari\nProcessing Record 3 of Set 2 : aleppo\nCity not found. Skipping...\nProcessing Record 4 of Set 2 : faanui\nProcessing Record 5 of Set 2 : vao\nProcessing Record 6 of Set 2 : georgetown\nProcessing Record 7 of Set 2 : marawi\nProcessing Record 8 of Set 2 : bluff\nProcessing Record 9 of Set 2 : diapaga\nCity not found. Skipping...\nProcessing Record 10 of Set 2 : buala\nProcessing Record 11 of Set 2 : busselton\nProcessing Record 12 of Set 2 : yellowknife\nProcessing Record 13 of Set 2 : castro\nProcessing Record 14 of Set 2 : isangel\nCity not found. Skipping...\nProcessing Record 15 of Set 2 : ulaanbaatar\nProcessing Record 16 of Set 2 : chino valley\nProcessing Record 17 of Set 2 : port elizabeth\nProcessing Record 18 of Set 2 : margate\nProcessing Record 19 of Set 2 : cape town\nProcessing Record 20 of Set 2 : vuktyl\nProcessing Record 21 of Set 2 : avarua\nProcessing Record 22 of Set 2 : ontario\nProcessing Record 23 of Set 2 : chuy\nProcessing Record 24 of Set 2 : kruisfontein\nProcessing Record 25 of Set 2 : arraial do cabo\nProcessing Record 26 of Set 2 : issoudun\nProcessing Record 27 of Set 2 : lahij\nProcessing Record 28 of Set 2 : bambous virieux\nProcessing Record 29 of Set 2 : trelew\nProcessing Record 30 of Set 2 : pilar\nProcessing Record 31 of Set 2 : kaitangata\nProcessing Record 32 of Set 2 : ushuaia\nCity not found. Skipping...\nProcessing Record 33 of Set 2 : patea\nProcessing Record 34 of Set 2 : lazaro cardenas\nProcessing Record 35 of Set 2 : nanortalik\nCity not found. Skipping...\nProcessing Record 36 of Set 2 : pangnirtung\nProcessing Record 37 of Set 2 : mar del plata\nProcessing Record 38 of Set 2 : la baule-escoublac\nProcessing Record 39 of Set 2 : garissa\nProcessing Record 40 of Set 2 : kiunga\nProcessing Record 41 of Set 2 : nikolskoye\nProcessing Record 42 of Set 2 : jamestown\nProcessing Record 43 of Set 2 : borova\nProcessing Record 44 of Set 2 : dingle\nProcessing Record 45 of Set 2 : dolores\nProcessing Record 46 of Set 2 : lorengau\nProcessing Record 47 of Set 2 : tymovskoye\nProcessing Record 48 of Set 2 : hohhot\nProcessing Record 49 of Set 2 : berliste\nProcessing Record 50 of Set 2 : dogondoutchi\nProcessing Record 0 of Set 3 : paripiranga\nProcessing Record 1 of Set 3 : ahipara\nProcessing Record 2 of Set 3 : payo\nProcessing Record 3 of Set 3 : sao joao da barra\nProcessing Record 4 of Set 3 : panjakent\nProcessing Record 5 of Set 3 : hakui\nProcessing Record 6 of Set 3 : luebo\nProcessing Record 7 of Set 3 : ostrovnoy\nProcessing Record 8 of Set 3 : sola\nProcessing Record 9 of Set 3 : saint george\nProcessing Record 10 of Set 3 : stephenville\nProcessing Record 11 of Set 3 : carnarvon\nProcessing Record 12 of Set 3 : atuona\nProcessing Record 13 of Set 3 : constitucion\nProcessing Record 14 of Set 3 : butaritari\nProcessing Record 15 of Set 3 : saldanha\nProcessing Record 16 of Set 3 : kurilsk\nProcessing Record 17 of Set 3 : nosy varika\nProcessing Record 18 of Set 3 : hualmay\nCity not found. Skipping...\nProcessing Record 19 of Set 3 : codrington\nProcessing Record 20 of Set 3 : indiaroba\nProcessing Record 21 of Set 3 : santa rosa\nProcessing Record 22 of Set 3 : te anau\nProcessing Record 23 of Set 3 : broome\nProcessing Record 24 of Set 3 : bilibino\nProcessing Record 25 of Set 3 : urengoy\nProcessing Record 26 of Set 3 : yabrud\nProcessing Record 27 of Set 3 : katsuura\nProcessing Record 28 of Set 3 : kapaa\nProcessing Record 29 of Set 3 : tasiilaq\nProcessing Record 30 of Set 3 : nicoya\nProcessing Record 31 of Set 3 : sorland\nProcessing Record 32 of Set 3 : qaanaaq\nProcessing Record 33 of Set 3 : zhigansk\nProcessing Record 34 of Set 3 : tongren\nProcessing Record 35 of Set 3 : ancud\nProcessing Record 36 of Set 3 : waipawa\nProcessing Record 37 of Set 3 : gat\nProcessing Record 38 of Set 3 : dwarka\nProcessing Record 39 of Set 3 : korla\nProcessing Record 40 of Set 3 : vila velha\nProcessing Record 41 of Set 3 : samandag\nProcessing Record 42 of Set 3 : peno\nProcessing Record 43 of Set 3 : terney\nProcessing Record 44 of Set 3 : andra\nProcessing Record 45 of Set 3 : lokosovo\nProcessing Record 46 of Set 3 : ornskoldsvik\nProcessing Record 47 of Set 3 : tuatapere\nProcessing Record 48 of Set 3 : hambantota\nProcessing Record 49 of Set 3 : ponta do sol\nProcessing Record 50 of Set 3 : lundazi\nProcessing Record 0 of Set 4 : hellvik\nProcessing Record 1 of Set 4 : nelson bay\nProcessing Record 2 of Set 4 : port hedland\nProcessing Record 3 of Set 4 : coroico\nProcessing Record 4 of Set 4 : thompson\nProcessing Record 5 of Set 4 : ilulissat\nProcessing Record 6 of Set 4 : mount isa\nProcessing Record 7 of Set 4 : tucupita\nCity not found. Skipping...\nProcessing Record 8 of Set 4 : khatanga\nProcessing Record 9 of Set 4 : vila franca do campo\nProcessing Record 10 of Set 4 : kodiak\nProcessing Record 11 of Set 4 : belyy yar\nProcessing Record 12 of Set 4 : milkovo\nProcessing Record 13 of Set 4 : xiongzhou\nProcessing Record 14 of Set 4 : guerrero negro\nProcessing Record 15 of Set 4 : kavieng\nProcessing Record 16 of Set 4 : pevek\nProcessing Record 17 of Set 4 : menongue\nProcessing Record 18 of Set 4 : sambava\nProcessing Record 19 of Set 4 : onega\nProcessing Record 20 of Set 4 : fairbanks\nProcessing Record 21 of Set 4 : isilkul\nProcessing Record 22 of Set 4 : savonlinna\nCity not found. Skipping...\nCity not found. Skipping...\nProcessing Record 23 of Set 4 : demba\nProcessing Record 24 of Set 4 : yorkton\nProcessing Record 25 of Set 4 : romitan\nProcessing Record 26 of Set 4 : kuvandyk\nProcessing Record 27 of Set 4 : sasovo\nProcessing Record 28 of Set 4 : carutapera\nCity not found. Skipping...\nProcessing Record 29 of Set 4 : baruun-urt\nProcessing Record 30 of Set 4 : fort nelson\nCity not found. Skipping...\nProcessing Record 31 of Set 4 : camacha\nProcessing Record 32 of Set 4 : pangai\nProcessing Record 33 of Set 4 : lagos\nProcessing Record 34 of Set 4 : san ramon\nProcessing Record 35 of Set 4 : borogontsy\nProcessing Record 36 of Set 4 : saskylakh\nProcessing Record 37 of Set 4 : nome\nProcessing Record 38 of Set 4 : ayan\nProcessing Record 39 of Set 4 : jimeta\nProcessing Record 40 of Set 4 : airai\nProcessing Record 41 of Set 4 : bengkulu\nProcessing Record 42 of Set 4 : mittagong\nProcessing Record 43 of Set 4 : conakry\nProcessing Record 44 of Set 4 : ayorou\nProcessing Record 45 of Set 4 : mesquite\nProcessing Record 46 of Set 4 : traverse city\nCity not found. Skipping...\nProcessing Record 47 of Set 4 : cidreira\nProcessing Record 48 of Set 4 : karasburg\nProcessing Record 49 of Set 4 : zhangye\nProcessing Record 50 of Set 4 : miri\nProcessing Record 0 of Set 5 : northam\nProcessing Record 1 of Set 5 : lagoa\nProcessing Record 2 of Set 5 : taltal\nProcessing Record 3 of Set 5 : tiznit\nProcessing Record 4 of Set 5 : flin flon\nProcessing Record 5 of Set 5 : lat yao\nProcessing Record 6 of Set 5 : hervey bay\nProcessing Record 7 of Set 5 : wuda\nProcessing Record 8 of Set 5 : novyy urengoy\nProcessing Record 9 of Set 5 : severobaykalsk\nProcessing Record 10 of Set 5 : lusambo\nProcessing Record 11 of Set 5 : luwuk\nProcessing Record 12 of Set 5 : bafq\nProcessing Record 13 of Set 5 : hithadhoo\nProcessing Record 14 of Set 5 : salym\nProcessing Record 15 of Set 5 : urucara\nProcessing Record 16 of Set 5 : bredasdorp\nProcessing Record 17 of Set 5 : nang rong\nProcessing Record 18 of Set 5 : coquimbo\nProcessing Record 19 of Set 5 : baykit\nProcessing Record 20 of Set 5 : lata\nProcessing Record 21 of Set 5 : baijiantan\nProcessing Record 22 of Set 5 : calbuco\nProcessing Record 23 of Set 5 : verkhoshizhemye\nProcessing Record 24 of Set 5 : san quintin\nProcessing Record 25 of Set 5 : egvekinot\nProcessing Record 26 of Set 5 : la carlota\nProcessing Record 27 of Set 5 : souillac\nProcessing Record 28 of Set 5 : umm kaddadah\nProcessing Record 29 of Set 5 : saint-pierre\nProcessing Record 30 of Set 5 : puerto escondido\nProcessing Record 31 of Set 5 : mahebourg\nProcessing Record 32 of Set 5 : cairns\nProcessing Record 33 of Set 5 : san patricio\nProcessing Record 34 of Set 5 : lakselv\nProcessing Record 35 of Set 5 : qasigiannguit\nProcessing Record 36 of Set 5 : beidao\nProcessing Record 37 of Set 5 : mananjary\nProcessing Record 38 of Set 5 : saint-philippe\nCity not found. Skipping...\nProcessing Record 39 of Set 5 : santa maria\nProcessing Record 40 of Set 5 : aklavik\nProcessing Record 41 of Set 5 : salina\nProcessing Record 42 of Set 5 : dalvik\nProcessing Record 43 of Set 5 : savannah bight\nProcessing Record 44 of Set 5 : takoradi\nProcessing Record 45 of Set 5 : muros\nCity not found. Skipping...\nProcessing Record 46 of Set 5 : bilma\nProcessing Record 47 of Set 5 : haverfordwest\nCity not found. Skipping...\nCity not found. Skipping...\nProcessing Record 48 of Set 5 : kutum\nProcessing Record 49 of Set 5 : ampanihy\nProcessing Record 50 of Set 5 : imbituba\nProcessing Record 0 of Set 6 : amapa\nProcessing Record 1 of Set 6 : laguna\nProcessing Record 2 of Set 6 : quatre cocos\nProcessing Record 3 of Set 6 : cabra\nProcessing Record 4 of Set 6 : itacoatiara\nProcessing Record 5 of Set 6 : karaton\nProcessing Record 6 of Set 6 : suhum\nProcessing Record 7 of Set 6 : iqaluit\nProcessing Record 8 of Set 6 : honiara\nProcessing Record 9 of Set 6 : jiayuguan\nProcessing Record 10 of Set 6 : poum\nProcessing Record 11 of Set 6 : usinsk\nProcessing Record 12 of Set 6 : acarau\nProcessing Record 13 of Set 6 : nguru\nCity not found. Skipping...\nProcessing Record 14 of Set 6 : ormara\nProcessing Record 15 of Set 6 : amli\nProcessing Record 16 of Set 6 : cherskiy\nProcessing Record 17 of Set 6 : kununurra\nCity not found. Skipping...\nProcessing Record 18 of Set 6 : areka\nProcessing Record 19 of Set 6 : sovetskiy\nProcessing Record 20 of Set 6 : zakamensk\nProcessing Record 21 of Set 6 : nara\nCity not found. Skipping...\nProcessing Record 22 of Set 6 : ahuimanu\nProcessing Record 23 of Set 6 : cayenne\nProcessing Record 24 of Set 6 : tura\nProcessing Record 25 of Set 6 : sur\nCity not found. Skipping...\nProcessing Record 26 of Set 6 : cartagena\nProcessing Record 27 of Set 6 : cabo san lucas\nProcessing Record 28 of Set 6 : lakes entrance\nProcessing Record 29 of Set 6 : lebu\nProcessing Record 30 of Set 6 : gamba\nProcessing Record 31 of Set 6 : tombouctou\nProcessing Record 32 of Set 6 : nova olinda do norte\nCity not found. Skipping...\nProcessing Record 33 of Set 6 : beckley\nProcessing Record 34 of Set 6 : penzance\nProcessing Record 35 of Set 6 : karratha\nProcessing Record 36 of Set 6 : najran\nProcessing Record 37 of Set 6 : utrecht\nCity not found. Skipping...\nProcessing Record 38 of Set 6 : gostyn\nProcessing Record 39 of Set 6 : chingirlau\nProcessing Record 40 of Set 6 : price\nProcessing Record 41 of Set 6 : wakema\nProcessing Record 42 of Set 6 : bindura\nProcessing Record 43 of Set 6 : atar\nProcessing Record 44 of Set 6 : ketchikan\nCity not found. Skipping...\nProcessing Record 45 of Set 6 : luanda\nProcessing Record 46 of Set 6 : elblag\nProcessing Record 47 of Set 6 : hasaki\nCity not found. Skipping...\nProcessing Record 48 of Set 6 : faya\nProcessing Record 49 of Set 6 : havelock\nProcessing Record 50 of Set 6 : aquiraz\nProcessing Record 0 of Set 7 : pacific grove\nProcessing Record 1 of Set 7 : linjiang\nProcessing Record 2 of Set 7 : miyako\nProcessing Record 3 of Set 7 : srednekolymsk\nProcessing Record 4 of Set 7 : adrar\nProcessing Record 5 of Set 7 : phan thiet\nProcessing Record 6 of Set 7 : cody\nProcessing Record 7 of Set 7 : esperance\nProcessing Record 8 of Set 7 : khor\nProcessing Record 9 of Set 7 : nyurba\nProcessing Record 10 of Set 7 : klaksvik\nProcessing Record 11 of Set 7 : manokwari\nCity not found. Skipping...\nProcessing Record 12 of Set 7 : zyryanka\nProcessing Record 13 of Set 7 : paita\nProcessing Record 14 of Set 7 : haines junction\nProcessing Record 15 of Set 7 : bassar\nProcessing Record 16 of Set 7 : benguela\nProcessing Record 17 of Set 7 : bowen\nProcessing Record 18 of Set 7 : velyka oleksandrivka\nCity not found. Skipping...\nProcessing Record 19 of Set 7 : anlu\nProcessing Record 20 of Set 7 : college\nProcessing Record 21 of Set 7 : christchurch\nProcessing Record 22 of Set 7 : vardo\nProcessing Record 23 of Set 7 : oussouye\nProcessing Record 24 of Set 7 : itarema\nProcessing Record 25 of Set 7 : newcastleton\nProcessing Record 26 of Set 7 : fontem\nProcessing Record 27 of Set 7 : yeppoon\nProcessing Record 28 of Set 7 : synya\nProcessing Record 29 of Set 7 : harrisonburg\nProcessing Record 30 of Set 7 : hofn\nProcessing Record 31 of Set 7 : kalaleh\nProcessing Record 32 of Set 7 : nouadhibou\nCity not found. Skipping...\nProcessing Record 33 of Set 7 : anadyr\nProcessing Record 34 of Set 7 : roma\nProcessing Record 35 of Set 7 : kozhva\nProcessing Record 36 of Set 7 : republic\nProcessing Record 37 of Set 7 : moron\nCity not found. Skipping...\nProcessing Record 38 of Set 7 : bella union\nProcessing Record 39 of Set 7 : asau\nProcessing Record 40 of Set 7 : bondo\nProcessing Record 41 of Set 7 : labuhan\nCity not found. Skipping...\nProcessing Record 42 of Set 7 : paamiut\nProcessing Record 43 of Set 7 : koutsouras\nProcessing Record 44 of Set 7 : lindi\nProcessing Record 45 of Set 7 : kirakira\nProcessing Record 46 of Set 7 : shawnee\nProcessing Record 47 of Set 7 : padang\nCity not found. Skipping...\nProcessing Record 48 of Set 7 : norton\nProcessing Record 49 of Set 7 : teya\nProcessing Record 50 of Set 7 : vzmorye\nProcessing Record 0 of Set 8 : ukiah\nProcessing Record 1 of Set 8 : san rafael\nProcessing Record 2 of Set 8 : villa sandino\nProcessing Record 3 of Set 8 : lakatoro\nProcessing Record 4 of Set 8 : namibe\nProcessing Record 5 of Set 8 : verkh-usugli\nProcessing Record 6 of Set 8 : rovaniemi\nProcessing Record 7 of Set 8 : waddan\nProcessing Record 8 of Set 8 : zaozerne\nProcessing Record 9 of Set 8 : matara\nProcessing Record 10 of Set 8 : buta\nProcessing Record 11 of Set 8 : jammalamadugu\nProcessing Record 12 of Set 8 : san cristobal\nProcessing Record 13 of Set 8 : elko\nProcessing Record 14 of Set 8 : anori\nProcessing Record 15 of Set 8 : kibakwe\nProcessing Record 16 of Set 8 : sao filipe\nProcessing Record 17 of Set 8 : severo-kurilsk\nProcessing Record 18 of Set 8 : gushikawa\nProcessing Record 19 of Set 8 : clyde river\nProcessing Record 20 of Set 8 : ikorodu\nProcessing Record 21 of Set 8 : benjamin aceval\nProcessing Record 22 of Set 8 : taoudenni\nCity not found. Skipping...\nProcessing Record 23 of Set 8 : lisala\nProcessing Record 24 of Set 8 : dapdap\nProcessing Record 25 of Set 8 : ulaangom\nProcessing Record 26 of Set 8 : flinders\nProcessing Record 27 of Set 8 : nynashamn\nProcessing Record 28 of Set 8 : chokurdakh\nProcessing Record 29 of Set 8 : avondale\nProcessing Record 30 of Set 8 : novooleksiyivka\nProcessing Record 31 of Set 8 : gizo\nProcessing Record 32 of Set 8 : lima\nProcessing Record 33 of Set 8 : povenets\nCity not found. Skipping...\nProcessing Record 34 of Set 8 : alpena\nProcessing Record 35 of Set 8 : geraldton\nProcessing Record 36 of Set 8 : nuuk\nProcessing Record 37 of Set 8 : loncoche\nProcessing Record 38 of Set 8 : garowe\nProcessing Record 39 of Set 8 : novouzensk\nProcessing Record 40 of Set 8 : danvers\nProcessing Record 41 of Set 8 : inta\nProcessing Record 42 of Set 8 : caravelas\nProcessing Record 43 of Set 8 : sernovodsk\nProcessing Record 44 of Set 8 : tutoia\nProcessing Record 45 of Set 8 : fonte boa\nProcessing Record 46 of Set 8 : fortuna\nProcessing Record 47 of Set 8 : hovd\nProcessing Record 48 of Set 8 : gambela\nProcessing Record 49 of Set 8 : pa sang\nProcessing Record 50 of Set 8 : vilcun\nProcessing Record 0 of Set 9 : little current\nProcessing Record 1 of Set 9 : aiken\nProcessing Record 2 of Set 9 : acton vale\nProcessing Record 3 of Set 9 : mantua\nProcessing Record 4 of Set 9 : naze\nProcessing Record 5 of Set 9 : keflavik\nProcessing Record 6 of Set 9 : bonthe\nProcessing Record 7 of Set 9 : birao\nProcessing Record 8 of Set 9 : aykhal\nCity not found. Skipping...\nProcessing Record 9 of Set 9 : nadym\nCity not found. Skipping...\nProcessing Record 10 of Set 9 : laurel\nProcessing Record 11 of Set 9 : henties bay\nCity not found. Skipping...\nProcessing Record 12 of Set 9 : sinop\nProcessing Record 13 of Set 9 : fukue\nProcessing Record 14 of Set 9 : florida\nProcessing Record 15 of Set 9 : martinsburg\nProcessing Record 16 of Set 9 : eureka\nProcessing Record 17 of Set 9 : kalevala\nProcessing Record 18 of Set 9 : lasa\nProcessing Record 19 of Set 9 : meadow lake\nProcessing Record 20 of Set 9 : mairi\nProcessing Record 21 of Set 9 : iranshahr\nProcessing Record 22 of Set 9 : belleville\nProcessing Record 23 of Set 9 : victor harbor\nProcessing Record 24 of Set 9 : general pico\nProcessing Record 25 of Set 9 : alofi\nProcessing Record 26 of Set 9 : tolaga bay\nProcessing Record 27 of Set 9 : tres picos\nProcessing Record 28 of Set 9 : tautira\nCity not found. Skipping...\nProcessing Record 29 of Set 9 : cozumel\nProcessing Record 30 of Set 9 : tilichiki\nProcessing Record 31 of Set 9 : pyay\nProcessing Record 32 of Set 9 : leningradskiy\nProcessing Record 33 of Set 9 : tarko-sale\nProcessing Record 34 of Set 9 : banda aceh\nProcessing Record 35 of Set 9 : izhma\nProcessing Record 36 of Set 9 : touros\nProcessing Record 37 of Set 9 : timizart\nProcessing Record 38 of Set 9 : yilan\nProcessing Record 39 of Set 9 : okhotsk\nProcessing Record 40 of Set 9 : syracuse\nProcessing Record 41 of Set 9 : namatanai\nCity not found. Skipping...\nProcessing Record 42 of Set 9 : sztum\nProcessing Record 43 of Set 9 : deputatskiy\nProcessing Record 44 of Set 9 : dali\nProcessing Record 45 of Set 9 : quimper\nProcessing Record 46 of Set 9 : kloulklubed\nProcessing Record 47 of Set 9 : chokwe\nProcessing Record 48 of Set 9 : nokia\nProcessing Record 49 of Set 9 : galveston\nProcessing Record 50 of Set 9 : changping\nProcessing Record 0 of Set 10 : chesterville\nProcessing Record 1 of Set 10 : coahuayana\nProcessing Record 2 of Set 10 : kachiry\nProcessing Record 3 of Set 10 : port lincoln\nProcessing Record 4 of Set 10 : ariquemes\nCity not found. Skipping...\nProcessing Record 5 of Set 10 : aksum\nProcessing Record 6 of Set 10 : goma\nProcessing Record 7 of Set 10 : mwinilunga\nProcessing Record 8 of Set 10 : darhan\nProcessing Record 9 of Set 10 : sao felix do xingu\nProcessing Record 10 of Set 10 : port-gentil\nProcessing Record 11 of Set 10 : apan\nProcessing Record 12 of Set 10 : yulara\nProcessing Record 13 of Set 10 : portland\nProcessing Record 14 of Set 10 : staraya poltavka\nProcessing Record 15 of Set 10 : aizpute\nProcessing Record 16 of Set 10 : rioja\nProcessing Record 17 of Set 10 : yumen\nProcessing Record 18 of Set 10 : pokhara\nProcessing Record 19 of Set 10 : sartana\nProcessing Record 20 of Set 10 : bubaque\nProcessing Record 21 of Set 10 : am timan\nProcessing Record 22 of Set 10 : tamparan\nProcessing Record 23 of Set 10 : kavaratti\nProcessing Record 24 of Set 10 : nanuque\nProcessing Record 25 of Set 10 : batemans bay\nProcessing Record 26 of Set 10 : muravlenko\nProcessing Record 27 of Set 10 : tessalit\nProcessing Record 28 of Set 10 : codajas\nProcessing Record 29 of Set 10 : arroio grande\nProcessing Record 30 of Set 10 : manjacaze\nProcessing Record 31 of Set 10 : martapura\nProcessing Record 32 of Set 10 : kirby\nProcessing Record 33 of Set 10 : santa cruz\nProcessing Record 34 of Set 10 : labytnangi\nProcessing Record 35 of Set 10 : okha\nProcessing Record 36 of Set 10 : yarmouth\nProcessing Record 37 of Set 10 : petatlan\nProcessing Record 38 of Set 10 : camacupa\nProcessing Record 39 of Set 10 : dunedin\nProcessing Record 40 of Set 10 : locri\nProcessing Record 41 of Set 10 : high rock\nCity not found. Skipping...\nProcessing Record 42 of Set 10 : chabahar\nProcessing Record 43 of Set 10 : hun\nProcessing Record 44 of Set 10 : asyut\nProcessing Record 45 of Set 10 : nantucket\nProcessing Record 46 of Set 10 : lompoc\nProcessing Record 47 of Set 10 : havoysund\nProcessing Record 48 of Set 10 : gondanglegi\nProcessing Record 49 of Set 10 : marienburg\nProcessing Record 50 of Set 10 : luancheng\nProcessing Record 0 of Set 11 : kashi\nProcessing Record 1 of Set 11 : artvin\nProcessing Record 2 of Set 11 : lethem\nProcessing Record 3 of Set 11 : safonovo\nProcessing Record 4 of Set 11 : kahului\nProcessing Record 5 of Set 11 : paraiso\nProcessing Record 6 of Set 11 : sobolevo\nProcessing Record 7 of Set 11 : bonfim\nProcessing Record 8 of Set 11 : vestmannaeyjar\nProcessing Record 9 of Set 11 : tefe\nProcessing Record 10 of Set 11 : costa rica\nProcessing Record 11 of Set 11 : batie\nProcessing Record 12 of Set 11 : mount gambier\nProcessing Record 13 of Set 11 : mukhen\nProcessing Record 14 of Set 11 : solnechnyy\nProcessing Record 15 of Set 11 : valparaiso\nProcessing Record 16 of Set 11 : katherine\nProcessing Record 17 of Set 11 : ngunguru\nProcessing Record 18 of Set 11 : palmer\nProcessing Record 19 of Set 11 : doka\nCity not found. Skipping...\nProcessing Record 20 of Set 11 : pryozerne\nProcessing Record 21 of Set 11 : alice springs\nProcessing Record 22 of Set 11 : diamantino\nProcessing Record 23 of Set 11 : bethel\nCity not found. Skipping...\nProcessing Record 24 of Set 11 : port moresby\nProcessing Record 25 of Set 11 : kamaishi\nProcessing Record 26 of Set 11 : ixtapa\nProcessing Record 27 of Set 11 : bogorodskoye\nProcessing Record 28 of Set 11 : wasilla\nProcessing Record 29 of Set 11 : khuzdar\nCity not found. Skipping...\nProcessing Record 30 of Set 11 : charters towers\nProcessing Record 31 of Set 11 : marysville\nProcessing Record 32 of Set 11 : road town\nProcessing Record 33 of Set 11 : bac lieu\nProcessing Record 34 of Set 11 : san ignacio\nProcessing Record 35 of Set 11 : pisco\nProcessing Record 36 of Set 11 : khasan\nProcessing Record 37 of Set 11 : amahai\nProcessing Record 38 of Set 11 : panzhihua\nProcessing Record 39 of Set 11 : bakau\nProcessing Record 40 of Set 11 : massakory\nProcessing Record 41 of Set 11 : santa barbara\nProcessing Record 42 of Set 11 : wattegama\nProcessing Record 43 of Set 11 : unity\nProcessing Record 44 of Set 11 : san policarpo\nProcessing Record 45 of Set 11 : ponta delgada\nProcessing Record 46 of Set 11 : san isidro\nProcessing Record 47 of Set 11 : warmbad\nCity not found. Skipping...\nCity not found. Skipping...\nProcessing Record 48 of Set 11 : pimenta bueno\nProcessing Record 49 of Set 11 : ust-kuyga\nProcessing Record 50 of Set 11 : gampaha\nCity not found. Skipping...\nProcessing Record 0 of Set 12 : pitkyaranta\nProcessing Record 1 of Set 12 : gornopravdinsk\nProcessing Record 2 of Set 12 : guangyuan\nProcessing Record 3 of Set 12 : booue\nProcessing Record 4 of Set 12 : kyaikto\nCity not found. Skipping...\nProcessing Record 5 of Set 12 : wamba\n"
],
[
"weather_dictionary = {\n \"City\": city,\n \"Cloudiness\": cloudiness,\n \"Country\": country,\n \"Date\": date,\n \"Humidity\": humidity,\n \"Lat\": latitude_list,\n \"Lng\": longitude_list,\n \"Max Temp\": maximum_temp,\n \"Wind Speed\": wind_speed\n}",
"_____no_output_____"
],
[
"weather_dataframe = pd.DataFrame(weather_dictionary)\nweather_dataframe.head(10)\nweather_dataframe.to_csv(\"weather_df.csv\", index=False)",
"_____no_output_____"
]
],
[
[
"#Plotting the Data \nUse proper labeling of the plots using plot titles (including date of analysis) and axes labels.\nSave the plotted figures as .pngs.\n",
"_____no_output_____"
]
],
[
[
"#Latitude vs. Temperature Plot\nplt.scatter(weather_dictionary[\"Lat\"], weather_dictionary[\"Max Temp\"], facecolor = \"lightblue\", edgecolor = \"black\")\nplt.title(\"City Latitude vs. Max Temperature (01/17/20)\")\nplt.xlabel(\"Latitude\")\nplt.ylabel(\"Maximum Temperature (F)\")\nplt.grid(linestyle='-', linewidth=1, alpha = 0.5)\n# Save the plotted figure as .pngs\nplt.savefig(\"Latitude vs Max Temperature.png\")\n",
"_____no_output_____"
]
],
[
[
"Observation- Latitude vs Max Temperature.png:\nAs the latitude increases, temperature drops and the maximum temp. is found around 0 latitude.",
"_____no_output_____"
]
],
[
[
"#Latitude vs. Humidity Plot\nplt.scatter(weather_dataframe[\"Lat\"], weather_dataframe[\"Humidity\"], marker='o', s=30, edgecolors= \"black\")\nplt.title(\"City Latitude vs Humidity\")\nplt.ylabel(\"Humidity Level (%)\")\nplt.xlabel(\"Latitude\")\nplt.grid()\nplt.savefig(\"Latitude vs Humidity.png\")",
"_____no_output_____"
]
],
[
[
"Observation- Latitude vs Humidity:\nAs the latitude gets higher,humidity gets higher too",
"_____no_output_____"
]
],
[
[
"#Latitude vs. Cloudiness Plot\nplt.scatter(weather_dataframe[\"Lat\"], weather_dataframe[\"Cloudiness\"], marker='o', s=30, edgecolors= \"black\")\nplt.title(\"City Latitude vs Cloudiness\")\nplt.ylabel(\"Cloudiness Level (%)\")\nplt.xlabel(\"Latitude\")\nplt.grid()\n# plt.show()\nplt.savefig(\"Latitude vs Cloudiness.png\")",
"_____no_output_____"
]
],
[
[
"Observation- Latitude vs Cloudiness: Cloudiness is all over the latitude.\n",
"_____no_output_____"
]
],
[
[
"#Latitude vs. Wind Speed Plot\nplt.scatter(weather_dataframe[\"Lat\"], weather_dataframe[\"Wind Speed\"], marker='o', s=30, edgecolors= \"black\")\nplt.title(\"City Latitude vs Wind Speed\")\nplt.ylabel(\"Wind Speed (mph)\")\nplt.xlabel(\"Latitude\")\nplt.grid()\nplt.savefig(\"Latitude vs Wind Speed.png\")",
"_____no_output_____"
]
],
[
[
"Observation- Latitude vs Wind Speed:Wind Speed is present across the latitude\n",
"_____no_output_____"
]
],
[
[
"#Linear Regression\n\n# Create Northern and Southern Hemisphere DataFrames\nnorthern_hemisphere = weather_dataframe.loc[weather_dataframe[\"Lat\"] >= 0]\nsouthern_hemisphere = weather_dataframe.loc[weather_dataframe[\"Lat\"] < 0]",
"_____no_output_____"
],
[
"#Northern Hemisphere - Max Temp vs. Latitude Linear Regression\n\n#Create a Scatter Plot for Lattitude vs Temperature of City\nx_values = northern_hemisphere['Lat']\ny_values = northern_hemisphere['Max Temp']\n(slope, intercept, rvalue, pvalue, stderr) = linregress(x_values, y_values)\nreg_values = x_values * slope + intercept\nline_eq = \"y = \" + str(round(slope,2)) + \"x + \" + str(round(intercept,2))\nplt.scatter(x_values,y_values)\nplt.plot(x_values,reg_values,\"red\")\nplt.annotate(line_eq,(5,10),fontsize=15,color=\"red\")\nplt.ylim(0,100)\nplt.xlim(0, 80)\n\nplt.ylabel(\"Max. Temp\")\nplt.xlabel(\"Latitude\")\nplt.savefig(\"North Max Temp vs Latitude Regression.png\")",
"_____no_output_____"
]
],
[
[
"\nObservation: negative correlation between latitude and Max Temperature in Northern Hemisphere",
"_____no_output_____"
]
],
[
[
"#Southern Hemisphere - Max Temp vs. Latitude Linear Regression\n\n#Create a Scatter Plot for Lattitude vs Temperature of City (Southern Hemisphere)\nx_values = southern_hemisphere['Lat']\ny_values = southern_hemisphere['Max Temp']\nreg_values = x_values * slope + intercept\nline_eq = \"y = \" + str(round(slope,2)) + \"x + \" + str(round(intercept,2))\nplt.scatter(x_values,y_values)\nplt.plot(x_values,reg_values,\"r-\")\nplt.annotate(line_eq,(6,10),fontsize=15,color=\"red\")\nplt.ylim(30, 100)\nplt.xlim(-60, 0, 10)\n\nplt.ylabel(\"Max. Temp\")\nplt.xlabel(\"Latitude\")\nplt.savefig(\"South Max Temp vs Latitude Regression.png\")",
"_____no_output_____"
],
[
"#Northern Hemisphere - Humidity (%) vs. Latitude Linear Regression\nx_values = northern_hemisphere['Lat']\ny_values = northern_hemisphere['Humidity']\nreg_values = x_values * slope + intercept\nline_eq = \"y = \" + str(round(slope,2)) + \"x + \" + str(round(intercept,2))\nplt.scatter(x_values,y_values)\nplt.plot(x_values,reg_values,\"r-\")\nplt.annotate(line_eq,(6,10),fontsize=15,color=\"red\")\nplt.ylabel(\"Humidity\")\nplt.xlabel(\"Latitude\")\nplt.savefig(\"North Humidity vs Latitude Linear Regression.png\")",
"_____no_output_____"
]
],
[
[
"Observation: Negative constant correlation\n",
"_____no_output_____"
]
],
[
[
"#Southern Hemisphere - Humidity (%) vs. Latitude Linear Regression\n\nx_values = southern_hemisphere['Lat']\ny_values = southern_hemisphere['Humidity']\nreg_values = x_values * slope + intercept\nline_eq = \"y = \" + str(round(slope,2)) + \"x + \" + str(round(intercept,2))\nplt.scatter(x_values,y_values)\nplt.plot(x_values,reg_values,\"r-\")\nplt.annotate(line_eq,(-25,10),fontsize=15,color=\"red\")\n\nplt.ylim(0, 100)\n\nplt.ylabel(\"Humidity\")\nplt.xlabel(\"Latitude\")\nplt.savefig(\"South Humidity vs Latitude Linear Regression.png\")",
"_____no_output_____"
],
[
"#Northern Hemisphere - Cloudiness (%) vs. Latitude Linear Regression\nx_values = northern_hemisphere['Lat']\ny_values = northern_hemisphere['Cloudiness']\nreg_values = x_values * slope + intercept\nline_eq = \"y = \" + str(round(slope,2)) + \"x + \" + str(round(intercept,2))\nplt.scatter(x_values,y_values)\nplt.plot(x_values,reg_values,\"r-\")\nplt.annotate(line_eq,(6,10),fontsize=15,color=\"red\")\nplt.ylabel(\"Cloudiness\")\nplt.xlabel(\"Latitude\")\nplt.savefig(\"North Cloudiness vs Latitude Linear Regression.png\")",
"_____no_output_____"
],
[
"#Southern Hemisphere - Cloudiness (%) vs. Latitude Linear Regression\nx_values = southern_hemisphere['Lat']\ny_values = southern_hemisphere['Cloudiness']\nreg_values = x_values * slope + intercept\nline_eq = \"y = \" + str(round(slope,2)) + \"x + \" + str(round(intercept,2))\nplt.scatter(x_values,y_values)\nplt.plot(x_values,reg_values,\"r-\")\nplt.annotate(line_eq,(-25,10),fontsize=15,color=\"red\")\nplt.ylabel(\"Cloudiness\")\nplt.xlabel(\"Latitude\")\nplt.savefig(\"South Cloudiness vs Latitude Linear Regression.png\")",
"_____no_output_____"
],
[
"#Northern Hemisphere - Wind Speed (mph) vs. Latitude Linear Regression\nx_values = northern_hemisphere['Lat']\ny_values = northern_hemisphere['Wind Speed']\n(slope, intercept, rvalue, pvalue, stderr) = linregress(x_values, y_values)\nreg_values = x_values * slope + intercept\nline_eq = \"y = \" + str(round(slope,2)) + \"x + \" + str(round(intercept,2))\nplt.scatter(x_values,y_values)\nplt.plot(x_values,reg_values,\"r-\")\nplt.annotate(line_eq,(45,22),fontsize=15,color=\"red\")\nplt.ylabel(\"Cloudiness\")\nplt.xlabel(\"Latitude\")\nplt.savefig(\"North Wind speed vs Latitude Linear Regression.png\")",
"_____no_output_____"
]
],
[
[
"Observation: low positive correlation",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
cbece18c2a3515ca46cd3c313797a77024f59c77
| 155,370 |
ipynb
|
Jupyter Notebook
|
playground/beat_toedtli/word2word_matching/evaluate_word2wordmatcher.ipynb
|
archity/rg_text_to_sound
|
acf54e680661d378dbe79ba25b6c966d1ff10f88
|
[
"MIT"
] | 1 |
2021-07-11T10:57:11.000Z
|
2021-07-11T10:57:11.000Z
|
playground/beat_toedtli/word2word_matching/evaluate_word2wordmatcher.ipynb
|
archity/rg_text_to_sound
|
acf54e680661d378dbe79ba25b6c966d1ff10f88
|
[
"MIT"
] | 2 |
2021-02-12T14:16:11.000Z
|
2021-10-03T10:58:46.000Z
|
playground/beat_toedtli/word2word_matching/evaluate_word2wordmatcher.ipynb
|
archity/rg_text_to_sound
|
acf54e680661d378dbe79ba25b6c966d1ff10f88
|
[
"MIT"
] | 7 |
2021-02-12T14:14:16.000Z
|
2021-05-22T15:21:31.000Z
| 127.666393 | 63,057 | 0.839351 |
[
[
[
"## Next Task: compute precision and recall\n\nthreshold 25: zoomy, sustain->thick, smooth (user results) \nzoomy, sustain -> dark, smooth (word2word matcher resuts) \nsmooth tp \ndark fp \nthik tn (fn?) \n\nprecision = tp/(tp+fp) \nrecall = tp/(tp+fn) \n\nfor one word, cant compute recall \nlater: tensorflow language models, Optimising (Kullback-Leibler) for the distribution",
"_____no_output_____"
],
[
"However, note: \n\n Let A and B be any sets with |A|=|B| (|.| being the set cardinality, i.e. number of elements in the set). It follows that\n fp = |B\\A∩B|=|B|-|A∩B| = |A|-|A∩B| = |A\\A∩B|=fn.\n It hence follows that\n precision = tp/(tp+fp)=tp/(tp+fn)=recall\n I understood your definition\n \"A is the set of words in our ground truth, when you apply a threshold to the sliders\n B is the set of words from the output of our words matcher\"\n in a way such that |A|=|B|",
"_____no_output_____"
]
],
[
[
"import sys\nimport ipdb\nimport pandas as pd\nimport numpy as np\nfrom tqdm import tqdm\nsys.path.append(r'C:\\Temp\\SoundOfAI\\rg_text_to_sound\\tts_pipeline\\src')\n\nfrom match_word_to_words import prepare_dataset,word_to_wordpair_estimator,word_to_words_matcher,prepare_dataset\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"df = pd.read_csv('text_to_qualities.csv')\ncolnames = df.columns\ndisplay(df.head(2))\ndf.shape",
"_____no_output_____"
],
[
"df = pd.read_csv('text_to_qualities.csv')\ndfnew[dfnew.description.str.match('\\'')]\ndfnew['description'] = dfnew.description.str.replace(\"'\",\"\")\ndfnew['description']=dfnew.description.str.lower().str.replace('(\\(not.*\\))','',regex=True)\ndfnew = dfnew[~dfnew.description.str.match('\\(.*\\)')]\ndfnew.head()",
"_____no_output_____"
],
[
"wordlist = dfnew.description\nunique_word_list = np.unique(wordlist).tolist()\nlen(wordlist),len(unique_word_list)",
"_____no_output_____"
]
],
[
[
"threshold 25: zoomy, sustain->thick, smooth (user results) \nzoomy, sustain -> dark, smooth (word2word matcher resuts) \nsmooth tp \ndark fp \nthik tn \n\nprecision = tp/(tp+fp) \nrecall = tp/(tp+fn) \n\nfor one word, cant compute recall",
"_____no_output_____"
],
[
"# word pair estimator",
"_____no_output_____"
]
],
[
[
"df_score",
"_____no_output_____"
],
[
"df_score = dfnew.iloc[:,1:]\n\ndescriptions = dfnew.iloc[:,0]\nwordpairnames = df_score.columns.tolist()\ndf_score.head()",
"_____no_output_____"
],
[
"target_word_pairs = [('bright', 'dark'), ('full', 'hollow'),( 'smooth', 'rough'), ('warm', 'metallic'), ('clear', 'muddy'), ('thin', 'thick'), ('pure', 'noisy'), ('rich', 'sparse'), ('soft', 'hard')]\nwordpairnames_to_wordpair_dict = {s:t for s,t in zip(wordpairnames,target_word_pairs)}\nwordpairnames_to_wordpair_dict",
"_____no_output_____"
],
[
"list(np.arange(49.8,50,0.1))\nA=set([1,2,3])\nB=set([3,4,5])\nAandB = A.intersection(B)\n\nB.difference(AandB)",
"_____no_output_____"
],
[
"def single_word_precision_recall(word,scorerow,threshold,w2wpe,wordpairnames_to_wordpair_dict):\n elems_above = scorerow[(scorerow>(100-threshold)) ]\n elems_below = scorerow[(scorerow<=threshold) ]\n words_above = [wordpairnames_to_wordpair_dict[wordpairname][1] for wordpairname in elems_above.index]\n words_below = [wordpairnames_to_wordpair_dict[wordpairname][0] for wordpairname in elems_below.index]\n A = set(words_above+words_below)\n opposite_pairs_beyond_threshold = elems_above.index.tolist()+elems_below.index.tolist()\n B = set([w2wpe.match_word_to_wordpair(word,ind)['closest word'] for ind in opposite_pairs_beyond_threshold])\n assert len(A)==len(B), 'This should never occurr!'\n \n AandB = set(A).intersection(B)\n tp = AandB\n fp = B.difference(AandB) # were found but shouldn't have been\n fn = A.difference(AandB) # were not found but should have been\n\n den = len(tp)+len(fp)\n if den==0:\n precision = np.NaN\n else:\n precision = len(tp)/den\n\n den = len(tp)+len(fn)\n if den==0:\n recall = np.NaN\n else:\n recall = len(tp)/den\n \n if precision!=recall and not np.isnan(precision): \n print('This should never occur!')\n print('word, A,B,AandB,tp,fp,fn,precision,recall')\n print(word, A,B,AandB,tp,fp,fn,precision,recall)\n return precision,recall,len(A)",
"_____no_output_____"
],
[
"lang_model='en_core_web_sm'\nw2wpe = word_to_wordpair_estimator()\nw2wpe.build(wordpairnames,target_word_pairs,lang_model=lang_model)\nw2wpe.match_word_to_wordpair('full','full_vs_hollow')",
"_____no_output_____"
],
[
"word = descriptions[0]\nscorerow = df_score.iloc[0,:]\nprec_50_list=[]\nNrRelevantWordpairList=[]\nfor word, (irow,scorerow) in tqdm(zip(descriptions, df_score.iterrows())):\n prec,rec,NrRelevantWordpairs = single_word_precision_recall(word,scorerow,10,w2wpe,wordpairnames_to_wordpair_dict)\n prec_50_list.append(prec)\n NrRelevantWordpairList.append(NrRelevantWordpairs)",
"668it [00:03, 184.66it/s]\n"
],
[
"pd.Series(prec_50_list).dropna()",
"_____no_output_____"
],
[
"\n\nlen(prec_50_list),np.mean(prec_50_list)",
"_____no_output_____"
],
[
"\n' '.join([f'{i:1.1f}' for i in thresholdlist])",
"_____no_output_____"
],
[
"def compute_accuracy(lang_model='en_core_web_lg',thresholdlist=None):\n w2wpe = word_to_wordpair_estimator()\n w2wpe.build(wordpairnames,target_word_pairs,lang_model=lang_model)\n if thresholdlist is None:\n thresholdlist = list(np.arange(0,50,2))+list(np.arange(45,50,0.5))+[50.]\n\n mean_accuracy_list = []\n nrrelevantlist = []\n for threshold in tqdm(thresholdlist):\n acc_list=[]\n NrRelevantWordpairList=[]\n for word, (irow,scorerow) in zip(descriptions, df_score.iterrows()):\n precision,recall,NrRelevantWordpairs = single_word_precision_recall(word,scorerow,threshold,w2wpe,wordpairnames_to_wordpair_dict)\n acc_list.append(precision)\n NrRelevantWordpairList.append(NrRelevantWordpairs)\n assert len(acc_list)>0, 'something is wrong...'\n meanAccuracyVal = pd.Series(acc_list).dropna().mean()\n NrRelevantVal = np.mean(NrRelevantWordpairList)\n\n mean_accuracy_list.append(meanAccuracyVal)\n nrrelevantlist.append(NrRelevantVal)\n return mean_accuracy_list,nrrelevantlist",
"_____no_output_____"
],
[
"%time\nlang_model1 = 'en_core_web_sm'\nlang_model2 = 'en_core_web_lg'\nmean_accuracy_list1,nrrelevantlist1 = compute_accuracy(lang_model=lang_model1)\nmean_accuracy_list2,nrrelevantlist2 = compute_accuracy(lang_model=lang_model2)",
"Wall time: 0 ns\n"
],
[
"lang_model3 = 'en_core_web_md'\nthresholdlist = list(np.arange(0,50,2))+list(np.arange(45,50,0.5))+[50.]\nmean_accuracy_list3,nrrelevantlist3 = compute_accuracy(lang_model=lang_model3,thresholdlist=thresholdlist)",
"100%|██████████████████████████████████████████████████████████████████████████████████| 36/36 [17:52<00:00, 29.80s/it]\n"
],
[
"from nltk.corpus import wordnet\n \n# Then, we're going to use the term \"program\" to find synsets like so:\nsyns = wordnet.synsets(\"program\")",
"_____no_output_____"
],
[
"if np.all(np.isclose(np.array(nrrelevantlist1),np.array(nrrelevantlist2))):\n nrrelevantlist = nrrelevantlist1",
"_____no_output_____"
],
[
"plt.figure(1,figsize=(15,7))\nplt.subplot(3,1,1)\nplt.plot(thresholdlist,mean_accuracy_list1,marker='o',label='Accuracy')\nplt.suptitle(f'Accuracy vs. Threshold\\nWords considered have (score <= threshold) or (score > 100-threshold)')\nplt.title(f'Accuracy of {lang_model1}')\nplt.ylabel('Accuracy')\nplt.legend()\n\nplt.subplot(2,1,2)\nplt.plot(thresholdlist,mean_accuracy_list2,marker='o',label='Accuracy')\nplt.title(f'Accuracy of {lang_model2}')\nplt.ylabel('Accuracy')\nplt.legend()\n\nplt.subplot(3,1,3)\nplt.plot(thresholdlist,nrrelevantlist,marker='o')\nplt.title('Average number of relevant sliders')\nplt.xlabel('threshold value')\nplt.ylabel('Nr of Sliders')\nplt.yticks(np.arange(1,10,2))\nplt.subplots_adjust(hspace=.6)",
"_____no_output_____"
],
[
"plt.figure(1,figsize=(15,7))\nplt.subplot(1,1,1)\nplt.plot(thresholdlist,mean_accuracy_list3,marker='o',label='Accuracy')\nplt.suptitle(f'Accuracy vs. Threshold\\nWords considered have (score <= threshold) or (score > 100-threshold)')\nplt.title(f'Accuracy of {lang_model3}')\nplt.ylabel('Accuracy')\nplt.legend()",
"_____no_output_____"
],
[
"plt.figure(1,figsize=(15,7))\nplt.subplot(2,1,1)\nplt.plot(thresholdlist,mean_accuracy_list1,marker='o',label=f'Accuracy of {lang_model1}')\nplt.plot(thresholdlist,mean_accuracy_list2,marker='o',label=f'Accuracy of {lang_model2}')\nplt.suptitle(f'Accuracy vs. Threshold\\nWords considered have (score <= threshold) or (score > 100-threshold)')\nplt.ylabel('Accuracy')\nplt.legend()\n\nplt.subplot(2,1,2)\n\nplt.plot(thresholdlist,nrrelevantlist,marker='o')\nplt.title('Average number of relevant sliders')\nplt.xlabel('threshold value')\nplt.ylabel('Nr of Sliders')\nplt.yticks(np.arange(1,10,2))\nplt.subplots_adjust(hspace=.6)\nplt.savefig('Accuracy_vs_Threshold.svg')",
"_____no_output_____"
],
[
"row\nlang_model = 'en_core_web_sm'\n\nw2wpe = word_to_wordpair_estimator()\nw2wpe.build(wordpairnames,target_word_pairs,lang_model=lang_model)\nprediction_dict = w2wpe.match_word_to_wordpair(word,ind)\nind,prediction_dict[]",
"_____no_output_____"
],
[
"ind,w2wpe.match_word_to_wordpair(word,ind)",
"_____no_output_____"
],
[
"\ndef compute_mean_acc(dfnew,df_score,thresholdmargin,threshold=50, required_confidence=0, lang_model='en_core_web_sm'):\n \"\"\"\n Take the opposite quality pairs for which the slider value is outside the 50+/- <thresholdmargin> band. \n Compute the accuracy in predicting the correct opposite-pair word for each such pair. \n threshold: where to split a score to lower or upper quality in pair: 50 is the most natural value.\n The prediction must be with a (minimum) < required_confidence > otherwise the prediction is deemed unsure.\n The returned accuracy is computed as \n accuracy = NrCorrect/(NrCorrect+NrWrong+NrUnsure)\n averaged over all words in <dfnew>.description\n \"\"\"\n \n w2wpe = word_to_wordpair_estimator()\n w2wpe.build(wordpairnames,target_word_pairs,lang_model=lang_model)\n acc_list = []\n unsure_list = []\n NrCorrect = 0\n NrWrong = 0\n NrUnsure = 0\n for word, (irow,scorerow) in zip(dfnew.description, df_score.iterrows()):\n #determine which opposite quality pairs will be correctly predicted as the first and second word in the word pair, respectively\n valid_qualities = scorerow[(scorerow > threshold+thresholdmargin )|(scorerow < threshold-thresholdmargin)]\n \n below_th = valid_qualities[valid_qualities<threshold].index.tolist()#first word in the word pair is correct\n above_th = valid_qualities[valid_qualities>threshold].index.tolist()#second word in the word pair is correct\n \n #word_pair_tuple = wordpairnames_to_wordpair_dict[word_pair]\n \n \n NrCorrect = 0\n NrWrong = 0\n NrUnsure = 0\n for word_pair in above_th:\n res = w2wpe.match_word_to_wordpair(word,word_pair) \n if res['slider value']>(threshold+required_confidence):# Add prediction threshold?\n NrCorrect+=1\n elif res['slider value']<(threshold-required_confidence):\n NrWrong+=1 \n else:\n NrUnsure+=1 #if required confidence was not reached\n for word_pair in below_th:\n res = w2wpe.match_word_to_wordpair(word,word_pair) \n if res['slider value']<(threshold-required_confidence):# Add prediction threshold?\n NrCorrect+=1\n elif res['slider value']>threshold+required_confidence:\n NrWrong+=1\n else:\n NrUnsure+=1 #if required confidence was not reached\n if len(below_th)+len(above_th)==0: continue\n accuracy = NrCorrect/(NrCorrect+NrWrong+NrUnsure)\n unsure_ratio = NrUnsure/(NrCorrect+NrWrong+NrUnsure) # the fraction of cases where the prediction did not reach the required confidence\n acc_list.append(accuracy)\n unsure_list.append(unsure_ratio)\n #resdict = {'NrCorrect':NrCorrect, 'NrWrong':NrWrong, 'NrUnsure':NrUnsure}\n mean_acc = np.mean(acc_list) #list of accuracies for each word, over all available sliders\n mean_unsure = np.mean(unsure_list)\n del w2wpe\n return mean_acc,mean_unsure",
"_____no_output_____"
],
[
"def f():\n ipdb.set_trace()\n return wordpair_matcher_dict['bright_vs_dark'].match_word_to_words('sunny')\nf()",
"_____no_output_____"
],
[
"y = np.array([np.where(row['bright_vs_dark']>=50,1,0) for row in rowlist])\ny.shape,yhat1.shape",
"_____no_output_____"
],
[
"yhat_binary = np.array([0 if yhatelem==target_word_pair[0] else 1 for yhatelem in yhat1])\nyhat_binary.shape",
"_____no_output_____"
],
[
"len(yhat),len(rowlist)",
"_____no_output_____"
],
[
"accuracy_score(y,yhat_binary)",
"_____no_output_____"
],
[
"yhat1",
"_____no_output_____"
],
[
"df_detailed = pd.DataFrame(index=wordlist)\ndf_detailed.head(7)",
"_____no_output_____"
],
[
"wordlist = [w for r,w in generate_training_examples(df)]\nrowlist = [r for r,w in generate_training_examples(df)]\n\nacc_scores=dict()\nfor target_word_pair,opposite_quality_pair in zip(target_word_pairs,colnames):\n y = np.array([np.where(row[opposite_quality_pair]>=50,1,0) for row in rowlist])\n \n print(target_word_pair,opposite_quality_pair)\n w2wm = word_to_words_matcher()\n w2wm.build(target_word_pair)\n yhat1 = np.array(f(wordlist,w2wm,variant=1))\n df_detailed[opposite_quality_pair] = yhat1\n yhat_binary = np.array([0 if yhatelem==target_word_pair[0] else 1 for yhatelem in yhat1])\n acc_score = accuracy_score(y,yhat_binary)\n print(f'{acc_score:1.3f}')\n acc_scores[opposite_quality_pair] = acc_score",
"_____no_output_____"
],
[
"print(df_detailed.shape)\ndf_detailed.to_excel('predicted_qualities.xlsx')\ndf_detailed.head(20)",
"_____no_output_____"
],
[
"pd.Series(acc_scores).plot.bar(ylabel='accuracy')\nplt.plot(plt.xlim(),[0.5,0.5],'--',c='k')\nplt.title(f'Accuracy of Spacy word vectors in predicting\\ntext_to_qualities.csv ({len(wordlist)} qualities)')\nplt.ylim(0,1)",
"_____no_output_____"
]
],
[
[
"## Next Task: compute precision and recall\n\nthreshold 25: zoomy, sustain->thick, smooth (user results) \nzoomy, sustain -> dark, smooth (word2word matcher resuts) \nsmooth tp \ndark fp \nthik tn \n\nprecision = tp/(tp+fp) \nrecall = tp/(tp+fn) \n\nfor one word, cant compute recall \nlater: tensorflow language models, Optimising (Kullback-Leibler) for the distribution",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
cbece2000aa23510d353a7331a5fb8bdc697dd47
| 3,469 |
ipynb
|
Jupyter Notebook
|
tensorflow_examples/lite/model_maker/demo/text_classification.ipynb
|
riyaj5246/skin-cancer-with-tflite
|
2a3fbd58deb0984c1224464638ec7f45f7e1fd9b
|
[
"Apache-2.0"
] | 6,484 |
2019-02-13T21:32:29.000Z
|
2022-03-31T20:50:20.000Z
|
tensorflow_examples/lite/model_maker/demo/text_classification.ipynb
|
riyaj5246/skin-cancer-with-tflite
|
2a3fbd58deb0984c1224464638ec7f45f7e1fd9b
|
[
"Apache-2.0"
] | 288 |
2019-02-13T22:56:03.000Z
|
2022-03-24T11:15:19.000Z
|
tensorflow_examples/lite/model_maker/demo/text_classification.ipynb
|
riyaj5246/skin-cancer-with-tflite
|
2a3fbd58deb0984c1224464638ec7f45f7e1fd9b
|
[
"Apache-2.0"
] | 7,222 |
2019-02-13T21:39:34.000Z
|
2022-03-31T22:23:54.000Z
| 38.544444 | 314 | 0.59729 |
[
[
[
"##### Copyright 2019 The TensorFlow Authors.",
"_____no_output_____"
]
],
[
[
"#@title Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# https://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.",
"_____no_output_____"
]
],
[
[
"# Text classification with TensorFlow Lite Model Maker with TensorFlow 2.0",
"_____no_output_____"
],
[
"<table class=\"tfo-notebook-buttons\" align=\"left\">\n <td>\n <a target=\"_blank\" href=\"https://www.tensorflow.org/lite/tutorials/model_maker_text_classification\"><img src=\"https://www.tensorflow.org/images/tf_logo_32px.png\" />View on TensorFlow.org</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://colab.research.google.com/github/tensorflow/tensorflow/blob/master/tensorflow/lite/g3doc/tutorials/model_maker_text_classification.ipynb\"><img src=\"https://www.tensorflow.org/images/colab_logo_32px.png\" />Run in Google Colab</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://github.com/tensorflow/tensorflow/blob/master/tensorflow/lite/g3doc/tutorials/model_maker_text_classification.ipynb\"><img src=\"https://www.tensorflow.org/images/GitHub-Mark-32px.png\" />View source on GitHub</a>\n </td>\n <td>\n <a href=\"https://storage.googleapis.com/tensorflow_docs/tensorflow/tensorflow/lite/g3doc/tutorials/model_maker_text_classification.ipynb\"><img src=\"https://www.tensorflow.org/images/download_logo_32px.png\" />Download notebook</a>\n </td>\n</table>",
"_____no_output_____"
],
[
"This notebook has been moved [here](https://www.tensorflow.org/lite/tutorials/model_maker_text_classification).",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
cbece4845f92fed2025983867ff97e7221b1d404
| 45,481 |
ipynb
|
Jupyter Notebook
|
.ipynb_checkpoints/Estudando com Movies On Streaming Platforms-checkpoint.ipynb
|
wander-asb/Analise-de-titulos-de-plataformas-de-streaming
|
791b6fb37500f5c031ae66061fae44ba210e1f05
|
[
"MIT"
] | null | null | null |
.ipynb_checkpoints/Estudando com Movies On Streaming Platforms-checkpoint.ipynb
|
wander-asb/Analise-de-titulos-de-plataformas-de-streaming
|
791b6fb37500f5c031ae66061fae44ba210e1f05
|
[
"MIT"
] | null | null | null |
.ipynb_checkpoints/Estudando com Movies On Streaming Platforms-checkpoint.ipynb
|
wander-asb/Analise-de-titulos-de-plataformas-de-streaming
|
791b6fb37500f5c031ae66061fae44ba210e1f05
|
[
"MIT"
] | null | null | null | 43.150854 | 12,168 | 0.568589 |
[
[
[
"## Questões:\n1. Qual plataforma possui mais títulos incluídos ao catálogo ?\n2. Após a visualização do número de títulos em cada plataforma, desejo saber quantos filmes e séries presentes na Netflix estão também vinculados à outras plataformas de streaming, e assim por diante.\n3. Rankear pela nota do IMDb, retirando os dados ausentes da coluna, qual plataforma é melhor para o cliente assinar.",
"_____no_output_____"
],
[
"### Importação de bibliotecas:",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport seaborn as sns",
"_____no_output_____"
]
],
[
[
"### Leitura de Arquivos",
"_____no_output_____"
]
],
[
[
"df_dados = pd.read_csv(\"MoviesOnStreamingPlatforms_updated.csv\")\ndf_dados.head()",
"_____no_output_____"
]
],
[
[
"### Descarte de colunas e Reindexação",
"_____no_output_____"
]
],
[
[
"#Apagar colunas que são desnecessárias utilizando a função .drop():\ndf_dados.drop(['Unnamed: 0','ID','Type'], axis = 1, inplace=True)\ndf_dados.head()",
"_____no_output_____"
]
],
[
[
"### Análise de valores faltantes",
"_____no_output_____"
]
],
[
[
"#Verificar possíveis dados faltantes\ndf_dados.isnull().sum()",
"_____no_output_____"
]
],
[
[
"- Em motivo de aprendizado maior, irei trabalhar com indexação entre minhas colunas úteis;\n- Decidir fazer isso por motivo de tentar entender o método, não necessariamente atrapalharia ou melhoraria o levantamento de respostas das minhas hipóteses",
"_____no_output_____"
]
],
[
[
"nome_colunas = ['Title','Year','Age','IMDb','Rotten Tomatoes','Netflix','Hulu','Prime Video','Disney+',\n 'Directors','Genres','Country','Language','Runtime']",
"_____no_output_____"
],
[
"#Criei uma lista com o nome das colunas de meu DataFrame, porém, em ordem que eu me sinta confortável\ndf_dados = df_dados.reindex(columns=nome_colunas)",
"_____no_output_____"
]
],
[
[
"### Seleção com loc e iloc\n- Para selecionar subconjuntos de linhas e colunas do meu DataFrame, utilizarei .loc e .iloc, na qual são reconhecidas pelos rótulos de eixo(**.loc**) e inteiro(**.iloc**).\n\n- Para entendermos a estrutura de seleção loc, vamos selecionar as 15 primeiras linhas e pedir apenas as colunas de título e diretors:\n",
"_____no_output_____"
]
],
[
[
"df_dados.loc[0:15, ['Title','Directors']]",
"_____no_output_____"
]
],
[
[
"### Qual plataforma possui mais títulos incluídos ao catálogo:",
"_____no_output_____"
]
],
[
[
"#Filmes vinculados à Netflix\nnetflix_data = df_dados.loc[df_dados['Netflix'] == 1]",
"_____no_output_____"
],
[
"#Filmes vinculados à Hulu\nhulu_data = df_dados.loc[df_dados['Hulu'] ==1]",
"_____no_output_____"
],
[
"#Filmes vinculados à Prime Video\nprime_data = df_dados.loc[df_dados['Prime Video'] ==1]",
"_____no_output_____"
],
[
"#Filmes vinculados à Disney+\ndisney_data = df_dados.loc[df_dados['Disney+'] ==1]",
"_____no_output_____"
]
],
[
[
"### Criação de listas para armazenar o número de títulos pertecentes a cada plataforma de stream:",
"_____no_output_____"
]
],
[
[
"# Eu poderia criar uma variável para cada filtro, mas decidir visualizar as listas durante o processo\nnumero_titulos = [netflix_data['Title'].count(), hulu_data['Title'].count(),\n prime_data['Title'].count() , disney_data['Title'].count()]\n\n# Lista de nomes de cada plataforma:\nnomes_plataformas = ['Netflix','Hulu','Prime Video','Disney+']",
"_____no_output_____"
]
],
[
[
"### Criação de gráfico de barras para comparação:",
"_____no_output_____"
]
],
[
[
"#Construção de plotagem do gráfico de barras:\nplt.bar(nomes_plataformas, numero_titulos, color='Blue')\n\n#Título do gráfico\nplt.title('Número de títulos x Plataformas')\n\n#A label para o eixo Y:\nplt.ylabel('Número de Títulos')\n\n#A label para o eixo X:\nplt.xlabel('Plataformas')\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"- A partir do gráfico acima, podemos observar que a plataforma que mais possui títulos é a Amazom Prime Video, com cerca de mais de 12.000 títulos na plataforma",
"_____no_output_____"
],
[
"### Após a visualização do número de títulos em cada plataforma, desejo saber quantos filmes e séries presentes na Netflix estão também vinculados à outras plataformas de streaming, e assim por diante:",
"_____no_output_____"
]
],
[
[
"netflix_data[['Netflix','Hulu','Prime Video','Disney+']].sum()",
"_____no_output_____"
]
],
[
[
"##### Na plataforma da Netflix, dos 3560 títulos, podemos perceber que :\n1. 25 títulos presentes na Hulu\n2. 345 títulos presentes na Prime Video\n3. 10 títulos presentes na Disney+",
"_____no_output_____"
]
],
[
[
"hulu_data[['Netflix','Hulu','Prime Video','Disney+']].sum()",
"_____no_output_____"
]
],
[
[
"##### Na plataforma da Hulu, dos 903 títulos, podemos perceber que :\n1. 25 títulos presentes na Netflix\n2. 241 títulos presentes na Prime Video\n3. 7 títulos presentes na Disney+",
"_____no_output_____"
]
],
[
[
"prime_data[['Netflix','Hulu','Prime Video','Disney+']].sum()",
"_____no_output_____"
]
],
[
[
"#### Na plataforma da Prime Video, dos 12354 títulos, podemos perceber que:\n1. 345 títulos presentes na Netflix\n2. 241 títulos presentes na Hulu\n3. 19 títulos presentes na Disney+",
"_____no_output_____"
]
],
[
[
"disney_data[['Netflix','Hulu','Prime Video','Disney+']].sum()",
"_____no_output_____"
]
],
[
[
"#### Na plataforma da Disney+, dos 564 títulos, podemos perceber que:\n1. 10 títulos presentes na Netflix\n2. 7 títulos presentes na Hulu\n3. 19 títulos presentes na Prime Video",
"_____no_output_____"
],
[
"### Rankear pela nota do IMDb, retirando os dados ausentes da coluna, qual plataforma é melhor para o cliente assinar:",
"_____no_output_____"
]
],
[
[
"print(\"Média de nota do IMDb dos títulos presentes na Netflix : {:.2f}\".format(netflix_data['IMDb'].dropna().mean()))\n\nprint(\"Média de nota do IMDb dos títulos presentes na Disney+ : {:.2f}\".format(disney_data['IMDb'].dropna().mean()))\n\nprint(\"Média de nota do IMDb dos títulos presentes na Hulu : {:.2f}\".format(hulu_data['IMDb'].dropna().mean()))\n\nprint(\"Média de nota do IMDb dos títulos presentes na Prime Video : {:.2f}\".format(prime_data['IMDb'].dropna().mean()))",
"Média de nota do IMDb dos títulos presentes na Netflix : 6.25\nMédia de nota do IMDb dos títulos presentes na Disney+ : 6.44\nMédia de nota do IMDb dos títulos presentes na Hulu : 6.14\nMédia de nota do IMDb dos títulos presentes na Prime Video : 5.77\n"
]
],
[
[
"### Análise de gêneros de títulos",
"_____no_output_____"
]
],
[
[
"netflix_data['Genres'].str.contains('Drama').dropna().sum()",
"_____no_output_____"
],
[
"print(\"Número de títulos relacionados ao gênero de drama na Netflix : {}\".format(netflix_data['Genres'].str.contains('Drama').dropna().sum()))\nprint(\"Número de títulos relacionados ao gênero de drama na Hulu : {}\".format(hulu_data['Genres'].str.contains('Drama').dropna().sum()))\nprint(\"Número de títulos relacionados ao gênero de drama na Disney+ : {}\".format(disney_data['Genres'].str.contains('Drama').dropna().sum()))\nprint(\"Número de títulos relacionados ao gênero de drama na Prime Video : {}\".format(prime_data['Genres'].str.contains('Drama').dropna().sum()))",
"Número de títulos relacionados ao gênero de drama na Netflix : 1501\nNúmero de títulos relacionados ao gênero de drama na Hulu : 438\nNúmero de títulos relacionados ao gênero de drama na Disney+ : 162\nNúmero de títulos relacionados ao gênero de drama na Prime Video : 5437\n"
],
[
"print(\"Número de títulos relacionados ao gênero de ação na Netflix : {}\"\n .format(netflix_data['Genres'].str.contains('Action').dropna().sum()))\nprint(\"Número de títulos relacionados ao gênero de ação na Hulu : {}\"\n .format(hulu_data['Genres'].str.contains('Action').dropna().sum()))\nprint(\"Número de títulos relacionados ao gênero de ação na Disney+ : {}\"\n .format(disney_data['Genres'].str.contains('Action').dropna().sum()))\nprint(\"Número de títulos relacionados ao gênero de ação na Prime Video : {}\"\n .format(prime_data['Genres'].str.contains('Action').dropna().sum()))",
"Número de títulos relacionados ao gênero de ação na Netflix : 545\nNúmero de títulos relacionados ao gênero de ação na Hulu : 129\nNúmero de títulos relacionados ao gênero de ação na Disney+ : 88\nNúmero de títulos relacionados ao gênero de ação na Prime Video : 2049\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
cbece83c78401bdfb08426a4394c09a343c3a991
| 216,570 |
ipynb
|
Jupyter Notebook
|
zf_embedding_learning.ipynb
|
danstowell/ml4bl
|
ec6041d773b9c57e4423d84eb049437715440a4f
|
[
"MIT"
] | null | null | null |
zf_embedding_learning.ipynb
|
danstowell/ml4bl
|
ec6041d773b9c57e4423d84eb049437715440a4f
|
[
"MIT"
] | null | null | null |
zf_embedding_learning.ipynb
|
danstowell/ml4bl
|
ec6041d773b9c57e4423d84eb049437715440a4f
|
[
"MIT"
] | 1 |
2022-03-07T14:21:14.000Z
|
2022-03-07T14:21:14.000Z
| 58.914581 | 322 | 0.446793 |
[
[
[
"import keras\nimport tensorflow as tf",
"Using TensorFlow backend.\n"
],
[
"import librosa\nimport numpy as np\nimport pandas\nimport pickle\nimport os\nfrom os import listdir\nfrom os.path import isfile, join\nimport matplotlib.pyplot as plt\nfrom mpl_toolkits.mplot3d import Axes3D\nimport h5py\nimport math\nimport random\nimport re\nimport csv\nfrom sklearn.manifold import TSNE, MDS",
"_____no_output_____"
],
[
"from keras.models import Model, load_model\nfrom keras.layers import Dropout, concatenate, Concatenate, Activation, Input, Dense, Conv2D, GRU, MaxPooling2D, MaxPooling1D, Flatten, Reshape, LeakyReLU, PReLU, BatchNormalization, Bidirectional, TimeDistributed, Lambda, GlobalMaxPool1D, GlobalMaxPool2D, GlobalAveragePooling2D, Multiply, GlobalAveragePooling2D\nfrom keras.optimizers import Adam, SGD\nimport keras.backend as K\nfrom keras import regularizers\nfrom keras.initializers import random_normal, glorot_uniform, glorot_normal\nimport tensorflow as tf\nfrom keras.callbacks import TensorBoard, ModelCheckpoint, LearningRateScheduler, ReduceLROnPlateau, Callback, EarlyStopping",
"_____no_output_____"
],
[
"path_mel = './ML4BL_ZF/melspecs/'\npath_files = './ML4BL_ZF/files/'\n\ntrain_triplet_file = 'train_triplets_50_70_single.pckl'\ntrain_gt_file = 'train_gt_50_70_single.pckl'\ntrain_cons_file = 'train_cons_50_70_single.pckl'\ntrain_trials_file = 'train_trials_50_70_single.pckl'\n\ntest_triplet_file = 'test_triplets_50_70_single.pckl'\ntest_gt_file = 'test_gt_50_70_single.pckl'\ntest_cons_file = 'test_cons_50_70_single.pckl'\ntest_trials_file = 'test_trials_50_70_single.pckl'",
"_____no_output_____"
],
[
"luscinia_triplets_file = 'luscinia_triplets_filtered.csv'",
"_____no_output_____"
],
[
"luscinia_triplets = []\nwith open(path_files+luscinia_triplets_file, 'r', newline='') as csvfile:\n csv_r = csv.reader(csvfile, delimiter=',')\n for row in csv_r:\n luscinia_triplets.append(row)",
"_____no_output_____"
],
[
"luscinia_triplets = luscinia_triplets[1:]\nluscinia_train_len = round(8*len(luscinia_triplets)/10)\nluscinia_val_len = len(luscinia_triplets) - luscinia_train_len",
"_____no_output_____"
],
[
"f = open(path_files+'mean_std_luscinia_pretraining.pckl', 'rb')\ntrain_dict = pickle.load(f)\nM_l = train_dict['mean']\nS_l = train_dict['std']\nf.close()",
"_____no_output_____"
],
[
"f = open(path_files+'training_setup_1_ordered_acc_single_cons_50_70_trials.pckl', 'rb')\ntrain_dict = pickle.load(f)\ntrain_keys = train_dict['train_keys']\ntraining_triplets = train_dict['train_triplets']\nval_keys = train_dict['val_keys']\nvalidation_triplets = train_dict['vali_triplets']\ntest_triplet = train_dict['test_triplets']\ntest_keys = train_dict['test_keys']\nM = train_dict['train_mean']\nS = train_dict['train_std']\nf.close()",
"_____no_output_____"
]
],
[
[
"# Network",
"_____no_output_____"
]
],
[
[
"def convBNpr(a, dilation, num_filters, kernel):\n c1 = Conv2D(filters=num_filters, kernel_size=kernel, strides=(1, 1), dilation_rate=dilation, padding='same', use_bias=False, kernel_initializer=glorot_uniform(seed=123), kernel_regularizer=regularizers.l2(1e-4))(a)\n c1 = BatchNormalization(axis=-1, momentum=0.99, epsilon=0.001)(c1)\n c1 = LeakyReLU(alpha=0.3)(c1)\n return c1\ndef createModelMatrix(emb_size, input_shape=(170, 150, 1)):\n a = Input(shape=(input_shape)) \n \n c = convBNpr(a, 1, 12, (3,3))\n c = convBNpr(c, 2, 32, (3,3))\n c = convBNpr(c, 4, 64, (3,3))\n c = convBNpr(c, 8, 128, (3,3))\n # attention with sigmoid\n a1 = Conv2D(filters=128, kernel_size=(1,1), strides=(1, 1), padding='same', activation = 'sigmoid', use_bias=True, kernel_regularizer=regularizers.l2(1e-4),kernel_initializer=glorot_uniform(seed=123))(c)\n\n \n # sum of sum of attention\n s = Lambda(lambda x: K.sum(K.sum(x,axis=1, keepdims=True), axis=2, keepdims=True))(a1)\n s = Lambda(lambda x: K.repeat_elements(x, 170, axis=1))(s)\n s = Lambda(lambda x: K.repeat_elements(x, 150, axis=2))(s)\n \n # probability matrix of attention\n p = Lambda(lambda x: x[0]/x[1])([a1,s])\n \n # inner product of attention and projection matrices\n m = Multiply()([c, p])\n \n # output\n out_sum = Lambda(lambda x: K.sum(K.sum(x, axis=1), axis=1))(m)\n \n # attention side\n d3 = Dense(emb_size*100, kernel_initializer=glorot_normal(seed=222), kernel_regularizer=regularizers.l2(1e-4),activation='relu')(out_sum)\n d3 = Dropout(.2, seed=222)(d3)\n d4 = Dense(emb_size*10, kernel_initializer=glorot_normal(seed=333), kernel_regularizer=regularizers.l2(1e-4),activation='relu')(d3)\n d4 = Dropout(.2, seed=333)(d4)\n d5 = Dense(emb_size, kernel_initializer=glorot_normal(seed=132), kernel_regularizer=regularizers.l2(1e-4))(d4)\n d5 = Dropout(.2, seed = 132)(d5)\n \n # maxpool side\n x = convBNpr(c, 1, 64, (3,3)) \n x = MaxPooling2D(pool_size=(2, 2))(x)\n x = convBNpr(x, 1, 32, (3,3))\n x = MaxPooling2D(pool_size=(2, 2))(x)\n x = convBNpr(x, 1, 12, (3,3))\n\n f = Flatten()(x)\n df1 = Dense(emb_size*100, kernel_initializer=glorot_normal(seed=456), kernel_regularizer=regularizers.l2(1e-4), activation='relu')(f)#\n df1 = Dropout(.2, seed=456)(df1)\n df2 = Dense(emb_size*10, kernel_initializer=glorot_normal(seed=654), kernel_regularizer=regularizers.l2(1e-4), activation='relu')(df1)#\n df2 = Dropout(.2, seed=654)(df2)\n df3 = Dense(emb_size, kernel_initializer=glorot_normal(seed=546), kernel_regularizer=regularizers.l2(1e-4))(df2)#\n df3 = Dropout(.2, seed=546)(df3)\n\n concat = Concatenate(axis=-1)([d5, df3])\n dd = Dense(emb_size, kernel_initializer=glorot_normal(seed=999), kernel_regularizer=regularizers.l2(1e-4))(concat)\n\n sph = Lambda(lambda x: K.l2_normalize(x,axis=1))(dd)\n \n # base model creation\n base_model = Model(a,sph) \n \n # triplet framework\n input_anchor = Input(shape=(input_shape))\n input_positive = Input(shape=(input_shape))\n input_negative = Input(shape=(input_shape)) \n \n net_anchor = base_model(input_anchor)\n net_positive = base_model(input_positive)\n net_negative = base_model(input_negative)\n \n base_model.summary()\n \n merged_vector = concatenate([net_anchor, net_positive, net_negative], axis=-1)\n \n model = Model([input_anchor, input_positive, input_negative], outputs=merged_vector)\n\n return model",
"_____no_output_____"
]
],
[
[
"# Functions",
"_____no_output_____"
]
],
[
[
"def masked_weighted_triplet_loss(margin, emb_size, m = 0 , w = 0, lh = 1):\n def lossFunction(y_true,y_pred):\n \n weight = y_true[:, 0] # acc\n cons = y_true[:, 1] # consistency\n trials = y_true[:, 2] # number of trials\n \n anchor = y_pred[:, 0:emb_size]\n positive = y_pred[:, emb_size:emb_size*2]\n negative = y_pred[:, emb_size*2:emb_size*3]\n\n # distance between the anchor and the positive\n pos_dist = K.sqrt(K.sum(K.square(anchor - positive), axis=1)) # l2 distance\n #pos_dist = K.sum(K.abs(anchor-positive), axis=1) # l1 distance\n\n # distance between the anchor and the negative\n neg_dist = K.sqrt(K.sum(K.square(anchor - negative), axis=1)) # l2 distance\n #neg_dist = K.sum(K.abs(anchor-negative), axis=1) # l1 distance\n\n loss_h = 0\n loss_l = 0\n \n if lh == 1:\n # DOES NOT WORK WITH MASKED LOSS\n # low-high margin loss\n p_c = K.square(neg_dist) - K.square(pos_dist) - margin \n p_i = K.square(neg_dist) - K.square(pos_dist)\n \n loss_1 = cons*(1-K.exp(p_c)) + (1-cons)*(1-K.exp(-K.abs(p_i)))\n \n if m != 0:\n # masked loss\n basic_loss = pos_dist - neg_dist + margin\n \n threshold = K.max(basic_loss) * m\n mask = 2 + margin - K.maximum(basic_loss, threshold) \n\n loss_1 = basic_loss * mask\n \n if w == 1:\n # weighted based on acc\n weighted_loss = weight*loss_1\n else:\n # non-weighted\n weighted_loss = loss_1\n \n loss = K.maximum(weighted_loss, 0.0)\n\n return loss\n return lossFunction",
"_____no_output_____"
],
[
"def discard_some_low(triplet_list, cons, acc):\n low_margin = []\n high_margin = []\n \n for i in range(len(triplet_list)):\n if float(triplet_list[i][-1]) < cons: # low margin\n if float(triplet_list[i][-2]) >= acc: # ACC \n low_margin.append(triplet_list[i])\n else: # high margin\n high_margin.append(triplet_list[i])\n \n random.seed(123)\n random.shuffle(low_margin)\n random.shuffle(high_margin)\n \n low_margin.extend(high_margin)\n \n return low_margin",
"_____no_output_____"
],
[
"def balance_input(triplet_list, cons, hi_balance = 6, lo_balance = 6):\n batchsize = hi_balance + lo_balance\n low_margin = []\n high_margin = []\n \n for i in range(len(triplet_list)):\n if float(triplet_list[i][-1]) < cons: # low margin\n low_margin.append(triplet_list[i])\n else: # high margin\n high_margin.append(triplet_list[i])\n \n random.seed(123)\n random.shuffle(low_margin)\n random.shuffle(high_margin)\n \n new_triplet_list = []\n maxlen = np.maximum(len(low_margin), len(high_margin))\n \n hi_start = 0\n lo_start = 0\n for i in range(0,int(maxlen/hi_balance)*batchsize,batchsize):\n for j in range(hi_start,hi_start+hi_balance,1):\n new_triplet_list.append(high_margin[np.mod(j,len(high_margin))])\n hi_start+=hi_balance\n for j in range(lo_start, lo_start+lo_balance,1):\n new_triplet_list.append(low_margin[np.mod(j,len(low_margin))])\n lo_start+=lo_balance\n \n return low_margin, high_margin, new_triplet_list",
"_____no_output_____"
]
],
[
[
"# Generators",
"_____no_output_____"
]
],
[
[
"def train_generator_mixed(triplet_list, M, S, luscinia_triplets, M_l, S_l, batchsize, lo, hi, lu, emb_size, path_mel):\n \n acc_gt = np.zeros((batchsize, emb_size))\n \n random.seed(123)\n random.shuffle(luscinia_triplets)\n \n while 1:\n \n anchors_input = np.empty((batchsize, 170, 150, 1))\n positives_input = np.empty((batchsize, 170, 150, 1))\n negatives_input = np.empty((batchsize, 170, 150, 1))\n \n imax = int(len(triplet_list)/(lo+hi))\n \n list_cnt = 0\n luscinia_cnt = 0\n \n for i in range(imax): \n for j in range(batchsize):\n \n if j < (lo+hi):\n triplet = triplet_list[list_cnt]\n list_cnt += 1\n \n tr_anc = triplet[3][:-4]+'.pckl'\n tr_pos = triplet[1][:-4]+'.pckl'\n tr_neg = triplet[2][:-4]+'.pckl'\n acc_gt[j][0] = float(triplet[-2]) # acc\n acc_gt[j][1] = 1 if float(triplet[-1])>=0.7 else 0 # cons\n acc_gt[j][2] = int(triplet[-3]) # number of trials\n\n else:\n triplet = luscinia_triplets[luscinia_cnt]\n luscinia_cnt += 1\n \n tr_anc = triplet[2][:-4]+'.pckl'\n tr_pos = triplet[0][:-4]+'.pckl'\n tr_neg = triplet[1][:-4]+'.pckl'\n acc_gt[j][0] = 1 # acc\n acc_gt[j][1] = 1 # cons\n acc_gt[j][2] = 1 # number of trials\n \n f = open(path_mel+tr_anc, 'rb')\n anc = pickle.load(f).T\n f.close()\n anc = (anc - M)/S\n anc = np.expand_dims(anc, axis=-1)\n \n f = open(path_mel+tr_pos, 'rb')\n pos = pickle.load(f).T\n f.close()\n pos = (pos - M)/S\n pos = np.expand_dims(pos, axis=-1)\n \n f = open(path_mel+tr_neg, 'rb')\n neg = pickle.load(f).T\n f.close()\n neg = (neg - M)/S\n neg = np.expand_dims(neg, axis=-1)\n \n anchors_input[j] = anc\n positives_input[j] = pos\n negatives_input[j] = neg\n \n yield [anchors_input, positives_input, negatives_input], acc_gt",
"_____no_output_____"
],
[
"def train_generator_luscinia(triplet_list, M, S, batchsize, emb_size, path_mel, ordered = True):\n \n acc_gt = np.zeros((batchsize, emb_size))\n \n random.seed(123)\n random.shuffle(triplet_list) \n \n while 1:\n \n anchors_input = np.empty((batchsize, 170, 150, 1))\n positives_input = np.empty((batchsize, 170, 150, 1))\n negatives_input = np.empty((batchsize, 170, 150, 1))\n \n imax = int(len(triplet_list)/batchsize)\n \n for i in range(imax): \n for j in range(batchsize):\n triplet = triplet_list[i*batchsize+j]\n \n tr_anc = triplet[2][:-4]+'.pckl'\n tr_pos = triplet[0][:-4]+'.pckl'\n tr_neg = triplet[1][:-4]+'.pckl'\n acc_gt[j][0] = 1 # acc\n acc_gt[j][1] = 1 # cons\n acc_gt[j][2] = 1 # number of trials\n \n f = open(path_mel+tr_anc, 'rb')\n anc = pickle.load(f).T\n f.close()\n anc = (anc - M)/S\n anc = np.expand_dims(anc, axis=-1)\n \n f = open(path_mel+tr_pos, 'rb')\n pos = pickle.load(f).T\n f.close()\n pos = (pos - M)/S\n pos = np.expand_dims(pos, axis=-1)\n \n f = open(path_mel+tr_neg, 'rb')\n neg = pickle.load(f).T\n f.close()\n neg = (neg - M)/S\n neg = np.expand_dims(neg, axis=-1)\n \n anchors_input[j] = anc\n positives_input[j] = pos\n negatives_input[j] = neg\n \n yield [anchors_input, positives_input, negatives_input], acc_gt",
"_____no_output_____"
],
[
"def train_generator(triplet_list, M, S, batchsize, emb_size, path_mel, ordered = True):\n \n acc_gt = np.zeros((batchsize, emb_size))\n \n random.seed(123)\n random.shuffle(triplet_list) \n \n while 1:\n \n anchors_input = np.empty((batchsize, 170, 150, 1))\n positives_input = np.empty((batchsize, 170, 150, 1))\n negatives_input = np.empty((batchsize, 170, 150, 1))\n \n imax = int(len(triplet_list)/batchsize)\n \n for i in range(imax): \n for j in range(batchsize):\n triplet = triplet_list[i*batchsize+j]\n \n tr_anc = triplet[3][:-4]+'.pckl'\n \n if ordered == False:\n if triplet[-1] == '0':\n tr_pos = triplet[1][:-4]+'.pckl'\n tr_neg = triplet[2][:-4]+'.pckl'\n else:\n tr_pos = triplet[2][:-4]+'.pckl'\n tr_neg = triplet[1][:-4]+'.pckl'\n else: \n tr_pos = triplet[1][:-4]+'.pckl'\n tr_neg = triplet[2][:-4]+'.pckl'\n acc_gt[j][0] = float(triplet[-2]) # acc\n acc_gt[j][1] = 1 if float(triplet[-1])>=0.7 else 0 # cons\n acc_gt[j][2] = int(triplet[-3]) # number of trials\n \n f = open(path_mel+tr_anc, 'rb')\n anc = pickle.load(f).T\n f.close()\n anc = (anc - M)/S\n anc = np.expand_dims(anc, axis=-1)\n \n f = open(path_mel+tr_pos, 'rb')\n pos = pickle.load(f).T\n f.close()\n pos = (pos - M)/S\n pos = np.expand_dims(pos, axis=-1)\n \n f = open(path_mel+tr_neg, 'rb')\n neg = pickle.load(f).T\n f.close()\n neg = (neg - M)/S\n neg = np.expand_dims(neg, axis=-1)\n \n anchors_input[j] = anc\n positives_input[j] = pos\n negatives_input[j] = neg\n \n yield [anchors_input, positives_input, negatives_input], acc_gt",
"_____no_output_____"
]
],
[
[
"# Training",
"_____no_output_____"
]
],
[
[
"emb_size=16\nmargin = 0.1\nm = 0\nlr = 1e-8\nadam = Adam(lr = lr)\n\ntriplet_model = createModelMatrix(emb_size=emb_size, input_shape=(170, 150, 1))\ntriplet_model.summary()\ntriplet_model.compile(loss=masked_weighted_triplet_loss(margin=margin, emb_size=emb_size, m=m, w = 0, lh = 1),optimizer=adam) ",
"Model: \"model\"\n__________________________________________________________________________________________________\nLayer (type) Output Shape Param # Connected to \n==================================================================================================\ninput_1 (InputLayer) [(None, 170, 150, 1) 0 \n__________________________________________________________________________________________________\nconv2d (Conv2D) (None, 170, 150, 12) 108 input_1[0][0] \n__________________________________________________________________________________________________\nbatch_normalization (BatchNorma (None, 170, 150, 12) 48 conv2d[0][0] \n__________________________________________________________________________________________________\nleaky_re_lu (LeakyReLU) (None, 170, 150, 12) 0 batch_normalization[0][0] \n__________________________________________________________________________________________________\nconv2d_1 (Conv2D) (None, 170, 150, 32) 3456 leaky_re_lu[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_1 (BatchNor (None, 170, 150, 32) 128 conv2d_1[0][0] \n__________________________________________________________________________________________________\nleaky_re_lu_1 (LeakyReLU) (None, 170, 150, 32) 0 batch_normalization_1[0][0] \n__________________________________________________________________________________________________\nconv2d_2 (Conv2D) (None, 170, 150, 64) 18432 leaky_re_lu_1[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_2 (BatchNor (None, 170, 150, 64) 256 conv2d_2[0][0] \n__________________________________________________________________________________________________\nleaky_re_lu_2 (LeakyReLU) (None, 170, 150, 64) 0 batch_normalization_2[0][0] \n__________________________________________________________________________________________________\nconv2d_3 (Conv2D) (None, 170, 150, 128 73728 leaky_re_lu_2[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_3 (BatchNor (None, 170, 150, 128 512 conv2d_3[0][0] \n__________________________________________________________________________________________________\nleaky_re_lu_3 (LeakyReLU) (None, 170, 150, 128 0 batch_normalization_3[0][0] \n__________________________________________________________________________________________________\nconv2d_5 (Conv2D) (None, 170, 150, 64) 73728 leaky_re_lu_3[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_4 (BatchNor (None, 170, 150, 64) 256 conv2d_5[0][0] \n__________________________________________________________________________________________________\nleaky_re_lu_4 (LeakyReLU) (None, 170, 150, 64) 0 batch_normalization_4[0][0] \n__________________________________________________________________________________________________\nmax_pooling2d (MaxPooling2D) (None, 85, 75, 64) 0 leaky_re_lu_4[0][0] \n__________________________________________________________________________________________________\nconv2d_6 (Conv2D) (None, 85, 75, 32) 18432 max_pooling2d[0][0] \n__________________________________________________________________________________________________\nconv2d_4 (Conv2D) (None, 170, 150, 128 16512 leaky_re_lu_3[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_5 (BatchNor (None, 85, 75, 32) 128 conv2d_6[0][0] \n__________________________________________________________________________________________________\nlambda (Lambda) (None, 1, 1, 128) 0 conv2d_4[0][0] \n__________________________________________________________________________________________________\nleaky_re_lu_5 (LeakyReLU) (None, 85, 75, 32) 0 batch_normalization_5[0][0] \n__________________________________________________________________________________________________\nlambda_1 (Lambda) (None, 170, 1, 128) 0 lambda[0][0] \n__________________________________________________________________________________________________\nmax_pooling2d_1 (MaxPooling2D) (None, 42, 37, 32) 0 leaky_re_lu_5[0][0] \n__________________________________________________________________________________________________\nlambda_2 (Lambda) (None, 170, 150, 128 0 lambda_1[0][0] \n__________________________________________________________________________________________________\nconv2d_7 (Conv2D) (None, 42, 37, 12) 3456 max_pooling2d_1[0][0] \n__________________________________________________________________________________________________\nlambda_3 (Lambda) (None, 170, 150, 128 0 conv2d_4[0][0] \n lambda_2[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_6 (BatchNor (None, 42, 37, 12) 48 conv2d_7[0][0] \n__________________________________________________________________________________________________\nmultiply (Multiply) (None, 170, 150, 128 0 leaky_re_lu_3[0][0] \n lambda_3[0][0] \n__________________________________________________________________________________________________\nleaky_re_lu_6 (LeakyReLU) (None, 42, 37, 12) 0 batch_normalization_6[0][0] \n__________________________________________________________________________________________________\nlambda_4 (Lambda) (None, 128) 0 multiply[0][0] \n__________________________________________________________________________________________________\nflatten (Flatten) (None, 18648) 0 leaky_re_lu_6[0][0] \n__________________________________________________________________________________________________\ndense (Dense) (None, 1600) 206400 lambda_4[0][0] \n__________________________________________________________________________________________________\ndense_3 (Dense) (None, 1600) 29838400 flatten[0][0] \n__________________________________________________________________________________________________\ndropout (Dropout) (None, 1600) 0 dense[0][0] \n__________________________________________________________________________________________________\ndropout_3 (Dropout) (None, 1600) 0 dense_3[0][0] \n__________________________________________________________________________________________________\ndense_1 (Dense) (None, 160) 256160 dropout[0][0] \n__________________________________________________________________________________________________\ndense_4 (Dense) (None, 160) 256160 dropout_3[0][0] \n__________________________________________________________________________________________________\ndropout_1 (Dropout) (None, 160) 0 dense_1[0][0] \n__________________________________________________________________________________________________\ndropout_4 (Dropout) (None, 160) 0 dense_4[0][0] \n__________________________________________________________________________________________________\ndense_2 (Dense) (None, 16) 2576 dropout_1[0][0] \n__________________________________________________________________________________________________\ndense_5 (Dense) (None, 16) 2576 dropout_4[0][0] \n__________________________________________________________________________________________________\ndropout_2 (Dropout) (None, 16) 0 dense_2[0][0] \n__________________________________________________________________________________________________\ndropout_5 (Dropout) (None, 16) 0 dense_5[0][0] \n__________________________________________________________________________________________________\nconcatenate (Concatenate) (None, 32) 0 dropout_2[0][0] \n dropout_5[0][0] \n__________________________________________________________________________________________________\ndense_6 (Dense) (None, 16) 528 concatenate[0][0] \n__________________________________________________________________________________________________\nlambda_5 (Lambda) (None, 16) 0 dense_6[0][0] \n==================================================================================================\nTotal params: 30,772,028\nTrainable params: 30,771,340\nNon-trainable params: 688\n__________________________________________________________________________________________________\nModel: \"model_1\"\n__________________________________________________________________________________________________\nLayer (type) Output Shape Param # Connected to \n==================================================================================================\ninput_2 (InputLayer) [(None, 170, 150, 1) 0 \n__________________________________________________________________________________________________\ninput_3 (InputLayer) [(None, 170, 150, 1) 0 \n__________________________________________________________________________________________________\ninput_4 (InputLayer) [(None, 170, 150, 1) 0 \n__________________________________________________________________________________________________\nmodel (Functional) (None, 16) 30772028 input_2[0][0] \n input_3[0][0] \n input_4[0][0] \n__________________________________________________________________________________________________\nconcatenate_1 (Concatenate) (None, 48) 0 model[0][0] \n model[1][0] \n model[2][0] \n==================================================================================================\nTotal params: 30,772,028\nTrainable params: 30,771,340\nNon-trainable params: 688\n__________________________________________________________________________________________________\n"
]
],
[
[
"# PRE (pretraining on Luscinia triplets)",
"_____no_output_____"
]
],
[
[
"lo = 6\nhi = 8\nlu = 10\nbatchsize = lo+hi+lu \n\ncpCallback = ModelCheckpoint('ZF_emb_'+str(emb_size)+'D_LUSCINIA_PRE_margin_loss_backup.h5', monitor='val_loss', save_best_only=True, save_weights_only=True, mode='min', save_freq='epoch')\nreduce_lr = ReduceLROnPlateau(monitor='val_loss', factor=0.5, patience=10, verbose=1, min_lr=1e-12)\nearlystop = EarlyStopping(monitor='val_loss', min_delta=0, patience=20, verbose=1, mode='auto')\n\nhistory = triplet_model.fit(train_generator_luscinia(luscinia_triplets[:int(luscinia_train_len/10)], M_l, S_l, batchsize, emb_size, path_mel),\n steps_per_epoch=int(int(luscinia_train_len/10)/batchsize), epochs=1000, verbose=1,\n validation_data=train_generator_luscinia(luscinia_triplets[luscinia_train_len:luscinia_train_len+200], M_l, S_l, batchsize, emb_size, path_mel),\n validation_steps=int(200/batchsize), callbacks=[cpCallback, reduce_lr, earlystop])\n",
"Epoch 1/1000\n348/348 [==============================] - 171s 346ms/step - loss: 0.5265 - val_loss: 0.4591\nEpoch 2/1000\n348/348 [==============================] - 119s 342ms/step - loss: 0.4536 - val_loss: 0.4486\nEpoch 3/1000\n348/348 [==============================] - 119s 342ms/step - loss: 0.4392 - val_loss: 0.4454\nEpoch 4/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.4334 - val_loss: 0.4415\nEpoch 5/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.4304 - val_loss: 0.4401\nEpoch 6/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.4283 - val_loss: 0.4373\nEpoch 7/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.4273 - val_loss: 0.4341\nEpoch 8/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.4250 - val_loss: 0.4324\nEpoch 9/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.4237 - val_loss: 0.4305\nEpoch 10/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.4225 - val_loss: 0.4294\nEpoch 11/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.4211 - val_loss: 0.4287\nEpoch 12/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.4205 - val_loss: 0.4278\nEpoch 13/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.4192 - val_loss: 0.4261\nEpoch 14/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.4182 - val_loss: 0.4254\nEpoch 15/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.4169 - val_loss: 0.4238\nEpoch 16/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.4155 - val_loss: 0.4229\nEpoch 17/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.4143 - val_loss: 0.4208\nEpoch 18/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.4128 - val_loss: 0.4198\nEpoch 19/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.4117 - val_loss: 0.4165\nEpoch 20/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.4103 - val_loss: 0.4160\nEpoch 21/1000\n348/348 [==============================] - 118s 341ms/step - loss: 0.4086 - val_loss: 0.4148\nEpoch 22/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.4069 - val_loss: 0.4140\nEpoch 23/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.4053 - val_loss: 0.4140\nEpoch 24/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.4038 - val_loss: 0.4105\nEpoch 25/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.4022 - val_loss: 0.4080\nEpoch 26/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.4006 - val_loss: 0.4055\nEpoch 27/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.3990 - val_loss: 0.4061\nEpoch 28/1000\n348/348 [==============================] - 118s 341ms/step - loss: 0.3972 - val_loss: 0.4034\nEpoch 29/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.3955 - val_loss: 0.4016\nEpoch 30/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.3937 - val_loss: 0.3982\nEpoch 31/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.3920 - val_loss: 0.3959\nEpoch 32/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.3904 - val_loss: 0.3936\nEpoch 33/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.3887 - val_loss: 0.3930\nEpoch 34/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.3867 - val_loss: 0.3898\nEpoch 35/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.3849 - val_loss: 0.3885\nEpoch 36/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.3829 - val_loss: 0.3864\nEpoch 37/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.3809 - val_loss: 0.3850\nEpoch 38/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.3789 - val_loss: 0.3829\nEpoch 39/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.3768 - val_loss: 0.3799\nEpoch 40/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.3747 - val_loss: 0.3797\nEpoch 41/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.3724 - val_loss: 0.3767\nEpoch 42/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.3701 - val_loss: 0.3738\nEpoch 43/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.3678 - val_loss: 0.3706\nEpoch 44/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.3654 - val_loss: 0.3684\nEpoch 45/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.3631 - val_loss: 0.3654\nEpoch 46/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.3608 - val_loss: 0.3624\nEpoch 47/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.3583 - val_loss: 0.3606\nEpoch 48/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.3560 - val_loss: 0.3587\nEpoch 49/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.3537 - val_loss: 0.3563\nEpoch 50/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.3515 - val_loss: 0.3542\nEpoch 51/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.3491 - val_loss: 0.3525\nEpoch 52/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.3467 - val_loss: 0.3510\nEpoch 53/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.3447 - val_loss: 0.3481\nEpoch 54/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.3423 - val_loss: 0.3459\nEpoch 55/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.3402 - val_loss: 0.3432\nEpoch 56/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.3381 - val_loss: 0.3413\nEpoch 57/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.3359 - val_loss: 0.3405\nEpoch 58/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.3335 - val_loss: 0.3379\nEpoch 59/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.3311 - val_loss: 0.3344\nEpoch 60/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.3287 - val_loss: 0.3322\nEpoch 61/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.3263 - val_loss: 0.3293\nEpoch 62/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.3237 - val_loss: 0.3266\nEpoch 63/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.3212 - val_loss: 0.3241\nEpoch 64/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.3187 - val_loss: 0.3216\nEpoch 65/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.3161 - val_loss: 0.3200\nEpoch 66/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.3136 - val_loss: 0.3164\nEpoch 67/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.3111 - val_loss: 0.3137\nEpoch 68/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.3085 - val_loss: 0.3119\nEpoch 69/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.3058 - val_loss: 0.3094\nEpoch 70/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.3031 - val_loss: 0.3067\nEpoch 71/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.3005 - val_loss: 0.3051\nEpoch 72/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.2979 - val_loss: 0.3013\nEpoch 73/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.2955 - val_loss: 0.2980\nEpoch 74/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.2933 - val_loss: 0.2962\nEpoch 75/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.2914 - val_loss: 0.2943\nEpoch 76/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.2894 - val_loss: 0.2921\nEpoch 77/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.2873 - val_loss: 0.2896\nEpoch 78/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.2853 - val_loss: 0.2877\nEpoch 79/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.2832 - val_loss: 0.2846\nEpoch 80/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.2810 - val_loss: 0.2818\nEpoch 81/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.2789 - val_loss: 0.2802\nEpoch 82/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.2768 - val_loss: 0.2783\nEpoch 83/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.2747 - val_loss: 0.2765\nEpoch 84/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.2724 - val_loss: 0.2739\nEpoch 85/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.2701 - val_loss: 0.2720\nEpoch 86/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.2677 - val_loss: 0.2700\nEpoch 87/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.2655 - val_loss: 0.2678\nEpoch 88/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.2633 - val_loss: 0.2673\nEpoch 89/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.2613 - val_loss: 0.2636\nEpoch 90/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.2593 - val_loss: 0.2633\nEpoch 91/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.2574 - val_loss: 0.2602\nEpoch 92/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.2555 - val_loss: 0.2584\nEpoch 93/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.2538 - val_loss: 0.2567\nEpoch 94/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.2522 - val_loss: 0.2559\nEpoch 95/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.2506 - val_loss: 0.2518\nEpoch 96/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.2490 - val_loss: 0.2513\nEpoch 97/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.2473 - val_loss: 0.2494\nEpoch 98/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.2455 - val_loss: 0.2474\nEpoch 99/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.2436 - val_loss: 0.2456\nEpoch 100/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.2418 - val_loss: 0.2454\nEpoch 101/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.2400 - val_loss: 0.2430\nEpoch 102/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.2382 - val_loss: 0.2421\nEpoch 103/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.2365 - val_loss: 0.2400\nEpoch 104/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.2346 - val_loss: 0.2379\nEpoch 105/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.2326 - val_loss: 0.2356\nEpoch 106/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.2305 - val_loss: 0.2336\nEpoch 107/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.2284 - val_loss: 0.2311\nEpoch 108/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.2262 - val_loss: 0.2286\nEpoch 109/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.2241 - val_loss: 0.2276\nEpoch 110/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.2220 - val_loss: 0.2255\nEpoch 111/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.2200 - val_loss: 0.2241\nEpoch 112/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.2179 - val_loss: 0.2212\nEpoch 113/1000\n348/348 [==============================] - 118s 340ms/step - loss: 0.2157 - val_loss: 0.2199\nEpoch 114/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.2135 - val_loss: 0.2170\nEpoch 115/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.2116 - val_loss: 0.2136\nEpoch 116/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.2100 - val_loss: 0.2119\nEpoch 117/1000\n348/348 [==============================] - 118s 341ms/step - loss: 0.2084 - val_loss: 0.2109\nEpoch 118/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.2070 - val_loss: 0.2103\nEpoch 119/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.2056 - val_loss: 0.2089\nEpoch 120/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.2041 - val_loss: 0.2081\nEpoch 121/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.2027 - val_loss: 0.2075\nEpoch 122/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.2013 - val_loss: 0.2060\nEpoch 123/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1998 - val_loss: 0.2047\nEpoch 124/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1985 - val_loss: 0.2024\nEpoch 125/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1972 - val_loss: 0.2012\nEpoch 126/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1960 - val_loss: 0.1994\nEpoch 127/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1947 - val_loss: 0.1975\nEpoch 128/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1933 - val_loss: 0.1960\nEpoch 129/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1919 - val_loss: 0.1944\nEpoch 130/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1904 - val_loss: 0.1927\nEpoch 131/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1890 - val_loss: 0.1917\nEpoch 132/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1876 - val_loss: 0.1912\nEpoch 133/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1863 - val_loss: 0.1896\nEpoch 134/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1852 - val_loss: 0.1905\nEpoch 135/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1838 - val_loss: 0.1888\nEpoch 136/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1825 - val_loss: 0.1872\nEpoch 137/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1813 - val_loss: 0.1845\nEpoch 138/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1800 - val_loss: 0.1838\nEpoch 139/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1788 - val_loss: 0.1828\nEpoch 140/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1775 - val_loss: 0.1808\nEpoch 141/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1762 - val_loss: 0.1796\nEpoch 142/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1748 - val_loss: 0.1779\nEpoch 143/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1734 - val_loss: 0.1770\nEpoch 144/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1722 - val_loss: 0.1735\nEpoch 145/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1711 - val_loss: 0.1730\nEpoch 146/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1702 - val_loss: 0.1720\nEpoch 147/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1693 - val_loss: 0.1706\nEpoch 148/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1684 - val_loss: 0.1696\nEpoch 149/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1674 - val_loss: 0.1682\nEpoch 150/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1663 - val_loss: 0.1683\nEpoch 151/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1653 - val_loss: 0.1662\nEpoch 152/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1642 - val_loss: 0.1651\nEpoch 153/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1631 - val_loss: 0.1639\nEpoch 154/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1618 - val_loss: 0.1628\nEpoch 155/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1605 - val_loss: 0.1617\nEpoch 156/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1591 - val_loss: 0.1595\nEpoch 157/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1580 - val_loss: 0.1589\nEpoch 158/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1571 - val_loss: 0.1579\nEpoch 159/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1563 - val_loss: 0.1576\nEpoch 160/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1555 - val_loss: 0.1570\nEpoch 161/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1548 - val_loss: 0.1562\nEpoch 162/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1540 - val_loss: 0.1553\nEpoch 163/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1533 - val_loss: 0.1543\nEpoch 164/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1525 - val_loss: 0.1545\nEpoch 165/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1516 - val_loss: 0.1530\nEpoch 166/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1507 - val_loss: 0.1523\nEpoch 167/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1498 - val_loss: 0.1518\nEpoch 168/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1488 - val_loss: 0.1523\nEpoch 169/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1480 - val_loss: 0.1506\nEpoch 170/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1471 - val_loss: 0.1496\nEpoch 171/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1463 - val_loss: 0.1492\nEpoch 172/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1455 - val_loss: 0.1484\nEpoch 173/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1447 - val_loss: 0.1465\nEpoch 174/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1438 - val_loss: 0.1452\nEpoch 175/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1429 - val_loss: 0.1441\nEpoch 176/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1419 - val_loss: 0.1438\nEpoch 177/1000\n348/348 [==============================] - 118s 341ms/step - loss: 0.1410 - val_loss: 0.1430\nEpoch 178/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1400 - val_loss: 0.1419\nEpoch 179/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1390 - val_loss: 0.1404\nEpoch 180/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1380 - val_loss: 0.1395\nEpoch 181/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1370 - val_loss: 0.1389\nEpoch 182/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1361 - val_loss: 0.1386\nEpoch 183/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1353 - val_loss: 0.1382\nEpoch 184/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1345 - val_loss: 0.1367\nEpoch 185/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1338 - val_loss: 0.1359\nEpoch 186/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1331 - val_loss: 0.1347\nEpoch 187/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1324 - val_loss: 0.1338\nEpoch 188/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1318 - val_loss: 0.1332\nEpoch 189/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1311 - val_loss: 0.1325\nEpoch 190/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1305 - val_loss: 0.1324\nEpoch 191/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1299 - val_loss: 0.1320\nEpoch 192/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1292 - val_loss: 0.1320\nEpoch 193/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1285 - val_loss: 0.1299\nEpoch 194/1000\n348/348 [==============================] - 118s 341ms/step - loss: 0.1278 - val_loss: 0.1300\nEpoch 195/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1272 - val_loss: 0.1293\nEpoch 196/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1265 - val_loss: 0.1293\nEpoch 197/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1259 - val_loss: 0.1283\nEpoch 198/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1254 - val_loss: 0.1269\nEpoch 199/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1248 - val_loss: 0.1266\nEpoch 200/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1243 - val_loss: 0.1260\nEpoch 201/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1237 - val_loss: 0.1254\nEpoch 202/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1231 - val_loss: 0.1252\nEpoch 203/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1224 - val_loss: 0.1243\nEpoch 204/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1217 - val_loss: 0.1238\nEpoch 205/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1210 - val_loss: 0.1230\nEpoch 206/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1202 - val_loss: 0.1222\nEpoch 207/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1194 - val_loss: 0.1212\nEpoch 208/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1187 - val_loss: 0.1209\nEpoch 209/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1178 - val_loss: 0.1202\nEpoch 210/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1170 - val_loss: 0.1195\nEpoch 211/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1162 - val_loss: 0.1183\nEpoch 212/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1155 - val_loss: 0.1176\nEpoch 213/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1149 - val_loss: 0.1167\nEpoch 214/1000\n348/348 [==============================] - 118s 341ms/step - loss: 0.1143 - val_loss: 0.1159\nEpoch 215/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1138 - val_loss: 0.1165\nEpoch 216/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1133 - val_loss: 0.1156\nEpoch 217/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1127 - val_loss: 0.1157\nEpoch 218/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1123 - val_loss: 0.1137\nEpoch 219/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1116 - val_loss: 0.1133\nEpoch 220/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1111 - val_loss: 0.1122\nEpoch 221/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1105 - val_loss: 0.1115\nEpoch 222/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1099 - val_loss: 0.1106\nEpoch 223/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1092 - val_loss: 0.1103\nEpoch 224/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1086 - val_loss: 0.1095\nEpoch 225/1000\n348/348 [==============================] - 118s 341ms/step - loss: 0.1081 - val_loss: 0.1090\nEpoch 226/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1075 - val_loss: 0.1084\nEpoch 227/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1069 - val_loss: 0.1081\nEpoch 228/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1063 - val_loss: 0.1075\nEpoch 229/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1057 - val_loss: 0.1077\nEpoch 230/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1050 - val_loss: 0.1067\nEpoch 231/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1044 - val_loss: 0.1061\nEpoch 232/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1037 - val_loss: 0.1054\nEpoch 233/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1031 - val_loss: 0.1045\nEpoch 234/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1025 - val_loss: 0.1038\nEpoch 235/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1018 - val_loss: 0.1032\nEpoch 236/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.1011 - val_loss: 0.1025\nEpoch 237/1000\n348/348 [==============================] - 118s 340ms/step - loss: 0.1004 - val_loss: 0.1018\nEpoch 238/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0997 - val_loss: 0.1012\nEpoch 239/1000\n348/348 [==============================] - 118s 341ms/step - loss: 0.0991 - val_loss: 0.1014\nEpoch 240/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0986 - val_loss: 0.1004\nEpoch 241/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0981 - val_loss: 0.1000\nEpoch 242/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0977 - val_loss: 0.0995\nEpoch 243/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0972 - val_loss: 0.1002\nEpoch 244/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0968 - val_loss: 0.0994\nEpoch 245/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0964 - val_loss: 0.0992\nEpoch 246/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0961 - val_loss: 0.0987\nEpoch 247/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0957 - val_loss: 0.0981\nEpoch 248/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0953 - val_loss: 0.0975\nEpoch 249/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0949 - val_loss: 0.0968\nEpoch 250/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0944 - val_loss: 0.0963\nEpoch 251/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0940 - val_loss: 0.0957\nEpoch 252/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0935 - val_loss: 0.0951\nEpoch 253/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0931 - val_loss: 0.0955\nEpoch 254/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0927 - val_loss: 0.0946\nEpoch 255/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0924 - val_loss: 0.0938\nEpoch 256/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0920 - val_loss: 0.0937\nEpoch 257/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0916 - val_loss: 0.0933\nEpoch 258/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0912 - val_loss: 0.0934\nEpoch 259/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0909 - val_loss: 0.0930\nEpoch 260/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0905 - val_loss: 0.0923\nEpoch 261/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0901 - val_loss: 0.0919\nEpoch 262/1000\n348/348 [==============================] - 118s 341ms/step - loss: 0.0897 - val_loss: 0.0919\nEpoch 263/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0894 - val_loss: 0.0922\nEpoch 264/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0890 - val_loss: 0.0919\nEpoch 265/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0887 - val_loss: 0.0912\nEpoch 266/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0883 - val_loss: 0.0908\nEpoch 267/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0879 - val_loss: 0.0902\nEpoch 268/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0875 - val_loss: 0.0898\nEpoch 269/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0871 - val_loss: 0.0898\nEpoch 270/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0866 - val_loss: 0.0897\nEpoch 271/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0862 - val_loss: 0.0892\nEpoch 272/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0857 - val_loss: 0.0887\nEpoch 273/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0853 - val_loss: 0.0886\nEpoch 274/1000\n348/348 [==============================] - 118s 341ms/step - loss: 0.0848 - val_loss: 0.0887\nEpoch 275/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0843 - val_loss: 0.0889\nEpoch 276/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0838 - val_loss: 0.0890\nEpoch 277/1000\n348/348 [==============================] - 118s 341ms/step - loss: 0.0833 - val_loss: 0.0863\nEpoch 278/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0828 - val_loss: 0.0859\nEpoch 279/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0824 - val_loss: 0.0853\nEpoch 280/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0819 - val_loss: 0.0846\nEpoch 281/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0814 - val_loss: 0.0842\nEpoch 282/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0811 - val_loss: 0.0844\nEpoch 283/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0807 - val_loss: 0.0843\nEpoch 284/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0803 - val_loss: 0.0839\nEpoch 285/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0799 - val_loss: 0.0836\nEpoch 286/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0795 - val_loss: 0.0829\nEpoch 287/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0791 - val_loss: 0.0827\nEpoch 288/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0787 - val_loss: 0.0825\nEpoch 289/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0783 - val_loss: 0.0822\nEpoch 290/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0779 - val_loss: 0.0817\nEpoch 291/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0774 - val_loss: 0.0821\nEpoch 292/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0771 - val_loss: 0.0802\nEpoch 293/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0766 - val_loss: 0.0801\nEpoch 294/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0763 - val_loss: 0.0797\nEpoch 295/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0760 - val_loss: 0.0794\nEpoch 296/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0756 - val_loss: 0.0791\nEpoch 297/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0753 - val_loss: 0.0782\nEpoch 298/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0749 - val_loss: 0.0778\nEpoch 299/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0746 - val_loss: 0.0774\nEpoch 300/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0743 - val_loss: 0.0770\nEpoch 301/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0739 - val_loss: 0.0766\nEpoch 302/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0735 - val_loss: 0.0762\nEpoch 303/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0730 - val_loss: 0.0757\nEpoch 304/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0726 - val_loss: 0.0750\nEpoch 305/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0721 - val_loss: 0.0745\nEpoch 306/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0716 - val_loss: 0.0740\nEpoch 307/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0712 - val_loss: 0.0734\nEpoch 308/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0708 - val_loss: 0.0727\nEpoch 309/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0706 - val_loss: 0.0724\nEpoch 310/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0702 - val_loss: 0.0719\nEpoch 311/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0699 - val_loss: 0.0715\nEpoch 312/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0696 - val_loss: 0.0712\nEpoch 313/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0692 - val_loss: 0.0719\nEpoch 314/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0690 - val_loss: 0.0715\nEpoch 315/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0686 - val_loss: 0.0710\nEpoch 316/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0683 - val_loss: 0.0704\nEpoch 317/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0680 - val_loss: 0.0703\nEpoch 318/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0677 - val_loss: 0.0702\nEpoch 319/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0674 - val_loss: 0.0698\nEpoch 320/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0670 - val_loss: 0.0695\nEpoch 321/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0667 - val_loss: 0.0688\nEpoch 322/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0664 - val_loss: 0.0693\nEpoch 323/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0662 - val_loss: 0.0693\nEpoch 324/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0660 - val_loss: 0.0689\nEpoch 325/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0658 - val_loss: 0.0686\nEpoch 326/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0656 - val_loss: 0.0691\nEpoch 327/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0654 - val_loss: 0.0696\nEpoch 328/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0652 - val_loss: 0.0693\nEpoch 329/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0650 - val_loss: 0.0685\nEpoch 330/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0647 - val_loss: 0.0682\nEpoch 331/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0644 - val_loss: 0.0689\nEpoch 332/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0641 - val_loss: 0.0688\nEpoch 333/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0639 - val_loss: 0.0672\nEpoch 334/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0636 - val_loss: 0.0672\nEpoch 335/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0633 - val_loss: 0.0674\nEpoch 336/1000\n348/348 [==============================] - 118s 341ms/step - loss: 0.0630 - val_loss: 0.0666\nEpoch 337/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0627 - val_loss: 0.0668\nEpoch 338/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0624 - val_loss: 0.0657\nEpoch 339/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0621 - val_loss: 0.0659\nEpoch 340/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0618 - val_loss: 0.0656\nEpoch 341/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0615 - val_loss: 0.0659\nEpoch 342/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0612 - val_loss: 0.0651\nEpoch 343/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0608 - val_loss: 0.0645\nEpoch 344/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0605 - val_loss: 0.0660\nEpoch 345/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0602 - val_loss: 0.0640\nEpoch 346/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0598 - val_loss: 0.0637\nEpoch 347/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0595 - val_loss: 0.0635\nEpoch 348/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0593 - val_loss: 0.0639\nEpoch 349/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0591 - val_loss: 0.0638\nEpoch 350/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0589 - val_loss: 0.0636\nEpoch 351/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0587 - val_loss: 0.0635\nEpoch 352/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0584 - val_loss: 0.0632\nEpoch 353/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0581 - val_loss: 0.0629\nEpoch 354/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0578 - val_loss: 0.0628\nEpoch 355/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0575 - val_loss: 0.0620\nEpoch 356/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0573 - val_loss: 0.0618\nEpoch 357/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0571 - val_loss: 0.0619\nEpoch 358/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0570 - val_loss: 0.0613\nEpoch 359/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0569 - val_loss: 0.0615\nEpoch 360/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0566 - val_loss: 0.0615\nEpoch 361/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0565 - val_loss: 0.0619\nEpoch 362/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0563 - val_loss: 0.0614\nEpoch 363/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0561 - val_loss: 0.0616\nEpoch 364/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0558 - val_loss: 0.0613\nEpoch 365/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0556 - val_loss: 0.0610\nEpoch 366/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0553 - val_loss: 0.0605\nEpoch 367/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0551 - val_loss: 0.0597\nEpoch 368/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0548 - val_loss: 0.0593\nEpoch 369/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0547 - val_loss: 0.0594\nEpoch 370/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0546 - val_loss: 0.0585\nEpoch 371/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0544 - val_loss: 0.0581\nEpoch 372/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0543 - val_loss: 0.0579\nEpoch 373/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0541 - val_loss: 0.0578\nEpoch 374/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0539 - val_loss: 0.0584\nEpoch 375/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0538 - val_loss: 0.0578\nEpoch 376/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0537 - val_loss: 0.0576\nEpoch 377/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0535 - val_loss: 0.0572\nEpoch 378/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0534 - val_loss: 0.0572\nEpoch 379/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0532 - val_loss: 0.0571\nEpoch 380/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0530 - val_loss: 0.0570\nEpoch 381/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0529 - val_loss: 0.0568\nEpoch 382/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0527 - val_loss: 0.0565\nEpoch 383/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0524 - val_loss: 0.0563\nEpoch 384/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0522 - val_loss: 0.0578\nEpoch 385/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0519 - val_loss: 0.0562\nEpoch 386/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0518 - val_loss: 0.0548\nEpoch 387/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0516 - val_loss: 0.0543\nEpoch 388/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0515 - val_loss: 0.0543\nEpoch 389/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0514 - val_loss: 0.0542\nEpoch 390/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0512 - val_loss: 0.0541\nEpoch 391/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0511 - val_loss: 0.0547\nEpoch 392/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0509 - val_loss: 0.0546\nEpoch 393/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0508 - val_loss: 0.0545\nEpoch 394/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0506 - val_loss: 0.0547\nEpoch 395/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0504 - val_loss: 0.0549\nEpoch 396/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0503 - val_loss: 0.0544\nEpoch 397/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0501 - val_loss: 0.0550\nEpoch 398/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0500 - val_loss: 0.0545\nEpoch 399/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0499 - val_loss: 0.0542\n\nEpoch 00399: ReduceLROnPlateau reducing learning rate to 4.999999987376214e-07.\nEpoch 400/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0497 - val_loss: 0.0541\nEpoch 401/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0497 - val_loss: 0.0534\nEpoch 402/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0496 - val_loss: 0.0535\nEpoch 403/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0496 - val_loss: 0.0538\nEpoch 404/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0495 - val_loss: 0.0537\nEpoch 405/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0494 - val_loss: 0.0537\nEpoch 406/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0493 - val_loss: 0.0536\nEpoch 407/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0492 - val_loss: 0.0527\nEpoch 408/1000\n348/348 [==============================] - 118s 340ms/step - loss: 0.0491 - val_loss: 0.0526\nEpoch 409/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0491 - val_loss: 0.0524\nEpoch 410/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0490 - val_loss: 0.0523\nEpoch 411/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0489 - val_loss: 0.0525\nEpoch 412/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0488 - val_loss: 0.0524\nEpoch 413/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0486 - val_loss: 0.0525\nEpoch 414/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0485 - val_loss: 0.0524\nEpoch 415/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0483 - val_loss: 0.0524\nEpoch 416/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0482 - val_loss: 0.0531\nEpoch 417/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0480 - val_loss: 0.0515\nEpoch 418/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0479 - val_loss: 0.0509\nEpoch 419/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0478 - val_loss: 0.0508\nEpoch 420/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0477 - val_loss: 0.0506\nEpoch 421/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0476 - val_loss: 0.0504\nEpoch 422/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0475 - val_loss: 0.0503\nEpoch 423/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0474 - val_loss: 0.0502\nEpoch 424/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0473 - val_loss: 0.0501\nEpoch 425/1000\n348/348 [==============================] - 118s 340ms/step - loss: 0.0471 - val_loss: 0.0499\nEpoch 426/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0470 - val_loss: 0.0497\nEpoch 427/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0468 - val_loss: 0.0497\nEpoch 428/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0467 - val_loss: 0.0492\nEpoch 429/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0466 - val_loss: 0.0492\nEpoch 430/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0464 - val_loss: 0.0492\nEpoch 431/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0463 - val_loss: 0.0491\nEpoch 432/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0462 - val_loss: 0.0490\nEpoch 433/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0460 - val_loss: 0.0491\nEpoch 434/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0459 - val_loss: 0.0489\nEpoch 435/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0458 - val_loss: 0.0490\nEpoch 436/1000\n348/348 [==============================] - 118s 341ms/step - loss: 0.0457 - val_loss: 0.0490\nEpoch 437/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0456 - val_loss: 0.0489\nEpoch 438/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0455 - val_loss: 0.0488\nEpoch 439/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0454 - val_loss: 0.0486\nEpoch 440/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0452 - val_loss: 0.0485\nEpoch 441/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0450 - val_loss: 0.0483\nEpoch 442/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0449 - val_loss: 0.0481\nEpoch 443/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0446 - val_loss: 0.0480\nEpoch 444/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0444 - val_loss: 0.0480\nEpoch 445/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0443 - val_loss: 0.0479\nEpoch 446/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0441 - val_loss: 0.0477\nEpoch 447/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0440 - val_loss: 0.0480\nEpoch 448/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0439 - val_loss: 0.0465\nEpoch 449/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0438 - val_loss: 0.0464\nEpoch 450/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0437 - val_loss: 0.0465\nEpoch 451/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0437 - val_loss: 0.0463\nEpoch 452/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0436 - val_loss: 0.0460\nEpoch 453/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0436 - val_loss: 0.0461\nEpoch 454/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0435 - val_loss: 0.0462\nEpoch 455/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0435 - val_loss: 0.0466\nEpoch 456/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0434 - val_loss: 0.0465\nEpoch 457/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0433 - val_loss: 0.0465\nEpoch 458/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0433 - val_loss: 0.0464\nEpoch 459/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0432 - val_loss: 0.0463\nEpoch 460/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0431 - val_loss: 0.0462\nEpoch 461/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0430 - val_loss: 0.0458\nEpoch 462/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0429 - val_loss: 0.0457\nEpoch 463/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0428 - val_loss: 0.0456\nEpoch 464/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0428 - val_loss: 0.0455\nEpoch 465/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0427 - val_loss: 0.0454\nEpoch 466/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0426 - val_loss: 0.0454\nEpoch 467/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0426 - val_loss: 0.0454\nEpoch 468/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0425 - val_loss: 0.0451\nEpoch 469/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0425 - val_loss: 0.0453\nEpoch 470/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0424 - val_loss: 0.0452\nEpoch 471/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0424 - val_loss: 0.0451\nEpoch 472/1000\n348/348 [==============================] - 118s 341ms/step - loss: 0.0423 - val_loss: 0.0450\nEpoch 473/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0422 - val_loss: 0.0445\nEpoch 474/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0421 - val_loss: 0.0445\nEpoch 475/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0420 - val_loss: 0.0444\nEpoch 476/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0419 - val_loss: 0.0444\nEpoch 477/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0418 - val_loss: 0.0443\nEpoch 478/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0417 - val_loss: 0.0444\nEpoch 479/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0416 - val_loss: 0.0447\nEpoch 480/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0416 - val_loss: 0.0447\nEpoch 481/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0415 - val_loss: 0.0447\nEpoch 482/1000\n348/348 [==============================] - 118s 340ms/step - loss: 0.0415 - val_loss: 0.0447\nEpoch 483/1000\n348/348 [==============================] - 118s 340ms/step - loss: 0.0414 - val_loss: 0.0447\nEpoch 484/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0414 - val_loss: 0.0449\nEpoch 485/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0413 - val_loss: 0.0448\nEpoch 486/1000\n348/348 [==============================] - 118s 341ms/step - loss: 0.0413 - val_loss: 0.0443\nEpoch 487/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0412 - val_loss: 0.0443\n\nEpoch 00487: ReduceLROnPlateau reducing learning rate to 2.499999993688107e-07.\nEpoch 488/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0412 - val_loss: 0.0440\nEpoch 489/1000\n348/348 [==============================] - 118s 340ms/step - loss: 0.0411 - val_loss: 0.0442\nEpoch 490/1000\n348/348 [==============================] - 118s 341ms/step - loss: 0.0411 - val_loss: 0.0441\nEpoch 491/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0411 - val_loss: 0.0442\nEpoch 492/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0410 - val_loss: 0.0442\nEpoch 493/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0410 - val_loss: 0.0442\nEpoch 494/1000\n348/348 [==============================] - 118s 341ms/step - loss: 0.0410 - val_loss: 0.0442\nEpoch 495/1000\n348/348 [==============================] - 118s 341ms/step - loss: 0.0409 - val_loss: 0.0441\nEpoch 496/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0409 - val_loss: 0.0444\nEpoch 497/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0408 - val_loss: 0.0444\nEpoch 498/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0408 - val_loss: 0.0444\n\nEpoch 00498: ReduceLROnPlateau reducing learning rate to 1.2499999968440534e-07.\nEpoch 499/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0407 - val_loss: 0.0443\nEpoch 500/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0407 - val_loss: 0.0442\nEpoch 501/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0407 - val_loss: 0.0441\nEpoch 502/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0407 - val_loss: 0.0440\nEpoch 503/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0406 - val_loss: 0.0440\nEpoch 504/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0406 - val_loss: 0.0440\nEpoch 505/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0406 - val_loss: 0.0439\nEpoch 506/1000\n348/348 [==============================] - 118s 340ms/step - loss: 0.0405 - val_loss: 0.0439\nEpoch 507/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0405 - val_loss: 0.0438\nEpoch 508/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0404 - val_loss: 0.0438\nEpoch 509/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0404 - val_loss: 0.0437\nEpoch 510/1000\n348/348 [==============================] - 118s 341ms/step - loss: 0.0403 - val_loss: 0.0431\nEpoch 511/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0403 - val_loss: 0.0430\nEpoch 512/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0402 - val_loss: 0.0430\nEpoch 513/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0402 - val_loss: 0.0430\nEpoch 514/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0402 - val_loss: 0.0429\nEpoch 515/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0401 - val_loss: 0.0429\nEpoch 516/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0401 - val_loss: 0.0429\nEpoch 517/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0400 - val_loss: 0.0429\nEpoch 518/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0400 - val_loss: 0.0430\nEpoch 519/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0400 - val_loss: 0.0430\nEpoch 520/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0399 - val_loss: 0.0430\nEpoch 521/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0399 - val_loss: 0.0430\nEpoch 522/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0399 - val_loss: 0.0432\nEpoch 523/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0399 - val_loss: 0.0431\n\nEpoch 00523: ReduceLROnPlateau reducing learning rate to 6.249999984220267e-08.\nEpoch 524/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0398 - val_loss: 0.0431\nEpoch 525/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0398 - val_loss: 0.0431\nEpoch 526/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0398 - val_loss: 0.0431\nEpoch 527/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0398 - val_loss: 0.0431\nEpoch 528/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0398 - val_loss: 0.0431\nEpoch 529/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0398 - val_loss: 0.0431\nEpoch 530/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0398 - val_loss: 0.0431\nEpoch 531/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0397 - val_loss: 0.0430\nEpoch 532/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0397 - val_loss: 0.0430\nEpoch 533/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0397 - val_loss: 0.0430\n\nEpoch 00533: ReduceLROnPlateau reducing learning rate to 3.1249999921101335e-08.\nEpoch 534/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0397 - val_loss: 0.0430\nEpoch 535/1000\n348/348 [==============================] - 119s 341ms/step - loss: 0.0397 - val_loss: 0.0429\nEpoch 00535: early stopping\n"
]
],
[
[
"# PRE trained (training on bird decisions after pretraining on Luscinia triplets)",
"_____no_output_____"
]
],
[
[
"# load pretrained model\ntriplet_model.load_weights('ZF_emb_'+str(emb_size)+'D_LUSCINIA_PRE_margin_loss_backup.h5')",
"_____no_output_____"
],
[
"lo = 6\nhi = 8\nlu = 10\nbatchsize = lo+hi+lu \n\ncpCallback = ModelCheckpoint('ZF_emb_'+str(emb_size)+'D_LUSCINIA_PRE_margin_loss_trained.h5', monitor='val_loss', save_best_only=True, save_weights_only=True, mode='min', save_freq='epoch')\nreduce_lr = ReduceLROnPlateau(monitor='val_loss', factor=0.5, patience=10, verbose=1, min_lr=1e-12)\nearlystop = EarlyStopping(monitor='val_loss', min_delta=0, patience=20, verbose=1, mode='auto')\n\ndis_tr_triplets = discard_some_low(training_triplets, 0.7, 0.7)\ndis_val_triplets = discard_some_low(validation_triplets, 0.7, 0.7)\n\nhistory = triplet_model.fit(train_generator(dis_tr_triplets, M, S, batchsize, emb_size, path_mel),\n steps_per_epoch=int(len(dis_tr_triplets)/batchsize), epochs=1000, verbose=1,\n validation_data=train_generator(dis_val_triplets, M,S, batchsize, emb_size, path_mel),\n validation_steps=int(len(dis_val_triplets)/batchsize), callbacks=[cpCallback, reduce_lr,earlystop])",
"Epoch 1/1000\n58/58 [==============================] - 28s 394ms/step - loss: 0.1178 - val_loss: 0.1106\nEpoch 2/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1201 - val_loss: 0.1079\nEpoch 3/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1196 - val_loss: 0.1073\nEpoch 4/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1166 - val_loss: 0.1070\nEpoch 5/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1183 - val_loss: 0.1068\nEpoch 6/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1158 - val_loss: 0.1066\nEpoch 7/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1166 - val_loss: 0.1064\nEpoch 8/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1157 - val_loss: 0.1062\nEpoch 9/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1146 - val_loss: 0.1060\nEpoch 10/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1154 - val_loss: 0.1058\nEpoch 11/1000\n58/58 [==============================] - 22s 382ms/step - loss: 0.1144 - val_loss: 0.1056\nEpoch 12/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1155 - val_loss: 0.1054\nEpoch 13/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1149 - val_loss: 0.1053\nEpoch 14/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1144 - val_loss: 0.1051\nEpoch 15/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1145 - val_loss: 0.1049\nEpoch 16/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1145 - val_loss: 0.1048\nEpoch 17/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1156 - val_loss: 0.1046\nEpoch 18/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1148 - val_loss: 0.1044\nEpoch 19/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1123 - val_loss: 0.1043\nEpoch 20/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1138 - val_loss: 0.1041\nEpoch 21/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1109 - val_loss: 0.1040\nEpoch 22/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1112 - val_loss: 0.1038\nEpoch 23/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1100 - val_loss: 0.1037\nEpoch 24/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1109 - val_loss: 0.1036\nEpoch 25/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1115 - val_loss: 0.1034\nEpoch 26/1000\n58/58 [==============================] - 22s 378ms/step - loss: 0.1109 - val_loss: 0.1033\nEpoch 27/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1100 - val_loss: 0.1032\nEpoch 28/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1115 - val_loss: 0.1030\nEpoch 29/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1100 - val_loss: 0.1029\nEpoch 30/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1113 - val_loss: 0.1028\nEpoch 31/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1081 - val_loss: 0.1026\nEpoch 32/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1111 - val_loss: 0.1025\nEpoch 33/1000\n58/58 [==============================] - 22s 378ms/step - loss: 0.1086 - val_loss: 0.1024\nEpoch 34/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1096 - val_loss: 0.1023\nEpoch 35/1000\n58/58 [==============================] - 22s 378ms/step - loss: 0.1096 - val_loss: 0.1022\nEpoch 36/1000\n58/58 [==============================] - 22s 378ms/step - loss: 0.1074 - val_loss: 0.1021\nEpoch 37/1000\n58/58 [==============================] - 22s 378ms/step - loss: 0.1076 - val_loss: 0.1020\nEpoch 38/1000\n58/58 [==============================] - 22s 378ms/step - loss: 0.1099 - val_loss: 0.1019\nEpoch 39/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1074 - val_loss: 0.1018\nEpoch 40/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1095 - val_loss: 0.1017\nEpoch 41/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1076 - val_loss: 0.1016\nEpoch 42/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1067 - val_loss: 0.1015\nEpoch 43/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1087 - val_loss: 0.1014\nEpoch 44/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1061 - val_loss: 0.1013\nEpoch 45/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1074 - val_loss: 0.1013\nEpoch 46/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1080 - val_loss: 0.1012\nEpoch 47/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1068 - val_loss: 0.1011\nEpoch 48/1000\n58/58 [==============================] - 22s 378ms/step - loss: 0.1071 - val_loss: 0.1010\nEpoch 49/1000\n58/58 [==============================] - 22s 378ms/step - loss: 0.1058 - val_loss: 0.1009\nEpoch 50/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1061 - val_loss: 0.1008\nEpoch 51/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1072 - val_loss: 0.1008\nEpoch 52/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1070 - val_loss: 0.1007\nEpoch 53/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1077 - val_loss: 0.1006\nEpoch 54/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1065 - val_loss: 0.1005\nEpoch 55/1000\n58/58 [==============================] - 22s 378ms/step - loss: 0.1061 - val_loss: 0.1005\nEpoch 56/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1070 - val_loss: 0.1004\nEpoch 57/1000\n58/58 [==============================] - 22s 378ms/step - loss: 0.1057 - val_loss: 0.1003\nEpoch 58/1000\n58/58 [==============================] - 22s 378ms/step - loss: 0.1057 - val_loss: 0.1003\nEpoch 59/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1042 - val_loss: 0.1002\nEpoch 60/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1059 - val_loss: 0.1001\nEpoch 61/1000\n58/58 [==============================] - 22s 378ms/step - loss: 0.1049 - val_loss: 0.1001\nEpoch 62/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1052 - val_loss: 0.1000\nEpoch 63/1000\n58/58 [==============================] - 22s 378ms/step - loss: 0.1046 - val_loss: 0.1000\nEpoch 64/1000\n58/58 [==============================] - 22s 378ms/step - loss: 0.1035 - val_loss: 0.0999\nEpoch 65/1000\n58/58 [==============================] - 22s 378ms/step - loss: 0.1027 - val_loss: 0.0998\nEpoch 66/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1037 - val_loss: 0.0998\nEpoch 67/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1054 - val_loss: 0.0997\nEpoch 68/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1029 - val_loss: 0.0997\nEpoch 69/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1045 - val_loss: 0.0996\nEpoch 70/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1042 - val_loss: 0.0996\nEpoch 71/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1043 - val_loss: 0.0995\nEpoch 72/1000\n58/58 [==============================] - 22s 381ms/step - loss: 0.1029 - val_loss: 0.0995\nEpoch 73/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1036 - val_loss: 0.0994\nEpoch 74/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1025 - val_loss: 0.0994\nEpoch 75/1000\n58/58 [==============================] - 22s 378ms/step - loss: 0.1024 - val_loss: 0.0994\nEpoch 76/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1038 - val_loss: 0.0993\nEpoch 77/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1029 - val_loss: 0.0993\nEpoch 78/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1022 - val_loss: 0.0992\nEpoch 79/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1040 - val_loss: 0.0992\nEpoch 80/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1027 - val_loss: 0.0991\nEpoch 81/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1037 - val_loss: 0.0991\nEpoch 82/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1027 - val_loss: 0.0991\nEpoch 83/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1029 - val_loss: 0.0990\nEpoch 84/1000\n58/58 [==============================] - 22s 378ms/step - loss: 0.1030 - val_loss: 0.0990\nEpoch 85/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1025 - val_loss: 0.0990\nEpoch 86/1000\n58/58 [==============================] - 22s 378ms/step - loss: 0.1031 - val_loss: 0.0989\nEpoch 87/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1026 - val_loss: 0.0989\nEpoch 88/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1033 - val_loss: 0.0989\nEpoch 89/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1024 - val_loss: 0.0988\nEpoch 90/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1021 - val_loss: 0.0988\nEpoch 91/1000\n58/58 [==============================] - 22s 378ms/step - loss: 0.1021 - val_loss: 0.0988\nEpoch 92/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1029 - val_loss: 0.0987\nEpoch 93/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1022 - val_loss: 0.0987\nEpoch 94/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1011 - val_loss: 0.0987\nEpoch 95/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1014 - val_loss: 0.0987\nEpoch 96/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1029 - val_loss: 0.0986\nEpoch 97/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1013 - val_loss: 0.0986\nEpoch 98/1000\n58/58 [==============================] - 22s 378ms/step - loss: 0.1015 - val_loss: 0.0986\nEpoch 99/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1007 - val_loss: 0.0986\nEpoch 100/1000\n58/58 [==============================] - 22s 378ms/step - loss: 0.1017 - val_loss: 0.0985\nEpoch 101/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1016 - val_loss: 0.0985\nEpoch 102/1000\n58/58 [==============================] - 22s 378ms/step - loss: 0.1021 - val_loss: 0.0985\nEpoch 103/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1010 - val_loss: 0.0985\nEpoch 104/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1006 - val_loss: 0.0984\nEpoch 105/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1004 - val_loss: 0.0984\nEpoch 106/1000\n58/58 [==============================] - 22s 378ms/step - loss: 0.1014 - val_loss: 0.0984\nEpoch 107/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1006 - val_loss: 0.0984\nEpoch 108/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1009 - val_loss: 0.0983\nEpoch 109/1000\n58/58 [==============================] - 22s 378ms/step - loss: 0.1006 - val_loss: 0.0983\nEpoch 110/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1008 - val_loss: 0.0983\nEpoch 111/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0997 - val_loss: 0.0983\nEpoch 112/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0998 - val_loss: 0.0983\nEpoch 113/1000\n58/58 [==============================] - 22s 378ms/step - loss: 0.1008 - val_loss: 0.0983\nEpoch 114/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1004 - val_loss: 0.0982\nEpoch 115/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1017 - val_loss: 0.0982\nEpoch 116/1000\n58/58 [==============================] - 22s 378ms/step - loss: 0.1003 - val_loss: 0.0982\nEpoch 117/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1007 - val_loss: 0.0982\nEpoch 118/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1003 - val_loss: 0.0981\nEpoch 119/1000\n58/58 [==============================] - 22s 378ms/step - loss: 0.1004 - val_loss: 0.0981\nEpoch 120/1000\n58/58 [==============================] - 22s 378ms/step - loss: 0.0989 - val_loss: 0.0981\nEpoch 121/1000\n58/58 [==============================] - 22s 378ms/step - loss: 0.0983 - val_loss: 0.0981\nEpoch 122/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1003 - val_loss: 0.0981\nEpoch 123/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1001 - val_loss: 0.0981\nEpoch 124/1000\n58/58 [==============================] - 22s 378ms/step - loss: 0.0995 - val_loss: 0.0981\nEpoch 125/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1001 - val_loss: 0.0980\nEpoch 126/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0999 - val_loss: 0.0980\nEpoch 127/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0980 - val_loss: 0.0980\nEpoch 128/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1004 - val_loss: 0.0980\nEpoch 129/1000\n58/58 [==============================] - 22s 378ms/step - loss: 0.1013 - val_loss: 0.0980\nEpoch 130/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1000 - val_loss: 0.0980\nEpoch 131/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0999 - val_loss: 0.0979\nEpoch 132/1000\n58/58 [==============================] - 22s 378ms/step - loss: 0.0995 - val_loss: 0.0979\nEpoch 133/1000\n58/58 [==============================] - 22s 381ms/step - loss: 0.1007 - val_loss: 0.0979\nEpoch 134/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0996 - val_loss: 0.0979\nEpoch 135/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0993 - val_loss: 0.0979\nEpoch 136/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1000 - val_loss: 0.0979\nEpoch 137/1000\n58/58 [==============================] - 22s 378ms/step - loss: 0.0986 - val_loss: 0.0979\nEpoch 138/1000\n58/58 [==============================] - 22s 378ms/step - loss: 0.1003 - val_loss: 0.0979\nEpoch 139/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0996 - val_loss: 0.0979\nEpoch 140/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1004 - val_loss: 0.0978\nEpoch 141/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0993 - val_loss: 0.0978\nEpoch 142/1000\n58/58 [==============================] - 22s 378ms/step - loss: 0.0987 - val_loss: 0.0978\nEpoch 143/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0992 - val_loss: 0.0978\nEpoch 144/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1000 - val_loss: 0.0978\nEpoch 145/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0981 - val_loss: 0.0978\nEpoch 146/1000\n58/58 [==============================] - 22s 378ms/step - loss: 0.1002 - val_loss: 0.0978\nEpoch 147/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0981 - val_loss: 0.0978\nEpoch 148/1000\n58/58 [==============================] - 22s 378ms/step - loss: 0.0983 - val_loss: 0.0978\nEpoch 149/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1006 - val_loss: 0.0978\nEpoch 150/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0989 - val_loss: 0.0977\nEpoch 151/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1002 - val_loss: 0.0977\nEpoch 152/1000\n58/58 [==============================] - 22s 378ms/step - loss: 0.0988 - val_loss: 0.0977\nEpoch 153/1000\n58/58 [==============================] - 22s 378ms/step - loss: 0.1004 - val_loss: 0.0977\nEpoch 154/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0983 - val_loss: 0.0977\nEpoch 155/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0982 - val_loss: 0.0977\nEpoch 156/1000\n58/58 [==============================] - 22s 378ms/step - loss: 0.0992 - val_loss: 0.0977\nEpoch 157/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0986 - val_loss: 0.0977\nEpoch 158/1000\n58/58 [==============================] - 22s 378ms/step - loss: 0.0990 - val_loss: 0.0977\nEpoch 159/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0999 - val_loss: 0.0977\n\nEpoch 00159: ReduceLROnPlateau reducing learning rate to 4.999999969612645e-09.\nEpoch 160/1000\n58/58 [==============================] - 22s 378ms/step - loss: 0.0988 - val_loss: 0.0977\nEpoch 161/1000\n58/58 [==============================] - 22s 378ms/step - loss: 0.0980 - val_loss: 0.0977\nEpoch 162/1000\n58/58 [==============================] - 22s 378ms/step - loss: 0.0997 - val_loss: 0.0977\nEpoch 163/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0999 - val_loss: 0.0977\nEpoch 164/1000\n58/58 [==============================] - 22s 378ms/step - loss: 0.0984 - val_loss: 0.0977\nEpoch 165/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0995 - val_loss: 0.0977\nEpoch 166/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0982 - val_loss: 0.0977\nEpoch 167/1000\n58/58 [==============================] - 22s 378ms/step - loss: 0.0984 - val_loss: 0.0977\nEpoch 168/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0978 - val_loss: 0.0977\nEpoch 169/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0990 - val_loss: 0.0977\n\nEpoch 00169: ReduceLROnPlateau reducing learning rate to 2.4999999848063226e-09.\nEpoch 170/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0984 - val_loss: 0.0977\nEpoch 171/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0989 - val_loss: 0.0977\nEpoch 172/1000\n58/58 [==============================] - 22s 378ms/step - loss: 0.0981 - val_loss: 0.0977\nEpoch 173/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0983 - val_loss: 0.0977\nEpoch 174/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0991 - val_loss: 0.0977\nEpoch 175/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.1000 - val_loss: 0.0977\nEpoch 176/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0989 - val_loss: 0.0976\nEpoch 177/1000\n58/58 [==============================] - 22s 378ms/step - loss: 0.0995 - val_loss: 0.0976\nEpoch 178/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0977 - val_loss: 0.0976\nEpoch 179/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0975 - val_loss: 0.0976\nEpoch 180/1000\n58/58 [==============================] - 22s 378ms/step - loss: 0.0981 - val_loss: 0.0976\nEpoch 181/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0998 - val_loss: 0.0976\nEpoch 182/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0979 - val_loss: 0.0976\nEpoch 183/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0982 - val_loss: 0.0976\nEpoch 184/1000\n58/58 [==============================] - 22s 378ms/step - loss: 0.0996 - val_loss: 0.0976\nEpoch 185/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0995 - val_loss: 0.0976\n\nEpoch 00185: ReduceLROnPlateau reducing learning rate to 1.2499999924031613e-09.\nEpoch 186/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0994 - val_loss: 0.0976\nEpoch 187/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0969 - val_loss: 0.0976\nEpoch 188/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0996 - val_loss: 0.0976\nEpoch 189/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0992 - val_loss: 0.0976\nEpoch 190/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0985 - val_loss: 0.0976\nEpoch 191/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0983 - val_loss: 0.0976\nEpoch 192/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0987 - val_loss: 0.0976\nEpoch 193/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0987 - val_loss: 0.0976\nEpoch 194/1000\n58/58 [==============================] - 22s 381ms/step - loss: 0.0975 - val_loss: 0.0976\nEpoch 195/1000\n58/58 [==============================] - 22s 378ms/step - loss: 0.0975 - val_loss: 0.0976\n\nEpoch 00195: ReduceLROnPlateau reducing learning rate to 6.249999962015806e-10.\nEpoch 196/1000\n58/58 [==============================] - 22s 378ms/step - loss: 0.1002 - val_loss: 0.0976\nEpoch 197/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0993 - val_loss: 0.0976\nEpoch 198/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0988 - val_loss: 0.0976\nEpoch 199/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0987 - val_loss: 0.0976\nEpoch 200/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0986 - val_loss: 0.0976\nEpoch 201/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0984 - val_loss: 0.0976\nEpoch 202/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0971 - val_loss: 0.0976\nEpoch 203/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0982 - val_loss: 0.0976\nEpoch 204/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0987 - val_loss: 0.0976\nEpoch 205/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0979 - val_loss: 0.0976\n\nEpoch 00205: ReduceLROnPlateau reducing learning rate to 3.124999981007903e-10.\nEpoch 206/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0972 - val_loss: 0.0976\nEpoch 207/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0993 - val_loss: 0.0976\nEpoch 208/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0991 - val_loss: 0.0976\nEpoch 209/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0989 - val_loss: 0.0976\nEpoch 210/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0974 - val_loss: 0.0976\nEpoch 211/1000\n58/58 [==============================] - 22s 378ms/step - loss: 0.0992 - val_loss: 0.0976\nEpoch 212/1000\n58/58 [==============================] - 22s 378ms/step - loss: 0.0996 - val_loss: 0.0976\nEpoch 213/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0987 - val_loss: 0.0976\nEpoch 214/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0971 - val_loss: 0.0976\nEpoch 215/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0994 - val_loss: 0.0976\n\nEpoch 00215: ReduceLROnPlateau reducing learning rate to 1.5624999905039516e-10.\nEpoch 216/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0989 - val_loss: 0.0976\nEpoch 217/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0989 - val_loss: 0.0976\nEpoch 218/1000\n58/58 [==============================] - 22s 378ms/step - loss: 0.1000 - val_loss: 0.0976\nEpoch 219/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0984 - val_loss: 0.0976\nEpoch 220/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0978 - val_loss: 0.0976\nEpoch 221/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0991 - val_loss: 0.0976\nEpoch 222/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0986 - val_loss: 0.0976\nEpoch 223/1000\n58/58 [==============================] - 22s 378ms/step - loss: 0.0976 - val_loss: 0.0976\nEpoch 224/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0997 - val_loss: 0.0976\nEpoch 225/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0981 - val_loss: 0.0976\n\nEpoch 00225: ReduceLROnPlateau reducing learning rate to 7.812499952519758e-11.\nEpoch 226/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0991 - val_loss: 0.0976\nEpoch 227/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0989 - val_loss: 0.0976\nEpoch 228/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0993 - val_loss: 0.0976\nEpoch 229/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0984 - val_loss: 0.0976\nEpoch 230/1000\n58/58 [==============================] - 22s 378ms/step - loss: 0.0988 - val_loss: 0.0976\nEpoch 231/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0987 - val_loss: 0.0976\nEpoch 232/1000\n58/58 [==============================] - 22s 378ms/step - loss: 0.0987 - val_loss: 0.0976\nEpoch 233/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0974 - val_loss: 0.0976\nEpoch 234/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0994 - val_loss: 0.0976\nEpoch 235/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0989 - val_loss: 0.0976\n\nEpoch 00235: ReduceLROnPlateau reducing learning rate to 3.906249976259879e-11.\nEpoch 236/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0991 - val_loss: 0.0976\nEpoch 237/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0974 - val_loss: 0.0976\nEpoch 238/1000\n58/58 [==============================] - 22s 378ms/step - loss: 0.0977 - val_loss: 0.0976\nEpoch 239/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0983 - val_loss: 0.0976\nEpoch 240/1000\n58/58 [==============================] - 22s 378ms/step - loss: 0.0996 - val_loss: 0.0976\nEpoch 241/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0983 - val_loss: 0.0976\nEpoch 242/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0988 - val_loss: 0.0976\nEpoch 243/1000\n58/58 [==============================] - 22s 378ms/step - loss: 0.0991 - val_loss: 0.0976\nEpoch 244/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0984 - val_loss: 0.0976\nEpoch 245/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0993 - val_loss: 0.0976\n\nEpoch 00245: ReduceLROnPlateau reducing learning rate to 1.9531249881299395e-11.\nEpoch 246/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0986 - val_loss: 0.0976\nEpoch 247/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0998 - val_loss: 0.0976\nEpoch 248/1000\n58/58 [==============================] - 22s 378ms/step - loss: 0.0993 - val_loss: 0.0976\nEpoch 249/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0986 - val_loss: 0.0976\nEpoch 250/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0993 - val_loss: 0.0976\nEpoch 251/1000\n58/58 [==============================] - 22s 378ms/step - loss: 0.0992 - val_loss: 0.0976\nEpoch 252/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0988 - val_loss: 0.0976\nEpoch 253/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0985 - val_loss: 0.0976\nEpoch 254/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0986 - val_loss: 0.0976\nEpoch 255/1000\n58/58 [==============================] - 22s 381ms/step - loss: 0.0983 - val_loss: 0.0976\n\nEpoch 00255: ReduceLROnPlateau reducing learning rate to 9.765624940649698e-12.\nEpoch 256/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0987 - val_loss: 0.0976\nEpoch 257/1000\n58/58 [==============================] - 22s 378ms/step - loss: 0.0984 - val_loss: 0.0976\nEpoch 258/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0983 - val_loss: 0.0976\nEpoch 259/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0984 - val_loss: 0.0976\nEpoch 260/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0988 - val_loss: 0.0976\nEpoch 261/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0988 - val_loss: 0.0976\nEpoch 262/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0982 - val_loss: 0.0976\nEpoch 263/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0987 - val_loss: 0.0976\nEpoch 264/1000\n58/58 [==============================] - 22s 378ms/step - loss: 0.0979 - val_loss: 0.0976\nEpoch 265/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0985 - val_loss: 0.0976\n\nEpoch 00265: ReduceLROnPlateau reducing learning rate to 4.882812470324849e-12.\nEpoch 266/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0990 - val_loss: 0.0976\nEpoch 267/1000\n58/58 [==============================] - 22s 378ms/step - loss: 0.0984 - val_loss: 0.0976\nEpoch 268/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0987 - val_loss: 0.0976\nEpoch 269/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0988 - val_loss: 0.0976\nEpoch 270/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0991 - val_loss: 0.0976\nEpoch 271/1000\n58/58 [==============================] - 22s 378ms/step - loss: 0.0977 - val_loss: 0.0976\nEpoch 272/1000\n58/58 [==============================] - 22s 377ms/step - loss: 0.0992 - val_loss: 0.0976\nEpoch 273/1000\n58/58 [==============================] - 22s 378ms/step - loss: 0.0972 - val_loss: 0.0976\nEpoch 00273: early stopping\n"
]
],
[
[
"# MIXED (training on both bird decisions and Luscinia triplets - w/o pretraining)",
"_____no_output_____"
]
],
[
[
"lo = 6\nhi = 8\nlu = 10\nbatchsize = lo+hi+lu #26\n\ncpCallback = ModelCheckpoint('ZF_emb_'+str(emb_size)+'D_LUSCINIA_MIXED_margin_loss.h5', monitor='val_loss', save_best_only=True, save_weights_only=True, mode='min', save_freq='epoch')\nreduce_lr = ReduceLROnPlateau(monitor='val_loss', factor=0.5, patience=10, verbose=1, min_lr=1e-12)\nearlystop = EarlyStopping(monitor='val_loss', min_delta=0, patience=20, verbose=1, mode='auto')\n\ndis_tr_triplets = discard_some_low(training_triplets, 0.7, 0.7)\ndis_val_triplets = discard_some_low(validation_triplets, 0.7, 0.7)\n\nlow_margin, high_margin, bal_training_triplets = balance_input(dis_tr_triplets, 0.7, hi_balance = hi, lo_balance = lo)\nvlow_margin, vhigh_margin, bal_val_triplets = balance_input(dis_val_triplets, 0.7, hi_balance = hi, lo_balance = lo)\n\n\nhistory = triplet_model.fit(train_generator_mixed(bal_training_triplets, M, S, luscinia_triplets[:luscinia_train_len],M_l, S_l, batchsize, lo, hi, lu, emb_size, path_mel),\n steps_per_epoch=int(len(bal_training_triplets)/(lo+hi)), epochs=1000, verbose=1,\n validation_data=train_generator_mixed(bal_val_triplets, M, S, luscinia_triplets[luscinia_train_len:],M_l, S_l, batchsize, lo, hi, lu, emb_size, path_mel),\n validation_steps=int(len(bal_val_triplets)/(lo+hi)), callbacks=[cpCallback, reduce_lr, earlystop])\n",
"Epoch 1/1000\n117/117 [==============================] - 50s 380ms/step - loss: 0.6301 - val_loss: 0.5736\nEpoch 2/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.6108 - val_loss: 0.5940\nEpoch 3/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.6125 - val_loss: 0.5691\nEpoch 4/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.5836 - val_loss: 0.5496\nEpoch 5/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.5807 - val_loss: 0.5335\nEpoch 6/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.5562 - val_loss: 0.5218\nEpoch 7/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.5476 - val_loss: 0.5129\nEpoch 8/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.5425 - val_loss: 0.5063\nEpoch 9/1000\n117/117 [==============================] - 44s 372ms/step - loss: 0.5374 - val_loss: 0.5008\nEpoch 10/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.5245 - val_loss: 0.4966\nEpoch 11/1000\n117/117 [==============================] - 44s 375ms/step - loss: 0.5251 - val_loss: 0.4928\nEpoch 12/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.5145 - val_loss: 0.4898\nEpoch 13/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.5166 - val_loss: 0.4878\nEpoch 14/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.5131 - val_loss: 0.4860\nEpoch 15/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.5089 - val_loss: 0.4847\nEpoch 16/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.5063 - val_loss: 0.4836\nEpoch 17/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.5037 - val_loss: 0.4827\nEpoch 18/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4987 - val_loss: 0.4817\nEpoch 19/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.5007 - val_loss: 0.4808\nEpoch 20/1000\n117/117 [==============================] - 44s 372ms/step - loss: 0.4962 - val_loss: 0.4801\nEpoch 21/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4969 - val_loss: 0.4796\nEpoch 22/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4932 - val_loss: 0.4791\nEpoch 23/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4944 - val_loss: 0.4787\nEpoch 24/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4937 - val_loss: 0.4782\nEpoch 25/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4924 - val_loss: 0.4778\nEpoch 26/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4897 - val_loss: 0.4774\nEpoch 27/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4895 - val_loss: 0.4769\nEpoch 28/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4886 - val_loss: 0.4766\nEpoch 29/1000\n117/117 [==============================] - 44s 372ms/step - loss: 0.4868 - val_loss: 0.4761\nEpoch 30/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4866 - val_loss: 0.4757\nEpoch 31/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4851 - val_loss: 0.4755\nEpoch 32/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4878 - val_loss: 0.4754\nEpoch 33/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4852 - val_loss: 0.4752\nEpoch 34/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4839 - val_loss: 0.4750\nEpoch 35/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4822 - val_loss: 0.4748\nEpoch 36/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4832 - val_loss: 0.4746\nEpoch 37/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4821 - val_loss: 0.4744\nEpoch 38/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4808 - val_loss: 0.4742\nEpoch 39/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4823 - val_loss: 0.4741\nEpoch 40/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4813 - val_loss: 0.4740\nEpoch 41/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4791 - val_loss: 0.4738\nEpoch 42/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4792 - val_loss: 0.4737\nEpoch 43/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4818 - val_loss: 0.4735\nEpoch 44/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4766 - val_loss: 0.4732\nEpoch 45/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4789 - val_loss: 0.4728\nEpoch 46/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4783 - val_loss: 0.4726\nEpoch 47/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4794 - val_loss: 0.4725\nEpoch 48/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4788 - val_loss: 0.4724\nEpoch 49/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4787 - val_loss: 0.4722\nEpoch 50/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4766 - val_loss: 0.4721\nEpoch 51/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4764 - val_loss: 0.4719\nEpoch 52/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4740 - val_loss: 0.4717\nEpoch 53/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4773 - val_loss: 0.4715\nEpoch 54/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4736 - val_loss: 0.4714\nEpoch 55/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4767 - val_loss: 0.4712\nEpoch 56/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4729 - val_loss: 0.4711\nEpoch 57/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4750 - val_loss: 0.4709\nEpoch 58/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4749 - val_loss: 0.4707\nEpoch 59/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4733 - val_loss: 0.4705\nEpoch 60/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4733 - val_loss: 0.4704\nEpoch 61/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4737 - val_loss: 0.4703\nEpoch 62/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4711 - val_loss: 0.4701\nEpoch 63/1000\n117/117 [==============================] - 44s 374ms/step - loss: 0.4718 - val_loss: 0.4699\nEpoch 64/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4717 - val_loss: 0.4698\nEpoch 65/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4714 - val_loss: 0.4697\nEpoch 66/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4717 - val_loss: 0.4696\nEpoch 67/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4712 - val_loss: 0.4695\nEpoch 68/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4688 - val_loss: 0.4694\nEpoch 69/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4709 - val_loss: 0.4693\nEpoch 70/1000\n117/117 [==============================] - 44s 372ms/step - loss: 0.4701 - val_loss: 0.4691\nEpoch 71/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4702 - val_loss: 0.4690\nEpoch 72/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4687 - val_loss: 0.4688\nEpoch 73/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4680 - val_loss: 0.4687\nEpoch 74/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4708 - val_loss: 0.4686\nEpoch 75/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4677 - val_loss: 0.4685\nEpoch 76/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4686 - val_loss: 0.4683\nEpoch 77/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4686 - val_loss: 0.4681\nEpoch 78/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4668 - val_loss: 0.4680\nEpoch 79/1000\n117/117 [==============================] - 44s 372ms/step - loss: 0.4689 - val_loss: 0.4679\nEpoch 80/1000\n117/117 [==============================] - 44s 372ms/step - loss: 0.4677 - val_loss: 0.4678\nEpoch 81/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4670 - val_loss: 0.4677\nEpoch 82/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4662 - val_loss: 0.4675\nEpoch 83/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4687 - val_loss: 0.4674\nEpoch 84/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4686 - val_loss: 0.4673\nEpoch 85/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4672 - val_loss: 0.4672\nEpoch 86/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4676 - val_loss: 0.4670\nEpoch 87/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4653 - val_loss: 0.4669\nEpoch 88/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4665 - val_loss: 0.4668\nEpoch 89/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4655 - val_loss: 0.4668\nEpoch 90/1000\n117/117 [==============================] - 44s 373ms/step - loss: 0.4644 - val_loss: 0.4667\nEpoch 91/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4660 - val_loss: 0.4665\nEpoch 92/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4638 - val_loss: 0.4665\nEpoch 93/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4642 - val_loss: 0.4664\nEpoch 94/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4650 - val_loss: 0.4663\nEpoch 95/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4649 - val_loss: 0.4663\nEpoch 96/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4621 - val_loss: 0.4662\nEpoch 97/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4648 - val_loss: 0.4661\nEpoch 98/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4645 - val_loss: 0.4660\nEpoch 99/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4650 - val_loss: 0.4659\nEpoch 100/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4636 - val_loss: 0.4658\nEpoch 101/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4631 - val_loss: 0.4657\nEpoch 102/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4628 - val_loss: 0.4656\nEpoch 103/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4617 - val_loss: 0.4655\nEpoch 104/1000\n117/117 [==============================] - 44s 373ms/step - loss: 0.4629 - val_loss: 0.4654\nEpoch 105/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4626 - val_loss: 0.4653\nEpoch 106/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4636 - val_loss: 0.4652\nEpoch 107/1000\n117/117 [==============================] - 44s 372ms/step - loss: 0.4617 - val_loss: 0.4651\nEpoch 108/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4626 - val_loss: 0.4650\nEpoch 109/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4619 - val_loss: 0.4649\nEpoch 110/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4626 - val_loss: 0.4648\nEpoch 111/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4623 - val_loss: 0.4647\nEpoch 112/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4624 - val_loss: 0.4646\nEpoch 113/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4619 - val_loss: 0.4645\nEpoch 114/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4601 - val_loss: 0.4644\nEpoch 115/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4605 - val_loss: 0.4644\nEpoch 116/1000\n117/117 [==============================] - 44s 373ms/step - loss: 0.4607 - val_loss: 0.4643\nEpoch 117/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4604 - val_loss: 0.4642\nEpoch 118/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4612 - val_loss: 0.4642\nEpoch 119/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4608 - val_loss: 0.4641\nEpoch 120/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4598 - val_loss: 0.4640\nEpoch 121/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4595 - val_loss: 0.4638\nEpoch 122/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4596 - val_loss: 0.4638\nEpoch 123/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4604 - val_loss: 0.4637\nEpoch 124/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4600 - val_loss: 0.4636\nEpoch 125/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4590 - val_loss: 0.4635\nEpoch 126/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4588 - val_loss: 0.4634\nEpoch 127/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4593 - val_loss: 0.4634\nEpoch 128/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4595 - val_loss: 0.4633\nEpoch 129/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4584 - val_loss: 0.4632\nEpoch 130/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4592 - val_loss: 0.4631\nEpoch 131/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4591 - val_loss: 0.4631\nEpoch 132/1000\n117/117 [==============================] - 44s 373ms/step - loss: 0.4580 - val_loss: 0.4630\nEpoch 133/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4585 - val_loss: 0.4629\nEpoch 134/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4585 - val_loss: 0.4628\nEpoch 135/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4584 - val_loss: 0.4628\nEpoch 136/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4580 - val_loss: 0.4627\nEpoch 137/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4571 - val_loss: 0.4627\nEpoch 138/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4576 - val_loss: 0.4626\nEpoch 139/1000\n117/117 [==============================] - 44s 374ms/step - loss: 0.4586 - val_loss: 0.4626\nEpoch 140/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4592 - val_loss: 0.4625\nEpoch 141/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4582 - val_loss: 0.4624\nEpoch 142/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4576 - val_loss: 0.4624\nEpoch 143/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4565 - val_loss: 0.4624\nEpoch 144/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4577 - val_loss: 0.4623\nEpoch 145/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4570 - val_loss: 0.4622\nEpoch 146/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4568 - val_loss: 0.4622\nEpoch 147/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4578 - val_loss: 0.4621\nEpoch 148/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4584 - val_loss: 0.4620\nEpoch 149/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4569 - val_loss: 0.4620\nEpoch 150/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4561 - val_loss: 0.4620\nEpoch 151/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4574 - val_loss: 0.4619\nEpoch 152/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4566 - val_loss: 0.4618\nEpoch 153/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4565 - val_loss: 0.4617\nEpoch 154/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4572 - val_loss: 0.4617\nEpoch 155/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4562 - val_loss: 0.4617\nEpoch 156/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4561 - val_loss: 0.4616\nEpoch 157/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4567 - val_loss: 0.4616\nEpoch 158/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4555 - val_loss: 0.4615\nEpoch 159/1000\n117/117 [==============================] - 44s 372ms/step - loss: 0.4556 - val_loss: 0.4615\nEpoch 160/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4550 - val_loss: 0.4614\nEpoch 161/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4560 - val_loss: 0.4613\nEpoch 162/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4537 - val_loss: 0.4613\nEpoch 163/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4563 - val_loss: 0.4612\nEpoch 164/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4560 - val_loss: 0.4612\nEpoch 165/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4554 - val_loss: 0.4611\nEpoch 166/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4555 - val_loss: 0.4611\nEpoch 167/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4553 - val_loss: 0.4611\nEpoch 168/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4542 - val_loss: 0.4610\nEpoch 169/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4536 - val_loss: 0.4610\nEpoch 170/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4554 - val_loss: 0.4610\nEpoch 171/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4552 - val_loss: 0.4610\nEpoch 172/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4540 - val_loss: 0.4609\nEpoch 173/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4537 - val_loss: 0.4609\nEpoch 174/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4537 - val_loss: 0.4609\nEpoch 175/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4548 - val_loss: 0.4608\nEpoch 176/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4540 - val_loss: 0.4608\nEpoch 177/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4533 - val_loss: 0.4608\nEpoch 178/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4519 - val_loss: 0.4607\nEpoch 179/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4537 - val_loss: 0.4607\nEpoch 180/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4531 - val_loss: 0.4606\nEpoch 181/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4534 - val_loss: 0.4606\nEpoch 182/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4535 - val_loss: 0.4606\nEpoch 183/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4538 - val_loss: 0.4605\nEpoch 184/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4541 - val_loss: 0.4605\nEpoch 185/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4522 - val_loss: 0.4604\nEpoch 186/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4516 - val_loss: 0.4604\nEpoch 187/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4528 - val_loss: 0.4604\nEpoch 188/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4530 - val_loss: 0.4603\nEpoch 189/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4539 - val_loss: 0.4603\nEpoch 190/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4531 - val_loss: 0.4602\nEpoch 191/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4521 - val_loss: 0.4602\nEpoch 192/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4514 - val_loss: 0.4602\nEpoch 193/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4517 - val_loss: 0.4601\nEpoch 194/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4522 - val_loss: 0.4601\nEpoch 195/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4523 - val_loss: 0.4601\nEpoch 196/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4512 - val_loss: 0.4600\nEpoch 197/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4513 - val_loss: 0.4599\nEpoch 198/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4523 - val_loss: 0.4599\nEpoch 199/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4506 - val_loss: 0.4599\nEpoch 200/1000\n117/117 [==============================] - 44s 374ms/step - loss: 0.4510 - val_loss: 0.4599\nEpoch 201/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4516 - val_loss: 0.4598\nEpoch 202/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4511 - val_loss: 0.4598\nEpoch 203/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4516 - val_loss: 0.4597\nEpoch 204/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4516 - val_loss: 0.4596\nEpoch 205/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4508 - val_loss: 0.4596\nEpoch 206/1000\n117/117 [==============================] - 44s 372ms/step - loss: 0.4518 - val_loss: 0.4595\nEpoch 207/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4523 - val_loss: 0.4594\nEpoch 208/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4516 - val_loss: 0.4594\nEpoch 209/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4505 - val_loss: 0.4594\nEpoch 210/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4507 - val_loss: 0.4593\nEpoch 211/1000\n117/117 [==============================] - 44s 372ms/step - loss: 0.4511 - val_loss: 0.4593\nEpoch 212/1000\n117/117 [==============================] - 44s 372ms/step - loss: 0.4505 - val_loss: 0.4592\nEpoch 213/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4506 - val_loss: 0.4592\nEpoch 214/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4512 - val_loss: 0.4592\nEpoch 215/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4513 - val_loss: 0.4592\nEpoch 216/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4505 - val_loss: 0.4591\nEpoch 217/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4494 - val_loss: 0.4591\nEpoch 218/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4499 - val_loss: 0.4591\nEpoch 219/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4493 - val_loss: 0.4591\nEpoch 220/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4494 - val_loss: 0.4591\nEpoch 221/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4496 - val_loss: 0.4590\nEpoch 222/1000\n117/117 [==============================] - 44s 372ms/step - loss: 0.4504 - val_loss: 0.4590\nEpoch 223/1000\n117/117 [==============================] - 44s 372ms/step - loss: 0.4495 - val_loss: 0.4590\nEpoch 224/1000\n117/117 [==============================] - 44s 372ms/step - loss: 0.4506 - val_loss: 0.4590\nEpoch 225/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4511 - val_loss: 0.4590\nEpoch 226/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4499 - val_loss: 0.4590\nEpoch 227/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4498 - val_loss: 0.4590\nEpoch 228/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4494 - val_loss: 0.4590\nEpoch 229/1000\n117/117 [==============================] - 44s 372ms/step - loss: 0.4497 - val_loss: 0.4590\n\nEpoch 00229: ReduceLROnPlateau reducing learning rate to 5.000000058430487e-08.\nEpoch 230/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4495 - val_loss: 0.4590\nEpoch 231/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4495 - val_loss: 0.4590\nEpoch 232/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4499 - val_loss: 0.4589\nEpoch 233/1000\n117/117 [==============================] - 44s 372ms/step - loss: 0.4493 - val_loss: 0.4589\nEpoch 234/1000\n117/117 [==============================] - 44s 372ms/step - loss: 0.4486 - val_loss: 0.4589\nEpoch 235/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4485 - val_loss: 0.4589\nEpoch 236/1000\n117/117 [==============================] - 44s 372ms/step - loss: 0.4501 - val_loss: 0.4589\nEpoch 237/1000\n117/117 [==============================] - 44s 372ms/step - loss: 0.4487 - val_loss: 0.4588\nEpoch 238/1000\n117/117 [==============================] - 44s 372ms/step - loss: 0.4483 - val_loss: 0.4588\nEpoch 239/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4486 - val_loss: 0.4588\nEpoch 240/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4492 - val_loss: 0.4588\nEpoch 241/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4488 - val_loss: 0.4587\nEpoch 242/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4491 - val_loss: 0.4587\nEpoch 243/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4485 - val_loss: 0.4587\nEpoch 244/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4480 - val_loss: 0.4587\nEpoch 245/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4484 - val_loss: 0.4587\nEpoch 246/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4482 - val_loss: 0.4587\nEpoch 247/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4498 - val_loss: 0.4587\nEpoch 248/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4490 - val_loss: 0.4586\nEpoch 249/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4489 - val_loss: 0.4586\nEpoch 250/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4477 - val_loss: 0.4586\nEpoch 251/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4478 - val_loss: 0.4586\nEpoch 252/1000\n117/117 [==============================] - 44s 374ms/step - loss: 0.4496 - val_loss: 0.4586\nEpoch 253/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4482 - val_loss: 0.4586\nEpoch 254/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4487 - val_loss: 0.4586\nEpoch 255/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4479 - val_loss: 0.4586\nEpoch 256/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4474 - val_loss: 0.4586\nEpoch 257/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4484 - val_loss: 0.4585\nEpoch 258/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4483 - val_loss: 0.4585\nEpoch 259/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4473 - val_loss: 0.4585\nEpoch 260/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4478 - val_loss: 0.4585\nEpoch 261/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4492 - val_loss: 0.4585\nEpoch 262/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4478 - val_loss: 0.4585\nEpoch 263/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4491 - val_loss: 0.4584\nEpoch 264/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4481 - val_loss: 0.4584\nEpoch 265/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4483 - val_loss: 0.4584\nEpoch 266/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4478 - val_loss: 0.4584\nEpoch 267/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4470 - val_loss: 0.4584\nEpoch 268/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4471 - val_loss: 0.4583\nEpoch 269/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4474 - val_loss: 0.4583\nEpoch 270/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4478 - val_loss: 0.4583\nEpoch 271/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4487 - val_loss: 0.4582\nEpoch 272/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4477 - val_loss: 0.4582\nEpoch 273/1000\n117/117 [==============================] - 44s 372ms/step - loss: 0.4478 - val_loss: 0.4582\nEpoch 274/1000\n117/117 [==============================] - 44s 372ms/step - loss: 0.4489 - val_loss: 0.4582\nEpoch 275/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4470 - val_loss: 0.4582\nEpoch 276/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4479 - val_loss: 0.4581\nEpoch 277/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4478 - val_loss: 0.4581\nEpoch 278/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4484 - val_loss: 0.4581\nEpoch 279/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4480 - val_loss: 0.4581\nEpoch 280/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4477 - val_loss: 0.4580\nEpoch 281/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4478 - val_loss: 0.4580\nEpoch 282/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4478 - val_loss: 0.4580\nEpoch 283/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4476 - val_loss: 0.4580\nEpoch 284/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4472 - val_loss: 0.4580\nEpoch 285/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4473 - val_loss: 0.4580\nEpoch 286/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4465 - val_loss: 0.4580\nEpoch 287/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4473 - val_loss: 0.4579\nEpoch 288/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4475 - val_loss: 0.4579\nEpoch 289/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4468 - val_loss: 0.4579\nEpoch 290/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4473 - val_loss: 0.4579\nEpoch 291/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4470 - val_loss: 0.4579\nEpoch 292/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4486 - val_loss: 0.4579\nEpoch 293/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4464 - val_loss: 0.4579\nEpoch 294/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4475 - val_loss: 0.4578\nEpoch 295/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4459 - val_loss: 0.4578\nEpoch 296/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4463 - val_loss: 0.4578\nEpoch 297/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4462 - val_loss: 0.4577\nEpoch 298/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4453 - val_loss: 0.4577\nEpoch 299/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4477 - val_loss: 0.4577\nEpoch 300/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4472 - val_loss: 0.4577\nEpoch 301/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4470 - val_loss: 0.4576\nEpoch 302/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4469 - val_loss: 0.4576\nEpoch 303/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4465 - val_loss: 0.4576\nEpoch 304/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4472 - val_loss: 0.4576\nEpoch 305/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4467 - val_loss: 0.4575\nEpoch 306/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4470 - val_loss: 0.4575\nEpoch 307/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4471 - val_loss: 0.4576\nEpoch 308/1000\n117/117 [==============================] - 44s 372ms/step - loss: 0.4473 - val_loss: 0.4576\nEpoch 309/1000\n117/117 [==============================] - 44s 372ms/step - loss: 0.4472 - val_loss: 0.4576\nEpoch 310/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4460 - val_loss: 0.4576\nEpoch 311/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4468 - val_loss: 0.4576\n\nEpoch 00311: ReduceLROnPlateau reducing learning rate to 2.5000000292152436e-08.\nEpoch 312/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4454 - val_loss: 0.4576\nEpoch 313/1000\n117/117 [==============================] - 44s 372ms/step - loss: 0.4471 - val_loss: 0.4576\nEpoch 314/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4464 - val_loss: 0.4576\nEpoch 315/1000\n117/117 [==============================] - 44s 372ms/step - loss: 0.4460 - val_loss: 0.4576\nEpoch 316/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4469 - val_loss: 0.4575\nEpoch 317/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4449 - val_loss: 0.4575\nEpoch 318/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4461 - val_loss: 0.4575\nEpoch 319/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4454 - val_loss: 0.4575\nEpoch 320/1000\n117/117 [==============================] - 44s 373ms/step - loss: 0.4467 - val_loss: 0.4575\nEpoch 321/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4461 - val_loss: 0.4575\nEpoch 322/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4471 - val_loss: 0.4575\nEpoch 323/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4465 - val_loss: 0.4575\nEpoch 324/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4463 - val_loss: 0.4575\nEpoch 325/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4457 - val_loss: 0.4575\nEpoch 326/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4466 - val_loss: 0.4574\nEpoch 327/1000\n117/117 [==============================] - 44s 372ms/step - loss: 0.4455 - val_loss: 0.4574\nEpoch 328/1000\n117/117 [==============================] - 44s 374ms/step - loss: 0.4446 - val_loss: 0.4574\nEpoch 329/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4458 - val_loss: 0.4574\nEpoch 330/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4467 - val_loss: 0.4574\nEpoch 331/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4459 - val_loss: 0.4574\nEpoch 332/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4455 - val_loss: 0.4574\nEpoch 333/1000\n117/117 [==============================] - 44s 373ms/step - loss: 0.4454 - val_loss: 0.4574\nEpoch 334/1000\n117/117 [==============================] - 44s 372ms/step - loss: 0.4457 - val_loss: 0.4574\nEpoch 335/1000\n117/117 [==============================] - 44s 372ms/step - loss: 0.4463 - val_loss: 0.4574\nEpoch 336/1000\n117/117 [==============================] - 44s 372ms/step - loss: 0.4456 - val_loss: 0.4574\nEpoch 337/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4460 - val_loss: 0.4573\nEpoch 338/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4454 - val_loss: 0.4573\nEpoch 339/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4443 - val_loss: 0.4573\nEpoch 340/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4450 - val_loss: 0.4573\n\nEpoch 00340: ReduceLROnPlateau reducing learning rate to 1.2500000146076218e-08.\nEpoch 341/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4451 - val_loss: 0.4573\nEpoch 342/1000\n117/117 [==============================] - 44s 372ms/step - loss: 0.4455 - val_loss: 0.4573\nEpoch 343/1000\n117/117 [==============================] - 44s 373ms/step - loss: 0.4463 - val_loss: 0.4573\nEpoch 344/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4457 - val_loss: 0.4573\nEpoch 345/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4466 - val_loss: 0.4573\nEpoch 346/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4460 - val_loss: 0.4573\nEpoch 347/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4462 - val_loss: 0.4573\nEpoch 348/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4462 - val_loss: 0.4573\nEpoch 349/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4461 - val_loss: 0.4573\nEpoch 350/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4457 - val_loss: 0.4573\nEpoch 351/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4444 - val_loss: 0.4573\nEpoch 352/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4452 - val_loss: 0.4573\nEpoch 353/1000\n117/117 [==============================] - 44s 372ms/step - loss: 0.4454 - val_loss: 0.4573\n\nEpoch 00353: ReduceLROnPlateau reducing learning rate to 6.250000073038109e-09.\nEpoch 354/1000\n117/117 [==============================] - 44s 372ms/step - loss: 0.4458 - val_loss: 0.4573\nEpoch 355/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4465 - val_loss: 0.4573\nEpoch 356/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4448 - val_loss: 0.4573\nEpoch 357/1000\n117/117 [==============================] - 44s 372ms/step - loss: 0.4467 - val_loss: 0.4573\nEpoch 358/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4459 - val_loss: 0.4573\nEpoch 359/1000\n117/117 [==============================] - 44s 373ms/step - loss: 0.4454 - val_loss: 0.4573\nEpoch 360/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4452 - val_loss: 0.4573\nEpoch 361/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4451 - val_loss: 0.4573\nEpoch 362/1000\n117/117 [==============================] - 44s 373ms/step - loss: 0.4455 - val_loss: 0.4573\nEpoch 363/1000\n117/117 [==============================] - 44s 372ms/step - loss: 0.4458 - val_loss: 0.4573\n\nEpoch 00363: ReduceLROnPlateau reducing learning rate to 3.1250000365190544e-09.\nEpoch 364/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4454 - val_loss: 0.4573\nEpoch 365/1000\n117/117 [==============================] - 44s 373ms/step - loss: 0.4455 - val_loss: 0.4573\nEpoch 366/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4457 - val_loss: 0.4573\nEpoch 367/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4452 - val_loss: 0.4573\nEpoch 368/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4446 - val_loss: 0.4573\nEpoch 369/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4455 - val_loss: 0.4573\nEpoch 370/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4456 - val_loss: 0.4573\nEpoch 371/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4462 - val_loss: 0.4573\nEpoch 372/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4442 - val_loss: 0.4573\nEpoch 373/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4453 - val_loss: 0.4573\n\nEpoch 00373: ReduceLROnPlateau reducing learning rate to 1.5625000182595272e-09.\nEpoch 374/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4461 - val_loss: 0.4573\nEpoch 375/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4457 - val_loss: 0.4573\nEpoch 376/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4450 - val_loss: 0.4573\nEpoch 377/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4444 - val_loss: 0.4573\nEpoch 378/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4462 - val_loss: 0.4573\nEpoch 379/1000\n117/117 [==============================] - 44s 372ms/step - loss: 0.4467 - val_loss: 0.4573\nEpoch 380/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4449 - val_loss: 0.4573\nEpoch 381/1000\n117/117 [==============================] - 44s 372ms/step - loss: 0.4461 - val_loss: 0.4573\nEpoch 382/1000\n117/117 [==============================] - 44s 373ms/step - loss: 0.4466 - val_loss: 0.4573\nEpoch 383/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4455 - val_loss: 0.4573\n\nEpoch 00383: ReduceLROnPlateau reducing learning rate to 7.812500091297636e-10.\nEpoch 384/1000\n117/117 [==============================] - 44s 373ms/step - loss: 0.4455 - val_loss: 0.4573\nEpoch 385/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4441 - val_loss: 0.4573\nEpoch 386/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4455 - val_loss: 0.4573\nEpoch 387/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4461 - val_loss: 0.4573\nEpoch 388/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4466 - val_loss: 0.4573\nEpoch 389/1000\n117/117 [==============================] - 44s 374ms/step - loss: 0.4459 - val_loss: 0.4573\nEpoch 390/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4444 - val_loss: 0.4573\nEpoch 391/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4470 - val_loss: 0.4573\nEpoch 392/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4460 - val_loss: 0.4573\nEpoch 393/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4447 - val_loss: 0.4573\n\nEpoch 00393: ReduceLROnPlateau reducing learning rate to 3.906250045648818e-10.\nEpoch 394/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4466 - val_loss: 0.4573\nEpoch 395/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4467 - val_loss: 0.4573\nEpoch 396/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4450 - val_loss: 0.4573\nEpoch 397/1000\n117/117 [==============================] - 44s 372ms/step - loss: 0.4450 - val_loss: 0.4573\nEpoch 398/1000\n117/117 [==============================] - 44s 372ms/step - loss: 0.4465 - val_loss: 0.4573\nEpoch 399/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4455 - val_loss: 0.4573\nEpoch 400/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4466 - val_loss: 0.4573\nEpoch 401/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4450 - val_loss: 0.4573\nEpoch 402/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.4461 - val_loss: 0.4573\nEpoch 00402: early stopping\n"
]
],
[
[
"# PRE + MIXED (training on both bird decisions and Luscinia triplets - w/ pretraining on Luscinia)",
"_____no_output_____"
]
],
[
[
"# load pre-trained model\ntriplet_model.load_weights('ZF_emb_'+str(emb_size)+'D_LUSCINIA_PRE_margin_loss_backup.h5')",
"_____no_output_____"
],
[
"lo = 6\nhi = 8\nlu = 10\nbatchsize = lo+hi+lu \n\ncpCallback = ModelCheckpoint('ZF_emb_'+str(emb_size)+'D_LUSCINIA_PRE_MIXED_margin_loss.h5', monitor='val_loss', save_best_only=True, save_weights_only=True, mode='min', save_freq='epoch')\nreduce_lr = ReduceLROnPlateau(monitor='val_loss', factor=0.5, patience=10, verbose=1, min_lr=1e-12)\nearlystop = EarlyStopping(monitor='val_loss', min_delta=0, patience=20, verbose=1, mode='auto')\n\ndis_tr_triplets = discard_some_low(training_triplets, 0.7, 0.7)\ndis_val_triplets = discard_some_low(validation_triplets, 0.7, 0.7)\n\nlow_margin, high_margin, bal_training_triplets = balance_input(dis_tr_triplets, 0.7, hi_balance = hi, lo_balance = lo)\nvlow_margin, vhigh_margin, bal_val_triplets = balance_input(dis_val_triplets, 0.7, hi_balance = hi, lo_balance = lo)\n\n\nhistory = triplet_model.fit(train_generator_mixed(bal_training_triplets, M, S, luscinia_triplets[:luscinia_train_len],M_l, S_l, batchsize, lo, hi, lu, emb_size, path_mel),\n steps_per_epoch=int(len(bal_training_triplets)/(lo+hi)), epochs=1000, verbose=1,\n validation_data=train_generator_mixed(bal_val_triplets, M, S, luscinia_triplets[luscinia_train_len:],M_l, S_l, batchsize, lo, hi, lu, emb_size, path_mel),\n validation_steps=int(len(bal_val_triplets)/(lo+hi)), callbacks=[cpCallback, reduce_lr, earlystop])\n",
"Epoch 1/1000\n117/117 [==============================] - 50s 380ms/step - loss: 0.0973 - val_loss: 0.0863\nEpoch 2/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.0958 - val_loss: 0.0856\nEpoch 3/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.0949 - val_loss: 0.0852\nEpoch 4/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.0937 - val_loss: 0.0849\nEpoch 5/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.0921 - val_loss: 0.0845\nEpoch 6/1000\n117/117 [==============================] - 44s 375ms/step - loss: 0.0927 - val_loss: 0.0842\nEpoch 7/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.0911 - val_loss: 0.0839\nEpoch 8/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.0919 - val_loss: 0.0836\nEpoch 9/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.0894 - val_loss: 0.0834\nEpoch 10/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.0896 - val_loss: 0.0831\nEpoch 11/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.0880 - val_loss: 0.0828\nEpoch 12/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.0874 - val_loss: 0.0826\nEpoch 13/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.0871 - val_loss: 0.0824\nEpoch 14/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.0871 - val_loss: 0.0822\nEpoch 15/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.0865 - val_loss: 0.0820\nEpoch 16/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.0837 - val_loss: 0.0818\nEpoch 17/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.0843 - val_loss: 0.0816\nEpoch 18/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.0842 - val_loss: 0.0815\nEpoch 19/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.0828 - val_loss: 0.0813\nEpoch 20/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.0832 - val_loss: 0.0812\nEpoch 21/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.0832 - val_loss: 0.0810\nEpoch 22/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.0824 - val_loss: 0.0809\nEpoch 23/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.0821 - val_loss: 0.0807\nEpoch 24/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.0799 - val_loss: 0.0806\nEpoch 25/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.0817 - val_loss: 0.0805\nEpoch 26/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.0806 - val_loss: 0.0804\nEpoch 27/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.0807 - val_loss: 0.0803\nEpoch 28/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.0788 - val_loss: 0.0801\nEpoch 29/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.0803 - val_loss: 0.0800\nEpoch 30/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.0802 - val_loss: 0.0799\nEpoch 31/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.0807 - val_loss: 0.0798\nEpoch 32/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.0788 - val_loss: 0.0797\nEpoch 33/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.0781 - val_loss: 0.0797\nEpoch 34/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.0802 - val_loss: 0.0796\nEpoch 35/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.0789 - val_loss: 0.0795\nEpoch 36/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.0791 - val_loss: 0.0794\nEpoch 37/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.0778 - val_loss: 0.0794\nEpoch 38/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.0779 - val_loss: 0.0793\nEpoch 39/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.0781 - val_loss: 0.0792\nEpoch 40/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.0777 - val_loss: 0.0792\nEpoch 41/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.0786 - val_loss: 0.0791\nEpoch 42/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.0780 - val_loss: 0.0791\nEpoch 43/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.0783 - val_loss: 0.0790\nEpoch 44/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.0779 - val_loss: 0.0790\nEpoch 45/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.0777 - val_loss: 0.0789\nEpoch 46/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.0765 - val_loss: 0.0789\nEpoch 47/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.0770 - val_loss: 0.0788\nEpoch 48/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.0763 - val_loss: 0.0788\nEpoch 49/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.0768 - val_loss: 0.0788\nEpoch 50/1000\n117/117 [==============================] - 44s 373ms/step - loss: 0.0769 - val_loss: 0.0787\nEpoch 51/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.0768 - val_loss: 0.0787\nEpoch 52/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.0758 - val_loss: 0.0787\nEpoch 53/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.0762 - val_loss: 0.0787\nEpoch 54/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.0760 - val_loss: 0.0786\nEpoch 55/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.0766 - val_loss: 0.0786\nEpoch 56/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.0753 - val_loss: 0.0786\nEpoch 57/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.0749 - val_loss: 0.0786\nEpoch 58/1000\n117/117 [==============================] - 44s 373ms/step - loss: 0.0753 - val_loss: 0.0785\nEpoch 59/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.0751 - val_loss: 0.0785\nEpoch 60/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.0754 - val_loss: 0.0785\nEpoch 61/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.0755 - val_loss: 0.0785\nEpoch 62/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.0752 - val_loss: 0.0785\nEpoch 63/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.0749 - val_loss: 0.0785\nEpoch 64/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.0750 - val_loss: 0.0785\nEpoch 65/1000\n117/117 [==============================] - 44s 373ms/step - loss: 0.0751 - val_loss: 0.0785\nEpoch 66/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.0750 - val_loss: 0.0785\nEpoch 67/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.0748 - val_loss: 0.0785\nEpoch 68/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.0756 - val_loss: 0.0784\nEpoch 69/1000\n117/117 [==============================] - 43s 372ms/step - loss: 0.0747 - val_loss: 0.0784\nEpoch 70/1000\n 66/117 [===============>..............] - ETA: 17s - loss: 0.0731"
]
],
[
[
"# Evaluation (for both ambiguous and unambiguous triplets - based on different distance margins)",
"_____no_output_____"
]
],
[
[
"def low_high_evaluation(path_mel, vhigh_margin, vlow_margin, triplet_model, margin = 0.00000001, max_margin = 0.0000001, step = 0.00000001):\n # pos, neg, anc\n while margin < max_margin:\n acc_cnt = 0\n high_cnt = 0\n low_cnt = 0\n for triplet in vhigh_margin:\n tr_pos = triplet[1][:-4]+'.pckl'\n tr_neg = triplet[2][:-4]+'.pckl'\n tr_anc = triplet[3][:-4]+'.pckl'\n\n f = open(path_mel+tr_anc, 'rb')\n anc = pickle.load(f).T\n f.close()\n anc = (anc - M)/S\n anc = np.expand_dims(anc, axis=0)\n anc = np.expand_dims(anc, axis=-1)\n\n f = open(path_mel+tr_pos, 'rb')\n pos = pickle.load(f).T\n f.close()\n pos = (pos - M)/S\n pos = np.expand_dims(pos, axis=0)\n pos = np.expand_dims(pos, axis=-1)\n\n f = open(path_mel+tr_neg, 'rb')\n neg = pickle.load(f).T\n f.close()\n neg = (neg - M)/S\n neg = np.expand_dims(neg, axis=0)\n neg = np.expand_dims(neg, axis=-1)\n\n y_pred = triplet_model.predict([anc, pos, neg])\n\n anchor1 = y_pred[:, 0:emb_size]\n positive1 = y_pred[:, emb_size:emb_size*2]\n negative1 = y_pred[:, emb_size*2:emb_size*3]\n\n pos_dist = np.sqrt(np.sum(np.square(anchor1 - positive1), axis=1))[0]\n neg_dist = np.sqrt(np.sum(np.square(anchor1 - negative1), axis=1))[0]\n\n if np.square(neg_dist) > np.square(pos_dist) + margin:\n acc_cnt += 1\n high_cnt += 1\n for triplet in vlow_margin:\n tr_pos = triplet[1][:-4]+'.pckl'\n tr_neg = triplet[2][:-4]+'.pckl'\n tr_anc = triplet[3][:-4]+'.pckl'\n\n f = open(path_mel+tr_anc, 'rb')\n anc = pickle.load(f).T\n f.close()\n anc = (anc - M)/S\n anc = np.expand_dims(anc, axis=0)\n anc = np.expand_dims(anc, axis=-1)\n\n f = open(path_mel+tr_pos, 'rb')\n pos = pickle.load(f).T\n f.close()\n pos = (pos - M)/S\n pos = np.expand_dims(pos, axis=0)\n pos = np.expand_dims(pos, axis=-1)\n\n f = open(path_mel+tr_neg, 'rb')\n neg = pickle.load(f).T\n f.close()\n neg = (neg - M)/S\n neg = np.expand_dims(neg, axis=0)\n neg = np.expand_dims(neg, axis=-1)\n\n y_pred = triplet_model.predict([anc, pos, neg])\n\n anchor1 = y_pred[:, 0:emb_size]\n positive1 = y_pred[:, emb_size:emb_size*2]\n negative1 = y_pred[:, emb_size*2:emb_size*3]\n\n pos_dist = np.sqrt(np.sum(np.square(anchor1 - positive1), axis=1))[0]\n neg_dist = np.sqrt(np.sum(np.square(anchor1 - negative1), axis=1))[0]\n\n if np.abs(np.square(pos_dist) - np.square(neg_dist)) <= margin:\n acc_cnt+=1\n low_cnt+=1\n print('MARGIN = ', margin)\n print('Macro-average Low-High margin accuracy: ',0.5*(high_cnt/(len(vhigh_margin)) + low_cnt/(len(vlow_margin)))*100, '%')\n print('Micro-average Low-High margin accuracy: ',(acc_cnt/(len(vhigh_margin)+len(vlow_margin)))*100, '%') \n print('High margin accuracy: ',(high_cnt/(len(vhigh_margin)))*100, '%') \n print('Low margin accuracy: ',(low_cnt/(len(vlow_margin)))*100, '%') \n margin += step\n \n return ",
"_____no_output_____"
],
[
"# Separate sets between low-margin (ambiguous) and high-margin (unambiguous) triplets\nlow_margin = pickle.load( open(path_files+'train_triplets_low_50_70_ACC70.pckl', 'rb'))\nvlow_margin = pickle.load(open(path_files+'val_triplets_low_50_70_ACC70.pckl', 'rb'))\nhigh_margin = pickle.load(open(path_files+'train_triplets_high_50_70_ACC70.pckl', 'rb'))\nvhigh_margin =pickle.load(open(path_files+'val_triplets_high_50_70_ACC70.pckl', 'rb'))\ntlow_margin =pickle.load(open(path_files+'test_triplets_low_50_70_ACC70.pckl', 'rb'))\nthigh_margin = pickle.load(open(path_files+'test_triplets_high_50_70_ACC70.pckl', 'rb'))",
"_____no_output_____"
],
[
"# run evaluation on a high margin and low margin set of the same split\nlow_high_evauation(path_mel, high_margin, low_margin, triplet_model, margin = 0.0, max_margin = 0.01, step = 0.005)",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
cbecf686a1f5369a3ec51ad6ac301be03ee2b419
| 71,907 |
ipynb
|
Jupyter Notebook
|
regularized-wasserstein-estimator/SGDforWassEst.ipynb
|
MarinBallu/regularized-wasserstein-estimator
|
aeb21778180a5f7b88789ac9640bf0aa90a07552
|
[
"MIT"
] | null | null | null |
regularized-wasserstein-estimator/SGDforWassEst.ipynb
|
MarinBallu/regularized-wasserstein-estimator
|
aeb21778180a5f7b88789ac9640bf0aa90a07552
|
[
"MIT"
] | null | null | null |
regularized-wasserstein-estimator/SGDforWassEst.ipynb
|
MarinBallu/regularized-wasserstein-estimator
|
aeb21778180a5f7b88789ac9640bf0aa90a07552
|
[
"MIT"
] | null | null | null | 138.816602 | 11,292 | 0.861599 |
[
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\nfrom solve import solve_dual_entropic",
"_____no_output_____"
],
[
"### INITIALIZE TEST\n\n# set seed\nnp.random.seed(0)\n\n# set dimension\nn_source = 30\nn_target = 30\n\n# set target measure\n#a = np.ones(n_source)\n#a = a/a.sum()\na = 1.5 + np.sin([1*(k / n_source) * 15 for k in range(n_source)]) \na = a / a.sum()\n\n# set prior measure\nb = np.ones(n_target)\nb = b/b.sum()\n\n# random distance matrix\n# rng = np.random.RandomState(0)\n# X_source = rng.randn(n_source, 2)\n# Y_target = rng.randn(n_target, 2)\n# M = ot.dist(X_source, Y_target)\n\n# discrete distance\n# M = (np.ones(n_source) - np.identity(n_source))\n\n# distance on the line\nX_source = np.array([k for k in range(n_source)])\nY_target = X_source\nM = abs(X_source[:,None] - Y_target[None, :])\n\n# normalize distance matrix (optional)\nM = M / (M.max() - M.min())\n\n# make distance matrix positive (mandatory!)\nM = M - M.min()\n\n# graph of matrix and target measure\nfig, ax = plt.subplots()\nprint(\"Cost matrix\")\nax.imshow(M, cmap=plt.cm.Blues)\nplt.axis('off')\nplt.show()\nfig.savefig('tmp/cost_matrix.pdf', bbox_inches='tight')\n\nprint(\"Target measure\")\nplt.bar(range(n_source), a)\nplt.show()",
"Cost matrix\n"
],
[
"from graphs import performance_graphs\n\n### PERFORMANCE TEST 1\n# set seed\nnp.random.seed(1)\n# set regularizer parameter\nreg1 = 0.1\nreg2 = 0.1\n# set learning rate (reg1 is close to optimal, using a bigger one might make it diverge)\nlr = reg1\n# set batch size (0 means use of full gradient in beta, while stochastic in alpha)\nbatch_size = 1\n# set algorithmic parameters (count between 10000 and 70000 iterations per seconds)\nnumItermax = 1000000\nmaxTime = 100\n\navg_alpha_list, avg_beta_list, time_list = solve_dual_entropic(a, b, M, reg1, reg2, numItermax, batch_size, lr, maxTime)\nperformance_graphs(a, b, M, reg1, reg2, avg_alpha_list, avg_beta_list, time_list)",
"Nb iter: 1000000\nTime: 26.3506757\nAverage iteration time: 2.63506757e-05\nTransportation matrix\n"
],
[
"### PERFORMANCE TEST 2\n# set seed\nnp.random.seed(1)\n# set regularizer parameter\nreg1 = 0.01\nreg2 = 0.01\n# set learning rate (reg1 is close to optimal, using a bigger one might make it diverge)\nlr = reg1\n# set batch size (0 means use of full gradient in beta, while stochastic in alpha)\nbatch_size = 1\n# set algorithmic parameters (count between 10000 and 70000 iterations per seconds)\nnumItermax = 1000000\nmaxTime = 100\n\navg_alpha_list, avg_beta_list, time_list = solve_dual_entropic(a, b, M, reg1, reg2, numItermax, batch_size, lr, maxTime)\nperformance_graphs(a, b, M, reg1, reg2, avg_alpha_list, avg_beta_list, time_list)",
"Nb iter: 1000000\nTime: 26.225920900000006\nAverage iteration time: 2.6225920900000005e-05\nTransportation matrix\n"
],
[
"from graphs import compare_results\n\n# PERFORMANCE: when regularization varies\nnumItermax = 1000000\nnp.random.seed(3)\nreg1 = 0.01\nreg2 = 0.01\nlr = reg1\navg_alpha_list1, avg_beta_list1, time_list1 = solve_dual_entropic(a, b, M, reg1, reg2, numItermax, batch_size, lr/8, maxTime)\navg_alpha_list2, avg_beta_list2, time_list2 = solve_dual_entropic(a, b, M, reg1, reg2, numItermax, batch_size, lr/4, maxTime)\navg_alpha_list3, avg_beta_list3, time_list3 = solve_dual_entropic(a, b, M, reg1, reg2, numItermax, batch_size, lr/2, maxTime)\navg_alpha_list4, avg_beta_list4, time_list4 = solve_dual_entropic(a, b, M, reg1, reg2, numItermax, batch_size, lr, maxTime)\navg_alpha_list5, avg_beta_list5, time_list5 = solve_dual_entropic(a, b, M, reg1, reg2, numItermax, batch_size, 2 * lr, maxTime)\nlist_results_alpha = [avg_alpha_list1, avg_alpha_list2, avg_alpha_list3, avg_alpha_list4, avg_alpha_list5]\nlist_results_beta = [avg_beta_list1, avg_beta_list2, avg_beta_list3, avg_beta_list4, avg_beta_list5]\ncompare_results(list_results_alpha, list_results_beta)",
"Nb iter: 1000000\nTime: 26.236947999999984\nAverage iteration time: 2.6236947999999984e-05\nNb iter: 1000000\nTime: 24.212753299999974\nAverage iteration time: 2.4212753299999976e-05\nNb iter: 1000000\nTime: 24.79928849999999\nAverage iteration time: 2.479928849999999e-05\nNb iter: 1000000\nTime: 25.77947119999999\nAverage iteration time: 2.577947119999999e-05\n"
],
[
"# PERFORMANCE: when dimension varies\n# This initializes the variables with sample gaussians\ndef generate_var(n_source, n_target):\n X_source = rng.randn(n_source, 3)\n Y_target = rng.randn(n_target, 3)\n M = ot.dist(X_source, Y_target)\n M = M / (M.max() - M.min())\n M = M - M.min()\n a = np.ones(n_source)/n_source\n b = np.ones(n_target)/n_target\n return X_source, Y_target, M, a, b\nbatch_size = 1\nnp.random.seed(4)\nreg1 = 0.01\nreg2 = 0.01\nlr = reg1\nnumItermax = 1000000\nlist_dimensions = [[30, 30], [100, 30], [30,100], [100, 100]]\nlist_results_alpha, list_results_beta = [], []\nrng = np.random.RandomState(0)\nfig, ax = plt.subplots()\nSetPlotRC()\nfor D in list_dimensions:\n n_source = D[0]\n n_target = D[1]\n X_source, Y_target, M, a, b = generate_var(n_source, n_target)\n avg_alpha_list, avg_beta_list, time_list = solve_dual_entropic(a, b, M, reg1, reg2, numItermax, batch_size, lr, maxTime)\n target_list = [dual_to_target(b, reg2, beta) for beta in avg_beta_list]\n norm0 = norm_grad_dual(a, b, target_list[0], M, reg1, avg_alpha_list[0], avg_beta_list[0])\n grad_norm_list = [norm_grad_dual(a, b, target_list[i], M, reg1, avg_alpha_list[i], avg_beta_list[i])/norm0 for i in range(len(avg_alpha_list))]\n print(\"Final gradient norm:\", grad_norm_list[-1])\n graph_loglog(grad_norm_list)\nlabels=['I = ' + str(D[0]) + ', J = ' + str(D[1]) for D in list_dimensions]\nplt.grid()\nplt.ylabel('Gradient norm', fontsize=12)\nplt.xlabel('Number of iterations', fontsize=12)\nplt.legend(labels)\nApplyFont(plt.gca())\nfig.savefig('tmp/dimension_sens.pdf', bbox_inches='tight')\nplt.show()",
"_____no_output_____"
],
[
"## PREPARE FOR ANIMATION\n\nimport matplotlib.pyplot as plt\nfrom matplotlib import animation\nfrom IPython.display import HTML\n\ntarget_list = [dual_to_target(b, reg2, beta) for beta in avg_beta_list[:50000]]\n\n#def transport_map(alpha, beta, M, reg1, a, b):\n# G = np.exp((alpha[:, None] + beta[None, :] - M) / reg1) * a[:, None] * b[None, :]\n# return G / G.sum()\n\n#transport_map_list = [transport_map(avg_alpha_list[i], avg_beta_list[i], M, reg1, a, b) for i in range(numItermax)]",
"_____no_output_____"
],
[
"# ANIMATION OF THE TARGET MEASURE\n\nfig=plt.figure()\n# Number of frames (100 frames per second)\nn = 1000\n\nk = int(len(target_list)) // n\nbarWidth = 0.4\n\nr1 = np.arange(n_source)\nr2 = [x + barWidth for x in r1]\nbarcollection = plt.bar(r1, target_list[0], width=barWidth)\nbarcollection1 = plt.bar(r2, target_list[-1], width=barWidth)\nplt.ylim(0, np.max(target_list[-1])*1.1)\n\ndef animate(t):\n y=target_list[t * k]\n for i, b in enumerate(barcollection):\n b.set_height(y[i])\n\nanim=animation.FuncAnimation(fig, animate, repeat=False, blit=False, frames=n,\n interval=10)\n\nHTML(anim.to_html5_video())",
"_____no_output_____"
],
[
"# ANIMATION OF THE TRANSPORT MAP\nfig=plt.figure()\n# Number of frames (100 frames per second)\nn = 500\n\nk = len(transport_map_list) // n\nims = []\nfor i in range(n):\n im = plt.imshow(transport_map_list[i*k], animated=True, cmap=plt.cm.Blues)\n ims.append([im])\nanim2 = animation.ArtistAnimation(fig, ims, repeat=False, blit=False,\n interval=10)\n\nHTML(anim2.to_html5_video())",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbed192e87db805723e493936c37422366751e9b
| 14,938 |
ipynb
|
Jupyter Notebook
|
notebooks/The ISB-CGC open-access TCGA tables in BigQuery.ipynb
|
deflaux/examples-Python
|
74ffb7cd953e514b83bb9191abef01c6c0fee43e
|
[
"Apache-2.0"
] | 47 |
2015-11-22T03:17:59.000Z
|
2022-02-02T10:37:59.000Z
|
notebooks/The ISB-CGC open-access TCGA tables in BigQuery.ipynb
|
deflaux/examples-Python
|
74ffb7cd953e514b83bb9191abef01c6c0fee43e
|
[
"Apache-2.0"
] | 2 |
2017-02-11T00:38:34.000Z
|
2018-07-12T00:23:03.000Z
|
notebooks/The ISB-CGC open-access TCGA tables in BigQuery.ipynb
|
deflaux/examples-Python
|
74ffb7cd953e514b83bb9191abef01c6c0fee43e
|
[
"Apache-2.0"
] | 22 |
2015-11-18T00:38:51.000Z
|
2022-01-13T23:20:06.000Z
| 81.628415 | 969 | 0.716562 |
[
[
[
"# The ISB-CGC open-access TCGA tables in Big-Query\n\nThe goal of this notebook is to introduce you to a new publicly-available, open-access dataset in BigQuery. This set of BigQuery tables was produced by the [ISB-CGC](http://www.isb-cgc.org) project, based on the open-access [TCGA](http://cancergenome.nih.gov/) data available at the TCGA [Data Portal](https://tcga-data.nci.nih.gov/tcga/). You will need to have access to a Google Cloud Platform (GCP) project in order to use BigQuery. If you don't already have one, you can sign up for a [free-trial](https://cloud.google.com/free-trial/) or contact [us](mailto://[email protected]) and become part of the community evaluation phase of our Cancer Genomics Cloud pilot. (You can find more information about this NCI-funded program [here](https://cbiit.nci.nih.gov/ncip/nci-cancer-genomics-cloud-pilots).)\n\nWe are not attempting to provide a thorough BigQuery or IPython tutorial here, as a wealth of such information already exists. Here are links to some resources that you might find useful: \n* [BigQuery](https://cloud.google.com/bigquery/what-is-bigquery), \n* the BigQuery [web UI](https://bigquery.cloud.google.com/) where you can run queries interactively, \n* [IPython](http://ipython.org/) (now known as [Jupyter](http://jupyter.org/)), and \n* [Cloud Datalab](https://cloud.google.com/datalab/) the recently announced interactive cloud-based platform that this notebook is being developed on. \n\nThere are also many tutorials and samples available on github (see, in particular, the [datalab](https://github.com/GoogleCloudPlatform/datalab) repo and the [Google Genomics]( https://github.com/googlegenomics) project).\n\nIn order to work with BigQuery, the first thing you need to do is import the [gcp.bigquery](http://googlecloudplatform.github.io/datalab/gcp.bigquery.html) package:",
"_____no_output_____"
]
],
[
[
"import gcp.bigquery as bq",
"_____no_output_____"
]
],
[
[
"The next thing you need to know is how to access the specific tables you are interested in. BigQuery tables are organized into datasets, and datasets are owned by a specific GCP project. The tables we are introducing in this notebook are in a dataset called **`tcga_201607_beta`**, owned by the **`isb-cgc`** project. A full table identifier is of the form `<project_id>:<dataset_id>.<table_id>`. Let's start by getting some basic information about the tables in this dataset:",
"_____no_output_____"
]
],
[
[
"d = bq.DataSet('isb-cgc:tcga_201607_beta')\nfor t in d.tables():\n print '%10d rows %12d bytes %s' \\\n % (t.metadata.rows, t.metadata.size, t.name.table_id)",
" 6322 rows 1729204 bytes Annotations\n 23797 rows 6382147 bytes Biospecimen_data\n 11160 rows 4201379 bytes Clinical_data\n 2646095 rows 333774244 bytes Copy_Number_segments\n3944304319 rows 445303830985 bytes DNA_Methylation_betas\n 382335670 rows 43164264006 bytes DNA_Methylation_chr1\n 197519895 rows 22301345198 bytes DNA_Methylation_chr10\n 235823572 rows 26623975945 bytes DNA_Methylation_chr11\n 198050739 rows 22359642619 bytes DNA_Methylation_chr12\n 97301675 rows 10986815862 bytes DNA_Methylation_chr13\n 123239379 rows 13913712352 bytes DNA_Methylation_chr14\n 124566185 rows 14064712239 bytes DNA_Methylation_chr15\n 179772812 rows 20296128173 bytes DNA_Methylation_chr16\n 234003341 rows 26417830751 bytes DNA_Methylation_chr17\n 50216619 rows 5669139362 bytes DNA_Methylation_chr18\n 211386795 rows 23862583107 bytes DNA_Methylation_chr19\n 279668485 rows 31577200462 bytes DNA_Methylation_chr2\n 86858120 rows 9805923353 bytes DNA_Methylation_chr20\n 35410447 rows 3997986812 bytes DNA_Methylation_chr21\n 70676468 rows 7978947938 bytes DNA_Methylation_chr22\n 201119616 rows 22705358910 bytes DNA_Methylation_chr3\n 159148744 rows 17968482285 bytes DNA_Methylation_chr4\n 195864180 rows 22113162401 bytes DNA_Methylation_chr5\n 290275524 rows 32772371379 bytes DNA_Methylation_chr6\n 240010275 rows 27097948808 bytes DNA_Methylation_chr7\n 164810092 rows 18607886221 bytes DNA_Methylation_chr8\n 81260723 rows 9173717922 bytes DNA_Methylation_chr9\n 98082681 rows 11072059468 bytes DNA_Methylation_chrX\n 2330426 rows 263109775 bytes DNA_Methylation_chrY\n 1867233 rows 207365611 bytes Protein_RPPA_data\n 5356089 rows 5715538107 bytes Somatic_Mutation_calls\n 5738048 rows 657855993 bytes mRNA_BCGSC_GA_RPKM\n 38299138 rows 4459086535 bytes mRNA_BCGSC_HiSeq_RPKM\n 44037186 rows 5116942528 bytes mRNA_BCGSC_RPKM\n 16794358 rows 1934755686 bytes mRNA_UNC_GA_RSEM\n 211284521 rows 24942992190 bytes mRNA_UNC_HiSeq_RSEM\n 228078879 rows 26877747876 bytes mRNA_UNC_RSEM\n 11997545 rows 2000881026 bytes miRNA_BCGSC_GA_isoform\n 4503046 rows 527101917 bytes miRNA_BCGSC_GA_mirna\n 90237323 rows 15289326462 bytes miRNA_BCGSC_HiSeq_isoform\n 28207741 rows 3381212265 bytes miRNA_BCGSC_HiSeq_mirna\n 102234868 rows 17290207488 bytes miRNA_BCGSC_isoform\n 32710787 rows 3908314182 bytes miRNA_BCGSC_mirna\n 26763022 rows 3265303352 bytes miRNA_Expression\n"
]
],
[
[
"These tables are based on the open-access TCGA data as of July 2016. The molecular data is all \"Level 3\" data, and is divided according to platform/pipeline. See [here](https://tcga-data.nci.nih.gov/tcga/tcgaDataType.jsp) for additional details regarding the TCGA data levels and data types.\n\nAdditional notebooks go into each of these tables in more detail, but here is an overview, in the same alphabetical order that they are listed in above and in the BigQuery web UI:\n\n\n- **Annotations**: This table contains the annotations that are also available from the interactive [TCGA Annotations Manager](https://tcga-data.nci.nih.gov/annotations/). Annotations can be associated with any type of \"item\" (*eg* Patient, Sample, Aliquot, etc), and a single item may have more than one annotation. Common annotations include \"Item flagged DNU\", \"Item is noncanonical\", and \"Prior malignancy.\" More information about this table can be found in the [TCGA Annotations](https://github.com/isb-cgc/examples-Python/blob/master/notebooks/TCGA%20Annotations.ipynb) notebook.\n\n\n- **Biospecimen_data**: This table contains information obtained from the \"biospecimen\" and \"auxiliary\" XML files in the TCGA Level-1 \"bio\" archives. Each row in this table represents a single \"biospecimen\" or \"sample\". Most participants in the TCGA project provided two samples: a \"primary tumor\" sample and a \"blood normal\" sample, but others provided normal-tissue, metastatic, or other types of samples. This table contains metadata about all of the samples, and more information about exploring this table and using this information to create your own custom analysis cohort can be found in the [Creating TCGA cohorts (part 1)](https://github.com/isb-cgc/examples-Python/blob/master/notebooks/Creating%20TCGA%20cohorts%20--%20part%201.ipynb) and [(part 2)](https://github.com/isb-cgc/examples-Python/blob/master/notebooks/Creating%20TCGA%20cohorts%20--%20part%202.ipynb) notebooks.\n\n\n- **Clinical_data**: This table contains information obtained from the \"clinical\" XML files in the TCGA Level-1 \"bio\" archives. Not all fields in the XML files are represented in this table, but any field which was found to be significantly filled-in for at least one tumor-type has been retained. More information about exploring this table and using this information to create your own custom analysis cohort can be found in the [Creating TCGA cohorts (part 1)](https://github.com/isb-cgc/examples-Python/blob/master/notebooks/Creating%20TCGA%20cohorts%20--%20part%201.ipynb) and [(part 2)](https://github.com/isb-cgc/examples-Python/blob/master/notebooks/Creating%20TCGA%20cohorts%20--%20part%202.ipynb) notebooks.\n\n\n- **Copy_Number_segments**: This table contains Level-3 copy-number segmentation results generated by The Broad Institute, from Genome Wide SNP 6 data using the CBS (Circular Binary Segmentation) algorithm. The values are base2 log(copynumber/2), centered on 0. More information about this data table can be found in the [Copy Number segments](https://github.com/isb-cgc/examples-Python/blob/master/notebooks/Copy%20Number%20segments.ipynb) notebook.\n\n\n- **DNA_Methylation_betas**: This table contains Level-3 summary measures of DNA methylation for each interrogated locus (beta values: M/(M+U)). This table contains data from two different platforms: the Illumina Infinium HumanMethylation 27k and 450k arrays. More information about this data table can be found in the [DNA Methylation](https://github.com/isb-cgc/examples-Python/blob/master/notebooks/DNA%20Methylation.ipynb) notebook. Note that individual chromosome-specific DNA Methylation tables are also available to cut down on the amount of data that you may need to query (depending on yoru use case). \n\n\n- **Protein_RPPA_data**: This table contains the normalized Level-3 protein expression levels based on each antibody used to probe the sample. More information about how this data was generated by the RPPA Core Facility at MD Anderson can be found [here](https://wiki.nci.nih.gov/display/TCGA/Protein+Array+Data+Format+Specification#ProteinArrayDataFormatSpecification-Expression-Protein), and more information about this data table can be found in the [Protein expression](https://github.com/isb-cgc/examples-Python/blob/master/notebooks/Protein%20expression.ipynb) notebook.\n\n\n- **Somatic_Mutation_calls**: This table contains annotated somatic mutation calls. All current MAF (Mutation Annotation Format) files were annotated using [Oncotator](http://onlinelibrary.wiley.com/doi/10.1002/humu.22771/abstract;jsessionid=15E7960BA5FEC21EE608E6D262390C52.f01t04) v1.5.1.0, and merged into a single table. More information about this data table can be found in the [Somatic Mutations](https://github.com/isb-cgc/examples-Python/blob/master/notebooks/Somatic%20Mutations.ipynb) notebook, including an example of how to use the [Tute Genomics annotations database in BigQuery](http://googlegenomics.readthedocs.org/en/latest/use_cases/annotate_variants/tute_annotation.html).\n\n\n- **mRNA_BCGSC_HiSeq_RPKM**: This table contains mRNAseq-based gene expression data produced by the [BC Cancer Agency](http://www.bcgsc.ca/). (For details about a very similar table, take a look at a [notebook](https://github.com/isb-cgc/examples-Python/blob/master/notebooks/UNC%20HiSeq%20mRNAseq%20gene%20expression.ipynb) describing the other mRNAseq gene expression table.)\n\n\n- **mRNA_UNC_HiSeq_RSEM**: This table contains mRNAseq-based gene expression data produced by [UNC Lineberger](https://unclineberger.org/). More information about this data table can be found in the [UNC HiSeq mRNAseq gene expression](https://github.com/isb-cgc/examples-Python/blob/master/notebooks/UNC%20HiSeq%20mRNAseq%20gene%20expression.ipynb) notebook.\n\n\n- **miRNA_expression**: This table contains miRNAseq-based expression data for mature microRNAs produced by the [BC Cancer Agency](http://www.bcgsc.ca/). More information about this data table can be found in the [microRNA expression](https://github.com/isb-cgc/examples-Python/blob/master/notebooks/BCGSC%20microRNA%20expression.ipynb) notebook.",
"_____no_output_____"
],
[
"### Where to start?\nWe suggest that you start with the two \"Creating TCGA cohorts\" notebooks ([part 1](https://github.com/isb-cgc/examples-Python/blob/master/notebooks/Creating%20TCGA%20cohorts%20--%20part%201.ipynb) and [part 2](https://github.com/isb-cgc/examples-Python/blob/master/notebooks/Creating%20TCGA%20cohorts%20--%20part%202.ipynb)) which describe and make use of the Clinical and Biospecimen tables. From there you can delve into the various molecular data tables as well as the Annotations table. For now these sample notebooks are intentionally relatively simple and do not do any analysis that integrates data from multiple tables but once you have a grasp of how to use the data, developing your own more complex analyses should not be difficult. You could even contribute an example back to our github repository! You are also welcome to submit bug reports, comments, and feature-requests as [github issues](https://github.com/isb-cgc/examples-Python/issues).",
"_____no_output_____"
],
[
"### A note about BigQuery tables and \"tidy data\"\nYou may be used to thinking about a molecular data table such as a gene-expression table as a matrix where the rows are genes and the columns are samples (or *vice versa*). These BigQuery tables instead use the [tidy data](https://cran.r-project.org/web/packages/tidyr/vignettes/tidy-data.html) approach, with each \"cell\" from the traditional data-matrix becoming a single row in the BigQuery table. A 10,000 gene x 500 sample matrix would therefore become a 5,000,000 row BigQuery table.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
cbed1bf42f0ba231fb8d6e8233484c56659a9b54
| 59,126 |
ipynb
|
Jupyter Notebook
|
src/models/Train_and_test.ipynb
|
K-Ellis/Turnover-Classification
|
871435fab21e42538cb4b7d191ae34263a7e9853
|
[
"BSD-3-Clause"
] | null | null | null |
src/models/Train_and_test.ipynb
|
K-Ellis/Turnover-Classification
|
871435fab21e42538cb4b7d191ae34263a7e9853
|
[
"BSD-3-Clause"
] | null | null | null |
src/models/Train_and_test.ipynb
|
K-Ellis/Turnover-Classification
|
871435fab21e42538cb4b7d191ae34263a7e9853
|
[
"BSD-3-Clause"
] | null | null | null | 165.619048 | 24,132 | 0.878987 |
[
[
[
"# %%writefile train.py\n# Import Packages\nimport pandas as pd\nimport numpy as np\nfrom sklearn.preprocessing import LabelEncoder\nimport xgboost as xgb\nfrom pre_ml_process import pre_ml_process\nimport pickle\nfrom plot_confusion_matrix import plot_confusion_matrix\nfrom sklearn.metrics import confusion_matrix, average_precision_score, precision_score, recall_score, roc_auc_score\nimport matplotlib.pyplot as plt\n%matplotlib inline\n\n# location of training data\ntraining_data_loc = input(\"Please input the training data dir:\") \n\n# Import Data\ndf_raw = pd.read_csv(training_data_loc, encoding=\"cp1252\")\n\n# Data cleaning\ndf = df_raw.dropna()\ndf = df.loc[df[\"f7\"] != \"#\"]\ndf[\"f7\"] = df[\"f7\"].astype(float)\n\n# f9 - remove the unknown record and binary encode the remaining two classes\ndf = df.loc[df[\"f9\"] != \"unknown\"]\nle_f9 = LabelEncoder()\ndf[\"f9\"] = le_f9.fit_transform(df[\"f9\"])\n\n# isolate the numerical columns\nnumerical_cols = df.dtypes[df.dtypes != object].index.tolist()\ndf_num = df[numerical_cols]\n\n# drop employee id primary key\ndf_num = df_num.drop(\"employee_id\", axis=1)\n\n# label encode string columns\ndef fit_label_encoders(df_in):\n fitted_label_encoders = {}\n for col in df_in.dtypes[df_in.dtypes == object].index.tolist():\n fitted_label_encoders[col] = LabelEncoder().fit(df_in[col])\n return fitted_label_encoders\n\nfitted_label_encoders = fit_label_encoders(df.drop(\"employee_id\", axis=1))\n\n# concat the label encoded dataframe with the baseline dataframe \ndef add_label_encoded(df_baseline, df_to_le, cols, fitted_label_encoders):\n df_out = df_baseline.copy()\n for col in cols:\n df_le = fitted_label_encoders[col].transform(df_to_le[col])\n df_out[col] = df_le\n return df_out\n\ndf_num_allLE = add_label_encoded(df_num, df, [\"f1\", \"f2\", \"f3\", \"f4\", \"f10\", \"f12\"], fitted_label_encoders)\n\n\nXGC=xgb.XGBClassifier(random_state=0, n_estimators=100) \n\n# parameters\nsplit_random_state=42\nxgb_fit_eval_metric=\"aucpr\"\ntrain_test_split_random_state=0\nRandomOverSampler_random_state=0\ntest_size=0.33\n\n# preprocessing\ndf_ignore, X, y, X_train, X_test, y_train, y_test, \\\n scaler, X_train_resample_scaled, y_train_resample, \\\n X_test_scaled, ros, poly_ignore = \\\n pre_ml_process(df_num_allLE, \n test_size,\n train_test_split_random_state,\n RandomOverSampler_random_state)\n\n# save scaler to file\npickle.dump(scaler, open(\"../../models/scaler.p\", \"wb\"))\n\n# Train with XGBoost Classifier\nclf_XG = XGC.fit(X_train_resample_scaled, y_train_resample, eval_metric=xgb_fit_eval_metric)\n\n# Model evaluation\n\n# Get test set predictions\ny_test_hat = clf_XG.predict(X_test_scaled)\ny_test_proba = clf_XG.predict_proba(X_test_scaled)[:,1]\n\n# Confusion Matrix\ndf_cm = confusion_matrix(y_test, y_test_hat, labels=[1, 0])\nplot_confusion_matrix(df_cm, \n target_names=[1, 0], \n title=\"%s Confusion Matrix\" % (type(clf_XG).__name__),\n normalize=True)\nplt.show()\n\n# Accuracy metrics\nap = average_precision_score(y_test, y_test_proba)\nps = precision_score(y_test, y_test_hat)\nrs = recall_score(y_test, y_test_hat)\nroc = roc_auc_score(y_test, y_test_hat)\n\nprint(\"average_precision_score = {:.3f}\".format(ap))\nprint(\"precision_score = {:.3f}\".format(ps))\nprint(\"recall_score = {:.3f}\".format(rs))\nprint(\"roc_auc_score = {:.3f}\".format(roc))\n\n# Feature Importances\ndf_feature_importances = pd.DataFrame(clf_XG.feature_importances_, columns=[\"Importance\"])\ncol_names = df_num_allLE.columns.tolist()\ncol_names.remove(\"has_left\")\ndf_feature_importances[\"Feature\"] = col_names\ndf_feature_importances.sort_values(\"Importance\", ascending=False, inplace=True)\ndf_feature_importances = df_feature_importances.round(4)\ndf_feature_importances = df_feature_importances.reset_index(drop=True)\nprint(df_feature_importances)\n\n# export trained model\npickle.dump(clf_XG, open(\"../../models/xgb_model.p\", \"wb\"))",
"Please input the training data dir: ../dataset.csv\n"
],
[
"# %%writefile test.py\n# import packages\nimport pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\n%matplotlib inline\nfrom sklearn.linear_model import LogisticRegression\nfrom sklearn.ensemble import RandomForestClassifier\nfrom sklearn.preprocessing import MinMaxScaler, LabelEncoder\nfrom sklearn.metrics import confusion_matrix, average_precision_score, precision_score, recall_score, roc_auc_score\nimport xgboost as xgb\nfrom sklearn.decomposition import PCA\nfrom pre_ml_process import pre_ml_process\nfrom plot_confusion_matrix import plot_confusion_matrix\nimport pickle\n\n# model location\nmodel_loc = input(\"Please input the trained model dir:\") \n\n# Import trained model\nclf = pickle.load(open(model_loc, \"rb\"))\n\n# Import Data\ndf_raw_loc = input(\"Please input the testing or prediction data dir:\") \ndf_raw = pd.read_csv(df_raw_loc, encoding=\"cp1252\")\n\n# Data cleaning\ndf = df_raw.dropna()\ndf = df.loc[df[\"f7\"] != \"#\"]\ndf[\"f7\"] = df[\"f7\"].astype(float)\n\n# f9 - remove the unknown record and binary encode the remaining two classes\ndf = df.loc[df[\"f9\"] != \"unknown\"]\nle_f9 = LabelEncoder()\ndf[\"f9\"] = le_f9.fit_transform(df[\"f9\"])\n\n# isolate the numerical columns\nnumerical_cols = df.dtypes[df.dtypes != object].index.tolist()\ndf_num = df[numerical_cols]\n\n# drop employee id primary key\ndf_num = df_num.drop(\"employee_id\", axis=1)\n\n# label encode string columns\ndef fit_label_encoders(df_in):\n fitted_label_encoders = {}\n for col in df_in.dtypes[df_in.dtypes == object].index.tolist():\n fitted_label_encoders[col] = LabelEncoder().fit(df_in[col])\n return fitted_label_encoders\n\nfitted_label_encoders = fit_label_encoders(df.drop(\"employee_id\", axis=1))\n\n# concat the label encoded dataframe with the baseline dataframe \ndef add_label_encoded(df_baseline, df_to_le, cols, fitted_label_encoders):\n df_out = df_baseline.copy()\n for col in cols:\n df_le = fitted_label_encoders[col].transform(df_to_le[col])\n df_out[col] = df_le\n return df_out\n\ndf_num_allLE = add_label_encoded(df_num, df, [\"f1\", \"f2\", \"f3\", \"f4\", \"f10\", \"f12\"], fitted_label_encoders)\n\n# Separate X and y\ny_col = \"has_left\"\ny = df_num_allLE[y_col]\nX = df_num_allLE.drop(y_col, axis=1)\nX = X.astype(float)\n\n# Scale predictors \nscaler = pickle.load(open(\"scaler.p\", \"rb\"))\nX_scaled = scaler.transform(X)\n\n# Get predictions\ny_hat = clf.predict(X_scaled)\ny_proba = clf.predict_proba(X_scaled)[:,1]\n\n# Confusion Matrix\ndf_cm = confusion_matrix(y, y_hat, labels=[1, 0])\nplot_confusion_matrix(df_cm, \n target_names=[1, 0], \n title=\"%s Confusion Matrix\" % (type(clf).__name__),\n normalize=True)\n\n# accuracy metrics\nap = average_precision_score(y, y_proba)\nps = precision_score(y, y_hat)\nrs = recall_score(y, y_hat)\nroc = roc_auc_score(y, y_hat)\n\nprint(\"average_precision_score = {:.3f}\".format(ap))\nprint(\"precision_score = {:.3f}\".format(ps))\nprint(\"recall_score = {:.3f}\".format(rs))\nprint(\"roc_auc_score = {:.3f}\".format(roc))\n\n# Feature Importances\ndf_feature_importances = pd.DataFrame(clf.feature_importances_, columns=[\"Importance\"])\ncol_names = df_num_allLE.columns.tolist()\ncol_names.remove(\"has_left\")\ndf_feature_importances[\"Feature\"] = col_names\ndf_feature_importances.sort_values(\"Importance\", ascending=False, inplace=True)\ndf_feature_importances = df_feature_importances.round(4)\ndf_feature_importances = df_feature_importances.reset_index(drop=True)\nprint(df_feature_importances)\n\n# concat test data with predictions\ndf_in_with_predictions = pd.concat([df_num_allLE, pd.Series(y_hat, name=\"y_hat\"), pd.Series(y_proba, name=\"y_hat_probability\")], axis=1)\n\n# Export predictions\ndf_in_with_predictions.to_csv(\"../../data/prediction/prediction_export.csv\", index=False)\n",
"Please input the trained model dir: xgb_model.p\nPlease input the testing data dir: ../dataset.csv\n"
]
]
] |
[
"code"
] |
[
[
"code",
"code"
]
] |
cbed25fb273a76ef06f68da6a2b05c96a8108869
| 139,366 |
ipynb
|
Jupyter Notebook
|
Notebooks/paper_analytics.ipynb
|
pranavmishra90/COVID-19
|
12cd58c4d312e62207c21bd8c6ea703852841c78
|
[
"MIT"
] | 1 |
2021-01-11T06:37:18.000Z
|
2021-01-11T06:37:18.000Z
|
Notebooks/paper_analytics.ipynb
|
pranavmishra90/COVID-19
|
12cd58c4d312e62207c21bd8c6ea703852841c78
|
[
"MIT"
] | null | null | null |
Notebooks/paper_analytics.ipynb
|
pranavmishra90/COVID-19
|
12cd58c4d312e62207c21bd8c6ea703852841c78
|
[
"MIT"
] | null | null | null | 1,151.785124 | 86,907 | 0.854642 |
[
[
[
"import pandas as pd\nimport numpy as np\nimport seaborn as sns\nfrom matplotlib import cbook as cbook\nfrom matplotlib import pyplot as plt\nfrom matplotlib import image as image",
"_____no_output_____"
],
[
"analytics = pd.read_csv('../Data/Paper_Analytics/analytics_medxriv_deductive_modeling.txt')\n\nanalytics['Year'] = pd.DatetimeIndex(analytics['Month']).year\nanalytics['Month'] = pd.DatetimeIndex(analytics['Month']).month",
"_____no_output_____"
],
[
"data = analytics.melt(id_vars=['Month'], value_vars=['Abstract', 'Pdf'])\n\n#Formatting the following figures\n\naxes_fontsize = 15\ntitle_fontsize = 20\nlabel_size = 10\nfig_dpi = 150\nfig_width = 8\nfig_height = 5\ntwitter_alpha = .5\n\nindia_color = \"#FF8F1C\"\nusa_color = \"#041E42\"\n\ndata_source = \"Data source: Medxriv\"\n\n\nwith cbook.get_sample_data('Twitter/twitter_watermark_black.png') as file:\n twitter = image.imread(file)\n\n#Plotting the figure\n\nfig1, ax1 = plt.subplots(figsize=(fig_width, fig_height), dpi = fig_dpi)\n\nfig1 = sns.lineplot(data = analytics, x=\"Month\", y='value', hue='var')\nplt.title('Distribution of Doubling Times')\nax1.set_xlabel(\"Doubling Time (Days)\")\nax1.set_ylabel(\"Probability\")\n\nfig1 = fig1.get_figure()\n\n\n#JHU Source\nplt.text(-2, -150, data_source, zorder=1, alpha=twitter_alpha, fontsize=6)\n\n#Twitter Watermark\n[twitter_width, twitter_height] = fig1.get_size_inches()*fig1.dpi\ntwitter_width = (twitter_width*.7).round(0)\ntwitter_height = (twitter_height*.8).round(0)\n\nfig1.figimage(twitter, twitter_width, twitter_height, zorder=4, alpha=twitter_alpha) \n\n\nfig1.savefig('../Data/Paper_Analytics/analytics.jpg', bbox_inches=\"tight\", pad_inches=0.3, transparent=False)",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code"
]
] |
cbed40e0c1f4d49b28e49e8ad35f92461168224e
| 22,170 |
ipynb
|
Jupyter Notebook
|
predict.ipynb
|
satish860/foodimageclassifier
|
12ce22e56f42de36f2b32d4344a57d20cf2fe089
|
[
"Apache-2.0"
] | null | null | null |
predict.ipynb
|
satish860/foodimageclassifier
|
12ce22e56f42de36f2b32d4344a57d20cf2fe089
|
[
"Apache-2.0"
] | null | null | null |
predict.ipynb
|
satish860/foodimageclassifier
|
12ce22e56f42de36f2b32d4344a57d20cf2fe089
|
[
"Apache-2.0"
] | null | null | null | 45.430328 | 118 | 0.480424 |
[
[
[
"from PIL import Image\nimport torchvision.transforms as tt\nimport torch\nfrom torchvision import models\nimport torch.nn as nn\nimport torch.nn.functional as F",
"_____no_output_____"
],
[
"stats = ((0.4914, 0.4822, 0.4465), (0.2023, 0.1994, 0.2010))\nvalid_tfms = tt.Compose([tt.Resize([224,224]),tt.ToTensor(), tt.Normalize(*stats)])",
"_____no_output_____"
],
[
"class Flatten(nn.Module):\n def forward(self,x):\n return torch.flatten(x,1)\n\nclass FoodImageClassifer(nn.Module):\n def __init__(self):\n super().__init__()\n mobilenet = models.mobilenet_v2(pretrained=True)\n self.body = mobilenet.features\n self.head = nn.Sequential(\n nn.Dropout(p=0.2),\n nn.Linear(1280,101))\n \n def forward(self,x):\n x = self.body(x)\n x = nn.functional.adaptive_avg_pool2d(x, (1, 1))\n x = torch.flatten(x, 1) \n return self.head(x)\n \n def freeze(self):\n for name,param in self.body.named_parameters():\n param.requires_grad = False",
"_____no_output_____"
],
[
"model = FoodImageClassifer()\nmodel.load_state_dict(torch.load('food_classifier.pth'))\nmodel.eval()",
"_____no_output_____"
],
[
"imgpath = 'food-101/images/apple_pie/3004621.jpg'\nimg = Image.open(imgpath)\nimg_ts=valid_tfms(img)\nbatch_t = torch.unsqueeze(img_ts, 0)\nout = model(batch_t)\nprob = torch.nn.functional.softmax(out, dim = 1)[0] * 100\n_, indices = torch.sort(out, descending = True)",
"_____no_output_____"
],
[
"indices",
"_____no_output_____"
],
[
" with open('classes.txt') as f:\n classes = [line.strip() for line in f.readlines()]",
"_____no_output_____"
],
[
"val = [(classes[idx], prob[idx].item()) for idx in indices[0][:5]]",
"_____no_output_____"
],
[
"val",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbed56818206652706ee18ee081a78efe91c3ec2
| 273,299 |
ipynb
|
Jupyter Notebook
|
code.ipynb
|
Bate90/Linear-Regression
|
0ab06d798db971749762c7784af8bbfcda00cd71
|
[
"Apache-2.0"
] | 1 |
2022-01-20T18:14:20.000Z
|
2022-01-20T18:14:20.000Z
|
code.ipynb
|
Bate90/Linear-Regression
|
0ab06d798db971749762c7784af8bbfcda00cd71
|
[
"Apache-2.0"
] | null | null | null |
code.ipynb
|
Bate90/Linear-Regression
|
0ab06d798db971749762c7784af8bbfcda00cd71
|
[
"Apache-2.0"
] | null | null | null | 586.478541 | 45,608 | 0.950486 |
[
[
[
"import pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt \nimport seaborn as sns\nimport statsmodels.api as sm\nimport patsy",
"_____no_output_____"
],
[
"# Data: https://archive.ics.uci.edu/ml/datasets/Bike+Sharing+Dataset\n# UCI citation:\n# Dua, D. and Graff, C. (2019). UCI Machine Learning Repository [http://archive.ics.uci.edu/ml]. Irvine, CA: University of California, School of Information and Computer Science.\n\n# Source:\n# # Hadi Fanaee-T \n\n# Laboratory of Artificial Intelligence and Decision Support (LIAAD), University of Porto \n# INESC Porto, Campus da FEUP \n# Rua Dr. Roberto Frias, 378 \n# 4200 - 465 Porto, Portugal \n\n# Original Source: http://capitalbikeshare.com/system-data \n\n\n\nbikes = pd.read_csv('bikes.csv')",
"_____no_output_____"
],
[
"# Fit model1\nmodel1 = sm.OLS.from_formula('cnt ~ temp + windspeed + holiday', data=bikes).fit()\n\n# Fit model2\nmodel2 = sm.OLS.from_formula('cnt ~ hum + season + weekday', data=bikes).fit()\n\n# Print R-squared for both models\nprint(model1.rsquared)\nprint(model2.rsquared)",
"0.41508122190306995\n0.3883324473437183\n"
],
[
"sns.scatterplot(x='temp', y='cnt', data = bikes)\nplt.show()",
"_____no_output_____"
],
[
"model1 = sm.OLS.from_formula('cnt ~ temp', data=bikes).fit()\nxs = pd.DataFrame({'temp': np.linspace(bikes.temp.min(), bikes.temp.max(), 100)})\nys = model1.predict(xs)\nsns.scatterplot(x='temp', y='cnt', data = bikes)\nplt.plot(xs, ys, color = 'black', linewidth=4)\nplt.show()",
"_____no_output_____"
],
[
"model2 = sm.OLS.from_formula('cnt ~ temp + np.power(temp, 2)', data=bikes).fit()\nxs = pd.DataFrame({'temp': np.linspace(bikes.temp.min(), bikes.temp.max(), 100)})\nys = model2.predict(xs)\nsns.scatterplot(x='temp', y='cnt', data = bikes)\nplt.plot(xs, ys, color = 'black', linewidth=4)\nplt.show()",
"_____no_output_____"
],
[
"model3 = sm.OLS.from_formula('cnt ~ temp + np.power(temp, 2) + np.power(temp, 3)', data=bikes).fit()\nxs = pd.DataFrame({'temp': np.linspace(bikes.temp.min(), bikes.temp.max(), 100)})\nys = model3.predict(xs)\nsns.scatterplot(x='temp', y='cnt', data = bikes)\nplt.plot(xs, ys, color = 'black', linewidth=4)\nplt.show()",
"_____no_output_____"
],
[
"model4 = sm.OLS.from_formula('cnt ~ temp + np.power(temp, 2) + np.power(temp, 3) + np.power(temp, 4) + np.power(temp, 5)', data=bikes).fit()\nxs = pd.DataFrame({'temp': np.linspace(bikes.temp.min(), bikes.temp.max(), 100)})\nys = model4.predict(xs)\nsns.scatterplot(x='temp', y='cnt', data = bikes)\nplt.plot(xs, ys, color = 'black', linewidth=4)\nplt.show()",
"_____no_output_____"
],
[
"model5 = sm.OLS.from_formula('cnt ~ temp + np.power(temp, 2) + np.power(temp, 3) + np.power(temp, 4) + np.power(temp, 5) + np.power(temp, 6) + np.power(temp, 7) + np.power(temp, 8) + np.power(temp, 9) + np.power(temp, 10)', data=bikes).fit()\nxs = pd.DataFrame({'temp': np.linspace(bikes.temp.min(), bikes.temp.max(), 100)})\nys = model5.predict(xs)\nsns.scatterplot(x='temp', y='cnt', data = bikes)\nplt.plot(xs, ys, color = 'black', linewidth=4)\nplt.show()",
"_____no_output_____"
],
[
"print(model1.rsquared)\nprint(model2.rsquared)\nprint(model3.rsquared)\nprint(model4.rsquared)\nprint(model5.rsquared)",
"0.3937487313729241\n0.4531790254908288\n0.4626662622590385\n0.4628445586311082\n0.47118613034158985\n"
],
[
"print(model1.rsquared_adj)\nprint(model2.rsquared_adj)\nprint(model3.rsquared_adj)\nprint(model4.rsquared_adj)\nprint(model5.rsquared_adj)",
"0.3929171109770022\n0.4516767700663531\n0.4597057457976558\n0.4591400383458055\n0.46458512503378724\n"
],
[
"from statsmodels.stats.anova import anova_lm\nanova_results = anova_lm(model1, model2, model3, model4, model5)\nprint(anova_results.round(2))",
" df_resid ssr df_diff ss_diff F Pr(>F)\n0 729.0 1.660847e+09 0.0 NaN NaN NaN\n1 728.0 1.498035e+09 1.0 1.628114e+08 81.03 0.00\n2 726.0 1.472045e+09 2.0 2.599062e+07 6.47 0.00\n3 725.0 1.471556e+09 1.0 4.884492e+05 0.24 0.62\n4 721.0 1.448704e+09 4.0 2.285203e+07 2.84 0.02\n"
],
[
"print(model1.llf)\nprint(model2.llf)\nprint(model3.llf)\nprint(model4.llf)\nprint(model5.llf)",
"-6386.767848313633\n-6349.0580904730305\n-6342.661081788197\n-6342.539782614866\n-6336.819342703741\n"
],
[
"print(model1.aic)\nprint(model2.aic)\nprint(model3.aic)\nprint(model4.aic)\nprint(model5.aic)",
"12777.535696627267\n12704.116180946061\n12695.322163576395\n12697.079565229733\n12693.638685407483\n"
],
[
"print(model1.bic)\nprint(model2.bic)\nprint(model3.bic)\nprint(model4.bic)\nprint(model5.bic)",
"12786.724523546765\n12717.89942132531\n12718.294230875144\n12724.646045988231\n12739.58282000498\n"
],
[
"# Set seed (don't change this)\nnp.random.seed(123)\n\n# Split bikes data\nindices = range(len(bikes))\ns = int(0.8*len(indices))\ntrain_ind = np.random.choice(indices, size = s, replace = False)\ntest_ind = list(set(indices) - set(train_ind))\nbikes_train = bikes.iloc[train_ind]\nbikes_test = bikes.iloc[test_ind]\n\n# Fit model1\nmodel1 = sm.OLS.from_formula('cnt ~ temp + atemp + hum', data=bikes_train).fit()\n\n# Fit model2\nmodel2 = sm.OLS.from_formula('cnt ~ season + windspeed + weekday', data=bikes_train).fit()\n\n# Calculate predicted cnt based on model1\nfitted1 = model1.predict(bikes_test)\n\n# Calculate predicted cnt based on model2\nfitted2 = model2.predict(bikes_test)\n\n# Calculate PRMSE for model1\ntrue = bikes_test.cnt\nprmse1 = np.mean((true-fitted1)**2)**.5\n\n# Calculate PRMSE for model2\nprmse2 = np.mean((true-fitted2)**2)**.5\n\n# Print PRMSE for both models\nprint(prmse1)\nprint(prmse2)",
"1460.39929474624\n1570.4805833457629\n"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbed62701409cab44fc3d087f96591fcd41e3775
| 46,155 |
ipynb
|
Jupyter Notebook
|
R/22_Transforms.ipynb
|
NanYoMy/SimpleITK-Notebooks
|
11f03ea1ee42849940a713367fabe3c1857502f8
|
[
"Apache-2.0"
] | 657 |
2015-02-20T13:50:28.000Z
|
2022-03-31T11:42:22.000Z
|
R/22_Transforms.ipynb
|
malekosh/SimpleITK-Notebooks
|
03137dbfc290b4438f6f7996eb7074b8eeaa990b
|
[
"Apache-2.0"
] | 100 |
2015-02-19T18:53:06.000Z
|
2022-03-29T15:20:39.000Z
|
R/22_Transforms.ipynb
|
malekosh/SimpleITK-Notebooks
|
03137dbfc290b4438f6f7996eb7074b8eeaa990b
|
[
"Apache-2.0"
] | 313 |
2015-01-29T20:23:36.000Z
|
2022-03-23T06:34:21.000Z
| 44.422522 | 494 | 0.630549 |
[
[
[
"<h1 align=\"center\">SimpleITK Spatial Transformations</h1>\n\n\n**Summary:**\n\n1. Points are represented by vector-like data types: Tuple, Numpy array, List.\n2. Matrices are represented by vector-like data types in row major order.\n3. Default transformation initialization as the identity transform.\n4. Angles specified in radians, distances specified in unknown but consistent units (nm,mm,m,km...).\n5. All global transformations **except translation** are of the form:\n$$T(\\mathbf{x}) = A(\\mathbf{x}-\\mathbf{c}) + \\mathbf{t} + \\mathbf{c}$$\n\n Nomenclature (when printing your transformation):\n\n * Matrix: the matrix $A$\n * Center: the point $\\mathbf{c}$\n * Translation: the vector $\\mathbf{t}$\n * Offset: $\\mathbf{t} + \\mathbf{c} - A\\mathbf{c}$\n6. Bounded transformations, BSplineTransform and DisplacementFieldTransform, behave as the identity transform outside the defined bounds.\n7. DisplacementFieldTransform:\n * Initializing the DisplacementFieldTransform using an image requires that the image's pixel type be sitk.sitkVectorFloat64.\n * Initializing the DisplacementFieldTransform using an image will \"clear out\" your image (your alias to the image will point to an empty, zero sized, image).\n8. Composite transformations are applied in stack order (first added, last applied).\n",
"_____no_output_____"
],
[
"# Transformation Types\nThis notebook introduces the transformation types supported by SimpleITK and illustrates how to \"promote\" transformations from a lower to higher parameter space (e.g. 3D translation to 3D rigid). \n\n\n| Class Name | Details|\n|:-------------|:---------|\n|[TranslationTransform](https://simpleitk.org/doxygen/latest/html/classitk_1_1simple_1_1TranslationTransform.html) | 2D or 3D, translation|\n|[VersorTransform](https://simpleitk.org/doxygen/latest/html/classitk_1_1simple_1_1VersorTransform.html)| 3D, rotation represented by a versor|\n|[VersorRigid3DTransform](https://simpleitk.org/doxygen/latest/html/classitk_1_1simple_1_1VersorRigid3DTransform.html)|3D, rigid transformation with rotation represented by a versor|\n|[Euler2DTransform](https://simpleitk.org/doxygen/latest/html/classitk_1_1simple_1_1Euler2DTransform.html)| 2D, rigid transformation with rotation represented by a Euler angle|\n|[Euler3DTransform](https://simpleitk.org/doxygen/latest/html/classitk_1_1simple_1_1Euler3DTransform.html)| 3D, rigid transformation with rotation represented by Euler angles|\n|[Similarity2DTransform](https://simpleitk.org/doxygen/latest/html/classitk_1_1simple_1_1Similarity2DTransform.html)| 2D, composition of isotropic scaling and rigid transformation with rotation represented by a Euler angle|\n|[Similarity3DTransform](https://simpleitk.org/doxygen/latest/html/classitk_1_1simple_1_1Similarity3DTransform.html) | 3D, composition of isotropic scaling and rigid transformation with rotation represented by a versor|\n|[ScaleTransform](https://simpleitk.org/doxygen/latest/html/classitk_1_1simple_1_1ScaleTransform.html)|2D or 3D, anisotropic scaling|\n|[ScaleVersor3DTransform](https://simpleitk.org/doxygen/latest/html/classitk_1_1simple_1_1ScaleVersor3DTransform.html)| 3D, rigid transformation and anisotropic scale is **added** to the rotation matrix part (not composed as one would expect)|\n|[ScaleSkewVersor3DTransform](https://simpleitk.org/doxygen/latest/html/classitk_1_1simple_1_1ScaleSkewVersor3DTransform.html#details)|3D, rigid transformation with anisotropic scale and skew matrices **added** to the rotation matrix part (not composed as one would expect) |\n|[AffineTransform](https://simpleitk.org/doxygen/latest/html/classitk_1_1simple_1_1AffineTransform.html)| 2D or 3D, affine transformation|\n|[BSplineTransform](https://simpleitk.org/doxygen/latest/html/classitk_1_1simple_1_1BSplineTransform.html)|2D or 3D, deformable transformation represented by a sparse regular grid of control points |\n|[DisplacementFieldTransform](https://simpleitk.org/doxygen/latest/html/classitk_1_1simple_1_1DisplacementFieldTransform.html)| 2D or 3D, deformable transformation represented as a dense regular grid of vectors|\n|[CompositeTransform](https://simpleitk.org/doxygen/latest/html/classitk_1_1simple_1_1CompositeTransform.html)| 2D or 3D, stack of transformations concatenated via composition, last added, first applied|\n|[Transform](https://simpleitk.org/doxygen/latest/html/classitk_1_1simple_1_1Transform.html#details) | 2D or 3D, parent/superclass for all transforms",
"_____no_output_____"
]
],
[
[
"library(SimpleITK)\n\nlibrary(scatterplot3d)\n\nOUTPUT_DIR <- \"Output\"\n\nprint(Version())",
"_____no_output_____"
]
],
[
[
"## Points in SimpleITK",
"_____no_output_____"
],
[
"### Utility functions\n\nA number of functions that deal with point data in a uniform manner.",
"_____no_output_____"
]
],
[
[
"# Format a point for printing, based on specified precision with trailing zeros. Uniform printing for vector-like data \n# (vector, array, list).\n# @param point (vector-like): nD point with floating point coordinates.\n# @param precision (int): Number of digits after the decimal point.\n# @return: String representation of the given point \"xx.xxx yy.yyy zz.zzz...\".\npoint2str <- function(point, precision=1)\n{\n precision_str <- sprintf(\"%%.%df\",precision)\n return(paste(lapply(point, function(x) sprintf(precision_str, x)), collapse=\", \"))\n}\n \n \n# Generate random (uniform withing bounds) nD point cloud. Dimension is based on the number of pairs in the \n# bounds input.\n# @param bounds (list(vector-like)): List where each vector defines the coordinate bounds.\n# @param num_points (int): Number of points to generate.\n# @return (matrix): Matrix whose columns are the set of points. \nuniform_random_points <- function(bounds, num_points)\n{\n return(t(sapply(bounds, function(bnd,n=num_points) runif(n, min(bnd),max(bnd)))))\n}\n \n\n# Distances between points transformed by the given transformation and their\n# location in another coordinate system. When the points are only used to evaluate\n# registration accuracy (not used in the registration) this is the target registration\n# error (TRE).\n# @param tx (SimpleITK transformation): Transformation applied to the points in point_list\n# @param point_data (matrix): Matrix whose columns are points which we transform using tx.\n# @param reference_point_data (matrix): Matrix whose columns are points to which we compare \n# the transformed point data. \n# @return (vector): Distances between the transformed points and the reference points.\ntarget_registration_errors <- function(tx, point_data, reference_point_data)\n{\n transformed_points_mat <- apply(point_data, MARGIN=2, tx$TransformPoint)\n return (sqrt(colSums((transformed_points_mat - reference_point_data)^2)))\n}\n \n \n# Check whether two transformations are \"equivalent\" in an arbitrary spatial region \n# either 3D or 2D, [x=(-10,10), y=(-100,100), z=(-1000,1000)]. This is just a sanity check, \n# as we are just looking at the effect of the transformations on a random set of points in\n# the region.\nprint_transformation_differences <- function(tx1, tx2)\n{\n if (tx1$GetDimension()==2 && tx2$GetDimension()==2)\n {\n bounds <- list(c(-10,10), c(-100,100))\n }\n else if(tx1$GetDimension()==3 && tx2$GetDimension()==3)\n {\n bounds <- list(c(-10,10), c(-100,100), c(-1000,1000))\n }\n else\n stop('Transformation dimensions mismatch, or unsupported transformation dimensionality')\n num_points <- 10\n point_data <- uniform_random_points(bounds, num_points)\n tx1_point_data <- apply(point_data, MARGIN=2, tx1$TransformPoint)\n differences <- target_registration_errors(tx2, point_data, tx1_point_data)\n cat(tx1$GetName(), \"-\", tx2$GetName(), \":\\tminDifference: \", \n toString(min(differences)), \" maxDifference: \",toString(max(differences))) \n}",
"_____no_output_____"
]
],
[
[
"In SimpleITK points can be represented by any vector-like data type. In R these include vector, array, and list. In general R will treat these data types differently, as illustrated by the print function below.",
"_____no_output_____"
]
],
[
[
"# SimpleITK points represented by vector-like data structures. \npoint_vector <- c(9.0, 10.531, 11.8341)\npoint_array <- array(c(9.0, 10.531, 11.8341),dim=c(1,3)) \npoint_list <- list(9.0, 10.531, 11.8341)\n\nprint(point_vector)\nprint(point_array)\nprint(point_list)\n\n# Uniform printing with specified precision.\nprecision <- 2\nprint(point2str(point_vector, precision))\nprint(point2str(point_array, precision))\nprint(point2str(point_list, precision))\n",
"_____no_output_____"
]
],
[
[
"## Global Transformations\nAll global transformations <i>except translation</i> are of the form:\n$$T(\\mathbf{x}) = A(\\mathbf{x}-\\mathbf{c}) + \\mathbf{t} + \\mathbf{c}$$\n\nIn ITK speak (when printing your transformation):\n<ul>\n<li>Matrix: the matrix $A$</li>\n<li>Center: the point $\\mathbf{c}$</li>\n<li>Translation: the vector $\\mathbf{t}$</li>\n<li>Offset: $\\mathbf{t} + \\mathbf{c} - A\\mathbf{c}$</li>\n</ul>",
"_____no_output_____"
],
[
"## TranslationTransform",
"_____no_output_____"
]
],
[
[
"# A 3D translation. Note that you need to specify the dimensionality, as the sitk TranslationTransform \n# represents both 2D and 3D translations.\ndimension <- 3 \noffset <- c(1,2,3) # offset can be any vector-like data \ntranslation <- TranslationTransform(dimension, offset)\nprint(translation)\ntranslation$GetOffset()",
"_____no_output_____"
],
[
"# Transform a point and use the inverse transformation to get the original back.\npoint <- c(10, 11, 12)\ntransformed_point <- translation$TransformPoint(point)\ntranslation_inverse <- translation$GetInverse()\ncat(paste0(\"original point: \", point2str(point), \"\\n\",\n \"transformed point: \", point2str(transformed_point), \"\\n\",\n \"back to original: \", point2str(translation_inverse$TransformPoint(transformed_point))))",
"_____no_output_____"
]
],
[
[
"## Euler2DTransform",
"_____no_output_____"
]
],
[
[
"point <- c(10, 11)\nrotation2D <- Euler2DTransform()\nrotation2D$SetTranslation(c(7.2, 8.4))\nrotation2D$SetAngle(pi/2.0)\ncat(paste0(\"original point: \", point2str(point), \"\\n\",\n \"transformed point: \", point2str(rotation2D$TransformPoint(point)),\"\\n\"))\n\n# Change the center of rotation so that it coincides with the point we want to\n# transform, why is this a unique configuration?\nrotation2D$SetCenter(point)\ncat(paste0(\"original point: \", point2str(point), \"\\n\",\n \"transformed point: \", point2str(rotation2D$TransformPoint(point)),\"\\n\"))",
"_____no_output_____"
]
],
[
[
"## VersorTransform",
"_____no_output_____"
]
],
[
[
"# Rotation only, parametrized by Versor (vector part of unit quaternion),\n# quaternion defined by rotation of theta around axis n: \n# q = [n*sin(theta/2), cos(theta/2)]\n \n# 180 degree rotation around z axis\n\n# Use a versor:\nrotation1 <- VersorTransform(c(0,0,1,0))\n\n# Use axis-angle:\nrotation2 <- VersorTransform(c(0,0,1), pi)\n\n# Use a matrix:\nrotation3 <- VersorTransform()\nrotation3$SetMatrix(c(-1, 0, 0, 0, -1, 0, 0, 0, 1))\n\npoint <- c(10, 100, 1000)\n\np1 <- rotation1$TransformPoint(point)\np2 <- rotation2$TransformPoint(point)\np3 <- rotation3$TransformPoint(point)\n\ncat(paste0(\"Points after transformation:\\np1=\", point2str(p1,15), \n \"\\np2=\", point2str(p2,15),\"\\np3=\", point2str(p3,15)))",
"_____no_output_____"
]
],
[
[
"We applied the \"same\" transformation to the same point, so why are the results slightly different for the second initialization method?\n \nThis is where theory meets practice. Using the axis-angle initialization method involves trigonometric functions which on a fixed precision machine lead to these slight differences. In many cases this is not an issue, but it is something to remember. From here on we will sweep it under the rug (printing with a more reasonable precision). ",
"_____no_output_____"
],
[
"## Translation to Rigid [3D]\nCopy the translational component.",
"_____no_output_____"
]
],
[
[
"dimension <- 3 \ntrans <- c(1,2,3) \ntranslation <- TranslationTransform(dimension, trans)\n\n# Only need to copy the translational component.\nrigid_euler <- Euler3DTransform()\nrigid_euler$SetTranslation(translation$GetOffset()) \nrigid_versor <- VersorRigid3DTransform()\nrigid_versor$SetTranslation(translation$GetOffset())\n\n# Sanity check to make sure the transformations are equivalent.\nbounds <- list(c(-10,10), c(-100,100), c(-1000,1000))\nnum_points <- 10\npoint_data <- uniform_random_points(bounds, num_points)\ntransformed_point_data <- apply(point_data, MARGIN=2, translation$TransformPoint) \n\n# Draw the original and transformed points.\nall_data <- cbind(point_data, transformed_point_data)\nxbnd <- range(all_data[1,])\nybnd <- range(all_data[2,])\nzbnd <- range(all_data[3,])\n\ns3d <- scatterplot3d(t(point_data), color = \"blue\", pch = 19, xlab='', ylab='', zlab='',\n xlim=xbnd, ylim=ybnd, zlim=zbnd)\ns3d$points3d(t(transformed_point_data), col = \"red\", pch = 17)\nlegend(\"topleft\", col= c(\"blue\", \"red\"), pch=c(19,17), legend = c(\"Original points\", \"Transformed points\"))\n\neuler_errors <- target_registration_errors(rigid_euler, point_data, transformed_point_data)\nversor_errors <- target_registration_errors(rigid_versor, point_data, transformed_point_data)\n\ncat(paste0(\"Euler\\tminError:\", point2str(min(euler_errors)),\" maxError: \", point2str(max(euler_errors)),\"\\n\"))\ncat(paste0(\"Versor\\tminError:\", point2str(min(versor_errors)),\" maxError: \", point2str(max(versor_errors)),\"\\n\"))",
"_____no_output_____"
]
],
[
[
"## Rotation to Rigid [3D]\nCopy the matrix or versor and <b>center of rotation</b>.",
"_____no_output_____"
]
],
[
[
"rotationCenter <- c(10, 10, 10)\nrotation <- VersorTransform(c(0,0,1,0), rotationCenter)\n\nrigid_euler <- Euler3DTransform()\nrigid_euler$SetMatrix(rotation$GetMatrix())\nrigid_euler$SetCenter(rotation$GetCenter())\n\nrigid_versor <- VersorRigid3DTransform()\nrigid_versor$SetRotation(rotation$GetVersor())\n#rigid_versor.SetCenter(rotation.GetCenter()) #intentional error\n\n# Sanity check to make sure the transformations are equivalent.\nbounds <- list(c(-10,10),c(-100,100), c(-1000,1000))\nnum_points = 10\npoint_data = uniform_random_points(bounds, num_points)\ntransformed_point_data <- apply(point_data, MARGIN=2, rotation$TransformPoint) \n \neuler_errors = target_registration_errors(rigid_euler, point_data, transformed_point_data)\nversor_errors = target_registration_errors(rigid_versor, point_data, transformed_point_data)\n\n# Draw the points transformed by the original transformation and after transformation\n# using the incorrect transformation, illustrate the effect of center of rotation.\nincorrect_transformed_point_data <- apply(point_data, 2, rigid_versor$TransformPoint) \n\nall_data <- cbind(transformed_point_data, incorrect_transformed_point_data)\nxbnd <- range(all_data[1,])\nybnd <- range(all_data[2,])\nzbnd <- range(all_data[3,])\ns3d <- scatterplot3d(t(transformed_point_data), color = \"blue\", pch = 19, xlab='', ylab='', zlab='',\n xlim=xbnd, ylim=ybnd, zlim=zbnd)\ns3d$points3d(t(incorrect_transformed_point_data), col = \"red\", pch = 17)\nlegend(\"topleft\", col= c(\"blue\", \"red\"), pch=c(19,17), legend = c(\"Original points\", \"Transformed points\"))\n\n\ncat(paste0(\"Euler\\tminError:\", point2str(min(euler_errors)),\" maxError: \", point2str(max(euler_errors)),\"\\n\"))\ncat(paste0(\"Versor\\tminError:\", point2str(min(versor_errors)),\" maxError: \", point2str(max(versor_errors)),\"\\n\"))",
"_____no_output_____"
]
],
[
[
"## Similarity [2D]\n\nWhen the center of the similarity transformation is not at the origin the effect of the transformation is not what most of us expect. This is readily visible if we limit the transformation to scaling: $T(\\mathbf{x}) = s\\mathbf{x}-s\\mathbf{c} + \\mathbf{c}$. Changing the transformation's center results in scale + translation.",
"_____no_output_____"
]
],
[
[
"# 2D square centered on (0,0)\npoints <- matrix(data=c(-1.0,-1.0, -1.0,1.0, 1.0,1.0, 1.0,-1.0), ncol=4, nrow=2) \n# Scale by 2 (center default is [0,0])\nsimilarity <- Similarity2DTransform();\nsimilarity$SetScale(2)\n\nscaled_points <- apply(points, MARGIN=2, similarity$TransformPoint) \n\n#Uncomment the following lines to change the transformations center and see what happens:\n#similarity$SetCenter(c(0,2))\n#scaled_points <- apply(points, 2, similarity$TransformPoint) \n\nplot(points[1,],points[2,], xlim=c(-10,10), ylim=c(-10,10), pch=19, col=\"blue\", xlab=\"\", ylab=\"\", las=1)\npoints(scaled_points[1,], scaled_points[2,], col=\"red\", pch=17)\nlegend('top', col= c(\"red\", \"blue\"), pch=c(17,19), legend = c(\"transformed points\", \"original points\"))",
"_____no_output_____"
]
],
[
[
"## Rigid to Similarity [3D]\nCopy the translation, center, and matrix or versor.",
"_____no_output_____"
]
],
[
[
"rotation_center <- c(100, 100, 100)\ntheta_x <- 0.0\ntheta_y <- 0.0\ntheta_z <- pi/2.0\ntranslation <- c(1,2,3)\n\nrigid_euler <- Euler3DTransform(rotation_center, theta_x, theta_y, theta_z, translation)\n\nsimilarity <- Similarity3DTransform()\nsimilarity$SetMatrix(rigid_euler$GetMatrix())\nsimilarity$SetTranslation(rigid_euler$GetTranslation())\nsimilarity$SetCenter(rigid_euler$GetCenter())\n\n# Apply the transformations to the same set of random points and compare the results\n# (see utility functions at top of notebook).\nprint_transformation_differences(rigid_euler, similarity)",
"_____no_output_____"
]
],
[
[
"## Similarity to Affine [3D]\nCopy the translation, center and matrix.",
"_____no_output_____"
]
],
[
[
"rotation_center <- c(100, 100, 100)\naxis <- c(0,0,1)\nangle <- pi/2.0\ntranslation <- c(1,2,3)\nscale_factor <- 2.0\nsimilarity <- Similarity3DTransform(scale_factor, axis, angle, translation, rotation_center)\n\naffine <- AffineTransform(3)\naffine$SetMatrix(similarity$GetMatrix())\naffine$SetTranslation(similarity$GetTranslation())\naffine$SetCenter(similarity$GetCenter())\n\n# Apply the transformations to the same set of random points and compare the results\n# (see utility functions at top of notebook).\nprint_transformation_differences(similarity, affine)",
"_____no_output_____"
]
],
[
[
"## Scale Transform\n\nJust as the case was for the similarity transformation above, when the transformations center is not at the origin, instead of a pure anisotropic scaling we also have translation ($T(\\mathbf{x}) = \\mathbf{s}^T\\mathbf{x}-\\mathbf{s}^T\\mathbf{c} + \\mathbf{c}$).",
"_____no_output_____"
]
],
[
[
"# 2D square centered on (0,0).\npoints <- matrix(data=c(-1.0,-1.0, -1.0,1.0, 1.0,1.0, 1.0,-1.0), ncol=4, nrow=2) \n\n# Scale by half in x and 2 in y.\nscale <- ScaleTransform(2, c(0.5,2));\n\nscaled_points <- apply(points, 2, scale$TransformPoint) \n\n#Uncomment the following lines to change the transformations center and see what happens:\n#scale$SetCenter(c(0,2))\n#scaled_points <- apply(points, 2, scale$TransformPoint) \n\nplot(points[1,],points[2,], xlim=c(-10,10), ylim=c(-10,10), pch=19, col=\"blue\", xlab=\"\", ylab=\"\", las=1)\npoints(scaled_points[1,], scaled_points[2,], col=\"red\", pch=17)\nlegend('top', col= c(\"red\", \"blue\"), pch=c(17,19), legend = c(\"transformed points\", \"original points\"))",
"_____no_output_____"
]
],
[
[
"## Scale Versor\n\nThis is not what you would expect from the name (composition of anisotropic scaling and rigid). This is:\n$$T(x) = (R+S)(\\mathbf{x}-\\mathbf{c}) + \\mathbf{t} + \\mathbf{c},\\;\\; \\textrm{where } S= \\left[\\begin{array}{ccc} s_0-1 & 0 & 0 \\\\ 0 & s_1-1 & 0 \\\\ 0 & 0 & s_2-1 \\end{array}\\right]$$ \n\nThere is no natural way of \"promoting\" the similarity transformation to this transformation.",
"_____no_output_____"
]
],
[
[
"scales <- c(0.5,0.7,0.9)\ntranslation <- c(1,2,3)\naxis <- c(0,0,1)\nangle <- 0.0\nscale_versor <- ScaleVersor3DTransform(scales, axis, angle, translation)\nprint(scale_versor)",
"_____no_output_____"
]
],
[
[
"## Scale Skew Versor\n\nAgain, not what you expect based on the name, this is not a composition of transformations. This is:\n$$T(x) = (R+S+K)(\\mathbf{x}-\\mathbf{c}) + \\mathbf{t} + \\mathbf{c},\\;\\; \\textrm{where } S = \\left[\\begin{array}{ccc} s_0-1 & 0 & 0 \\\\ 0 & s_1-1 & 0 \\\\ 0 & 0 & s_2-1 \\end{array}\\right]\\;\\; \\textrm{and } K = \\left[\\begin{array}{ccc} 0 & k_0 & k_1 \\\\ k_2 & 0 & k_3 \\\\ k_4 & k_5 & 0 \\end{array}\\right]$$ \n\nIn practice this is an over-parametrized version of the affine transform, 15 (scale, skew, versor, translation) vs. 12 parameters (matrix, translation).",
"_____no_output_____"
]
],
[
[
"scale <- c(2,2.1,3)\nskew <- 0:1/6.0:1 #six equally spaced values in[0,1], an arbitrary choice\ntranslation <- c(1,2,3)\nversor <- c(0,0,0,1.0)\nscale_skew_versor <- ScaleSkewVersor3DTransform(scale, skew, versor, translation)\nprint(scale_skew_versor)",
"_____no_output_____"
]
],
[
[
"## Bounded Transformations\n\nSimpleITK supports two types of bounded non-rigid transformations, BSplineTransform (sparse representation) and \tDisplacementFieldTransform (dense representation).\n\nTransforming a point that is outside the bounds will return the original point - identity transform.",
"_____no_output_____"
]
],
[
[
"#\n# This function displays the effects of the deformable transformation on a grid of points by scaling the\n# initial displacements (either of control points for BSpline or the deformation field itself). It does\n# assume that all points are contained in the range(-2.5,-2.5), (2.5,2.5) - for display.\n#\ndisplay_displacement_scaling_effect <- function(s, original_x_mat, original_y_mat, tx, original_control_point_displacements)\n{\n if(tx$GetDimension()!=2)\n stop('display_displacement_scaling_effect only works in 2D')\n\n tx$SetParameters(s*original_control_point_displacements)\n transformed_points <- mapply(function(x,y) tx$TransformPoint(c(x,y)), original_x_mat, original_y_mat)\n \n plot(original_x_mat,original_y_mat, xlim=c(-2.5,2.5), ylim=c(-2.5,2.5), pch=19, col=\"blue\", xlab=\"\", ylab=\"\", las=1)\n points(transformed_points[1,], transformed_points[2,], col=\"red\", pch=17)\n legend('top', col= c(\"red\", \"blue\"), pch=c(17,19), legend = c(\"transformed points\", \"original points\"))\n}",
"_____no_output_____"
]
],
[
[
"## BSpline\nUsing a sparse set of control points to control a free form deformation.",
"_____no_output_____"
]
],
[
[
"# Create the transformation (when working with images it is easier to use the BSplineTransformInitializer function\n# or its object oriented counterpart BSplineTransformInitializerFilter).\ndimension <- 2\nspline_order <- 3\ndirection_matrix_row_major <- c(1.0,0.0,0.0,1.0) # identity, mesh is axis aligned\norigin <- c(-1.0,-1.0) \ndomain_physical_dimensions <- c(2,2)\n\nbspline <- BSplineTransform(dimension, spline_order)\nbspline$SetTransformDomainOrigin(origin)\nbspline$SetTransformDomainDirection(direction_matrix_row_major)\nbspline$SetTransformDomainPhysicalDimensions(domain_physical_dimensions)\nbspline$SetTransformDomainMeshSize(c(4,3))\n\n# Random displacement of the control points.\noriginalControlPointDisplacements <- runif(length(bspline$GetParameters()))\nbspline$SetParameters(originalControlPointDisplacements)\n\n# Apply the bspline transformation to a grid of points \n# starting the point set exactly at the origin of the bspline mesh is problematic as\n# these points are considered outside the transformation's domain,\n# remove epsilon below and see what happens.\nnumSamplesX = 10\nnumSamplesY = 20\n\neps <- .Machine$double.eps\n\ncoordsX <- seq(origin[1] + eps,\n origin[1] + domain_physical_dimensions[1],\n (domain_physical_dimensions[1]-eps)/(numSamplesX-1))\ncoordsY <- seq(origin[2] + eps,\n origin[2] + domain_physical_dimensions[2],\n (domain_physical_dimensions[2]-eps)/(numSamplesY-1))\n# next two lines equivalent to Python's/MATLAB's meshgrid \nXX <- outer(coordsY*0, coordsX, \"+\")\nYY <- outer(coordsY, coordsX*0, \"+\") \n\ndisplay_displacement_scaling_effect(0.0, XX, YY, bspline, originalControlPointDisplacements)\n\n#uncomment the following line to see the effect of scaling the control point displacements \n# on our set of points (we recommend keeping the scaling in the range [-1.5,1.5] due to display bounds) \n#display_displacement_scaling_effect(0.5, XX, YY, bspline, originalControlPointDisplacements)",
"_____no_output_____"
]
],
[
[
"## DisplacementField\n\nA dense set of vectors representing the displacement inside the given domain. The most generic representation of a transformation.",
"_____no_output_____"
]
],
[
[
"# Create the displacement field. \n \n# When working with images the safer thing to do is use the image based constructor,\n# DisplacementFieldTransform(my_image), all the fixed parameters will be set correctly and the displacement\n# field is initialized using the vectors stored in the image. SimpleITK requires that the image's pixel type be \n# \"sitkVectorFloat64\".\ndisplacement <- DisplacementFieldTransform(2)\nfield_size <- c(10,20)\nfield_origin <- c(-1.0,-1.0) \nfield_spacing <- c(2.0/9.0,2.0/19.0) \nfield_direction <- c(1,0,0,1) # direction cosine matrix (row major order) \n\n# Concatenate all the information into a single list\ndisplacement$SetFixedParameters(c(field_size, field_origin, field_spacing, field_direction))\n# Set the interpolator, either sitkLinear which is default or nearest neighbor\ndisplacement$SetInterpolator(\"sitkNearestNeighbor\")\n\noriginalDisplacements <- runif(length(displacement$GetParameters()))\ndisplacement$SetParameters(originalDisplacements)\n\ncoordsX <- seq(field_origin[1],\n field_origin[1]+(field_size[1]-1)*field_spacing[1],\n field_spacing[1])\ncoordsY <- seq(field_origin[2],\n field_origin[2]+(field_size[2]-1)*field_spacing[2],\n field_spacing[2])\n\n# next two lines equivalent to Python's/MATLAB's meshgrid \nXX <- outer(coordsY*0, coordsX, \"+\")\nYY <- outer(coordsY, coordsX*0, \"+\") \n\ndisplay_displacement_scaling_effect(0.0, XX, YY, displacement, originalDisplacements)\n\n#uncomment the following line to see the effect of scaling the control point displacements \n# on our set of points (we recommend keeping the scaling in the range [-1.5,1.5] due to display bounds) \n#display_displacement_scaling_effect(0.5, XX, YY, displacement, originalDisplacements)",
"_____no_output_____"
]
],
[
[
"Displacement field transform created from an image. Remember that SimpleITK will clear the image you provide, as shown in the cell below.",
"_____no_output_____"
]
],
[
[
"displacement_image <- Image(c(64,64), \"sitkVectorFloat64\")\n\n# The only point that has any displacement is at physical SimpleITK index (0,0), R index (1,1)\ndisplacement <- c(0.5,0.5)\n# Note that SimpleITK indexing starts at zero.\ndisplacement_image$SetPixel(c(0,0), displacement)\n\ncat('Original displacement image size: ',point2str(displacement_image$GetSize()),\"\\n\")\n\ndisplacement_field_transform <- DisplacementFieldTransform(displacement_image)\n\ncat(\"After using the image to create a transform, displacement image size: \",\n point2str(displacement_image$GetSize()), \"\\n\")\n\n# Check that the displacement field transform does what we expect.\ncat(\"Expected result: \",point2str(displacement),\n \"\\nActual result: \", displacement_field_transform$TransformPoint(c(0,0)),\"\\n\")",
"_____no_output_____"
]
],
[
[
"## CompositeTransform\n\nThis class represents a composition of transformations, multiple transformations applied one after the other. \n\nThe choice of whether to use a composite transformation or compose transformations on your own has subtle differences in the registration framework.\n\nBelow we represent the composite transformation $T_{affine}(T_{rigid}(x))$ in two ways: (1) use a composite transformation to contain the two; (2) combine the two into a single affine transformation. We can use both as initial transforms (SetInitialTransform) for the registration framework (ImageRegistrationMethod). The difference is that in the former case the optimized parameters belong to the rigid transformation and in the later they belong to the combined-affine transformation. ",
"_____no_output_____"
]
],
[
[
"# Create a composite transformation: T_affine(T_rigid(x)).\nrigid_center <- c(100,100,100)\ntheta_x <- 0.0\ntheta_y <- 0.0\ntheta_z <- pi/2.0\nrigid_translation <- c(1,2,3)\nrigid_euler <- Euler3DTransform(rigid_center, theta_x, theta_y, theta_z, rigid_translation)\n\naffine_center <- c(20, 20, 20)\naffine_translation <- c(5,6,7) \n\n# Matrix is represented as a vector-like data in row major order.\naffine_matrix <- runif(9) \naffine <- AffineTransform(affine_matrix, affine_translation, affine_center)\n\n# Using the composite transformation we just add them in (stack based, first in - last applied).\ncomposite_transform <- CompositeTransform(affine)\ncomposite_transform$AddTransform(rigid_euler)\n\n# Create a single transform manually. this is a recipe for compositing any two global transformations\n# into an affine transformation, T_0(T_1(x)):\n# A = A=A0*A1\n# c = c1\n# t = A0*[t1+c1-c0] + t0+c0-c1\nA0 <- t(matrix(affine$GetMatrix(), 3, 3))\nc0 <- affine$GetCenter()\nt0 <- affine$GetTranslation()\n\nA1 <- t(matrix(rigid_euler$GetMatrix(), 3, 3))\nc1 <- rigid_euler$GetCenter()\nt1 <- rigid_euler$GetTranslation()\n\ncombined_mat <- A0%*%A1\ncombined_center <- c1\ncombined_translation <- A0 %*% (t1+c1-c0) + t0+c0-c1\ncombined_affine <- AffineTransform(c(t(combined_mat)), combined_translation, combined_center)\n\n# Check if the two transformations are \"equivalent\".\ncat(\"Apply the two transformations to the same point cloud:\\n\")\nprint_transformation_differences(composite_transform, combined_affine)\n\ncat(\"\\nTransform parameters:\\n\")\ncat(paste(\"\\tComposite transform: \", point2str(composite_transform$GetParameters(),2),\"\\n\"))\ncat(paste(\"\\tCombined affine: \", point2str(combined_affine$GetParameters(),2),\"\\n\"))\n\ncat(\"Fixed parameters:\\n\")\ncat(paste(\"\\tComposite transform: \", point2str(composite_transform$GetFixedParameters(),2),\"\\n\"))\ncat(paste(\"\\tCombined affine: \", point2str(combined_affine$GetFixedParameters(),2),\"\\n\"))",
"_____no_output_____"
]
],
[
[
"Composite transforms enable a combination of a global transformation with multiple local/bounded transformations. This is useful if we want to apply deformations only in regions that deform while other regions are only effected by the global transformation.\n\nThe following code illustrates this, where the whole region is translated and subregions have different deformations.",
"_____no_output_____"
]
],
[
[
"# Global transformation.\ntranslation <- TranslationTransform(2, c(1.0,0.0))\n\n# Displacement in region 1.\ndisplacement1 <- DisplacementFieldTransform(2)\nfield_size <- c(10,20)\nfield_origin <- c(-1.0,-1.0) \nfield_spacing <- c(2.0/9.0,2.0/19.0) \nfield_direction <- c(1,0,0,1) # direction cosine matrix (row major order) \n\n# Concatenate all the information into a single list.\ndisplacement1$SetFixedParameters(c(field_size, field_origin, field_spacing, field_direction))\ndisplacement1$SetParameters(rep(1.0, length(displacement1$GetParameters())))\n\n# Displacement in region 2.\ndisplacement2 <- DisplacementFieldTransform(2)\nfield_size <- c(10,20)\nfield_origin <- c(1.0,-3) \nfield_spacing <- c(2.0/9.0,2.0/19.0) \nfield_direction <- c(1,0,0,1) #direction cosine matrix (row major order) \n\n# Concatenate all the information into a single list.\ndisplacement2$SetFixedParameters(c(field_size, field_origin, field_spacing, field_direction))\ndisplacement2$SetParameters(rep(-1.0, length(displacement2$GetParameters())))\n\n# Composite transform which applies the global and local transformations.\ncomposite <- CompositeTransform(translation)\ncomposite$AddTransform(displacement1)\ncomposite$AddTransform(displacement2)\n\n# Apply the composite transformation to points in ([-1,-3],[3,1]) and \n# display the deformation using a quiver plot.\n \n# Generate points.\nnumSamplesX <- 10\nnumSamplesY <- 10\ncoordsX <- seq(-1.0, 3.0, 4.0/(numSamplesX-1))\ncoordsY <- seq(-3.0, 1.0, 4.0/(numSamplesY-1))\n# next two lines equivalent to Python's/MATLAB's meshgrid \noriginal_x_mat <- outer(coordsY*0, coordsX, \"+\")\noriginal_y_mat <- outer(coordsY, coordsX*0, \"+\") \n\n# Transform points and plot.\noriginal_points <- mapply(function(x,y) c(x,y), original_x_mat, original_y_mat)\ntransformed_points <- mapply(function(x,y) composite$TransformPoint(c(x,y)), original_x_mat, original_y_mat)\nplot(0,0,xlim=c(-1.0,3.0), ylim=c(-3.0,1.0), las=1)\narrows(original_points[1,], original_points[2,], transformed_points[1,], transformed_points[2,])",
"_____no_output_____"
]
],
[
[
"## Transform\n\nThis class represents a generic transform and is the return type from the registration framework (if not done in place). Underneath the generic facade is one of the actual classes. To find out who is hiding under the hood we can query the transform to obtain the [TransformEnum](https://simpleitk.org/doxygen/latest/html/namespaceitk_1_1simple.html#a527cb966ed81d0bdc65999f4d2d4d852).\n\nWe can then downcast the generic transform to its actual type and obtain access to the relevant methods. Note that attempting to access the method will fail but not invoke an exception so we cannot use `try`, `tryCatch`.",
"_____no_output_____"
]
],
[
[
"tx <- Transform(TranslationTransform(2,c(1.0,0.0)))\nif(tx$GetTransformEnum() == 'sitkTranslation') {\n translation = TranslationTransform(tx)\n cat(paste(c('Translation is:', translation$GetOffset()), collapse=' '))\n}",
"_____no_output_____"
]
],
[
[
"## Writing and Reading\n\nThe SimpleITK.ReadTransform() returns a SimpleITK.Transform . The content of the file can be any of the SimpleITK transformations or a composite (set of transformations). \n\n**Details of note**:\n1. When read from file, the type of the returned transform is the generic `Transform`. We can then obtain the \"true\" transform type via the `Downcast` method.\n2. Writing of nested composite transforms is not supported, you will need to \"flatten\" the transform before writing it to file.",
"_____no_output_____"
]
],
[
[
"# Create a 2D rigid transformation, write it to disk and read it back.\nbasic_transform <- Euler2DTransform()\nbasic_transform$SetTranslation(c(1,2))\nbasic_transform$SetAngle(pi/2.0)\n\nfull_file_name <- file.path(OUTPUT_DIR, \"euler2D.tfm\")\n\nWriteTransform(basic_transform, full_file_name)\n\n# The ReadTransform function returns a SimpleITK Transform no matter the type of the transform \n# found in the file (global, bounded, composite).\nread_result <- ReadTransform(full_file_name)\ncat(paste(\"Original type: \",basic_transform$GetName(),\"\\nType after reading: \", read_result$GetName(),\"\\n\"))\nprint_transformation_differences(basic_transform, read_result)\n\n\n# Create a composite transform then write and read.\ndisplacement <- DisplacementFieldTransform(2)\nfield_size <- c(10,20)\nfield_origin <- c(-10.0,-100.0) \nfield_spacing <- c(20.0/(field_size[1]-1),200.0/(field_size[2]-1)) \nfield_direction <- c(1,0,0,1) #direction cosine matrix (row major order)\n\n# Concatenate all the information into a single list.\ndisplacement$SetFixedParameters(c(field_size, field_origin, field_spacing, field_direction))\ndisplacement$SetParameters(runif(length(displacement$GetParameters())))\n\ncomposite_transform <- Transform(basic_transform)\ncomposite_transform$AddTransform(displacement)\n\nfull_file_name <- file.path(OUTPUT_DIR, \"composite.tfm\")\n\nWriteTransform(composite_transform, full_file_name)\nread_result <- ReadTransform(full_file_name)\ncat(\"\\n\")\nprint_transformation_differences(composite_transform, read_result) ",
"_____no_output_____"
],
[
"x_translation <- TranslationTransform(2,c(1,0))\ny_translation <- TranslationTransform(2,c(0,1))\n# Create composite transform with the x_translation repeated 3 times\ncomposite_transform1 <- CompositeTransform(x_translation)\ncomposite_transform1$AddTransform(x_translation)\ncomposite_transform1$AddTransform(x_translation)\n# Create a nested composite transform\ncomposite_transform <- CompositeTransform(y_translation)\ncomposite_transform$AddTransform(composite_transform1)\ncat(paste0('Nested composite transform contains ',composite_transform$GetNumberOfTransforms(), ' transforms.\\n'))\n\n# We cannot write nested composite transformations, so we \n# flatten it (unravel the nested part)\ncomposite_transform$FlattenTransform()\ncat(paste0('Nested composite transform after flattening contains ',composite_transform$GetNumberOfTransforms(), ' transforms.\\n'))\nfull_file_name <- file.path(OUTPUT_DIR, \"composite.tfm\")\nWriteTransform(composite_transform, full_file_name)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
cbed952aebf00abdc22563c81bdbe45c57164fe8
| 110,114 |
ipynb
|
Jupyter Notebook
|
analysis/Analysis Notebook/Analysis Notebook.ipynb
|
data301-2020-winter1/course-project-group_6022
|
892e9f42c757e53094ea07185136393aa4353d5d
|
[
"MIT"
] | null | null | null |
analysis/Analysis Notebook/Analysis Notebook.ipynb
|
data301-2020-winter1/course-project-group_6022
|
892e9f42c757e53094ea07185136393aa4353d5d
|
[
"MIT"
] | 1 |
2020-11-26T22:09:39.000Z
|
2020-11-26T22:09:39.000Z
|
analysis/Analysis Notebook/Analysis Notebook.ipynb
|
data301-2020-winter1/course-project-group_6022
|
892e9f42c757e53094ea07185136393aa4353d5d
|
[
"MIT"
] | null | null | null | 95.08981 | 26,432 | 0.746172 |
[
[
[
"### Research Question 1\n<p><b>What is the exact demographic most at risk for covid19 in Toronto?</b></p> \nTo find this we'll need to produce graphs of Age, Gender, and if they were hospitalized.\n",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nimport pandas_profiling\nimport sys, os\nimport seaborn as sns\nfrom matplotlib import pyplot as plt\nsys.path.insert(0, os.path.abspath('..'))\nfrom scripts import project_functions as sc\npath = \"\\\\analysis\\\\Analysis Notebook\" \n#this formats everything quickly. \ndf = sc.ez_format(path)\ndf",
"_____no_output_____"
],
[
"#create a new data frame of values we care about\nndf = df.groupby([\"Gender\", \"Age_Group\", \"Ever_Hospitalized\"]).size().reset_index()\nndf = ndf.rename(columns = {0:'D_Count'})\nndf = ndf.sort_values(by = 'D_Count', ascending = False)\n#drop those who were not hosplitalized, and then drop the hopitalized column. \nndf = ndf.loc[ndf[\"Ever_Hospitalized\"].str.contains(\"Yes\")]\nndf = ndf.drop([\"Ever_Hospitalized\"], axis = 1)\nndf = ndf.loc[~df[\"Gender\"].str.contains(\"UNKNOWN\")]\n#create a demographic column, combine gender and agegroup together\nndf.insert( 2,column = 'Demographic', value = (ndf['Gender'][:] + \" \" + ndf['Age_Group'][:]))\norder = ndf['Demographic'].to_list()\nndf = ndf.reset_index(drop = True)\nndf",
"_____no_output_____"
],
[
"import seaborn as sns\nfrom matplotlib import pyplot as plt\nplt.figure(figsize=(6,5))\nsns.set_context(\"notebook\")\nsns.barplot(x ='D_Count', y = 'Demographic', data= ndf, order= order, palette = 'viridis_r')\nsns.despine(top = True, right = True, left = False, bottom = True)\nplt.title(\"Demographic by Hospitalization Cases\")\nplt.xlabel(\"Amount of Cases resulting in Hospitalization\")",
"_____no_output_____"
],
[
"#quickly draw up a new dataframe. \ndf = sc.ez_format(path)\ndf = df.loc[df[\"Ever_Hospitalized\"].str.contains(\"Yes\")]\ndf = df.loc[~df[\"Gender\"].str.contains(\"UNKNOWN\")]\ndf = df.reset_index(drop = True)\nls = df['Age_Group'].value_counts().to_dict()\nplt.figure(figsize=(7,5))\n\"\"\"\nUse the seaborn count plot to visualize our data. Personally, graph 1 is easier to look at than graph 2,\nbut I also want to go for style points. \n\"\"\"\nsns.countplot(x= 'Gender', hue = \"Age_Group\", data=df, hue_order = ls, dodge = True, palette = 'viridis_r' )\nsns.despine(top = True, right = True, left = False)\nplt.title(\"Demographic by Hospitalization Cases\")\nplt.xlabel(\"Gender\")\nplt.legend(loc='upper right')\n\n",
"_____no_output_____"
]
],
[
[
"### Result\nBy our graph we can see that senior males are most succeptable to the virus. This is intriuging becase our data set actually has \nmore females than males. My initial hypothesis was that the most succeptable demographic would be female, since there was over 1000 more female cases.\nHere's the image of that graph that from EDA_MattKuelker.ipynb. (If you still can't find it, it's in output.png here) \n\n\n\nNot only that, but it apears that across the board, elderly men are most at risk for the virus, despite being a less prominent group in the data set. \nAs for those least affected, 90+ is to be expected, since most people don't live that long. I am surpised that there was a trangender case that was hospitalized, considering that there are not that many trans people compared to cis gendered people. \n\nFrom this data set we can also guess that females <19 are the least to be affected. Not only are there few hospitializations, but also their male <19 counterparts are also at the bottom of the case count. ",
"_____no_output_____"
],
[
"### Research Question 2\n<p><b>Do women have a higher spread via physical contact than men? </b></p> \n<i>Women are scientifically proven to have closer relationships than males. Let's see if we can draw a correllation between physical contact and cases for gender. </i>\n\nHow are we going to prove this? Well first to visualize our data lets draw up a countplot via gender, than see if we can draw a correllation. ",
"_____no_output_____"
]
],
[
[
"path =\"\\\\analysis\\\\Analysis Notebook\"\n#this formats everything quickly. \ndf = sc.ez_format(path)\n\ndf.head(5)\n",
"_____no_output_____"
]
],
[
[
"We're going to be filtering out pending, unkown, and N/A / outbreak associated, since those do not directly tell us how exactly people got covid 19. We'll also only focus on males and females since there isn't enough transgender cases to add draw any meaningful conclusions. ",
"_____no_output_____"
]
],
[
[
"df = df[~df['Source_of_Infection'].isin(['N/A - Outbreak associated','Pending','Unknown/Missing'])]\ndf = df.loc[df[\"Gender\"].str.contains(\"MALE\")] #We're dropping all other genders and whatnots from the dataset beccause they are minscule in count. \nls = df['Source_of_Infection'].value_counts().to_dict()\nplt.figure(figsize=(6,5))\nsns.set_context(\"notebook\")\nsns.countplot(y = \"Source_of_Infection\", data =df, order = ls, hue = \"Gender\", palette = \"Blues_r\")\nplt.legend(loc = \"center right\")\nplt.ylabel(\"Source of Infection\")\nplt.xlabel(\"Count\")\nsns.despine()",
"_____no_output_____"
]
],
[
[
"Look at this! After some decent filtering it turns out my hypothesis is completely wrong, and the most notable desparity between male and female is actually community contact. Turns out the demographic with the highest infection count in community contact was male. \n\nLet's modify our question then and ask ourselves what the age demographic is of males who were infected via community contact. My guess is bachelor men in their 20's and seniors in elderly homes, since we had proven in our research question above older men were most at risk for the virus. \n\n<p><b>New Question 2: What is the demographic of these males infected in community? </b></p> \n\nTime for some filtering. ",
"_____no_output_____"
]
],
[
[
"df = sc.ez_format(path)\ndf = df.loc[df[\"Gender\"].str.contains(\"MALE\")] #This almost got me. Always anything containing male will be kept.... which means we'd still have females in our dataset. \ndf = df.loc[~df[\"Gender\"].str.contains(\"FEMALE\")] \ndf = df.loc[df[\"Source_of_Infection\"].str.contains(\"Community\")]\ndf\nls = df['Age_Group'].value_counts(ascending = True).to_dict()\nplt.figure(figsize=(6,5))\nsns.set_context(\"notebook\")\nsns.countplot(x = \"Age_Group\", data = df, order = ls, palette = \"Spectral\")\nplt.ylabel(\"Males Infected\")\nplt.xlabel(\"Community Contact Cases\")\nplt.title(\"Infected Males via Community Contact\")\nsns.despine()\n\n",
"_____no_output_____"
]
],
[
[
"### Conclusion\nAs I had hypothesized, Males in their 20's had a large amount of ... however the largest group wasn't elderly men but men in their 50's. Likely however we can conclude that community contact then isn't caused exclusivley by males living together in the same residence.",
"_____no_output_____"
]
],
[
[
"df = pd.read_csv(sc.autopath(path))\ndf",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cbed975f6894ba2ae740d0e3cf1d3b9afe0091cc
| 4,713 |
ipynb
|
Jupyter Notebook
|
HubSpot/HubSpot_Get_contacts_associated_to_deal.ipynb
|
krajai/testt
|
3aaf5fd7fe85e712c8c1615852b50f9ccb6737e5
|
[
"BSD-3-Clause"
] | 1 |
2022-03-24T07:46:45.000Z
|
2022-03-24T07:46:45.000Z
|
HubSpot/HubSpot_Get_contacts_associated_to_deal.ipynb
|
PZawieja/awesome-notebooks
|
8ae86e5689749716e1315301cecdad6f8843dcf8
|
[
"BSD-3-Clause"
] | null | null | null |
HubSpot/HubSpot_Get_contacts_associated_to_deal.ipynb
|
PZawieja/awesome-notebooks
|
8ae86e5689749716e1315301cecdad6f8843dcf8
|
[
"BSD-3-Clause"
] | null | null | null | 19.970339 | 301 | 0.504774 |
[
[
[
"<img width=\"10%\" alt=\"Naas\" src=\"https://landen.imgix.net/jtci2pxwjczr/assets/5ice39g4.png?w=160\"/>",
"_____no_output_____"
],
[
"# HubSpot - Get contacts associated to deal\n<a href=\"https://app.naas.ai/user-redirect/naas/downloader?url=https://raw.githubusercontent.com/jupyter-naas/awesome-notebooks/master/HubSpot/HubSpot_Get_contacts_associated_to_deal.ipynb\" target=\"_parent\"><img src=\"https://naasai-public.s3.eu-west-3.amazonaws.com/open_in_naas.svg\"/></a>",
"_____no_output_____"
],
[
"**Tags:** #hubspot #crm #sales #deal #contact #naas_drivers #snippet #dataframe",
"_____no_output_____"
],
[
"**Author:** [Florent Ravenel](https://www.linkedin.com/in/florent-ravenel/)",
"_____no_output_____"
],
[
"## Input",
"_____no_output_____"
],
[
"### Import library",
"_____no_output_____"
]
],
[
[
"from naas_drivers import hubspot",
"_____no_output_____"
]
],
[
[
"### Setup your HubSpot\n👉 Access your [HubSpot API key](https://knowledge.hubspot.com/integrations/how-do-i-get-my-hubspot-api-key)",
"_____no_output_____"
]
],
[
[
"HS_API_KEY = 'YOUR_HUBSPOT_API_KEY'",
"_____no_output_____"
]
],
[
[
"### Enter deal ID",
"_____no_output_____"
]
],
[
[
"deal_id = '3452242690'",
"_____no_output_____"
]
],
[
[
"## Model",
"_____no_output_____"
],
[
"### Get association",
"_____no_output_____"
]
],
[
[
"result = hubspot.connect(HS_API_KEY).associations.get('deal', \n deal_id,\n 'contact')",
"_____no_output_____"
]
],
[
[
"## Output",
"_____no_output_____"
],
[
"### Display result",
"_____no_output_____"
]
],
[
[
"result",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
]
] |
cbed9ac3938afe219bf2ab7f6ecd09cb237a4777
| 3,866 |
ipynb
|
Jupyter Notebook
|
Trabalhando String.ipynb
|
marcelobentes/Python
|
6a1e280f6c11d9c3b5c09836c7d2b455ab1d109c
|
[
"MIT"
] | null | null | null |
Trabalhando String.ipynb
|
marcelobentes/Python
|
6a1e280f6c11d9c3b5c09836c7d2b455ab1d109c
|
[
"MIT"
] | null | null | null |
Trabalhando String.ipynb
|
marcelobentes/Python
|
6a1e280f6c11d9c3b5c09836c7d2b455ab1d109c
|
[
"MIT"
] | null | null | null | 29.738462 | 122 | 0.536989 |
[
[
[
"print(ord('a'))\nprint(chr(97))\nprint(chr(55))\n\nstr1 = 'Uma string!'\nstr2 = \"Outra string!\"\nstr3 = \"\"\"Mais uma string!\"\"\"\n\nopcao = input('Tecle \"S\" para SIM e \"N\" para não: ')\nopcao = input(\"Tecle 'S' para SIM e 'N' para não:\")\nopcao = input(\"\"\"Tecle 'S' para SIM e 'N' para não:\"\"\")\n\n#acessando o indice de uma String\n\nx = \"Vamos lá!\"\nprint(\"tipo da variavel: \",type(x))#retorna o tipo da variavel\nprint(x[2])#acessando o indice da String\nprint(len(x))\nprimeira_parte = x[ : len(x) // 2]#divindo o fatiamento do tamanho da String\nprint(primeira_parte)\n\n#passando um String para maiuscula\nmsg_MAIUSCULA = x.upper()\nprint(msg_MAIUSCULA)\n\n#passando um String para minuscula\nmsg_minuscula = x.lower()\nprint(msg_minuscula)\n\n#contando quantas vezes a palavra se repete\nquantos = \"\"\"João amava Teresa que amava Raimundo\nque amava Maria que amava Joaquim que amava Lili\nque não amava ninguém. \"\"\".count(\"amava\")\nprint(quantos) # vai imprimir o valor 6\n\n#substituindo a palavra\nquantos = \"\"\"João amava Teresa que amava Raimundo\nque amava Maria que amava Joaquim que amava Lili\nque não amava ninguém. \"\"\".replace(\"amava\", \"Menicava\")\nprint(quantos) \n\n#Utilizando o operador in, é possível verificar se existe uma dada palavra em uma string, como no exemplo abaixo:\n\ntexto = \"\"\"João amava Teresa que amava Raimundo\nque amava Maria que amava Joaquim que amava Lili\nque não amava ninguém. \"\"\"\nif \"Paulo\" in texto:\n print(\"Paulo está na lista de pessoas.\")\nelse:\n print(\"Paulo não está na lista de pessoas.\")\n \n#Usando o format() \nnome = input('Digite seu nome: ')\nidade = int(input('Digite sua idade: '))\naltura = float(input('Digite sua altura: '))\n \nformato_msg = 'Olá, {}! Sua idade é {} anos e sua altura é {}m. '\nmsg = formato_msg.format(nome, idade, altura)\nprint(msg)",
"97\na\n7\nTecle \"S\" para SIM e \"N\" para não: s\nTecle 'S' para SIM e 'N' para não:s\nTecle 'S' para SIM e 'N' para não:s\ntipo da variavel: <class 'str'>\nm\n9\nVamo\nVAMOS LÁ!\nvamos lá!\n6\nJoão Menicava Teresa que Menicava Raimundo\nque Menicava Maria que Menicava Joaquim que Menicava Lili\nque não Menicava ninguém. \nPaulo não está na lista de pessoas.\nDigite seu nome: Marcelo\nDigite sua idade: 36\nDigite sua altura: 1.69\nOlá, Marcelo! Sua idade é 36 anos e sua altura é 1.69m. \n"
]
]
] |
[
"code"
] |
[
[
"code"
]
] |
cbedaa596a16c9d9329a1a53a102e4f5d8d62437
| 222,344 |
ipynb
|
Jupyter Notebook
|
task2/Untitled.ipynb
|
alexsubota/OpenDataScience
|
b490a60ed87b551d478348781c7df888ed01558c
|
[
"MIT"
] | null | null | null |
task2/Untitled.ipynb
|
alexsubota/OpenDataScience
|
b490a60ed87b551d478348781c7df888ed01558c
|
[
"MIT"
] | null | null | null |
task2/Untitled.ipynb
|
alexsubota/OpenDataScience
|
b490a60ed87b551d478348781c7df888ed01558c
|
[
"MIT"
] | null | null | null | 244.334066 | 33,612 | 0.897474 |
[
[
[
"# подгружаем все нужные пакеты\nimport pandas as pd\nimport numpy as np\n\n# игнорируем warnings\nimport warnings\nwarnings.filterwarnings(\"ignore\")\n\nimport seaborn as sns\n\nimport matplotlib\nimport matplotlib.pyplot as plt\nimport matplotlib.ticker\n%matplotlib inline\n\n# настройка внешнего вида графиков в seaborn\nsns.set_context(\n \"notebook\", \n font_scale = 1.5, \n rc = { \n \"figure.figsize\" : (12, 9), \n \"axes.titlesize\" : 18 \n }\n)",
"_____no_output_____"
],
[
"train = pd.read_csv('mlbootcamp5_train.csv', sep=';',\n index_col='id')",
"_____no_output_____"
],
[
"print('Размер датасета: ', train.shape)\ntrain.head()",
"Размер датасета: (70000, 12)\n"
],
[
"train_uniques = pd.melt(frame=train, value_vars=['gender','cholesterol', \n 'gluc', 'smoke', 'alco', \n 'active', 'cardio'])\ntrain_uniques = pd.DataFrame(train_uniques.groupby(['variable', \n 'value'])['value'].count()) \\\n .sort_index(level=[0, 1]) \\\n .rename(columns={'value': 'count'}) \\\n .reset_index()\n \nsns.factorplot(x='variable', y='count', hue='value', \n data=train_uniques, kind='bar', size=12);",
"_____no_output_____"
],
[
"train_uniques = pd.melt(frame=train, value_vars=['gender','cholesterol', \n 'gluc', 'smoke', 'alco', \n 'active'], \n id_vars=['cardio'])\ntrain_uniques = pd.DataFrame(train_uniques.groupby(['variable', 'value', \n 'cardio'])['value'].count()) \\\n .sort_index(level=[0, 1]) \\\n .rename(columns={'value': 'count'}) \\\n .reset_index()\n \nsns.factorplot(x='variable', y='count', hue='value', \n col='cardio', data=train_uniques, kind='bar', size=9);",
"_____no_output_____"
],
[
"for c in train.columns:\n n = train[c].nunique()\n print(c)\n \n if n <= 3:\n print(n, sorted(train[c].value_counts().to_dict().items()))\n else:\n print(n)\n print(10 * '-')",
"age\n8076\n----------\ngender\n2 [(1, 45530), (2, 24470)]\n----------\nheight\n109\n----------\nweight\n287\n----------\nap_hi\n153\n----------\nap_lo\n157\n----------\ncholesterol\n3 [(1, 52385), (2, 9549), (3, 8066)]\n----------\ngluc\n3 [(1, 59479), (2, 5190), (3, 5331)]\n----------\nsmoke\n2 [(0, 63831), (1, 6169)]\n----------\nalco\n2 [(0, 66236), (1, 3764)]\n----------\nactive\n2 [(0, 13739), (1, 56261)]\n----------\ncardio\n2 [(0, 35021), (1, 34979)]\n----------\n"
],
[
"train.head()",
"_____no_output_____"
],
[
"corrTrain = train.corr()\nsns.heatmap(corrTrain);",
"_____no_output_____"
],
[
"#choresterol gluc",
"_____no_output_____"
],
[
"train2 = train.melt(id_vars=['height'], value_vars=['gender'])",
"_____no_output_____"
],
[
"sns.violinplot(x='value', y='height', data=train2);",
"_____no_output_____"
],
[
"_, axes = plt.subplots(1, 2, sharey=True, figsize=(16,6))\nsns.kdeplot(train2[train2['value'] == 1]['height'], ax = axes[0])\nsns.kdeplot(train2[train2['value'] == 2]['height'], ax = axes[1])",
"_____no_output_____"
],
[
"corrTrain = train.corr(method = 'spearman')",
"_____no_output_____"
],
[
"sns.heatmap(corrTrain);",
"_____no_output_____"
],
[
"#ap_hi ap_ho\n#Природа данных",
"_____no_output_____"
],
[
"#g = sns.jointplot(corrTrain['ap_hi'], corrTrain['ap_lo'], data = filtered)\ndf4=train.copy()[['ap_hi','ap_lo']]\ndf4=df4[(df4['ap_hi']>0) & (df4['ap_lo']>0)]\ndf4['l'+'ap_hi']=df4['ap_hi'].apply(np.log1p)\ndf4['l'+'ap_lo']=df4['ap_lo'].apply(np.log1p)\ng=sns.jointplot(x='l'+'ap_hi',y='l'+'ap_lo',data=df4, dropna=True)\n\n#\"\"\"Сетка\"\"\"\ng.ax_joint.grid(True) \n\n#\"\"\"Преобразуем логарифмические значения на шкалах в реальные\"\"\"\ng.ax_joint.yaxis.set_major_formatter(matplotlib.ticker.FuncFormatter(lambda x, pos: str(round(int(np.exp(x))))))\ng.ax_joint.xaxis.set_major_formatter(matplotlib.ticker.FuncFormatter(lambda x, pos: str(round(int(np.exp(x))))))",
"_____no_output_____"
],
[
"#3??",
"_____no_output_____"
],
[
"train['age_years'] = (train['age'] // 365.25).astype(int)",
"_____no_output_____"
],
[
"train.head()",
"_____no_output_____"
],
[
"plt.subplots(figsize = (18,10))\nsns.countplot(x = 'age_years', hue = 'cardio', data = train)",
"_____no_output_____"
],
[
"#53",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbedb02a727a104d51318ab75473e5bc123165db
| 168,446 |
ipynb
|
Jupyter Notebook
|
Implementations/unsupervised/.ipynb_checkpoints/K-means - Empty-checkpoint.ipynb
|
teh67/teh67.github.io
|
006f8fbf2383802c9e5ce9217c69478b3af3de24
|
[
"CC-BY-3.0"
] | 1 |
2021-09-25T22:15:37.000Z
|
2021-09-25T22:15:37.000Z
|
Implementations/unsupervised/.ipynb_checkpoints/K-means - Empty-checkpoint.ipynb
|
openCAML/openCAML.github.io
|
26df37fd0b9daa2c40f9615e44b8ec63e1322302
|
[
"CC-BY-3.0",
"BSD-3-Clause"
] | null | null | null |
Implementations/unsupervised/.ipynb_checkpoints/K-means - Empty-checkpoint.ipynb
|
openCAML/openCAML.github.io
|
26df37fd0b9daa2c40f9615e44b8ec63e1322302
|
[
"CC-BY-3.0",
"BSD-3-Clause"
] | null | null | null | 323.31286 | 42,288 | 0.925412 |
[
[
[
"# K-means clustering\n\nWhen working with large datasets it can be helpful to group similar observations together. This process, known as clustering, is one of the most widely used in Machine Learning and is often used when our dataset comes without pre-existing labels. \n\nIn this notebook we're going to implement the classic K-means algorithm, the simplest and most widely used clustering method. Once we've implemented it we'll use it to split a dataset into groups and see how our clustering compares to the 'true' labelling.",
"_____no_output_____"
],
[
"## Import Modules",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport random\nimport pandas as pd\nimport matplotlib.pyplot as plt\nfrom scipy.stats import multivariate_normal\n",
"_____no_output_____"
]
],
[
[
"## Generate Dataset",
"_____no_output_____"
]
],
[
[
"modelParameters = {'mu':[[-2,1], [0.5, -1], [0,1]],\n 'pi':[0.2, 0.35, 0.45],\n 'sigma':0.4,\n 'n':200} \n\n#Check that pi sums to 1\n\nif np.sum(modelParameters['pi']) != 1:\n print('Mixture weights must sum to 1!')\n \ndata = []\n\n#determine which mixture each point belongs to\n\ndef generateLabels(n, pi):\n #Generate n realisations of a categorical distribution given the parameters pi\n unif = np.random.uniform(size = n) #Generate uniform random variables\n labels = [(u < np.cumsum(pi)).argmax() for u in unif] #assign cluster\n return labels\n\n#Given the labels, generate from the corresponding normal distribution\n\ndef generateMixture(labels, params):\n \n normalSamples = []\n for label in labels:\n \n #Select Parameters\n mu = params['mu'][label]\n Sigma = np.diag([params['sigma']**2]*len(mu))\n \n \n #sample from multivariate normal\n samp = np.random.multivariate_normal(mean = mu, cov = Sigma, size = 1)\n normalSamples.append(samp)\n \n normalSamples = np.reshape(normalSamples, (len(labels), len(params['mu'][0])))\n \n return normalSamples\n ",
"_____no_output_____"
],
[
"labels = generateLabels(100, modelParameters['pi']) #labels - (in practice we don't actually know what these are!)\nX = generateMixture(labels, modelParameters) #features - (we do know what these are)",
"_____no_output_____"
]
],
[
[
"# Quickly plot the data so we know what it looks like",
"_____no_output_____"
]
],
[
[
"plt.figure(figsize=(10,6))\nplt.scatter(X[:,0], X[:,1],c = labels)\nplt.show()",
"_____no_output_____"
]
],
[
[
"When doing K-means clustering, our goal is to sort the data into 3 clusters using the data $X$. When we're doing clustering we don't have access to the colour (label) of each point, so the data we're actually given would look like this:",
"_____no_output_____"
]
],
[
[
"plt.figure(figsize=(10,6))\nplt.scatter(X[:,0], X[:,1])\nplt.title('Example data - no labels')\nplt.show()",
"_____no_output_____"
]
],
[
[
"If we inspect the data we can still see that the data are roughly made up by 3 groups, one in the top left corner, one in the top right corner and one in the bottom right corner",
"_____no_output_____"
],
[
"## How does K-means work?\n\nThe K in K-means represents the number of clusters, K, that we will sort the data into.\n\nLet's imagine we had already sorted the data into K clusters (like in the first plot above) and were trying to decide what the label of a new point should be. It would make sense to assign it to the cluster which it is closest to.\n\nBut how do we define 'closest to'? One way would be to give it the same label as the point that is closest to it (a 'nearest neighbour' approach), but a more robust way would be to determine where the 'middle' of each cluster was and assign the new point to the cluster with the closest middle. We call this 'middle' the Cluster Centroid and we calculate it be taking the average of all the points in the cluster.",
"_____no_output_____"
],
[
"That's all very well and good if we already have the clusters in place, but the whole point of the algorithm is to find out what the clusters are!\n\nTo find the clusters, we do the following:\n\n1. Randomly initialise K Cluster Centroids\n2. Assign each point to the Cluster Centroid that it is closest to.\n3. Update the Cluster Centroids as the average of all points currently assigned to that centroid\n4. Repeat steps 2-3 until convergence\n\n### Why does K-means work?\n\nOur aim is to find K Cluster Centroids such that the overall distance between each datapoint and its Cluster Centroid is minimised. That is, we want to choose cluster centroids $C = \\{C_1,...,C_K\\}$ such that the error function:\n\n$$E(C) = \\sum_{i=1}^n ||x_i-C_{x_i}||^2$$\n\nis minimised, where $C_{x_i}$ is the Cluster Centroid associated with the ith observation and $||x_i-C_{x_i}||$ is the Euclidean distance between the ith observation and associated Cluster Centroid. \n\n\n\n\n\nNow assume after $m$ iterations of the algorithm, the current value of $E(C)$ was $\\alpha$. By carrying out step 2, we make sure that each point is assigned to the nearest cluster centroid - by doing this, either $\\alpha$ stays the same (every point was already assigned to the closest centroid) or $\\alpha$ gets smaller (one or more points is moved to a nearer centroid and hence the total distance is reduced). Similarly with step 3, by changing the centroid to be the average of all points in the cluster, we minimise the total distance associated with that cluster, meaning $\\alpha$ can either stay the same or go down.\n\nIn this way we see that as we run the algorithm $E(C)$ is non-increasing, so by continuing to run the algorithm our results can't get worse - hopefully if we run it for long enough then the results will be sensible!",
"_____no_output_____"
]
],
[
[
"class KMeans:\n \n def __init__(self, data, K):\n \n self.data = data #dataset with no labels\n self.K = K #Number of clusters to sort the data into\n \n #Randomly initialise Centroids\n \n self.Centroids = np.random.normal(0,1,(self.K, self.data.shape[1])) #If the data has p features then should be a K x p array\n \n def closestCentroid(self, x):\n #Takes a single example and returns the index of the closest centroid\n #Recall centroids are saved as self.Centroids\n \n pass\n \n def assignToCentroid(self):\n #Want to assign each observation to a centroid by passing each observation to the function closestCentroid\n \n pass\n \n\n def updateCentroids(self):\n #Now based on the current cluster assignments (stored in self.assignments) update the Centroids\n \n pass\n \n def runKMeans(self, tolerance = 0.00001):\n #When the improvement between two successive evaluations of our error function is less than tolerance, we stop\n \n change = 1000 #Initialise change to be a big number\n numIterations = 0\n \n self.CentroidStore = [np.copy(self.Centroids)] #We want to be able to keep track of how the centroids evolved over time\n \n #while change > tolerance:\n #Code goes here...\n \n print(f'K-means Algorithm converged in {numIterations} steps')",
"_____no_output_____"
],
[
"myKM = KMeans(X,3)",
"_____no_output_____"
],
[
"myKM.runKMeans()",
"K-means Algorithm converged in 4 steps\n"
]
],
[
[
"## Let's plot the results",
"_____no_output_____"
]
],
[
[
"c = [0,1,2]*len(myKM.CentroidStore)\nplt.figure(figsize=(10,6))\nplt.scatter(np.array(myKM.CentroidStore).reshape(-1,2)[:,0], np.array(myKM.CentroidStore).reshape(-1,2)[:,1],c=np.array(c), s = 200, marker = '*')\nplt.scatter(X[:,0], X[:,1], s = 12)\nplt.title('Example data from a mixture of Gaussians - Cluster Centroid traces')\nplt.show()",
"_____no_output_____"
]
],
[
[
"The stars of each colour above represents the trajectory of each cluster centroid as the algorithm progressed. Starting from a random initialisation, the centroids raplidly converged to a separate cluster, which is encouraging.\n\nNow let's plot the data with the associated labels that we've assigned to them.",
"_____no_output_____"
]
],
[
[
"plt.figure(figsize=(10,6))\nplt.scatter(X[:,0], X[:,1], s = 20, c = myKM.assignments)\nplt.scatter(np.array(myKM.Centroids).reshape(-1,2)[:,0], np.array(myKM.Centroids).reshape(-1,2)[:,1], s = 200, marker = '*', c = 'red')\nplt.title('Example data from a mixture of Gaussians - Including Cluster Centroids')\nplt.show()",
"_____no_output_____"
]
],
[
[
"The plot above shows the final clusters (with red Cluster Centroids) assigned by the model, which should be pretty close to the 'true' clusters at the top of the page. Note: It's possible that although the clusters are the same the labels might be different - remember that K-means isn't supposed to identify the correct label, it's supposed to group the data in clusters which in reality share the same labels.\n\n",
"_____no_output_____"
],
[
"The data we've worked with in this notebook had an underlying structure that made it easy for K-means to identify distinct clusters. However let's look at an example where K-means doesn't perform so well\n\n\n## The sting in the tail - A more complex data structure",
"_____no_output_____"
]
],
[
[
"theta = np.linspace(0, 2*np.pi, 100)\n\nr = 15\n\nx1 = r*np.cos(theta)\nx2 = r*np.sin(theta)\n\n#Perturb the values in the circle\nx1 = x1 + np.random.normal(0,2,x1.shape[0])\nx2 = x2 + np.random.normal(0,2,x2.shape[0])\nz1 = np.random.normal(0,3,x1.shape[0])\nz2 = np.random.normal(0,3,x2.shape[0])\nx1 = np.array([x1,z1]).reshape(-1)\nx2 = np.array([x2,z2]).reshape(-1)\n\nplt.scatter(x1,x2)\nplt.show()",
"_____no_output_____"
]
],
[
[
"It might be the case that the underlying generative structure that we want to capture is that the 'outer ring' in the plot corresponds to a certain kind of process and the 'inner circle' corresponds to another.",
"_____no_output_____"
]
],
[
[
"#Get data in the format we want\nnewX = []\nfor i in range(x1.shape[0]):\n newX.append([x1[i], x2[i]])\n \nnewX = np.array(newX)",
"_____no_output_____"
],
[
"#Run KMeans\nmyNewKM = KMeans(newX,2)\nmyNewKM.runKMeans()",
"K-means Algorithm converged in 36 steps\n"
],
[
"plt.figure(figsize=(10,6))\nplt.scatter(newX[:,0], newX[:,1], s = 20, c = np.array(myNewKM.assignments))\nplt.scatter(np.array(myNewKM.Centroids).reshape(-1,2)[:,0], np.array(myNewKM.Centroids).reshape(-1,2)[:,1], s = 200, marker = '*', c = 'red')\nplt.title('Assigned K-Means labels for Ring data ')\nplt.show()",
"_____no_output_____"
]
],
[
[
"The above plot indicates that K-means isn't able to identify the ring-like structure that we mentioned above. The clustering it has performed is perfectly valid - remember in K-means' world, labels don't exist and this is a legitmate clustering of the data! However if we were to use this clustering our subsequent analyses might be negatively impacted. ",
"_____no_output_____"
],
[
"In a future post we'll implement a method which is capable of capturing non-linear relationships more effectively (the Gaussian Mixture Model).",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
]
] |
cbedc0cc4e42b88d1c9f1a952ca3e77efb9e9293
| 1,025,505 |
ipynb
|
Jupyter Notebook
|
intro-to-pytorch/Part 8 - Transfer Learning (Exercises).ipynb
|
gabriel301/deep-learning-v2-pytorch
|
a89f5823b572f2bd38da216d205997551f2c797f
|
[
"MIT"
] | null | null | null |
intro-to-pytorch/Part 8 - Transfer Learning (Exercises).ipynb
|
gabriel301/deep-learning-v2-pytorch
|
a89f5823b572f2bd38da216d205997551f2c797f
|
[
"MIT"
] | null | null | null |
intro-to-pytorch/Part 8 - Transfer Learning (Exercises).ipynb
|
gabriel301/deep-learning-v2-pytorch
|
a89f5823b572f2bd38da216d205997551f2c797f
|
[
"MIT"
] | null | null | null | 466.775148 | 138,084 | 0.909488 |
[
[
[
"# Transfer Learning\n\nIn this notebook, you'll learn how to use pre-trained networks to solved challenging problems in computer vision. Specifically, you'll use networks trained on [ImageNet](http://www.image-net.org/) [available from torchvision](http://pytorch.org/docs/0.3.0/torchvision/models.html). \n\nImageNet is a massive dataset with over 1 million labeled images in 1000 categories. It's used to train deep neural networks using an architecture called convolutional layers. I'm not going to get into the details of convolutional networks here, but if you want to learn more about them, please [watch this](https://www.youtube.com/watch?v=2-Ol7ZB0MmU).\n\nOnce trained, these models work astonishingly well as feature detectors for images they weren't trained on. Using a pre-trained network on images not in the training set is called transfer learning. Here we'll use transfer learning to train a network that can classify our cat and dog photos with near perfect accuracy.\n\nWith `torchvision.models` you can download these pre-trained networks and use them in your applications. We'll include `models` in our imports now.",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\n%config InlineBackend.figure_format = 'retina'\n\nimport matplotlib.pyplot as plt\n\nimport torch\nfrom torch import nn\nfrom torch import optim\nimport torch.nn.functional as F\nfrom torchvision import datasets, transforms, models",
"_____no_output_____"
]
],
[
[
"Most of the pretrained models require the input to be 224x224 images. Also, we'll need to match the normalization used when the models were trained. Each color channel was normalized separately, the means are `[0.485, 0.456, 0.406]` and the standard deviations are `[0.229, 0.224, 0.225]`.",
"_____no_output_____"
]
],
[
[
"data_dir = './Cat_Dog_data'\n\n# TODO: Define transforms for the training data and testing data\ntrain_transforms =\n\ntest_transforms =\n\n# Pass transforms in here, then run the next cell to see how the transforms look\ntrain_data = datasets.ImageFolder(data_dir + '/train', transform=train_transforms)\ntest_data = datasets.ImageFolder(data_dir + '/test', transform=test_transforms)\n\ntrainloader = torch.utils.data.DataLoader(train_data, batch_size=64, shuffle=True)\ntestloader = torch.utils.data.DataLoader(test_data, batch_size=64)",
"_____no_output_____"
]
],
[
[
"We can load in a model such as [DenseNet](http://pytorch.org/docs/0.3.0/torchvision/models.html#id5). Let's print out the model architecture so we can see what's going on.",
"_____no_output_____"
]
],
[
[
"model = models.densenet121(pretrained=True)\nmodel",
"/home/gabriel/anaconda3/envs/pytorchChallenge/lib/python3.6/site-packages/torchvision-0.2.1-py3.6.egg/torchvision/models/densenet.py:212: UserWarning: nn.init.kaiming_normal is now deprecated in favor of nn.init.kaiming_normal_.\n"
]
],
[
[
"This model is built out of two main parts, the features and the classifier. The features part is a stack of convolutional layers and overall works as a feature detector that can be fed into a classifier. The classifier part is a single fully-connected layer `(classifier): Linear(in_features=1024, out_features=1000)`. This layer was trained on the ImageNet dataset, so it won't work for our specific problem. That means we need to replace the classifier, but the features will work perfectly on their own. In general, I think about pre-trained networks as amazingly good feature detectors that can be used as the input for simple feed-forward classifiers.",
"_____no_output_____"
]
],
[
[
"# Freeze parameters so we don't backprop through them\nfor param in model.parameters():\n param.requires_grad = False\n\nfrom collections import OrderedDict\nclassifier = nn.Sequential(OrderedDict([\n ('fc1', nn.Linear(1024, 500)),\n ('relu', nn.ReLU()),\n ('fc2', nn.Linear(500, 2)),\n ('output', nn.LogSoftmax(dim=1))\n ]))\n \nmodel.classifier = classifier",
"_____no_output_____"
]
],
[
[
"With our model built, we need to train the classifier. However, now we're using a **really deep** neural network. If you try to train this on a CPU like normal, it will take a long, long time. Instead, we're going to use the GPU to do the calculations. The linear algebra computations are done in parallel on the GPU leading to 100x increased training speeds. It's also possible to train on multiple GPUs, further decreasing training time.\n\nPyTorch, along with pretty much every other deep learning framework, uses [CUDA](https://developer.nvidia.com/cuda-zone) to efficiently compute the forward and backwards passes on the GPU. In PyTorch, you move your model parameters and other tensors to the GPU memory using `model.to('cuda')`. You can move them back from the GPU with `model.to('cpu')` which you'll commonly do when you need to operate on the network output outside of PyTorch. As a demonstration of the increased speed, I'll compare how long it takes to perform a forward and backward pass with and without a GPU.",
"_____no_output_____"
]
],
[
[
"import time",
"_____no_output_____"
],
[
"for device in ['cpu', 'cuda']:\n\n criterion = nn.NLLLoss()\n # Only train the classifier parameters, feature parameters are frozen\n optimizer = optim.Adam(model.classifier.parameters(), lr=0.001)\n\n model.to(device)\n\n for ii, (inputs, labels) in enumerate(trainloader):\n\n # Move input and label tensors to the GPU\n inputs, labels = inputs.to(device), labels.to(device)\n\n start = time.time()\n\n outputs = model.forward(inputs)\n loss = criterion(outputs, labels)\n loss.backward()\n optimizer.step()\n\n if ii==3:\n break\n \n print(f\"Device = {device}; Time per batch: {(time.time() - start)/3:.3f} seconds\")",
"Device = cpu; Time per batch: 5.765 seconds\nDevice = cuda; Time per batch: 0.012 seconds\n"
]
],
[
[
"You can write device agnostic code which will automatically use CUDA if it's enabled like so:\n```python\n# at beginning of the script\ndevice = torch.device(\"cuda:0\" if torch.cuda.is_available() else \"cpu\")\n\n...\n\n# then whenever you get a new Tensor or Module\n# this won't copy if they are already on the desired device\ninput = data.to(device)\nmodel = MyModule(...).to(device)\n```\n\nFrom here, I'll let you finish training the model. The process is the same as before except now your model is much more powerful. You should get better than 95% accuracy easily.\n\n>**Exercise:** Train a pretrained models to classify the cat and dog images. Continue with the DenseNet model, or try ResNet, it's also a good model to try out first. Make sure you are only training the classifier and the parameters for the features part are frozen.",
"_____no_output_____"
]
],
[
[
"class MyClassifier(nn.Module):\n def __init__(self):\n super().__init__()\n self.fc1 = nn.Linear(1024,512)\n self.fc2 = nn.Linear(512,256)\n self.output = nn.Linear(256,2)\n self.dropout = nn.Dropout(0.2)\n #self.fc1 = nn.Linear(32,32)\n \n def forward(self,x):\n x = self.dropout(F.relu(self.fc1(x)))\n x = self.dropout(F.relu(self.fc2(x)))\n #x = self.dropout(F.relu(self.fc3(x)))\n #x = self.dropout(F.relu(self.fc4(x)))\n x = F.log_softmax(self.output(x),dim=1)\n return x",
"_____no_output_____"
],
[
"## TODO: Use a pretrained model to classify the cat and dog images\ndevice = torch.device(\"cuda:0\" if torch.cuda.is_available() else \"cpu\")",
"_____no_output_____"
],
[
"# Only train the classifier parameters, feature parameters are frozen\n# Freeze parameters so we don't backprop through them\nfor param in model.parameters():\n param.requires_grad = False\n\n\n\nmodel.classifier = MyClassifier()\nmodel.to(device)\n\ncriterion = nn.NLLLoss()\noptimizer = optim.Adam(model.classifier.parameters(), lr=0.001)\n\nepochs = 1\ntrain_losses,test_losses = [],[]\nsteps = 0\nprint_every = 5\n\nstart = time.time()\nfor epoch in range(epochs):\n currentLoss = 0\n steps = 0\n for inputs, labels in trainloader:\n steps += 1\n # Move input and label tensors to the GPU\n inputs, labels = inputs.to(device), labels.to(device)\n \n optimizer.zero_grad()\n outputs = model.forward(inputs)\n loss = criterion(outputs, labels)\n loss.backward()\n optimizer.step()\n currentLoss += loss.item()\n \n if steps % print_every == 0:\n ## TODO: Implement the validation pass and print out the validation accuracy\n test_loss = 0\n accuracy = 0\n\n # Turn off gradients for validation, saves memory and computations\n with torch.no_grad():\n model.eval()\n for images, labels in testloader:\n images,labels = images.to(device),labels.to(device)\n log_ps = model(images)\n test_loss += criterion(log_ps, labels)\n\n ps = torch.exp(log_ps)\n top_p, top_class = ps.topk(1, dim=1)\n equals = top_class == labels.view(*top_class.shape)\n accuracy += torch.mean(equals.type(torch.FloatTensor))\n model.train()\n train_losses.append(currentLoss/len(trainloader))\n test_losses.append(test_loss/len(testloader))\n\n print(\"Epoch: {}/{}.. \".format(epoch+1, epochs),\n \"Step: {}/{}.. \".format(steps, len(trainloader)),\n \"Training Loss: {:.3f}.. \".format(currentLoss/len(trainloader)),\n \"Test Loss: {:.3f}.. \".format(test_loss/len(testloader)),\n \"Test Accuracy: {:.3f}\".format(accuracy/len(testloader))) \nend = time.time()\nprint(f\"Device = {device}; Trainning Time: {(end-start)/60:.3f} minutes\")",
"Epoch: 1/1.. Step: 5/352.. Training Loss: 0.008.. Test Loss: 0.301.. Test Accuracy: 0.884\nEpoch: 1/1.. Step: 10/352.. Training Loss: 0.014.. Test Loss: 0.155.. Test Accuracy: 0.950\nEpoch: 1/1.. Step: 15/352.. Training Loss: 0.021.. Test Loss: 0.142.. Test Accuracy: 0.959\nEpoch: 1/1.. Step: 20/352.. Training Loss: 0.027.. Test Loss: 0.148.. Test Accuracy: 0.961\nEpoch: 1/1.. Step: 25/352.. Training Loss: 0.032.. Test Loss: 0.215.. Test Accuracy: 0.920\nEpoch: 1/1.. Step: 30/352.. Training Loss: 0.036.. Test Loss: 0.118.. Test Accuracy: 0.974\nEpoch: 1/1.. Step: 35/352.. Training Loss: 0.039.. Test Loss: 0.084.. Test Accuracy: 0.974\nEpoch: 1/1.. Step: 40/352.. Training Loss: 0.042.. Test Loss: 0.063.. Test Accuracy: 0.978\nEpoch: 1/1.. Step: 45/352.. Training Loss: 0.046.. Test Loss: 0.064.. Test Accuracy: 0.977\nEpoch: 1/1.. Step: 50/352.. Training Loss: 0.047.. Test Loss: 0.062.. Test Accuracy: 0.978\nEpoch: 1/1.. Step: 55/352.. Training Loss: 0.050.. Test Loss: 0.050.. Test Accuracy: 0.980\nEpoch: 1/1.. Step: 60/352.. Training Loss: 0.053.. Test Loss: 0.052.. Test Accuracy: 0.980\nEpoch: 1/1.. Step: 65/352.. Training Loss: 0.056.. Test Loss: 0.067.. Test Accuracy: 0.974\nEpoch: 1/1.. Step: 70/352.. Training Loss: 0.058.. Test Loss: 0.067.. Test Accuracy: 0.976\nEpoch: 1/1.. Step: 75/352.. Training Loss: 0.060.. Test Loss: 0.151.. Test Accuracy: 0.939\nEpoch: 1/1.. Step: 80/352.. Training Loss: 0.066.. Test Loss: 0.054.. Test Accuracy: 0.982\nEpoch: 1/1.. Step: 85/352.. Training Loss: 0.070.. Test Loss: 0.101.. Test Accuracy: 0.973\nEpoch: 1/1.. Step: 90/352.. Training Loss: 0.073.. Test Loss: 0.102.. Test Accuracy: 0.965\nEpoch: 1/1.. Step: 95/352.. Training Loss: 0.076.. Test Loss: 0.079.. Test Accuracy: 0.975\nEpoch: 1/1.. Step: 100/352.. Training Loss: 0.078.. Test Loss: 0.053.. Test Accuracy: 0.982\nEpoch: 1/1.. Step: 105/352.. Training Loss: 0.080.. Test Loss: 0.047.. Test Accuracy: 0.982\nEpoch: 1/1.. Step: 110/352.. Training Loss: 0.082.. Test Loss: 0.046.. Test Accuracy: 0.982\nEpoch: 1/1.. Step: 115/352.. Training Loss: 0.084.. Test Loss: 0.045.. Test Accuracy: 0.984\nEpoch: 1/1.. Step: 120/352.. Training Loss: 0.087.. Test Loss: 0.052.. Test Accuracy: 0.979\nEpoch: 1/1.. Step: 125/352.. Training Loss: 0.090.. Test Loss: 0.046.. Test Accuracy: 0.983\nEpoch: 1/1.. Step: 130/352.. Training Loss: 0.091.. Test Loss: 0.061.. Test Accuracy: 0.979\nEpoch: 1/1.. Step: 135/352.. Training Loss: 0.094.. Test Loss: 0.057.. Test Accuracy: 0.978\nEpoch: 1/1.. Step: 140/352.. Training Loss: 0.096.. Test Loss: 0.046.. Test Accuracy: 0.984\nEpoch: 1/1.. Step: 145/352.. Training Loss: 0.099.. Test Loss: 0.044.. Test Accuracy: 0.984\nEpoch: 1/1.. Step: 150/352.. Training Loss: 0.100.. Test Loss: 0.042.. Test Accuracy: 0.983\nEpoch: 1/1.. Step: 155/352.. Training Loss: 0.103.. Test Loss: 0.073.. Test Accuracy: 0.975\nEpoch: 1/1.. Step: 160/352.. Training Loss: 0.105.. Test Loss: 0.060.. Test Accuracy: 0.980\nEpoch: 1/1.. Step: 165/352.. Training Loss: 0.108.. Test Loss: 0.044.. Test Accuracy: 0.983\nEpoch: 1/1.. Step: 170/352.. Training Loss: 0.111.. Test Loss: 0.088.. Test Accuracy: 0.966\nEpoch: 1/1.. Step: 175/352.. Training Loss: 0.113.. Test Loss: 0.055.. Test Accuracy: 0.982\nEpoch: 1/1.. Step: 180/352.. Training Loss: 0.116.. Test Loss: 0.069.. Test Accuracy: 0.975\nEpoch: 1/1.. Step: 185/352.. Training Loss: 0.118.. Test Loss: 0.061.. Test Accuracy: 0.979\nEpoch: 1/1.. Step: 190/352.. Training Loss: 0.121.. Test Loss: 0.048.. Test Accuracy: 0.983\nEpoch: 1/1.. Step: 195/352.. Training Loss: 0.123.. Test Loss: 0.045.. Test Accuracy: 0.984\nEpoch: 1/1.. Step: 200/352.. Training Loss: 0.126.. Test Loss: 0.069.. Test Accuracy: 0.976\nEpoch: 1/1.. Step: 205/352.. Training Loss: 0.128.. Test Loss: 0.043.. Test Accuracy: 0.984\nEpoch: 1/1.. Step: 210/352.. Training Loss: 0.131.. Test Loss: 0.043.. Test Accuracy: 0.984\nEpoch: 1/1.. Step: 215/352.. Training Loss: 0.133.. Test Loss: 0.040.. Test Accuracy: 0.984\nEpoch: 1/1.. Step: 220/352.. Training Loss: 0.135.. Test Loss: 0.047.. Test Accuracy: 0.983\nEpoch: 1/1.. Step: 225/352.. Training Loss: 0.138.. Test Loss: 0.040.. Test Accuracy: 0.983\nEpoch: 1/1.. Step: 230/352.. Training Loss: 0.140.. Test Loss: 0.041.. Test Accuracy: 0.983\nEpoch: 1/1.. Step: 235/352.. Training Loss: 0.142.. Test Loss: 0.039.. Test Accuracy: 0.983\nEpoch: 1/1.. Step: 240/352.. Training Loss: 0.144.. Test Loss: 0.041.. Test Accuracy: 0.984\nEpoch: 1/1.. Step: 245/352.. Training Loss: 0.145.. Test Loss: 0.041.. Test Accuracy: 0.984\nEpoch: 1/1.. Step: 250/352.. Training Loss: 0.149.. Test Loss: 0.060.. Test Accuracy: 0.979\nEpoch: 1/1.. Step: 255/352.. Training Loss: 0.152.. Test Loss: 0.062.. Test Accuracy: 0.978\nEpoch: 1/1.. Step: 260/352.. Training Loss: 0.155.. Test Loss: 0.054.. Test Accuracy: 0.984\nEpoch: 1/1.. Step: 265/352.. Training Loss: 0.157.. Test Loss: 0.068.. Test Accuracy: 0.975\nEpoch: 1/1.. Step: 270/352.. Training Loss: 0.159.. Test Loss: 0.042.. Test Accuracy: 0.984\nEpoch: 1/1.. Step: 275/352.. Training Loss: 0.161.. Test Loss: 0.051.. Test Accuracy: 0.981\nEpoch: 1/1.. Step: 280/352.. Training Loss: 0.163.. Test Loss: 0.041.. Test Accuracy: 0.984\nEpoch: 1/1.. Step: 285/352.. Training Loss: 0.165.. Test Loss: 0.040.. Test Accuracy: 0.984\nEpoch: 1/1.. Step: 290/352.. Training Loss: 0.168.. Test Loss: 0.039.. Test Accuracy: 0.984\nEpoch: 1/1.. Step: 295/352.. Training Loss: 0.171.. Test Loss: 0.043.. Test Accuracy: 0.985\nEpoch: 1/1.. Step: 300/352.. Training Loss: 0.173.. Test Loss: 0.047.. Test Accuracy: 0.984\nEpoch: 1/1.. Step: 305/352.. Training Loss: 0.175.. Test Loss: 0.045.. Test Accuracy: 0.985\nEpoch: 1/1.. Step: 310/352.. Training Loss: 0.177.. Test Loss: 0.042.. Test Accuracy: 0.984\nEpoch: 1/1.. Step: 315/352.. Training Loss: 0.179.. Test Loss: 0.038.. Test Accuracy: 0.985\nEpoch: 1/1.. Step: 320/352.. Training Loss: 0.180.. Test Loss: 0.038.. Test Accuracy: 0.985\nEpoch: 1/1.. Step: 325/352.. Training Loss: 0.183.. Test Loss: 0.049.. Test Accuracy: 0.981\nEpoch: 1/1.. Step: 330/352.. Training Loss: 0.186.. Test Loss: 0.037.. Test Accuracy: 0.984\nEpoch: 1/1.. Step: 335/352.. Training Loss: 0.187.. Test Loss: 0.044.. Test Accuracy: 0.984\nEpoch: 1/1.. Step: 340/352.. Training Loss: 0.190.. Test Loss: 0.038.. Test Accuracy: 0.986\nEpoch: 1/1.. Step: 345/352.. Training Loss: 0.192.. Test Loss: 0.040.. Test Accuracy: 0.986\nEpoch: 1/1.. Step: 350/352.. Training Loss: 0.195.. Test Loss: 0.037.. Test Accuracy: 0.985\nDevice = cuda:0; Trainning Time: 40.329 minutes\n"
],
[
"plt.plot(train_losses, label='Training loss')\nplt.plot(test_losses, label='Validation loss')\nplt.legend(frameon=False)",
"_____no_output_____"
],
[
"model",
"_____no_output_____"
],
[
"#Save the classifier\n\ntorch.save(model, 'checkpoint_dog_cat.pth')",
"/home/gabriel/anaconda3/envs/pytorchChallenge/lib/python3.6/site-packages/torch/serialization.py:241: UserWarning: Couldn't retrieve source code for container of type MyClassifier. It won't be checked for correctness upon loading.\n \"type \" + obj.__name__ + \". It won't be checked \"\n"
],
[
"#Load the Classifier\ncls = torch.load('checkpoint_dog_cat.pth')\nprint(cls)",
"DenseNet(\n (features): Sequential(\n (conv0): Conv2d(3, 64, kernel_size=(7, 7), stride=(2, 2), padding=(3, 3), bias=False)\n (norm0): BatchNorm2d(64, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu0): ReLU(inplace)\n (pool0): MaxPool2d(kernel_size=3, stride=2, padding=1, dilation=1, ceil_mode=False)\n (denseblock1): _DenseBlock(\n (denselayer1): _DenseLayer(\n (norm1): BatchNorm2d(64, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu1): ReLU(inplace)\n (conv1): Conv2d(64, 128, kernel_size=(1, 1), stride=(1, 1), bias=False)\n (norm2): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu2): ReLU(inplace)\n (conv2): Conv2d(128, 32, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n )\n (denselayer2): _DenseLayer(\n (norm1): BatchNorm2d(96, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu1): ReLU(inplace)\n (conv1): Conv2d(96, 128, kernel_size=(1, 1), stride=(1, 1), bias=False)\n (norm2): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu2): ReLU(inplace)\n (conv2): Conv2d(128, 32, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n )\n (denselayer3): _DenseLayer(\n (norm1): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu1): ReLU(inplace)\n (conv1): Conv2d(128, 128, kernel_size=(1, 1), stride=(1, 1), bias=False)\n (norm2): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu2): ReLU(inplace)\n (conv2): Conv2d(128, 32, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n )\n (denselayer4): _DenseLayer(\n (norm1): BatchNorm2d(160, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu1): ReLU(inplace)\n (conv1): Conv2d(160, 128, kernel_size=(1, 1), stride=(1, 1), bias=False)\n (norm2): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu2): ReLU(inplace)\n (conv2): Conv2d(128, 32, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n )\n (denselayer5): _DenseLayer(\n (norm1): BatchNorm2d(192, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu1): ReLU(inplace)\n (conv1): Conv2d(192, 128, kernel_size=(1, 1), stride=(1, 1), bias=False)\n (norm2): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu2): ReLU(inplace)\n (conv2): Conv2d(128, 32, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n )\n (denselayer6): _DenseLayer(\n (norm1): BatchNorm2d(224, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu1): ReLU(inplace)\n (conv1): Conv2d(224, 128, kernel_size=(1, 1), stride=(1, 1), bias=False)\n (norm2): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu2): ReLU(inplace)\n (conv2): Conv2d(128, 32, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n )\n )\n (transition1): _Transition(\n (norm): BatchNorm2d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu): ReLU(inplace)\n (conv): Conv2d(256, 128, kernel_size=(1, 1), stride=(1, 1), bias=False)\n (pool): AvgPool2d(kernel_size=2, stride=2, padding=0)\n )\n (denseblock2): _DenseBlock(\n (denselayer1): _DenseLayer(\n (norm1): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu1): ReLU(inplace)\n (conv1): Conv2d(128, 128, kernel_size=(1, 1), stride=(1, 1), bias=False)\n (norm2): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu2): ReLU(inplace)\n (conv2): Conv2d(128, 32, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n )\n (denselayer2): _DenseLayer(\n (norm1): BatchNorm2d(160, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu1): ReLU(inplace)\n (conv1): Conv2d(160, 128, kernel_size=(1, 1), stride=(1, 1), bias=False)\n (norm2): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu2): ReLU(inplace)\n (conv2): Conv2d(128, 32, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n )\n (denselayer3): _DenseLayer(\n (norm1): BatchNorm2d(192, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu1): ReLU(inplace)\n (conv1): Conv2d(192, 128, kernel_size=(1, 1), stride=(1, 1), bias=False)\n (norm2): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu2): ReLU(inplace)\n (conv2): Conv2d(128, 32, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n )\n (denselayer4): _DenseLayer(\n (norm1): BatchNorm2d(224, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu1): ReLU(inplace)\n (conv1): Conv2d(224, 128, kernel_size=(1, 1), stride=(1, 1), bias=False)\n (norm2): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu2): ReLU(inplace)\n (conv2): Conv2d(128, 32, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n )\n (denselayer5): _DenseLayer(\n (norm1): BatchNorm2d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu1): ReLU(inplace)\n (conv1): Conv2d(256, 128, kernel_size=(1, 1), stride=(1, 1), bias=False)\n (norm2): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu2): ReLU(inplace)\n (conv2): Conv2d(128, 32, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n )\n (denselayer6): _DenseLayer(\n (norm1): BatchNorm2d(288, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu1): ReLU(inplace)\n (conv1): Conv2d(288, 128, kernel_size=(1, 1), stride=(1, 1), bias=False)\n (norm2): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu2): ReLU(inplace)\n (conv2): Conv2d(128, 32, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n )\n (denselayer7): _DenseLayer(\n (norm1): BatchNorm2d(320, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu1): ReLU(inplace)\n (conv1): Conv2d(320, 128, kernel_size=(1, 1), stride=(1, 1), bias=False)\n (norm2): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu2): ReLU(inplace)\n (conv2): Conv2d(128, 32, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n )\n (denselayer8): _DenseLayer(\n (norm1): BatchNorm2d(352, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu1): ReLU(inplace)\n (conv1): Conv2d(352, 128, kernel_size=(1, 1), stride=(1, 1), bias=False)\n (norm2): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu2): ReLU(inplace)\n (conv2): Conv2d(128, 32, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n )\n (denselayer9): _DenseLayer(\n (norm1): BatchNorm2d(384, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu1): ReLU(inplace)\n (conv1): Conv2d(384, 128, kernel_size=(1, 1), stride=(1, 1), bias=False)\n (norm2): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu2): ReLU(inplace)\n (conv2): Conv2d(128, 32, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n )\n (denselayer10): _DenseLayer(\n (norm1): BatchNorm2d(416, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu1): ReLU(inplace)\n (conv1): Conv2d(416, 128, kernel_size=(1, 1), stride=(1, 1), bias=False)\n (norm2): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu2): ReLU(inplace)\n (conv2): Conv2d(128, 32, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n )\n (denselayer11): _DenseLayer(\n (norm1): BatchNorm2d(448, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu1): ReLU(inplace)\n (conv1): Conv2d(448, 128, kernel_size=(1, 1), stride=(1, 1), bias=False)\n (norm2): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu2): ReLU(inplace)\n (conv2): Conv2d(128, 32, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n )\n (denselayer12): _DenseLayer(\n (norm1): BatchNorm2d(480, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu1): ReLU(inplace)\n (conv1): Conv2d(480, 128, kernel_size=(1, 1), stride=(1, 1), bias=False)\n (norm2): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu2): ReLU(inplace)\n (conv2): Conv2d(128, 32, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n )\n )\n (transition2): _Transition(\n (norm): BatchNorm2d(512, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu): ReLU(inplace)\n (conv): Conv2d(512, 256, kernel_size=(1, 1), stride=(1, 1), bias=False)\n (pool): AvgPool2d(kernel_size=2, stride=2, padding=0)\n )\n (denseblock3): _DenseBlock(\n (denselayer1): _DenseLayer(\n (norm1): BatchNorm2d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu1): ReLU(inplace)\n (conv1): Conv2d(256, 128, kernel_size=(1, 1), stride=(1, 1), bias=False)\n (norm2): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu2): ReLU(inplace)\n (conv2): Conv2d(128, 32, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n )\n (denselayer2): _DenseLayer(\n (norm1): BatchNorm2d(288, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu1): ReLU(inplace)\n (conv1): Conv2d(288, 128, kernel_size=(1, 1), stride=(1, 1), bias=False)\n (norm2): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu2): ReLU(inplace)\n (conv2): Conv2d(128, 32, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n )\n (denselayer3): _DenseLayer(\n (norm1): BatchNorm2d(320, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu1): ReLU(inplace)\n (conv1): Conv2d(320, 128, kernel_size=(1, 1), stride=(1, 1), bias=False)\n (norm2): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu2): ReLU(inplace)\n (conv2): Conv2d(128, 32, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n )\n (denselayer4): _DenseLayer(\n (norm1): BatchNorm2d(352, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu1): ReLU(inplace)\n (conv1): Conv2d(352, 128, kernel_size=(1, 1), stride=(1, 1), bias=False)\n (norm2): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu2): ReLU(inplace)\n (conv2): Conv2d(128, 32, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n )\n (denselayer5): _DenseLayer(\n (norm1): BatchNorm2d(384, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu1): ReLU(inplace)\n (conv1): Conv2d(384, 128, kernel_size=(1, 1), stride=(1, 1), bias=False)\n (norm2): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu2): ReLU(inplace)\n (conv2): Conv2d(128, 32, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n )\n (denselayer6): _DenseLayer(\n (norm1): BatchNorm2d(416, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu1): ReLU(inplace)\n (conv1): Conv2d(416, 128, kernel_size=(1, 1), stride=(1, 1), bias=False)\n (norm2): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu2): ReLU(inplace)\n (conv2): Conv2d(128, 32, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n )\n (denselayer7): _DenseLayer(\n (norm1): BatchNorm2d(448, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu1): ReLU(inplace)\n (conv1): Conv2d(448, 128, kernel_size=(1, 1), stride=(1, 1), bias=False)\n (norm2): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu2): ReLU(inplace)\n (conv2): Conv2d(128, 32, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n )\n (denselayer8): _DenseLayer(\n (norm1): BatchNorm2d(480, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu1): ReLU(inplace)\n (conv1): Conv2d(480, 128, kernel_size=(1, 1), stride=(1, 1), bias=False)\n (norm2): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu2): ReLU(inplace)\n (conv2): Conv2d(128, 32, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n )\n (denselayer9): _DenseLayer(\n (norm1): BatchNorm2d(512, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu1): ReLU(inplace)\n (conv1): Conv2d(512, 128, kernel_size=(1, 1), stride=(1, 1), bias=False)\n (norm2): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu2): ReLU(inplace)\n (conv2): Conv2d(128, 32, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n )\n (denselayer10): _DenseLayer(\n (norm1): BatchNorm2d(544, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu1): ReLU(inplace)\n (conv1): Conv2d(544, 128, kernel_size=(1, 1), stride=(1, 1), bias=False)\n (norm2): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu2): ReLU(inplace)\n (conv2): Conv2d(128, 32, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n )\n (denselayer11): _DenseLayer(\n (norm1): BatchNorm2d(576, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu1): ReLU(inplace)\n (conv1): Conv2d(576, 128, kernel_size=(1, 1), stride=(1, 1), bias=False)\n (norm2): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu2): ReLU(inplace)\n (conv2): Conv2d(128, 32, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n )\n (denselayer12): _DenseLayer(\n (norm1): BatchNorm2d(608, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu1): ReLU(inplace)\n (conv1): Conv2d(608, 128, kernel_size=(1, 1), stride=(1, 1), bias=False)\n (norm2): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu2): ReLU(inplace)\n (conv2): Conv2d(128, 32, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n )\n (denselayer13): _DenseLayer(\n (norm1): BatchNorm2d(640, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu1): ReLU(inplace)\n (conv1): Conv2d(640, 128, kernel_size=(1, 1), stride=(1, 1), bias=False)\n (norm2): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu2): ReLU(inplace)\n (conv2): Conv2d(128, 32, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n )\n (denselayer14): _DenseLayer(\n (norm1): BatchNorm2d(672, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu1): ReLU(inplace)\n (conv1): Conv2d(672, 128, kernel_size=(1, 1), stride=(1, 1), bias=False)\n (norm2): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu2): ReLU(inplace)\n (conv2): Conv2d(128, 32, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n )\n (denselayer15): _DenseLayer(\n (norm1): BatchNorm2d(704, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu1): ReLU(inplace)\n (conv1): Conv2d(704, 128, kernel_size=(1, 1), stride=(1, 1), bias=False)\n (norm2): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu2): ReLU(inplace)\n (conv2): Conv2d(128, 32, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n )\n (denselayer16): _DenseLayer(\n (norm1): BatchNorm2d(736, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu1): ReLU(inplace)\n (conv1): Conv2d(736, 128, kernel_size=(1, 1), stride=(1, 1), bias=False)\n (norm2): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu2): ReLU(inplace)\n (conv2): Conv2d(128, 32, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n )\n (denselayer17): _DenseLayer(\n (norm1): BatchNorm2d(768, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu1): ReLU(inplace)\n (conv1): Conv2d(768, 128, kernel_size=(1, 1), stride=(1, 1), bias=False)\n (norm2): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu2): ReLU(inplace)\n (conv2): Conv2d(128, 32, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n )\n (denselayer18): _DenseLayer(\n (norm1): BatchNorm2d(800, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu1): ReLU(inplace)\n (conv1): Conv2d(800, 128, kernel_size=(1, 1), stride=(1, 1), bias=False)\n (norm2): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu2): ReLU(inplace)\n (conv2): Conv2d(128, 32, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n )\n (denselayer19): _DenseLayer(\n (norm1): BatchNorm2d(832, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu1): ReLU(inplace)\n (conv1): Conv2d(832, 128, kernel_size=(1, 1), stride=(1, 1), bias=False)\n (norm2): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu2): ReLU(inplace)\n (conv2): Conv2d(128, 32, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n )\n (denselayer20): _DenseLayer(\n (norm1): BatchNorm2d(864, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu1): ReLU(inplace)\n (conv1): Conv2d(864, 128, kernel_size=(1, 1), stride=(1, 1), bias=False)\n (norm2): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu2): ReLU(inplace)\n (conv2): Conv2d(128, 32, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n )\n (denselayer21): _DenseLayer(\n (norm1): BatchNorm2d(896, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu1): ReLU(inplace)\n (conv1): Conv2d(896, 128, kernel_size=(1, 1), stride=(1, 1), bias=False)\n (norm2): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu2): ReLU(inplace)\n (conv2): Conv2d(128, 32, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n )\n (denselayer22): _DenseLayer(\n (norm1): BatchNorm2d(928, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu1): ReLU(inplace)\n (conv1): Conv2d(928, 128, kernel_size=(1, 1), stride=(1, 1), bias=False)\n (norm2): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu2): ReLU(inplace)\n (conv2): Conv2d(128, 32, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n )\n (denselayer23): _DenseLayer(\n (norm1): BatchNorm2d(960, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu1): ReLU(inplace)\n (conv1): Conv2d(960, 128, kernel_size=(1, 1), stride=(1, 1), bias=False)\n (norm2): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu2): ReLU(inplace)\n (conv2): Conv2d(128, 32, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n )\n (denselayer24): _DenseLayer(\n (norm1): BatchNorm2d(992, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu1): ReLU(inplace)\n (conv1): Conv2d(992, 128, kernel_size=(1, 1), stride=(1, 1), bias=False)\n (norm2): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu2): ReLU(inplace)\n (conv2): Conv2d(128, 32, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n )\n )\n (transition3): _Transition(\n (norm): BatchNorm2d(1024, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu): ReLU(inplace)\n (conv): Conv2d(1024, 512, kernel_size=(1, 1), stride=(1, 1), bias=False)\n (pool): AvgPool2d(kernel_size=2, stride=2, padding=0)\n )\n (denseblock4): _DenseBlock(\n (denselayer1): _DenseLayer(\n (norm1): BatchNorm2d(512, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu1): ReLU(inplace)\n (conv1): Conv2d(512, 128, kernel_size=(1, 1), stride=(1, 1), bias=False)\n (norm2): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu2): ReLU(inplace)\n (conv2): Conv2d(128, 32, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n )\n (denselayer2): _DenseLayer(\n (norm1): BatchNorm2d(544, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu1): ReLU(inplace)\n (conv1): Conv2d(544, 128, kernel_size=(1, 1), stride=(1, 1), bias=False)\n (norm2): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu2): ReLU(inplace)\n (conv2): Conv2d(128, 32, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n )\n (denselayer3): _DenseLayer(\n (norm1): BatchNorm2d(576, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu1): ReLU(inplace)\n (conv1): Conv2d(576, 128, kernel_size=(1, 1), stride=(1, 1), bias=False)\n (norm2): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu2): ReLU(inplace)\n (conv2): Conv2d(128, 32, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n )\n (denselayer4): _DenseLayer(\n (norm1): BatchNorm2d(608, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu1): ReLU(inplace)\n (conv1): Conv2d(608, 128, kernel_size=(1, 1), stride=(1, 1), bias=False)\n (norm2): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu2): ReLU(inplace)\n (conv2): Conv2d(128, 32, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n )\n (denselayer5): _DenseLayer(\n (norm1): BatchNorm2d(640, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu1): ReLU(inplace)\n (conv1): Conv2d(640, 128, kernel_size=(1, 1), stride=(1, 1), bias=False)\n (norm2): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu2): ReLU(inplace)\n (conv2): Conv2d(128, 32, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n )\n (denselayer6): _DenseLayer(\n (norm1): BatchNorm2d(672, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu1): ReLU(inplace)\n (conv1): Conv2d(672, 128, kernel_size=(1, 1), stride=(1, 1), bias=False)\n (norm2): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu2): ReLU(inplace)\n (conv2): Conv2d(128, 32, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n )\n (denselayer7): _DenseLayer(\n (norm1): BatchNorm2d(704, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu1): ReLU(inplace)\n (conv1): Conv2d(704, 128, kernel_size=(1, 1), stride=(1, 1), bias=False)\n (norm2): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu2): ReLU(inplace)\n (conv2): Conv2d(128, 32, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n )\n (denselayer8): _DenseLayer(\n (norm1): BatchNorm2d(736, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu1): ReLU(inplace)\n (conv1): Conv2d(736, 128, kernel_size=(1, 1), stride=(1, 1), bias=False)\n (norm2): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu2): ReLU(inplace)\n (conv2): Conv2d(128, 32, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n )\n (denselayer9): _DenseLayer(\n (norm1): BatchNorm2d(768, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu1): ReLU(inplace)\n (conv1): Conv2d(768, 128, kernel_size=(1, 1), stride=(1, 1), bias=False)\n (norm2): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu2): ReLU(inplace)\n (conv2): Conv2d(128, 32, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n )\n (denselayer10): _DenseLayer(\n (norm1): BatchNorm2d(800, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu1): ReLU(inplace)\n (conv1): Conv2d(800, 128, kernel_size=(1, 1), stride=(1, 1), bias=False)\n (norm2): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu2): ReLU(inplace)\n (conv2): Conv2d(128, 32, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n )\n (denselayer11): _DenseLayer(\n (norm1): BatchNorm2d(832, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu1): ReLU(inplace)\n (conv1): Conv2d(832, 128, kernel_size=(1, 1), stride=(1, 1), bias=False)\n (norm2): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu2): ReLU(inplace)\n (conv2): Conv2d(128, 32, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n )\n (denselayer12): _DenseLayer(\n (norm1): BatchNorm2d(864, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu1): ReLU(inplace)\n (conv1): Conv2d(864, 128, kernel_size=(1, 1), stride=(1, 1), bias=False)\n (norm2): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu2): ReLU(inplace)\n (conv2): Conv2d(128, 32, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n )\n (denselayer13): _DenseLayer(\n (norm1): BatchNorm2d(896, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu1): ReLU(inplace)\n (conv1): Conv2d(896, 128, kernel_size=(1, 1), stride=(1, 1), bias=False)\n (norm2): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu2): ReLU(inplace)\n (conv2): Conv2d(128, 32, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n )\n (denselayer14): _DenseLayer(\n (norm1): BatchNorm2d(928, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu1): ReLU(inplace)\n (conv1): Conv2d(928, 128, kernel_size=(1, 1), stride=(1, 1), bias=False)\n (norm2): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu2): ReLU(inplace)\n (conv2): Conv2d(128, 32, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n )\n (denselayer15): _DenseLayer(\n (norm1): BatchNorm2d(960, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu1): ReLU(inplace)\n (conv1): Conv2d(960, 128, kernel_size=(1, 1), stride=(1, 1), bias=False)\n (norm2): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu2): ReLU(inplace)\n (conv2): Conv2d(128, 32, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n )\n (denselayer16): _DenseLayer(\n (norm1): BatchNorm2d(992, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu1): ReLU(inplace)\n (conv1): Conv2d(992, 128, kernel_size=(1, 1), stride=(1, 1), bias=False)\n (norm2): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (relu2): ReLU(inplace)\n (conv2): Conv2d(128, 32, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n )\n )\n (norm5): BatchNorm2d(1024, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n )\n (classifier): MyClassifier(\n (fc1): Linear(in_features=1024, out_features=512, bias=True)\n (fc2): Linear(in_features=512, out_features=256, bias=True)\n (output): Linear(in_features=256, out_features=2, bias=True)\n (dropout): Dropout(p=0.2)\n )\n)\n"
],
[
"# Import helper module (should be in the repo)\nimport helper\ncross_data = datasets.ImageFolder(data_dir + '/cross/', transform=test_transforms)\n\ncrossloader = torch.utils.data.DataLoader(cross_data, batch_size=64,shuffle=True)\n# Test out your network!\n\n#cls.eval()\n\ndataiter = iter(crossloader)\nimages, labels = dataiter.next()\nimages, labels = images.to(device),labels.to(device)\n\n\n# Calculate the class probabilities (softmax) for img\nwith torch.no_grad():\n output = cls(images)\n\nps = torch.exp(output)\nfor i in range(len(images)):\n index = torch.argmax(ps[i])\n if index == 0:\n pred = \"Cat\"\n else:\n pred = \"Dog\"\n print(\"Predicted: \" + str(pred))\n helper.imshow(images[i].cpu(), normalize=True)\n\n",
"Predicted: Cat\nPredicted: Cat\nPredicted: Cat\nPredicted: Cat\nPredicted: Cat\nPredicted: Cat\nPredicted: Dog\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbedc732e04cf496f82d5a0b8430cd3ee90ad366
| 69,702 |
ipynb
|
Jupyter Notebook
|
tutorials/W0D5_Statistics/student/W0D5_Tutorial2.ipynb
|
bgalbraith/course-content
|
3db3bbba0fee7af1def2a67e34be073c43434f4a
|
[
"CC-BY-4.0",
"BSD-3-Clause"
] | null | null | null |
tutorials/W0D5_Statistics/student/W0D5_Tutorial2.ipynb
|
bgalbraith/course-content
|
3db3bbba0fee7af1def2a67e34be073c43434f4a
|
[
"CC-BY-4.0",
"BSD-3-Clause"
] | 1 |
2021-06-16T05:41:08.000Z
|
2021-06-16T05:41:08.000Z
|
tutorials/W0D5_Statistics/student/W0D5_Tutorial2.ipynb
|
bgalbraith/course-content
|
3db3bbba0fee7af1def2a67e34be073c43434f4a
|
[
"CC-BY-4.0",
"BSD-3-Clause"
] | null | null | null | 55.406995 | 14,390 | 0.683036 |
[
[
[
"<a href=\"https://colab.research.google.com/github/NeuromatchAcademy/course-content/blob/master/tutorials/W0D5_Statistics/student/W0D5_Tutorial2.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# Tutorial 2: Statistical Inference\n**Week 0, Day 5: Statistics**\n\n**By Neuromatch Academy**\n\n__Content creators:__ Ulrik Beierholm\n\nIf an editor really did a lot of content creation add \"with help from Name Surname\" to the above\n\n__Content reviewers:__ Ethan Cheng, Manisha Sinha\n\nName Surname, Name Surname. This includes both reviewers and editors. Add reviewers first then editors (paper-like seniority :) ).\n",
"_____no_output_____"
],
[
"---\n#Tutorial Objectives\n\nThis tutorial builds on Tutorial 1 by explaining how to do inference through inverting the generative process.\n\nBy completing the exercises in this tutorial, you should:\n* understand what the likelihood function is, and have some intuition of why it is important\n* know how to summarise the Gaussian distribution using mean and variance \n* know how to maximise a likelihood function\n* be able to do simple inference in both classical and Bayesian ways\n* (Optional) understand how Bayes Net can be used to model causal relationships",
"_____no_output_____"
]
],
[
[
"#@markdown Tutorial slides (to be added)\n# you should link the slides for all tutorial videos here (we will store pdfs on osf)\n\nfrom IPython.display import HTML\nHTML('<iframe src=\"https://mfr.ca-1.osf.io/render?url=https://osf.io/kaq2x/?direct%26mode=render%26action=download%26mode=render\" frameborder=\"0\" width=\"960\" height=\"569\" allowfullscreen=\"true\" mozallowfullscreen=\"true\" webkitallowfullscreen=\"true\"></iframe>')",
"_____no_output_____"
]
],
[
[
"---\n# Setup\nMake sure to run this before you get started",
"_____no_output_____"
]
],
[
[
"# Imports\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport scipy as sp\nfrom numpy.random import default_rng # a default random number generator\nfrom scipy.stats import norm # the normal probability distribution",
"_____no_output_____"
],
[
"#@title Figure settings\nimport ipywidgets as widgets # interactive display\nfrom ipywidgets import interact, fixed, HBox, Layout, VBox, interactive, Label, interact_manual\n%config InlineBackend.figure_format = 'retina'\n# plt.style.use(\"https://raw.githubusercontent.com/NeuromatchAcademy/course-content/master/nma.mplstyle\")\nplt.style.use(\"https://raw.githubusercontent.com/NeuromatchAcademy/course-content/NMA2020/nma.mplstyle\")",
"_____no_output_____"
],
[
"#@title Plotting & Helper functions\n\ndef plot_hist(data, xlabel, figtitle = None, num_bins = None):\n \"\"\" Plot the given data as a histogram.\n\n Args:\n data (ndarray): array with data to plot as histogram\n xlabel (str): label of x-axis\n figtitle (str): title of histogram plot (default is no title)\n num_bins (int): number of bins for histogram (default is 10)\n\n Returns:\n count (ndarray): number of samples in each histogram bin\n bins (ndarray): center of each histogram bin\n \"\"\"\n fig, ax = plt.subplots()\n ax.set_xlabel(xlabel)\n ax.set_ylabel('Count')\n if num_bins is not None:\n count, bins, _ = plt.hist(data, max(data), bins = num_bins)\n else:\n count, bins, _ = plt.hist(data, max(data)) # 10 bins default\n if figtitle is not None:\n fig.suptitle(figtitle, size=16)\n plt.show()\n return count, bins\n\ndef plot_gaussian_samples_true(samples, xspace, mu, sigma, xlabel, ylabel):\n \"\"\" Plot a histogram of the data samples on the same plot as the gaussian\n distribution specified by the give mu and sigma values.\n\n Args:\n samples (ndarray): data samples for gaussian distribution\n xspace (ndarray): x values to sample from normal distribution\n mu (scalar): mean parameter of normal distribution\n sigma (scalar): variance parameter of normal distribution\n xlabel (str): the label of the x-axis of the histogram\n ylabel (str): the label of the y-axis of the histogram\n\n Returns:\n Nothing.\n \"\"\"\n fig, ax = plt.subplots()\n ax.set_xlabel(xlabel)\n ax.set_ylabel(ylabel)\n # num_samples = samples.shape[0]\n\n count, bins, _ = plt.hist(samples, density=True) # probability density function\n\n plt.plot(xspace, norm.pdf(xspace, mu, sigma),'r-')\n plt.show()\n\ndef plot_likelihoods(likelihoods, mean_vals, variance_vals):\n \"\"\" Plot the likelihood values on a heatmap plot where the x and y axes match\n the mean and variance parameter values the likelihoods were computed for.\n\n Args:\n likelihoods (ndarray): array of computed likelihood values\n mean_vals (ndarray): array of mean parameter values for which the\n likelihood was computed\n variance_vals (ndarray): array of variance parameter values for which the\n likelihood was computed\n\n Returns:\n Nothing.\n \"\"\"\n fig, ax = plt.subplots()\n im = ax.imshow(likelihoods)\n\n cbar = ax.figure.colorbar(im, ax=ax)\n cbar.ax.set_ylabel('log likelihood', rotation=-90, va=\"bottom\")\n\n ax.set_xticks(np.arange(len(mean_vals)))\n ax.set_yticks(np.arange(len(variance_vals)))\n ax.set_xticklabels(mean_vals)\n ax.set_yticklabels(variance_vals)\n ax.set_xlabel('Mean')\n ax.set_ylabel('Variance')\n\ndef posterior_plot(x, likelihood=None, prior=None, posterior_pointwise=None, ax=None):\n \"\"\"\n Plots normalized Gaussian distributions and posterior.\n\n Args:\n x (numpy array of floats): points at which the likelihood has been evaluated\n auditory (numpy array of floats): normalized probabilities for auditory likelihood evaluated at each `x`\n visual (numpy array of floats): normalized probabilities for visual likelihood evaluated at each `x`\n posterior (numpy array of floats): normalized probabilities for the posterior evaluated at each `x`\n ax: Axis in which to plot. If None, create new axis.\n\n Returns:\n Nothing.\n \"\"\"\n if likelihood is None:\n likelihood = np.zeros_like(x)\n\n if prior is None:\n prior = np.zeros_like(x)\n\n if posterior_pointwise is None:\n posterior_pointwise = np.zeros_like(x)\n\n if ax is None:\n fig, ax = plt.subplots()\n\n ax.plot(x, likelihood, '-C1', LineWidth=2, label='Auditory')\n ax.plot(x, prior, '-C0', LineWidth=2, label='Visual')\n ax.plot(x, posterior_pointwise, '-C2', LineWidth=2, label='Posterior')\n ax.legend()\n ax.set_ylabel('Probability')\n ax.set_xlabel('Orientation (Degrees)')\n plt.show()\n\n return ax\n\ndef plot_classical_vs_bayesian_normal(num_points, mu_classic, var_classic,\n mu_bayes, var_bayes):\n \"\"\" Helper function to plot optimal normal distribution parameters for varying\n observed sample sizes using both classic and Bayesian inference methods.\n\n Args:\n num_points (int): max observed sample size to perform inference with\n mu_classic (ndarray): estimated mean parameter for each observed sample size\n using classic inference method\n var_classic (ndarray): estimated variance parameter for each observed sample size\n using classic inference method\n mu_bayes (ndarray): estimated mean parameter for each observed sample size\n using Bayesian inference method\n var_bayes (ndarray): estimated variance parameter for each observed sample size\n using Bayesian inference method\n\n Returns:\n Nothing.\n \"\"\"\n xspace = np.linspace(0, num_points, num_points)\n fig, ax = plt.subplots()\n ax.set_xlabel('n data points')\n ax.set_ylabel('mu')\n plt.plot(xspace, mu_classic,'r-', label = \"Classical\")\n plt.plot(xspace, mu_bayes,'b-', label = \"Bayes\")\n plt.legend()\n plt.show()\n\n fig, ax = plt.subplots()\n ax.set_xlabel('n data points')\n ax.set_ylabel('sigma^2')\n plt.plot(xspace, var_classic,'r-', label = \"Classical\")\n plt.plot(xspace, var_bayes,'b-', label = \"Bayes\")\n plt.legend()\n plt.show()",
"_____no_output_____"
]
],
[
[
"---\n# Section 1: Statistical Inference and Likelihood",
"_____no_output_____"
]
],
[
[
"#@title Video 4: Inference\nfrom IPython.display import YouTubeVideo\nvideo = YouTubeVideo(id=\"765S2XKYoJ8\", width=854, height=480, fs=1)\nprint(\"Video available at https://youtu.be/\" + video.id)\nvideo",
"_____no_output_____"
]
],
[
[
"A generative model (such as the Gaussian distribution from the previous tutorial) allows us to make prediction about outcomes. \n\nHowever, after we observe $n$ data points, we can also evaluate our model (and any of its associated parameters) by calculating the **likelihood** of our model having generated each of those data points $x_i$.\n\n$$P(x_i|\\mu,\\sigma)=\\mathcal{N}(x_i,\\mu,\\sigma)$$\n\nFor all data points $\\mathbf{x}=(x_1, x_2, x_3, ...x_n) $ we can then calculate the likelihood for the whole dataset by computing the product of the likelihood for each single data point.\n\n$$P(\\mathbf{x}|\\mu,\\sigma)=\\prod_{i=1}^n \\mathcal{N}(x_i,\\mu,\\sigma)$$\n\nAs a function of the parameters (when the data points $x$ are fixed), this is referred to as the **likelihood function**, $L(\\mu,\\sigma)$.\n\nIn the last tutorial we reviewed how the data was generated given the selected parameters of the generative process. If we do not know the parameters $\\mu$, $\\sigma$ that generated the data, we can ask which parameter values (given our model) gives the best (highest) likelihood. \n",
"_____no_output_____"
],
[
"## Exercise 1A: Likelihood, mean and variance\n\n\nWe can use the likelihood to find the set of parameters that are most likely to have generated the data (given the model we are using). That is, we want to infer the parameters that gave rise to the data we observed. We will try a couple of ways of doing statistical inference.\n\nIn the following exercise, we will sample from the Gaussian distribution (again), plot a histogram and the Gaussian probability density function, and calculate some statistics from the samples.\n\nSpecifically we will calculate:\n\n* Likelihood\n* Mean\n* Standard deviation\n\nStatistical moments are defined based on the expectations. The first moment is the expected value, i.e. the mean, the second moment is the expected squared value, i.e. variance, and so on.\n\nThe special thing about the Gaussian is that mean and standard deviation of the random sample can effectively approximate the two parameters of a Gaussian, $\\mu, \\sigma$.\n\nHence using the sample mean, $\\bar{x}=\\frac{1}{n}\\sum_i x_i$, and variance, $\\bar{\\sigma}^2=\\frac{1}{n} \\sum_i (x_i-\\bar{x})^2 $ should give us the best/maximum likelihood, $L(\\bar{x},\\bar{\\sigma}^2)$.\n\nLet's see if that actually works. If we search through different combinations of $\\mu$ and $\\sigma$ values, do the sample mean and variance values give us the maximum likelihood (of observing our data)?\n\nYou need to modify two lines below to generate the data from a normal distribution $N(5, 1)$, and plot the theoretical distribution. Note that we are reusing functions from tutorial 1, so review that tutorial if needed. Then you will use this random sample to calculate the likelihood for a variety of potential mean and variance parameter values. For this tutorial we have chosen a variance parameter of 1, meaning the standard deviation is also 1 in this case. Most of our functions take the standard deviation sigma as a parameter, so we will write $\\sigma = 1$.\n\n(Note that in practice computing the sample variance like this $$\\bar{\\sigma}^2=\\frac{1}{(n-1)} \\sum_i (x_i-\\bar{x})^2 $$ is actually better, take a look at any statistics textbook for an explanation of this.)",
"_____no_output_____"
]
],
[
[
"def generate_normal_samples(mu, sigma, num_samples):\n \"\"\" Generates a desired number of samples from a normal distribution,\n Normal(mu, sigma).\n\n Args:\n mu (scalar): mean parameter of the normal distribution\n sigma (scalar): standard deviation parameter of the normal distribution\n num_samples (int): number of samples drawn from normal distribution\n\n Returns:\n sampled_values (ndarray): a array of shape (samples, ) containing the samples\n \"\"\"\n random_num_generator = default_rng(0)\n sampled_values = random_num_generator.normal(mu, sigma, num_samples)\n return sampled_values\n\ndef compute_likelihoods_normal(x, mean_vals, variance_vals):\n \"\"\" Computes the log-likelihood values given a observed data sample x, and\n potential mean and variance values for a normal distribution\n\n Args:\n x (ndarray): 1-D array with all the observed data\n mean_vals (ndarray): 1-D array with all potential mean values to\n compute the likelihood function for\n variance_vales (ndarray): 1-D array with all potential variance values to\n compute the likelihood function for\n\n Returns:\n likelihood (ndarray): 2-D array of shape (number of mean_vals,\n number of variance_vals) for which the likelihood\n of the observed data was computed\n \"\"\"\n # Initialise likelihood collection array\n likelihood = np.zeros((mean_vals.shape[0], variance_vals.shape[0]))\n\n # Compute the likelihood for observing the gvien data x assuming\n # each combination of mean and variance values\n for idxMean in range(mean_vals.shape[0]):\n for idxVar in range(variance_vals.shape[0]):\n likelihood[idxVar,idxMean]= sum(np.log(norm.pdf(x, mean_vals[idxMean],\n variance_vals[idxVar])))\n\n return likelihood\n\n###################################################################\n## TODO for students: Generate 1000 random samples from a normal distribution\n## with mu = 5 and sigma = 1\n# Fill out the following then remove\nraise NotImplementedError(\"Student exercise: need to generate samples\")\n###################################################################\n\n# Generate data\nmu = 5\nsigma = 1 # since variance = 1, sigma = 1\nx = ...\n\n# You can calculate mean and variance through either numpy or scipy\nprint(\"This is the sample mean as estimated by numpy: \" + str(np.mean(x)))\nprint(\"This is the sample standard deviation as estimated by numpy: \" + str(np.std(x)))\n# or\nmeanX, varX = sp.stats.norm.stats(x)\nprint(\"This is the sample mean as estimated by scipy: \" + str(meanX[0]))\nprint(\"This is the sample standard deviation as estimated by scipy: \" + str(varX[0]))\n\n###################################################################\n## TODO for students: Use the given function to compute the likelihood for\n## a variety of mean and variance values\n# Fill out the following then remove\nraise NotImplementedError(\"Student exercise: need to compute likelihoods\")\n###################################################################\n\n# Let's look through possible mean and variance values for the highest likelihood\n# using the compute_likelihood function\nmeanTest = np.linspace(1, 10, 10) # potential mean values to try\nvarTest = np.array([0.7, 0.8, 0.9, 1, 1.2, 1.5, 2, 3, 4, 5]) # potential variance values to try\nlikelihoods = ...\n\n# Uncomment once you've generated the samples and compute likelihoods\n# xspace = np.linspace(0, 10, 100)\n# plot_gaussian_samples_true(x, xspace, mu, sigma, \"x\", \"Count\")\n# plot_likelihoods(likelihoods, meanTest, varTest)",
"_____no_output_____"
]
],
[
[
"[*Click for solution*](https://github.com/NeuromatchAcademy/course-content/tree/master//tutorials/W0D5_Statistics/solutions/W0D5_Tutorial2_Solution_7687f6b1.py)\n\n*Example output:*\n\n<img alt='Solution hint' align='left' width=560 height=416 src=https://raw.githubusercontent.com/NeuromatchAcademy/course-content/master/tutorials/W0D5_Statistics/static/W0D5_Tutorial2_Solution_7687f6b1_1.png>\n\n<img alt='Solution hint' align='left' width=534 height=414 src=https://raw.githubusercontent.com/NeuromatchAcademy/course-content/master/tutorials/W0D5_Statistics/static/W0D5_Tutorial2_Solution_7687f6b1_2.png>\n\n",
"_____no_output_____"
],
[
"The top figure shows hopefully a nice fit between the histogram and the distribution that generated the data. So far so good.\n\nUnderneath you should see the sample mean and variance values, which are close to the true values (that we happen to know here).\n\nIn the heatmap we should be able to see that the mean and variance parameters values yielding the highest likelihood (yellow) corresponds to (roughly) the combination of the calculated sample mean and variance from the dataset.\nBut it can be hard to see from such a rough **grid-search** simulation, as it is only as precise as the resolution of the grid we are searching. \n\nImplicitly, by looking for the parameters that give the highest likelihood, we have been searching for the **maximum likelihood** estimate.\n$$(\\hat{\\mu},\\hat{\\sigma})=argmax_{\\mu,\\sigma}L(\\mu,\\sigma)=argmax_{\\mu,\\sigma} \\prod_{i=1}^n \\mathcal{N}(x_i,\\mu,\\sigma)$$.\n\nFor a simple Gaussian this can actually be done analytically (you have likely already done so yourself), using the statistical moments: mean and standard deviation (variance). \n\nIn next section we will look at other ways of inferring such parameter variables.",
"_____no_output_____"
],
[
"## Interactive Demo: Maximum likelihood inference",
"_____no_output_____"
],
[
"We want to do inference on this data set, i.e. we want to infer the parameters that most likely gave rise to the data given our model. Intuitively that means that we want as good as possible a fit between the observed data and the probability distribution function with the best inferred parameters. \n\nFor now, just try to see how well you can fit the probability distribution to the data by using the demo sliders to control the mean and standard deviation parameters of the distribution.",
"_____no_output_____"
]
],
[
[
"#@title\n\n#@markdown Make sure you execute this cell to enable the widget and fit by hand!\nvals = generate_normal_samples(mu, sigma, 1000)\ndef plotFnc(mu,sigma):\n #prepare to plot\n fig, ax = plt.subplots()\n ax.set_xlabel('x')\n ax.set_ylabel('probability')\n loglikelihood= sum(np.log(norm.pdf(vals,mu,sigma)))\n\n #calculate histogram\n count, bins, ignored = plt.hist(vals,density=True)\n x = np.linspace(0,10,100)\n #plot\n plt.plot(x, norm.pdf(x,mu,sigma),'r-')\n plt.show()\n print(\"The log-likelihood for the selected parameters is: \" + str(loglikelihood))\n\n#interact(plotFnc, mu=5.0, sigma=2.1);\n#interact(plotFnc, mu=widgets.IntSlider(min=0.0, max=10.0, step=1, value=4.0),sigma=widgets.IntSlider(min=0.1, max=10.0, step=1, value=4.0));\ninteract(plotFnc, mu=(0.0, 10.0, 0.1),sigma=(0.1, 10.0, 0.1));",
"_____no_output_____"
]
],
[
[
"Did you notice the number below the plot? That is the summed log-likelihood, which increases (becomes less negative) as the fit improves. The log-likelihood should be greatest when $\\mu$ = 5 and $\\sigma$ = 1.\n\nBuilding upon what we did in the previous exercise, we want to see if we can do inference on observed data in a bit more principled way.\n",
"_____no_output_____"
],
[
"## Exercise 1B: Maximum Likelihood Estimation\n\nLet's again assume that we have a data set, $\\mathbf{x}$, assumed to be generated by a normal distribution (we actually generate it ourselves in line 1, so we know how it was generated!).\nWe want to maximise the likelihood of the parameters $\\mu$ and $\\sigma^2$. We can do so using a couple of tricks:\n\n* Using a log transform will not change the maximum of the function, but will allow us to work with very small numbers that could lead to problems with machine precision.\n* Maximising a function is the same as minimising the negative of a function, allowing us to use the minimize optimisation provided by scipy.\n\nIn the code below, insert the missing line (see the `compute_likelihoods_normal` function from previous exercise), with the mean as theta[0] and variance as theta[1].\n",
"_____no_output_____"
]
],
[
[
"mu = 5\nsigma = 1\n\n# Generate 1000 random samples from a Gaussian distribution\ndataX = generate_normal_samples(mu, sigma, 1000)\n\n# We define the function to optimise, the negative log likelihood\ndef negLogLike(theta):\n \"\"\" Function for computing the negative log-likelihood given the observed data\n and given parameter values stored in theta.\n\n Args:\n dataX (ndarray): array with observed data points\n theta (ndarray): normal distribution parameters (mean is theta[0],\n variance is theta[1])\n\n Returns:\n Calculated negative Log Likelihood value!\n \"\"\"\n ###################################################################\n ## TODO for students: Compute the negative log-likelihood value for the\n ## given observed data values and parameters (theta)\n # Fill out the following then remove\n raise NotImplementedError(\"Student exercise: need to compute the negative \\\n log-likelihood value\")\n ###################################################################\n return ...\n\n# Define bounds, var has to be positive\nbnds = ((None, None), (0, None))\n\n# Optimize with scipy!\n# Uncomment once function above is implemented\n# optimal_parameters = sp.optimize.minimize(negLogLike, (2, 2), bounds = bnds)\n# print(\"The optimal mean estimate is: \" + str(optimal_parameters.x[0]))\n# print(\"The optimal variance estimate is: \" + str(optimal_parameters.x[1]))\n\n# optimal_parameters contains a lot of information about the optimization,\n# but we mostly want the mean and variance",
"_____no_output_____"
]
],
[
[
"[*Click for solution*](https://github.com/NeuromatchAcademy/course-content/tree/master//tutorials/W0D5_Statistics/solutions/W0D5_Tutorial2_Solution_29984e0b.py)\n\n",
"_____no_output_____"
],
[
"These are the approximations of the parameters that maximise the likelihood ($\\mu$ ~ 5.281 and $\\sigma$ ~ 1.170)\n\nCompare these values to the first and second moment (sample mean and variance) from the previous exercise, as well as to the true values (which we only know because we generated the numbers!). Consider the relationship discussed about statistical moments and maximising likelihood.\n\nGo back to the previous exercise and modify the mean and standard deviation values used to generate the observed data $x$, and verify that the values still work out.",
"_____no_output_____"
],
[
"[*Click for solution*](https://github.com/NeuromatchAcademy/course-content/tree/master//tutorials/W0D5_Statistics/solutions/W0D5_Tutorial2_Solution_b0145e28.py)\n\n",
"_____no_output_____"
],
[
"---\n# Section 2: Bayesian Inference",
"_____no_output_____"
]
],
[
[
"#@title Video 5: Bayes\nfrom IPython.display import YouTubeVideo\nvideo = YouTubeVideo(id=\"12tk5FsVMBQ\", width=854, height=480, fs=1)\nprint(\"Video available at https://youtu.be/\" + video.id)\nvideo",
"_____no_output_____"
]
],
[
[
"\nFor Bayesian inference we do not focus on the likelihood function $L(y)=P(x|y)$, but instead focus on the posterior distribution: \n\n$$P(y|x)=\\frac{P(x|y)P(y)}{P(x)}$$\n\nwhich is composed of the likelihood function $P(x|y)$, the prior $P(y)$ and a normalising term $P(x)$ (which we will ignore for now).\n\nWhile there are other advantages to using Bayesian inference (such as the ability to derive Bayesian Nets, see optional bonus task below), we will first mostly focus on the role of the prior in inference.",
"_____no_output_____"
],
[
"## Exercise 2A: Performing Bayesian inference\n\nIn the above sections we performed inference using maximum likelihood, i.e. finding the parameters that maximised the likelihood of a set of parameters, given the model and data.\n\nWe will now repeat the inference process, but with an added Bayesian prior, and compare it to the classical inference process we did before (Section 1). When using conjugate priors we can just update the parameter values of the distributions (here Gaussian distributions). \n\nFor the prior we start by guessing a mean of 6 (mean of observed data points 5 and 7) and variance of 1 (variance of 5 and 7). This is a simplified way of applying a prior, that allows us to just add these 2 values (pseudo-data) to the real data.\n\nIn the code below, complete the missing lines.",
"_____no_output_____"
]
],
[
[
"def classic_vs_bayesian_normal(mu, sigma, num_points, prior):\n \"\"\" Compute both classical and Bayesian inference processes over the range of\n data sample sizes (num_points) for a normal distribution with parameters\n mu, sigma for comparison.\n\n Args:\n mu (scalar): the mean parameter of the normal distribution\n sigma (scalar): the standard deviation parameter of the normal distribution\n num_points (int): max number of points to use for inference\n prior (ndarray): prior data points for Bayesian inference\n\n Returns:\n mean_classic (ndarray): estimate mean parameter via classic inference\n var_classic (ndarray): estimate variance parameter via classic inference\n mean_bayes (ndarray): estimate mean parameter via Bayesian inference\n var_bayes (ndarray): estimate variance parameter via Bayesian inference\n \"\"\"\n\n # Initialize the classical and Bayesian inference arrays that will estimate\n # the normal parameters given a certain number of randomly sampled data points\n mean_classic = np.zeros(num_points)\n var_classic = np.zeros(num_points)\n\n mean_bayes = np.zeros(num_points)\n var_bayes = np.zeros(num_points)\n\n for nData in range(num_points):\n\n ###################################################################\n ## TODO for students: Complete classical inference for increasingly\n ## larger sets of random data points\n # Fill out the following then remove\n raise NotImplementedError(\"Student exercise: need to code classical inference\")\n ###################################################################\n\n # Randomly sample nData + 1 number of points\n x = ...\n # Compute the mean of those points and set the corresponding array entry to this value\n mean_classic[nData] = ...\n # Compute the variance of those points and set the corresponding array entry to this value\n var_classic[nData] = ...\n\n # Bayesian inference with the given prior is performed below for you\n xsupp = np.hstack((x, prior))\n mean_bayes[nData] = np.mean(xsupp)\n var_bayes[nData] = np.var(xsupp)\n\n return mean_classic, var_classic, mean_bayes, var_bayes\n\n# Set normal distribution parameters, mu and sigma\nmu = 5\nsigma = 1\n\n# Set the prior to be two new data points, 5 and 7, and print the mean and variance\nprior = np.array((5, 7))\nprint(\"The mean of the data comprising the prior is: \" + str(np.mean(prior)))\nprint(\"The variance of the data comprising the prior is: \" + str(np.var(prior)))\n\n# Uncomment once the function above is completed\n# mean_classic, var_classic, mean_bayes, var_bayes = classic_vs_bayesian_normal(mu, sigma, 30, prior)\n# plot_classical_vs_bayesian_normal(30, mean_classic, var_classic, mean_bayes, var_bayes)",
"_____no_output_____"
]
],
[
[
"[*Click for solution*](https://github.com/NeuromatchAcademy/course-content/tree/master//tutorials/W0D5_Statistics/solutions/W0D5_Tutorial2_Solution_4cfc70ca.py)\n\n*Example output:*\n\n<img alt='Solution hint' align='left' width=560 height=416 src=https://raw.githubusercontent.com/NeuromatchAcademy/course-content/master/tutorials/W0D5_Statistics/static/W0D5_Tutorial2_Solution_4cfc70ca_1.png>\n\n<img alt='Solution hint' align='left' width=560 height=416 src=https://raw.githubusercontent.com/NeuromatchAcademy/course-content/master/tutorials/W0D5_Statistics/static/W0D5_Tutorial2_Solution_4cfc70ca_2.png>\n\n",
"_____no_output_____"
],
[
"Hopefully you can see that the blue line stays a little closer to the true values ($\\mu=5$, $\\sigma^2=1$). Having a simple prior in the Bayesian inference process (blue) helps to regularise the inference of the mean and variance parameters when you have very little data, but has little effect with large data. You can see that as the number of data points (x-axis) increases, both inference processes (blue and red lines) get closer and closer together, i.e. their estimates for the true parameters converge as sample size increases.",
"_____no_output_____"
],
[
"## Think! 2A: Bayesian Brains\nIt should be clear how Bayesian inference can help you when doing data analysis. But consider whether the brain might be able to benefit from this too. If the brain needs to make inferences about the world, would it be useful to do regularisation on the input? ",
"_____no_output_____"
],
[
"[*Click for solution*](https://github.com/NeuromatchAcademy/course-content/tree/master//tutorials/W0D5_Statistics/solutions/W0D5_Tutorial2_Solution_daa12602.py)\n\n",
"_____no_output_____"
],
[
"## Exercise 2B: Finding the posterior computationally\n***(Exercise moved from NMA2020 Bayes day, all credit to original creators!)***\n\nImagine an experiment where participants estimate the location of a noise-emitting object. To estimate its position, the participants can use two sources of information: \n 1. new noisy auditory information (the likelihood)\n 2. prior visual expectations of where the stimulus is likely to come from (visual prior). \n\nThe auditory and visual information are both noisy, so participants will combine these sources of information to better estimate the position of the object.\n\nWe will use Gaussian distributions to represent the auditory likelihood (in red), and a Gaussian visual prior (expectations - in blue). Using Bayes rule, you will combine them into a posterior distribution that summarizes the probability that the object is in each possible location. \n\nWe have provided you with a ready-to-use plotting function, and a code skeleton.\n\n* You can use `my_gaussian` from Tutorial 1 (also included below), to generate an auditory likelihood with parameters $\\mu$ = 3 and $\\sigma$ = 1.5\n* Generate a visual prior with parameters $\\mu$ = -1 and $\\sigma$ = 1.5\n* Calculate the posterior using pointwise multiplication of the likelihood and prior. Don't forget to normalize so the posterior adds up to 1\n* Plot the likelihood, prior and posterior using the predefined function `posterior_plot`\n\n",
"_____no_output_____"
]
],
[
[
"def my_gaussian(x_points, mu, sigma):\n \"\"\" Returns normalized Gaussian estimated at points `x_points`, with parameters:\n mean `mu` and standard deviation `sigma`\n\n Args:\n x_points (ndarray of floats): points at which the gaussian is evaluated\n mu (scalar): mean of the Gaussian\n sigma (scalar): standard deviation of the gaussian\n\n Returns:\n (numpy array of floats) : normalized Gaussian evaluated at `x`\n \"\"\"\n px = 1/(2*np.pi*sigma**2)**1/2 *np.exp(-(x_points-mu)**2/(2*sigma**2))\n\n # as we are doing numerical integration we may have to remember to normalise\n # taking into account the stepsize (0.1)\n px = px/(0.1*sum(px))\n return px\n\ndef compute_posterior_pointwise(prior, likelihood):\n \"\"\" Compute the posterior probability distribution point-by-point using Bayes\n Rule.\n\n Args:\n prior (ndarray): probability distribution of prior\n likelihood (ndarray): probability distribution of likelihood\n\n Returns:\n posterior (ndarray): probability distribution of posterior\n \"\"\"\n ##############################################################################\n # TODO for students: Write code to compute the posterior from the prior and\n # likelihood via pointwise multiplication. (You may assume both are defined\n # over the same x-axis)\n #\n # Comment out the line below to test your solution\n raise NotImplementedError(\"Finish the simulation code first\")\n ##############################################################################\n\n posterior = ...\n\n return posterior\n\ndef localization_simulation(mu_auditory = 3.0, sigma_auditory = 1.5,\n mu_visual = -1.0, sigma_visual = 1.5):\n \"\"\" Perform a sound localization simulation with an auditory prior.\n\n Args:\n mu_auditory (float): mean parameter value for auditory prior\n sigma_auditory (float): standard deviation parameter value for auditory\n prior\n mu_visual (float): mean parameter value for visual likelihood distribution\n sigma_visual (float): standard deviation parameter value for visual\n likelihood distribution\n\n Returns:\n x (ndarray): range of values for which to compute probabilities\n auditory (ndarray): probability distribution of the auditory prior\n visual (ndarray): probability distribution of the visual likelihood\n posterior_pointwise (ndarray): posterior probability distribution\n \"\"\"\n ##############################################################################\n ## Using the x variable below,\n ## create a gaussian called 'auditory' with mean 3, and std 1.5\n ## create a gaussian called 'visual' with mean -1, and std 1.5\n #\n #\n ## Comment out the line below to test your solution\n raise NotImplementedError(\"Finish the simulation code first\")\n ###############################################################################\n x = np.arange(-8, 9, 0.1)\n\n auditory = ...\n visual = ...\n posterior = compute_posterior_pointwise(auditory, visual)\n\n return x, auditory, visual, posterior\n\n\n# Uncomment the lines below to plot the results\n# x, auditory, visual, posterior_pointwise = localization_simulation()\n# _ = posterior_plot(x, auditory, visual, posterior_pointwise)",
"_____no_output_____"
]
],
[
[
"[*Click for solution*](https://github.com/NeuromatchAcademy/course-content/tree/master//tutorials/W0D5_Statistics/solutions/W0D5_Tutorial2_Solution_ab4b98de.py)\n\n*Example output:*\n\n<img alt='Solution hint' align='left' width=560 height=416 src=https://raw.githubusercontent.com/NeuromatchAcademy/course-content/master/tutorials/W0D5_Statistics/static/W0D5_Tutorial2_Solution_ab4b98de_1.png>\n\n",
"_____no_output_____"
],
[
"Combining the the visual and auditory information could help the brain get a better estimate of the location of an audio-visual object, with lower variance. For this specific example we did not use a Bayesian prior for simplicity, although it would be a good idea in a practical modeling study.\n\n**Main course preview:** On Week 3 Day 1 (W3D1) there will be a whole day devoted to examining whether the brain uses Bayesian inference. Is the brain Bayesian?!",
"_____no_output_____"
],
[
"---\n# Summary\n",
"_____no_output_____"
]
],
[
[
"#@title Video 6: Outro\nfrom IPython.display import YouTubeVideo\nvideo = YouTubeVideo(id= \"BL5qNdZS-XQ\", width=854, height=480, fs=1)\nprint(\"Video available at https://youtu.be/\" + video.id)\nvideo",
"_____no_output_____"
]
],
[
[
"\nHaving done the different exercises you should now:\n* understand what the likelihood function is, and have some intuition of why it is important\n* know how to summarise the Gaussian distribution using mean and variance \n* know how to maximise a likelihood function\n* be able to do simple inference in both classical and Bayesian ways",
"_____no_output_____"
],
[
"---\n# Bonus\n\nFor more reading on these topics see:\nTextbook",
"_____no_output_____"
],
[
"\n\n## Extra exercise: Bayes Net\nIf you have the time, here is another extra exercise.\n\nBayes Net, or Bayesian Belief Networks, provide a way to make inferences about multiple levels of information, which would be very difficult to do in a classical frequentist paradigm.\n\nWe can encapsulate our knowledge about causal relationships and use this to make inferences about hidden properties.",
"_____no_output_____"
],
[
"We will try a simple example of a Bayesian Net (aka belief network). Imagine that you have a house with an unreliable sprinkler system installed for watering the grass. This is set to water the grass independently of whether it has rained that day. We have three variables, rain ($r$), sprinklers ($s$) and wet grass ($w$). Each of these can be true (1) or false (0). See the graphical model representing the relationship between the variables.",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"There is a table below describing all the relationships between $w, r$, and s$.\n\nObviously the grass is more likely to be wet if either the sprinklers were on or it was raining. On any given day the sprinklers have probability 0.25 of being on, $P(s = 1) = 0.25$, while there is a probability 0.1 of rain, $P (r = 1) = 0.1$. The table then lists the conditional probabilities for the given being wet, given a rain and sprinkler condition for that day.",
"_____no_output_____"
],
[
"\\begin{array}{|l | l || ll |} \\hline\nr &s&P(w=0|r,s) &P(w=1|r,s)$\\\\ \\hline\n0& 0 &0.999 &0.001\\\\\n0& 1 &0.1& 0.9\\\\\n1& 0 &0.01 &0.99\\\\\n1& 1& 0.001 &0.999\\\\ \\hline\n\\end{array}\n",
"_____no_output_____"
],
[
"\nYou come home and find that the the grass is wet, what is the probability the sprinklers were on today (you do not know if it was raining)?\n\nWe can start by writing out the joint probability:\n$P(r,w,s)=P(w|r,s)P(r)P(s)$\n\nThe conditional probability is then:\n\n$\nP(s|w)=\\frac{\\sum_{r} P(w|s,r)P(s) P(r)}{P(w)}=\\frac{P(s) \\sum_{r} P(w|s,r) P(r)}{P(w)}\n$\n\nNote that we are summing over all possible conditions for $r$ as we do not know if it was raining. Specifically, we want to know the probability of sprinklers having been on given the wet grass, $P(s=1|w=1)$:\n\n$\nP(s=1|w=1)=\\frac{P(s = 1)( P(w = 1|s = 1, r = 1) P(r = 1)+ P(w = 1|s = 1,r = 0) P(r = 0))}{P(w = 1)} \n$\n\nwhere\n\n\\begin{eqnarray}\nP(w=1)=P(s=1)( P(w=1|s=1,r=1 ) P(r=1) &+ P(w=1|s=1,r=0) P(r=0))\\\\\n+P(s=0)( P(w=1|s=0,r=1 ) P(r=1) &+ P(w=1|s=0,r=0) P(r=0))\\\\\n\\end{eqnarray}\n\nThis code has been written out below, you just need to insert the right numbers from the table.",
"_____no_output_____"
]
],
[
[
"##############################################################################\n# TODO for student: Write code to insert the correct conditional probabilities\n# from the table; see the comments to match variable with table entry.\n# Comment out the line below to test your solution\nraise NotImplementedError(\"Finish the simulation code first\")\n##############################################################################\n\nPw1r1s1 = ... # the probability of wet grass given rain and sprinklers on\nPw1r1s0 = ... # the probability of wet grass given rain and sprinklers off\nPw1r0s1 = ... # the probability of wet grass given no rain and sprinklers on\nPw1r0s0 = ... # the probability of wet grass given no rain and sprinklers off\nPs = ... # the probability of the sprinkler being on\nPr = ... # the probability of rain that day\n\n\n# Uncomment once variables are assigned above\n# A= Ps * (Pw1r1s1 * Pr + (Pw1r0s1) * (1 - Pr))\n# B= (1 - Ps) * (Pw1r1s0 *Pr + (Pw1r0s0) * (1 - Pr))\n# print(\"Given that the grass is wet, the probability the sprinkler was on is: \" +\n# str(A/(A + B)))",
"_____no_output_____"
]
],
[
[
"[*Click for solution*](https://github.com/NeuromatchAcademy/course-content/tree/master//tutorials/W0D5_Statistics/solutions/W0D5_Tutorial2_Solution_204db048.py)\n\n",
"_____no_output_____"
],
[
"The probability you should get is about 0.7522.\n\nYour neighbour now tells you that it was indeed \nraining today, $P (r = 1) = 1$, so what is now the probability the sprinklers were on? Try changing the numbers above.\n\n",
"_____no_output_____"
],
[
"## Think! Bonus: Causality in the Brain\n\nIn a causal stucture this is the correct way to calculate the probabilities. Do you think this is how the brain solves such problems? Would it be different for task involving novel stimuli (e.g. for someone with no previous exposure to sprinklers), as opposed to common stimuli?\n\n**Main course preview:** On W3D5 we will discuss causality further!",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
cbedc8f938439bcd1c54f3e046a0ecfe9f25e399
| 34,997 |
ipynb
|
Jupyter Notebook
|
Selenium/Selenium.ipynb
|
JuanManiglia/WebScraping
|
afe0af0831ae92c3ca7bf2d52ca59d91940ebcdd
|
[
"MIT"
] | null | null | null |
Selenium/Selenium.ipynb
|
JuanManiglia/WebScraping
|
afe0af0831ae92c3ca7bf2d52ca59d91940ebcdd
|
[
"MIT"
] | null | null | null |
Selenium/Selenium.ipynb
|
JuanManiglia/WebScraping
|
afe0af0831ae92c3ca7bf2d52ca59d91940ebcdd
|
[
"MIT"
] | null | null | null | 26.214981 | 735 | 0.576078 |
[
[
[
"# Web Scraping: Selenium",
"_____no_output_____"
],
[
"A menudo, los datos están disponibles públicamente para nosotros, pero no en una forma que sea fácilmente utilizable. Ahí es donde entra en juego el web scraping, podemos usar web scraping para obtener nuestros datos deseados en un formato conveniente que luego se puede usar. a continuación, mostraré cómo se puede extraer información de interés de un sitio web usando el paquete Selenium en Python. Selenium nos permite manejar una ventana del navegador e interactuar con el sitio web mediante programación. \n\nSelenium también tiene varios métodos que facilitan la extracción de datos.\nEn este Jupyter Notebook vamos a usar Python 3 en Windows.\n\nEn primer lugar, tendremos que descargar un controlador.\n\nUsaremos ChromeDriver para Google Chrome. Para obtener una lista completa de controladores y plataformas compatibles, consulte [Selenium](https://www.selenium.dev/downloads/). Si desea utilizar Google Chrome, diríjase a [chrome](https://chromedriver.chromium.org/) y descargue el controlador que corresponde a su versión actual de Google Chrome.\n\nComo saber cual es la version de chrome que utilizo simple utilizamos pegamos el siguiente enlace en la barra de chrome chrome://settings/help\n\nAntes de comenzar se preguntaran si ya se BeautifulSoup cual es la diferencia con Selenium.\n\nA diferencia BeautifulSoup, Selenium no trabaja con el texto fuente en HTML de la web en cuestión, sino que carga la página en un navegador sin interfaz de usuario. El navegador interpreta entonces el código fuente de la página y crea, a partir de él, un Document Object Model (modelo de objetos de documento o DOM). Esta interfaz estandarizada permite poner a prueba las interacciones de los usuarios. De esta forma se consigue, por ejemplo, simular clics y rellenar formularios automáticamente. Los cambios en la web que resultan de dichas acciones se reflejan en el DOM. La estructura del proceso de web scraping con Selenium es la siguiente:\n\nURL → Solicitud HTTP → HTML → Selenium → DOM\n\n",
"_____no_output_____"
],
[
"## Comencemos importando las bibliotecas que usaremos:",
"_____no_output_____"
]
],
[
[
"from selenium import webdriver\nimport urllib3 # urllib3 es un cliente HTTP potente y fácil de usar para Python.\nimport re # Expresiones regulares \nimport time\nimport pandas as pd",
"_____no_output_____"
]
],
[
[
"El objeto driver es con el que trabajaremos a partir de ahora",
"_____no_output_____"
]
],
[
[
"# especificamos el path hasta nuestro driver recién descargado:\nchrome_driver_path = 'chromedriver.exe'\noptions = webdriver.ChromeOptions()",
"_____no_output_____"
],
[
"# Creamos el driver con el que nos vamos a manejar en la sesión de scrapeo:\ndriver = webdriver.Chrome(executable_path = chrome_driver_path, options = options)",
"_____no_output_____"
],
[
"# indicamos la URL de la página web a la que queremos acceder:\nurl = 'https://insolvencyinsider.ca/filing/'\n# el objeto driver nos va a permitir alterar el estado del la página\ndriver.get(url)",
"_____no_output_____"
]
],
[
[
"Ahora si queremos hacer click en el boton de \"Load more\"..",
"_____no_output_____"
],
[
"Selenium proporciona varios métodos para localizar elementos en la página web. Usaremos el método find_element_by_xpath() para crear un objeto de botón, con el que luego podremos interactuar:",
"_____no_output_____"
],
[
"/html/body/div[2]/div/main/div/div/div/button",
"_____no_output_____"
]
],
[
[
"loadMore = driver.find_element_by_xpath(xpath=\"\")",
"_____no_output_____"
]
],
[
[
"Antes de continuar, necesitaremos saber cuántas páginas hay para saber cuántas veces debemos hacer clic en el botón. Necesitaremos una forma de extraer el código fuente del sitio web. Afortunadamente, este proceso es relativamente sencillo con las bibliotecas urllib3 y re.",
"_____no_output_____"
]
],
[
[
"url = \"https://insolvencyinsider.ca/filing/\"\nhttp = urllib3.PoolManager()\nr = http.request(\"GET\", url)\ntext = str(r.data)\n",
"_____no_output_____"
]
],
[
[
"```text``` ahora es una cadena. Ahora, necesitamos una forma de extraer total_pages de nuestra cadena de texto. Imprima texto para ver cómo podemos extraerlo usando RegEx con el paquete re. Podemos totalizar_páginas así:",
"_____no_output_____"
]
],
[
[
"totalPagesObj = re.search(pattern='\"total_pages\":\\d+', string=text)\ntotalPagesStr = totalPagesObj.group(0)\ntotalPages = int((re.search(pattern=\"\\d+\", string=totalPagesStr)).group(0))",
"_____no_output_____"
]
],
[
[
"El método de búsqueda toma un patrón y una cadena. En este caso nuestro patrón es '\"total_pages\":\\d+' . Si no está familiarizado con RegEx, todo esto significa que estamos buscando la cadena \"total_pages\": con dos o más dígitos después de los dos puntos. \\d se refiere a un dígito entre 0 y 9, mientras que + indica que Python debe buscar una o más de las expresiones regulares anteriores. Puedes leer más sobre el paquete re aquí. El método search() devuelve un objeto Match. re proporciona el método group() que devuelve uno o más subgrupos de la coincidencia. Pasamos 0 como argumento para indicar que queremos el parche completo. La tercera línea simplemente extrae el entero correspondiente a total_pages de la cadena.",
"_____no_output_____"
]
],
[
[
"print(totalPagesObj)\nprint(totalPagesStr)\nprint(totalPages)",
"_____no_output_____"
]
],
[
[
"Con eso completo, ahora podemos cargar todas las páginas de Insolvency Insider. Podemos hacer clic en el botón Cargar más accediendo al método click() del objeto. Esperamos tres segundos entre clics para no sobrecargar el sitio web.",
"_____no_output_____"
],
[
"Recuerde que el total de las páginas son 88 pero comenzamos en 0 asi que es 88-1",
"_____no_output_____"
]
],
[
[
"for i in range(totalPages-1):\n loadMore.click()\n time.sleep(3)",
"_____no_output_____"
]
],
[
[
"Una vez que ejecute esto, debería ver que se hace clic en el botón Cargar más y que se cargan las páginas restantes.\nUna vez que se carga cada página, podemos comenzar a raspar el contenido. Ahora, eliminar ciertos elementos como el nombre de presentación, la fecha y la hiperreferencia es bastante sencillo. Podemos usar los métodos find_elements_by_class_name() y find_elements_by_xpath() de Selenium (importante la ```s``` extra después de element):",
"_____no_output_____"
],
[
"filing-name\nfiling-date\n//*[@id='content']/div[2]/div/div[1]/h3/a",
"_____no_output_____"
]
],
[
[
"filingNamesElements = driver.find_elements_by_class_name(\"\")\n\nfilingDateElements = driver.find_elements_by_class_name(\"\")\n\nfilingHrefElements = driver.find_elements_by_xpath(\"\")",
"_____no_output_____"
]
],
[
[
"También nos gustaría conocer los metadatos de presentación, es decir, el tipo de archivo, el sector de la empresa y el lugar en la que operan. Extraer estos datos requiere un poco más de trabajo.",
"_____no_output_____"
],
[
"//*[@id='content']/div[2]/div[%d]/div[2]/div[1]",
"_____no_output_____"
]
],
[
[
"filingMetas = []\nfor i in range(len(filingNamesElements) + 1):\n filingMetai = driver.find_elements_by_xpath((\"\" %(i)))\n for element in filingMetai:\n filingMetaTexti = element.text\n filingMetas.append(filingMetaTexti)",
"_____no_output_____"
]
],
[
[
"De cada elemento de la presentación de Metas podemos extraer el tipo de presentación, la industria y la provincia, así:",
"_____no_output_____"
],
[
"********",
"_____no_output_____"
]
],
[
[
"metaDict = {\"Filing Type\": [], \"Industry\": [], \"Province\": []}\nfor filing in filingMetas:\n filingSplit = filing.split(\"\\n\")\n \n for item in filingSplit:\n itemSplit = item.split(\":\")\n\n \n if itemSplit[0] == \"Filing Type\":\n metaDict[\"Filing Type\"].append(itemSplit[1])\n elif itemSplit[0] == \"Industry\":\n metaDict[\"Industry\"].append(itemSplit[1])\n elif itemSplit[0] == \"Province\":\n metaDict[\"Province\"].append(itemSplit[1])\n \n if \"Filing Type\" not in filing:\n metaDict[\"Filing Type\"].append(\"NA\")\n elif \"Industry\" not in filing:\n metaDict[\"Industry\"].append(\"NA\")\n elif \"Province\" not in filing:\n metaDict[\"Province\"].append(\"NA\")",
"_____no_output_____"
],
[
"for key in metaDict:\n print(len(metaDict[key]))",
"_____no_output_____"
]
],
[
[
"*********",
"_____no_output_____"
],
[
"Ahora, todavía tenemos que poner nuestros nombres y fechas de presentación en las listas. Hacemos esto agregando el texto de cada elemento a una lista usando el método text() de antes:",
"_____no_output_____"
]
],
[
[
"filingName = []\nfilingDate = []\nfilingLink = []\n# para cada elemento en la lista de elementos de nombre de archivo, agrega el\n# texto del elemento a la lista de nombres de archivo.\nfor element in filingNamesElements:\n filingName.append(element.text)\n# para cada elemento en la lista de elementos de la fecha de presentación, agrega el\n# texto del elemento a la lista de fechas de presentación.\nfor element in filingDateElements:\n filingDate.append(element.text)\nfor link in filingHrefElements:\n if link.get_attribute(\"href\"):\n filingLink.append(link.get_attribute(\"href\"))",
"_____no_output_____"
]
],
[
[
"Una vez que tengamos eso, estamos listos para poner todo en un diccionario y luego crear un DataFrame de pandas:",
"_____no_output_____"
]
],
[
[
"# Crea un diccionario final con nombres y fechas de archivo.\nfullDict = {\n \"Filing Name\": filingName,\n \"Filing Date\": filingDate, \n \"Filing Type\": metaDict[\"Filing Type\"],\n \"Industry\": metaDict[\"Industry\"],\n \"Province\": metaDict[\"Province\"],\n \"Link\": filingLink\n}\n# Crea un DataFrame.\ndf = pd.DataFrame(fullDict)\ndf[\"Filing Date\"] = pd.to_datetime(df[\"Filing Date\"], infer_datetime_format=True)",
"_____no_output_____"
],
[
"df",
"_____no_output_____"
]
],
[
[
"------------------------",
"_____no_output_____"
],
[
"# Ahora algo más visual",
"_____no_output_____"
]
],
[
[
"driver = webdriver.Chrome(executable_path = chrome_driver_path, options = options)",
"_____no_output_____"
],
[
"# indicamos la URL de la página web a la que queremos acceder:\nurl = 'https://www.filmaffinity.com/es/main.html'\n# el objeto driver nos va a permitir alterar el estado del la página\ndriver.get(url)",
"_____no_output_____"
]
],
[
[
"La página de Filmaffinity se ha abierto",
"_____no_output_____"
],
[
"Pero....",
"_____no_output_____"
],
[
"Nos hemos encontrado con un pop-up que nos pide aceptar cookies",
"_____no_output_____"
],
[
"1. Buscamos el botón\n2. Hacemos click en el botón",
"_____no_output_____"
],
[
"Vamos a quitar el boton para seguir",
"_____no_output_____"
]
],
[
[
"elements_by_tag = driver.find_elements_by_tag_name('button')\nelements_by_class_name = driver.find_elements_by_class_name('css-v43ltw')\nelement_by_xpath = driver.find_element_by_xpath('/html/body/div[1]/div/div/div/div[2]/div/button[2]')",
"_____no_output_____"
]
],
[
[
"Una vez tenemos los elementos podemos hacer varias cosas con ellos",
"_____no_output_____"
],
[
"Podemos extraer todos los atributos que tenga",
"_____no_output_____"
]
],
[
[
"dir(element_by_xpath)\n# obtenemos todos sus métodos y atributos:",
"_____no_output_____"
]
],
[
[
"Podemos evaluar que tipo de elemento es (tag)",
"_____no_output_____"
]
],
[
[
"element_by_xpath.tag_name",
"_____no_output_____"
]
],
[
[
"Podemos sacar el valor que tiene (el texto)",
"_____no_output_____"
]
],
[
[
"element_by_xpath.text",
"_____no_output_____"
],
[
"for i in range(0,len(elements_by_tag)):\n print(elements_by_tag[i].text)",
"_____no_output_____"
]
],
[
[
"Incluso podemos guardar una imagen del elemento",
"_____no_output_____"
]
],
[
[
"type(element_by_xpath)\n# Vemos que es tipo 'WebElement' y en la documentación podremos encontrar sus métodos",
"_____no_output_____"
],
[
"# guardamos como 'mi_imagen.png' la imagen asociada al xpath\nelement_by_xpath.screenshot('mi_imagen.png')",
"_____no_output_____"
]
],
[
[
"Evaluamos que elementos hemos encontrado por el tag:",
"_____no_output_____"
]
],
[
[
"for index, element in enumerate(elements_by_tag):\n print('Elemento:', index)\n print('Texto del elemento',index, 'es', element.text)\n print('El tag del elemento',index, 'es', element.tag_name)\n element.screenshot('mi_imagen'+str(index)+'.png')",
"_____no_output_____"
]
],
[
[
"Basta de tonterias seguimos",
"_____no_output_____"
],
[
"Instanciamos el elemento del tag [2] en la variable boton aceptar",
"_____no_output_____"
]
],
[
[
"boton_aceptar = elements_by_tag[2]",
"_____no_output_____"
]
],
[
[
"Si el elemento es interactivo podremos hacer más cosas además de las anteriores. Por ejemplo: hacer click",
"_____no_output_____"
]
],
[
[
"boton_aceptar.click()",
"_____no_output_____"
]
],
[
[
"Buscamos una película por título",
"_____no_output_____"
]
],
[
[
"from selenium.webdriver.common.keys import Keys",
"_____no_output_____"
]
],
[
[
"/html/body/div[2]/div[1]/div/div[2]/form/div/input",
"_____no_output_____"
]
],
[
[
"buscador = driver.find_element_by_xpath('')",
"_____no_output_____"
],
[
"buscador.send_keys('')",
"_____no_output_____"
],
[
"buscador.clear()",
"_____no_output_____"
],
[
"# una vez escrita la búsqueda deberíamos poder activarla:\nbuscador.send_keys(Keys.ENTER)",
"_____no_output_____"
],
[
"# volvemos a la página anterior\ndriver.back()",
"_____no_output_____"
]
],
[
[
"### Vamos a buscar todas las películas que se estrenan el próximo viernes",
"_____no_output_____"
],
[
"1. Cogemos los containers que hay en la zona lateral",
"_____no_output_____"
]
],
[
[
"menu_lateral = driver.find_element_by_id('lsmenu')\nmenu_lateral",
"_____no_output_____"
],
[
"mis_secciones = menu_lateral.find_elements_by_tag_name('a')",
"_____no_output_____"
]
],
[
[
"2. Vemos con cuál nos tenemos que quedar",
"_____no_output_____"
]
],
[
[
"for a in mis_secciones:\n if a.text == 'Próximos estrenos':\n a.click()\n break",
"_____no_output_____"
]
],
[
[
"Accedemos al container central, en el que aparecen los estrenos por semana que queremos ver, exactamente igual que hemos hecho antes",
"_____no_output_____"
]
],
[
[
"cajon_central = driver.find_elements_by_id('main-wrapper-rdcat')",
"_____no_output_____"
],
[
"type(cajon_central)",
"_____no_output_____"
],
[
"for semana in cajon_central:\n print(semana.find_element_by_tag_name('div').text)\n print(semana.find_element_by_tag_name('div').get_attribute('id'))",
"_____no_output_____"
],
[
"for semana in cajon_central:\n fecha = semana.find_element_by_tag_name('div').get_attribute('id')\n if fecha == '2022-02-25':\n break",
"_____no_output_____"
]
],
[
[
"Buscamos cómo acceder a las películas",
"_____no_output_____"
]
],
[
[
"caratulas = semana.find_elements_by_class_name('')\nlista_pelis = []\nfor peli in caratulas:\n lista_pelis.append(peli.find_element_by_tag_name('a').get_attribute('href'))",
"_____no_output_____"
],
[
"lista_pelis",
"_____no_output_____"
]
],
[
[
"Una vez tenemos todas las urls vamos a ver qué hacemos con cada una de ellas",
"_____no_output_____"
]
],
[
[
"# Accedemos a la página de la primera pelicula\ndriver.get(lista_pelis[0])",
"_____no_output_____"
]
],
[
[
"Vamos a ver el proceso que deberíamos hacer con cada una de las películas:",
"_____no_output_____"
],
[
"1. Sacamos toda la información que nos interesa",
"_____no_output_____"
]
],
[
[
"# titulo, nota, numero de votos y ficha técnica\ntitulo = driver.find_element_by_xpath('/html/body/div[4]/table/tbody/tr/td[2]/div[1]/div[4]/h1/span').text\nnota = driver.find_element_by_xpath('/html/body/div[4]/table/tbody/tr/td[2]/div[1]/div[4]/div/div[2]/div[2]/div[1]/div[2]/div[1]').text\nvotos = driver.find_element_by_xpath('/html/body/div[4]/table/tbody/tr/td[2]/div[1]/div[4]/div/div[2]/div[2]/div[1]/div[2]/div[2]/span').text\nficha = driver.find_element_by_xpath('/html/body/div[4]/table/tbody/tr/td[2]/div[1]/div[4]/div/div[3]/dl[1]')",
"_____no_output_____"
],
[
"titulo",
"_____no_output_____"
]
],
[
[
"2. Creamos una lista a partir de la ficha técnica",
"_____no_output_____"
]
],
[
[
"# Los nombres estan con tag = 'dt' y los valores con 'dd'\nficha_names = []\nficha_values = []\n\nfor name in ficha.find_elements_by_tag_name('dt'):\n ficha_names.append(name.text)\nfor value in ficha.find_elements_by_tag_name('dd'):\n ficha_values.append(value.text)",
"_____no_output_____"
],
[
"ficha_values",
"_____no_output_____"
]
],
[
[
"3. Creamos un dataframe con la info",
"_____no_output_____"
]
],
[
[
"columns = ['Titulo', 'Nota', 'Votos']\ncolumns.extend(ficha_names)\nlen(columns)",
"_____no_output_____"
],
[
"values = [titulo, nota, votos]\nvalues.extend(ficha_values)\nlen(values)",
"_____no_output_____"
],
[
"pd.DataFrame([values],columns=columns)",
"_____no_output_____"
]
],
[
[
"Ahora vamos a crear una función que nos haga todo esto para cada una de las películas:",
"_____no_output_____"
]
],
[
[
"def sacar_info(driver):\n \n titulo = driver.find_element_by_xpath('/html/body/div[4]/table/tbody/tr/td[2]/div[1]/div[4]/h1/span').text\n try:\n nota = driver.find_element_by_xpath('/html/body/div[4]/table/tbody/tr/td[2]/div[1]/div[4]/div/div[2]/div[2]/div[1]/div[2]').text\n votos = driver.find_element_by_xpath('/html/body/div[4]/table/tbody/tr/td[2]/div[1]/div[4]/div/div[2]/div[2]/div[1]/div[2]/div[2]').text\n except:\n nota = None\n votos = None\n ficha = driver.find_element_by_xpath('/html/body/div[4]/table/tbody/tr/td[2]/div[1]/div[4]/div/div[3]/dl[1]')\n \n return titulo, nota, votos, ficha\n\ndef sacar_ficha(ficha):\n \n ficha_names = []\n ficha_values = []\n\n for name in ficha.find_elements_by_tag_name('dt'):\n ficha_names.append(name.text)\n for value in ficha.find_elements_by_tag_name('dd'):\n ficha_values.append(value.text)\n \n return ficha_names, ficha_values\n\ndef montar_df(ficha_names, ficha_values, titulo, nota, votos):\n \n columns = ['Titulo', 'Nota', 'Votos']\n columns.extend(ficha_names)\n values = [titulo, nota, votos]\n values.extend(ficha_values)\n \n return pd.DataFrame([values], columns = columns)\n \ndef nueva_pelicula(driver):\n \n titulo, nota, votos, ficha = sacar_info(driver)\n ficha_names, ficha_values = sacar_ficha(ficha)\n df_peli = montar_df(ficha_names, ficha_values, titulo, nota, votos)\n \n return df_peli\n",
"_____no_output_____"
]
],
[
[
"Vamos a ver cómo nos podemos mover entre ventanas del navegador\n\nAbrir nueva ventana:",
"_____no_output_____"
]
],
[
[
"driver.execute_script('window.open(\"\");')",
"_____no_output_____"
]
],
[
[
"Movernos a otra ventana",
"_____no_output_____"
]
],
[
[
"driver.switch_to.window(driver.window_handles[0])",
"_____no_output_____"
]
],
[
[
"Cerrar ventana",
"_____no_output_____"
]
],
[
[
"driver.close()",
"_____no_output_____"
]
],
[
[
"Una vez cerramos la ventana tenemos que indicarle a qué ventana tiene que ir",
"_____no_output_____"
]
],
[
[
"driver.switch_to.window(driver.window_handles[-1])",
"_____no_output_____"
]
],
[
[
"Sabiendo cómo podemos movernos por entre las ventanas y sabiendo cómo extraer de cada página toda la información que necesitamos vamos a crear nuestro dataframe:",
"_____no_output_____"
]
],
[
[
"# para abrir todos los links en lista_pelis\nfor link in lista_pelis:\n driver.execute_script('window.open(\"'+link+'\");')\n driver.switch_to.window(driver.window_handles[-1])\n driver.get(link)",
"_____no_output_____"
],
[
"# Creamos un dataframe con todas las pelis que se estrenan la próxima semana:\ndf_peliculas = pd.DataFrame()\n\nfor link in lista_pelis:\n driver.execute_script('window.open(\"\");')\n driver.switch_to.window(driver.window_handles[-1])\n driver.get(link)\n nueva_peli = nueva_pelicula(driver)\n df_peliculas = df_peliculas.append(nueva_peli)",
"_____no_output_____"
],
[
"df_peliculas.info()",
"_____no_output_____"
],
[
"df_peliculas",
"_____no_output_____"
]
],
[
[
"Ya tenemos un dataframe con todas las películas que se van a estrenar el próximo viernes",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
cbedcdcb2f71bd703afc20aaa7f0a9276b5674df
| 50,356 |
ipynb
|
Jupyter Notebook
|
2019/GEOS 657-Lab9-InSARTimeSeriesAnalysis.ipynb
|
uafgeoteach/GEOS657_MRS
|
682d9d936e058c692d3f3f1492c243e569cd0f6f
|
[
"BSD-3-Clause"
] | 5 |
2021-11-02T04:02:17.000Z
|
2022-03-22T20:44:53.000Z
|
2019/GEOS 657-Lab9-InSARTimeSeriesAnalysis.ipynb
|
uafgeoteach/GEOS657_MRS
|
682d9d936e058c692d3f3f1492c243e569cd0f6f
|
[
"BSD-3-Clause"
] | null | null | null |
2019/GEOS 657-Lab9-InSARTimeSeriesAnalysis.ipynb
|
uafgeoteach/GEOS657_MRS
|
682d9d936e058c692d3f3f1492c243e569cd0f6f
|
[
"BSD-3-Clause"
] | 4 |
2021-11-30T16:12:46.000Z
|
2022-03-22T20:02:33.000Z
| 37.60717 | 722 | 0.593534 |
[
[
[
"<img src=\"NotebookAddons/blackboard-banner.png\" width=\"100%\" />\n<font face=\"Calibri\">\n<br>\n<font size=\"7\"> <b> GEOS 657: Microwave Remote Sensing<b> </font>\n\n<font size=\"5\"> <b>Lab 9: InSAR Time Series Analysis using GIAnT within Jupyter Notebooks</b> </font>\n\n<br>\n<font size=\"4\"> <b> Franz J Meyer & Joshua J C Knicely; University of Alaska Fairbanks</b> <br>\n<img src=\"NotebookAddons/UAFLogo_A_647.png\" width=\"170\" align=\"right\" /><font color='rgba(200,0,0,0.2)'> <b>Due Date: </b>NONE</font>\n</font>\n\n<font size=\"3\"> This Lab is part of the UAF course <a href=\"https://radar.community.uaf.edu/\" target=\"_blank\">GEOS 657: Microwave Remote Sensing</a>. The primary goal of this lab is to demonstrate how to process InSAR data, specifically interferograms, using the Generic InSAR Analysis Toolbox (<a href=\"http://earthdef.caltech.edu/projects/giant/wiki\" target=\"_blank\">GIAnT</a>) in the framework of *Jupyter Notebooks*.<br>\n\n<b>Our specific objectives for this lab are to:</b>\n\n- Learn how to prepare data for GIAnT. \n- Use GIAnT to create maps of surface deformation. \n - Understand its capabilities. \n - Understand its limitations. \n</font>\n\n<br>\n<font face=\"Calibri\">\n\n<font size=\"5\"> <b> Target Description </b> </font>\n\n<font size=\"3\"> In this lab, we will analyze the volcano Sierra Negra. This is a highly active volcano on the Galapagos hotpsot. The most recent eruption occurred from 29 June to 23 August 2018. The previous eruption occurred in October 2005, prior to the launch of the Sentinel-1 satellites, which will be the source of data we use for this lab. We will be looking at the deformation that occurred prior to the volcano's 2018 eruption. </font>\n\n<font size=\"4\"> <font color='rgba(200,0,0,0.2)'> <b>THIS NOTEBOOK INCLUDES NO HOMEWORK ASSIGNMENTS.</b></font> <br>\n\nContact me at [email protected] should you run into any problems.\n</font>",
"_____no_output_____"
]
],
[
[
"import url_widget as url_w\nnotebookUrl = url_w.URLWidget()\ndisplay(notebookUrl)",
"_____no_output_____"
],
[
"from IPython.display import Markdown\nfrom IPython.display import display\n\nnotebookUrl = notebookUrl.value\nuser = !echo $JUPYTERHUB_USER\nenv = !echo $CONDA_PREFIX\nif env[0] == '':\n env[0] = 'Python 3 (base)'\nif env[0] != '/home/jovyan/.local/envs/insar_analysis':\n display(Markdown(f'<text style=color:red><strong>WARNING:</strong></text>'))\n display(Markdown(f'<text style=color:red>This notebook should be run using the \"insar_analysis\" conda environment.</text>'))\n display(Markdown(f'<text style=color:red>It is currently using the \"{env[0].split(\"/\")[-1]}\" environment.</text>'))\n display(Markdown(f'<text style=color:red>Select \"insar_analysis\" from the \"Change Kernel\" submenu of the \"Kernel\" menu.</text>'))\n display(Markdown(f'<text style=color:red>If the \"insar_analysis\" environment is not present, use <a href=\"{notebookUrl.split(\"/user\")[0]}/user/{user[0]}/notebooks/conda_environments/Create_OSL_Conda_Environments.ipynb\"> Create_OSL_Conda_Environments.ipynb </a> to create it.</text>'))\n display(Markdown(f'<text style=color:red>Note that you must restart your server after creating a new environment before it is usable by notebooks.</text>'))",
"_____no_output_____"
]
],
[
[
"<font face='Calibri'><font size='5'><b>Overview</b></font>\n<br>\n<font size='3'><b>About GIAnT</b>\n<br>\nGIAnT is a Python framework that allows rapid time series analysis of low amplitude deformation signals. It allows users to use multiple time series analysis technqiues: Small Baseline Subset (SBAS), New Small Baseline Subset (N-SBAS), and Multiscale InSAR Time-Series (MInTS). As a part of this, it includes the ability to correct for atmospheric delays by assuming a spatially uniform stratified atmosphere. \n<br><br>\n<b>Limitations</b>\n<br>\nGIAnT has a number of limitations that are important to keep in mind as these can affect its effectiveness for certain applications. It implements the simplest time-series inversion methods. Its single coherence threshold is very conservative in terms of pixel selection. It does not include any consistency checks for unwrapping errors. It has a limited dictionary of temporal model functions. It cannot correct for atmospheric effects due to differing surface elevations. \n<br><br>\n<b>Steps to use GIAnT</b><br>\nAlthough GIAnT is an incredibly powerful tool, it requires very specific input. Because of the input requirements, the majority of one's effort goes to getting the data into a form that GIAnT can manipulate and to creating files that tell GIAnT what to do. The general steps to use GIAnT are below. \n\n- Download Data\n- Identify Area of Interest\n- Subset (Crop) Data to Area of Interest\n- Prepare Data for GIAnT\n - Adjust file names\n - Remove potentially disruptive default values (optional)\n - Convert data from '.tiff' to '.flt' format\n- Create Input Files for GIAnT\n - Create 'ifg.list'\n - Create 'date.mli.par'\n - Make prepxml_SBAS.py\n - Run prepxml_SBAS.py\n - Make userfn.py\n- Run GIAnT\n - PrepIgramStack.py*\n - ProcessStack.py\n - SBASInvert.py\n - SBASxval.py\n- Data Visualization\n\n<br>\nThe steps from PrepIgramStack.py and above have been completed for you in order to save disk space and computation time. This allows us to concentrate on the usage of GIAnT and data visualization. Some of the code to create the prepatory files (e.g., 'ifg.list', 'date.mli.par', etc.) have been incldued for your potential use. More information about GIAnT can be found here: (<a href=\"http://earthdef.caltech.edu/projects/giant/wiki\" target=\"_blank\">http://earthdef.caltech.edu/projects/giant/wiki</a>).",
"_____no_output_____"
],
[
"<hr>\n<font face=\"Calibri\" size=\"5\" color=\"darkred\"> <b>Important Note about JupyterHub</b> </font>\n<br><br>\n<font face=\"Calibri\" size=\"3\"> <b>Your JupyterHub server will automatically shutdown when left idle for more than 1 hour. Your notebooks will not be lost but you will have to restart their kernels and re-run them from the beginning. You will not be able to seamlessly continue running a partially run notebook.</b> </font>\n",
"_____no_output_____"
],
[
"<font face='Calibri'><font size='5'><b>0. Import Python Libraries:</b></font><br><br>\n<font size='3'><b>Import the Python libraries and modules we will need to run this lab:</b></font>",
"_____no_output_____"
]
],
[
[
"%%capture\nfrom datetime import date\nimport glob\nimport h5py # for is_hdf5\nimport os\nimport shutil\n\nfrom osgeo import gdal\nimport matplotlib.pyplot as plt\nimport matplotlib.animation\nfrom matplotlib import rc\nimport numpy as np\n\nfrom IPython.display import HTML\n\nimport opensarlab_lib as asfn\nasfn.jupytertheme_matplotlib_format()",
"_____no_output_____"
]
],
[
[
"<font face='Calibri'><font size='5'><b>1. Transfer data to a local directory</b></font><br>\n <font size='3'>The data cube (referred to as a stack in the GIAnT documentation and code) and several other needed files have been created and stored in the GIAnT server. We will download this data to a local directory and unzip it. </font></font>",
"_____no_output_____"
],
[
"<font face=\"Calibri\" size=\"3\"> Before we download anything, <b>create a working directory for this analysis and change into it:</b> </font>",
"_____no_output_____"
]
],
[
[
"path = f\"{os.getcwd()}/2019/lab_9_data\"\nif not os.path.exists(path):\n os.makedirs(path)\nos.chdir(path)\nprint(f\"Current working directory: {os.getcwd()}\")",
"_____no_output_____"
]
],
[
[
"<font face = 'Calibri' size='3'>First step is to find the zip file and download it to a local directory. This zip file has been placed in the S3 bucket for this class.\n<br><br>\n<b>Display the contents of the S3 bucket:</b></font>",
"_____no_output_____"
]
],
[
[
"!aws s3 ls --region=us-west-2 --no-sign-request s3://asf-jupyter-data-west/",
"_____no_output_____"
]
],
[
[
"<font face = 'Calibri' size='3'><b>Copy the desired file ('Lab9Files.zip') to your data directory:</b></font>",
"_____no_output_____"
]
],
[
[
"!aws s3 cp --region=us-west-2 --no-sign-request s3://asf-jupyter-data-west/Lab9Files.zip .",
"_____no_output_____"
]
],
[
[
"<font face='Calibri'><font size='3'><b>Create the directories where we will perform the GIAnT analysis and store the data:</b></font>",
"_____no_output_____"
]
],
[
[
"stack_path = f\"{os.getcwd()}/Stack\" # directory GIAnT prefers to access and store data steps. \nif not os.path.exists(stack_path):\n os.makedirs(stack_path)",
"_____no_output_____"
]
],
[
[
"<font face='Calibri'><font size='3'><b>Extract the zipped file to path and delete it:</b></font>",
"_____no_output_____"
]
],
[
[
"zipped = 'Lab9Files.zip'\nasfn.asf_unzip(path, zipped)\nif os.path.exists(zipped):\n os.remove(zipped)",
"_____no_output_____"
]
],
[
[
"<font face='Calibri' size='3'>The files have been extracted and placed in a folder called 'Lab9Files'. <b>Move the amplitude image, data.xml, date.mli.par, and sbas.xml files to path and RAW-STACK.h5 to stack_path:</b></font>",
"_____no_output_____"
]
],
[
[
"temp_dir = f\"{path}/Lab9Files\"\nif not os.path.exists(f\"{stack_path}/RAW-STACK.h5\"):\n shutil.move(f\"{temp_dir}/RAW-STACK.h5\", stack_path) \nfiles = glob.glob(f\"{temp_dir}/*.*\")\nfor file in files:\n if os.path.exists(file):\n shutil.move(file, path)\nif os.path.exists(temp_dir):\n os.rmdir(temp_dir)",
"_____no_output_____"
]
],
[
[
"<font face='Calibri'><font size='5'><b>2. Create Input Files And Code for GIAnT</b></font>\n <br>\n <font size ='3'>The code below shows how to create the input files and specialty code that GIAnT requires. For this lab, 'ifg.list' is not needed, 'date.mli.par' has already been provided, 'prepxml_SBAS.py' is not needed as the 'sbas.xml' and 'data.xml' files it would create have already been provided, and 'userfn.py' is not needed as we are skipping the step in which it would be used. <br>The files that would be created are listed below. \n <br>\n \n- ifg.list\n - List of the interferogram properties including master and slave date, perpendicular baseline, and sensor. \n- date.mli.par\n - File from which GIAnT pulls requisite information about the sensor. \n - This is specifically for GAMMA files. When using other interferogram processing techniques, an alternate file is required. \n- prepxml_SBAS.py\n - Python function to create an xml file that specifies the processing options to GIAnT. \n - This must be modified by the user for their particular application. \n- userfn.py\n - Python function to map the interferogram dates to a phyiscal file on disk. \n - This must be modified by the user for their particular application. \n </font>\n </font>",
"_____no_output_____"
],
[
"<font face='Calibri' size='4'> <b>2.1 Create 'ifg.list' File </b> </font> </font>\n<br>\n<font face='Calibri' size='3'> This will create simple 4 column text file will communicate network information to GIAnT. It will be created within the <b>GIAnT</b> folder.\n<br><br>\n<b>This step has already been done, so we will not actually create the 'ifg.list' file. This code is displayed for your potential future use.</b></font>",
"_____no_output_____"
]
],
[
[
"\"\"\"\n# Get one of each file name. This assumes the unwrapped phase geotiff has been converted to a '.flt' file\nfiles = [f for f in os.listdir(datadirectory) if f.endswith('_unw_phase.flt')] \n\n# Get all of the master and slave dates. \nmasterDates,slaveDates = [],[]\nfor file in files:\n masterDates.append(file[0:8])\n slaveDates.append(file[9:17])\n# Sort the dates according to the master dates. \nmaster_dates,sDates = (list(t) for t in zip(*sorted(zip(masterDates,slaveDates))))\n\nwith open( os.path.join('GIAnT', 'ifg.list'), 'w') as fid:\n for i in range(len(master_dates)):\n masterDate = master_dates[i] # pull out master Date (first set of numbers)\n slaveDate = sDates[i] # pull out slave Date (second set of numbers)\n bperp = '0.0' # according to JPL notebooks\n sensor = 'S1' # according to JPL notebooks\n fid.write(f'{masterDate} {slaveDate} {bperp} {sensor}\\n') # write values to the 'ifg.list' file. \n\"\"\"",
"_____no_output_____"
]
],
[
[
"<font face='Calibri'><font size='3'>You may notice that the code above sets the perpendicular baseline to a value of 0.0 m. This is not the true perpendicular baseline. That value can be found in metadata file (titled '$<$master timestamp$>$_$<$slave timestamp$>$.txt') that comes with the original interferogram. Generally, we would want the true baseline for each interferogram. However, since Sentinel-1 has such a short baseline, a value of 0.0 m is sufficient for our purposes. </font></font>",
"_____no_output_____"
],
[
"<font face='Calibri' size='4'> <b>2.2 Create 'date.mli.par' File </b></font> \n<br>\n<font face='Calibri' size='3'>As we are using GAMMA products, we must create a 'date.mli.par' file from which GIAnT will pull necessary information. If another processing technique is used to create the interferograms, an alterante file name and file inputs are required.\n<br><br>\n<b>Again, this step has already been completed and the code is only displayed for your potential future use.</b></font>",
"_____no_output_____"
]
],
[
[
"\"\"\"\n# Create file 'date.mli.par'\n\n# Get file names\nfiles = [f for f in os.listdir(datadirectory) if f.endswith('_unw_phase.flt')]\n\n# Get WIDTH (xsize) and FILE_LENGTH (ysize) information\nds = gdal.Open(datadirectory+files[0], gdal.GA_ReadOnly)\ntype(ds)\n\nnLines = ds.RasterYSize\nnPixels = ds.RasterXSize\n\ntrans = ds.GetGeoTransform()\nds = None\n\n# Get the center line UTC time stamp; can also be found inside <date>_<date>.txt file and hard coded\ndirName = os.listdir('ingrams')[0] # get original file name (any file can be used; the timestamps are different by a few seconds)\nvals = dirName.split('-') # break file name into parts using the separator '-'\ntstamp = vals[2][9:16] # extract the time stamp from the 2nd datetime (could be the first)\nc_l_utc = int(tstamp[0:2])*3600 + int(tstamp[2:4])*60 + int(tstamp[4:6])\n\nrfreq = 299792548.0 / 0.055465763 # radar frequency; speed of light divided by radar wavelength of Sentinel1 in meters\n\n# write the 'date.mli.par' file\nwith open(os.path.join(path, 'date.mli.par'), 'w') as fid:\n # Method 1\n fid.write(f'radar_frequency: {rfreq} \\n') # when using GAMMA products, GIAnT requires the radar frequency. Everything else is in wavelength (m) \n fid.write(f'center_time: {c_l_utc} \\n') # Method from Tom Logan's prepGIAnT code; can also be found inside <date>_<date>.txt file and hard coded\n fid.write( 'heading: -11.9617913 \\n') # inside <date>_<date>.txt file; can be hardcoded or set up so code finds it. \n fid.write(f'azimuth_lines: {nLines} \\n') # number of lines in direction of the satellite's flight path\n fid.write(f'range_samples: {nPixels} \\n') # number of pixels in direction perpendicular to satellite's flight path\n fid.close() # close the file\n\"\"\"",
"_____no_output_____"
]
],
[
[
"<font face='Calibri'><font size='4'><b>2.3 Make prepxml_SBAS.py</b> </font>\n<br>\n<font size='3'>We will create a prepxml_SBAS.py function and put it into our GIAnT working directory. Again, this is shown for anyone that may want to use GIAnT on their own.<br>If we do wish to change 'sbas.xml' or 'data.xml', this can be done by creating and running a new 'prepxml_SBAS.py'. </font>\n</font>",
"_____no_output_____"
],
[
"<font face='Calibri'> <font size='3'><b>2.3.1 Necessary prepxml_SBAS.py edits</b></font>\n<br>\n<font size='3'> GIAnT comes with an example prepxml_SBAS.py, but requries significant edits for our purposes. These alterations have already been made, so we don't have to do anything now, but it is good to know the kinds of things that have to be altered. The details of some of these options can be found in the GIAnT documentation. The rest must be found in the GIAnT processing files themselves, most notably the tsxml.py and tsio.py functions. <br>The following alterations were made:\n<br>\n- Changed 'example' ► 'date.mli.par'\n- Removed 'xlim', 'ylim', 'ref_x_lim', and 'ref_y_lim'\n - These are used for clipping the files in GIAnT. As we have already done this, it is not necessary. \n- Removed latfile='lat.map' and lonfile='lon.map'\n - These are optional inputs for the latitude and longitude maps. \n- Removed hgtfile='hgt.map'\n - This is an optional altitude file for the sensor. \n- Removed inc=21.\n - This is the optional incidence angle information. \n - It can be a constant float value or incidence angle file. \n - For Sentinel1, it varies from 29.1-46.0°.\n- Removed masktype='f4'\n - This is the mask designation. \n - We are not using any masks for this. \n- Changed unwfmt='RMG' ► unwfmt='GRD'\n - Read data using GDAL. \n- Removed demfmt='RMG'\n- Changed corfmt='RMG' ► corfmt='GRD'\n - Read data using GDAL. \n- Changed nvalid=30 -> nvalid=1\n - This is the minimum number of interferograms in which a pixel must be coherent. A particular pixel will be included only if its coherence is above the coherence threshold, cohth, in more than nvalid number of interferograms. \n- Removed atmos='ECMWF'\n - This is an amtospheric correction command. It depends on a library called 'pyaps' developed for GIAnT. This library has not been installed yet. \n- Changed masterdate='19920604' ► masterdate='20161119'\n - Use our actual masterdate. \n - I simply selected the earliest date as the masterdate. \n\n</font>",
"_____no_output_____"
],
[
"<font face='Calibri' size='3'>Defining a reference region is a potentially important step. This is a region at which there should be no deformation. For a volcano, this should be some significant distance away from the volcano. GIAnT has the ability to automatically select a reference region which we will use for this exercise. <br>Below is an example of how the reference region would be defined. If we look at the prepxml_SBAS.py code below, ref_x_lim and ref_y_lim, the pixel based location of the reference region, is within the code, but has been commented out. \n<br><br>\n<b>Define reference region:</b></font>",
"_____no_output_____"
]
],
[
[
"ref_x_lim, ref_y_lim = [0, 10], [95, 105]",
"_____no_output_____"
]
],
[
[
"<font face='Calibri' size='3'>Below is an example of how the reference region would be defined. Look at the prepxml_SBAS.py code below. Note that ref_x_lim and ref_y_lim (the pixel based location of the reference region) are within the code.\n<br><br>\n<b>This has already been completed but the code is here as an example script for creating XML files for use with the SBAS processing chain.</b></font>",
"_____no_output_____"
]
],
[
[
"'''\n#!/usr/bin/env python\n\nimport tsinsar as ts\nimport argparse\nimport numpy as np\n\ndef parse():\n parser= argparse.ArgumentParser(description='Preparation of XML files for setting up the processing chain. Check tsinsar/tsxml.py for details on the parameters.')\n parser.parse_args()\n\nparse()\ng = ts.TSXML('data')\ng.prepare_data_xml(\n 'date.mli.par', proc='GAMMA', \n #ref_x_lim = [{1},{2}], ref_y_lim=[{3},{4}],\n inc = 21., cohth=0.10, \n unwfmt='GRD', corfmt='GRD', chgendian='True', endianlist=['UNW','COR'])\ng.writexml('data.xml')\n\n\ng = ts.TSXML('params')\ng.prepare_sbas_xml(nvalid=1, netramp=True, demerr=False, uwcheck=False, regu=True, masterdate='{5}', filt=1.0)\ng.writexml('sbas.xml')\n\n\n############################################################\n# Program is part of GIAnT v1.0 #\n# Copyright 2012, by the California Institute of Technology#\n# Contact: [email protected] #\n############################################################\n\n'''",
"_____no_output_____"
]
],
[
[
"<font face='Calibri' size='3'><b>Set the master date and create a script for creating XML files for use with the SBAS processing chain: </b></font>",
"_____no_output_____"
]
],
[
[
"#files = [f for f in os.listdir(datadirectory) if f.endswith('_unw_phase.flt')]\n#master_date = min([files[i][0:8] for i in range(len(files))], key=int)\n\nmaster_date = '20161119'\n\nprepxml_SBAS_Template = '''\n#!/usr/bin/env python\n\"\"\"Example script for creating XML files for use with the SBAS processing chain. This script is supposed to be copied to the working directory and modified as needed.\"\"\"\n\n\nimport tsinsar as ts\nimport argparse\nimport numpy as np\n\ndef parse():\n parser= argparse.ArgumentParser(description='Preparation of XML files for setting up the processing chain. Check tsinsar/tsxml.py for details on the parameters.')\n parser.parse_args()\n\nparse()\ng = ts.TSXML('data')\ng.prepare_data_xml(\n 'date.mli.par', proc='GAMMA', \n #ref_x_lim = [{1},{2}], ref_y_lim=[{3},{4}],\n inc = 21., cohth=0.10, \n unwfmt='GRD', corfmt='GRD', chgendian='True', endianlist=['UNW','COR'])\ng.writexml('data.xml')\n\n\ng = ts.TSXML('params')\ng.prepare_sbas_xml(nvalid=1, netramp=True, demerr=False, uwcheck=False, regu=True, masterdate='{5}', filt=1.0)\ng.writexml('sbas.xml')\n\n\n############################################################\n# Program is part of GIAnT v1.0 #\n# Copyright 2012, by the California Institute of Technology#\n# Contact: [email protected] #\n############################################################\n\n'''\nwith open(os.path.join(path,'prepxml_SBAS.py'), 'w') as fid:\n fid.write(prepxml_SBAS_Template.format(path,ref_x_lim[0],ref_x_lim[1],ref_y_lim[0],ref_y_lim[1],master_date))",
"_____no_output_____"
]
],
[
[
"<font face='Calibri'><font size='3'>To create a new 'sbas.xml' and 'data.xml' file, we would modify the above code to give new parameters and to write to the appropriate folder (e.g., to change the time filter from 1 year to none and to write to the directory in which we are working; 'filt=1.0' -> 'filt=0.0'; and 'os.path.join(path,'prepxml_SBAS.py') -> 'prepxml_SBAS.py' OR '%cd ~' into your home directory). Then we would run it below. </font></font>",
"_____no_output_____"
],
[
"<font face='Calibri' size='4'> <b>2.4 Run prepxml_SBAS.py </b> </font>\n<br>\n<font face='Calibri' size='3'> Here we run <b>prepxml_SBAS.py</b> to create the 2 needed files</font>\n\n- data.xml \n- sbas.xml\n\n<font face='Calibri' size='3'> To use MinTS, we would run <b>prepxml_MinTS.py</b> to create</font>\n\n- data.xml\n- mints.xml\n \n<font face='Calibri' size='3'> These files are needed by <b>PrepIgramStack.py</b>. \n<br>\nWe must first switch to the GIAnT folder in which <b>prepxml_SBAS.py</b> is contained, then call it. Otherwise, <b>prepxml_SBAS.py</b> will not be able to find the file 'date.mli.par', which holds necessary processing information. \n<br><br>\n<b>Create a variable holding the general path to the GIAnT code base and download GIAnT from the `asf-jupyter-data-west` S3 bucket, if not present.</b>\n <br>\n GIAnT is no longer supported (Python 2). This unofficial version of GIAnT has been partially ported to Python 3 to run this notebook. Only the portions of GIAnT used in this notebook have been tested.\n</font> ",
"_____no_output_____"
]
],
[
[
"giant_path = \"/home/jovyan/.local/GIAnT/SCR\"\n\nif not os.path.exists(\"/home/jovyan/.local/GIAnT\"):\n download_path = 's3://asf-jupyter-data-west/GIAnT_5_21.zip'\n output_path = f\"/home/jovyan/.local/{os.path.basename(download_path)}\"\n !aws --region=us-west-2 --no-sign-request s3 cp $download_path $output_path\n if os.path.isfile(output_path):\n !unzip $output_path -d /home/jovyan/.local/\n os.remove(output_path)",
"_____no_output_____"
]
],
[
[
"<font face='Calibri' size='3'><b>Run prepxml_SBAS.py and check the output to confirm that your input values are correct:</b></font>",
"_____no_output_____"
]
],
[
[
"# !python $giant_path/prepxml_SBAS.py # this has already been done. data.xml and sbas.xml already exist",
"_____no_output_____"
]
],
[
[
"<font face='Calibri' size='3'><b>Make sure the two requisite xml files (data.xml and sbas.xml) were produced after running prepxml_SBAS.py.</b></font>\n<br><br>\n<font face='Calibri' size='3'><b>Display the contents of data.xml:</b></font>",
"_____no_output_____"
]
],
[
[
"if os.path.exists('data.xml'):\n !cat data.xml ",
"_____no_output_____"
]
],
[
[
"<font face='Calibri' size='3'><b>Display the contents of sbas.xml:</b></font>",
"_____no_output_____"
]
],
[
[
"if os.path.exists('sbas.xml'):\n !cat sbas.xml",
"_____no_output_____"
]
],
[
[
"<font face='Calibri'><font size='4'><b>2.5 Create userfn.py</b></font>\n<br>\n<font size='3'>Before running the next piece of code, <b>PrepIgramStack.py</b>, we must create a python file called <b>userfn.py</b>. This file maps the interferogram dates to a physical file on disk. This python file must be in our working directory, <b>/GIAnT</b>. We can create this file from within the notebook using python. \n<br><br>\n<b>Again, this step has already been preformed and is unnecessary, but the code is provided as an example.</b></font>",
"_____no_output_____"
]
],
[
[
"userfnTemplate = \"\"\"\n#!/usr/bin/env python\nimport os \n\ndef makefnames(dates1, dates2, sensor):\n dirname = '{0}'\n root = os.path.join(dirname, dates1+'-'+dates2)\n #unwname = root+'_unw_phase.flt' # for potentially disruptive default values kept. \n unwname = root+'_unw_phase_no_default.flt' # for potentially disruptive default values removed. \n corname = root+'_corr.flt'\n return unwname, corname\n\"\"\"\n\nwith open('userfn.py', 'w') as fid:\n fid.write(userfnTemplate.format(path))",
"_____no_output_____"
]
],
[
[
"<font face='Calibri'><font size='5'><b>3. Run GIAnT</b></font>\n <br>\n <font size='3'>We have now created all of the necessary files to run GIAnT. The full GIAnT process requires 3 function calls.\n- PrepIgramStack.py\n - After PrepIgramStack.py, we will actually start running GIAnT. \n- ProcessStack.py\n- SBASInvert.py\n- SBASxval.py\n - This 4th function call is not necessary and we will skip it, but provides some error estimation that can be useful.",
"_____no_output_____"
],
[
"<font face='Calibri' size='4'> <b>3.1 Run PrepIgramStack.py </b> </font>\n<br>\n<font face='Calibri' size='3'> Here we would run <b>PrepIgramStack.py</b> to create the files for GIAnT. This would read in the input data and the files we previously created and output an HDF5 file. As we do not actually need to call this, it is currently set up to display some help information.<br>\nInputs: \n- ifg.list\n- data.xml\n- sbas.xml \n- interferograms\n- coherence files \n\nOutputs:\n- RAW-STACK.h5\n- PNG previews under 'GIAnT/Figs/Igrams'\n \n</font>\n<br>\n<font size='3'><b>Display some help information for PrepIgramStack.py:</b></font>",
"_____no_output_____"
]
],
[
[
"!python $giant_path/PrepIgramStack.py -h",
"_____no_output_____"
]
],
[
[
"<font size='3'><b>Run PrepIgramStack.py (in our case, this has already been done):</b></font>",
"_____no_output_____"
]
],
[
[
"#!python $giant_path/PrepIgramStack.py",
"_____no_output_____"
]
],
[
[
"<hr>\n<font face='Calibri'><font size='3'>PrepIgramStack.py creates a file called 'RAW-STACK.h5'.\n<br><br>\n<b>Verify that RAW-STACK.h5 is an HDF5 file as required by the rest of GIAnT.</b></font>",
"_____no_output_____"
]
],
[
[
"raw_h5 = f\"{stack_path}/RAW-STACK.h5\"\nif not h5py.is_hdf5(raw_h5):\n print(f\"Not an HDF5 file: {raw_h5}\")\nelse:\n print(f\"Confirmed: {raw_h5} is an HDF5 file.\")",
"_____no_output_____"
]
],
[
[
"<font face='Calibri' size='4'> <b>3.2 Run ProcessStack.py </b> </font>\n<br>\n<font face='Calibri' size='3'> This seems to be an optional step. Does atmospheric corrections and estimation of orbit residuals. <br>\nInputs:\n\n- HDF5 files from PrepIgramStack.py, RAW-STACK.h5\n- data.xml \n- sbas.xml\n- GPS Data (optional; we don't have this)\n- Weather models (downloaded automatically)\n\nOutputs: \n\n- HDF5 files, PROC-STACK.h5\n \nThese files are then fed into SBAS. \n</font> \n<br><br>\n<font face='Calibri' size='3'><b>Display the help information for ProcessStack.py:</b></font>",
"_____no_output_____"
]
],
[
[
"!python $giant_path/ProcessStack.py -h",
"_____no_output_____"
]
],
[
[
"<font face='Calibri' size='3'><b>Run ProcessStack.py:</b></font>",
"_____no_output_____"
]
],
[
[
"!python $giant_path/ProcessStack.py",
"_____no_output_____"
]
],
[
[
"<hr>\n<font face='Calibri'><font size='3'>ProcessStack.py creates a file called 'PROC-STACK.h5'.\n<br><br>\n<b>Verify that PROC-STACK.h5 is an HDF5 file as required by the rest of GIAnT:</b></font>",
"_____no_output_____"
]
],
[
[
"proc_h5 = f\"{stack_path}/PROC-STACK.h5\"\nif not h5py.is_hdf5(proc_h5):\n print(f\"Not an HDF5 file: {proc_h5}\")\nelse:\n print(f\"Confirmed: {proc_h5} is an HDF5 file.\")",
"_____no_output_____"
]
],
[
[
"<font face='Calibri' size='4'> <b>3.3 Run SBASInvert.py </b></font>\n<br>\n<font face='Calibri' size='3'> Actually do the time series. \n \nInputs\n\n- HDF5 file, PROC-STACK.h5\n- data.xml\n- sbas.xml\n\nOutputs\n\n- HDF5 file: LS-PARAMS.h5\n\n<b>Display the help information for SBASInvert.py:</b>\n</font>\n",
"_____no_output_____"
]
],
[
[
"!python $giant_path/SBASInvert.py -h",
"_____no_output_____"
]
],
[
[
"<font face='Calibri' size='3'><b>Run SBASInvert.py:</b></font>",
"_____no_output_____"
]
],
[
[
"!python $giant_path/SBASInvert.py",
"_____no_output_____"
]
],
[
[
"<hr>\n<font face='Calibri'><font size='3'>SBASInvert.py creates a file called 'LS-PARAMS.h5'.\n<br><br>\n<b>Verify that LS-PARAMS.h5 is an HDF5 file as required by the rest of GIAnT:</b></font>",
"_____no_output_____"
]
],
[
[
"params_h5 = f\"{stack_path}/LS-PARAMS.h5\"\nif not h5py.is_hdf5(params_h5):\n print(f\"Not an HDF5 file: {params_h5}\")\nelse:\n print(f\"Confirmed: {params_h5} is an HDF5 file.\")",
"_____no_output_____"
]
],
[
[
"<font face='Calibri' size='4'> <b>3.4 Run SBASxval.py </b></font>\n<br>\n<font face='Calibri' size='3'> Get an uncertainty estimate for each pixel and epoch using a Jacknife test. We are skipping this function as we won't be doing anything with its output and it takes a significant amount of time to run relative to the other GIAnT functions.\n \nInputs: \n\n- HDF5 files, PROC-STACK.h5\n- data.xml\n- sbas.xml\n\nOutputs:\n\n- HDF5 file, LS-xval.h5\n\n<br>\n<b>Display the help information for SBASxval.py:</b></font>",
"_____no_output_____"
]
],
[
[
"#!python $giant_path/SBASxval.py -h",
"_____no_output_____"
]
],
[
[
"<font face='Calibri' size='3'><b>Run SBASxval.py:</b></font>",
"_____no_output_____"
]
],
[
[
"#!python $giant_path/SBASxval.py",
"_____no_output_____"
]
],
[
[
"<hr>\n<font face='Calibri'><font size='3'>SBASxval.py creates a file called 'LS-xval.h5'.\n<br><br>\n<b>Verify that LS-xval.h5 is an HDF5 file as required by the rest of GIAnT:</b></font>",
"_____no_output_____"
]
],
[
[
"'''\nxval_h5 = f\"{stack_path}/LS-xval.h5\"\nif not h5py.is_hdf5(xval_h5):\n print(f\"Not an HDF5 file: {xval_h5}\")\nelse:\n print(f\"Confirmed: {xval_h5} is an HDF5 file.\")\n'''",
"_____no_output_____"
]
],
[
[
"<font face='Calibri' size='5'><b>4. Data Visualization</b></font>\n<br>\n<font face='Calibri' size='3'>Now we visualize the data. This is largely copied from Lab 4.\n<br><br>\n<b>Create a directory in which to store our plots and move into it:</b></font>",
"_____no_output_____"
]
],
[
[
"plot_dir = f\"{path}/plots\"\nif not os.path.exists(plot_dir):\n os.makedirs(plot_dir)\nif os.path.exists(plot_dir):\n os.chdir(plot_dir)\nprint(f\"Current Working Directory: {os.getcwd()}\")",
"_____no_output_____"
]
],
[
[
"<font face='Calibri' size='3'><b>Load the stack produced by GIAnT and read it into an array so we can manipulate and display it:</b></font>",
"_____no_output_____"
]
],
[
[
"f = h5py.File(params_h5, 'r')",
"_____no_output_____"
]
],
[
[
"<font face='Calibri' size='3'><b>List all groups ('key's) within the HDF5 file that has been loaded into the object 'f'</b></font>",
"_____no_output_____"
]
],
[
[
"print(\"Keys: %s\" %f.keys())",
"_____no_output_____"
]
],
[
[
"<font face='Calibri' size='3'>Details on what each of these keys means can be found in the GIAnT documentation. For now, the only keys with which we are concerned are <b>'recons'</b> (the filtered time series of each pixel) and <b>'dates'</b> (the dates of acquisition). It is important to note that the dates are given in a type of Julian Day number called Rata Die number. This will have to be converted later, but this can easily be done via one of several different methods in Python.</font>\n<br><br>\n<font face='Calibri' size='3'><b>Get our data from the stack:</b></font>",
"_____no_output_____"
]
],
[
[
"data_cube = f['recons'][()]",
"_____no_output_____"
]
],
[
[
"<font face='Calibri' size='3'><b>Get the dates for each raster from the stack:</b></font>",
"_____no_output_____"
]
],
[
[
"dates = list(f['dates']) # these dates appear to be given in Rata Die style: floor(Julian Day Number - 1721424.5). \nif data_cube.shape[0] is not len(dates):\n print('Problem:')\n print('Number of rasters in data_cube: ',data_cube.shape[0])\n print('Number of dates: ',len(dates))",
"_____no_output_____"
]
],
[
[
"<font face='Calibri' size='3'><b>Plot and save amplitude image with transparency determined by alpha (SierraNegra-dBScaled-AmplitudeImage.png):</b></font>",
"_____no_output_____"
]
],
[
[
"plt.rcParams.update({'font.size': 14})\nradar_tiff = f\"{path}/20161119-20170106_amp.tiff\"\nradar=gdal.Open(radar_tiff)\nim_radar = radar.GetRasterBand(1).ReadAsArray()\nradar = None\ndbplot = np.ma.log10(im_radar)\nvmin=np.percentile(dbplot,3)\nvmax=np.percentile(dbplot,97)\nfig = plt.figure(figsize=(18,10)) # Initialize figure with a size\nax1 = fig.add_subplot(111) # 221 determines: 2 rows, 2 plots, first plot\nax1.imshow(dbplot, cmap='gray',vmin=vmin,vmax=vmax,alpha=1);\nplt.title('Example dB-scaled SAR Image for Ifgrm 20161119-20170106')\nplt.grid()\nplt.savefig('SierraNegra-dBScaled-AmplitudeImage.png',dpi=200,transparent='false')",
"_____no_output_____"
]
],
[
[
"<font face='Calibri' size='3'><b>Display and save an overlay of the clipped deformation map and amplitude image (SierraNegra-DeformationComposite.png):</b></font>",
"_____no_output_____"
]
],
[
[
"# We will define a short function that can plot an overaly of our radar image and deformation map. \ndef defNradar_plot(deformation, radar):\n fig = plt.figure(figsize=(18, 10))\n ax = fig.add_subplot(111)\n vmin = np.percentile(radar, 3)\n vmax = np.percentile(radar, 97)\n ax.imshow(radar, cmap='gray', vmin=vmin, vmax=vmax)\n fin_plot = ax.imshow(deformation, cmap='RdBu', vmin=-50.0, vmax=50.0, alpha=0.75)\n fig.colorbar(fin_plot, fraction=0.24, pad=0.02)\n ax.set(title=\"Integrated Defo [mm] Overlain on Clipped db-Scaled Amplitude Image\")\n plt.grid()\n \n# Get deformation map and radar image we wish to plot\ndeformation = data_cube[data_cube.shape[0]-1]\n\n# Call function to plot an overlay of our deformation map and radar image.\ndefNradar_plot(deformation, dbplot)\nplt.savefig('SierraNegra-DeformationComposite.png', dpi=200, transparent='false')",
"_____no_output_____"
]
],
[
[
"<font face='Calibri' size='3'><b>Convert from Rata Die number (similar to Julian Day number) contained in 'dates' to Gregorian date:</b></font>",
"_____no_output_____"
]
],
[
[
"tindex = []\nfor d in dates:\n tindex.append(date.fromordinal(int(d)))",
"_____no_output_____"
]
],
[
[
"<font face='Calibri' size='3'><b>Create an animation of the deformation</b></font>",
"_____no_output_____"
]
],
[
[
"%%capture\nfig = plt.figure(figsize=(14, 8))\nax = fig.add_subplot(111)\nax.axis('off')\nvmin=np.percentile(data_cube.flatten(), 5)\nvmax=np.percentile(data_cube.flatten(), 95)\n\n\nim = ax.imshow(data_cube[0], cmap='RdBu', vmin=-50.0, vmax=50.0)\nax.set_title(\"Animation of Deformation Time Series - Sierra Negra, Galapagos\")\nfig.colorbar(im)\nplt.grid()\n\ndef animate(i):\n ax.set_title(\"Date: {}\".format(tindex[i]))\n im.set_data(data_cube[i])\n \nani = matplotlib.animation.FuncAnimation(fig, animate, frames=data_cube.shape[0], interval=400)",
"_____no_output_____"
]
],
[
[
"<font face=\"Calibri\" size=\"3\"><b>Configure matplotlib's RC settings for the animation:</b></font> ",
"_____no_output_____"
]
],
[
[
"rc('animation', embed_limit=10.0**9)",
"_____no_output_____"
]
],
[
[
"<font face=\"Calibri\" size=\"3\"><b>Create a javascript animation of the time-series running inline in the notebook:</b></font> ",
"_____no_output_____"
]
],
[
[
"HTML(ani.to_jshtml())",
"_____no_output_____"
]
],
[
[
"<font face=\"Calibri\" size=\"3\"><b>Save the animation as a 'gif' file (SierraNegraDeformationTS.gif):</b></font> ",
"_____no_output_____"
]
],
[
[
"ani.save('SierraNegraDeformationTS.gif', writer='pillow', fps=2)",
"_____no_output_____"
]
],
[
[
"<font face='Calibri'><font size='5'><b>5. Alter the time filter parameter</b></font><br>\n <font size='3'>Looking at the video above, you may notice that the deformation has a very smoothed appearance. This may be because of our time filter which is currently set to 1 year ('filt=1.0' in the prepxml_SBAS.py code). Let's repeat the lab from there with 2 different time filters. <br>First, using no time filter ('filt=0.0') and then using a 1 month time filter ('filt=0.082'). Change the output file name for anything you want saved (e.g., 'SierraNegraDeformationTS.gif' to 'YourDesiredFileName.gif'). Otherwise, it will be overwritten. <br><br>How did these changes affect the output time series?<br>How might we figure out the right filter length?<br>What does this say about the parameters we select?",
"_____no_output_____"
],
[
"<font face='Calibri'><font size='5'><b>6. Clear data (optional)</b></font>\n <br>\n <font size='3'>This lab has produced a large quantity of data. If you look at this notebook in your home directory, it should now be ~13 MB. This can take a long time to load in a Jupyter Notebook. It may be useful to clear the cell outputs. <br>To clear the cell outputs, go Cell->All Output->Clear. This will clear the outputs of the Jupyter Notebook and restore it to its original size of ~60 kB. This will not delete any of the files we have created. </font>\n </font>",
"_____no_output_____"
],
[
"<font face=\"Calibri\" size=\"2\"> <i>GEOS 657-Lab9-InSARTimeSeriesAnalysis.ipynb - Version 1.2.0 - April 2021\n <br>\n <b>Version Changes:</b>\n <ul>\n <li>from osgeo import gdal</li>\n <li>namespace asf_notebook</li>\n </ul>\n </i>\n</font>",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
cbeddbaace31b8054ddeb0cd871df9dbc70649e8
| 181,554 |
ipynb
|
Jupyter Notebook
|
apartments_graz_predictions.ipynb
|
keller-kirill/graz-apartments-analytics
|
30153dd093a75cb95a253627f41584b7503b8c70
|
[
"MIT"
] | null | null | null |
apartments_graz_predictions.ipynb
|
keller-kirill/graz-apartments-analytics
|
30153dd093a75cb95a253627f41584b7503b8c70
|
[
"MIT"
] | null | null | null |
apartments_graz_predictions.ipynb
|
keller-kirill/graz-apartments-analytics
|
30153dd093a75cb95a253627f41584b7503b8c70
|
[
"MIT"
] | null | null | null | 95.605055 | 137,648 | 0.828095 |
[
[
[
"# Linear regression",
"_____no_output_____"
],
[
"## Problem\nBuild a model and predict pricing of the apartment rent in Graz based on data in the ad\n\n## Goals\n\n- Manually write linear regression algorithm\n - Gradient descent function\n - Cost function implementation\n - Normal equation function\n - Feature enumeration and normalization function\n- Use libraries and compare it with manual result\n\n## Data description\n\n| Feature | Variable Type | Variable | Value Type |\n|---------|--------------|---------------|------------|\n| Area | Objective Feature | area | float (square meters) |\n| Rooms number | Objective Feature | rooms | string |\n| Zip code | Objective Feature | zip | string |\n| District | Objective Feature | district | string |\n| Is the ad private| Objective Feature | is_private | boolean |\n| Is the flat in the city center | Objective Feature | center | boolean |\n| Pricing of the ad | Target Variable | price | float |",
"_____no_output_____"
],
[
"## Read data from pickle",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport matplotlib.pyplot as plt\nimport numpy as np\nimport matplotlib\n\n%matplotlib inline\n%config InlineBackend.figure_format = 'retina'\n\nmatplotlib.rcParams.update({'font.size': 16})\n\ndf = pd.read_pickle('apartmetns.pkl')",
"_____no_output_____"
]
],
[
[
"## Data selection",
"_____no_output_____"
]
],
[
[
"# We will make a copy of the dataset\nX = df.loc[:, ~df.columns.isin(['price', 'advertiser', 'link-href', 'is_private', 'zip', 'district'])]\ny = df['price']",
"_____no_output_____"
],
[
"X",
"_____no_output_____"
],
[
"y.head()",
"_____no_output_____"
]
],
[
[
"## Categorical features enumeration",
"_____no_output_____"
]
],
[
[
"def cats_to_codes(df, feature, ordered=None):\n return dict(df[feature].value_counts().astype('category').cat.codes)",
"_____no_output_____"
],
[
"def codes_to_cats(feature, code_dict):\n return {value:key for key, value in code_dict.items()}",
"_____no_output_____"
]
],
[
[
"zip_codes = cats_to_codes(X, 'zip')\nX.zip = X.zip.map(zip_codes)",
"_____no_output_____"
]
],
[
[
"X.head()",
"_____no_output_____"
],
[
"rooms_codes = cats_to_codes(X, 'rooms')\nX.rooms = X.rooms.map(rooms_codes)",
"/opt/miniconda3/envs/mlenv/lib/python3.8/site-packages/pandas/core/generic.py:5168: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\n self[name] = value\n"
],
[
"X.head()",
"_____no_output_____"
]
],
[
[
"district_codes = cats_to_codes(X, 'district')\nX.district = X.district.map(district_codes)",
"_____no_output_____"
]
],
[
[
"#X.is_private = X.is_private.astype('int')\nX.center = X.center.astype('int')",
"/opt/miniconda3/envs/mlenv/lib/python3.8/site-packages/pandas/core/generic.py:5168: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\n self[name] = value\n"
],
[
"X",
"_____no_output_____"
]
],
[
[
"## Feature normalization",
"_____no_output_____"
]
],
[
[
"mean_X = np.mean(X)\nstd_X = np.std(X)",
"_____no_output_____"
],
[
"def normalize(x, mean, std):\n return (x - mean)/std",
"_____no_output_____"
],
[
"for feat in X.columns:\n X[feat] = (X[feat] - mean_X[feat])/std_X[feat]",
"<ipython-input-14-3033a60703a8>:2: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\n X[feat] = (X[feat] - mean_X[feat])/std_X[feat]\n"
],
[
"X",
"_____no_output_____"
]
],
[
[
"## First hyphotesis\n$$\nh_\\theta(x) = \\theta_0 x_0 + \\theta_1 x_1 + \\theta_2 x_2 + \\theta_3 x_3 + \n\\theta_4 x_4 + \\theta_5 x_5 + \\theta_6 x_6 \n$$",
"_____no_output_____"
],
[
"### Adding of intercept term x0",
"_____no_output_____"
]
],
[
[
"X.insert(loc=0, column='x0', value=np.ones(len(X)))",
"_____no_output_____"
],
[
"X",
"_____no_output_____"
]
],
[
[
"### Conversion of X and y to numpy arrays ",
"_____no_output_____"
]
],
[
[
"X = X.to_numpy()\ny = y.to_numpy().reshape((-1, 1))",
"_____no_output_____"
],
[
"X.shape, y.shape",
"_____no_output_____"
],
[
"def computeCost(X, y, theta):\n m = len(y) \n J = 1/(2*m) * np.sum(np.power(np.subtract(X.dot(theta), y), 2))\n return J",
"_____no_output_____"
],
[
"theta = np.zeros((X.shape[1], 1))",
"_____no_output_____"
],
[
"theta.shape",
"_____no_output_____"
],
[
"computeCost(X, y, theta)",
"_____no_output_____"
],
[
"def gradientDescent(X, y, theta, alpha, num_iters):\n m = len(y)\n J_history = np.zeros((num_iters, 1))\n for i in range(num_iters):\n error = X.dot(theta) - y\n theta = theta - (alpha/m) * X.T.dot(error)\n J_history[i] = computeCost(X, y, theta)\n return theta, J_history",
"_____no_output_____"
],
[
"new_theta, J_history = gradientDescent(X, y, theta, 0.1, 100)",
"_____no_output_____"
],
[
"computeCost(X, y, new_theta)",
"_____no_output_____"
],
[
"plt.figure(figsize=(10,7));\n\nalphas = np.linspace(0.001, 0.3, 5)\nfor a in alphas:\n _, J_history = gradientDescent(X, y, theta, a, 50)\n plt.plot(np.arange(J_history.shape[0]), J_history, label=(r'$\\alpha$ = {:1.3f}'.format(a) \\\n + '\\n J = {}'.format(int(J_history[-1][0]))))\n\nplt.legend()\nplt.xlabel('Number of iterations');\nplt.ylabel(r'Cost function J($\\theta$)')\nplt.title(\"Gradient descent\");",
"_____no_output_____"
]
],
[
[
"### Quick test for hyphothesis",
"_____no_output_____"
]
],
[
[
"new_theta, J_history = gradientDescent(X, y, theta, 0.1, 100)",
"_____no_output_____"
],
[
"new_theta",
"_____no_output_____"
],
[
"mean_X, std_X",
"_____no_output_____"
],
[
"f = [44, 1, 1]\nx = []\n\nfor i in range(mean_X.shape[0]):\n x.append(normalize(f[i], mean_X[i], std_X[i]))",
"_____no_output_____"
],
[
"x = [1,] + x",
"_____no_output_____"
],
[
"x = np.array(x)",
"_____no_output_____"
],
[
"new_theta.T.dot(x)",
"_____no_output_____"
]
],
[
[
"Looks valid",
"_____no_output_____"
],
[
"## Model validation",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import train_test_split\ntrain_X, val_X, train_y, val_y = train_test_split(X, y, random_state=1)",
"_____no_output_____"
],
[
"def predict(X, theta):\n return X.dot(theta)",
"_____no_output_____"
],
[
"theta",
"_____no_output_____"
],
[
"new_theta, _ = gradientDescent(train_X, train_y, theta, 0.1, 100)",
"_____no_output_____"
],
[
"val_predictions = predict(val_X, new_theta)",
"_____no_output_____"
],
[
"print(val_predictions[:10])",
"[[ 768.22493417]\n [ 577.35782165]\n [ 851.11817124]\n [ 692.52226332]\n [1014.75103101]\n [ 448.74502025]\n [ 547.08664618]\n [1305.48778157]\n [ 653.0604925 ]\n [ 490.80205961]]\n"
],
[
"print(val_y[:10])",
"[[ 895. ]\n [ 550. ]\n [ 745. ]\n [ 728.43]\n [ 999.24]\n [ 400. ]\n [ 396.5 ]\n [1453.63]\n [ 545. ]\n [ 495. ]]\n"
]
],
[
[
"### Calculation of the Mean Absolute Error in Validation Data",
"_____no_output_____"
]
],
[
[
"from sklearn.metrics import mean_absolute_error\nval_mae = mean_absolute_error(val_y, val_predictions)",
"_____no_output_____"
],
[
"val_mae",
"_____no_output_____"
]
],
[
[
"## Normal equation",
"_____no_output_____"
],
[
"$$\n\\theta = (X^T X)^{-1}X^T \\bar{y}\n$$",
"_____no_output_____"
]
],
[
[
"def normalEqn(X, y):\n m = len(y)\n theta = np.linalg.pinv(X.T.dot(X)).dot(X.T).dot(y)\n return theta",
"_____no_output_____"
],
[
"norm_theta = normalEqn(X, y)",
"_____no_output_____"
]
],
[
[
"Should be more accurate",
"_____no_output_____"
]
],
[
[
"train_X, val_X, train_y, val_y = train_test_split(X, y, random_state=1)",
"_____no_output_____"
],
[
"val_predictions = predict(val_X, norm_theta)",
"_____no_output_____"
],
[
"val_mae = mean_absolute_error(val_y, val_predictions)\nval_mae",
"_____no_output_____"
]
],
[
[
"## Predicting prices usign scikit linear regressor",
"_____no_output_____"
]
],
[
[
"X",
"_____no_output_____"
],
[
"X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.25, random_state=1)",
"_____no_output_____"
],
[
"from sklearn.linear_model import LinearRegression\n\nregressor = LinearRegression(fit_intercept=False, n_jobs=-1)\nregressor.fit(X_train, y_train)",
"_____no_output_____"
],
[
"print('Weight coefficients: ', regressor.coef_)",
"Weight coefficients: [[697.93071978 177.18853113 -6.41221045 -6.80762635]]\n"
],
[
"y_pred_train = regressor.predict(X_test)",
"_____no_output_____"
],
[
"y_pred_train[0:10]",
"_____no_output_____"
],
[
"val_mae = mean_absolute_error(y_test, y_pred_train)\nval_mae",
"_____no_output_____"
],
[
"error = y_pred_train.sum() / y_test.sum() - 1",
"_____no_output_____"
],
[
"print(\"Percentage error: {:.2f}%\".format(error*100))",
"Percentage error: 0.83%\n"
],
[
"from sklearn.linear_model import RidgeCV",
"_____no_output_____"
],
[
"model = RidgeCV(fit_intercept=False, cv=5)",
"_____no_output_____"
],
[
"model.fit(X_train, y_train)",
"_____no_output_____"
],
[
"model.best_score_",
"_____no_output_____"
],
[
"model.alpha_",
"_____no_output_____"
],
[
"model.coef_",
"_____no_output_____"
],
[
"error = model.predict(X_test).sum() / y_test.sum() - 1\nprint(\"Percentage error: {:.2f}%\".format(error*100))",
"Percentage error: 0.79%\n"
],
[
"from sklearn.linear_model import LassoCV",
"_____no_output_____"
],
[
"model = LassoCV(fit_intercept=False, cv=5)\nmodel.fit(X_train, y_train.reshape(-1))",
"_____no_output_____"
],
[
"model.coef_",
"_____no_output_____"
],
[
"error = model.predict(X_test).sum() / y_test.sum() - 1\nprint(\"Percentage error: {:.2f}%\".format(error*100))",
"Percentage error: 0.73%\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbede37c8a4acef0b6efb6423bd5b2cf553d037c
| 21,012 |
ipynb
|
Jupyter Notebook
|
module-1/List-Comprehension/your-code/main.ipynb
|
gracegm/daft-miami-1019-labs
|
4627491e1772b0593574f03e928e53fc1d0170b2
|
[
"MIT"
] | null | null | null |
module-1/List-Comprehension/your-code/main.ipynb
|
gracegm/daft-miami-1019-labs
|
4627491e1772b0593574f03e928e53fc1d0170b2
|
[
"MIT"
] | null | null | null |
module-1/List-Comprehension/your-code/main.ipynb
|
gracegm/daft-miami-1019-labs
|
4627491e1772b0593574f03e928e53fc1d0170b2
|
[
"MIT"
] | null | null | null | 51.626536 | 3,281 | 0.605606 |
[
[
[
"# List Comprehensions\n\nComplete the following set of exercises to solidify your knowledge of list comprehensions.",
"_____no_output_____"
]
],
[
[
"import os;",
"_____no_output_____"
]
],
[
[
"#### 1. Use a list comprehension to create and print a list of consecutive integers starting with 1 and ending with 50.",
"_____no_output_____"
]
],
[
[
"lst = [i for i in range(1,51)]\nprint(lst)",
"[1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50]\n"
]
],
[
[
"#### 2. Use a list comprehension to create and print a list of even numbers starting with 2 and ending with 200.",
"_____no_output_____"
]
],
[
[
"lst = [i for i in range(2,202) if i >= 2]\nfor num in lst:\n if num % 2 == 0: \n print(num, end = \" \") ",
"2 4 6 8 10 12 14 16 18 20 22 24 26 28 30 32 34 36 38 40 42 44 46 48 50 52 54 56 58 60 62 64 66 68 70 72 74 76 78 80 82 84 86 88 90 92 94 96 98 100 102 104 106 108 110 112 114 116 118 120 122 124 126 128 130 132 134 136 138 140 142 144 146 148 150 152 154 156 158 160 162 164 166 168 170 172 174 176 178 180 182 184 186 188 190 192 194 196 198 200 "
]
],
[
[
"#### 3. Use a list comprehension to create and print a list containing all elements of the 10 x 4 array below.",
"_____no_output_____"
]
],
[
[
"a = [[0.84062117, 0.48006452, 0.7876326 , 0.77109654],\n [0.44409793, 0.09014516, 0.81835917, 0.87645456],\n [0.7066597 , 0.09610873, 0.41247947, 0.57433389],\n [0.29960807, 0.42315023, 0.34452557, 0.4751035 ],\n [0.17003563, 0.46843998, 0.92796258, 0.69814654],\n [0.41290051, 0.19561071, 0.16284783, 0.97016248],\n [0.71725408, 0.87702738, 0.31244595, 0.76615487],\n [0.20754036, 0.57871812, 0.07214068, 0.40356048],\n [0.12149553, 0.53222417, 0.9976855 , 0.12536346],\n [0.80930099, 0.50962849, 0.94555126, 0.33364763]];\n\nlst = []\n\n\nfor i in range(len(a[0])):\n lst_row = []\n\n for row in a:\n lst_row.append(row[i])\n lst.append(lst_row)\n\nprint(lst)\n\n",
"[[0.84062117, 0.44409793, 0.7066597, 0.29960807, 0.17003563, 0.41290051, 0.71725408, 0.20754036, 0.12149553, 0.80930099], [0.48006452, 0.09014516, 0.09610873, 0.42315023, 0.46843998, 0.19561071, 0.87702738, 0.57871812, 0.53222417, 0.50962849], [0.7876326, 0.81835917, 0.41247947, 0.34452557, 0.92796258, 0.16284783, 0.31244595, 0.07214068, 0.9976855, 0.94555126], [0.77109654, 0.87645456, 0.57433389, 0.4751035, 0.69814654, 0.97016248, 0.76615487, 0.40356048, 0.12536346, 0.33364763]]\n"
],
[
"a = [[0.84062117, 0.48006452, 0.7876326 , 0.77109654],\n [0.44409793, 0.09014516, 0.81835917, 0.87645456],\n [0.7066597 , 0.09610873, 0.41247947, 0.57433389],\n [0.29960807, 0.42315023, 0.34452557, 0.4751035 ],\n [0.17003563, 0.46843998, 0.92796258, 0.69814654],\n [0.41290051, 0.19561071, 0.16284783, 0.97016248],\n [0.71725408, 0.87702738, 0.31244595, 0.76615487],\n [0.20754036, 0.57871812, 0.07214068, 0.40356048],\n [0.12149553, 0.53222417, 0.9976855 , 0.12536346],\n [0.80930099, 0.50962849, 0.94555126, 0.33364763]];\n\na = [int(i) for i in a] \nprint (\"Modified list is : \" + str(a)) ",
"_____no_output_____"
]
],
[
[
"#### 4. Add a condition to the list comprehension above so that only values greater than or equal to 0.5 are printed.",
"_____no_output_____"
]
],
[
[
"lst = [[0.84062117, 0.44409793, 0.7066597, 0.29960807, 0.17003563, 0.41290051, 0.71725408, 0.20754036, 0.12149553, 0.80930099], [0.48006452, 0.09014516, 0.09610873, 0.42315023, 0.46843998, 0.19561071, 0.87702738, 0.57871812, 0.53222417, 0.50962849], [0.7876326, 0.81835917, 0.41247947, 0.34452557, 0.92796258, 0.16284783, 0.31244595, 0.07214068, 0.9976855, 0.94555126], [0.77109654, 0.87645456, 0.57433389, 0.4751035, 0.69814654, 0.97016248, 0.76615487, 0.40356048, 0.12536346, 0.33364763]]\n\nlst_new = [ x for x in range(lst) if x >= 0.5]\nprint(lst_new)\n",
"_____no_output_____"
],
[
"lst = [i for i in range(lst) if i >= 0.5]\nprint(lst)",
"_____no_output_____"
]
],
[
[
"#### 5. Use a list comprehension to create and print a list containing all elements of the 5 x 2 x 3 array below.",
"_____no_output_____"
]
],
[
[
"b = [[[0.55867166, 0.06210792, 0.08147297],\n [0.82579068, 0.91512478, 0.06833034]],\n\n [[0.05440634, 0.65857693, 0.30296619],\n [0.06769833, 0.96031863, 0.51293743]],\n\n [[0.09143215, 0.71893382, 0.45850679],\n [0.58256464, 0.59005654, 0.56266457]],\n\n [[0.71600294, 0.87392666, 0.11434044],\n [0.8694668 , 0.65669313, 0.10708681]],\n\n [[0.07529684, 0.46470767, 0.47984544],\n [0.65368638, 0.14901286, 0.23760688]]];\n\n\nlst_new = []\n\n\n# done in a long format-\n# for i in range(len(b[0])):\n# don't have to describe range with 0\n\nfor i in b:\n# lst_row = []\n for row in i:\n lst_new.append(row)\n# lst.append(lst_row)\n\nprint(lst_new)\n\n\n# or in a shorthand way-\n\nlst_new_2 = [row for i in b for row in i]\nprint(lst_new_2)",
"[[0.55867166, 0.06210792, 0.08147297], [0.82579068, 0.91512478, 0.06833034], [0.05440634, 0.65857693, 0.30296619], [0.06769833, 0.96031863, 0.51293743], [0.09143215, 0.71893382, 0.45850679], [0.58256464, 0.59005654, 0.56266457], [0.71600294, 0.87392666, 0.11434044], [0.8694668, 0.65669313, 0.10708681], [0.07529684, 0.46470767, 0.47984544], [0.65368638, 0.14901286, 0.23760688]]\n[[0.55867166, 0.06210792, 0.08147297], [0.82579068, 0.91512478, 0.06833034], [0.05440634, 0.65857693, 0.30296619], [0.06769833, 0.96031863, 0.51293743], [0.09143215, 0.71893382, 0.45850679], [0.58256464, 0.59005654, 0.56266457], [0.71600294, 0.87392666, 0.11434044], [0.8694668, 0.65669313, 0.10708681], [0.07529684, 0.46470767, 0.47984544], [0.65368638, 0.14901286, 0.23760688]]\n"
]
],
[
[
"#### 6. Add a condition to the list comprehension above so that the last value in each subarray is printed, but only if it is less than or equal to 0.5.",
"_____no_output_____"
]
],
[
[
"b = [[[0.55867166, 0.06210792, 0.08147297],\n [0.82579068, 0.91512478, 0.06833034]],\n\n [[0.05440634, 0.65857693, 0.30296619],\n [0.06769833, 0.96031863, 0.51293743]],\n\n [[0.09143215, 0.71893382, 0.45850679],\n [0.58256464, 0.59005654, 0.56266457]],\n\n [[0.71600294, 0.87392666, 0.11434044],\n [0.8694668 , 0.65669313, 0.10708681]],\n\n [[0.07529684, 0.46470767, 0.47984544],\n [0.65368638, 0.14901286, 0.23760688]]];\n\nfor x in b:\n for y in x:\n if y[-1] <= .5:\n print(y[-1])",
"0.08147297\n0.06833034\n0.30296619\n0.45850679\n0.11434044\n0.10708681\n0.47984544\n0.23760688\n"
]
],
[
[
"#### 7. Use a list comprehension to select and print the names of all CSV files in the */data* directory.",
"_____no_output_____"
]
],
[
[
"import csv \n\nimport os\n\n\nfile_list = [x for x in os.listdir('./data') if x.endswith('.csv')]\ndata_sets = [pd.read_csv(os.path.join('./data', x)) for x in file_list]\ndata = pd.concat(data_sets, axis=0)\n",
"_____no_output_____"
]
],
[
[
"### Bonus",
"_____no_output_____"
],
[
"Try to solve these katas using list comprehensions.",
"_____no_output_____"
],
[
"**Easy**\n- [Insert values](https://www.codewars.com/kata/invert-values)\n- [Sum Square(n)](https://www.codewars.com/kata/square-n-sum)\n- [Digitize](https://www.codewars.com/kata/digitize)\n- [List filtering](https://www.codewars.com/kata/list-filtering)\n- [Arithmetic list](https://www.codewars.com/kata/541da001259d9ca85d000688)\n\n**Medium**\n- [Multiples of 3 or 5](https://www.codewars.com/kata/514b92a657cdc65150000006)\n- [Count of positives / sum of negatives](https://www.codewars.com/kata/count-of-positives-slash-sum-of-negatives)\n- [Categorize new member](https://www.codewars.com/kata/5502c9e7b3216ec63c0001aa)\n\n**Advanced**\n- [Queue time counter](https://www.codewars.com/kata/queue-time-counter)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
cbede399dd127bc9229f7d84d5ab10e5285f0121
| 46,056 |
ipynb
|
Jupyter Notebook
|
attention/Attention_Basics_Solution.ipynb
|
prokokok/deep-learning-v2-pytorch
|
5f04298aab7d51873b59e8720122def5673f1815
|
[
"MIT"
] | null | null | null |
attention/Attention_Basics_Solution.ipynb
|
prokokok/deep-learning-v2-pytorch
|
5f04298aab7d51873b59e8720122def5673f1815
|
[
"MIT"
] | null | null | null |
attention/Attention_Basics_Solution.ipynb
|
prokokok/deep-learning-v2-pytorch
|
5f04298aab7d51873b59e8720122def5673f1815
|
[
"MIT"
] | null | null | null | 105.633028 | 11,300 | 0.867487 |
[
[
[
"# [SOLUTION] Attention Basics\nIn this notebook, we look at how attention is implemented. We will focus on implementing attention in isolation from a larger model. That's because when implementing attention in a real-world model, a lot of the focus goes into piping the data and juggling the various vectors rather than the concepts of attention themselves.\n\nWe will implement attention scoring as well as calculating an attention context vector.\n\n## Attention Scoring\n### Inputs to the scoring function\nLet's start by looking at the inputs we'll give to the scoring function. We will assume we're in the first step in the decoding phase. The first input to the scoring function is the hidden state of decoder (assuming a toy RNN with three hidden nodes -- not usable in real life, but easier to illustrate):",
"_____no_output_____"
]
],
[
[
"dec_hidden_state = [5,1,20]",
"_____no_output_____"
]
],
[
[
"Let's visualize this vector:",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport seaborn as sns\n\n# Let's visualize our decoder hidden state\nplt.figure(figsize=(1.5, 4.5))\nsns.heatmap(np.transpose(np.matrix(dec_hidden_state)), annot=True, cmap=sns.light_palette(\"purple\", as_cmap=True), linewidths=1)",
"_____no_output_____"
]
],
[
[
"Our first scoring function will score a single annotation (encoder hidden state), which looks like this:",
"_____no_output_____"
]
],
[
[
"annotation = [3,12,45] #e.g. Encoder hidden state",
"_____no_output_____"
],
[
"# Let's visualize the single annotation\nplt.figure(figsize=(1.5, 4.5))\nsns.heatmap(np.transpose(np.matrix(annotation)), annot=True, cmap=sns.light_palette(\"orange\", as_cmap=True), linewidths=1)",
"_____no_output_____"
]
],
[
[
"### IMPLEMENT: Scoring a Single Annotation\nLet's calculate the dot product of a single annotation. NumPy's [dot()](https://docs.scipy.org/doc/numpy/reference/generated/numpy.dot.html) is a good candidate for this operation",
"_____no_output_____"
]
],
[
[
"def single_dot_attention_score(dec_hidden_state, enc_hidden_state):\n # TODO: return the dot product of the two vectors\n return np.dot(dec_hidden_state, enc_hidden_state)\n \nsingle_dot_attention_score(dec_hidden_state, annotation)",
"_____no_output_____"
]
],
[
[
"\n### Annotations Matrix\nLet's now look at scoring all the annotations at once. To do that, here's our annotation matrix:",
"_____no_output_____"
]
],
[
[
"annotations = np.transpose([[3,12,45], [59,2,5], [1,43,5], [4,3,45.3]])",
"_____no_output_____"
]
],
[
[
"And it can be visualized like this (each column is a hidden state of an encoder time step):",
"_____no_output_____"
]
],
[
[
"# Let's visualize our annotation (each column is an annotation)\nax = sns.heatmap(annotations, annot=True, cmap=sns.light_palette(\"orange\", as_cmap=True), linewidths=1)",
"_____no_output_____"
]
],
[
[
"### IMPLEMENT: Scoring All Annotations at Once\nLet's calculate the scores of all the annotations in one step using matrix multiplication. Let's continue to us the dot scoring method\n\n<img src=\"images/scoring_functions.png\" />\n\nTo do that, we'll have to transpose `dec_hidden_state` and [matrix multiply](https://docs.scipy.org/doc/numpy/reference/generated/numpy.matmul.html) it with `annotations`.",
"_____no_output_____"
]
],
[
[
"def dot_attention_score(dec_hidden_state, annotations):\n # TODO: return the product of dec_hidden_state transpose and enc_hidden_states\n return np.matmul(np.transpose(dec_hidden_state), annotations)\n \nattention_weights_raw = dot_attention_score(dec_hidden_state, annotations)\nattention_weights_raw",
"_____no_output_____"
]
],
[
[
"Looking at these scores, can you guess which of the four vectors will get the most attention from the decoder at this time step?\n\n## Softmax\nNow that we have our scores, let's apply softmax:\n<img src=\"images/softmax.png\" />",
"_____no_output_____"
]
],
[
[
"def softmax(x):\n x = np.array(x, dtype=np.float128)\n e_x = np.exp(x)\n return e_x / e_x.sum(axis=0) \n\nattention_weights = softmax(attention_weights_raw)\nattention_weights",
"_____no_output_____"
]
],
[
[
"Even when knowing which annotation will get the most focus, it's interesting to see how drastic softmax makes the end score become. The first and last annotation had the respective scores of 927 and 929. But after softmax, the attention they'll get is 0.119 and 0.880 respectively.\n\n# Applying the scores back on the annotations\nNow that we have our scores, let's multiply each annotation by its score to proceed closer to the attention context vector. This is the multiplication part of this formula (we'll tackle the summation part in the latter cells)\n\n<img src=\"images/Context_vector.png\" />",
"_____no_output_____"
]
],
[
[
"def apply_attention_scores(attention_weights, annotations):\n # TODO: Multiple the annotations by their weights\n return attention_weights * annotations\n\napplied_attention = apply_attention_scores(attention_weights, annotations)\napplied_attention",
"_____no_output_____"
]
],
[
[
"Let's visualize how the context vector looks now that we've applied the attention scores back on it:",
"_____no_output_____"
]
],
[
[
"# Let's visualize our annotations after applying attention to them\nax = sns.heatmap(applied_attention, annot=True, cmap=sns.light_palette(\"orange\", as_cmap=True), linewidths=1)",
"_____no_output_____"
]
],
[
[
"Contrast this with the raw annotations visualized earlier in the notebook, and we can see that the second and third annotations (columns) have been nearly wiped out. The first annotation maintains some of its value, and the fourth annotation is the most pronounced.\n\n# Calculating the Attention Context Vector\nAll that remains to produce our attention context vector now is to sum up the four columns to produce a single attention context vector\n",
"_____no_output_____"
]
],
[
[
"def calculate_attention_vector(applied_attention):\n return np.sum(applied_attention, axis=1)\n\nattention_vector = calculate_attention_vector(applied_attention)\nattention_vector",
"_____no_output_____"
],
[
"# Let's visualize the attention context vector\nplt.figure(figsize=(1.5, 4.5))\nsns.heatmap(np.transpose(np.matrix(attention_vector)), annot=True, cmap=sns.light_palette(\"Blue\", as_cmap=True), linewidths=1)",
"_____no_output_____"
]
],
[
[
"Now that we have the context vector, we can concatenate it with the hidden state and pass it through a hidden layer to produce the the result of this decoding time step.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
cbedf7feb76a972e2fe356dc69a75646308a7b44
| 171,741 |
ipynb
|
Jupyter Notebook
|
Pre_trained_BERT_contextualized_word_embeddings.ipynb
|
Sudhir22/bert
|
b1e58d2a316d91f47530817780d1184b181b3953
|
[
"Apache-2.0"
] | null | null | null |
Pre_trained_BERT_contextualized_word_embeddings.ipynb
|
Sudhir22/bert
|
b1e58d2a316d91f47530817780d1184b181b3953
|
[
"Apache-2.0"
] | null | null | null |
Pre_trained_BERT_contextualized_word_embeddings.ipynb
|
Sudhir22/bert
|
b1e58d2a316d91f47530817780d1184b181b3953
|
[
"Apache-2.0"
] | null | null | null | 80.629577 | 2,096 | 0.556524 |
[
[
[
"<a href=\"https://colab.research.google.com/github/Sudhir22/bert/blob/master/Pre_trained_BERT_contextualized_word_embeddings.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"!rm -rf bert\n!git clone https://github.com/google-research/bert",
"Cloning into 'bert'...\nremote: Enumerating objects: 340, done.\u001b[K\nReceiving objects: 0% (1/340) \rReceiving objects: 1% (4/340) \rReceiving objects: 2% (7/340) \rReceiving objects: 3% (11/340) \rReceiving objects: 4% (14/340) \rReceiving objects: 5% (17/340) \rReceiving objects: 6% (21/340) \rReceiving objects: 7% (24/340) \rReceiving objects: 8% (28/340) \rReceiving objects: 9% (31/340) \rReceiving objects: 10% (34/340) \rReceiving objects: 11% (38/340) \rReceiving objects: 12% (41/340) \rReceiving objects: 13% (45/340) \rReceiving objects: 14% (48/340) \rReceiving objects: 15% (51/340) \rReceiving objects: 16% (55/340) \rReceiving objects: 17% (58/340) \rReceiving objects: 18% (62/340) \rReceiving objects: 19% (65/340) \rReceiving objects: 20% (68/340) \rReceiving objects: 21% (72/340) \rReceiving objects: 22% (75/340) \rReceiving objects: 23% (79/340) \rReceiving objects: 24% (82/340) \rReceiving objects: 25% (85/340) \rReceiving objects: 26% (89/340) \rReceiving objects: 27% (92/340) \rReceiving objects: 28% (96/340) \rReceiving objects: 29% (99/340) \rReceiving objects: 30% (102/340) \rReceiving objects: 31% (106/340) \rReceiving objects: 32% (109/340) \rReceiving objects: 33% (113/340) \rReceiving objects: 34% (116/340) \rReceiving objects: 35% (119/340) \rReceiving objects: 36% (123/340) \rReceiving objects: 37% (126/340) \rReceiving objects: 38% (130/340) \rReceiving objects: 39% (133/340) \rReceiving objects: 40% (136/340) \rReceiving objects: 41% (140/340) \rReceiving objects: 42% (143/340) \rReceiving objects: 43% (147/340) \rReceiving objects: 44% (150/340) \rReceiving objects: 45% (153/340) \rReceiving objects: 46% (157/340) \rReceiving objects: 47% (160/340) \rremote: Total 340 (delta 0), reused 0 (delta 0), pack-reused 340\u001b[K\nReceiving objects: 48% (164/340) \rReceiving objects: 49% (167/340) \rReceiving objects: 50% (170/340) \rReceiving objects: 51% (174/340) \rReceiving objects: 52% (177/340) \rReceiving objects: 53% (181/340) \rReceiving objects: 54% (184/340) \rReceiving objects: 55% (187/340) \rReceiving objects: 56% (191/340) \rReceiving objects: 57% (194/340) \rReceiving objects: 58% (198/340) \rReceiving objects: 59% (201/340) \rReceiving objects: 60% (204/340) \rReceiving objects: 61% (208/340) \rReceiving objects: 62% (211/340) \rReceiving objects: 63% (215/340) \rReceiving objects: 64% (218/340) \rReceiving objects: 65% (221/340) \rReceiving objects: 66% (225/340) \rReceiving objects: 67% (228/340) \rReceiving objects: 68% (232/340) \rReceiving objects: 69% (235/340) \rReceiving objects: 70% (238/340) \rReceiving objects: 71% (242/340) \rReceiving objects: 72% (245/340) \rReceiving objects: 73% (249/340) \rReceiving objects: 74% (252/340) \rReceiving objects: 75% (255/340) \rReceiving objects: 76% (259/340) \rReceiving objects: 77% (262/340) \rReceiving objects: 78% (266/340) \rReceiving objects: 79% (269/340) \rReceiving objects: 80% (272/340) \rReceiving objects: 81% (276/340) \rReceiving objects: 82% (279/340) \rReceiving objects: 83% (283/340) \rReceiving objects: 84% (286/340) \rReceiving objects: 85% (289/340) \rReceiving objects: 86% (293/340) \rReceiving objects: 87% (296/340) \rReceiving objects: 88% (300/340) \rReceiving objects: 89% (303/340) \rReceiving objects: 90% (306/340) \rReceiving objects: 91% (310/340) \rReceiving objects: 92% (313/340) \rReceiving objects: 93% (317/340) \rReceiving objects: 94% (320/340) \rReceiving objects: 95% (323/340) \rReceiving objects: 96% (327/340) \rReceiving objects: 97% (330/340) \rReceiving objects: 98% (334/340) \rReceiving objects: 99% (337/340) \rReceiving objects: 100% (340/340) \rReceiving objects: 100% (340/340), 300.28 KiB | 3.90 MiB/s, done.\nResolving deltas: 0% (0/185) \rResolving deltas: 1% (2/185) \rResolving deltas: 2% (5/185) \rResolving deltas: 3% (7/185) \rResolving deltas: 4% (9/185) \rResolving deltas: 6% (12/185) \rResolving deltas: 8% (15/185) \rResolving deltas: 10% (19/185) \rResolving deltas: 11% (22/185) \rResolving deltas: 20% (37/185) \rResolving deltas: 21% (39/185) \rResolving deltas: 30% (57/185) \rResolving deltas: 31% (59/185) \rResolving deltas: 40% (74/185) \rResolving deltas: 44% (82/185) \rResolving deltas: 45% (84/185) \rResolving deltas: 48% (89/185) \rResolving deltas: 90% (167/185) \rResolving deltas: 100% (185/185) \rResolving deltas: 100% (185/185), done.\n"
],
[
"%tensorflow_version 1.x",
"TensorFlow is already loaded. Please restart the runtime to change versions.\n"
],
[
"import sys\n\nsys.path.append('bert/')\n\nfrom __future__ import absolute_import\nfrom __future__ import division\nfrom __future__ import print_function\n\nimport codecs\nimport collections\nimport json\nimport re\nimport os\nimport pprint\nimport numpy as np\nimport tensorflow as tf\n\nimport modeling\nimport tokenization",
"_____no_output_____"
],
[
"tf.__version__",
"_____no_output_____"
],
[
"assert 'COLAB_TPU_ADDR' in os.environ, 'ERROR: Not connected to a TPU runtime; please see the first cell in this notebook for instructions!'\nTPU_ADDRESS = 'grpc://' + os.environ['COLAB_TPU_ADDR']\nprint('TPU address is', TPU_ADDRESS)\n\nfrom google.colab import auth\nauth.authenticate_user()\nwith tf.Session(TPU_ADDRESS) as session:\n print('TPU devices:')\n pprint.pprint(session.list_devices())\n\n # Upload credentials to TPU.\n with open('/content/adc.json', 'r') as f:\n auth_info = json.load(f)\n tf.contrib.cloud.configure_gcs(session, credentials=auth_info)\n # Now credentials are set for all future sessions on this TPU.",
"TPU address is grpc://10.73.133.122:8470\nTPU devices:\n[_DeviceAttributes(/job:tpu_worker/replica:0/task:0/device:CPU:0, CPU, -1, 4991885305906145687),\n _DeviceAttributes(/job:tpu_worker/replica:0/task:0/device:XLA_CPU:0, XLA_CPU, 17179869184, 15247344399970103192),\n _DeviceAttributes(/job:tpu_worker/replica:0/task:0/device:TPU:0, TPU, 17179869184, 1415904661961376636),\n _DeviceAttributes(/job:tpu_worker/replica:0/task:0/device:TPU:1, TPU, 17179869184, 10077710641892051921),\n _DeviceAttributes(/job:tpu_worker/replica:0/task:0/device:TPU:2, TPU, 17179869184, 14609532431765640584),\n _DeviceAttributes(/job:tpu_worker/replica:0/task:0/device:TPU:3, TPU, 17179869184, 13700206246418090601),\n _DeviceAttributes(/job:tpu_worker/replica:0/task:0/device:TPU:4, TPU, 17179869184, 6990872653339197783),\n _DeviceAttributes(/job:tpu_worker/replica:0/task:0/device:TPU:5, TPU, 17179869184, 10895862202158335490),\n _DeviceAttributes(/job:tpu_worker/replica:0/task:0/device:TPU:6, TPU, 17179869184, 14707221350351964306),\n _DeviceAttributes(/job:tpu_worker/replica:0/task:0/device:TPU:7, TPU, 17179869184, 6598222520871302844),\n _DeviceAttributes(/job:tpu_worker/replica:0/task:0/device:TPU_SYSTEM:0, TPU_SYSTEM, 8589934592, 8017795357732511371)]\nWARNING:tensorflow:\nThe TensorFlow contrib module will not be included in TensorFlow 2.0.\nFor more information, please see:\n * https://github.com/tensorflow/community/blob/master/rfcs/20180907-contrib-sunset.md\n * https://github.com/tensorflow/addons\n * https://github.com/tensorflow/io (for I/O related ops)\nIf you depend on functionality not listed there, please file an issue.\n\n"
],
[
"# Available pretrained model checkpoints:\n# uncased_L-12_H-768_A-12: uncased BERT base model\n# uncased_L-24_H-1024_A-16: uncased BERT large model\n# cased_L-12_H-768_A-12: cased BERT large model\nBERT_MODEL = 'multi_cased_L-12_H-768_A-12' #@param {type:\"string\"}\nBERT_PRETRAINED_DIR = 'gs://cloud-tpu-checkpoints/bert/' + BERT_MODEL\nprint('***** BERT pretrained directory: {} *****'.format(BERT_PRETRAINED_DIR))\n!gsutil ls $BERT_PRETRAINED_DIR",
"***** BERT pretrained directory: gs://cloud-tpu-checkpoints/bert/multi_cased_L-12_H-768_A-12 *****\nCommandException: One or more URLs matched no objects.\n"
],
[
"LAYERS = [-1,-2,-3,-4]\nNUM_TPU_CORES = 8\nMAX_SEQ_LENGTH = 87\nBERT_CONFIG = BERT_PRETRAINED_DIR + '/bert_config.json'\nCHKPT_DIR = BERT_PRETRAINED_DIR + '/bert_model.ckpt'\nVOCAB_FILE = BERT_PRETRAINED_DIR + '/vocab.txt'\nINIT_CHECKPOINT = BERT_PRETRAINED_DIR + '/bert_model.ckpt'\nBATCH_SIZE = 128",
"_____no_output_____"
],
[
"class InputExample(object):\n\n def __init__(self, unique_id, text_a, text_b=None):\n self.unique_id = unique_id\n self.text_a = text_a\n self.text_b = text_b\n",
"_____no_output_____"
],
[
"class InputFeatures(object):\n \"\"\"A single set of features of data.\"\"\"\n\n def __init__(self, unique_id, tokens, input_ids, input_mask, input_type_ids):\n self.unique_id = unique_id\n self.tokens = tokens\n self.input_ids = input_ids\n self.input_mask = input_mask\n self.input_type_ids = input_type_ids",
"_____no_output_____"
],
[
"def input_fn_builder(features, seq_length):\n \"\"\"Creates an `input_fn` closure to be passed to TPUEstimator.\"\"\"\n\n all_unique_ids = []\n all_input_ids = []\n all_input_mask = []\n all_input_type_ids = []\n\n for feature in features:\n all_unique_ids.append(feature.unique_id)\n all_input_ids.append(feature.input_ids)\n all_input_mask.append(feature.input_mask)\n all_input_type_ids.append(feature.input_type_ids)\n\n def input_fn(params):\n \"\"\"The actual input function.\"\"\"\n batch_size = params[\"batch_size\"]\n\n num_examples = len(features)\n\n # This is for demo purposes and does NOT scale to large data sets. We do\n # not use Dataset.from_generator() because that uses tf.py_func which is\n # not TPU compatible. The right way to load data is with TFRecordReader.\n d = tf.data.Dataset.from_tensor_slices({\n \"unique_ids\":\n tf.constant(all_unique_ids, shape=[num_examples], dtype=tf.int32),\n \"input_ids\":\n tf.constant(\n all_input_ids, shape=[num_examples, seq_length],\n dtype=tf.int32),\n \"input_mask\":\n tf.constant(\n all_input_mask,\n shape=[num_examples, seq_length],\n dtype=tf.int32),\n \"input_type_ids\":\n tf.constant(\n all_input_type_ids,\n shape=[num_examples, seq_length],\n dtype=tf.int32),\n })\n\n d = d.batch(batch_size=batch_size, drop_remainder=False)\n return d\n\n return input_fn",
"_____no_output_____"
],
[
"def model_fn_builder(bert_config, init_checkpoint, layer_indexes, use_tpu,\n use_one_hot_embeddings):\n \"\"\"Returns `model_fn` closure for TPUEstimator.\"\"\"\n\n def model_fn(features, labels, mode, params): # pylint: disable=unused-argument\n \"\"\"The `model_fn` for TPUEstimator.\"\"\"\n\n unique_ids = features[\"unique_ids\"]\n input_ids = features[\"input_ids\"]\n input_mask = features[\"input_mask\"]\n input_type_ids = features[\"input_type_ids\"]\n\n model = modeling.BertModel(\n config=bert_config,\n is_training=False,\n input_ids=input_ids,\n input_mask=input_mask,\n token_type_ids=input_type_ids,\n use_one_hot_embeddings=use_one_hot_embeddings)\n\n if mode != tf.estimator.ModeKeys.PREDICT:\n raise ValueError(\"Only PREDICT modes are supported: %s\" % (mode))\n\n tvars = tf.trainable_variables()\n scaffold_fn = None\n (assignment_map,\n initialized_variable_names) = modeling.get_assignment_map_from_checkpoint(\n tvars, init_checkpoint)\n if use_tpu:\n\n def tpu_scaffold():\n tf.train.init_from_checkpoint(init_checkpoint, assignment_map)\n return tf.train.Scaffold()\n\n scaffold_fn = tpu_scaffold\n else:\n tf.train.init_from_checkpoint(init_checkpoint, assignment_map)\n\n tf.logging.info(\"**** Trainable Variables ****\")\n for var in tvars:\n init_string = \"\"\n if var.name in initialized_variable_names:\n init_string = \", *INIT_FROM_CKPT*\"\n tf.logging.info(\" name = %s, shape = %s%s\", var.name, var.shape,\n init_string)\n\n all_layers = model.get_all_encoder_layers()\n\n predictions = {\n \"unique_id\": unique_ids,\n }\n\n for (i, layer_index) in enumerate(layer_indexes):\n predictions[\"layer_output_%d\" % i] = all_layers[layer_index]\n\n output_spec = tf.contrib.tpu.TPUEstimatorSpec(\n mode=mode, predictions=predictions, scaffold_fn=scaffold_fn)\n return output_spec\n\n return model_fn",
"_____no_output_____"
],
[
"def convert_examples_to_features(examples, seq_length, tokenizer):\n \"\"\"Loads a data file into a list of `InputBatch`s.\"\"\"\n\n features = []\n for (ex_index, example) in enumerate(examples):\n tokens_a = tokenizer.tokenize(example.text_a)\n\n tokens_b = None\n if example.text_b:\n tokens_b = tokenizer.tokenize(example.text_b)\n\n if tokens_b:\n # Modifies `tokens_a` and `tokens_b` in place so that the total\n # length is less than the specified length.\n # Account for [CLS], [SEP], [SEP] with \"- 3\"\n _truncate_seq_pair(tokens_a, tokens_b, seq_length - 3)\n else:\n # Account for [CLS] and [SEP] with \"- 2\"\n if len(tokens_a) > seq_length - 2:\n tokens_a = tokens_a[0:(seq_length - 2)]\n\n # The convention in BERT is:\n # (a) For sequence pairs:\n # tokens: [CLS] is this jack ##son ##ville ? [SEP] no it is not . [SEP]\n # type_ids: 0 0 0 0 0 0 0 0 1 1 1 1 1 1\n # (b) For single sequences:\n # tokens: [CLS] the dog is hairy . [SEP]\n # type_ids: 0 0 0 0 0 0 0\n #\n # Where \"type_ids\" are used to indicate whether this is the first\n # sequence or the second sequence. The embedding vectors for `type=0` and\n # `type=1` were learned during pre-training and are added to the wordpiece\n # embedding vector (and position vector). This is not *strictly* necessary\n # since the [SEP] token unambiguously separates the sequences, but it makes\n # it easier for the model to learn the concept of sequences.\n #\n # For classification tasks, the first vector (corresponding to [CLS]) is\n # used as as the \"sentence vector\". Note that this only makes sense because\n # the entire model is fine-tuned.\n tokens = []\n input_type_ids = []\n tokens.append(\"[CLS]\")\n input_type_ids.append(0)\n for token in tokens_a:\n tokens.append(token)\n input_type_ids.append(0)\n tokens.append(\"[SEP]\")\n input_type_ids.append(0)\n\n if tokens_b:\n for token in tokens_b:\n tokens.append(token)\n input_type_ids.append(1)\n tokens.append(\"[SEP]\")\n input_type_ids.append(1)\n\n input_ids = tokenizer.convert_tokens_to_ids(tokens)\n\n # The mask has 1 for real tokens and 0 for padding tokens. Only real\n # tokens are attended to.\n input_mask = [1] * len(input_ids)\n\n # Zero-pad up to the sequence length.\n while len(input_ids) < seq_length:\n input_ids.append(0)\n input_mask.append(0)\n input_type_ids.append(0)\n\n assert len(input_ids) == seq_length\n assert len(input_mask) == seq_length\n assert len(input_type_ids) == seq_length\n\n if ex_index < 5:\n tf.logging.info(\"*** Example ***\")\n tf.logging.info(\"unique_id: %s\" % (example.unique_id))\n tf.logging.info(\"tokens: %s\" % \" \".join(\n [tokenization.printable_text(x) for x in tokens]))\n tf.logging.info(\"input_ids: %s\" % \" \".join([str(x) for x in input_ids]))\n tf.logging.info(\"input_mask: %s\" % \" \".join([str(x) for x in input_mask]))\n tf.logging.info(\n \"input_type_ids: %s\" % \" \".join([str(x) for x in input_type_ids]))\n\n features.append(\n InputFeatures(\n unique_id=example.unique_id,\n tokens=tokens,\n input_ids=input_ids,\n input_mask=input_mask,\n input_type_ids=input_type_ids))\n return features",
"_____no_output_____"
],
[
"def _truncate_seq_pair(tokens_a, tokens_b, max_length):\n \"\"\"Truncates a sequence pair in place to the maximum length.\"\"\"\n\n # This is a simple heuristic which will always truncate the longer sequence\n # one token at a time. This makes more sense than truncating an equal percent\n # of tokens from each, since if one sequence is very short then each token\n # that's truncated likely contains more information than a longer sequence.\n while True:\n total_length = len(tokens_a) + len(tokens_b)\n if total_length <= max_length:\n break\n if len(tokens_a) > len(tokens_b):\n tokens_a.pop()\n else:\n tokens_b.pop()",
"_____no_output_____"
],
[
"def read_sequence(input_sentences):\n examples = []\n unique_id = 0\n for sentence in input_sentences:\n line = tokenization.convert_to_unicode(sentence)\n examples.append(InputExample(unique_id=unique_id, text_a=line))\n unique_id += 1\n return examples\n ",
"_____no_output_____"
],
[
"def get_features(input_text, dim=768):\n# tf.logging.set_verbosity(tf.logging.INFO)\n\n layer_indexes = LAYERS\n\n bert_config = modeling.BertConfig.from_json_file(BERT_CONFIG)\n\n tokenizer = tokenization.FullTokenizer(\n vocab_file=VOCAB_FILE, do_lower_case=True)\n\n is_per_host = tf.contrib.tpu.InputPipelineConfig.PER_HOST_V2\n tpu_cluster_resolver = tf.contrib.cluster_resolver.TPUClusterResolver(TPU_ADDRESS)\n run_config = tf.contrib.tpu.RunConfig(\n cluster=tpu_cluster_resolver,\n tpu_config=tf.contrib.tpu.TPUConfig(\n num_shards=NUM_TPU_CORES,\n per_host_input_for_training=is_per_host))\n\n examples = read_sequence(input_text)\n\n features = convert_examples_to_features(\n examples=examples, seq_length=MAX_SEQ_LENGTH, tokenizer=tokenizer)\n\n unique_id_to_feature = {}\n for feature in features:\n unique_id_to_feature[feature.unique_id] = feature\n\n model_fn = model_fn_builder(\n bert_config=bert_config,\n init_checkpoint=INIT_CHECKPOINT,\n layer_indexes=layer_indexes,\n use_tpu=True,\n use_one_hot_embeddings=True)\n\n # If TPU is not available, this will fall back to normal Estimator on CPU\n # or GPU.\n estimator = tf.contrib.tpu.TPUEstimator(\n use_tpu=True,\n model_fn=model_fn,\n config=run_config,\n predict_batch_size=BATCH_SIZE,\n train_batch_size=BATCH_SIZE)\n\n input_fn = input_fn_builder(\n features=features, seq_length=MAX_SEQ_LENGTH)\n\n # Get features\n for result in estimator.predict(input_fn, yield_single_examples=True):\n unique_id = int(result[\"unique_id\"])\n feature = unique_id_to_feature[unique_id]\n output = collections.OrderedDict()\n for (i, token) in enumerate(feature.tokens):\n layers = []\n for (j, layer_index) in enumerate(layer_indexes):\n layer_output = result[\"layer_output_%d\" % j]\n layer_output_flat = np.array([x for x in layer_output[i:(i + 1)].flat])\n layers.append(layer_output_flat)\n output[token] = sum(layers)[:dim]\n \n return output",
"_____no_output_____"
],
[
"embeddings = get_features([\"This is a test\"])\nprint(embeddings)",
"INFO:tensorflow:*** Example ***\nINFO:tensorflow:unique_id: 0\nINFO:tensorflow:tokens: [CLS] this is a test [SEP]\nINFO:tensorflow:input_ids: 101 2023 2003 1037 3231 102 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0\nINFO:tensorflow:input_mask: 1 1 1 1 1 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0\nINFO:tensorflow:input_type_ids: 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0\nWARNING:tensorflow:Estimator's model_fn (<function model_fn_builder.<locals>.model_fn at 0x7f34d72d1840>) includes params argument, but params are not passed to Estimator.\nWARNING:tensorflow:Using temporary folder as model directory: /tmp/tmpiyk27bc2\nINFO:tensorflow:Using config: {'_model_dir': '/tmp/tmpiyk27bc2', '_tf_random_seed': None, '_save_summary_steps': 100, '_save_checkpoints_steps': None, '_save_checkpoints_secs': 600, '_session_config': allow_soft_placement: true\ncluster_def {\n job {\n name: \"worker\"\n tasks {\n key: 0\n value: \"10.73.133.122:8470\"\n }\n }\n}\nisolate_session_state: true\n, '_keep_checkpoint_max': 5, '_keep_checkpoint_every_n_hours': 10000, '_log_step_count_steps': None, '_train_distribute': None, '_device_fn': None, '_protocol': None, '_eval_distribute': None, '_experimental_distribute': None, '_experimental_max_worker_delay_secs': None, '_session_creation_timeout_secs': 7200, '_service': None, '_cluster_spec': <tensorflow.python.training.server_lib.ClusterSpec object at 0x7f34d6c65908>, '_task_type': 'worker', '_task_id': 0, '_global_id_in_cluster': 0, '_master': 'grpc://10.73.133.122:8470', '_evaluation_master': 'grpc://10.73.133.122:8470', '_is_chief': True, '_num_ps_replicas': 0, '_num_worker_replicas': 1, '_tpu_config': TPUConfig(iterations_per_loop=2, num_shards=8, num_cores_per_replica=None, per_host_input_for_training=3, tpu_job_name=None, initial_infeed_sleep_secs=None, input_partition_dims=None, eval_training_input_configuration=2, experimental_host_call_every_n_steps=1), '_cluster': <tensorflow.python.distribute.cluster_resolver.tpu_cluster_resolver.TPUClusterResolver object at 0x7f34d717b898>}\nINFO:tensorflow:_TPUContext: eval_on_tpu True\nINFO:tensorflow:Could not find trained model in model_dir: /tmp/tmpiyk27bc2, running initialization to predict.\nINFO:tensorflow:Querying Tensorflow master (grpc://10.73.133.122:8470) for TPU system metadata.\nINFO:tensorflow:Found TPU system:\nINFO:tensorflow:*** Num TPU Cores: 8\nINFO:tensorflow:*** Num TPU Workers: 1\nINFO:tensorflow:*** Num TPU Cores Per Worker: 8\nINFO:tensorflow:*** Available Device: _DeviceAttributes(/job:worker/replica:0/task:0/device:CPU:0, CPU, -1, 4991885305906145687)\nINFO:tensorflow:*** Available Device: _DeviceAttributes(/job:worker/replica:0/task:0/device:TPU:0, TPU, 17179869184, 1415904661961376636)\nINFO:tensorflow:*** Available Device: _DeviceAttributes(/job:worker/replica:0/task:0/device:TPU:1, TPU, 17179869184, 10077710641892051921)\nINFO:tensorflow:*** Available Device: _DeviceAttributes(/job:worker/replica:0/task:0/device:TPU:2, TPU, 17179869184, 14609532431765640584)\nINFO:tensorflow:*** Available Device: _DeviceAttributes(/job:worker/replica:0/task:0/device:TPU:3, TPU, 17179869184, 13700206246418090601)\nINFO:tensorflow:*** Available Device: _DeviceAttributes(/job:worker/replica:0/task:0/device:TPU:4, TPU, 17179869184, 6990872653339197783)\nINFO:tensorflow:*** Available Device: _DeviceAttributes(/job:worker/replica:0/task:0/device:TPU:5, TPU, 17179869184, 10895862202158335490)\nINFO:tensorflow:*** Available Device: _DeviceAttributes(/job:worker/replica:0/task:0/device:TPU:6, TPU, 17179869184, 14707221350351964306)\nINFO:tensorflow:*** Available Device: _DeviceAttributes(/job:worker/replica:0/task:0/device:TPU:7, TPU, 17179869184, 6598222520871302844)\nINFO:tensorflow:*** Available Device: _DeviceAttributes(/job:worker/replica:0/task:0/device:TPU_SYSTEM:0, TPU_SYSTEM, 8589934592, 8017795357732511371)\nINFO:tensorflow:*** Available Device: _DeviceAttributes(/job:worker/replica:0/task:0/device:XLA_CPU:0, XLA_CPU, 17179869184, 15247344399970103192)\nINFO:tensorflow:Calling model_fn.\nWARNING:tensorflow:Entity <function _InputsWithStoppingSignals.insert_stopping_signal.<locals>._map_fn at 0x7f34d7300c80> could not be transformed and will be executed as-is. Please report this to the AutoGraph team. When filing the bug, set the verbosity to 10 (on Linux, `export AUTOGRAPH_VERBOSITY=10`) and attach the full output. Cause: module 'gast' has no attribute 'Num'\nWARNING: Entity <function _InputsWithStoppingSignals.insert_stopping_signal.<locals>._map_fn at 0x7f34d7300c80> could not be transformed and will be executed as-is. Please report this to the AutoGraph team. When filing the bug, set the verbosity to 10 (on Linux, `export AUTOGRAPH_VERBOSITY=10`) and attach the full output. Cause: module 'gast' has no attribute 'Num'\nWARNING:tensorflow:Entity <function _InputsWithStoppingSignals.insert_stopping_signal.<locals>._map_fn at 0x7f34d730fbf8> could not be transformed and will be executed as-is. Please report this to the AutoGraph team. When filing the bug, set the verbosity to 10 (on Linux, `export AUTOGRAPH_VERBOSITY=10`) and attach the full output. Cause: module 'gast' has no attribute 'Num'\nWARNING: Entity <function _InputsWithStoppingSignals.insert_stopping_signal.<locals>._map_fn at 0x7f34d730fbf8> could not be transformed and will be executed as-is. Please report this to the AutoGraph team. When filing the bug, set the verbosity to 10 (on Linux, `export AUTOGRAPH_VERBOSITY=10`) and attach the full output. Cause: module 'gast' has no attribute 'Num'\nWARNING:tensorflow:Entity <function _InputsWithStoppingSignals.__init__.<locals>._set_mask at 0x7f34d75d4378> could not be transformed and will be executed as-is. Please report this to the AutoGraph team. When filing the bug, set the verbosity to 10 (on Linux, `export AUTOGRAPH_VERBOSITY=10`) and attach the full output. Cause: module 'gast' has no attribute 'Num'\nWARNING: Entity <function _InputsWithStoppingSignals.__init__.<locals>._set_mask at 0x7f34d75d4378> could not be transformed and will be executed as-is. Please report this to the AutoGraph team. When filing the bug, set the verbosity to 10 (on Linux, `export AUTOGRAPH_VERBOSITY=10`) and attach the full output. Cause: module 'gast' has no attribute 'Num'\nINFO:tensorflow:**** Trainable Variables ****\nINFO:tensorflow: name = bert/embeddings/word_embeddings:0, shape = (30522, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/embeddings/token_type_embeddings:0, shape = (2, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/embeddings/position_embeddings:0, shape = (512, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/embeddings/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/embeddings/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_0/attention/self/query/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_0/attention/self/query/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_0/attention/self/key/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_0/attention/self/key/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_0/attention/self/value/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_0/attention/self/value/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_0/attention/output/dense/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_0/attention/output/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_0/attention/output/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_0/attention/output/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_0/intermediate/dense/kernel:0, shape = (768, 3072), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_0/intermediate/dense/bias:0, shape = (3072,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_0/output/dense/kernel:0, shape = (3072, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_0/output/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_0/output/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_0/output/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_1/attention/self/query/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_1/attention/self/query/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_1/attention/self/key/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_1/attention/self/key/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_1/attention/self/value/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_1/attention/self/value/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_1/attention/output/dense/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_1/attention/output/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_1/attention/output/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_1/attention/output/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_1/intermediate/dense/kernel:0, shape = (768, 3072), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_1/intermediate/dense/bias:0, shape = (3072,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_1/output/dense/kernel:0, shape = (3072, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_1/output/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_1/output/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_1/output/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_2/attention/self/query/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_2/attention/self/query/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_2/attention/self/key/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_2/attention/self/key/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_2/attention/self/value/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_2/attention/self/value/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_2/attention/output/dense/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_2/attention/output/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_2/attention/output/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_2/attention/output/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_2/intermediate/dense/kernel:0, shape = (768, 3072), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_2/intermediate/dense/bias:0, shape = (3072,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_2/output/dense/kernel:0, shape = (3072, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_2/output/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_2/output/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_2/output/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_3/attention/self/query/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_3/attention/self/query/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_3/attention/self/key/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_3/attention/self/key/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_3/attention/self/value/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_3/attention/self/value/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_3/attention/output/dense/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_3/attention/output/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_3/attention/output/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_3/attention/output/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_3/intermediate/dense/kernel:0, shape = (768, 3072), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_3/intermediate/dense/bias:0, shape = (3072,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_3/output/dense/kernel:0, shape = (3072, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_3/output/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_3/output/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_3/output/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_4/attention/self/query/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_4/attention/self/query/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_4/attention/self/key/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_4/attention/self/key/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_4/attention/self/value/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_4/attention/self/value/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_4/attention/output/dense/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_4/attention/output/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_4/attention/output/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_4/attention/output/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_4/intermediate/dense/kernel:0, shape = (768, 3072), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_4/intermediate/dense/bias:0, shape = (3072,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_4/output/dense/kernel:0, shape = (3072, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_4/output/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_4/output/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_4/output/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_5/attention/self/query/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_5/attention/self/query/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_5/attention/self/key/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_5/attention/self/key/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_5/attention/self/value/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_5/attention/self/value/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_5/attention/output/dense/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_5/attention/output/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_5/attention/output/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_5/attention/output/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_5/intermediate/dense/kernel:0, shape = (768, 3072), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_5/intermediate/dense/bias:0, shape = (3072,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_5/output/dense/kernel:0, shape = (3072, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_5/output/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_5/output/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_5/output/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_6/attention/self/query/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_6/attention/self/query/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_6/attention/self/key/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_6/attention/self/key/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_6/attention/self/value/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_6/attention/self/value/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_6/attention/output/dense/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_6/attention/output/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_6/attention/output/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_6/attention/output/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_6/intermediate/dense/kernel:0, shape = (768, 3072), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_6/intermediate/dense/bias:0, shape = (3072,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_6/output/dense/kernel:0, shape = (3072, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_6/output/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_6/output/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_6/output/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_7/attention/self/query/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_7/attention/self/query/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_7/attention/self/key/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_7/attention/self/key/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_7/attention/self/value/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_7/attention/self/value/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_7/attention/output/dense/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_7/attention/output/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_7/attention/output/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_7/attention/output/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_7/intermediate/dense/kernel:0, shape = (768, 3072), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_7/intermediate/dense/bias:0, shape = (3072,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_7/output/dense/kernel:0, shape = (3072, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_7/output/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_7/output/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_7/output/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_8/attention/self/query/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_8/attention/self/query/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_8/attention/self/key/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_8/attention/self/key/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_8/attention/self/value/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_8/attention/self/value/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_8/attention/output/dense/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_8/attention/output/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_8/attention/output/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_8/attention/output/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_8/intermediate/dense/kernel:0, shape = (768, 3072), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_8/intermediate/dense/bias:0, shape = (3072,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_8/output/dense/kernel:0, shape = (3072, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_8/output/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_8/output/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_8/output/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_9/attention/self/query/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_9/attention/self/query/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_9/attention/self/key/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_9/attention/self/key/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_9/attention/self/value/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_9/attention/self/value/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_9/attention/output/dense/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_9/attention/output/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_9/attention/output/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_9/attention/output/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_9/intermediate/dense/kernel:0, shape = (768, 3072), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_9/intermediate/dense/bias:0, shape = (3072,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_9/output/dense/kernel:0, shape = (3072, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_9/output/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_9/output/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_9/output/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_10/attention/self/query/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_10/attention/self/query/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_10/attention/self/key/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_10/attention/self/key/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_10/attention/self/value/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_10/attention/self/value/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_10/attention/output/dense/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_10/attention/output/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_10/attention/output/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_10/attention/output/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_10/intermediate/dense/kernel:0, shape = (768, 3072), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_10/intermediate/dense/bias:0, shape = (3072,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_10/output/dense/kernel:0, shape = (3072, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_10/output/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_10/output/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_10/output/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_11/attention/self/query/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_11/attention/self/query/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_11/attention/self/key/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_11/attention/self/key/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_11/attention/self/value/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_11/attention/self/value/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_11/attention/output/dense/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_11/attention/output/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_11/attention/output/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_11/attention/output/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_11/intermediate/dense/kernel:0, shape = (768, 3072), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_11/intermediate/dense/bias:0, shape = (3072,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_11/output/dense/kernel:0, shape = (3072, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_11/output/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_11/output/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_11/output/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/pooler/dense/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/pooler/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow:Done calling model_fn.\nINFO:tensorflow:TPU job name worker\nINFO:tensorflow:Graph was finalized.\nINFO:tensorflow:Running local_init_op.\nINFO:tensorflow:Done running local_init_op.\nINFO:tensorflow:Init TPU system\nINFO:tensorflow:Initialized TPU in 8 seconds\nINFO:tensorflow:Starting infeed thread controller.\nINFO:tensorflow:Starting outfeed thread controller.\nINFO:tensorflow:Initialized dataset iterators in 0 seconds\nINFO:tensorflow:Enqueue next (1) batch(es) of data to infeed.\nINFO:tensorflow:Dequeue next (1) batch(es) of data from outfeed.\nINFO:tensorflow:Outfeed finished for iteration (0, 0)\nINFO:tensorflow:Enqueue next (1) batch(es) of data to infeed.\nINFO:tensorflow:Dequeue next (1) batch(es) of data from outfeed.\nINFO:tensorflow:Stop infeed thread controller\nINFO:tensorflow:Shutting down InfeedController thread.\nINFO:tensorflow:InfeedController received shutdown signal, stopping.\nINFO:tensorflow:Infeed thread finished, shutting down.\nINFO:tensorflow:infeed marked as finished\nINFO:tensorflow:Stop output thread controller\nINFO:tensorflow:Shutting down OutfeedController thread.\nINFO:tensorflow:OutfeedController received shutdown signal, stopping.\nINFO:tensorflow:Outfeed thread finished, shutting down.\nINFO:tensorflow:outfeed marked as finished\nINFO:tensorflow:Shutdown TPU system.\nINFO:tensorflow:prediction_loop marked as finished\nINFO:tensorflow:prediction_loop marked as finished\nOrderedDict([('[CLS]', array([-5.87150991e-01, 9.16414261e-01, -1.34775972e+00, -1.99347770e+00,\n -2.04564190e+00, -2.73920655e+00, 1.31017947e+00, 8.94557774e-01,\n 1.79879177e+00, -2.54441857e+00, -2.46549666e-01, -3.13557088e-01,\n -2.10572779e-01, 1.60245299e-02, -1.53711855e-01, -1.28177464e+00,\n -3.68787289e-01, 1.53126359e+00, -1.05414271e-01, -1.94623184e+00,\n -1.86052239e+00, -6.47736847e-01, -1.85206962e+00, -2.37619138e+00,\n 4.65067595e-01, -1.45155859e+00, -1.20392752e+00, -1.20901018e-01,\n -3.85949314e-02, 1.58377898e+00, 2.11463881e+00, 7.57320523e-01,\n -1.87653315e+00, 5.32531500e-01, 1.46711528e+00, 7.95631558e-02,\n -7.32481003e-01, -6.99452311e-02, -1.80791259e+00, 1.15414321e+00,\n -2.03683305e+00, -4.20604467e-01, 5.72056234e-01, -1.25879240e+00,\n -1.63536429e+00, -5.64718902e-01, -1.66696320e+01, 1.14634621e+00,\n 1.10821891e+00, -3.39787817e+00, -1.39261246e-01, 8.68356705e-01,\n 7.90362179e-01, 2.81426692e+00, 2.46805644e+00, 1.03919315e+00,\n -1.41334021e+00, -2.75762588e-01, -1.91190541e+00, -6.29536867e-01,\n -1.14416265e+00, 2.62927294e-01, 2.39279941e-01, -4.67865586e-01,\n 2.06246272e-01, 1.43070364e+00, 1.89287961e+00, 3.52403045e-01,\n -2.00437260e+00, 2.56410456e+00, -3.03069162e+00, 1.23118234e+00,\n 4.33506608e-01, 1.06575966e+00, -8.21534991e-02, -7.49483705e-04,\n -5.97475529e-01, 8.12771022e-02, -9.45651889e-01, 7.65816197e-02,\n -7.06242025e-01, 1.81753516e+00, 5.94962597e-01, -1.01607096e+00,\n 6.63127363e-01, 4.26659763e-01, -1.36161375e+00, -3.41837263e+00,\n -7.14064598e-01, 1.50662076e+00, -7.77077496e-01, 2.04711914e+00,\n 2.60857433e-01, 1.95579231e-01, 1.31602240e+00, -7.36657143e-01,\n -3.84083390e-01, -1.18201399e+00, 2.22382832e+00, -4.96253431e-01,\n -9.52993870e-01, -1.61791992e+00, -4.79695648e-01, -1.23452127e+00,\n -6.68061614e-01, 1.76494401e-02, -1.78336859e-01, 1.60516691e+00,\n 1.73195362e-01, -2.78933644e+00, 1.93503714e+00, 1.97558105e-01,\n -8.25254992e-02, -1.86251903e+00, -1.77632785e+00, 3.84783030e+00,\n 1.17947960e+00, 1.70811415e+00, 2.17735600e+00, -1.59811664e+00,\n -2.54235172e+00, -3.42736185e-01, -4.91923034e-01, -5.04935443e-01,\n 2.92190742e+00, 9.53414977e-01, 5.41207850e-01, -1.16975343e+00,\n 4.48427141e-01, 1.49893987e+00, 1.71559918e+00, 1.33158290e+00,\n 1.40566087e+00, 2.95115381e-01, -1.89850450e-01, 1.65237498e+00,\n 9.33347106e-01, -1.83930135e+00, -2.27226090e+00, 3.07688117e-01,\n 9.80433643e-01, 3.53047132e-01, -6.19372272e+00, 1.07157660e+00,\n 2.25400829e+00, -6.23895049e-01, -2.09311032e+00, -7.60081470e-01,\n 1.25285935e+00, -6.03778124e-01, 1.76023602e-01, 1.69679737e+00,\n -1.87517488e+00, 1.24070382e+00, -5.26095748e-01, -2.20557070e+00,\n -8.12207043e-01, -1.45132875e+00, 2.01579094e+00, 1.45263577e+00,\n -8.89456391e-01, -9.39521909e-01, 3.15750170e+00, -1.83740807e+00,\n 3.26807320e-01, -1.52365005e+00, 1.33625638e+00, 2.84647369e+00,\n 1.26503944e+00, -2.50071430e+00, -1.17745888e+00, 4.30200249e-02,\n 2.80416393e+00, -6.40816927e-01, -4.34664667e-01, 5.36592603e-01,\n 6.65054560e-01, 1.43984675e+00, 5.31069756e-01, 1.30182302e+00,\n -7.47437358e-01, 9.99816895e-01, 3.12953174e-01, -6.13616168e-01,\n 1.76342523e+00, -1.26698554e-01, 1.18118894e+00, -1.58426535e+00,\n -5.08319587e-02, 1.46625745e+00, -4.60047722e-01, 1.00126338e+00,\n 4.11811203e-01, -2.85992789e+00, 1.65475082e+00, -8.68700445e-01,\n -5.47131360e-01, -1.26741755e+00, 1.06084883e+00, -4.36745018e-01,\n -3.48072976e-01, 6.28414333e-01, -3.14913130e+00, -1.11696053e+00,\n 5.67448795e-01, 1.07202044e+01, 1.12433171e+00, -1.77822217e-01,\n 2.07338715e+00, 1.65330732e+00, -1.53731608e+00, 2.10988736e+00,\n -2.27002501e+00, -1.37826586e+00, -3.62643242e-01, 1.01898634e+00,\n -3.61781418e-02, 2.48204991e-01, 6.72576249e-01, 1.51021600e+00,\n 1.69167805e+00, 1.61153829e+00, -2.96090245e-01, -6.52378917e-01,\n -1.63595295e+00, 7.87445545e-01, 1.65368819e+00, 6.46828532e-01,\n 1.00242138e+00, -8.27019024e+00, 4.86654639e-01, 8.68731797e-01,\n -1.27056766e+00, 1.12306952e+00, -7.74196386e-01, -1.54052401e+00,\n 2.26118040e+00, 6.68719858e-02, 1.57762277e+00, -8.89992535e-01,\n -1.19809794e+00, 1.96297276e+00, 2.13182497e+00, 2.04498529e+00,\n 1.77054095e+00, 1.21573758e+00, 1.63002992e+00, -6.84165657e-01,\n 5.68361104e-01, -5.27800202e-01, 1.38243675e+00, -2.41504645e+00,\n 2.40301549e-01, -1.43389678e+00, -2.71262360e+00, 3.57950926e-01,\n -8.15853298e-01, -1.11494744e+00, -9.39794779e-01, 3.73668462e-01,\n -6.39853776e-01, 5.69956899e-01, -6.51559532e-02, 2.62185931e+00,\n -9.96891439e-01, -4.32568461e-01, 8.87186825e-01, -6.18574083e-01,\n 3.19231844e+00, -1.42100954e+00, -1.26737761e+00, -8.75054359e-01,\n -1.74383068e+00, -4.91304827e+00, 1.83709550e+00, -5.92774451e-01,\n 1.56971717e+00, 3.34292746e+00, 1.53008533e+00, 2.53206849e-01,\n -2.26664636e-02, -9.53170300e-01, -2.25510311e+00, 8.32451701e-01,\n 3.58676529e+00, 7.03689575e-01, 8.22514176e-01, -3.34593201e+00,\n 2.23384356e+00, 2.14299262e-01, -2.21936679e+00, 3.77049059e-01,\n -7.15882421e-01, 9.29809332e-01, 4.13222599e+00, -1.27233124e+00,\n 1.58235598e+00, 6.05476141e-01, -9.50400710e-01, -3.26015902e+00,\n -9.44414377e-01, 1.05015469e+00, 5.28346956e-01, -1.24074960e+00,\n -9.67255414e-01, 1.18260361e-01, -9.20743465e-01, -1.49569392e+00,\n -3.63394318e+01, 2.24724621e-01, 2.98433542e-01, -1.41793871e+00,\n -3.41986597e-01, -2.55600858e+00, 4.11405385e-01, 6.75159812e-01,\n -1.01440825e-01, -2.16468751e-01, 2.88538963e-01, -1.21148705e+00,\n -1.48365223e+00, -6.48296237e-01, 1.23553562e+00, -2.26408172e+00,\n 1.63568437e+00, 1.43106833e-01, 8.60240579e-01, -6.42487526e-01,\n -2.01840067e+00, 3.00677329e-01, 6.36345744e-02, 1.29079968e-01,\n -5.04900515e-01, -8.70016336e-01, -2.41178989e+00, 5.09477615e-01,\n 1.61661232e+00, -1.40061605e+00, -9.39925730e-01, -1.66604090e+00,\n -7.04529881e-01, -1.12063146e+00, -2.64631987e+00, -1.76401901e+00,\n 4.82527643e-01, -4.69870627e-01, 3.71675944e+00, -4.39115077e-01,\n -4.61548895e-01, 3.72160584e-01, 1.80848765e+00, 1.18305993e+00,\n 3.74418229e-01, -3.26664120e-01, 3.32347512e-01, 5.17988563e-01,\n 5.83628297e-01, 8.17077875e-01, -2.54582167e-02, 1.54723488e-02,\n 5.71445084e+00, -2.34178543e+00, 1.85085344e+00, -6.32615030e-01,\n 2.25359774e+00, 1.52598834e+00, 1.79935801e+00, -1.11094201e+00,\n 2.66553736e+00, -6.94558442e-01, 2.51942921e+00, 3.98118615e-01,\n -2.68395066e-01, -9.69636679e-01, 1.65088773e+00, -9.17464614e-01,\n -3.73154134e-01, -7.29087651e-01, 1.20044971e+00, 1.36977756e+00,\n 9.10005689e-01, -1.15775700e+01, -2.11749816e+00, 1.73276082e-01,\n -1.46296573e+00, 6.18114829e-01, 1.79159737e+00, -4.46071059e-01,\n -1.79424274e+00, -1.10926199e+00, -1.50003481e+00, -3.62349421e-01,\n -1.33447945e+00, -4.13753361e-01, -1.35755822e-01, -2.99708247e-02,\n -2.40106195e-01, 1.83773446e+00, 7.58728504e-01, 1.63628101e+00,\n 5.63682675e-01, 1.95888042e+00, -4.86234426e-01, 6.65245056e-01,\n 5.28533340e-01, -2.50786161e+00, 4.42728072e-01, 7.19163597e-01,\n -1.49332881e+00, -2.27842307e+00, -1.58049488e+00, 1.69352794e+00,\n -1.07663131e+00, -1.04310560e+00, -8.12672138e-01, 3.57156491e+00,\n -6.96689487e-01, 1.79493916e+00, 1.36431956e+00, 1.45777392e+00,\n 2.08307624e+00, 1.61071312e+00, 1.67565846e+00, 1.42075956e+00,\n -9.33076024e-01, 1.94427609e+00, -4.85617042e-01, -6.90784991e-01,\n 1.94864774e+00, -1.11848676e+00, 1.55980206e+00, -2.74368525e-02,\n -6.48513436e-03, -9.99986231e-02, -9.34252143e-01, -1.60313129e+00,\n -1.81533849e+00, 2.24116966e-02, 8.71294796e-01, 2.45068264e+00,\n -1.22386384e+00, -9.88404691e-01, -1.60910642e+00, -1.54355526e-01,\n -9.76397753e-01, 2.88214254e+00, -8.56772959e-01, 2.26140571e+00,\n 2.02857351e+00, 5.81454992e-01, -1.58725071e+00, 2.61525679e+00,\n -7.40779877e-01, 3.83542001e-01, -1.42914009e+00, -5.31249195e-02,\n 1.18680072e+00, 2.84455776e+00, 2.65889478e+00, -1.63796437e+00,\n 1.49737465e+00, 1.15662909e+00, 6.80022463e-02, -2.36854345e-01,\n -6.37532324e-02, -2.59194827e+00, -1.32615983e-01, -3.85613471e-01,\n 8.44255984e-01, -8.12782645e-01, -1.00758953e+01, -2.70959526e-01,\n 1.87327468e+00, -5.09644091e-01, -1.40541613e-01, -5.59402853e-02,\n -1.23355329e+00, 1.71531546e+00, 3.49223614e-01, -1.63709924e-01,\n 7.78623998e-01, 5.97101688e-01, 6.86114907e-01, -7.83254266e-01,\n 2.24757338e+00, -7.54355848e-01, 2.24270725e+00, -1.22296262e+00,\n 2.11074615e+00, 1.56837654e+00, -1.18844414e+00, 1.54504216e+00,\n -1.31026793e+00, -9.08147156e-01, -9.43682611e-01, -7.79524624e-01,\n -2.95606762e-01, 1.18126726e+00, -1.94547582e+00, -7.48109102e-01,\n 2.64530206e+00, -3.28122759e+00, -1.10137796e+00, -1.31490302e+00,\n 4.81687725e-01, 8.10500205e-01, 1.21029329e+00, 3.26501876e-01,\n 1.11194730e+00, 1.53131497e+00, 2.79272616e-01, 3.18860531e+00,\n 1.88937932e-01, 7.67825007e-01, 1.42330170e+00, 1.06220174e+00,\n 3.39529872e-01, -1.22175789e+00, 2.04732656e+00, -6.81114316e-01,\n -2.64645958e+00, -2.40567470e+00, -2.34359908e+00, -6.99413002e-01,\n -8.08843255e-01, 6.36797667e-01, -1.14798680e-01, -1.56272268e+00,\n -1.67161024e+00, -8.07896078e-01, 2.49541044e+00, -1.34053767e+00,\n 1.27311730e+00, -4.69142288e-01, -6.07361019e-01, -4.14114475e+00,\n -9.39035773e-01, -1.07580578e+00, -1.56427884e+00, -2.30472490e-01,\n 6.66219234e-01, -6.72831476e-01, -6.34416714e-02, -7.32083440e-01,\n -2.60582709e+00, 3.16968632e+00, 7.69653320e-01, 1.89172566e-01,\n 1.03248060e+00, -2.16062832e+00, 1.21445701e-01, -8.68923664e-01,\n -1.27871168e+00, -1.55026925e+00, 4.21189904e-01, -4.67507005e-01,\n -9.76860166e-01, 2.36488605e+00, -1.18901181e+00, -1.31253886e+00,\n -2.49230433e+00, -1.55409241e+00, 2.93542910e+00, -4.50974643e-01,\n 1.89961779e+00, 7.48454571e-01, 2.07556152e+00, -5.60104489e-01,\n -1.13896060e+00, 9.16581511e-01, -1.28092074e+00, 6.05016530e-01,\n -1.67907000e-01, 8.97754580e-02, 1.44320083e+00, 3.60537958e+00,\n 2.34955263e+00, 5.59324145e-01, -1.46051013e+00, -1.00028539e+00,\n -2.17396811e-01, 2.25092220e+00, -1.22809160e+00, -6.98592067e-01,\n -6.40538335e-01, 3.40050966e-01, 8.97307634e-01, 1.10488570e+00,\n 4.02577209e+00, 8.94767702e-01, -1.55718178e-01, -3.31113398e-01,\n 2.36069530e-01, 1.53828418e+00, -4.79909241e-01, 6.52987003e-01,\n -1.30946112e+00, 1.18409181e+00, -1.20681345e-01, -1.44584608e+00,\n 3.43077958e-01, -5.43195233e-02, 5.77160358e-01, 1.17709303e+00,\n -1.13257718e+00, 9.07306731e-01, -3.39235210e+00, -1.42154384e+00,\n -1.29177499e+00, 3.45986271e+00, 9.81493294e-02, -7.89033294e-01,\n 4.16693777e-01, 1.53297710e+00, -1.27547312e+00, -5.97906411e-01,\n -1.74597979e+00, 5.21436930e-01, -5.28528094e-01, -6.41765654e-01,\n 6.19217396e-01, -4.40612704e-01, -4.11981225e-01, 1.66972959e+00,\n -1.23518479e+00, -7.97375590e-02, -1.10956788e+00, 1.60628211e+00,\n 8.16543341e-01, 4.15315956e-01, 2.62563658e+00, -1.39390075e+00,\n -1.74842334e+00, 2.12195134e+00, -7.67391920e-01, -1.89208412e+00,\n -1.55518174e+00, 5.90426862e-01, 1.89484990e+00, -2.41832566e+00,\n 6.95349395e-01, 7.64952719e-01, -2.98891127e-01, -1.22985625e+00,\n 8.91265810e-01, -2.20669955e-02, -2.54415369e+00, 2.36745429e+00,\n -3.74132305e-01, -6.60323143e-01, 1.49576950e+00, 1.10131943e+00,\n 1.60291016e+00, -6.10825658e-01, 2.64988005e-01, -2.68210053e-01,\n -2.05898929e+00, 1.40765917e+00, 1.62395149e-01, 3.03424549e+00,\n 6.02984786e-01, 3.76291871e-01, 2.39166403e+00, -1.64612547e-01,\n 2.86073780e+00, -4.19103622e-01, -1.01964319e+00, -5.84569693e+00,\n 4.94367957e-01, 1.38775468e+00, 2.98384875e-01, -1.00001514e+00,\n 1.92587090e+00, 1.68436646e-01, -1.42610884e+00, -3.56228977e-01,\n -6.14142567e-02, 3.32186222e-01, 9.66554523e-01, 1.29591155e+00,\n 1.42639744e+00, 1.55939364e+00, 1.24038029e+00, -1.24705577e+00,\n -3.52879286e+00, -2.70000160e-01, -1.92006207e+00, 5.75176597e-01,\n -4.12145197e-01, -9.47761893e-01, -5.12822986e-01, 1.19601309e-01,\n 5.76616079e-03, -4.34519976e-01, -2.70797133e+00, -3.65775168e-01,\n -3.90139818e-02, -1.22729015e+00, -9.13863182e-01, 2.22465014e+00,\n -1.21527791e+00, -4.67408717e-01, 9.73998010e-01, -3.17882121e-01,\n -6.12214148e-01, 5.46795964e-01, 8.02254915e-01, -1.20154214e+00,\n 9.12286043e-01, -1.88789546e+00, -8.58837366e-01, -1.19406509e+00,\n -7.68777132e-01, 1.84594655e+00, -1.47514927e+00, 1.01477575e+00,\n -9.04510498e-01, 3.67100567e-01, 4.45615679e-01, -2.79527259e+00,\n 3.75208795e-01, 2.46652842e+00, -1.57931066e+00, -6.66242063e-01,\n 1.77684677e+00, 3.60953659e-01, 2.20948935e+00, 2.01143846e-01,\n -1.60177469e+00, -6.72536612e-01, 2.65313119e-01, 1.60263324e+00,\n -2.38276625e+00, -4.27407622e-01, 3.19540024e-01, -9.34923530e-01,\n 5.17068267e-01, -2.14932114e-01, 6.52135551e-01, 4.23134625e-01,\n -3.72015953e-01, 3.32559013e+00, -7.18798399e-01, 1.04962081e-01,\n 1.22510147e+00, -1.33977517e-01, 1.56305027e+00, 7.73974419e-01,\n -2.21012354e+00, 9.96462047e-01, -2.54472160e+00, -4.03058082e-01,\n -1.02060823e+01, 8.18396807e-02, -2.06599402e+00, 4.85000312e-01,\n 4.12496150e-01, -8.72230008e-02, -2.10239506e+00, -5.08403063e-01,\n -4.29701358e-02, -1.45416749e+00, -8.51665139e-01, 2.39043131e-01,\n 1.49118766e-01, -1.23436666e+00, -7.36167252e-01, 2.56243086e+00],\n dtype=float32)), ('this', array([-2.06151748e+00, -6.60988212e-01, 9.01244402e-01, -1.64779997e+00,\n 2.63820267e+00, 7.90307224e-01, -1.84054875e+00, 2.22880268e+00,\n -1.61736691e+00, -1.39133662e-01, -1.87350251e-02, -3.90823126e-01,\n 1.04033315e+00, 2.88460821e-01, 1.58469427e+00, 2.77557635e+00,\n -1.86856842e+00, 8.16558659e-01, -9.06831086e-01, -1.36792850e+00,\n 2.75060534e+00, 1.19425690e+00, -2.02801275e+00, -3.81437957e-01,\n 4.18794537e+00, -1.53663850e+00, -1.19022751e+00, 2.74695426e-01,\n -4.84578639e-01, -4.87662613e-01, 2.34219342e-01, 1.85535014e+00,\n 1.50280881e+00, -6.49968535e-02, -1.21994829e+00, -7.72967041e-01,\n 4.62442160e-01, -2.82695389e+00, -2.54010105e+00, 1.70868361e+00,\n 3.21304590e-01, -1.46408975e-01, 2.51152420e+00, -2.70429611e-01,\n -1.05066454e+00, -3.80472243e-01, 1.47800136e+00, -1.87119770e+00,\n -1.01100850e+00, -2.36425328e+00, -4.23852825e+00, 2.10366696e-01,\n -9.48942721e-01, 1.59495711e-01, -4.83654404e+00, 1.06716323e+00,\n 2.56783068e-01, -2.30803800e+00, -3.24087799e-01, -8.39685082e-01,\n 2.13124895e+00, -2.80119121e-01, 2.33581871e-01, 1.48563313e+00,\n 1.77941394e+00, 1.59973872e+00, 7.69077301e-01, 1.65162563e+00,\n -1.75667989e+00, -1.39260268e+00, -1.32475162e+00, -3.57547426e+00,\n 8.38259816e-01, 3.20036739e-01, 1.65423894e+00, 9.07088637e-01,\n -5.92350483e-01, -3.51595759e-01, -2.28519469e-01, -3.68335724e+00,\n 6.41822517e-01, 4.79975510e+00, 1.64354455e+00, -4.33025151e-01,\n 8.82280707e-01, 9.44206417e-01, -1.49712968e+00, 3.30670178e-01,\n -3.17750263e+00, 2.39856482e+00, -1.41454518e-01, -2.03954387e+00,\n -2.14461517e+00, 3.48352730e-01, 3.03874373e-01, -4.15034533e+00,\n -1.85126090e+00, -2.97060061e+00, 5.50660014e-01, -3.63028860e+00,\n -2.55361700e+00, -6.23918247e+00, -4.85539615e-01, 1.03205538e+00,\n 9.13876474e-01, -6.13795996e-01, 9.32011843e-01, 3.65330124e+00,\n 2.88257027e+00, -3.07537746e+00, 4.80245233e-01, 1.53483796e+00,\n 5.00141501e-01, -1.88190520e-01, -2.35021710e+00, 2.70690584e+00,\n -5.24845004e-01, 1.89043951e+00, 2.67473960e+00, -1.78898811e+00,\n -3.88324738e+00, 5.32040656e-01, 1.26099753e+00, 1.87876761e+00,\n 5.24927139e-01, 8.52876723e-01, 1.76951289e+00, 1.54487979e+00,\n 6.09962821e-01, -2.67511988e+00, 1.16187131e+00, 3.80747914e+00,\n 3.29138994e+00, -1.97444141e-01, -5.34155190e-01, -1.64079773e+00,\n -5.12093425e-01, -3.50186801e+00, -3.29415560e+00, -1.08402669e+00,\n 2.74042940e+00, 2.21246883e-01, 3.60517025e+00, 4.43118155e-01,\n 6.84380949e-01, -1.60800838e+00, -2.58211064e+00, -8.89470220e-01,\n -4.10495281e-01, -3.16021800e-01, 5.79772413e-01, 1.51941693e+00,\n -1.35226178e+00, -2.18451357e+00, -7.22791195e-01, -9.68789101e-01,\n -2.31947708e+00, 2.29124784e+00, 9.16360259e-01, 2.20985746e+00,\n 1.23639345e+00, 1.20288622e+00, 3.27279472e+00, 3.12709951e+00,\n 1.92695677e+00, -1.81729913e+00, -1.04907835e+00, -8.40046406e-01,\n 1.16908216e+00, -3.25442648e+00, 1.05546737e+00, -3.30256629e+00,\n 2.53007746e+00, 1.42755699e+00, -8.77801001e-01, -5.30875176e-02,\n 1.24621534e+00, -1.54841471e+00, 1.88082767e+00, 1.75026238e+00,\n -1.81591284e+00, -1.99626553e+00, -2.96949935e+00, -7.08848417e-01,\n 1.47713661e+00, 5.06355166e-01, -5.06473899e-01, -4.45754147e+00,\n -2.09264159e-01, 5.33231831e+00, -1.17951536e+00, 3.01163578e+00,\n 1.17991531e+00, -4.22484064e+00, 1.53453100e+00, -2.54382801e+00,\n -2.24179363e+00, -1.18559587e+00, 2.19983554e+00, -1.89659202e+00,\n 6.82720006e-01, 2.26982522e+00, -1.78928912e-01, -5.12818396e-02,\n 5.02129972e-01, 5.28296757e+00, 1.66096711e+00, -9.50530052e-01,\n 7.29229867e-01, -8.20072055e-01, -3.74128997e-01, 6.42508459e+00,\n -1.48333883e+00, -5.93578219e-01, 1.33169460e+00, 2.10809398e+00,\n -2.54307652e+00, 9.87953663e-01, 1.02637303e+00, 1.80206239e+00,\n 1.17413366e+00, -1.70701230e+00, -1.35935068e+00, 1.95292485e+00,\n -1.21594071e+00, 4.50632334e+00, 2.52183056e+00, 1.25438011e+00,\n 1.50238872e+00, -6.61006212e-01, -9.39444423e-01, -2.09089935e-01,\n -1.00152659e+00, -2.29346603e-01, -3.45885724e-01, 4.59669065e+00,\n 9.13417339e-01, -5.81251085e-01, -9.51805413e-02, 1.61924613e+00,\n -1.54568148e+00, 1.46689951e+00, -3.02519977e-01, 3.04833603e+00,\n -3.28942871e+00, -7.35817790e-01, -2.84884036e-01, -2.49692130e+00,\n -1.43487263e+00, -9.72960711e-01, -1.88861251e+00, -8.51065338e-01,\n -4.18707430e-01, -1.70045507e+00, -4.95572042e+00, -5.00460118e-02,\n 4.77657169e-02, 1.92613924e+00, 5.19203782e-01, 8.97293210e-01,\n 1.45387661e+00, 1.55786967e+00, 1.20036674e+00, -1.37592554e-01,\n -1.04275787e+00, 6.28491879e-01, -2.63064837e+00, 2.17257094e+00,\n 1.78405988e+00, 1.03657079e+00, -3.07841587e+00, -2.62570977e+00,\n -9.77001309e-01, -1.08873332e+00, -2.94658685e+00, -1.70976520e+00,\n -3.43563855e-02, 2.21203268e-02, -1.73493803e+00, -3.52413923e-01,\n -2.84776717e-01, 2.63769698e+00, 4.94865388e-01, 8.57661963e-01,\n 2.87633419e+00, -1.73457980e+00, 7.14749932e-01, -4.97826576e-01,\n -5.29348999e-02, 9.26089287e-02, -3.24941516e+00, 2.47544813e+00,\n -2.24171090e+00, -5.36972761e+00, 4.02708769e+00, -1.27829897e+00,\n 2.29108572e+00, 6.04518533e-01, -2.72756672e+00, -3.39956260e+00,\n -1.07763410e+00, -2.58415079e+00, 1.79191434e+00, 1.16855919e-01,\n -1.71414936e+00, 2.21213484e+00, -1.56503534e+00, -1.55671310e+00,\n -2.70496445e+01, 1.35250136e-01, -1.02360201e+00, -7.60923743e-01,\n -1.64519048e+00, 2.02327639e-01, -4.19679612e-01, 8.99103642e-01,\n -1.33351183e+00, -6.34252310e-01, -1.39443660e+00, -2.85758400e+00,\n -1.91844308e+00, 1.20011091e+00, -1.16089061e-01, -1.04893303e+00,\n 1.29639578e+00, 3.15334344e+00, 3.00656199e-01, 4.60581303e+00,\n -1.21655333e+00, -1.54574561e+00, -1.24810815e-01, -2.03697300e+00,\n -5.13954353e+00, 1.00324464e+00, -6.25430703e-01, 1.64058101e+00,\n -1.55852878e+00, 1.82162523e+00, 4.78050560e-01, -1.29992819e+00,\n -6.80713177e-01, 1.81918097e+00, -2.34114766e+00, 7.23255932e-01,\n 4.51537430e-01, -2.11234760e+00, 1.24817991e+00, 9.02195811e-01,\n -7.27035284e-01, -9.97290194e-01, 2.07151961e+00, 2.70721650e+00,\n -7.09761143e-01, 8.02284718e-01, -1.12379670e+00, -1.56643569e+00,\n -4.34926480e-01, 9.80157852e-01, -1.21399617e+00, -2.31257010e+00,\n 5.54166377e-01, -5.16683519e-01, -3.07023108e-01, 1.28092170e+00,\n -3.94111097e-01, -2.30953634e-01, 1.86944056e+00, 5.76802492e-01,\n 1.94921970e+00, -3.93669987e+00, -2.67622948e+00, 2.92488194e+00,\n -1.36228934e-01, -4.11785841e+00, -4.04014289e-01, -1.85826051e+00,\n -8.33527803e-01, 1.73215389e+00, -1.65354443e+00, 1.36537945e+00,\n 3.59903002e+00, -1.28343201e+01, -1.90786874e+00, -1.22698355e+00,\n 1.94380093e+00, 3.61065358e-01, 5.39731312e+00, -3.93930554e-01,\n -1.69107091e+00, -3.24742627e+00, 2.34137726e+00, 6.73783302e-01,\n 1.87343633e+00, -4.75728929e-01, 1.63546622e+00, -1.22131240e+00,\n -1.27176452e+00, -2.97526836e-01, -3.17002010e+00, 3.13992691e+00,\n -8.09281170e-01, 2.12504888e+00, -5.68572760e-01, 1.51250586e-01,\n 1.29395270e+00, -2.65934229e+00, -2.48478800e-01, 2.42425489e+00,\n 2.62182999e+00, -3.97840858e-01, 3.11840796e+00, 2.18603778e+00,\n 4.76051426e+00, -9.93168712e-01, -2.96750617e+00, 3.88353634e+00,\n -5.30234575e-01, 2.71193576e+00, -2.06041813e+00, -8.07008982e-01,\n -1.69010961e+00, -4.61568773e-01, -3.08530211e+00, 2.63426852e+00,\n -1.67073369e+00, 1.71168435e+00, 1.75999367e+00, -2.47717905e+00,\n -7.03588352e-02, 1.74219120e+00, -1.75053537e+00, -2.74892139e+00,\n -2.51376104e+00, -1.04232430e-01, -2.31501913e+00, 2.52336431e+00,\n -3.35380840e+00, 2.06405735e+00, -1.96217167e+00, 2.19019961e+00,\n -1.66168141e+00, 2.82280743e-01, 1.36640000e+00, -1.23139128e-01,\n -8.19660127e-01, 5.64721203e+00, 4.21977901e+00, -3.76158118e-01,\n 2.78726935e-01, 2.77557111e+00, 1.00555301e+00, 2.45483780e+00,\n 1.57262874e+00, 2.57323456e+00, -5.57377338e-01, -2.10861635e+00,\n -4.70240563e-02, 9.85036254e-01, 1.91405430e-01, -2.00530672e+00,\n 1.31374693e+00, -1.16779160e+00, 9.96227384e-01, 4.22857821e-01,\n 2.40403366e+00, -8.70058954e-01, 3.58269334e-01, -3.39673924e+00,\n 1.04052687e+00, -1.40798354e+00, -8.07139754e-01, -1.46334088e+00,\n 3.04123139e+00, 7.59105325e-01, 2.32768744e-01, 4.43320647e-02,\n -2.46330428e+00, -4.19368172e+00, -1.74058986e+00, 1.08369493e+00,\n 9.82749611e-02, -2.13859940e+00, 1.63009632e+00, -2.23901892e+00,\n -1.02638036e-01, -1.00240552e+00, 1.02284265e+00, 1.02040243e+00,\n 8.34562421e-01, 1.84578300e+00, 1.92636895e+00, 6.56498194e-01,\n 1.44521129e+00, -7.66581893e-01, -4.33097929e-01, -6.48968637e-01,\n 3.87490571e-01, 1.58110428e+00, -9.91709590e-01, 6.86446190e-01,\n -8.74638677e-01, -2.56020904e+00, -2.76195002e+00, -2.55254412e+00,\n 4.43155646e-01, 3.14474225e-01, 2.06457090e+00, 2.17746639e+00,\n 7.45012939e-01, 2.84237242e+00, -9.50627446e-01, -1.29374290e+00,\n -5.34832060e-01, 2.57555938e+00, -9.13931966e-01, 8.13854039e-01,\n 8.47411990e-01, -1.16847730e+00, -8.36292863e-01, 4.65259552e-01,\n -2.09995914e+00, -4.21928883e+00, -1.82238424e+00, -2.59686470e+00,\n -2.12247705e+00, 2.63694859e+00, -2.75659800e+00, -1.82731581e+00,\n -7.70584345e-01, 2.55915880e-01, 8.25502753e-01, -1.94935828e-01,\n -9.47885931e-01, -3.25217175e+00, 3.17397451e+00, -3.87598586e+00,\n 2.74548578e+00, -1.05592296e-01, -1.49303675e+00, -4.11066341e+00,\n 1.16900837e+00, 1.23952913e+00, -1.32276464e+00, -4.34994602e+00,\n -3.28251052e+00, 3.55913967e-01, 3.65062416e-01, -1.25570667e+00,\n 1.13799775e+00, -4.14410305e+00, 1.70531225e+00, -6.12771869e-01,\n -2.99816847e+00, -4.01915073e+00, 4.43169147e-01, -1.15466774e+00,\n 4.18475688e-01, 2.25445580e+00, -8.46850276e-02, 3.24470139e+00,\n -7.38708019e-01, -4.18693036e-01, -9.95072722e-02, -2.30658793e+00,\n 2.79425049e+00, 1.07988417e+00, 4.84989285e-02, 2.05558848e+00,\n 8.68896842e-01, 1.52186966e+00, 2.78109932e+00, 3.93874955e+00,\n -2.28132653e+00, 1.57945466e+00, -1.65724844e-01, -7.21502185e-01,\n 3.47642332e-01, 1.68776512e+00, -3.64121675e+00, 2.49744654e+00,\n 8.47951770e-01, 1.85247636e+00, 2.69540453e+00, -8.72950077e-01,\n -8.24803233e-01, 4.00232151e-02, -1.76669168e+00, 2.41407424e-01,\n 8.14830959e-01, 4.90098476e-01, -3.60848761e+00, -5.59456110e-01,\n -1.80803645e+00, 9.04515386e-01, -1.73901165e+00, -6.95377350e-01,\n 2.42816091e+00, -7.99551845e-01, -2.45431113e+00, -1.22833326e-01,\n 1.43067575e+00, 1.91327333e+00, 3.99058223e-01, 4.27168846e-01,\n 2.45602155e+00, 1.24232602e+00, 6.68387949e-01, -2.88281739e-02,\n -6.09493136e-01, 1.89938998e+00, -5.86492896e-01, 8.05594981e-01,\n -3.41339350e-01, 2.13022614e+00, 2.71380401e+00, -2.30834186e-02,\n -9.23136592e-01, 1.89873302e+00, -1.33142412e+00, 1.32467556e+00,\n 9.07860518e-01, -9.43488002e-01, 9.33475733e-01, 1.71026778e+00,\n 3.44864416e+00, -1.37888968e+00, -7.88728237e-01, 2.48014736e+00,\n -2.51914072e+00, 1.38224423e-01, 1.19576901e-01, -2.00510073e+00,\n -2.38182044e+00, -2.68934965e-01, -2.57933497e+00, -1.50733745e+00,\n -3.92813712e-01, -1.44191384e+00, 4.44592535e-02, -1.00061333e+00,\n -2.39194250e+00, 3.17540884e+00, -1.21450126e-01, 2.64450645e+00,\n -6.86704278e-01, -2.13251615e+00, 2.00330257e+00, 1.67906976e+00,\n -2.57708788e+00, -1.29647374e-01, 2.45424724e+00, 1.80543482e+00,\n -1.58560300e+00, -1.39036131e+00, -3.35764438e-01, 4.32217538e-01,\n -8.01830411e-01, 1.24492753e+00, 2.65466928e+00, 1.35145545e+00,\n 1.53817892e+00, -1.45809484e+00, -3.83151054e-01, 1.87781084e+00,\n 2.22314429e+00, 2.12585354e+00, 2.24408507e+00, 1.98697507e-01,\n 2.17358732e+00, 2.45930821e-01, 8.51337552e-01, -1.01808202e+00,\n -1.06195256e-01, 1.36613560e+00, 2.04582858e+00, 1.98897398e+00,\n 7.54615128e-01, -1.02346158e+00, -4.97204006e-01, 2.39321280e+00,\n 1.64808202e+00, 2.47932291e+00, -1.68948889e+00, 4.50206280e-01,\n -5.32100439e-01, 4.54317212e-01, -7.88925290e-01, -1.56387985e+00,\n 1.12469935e+00, 3.13415498e-01, 7.45716393e-01, -5.03132701e-01,\n -2.24132597e-01, -1.49285126e+00, -8.29637527e-01, 2.79790461e-02,\n 1.50920063e-01, -4.73168671e-01, 1.08686340e+00, 8.99161935e-01,\n -2.80048996e-01, -9.20594335e-01, 2.11123848e+00, -4.68289346e-01,\n -1.74462426e+00, -5.69986880e-01, 1.46353030e+00, -2.70267534e+00,\n 1.27518535e+00, 1.56568885e-01, -4.53318059e-02, -2.10661578e+00,\n -1.70757425e+00, 9.95355248e-02, -1.08474374e+00, 2.61613488e+00,\n -1.73607445e+00, -3.13704038e+00, 1.65696049e+00, 5.50683737e-01,\n 5.93220353e-01, -1.98653293e+00, -3.91659451e+00, 1.12145340e+00,\n 5.91378689e-01, -2.53870630e+00, -1.83522308e+00, 7.55941570e-01,\n -2.96283937e+00, -3.49579120e+00, -3.23328137e-01, 4.04899883e+00,\n 3.22775662e-01, -2.84158468e+00, -3.89616281e-01, -1.73561946e-01,\n -2.85589695e+00, -2.47964048e+00, 1.66222584e+00, 2.56132102e+00,\n -2.69972175e-01, -2.15884134e-01, -7.68641949e-01, -4.07402039e+00,\n 1.39365733e+00, -1.82120502e+00, 1.85946596e+00, -1.66499305e+00,\n -5.76051995e-02, 4.73632240e+00, -1.45582652e+00, 1.33207333e+00,\n -1.20725536e+00, -1.23040473e+00, -7.56453753e-01, -3.10782909e+00,\n 3.03737712e+00, 1.29491687e+00, 2.51569843e+00, -3.99123430e-01,\n 8.16602349e-01, -1.91904163e+00, -3.65072775e+00, 4.74868679e+00,\n -3.15397114e-01, -2.41786265e+00, 5.58235884e-01, 2.19366431e+00],\n dtype=float32)), ('is', array([-1.49126601e+00, -9.43653047e-01, 2.48240852e+00, -7.37955689e-01,\n -2.00913048e+00, -7.77409256e-01, -1.95818233e+00, 1.80954468e+00,\n -1.35973394e+00, -3.45780253e+00, 3.31536412e-01, -8.36897016e-01,\n -1.33736169e+00, 1.12518990e+00, -1.52375555e+00, 1.41726255e+00,\n -1.09342480e+00, -1.01838601e+00, 1.48238039e+00, -1.35435462e+00,\n 1.07505083e+00, 2.85449600e+00, -2.39840078e+00, -1.19180083e+00,\n 8.19160461e-01, 1.45340610e+00, -2.74805129e-01, -2.52085567e+00,\n -2.64881492e-01, 1.11394727e+00, 1.97445226e+00, 9.85239208e-01,\n 1.57252407e+00, -2.27361870e+00, -2.21182942e+00, -1.90472674e+00,\n 1.23033166e-01, -2.27698517e+00, -4.56972408e+00, 5.48222184e-01,\n -1.11670649e+00, -1.59575176e+00, 1.45195687e+00, 5.70956647e-01,\n -1.16829026e+00, 2.77093142e-01, 3.30588627e+00, -7.16315269e-01,\n 1.50581646e+00, -1.80108762e+00, -3.96876907e+00, -2.29787207e+00,\n 2.07470441e+00, 2.16017127e+00, -1.49040055e+00, 2.49467683e+00,\n -3.60778928e-01, -3.01101255e+00, -5.91381431e-01, -7.70706952e-01,\n 6.22554898e-01, -4.17888939e-01, 7.10910201e-01, 1.89667916e+00,\n 2.39778304e+00, 2.27681494e+00, -4.29660857e-01, 6.22181147e-02,\n -1.26079381e+00, 1.54760265e+00, 9.07531619e-01, -1.88656056e+00,\n 3.53369999e+00, 2.57900685e-01, -1.39359975e+00, 5.86659729e-01,\n -1.08971608e+00, -9.39617157e-01, 2.97544765e+00, -2.81365061e+00,\n -6.18298829e-01, 5.11932945e+00, -4.93881628e-02, 3.04182887e-01,\n 6.76466048e-01, -4.11224222e+00, -1.07441998e+00, -8.34418774e-01,\n -4.03038216e+00, -8.23588014e-01, 1.39967608e+00, -2.15924954e+00,\n 4.45227653e-01, -1.20315921e+00, 7.59126902e-01, -5.21934557e+00,\n -7.47040093e-01, -2.55360198e+00, -7.08164722e-02, -3.57310128e+00,\n -1.87825692e+00, -5.10245371e+00, 1.88420987e+00, -5.00296891e-01,\n -1.91885740e-01, -7.08465219e-01, -2.33401537e+00, 5.20400476e+00,\n 3.47468257e+00, -3.13773966e+00, 1.51250303e+00, 2.04615164e+00,\n 1.28231764e-01, 2.71312833e-01, -1.13982713e+00, 2.15454674e+00,\n -2.81179160e-01, 2.45237336e-01, 1.91016173e+00, -1.97350562e+00,\n -1.12345207e+00, -1.16771054e+00, -5.97940087e-01, 2.18151951e+00,\n -4.63656843e-01, 2.22407055e+00, -1.71341300e+00, 1.23503745e+00,\n -4.72254276e-01, -2.34765434e+00, 2.65903711e+00, 9.03169513e-01,\n 3.02865028e+00, -2.17571330e+00, 3.56160450e+00, 5.79993248e-01,\n 1.24466598e-01, -1.60528421e+00, -5.67695904e+00, -9.15822536e-02,\n 1.95025897e+00, 1.52327561e+00, 3.06210947e+00, -1.43937135e+00,\n -1.46194696e+00, -2.18500996e+00, -3.61036277e+00, 5.78358710e-01,\n 1.82440841e+00, 3.89762402e-01, -5.62306166e-01, 1.77419233e+00,\n 4.87029433e-01, -2.38311648e+00, 1.10754371e+00, -2.24959159e+00,\n -8.95443201e-01, 2.15968323e+00, 9.50986028e-01, 5.07258892e-01,\n 4.15195286e-01, 5.12130260e-01, 1.17998409e+00, 1.61869705e+00,\n 3.36331040e-01, -1.96278840e-01, -5.34239471e-01, 9.95529413e-01,\n 2.41796541e+00, -3.73321462e+00, 8.08570027e-01, -4.36797529e-01,\n -1.13277987e-01, -1.33749098e-02, 2.26724792e+00, 1.02044749e+00,\n -1.84577835e+00, 1.34707704e-01, 4.40607882e+00, 1.64789248e+00,\n -9.85833883e-01, -2.25869489e+00, -1.48049164e+00, -3.63306594e+00,\n -2.34360427e-01, -7.53898621e-02, -7.65656352e-01, -9.10130441e-01,\n -1.00085330e+00, 3.65413451e+00, 3.04004341e-01, 3.50726604e+00,\n 1.86765265e+00, -2.45345783e+00, -3.99615079e-01, -2.40356970e+00,\n -1.91600585e+00, -2.47108221e+00, -6.11715853e-01, -2.57968593e+00,\n 1.21236062e+00, 1.89139783e+00, 1.60503614e+00, -1.20580947e+00,\n -1.57958877e+00, 3.62749791e+00, 2.36892533e+00, -1.10803342e+00,\n -1.44388807e+00, 6.07312202e-01, 1.04578364e+00, 5.15290308e+00,\n -1.07205600e-01, -1.91798657e-01, 2.13868260e+00, 2.75123739e+00,\n 1.13493586e+00, -1.51974416e+00, 7.95320511e-01, 2.15098763e+00,\n 2.37411332e+00, -3.44415808e+00, -4.42123175e+00, 1.50133908e-01,\n -1.73098135e+00, 1.23942232e+00, 6.92882001e-01, 1.76000178e+00,\n 1.74554372e+00, -2.10563612e+00, -3.43099892e-01, 7.16276348e-01,\n 2.45144397e-01, 2.16568089e+00, 9.40388203e-01, 3.79211712e+00,\n 6.42793179e-02, 1.29858398e+00, 2.59019709e+00, 1.80356729e+00,\n 8.14835608e-01, 3.00816488e+00, 4.53192520e+00, 6.03188634e-01,\n 3.15965712e-01, -8.69524956e-01, -4.73948061e-01, -2.08995533e+00,\n -2.56012893e+00, -1.69470692e+00, -2.28035760e+00, -3.84858322e+00,\n -4.09252256e-01, -2.28163674e-02, -3.80629992e+00, -5.00131369e-01,\n -1.98133612e+00, -7.24980235e-02, 3.50680470e-01, 8.79511833e-01,\n -2.20601544e-01, 1.51045763e+00, -1.08350134e+00, -3.63689214e-01,\n -8.67066681e-01, 4.39716697e-01, 3.47551405e-01, 1.75882077e+00,\n 5.04951572e+00, 1.12705040e+00, -3.89148831e-01, -1.74457264e+00,\n 1.73626411e+00, 5.35147667e-01, -6.78079486e-01, -3.86440605e-01,\n -1.02311218e+00, -5.48225820e-01, -2.58845687e+00, 1.15553606e+00,\n 2.60133791e+00, -1.78057745e-01, 2.55747914e-01, -1.10384691e+00,\n 1.85323584e+00, -8.90728951e-01, -8.13649118e-01, -5.01150727e-01,\n -1.98465598e+00, -2.01095200e+00, -3.77061677e+00, 3.23753953e+00,\n 3.58389020e-01, -7.20858288e+00, 4.78798962e+00, -2.43055630e+00,\n 7.50351787e-01, 2.40659189e+00, -3.44288898e+00, -6.83531523e-01,\n -4.35844213e-01, -3.51787734e+00, 1.17401755e+00, -1.86902642e+00,\n -5.54093838e-01, -1.28015256e+00, -1.51746726e+00, 7.95142055e-01,\n -2.99072857e+01, -2.42203379e+00, 3.49145621e-01, -2.00196004e+00,\n -1.25928640e-01, 1.98652312e-01, -2.88321614e-01, 1.30309200e+00,\n -2.23386145e+00, 8.85055423e-01, -1.02029383e+00, -2.84918928e+00,\n -2.72842360e+00, 1.28254521e+00, 2.05543780e+00, -4.28827715e+00,\n -8.60526621e-01, 1.07620239e+00, 1.83247119e-01, 7.42765951e+00,\n -3.92079210e+00, -1.71073949e+00, 6.02431774e-01, -2.93414807e+00,\n -2.36623025e+00, 2.54542542e+00, 1.25756562e-02, 8.69601727e-01,\n -2.45497894e+00, -1.83600688e+00, -2.26969576e+00, 2.22250104e-01,\n 1.53597856e+00, 2.49344468e+00, -3.14198554e-01, -7.72304535e-02,\n -1.74641371e-01, -2.26883173e+00, 2.34960389e+00, -1.80539739e+00,\n -7.33242035e-01, -1.18008220e+00, 1.43859124e+00, 1.06047642e+00,\n 1.37566626e+00, -1.43849611e-01, 4.36722636e-01, -2.95693564e+00,\n 1.34736454e+00, 4.07657957e+00, -2.94853640e+00, -2.02462530e+00,\n -2.70367086e-01, -2.48207378e+00, 1.31244004e+00, 8.07296932e-01,\n 3.25785398e+00, 3.00726265e-01, 3.59229326e+00, -1.42439079e+00,\n 1.11430717e+00, -3.21023822e+00, -2.59298563e+00, 5.88242197e+00,\n -6.05805933e-01, 1.58903822e-01, -4.66701651e+00, -2.60726786e+00,\n 1.41685736e+00, 9.85289633e-01, 4.07405764e-01, 2.25500154e+00,\n 1.35790801e+00, -1.20320034e+01, -1.92002869e+00, -1.30287111e+00,\n 2.83787179e+00, -2.10869044e-01, 7.10515201e-01, 1.03971767e+00,\n -3.06053734e+00, -4.46737099e+00, 1.23164341e-01, 2.32203889e+00,\n -3.26915741e-01, -1.01978429e-01, 2.96106339e-02, 5.63665509e-01,\n 2.57334471e-01, 1.33485699e+00, 2.20361918e-01, 3.11618161e+00,\n -3.42092752e-01, 2.25000143e+00, -1.97125244e+00, 7.48440504e-01,\n 2.36628342e+00, -1.74806499e+00, 7.18164384e-01, 2.28968453e+00,\n 4.81662512e+00, 2.59959757e-01, 1.38929701e+00, 4.53423500e+00,\n 6.20340061e+00, 2.50169420e+00, -3.61102962e+00, 2.00748873e+00,\n 2.46403670e+00, 1.27324796e+00, 9.98339295e-01, -1.68876052e-01,\n -1.46798420e+00, -2.86310911e-03, -3.66906238e+00, 2.18176222e+00,\n -2.01064014e+00, 2.16561055e+00, 1.11374581e+00, -2.94069457e+00,\n -1.02281451e+00, -4.30156529e-01, -1.44194651e+00, -3.43218303e+00,\n -4.75802040e+00, -8.16402435e-01, -1.45620608e+00, 7.55139470e-01,\n -1.80856085e+00, 1.64795434e+00, 5.69868833e-02, 4.42695189e+00,\n 1.43387949e+00, 1.02167571e+00, -8.19991469e-01, 8.42229500e-02,\n -1.73456407e+00, 5.18062401e+00, -3.80029887e-01, -1.22410335e-01,\n 5.01329660e-01, 2.67164874e+00, -1.97124910e+00, -1.09700918e+00,\n -2.37445068e+00, 4.70610285e+00, 6.33587539e-01, -8.84274602e-01,\n 1.32701683e+00, -1.07883596e+00, -2.81650400e+00, 2.01244503e-01,\n 2.87770271e+00, 1.90154099e+00, -7.14639366e-01, -2.45423168e-02,\n 3.44006091e-01, -1.10941374e+00, -1.55289268e+00, -5.61483526e+00,\n 2.18294954e+00, 2.20178440e-01, 4.88469601e-01, 1.18314528e+00,\n 1.38093257e+00, 1.02358675e+00, -8.11505139e-01, 9.37226117e-01,\n -2.45631361e+00, -2.53804755e+00, -1.03802848e+00, 1.30918074e+00,\n -9.71696615e-01, -1.03270805e+00, 3.17397738e+00, -4.49946105e-01,\n -1.21611786e+00, -1.88782299e+00, 1.44047546e+00, 5.84089100e-01,\n 9.64816928e-01, 7.55962431e-02, 4.92330790e-01, -1.15164304e+00,\n -1.04969549e+00, 5.28398871e-01, 2.94067591e-01, 3.74340594e-01,\n 6.37677252e-01, -2.35908881e-01, -4.10497332e+00, 3.94325286e-01,\n -7.34709680e-01, -1.18321383e+00, -5.77900827e-01, -3.33582711e+00,\n 3.78584027e-01, 4.55937058e-01, -1.29973519e+00, 1.45043671e+00,\n 1.00099945e+00, 4.09049988e-02, -1.41216487e-01, -6.54326975e-02,\n -7.28478432e-01, 2.25739908e+00, -1.41448832e+00, -1.01417816e+00,\n -8.09604168e-01, -2.41343474e+00, 8.41779351e-01, -1.53401166e-01,\n 1.63395178e+00, -1.91967726e+00, -6.95088923e-01, -9.82195735e-01,\n 1.82589173e+00, 1.62946534e+00, -4.40049934e+00, 1.56085283e-01,\n -1.14990306e+00, -7.28949666e-01, 2.40279174e+00, -7.32653856e-01,\n 7.18039274e-01, -2.50960946e+00, 6.99720740e-01, -3.61363339e+00,\n 1.82613850e+00, -1.12793505e+00, -5.01489937e-01, -2.58955312e+00,\n -4.15890247e-01, 3.83580983e-01, -3.52736568e+00, -7.00485766e-01,\n -6.30514324e-01, 3.36504668e-01, 1.38212359e+00, -9.04867291e-01,\n -1.22168589e+00, -3.28530765e+00, -7.35800982e-01, -1.40724349e+00,\n 5.31000853e-01, -5.36806440e+00, 1.63398635e+00, -1.76243949e+00,\n -1.09221053e+00, 1.46734238e+00, 1.01785636e+00, 2.76797676e+00,\n 8.53842497e-01, -2.34179544e+00, -1.61211401e-01, -3.14831996e+00,\n -1.98748386e+00, 1.44071829e+00, 2.94050217e-01, 7.67786801e-02,\n 8.66885424e-01, 2.61335182e+00, -6.03698015e-01, 1.05699074e+00,\n -5.19798696e-01, 8.16070065e-02, 3.52043533e+00, 1.84486961e+00,\n 1.53232181e+00, 1.68656707e+00, -3.39691687e+00, 2.11678505e+00,\n 2.23781013e+00, 1.77320397e+00, 2.14734387e+00, -2.16879058e+00,\n -1.29038298e+00, 1.28758621e+00, 1.04954791e+00, -2.35532045e+00,\n -1.18120563e+00, -1.05894232e+00, -3.79061818e+00, 9.75292325e-01,\n 1.75827503e-01, 7.31311619e-01, 1.19312537e+00, 2.60286617e+00,\n 8.69498432e-01, -6.74136817e-01, -3.19753623e+00, -3.91345918e-01,\n -2.57603216e+00, -3.93398094e+00, 5.72923899e-01, -8.74991655e-01,\n 3.93119621e+00, 1.71358085e+00, 3.35435092e-01, -3.02256250e+00,\n 2.48699144e-01, 3.35746098e+00, 3.40357971e+00, -8.92864466e-02,\n -3.59576607e+00, 2.24764800e+00, -2.29200900e-01, 6.38916731e-01,\n -1.38058102e+00, 1.61436462e+00, -1.08875895e+00, -1.85471058e-01,\n 1.57091796e+00, 2.48134643e-01, 2.42971802e+00, -9.94265556e-01,\n 3.17785054e-01, -2.21412468e+00, -9.57366586e-01, 2.75902510e+00,\n -2.14359665e+00, -2.19539595e+00, 2.86046505e+00, -8.48217964e-01,\n -1.88282049e+00, 1.56468999e+00, -1.57652080e+00, -3.27210593e+00,\n -3.99587244e-01, -1.34626985e-01, 4.65337932e-01, -1.31918824e+00,\n -2.08976507e+00, 1.43362856e+00, -2.73568928e-02, -1.37924087e+00,\n 1.32718325e+00, -3.97269177e+00, 5.39630699e+00, 2.76890337e-01,\n -1.59833145e+00, -1.55421352e+00, 2.02858353e+00, 2.57484579e+00,\n -9.50849473e-01, -2.16783834e+00, 5.79418480e-01, -1.62456524e+00,\n -1.28814948e+00, 3.64231539e+00, 1.21584487e+00, -4.92766052e-02,\n 1.50074923e+00, 5.10802865e-02, 1.33486462e+00, -2.93035150e+00,\n 1.34032035e+00, 3.45545387e+00, -4.10508215e-01, 1.26751125e-01,\n 3.10301304e+00, 1.80074084e+00, 2.55082941e+00, -5.68139613e-01,\n -6.91802263e-01, 3.13867402e+00, 1.23244393e+00, 3.51930523e+00,\n 1.22267628e+00, -1.91458130e+00, -2.41045803e-01, 1.19508278e+00,\n 2.54850006e+00, 1.24378335e+00, 3.35205436e-01, 2.60172272e+00,\n -1.40075564e+00, -1.33622658e+00, -1.91406202e+00, -6.29880309e-01,\n 2.15883899e+00, -6.64399415e-02, 3.29563856e-01, -1.49226284e+00,\n 1.40853775e+00, -2.40254712e+00, -4.64376748e-01, -4.19757032e+00,\n 6.07549012e-01, -4.42348421e-01, -5.16312957e-01, 1.93001544e+00,\n -5.51427722e-01, -2.26106834e+00, 2.04624653e+00, -1.44312882e+00,\n -9.24660683e-01, -1.10053554e-01, 1.07343078e+00, -1.07876986e-01,\n -4.65283334e-01, -2.42567515e+00, -7.49818504e-01, 9.52145338e-01,\n -3.26553851e-01, -3.10787022e-01, -1.50825381e+00, 2.70121312e+00,\n -1.17554486e+00, -2.91936541e+00, 2.12064099e+00, 1.53117442e+00,\n -4.80946362e-01, -2.86469907e-01, -1.48246002e+00, 2.37165618e+00,\n 3.52479219e-02, -3.22444153e+00, -3.05905151e+00, -7.33895898e-01,\n -4.23499203e+00, -3.93101215e+00, -5.76530218e-01, 2.87378001e+00,\n 4.51699197e-01, -1.29670966e+00, 2.01036215e-01, 3.49831998e-01,\n 3.87923360e-01, -3.75383198e-01, 2.42779160e+00, 2.94953537e+00,\n -1.38986540e+00, 4.33640718e+00, -4.76655483e-01, -4.83209085e+00,\n -2.08360702e-01, 2.74711180e+00, 1.20358467e+00, 3.73331875e-01,\n 8.80917311e-01, 4.42702866e+00, 6.54141486e-01, -1.17656231e+00,\n -4.44818735e-01, 1.49961746e+00, -1.38378274e+00, 2.20370150e+00,\n 1.74790633e+00, 1.75777316e+00, 1.81595385e+00, -3.65627468e-01,\n 1.33242488e+00, -2.46422815e+00, -1.26262796e+00, 1.32544804e+00,\n 1.56579089e+00, -1.26347923e+00, 3.84304732e-01, 4.71417189e+00],\n dtype=float32)), ('a', array([ 5.07802844e-01, -1.06117237e+00, 4.41133785e+00, -1.17083825e-01,\n 3.69355381e-01, -1.96878731e+00, 1.22007346e+00, 6.44900560e-01,\n -1.90829778e+00, -8.80424321e-01, -1.91740125e-01, -4.48422813e+00,\n -2.94252336e-01, 1.23842001e+00, -8.20129097e-01, 4.62310016e-01,\n -6.71495497e-01, -2.55870819e+00, 3.16150141e+00, -2.39068580e+00,\n -9.16247845e-01, 3.58080536e-01, -4.86178780e+00, 1.29958123e-01,\n 1.25049412e+00, 3.76713610e+00, -1.99166998e-01, -1.20508134e+00,\n 7.62005925e-01, 4.72482204e-01, 2.55354929e+00, 3.69455159e-01,\n -4.35217321e-01, -2.67912769e+00, 5.93520522e-01, -3.08449841e+00,\n -1.12360549e+00, -2.67890787e+00, -2.35526967e+00, 2.12023640e+00,\n 1.74699140e+00, -7.41613030e-01, 3.95051527e+00, -1.98433268e+00,\n -3.50120163e+00, -2.51622736e-01, -4.70171452e-01, -2.29992628e+00,\n -1.21117032e+00, -3.26710391e+00, -2.64911175e+00, -2.62089968e+00,\n 8.49430040e-02, 3.45292687e-01, -1.21689034e+00, 7.39157915e-01,\n -1.37094975e+00, -3.66611767e+00, -3.22945625e-01, -4.91381586e-02,\n 2.49619770e+00, 1.08188200e+00, 1.12249601e+00, 7.21617937e-01,\n 3.04453087e+00, 2.04524064e+00, 1.39251125e+00, -1.07958305e+00,\n -2.78507543e+00, 2.64367402e-01, -1.30487835e+00, -3.08552909e+00,\n 7.89369941e-01, 2.52710581e-01, -3.10505271e-01, -1.57924914e+00,\n 2.15336576e-01, -1.15986633e+00, 1.14758956e+00, -1.94586039e+00,\n 1.68117210e-01, 9.11324263e-01, -1.53639305e+00, 2.91550851e+00,\n -1.73234165e+00, -6.43028438e-01, -2.13933229e+00, 1.90026927e+00,\n -4.41903543e+00, -3.59838915e+00, 5.59211671e-01, -7.28084087e-01,\n -2.23688459e+00, -1.77448952e+00, 9.47833836e-01, -1.48217678e+00,\n -7.39580870e-01, -3.38110781e+00, 4.52060550e-01, -3.91360283e+00,\n 9.02087390e-01, -3.34385204e+00, 2.17929602e+00, 3.55464602e+00,\n 1.64799213e-01, -2.98888874e+00, -9.81497347e-01, 7.39489317e-01,\n 3.39735031e-01, -6.04395032e-01, 4.59147900e-01, 3.23970318e+00,\n 1.01950121e+00, 3.63660216e-01, -1.97952116e+00, 4.04764986e+00,\n -2.89851189e+00, 1.72316039e+00, 2.63089848e+00, -1.40337014e+00,\n 8.24954987e-01, -3.96322131e-01, -8.23990524e-01, -4.58814323e-01,\n 6.78181171e-01, 2.26164985e+00, -2.93313909e+00, 2.52867556e+00,\n -2.69507265e+00, -1.61709809e+00, 1.54914951e+00, 4.62550819e-01,\n 2.05633616e+00, -1.15216374e+00, 4.43822527e+00, -5.75630724e-01,\n 3.95617545e-01, -3.90297222e+00, -5.32317829e+00, 2.28882730e-02,\n -1.93847144e+00, -1.69649816e+00, 1.82130408e+00, -2.82435822e+00,\n 2.48808432e+00, -2.62913418e+00, -5.69189131e-01, -1.63734996e+00,\n 1.93988514e+00, 3.49726439e+00, 4.96956259e-02, 2.58031869e+00,\n -2.63546944e+00, -3.18895555e+00, 5.05302072e-01, -1.42982805e+00,\n -3.07441616e+00, 2.30720568e+00, -1.83655763e+00, 1.44350350e+00,\n 1.79443610e+00, 2.58027267e+00, 2.40159035e+00, 7.52484202e-01,\n 1.38622928e+00, -4.45992351e-01, 2.60981560e+00, 4.05805779e+00,\n 3.63905430e-01, -3.56114179e-01, -1.61782146e+00, -1.89434052e-01,\n 2.72379684e+00, 1.87250268e+00, -1.42823720e+00, 1.71746516e+00,\n 7.47376919e-01, -5.99662215e-03, 1.84485018e+00, -2.52807021e-01,\n -9.97238457e-01, -5.90428531e-01, 9.19176042e-02, -2.21798182e+00,\n -1.07128167e+00, -1.18757975e+00, 1.05657436e-01, 1.88352168e-03,\n 2.18691945e+00, 4.33225489e+00, -1.00709224e+00, -3.20988238e-01,\n -3.45059097e-01, -1.43129140e-01, -8.93366814e-01, -8.27056289e-01,\n -1.86295247e+00, -3.34881186e-01, 1.23703182e+00, -8.19636226e-01,\n 6.89537644e-01, 4.89349425e-01, 2.87305212e+00, -2.58215761e+00,\n -3.20013475e+00, 1.91823447e+00, 1.51000750e+00, -8.54956150e-01,\n -3.32648969e+00, 3.15958047e+00, -1.55761981e+00, 4.43540955e+00,\n 7.69676805e-01, -1.24326539e+00, 1.30096889e+00, 3.51839590e+00,\n -9.51706767e-01, 2.29673699e-01, 1.63190985e+00, 1.52527463e+00,\n 3.47037864e+00, -1.24515332e-01, -2.07874608e+00, -3.54705125e-01,\n -4.37284291e-01, 3.34935999e+00, 3.01289368e+00, -3.11615199e-01,\n 7.86303711e+00, -1.27987528e+00, 8.01652670e-01, 4.28349167e-01,\n -5.72625771e-02, 5.34797955e+00, 1.05822182e+00, 3.04552841e+00,\n -2.04093561e-01, 2.43633911e-01, 2.15612173e+00, 3.41290593e-01,\n -1.36016399e-01, 1.78708136e+00, -4.35770839e-01, 2.23096085e+00,\n 1.18515706e+00, -2.34348595e-01, -1.42986584e+00, -8.86536464e-02,\n 8.53213489e-01, -3.59568739e+00, -2.75205421e+00, -5.04335976e+00,\n 1.11166179e-01, -1.06448388e+00, -3.52669382e+00, -1.38224888e+00,\n -2.25509620e+00, -3.93562007e+00, 2.79908717e-01, 2.92531347e+00,\n -6.31487012e-01, 3.57270420e-01, -9.98343825e-01, -2.57038546e+00,\n 6.29045606e-01, 2.66126776e+00, 7.26686716e-02, 7.27616549e-02,\n 2.62545824e+00, -1.23612285e+00, 3.08900774e-01, -1.72362924e+00,\n -8.05987954e-01, 1.67993188e+00, -4.41004372e+00, 7.81975091e-01,\n -1.72089052e+00, -2.13537788e+00, -1.76450133e+00, 2.89367175e+00,\n 2.14982939e+00, -9.75397527e-02, -2.13093948e+00, 1.58561468e-02,\n 3.22913051e+00, -1.87988353e+00, -1.90858531e+00, 2.99272490e+00,\n -2.14168692e+00, -5.64497352e-01, -2.43782949e+00, 2.09602976e+00,\n -4.97858524e-02, -5.31659889e+00, 3.40418720e+00, -1.12013662e+00,\n 1.96556652e+00, 2.23409629e+00, -1.72265005e+00, -8.07241261e-01,\n -5.67136467e-01, -1.01025116e+00, -2.94456911e+00, 1.52192259e+00,\n 2.00670075e+00, -5.77446043e-01, -2.95572472e+00, -2.50309300e+00,\n -3.40872650e+01, -3.88903528e-01, 5.09424925e-01, -3.71284628e+00,\n 3.13348234e-01, 1.40375626e+00, -3.42622566e+00, -4.53453660e-01,\n -2.06281996e+00, 2.50510037e-01, -4.01020050e+00, -5.15466630e-01,\n -3.66351008e-03, 6.01433396e-01, 1.26050258e+00, -3.58434057e+00,\n -3.14445114e+00, -2.79902611e-02, 2.55207014e+00, 2.72946715e+00,\n -2.24160552e-01, 6.47100091e-01, -6.03894770e-01, -4.88319159e+00,\n -2.22214103e+00, 4.12485218e+00, -4.32007837e+00, 1.85806513e-01,\n -7.28859186e-01, 8.93635511e-01, -1.99701977e+00, -7.77530670e-03,\n 1.50366592e+00, 3.30237269e-01, -1.65977979e+00, 1.60207808e+00,\n 4.48381305e-01, -3.30118203e+00, 1.12954330e+00, -1.98948994e-01,\n -6.07776284e-01, -6.79933667e-01, 2.13775086e+00, 5.05365229e+00,\n 8.80399585e-01, -3.43154430e-01, -5.60864806e-01, 1.33308887e+00,\n -1.10947263e+00, 2.65330505e+00, -1.96705842e+00, -1.50493264e+00,\n 8.41777563e-01, -2.12496495e+00, 1.06873441e+00, 6.87127888e-01,\n 5.25961733e+00, 9.70314860e-01, 3.54917908e+00, -7.41479993e-01,\n -2.94973612e+00, -4.85063267e+00, 1.41322637e+00, 6.02266264e+00,\n 6.82046294e-01, -9.50667202e-01, 2.34019458e-01, -5.08324671e+00,\n 1.76081693e+00, -9.41113949e-01, -1.05289364e+00, 2.22720075e+00,\n 1.78534257e+00, -1.10745964e+01, -4.38632756e-01, -3.95092010e+00,\n -1.10130298e+00, 7.61848986e-02, 2.57536101e+00, -9.75444853e-01,\n -2.10465693e+00, -2.03837109e+00, 9.32009935e-01, -2.58459866e-01,\n 1.78992426e+00, 2.39304876e+00, -5.18357158e-01, 1.58856404e+00,\n 1.76648664e+00, 1.11896563e+00, 2.79214978e-02, 5.51461649e+00,\n 1.66734719e+00, 1.56763470e+00, -1.08275199e+00, 6.82215452e-01,\n 3.13286686e+00, -1.44376016e+00, 2.61841655e+00, 2.43181682e+00,\n 2.05461645e+00, -1.72297740e+00, 1.30390644e+00, 2.88440895e+00,\n 2.18690109e+00, -1.09910583e+00, -4.71469545e+00, 3.72804785e+00,\n 2.25670862e+00, 2.77444339e+00, 2.70471358e+00, 6.03573501e-01,\n -9.40879881e-01, -6.86792493e-01, -2.14993620e+00, -2.56840765e-01,\n -9.94733214e-01, 1.95555925e-01, 2.40341353e+00, 7.04857230e-01,\n 5.46656370e-01, 1.14875436e+00, -1.14626682e+00, -4.46563625e+00,\n -2.22677779e+00, -1.12981141e-01, -2.37440062e+00, 1.97492909e+00,\n -2.31417704e+00, -8.36191416e-01, -2.32006836e+00, 4.01829243e+00,\n 1.55427909e+00, -1.22357666e+00, -7.59840965e-01, -9.67922449e-01,\n -2.07545590e+00, 5.18809080e+00, 1.61542106e+00, -4.74571139e-01,\n -1.26539969e+00, -7.34522104e-01, 5.00223994e-01, -2.58528233e+00,\n -1.81103826e+00, -1.88354462e-01, -1.21749103e-01, -1.60647023e+00,\n 1.83811545e-01, -2.54272413e+00, -1.95348275e+00, -5.21178782e-01,\n 3.59991503e+00, 1.51090324e+00, -1.28655958e+00, -1.44162464e+00,\n 4.55789804e+00, -1.58368540e+00, -1.89961684e+00, -2.32443953e+00,\n 1.17231905e-02, -6.03009880e-01, 3.73146415e+00, 2.91737646e-01,\n 1.38632107e+00, -3.24780792e-01, -1.19417465e+00, 9.12755072e-01,\n -3.50191236e+00, -3.53854895e+00, -3.02501297e+00, 1.12756145e+00,\n 1.39822900e-01, -2.44987822e+00, 2.43716383e+00, 1.78301072e+00,\n 5.77345848e-01, -1.00931656e+00, -1.38846147e+00, -2.19917059e+00,\n 1.98742557e+00, 1.47544742e+00, 3.81716037e+00, -6.35063350e-01,\n -2.97075450e-01, -3.58690917e-01, -2.20702887e+00, 2.05800200e+00,\n 7.39620328e-02, -5.86174548e-01, -1.27619827e+00, 1.70883048e+00,\n -2.66930866e+00, -4.07645583e-01, -1.11055362e+00, 7.86311567e-01,\n 2.43980217e+00, 2.22618327e-01, -1.18055046e+00, 2.48940721e-01,\n -4.89941448e-01, 5.94013214e-01, -1.36220455e-01, -1.49416316e+00,\n 6.51035011e-01, -5.12503564e-01, -1.78989553e+00, -1.09263206e+00,\n -9.76574302e-01, -2.54426908e+00, 1.56370020e+00, -1.55812192e+00,\n 1.17911470e+00, -3.28970242e+00, 3.68504703e-01, -6.61712408e-01,\n 9.97716129e-01, 1.88261294e+00, -2.78714108e+00, 1.68466747e-01,\n 3.55342078e+00, -1.59390056e+00, -1.00930166e+00, -5.26037365e-02,\n 6.21717721e-02, -9.21564817e-01, -2.48046923e+00, -1.58819437e+00,\n 2.64276767e+00, 2.85040438e-01, 2.09645009e+00, -3.56618643e+00,\n -1.16459990e+00, 1.16243958e+00, -8.69494259e-01, -2.71303058e-02,\n -3.44445848e+00, 5.97154617e-01, -1.75839275e-01, 5.49545586e-02,\n -2.31434727e+00, -1.39014959e+00, -4.36397934e+00, -6.43653393e-01,\n 2.57969022e+00, -5.65755606e+00, 2.35415936e+00, -1.94090855e+00,\n -1.06177473e+00, 5.73748946e-01, -1.47189558e+00, 1.09320879e+00,\n 1.10538101e+00, -3.65148044e+00, 6.66398406e-01, -6.18180990e-01,\n -1.71033609e+00, 3.18805575e+00, -1.36622608e-01, 3.14431858e+00,\n 1.16603702e-01, 1.61620259e+00, 1.12834990e+00, 1.66340411e+00,\n -9.73834753e-01, -1.63568699e+00, 4.05063200e+00, 1.02819049e+00,\n -1.92326754e-01, 3.92267418e+00, -2.95773029e+00, 3.63398123e+00,\n 1.78934306e-01, 2.40313959e+00, 1.83033335e+00, -1.71921349e+00,\n -4.49493349e-01, -4.02924657e-01, -9.45420861e-01, 1.43751681e+00,\n -1.52871394e+00, -3.50116432e-01, -4.77039278e-01, -8.08803678e-01,\n 2.67264271e+00, 3.91319335e-01, 4.09246206e-01, -1.02901125e+00,\n 4.94470507e-01, -2.06567407e+00, -3.29359341e+00, 1.26669192e+00,\n -1.99827135e+00, -1.09899294e+00, -4.71857727e-01, -1.28315598e-01,\n -2.38229007e-01, 8.01404715e-01, 8.84505033e-01, -2.79429615e-01,\n -6.91534281e-01, 2.48864412e+00, 2.53036070e+00, 1.14327919e+00,\n -3.78893161e+00, 2.31249833e+00, -1.61336541e+00, -2.89638615e+00,\n -3.47799271e-01, 3.16783047e+00, -1.54907310e+00, -1.55172539e+00,\n 2.60501242e+00, 1.06240475e+00, -2.97969818e-01, 9.11554039e-01,\n 2.49240923e+00, -4.97940987e-01, -1.99682307e+00, 1.78390229e+00,\n -1.32283962e+00, -1.06370008e+00, 5.15147328e-01, -2.31077647e+00,\n -6.70537055e-01, 2.36643624e+00, 1.30152059e+00, -3.69527984e+00,\n -9.86149549e-01, 3.03048879e-01, 7.82625377e-01, 1.21344936e+00,\n -1.19349408e+00, -2.34933186e+00, 1.91485792e-01, -9.61004138e-01,\n -1.79026401e+00, -4.22141552e+00, 5.46995687e+00, 3.08210850e+00,\n -7.32132733e-01, -1.09064054e+00, 3.63925910e+00, 1.76205671e+00,\n -1.32313418e+00, -3.50435543e+00, 2.45584464e+00, -7.95406759e-01,\n -3.65489388e+00, 4.22102451e+00, -8.90663922e-01, 8.43321621e-01,\n 1.03537202e+00, -1.41281724e+00, 1.30674362e+00, -1.48225832e+00,\n 9.56342578e-01, 2.39404464e+00, 4.14453840e+00, 2.20684886e-01,\n 4.55114067e-01, 1.52212572e+00, 2.73236656e+00, -4.03751493e-01,\n 1.37280321e+00, 1.78054559e+00, -7.08067536e-01, -5.55735648e-01,\n -5.83247304e-01, -1.09083414e+00, 3.95735145e-01, 1.48730338e-01,\n 5.17477751e+00, 5.46011019e+00, 1.33546066e+00, -6.29849136e-01,\n 5.28645098e-01, 1.68083215e+00, 7.65509248e-01, 1.10275769e+00,\n -3.48606586e-01, 2.02490807e+00, -3.29611093e-01, -1.07311368e+00,\n -9.20139074e-01, 5.18852532e-01, -7.53180087e-01, -1.68931723e+00,\n -6.60301208e-01, -2.77323246e+00, -5.31252474e-02, 2.01806021e+00,\n -2.44604111e+00, -2.72015381e+00, 3.57080531e+00, -2.64392793e-01,\n 8.90464902e-01, 6.57258213e-01, 1.99155200e+00, -2.11901236e+00,\n 1.64608383e+00, -1.03352833e+00, 7.23861575e-01, 2.97336012e-01,\n -2.02465057e+00, -1.23384595e+00, 9.09818888e-01, 2.12584972e+00,\n -3.65417433e+00, -1.75048709e+00, 3.22526288e+00, 3.88889432e-01,\n 2.81787306e-01, 2.38002348e+00, -1.96070552e+00, 5.57268322e-01,\n 1.57684326e+00, -6.46983802e-01, -8.87143433e-01, 7.76874661e-01,\n -3.64990401e+00, -2.13031363e+00, 1.47392964e+00, 3.33579755e+00,\n -6.97958112e-01, -3.91470766e+00, 3.56959105e-01, -6.09571874e-01,\n -1.59522939e+00, 2.55956650e-01, -1.42578438e-01, 1.15956616e+00,\n -1.03169394e+00, 2.50078917e+00, 1.26865298e-01, -4.29148293e+00,\n -1.64948845e+00, -4.68269527e-01, -1.75706357e-01, -2.79768407e-01,\n 8.80624533e-01, 2.49166012e+00, 1.34315765e+00, -1.40455449e+00,\n -2.67545915e+00, 1.76955140e+00, 2.61269271e-01, 6.96819007e-01,\n -1.66124344e-01, 4.86076295e-01, -6.61207080e-01, 3.98863316e-01,\n 2.42528129e+00, -2.72965026e+00, -1.36617529e+00, 2.68732762e+00,\n 7.67819881e-01, 2.14403525e-01, 3.94346774e-01, 6.74068975e+00],\n dtype=float32)), ('test', array([ 6.7454159e-01, -2.9485202e+00, -9.3779439e-01, -5.6110913e-01,\n 1.6011660e+00, 2.4794657e+00, -8.1066269e-01, -1.9208476e-01,\n -8.3791518e-01, -7.6063937e-01, -6.4914387e-01, 2.5206871e+00,\n 1.6188793e+00, -1.3911190e+00, -4.9176180e-01, -2.0117500e+00,\n -2.3552936e-01, 8.2505262e-01, 4.9045534e+00, 1.1899111e+00,\n 1.2878735e+00, 1.0135154e+00, -1.5534127e+00, -1.1296377e+00,\n -2.1283522e-01, 1.2046744e+00, -1.4018006e+00, -2.2609085e-01,\n -3.0025188e-02, 1.3588805e+00, 1.1163551e+00, 1.6365018e+00,\n 2.2707908e+00, 4.3582525e+00, -4.7298951e+00, -1.9903003e+00,\n 1.7178652e+00, 1.7830758e+00, -4.4581380e+00, 2.3848023e-01,\n -7.7810258e-02, -1.9992583e+00, 2.3104734e+00, -1.9359531e+00,\n -1.0847528e+00, -8.4899110e-01, 7.4549294e-01, -3.0071647e+00,\n -1.8140261e+00, -2.2022645e+00, -3.8446991e+00, 1.7053132e+00,\n -2.0258565e+00, 1.7132781e+00, -7.5042576e-01, -1.6629399e+00,\n -3.3699852e-01, -1.1778991e+00, -2.0571127e+00, -1.6935511e+00,\n 1.9343424e+00, 1.2101406e+00, -8.2859969e-01, -3.9708874e-01,\n 3.0229936e+00, -3.0691069e-01, 3.0227385e+00, 1.6994691e+00,\n -2.1029420e+00, 7.9853350e-01, -4.7545843e+00, 3.6953602e+00,\n 1.9907160e-01, -1.9756621e+00, -1.1531063e+00, 2.9534793e-01,\n -6.2364006e-01, 8.2998216e-02, 1.3671582e+00, -4.8110747e-01,\n -8.0155671e-01, 3.0010507e+00, 9.3118513e-01, 6.5583789e-01,\n 3.3422366e-02, -1.7676616e+00, 8.7482840e-02, -1.6947997e+00,\n -2.7512437e-01, 2.3508072e-03, 8.8666379e-03, -8.1585532e-01,\n -2.6670227e+00, -1.7784169e+00, 2.3081000e+00, 5.7218331e-01,\n -2.6416199e+00, -1.0310897e+00, 2.0310845e+00, -4.7691588e+00,\n -2.7874718e+00, -3.1060390e+00, -1.7195554e+00, 2.3633847e+00,\n -1.5312880e+00, -1.7269542e+00, -1.3894429e+00, -2.2982191e-02,\n -1.6309316e+00, -8.2867861e-01, 1.3217248e+00, 2.7352393e+00,\n 1.2000694e+00, -1.4446099e+00, -1.6270702e+00, 2.9784577e+00,\n -3.5486406e-01, -2.2850308e-01, 4.0588379e+00, 5.2585852e-01,\n -9.7687805e-01, 8.3297765e-01, -1.7562144e-01, 2.7430701e+00,\n 5.7804936e-01, 1.9571285e+00, -1.3075140e+00, -2.2691697e-01,\n -2.4482553e+00, 8.0450487e-01, 6.2241292e-01, 3.9451001e+00,\n 1.3857167e+00, 1.1599030e+00, -1.9921854e+00, -2.4505224e+00,\n 1.8359181e-01, -6.9700670e-01, -2.2796805e+00, -2.4446363e+00,\n 1.6709605e+00, 2.0261898e+00, -4.7978556e-01, 6.6757298e-01,\n 1.0969636e+00, -4.7542506e-01, -4.8633795e+00, -1.6106855e+00,\n -1.7652805e+00, 1.3570098e+00, -1.1417339e+00, 3.9819660e+00,\n -2.3680239e+00, -8.7991381e-01, -1.3387605e+00, 7.6862955e-01,\n -3.3904150e+00, -5.2760363e-01, -1.3698745e+00, 2.4581778e+00,\n 3.1759341e+00, -2.2602688e-01, 7.8396273e-01, 3.0825744e+00,\n 3.4668574e-01, 2.3707359e+00, 2.8294353e+00, 3.3378835e+00,\n 3.4534195e-01, 5.3361368e-01, 7.9951212e-02, -5.1760733e-01,\n 1.5857354e+00, 3.6554480e-01, -3.4956665e+00, 1.3950533e+00,\n 1.0521983e+00, -1.6769440e+00, 6.0511404e-01, -4.6315768e-01,\n -2.1440337e+00, -2.6197124e-01, -2.4589000e+00, -5.8522981e-01,\n -1.0816635e+00, 1.0202338e+00, 2.6896534e+00, -3.4616728e+00,\n 1.4572464e+00, -2.4800055e+00, -3.0777216e+00, -3.3052940e+00,\n -2.8589604e+00, -3.2933536e+00, -1.2403674e+00, -7.1323931e-01,\n -1.7342139e+00, 6.5414941e-01, 9.1691226e-01, 4.7522777e-01,\n 3.5224752e+00, -7.1317625e-01, -1.2755244e+00, 2.5180531e+00,\n 9.1673195e-02, 1.0665349e+00, 2.4363656e+00, 1.8633458e-01,\n -1.1934663e+00, 1.5239868e+00, -1.6552957e+00, 4.0155096e+00,\n -8.2887244e-01, 8.2592320e-01, 1.9623172e+00, -3.5699213e-01,\n -5.5807519e-01, 1.0552856e+00, 7.7925986e-01, 9.1124749e-01,\n 2.1137004e+00, -3.3382478e+00, -1.8599565e+00, -1.0191116e+00,\n -3.1507349e+00, 1.6007267e+00, 2.5802016e+00, -2.6373525e+00,\n 3.3852406e+00, -1.6202480e+00, 2.0115142e+00, -2.8847535e+00,\n 1.3671528e+00, -8.0547184e-01, 1.8455622e+00, 2.4308660e+00,\n 2.2670443e+00, -8.5782528e-02, -2.3252585e+00, 1.7226653e-01,\n -2.0220950e+00, 3.1533876e-01, 1.0265582e+00, 1.4944217e+00,\n -5.7553989e-01, 7.9426485e-01, 3.0104668e+00, -1.5131805e+00,\n 9.6806085e-01, -1.3706075e+00, -1.2335411e+00, -6.2647681e+00,\n 1.4137797e+00, 1.9937580e+00, -5.1735840e+00, -1.6724211e+00,\n -1.7919513e+00, -3.3046021e+00, 2.9210396e+00, 2.0421333e+00,\n -4.6221008e+00, 1.8205376e+00, 3.3740339e+00, -1.9418465e-01,\n 1.8280522e+00, 6.7915863e-01, 3.9480040e+00, 1.9717607e+00,\n 3.5077696e+00, 8.0250168e-01, 1.2862362e-01, -2.0225592e+00,\n -1.4668193e-01, -9.7597980e-01, -3.0131903e+00, 1.0988512e+00,\n 5.6748724e-01, 1.1645736e+00, -1.7243446e-01, 4.9036641e+00,\n 1.8574834e-02, 2.3444030e-01, -1.8271809e+00, 1.5183340e+00,\n 6.5318646e+00, -2.0813355e+00, -8.4424019e-01, -3.5965118e+00,\n 8.8247812e-01, -1.2848207e+00, -1.1688627e+00, 3.7072713e+00,\n 6.0729563e-01, 1.2935691e-01, 5.2924805e+00, 9.3604338e-01,\n -2.1904975e-02, -4.5124367e-03, 5.4956537e-01, -2.0142798e+00,\n -2.7123289e+00, 2.7048712e+00, 3.9228806e+00, 9.7608829e-01,\n 2.8662477e+00, 1.0380677e+00, -1.5654715e+00, 6.0412198e-02,\n -2.7979744e+01, 3.4949207e-01, 2.1690545e+00, -1.5722148e+00,\n 5.8181190e-01, -1.9456251e+00, -2.8152363e+00, 7.4750572e-01,\n -4.6599889e+00, 1.6202610e+00, 2.4890316e-01, -2.2398655e-01,\n 9.7433180e-02, -2.7999571e-01, -7.2192937e-02, -1.5432613e+00,\n -1.9225668e+00, -1.9701161e+00, -2.2956483e-01, 4.0906286e+00,\n -1.9643922e+00, 5.3120816e-01, 1.6076261e+00, -2.5221786e+00,\n -3.4437079e+00, 1.4655004e+00, -1.6293845e-01, 2.8352537e+00,\n -3.0255907e+00, -9.0846360e-01, -1.6899428e+00, -1.3585156e+00,\n 1.7556839e+00, 3.2594686e+00, 5.3100133e-01, -1.5271184e+00,\n 7.1964949e-01, -7.4639630e-01, 3.5488915e+00, -1.1783072e-01,\n 1.0882401e+00, -1.2461210e+00, -4.6852857e-02, 5.7202821e+00,\n -3.4170258e-01, -2.4987847e-01, -6.7338365e-01, 1.1053495e+00,\n 3.2957518e-01, -7.4288285e-01, 1.3156901e-01, -2.3930037e-01,\n 2.7488020e-01, -2.8307202e+00, -1.4558913e+00, -2.9550189e-01,\n 3.1637490e-01, 1.5424910e+00, 1.0725863e+00, -2.0835261e+00,\n -3.8241118e-01, -2.1353705e+00, 4.7322342e-01, 3.7512918e+00,\n -5.2364719e-01, 2.7699035e-01, -2.8344598e+00, -7.9719657e-01,\n 1.5753702e+00, -1.4336437e-02, 1.7369319e-01, -3.0729508e+00,\n 2.8352597e+00, -1.2550900e+01, -2.6994975e+00, 2.9342353e+00,\n -7.6706910e-01, -2.7937555e-01, -1.9705291e-01, -1.9438813e+00,\n 8.3813316e-01, -1.6667647e+00, 2.9747885e-01, 1.3111782e-01,\n 2.1675141e+00, 9.1723037e-01, -1.6112453e-01, -5.2701063e+00,\n 1.0086018e+00, 6.4099848e-01, -1.0795178e+00, 9.5995545e-01,\n 4.7536759e+00, -6.2577897e-01, 2.5910573e+00, 1.0443070e+00,\n 1.0670985e+00, -4.2491758e-01, -1.1450024e+00, 4.3415743e-01,\n -1.3025869e+00, -3.7637930e+00, -5.0411785e-01, 8.2908064e-02,\n 1.6712284e-01, -2.2470644e+00, -5.3389983e+00, 2.4554851e+00,\n 4.2048725e-01, 1.4125308e+00, -6.9314545e-01, 2.1806238e+00,\n -3.8111252e-01, 1.0594931e-01, -1.7724484e+00, -5.0749266e-01,\n -1.5291820e+00, 3.4449261e-01, 6.6106427e-01, 9.7719741e-01,\n -1.4101398e+00, 1.0587924e+00, -1.3905164e+00, -4.5856011e-01,\n 4.3098798e-01, -2.2400534e+00, -3.1165771e+00, -2.5050352e+00,\n -6.4245653e-01, -6.7435640e-01, 5.5712372e-01, 2.3543060e+00,\n -6.3309282e-01, -1.8685431e+00, -3.3579215e-01, -3.0565721e-01,\n -2.0926437e+00, 3.9096832e+00, -7.4025404e-01, 2.3010716e+00,\n 4.0156527e+00, -5.0853783e-01, 2.2318130e+00, -1.0117307e+00,\n 1.1005436e+00, -1.4936535e+00, 3.3458251e-01, -7.4931216e-01,\n -1.6413043e+00, 1.2961667e+00, 3.2943654e+00, -1.6017804e+00,\n 1.5648829e+00, 2.8548667e+00, 2.2776265e+00, 3.2134330e+00,\n 3.6694341e+00, -3.6612396e+00, 9.1231167e-01, -2.7845681e+00,\n 5.8567369e-01, 1.6318277e-01, 1.3142579e+00, -2.0883843e-01,\n 2.7095246e+00, 1.8372931e+00, -1.8416164e+00, 7.6144511e-01,\n -8.6211324e-01, -2.0925560e+00, 4.9880201e-01, -2.3730643e+00,\n -8.5365713e-01, 3.6383100e+00, 1.7978468e+00, -2.2195873e+00,\n -4.7145754e-02, -2.1318762e+00, -1.2122815e+00, 2.0655005e+00,\n 1.3078371e+00, 2.4056952e-01, -2.0961735e-01, -1.2023876e+00,\n -1.5414235e-01, 2.3296189e+00, -1.4581020e+00, -3.5422748e-01,\n 3.3555022e-01, 2.7417049e+00, -4.4721091e-01, -3.6386222e-01,\n 1.0500710e+00, 1.6650391e+00, 1.0505176e+00, -7.3722863e-01,\n 1.1099629e+00, 3.4338884e+00, 2.2211311e+00, 1.8710246e+00,\n -1.2937369e+00, -4.2000465e+00, 1.7472847e+00, -7.7561635e-01,\n 1.8234248e-01, 2.9546952e-01, -1.3764184e+00, 2.3141527e+00,\n -2.7311432e+00, -1.3836352e+00, 1.6433214e+00, -2.5646377e+00,\n -3.6945858e+00, -3.9826958e+00, 2.0479882e+00, 3.0926204e+00,\n 1.0302339e+00, 5.3717947e-01, 5.2819753e-01, -1.8396453e+00,\n -1.7234549e-01, -1.8522911e+00, 2.1416790e+00, -1.4160035e+00,\n -3.5686210e-01, -2.6201816e+00, 1.9618342e+00, -3.6884031e+00,\n -2.7682939e+00, -8.9253902e-01, 3.6373088e-01, 2.2010601e+00,\n -1.5600955e+00, 1.7532727e-01, -9.2840153e-01, -2.7775881e+00,\n -4.6001291e+00, 2.0467737e+00, -5.5051696e-01, -1.4272431e+00,\n -9.3356025e-01, -1.4253471e+00, -5.0033751e+00, 1.2293327e+00,\n -3.7727678e-01, -5.7396752e-01, -1.5653009e+00, 9.0757251e-01,\n 2.4195855e+00, 4.1939383e+00, -1.8919008e+00, 7.9781222e-01,\n 3.6484382e-01, -2.4352460e+00, 1.2266825e+00, -3.2968292e+00,\n 2.3333864e+00, -1.1053841e+00, -2.3256049e-01, 3.7436435e+00,\n -1.9000156e+00, 1.0733070e+00, -7.6266372e-01, 2.6880543e+00,\n -1.3394560e+00, 1.0983888e+00, -1.6400521e+00, 4.2876032e-01,\n 1.7290025e+00, 2.7860694e+00, -2.9117098e+00, -8.0596870e-01,\n -2.6325148e-01, 2.0402193e+00, -2.9534655e+00, -5.0605452e-01,\n 7.5317931e-01, -3.1212645e+00, -3.5827044e-01, 3.3229139e+00,\n -1.3173406e+00, -2.6654172e-01, 2.1813560e+00, 1.2030385e+00,\n -1.1393309e+00, 2.2912612e+00, -2.4206247e+00, -2.5461216e+00,\n -1.6820468e+00, -3.6278114e+00, 1.6799066e+00, -6.0671437e-01,\n 2.1508217e-01, 8.0482394e-01, 1.1128571e+00, 3.7576594e+00,\n -1.3334596e+00, 1.4441613e+00, 7.0231605e-01, -7.7842307e-01,\n -2.1327078e+00, 1.7863853e+00, 7.2396028e-01, -6.1425591e-01,\n -2.3452106e+00, 1.7802800e+00, -4.5547342e-01, -1.2603570e+00,\n -5.7058239e-01, 4.1208634e-01, -3.9934391e-01, 2.6481837e-01,\n -2.6431763e-01, 9.7862750e-01, -3.7226193e+00, -4.6707407e-02,\n 1.5567477e+00, -2.5732207e+00, 2.6971713e-01, 4.8541842e+00,\n -1.9882680e+00, 3.5352120e-01, 2.7531719e+00, -2.3147726e+00,\n -1.1406830e-01, 2.2577653e+00, -9.8775107e-01, -7.3283195e-01,\n 3.2155073e-01, 1.2584739e+00, 7.8512627e-01, 1.3955966e+00,\n 1.6924717e+00, 3.1141372e+00, 2.2362034e+00, 3.4651366e-01,\n 2.1716623e+00, -7.0629317e-01, -7.0598114e-01, 1.4169483e+00,\n -3.1333500e-01, -2.6284633e+00, 3.0263585e-01, 7.9585159e-01,\n -1.2160642e+00, -2.7030520e+00, -1.2417159e+00, -1.5634908e-01,\n -2.6419582e+00, 4.7861786e+00, -1.9751813e+00, 5.0474286e-01,\n -3.1389322e+00, -2.6988220e-01, -5.5410075e-01, -1.9638355e+00,\n 2.6873729e-01, -8.1858253e-01, -3.2008545e+00, 1.6037093e+00,\n 3.5142539e+00, -9.3081564e-01, 1.1142590e+00, -5.4520124e-01,\n -6.9684625e-01, 1.8266375e+00, 1.1813836e+00, -1.8361135e+00,\n -8.1695926e-01, -2.0470436e+00, -2.6212220e+00, 2.4359646e+00,\n 2.5919704e+00, 4.4629083e+00, 1.6344283e+00, -5.6113893e-01,\n 2.9651020e+00, 4.5486015e-01, -2.1389031e+00, -1.9014281e+00,\n 1.3523400e+00, 8.0850285e-01, -1.3208122e+00, 5.0747514e-01,\n 4.7916117e+00, -2.4160296e-02, 2.3206816e+00, 1.5962381e+00,\n 4.5423156e-01, -2.1983457e+00, -4.1315570e-01, -9.4483815e-02,\n -4.7505967e-02, -2.6341305e+00, 3.6207583e+00, -1.1999235e+00,\n -1.2500976e+00, -1.2757815e+00, 2.3853219e+00, -3.7675264e-01,\n -1.2696260e-01, -2.2368083e+00, -6.1488390e-01, -2.1710598e+00,\n -3.1741631e+00, 6.9366455e-02, -1.2309127e+00, 2.4059958e+00,\n -3.8937249e+00, -1.9036443e+00, 2.7996178e+00, 1.3424644e+00,\n -6.5701127e-01, 2.3665893e+00, -2.5992389e+00, -1.1981647e+00,\n 1.9235219e+00, -1.5745518e+00, 2.5923760e+00, -2.7526841e+00,\n -3.3179617e+00, -2.3521174e-01, -1.8332132e+00, -5.3201962e-01,\n 2.0913906e+00, -7.0595036e+00, -2.0157035e-01, -2.0286736e+00,\n 5.8290410e-01, 2.0489588e+00, 2.8443568e+00, 7.9657972e-01,\n 8.1662977e-01, 5.8368473e+00, -7.7982366e-01, -3.3870583e+00,\n 1.2299931e+00, -2.7636406e+00, 1.8091664e+00, -8.8804114e-01,\n -2.2952509e+00, 1.3853090e+00, 1.3686513e+00, -1.9734201e+00,\n -5.8277565e-01, 9.5532954e-01, -2.5400834e+00, 4.8777759e-02,\n 1.2867297e+00, 1.5484427e+00, 2.5072465e+00, -2.1705463e+00,\n 1.0178548e+00, -2.7823870e+00, -1.3135016e+00, 1.4083505e-02,\n 2.1600375e+00, 1.2855756e-01, -5.0620764e-01, -2.5290141e+00],\n dtype=float32)), ('[SEP]', array([ 7.47277200e-01, 1.19544253e-01, -2.69795090e-01, 4.65479672e-01,\n -7.22580433e-01, -7.14895427e-01, 2.82050461e-01, -9.72757220e-01,\n 7.98530698e-01, 1.57996044e-01, 2.06953719e-01, -4.15753901e-01,\n 3.69492382e-01, -3.30903351e-01, -6.45951927e-01, -3.03770036e-01,\n -7.99396038e-02, -3.11046541e-01, 1.63409635e-01, -2.50373840e-01,\n 1.33972168e-02, 6.13581687e-02, 4.80295807e-01, -2.87004054e-01,\n 1.23746045e-01, -2.39073932e-02, -1.82742327e-01, -2.11694688e-02,\n -3.77651870e-01, -8.63070726e-01, -7.34691739e-01, -6.24354899e-01,\n 7.68505931e-02, 7.11904764e-01, 2.25533441e-01, -6.41036704e-02,\n 2.92308301e-01, -4.29102033e-01, -4.06768024e-01, -7.01638609e-02,\n -5.31484723e-01, 3.67732048e-01, -2.60485172e-01, 1.99946567e-01,\n 1.87113255e-01, -4.68064845e-01, 3.47647667e-01, 3.39790463e-01,\n -9.07869563e-02, 5.28540909e-01, -1.47728175e-02, 3.28741491e-01,\n -2.00101003e-01, 5.14364660e-01, 1.24816768e-01, 2.15644404e-01,\n 5.21012783e-01, -4.48380649e-01, 2.46154994e-01, 3.99183780e-01,\n 1.35531500e-01, 2.83468783e-01, -1.36227399e-01, -3.87113094e-02,\n 4.18502867e-01, 4.57278863e-02, -3.36264521e-02, -6.93121731e-01,\n -6.27542615e-01, -1.61436304e-01, 6.05547428e-03, -1.11898327e+00,\n 5.70075214e-01, -2.08351761e-02, 2.00511113e-01, 4.02795345e-01,\n -4.79098499e-01, 8.98899078e-01, 1.28819630e-01, -1.22114271e-02,\n -2.08447486e-01, 8.41861218e-03, -1.07850820e-01, -3.46357748e-03,\n 7.18200982e-01, -1.25811230e-02, -6.00567907e-02, 2.08290145e-01,\n -1.55712605e-01, -3.31124783e-01, 4.29554224e-01, 3.69922101e-01,\n -4.87964153e-02, -1.73442364e-02, -1.02721281e-01, 3.94879848e-01,\n -3.42686921e-01, 2.35362455e-01, -4.39764857e-02, -2.05473557e-01,\n 2.52509892e-01, -1.23153642e-01, 1.30239755e-01, 8.72043967e-01,\n -6.53034300e-02, -1.95996016e-01, 2.92918593e-01, 6.08742833e-01,\n 7.61048198e-01, 8.51544619e-01, 1.10713315e+00, 3.57210338e-01,\n 3.79045665e-01, -1.29406676e-02, -3.68514478e-01, -4.49868321e-01,\n 5.29984176e-01, 2.11055130e-01, 7.31787562e-01, 5.67774773e-02,\n -5.39311469e-01, -7.96676636e-01, 2.08583236e-01, 3.40144134e+00,\n -4.93464172e-02, 1.18327893e-01, 6.52801618e-02, -1.06249046e+00,\n -1.89788789e-01, -5.41747272e-01, -5.16198099e-01, 5.01799583e-01,\n 4.70110595e-01, 9.42048430e-01, 1.21226296e-01, 4.01262939e-01,\n -3.49941194e-01, 2.98638374e-01, -7.18815088e-01, 2.96448827e-01,\n -6.34001940e-02, 9.73003626e-01, 5.81524253e-01, -1.54853392e+00,\n -2.55737454e-02, 6.53061390e-01, 7.50534296e-01, 3.34834903e-02,\n 5.96073031e-01, -4.84697998e-01, 8.43108535e-01, -3.98649156e-01,\n -3.65941882e-01, 3.48592177e-03, -5.76770365e-01, -5.07139981e-01,\n -1.40983313e-01, 2.24765725e-02, 6.65667057e-01, 7.68852413e-01,\n 3.92320365e-01, 5.01994371e-01, 6.23312831e-01, 1.09278820e-01,\n -3.96356910e-01, 2.01923564e-01, -1.29441094e+00, -1.95931196e-01,\n 6.70691848e-01, 5.40560186e-01, -5.02358973e-01, -3.14169109e-01,\n -4.17603165e-01, -1.66797787e-02, -5.21470070e-01, 3.52788091e-01,\n 9.72120017e-02, -1.23987339e-01, -3.26862514e-01, -9.92481291e-01,\n -1.46224356e+01, -7.77860403e-01, 8.66041034e-02, -4.49697524e-02,\n -6.58191890e-02, -1.51695937e-01, -1.00449562e+00, -7.49273524e-02,\n -2.50288844e-03, -1.15428269e+00, 3.27301592e-01, 7.23046958e-02,\n -7.38146544e-01, 3.55511546e-01, 2.69556075e-01, -2.17322677e-01,\n -1.51871920e-01, 1.75427794e-02, -4.69003946e-01, 1.78998262e-01,\n 2.50537574e-01, -4.33816314e-01, -3.17495286e-01, 5.01106560e-01,\n -2.39633605e-01, -1.63334024e+00, 3.99657786e-01, -3.58009338e-01,\n 1.49196871e-02, 1.63568228e-01, -1.11279929e+00, -7.92719126e-02,\n 1.66915983e-01, -8.36365342e-01, 5.94085932e-01, -2.83981532e-01,\n 3.17858636e-01, -4.02517617e-01, -8.62414658e-01, -4.34086621e-01,\n -8.40050399e-01, -5.54817200e-01, -2.09124923e-01, -3.39545369e-01,\n 7.26818860e-01, -2.69183588e+00, 6.79075062e-01, 1.01200068e+00,\n 7.06579328e-01, 1.58233076e-01, -2.28230715e-01, 3.01975399e-01,\n 4.33452338e-01, 3.22976172e-01, 4.75912184e-01, -6.12596050e-02,\n 1.51240796e-01, -7.11405352e-02, -5.99880099e-01, -1.33344382e-01,\n 5.90555519e-02, 4.86671776e-01, 4.55638707e-01, -2.19054624e-01,\n -6.77810669e-01, -9.51805174e-01, -1.02408260e-01, 7.83316493e-01,\n 1.01495242e+00, 3.64032388e-03, -4.24309462e-01, -1.00062835e+00,\n 3.22966650e-02, -5.62163532e-01, 3.73830587e-01, 8.12983096e-01,\n 4.24623489e-03, 2.73465037e-01, 1.23026931e+00, -2.86635250e-01,\n 4.70842779e-01, 7.86568701e-01, 2.14118257e-01, 1.14284702e-01,\n -5.16113579e-01, 1.15759027e+00, 2.80978501e-01, 4.09993351e-01,\n -2.42483690e-01, 3.15053463e-01, 2.12339818e-01, -3.87657136e-01,\n -2.35158995e-01, 5.18051386e-01, 9.90481600e-02, -9.41838622e-01,\n 3.67467731e-01, 1.09189637e-02, 3.67206812e-01, -1.45973172e-02,\n -5.34822583e-01, -3.77859771e-02, -3.22057530e-02, 9.56173539e-01,\n 6.57714844e-01, -5.70637763e-01, -1.16847491e+00, -3.58010009e-02,\n 3.60460401e-01, -7.47398257e-01, -1.74480811e-01, -3.10095638e-01,\n 3.05545807e-01, 3.28680992e-01, -1.17556714e-02, 1.71635337e-02,\n 7.96749055e-01, -9.61633027e-02, -8.24414551e-01, -2.86415100e-01,\n 4.15685236e-01, -6.77867889e-01, -6.61213323e-02, -1.38597801e-01,\n -8.17664504e-01, -9.06744897e-02, 1.60091117e-01, 3.04878831e-01,\n 5.04516506e+00, -7.66995400e-02, 6.63899407e-02, -7.69001901e-01,\n 5.71159534e-02, 3.03661656e-02, 8.07254538e-02, -1.80927977e-01,\n -5.51925957e-01, -4.29255486e-01, 7.53005221e-03, -7.27976084e-01,\n 3.12609971e-01, 2.22065866e-01, -3.48325014e-01, 1.08633173e+00,\n 2.32963376e-02, -4.04967099e-01, -6.88609898e-01, -5.83201408e-01,\n 2.37168446e-02, 3.62407565e-01, 1.04253307e-01, -4.19051766e-01,\n 6.33383870e-01, -5.78164935e-01, -6.73843205e-01, 2.80672275e-02,\n 1.22984543e-01, 1.05578408e-01, -4.27697003e-01, -7.50363350e-01,\n -3.93532127e-01, -7.81694591e-01, 6.33218139e-03, 3.37018490e-01,\n -3.00740659e-01, 4.17670488e-01, 5.79307377e-01, -1.25942454e-01,\n -6.71937615e-02, 9.55632985e-01, 2.49431059e-01, -8.85666072e-01,\n -9.92519915e-01, -3.64751756e-01, -6.82361364e-01, -2.38570005e-01,\n -5.64868152e-02, -2.21540146e-02, 2.57586330e-01, -2.60549247e-01,\n -4.37264323e-01, 2.78154820e-01, -7.98489869e-01, -3.92585337e-01,\n 1.18384756e-01, 2.70041466e-01, 1.21253319e-02, -4.06758189e-01,\n 6.30180955e-01, -7.32773423e-01, 6.42872572e-01, 3.77312481e-01,\n -4.07330811e-01, 4.17000830e-01, 4.44727182e-01, 4.47301060e-01,\n -3.25813174e-01, 1.47019714e-01, -5.38453534e-02, -6.36136413e-01,\n -7.56918669e-01, -1.04030876e+01, -5.39244771e-01, 3.77115667e-01,\n -9.39559519e-01, 3.56085747e-01, 9.52926159e-01, -1.17427446e-01,\n 3.47996682e-01, -6.32461831e-02, -4.73790169e-02, 2.55489737e-01,\n -2.57051885e-01, 6.27693012e-02, 9.07871500e-02, 3.63426320e-02,\n 6.91977143e-01, -4.88751262e-01, 6.28802553e-02, 5.66762149e-01,\n 4.96241376e-02, -1.33674696e-01, 3.76361683e-02, 6.77354336e-01,\n 6.32003367e-01, 2.54655600e-01, 3.37713867e-01, 3.28810692e-01,\n -4.37369883e-01, -5.98554239e-02, 8.04426193e-01, -1.64897382e-01,\n -1.34422675e-01, 5.13696492e-01, -8.42697024e-01, -3.83413494e-01,\n -6.36178374e-01, -2.24019885e-01, 7.98433721e-02, -5.28987586e-01,\n 4.05215979e-01, 7.75448456e-02, -4.91976678e-01, -4.74331647e-01,\n -2.82053232e-01, 7.85411954e-01, 4.39621210e-02, 5.03493071e-01,\n 8.65300536e-01, 3.14397454e-01, 6.80294335e-01, 2.85041332e-01,\n -4.34756130e-02, -2.64850825e-01, 4.84166443e-01, -4.96896267e-01,\n 2.49236345e-01, -1.84241295e-01, 3.53078365e-01, 2.61592239e-01,\n -1.91861078e-01, -6.51381671e-01, -6.31134868e-01, 1.01643765e+00,\n 9.42196488e-01, 1.50000310e+00, 7.92521894e-01, 4.27157462e-01,\n 1.45513207e-01, 3.73194307e-01, -8.39729667e-01, -1.51113018e-01,\n -1.93717673e-01, 8.45905960e-01, -7.03491509e-01, 3.52909416e-01,\n 8.94982338e-01, 3.15302014e-02, 7.13599443e-01, 2.63230681e-01,\n 3.85412186e-01, -7.87497640e-01, -3.92595381e-02, -8.49693418e-01,\n -1.22719407e-02, 1.85883909e-01, -1.25940108e+00, 1.93139717e-01,\n -5.76790929e-01, -1.43338025e-01, 7.04292655e-01, -7.84686804e-02,\n -6.59248173e-01, -1.00539410e+00, 3.94983530e-01, -5.36350846e-01,\n -4.63372886e-01, 2.79267967e-01, 4.52531457e-01, -6.93497211e-02,\n 7.56990612e-01, -1.77658409e-01, -2.07795650e-01, 7.17090666e-01,\n 2.14408070e-01, -3.04786503e-01, 1.17867142e-02, -3.49925280e-01,\n 2.57581919e-01, -9.51970279e-01, 2.08498240e-02, -2.67667919e-01,\n 3.16156119e-01, -1.47699952e-01, -2.85970330e-01, -5.73386908e-01,\n 5.30821443e-01, -1.29520789e-01, 4.87134516e-01, -9.47579205e-01,\n -8.96620393e-01, 1.33827366e-02, -3.46674323e-01, 6.55562401e-01,\n 5.89349926e-01, -5.92327356e-01, 5.38228095e-01, 1.76440999e-01,\n 7.01240480e-01, 2.75054425e-01, -6.42431080e-01, 7.18982890e-04,\n -5.09306192e-01, 8.95830393e-02, -6.04627393e-02, -2.95443058e-01,\n -4.00094151e-01, -6.74588144e-01, 4.21255022e-01, -4.60031152e-01,\n -1.99148193e-01, -4.75323498e-01, 5.36219776e-01, 4.48370039e-01,\n 4.27922398e-01, -2.25051850e-01, 1.74733460e+00, -2.42062137e-01,\n -7.28633165e-01, -6.56704664e-01, -5.55008650e-04, 4.13772851e-01,\n 2.54790127e-01, -1.03530705e+00, -9.41522181e-01, 2.46725500e-01,\n -4.67434973e-01, 2.64016449e-01, -3.86801362e-02, 1.25489724e+00,\n -8.40474427e-01, -6.69294447e-02, 8.22278559e-01, -7.63152353e-03,\n 7.51231462e-02, 6.46436274e-01, 2.68672287e-01, -1.80111617e-01,\n -8.39132011e-01, -3.30313265e-01, -1.42777324e-01, -2.67261624e-01,\n -5.50760925e-02, -2.72957444e-01, 2.98006982e-01, 1.33635688e+00,\n 2.27536093e-02, -1.17700875e+00, -3.47259641e-01, -4.58708704e-02,\n 4.37102944e-01, 1.79467157e-01, -5.28155863e-01, -4.26109612e-01,\n 7.11461753e-02, -9.33481678e-02, -9.98666361e-02, -6.00276887e-01,\n 3.38462114e-01, 1.91928595e-01, 9.16126430e-01, -7.12594271e-01,\n 1.16142482e-01, 2.90572226e-01, 3.96926105e-01, -7.52364025e-02,\n -8.83707404e-02, 5.05500197e-01, -1.40293315e-01, 6.27462387e-01,\n 1.12909526e-01, 1.26804203e-01, -9.06096339e-01, 4.52533066e-01,\n 4.17275041e-01, -1.88256100e-01, -9.47703362e-01, 1.26599476e-01,\n -6.66880757e-02, 5.43238580e-01, -5.49229860e-01, -4.18547511e-01,\n -2.34864473e-01, 1.16352834e-01, 1.70271948e-01, 5.19884884e-01,\n -1.31784603e-01, -5.82413554e-01, 4.49904323e-01, -1.23372376e-01,\n -3.05623174e-01, 1.30007654e-01, -2.84432292e-01, 2.10333526e-01,\n -4.64999139e-01, 7.60375738e-01, -4.48388249e-01, -4.58142370e-01,\n 3.02865356e-02, 8.68064702e-01, 6.88253045e-02, 5.42488024e-02,\n 1.94935203e-02, -1.91484168e-01, 7.27310956e-01, 8.87250826e-02,\n -1.70166045e-01, -1.52007967e-01, -6.12322450e-01, 7.42658079e-01,\n 4.78442609e-02, 5.83464146e-01, -1.53904986e+00, -1.28770620e-01,\n 4.11456972e-02, -7.85531759e-01, -1.85755521e-01, 1.12253225e+00,\n 1.37069449e-03, -8.75927657e-02, -9.41453055e-02, -5.48875630e-01,\n -1.21809617e-01, -1.14224650e-01, -6.24941170e-01, -5.05941808e-01,\n -7.77410924e-01, -2.03377962e-01, 7.60520279e-01, 9.93452132e-01,\n 2.36683786e-01, 4.01045948e-01, 5.59971109e-02, -7.67491981e-02,\n 4.86585438e-01, 7.73757577e-01, -1.89116195e-01, -1.15080905e+00,\n 2.00615637e-03, 1.50798082e-01, -3.96724164e-01, 2.07813725e-01,\n -1.68358251e-01, -9.86884311e-02, 2.81797200e-01, 3.93666774e-02,\n -2.10500926e-01, -3.15205097e-01, 2.49992818e-01, -1.12620763e-01,\n -5.38319945e-01, 4.53690857e-01, -6.33629084e-01, 3.78209472e-01,\n 3.70509744e-01, 6.28506184e-01, 1.91421453e-02, -1.21532366e-01,\n -2.95967907e-01, -7.64577448e-01, -1.06140304e+00, -4.03031588e-01,\n -3.19335833e-02, 9.57063079e-01, 3.45315337e-01, -1.46200016e-01,\n -1.06994057e+00, 1.68559730e-01, 5.39638996e-01, 7.35182822e-01,\n -3.15263569e-01, -5.74359238e-01, -2.96879172e-01, 5.23734808e-01,\n -4.70534086e-01, 5.01377106e-01, -8.27666342e-01, 1.56010121e-01,\n 8.20634305e-01, 1.91381723e-01, -6.39304519e-02, 1.05225265e-01,\n 1.74418122e-01, -5.72201252e-01, -3.24726909e-01, -2.95729816e-01,\n 3.23628247e-01, 1.03967562e-01, 1.41976178e-01, -9.01225984e-01,\n -2.40003586e-01, 3.29296052e-01, 8.98381114e-01, 3.16089541e-01,\n 2.00824425e-01, -8.04099292e-02, 3.75158668e-01, 4.59516257e-01,\n 9.16634679e-01, -5.42102873e-01, -5.78891277e-01, 3.66972536e-01,\n -5.77074736e-02, -2.43312374e-01, 1.20764291e+00, -3.96294087e-01,\n 8.42844188e-01, -6.44075036e-01, -9.63378489e-01, -1.32573515e-01,\n -5.31032324e+00, 2.05761448e-01, -3.18816632e-01, -3.25189352e-01,\n -1.23869739e-01, 5.24408519e-01, 2.70734150e-02, -4.40607756e-01,\n -4.95121896e-01, -1.97602078e-01, 3.89996529e-01, 2.51638114e-01,\n -3.32608849e-01, -4.37130630e-02, -2.86759138e-01, 9.02597725e-01,\n -2.84475058e-01, -4.34994936e-01, 6.41452491e-01, 1.80947438e-01,\n 3.77803355e-01, 1.64125830e-01, 4.85077083e-01, -4.23475623e-01,\n -5.27139641e-02, -3.06918055e-01, -1.59317881e-01, -7.20196664e-01,\n -2.59837747e-01, 2.00399607e-02, 6.52360320e-01, 1.68055400e-01,\n 8.51624787e-01, -5.15118599e-01, 1.11459529e+00, -1.21983491e-01,\n 4.09144759e-02, -4.18154776e-01, 1.54754952e-01, 1.57715812e-01,\n -3.98643106e-01, -6.09316289e-01, -1.55087322e-01, -4.41018939e-01,\n 1.76068574e-01, 1.76927492e-01, -1.08204603e+00, -1.86846420e-01],\n dtype=float32))])\n"
],
[
"",
"_____no_output_____"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbedf955de74e623a6ac1e83af5bb96507741304
| 82,243 |
ipynb
|
Jupyter Notebook
|
doc/ipython_notebooks/Degree.ipynb
|
daniel-ge/brainx
|
121f4b7d132a1e0866c5dbfe81f3426a91e32893
|
[
"BSD-3-Clause"
] | 28 |
2015-02-05T00:05:48.000Z
|
2021-11-14T03:31:25.000Z
|
doc/ipython_notebooks/Degree.ipynb
|
daniel-ge/brainx
|
121f4b7d132a1e0866c5dbfe81f3426a91e32893
|
[
"BSD-3-Clause"
] | 2 |
2015-06-01T20:37:11.000Z
|
2015-06-05T22:31:07.000Z
|
doc/ipython_notebooks/Degree.ipynb
|
daniel-ge/brainx
|
121f4b7d132a1e0866c5dbfe81f3426a91e32893
|
[
"BSD-3-Clause"
] | 19 |
2015-03-23T01:38:06.000Z
|
2021-08-06T04:07:40.000Z
| 451.884615 | 49,667 | 0.926535 |
[
[
[
"empty"
]
]
] |
[
"empty"
] |
[
[
"empty"
]
] |
cbee0b455de644142fb73d5a9ebe828997bc4bab
| 9,378 |
ipynb
|
Jupyter Notebook
|
notebooks/RDF_Python_analytics.ipynb
|
SLIPO-EU/tutorial
|
15f4e0bc8c3ef2b937daa23e7b759ecf44fce5d7
|
[
"Apache-2.0"
] | 1 |
2020-11-03T14:59:01.000Z
|
2020-11-03T14:59:01.000Z
|
notebooks/RDF_Python_analytics.ipynb
|
SLIPO-EU/tutorial
|
15f4e0bc8c3ef2b937daa23e7b759ecf44fce5d7
|
[
"Apache-2.0"
] | null | null | null |
notebooks/RDF_Python_analytics.ipynb
|
SLIPO-EU/tutorial
|
15f4e0bc8c3ef2b937daa23e7b759ecf44fce5d7
|
[
"Apache-2.0"
] | 1 |
2021-07-16T08:33:36.000Z
|
2021-07-16T08:33:36.000Z
| 26.717949 | 176 | 0.523459 |
[
[
[
"# RDF graph processing against the integrated POIs",
"_____no_output_____"
],
[
"#### Auxiliary function to format SPARQL query results as a data frame:",
"_____no_output_____"
]
],
[
[
"import pandas as pds\n\ndef sparql_results_frame(qres):\n\n cols = qres.vars\n\n out = []\n for row in qres:\n item = []\n for c in cols:\n item.append(row[c])\n out.append(item)\n\n pds.set_option('display.max_colwidth', 0)\n \n return pds.DataFrame(out, columns=cols)",
"_____no_output_____"
]
],
[
[
"#### Create an **RDF graph** with the triples resulting from data integration:",
"_____no_output_____"
]
],
[
[
"from rdflib import Graph,URIRef\n\ng = Graph()\ng.parse('./output/integrated.nt', format=\"nt\")\n\n# Get graph size (in number of statements)\nlen(g)",
"_____no_output_____"
]
],
[
[
"#### Number of statements per predicate:",
"_____no_output_____"
]
],
[
[
"# SPARQL query is used to retrieve the results from the graph\nqres = g.query(\n \"\"\"SELECT ?p (COUNT(*) AS ?cnt) {\n ?s ?p ?o .\n } GROUP BY ?p ORDER BY DESC(?cnt)\"\"\")\n\n# display unformatted query results\n#for row in qres:\n# print(\"%s %s\" % row)\n \n# display formatted query results\nsparql_results_frame(qres)",
"_____no_output_____"
]
],
[
[
"#### Identify POIs having _**name**_ similar to a user-specified one:",
"_____no_output_____"
]
],
[
[
"# SPARQL query is used to retrieve the results from the graph\nqres = g.query(\n \"\"\"PREFIX slipo: <http://slipo.eu/def#>\n PREFIX provo: <http://www.w3.org/ns/prov#>\n SELECT DISTINCT ?poiURI ?title\n WHERE { ?poiURI slipo:name ?n .\n ?n slipo:nameValue ?title .\n FILTER regex(?title, \"^Achilleio\", \"i\") \n }\n\"\"\")\n\n# display query results\nsparql_results_frame(qres)",
"_____no_output_____"
]
],
[
[
"#### **Fusion action** regarding a specific POI:",
"_____no_output_____"
]
],
[
[
"# SPARQL query is used to retrieve the results from the graph\nqres = g.query(\n \"\"\"PREFIX slipo: <http://slipo.eu/def#>\n PREFIX provo: <http://www.w3.org/ns/prov#>\n SELECT ?prov ?defaultAction ?conf\n WHERE { ?poiURI provo:wasDerivedFrom ?prov .\n ?poiURI slipo:name ?n .\n ?n slipo:nameValue ?title .\n ?poiURI slipo:address ?a .\n ?a slipo:street ?s .\n ?prov provo:default-fusion-action ?defaultAction .\n ?prov provo:fusion-confidence ?conf .\n FILTER regex(?title, \"Achilleio\", \"i\")\n }\n\"\"\")\n\nprint(\"Query returned %d results.\" % len(qres) ) \n\n# display query results\nsparql_results_frame(qres)",
"_____no_output_____"
]
],
[
[
"#### **Pair of original POIs** involved in this fusion:",
"_____no_output_____"
]
],
[
[
"# SPARQL query is used to retrieve the results from the graph\nqres = g.query(\n \"\"\"PREFIX slipo: <http://slipo.eu/def#>\n PREFIX provo: <http://www.w3.org/ns/prov#>\n SELECT ?leftURI ?rightURI ?conf\n WHERE { <http://www.provbook.org/d494ddbd-9a98-39b0-bec9-0477636c42f7> provo:left-uri ?leftURI .\n <http://www.provbook.org/d494ddbd-9a98-39b0-bec9-0477636c42f7> provo:right-uri ?rightURI .\n <http://www.provbook.org/d494ddbd-9a98-39b0-bec9-0477636c42f7> provo:fusion-confidence ?conf .\n }\n\"\"\")\n \nprint(\"Query returned %d results.\" % len(qres))\n\n# display pair of POI URIs along with the fusion confidence\nsparql_results_frame(qres)",
"_____no_output_____"
]
],
[
[
"#### Values per attribute **before and after fusion** regarding this POI:",
"_____no_output_____"
]
],
[
[
"# SPARQL query is used to retrieve the results from the graph\nqres = g.query(\n \"\"\"PREFIX slipo: <http://slipo.eu/def#>\n PREFIX provo: <http://www.w3.org/ns/prov#>\n SELECT DISTINCT ?valLeft ?valRight ?valFused\n WHERE { ?poiURI provo:wasDerivedFrom <http://www.provbook.org/d494ddbd-9a98-39b0-bec9-0477636c42f7> .\n ?poiURI provo:appliedAction ?action .\n ?action provo:attribute ?attr .\n ?action provo:left-value ?valLeft .\n ?action provo:right-value ?valRight .\n ?action provo:fused-value ?valFused .\n }\n\"\"\")\n \nprint(\"Query returned %d results.\" % len(qres)) \n\n# print query results\nsparql_results_frame(qres)",
"_____no_output_____"
]
],
[
[
"# POI Analytics\n\n#### Once integrated POI data has been saved locally, analysis can be perfomed using tools like **pandas** _DataFrames_, **geopandas** _GeoDataFrames_ or other libraries.",
"_____no_output_____"
],
[
"#### Unzip exported CSV file with the results of data integration:",
"_____no_output_____"
]
],
[
[
"import os\nimport zipfile\n\nwith zipfile.ZipFile('./output/corfu-integrated-pois.zip','r') as zip_ref:\n zip_ref.extractall(\"./output/\")\n \nos.rename('./output/points.csv', './output/corfu_pois.csv')",
"_____no_output_____"
]
],
[
[
"#### Load CSV data in a _DataFrame_:",
"_____no_output_____"
]
],
[
[
"import pandas as pd\n\npois = pd.read_csv('./output/corfu_pois.csv', delimiter='|', error_bad_lines=False)\n\n# Geometries in the exported CSV file are listed in Extended Well-Known Text (EWKT)\n# Since shapely does not support EWKT, update the geometry by removing the SRID value from EWKT\npois['the_geom'] = pois['the_geom'].apply(lambda x: x.split(';')[1])\n\npois.head()",
"_____no_output_____"
]
],
[
[
"#### Create a _GeoDataFrame_:",
"_____no_output_____"
]
],
[
[
"import geopandas\nfrom shapely import wkt\n\npois['the_geom'] = pois['the_geom'].apply(wkt.loads)\n\ngdf = geopandas.GeoDataFrame(pois, geometry='the_geom')",
"_____no_output_____"
]
],
[
[
"#### Display the location of the exported POIs on a **simplified plot** using _matplotlib_:",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\n\nimport matplotlib.pyplot as plt\n\nworld = geopandas.read_file(geopandas.datasets.get_path('naturalearth_lowres'))\n\n# Restrict focus to Germany:\nax = world[world.name == 'Greece'].plot(\n color='white', edgecolor='black')\n\n# Plot the contents of the GeoDataFrame in blue dots:\ngdf.plot(ax=ax, color='blue')\n\nplt.show()",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cbee0d5cf3751fc4b47f3610321eac74f25abfc6
| 19,871 |
ipynb
|
Jupyter Notebook
|
notebooks/drafts/data_cleaning_at2.ipynb
|
NB-01/twitter
|
ba755bbc4da409bf616039fe5a75817516d37771
|
[
"MIT"
] | null | null | null |
notebooks/drafts/data_cleaning_at2.ipynb
|
NB-01/twitter
|
ba755bbc4da409bf616039fe5a75817516d37771
|
[
"MIT"
] | null | null | null |
notebooks/drafts/data_cleaning_at2.ipynb
|
NB-01/twitter
|
ba755bbc4da409bf616039fe5a75817516d37771
|
[
"MIT"
] | null | null | null | 52.018325 | 1,488 | 0.633033 |
[
[
[
"%load_ext lab_black",
"_____no_output_____"
],
[
"# Import all the necessary packages\nimport pandas as pd\nimport numpy as np\n\nimport statsmodels.api as sm\nimport scipy.stats as st\n\nimport matplotlib.pyplot as plt\nfrom matplotlib import rcParams\nimport seaborn as sns\n\nimport json\nimport requests\nimport yaml\nfrom twython import Twython\nfrom twython import Twython, TwythonError\n\nfrom pandas import json_normalize\n\nfrom datetime import datetime\nfrom dateutil import parser",
"_____no_output_____"
],
[
"dorsey = pd.read_csv(\"dorsey.csv\")\ntwitter = pd.read_csv(\"TWTR.csv\")",
"_____no_output_____"
],
[
"dorsey.head(5)",
"_____no_output_____"
],
[
"columns = [\"created_at\", \"id\", \"full_text\", \"retweet_count\", \"favorite_count\"]\ndorsey = dorsey[columns]\ndorsey[\"created_at\"] = pd.to_datetime(dorsey[\"created_at\"]).dt.date",
"_____no_output_____"
],
[
"dorsey.head()",
"_____no_output_____"
],
[
"dorsey.dtypes",
"_____no_output_____"
],
[
"import re\n\n# Clean The Data\ndef cleantext(text):\n text = re.sub(r\"@[A-Za-z0-9]+\", \"\", text) # Remove Mentions\n text = re.sub(r\"#\", \"\", text) # Remove Hashtags Symbol\n text = re.sub(r\"RT[\\s]+\", \"\", text) # Remove Retweets\n text = re.sub(r\"https?:\\/\\/\\S+\", \"\", text) # Remove The Hyper Link\n\n return text\n\n\n# Clean The Text\ndorsey[\"full_text\"] = dorsey[\"full_text\"].apply(cleantext)\ndorsey.head()",
"_____no_output_____"
],
[
"from textblob import TextBlob\n\n# Get The Subjectivity\ndef sentiment_analysis(ds):\n sentiment = TextBlob(ds).sentiment\n return pd.Series([sentiment.subjectivity, sentiment.polarity])\n\n\n# Adding Subjectivity & Polarity\ndorsey[[\"subjectivity\", \"polarity\"]] = dorsey[\"full_text\"].apply(sentiment_analysis)\ndorsey.head()",
"_____no_output_____"
],
[
"df_a = (\n dorsey.groupby([\"created_at\"])\n .agg(\n agg_string=(\"full_text\", \"sum\"),\n agg_retweet=(\"retweet_count\", \"sum\"),\n agg_count=(\"favorite_count\", \"sum\"),\n subjectivity_mean=(\"subjectivity\", \"mean\"),\n polarity_mean=(\"polarity\", \"mean\"),\n )\n .reset_index()\n)\ndf_a.head()",
"_____no_output_____"
],
[
"from wordcloud import WordCloud\n\nallwords = \" \".join([twts for twts in dorsey[\"full_text\"]])\nwordCloud = WordCloud(\n width=1000, height=1000, random_state=21, max_font_size=119\n).generate(allwords)\nplt.figure(figsize=(20, 20), dpi=80)\nplt.imshow(wordCloud, interpolation=\"bilinear\")\nplt.axis(\"off\")\nplt.show()",
"_____no_output_____"
],
[
"sentiment = TextBlob(\"not sure anyone has heard but, I resigned from Twitter\").sentiment\nprint(sentiment)",
"Sentiment(polarity=-0.25, subjectivity=0.8888888888888888)\n"
],
[
"df_a.columns = [\n \"Date\",\n \"agg_text\",\n \"agg_retweet\",\n \"agg_count\",\n \"subjectivity\",\n \"polarity\",\n]",
"_____no_output_____"
],
[
"d = df_a.iloc[:][\"Date\"]\nd",
"_____no_output_____"
],
[
"df = pd.merge(twitter, df_a, on=\"Date\", how=\"outer\")",
"_____no_output_____"
],
[
"df",
"_____no_output_____"
],
[
"df_a",
"_____no_output_____"
],
[
"twitter[\"Date\"] = pd.to_datetime(twitter[\"Date\"]).dt.date",
"_____no_output_____"
],
[
"twitter.dtypes",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbee184219486ca256cb7e847247bcb69d834cb4
| 16,252 |
ipynb
|
Jupyter Notebook
|
solutions_do_not_open/Lab_10_DL Keras Intro Shallow Models_solution.ipynb
|
differentmatt/ztdl-5-day-bootcamp
|
66a300ed73857dd9b4e1b2fa6aec54e1f87b6615
|
[
"MIT"
] | 1 |
2018-09-23T05:40:20.000Z
|
2018-09-23T05:40:20.000Z
|
solutions_do_not_open/Lab_10_DL Keras Intro Shallow Models_solution.ipynb
|
differentmatt/ztdl-5-day-bootcamp
|
66a300ed73857dd9b4e1b2fa6aec54e1f87b6615
|
[
"MIT"
] | null | null | null |
solutions_do_not_open/Lab_10_DL Keras Intro Shallow Models_solution.ipynb
|
differentmatt/ztdl-5-day-bootcamp
|
66a300ed73857dd9b4e1b2fa6aec54e1f87b6615
|
[
"MIT"
] | null | null | null | 22.081522 | 489 | 0.535565 |
[
[
[
"# Keras Intro: Shallow Models\n\nKeras Documentation: https://keras.io\n\nIn this notebook we explore how to use Keras to implement 2 traditional Machine Learning models:\n- **Linear Regression** to predict continuous data\n- **Logistic Regression** to predict categorical data",
"_____no_output_____"
],
[
"## Linear Regression",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\nimport matplotlib.pyplot as plt\nimport pandas as pd\nimport numpy as np",
"_____no_output_____"
]
],
[
[
"### 0. Load data",
"_____no_output_____"
]
],
[
[
"df = pd.read_csv('../data/weight-height.csv')",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
],
[
"df.plot(kind='scatter',\n x='Height',\n y='Weight',\n title='Weight and Height in adults')",
"_____no_output_____"
]
],
[
[
"### 1. Create Train/Test split",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import train_test_split",
"_____no_output_____"
],
[
"X = df[['Height']].values\ny = df['Weight'].values\n\nX_train, X_test, y_train, y_test = train_test_split(X, y, \n test_size = 0.3, random_state=0)",
"_____no_output_____"
]
],
[
[
"### 2. Train Linear Regression Model",
"_____no_output_____"
]
],
[
[
"from keras.models import Sequential\nfrom keras.layers import Dense\nfrom keras.optimizers import Adam, SGD",
"_____no_output_____"
],
[
"model = Sequential()",
"_____no_output_____"
],
[
"model.add(Dense(1, input_shape=(1,)))",
"_____no_output_____"
],
[
"model.summary()",
"_____no_output_____"
],
[
"model.compile(Adam(lr=0.9), 'mean_squared_error')",
"_____no_output_____"
],
[
"model.fit(X_train, y_train, epochs=40)",
"_____no_output_____"
]
],
[
[
"### 3. Evaluate Model Performance",
"_____no_output_____"
]
],
[
[
"from sklearn.metrics import r2_score",
"_____no_output_____"
],
[
"y_train_pred = model.predict(X_train).ravel()\ny_test_pred = model.predict(X_test).ravel()",
"_____no_output_____"
],
[
"print(\"The R2 score on the Train set is:\\t{:0.3f}\".format(r2_score(y_train, y_train_pred)))\nprint(\"The R2 score on the Test set is:\\t{:0.3f}\".format(r2_score(y_test, y_test_pred)))",
"_____no_output_____"
],
[
"df.plot(kind='scatter',\n x='Height',\n y='Weight',\n title='Weight and Height in adults')\nplt.plot(X_test, y_test_pred, color='red')",
"_____no_output_____"
],
[
"W, B = model.get_weights()",
"_____no_output_____"
],
[
"W",
"_____no_output_____"
],
[
"B",
"_____no_output_____"
]
],
[
[
"# Classification",
"_____no_output_____"
],
[
"### 0. Load Data",
"_____no_output_____"
]
],
[
[
"df = pd.read_csv('../data/user_visit_duration.csv')",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
],
[
"df.plot(kind='scatter', x='Time (min)', y='Buy')",
"_____no_output_____"
]
],
[
[
"### 1. Create Train/Test split",
"_____no_output_____"
]
],
[
[
"X = df[['Time (min)']].values\ny = df['Buy'].values",
"_____no_output_____"
],
[
"X_train, X_test, y_train, y_test = train_test_split(X, y, \n test_size = 0.3, random_state=0)",
"_____no_output_____"
]
],
[
[
"### 2. Train Logistic Regression Model",
"_____no_output_____"
]
],
[
[
"model = Sequential()",
"_____no_output_____"
],
[
"model.add(Dense(1, input_shape=(1,), activation='sigmoid'))",
"_____no_output_____"
],
[
"model.summary()",
"_____no_output_____"
],
[
"model.compile(SGD(lr=0.5), 'binary_crossentropy', metrics=['accuracy'])",
"_____no_output_____"
],
[
"model.fit(X_train, y_train, epochs=40)",
"_____no_output_____"
],
[
"ax = df.plot(kind='scatter', x='Time (min)', y='Buy',\n title='Purchase behavior VS time spent on site')\n\nt = np.linspace(0, 4)\nax.plot(t, model.predict(t), color='orange')\n\nplt.legend(['model', 'data'])",
"_____no_output_____"
]
],
[
[
"### 3. Evaluate Model Performance",
"_____no_output_____"
],
[
"#### Accuracy",
"_____no_output_____"
]
],
[
[
"from sklearn.metrics import accuracy_score",
"_____no_output_____"
],
[
"y_train_pred = model.predict_classes(X_train)\ny_test_pred = model.predict_classes(X_test)",
"_____no_output_____"
],
[
"print(\"The train accuracy score is {:0.3f}\".format(accuracy_score(y_train, y_train_pred)))\nprint(\"The test accuracy score is {:0.3f}\".format(accuracy_score(y_test, y_test_pred)))",
"_____no_output_____"
]
],
[
[
"#### Confusion Matrix & Classification Report",
"_____no_output_____"
]
],
[
[
"from sklearn.metrics import confusion_matrix",
"_____no_output_____"
],
[
"confusion_matrix(y_test, y_test_pred)",
"_____no_output_____"
],
[
"def pretty_confusion_matrix(y_true, y_pred, labels=[\"False\", \"True\"]):\n cm = confusion_matrix(y_true, y_pred)\n pred_labels = ['Predicted '+ l for l in labels]\n df = pd.DataFrame(cm, index=labels, columns=pred_labels)\n return df",
"_____no_output_____"
],
[
"pretty_confusion_matrix(y_test, y_test_pred, ['Not Buy', 'Buy'])",
"_____no_output_____"
],
[
"from sklearn.metrics import classification_report",
"_____no_output_____"
],
[
"print(classification_report(y_test, y_test_pred))",
"_____no_output_____"
]
],
[
[
"## Exercise",
"_____no_output_____"
],
[
"You've just been hired at a real estate investment firm and they would like you to build a model for pricing houses. You are given a dataset that contains data for house prices and a few features like number of bedrooms, size in square feet and age of the house. Let's see if you can build a model that is able to predict the price. In this exercise we extend what we have learned about linear regression to a dataset with more than one feature. Here are the steps to complete it:\n\n1. Load the dataset ../data/housing-data.csv\n- create 2 variables called X and y: X shall be a matrix with 3 columns (sqft,bdrms,age) and y shall be a vector with 1 column (price)\n- create a linear regression model in Keras with the appropriate number of inputs and output\n- split the data into train and test with a 20% test size, use `random_state=0` for consistency with classmates\n- train the model on the training set and check its accuracy on training and test set\n- how's your model doing? Is the loss decreasing?\n- try to improve your model with these experiments:\n - normalize the input features:\n - divide sqft by 1000\n - divide age by 10\n - divide price by 100000\n - use a different value for the learning rate of your model\n - use a different optimizer\n- once you're satisfied with training, check the R2score on the test set",
"_____no_output_____"
]
],
[
[
"# Load the dataset ../data/housing-data.csv\ndf = pd.read_csv('../data/housing-data.csv')\ndf.head()",
"_____no_output_____"
],
[
"df.columns",
"_____no_output_____"
],
[
"# create 2 variables called X and y:\n# X shall be a matrix with 3 columns (sqft,bdrms,age)\n# and y shall be a vector with 1 column (price)\nX = df[['sqft', 'bdrms', 'age']].values\ny = df['price'].values",
"_____no_output_____"
],
[
"# create a linear regression model in Keras\n# with the appropriate number of inputs and output\nmodel = Sequential()\nmodel.add(Dense(1, input_shape=(3,)))\nmodel.compile(Adam(lr=0.8), 'mean_squared_error')",
"_____no_output_____"
],
[
"# split the data into train and test with a 20% test size\nX_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=0)",
"_____no_output_____"
],
[
"# train the model on the training set and check its accuracy on training and test set\n# how's your model doing? Is the loss decreasing?\nmodel.fit(X_train, y_train, epochs=50)",
"_____no_output_____"
],
[
"# check the R2score on training and test set (probably very bad)\n\ny_train_pred = model.predict(X_train)\ny_test_pred = model.predict(X_test)\n\nprint(\"The R2 score on the Train set is:\\t{:0.3f}\".format(r2_score(y_train, y_train_pred)))\nprint(\"The R2 score on the Test set is:\\t{:0.3f}\".format(r2_score(y_test, y_test_pred)))",
"_____no_output_____"
],
[
"# try to improve your model with these experiments:\n# - normalize the input features with one of the rescaling techniques mentioned above\n# - use a different value for the learning rate of your model\n# - use a different optimizer\ndf['sqft1000'] = df['sqft']/1000.0\ndf['age10'] = df['age']/10.0\ndf['price100k'] = df['price']/1e5",
"_____no_output_____"
],
[
"X = df[['sqft1000', 'bdrms', 'age10']].values\ny = df['price100k'].values",
"_____no_output_____"
],
[
"X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=0)",
"_____no_output_____"
],
[
"model = Sequential()\nmodel.add(Dense(1, input_dim=3))\nmodel.compile(Adam(lr=0.1), 'mean_squared_error')\nmodel.fit(X_train, y_train, epochs=50)",
"_____no_output_____"
],
[
"# once you're satisfied with training, check the R2score on the test set\n\ny_train_pred = model.predict(X_train)\ny_test_pred = model.predict(X_test)\n\nprint(\"The R2 score on the Train set is:\\t{:0.3f}\".format(r2_score(y_train, y_train_pred)))\nprint(\"The R2 score on the Test set is:\\t{:0.3f}\".format(r2_score(y_test, y_test_pred)))",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbee37271f059a8094e1d010755cd592a0fc7dd7
| 42,591 |
ipynb
|
Jupyter Notebook
|
Logistic Regression.ipynb
|
reata/MachineLearning
|
f1082ccbe79d65008ac6bcefe9e184a090eb91bb
|
[
"MIT"
] | null | null | null |
Logistic Regression.ipynb
|
reata/MachineLearning
|
f1082ccbe79d65008ac6bcefe9e184a090eb91bb
|
[
"MIT"
] | null | null | null |
Logistic Regression.ipynb
|
reata/MachineLearning
|
f1082ccbe79d65008ac6bcefe9e184a090eb91bb
|
[
"MIT"
] | 2 |
2018-10-08T16:05:27.000Z
|
2020-12-14T14:58:23.000Z
| 159.516854 | 21,018 | 0.865183 |
[
[
[
"# 分类和逻辑回归 Classification and Logistic Regression\n\n引入科学计算和绘图相关包:",
"_____no_output_____"
]
],
[
[
"import numpy as np\nfrom sklearn import linear_model, datasets\n\nimport matplotlib.pyplot as plt\nimport seaborn as sns\n\nsns.set_style('whitegrid')\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"分类和回归的唯一区别在于,分类问题中我们希望预测的目标变量 $y$ 只会取少数几个离散值。本节我们将主要关注**二元分类 binary classification**问题,$y$ 在二元分类中只会取 $0, 1$ 两个值。$0$ 也被称为**反类 negative class**,$1$ 也被称为**正类 positive class**,它们有时也使用符号 $-, +$ 来标识。给定 $x^{(i)}$,对应的 $y^{(i)}$ 也被称为训练样本的**标签 label**。\n\n本节包括以下内容:\n\n1. 逻辑回归 Logistic regression\n2. 感知器 The perceptron learning algorithm\n3. 牛顿法:最大化 $\\ell(\\theta)$ 的另一种算法 Newton's Method: Another algorithm for maximizing $\\ell(\\theta)$",
"_____no_output_____"
],
[
"## 1. 逻辑回归 Logistic Regression\n\n在逻辑回归中,我们的假设函数 $h_\\theta(x)$ 的形式为:\n$$ h_\\theta(x) = g(\\theta^Tx) = \\frac{1}{1+e^{-\\theta^Tx}}, $$\n其中\n$$ g(z) = \\frac{1}{1+e^{-z}} $$\n被称为**logistic函数**或**sigmoid函数**,$g(z)$ 的形式如下:",
"_____no_output_____"
]
],
[
[
"x = np.arange(-10., 10., 0.2)\ny = 1 / (1 + np.e ** (-x))\nplt.plot(x, y)\nplt.title(' Logistic Function ')\nplt.show()",
"_____no_output_____"
]
],
[
[
"当 $z \\rightarrow \\infty$ 时,$g(z) \\rightarrow 1$;当 $z \\rightarrow -\\infty$ 时,$g(z) \\rightarrow 0$;$g(z)$ 的值域为 $(0, 1)$。我们保留令 $x_0 = 1$ 的习惯,$\\theta^Tx = \\theta_0 + \\sum_{j=1}^n \\theta_jx_j$。\n\n之后在讲广义线性模型时,我们会介绍sigmoid函数的由来。暂时我们只是将其作为给定条件。sigmoid函数的导数有一个十分有用的性质,后续在推导时会用到:\n$$\n\\begin{split}\ng'(z) &= \\frac{d}{dz}\\frac{1}{1+e^{-z}} \\\\\n&= \\frac{1}{(1+e^{-z})^2}e^{-z} \\\\\n&= \\frac{1}{(1+e^{-z})} \\cdot (1 - \\frac{1}{(1+e^{-z})}) \\\\\n&= g(z) \\cdot (1-g(z))\n\\end{split}\n$$\n\n在线性回归的概率诠释中,根据一定的假设,我们通过最大似然估计法计算 $\\theta$。类似地,在逻辑回归中,我们也采用同样的策略,假设:\n$$ P(y=1|x;\\theta) = h_\\theta(x) $$\n$$ P(y=0|x;\\theta) = 1 - h_\\theta(x) $$\n这两个假设可以合并为:\n$$ P(y|x;\\theta) = (h_\\theta(x))^y(1-h_\\theta(x))^{1-y} $$\n\n继续假设 $m$ 个训练样本相互独立,似然函数因此可以写成:\n$$\n\\begin{split}\nL(\\theta) & = p(y|X; \\theta) \\\\\n& = \\prod_{i=1}^{m} p(y^{(i)}|x^{(i)}; \\theta) \\\\\n& = \\prod_{i=1}^{m} (h_\\theta(x^{(i)}))^{y^{(i)}}(1-h_\\theta(x^{(i)}))^{1-y^{(i)}}\n\\end{split}\n$$\n\n相应的对数似然函数可以写成:\n$$\n\\begin{split}\n\\ell(\\theta) &= logL(\\theta) \\\\\n&= \\sum_{i=1}^m (y^{(i)}logh(x^{(i)})+(1-y^{(i)})log(1-h(x^{(i)})))\n\\end{split}\n$$\n\n为了求解对数似然函数的最大值,我们可以采用梯度上升的算法 $\\theta = \\theta + \\alpha\\nabla_\\theta\\ell(\\theta)$,其中偏导为:\n$$\n\\begin{split}\n\\frac{\\partial}{\\partial\\theta_j}\\ell(\\theta) &= \\sum(y\\frac{1}{g(\\theta^Tx)} - (1-y)\\frac{1}{1-g(\\theta^Tx)})\\frac{\\partial}{\\partial\\theta_j}g(\\theta^Tx) \\\\\n&= \\sum(y\\frac{1}{g(\\theta^Tx)} - (1-y)\\frac{1}{1-g(\\theta^Tx)})g(\\theta^Tx)(1-g(\\theta^Tx))\\frac{\\partial}{\\partial\\theta_j}\\theta^Tx \\\\\n&= \\sum(y(1-g(\\theta^Tx))-(1-y)g(\\theta^Tx))x_j \\\\\n&= \\sum(y-h_\\theta(x))x_j\n\\end{split}\n$$\n\n而对于每次迭代只使用单个样本的随机梯度上升算法而言:\n$$ \\theta_j = \\theta_j + \\alpha(y^{(i)}-h_\\theta(x^{(i)}))x^{(i)}_j = \\theta_j - \\alpha(h_\\theta(x^{(i)}) - y^{(i)})x_j^{(i)}$$\n\n可以看到,除去假设函数 $h_\\theta(x)$ 本身不同之外,逻辑回归和线性回归的梯度更新是十分类似的。广义线性模型将会解释这里的“巧合”。",
"_____no_output_____"
],
[
"## 2. 感知器 The Perceptron Learning Algorithm\n\n在逻辑回归中,我们通过sigmoid函数,使得最终的目标变量落在 $(0, 1)$ 的区间内,并假设目标变量的值就是其为正类的概率。\n\n设想我们使目前变量严格地取 $0$ 或 $1$:\n$$ g(z) =\\left\\{\n\\begin{aligned}\n1 & , z \\geq 0 \\\\\n0 & , z < 0\n\\end{aligned}\n\\right.\n$$\n\n和之前一样,我们令 $h_\\theta(x) = g(\\theta^Tx)$,并根据以下规则更新:\n$$ \\theta_j = \\theta_j + \\alpha(y^{(i)} - h_\\theta(x^{(i)}))x_j^{(i)} $$\n这个算法被称为**感知器学习算法 perceptron learning algorithm**。\n\n感知器在上世纪60年代一直被视作大脑中单个神经元的粗略模拟。但注意,尽管感知器和逻辑回归的形式非常相似,但由于 $g(z)$ 无法使用概率假设来描述,因而也无法使用最大似然估计法进行参数估计。实际上感知器和线性模型是完全不同的算法类型,它是神经网络算法的起源,之后我们会回到这个话题。",
"_____no_output_____"
],
[
"## 3. 牛顿法:最大化 $\\ell(\\theta)$ 的另一种算法 Newton's Method: Another algorithm for maximizing $\\ell(\\theta)$\n\n回到逻辑回归,为了求解其对数似然函数的最大值,除了梯度上升算法外,这里介绍通过牛顿法进行求解。\n\n牛顿法主要用来求解方程的根。设想有一个函数 $f: \\mathbb{R} \\rightarrow \\mathbb{R}$,我们希望找到一个值 $\\theta$ 使得 $f(\\theta)=0$。牛顿法的迭代过程如下:\n$$ \\theta = \\theta - \\frac{f(\\theta)}{f'(\\theta)} $$",
"_____no_output_____"
]
],
[
[
"f = lambda x: x ** 2\nf_prime = lambda x: 2 * x\nimprove_x = lambda x: x - f(x) / f_prime(x)\n\nx = np.arange(0, 3, 0.2)\nx0 = 2\ntangent0 = lambda x: f_prime(x0) * (x - x0) + f(x0)\nx1 = improve_x(x0)\ntangent1 = lambda x: f_prime(x1) * (x - x1) + f(x1)\n\nplt.plot(x, f(x), label=\"y=x^2\")\nplt.plot(x, np.zeros_like(x), label=\"x axis\")\nplt.plot(x, tangent0(x), label=\"y=4x-4\")\nplt.plot(x, tangent1(x), label=\"y=2x-1\")\nplt.legend(loc=\"best\")\nplt.show()",
"_____no_output_____"
]
],
[
[
"牛顿法的迭代过程可以非常直观地用上图解释,我们需要求解 $y = x^2$ 的根,函数如蓝线所示。假设我们设置的初始点 $x_0 = 2$,对这个点求导做切线,如绿线所示,绿线和 $x$ 轴的交点就是我们第一轮迭代的结果 $x_1 = 1$。继续这个过程,得到切线用红色表示,红线与 $x$ 轴为第二轮迭代结果 $x_2 = 0.5$。重复迭代可以到达 $x=0$;\n\n对于我们的对数似然函数来说,要求解其最大值,也就是要求解 $\\ell'(\\theta) = 0$:\n$$ \\theta = \\theta - \\frac{\\ell'(\\theta)}{\\ell''(\\theta)} $$\n\n注意这里的充要关系,在最大值点一阶导数必然为0,反之则未必,所以我们求解的,实际上也可能是局部/全局最小值点,或者鞍点。\n\n最后,在逻辑回归中,$\\theta$ 是一个向量,所以我们需要据此扩展牛顿法。牛顿法在高维空间中的,也称为**牛顿-拉弗森法 Newton-Raphson method**:\n$$ \\theta = \\theta - H^{-1}\\nabla_\\theta\\ell(\\theta) $$\n\n这里,$\\nabla_\\theta\\ell(\\theta)$ 是 $\\ell(\\theta)$ 针对向量 $\\theta$ 的偏导。$H$ 是一个 $n \\times n$ 的矩阵(加上截距项,实际是 $n+1 \\times n+1$),称为**海森矩阵 Hessian Matrix**:\n$$ H_{ij} = \\frac{\\partial^2\\ell(\\theta)}{\\partial\\theta_i\\partial\\theta_j} $$\n\n通常牛顿法会比(批量)梯度下降在更短的迭代次数内收敛。同时,一次牛顿法迭代过程,由于要对海森矩阵求逆,会比梯度下降的一次迭代慢。只要 $n$ 的值不至于过大,牛顿法总的来说会比梯度下降收敛快得多。\n\n应用牛顿法解最大似然函数,也被称为**费舍尔评分 Fisher's scoring**。",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
cbee40ca9f2feb5ca223df98c3b5287e4f8a0377
| 3,663 |
ipynb
|
Jupyter Notebook
|
homework.ipynb
|
KarenTadevosyan/Econometrics2022
|
3d85661db8ae574c2a44c5e9cd159177eca8eaa5
|
[
"MIT"
] | null | null | null |
homework.ipynb
|
KarenTadevosyan/Econometrics2022
|
3d85661db8ae574c2a44c5e9cd159177eca8eaa5
|
[
"MIT"
] | null | null | null |
homework.ipynb
|
KarenTadevosyan/Econometrics2022
|
3d85661db8ae574c2a44c5e9cd159177eca8eaa5
|
[
"MIT"
] | null | null | null | 16.065789 | 77 | 0.441168 |
[
[
[
"str1 = \"This is my homework\"\nstr1",
"_____no_output_____"
],
[
"str1.upper()",
"_____no_output_____"
],
[
"str1.lower()",
"_____no_output_____"
],
[
"str1.replace(\"my\",\"a\")",
"_____no_output_____"
],
[
"str1.find(\"my\")",
"_____no_output_____"
],
[
"len(str1)",
"_____no_output_____"
],
[
"print(str1[11:19])",
"homework\n"
],
[
"print(str1[::4])",
"T moo\n"
],
[
"print(str1[-10:-4])",
"y home\n"
],
[
"str2 = \"1, 2, 3, 4, 5\"\nstr2\nprint(str2[::-1])",
"5 ,4 ,3 ,2 ,1\n"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbee4437af7e03f6594d8285c76eaddd0c40ca17
| 10,307 |
ipynb
|
Jupyter Notebook
|
.ipynb_checkpoints/iris_snippets_debug-checkpoint.ipynb
|
petercunning/notebook
|
5b26f2dc96bcb36434542b397de6ca5fa3b61a0a
|
[
"MIT"
] | 32 |
2015-01-07T01:48:05.000Z
|
2022-03-02T07:07:42.000Z
|
.ipynb_checkpoints/iris_snippets_debug-checkpoint.ipynb
|
petercunning/notebook
|
5b26f2dc96bcb36434542b397de6ca5fa3b61a0a
|
[
"MIT"
] | 1 |
2015-04-13T21:00:18.000Z
|
2015-04-13T21:00:18.000Z
|
.ipynb_checkpoints/iris_snippets_debug-checkpoint.ipynb
|
petercunning/notebook
|
5b26f2dc96bcb36434542b397de6ca5fa3b61a0a
|
[
"MIT"
] | 30 |
2015-01-28T09:31:29.000Z
|
2022-03-07T03:08:28.000Z
| 35.912892 | 99 | 0.485786 |
[
[
[
"empty"
]
]
] |
[
"empty"
] |
[
[
"empty"
]
] |
cbee526742169a71edfa2844c4fd13ba16647d63
| 76,016 |
ipynb
|
Jupyter Notebook
|
AI/Tree Data/Deep Learning - TreeData.ipynb
|
pierpaolo28/Alleviate-Children-s-Health-Issues-through-Games-and-Machine-Learning
|
2d6ffd637d36fbf18b94d149fd5c84da90d58fad
|
[
"Apache-2.0"
] | 4 |
2020-06-07T17:39:35.000Z
|
2021-12-14T11:53:26.000Z
|
AI/Tree Data/Deep Learning - TreeData.ipynb
|
pierpaolo28/Alleviate-Children-s-Health-Issues-through-Games-and-Machine-Learning
|
2d6ffd637d36fbf18b94d149fd5c84da90d58fad
|
[
"Apache-2.0"
] | null | null | null |
AI/Tree Data/Deep Learning - TreeData.ipynb
|
pierpaolo28/Alleviate-Children-s-Health-Issues-through-Games-and-Machine-Learning
|
2d6ffd637d36fbf18b94d149fd5c84da90d58fad
|
[
"Apache-2.0"
] | 1 |
2021-07-26T07:28:54.000Z
|
2021-07-26T07:28:54.000Z
| 161.73617 | 58,772 | 0.882367 |
[
[
[
"import pandas as pd\nimport numpy as np\nimport pickle\nimport matplotlib.pyplot as plt\nfrom scipy import stats\nimport tensorflow as tf\nimport seaborn as sns\nfrom pylab import rcParams\nfrom sklearn import metrics\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.preprocessing import StandardScaler\n\n%matplotlib inline\n\nsns.set(style='whitegrid', palette='muted', font_scale=1.5)\n\nrcParams['figure.figsize'] = 14, 8\n\nRANDOM_SEED = 42",
"C:\\Apps\\Anaconda3\\lib\\site-packages\\h5py\\__init__.py:36: FutureWarning: Conversion of the second argument of issubdtype from `float` to `np.floating` is deprecated. In future, it will be treated as `np.float64 == np.dtype(float).type`.\n from ._conv import register_converters as _register_converters\n"
],
[
"df=pd.read_csv('TreeData.csv')\n# df.head(22)\ndf.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 381000 entries, 0 to 380999\nColumns: 129 entries, CH1 to Label\ndtypes: float64(129)\nmemory usage: 375.0 MB\n"
],
[
"N_TIME_STEPS = 250\nN_FEATURES = 128 #128\nstep = 10 # 20\nsegments = []\nfor i in range(0, len(df) - N_TIME_STEPS, step):\n ch = []\n for j in range(0, N_FEATURES):\n ch.append(df.iloc[:, j].values[i: i + N_TIME_STEPS])\n segments.append(ch)",
"_____no_output_____"
],
[
"labels = []\nfor i in range(0, len(df) - N_TIME_STEPS, step):\n label = stats.mode(df['Label'][i: i + N_TIME_STEPS])[0][0]\n labels.append(label)\nlabelsl = np.asarray(pd.get_dummies(labels), dtype = np.float32)\n#print(labelsl)",
"_____no_output_____"
],
[
"reshaped_segments = np.asarray(segments, dtype= np.float32).reshape(-1, N_TIME_STEPS, N_FEATURES)\nX_train, X_test, y_train, y_test = train_test_split(\n reshaped_segments, labelsl, test_size=0.2, random_state=RANDOM_SEED)",
"_____no_output_____"
],
[
"print(np.array(segments).shape, reshaped_segments.shape, labelsl[0], len(X_train), len(X_test))",
"(38075, 128, 250) (38075, 250, 128) [1. 0.] 30460 7615\n"
]
],
[
[
"# Building the model",
"_____no_output_____"
]
],
[
[
"N_CLASSES = 2\nN_HIDDEN_UNITS = 64",
"_____no_output_____"
],
[
"# https://medium.com/@curiousily/human-activity-recognition-using-lstms-on-android-tensorflow-for-hackers-part-vi-492da5adef64\ndef create_LSTM_model(inputs):\n W = {\n 'hidden': tf.Variable(tf.random_normal([N_FEATURES, N_HIDDEN_UNITS])),\n 'output': tf.Variable(tf.random_normal([N_HIDDEN_UNITS, N_CLASSES]))\n }\n biases = {\n 'hidden': tf.Variable(tf.random_normal([N_HIDDEN_UNITS], mean=1.0)),\n 'output': tf.Variable(tf.random_normal([N_CLASSES]))\n }\n \n X = tf.transpose(inputs, [1, 0, 2])\n X = tf.reshape(X, [-1, N_FEATURES])\n hidden = tf.nn.relu(tf.matmul(X, W['hidden']) + biases['hidden'])\n hidden = tf.split(hidden, N_TIME_STEPS, 0)\n\n # Stack 2 LSTM layers\n lstm_layers = [tf.contrib.rnn.BasicLSTMCell(N_HIDDEN_UNITS, forget_bias=1.0) for _ in range(2)]\n lstm_layers = tf.contrib.rnn.MultiRNNCell(lstm_layers)\n\n outputs, _ = tf.contrib.rnn.static_rnn(lstm_layers, hidden, dtype=tf.float32)\n\n # Get output for the last time step\n lstm_last_output = outputs[-1]\n\n return tf.matmul(lstm_last_output, W['output']) + biases['output']",
"_____no_output_____"
],
[
"tf.reset_default_graph()\n\nX = tf.placeholder(tf.float32, [None, N_TIME_STEPS, N_FEATURES], name=\"input\")\nY = tf.placeholder(tf.float32, [None, N_CLASSES])",
"_____no_output_____"
],
[
"pred_Y = create_LSTM_model(X)\n\npred_softmax = tf.nn.softmax(pred_Y, name=\"y_\")",
"WARNING:tensorflow:From C:\\Apps\\Anaconda3\\lib\\site-packages\\tensorflow\\python\\framework\\op_def_library.py:263: colocate_with (from tensorflow.python.framework.ops) is deprecated and will be removed in a future version.\nInstructions for updating:\nColocations handled automatically by placer.\n\nWARNING: The TensorFlow contrib module will not be included in TensorFlow 2.0.\nFor more information, please see:\n * https://github.com/tensorflow/community/blob/master/rfcs/20180907-contrib-sunset.md\n * https://github.com/tensorflow/addons\nIf you depend on functionality not listed there, please file an issue.\n\nWARNING:tensorflow:From <ipython-input-8-c8ae7df74e6e>:17: BasicLSTMCell.__init__ (from tensorflow.python.ops.rnn_cell_impl) is deprecated and will be removed in a future version.\nInstructions for updating:\nThis class is equivalent as tf.keras.layers.LSTMCell, and will be replaced by that in Tensorflow 2.0.\nWARNING:tensorflow:From <ipython-input-8-c8ae7df74e6e>:18: MultiRNNCell.__init__ (from tensorflow.python.ops.rnn_cell_impl) is deprecated and will be removed in a future version.\nInstructions for updating:\nThis class is equivalent as tf.keras.layers.StackedRNNCells, and will be replaced by that in Tensorflow 2.0.\nWARNING:tensorflow:From <ipython-input-8-c8ae7df74e6e>:20: static_rnn (from tensorflow.python.ops.rnn) is deprecated and will be removed in a future version.\nInstructions for updating:\nPlease use `keras.layers.RNN(cell, unroll=True)`, which is equivalent to this API\n"
],
[
"L2_LOSS = 0.0015\n\nl2 = L2_LOSS * \\\n sum(tf.nn.l2_loss(tf_var) for tf_var in tf.trainable_variables())\n\nloss = tf.reduce_mean(tf.nn.softmax_cross_entropy_with_logits_v2(logits = pred_Y, labels = Y)) + l2",
"_____no_output_____"
],
[
"LEARNING_RATE = 0.0025\n\noptimizer = tf.train.AdamOptimizer(learning_rate=LEARNING_RATE).minimize(loss)\n\ncorrect_pred = tf.equal(tf.argmax(pred_softmax, 1), tf.argmax(Y, 1))\naccuracy = tf.reduce_mean(tf.cast(correct_pred, dtype=tf.float32))",
"_____no_output_____"
]
],
[
[
"# Training",
"_____no_output_____"
]
],
[
[
"N_EPOCHS = 50 # 50\nBATCH_SIZE = 1024 # 1024",
"_____no_output_____"
],
[
"# https://medium.com/@curiousily/human-activity-recognition-using-lstms-on-android-tensorflow-for-hackers-part-vi-492da5adef64\nsaver = tf.train.Saver()\n\nhistory = dict(train_loss=[], \n train_acc=[], \n test_loss=[], \n test_acc=[])\n\nsess=tf.InteractiveSession()\nsess.run(tf.global_variables_initializer())\n\ntrain_count = len(X_train)\n\nfor i in range(1, N_EPOCHS + 1):\n for start, end in zip(range(0, train_count, BATCH_SIZE),\n range(BATCH_SIZE, train_count + 1,BATCH_SIZE)):\n sess.run(optimizer, feed_dict={X: X_train[start:end],\n Y: y_train[start:end]})\n\n _, acc_train, loss_train = sess.run([pred_softmax, accuracy, loss], feed_dict={\n X: X_train, Y: y_train})\n\n _, acc_test, loss_test = sess.run([pred_softmax, accuracy, loss], feed_dict={\n X: X_test, Y: y_test})\n\n history['train_loss'].append(loss_train)\n history['train_acc'].append(acc_train)\n history['test_loss'].append(loss_test)\n history['test_acc'].append(acc_test)\n\n# if i != 1 and i % 10 != 0:\n# continue\n\n print(f'epoch: {i} test accuracy: {acc_test} loss: {loss_test}')\n \npredictions, acc_final, loss_final = sess.run([pred_softmax, accuracy, loss], feed_dict={X: X_test, Y: y_test})\n\nprint()\nprint(f'final results: accuracy: {acc_final} loss: {loss_final}')",
"epoch: 1 test accuracy: 0.5491792559623718 loss: 6.679009914398193\nepoch: 2 test accuracy: 0.598161518573761 loss: 6.081445693969727\nepoch: 3 test accuracy: 0.6346684098243713 loss: 5.547723770141602\nepoch: 4 test accuracy: 0.6502954959869385 loss: 5.051041126251221\nepoch: 5 test accuracy: 0.637032151222229 loss: 4.660353660583496\nepoch: 6 test accuracy: 0.6844385862350464 loss: 4.257678031921387\nepoch: 7 test accuracy: 0.6764280796051025 loss: 3.9191951751708984\nepoch: 8 test accuracy: 0.6803677082061768 loss: 3.6233041286468506\nepoch: 9 test accuracy: 0.6608010530471802 loss: 3.3527159690856934\nepoch: 10 test accuracy: 0.700590968132019 loss: 3.070573329925537\nepoch: 11 test accuracy: 0.6345371007919312 loss: 2.908764362335205\nepoch: 12 test accuracy: 0.6833880543708801 loss: 2.6737332344055176\nepoch: 13 test accuracy: 0.7047931551933289 loss: 2.4761414527893066\nepoch: 14 test accuracy: 0.6970453262329102 loss: 2.3195760250091553\nepoch: 15 test accuracy: 0.6593565344810486 loss: 2.203551769256592\nepoch: 16 test accuracy: 0.6915298700332642 loss: 2.0374956130981445\nepoch: 17 test accuracy: 0.684175968170166 loss: 1.9220277070999146\nepoch: 18 test accuracy: 0.6567301154136658 loss: 1.8521720170974731\nepoch: 19 test accuracy: 0.7024294137954712 loss: 1.6897430419921875\nepoch: 20 test accuracy: 0.6856204867362976 loss: 1.6988005638122559\nepoch: 21 test accuracy: 0.7348654270172119 loss: 1.484269142150879\nepoch: 22 test accuracy: 0.7506237626075745 loss: 1.3973190784454346\nepoch: 23 test accuracy: 0.6601444482803345 loss: 1.5000097751617432\nepoch: 24 test accuracy: 0.743926465511322 loss: 1.2885267734527588\nepoch: 25 test accuracy: 0.7553513050079346 loss: 1.2124427556991577\nepoch: 26 test accuracy: 0.7523308992385864 loss: 1.1660268306732178\nepoch: 27 test accuracy: 0.7615233063697815 loss: 1.1193861961364746\nepoch: 28 test accuracy: 0.7661194801330566 loss: 1.072493076324463\nepoch: 29 test accuracy: 0.7684832811355591 loss: 1.051013469696045\nepoch: 30 test accuracy: 0.7586342692375183 loss: 1.0569052696228027\nepoch: 31 test accuracy: 0.767432689666748 loss: 0.980268120765686\nepoch: 32 test accuracy: 0.7712409496307373 loss: 0.9483214020729065\nepoch: 33 test accuracy: 0.759290874004364 loss: 0.9309238195419312\nepoch: 34 test accuracy: 0.7585029602050781 loss: 0.921152651309967\nepoch: 35 test accuracy: 0.7808273434638977 loss: 0.885219931602478\nepoch: 36 test accuracy: 0.7344714403152466 loss: 0.9116696119308472\nepoch: 37 test accuracy: 0.7508863806724548 loss: 0.87147057056427\nepoch: 38 test accuracy: 0.7737360596656799 loss: 0.8312199711799622\nepoch: 39 test accuracy: 0.7739986777305603 loss: 0.816388726234436\nepoch: 40 test accuracy: 0.7783322334289551 loss: 0.8092168569564819\nepoch: 41 test accuracy: 0.7725541591644287 loss: 0.7965925931930542\nepoch: 42 test accuracy: 0.7961917519569397 loss: 0.7669544219970703\nepoch: 43 test accuracy: 0.7764937877655029 loss: 0.7741450071334839\nepoch: 44 test accuracy: 0.7929087281227112 loss: 0.7412621974945068\nepoch: 45 test accuracy: 0.7948785424232483 loss: 0.7452307939529419\nepoch: 46 test accuracy: 0.7826657891273499 loss: 0.7869150638580322\nepoch: 47 test accuracy: 0.769271194934845 loss: 0.809227466583252\nepoch: 48 test accuracy: 0.791070282459259 loss: 0.7862563133239746\nepoch: 49 test accuracy: 0.7641497254371643 loss: 0.8093451261520386\nepoch: 50 test accuracy: 0.784635603427887 loss: 0.7785685062408447\n\nfinal results: accuracy: 0.784635603427887 loss: 0.7785685062408447\n"
]
],
[
[
"# Evaluation",
"_____no_output_____"
]
],
[
[
"# https://medium.com/@curiousily/human-activity-recognition-using-lstms-on-android-tensorflow-for-hackers-part-vi-492da5adef64\nplt.figure(figsize=(12, 8))\n\nplt.plot(np.array(history['train_loss']), \"r--\", label=\"Train loss\")\nplt.plot(np.array(history['train_acc']), \"g--\", label=\"Train accuracy\")\n\nplt.plot(np.array(history['test_loss']), \"r-\", label=\"Test loss\")\nplt.plot(np.array(history['test_acc']), \"g-\", label=\"Test accuracy\")\n\nplt.title(\"Training session's progress over iterations\")\nplt.legend(loc='upper right', shadow=True)\nplt.ylabel('Training Progress (Loss or Accuracy values)')\nplt.xlabel('Training Epoch')\nplt.ylim(0)\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"# Saving Model",
"_____no_output_____"
]
],
[
[
"import os\n\nfile_info = [N_HIDDEN_UNITS, BATCH_SIZE, N_EPOCHS]\n\ndirname = os.path.dirname(\"nhid-{}_bat-{}_nepoc-{}/dumps/\".format(*file_info))\nif not os.path.exists(dirname):\n os.makedirs(dirname)\ndirname = os.path.dirname(\"nhid-{}_bat-{}_nepoc-{}/logs/\".format(*file_info))\nif not os.path.exists(dirname):\n os.makedirs(dirname)\n\npickle.dump(predictions, open(\"nhid-{}_bat-{}_nepoc-{}/dumps/predictions.p\".format(*file_info), \"wb\"))\npickle.dump(history, open(\"nhid-{}_bat-{}_nepoc-{}/dumps/history.p\".format(*file_info), \"wb\"))\ntf.train.write_graph(sess.graph, \"nhid-{}_bat-{}_nepoc-{}/logs\".format(*file_info), 'har.pbtxt') \nsaver.save(sess, 'nhid-{}_bat-{}_nepoc-{}/logs/har.ckpt'.format(*file_info))\n\nwriter = tf.summary.FileWriter('nhid-{}_bat-{}_nepoc-{}/logs'.format(*file_info))\nwriter.add_graph(sess.graph)",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cbee5dd96658210c56e51d6bfdd6ba9c26895488
| 921,643 |
ipynb
|
Jupyter Notebook
|
Solution_VAST.ipynb
|
LasTAD/VAST-2017-MC-1
|
a49d9d95e684ea9d73cbd699ab0a825bfe007439
|
[
"MIT"
] | 1 |
2020-04-08T16:52:49.000Z
|
2020-04-08T16:52:49.000Z
|
Solution_VAST.ipynb
|
LasTAD/VAST-2017-MC-1
|
a49d9d95e684ea9d73cbd699ab0a825bfe007439
|
[
"MIT"
] | null | null | null |
Solution_VAST.ipynb
|
LasTAD/VAST-2017-MC-1
|
a49d9d95e684ea9d73cbd699ab0a825bfe007439
|
[
"MIT"
] | null | null | null | 88.619519 | 93,550 | 0.486737 |
[
[
[
"# VAST 2017 MC-1",
"_____no_output_____"
],
[
"## Задание\nПриродный заповедник Бунсонг Лекагуль используется местными жителями и туристами для однодневных поездок, ночевок в кемпингах, а иногда и просто для доступа к основным магистралям на противоположных сторонах заповедника.\n\nВходные кабинки заповедника контролируются с целью получения дохода, а также мониторинга использования. Транспортные средства, въезжающие в заповедник и выезжающие из него, должны платить пошлину в зависимости от количества осей (личный автомобиль, развлекательный прицеп, полуприцеп и т.д.).\n\nЭто создает поток данных с отметками времени входа / выхода и типом транспортного средства. Есть также другие места в части, которые регистрируют трафик, проходящий через заповедник. Путешествуя по различным частям заповедника, Митч заметил странное поведение транспортных средств, которое, по его мнению, не соответствует видам посетителей парка, которых он ожидал. Если бы Митч мог каким-то образом анализировать поведение автомобилей в парке с течением времени, это сможет помочь ему в его расследовании.\n\n### Пример исходных данных\n\n### Необходимые импорты",
"_____no_output_____"
]
],
[
[
"import sqlite3\nimport numpy as np\nimport matplotlib.pyplot as plt\nfrom sklearn.cluster import KMeans\nfrom SOM import SOM\nimport seaborn as sb",
"_____no_output_____"
]
],
[
[
"## Подготовка данных\n\nС помощью языка программирования Python распарсили данные из таблицы, затем объединили в группы сенсоры по Car-Id, тем самым нашли все датчики, которые проехало данное ТС, его путь.",
"_____no_output_____"
]
],
[
[
"data_set = open(\"Data/Lekagul Sensor Data.csv\", \"r\")\ndata = data_set.readlines()\ndata_set.close()\ntraces = []\ngates = set()\nfor line in data:\n args = line.split(\";\")\n gates.add(args[3])\n traces.append(args)\ngates = sorted(gates)\ngroupedTraces = []\nfor t in traces:\n if t[1] in groupedTraces:\n groupedTraces[t[1]].append(t)\n else:\n groupedTraces[t[1]] = [t]",
"_____no_output_____"
]
],
[
[
"Вычленяем типа автомобиля из списка путей.",
"_____no_output_____"
]
],
[
[
"target = []\nfor x in groupedTraces:\n target.append(groupedTraces[x][1][2])\ntargets = []\nfor rec in target:\n if rec == '2P':\n targets.append('7')\n else:\n targets.append(str(rec))\nprint(targets)",
"['4', '1', '4', '4', '3', '1', '5', '1', '2', '3', '1', '1', '1', '3', '7', '2', '1', '1', '3', '1', '1', '2', '1', '3', '3', '1', '2', '1', '1', '1', '7', '5', '7', '7', '4', '2', '2', '3', '2', '4', '4', '5', '1', '1', '3', '1', '3', '2', '7', '6', '1', '5', '1', '1', '5', '5', '3', '1', '3', '3', '1', '1', '3', '3', '2', '2', '1', '3', '7', '7', '3', '2', '2', '5', '2', '5', '2', '1', '2', '2', '2', '2', '3', '2', '3', '2', '3', '5', '3', '1', '1', '1', '2', '1', '1', '7', '7', '1', '2', '4', '2', '1', '1', '1', '3', '1', '4', '1', '1', '1', '1', '3', '7', '5', '1', '3', '1', '2', '4', '4', '3', '2', '1', '6', '3', '1', '6', '1', '1', '3', '1', '7', '7', '3', '2', '2', '1', '1', '2', '4', '7', '1', '4', '2', '5', '2', '7', '1', '2', '2', '4', '5', '1', '4', '1', '2', '4', '2', '1', '7', '1', '1', '1', '2', '1', '2', '1', '2', '2', '7', '2', '2', '1', '3', '1', '3', '3', '7', '2', '1', '1', '2', '4', '1', '3', '1', '2', '2', '4', '2', '4', '2', '5', '5', '1', '1', '1', '4', '4', '1', '2', '2', '7', '2', '1', '1', '2', '1', '3', '2', '3', '2', '2', '7', '1', '3', '4', '1', '1', '3', '1', '1', '1', '1', '2', '1', '2', '6', '2', '3', '1', '4', '2', '1', '1', '2', '1', '3', '4', '1', '2', '1', '2', '4', '1', '1', '2', '6', '3', '2', '3', '1', '3', '2', '6', '1', '7', '3', '4', '1', '1', '2', '1', '4', '4', '1', '2', '4', '1', '1', '1', '3', '2', '3', '1', '2', '1', '3', '7', '2', '3', '3', '7', '2', '1', '4', '1', '1', '3', '2', '7', '3', '1', '7', '1', '2', '1', '1', '1', '4', '2', '2', '2', '2', '5', '2', '2', '2', '1', '1', '3', '2', '1', '2', '1', '3', '1', '1', '5', '2', '3', '2', '1', '1', '2', '3', '5', '1', '4', '1', '2', '1', '1', '4', '1', '1', '7', '1', '2', '1', '2', '2', '2', '7', '5', '1', '2', '2', '7', '2', '2', '2', '4', '5', '2', '2', '5', '2', '2', '4', '5', '5', '4', '1', '2', '2', '2', '1', '7', '2', '2', '2', '7', '7', '2', '2', '1', '1', '1', '1', '1', '1', '1', '7', '6', '3', '2', '1', '1', '2', '5', '4', '2', '1', '2', '1', '1', '1', '1', '3', '3', '4', '3', '1', '2', '7', '6', '2', '1', '2', '5', '3', '1', '2', '3', '1', '1', '1', '1', '3', '7', '7', '3', '4', '1', '2', '4', '7', '2', '4', '1', '1', '3', '2', '1', '1', '1', '4', '3', '2', '3', '2', '3', '5', '6', '4', '4', '3', '1', '3', '1', '7', '1', '7', '5', '6', '3', '2', '1', '1', '1', '7', '1', '7', '1', '4', '2', '2', '2', '3', '2', '1', '2', '2', '3', '1', '6', '2', '3', '2', '4', '4', '4', '2', '6', '3', '2', '1', '1', '1', '1', '7', '2', '2', '1', '7', '5', '1', '1', '3', '2', '2', '2', '1', '3', '5', '2', '1', '4', '7', '2', '1', '2', '1', '4', '2', '1', '2', '1', '4', '3', '5', '2', '4', '4', '3', '3', '5', '5', '2', '1', '3', '1', '1', '1', '4', '7', '2', '1', '1', '3', '3', '2', '1', '1', '1', '7', '2', '5', '1', '7', '1', '1', '2', '5', '3', '3', '1', '2', '2', '6', '3', '1', '3', '5', '5', '2', '4', '1', '1', '5', '4', '2', '2', '2', '3', '5', '1', '5', '2', '3', '1', '2', '2', '4', '3', '2', '2', '7', '1', '1', '3', '7', '2', '4', '3', '2', '1', '2', '7', '2', '1', '3', '7', '3', '2', '3', '1', '1', '1', '1', '2', '1', '2', '2', '3', '4', '6', '3', '2', '1', '2', '4', '1', '3', '2', '7', '2', '3', '2', '2', '1', '1', '6', '1', '1', '2', '2', '2', '1', '2', '1', '2', '3', '2', '6', '1', '1', '1', '7', '1', '1', '2', '2', '3', '2', '1', '1', '1', '2', '5', '3', '4', '1', '3', '3', '2', '1', '3', '1', '1', '7', '1', '2', '5', '1', '2', '2', '4', '2', '2', '2', '7', '1', '4', '1', '7', '2', '2', '1', '1', '1', '2', '1', '3', '3', '2', '2', '2', '4', '4', '2', '2', '3', '5', '2', '4', '2', '1', '2', '5', '7', '2', '3', '1', '6', '1', '1', '2', '1', '1', '1', '3', '2', '1', '2', '1', '2', '1', '2', '2', '5', '2', '5', '1', '2', '2', '1', '2', '4', '1', '3', '7', '5', '2', '2', '1', '3', '2', '4', '2', '5', '3', '2', '1', '7', '4', '3', '2', '4', '1', '1', '4', '3', '1', '1', '1', '2', '1', '2', '3', '2', '2', '4', '1', '3', '2', '1', '1', '3', '1', '1', '3', '1', '1', '3', '2', '7', '3', '1', '1', '1', '2', '6', '1', '5', '2', '6', '1', '1', '2', '2', '2', '4', '3', '1', '7', '1', '1', '3', '2', '2', '2', '6', '5', '1', '1', '4', '1', '1', '3', '2', '7', '7', '2', '4', '7', '4', '3', '2', '3', '2', '2', '2', '1', '2', '1', '3', '4', '2', '7', '1', '3', '1', '2', '1', '1', '1', '4', '2', '1', '1', '1', '2', '1', '1', '2', '1', '1', '1', '3', '2', '2', '3', '7', '1', '1', '2', '2', '1', '2', '4', '3', '1', '1', '1', '1', '1', '1', '1', '1', '5', '2', '7', '2', '2', '3', '1', '2', '1', '1', '1', '5', '2', '2', '1', '7', '2', '1', '4', '1', '3', '4', '2', '3', '2', '4', '1', '2', '2', '5', '6', '1', '7', '1', '2', '3', '1', '2', '3', '2', '3', '1', '2', '1', '2', '2', '3', '2', '2', '3', '2', '7', '1', '4', '1', '7', '2', '1', '1', '3', '1', '1', '1', '2', '4', '5', '1', '1', '3', '4', '2', '1', '1', '1', '2', '1', '7', '1', '1', '1', '1', '3', '4', '2', '3', '1', '3', '1', '4', '2', '3', '1', '7', '4', '5', '1', '7', '1', '1', '1', '2', '1', '1', '1', '1', '3', '1', '4', '4', '3', '3', '4', '1', '1', '4', '4', '2', '1', '7', '1', '1', '1', '7', '1', '3', '7', '7', '1', '1', '2', '2', '1', '2', '3', '2', '5', '2', '2', '5', '2', '3', '1', '4', '1', '3', '1', '2', '4', '1', '1', '4', '1', '4', '1', '3', '3', '2', '1', '1', '1', '5', '2', '3', '2', '4', '1', '1', '7', '1', '7', '1', '2', '1', '2', '1', '2', '2', '3', '1', '3', '1', '1', '7', '1', '3', '3', '1', '3', '1', '1', '3', '3', '3', '3', '1', '3', '1', '1', '2', '1', '2', '4', '2', '7', '3', '2', '7', '2', '7', '1', '1', '1', '1', '1', '1', '2', '3', '1', '2', '1', '1', '1', '3', '3', '3', '4', '1', '2', '3', '7', '2', '7', '1', '2', '1', '5', '1', '5', '3', '4', '7', '2', '1', '1', '3', '6', '1', '1', '6', '1', '4', '7', '1', '1', '1', '4', '3', '2', '3', '2', '1', '1', '4', '1', '2', '2', '1', '2', '3', '5', '4', '3', '2', '1', '3', '2', '7', '1', '2', '4', '1', '2', '1', '2', '2', '3', '1', '4', '2', '1', '2', '2', '2', '2', '1', '7', '3', '4', '7', '7', '2', '1', '1', '3', '3', '2', '1', '4', '2', '1', '3', '5', '4', '4', '4', '2', '1', '3', '3', '3', '2', '7', '3', '2', '2', '2', '1', '5', '2', '3', '1', '2', '1', '2', '2', '5', '2', '1', '1', '7', '7', '3', '2', '1', '3', '3', '1', '1', '3', '2', '1', '6', '1', '5', '2', '1', '2', '1', '7', '2', '5', '5', '2', '7', '1', '1', '1', '1', '3', '6', '1', '1', '2', '2', '5', '2', '3', '7', '2', '1', '1', '1', '1', '5', '2', '2', '1', '4', '1', '5', '1', '2', '2', '3', '4', '6', '1', '2', '2', '3', '1', '1', '2', '3', '2', '1', '1', '3', '2', '2', '2', '1', '1', '1', '1', '2', '1', '2', '1', '2', '2', '3', '5', '1', '1', '3', '3', '7', '1', '1', '4', '3', '1', '1', '1', '5', '1', '1', '3', '2', '2', '2', '1', '1', '1', '1', '2', '1', '3', '1', '7', '1', '2', '2', '5', '4', '6', '2', '4', '4', '1', '2', '3', '4', '4', '1', '1', '1', '4', '5', '3', '3', '2', '1', '3', '3', '1', '1', '2', '2', '1', '2', '2', '1', '7', '2', '1', '1', '1', '1', '3', '2', '1', '1', '1', '3', '2', '7', '1', '7', '3', '2', '2', '3', '1', '2', '1', '2', '3', '2', '2', '1', '2', '5', '2', '3', '1', '1', '7', '1', '1', '3', '3', '3', '2', '1', '1', '4', '1', '1', '3', '1', '2', '1', '3', '2', '1', '1', '2', '1', '3', '3', '3', '4', '3', '4', '5', '5', '1', '4', '1', '4', '1', '4', '1', '3', '2', '1', '2', '2', '2', '5', '1', '1', '1', '2', '1', '1', '1', '1', '1', '6', '1', '1', '1', '3', '1', '2', '7', '2', '3', '3', '3', '2', '1', '3', '5', '1', '1', '2', '1', '2', '3', '2', '1', '1', '3', '1', '2', '1', '1', '1', '3', '1', '7', '1', '3', '2', '1', '3', '1', '2', '1', '1', '3', '3', '1', '1', '3', '2', '3', '5', '3', '2', '5', '1', '2', '1', '5', '1', '2', '2', '1', '3', '1', '2', '1', '1', '1', '3', '2', '6', '1', '2', '1', '7', '3', '1', '1', '1', '3', '3', '2', '2', '3', '1', '3', '4', '1', '3', '1', '1', '1', '2', '1', '4', '2', '1', '2', '3', '2', '2', '3', '1', '3', '1', '4', '1', '1', '3', '2', '1', '1', '5', '7', '1', '2', '3', '7', '1', '3', '1', '4', '1', '3', '1', '3', '2', '6', '3', '4', '2', '2', '1', '2', '1', '1', '2', '5', '1', '2', '1', '6', '5', '5', '6', '3', '1', '1', '2', '3', '3', '4', '2', '1', '1', '1', '3', '3', '4', '2', '2', '2', '1', '2', '1', '2', '3', '2', '2', '1', '1', '1', '3', '1', '1', '2', '2', '2', '2', '2', '2', '1', '2', '7', '3', '7', '3', '1', '1', '1', '1', '2', '7', '2', '6', '1', '3', '1', '2', '6', '3', '1', '1', '2', '2', '1', '3', '1', '2', '1', '3', '4', '1', '1', '6', '1', '1', '1', '1', '2', '3', '6', '3', '2', '1', '1', '1', '2', '2', '1', '1', '3', '5', '2', '1', '1', '2', '1', '2', '3', '2', '3', '1', '2', '2', '1', '4', '1', '2', '2', '1', '1', '1', '7', '1', '3', '1', '3', '2', '1', '1', '1', '1', '2', '2', '3', '1', '7', '2', '1', '2', '1', '2', '1', '1', '1', '1', '1', '5', '1', '2', '2', '2', '3', '2', '1', '1', '2', '1', '1', '3', '3', '2', '2', '1', '1', '2', '3', '1', '2', '1', '1', '1', '4', '3', '2', '1', '3', '2', '7', '2', '3', '2', '3', '3', '2', '1', '2', '2', '2', '1', '1', '3', '2', '1', '3', '1', '4', '2', '4', '2', '1', '1', '1', '2', '1', '2', '1', '2', '1', '2', '6', '2', '1', '6', '2', '4', '5', '2', '5', '2', '1', '5', '3', '1', '2', '2', '1', '2', '1', '2', '1', '2', '3', '7', '1', '1', '1', '1', '1', '3', '1', '1', '1', '1', '1', '1', '1', '2', '1', '5', '1', '4', '2', '4', '1', '1', '3', '2', '1', '1', '3', '2', '2', '2', '2', '2', '2', '1', '3', '1', '2', '1', '2', '1', '7', '2', '2', '3', '7', '1', '2', '2', '1', '1', '1', '2', '2', '1', '1', '3', '3', '3', '7', '1', '3', '1', '1', '1', '1', '1', '2', '2', '1', '2', '2', '2', '2', '5', '3', '2', '4', '1', '3', '1', '1', '1', '1', '2', '2', '2', '4', '2', '2', '1', '4', '1', '1', '1', '1', '4', '1', '1', '6', '1', '2', '2', '1', '3', '2', '4', '1', '6', '1', '3', '1', '5', '1', '1', '1', '2', '1', '1', '3', '4', '1', '3', '1', '1', '1', '1', '1', '2', '1', '2', '2', '1', '2', '3', '2', '2', '1', '3', '1', '1', '2', '1', '2', '3', '2', '2', '1', '1', '7', '1', '2', '1', '1', '3', '3', '1', '7', '1', '1', '1', '1', '3', '2', '1', '3', '3', '1', '1', '1', '2', '6', '4', '5', '5', '4', '1', '5', '3', '2', '1', '4', '6', '2', '3', '1', '1', '2', '1', '3', '4', '2', '1', '1', '2', '2', '1', '3', '3', '2', '3', '2', '1', '2', '3', '1', '1', '2', '1', '1', '2', '2', '1', '5', '2', '3', '2', '1', '5', '3', '3', '1', '2', '2', '1', '1', '2', '1', '3', '1', '1', '3', '1', '1', '1', '2', '2', '1', '2', '1', '7', '1', '1', '3', '3', '3', '1', '1', '3', '2', '1', '1', '2', '5', '1', '4', '1', '1', '2', '1', '3', '6', '1', '4', '3', '2', '2', '1', '1', '2', '2', '1', '1', '3', '2', '2', '2', '2', '2', '2', '1', '2', '4', '2', '1', '3', '1', '1', '3', '1', '2', '1', '1', '2', '7', '2', '4', '4', '1', '1', '1', '1', '3', '1', '1', '1', '1', '1', '1', '1', '3', '3', '3', '2', '2', '7', '3', '2', '1', '5', '6', '3', '3', '3', '2', '2', '2', '2', '1', '3', '1', '7', '3', '4', '1', '2', '7', '3', '2', '1', '1', '1', '1', '4', '4', '2', '1', '1', '3', '1', '1', '1', '1', '2', '1', '1', '2', '3', '2', '3', '7', '1', '1', '1', '3', '3', '1', '2', '1', '1', '5', '2', '1', '1', '3', '1', '2', '1', '1', '1', '2', '3', '5', '3', '2', '3', '2', '1', '1', '1', '3', '1', '2', '1', '1', '1', '4', '2', '1', '3', '1', '2', '3', '1', '1', '2', '3', '1', '3', '3', '2', '6', '3', '3', '2', '1', '1', '2', '2', '1', '3', '2', '2', '1', '1', '1', '2', '3', '1', '1', '3', '1', '1', '1', '1', '1', '6', '1', '1', '2', '2', '2', '1', '1', '2', '3', '2', '3', '7', '1', '3', '1', '1', '1', '3', '1', '2', '1', '2', '2', '1', '3', '7', '1', '1', '1', '1', '1', '3', '1', '1', '1', '1', '1', '1', '1', '2', '2', '1', '1', '1', '2', '1', '2', '1', '1', '1', '1', '2', '7', '1', '1', '2', '1', '3', '1', '1', '1', '2', '1', '6', '1', '5', '4', '1', '2', '2', '5', '1', '1', '6', '3', '4', '2', '3', '2', '3', '2', '2', '1', '3', '1', '2', '1', '2', '3', '1', '2', '3', '2', '1', '6', '1', '1', '1', '1', '2', '1', '1', '1', '1', '3', '2', '3', '1', '1', '1', '2', '3', '1', '1', '3', '1', '1', '1', '1', '2', '1', '1', '5', '2', '2', '1', '1', '1', '3', '2', '1', '1', '1', '3', '1', '1', '1', '1', '2', '2', '3', '2', '3', '6', '2', '1', '1', '2', '7', '3', '2', '1', '2', '3', '4', '1', '1', '1', '5', '1', '1', '1', '3', '1', '2', '1', '1', '2', '2', '3', '2', '3', '3', '2', '5', '3', '1', '4', '2', '1', '1', '6', '1', '1', '6', '2', '3', '1', '1', '2', '1', '3', '2', '7', '3', '3', '1', '2', '1', '1', '1', '1', '3', '3', '1', '1', '1', '2', '1', '1', '1', '1', '1', '1', '7', '2', '3', '2', '1', '3', '1', '3', '3', '2', '2', '6', '1', '1', '1', '2', '1', '5', '1', '1', '1', '2', '1', '1', '2', '2', '5', '1', '2', '1', '3', '1', '1', '1', '2', '1', '7', '1', '3', '1', '1', '2', '2', '2', '6', '3', '6', '4', '2', '3', '1', '2', '4', '6', '1', '6', '2', '3', '2', '1', '4', '2', '2', '3', '2', '7', '1', '1', '2', '7', '7', '3', '1', '2', '1', '2', '1', '2', '1', '2', '1', '1', '2', '2', '3', '2', '1', '3', '2', '2', '2', '3', '1', '3', '1', '3', '4', '5', '2', '1', '5', '2', '1', '4', '7', '1', '1', '1', '1', '4', '1', '2', '1', '1', '2', '4', '1', '1', '2', '1', '1', '3', '1', '1', '2', '1', '1', '2', '1', '1', '1', '1', '3', '2', '1', '1', '4', '3', '1', '1', '2', '1', '1', '1', '2', '3', '2', '1', '5', '1', '2', '1', '1', '4', '3', '2', '1', '7', '3', '1', '1', '1', '2', '2', '2', '1', '2', '7', '3', '3', '1', '1', '1', '1', '5', '1', '2', '7', '1', '3', '4', '2', '2', '5', '5', '1', '1', '1', '7', '2', '1', '1', '1', '1', '5', '1', '5', '1', '1', '1', '3', '2', '1', '2', '2', '1', '3', '2', '3', '2', '4', '3', '1', '1', '1', '1', '3', '3', '3', '1', '1', '1', '2', '2', '1', '1', '2', '2', '6', '1', '3', '2', '1', '1', '2', '5', '3', '1', '1', '2', '1', '2', '5', '2', '5', '2', '4', '2', '2', '1', '1', '5', '2', '1', '1', '2', '2', '2', '1', '1', '1', '1', '1', '1', '6', '6', '1', '1', '3', '3', '1', '1', '1', '2', '1', '2', '1', '3', '1', '1', '2', '2', '1', '3', '2', '3', '1', '1', '1', '3', '3', '2', '1', '2', '1', '3', '2', '1', '1', '1', '6', '2', '1', '1', '2', '1', '2', '2', '5', '1', '2', '1', '2', '2', '3', '1', '1', '1', '3', '1', '3', '2', '7', '3', '3', '1', '1', '1', '4', '3', '4', '1', '2', '2', '1', '3', '1', '4', '2', '6', '1', '1', '1', '2', '1', '4', '2', '2', '2', '2', '3', '2', '1', '2', '3', '2', '5', '2', '5', '1', '1', '3', '1', '3', '2', '1', '1', '1', '4', '2', '3', '1', '3', '3', '1', '1', '2', '1', '3', '1', '1', '1', '2', '2', '7', '1', '2', '2', '1', '4', '3', '1', '1', '2', '3', '2', '1', '3', '1', '1', '1', '1', '1', '2', '1', '1', '2', '4', '2', '1', '2', '3', '2', '1', '2', '2', '1', '2', '6', '1', '1', '4', '5', '2', '5', '2', '3', '2', '5', '3', '6', '3', '1', '4', '2', '2', '1', '3', '2', '3', '2', '1', '1', '2', '2', '1', '2', '1', '1', '2', '2', '3', '1', '3', '2', '3', '1', '1', '1', '3', '3', '3', '1', '1', '1', '2', '1', '1', '2', '2', '2', '5', '3', '2', '1', '2', '1', '2', '1', '1', '1', '2', '3', '1', '3', '3', '7', '1', '1', '1', '3', '1', '1', '4', '2', '1', '1', '2', '2', '1', '1', '1', '7', '2', '7', '3', '3', '1', '3', '1', '1', '1', '5', '4', '2', '1', '1', '2', '3', '6', '5', '2', '6', '6', '1', '2', '3', '2', '3', '2', '1', '3', '1', '3', '1', '1', '2', '1', '1', '1', '3', '1', '1', '1', '3', '1', '2', '2', '2', '1', '1', '4', '1', '1', '1', '1', '1', '1', '3', '3', '3', '1', '2', '1', '2', '3', '2', '2', '1', '1', '1', '2', '2', '1', '3', '3', '2', '2', '2', '1', '1', '3', '2', '2', '1', '1', '1', '7', '1', '2', '2', '2', '2', '1', '1', '1', '2', '1', '3', '7', '1', '2', '3', '2', '3', '1', '2', '1', '2', '2', '7', '3', '1', '1', '3', '2', '1', '2', '1', '1', '1', '2', '2', '1', '6', '3', '1', '3', '1', '4', '2', '3', '3', '3', '3', '1', '1', '2', '3', '1', '1', '2', '1', '2', '1', '1', '1', '2', '1', '1', '7', '1', '1', '4', '2', '2', '1', '1', '3', '1', '3', '2', '3', '3', '7', '2', '1', '2', '1', '1', '1', '1', '2', '2', '3', '2', '1', '3', '1', '1', '1', '1', '3', '2', '3', '3', '2', '2', '3', '1', '1', '1', '3', '3', '3', '1', '3', '1', '1', '1', '1', '2', '7', '2', '1', '4', '1', '2', '1', '3', '2', '1', '1', '1', '3', '2', '1', '1', '2', '3', '4', '1', '1', '1', '2', '7', '1', '1', '2', '1', '2', '2', '3', '1', '3', '1', '1', '3', '1', '6', '1', '1', '1', '1', '3', '3', '5', '2', '4', '2', '1', '1', '1', '2', '3', '1', '1', '2', '2', '3', '1', '3', '1', '2', '1', '2', '2', '1', '7', '1', '4', '2', '3', '2', '3', '1', '3', '2', '4', '2', '1', '1', '1', '2', '1', '7', '3', '1', '1', '4', '3', '1', '1', '2', '2', '2', '1', '1', '1', '2', '3', '1', '1', '2', '1', '2', '3', '1', '2', '2', '1', '1', '1', '2', '1', '1', '3', '1', '2', '3', '1', '2', '1', '3', '1', '4', '5', '1', '4', '1', '4', '3', '4', '2', '2', '1', '2', '2', '2', '5', '5', '1', '3', '1', '1', '1', '1', '2', '2', '2', '3', '2', '1', '2', '3', '2', '1', '4', '2', '1', '2', '1', '2', '1', '1', '1', '1', '7', '7', '2', '2', '2', '3', '1', '1', '1', '1', '7', '2', '1', '3', '1', '3', '3', '2', '1', '1', '1', '1', '1', '3', '1', '2', '2', '1', '1', '2', '2', '1', '1', '3', '1', '2', '3', '5', '1', '2', '4', '3', '2', '1', '2', '3', '7', '1', '4', '1', '1', '2', '1', '2', '1', '5', '1', '1', '1', '2', '1', '4', '3', '5', '5', '2', '3', '2', '1', '6', '6', '1', '5', '1', '1', '3', '1', '1', '4', '4', '2', '1', '2', '1', '2', '2', '1', '2', '2', '1', '1', '2', '1', '1', '1', '3', '7', '1', '3', '3', '1', '1', '3', '1', '1', '1', '2', '1', '1', '3', '2', '1', '3', '1', '1', '3', '3', '1', '1', '2', '1', '1', '1', '1', '3', '2', '1', '2', '4', '2', '1', '2', '1', '3', '2', '1', '1', '3', '2', '2', '1', '1', '2', '2', '1', '1', '1', '1', '3', '1', '3', '3', '2', '3', '1', '1', '2', '6', '3', '1', '1', '2', '2', '2', '1', '1', '1', '1', '2', '2', '3', '2', '2', '3', '4', '2', '1', '4', '1', '6', '2', '1', '4', '6', '2', '2', '1', '3', '1', '2', '3', '1', '3', '3', '1', '1', '3', '3', '3', '2', '1', '2', '1', '2', '1', '2', '1', '1', '2', '1', '1', '2', '1', '2', '1', '1', '1', '2', '1', '1', '1', '1', '5', '3', '2', '2', '7', '3', '1', '6', '1', '3', '2', '1', '1', '1', '1', '1', '1', '1', '2', '3', '7', '1', '2', '1', '1', '3', '1', '7', '4', '2', '4', '6', '2', '3', '2', '1', '2', '3', '2', '2', '2', '3', '2', '3', '1', '2', '4', '1', '3', '1', '5', '3', '2', '2', '1', '1', '3', '4', '2', '1', '2', '3', '4', '1', '1', '2', '3', '1', '2', '1', '2', '3', '2', '1', '2', '1', '3', '1', '2', '2', '2', '1', '2', '1', '2', '1', '2', '1', '1', '1', '3', '1', '2', '7', '4', '3', '1', '1', '1', '1', '2', '1', '2', '1', '1', '1', '1', '1', '2', '2', '3', '1', '1', '1', '1', '3', '2', '1', '2', '1', '1', '2', '2', '1', '1', '1', '3', '2', '5', '5', '2', '4', '1', '3', '2', '1', '3', '1', '6', '4', '6', '1', '2', '1', '5', '2', '3', '3', '2', '2', '3', '3', '7', '3', '1', '3', '1', '2', '1', '1', '2', '1', '1', '1', '4', '3', '1', '1', '1', '3', '2', '2', '2', '1', '1', '1', '1', '1', '1', '1', '2', '7', '2', '1', '1', '1', '1', '3', '1', '1', '1', '1', '2', '7', '2', '1', '2', '1', '3', '2', '1', '6', '1', '2', '1', '7', '1', '1', '1', '1', '3', '1', '2', '3', '2', '1', '1', '1', '1', '1', '1', '3', '6', '6', '1', '3', '2', '2', '1', '4', '4', '2', '3', '2', '3', '3', '1', '2', '3', '1', '3', '3', '3', '1', '2', '3', '1', '2', '3', '3', '1', '5', '7', '3', '1', '1', '2', '2', '1', '1', '1', '1', '3', '3', '1', '4', '2', '3', '3', '2', '2', '1', '1', '2', '2', '1', '1', '1', '2', '1', '1', '4', '3', '1', '2', '5', '3', '1', '3', '7', '3', '7', '2', '2', '2', '2', '1', '2', '1', '1', '3', '1', '3', '1', '1', '2', '2', '4', '2', '2', '7', '6', '1', '3', '2', '6', '1', '2', '2', '4', '3', '1', '2', '3', '3', '1', '3', '1', '3', '1', '1', '1', '3', '3', '2', '1', '2', '1', '2', '2', '1', '1', '1', '2', '2', '1', '2', '3', '1', '3', '4', '4', '3', '1', '1', '1', '2', '2', '3', '1', '2', '3', '2', '5', '2', '1', '1', '1', '1', '7', '1', '1', '1', '2', '2', '1', '1', '1', '2', '2', '2', '2', '2', '3', '1', '1', '3', '2', '3', '3', '1', '1', '2', '1', '2', '1', '1', '1', '1', '1', '1', '1', '6', '4', '2', '5', '5', '2', '1', '2', '3', '3', '1', '1', '1', '1', '2', '1', '2', '2', '2', '2', '1', '3', '1', '2', '3', '2', '2', '1', '2', '1', '1', '1', '7', '1', '1', '1', '2', '2', '1', '1', '1', '2', '2', '3', '1', '1', '1', '3', '1', '2', '2', '2', '2', '7', '1', '1', '3', '2', '2', '1', '1', '1', '1', '1', '3', '2', '2', '3', '1', '1', '3', '1', '1', '1', '1', '3', '1', '2', '2', '2', '1', '2', '1', '1', '3', '2', '4', '1', '1', '3', '2', '3', '4', '2', '5', '2', '2', '1', '4', '5', '3', '2', '1', '2', '5', '2', '1', '2', '3', '6', '3', '3', '1', '3', '1', '3', '1', '3', '3', '1', '2', '1', '1', '2', '1', '2', '1', '1', '5', '1', '1', '1', '2', '3', '1', '2', '1', '1', '1', '1', '3', '3', '2', '3', '1', '4', '4', '1', '3', '3', '6', '4', '2', '1', '2', '1', '1', '1', '1', '2', '1', '1', '1', '2', '2', '1', '2', '2', '2', '2', '1', '1', '1', '2', '1', '1', '1', '2', '3', '3', '2', '3', '2', '1', '1', '2', '1', '3', '1', '2', '4', '2', '1', '2', '1', '1', '1', '2', '3', '3', '1', '1', '2', '1', '1', '1', '1', '3', '3', '2', '3', '3', '2', '1', '1', '3', '7', '1', '7', '1', '3', '3', '1', '1', '2', '2', '2', '1', '2', '3', '2', '3', '2', '1', '3', '3', '3', '2', '3', '4', '6', '6', '4', '2', '3', '1', '1', '1', '1', '1', '1', '1', '1', '3', '1', '1', '1', '1', '1', '2', '1', '1', '1', '1', '3', '2', '1', '1', '2', '2', '2', '1', '1', '3', '7', '3', '2', '1', '1', '2', '2', '2', '3', '1', '3', '3', '2', '2', '2', '1', '6', '1', '2', '3', '3', '1', '1', '1', '3', '3', '3', '1', '1', '1', '3', '1', '1', '1', '1', '4', '1', '1', '1', '1', '1', '2', '1', '1', '1', '1', '2', '2', '1', '2', '2', '2', '2', '2', '1', '2', '7', '1', '4', '2', '1', '1', '1', '1', '2', '2', '2', '1', '1', '1', '2', '2', '1', '2', '5', '2', '1', '5', '1', '5', '1', '5', '2', '3', '2', '6', '2', '3', '2', '1', '1', '2', '2', '2', '3', '1', '2', '2', '2', '3', '1', '1', '3', '2', '3', '1', '2', '1', '3', '1', '1', '1', '1', '3', '1', '2', '2', '3', '3', '1', '3', '3', '3', '1', '1', '1', '1', '6', '2', '2', '2', '4', '4', '1', '2', '4', '1', '3', '2', '1', '2', '2', '1', '2', '1', '1', '1', '3', '1', '1', '3', '3', '1', '2', '2', '1', '1', '2', '1', '2', '2', '2', '1', '1', '3', '2', '3', '2', '2', '1', '3', '1', '1', '1', '1', '2', '3', '2', '1', '2', '1', '2', '3', '2', '2', '1', '2', '1', '2', '2', '3', '1', '1', '2', '1', '2', '3', '2', '1', '3', '1', '1', '1', '7', '1', '2', '2', '4', '3', '1', '1', '3', '1', '3', '1', '1', '1', '3', '2', '1', '2', '1', '1', '1', '1', '1', '1', '1', '7', '3', '3', '1', '1', '2', '1', '2', '1', '4', '3', '5', '5', '1', '1', '5', '1', '3', '2', '2', '5', '1', '2', '4', '3', '1', '3', '2', '2', '2', '1', '2', '1', '3', '2', '1', '2', '3', '1', '1', '1', '1', '1', '2', '1', '2', '3', '1', '2', '1', '1', '3', '1', '2', '1', '5', '1', '2', '2', '1', '1', '1', '1', '1', '1', '3', '2', '1', '2', '2', '3', '2', '1', '1', '2', '1', '1', '2', '2', '1', '1', '1', '3', '1', '2', '3', '1', '3', '1', '2', '2', '2', '1', '1', '2', '1', '1', '3', '1', '3', '1', '1', '2', '2', '1', '3', '1', '3', '7', '3', '1', '2', '2', '3', '2', '1', '3', '1', '2', '1', '1', '1', '1', '2', '3', '2', '1', '3', '2', '7', '1', '1', '1', '1', '3', '2', '1', '3', '1', '2', '2', '3', '2', '2', '1', '3', '1', '1', '2', '3', '1', '1', '1', '3', '1', '5', '1', '3', '2', '1', '1', '2', '2', '1', '4', '2', '2', '1', '3', '3', '1', '2', '1', '2', '3', '2', '3', '1', '1', '2', '1', '2', '1', '2', '3', '1', '5', '4', '4', '2', '2', '3', '5', '1', '1', '4', '3', '1', '1', '1', '1', '1', '1', '2', '6', '3', '2', '2', '1', '1', '2', '1', '2', '1', '1', '1', '1', '3', '5', '3', '7', '1', '2', '1', '1', '3', '2', '2', '1', '1', '2', '2', '1', '3', '2', '2', '1', '2', '1', '1', '3', '7', '3', '3', '1', '1', '3', '1', '2', '1', '2', '2', '3', '2', '1', '1', '3', '1', '3', '2', '1', '1', '1', '1', '2', '1', '3', '1', '3', '3', '3', '2', '2', '3', '2', '2', '1', '2', '1', '1', '2', '3', '3', '1', '1', '3', '1', '1', '3', '1', '1', '1', '1', '2', '2', '2', '1', '3', '1', '3', '2', '2', '1', '3', '2', '1', '1', '1', '3', '1', '1', '2', '1', '1', '1', '3', '2', '1', '1', '2', '2', '3', '1', '2', '1', '1', '3', '1', '2', '3', '4', '2', '1', '2', '3', '3', '2', '2', '1', '2', '7', '1', '1', '5', '1', '1', '1', '2', '3', '1', '1', '3', '5', '1', '2', '4', '1', '4', '5', '1', '5', '5', '5', '1', '1', '4', '5', '5', '5', '3', '2', '1', '1', '3', '1', '3', '2', '3', '1', '2', '3', '2', '3', '2', '2', '2', '2', '1', '1', '1', '2', '1', '1', '1', '5', '2', '2', '1', '1', '3', '1', '4', '3', '1', '3', '1', '1', '1', '1', '1', '1', '3', '3', '2', '1', '3', '2', '1', '2', '3', '2', '1', '3', '1', '7', '1', '2', '2', '2', '2', '1', '2', '3', '1', '5', '2', '3', '1', '1', '2', '2', '2', '3', '1', '4', '4', '1', '2', '1', '3', '3', '1', '3', '1', '2', '3', '2', '1', '1', '2', '3', '2', '5', '2', '1', '1', '1', '4', '2', '4', '6', '2', '4', '1', '3', '3', '2', '1', '2', '1', '7', '1', '1', '7', '1', '2', '1', '1', '1', '1', '2', '1', '1', '2', '2', '3', '3', '2', '1', '2', '3', '1', '2', '2', '7', '1', '1', '4', '1', '1', '2', '1', '3', '4', '1', '2', '1', '3', '3', '1', '2', '2', '1', '3', '3', '3', '2', '1', '1', '1', '1', '1', '2', '3', '1', '2', '1', '1', '1', '2', '1', '3', '3', '1', '2', '2', '1', '3', '1', '3', '3', '2', '1', '5', '1', '5', '2', '1', '1', '1', '1', '1', '2', '3', '1', '1', '2', '2', '3', '1', '2', '2', '3', '5', '2', '4', '1', '1', '2', '1', '1', '3', '2', '1', '1', '2', '3', '3', '3', '3', '3', '1', '3', '1', '1', '1', '3', '2', '1', '2', '3', '1', '2', '3', '1', '1', '3', '1', '1', '1', '3', '1', '2', '1', '1', '3', '3', '2', '4', '1', '1', '1', '1', '2', '3', '2', '1', '3', '3', '2', '2', '2', '2', '7', '3', '1', '3', '2', '1', '1', '3', '2', '1', '1', '3', '1', '3', '1', '2', '2', '3', '1', '3', '2', '1', '2', '4', '1', '5', '1', '2', '1', '7', '3', '1', '7', '2', '3', '1', '1', '2', '3', '1', '1', '1', '2', '1', '1', '3', '1', '1', '1', '3', '1', '2', '2', '1', '2', '2', '1', '1', '6', '1', '1', '1', '1', '2', '3', '1', '2', '1', '1', '1', '2', '2', '2', '2', '3', '5', '4', '3', '3', '1', '3', '1', '1', '1', '1', '2', '2', '2', '3', '1', '1', '1', '1', '2', '2', '2', '2', '2', '2', '3', '1', '1', '3', '2', '1', '1', '1', '1', '1', '2', '1', '1', '1', '3', '2', '2', '2', '1', '5', '1', '1', '3', '1', '1', '2', '1', '1', '3', '1', '1', '1', '3', '2', '1', '1', '1', '1', '1', '6', '1', '1', '1', '3', '3', '1', '4', '1', '2', '1', '1', '3', '1', '2', '2', '2', '3', '3', '2', '2', '2', '7', '1', '1', '1', '1', '1', '2', '1', '2', '2', '1', '3', '1', '3', '1', '6', '1', '3', '1', '3', '2', '1', '1', '1', '4', '4', '2', '1', '1', '1', '5', '1', '1', '2', '4', '5', '3', '3', '4', '1', '2', '1', '2', '2', '2', '1', '2', '1', '1', '1', '2', '5', '2', '2', '3', '1', '1', '1', '1', '1', '1', '1', '3', '3', '2', '1', '1', '3', '2', '2', '1', '3', '2', '1', '2', '2', '1', '1', '2', '1', '2', '2', '1', '7', '1', '1', '1', '1', '6', '1', '1', '2', '1', '2', '3', '1', '2', '4', '1', '3', '2', '1', '1', '1', '1', '2', '2', '1', '1', '2', '1', '3', '5', '2', '1', '1', '1', '4', '1', '2', '3', '2', '2', '4', '3', '3', '2', '3', '1', '2', '2', '1', '2', '1', '2', '1', '1', '1', '1', '1', '2', '3', '1', '2', '2', '2', '1', '3', '2', '6', '2', '3', '2', '1', '1', '1', '2', '1', '2', '1', '1', '1', '4', '1', '2', '3', '1', '1', '1', '1', '1', '1', '4', '1', '3', '5', '1', '2', '2', '2', '6', '4', '1', '2', '5', '5', '4', '5', '2', '2', '4', '1', '5', '1', '2', '1', '2', '1', '3', '2', '2', '1', '1', '2', '1', '1', '1', '1', '3', '2', '4', '1', '2', '1', '5', '1', '1', '1', '1', '1', '7', '5', '1', '2', '3', '1', '1', '3', '2', '1', '1', '1', '1', '3', '3', '1', '2', '2', '3', '3', '2', '3', '3', '1', '1', '1', '1', '1', '2', '1', '1', '1', '3', '1', '2', '1', '1', '3', '3', '3', '1', '1', '1', '2', '1', '3', '1', '1', '2', '2', '2', '1', '3', '1', '1', '1', '2', '3', '1', '1', '2', '1', '1', '1', '1', '3', '7', '2', '3', '2', '7', '1', '1', '1', '1', '2', '3', '1', '2', '1', '1', '2', '2', '1', '1', '3', '1', '1', '3', '1', '3', '3', '2', '1', '1', '1', '1', '1', '2', '3', '4', '1', '1', '1', '1', '7', '1', '1', '1', '4', '3', '1', '1', '1', '1', '2', '2', '1', '2', '1', '3', '2', '2', '1', '2', '3', '2', '2', '1', '1', '1', '3', '1', '1', '1', '5', '3', '2', '1', '3', '2', '2', '1', '1', '1', '4', '2', '3', '6', '1', '2', '5', '4', '1', '4', '2', '2', '3', '2', '2', '1', '1', '1', '1', '1', '1', '1', '3', '3', '2', '1', '2', '2', '3', '1', '1', '1', '2', '2', '1', '3', '4', '3', '1', '1', '1', '1', '1', '1', '2', '3', '1', '1', '1', '3', '1', '3', '2', '1', '3', '1', '1', '4', '3', '3', '1', '1', '3', '1', '3', '1', '1', '2', '2', '1', '2', '3', '1', '1', '1', '3', '2', '2', '2', '1', '3', '1', '1', '1', '2', '1', '2', '3', '1', '1', '1', '1', '3', '3', '3', '2', '2', '1', '1', '1', '2', '1', '2', '1', '3', '1', '2', '3', '1', '1', '1', '7', '3', '1', '3', '1', '1', '1', '1', '2', '3', '3', '3', '1', '1', '1', '2', '2', '1', '1', '1', '1', '3', '6', '1', '3', '2', '4', '5', '1', '2', '1', '2', '3', '1', '1', '2', '4', '1', '3', '3', '1', '1', '1', '1', '3', '2', '4', '2', '1', '1', '1', '7', '1', '1', '1', '2', '1', '1', '1', '1', '2', '3', '2', '1', '2', '4', '2', '1', '3', '2', '2', '2', '3', '1', '2', '2', '1', '5', '6', '1', '1', '1', '2', '1', '1', '2', '2', '3', '5', '2', '1', '2', '2', '3', '1', '3', '2', '1', '1', '3', '2', '1', '2', '1', '6', '1', '3', '1', '3', '1', '2', '2', '1', '1', '1', '2', '3', '1', '2', '2', '2', '1', '1', '2', '2', '1', '1', '4', '7', '4', '5', '1', '1', '2', '2', '1', '5', '1', '4', '3', '4', '5', '4', '4', '3', '5', '4', '4', '4', '3', '3', '4', '4', '5', '6', '3', '1', '5', '1', '3', '2', '1', '2', '2', '1', '2', '1', '1', '1', '2', '3', '3', '1', '1', '4', '1', '2', '2', '1', '1', '1', '1', '3', '1', '1', '6', '1', '3', '1', '1', '3', '3', '1', '4', '1', '3', '3', '1', '1', '1', '1', '2', '7', '1', '1', '1', '1', '3', '1', '3', '1', '1', '2', '1', '4', '1', '1', '2', '1', '2', '1', '2', '1', '5', '1', '1', '1', '1', '3', '4', '2', '3', '3', '5', '2', '1', '3', '2', '2', '1', '2', '1', '1', '1', '2', '1', '3', '5', '1', '1', '6', '2', '1', '2', '2', '3', '2', '4', '1', '5', '2', '1', '2', '2', '3', '4', '2', '5', '2', '3', '1', '1', '1', '2', '3', '1', '1', '2', '2', '2', '3', '1', '4', '3', '1', '2', '1', '1', '1', '2', '2', '1', '2', '2', '1', '1', '1', '2', '3', '3', '2', '1', '2', '1', '3', '7', '2', '5', '2', '2', '1', '3', '2', '1', '1', '1', '1', '2', '1', '2', '3', '1', '1', '2', '2', '1', '2', '3', '1', '1', '3', '1', '1', '2', '2', '1', '2', '2', '4', '2', '3', '3', '3', '4', '4', '1', '2', '4', '1', '2', '1', '1', '3', '5', '5', '2', '1', '4', '3', '3', '1', '3', '3', '1', '2', '1', '2', '1', '3', '1', '2', '3', '1', '1', '1', '1', '3', '1', '2', '1', '3', '1', '1', '2', '3', '2', '1', '1', '2', '1', '1', '1', '3', '2', '1', '1', '1', '1', '2', '3', '2', '2', '1', '1', '1', '1', '2', '1', '2', '7', '1', '1', '1', '4', '1', '1', '3', '1', '2', '2', '2', '7', '1', '1', '2', '1', '1', '3', '1', '2', '3', '1', '1', '1', '1', '1', '1', '1', '2', '1', '2', '5', '4', '2', '2', '1', '1', '2', '2', '2', '4', '3', '1', '3', '1', '3', '1', '1', '3', '4', '3', '4', '1', '1', '2', '1', '1', '3', '2', '1', '3', '2', '2', '1', '3', '2', '1', '3', '1', '3', '1', '1', '1', '3', '1', '1', '2', '3', '3', '1', '1', '1', '3', '3', '2', '1', '2', '2', '1', '2', '3', '2', '1', '1', '2', '1', '2', '1', '3', '1', '3', '3', '2', '1', '1', '3', '1', '1', '3', '1', '2', '1', '1', '2', '2', '1', '1', '2', '1', '1', '1', '2', '1', '3', '1', '1', '1', '1', '1', '1', '1', '2', '1', '5', '1', '2', '3', '1', '3', '1', '7', '1', '1', '1', '1', '2', '1', '1', '2', '3', '1', '2', '1', '3', '1', '2', '2', '1', '1', '7', '3', '3', '1', '1', '3', '2', '1', '3', '2', '2', '1', '1', '2', '1', '1', '1', '1', '3', '3', '3', '2', '1', '2', '1', '1', '3', '1', '1', '2', '2', '1', '2', '1', '2', '1', '4', '1', '7', '3', '1', '1', '1', '1', '1', '1', '2', '1', '1', '4', '2', '2', '1', '6', '2', '1', '3', '1', '2', '4', '2', '2', '1', '2', '1', '2', '3', '2', '3', '5', '1', '4', '1', '2', '1', '1', '2', '1', '3', '1', '4', '1', '1', '3', '2', '2', '3', '2', '2', '2', '3', '2', '3', '3', '2', '1', '1', '1', '1', '1', '2', '1', '7', '1', '1', '1', '2', '3', '2', '3', '1', '1', '2', '1', '1', '1', '1', '1', '1', '4', '1', '2', '1', '1', '2', '2', '1', '3', '1', '3', '1', '1', '2', '2', '4', '2', '7', '1', '1', '3', '3', '2', '1', '1', '1', '3', '1', '1', '3', '2', '1', '3', '1', '1', '2', '5', '3', '2', '3', '1', '2', '1', '2', '3', '2', '1', '1', '1', '4', '3', '2', '1', '1', '1', '3', '1', '2', '1', '2', '3', '1', '2', '1', '1', '5', '2', '2', '2', '1', '5', '2', '4', '2', '4', '4', '5', '4', '1', '6', '2', '1', '1', '7', '3', '1', '1', '1', '3', '1', '3', '2', '2', '1', '3', '1', '5', '4', '3', '3', '1', '1', '1', '2', '2', '2', '1', '2', '1', '1', '2', '3', '1', '3', '1', '2', '5', '2', '3', '2', '1', '1', '2', '1', '1', '2', '2', '2', '2', '5', '1', '1', '3', '3', '3', '5', '2', '2', '1', '1', '3', '1', '1', '1', '3', '1', '1', '1', '2', '3', '1', '3', '1', '2', '1', '2', '2', '1', '3', '1', '3', '3', '7', '1', '2', '3', '4', '3', '2', '2', '1', '2', '1', '1', '2', '2', '2', '2', '7', '1', '2', '1', '2', '1', '2', '1', '2', '1', '1', '3', '2', '3', '1', '3', '3', '1', '4', '2', '2', '4', '5', '1', '1', '2', '1', '1', '4', '3', '5', '1', '1', '1', '4', '5', '3', '2', '3', '2', '5', '5', '1', '1', '5', '1', '2', '2', '1', '1', '1', '1', '1', '2', '3', '3', '3', '3', '2', '2', '1', '1', '1', '2', '1', '1', '7', '2', '3', '1', '3', '3', '2', '3', '2', '2', '2', '2', '1', '2', '1', '3', '1', '2', '2', '3', '1', '1', '3', '1', '1', '2', '2', '1', '1', '1', '1', '4', '7', '6', '1', '1', '1', '1', '1', '2', '1', '1', '2', '2', '1', '3', '1', '1', '3', '1', '1', '3', '1', '2', '1', '1', '1', '3', '5', '1', '1', '1', '4', '3', '1', '3', '1', '1', '2', '1', '2', '3', '1', '3', '4', '5', '2', '1', '4', '4', '3', '2', '6', '5', '1', '2', '2', '1', '4', '4', '2', '1', '4', '1', '2', '2', '2', '1', '1', '1', '3', '1', '3', '5', '3', '3', '2', '1', '1', '1', '2', '3', '3', '1', '1', '1', '2', '1', '4', '1', '3', '1', '7', '1', '2', '1', '3', '1', '3', '1', '1', '3', '2', '4', '1', '1', '2', '1', '3', '1', '1', '3', '1', '5', '3', '7', '3', '2', '2', '1', '1', '1', '3', '1', '3', '4', '1', '1', '1', '1', '1', '2', '1', '2', '7', '2', '1', '3', '1', '1', '1', '1', '2', '1', '1', '2', '2', '1', '3', '2', '2', '3', '2', '5', '4', '6', '1', '2', '3', '4', '4', '5', '5', '1', '5', '6', '4', '1', '5', '1', '3', '1', '2', '1', '1', '5', '3', '2', '2', '3', '3', '2', '2', '3', '1', '1', '1', '1', '2', '3', '2', '6', '1', '2', '2', '2', '1', '1', '2', '3', '1', '1', '1', '1', '3', '2', '2', '1', '1', '2', '1', '3', '1', '2', '4', '1', '1', '2', '1', '2', '1', '1', '1', '1', '1', '2', '3', '1', '1', '1', '1', '1', '1', '1', '3', '2', '2', '1', '7', '3', '7', '1', '1', '1', '3', '1', '3', '1', '1', '2', '2', '2', '1', '1', '1', '2', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '3', '3', '1', '4', '2', '1', '1', '1', '4', '1', '2', '1', '3', '1', '1', '1', '3', '6', '5', '4', '4', '2', '2', '2', '2', '4', '3', '6', '6', '3', '1', '2', '5', '1', '2', '1', '1', '4', '1', '1', '1', '1', '1', '3', '2', '2', '2', '1', '1', '3', '1', '3', '2', '1', '3', '2', '1', '3', '5', '1', '1', '3', '3', '1', '3', '3', '2', '1', '1', '1', '1', '1', '1', '1', '1', '3', '2', '1', '1', '2', '1', '1', '2', '2', '1', '1', '1', '1', '2', '7', '1', '3', '1', '1', '3', '3', '1', '1', '1', '1', '1', '2', '1', '1', '3', '1', '1', '3', '1', '3', '1', '3', '1', '2', '2', '1', '1', '2', '1', '1', '3', '2', '1', '3', '3', '1', '4', '1', '1', '1', '1', '7', '1', '2', '1', '2', '1', '2', '3', '2', '2', '1', '1', '1', '1', '1', '5', '1', '1', '2', '1', '4', '2', '2', '2', '1', '2', '1', '6', '5', '4', '2', '6', '6', '1', '1', '4', '3', '2', '1', '1', '1', '4', '1', '2', '1', '1', '1', '1', '3', '1', '2', '1', '1', '1', '2', '1', '2', '1', '1', '1', '1', '2', '1', '6', '1', '1', '1', '1', '2', '1', '3', '3', '2', '2', '1', '2', '3', '1', '1', '1', '1', '1', '1', '1', '1', '2', '1', '5', '2', '2', '3', '1', '1', '1', '1', '1', '1', '1', '3', '2', '2', '1', '7', '1', '2', '7', '3', '1', '3', '1', '1', '2', '1', '1', '3', '1', '1', '2', '7', '3', '3', '2', '3', '1', '1', '3', '1', '3', '2', '4', '3', '1', '1', '1', '2', '2', '3', '1', '3', '1', '1', '2', '3', '1', '2', '2', '2', '3', '1', '1', '3', '1', '2', '3', '2', '1', '2', '3', '2', '1', '3', '1', '2', '2', '1', '1', '1', '1', '1', '1', '3', '2', '1', '3', '1', '3', '3', '3', '2', '5', '4', '2', '2', '5', '1', '6', '6', '3', '4', '4', '1', '1', '3', '2', '4', '6', '3', '4', '5', '1', '5', '5', '1', '1', '1', '1', '2', '1', '2', '1', '4', '1', '4', '2', '2', '3', '2', '2', '1', '1', '2', '1', '3', '3', '1', '2', '2', '1', '1', '2', '1', '1', '1', '2', '1', '1', '1', '3', '2', '3', '2', '1', '3', '3', '3', '2', '2', '1', '3', '3', '2', '1', '1', '1', '1', '3', '2', '7', '2', '1', '1', '2', '1', '2', '1', '1', '1', '3', '3', '1', '1', '1', '1', '3', '2', '2', '2', '3', '5', '3', '2', '2', '2', '2', '1', '1', '1', '2', '1', '1', '1', '4', '1', '2', '3', '2', '2', '1', '2', '1', '1', '2', '1', '2', '1', '2', '1', '7', '1', '3', '1', '3', '2', '1', '3', '1', '3', '2', '1', '2', '1', '2', '5', '1', '2', '1', '1', '2', '1', '6', '1', '2', '4', '4', '2', '1', '3', '2', '2', '1', '1', '1', '1', '1', '1', '1', '1', '1', '7', '3', '1', '1', '1', '1', '1', '1', '2', '3', '3', '2', '3', '2', '1', '3', '3', '2', '2', '1', '2', '3', '1', '2', '3', '1', '1', '3', '2', '7', '2', '1', '3', '1', '1', '3', '2', '1', '4', '1', '1', '2', '1', '2', '3', '3', '1', '2', '1', '1', '2', '3', '2', '1', '1', '2', '3', '1', '1', '3', '1', '3', '1', '3', '1', '7', '1', '1', '2', '2', '2', '3', '1', '1', '1', '1', '1', '1', '2', '3', '1', '2', '2', '1', '2', '1', '5', '1', '2', '1', '3', '3', '1', '5', '3', '6', '2', '3', '1', '7', '1', '1', '1', '1', '3', '2', '1', '3', '2', '1', '1', '2', '1', '2', '1', '5', '1', '3', '4', '2', '2', '1', '1', '3', '1', '3', '2', '5', '2', '3', '2', '1', '3', '3', '1', '2', '3', '3', '5', '1', '1', '2', '5', '2', '2', '3', '1', '2', '1', '2', '1', '7', '1', '2', '7', '2', '1', '3', '1', '1', '2', '2', '2', '1', '1', '6', '3', '1', '1', '1', '2', '7', '3', '1', '1', '1', '1', '2', '1', '1', '2', '1', '3', '3', '1', '2', '1', '3', '1', '1', '1', '2', '1', '2', '1', '4', '1', '1', '1', '2', '3', '4', '5', '3', '1', '1', '1', '2', '2', '2', '1', '1', '2', '4', '2', '2', '2', '3', '1', '1', '5', '1', '2', '4', '2', '3', '1', '3', '2', '3', '3', '1', '1', '1', '1', '1', '1', '3', '1', '3', '3', '1', '3', '1', '3', '3', '1', '2', '1', '1', '1', '3', '1', '3', '1', '1', '1', '3', '1', '2', '1', '2', '1', '1', '1', '1', '3', '3', '1', '1', '2', '1', '3', '1', '3', '1', '2', '3', '1', '1', '2', '2', '1', '1', '2', '1', '2', '1', '3', '3', '1', '1', '1', '1', '1', '1', '2', '2', '1', '1', '1', '7', '2', '2', '2', '3', '1', '3', '3', '1', '3', '1', '1', '3', '3', '4', '2', '1', '6', '4', '1', '4', '2', '5', '1', '2', '3', '2', '1', '4', '1', '1', '4', '5', '3', '4', '2', '1', '1', '1', '3', '2', '1', '1', '1', '3', '1', '2', '1', '1', '1', '1', '1', '2', '6', '1', '2', '7', '1', '2', '3', '2', '2', '2', '1', '2', '1', '1', '1', '3', '1', '1', '1', '1', '1', '3', '1', '3', '1', '2', '2', '3', '2', '1', '3', '1', '2', '1', '2', '3', '1', '2', '1', '1', '1', '3', '1', '3', '2', '1', '1', '1', '2', '2', '3', '2', '2', '2', '2', '3', '3', '1', '7', '4', '7', '1', '1', '2', '1', '1', '1', '1', '1', '4', '7', '2', '1', '5', '1', '1', '5', '2', '2', '2', '3', '4', '2', '2', '5', '2', '1', '1', '2', '1', '4', '3', '1', '4', '6', '4', '2', '1', '2', '5', '6', '3', '1', '2', '1', '4', '6', '2', '2', '1', '3', '5', '1', '1', '1', '1', '2', '2', '1', '1', '1', '1', '3', '1', '2', '2', '3', '1', '1', '3', '3', '1', '2', '1', '3', '1', '3', '1', '2', '1', '1', '3', '2', '2', '2', '1', '3', '5', '3', '1', '3', '1', '3', '1', '2', '2', '3', '1', '2', '3', '1', '1', '1', '4', '1', '3', '1', '1', '2', '1', '2', '1', '2', '2', '1', '1', '3', '5', '1', '1', '1', '4', '3', '2', '6', '3', '7', '1', '1', '2', '2', '1', '1', '1', '1', '2', '3', '3', '1', '1', '1', '1', '3', '3', '1', '4', '2', '2', '2', '3', '1', '1', '3', '3', '2', '1', '4', '2', '1', '2', '1', '1', '1', '1', '1', '1', '2', '4', '2', '2', '2', '2', '6', '6', '1', '1', '1', '5', '2', '1', '5', '2', '1', '2', '3', '4', '1', '4', '1', '1', '3', '2', '1', '3', '1', '1', '2', '2', '1', '3', '1', '2', '1', '2', '7', '3', '2', '2', '1', '1', '2', '1', '2', '5', '1', '1', '1', '1', '2', '1', '1', '3', '1', '1', '1', '7', '1', '1', '2', '2', '1', '2', '3', '1', '1', '2', '1', '3', '1', '1', '1', '1', '7', '2', '2', '2', '1', '3', '3', '2', '1', '2', '5', '3', '1', '1', '1', '1', '2', '3', '2', '2', '2', '3', '1', '2', '1', '1', '3', '1', '1', '3', '1', '1', '1', '1', '1', '1', '1', '1', '2', '2', '2', '1', '1', '2', '1', '2', '2', '2', '4', '1', '2', '3', '1', '1', '3', '1', '3', '1', '2', '2', '2', '3', '5', '3', '2', '4', '1', '1', '1', '3', '1', '5', '1', '2', '3', '2', '2', '3', '1', '2', '1', '2', '1', '1', '1', '2', '3', '1', '2', '2', '1', '2', '3', '2', '6', '2', '5', '1', '2', '3', '3', '2', '1', '4', '6', '6', '3', '5', '4', '2', '1', '2', '1', '3', '4', '1', '1', '1', '7', '2', '1', '2', '2', '1', '2', '1', '3', '1', '2', '3', '1', '1', '3', '1', '2', '1', '2', '1', '1', '3', '3', '1', '1', '2', '2', '1', '3', '3', '1', '1', '1', '3', '1', '1', '5', '3', '2', '1', '2', '2', '1', '1', '1', '2', '5', '1', '1', '1', '1', '1', '5', '2', '5', '2', '2', '3', '1', '1', '1', '1', '1', '3', '1', '2', '1', '3', '1', '1', '2', '1', '3', '3', '1', '1', '2', '2', '1', '7', '1', '1', '2', '1', '1', '2', '2', '2', '1', '1', '2', '2', '2', '1', '2', '1', '1', '1', '1', '2', '4', '4', '4', '1', '3', '2', '2', '1', '3', '5', '3', '2', '1', '1', '3', '4', '4', '1', '3', '1', '2', '3', '4', '2', '2', '2', '1', '1', '1', '3', '3', '1', '3', '1', '1', '3', '1', '1', '1', '2', '1', '1', '3', '2', '3', '1', '1', '1', '1', '1', '3', '1', '1', '2', '1', '1', '3', '1', '1', '2', '1', '3', '1', '1', '1', '1', '4', '2', '1', '4', '1', '1', '1', '1', '1', '1', '1', '1', '2', '1', '6', '1', '2', '6', '2', '1', '1', '1', '2', '1', '1', '1', '2', '1', '2', '1', '1', '1', '1', '2', '5', '1', '1', '2', '1', '3', '7', '2', '1', '2', '1', '1', '2', '2', '1', '3', '3', '2', '1', '1', '1', '3', '3', '1', '3', '1', '1', '2', '1', '3', '1', '1', '3', '1', '1', '3', '4', '4', '3', '4', '2', '2', '4', '1', '5', '4', '5', '1', '4', '6', '4', '1', '1', '2', '1', '1', '3', '2', '3', '3', '3', '2', '1', '1', '1', '1', '1', '2', '1', '1', '2', '3', '1', '3', '2', '3', '1', '5', '2', '3', '5', '3', '4', '1', '1', '1', '2', '2', '2', '2', '3', '1', '2', '1', '1', '3', '1', '1', '1', '1', '1', '5', '3', '3', '1', '1', '2', '1', '1', '3', '4', '1', '1', '2', '5', '1', '3', '1', '1', '1', '3', '1', '3', '1', '2', '2', '1', '3', '3', '7', '1', '1', '2', '3', '3', '1', '2', '1', '2', '2', '4', '3', '1', '5', '2', '5', '5', '1', '4', '5', '4', '1', '2', '2', '1', '1', '1', '1', '2', '1', '2', '1', '1', '1', '1', '1', '2', '1', '1', '5', '4', '1', '2', '3', '1', '2', '2', '1', '1', '2', '5', '1', '1', '1', '2', '2', '3', '2', '1', '2', '3', '2', '3', '1', '4', '1', '1', '2', '2', '3', '1', '2', '1', '4', '7', '2', '3', '2', '2', '1', '1', '4', '7', '1', '3', '1', '3', '3', '1', '3', '2', '3', '1', '1', '1', '3', '1', '1', '3', '1', '3', '5', '1', '1', '3', '1', '1', '1', '1', '3', '2', '1', '7', '2', '3', '2', '5', '4', '1', '2', '2', '5', '3', '1', '4', '4', '4', '1', '1', '3', '3', '1', '2', '1', '6', '1', '1', '1', '3', '5', '3', '3', '1', '4', '2', '2', '2', '2', '1', '3', '1', '2', '1', '1', '3', '2', '1', '3', '3', '1', '2', '1', '1', '1', '1', '1', '7', '3', '4', '1', '2', '2', '2', '1', '2', '1', '4', '1', '1', '1', '2', '2', '2', '1', '1', '1', '1', '4', '3', '1', '3', '3', '2', '1', '1', '1', '2', '1', '2', '1', '4', '2', '2', '3', '1', '3', '2', '2', '2', '1', '1', '2', '5', '2', '1', '2', '3', '1', '2', '1', '2', '1', '1', '1', '1', '3', '1', '3', '1', '2', '7', '1', '3', '1', '1', '1', '1', '1', '4', '1', '2', '1', '1', '1', '3', '1', '1', '3', '6', '1', '3', '2', '3', '2', '1', '2', '1', '2', '1', '1', '1', '1', '1', '1', '7', '1', '3', '7', '1', '1', '4', '3', '2', '1', '1', '7', '3', '1', '3', '1', '2', '2', '2', '2', '3', '1', '4', '2', '1', '4', '2', '3', '1', '1', '3', '1', '1', '3', '1', '3', '3', '1', '1', '6', '1', '1', '6', '4', '4', '1', '1', '2', '4', '1', '4', '6', '1', '6', '1', '5', '1', '2', '2', '1', '1', '1', '1', '2', '1', '1', '3', '3', '2', '5', '1', '1', '2', '3', '1', '1', '1', '3', '1', '1', '2', '2', '2', '2', '5', '2', '1', '2', '1', '1', '2', '1', '3', '2', '3', '1', '1', '1', '3', '1', '1', '2', '3', '1', '3', '2', '2', '2', '2', '1', '1', '2', '2', '2', '1', '1', '4', '4', '1', '1', '1', '1', '3', '1', '1', '3', '1', '1', '2', '5', '2', '1', '2', '1', '2', '1', '1', '2', '7', '3', '2', '2', '2', '2', '1', '1', '2', '1', '2', '2', '1', '3', '3', '1', '1', '2', '2', '2', '1', '3', '1', '1', '1', '2', '2', '4', '1', '2', '1', '3', '3', '1', '2', '3', '3', '1', '2', '3', '1', '2', '3', '1', '2', '2', '2', '3', '4', '2', '2', '5', '2', '2', '6', '5', '5', '3', '5', '2', '1', '4', '1', '1', '1', '1', '2', '1', '1', '1', '1', '1', '1', '1', '1', '1', '3', '1', '2', '1', '3', '1', '1', '2', '2', '5', '3', '2', '2', '1', '1', '1', '1', '1', '4', '1', '1', '3', '2', '2', '5', '1', '2', '1', '1', '2', '2', '3', '1', '1', '7', '1', '1', '1', '1', '1', '2', '1', '3', '2', '1', '1', '1', '2', '6', '3', '5', '2', '1', '1', '3', '1', '3', '2', '7', '1', '1', '5', '2', '1', '2', '1', '1', '1', '6', '1', '1', '1', '1', '3', '2', '1', '2', '1', '1', '1', '1', '3', '2', '1', '2', '1', '1', '1', '2', '5', '3', '1', '3', '3', '1', '4', '4', '2', '1', '3', '3', '1', '1', '1', '1', '2', '2', '2', '3', '1', '7', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '3', '1', '4', '1', '1', '3', '1', '2', '1', '1', '3', '1', '1', '2', '1', '1', '1', '2', '2', '1', '2', '2', '2', '2', '1', '1', '1', '1', '1', '3', '1', '5', '3', '1', '1', '3', '2', '1', '1', '2', '1', '3', '4', '2', '1', '1', '1', '2', '3', '7', '1', '1', '1', '1', '3', '1', '3', '1', '1', '2', '3', '4', '1', '1', '1', '2', '1', '7', '3', '1', '2', '3', '3', '1', '1', '3', '3', '2', '1', '1', '1', '3', '1', '2', '5', '5', '2', '4', '4', '2', '5', '4', '4', '6', '5', '1', '1', '1', '2', '1', '4', '4', '2', '2', '3', '1', '1', '1', '1', '2', '3', '1', '1', '2', '3', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '1', '2', '3', '1', '1', '1', '2', '1', '1', '2', '1', '2', '3', '3', '2', '1', '1', '2', '3', '2', '1', '7', '3', '1', '7', '1', '3', '4', '2', '1', '3', '1', '3', '1', '2', '2', '2', '4', '1', '1', '1', '1', '7', '3', '1', '3', '3', '1', '1', '1', '1', '1', '3', '2', '1', '1', '2', '1', '2', '1', '1', '1', '1', '1', '1', '1', '3', '5', '1', '2', '7', '2', '3', '3', '3', '2', '1', '3', '2', '5', '1', '2', '3', '6', '5', '1', '1', '1', '3', '6', '2', '2', '2', '1', '1', '2', '4', '3', '7', '1', '1', '3', '2', '1', '1', '1', '7', '1', '3', '2', '1', '1', '2', '1', '2', '2', '1', '2', '1', '1', '2', '6', '1', '1', '2', '1', '1', '1', '1', '1', '1', '3', '5', '1', '1', '2', '1', '7', '1', '3', '2', '3', '1', '1', '1', '3', '7', '3', '1', '1', '1', '1', '2', '1', '3', '2', '1', '1', '1', '1', '3', '1', '2', '3', '2', '2', '4', '3', '2', '2', '4', '3', '2', '6', '4', '4', '2', '4', '2', '2', '1', '1', '3', '3', '5', '6', '2', '1', '2', '6', '1', '5', '3', '7', '1', '4', '1', '1', '1', '1', '1', '1', '2', '3', '1', '1', '1', '5', '1', '2', '3', '1', '3', '3', '1', '1', '4', '1', '1', '2', '3', '1', '4', '1', '1', '1', '5', '1', '1', '2', '1', '7', '1', '2', '2', '1', '5', '1', '1', '2', '1', '2', '2', '1', '1', '1', '2', '1', '2', '2', '1', '1', '1', '1', '3', '1', '3', '1', '3', '1', '2', '1', '1', '1', '3', '1', '3', '3', '4', '1', '4', '2', '4', '2', '3', '4', '1', '1', '2', '5', '3', '1', '2', '1', '1', '1', '3', '3', '3', '7', '1', '2', '4', '1', '3', '1', '1', '2', '1', '1', '3', '1', '1', '1', '2', '1', '2', '1', '2', '3', '1', '5', '1', '1', '1', '1', '3', '2', '1', '1', '3', '1', '1', '3', '2', '1', '2', '2', '1', '1', '3', '2', '2', '2', '2', '2', '3', '2', '2', '1', '1', '1', '4', '1', '2', '4', '3', '4', '2', '2', '3', '2', '3', '1', '2', '1', '2', '1', '1', '5', '1', '1', '1', '1', '1', '1', '1', '2', '1', '2', '7', '1', '1', '2', '1', '3', '2', '2', '1', '3', '1', '1', '4', '3', '1', '2', '2', '4', '3', '1', '2', '3', '1', '3', '2', '1', '2', '2', '1', '2', '2', '2', '2', '1', '1', '1', '1', '1', '2', '1', '3', '1', '1', '1', '2', '1', '1', '2', '2', '3', '3', '3', '2', '1', '3', '3', '1', '2', '1', '2', '1', '2', '1', '3', '3', '3', '2', '2', '1', '2', '1', '1', '1', '1', '1', '1', '2', '2', '1', '1', '3', '2', '2', '1', '1', '2', '2', '2', '7', '1', '3', '3', '1', '3', '1', '2', '6', '2', '2', '3', '1', '2', '1', '1', '2', '4', '3', '1', '2', '1', '2', '1', '6', '4', '3', '4', '2', '3', '4', '3', '3', '1', '3', '3', '1', '1', '7', '1', '3', '3', '1', '1', '2', '1', '1', '2', '5', '3', '1', '1', '1', '3', '2', '2', '2', '1', '5', '2', '2', '1', '1', '3', '1', '1', '1', '1', '2', '1', '1', '3', '2', '1', '3', '2', '1', '1', '1', '3', '3', '3', '1', '2', '1', '1', '1', '3', '1', '3', '1', '3', '1', '1', '1', '1', '3', '3', '3', '1', '3', '1', '1', '1', '4', '1', '1', '2', '2', '3', '3', '1', '1', '1', '3', '1', '1', '7', '1', '1', '1', '2', '2', '1', '2', '1', '2', '1', '3', '1', '1', '4', '1', '2', '2', '2', '1', '5', '3', '3', '2', '5', '1', '2', '3', '1', '5', '2', '2', '3', '2', '1', '1', '1', '1', '3', '3', '1', '1', '1', '3', '3', '1', '1', '5', '3', '3', '2', '1', '3', '1', '1', '1', '7', '2', '1', '2', '2', '1', '2', '1', '3', '1', '1', '3', '1', '2', '2', '3', '1', '1', '7', '1', '1', '1', '1', '1', '1', '7', '3', '1', '3', '2', '1', '1', '1', '2', '1', '3', '1', '2', '7', '1', '1', '3', '2', '1', '1', '1', '2', '1', '1', '1', '1', '1', '1', '4', '2', '1', '2', '2', '1', '2', '1', '1', '2', '2', '3', '3', '2', '1', '2', '1', '5', '2', '1', '3', '5', '1', '5', '2', '1', '1', '3', '3', '4', '2', '1', '4', '5', '1', '1', '1', '1', '1', '2', '7', '3', '1', '3', '1', '3', '3', '1', '3', '3', '2', '2', '7', '3', '1', '1', '1', '1', '2', '3', '3', '2', '1', '1', '3', '4', '1', '1', '3', '1', '1', '2', '1', '3', '1', '1', '1', '2', '1', '2', '2', '3', '7', '5', '1', '3', '4', '1', '1', '5', '1', '1', '2', '2', '2', '1', '2', '3', '4', '2', '1', '3', '1', '1', '1', '2', '4', '5', '2', '1', '1', '5', '4', '2', '2', '3', '1', '1', '1', '1', '3', '2', '4', '4', '2', '3', '3', '2', '1', '2', '4', '1', '1', '1', '2', '1', '2', '3', '7', '1', '1', '2', '1', '1', '2', '2', '2', '1', '3', '1', '2', '1', '1', '2', '1', '1', '2', '3', '1', '1', '2', '2', '3', '2', '1', '2', '3', '1', '3', '2', '1', '1', '5', '1', '2', '1', '1', '2', '2', '3', '1', '5', '1', '1', '3', '1', '1', '1', '1', '3', '1', '1', '6', '1', '1', '1', '1', '1', '2', '2', '2', '2', '1', '1', '1', '2', '2', '1', '1', '1', '4', '1', '2', '1', '1', '3', '1', '2', '2', '2', '1', '4', '1', '1', '2', '3', '1', '3', '3', '4', '3', '4', '2', '4', '4', '4', '1', '1', '5', '1', '1', '4', '2', '1', '1', '6', '4', '1', '7', '1', '1', '1', '2', '2', '1', '1', '1', '1', '2', '1', '1', '3', '1', '4', '1', '1', '1', '1', '1', '3', '3', '2', '1', '1', '1', '1', '4', '1', '3', '1', '1', '1', '1', '2', '1', '1', '1', '1', '1', '1', '1', '1', '1', '5', '2', '2', '3', '2', '2', '1', '4', '3', '4', '1', '1', '1', '3', '3', '2', '1', '3', '1', '7', '2', '2', '2', '2', '1', '1', '1', '5', '2', '1', '3', '4', '1', '4', '3', '6', '4', '3', '1', '1', '1', '1', '2', '3', '2', '1', '2', '3', '1', '1', '1', '1', '1', '2', '1', '1', '1', '1', '1', '1', '1', '1', '3', '3', '2', '2', '3', '3', '4', '2', '1', '3', '1', '1', '2', '3', '6', '2', '3', '1', '2', '1', '1', '3', '2', '1', '2', '2', '1', '1', '7', '1', '2', '3', '6', '2', '2', '2', '6', '1', '3', '1', '1', '1', '2', '2', '1', '3', '1', '2', '1', '7', '1', '3', '3', '4', '2', '2', '1', '3', '4', '2', '2', '6', '6', '1', '5', '4', '6', '2', '6', '1', '1', '1', '3', '2', '3', '1', '4', '7', '1', '2', '6', '1', '2', '1', '3', '1', '2', '3', '4', '2', '2', '2', '1', '2', '1', '1', '1', '7', '1', '2', '2', '1', '2', '2', '1', '1', '1', '1', '1', '2', '2', '2', '1', '3', '1', '1', '3', '2', '2', '3', '3', '1', '2', '1', '1', '7', '2', '1', '5', '3', '1', '2', '1', '1', '2', '1', '2', '1', '3', '1', '3', '4', '1', '1', '3', '2', '3', '2', '1', '2', '1', '3', '1', '1', '3', '2', '1', '1', '2', '1', '1', '4', '3', '7', '3', '2', '1', '2', '1', '1', '1', '5', '4', '1', '4', '4', '1', '1', '3', '1', '1', '3', '1', '1', '1', '2', '1', '1', '1', '2', '1', '3', '1', '2', '1', '1', '1', '3', '3', '1', '7', '4', '1', '3', '1', '1', '1', '1', '1', '3', '2', '1', '1', '2', '6', '1', '3', '1', '2', '4', '2', '1', '1', '1', '2', '1', '2', '2', '1', '2', '2', '3', '7', '1', '3', '3', '3', '2', '1', '2', '1', '1', '1', '3', '2', '2', '2', '1', '1', '1', '2', '2', '1', '1', '2', '1', '2', '1', '1', '1', '4', '1', '3', '2', '7', '1', '2', '4', '1', '3', '1', '2', '4', '2', '1', '2', '2', '1', '3', '1', '3', '2', '2', '2', '2', '1', '3', '2', '2', '4', '4', '1', '2', '2', '2', '5', '1', '2', '4', '1', '2', '3', '1', '3', '1', '4', '3', '1', '1', '3', '1', '1', '1', '1', '3', '1', '2', '2', '2', '1', '3', '3', '1', '2', '1', '2', '1', '5', '2', '1', '3', '4', '1', '5', '2', '1', '4', '1', '2', '1', '3', '5', '1', '1', '2', '2', '1', '7', '3', '2', '1', '1', '2', '1', '1', '3', '1', '1', '3', '2', '1', '3', '1', '3', '1', '2', '1', '2', '2', '1', '2', '2', '3', '1', '2', '5', '3', '1', '1', '1', '2', '2', '5', '1', '2', '1', '1', '3', '1', '2', '4', '3', '3', '2', '1', '3', '1', '2', '2', '1', '2', '1', '4', '2', '3', '1', '1', '1', '2', '2', '3', '1', '1', '2', '2', '3', '3', '1', '1', '1', '2', '2', '4', '2', '1', '1', '1', '1', '5', '3', '4', '2', '3', '2', '2', '2', '1', '1', '2', '1', '1', '1', '5', '3', '1', '4', '5', '3', '2', '2', '3', '7', '1', '1', '5', '1', '2', '1', '1', '1', '1', '3', '1', '3', '1', '2', '2', '3', '2', '1', '1', '1', '1', '1', '3', '1', '2', '2', '1', '2', '1', '1', '3', '3', '1', '2', '1', '2', '5', '3', '1', '1', '3', '3', '2', '2', '1', '3', '1', '1', '4', '7', '2', '2', '1', '1', '1', '2', '2', '1', '3', '1', '2', '1', '1', '1', '1', '2', '1', '3', '1', '3', '1', '3', '1', '2', '1', '4', '2', '4', '4', '3', '5', '4', '1', '4', '3', '3', '1', '1', '2', '1', '1', '5', '2', '1', '1', '1', '1', '1', '2', '2', '2', '7', '2', '1', '1', '3', '2', '2', '7', '1', '1', '1', '1', '1', '1', '1', '1', '2', '1', '2', '3', '2', '1', '2', '2', '1', '1', '2', '1', '2', '3', '2', '2', '1', '4', '4', '1', '1', '1', '1', '1', '1', '3', '2', '1', '1', '3', '1', '2', '1', '2', '2', '1', '1', '2', '3', '7', '5', '1', '4', '1', '1', '2', '2', '2', '1', '3', '2', '3', '2', '1', '2', '3', '1', '3', '2', '3', '5', '1', '4', '4', '2', '3', '1', '1', '1', '3', '1', '2', '1', '1', '4', '2', '7', '1', '1', '1', '1', '3', '1', '3', '1', '1', '1', '2', '2', '5', '3', '3', '2', '2', '1', '3', '2', '1', '4', '1', '3', '3', '5', '1', '2', '2', '1', '1', '3', '1', '3', '3', '1', '1', '1', '3', '7', '1', '1', '1', '2', '7', '1', '1', '2', '3', '1', '2', '4', '1', '3', '1', '2', '4', '2', '1', '2', '1', '7', '1', '2', '1', '1', '3', '2', '1', '1', '3', '3', '1', '1', '1', '3', '1', '1', '1', '1', '2', '2', '4', '2', '1', '3', '2', '3', '3', '5', '2', '5', '4', '5', '3', '2', '2', '5', '1', '2', '1', '2', '2', '1', '1', '1', '3', '1', '1', '2', '7', '2', '2', '2', '1', '1', '1', '2', '3', '2', '2', '2', '1', '3', '1', '4', '1', '2', '1', '1', '1', '1', '2', '2', '1', '1', '1', '2', '7', '2', '2', '2', '1', '1', '3', '6', '2', '1', '1', '1', '3', '4', '1', '2', '1', '2', '3', '1', '1', '3', '2', '3', '1', '1', '1', '2', '4', '1', '2', '2', '1', '1', '1', '3', '2', '1', '1', '1', '1', '1', '2', '1', '2', '3', '1', '1', '1', '3', '3', '2', '2', '2', '2', '1', '4', '1', '3', '4', '3', '4', '1', '1', '1', '1', '2', '1', '1', '1', '1', '2', '3', '1', '4', '2', '1', '2', '1', '5', '2', '1', '1', '1', '1', '1', '2', '1', '1', '3', '1', '1', '7', '4', '3', '1', '2', '7', '2', '1', '1', '3', '3', '1', '1', '2', '1', '1', '2', '1', '2', '1', '1', '2', '1', '1', '3', '2', '2', '2', '3', '1', '1', '1', '1', '1', '1', '2', '4', '1', '3', '1', '1', '2', '2', '1', '3', '1', '3', '2', '3', '3', '2', '1', '2', '2', '3', '2', '1', '1', '3', '1', '1', '2', '2', '1', '1', '1', '3', '5', '3', '1', '3', '1', '5', '3', '5', '2', '2', '3', '1', '2', '2', '1', '1', '1', '1', '2', '2', '1', '3', '1', '1', '1', '2', '1', '2', '1', '2', '1', '2', '3', '2', '2', '1', '2', '1', '2', '4', '2', '1', '1', '2', '1', '2', '1', '2', '3', '2', '1', '2', '1', '1', '1', '1', '7', '3', '3', '1', '1', '1', '2', '2', '1', '1', '3', '2', '1', '2', '1', '1', '2', '1', '1', '3', '1', '4', '1', '2', '3', '2', '2', '2', '1', '1', '2', '3', '3', '1', '2', '1', '3', '1', '1', '2', '1', '2', '1', '1', '2', '1', '2', '2', '2', '3', '1', '3', '1', '2', '4', '4', '6', '2', '4', '6', '3', '5', '4', '1', '1', '1', '3', '1', '1', '1', '2', '3', '1', '1', '1', '2', '3', '7', '1', '3', '3', '1', '2', '1', '3', '2', '1', '1', '1', '1', '2', '2', '1', '1', '1', '2', '3', '1', '2', '1', '1', '1', '2', '3', '1', '1', '5', '1', '3', '1', '7', '1', '7', '1', '1', '1', '1', '2', '3', '1', '1', '2', '4', '2', '2', '3', '1', '1', '1', '1', '2', '3', '7', '1', '1', '2', '1', '3', '2', '1', '2', '5', '3', '1', '3', '3', '2', '2', '3', '1', '1', '1', '2', '2', '1', '1', '1', '2', '1', '3', '1', '1', '1', '2', '2', '1', '1', '3', '2', '2', '3', '3', '3', '1', '3', '2', '5', '2', '5', '4', '1', '2', '1', '2', '2', '4', '3', '6', '1', '3', '2', '3', '1', '1', '1', '1', '1', '2', '1', '3', '1', '4', '1', '2', '1', '2', '1', '3', '1', '1', '1', '1', '1', '1', '1', '1', '1', '4', '1', '2', '4', '1', '1', '1', '1', '1', '1', '1', '2', '3', '1', '2', '2', '2', '1', '3', '1', '7', '3', '1', '1', '7', '1', '1', '2', '1', '1', '1', '7', '1', '1', '2', '1', '4', '1', '3', '1', '2', '1', '2', '1', '4', '1', '1', '3', '2', '1', '1', '1', '1', '4', '3', '1', '2', '2', '1', '1', '6', '1', '4', '5', '5', '4', '5', '4', '2', '3', '1', '2', '1', '2', '5', '3', '5', '2', '1', '1', '4', '1', '3', '3', '5', '2', '1', '1', '3', '1', '3', '3', '1', '1', '1', '4', '1', '1', '1', '2', '3', '2', '1', '1', '1', '1', '1', '1', '1', '1', '1', '3', '6', '3', '1', '1', '1', '1', '1', '3', '2', '1', '1', '1', '7', '3', '1', '2', '2', '1', '1', '3', '5', '2', '2', '7', '1', '1', '3', '2', '1', '1', '1', '2', '2', '2', '5', '2', '4', '1', '4', '2', '1', '3', '2', '2', '1', '5', '4', '4', '2', '3', '1', '1', '2', '5', '3', '3', '4', '1', '1', '1', '1', '1', '3', '1', '2', '2', '1', '2', '2', '2', '7', '3', '1', '1', '1', '3', '2', '1', '5', '1', '3', '2', '7', '3', '1', '4', '1', '1', '1', '1', '2', '3', '3', '1', '1', '1', '7', '3', '6', '1', '3', '2', '2', '2', '1', '1', '2', '2', '1', '1', '2', '1', '3', '6', '2', '1', '1', '3', '1', '1', '2', '6', '2', '2', '1', '1', '3', '6', '1', '1', '1', '2', '3', '1', '7', '2', '1', '2', '3', '1', '3', '1', '1', '3', '1', '2', '6', '2', '3', '3', '2', '1', '4', '3', '1', '2', '1', '1', '4', '7', '1', '1', '1', '4', '3', '1', '2', '2', '4', '5', '2', '1', '4', '1', '1', '2', '2', '3', '2', '1', '5', '2', '1', '3', '6', '1', '2', '1', '3', '2', '1', '1', '1', '1', '2', '2', '2', '2', '1', '1', '2', '1', '1', '1', '1', '1', '3', '4', '1', '1', '3', '1', '3', '2', '1', '2', '1', '3', '1', '1', '7', '2', '2', '1', '1', '2', '6', '2', '4', '3', '2', '1', '2', '1', '3', '2', '2', '2', '4', '1', '1', '2', '2', '1', '3', '1', '1', '1', '1', '1', '1', '1', '1', '3', '2', '3', '7', '1', '3', '1', '1', '3', '1', '4', '1', '1', '1', '2', '3', '1', '1', '2', '1', '1', '2', '1', '1', '5', '1', '4', '4', '6', '1', '2', '2', '3', '4', '1', '2', '1', '2', '1', '1', '3', '2', '1', '1', '5', '2', '4', '6', '4', '3', '1', '3', '1', '1', '1', '7', '2', '3', '1', '2', '2', '1', '2', '2', '1', '2', '1', '1', '3', '1', '1', '1', '3', '1', '2', '2', '2', '3', '1', '3', '2', '1', '2', '7', '4', '1', '1', '1', '1', '2', '3', '1', '2', '1', '4', '7', '3', '2', '2', '3', '4', '1', '4', '1', '2', '1', '1', '3', '2', '2', '1', '2', '2', '1', '3', '2', '1', '3', '6', '1', '1', '2', '1', '4', '1', '5', '1', '6', '2', '1', '3', '2', '2', '6', '1', '3', '1', '1', '2', '1', '3', '1', '1', '2', '1', '3', '4', '3', '3', '1', '1', '1', '7', '1', '1', '1', '2', '1', '1', '1', '1', '2', '5', '3', '1', '1', '1', '3', '7', '2', '1', '1', '1', '1', '1', '2', '7', '5', '1', '1', '1', '1', '3', '1', '2', '1', '1', '1', '1', '2', '2', '1', '4', '4', '2', '2', '2', '2', '3', '4', '2', '1', '2', '4', '4', '4', '2', '2', '5', '1', '5', '5', '6', '1', '1', '3', '4', '4', '1', '1', '1', '3', '3', '3', '2', '1', '3', '2', '1', '2', '2', '1', '2', '1', '3', '1', '3', '1', '2', '3', '1', '7', '3', '2', '1', '4', '2', '2', '1', '2', '6', '2', '2', '5', '4', '3', '1', '7', '1', '2', '2', '2', '1', '5', '7', '3', '1', '3', '1', '2', '1', '2', '7', '2', '2', '1', '2', '5', '2', '1', '3', '1', '4', '2', '2', '1', '3', '3', '1', '6', '1', '5', '2', '1', '2', '1', '1', '2', '2', '1', '3', '2', '1', '4', '2', '3', '2', '2', '1', '6', '1', '1', '1', '3', '1', '1', '4', '2', '2', '2', '3', '3', '5', '2', '1', '2', '1', '1', '1', '1', '1', '2', '2', '1', '2', '1', '2', '1', '2', '3', '1', '1', '3', '3', '1', '1', '7', '5', '2', '1', '2', '1', '4', '2', '2', '2', '6', '1', '3', '4', '2', '1', '3', '3', '4', '1', '2', '5', '1', '5', '2', '1', '5', '3', '3', '6', '1', '6', '5', '1', '1', '3', '1', '3', '1', '3', '1', '2', '7', '2', '1', '2', '2', '5', '1', '2', '2', '1', '5', '3', '2', '1', '1', '3', '2', '3', '2', '3', '1', '1', '1', '3', '1', '3', '2', '1', '2', '1', '7', '1', '3', '7', '2', '3', '1', '3', '2', '1', '1', '4', '4', '5', '1', '2', '2', '4', '1', '4', '1', '2', '1', '1', '1', '2', '2', '2', '5', '1', '1', '1', '2', '2', '3', '1', '2', '1', '3', '1', '2', '2', '1', '3', '7', '2', '1', '7', '1', '2', '3', '4', '2', '3', '3', '1', '2', '2', '2', '3', '2', '1', '3', '1', '2', '3', '3', '5', '4', '1', '1', '1', '1', '2', '1', '2', '1', '1', '1', '6', '4', '1', '1', '1', '4', '2', '4', '1', '4', '5', '5', '2', '2', '1', '2', '2', '1', '3', '3', '1', '1', '1', '2', '3', '1', '3', '2', '2', '1', '3', '1', '2', '3', '1', '2', '1', '1', '2', '1', '1', '3', '1', '1', '2', '1', '1', '2', '1', '1', '4', '6', '1', '2', '2', '1', '6', '3', '2', '5', '2', '2', '1', '3', '4', '2', '1', '1', '2', '1', '3', '7', '1', '3', '2', '1', '1', '1', '1', '1', '1', '2', '2', '1', '2', '3', '6', '6', '2', '3', '4', '1', '6', '1', '1', '3', '6', '6', '3', '3', '4', '1', '2', '2', '2', '4', '2', '4', '3', '1', '1', '1', '1', '1', '2', '2', '2', '1', '2', '2', '3', '4', '3', '1', '2', '2', '1', '3', '2', '2', '1', '3', '3', '3', '1', '2', '1', '2', '2', '1', '3', '2', '1', '1', '3', '1', '2', '3', '7', '2', '1', '1', '1', '1', '1', '1', '1', '1', '2', '1', '7', '2', '1', '3', '2', '1', '1', '1', '1', '2', '1', '1', '1', '2', '1', '1', '2', '1', '2', '1', '3', '2', '2', '1', '2', '1', '4', '1', '6', '4', '2', '1', '3', '2', '2', '2', '2', '1', '6', '2', '1', '5', '4', '2', '1', '1', '1', '2', '2', '1', '1', '1', '1', '7', '1', '1', '1', '1', '3', '1', '1', '3', '1', '7', '2', '2', '2', '1', '2', '1', '7', '1', '1', '3', '1', '1', '1', '2', '7', '1', '2', '1', '1', '2', '3', '1', '1', '3', '1', '1', '2', '3', '1', '3', '4', '1', '4', '1', '1', '3', '2', '1', '1', '1', '2', '4', '5', '1', '1', '3', '1', '1', '1', '3', '2', '1', '1', '2', '2', '4', '2', '2', '4', '5', '4', '1', '2', '1', '1', '5', '1', '3', '1', '1', '2', '3', '3', '3', '3', '1', '2', '3', '2', '1', '1', '2', '3', '1', '2', '3', '2', '1', '1', '2', '2', '1', '1', '1', '2', '1', '2', '1', '3', '1', '1', '1', '1', '3', '1', '1', '1', '3', '1', '3', '3', '1', '1', '1', '7', '2', '2', '7', '1', '1', '2', '3', '2', '2', '6', '1', '2', '1', '6', '1', '4', '4', '6', '4', '2', '1', '1', '1', '2', '7', '1', '1', '2', '1', '2', '1', '7', '2', '2', '2', '2', '1', '2', '4', '1', '1', '5', '1', '5', '1', '1', '1', '1', '1', '3', '1', '2', '1', '3', '3', '3', '2', '3', '1', '1', '1', '1', '2', '1', '3', '2', '3', '3', '2', '1', '4', '1', '3', '3', '2', '1', '3', '1', '3', '3', '1', '5', '1', '5', '1', '7', '1', '5', '1', '5', '1', '1', '5', '1', '3', '2', '1', '2', '1', '1', '6', '1', '2', '2', '1', '1', '2', '1', '3', '1', '1', '2', '3', '1', '6', '2', '2', '1', '2', '1', '1', '2', '1', '1', '1', '2', '2', '1', '1', '1', '5', '1', '2', '1', '1', '2', '4', '1', '2', '2', '2', '3', '3', '2', '2', '7', '1', '1', '4', '2', '2', '2', '3', '3', '1', '2', '1', '2', '1', '2', '7', '2', '3', '3', '4', '3', '2', '1', '3', '1', '4', '1', '1', '2', '1', '1', '6', '2', '2', '4', '1', '4', '1', '3', '2', '2', '2', '1', '3', '1', '1', '1', '1', '3', '1', '2', '1', '3', '1', '2', '1', '2', '1', '1', '3', '3', '2', '2', '2', '5', '1', '3', '1', '1', '2', '1', '5', '2', '2', '1', '3', '3', '1', '1', '2', '2', '3', '1', '1', '2', '1', '2', '7', '1', '2', '2', '3', '1', '1', '2', '2', '1', '3', '3', '3', '3', '5', '2', '2', '3', '1', '3', '3', '2', '3', '3', '1', '3', '1', '2', '1', '1', '1', '2', '2', '1', '2', '1', '3', '2', '2', '3', '1', '3', '1', '7', '1', '3', '1', '3', '3', '1', '1', '1', '5', '3', '3', '2', '2', '7', '3', '3', '2', '2', '1', '1', '7', '2', '6', '7', '3', '2', '3', '3', '2', '2', '1', '1', '1', '1', '1', '1', '1', '3', '3', '5', '3', '5', '3', '3', '2', '3', '3', '1', '1', '1', '5', '3', '1', '3', '3', '1', '2', '2', '3', '2', '5', '1', '5', '4', '3', '2', '1', '5', '1', '2', '2', '3', '4', '2', '1', '2', '1', '2', '1', '1', '7', '7', '3', '2', '1', '1', '4', '1', '1', '3', '2', '3', '3', '2', '3', '3', '1', '1', '1', '4', '3', '2', '2', '2', '1', '2', '1', '3', '2', '1', '3', '1', '2', '3', '2', '3', '4', '1', '3', '1', '1', '2', '2', '3', '1', '1', '1', '1', '2', '3', '3', '4', '2', '2', '1', '2', '3', '1', '2', '1', '1', '2', '1', '2', '1', '2', '2', '2', '3', '1', '3', '1', '1', '1', '1', '2', '1', '1', '4', '2', '4', '6', '1', '2', '1', '1', '1', '2', '5', '2', '1', '6', '3', '4', '1', '4', '1', '2', '3', '1', '1', '7', '2', '1', '1', '2', '1', '2', '2', '2', '3', '2', '3', '2', '3', '5', '2', '2', '3', '1', '2', '2', '2', '1', '4', '4', '3', '7', '2', '3', '2', '7', '1', '4', '3', '3', '2', '1', '1', '1', '7', '5', '1', '1', '1', '3', '1', '1', '1', '2', '3', '2', '2', '1', '1', '3', '3', '2', '1', '3', '1', '2', '2', '1', '1', '6', '2', '2', '3', '4', '5', '4', '5', '2', '5', '1', '2', '5', '3', '1', '4', '4', '2', '1', '2', '3', '1', '6', '2', '3', '1', '1', '3', '3', '2', '1', '1', '1', '3', '1', '1', '1', '3', '7', '5', '3', '2', '1', '3', '3', '1', '3', '1', '4', '4', '5', '1', '1', '1', '1', '1', '2', '1', '2', '1', '2', '3', '3', '1', '5', '1', '2', '1', '3', '4', '2', '1', '2', '1', '1', '4', '5', '1', '1', '5', '2', '4', '1', '1', '1', '1', '3', '1', '3', '1', '2', '4', '3', '7', '3', '2', '1', '3', '2', '3', '1', '2', '3', '1', '1', '7', '3', '1', '1', '7', '1', '3', '1', '3', '1', '3', '2', '3', '1', '1', '1', '2', '3', '1', '1', '5', '1', '2', '2', '3', '1', '2', '3', '1', '1', '1', '1', '1', '2', '2', '1', '3', '1', '2', '6', '1', '2', '3', '1', '3', '3', '4', '2', '3', '1', '3', '2', '2', '1', '1', '1', '1', '1', '1', '3', '1', '1', '2', '1', '1', '3', '2', '3', '1', '1', '1', '2', '3', '2', '1', '2', '2', '2', '3', '6', '2', '5', '3', '4', '1', '1', '1', '7', '3', '5', '2', '2', '1', '1', '1', '2', '1', '4', '1', '2', '1', '2', '2', '3', '4', '1', '2', '3', '2', '2', '4', '3', '3', '3', '3', '2', '4', '3', '3', '1', '2', '2', '2', '2', '4', '1', '4', '1', '1', '3', '2', '1', '2', '3', '1', '2', '3', '4', '2', '2', '3', '3', '2', '5', '1', '4', '2', '1', '2', '2', '1', '1', '3', '2', '1', '3', '4', '1', '1', '2', '2', '3', '4', '2', '1', '2', '1', '2', '2', '2', '5', '1', '3', '1', '1', '1', '1', '3', '3', '2', '3', '2', '4', '7', '1', '1', '2', '3', '1', '1', '2', '1', '5', '2', '2', '2', '1', '1', '2', '4', '2', '4', '2', '1', '1', '6', '3', '5', '1', '1', '1', '1', '1', '3', '1', '1', '2', '1', '1', '1', '1', '2', '2', '3', '2', '3', '2', '2', '1', '4', '2', '1', '1', '1', '2', '7', '3', '1', '2', '2', '2', '1', '1', '2', '2', '1', '2', '3', '2', '1', '4', '1', '1', '2', '1', '1', '1', '2', '3', '1', '1', '3', '1', '2', '1', '2', '3', '2', '1', '7', '1', '2', '2', '3', '1', '4', '2', '2', '1', '6', '1', '3', '2', '3', '3', '2', '1', '1', '2', '2', '6', '2', '2', '1', '6', '5', '3', '2', '1', '3', '2', '2', '3', '6', '3', '1', '1', '1', '3', '3', '1', '2', '3', '1', '3', '3', '1', '4', '3', '2', '1', '2', '1', '3', '2', '1', '1', '1', '3', '2', '4', '2', '1', '1', '2', '1', '3', '3', '3', '1', '2', '2', '1', '2', '1', '3', '1', '2', '3', '3', '3', '2', '2', '1', '1', '5', '2', '1', '2', '1', '1', '1', '1', '2', '1', '1', '1', '1', '1', '3', '1', '1', '7', '1', '2', '1', '6', '1', '2', '4', '5', '2', '1', '2', '4', '1', '3', '2', '6', '2', '1', '2', '2', '2', '3', '3', '7', '3', '4', '1', '3', '2', '1', '3', '1', '1', '1', '1', '3', '2', '3', '1', '1', '7', '5', '2', '2', '1', '3', '2', '1', '1', '1', '1', '5', '1', '7', '2', '2', '1', '3', '1', '2', '1', '2', '1', '1', '1', '1', '1', '1', '2', '2', '2', '1', '3', '2', '2', '1', '2', '1', '1', '3', '1', '3', '1', '1', '3', '4', '1', '2', '6', '1', '2', '3', '2', '3', '2', '1', '1', '1', '1', '3', '1', '1', '2', '3', '1', '3', '3', '7', '2', '1', '1', '2', '2', '2', '1', '7', '2', '1', '4', '2', '1', '2', '1', '7', '2', '1', '2', '1', '1', '2', '2', '2', '2', '2', '3', '7', '5', '1', '1', '1', '3', '1', '3', '1', '2', '2', '2', '1', '3', '4', '2', '1', '2', '2', '1', '5', '3', '1', '2', '1', '5', '6', '2', '3', '2', '1', '2', '2', '1', '3', '1', '4', '2', '2', '1', '3', '1', '1', '3', '2', '2', '2', '3', '3', '1', '2', '1', '1', '2', '5', '1', '2', '1', '2', '1', '3', '1', '1', '1', '1', '1', '1', '1', '3', '1', '1', '1', '1', '1', '1', '1', '2', '4', '1', '2', '2', '3', '2', '1', '2', '1', '2', '1', '1', '5', '1', '2', '2', '1', '1', '1', '1', '2', '1', '1', '3', '1', '7', '1', '4', '1', '3', '1', '4', '1', '6', '5', '1', '2', '1', '3', '2', '7', '1', '2', '2', '1', '3', '2', '1', '4', '1', '1', '6', '2', '1', '2', '1', '1', '3', '3', '2', '1', '1', '2', '2', '1', '7', '1', '3', '3', '3', '1', '2', '7', '2', '2', '2', '1', '1', '1', '3', '1', '1', '3', '2', '2', '1', '1', '2', '3', '1', '3', '7', '4', '2', '1', '3', '1', '1', '2', '6', '1', '5', '1', '4', '3', '3', '2', '4', '2', '4', '1', '7', '3', '1', '1', '1', '3', '1', '1', '6', '7', '3', '1', '1', '1', '7', '3', '2', '2', '4', '1', '3', '1', '1', '6', '2', '1', '1', '2', '1', '5', '3', '1', '2', '7', '2', '3', '1', '3', '1', '3', '3', '5', '2', '1', '1', '4', '1', '4', '1', '4', '5', '2', '6', '2', '1', '3', '7', '7', '3', '5', '2', '2', '1', '1', '2', '1', '1', '6', '2', '1', '5', '2', '5', '6', '1', '4', '4', '1', '1', '4', '3', '2', '2', '4', '5', '2', '3', '1', '3', '1', '5', '3', '7', '1', '3', '4', '1', '5', '1', '6', '7', '1', '1', '1', '1', '5', '5', '2', '2', '5', '2', '2', '2', '3', '3', '1', '4', '1', '1', '1', '6', '2', '1', '4', '1', '1', '1', '1', '1', '7', '3', '1', '4', '3', '4', '3', '3', '3', '7', '2', '3', '1', '3', '2', '2', '1', '4', '2', '1', '4', '1', '1', '1', '3', '2', '2', '2', '2', '1', '4', '2', '1', '2', '2', '2', '2', '1', '5', '2', '1', '1', '2', '3', '7', '4', '1', '2', '3', '1', '5', '2', '1', '4', '6', '5', '4', '1', '4', '3', '1', '6', '3', '1', '7', '1', '2', '7', '2', '7', '1', '2', '3', '2', '1', '2', '3', '2', '4', '1', '1', '6', '1', '1', '2', '1', '1', '7', '1', '2', '2', '4', '1', '2', '4', '1', '4', '1', '2', '2', '1', '1', '2', '3', '3', '1', '2', '3', '3', '1', '7', '1', '1', '3', '3', '3', '1', '1', '3', '1', '4', '3', '3', '5', '3', '3', '1', '1', '3', '2', '3', '1', '2', '2', '2', '1', '2', '4', '5', '7', '1', '4', '2', '1', '3', '3', '4', '1', '5', '1', '1', '3', '7', '5', '5', '2', '1', '2', '2', '3', '4', '1', '4', '2', '1', '1', '1', '2', '2', '2', '1', '3', '2', '1', '7', '5', '7', '7', '1', '1', '2', '2', '2', '4', '2', '2', '6', '5', '4', '1', '2', '7', '2', '2', '7', '3', '2', '1', '7', '2', '1', '3', '2', '2', '3', '3', '7', '3', '2', '6', '1', '2', '1', '1', '2', '1', '6', '1', '4', '3', '1', '1', '3', '5', '1', '6', '5', '3', '3', '7', '2', '7', '1', '7', '2', '1', '2', '5', '3', '4', '4', '3', '2', '1', '1', '2', '2', '1', '3', '2', '1', '5', '3', '4', '3', '1', '7', '7', '7', '3', '2', '7', '3', '1', '3', '4', '3', '2', '1', '2', '2', '1', '2', '3', '3', '6', '6', '1', '1', '4', '1', '7', '2', '2', '7', '2', '1', '1', '3', '2', '3', '1', '2', '2', '1', '7', '6', '2', '2', '1', '7', '2', '1', '2', '2', '3', '1', '7', '1', '6', '1', '1', '1', '7', '7', '3', '1', '1', '6', '3', '2', '1', '2', '1', '2', '2', '5', '1', '7', '6', '2', '3', '2', '2', '1', '4', '7', '2', '7', '3', '2', '2', '1', '1', '1', '7', '1', '3', '2', '3', '3', '1', '2', '1', '2', '2', '2', '1', '4', '2', '6', '2', '2', '1', '2', '1', '4', '7', '1', '1', '7', '7', '4', '4', '1', '2', '4', '1', '4', '4', '4', '3', '7', '1', '4', '1', '4', '1', '1', '1', '5', '1', '7', '7', '4', '4', '3', '5', '5', '2', '1', '4', '5', '3', '1', '4', '4', '2', '3', '7', '2', '1', '2', '2', '1', '1', '1', '4', '7', '2', '2', '5', '5', '2', '5', '1', '5', '3', '3', '3', '3', '4', '3', '5', '1', '3', '1', '1', '2', '1', '1', '1', '3', '3', '1', '7', '7', '4', '4', '2', '7', '1', '3', '7', '2', '3', '3', '1', '2', '4', '5', '2', '2', '3', '2', '4', '1', '5', '3', '3', '2', '1', '1', '2', '1', '3', '7', '4', '7', '5', '2', '3', '1', '2', '3', '2', '6', '3', '2', '3', '4', '2', '1', '2', '3', '1', '1', '2', '2', '2', '4', '2', '1', '2', '3', '1', '1', '7', '2', '1', '5', '3', '3', '2', '2', '2', '1', '4', '2', '4', '3', '1', '3', '4', '4', '1', '4', '5', '3', '3', '1', '2', '2', '3', '1', '2', '2', '2', '1', '7', '2', '2', '1', '2', '1', '1', '4', '7', '3', '2', '5', '4', '1', '1', '5', '5', '5', '3', '2', '4', '1', '1', '4', '1', '2', '7', '3', '4', '1', '5', '3', '2', '3', '7', '1', '2', '6', '2', '3', '1', '5', '1', '6', '2', '6', '6', '4', '1', '2', '4', '4', '2', '1', '5', '1', '1', '3', '4', '5', '4', '7', '4', '1', '4', '1', '2', '7', '7', '1', '3', '3', '1', '2', '3', '2', '1', '4', '1', '1', '2', '2', '1', '2', '2', '3', '1', '4', '7', '6', '1', '7', '7', '2', '2', '2', '4', '4', '3', '1', '6', '1', '3', '1', '1', '1', '4', '2', '2', '3', '1', '3', '1', '1', '6', '7', '2', '1', '7', '1', '2', '5', '1', '3', '2', '2', '3', '3', '1', '1', '1', '5', '3', '2', '1', '5', '1', '2', '4', '4', '1', '1', '1', '1', '5', '4', '5', '6', '1', '3', '7', '2', '1', '1', '1', '1', '1', '6', '3', '3', '1', '1', '5', '1', '3', '2', '1', '7', '1', '1', '1', '2', '1', '1', '7', '1', '7', '1', '2', '3', '1', '7', '1', '3', '4', '2', '3', '2', '2', '4', '6', '1', '5', '2', '4', '5', '1', '4', '1', '2', '7', '7', '1', '1', '1', '6', '3', '1', '3', '1', '2', '2', '6', '1', '1', '2', '5', '2', '7', '4', '3', '2', '4', '1', '1', '1', '3', '1', '3', '7', '4', '4', '7', '5', '1', '3', '2', '2', '3', '7', '1', '6', '3', '2', '5', '2', '2', '7', '2', '2', '7', '4', '5', '5', '2', '2', '7', '3', '5', '4', '3', '1', '3', '7', '7', '1', '1', '3', '7', '3', '2', '1', '1', '7', '4', '3', '4', '1', '1', '1', '2', '2', '5', '1', '7', '1', '3', '7', '7', '1', '6', '7', '2', '6', '2', '2', '2', '1', '2', '7', '7', '1', '2', '2', '2', '1', '2', '1', '7', '4', '7', '7', '7', '2', '3', '6', '3', '2', '1', '2', '2', '1', '1', '1', '7', '7', '5', '7', '1', '7', '4', '4', '4', '4', '1', '4', '1', '1', '1', '7', '4', '2', '2', '7', '7', '2', '1', '6', '3', '1', '3', '2', '7', '7', '2', '4', '1', '2', '1', '3', '5', '2', '1', '7', '1', '3', '3', '7', '7', '3', '4', '3', '7', '2', '4', '2', '6', '2', '1', '1', '7', '3', '7', '7', '2', '5', '2', '7', '7', '1', '7', '3', '5', '4', '2', '1', '7', '5', '2', '2', '2', '2', '7', '3', '2', '1', '7', '7', '4', '7', '5', '1', '4', '3', '4', '5', '7', '1', '3', '5', '2', '4', '5', '6', '2', '6', '3', '3', '7', '7', '4', '1', '2', '5', '2', '4', '3', '7', '2', '7', '3', '4', '1', '5', '5', '7', '2', '7', '6', '2', '7', '7', '5', '3', '2', '2', '6', '1', '4', '7', '7', '1', '7', '2', '1', '6', '3', '1', '2', '6', '2', '1', '2', '1', '1', '7', '2', '3', '7', '7', '2', '6', '6', '2', '3', '2', '3', '1', '7', '7', '2', '7', '2', '1', '2', '4', '1', '2', '2', '7', '7', '2', '7', '5', '7', '6', '2', '3', '3', '6', '7', '3', '7', '2', '1', '7', '7', '1', '5', '7', '1', '4', '3', '4', '1', '3', '3', '2', '1', '1', '4', '4', '1', '7', '3', '7', '3', '2', '3', '7', '1', '5', '4', '1', '7', '1', '2', '3', '1', '7', '2', '7', '4', '4', '7', '1', '4', '3', '4', '5', '2', '1', '7', '7', '7', '4', '4', '5', '2', '1', '4', '1', '1', '4', '7', '1', '7', '1', '1', '6', '5', '1', '7', '7', '4', '5', '2', '2', '2', '7', '7', '3', '7', '7', '1', '1', '1', '5', '3', '4', '7', '4', '6', '1', '3', '1', '3', '7', '7', '4', '1', '3', '7', '7', '6', '2', '3', '7', '4', '1', '2', '7', '5', '7', '3', '7', '5', '7', '1', '1', '1', '6', '3', '7', '7', '1', '6', '2', '1', '1', '7', '2', '2', '7', '4', '7', '5', '3', '2', '7', '5', '7', '1', '1', '4', '7', '7', '1', '1', '1', '3', '2', '7', '7', '1', '7', '7', '1', '6', '2', '7', '3', '7', '2', '7', '3', '4', '6', '2', '7', '1', '7', '1', '2', '3', '1', '2', '2', '7', '7', '1', '7', '7', '1', '2', '7', '7', '7', '1', '6', '6', '6', '2', '2', '4', '7', '7', '1', '3', '7', '5', '4', '3', '3', '7', '2', '7', '7', '4', '4', '1', '2', '7', '2', '2', '7', '7', '7', '2', '2', '2', '7', '2', '1', '5', '7', '2', '7', '7', '3', '3', '2', '4', '2', '2', '7', '1', '1', '7', '3', '7', '7', '3', '6', '1', '2', '1', '2', '7', '5', '1', '7', '3', '3', '7', '5', '1', '7', '2', '7', '6', '1', '7', '1', '3', '7', '7', '2', '4', '5', '3', '1', '2', '7', '1', '2', '7', '3', '2', '3', '7', '1', '2', '7', '4', '1', '1', '2', '3', '7', '7', '1', '7', '7', '1', '4', '3', '7', '3', '4', '4', '7', '1', '3', '3', '3', '1', '7', '7', '7', '3', '7', '7', '7', '3', '7', '2', '2', '1', '6', '2', '1', '7', '2', '7', '7', '1', '2', '4', '7', '2', '2', '4', '4', '7', '7', '2', '7', '1', '5', '2', '4', '7', '7', '2', '7', '1', '1', '1', '6', '6', '1', '7', '7', '7', '1', '7', '1', '4', '7', '7', '3', '5', '7', '2', '7', '2', '4', '3', '7', '6', '1', '3', '6', '7', '3', '7', '1', '7', '6', '4', '4', '7', '5', '7', '1', '7', '7', '5', '2', '7', '2', '7', '2', '2', '7', '3', '2', '7', '7', '1', '7', '2', '7', '2', '1', '7', '7', '1', '7', '6', '1', '3', '1', '7', '7', '7', '7', '1', '3', '1', '7', '2', '2', '2', '4', '2', '7', '7', '1', '4', '7', '2', '1', '7', '1', '1', '2', '3', '2', '5', '1', '7', '1', '7', '7', '2', '4', '1', '2', '2', '7', '7', '6', '7', '7', '1', '3', '4', '3', '7', '1', '3', '4', '3', '7', '2', '7', '4', '1', '7', '6', '4', '7', '2', '4', '1', '7', '7', '7', '1', '1', '7', '2', '7', '5', '7', '2', '7', '1', '7', '3', '1', '3', '3', '7', '7', '7', '2', '4', '1', '7', '1', '3', '1', '7', '1', '5', '1', '4', '2', '2', '3', '7', '3', '7', '2', '1', '1', '7', '3', '7', '7', '4', '1', '5', '2', '1', '1', '7', '3', '1', '3', '3', '1', '1', '3', '4', '4', '1', '1', '5', '1', '2', '5', '1', '2', '2', '1', '7', '4', '2', '2', '4', '3', '4', '4', '7', '1', '1', '3', '5', '1', '1', '1', '2', '1', '1', '6', '6', '7', '5', '1', '7', '7', '7', '4', '6', '2', '5', '1', '3', '1', '1', '3', '2', '2', '3', '1', '1', '7', '2', '2', '7', '1', '2', '1', '1', '2', '2', '3', '7', '3', '3', '3', '1', '7', '2', '3', '1', '2', '3', '4', '3', '7', '1', '1', '2', '1', '5', '4', '4', '7', '3', '1', '2', '4', '4', '4', '6', '1', '1', '3', '7', '4', '1', '2', '2', '2', '1', '5', '4', '7', '2', '4', '3', '4', '4', '3', '2', '1', '1', '1', '3', '7', '4', '1', '2', '5', '2', '5', '3', '2', '2', '1', '5', '1', '3', '4', '1', '4', '7', '2', '4', '2', '1', '6', '3', '1', '7', '1', '1', '1', '1', '1', '1', '5', '7', '6', '1', '2', '6', '1', '5', '1', '2', '7', '5', '1', '1', '1', '1', '4', '2', '5', '5', '7', '7', '1', '3', '1', '1', '1', '4', '1', '7', '2', '6', '2', '5', '5', '3', '5', '5', '7', '3', '3', '7', '7', '5', '1', '4', '2', '5', '4', '1', '3', '7', '3', '1', '7', '5', '3', '7', '7', '3', '1', '2', '1', '2', '5', '5', '2', '1', '4', '2', '2', '3', '4', '2', '2', '2', '7', '2', '4', '4', '7', '4', '7', '3', '6', '1', '6', '4', '1', '3', '4', '3', '3', '7', '7', '6', '1', '7', '5', '4', '4', '4', '1', '1', '2', '2', '4', '3', '1', '4', '3', '7', '2', '2', '2', '5', '2', '1', '5', '4', '7', '2', '1', '6', '4', '2', '1', '4', '7', '3', '3', '7', '7', '7', '2', '2', '1', '5', '5', '4', '2', '1', '6', '4', '3', '2', '3', '6', '5', '3', '7', '2', '1', '2', '1', '2', '5', '3', '1', '1', '4', '5', '7', '7', '3', '5', '4', '1', '1', '1', '1', '2', '7', '3', '2', '4', '7', '1', '3', '3', '2', '5', '7', '4', '2', '1', '4', '1', '4', '6', '4', '1', '4', '3', '2', '7', '2', '2', '5', '7', '7', '2', '1', '2', '7', '1', '4', '1', '4', '6', '7', '4', '1', '4', '2', '2', '5', '6', '1', '3', '5', '7', '2', '4', '6', '3', '7', '1', '3', '7', '1', '7', '5', '3', '2', '2', '1', '2', '1', '3', '1', '3', '2', '5', '3', '3', '7', '1', '1', '1', '4', '4', '3', '3', '2', '1', '7', '7', '6', '1', '1', '7', '5', '1', '1', '1', '7', '1', '4', '1', '4', '1', '2', '2', '3', '5', '5', '1', '7', '5', '3', '1', '2', '3', '2', '6', '4', '7', '2', '3', '7', '5', '3', '4', '3', '3', '1', '2', '4', '4', '2', '3', '1', '1', '1', '1', '2', '1', '1', '2', '7', '7', '1', '2', '7', '2', '2', '2', '5', '1', '2', '5', '3', '4', '1', '5', '2', '1', '7', '2', '1', '3', '4', '3', '4', '2', '4', '7', '7', '4', '7', '1', '1', '1', '1', '1', '2', '2', '5', '1', '3', '3', '6', '1', '2', '7', '4', '2', '3', '3', '2', '3', '2', '6', '2', '1', '1', '2', '6', '1', '2', '4', '4', '2', '3', '2', '1', '1', '2', '7', '2', '4', '5', '2', '4', '6', '3', '2', '1', '7', '1', '7', '3', '4', '7', '1', '5', '4', '1', '1', '5', '5', '3', '6', '2', '7', '5', '1', '2', '2', '7', '7', '2', '1', '7', '2', '2', '3', '3', '1', '2', '1', '2', '1', '3', '2', '2', '1', '7', '1', '7', '7', '5', '5', '4', '5', '2', '5', '3', '5', '3', '2', '1', '2', '1', '5', '2', '1', '5', '7', '3', '7', '1', '2', '3', '7', '1', '7', '1', '4', '2', '1', '1', '7', '6', '4', '1', '2', '1', '4', '5', '7', '3', '3', '2', '7', '1', '7', '7', '2', '1', '2', '2', '1', '2', '4', '3', '4', '7', '1', '3', '2', '2', '1', '2', '7', '1', '1', '1', '1', '6', '3', '2', '1', '7', '5', '1', '3', '2', '6', '2', '7', '7', '2', '1', '2', '2', '3', '4', '7', '7', '7', '1', '1', '2', '3', '2', '1', '3', '4', '5', '4', '4', '4', '7', '4', '7', '7', '1', '1', '7', '1', '2', '2', '1', '5', '2', '1', '2', '6', '2', '4', '2', '1', '4', '2', '2', '1', '7', '7', '2', '1', '1', '3', '1', '1', '5', '5', '4', '4', '2', '5', '6', '3', '1', '7', '4', '2', '5', '2', '4', '4', '7', '7', '1', '2', '4', '1', '3', '3', '1', '5', '2', '7', '5', '3', '3', '4', '7', '7', '7', '5', '1', '1', '1', '2', '1', '5', '7', '7', '6', '7', '4', '1', '2', '5', '7', '1', '1', '1', '2', '3', '1', '1', '7', '1', '3', '7', '2', '3', '7', '7', '2', '4', '2', '3', '3', '1', '1', '3', '2', '4', '7', '1', '7', '5', '7', '2', '3', '4', '3', '2', '1', '6', '2', '1', '2', '1', '2', '4', '1', '7', '4', '2', '7', '3', '2', '4', '3', '2', '1', '1', '2', '1', '2', '7', '3', '1', '4', '4', '7', '2', '7', '1', '2', '1', '4', '2', '6', '6', '3', '2', '1', '4', '7', '4', '2', '2', '4', '4', '2', '5', '1', '1', '7', '1', '7', '4', '7', '7', '1', '1', '2', '5', '1', '4', '4', '1', '4', '2', '1', '2', '4', '2', '7', '4', '2', '6', '2', '1', '1', '3', '4', '1', '2', '1', '4', '2', '5', '2', '6', '2', '1', '1', '7', '2', '2', '1', '1', '2', '1', '4', '1', '4', '1', '4', '2', '1', '3', '1', '7', '1', '7', '1', '1', '4', '3', '1', '2', '3', '1', '4', '7', '2', '2', '1', '2', '1', '2', '1', '6', '1', '7', '7', '4', '5', '1', '3', '1', '7', '2', '1', '1', '5', '2', '7', '2', '7', '4', '2', '1', '2', '1', '1', '1', '2', '2', '2', '7', '4', '7', '1', '1', '1', '1', '5', '1', '5', '3', '4', '6', '4', '6', '5', '1', '7', '4', '3', '1', '7', '5', '3', '1', '3', '5', '5', '1', '5', '7', '2', '7', '1', '2', '2', '2', '3', '1', '2', '1', '6', '2', '4', '4', '1', '5', '3', '5', '2', '2', '4', '4', '2', '1', '7', '3', '1', '1', '1', '4', '4', '5', '1', '5', '1', '6', '3', '4', '5', '3', '2', '2', '1', '3', '5', '7', '1', '1', '1', '7', '7', '2', '5', '6', '3', '2', '4', '1', '4', '2', '7', '5', '2', '7', '2', '3', '1', '1', '1', '4', '3', '2', '2', '3', '4', '2', '6', '7', '1', '3', '2', '2', '5', '7', '5', '7', '2', '7', '2', '1', '2', '3', '7', '3', '1', '6', '7', '1', '2', '6', '7', '2', '7', '4', '4', '3', '4', '4', '2', '2', '3', '1', '1', '3', '1', '6', '3', '2', '3', '1', '4', '3', '2', '7', '4', '7', '1', '7', '2', '1', '6', '1', '5', '1', '2', '6', '1', '2', '2', '1', '3', '1', '6', '7', '7', '7', '1', '7', '2', '4', '2', '1', '4', '6', '1', '1', '1', '2', '2', '2', '3', '2', '2', '4', '7', '1', '2', '2', '6', '3', '4', '6', '2', '2', '1', '4', '2', '1', '1', '7', '4', '7', '2', '1', '2', '5', '3', '1', '2', '1', '5', '3', '2', '4', '2', '3', '4', '2', '1', '3', '7', '2', '4', '2', '3', '2', '1', '2', '5', '4', '4', '1', '3', '6', '1', '3', '1', '1', '2', '7', '1', '7', '4', '1', '2', '4', '2', '2', '2', '2', '1', '1', '1', '3', '3', '2', '1', '2', '2', '2', '1', '7', '2', '6', '2', '5', '1', '1', '5', '7', '7', '3', '3', '4', '3', '1', '1', '2', '4', '3', '6', '1', '1', '1', '5', '3', '4', '7', '1', '4', '1', '5', '6', '1', '2', '4', '3', '1', '3', '2', '1', '2', '2', '1', '7', '3', '1', '5', '5', '1', '2', '1', '7', '2', '2', '2', '1', '1', '1', '1', '3', '5', '6', '1', '2', '3', '1', '4', '1', '5', '1', '1', '7', '2', '1', '1', '1', '4', '3', '4', '1', '1', '1', '1', '2', '4', '3', '3', '4', '1', '7', '3', '1', '3', '6', '5', '7', '3', '1', '1', '7', '1', '1', '1', '1', '1', '1', '1', '6', '1', '7', '7', '2', '7', '5', '7', '3', '4', '1', '6', '5', '2', '2', '4', '1', '1', '1', '1', '3', '2', '1', '2', '3', '2', '7', '1', '7', '5', '4', '2', '2', '3', '2', '1', '3', '4', '1', '1', '2', '2', '2', '7', '7', '1', '2', '4', '5', '1', '2', '1', '1', '1', '1', '4', '2', '1', '2', '2', '6', '2', '2', '4', '2', '1', '4', '2', '7', '5', '2', '2', '2', '2', '3', '1', '1', '2', '1', '7', '2', '5', '1', '2', '6', '1', '2', '6', '4', '5', '1', '5', '3', '3', '7', '2', '5', '3', '2', '2', '1', '4', '4', '4', '2', '3', '1', '2', '1', '3', '5', '3', '1', '2', '4', '1', '4', '4', '3', '1', '4', '4', '7', '3', '7', '3', '2', '2', '7', '2', '3', '5', '2', '1', '3', '1', '2', '1', '5', '4', '1', '5', '3', '7', '4', '2', '6', '4', '1', '1', '1', '7', '2', '2', '3', '4', '7', '2', '5', '2', '6', '1', '2', '1', '7', '4', '3', '1', '7', '1', '4', '4', '1', '2', '1', '2', '1', '2', '1', '2', '5', '1', '2', '7', '2', '1', '7', '7', '1', '1', '4', '2', '1', '3', '2', '1', '2', '2', '7', '1', '1', '4', '5', '7', '1', '1', '2', '2', '2', '3', '3', '2', '3', '5', '2', '2', '4', '3', '4', '7', '2', '2', '4', '1', '4', '6', '1', '1', '2', '1', '1', '2', '2', '6', '1', '4', '1', '3', '3', '4', '7', '7', '7', '3', '3', '4', '1', '2', '7', '5', '1', '1', '5', '1', '2', '1', '3', '1', '2', '7', '2', '1', '4', '2', '4', '2', '7', '5', '2', '4', '2', '2', '2', '2', '1', '2', '5', '3', '1', '6', '2', '2', '2', '2', '3', '2', '1', '1', '3', '7', '1', '2', '1', '3', '3', '1', '2', '3', '3', '1', '3', '3', '3', '1', '2', '3', '3', '2', '2', '2', '1', '2', '2', '2', '1', '2', '1', '1', '4', '1', '5', '3', '2', '1', '6', '5', '2', '1', '7', '1', '1', '2', '2', '1', '5', '6', '2', '1', '5', '7', '3', '3', '2', '2', '1', '1', '2', '7', '3', '1', '2', '2', '1', '1', '2', '4', '1', '3', '3', '1', '5', '1', '4', '1', '1', '4', '1', '2', '4', '5', '2', '1', '7', '1', '1', '2', '1', '2', '3', '1', '5', '5', '6', '1', '2', '3', '1', '1', '1', '1', '1', '3', '2', '7', '1', '2', '5', '1', '3', '1', '4', '3', '5', '4', '2', '3', '3', '1', '2', '4', '1', '1', '1', '4', '1', '4', '3', '1', '7', '7', '1', '4', '2', '3', '2', '1', '7', '3', '1', '2', '1', '1', '3', '1', '3', '1', '5', '4', '6', '1', '2', '3', '2', '1', '5', '3', '2', '5', '1', '4', '2', '1', '1', '1', '1', '1', '2', '5', '1', '1', '2', '4', '4', '1', '1', '3', '3', '1', '1', '1', '1', '3', '7', '1', '2', '3', '4', '1', '1', '2', '4', '2', '1', '1', '2', '1', '1', '6', '5', '1', '2', '1', '1', '3', '3', '2', '2', '1', '1', '3', '3', '2', '1', '7', '7', '2', '1', '2', '4', '1', '1', '7', '7', '2', '2', '1', '4', '2', '2', '2', '1', '5', '1', '4', '4', '5', '6', '5', '3', '5', '6', '4', '3', '2', '2', '1', '2', '1', '1', '1', '2', '1', '7', '1', '1', '5', '2', '3', '3', '3', '2', '2', '1', '7', '1', '3', '2', '2', '1', '2', '1', '3', '7', '1', '4', '3', '1', '2', '4', '1', '4', '2', '3', '4', '6', '1', '3', '4', '2', '1', '2', '1', '1', '2', '1', '3', '1', '2', '2', '2', '4', '2', '1', '2', '7', '3', '1', '3', '1', '1', '3', '7', '1', '3', '4', '1', '1', '1', '5', '3', '3', '2', '2', '3', '2', '1', '1', '5', '4', '5', '7', '5', '4', '2', '4', '3', '3', '1', '4', '4', '1', '1', '1', '5', '2', '1', '1', '3', '1', '5', '1', '4', '5', '1', '5', '5', '3', '2', '1', '4', '6', '2', '2', '3', '1', '2', '1', '3', '2', '1', '4', '7', '5', '2', '1', '1', '5', '5', '7', '3', '3', '6', '2', '3', '1', '3', '6', '1', '3', '1', '1', '2', '2', '7', '2', '7', '2', '1', '1', '2', '1', '4', '5', '1', '2', '2', '2', '2', '1', '1', '6', '4', '4', '2', '2', '2', '4', '1', '1', '4', '1', '5', '1', '1', '2', '1', '1', '2', '1', '4', '2', '7', '7', '2', '5', '1', '3', '5', '1', '1', '1', '2', '2', '1', '7', '1', '2', '3', '1', '2', '7', '2', '4', '1', '2', '2', '4', '5', '5', '6', '2', '6', '4', '4', '1', '1', '6', '1', '5', '3', '1', '1', '2', '4', '3', '2', '1', '1', '2', '4', '1', '2', '7', '7', '2', '7', '4', '3', '3', '2', '1', '1', '5', '2', '2', '4', '2', '1', '4', '6', '1', '1', '1', '4', '5', '1', '1', '1', '1', '1', '7', '1', '1', '1', '2', '7', '1', '1', '1', '1', '4', '1', '1', '3', '1', '3', '3', '1', '3', '1', '2', '2', '2', '7', '1', '7', '1', '1', '1', '1', '2', '2', '1', '4', '1', '5', '1', '1', '3', '5', '3', '4', '1', '1', '1', '1', '2', '1', '1', '1', '1', '2', '2', '2', '3', '1', '1', '1', '3', '7', '1', '1', '4', '1', '2', '2', '3', '1', '3', '2', '3', '3', '1', '5', '1', '4', '4', '1', '2', '4', '2', '1', '5', '6', '4', '1', '1', '5', '1', '4', '1', '3', '2', '2', '5', '1', '1', '1', '6', '1', '2', '1', '3', '3', '1', '1', '1', '1', '1', '2', '1', '7', '5', '2', '1', '2', '5', '2', '1', '4', '1', '3', '2', '3', '3', '1', '2', '3', '3', '2', '1', '2', '1', '4', '3', '2', '1', '2', '7', '1', '1', '4', '3', '1', '3', '4', '1', '1', '1', '6', '3', '1', '3', '2', '1', '4', '2', '6', '3', '3', '5', '6', '1', '2', '2', '2', '2', '3', '1', '1', '3', '2', '2', '1', '1', '1', '1', '1', '1', '3', '2', '2', '5', '2', '6', '1', '1', '2', '2', '7', '1', '1', '1', '1', '1', '4', '1', '4', '2', '2', '1', '1', '2', '2', '2', '1', '2', '1', '2', '7', '1', '4', '1', '2', '1', '1', '3', '7', '7', '2', '4', '4', '1', '1', '4', '7', '2', '1', '2', '4', '2', '5', '4', '2', '4', '5', '2', '3', '4', '3', '1', '1', '1', '3', '2', '2', '3', '2', '1', '1', '2', '1', '1', '7', '5', '2', '7', '2', '1', '5', '1', '2', '4', '3', '6', '7', '4', '7', '2', '5', '2', '4', '2', '5', '2', '6', '4', '2', '4', '4', '2', '4', '6', '2', '1', '1', '5', '3', '1', '3', '7', '3', '1', '7', '4', '7', '1', '1', '1', '5', '4', '1', '3', '2', '3', '2', '1', '1', '1', '2', '1', '1', '1', '5', '1', '3', '4', '1', '7', '1', '1', '5', '1', '4', '2', '4', '5', '5', '3', '4', '3', '3', '3', '4', '1', '1', '4', '1', '2', '1', '5', '1', '1', '1', '1', '4', '1', '7', '7', '1', '2', '3', '1', '5', '5', '1', '2', '1', '6', '2', '1', '1', '2', '1', '1', '1', '4', '3', '1', '7', '2', '4', '1', '3', '4', '5', '2', '2', '5', '1', '5', '4', '3', '4', '1', '1', '2', '5', '5', '1', '3', '1', '1', '3', '1', '7', '5', '5', '1', '4', '6', '2', '3', '1', '4', '3', '1', '2', '1', '1', '1', '1', '1', '1', '4', '1', '1', '4', '4', '5', '2', '3', '2', '3', '1', '1', '4', '1', '1', '1', '2', '4', '5', '2', '1', '1', '1', '1', '2', '1', '4', '2', '1', '1', '2', '1', '1', '2', '1', '2', '3', '7', '2', '1', '5', '1', '3', '4', '1', '4', '4', '5', '5', '4', '6', '2', '1', '3', '4', '1', '1', '1', '1', '2', '3', '1', '2', '1', '3', '1', '1', '2', '5', '1', '5', '3', '1', '1', '2', '4', '1', '5', '1', '1', '1', '7', '3', '3', '3', '2', '3', '1', '4', '6', '1', '1', '4', '2', '1', '2', '2', '4', '3', '3', '1', '2', '2', '1', '3', '1', '5', '1', '2', '3', '1', '1', '6', '1', '7', '2', '1', '2', '7', '7', '6', '2', '1', '3', '2', '2', '2', '2', '2', '1', '2', '1', '2', '5', '1', '3', '4', '1', '3', '3', '5', '4', '1', '1', '1', '4', '3', '2', '1', '1', '5', '4', '3', '1', '2', '2', '4', '1', '1', '1', '1', '7', '1', '1', '4', '5', '4', '2', '2', '1', '2', '4', '2', '5', '4', '2', '1', '3', '2', '1', '3', '1', '3', '2', '2', '2', '1', '1', '1', '2', '2', '2', '2', '1', '3', '7', '3', '1', '2', '3', '3', '1', '1', '2', '3', '2', '1', '1', '1', '3', '1', '4', '1', '2', '2', '2', '1', '2', '3', '1', '4', '2', '1', '2', '5', '2', '5', '6', '1', '1', '1', '1', '1', '4', '1', '2', '3', '4', '3', '1', '1', '5', '1', '1', '1', '3', '4', '1', '3', '2', '2', '2', '7', '1', '3', '1', '2', '2', '1', '5', '3', '1', '3', '1', '5', '4', '2', '3', '1', '1', '2', '2', '5', '1', '5', '1', '4', '4', '4', '5', '4', '5', '1', '7', '2', '1', '3', '1', '3', '2', '1', '3', '2', '3', '1', '7', '2', '2', '1', '7', '1', '6', '1', '1', '1', '2', '1', '3', '5', '1', '2', '3', '1', '2', '3', '1', '2', '4', '1', '3', '1', '2', '5', '1', '7', '1', '7', '2', '3', '7', '6', '2', '1', '1', '2', '2', '1', '5', '1', '1', '1', '6', '1', '1', '5', '4', '4', '2', '5', '5', '1', '3', '4', '5', '3', '3', '2', '4', '1', '2', '7', '1', '1', '1', '7', '5', '1', '1', '1', '4', '2', '5', '5', '1', '1', '2', '1', '1', '2', '2', '1', '4', '2', '1', '1', '5', '2', '3', '1', '5', '6', '3', '2', '5', '1', '1', '3', '1', '2', '1', '6']\n"
]
],
[
[
"Отнормировали датчики по общему количеству пройденных датчиков (если транспортное средство никак не взаимодействовало с сенсором - присваиваем значение 0). В итоге получаем вектор, где координатами являются сенсоры со значениями [0, 1].",
"_____no_output_____"
]
],
[
[
"vectors = []\nfor name, gt in groupedTraces.items():\n groupGates = np.zeros(len(gates))\n for t in gt:\n groupGates[gates.index(t[3])] += 1\n vector = []\n for rec in groupGates:\n vector.append(str(rec))\n # vectors.write(\";\")\n vectors.append(vector)\n\nfor vector in vectors:\n print(vector)",
"_____no_output_____"
]
],
[
[
"## Заключение\n### SOM\nПо итогу работы с алгоритмом SOM можно выделить следующие плюсы и минусы.\n\nПлюсы:\n1. Алгоритм является инструментом снижения размерности и визуализации данных;\n2. Отличная видимость кластеров на карте отображения сработавших нейронов совместно с точками данных;\n3. Отличная видимость аномалий при любых параметрах, что позволило нам быстро найти ответ на главный вопрос челленджа;\n4. Сравнивая интуитивное разбиение и разбиение алгоритмом k-means, получили небольшую погрешность. Это говорит о том, что карта SOM вполне подходит для определения кластеров в данных.\n\nНезначительные минусы: ресурсозатратность, что подразумевает длительную работу алгоритма; ограниченный выбор форм визуализации результата работы алгоритма (SOM-карта и U-матрица).\n\nЛучшая репрезентативность визуализации была достигнута при параметрах размерность карты: 20х20, количество поколений: 10000, инициализация: PCA.\n\nМы смогли выявить аномалию заметив, что 4-осный грузовик проходят по мрашрутам грузовиков рейнджеров, несмотря на то, что у него нет на это разрешения. В визуализации в раскраске грузовиков рейнджеров отмечены вкрапления цвета 4-осных грузовиков.\n",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
cbee7696ab8433f41de27c5e1f862ab47ad779b3
| 905,661 |
ipynb
|
Jupyter Notebook
|
notebooks/BLS_2019.ipynb
|
asfox/PSC290_WQ2020
|
22a7bf2d65c8a5706d4c09168eb7324bc669d3fb
|
[
"BSD-3-Clause"
] | null | null | null |
notebooks/BLS_2019.ipynb
|
asfox/PSC290_WQ2020
|
22a7bf2d65c8a5706d4c09168eb7324bc669d3fb
|
[
"BSD-3-Clause"
] | null | null | null |
notebooks/BLS_2019.ipynb
|
asfox/PSC290_WQ2020
|
22a7bf2d65c8a5706d4c09168eb7324bc669d3fb
|
[
"BSD-3-Clause"
] | null | null | null | 860.076923 | 275,080 | 0.945885 |
[
[
[
"# HOMEWORK NOTES\n\n- __Do the assignment or don't.__ \n\n - If you run into issues, ask questions. \n - If you run into issues at the last minue, explain what the issue is in your homework. \n\n\n- __Read the instructions.__\n\n - For example, In this assignment the rules were to load *more than 10 variables, stored across multiple files*, and *Organize/munge the data into a single Pandas DataFrame*. Some of you did not do this. \n\n\n- __If you use a custom dataset, you MUST include it in your submission.__\n\n\n- The more feedback you can give when editing code, the better off we all are. \n\n\n\n\n\n",
"_____no_output_____"
],
[
"# It's a good idea to import things at the top of the file.\n\nThis isn't strictly necessary, but it will make your life better in the long run, as it will help you know what packages you need installed in order to run something. ",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nimport numpy as np\n\nimport itertools \n\n%matplotlib inline\n",
"_____no_output_____"
]
],
[
[
"# Start by reading in files",
"_____no_output_____"
]
],
[
[
"series_file = 'bls/bls_series.csv'\nrecords_file = 'bls/bls_records.csv'\n",
"_____no_output_____"
],
[
"series = pd.read_csv(series_file)\nrecords = pd.read_csv(records_file, parse_dates=[2])\n\n",
"_____no_output_____"
],
[
"print( records.head() )\nprint( type( records['blsid'][0] ))\n\nrecords.sort_values('period', inplace=True)\n# functionally equal to: records = records.sort_values('period')\n\nprint( records.head() )",
" series_id blsid period value footnote\n0 1 LAUST280000000000006 2008-03-01 1312045.0 NaN\n1 1 LAUST280000000000006 2008-01-01 1313585.0 NaN\n2 1 LAUST280000000000006 2008-02-01 1311059.0 NaN\n3 1 LAUST280000000000006 2002-07-01 1294242.0 NaN\n4 1 LAUST280000000000006 2007-12-01 1330529.0 NaN\n<class 'str'>\n series_id blsid period value footnote\n242450 1333 SMU01000002000000001 2000-01-01 101.1 NaN\n143064 787 SMS20000002000000001 2000-01-01 66.5 NaN\n14014 78 LAUST240000000000006 2000-01-01 2763818.0 NaN\n158215 870 SMS41000004000000001 2000-01-01 321.3 NaN\n149426 822 SMU49000000000000001 2000-01-01 1044.9 NaN\n"
],
[
"print(series.head())",
" id blsid title \\\n0 1613 LNS12300000 Employment-Population Ratio \n1 1615 LNS12600000 Employed, Usually Work Part Time \n2 1616 LNU02036012 Employment Level, Nonag. Industries, With a Jo... \n3 1617 LNS14027689 Unemployment Rate - 25 Years & Over, Some Coll... \n4 1618 LNS13327709 Alternative measure of labor underutilization U-6 \n\n source is_primary delta_id is_delta is_adjusted \n0 CPS t 1686.0 f f \n1 CPS t 1688.0 f f \n2 CPS t 1689.0 f f \n3 CPS t 1690.0 f f \n4 CPS t 1691.0 f f \n"
]
],
[
[
"# Merge Files\n\n\nThis is where the magic happens.",
"_____no_output_____"
]
],
[
[
"df = records.merge(series, on='blsid', how='inner')\n\n# df = records.merge(series, left_on='blsid', right_on='blsid')\n# df = pd.merge( left=records, right=series, left_on='blsid', right_on='blsid')",
"_____no_output_____"
],
[
"df.sample(n=5)['title'].values\n",
"_____no_output_____"
],
[
"print( len( df['title'].str.contains('unemployment') ) )\nprint( len( df['title'] ) )\n\ndf.loc[ df['title'].str.contains('unemployment'),'title' ].unique()",
"609207\n609207\n"
],
[
"\nselected_columns = df['title'].str.contains('seasonally adjusted - unemployment$') & ~df['title'].str.contains('not seasonally')\n\n\nprint ( df.loc[ selected_columns, 'title'].sample(n=5))\n\n",
"234728 California, seasonally adjusted - unemployment\n264367 Texas, seasonally adjusted - unemployment\n84271 Illinois, seasonally adjusted - unemployment\n191489 Pennsylvania, seasonally adjusted - unemployment\n41160 Maryland, seasonally adjusted - unemployment\nName: title, dtype: object\n"
],
[
"df.loc[selected_columns].groupby('period')['value'].mean().plot()\n\n",
"_____no_output_____"
],
[
"# using non-pandas plot function... \nf, axarr = plt.subplots(1,2)\naxarr[0].plot( df.loc[selected_columns].groupby('period')['value'].mean(), 'r' )\naxarr[1].plot( df.loc[selected_columns].groupby('period')['value'].sum(), 'b')\n",
"_____no_output_____"
],
[
"# using seaborn lineplot\n\nsns.lineplot( x='period', y='value', data=df.loc[selected_columns])\n\n",
"_____no_output_____"
],
[
"# using seaborn regplot\nsns.relplot(x=\"period\", y=\"value\", hue=\"title\", kind=\"line\", legend=\"full\", data=df.loc[selected_columns] )",
"_____no_output_____"
],
[
"# regplot sorted by title... \nsns.relplot(x=\"period\", y=\"value\", hue=\"title\", kind=\"line\", legend=\"full\", data=df.loc[selected_columns].sort_values('title') )",
"_____no_output_____"
],
[
"# select unenplotment *rate*\nselected_columns_rate = df['title'].str.contains('seasonally adjusted - unemployment rate$') & ~df['title'].str.contains('not seasonally')\nsns.relplot(x=\"period\", y=\"value\", hue=\"title\", kind=\"line\", legend=\"full\", data=df.loc[selected_columns_rate].sort_values('title') )\n\n\n",
"_____no_output_____"
],
[
"# for s in df.loc[selected_columns_rate, 'title'].unique():\n# print( s )",
"_____no_output_____"
],
[
"# in order to get matplotlib to deal with dates, correctly. \nimport matplotlib.dates as mdates\n\nsns.set_style('white')\n\n# get a list of variables to use...\nstate_unemployment_vars = df.loc[ df['title'].str.contains(', not seasonally adjusted - unemployment rate$') ]\nstate_unemployment_vars = state_unemployment_vars.sort_values('title', ascending=False)\n\n# get a list of bslids for the unenployment rate\nstate_unenployment_ids = state_unemployment_vars['blsid'].unique()\n\n# create a pivoted dataframe of unemployment. \nunemployment_df = df.pivot(index='period', columns='blsid', values='value').copy()\n\n# select a colormap\nmy_cmap = sns.cubehelix_palette(n_colors=10, as_cmap=True )\n\n# here is where we use mdates to set the x axis.\nxlim = mdates.date2num([min(unemployment_df.index), max(unemployment_df.index)])\n\n# create figure\nf,ax = plt.subplots(1,1,figsize=(10,10))\n\n# # create axis. \nax.imshow( unemployment_df[state_unenployment_ids].transpose(), \n extent=[xlim[0], xlim[1], 0,len(state_unenployment_ids)], \n aspect='auto', interpolation='none',\n origin='lower', cmap=my_cmap,\n vmin=2, vmax=13)\n\n# set the x-axis to be a date.\nax.xaxis_date()\n\n# get the list of titles\nmy_titles = state_unemployment_vars['title'].unique()\n\n# turn the list of titles into a list of states.\nmy_states = [title.replace(', not seasonally adjusted - unemployment rate','') for title in my_titles]\n\n# # make sure every row has a y-tick. \nax.set_yticks([x+.5 for x in range(0,len(state_unenployment_ids))])\n\n# # set the name of the y_ticks to states.\nax.set_yticklabels( my_states ) ;\n\n\n",
"_____no_output_____"
],
[
"# custom legend\n# x-ticks on big plot\n\n",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbee7eeffd6fd232392efe19925ee1be15af1695
| 243,987 |
ipynb
|
Jupyter Notebook
|
Search for genders - Part 2 - Feature Engineering.ipynb
|
patrickphat/gfg_challenge
|
95579c90e9768545f6a7344e8e6c323f28a6e049
|
[
"Apache-2.0"
] | null | null | null |
Search for genders - Part 2 - Feature Engineering.ipynb
|
patrickphat/gfg_challenge
|
95579c90e9768545f6a7344e8e6c323f28a6e049
|
[
"Apache-2.0"
] | null | null | null |
Search for genders - Part 2 - Feature Engineering.ipynb
|
patrickphat/gfg_challenge
|
95579c90e9768545f6a7344e8e6c323f28a6e049
|
[
"Apache-2.0"
] | null | null | null | 139.74055 | 140,676 | 0.803891 |
[
[
[
"import pandas as pd\nimport json\nimport seaborn as sns\nimport matplotlib.pyplot as plt\n\npd.set_option('display.max_columns', 50)",
"_____no_output_____"
]
],
[
[
"So in the last time we're able to \"softly\" label ~54% of the dataset based on 2 features which are `male_items` and `female_items`. However, other features might also contribute to the gender, however other features is not clear in a way that we can intepret, we don't even have labels! This is where deep learning shines. \nLater, we will build a representation model on contrastive learning \"softly\" labeled data. Then use that to predict the other half uncertainty.\n\nFor now we're preparing some useful features for the model.",
"_____no_output_____"
],
[
"# 1. Load data",
"_____no_output_____"
]
],
[
[
"with open(\"clean_data.csv\", \"r\") as f:\n df = pd.read_csv(f)\nwith open(\"partial_labels.csv\",\"r\") as f:\n partial_labels_df = pd.read_csv(f)",
"_____no_output_____"
],
[
"partial_labels_df",
"_____no_output_____"
],
[
"# Merge df with label\nmain_df = pd.merge(df, partial_labels_df, how=\"right\", left_on=\"customer_id\", right_on=\"customer_id\")\n\n# Drop redundant key label after merged\nmain_df.orders",
"_____no_output_____"
],
[
"main_df.keys()",
"_____no_output_____"
]
],
[
[
"# 2. Feature engineering",
"_____no_output_____"
]
],
[
[
"# skip visualize due to categorical object\nskip_columns = ['is_newsletter_subscriber','customer_id','female_flag']\ntop_n = 5\ni = 0\nfor column in main_df:\n if i == top_n:\n break\n elif column in skip_columns:\n continue\n print(f\"Column {column}\")\n plt.figure()\n main_df.boxplot([column])\n i += 1",
"Column days_since_first_order\nColumn days_since_last_order\nColumn orders\nColumn items\nColumn returns\n"
]
],
[
[
"From few boxplots we can see that the outliers deviates from the mean a lot. This is due to the nature of accumulative values.\n\nFor example:\n- returns is accumulative value since because it rises after customer return an order but do not decrease by any mean.\n- this kind of feature is not useful because it does not reflect users buying habit. \n- to reflect accurately the user habit we should relate it to the another \"time span\" variable. For example, returns per order, or returns per items, etc.\n ",
"_____no_output_____"
],
[
"Here we introduce some features normalized with the \"time span\" variable (for example `orders` or `items`) to reflect correctly the users behaviors",
"_____no_output_____"
]
],
[
[
"from utils.data_composer import feature_engineering",
"_____no_output_____"
],
[
"main_df = feature_engineering(main_df)\n# Get engineered data\nengineered_df = main_df.iloc[:,33:]\n\n# Also append column \"devices\" and \"coupon_discount_applied\" into\nengineered_df = pd.concat([engineered_df, main_df.loc[:,[\"coupon_discount_applied\",\"devices\",\"customer_id\"]]],axis=1)",
"_____no_output_____"
],
[
"engineered_df.describe()",
"_____no_output_____"
],
[
"engineered_df",
"_____no_output_____"
],
[
"# df_ = df.copy()\n# df_ = df_.drop([\"days_since_first_order\",\"customer_id\",\"days_since_last_order\"], axis=1)\nsns.set(rc={'figure.figsize':(11.7,8.27)})\n\ncorr = engineered_df.drop(\"female_flag\",axis=1).corr()\nax = sns.heatmap(\n corr, \n vmin=-1, vmax=1, center=0,\n cmap=sns.diverging_palette(20, 220, n=200),\n square=True\n)\nax.set_xticklabels(\n ax.get_xticklabels(),\n rotation=90,\n horizontalalignment='right'\n);",
"_____no_output_____"
],
[
"\nc = corr\n\ns = c.unstack()\nso = s.sort_values(kind=\"quicksort\", ascending=False, key=lambda value: value.abs())\nso.head(20)\n\n# Remove self correlated\nfor index in so.index:\n if index[0] == index[1]:\n so.drop(index,inplace=True)\n \n# Remove duplicate pair\nalready_paired = []\n\nfor index in so.index:\n if index[0] == index[1]:\n so.drop(index,inplace=True)\n elif (index[1], index[0]) in already_paired:\n so.drop(index, inplace=True)\n already_paired.append(index)",
"_____no_output_____"
],
[
"so.head(32)",
"_____no_output_____"
],
[
"s.sort_values(kind=\"quicksort\", ascending=False)",
"_____no_output_____"
],
[
"s.index[0]",
"_____no_output_____"
]
],
[
[
"Okay so we have most data scaled between 0 and 1. That's a great signal. For other column, outlier might be a problem. But no worry, we will use RobustScaler to scale it down without being suffered from largely deviated outliers.",
"_____no_output_____"
]
],
[
[
"engineered_df.to_csv(\"modeling_data.csv\",index=False)",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
cbee8ef23fd194fb8849c49d43ee10a93b402999
| 433,625 |
ipynb
|
Jupyter Notebook
|
ipynb/Ames Housing Prices-Copy3.ipynb
|
Mazharul-Hossain/DSP-COMP6993
|
9770d638d551fda4e15bfd1a7ede2e319f987998
|
[
"MIT"
] | null | null | null |
ipynb/Ames Housing Prices-Copy3.ipynb
|
Mazharul-Hossain/DSP-COMP6993
|
9770d638d551fda4e15bfd1a7ede2e319f987998
|
[
"MIT"
] | null | null | null |
ipynb/Ames Housing Prices-Copy3.ipynb
|
Mazharul-Hossain/DSP-COMP6993
|
9770d638d551fda4e15bfd1a7ede2e319f987998
|
[
"MIT"
] | null | null | null | 220.226003 | 40,908 | 0.859867 |
[
[
[
"from __future__ import absolute_import, division, print_function",
"_____no_output_____"
],
[
"import numpy, os, pandas ",
"_____no_output_____"
],
[
"import tensorflow",
"_____no_output_____"
],
[
"from tensorflow import keras",
"_____no_output_____"
],
[
"print(tensorflow.__version__)",
"1.11.0\n"
],
[
"AmesHousing = pandas.read_excel('../data/AmesHousing.xls')\nAmesHousing.head(10)",
"_____no_output_____"
],
[
"cd ..",
"D:\\UofMemphis\\Fall-18\\COMP6993\\DSP-COMP6993\n"
],
[
"from libpy import NS_dp",
"_____no_output_____"
],
[
"from sklearn.model_selection import train_test_split",
"_____no_output_____"
]
],
[
[
"We use our own function to clean Data",
"_____no_output_____"
]
],
[
[
"df = NS_dp.clean_Ames_Housing(AmesHousing)",
"E:\\Anaconda3\\lib\\site-packages\\sklearn\\preprocessing\\data.py:323: DataConversionWarning: Data with input dtype int64, float64 were all converted to float64 by MinMaxScaler.\n return self.partial_fit(X, y)\n"
],
[
"data, labels = df.iloc[ : , 2: ].drop( columns=[ \"SalePrice\" ] ), df[ \"SalePrice\" ]",
"_____no_output_____"
],
[
"train_data, test_data, train_labels, test_labels = train_test_split(data, labels, test_size=0.2)",
"_____no_output_____"
],
[
"from libpy import FS",
"_____no_output_____"
],
[
"# train_data, train_labels, test_data, test_labels = FS.feature_select(df)",
"_____no_output_____"
],
[
"print(\"Training set: {}\".format(train_data.shape)) # 1607 examples, ** features\nprint(\"Testing set: {}\".format(test_data.shape)) # 1071 examples, 13 features",
"Training set: (2142, 210)\nTesting set: (536, 210)\n"
],
[
"train_data.sample(10)",
"_____no_output_____"
]
],
[
[
"### Model building",
"_____no_output_____"
],
[
"Here we created a neural network of our own. Trained it and ecaluted it's score to to measure performance. Later we plot our results.",
"_____no_output_____"
],
[
"### Train a model",
"_____no_output_____"
]
],
[
[
"from libpy import KR",
"_____no_output_____"
]
],
[
[
"Here we use default 64 node Neural Network",
"_____no_output_____"
]
],
[
[
"model = KR.build_model(train_data)\nmodel.summary()",
"_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\ndense (Dense) (None, 64) 13504 \n_________________________________________________________________\ndense_1 (Dense) (None, 64) 4160 \n_________________________________________________________________\ndense_2 (Dense) (None, 64) 4160 \n_________________________________________________________________\ndense_3 (Dense) (None, 1) 65 \n=================================================================\nTotal params: 21,889\nTrainable params: 21,889\nNon-trainable params: 0\n_________________________________________________________________\n"
],
[
"history, model = KR.train_model( model, train_data, train_labels )",
"\n....................................................................................................\n................................................................................................."
]
],
[
[
"### Plot",
"_____no_output_____"
],
[
"Here we plot our model performance. ",
"_____no_output_____"
]
],
[
[
"from matplotlib import pyplot",
"_____no_output_____"
],
[
"from libpy import DNN_plot",
"_____no_output_____"
],
[
"DNN_plot.plot_history(history)",
"_____no_output_____"
]
],
[
[
"We shown our Training vs Validation loss. Here we used tf.losses.mean_squared_error (mse) as loss aprameter and mean_absolute_error (mae) to plot our Training performaance",
"_____no_output_____"
],
[
"In below, we calculate the mae score to find test accuracy, aka our model's accuracy",
"_____no_output_____"
]
],
[
[
"[loss, mae] = model.evaluate(test_data, test_labels, verbose=0)\n\nprint(\"Testing set Mean Abs Error: ${:7.2f}\".format( mae ))",
"Testing set Mean Abs Error: $21458.43\n"
],
[
"test_predictions = model.predict(test_data).flatten()",
"_____no_output_____"
]
],
[
[
"Here we can see the Regression model ",
"_____no_output_____"
]
],
[
[
"DNN_plot.plot_predict( test_labels, test_predictions )",
"_____no_output_____"
]
],
[
[
"We can check how much error we get",
"_____no_output_____"
]
],
[
[
"DNN_plot.plot_predict_error(test_labels, test_predictions)",
"_____no_output_____"
]
],
[
[
"### Experiment Depth of Neural Network",
"_____no_output_____"
],
[
"We want to check with increase of Hidden layer, does our model performs better? We increased up to 7 hidden layer",
"_____no_output_____"
]
],
[
[
"from libpy import CV",
"_____no_output_____"
],
[
"depths = []\nscores_mae = []\n\nfor i in range( 7 ):\n model = KR.build_model(train_data, depth=i) \n \n history, model = KR.train_model( model, train_data, train_labels )\n model.summary()\n \n DNN_plot.plot_history(history)\n \n [loss, mae] = model.evaluate(test_data, test_labels, verbose=0)\n print(\"Testing set Mean Abs Error: ${:7.2f}\".format( mae ))\n \n test_predictions = model.predict(test_data).flatten()\n DNN_plot.plot_predict( test_labels, test_predictions )\n \n depths.append( i+2 )\n scores_mae.append( mae) ",
"\n.\n.\n.\n.\n.\n.\n......................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................\n.\n.\n.\n.\n.\n.\n......................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................\n.\n.\n.\n.\n.\n.\n......................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................\n.\n.\n.\n.\n.\n.\n......................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................\n.\n.\n.\n.\n.\n.\n......................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\ndense_24 (Dense) (None, 64) 13504 \n_________________________________________________________________\ndense_25 (Dense) (None, 1) 65 \n=================================================================\nTotal params: 13,569\nTrainable params: 13,569\nNon-trainable params: 0\n_________________________________________________________________\n"
],
[
"CV.plot_any( depths, scores_mae, xlabel='Depth', ylabel='Mean Abs Error [1000$]' )",
"_____no_output_____"
]
],
[
[
"### Experiment Overfitting",
"_____no_output_____"
],
[
"In this part, we try multiple neural network model, with various node number to check Overfitting vs Underfitting ",
"_____no_output_____"
]
],
[
[
"model_16 = KR.build_model(train_data, units=16)\nhistory_16, model_16 = KR.train_model( model_16, train_data, train_labels )\nmodel_16.summary()",
"_____no_output_____"
],
[
"loss, acc = model_16.evaluate( train_data, train_labels )\nprint(\"Trained model, accuracy: {:5.2f}%\".format( acc ))",
"_____no_output_____"
],
[
"model_32 = KR.build_model(train_data, units=32)\nhistory_32, model_32 = KR.train_model( model_32, train_data, train_labels )\nmodel_32.summary()",
"_____no_output_____"
],
[
"loss, acc = model_32.evaluate( train_data, train_labels )\nprint(\"Trained model, accuracy: {:5.2f}%\".format( acc))",
"_____no_output_____"
],
[
"model_48 = KR.build_model(train_data, units=48)\nhistory_48, model_48 = KR.train_model( model_48, train_data, train_labels )\nmodel_48.summary()",
"_____no_output_____"
],
[
"loss, acc = model_48.evaluate( train_data, train_labels )\nprint(\"Trained model, accuracy: {:5.2f}%\".format( acc))",
"_____no_output_____"
],
[
"model_64 = KR.build_model( train_data, units=64 )\nhistory_64, model_64 = KR.train_model( model_64, train_data, train_labels )\nmodel_64.summary()",
"_____no_output_____"
],
[
"loss, acc = model_64.evaluate( train_data, train_labels )\nprint(\"Trained model, accuracy: {:5.2f}%\".format( acc))",
"_____no_output_____"
],
[
"model_128 = KR.build_model( train_data, units=128)\nhistory_128, model_128 = KR.train_model( model_128, train_data, train_labels )\nmodel_128.summary()",
"_____no_output_____"
],
[
"loss, acc = model_128.evaluate( train_data, train_labels )\nprint(\"Trained model, accuracy: {:5.2f}%\".format( acc))",
"_____no_output_____"
],
[
"model_512 = KR.build_model(train_data, units=512)\nhistory_512, model_512 = KR.train_model( model_512, train_data, train_labels )\nmodel_512.summary()",
"_____no_output_____"
],
[
"loss, acc = model_512.evaluate( train_data, train_labels )\nprint(\"Trained model, accuracy: {:5.2f}%\".format( acc))",
"_____no_output_____"
],
[
"DNN_plot.plot_compare_history( [\n ('history_16', history_16 ),\n ('history_32', history_32 ),\n ('history_48', history_48 ),\n ('history_64', history_64 ),\n ('history_128', history_128 ),\n ('history_512', history_512 )\n] )",
"_____no_output_____"
]
],
[
[
"In our case, Validation and Training loss corrosponds to each other, all the models does not face any Overfitting or Underfitting. As we used EarlyStopping to stop training when val_loss stops to update. At the same time we used keras.regularizers.l2 to regularize our model.",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
cbeeaf067c186e658a6f0155dbf0ba567f50b5ae
| 4,874 |
ipynb
|
Jupyter Notebook
|
notebooks/elevation_indices.ipynb
|
Jiangchao3/pyflwdir
|
4fb72b6a2e60faae9b9d87b996aabce67b840b7e
|
[
"MIT"
] | 20 |
2021-09-27T19:19:39.000Z
|
2022-03-23T22:44:20.000Z
|
notebooks/elevation_indices.ipynb
|
Jiangchao3/pyflwdir
|
4fb72b6a2e60faae9b9d87b996aabce67b840b7e
|
[
"MIT"
] | 19 |
2021-09-24T20:48:49.000Z
|
2022-03-07T09:34:41.000Z
|
notebooks/elevation_indices.ipynb
|
Jiangchao3/pyflwdir
|
4fb72b6a2e60faae9b9d87b996aabce67b840b7e
|
[
"MIT"
] | 1 |
2022-01-18T07:25:17.000Z
|
2022-01-18T07:25:17.000Z
| 32.493333 | 655 | 0.60238 |
[
[
[
"# Elevation indices",
"_____no_output_____"
],
[
"Here we assume that flow directions are known. We read the flow direction raster data, including meta-data, using [rasterio](https://rasterio.readthedocs.io/en/latest/) and parse it to a pyflwdir `FlwDirRaster` object, see earlier examples for more background.",
"_____no_output_____"
]
],
[
[
"# import pyflwdir, some dependencies and convenience methods\nimport numpy as np\nimport rasterio\nimport pyflwdir\n\n# local convenience methods (see utils.py script in notebooks folder)\nfrom utils import quickplot, plt # data specific quick plot method\n\n# read and parse flow direciton data\nwith rasterio.open(\"rhine_d8.tif\", \"r\") as src:\n flwdir = src.read(1)\n crs = src.crs\n extent = np.array(src.bounds)[[0, 2, 1, 3]]\n flw = pyflwdir.from_array(\n flwdir,\n ftype=\"d8\",\n transform=src.transform,\n latlon=crs.is_geographic,\n cache=True,\n )\n# read elevation data\nwith rasterio.open(\"rhine_elv0.tif\", \"r\") as src:\n elevtn = src.read(1)",
"_____no_output_____"
]
],
[
[
"## height above nearest drain (HAND)",
"_____no_output_____"
],
[
"The [hand()](reference.rst#pyflwdir.FlwdirRaster.hand) method uses drainage-normalized topography and flowpaths to delineate the relative vertical distances (drop) to the nearest river (drain) as a proxy for the potential extent of flooding ([Nobre et al. 2016](https://doi.org/10.1002/hyp.10581)). The pyflwdir implementation requires stream mask `drain` and elevation raster `elevtn`. The stream mask is typically determined based on a threshold on [upstream_area()](reference.rst#pyflwdir.FlwdirRaster.upstream_area) or [stream_order()](reference.rst#pyflwdir.FlwdirRaster.stream_order), but can also be set from rasterizing a vector stream file.",
"_____no_output_____"
]
],
[
[
"# first we derive the upstream area map\nuparea = flw.upstream_area(\"km2\")",
"_____no_output_____"
],
[
"# HAND based on streams defined by a minimal upstream area of 1000 km2\nhand = flw.hand(drain=uparea > 1000, elevtn=elevtn)\n# plot\nax = quickplot(title=\"Height above nearest drain (HAND)\")\nim = ax.imshow(\n np.ma.masked_equal(hand, -9999),\n extent=extent,\n cmap=\"gist_earth_r\",\n alpha=0.5,\n vmin=0,\n vmax=150,\n)\nfig = plt.gcf()\ncax = fig.add_axes([0.82, 0.37, 0.02, 0.12])\nfig.colorbar(im, cax=cax, orientation=\"vertical\")\ncax.set_ylabel(\"HAND [m]\")\nplt.savefig(\"hand.png\")",
"_____no_output_____"
]
],
[
[
"## Floodplains",
"_____no_output_____"
],
[
"The [floodplains()](reference.rst#pyflwdir.FlwdirRaster.floodplains) method delineates geomorphic floodplain boundaries based on a power-law relation between upstream area and a maximum HAND contour as developed by [Nardi et al (2019)](http://www.doi.org/10.1038/sdata.2018.309). Here, streams are defined based on a minimum upstream area threshold `upa_min` and floodplains on the scaling parameter `b` of the power-law relationship.",
"_____no_output_____"
]
],
[
[
"floodplains = flw.floodplains(elevtn=elevtn, uparea=uparea, upa_min=1000)\n# plot\nfloodmap = (floodplains, -1, dict(cmap=\"Blues\", alpha=0.5, vmin=0))\nax = quickplot(\n raster=floodmap, title=\"Geomorphic floodplains\", filename=\"flw_floodplain\"\n)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
]
] |
cbeecc0b5072ba6ed02483d39d0be08e9f798653
| 46,354 |
ipynb
|
Jupyter Notebook
|
assignments/00_Basics/D1-exercises.ipynb
|
luschu/P1.4_seed
|
1c44f6f54f0544faac97cc73ddd447f652a1ef71
|
[
"CC-BY-4.0"
] | null | null | null |
assignments/00_Basics/D1-exercises.ipynb
|
luschu/P1.4_seed
|
1c44f6f54f0544faac97cc73ddd447f652a1ef71
|
[
"CC-BY-4.0"
] | null | null | null |
assignments/00_Basics/D1-exercises.ipynb
|
luschu/P1.4_seed
|
1c44f6f54f0544faac97cc73ddd447f652a1ef71
|
[
"CC-BY-4.0"
] | null | null | null | 31.026774 | 381 | 0.554364 |
[
[
[
"# Pure Python evaluation of vector norms\n\nGenerate a list of random floats of a given dimension (dim), and store its result in the variable `vec`.",
"_____no_output_____"
]
],
[
[
"# This is used for plots and numpy\n%pylab inline \n\n#import random\ndim = int(1000)\n\n# YOUR CODE HERE\n#vec = []\n#[vec.append(random.random() for i in range(dim))]\n#print(vec)\nvec = random.random(dim)\nvec\n#raise NotImplementedError()",
"Populating the interactive namespace from numpy and matplotlib\n"
],
[
"from numpy.testing import *\n\nassert_equal(type(vec), list)\nassert_equal(len(vec), dim)\nfor ob in vec:\n assert_equal(type(ob), float)",
"_____no_output_____"
]
],
[
[
"Write a function that evaluates the $l_p$ norm of a vector in $R^d$. We remind:\n$$\n\\|v \\|_{p} := \\left(\\sum_i (v_i)^p\\right)^{1/p}\n$$\n\nthe function should take as arguments a `list`, containing your $R^d$ vector, and a number `p` in the range $[1, \\infty]$, indicating the exponent of the norm. \n\n**Note:** an infinite float number is given by `float(\"inf\")`.\n\nThrow an assertion (look it up on google!) if the exponent is not in the range you expect.",
"_____no_output_____"
]
],
[
[
"def p_norm(vector,p):\n # YOUR CODE HERE\n raise NotImplementedError()",
"_____no_output_____"
],
[
"assert_equal(p_norm(range(10),1), 45.0)\nassert_equal(p_norm([3,4], 2), 5.0)",
"_____no_output_____"
],
[
"assert_equal(p_norm([-1,-.5,.5], float(\"inf\")), 1)",
"_____no_output_____"
],
[
"assert_raises(AssertionError, p_norm, [2,3], 0)\nassert_raises(AssertionError, p_norm, [2,3], -1)",
"_____no_output_____"
]
],
[
[
"# Playing with condition numbers\n\n\nIn this exercise you will have to figure out what are the optimal\nvalues of the stepping interval when approximating derivatives using\nthe finite difference method. See here_ for a short introduction on\nhow to run these programs on SISSA machines.\n\n## 1. Finite differences\nWrite a program to compute the finite difference (`FD`)\napproximation of the derivative of a function `f`, computed at\npoint `x`, using a stepping of size `h`. Recall the definition of\napproximate derivative:\n\n$$\nFD(f,x,h) := \\frac{f(x+h)-f(x)}{h}\n$$",
"_____no_output_____"
]
],
[
[
"def FD(f, x, h):\n # YOUR CODE HERE\n raise NotImplementedError()",
"_____no_output_____"
],
[
"assert_equal(FD(lambda x: x, 0, .125), 1.0)",
"_____no_output_____"
]
],
[
[
"## 2. Compute FD\n\nEvaluate this function for the derivative of `sin(x)` evaluated at `x=1`, for values of `h` equal to `1e-i`, with `i=0,...,20`. Store the values of the finite differences in the list `fd1`.",
"_____no_output_____"
]
],
[
[
"# YOUR CODE HERE\nraise NotImplementedError()",
"_____no_output_____"
],
[
"assert_equal(len(fd1), 21)\nexpected = [0.067826442017785205, 0.49736375253538911, 0.53608598101186899, 0.5398814803603269, 0.54026023141862112, 0.54029809850586474, 0.54030188512133037, 0.54030226404044868, 0.54030229179602429, 0.54030235840940577, 0.54030224738710331, 0.54030113716407868, 0.54034554608506369, 0.53956838996782608, 0.53290705182007514, 0.55511151231257827, 0.0, 0.0, 0.0, 0.0, 0.0]\nassert_almost_equal(fd1,expected,decimal=4)",
"_____no_output_____"
]
],
[
[
"## 3. Error plots\n\nPlot the error, defined as `abs(FD-cos(1.0))` where `FD` is your approximation, in `loglog` format and explain what you see. A good way to emphasize the result is to give the option `'-o'` to the plot command.",
"_____no_output_____"
]
],
[
[
"# YOUR CODE HERE\nraise NotImplementedError()",
"_____no_output_____"
]
],
[
[
"YOUR ANSWER HERE",
"_____no_output_____"
],
[
"## 4. Error plots base 2\nRepeate step 2 and 3 above, but using powers of `2` instead of powers of `10`, i.e., using `h` equal to `2**(-i)` for `i=1,...,60`. Do you see differences? How do you explain these differences? Shortly comment. A good way to emphasize the result is to give the option `'-o'` to the plot command.",
"_____no_output_____"
],
[
"YOUR ANSWER HERE",
"_____no_output_____"
],
[
"YOUR ANSWER HERE",
"_____no_output_____"
],
[
"## 5. Central Finite Differences\nWrite a function that computes the central finite difference approximation (`CFD`), defined as \n\n$$\nCFD(f,x,h) := \\frac{f(x+h)-f(x-h)}{2h}\n$$\n",
"_____no_output_____"
]
],
[
[
"def CFD(f, x, h):\n # YOUR CODE HERE\n raise NotImplementedError()",
"_____no_output_____"
],
[
"assert_equal(CFD(lambda x: x**2, 0.0, .5), 0.0)\nassert_equal(CFD(lambda x: x**2, 1.0, .5), 2.0)",
"_____no_output_____"
]
],
[
[
"## 6. Error plots for CFD\n\nRepeat steps 2., 3. and 4. and explain what you see. What is the *order* of the approximation 1. and what is the order of the approximation 5.? What's the order of the cancellation errors? ",
"_____no_output_____"
]
],
[
[
"# YOUR CODE HERE\nraise NotImplementedError()",
"_____no_output_____"
]
],
[
[
"YOUR ANSWER HERE",
"_____no_output_____"
],
[
"# Numpy\n\nNumpy provides a very powerful array container. The first line of this ipython notebook has imported all of numpy functionalities in your notebook, just as if you typed::\n\n from numpy import *\n \nCreate a numpy array whith entries that range form 0 to 64. Use the correct numpy function to do so. Call it `x`. ",
"_____no_output_____"
]
],
[
[
"# YOUR CODE HERE\nraise NotImplementedError()",
"_____no_output_____"
],
[
"assert_equal(type(x), ndarray)\nassert_equal(len(x), 64)\nfor i in xrange(64):\n assert_equal(x[i], float(i))",
"_____no_output_____"
]
],
[
[
"Reshape the one dimensional array, to become a 4 rows 2 dimensional array, let numpy evaluate the correct number of culumns. Call it `y`.",
"_____no_output_____"
]
],
[
[
"# YOUR CODE HERE\nraise NotImplementedError()",
"_____no_output_____"
],
[
"assert_equal(shape(y), (4,16))",
"_____no_output_____"
]
],
[
[
"Get the following *slices* of `y`:\n\n* All the rows and the first three colums. Name it `sl1`.\n* All the colums and the first three rows. Name it `sl2`.\n* Third to sixth (included) columns and all the rows. Name it `sl3`.\n* The last three columns and all the rows. Name it `sl4`.",
"_____no_output_____"
]
],
[
[
"# YOUR CODE HERE\nraise NotImplementedError()",
"_____no_output_____"
],
[
"assert_equal(sl1,[[0,1,2],[16,17,18],[32,33,34],[48,49,50]])\nassert_equal(sl2,[[0,1,2,3,4,5,6,7,8,9,10,11,12,13,14,15],[16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31],[32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47]])\nassert_equal(sl3,[[3,4,5,6],[19,20,21,22],[35,36,37,38],[51,52,53,54]])\nassert_equal(sl4,[[13,14,15],[29,30,31],[45,46,47],[61,62,63]])",
"_____no_output_____"
]
],
[
[
"Now reshape the array, as if you wanted to feed it to a fortran routine. Call it `z`.",
"_____no_output_____"
]
],
[
[
"# YOUR CODE HERE\nraise NotImplementedError()",
"_____no_output_____"
]
],
[
[
"Comment on the result, what has changed with respect to `y`? ",
"_____no_output_____"
],
[
"YOUR ANSWER HERE",
"_____no_output_____"
],
[
"Set the fourth element of `x` to 666666, and print `x`, `y`, `z`. Comment on the result",
"_____no_output_____"
]
],
[
[
"# YOUR CODE HERE\nraise NotImplementedError()",
"_____no_output_____"
]
],
[
[
"YOUR ANSWER HERE",
"_____no_output_____"
],
[
"## Arrays and Matrices\n\nDefine 2 arrays, `A` of dimensions (2,3) and `B` of dimension (3,4).\n\n* Perform the operation `C = A.dot(B)`. Comment the result, or the error you get.",
"_____no_output_____"
]
],
[
[
"# YOUR CODE HERE\nraise NotImplementedError()",
"_____no_output_____"
],
[
"assert_equal(A.shape,(2,3))\nassert_equal(B.shape,(3,4))\nassert_equal(C.shape,(2,4))\nexpected = sum(A[1,:]*B[:,2])\nassert_equal(C[1,2],expected)",
"_____no_output_____"
]
],
[
[
"YOUR ANSWER HERE",
"_____no_output_____"
],
[
"* Perform the operation `C = A*(B)`. Comment the result, or the error you get.",
"_____no_output_____"
]
],
[
[
"C = A*B",
"_____no_output_____"
]
],
[
[
"YOUR ANSWER HERE",
"_____no_output_____"
],
[
" * Convert A and B, from arrays to matrices and perform `A*B`. Comment the result.",
"_____no_output_____"
]
],
[
[
"# YOUR CODE HERE\nraise NotImplementedError()",
"_____no_output_____"
],
[
"assert_equal(type(A),numpy.matrixlib.defmatrix.matrix)\nassert_equal(type(B),numpy.matrixlib.defmatrix.matrix)\nassert_equal(type(C),numpy.matrixlib.defmatrix.matrix)\nassert_equal(A.shape,(2,3))\nassert_equal(B.shape,(3,4))\nassert_equal(C.shape,(2,4))\nexpected = sum(A[1,:]*B[:,2])\nassert_equal(C[1,2],expected)",
"_____no_output_____"
]
],
[
[
"YOUR ANSWER HERE",
"_____no_output_____"
],
[
"# Playing with polynomials\n\nThe polynomial `(1-x)^6` can be expanded to::\n \n x^6 - 6*x^5 + 15*x^4 - 20*x^3 + 15*x^2 - 6*x + 1\n\n The two forms above are equivalent from a mathematical point of\n view, but may yield different results in a computer machine.\n\n Compute and plot the values of this polynomial, using each of the\n two forms, for 101 equally spaced points in the interval\n `[0.995,1.005]`, i.e., with a spacing of 0.0001 (use linspace).\n \n Can you explain this behavior?\n",
"_____no_output_____"
]
],
[
[
"# YOUR CODE HERE\nraise NotImplementedError()",
"_____no_output_____"
]
],
[
[
"YOUR ANSWER HERE",
"_____no_output_____"
],
[
"**Playing with interpolation in python**\n\n1. Given a set of $n+1$ points $x_i$ as input (either a list of floats, or a numpy array of floats), construct a function `lagrange_basis(xi,i,x)` that returns the $i$-th Lagrange\npolynomial associated to $x_i$, evaluated at $x$. The $i$-th Lagrange polynomial is defined as polynomial of degree $n$ such that $l_i(x_j) = \\delta_{ij}$, where $\\delta$ is one if $i == j$ and zero otherwise.\n \nRecall the mathematical definition of the $l_i(x)$ polynomials:\n\n$$\nl_i(x) := \\prod_{j=0, j\\neq i}^{n} \\frac{x-x_j}{x_i-x_j}\n$$\n",
"_____no_output_____"
]
],
[
[
"def lagrange_basis(xi, i, x):\n # YOUR CODE HERE\n raise NotImplementedError()",
"_____no_output_____"
],
[
"x = linspace(0,1,5)\nd = 3\nxi = linspace(0,1,d)\n\nassert_equal(list(lagrange_basis(xi, 0, x)),[1.0, 0.375, -0.0, -0.125, 0.0])\nassert_equal(list(lagrange_basis(xi, 1, x)),[0.0, 0.75, 1.0, 0.75, -0.0])\nassert_equal(list(lagrange_basis(xi, 2, x)),[-0.0, -0.125, 0.0, 0.375, 1.0])\n\nassert_raises(AssertionError, lagrange_basis, xi, -1, x)\nassert_raises(AssertionError, lagrange_basis, xi, 10, x)",
"_____no_output_____"
]
],
[
[
"Construct the function `lagrange_interpolation(xi,g)` that, given the set of interpolation points `xi` and a function `g`, it returns **another function** that when evaluated at **x** returns the Lagrange interpolation polynomial of `g` defined as \n\n$$\n \\mathcal{L} g(x) := \\sum_{i=0}^n g(x_i) l_i(x)\n$$\n\nYou could use this function as follows::\n\n Lg = lagrange_interpolation(xi, g)\n xi = linspace(0,1,101)\n plot(x, g(x))\n plot(x, Lg(x))\n plot(xi, g(xi), 'or')",
"_____no_output_____"
]
],
[
[
"def lagrange_interpolation(xi,f):\n # YOUR CODE HERE\n raise NotImplementedError()",
"_____no_output_____"
],
[
"# Check for polynomials. This should be **exact**\ng = lambda x: x**3+x**2\n\nxi = linspace(0,1,4)\nLg = lagrange_interpolation(xi, g)\n\nx = linspace(0,1,1001)\n\nassert p_norm(g(x) - Lg(x),float('inf')) < 1e-15, 'This should be zero...'",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
cbeed8888e2e4584654e60ac579f08b0a5e0750c
| 38,980 |
ipynb
|
Jupyter Notebook
|
notebooks/000. numpy.ipynb
|
gillouche/exploratory-data-analysis
|
b2e2361ffd2eccbdbcfe72b9f223a7f06cc9d064
|
[
"MIT"
] | null | null | null |
notebooks/000. numpy.ipynb
|
gillouche/exploratory-data-analysis
|
b2e2361ffd2eccbdbcfe72b9f223a7f06cc9d064
|
[
"MIT"
] | 2 |
2021-06-08T21:43:25.000Z
|
2021-12-13T20:41:55.000Z
|
notebooks/000. numpy.ipynb
|
gillouche/exploratory-data-analysis
|
b2e2361ffd2eccbdbcfe72b9f223a7f06cc9d064
|
[
"MIT"
] | null | null | null | 17.582318 | 107 | 0.421036 |
[
[
[
"# Numpy",
"_____no_output_____"
]
],
[
[
"import numpy as np",
"_____no_output_____"
]
],
[
[
"## Create numpy arrays",
"_____no_output_____"
]
],
[
[
"np.array([1, 2]).shape",
"_____no_output_____"
],
[
"np.array([ [1, 2], [3, 4] ]).shape",
"_____no_output_____"
],
[
"np.array([ [1, 2], [3, 4], [5, 6] ]).shape",
"_____no_output_____"
],
[
"np.zeros((2, 2))",
"_____no_output_____"
],
[
"np.ones((2, 2))",
"_____no_output_____"
],
[
"np.full((2, 2), 5)",
"_____no_output_____"
],
[
"np.eye(3)",
"_____no_output_____"
]
],
[
[
"## Generate data",
"_____no_output_____"
]
],
[
[
"np.random.random()",
"_____no_output_____"
],
[
"np.random.randint(0, 10)",
"_____no_output_____"
],
[
"lower_bound_value = 0\nupper_bound_value = 100\nnum_rows = 1000\nnum_cols = 50\nA = np.random.randint(lower_bound_value, upper_bound_value, size=(num_rows, num_cols))\nA",
"_____no_output_____"
],
[
"A.shape",
"_____no_output_____"
],
[
"A.min()",
"_____no_output_____"
],
[
"A.max()",
"_____no_output_____"
],
[
"v = np.random.uniform(size=4)\nv",
"_____no_output_____"
],
[
"np.random.choice(v)",
"_____no_output_____"
],
[
"np.random.choice(10, size=(3, 3))",
"_____no_output_____"
],
[
"np.random.normal(size=4)",
"_____no_output_____"
],
[
"# gaussian (normal) distribution, mean = 0 and variance = 1\nnp.random.randn(2, 3)",
"_____no_output_____"
]
],
[
[
"## Numpy operations",
"_____no_output_____"
]
],
[
[
"array = np.array([ [1, 2, 3], [4, 5, 6], [7, 8, 9] ])\narray",
"_____no_output_____"
],
[
"array[:]",
"_____no_output_____"
],
[
"array[0]",
"_____no_output_____"
],
[
"array[2]",
"_____no_output_____"
],
[
"array[:, 0]",
"_____no_output_____"
],
[
"array[:, 2:]",
"_____no_output_____"
],
[
"array[1, 1]",
"_____no_output_____"
],
[
"array[-1, -1]",
"_____no_output_____"
],
[
"array_2 = np.concatenate([array, np.array([ [10, 11, 12] ])])\narray_2",
"_____no_output_____"
],
[
"array_2[0, 0] = 0\narray_2",
"_____no_output_____"
]
],
[
[
"## Vectors, Matrices arithmetic and linear systems",
"_____no_output_____"
]
],
[
[
"array_1 = np.array([1, 2])\narray_1",
"_____no_output_____"
],
[
"array_2 = np.array([3, 4])\narray_2",
"_____no_output_____"
],
[
"array_1 + array_2",
"_____no_output_____"
],
[
"array_1 - array_2",
"_____no_output_____"
],
[
"array_1 * 2",
"_____no_output_____"
],
[
"array_1 ** 3",
"_____no_output_____"
],
[
"array_1 * array_2",
"_____no_output_____"
],
[
"np.dot(array_1, array_2)",
"_____no_output_____"
],
[
"mat_1 = np.array([ [1, 2, 3], [4, 5, 6] ])\nmat_1",
"_____no_output_____"
],
[
"mat_1.T",
"_____no_output_____"
],
[
"mat_2 = np.array([ [1, 2, 3], [4, 5, 6] ])\nmat_2",
"_____no_output_____"
],
[
"mat_1 * 10",
"_____no_output_____"
],
[
"mat_1 * mat_2",
"_____no_output_____"
],
[
"np.dot(mat_1, mat_2.T)",
"_____no_output_____"
],
[
"mat_1 @ mat_2.T",
"_____no_output_____"
],
[
"np.linalg.inv(np.eye(3))",
"_____no_output_____"
],
[
"mat_3 = np.matrix([ [1, 2], [3, 4] ])\nmat_3",
"_____no_output_____"
],
[
"np.linalg.det(mat_3)",
"_____no_output_____"
],
[
"np.linalg.inv(mat_3)",
"_____no_output_____"
],
[
"np.linalg.inv(mat_3).dot(mat_3)",
"_____no_output_____"
],
[
"np.trace(mat_3)",
"_____no_output_____"
],
[
"np.diag(mat_3)",
"_____no_output_____"
],
[
"np.diag([1, 4])",
"_____no_output_____"
]
],
[
[
"$$ a^T b = \\vert\\vert a \\vert\\vert \\vert\\vert b \\vert\\vert \\cos(\\theta) $$\n\n$$ \\cos \\theta_{ab} = \\frac{a^T b}{ \\vert\\vert a \\vert\\vert \\vert\\vert b \\vert\\vert} $$\n\n$$ \\vert\\vert a \\vert\\vert = \\sqrt{ \\sum_{d=1}^{D} a^2_{d} } $$ ",
"_____no_output_____"
]
],
[
[
"a = np.array([1, 2])\nb = np.array([3, 4])",
"_____no_output_____"
],
[
"a_mag = np.sqrt((a * a).sum())\na_mag",
"_____no_output_____"
],
[
"np.linalg.norm(a)",
"_____no_output_____"
],
[
"cos_theta = a.dot(b) / (np.linalg.norm(a) * np.linalg.norm(b))\ncos_theta",
"_____no_output_____"
],
[
"angle = np.arccos(cos_theta)\nangle",
"_____no_output_____"
]
],
[
[
"## Eigen vectors and eigen values",
"_____no_output_____"
]
],
[
[
"A = np.matrix([ [1, 2], [3, 4] ])",
"_____no_output_____"
],
[
"eig_values, eig_vectors = np.linalg.eig(A)",
"_____no_output_____"
],
[
"eig_values",
"_____no_output_____"
],
[
"eig_vectors",
"_____no_output_____"
],
[
"eig_vectors[:, 0] * eig_values[0]",
"_____no_output_____"
],
[
"A @ eig_vectors[:, 0]",
"_____no_output_____"
],
[
"# not true true because of numerical precision, need to use np.allclose\neig_vectors[:, 0] * eig_values[0] == A @ eig_vectors[:, 0]",
"_____no_output_____"
],
[
"np.allclose(eig_vectors[:, 0] * eig_values[0], A @ eig_vectors[:, 0])",
"_____no_output_____"
],
[
"# check all\nnp.allclose(eig_vectors @ np.diag(eig_values), A @ eig_vectors)",
"_____no_output_____"
]
],
[
[
"## Broadcasting\n\nperform arithmetic operations on different shaped arrays\n\nsmaller array is broadcast across the larger array to ensure shape consistency\n\nRules:\n\n* one dimension (either column or row) should have the same dimension for both arrays\n* the lower dimension array should be a 1d array",
"_____no_output_____"
]
],
[
[
"mat_1 = np.arange(20).reshape(5, 4)\nmat_1",
"_____no_output_____"
],
[
"mat_2 = np.arange(5) # 1x5 cannot be added to mat_1\nmat_3 = mat_2.reshape(5, 1)\nmat_3",
"_____no_output_____"
],
[
"mat_1 + mat_3",
"_____no_output_____"
],
[
"mat_1 * mat_3",
"_____no_output_____"
]
],
[
[
"## Solve equations",
"_____no_output_____"
]
],
[
[
"mat_1 = np.array([ [2, 1], [1, -1] ])\nmat_1",
"_____no_output_____"
],
[
"array = np.array([4, -1])\narray",
"_____no_output_____"
],
[
"%%time\nnp.linalg.inv(mat_1).dot(array)",
"CPU times: user 121 µs, sys: 155 µs, total: 276 µs\nWall time: 215 µs\n"
],
[
"%%time\nnp.linalg.solve(mat_1, array)",
"CPU times: user 89 µs, sys: 114 µs, total: 203 µs\nWall time: 94.7 µs\n"
],
[
"inv_mat_1 = np.linalg.inv(mat_1)\ninv_mat_1",
"_____no_output_____"
],
[
"inv_mat_1.dot(array)",
"_____no_output_____"
],
[
"mat_1 = np.array([ [1, 2, 3], [4, 5, 2], [2, 8, 5] ])\nmat_1",
"_____no_output_____"
],
[
"array = np.array([5, 10, 15])\narray",
"_____no_output_____"
],
[
"np.linalg.solve(mat_1, array)",
"_____no_output_____"
]
],
[
[
"## Statistical operations",
"_____no_output_____"
]
],
[
[
"mat_1 = np.array([ [1, 2, 3, 4], [3, 4, 5, 6], [7, 8, 9, 6], [12, 7, 10, 9], [2, 11, 8, 10] ])\nmat_1",
"_____no_output_____"
],
[
"mat_1.sum()",
"_____no_output_____"
],
[
"np.sum(mat_1)",
"_____no_output_____"
],
[
"mat_1.sum(axis=0) # column wise sum",
"_____no_output_____"
],
[
"mat_1.sum(axis=1) # row wise sum",
"_____no_output_____"
],
[
"mat_1.mean()",
"_____no_output_____"
],
[
"mat_1.mean(axis=0)",
"_____no_output_____"
],
[
"mat_1.mean(axis=1)",
"_____no_output_____"
],
[
"np.median(mat_1)",
"_____no_output_____"
],
[
"np.median(mat_1, axis=0)",
"_____no_output_____"
],
[
"np.std(mat_1, axis=1)",
"_____no_output_____"
],
[
"np.std(mat_1)",
"_____no_output_____"
],
[
"# percentile: value below which a given percentage of observations can be found\npercentile = [25, 50, 75]\nfor p in percentile:\n print(f'Percentile {p}: {np.percentile(mat_1, p, axis=1)}')",
"Percentile 25: [1.75 3.75 6.75 8.5 6.5 ]\nPercentile 50: [2.5 4.5 7.5 9.5 9. ]\nPercentile 75: [ 3.25 5.25 8.25 10.5 10.25]\n"
],
[
"R = np.random.randn(10_000)",
"_____no_output_____"
],
[
"R.mean()",
"_____no_output_____"
],
[
"R.var()",
"_____no_output_____"
],
[
"R.std()",
"_____no_output_____"
],
[
"np.sqrt(R.var())",
"_____no_output_____"
],
[
"R = np.random.randn(10_000, 3)\nR.mean(axis=0).shape",
"_____no_output_____"
],
[
"R.mean(axis=1).shape",
"_____no_output_____"
],
[
"np.cov(R).shape",
"_____no_output_____"
],
[
"np.cov(R.T)",
"_____no_output_____"
],
[
"np.cov(R, rowvar=False)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbeed9f1ec2bdb42db86f3419eae4d5ef81ee51d
| 466,891 |
ipynb
|
Jupyter Notebook
|
urban-heat-islands.ipynb
|
thomasdubdub/urban-heat-islands
|
ff0dfd2a111fc824d27b826b333fed544c0011d2
|
[
"MIT"
] | null | null | null |
urban-heat-islands.ipynb
|
thomasdubdub/urban-heat-islands
|
ff0dfd2a111fc824d27b826b333fed544c0011d2
|
[
"MIT"
] | null | null | null |
urban-heat-islands.ipynb
|
thomasdubdub/urban-heat-islands
|
ff0dfd2a111fc824d27b826b333fed544c0011d2
|
[
"MIT"
] | 1 |
2020-03-10T01:26:31.000Z
|
2020-03-10T01:26:31.000Z
| 1,978.351695 | 459,976 | 0.959791 |
[
[
[
"# Visualize Urban Heat Islands (UHI) in Toulouse - France",
"_____no_output_____"
],
[
"#### <br> Data from meteo stations can be downloaded on the French open source portal https://www.data.gouv.fr/",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\nimport matplotlib.pyplot as plt\nimport pandas as pd\nimport string\nfrom glob import glob\nfrom matplotlib.dates import DateFormatter\nfrom ipyleaflet import Map, Marker, basemaps, basemap_to_tiles\nfrom ipywidgets import Layout",
"_____no_output_____"
],
[
"met_files_folder = 'station-meteo' # Folder where to put meteo data files\nlegend_file = 'stations-meteo-en-place.csv' # File listing all meteo stations\nstart_date = '2019-06-27'\nend_date = '2019-06-27'\ntoulouse_center = (43.60426, 1.44367)\ndefault_zoom = 12",
"_____no_output_____"
]
],
[
[
"#### <br> Parse file listing all met stations ",
"_____no_output_____"
]
],
[
[
"leg = pd.read_csv(legend_file, sep=';')\n\ndef get_legend(id):\n return leg.loc[leg['FID']==id]['Nom_Station'].values[0]\n\ndef get_lon(id):\n return leg.loc[leg['FID']==id]['x'].values[0]\n\ndef get_lat(id):\n return leg.loc[leg['FID']==id]['y'].values[0]",
"_____no_output_____"
]
],
[
[
"#### <br> Build a Pandas dataframe from a met file",
"_____no_output_____"
]
],
[
[
"def get_table(file):\n df = pd.read_csv(file, sep=';')\n df.columns = list(string.ascii_lowercase)[:17]\n df['id'] = df['b']\n df['annee'] = df['e'] + 2019\n df['heure'] = (df['f'] - 1) * 15 // 60\n df['minute'] = 1 + (df['f'] - 1) * 15 % 60\n df = df.loc[df['g'] > 0] # temperature field null\n df['temperature'] = df['g'] - 50 + df['h'] / 10\n df['pluie'] = df['j'] * 0.2 # auget to mm\n df['vent_dir'] = df['k'] * 2\n df['vent_force'] = df['l'] # en dessous de 80 pareil au dessus / 2 ?\n df['pression'] = df['m'] + 900\n df['vent_max_dir'] = df['n'] * 22.5\n df['vent_max_force'] = df['o'] # en dessous de 80 pareil au dessus / 2 ?\n df['pluie_plus_intense'] = df['p'] * 0.2\n df['date'] = df['annee'].map(str) + '-' + df['c'].map(str) + '-' + df['d'].map(str) \\\n + ':' + df['heure'].map(str) + '-' + df['minute'].map(str)\n df['date'] = pd.to_datetime(df['date'], format='%Y-%m-%d:%H-%M')\n df = df[['date','id','temperature','pression','pluie','pluie_plus_intense','vent_dir', \\\n 'vent_force','vent_max_dir','vent_max_force']]\n df.set_index('date', inplace=True)\n df = df.loc[start_date:end_date]\n return df",
"_____no_output_____"
]
],
[
[
"#### <br> Parse met files (in met_files_folder)",
"_____no_output_____"
]
],
[
[
"table_list = []\nfor file in glob(met_files_folder + '/*.csv'):\n table_list.append(get_table(file))\ntables = [table for table in table_list if not table.empty]\nlegs = [get_legend(table['id'].iloc[0]) for table in tables]\nlats = [get_lat(table['id'].iloc[0]) for table in tables]\nlongs = [get_lon(table['id'].iloc[0]) for table in tables]\nprint('Number of meteo stations with available recordings for this time period: {}'.format(len(legs)))\nprint(legs)",
"Number of meteo stations with available recordings for this time period: 21\n['Nakache', 'Toulouse_Cote_Pavee', 'Toulouse_Paul_Sabatier', 'Toulouse_Carmes', 'Toulouse_parc_japonais', 'Basso_Cambo', 'Toulouse_Lardenne', 'Marengo', 'Montaudran', 'Busca', 'La_Salade', 'Pech_David', 'Avenue_Grde_Bretagne', 'Soupetard', 'Toulouse_parc_Jardin_Plantes', 'Meteopole', 'Toulouse_Cyprien', 'Castelginest', 'Toulouse_Canceropole', 'Valade', 'Colomiers_ZA_Perget']\n"
]
],
[
[
"#### <br> Plot all met stations around Toulouse",
"_____no_output_____"
]
],
[
[
"m = Map(center=toulouse_center, zoom=default_zoom, layout=Layout(width='100%', height='500px'))\nfor i in range(len(legs)):\n m.add_layer(Marker(location=(lats[i], longs[i]), draggable=False, title=legs[i]))\nm",
"_____no_output_____"
]
],
[
[
"#### <br> Plot temperature chart for all met stations",
"_____no_output_____"
]
],
[
[
"ax = tables[0]['temperature'].plot(grid=True, figsize=[25,17])\nfor i in range(1, len(tables)):\n tables[i]['temperature'].plot(grid=True, ax=ax)\nax.legend(legs)\nax.xaxis.set_major_formatter(DateFormatter('%H:%M'))\nax.set_xlabel('Temperatures of ' + start_date)\nplt.savefig('temperatures.png')",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.