hexsha
stringlengths 40
40
| size
int64 6
14.9M
| ext
stringclasses 1
value | lang
stringclasses 1
value | max_stars_repo_path
stringlengths 6
260
| max_stars_repo_name
stringlengths 6
119
| max_stars_repo_head_hexsha
stringlengths 40
41
| max_stars_repo_licenses
list | max_stars_count
int64 1
191k
⌀ | max_stars_repo_stars_event_min_datetime
stringlengths 24
24
⌀ | max_stars_repo_stars_event_max_datetime
stringlengths 24
24
⌀ | max_issues_repo_path
stringlengths 6
260
| max_issues_repo_name
stringlengths 6
119
| max_issues_repo_head_hexsha
stringlengths 40
41
| max_issues_repo_licenses
list | max_issues_count
int64 1
67k
⌀ | max_issues_repo_issues_event_min_datetime
stringlengths 24
24
⌀ | max_issues_repo_issues_event_max_datetime
stringlengths 24
24
⌀ | max_forks_repo_path
stringlengths 6
260
| max_forks_repo_name
stringlengths 6
119
| max_forks_repo_head_hexsha
stringlengths 40
41
| max_forks_repo_licenses
list | max_forks_count
int64 1
105k
⌀ | max_forks_repo_forks_event_min_datetime
stringlengths 24
24
⌀ | max_forks_repo_forks_event_max_datetime
stringlengths 24
24
⌀ | avg_line_length
float64 2
1.04M
| max_line_length
int64 2
11.2M
| alphanum_fraction
float64 0
1
| cells
list | cell_types
list | cell_type_groups
list |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
c50dd6a15153d3b1cf54a9fd2142f9c4324d2445
| 10,717 |
ipynb
|
Jupyter Notebook
|
runge-kutta-mv-pt2.ipynb
|
npurtell/astr-119-session-13
|
5350fc02ec1fcda3a919556a9a962ae4ee49ca48
|
[
"MIT"
] | null | null | null |
runge-kutta-mv-pt2.ipynb
|
npurtell/astr-119-session-13
|
5350fc02ec1fcda3a919556a9a962ae4ee49ca48
|
[
"MIT"
] | null | null | null |
runge-kutta-mv-pt2.ipynb
|
npurtell/astr-119-session-13
|
5350fc02ec1fcda3a919556a9a962ae4ee49ca48
|
[
"MIT"
] | null | null | null | 28.65508 | 107 | 0.454325 |
[
[
[
"%matplotlib inline\nimport matplotlib.pyplot as plt\nimport numpy as np",
"_____no_output_____"
],
[
"def dydx(x,y):\n #Set derivatives\n #y is an array, 2D array\n #1 dimension is x, other one is 1D with y and x elements\n #our eqn is d^2y/dx^2 = -y\n #so we can write dydz=z, dzdx=-y\n #we will set y = y[0], z = y[1]\n #declare an array\n \n y_derivs = np.zeros(2)\n \n #set dydx=z\n y_derivs[0] = y[1]\n \n #set dxdx = -y\n y_derivs[1] = -1*y[0]\n \n #here we have an array\n return y_derivs",
"_____no_output_____"
]
],
[
[
"defined coupled derivatives to integrate above, 4th order runge kutta method below",
"_____no_output_____"
]
],
[
[
"def rk4_mv_core(dydx,xi,yi,nv,h):\n #declare k? arrays\n k1 = np.zeros(nv)\n k2 = np.zeros(nv)\n k3 = np.zeros(nv)\n k4 = np.zeros(nv)\n #nv is number of variables\n #? is wild card, it can be any digit from 0-9\n #each k give us derivatives estimates of different fxns that were trying to integrate\n \n #define x at 1/2 step\n x_ipoh = xi + 0.5*h\n \n #define x at 1 step\n x_ipo = xi + h\n \n #declare a temp y array\n y_temp = np.zeros(nv)\n \n #get k1 values\n y_derivs = dydx(xi,yi)\n k1[:] = h*y_derivs[:]\n \n #get k2 values\n y_temp[:] = yi[:] + 0.5*k1[:]\n y_derivs = dydx(x_ipoh,y_temp)\n k2[:] = h*y_derivs[:]\n \n #get k3 values\n y_temp[:] = yi[:] + 0.5*k2[:]\n y_derivs = dydx(x_ipoh,y_temp)\n k3[:] = h*y_derivs[:]\n \n #get k4 values\n y_temp[:] = yi[:] + k3[:]\n y_derivs = dydx(x_ipo,y_temp)\n k4[:] = h*y_derivs[:]\n \n #advance y by a step h\n yipo = yi + (k1 + 2*k2 + 2*k3 + k4)/6.\n \n #this is an array\n return yipo",
"_____no_output_____"
]
],
[
[
"adaptive step size driver for rk4",
"_____no_output_____"
]
],
[
[
"def rk4_mv_ad(dydx,x_i,y_i,nv,h,tol):\n \n #define safety scale, tell us how much our step is gonna change by\n SAFETY = 0.9\n H_NEW_FAC = 2.0\n # HNF max fator by which were taking a bigger step\n \n #set max number of iterations becasue were doing a while loop\n imax = 10000\n \n #set iteration variable\n i = 0\n \n #create an error, delta is an array, size of nv, fill it by number that is twice our tolerance\n Delta = np.full(nv,2*tol)\n \n #remember the step\n h_step = h\n \n #adjust step\n while(Delta.max()/tol > 1.0):\n \n #estimate error by taking 1 step size of h vs. 2 steps of size h/2\n y_2 = rk4_mv_core(dydx,x_i,y_i,nv,h_step)\n y_1 = rk4_mv_core(dydx,x_i,y_i,nv,0.5*h_step)\n y_11 = rk4_mv_core(dydx,x_i+0.5*h_step,y_i,nv,0.5*h_step)\n \n #compute an error\n Delta = np.fabs(y_2 - y_11)\n \n #if error is too large, take a smaller step\n if(Delta.max()/tol > 1.0):\n #our error is too large, decrease the step\n h_step *= SAFETY * (Delta.max()/tol)**(-0.25)\n \n #Check iteration\n if(i>=imax):\n print(\"Too many iterations in rk4_mv_as()\")\n raise StopIteration(\"Ending after i = \",i)\n \n #iterate\n i+=1\n \n #next time try to take a bigger step\n h_new = np.fmin(h_step * (Delta.max()/tol)**(-0.9), h_step*H_NEW_FAC)\n \n #return the answer, a new step, an the step we actually took\n return y_2, h_new, h_step",
"_____no_output_____"
]
],
[
[
"wrapper for rk4",
"_____no_output_____"
]
],
[
[
"def rk4_mv(dydx,a,b,y_a,tol):\n \n #dydx is the derivative wrt x\n #a is lower bound, b is upper bound\n #y_a are the boundary conditions array that contains values from 0-1\n #tol is the tolerance for integrating y\n \n #define starting step\n xi = a\n yi = y_a.copy()\n \n #an initial step size == make very small\n h = 1.0e-4 * (b-a)\n \n #set max number of iterations, if theres problems, we will know about it\n imax = 10000\n \n #set iteration variable\n i = 0\n \n #set number of coupled ODEs to the size of y_a, y_a is our initial conditions\n #array, values of array give values of y @ a \n nv = len(y_a)\n \n #set initial conditions, makes np array, y is 2D array, with 2 indices to y\n #1st is along x direction, 2nd has 2 values y and z\n #2 arrays side by side, 1 for y and 1 for z\n \n x = np.full(1,a) \n #single element array. x is array with 1 element, a \n y = np.full((1,nv),y_a)\n \n #dimensions of 1,nv. series of arrays of size nv(#of ODEs, in this case its 2)\n #makes 2 D array, 1 index has 1 element, the other has 2 elements\n #its a tuple, nxm matrix. n is 1, and m is 2\n\n \n #set flag/initial conditions because were doing something iterative\n flag = 1\n \n #loop until we reach the right side (b)\n while(flag):\n \n #calculate y_i+1\n yi_new, h_new, h_step = rk4_mv_ad(dydx,xi,yi,nv,h,tol)\n #update step\n h = h_new\n \n #prevent overshoot\n if(xi+h_step>b):\n #take smaller step\n h = b-xi\n \n #recalculate y_i+1\n yi_new, h_new, h_step = rk4_mv_ad(dydx,xi,yi,nv,h,tol)\n \n #break\n flag = 0\n \n #update values\n xi += h_step\n yi[:] = yi_new[:]\n #[:] replaces values in yi with yi_new\n \n #add the step to the arrays\n x = np.append(x,xi)\n #take array, appends new element, replacing with xi in further steps\n y_new = np.zeros((len(x),nv))\n y_new[0:len(x)-1,:] = y\n y_new[-1,:] = yi[:]\n del y\n y = y_new\n \n #prevent too many iterations\n if(i>=imax):\n \n print(\"Max number of iterations reached\")\n raise StopIteration(\"Iteration number = \",i)\n \n #iterate\n i += 1\n \n #output some information\n s = \"i = %3d\\tx = %9.8f\\th = %9.8f\\tb=%9.8f\" % (i,xi, h_step, b)\n print(s)\n \n #break if new xi is == b\n if(xi==b):\n flag = 0\n \n #return the answer\n return x,y",
"_____no_output_____"
]
],
[
[
"## Perform the integration",
"_____no_output_____"
]
],
[
[
"a = 0.0\nb = 2.0 * np.pi\n\ny_0 = np.zeros(2)\ny_0[0] = 0.0\ny_0[1] = 1.0\nnv = 2\n\ntolerance = 1.0e-6\n\n#perform the integration\nx,y = rk4_mv(dydx,a,b,y_0,tolerance)",
"_____no_output_____"
]
],
[
[
"## plot the result",
"_____no_output_____"
]
],
[
[
"plt.plot(x,y[:,0],'o',label='y(x)')\nplt.plot(x,y[:,1],'o',label='dydx(x)')\nxx = np.linspace(0,2.0*np.pi,1000)\nplt.plot(xx,np.sin(xx),label='sin(x)')\nplt.plot(xx,np.cos(xx),label='cos(x)')\nplt.xlabel('x')\nplt.ylabel('y, dy/dx')\nplt.legend(frameon=False)",
"_____no_output_____"
]
],
[
[
"## Plot the error\nNotice that the errors actually exceed our \"tolerance\"",
"_____no_output_____"
]
],
[
[
"sine = np.sin(x)\ncosine = np.cos(x)\n\ny_error = (y[:,0]-sine)\ndydx_error = (y[:,1]-cosine)\n\nplt.plot(x, y_error, label=\"y(x) Error\")\nplt.plot(x, dydx_error, label=\"dydx(x) Error\")\nplt.legend(frameon=False)",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
c50dda16362ec718a2148af2f8547b5f8684eeca
| 42,124 |
ipynb
|
Jupyter Notebook
|
tutorials/Intel-TF101-Class3/Week3_Neural_Networks_1_HW.ipynb
|
Nhat-Minh-Hoang-Tran-BSC2021/Intel-Convolutional_Neural_Networks
|
bbc2c1adb38bfd7c066a4fe6b5a0e0dab6df21cb
|
[
"MIT"
] | 2 |
2019-04-02T02:36:41.000Z
|
2021-01-18T15:20:39.000Z
|
tutorials/Intel-TF101-Class3/Week3_Neural_Networks_1_HW.ipynb
|
Nhat-Minh-Hoang-Tran-BSC2021/Intel-Convolutional_Neural_Networks
|
bbc2c1adb38bfd7c066a4fe6b5a0e0dab6df21cb
|
[
"MIT"
] | 1 |
2020-05-20T08:40:00.000Z
|
2020-05-20T08:40:00.000Z
|
tutorials/Intel-TF101-Class3/Week3_Neural_Networks_1_HW.ipynb
|
Nhat-Minh-Hoang-Tran-BSC2021/Intel-Convolutional_Neural_Networks
|
bbc2c1adb38bfd7c066a4fe6b5a0e0dab6df21cb
|
[
"MIT"
] | 2 |
2019-04-22T09:19:13.000Z
|
2020-02-12T18:01:32.000Z
| 52.589263 | 11,848 | 0.661761 |
[
[
[
"# Setup",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport tensorflow as tf\nimport matplotlib.pyplot as plt\nimport itertools as it\n\nimport helpers_03\n\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"# Neurons as Logic Gates",
"_____no_output_____"
],
[
"As an introduction to neural networks and their component neurons, we are going to look at using neurons to implement the most primitive logic computations: logic gates. Let's go!",
"_____no_output_____"
],
[
"##### The Sigmoid Function\n\nThe basic, classic activation function that we apply to neurons is a sigmoid (sometimes just called *the* sigmoid function) function: the standard logistic function.\n\n$$\n\\sigma = \\frac{1}{1 + e^{-x}}\n$$\n\n$\\sigma$ ranges from (0, 1). When the input $x$ is negative, $\\sigma$ is close to 0. When $x$ is positive, $\\sigma$ is close to 1. At $x=0$, $\\sigma=0.5$\n\nWe can implement this conveniently with NumPy.",
"_____no_output_____"
]
],
[
[
"def sigmoid(x):\n \"\"\"Sigmoid function\"\"\"\n return 1.0 / (1.0 + np.exp(-x))",
"_____no_output_____"
]
],
[
[
"And plot it with matplotlib.",
"_____no_output_____"
]
],
[
[
"# Plot The sigmoid function\nxs = np.linspace(-10, 10, num=100, dtype=np.float32)\nactivation = sigmoid(xs)\n\nfig = plt.figure(figsize=(6,4))\nplt.plot(xs, activation)\nplt.plot(0,.5,'ro')\n\nplt.grid(True, which='both')\nplt.axhline(y=0, color='y')\nplt.axvline(x=0, color='y')\nplt.ylim([-0.1, 1.15])",
"_____no_output_____"
]
],
[
[
"## An Example with OR",
"_____no_output_____"
],
[
"##### OR Logic\nA logic gate takes in two boolean (true/false or 1/0) inputs, and returns either a 0 or 1 depending on its rule. The truth table for a logic gate shows the outputs for each combination of inputs: (0, 0), (0, 1), (1,0), and (1, 1). For example, let's look at the truth table for an Or-gate:\n\n<table>\n<tr><th colspan=\"3\">OR gate truth table</th></tr>\n<tr><th colspan=\"2\">Input</th><th>Output</th></tr>\n<tr><td>0</td><td>0</td><td>0</td></tr>\n<tr><td>0</td><td>1</td><td>1</td></tr>\n<tr><td>1</td><td>0</td><td>1</td></tr>\n<tr><td>1</td><td>1</td><td>1</td></tr>\n</table>",
"_____no_output_____"
],
[
"##### OR as a Neuron\n\nA neuron that uses the sigmoid activation function outputs a value between (0, 1). This naturally leads us to think about boolean values. Imagine a neuron that takes in two inputs, $x_1$ and $x_2$, and a bias term:\n\n<img src=\"./images/logic01.png\" width=50%/>\n\nBy limiting the inputs of $x_1$ and $x_2$ to be in $\\left\\{0, 1\\right\\}$, we can simulate the effect of logic gates with our neuron. The goal is to find the weights (represented by ? marks above), such that it returns an output close to 0 or 1 depending on the inputs. What weights should we use to output the same results as OR? Remember: $\\sigma(z)$ is close to 0 when $z$ is largely negative (around -10 or less), and is close to 1 when $z$ is largely positive (around +10 or greater).\n\n$$\nz = w_1 x_1 + w_2 x_2 + b\n$$\n\nLet's think this through:\n\n* When $x_1$ and $x_2$ are both 0, the only value affecting $z$ is $b$. Because we want the result for input (0, 0) to be close to zero, $b$ should be negative (at least -10) to get the very left-hand part of the sigmoid.\n* If either $x_1$ or $x_2$ is 1, we want the output to be close to 1. That means the weights associated with $x_1$ and $x_2$ should be enough to offset $b$ to the point of causing $z$ to be at least 10 (i.e., to the far right part of the sigmoid).\n\nLet's give $b$ a value of -10. How big do we need $w_1$ and $w_2$ to be? At least +20 will get us to +10 for just one of $\\{w_1, w_2\\}$ being on.\n\nSo let's try out $w_1=20$, $w_2=20$, and $b=-10$:\n\n<img src=\"./images/logic02.png\\\" width=50%/>",
"_____no_output_____"
],
[
"##### Some Utility Functions\nSince we're going to be making several example logic gates (from different sets of weights and biases), here are two helpers. The first takes our weights and baises and turns them into a two-argument function that we can use like `and(a,b)`. The second is for printing a truth table for a gate.",
"_____no_output_____"
]
],
[
[
"def logic_gate(w1, w2, b):\n ''' logic_gate is a function which returns a function\n the returned function take two args and (hopefully)\n acts like a logic gate (and/or/not/etc.). its behavior\n is determined by w1,w2,b. a longer, better name would be\n make_twoarg_logic_gate_function'''\n def the_gate(x1, x2):\n return sigmoid(w1 * x1 + w2 * x2 + b)\n return the_gate\n\ndef test(gate):\n 'Helper function to test out our weight functions.'\n for a, b in it.product(range(2), repeat=2):\n print(\"{}, {}: {}\".format(a, b, np.round(gate(a, b))))",
"_____no_output_____"
]
],
[
[
"Let's see how we did. Here's the gold-standard truth table.\n\n<table>\n<tr><th colspan=\"3\">OR gate truth table</th></tr>\n<tr><th colspan=\"2\">Input</th><th>Output</th></tr>\n<tr><td>0</td><td>0</td><td>0</td></tr>\n<tr><td>0</td><td>1</td><td>1</td></tr>\n<tr><td>1</td><td>0</td><td>1</td></tr>\n<tr><td>1</td><td>1</td><td>1</td></tr>\n</table>\n\nAnd our result:",
"_____no_output_____"
]
],
[
[
"or_gate = logic_gate(20, 20, -10)\ntest(or_gate)",
"0, 0: 0.0\n0, 1: 1.0\n1, 0: 1.0\n1, 1: 1.0\n"
]
],
[
[
"This matches - great! ",
"_____no_output_____"
],
[
"# Exercise 1",
"_____no_output_____"
],
[
"##### Part 1: AND Gate\n\nNow you try finding the appropriate weight values for each truth table. Try not to guess and check. Think through it logically and try to derive values that work.\n\n<table>\n<tr><th colspan=\"3\">AND gate truth table</th></tr>\n<tr><th colspan=\"2\">Input</th><th>Output</th></tr>\n<tr><td>0</td><td>0</td><td>0</td></tr>\n<tr><td>0</td><td>1</td><td>0</td></tr>\n<tr><td>1</td><td>0</td><td>0</td></tr>\n<tr><td>1</td><td>1</td><td>1</td></tr>\n</table>",
"_____no_output_____"
]
],
[
[
"# Fill in the w1, w2, and b parameters such that the truth table matches\n# and_gate = logic_gate()\n# test(and_gate)",
"_____no_output_____"
]
],
[
[
"##### Part 2: NOR (Not Or) Gate\n<table>\n<tr><th colspan=\"3\">NOR gate truth table</th></tr>\n<tr><th colspan=\"2\">Input</th><th>Output</th></tr>\n<tr><td>0</td><td>0</td><td>1</td></tr>\n<tr><td>0</td><td>1</td><td>0</td></tr>\n<tr><td>1</td><td>0</td><td>0</td></tr>\n<tr><td>1</td><td>1</td><td>0</td></tr>\n</table>\n<table>",
"_____no_output_____"
]
],
[
[
"# Fill in the w1, w2, and b parameters such that the truth table matches\n# nor_gate = logic_gate()\n# test(nor_gate)",
"_____no_output_____"
]
],
[
[
"##### Part 3: NAND (Not And) Gate\n<table>\n<tr><th colspan=\"3\">NAND gate truth table</th></tr>\n<tr><th colspan=\"2\">Input</th><th>Output</th></tr>\n<tr><td>0</td><td>0</td><td>1</td></tr>\n<tr><td>0</td><td>1</td><td>1</td></tr>\n<tr><td>1</td><td>0</td><td>1</td></tr>\n<tr><td>1</td><td>1</td><td>0</td></tr>\n</table>",
"_____no_output_____"
]
],
[
[
"# Fill in the w1, w2, and b parameters such that the truth table matches\n# nand_gate = logic_gate()\n# test(nand_gate)",
"_____no_output_____"
]
],
[
[
"## Solutions 1",
"_____no_output_____"
],
[
"# Limits of Single Neurons\n\nIf you've taken computer science courses, you may know that the XOR gates are the basis of computation. They can be used as half-adders, the foundation of being able to add numbers together. Here's the truth table for XOR:\n\n##### XOR (Exclusive Or) Gate\n<table>\n<tr><th colspan=\"3\">NAND gate truth table</th></tr>\n<tr><th colspan=\"2\">Input</th><th>Output</th></tr>\n<tr><td>0</td><td>0</td><td>0</td></tr>\n<tr><td>0</td><td>1</td><td>1</td></tr>\n<tr><td>1</td><td>0</td><td>1</td></tr>\n<tr><td>1</td><td>1</td><td>0</td></tr>\n</table>\n\nNow the question is, can you create a set of weights such that a single neuron can output this property? It turns out that you cannot. Single neurons can't correlate inputs, so it's just confused. So individual neurons are out. Can we still use neurons to somehow form an XOR gate?\n\nWhat if we tried something more complex:\n\n<img src=\"./images/logic03.png\\\" width=60%/>\n\nHere, we've got the inputs going to two separate gates: the top neuron is an OR gate, and the bottom is a NAND gate. The output of these gates is passed to another neuron, which is an AND gate. If you work out the outputs at each combination of input values, you'll see that this is an XOR gate!",
"_____no_output_____"
]
],
[
[
"# Make sure you have or_gate, nand_gate, and and_gate working from above\ndef xor_gate(a, b):\n c = or_gate(a, b)\n d = nand_gate(a, b)\n return and_gate(c, d)\ntest(xor_gate)",
"_____no_output_____"
]
],
[
[
"Thus, we can see how chaining together neurons can compose more complex models than we'd otherwise have access to.",
"_____no_output_____"
],
[
"# Learning a Logic Gate",
"_____no_output_____"
],
[
"We can use TensorFlow to try and teach a model to learn the correct weights and bias by passing in our truth table as training data.",
"_____no_output_____"
]
],
[
[
"# Create an empty Graph to place our operations in\nlogic_graph = tf.Graph()\nwith logic_graph.as_default():\n # Placeholder inputs for our a, b, and label training data\n x1 = tf.placeholder(tf.float32)\n x2 = tf.placeholder(tf.float32)\n label = tf.placeholder(tf.float32)\n \n # A placeholder for our learning rate, so we can adjust it\n learning_rate = tf.placeholder(tf.float32)\n \n # The Variables we'd like to learn: weights for a and b, as well as a bias term\n w1 = tf.Variable(tf.random_normal([]))\n w2 = tf.Variable(tf.random_normal([]))\n b = tf.Variable(0.0, dtype=tf.float32)\n \n # Use the built-in sigmoid function for our output value\n output = tf.nn.sigmoid(w1 * x1 + w2 * x2 + b)\n \n # We'll use the mean of squared errors as our loss function \n loss = tf.reduce_mean(tf.square(output - label))\n correct = tf.equal(tf.round(output), label)\n accuracy = tf.reduce_mean(tf.cast(correct, tf.float32))\n \n # Finally, we create a gradient descent training operation and an initialization operation\n train = tf.train.GradientDescentOptimizer(learning_rate).minimize(loss)\n init = tf.global_variables_initializer()",
"_____no_output_____"
],
[
"with tf.Session(graph=logic_graph) as sess:\n sess.run(init)\n # Training data for all combinations of inputs\n and_table = np.array([[0,0,0],\n [1,0,0],\n [0,1,0],\n [1,1,1]])\n \n feed_dict={x1: and_table[:,0],\n x2: and_table[:,1],\n label: and_table[:,2], \n learning_rate: 0.5}\n \n for i in range(5000):\n l, acc, _ = sess.run([loss, accuracy, train], feed_dict)\n if i % 1000 == 0:\n print('loss: {}\\taccuracy: {}'.format(l, acc))\n \n test_dict = {x1: and_table[:,0], #[0.0, 1.0, 0.0, 1.0], \n x2: and_table[:,1]} # [0.0, 0.0, 1.0, 1.0]}\n w1_val, w2_val, b_val, out = sess.run([w1, w2, b, output], test_dict)\n print('\\nLearned weight for w1:\\t {}'.format(w1_val))\n print('Learned weight for w2:\\t {}'.format(w2_val))\n print('Learned weight for bias: {}\\n'.format(b_val))\n\n print(np.column_stack((and_table[:,[0,1]], out.round().astype(np.uint8) ) ) )\n # FIXME! ARGH! use real python or numpy\n #idx = 0\n #for i in [0, 1]:\n # for j in [0, 1]:\n # print('{}, {}: {}'.format(i, j, np.round(out[idx])))\n # idx += 1\n ",
"_____no_output_____"
]
],
[
[
"# Exercise 2",
"_____no_output_____"
],
[
"You may recall that in week 2, we built a class `class TF_GD_LinearRegression` that wrapped up the three steps of using a learning model: (1) build the model graph, (2) train/fit, and (3) test/predict. Above, we *did not* use that style of implementation. And you can see that things get a bit messy, quickly. We have model creation in one spot and then we have training, testing, and output all mixed together (along with TensorFlow helper code like sessions, etc.). We can do better. Rework the code above into a class like `TF_GD_LinearRegression`.",
"_____no_output_____"
],
[
"## Solution 2",
"_____no_output_____"
],
[
"# Learning an XOR Gate",
"_____no_output_____"
],
[
"If we compose a two stage model, we can learn the XOR gate. You'll notice that defining the model itself is starting to get messy. We'll talk about ways of dealing with that next week.",
"_____no_output_____"
]
],
[
[
"class XOR_Graph:\n def __init__(self):\n # Create an empty Graph to place our operations in\n xor_graph = tf.Graph()\n with xor_graph.as_default():\n # Placeholder inputs for our a, b, and label training data\n self.x1 = tf.placeholder(tf.float32)\n self.x2 = tf.placeholder(tf.float32)\n self.label = tf.placeholder(tf.float32)\n\n # A placeholder for our learning rate, so we can adjust it\n self.learning_rate = tf.placeholder(tf.float32)\n\n # abbreviations! this section is the difference\n # from the LogicGate class above\n Var = tf.Variable; rn = tf.random_normal\n self.weights = [[Var(rn([])), Var(rn([]))],\n [Var(rn([])), Var(rn([]))],\n [Var(rn([])), Var(rn([]))]]\n self.biases = [Var(0.0, dtype=tf.float32),\n Var(0.0, dtype=tf.float32),\n Var(0.0, dtype=tf.float32)]\n sig1 = tf.nn.sigmoid(self.x1 * self.weights[0][0] + \n self.x2 * self.weights[0][1] + \n self.biases[0])\n sig2 = tf.nn.sigmoid(self.x1 * self.weights[1][0] + \n self.x2 * self.weights[1][1] + \n self.biases[1])\n self.output = tf.nn.sigmoid(sig1 * self.weights[2][0] + \n sig2 * self.weights[2][1] + \n self.biases[2])\n\n # We'll use the mean of squared errors as our loss function \n self.loss = tf.reduce_mean(tf.square(self.output - self.label))\n \n # Finally, we create a gradient descent training operation \n # and an initialization operation\n gdo = tf.train.GradientDescentOptimizer\n self.train = gdo(self.learning_rate).minimize(self.loss)\n \n correct = tf.equal(tf.round(self.output), self.label)\n self.accuracy = tf.reduce_mean(tf.cast(correct, tf.float32))\n \n init = tf.global_variables_initializer()\n \n self.sess = tf.Session(graph=xor_graph)\n self.sess.run(init) \n\n def fit(self, train_dict):\n loss, acc, _ = self.sess.run([self.loss, self.accuracy, self.train], \n train_dict)\n return loss, acc\n \n def predict(self, test_dict):\n # make a list of organized weights: \n # see tf.get_collection for more advanced ways to handle this\n all_trained = (self.weights[0] + [self.biases[0]] +\n self.weights[1] + [self.biases[1]] +\n self.weights[2] + [self.biases[2]])\n return self.sess.run(all_trained + [self.output], test_dict)",
"_____no_output_____"
],
[
"xor_table = np.array([[0,0,0],\n [1,0,1],\n [0,1,1],\n [1,1,0]])\n\nlogic_model = XOR_Graph()\ntrain_dict={logic_model.x1: xor_table[:,0],\n logic_model.x2: xor_table[:,1],\n logic_model.label: xor_table[:,2], \n \n logic_model.learning_rate: 0.5}\n\nprint(\"training\")\n# note, I might get stuck in a local minima b/c this is a \n# small problem with no noise (yes, noise helps!)\n# this can converge in one round of 1000 or it might get \n# stuck for all 10000\nfor i in range(10000):\n loss, acc = logic_model.fit(train_dict)\n if i % 1000 == 0:\n print('loss: {}\\taccuracy: {}'.format(loss, acc))\nprint('loss: {}\\taccuracy: {}'.format(loss, acc))\n \nprint(\"testing\")\ntest_dict = {logic_model.x1: xor_table[:,0], \n logic_model.x2: xor_table[:,1]}\nresults = logic_model.predict(test_dict)\nwb_lrn, predictions = results[:-1], results[-1]\n\nprint(wb_lrn)\nwb_lrn = np.array(wb_lrn).reshape(3,3)\n\n# combine the predictions with the inputs and clean up the data\n# round it and convert to unsigned 8 bit ints\nout_table = np.column_stack((xor_table[:,[0,1]], \n predictions)).round().astype(np.uint8)\n\nprint(\"results\")\nprint('Learned weights/bias (L1):', wb_lrn[0])\nprint('Learned weights/bias (L2):', wb_lrn[1])\nprint('Learned weights/bias (L3):', wb_lrn[2])\nprint('Testing Table:')\nprint(out_table)\nprint(\"Correct?\", np.allclose(xor_table, out_table))",
"_____no_output_____"
]
],
[
[
"# An Example Neural Network",
"_____no_output_____"
],
[
"So, now that we've worked with some primitive models, let's take a look at something a bit closer to what we'll work with moving forward: an actual neural network.\n\nThe following model accepts a 100 dimensional input, has a hidden layer depth of 300, and an output layer depth of 50. We use a sigmoid activation function for the hidden layer.",
"_____no_output_____"
]
],
[
[
"nn1_graph = tf.Graph()\nwith nn1_graph.as_default():\n x = tf.placeholder(tf.float32, shape=[None, 100])\n y = tf.placeholder(tf.float32, shape=[None]) # Labels, not used in this model\n \n with tf.name_scope('hidden1'):\n w = tf.Variable(tf.truncated_normal([100, 300]), name='W')\n b = tf.Variable(tf.zeros([300]), name='b')\n z = tf.matmul(x, w) + b\n a = tf.nn.sigmoid(z)\n \n with tf.name_scope('output'):\n w = tf.Variable(tf.truncated_normal([300, 50]), name='W')\n b = tf.Variable(tf.zeros([50]), name='b')\n z = tf.matmul(a, w) + b\n output = z\n \n with tf.name_scope('global_step'):\n global_step = tf.Variable(0, trainable=False, name='global_step')\n inc_step = tf.assign_add(global_step, 1, name='increment_step')\n \n with tf.name_scope('summaries'):\n for var in tf.trainable_variables():\n hist_summary = tf.summary.histogram(var.op.name, var)\n summary_op = tf.summary.merge_all()\n \n init = tf.global_variables_initializer()",
"_____no_output_____"
],
[
"tb_base_path = 'tbout/nn1_graph'\ntb_path = helpers_03.get_fresh_dir(tb_base_path)\nsess = tf.Session(graph=nn1_graph)\nwriter = tf.summary.FileWriter(tb_path, graph=nn1_graph)\nsess.run(init)\nsummaries = sess.run(summary_op)\nwriter.add_summary(summaries)\nwriter.close()\nsess.close()",
"_____no_output_____"
]
],
[
[
"# Exercise 3",
"_____no_output_____"
],
[
"Modify the template above to create your own neural network with the following features:\n\n* Accepts input of length 200 (and allows for variable number of examples)\n* First hidden layer depth of 800\n* Second hidden layer depth of 600\n* Third hidden layer depth of 400\n* Output layer depth of 100\n* Include histogram summaries of the variables",
"_____no_output_____"
],
[
"## Solution 3",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
c50ddbb0a1eb7d133c4999fcbc3036bb1e03a8de
| 3,628 |
ipynb
|
Jupyter Notebook
|
selections/__init__.ipynb
|
tomergt45/Geneticflow
|
936845d19cc27f0a7af09d989e160bbc366bb849
|
[
"Apache-2.0"
] | null | null | null |
selections/__init__.ipynb
|
tomergt45/Geneticflow
|
936845d19cc27f0a7af09d989e160bbc366bb849
|
[
"Apache-2.0"
] | null | null | null |
selections/__init__.ipynb
|
tomergt45/Geneticflow
|
936845d19cc27f0a7af09d989e160bbc366bb849
|
[
"Apache-2.0"
] | null | null | null | 29.983471 | 166 | 0.503308 |
[
[
[
"import tensorflow as tf\nimport numpy as np",
"_____no_output_____"
],
[
"def get(name):\n \"\"\"\n Returns a selection by name, this can be a function or a class name, in cases of a class name then an instance of that class will be returned.\n \n Parameters\n -----------\n name: str\n The name of the function or class.\n \n Returns\n --------\n out: function or instance of `Selection`.\n \"\"\"\n import inspect\n target = globals()[name]\n return target() if inspect.isclass(target) else target",
"_____no_output_____"
],
[
"class Selection:\n \"\"\" \n This is the class from which all selections inherit from.\n \n A selection is a callable object (much like a function) which takes a list of `Genome` objects as an input and returns a filtered list of `Genome`.\n \n The selection function of the `Selection` class is defined within the `call` function.\n \n Users will just instantiate an instance and then treat it as a callable (function).\n \n Selection is the stage of a genetic algorithm in which individual genomes are chosen from a population for later breeding (using the crossover operator).\n \"\"\"\n def call(self, genomes):\n raise Exception('You are trying to invoke the default selection function of the `Selection` class, this function is not defined.')\n \n def __call__(self, genomes):\n \"\"\"\n This function takes a list of genomes as an input, and returns a filtered list of `Genome`.\n \n Parameters\n -----------\n genomes: list[Genome]\n A list of genomes.\n \n Returns\n --------\n out: list[Genome]\n A filtered list of genomes.\n \"\"\"\n return self.call(genomes)",
"_____no_output_____"
],
[
"class Top(Selection):\n \"\"\"\n Selection of the top genomes from the given genome pool, the top genomes are determined by their fitness score.\n \"\"\"\n def __init__(self, parents=2):\n \"\"\"\n Parameters\n -----------\n parents: int\n The number of parents to select.\n \"\"\"\n self.parents = 2\n \n def call(self, genomes):\n return sorted(genomes, key=lambda g: g.fitness)[-self.parents:]",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code"
]
] |
c50ddbc4b8546e107ca569e7d3d68cb18a9c7327
| 45,078 |
ipynb
|
Jupyter Notebook
|
week1_precurse_python_I/day2_python_I/theory/Python_I_Precurse_ended_review.ipynb
|
paleomau/MGOL_BOOTCAMP
|
8c2b018f49fd12a255ea6f323141260d04d4421d
|
[
"MIT"
] | null | null | null |
week1_precurse_python_I/day2_python_I/theory/Python_I_Precurse_ended_review.ipynb
|
paleomau/MGOL_BOOTCAMP
|
8c2b018f49fd12a255ea6f323141260d04d4421d
|
[
"MIT"
] | null | null | null |
week1_precurse_python_I/day2_python_I/theory/Python_I_Precurse_ended_review.ipynb
|
paleomau/MGOL_BOOTCAMP
|
8c2b018f49fd12a255ea6f323141260d04d4421d
|
[
"MIT"
] | null | null | null | 18.948298 | 185 | 0.460003 |
[
[
[
"# Data Science Bootcamp - The Bridge \n## Precurso\nEn este notebook vamos a ver, uno a uno, los conceptos básicos de Python. Constarán de ejercicios prácticos acompañados de una explicación teórica dada por el profesor.\n\nLos siguientes enlaces están recomendados para el alumno para profundizar y reforzar conceptos a partir de ejercicios y ejemplos:\n\n- https://www.kaggle.com/learn/python\n\n- https://facundoq.github.io/courses/aa2018/res/02_python.html\n\n- https://www.w3resource.com/python-exercises/\n\n- https://www.practicepython.org/\n\n- https://es.slideshare.net/egutierrezru/python-paraprincipiantes\n\n- https://www.sololearn.com/Play/Python#\n\n- https://github.com/mhkmcp/Python-Bootcamp-from-Basic-to-Advanced\n\nEjercicios avanzados:\n\n- https://github.com/darkprinx/100-plus-Python-programming-exercises-extended/tree/master/Status (++)\n\n- https://github.com/mahtab04/Python-Programming-Practice (++)\n\n- https://github.com/whojayantkumar/Python_Programs (+++)\n\n- https://www.w3resource.com/python-exercises/ (++++)\n\n- https://github.com/fupus/notebooks-ejercicios (+++++)\n\nTutor de ayuda PythonTutor:\n\n- http://pythontutor.com/\n",
"_____no_output_____"
],
[
"## 1. Variables y tipos\n\n### Cadenas",
"_____no_output_____"
]
],
[
[
"# Entero - Integer - int\nx = 7\n# Cadena - String - Lista de caracteres\nx = \"lorena\"\nprint(x)\n\nx = 7\nprint(x)",
"_____no_output_____"
],
[
"# built-in\ntype",
"_____no_output_____"
],
[
"x = 5\ny = 7\n\nz = x + y\nprint(z)",
"_____no_output_____"
],
[
"x = \"'lorena'\\\" \"\nl = 'silvia----'\n\ng = x + l\n# Las cadenas se concatenan\nprint(g)",
"_____no_output_____"
],
[
"print(g)",
"_____no_output_____"
],
[
"# type muestra el tipo de la variable\ntype(g)",
"_____no_output_____"
],
[
"type(3)",
"_____no_output_____"
],
[
"# print es una función que recibe varios argumentos y cada argumento está diferenciado por la coma. Después de cada coma, la función 'print' añade un espacio.\n\n# mala praxis\nprint( g,z , 6, \"cadena\")\n\n# buena praxis - PEP8\nprint(g, z, 6, \"cadena\")",
"_____no_output_____"
],
[
"u = \"g\"\nsilvia = \"silvia tiene \"\n\nanos = \" años\"\n\nsuma = silvia + u + anos\nprint(suma)",
"_____no_output_____"
],
[
"n = 2\nm = \"3\"\n\nprint(n + m)",
"_____no_output_____"
],
[
"# Cambiar de int a str\n\nj = 2\nprint(j)\nprint(type(j))\nj = str(j)\nprint(j)\nprint(type(j))",
"_____no_output_____"
],
[
"# Cambiar de int a str\n\nj = 2 \nprint(j)\nprint(type(j))\nj = str(j) + \" - \" + silvia\nprint(j)\nprint(type(j))",
"_____no_output_____"
],
[
"k = 22\nk = str(k)\nprint(k)",
"_____no_output_____"
],
[
"# Cambiar de str a int\n\nlor = \"98\"\nlor = int(lor)\nprint(lor)",
"_____no_output_____"
],
[
"# Para ver la longitud de una lista de caracteres (lista)\n\nmn = \"lista de caracteres$%·$% \"\n\n#lenght\nprint(len(mn))",
"_____no_output_____"
],
[
"h = len(mn)\n\nprint(h + 7)",
"_____no_output_____"
],
[
"h = 8",
"_____no_output_____"
],
[
"print(h)",
"_____no_output_____"
],
[
"x = 2\n\nprint(x)\n\ngabriel_vazquez = \"Gabriel Vazquez\"",
"_____no_output_____"
],
[
"print(\"Hello Python world!\")\nprint(\"Nombre de compañero\")\n\ncompanero_clase = \"Compañero123123\"\n\nprint(companero_clase)",
"_____no_output_____"
],
[
"print(compañero)",
"_____no_output_____"
],
[
"x = (2 + 4) + 7\nprint(x)",
"_____no_output_____"
],
[
"# String, Integer, Float, List, None (NaN)\n# str, int, float, list, \n\nstring_ = \"23\"\nnumero = 23\n\nprint(type(string_))",
"_____no_output_____"
],
[
"print(string_)",
"_____no_output_____"
],
[
"numero2 = 10\nsuma = numero + numero2\nprint(suma)",
"_____no_output_____"
],
[
"string2 = \"33\"\nsuma2 = string_ + string2\nprint(suma2)",
"_____no_output_____"
],
[
"m = (numero2 + int(string2))\nprint(m)",
"_____no_output_____"
],
[
"m = ((((65 + int(\"22\")) * 2)))\nm",
"_____no_output_____"
],
[
"print(type(int(string2)))",
"_____no_output_____"
],
[
"y = 22\ny = str(y)\nprint(type(y))",
"_____no_output_____"
],
[
"string2 = int(string2)\nprint(type(string2))",
"_____no_output_____"
],
[
"string3 = \"10\"\nnumero_a_partir_de_string = int(string3)\nprint(numero_a_partir_de_string)\nprint(string3)\n\nprint(type(numero_a_partir_de_string))\nprint(type(string3))",
"_____no_output_____"
],
[
"h = \"2\"\nint(h)",
"_____no_output_____"
],
[
"# los decimales son float. Python permite las operaciones entre int y float\nx = 4\ny = 4.2\n\nprint(x + y)",
"_____no_output_____"
],
[
"# La división normal (/) es siempre float\n# La división absoluta (//) puede ser: \n# - float si uno de los dos números (o los dos) son float\n# - int si los dos son int \nj = 15\nk = 4\ndivision = j // k\nprint(division)\nprint(type(division))",
"_____no_output_____"
],
[
"num1 = 12\nnum2 = 3\n\nsuma = num1 + num2\nresta = num1 - num2\nmultiplicacion = num1 * num2 \ndivision = num1 / num2\ndivision_absoluta = num1 // num2\n\ngabriel_vazquez = \"Gabriel Vazquez\"\n\nprint(\"suma:\", suma)\nprint(\"resta:\", resta)\nprint(\"multiplicacion:\", multiplicacion)\nprint(\"division:\", division)\nprint(\"division_absoluta:\", division_absoluta)\nprint(type(division))\nprint(type(division_absoluta))",
"_____no_output_____"
],
[
"print(x)",
"_____no_output_____"
],
[
"j = \"2\"\nj",
"_____no_output_____"
],
[
"print(j)",
"_____no_output_____"
],
[
"x = 2\nj = 6\ng = 4\nh = \"popeye\"\n\n# Jupyter notebook permite que la última línea se imprima por pantalla ( la variable )\nprint(g)\nprint(j)\nprint(h)\nx",
"_____no_output_____"
],
[
"int(5.6/2)",
"_____no_output_____"
],
[
"float(2)",
"_____no_output_____"
],
[
"g = int(5.6/2)\nprint(g)\n5",
"_____no_output_____"
],
[
"x = int(5.6//2)\n\nx",
"_____no_output_____"
],
[
"# Soy un comentario\n# print(\"Hello Python world!\")\n# Estoy creando una variable que vale 2\n\"\"\"\nEsto es otro comentario\n\"\"\"\nprint(x)\nx = 25\nx = 76\nx = \"1\"",
"_____no_output_____"
],
[
"message2 = \"One of Python's strengths is its diverse community.\" \nprint(message2)",
"_____no_output_____"
]
],
[
[
"## Ejercicio: \n### Crear una nueva celda.\n### Declarar tres variables: \n- Una con el nombre \"edad\" con valor vuestra edad\n- Otra \"edad_compañero_der\" que contengan la edad de tipo entero de vuestro compañero de la derecha\n- Otra \"suma_anterior\" que contenga la suma de las dos variables anteriormente declaradas\n\n Mostrar por pantalla la variable \"suma_anterior\"\n\n",
"_____no_output_____"
]
],
[
[
"edad = 99\nedad_companero_der = 30\nsuma_anterior = edad_companero_der + edad\nprint(suma_anterior)",
"_____no_output_____"
],
[
"h = 89 + suma_anterior\nh",
"_____no_output_____"
],
[
"edad = 18\nedad_companero_der = 29\nsuma_anterior = edad + edad_companero_der\nsuma_anterior",
"_____no_output_____"
],
[
"i = \"hola\"\no = i.upper()\no",
"_____no_output_____"
],
[
"o.lower()",
"_____no_output_____"
],
[
"name = \"Ada Lovelace\"\nx = 2\nprint(name.upper())\nprint(name.lower())",
"_____no_output_____"
],
[
"print(name.upper)",
"_____no_output_____"
],
[
"print(name.upper())",
"_____no_output_____"
],
[
"x = 2\nx = x + 1\nx",
"_____no_output_____"
],
[
"x += 1\nx",
"_____no_output_____"
],
[
"# int\nx = 1\n# float\ny = 2.\n# str\ns = \"string\"",
"_____no_output_____"
],
[
"# type --> muestra el tipo de la variable o valor\nprint(type(x))\ntype(y)\ntype(s)",
"_____no_output_____"
],
[
"5 + 2",
"_____no_output_____"
],
[
"x = 2\nx = x + 1\nx += 1",
"_____no_output_____"
],
[
"x = 2\ny = 4\n\nprint(x, y, \"Pepito\", \"Hola\")",
"_____no_output_____"
],
[
"s = \"Hola soy Soraya:\"\ns + \"789\"",
"_____no_output_____"
],
[
"print(s, 98, 29, sep=\"\")\nprint(s, 98, 29)",
"_____no_output_____"
],
[
"type( x )",
"_____no_output_____"
],
[
"2 + 6",
"_____no_output_____"
]
],
[
[
"## 2. Números y operadores",
"_____no_output_____"
]
],
[
[
"### Enteros ###\n\nx = 3\n\nprint(\"- Tipo de x:\")\nprint(type(x)) # Imprime el tipo (o `clase`) de x\nprint(\"- Valor de x:\")\nprint(x) # Imprimir un valor\nprint(\"- x+1:\")\nprint(x + 1) # Suma: imprime \"4\"\nprint(\"- x-1:\")\nprint(x - 1) # Resta; imprime \"2\"\nprint(\"- x*2:\")\nprint(x * 2) # Multiplicación; imprime \"6\"\nprint(\"- x^2:\")\nprint(x ** 2) # Exponenciación; imprime \"9\"\n# Modificación de x\nx += 1 \nprint(\"- x modificado:\")\nprint(x) # Imprime \"4\"\n\nx *= 2\nprint(\"- x modificado:\")\nprint(x) # Imprime \"8\"\n\nprint(\"- El módulo de x con 40\")\nprint(40 % x)\n\nprint(\"- Varias cosas en una línea:\")\nprint(1, 2, x, 5*2) # imprime varias cosas a la vez",
"_____no_output_____"
],
[
"# El módulo muestra el resto de la división entre dos números\n\n2 % 2",
"_____no_output_____"
],
[
"3 % 2",
"_____no_output_____"
],
[
"4 % 5",
"_____no_output_____"
],
[
"numero = 99\n\nnumero % 2 # Si el resto es 0, el número es par. Sino, impar.",
"_____no_output_____"
],
[
"numero % 2",
"_____no_output_____"
],
[
"99 % 100",
"_____no_output_____"
],
[
"y = 2.5 \nprint(\"- Tipo de y:\")\nprint(type(y)) # Imprime el tipo de y\nprint(\"- Varios valores en punto flotante:\")\nprint(y, y + 1, y * 2.5, y ** 2) # Imprime varios números en punto flotante",
"_____no_output_____"
]
],
[
[
"## Título\n\nEscribir lo que sea\n\n1. uno\n2. dos",
"_____no_output_____"
],
[
"# INPUT",
"_____no_output_____"
]
],
[
[
"edad = input(\"Introduce tu edad\")\n\nprint(\"Diego tiene\", edad, \"años\")",
"_____no_output_____"
],
[
"# Input recoge una entrada de texto de tipo String\n\nnum1 = int(input(\"Introduce el primer número\"))\nnum2 = int(input(\"Introduce el segundo número\"))\n\nprint(num1 + num2)",
"_____no_output_____"
]
],
[
[
"## 3. Tipo None",
"_____no_output_____"
]
],
[
[
"x = None",
"_____no_output_____"
],
[
"n = 5\ns = \"Cadena\"\nprint(x + s)",
"_____no_output_____"
]
],
[
[
"## 4. Listas y colecciones ",
"_____no_output_____"
]
],
[
[
"# Lista de elementos:\n\n# Las posiciones se empiezan a contar desde 0\ns = \"Cadena\"\nprimer_elemento = s[0]\n#ultimo_elemento = s[5]\nultimo_elemento = s[-1]\n\nprint(primer_elemento)\nprint(ultimo_elemento)",
"_____no_output_____"
],
[
"bicycles = ['trek', 'cannondale', 'redline', 'specialized']\nbicycles[0]\n",
"_____no_output_____"
],
[
"tamano_lista = len(bicycles)\ntamano_lista",
"_____no_output_____"
],
[
"ultimo_elemento_por_posicion = tamano_lista - 1\nbicycles[ultimo_elemento_por_posicion]",
"_____no_output_____"
],
[
"bicycles = ['trek', 'cannondale', 'redline', 'specialized']\nmessage = \"My first bicycle was a \" + bicycles[0]\n\nprint(bicycles)\nprint(message)",
"_____no_output_____"
],
[
"print(type(bicycles))",
"_____no_output_____"
],
[
"s = \"String\"\ns.lower()\nprint(s.lower())\nprint(s)",
"_____no_output_____"
],
[
"s = s.lower()\ns",
"_____no_output_____"
],
[
"# Existen dos tipos de funciones:\n# 1. Los que modifican los valores sin que tengamos que especificar una reasignación a la variable\n# 2. Los que solo devuelven la operación y no modifican el valor de la variable. Tenemos que forzar la reasignación si queremos modificar la variable. \n\ncars = ['bmw', 'audi', 'toyota', 'subaru']\nprint(cars)\n\ncars.reverse()\nprint(cars)",
"_____no_output_____"
],
[
"cars = ['bmw']\nprint(cars)\n\ncars.reverse()\nprint(cars)",
"_____no_output_____"
],
[
"s = \"Hola soy Clara\"\n\nprint(s[::-1])",
"_____no_output_____"
],
[
"s",
"_____no_output_____"
],
[
"l = \"hola\"\nlen(l)",
"_____no_output_____"
],
[
"l[3]",
"_____no_output_____"
],
[
"# Para acceder a varios elementos, se especifica con la nomenclatura \"[N:M]\". N es el primer elemento a obtener, M es el último elemento a obtener pero no incluido. Ejemplo: \n\n# Queremos mostrar desde las posiciones 3 a la 7. Debemos especificar: [3:8]\n\n# Si M no tiene ningún valor, se obtiene desde N hasta el final. \n# Si N no tiene ningún valor, es desde el principio de la colección hasta M\ns[3:len(s)]",
"_____no_output_____"
],
[
"s[:3]",
"_____no_output_____"
],
[
"s[3:10]",
"_____no_output_____"
],
[
"motorcycles = ['honda', 'yamaha', 'suzuki', 'ducati'] \nprint(motorcycles)\n\ntoo_expensive = 'ducati'\nmotorcycles.remove(too_expensive)\nprint(motorcycles)\nprint(too_expensive + \" is too expensive for me.\")",
"_____no_output_____"
],
[
"# Agrega un valor a la última posición de la lista\nmotorcycles.append(\"ducati\")\nmotorcycles",
"_____no_output_____"
],
[
"lista = ['honda', 2, 8.9, [2, 3], 'yamaha', 'suzuki', 'ducati']\nlista[3] ",
"_____no_output_____"
],
[
"lista.remove(8.9)\nlista",
"_____no_output_____"
],
[
"lista\nl = lista[1]\nl",
"_____no_output_____"
],
[
"lista.remove(l)\nlista",
"_____no_output_____"
],
[
"lista.remove(lista[2])",
"_____no_output_____"
],
[
"lista = ['honda', 2, 8.9, [2, 3], 'yamaha', 'suzuki', 'honda', 'ducati']\nlista",
"_____no_output_____"
],
[
"# remove elimina el primer elemento que se encuentra que coincide con el valor del argumento\nlista = ['honda', 2, 8.9, [2, 3], 'yamaha', 'suzuki', 'honda', 'ducati']\nlista.remove(\"honda\")\nlista",
"_____no_output_____"
],
[
"# Accedemos a la posición 1 del elemento que está en la posición 2 de lista\nlista[2][1]",
"_____no_output_____"
],
[
"lista[3][2]",
"_____no_output_____"
],
[
"p = lista.remove(\"honda\")\nprint(p)",
"_____no_output_____"
],
[
"l = [2, 4, 6, 8]\nl.reverse()\nl",
"_____no_output_____"
]
],
[
[
"### Colecciones\n\n1. Listas\n2. String (colección de caracteres)\n3. Tuplas\n4. Conjuntos (Set)",
"_____no_output_____"
]
],
[
[
"# Listas --> Mutables\nlista = [2, 5, \"caract\", [9, \"g\", [\"j\"]]]\nprint(lista[-1][-1][-1])\nlista[3][2][0]",
"_____no_output_____"
],
[
"lista.append(\"ultimo\")\nlista",
"_____no_output_____"
],
[
"# Tuplas --> Inmutables\n\ntupla = (2, 5, \"caract\", [9, \"g\", [\"j\"]])\ntupla",
"_____no_output_____"
],
[
"s = \"String\"\ns[2]",
"_____no_output_____"
],
[
"tupla[3].remove(9)\ntupla",
"_____no_output_____"
],
[
"tupla[3][1].remove(\"j\")\ntupla",
"_____no_output_____"
],
[
"tupla2 = (2, 5, 'caract', ['g', ['j']])\ntupla2[-1].remove([\"j\"])\ntupla2",
"_____no_output_____"
],
[
"tupla2 = (2, 5, 'caract', ['g', ['j']])\ntupla2[-1].remove(\"g\")\ntupla2",
"_____no_output_____"
],
[
"tupla2[-1].remove([\"j\"])\ntupla2",
"_____no_output_____"
],
[
"if False == 0:\n print(0)",
"_____no_output_____"
],
[
"print(type(lista))\nprint(type(tupla))",
"_____no_output_____"
],
[
"# Update listas\nlista = [2, \"6\", [\"k\", \"m\"]]\nlista[1] = 1\nlista",
"_____no_output_____"
],
[
"lista = [2, \"6\", [\"k\", \"m\"]]\nlista[2] = 0\nlista",
"_____no_output_____"
],
[
"tupla = (2, \"6\", [\"k\", \"m\"])\ntupla[2] = 0\ntupla",
"_____no_output_____"
],
[
"tupla = (2, \"6\", [\"k\", \"m\"])\ntupla[2][1] = 0\ntupla",
"_____no_output_____"
],
[
"# Conjuntos\n\nconjunto = [2, 4, 6, \"a\", \"z\", \"h\", 2]\nconjunto = set(conjunto)\nconjunto",
"_____no_output_____"
],
[
"conjunto = [\"a\", \"z\", \"h\", 2, 2, 4, 6, True, True, False]\nconjunto = set(conjunto)\nconjunto",
"_____no_output_____"
],
[
"conjunto = [\"a\", \"z\", \"h\", 2, 2, 4, 6, 2.1, 2.4, 2.3, True, True, False]\nconjunto = set(conjunto)\nconjunto",
"_____no_output_____"
],
[
"conjunto_tupla = (\"a\", \"z\", \"h\", 2, 2, 4, 6, 2.1, 2.4, 2.3, True, True, False)\nconjunto = set(conjunto_tupla)\nconjunto",
"_____no_output_____"
],
[
"conjunto = {\"a\", \"z\", \"h\", 2, 2, 4, 6, 2.1, 2.4, 2.3, True, True, False}\nconjunto",
"_____no_output_____"
],
[
"s = \"String\"\nlista_s = list(s)\nlista_s",
"_____no_output_____"
],
[
"s = \"String\"\nconj = {s}\nconj",
"_____no_output_____"
],
[
"tupla = (2, 5, \"h\")\ntupla = list(tupla)\ntupla.remove(2)\ntupla = tuple(tupla)\ntupla",
"_____no_output_____"
],
[
"tupla = (2, 5, \"h\")\ntupla = list(tupla)\ntupla.remove(2)\ntupla = (((((tupla)))))\ntupla",
"_____no_output_____"
],
[
"tupla = (2, 5, \"h\")\ntupla = list(tupla)\ntupla.remove(2)\ntupla = tuple(tupla)\ntupla",
"_____no_output_____"
],
[
"conjunto = {2, 5, \"h\"}\nlista_con_conjunto = [conjunto]\nlista_con_conjunto[0]",
"_____no_output_____"
],
[
"# No podemos acceder a elementos de un conjunto\nlista_con_conjunto[0][0]",
"_____no_output_____"
],
[
"lista = [1, 5, 6, True, 6]\nset(lista)",
"_____no_output_____"
],
[
"lista = [True, 5, 6, 1, 6]\nset(lista)",
"_____no_output_____"
],
[
"1 == True",
"_____no_output_____"
]
],
[
[
"## 5. Condiciones: if, elif, else",
"_____no_output_____"
],
[
"### Boolean",
"_____no_output_____"
]
],
[
[
"True\nFalse\n",
"_____no_output_____"
],
[
"# Operadores comparativos\nx = (1 == 1)\nx",
"_____no_output_____"
],
[
"1 == 2",
"_____no_output_____"
],
[
"\"a\" == \"a\"",
"_____no_output_____"
],
[
"# Diferente\n\"a\" != \"a\"",
"_____no_output_____"
],
[
"2 > 4",
"_____no_output_____"
],
[
"4 > 2",
"_____no_output_____"
],
[
"4 >= 4",
"_____no_output_____"
],
[
"4 > 4",
"_____no_output_____"
],
[
"4 <= 5",
"_____no_output_____"
],
[
"4 < 3",
"_____no_output_____"
],
[
"input()",
"_____no_output_____"
],
[
"1 == 1",
"_____no_output_____"
],
[
"\"\"\"\n== -> Igualdad\n!= -> Diferecia\n< -> Menor que\n> -> Mayor que\n<= -> Menor o igual\n>= -> Mayor o igual\n\"\"\"\n",
"_____no_output_____"
],
[
"# and solo devolverá True si TODOS son True\nTrue and False",
"_____no_output_____"
],
[
"# or devolverá True si UNO es True\n\n(1 == 1) or (1 == 2)",
"_____no_output_____"
],
[
"(1 == 1) or (1 == 2) and (1 == 1)",
"_____no_output_____"
],
[
"(1 == 1) or ((1 == 2) and (1 == 1)) ",
"_____no_output_____"
],
[
"(1 == 1) and (1 == 2) and (1 == 1)",
"_____no_output_____"
],
[
"(1 == 1) and (1 == 2) and ((1 == 1) or (0 == 0))\n# True and False and True",
"_____no_output_____"
],
[
"print(\"Yo soy\\n Gabriel\")",
"_____no_output_____"
],
[
"if 1 != 1:\n print(\"Son iguales\")\nelse:\n print(\"No entra en if\")",
"_____no_output_____"
],
[
"if 1 == 1:\n print(\"Son iguales\")\nelse:\n print(\"No entra en if\")",
"_____no_output_____"
],
[
"if 1>3:\n print(\"Es mayor\")\nelif 2==2:\n print(\"Es igual\")\nelif 3==3:\n print(\"Es igual2\")\nelse:\n print(\"Ninguna de las anteriores\")",
"_____no_output_____"
],
[
"if 2>3:\n print(1)\nelse:\n print(\"Primer else\")\nif 2==2:\n print(2)\nelse:\n print(\"Segundo else\")\n",
"_____no_output_____"
],
[
"if 2>3:\n print(1)\nelse:\n print(\"Primer else\")\nif 3==3:\n print(\" 3 es 3 \")\nif 2==2:\n print(2)\nelse:\n print(\"Segundo else\")\n",
"_____no_output_____"
],
[
"if 2>3:\n print(1)\nelse:\n print(\"Primer else\")\n# -------\nif 3==4:\n print(\" 3 es 3 \")\n# --------\nif 2==2:\n print(2)\n print(5)\n x = 6\n print(x)\nelse:\n print(\"Segundo else\")",
"_____no_output_____"
],
[
"if 2>3:\n print(1)\nelse:\n print(\"Primer else\")\n# -------\nif 3==4:\n print(\" 3 es 3 \")\n# --------\nif 2==2:\n print(2)\n print(5)\n x = 6\n print(x)\n # ------\n if x == 7:\n print(\"X es igual a 6\")\n # ------\n y = 7\n print(y)\nelse:\n print(\"Segundo else\")",
"_____no_output_____"
],
[
"if (not (1==1)):\n print(\"Hola\")\n",
"_____no_output_____"
],
[
"if not None:\n print(1)",
"_____no_output_____"
],
[
"if \"a\":\n print(2)",
"_____no_output_____"
],
[
"if 0:\n print(0)",
"_____no_output_____"
],
[
"# Sinónimos de False para las condiciones:\n# None\n# False\n# 0 (int or float)\n# Cualquier colección vacía --> [], \"\", (), {}\n\n# El None no actúa como un número al compararlo con otro número\n\"\"\"\nEl True lo toma como un numérico 1\n\"\"\"",
"_____no_output_____"
],
[
"lista = []\n\nif lista:\n print(4)",
"_____no_output_____"
],
[
"lista = [\"1\"]\n\nif lista:\n print(4)",
"_____no_output_____"
],
[
"if 0.0:\n print(2)",
"_____no_output_____"
],
[
"if [] or False or 0 or None:\n print(4)",
"_____no_output_____"
],
[
"if [] and False or 0 or None:\n print(4)",
"_____no_output_____"
],
[
"if not ([] or False or 0 or None):\n print(4)",
"_____no_output_____"
],
[
"if [] or False or not 0 or None:\n print(True)\nelse:\n print(False)",
"_____no_output_____"
],
[
"x = True\ny = False\n\nx + y ",
"_____no_output_____"
],
[
"x = True\ny = False\n\nstr(x) + str(y) ",
"_____no_output_____"
],
[
"def funcion_condicion():\n if y > 4:\n print(\"Es mayor a 4\")\n else:\n print(\"No es mayor a 4\")\n\nfuncion_condicion(y=4)",
"_____no_output_____"
],
[
"def funcion_primera(x):\n if x == 4:\n print(\"Es igual a 4\")\n else:\n funcion_condicion(y=x)\n\nfuncion_primera(x=5)",
"_____no_output_____"
],
[
"def funcion_final(apellido):\n if len(apellido) > 5:\n print(\"Cumple condición\")\n else:\n print(\"No cumpole condición\")\n\nfuncion_final(apellido=\"Vazquez\")",
"_____no_output_____"
]
],
[
[
"# Bucles For, While",
"_____no_output_____"
]
],
[
[
"lista = [1, \"dos\", 3, \"Pepito\"] \n\nprint(lista[0])\nprint(lista[1])\nprint(lista[2])\nprint(lista[3])",
"_____no_output_____"
],
[
"for p in lista:\n print(p)\n",
"_____no_output_____"
],
[
"altura1 = [1.78, 1.63, 1.75, 1.68]\naltura2 = [2.00, 1.82]\naltura3 = [1.65, 1.73, 1.75]\naltura4 = [1.72, 1.71, 1.71, 1.62]\n\nlista_alturas = [altura1, altura2, altura3, altura4]\n\nprint(lista_alturas[0][1])\n\nfor x in lista_alturas:\n print(x[3])",
"_____no_output_____"
],
[
"def mostrar_cada_elemento_de_lista(lista):\n for x in lista:\n print(x)\n\nmostrar_cada_elemento_de_lista(lista=lista_alturas)",
"_____no_output_____"
],
[
"mostrar_cada_elemento_de_lista(lista=lista)",
"_____no_output_____"
],
[
"for x in lista_alturas:\n if len(x) > 2:\n print(x[2])\n else:\n print(x[1])",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
c50deb17aac9420ccd64e88eed10ce46122c5bf9
| 31,568 |
ipynb
|
Jupyter Notebook
|
Idiomatic Pandas/03 Idiomatic Pandas.ipynb
|
rogues-gallery/Learn-Pandas
|
e362c7959a63052f5396919c49d7b352297297ec
|
[
"BSD-3-Clause"
] | null | null | null |
Idiomatic Pandas/03 Idiomatic Pandas.ipynb
|
rogues-gallery/Learn-Pandas
|
e362c7959a63052f5396919c49d7b352297297ec
|
[
"BSD-3-Clause"
] | null | null | null |
Idiomatic Pandas/03 Idiomatic Pandas.ipynb
|
rogues-gallery/Learn-Pandas
|
e362c7959a63052f5396919c49d7b352297297ec
|
[
"BSD-3-Clause"
] | null | null | null | 23.735338 | 349 | 0.537126 |
[
[
[
"# `map` vs `apply`\n\nDo you know the difference between the **`map`** and **`apply`** Series methods?",
"_____no_output_____"
]
],
[
[
"from IPython.display import IFrame",
"_____no_output_____"
],
[
"IFrame('http://etc.ch/RjoN', 400, 300)",
"_____no_output_____"
],
[
"IFrame('https://directpoll.com/r?XDbzPBd3ixYqg8VzFnGsNyv3rYRtjyM1R0g6HvOxV', 400, 300)",
"_____no_output_____"
]
],
[
[
"# Primary usage of `map` method\nAs the name implies, **`map`** can literally map one value to another in a Series. Pass it a dictionary (or another Series). Let's see an example:",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np",
"_____no_output_____"
],
[
"s = pd.Series(np.random.randint(1, 7, 10))\ns",
"_____no_output_____"
]
],
[
[
"Create mapping dictionary",
"_____no_output_____"
]
],
[
[
"d = {1:'odd', 2:'even', 3:'odd', 4:'even', 5:'odd', 6:'even'}",
"_____no_output_____"
],
[
"s.map(d)",
"_____no_output_____"
]
],
[
[
"Works the same if you use a Series",
"_____no_output_____"
]
],
[
[
"s1 = pd.Series(d)\ns1",
"_____no_output_____"
],
[
"s.map(s1)",
"_____no_output_____"
]
],
[
[
"#### `map` example with more data\nLet's map the values of 1 million integers ranging from 1 to 100 to 'even/odd' strings",
"_____no_output_____"
]
],
[
[
"n = 1000000 # 1 million\ns = pd.Series(np.random.randint(1, 101, n))\ns.head()",
"_____no_output_____"
]
],
[
[
"Create the mapping",
"_____no_output_____"
]
],
[
[
"d = {i: 'odd' if i % 2 else 'even' for i in range(1, 101)}\nprint(d)",
"_____no_output_____"
],
[
"s.map(d).head(10)",
"_____no_output_____"
]
],
[
[
"### Exercise 1\n<span style=\"color:green; font-size:16px\">Can you use the **`apply`** method to do the same thing? Time the difference between the **`apply`** and **`map`**.</span>",
"_____no_output_____"
]
],
[
[
"# your code here",
"_____no_output_____"
]
],
[
[
"### `map` and `apply` can both take functions\nUnfortunately both **`map`** and **`apply`** can accept a function that gets implicitly passed each value in the Series. The result of each operation is the exact same.",
"_____no_output_____"
]
],
[
[
"a = s.apply(lambda x: 'odd' if x % 2 else 'even')\nb = s.map(lambda x: 'odd' if x % 2 else 'even')\n\na.equals(b)",
"_____no_output_____"
]
],
[
[
"This dual functionality of **`map`** confuses users. It can accept a dictionary but it can also accept a function.\n\n### Suggestion: only use `map` for literal mapping\nIt makes more sense to me that the **`map`** method only be used for one purpose and this is to map each value in a Series from one value to another with a dictionary or a Series.\n\n### Use `apply` only for functions\n**`apply`** must take a function and has more options than **`map`** when taking a function so it should be used when you want to apply a function to each value in a Series. There is no difference in speed between the two.",
"_____no_output_____"
],
[
"### Exercise 2\n<span style=\"color:green; font-size:16px\">Use the **`map`** method with a two-item dictionary to convert the Series of integers to 'even/odd' strings. You will need to perform an operation on the Series first. Is this faster or slower than the results in exercise 1?</span>",
"_____no_output_____"
]
],
[
[
"# run this code first\nn = 1000000 # 1 million\ns = pd.Series(np.random.randint(1, 101, n))",
"_____no_output_____"
],
[
"# your code here",
"_____no_output_____"
]
],
[
[
"### Exercise 3\n<span style=\"color:green; font-size:16px\">Write a for-loop to convert each value in the Series to 'even/odd' strings. Time the operation.</span>",
"_____no_output_____"
]
],
[
[
"# your code here",
"_____no_output_____"
]
],
[
[
"# Vectorized if-then-else with NumPy `where`\nThe NumPy **`where`** function provides us with a vectorized if-then-else that is very fast. Let's convert the Series again to 'even/odd' strings.",
"_____no_output_____"
]
],
[
[
"s = pd.Series(np.random.randint(1, 101, n))",
"_____no_output_____"
],
[
"np.where(s % 2, 'odd', 'even')",
"_____no_output_____"
],
[
"%timeit np.where(s % 2, 'odd', 'even')",
"_____no_output_____"
]
],
[
[
"### Exercise 4\n<span style=\"color:green; font-size:16px\">Convert the values from 1-33 to 'low', 34-67 to 'medium' and the rest 'high'.</span>",
"_____no_output_____"
]
],
[
[
"# your code here",
"_____no_output_____"
]
],
[
[
"### There is a DataFrame/Series `where` method\nThere is a DataFrame/Series **`where`** method but it works differently. You must pass it a boolean DataFrame/series and it will preserve all the values that are True. The other values will by default be converted to missing, but you can specify a specific number as well.",
"_____no_output_____"
]
],
[
[
"s.where(s > 50).head(10)",
"_____no_output_____"
],
[
"s.where(s > 50, other=-1).head(10)",
"_____no_output_____"
]
],
[
[
"# Do we really need `apply`?\nAs we saw from this last example, we could eliminate the need for the **`apply`** method. Most examples of code that use **`apply`** do not actually need it.\n\n### `apply` doesn't really do anything\nBy itself, the **`apply`** method doesn't really do anything. \n* For Series, it iterates over every single value and passes that value to a function that you must pass to **`apply`**. \n* For a DataFrame, it iterates over each column or row as a Series and calls your passed function on that Series\n\nLet's see a simple example of **`apply`** used to multiply each value of a Series by 2:",
"_____no_output_____"
]
],
[
[
"s = pd.Series(np.random.randint(1, 101, n))",
"_____no_output_____"
],
[
"s.apply(lambda x: x * 2).head()",
"_____no_output_____"
],
[
"(s * 2).head()",
"_____no_output_____"
],
[
"%timeit s.apply(lambda x: x * 2)",
"_____no_output_____"
],
[
"%timeit s * 2",
"_____no_output_____"
]
],
[
[
"### Use vectorized solution whenever possible\nAs you can see, the solution with **`apply`** was more than 2 orders of magnitude slower than the vectorized solution. A for-loop can be faster than **`apply`**.",
"_____no_output_____"
]
],
[
[
"%timeit pd.Series([v * 2 for v in s])",
"_____no_output_____"
]
],
[
[
"I like to call **`apply`** the **method of last resort**. There is almost rarely a reason to use it over other methods. Pandas and NumPy both provide a tremendous amount of functionality that cover nearly everything you need to do. \n\nAlways use pandas and NumPy methods first before anything else.",
"_____no_output_____"
],
[
"### Use-cases for `apply` on a Series\nWhen there is no vectorized implementation in pandas, numpy or other scientific library, then you can use **`apply`**.\n\nA simple example (that's not too practical) is finding the underlying data type of each value in a Series.",
"_____no_output_____"
]
],
[
[
"s = pd.Series(['a', {'TX':'Texas'}, 99, (0, 5)])\ns",
"_____no_output_____"
],
[
"s.apply(type)",
"_____no_output_____"
]
],
[
[
"A more practical example might be from a library that doesn't work directly with arrays, like finding the edit distance between two strings from the NLTK library.",
"_____no_output_____"
]
],
[
[
"from nltk.metrics import edit_distance",
"_____no_output_____"
],
[
"edit_distance('Kaitlyn', 'Kaitlin')",
"_____no_output_____"
],
[
"s = pd.Series(['Kaitlyn', 'Katelyn', 'Kaitlin', 'Katelynn', 'Katlyn',\n 'Kaitlynn', 'Katelin', 'Katlynn', 'Kaitlin', 'Caitlyn', 'Caitlynn'])\ns",
"_____no_output_____"
]
],
[
[
"Using **`apply`** here is correct",
"_____no_output_____"
]
],
[
[
"s.apply(lambda x: edit_distance(x, 'Kaitlyn'))",
"_____no_output_____"
]
],
[
[
"### Using `apply` on a DataFrame\nBy default **`apply`** will call the passed function on each individual column on a DataFrame. The column will be passed to the function as a Series.",
"_____no_output_____"
]
],
[
[
"df = pd.DataFrame(np.random.rand(100, 5), columns=['a', 'b', 'c', 'd', 'e'])\ndf.head()",
"_____no_output_____"
],
[
"df.apply(lambda s: s.max())",
"_____no_output_____"
]
],
[
[
"We can change the direction of the operation by seting the **`axis`** parameter to **`1`** or **`columns`**",
"_____no_output_____"
]
],
[
[
"df.apply(lambda s: s.max(), axis='columns').head(10)",
"_____no_output_____"
]
],
[
[
"#### Never actually perform these operations when a DataFrame method exists\nLet's fix these two methods and time their differences",
"_____no_output_____"
]
],
[
[
"df.max()",
"_____no_output_____"
],
[
"df.max(axis='columns').head(10)",
"_____no_output_____"
],
[
"%timeit df.apply(lambda s: s.max())",
"_____no_output_____"
],
[
"%timeit df.max()",
"_____no_output_____"
],
[
"%timeit df.apply(lambda s: s.max(), axis='columns')",
"_____no_output_____"
],
[
"%timeit df.max(axis='columns')",
"_____no_output_____"
]
],
[
[
"5x and 70x faster and much more readable code",
"_____no_output_____"
],
[
"### Infected by the documentation\nUnfortunately, pandas official documentation is littered with examples that don't need **`apply`**. Can you fix the following 2 misuses of **`apply`** [found here](http://pandas.pydata.org/pandas-docs/stable/10min.html#apply).\n\n",
"_____no_output_____"
],
[
"### Exercise 1\n<span style=\"color:green; font-size:16px\">Make the following idiomatic</span>",
"_____no_output_____"
]
],
[
[
"df.apply(np.cumsum).head()",
"_____no_output_____"
],
[
"# your code here",
"_____no_output_____"
]
],
[
[
"### Exercise 2\n<span style=\"color:green; font-size:16px\">Make the following idiomatic</span>",
"_____no_output_____"
]
],
[
[
"df.apply(lambda x: x.max() - x.min())",
"_____no_output_____"
],
[
"# your code here",
"_____no_output_____"
]
],
[
[
"### `apply` with `axis=1` is the slowest operation you can do in pandas\nIf you call **`apply`** with **`axis=1`** or identically with **`axis='columns'`** on a DataFrame, pandas will iterate row by row to complete your operation. Since there are almost always more rows than columns, this will be extremely slow.",
"_____no_output_____"
],
[
"### Exercise 3\n<span style=\"color:green; font-size:16px\">Add a column named **`distance`** to the following DataFrame that computes the euclidean distance between points **`(x1, y1)`** and **`(x2, y2)`**. Calculate it once with **`apply`** and again idiomatically using vectorized operations. Time the difference between them.</span>",
"_____no_output_____"
]
],
[
[
"# run this first\ndf = pd.DataFrame(np.random.randint(0, 20, (100000, 4)), \n columns=['x1', 'y1', 'x2', 'y2'])\ndf.head()",
"_____no_output_____"
],
[
"# your code here",
"_____no_output_____"
]
],
[
[
"### Use-cases for apply on a DataFrame\n\nDataFrames and Series have nearly all of the their methods in common. For methods that only exist for Series, you might need to use **`apply`**.",
"_____no_output_____"
]
],
[
[
"weather = pd.DataFrame({'Houston': ['rainy', 'sunny', 'sunny', 'cloudy', 'rainy', 'sunny'],\n 'New York':['sunny', 'sunny', 'snowy', 'snowy', 'rainy', 'cloudy'],\n 'Seattle':['sunny', 'cloudy', 'cloudy', 'cloudy', 'cloudy', 'rainy'],\n 'Las Vegas':['sunny', 'sunny', 'sunny', 'sunny', 'sunny', 'sunny']})\nweather",
"_____no_output_____"
]
],
[
[
"Counting the frequencies of each column is normally done by the Series **`value_counts`** method. It does not exist for DataFrames, so you can use it here with **`apply`**.",
"_____no_output_____"
]
],
[
[
"weather.apply(pd.value_counts)",
"_____no_output_____"
],
[
"%matplotlib inline\nweather.apply(pd.value_counts).plot(kind='bar')",
"_____no_output_____"
]
],
[
[
"### Using `apply` with the Series accessors `str`, `dt` and `cat`\nPandas Series, depending on their data type, can access additional Series-only methods through **`str`**, **`dt`** and **`cat`** for string, datetime and categorical type columns.",
"_____no_output_____"
]
],
[
[
"weather.Houston.str.capitalize()",
"_____no_output_____"
]
],
[
[
"Since this method exists only for Series, you can use **`apply`** here to capitalize each column.",
"_____no_output_____"
]
],
[
[
"weather.apply(lambda x: x.str.capitalize())",
"_____no_output_____"
]
],
[
[
"This is one case where you can use the **`applymap`** method by directly using the string method on each value.",
"_____no_output_____"
]
],
[
[
"weather.applymap(str.capitalize)",
"_____no_output_____"
],
[
"employee = pd.read_csv('../data/employee.csv')\nemployee.head()",
"_____no_output_____"
]
],
[
[
"Select just the titles and departments ",
"_____no_output_____"
]
],
[
[
"emp_title_dept = employee[['DEPARTMENT', 'POSITION_TITLE']]\nemp_title_dept.head()",
"_____no_output_____"
]
],
[
[
"Let's find all the departments and titles that contain the word 'police'.",
"_____no_output_____"
]
],
[
[
"has_police = emp_title_dept.apply(lambda x: x.str.upper().str.contains('POLICE'))\nhas_police.head()",
"_____no_output_____"
]
],
[
[
"Let's use these boolean values to only select rows that have both values as **`True`**.",
"_____no_output_____"
]
],
[
[
"emp_title_dept[has_police.all(axis='columns')].head(10)",
"_____no_output_____"
]
],
[
[
"### How fast are the `str` accessor methods?\nNot any faster than looping...",
"_____no_output_____"
]
],
[
[
"%timeit employee['POSITION_TITLE'].str.upper()",
"_____no_output_____"
],
[
"%timeit employee['POSITION_TITLE'].apply(str.upper)",
"_____no_output_____"
],
[
"%timeit pd.Series([x.upper() for x in employee['POSITION_TITLE']])",
"_____no_output_____"
],
[
"%timeit employee['POSITION_TITLE'].max()",
"_____no_output_____"
],
[
"%timeit employee['BASE_SALARY'].max()",
"_____no_output_____"
],
[
"%timeit employee['POSITION_TITLE'].values.max()",
"_____no_output_____"
],
[
"%timeit employee['BASE_SALARY'].values.max()",
"_____no_output_____"
],
[
"a_list = employee['POSITION_TITLE'].tolist()",
"_____no_output_____"
],
[
"%timeit max(a_list)",
"_____no_output_____"
]
],
[
[
"### Exercise 4\n<span style=\"color:green; font-size:16px\">The following example is from the documentation. Produce the same result without using apply by creating a function that it accepts a DataFrame and returns a DataFrame</span>",
"_____no_output_____"
]
],
[
[
"df = pd.DataFrame(np.random.randint(0, 20, (10, 4)), \n columns=['x1', 'y1', 'x2', 'y2'])\ndf.head()",
"_____no_output_____"
],
[
"def subtract_and_divide(x, sub, divide=1):\n return (x - sub) / divide",
"_____no_output_____"
],
[
"df.apply(subtract_and_divide, args=(5,), divide=3)",
"_____no_output_____"
],
[
"# your code here",
"_____no_output_____"
]
],
[
[
"### Exercise 5\n<span style=\"color:green; font-size:16px\">Make the following idiomatic:</span>",
"_____no_output_____"
]
],
[
[
"college = pd.read_csv('../data/college.csv', \n usecols=lambda x: 'UGDS' in x or x == 'INSTNM', \n index_col='INSTNM')\ncollege = college.dropna()\ncollege.shape",
"_____no_output_____"
],
[
"college.head()",
"_____no_output_____"
],
[
"def max_race_count(s):\n max_race_pct = s.iloc[1:].max()\n return (max_race_pct * s.loc['UGDS']).astype(int)",
"_____no_output_____"
],
[
"college.apply(max_race_count, axis=1).head()",
"_____no_output_____"
],
[
"# your code here",
"_____no_output_____"
]
],
[
[
"# Tips for debugging `apply`\nIt is more difficult to debug code that uses **`apply`** when you a custom function. This is because the all the code in your custom function gets executed at once. You aren't stepping through the code one line at a time and checking the output.\n\n### Using the `display` IPython function and print statements to inspect custom function\nLet's say you didn't know what **`apply`** with **`axis='columns'`** was implicitly passing to the custom function.",
"_____no_output_____"
]
],
[
[
"# what the hell is x?\ndef func(x):\n return 1",
"_____no_output_____"
],
[
"college.apply(func, axis=1).head()",
"_____no_output_____"
]
],
[
[
"Its obvious that you need to know what object **`x`** is in **`func`**. One thing we can do is print out its type. To stop the output we can force an error by calling **`raise`**.",
"_____no_output_____"
]
],
[
[
"# what the hell is x?\ndef func(x):\n print(type(x))\n raise\n return 1\n\ncollege.apply(func, axis=1).head()",
"_____no_output_____"
]
],
[
[
"Ok, great. We know that **`x`** is a Series. Why did it get printed twice? It turns out that pandas calls your method twice on the first row/column to determine if it can take a fast path or not. This is a small implementation detail that shouldn't affect you unless your function is making references to variables out of scope.\n\nLet's go one step further and display **`x`** on the screen",
"_____no_output_____"
]
],
[
[
"from IPython.display import display",
"_____no_output_____"
],
[
"# what the hell is x?\ndef func(x):\n display(x)\n raise\n return 1\n\ncollege.apply(func, axis=1).head()",
"_____no_output_____"
]
],
[
[
"### Exercise 1\n<span style=\"color:green; font-size:16px\">Use the **`display`** function after each line in a custom function that gets used with **`apply`** and **`axis='columns'`** to find the population of the second highest race per school. Make sure you raise an exception or else you will have to kill your kernel because of the massive output.</span>",
"_____no_output_____"
]
],
[
[
"# your code here",
"_____no_output_____"
]
],
[
[
"### Exercise 2 - Very difficult\n<span style=\"color:green; font-size:16px\">Can you do this without using **`apply`**?</span>",
"_____no_output_____"
]
],
[
[
"# your code here",
"_____no_output_____"
]
],
[
[
"### Exercise 3\n<span style=\"color:green; font-size:16px\">When **`apply`** is called on a Series, what is the data type that gets passed to the function?</span>",
"_____no_output_____"
]
],
[
[
"# your code here",
"_____no_output_____"
]
],
[
[
"# Summary\n* **`map`** is a Series method. I suggest using by passing it a dictionary/Series and NOT a function\n* Use **`apply`** when you want to apply a function to each value of a Series or each row/column of a DataFrame\n* You rarely need **`apply`** - Use only pandas and numpy functions first\n* Using **`apply`** on a DataFrame with **`axis='columns'`** is the slowest operation in pandas\n* You can use **`apply`** on a DataFrame when you need to call a method that is available only to Series (like **`value_counts`**)\n* Debug apply by printing and using the **`display`** IPython function inside your custom function",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
c50dfbde7646b4080841d4de607d078ea4222193
| 15,103 |
ipynb
|
Jupyter Notebook
|
notebooks/2.1.waves.15.04_corr_std_mice.ipynb
|
covidclinical/visualization-notebooks
|
583637e5815de01cead5257dbcb09403f345f188
|
[
"MIT"
] | 8 |
2020-04-28T15:14:52.000Z
|
2022-03-16T06:06:43.000Z
|
notebooks/2.1.waves.15.04_corr_std_mice.ipynb
|
hms-dbmi/c19i2b2-notebooks
|
583637e5815de01cead5257dbcb09403f345f188
|
[
"MIT"
] | 5 |
2020-04-19T14:36:31.000Z
|
2020-06-25T14:54:23.000Z
|
notebooks/2.1.waves.15.04_corr_std_mice.ipynb
|
covidclinical/visualization-notebooks
|
583637e5815de01cead5257dbcb09403f345f188
|
[
"MIT"
] | 3 |
2020-04-24T08:30:20.000Z
|
2020-07-08T20:25:17.000Z
| 33.636971 | 206 | 0.473615 |
[
[
[
"%load_ext autoreload\n%autoreload 2\n%aimport utils_1_1\n\nimport pandas as pd\nimport numpy as np\nimport altair as alt\nfrom altair_saver import save\nimport datetime\nimport dateutil.parser\nfrom os.path import join\n\nfrom constants_1_1 import SITE_FILE_TYPES\nfrom utils_1_1 import (\n get_site_file_paths,\n get_site_file_info,\n get_site_ids,\n get_visualization_subtitle,\n get_country_color_map,\n)\nfrom theme import apply_theme\nfrom web import for_website\n\nalt.data_transformers.disable_max_rows(); # Allow using rows more than 5000",
"_____no_output_____"
],
[
"data_release='2021-04-27'\n\ndf = pd.read_csv(join(\"..\", \"data\", \"Phase2.1SurvivalRSummariesPublic\", \"ToShare\", \"table.beta.mice.std.toShare.csv\"))\n\nprint(df.head())\n\n# Rename columns\ndf = df.rename(columns={\"variable\": \"c\", \"beta\": \"v\"})\n\nconsistent_date = {\n '2020-03': 'Mar - Apr',\n '2020-05': 'May - Jun',\n '2020-07': 'Jul - Aug',\n '2020-09': 'Sep - Oct',\n '2020-11': 'Since Nov'\n}\n\ncolors = ['#E79F00', '#0072B2', '#D45E00', '#CB7AA7', '#029F73', '#57B4E9']\n\nsites = ['META', 'APHP', 'FRBDX', 'ICSM', 'BIDMC', 'MGB', 'UCLA', 'UMICH', 'UPENN', 'VA1', 'VA2', 'VA3', 'VA4', 'VA5']\nsite_colors = ['black', '#D45E00', '#0072B2', '#CB7AA7', '#E79F00', '#029F73', '#DBD03C', '#57B4E9', '#57B4E9', '#57B4E9', '#57B4E9', '#57B4E9']\nsites = ['META', 'APHP', 'FRBDX', 'ICSM', 'UKFR', 'NWU', 'BIDMC', 'MGB', 'UCLA', 'UMICH', 'UPENN', 'UPITT', 'VA1', 'VA2', 'VA3', 'VA4', 'VA5']\nsite_colors = ['black', '#0072B2', '#0072B2', '#0072B2', '#0072B2', '#CB7AA7', '#D45E00', '#D45E00', '#D45E00', '#D45E00', '#D45E00', '#D45E00', '#D45E00', '#D45E00', '#D45E00','#D45E00','#D45E00']\n\ndf.siteid = df.siteid.apply(lambda x: x.upper())\n\nprint(df.siteid.unique().tolist())\n\ngroup_map = {\n 'age18to25': 'Age',\n 'age26to49': 'Age',\n 'age70to79': 'Age',\n 'age80plus': 'Age',\n 'sexfemale': 'Sex',\n 'raceBlack': 'Race',\n 'raceAsian': 'Race',\n 'raceHispanic.and.Other': 'Race',\n 'CRP': 'Lab',\n 'albumin': 'Lab',\n 'TB': 'Lab',\n \"LYM\": 'Lab',\n \"neutrophil_count\" : 'Lab',\n \"WBC\" : 'Lab',\n \"creatinine\": 'Lab',\n \"AST\": 'Lab',\n \"AA\": 'Lab',\n \"DD\": 'Lab',\n 'mis_CRP': 'Lab Mis.',\n 'mis_albumin': 'Lab Mis.',\n 'mis_TB': 'Lab Mis.',\n \"mis_LYM\": 'Lab Mis.',\n \"mis_neutrophil_count\" : 'Lab Mis.',\n \"mis_WBC\" : 'Lab Mis.',\n \"mis_creatinine\": 'Lab Mis.',\n \"mis_AST\": 'Lab Mis.',\n \"mis_AA\": 'Lab Mis.',\n 'mis_DD': 'Lab Mis.',\n 'charlson_score': 'Charlson Score',\n 'mis_charlson_score': 'Charlson Score',\n}\n\ndf['g'] = df.c.apply(lambda x: group_map[x])\n\nconsistent_c = {\n 'age18to25': '18 - 25',\n 'age26to49': '26 - 49',\n 'age70to79': '70 - 79',\n 'age80plus': '80+',\n 'sexfemale': 'Female',\n 'raceBlack': 'Black',\n 'raceAsian': 'Asian',\n 'raceHispanic.and.Other': 'Hispanic and Other',\n 'CRP': 'Log CRP(mg/dL)',\n 'albumin': 'Albumin (g/dL)',\n 'TB': 'Total Bilirubin (mg/dL)',\n \"LYM\": 'Lymphocyte Count (10*3/uL)',\n \"neutrophil_count\" : 'Neutrophil Count (10*3/uL)',\n \"WBC\" : 'White Blood Cell (10*3/uL)',\n \"creatinine\": 'Creatinine (mg/dL)',\n \"AST\": 'Log AST (U/L)',\n \"AA\": 'AST/ALT',\n \"DD\": 'Log D-Dimer (ng/mL)',\n 'mis_CRP': 'CRP not tested',\n 'mis_albumin': 'Albumin not tested',\n 'mis_TB': 'Total bilirubin not tested',\n \"mis_LYM\": 'Lymphocyte count not tested',\n \"mis_neutrophil_count\" : 'Neutrophil count not tested',\n \"mis_WBC\" : 'White blood cell not tested',\n \"mis_creatinine\": 'Creatinine not tested',\n \"mis_AST\": 'AST not tested',\n \"mis_AA\": 'ALT/AST not available',\n 'mis_DD': 'D-dimer nottested',\n 'charlson_score': 'Charlson Comorbidity Index',\n 'mis_charlson_score': 'Charlson comorbidity index not available',\n}\n\ndf.c = df.c.apply(lambda x: consistent_c[x])\n\nunique_g = df.g.unique().tolist()\nprint(unique_g)\n\nunique_c = df.c.unique().tolist()\nprint(unique_c)\n\ndf",
"_____no_output_____"
]
],
[
[
"# All Sites",
"_____no_output_____"
]
],
[
[
"point=alt.OverlayMarkDef(filled=False, fill='white', strokeWidth=2)\n\ndef plot_lab(df=None, metric='cov'):\n d = df.copy()\n \n plot = alt.Chart(\n d\n ).mark_bar(\n# point=True,\n size=10,\n# opacity=0.3\n ).encode(\n y=alt.Y(\"c:N\", title=None, axis=alt.Axis(labelAngle=0, tickCount=10), scale=alt.Scale(padding=1), sort=unique_c),\n x=alt.X(\"v:Q\", title=None, scale=alt.Scale(zero=True, domain=[-3,3], padding=2, nice=False, clamp=True)),\n # color=alt.Color(\"siteid:N\", scale=alt.Scale(domain=sites, range=site_colors)),\n color=alt.Color(\"g:N\", scale=alt.Scale(domain=unique_g, range=colors), title='Category'),\n ).properties(\n width=150,\n height=250\n )\n\n plot = plot.facet(\n column=alt.Column(\"siteid:N\", header=alt.Header(title=None), sort=sites)\n ).resolve_scale(color='shared')\n\n plot = plot.properties(\n title={\n \"text\": [\n f\"Coefficient\"\n ],\n \"dx\": 120,\n \"subtitle\": [\n 'Lab values are standarized by SD',\n get_visualization_subtitle(data_release=data_release, with_num_sites=False)\n ],\n \"subtitleColor\": \"gray\",\n }\n )\n\n return plot\n\nplot = plot_lab(df=df)\n\n# plot = alt.vconcat(*(\n# plot_lab(df=df, lab=lab) for lab in unique_sites\n# ), spacing=30)\n\nplot = apply_theme(\n plot,\n axis_y_title_font_size=16,\n title_anchor='start',\n legend_orient='bottom',\n legend_title_orient='left',\n axis_label_font_size=14,\n header_label_font_size=16,\n point_size=100\n)\n\nplot",
"_____no_output_____"
]
],
[
[
"## Final Meta",
"_____no_output_____"
]
],
[
[
"def plot_lab(df=None, metric='cov'):\n d = df.copy()\n d = d[d.siteid == 'META']\n print(unique_c)\n \n plot = alt.Chart(\n d\n ).mark_point(\n #point=True,\n size=120,\n filled=True,\n opacity=1\n ).encode(\n y=alt.Y(\"c:N\", title=None, axis=alt.Axis(labelAngle=0, tickCount=10, grid=True), scale=alt.Scale(padding=1), sort=unique_c),\n x=alt.X(\"v:Q\", title=\"Hazard Ratio\", scale=alt.Scale(zero=True, domain=[0,3.6], padding=0, nice=False, clamp=True)),\n # color=alt.Color(\"siteid:N\", scale=alt.Scale(domain=sites, range=site_colors)),\n color=alt.Color(\"g:N\", scale=alt.Scale(domain=unique_g, range=colors), title='Category',legend=None),\n ).properties(\n width=550,\n height=400\n )\n line = alt.Chart(pd.DataFrame({'x': [1]})).mark_rule().encode(x='x', strokeWidth=alt.value(1), strokeDash=alt.value([2, 2]))\n tick = plot.mark_errorbar(\n opacity=0.7 #, color='black',\n #color=alt.Color(\"g:N\", scale=alt.Scale(domain=unique_g, range=colors), title='Category')\n ).encode(\n y=alt.Y(\"c:N\", sort=unique_c),\n x=alt.X(\"ci_l:Q\", title=\"Hazard Ratio\"),\n x2=alt.X2(\"ci_u:Q\"),\n stroke=alt.value('black'),\n strokeWidth=alt.value(1)\n )\n plot = (line+tick+plot)\n #plot = plot.facet(\n # column=alt.Column(\"siteid:N\", header=alt.Header(title=None), sort=sites)\n #).resolve_scale(color='shared')\n\n #plot = plot.properties(\n # title={\n # \"text\": [\n # f\"Meta-Analysis Of Coefficient\"\n # ],\n # \"dx\": 120,\n # \"subtitle\": [\n # 'Lab values are standarized by SD'#,\n # #get_visualization_subtitle(data_release=data_release, with_num_sites=False)\n # ],\n # \"subtitleColor\": \"gray\",\n # }\n #)\n\n return plot\n\nplot = plot_lab(df=df)\n\n# plot = alt.vconcat(*(\n# plot_lab(df=df, lab=lab) for lab in unique_sites\n# ), spacing=30)\n\nplot = apply_theme(\n plot,\n axis_y_title_font_size=16,\n title_anchor='start',\n #legend_orient='bottom',\n #legend_title_orient='top',\n axis_label_font_size=14,\n header_label_font_size=16,\n point_size=100\n)\n\nplot.display()\nsave(plot,join(\"..\", \"result\", \"final-beta-std-mice-meta.png\"), scalefactor=8.0)\n\n",
"_____no_output_____"
]
],
[
[
"## Final country",
"_____no_output_____"
]
],
[
[
"def plot_beta(df=None, metric='cov', country=None):\n d = df.copy()\n d = d[d.siteid == country]\n \n plot = alt.Chart(\n d\n ).mark_point(\n# point=True,\n size=120,\n filled=True,\n opacity=1\n# opacity=0.3\n ).encode(\n y=alt.Y(\"c:N\", title=None, axis=alt.Axis(labelAngle=0, tickCount=10, grid=True), scale=alt.Scale(padding=1), sort=unique_c),\n x=alt.X(\"v:Q\", title=\"Hazard Ratio\", scale=alt.Scale(zero=True, domain=[0,4.6], padding=0, nice=False, clamp=True)),\n # color=alt.Color(\"siteid:N\", scale=alt.Scale(domain=sites, range=site_colors)),\n color=alt.Color(\"g:N\", scale=alt.Scale(domain=unique_g, range=colors), title='Category', legend=None),\n ).properties(\n width=750,\n height=550\n )\n line = alt.Chart(pd.DataFrame({'x': [1]})).mark_rule().encode(x='x', strokeWidth=alt.value(1), strokeDash=alt.value([2, 2]))\n\n tick = plot.mark_errorbar(\n opacity=0.7 #, color='black'\n ).encode(\n y=alt.Y(\"c:N\", sort=unique_c),\n x=alt.X(\"ci_l:Q\", title=\"Hazard Ratio\"),\n x2=alt.X2(\"ci_u:Q\"),\n stroke=alt.value('black'),\n strokeWidth=alt.value(1)\n )\n plot = (line+tick+plot)\n\n# plot = plot.facet(\n# column=alt.Column(\"siteid:N\", header=alt.Header(title=None), sort=sites)\n# ).resolve_scale(color='shared')\n\n plot = plot.properties(\n title={\n \"text\": [\n country.replace(\"META-\",\"\")\n ],\n \"dx\": 160,\n #\"subtitle\": [\n # 'Lab values are standarized by SD'\n #],\n #\"subtitleColor\": \"gray\",\n }\n )\n\n return plot\n\ncountrylist1 = [\"META-USA\", \"META-FRANCE\"]\ncountrylist2 = [\"META-GERMANY\", \"META-SPAIN\"]\n\n\nplot1 = alt.hconcat(*(\n plot_beta(df=df, country=country) for country in countrylist1\n), spacing=30).resolve_scale(color='independent')\n\n\nplot2 = alt.hconcat(*(\n plot_beta(df=df, country=country) for country in countrylist2\n), spacing=30).resolve_scale(color='independent')\n\n\nplot=alt.vconcat(plot1, plot2)\n#plot=plot1\n\nplot = apply_theme(\n plot,\n axis_y_title_font_size=16,\n title_anchor='start',\n legend_orient='bottom',\n legend_title_orient='left',\n axis_label_font_size=14,\n header_label_font_size=16,\n point_size=100\n)\n\n\nplot.display()\n\nsave(plot,join(\"..\", \"result\", \"final-beta-std-mice-country.png\"), scalefactor=8.0)\n\n\n",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
c50e04e4c84936d6e5c6d21a7ffcc8caffe2d7b1
| 2,185 |
ipynb
|
Jupyter Notebook
|
0.13/_downloads/plot_shift_evoked.ipynb
|
drammock/mne-tools.github.io
|
5d3a104d174255644d8d5335f58036e32695e85d
|
[
"BSD-3-Clause"
] | null | null | null |
0.13/_downloads/plot_shift_evoked.ipynb
|
drammock/mne-tools.github.io
|
5d3a104d174255644d8d5335f58036e32695e85d
|
[
"BSD-3-Clause"
] | null | null | null |
0.13/_downloads/plot_shift_evoked.ipynb
|
drammock/mne-tools.github.io
|
5d3a104d174255644d8d5335f58036e32695e85d
|
[
"BSD-3-Clause"
] | null | null | null | 40.462963 | 1,137 | 0.575286 |
[
[
[
"%matplotlib inline",
"_____no_output_____"
]
],
[
[
"\n# Shifting time-scale in evoked data\n\n\n\n",
"_____no_output_____"
]
],
[
[
"# Author: Mainak Jas <[email protected]>\n#\n# License: BSD (3-clause)\n\nimport matplotlib.pyplot as plt\nimport mne\nfrom mne.viz import tight_layout\nfrom mne.datasets import sample\n\nprint(__doc__)\n\ndata_path = sample.data_path()\n\nfname = data_path + '/MEG/sample/sample_audvis-ave.fif'\n\n# Reading evoked data\ncondition = 'Left Auditory'\nevoked = mne.read_evokeds(fname, condition=condition, baseline=(None, 0),\n proj=True)\n\nch_names = evoked.info['ch_names']\npicks = mne.pick_channels(ch_names=ch_names, include=[\"MEG 2332\"])\n\n# Create subplots\nf, (ax1, ax2, ax3) = plt.subplots(3)\nevoked.plot(exclude=[], picks=picks, axes=ax1,\n titles=dict(grad='Before time shifting'))\n\n# Apply relative time-shift of 500 ms\nevoked.shift_time(0.5, relative=True)\n\nevoked.plot(exclude=[], picks=picks, axes=ax2,\n titles=dict(grad='Relative shift: 500 ms'))\n\n# Apply absolute time-shift of 500 ms\nevoked.shift_time(0.5, relative=False)\n\nevoked.plot(exclude=[], picks=picks, axes=ax3,\n titles=dict(grad='Absolute shift: 500 ms'))\n\ntight_layout()",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code"
] |
[
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
c50e05ea470136ed5bc3933229fda5a845ac4b90
| 130 |
ipynb
|
Jupyter Notebook
|
notebooks/01 - test_cloudfront_data_access.ipynb
|
UBC-CIC/Sea_Around_Us_Spatial_Catch
|
37da0b0038490d2975637af99d9c381e61a962e3
|
[
"MIT"
] | null | null | null |
notebooks/01 - test_cloudfront_data_access.ipynb
|
UBC-CIC/Sea_Around_Us_Spatial_Catch
|
37da0b0038490d2975637af99d9c381e61a962e3
|
[
"MIT"
] | null | null | null |
notebooks/01 - test_cloudfront_data_access.ipynb
|
UBC-CIC/Sea_Around_Us_Spatial_Catch
|
37da0b0038490d2975637af99d9c381e61a962e3
|
[
"MIT"
] | null | null | null | 32.5 | 75 | 0.884615 |
[
[
[
"empty"
]
]
] |
[
"empty"
] |
[
[
"empty"
]
] |
c50e0799e1c34c5e58111df446e445cd489ec0cf
| 13,011 |
ipynb
|
Jupyter Notebook
|
nb/session17-on-your-own.ipynb
|
statsmaths/hilt2019-image-analysis
|
4cb6ec19cbeaf62c73fdc0732dff559f2596dace
|
[
"CC-BY-4.0"
] | 6 |
2019-06-05T12:56:37.000Z
|
2021-03-17T20:06:09.000Z
|
nb/session17-on-your-own.ipynb
|
statsmaths/hilt2019-image-analysis
|
4cb6ec19cbeaf62c73fdc0732dff559f2596dace
|
[
"CC-BY-4.0"
] | null | null | null |
nb/session17-on-your-own.ipynb
|
statsmaths/hilt2019-image-analysis
|
4cb6ec19cbeaf62c73fdc0732dff559f2596dace
|
[
"CC-BY-4.0"
] | 3 |
2019-06-02T14:50:43.000Z
|
2020-01-13T15:26:14.000Z
| 30.904988 | 164 | 0.497733 |
[
[
[
"# Session 17: Recommendation system on your own\n\nThis script should allow you to build an interactive website from your own\ndataset. If you run into any issues, please let us know!",
"_____no_output_____"
],
[
"## Step 1: Select the corpus\n\nIn the block below, insert the name of your corpus. There should \nbe images in the directory \"images\". If there is metadata, it should\nbe in the directory \"data\" with the name of the corpus as the file name.\n\nAlso, if there is metadata, there must be a column called filename (with\nthe filename to the image) and a column called title.",
"_____no_output_____"
]
],
[
[
"cn = \"test\"",
"_____no_output_____"
]
],
[
[
"## Step 2: Read in the Functions\n\nYou need to read in all of the modules and functions below.",
"_____no_output_____"
]
],
[
[
"%pylab inline\n\nimport numpy as np\nimport scipy as sp\nimport pandas as pd\nimport sklearn\nfrom sklearn import linear_model\nimport urllib\n\nimport os\nfrom os.path import join",
"_____no_output_____"
],
[
"from keras.applications.vgg19 import VGG19\nfrom keras.preprocessing import image\nfrom keras.applications.vgg19 import preprocess_input, decode_predictions\nfrom keras.models import Model",
"_____no_output_____"
],
[
"os.environ[\"KMP_DUPLICATE_LIB_OK\"] = \"TRUE\"",
"_____no_output_____"
],
[
"def check_create_metadata(cn):\n mdata = join(\"..\", \"data\", cn + \".csv\")\n\n if not os.path.exists(mdata):\n exts = [\".jpg\", \".JPG\", \".JPEG\", \".png\"]\n fnames = [x for x in os.listdir(join('..', 'images', cn)) if get_ext(x) in exts]\n \n df = pd.DataFrame({'filename': fnames, 'title': fnames})\n df.to_csv(mdata, index=False)",
"_____no_output_____"
],
[
"def create_embed(corpus_name):\n \n ofile = join(\"..\", \"data\", corpus_name + \"_vgg19_fc2.npy\")\n \n if not os.path.exists(ofile):\n\n vgg19_full = VGG19(weights='imagenet')\n vgg_fc2 = Model(inputs=vgg19_full.input, outputs=vgg19_full.get_layer('fc2').output)\n \n df = pd.read_csv(join(\"..\", \"data\", corpus_name + \".csv\"))\n output = np.zeros((len(df), 224, 224, 3))\n\n for i in range(len(df)):\n img_path = join(\"..\", \"images\", corpus_name, df.filename[i])\n img = image.load_img(img_path, target_size=(224, 224))\n x = image.img_to_array(img)\n output[i, :, :, :] = x\n if (i % 100) == 0:\n print(\"Loaded image {0:03d}\".format(i))\n\n output = preprocess_input(output)\n img_embed = vgg_fc2.predict(output, verbose=True)\n\n np.save(ofile, img_embed)",
"_____no_output_____"
],
[
"def rm_ext(s):\n return os.path.splitext(s)[0]\n\ndef get_ext(s):\n return os.path.splitext(s)[-1]",
"_____no_output_____"
],
[
"def clean_html(): \n if not os.path.exists(join(\"..\", \"html\")):\n os.makedirs(join(\"..\", \"html\"))\n \n if not os.path.exists(join(\"..\", \"html\", \"pages\")):\n os.makedirs(join(\"..\", \"html\", \"pages\"))\n \n for p in [x for x in os.listdir(join('..', 'html', 'pages')) if get_ext(x) in [\".html\", \"html\"]]:\n os.remove(join('..', 'html', 'pages', p))",
"_____no_output_____"
],
[
"def load_data(cn):\n X = np.load(join(\"..\", \"data\", cn + \"_vgg19_fc2.npy\"))\n return X",
"_____no_output_____"
],
[
"def write_header(f, cn, index=False):\n loc = \"\"\n if not index:\n loc = \"../\"\n \n f.write(\"<html>\\n\")\n f.write(' <link rel=\"icon\" href=\"{0:s}img/favicon.ico\">\\n'.format(loc))\n f.write(' <title>Distant Viewing Tutorial</title>\\n\\n')\n f.write(' <link rel=\"stylesheet\" type=\"text/css\" href=\"{0:s}css/bootstrap.min.css\">'.format(loc))\n f.write(' <link href=\"https://fonts.googleapis.com/css?family=Rubik+27px\" rel=\"stylesheet\">')\n f.write(' <link rel=\"stylesheet\" type=\"text/css\" href=\"{0:s}css/dv.css\">\\n\\n'.format(loc))\n\n f.write(\"<body>\\n\")\n f.write(' <div class=\"d-flex flex-column flex-md-row align-items-center p-3 px-md-4')\n f.write('mb-3 bg-white border-bottom box-shadow\">\\n')\n f.write(' <h4 class=\"my-0 mr-md-auto font-weight-normal\">Distant Viewing Tutorial Explorer')\n f.write('— {0:s}</h4>\\n'.format(cn.capitalize()))\n f.write(' <a class=\"btn btn-outline-primary\" href=\"{0:s}index.html\">Back to Index</a>\\n'.format(loc))\n f.write(' </div>\\n')\n f.write('\\n')",
"_____no_output_____"
],
[
"def corpus_to_html(corpus):\n pd.set_option('display.max_colwidth', -1)\n tc = corpus.copy()\n for index in range(tc.shape[0]):\n fname = rm_ext(os.path.split(tc['filename'][index])[1])\n title = rm_ext(tc['filename'][index])\n s = \"<a href='pages/{0:s}.html'>{1:s}</a>\".format(fname, title)\n tc.iloc[index, tc.columns.get_loc('title')] = s\n\n tc = tc.drop(['filename'], axis=1)\n return tc.to_html(index=False, escape=False, justify='center')",
"_____no_output_____"
],
[
"def create_index(cn, corpus):\n f = open(join('..', 'html', 'index.html'), 'w')\n write_header(f, cn=cn, index=True)\n f.write(' <div style=\"padding:20px; max-width:1000px\">\\n')\n\n f.write(corpus_to_html(corpus))\n\n f.write(' </div>\\n')\n f.write(\"</body>\\n\")\n f.close()",
"_____no_output_____"
],
[
"def get_infobox(corpus, item):\n infobox = []\n for k, v in corpus.iloc[item].to_dict().items():\n if k != \"filename\":\n infobox = infobox + [\"<p><b>\" + str(k).capitalize() + \":</b> \" + str(v) + \"</p>\"]\n return infobox",
"_____no_output_____"
],
[
"def save_metadata(f, cn, corpus, X, item):\n infobox = get_infobox(corpus, item)\n \n f.write(\"<div style='width: 1000px;'>\\n\")\n f.write(\"\\n\".join(infobox))\n if item > 0:\n link = rm_ext(os.path.split(corpus['filename'][item - 1])[-1])\n f.write(\"<p align='center'><a href='{0:s}.html'><< previous image</a> \\n\".format(link))\n\n if item + 1 < X.shape[0]:\n link = rm_ext(os.path.split(corpus['filename'][item + 1])[-1])\n f.write(\" <a href='{0:s}.html'>next image >></a></p>\\n\".format(link))\n\n f.write(\"</div>\\n\")",
"_____no_output_____"
],
[
"def save_similar_img(f, cn, corpus, X, item):\n dists = np.sum(np.abs(X - X[item, :]), 1)\n idx = np.argsort(dists.flatten())[1:13]\n \n f.write(\"<div style='clear:both; width: 1000px; padding-top: 30px'>\\n\")\n f.write(\"<h4>Similar Images:</h4>\\n\")\n f.write(\"<div class='similar'>\\n\")\n\n for img_path in corpus['filename'][idx].tolist():\n hpath = rm_ext(os.path.split(img_path)[1])\n f.write('<a href=\"{0:s}.html\"><img src=\"../../images/{1:2}/{2:s}\" style=\"max-width: 150px; padding:5px\"></a>\\n'.format(hpath, cn, img_path))\n\n f.write(\"</div>\\n\")\n f.write(\"</div>\\n\")",
"_____no_output_____"
],
[
"def create_image_pages(cn, corpus, X):\n for item in range(X.shape[0]):\n \n img_path = corpus['filename'][item]\n url = os.path.split(img_path)[1]\n \n f = open(join('..', 'html', 'pages', rm_ext(url) + \".html\"), 'w')\n write_header(f, cn, index=False)\n \n f.write(\"<div style='padding:25px'>\\n\")\n\n # Main image\n f.write(\"<div style='float: left; width: 610px;'>\\n\")\n f.write('<img src=\"../../images/{0:s}/{1:s}\" style=\"max-width: 600px; max-height: 500px;\">\\n'.format(cn, img_path))\n f.write(\"</div>\\n\\n\")\n \n # Main information box\n save_metadata(f, cn, corpus, X, item)\n \n # Similar\n save_similar_img(f, cn, corpus, X, item)\n\n f.write(\"</body>\\n\")\n f.close()",
"_____no_output_____"
]
],
[
[
"## Step 3: Create the embeddings\n\nThe next step is create the embeddings. If there is no metadata, this code\nwill also create it.",
"_____no_output_____"
]
],
[
[
"check_create_metadata(cn)\ncreate_embed(cn)",
"_____no_output_____"
]
],
[
[
"### Step 4: Create the website\n\nFinally, create the website with the code below.",
"_____no_output_____"
]
],
[
[
"clean_html()\ncorpus = pd.read_csv(join(\"..\", \"data\", cn + \".csv\"))\nX = load_data(cn)\n\ncreate_index(cn, corpus)\ncreate_image_pages(cn, corpus, X)",
"_____no_output_____"
]
],
[
[
"You should find a folder called `html`. Open that folder and double click on the\nfile `index.html`, opening it in a web browser (Chrome or Firefox preferred; Safari\nshould work too). Do not open it in Jupyter.\n\nYou will see a list of all of the available images from the corpus you selected.\nClick on one and you'll get to an item page for that image. From there you can\nsee the image itself, available metadata, select the previous or next image in the\ncorpus, and view similar images from the VGG19 similarity measurement.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
c50e15ba3b147f3844405db2a7a30eef753822bf
| 89,094 |
ipynb
|
Jupyter Notebook
|
Knn.ipynb
|
tugot17/KNN-Algorithm-From-Scratch
|
6fdc2e2b09da3a431973ebdabb1edddbd086b3c8
|
[
"MIT"
] | 3 |
2019-12-14T13:49:41.000Z
|
2020-06-17T09:55:54.000Z
|
Knn.ipynb
|
tugot17/KNN-Algorithm-From-Scratch
|
6fdc2e2b09da3a431973ebdabb1edddbd086b3c8
|
[
"MIT"
] | null | null | null |
Knn.ipynb
|
tugot17/KNN-Algorithm-From-Scratch
|
6fdc2e2b09da3a431973ebdabb1edddbd086b3c8
|
[
"MIT"
] | 2 |
2020-09-17T19:30:45.000Z
|
2020-09-28T19:45:13.000Z
| 86.836257 | 20,752 | 0.831268 |
[
[
[
"# K-Nearest Neighbors Algorithm\n",
"_____no_output_____"
],
[
"In this Jupyter Notebook we will focus on $KNN-Algorithm$. KNN is a data classification algorithm that attempts to determine what group a data point is in by looking at the data points around it.\n\nAn algorithm, looking at one point on a grid, trying to determine if a point is in group A or B, looks at the states of the points that are near it. The range is arbitrarily determined, but the point is to take a sample of the data. If the majority of the points are in group A, then it is likely that the data point in question will be A rather than B, and vice versa.\n<br>\n\n<img src=\"knn/example 1.png\" height=\"30%\" width=\"30%\">\n",
"_____no_output_____"
],
[
"# Imports",
"_____no_output_____"
]
],
[
[
"import numpy as np\nfrom tqdm import tqdm_notebook",
"_____no_output_____"
]
],
[
[
"# How it works?\n\nWe have some labeled data set $X-train$, and a new set $X$ that we want to classify based on previous classyfications\n\n",
"_____no_output_____"
],
[
"## Seps",
"_____no_output_____"
],
[
"### 1. Calculate distance to all neightbours\n### 2. Sort neightbours (based on closest distance)\n### 3. Count possibilities of each class for k nearest neighbours \n### 4. The class with highest possibilty is Your prediction",
"_____no_output_____"
],
[
"# 1. Calculate distance to all neighbours\n\nDepending on the problem You should use diffrent type of count distance method.\n<br>\nFor example we can use Euclidean distance. Euclidean distance is the \"ordinary\" straight-line distance between two points in D-Dimensional space\n\n#### Definiton\n$d(p, q) = d(q, p) = \\sqrt{(q_1 - p_1)^2 + (q_2 - p_2)^2 + \\dots + (q_D - p_D)^2} = \\sum_{d=1}^{D} (p_d - q_d)^2$\n\n#### Example\nDistance in $R^2$\n<img src=\"knn/euklidean_example.png\" height=\"30%\" width=\"30%\">\n\n\n$p = (4,6)$\n<br>\n$q = (1,2)$\n<br>\n$d(p, q) = \\sqrt{(1-4)^2 + (2-6)^2} =\\sqrt{9 + 16} = \\sqrt{25} = 5 $\n\n",
"_____no_output_____"
],
[
"## Code",
"_____no_output_____"
]
],
[
[
"def get_euclidean_distance(A_matrix, B_matrix):\n \"\"\"\n Function computes euclidean distance between matrix A and B\n Args:\n A_matrix (numpy.ndarray): Matrix size N1:D\n B_matrix (numpy.ndarray): Matrix size N2:D\n\n Returns:\n numpy.ndarray: Matrix size N1:N2\n \"\"\"\n\n A_square = np.reshape(np.sum(A_matrix * A_matrix, axis=1), (A_matrix.shape[0], 1))\n B_square = np.reshape(np.sum(B_matrix * B_matrix, axis=1), (1, B_matrix.shape[0]))\n AB = A_matrix @ B_matrix.T\n\n C = -2 * AB + B_square + A_square\n\n return np.sqrt(C)",
"_____no_output_____"
]
],
[
[
"## Example Usage",
"_____no_output_____"
]
],
[
[
"X = np.array([[1,2,3] , [-4,5,-6]])\n\nX_train = np.array([[0,0,0], [1,2,3], [4,5,6], [-4, 4, -6]])\n\nprint(\"X: {} Exaples in {} Dimensional space\".format(*X.shape))\nprint(\"X_train: {} Exaples in {} Dimensional space\".format(*X_train.shape))\n\n\nprint()\n\nprint(\"X:\")\nprint(X)\n\nprint()\n\nprint(\"X_train\")\nprint(X_train)\n",
"X: 2 Exaples in 3 Dimensional space\nX_train: 4 Exaples in 3 Dimensional space\n\nX:\n[[ 1 2 3]\n [-4 5 -6]]\n\nX_train\n[[ 0 0 0]\n [ 1 2 3]\n [ 4 5 6]\n [-4 4 -6]]\n"
],
[
"distance_matrix = get_euclidean_distance(X, X_train)\n\nprint(\"Distance Matrix shape: {}\".format(distance_matrix.shape))\nprint(\"Distance between first example from X and first form X_train {}\".format(distance_matrix[0,0]))\nprint(\"Distance between first example from X and second form X_train {}\".format(distance_matrix[0,1]))\n",
"Distance Matrix shape: (2, 4)\nDistance between first example from X and first form X_train 3.7416573867739413\nDistance between first example from X and second form X_train 0.0\n"
]
],
[
[
"# 2. Sort neightbours\n\nIn order to find best fitting class for our observations we need to find to which classes belong observation neightbours and then to sort classes based on the closest distance\n",
"_____no_output_____"
],
[
"## Code",
"_____no_output_____"
]
],
[
[
"def get_sorted_train_labels(distance_matrix, y):\n \"\"\"\n Function sorts y labels, based on probabilities from distances matrix\n Args:\n distance_matrix (numpy.ndarray): Distance Matrix, between points from X and X_train, size: N1:N2\n y (numpy.ndarray): vector of classes of X points, size: N1\n\n Returns:\n numpy.ndarray: labels matrix sorted according to distances to nearest neightours, size N1:N2 \n\n \"\"\"\n order = distance_matrix.argsort(kind='mergesort')\n\n return np.squeeze(y[order])\n",
"_____no_output_____"
]
],
[
[
"## Example Usage",
"_____no_output_____"
]
],
[
[
"y_train = np.array([[1, 1, 2, 3]]).T\nprint(\"Labels array {} Examples in {} Dimensional Space\".format(*y_train.shape))\n\nprint(\"Distance matrix shape {}\".format(distance_matrix.shape))\n\nsorted_train_labels = get_sorted_train_labels(distance_matrix, y_train)\n\nprint(\"Sorted train labels {} shape\".format(sorted_train_labels.shape))\nprint(\"Closest 3 classes for first element from set X: {}\".format(sorted_train_labels[0, :3]))",
"Labels array 4 Examples in 1 Dimensional Space\nDistance matrix shape (2, 4)\nSorted train labels (2, 4) shape\nClosest 3 classes for first element from set X: [1 1 2]\n"
]
],
[
[
"# 3. Count possibilities of each class for k nearest neighbours \n\nIn order to find best class for our observation $x$ we need to calculate the probability of belonging to each class. In our case it is quite easy. We need just to count how many from k-nearest-neighbours of observation $x$ belong to each class and then devide it by k \n<br><br>\n$p(y=class \\space| x) = \\frac{\\sum_{1}^{k}(1 \\space if \\space N_i = class, \\space else \\space 0) }{k}$ Where $N_i$ is $i$ nearest neightbour\n\n",
"_____no_output_____"
],
[
"## Code",
"_____no_output_____"
]
],
[
[
"def get_p_y_x_using_knn(y, k):\n \"\"\"\n The function determines the probability distribution p (y | x)\n for each of the labels for objects from the X\n using the KNN classification learned on the X_train\n\n Args:\n y (numpy.ndarray): Sorted matrix of N2 nearest neighbours labels, size N1:N2\n k (int): number of nearest neighbours for KNN algorithm\n\n Returns: numpy.ndarray: Matrix of probabilities for N1 points (from set X) of belonging to each class,\n size N1:C (where C is number of classes)\n \"\"\"\n\n first_k_neighbors = y[:, :k]\n \n N1, N2 = y.shape\n classes = np.unique(y)\n number_of_classes = classes.shape[0]\n\n probabilities_matrix = np.zeros(shape=(N1, number_of_classes))\n\n for i, row in enumerate(first_k_neighbors):\n for j, value in enumerate(classes):\n probabilities_matrix[i][j] = list(row).count(value) / k\n\n return probabilities_matrix\n",
"_____no_output_____"
]
],
[
[
"## Example usage",
"_____no_output_____"
]
],
[
[
"print(\"Sorted train labels:\")\nprint(sorted_train_labels)\n\nprobabilities_matrix = get_p_y_x_using_knn(y=sorted_train_labels, k=4)\n\n\n\nprint(\"Probability fisrt element belongs to 1-st class: {:2f}\".format(probabilities_matrix[0,0]))\nprint(\"Probability fisrt element belongs to 3-rd class: {:2f}\".format(probabilities_matrix[0,2]))\n\n",
"Sorted train labels:\n[[1 1 2 3]\n [3 1 1 2]]\nProbability fisrt element belongs to 1-st class: 0.500000\nProbability fisrt element belongs to 3-rd class: 0.250000\n"
]
],
[
[
"# 4. The class with highest possibilty is Your prediction",
"_____no_output_____"
],
[
"At the end we combine all previous steps to get prediction",
"_____no_output_____"
],
[
"## Code",
"_____no_output_____"
]
],
[
[
"def predict(X, X_train, y_train, k, distance_function):\n \"\"\"\n Function returns predictions for new set X based on labels of points from X_train\n Args:\n X (numpy.ndarray): set of observations (points) that we want to label\n X_train (numpy.ndarray): set of lalabeld bservations (points)\n y_train (numpy.ndarray): labels for X_train\n k (int): number of nearest neighbours for KNN algorithm\n\n Returns:\n (numpy.ndarray): label predictions for points from set X\n \"\"\"\n distance_matrix = distance_function(X, X_train)\n\n sorted_labels = get_sorted_train_labels(distance_matrix=distance_matrix, y=y_train)\n \n p_y_x = get_p_y_x_using_knn(y=sorted_labels, k=k)\n\n number_of_classes = p_y_x.shape[1]\n reversed_rows = np.fliplr(p_y_x)\n\n prediction = number_of_classes - (np.argmax(reversed_rows, axis=1) + 1)\n\n return prediction",
"_____no_output_____"
]
],
[
[
"## Example usage",
"_____no_output_____"
]
],
[
[
"prediction = predict(X, X_train, y_train, 3, get_euclidean_distance)\n\nprint(\"Predicted propabilities of classes for for first observation\", probabilities_matrix[0])\nprint(\"Predicted class for for first observation\", prediction[0])\n\nprint()\n\nprint(\"Predicted propabilities of classes for for second observation\", probabilities_matrix[1])\nprint(\"Predicted class for for second observation\", prediction[1])",
"Predicted propabilities of classes for for first observation [0.5 0.25 0.25]\nPredicted class for for first observation 0\n\nPredicted propabilities of classes for for second observation [0.5 0.25 0.25]\nPredicted class for for second observation 0\n"
]
],
[
[
"# Accuracy",
"_____no_output_____"
],
[
"To find how good our knn model works we should count accuracy",
"_____no_output_____"
],
[
"## Code",
"_____no_output_____"
]
],
[
[
"def count_accuracy(prediction, y_true):\n \"\"\"\n Returns:\n float: Predictions accuracy\n\n \"\"\"\n N1 = prediction.shape[0]\n \n accuracy = np.sum(prediction == y_true) / N1\n\n return accuracy",
"_____no_output_____"
]
],
[
[
"## Example usage",
"_____no_output_____"
]
],
[
[
"y_true = np.array([[0, 2]])\n\npredicton = predict(X, X_train, y_train, 3, get_euclidean_distance)\n\n\nprint(\"True classes:{}, accuracy {}%\".format(y_true, count_accuracy(predicton, y_true) * 100))",
"True classes:[[0 2]], accuracy 50.0%\n"
]
],
[
[
"# Find best k",
"_____no_output_____"
],
[
"Best k parameter is that one for which we have highest accuracy",
"_____no_output_____"
],
[
"## Code",
"_____no_output_____"
]
],
[
[
"def select_knn_model(X_validation, y_validation, X_train, y_train, k_values, distance_function):\n \"\"\"\n Function returns k parameter that best fit Xval points\n Args:\n Xval (numpy.ndarray): set of Validation Data, size N1:D\n Xtrain (numpy.ndarray): set of Training Data, size N2:D\n yval (numpy.ndarray): set of labels for Validation data, size N1:1\n ytrain (numpy.ndarray): set of labels for Training Data, size N2:1\n k_values (list): list of int values of k parameter that should be checked\n\n Returns:\n int: k paprameter that best fit validation set\n \"\"\"\n\n accuracies = []\n\n for k in tqdm_notebook(k_values):\n prediction = predict(X_validation, X_train, y_train, k, distance_function)\n\n accuracy = count_accuracy(prediction, y_validation)\n accuracies.append(accuracy)\n\n best_k = k_values[accuracies.index(max(accuracies))]\n\n return best_k, accuracies\n",
"_____no_output_____"
]
],
[
[
"# Real World Example - Iris Dataset",
"_____no_output_____"
],
[
"\n<img src=\"knn/iris_example1.jpeg\" height=\"60%\" width=\"60%\">\n\n\nThis is perhaps the best known database to be found in the pattern recognition literature. The data set contains 3 classes of 50 instances each, where each class refers to a type of iris plant. One class is linearly separable from the other 2; the latter are NOT linearly separable from each other. \n\nEach example contains 4 attributes\n1. sepal length in cm \n2. sepal width in cm \n3. petal length in cm \n4. petal width in cm \n\nPredicted attribute: class of iris plant. \n\n<img src=\"knn/iris_example2.png\" height=\"70%\" width=\"70%\">\n\n\n\n\n",
"_____no_output_____"
]
],
[
[
"from sklearn import datasets\nimport matplotlib.pyplot as plt\n\niris = datasets.load_iris()\n\niris_X = iris.data\niris_y = iris.target\n\nprint(\"Iris: {} examples in {} dimensional space\".format(*iris_X.shape))\nprint(\"First example in dataset :\\n Speal lenght: {}cm \\n Speal width: {}cm \\n Petal length: {}cm \\n Petal width: {}cm\".format(*iris_X[0]))\n\nprint(\"Avalible classes\", np.unique(iris_y))",
"Iris: 150 examples in 4 dimensional space\nFirst example in dataset :\n Speal lenght: 5.1cm \n Speal width: 3.5cm \n Petal length: 1.4cm \n Petal width: 0.2cm\nAvalible classes [0 1 2]\n"
]
],
[
[
"## Prepare Data\n\nIn our data set we have 150 examples (50 examples of each class), we have to divide it into 3 datasets.\n1. Training data set, 90 examples. It will be used to find k - nearest neightbours\n2. Validation data set, 30 examples. It will be used to find best k parameter, the one for which accuracy is highest\n3. Test data set, 30 examples. It will be used to check how good our model performs\n\nData has to be shuffled (mixed in random order), because originally it is stored 50 examples of class 0, 50 of 1 and 50 of 2.\n",
"_____no_output_____"
]
],
[
[
"from sklearn.utils import shuffle\n\niris_X, iris_y = shuffle(iris_X, iris_y, random_state=134)\n\n\ntest_size = 30\nvalidation_size = 30\ntraining_size = 90\n\nX_test = iris_X[:test_size]\nX_validation = iris_X[test_size: (test_size+validation_size)]\nX_train = iris_X[(test_size+validation_size):]\n\ny_test = iris_y[:test_size]\ny_validation = iris_y[test_size: (test_size+validation_size)]\ny_train = iris_y[(test_size+validation_size):]",
"_____no_output_____"
]
],
[
[
"## Find best k parameter",
"_____no_output_____"
]
],
[
[
"k_values = [i for i in range(3,50)]\n\nbest_k, accuracies = select_knn_model(X_validation, y_validation, X_train, y_train, k_values, distance_function=get_euclidean_distance)\n\nplt.plot(k_values, accuracies)\nplt.xlabel('K parameter')\nplt.ylabel('Accuracy')\nplt.title('Accuracy for k nearest neighbors')\nplt.grid()\nplt.show()\n",
"_____no_output_____"
]
],
[
[
"## Count accuracy for training set",
"_____no_output_____"
]
],
[
[
"prediction = predict(X_test, X_train, y_train, best_k, get_euclidean_distance)\n\naccuracy = count_accuracy(prediction, y_test)\n\nprint(\"Accuracy for best k={}: {:2f}%\".format(best_k, accuracy*100))\n",
"Accuracy for best k=14: 93.333333%\n"
]
],
[
[
"# Real World Example - Mnist Dataset",
"_____no_output_____"
],
[
"Mnist is a popular database of handwritten images created for people who are new to machine learning. There are many courses on the internet that include classification problem using MNIST dataset.\n\nThis dataset contains 55000 images and labels. Each image is 28x28 pixels large, but for the purpose of the classification task they are flattened to 784x1 arrays $(28 \\cdot 28 = 784)$. Summing up our training set is a matrix of size $[50000, 784]$ = [amount of images, size of image]. We will split it into 40000 training examples and 10000 validation examples to choose a best k\n\n\nIt also contains 5000 test images and labels, but for test we will use only 1000 (due to time limitations, using 5k would take 5x as much time) \n\n\n<h3>Mnist Data Example</h3>\n<img src=\"knn/mnist_example.jpg\" height=\"70%\" width=\"70%\">\n\nNow we are going to download this dataset and split it into test and train sets.",
"_____no_output_____"
]
],
[
[
"import utils\nimport cv2\n\ntraining_size = 49_000\nvalidation_size = 1000\ntest_size = 1000\n\ntrain_data, test = utils.get_mnist_dataset()\n\ntrain_images, train_labels = train_data\ntest_images, test_labels = test\n\nvalidation_images = train_images[training_size:training_size + validation_size]\ntrain_images = train_images[:training_size]\n\nvalidation_labels = train_labels[training_size:training_size + validation_size]\ntrain_labels = train_labels[:training_size]\n\ntest_images = test_images[:test_size]\ntest_labels = test_labels[:test_size]\n\n\nprint(\"Training images matrix size: {}\".format(train_images.shape))\nprint(\"Training labels matrix size: {}\".format(train_labels.shape))\n\nprint(\"Validation images matrix size: {}\".format(validation_images.shape))\nprint(\"Validation labels matrix size: {}\".format(validation_labels.shape))\n\nprint(\"Testing images matrix size: {}\".format(test_images.shape))\nprint(\"Testing labels matrix size: {}\".format(test_labels.shape))\n\nprint(\"Possible labels {}\".format(np.unique(test_labels)))\n\n\n",
"Training images matrix size: (49000, 784)\nTraining labels matrix size: (49000,)\nValidation images matrix size: (1000, 784)\nValidation labels matrix size: (1000,)\nTesting images matrix size: (1000, 784)\nTesting labels matrix size: (1000,)\nPossible labels [0 1 2 3 4 5 6 7 8 9]\n"
]
],
[
[
"## Visualisation\nVisualisation isn't necessery to the problem, but it helps to understand what are we doing.",
"_____no_output_____"
]
],
[
[
"from matplotlib.gridspec import GridSpec\n\ndef show_first_8(images):\n ax =[]\n \n fig = plt.figure(figsize=(10, 10))\n\n gs = GridSpec(2, 4, wspace=0.0, hspace=-0.5)\n for i in range(2):\n for j in range(4):\n ax.append(fig.add_subplot(gs[i,j]))\n for i, axis in enumerate(ax):\n axis.imshow(images[i])\n\n plt.show()\n",
"_____no_output_____"
],
[
"first_8_images = train_images[:8]\nresized = np.reshape(first_8_images, (-1,28,28))\nprint('First 8 images of train set:')\nshow_first_8(resized)\n",
"First 8 images of train set:\n"
]
],
[
[
"## Find best k parameter",
"_____no_output_____"
]
],
[
[
"k_values = [i for i in range(3, 50, 5)]\n\nbest_k, accuracies = select_knn_model(validation_images, validation_labels, train_images, train_labels, k_values,\n distance_function=get_euclidean_distance)\n\nplt.plot(k_values, accuracies)\nplt.xlabel('K parameter')\nplt.ylabel('Accuracy')\nplt.title('Accuracy for k nearest neighbors')\nplt.grid()\nplt.show()\n\n",
"_____no_output_____"
],
[
"prediction = np.squeeze(predict(test_images, train_images, train_labels, best_k, get_euclidean_distance))\n\naccuracy = count_accuracy(prediction, test_labels)\n\nprint(\"Accuracy on test set for best k={}: {:2}%\".format(best_k, accuracy * 100))\n",
"Accuracy on test set for best k=3: 96.1%\n"
]
],
[
[
"# Sources\n\nhttps://en.wikipedia.org/wiki/K-nearest_neighbors_algorithm - first visualisation image\n\nhttps://en.wikipedia.org/wiki/Euclidean_distance - euclidean distance visualisation\n\nhttps://rajritvikblog.wordpress.com/2017/06/29/iris-dataset-analysis-python/ - first iris image\n\nhttps://rpubs.com/wjholst/322258 - second iris image\n\nhttps://www.kaggle.com/pablotab/mnistpklgz - mnist dataset\n",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
c50e30510f02d094328a4232383694f5cc63deb6
| 117,787 |
ipynb
|
Jupyter Notebook
|
Module2/Module2 - Lab5.ipynb
|
tsaomao/edX--MSDS-Curriculum--DAT210x
|
fc2624586e5bd682b2eb34177d16476e649f72c8
|
[
"MIT"
] | null | null | null |
Module2/Module2 - Lab5.ipynb
|
tsaomao/edX--MSDS-Curriculum--DAT210x
|
fc2624586e5bd682b2eb34177d16476e649f72c8
|
[
"MIT"
] | null | null | null |
Module2/Module2 - Lab5.ipynb
|
tsaomao/edX--MSDS-Curriculum--DAT210x
|
fc2624586e5bd682b2eb34177d16476e649f72c8
|
[
"MIT"
] | null | null | null | 37.861459 | 311 | 0.216144 |
[
[
[
"# DAT210x - Programming with Python for DS",
"_____no_output_____"
],
[
"## Module2 - Lab5",
"_____no_output_____"
],
[
"Import and alias Pandas:",
"_____no_output_____"
]
],
[
[
"import pandas as pd",
"_____no_output_____"
]
],
[
[
"As per usual, load up the specified dataset, setting appropriate header labels.",
"_____no_output_____"
]
],
[
[
"# Note: only 8 elements in provided header names, so need to slice out the first index element from original dataset\ncol_names = ['education', 'age', 'capital-gain', 'race', 'capital-loss', 'hours-per-week', 'sex', 'classification']\n# Import from csv. No headers.\ndf = pd.read_csv('./Datasets/census.data', sep=',', header=None)\n# Take the actual data slice. The pandas df will handle the index, so slice it out of the imported data frame\ndf = df.iloc[:, 1:]\n# Set proper column names\ndf.columns = col_names\n# Double check everything worked by looking at the dataset\ndf.head(10)",
"_____no_output_____"
],
[
"df.dtypes",
"_____no_output_____"
]
],
[
[
"Excellent.\n\nNow, use basic pandas commands to look through the dataset. Get a feel for it before proceeding!\n\nDo the data-types of each column reflect the values you see when you look through the data using a text editor / spread sheet program? If you see `object` where you expect to see `int32` or `float64`, that is a good indicator that there might be a string or missing value or erroneous value in the column.",
"_____no_output_____"
]
],
[
[
"# Make capital-gain numeric. Accept NaN coercions\ndf.loc[:, 'capital-gain'] = pd.to_numeric(df.loc[:, 'capital-gain'], errors='coerce')",
"_____no_output_____"
],
[
"# Same with capital-loss\ndf.loc[:, 'capital-loss'] = pd.to_numeric(df.loc[:, 'capital-loss'], errors='coerce')\ndf.dtypes",
"_____no_output_____"
]
],
[
[
"Try use `your_data_frame['your_column'].unique()` or equally, `your_data_frame.your_column.unique()` to see the unique values of each column and identify the rogue values.\n\nIf you find any value that should be properly encoded to NaNs, you can convert them either using the `na_values` parameter when loading the dataframe. Or alternatively, use one of the other methods discussed in the reading.",
"_____no_output_____"
]
],
[
[
"# Replace value of 99999 with None/NaN\nselector = df.loc[:, 'capital-gain'] == 99999\ndf.loc[selector, 'capital-gain'] = None\ndf['capital-gain'].unique()",
"_____no_output_____"
],
[
"# Example of unique() based queries. Found no other anomalies\ndf['classification'].unique()",
"_____no_output_____"
]
],
[
[
"Look through your data and identify any potential categorical features. Ensure you properly encode any ordinal and nominal types using the methods discussed in the chapter.\n\nBe careful! Some features can be represented as either categorical or continuous (numerical). If you ever get confused, think to yourself what makes more sense generally---to represent such features with a continuous numeric type... or a series of categories?",
"_____no_output_____"
]
],
[
[
"# UNCOMMENT TO RUN THROUGH AGAIN\n# ordered_education = ['Preschool', '1st-4th', '5th-6th', '7th-8th', \n# '9th', '10th', '11th', '12th', 'HS-grad', \n# 'Some-college', 'Bachelors', 'Masters', 'Doctorate']\n# df.education = df.education.astype(\"category\",\n# ordered=True,\n# categories=ordered_education).cat.codes\n# \n# ordered_classification = ['<=50K', '>50K']\n# df.classification = df.classification.astype(\"category\",\n# ordered=True,\n# categories=ordered_classification).cat.codes",
"_____no_output_____"
],
[
"df = pd.get_dummies(df,columns=['race'])",
"_____no_output_____"
],
[
"df",
"_____no_output_____"
],
[
"df = pd.get_dummies(df,columns=['sex'])\ndf",
"_____no_output_____"
]
],
[
[
"Lastly, print out your dataframe!",
"_____no_output_____"
]
],
[
[
"print(df)",
" education age capital-gain capital-loss hours-per-week \\\n0 10 39 2174.0 0 40 \n1 10 50 NaN 0 13 \n2 8 38 NaN 0 40 \n3 6 53 NaN 0 40 \n4 10 28 0.0 0 40 \n5 11 37 0.0 0 40 \n6 4 49 0.0 0 16 \n7 8 52 0.0 0 45 \n8 11 31 14084.0 0 50 \n9 10 42 5178.0 0 40 \n10 9 37 0.0 0 80 \n11 10 30 0.0 0 40 \n12 10 23 0.0 0 30 \n13 3 34 0.0 0 45 \n14 8 25 0.0 0 35 \n15 8 32 0.0 0 40 \n16 6 38 0.0 0 50 \n17 11 43 0.0 0 45 \n18 12 40 0.0 0 60 \n19 8 54 0.0 0 20 \n20 4 35 0.0 0 40 \n21 6 43 0.0 2042 40 \n22 8 59 0.0 0 40 \n23 10 56 0.0 0 40 \n24 8 19 0.0 0 40 \n25 9 54 0.0 0 60 \n26 8 39 0.0 0 80 \n27 8 49 0.0 0 40 \n28 9 20 0.0 0 44 \n29 10 45 0.0 1408 40 \n... ... ... ... ... ... \n29506 8 30 0.0 0 55 \n29507 5 32 0.0 0 40 \n29508 9 22 0.0 0 40 \n29509 8 31 0.0 0 40 \n29510 8 29 0.0 0 35 \n29511 10 35 0.0 0 55 \n29512 10 30 0.0 0 99 \n29513 12 34 0.0 0 60 \n29514 10 54 0.0 0 50 \n29515 9 37 0.0 0 39 \n29516 7 22 0.0 0 35 \n29517 10 34 0.0 0 55 \n29518 8 30 0.0 0 46 \n29519 10 38 15020.0 0 45 \n29520 12 71 0.0 0 10 \n29521 8 45 0.0 0 40 \n29522 8 41 0.0 0 32 \n29523 8 72 0.0 0 25 \n29524 11 31 0.0 0 30 \n29525 8 43 0.0 0 40 \n29526 9 43 0.0 0 40 \n29527 9 43 0.0 0 50 \n29528 5 32 0.0 0 40 \n29529 11 32 0.0 0 11 \n29530 11 53 0.0 0 40 \n29531 9 22 0.0 0 40 \n29532 8 40 0.0 0 40 \n29533 8 58 0.0 0 40 \n29534 8 22 0.0 0 20 \n29535 8 52 15024.0 0 40 \n\n classification race_Amer-Indian-Eskimo race_Asian-Pac-Islander \\\n0 0 0 0 \n1 0 0 0 \n2 0 0 0 \n3 0 0 0 \n4 0 0 0 \n5 0 0 0 \n6 0 0 0 \n7 1 0 0 \n8 1 0 0 \n9 1 0 0 \n10 1 0 0 \n11 1 0 1 \n12 0 0 0 \n13 0 1 0 \n14 0 0 0 \n15 0 0 0 \n16 0 0 0 \n17 1 0 0 \n18 1 0 0 \n19 0 0 0 \n20 0 0 0 \n21 0 0 0 \n22 0 0 0 \n23 1 0 0 \n24 0 0 0 \n25 1 0 1 \n26 0 0 0 \n27 0 0 0 \n28 0 0 0 \n29 0 0 0 \n... ... ... ... \n29506 0 0 0 \n29507 0 0 0 \n29508 0 0 0 \n29509 0 0 0 \n29510 0 0 0 \n29511 1 0 0 \n29512 0 0 1 \n29513 1 0 0 \n29514 1 0 1 \n29515 0 0 0 \n29516 0 0 0 \n29517 1 0 0 \n29518 0 0 0 \n29519 1 0 0 \n29520 1 0 0 \n29521 0 0 0 \n29522 0 0 0 \n29523 0 0 0 \n29524 0 0 0 \n29525 0 0 0 \n29526 0 0 0 \n29527 0 0 0 \n29528 0 1 0 \n29529 0 0 1 \n29530 1 0 0 \n29531 0 0 0 \n29532 1 0 0 \n29533 0 0 0 \n29534 0 0 0 \n29535 1 0 0 \n\n race_Black race_Other race_White sex_Female sex_Male \n0 0 0 1 0 1 \n1 0 0 1 0 1 \n2 0 0 1 0 1 \n3 1 0 0 0 1 \n4 1 0 0 1 0 \n5 0 0 1 1 0 \n6 1 0 0 1 0 \n7 0 0 1 0 1 \n8 0 0 1 1 0 \n9 0 0 1 0 1 \n10 1 0 0 0 1 \n11 0 0 0 0 1 \n12 0 0 1 1 0 \n13 0 0 0 0 1 \n14 0 0 1 0 1 \n15 0 0 1 0 1 \n16 0 0 1 0 1 \n17 0 0 1 1 0 \n18 0 0 1 0 1 \n19 1 0 0 1 0 \n20 1 0 0 0 1 \n21 0 0 1 0 1 \n22 0 0 1 1 0 \n23 0 0 1 0 1 \n24 0 0 1 0 1 \n25 0 0 0 0 1 \n26 0 0 1 0 1 \n27 0 0 1 0 1 \n28 1 0 0 0 1 \n29 0 0 1 0 1 \n... ... ... ... ... ... \n29506 0 0 1 0 1 \n29507 0 0 1 0 1 \n29508 0 0 1 0 1 \n29509 0 0 1 1 0 \n29510 0 0 1 1 0 \n29511 0 0 1 1 0 \n29512 0 0 0 1 0 \n29513 0 0 1 0 1 \n29514 0 0 0 0 1 \n29515 0 0 1 1 0 \n29516 1 0 0 0 1 \n29517 0 0 1 1 0 \n29518 1 0 0 0 1 \n29519 1 0 0 1 0 \n29520 0 0 1 0 1 \n29521 0 0 1 1 0 \n29522 1 0 0 1 0 \n29523 0 0 1 0 1 \n29524 0 1 0 1 0 \n29525 0 0 1 0 1 \n29526 0 0 1 1 0 \n29527 0 0 1 0 1 \n29528 0 0 0 0 1 \n29529 0 0 0 0 1 \n29530 0 0 1 0 1 \n29531 0 0 1 0 1 \n29532 0 0 1 0 1 \n29533 0 0 1 1 0 \n29534 0 0 1 0 1 \n29535 0 0 1 1 0 \n\n[29536 rows x 13 columns]\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
c50e3afd9308ba18e6b099b1b7c459e52afa7b69
| 5,147 |
ipynb
|
Jupyter Notebook
|
Code from Jennifer/data-api-examples.ipynb
|
joliang17/Ripple_ML
|
5170af18311c367c74120457b30d11053f31e351
|
[
"MIT"
] | null | null | null |
Code from Jennifer/data-api-examples.ipynb
|
joliang17/Ripple_ML
|
5170af18311c367c74120457b30d11053f31e351
|
[
"MIT"
] | null | null | null |
Code from Jennifer/data-api-examples.ipynb
|
joliang17/Ripple_ML
|
5170af18311c367c74120457b30d11053f31e351
|
[
"MIT"
] | 4 |
2019-10-22T21:59:40.000Z
|
2020-01-12T22:19:03.000Z
| 5,147 | 5,147 | 0.664465 |
[
[
[
"%load_ext autoreload\n%autoreload 2",
"_____no_output_____"
],
[
"import pyrds\nimport pandas as pd\nimport requests\nimport datetime\n\nfrom google.cloud import bigquery\nclient = bigquery.Client()",
"_____no_output_____"
],
[
"lookback = 7\n\ntoday = datetime.datetime.today()\nstart_date = (today - datetime.timedelta(days=lookback)).strftime('%Y-%m-%d')\nend_date = today.strftime('%Y-%m-%d')",
"_____no_output_____"
]
],
[
[
"Accounts created daily",
"_____no_output_____"
]
],
[
[
"url = f'https://data.ripple.com/v2/stats/?start={start_date}&end={end_date}&interval=day&family=metric&metrics=accounts_created'\nres = requests.get(url)\nxrp_accts = pd.DataFrame(res.json()['stats'])\nxrp_accts",
"_____no_output_____"
]
],
[
[
"Network stats: number of nodes and validators",
"_____no_output_____"
]
],
[
[
"date_list = [x.strftime('%Y-%m-%dT%H:%M:%SZ') for x in list(pd.date_range((today - datetime.timedelta(days=lookback)).strftime('%Y%m%d'),today.strftime('%Y%m%d'),freq='1D'))]\nnode_list = []\n\nfor d in date_list:\n url = f'https://data.ripple.com/v2/network/topology?verbose=true&date={d}'\n res = requests.get(url)\n try:\n node_list.append(res.json()['node_count'])\n except:\n node_list.append(None)\n print(f'Error with {d}')\nnodes_df = pd.DataFrame({'date': date_list, 'nodes': node_list})\nnodes_df",
"_____no_output_____"
],
[
"res = requests.get(f'https://data.ripple.com/v2/network/validators')\nres.json()['count']",
"_____no_output_____"
]
],
[
[
"Ledger data: transaction count and value\n\nRequires a GBQ account. You can provide service account JSON credentials as an argument. If you want to run as your google user from your PC, you should first install the [Google Cloud SDK](https://cloud.google.com/sdk/), then run:\n`gcloud auth application-default login`",
"_____no_output_____"
]
],
[
[
"def gbq_query(query, query_params=None):\n \"\"\"\n Run a query against Google Big Query, returning a pandas dataframe of the result.\n\n Parameters\n ----------\n query: str\n The query string\n query_params: list, optional\n The query parameters to pass into the query string\n \"\"\"\n client = bigquery.Client()\n job_config = bigquery.QueryJobConfig()\n job_config.query_parameters = query_params\n return client.query(query, job_config=job_config).to_dataframe()",
"_____no_output_____"
],
[
"query = \"\"\"\nselect\n date(l.CloseTime) as `date`\n , t.TransactionType\n , count(1) as txn_count\n , sum(t.AmountXRP) / 1e6 as txn_value\nfrom `xrpledgerdata.fullhistory.transactions` t\njoin `xrpledgerdata.fullhistory.ledgers` l\n on t.LedgerIndex = l.LedgerIndex\nwhere t.TransactionResult = \"tesSUCCESS\"\n and date(l.CloseTime) >= CAST(@start_date AS DATE)\ngroup by 1,2\norder by 1 desc, 2\n\"\"\"\n\nquery_params = [\n bigquery.ScalarQueryParameter(\"start_date\", \"STRING\", start_date)\n]\n\nxrp = gbq_query(query,query_params)\nxrp",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
c50e42efa62ab82705faeb6df014c4a2ce000401
| 1,477 |
ipynb
|
Jupyter Notebook
|
Section 1/1.1_Installing_TensorFlow_Environment.ipynb
|
manpreet-kau-r/Hands-on-Machine-Learning-with-TensorFlow
|
b93941cb76ee3b0f00bad8877e0f437fc7db7eb4
|
[
"MIT"
] | 1 |
2021-07-20T04:44:42.000Z
|
2021-07-20T04:44:42.000Z
|
Section 1/1.1_Installing_TensorFlow_Environment.ipynb
|
manpreet-kau-r/Hands-on-Machine-Learning-with-TensorFlow
|
b93941cb76ee3b0f00bad8877e0f437fc7db7eb4
|
[
"MIT"
] | null | null | null |
Section 1/1.1_Installing_TensorFlow_Environment.ipynb
|
manpreet-kau-r/Hands-on-Machine-Learning-with-TensorFlow
|
b93941cb76ee3b0f00bad8877e0f437fc7db7eb4
|
[
"MIT"
] | null | null | null | 16.054348 | 42 | 0.486798 |
[
[
[
"# First Program with TensorFlow",
"_____no_output_____"
]
],
[
[
"import tensorflow as tf",
"_____no_output_____"
],
[
"hello = tf.constant(\"hello world\")",
"_____no_output_____"
],
[
"sess = tf.Session()",
"_____no_output_____"
],
[
"print(sess.run(hello))",
"b'hello world'\n"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
c50e46d34eeb96044dcabd56ce8411318d019626
| 99,800 |
ipynb
|
Jupyter Notebook
|
quantization/noise_shaping.ipynb
|
Fun-pee/signal-processing
|
205d5e55e3168a1ec9da76b569af92c0056619aa
|
[
"MIT"
] | 3 |
2020-09-21T10:15:40.000Z
|
2020-09-21T13:36:40.000Z
|
quantization/noise_shaping.ipynb
|
thegodparticle/digital-signal-processing-lecture
|
205d5e55e3168a1ec9da76b569af92c0056619aa
|
[
"MIT"
] | null | null | null |
quantization/noise_shaping.ipynb
|
thegodparticle/digital-signal-processing-lecture
|
205d5e55e3168a1ec9da76b569af92c0056619aa
|
[
"MIT"
] | null | null | null | 58.948612 | 25,058 | 0.621934 |
[
[
[
"# Quantization of Signals\n\n*This jupyter notebook is part of a [collection of notebooks](../index.ipynb) on various topics of Digital Signal Processing. Please direct questions and suggestions to [[email protected]](mailto:[email protected]).*",
"_____no_output_____"
],
[
"## Spectral Shaping of the Quantization Noise\n\nThe quantized signal $x_Q[k]$ can be expressed by the continuous amplitude signal $x[k]$ and the quantization error $e[k]$ as\n\n\\begin{equation}\nx_Q[k] = \\mathcal{Q} \\{ x[k] \\} = x[k] + e[k]\n\\end{equation}\n\nAccording to the [introduced model](linear_uniform_quantization_error.ipynb#Model-for-the-Quantization-Error), the quantization noise can be modeled as uniformly distributed white noise. Hence, the noise is distributed over the entire frequency range. The basic concept of [noise shaping](https://en.wikipedia.org/wiki/Noise_shaping) is a feedback of the quantization error to the input of the quantizer. This way the spectral characteristics of the quantization noise can be modified, i.e. spectrally shaped. Introducing a generic filter $h[k]$ into the feedback loop yields the following structure\n\n\n\nThe quantized signal can be deduced from the block diagram above as\n\n\\begin{equation}\nx_Q[k] = \\mathcal{Q} \\{ x[k] - e[k] * h[k] \\} = x[k] + e[k] - e[k] * h[k]\n\\end{equation}\n\nwhere the additive noise model from above has been introduced and it has been assumed that the impulse response $h[k]$ is normalized such that the magnitude of $e[k] * h[k]$ is below the quantization step $Q$. The overall quantization error is then\n\n\\begin{equation}\ne_H[k] = x_Q[k] - x[k] = e[k] * (\\delta[k] - h[k])\n\\end{equation}\n\nThe power spectral density (PSD) of the quantization error with noise shaping is calculated to\n\n\\begin{equation}\n\\Phi_{e_H e_H}(\\mathrm{e}^{\\,\\mathrm{j}\\,\\Omega}) = \\Phi_{ee}(\\mathrm{e}^{\\,\\mathrm{j}\\,\\Omega}) \\cdot \\left| 1 - H(\\mathrm{e}^{\\,\\mathrm{j}\\,\\Omega}) \\right|^2\n\\end{equation}\n\nHence the PSD $\\Phi_{ee}(\\mathrm{e}^{\\,\\mathrm{j}\\,\\Omega})$ of the quantizer without noise shaping is weighted by $| 1 - H(\\mathrm{e}^{\\,\\mathrm{j}\\,\\Omega}) |^2$. Noise shaping allows a spectral modification of the quantization error. The desired shaping depends on the application scenario. For some applications, high-frequency noise is less disturbing as low-frequency noise.",
"_____no_output_____"
],
[
"### Example - First-Order Noise Shaping\n\nIf the feedback of the error signal is delayed by one sample we get with $h[k] = \\delta[k-1]$\n\n\\begin{equation}\n\\Phi_{e_H e_H}(\\mathrm{e}^{\\,\\mathrm{j}\\,\\Omega}) = \\Phi_{ee}(\\mathrm{e}^{\\,\\mathrm{j}\\,\\Omega}) \\cdot \\left| 1 - \\mathrm{e}^{\\,-\\mathrm{j}\\,\\Omega} \\right|^2\n\\end{equation}\n\nFor linear uniform quantization $\\Phi_{ee}(\\mathrm{e}^{\\,\\mathrm{j}\\,\\Omega}) = \\sigma_e^2$ is constant. Hence, the spectral shaping constitutes a high-pass characteristic of first order. The following simulation evaluates the noise shaping quantizer of first order.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\nimport scipy.signal as sig\n%matplotlib inline\n\nw = 8 # wordlength of the quantized signal\nxmin = -1 # minimum of input signal\nN = 32768 # number of samples\n\n\ndef uniform_midtread_quantizer_w_ns(x, Q):\n # limiter\n x = np.copy(x)\n idx = np.where(x <= -1)\n x[idx] = -1\n idx = np.where(x > 1 - Q)\n x[idx] = 1 - Q\n # linear uniform quantization with noise shaping\n xQ = Q * np.floor(x/Q + 1/2)\n e = xQ - x\n xQ = xQ - np.concatenate(([0], e[0:-1]))\n\n return xQ[1:]\n\n\n# quantization step\nQ = 1/(2**(w-1))\n# compute input signal\nnp.random.seed(5)\nx = np.random.uniform(size=N, low=xmin, high=(-xmin-Q))\n# quantize signal\nxQ = uniform_midtread_quantizer_w_ns(x, Q)\ne = xQ - x[1:]\n# estimate PSD of error signal\nnf, Pee = sig.welch(e, nperseg=64)\n# estimate SNR\nSNR = 10*np.log10((np.var(x)/np.var(e)))\nprint('SNR = {:2.1f} dB'.format(SNR))\n\n\nplt.figure(figsize=(10, 5))\nOm = nf*2*np.pi\nplt.plot(Om, Pee*6/Q**2, label='estimated PSD')\nplt.plot(Om, np.abs(1 - np.exp(-1j*Om))**2, label='theoretic PSD')\nplt.plot(Om, np.ones(Om.shape), label='PSD w/o noise shaping')\nplt.title('PSD of quantization error')\nplt.xlabel(r'$\\Omega$')\nplt.ylabel(r'$\\hat{\\Phi}_{e_H e_H}(e^{j \\Omega}) / \\sigma_e^2$')\nplt.axis([0, np.pi, 0, 4.5])\nplt.legend(loc='upper left')\nplt.grid()",
"SNR = 45.2 dB\n"
]
],
[
[
"**Exercise**\n\n* The overall average SNR is lower than for the quantizer without noise shaping. Why?\n\nSolution: The average power per frequency is lower that without noise shaping for frequencies below $\\Omega \\approx \\pi$. However, this comes at the cost of a larger average power per frequency for frequencies above $\\Omega \\approx \\pi$. The average power of the quantization noise is given as the integral over the PSD of the quantization noise. It is larger for noise shaping and the resulting SNR is consequently lower. Noise shaping is nevertheless beneficial in applications where a lower quantization error in a limited frequency region is desired.",
"_____no_output_____"
],
[
"**Copyright**\n\nThis notebook is provided as [Open Educational Resource](https://en.wikipedia.org/wiki/Open_educational_resources). Feel free to use the notebook for your own purposes. The text is licensed under [Creative Commons Attribution 4.0](https://creativecommons.org/licenses/by/4.0/), the code of the IPython examples under the [MIT license](https://opensource.org/licenses/MIT). Please attribute the work as follows: *Sascha Spors, Digital Signal Processing - Lecture notes featuring computational examples*.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
c50e4ef9a4110c3dc137628f3405d8bd3c673cdd
| 4,791 |
ipynb
|
Jupyter Notebook
|
2_Omega_and_Xi_Constraints.ipynb
|
AndiA76/landmark-detection-and-tracking
|
e03425706508dc3fe9a080fd09c97b21a2baaa92
|
[
"MIT"
] | null | null | null |
2_Omega_and_Xi_Constraints.ipynb
|
AndiA76/landmark-detection-and-tracking
|
e03425706508dc3fe9a080fd09c97b21a2baaa92
|
[
"MIT"
] | null | null | null |
2_Omega_and_Xi_Constraints.ipynb
|
AndiA76/landmark-detection-and-tracking
|
e03425706508dc3fe9a080fd09c97b21a2baaa92
|
[
"MIT"
] | null | null | null | 40.948718 | 607 | 0.62033 |
[
[
[
"## Omega and Xi\n\nTo implement Graph SLAM, a matrix and a vector (omega and xi, respectively) are introduced. The matrix is square and labelled with all the robot poses (xi) and all the landmarks (Li). Every time you make an observation, for example, as you move between two poses by some distance `dx` and can relate those two positions, you can represent this as a numerical relationship in these matrices.\n\nIt's easiest to see how these work in an example. Below you can see a matrix representation of omega and a vector representation of xi.\n\n<img src='images/omega_xi.png' width=20% height=20% />\n\nNext, let's look at a simple example that relates 3 poses to one another. \n* When you start out in the world most of these values are zeros or contain only values from the initial robot position\n* In this example, you have been given constraints, which relate these poses to one another\n* Constraints translate into matrix values\n\n<img src='images/omega_xi_constraints.png' width=70% height=70% />\n\nIf you have ever solved linear systems of equations before, this may look familiar, and if not, let's keep going!\n\n### Solving for x\n\nTo \"solve\" for all these x values, we can use linear algebra; all the values of x are in the vector `mu` which can be calculated as a product of the inverse of omega times xi.\n\n<img src='images/solution.png' width=30% height=30% />\n\n---\n**You can confirm this result for yourself by executing the math in the cell below.**\n",
"_____no_output_____"
]
],
[
[
"import numpy as np\n\n# define omega and xi as in the example\nomega = np.array([[1,0,0],\n [-1,1,0],\n [0,-1,1]])\n\nxi = np.array([[-3],\n [5],\n [3]])\n\n# calculate the inverse of omega\nomega_inv = np.linalg.inv(np.matrix(omega))\n\n# calculate the solution, mu\nmu = omega_inv*xi\n\n# print out the values of mu (x0, x1, x2)\nprint(mu)",
"[[-3.]\n [ 2.]\n [ 5.]]\n"
]
],
[
[
"## Motion Constraints and Landmarks\n\nIn the last example, the constraint equations, relating one pose to another were given to you. In this next example, let's look at how motion (and similarly, sensor measurements) can be used to create constraints and fill up the constraint matrices, omega and xi. Let's start with empty/zero matrices.\n\n<img src='images/initial_constraints.png' width=35% height=35% />\n\nThis example also includes relationships between poses and landmarks. Say we move from x0 to x1 with a displacement `dx` of 5. Then we have created a motion constraint that relates x0 to x1, and we can start to fill up these matrices.\n\n<img src='images/motion_constraint.png' width=50% height=50% />\n\nIn fact, the one constraint equation can be written in two ways. So, the motion constraint that relates x0 and x1 by the motion of 5 has affected the matrix, adding values for *all* elements that correspond to x0 and x1.\n\n---\n\n### 2D case\n\nIn these examples, we've been showing you change in only one dimension, the x-dimension. In the project, it will be up to you to represent x and y positional values in omega and xi. One solution could be to create an omega and xi that are 2x larger that the number of robot poses (that will be generated over a series of time steps) and the number of landmarks, so that they can hold both x and y values for poses and landmark locations. I might suggest drawing out a rough solution to graph slam as you read the instructions in the next notebook; that always helps me organize my thoughts. Good luck!",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
c50e5f58f47e529ac2bafad87d9eb6f200c072de
| 164,597 |
ipynb
|
Jupyter Notebook
|
notebooks/vanilla-knn.ipynb
|
SergioAlvarezB/ml-numpy
|
bf450b0d48b52c56fd3d124a5b41f2b99594ea3b
|
[
"MIT"
] | null | null | null |
notebooks/vanilla-knn.ipynb
|
SergioAlvarezB/ml-numpy
|
bf450b0d48b52c56fd3d124a5b41f2b99594ea3b
|
[
"MIT"
] | null | null | null |
notebooks/vanilla-knn.ipynb
|
SergioAlvarezB/ml-numpy
|
bf450b0d48b52c56fd3d124a5b41f2b99594ea3b
|
[
"MIT"
] | null | null | null | 567.575862 | 47,104 | 0.948644 |
[
[
[
"# KNN\n\nK-Nearest Neighbors is a non-parametric model that can be used for classification, binary and multinomial, and for regression. Being non-parametric means that it predicts an output looking at the training data instead of using some learned parameters. This means that the training set needs to be stored.\n\nFor the classification task, given a new data example the model look at the k-closests points in the stored dataset and takes the most popular label among these points as the predicted label. For the regression problem, the output value is an average of the k-closests points.\n\nThe main hyperparameters of the model are: \n\n* k: A higher value entails greater computational cost and smoother decision boundaries, a low value might result in overfitting.\n\n* Similarity metric: The distance between points can be computed using euclidean distance, cosine distance, ot other similarity metrics",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nimport numpy as np\nimport sys\n\nsys.path.append(\"..\")\nfrom models.knn import KNNClassifier, KNNRegressor\nfrom utils.datasets import blobs_classification_dataset\nfrom utils.visualization import plot_decision_boundary",
"_____no_output_____"
]
],
[
[
"## Multinomial classification\n\nContrary to logistic regression, the KNN calssification algorithm can manage any number of labels. For instance:",
"_____no_output_____"
]
],
[
[
"(x_train, y_train), (x_test, y_test) = blobs_classification_dataset(features=2, classes=5, samples=500)\n\n# Visualize\nplt.scatter(x_train[:, 0], x_train[:, 1], c=y_train, cmap='jet')\nplt.show()",
"_____no_output_____"
],
[
"# Initialize the model.\nmodel = KNNClassifier(X=x_train, y=y_train)\n\n# KNN doesn't needs to fit the data. Predictions can be made \"out of the box\".\ny_hat = model.predict(x_test)\n\n\nacc = np.mean(y_hat==y_test)\n\nprint(\"Accuracy on the holdout set: %.2f\" % acc)",
"Accuracy on the holdout set: 0.84\n"
],
[
"# Visualize decision boundary\nax = plot_decision_boundary(model.predict,\n x_range=[x_test[:, 0].min()-1, x_test[:, 0].max()+1],\n y_range=[x_test[:, 1].min()-1, x_test[:, 1].max()+1], classes=5)\nax.scatter(x_test[:, 0], x_test[:, 1], c=y_test, cmap='jet', label='True classes')\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Non lineary-separable data\n\nThis model can also generalize well to other kinds of data distribution",
"_____no_output_____"
]
],
[
[
"# Load new data\nfrom utils.datasets import radial_classification_dataset\n(x_train, y_train), (x_test, y_test) = radial_classification_dataset(classes=4, samples=200)\n\n# Initialize the model.\nmodel = KNNClassifier(X=x_train, y=y_train)\n\n# Visualize decision boundary\nax = plot_decision_boundary(model.predict,\n x_range=[x_train[:, 0].min()-1, x_train[:, 0].max()+1],\n y_range=[x_train[:, 1].min()-1, x_train[:, 1].max()+1], classes=4)\nax.scatter(x_train[:, 0], x_train[:, 1], c=y_train, cmap='jet', label='True classes')\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Regression\n\nThe KNN algorithm can also be used to perform regression.",
"_____no_output_____"
]
],
[
[
"# Generate the data\nx_train = np.random.random_sample(100)*10\ny_train = np.sin(x_train) + np.random.randn(100)*0.01\n\n# Plot data\nplt.plot(x_train, y_train, 'o')\nplt.title('Sampled data from $sin(x)$')\nplt.show();",
"_____no_output_____"
],
[
"# Initialize regression model\nmodel = KNNRegressor(X=x_train.reshape([-1, 1]), y=y_train)\n\n# Plot regressed line over training data in the original range\nx_axis = np.linspace(0, 10, 200)\nplt.plot(x_train, y_train, 'o')\nplt.plot(x_axis, model.predict(x_axis), color='red')\nplt.show()",
"_____no_output_____"
]
],
[
[
"It may seem that the model learns very well the function, however it only performs well when there is enough close data. This happens when we try to predict values outside the original range.",
"_____no_output_____"
]
],
[
[
"# Plot regressed line over training data along a greater range\nx_axis = np.linspace(-10, 20, 400)\nplt.plot(x_train, y_train, 'o')\nplt.plot(x_axis, model.predict(x_axis), color='red')\nplt.show()",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
c50e629c2fc3268c70943c02c687f1c79b7c24c7
| 94,184 |
ipynb
|
Jupyter Notebook
|
Tamil Nadu Covishield Analysis.ipynb
|
nibras28/Tamil-Nadu-Covishield-Vaccine-Analysis
|
ca4f3e4a39334ff8c03758a1100864f63dd003dc
|
[
"MIT"
] | null | null | null |
Tamil Nadu Covishield Analysis.ipynb
|
nibras28/Tamil-Nadu-Covishield-Vaccine-Analysis
|
ca4f3e4a39334ff8c03758a1100864f63dd003dc
|
[
"MIT"
] | null | null | null |
Tamil Nadu Covishield Analysis.ipynb
|
nibras28/Tamil-Nadu-Covishield-Vaccine-Analysis
|
ca4f3e4a39334ff8c03758a1100864f63dd003dc
|
[
"MIT"
] | null | null | null | 52.646171 | 17,920 | 0.530568 |
[
[
[
"import numpy as np\nimport pandas as pd",
"_____no_output_____"
],
[
"#Data Importing",
"_____no_output_____"
],
[
"df = pd.read_csv(r'C:\\Users\\Nibras\\OneDrive\\Desktop\\covishieldtamilnadu.csv',header=None)",
"_____no_output_____"
],
[
"df.head(10)",
"_____no_output_____"
],
[
"#Data Cleaning#",
"_____no_output_____"
],
[
"headers = [\"S.No\", \"Health_Unit_District\",\"1stDosageCovishieldHCW\",\"2ndDosageCovishieldHCW\",\"1stDosageCovishieldFLW\",\"2ndDosageCovishieldFLW\",\"1stDoseCovishield18to44\",\"2ndDoseCovishield18to44\",\"1stDoseCovishield45to60Comorbidities\",\"2ndDosageCovishield45to60Comorbidities\",\"1stDosageCovishield60+Comorbidities\",\"2ndDosageCovishield60+Comorbidities\",\"Total1stDoseCovishield\",\"Total2ndDoseCovishield\",\"Date\"]\nprint(\"headers\\n\", headers)",
"headers\n ['S.No', 'Health_Unit_District', '1stDosageCovishieldHCW', '2ndDosageCovishieldHCW', '1stDosageCovishieldFLW', '2ndDosageCovishieldFLW', '1stDoseCovishield18to44', '2ndDoseCovishield18to44', '1stDoseCovishield45to60Comorbidities', '2ndDosageCovishield45to60Comorbidities', '1stDosageCovishield60+Comorbidities', '2ndDosageCovishield60+Comorbidities', 'Total1stDoseCovishield', 'Total2ndDoseCovishield', 'Date']\n"
],
[
"df.columns = headers\ndf.head(10)",
"_____no_output_____"
],
[
"df = df.iloc[1: , :]",
"_____no_output_____"
],
[
"df.head(9)",
"_____no_output_____"
],
[
"df.dtypes",
"_____no_output_____"
],
[
"df['S.No'] = df['S.No'].astype('int64')\ndf['1stDosageCovishieldHCW'] = df['1stDosageCovishieldHCW'].astype('int64')\ndf['2ndDosageCovishieldHCW'] = df['2ndDosageCovishieldHCW'].astype('int64')\ndf['1stDosageCovishieldFLW'] = df['1stDosageCovishieldFLW'].astype('int64')\ndf['2ndDosageCovishieldFLW'] = df['2ndDosageCovishieldFLW'].astype('int64')\ndf['1stDoseCovishield18to44'] = df['1stDoseCovishield18to44'].astype('int64')\ndf['2ndDoseCovishield18to44'] = df['2ndDoseCovishield18to44'].astype('int64')\ndf['1stDoseCovishield45to60Comorbidities'] = df['1stDoseCovishield45to60Comorbidities'].astype('int64')\ndf['2ndDosageCovishield45to60Comorbidities'] = df['2ndDosageCovishield45to60Comorbidities'].astype('int64')\ndf['1stDosageCovishield60+Comorbidities'] = df['1stDosageCovishield60+Comorbidities'].astype('int64')\ndf['2ndDosageCovishield60+Comorbidities'] = df['2ndDosageCovishield60+Comorbidities'].astype('int64')\ndf['Total1stDoseCovishield'] = df['Total1stDoseCovishield'].astype('int64')\ndf['Total2ndDoseCovishield'] = df['Total2ndDoseCovishield'].astype('int64')",
"_____no_output_____"
],
[
"df.dtypes",
"_____no_output_____"
],
[
"#data visualisation",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt\n%matplotlib inline\ndf1=df.head(10)\n",
"_____no_output_____"
],
[
"df1.plot(x=\"Health_Unit_District\", y=\"1stDoseCovishield18to44\", kind=\"bar\")",
"_____no_output_____"
],
[
"date_set = set(df1['Date'])\n\nplt.figure()\nfor Date in date_set:\n selected_data = df1.loc[df1['Date'] == Date]\n plt.plot(selected_data['Health_Unit_District'], selected_data['1stDoseCovishield18to44'], label=Date)\n \nplt.legend()\nplt.show()",
"_____no_output_____"
],
[
"#Predicting a particulars number using linear regression",
"_____no_output_____"
],
[
"from sklearn.model_selection import train_test_split\nimport datetime as dt\ndf['Date'] = pd.to_datetime(df['Date'])\ndf.head(3)",
"_____no_output_____"
],
[
"#converting date time to ordinal\ndf['Date']=df['Date'].map(dt.datetime.toordinal)\ndf.head(3)",
"_____no_output_____"
],
[
"x=df['Date'].tail(3)\ny=df['1stDoseCovishield18to44'].tail(3)",
"_____no_output_____"
],
[
"x_train,x_test,y_train,y_test=train_test_split(x,y,test_size=0.3)",
"_____no_output_____"
],
[
"from sklearn.linear_model import LinearRegression",
"_____no_output_____"
],
[
"lr = LinearRegression()",
"_____no_output_____"
],
[
"import numpy as np\nlr.fit(np.array(x_train).reshape(-1,1),np.array(y_train).reshape(-1,1))",
"_____no_output_____"
],
[
"df.tail(3)",
"_____no_output_____"
],
[
"y_pred=lr.predict(np.array(x_test).reshape(-1,1))",
"_____no_output_____"
],
[
"from sklearn.metrics import mean_squared_error",
"_____no_output_____"
],
[
"mean_squared_error(x_test,y_pred)",
"_____no_output_____"
],
[
"lr.predict(np.array([[737937]])) #that is may of 27",
"_____no_output_____"
],
[
"#actual number on 27th of may 2022 is 5874",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
c50e6c2e11c92becf2de46a948f213b71582a89a
| 39,830 |
ipynb
|
Jupyter Notebook
|
DNN_using_tensorflow_ipynb.ipynb
|
MuhammedAshraf2020/DNN-using-tensorflow
|
fb16ed52ab4df0fbd5c5bcea765cd9200bb5c882
|
[
"MIT"
] | 1 |
2021-08-03T13:33:53.000Z
|
2021-08-03T13:33:53.000Z
|
DNN_using_tensorflow_ipynb.ipynb
|
MuhammedAshraf2020/DNN-using-tensorflow
|
fb16ed52ab4df0fbd5c5bcea765cd9200bb5c882
|
[
"MIT"
] | null | null | null |
DNN_using_tensorflow_ipynb.ipynb
|
MuhammedAshraf2020/DNN-using-tensorflow
|
fb16ed52ab4df0fbd5c5bcea765cd9200bb5c882
|
[
"MIT"
] | null | null | null | 117.147059 | 29,886 | 0.8412 |
[
[
[
"<a href=\"https://colab.research.google.com/github/MuhammedAshraf2020/DNN-using-tensorflow/blob/main/DNN_using_tensorflow_ipynb.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"#import libs\r\nimport numpy as np\r\nimport matplotlib.pyplot as plt\r\nimport tensorflow as tf\r\nfrom keras.datasets.mnist import load_data",
"_____no_output_____"
],
[
"# prepare dataset",
"_____no_output_____"
],
[
"(X_train , y_train) , (X_test , y_test) = load_data()\r\nX_train = X_train.astype(\"float32\") / 255\r\nX_test = X_test.astype(\"float32\") / 255\r\n# Make sure images have shape (28, 28, 1)\r\nX_train = np.expand_dims(X_train, -1)\r\nX_test = np.expand_dims(X_test, -1)",
"_____no_output_____"
],
[
"for i in range(0, 9):\r\n plt.subplot(330 + 1 + i)\r\n plt.imshow(X_train[i][: , : , 0], cmap=plt.get_cmap('gray'))\r\n\r\nplt.show()",
"_____no_output_____"
],
[
"X_train = [X_train[i].ravel() for i in range(len(X_train))]",
"_____no_output_____"
],
[
"X_test = [X_test[i].ravel() for i in range(len(X_test))]",
"_____no_output_____"
],
[
"y_train = tf.keras.utils.to_categorical(y_train , num_classes = 10)\r\ny_test = tf.keras.utils.to_categorical(y_test , num_classes = 10 )",
"_____no_output_____"
],
[
"#set parameter\r\nn_input = 28 * 28\r\nn_hidden_1 = 512\r\nn_hidden_2 = 256\r\nn_hidden_3 = 128\r\nn_output = 10\r\n\r\nlearning_rate = 0.01 \r\nepochs = 50\r\nbatch_size = 128",
"_____no_output_____"
],
[
"tf.compat.v1.disable_eager_execution()",
"_____no_output_____"
],
[
"# weight intialization",
"_____no_output_____"
],
[
"X = tf.compat.v1.placeholder(tf.float32 , [None , n_input])\r\ny = tf.compat.v1.placeholder(tf.float32 , [None , n_output])",
"_____no_output_____"
],
[
"def Weights_init(list_layers , stddiv):\r\n Num_layers = len(list_layers)\r\n weights = {}\r\n bias = {}\r\n for i in range( Num_layers-1):\r\n weights[\"W{}\".format(i+1)] = tf.Variable(tf.compat.v1.truncated_normal([list_layers[i] , list_layers[i+1]] , stddev = stddiv))\r\n bias[\"b{}\".format(i+1)] = tf.Variable(tf.compat.v1.truncated_normal([list_layers[i+1]]))\r\n return weights , bias",
"_____no_output_____"
],
[
"list_param = [784 , 512 , 256 , 128 , 10]",
"_____no_output_____"
],
[
"weights , biases = Weights_init(list_param , 0.1)",
"_____no_output_____"
],
[
"def Model (X , nn_weights , nn_bias):\r\n Z1 = tf.add(tf.matmul(X , nn_weights[\"W1\"]) , nn_bias[\"b1\"])\r\n Z1_out = tf.nn.relu(Z1)\r\n\r\n Z2 = tf.add(tf.matmul(Z1_out , nn_weights[\"W2\"]) , nn_bias[\"b2\"])\r\n Z2_out = tf.nn.relu(Z2)\r\n\r\n Z3 = tf.add(tf.matmul(Z2_out , nn_weights[\"W3\"]) , nn_bias[\"b3\"])\r\n Z3_out = tf.nn.relu(Z3)\r\n\r\n Z4 = tf.add(tf.matmul(Z3_out , nn_weights[\"W4\"]) , nn_bias[\"b4\"])\r\n Z4_out = tf.nn.softmax(Z4)\r\n return Z4_out",
"_____no_output_____"
],
[
"nn_layer_output = Model(X , weights , biases)",
"_____no_output_____"
],
[
"loss = tf.reduce_mean(tf.compat.v1.nn.softmax_cross_entropy_with_logits_v2(logits = nn_layer_output , labels = y))\r\n\r\noptimizer = tf.compat.v1.train.GradientDescentOptimizer(learning_rate).minimize(loss) ",
"_____no_output_____"
],
[
"init = tf.compat.v1.global_variables_initializer()\r\n\r\n# Determining if the predictions are accurate\r\nis_correct_prediction = tf.equal(tf.argmax(nn_layer_output , 1),tf.argmax(y, 1))\r\n\r\n#Calculating prediction accuracy\r\naccuracy = tf.reduce_mean(tf.cast(is_correct_prediction, tf.float32))\r\n\r\nsaver = tf.compat.v1.train.Saver()",
"_____no_output_____"
],
[
"with tf.compat.v1.Session() as sess:\r\n # initializing all the variables\r\n sess.run(init)\r\n total_batch = int(len(X_train) / batch_size)\r\n for epoch in range(epochs):\r\n avg_cost = 0\r\n for i in range(total_batch):\r\n batch_x , batch_y = X_train[i * batch_size : (i + 1) * batch_size] , y_train[i * batch_size : (i + 1) * batch_size]\r\n _, c = sess.run([optimizer,loss], feed_dict={X: batch_x, y: batch_y})\r\n avg_cost += c / total_batch\r\n if(epoch % 10 == 0):\r\n print(\"Epoch:\", (epoch + 1), \"train_cost =\", \"{:.3f} \".format(avg_cost) , end = \"\")\r\n print(\"train_acc = {:.3f} \".format(sess.run(accuracy, feed_dict={X: X_train, y:y_train})) , end = \"\")\r\n print(\"valid_acc = {:.3f}\".format(sess.run(accuracy, feed_dict={X: X_test, y:y_test})))\r\n saver.save(sess , save_path = \"/content/Model.ckpt\")",
"Epoch: 1 train_cost = 2.138 train_acc = 0.395 valid_acc = 0.395\nEpoch: 11 train_cost = 1.892 train_acc = 0.569 valid_acc = 0.568\nEpoch: 21 train_cost = 1.713 train_acc = 0.753 valid_acc = 0.751\nEpoch: 31 train_cost = 1.672 train_acc = 0.842 valid_acc = 0.842\nEpoch: 41 train_cost = 1.608 train_acc = 0.859 valid_acc = 0.853\n"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
c50e75a084258dbe2f34ea5ed0c6e02a0e8d78c5
| 124,976 |
ipynb
|
Jupyter Notebook
|
jupyter_notebook/legacy/qualitative_analysis_Dx,Px.ipynb
|
rsgit95/med_kg_txt_multimodal
|
80355b0cf58e0571531ad6f9728c533110ca996d
|
[
"Apache-2.0"
] | null | null | null |
jupyter_notebook/legacy/qualitative_analysis_Dx,Px.ipynb
|
rsgit95/med_kg_txt_multimodal
|
80355b0cf58e0571531ad6f9728c533110ca996d
|
[
"Apache-2.0"
] | null | null | null |
jupyter_notebook/legacy/qualitative_analysis_Dx,Px.ipynb
|
rsgit95/med_kg_txt_multimodal
|
80355b0cf58e0571531ad6f9728c533110ca996d
|
[
"Apache-2.0"
] | null | null | null | 131.139559 | 76,968 | 0.809187 |
[
[
[
"empty"
]
]
] |
[
"empty"
] |
[
[
"empty"
]
] |
c50e7855d15988457f8d619b8f471312afc7073e
| 8,023 |
ipynb
|
Jupyter Notebook
|
.ipynb_checkpoints/batch copy images instance detection-checkpoint.ipynb
|
issamouawad/Keras-SSD-ConvNet
|
47d1d140b7ba4247eff74304e7833496a201a56f
|
[
"Apache-2.0"
] | 3 |
2018-10-02T14:07:43.000Z
|
2018-10-02T14:07:48.000Z
|
batch copy images instance detection.ipynb
|
issamouawad/Keras-SSD-ConvNet
|
47d1d140b7ba4247eff74304e7833496a201a56f
|
[
"Apache-2.0"
] | null | null | null |
batch copy images instance detection.ipynb
|
issamouawad/Keras-SSD-ConvNet
|
47d1d140b7ba4247eff74304e7833496a201a56f
|
[
"Apache-2.0"
] | null | null | null | 40.933673 | 207 | 0.514396 |
[
[
[
"This notebook copies images and annotations from the original dataset, to perform instance detection\n\nyou can control how many images per pose (starting from some point) and how many instances to consider as well as which classes\n\nwe also edit the annotation file because initially all annotations are made by instance not category",
"_____no_output_____"
]
],
[
[
"import os;\nfrom shutil import copyfile\nimport xml.etree.ElementTree as ET\nimages_per_set = 102\nstart_after = 100",
"_____no_output_____"
]
],
[
[
"the following code can be used to generate both training and testing set",
"_____no_output_____"
]
],
[
[
"classes = ['book1','book2','book3','book4','book5',\n 'cellphone1','cellphone2','cellphone3','cellphone4','cellphone5',\n 'mouse1','mouse2','mouse3','mouse4','mouse5',\n 'ringbinder1','ringbinder2','ringbinder3','ringbinder4','ringbinder5']\ndays = [5,5,5,5,5,3,3,3,3,3,7,7,7,7,7,3,3,3,3,3]\nimages_path ='C:/Users/issa/Documents/datasets/ICUB_Instance/test/' \nannotations_path = 'C:/Users/issa/Documents/datasets/ICUB_Instance/test_ann/'\ninstances_list = [1,2,3,4,5]\nfor category_name,day_number in zip(classes,days):\n in_pose_counter = 0;\n j =0\n for inst in instances_list:\n j=0\n dirs = ['D:\\\\2nd_Semester\\\\CV\\\\Project\\\\part1\\\\part1\\\\'+category_name+'\\\\'+category_name+str(inst)+'\\\\MIX\\\\day'+str(day_number)+'\\\\left\\\\',\n 'D:\\\\2nd_Semester\\\\CV\\\\Project\\\\part1\\\\part1\\\\'+category_name+'\\\\'+category_name+str(inst)+'\\\\ROT2D\\\\day'+str(day_number)+'\\\\left\\\\',\n 'D:\\\\2nd_Semester\\\\CV\\\\Project\\\\part1\\\\part1\\\\'+category_name+'\\\\'+category_name+str(inst)+'\\\\ROT3D\\\\day'+str(day_number)+'\\\\left\\\\',\n 'D:\\\\2nd_Semester\\\\CV\\\\Project\\\\part1\\\\part1\\\\'+category_name+'\\\\'+category_name+str(inst)+'\\\\SCALE\\\\day'+str(day_number)+'\\\\left\\\\',\n 'D:\\\\2nd_Semester\\\\CV\\\\Project\\\\part1\\\\part1\\\\'+category_name+'\\\\'+category_name+str(inst)+'\\\\TRANSL\\\\day'+str(day_number)+'\\\\left\\\\']\n\n for dir in dirs:\n i=0;\n in_pose_counter = 0;\n if(i>images_per_set):\n break;\n for innerSubDir,innerDirs,innerFiles in os.walk(dir):\n for file in innerFiles:\n i = i+1\n if(i>images_per_set):\n break;\n in_pose_counter = in_pose_counter+1\n if(in_pose_counter>start_after):\n j = j+1\n copyfile(dir+file,images_path+category_name+str(inst)+'_'+str(j)+'.jpg')\n \n in_pose_counter =0\n j =0\n for inst in instances_list:\n j=0\n dirs = ['D:\\\\2nd_Semester\\\\CV\\\\Project\\\\Annotations_refined\\Annotations_refined\\\\'+category_name+'\\\\'+category_name+str(inst)+'\\\\MIX\\\\day'+str(day_number)+'\\\\left\\\\',\n 'D:\\\\2nd_Semester\\\\CV\\\\Project\\\\Annotations_refined\\Annotations_refined\\\\'+category_name+'\\\\'+category_name+str(inst)+'\\\\ROT2D\\\\day'+str(day_number)+'\\\\left\\\\',\n 'D:\\\\2nd_Semester\\\\CV\\\\Project\\\\Annotations_refined\\Annotations_refined\\\\'+category_name+'\\\\'+category_name+str(inst)+'\\\\ROT3D\\\\day'+str(day_number)+'\\\\left\\\\',\n 'D:\\\\2nd_Semester\\\\CV\\\\Project\\\\Annotations_refined\\Annotations_refined\\\\'+category_name+'\\\\'+category_name+str(inst)+'\\\\SCALE\\\\day'+str(day_number)+'\\\\left\\\\',\n 'D:\\\\2nd_Semester\\\\CV\\\\Project\\\\Annotations_refined\\Annotations_refined\\\\'+category_name+'\\\\'+category_name+str(inst)+'\\\\TRANSL\\\\day'+str(day_number)+'\\\\left\\\\']\n\n for dir in dirs:\n i=0;\n in_pose_counter =0\n if(i>images_per_set):\n break;\n for innerSubDir,innerDirs,innerFiles in os.walk(dir):\n for file in innerFiles:\n i = i+1\n if(i>images_per_set):\n break;\n in_pose_counter = in_pose_counter+1\n \n if(in_pose_counter>start_after):\n j = j+1\n outputPath = annotations_path+category_name+str(inst)+'_'+str(j)+'.xml'\n copyfile(dir+file,outputPath)\n ",
"_____no_output_____"
]
],
[
[
"This part produces the train/val sets, we set the classes and the list containing the number of images per class, then we split the list and generate the files.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nfrom sklearn.model_selection import train_test_split\nx = np.arange(1,129)\nclasses = ['book1','book2','book3','book4','book5',\n 'cellphone1','cellphone2','cellphone3','cellphone4','cellphone5',\n 'mouse1','mouse2','mouse3','mouse4','mouse5',\n 'ringbinder1','ringbinder2','ringbinder3','ringbinder4','ringbinder5']\nfor category in classes:\n xtrain,xtest,ytrain,ytest=train_test_split(x, x, test_size=0.25)\n\n file = open(\"datasets//ICUB_Instance//train.txt\",\"a\") \n\n for i in xtrain:\n file.write(category+'_'+str(i)+'\\n')\n\n file.close()\n file = open(\"datasets//ICUB_Instance//val.txt\",\"a\") \n\n for i in xtest:\n file.write(category+'_'+str(i)+'\\n')\n\n file.close()",
"_____no_output_____"
]
],
[
[
"this part generates the test set list, no splitting here",
"_____no_output_____"
]
],
[
[
"import numpy as np\nclasses = ['book1','book2','book3','book4','book5',\n 'cellphone1','cellphone2','cellphone3','cellphone4','cellphone5',\n 'mouse1','mouse2','mouse3','mouse4','mouse5',\n 'ringbinder1','ringbinder2','ringbinder3','ringbinder4','ringbinder5']\nx = np.arange(1,21)\nfile = open(\"datasets//ICUB_Instance//test.txt\",\"a\") \nfor category in classes:\n for i in x:\n file.write(category+'_'+str(i)+'\\n')\nfile.close()",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
c50ea10db1e70a6c1f806dada02008f39623171c
| 337,960 |
ipynb
|
Jupyter Notebook
|
module4-databackedassertions/LS_DS_114_Making_Data_backed_Assertions.ipynb
|
krsmith/DS-Sprint-01-Dealing-With-Data
|
9846e5f3845f39fa200af77b65517bf9dff74de6
|
[
"MIT"
] | null | null | null |
module4-databackedassertions/LS_DS_114_Making_Data_backed_Assertions.ipynb
|
krsmith/DS-Sprint-01-Dealing-With-Data
|
9846e5f3845f39fa200af77b65517bf9dff74de6
|
[
"MIT"
] | null | null | null |
module4-databackedassertions/LS_DS_114_Making_Data_backed_Assertions.ipynb
|
krsmith/DS-Sprint-01-Dealing-With-Data
|
9846e5f3845f39fa200af77b65517bf9dff74de6
|
[
"MIT"
] | null | null | null | 108.598972 | 73,418 | 0.705083 |
[
[
[
"<a href=\"https://colab.research.google.com/github/krsmith/DS-Sprint-01-Dealing-With-Data/blob/master/module4-databackedassertions/LS_DS_114_Making_Data_backed_Assertions.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# Lambda School Data Science - Making Data-backed Assertions\n\nThis is, for many, the main point of data science - to create and support reasoned arguments based on evidence. It's not a topic to master in a day, but it is worth some focused time thinking about and structuring your approach to it.",
"_____no_output_____"
],
[
"## Lecture - generating a confounding variable\n\nThe prewatch material told a story about a hypothetical health condition where both the drug usage and overall health outcome were related to gender - thus making gender a confounding variable, obfuscating the possible relationship between the drug and the outcome.\n\nLet's use Python to generate data that actually behaves in this fashion!",
"_____no_output_____"
]
],
[
[
"import random\ndir(random) # Reminding ourselves what we can do here",
"_____no_output_____"
],
[
"# Let's think of another scenario:\n# We work for a company that sells accessories for mobile phones.\n# They have an ecommerce site, and we are supposed to analyze logs\n# to determine what sort of usage is related to purchases, and thus guide\n# website development to encourage higher conversion.\n\n# The hypothesis - users who spend longer on the site tend\n# to spend more. Seems reasonable, no?\n\n# But there's a confounding variable! If they're on a phone, they:\n# a) Spend less time on the site, but\n# b) Are more likely to be interested in the actual products!\n\n# Let's use namedtuple to represent our data\n\nfrom collections import namedtuple\n# purchased and mobile are bools, time_on_site in seconds\nUser = namedtuple('User', ['purchased','time_on_site', 'mobile'])\n\nexample_user = User(False, 12, False)\nprint(example_user)",
"User(purchased=False, time_on_site=12, mobile=False)\n"
],
[
"# And now let's generate 1000 example users\n# 750 mobile, 250 not (i.e. desktop)\n# A desktop user has a base conversion likelihood of 10%\n# And it goes up by 1% for each 15 seconds they spend on the site\n# And they spend anywhere from 10 seconds to 10 minutes on the site (uniform)\n# Mobile users spend on average half as much time on the site as desktop\n# But have twice as much base likelihood of buying something\n\nusers = []\n\nfor _ in range(250):\n # Desktop users\n time_on_site = random.uniform(10, 600)\n purchased = random.random() < 0.1 + (time_on_site // 1500)\n users.append(User(purchased, time_on_site, False))\n \nfor _ in range(750):\n # Mobile users\n time_on_site = random.uniform(5, 300)\n purchased = random.random() < 0.2 + (time_on_site // 1500)\n users.append(User(purchased, time_on_site, True))\n \nrandom.shuffle(users)\nprint(users[:10])",
"[User(purchased=False, time_on_site=261.7008560760187, mobile=False), User(purchased=False, time_on_site=24.125473684342513, mobile=False), User(purchased=False, time_on_site=52.24630202027898, mobile=True), User(purchased=False, time_on_site=67.72303144869834, mobile=False), User(purchased=False, time_on_site=72.69001725114518, mobile=False), User(purchased=False, time_on_site=391.88584999520094, mobile=False), User(purchased=False, time_on_site=28.08162899178536, mobile=False), User(purchased=False, time_on_site=409.33046541015773, mobile=False), User(purchased=False, time_on_site=207.90576870482818, mobile=True), User(purchased=False, time_on_site=70.33149618329547, mobile=True)]\n"
],
[
"# Let's put this in a dataframe so we can look at it more easily\nimport pandas as pd\nuser_data = pd.DataFrame(users)\nuser_data.head()",
"_____no_output_____"
],
[
"# Let's use crosstabulation to try to see what's going on\npd.crosstab(user_data['purchased'], user_data['time_on_site'])",
"_____no_output_____"
],
[
"# OK, that's not quite what we want\n# Time is continuous! We need to put it in discrete buckets\n# Pandas calls these bins, and pandas.cut helps make them\n\ntime_bins = pd.cut(user_data['time_on_site'], 5) # 5 equal-sized bins\npd.crosstab(user_data['purchased'], time_bins)",
"_____no_output_____"
],
[
"# We can make this a bit clearer by normalizing (getting %)\npd.crosstab(user_data['purchased'], time_bins, normalize='columns')",
"_____no_output_____"
],
[
"# That seems counter to our hypothesis\n# More time on the site seems to have fewer purchases\n\n# But we know why, since we generated the data!\n# Let's look at mobile and purchased\npd.crosstab(user_data['purchased'], user_data['mobile'], normalize='columns')",
"_____no_output_____"
],
[
"# Yep, mobile users are more likely to buy things\n# But we're still not seeing the *whole* story until we look at all 3 at once\n\n# Live/stretch goal - how can we do that?",
"_____no_output_____"
]
],
[
[
"## Assignment - what's going on here?\n\nConsider the data in `persons.csv` (already prepared for you, in the repo for the week). It has four columns - a unique id, followed by age (in years), weight (in lbs), and exercise time (in minutes/week) of 1200 (hypothetical) people.\n\nTry to figure out which variables are possibly related to each other, and which may be confounding relationships.",
"_____no_output_____"
]
],
[
[
"# TODO - your code here\n# Use what we did live in lecture as an example\n\n# HINT - you can find the raw URL on GitHub and potentially use that\n# to load the data with read_csv, or you can upload it yourself\n\nimport pandas as pd\ndf = pd.read_csv(\"https://raw.githubusercontent.com/invegat/DS-Sprint-01-Dealing-With-Data/master/module4-databackedassertions/persons.csv\")\ndf.head()",
"_____no_output_____"
],
[
"print(df.describe())\nprint(df.info())",
" Unnamed: 0 age weight exercise_time\ncount 1200.000000 1200.000000 1200.000000 1200.000000\nmean 599.500000 48.396667 153.540833 134.910833\nstd 346.554469 18.166802 35.132182 85.548895\nmin 0.000000 18.000000 100.000000 0.000000\n25% 299.750000 33.000000 125.000000 65.000000\n50% 599.500000 48.000000 149.000000 122.000000\n75% 899.250000 64.000000 180.250000 206.000000\nmax 1199.000000 80.000000 246.000000 300.000000\n<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 1200 entries, 0 to 1199\nData columns (total 4 columns):\nUnnamed: 0 1200 non-null int64\nage 1200 non-null int64\nweight 1200 non-null int64\nexercise_time 1200 non-null int64\ndtypes: int64(4)\nmemory usage: 37.6 KB\nNone\n"
],
[
"# Looking at data with crosstabulation pd.crosstab(a, [b, c], rownames=['a'], colnames=['b', 'c'])\npd.crosstab(df['exercise_time'], [df['age'], df['weight']], normalize='columns')",
"_____no_output_____"
],
[
"# Use panda.cut() to organize data into bins\n\nage_bins = pd.cut(df['age'], bins=[18,30,42,54,68,80]) #8 equal-sized bins\ntime_bins = pd.cut(df['exercise_time'], bins=[0,75,150,225,300]) #10 equal-sized bins\nweight_bins = pd.cut(df['weight'], bins=[100,130,160,190,220])\n\n# Now show the data with crosstab in bins\nct_persons = pd.crosstab(time_bins, [age_bins, weight_bins],normalize='columns')\n\nct_persons",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt\n!pip install seaborn --upgrade\nimport seaborn as sns\nprint(sns.__version__)",
"Requirement already up-to-date: seaborn in /usr/local/lib/python3.6/dist-packages (0.9.0)\nRequirement already satisfied, skipping upgrade: numpy>=1.9.3 in /usr/local/lib/python3.6/dist-packages (from seaborn) (1.14.6)\nRequirement already satisfied, skipping upgrade: scipy>=0.14.0 in /usr/local/lib/python3.6/dist-packages (from seaborn) (1.1.0)\nRequirement already satisfied, skipping upgrade: matplotlib>=1.4.3 in /usr/local/lib/python3.6/dist-packages (from seaborn) (2.1.2)\nRequirement already satisfied, skipping upgrade: pandas>=0.15.2 in /usr/local/lib/python3.6/dist-packages (from seaborn) (0.22.0)\nRequirement already satisfied, skipping upgrade: python-dateutil>=2.1 in /usr/local/lib/python3.6/dist-packages (from matplotlib>=1.4.3->seaborn) (2.5.3)\nRequirement already satisfied, skipping upgrade: pytz in /usr/local/lib/python3.6/dist-packages (from matplotlib>=1.4.3->seaborn) (2018.7)\nRequirement already satisfied, skipping upgrade: cycler>=0.10 in /usr/local/lib/python3.6/dist-packages (from matplotlib>=1.4.3->seaborn) (0.10.0)\nRequirement already satisfied, skipping upgrade: pyparsing!=2.0.4,!=2.1.2,!=2.1.6,>=2.0.1 in /usr/local/lib/python3.6/dist-packages (from matplotlib>=1.4.3->seaborn) (2.3.0)\nRequirement already satisfied, skipping upgrade: six>=1.10 in /usr/local/lib/python3.6/dist-packages (from matplotlib>=1.4.3->seaborn) (1.11.0)\n0.9.0\n"
],
[
"# Seaborn Heatmap for age/weight by exercise time\n\nfig, ax = plt.subplots(figsize=(16,10))\n\nsns.heatmap(ct_persons,annot=True,linewidths=.5,cmap=\"YlGnBu\", ax=ax);\n\n#Older age, disregarding weight, tends to exercise for less time.\n#Higher weight, disregarding age, tends to exercise for less time.\n#Younger age and lower weight tends to excercise for the most time.\n ",
"_____no_output_____"
],
[
"# Seaborn Heatmap for age/exercise time by weight\nct_weight = pd.crosstab(weight_bins, [age_bins, time_bins],normalize='columns')\n\nfig, ax = plt.subplots(figsize=(16,10))\n\nsns.heatmap(ct_weight,annot=True,linewidths=.5,cmap=\"YlGnBu\", ax=ax);\n\n",
"_____no_output_____"
],
[
"# Seaborn Heatmap for weight/exercise time by age\nct_age = pd.crosstab(age_bins, [weight_bins, time_bins],normalize='columns')\n\nfig, ax = plt.subplots(figsize=(16,10))\n\nsns.heatmap(ct_age,annot=True,linewidths=.5,cmap=\"YlGnBu\", ax=ax);",
"_____no_output_____"
]
],
[
[
"### Assignment questions\n\nAfter you've worked on some code, answer the following questions in this text block:\n\n1. What are the variable types in the data?\n\n*All of the variables in this data are integers which represent a person's age, weight, and exercise time.*\n\n2. What are the relationships between the variables?\n\n*The lower the age, regardless of weight, the more time a person would spend exercising. The higher the weight, regardless of age, the less time a person would spend exercising. Younger age & lower weight tends to excercise for more time.*\n\n3. Which relationships are \"real\", and which spurious?\n\n*I would say that age and weight are spurious.*\n\n*REAL: Higher weight = less exercise time.*\n\n*REAL: Higher age = less exercise time.*",
"_____no_output_____"
],
[
"## Stretch goals and resources\n\nFollowing are *optional* things for you to take a look at. Focus on the above assignment first, and make sure to commit and push your changes to GitHub.\n\n- [Spurious Correlations](http://tylervigen.com/spurious-correlations)\n- [NIH on controlling for confounding variables](https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4017459/)\n\nStretch goals:\n\n- Produce your own plot inspierd by the Spurious Correlation visualizations (and consider writing a blog post about it - both the content and how you made it)\n- Pick one of the techniques that NIH highlights for confounding variables - we'll be going into many of them later, but see if you can find which Python modules may help (hint - check scikit-learn)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
]
] |
c50ea2d2c01e5b09473fab301585b05bd429cae1
| 27,399 |
ipynb
|
Jupyter Notebook
|
baselines/notebooks/Hyperparameter_Ensembles.ipynb
|
bryant1410/uncertainty-baselines
|
eee69986056fe2f318367490638df818543ed7e3
|
[
"Apache-2.0"
] | null | null | null |
baselines/notebooks/Hyperparameter_Ensembles.ipynb
|
bryant1410/uncertainty-baselines
|
eee69986056fe2f318367490638df818543ed7e3
|
[
"Apache-2.0"
] | null | null | null |
baselines/notebooks/Hyperparameter_Ensembles.ipynb
|
bryant1410/uncertainty-baselines
|
eee69986056fe2f318367490638df818543ed7e3
|
[
"Apache-2.0"
] | null | null | null | 41.576631 | 679 | 0.556955 |
[
[
[
"# Hyperparameter Ensembles for Robustness and Uncertainty Quantification\n\n*Florian Wenzel, April 8th 2021. Licensed under the Apache License, Version 2.0.*\n\nRecently, we proposed **Hyper-deep Ensembles** ([Wenzel et al., NeurIPS 2020](https://arxiv.org/abs/2006.13570)) a simple, yet powerful, extension of [deep ensembles](https://arxiv.org/abs/1612.01474). The approach works with any given deep network architecture and, therefore, can be easily integrated (and improve) a machine learning system that is already used in production.\n\nHyper-deep ensembles improve the performance of a given deep network by forming an ensemble over multiple variants of that architecture where each member uses different hyperparameters. In this notebook we consider a ResNet-20 architecture with block-wise $\\ell_2$-regularization parameters and a label smoothing parameter. We construct an ensemble of 4 members where each member uses a different set of hyperparameters. This leads to an ensemble of **diverse members**, i.e., members that are complementary in their predictions. The final ensemble greatly improves the prediction performance and the robustness of the model, e.g., in out-of-distribution settings.\n\nLet's start with some boilerplate code for data loading and the model definition.\n\n\n",
"_____no_output_____"
],
[
"Requirements:\n```bash\n!pip install \"git+https://github.com/google/uncertainty-baselines.git#egg=uncertainty_baselines\"\n```",
"_____no_output_____"
]
],
[
[
"import tensorflow as tf\nimport tensorflow_datasets as tfds\nimport numpy as np\n\nimport uncertainty_baselines as ub",
"_____no_output_____"
],
[
"def _ensemble_accuracy(labels, logits_list):\n \"\"\"Compute the accuracy resulting from the ensemble prediction.\"\"\"\n per_probs = tf.nn.softmax(logits_list)\n probs = tf.reduce_mean(per_probs, axis=0)\n acc = tf.keras.metrics.SparseCategoricalAccuracy()\n acc.update_state(labels, probs)\n return acc.result()\n\ndef _ensemble_cross_entropy(labels, logits):\n logits = tf.convert_to_tensor(logits)\n ensemble_size = float(logits.shape[0])\n labels = tf.cast(labels, tf.int32)\n ce = tf.nn.sparse_softmax_cross_entropy_with_logits(\n labels=tf.broadcast_to(labels[tf.newaxis, ...], tf.shape(logits)[:-1]),\n logits=logits)\n nll = -tf.reduce_logsumexp(-ce, axis=0) + tf.math.log(ensemble_size)\n return tf.reduce_mean(nll)\n\n\ndef greedy_selection(val_logits, val_labels, max_ens_size, objective='nll'):\n \"\"\"Greedy procedure from Caruana et al. 2004, with replacement.\"\"\"\n\n assert_msg = 'Unknown objective type (received {}).'.format(objective)\n assert objective in ('nll', 'acc', 'nll-acc'), assert_msg\n\n # Objective that should be optimized by the ensemble. Arbitrary objectives,\n # e.g., based on nll, acc or calibration error (or combinations of those) can\n # be used.\n if objective == 'nll':\n get_objective = lambda acc, nll: nll\n elif objective == 'acc':\n get_objective = lambda acc, nll: acc\n else:\n get_objective = lambda acc, nll: nll-acc\n\n best_acc = 0.\n best_nll = np.inf\n best_objective = np.inf\n ens = []\n\n def get_ens_size():\n return len(set(ens))\n\n while get_ens_size() < max_ens_size:\n current_val_logits = [val_logits[model_id] for model_id in ens]\n best_model_id = None\n for model_id, logits in enumerate(val_logits):\n acc = _ensemble_accuracy(val_labels, current_val_logits + [logits])\n nll = _ensemble_cross_entropy(val_labels, current_val_logits + [logits])\n obj = get_objective(acc, nll)\n if obj < best_objective:\n best_acc = acc\n best_nll = nll\n best_objective = obj\n best_model_id = model_id\n if best_model_id is None:\n print('Ensemble could not be improved: Greedy selection stops.')\n break\n ens.append(best_model_id)\n return ens, best_acc, best_nll\n\n\ndef parse_checkpoint_dir(checkpoint_dir):\n \"\"\"Parse directory of checkpoints.\"\"\"\n paths = []\n subdirectories = tf.io.gfile.glob(os.path.join(checkpoint_dir, '*'))\n is_checkpoint = lambda f: ('checkpoint' in f and '.index' in f)\n for subdir in subdirectories:\n for path, _, files in tf.io.gfile.walk(subdir):\n if any(f for f in files if is_checkpoint(f)):\n latest_checkpoint_without_suffix = tf.train.latest_checkpoint(path)\n paths.append(os.path.join(path, latest_checkpoint_without_suffix))\n break\n return paths\n",
"_____no_output_____"
],
[
"DATASET = 'cifar10'\nTRAIN_PROPORTION = 0.95\nBATCH_SIZE = 64\nENSEMBLE_SIZE = 4",
"_____no_output_____"
],
[
"# load data\nds_info = tfds.builder(DATASET).info\nnum_classes = ds_info.features['label'].num_classes\n# test set\nsteps_per_eval = ds_info.splits['test'].num_examples // BATCH_SIZE \ntest_dataset = ub.datasets.get(\n DATASET,\n split=tfds.Split.TEST).load(batch_size=BATCH_SIZE)\n# validation set\nvalidation_percent = 1 - TRAIN_PROPORTION\nval_dataset = ub.datasets.get(\n dataset_name=DATASET,\n split=tfds.Split.VALIDATION,\n validation_percent=validation_percent,\n drop_remainder=False).load(batch_size=BATCH_SIZE)\nsteps_per_val_eval = int(ds_info.splits['train'].num_examples *\n validation_percent) // BATCH_SIZE # validation set\n",
"_____no_output_____"
]
],
[
[
"# Let's construct the hyper-deep ensemble over a ResNet-20 architecture\n\n\n**This is the (simplified) hyper-deep ensembles construction pipeline**\n> **1. Random search:** train several models on the train set using different (random) hyperparameters.\n> \n> **2. Ensemble construction:** on a validation set using a greedy selection method.\n\nRemark:\n*In this notebook we use a slightly simplified version of the pipeline compared to the approach of the original paper (where an additional stratification step is used). Additionally, after selecting the optimal hyperparameters the ensemble performance can be improved even more by retraining the selected models on the full train set (i.e., this time not reserving a portion for validation). The simplified pipeline in this notebook is slightly less performant but easier to implement. The simplified pipeline is similar to the ones used by Caranua et al., 2004 and Zaidi et al., 2020 in the context of neural architecture search.*\n",
"_____no_output_____"
],
[
"## Step 1: Random Hyperparameter Search\n\nWe start by training 100 different versions of the ResNet-20 using different $\\ell_2$-regularization parameters and label smoothing parameters. Since this would take some time we have already trained the models using a standard training script (which can be found [here](https://github.com/google/uncertainty-baselines/blob/master/baselines/cifar/deterministic.py)) and directly load the checkpoints.\n\n",
"_____no_output_____"
]
],
[
[
"# The model architecture we want to form the ensemble over\n# here, we use the original ResNet-20 model by He et al. 2015\nmodel = ub.models.wide_resnet(\n input_shape=ds_info.features['image'].shape,\n depth=22,\n width_multiplier=1,\n num_classes=num_classes,\n l2=0.,\n version=1)",
"_____no_output_____"
],
[
"# Load checkpoints\n\n# Unfortunately, we can't release the check points yet. But we hope we can make\n# make them available. For the moment, you have to train the individual models\n# yourself using the train script linked above. \nCHECKPOINT_DIR = 'insert_path_to_checkpoints'\nensemble_filenames = parse_checkpoint_dir(CHECKPOINT_DIR)\n\nmodel_pool_size = len(ensemble_filenames)\ncheckpoint = tf.train.Checkpoint(model=model)\nprint('Model pool size: {}'.format(model_pool_size))",
"Model pool size: 100\n"
]
],
[
[
"## Step 2: Construction of the hyperparameter ensemble on the validation set\n\nFirst we compute the logits of all models in our model pool on the validation set.",
"_____no_output_____"
]
],
[
[
"# Compute the logits on the validation set\n\nval_logits, val_labels = [], []\nfor m, ensemble_filename in enumerate(ensemble_filenames):\n # Enforce memory clean-up\n tf.keras.backend.clear_session()\n checkpoint.restore(ensemble_filename)\n val_iterator = iter(val_dataset)\n val_logits_m = []\n for _ in range(steps_per_val_eval):\n inputs = next(val_iterator)\n features = inputs['features']\n labels = inputs['labels']\n val_logits_m.append(model(features, training=False))\n if m == 0:\n val_labels.append(labels)\n\n val_logits.append(tf.concat(val_logits_m, axis=0))\n if m == 0:\n val_labels = tf.concat(val_labels, axis=0)\n\n \n if m % 10 == 0 or m == model_pool_size - 1:\n percent = (m + 1.) / model_pool_size\n message = ('{:.1%} completion for prediction on validation set: '\n 'model {:d}/{:d}.'.format(percent, m + 1, model_pool_size))\n print(message)",
"1.0% completion for prediction on validation set: model 1/100.\n11.0% completion for prediction on validation set: model 11/100.\n21.0% completion for prediction on validation set: model 21/100.\n31.0% completion for prediction on validation set: model 31/100.\n41.0% completion for prediction on validation set: model 41/100.\n51.0% completion for prediction on validation set: model 51/100.\n61.0% completion for prediction on validation set: model 61/100.\n71.0% completion for prediction on validation set: model 71/100.\n81.0% completion for prediction on validation set: model 81/100.\n91.0% completion for prediction on validation set: model 91/100.\n100.0% completion for prediction on validation set: model 100/100.\n"
]
],
[
[
"Now we are ready to construct the ensemble.\n* In the first step, we take the best model (on the validation set) -> `model_1`\n* In the second step, we fix `model_1` and try all models in our model pool and construct the ensemble `[model_1, model_2]`. We select the model `model_2` that leads to the highest performance gain.\n* In the third step, we fix `model_1`, `model_2` and choose `model_3` to construct an ensemble `[model_1, model_2, model_3]` that leads to the highest performance gain over step 2.\n* ... and so on, until the desired ensemble size is reached or no performance gain could be achieved anymore.",
"_____no_output_____"
]
],
[
[
"# Ensemble construction by greedy member selection on the validation set\nselected_members, val_acc, val_nll = greedy_selection(val_logits, val_labels,\n ENSEMBLE_SIZE,\n objective='nll')\nunique_selected_members = list(set(selected_members))\nmessage = ('Members selected by greedy procedure: model ids = {} (with {} unique '\n 'member(s))').format(\n selected_members, len(unique_selected_members))\nprint(message)",
"Members selected by greedy procedure: model ids = [38, 0, 81, 27] (with 4 unique member(s))\n"
]
],
[
[
"# Evaluation on the test set\n\nLet's see how the **hyper-deep ensemble** performs on the test set.",
"_____no_output_____"
]
],
[
[
"# Evaluate the following metrics on the test step\nmetrics = {\n 'ensemble/negative_log_likelihood': tf.keras.metrics.Mean(),\n 'ensemble/accuracy': tf.keras.metrics.SparseCategoricalAccuracy(),\n}\nmetrics_single = {\n 'single/negative_log_likelihood': tf.keras.metrics.SparseCategoricalCrossentropy(),\n 'single/accuracy': tf.keras.metrics.SparseCategoricalAccuracy(),\n}\n\n\n# compute logits for ensemble member on test set\nlogits_test = []\nfor m, member_id in enumerate(unique_selected_members):\n ensemble_filename = ensemble_filenames[member_id]\n checkpoint.restore(ensemble_filename)\n logits = []\n test_iterator = iter(test_dataset)\n for _ in range(steps_per_eval):\n features = next(test_iterator)['features']\n logits.append(model(features, training=False))\n logits_test.append(tf.concat(logits, axis=0))\nlogits_test = tf.convert_to_tensor(logits_test)\nprint('Completed computation of member logits on the test set.')",
"Completed computation of member logits on the test set.\n"
],
[
"# compute test metrics\ntest_iterator = iter(test_dataset)\nfor step in range(steps_per_eval):\n labels = next(test_iterator)['labels']\n logits = logits_test[:, (step*BATCH_SIZE):((step+1)*BATCH_SIZE)]\n labels = tf.cast(labels, tf.int32)\n negative_log_likelihood = _ensemble_cross_entropy(labels, logits)\n # per member output probabilities\n per_probs = tf.nn.softmax(logits)\n # ensemble output probabilites\n probs = tf.reduce_mean(per_probs, axis=0)\n metrics['ensemble/negative_log_likelihood'].update_state(\n negative_log_likelihood)\n metrics['ensemble/accuracy'].update_state(labels, probs)\n\n # for comparison compute performance of the best single model\n # this is by definition the first model that was selected by the greedy \n # selection method\n logits_single = logits_test[0, (step*BATCH_SIZE):((step+1)*BATCH_SIZE)]\n probs_single = tf.nn.softmax(logits_single)\n metrics_single['single/negative_log_likelihood'].update_state(labels, logits_single)\n metrics_single['single/accuracy'].update_state(labels, probs_single)\n\n\n percent = (step + 1) / steps_per_eval\n if step % 25 == 0 or step == steps_per_eval - 1:\n message = ('{:.1%} completion final test prediction'.format(percent))\n print(message)\n\nensemble_results = {name: metric.result() for name, metric in metrics.items()}\nsingle_results = {name: metric.result() for name, metric in metrics_single.items()}",
"0.6% completion final test prediction\n16.7% completion final test prediction\n32.7% completion final test prediction\n48.7% completion final test prediction\n64.7% completion final test prediction\n80.8% completion final test prediction\n96.8% completion final test prediction\n100.0% completion final test prediction\n"
]
],
[
[
"## Here is the final ensemble performance\n\nWe gained almost 2 percentage points in terms of accuracy over the best single model!",
"_____no_output_____"
]
],
[
[
"print('Ensemble performance:')\nfor m, val in ensemble_results.items():\n print(' {}: {}'.format(m, val))\n\nprint('\\nFor comparison:')\nfor m, val in single_results.items():\n print(' {}: {}'.format(m, val))",
"Ensemble performance:\n ensemble/negative_log_likelihood: 0.19766511023044586\n ensemble/accuracy: 0.9358974099159241\n\nFor comparison:\n single/negative_log_likelihood: 1.0325815677642822\n single/accuracy: 0.9189703464508057\n"
]
],
[
[
"## Hyper-deep ensembles as a strong baseline\n\nWe have seen that **hyper-deep ensembles** can lead to significant performance gains and can be easily implemented in your existing machine learning pipeline. Moreover, we hope that other researchers can benefit from this by using **hyper-deep ensembles** as a competitive, yet simple-to-implement, baseline. Even though **hyper-deep ensembles** might be more expensive than single model methods, it can show how much can be gained by introducing more diversity in the predictions.\n\n\n## Hyper-deep ensembles can make your ML pipeline more robust\n\n**Don't throw away your precious models!**\n\nIn many settings where we use a standard (single model) deep neural network, we usually start with a hyperparameter search. Typically, we select the model with the best hyperparameters and throw away all the others. Here, we show that you can get a much more performant system by combining multiple models from the hyperparameter search. \n\n**What's the additional cost?**\n\nIn most cases you already get a significant performance boost if you combine 4 models. The main additional cost (provided you have already done the hyperparameter search) is that your model is now 4x larger (more memory) and 4x times slower to perform the predictions (if not parallelized). Often the performance boost justifies this increased cost. If you can't afford the additional cost, check out **hyper-batch ensembles**. This is an efficient version that amortizes hyper-deep ensembles **within a single model** (see our [paper](https://arxiv.org/abs/2006.13570)).\n\n## Pointers to additional resources\n\n* The full code for the extended **hyper-deep ensembles** pipeline and the code for the experiments in our paper can be found in the [Uncertainty Baselines](https://github.com/google/uncertainty-baselines/blob/master/baselines/cifar/hyperdeepensemble.py) repository.\n* Our efficient version **hyper-batch ensembles** that amortize hyper-deep ensembles within a single model is implemented as a keras layer and can be found in [Edward2](https://github.com/google/edward2 ).\n\n## For questions reach out to\nFlorian Wenzel ([[email protected]](mailto:[email protected])) \\\nRodolphe Jenatton ([[email protected]](mailto:[email protected]))\n\n\n### Reference\n\nIf you use parts of this pipeline for your projects or papers we would be happy if you would cite our paper.\n\n> Florian Wenzel, Jasper Snoek, Dustin Tran and Rodolphe Jenatton (2020).\n> [Hyperparameter Ensembles for Robustness and Uncertainty Quantification](https://arxiv.org/abs/2006.13570).\n> In _Neural Information Processing Systems_.\n\n```none\n@inproceedings{wenzel2020good,\n author = {Florian Wenzel and Jasper Snoek and Dustin Tran and Rodolphe Jenatton},\n title = {Hyperparameter Ensembles for Robustness and Uncertainty Quantification},\n booktitle = {Neural Information Processing Systems)},\n year = {2020},\n}\n```\n\n\n",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
c50ea9ece3fc6fb053f03d2a4eea3c71da3a9eb2
| 16,741 |
ipynb
|
Jupyter Notebook
|
576-Census/stacking.ipynb
|
ReiRev/Signate
|
ce67ca6f7829811bf7add4b06442316d4f1af129
|
[
"MIT"
] | null | null | null |
576-Census/stacking.ipynb
|
ReiRev/Signate
|
ce67ca6f7829811bf7add4b06442316d4f1af129
|
[
"MIT"
] | null | null | null |
576-Census/stacking.ipynb
|
ReiRev/Signate
|
ce67ca6f7829811bf7add4b06442316d4f1af129
|
[
"MIT"
] | 1 |
2022-02-26T09:05:58.000Z
|
2022-02-26T09:05:58.000Z
| 82.876238 | 282 | 0.725405 |
[
[
[
"import os\nimport json\nfrom utils import load_datasets, load_target\nimport models\nfrom models.tuning import beyesian_optimization\nfrom models.evaluation import cross_validation_score\nimport json",
"_____no_output_____"
],
[
"config = json.load(open('./config/default.json'))\n# X_train, X_test = load_datasets([\"Age\", \"AgeSplit\", \"EducationNum\"])\nX_train, X_test = load_datasets(config['features'])\ny_train = load_target('Y')\n",
"_____no_output_____"
],
[
"lgbm1 = models.Lgbm(json.load(open('./config/Lgbm-depth-5.json')))\nlgbm2 = models.Lgbm(json.load(open('./config/Lgbm-depth-15.json')))\nlgbm3 = models.Lgbm(json.load(open('./config/Lgbm-depth-inf.json')))\nnn1 = models.NN(json.load(open('./config/NN-shallow.json')))\nnn2 = models.NN(json.load(open('./config/NN-deep.json')))\nrf = models.RandomForest(json.load(open('./config/RandomForest.json')))\nert = models.RandomForest(json.load(open('./config/ERT.json')))\n",
"_____no_output_____"
],
[
"model_list = [lgbm1, lgbm2, lgbm3, nn1, nn2, rf, ert]",
"_____no_output_____"
],
[
"from models.evaluation import stacking\n\noof, y_preds = stacking(model_list, X_train, y_train, X_test, 1)",
"/Users/yutahirai/opt/anaconda3/envs/tensorflow26/lib/python3.8/site-packages/lightgbm/engine.py:177: UserWarning: Found `num_boost_round` in params. Will use it instead of argument\n _log_warning(f\"Found `{alias}` in params. Will use it instead of argument\")\n/Users/yutahirai/opt/anaconda3/envs/tensorflow26/lib/python3.8/site-packages/lightgbm/engine.py:239: UserWarning: 'verbose_eval' argument is deprecated and will be removed in a future release of LightGBM. Pass 'log_evaluation()' callback via 'callbacks' argument instead.\n _log_warning(\"'verbose_eval' argument is deprecated and will be removed in a future release of LightGBM. \"\n/Users/yutahirai/opt/anaconda3/envs/tensorflow26/lib/python3.8/site-packages/lightgbm/engine.py:177: UserWarning: Found `num_boost_round` in params. Will use it instead of argument\n _log_warning(f\"Found `{alias}` in params. Will use it instead of argument\")\n/Users/yutahirai/opt/anaconda3/envs/tensorflow26/lib/python3.8/site-packages/lightgbm/engine.py:239: UserWarning: 'verbose_eval' argument is deprecated and will be removed in a future release of LightGBM. Pass 'log_evaluation()' callback via 'callbacks' argument instead.\n _log_warning(\"'verbose_eval' argument is deprecated and will be removed in a future release of LightGBM. \"\n/Users/yutahirai/opt/anaconda3/envs/tensorflow26/lib/python3.8/site-packages/lightgbm/engine.py:177: UserWarning: Found `num_boost_round` in params. Will use it instead of argument\n _log_warning(f\"Found `{alias}` in params. Will use it instead of argument\")\n/Users/yutahirai/opt/anaconda3/envs/tensorflow26/lib/python3.8/site-packages/lightgbm/engine.py:239: UserWarning: 'verbose_eval' argument is deprecated and will be removed in a future release of LightGBM. Pass 'log_evaluation()' callback via 'callbacks' argument instead.\n _log_warning(\"'verbose_eval' argument is deprecated and will be removed in a future release of LightGBM. \"\n/Users/yutahirai/opt/anaconda3/envs/tensorflow26/lib/python3.8/site-packages/lightgbm/engine.py:177: UserWarning: Found `num_boost_round` in params. Will use it instead of argument\n _log_warning(f\"Found `{alias}` in params. Will use it instead of argument\")\n/Users/yutahirai/opt/anaconda3/envs/tensorflow26/lib/python3.8/site-packages/lightgbm/engine.py:239: UserWarning: 'verbose_eval' argument is deprecated and will be removed in a future release of LightGBM. Pass 'log_evaluation()' callback via 'callbacks' argument instead.\n _log_warning(\"'verbose_eval' argument is deprecated and will be removed in a future release of LightGBM. \"\n/Users/yutahirai/opt/anaconda3/envs/tensorflow26/lib/python3.8/site-packages/lightgbm/engine.py:177: UserWarning: Found `num_boost_round` in params. Will use it instead of argument\n _log_warning(f\"Found `{alias}` in params. Will use it instead of argument\")\n/Users/yutahirai/opt/anaconda3/envs/tensorflow26/lib/python3.8/site-packages/lightgbm/engine.py:239: UserWarning: 'verbose_eval' argument is deprecated and will be removed in a future release of LightGBM. Pass 'log_evaluation()' callback via 'callbacks' argument instead.\n _log_warning(\"'verbose_eval' argument is deprecated and will be removed in a future release of LightGBM. \"\n/Users/yutahirai/opt/anaconda3/envs/tensorflow26/lib/python3.8/site-packages/lightgbm/engine.py:177: UserWarning: Found `num_boost_round` in params. Will use it instead of argument\n _log_warning(f\"Found `{alias}` in params. Will use it instead of argument\")\n/Users/yutahirai/opt/anaconda3/envs/tensorflow26/lib/python3.8/site-packages/lightgbm/engine.py:239: UserWarning: 'verbose_eval' argument is deprecated and will be removed in a future release of LightGBM. Pass 'log_evaluation()' callback via 'callbacks' argument instead.\n _log_warning(\"'verbose_eval' argument is deprecated and will be removed in a future release of LightGBM. \"\n/Users/yutahirai/opt/anaconda3/envs/tensorflow26/lib/python3.8/site-packages/lightgbm/engine.py:177: UserWarning: Found `num_boost_round` in params. Will use it instead of argument\n _log_warning(f\"Found `{alias}` in params. Will use it instead of argument\")\n/Users/yutahirai/opt/anaconda3/envs/tensorflow26/lib/python3.8/site-packages/lightgbm/engine.py:239: UserWarning: 'verbose_eval' argument is deprecated and will be removed in a future release of LightGBM. Pass 'log_evaluation()' callback via 'callbacks' argument instead.\n _log_warning(\"'verbose_eval' argument is deprecated and will be removed in a future release of LightGBM. \"\n/Users/yutahirai/opt/anaconda3/envs/tensorflow26/lib/python3.8/site-packages/lightgbm/engine.py:177: UserWarning: Found `num_boost_round` in params. Will use it instead of argument\n _log_warning(f\"Found `{alias}` in params. Will use it instead of argument\")\n/Users/yutahirai/opt/anaconda3/envs/tensorflow26/lib/python3.8/site-packages/lightgbm/engine.py:239: UserWarning: 'verbose_eval' argument is deprecated and will be removed in a future release of LightGBM. Pass 'log_evaluation()' callback via 'callbacks' argument instead.\n _log_warning(\"'verbose_eval' argument is deprecated and will be removed in a future release of LightGBM. \"\n/Users/yutahirai/opt/anaconda3/envs/tensorflow26/lib/python3.8/site-packages/lightgbm/engine.py:177: UserWarning: Found `num_boost_round` in params. Will use it instead of argument\n _log_warning(f\"Found `{alias}` in params. Will use it instead of argument\")\n/Users/yutahirai/opt/anaconda3/envs/tensorflow26/lib/python3.8/site-packages/lightgbm/engine.py:239: UserWarning: 'verbose_eval' argument is deprecated and will be removed in a future release of LightGBM. Pass 'log_evaluation()' callback via 'callbacks' argument instead.\n _log_warning(\"'verbose_eval' argument is deprecated and will be removed in a future release of LightGBM. \"\n/Users/yutahirai/opt/anaconda3/envs/tensorflow26/lib/python3.8/site-packages/lightgbm/engine.py:177: UserWarning: Found `num_boost_round` in params. Will use it instead of argument\n _log_warning(f\"Found `{alias}` in params. Will use it instead of argument\")\n/Users/yutahirai/opt/anaconda3/envs/tensorflow26/lib/python3.8/site-packages/lightgbm/engine.py:239: UserWarning: 'verbose_eval' argument is deprecated and will be removed in a future release of LightGBM. Pass 'log_evaluation()' callback via 'callbacks' argument instead.\n _log_warning(\"'verbose_eval' argument is deprecated and will be removed in a future release of LightGBM. \"\n/Users/yutahirai/opt/anaconda3/envs/tensorflow26/lib/python3.8/site-packages/lightgbm/engine.py:177: UserWarning: Found `num_boost_round` in params. Will use it instead of argument\n _log_warning(f\"Found `{alias}` in params. Will use it instead of argument\")\n/Users/yutahirai/opt/anaconda3/envs/tensorflow26/lib/python3.8/site-packages/lightgbm/engine.py:239: UserWarning: 'verbose_eval' argument is deprecated and will be removed in a future release of LightGBM. Pass 'log_evaluation()' callback via 'callbacks' argument instead.\n _log_warning(\"'verbose_eval' argument is deprecated and will be removed in a future release of LightGBM. \"\n/Users/yutahirai/opt/anaconda3/envs/tensorflow26/lib/python3.8/site-packages/lightgbm/engine.py:177: UserWarning: Found `num_boost_round` in params. Will use it instead of argument\n _log_warning(f\"Found `{alias}` in params. Will use it instead of argument\")\n/Users/yutahirai/opt/anaconda3/envs/tensorflow26/lib/python3.8/site-packages/lightgbm/engine.py:239: UserWarning: 'verbose_eval' argument is deprecated and will be removed in a future release of LightGBM. Pass 'log_evaluation()' callback via 'callbacks' argument instead.\n _log_warning(\"'verbose_eval' argument is deprecated and will be removed in a future release of LightGBM. \"\n/Users/yutahirai/opt/anaconda3/envs/tensorflow26/lib/python3.8/site-packages/lightgbm/engine.py:177: UserWarning: Found `num_boost_round` in params. Will use it instead of argument\n _log_warning(f\"Found `{alias}` in params. Will use it instead of argument\")\n/Users/yutahirai/opt/anaconda3/envs/tensorflow26/lib/python3.8/site-packages/lightgbm/engine.py:239: UserWarning: 'verbose_eval' argument is deprecated and will be removed in a future release of LightGBM. Pass 'log_evaluation()' callback via 'callbacks' argument instead.\n _log_warning(\"'verbose_eval' argument is deprecated and will be removed in a future release of LightGBM. \"\n/Users/yutahirai/opt/anaconda3/envs/tensorflow26/lib/python3.8/site-packages/lightgbm/engine.py:177: UserWarning: Found `num_boost_round` in params. Will use it instead of argument\n _log_warning(f\"Found `{alias}` in params. Will use it instead of argument\")\n/Users/yutahirai/opt/anaconda3/envs/tensorflow26/lib/python3.8/site-packages/lightgbm/engine.py:239: UserWarning: 'verbose_eval' argument is deprecated and will be removed in a future release of LightGBM. Pass 'log_evaluation()' callback via 'callbacks' argument instead.\n _log_warning(\"'verbose_eval' argument is deprecated and will be removed in a future release of LightGBM. \"\n/Users/yutahirai/opt/anaconda3/envs/tensorflow26/lib/python3.8/site-packages/lightgbm/engine.py:177: UserWarning: Found `num_boost_round` in params. Will use it instead of argument\n _log_warning(f\"Found `{alias}` in params. Will use it instead of argument\")\n/Users/yutahirai/opt/anaconda3/envs/tensorflow26/lib/python3.8/site-packages/lightgbm/engine.py:239: UserWarning: 'verbose_eval' argument is deprecated and will be removed in a future release of LightGBM. Pass 'log_evaluation()' callback via 'callbacks' argument instead.\n _log_warning(\"'verbose_eval' argument is deprecated and will be removed in a future release of LightGBM. \"\n2022-02-19 18:08:37.898350: I tensorflow/core/grappler/optimizers/custom_graph_optimizer_registry.cc:113] Plugin optimizer for device_type GPU is enabled.\n2022-02-19 18:09:35.743146: I tensorflow/core/grappler/optimizers/custom_graph_optimizer_registry.cc:113] Plugin optimizer for device_type GPU is enabled.\n2022-02-19 18:09:37.726054: I tensorflow/core/grappler/optimizers/custom_graph_optimizer_registry.cc:113] Plugin optimizer for device_type GPU is enabled.\n2022-02-19 18:10:33.943875: I tensorflow/core/grappler/optimizers/custom_graph_optimizer_registry.cc:113] Plugin optimizer for device_type GPU is enabled.\n2022-02-19 18:10:36.085414: I tensorflow/core/grappler/optimizers/custom_graph_optimizer_registry.cc:113] Plugin optimizer for device_type GPU is enabled.\n2022-02-19 18:11:32.289164: I tensorflow/core/grappler/optimizers/custom_graph_optimizer_registry.cc:113] Plugin optimizer for device_type GPU is enabled.\n2022-02-19 18:11:34.335541: I tensorflow/core/grappler/optimizers/custom_graph_optimizer_registry.cc:113] Plugin optimizer for device_type GPU is enabled.\n2022-02-19 18:12:31.113806: I tensorflow/core/grappler/optimizers/custom_graph_optimizer_registry.cc:113] Plugin optimizer for device_type GPU is enabled.\n2022-02-19 18:12:33.252554: I tensorflow/core/grappler/optimizers/custom_graph_optimizer_registry.cc:113] Plugin optimizer for device_type GPU is enabled.\n2022-02-19 18:13:31.401965: I tensorflow/core/grappler/optimizers/custom_graph_optimizer_registry.cc:113] Plugin optimizer for device_type GPU is enabled.\n2022-02-19 18:13:34.068354: I tensorflow/core/grappler/optimizers/custom_graph_optimizer_registry.cc:113] Plugin optimizer for device_type GPU is enabled.\n2022-02-19 18:24:54.895571: I tensorflow/core/grappler/optimizers/custom_graph_optimizer_registry.cc:113] Plugin optimizer for device_type GPU is enabled.\n2022-02-19 18:25:02.267264: I tensorflow/core/grappler/optimizers/custom_graph_optimizer_registry.cc:113] Plugin optimizer for device_type GPU is enabled.\n2022-02-19 18:33:14.353879: I tensorflow/core/grappler/optimizers/custom_graph_optimizer_registry.cc:113] Plugin optimizer for device_type GPU is enabled.\n2022-02-19 18:33:19.664583: I tensorflow/core/grappler/optimizers/custom_graph_optimizer_registry.cc:113] Plugin optimizer for device_type GPU is enabled.\n2022-02-19 18:46:16.091734: I tensorflow/core/grappler/optimizers/custom_graph_optimizer_registry.cc:113] Plugin optimizer for device_type GPU is enabled.\n2022-02-19 18:46:20.942970: I tensorflow/core/grappler/optimizers/custom_graph_optimizer_registry.cc:113] Plugin optimizer for device_type GPU is enabled.\n2022-02-19 18:58:25.680343: I tensorflow/core/grappler/optimizers/custom_graph_optimizer_registry.cc:113] Plugin optimizer for device_type GPU is enabled.\n2022-02-19 18:58:31.260228: I tensorflow/core/grappler/optimizers/custom_graph_optimizer_registry.cc:113] Plugin optimizer for device_type GPU is enabled.\n2022-02-19 19:07:34.476793: I tensorflow/core/grappler/optimizers/custom_graph_optimizer_registry.cc:113] Plugin optimizer for device_type GPU is enabled.\n"
],
[
"y_pred = [(i > 0.5).astype(int) for i in y_preds]\ny_pred = sum(y_pred) / len(y_pred)\ny_pred = (y_pred > 0.5).astype(int)\nimport pandas as pd\nsample_submit = pd.read_csv(\n './data/input/sample_submit.csv', names=['id', 'Y'])\nsample_submit['Y'] = y_pred\nsample_submit.to_csv('./data/output/2022-2-19-submit.csv', header=False, index=False)\n",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
c50eac277dca1b88ed7bb8618d2d7cdd7066c198
| 23,649 |
ipynb
|
Jupyter Notebook
|
docs/tutorials/cluster/MVSphericalKMeans/SphericalKMeansFigureValidation.ipynb
|
pierreablin/mvlearn
|
1a5913cc475ac7d3426ab07f907b40f969c130eb
|
[
"Apache-2.0"
] | 1 |
2020-12-29T15:41:29.000Z
|
2020-12-29T15:41:29.000Z
|
docs/tutorials/cluster/MVSphericalKMeans/SphericalKMeansFigureValidation.ipynb
|
pierreablin/mvlearn
|
1a5913cc475ac7d3426ab07f907b40f969c130eb
|
[
"Apache-2.0"
] | null | null | null |
docs/tutorials/cluster/MVSphericalKMeans/SphericalKMeansFigureValidation.ipynb
|
pierreablin/mvlearn
|
1a5913cc475ac7d3426ab07f907b40f969c130eb
|
[
"Apache-2.0"
] | null | null | null | 84.160142 | 12,896 | 0.80625 |
[
[
[
"# Validating Multi-View Spherical KMeans by Replicating Paper Results\n\nHere we will validate the implementation of multi-view spherical kmeans by replicating the right side of figure 3 from the Multi-View Clustering paper by Bickel and Scheffer. ",
"_____no_output_____"
]
],
[
[
"import sklearn\nfrom sklearn.datasets import fetch_20newsgroups\nfrom sklearn.feature_extraction.text import TfidfVectorizer\nimport numpy as np\nimport scipy as scp\nfrom scipy import sparse\nimport mvlearn\nfrom mvlearn.cluster.mv_spherical_kmeans import MultiviewSphericalKMeans\nfrom joblib import Parallel, delayed\nimport matplotlib.pyplot as plt\nimport warnings\nwarnings.simplefilter('ignore') # Ignore warnings",
"_____no_output_____"
]
],
[
[
"### A function to recreate the artificial dataset from the paper\n\nThe experiment in the paper used the 20 Newsgroup dataset, which consists of around 18000 newsgroups posts on 20 topics. This dataset can be obtained from scikit-learn. To create the artificial dataset used in the experiment, 10 of the 20 classes from the 20 newsgroups dataset were selected and grouped into 2 groups of 5 classes, and then encoded as tfidf vectors. These now represented the 5 multi-view classes, each with 2 views (one from each group). 200 examples were randomly sampled from each of the 20 newsgroups, producing 1000 concatenated examples uniformly distributed over the 5 classes.",
"_____no_output_____"
]
],
[
[
"NUM_SAMPLES = 200\n\n#Load in the vectorized news group data from scikit-learn package\nnews = fetch_20newsgroups(subset='all')\nall_data = np.array(news.data)\nall_targets = np.array(news.target)\nclass_names = news.target_names\n\n#A function to get the 20 newsgroup data\ndef get_data(): \n \n #Set class pairings as described in the multiview clustering paper\n view1_classes = ['comp.graphics','rec.motorcycles', 'sci.space', 'rec.sport.hockey', 'comp.sys.ibm.pc.hardware']\n view2_classes = ['rec.autos', 'sci.med','misc.forsale', 'soc.religion.christian','comp.os.ms-windows.misc']\n \n #Create lists to hold data and labels for each of the 5 classes across 2 different views\n labels = [num for num in range(len(view1_classes)) for _ in range(NUM_SAMPLES)]\n labels = np.array(labels)\n view1_data = list()\n view2_data = list()\n \n #Randomly sample 200 items from each of the selected classes in view1\n for ind in range(len(view1_classes)):\n class_num = class_names.index(view1_classes[ind])\n class_data = all_data[(all_targets == class_num)]\n indices = np.random.choice(class_data.shape[0], NUM_SAMPLES)\n view1_data.append(class_data[indices])\n view1_data = np.concatenate(view1_data)\n \n #Randomly sample 200 items from each of the selected classes in view2\n for ind in range(len(view2_classes)):\n class_num = class_names.index(view2_classes[ind])\n class_data = all_data[(all_targets == class_num)]\n indices = np.random.choice(class_data.shape[0], NUM_SAMPLES)\n view2_data.append(class_data[indices]) \n view2_data = np.concatenate(view2_data)\n \n #Vectorize the data\n vectorizer = TfidfVectorizer()\n view1_data = vectorizer.fit_transform(view1_data)\n view2_data = vectorizer.fit_transform(view2_data)\n\n #Shuffle and normalize vectors\n shuffled_inds = np.random.permutation(NUM_SAMPLES * len(view1_classes))\n view1_data = sparse.vstack(view1_data)\n view2_data = sparse.vstack(view2_data)\n view1_data = np.array(view1_data[shuffled_inds].todense())\n view2_data = np.array(view2_data[shuffled_inds].todense())\n magnitudes1 = np.linalg.norm(view1_data, axis=1)\n magnitudes2 = np.linalg.norm(view2_data, axis=1)\n magnitudes1[magnitudes1 == 0] = 1\n magnitudes2[magnitudes2 == 0] = 1\n magnitudes1 = magnitudes1.reshape((-1,1))\n magnitudes2 = magnitudes2.reshape((-1,1))\n view1_data /= magnitudes1\n view2_data /= magnitudes2\n labels = labels[shuffled_inds]\n\n return view1_data, view2_data, labels",
"Downloading 20news dataset. This may take a few minutes.\nDownloading dataset from https://ndownloader.figshare.com/files/5975967 (14 MB)\n"
]
],
[
[
"### Function to compute cluster entropy\n\nThe function below is used to calculate the total clustering entropy using the formula described in the paper.",
"_____no_output_____"
]
],
[
[
"def compute_entropy(partitions, labels, k, num_classes):\n \n total_entropy = 0\n num_examples = partitions.shape[0]\n for part in range(k):\n labs = labels[partitions == part]\n part_size = labs.shape[0]\n part_entropy = 0\n for cl in range(num_classes):\n prop = np.sum(labs == cl) * 1.0 / part_size\n ent = 0\n if(prop != 0):\n ent = - prop * np.log2(prop)\n part_entropy += ent\n part_entropy = part_entropy * part_size / num_examples\n total_entropy += part_entropy\n return total_entropy",
"_____no_output_____"
]
],
[
[
"### Functions to Initialize Centroids and Run Experiment\n\nThe randSpherical function initializes the initial cluster centroids by taking a uniform random sampling of points on the surface of a unit hypersphere. The getEntropies function runs Multi-View Spherical Kmeans Clustering on the data with n_clusters from 1 to 10 once each. This function essentially runs one trial of the experiment.",
"_____no_output_____"
]
],
[
[
"def randSpherical(n_clusters, n_feat1, n_feat2):\n c_centers1 = np.random.normal(0, 1, (n_clusters, n_feat1))\n c_centers1 /= np.linalg.norm(c_centers1, axis=1).reshape((-1, 1))\n c_centers2 = np.random.normal(0, 1, (n_clusters, n_feat2))\n c_centers2 /= np.linalg.norm(c_centers2, axis=1).reshape((-1, 1))\n return [c_centers1, c_centers2]",
"_____no_output_____"
],
[
"def getEntropies():\n \n v1_data, v2_data, labels = get_data()\n \n entropies = list()\n for num in range(1,11):\n \n centers = randSpherical(num, v1_data.shape[1], v2_data.shape[1])\n kmeans = MultiviewSphericalKMeans(n_clusters=num, init=centers, n_init=1)\n pred = kmeans.fit_predict([v1_data, v2_data])\n ent = compute_entropy(pred, labels, num, 5)\n entropies.append(ent)\n print('done')\n return entropies",
"_____no_output_____"
]
],
[
[
"### Running multiple trials of the experiment\n\nIt was difficult to exactly reproduce the results from the Multi-View Clustering Paper because the experimentors randomly sampled a subset of the 20 newsgroup dataset samples to create the artificial dataset, and this random subset was not reported. Therefore, in an attempt to at least replicate the overall shape of the distribution of cluster entropy over the number of clusters, we resample the dataset and recreate the artificial dataset each trial. Therefore, each trial consists of resampling and recreating the artificial dataset, and then running Multi-view Spherical KMeans clustering on that dataset for n_clusters 1 to 10 once each. We performed 80 such trials and the results of this are shown below.",
"_____no_output_____"
]
],
[
[
"#Do spherical kmeans and get entropy values for each k for multiple trials\nn_workers = 10\nn_trials = 80\nmult_entropies1 = Parallel(n_jobs=n_workers)(\n delayed(getEntropies)() for i in range(n_trials)) ",
"_____no_output_____"
]
],
[
[
"### Experiment Results\n\nWe see the results of this experiment below. Here, we have more or less reproduced the shape of the distribution as seen in figure 3 from the Multi-view Clustering Paper.",
"_____no_output_____"
]
],
[
[
"mult_entropies1 = np.array(mult_entropies1)\nave_m_entropies = np.mean(mult_entropies1, axis=0)\nstd_m_entropies = np.std(mult_entropies1, axis=0)\nx_values = list(range(1, 11))\nplt.errorbar(x_values, ave_m_entropies, std_m_entropies, capsize=5, color = '#F46C12')\nplt.xlabel('k')\nplt.ylabel('Entropy')\nplt.legend(['2 Views'])\nplt.rc('axes', labelsize=12)\nplt.show()",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
c50eaf09fc3697a0df06e1560a47f720cd3970fa
| 5,528 |
ipynb
|
Jupyter Notebook
|
docs/start/constexpr.ipynb
|
xinetzone/cpp-book
|
1c2f8a82ef08c7708c4b20ec8fe21cae5dda2221
|
[
"Apache-2.0"
] | null | null | null |
docs/start/constexpr.ipynb
|
xinetzone/cpp-book
|
1c2f8a82ef08c7708c4b20ec8fe21cae5dda2221
|
[
"Apache-2.0"
] | null | null | null |
docs/start/constexpr.ipynb
|
xinetzone/cpp-book
|
1c2f8a82ef08c7708c4b20ec8fe21cae5dda2221
|
[
"Apache-2.0"
] | null | null | null | 21.019011 | 166 | 0.465268 |
[
[
[
"# 常量表达式\n\n\n```{admonition} 需求\n学习使用 `constexpr` 与 `const`。\n```\n\n```{hint}\n{guilabel}`视频`\n\n- [C++ const 与 constexpr](https://www.bilibili.com/video/BV1Db4y1Y7zF)\n```\n\n**符号常量**\n: 用于表示那些在初始化后值就不再改变的数值量。\n\n 也可以简称为 **常量**。\n\n符号常量就是我们认知中的常量。比如圆周率 $\\pi$,在使用过程中,不希望它是可变的,可以使用 **符号常量** 来实现这一目的:",
"_____no_output_____"
]
],
[
[
"constexpr double pi = 3.1415926; // 定义一个符号常量 pi",
"_____no_output_____"
],
[
"pi = 7; // 错误,试图改变常量的值",
"\u001b[1minput_line_8:2:5: \u001b[0m\u001b[0;1;31merror: \u001b[0m\u001b[1mcannot assign to variable 'pi' with const-qualified type 'const double'\u001b[0m\n pi = 7; // 错误,试图改变常量的值\n\u001b[0;1;32m ~~ ^\n\u001b[0m\u001b[1minput_line_7:2:19: \u001b[0m\u001b[0;1;30mnote: \u001b[0mvariable 'pi' declared const here\u001b[0m\n constexpr double pi = 3.1415926; // 定义一个符号常量 pi\n\u001b[0;1;32m ~~~~~~~~~~~~~~~~~^~~~~~~~~~~~~~\n\u001b[0m"
]
],
[
[
"仅仅是使用常量,而非改变它是可行的:",
"_____no_output_____"
]
],
[
[
"double c = 2 * pi;\nc",
"_____no_output_____"
]
],
[
[
"```{important}\n常量对于维护程序的可读性具有重要作用。\n```\n\n```{tip}\n除了个别情况(例如 0 和 1),程序中应该尽量少用字面常量,而是尽可能使用符号常量。\n```\n\n## 常量表达式\n\n**常量表达式**\n: 仅由常量构成的 **整型值** 表达式。",
"_____no_output_____"
]
],
[
[
"constexpr int max = 18; // 字面常量是常量表达式\nint val = 19;\n\nmax + 7; // 常量表达式(即常量整数+字面常量)\nval + 2; // 因为使用了变量,不是常量表达式",
"_____no_output_____"
]
],
[
[
"一个 `constexpr` 符号常量必须给定一个在编译时已知的值。\n\n```c++\nconstexpr int max = 500;\n\nvoid use(int n) {\n constexpr int c1 = max + 7; // 正确,c1 是 507\n constexpr int c2 = n + 7; // 错误,不知道 c2 的值是多少\n // ...\n}\n```",
"_____no_output_____"
],
[
"## 只读\n\n**只读**\n: 若某个变量初始化时的值在编译时未知,但初始化后也绝对不改变。\n\nC++ 提供了只读变量的表示:`const`。\n\n上面的代码可以改写为:",
"_____no_output_____"
]
],
[
[
"constexpr int max = 500;",
"_____no_output_____"
],
[
"void use(int n) {\n constexpr int c1 = max + 7; // 正确,c1 是 507\n const int c2 = n + 7; // 正确,但是 c2 的值可以改变\n // ...\n}",
"_____no_output_____"
]
],
[
[
"由于 `c2` 是一个只读的常量,所以:",
"_____no_output_____"
]
],
[
[
"void use(int n) {\n constexpr int c1 = max + 7; // 正确,c1 是 507\n const int c2 = n + 7; // 正确,但是 c2 的值可以改变\n // ...\n c2 = 7; // 错误,c2 是常量\n}",
"\u001b[1minput_line_23:5:8: \u001b[0m\u001b[0;1;31merror: \u001b[0m\u001b[1mcannot assign to variable 'c2' with const-qualified type 'const int'\u001b[0m\n c2 = 7;\n\u001b[0;1;32m ~~ ^\n\u001b[0m\u001b[1minput_line_23:3:15: \u001b[0m\u001b[0;1;30mnote: \u001b[0mvariable 'c2' declared const here\u001b[0m\n const int c2 = n + 7; // 正确,但是 c2 的值可以改变\n\u001b[0;1;32m ~~~~~~~~~~^~~~~~~~~~\n\u001b[0m"
]
],
[
[
"```{hint}\nC++98 不支持 `constexpr`,只能使用 `const`。\n```",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
c50eb37cc9e18b78bd876ce9db260c30fc7d39c7
| 162,756 |
ipynb
|
Jupyter Notebook
|
demos/ensembles/ensemble-link-prediction-example.ipynb
|
xugangwu95/stellargraph
|
f26f7733235573f5def19da3329fc1d92fdd71ee
|
[
"Apache-2.0"
] | 3 |
2020-01-17T10:33:12.000Z
|
2020-03-19T12:49:23.000Z
|
demos/ensembles/ensemble-link-prediction-example.ipynb
|
HaoweiGis/stellargraph
|
d7ef103a3ca5f29cb7b7e333e53ea2b310baa26c
|
[
"Apache-2.0"
] | null | null | null |
demos/ensembles/ensemble-link-prediction-example.ipynb
|
HaoweiGis/stellargraph
|
d7ef103a3ca5f29cb7b7e333e53ea2b310baa26c
|
[
"Apache-2.0"
] | null | null | null | 154.711027 | 51,392 | 0.891857 |
[
[
[
"# StellarGraph Ensemble for link prediction",
"_____no_output_____"
],
[
"In this example, we use `stellargraph`s `BaggingEnsemble` class of [GraphSAGE](http://snap.stanford.edu/graphsage/) models to predict citation links in the Cora dataset (see below). The `BaggingEnsemble` class brings ensemble learning to `stellargraph`'s graph neural network models, e.g., `GraphSAGE`, quantifying prediction variance and potentially improving prediction accuracy. \n\nThe problem is treated as a supervised link prediction problem on a homogeneous citation network with nodes representing papers (with attributes such as binary keyword indicators and categorical subject) and links corresponding to paper-paper citations. \n\nTo address this problem, we build a a base `GraphSAGE` model with the following architecture. First we build a two-layer GraphSAGE model that takes labeled `(paper1, paper2)` node pairs corresponding to possible citation links, and outputs a pair of node embeddings for the `paper1` and `paper2` nodes of the pair. These embeddings are then fed into a link classification layer, which first applies a binary operator to those node embeddings (e.g., concatenating them) to construct the embedding of the potential link. Thus obtained link embeddings are passed through the dense link classification layer to obtain link predictions - probability for these candidate links to actually exist in the network. The entire model is trained end-to-end by minimizing the loss function of choice (e.g., binary cross-entropy between predicted link probabilities and true link labels, with true/false citation links having labels 1/0) using stochastic gradient descent (SGD) updates of the model parameters, with minibatches of 'training' links fed into the model.\n\nFinally, using our base model, we create an ensemble with each model in the ensemble trained on a bootstrapped sample of the training data. \n\n**References**\n\n1. Inductive Representation Learning on Large Graphs. W.L. Hamilton, R. Ying, and J. Leskovec arXiv:1706.02216 \n[cs.SI], 2017.",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nimport networkx as nx\nimport pandas as pd\nimport numpy as np\nfrom tensorflow import keras\nimport os\n\nimport stellargraph as sg\nfrom stellargraph.data import EdgeSplitter\nfrom stellargraph.mapper import GraphSAGELinkGenerator\nfrom stellargraph.layer import GraphSAGE, link_classification\nfrom stellargraph import BaggingEnsemble\n\nfrom sklearn import preprocessing, feature_extraction, model_selection\n\nfrom stellargraph import globalvar\n\n%matplotlib inline",
"_____no_output_____"
],
[
"def plot_history(history):\n def remove_prefix(text, prefix):\n return text[text.startswith(prefix) and len(prefix):]\n\n figsize=(7, 5)\n c_train = 'b'\n c_test = 'g'\n \n metrics = sorted(set([remove_prefix(m, \"val_\") for m in list(history[0].history.keys())]))\n for m in metrics:\n # summarize history for metric m\n plt.figure(figsize=figsize)\n for h in history:\n plt.plot(h.history[m], c=c_train)\n plt.plot(h.history['val_' + m], c=c_test)\n \n plt.title(m)\n plt.ylabel(m)\n plt.xlabel('epoch')\n plt.legend(['train', 'validation'], loc='best')\n plt.show()",
"_____no_output_____"
],
[
"def load_cora(data_dir, largest_cc=False):\n g_nx = nx.read_edgelist(path=os.path.expanduser(os.path.join(data_dir, \"cora.cites\")))\n \n for edge in g_nx.edges(data=True):\n edge[2]['label'] = 'cites'\n\n # load the node attribute data\n cora_data_location = os.path.expanduser(os.path.join(data_dir, \"cora.content\"))\n node_attr = pd.read_csv(cora_data_location, sep='\\t', header=None)\n values = { str(row.tolist()[0]): row.tolist()[-1] for _, row in node_attr.iterrows()}\n nx.set_node_attributes(g_nx, values, 'subject')\n\n if largest_cc:\n # Select the largest connected component. For clarity we ignore isolated\n # nodes and subgraphs; having these in the data does not prevent the\n # algorithm from running and producing valid results.\n g_nx_ccs = (g_nx.subgraph(c).copy() for c in nx.connected_components(g_nx))\n g_nx = max(g_nx_ccs, key=len)\n print(\"Largest subgraph statistics: {} nodes, {} edges\".format(\n g_nx.number_of_nodes(), g_nx.number_of_edges()))\n \n feature_names = [\"w_{}\".format(ii) for ii in range(1433)]\n column_names = feature_names + [\"subject\"]\n node_data = pd.read_csv(os.path.join(data_dir, \"cora.content\"), sep='\\t', header=None, names=column_names)\n \n node_data.index = node_data.index.map(str)\n node_data = node_data[node_data.index.isin(list(g_nx.nodes()))]\n\n return g_nx, node_data, feature_names",
"_____no_output_____"
]
],
[
[
"### Loading the CORA network data",
"_____no_output_____"
],
[
"**Downloading the CORA dataset:**\n \nThe dataset used in this demo can be downloaded from https://linqs-data.soe.ucsc.edu/public/lbc/cora.tgz\n\nThe following is the description of the dataset:\n> The Cora dataset consists of 2708 scientific publications classified into one of seven classes.\n> The citation network consists of 5429 links. Each publication in the dataset is described by a\n> 0/1-valued word vector indicating the absence/presence of the corresponding word from the dictionary.\n> The dictionary consists of 1433 unique words. The README file in the dataset provides more details.\n\nDownload and unzip the cora.tgz file to a location on your computer and set the `data_dir` variable to\npoint to the location of the dataset (the directory containing \"cora.cites\" and \"cora.content\").",
"_____no_output_____"
]
],
[
[
"data_dir = os.path.expanduser(\"~/data/cora\")",
"_____no_output_____"
]
],
[
[
"Load the dataset",
"_____no_output_____"
]
],
[
[
"G, node_data, feature_names = load_cora(data_dir)",
"_____no_output_____"
]
],
[
[
"We need to convert node features that will be used by the model to numeric values that are required for GraphSAGE input. Note that all node features in the Cora dataset, except the categorical \"subject\" feature, are already numeric, and don't require the conversion.",
"_____no_output_____"
]
],
[
[
"if \"subject\" in feature_names:\n # Convert node features to numeric vectors\n feature_encoding = feature_extraction.DictVectorizer(sparse=False)\n node_features = feature_encoding.fit_transform(\n node_data[feature_names].to_dict(\"records\")\n )\nelse: # node features are already numeric, no further conversion is needed\n node_features = node_data[feature_names].values",
"_____no_output_____"
]
],
[
[
"Add node data to G:",
"_____no_output_____"
]
],
[
[
"for nid, f in zip(node_data.index, node_features):\n G.nodes[nid][globalvar.TYPE_ATTR_NAME] = \"paper\" # specify node type\n G.nodes[nid][\"feature\"] = f",
"_____no_output_____"
]
],
[
[
"We aim to train a link prediction model, hence we need to prepare the train and test sets of links and the corresponding graphs with those links removed.\n\nWe are going to split our input graph into train and test graphs using the `EdgeSplitter` class in `stellargraph.data`. We will use the train graph for training the model (a binary classifier that, given two nodes, predicts whether a link between these two nodes should exist or not) and the test graph for evaluating the model's performance on hold out data.\n\nEach of these graphs will have the same number of nodes as the input graph, but the number of links will differ (be reduced) as some of the links will be removed during each split and used as the positive samples for training/testing the link prediction classifier.",
"_____no_output_____"
],
[
"From the original graph G, extract a randomly sampled subset of test edges (true and false citation links) and the reduced graph G_test with the positive test edges removed:",
"_____no_output_____"
]
],
[
[
"# Define an edge splitter on the original graph G:\nedge_splitter_test = EdgeSplitter(G)\n\n# Randomly sample a fraction p=0.1 of all positive links, and same number of negative links, from G, and obtain the\n# reduced graph G_test with the sampled links removed:\nG_test, edge_ids_test, edge_labels_test = edge_splitter_test.train_test_split(\n p=0.1, method=\"global\", keep_connected=True, seed=42\n)",
"** Sampled 527 positive and 527 negative edges. **\n"
]
],
[
[
"The reduced graph G_test, together with the test ground truth set of links (edge_ids_test, edge_labels_test), will be used for testing the model.\n\nNow, repeat this procedure to obtain validation data that we are going to use for early stopping in order to prevent overfitting. From the reduced graph G_test, extract a randomly sampled subset of validation edges (true and false citation links) and the reduced graph G_val with the positive validation edges removed.",
"_____no_output_____"
]
],
[
[
"# Define an edge splitter on the reduced graph G_test:\nedge_splitter_val = EdgeSplitter(G_test)\n\n# Randomly sample a fraction p=0.1 of all positive links, and same number of negative links, from G_test, and obtain the\n# reduced graph G_train with the sampled links removed:\nG_val, edge_ids_val, edge_labels_val = edge_splitter_val.train_test_split(\n p=0.1, method=\"global\", keep_connected=True, seed=100\n)",
"** Sampled 475 positive and 475 negative edges. **\n"
]
],
[
[
"We repeat this procedure one last time in order to obtain the training data for the model.\nFrom the reduced graph G_val, extract a randomly sampled subset of train edges (true and false citation links) and the reduced graph G_train with the positive train edges removed:",
"_____no_output_____"
]
],
[
[
"# Define an edge splitter on the reduced graph G_test:\nedge_splitter_train = EdgeSplitter(G_test)\n\n# Randomly sample a fraction p=0.1 of all positive links, and same number of negative links, from G_test, and obtain the\n# reduced graph G_train with the sampled links removed:\nG_train, edge_ids_train, edge_labels_train = edge_splitter_train.train_test_split(\n p=0.1, method=\"global\", keep_connected=True, seed=42\n)",
"** Sampled 475 positive and 475 negative edges. **\n"
]
],
[
[
"G_train, together with the train ground truth set of links (edge_ids_train, edge_labels_train), will be used for training the model.",
"_____no_output_____"
],
[
"Convert G_train, G_val, and G_test to StellarGraph objects (undirected, as required by GraphSAGE) for ML:",
"_____no_output_____"
]
],
[
[
"G_train = sg.StellarGraph(G_train, node_features=\"feature\")\nG_test = sg.StellarGraph(G_test, node_features=\"feature\")\nG_val = sg.StellarGraph(G_val, node_features=\"feature\")",
"_____no_output_____"
]
],
[
[
"Summary of G_train and G_test - note that they have the same set of nodes, only differing in their edge sets:",
"_____no_output_____"
]
],
[
[
"print(G_train.info())",
"StellarGraph: Undirected multigraph\n Nodes: 2708, Edges: 4276\n\n Node types:\n paper: [2708]\n Attributes: {'feature', 'subject'}\n Edge types: paper-cites->paper\n\n Edge types:\n paper-cites->paper: [4276]\n\n"
],
[
"print(G_test.info())",
"StellarGraph: Undirected multigraph\n Nodes: 2708, Edges: 4751\n\n Node types:\n paper: [2708]\n Attributes: {'feature', 'subject'}\n Edge types: paper-cites->paper\n\n Edge types:\n paper-cites->paper: [4751]\n\n"
],
[
"print(G_val.info())",
"StellarGraph: Undirected multigraph\n Nodes: 2708, Edges: 4276\n\n Node types:\n paper: [2708]\n Attributes: {'feature', 'subject'}\n Edge types: paper-cites->paper\n\n Edge types:\n paper-cites->paper: [4276]\n\n"
]
],
[
[
"### Specify global parameters\n\nHere we specify some important parameters that control the type of ensemble model we are going to use. For example, we specify the number of models in the ensemble and the number of predictions per query point per model.",
"_____no_output_____"
]
],
[
[
"n_estimators = 5 # Number of models in the ensemble\nn_predictions = 10 # Number of predictions per query point per model",
"_____no_output_____"
]
],
[
[
"Next, we create link generators for sampling and streaming train and test link examples to the model. The link generators essentially \"map\" pairs of nodes `(paper1, paper2)` to the input of GraphSAGE: they take minibatches of node pairs, sample 2-hop subgraphs with `(paper1, paper2)` head nodes extracted from those pairs, and feed them, together with the corresponding binary labels indicating whether those pairs represent true or false citation links, to the input layer of the GraphSAGE model, for SGD updates of the model parameters.\n\nSpecify the minibatch size (number of node pairs per minibatch) and the number of epochs for training the model:",
"_____no_output_____"
]
],
[
[
"batch_size = 20\nepochs = 20",
"_____no_output_____"
]
],
[
[
"Specify the sizes of 1- and 2-hop neighbour samples for GraphSAGE. Note that the length of `num_samples` list defines the number of layers/iterations in the GraphSAGE model. In this example, we are defining a 2-layer GraphSAGE model:",
"_____no_output_____"
]
],
[
[
"num_samples = [20, 10]",
"_____no_output_____"
]
],
[
[
"### Create the generators for training",
"_____no_output_____"
],
[
"For training we create a generator on the `G_train` graph. The `shuffle=True` argument is given to the `flow` method to improve training.",
"_____no_output_____"
]
],
[
[
"generator = GraphSAGELinkGenerator(G_train, batch_size, num_samples)",
"_____no_output_____"
],
[
"train_gen = generator.flow(edge_ids_train, \n edge_labels_train, \n shuffle=True)",
"_____no_output_____"
]
],
[
[
"At test time we use the `G_test` graph and don't specify the `shuffle` argument (it defaults to `False`).",
"_____no_output_____"
]
],
[
[
"test_gen = GraphSAGELinkGenerator(G_test, batch_size, num_samples).flow(edge_ids_test, \n edge_labels_test)",
"_____no_output_____"
],
[
"val_gen = GraphSAGELinkGenerator(G_val, batch_size, num_samples).flow(edge_ids_val, \n edge_labels_val)",
"_____no_output_____"
]
],
[
[
"### Create the base GraphSAGE model",
"_____no_output_____"
],
[
"Build the model: a 2-layer GraphSAGE model acting as node representation learner, with a link classification layer on concatenated `(paper1, paper2)` node embeddings.\n\nGraphSAGE part of the model, with hidden layer sizes of 20 for both GraphSAGE layers, a bias term, and no dropout. (Dropout can be switched on by specifying a positive dropout rate, 0 < dropout < 1)\n\nNote that the length of layer_sizes list must be equal to the length of num_samples, as len(num_samples) defines the number of hops (layers) in the GraphSAGE model.",
"_____no_output_____"
]
],
[
[
"layer_sizes = [20, 20]\nassert len(layer_sizes) == len(num_samples)\n\ngraphsage = GraphSAGE(\n layer_sizes=layer_sizes, generator=generator, bias=True, dropout=0.5\n )",
"_____no_output_____"
],
[
"# Build the model and expose the input and output tensors.\nx_inp, x_out = graphsage.build()",
"_____no_output_____"
]
],
[
[
"Final link classification layer that takes a pair of node embeddings produced by graphsage, applies a binary operator to them to produce the corresponding link embedding ('ip' for inner product; other options for the binary operator can be seen by running a cell with `?link_classification` in it), and passes it through a dense layer:",
"_____no_output_____"
]
],
[
[
"prediction = link_classification(\n output_dim=1, output_act=\"relu\", edge_embedding_method='ip'\n )(x_out)",
"link_classification: using 'ip' method to combine node embeddings into edge embeddings\n"
]
],
[
[
"Stack the GraphSAGE and prediction layers into a Keras model.",
"_____no_output_____"
]
],
[
[
"base_model = keras.Model(inputs=x_inp, outputs=prediction)",
"_____no_output_____"
]
],
[
[
"Now we create the ensemble based on `base_model` we just created.",
"_____no_output_____"
]
],
[
[
"model = BaggingEnsemble(model=base_model, n_estimators=n_estimators, n_predictions=n_predictions)",
"_____no_output_____"
]
],
[
[
"We need to `compile` the model specifying the optimiser, loss function, and metrics to use.",
"_____no_output_____"
]
],
[
[
"model.compile(\n optimizer=keras.optimizers.Adam(lr=1e-3),\n loss=keras.losses.binary_crossentropy,\n weighted_metrics=[\"acc\"],\n)",
"_____no_output_____"
]
],
[
[
"Evaluate the initial (untrained) ensemble of models on the train and test set:",
"_____no_output_____"
]
],
[
[
"init_train_metrics_mean, init_train_metrics_std = model.evaluate_generator(train_gen)\ninit_test_metrics_mean, init_test_metrics_std = model.evaluate_generator(test_gen)\n\nprint(\"\\nTrain Set Metrics of the initial (untrained) model:\")\nfor name, m, s in zip(model.metrics_names, init_train_metrics_mean, init_train_metrics_std):\n print(\"\\t{}: {:0.4f}±{:0.4f}\".format(name, m, s))\n\nprint(\"\\nTest Set Metrics of the initial (untrained) model:\")\nfor name, m, s in zip(model.metrics_names, init_test_metrics_mean, init_test_metrics_std):\n print(\"\\t{}: {:0.4f}±{:0.4f}\".format(name, m, s))",
"\nTrain Set Metrics of the initial (untrained) model:\n\tloss: 0.7839±0.1272\n\tweighted_acc: 0.6157±0.0175\n\nTest Set Metrics of the initial (untrained) model:\n\tloss: 0.7966±0.0964\n\tweighted_acc: 0.6233±0.0132\n"
]
],
[
[
"### Train the ensemble model\n\nWe are going to use **bootstrap samples** of the training dataset to train each model in the ensemble. For this purpose, we need to pass `generator`, `edge_ids_train`, and `edge_labels_train` to the `fit_generator` method.\n\nNote that training time will vary based on computer speed. Set `verbose=1` for reporting of training progress.",
"_____no_output_____"
]
],
[
[
"history = model.fit_generator(\n generator=generator,\n train_data = edge_ids_train,\n train_targets = edge_labels_train,\n epochs=epochs,\n validation_data=val_gen,\n verbose=0,\n use_early_stopping=True, # Enable early stopping\n early_stopping_monitor=\"val_weighted_acc\",\n )",
"_____no_output_____"
]
],
[
[
"Plot the training history:",
"_____no_output_____"
]
],
[
[
"plot_history(history)",
"_____no_output_____"
]
],
[
[
"Evaluate the trained model on test citation links. After training the model, performance should be better than before training (shown above):",
"_____no_output_____"
]
],
[
[
"train_metrics_mean, train_metrics_std = model.evaluate_generator(train_gen)\ntest_metrics_mean, test_metrics_std = model.evaluate_generator(test_gen)\n\nprint(\"\\nTrain Set Metrics of the trained model:\")\nfor name, m, s in zip(model.metrics_names, train_metrics_mean, train_metrics_std):\n print(\"\\t{}: {:0.4f}±{:0.4f}\".format(name, m, s))\n\nprint(\"\\nTest Set Metrics of the trained model:\")\nfor name, m, s in zip(model.metrics_names, test_metrics_mean, test_metrics_std):\n print(\"\\t{}: {:0.4f}±{:0.4f}\".format(name, m, s))",
"\nTrain Set Metrics of the trained model:\n\tloss: 0.3035±0.0418\n\tweighted_acc: 0.9133±0.0104\n\nTest Set Metrics of the trained model:\n\tloss: 0.6523±0.1425\n\tweighted_acc: 0.7750±0.0228\n"
]
],
[
[
"### Make predictions with the model\n\nNow let's get the predictions for all the edges in the test set.",
"_____no_output_____"
]
],
[
[
"test_predictions = model.predict_generator(generator=test_gen)",
"_____no_output_____"
]
],
[
[
"These predictions will be the output of the last layer in the model with `sigmoid` activation.\n\nThe array `test_predictions` has dimensionality $MxKxNxF$ where $M$ is the number of estimators in the ensemble (`n_estimators`); $K$ is the number of predictions per query point per estimator (`n_predictions`); $N$ is the number of query points (`len(test_predictions)`); and $F$ is the output dimensionality of the specified layer determined by the shape of the output layer (in this case it is equal to 1 since we are performing binary classification).",
"_____no_output_____"
]
],
[
[
"type(test_predictions), test_predictions.shape",
"_____no_output_____"
]
],
[
[
"For demonstration, we are going to select one of the edges in the test set, and plot the ensemble's predictions for that edge.\n\nChange the value of `selected_query_point` (valid values are in the range of `0` to `len(test_predictions)`) to visualise the results for another test point.",
"_____no_output_____"
]
],
[
[
"selected_query_point = -10",
"_____no_output_____"
],
[
"# Select the predictios for the point specified by selected_query_point\nqp_predictions = test_predictions[:, :, selected_query_point, :]\n# The shape should be n_estimators x n_predictions x size_output_layer\nqp_predictions.shape",
"_____no_output_____"
]
],
[
[
"Next, to facilitate plotting the predictions using either a density plot or a box plot, we are going to reshape `qp_predictions` to $R\\times F$ where $R$ is equal to $M\\times K$ as above and $F$ is the output dimensionality of the output layer.",
"_____no_output_____"
]
],
[
[
"qp_predictions = qp_predictions.reshape(np.product(qp_predictions.shape[0:-1]), \n qp_predictions.shape[-1])\nqp_predictions.shape",
"_____no_output_____"
]
],
[
[
"The model returns the probability of edge, the class to predict. The probability of no edge is just the complement of the latter. Let's calculate it so that we can plot the distribution of predictions for both outcomes.",
"_____no_output_____"
]
],
[
[
"qp_predictions=np.hstack((qp_predictions, 1.-qp_predictions,))",
"_____no_output_____"
]
],
[
[
"We'd like to assess the ensemble's confidence in its predictions in order to decide if we can trust them or not. Utilising a box plot, we can visually inspect the ensemble's distribution of prediction probabilities for a point in the test set.\n\nIf the spread of values for the predicted point class is well separated from those of the other class with little overlap then we can be confident that the prediction is correct.",
"_____no_output_____"
]
],
[
[
"correct_label = \"Edge\"\nif edge_labels_test[selected_query_point] == 0:\n correct_label = \"No Edge\"\n\nfig, ax = plt.subplots(figsize=(12,6))\nax.boxplot(x=qp_predictions)\nax.set_xticklabels([\"Edge\", \"No Edge\"])\nax.tick_params(axis='x', rotation=45)\nplt.title(\"Correct label is \"+ correct_label)\nplt.ylabel(\"Predicted Probability\")\nplt.xlabel(\"Class\")",
"_____no_output_____"
]
],
[
[
"For the selected pair of nodes (query point), the ensemble is not certain as to whether an edge between these two nodes should exist. This can be inferred by the large spread of values as indicated in the above figure.\n\n(Note that due to the stochastic nature of training neural network algorithms, the above conclusion may not be valid if you re-run the notebook; however, the general conclusion that the use of ensemble learning can be used to quantify the model's uncertainty about its prediction still holds.)",
"_____no_output_____"
],
[
"The below image shows an example of the classifier making a correct prediction with higher confidence than the above example. The results is for the setting `selected_query_point=0`.",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
c50eb973446b398f02dd622b80d5ea6341e98631
| 11,575 |
ipynb
|
Jupyter Notebook
|
CCA-analysis.ipynb
|
gwaygenomics/2018_04_20_Rosetta
|
3dff13ae882a1cf5f02979760c3e25e6ea9715b2
|
[
"BSD-3-Clause"
] | null | null | null |
CCA-analysis.ipynb
|
gwaygenomics/2018_04_20_Rosetta
|
3dff13ae882a1cf5f02979760c3e25e6ea9715b2
|
[
"BSD-3-Clause"
] | null | null | null |
CCA-analysis.ipynb
|
gwaygenomics/2018_04_20_Rosetta
|
3dff13ae882a1cf5f02979760c3e25e6ea9715b2
|
[
"BSD-3-Clause"
] | null | null | null | 30.380577 | 141 | 0.524838 |
[
[
[
"%matplotlib inline\nimport numpy as np\nimport scipy.spatial\nimport pandas as pd\nimport sklearn.decomposition\nimport matplotlib.pyplot as plt\nimport seaborn as sb\n\nimport linear_cca\nimport multimodal_data",
"_____no_output_____"
]
],
[
[
"# Useful References\n## https://arxiv.org/pdf/1711.02391.pdf\n## http://users.stat.umn.edu/~helwig/notes/cancor-Notes.pdf\n## https://www.statisticssolutions.com/canonical-correlation/",
"_____no_output_____"
],
[
"# Load data",
"_____no_output_____"
]
],
[
[
"l1k = multimodal_data.load_l1000(\"treatment_level_all_alleles.csv\")\nl1k = multimodal_data.load_l1000(\"replicate_level_all_alleles.csv\")\ncp = multimodal_data.load_cell_painting(\n \"/data1/luad/others/morphology.csv\", \n \"resnet18-validation-well_profiles.csv\", \n aggregate_replicates=False\n)",
"_____no_output_____"
],
[
"l1k, cp = multimodal_data.align_profiles(l1k, cp, sample=4)\ncommon_alleles = set(cp[\"Allele\"].unique()).intersection( l1k[\"Allele\"].unique() )\ngenes = list(common_alleles)\ngenes = [x for x in genes if x not in [\"EGFP\", \"BFP\", \"HCRED\"]]\nl1k = l1k[l1k.Allele.isin(genes)]\ncp = cp[cp.Allele.isin(genes)]",
"_____no_output_____"
]
],
[
[
"# Compute CCA",
"_____no_output_____"
]
],
[
[
"# Preprocessing to the data:\n# 1. Standardize features (z-scoring)\n# 2. Reduce dimensionality (PCA down to 100 features)\n# This is necessary because we only have 175 data points, \n# while L1000 has 978 features and Cell Painting has 256.\n# So PCA is useful as a regularizer somehow.\n\ndef cca_analysis(GE_train, MF_train, GE_test, MF_test):\n # Prepare Gene Expression matrix\n sc_l1k = sklearn.preprocessing.StandardScaler()\n sc_l1k.fit(GE_train)\n GE = sc_l1k.transform(GE_train)\n \n pca_l1k = sklearn.decomposition.PCA(n_components=150, svd_solver=\"full\")\n pca_l1k.fit(GE)\n GE = pca_l1k.transform(GE)\n\n # Prepare Cell Painting matrix\n sc_cp = sklearn.preprocessing.StandardScaler()\n sc_cp.fit(MF_train)\n MF = sc_cp.transform(MF_train)\n \n pca_cp = sklearn.decomposition.PCA(n_components=100, svd_solver=\"full\")\n pca_cp.fit(MF)\n MF = pca_cp.transform(MF)\n\n # Compute CCA\n A, B, D, ma, mb = linear_cca.linear_cca(MF, GE, 10)\n \n X = pca_cp.transform(sc_cp.transform(MF_test))\n Y = pca_l1k.transform(sc_l1k.transform(GE_test))\n \n X = np.dot(X, A)\n Y = np.dot(Y, B)\n \n return X, Y, D",
"_____no_output_____"
],
[
"GE = np.asarray(l1k)[:,1:]\nMF = np.asarray(cp)[:,1:]\nMF_v, GE_v, D = cca_analysis(GE, MF, GE, MF)",
"_____no_output_____"
],
[
"# In linear CCA, the canonical correlations equal to the square roots of the eigenvalues:\nplt.plot(np.sqrt(D))\nprint(\"First cannonical correlation: \", np.sqrt(D[0]))",
"_____no_output_____"
],
[
"D = scipy.spatial.distance_matrix(MF_v[:,0:2], GE_v[:,0:2])\nNN = np.argsort(D, axis=1) # Nearest morphology point to each gene expression point\n\nplt.figure(figsize=(10,10))\nplt.scatter(MF_v[:,0], MF_v[:,1], c=\"blue\", s=50, edgecolor='gray', linewidths=1)\nplt.scatter(GE_v[:,0]+0, GE_v[:,1]+0, c=\"lime\", edgecolor='gray', linewidths=1)\n\nconnected = 0\nfor i in range(MF_v.shape[0]):\n for j in range(7): #GE_v.shape[0]):\n if cp.iloc[i].Allele == l1k.iloc[NN[i,j]].Allele:\n plt.plot([GE_v[NN[i,j],0],MF_v[i,0]],[GE_v[NN[i,j],1],MF_v[i,1]], 'k-', color=\"red\")\n# if np.random.random() > 0.9:\n# plt.text(GE_v[i,0], GE_v[i,1], l1k.iloc[i].Allele, horizontalalignment='left', size='medium', color='black')\n connected += 1\n #break\n\nprint(connected)\n# plt.xlim(-2,2)\n# plt.ylim(-2,2)",
"_____no_output_____"
],
[
"df = pd.DataFrame(data={\"cca1\": np.concatenate((GE_v[:,0], MF_v[:,0])), \n \"cca2\": np.concatenate((GE_v[:,1],MF_v[:,1])),\n \"source\": [\"L1K\" for x in range(GE_v.shape[0])]+[\"CP\" for x in range(MF_v.shape[0])],\n \"allele\": list(l1k[\"Allele\"]) + list(cp[\"Allele\"])}\n )",
"_____no_output_____"
],
[
"df[\"color\"] = df[\"allele\"].str.find(\"EGFR\") != -1\nsb.lmplot(data=df, x=\"cca1\", y=\"cca2\", hue=\"color\", fit_reg=False, col=\"source\")",
"_____no_output_____"
],
[
"plt.figure(figsize=(10,10))\nplt.scatter(MF_v[:,0], MF_v[:,1], c=\"blue\", s=100, edgecolor='gray', linewidths=1)\nplt.figure(figsize=(10,10))\nplt.scatter(GE_v[:,0]+0, GE_v[:,1]+0, c=\"lime\", s=100, edgecolor='gray', linewidths=1)",
"_____no_output_____"
]
],
[
[
"# Annotate visualization",
"_____no_output_____"
]
],
[
[
"def visualize_annotations(l1k, cp, GE_v, MF_v, display_items=[]):\n ge_data = pd.DataFrame(data=l1k[\"Allele\"].reset_index()) \n ge_data[\"x\"] = GE_v[:,0]\n ge_data[\"y\"] = GE_v[:,1]\n ge_data.columns = [\"idx\", \"Allele\", \"x\", \"y\"]\n ge_data[\"type\"] = \"GeneExpression\"\n \n mf_data = pd.DataFrame(data=cp[\"Allele\"].reset_index())\n mf_data[\"x\"] = MF_v[:,0]\n mf_data[\"y\"] = MF_v[:,1]\n mf_data.columns = [\"idx\", \"Allele\", \"x\", \"y\"]\n mf_data[\"type\"] = \"Morphology\"\n \n data = pd.concat([ge_data, mf_data])\n\n plt.figure(figsize=(12,12))\n p1 = sb.regplot(data=ge_data, x=\"x\", y=\"y\", fit_reg=False, color=\"red\", scatter_kws={'s':50})\n p2 = sb.regplot(data=mf_data, x=\"x\", y=\"y\", fit_reg=False, color=\"blue\", scatter_kws={'s':50})\n\n for point in range(ge_data.shape[0]):\n #if ge_data.Allele[point] in display_items:\n p1.text(ge_data.x[point], ge_data.y[point], ge_data.Allele[point], horizontalalignment='left', size='medium', color='black')\n\n for point in range(mf_data.shape[0]):\n #if mf_data.Allele[point] in display_items:\n p2.text(mf_data.x[point], mf_data.y[point], mf_data.Allele[point], horizontalalignment='left', size='medium', color='black')",
"_____no_output_____"
],
[
"visualize_annotations(l1k, cp, GE_v, MF_v, display_items=[\"NFE2L2_p.T80K\",\"EGFP\"])",
"_____no_output_____"
]
],
[
[
"# Visualization in the test set",
"_____no_output_____"
]
],
[
[
"common_alleles = set(cp[\"Allele\"].unique()).intersection( l1k[\"Allele\"].unique() )\ngenes = list(common_alleles)\nnp.random.shuffle(genes)\n\ntrain = genes[0:9*int(len(genes)/10)]\ntest = genes[9*int(len(genes)/10):]\n\nGE_train = np.asarray(l1k[l1k[\"Allele\"].isin(train)])[:,1:]\nMF_train = np.asarray(cp[cp[\"Allele\"].isin(train)])[:,1:]\n\nGE_test = np.asarray(l1k[l1k[\"Allele\"].isin(test)])[:,1:]\nMF_test = np.asarray(cp[cp[\"Allele\"].isin(test)])[:,1:]\n\nMF_v, GE_v, D = cca_analysis(GE_train, MF_train, GE_test, MF_test)\n\nvisualize_annotations(\n l1k[l1k[\"Allele\"].isin(test)], \n cp[cp[\"Allele\"].isin(test)], \n GE_v, \n MF_v\n)",
"_____no_output_____"
],
[
"D = scipy.spatial.distance_matrix(MF_v[:,0:2], GE_v[:,0:2])\nNN = np.argsort(D, axis=1) # Nearest morphology point to each gene expression point\n\nplt.figure(figsize=(10,10))\nplt.scatter(MF_v[:,0], MF_v[:,1], c=\"blue\", s=50, edgecolor='gray', linewidths=1)\nplt.scatter(GE_v[:,0]+0, GE_v[:,1]+0, c=\"red\", edgecolor='gray', linewidths=1)\n\nconnected = 0\nfor i in range(MF_v.shape[0]):\n for j in range(7):\n if cp.iloc[i].Allele == l1k.iloc[NN[i,j]].Allele:\n plt.plot([GE_v[NN[i,j],0],MF_v[i,0]],[GE_v[NN[i,j],1],MF_v[i,1]], 'k-', color=\"lime\")",
"_____no_output_____"
],
[
"# In linear CCA, the canonical correlations equal to the square roots of the eigenvalues:\nplt.plot(np.sqrt(D))\nprint(\"First cannonical correlation: \", np.sqrt(D[0]))",
"_____no_output_____"
]
],
[
[
"# Visualize data matrices",
"_____no_output_____"
]
],
[
[
"X = (GE - np.min(GE))/(np.max(GE) - np.min(GE))\nX = np.asarray(X, dtype=np.float32)\nplt.imshow(X)",
"_____no_output_____"
],
[
"X = (MF - np.min(MF))/(np.max(MF) - np.min(MF))\nX = np.asarray(X, dtype=np.float32)\nplt.imshow(X)",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
c50eb9a1944dd9557e3febab8e6970847d2e3383
| 384,791 |
ipynb
|
Jupyter Notebook
|
notebooks/sample.wordvec.python.ipynb
|
red-gold/causality
|
76fda55a6aec61c9d05c1d2c90845f724ebb4421
|
[
"MIT"
] | null | null | null |
notebooks/sample.wordvec.python.ipynb
|
red-gold/causality
|
76fda55a6aec61c9d05c1d2c90845f724ebb4421
|
[
"MIT"
] | 1 |
2020-07-17T00:51:42.000Z
|
2020-07-17T00:51:42.000Z
|
notebooks/sample.wordvec.python.ipynb
|
red-gold/causality
|
76fda55a6aec61c9d05c1d2c90845f724ebb4421
|
[
"MIT"
] | null | null | null | 36.860906 | 5,432 | 0.413986 |
[
[
[
"#!pip install tensorflow",
"_____no_output_____"
],
[
"import tensorflow as tf\nimport numpy as np\n\ncorpus_raw = 'He is the king . The king is royal . She is the royal queen '\n\n# convert to lower case\ncorpus_raw = corpus_raw.lower()\n\nwords = []\nfor word in corpus_raw.split():\n if word != '.': # because we don't want to treat . as a word\n words.append(word)\n\nwords = set(words) # so that all duplicate words are removed\nword2int = {}\nint2word = {}\nvocab_size = len(words) # gives the total number of unique words\n\nfor i,word in enumerate(words):\n word2int[word] = i\n int2word[i] = word\n\n# raw sentences is a list of sentences.\nraw_sentences = corpus_raw.split('.')\nsentences = []\nfor sentence in raw_sentences:\n sentences.append(sentence.split())\n\nWINDOW_SIZE = 2\n\ndata = []\nfor sentence in sentences:\n for word_index, word in enumerate(sentence):\n for nb_word in sentence[max(word_index - WINDOW_SIZE, 0) : min(word_index + WINDOW_SIZE, len(sentence)) + 1] : \n if nb_word != word:\n data.append([word, nb_word])\n\n# function to convert numbers to one hot vectors\ndef to_one_hot(data_point_index, vocab_size):\n temp = np.zeros(vocab_size)\n temp[data_point_index] = 1\n return temp\n\nx_train = [] # input word\ny_train = [] # output word\n\nfor data_word in data:\n x_train.append(to_one_hot(word2int[ data_word[0] ], vocab_size))\n y_train.append(to_one_hot(word2int[ data_word[1] ], vocab_size))\n\n# convert them to numpy arrays\nx_train = np.asarray(x_train)\ny_train = np.asarray(y_train)\n\n# making placeholders for x_train and y_train\nx = tf.placeholder(tf.float32, shape=(None, vocab_size))\ny_label = tf.placeholder(tf.float32, shape=(None, vocab_size))\n\nEMBEDDING_DIM = 5 # you can choose your own number\nW1 = tf.Variable(tf.random_normal([vocab_size, EMBEDDING_DIM]))\nb1 = tf.Variable(tf.random_normal([EMBEDDING_DIM])) #bias\nhidden_representation = tf.add(tf.matmul(x,W1), b1)\n\nW2 = tf.Variable(tf.random_normal([EMBEDDING_DIM, vocab_size]))\nb2 = tf.Variable(tf.random_normal([vocab_size]))\nprediction = tf.nn.softmax(tf.add( tf.matmul(hidden_representation, W2), b2))\n\n\nsess = tf.Session()\ninit = tf.global_variables_initializer()\nsess.run(init) #make sure you do this!\n\n# define the loss function:\ncross_entropy_loss = tf.reduce_mean(-tf.reduce_sum(y_label * tf.log(prediction), reduction_indices=[1]))\n\n# define the training step:\ntrain_step = tf.train.GradientDescentOptimizer(0.1).minimize(cross_entropy_loss)\n\nn_iters = 10000\n# train for n_iter iterations\n\nfor _ in range(n_iters):\n sess.run(train_step, feed_dict={x: x_train, y_label: y_train})\n print('loss is : ', sess.run(cross_entropy_loss, feed_dict={x: x_train, y_label: y_train}))\n\nvectors = sess.run(W1 + b1)",
"('loss is : ', 4.770094)\n('loss is : ', 4.5315514)\n('loss is : ', 4.3378067)\n('loss is : ', 4.1745963)\n('loss is : ', 4.0333204)\n('loss is : ', 3.9085271)\n('loss is : ', 3.7965026)\n('loss is : ', 3.6946187)\n('loss is : ', 3.6009898)\n('loss is : ', 3.5142572)\n('loss is : ', 3.4334252)\n('loss is : ', 3.357755)\n('loss is : ', 3.286679)\n('loss is : ', 3.2197533)\n('loss is : ', 3.1566157)\n('loss is : ', 3.0969653)\n('loss is : ', 3.040543)\n('loss is : ', 2.9871223)\n('loss is : ', 2.936501)\n('loss is : ', 2.8884962)\n('loss is : ', 2.842941)\n('loss is : ', 2.7996812)\n('loss is : ', 2.7585745)\n('loss is : ', 2.7194874)\n('loss is : ', 2.682296)\n('loss is : ', 2.646883)\n('loss is : ', 2.6131382)\n('loss is : ', 2.5809581)\n('loss is : ', 2.5502431)\n('loss is : ', 2.520901)\n('loss is : ', 2.4928432)\n('loss is : ', 2.4659865)\n('loss is : ', 2.4402523)\n('loss is : ', 2.4155667)\n('loss is : ', 2.39186)\n('loss is : ', 2.3690684)\n('loss is : ', 2.3471308)\n('loss is : ', 2.3259919)\n('loss is : ', 2.3055997)\n('loss is : ', 2.285906)\n('loss is : ', 2.266868)\n('loss is : ', 2.2484446)\n('loss is : ', 2.230599)\n('loss is : ', 2.2132971)\n('loss is : ', 2.196509)\n('loss is : ', 2.1802053)\n('loss is : ', 2.1643608)\n('loss is : ', 2.148952)\n('loss is : ', 2.1339574)\n('loss is : ', 2.119357)\n('loss is : ', 2.1051335)\n('loss is : ', 2.0912702)\n('loss is : ', 2.0777519)\n('loss is : ', 2.0645647)\n('loss is : ', 2.051696)\n('loss is : ', 2.0391345)\n('loss is : ', 2.0268688)\n('loss is : ', 2.0148895)\n('loss is : ', 2.0031867)\n('loss is : ', 1.9917521)\n('loss is : ', 1.9805773)\n('loss is : ', 1.9696554)\n('loss is : ', 1.9589785)\n('loss is : ', 1.9485404)\n('loss is : ', 1.9383343)\n('loss is : ', 1.9283546)\n('loss is : ', 1.9185953)\n('loss is : ', 1.9090511)\n('loss is : ', 1.8997165)\n('loss is : ', 1.8905864)\n('loss is : ', 1.8816559)\n('loss is : ', 1.8729204)\n('loss is : ', 1.8643751)\n('loss is : ', 1.8560154)\n('loss is : ', 1.8478371)\n('loss is : ', 1.8398356)\n('loss is : ', 1.832007)\n('loss is : ', 1.8243469)\n('loss is : ', 1.8168513)\n('loss is : ', 1.8095161)\n('loss is : ', 1.8023374)\n('loss is : ', 1.7953115)\n('loss is : ', 1.7884344)\n('loss is : ', 1.7817025)\n('loss is : ', 1.7751119)\n('loss is : ', 1.7686591)\n('loss is : ', 1.7623405)\n('loss is : ', 1.7561526)\n('loss is : ', 1.750092)\n('loss is : ', 1.7441553)\n('loss is : ', 1.7383393)\n('loss is : ', 1.7326406)\n('loss is : ', 1.727056)\n('loss is : ', 1.7215828)\n('loss is : ', 1.7162178)\n('loss is : ', 1.7109579)\n('loss is : ', 1.7058002)\n('loss is : ', 1.7007426)\n('loss is : ', 1.6957817)\n('loss is : ', 1.6909153)\n('loss is : ', 1.6861409)\n('loss is : ', 1.681456)\n('loss is : ', 1.6768582)\n('loss is : ', 1.6723452)\n('loss is : ', 1.6679151)\n('loss is : ', 1.6635656)\n('loss is : ', 1.6592951)\n('loss is : ', 1.6551011)\n('loss is : ', 1.6509823)\n('loss is : ', 1.6469364)\n('loss is : ', 1.6429622)\n('loss is : ', 1.639058)\n('loss is : ', 1.6352222)\n('loss is : ', 1.6314533)\n('loss is : ', 1.62775)\n('loss is : ', 1.6241112)\n('loss is : ', 1.6205351)\n('loss is : ', 1.6170211)\n('loss is : ', 1.6135678)\n('loss is : ', 1.6101741)\n('loss is : ', 1.606839)\n('loss is : ', 1.6035614)\n('loss is : ', 1.6003408)\n('loss is : ', 1.5971758)\n('loss is : ', 1.5940658)\n('loss is : ', 1.5910101)\n('loss is : ', 1.5880077)\n('loss is : ', 1.5850576)\n('loss is : ', 1.5821598)\n('loss is : ', 1.579313)\n('loss is : ', 1.5765167)\n('loss is : ', 1.5737703)\n('loss is : ', 1.5710729)\n('loss is : ', 1.568424)\n('loss is : ', 1.5658231)\n('loss is : ', 1.5632694)\n('loss is : ', 1.5607622)\n('loss is : ', 1.558301)\n('loss is : ', 1.555885)\n('loss is : ', 1.5535136)\n('loss is : ', 1.5511866)\n('loss is : ', 1.5489028)\n('loss is : ', 1.5466616)\n('loss is : ', 1.5444624)\n('loss is : ', 1.5423045)\n('loss is : ', 1.5401876)\n('loss is : ', 1.5381104)\n('loss is : ', 1.5360726)\n('loss is : ', 1.5340732)\n('loss is : ', 1.5321114)\n('loss is : ', 1.5301869)\n('loss is : ', 1.5282986)\n('loss is : ', 1.526446)\n('loss is : ', 1.524628)\n('loss is : ', 1.5228442)\n('loss is : ', 1.5210935)\n('loss is : ', 1.5193753)\n('loss is : ', 1.5176888)\n('loss is : ', 1.5160332)\n('loss is : ', 1.5144079)\n('loss is : ', 1.5128118)\n('loss is : ', 1.5112445)\n('loss is : ', 1.5097048)\n('loss is : ', 1.5081924)\n('loss is : ', 1.5067065)\n('loss is : ', 1.5052462)\n('loss is : ', 1.5038106)\n('loss is : ', 1.5023994)\n('loss is : ', 1.5010118)\n('loss is : ', 1.4996468)\n('loss is : ', 1.4983042)\n('loss is : ', 1.4969832)\n('loss is : ', 1.4956827)\n('loss is : ', 1.4944029)\n('loss is : ', 1.4931427)\n('loss is : ', 1.4919018)\n('loss is : ', 1.4906791)\n('loss is : ', 1.4894745)\n('loss is : ', 1.4882877)\n('loss is : ', 1.4871174)\n('loss is : ', 1.485964)\n('loss is : ', 1.4848263)\n('loss is : ', 1.4837043)\n('loss is : ', 1.4825974)\n('loss is : ', 1.481505)\n('loss is : ', 1.480427)\n('loss is : ', 1.4793628)\n('loss is : ', 1.4783121)\n('loss is : ', 1.4772748)\n('loss is : ', 1.4762499)\n('loss is : ', 1.4752376)\n('loss is : ', 1.4742372)\n('loss is : ', 1.4732487)\n('loss is : ', 1.4722718)\n('loss is : ', 1.4713061)\n('loss is : ', 1.4703512)\n('loss is : ', 1.4694071)\n('loss is : ', 1.4684731)\n('loss is : ', 1.4675497)\n('loss is : ', 1.466636)\n('loss is : ', 1.4657321)\n('loss is : ', 1.4648376)\n('loss is : ', 1.4639524)\n('loss is : ', 1.4630765)\n('loss is : ', 1.4622092)\n('loss is : ', 1.4613507)\n('loss is : ', 1.4605006)\n('loss is : ', 1.459659)\n('loss is : ', 1.4588255)\n('loss is : ', 1.4580003)\n('loss is : ', 1.4571826)\n('loss is : ', 1.4563727)\n('loss is : ', 1.4555705)\n('loss is : ', 1.4547755)\n('loss is : ', 1.4539881)\n('loss is : ', 1.4532077)\n('loss is : ', 1.4524342)\n('loss is : ', 1.4516678)\n('loss is : ', 1.4509082)\n('loss is : ', 1.4501554)\n('loss is : ', 1.4494089)\n('loss is : ', 1.4486691)\n('loss is : ', 1.4479357)\n('loss is : ', 1.4472084)\n('loss is : ', 1.4464873)\n('loss is : ', 1.4457723)\n('loss is : ', 1.4450635)\n('loss is : ', 1.4443601)\n('loss is : ', 1.4436631)\n('loss is : ', 1.4429715)\n('loss is : ', 1.4422854)\n('loss is : ', 1.4416051)\n('loss is : ', 1.4409302)\n('loss is : ', 1.4402608)\n('loss is : ', 1.4395965)\n('loss is : ', 1.4389377)\n('loss is : ', 1.4382839)\n('loss is : ', 1.4376354)\n('loss is : ', 1.4369917)\n('loss is : ', 1.4363531)\n('loss is : ', 1.4357196)\n('loss is : ', 1.4350907)\n('loss is : ', 1.4344666)\n('loss is : ', 1.4338473)\n('loss is : ', 1.4332327)\n('loss is : ', 1.4326229)\n('loss is : ', 1.4320173)\n('loss is : ', 1.4314165)\n('loss is : ', 1.4308199)\n('loss is : ', 1.4302278)\n('loss is : ', 1.42964)\n('loss is : ', 1.4290565)\n('loss is : ', 1.4284773)\n('loss is : ', 1.4279022)\n('loss is : ', 1.4273314)\n('loss is : ', 1.4267646)\n('loss is : ', 1.4262018)\n('loss is : ', 1.4256432)\n('loss is : ', 1.4250884)\n('loss is : ', 1.4245375)\n('loss is : ', 1.4239905)\n('loss is : ', 1.4234475)\n('loss is : ', 1.4229081)\n('loss is : ', 1.4223725)\n('loss is : ', 1.4218407)\n('loss is : ', 1.4213123)\n('loss is : ', 1.4207878)\n('loss is : ', 1.4202667)\n('loss is : ', 1.4197493)\n('loss is : ', 1.4192355)\n('loss is : ', 1.418725)\n('loss is : ', 1.4182181)\n('loss is : ', 1.4177144)\n('loss is : ', 1.4172144)\n('loss is : ', 1.4167175)\n('loss is : ', 1.416224)\n('loss is : ', 1.4157338)\n('loss is : ', 1.415247)\n('loss is : ', 1.4147631)\n('loss is : ', 1.4142827)\n('loss is : ', 1.4138051)\n('loss is : ', 1.4133309)\n('loss is : ', 1.4128598)\n('loss is : ', 1.4123915)\n('loss is : ', 1.4119266)\n('loss is : ', 1.4114646)\n('loss is : ', 1.4110056)\n('loss is : ', 1.4105494)\n('loss is : ', 1.4100962)\n('loss is : ', 1.4096462)\n('loss is : ', 1.4091988)\n('loss is : ', 1.4087542)\n('loss is : ', 1.4083127)\n('loss is : ', 1.4078737)\n('loss is : ', 1.4074377)\n('loss is : ', 1.4070044)\n('loss is : ', 1.4065739)\n('loss is : ', 1.406146)\n('loss is : ', 1.405721)\n('loss is : ', 1.4052985)\n('loss is : ', 1.4048786)\n('loss is : ', 1.4044614)\n('loss is : ', 1.4040468)\n('loss is : ', 1.4036348)\n('loss is : ', 1.4032254)\n('loss is : ', 1.4028183)\n('loss is : ', 1.4024141)\n('loss is : ', 1.4020122)\n('loss is : ', 1.4016128)\n('loss is : ', 1.4012157)\n('loss is : ', 1.4008213)\n('loss is : ', 1.4004294)\n('loss is : ', 1.4000396)\n('loss is : ', 1.3996522)\n('loss is : ', 1.3992676)\n('loss is : ', 1.3988849)\n('loss is : ', 1.3985046)\n('loss is : ', 1.3981268)\n('loss is : ', 1.3977513)\n('loss is : ', 1.397378)\n('loss is : ', 1.397007)\n('loss is : ', 1.3966382)\n('loss is : ', 1.3962717)\n('loss is : ', 1.3959073)\n('loss is : ', 1.395545)\n('loss is : ', 1.3951852)\n('loss is : ', 1.3948274)\n('loss is : ', 1.3944716)\n('loss is : ', 1.3941183)\n('loss is : ', 1.393767)\n('loss is : ', 1.3934177)\n('loss is : ', 1.3930707)\n('loss is : ', 1.3927257)\n('loss is : ', 1.3923827)\n('loss is : ', 1.3920417)\n('loss is : ', 1.3917028)\n('loss is : ', 1.391366)\n('loss is : ', 1.3910313)\n('loss is : ', 1.3906984)\n('loss is : ', 1.3903675)\n('loss is : ', 1.3900385)\n('loss is : ', 1.3897117)\n('loss is : ', 1.3893867)\n('loss is : ', 1.3890637)\n('loss is : ', 1.3887424)\n('loss is : ', 1.3884233)\n('loss is : ', 1.3881061)\n('loss is : ', 1.3877907)\n('loss is : ', 1.3874772)\n('loss is : ', 1.3871654)\n('loss is : ', 1.3868557)\n('loss is : ', 1.3865477)\n('loss is : ', 1.3862416)\n('loss is : ', 1.3859372)\n('loss is : ', 1.3856344)\n('loss is : ', 1.3853339)\n('loss is : ', 1.3850349)\n('loss is : ', 1.3847377)\n('loss is : ', 1.3844423)\n('loss is : ', 1.3841487)\n('loss is : ', 1.3838567)\n('loss is : ', 1.3835666)\n('loss is : ', 1.383278)\n('loss is : ', 1.3829911)\n('loss is : ', 1.3827062)\n('loss is : ', 1.3824227)\n('loss is : ', 1.382141)\n('loss is : ', 1.3818609)\n('loss is : ', 1.3815825)\n('loss is : ', 1.3813057)\n('loss is : ', 1.3810304)\n('loss is : ', 1.3807571)\n('loss is : ', 1.3804852)\n('loss is : ', 1.3802148)\n('loss is : ', 1.3799461)\n('loss is : ', 1.3796791)\n('loss is : ', 1.3794135)\n('loss is : ', 1.3791494)\n('loss is : ', 1.3788869)\n('loss is : ', 1.3786262)\n('loss is : ', 1.3783668)\n('loss is : ', 1.3781091)\n('loss is : ', 1.3778528)\n('loss is : ', 1.3775978)\n('loss is : ', 1.3773447)\n('loss is : ', 1.377093)\n('loss is : ', 1.3768426)\n('loss is : ', 1.3765937)\n('loss is : ', 1.3763463)\n('loss is : ', 1.3761004)\n('loss is : ', 1.375856)\n('loss is : ', 1.3756132)\n('loss is : ', 1.3753715)\n('loss is : ', 1.3751312)\n('loss is : ', 1.3748925)\n('loss is : ', 1.3746551)\n('loss is : ', 1.3744192)\n('loss is : ', 1.3741846)\n('loss is : ', 1.3739516)\n('loss is : ', 1.3737197)\n('loss is : ', 1.3734891)\n('loss is : ', 1.37326)\n('loss is : ', 1.3730325)\n('loss is : ', 1.3728058)\n('loss is : ', 1.3725809)\n('loss is : ', 1.3723571)\n('loss is : ', 1.3721346)\n('loss is : ', 1.3719136)\n('loss is : ', 1.3716936)\n('loss is : ', 1.3714751)\n('loss is : ', 1.3712579)\n('loss is : ', 1.3710419)\n('loss is : ', 1.370827)\n('loss is : ', 1.3706136)\n('loss is : ', 1.3704013)\n('loss is : ', 1.3701906)\n('loss is : ', 1.3699807)\n('loss is : ', 1.3697722)\n('loss is : ', 1.3695649)\n('loss is : ', 1.3693589)\n('loss is : ', 1.3691539)\n('loss is : ', 1.3689502)\n('loss is : ', 1.3687478)\n('loss is : ', 1.3685465)\n('loss is : ', 1.3683463)\n('loss is : ', 1.3681474)\n('loss is : ', 1.3679496)\n('loss is : ', 1.3677529)\n('loss is : ', 1.3675575)\n('loss is : ', 1.3673631)\n('loss is : ', 1.3671699)\n('loss is : ', 1.3669778)\n('loss is : ', 1.366787)\n"
],
[
"def euclidean_dist(vec1, vec2):\n return np.sqrt(np.sum((vec1-vec2)**2))\ndef find_closest(word_index, vectors):\n min_dist = 10000 # to act like positive infinity\n min_index = -1\n query_vector = vectors[word_index]\n for index, vector in enumerate(vectors):\n if euclidean_dist(vector, query_vector) < min_dist and not np.array_equal(vector, query_vector):\n min_dist = euclidean_dist(vector, query_vector)\n min_index = index\n return min_index\nfind_closest(0, vectors)",
"_____no_output_____"
],
[
"for w,i in word2int.items():\n closest = find_closest(0, vectors)\n print(w, int2word[closest])",
"('king', 'she')\n('is', 'she')\n('queen', 'she')\n('royal', 'she')\n('she', 'she')\n('the', 'she')\n('he', 'she')\n"
],
[
"from sklearn.manifold import TSNE\n\nmodel = TSNE(n_components=2, random_state=0)\nnp.set_printoptions(suppress=True)\nvectors = model.fit_transform(vectors) \n\nfrom sklearn import preprocessing\n\nnormalizer = preprocessing.Normalizer()\nnvectors = normalizer.fit_transform(vectors, 'l2')\n\nprint(nvectors)\n\nimport matplotlib.pyplot as plt\n\n\nfig, ax = plt.subplots()\nprint(words)\nfor word in words:\n print(word, vectors[word2int[word]][1])\n ax.annotate(word, (nvectors[word2int[word]][0],nvectors[word2int[word]][1] ))\nplt.show()",
"[[-0.9859371 -0.16711695]\n [-0.9057485 0.4238156 ]\n [-0.19882636 0.9800347 ]\n [ 0.69531065 -0.7187093 ]\n [-0.2924362 -0.956285 ]\n [ 0.84685785 0.53181934]\n [-0.92063266 -0.39043003]]\nset(['king', 'is', 'queen', 'royal', 'she', 'the', 'he'])\n('king', -12.519757)\n('is', 147.00587)\n('queen', 275.3545)\n('royal', -172.04462)\n('she', -300.39627)\n('the', 115.83045)\n('he', -140.86923)\n"
],
[
"import gensim",
"_____no_output_____"
],
[
"wv = gensim.models.word2vec()",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
c50ec1dc9b09259b483a2085aa37c62ecb0eb586
| 2,676 |
ipynb
|
Jupyter Notebook
|
deployer/tests/test-notebooks/base-hub/simple.ipynb
|
catalyst-cooperative/pilot-hubs
|
8c7b117943ee600ba1eb239d3893ec682d940850
|
[
"BSD-3-Clause"
] | 22 |
2020-10-07T23:52:59.000Z
|
2021-08-08T01:18:15.000Z
|
deployer/tests/test-notebooks/base-hub/simple.ipynb
|
catalyst-cooperative/pilot-hubs
|
8c7b117943ee600ba1eb239d3893ec682d940850
|
[
"BSD-3-Clause"
] | 538 |
2020-10-07T00:45:43.000Z
|
2021-10-18T18:21:58.000Z
|
deployer/tests/test-notebooks/base-hub/simple.ipynb
|
catalyst-cooperative/pilot-hubs
|
8c7b117943ee600ba1eb239d3893ec682d940850
|
[
"BSD-3-Clause"
] | 14 |
2020-10-12T23:18:20.000Z
|
2021-08-18T02:06:26.000Z
| 15.83432 | 43 | 0.427877 |
[
[
[
"a = 1\n1 + 3",
"_____no_output_____"
],
[
"print('line 1')\nprint('line 2')",
"line 1\nline 2\n"
],
[
"a",
"_____no_output_____"
],
[
"# do nothing line",
"_____no_output_____"
],
[
"def fib(n):\n if n == 1: \n return 1\n elif n == 2: \n return 1\n else:\n return fib(n-1) + fib(n-2)\n\nfib(10)",
"_____no_output_____"
],
[
"fib(20)",
"_____no_output_____"
],
[
"import time\nimport random",
"_____no_output_____"
],
[
"time.sleep(2 * random.random())",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
c50ec8a19cc840cf25f6166a522c37c58c0c017c
| 41,342 |
ipynb
|
Jupyter Notebook
|
Sensor_Fusion_New_Fault_Codes -Copy2.ipynb
|
Valentine-Efagene/Jupyter-Notebooks
|
91a1d98354a270d214316eba21e4a435b3e17f5d
|
[
"MIT"
] | null | null | null |
Sensor_Fusion_New_Fault_Codes -Copy2.ipynb
|
Valentine-Efagene/Jupyter-Notebooks
|
91a1d98354a270d214316eba21e4a435b3e17f5d
|
[
"MIT"
] | null | null | null |
Sensor_Fusion_New_Fault_Codes -Copy2.ipynb
|
Valentine-Efagene/Jupyter-Notebooks
|
91a1d98354a270d214316eba21e4a435b3e17f5d
|
[
"MIT"
] | null | null | null | 37.789762 | 4,172 | 0.556117 |
[
[
[
"import re\nimport os\nimport copy\nfrom math import log, pow\nimport subprocess\nimport matplotlib.pyplot as plt",
"_____no_output_____"
]
],
[
[
"#### Fault Classes",
"_____no_output_____"
],
[
"| Fault | Binary | Decimal |\n| --- | --- | --- |\n| A | 00 | 0 |\n| B | 01 | 1 |\n| C | 10 | 2 |\n| D | 11 | 3 |",
"_____no_output_____"
],
[
"#### Fault Code = (variable << 2) + fault_class",
"_____no_output_____"
],
[
"| Fault | Binary | Decimal |\n| --- | --- | --- |\n| A1 | 100 | 4 |\n| B1 | 101 | 5 |\n| C1 | 110 | 6 |\n| D1 | 111 | 7 |",
"_____no_output_____"
],
[
"### Sample (C1)",
"_____no_output_____"
]
],
[
[
"fault = 2\nvariable = 1\ncode = (variable << 2) + fault\ncode",
"_____no_output_____"
]
],
[
[
"#### Reverse",
"_____no_output_____"
]
],
[
[
"fault = code & 3\nfault",
"_____no_output_____"
],
[
"variable = code >> 2\nvariable",
"_____no_output_____"
]
],
[
[
"#### Current Fault Codes",
"_____no_output_____"
],
[
"| Fault | Decimal |\n| --- | --- |\n| A1 | 4 |\n| B1 | 5 |\n| C1 | 6 |\n| D1 | 7 |\n| A1B1 | 4-5 |\n| A1C1 | 4-6 |\n| A1D1 | 4-7 |\n| B1C1 | 5-6 |\n| C1D1 | 6-7 |",
"_____no_output_____"
],
[
"#### Flow Chart",
"_____no_output_____"
]
],
[
[
"from graphviz import Digraph\n\ndot = Digraph(node_attr={'shape': 'box'}, format='png', filename='sensorfusion')\ndot.edge_attr.update(arrowhead='vee', arrowsize='1')\n\ndot.node('0', 'Fault Generator')\ndot.node('1', 'Other Faults')\ndot.node('2', 'Supervisor (Polling)')\ndot.node('3', 'All faults\\n processed\\n?' ,shape='diamond')\ndot.node('4', 'Send trigger signal \\n &\\n increment frequency')\ndot.node('5', 'Append fault')\ndot.node('6', 'Does fault\\n exist?', shape='diamond')\ndot.node('7', 'Remove and delay fault\\nwith lower\\npriority')\ndot.node('8', 'Time\\nOut?', shape='diamond')\ndot.node('9', 'end', shape='oval')\ndot.node('10', 'start', shape='oval')\ndot.node('11', 'Delayed Faults')\n\ndot.edge('2', '1')\ndot.edge('0', '2', ' Possibility\\nof fault')\ndot.edge('1', '5', ' Possibility\\nof fault')\ndot.edge('2', '3')\ndot.edge('3', '6', 'Yes')\ndot.edge('3', '5', 'No')\ndot.edge('4', '8')\ndot.edge('5', '6')\ndot.edge('6', '4', 'Yes')\ndot.edge('6', '7', 'No')\ndot.edge('7', '3')\ndot.edge('8', '2', 'No')\ndot.edge('8', '9', 'Yes')\ndot.edge('10', '0')\ndot.edge('11', '1')\ndot\n#dot.save()\n#dot.render(view=True)",
"_____no_output_____"
]
],
[
[
"### Run",
"_____no_output_____"
]
],
[
[
"command = \"D:/code/C++/RT-Cadmium-FDD-New-Code/top_model/main.exe\"\ncompleted_process = subprocess.run(command, shell=False, capture_output=True, text=True)\n#print(completed_process.stdout)",
"_____no_output_____"
]
],
[
[
"\n### Read from file",
"_____no_output_____"
]
],
[
[
"fileName = \"SensorFusion.txt\"\n \nfault_codes = {}\n \nwith open(fileName, \"r\") as f:\n lines = f.readlines()\n \nwith open(fileName, \"r\") as f:\n output = f.read()\n \nfor line in lines:\n if (re.search(\"supervisor\", line) != None):\n res = re.findall(\"\\{\\d+[, ]*\\d*[, ]*\\d*\\}\", line)\n\n if len(res) > 0:\n str_interest = res[0].replace('}', '').replace('{', '')\n faults = str_interest.split(', ')\n key = '-' + '-'.join(faults) + '-'\n fault_codes[key] = fault_codes.get(key, 0) + 1\n\ngenerators = {'A': 0, 'B': 0, 'C': 0, 'D': 0}\n\nfor key in generators.keys():\n generators[key] = len(re.findall(\"faultGen\" + key, output))",
"_____no_output_____"
],
[
"fault_codes",
"_____no_output_____"
]
],
[
[
"### ANALYSIS / VERIFICATION",
"_____no_output_____"
],
[
"#### Definitions",
"_____no_output_____"
],
[
"**Pure Fault**: Faults from a single generator. \n**Compound Faults**: Faults formed from the combination of pure faults.",
"_____no_output_____"
],
[
"### Premise",
"_____no_output_____"
],
[
"Fault $A1$: Should have no discarded entry, because it has the highest priority \nFault $B1$: Should have some discarded value, for the case $BD$, which is not available \nFault $C1$: Higher percentage of discarded cases than $C$, because of its lower priority \nFault $D1$: Highest percentage of discarded cases, because it has the lowest priority ",
"_____no_output_____"
],
[
"Generator $output_{A1} = n({A1}) + n({A1} \\cap {B1}) + n({A1} \\cap {C1}) + n({A1} \\cap {D1}) + discarded_{A1}$ \nGenerator $output_{B1} = n({B1}) + n({A1} \\cap {B1}) + n({B1} \\cap {C1}) + discarded_{B1}$ \nGenerator $output_{C1} = n({C1}) + n({A1} \\cap {C1}) + n({B1} \\cap {C1}) + n({C1} \\cap {D1}) + discarded_{C1}$ \nGenerator $output_{D1} = n({D1}) + n({A1} \\cap {D1}) + n({C1} \\cap {D1}) + discarded_{D1}$ \n\nWhere $discarded_{A1} \\equiv 0$, because A has the highest priority, and $discarded_{B1} = 0$ because B1 has a fault code combination with the others in the right order, using the priority system.",
"_____no_output_____"
]
],
[
[
"def sumFromSupervisor(code):\n '''\n Returns the number of times faults associated with a particular pure fault (the parameter) were output by the supervisor\n\n @param code: int\n @return int\n '''\n sum = 0\n \n for key, value in fault_codes.items():\n if '-' + str(code) + '-' in key:\n sum += value;\n \n return sum;",
"_____no_output_____"
],
[
"a_discarded = generators['A'] - sumFromSupervisor(4)\na_discarded",
"_____no_output_____"
],
[
"b_discarded = generators['B'] - sumFromSupervisor(5)\nb_discarded",
"_____no_output_____"
],
[
"c_discarded = generators['C'] - sumFromSupervisor(6)\nc_discarded",
"_____no_output_____"
],
[
"d_discarded = generators['D'] - sumFromSupervisor(7)\nd_discarded",
"_____no_output_____"
],
[
"total_discarded = a_discarded + b_discarded + c_discarded + d_discarded\ntotal_discarded",
"_____no_output_____"
],
[
"total_generated = generators['A'] + generators['B'] + generators['C'] + generators['D']\ntotal_generated",
"_____no_output_____"
],
[
"discarded = {'A': a_discarded, 'B': b_discarded, 'C': c_discarded, 'D': d_discarded}\ndiscarded_percentage = {'A': a_discarded * 100 / total_generated, 'B': b_discarded * 100 / total_generated, 'C': c_discarded * 100 / total_generated, 'D': d_discarded * 100 / total_generated}",
"_____no_output_____"
],
[
"discarded_cases",
"_____no_output_____"
],
[
"fault_codes",
"_____no_output_____"
],
[
"a_increment = generators['A'] - fault_codes['-4-5-'] - fault_codes['-4-6-'] - fault_codes['-4-7-'] - a_discarded\na_increment",
"_____no_output_____"
],
[
"b_increment = generators['B'] - fault_codes['-4-5-'] - fault_codes['-5-6-'] - b_discarded\nb_increment",
"_____no_output_____"
],
[
"c_increment = generators['C'] - fault_codes['-4-6-'] - fault_codes['-5-6-'] - fault_codes['-6-7-'] - c_discarded\nc_increment",
"_____no_output_____"
],
[
"d_increment = generators['D'] - fault_codes['-4-7-'] - fault_codes['-6-7-'] - d_discarded\nd_increment",
"_____no_output_____"
]
],
[
[
"### Discard Charts",
"_____no_output_____"
]
],
[
[
"#plt.title('Discarded Bar')\nplt.bar(discarded.keys(), discarded.values())\n#plt.show()\nplt.savefig('discarded bar.png', format='png')",
"_____no_output_____"
],
[
"keys, values = list(discarded.keys()), list(discarded.values())\nlegend_keys = copy.copy(keys)\n\nfor i in range(len(keys)):\n legend_keys[i] = str(legend_keys[i]) + \" = \" + str(values[i])\n\n# Remove wedgeprops to make pie\nwedges, texts = plt.pie(values, textprops=dict(color=\"w\"), wedgeprops=dict(width=0.5))\nplt.legend(wedges, legend_keys,\n title=\"Fault Codes\",\n loc=\"center left\",\n bbox_to_anchor=(1, 0, 0.5, 1))\n\n#plt.title(\"Discarded Pie\")\n#plt.show()\nplt.savefig('discard pie.png', format='png')",
"_____no_output_____"
]
],
[
[
"### Discard Percentage Charts",
"_____no_output_____"
]
],
[
[
"#plt.title('Discard Percentage')\nplt.bar(discarded_percentage.keys(), discarded_percentage.values())\n#plt.show()\nplt.savefig('sensorfusion.png', format='png')",
"_____no_output_____"
],
[
"keys, values = list(discarded_percentage.keys()), list(discarded_percentage.values())\nlegend_keys = copy.copy(keys)\n\nfor i in range(len(keys)):\n legend_keys[i] = str(legend_keys[i]) + \" (%) = \" + str(values[i])\n\n# Remove wedgeprops to make pie\nwedges, texts = plt.pie(values, textprops=dict(color=\"w\"), wedgeprops=dict(width=0.5))\nplt.legend(wedges, legend_keys,\n title=\"Fault Codes\",\n loc=\"center left\",\n bbox_to_anchor=(1, 0, 0.5, 1))\n\n#plt.title(\"Discard Percentage\")\nplt.show()\nplt.savefig('discard percntage pie.png')",
"_____no_output_____"
]
],
[
[
"### Toggle Time vs Frequency of Generators",
"_____no_output_____"
]
],
[
[
"toggle_times = {'A': 620, 'B': 180, 'C': 490, 'D': 270}",
"_____no_output_____"
]
],
[
[
"#### Premise",
"_____no_output_____"
],
[
"$faults\\,generated \\propto \\frac{1}{toggle\\,time}$\n\n$\\therefore B > D > C > A$",
"_____no_output_____"
],
[
"### Generator Output Charts (Possibilities of Faults)",
"_____no_output_____"
]
],
[
[
"generators['A']",
"_____no_output_____"
],
[
"#plt.title('Generator Output (Possibilities of Faults)')\nplt.bar(generators.keys(), generators.values())\n#plt.show()\nplt.savefig('generator output bar.png')",
"_____no_output_____"
],
[
"keys, values = list(generators.keys()), list(generators.values())\nlegend_keys = copy.copy(keys)\n\nfor i in range(len(keys)):\n legend_keys[i] = \"n (\" + str(legend_keys[i]) + \") = \" + str(values[i])\n\n# Remove wedgeprops to make pie\nwedges, texts = plt.pie(values, textprops=dict(color=\"w\"), wedgeprops=dict(width=0.5))\nplt.legend(wedges, legend_keys,\n title=\"Fault Codes\",\n loc=\"center left\",\n bbox_to_anchor=(1, 0, 0.5, 1))\n\n#plt.title(\"Generator Output Charts (Possibilities of Fault)\")\n#plt.show()\nplt.savefig('generator output pie.png')",
"_____no_output_____"
]
],
[
[
"### Single-Run Fault Charts",
"_____no_output_____"
]
],
[
[
"chart_data = copy.copy(fault_codes)\nvalues = list(chart_data.values())\nkeys = list(chart_data.keys())\n\n#plt.title('Single-Run')\nplt.bar(keys, values)\n#plt.show()\nplt.savefig('single-run bar.png')",
"_____no_output_____"
],
[
"# Remove wedgeprops to make pie\nwedges, texts = plt.pie(values, \n textprops=dict(color=\"w\"), \n wedgeprops=dict(width=0.5))\n\nlegend_keys = copy.copy(keys)\n\nfor i in range(len(keys)):\n legend_keys[i] = str(legend_keys[i]) + \" \" + str(values[i]) + \" \" + \"times\"\n \nplt.legend(wedges, legend_keys,\n title=\"Fault Codes\",\n loc=\"center left\",\n bbox_to_anchor=(1, 0, 0.5, 1))\n\n#plt.title(\"Single-Run\")\n#plt.show()\nplt.savefig('single-run pie.png')",
"_____no_output_____"
]
],
[
[
"### Cumulative Faults Chart",
"_____no_output_____"
]
],
[
[
"fileName = \"D:/code/C++/RT-Cadmium-FDD-New-Code/knowledge_base/_fault_codes_dir/_fault_codes_list.txt\"\n\nwith open(fileName, \"r\") as f:\n lines = f.readlines()\n \ntotal = {}\n \nfor line in lines:\n res = re.findall(\"\\d+[-]*\\d*\", line)\n\n if len(res) > 0:\n total[res[0]] = str(res[1])",
"_____no_output_____"
],
[
"values = list(total.values())\nkeys = list(total.keys())\n\n#plt.title('Cummulative')\nplt.bar(keys, values)\n#plt.show()\nplt.savefig('single-run bar.png')",
"_____no_output_____"
],
[
"values = list(total.values())\nkeys = list(total.keys())\nlegend_keys = copy.copy(keys)\n\nfor i in range(len(keys)):\n legend_keys[i] = str(legend_keys[i]) + \" \" + str(values[i]) + \" \" + 'times'\n\n# Remove wedgeprops to make pie\nwedges, texts = plt.pie(values, textprops=dict(color=\"w\"), wedgeprops=dict(width=0.5))\nplt.legend(wedges, legend_keys,\n title=\"Fault Codes\",\n loc=\"center left\",\n bbox_to_anchor=(1, 0, 0.5, 1))\n\n#plt.title(\"Cumulative\")\n#plt.show()\nplt.savefig('cumulative pie.png')",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
c50eea9f40548becb7108babdf7371660cc50da8
| 479,292 |
ipynb
|
Jupyter Notebook
|
source/examples/basics/geocoding/geocoding_level_state_us.ipynb
|
JetBrains/lets-plot-docs
|
73583bce5308d34b341d9f8a7249ccb34a95f504
|
[
"MIT"
] | 2 |
2021-06-02T10:24:24.000Z
|
2021-11-08T09:50:22.000Z
|
source/examples/basics/geocoding/geocoding_level_state_us.ipynb
|
JetBrains/lets-plot-docs
|
73583bce5308d34b341d9f8a7249ccb34a95f504
|
[
"MIT"
] | 13 |
2021-05-25T19:49:50.000Z
|
2022-03-22T12:30:29.000Z
|
source/examples/basics/geocoding/geocoding_level_state_us.ipynb
|
JetBrains/lets-plot-docs
|
73583bce5308d34b341d9f8a7249ccb34a95f504
|
[
"MIT"
] | 4 |
2021-01-19T12:26:21.000Z
|
2022-03-19T07:47:52.000Z
| 2,648.022099 | 472,716 | 0.729676 |
[
[
[
"from lets_plot.geo_data import *\nfrom lets_plot import *\nLetsPlot.setup_html()",
"The geodata is provided by © OpenStreetMap contributors and is made available here under the Open Database License (ODbL).\n"
],
[
"gdf = geocode_states().countries('US-48').inc_res().get_boundaries()\n\nggplot() + \\\n geom_map(data=gdf, color='white', fill='#636363', \\\n tooltips=layer_tooltips().line('@{found name}')) + \\\n theme(axis_title='blank', axis_text='blank', axis_ticks='blank', axis_line='blank') + \\\n ggsize(800, 600) + ggtitle('US States') + \\\n theme(panel_grid='blank')",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code"
]
] |
c50eebfb232807d3c3a923dec4dbeef8a4308513
| 14,382 |
ipynb
|
Jupyter Notebook
|
Writeup.ipynb
|
robingh42/wikipedia_pathways
|
cfe8bf0eecc0a5d4c546e27f65afa1e710dd42cb
|
[
"MIT"
] | null | null | null |
Writeup.ipynb
|
robingh42/wikipedia_pathways
|
cfe8bf0eecc0a5d4c546e27f65afa1e710dd42cb
|
[
"MIT"
] | null | null | null |
Writeup.ipynb
|
robingh42/wikipedia_pathways
|
cfe8bf0eecc0a5d4c546e27f65afa1e710dd42cb
|
[
"MIT"
] | null | null | null | 40.512676 | 557 | 0.635586 |
[
[
[
"# Six Degrees Of Wikipedia\n**A Project by Robin Graham-Hayes**",
"_____no_output_____"
]
],
[
[
"# Run this code to import the nessisary files and libraries\nimport helpers\nimport pathways\nimport wikipedia as wiki",
"_____no_output_____"
]
],
[
[
"## Introduction",
"_____no_output_____"
],
[
"In the Wikipedia game the goal is to get from one Wikipedia page to another in the shortest amount of clicks. You can only use the links in the main article. Talk: pages and other meta Wikipedia pages are un interesting and outside of the heart of the game. \n\nMy project is a deviation of this game. Its goal is to find all the valid pathways between the two pages. It can realy be interesting to find unintuitive pathways between pages. A pathway such as: `| Toothpaste ---> Apricot ---> Alexander the Great |` demostrates how pathways can be unintuitive without the specific context the the articles give. Without this program a lot of these short weird pathways would be hard to find. ",
"_____no_output_____"
],
[
"## Methodology",
"_____no_output_____"
],
[
"#### The Wikipedia API\nThe first problem I had to solve was how to interface with wikipedia pages. I could use BeautifulSoup but I was able to find a nice Wikipedia API that made interfacing with wikipedia pretty simple. I used the bash command `pip install wikipedia` to install the API and ran some basic tests:",
"_____no_output_____"
]
],
[
[
"mars = wiki.page(\"Mars (planet)\")\nmars_links = mars.links\nprint(\"Here is a sample of the links:\", mars_links[20:25])",
"_____no_output_____"
]
],
[
[
"Now I have a basic way to get all the links in a page. In addition the Wikipedia API will throw errors if the page does not exist of if it is a disambiguation page which is useful for interacting with broken links and the alternitive format of a disambiguation page.",
"_____no_output_____"
],
[
"#### Depth First Searches\nThe next problem I had to tackle is how to actualy search through wikipedia pages. I knew I needed to specify a maximum depth to search, otherwise the program could end up thousands of pages deep in one pathway it needs to check. I also know that I dont know the precise amount of times I will have to iterate through a search. Following that a recursive depth fisrt search made the most sense for my purpose.\n\n*The Folowing graphic depicts the basic pattern of a depth first search:*",
"_____no_output_____"
],
[
" A \n / | \\ \n B E G \n /\\ | \\ \n C D F H ",
"_____no_output_____"
],
[
"The program starts by searching the first page. It will then go on to serch through the first link, and then through all the following links untill it reaches its max depth in the search. After that it will start going back through the previous branches that were not included in the first deep search. In the above graphic, the search pattern is depicted in alphebetical order.",
"_____no_output_____"
],
[
"#### The Algorithm\n\nTo create the search helpers `helper.get_pathway()` there are some checks that need to happen first. First we need to check if the file has the end page as a link in the article.",
"_____no_output_____"
]
],
[
[
"print(helpers.has_end(mars.title, wiki.page(\"The Moon\")))\nprint(helpers.has_end(mars.title, wiki.page(\"Salt\")))",
"_____no_output_____"
]
],
[
[
"Next we need to check if the max depth has been reached, since we are checking the links before hand this check will be one less than the given number.\n\nAfter those checks we start checking each link in the page recursivly to see if the end page present in that next page. During this step we also check wether the link we are about to search is in the existing trail and if it is we skip it to avoid entering a infinite loop.",
"_____no_output_____"
],
[
"#### Slow downs\n\nOnce I got the algorithm running, I ran into the problem that accessing thousands and thousands of webpages end up taking a lot of time. To reduce the time I had to limit the maximum depth of my tests to four chains deep. I also rand into the realization the the wikipedia API was pulling all of the links including all of the hundreds of links in the navigation boxes at the bottom of the page. These are typicaly not included in the wikipedia game and it is both uninteresting to look at those paths and significantly slow down the searches.",
"_____no_output_____"
],
[
"#### Getting the links\n\n",
"_____no_output_____"
],
[
"I quickly learned that even with a more robust Wikipedia API there was no way to distinguish links in the navigation boxes from the main links in the article. I slowly developed a way to parse the HTML retrived by the wikipedia API with BeautifulSoup. \nDoing this took a few steps, and is mostly contained in the function `helpers.parse_links()` I had to first seperate the navigation box from the main part of the article. I was able to do this using string indexing, where I found the first index of the navigation box, and cut off all the lines after that index. The next step was to pull only the wikipedia links from the page. This was fairly simple with BeautifulSoup as the search criteria was fairly limited, all the wiki links on the page had a title and no class except disambiguation pages.\n\nBeautifulSoup allows me to use the following functions to find regular links:\n\n`content_soup.find_all(\n name=\"a\", attrs={\"class\": None, \"title\": re.compile(\".\")})`\n\nAs well as finding all the disambiguation links:\n\n`content_soup.find_all(\n name=\"a\", attrs={\"class\": \"mw-disambig\", \"title\": re.compile(\".\")})`\n \nWe then only have to sort out all the meta pages. The function `helpers.get_titles()` which takes the ResultsSet object from `helpers.parse_links()` and only returns the titles that are from articles that are regular wikipedia pages.",
"_____no_output_____"
],
[
"All of this was then simplified into the function `helpers.get_links()` which parses the links of any page title given to it.",
"_____no_output_____"
]
],
[
[
"print(f\"The page {mars.title} has {len(mars_links)} links\")\nprint(f\"The page {mars.title} has {len(helpers.get_links(mars.title))} links\")",
"_____no_output_____"
]
],
[
[
"As you can see above this significantly reduces the number of links the program has to sort through.",
"_____no_output_____"
],
[
"#### Saving to files\n\nTo further save time I decided to save information localy. I created the function `helpers.save_links()` to save all the links to a file witht the title of the article and each link of the page listed on seperate lines of the file. The function `helpers.read_links()` was created to read the links from the file. Both of these functions were added to `helpers.get_links()` and significantly sped up the runtime of the search.\n",
"_____no_output_____"
],
[
"#### Making an Interactive Script\n",
"_____no_output_____"
],
[
"The result of all of these functions was `pathways.find_paths()` however I wanted to provide a way for any user to interact with the program and not need to know the ins and outs of how `pathways.find_paths()` works. I created the python script `Six_Degrees_of_Wiki.py` to do just that.\n\nBy running `python Six_Degrees_of_Wiki.py` in a bash terminal you will be prompted for the strarting article and if you would like to use the default arguments for the max steps and ending page. If you don't it will prompt you for those as well.",
"_____no_output_____"
],
[
"## Results",
"_____no_output_____"
],
[
"The results collected from the main function call are filterd through `helpers.plot_all_paths()` to format them in a more visualy and are then saved to a file. \n\nFor example the following function call produces a file containing the following text:",
"_____no_output_____"
]
],
[
[
"We found 4 paths from Toothpaste to Alexander the Great!\nThe shortest path(s) are 3 links long.\nThe first short path is: | Toothpaste ---> Egyptians ---> Alexander the Great |\n\n-------------------------------\n\n| Toothpaste ---> Egyptians ---> Alexander the Great |\n| Toothpaste ---> Ancient Greece ---> Alexander the Great |\n| Toothpaste ---> Ancient Rome ---> Alexander the Great |\n| Toothpaste ---> Apricot ---> Alexander the Great |",
"_____no_output_____"
]
],
[
[
"We can call `pathways.find_paths` to see the full out put of the function call:",
"_____no_output_____"
]
],
[
[
"# The following code will produce ~125 lines of text\n# to show you the process the search follows\npathways.find_paths(\"Toothpaste\", \"Alexander the Great\", 2)",
"_____no_output_____"
]
],
[
[
"If you use `python Six_Degrees_of_Wiki.py` to run the same search as the function call above does you would get the following terminal output:",
"_____no_output_____"
]
],
[
[
"Welcome to *Six Degrees of Wikipedia* \nOur goal is to find the links inbetween two pages on Wikipedia.\nAs the search runs, files will be saved localy to speed up later runs. \nThis might take some time to run initialy, sorry. \n\n-------------------------------\n\n What page would you like to start at: Toothpaste\n\nThe default end is Philosophy and the maximum number of chains is 3\n\n Would you like to keep this? (y/n): N\n\n What page would you like to end at: Alexander the Great\n\n How many links between articles do you want: 2\n\n-------------------------------\n\nNot a path: ['Toothpaste', 'Procter & Gamble'] \nNot a path: ['Toothpaste', 'Eucalyptus oil']\n**Pathway 1**: | Toothpaste ---> Egyptians ---> Alexander the Great | \nNot a path: ['Toothpaste', 'Toothbrush']\nNot a path: ['Toothpaste', 'Hydroxyapatite'] \nNot a path: ['Toothpaste', 'Tooth enamel']\nNot a path: ['Toothpaste', 'Colgate-Palmolive']\nNot a path: ['Toothpaste', 'Viscosity']\nNot a path: ['Toothpaste', 'Chloroform']\n**Pathway 2**: | Toothpaste ---> Ancient Greece ---> Alexander the Great |\nNot a path: ['Toothpaste', 'Xylitol']\nNot a path: ['Toothpaste', 'Newell Sill Jenkins']\n...",
"_____no_output_____"
]
],
[
[
"## Conclusion",
"_____no_output_____"
],
[
"I learned a lot through this project, I got alot more used to using an API to request data. I also learned a lot about BeautifulSoup, though I still have a lot to learn about using BeautifulSoup. Also learning the limits of programs and how a large program will actualy take time to run, as well as learning how to optimise a program to increase runtime efficiency. If you can avoid a try-except call without risking a error being thrown, that can be extreamly useful as they tend to add a lot of time when call repeatedly.",
"_____no_output_____"
],
[
"I think there are some really interesting paths between wikipedia pages that are for the most part unintuitive. Who knew you could get from Ketchup to The Bible with the path: `| Ketchup --> Salt --> Bible |` or from Toothpaste to Alexander the Great through apricot: `| Toothpaste ---> Apricot ---> Alexander the Great |`\n\nI think this project highlights how important context is. If you are just told something or given some information without context how would you know what to do with it. If I was playing the wikipedia game I would never click on Apricot to get to Alexander the Great. Things are not ment to existt in this world without any context. A student is not defined by their SAT scores, you need to consider the bigger picture.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"raw",
"markdown",
"code",
"markdown",
"raw",
"markdown"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"raw"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"raw"
],
[
"markdown",
"markdown",
"markdown"
]
] |
c50f0ea7126a597f3d363551dd8d5f6679beb3a7
| 159,787 |
ipynb
|
Jupyter Notebook
|
old-notes/old-ai/exam/k-means/3-final-results-with-plot.ipynb
|
mithi/algorithm-playground
|
ee35df7e343544b56c145b3912eca263c566d819
|
[
"MIT"
] | 85 |
2017-12-19T19:51:51.000Z
|
2021-05-26T20:00:39.000Z
|
old-notes/old-ai/exam/k-means/3-final-results-with-plot.ipynb
|
mithi/algorithm-playground
|
ee35df7e343544b56c145b3912eca263c566d819
|
[
"MIT"
] | 1 |
2019-01-02T07:00:40.000Z
|
2019-01-02T07:00:40.000Z
|
old-notes/old-ai/exam/k-means/3-final-results-with-plot.ipynb
|
mithi/algorithm-playground
|
ee35df7e343544b56c145b3912eca263c566d819
|
[
"MIT"
] | 34 |
2018-03-29T11:51:53.000Z
|
2020-11-17T08:24:51.000Z
| 581.043636 | 79,848 | 0.944151 |
[
[
[
" # `kmeans(data)`\n\n#### `def kmeans_more(data, nk=10, niter=100)`\n - `returns 3 items : best_k, vector of corresponding labels for each given sample, centroids for each cluster`\n\n#### `def kmeans(data, nk=10, niter=100)`\n - `returns 2 items: best_k, vector of corresponding labels for each given sample`\n\n# Requirements\n- where data is an MxN numpy array\n- This should return\n - an integer K, which should be programmatically identified\n - a vector of length M containing the cluster labels\n- `nk` is predefined as 10, which is the max number of clusters our program will test. So given a data set, the best k would be less than or equal to nk but greater than 1. \n- `niter` is the number of iterations before our algorithm \"gives up\", if it doesn't converge to a centroid after 100 iterations, it will just use the centroids it has computed the most recently\n- `kmeans_more()` is just `kmeans` but also returns the set of centroids. This is useful for visualization or plotting purposes. ",
"_____no_output_____"
]
],
[
[
"# x_kmeans returns error per k\n# kmeans returns k and data labels\nfrom KMeans import kmeans, kmeans_more, get_angle_between_3points\n\n# A list of four sets of 2d points\nfrom oldsamplesgen import gen_set1\n# helper plotting functions visualize what kmeans is doing\nfrom kmeansplottinghelper import initial_plots, colored_plots, eval_plots\n\nimport numpy as np\nimport matplotlib.pyplot as plt\n\n\n%matplotlib inline",
"_____no_output_____"
],
[
"# Load 4 data sets of 2d points with clusters [2, 3, 4, 5] respectively \npointset = gen_set1()",
"_____no_output_____"
],
[
"# let's get one of them to test for our k means \n\nsamples = pointset[3]\n# Make sure to shuffle the data, as they sorted by label\nnp.random.shuffle(samples)\n\nprint()\nprint(\"(M x N) row = M (number of samples) columns = N (number of features per sample)\")\nprint(\"Shape of array:\", samples.shape)\n\nprint()\nprint(\"Which means there are\", samples.shape[0], \"samples and\", samples.shape[1], \"features per sample\")\n\nprint()\nprint(\"Let's run our kmeans implementation\")\n\n#----------------------------------------------\nk, labels = kmeans(samples)\n#----------------------------------------------\n\nprint()\nprint()\nprint(\"Proposed number of clusters:\", k)\n\nprint(\"Labels shape:\")\nprint(labels.shape)\n\nprint(\"Print all the labels:\")\nprint(labels)",
"\n(M x N) row = M (number of samples) columns = N (number of features per sample)\nShape of array: (315, 2)\n\nWhich means there are 315 samples and 2 features per sample\n\nLet's run our kmeans implementation\n>>>>>>>>>>\n\nProposed number of clusters: 5\nLabels shape:\n(315,)\nPrint all the labels:\n[4 2 0 1 3 3 4 1 2 2 3 4 3 4 1 4 3 1 4 0 0 3 4 3 3 3 1 3 0 2 1 2 1 0 1 2 4\n 0 0 1 0 4 3 0 1 0 4 1 3 1 1 2 3 2 0 0 2 4 2 1 4 1 2 0 4 1 0 0 2 1 2 4 1 1\n 2 1 1 2 4 1 4 2 1 3 4 2 4 3 1 1 3 4 2 4 4 1 3 1 2 1 1 2 4 4 4 0 0 3 3 3 3\n 3 0 1 4 4 4 1 3 3 1 3 1 4 1 1 1 2 0 1 1 1 3 4 3 2 1 4 2 0 4 3 3 3 1 4 0 1\n 3 1 3 4 1 1 4 3 1 0 3 0 1 2 4 3 4 0 0 1 0 4 1 3 1 3 1 1 4 3 0 1 0 1 4 1 0\n 2 3 3 1 4 2 2 2 1 1 2 3 0 0 4 3 3 0 1 1 0 3 1 0 2 1 1 0 1 4 0 3 0 3 4 0 3\n 3 1 1 4 1 4 0 3 3 3 3 4 1 3 0 0 3 3 2 3 1 1 0 0 4 3 0 4 1 0 1 0 2 4 0 4 4\n 0 1 4 0 0 3 1 1 1 4 0 4 1 2 1 1 1 4 3 3 0 1 3 2 1 2 3 2 2 3 2 4 3 3 3 1 4\n 1 4 3 1 3 0 1 2 3 1 4 0 0 3 4 1 2 1 1]\n"
],
[
"# The synthetic dataset looks like this\n# They look like this\ninitial_plots(pointset)",
"_____no_output_____"
],
[
"# Plot a kmeans implementation given 4 sets of points\ndef plot_sample_kmeans_more(pointset):\n idata, ilabels, icentroids, inclusters = [], [], [], []\n\n for points in pointset:\n data = points\n np.random.shuffle(data)\n nclusters, labels, centroids = kmeans_more(data) \n idata.append(data)\n ilabels.append(labels)\n icentroids.append(centroids)\n inclusters.append(nclusters)\n\n colored_plots(idata, ilabels, icentroids, inclusters)",
"_____no_output_____"
],
[
"# returns the set the evaluated ks for each set\ndef test_final_kmeans(pointset):\n ks = []\n \n for i, points in enumerate(pointset):\n data = pointset[i]\n #Make sure to shuffle the data, as they sorted by label\n np.random.shuffle(data)\n k, _ = kmeans(data)\n ks.append(k)\n return ks",
"_____no_output_____"
],
[
"ks = test_final_kmeans(pointset)\nprint()\n# Should be [2, 3, 4, 5]\nprint(\"Proposed k for each set:\", ks)",
">>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>\nProposed k for each set: [2, 3, 4, 5]\n"
],
[
"plot_sample_kmeans_more(pointset)",
">>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>"
],
[
"# test if our \"compute angle between three points\" function is working\na = get_angle_between_3points([1, 2], [1, 1], [2, 1])\nb = get_angle_between_3points([1, 1], [2, 1], [3, 1])\n\nassert a, 90.0\nassert b, 180.0",
"_____no_output_____"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
c50f10aef9715e9487e18bd0268ed80188ad356b
| 56,602 |
ipynb
|
Jupyter Notebook
|
darkhistory/electrons/.ipynb_checkpoints/ion_heat_transfer_simplified-checkpoint.ipynb
|
wenzerq/DarkHistory
|
20aa55a46ee4981e09c3ae479f513c5df2949c71
|
[
"MIT"
] | 8 |
2019-04-23T02:09:40.000Z
|
2022-02-15T21:11:25.000Z
|
darkhistory/electrons/.ipynb_checkpoints/ion_heat_transfer_simplified-checkpoint.ipynb
|
wenzerq/DarkHistory
|
20aa55a46ee4981e09c3ae479f513c5df2949c71
|
[
"MIT"
] | 1 |
2017-09-29T17:36:45.000Z
|
2017-09-29T17:36:45.000Z
|
darkhistory/electrons/.ipynb_checkpoints/ion_heat_transfer_simplified-checkpoint.ipynb
|
wenzerq/DarkHistory
|
20aa55a46ee4981e09c3ae479f513c5df2949c71
|
[
"MIT"
] | 10 |
2019-04-23T16:29:58.000Z
|
2022-02-05T01:25:31.000Z
| 66.434272 | 15,418 | 0.766969 |
[
[
[
"%load_ext autoreload",
"_____no_output_____"
],
[
"%autoreload\n\nimport numpy as np\nimport matplotlib\nimport matplotlib.pyplot as plt\n\nimport math\nimport sys\nsys.path.append(\"..\")\nimport physics\nsys.path.append(\"../..\")\nfrom spec.spectrum import *\nimport spec.spectools as spectools\nimport xsecs",
"_____no_output_____"
],
[
"class Rates(object):\n \n def __init__(self, E_spec, n, den=[1,1,1], dNdW=np.zeros((2,1)), rates=np.zeros(4)):\n self.energy = E_spec.eng[n]\n self.n = n\n self.dNdE = E_spec.dNdE[n]\n self.den = den\n self.rates = rates\n self.v = np.sqrt(2*np.array([E_spec.eng[n]])/physics.me)*physics.c #units?\n self.mult = self.den*self.v\n self.dNdW = np.zeros((2, self.n ))\n \n def ion_dNdW_calc_H(self): #uses new integration method\n '''Fills *self.dNdW[0,:]* with the discretized singly differential xsec in rate form\n '''\n eng_temp = E_spec.eng[0:self.n]\n ion_s_rates = xsecs.ionize_s_cs_H_2(self.energy, eng_temp) #possible problem with np type\n self.dNdW[0] = ion_s_rates *self.mult[0] #dNdE? ;also, [0,:]?\n return self.dNdW\n \n def ion_rate_calc(self):\n '''Fills *self.rate[1:3]* vector by calculating total xsec and then converting to rate\n '''\n ion_rates = xsecs.ionize_cs(self.energy*np.ones(3),np.array([1,2,3]))*self.mult\n self.rates[1:4] = ion_rates\n return self.rates\n \n def heat_rate_calc(self, x_e, rs):\n '''Fills *self.rate[0]* vector with fraction going to heating\n x_e and rs...\n '''\n dE = xsecs.heating_dE(self.energy, x_e, rs, nH=physics.nH)\n delta_dNdE = np.zeros(len(E_spec.dNdE))\n np.put(delta_dNdE, self.n, self.dNdE)\n delta = Spectrum(E_spec.eng, delta_dNdE, rs)\n shift_delta_eng = E_spec.eng+dE\n delta.shift_eng(shift_delta_eng)\n delta.rebin(E_spec.eng)\n \n heating_frac = delta.dNdE[self.n]/self.dNdE\n self.rates[0] = 1-heating_frac #units? \n return(self.rates)\n \n def E_loss(self):\n '''loss fraction\n '''\n E_loss_ion=13.6*self.rates[1]\n E_loss_heat=(E_spec.eng[self.n]-E_spec.eng[self.n-1])*self.rates[0]\n E_frac = E_loss_ion/E_loss_heat\n return(E_frac)\n \n def ion_int_calc(self):\n '''gives total ionization rate\n '''\n bin_width = get_log_bin_width(E_spec.eng[0:self.n])\n integ = 0\n for i in range(self.n):\n integ += self.dNdW[0,i-1]*bin_width[i]*E_spec.eng[i]\n return integ\n \n ",
"_____no_output_____"
],
[
"def electron_low_e(E_spec, rs, ion_frac=[0.01,0.01,0.01], den=[1,1,1], dt=1 ,all_outputs=False):\n \n N = len(E_spec.eng)\n den[0]=(physics.nH*(1-ion_frac[0]))*(rs)**3 #units?\n \n R = np.zeros((2,N))\n R[1,0] = 1\n R[1,1] = 1\n R[1,2] = 1\n \n for n in range(3,N):\n e_rates = Rates(E_spec, n, den)\n e_rates.ion_rate_calc()\n e_rates.heat_rate_calc(ion_frac[0], rs)\n e_rates.ion_dNdW_calc_H() \n \n delta_E_spec = np.ediff1d(E_spec.eng)[0:(n)] #bin widths\n \n discrete_dN_dEdt_i = e_rates.dNdW[0]\n \n h_init=np.zeros(n)\n h_init[n-2] = e_rates.rates[0]\n h_init[n-1] = 1 - e_rates.rates[0]\n discrete_dN_dEdt_h = h_init/delta_E_spec\n \n \n R_in = ((13.6*e_rates.rates[1]) + np.sum(E_spec.eng[0:n-1]*discrete_dN_dEdt_i[0:n-1]*R[0,0:n-1]*delta_E_spec[0:n-1]) \\\n + np.sum(E_spec.eng[0:n-1]*discrete_dN_dEdt_h[0:n-1]*R[0,0:n-1]*delta_E_spec[0:n-1])) \\\n /(e_rates.energy*(np.sum(discrete_dN_dEdt_i[0:n-1])+np.sum(discrete_dN_dEdt_h[0:n-1])))\n \n \n R_hn = ((e_rates.energy*e_rates.rates[0]-np.sum(E_spec.eng[0:n-1]*discrete_dN_dEdt_h[0:n-1]*delta_E_spec[0:n-1])) \\\n + np.sum(E_spec.eng[0:n-1]*discrete_dN_dEdt_i[0:n-1]*R[1,0:n-1]*delta_E_spec[0:n-1]) \\\n + np.sum(E_spec.eng[0:n-1]*discrete_dN_dEdt_h[0:n-1]*R[1,0:n-1]*delta_E_spec[0:n-1])) \\\n /(e_rates.energy*(np.sum(discrete_dN_dEdt_i[0:n-1])+np.sum(discrete_dN_dEdt_h[0:n-1])))\n \n\n R[0,n] = R_in/(R_in+R_hn)\n R[1,n] = R_hn/(R_in+R_hn)\n \n if n==100 or n == 325 or n == 400:\n print('energy')\n print(e_rates.energy)\n print('rs')\n print(rs)\n print('ion')\n print(13.6*e_rates.rates[1])\n print(np.sum(E_spec.eng[0:n-1]*discrete_dN_dEdt_i[0:n-1]*R[0,0:n-1]*delta_E_spec[0:n-1]))\n print(np.sum(E_spec.eng[0:n-1]*discrete_dN_dEdt_h[0:n-1]*R[0,0:n-1]*delta_E_spec[0:n-1]))\n print('heat')\n print(e_rates.energy*e_rates.rates[0]-np.sum(E_spec.eng[0:n-1]*discrete_dN_dEdt_h[0:n-1]*delta_E_spec[0:n-1]))\n print(np.sum(E_spec.eng[0:n-1]*discrete_dN_dEdt_i[0:n-1]*R[1,0:n-1]*delta_E_spec[0:n-1]))\n print(np.sum(E_spec.eng[0:n-1]*discrete_dN_dEdt_h[0:n-1]*R[1,0:n-1]*delta_E_spec[0:n-1]))\n print('denominator')\n print((e_rates.energy*(np.sum(discrete_dN_dEdt_i[0:n-1])+np.sum(discrete_dN_dEdt_h[0:n-1]))))\n \n #R[0,n] = R_in\n #R[1,n] = R_hn\n \n #print(n, e_rates.energy,R_in,R_hn)\n #print(e_rates.energy*e_rates.rates[0], np.sum(E_spec.eng[0:n-1]*discrete_dN_dEdt_h[0:n-1]*delta_E_spec[0:n-1]), np.sum(E_spec.eng[0:n-1]*discrete_dN_dEdt_i[0:n-1]*R[1,0:n-1]*delta_E_spec[0:n-1]),np.sum(E_spec.eng[0:n-1]*discrete_dN_dEdt_h[0:n-1]*R[1,0:n-1]*delta_E_spec[0:n-1]) )\n \n \n return R\n ",
"_____no_output_____"
]
],
[
[
"<h1>Testing specific spectra:</h1>",
"_____no_output_____"
]
],
[
[
"eng1 = np.logspace(-4.,4.,num = 500)\n#dNdE1 = np.logspace(0.,5.,num = 500)\ndNdE1 = np.ones(500)\nrs=1\nE_spec = Spectrum(eng1,dNdE1,rs)",
"_____no_output_____"
],
[
"%%capture\nresults_ion_frac_0 = electron_low_e(E_spec,10**3)",
"_____no_output_____"
],
[
"%%capture\nresults_ion_frac_1 = electron_low_e(E_spec,10**1)",
"_____no_output_____"
],
[
"x=np.linspace(10,100,num = 10)\ny=np.zeros((10,1))\n\nfor k,rs in enumerate(x):\n y[k] = electron_low_e(E_spec, rs)[1,400]\n print(k)\n\nplt.plot(x,y, 'r')\nplt.show()\n#heat rate versus redshift",
"energy\n0.00401057288086\nrs\n10.0\nion\n0.0\n0.0\n0.0\nheat\n4.49069931844e-10\n0.0\n5.86068195652e-09\ndenominator\n4.50426751939e-05\nenergy\n16.2338243161\nrs\n10.0\nion\n9.29859673896e-12\n0.0\n5.69901148766e-12\nheat\n7.09281494106e-12\n0.0\n8.68672673887e-11\ndenominator\n1.75757715572e-10\nenergy\n258.71740786\nrs\n10.0\nion\n8.48832706949e-11\n3.63816528721e-13\n2.67247198591e-11\nheat\n2.09332920564e-12\n8.26313923059e-15\n5.94714898379e-13\ndenominator\n3.36131452709e-12\n0\nenergy\n0.00401057288086\nrs\n20.0\nion\n0.0\n0.0\n0.0\nheat\n3.59255944252e-09\n0.0\n4.68854554925e-08\ndenominator\n0.000360341400325\nenergy\n16.2338243161\nrs\n20.0\nion\n7.43887739116e-11\n0.0\n4.5338100279e-11\nheat\n5.64811006507e-11\n0.0\n6.91780428474e-10\ndenominator\n1.39958384729e-09\nenergy\n258.71740786\nrs\n20.0\nion\n6.79066165559e-10\n2.90988872684e-12\n1.87190242516e-10\nheat\n1.46512601727e-11\n6.67486167714e-14\n4.01912165084e-12\ndenominator\n2.36325062979e-11\n1\nenergy\n0.00401057288086\nrs\n30.0\nion\n0.0\n0.0\n0.0\nheat\n1.21248881232e-08\n0.0\n1.58238412349e-07\ndenominator\n0.00121615222657\nenergy\n16.2338243161\nrs\n30.0\nion\n2.51062111952e-10\n0.0\n1.53041704116e-10\nheat\n1.90590267703e-10\n0.0\n2.33429682338e-09\ndenominator\n4.72276667867e-09\nenergy\n258.71740786\nrs\n30.0\nion\n2.29184830876e-09\n9.82093322274e-12\n6.15055300374e-10\nheat\n4.81445274629e-11\n2.25217811943e-13\n1.32650199122e-11\ndenominator\n7.77329881949e-11\n2\nenergy\n0.00401057288086\nrs\n40.0\nion\n0.0\n0.0\n0.0\nheat\n2.87404755528e-08\n0.0\n3.75083644106e-07\ndenominator\n0.00288273120387\nenergy\n16.2338243161\nrs\n40.0\nion\n5.95110191293e-10\n0.0\n3.62741216396e-10\nheat\n4.51715530321e-10\n0.0\n5.53246768161e-09\ndenominator\n1.11933682688e-08\nenergy\n258.71740786\nrs\n40.0\nion\n5.43252932447e-09\n2.32789566952e-11\n1.44409018846e-09\nheat\n1.13039777104e-10\n5.34142053715e-13\n3.11592884442e-11\ndenominator\n1.82575816008e-10\n3\nenergy\n0.00401057288086\nrs\n50.0\nion\n0.0\n0.0\n0.0\nheat\n5.61337413101e-08\n0.0\n7.32585242342e-07\ndenominator\n0.00563033438215\nenergy\n16.2338243161\nrs\n50.0\nion\n1.16232459237e-09\n0.0\n7.08551262022e-10\nheat\n8.82349000658e-10\n0.0\n1.08067306592e-08\ndenominator\n2.18643297451e-08\nenergy\n258.71740786\nrs\n50.0\nion\n1.06104088369e-08\n4.54684719348e-11\n2.82140875245e-09\nheat\n2.20848275462e-10\n1.04148655916e-12\n6.08182935948e-11\ndenominator\n3.56698282668e-10\n4\nenergy\n0.00401057288086\nrs\n60.0\nion\n0.0\n0.0\n0.0\nheat\n9.69991049822e-08\n0.0\n1.26590729874e-06\ndenominator\n0.00972921781219\nenergy\n16.2338243161\nrs\n60.0\nion\n2.00849689561e-09\n0.0\n1.22432532447e-09\nheat\n1.52465415732e-09\n0.0\n1.8673495652e-08\ndenominator\n3.77804487999e-08\nenergy\n258.71740786\nrs\n60.0\nion\n1.83347864701e-08\n7.85691394847e-11\n4.88059175148e-09\nheat\n3.82032580029e-10\n1.80006879281e-12\n1.05205091751e-10\ndenominator\n6.17007086871e-10\n5\nenergy\n0.00401057288086\nrs\n70.0\nion\n0.0\n0.0\n0.0\nheat\n1.54030986152e-07\n0.0\n2.01021390495e-06\ndenominator\n0.0154496375443\nenergy\n16.2338243161\nrs\n70.0\nion\n3.18941868146e-09\n0.0\n1.94422231336e-09\nheat\n2.42112388209e-09\n0.0\n2.96531660756e-08\ndenominator\n5.99946856317e-08\nenergy\n258.71740786\nrs\n70.0\nion\n2.91149618484e-08\n1.2476629885e-10\n7.7555667274e-09\nheat\n6.07061381101e-10\n2.85702725766e-12\n1.67015993989e-10\ndenominator\n9.80418462954e-10\n6\nenergy\n0.00401057288086\nrs\n80.0\nion\n0.0\n0.0\n0.0\nheat\n2.29923804402e-07\n0.0\n3.00066915258e-06\ndenominator\n0.0230618496289\nenergy\n16.2338243161\nrs\n80.0\nion\n4.76088153035e-09\n0.0\n2.90214741095e-09\nheat\n3.61398591799e-09\n0.0\n4.42629388234e-08\ndenominator\n8.95534303845e-08\nenergy\n258.71740786\nrs\n80.0\nion\n4.34602345958e-08\n1.86236606384e-10\n1.15663287288e-08\nheat\n9.05366925705e-10\n4.26818360706e-12\n2.49353482937e-10\ndenominator\n1.4622371223e-09\n7\nenergy\n0.00401057288086\nrs\n90.0\nion\n0.0\n0.0\n0.0\nheat\n3.27371979312e-07\n0.0\n4.27243713323e-06\ndenominator\n0.0328361101159\nenergy\n16.2338243161\nrs\n90.0\nion\n6.7786770227e-09\n0.0\n4.13217189892e-09\nheat\n5.14573327507e-09\n0.0\n6.30233066129e-08\ndenominator\n1.27509646436e-07\nenergy\n258.71740786\nrs\n90.0\nion\n6.18799043366e-08\n2.65170476868e-10\n1.64738179466e-08\nheat\n1.28949079067e-09\n6.07560106852e-12\n3.54953864004e-10\ndenominator\n2.08260234515e-09\n8\nenergy\n0.00401057288086\nrs\n100.0\nion\n0.0\n0.0\n0.0\nheat\n4.49069930473e-07\n0.0\n5.86068193863e-06\ndenominator\n0.0450426750564\nenergy\n16.2338243161\nrs\n100.0\nion\n9.29859673896e-09\n0.0\n5.66825396306e-09\nheat\n7.05859485079e-09\n0.0\n8.64514284004e-08\ndenominator\n1.74909752536e-07\nenergy\n258.71740786\nrs\n100.0\nion\n8.48832706949e-08\n3.63745631566e-10\n2.25979394811e-08\nheat\n1.7688672673e-09\n8.33403638513e-12\n4.87036247184e-10\ndenominator\n2.85682202144e-09\n9\n"
],
[
"np.set_printoptions(threshold = np.nan)\n#print(np.transpose([E_spec.eng, results_ion_frac_0[0,:],results_ion_frac_0[1,:]]))\nplt.plot(E_spec.eng, results_ion_frac_0[0,:], 'r') #10**3\nplt.plot(E_spec.eng, results_ion_frac_1[0,:], 'r--') #10**1\nplt.plot(E_spec.eng, results_ion_frac_0[1,:], 'b')\nplt.plot(E_spec.eng, results_ion_frac_1[1,:], 'b--')\nplt.xscale('log')\nplt.yscale('log')\nplt.show()",
"_____no_output_____"
],
[
"#%%capture\nresults_ion_frac_t = electron_low_e(E_spec,1)",
"energy\n0.00401057288086\nrs\n1\nion\n0.0\n0.0\n0.0\nheat\n4.49071152785e-13\n0.0\n5.86069789068e-12\ndenominator\n4.5042797657e-08\n"
],
[
"plt.plot(E_spec.eng, results_ion_frac_t[0,:], 'r') #10**3\nplt.plot(E_spec.eng, results_ion_frac_t[1,:], 'b')\nplt.xscale('log')\nplt.yscale('log')\nplt.show()",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code"
] |
[
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
c50f137f0a95b8434c68f5b0636d64c55554eaec
| 4,823 |
ipynb
|
Jupyter Notebook
|
analysis/predictions/aparent_predict_native_pairs_data.ipynb
|
876lkj/APARENT
|
5c8b9c038a46b129b5e0e5ce1453c4725b62322e
|
[
"MIT"
] | 20 |
2019-04-23T20:35:23.000Z
|
2022-02-02T02:07:06.000Z
|
analysis/predictions/aparent_predict_native_pairs_data.ipynb
|
lafleur1/aparentGenomeTesting
|
e945d36e63d207d23b1508a4d1b7e5f63d66e304
|
[
"MIT"
] | 6 |
2019-10-14T16:35:00.000Z
|
2021-03-24T17:55:07.000Z
|
analysis/predictions/aparent_predict_native_pairs_data.ipynb
|
lafleur1/aparentGenomeTesting
|
e945d36e63d207d23b1508a4d1b7e5f63d66e304
|
[
"MIT"
] | 11 |
2019-06-10T08:53:57.000Z
|
2021-01-25T00:54:59.000Z
| 28.040698 | 261 | 0.594029 |
[
[
[
"from __future__ import print_function\nimport keras\nfrom keras.models import Sequential, Model, load_model\n\nimport tensorflow as tf\n\nimport pandas as pd\n\nimport os\nimport pickle\nimport numpy as np\n\nimport scipy.sparse as sp\nimport scipy.io as spio\n\nimport isolearn.io as isoio\nimport isolearn.keras as iso\n\nfrom aparent.data.aparent_data_native_pairs import load_data\n",
"Using TensorFlow backend.\n"
],
[
"#Load native pair-wise APA data\n\nfile_path = '../../data/prepared_data/apa_leslie_apadb_pair_data/'\nnative_gens = load_data(batch_size=1, file_path=file_path)\n",
"Pair-wise Native APA (APADB + Leslie) size = 29756\nTraining set size = 0\nValidation set size = 0\nTest set size = 29756\n"
],
[
"#Load APADB-tuned APARENT model\n\n#model_name = 'aparent_apadb_fitted'\nmodel_name = 'aparent_apadb_fitted_legacy_pasaligned_padded'\n\nsave_dir = os.path.join(os.getcwd(), '../../saved_models/legacy_models')\nmodel_path = os.path.join(save_dir, model_name + '.h5')\n\napadb_model = load_model(model_path)\n",
"WARNING:tensorflow:From /home/johli/anaconda3/envs/aparent/lib/python3.6/site-packages/tensorflow/python/framework/op_def_library.py:263: colocate_with (from tensorflow.python.framework.ops) is deprecated and will be removed in a future version.\nInstructions for updating:\nColocations handled automatically by placer.\nWARNING:tensorflow:From /home/johli/anaconda3/envs/aparent/lib/python3.6/site-packages/keras/backend/tensorflow_backend.py:3445: calling dropout (from tensorflow.python.ops.nn_ops) with keep_prob is deprecated and will be removed in a future version.\nInstructions for updating:\nPlease use `rate` instead of `keep_prob`. Rate should be set to `rate = 1 - keep_prob`.\n"
],
[
"#Predict from test data generator\n\niso_pred, cut_prox, cut_dist = apadb_model.predict_generator(native_gens['all'], workers=4, use_multiprocessing=True)\n\n#Calculate isoform logits\niso_pred = np.clip(np.ravel(iso_pred), 10**-5, 1. - 10**-5)\nlogodds_pred = np.log(iso_pred / (1. - iso_pred))\n",
"/home/johli/anaconda3/envs/aparent/lib/python3.6/site-packages/ipykernel_launcher.py:7: RuntimeWarning: divide by zero encountered in true_divide\n import sys\n"
],
[
"#Copy the test set dataframe and store isoform predictions\n\nnative_df = native_gens['all'].sources['df'].reset_index().copy()\n\nnative_df['iso_pred'] = iso_pred\nnative_df['logodds_pred'] = logodds_pred\n\nnative_df = native_df[['gene_id', 'iso_pred', 'logodds_pred']]\n",
"_____no_output_____"
],
[
"#Dump prediction dataframe and cut probability matrices\n\nisoio.dump({'native_df' : native_df, 'cut_prox' : sp.csr_matrix(cut_prox), 'cut_dist' : sp.csr_matrix(cut_dist)}, 'apa_leslie_apadb_pair_data/' + model_name + '_predictions')",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
c50f1644c7f351b68e8cf6a7538dbd8dbd35ed2b
| 29,060 |
ipynb
|
Jupyter Notebook
|
notebooks/pipelines/02-savor_data_pipelines_csv.ipynb
|
tobias-fyi/savor_data
|
b06cbef44f23048fa01f9ad7fb36b8fa715d63b2
|
[
"MIT"
] | null | null | null |
notebooks/pipelines/02-savor_data_pipelines_csv.ipynb
|
tobias-fyi/savor_data
|
b06cbef44f23048fa01f9ad7fb36b8fa715d63b2
|
[
"MIT"
] | null | null | null |
notebooks/pipelines/02-savor_data_pipelines_csv.ipynb
|
tobias-fyi/savor_data
|
b06cbef44f23048fa01f9ad7fb36b8fa715d63b2
|
[
"MIT"
] | null | null | null | 48.840336 | 4,933 | 0.501927 |
[
[
[
"# Savor Data\n\n> Taking advantage of my own big data.\n\nA data-driven project by [Tobias Reaper](https://github.com/tobias-fyi/)\n\n## Part 2: CSV Pipeline\n\nHere are the general steps in the pipeline:\n\n1. Load CSV data exported from Airtable's GUI\n2. Apply any needed transformations\n * Fixing column datatypes\n3. Insert into local Postgres database",
"_____no_output_____"
],
[
"---\n---\n\n## Load CSV Data",
"_____no_output_____"
]
],
[
[
"# === Some initial imports and config === #\n%load_ext autoreload\n%autoreload\n\nfrom os import environ\nfrom pprint import pprint\n\nimport pandas as pd\nimport janitor\n\npd.options.display.max_rows = 100\npd.options.display.max_columns = 50",
"The autoreload extension is already loaded. To reload it, use:\n %reload_ext autoreload\n"
],
[
"# === Set up environment variables === #\nfrom dotenv import load_dotenv\nfrom pathlib import Path\n\nenv_path = Path.cwd().parents[0] / \".env\"\nload_dotenv(dotenv_path=env_path)\n\n# === Airtable keys === #\nbase_key = environ.get(\"AIRTABLE_BASE_KEY\")\napi_key = environ.get(\"AIRTABLE_API_KEY\")",
"_____no_output_____"
],
[
"# === engage_log === #\ntable_name = \"engage_log\"\nairtable = Airtable(base_key, table_name, api_key=api_key)\nprint(airtable)",
"<Airtable table:engage_log>\n"
],
[
"# === Get all engagement records, sorted by time_in === #\nengage_log_records = airtable.get_all(sort=[\"time_in\"])\npprint(engage_log_records[0])",
"{'createdTime': '2020-09-07T20:14:40.000Z',\n 'fields': {'created': '2020-09-07T20:14:40.000Z',\n 'duration': {'specialValue': 'NaN'},\n 'id_num': 12344,\n 'mental': ['recVFgOi7povUvYjF', 'recPkaZr3nxxYyS37'],\n 'mental_note': 'Savor Data EDA and Viz - from CSV ::\\n\\n',\n 'modified': '2020-09-07T20:14:54.000Z',\n 'name': '12344-Sit-Cod',\n 'physical': ['recAEH30q7XSj0DCG'],\n 'project_location': ['recioKUrLNgcouGZW'],\n 'project_log': ['recpZPk1EhkKVds9P'],\n 'subloc': ['recSSXv8D15gISw4Y'],\n 'tags': ['recRjlzORPPL9M6qe',\n 'recuxV21zzIxFJUCc',\n 'rec6NWSQTIDn6rBRY',\n 'recbDqHY61m2rocNM']},\n 'id': 'receSicSHvRJm4DDI'}\n"
],
[
"# === mental === #\ntable_name = \"mental\"\nairtable = Airtable(base_key, table_name, api_key)\nmental_records = airtable.get_all()\n# Output is weird due to how airtable connects tables\n# pprint(mental_records[0])\n# We only need the `airtable_id` and `name`",
"_____no_output_____"
],
[
"# === physical === #\ntable_name = \"physical\"\nairtable = Airtable(base_key, table_name, api_key)\nphysical_records = airtable.get_all()\n# Output is weird due to how airtable connects tables\n# pprint(physical_records[0])\n# We only need the `airtable_id` and `name`",
"_____no_output_____"
]
],
[
[
"### Primary Keys\n\nI can't simply put the `fields` item into a dataframe and call it a day, because of the primary key / identifier that Airtable assigns on the back end of each record — e.g. `rec8GZsE62hEBtAst`. I'll need it to link records from different tables, but it's not included in the `fields`; it doesn't get brought into the dataframe without some extra processing.",
"_____no_output_____"
]
],
[
[
"def extract_and_concat_airtable_data(records: dict) -> pd.DataFrame:\n \"\"\"Extracts fields from the airtable data and concatenates them with airtable id.\n Uses pyjanitor to clean up column names.\n \"\"\"\n df = ( # Load and clean/fix names\n pd.DataFrame.from_records(records)\n .clean_names()\n .rename_column(\"id\", \"airtable_id\")\n )\n df2 = pd.concat( # Extract `fields` and concat to `airtable_id`\n [df[\"airtable_id\"], df[\"fields\"].apply(pd.Series)], axis=1\n )\n return df2",
"_____no_output_____"
],
[
"# === Use function to load and do initial transformations === #\ndf_engage_1 = \ndf_engage_1.shape",
"_____no_output_____"
],
[
"# === Write to CSV to save on API calls === #\ndf_engage_2.to_csv(\"../assets/data_/20-09-06-engage_log.csv\", index=False)",
"_____no_output_____"
],
[
"# === Test out loading from csv === #\ndf_engage_2 = pd.read_csv(\"../assets/data_/20-09-06-engage_log.csv\")",
"_____no_output_____"
],
[
"df_engage_2.head(3)",
"_____no_output_____"
],
[
"df_engage_2.tail()",
"_____no_output_____"
]
],
[
[
"---\n\n## Transform\n\n* Column data types\n * [ ] Date columns",
"_____no_output_____"
]
],
[
[
"df_engage_2.dtypes",
"_____no_output_____"
],
[
"pd.to_datetime(df_engage_2[\"time_in\"])",
"_____no_output_____"
],
[
"# === Automate datetime conversion in pipeline === #\ndatetime_cols = [\n \"time_in\",\n \"time_out\",\n \"created\",\n \"modified\",\n \"date\",\n]\n\ndef convert_datetime_cols(data: pd.DataFrame, dt_cols: list) -> pd.DataFrame:\n \"\"\"If datetime columns exist in dataframe, convert them to datetime.\"\"\"\n for col in dt_cols:\n if col in data.columns:\n data[col] = pd.to_datetime(data[col])\n return data",
"_____no_output_____"
],
[
"df_engage_2.iloc[12268, 8]",
"_____no_output_____"
]
],
[
[
"---\n\n## Postgres\n\nInserting the extracted data into a local Postgres instance using SQLAlchemy.\n\nThe SQLAlchemy `create_engine` function uses the following connection string format:\n\n dialect+driver://username:password@host:port/database",
"_____no_output_____"
]
],
[
[
"# === Set up connection to postgres db === #\nfrom sqlalchemy import create_engine\nfrom sqlalchemy.orm import sessionmaker\n\npg_user = environ.get(\"PG_USER\")\npg_pass = environ.get(\"PG_PASS\")\ndb_uri = f\"postgresql+psycopg2://{pg_user}:{pg_pass}@localhost:5432/savor\"\nengine = create_engine(uri, echo=True)\n\n# Instantiate new session\nSession = sessionmaker(bind=engine)\nsession = Session()",
"_____no_output_____"
]
],
[
[
"### Creating the tables\n\nThe first time this pipeline is run, the tables will have to be created in the Postgres database.\n\n",
"_____no_output_____"
]
],
[
[
"# === Define the declarative base class === #\nfrom sqlalchemy.ext.declarative import declarative_base\n\nBase = declarative_base()",
"_____no_output_____"
],
[
"# === Define the data model === #\nfrom sqlalchemy import Column, Integer, String \n\nclass EngageLog(Base):\n __tablename__ = \"engage_log\"\n \n ",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
c50f2e830446f25d460bb3f44b5bb6bcabff802d
| 50,741 |
ipynb
|
Jupyter Notebook
|
r/lecture1-eventlogs.ipynb
|
fmannhardt/course-processmining-intro
|
5857f18b7bb43546fb36161555429340dc254f10
|
[
"MIT"
] | 4 |
2021-04-22T22:04:49.000Z
|
2021-12-01T10:35:34.000Z
|
r/lecture1-eventlogs.ipynb
|
fmannhardt/course-processmining-intro
|
5857f18b7bb43546fb36161555429340dc254f10
|
[
"MIT"
] | null | null | null |
r/lecture1-eventlogs.ipynb
|
fmannhardt/course-processmining-intro
|
5857f18b7bb43546fb36161555429340dc254f10
|
[
"MIT"
] | 2 |
2021-04-21T08:08:12.000Z
|
2021-11-16T10:51:31.000Z
| 60.405952 | 14,078 | 0.71388 |
[
[
[
"# Applied Process Mining Module\n\nThis notebook is part of an Applied Process Mining module. The collection of notebooks is a *living document* and subject to change. \n\n# Lecture 1 - 'Event Logs and Process Visualization' (R / bupaR)",
"_____no_output_____"
],
[
"## Setup\n\n<img src=\"http://bupar.net/images/logo_text.PNG\" alt=\"bupaR\" style=\"width: 200px;\"/>\n\nIn this notebook, we are going to need the `tidyverse` and the `bupaR` packages.",
"_____no_output_____"
]
],
[
[
"## Perform the commented out commands below in a separate R session\n# install.packages(\"tidyverse\")\n# install.packages(\"bupaR\")",
"_____no_output_____"
],
[
"# for larger and readable plots\noptions(jupyter.plot_scale=1.25)",
"_____no_output_____"
],
[
"# the initial execution of these may give you warnings that you can safely ignore\nlibrary(tidyverse)\nlibrary(bupaR)\nlibrary(processanimateR)",
"-- \u001b[1mAttaching packages\u001b[22m ----------------------------------------------------------------------------------------------- tidyverse 1.3.1 --\n\n\u001b[32mv\u001b[39m \u001b[34mggplot2\u001b[39m 3.3.3 \u001b[32mv\u001b[39m \u001b[34mpurrr \u001b[39m 0.3.4\n\u001b[32mv\u001b[39m \u001b[34mtibble \u001b[39m 3.1.0 \u001b[32mv\u001b[39m \u001b[34mdplyr \u001b[39m 1.0.5\n\u001b[32mv\u001b[39m \u001b[34mtidyr \u001b[39m 1.1.3 \u001b[32mv\u001b[39m \u001b[34mstringr\u001b[39m 1.4.0\n\u001b[32mv\u001b[39m \u001b[34mreadr \u001b[39m 1.4.0 \u001b[32mv\u001b[39m \u001b[34mforcats\u001b[39m 0.5.1\n\n-- \u001b[1mConflicts\u001b[22m -------------------------------------------------------------------------------------------------- tidyverse_conflicts() --\n\u001b[31mx\u001b[39m \u001b[34mdplyr\u001b[39m::\u001b[32mfilter()\u001b[39m masks \u001b[34mstats\u001b[39m::filter()\n\u001b[31mx\u001b[39m \u001b[34mdplyr\u001b[39m::\u001b[32mlag()\u001b[39m masks \u001b[34mstats\u001b[39m::lag()\n\n\nAttaching package: 'bupaR'\n\n\nThe following object is masked from 'package:stats':\n\n filter\n\n\nThe following object is masked from 'package:utils':\n\n timestamp\n\n\n"
]
],
[
[
"## Event Logs\n\nThis part introduces event logs and their unique properties that provide the basis for any Process Mining method. Together with `bupaR` several event logs are distributed that can be loaded without further processing. \nIn this lecture we are going to make use of the following datasets:\n\n* Patients, a synthetically generated example event log in a hospital setting.\n* Sepsis, a real-life event log taken from a Dutch hospital. The event log is publicly available here: https://doi.org/10.4121/uuid:915d2bfb-7e84-49ad-a286-dc35f063a460 and has been used in many Process Mining related publications.",
"_____no_output_____"
],
[
"### Exploring Event Data\n\nLet us first explore the event data without any prior knowledge about event log structure or properties. We convert the `patients` event log below to a standard `tibble` (https://tibble.tidyverse.org/) and inspect the first rows.",
"_____no_output_____"
]
],
[
[
"patients %>%\n as_tibble() %>%\n head()",
"_____no_output_____"
]
],
[
[
"The most important ingredient of an event log is the timestamps column `time`. This allows us to establish a sequence of events.",
"_____no_output_____"
]
],
[
[
"patients %>% \n filter(time < '2017-01-31') %>% \n ggplot(aes(time, \"Event\")) + \n geom_point() + \n theme_bw()",
"_____no_output_____"
],
[
"patients %>%\n as_tibble() %>% \n distinct(handling)",
"_____no_output_____"
],
[
"patients %>%\n as_tibble() %>% \n distinct(patient) %>% \n head()",
"_____no_output_____"
],
[
"patients %>%\n as_tibble() %>% \n count(patient) %>% \n head()",
"_____no_output_____"
],
[
"patients %>% \n filter(time < '2017-01-31') %>% \n ggplot(aes(time, patient, color = handling)) + \n geom_point() + \n theme_bw()",
"_____no_output_____"
],
[
"patients %>% \n as_tibble() %>% \n arrange(patient, time) %>% \n head()",
"_____no_output_____"
]
],
[
[
"### Further resources",
"_____no_output_____"
],
[
"* [XES Standard](http://xes-standard.org/)\n* [Creating event logs from CSV files in bupaR](http://bupar.net/creating_eventlogs.html)\n* [Changing the case, activity notiions in bupaR](http://bupar.net/mapping.html)",
"_____no_output_____"
],
[
"### Reflection Questions\n\n* What could be the reason a column `.order` is included in this dataset?\n* How could the column `employee` be used?\n* What is the use of the column `handling_id` and in which situation is it required?",
"_____no_output_____"
],
[
"## Basic Process Visualization",
"_____no_output_____"
],
[
"### Set of Traces",
"_____no_output_____"
]
],
[
[
"patients %>% \n trace_explorer(coverage = 1.0, .abbreviate = T) # abbreviated here due to poor Jupyter notebook output scaling",
"_____no_output_____"
]
],
[
[
"### Dotted Chart",
"_____no_output_____"
]
],
[
[
"patients %>%\n filter(time < '2017-01-31') %>% \n dotted_chart(add_end_events = T)",
"_____no_output_____"
],
[
"patients %>% \n dotted_chart(\"relative\", add_end_events = T)",
"_____no_output_____"
]
],
[
[
"We can also use `plotly` to get an interactive visualization:",
"_____no_output_____"
]
],
[
[
"patients %>% \n plotly_dotted_chart(\"relative\", add_end_events = T)",
"_____no_output_____"
],
[
"sepsis %>% \n dotted_chart(\"relative_day\",\n sort = \"start_day\", \n units = \"hours\")",
"_____no_output_____"
]
],
[
[
"Check out other process visualization options using bupaR:\n\n* [Further Dotted Charts](http://bupar.net/dotted_chart.html)\n* [Exploring Time, Resources, Structuredness](http://bupar.net/exploring.html)",
"_____no_output_____"
],
[
"## Process Map Visualization",
"_____no_output_____"
]
],
[
[
"patients %>% \n precedence_matrix() %>% \n plot()",
"_____no_output_____"
],
[
"patients %>% \n process_map()",
"_____no_output_____"
],
[
"patients %>% \n process_map(type = performance(units = \"hours\"))",
"_____no_output_____"
]
],
[
[
"#### Challenge 1\nUse some other attribute to be shown in the `patients` dataset.",
"_____no_output_____"
]
],
[
[
"#patients %>% \n# process_map(type = custom(...))",
"_____no_output_____"
],
[
"patients %>% \n animate_process(mode = \"relative\")",
"_____no_output_____"
]
],
[
[
"#### Challenge 2\nReproduce the example shown on the lecture slides by animating some other attribute from the `traffic_fines` dataset.",
"_____no_output_____"
]
],
[
[
"traffic_fines %>% \n head()",
"_____no_output_____"
],
[
"traffic_fines %>% \n # WARNING: don't animate the full log in Jupyter (at least not on Firefox - it will really slow down your browser the library does not scale well)\n bupaR::sample_n(1000) %>%\n edeaR::filter_trace_frequency(percentage=0.95) %>%\n animate_process(mode = \"relative\")",
"_____no_output_____"
],
[
"# traffic_fines %>% ",
"_____no_output_____"
]
],
[
[
"## Real-life Processes",
"_____no_output_____"
]
],
[
[
"sepsis %>% \n precedence_matrix() %>% \n plot()",
"_____no_output_____"
]
],
[
[
"# Exercises - 1st Hands-on Session\n\nIn the first hands-on session, you are going to explore a real-life dataset (see the Assignment notebook) and apply what was presented in the lecture about event logs and basic process mining visualizations. The objective is to explore your dataset and as an event log and with the learned process mining visualizations in mind.\n\n* Analyse basic properties of the the process (business process or other process) that has generated it. \n * What are possible case notions / what is the or what are the case identifiers?\n * What are the activities? Are all activities on the same abstraction level? Can activities be derived from other data?\n * Can activities or actions be derived from other (non-activity) data?\n* Discovery a map of the process (or a sub-process) behind it.\n * Are there multiple processes that can be discovered?\n * What is the effect of taking a subset of the data?\n\n*Hint*: You may use/copy the code from this notebook to have a starting point. ",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
c50f48aa003e6f1b3200a5aee4553efa5bb5a066
| 24,071 |
ipynb
|
Jupyter Notebook
|
site/ja/tutorials/keras/regression.ipynb
|
ilyaspiridonov/docs-l10n
|
a061a44e40d25028d0a4458094e48ab717d3565c
|
[
"Apache-2.0"
] | 1 |
2021-09-23T09:56:29.000Z
|
2021-09-23T09:56:29.000Z
|
site/ja/tutorials/keras/regression.ipynb
|
ilyaspiridonov/docs-l10n
|
a061a44e40d25028d0a4458094e48ab717d3565c
|
[
"Apache-2.0"
] | null | null | null |
site/ja/tutorials/keras/regression.ipynb
|
ilyaspiridonov/docs-l10n
|
a061a44e40d25028d0a4458094e48ab717d3565c
|
[
"Apache-2.0"
] | 1 |
2020-06-23T07:43:49.000Z
|
2020-06-23T07:43:49.000Z
| 27.859954 | 429 | 0.493374 |
[
[
[
"##### Copyright 2018 The TensorFlow Authors.",
"_____no_output_____"
]
],
[
[
"#@title Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# https://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.",
"_____no_output_____"
],
[
"#@title MIT License\n#\n# Copyright (c) 2017 François Chollet\n#\n# Permission is hereby granted, free of charge, to any person obtaining a\n# copy of this software and associated documentation files (the \"Software\"),\n# to deal in the Software without restriction, including without limitation\n# the rights to use, copy, modify, merge, publish, distribute, sublicense,\n# and/or sell copies of the Software, and to permit persons to whom the\n# Software is furnished to do so, subject to the following conditions:\n#\n# The above copyright notice and this permission notice shall be included in\n# all copies or substantial portions of the Software.\n#\n# THE SOFTWARE IS PROVIDED \"AS IS\", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR\n# IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,\n# FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL\n# THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER\n# LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING\n# FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER\n# DEALINGS IN THE SOFTWARE.",
"_____no_output_____"
]
],
[
[
"# 回帰:燃費を予測する ",
"_____no_output_____"
],
[
"<table class=\"tfo-notebook-buttons\" align=\"left\">\n <td>\n <a target=\"_blank\" href=\"https://www.tensorflow.org/tutorials/keras/regression\"><img src=\"https://www.tensorflow.org/images/tf_logo_32px.png\" />View on TensorFlow.org</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://colab.research.google.com/github/tensorflow/docs-l10n/blob/master/site/ja/tutorials/keras/regression.ipynb\"><img src=\"https://www.tensorflow.org/images/colab_logo_32px.png\" />Run in Google Colab</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://github.com/tensorflow/docs-l10n/blob/master/site/ja/tutorials/keras/regression.ipynb\"><img src=\"https://www.tensorflow.org/images/GitHub-Mark-32px.png\" />View source on GitHub</a>\n </td>\n <td>\n <a href=\"https://storage.googleapis.com/tensorflow_docs/docs-l10n/site/ja/tutorials/keras/regression.ipynb\"><img src=\"https://www.tensorflow.org/images/download_logo_32px.png\" />Download notebook</a>\n </td>\n</table>",
"_____no_output_____"
],
[
"Note: これらのドキュメントは私たちTensorFlowコミュニティが翻訳したものです。コミュニティによる 翻訳は**ベストエフォート**であるため、この翻訳が正確であることや[英語の公式ドキュメント](https://www.tensorflow.org/?hl=en)の 最新の状態を反映したものであることを保証することはできません。 この翻訳の品質を向上させるためのご意見をお持ちの方は、GitHubリポジトリ[tensorflow/docs](https://github.com/tensorflow/docs)にプルリクエストをお送りください。 コミュニティによる翻訳やレビューに参加していただける方は、 [[email protected] メーリングリスト](https://groups.google.com/a/tensorflow.org/forum/#!forum/docs-ja)にご連絡ください。",
"_____no_output_____"
],
[
"回帰問題では、価格や確率といった連続的な値の出力を予測することが目的となります。これは、分類問題の目的が、(たとえば、写真にリンゴが写っているかオレンジが写っているかといった)離散的なラベルを予測することであるのとは対照的です。\n\nこのノートブックでは、古典的な[Auto MPG](https://archive.ics.uci.edu/ml/datasets/auto+mpg)データセットを使用し、1970年代後半から1980年台初めの自動車の燃費を予測するモデルを構築します。この目的のため、モデルにはこの時期の多数の自動車の仕様を読み込ませます。仕様には、気筒数、排気量、馬力、重量などが含まれています。\n\nこのサンプルでは`tf.keras` APIを使用しています。詳細は[このガイド](https://www.tensorflow.org/guide/keras)を参照してください。",
"_____no_output_____"
]
],
[
[
"# ペアプロットのためseabornを使用します\n!pip install seaborn",
"_____no_output_____"
],
[
"import pathlib\n\nimport matplotlib.pyplot as plt\nimport pandas as pd\nimport seaborn as sns\n\nimport tensorflow as tf\n\nfrom tensorflow import keras\nfrom tensorflow.keras import layers\n\nprint(tf.__version__)",
"_____no_output_____"
]
],
[
[
"## Auto MPG データセット\n\nこのデータセットは[UCI Machine Learning Repository](https://archive.ics.uci.edu/ml/)から入手可能です。",
"_____no_output_____"
],
[
"### データの取得\n\nまず、データセットをダウンロードします。",
"_____no_output_____"
]
],
[
[
"dataset_path = keras.utils.get_file(\"auto-mpg.data\", \"https://archive.ics.uci.edu/ml/machine-learning-databases/auto-mpg/auto-mpg.data\")\ndataset_path",
"_____no_output_____"
]
],
[
[
"pandasを使ってデータをインポートします。",
"_____no_output_____"
]
],
[
[
"column_names = ['MPG','Cylinders','Displacement','Horsepower','Weight',\n 'Acceleration', 'Model Year', 'Origin'] \nraw_dataset = pd.read_csv(dataset_path, names=column_names,\n na_values = \"?\", comment='\\t',\n sep=\" \", skipinitialspace=True)\n\ndataset = raw_dataset.copy()\ndataset.tail()",
"_____no_output_____"
]
],
[
[
"### データのクレンジング\n\nこのデータには、いくつか欠損値があります。",
"_____no_output_____"
]
],
[
[
"dataset.isna().sum()",
"_____no_output_____"
]
],
[
[
"この最初のチュートリアルでは簡単化のためこれらの行を削除します。",
"_____no_output_____"
]
],
[
[
"dataset = dataset.dropna()",
"_____no_output_____"
]
],
[
[
"`\"Origin\"`の列は数値ではなくカテゴリーです。このため、ワンホットエンコーディングを行います。",
"_____no_output_____"
]
],
[
[
"origin = dataset.pop('Origin')",
"_____no_output_____"
],
[
"dataset['USA'] = (origin == 1)*1.0\ndataset['Europe'] = (origin == 2)*1.0\ndataset['Japan'] = (origin == 3)*1.0\ndataset.tail()",
"_____no_output_____"
]
],
[
[
"### データを訓練用セットとテスト用セットに分割\n\nデータセットを訓練用セットとテスト用セットに分割しましょう。\n\nテスト用データセットは、作成したモデルの最終評価に使用します。",
"_____no_output_____"
]
],
[
[
"train_dataset = dataset.sample(frac=0.8,random_state=0)\ntest_dataset = dataset.drop(train_dataset.index)",
"_____no_output_____"
]
],
[
[
"### データの観察\n\n訓練用セットのいくつかの列の組み合わせの同時分布を見てみましょう。",
"_____no_output_____"
]
],
[
[
"sns.pairplot(train_dataset[[\"MPG\", \"Cylinders\", \"Displacement\", \"Weight\"]], diag_kind=\"kde\")",
"_____no_output_____"
]
],
[
[
"全体の統計値も見てみましょう。",
"_____no_output_____"
]
],
[
[
"train_stats = train_dataset.describe()\ntrain_stats.pop(\"MPG\")\ntrain_stats = train_stats.transpose()\ntrain_stats",
"_____no_output_____"
]
],
[
[
"### ラベルと特徴量の分離\n\nラベル、すなわち目的変数を特徴量から切り離しましょう。このラベルは、モデルに予測させたい数量です。",
"_____no_output_____"
]
],
[
[
"train_labels = train_dataset.pop('MPG')\ntest_labels = test_dataset.pop('MPG')",
"_____no_output_____"
]
],
[
[
"### データの正規化\n\n上の`train_stats`のブロックをもう一度見て、それぞれの特徴量の範囲がどれほど違っているかに注目してください。",
"_____no_output_____"
],
[
"スケールや値の範囲が異なる特徴量を正規化するのはよい習慣です。特徴量の正規化なしでもモデルは収束する**かもしれませんが**、モデルの訓練はより難しくなり、結果として得られたモデルも入力で使われる単位に依存することになります。\n\n注:(正規化に使用する)統計量は意図的に訓練用データセットだけを使って算出していますが、これらはテスト用データセットの正規化にも使うことになります。テスト用のデータセットを、モデルの訓練に使用した分布とおなじ分布に射影する必要があるのです。",
"_____no_output_____"
]
],
[
[
"def norm(x):\n return (x - train_stats['mean']) / train_stats['std']\nnormed_train_data = norm(train_dataset)\nnormed_test_data = norm(test_dataset)",
"_____no_output_____"
]
],
[
[
"この正規化したデータを使ってモデルを訓練することになります。\n\n注意:ここで入力の正規化に使った統計量(平均と標準偏差)は、さきほど実施したワンホットエンコーディングとともに、モデルに供給するほかのどんなデータにも適用する必要があります。テスト用データセットだけでなく、モデルをプロダクション環境で使用する際の生のデータについても同様です。",
"_____no_output_____"
],
[
"## モデル",
"_____no_output_____"
],
[
"### モデルの構築\n\nそれではモデルを構築しましょう。ここでは、2つの全結合の隠れ層と、1つの連続値を返す出力層からなる、`Sequential`モデルを使います。モデルを構築するステップは`build_model`という1つの関数の中に組み込みます。あとから2つ目のモデルを構築するためです。",
"_____no_output_____"
]
],
[
[
"def build_model():\n model = keras.Sequential([\n layers.Dense(64, activation='relu', input_shape=[len(train_dataset.keys())]),\n layers.Dense(64, activation='relu'),\n layers.Dense(1)\n ])\n\n optimizer = tf.keras.optimizers.RMSprop(0.001)\n\n model.compile(loss='mse',\n optimizer=optimizer,\n metrics=['mae', 'mse'])\n return model",
"_____no_output_____"
],
[
"model = build_model()",
"_____no_output_____"
]
],
[
[
"### モデルの検証\n\n`.summary`メソッドを使って、モデルの簡単な説明を表示します。",
"_____no_output_____"
]
],
[
[
"model.summary()",
"_____no_output_____"
]
],
[
[
"では、モデルを試してみましょう。訓練用データのうち`10`個のサンプルからなるバッチを取り出し、それを使って`model.predict`メソッドを呼び出します。",
"_____no_output_____"
]
],
[
[
"example_batch = normed_train_data[:10]\nexample_result = model.predict(example_batch)\nexample_result",
"_____no_output_____"
]
],
[
[
"うまく動作しているようです。予定どおりの型と形状の出力が得られています。",
"_____no_output_____"
],
[
"### モデルの訓練\n\nモデルを1000エポック訓練し、訓練と検証の正解率を`history`オブジェクトに記録します。",
"_____no_output_____"
]
],
[
[
"# エポックが終わるごとにドットを一つ出力することで進捗を表示\nclass PrintDot(keras.callbacks.Callback):\n def on_epoch_end(self, epoch, logs):\n if epoch % 100 == 0: print('')\n print('.', end='')\n\nEPOCHS = 1000\n\nhistory = model.fit(\n normed_train_data, train_labels,\n epochs=EPOCHS, validation_split = 0.2, verbose=0,\n callbacks=[PrintDot()])",
"_____no_output_____"
]
],
[
[
"`history`オブジェクトに保存された数値を使ってモデルの訓練の様子を可視化します。",
"_____no_output_____"
]
],
[
[
"hist = pd.DataFrame(history.history)\nhist['epoch'] = history.epoch\nhist.tail()",
"_____no_output_____"
],
[
"def plot_history(history):\n hist = pd.DataFrame(history.history)\n hist['epoch'] = history.epoch\n \n plt.figure()\n plt.xlabel('Epoch')\n plt.ylabel('Mean Abs Error [MPG]')\n plt.plot(hist['epoch'], hist['mae'],\n label='Train Error')\n plt.plot(hist['epoch'], hist['val_mae'],\n label = 'Val Error')\n plt.ylim([0,5])\n plt.legend()\n \n plt.figure()\n plt.xlabel('Epoch')\n plt.ylabel('Mean Square Error [$MPG^2$]')\n plt.plot(hist['epoch'], hist['mse'],\n label='Train Error')\n plt.plot(hist['epoch'], hist['val_mse'],\n label = 'Val Error')\n plt.ylim([0,20])\n plt.legend()\n plt.show()\n\n\nplot_history(history)",
"_____no_output_____"
]
],
[
[
"このグラフを見ると、検証エラーは100エポックを過ぎたあたりで改善が見られなくなり、むしろ悪化しているようです。検証スコアの改善が見られなくなったら自動的に訓練を停止するように、`model.fit`メソッド呼び出しを変更します。ここでは、エポック毎に訓練状態をチェックする*EarlyStopping*コールバックを使用します。設定したエポック数の間に改善が見られない場合、訓練を自動的に停止します。\n\nこのコールバックについての詳細は[ここ](https://www.tensorflow.org/api_docs/python/tf/keras/callbacks/EarlyStopping)を参照ください。",
"_____no_output_____"
]
],
[
[
"model = build_model()\n\n# patience は改善が見られるかを監視するエポック数を表すパラメーター\nearly_stop = keras.callbacks.EarlyStopping(monitor='val_loss', patience=10)\n\nhistory = model.fit(normed_train_data, train_labels, epochs=EPOCHS,\n validation_split = 0.2, verbose=0, callbacks=[early_stop, PrintDot()])\n\nplot_history(history)",
"_____no_output_____"
]
],
[
[
"検証用データセットでのグラフを見ると、平均誤差は+/- 2 MPG(マイル/ガロン)前後です。これはよい精度でしょうか?その判断はおまかせします。\n\nモデルの訓練に使用していない**テスト用**データセットを使って、モデルがどれくらい汎化できているか見てみましょう。これによって、モデルが実際の現場でどれくらい正確に予測できるかがわかります。",
"_____no_output_____"
]
],
[
[
"loss, mae, mse = model.evaluate(normed_test_data, test_labels, verbose=2)\n\nprint(\"Testing set Mean Abs Error: {:5.2f} MPG\".format(mae))",
"_____no_output_____"
]
],
[
[
"### モデルを使った予測\n\n最後に、テストデータを使ってMPG値を予測します。",
"_____no_output_____"
]
],
[
[
"test_predictions = model.predict(normed_test_data).flatten()\n\nplt.scatter(test_labels, test_predictions)\nplt.xlabel('True Values [MPG]')\nplt.ylabel('Predictions [MPG]')\nplt.axis('equal')\nplt.axis('square')\nplt.xlim([0,plt.xlim()[1]])\nplt.ylim([0,plt.ylim()[1]])\n_ = plt.plot([-100, 100], [-100, 100])\n",
"_____no_output_____"
]
],
[
[
"そこそこよい予測ができているように見えます。誤差の分布を見てみましょう。",
"_____no_output_____"
]
],
[
[
"error = test_predictions - test_labels\nplt.hist(error, bins = 25)\nplt.xlabel(\"Prediction Error [MPG]\")\n_ = plt.ylabel(\"Count\")",
"_____no_output_____"
]
],
[
[
"とても正規分布には見えませんが、サンプル数が非常に小さいからだと考えられます。",
"_____no_output_____"
],
[
"## 結論\n\nこのノートブックでは、回帰問題を扱うためのテクニックをいくつか紹介しました。\n\n* 平均二乗誤差(MSE: Mean Squared Error)は回帰問題に使われる一般的な損失関数です(分類問題には異なる損失関数が使われます)。\n* 同様に、回帰問題に使われる評価指標も分類問題とは異なります。回帰問題の一般的な評価指標は平均絶対誤差(MAE: Mean Absolute Error)です。\n* 入力数値特徴量の範囲が異なっている場合、特徴量ごとにおなじ範囲に正規化するべきです。\n* 訓練用データが多くない場合、過学習を避けるために少ない隠れ層をもつ小さいネットワークを使うというのがよい方策の1つです。\n* Early Stoppingは過学習を防止するための便利な手法の一つです。",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
c50f5db1b246aa9789731cee315ae1e9e0f30aae
| 644,141 |
ipynb
|
Jupyter Notebook
|
examples/methods_paper_plots/fig_6_max_R/make_fig_6.ipynb
|
TomWagg/COMPAS
|
b4898710f8c2fbe3bcca0730cd84f738bfe8fe96
|
[
"MIT"
] | null | null | null |
examples/methods_paper_plots/fig_6_max_R/make_fig_6.ipynb
|
TomWagg/COMPAS
|
b4898710f8c2fbe3bcca0730cd84f738bfe8fe96
|
[
"MIT"
] | null | null | null |
examples/methods_paper_plots/fig_6_max_R/make_fig_6.ipynb
|
TomWagg/COMPAS
|
b4898710f8c2fbe3bcca0730cd84f738bfe8fe96
|
[
"MIT"
] | null | null | null | 1,450.768018 | 332,420 | 0.954924 |
[
[
[
"<div style=\"text-align:center;\">\n <h1 style=\"font-size: 50px; margin: 0px; margin-bottom: 5px;\">Maximum Radial Extent Plot</h1>\n <h2 style=\"margin:0px; margin-bottom: 5px;\">COMPAS methods paper Figure 6</h2>\n <p style=\"text-align:center;\">A notebook for reproducing the maximum radial extent plot in the COMPAS methods paper.</p>\n</div>",
"_____no_output_____"
],
[
"<img src=\"https://compas.science/images/COMPAS_CasA.png\" style=\"width:50%; display:block; margin:auto; margin-bottom:20px\">",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport h5py as h5\nimport matplotlib.pyplot as plt\nimport astropy.constants as consts\nimport matplotlib\nimport astropy.units as u",
"_____no_output_____"
],
[
"# make the plots pretty\n%config InlineBackend.figure_format = 'retina'\nplt.rc('font', family='serif')\nfs = 24\n\nparams = {'legend.fontsize': fs,\n 'axes.labelsize': fs,\n 'xtick.labelsize':0.7*fs,\n 'ytick.labelsize':0.7*fs}\nplt.rcParams.update(params)",
"_____no_output_____"
]
],
[
[
"# Get the stellar types\nFirst we can import the stellar types array to use the same colour palette as the other plots.",
"_____no_output_____"
]
],
[
[
"import sys\nsys.path.append(\"../\")\nfrom stellar_types import stellar_types",
"_____no_output_____"
]
],
[
[
"# Data functions\nFunctions for getting variables from COMPAS detailed output files and gathering necessary information for maximum radius plots.",
"_____no_output_____"
]
],
[
[
"def get_detailed_output_vars(compas_file, variables):\n \"\"\" Return a list of variables from a COMPAS detailed output file \"\"\"\n with h5.File(compas_file, \"r\") as compas:\n var_list = [compas[variables[i]][...].squeeze() for i in range(len(variables))]\n return var_list",
"_____no_output_____"
],
[
"def gather_maximum_radius_data(folder, files, max_ev_time=13700.0):\n \"\"\" Get the range of masses, corresponding maximum radii as well as R_ZAMS \"\"\"\n n_masses = len(files)\n masses = np.zeros(n_masses)\n R_ZAMS = np.zeros(n_masses)\n maximum_radius = np.zeros((len(stellar_types) - 1, n_masses))\n for i in range(len(files)):\n file_path = \"{}/Detailed_Output/BSE_Detailed_Output_{}.h5\".format(folder, files[i])\n m_1, time, stellar_type, radius = get_detailed_output_vars(file_path, [\"Mass(1)\",\n \"Time\",\n \"Stellar_Type(1)\",\n \"Radius(1)\"])\n \n # change MS < 0.7 to just MS\n stellar_type[stellar_type == 0] = 1\n\n # store the mass\n masses[i] = m_1[0]\n \n # limit evolution time\n time_limit = time < max_ev_time\n stellar_type = stellar_type[time_limit]\n radius = radius[time_limit]\n \n # find maximum radius for each stellar type\n for st in range(1, len(stellar_types)):\n radius_at_st = radius[stellar_type == st]\n if len(radius_at_st) > 0:\n maximum_radius[st - 1][i] = np.max(radius_at_st)\n \n # store ZAMS radius\n if st == 1:\n R_ZAMS[i] = radius_at_st[0]\n \n return masses, maximum_radius, R_ZAMS",
"_____no_output_____"
],
[
"max_r_masses, maximum_radius_solar, R_ZAMS_solar = gather_maximum_radius_data(\"COMPAS_Output\", range(500))",
"_____no_output_____"
],
[
"max_r_masses, maximum_radius_lowz, R_ZAMS_lowz = gather_maximum_radius_data(\"COMPAS_Output\", range(500, 1000))",
"_____no_output_____"
]
],
[
[
"# Important transition masses\nThe following functions give the important transition masses from Hurley+2000",
"_____no_output_____"
]
],
[
[
"def m_hook(zeta):\n return 1.0185 + 0.16015 * zeta + 0.0892 * zeta**2\n\ndef m_helium_flash(zeta):\n return 1.995 + 0.25 * zeta + 0.087 * zeta**2\n\ndef m_FGB(Z):\n return (13.048 * (Z / 0.02)**0.06) / (1 + 0.0012 * (0.02 / Z)**1.27)",
"_____no_output_____"
]
],
[
[
"# Plotting function",
"_____no_output_____"
]
],
[
[
"def plot_max_R(masses, R_ZAMS, max_R, Z, mass_label_list,\n mass_label_loc=0.35, zloc=(0.98, 0.02), stellar_type_list=True,\n fig=None, ax=None, show=True):\n \"\"\" plot the maximum radius of each stellar type\n \n Parameters\n ----------\n masses : `float/array`\n Mass of each star\n\n R_ZAMS : `float/array`\n Radius at ZAMS (corresponding to each mass)\n\n max_R : `float/array`\n Maximum radius for stellar types 1-9 (corresponding to each mass)\n\n Z : `float`\n Metallicity of the stars\n\n mass_label_list : `tuple/array`\n A list of mass labels in the form (mass, label)\n\n mass_label_loc : `float`\n Y value of each mass label\n\n zloc : `tuple`\n Where to put the metallicity annotation\n\n stellar_type_list : `boolean`\n Whether to include the list of stellar types\n\n fig : `figure`, optional\n Matplotlib figure to, by default None\n \n ax : `axis`, optional\n Matplotlib axis to use, by default None\n \n show : `bool`, optional\n Whether to immediately show the plot or just return it, by default True\n \n \"\"\"\n if fig is None or ax is None:\n fig, ax = plt.subplots(1, figsize=(10, 8))\n\n # plot the ZAMS radius\n ax.plot(masses, R_ZAMS, color=\"grey\", label=\"ZAMS\", lw=2, zorder=10)\n\n # work out the top of the maximum radius plot\n top = np.maximum(np.maximum(max_R[3], max_R[4]), max_R[5])\n \n # fill the areas of case A,B,C mass transfer\n mask = masses > mass_label_list[0][0]\n ax.fill_between(masses[mask], np.zeros(len(R_ZAMS))[mask], top[mask], color=\"white\", zorder=2)\n ax.fill_between(masses[mask], R_ZAMS[mask], max_R[0][mask], color=stellar_types[1][\"colour\"], alpha=0.1, zorder=3)\n ax.fill_between(masses[mask], max_R[0][mask], max_R[3][mask], color=stellar_types[2][\"colour\"], alpha=0.1, zorder=3)\n ax.fill_between(masses[mask], max_R[3][mask], top[mask], color=stellar_types[4][\"colour\"], alpha=0.1, zorder=3) \n \n # plot each maximum radius track\n for st in range(1, 10):\n # for most of them only plot the line when it is above its predecessor\n if st < 7:\n mask = max_R[st - 1] > max_R[st - 2]\n # but for Helium stars plot everything\n else:\n mask = np.repeat(True, len(max_R[st - 1]))\n \n ax.plot(masses[mask], max_R[st - 1][mask], color=stellar_types[st][\"colour\"], label=stellar_types[st][\"short\"], lw=2, zorder=5)\n\n ax.set_xscale(\"log\")\n ax.set_yscale(\"log\")\n\n ax.set_ylabel(r\"$R_{\\rm max} \\ [\\rm R_{\\odot}]$\")\n ax.set_xlabel(r\"$M_{\\rm ZAMS} \\ [\\rm M_{\\odot}]$\")\n\n ax.set_xlim((np.min(masses), np.max(masses)))\n\n ax.annotate(r\"$Z = {}$\".format(Z), xy=zloc, xycoords=\"axes fraction\", ha=\"right\", va=\"bottom\", fontsize=0.7 * fs)\n\n # add mass limits\n for mass, label in mass_label_list:\n ax.axvline(mass, color=\"grey\", linestyle=\"dotted\", lw=1, zorder=1)\n ax.annotate(label, (mass * 0.98, mass_label_loc), fontsize=0.6 * fs, rotation=90, ha=\"right\", va=\"top\", color=\"grey\")\n\n # add a stellar type list\n if stellar_type_list:\n spacing = 0.05\n for st in range(1, 10):\n ax.annotate(stellar_types[st][\"short\"], xy=(1.01, spacing * (10 - st - 0.7)), xycoords=\"axes fraction\", color=stellar_types[st][\"colour\"], fontsize=0.7*fs, weight=\"bold\")\n ax.annotate(\"ZAMS\", xy=(1.01, spacing * (10 - 0.7)), xycoords=\"axes fraction\", color=\"grey\", fontsize=0.7*fs, weight=\"bold\")\n\n if show:\n plt.show()\n return fig, ax",
"_____no_output_____"
]
],
[
[
"# Recreate the plot from the paper",
"_____no_output_____"
]
],
[
[
"# create a two panel plot\nfig, axes = plt.subplots(1, 2, figsize=(20, 8))\nfig.subplots_adjust(wspace=0.0)\n\n# make the solar plot (with no stellar type list)\nZ = 0.0142\nzeta = np.log10(Z / 0.02)\nmass_label_list = [(0.92, r\"$M_{\\rm HT}$\"), (m_helium_flash(zeta), r\"$M_{\\rm HeF}$\"), (7.8, r\"$M_{\\rm SN}$\"), (m_FGB(Z), r\"$M_{\\rm FGB}$\"), (27, r\"$M_{\\rm WR}$\")]\nplot_max_R(max_r_masses, R_ZAMS_solar, maximum_radius_solar, Z, mass_label_list, mass_label_loc=6e3, zloc=(0.98, 0.02), fig=fig, ax=axes[0], show=False, stellar_type_list=False)\n\n# make the low Z plot (but with a stellar type list)\nZ = 0.001\nzeta = np.log10(Z / 0.02)\nmass_label_list = [(0.804, r\"$M_{\\rm HT}$\"), (m_helium_flash(zeta), r\"$M_{\\rm HeF}$\"), (6.3, r\"$M_{\\rm SN}$\"), (m_FGB(Z), r\"$M_{\\rm FGB}$\"), (37, r\"$M_{\\rm WR}$\")]\nplot_max_R(max_r_masses, R_ZAMS_lowz, maximum_radius_lowz, Z, mass_label_list, mass_label_loc=6e3, zloc=(0.98, 0.02), fig=fig, ax=axes[1], show=False)\n\n# make sure the scale is the same\nylims = (np.min([axes[0].get_ylim()[0], axes[1].get_ylim()[0]]),\n np.max([axes[0].get_ylim()[1], axes[1].get_ylim()[1]]))\nfor ax in axes:\n ax.set_ylim(ylims)\n \n# hide the yaxis stuff for the right panel\naxes[1].set_axisbelow(False)\naxes[1].tick_params(axis=\"y\", which=\"both\", left=True, right=True, direction=\"in\")\naxes[1].yaxis.set_ticklabels([])\naxes[1].set_ylabel(\"\")\n\nplt.savefig(\"maximum_radius.pdf\", format=\"pdf\", bbox_inches=\"tight\")\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"# Alternate version with split panels",
"_____no_output_____"
]
],
[
[
"fig, axes = plt.subplots(1, 2, figsize=(20, 8))\n\nZ = 0.0142\nzeta = np.log10(Z / 0.02)\nmass_label_list = [(0.92, r\"$M_{\\rm HT}$\"), (m_helium_flash(zeta), r\"$M_{\\rm HeF}$\"), (7.8, r\"$M_{\\rm SN}$\"), (m_FGB(Z), r\"$M_{\\rm FGB}$\"), (27, r\"$M_{\\rm WR}$\")]\nplot_max_R(max_r_masses, R_ZAMS_solar, maximum_radius_solar, Z, mass_label_list, mass_label_loc=6e3, zloc=(0.98, 0.02), fig=fig, ax=axes[0], show=False, stellar_type_list=False)\n\nZ = 0.001\nzeta = np.log10(Z / 0.02)\nmass_label_list = [(0.82, r\"$M_{\\rm HT}$\"), (m_helium_flash(zeta), r\"$M_{\\rm HeF}$\"), (6.3, r\"$M_{\\rm SN}$\"), (m_FGB(Z), r\"$M_{\\rm FGB}$\"), (37, r\"$M_{\\rm WR}$\")]\nplot_max_R(max_r_masses, R_ZAMS_lowz, maximum_radius_lowz, Z, mass_label_list, mass_label_loc=6e3, zloc=(0.98, 0.02), fig=fig, ax=axes[1], show=False, stellar_type_list=False)\n\n# make sure the scale is the same\nylims = (np.min([axes[0].get_ylim()[0], axes[1].get_ylim()[0]]),\n np.max([axes[0].get_ylim()[1], axes[1].get_ylim()[1]]))\nfor ax in axes:\n ax.set_ylim(ylims)\n\naxes[1].yaxis.set_tick_params(labelleft=True)\n\nplt.show()",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
c50f7038dd50844c03aab5ce830f1b1328a2a3da
| 102,769 |
ipynb
|
Jupyter Notebook
|
4-Largest palindrome product.ipynb
|
vss-py/TDD-ProjectEuler
|
cc8324f6a7ffa4b734739f5ebc31117f0fc75c4b
|
[
"MIT"
] | null | null | null |
4-Largest palindrome product.ipynb
|
vss-py/TDD-ProjectEuler
|
cc8324f6a7ffa4b734739f5ebc31117f0fc75c4b
|
[
"MIT"
] | null | null | null |
4-Largest palindrome product.ipynb
|
vss-py/TDD-ProjectEuler
|
cc8324f6a7ffa4b734739f5ebc31117f0fc75c4b
|
[
"MIT"
] | null | null | null | 24.527208 | 2,553 | 0.406718 |
[
[
[
"* # Largest palindrome product",
"_____no_output_____"
],
[
"A palindromic number reads the same both ways. The largest palindrome made from the product of two 2-digit numbers is 9009 = 91 × 99.\n\nFind the largest palindrome made from the product of two 3-digit numbers.",
"_____no_output_____"
],
[
"### Let's break the trouble in more steps, at least meanwhile:\n* Find palindromic numbers;\n* Find palindromic numbers from the product of two 3-digit numbers;\n* If the term is even, append this in one list;\n* Set the interval of the list;\n* Find the sum of all members of this list.",
"_____no_output_____"
]
],
[
[
"\"\"\"so, at the first moment we must check if the input number is a \npalindromic number or not:\"\"\"\nif __name__ == '__main__':\n assert palindromic(11) == True",
"_____no_output_____"
],
[
"def palindromic(n):\n return True\n\n\nif __name__ == '__main__':\n assert palindromic(11) == True\n assert palindromic(22) == True\n assert palindromic(33) == True\n assert palindromic(21) == False",
"_____no_output_____"
],
[
"def palindromic(n):\n if n == 21:\n return False\n return True\n\n\nif __name__ == '__main__':\n assert palindromic(11) == True\n assert palindromic(22) == True\n assert palindromic(33) == True\n assert palindromic(21) == False\n assert palindromic(12) == False\n assert palindromic(10) == False",
"_____no_output_____"
],
[
"def palindromic(n):\n if n == 21 or n == 12 or n == 10:\n return False\n return True\n\n\nif __name__ == '__main__':\n assert palindromic(11) == True\n assert palindromic(22) == True\n assert palindromic(33) == True\n assert palindromic(21) == False\n assert palindromic(12) == False\n assert palindromic(10) == False\n\"\"\"Now, I must to refactor this.\"\"\"",
"_____no_output_____"
],
[
"def palindromic(n):\n for m in n:\n if m[0] == m[1]:\n return True\n return False\n \"\"\"if n == 21 or n == 12 or n == 10:\n return False\n return True\"\"\"\n\n\nif __name__ == '__main__':\n assert palindromic(11) == True\n assert palindromic(22) == True\n assert palindromic(33) == True\n assert palindromic(21) == False\n assert palindromic(12) == False\n assert palindromic(10) == False",
"_____no_output_____"
],
[
"def palindromic(n):\n a = str(n)\n for m in a:\n if m[0] == m[1]:\n return True\n return False\n \"\"\"if n == 21 or n == 12 or n == 10:\n return False\n return True\"\"\"\n\n\nif __name__ == '__main__':\n assert palindromic(11) == True\n assert palindromic(22) == True\n assert palindromic(33) == True\n assert palindromic(21) == False\n assert palindromic(12) == False\n assert palindromic(10) == False",
"_____no_output_____"
],
[
"palindromic(11)",
"_____no_output_____"
],
[
"def palindromic(n):\n a = str(n)\n return(a)\n \"\"\"for m in a:\n if m[0] == m[1]:\n return True\n return False\"\"\"\n \"\"\"if n == 21 or n == 12 or n == 10:\n return False\n return True\"\"\"\n\n\nif __name__ == '__main__':\n assert type(palindromic(11)) == str\n assert palindromic(11) == '11'\n \"\"\"\n assert palindromic(11) == True\n assert palindromic(22) == True\n assert palindromic(33) == True\n assert palindromic(21) == False\n assert palindromic(12) == False\n assert palindromic(10) == False\"\"\"",
"_____no_output_____"
],
[
"#test:\ndef palindromic(n):\n a = str(n)\n return(a[0])\npalindromic(452)\n#Ok",
"_____no_output_____"
],
[
"#one more test attachment:\ndef palindromic(n):\n a = str(n)\n return(a[0:])\npalindromic(452)\n#Ok",
"_____no_output_____"
],
[
"def palindromic(n):\n a = str(n)\n if a[0] == a[-1]:\n return True\n return False\n \"\"\"for m in a:\n if m[0] == m[1]:\n return True\n return False\"\"\"\n \"\"\"if n == 21 or n == 12 or n == 10:\n return False\n return True\"\"\"\n\n\nif __name__ == '__main__':\n assert palindromic(11) == True\n assert palindromic(22) == True\n assert palindromic(33) == True\n assert palindromic(12) == False\n assert palindromic(21) == False\n assert palindromic(31) == False\n \"\"\"assert type(palindromic(11)) == str\n assert palindromic(11) == '11'\n assert palindromic(11) == True\n assert palindromic(22) == True\n assert palindromic(33) == True\n assert palindromic(21) == False\n assert palindromic(12) == False\n assert palindromic(10) == False\"\"\"",
"_____no_output_____"
],
[
"def palindromic(n):\n a = str(n)\n if a[0] == a[-1]:\n return True\n return False\n\n\nif __name__ == '__main__':\n assert palindromic(1) == False\n assert palindromic(2) == False\n assert palindromic(3) == False\n assert palindromic(11) == True\n assert palindromic(22) == True\n assert palindromic(33) == True\n assert palindromic(12) == False\n assert palindromic(21) == False\n assert palindromic(31) == False",
"_____no_output_____"
],
[
"def palindromic(n):\n a = str(n)\n if a == '1':\n return False\n if a[0] == a[-1]:\n return True\n return False\n\n\nif __name__ == '__main__':\n assert palindromic(1) == False\n assert palindromic(2) == False\n assert palindromic(3) == False\n assert palindromic(11) == True\n assert palindromic(22) == True\n assert palindromic(33) == True\n assert palindromic(12) == False\n assert palindromic(21) == False\n assert palindromic(31) == False",
"_____no_output_____"
],
[
"def palindromic(n):\n a = str(n)\n if a == '1' or a == '2' or a == '3':\n return False\n if a[0] == a[-1]:\n return True\n return False\n\n\nif __name__ == '__main__':\n assert palindromic(1) == False\n assert palindromic(2) == False\n assert palindromic(3) == False\n assert palindromic(11) == True\n assert palindromic(22) == True\n assert palindromic(33) == True\n assert palindromic(12) == False\n assert palindromic(21) == False\n assert palindromic(31) == False\n\"\"\"Let's refactor this\"\"\"",
"_____no_output_____"
],
[
"def palindromic(n):\n a = str(n)\n if len(a) == 1:\n return False\n if a == '1' or a == '2' or a == '3':\n return False\n if a[0] == a[-1]:\n return True\n return False\n\n\nif __name__ == '__main__':\n assert palindromic(1) == False\n assert palindromic(2) == False\n assert palindromic(3) == False\n assert palindromic(4) == False\n assert palindromic(5) == False\n assert palindromic(0) == False\n assert palindromic(11) == True\n assert palindromic(22) == True\n assert palindromic(33) == True\n assert palindromic(12) == False\n assert palindromic(21) == False\n assert palindromic(31) == False",
"_____no_output_____"
],
[
"def palindromic(n):\n a = str(n)\n if len(a) == 1:\n return False\n if a[0] == a[-1]:\n return True\n return False\n\n\nif __name__ == '__main__':\n assert palindromic(1) == False\n assert palindromic(2) == False\n assert palindromic(3) == False\n assert palindromic(4) == False\n assert palindromic(5) == False\n assert palindromic(0) == False\n assert palindromic(11) == True\n assert palindromic(22) == True\n assert palindromic(33) == True\n assert palindromic(12) == False\n assert palindromic(21) == False\n assert palindromic(31) == False\n \n\"\"\"\n\nFinally I checked if the number is a palindromic number!\n\n\"\"\"",
"_____no_output_____"
]
],
[
[
"### Now, I must to begin the next step. In this case, I must to find a number by the product of two 3-digit numbers",
"_____no_output_____"
]
],
[
[
"\"\"\"To get all the possible 3-digit numbers:\"\"\"\nfor i in range(100, 999):\n print(i)",
"100\n101\n102\n103\n104\n105\n106\n107\n108\n109\n110\n111\n112\n113\n114\n115\n116\n117\n118\n119\n120\n121\n122\n123\n124\n125\n126\n127\n128\n129\n130\n131\n132\n133\n134\n135\n136\n137\n138\n139\n140\n141\n142\n143\n144\n145\n146\n147\n148\n149\n150\n151\n152\n153\n154\n155\n156\n157\n158\n159\n160\n161\n162\n163\n164\n165\n166\n167\n168\n169\n170\n171\n172\n173\n174\n175\n176\n177\n178\n179\n180\n181\n182\n183\n184\n185\n186\n187\n188\n189\n190\n191\n192\n193\n194\n195\n196\n197\n198\n199\n200\n201\n202\n203\n204\n205\n206\n207\n208\n209\n210\n211\n212\n213\n214\n215\n216\n217\n218\n219\n220\n221\n222\n223\n224\n225\n226\n227\n228\n229\n230\n231\n232\n233\n234\n235\n236\n237\n238\n239\n240\n241\n242\n243\n244\n245\n246\n247\n248\n249\n250\n251\n252\n253\n254\n255\n256\n257\n258\n259\n260\n261\n262\n263\n264\n265\n266\n267\n268\n269\n270\n271\n272\n273\n274\n275\n276\n277\n278\n279\n280\n281\n282\n283\n284\n285\n286\n287\n288\n289\n290\n291\n292\n293\n294\n295\n296\n297\n298\n299\n300\n301\n302\n303\n304\n305\n306\n307\n308\n309\n310\n311\n312\n313\n314\n315\n316\n317\n318\n319\n320\n321\n322\n323\n324\n325\n326\n327\n328\n329\n330\n331\n332\n333\n334\n335\n336\n337\n338\n339\n340\n341\n342\n343\n344\n345\n346\n347\n348\n349\n350\n351\n352\n353\n354\n355\n356\n357\n358\n359\n360\n361\n362\n363\n364\n365\n366\n367\n368\n369\n370\n371\n372\n373\n374\n375\n376\n377\n378\n379\n380\n381\n382\n383\n384\n385\n386\n387\n388\n389\n390\n391\n392\n393\n394\n395\n396\n397\n398\n399\n400\n401\n402\n403\n404\n405\n406\n407\n408\n409\n410\n411\n412\n413\n414\n415\n416\n417\n418\n419\n420\n421\n422\n423\n424\n425\n426\n427\n428\n429\n430\n431\n432\n433\n434\n435\n436\n437\n438\n439\n440\n441\n442\n443\n444\n445\n446\n447\n448\n449\n450\n451\n452\n453\n454\n455\n456\n457\n458\n459\n460\n461\n462\n463\n464\n465\n466\n467\n468\n469\n470\n471\n472\n473\n474\n475\n476\n477\n478\n479\n480\n481\n482\n483\n484\n485\n486\n487\n488\n489\n490\n491\n492\n493\n494\n495\n496\n497\n498\n499\n500\n501\n502\n503\n504\n505\n506\n507\n508\n509\n510\n511\n512\n513\n514\n515\n516\n517\n518\n519\n520\n521\n522\n523\n524\n525\n526\n527\n528\n529\n530\n531\n532\n533\n534\n535\n536\n537\n538\n539\n540\n541\n542\n543\n544\n545\n546\n547\n548\n549\n550\n551\n552\n553\n554\n555\n556\n557\n558\n559\n560\n561\n562\n563\n564\n565\n566\n567\n568\n569\n570\n571\n572\n573\n574\n575\n576\n577\n578\n579\n580\n581\n582\n583\n584\n585\n586\n587\n588\n589\n590\n591\n592\n593\n594\n595\n596\n597\n598\n599\n600\n601\n602\n603\n604\n605\n606\n607\n608\n609\n610\n611\n612\n613\n614\n615\n616\n617\n618\n619\n620\n621\n622\n623\n624\n625\n626\n627\n628\n629\n630\n631\n632\n633\n634\n635\n636\n637\n638\n639\n640\n641\n642\n643\n644\n645\n646\n647\n648\n649\n650\n651\n652\n653\n654\n655\n656\n657\n658\n659\n660\n661\n662\n663\n664\n665\n666\n667\n668\n669\n670\n671\n672\n673\n674\n675\n676\n677\n678\n679\n680\n681\n682\n683\n684\n685\n686\n687\n688\n689\n690\n691\n692\n693\n694\n695\n696\n697\n698\n699\n700\n701\n702\n703\n704\n705\n706\n707\n708\n709\n710\n711\n712\n713\n714\n715\n716\n717\n718\n719\n720\n721\n722\n723\n724\n725\n726\n727\n728\n729\n730\n731\n732\n733\n734\n735\n736\n737\n738\n739\n740\n741\n742\n743\n744\n745\n746\n747\n748\n749\n750\n751\n752\n753\n754\n755\n756\n757\n758\n759\n760\n761\n762\n763\n764\n765\n766\n767\n768\n769\n770\n771\n772\n773\n774\n775\n776\n777\n778\n779\n780\n781\n782\n783\n784\n785\n786\n787\n788\n789\n790\n791\n792\n793\n794\n795\n796\n797\n798\n799\n800\n801\n802\n803\n804\n805\n806\n807\n808\n809\n810\n811\n812\n813\n814\n815\n816\n817\n818\n819\n820\n821\n822\n823\n824\n825\n826\n827\n828\n829\n830\n831\n832\n833\n834\n835\n836\n837\n838\n839\n840\n841\n842\n843\n844\n845\n846\n847\n848\n849\n850\n851\n852\n853\n854\n855\n856\n857\n858\n859\n860\n861\n862\n863\n864\n865\n866\n867\n868\n869\n870\n871\n872\n873\n874\n875\n876\n877\n878\n879\n880\n881\n882\n883\n884\n885\n886\n887\n888\n889\n890\n891\n892\n893\n894\n895\n896\n897\n898\n899\n900\n901\n902\n903\n904\n905\n906\n907\n908\n909\n910\n911\n912\n913\n914\n915\n916\n917\n918\n919\n920\n921\n922\n923\n924\n925\n926\n927\n928\n929\n930\n931\n932\n933\n934\n935\n936\n937\n938\n939\n940\n941\n942\n943\n944\n945\n946\n947\n948\n949\n950\n951\n952\n953\n954\n955\n956\n957\n958\n959\n960\n961\n962\n963\n964\n965\n966\n967\n968\n969\n970\n971\n972\n973\n974\n975\n976\n977\n978\n979\n980\n981\n982\n983\n984\n985\n986\n987\n988\n989\n990\n991\n992\n993\n994\n995\n996\n997\n998\n"
],
[
"for i in range(100, 999):\n for j in range(100, 999):\n if i == j:\n n = i * j\n print(n)\n",
"10000\n10201\n10404\n10609\n10816\n11025\n11236\n11449\n11664\n11881\n12100\n12321\n12544\n12769\n12996\n13225\n13456\n13689\n13924\n14161\n14400\n14641\n14884\n15129\n15376\n15625\n15876\n16129\n16384\n16641\n16900\n17161\n17424\n17689\n17956\n18225\n18496\n18769\n19044\n19321\n19600\n19881\n20164\n20449\n20736\n21025\n21316\n21609\n21904\n22201\n22500\n22801\n23104\n23409\n23716\n24025\n24336\n24649\n24964\n25281\n25600\n25921\n26244\n26569\n26896\n27225\n27556\n27889\n28224\n28561\n28900\n29241\n29584\n29929\n30276\n30625\n30976\n31329\n31684\n32041\n32400\n32761\n33124\n33489\n33856\n34225\n34596\n34969\n35344\n35721\n36100\n36481\n36864\n37249\n37636\n38025\n38416\n38809\n39204\n39601\n40000\n40401\n40804\n41209\n41616\n42025\n42436\n42849\n43264\n43681\n44100\n44521\n44944\n45369\n45796\n46225\n46656\n47089\n47524\n47961\n48400\n48841\n49284\n49729\n50176\n50625\n51076\n51529\n51984\n52441\n52900\n53361\n53824\n54289\n54756\n55225\n55696\n56169\n56644\n57121\n57600\n58081\n58564\n59049\n59536\n60025\n60516\n61009\n61504\n62001\n62500\n63001\n63504\n64009\n64516\n65025\n65536\n66049\n66564\n67081\n67600\n68121\n68644\n69169\n69696\n70225\n70756\n71289\n71824\n72361\n72900\n73441\n73984\n74529\n75076\n75625\n76176\n76729\n77284\n77841\n78400\n78961\n79524\n80089\n80656\n81225\n81796\n82369\n82944\n83521\n84100\n84681\n85264\n85849\n86436\n87025\n87616\n88209\n88804\n89401\n90000\n90601\n91204\n91809\n92416\n93025\n93636\n94249\n94864\n95481\n96100\n96721\n97344\n97969\n98596\n99225\n99856\n100489\n101124\n101761\n102400\n103041\n103684\n104329\n104976\n105625\n106276\n106929\n107584\n108241\n108900\n109561\n110224\n110889\n111556\n112225\n112896\n113569\n114244\n114921\n115600\n116281\n116964\n117649\n118336\n119025\n119716\n120409\n121104\n121801\n122500\n123201\n123904\n124609\n125316\n126025\n126736\n127449\n128164\n128881\n129600\n130321\n131044\n131769\n132496\n133225\n133956\n134689\n135424\n136161\n136900\n137641\n138384\n139129\n139876\n140625\n141376\n142129\n142884\n143641\n144400\n145161\n145924\n146689\n147456\n148225\n148996\n149769\n150544\n151321\n152100\n152881\n153664\n154449\n155236\n156025\n156816\n157609\n158404\n159201\n160000\n160801\n161604\n162409\n163216\n164025\n164836\n165649\n166464\n167281\n168100\n168921\n169744\n170569\n171396\n172225\n173056\n173889\n174724\n175561\n176400\n177241\n178084\n178929\n179776\n180625\n181476\n182329\n183184\n184041\n184900\n185761\n186624\n187489\n188356\n189225\n190096\n190969\n191844\n192721\n193600\n194481\n195364\n196249\n197136\n198025\n198916\n199809\n200704\n201601\n202500\n203401\n204304\n205209\n206116\n207025\n207936\n208849\n209764\n210681\n211600\n212521\n213444\n214369\n215296\n216225\n217156\n218089\n219024\n219961\n220900\n221841\n222784\n223729\n224676\n225625\n226576\n227529\n228484\n229441\n230400\n231361\n232324\n233289\n234256\n235225\n236196\n237169\n238144\n239121\n240100\n241081\n242064\n243049\n244036\n245025\n246016\n247009\n248004\n249001\n250000\n251001\n252004\n253009\n254016\n255025\n256036\n257049\n258064\n259081\n260100\n261121\n262144\n263169\n264196\n265225\n266256\n267289\n268324\n269361\n270400\n271441\n272484\n273529\n274576\n275625\n276676\n277729\n278784\n279841\n280900\n281961\n283024\n284089\n285156\n286225\n287296\n288369\n289444\n290521\n291600\n292681\n293764\n294849\n295936\n297025\n298116\n299209\n300304\n301401\n302500\n303601\n304704\n305809\n306916\n308025\n309136\n310249\n311364\n312481\n313600\n314721\n315844\n316969\n318096\n319225\n320356\n321489\n322624\n323761\n324900\n326041\n327184\n328329\n329476\n330625\n331776\n332929\n334084\n335241\n336400\n337561\n338724\n339889\n341056\n342225\n343396\n344569\n345744\n346921\n348100\n349281\n350464\n351649\n352836\n354025\n355216\n356409\n357604\n358801\n360000\n361201\n362404\n363609\n364816\n366025\n367236\n368449\n369664\n370881\n372100\n373321\n374544\n375769\n376996\n378225\n379456\n380689\n381924\n383161\n384400\n385641\n386884\n388129\n389376\n390625\n391876\n393129\n394384\n395641\n396900\n398161\n399424\n400689\n401956\n403225\n404496\n405769\n407044\n408321\n409600\n410881\n412164\n413449\n414736\n416025\n417316\n418609\n419904\n421201\n422500\n423801\n425104\n426409\n427716\n429025\n430336\n431649\n432964\n434281\n435600\n436921\n438244\n439569\n440896\n442225\n443556\n444889\n446224\n447561\n448900\n450241\n451584\n452929\n454276\n455625\n456976\n458329\n459684\n461041\n462400\n463761\n465124\n466489\n467856\n469225\n470596\n471969\n473344\n474721\n476100\n477481\n478864\n480249\n481636\n483025\n484416\n485809\n487204\n488601\n490000\n491401\n492804\n494209\n495616\n497025\n498436\n499849\n501264\n502681\n504100\n505521\n506944\n508369\n509796\n511225\n512656\n514089\n515524\n516961\n518400\n519841\n521284\n522729\n524176\n525625\n527076\n528529\n529984\n531441\n532900\n534361\n535824\n537289\n538756\n540225\n541696\n543169\n544644\n546121\n547600\n549081\n550564\n552049\n553536\n555025\n556516\n558009\n559504\n561001\n562500\n564001\n565504\n567009\n568516\n570025\n571536\n573049\n574564\n576081\n577600\n579121\n580644\n582169\n583696\n585225\n586756\n588289\n589824\n591361\n592900\n594441\n595984\n597529\n599076\n600625\n602176\n603729\n605284\n606841\n608400\n609961\n611524\n613089\n614656\n616225\n617796\n619369\n620944\n622521\n624100\n625681\n627264\n628849\n630436\n632025\n633616\n635209\n636804\n638401\n640000\n641601\n643204\n644809\n646416\n648025\n649636\n651249\n652864\n654481\n656100\n657721\n659344\n660969\n662596\n664225\n665856\n667489\n669124\n670761\n672400\n674041\n675684\n677329\n678976\n680625\n682276\n683929\n685584\n687241\n688900\n690561\n692224\n693889\n695556\n697225\n698896\n700569\n702244\n703921\n705600\n707281\n708964\n710649\n712336\n714025\n715716\n717409\n719104\n720801\n722500\n724201\n725904\n727609\n729316\n731025\n732736\n734449\n736164\n737881\n739600\n741321\n743044\n744769\n746496\n748225\n749956\n751689\n753424\n755161\n756900\n758641\n760384\n762129\n763876\n765625\n767376\n769129\n770884\n772641\n774400\n776161\n777924\n779689\n781456\n783225\n784996\n786769\n788544\n790321\n792100\n793881\n795664\n797449\n799236\n801025\n802816\n804609\n806404\n808201\n810000\n811801\n813604\n815409\n817216\n819025\n820836\n822649\n824464\n826281\n828100\n829921\n831744\n833569\n835396\n837225\n839056\n840889\n842724\n844561\n846400\n848241\n850084\n851929\n853776\n855625\n857476\n859329\n861184\n863041\n864900\n866761\n868624\n870489\n872356\n874225\n876096\n877969\n879844\n881721\n883600\n885481\n887364\n889249\n891136\n893025\n894916\n896809\n898704\n900601\n902500\n904401\n906304\n908209\n910116\n912025\n913936\n915849\n917764\n919681\n921600\n923521\n925444\n927369\n929296\n931225\n933156\n935089\n937024\n938961\n940900\n942841\n944784\n946729\n948676\n950625\n952576\n954529\n956484\n958441\n960400\n962361\n964324\n966289\n968256\n970225\n972196\n974169\n976144\n978121\n980100\n982081\n984064\n986049\n988036\n990025\n992016\n994009\n996004\n"
],
[
"\"\"\"I am doing this for I can see how my loop is doing. Therefore, I must to do it in \nunderstable way\"\"\"\nfor i in range(100, 999):\n for j in range(100, 150):\n if i == j:\n n = i * j\n print(n)\n print(i, j)",
"10000\n100 100\n10201\n101 101\n10404\n102 102\n10609\n103 103\n10816\n104 104\n11025\n105 105\n11236\n106 106\n11449\n107 107\n11664\n108 108\n11881\n109 109\n12100\n110 110\n12321\n111 111\n12544\n112 112\n12769\n113 113\n12996\n114 114\n13225\n115 115\n13456\n116 116\n13689\n117 117\n13924\n118 118\n14161\n119 119\n14400\n120 120\n14641\n121 121\n14884\n122 122\n15129\n123 123\n15376\n124 124\n15625\n125 125\n15876\n126 126\n16129\n127 127\n16384\n128 128\n16641\n129 129\n16900\n130 130\n17161\n131 131\n17424\n132 132\n17689\n133 133\n17956\n134 134\n18225\n135 135\n18496\n136 136\n18769\n137 137\n19044\n138 138\n19321\n139 139\n19600\n140 140\n19881\n141 141\n20164\n142 142\n20449\n143 143\n20736\n144 144\n21025\n145 145\n21316\n146 146\n21609\n147 147\n21904\n148 148\n22201\n149 149\n"
],
[
"\"\"\"So, let's see what numbers is a palindromic number:\"\"\"\nfor i in range(100, 999):\n for j in range(100, 150):\n if i == j:\n n = i * j\n n = str(n)\n if n[0] == n[-1]:\n print(n)\n print(i, j)",
"10201\n101 101\n11881\n109 109\n12321\n111 111\n14161\n119 119\n14641\n121 121\n16641\n129 129\n17161\n131 131\n19321\n139 139\n19881\n141 141\n"
]
],
[
[
"### Now I could see this way didn't work, because this 'rule' works only for the 2-digit numbers. So, I must change my 'rule'.",
"_____no_output_____"
]
],
[
[
"\"\"\"As I am working with number as a string, I can do this way:\"\"\"\nn = 12345\nd = str(n)\nprint('Direct:' + d)\nr = str(n)\nprint('Reverse:' + r[::-1])",
"Direct:12345\nReverse:54321\n"
],
[
"\"\"\"Now, let's do the test:\"\"\"\n\nif __name__ == '__main__':\n assert palindromic2(121) == True",
"_____no_output_____"
],
[
"def palindromic2(n):\n return True\n \n\nif __name__ == '__main__':\n assert palindromic2(121) == True",
"_____no_output_____"
],
[
"def palindromic2(n):\n return True\n \n\nif __name__ == '__main__':\n assert palindromic2(121) == True\n assert palindromic2(131) == True\n assert palindromic2(141) == True\n assert palindromic2(314) == False",
"_____no_output_____"
],
[
"def palindromic2(n):\n d = str(n)\n #print('Direct:' + d)\n r = str(n)\n r = r[::-1]\n #print('Reverse:' + r[::-1])\n if d == r:\n return True\n else:\n return False\n\nif __name__ == '__main__':\n assert palindromic2(121) == True\n assert palindromic2(131) == True\n assert palindromic2(141) == True\n assert palindromic2(314) == False",
"_____no_output_____"
],
[
"def palindromic2(n):\n d = str(n)\n r = str(n)\n r = r[::-1]\n if d == r:\n return True\n else:\n return False\n\nif __name__ == '__main__':\n assert palindromic2(121) == True\n assert palindromic2(131) == True\n assert palindromic2(141) == True\n assert palindromic2(314) == False\n assert palindromic2(10) == False\n assert palindromic2(1) == False",
"_____no_output_____"
],
[
"def palindromic2(n):\n d = str(n)\n r = str(n)\n r = r[::-1]\n if len(d) == 1:\n return False\n if d == r:\n return True\n else:\n return False\n\nif __name__ == '__main__':\n assert palindromic2(121) == True\n assert palindromic2(131) == True\n assert palindromic2(141) == True\n assert palindromic2(314) == False\n assert palindromic2(10) == False\n assert palindromic2(1) == False",
"_____no_output_____"
]
],
[
[
"### So, I must to return for the loop. Now, I am going to check the given result: 9009 = 91 × 99.\n\n",
"_____no_output_____"
]
],
[
[
"\"\"\"def palindromic2(n):\n d = str(n)\n r = str(n)\n r = r[::-1]\n if len(d) == 1:\n return False\n if d == r:\n return True\n else:\n return False\"\"\"\nlista = [] \nfor i in range(1, 100):\n for j in range(1, 100):\n n = i * j\n n = str(n)\n r = n[::-1]\n if r == n:\n m = int(n)\n lista.append(m)\nprint(max(lista))",
"9009\n"
]
],
[
[
"### At this point, I don't know how to test directly, therefore I must do it without a assert. But I know what I hope to see.",
"_____no_output_____"
]
],
[
[
"def palindromic3(n):\n d = str(n)\n r = str(n)\n r = r[::-1]\n if len(d) == 1:\n return False\n if d == r:\n return True\n else:\n return False\n\nlista = [] \nfor i in range(1, 100):\n for j in range(1, 100):\n n = i * j\n n = str(n)\n r = n[::-1]\n if r == n:\n if palindromic3(r) == True:\n lista.append(r)\nprint(lista)",
"['11', '22', '33', '44', '55', '66', '77', '88', '99', '22', '44', '66', '88', '33', '66', '99', '111', '141', '171', '222', '252', '282', '44', '88', '212', '232', '252', '272', '292', '55', '66', '222', '252', '282', '414', '444', '474', '77', '161', '252', '343', '434', '525', '595', '616', '686', '88', '232', '272', '424', '464', '616', '656', '696', '99', '171', '252', '333', '414', '585', '666', '747', '828', '11', '22', '33', '44', '55', '66', '77', '88', '99', '121', '242', '363', '484', '616', '737', '858', '979', '1001', '252', '444', '636', '696', '828', '888', '494', '585', '676', '767', '858', '949', '1001', '252', '434', '616', '686', '868', '525', '555', '585', '272', '464', '656', '848', '272', '323', '595', '646', '969', '252', '414', '666', '828', '171', '323', '494', '646', '969', '1881', '252', '525', '777', '22', '44', '66', '88', '242', '484', '616', '858', '2002', '2112', '161', '414', '575', '828', '989', '1771', '696', '888', '2112', '525', '575', '494', '676', '858', '2002', '999', '252', '616', '868', '2772', '232', '464', '696', '2552', '434', '868', '2112', '33', '66', '99', '363', '858', '1221', '1551', '1881', '2112', '2442', '2772', '3003', '272', '646', '2992', '525', '595', '252', '828', '2772', '111', '222', '333', '444', '555', '666', '777', '888', '999', '1221', '2442', '3663', '494', '646', '585', '858', '3003', '656', '252', '2772', '989', '44', '88', '484', '616', '2112', '2332', '2552', '2772', '2992', '4004', '4224', '585', '414', '828', '4554', '141', '282', '1551', '2112', '4224', '343', '686', '3773', '969', '676', '4004', '212', '424', '636', '848', '2332', '4664', '55', '5005', '5115', '5225', '5335', '5445', '616', '171', '969', '1881', '232', '464', '696', '2552', '767', '434', '868', '4774', '252', '2772', '2112', '4224', '6336', '585', '5005', '66', '858', '2112', '2442', '2772', '4224', '4554', '4884', '6006', '6336', '737', '272', '2992', '414', '828', '4554', '6336', '292', '949', '7227', '222', '444', '666', '888', '2442', '4884', '525', '5775', '77', '616', '1001', '1771', '2002', '2772', '3003', '3773', '4004', '4774', '5005', '5775', '6006', '6776', '7007', '858', '6006', '474', '656', '8118', '747', '252', '2772', '595', '696', '88', '616', '2112', '2552', '2992', '4224', '4664', '6336', '6776', '8008', '8448', '979', '1001', '2002', '3003', '4004', '5005', '6006', '7007', '8008', '9009', '828', '5115', '282', '5225', '2112', '4224', '6336', '8448', '5335', '686', '99', '1881', '2772', '3663', '4554', '5445', '6336', '7227', '8118', '9009']\n"
]
],
[
[
"### As it works very well, I can expect to see the number given by the statement.",
"_____no_output_____"
]
],
[
[
"def palindromic3(n):\n d = str(n)\n r = str(n)\n r = r[::-1]\n if len(d) == 1:\n return False\n if d == r:\n return True\n else:\n return False\n\ndef palindromic4():\n lista = [] \n for i in range(1, 100):\n for j in range(1, 100):\n n = i * j\n n = str(n)\n r = n[::-1]\n if r == n:\n if palindromic3(r) == True:\n lista.append(r)\n return(max(lista))\n \nif __name__ == '__main__':\n assert palindromic4() == 9009",
"_____no_output_____"
],
[
"palindromic4()\n\"\"\"So, I must to fix this:\"\"\"",
"_____no_output_____"
],
[
"def palindromic3(n):\n d = str(n)\n r = str(n)\n r = r[::-1]\n if len(d) == 1:\n return False\n if d == r:\n return True\n else:\n return False\n\ndef palindromic4():\n lista = [] \n for i in range(1, 100):\n for j in range(1, 100):\n n = i * j\n n = str(n)\n r = n[::-1]\n if r == n:\n if palindromic3(r) == True:\n lista.append(r)\n return(lista[-1])\n \nif __name__ == '__main__':\n assert palindromic4() == 9009",
"_____no_output_____"
],
[
"palindromic4()\n\"\"\"This value is a string value, not a numeric value\"\"\"",
"_____no_output_____"
],
[
"def palindromic3(n):\n d = str(n)\n r = str(n)\n r = r[::-1]\n if len(d) == 1:\n return False\n if d == r:\n return True\n else:\n return False\n\ndef palindromic4():\n lista = [] \n for i in range(1, 100):\n for j in range(1, 100):\n n = i * j\n n = str(n)\n r = n[::-1]\n if r == n:\n if palindromic3(r) == True:\n lista.append(r)\n return(int(lista[-1]))\n \nif __name__ == '__main__':\n assert palindromic4() == 9009\n\"\"\"I have two possibilities so fix this: Or I set my assert to hope a str value, \nor I set my function to return a int value.\"\"\"",
"_____no_output_____"
],
[
"#refactoring\ndef palindromic3(n):\n d = str(n)\n r = str(n)\n r = r[::-1]\n if len(d) == 1:\n return False\n if d == r:\n return True\n else:\n return False\n\ndef palindromic4():\n lista = [] \n for i in range(1, 100):\n for j in range(1, 100):\n n = i * j\n n = str(n)\n r = n[::-1]\n #if r == n: here\n if palindromic3(r) == True:\n lista.append(r)\n return(int(lista[-1]))\n \nif __name__ == '__main__':\n assert palindromic4() == 9009",
"_____no_output_____"
],
[
"palindromic4()",
"_____no_output_____"
]
],
[
[
"### As I could solve this problem to a product of two 2-digit numbers, I must to solve this problem to a product of two 3-digit numbers. ",
"_____no_output_____"
]
],
[
[
"def palindromic3(n):\n d = str(n)\n r = str(n)\n r = r[::-1]\n if len(d) == 1 or len(d) == 2:\n return False\n if d == r:\n return True\n else:\n return False\n\ndef palindromic4():\n lista = [] \n for i in range(100, 1001):\n for j in range(100, 1001):\n n = i * j\n n = str(n)\n if palindromic3(n) == True:\n lista.append(n)\n return(lista)\n ",
"_____no_output_____"
],
[
"palindromic4()",
"_____no_output_____"
]
],
[
[
"#### Now I could see that the last value isn't a higher value inside the list",
"_____no_output_____"
]
],
[
[
"def palindromic3(n):\n d = str(n)\n r = str(n)\n r = r[::-1]\n if len(d) == 1 or len(d) == 2:\n return False\n if d == r:\n return True\n else:\n return False\n\ndef palindromic4():\n lista = [] \n for i in range(100, 1001):\n for j in range(100, 1001):\n n = i * j\n n = str(n)\n if palindromic3(n) == True:\n n = int(n)#add this\n lista.append(n)#and this\n return(max(lista))#now the function can return a highest value from the list",
"_____no_output_____"
],
[
"palindromic4()",
"_____no_output_____"
]
],
[
[
"## Now, I only should to refact my code to make it more simple and beautifull, as the python's zen tell us. ",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
c50f79912a40372e239fcae127a2c8ec74ef6923
| 184,262 |
ipynb
|
Jupyter Notebook
|
aruco.ipynb
|
HaraDev001/Python3D
|
5c81096237f4bd94f5c312997338e12ab91ce4aa
|
[
"MIT"
] | null | null | null |
aruco.ipynb
|
HaraDev001/Python3D
|
5c81096237f4bd94f5c312997338e12ab91ce4aa
|
[
"MIT"
] | null | null | null |
aruco.ipynb
|
HaraDev001/Python3D
|
5c81096237f4bd94f5c312997338e12ab91ce4aa
|
[
"MIT"
] | null | null | null | 376.813906 | 61,084 | 0.929622 |
[
[
[
"# カメラの位置姿勢を求める",
"_____no_output_____"
]
],
[
[
"import cv2\nimport numpy as np\nimport matplotlib.pyplot as plt\nfrom skvideo.io import vread\nimport moviepy.editor as mpy\nfrom tqdm import tqdm\nfrom mpl_toolkits.mplot3d import axes3d, Axes3D\nfrom IPython.display import Image\n\n\ndef npy_to_gif(npy, filename):\n clip = mpy.ImageSequenceClip(list(npy), fps=10)\n clip.write_gif(filename)",
"_____no_output_____"
]
],
[
[
"### 1. 素材",
"_____no_output_____"
]
],
[
[
"vid = vread(\"src/shisa.mp4\")\nprint(vid.shape)",
"(524, 1920, 1080, 3)\n"
]
],
[
[
"### 2. アルコマーカー",
"_____no_output_____"
]
],
[
[
"aruco_dict = cv2.aruco.getPredefinedDictionary(cv2.aruco.DICT_4X4_50)\naruco = cv2.aruco.drawMarker(aruco_dict, 0, 256)\nplt.figure(figsize=(3,3)); plt.imshow(aruco); plt.show()\n# cv2.imwrite(\"aruco.png\", aruco)",
"_____no_output_____"
]
],
[
[
"### 3. カメラの設定",
"_____no_output_____"
]
],
[
[
"marker_length = 0.07 # [m] ### 注意!\nmtx = np.load(\"camera/mtx.npy\")\ndist = np.load(\"camera/dist.npy\")\nprint(mtx); print(dist)",
"[[1.83882882e+03 0.00000000e+00 5.20791172e+02]\n [0.00000000e+00 1.82803295e+03 9.76365659e+02]\n [0.00000000e+00 0.00000000e+00 1.00000000e+00]]\n[[ 2.97906356e-01 -4.84831541e-01 4.55397225e-03 8.49742572e-04\n -1.61521682e+00]]\n"
]
],
[
[
"### 4. マーカーの検出",
"_____no_output_____"
]
],
[
[
"frame = vid[0]\nframe = frame[...,::-1] # BGR2RGB\nframe = cv2.resize(frame, (360, 640))\n\ncorners, ids, rejectedImgPoints = cv2.aruco.detectMarkers(frame, aruco_dict)\nrvec, tvec, _ = cv2.aruco.estimatePoseSingleMarkers(corners, marker_length, mtx, dist)\n\n# ---- 描画\nframe = cv2.aruco.drawDetectedMarkers(frame, corners, ids)\nframe = cv2.aruco.drawAxis(frame, mtx, dist, rvec, tvec, marker_length/2)\n# ----\n\nplt.imshow(frame[...,::-1]); plt.show()",
"_____no_output_____"
]
],
[
[
"### 5. カメラの位置姿勢の計算",
"_____no_output_____"
]
],
[
[
"# def eulerAnglesToRotationMatrix(euler):\n# R_x = np.array([[ 1, 0, 0],\n# [ 0, np.cos(euler[0]), -np.sin(euler[0])],\n# [ 0, np.sin(euler[0]), np.cos(euler[0])]])\n# R_y = np.array([[ np.cos(euler[1]), 0, np.sin(euler[1])],\n# [ 0, 1, 0],\n# [-np.sin(euler[1]), 0, np.cos(euler[1])]])\n# R_z = np.array([[ np.cos(euler[2]), -np.sin(euler[2]), 0],\n# [ np.sin(euler[2]), np.cos(euler[2]), 0],\n# [ 0, 0, 1]])\n# R = np.dot(R_z, np.dot(R_y, R_x))\n# return R",
"_____no_output_____"
],
[
"XYZ = []\nRPY = []\nV_x = []\nV_y = []\nV_z = []\n\nfor frame in vid[:500:25]: # 全部処理すると重いので…\n frame = frame[...,::-1] # BGR2RGB\n frame = cv2.resize(frame, (360, 640))\n corners, ids, _ = cv2.aruco.detectMarkers(frame, aruco_dict)\n\n rvec, tvec, _ = cv2.aruco.estimatePoseSingleMarkers(corners, marker_length, mtx, dist)\n\n R = cv2.Rodrigues(rvec)[0] # 回転ベクトル -> 回転行列\n R_T = R.T\n T = tvec[0].T\n\n xyz = np.dot(R_T, - T).squeeze()\n XYZ.append(xyz)\n\n rpy = np.deg2rad(cv2.RQDecomp3x3(R_T)[0])\n RPY.append(rpy)\n # print(rpy)\n\n # rpy = cv2.decomposeProjectionMatrix(np.hstack([R_T, -T]))[6] # [0~5]は使わない\n # rpy = np.deg2rad(rpy.squeeze())\n # print(rpy)\n\n # r = np.arctan2(-R_T[2][1], R_T[2][2])\n # p = np.arcsin(R_T[2][0])\n # y = np.arctan2(-R_T[1][0], R_T[0][0])\n # rpy = - np.array([r, p, y])\n # print(rpy)\n\n # from scipy.spatial.transform import Rotation\n # diff = eulerAnglesToRotationMatrix(rpy) - R_T\n # print(diff.astype(np.float16))\n # diff = Rotation.from_euler('xyz', rpy).as_matrix() - R_T\n # print(diff.astype(np.float16))\n \n V_x.append(np.dot(R_T, np.array([1,0,0])))\n V_y.append(np.dot(R_T, np.array([0,1,0])))\n V_z.append(np.dot(R_T, np.array([0,0,1])))\n\n # ---- 描画\n # cv2.aruco.drawDetectedMarkers(frame, corners, ids, (0,255,255))\n # cv2.aruco.drawAxis(frame, mtx, dist, rvec, tvec, marker_length/2)\n # cv2.imshow('frame', frame)\n # cv2.waitKey(1)\n # ----\n\ncv2.destroyAllWindows()",
"_____no_output_____"
],
[
"def plot_all_frames(elev=90, azim=270):\n frames = []\n\n for t in tqdm(range(len(XYZ))):\n fig = plt.figure(figsize=(4,3))\n ax = Axes3D(fig)\n ax.view_init(elev=elev, azim=azim)\n ax.set_xlim(-2, 2); ax.set_ylim(-2, 2); ax.set_zlim(-2, 2)\n ax.set_xlabel(\"x\"); ax.set_ylabel(\"y\"); ax.set_zlabel(\"z\")\n\n x, y, z = XYZ[t]\n ux, vx, wx = V_x[t]\n uy, vy, wy = V_y[t]\n uz, vz, wz = V_z[t]\n\n # draw marker\n ax.scatter(0, 0, 0, color=\"k\")\n ax.quiver(0, 0, 0, 1, 0, 0, length=1, color=\"r\")\n ax.quiver(0, 0, 0, 0, 1, 0, length=1, color=\"g\")\n ax.quiver(0, 0, 0, 0, 0, 1, length=1, color=\"b\")\n ax.plot([-1,1,1,-1,-1], [-1,-1,1,1,-1], [0,0,0,0,0], color=\"k\", linestyle=\":\")\n\n # draw camera\n if t < 5:\n ax.quiver(x, y, z, ux, vx, wx, length=0.5, color=\"k\")\n ax.quiver(x, y, z, uy, vy, wy, length=0.5, color=\"k\")\n ax.quiver(x, y, z, uz, vz, wz, length=0.5, color=\"k\")\n else:\n ax.quiver(x, y, z, ux, vx, wx, length=0.5, color=\"r\")\n ax.quiver(x, y, z, uy, vy, wy, length=0.5, color=\"g\")\n ax.quiver(x, y, z, uz, vz, wz, length=0.5, color=\"b\")\n\n # save for animation\n fig.canvas.draw()\n frames.append(np.array(fig.canvas.renderer.buffer_rgba()))\n plt.close()\n\n return frames",
"_____no_output_____"
],
[
"frames = plot_all_frames(elev=105, azim=270)\nnpy_to_gif(frames, \"src/sample1.gif\"); Image(url='src/sample1.gif')",
"100%|██████████| 20/20 [00:02<00:00, 6.68it/s]\nt: 33%|███▎ | 7/21 [00:00<00:00, 66.75it/s, now=None]"
],
[
"frames = plot_all_frames(elev=165, azim=270)\nnpy_to_gif(frames, \"src/sample2.gif\"); Image(url='src/sample2.gif')",
"100%|██████████| 20/20 [00:03<00:00, 6.52it/s]\nt: 38%|███▊ | 8/21 [00:00<00:00, 74.57it/s, now=None]"
],
[
"plt.title(\"xyz\"); plt.plot(XYZ); plt.show() # 青:x, 橙:y, 緑:z\nplt.title(\"rpy\"); plt.plot(RPY); plt.show() # 青:r, 橙:p, 緑:y\nplt.title(\"(v_x)\"); plt.plot(V_x); plt.show()\nplt.title(\"(v_y)\"); plt.plot(V_y); plt.show()\nplt.title(\"(v_z)\"); plt.plot(V_z); plt.show()",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
c50f7aa5225cec98f4ea92d4cd27a20c4c2e611a
| 16,080 |
ipynb
|
Jupyter Notebook
|
notebooks/aot_autograd_optimizations.ipynb
|
zdevito/functorch
|
117477235bb0a12bd8f79ae45be3f3a759d74c60
|
[
"BSD-3-Clause"
] | null | null | null |
notebooks/aot_autograd_optimizations.ipynb
|
zdevito/functorch
|
117477235bb0a12bd8f79ae45be3f3a759d74c60
|
[
"BSD-3-Clause"
] | null | null | null |
notebooks/aot_autograd_optimizations.ipynb
|
zdevito/functorch
|
117477235bb0a12bd8f79ae45be3f3a759d74c60
|
[
"BSD-3-Clause"
] | null | null | null | 39.703704 | 571 | 0.624316 |
[
[
[
"# AOT Autograd - How to use and optimize?\n\n<a href=\"https://colab.research.google.com/github/pytorch/functorch/blob/main/notebooks/colab/aot_autograd_optimizations.ipynb\">\n <img style=\"width: auto\" src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/>\n</a>\n\n## Background\nIn this tutorial, we will learn how to use AOT Autograd to speedup training of deep learning models.\n\nFor background, AOT Autograd is a toolkit to assist developers in accelerating training on PyTorch. Broadly, it has two key features\n* AOT Autograd traces the forward and backward graph ahead of time. Presence of forward and backward graph ahead of time facilitates joint graph optimizations such as recomputation or activation checkpointing.\n* AOT Autograd provides simple mechanisms to compile the extracted forward and backward graphs through deep learning compilers, such as NVFuser, NNC, TVM and others.\n",
"_____no_output_____"
],
[
"\n## What will you learn?\nIn this tutorial, we will look at how AOT Autograd can be used, in conjunction with backend compilers, to accelerate the training of PyTorch models. More specifically, you will learn\n* How to use AOT Autograd?\n* How AOT Autograd uses backend compilers to perform operation fusion?\n* How AOT Autograd enables training-specific optimizations such as Recomputation?\n\nSo, lets get started.\n",
"_____no_output_____"
],
[
"## Setup\n\nLet's setup a simple model.\n",
"_____no_output_____"
]
],
[
[
"import torch\n\ndef fn(a, b, c, d):\n x = a + b + c + d\n return x.cos().cos()",
"_____no_output_____"
],
[
"# Test that it works\na, b, c, d = [torch.randn(2, 4, requires_grad=True) for _ in range(4)]\nref = fn(a, b, c, d)\nloss = ref.sum()\nloss.backward()",
"_____no_output_____"
]
],
[
[
"# Use AOT Autograd\n\nNow, lets use AOT Autograd and look at the extracted forward and backward graphs. Internally, AOT uses `__torch_dispatch__` based tracing mechanism to extract forward and backward graphs, and wraps them in `torch.Fx` GraphModule containers. Note that AOT Autograd tracing is different from the usual Fx symbolic tracing. AOT Autograd uses Fx GraphModule just to represent the traced graphs (and not for tracing).\n\nAOT Autograd then sends these forward and backward graphs to the user supplied compilers. So, lets write a compiler that just prints the graph.",
"_____no_output_____"
]
],
[
[
"from functorch.compile import aot_function\n\n# The compiler_fn is called after the forward and backward graphs are extracted.\n# Here, we just print the code in the compiler_fn. Return of this function is a callable.\ndef compiler_fn(fx_module: torch.fx.GraphModule, _):\n print(fx_module.code)\n return fx_module\n\n# Pass on the compiler_fn to the aot_function API\naot_print_fn = aot_function(fn, fw_compiler=compiler_fn, bw_compiler=compiler_fn)\n\n# Run the aot_print_fn once to trigger the compilation and print the graphs\nres = aot_print_fn(a, b, c, d)\nassert torch.allclose(ref, res)\n\nfrom functorch.compile import clear_compile_cache\nclear_compile_cache()",
"\n\n\ndef forward(self, primals_1, primals_2, primals_3, primals_4):\n add = torch.ops.aten.add(primals_1, primals_2); primals_1 = primals_2 = None\n add_1 = torch.ops.aten.add(add, primals_3); add = primals_3 = None\n add_2 = torch.ops.aten.add(add_1, primals_4); add_1 = primals_4 = None\n cos = torch.ops.aten.cos(add_2)\n cos_1 = torch.ops.aten.cos(cos)\n return [cos_1, add_2, cos]\n \n\n\n\ndef forward(self, add_2, cos, tangents_1):\n sin = torch.ops.aten.sin(cos); cos = None\n neg = torch.ops.aten.neg(sin); sin = None\n mul = torch.ops.aten.mul(tangents_1, neg); tangents_1 = neg = None\n sin_1 = torch.ops.aten.sin(add_2); add_2 = None\n neg_1 = torch.ops.aten.neg(sin_1); sin_1 = None\n mul_1 = torch.ops.aten.mul(mul, neg_1); mul = neg_1 = None\n return [mul_1, mul_1, mul_1, mul_1]\n \n"
]
],
[
[
"The above code prints the Fx graph for the forward and backward graph. You can see that in addition to the original input of the forward pass, the forward graph outputs some additional tensors. These tensors are saved for the backward pass for gradient calculation. We will come back to these later while talking about recomputation.",
"_____no_output_____"
],
[
"## Operator Fusion\nNow that we understand how to use AOT Autograd to print forward and backward graphs, let us use AOT Autograd to use some actual deep learning compiler. In this tutorial, we use PyTorch Neural Network Compiler (NNC) to perform pointwise operator fusion for CPU devices. For CUDA devices, a suitable alternative is NvFuser. So, lets use NNC",
"_____no_output_____"
]
],
[
[
"# AOT Autograd has a suite of already integrated backends. Lets import the NNC compiler backend - ts_compile\nfrom functorch.compile import ts_compile\n\n# Lets compile the forward and backward through ts_compile.\naot_nnc_fn = aot_function(fn, fw_compiler=ts_compile, bw_compiler=ts_compile)\n\n# Correctness checking. Lets clone the input so that we can check grads.\ncloned_inputs = [x.clone().detach().requires_grad_(True) for x in (a, b, c, d)]\ncloned_a, cloned_b, cloned_c, cloned_d = cloned_inputs\n\nres = aot_nnc_fn(*cloned_inputs)\nloss = res.sum()\nloss.backward()\nassert torch.allclose(ref, res)\nassert torch.allclose(a.grad, cloned_a.grad)\nassert torch.allclose(b.grad, cloned_b.grad)\nassert torch.allclose(c.grad, cloned_c.grad)\nassert torch.allclose(d.grad, cloned_d.grad)",
"_____no_output_____"
]
],
[
[
"Lets benchmark the original and AOT Autograd + NNC compiled function.",
"_____no_output_____"
]
],
[
[
"# Lets write a function to benchmark the forward and backward pass\nimport time\nimport statistics\n\ndef bench(fn, args, prefix):\n warmup = 10\n iterations = 100\n\n for _ in range(warmup):\n ref = fn(*args)\n ref.sum().backward()\n \n fw_latencies = []\n bw_latencies = []\n for _ in range(iterations):\n for arg in args:\n arg.grad = None\n\n fw_begin = time.perf_counter()\n ref = fn(*args)\n fw_end = time.perf_counter()\n\n loss = ref.sum() \n\n bw_begin = time.perf_counter()\n loss.backward()\n bw_end = time.perf_counter()\n\n fw_latencies.append(fw_end - fw_begin)\n bw_latencies.append(bw_end - bw_begin)\n \n avg_fw_latency = statistics.mean(fw_latencies) * 10**6\n avg_bw_latency = statistics.mean(bw_latencies) * 10**6\n print(prefix, \"Fwd = \" + str(avg_fw_latency) + \" us\", \"Bwd = \" + str(avg_bw_latency) + \" us\", sep=', ')\n",
"_____no_output_____"
],
[
"large_inputs = [torch.randn(1024, 2048, requires_grad=True) for _ in range(4)]\n\n# Benchmark the Eager and AOT Autograd functions\nbench(fn, large_inputs, \"Eager\")\nbench(aot_nnc_fn, large_inputs, \"AOT\")",
"Eager, Fwd = 982.6959593920038 us, Bwd = 1899.7003795811906 us\nAOT, Fwd = 734.2723174951971 us, Bwd = 831.1696897726506 us\n"
]
],
[
[
"With the help of NNC, AOT Autograd speeds up both the forward and backward pass. If we look at the printed graphs earlier, all the operators are pointwise. The pointwise operators are memory bandwidth bound, and thus benefit from operator fusion. Looking closely at the numbers, the backward pass gets higher speedup. This is because forward pass has to output some intermediate tensors for gradient calculation for the backward pass, preventing it from saving some memory reads and writes. However, such restriction does not exist in the backward graph.",
"_____no_output_____"
],
[
"## Recomputation (aka Activation Checkpointing)\nRecomputation (often called activation checkpointing) is a technique in which, instead of saving some activations for use in backwards, we recompute them **during** the backwards pass. Recomputing saves memory, but we incur performance overhead.\n\nHowever, in the presence of fusing compiler, we can do better that that. We can recompute the fusion-friendly operators to save memory, and then rely on the fusing compiler to fuse the recomputed operators. This reduces both memory and runtime. Please refer to this [discuss post](https://dev-discuss.pytorch.org/t/min-cut-optimal-recomputation-i-e-activation-checkpointing-with-aotautograd/467) for more details.\n\nHere, we use AOT Autograd with NNC to perform similar type of recomputation. At the end of `__torch_dispatch__` tracing, AOT Autograd has a forward graph and joint forward-backward graph. AOT Autograd then uses a partitioner to isolate the forward and backward graph. In the example above, we used a default partitioner. For this experiment, we will use another partitioner called `min_cut_rematerialization_partition` to perform smarter fusion-aware recomputation. The partitioner is configurable and one can write their own partitioner to plug it in AOT Autograd.",
"_____no_output_____"
]
],
[
[
"from functorch.compile import min_cut_rematerialization_partition\n\n# Lets set up the partitioner. Also set the fwd and bwd compilers to the printer function that we used earlier.\n# This will show us how the recomputation has modified the graph.\naot_fn = aot_function(fn, fw_compiler=compiler_fn, bw_compiler=compiler_fn, partition_fn=min_cut_rematerialization_partition)\nres = aot_fn(a, b, c, d)",
"\n\n\ndef forward(self, primals_1, primals_2, primals_3, primals_4):\n add = torch.ops.aten.add(primals_1, primals_2); primals_1 = primals_2 = None\n add_1 = torch.ops.aten.add(add, primals_3); add = primals_3 = None\n add_2 = torch.ops.aten.add(add_1, primals_4); add_1 = primals_4 = None\n cos = torch.ops.aten.cos(add_2)\n cos_1 = torch.ops.aten.cos(cos); cos = None\n return [cos_1, add_2]\n \n\n\n\ndef forward(self, add_2, tangents_1):\n cos = torch.ops.aten.cos(add_2)\n sin = torch.ops.aten.sin(cos); cos = None\n neg = torch.ops.aten.neg(sin); sin = None\n mul = torch.ops.aten.mul(tangents_1, neg); tangents_1 = neg = None\n sin_1 = torch.ops.aten.sin(add_2); add_2 = None\n neg_1 = torch.ops.aten.neg(sin_1); sin_1 = None\n mul_1 = torch.ops.aten.mul(mul, neg_1); mul = neg_1 = None\n return [mul_1, mul_1, mul_1, mul_1]\n \n"
]
],
[
[
"We can see that compared to default partitioner, forward pass now outputs fewer tensors, and recomputes some operations in the backward pass. Let us try NNC compiler now to perform operator fusions (note that we also have a wrapper function - `memory_efficient_fusion` which internally uses `min_cut_rematerialization_partition` and Torchscript compiler to achieve the same effect as following code).",
"_____no_output_____"
]
],
[
[
"\n# Lets set up the partitioner and NNC compiler.\naot_recompute_nnc_fn = aot_function(fn, fw_compiler=ts_compile, bw_compiler=ts_compile, partition_fn=min_cut_rematerialization_partition)\n\n# Correctness checking. Lets clone the input so that we can check grads.\ncloned_inputs = [x.clone().detach().requires_grad_(True) for x in (a, b, c, d)]\ncloned_a, cloned_b, cloned_c, cloned_d = cloned_inputs\n\nres = aot_recompute_nnc_fn(*cloned_inputs)\nloss = res.sum()\nloss.backward()\nassert torch.allclose(ref, res)\nassert torch.allclose(a.grad, cloned_a.grad)\nassert torch.allclose(b.grad, cloned_b.grad)\nassert torch.allclose(c.grad, cloned_c.grad)\nassert torch.allclose(d.grad, cloned_d.grad)",
"_____no_output_____"
]
],
[
[
"Finally, lets benchmark the different functions",
"_____no_output_____"
]
],
[
[
"bench(fn, large_inputs, \"Eager\")\nbench(aot_nnc_fn, large_inputs, \"AOT\")\nbench(aot_recompute_nnc_fn, large_inputs, \"AOT_Recomp\")",
"Eager, Fwd = 740.7676504226401 us, Bwd = 1560.5240693548694 us\nAOT, Fwd = 713.8530415249988 us, Bwd = 909.1200679540634 us\nAOT_Recomp, Fwd = 712.2249767417088 us, Bwd = 791.4606417762116 us\n"
]
],
[
[
"We observe that both forward and backward latency improve over the default partitioner (and a lot better than eager). Fewer outputs in the forward pass and fewer inputs in the backward pass, along with fusion, allows better memory bandwidth utilization leading to further speedups.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
c50f7fdd1afd6a37b5e761a0eb6a7e5771d1bf38
| 2,197 |
ipynb
|
Jupyter Notebook
|
examples/archive/mudensity/prototyping/prototype7.ipynb
|
lipteck/pymedphys
|
6e8e2b5db8173eafa6006481ceeca4f4341789e0
|
[
"Apache-2.0"
] | 2 |
2020-02-04T03:21:20.000Z
|
2020-04-11T14:17:53.000Z
|
prototyping/archive/mudensity/prototyping/prototype7.ipynb
|
SimonBiggs/pymedphys
|
83f02eac6549ac155c6963e0a8d1f9284359b652
|
[
"Apache-2.0"
] | 6 |
2020-10-06T15:36:46.000Z
|
2022-02-27T05:15:17.000Z
|
prototyping/archive/mudensity/prototyping/prototype7.ipynb
|
SimonBiggs/pymedphys
|
83f02eac6549ac155c6963e0a8d1f9284359b652
|
[
"Apache-2.0"
] | 1 |
2020-12-20T14:14:00.000Z
|
2020-12-20T14:14:00.000Z
| 20.726415 | 74 | 0.527082 |
[
[
[
"import os\nfrom glob import glob\n\nimport numpy as np\nimport matplotlib.pyplot as plt\n\nfrom decode_trf import delivery_data_from_logfile\nfrom mu_density import calc_mu_density",
"_____no_output_____"
],
[
"grid_resolution = 5/3 # mm",
"_____no_output_____"
],
[
"test_files = glob(os.path.join(\n \"S:\\\\Physics\\\\Programming\\\\data\\\\LinacLogFiles\",\n 'indexed', '*', '012194*', 'clinical', '*_VMAT', '*', '*.trf'\n))\n\ndelivery_data = delivery_data_from_logfile(test_files[0])",
"_____no_output_____"
],
[
"mu = np.array(delivery_data.monitor_units)\nmlc = np.swapaxes(delivery_data.mlc, 0, 2)\njaw = np.swapaxes(delivery_data.jaw, 0, 1)",
"_____no_output_____"
],
[
"grid_xx, grid_yy, mu_density = calc_mu_density(\n mu, mlc, jaw, grid_resolution=grid_resolution)",
"_____no_output_____"
],
[
"plt.pcolormesh(\n grid_xx, \n grid_yy,\n mu_density)\nplt.colorbar()",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
c50f848995a912d54290b072dcf24f71223f196c
| 5,578 |
ipynb
|
Jupyter Notebook
|
chts-attach-network-nodes.ipynb
|
ual/chts-attach-network
|
4c674fde319981aa807db1c4358d9a2c055e5fac
|
[
"BSD-3-Clause"
] | null | null | null |
chts-attach-network-nodes.ipynb
|
ual/chts-attach-network
|
4c674fde319981aa807db1c4358d9a2c055e5fac
|
[
"BSD-3-Clause"
] | null | null | null |
chts-attach-network-nodes.ipynb
|
ual/chts-attach-network
|
4c674fde319981aa807db1c4358d9a2c055e5fac
|
[
"BSD-3-Clause"
] | 1 |
2018-03-16T22:43:13.000Z
|
2018-03-16T22:43:13.000Z
| 25.354545 | 388 | 0.546074 |
[
[
[
"# Attach nearest network nodes to CHTS homes and workplaces",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nfrom sklearn.neighbors import BallTree",
"_____no_output_____"
],
[
"# identify bay area counties by fips code\nbayarea = {'Alameda':'001',\n 'Contra Costa':'013',\n 'Marin':'041',\n 'Napa':'055',\n 'San Francisco':'075',\n 'San Mateo':'081',\n 'Santa Clara':'085',\n 'Solano':'095',\n 'Sonoma':'097'}\n\nstate_counties = ['06{}'.format(county) for county in bayarea.values()]",
"_____no_output_____"
]
],
[
[
"## Load persons home and work data",
"_____no_output_____"
]
],
[
[
"# load persons and workplace data\npersons = pd.read_csv('data/chts/LookUp_PER.csv', dtype={'WBLOCK':str})\npersons = persons[['SAMPN', 'PERNO', 'WXCORD', 'WYCORD', 'WBLOCK']]\nmask = persons['WBLOCK'].str.slice(0, 5).isin(state_counties)\npersons = persons[mask]",
"_____no_output_____"
],
[
"# load homes data\nhomes = pd.read_csv('data/chts/LookUp_Home.csv', dtype={'HBLOCK':str})\nhomes = homes[['SAMPN', 'HXCORD', 'HYCORD', 'HBLOCK']]\nmask = homes['HBLOCK'].str.slice(0, 5).isin(state_counties)\nhomes = homes[mask]",
"_____no_output_____"
],
[
"# merge homes and workplaces for each person\ndf = pd.merge(persons, homes, how='inner', on='SAMPN').dropna()\ndf = df.set_index(['SAMPN', 'PERNO'])\nassert df.index.is_unique",
"_____no_output_____"
]
],
[
[
"## Find nearest network node to home and work locations\n\nFor fast nearest-neighbor search, use a ball tree with the haversine distance metric. This will be more accurate than using Euclidean distance at this spatial scale, and faster than projecting data to a metric projection and then using Euclidean distance. If minor inaccuracy is acceptible in the name of greater speed, just use scipy's basic cKDTree and Euclidean distance instead.",
"_____no_output_____"
]
],
[
[
"# load the network nodes\nnodes = pd.read_csv('data/network/bay_area_tertiary_strongly_nodes.csv')\nnodes = nodes.set_index('osmid')\nassert nodes.index.is_unique",
"_____no_output_____"
],
[
"# haversine requires data in form of [lat, lng] and inputs/outputs in units of radians\nnodes_rad = np.deg2rad(nodes[['y', 'x']])\nhomes_rad = np.deg2rad(df[['HYCORD', 'HXCORD']])\nworks_rad = np.deg2rad(df[['WYCORD', 'WXCORD']])",
"_____no_output_____"
],
[
"# build the tree for fast nearest-neighbor search\ntree = BallTree(nodes_rad, metric='haversine')",
"_____no_output_____"
],
[
"# query the tree for nearest node to each home\nidx = tree.query(homes_rad, return_distance=False)\ndf['HNODE'] = nodes.iloc[idx[:,0]].index",
"_____no_output_____"
],
[
"# query the tree for nearest node to each workplace\nidx = tree.query(works_rad, return_distance=False)\ndf['WNODE'] = nodes.iloc[idx[:,0]].index",
"_____no_output_____"
]
],
[
[
"## Save to disk",
"_____no_output_____"
]
],
[
[
"# how many home and work locations are so close that they resolve to same network node?\nlen(df[df['HNODE']==df['WNODE']])",
"_____no_output_____"
],
[
"# save ODs to disk\ndf_save = df[['HNODE', 'WNODE']].rename(columns={'HNODE':'orig', 'WNODE':'dest'})\ndf_save.to_csv('data/od.csv', index=True, encoding='utf-8')",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
c50f8612cfb329ce6847b3d496c0cf9543eb1313
| 159,117 |
ipynb
|
Jupyter Notebook
|
docs/tutorial/eaton_well.ipynb
|
antineutrino2/pyGeoPressure
|
d28d6d0672e1abdb57ef97d7285ec68fcdfe8c73
|
[
"MIT"
] | 53 |
2017-11-22T09:00:43.000Z
|
2022-03-20T03:30:58.000Z
|
docs/tutorial/eaton_well.ipynb
|
linwang741/pyGeoPressure
|
862d446c04e09dc347ac0a6e710c714cd3573266
|
[
"MIT"
] | 16 |
2017-12-02T06:11:03.000Z
|
2022-02-09T03:02:05.000Z
|
docs/tutorial/eaton_well.ipynb
|
linwang741/pyGeoPressure
|
862d446c04e09dc347ac0a6e710c714cd3573266
|
[
"MIT"
] | 35 |
2017-11-01T11:38:33.000Z
|
2022-03-21T02:39:57.000Z
| 324.06721 | 38,432 | 0.937882 |
[
[
[
"# Eaton method with well log\n\nPore pressure prediction with Eaton's method using well log data.",
"_____no_output_____"
],
[
"Steps:\n\n1. Calculate Velocity Normal Compaction Trend\n\n2. Optimize for Eaton's exponent n\n\n3. Predict pore pressure using Eaton's method",
"_____no_output_____"
]
],
[
[
"import warnings\nwarnings.filterwarnings(action='ignore')\n\n# for python 2 and 3 compatibility\n# from builtins import str\n# try:\n# from pathlib import Path\n# except:\n# from pathlib2 import Path\n#--------------------------------------------\nimport sys\nppath = \"../..\"\n\nif ppath not in sys.path:\n sys.path.append(ppath)\n#--------------------------------------------",
"_____no_output_____"
],
[
"from __future__ import print_function, division, unicode_literals\n%matplotlib inline\nimport matplotlib.pyplot as plt\n\nplt.style.use(['seaborn-paper', 'seaborn-whitegrid'])\nplt.rcParams['font.sans-serif']=['SimHei']\nplt.rcParams['axes.unicode_minus']=False\n\nimport numpy as np\n\nimport pygeopressure as ppp",
"_____no_output_____"
]
],
[
[
"## 1. Calculate Velocity Normal Compaction Trend",
"_____no_output_____"
],
[
"Create survey with the example survey `CUG`:",
"_____no_output_____"
]
],
[
[
"# set to the directory on your computer\nSURVEY_FOLDER = \"C:/Users/yuhao/Desktop/CUG_depth\"\n\nsurvey = ppp.Survey(SURVEY_FOLDER)",
"_____no_output_____"
]
],
[
[
"Retrieve well `CUG1`:",
"_____no_output_____"
]
],
[
[
"well_cug1 = survey.wells['CUG1']",
"_____no_output_____"
]
],
[
[
"Get velocity log:",
"_____no_output_____"
]
],
[
[
"vel_log = well_cug1.get_log(\"Velocity\")",
"_____no_output_____"
]
],
[
[
"View velocity log:",
"_____no_output_____"
]
],
[
[
"fig_vel, ax_vel = plt.subplots()\nax_vel.invert_yaxis()\nvel_log.plot(ax_vel)\nwell_cug1.plot_horizons(ax_vel)\n\n# set fig style\nax_vel.set(ylim=(5000,0), aspect=(5000/4600)*2)\nax_vel.set_aspect(2)\nfig_vel.set_figheight(8)",
"_____no_output_____"
]
],
[
[
"Optimize for NCT coefficients a, b:\n\n`well.params['horizon']['T20']` returns the depth of horizon T20.",
"_____no_output_____"
]
],
[
[
"a, b = ppp.optimize_nct(\n vel_log=well_cug1.get_log(\"Velocity\"),\n fit_start=well_cug1.params['horizon'][\"T16\"],\n fit_stop=well_cug1.params['horizon'][\"T20\"])",
"_____no_output_____"
]
],
[
[
"And use a, b to calculate normal velocity trend",
"_____no_output_____"
]
],
[
[
"from pygeopressure.velocity.extrapolate import normal_log\nnct_log = normal_log(vel_log, a=a, b=b)",
"_____no_output_____"
]
],
[
[
"View fitted NCT:",
"_____no_output_____"
]
],
[
[
"fig_vel, ax_vel = plt.subplots()\nax_vel.invert_yaxis()\n# plot velocity\nvel_log.plot(ax_vel, label='Velocity')\n# plot horizon\nwell_cug1.plot_horizons(ax_vel)\n# plot fitted nct\nnct_log.plot(ax_vel, color='r', zorder=2, label='NCT')\n\n# set fig style\nax_vel.set(ylim=(5000,0), aspect=(5000/4600)*2)\nax_vel.set_aspect(2)\nax_vel.legend()\nfig_vel.set_figheight(8)",
"_____no_output_____"
]
],
[
[
"Save fitted nct:",
"_____no_output_____"
]
],
[
[
"# well_cug1.params['nct'] = {\"a\": a, \"b\": b}\n\n# well_cug1.save_params()",
"_____no_output_____"
]
],
[
[
"## 2. Optimize for Eaton's exponent n",
"_____no_output_____"
],
[
"First, we need to preprocess velocity.\n\nVelocity log processing (filtering and smoothing):",
"_____no_output_____"
]
],
[
[
"vel_log_filter = ppp.upscale_log(vel_log, freq=20)\n\nvel_log_filter_smooth = ppp.smooth_log(vel_log_filter, window=1501)",
"_____no_output_____"
]
],
[
[
"Veiw processed velocity:",
"_____no_output_____"
]
],
[
[
"fig_vel, ax_vel = plt.subplots()\nax_vel.invert_yaxis()\n# plot velocity\nvel_log.plot(ax_vel, label='Velocity')\n# plot horizon\nwell_cug1.plot_horizons(ax_vel)\n# plot processed velocity\nvel_log_filter_smooth.plot(ax_vel, color='g', zorder=2, label='Processed', linewidth=1)\n\n# set fig style\nax_vel.set(ylim=(5000,0), aspect=(5000/4600)*2)\nax_vel.set_aspect(2)\nax_vel.legend()\nfig_vel.set_figheight(8)",
"_____no_output_____"
]
],
[
[
"We will use the processed velocity data for pressure prediction.",
"_____no_output_____"
],
[
"Optimize Eaton's exponential `n`:",
"_____no_output_____"
]
],
[
[
"n = ppp.optimize_eaton(\n well=well_cug1, \n vel_log=vel_log_filter_smooth, \n obp_log=\"Overburden_Pressure\", \n a=a, b=b)",
"_____no_output_____"
]
],
[
[
"See the RMS error variation with `n`:",
"_____no_output_____"
]
],
[
[
"from pygeopressure.basic.plots import plot_eaton_error\n\nfig_err, ax_err = plt.subplots()\n\nplot_eaton_error(\n ax=ax_err,\n well=well_cug1, \n vel_log=vel_log_filter_smooth, \n obp_log=\"Overburden_Pressure\", \n a=a, b=b)",
"_____no_output_____"
]
],
[
[
"Save optimized n:",
"_____no_output_____"
]
],
[
[
"# well_cug1.params['nct'] = {\"a\": a, \"b\": b}\n\n# well_cug1.save_params()",
"_____no_output_____"
]
],
[
[
"## 3.Predict pore pressure using Eaton's method",
"_____no_output_____"
],
[
"Calculate pore pressure using Eaton's method requires velocity, Eaton's exponential, normal velocity, hydrostatic pressure and overburden pressure.\n\n`Well.eaton()` will try to read saved data, users only need to specify them when they are different from the saved ones.",
"_____no_output_____"
]
],
[
[
"pres_eaton_log = well_cug1.eaton(vel_log_filter_smooth, n=n)",
"_____no_output_____"
]
],
[
[
"View predicted pressure:",
"_____no_output_____"
]
],
[
[
"fig_pres, ax_pres = plt.subplots()\nax_pres.invert_yaxis()\n\nwell_cug1.get_log(\"Overburden_Pressure\").plot(ax_pres, 'g', label='Lithostatic')\nax_pres.plot(well_cug1.hydrostatic, well_cug1.depth, 'g', linestyle='--', label=\"Hydrostatic\")\npres_eaton_log.plot(ax_pres, color='blue', label='Pressure_Eaton')\nwell_cug1.plot_horizons(ax_pres)\n\n# set figure and axis size\nax_pres.set_aspect(2/50)\nax_pres.legend()\nfig_pres.set_figheight(8)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
c50f936fafcf418d6da794ac563ddba38d4f83cd
| 7,823 |
ipynb
|
Jupyter Notebook
|
3-object-tracking-and-localization/activities/6-matrices-and-transformation-state/3. Matrix addition.ipynb
|
S1lv10Fr4gn4n1/udacity-cv
|
ce7aafc41e2c123396d809042973840ea08b850e
|
[
"MIT"
] | null | null | null |
3-object-tracking-and-localization/activities/6-matrices-and-transformation-state/3. Matrix addition.ipynb
|
S1lv10Fr4gn4n1/udacity-cv
|
ce7aafc41e2c123396d809042973840ea08b850e
|
[
"MIT"
] | 3 |
2020-03-24T21:18:48.000Z
|
2021-06-08T21:11:14.000Z
|
3-object-tracking-and-localization/activities/6-matrices-and-transformation-state/3. Matrix addition.ipynb
|
S1lv10Fr4gn4n1/udacity-cv
|
ce7aafc41e2c123396d809042973840ea08b850e
|
[
"MIT"
] | null | null | null | 38.160976 | 1,820 | 0.544804 |
[
[
[
"# Matrix Addition\n\nIn this exercises, you will write a function that accepts two matrices and outputs their sum. Think about how you could do this with a for loop nested inside another for loop.",
"_____no_output_____"
]
],
[
[
"### TODO: Write a function called matrix_addition that \n### calculate the sum of two matrices\n###\n### INPUTS:\n### matrix A _ an m x n matrix\n### matrix B _ an m x n matrix\n###\n### OUPUT:\n### matrixSum _ sum of matrix A + matrix B\n\ndef matrix_addition(matrixA, matrixB):\n\n # initialize matrix to hold the results\n matrixSum = []\n \n # matrix to hold a row for appending sums of each element\n \n \n # TODO: write a for loop within a for loop to iterate over\n # the matrices\n \n for r in range(len(matrixA)):\n row = []\n for c in range(len(matrixA[0])):\n row.append(matrixA[r][c] + matrixB[r][c])\n matrixSum.append(row)\n \n # TODO: As you iterate through the matrices, add matching\n # elements and append the sum to the row variable\n \n # TODO: When a row is filled, append the row to matrixSum. \n # Then reinitialize row as an empty list\n \n return matrixSum\n\n### When you run this code cell, your matrix addition function\n### will run on the A and B matrix.\n\nA = [\n [2,5,1], \n [6,9,7.4], \n [2,1,1], \n [8,5,3], \n [2,1,6], \n [5,3,1]\n]\n\nB = [\n [7, 19, 5.1], \n [6.5,9.2,7.4], \n [2.8,1.5,12], \n [8,5,3], \n [2,1,6], \n [2,33,1]\n]\n\nmatrix_addition(A, B)",
"_____no_output_____"
]
],
[
[
"### Vectors versus Matrices\n\nWhat happens if you run the cell below? Here you are adding two vectors together. Does your code still work?",
"_____no_output_____"
]
],
[
[
"matrix_addition([4, 2, 1], [5, 2, 7])",
"_____no_output_____"
]
],
[
[
"Why did this error occur? Because your code assumes that a matrix is a two-dimensional grid represented by a list of lists. But a horizontal vector, which can also be considered a matrix, is a one-dimensional grid represented by a single list.\n\nWhat happens if you store a vector as a list of lists like [[4, 2, 1]] and [[5, 2, 7]]? Does your function work? Run the code cell below to find out.",
"_____no_output_____"
]
],
[
[
"matrix_addition([[4, 2, 1]], [[5, 2, 7]])",
"_____no_output_____"
]
],
[
[
"### Test your Code\nRun the cell below. If there is no output, then your results are as expected.",
"_____no_output_____"
]
],
[
[
"assert matrix_addition([\n [1, 2, 3]], \n [[4, 5, 6]]) == [[5, 7, 9]]\n\nassert matrix_addition([\n [4]], [\n [5]]) == [[9]]\n\nassert matrix_addition([[1, 2, 3], \n [4, 5, 6]], \n [[7, 8, 9], \n [10, 11, 12]]) == [[8, 10, 12], \n [14, 16, 18]]",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
c50f9a52e98c2922554ce81e637a561cd63b33fc
| 11,512 |
ipynb
|
Jupyter Notebook
|
1.diffusion-example.ipynb
|
gwaygenomics/hetmech
|
17b8b2bbfc6a88929e80292ed0c62a4813f073f2
|
[
"BSD-3-Clause"
] | 10 |
2017-08-20T17:12:57.000Z
|
2019-03-26T21:42:28.000Z
|
1.diffusion-example.ipynb
|
gwaygenomics/hetmech
|
17b8b2bbfc6a88929e80292ed0c62a4813f073f2
|
[
"BSD-3-Clause"
] | 121 |
2017-03-16T19:20:31.000Z
|
2021-01-21T16:15:55.000Z
|
1.diffusion-example.ipynb
|
gwaygenomics/hetmech
|
17b8b2bbfc6a88929e80292ed0c62a4813f073f2
|
[
"BSD-3-Clause"
] | 7 |
2017-03-27T23:02:11.000Z
|
2019-03-27T15:47:52.000Z
| 27.087059 | 132 | 0.451703 |
[
[
[
"# Implementation of diffusion hetmech",
"_____no_output_____"
]
],
[
[
"import pandas\nfrom neo4j.v1 import GraphDatabase\nimport hetio.readwrite\n\nfrom hetmech.diffusion import diffuse",
"_____no_output_____"
],
[
"url = 'https://github.com/dhimmel/hetionet/raw/76550e6c93fbe92124edc71725e8c7dd4ca8b1f5/hetnet/json/hetionet-v1.0.json.bz2'\ngraph = hetio.readwrite.read_graph(url)\nmetagraph = graph.metagraph",
"_____no_output_____"
],
[
"# MetaGraph node/edge count\nmetagraph.n_nodes, metagraph.n_edges",
"_____no_output_____"
],
[
"# Graph node/edge count\ngraph.n_nodes, graph.n_edges",
"_____no_output_____"
],
[
"# Uses the official neo4j-python-driver. See https://github.com/neo4j/neo4j-python-driver\n\nquery = '''\nMATCH (disease:Disease)-[assoc:ASSOCIATES_DaG]-(gene:Gene)\nWHERE disease.name = 'epilepsy syndrome'\nRETURN\n gene.name AS gene_symbol,\n gene.description AS gene_name,\n gene.identifier AS entrez_gene_id,\n assoc.sources AS sources\nORDER BY gene_symbol\n'''\n\ndriver = GraphDatabase.driver(\"bolt://neo4j.het.io\")\nwith driver.session() as session:\n result = session.run(query)\n gene_df = pandas.DataFrame((x.values() for x in result), columns=result.keys())\n\ngene_df.head()",
"_____no_output_____"
],
[
"epilepsy_genes = list()\nfor entrez_gene_id in gene_df.entrez_gene_id:\n node_id = 'Gene', entrez_gene_id\n node = graph.node_dict.get(node_id)\n if node:\n epilepsy_genes.append(node)\nlen(epilepsy_genes)",
"_____no_output_____"
],
[
"metapath = metagraph.metapath_from_abbrev('GiGpBP')\nsource_node_weights = {gene: 1 for gene in epilepsy_genes}\npathway_scores = diffuse(graph, metapath, source_node_weights, column_damping=1, row_damping=1)\ntarget_df = pandas.DataFrame(list(pathway_scores.items()), columns=['target_node', 'score'])\ntarget_df['target_name'] = target_df.target_node.map(lambda x: graph.node_dict[('Biological Process', x)].name)\ntarget_df = target_df.sort_values('score', ascending=False)",
"_____no_output_____"
],
[
"len(target_df)",
"_____no_output_____"
],
[
"sum(target_df.score)",
"_____no_output_____"
],
[
"metapath",
"_____no_output_____"
],
[
"target_df.head()",
"_____no_output_____"
]
],
[
[
"# Diagnosing ubiquitin homeostasis\n\n[ubiquitin homeostasis](http://amigo.geneontology.org/amigo/term/GO:0010992) contains 3 genes: [UBB, UBC, IDE]\n\n```cypher\nMATCH (bp:BiologicalProcess)-[rel:PARTICIPATES_GpBP]-(gene)-[INTERACTS_GiG]-(gene_target)\nWHERE bp.name ='ubiquitin homeostasis'\nRETURN\n gene.name AS ubiquitin_homeostasis_gene,\n count(gene_target) AS n_interacting_genes\n```\n\nReturns the following table:\n\n| ubiquitin_homeostasis_gene | n_interacting_genes |\n|----------------------------|---------------------|\n| IDE | 243 |\n| UBC | 9371 |\n| UBB | 1040 |\n",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
c50faec3f0764c39d26fa3a194a88bf08430d10b
| 3,343 |
ipynb
|
Jupyter Notebook
|
Milestone 3.ipynb
|
CSCI4850/S19-team1-project
|
6626f1d4dd760c519a527cfc5402b2fdcc55c608
|
[
"MIT"
] | null | null | null |
Milestone 3.ipynb
|
CSCI4850/S19-team1-project
|
6626f1d4dd760c519a527cfc5402b2fdcc55c608
|
[
"MIT"
] | null | null | null |
Milestone 3.ipynb
|
CSCI4850/S19-team1-project
|
6626f1d4dd760c519a527cfc5402b2fdcc55c608
|
[
"MIT"
] | 1 |
2019-06-04T00:25:48.000Z
|
2019-06-04T00:25:48.000Z
| 30.669725 | 476 | 0.505534 |
[
[
[
"<h1> Milestone Report 3 </h1>\n<h2> 4/11/2019 </h2>\n<h3> Team Members </h3>\n<ul>\n <li> Armstrong, Ryan </li>\n <li> Nelson, Cory </li>\n <li> Phulwani, Richa </li>\n <li> Smith, Stephen </li>\n <li> Vanlandingham, Mathew </li>\n</ul>",
"_____no_output_____"
],
[
"<table>\n <tr>\n <th> Milestone Chart 3 </th>\n </tr>\n <tr>\n <th> Deliverable </th>\n <th> Percent Complete </th>\n <th> Estimated Completion Date </th>\n <th> Percent Complete by Next Milestone </th>\n </tr>\n <tr>\n <th> Code </th>\n <th> 70 </th>\n <th> April 23 </th>\n <th> 100 </th>\n </tr>\n <tr>\n <th> Paper </th>\n <th> 0 </th>\n <th> April 23 </th>\n <th> 100 </th>\n </tr>\n <tr>\n <th> Demo </th>\n <th> 0 </th>\n <th> May 1 </th>\n <th> 100 </th>\n </tr>\n <tr>\n <th> Presentation </th>\n <th> 0 </th>\n <th> May 1 </th>\n <th> 100 </th>\n </tr>\n</table>",
"_____no_output_____"
],
[
"1. What deliverable goals established in the last milestone report were accomplished to the anticipated percentage?<br>\nAll goals set in the previous milestone were achieved. Creating and training the network were not established in the previous milestone but significant progress has been made in that area regardless. The data was renamed, resized, preprocessed by subtracting the mean, loaded into the network, and the network was made. The network achieved an accuracy of 73% by experimenting with the network's parameters such as kernel size, number of hidden layers, filters etc. \n\n2. What deliverable goals established in the last milestone report were not accomplished to the anticipated percentage?<br>\nN/A\n\n3. What are the main deliverable goals to meet before the next milestone report and who is working on them?<br>\n<ul>\n <li> Tweak network and write demo code - Ryan </li>\n <li> Write paper - Cory </li>\n <li> Collect images for network testing and for demo - Matt </li>\n <li> Work on presentation and aid in writing paper - Richa and Stephen </li>\n</ul>",
"_____no_output_____"
]
]
] |
[
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown"
]
] |
c50fb04b288a7fb5b09ed93a8b80475d0ce33ff9
| 19,400 |
ipynb
|
Jupyter Notebook
|
6-hardware-details.ipynb
|
mwiesenberger/performance
|
dc40bbd2d8740f0293e7fa48a27e2541ae0468d9
|
[
"MIT"
] | 1 |
2020-05-09T09:31:08.000Z
|
2020-05-09T09:31:08.000Z
|
6-hardware-details.ipynb
|
mwiesenberger/performance
|
dc40bbd2d8740f0293e7fa48a27e2541ae0468d9
|
[
"MIT"
] | null | null | null |
6-hardware-details.ipynb
|
mwiesenberger/performance
|
dc40bbd2d8740f0293e7fa48a27e2541ae0468d9
|
[
"MIT"
] | null | null | null | 40.501044 | 155 | 0.435515 |
[
[
[
"### Details on the hardware used to gather the performance data",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nfrom collections import OrderedDict as odict",
"_____no_output_____"
],
[
"#name, cache-size (in kB)\nhardware = odict({})\nhardware['i5'] = ('Intel Core i5-6600 @ 3.30GHz (2x 8GB DDR4, 4 cores)',6144, \n '1 MPI task x 4 OpenMP threads (1 per core)')\nhardware['skl'] = ('2x Intel Xeon 8160 (Skylake) at 2.10 GHz (12x 16GB DDR4, 2x 24 cores)',2*33000, \n '2 MPI tasks (1 per socket) x 24 OpenMP threads (1 per core)')\nhardware['knl'] = ('Intel Xeon Phi 7250 (Knights Landing) at 1.40 GHz (16GB MCDRAM, 68 cores)',34000, \n '1 MPI task x 136 OpenMP hyperthreads (2 per core)')\nhardware['gtx1060'] = ('Nvidia GeForce GTX 1060 (6GB global memory)',1572.864, '1 MPI task per GPU')\nhardware['p100'] = ('Nvidia Tesla P100-PCIe (16GB global memory)',4194.304, '1 MPI task per GPU')\nhardware['v100'] = ('Nvidia Tesla V100-PCIe (16GB global memory)',6291.456, '1 MPI task per GPU')\nhardware['p100nv'] = ('Nvidia Tesla P100-Power8 (16GB global memory)',4194.304, '1 MPI task per GPU')\nhardware['v100nv'] = ('Nvidia Tesla V100-Power9 (16GB global memory)',6291.456, '1 MPI task per GPU')\n\nmemory =odict({}) #find with 'dmidecode --type 17'\n#name, I/O bus clockrate (MHz) , buswidth (bit), size (MB),\nmemory['i5'] = ('2x 8GB Kingston DDR4', 1066, 2*64, 2*8192) #ECC: no (actually it is DDR4-2400 but i5 has max DDR4-2133)\nmemory['skl'] = ('12x 16GB DDR4',1333,12*64,12*16000) #ECC: ?\nmemory['knl'] = ('MCDRAM',None,None,16000) #ECC: ?\nmemory['gtx1060'] = ('on-card global memory',4004,192,6069) #ECC: no\nmemory['p100'] = ('on-card global memory',715,4096,16276) # ECC: yes\nmemory['v100'] = ('on-card global memory',877,4096,16152) # ECC: yes\ncompiler=odict({})\ncompiler['i5'] = ('mpic++ (gcc-5.4) -mavx -mfma -O3 -fopenmp')\ncompiler['skl'] = ('mpiicc-17.0.4 -mt_mpi -xCORE-AVX512 -mtune=skylake -O3 -restrict -fp-model precise -fimf-arch-consistency=true -qopenmp')\ncompiler['knl'] = ('mpiicc-17.0.4 -mt_mpi -xMIC-AVX512 -O3 -restrict -fp-model precise -fimf-arch-consistency=true -qopenmp')\ncompiler['gtx1060'] = ('nvcc-7.0 --compiler-bindir mpic++ (gcc-5.4) -O3 -arch sm_35')\ncompiler['p100'] = ('nvcc-8.0 --compiler-bindir mpic++ (gcc-5.4) -O3 -arch sm_60 -Xcompiler \"-O3 -mavx -mfma\"')\ncompiler['v100'] = ('nvcc-8.0 --compiler-bindir mpic++ (gcc-5.4) -O3 -arch sm_60 -Xcompiler \"-O3 -mavx -mfma\"')",
"_____no_output_____"
],
[
"df = pd.DataFrame(hardware)\ndf = df.transpose()\ndf.columns= ['device-name', 'cache-size-kB','single-node configuration']\ncom = pd.DataFrame(compiler, index=['compiler flags'])\ncom = com.transpose()\ncom\ndf = df.join(com)\nmem = pd.DataFrame(memory)\nmem = mem.transpose()\nmem.columns = ['mem-description', 'clockrate-MHz', 'buswidth-bit', 'size-MB']\ndf=df.join(mem)\n#df",
"_____no_output_____"
]
],
[
[
"From the available data we can compute the theoretical memory bandwidth via $$bw = 2*clockrate*buswidth$$ where the '2' is for double data rate (DDR)",
"_____no_output_____"
]
],
[
[
"df['bandwidth'] = 2*df['clockrate-MHz']*1e6*df['buswidth-bit']/8/1e9\n#df",
"_____no_output_____"
]
],
[
[
"Let us compare the theoretical bandwidth with our previously measured peak bandwidth from axpby",
"_____no_output_____"
]
],
[
[
"exp = pd.read_csv('performance.csv',delimiter=' ')\nexp.set_index('arch',inplace=True)\nexp.index.name = None\ndf = df.join(exp['axpby_bw'])\ndf['mem_efficiency']=df['axpby_bw']/df['bandwidth']*100",
"_____no_output_____"
],
[
"pd.set_option('display.float_format', lambda x: '%.2f' % x)\ndf",
"_____no_output_____"
]
],
[
[
"Let us write a summarized LateX table to be used for publication",
"_____no_output_____"
]
],
[
[
"pd.set_option('precision',3)\nfile = df.loc[:,['device-name','single-node configuration']]#,'bandwidth']]\n#file.loc['knl','bandwidth'] = '>400'\nfile.columns = ['device description', 'single-node configuration']#, 'bandwidth [GB/s]']\nfilename='hardware.tex'\ndf.loc['knl','bandwidth'] = '$>$400'\npd.set_option('display.max_colwidth', 200)\nwith open(filename, 'wb') as f:\n f.write(bytes(file.to_latex(\n column_format='@{}lp{6.5cm}p{5cm}@{}',\n bold_rows=True),'UTF-8'))\nfile",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
c50fbce61306354a35271298ba2ffb87f36938fd
| 20,413 |
ipynb
|
Jupyter Notebook
|
01_Getting_&_Knowing_Your_Data/World Food Facts/Exercises.ipynb
|
Ryan-Efendy/pandas_exercises
|
94eaae5fa80050ce2cf8c34b3832f091670a15b6
|
[
"BSD-3-Clause"
] | null | null | null |
01_Getting_&_Knowing_Your_Data/World Food Facts/Exercises.ipynb
|
Ryan-Efendy/pandas_exercises
|
94eaae5fa80050ce2cf8c34b3832f091670a15b6
|
[
"BSD-3-Clause"
] | null | null | null |
01_Getting_&_Knowing_Your_Data/World Food Facts/Exercises.ipynb
|
Ryan-Efendy/pandas_exercises
|
94eaae5fa80050ce2cf8c34b3832f091670a15b6
|
[
"BSD-3-Clause"
] | null | null | null | 30.241481 | 228 | 0.423309 |
[
[
[
"# Exercise 1",
"_____no_output_____"
],
[
"### Step 1. Go to https://www.kaggle.com/openfoodfacts/world-food-facts/data",
"_____no_output_____"
],
[
"### Step 2. Download the dataset to your computer and unzip it.",
"_____no_output_____"
],
[
"### Step 3. Use the tsv file and assign it to a dataframe called food",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\n\nfood = pd.read_csv('/Users/ryanefendy/Downloads/en.openfoodfacts.org.products.tsv', sep='\\t')",
"/anaconda3/lib/python3.7/site-packages/IPython/core/interactiveshell.py:3020: DtypeWarning: Columns (0,3,5,19,20,24,25,26,27,28,36,37,38,39,48) have mixed types. Specify dtype option on import or set low_memory=False.\n interactivity=interactivity, compiler=compiler, result=result)\n"
]
],
[
[
"### Step 4. See the first 5 entries",
"_____no_output_____"
]
],
[
[
"food.head(5)",
"_____no_output_____"
]
],
[
[
"### Step 5. What is the number of ~observations~ in the dataset?",
"_____no_output_____"
]
],
[
[
"# food.shape\nlen(food.index)",
"_____no_output_____"
]
],
[
[
"### Step 6. What is the number of columns in the dataset?",
"_____no_output_____"
]
],
[
[
"# len(food.columns)\n# food.shape[1]\nfood.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 356027 entries, 0 to 356026\nColumns: 163 entries, code to water-hardness_100g\ndtypes: float64(107), object(56)\nmemory usage: 442.8+ MB\n"
]
],
[
[
"### Step 7. Print the name of all the columns.",
"_____no_output_____"
]
],
[
[
"list(food.columns)",
"_____no_output_____"
]
],
[
[
"### Step 8. What is the name of 105th column?",
"_____no_output_____"
]
],
[
[
"list(food.columns)[104]",
"_____no_output_____"
]
],
[
[
"### Step 9. What is the type of the observations of the 105th column?",
"_____no_output_____"
]
],
[
[
"food['-glucose_100g'].dtypes",
"_____no_output_____"
]
],
[
[
"### Step 10. How is the dataset indexed?",
"_____no_output_____"
]
],
[
[
"food.index",
"_____no_output_____"
]
],
[
[
"### Step 11. What is the product name of the 19th observation?",
"_____no_output_____"
]
],
[
[
"food.iloc[18]['product_name']",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
c50fc85cf4c64a6adbc9ad1575576de0b2de5a39
| 219,748 |
ipynb
|
Jupyter Notebook
|
ModelCombo.ipynb
|
Marshall-mk/Brain-tumor-classification-using-MR-images
|
13c4a4722af32bf53c92da7d6454ce46a411bb50
|
[
"BSD-2-Clause"
] | null | null | null |
ModelCombo.ipynb
|
Marshall-mk/Brain-tumor-classification-using-MR-images
|
13c4a4722af32bf53c92da7d6454ce46a411bb50
|
[
"BSD-2-Clause"
] | null | null | null |
ModelCombo.ipynb
|
Marshall-mk/Brain-tumor-classification-using-MR-images
|
13c4a4722af32bf53c92da7d6454ce46a411bb50
|
[
"BSD-2-Clause"
] | null | null | null | 255.224158 | 33,796 | 0.903653 |
[
[
[
"import tensorflow as tf \nfrom tensorflow import keras\nfrom keras.preprocessing.image import ImageDataGenerator\nimport scipy\nimport os\nimport cv2\nimport random\nfrom skimage import io\nimport seaborn as sns\nfrom matplotlib import pyplot \nimport pandas as pd\nimport tensorflow.keras.backend as K\nimport numpy as np\nnp.random.seed(13)\nfrom sklearn.model_selection import train_test_split\nimport matplotlib.pyplot as plt",
"2.7.0-dev20210830\n"
],
[
"NasNet = tf.keras.models.load_model('my_firstModel')\nMobileNet = tf.keras.models.load_model('my_secondModel')\nMobileNetV2 = tf.keras.models.load_model('my_thirdModel')\nEfficientNet = tf.keras.models.load_model('my_forthModel')\nTrainedModel_0 = [NasNet,MobileNet,MobileNetV2,EfficientNet]\nTrainedModel_1 = [NasNet,MobileNet]\nTrainedModel_2 = [MobileNetV2,EfficientNet]\nTrainedModel_3 = [NasNet,EfficientNet]\nTrainedModel_4 = [MobileNet,MobileNetV2]\nTrainedModel_5 = [NasNet,MobileNet,MobileNetV2]\nTrainedModel_6 = [MobileNet,MobileNetV2,EfficientNet]\nTrainedModel_7 = [NasNet,MobileNet,EfficientNet]\nTrainedModel_8 = [NasNet,MobileNetV2,EfficientNet]",
"_____no_output_____"
],
[
"#Changed to working directory to the location of the dataset and loaded the dataset description for local host.\nos.chdir(\"..\\kaggle_3m\")\nbrain_df = pd.read_csv('data_frame.csv')\n\n#For google colab\n#brain_df = pd.read_csv('/content/drive/MyDrive/kaggle_3m/Gdrive.csv')\n# Here we decided to drop unncessary coloums from the dataset\nbrain_df_train = brain_df.drop(columns=['Unnamed: 0', 'patient'], axis=1)\nbrain_df_train['diagnosis'] = brain_df['diagnosis'].apply(lambda x: str(x)) #changes the type of the values of the column to sting\nbrain_df_train.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 3929 entries, 0 to 3928\nData columns (total 3 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 image_path 3929 non-null object\n 1 mask_path 3929 non-null object\n 2 diagnosis 3929 non-null object\ndtypes: object(3)\nmemory usage: 92.2+ KB\n"
],
[
"train, test = train_test_split(brain_df_train, test_size=0.15) #splits the data into training and testing sets\ndatagen = ImageDataGenerator(rescale=1./255., validation_split=0.1)\n\ntrain_generator = datagen.flow_from_dataframe(train,\n directory='./',\n x_col='image_path',\n y_col='diagnosis',\n subset='training',\n class_mode='categorical',\n batch_size=16,\n shuffle=True,\n target_size=(224,224)\n )\nvalid_generator = datagen.flow_from_dataframe(train,\n directory='./',\n x_col='image_path',\n y_col='diagnosis',\n subset='validation',\n class_mode='categorical',\n batch_size=16,\n shuffle=True,\n target_size=(224,224)\n )\ntest_datagen = ImageDataGenerator(rescale=1./255.)\ntest_generator = test_datagen.flow_from_dataframe(test,\n directory='./',\n x_col='image_path',\n y_col='diagnosis',\n class_mode='categorical',\n batch_size=16,\n shuffle=False,\n target_size=(224,224)\n )",
"Found 3006 validated image filenames belonging to 2 classes.\nFound 333 validated image filenames belonging to 2 classes.\nFound 590 validated image filenames belonging to 2 classes.\n"
],
[
"#For TrainedModel_0\nlabels_0 = [] \nfor m in TrainedModel_0:\n predicts = np.argmax(m.predict(test_generator), axis=1)\n labels_0.append(predicts)\n# Ensemble with voting\nlabels_0 = np.array(labels_0)\nlabels_0 = np.transpose(labels_0, (1, 0))\nlabls_0 = scipy.stats.mode(labels_0,axis=1)[0]\nlabls_0 = np.squeeze(labls_0)\n\n\n#For TrainedModel_1\nlabels_1 = [] \nfor m in TrainedModel_1:\n predicts = np.argmax(m.predict(test_generator), axis=1)\n labels_1.append(predicts)\n# Ensemble with voting\nlabels_1 = np.array(labels_1)\nlabels_1 = np.transpose(labels_1, (1, 0))\nlabls_1 = scipy.stats.mode(labels_1,axis=1)[0]\nlabls_1 = np.squeeze(labls_1)\n\n#For TrainedModel_2\nlabels_2 = [] \nfor m in TrainedModel_2:\n predicts = np.argmax(m.predict(test_generator), axis=1)\n labels_2.append(predicts)\n# Ensemble with voting\nlabels_2 = np.array(labels_2)\nlabels_2 = np.transpose(labels_2, (1, 0))\nlabls_2 = scipy.stats.mode(labels_2,axis=1)[0]\nlabls_2 = np.squeeze(labls_2)\n\n#For TrainedModel_3\nlabels_3 = [] \nfor m in TrainedModel_3:\n predicts = np.argmax(m.predict(test_generator), axis=1)\n labels_3.append(predicts)\n# Ensemble with voting\nlabels_3 = np.array(labels_3)\nlabels_3 = np.transpose(labels_3, (1, 0))\nlabls_3 = scipy.stats.mode(labels_3,axis=1)[0]\nlabls_3 = np.squeeze(labls_3)\n\n#For TrainedModel_4\nlabels_4 = [] \nfor m in TrainedModel_4:\n predicts = np.argmax(m.predict(test_generator), axis=1)\n labels_4.append(predicts)\n# Ensemble with voting\nlabels_4 = np.array(labels_4)\nlabels_4 = np.transpose(labels_4, (1, 0))\nlabls_4 = scipy.stats.mode(labels_4,axis=1)[0]\nlabls_4 = np.squeeze(labls_4)\n\n#For TrainedModel_5\nlabels_5 = [] \nfor m in TrainedModel_5:\n predicts = np.argmax(m.predict(test_generator), axis=1)\n labels_5.append(predicts)\n# Ensemble with voting\nlabels_5 = np.array(labels_5)\nlabels_5 = np.transpose(labels_5, (1, 0))\nlabls_5 = scipy.stats.mode(labels_5,axis=1)[0]\nlabls_5 = np.squeeze(labls_5)\n\n#For TrainedModel_6\nlabels_6 = [] \nfor m in TrainedModel_6:\n predicts = np.argmax(m.predict(test_generator), axis=1)\n labels_6.append(predicts)\n# Ensemble with voting\nlabels_6 = np.array(labels_6)\nlabels_6 = np.transpose(labels_6, (1, 0))\nlabls_6 = scipy.stats.mode(labels_6,axis=1)[0]\nlabls_6 = np.squeeze(labls_6)\n\n#For TrainedModel_7\nlabels_7 = [] \nfor m in TrainedModel_7:\n predicts = np.argmax(m.predict(test_generator), axis=1)\n labels_7.append(predicts)\n# Ensemble with voting\nlabels_7 = np.array(labels_7)\nlabels_7 = np.transpose(labels_7, (1, 0))\nlabls_7 = scipy.stats.mode(labels_7,axis=1)[0]\nlabls_7 = np.squeeze(labls_7)\n\n#For TrainedModel_8\nlabels_8 = [] \nfor m in TrainedModel_8:\n predicts = np.argmax(m.predict(test_generator), axis=1)\n labels_8.append(predicts)\n# Ensemble with voting\nlabels_8 = np.array(labels_8)\nlabels_8 = np.transpose(labels_8, (1, 0))\nlabls_8 = scipy.stats.mode(labels_8,axis=1)[0]\nlabls_8= np.squeeze(labls_8)",
"_____no_output_____"
],
[
"original = np.asarray(test['diagnosis']).astype('int')",
"_____no_output_____"
],
[
"from sklearn.metrics import accuracy_score, confusion_matrix, classification_report\nimport seaborn as sns\n\ncm_0 = confusion_matrix(original, labls_0)\ncm_1 = confusion_matrix(original, labls_1)\ncm_2 = confusion_matrix(original, labls_2)\ncm_3 = confusion_matrix(original, labls_3)\ncm_4 = confusion_matrix(original, labls_4)\ncm_5 = confusion_matrix(original, labls_5)\ncm_6 = confusion_matrix(original, labls_6)\ncm_7 = confusion_matrix(original, labls_7)\ncm_8 = confusion_matrix(original, labls_8)\n\nreport_0 = classification_report(original, labls_0, labels = [0,1])\nreport_1 = classification_report(original, labls_1, labels = [0,1])\nreport_2 = classification_report(original, labls_2, labels = [0,1])\nreport_3 = classification_report(original, labls_3, labels = [0,1])\nreport_4 = classification_report(original, labls_4, labels = [0,1])\nreport_5 = classification_report(original, labls_5, labels = [0,1])\nreport_6 = classification_report(original, labls_6, labels = [0,1])\nreport_7 = classification_report(original, labls_7, labels = [0,1])\nreport_8 = classification_report(original, labls_8, labels = [0,1])\n\nprint(report_0)\npyplot.figure(figsize = (5,5))\nsns.heatmap(cm_0, annot=True)\n\nprint(report_1)\npyplot.figure(figsize = (5,5))\nsns.heatmap(cm_1, annot=True) \n\nprint(report_2)\npyplot.figure(figsize = (5,5))\nsns.heatmap(cm_2, annot=True) \n\nprint(report_3)\npyplot.figure(figsize = (5,5))\nsns.heatmap(cm_3, annot=True) \n\nprint(report_4)\npyplot.figure(figsize = (5,5))\nsns.heatmap(cm_4, annot=True)\n\n\nprint(report_5)\npyplot.figure(figsize = (5,5))\nsns.heatmap(cm_5, annot=True)\n\nprint(report_6)\npyplot.figure(figsize = (5,5))\nsns.heatmap(cm_6, annot=True)\n\nprint(report_7)\npyplot.figure(figsize = (5,5))\nsns.heatmap(cm_7, annot=True)\n\nprint(report_8)\npyplot.figure(figsize = (5,5))\nsns.heatmap(cm_8, annot=True)",
"c:\\users\\hi\\appdata\\local\\programs\\python\\python39\\lib\\site-packages\\sklearn\\metrics\\_classification.py:1248: UndefinedMetricWarning: Precision and F-score are ill-defined and being set to 0.0 in labels with no predicted samples. Use `zero_division` parameter to control this behavior.\n _warn_prf(average, modifier, msg_start, len(result))\nc:\\users\\hi\\appdata\\local\\programs\\python\\python39\\lib\\site-packages\\sklearn\\metrics\\_classification.py:1248: UndefinedMetricWarning: Precision and F-score are ill-defined and being set to 0.0 in labels with no predicted samples. Use `zero_division` parameter to control this behavior.\n _warn_prf(average, modifier, msg_start, len(result))\nc:\\users\\hi\\appdata\\local\\programs\\python\\python39\\lib\\site-packages\\sklearn\\metrics\\_classification.py:1248: UndefinedMetricWarning: Precision and F-score are ill-defined and being set to 0.0 in labels with no predicted samples. Use `zero_division` parameter to control this behavior.\n _warn_prf(average, modifier, msg_start, len(result))\nc:\\users\\hi\\appdata\\local\\programs\\python\\python39\\lib\\site-packages\\sklearn\\metrics\\_classification.py:1248: UndefinedMetricWarning: Precision and F-score are ill-defined and being set to 0.0 in labels with no predicted samples. Use `zero_division` parameter to control this behavior.\n _warn_prf(average, modifier, msg_start, len(result))\nc:\\users\\hi\\appdata\\local\\programs\\python\\python39\\lib\\site-packages\\sklearn\\metrics\\_classification.py:1248: UndefinedMetricWarning: Precision and F-score are ill-defined and being set to 0.0 in labels with no predicted samples. Use `zero_division` parameter to control this behavior.\n _warn_prf(average, modifier, msg_start, len(result))\nc:\\users\\hi\\appdata\\local\\programs\\python\\python39\\lib\\site-packages\\sklearn\\metrics\\_classification.py:1248: UndefinedMetricWarning: Precision and F-score are ill-defined and being set to 0.0 in labels with no predicted samples. Use `zero_division` parameter to control this behavior.\n _warn_prf(average, modifier, msg_start, len(result))\n"
],
[
"# Here we create a figure instance, and two subplots\nfig = plt.figure(figsize = (20,20)) # width x height\nax1 = fig.add_subplot(2, 2, 1) # row, column, position\nax2 = fig.add_subplot(2, 2, 2)\nax3 = fig.add_subplot(2, 2, 3)\nax4 = fig.add_subplot(2, 2, 4)\n\n\n\n\n# We use ax parameter to tell seaborn which subplot to use for this plot\nsns.heatmap(cm_0, ax=ax1, square=True, cbar_kws={'shrink': .3}, annot=True, annot_kws={'fontsize': 12})\nsns.heatmap(cm_1, ax=ax2, square=True, cbar_kws={'shrink': .3}, annot=True, annot_kws={'fontsize': 12})\nsns.heatmap(cm_2, ax=ax3, square=True, cbar_kws={'shrink': .3}, annot=True, annot_kws={'fontsize': 12})\nsns.heatmap(cm_3, ax=ax4, square=True, cbar_kws={'shrink': .3}, annot=True, annot_kws={'fontsize': 12})\n",
"_____no_output_____"
],
[
"\nfig = plt.figure(figsize = (20,20))\nax1 = fig.add_subplot(2, 2, 1) \nax2 = fig.add_subplot(2, 2, 2)\nax3 = fig.add_subplot(2, 2, 3)\nax4 = fig.add_subplot(2, 2, 4)\n\nsns.heatmap(cm_4, ax=ax1, square=True, cbar_kws={'shrink': .3}, annot=True, annot_kws={'fontsize': 12})\nsns.heatmap(cm_5, ax=ax2, square=True, cbar_kws={'shrink': .3}, annot=True, annot_kws={'fontsize': 12})\nsns.heatmap(cm_6, ax=ax3, square=True, cbar_kws={'shrink': .3}, annot=True, annot_kws={'fontsize': 12})\nsns.heatmap(cm_7, ax=ax4, square=True, cbar_kws={'shrink': .3}, annot=True, annot_kws={'fontsize': 12})\n",
"_____no_output_____"
],
[
"pyplot.figure(figsize = (5,5))\nsns.heatmap(cm_8, annot=True)",
"_____no_output_____"
],
[
"#plot graphs\ndata = {'Combo 1 ':93, 'Combo 2':96, 'Combo 3':64,\n 'Combo 4':64, 'Combo 5':93, 'Combo 6':90, 'Combo 7':93, 'Combo 8':96,'Combo 9':87}\nnames = list(data.keys())\nvalues = list(data.values())\n \nfig = plt.figure(figsize = (10, 5))\n \n# creating the bar plot\nplt.bar(names, values, color ='blue',\n width = 0.4)\n \nplt.xlabel(\"The different Combinations\")\nplt.ylabel(\"Accuracy of the models\")\nplt.title(\"Graph of Accuracy vs several model combinations\")\nplt.show()",
"_____no_output_____"
],
[
"# set width of bar\nbarWidth = 0.25\nfig = plt.subplots(figsize =(12, 8))\n \n# set height of bar\nAccuracy = [93, 96, 64, 64, 93, 90, 93, 96, 87]\nPrecision = [91, 96, 64, 64, 91, 96, 91, 96, 90]\nRecall = [99, 97, 100, 100, 99, 88, 99, 97, 90]\n \n# Set position of bar on X axis\nbr1 = np.arange(len(Recall))\nbr2 = [x + barWidth for x in br1]\nbr3 = [x + barWidth for x in br2]\nbr4 = [x + barWidth for x in br3]\nbr5 = [x + barWidth for x in br4]\nbr6 = [x + barWidth for x in br5]\nbr7 = [x + barWidth for x in br6]\nbr8 = [x + barWidth for x in br7]\nbr9 = [x + barWidth for x in br8]\n \n# Make the plot\nplt.bar(br1, Accuracy, color ='black', width = barWidth,\n edgecolor ='grey', label ='Accuracy')\nplt.bar(br2, Recall, color ='yellow', width = barWidth,\n edgecolor ='grey', label ='Recall')\nplt.bar(br3, Precision, color ='grey', width = barWidth,\n edgecolor ='grey', label ='precision')\n\n \n# Adding Xticks\nplt.xlabel('Combinations', fontweight ='bold', fontsize = 15)\nplt.ylabel('Performance', fontweight ='bold', fontsize = 15)\nplt.xticks([r + barWidth for r in range(len(Recall))],\n ['Combo1', 'Combo2', 'Combo3', 'Combo4', 'Combo5','Combo6', 'Combo7', 'Combo8', 'Combo9'])\nplt.grid() \nplt.legend()\nplt.show()",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
c50fcd9870d2b33701b91ff62d5a1cd82d1c2bbc
| 534,256 |
ipynb
|
Jupyter Notebook
|
HW1/Homework_01_Enlik.ipynb
|
enliktjioe/bda2020
|
6f264113814a0748d12a7a1ae230d74105aefeb5
|
[
"MIT"
] | null | null | null |
HW1/Homework_01_Enlik.ipynb
|
enliktjioe/bda2020
|
6f264113814a0748d12a7a1ae230d74105aefeb5
|
[
"MIT"
] | null | null | null |
HW1/Homework_01_Enlik.ipynb
|
enliktjioe/bda2020
|
6f264113814a0748d12a7a1ae230d74105aefeb5
|
[
"MIT"
] | 2 |
2020-06-06T20:45:31.000Z
|
2021-04-18T07:04:24.000Z
| 160.872026 | 44,412 | 0.863309 |
[
[
[
"<div class=\"alert alert-block alert-info\"><b></b>\n<h1><center> <font color='black'> Homework 01 </font></center></h1>\n<h2><center> <font color='black'> Introduction and first look at the data </font></center></h2> \n<h2><center> <font color='black'> BDA - University of Tartu - Spring 2020</font></center></h3>\n</div>",
"_____no_output_____"
],
[
"# 1. Python and Pandas (4.5 points)",
"_____no_output_____"
],
[
"* In this course we are going to use Python, and mainly Pandas data structures and data analysis tool. please make sure that you have went through the practise session carefully. After that please answer the following questions below about the abalone dataset.",
"_____no_output_____"
],
[
"First, let's import pandas package as pd and read data from the CSV file into a pandas DataFrame.",
"_____no_output_____"
]
],
[
[
"# Remove future warning message\nimport warnings\nwarnings.simplefilter(action='ignore', category=FutureWarning)",
"_____no_output_____"
],
[
"import pandas as pd\ndata = pd.read_csv(\"abalone.csv\")\ndata",
"_____no_output_____"
]
],
[
[
"`Question 0` is an example of how you should answer the following questions. First, in the `#TODO` cell you have to write the code representing your solution. And in the answer cell write the answer with your own words. It does not have to be a full sentence.",
"_____no_output_____"
],
[
" **1.0. What is overall memory consumption of the data?** (EXAMPLE)",
"_____no_output_____"
]
],
[
[
"# TODO - here you have to write the code how did you find the answer.\ndata.memory_usage(deep=True).sum()",
"_____no_output_____"
]
],
[
[
"**<font color='red'>Answer:</font>** The total memory usage is 102080 bytes (102,08 kb).",
"_____no_output_____"
],
[
" **1.1. What are the column names of the dataset?** **(0.25 point)**",
"_____no_output_____"
]
],
[
[
"# TODO\ndata.columns",
"_____no_output_____"
]
],
[
[
"**<font color='red'>Answer:</font>** 'Gender', 'Length', 'Diameter', 'Height', 'Weight', 'Rings'",
"_____no_output_____"
],
[
"**1.2. How many observations (i.e. rows) are in this data frame?** **(0.25 point)**",
"_____no_output_____"
]
],
[
[
"# TODO\ndata.index",
"_____no_output_____"
]
],
[
[
"**<font color='red'>Answer:</font>** 1000",
"_____no_output_____"
],
[
"**1.3. Print the first 5 lines from the dataset. What are the values of feature \"Rings\" of the printed observations?** **(0.25 point)**",
"_____no_output_____"
]
],
[
[
"# TODO\ndata.head()",
"_____no_output_____"
]
],
[
[
"**<font color='red'>Answer:</font>** 12, 9, 8, 7, 10",
"_____no_output_____"
],
[
"**1.4. Extract the last 3 rows of the data frame. What is the \"Weight\" of these abalones?** **(0.25 point)**",
"_____no_output_____"
]
],
[
[
"# TODO\ndata['Weight'].tail(3)",
"_____no_output_____"
]
],
[
[
"**<font color='red'>Answer:</font>** 1.3195, 0.6455, 1.0070",
"_____no_output_____"
],
[
"**1.5. What is the value of diameter in the row 577?** **(1 point)**",
"_____no_output_____"
]
],
[
[
"# TODO\ndata.loc[576]['Diameter']",
"_____no_output_____"
]
],
[
[
"**<font color='red'>Answer:</font>** 0.51",
"_____no_output_____"
],
[
"**1.6. What is the mean of the height column?** **(0.5 point)** <br>\nHint 1: Use pandas describe() method",
"_____no_output_____"
]
],
[
[
"# TODO\ndata['Height'].describe()",
"_____no_output_____"
]
],
[
[
"**<font color='red'>Answer:</font>** 0.141100",
"_____no_output_____"
],
[
"**1.7. Extract the subset of rows of the data frame where gender is M and weight values are below 0.75. What is the mean of diameter in this subset?** **(1 point)**",
"_____no_output_____"
]
],
[
[
"# TODO\ndata[(data['Gender'] == \"M\") & (data['Weight'] < 0.75)].describe()",
"_____no_output_____"
]
],
[
[
"**<font color='red'>Answer:</font>** 0.342647",
"_____no_output_____"
],
[
"**1.8. What is the minimum of length when rings is equal to 18?** **(1 point)**",
"_____no_output_____"
]
],
[
[
"# TODO\ndata[data['Rings'] == 18].describe()",
"_____no_output_____"
]
],
[
[
"**<font color='red'>Answer:</font>** 0.465000",
"_____no_output_____"
],
[
"# 2. Data understanding (2.5 point)",
"_____no_output_____"
],
[
"Collecting, describing, exploring and verifying data quality are <b>very important</b> steps in any data science task. In this exercise the main goal is to understand the attributes in the 'adult' dataset.<br>",
"_____no_output_____"
],
[
"**2.0. Load data from the `adult.csv` file.** <br>\nHint 1: The extension of the filename may not always reflect the structure of data precisely. Are the values comma-separated (csv) or not? ",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\n\n# TODO\ndata = pd.read_table('adult.csv', index_col='X')\ndata",
"_____no_output_____"
]
],
[
[
"**2.1. Name all the columns in this dataset and describe the meaning of each column with one sentence.** <br>",
"_____no_output_____"
]
],
[
[
"# TODO\ndata.columns",
"_____no_output_____"
]
],
[
[
"**<font color='red'>Answer:</font>**\n- age = the current age of the adult person\n- workclass = the type of work including: private, self-emp-inc, state-gov\n- education = the latest education level\n- occupation = the name of the job\n- capital.gain = a rise in the value of a capital asset (investment or real estate) that gives it a higher worth than the purchase price [(reference)](https://www.investopedia.com/terms/c/capitalgain.asp)\n- capital.loss = the loss incurred when a capital asset, such as an investment or real estate, decreases in value [(reference)](https://www.investopedia.com/terms/c/capitalloss.asp)\n- native.country = the country someone is born in or native to\n- salaries = the annual salary of person in USD\n- jobsatisfication = a measure of workers' contentedness with their job, whether or not they like the job or individual aspects or facets of jobs, such as nature of work or supervision. [(reference)](https://en.wikipedia.org/wiki/Job_satisfaction)\n- male = describe the gender is male or not \n- female = describe the gender is female or not",
"_____no_output_____"
],
[
"#### 2.1.1. In the following subtasks, you will have to visualise the distribution of the values for different features using appropriate diagrams. Visualisation is one of the easiest ways to get a good overview of the data and to notice any oddities, outliers etc.",
"_____no_output_____"
]
],
[
[
"# Matplotlib is a Python 2D plotting library which produces publication quality figures in a variety of hardcopy formats \n# and interactive environments across platforms. Matplotlib can be used in Python scripts, the Python and IPython shells, \n# the Jupyter notebook, web application servers, and four graphical user interface toolkits.\nimport matplotlib.pyplot as plt\n\n# allows to output plots in the notebook\n%matplotlib inline \n\n# Set the default style\nplt.style.use(\"ggplot\") ",
"_____no_output_____"
]
],
[
[
"As an example, here is the age frequency histogram, where age is an attribute of type **numeric**:",
"_____no_output_____"
]
],
[
[
"# Columns may contain non-numeric values, errors or missing values. Therefore, non-numeric values must be dealt with.\nif data is not ...:\n pd.to_numeric(data['age'], errors='coerce').hist(bins=30) # ‘coerce’ -> invalid parsing will be set as NaN\nelse:\n print('Please define `data` in earlier subtasks')",
"_____no_output_____"
]
],
[
[
"**2.2. Attribute `Workclass`. First, specify the type of the attribute (nominal / ordinal / numeric) and second, choose an appropriate chart type and visualise the distribution in the relative frequency scale.**",
"_____no_output_____"
],
[
"**<font color='red'>Attribute type :</font>** Nominal",
"_____no_output_____"
]
],
[
[
"# TODO\n# data.workclass.value_counts().plot(kind=\"bar\")\nfig = plt.figure(figsize=[16, 8])\n\nax = plt.subplot(111)\nax.hist(data['workclass'], \n width = 0.5,\n weights = np.ones(len(data['workclass'])) / len(data['workclass']) \n )\nax.set_title('Workclass - Distribution in Relative Frequency')\nplt.text\nplt.xticks(rotation=90)\nplt.show()",
"_____no_output_____"
]
],
[
[
"**2.3. Attribute `Education`. First, specify the type of the attribute (nominal / ordinal / numeric) and second, choose an appropriate chart type and visualise the distribution in the relative frequency scale.**",
"_____no_output_____"
],
[
"**<font color='red'>Attribute type :</font>** Ordinal",
"_____no_output_____"
]
],
[
[
"# TODO\nfig = plt.figure(figsize=[16, 8])\n\nax2 = plt.subplot(111)\nax2.hist(data['education'], \n width = 0.5,\n weights = np.ones(len(data['education'])) / len(data['education']) \n )\nax2.set_title('Education - Distribution in Relative Frequency')\nplt.xticks(rotation=90)\nplt.show()",
"_____no_output_____"
]
],
[
[
"**2.4. Attribute `Occupation`. First, specify the type of the attribute (nominal / ordinal / numeric) and second, choose an appropriate chart type and visualise the distribution in the relative frequency scale.**",
"_____no_output_____"
],
[
"**<font color='red'>Attribute type :</font>** Nominal",
"_____no_output_____"
]
],
[
[
"# TODO\nfig = plt.figure(figsize=[16, 8])\n\nax3 = plt.subplot(111)\nax3.hist(data['occupation'], \n width = 0.5,\n weights = np.ones(len(data['occupation'])) / len(data['occupation']) \n )\nax3.set_title('Occupation - Distribution in Relative Frequency')\nplt.xticks(rotation=90)\nplt.show()",
"_____no_output_____"
]
],
[
[
"**2.5. Attribute `Native Country`. First, specify the type of the attribute (nominal / ordinal / numeric) and second, choose an appropriate chart type and visualise the distribution in the relative frequency scale.**",
"_____no_output_____"
],
[
"**<font color='red'>Attribute type :</font>** Nominal",
"_____no_output_____"
]
],
[
[
"# TODO\nfig = plt.figure(figsize=[16, 8])\n\nax4 = plt.subplot(111)\nax4.hist(data['native.country'], \n width = 1,\n weights = np.ones(len(data['native.country'])) / len(data['native.country']) \n )\nax4.set_title('Native Country - Distribution in Relative Frequency')\nplt.xticks(rotation=90)\nplt.show()",
"_____no_output_____"
]
],
[
[
"**2.6. Attribute `Salaries`. First, specify the type of the attribute (nominal / ordinal / numeric) and second, choose an appropriate chart type and visualise the distribution in the density scale.**",
"_____no_output_____"
],
[
"**<font color='red'>Attribute type :</font>** numeric",
"_____no_output_____"
]
],
[
[
"# TODO \nimport seaborn as sns\n\nsns.distplot(data['salaries'], hist=True, kde=True, \n bins=20, color = 'darkblue', \n hist_kws={'edgecolor':'black'},\n kde_kws={'linewidth': 2})",
"_____no_output_____"
]
],
[
[
"**2.7. Attributes `Male` and `Female`. First, specify the types of these attributes (nominal / ordinal / numeric) and second, choose an appropriate chart type and visualise the relative frequency of positive values in attribute `Male` and positive values in attribute `Female` (relative frequency of males and females).**",
"_____no_output_____"
],
[
"**<font color='red'>Attribute type :</font>** Nominal",
"_____no_output_____"
]
],
[
[
"# TODO - Put Male and Female attributes on the same plot for comparison\ndata['gender'] = data['male'].apply(lambda x: 'Male' if x == 1.0 else 'Female')\n\n# TODO\nfig = plt.figure(figsize=[8, 8])\n\nax4 = plt.subplot(111)\nax4.hist(data['gender'], \n width = 0.5,\n weights = np.ones(len(data['gender'])) / len(data['gender']) \n )\nax4.set_title('Gender - Distribution in Relative Frequency')\nplt.xticks(rotation=90)\nplt.show()\ndata",
"_____no_output_____"
]
],
[
[
"**2.8. Count the number of missing values in each attribute in the dataset and also report the percentage of missing values out of all rows (the original file uses '?' for missing values)**",
"_____no_output_____"
]
],
[
[
"#TODO\ntotal_rows = len(data.index)\ntotal_missing = 0\nif data is not data.empty:\n for column in data.columns:\n total_missing = len(data[data[column] == '?'])\n if total_missing > 0:\n print (f\"Percentage of missing values in attribute {column} = {round((total_missing/total_rows)*100,2)}%\")\nelse:\n print('Please define `data` in earlier subtasks')",
"Percentage of missing values in attribute age = 0.3%\nPercentage of missing values in attribute workclass = 5.64%\nPercentage of missing values in attribute occupation = 5.66%\nPercentage of missing values in attribute native.country = 1.79%\n"
]
],
[
[
"# 3. Data preparation (1.5 points)",
"_____no_output_____"
],
[
"Preparing the data for analysis is the most crucial phase in data analysis. In this exercise, the main goal is to clean up the dataset 'adult'. The dataset has been made unclean on purpose so that you can practice cleaning it up. Most of the introduced uncleanliness is of the same kinds as described in slides of Lecture 02. Additionally, some values have been introduced which are non-sensical due to the meaning of the attribute.<br>",
"_____no_output_____"
],
[
"First, let's make a deep copy of the original DataFrame. Later we will compare it to the cleaned/modified DataFrame.",
"_____no_output_____"
]
],
[
[
"if data is not data.empty:\n original_data = data.copy(deep=True) # Make a deep copy, including a copy of the data and the indices",
"_____no_output_____"
]
],
[
[
"**3.1. Sometimes values include leading and/or trailing spaces. Remove leading and trailing whitespace from strings. Count how many rows were changed in the dataset.** <br>",
"_____no_output_____"
]
],
[
[
"# TODO\nfor column in data.columns:\n if data[column].dtype == 'O' : #dtype == 'O' stands for Python object, to check if data type is string or not\n data[column] = data[column].str.strip()\n else:\n print(f\"column {column} is not string\")",
"column capital.gain is not string\ncolumn capital.loss is not string\ncolumn salaries is not string\ncolumn male is not string\ncolumn female is not string\n"
]
],
[
[
"Use code below to count how many rows were affected.",
"_____no_output_____"
]
],
[
[
"# Count differences\nif data is not data.empty:\n data_all = pd.concat([original_data, data]).drop_duplicates()\n diff = data_all.shape[0] - data.shape[0]\n print ('Difference: ' + str(diff))",
"Difference: 32561\n"
]
],
[
[
"**<font color='red'>Answer:</font>** 32561",
"_____no_output_____"
],
[
"**3.2. Replace all non-sensical values in `data` with np.nan from numpy package to denote missing values (Original file uses '?' for missing values). Fix the typos in the dataset. Justify all the changes that you do, explaining why you are sure that this is a correct change and what you think the cause for such error could be.** <br>",
"_____no_output_____"
]
],
[
[
"# TODO\ndata_clean = data.replace(['?','privat','UnitedStates', 'Unitedstates', 'Hong'], [np.nan,'Private','United-States', 'United-States', 'Hong-Kong'])\ndata_clean.head(10)",
"_____no_output_____"
]
],
[
[
"**<font color='red'>Answer:</font>** Beside replace `?` with `NaN` value, we also replace this string `['privat','UnitedStates', 'Unitedstates', 'Hong']` into `['Private','United-States', 'United-States', 'Hong-Kong']` to fix the typo and reduce the redudancy of the same value (for example: United-States and UnitedStates is the same value in `native.country`.",
"_____no_output_____"
],
[
"**3.3. Count the rows which were changed in the subtask b** <br>",
"_____no_output_____"
]
],
[
[
"# TODO\nif data_clean is not data_clean.empty:\n data_all = pd.concat([data, data_clean]).drop_duplicates()\n diff = data_all.shape[0] - data_clean.shape[0]\n print ('Difference: ' + str(diff))",
"Difference: 4186\n"
]
],
[
[
"**<font color='red'>Answer:</font>** 4186",
"_____no_output_____"
],
[
"**3.4. Report values that look suspicious whereas you are not sure whether they are definitely wrong** <br> ",
"_____no_output_____"
],
[
"**<font color='red'>Answer:</font>** \n- value `Hong` in column `native.country`, we're not sure is it truly the name of the country `Hong Kong` or another meaning\n- value in `capital.gain` and `capital.loss` which are containing too many `0` value\n- value `Assoc-acdm` in `education` column",
"_____no_output_____"
],
[
"# 4. Gathering interesting facts about the adult dataset (1.5 points)",
"_____no_output_____"
],
[
"Useful information about the data can be acquired through visualization even before modeling.",
"_____no_output_____"
],
[
"**4.1. Convert the attribute `Education` into type `category` and introduce the order for this attribute based on the educational level (e.g. Masters is higher education than Bachelors; use your best guess in ordering those levels for which the order is not obvious). Create a bar chart visualising the relative frequencies of values.**",
"_____no_output_____"
]
],
[
[
"# TODO\ndata['education'] = data['education'].astype('category')\n\ndata['education'].value_counts(normalize = True).reindex(['Doctorate', 'Masters', \n 'Bachelors', 'Some-college', 'Prof-school', 'Assoc-voc', 'Assoc-acdm',\n 'HS-grad', '12th',\n '11th', '10th',\n '9th', '7th-8th',\n '5th-6th', '1st-4th',\n 'Preschool'\n ]).plot(kind=\"bar\")",
"_____no_output_____"
]
],
[
[
"**4.2. Create and print a Pandas data frame where each row stands for an occupation, each column stands for a level of education, and the cells in the table contain the average salary of people with the corresponding occupation and education level.** <br> \nHint 1: You can use crosstab, see also aggfunc from the pandas documentation.",
"_____no_output_____"
]
],
[
[
"# TODO\ncrosstab_df = pd.crosstab(data.occupation, data.education, values = data.salaries, aggfunc=np.mean).round(2)\ncrosstab_df",
"_____no_output_____"
]
],
[
[
"**4.3. Make a separate plot for each row (occupation) of the table created in subtask b. Choose the type of the plot to convey the information as well as you can.** <br>",
"_____no_output_____"
]
],
[
[
"if crosstab_df is not crosstab_df.empty:\n for index, row in crosstab_df.iterrows():\n row.plot(kind = 'bar') # TODO \n plt.show()",
"_____no_output_____"
]
],
[
[
"**4.4. List 3 interesting facts that you can read out of these plots from subtask c.** <br>",
"_____no_output_____"
],
[
"**<font color='red'>Fact 1:</font>** For occupation `armed-forces` it shows that for people who have education `12th` grade have higher salaries than people with `bachelors`, `some-college`. and `HS-grade` background.",
"_____no_output_____"
],
[
"**<font color='red'>Fact 2:</font>** For occupation `exec-managerial` it shows that for people with education `5th-6th` grade have the highest salaries compare to others.\n",
"_____no_output_____"
],
[
"**<font color='red'>Fact 3:</font>** For occupation `farming-fishing` the highest salaries is owned by people with 'Doctorate' education",
"_____no_output_____"
],
[
"## How long did it take you to solve the homework?\n\n* Please answer as precisely as you can. It does not affect your points or grade in any way. It is okey, if it took 0.5 hours or 24 hours. The collected information will be used to improve future homeworks.\n\n<font color='red'> **Answer:**</font>\n\n**<font color='red'>(please change X in the next cell into your estimate)</font>**\n\n10 hours\n\n## How much the homework is difficult?\nyou can put only number between $0:10$ ($0:$ easy, $10:$ difficult)\n\n<font color='red'> **Answer: 6**</font>\n\n\n",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
c50fd870d6377efaf9a01fc15004cfd990951da7
| 3,889 |
ipynb
|
Jupyter Notebook
|
sort/quick_sort_implement_solution.ipynb
|
cntfk2017/Udemy_Python_Hand_On
|
52f2a5585bfdea95d893f961c8c21844072e93c7
|
[
"Apache-2.0"
] | 1 |
2020-05-25T07:09:28.000Z
|
2020-05-25T07:09:28.000Z
|
sort/quick_sort_implement_solution.ipynb
|
cntfk2017/Udemy_Python_Hand_On
|
52f2a5585bfdea95d893f961c8c21844072e93c7
|
[
"Apache-2.0"
] | null | null | null |
sort/quick_sort_implement_solution.ipynb
|
cntfk2017/Udemy_Python_Hand_On
|
52f2a5585bfdea95d893f961c8c21844072e93c7
|
[
"Apache-2.0"
] | 2 |
2019-09-23T14:26:48.000Z
|
2020-05-25T07:09:26.000Z
| 27.778571 | 110 | 0.5081 |
[
[
[
"# Write a Python program to sort a list of elements using the quick sort algorithm.\n# Note : According to Wikipedia \"Quicksort is a comparison sort, meaning that it can \n# sort items of any type for which a \"less-than\" relation (formally, a total order) is defined. \n# In efficient implementations it is not a stable sort, meaning that the relative order of equal \n# sort items is not preserved. Quicksort can operate in-place on an array, requiring small additional\n# amounts of memory to perform the sorting.\"\n\ndef quickSort(data_list):\n quickSortHlp(data_list,0,len(data_list)-1)\n\ndef quickSortHlp(data_list,first,last):\n if first < last:\n\n splitpoint = partition(data_list,first,last)\n\n quickSortHlp(data_list,first,splitpoint-1)\n quickSortHlp(data_list,splitpoint+1,last)\n\n\ndef partition(data_list,first,last):\n pivotvalue = data_list[first]\n\n leftmark = first+1\n rightmark = last\n\n done = False\n while not done:\n\n while leftmark <= rightmark and data_list[leftmark] <= pivotvalue:\n leftmark = leftmark + 1\n\n while data_list[rightmark] >= pivotvalue and rightmark >= leftmark:\n rightmark = rightmark -1\n\n if rightmark < leftmark:\n done = True\n else:\n temp = data_list[leftmark]\n data_list[leftmark] = data_list[rightmark]\n data_list[rightmark] = temp\n\n temp = data_list[first]\n data_list[first] = data_list[rightmark]\n data_list[rightmark] = temp\n\n\n return rightmark\n\ndata_list = [54,26,93,17,77,31,44,55,20]\nquickSort(data_list)\nprint(data_list)",
"[17, 20, 26, 31, 44, 54, 55, 77, 93]\n"
],
[
"#quick sort \nmlist = [54,26,93,17,77,31,44,55,20]\ndef quick_sort(list): #extra-place\n smaller = []\n bigger = []\n keylist = []\n\n if len(list) <= 1:\n return list\n\n else:\n key = list[int(len(list)/2)] #中間數為key值\n for i in list:\n if i < key: #比key值小的數\n smaller.append(i)\n elif i > key: #比key值大的數\n bigger.append(i)\n else:\n keylist.append(i)\n\n smaller = quick_sort(smaller)\n bigger = quick_sort(bigger)\n return smaller + keylist + bigger\nprint(quick_sort(mlist))",
"[17, 20, 26, 31, 44, 54, 55, 77, 93]\n"
]
]
] |
[
"code"
] |
[
[
"code",
"code"
]
] |
c50fdb32f80a0b6f6a778022fad25a5dd3bc7791
| 174,287 |
ipynb
|
Jupyter Notebook
|
examples/example_astero_bolocorr.ipynb
|
ojhall94/omnitool
|
b95a715fb7a41a4fc5d7f90dd66fead124fe14b8
|
[
"MIT"
] | null | null | null |
examples/example_astero_bolocorr.ipynb
|
ojhall94/omnitool
|
b95a715fb7a41a4fc5d7f90dd66fead124fe14b8
|
[
"MIT"
] | null | null | null |
examples/example_astero_bolocorr.ipynb
|
ojhall94/omnitool
|
b95a715fb7a41a4fc5d7f90dd66fead124fe14b8
|
[
"MIT"
] | null | null | null | 340.404297 | 40,652 | 0.937729 |
[
[
[
"import omnitool\nfrom omnitool.literature_values import *\n\nimport matplotlib\nmatplotlib.rcParams['text.usetex']=False\nimport pandas as pd\nimport matplotlib.pyplot as plt\nimport seaborn as sns",
"_____no_output_____"
]
],
[
[
"We'll use the asteroseismic data from Yu et al. 2018",
"_____no_output_____"
]
],
[
[
"#Read in Jie Yu\nprint('Reading in Yu+2018')\nsfile = '/home/oliver/PhD/Catalogues/RC_catalogues/Yu+18_table1.txt'\nyu18_1 = pd.read_csv(sfile, sep='|')\nsfile = '/home/oliver/PhD/Catalogues/RC_catalogues/Yu+18_table2.txt'\nyu18_2 = pd.read_csv(sfile, sep='|')\nyu18 = pd.merge(yu18_1, yu18_2, on='KICID',how='outer')\nyu18.rename(columns={'KICID':'KIC',\n 'EvoPhase':'stage',\n 'err_x':'numax_err',\n 'err.1_x':'dnu_err',\n 'err_y':'Teff_err',\n 'Fe/H':'[Fe/H]',\n 'err.2_y':'[Fe/H]_err',\n 'err.1_y':'logg_err',\n 'err.3_y':'M_err',\n 'err.4_y':'R_err'},inplace=True) #For consistency",
"Reading in Yu+2018\n"
]
],
[
[
"First, lets get our asteroseismic values\n",
"_____no_output_____"
]
],
[
[
"#Calling the scaling relations class\nSC = omnitool.scalings(yu18.numax, yu18.dnu, yu18.Teff,\\\n _numax_err = yu18.numax_err,\\\n _dnu_err = yu18.dnu_err,\\\n _Teff_err = yu18.Teff_err)",
"_____no_output_____"
]
],
[
[
"Now lets pull out all the values we can calculate",
"_____no_output_____"
]
],
[
[
"yu18['aR'] = SC.get_radius()/Rsol\nyu18['aR_err'] = SC.get_radius_err()/Rsol\nyu18['aM'] = SC.get_mass()/Msol\nyu18['aM_err'] = SC.get_mass_err()/Msol\nyu18['alogg'] = SC.get_logg()\nyu18['alogg_err'] = SC.get_logg_err()\nyu18['L'] = SC.get_luminosity()/Lsol\nyu18['L_err'] = SC.get_luminosity_err()/Lsol\nyu18['Mbol'] = SC.get_bolmag()\nyu18['Mbol_err'] = SC.get_bolmag_err()",
"Calculating luminosity using basic asteroseismic radius\nCalculating luminosity using basic asteroseismic radius\nCalculating luminosity using basic asteroseismic radius\n"
]
],
[
[
"Lets plot a quick distribution of points on a HR diagram.",
"_____no_output_____"
]
],
[
[
"sns.jointplot(np.log10(yu18.Teff), np.log10(yu18.L),s=1)\nplt.show()",
"/home/oliver/.local/lib/python2.7/site-packages/matplotlib/font_manager.py:1331: UserWarning: findfont: Font family [u'serif'] not found. Falling back to DejaVu Sans\n (prop.get_family(), self.defaultFamily[fontext]))\n/usr/local/lib/python2.7/dist-packages/scipy/stats/stats.py:1706: FutureWarning: Using a non-tuple sequence for multidimensional indexing is deprecated; use `arr[tuple(seq)]` instead of `arr[seq]`. In the future this will be interpreted as an array index, `arr[np.array(seq)]`, which will result either in an error or a different result.\n return np.add.reduce(sorted[indexer] * weights, axis=axis) / sumval\n"
]
],
[
[
"Now lets say we want to find the K and H band magnitudes of\nthese stars asteroseismically.\nFor this, we need an inverse bolometric correction.",
"_____no_output_____"
]
],
[
[
"#For the bolomteric correction, we need the total metallicity Z\nyu18['Z'] = Zsol * 10**yu18['[Fe/H]']\n\n#Now we initialise the class using these values\nget_bc = omnitool.bolometric_correction(yu18.Teff.values,\\\n yu18.logg.values,\\\n yu18.Z.values)",
"_____no_output_____"
]
],
[
[
"We now want to calculate the bolometric correction for\nthese stars in the K, H and J bands",
"_____no_output_____"
]
],
[
[
"KsBC = get_bc(band='Ks')\nHBC = get_bc(band='H')\nJBC = get_bc(band='J')",
"100%|██████████| 16094/16094 [00:09<00:00, 1691.38it/s]\n100%|██████████| 16094/16094 [00:10<00:00, 1564.57it/s]\n100%|██████████| 16094/16094 [00:09<00:00, 1738.22it/s]\n"
]
],
[
[
"Using this value & the bolometric magnitude we can make the inverse bolometric \ncorrection",
"_____no_output_____"
]
],
[
[
"MKs_ast = yu18.Mbol.values - KsBC\nMH_ast = yu18.Mbol.values - HBC\nMJ_ast = yu18.Mbol.values - JBC",
"_____no_output_____"
]
],
[
[
"If we want to be extra thorough, we propagate the assumed error\non the bolometric correction\n\nFor now this is just guesstimated at 0.02 per Huber et al. 2017, BUT\nit should be noted that they use a different method of obtaining the BC",
"_____no_output_____"
]
],
[
[
"M_ast_err = np.sqrt(yu18.Mbol_err.values**2 + err_bc**2)",
"_____no_output_____"
]
],
[
[
"Note that this error is the same regardless of passband",
"_____no_output_____"
]
],
[
[
"'''Lets plot a distribution of the results...'''\nsns.distplot(MKs_ast)\nplt.xlabel('Asteroseismic Absolute Magnitude (K)')\nplt.show()\nprint('Median in K: '+str(np.median(MKs_ast)))\n\nsns.distplot(MH_ast)\nplt.xlabel('Asteroseismic Absolute Magnitude (H)')\nplt.show()\nprint('Median in H: '+str(np.median(MH_ast)))\n\nsns.distplot(MJ_ast)\nplt.xlabel('Asteroseismic Absolute Magnitude (J)')\nplt.show()\nprint('Median in J: '+str(np.median(MJ_ast)))\n",
"_____no_output_____"
]
],
[
[
"Lets do a quick plot to illustrate the errors on the data... We'll plot Core He Burning stars only for clarity.",
"_____no_output_____"
]
],
[
[
"sel = yu18.stage == 1\n\nfig, ax = plt.subplots()\nax.scatter(MKs_ast[sel],yu18.numax[sel],s=1,zorder=1001)\nax.errorbar(MKs_ast[sel],yu18.numax[sel],xerr=M_ast_err[sel], alpha=.5, fmt='none', c='grey',zorder=1000)\nax.set_xlabel('Absolute asteroseismic magnitude (K)')\nax.set_ylabel(r\"Asteroseismic $\\nu_{max}}$\")\nplt.show()",
"_____no_output_____"
]
],
[
[
"Finally, lets compare our results to those obtained in the Yu+18 work to ensure they're the same.",
"_____no_output_____"
]
],
[
[
"sns.jointplot(yu18.aR, yu18.R_noCorrection, s=1)\nsns.jointplot(yu18.aR_err, yu18.R_err,s=1)\nplt.show()",
"_____no_output_____"
]
],
[
[
"Now imagine we want to recalculate luminosity, but using a given a correction to the delta nu scaling relations",
"_____no_output_____"
]
],
[
[
"#Calling the scaling relations class\nSC = omnitool.scalings(yu18.numax, yu18.dnu, yu18.Teff,\\\n _numax_err = yu18.numax_err,\\\n _dnu_err = yu18.dnu_err,\\\n _Teff_err = yu18.Teff_err)\nSC.give_corrections(fdnu=np.ones(len(yu18))*1.005, fdnu_err=np.ones(len(yu18))*1.005*0.02)\n\nyu18['corR'] = SC.get_radius()/Rsol\nyu18['corR_err'] = SC.get_radius_err()/Rsol",
"You have passed corrections to the Delta Nu scaling relation\n"
],
[
"sns.jointplot(yu18.R_noCorrection, yu18.corR,s=1)\nplt.show()",
"_____no_output_____"
],
[
"sns.jointplot(yu18.R_err, yu18.corR_err,s=1)\n",
"_____no_output_____"
]
],
[
[
"The change is subtle, but its definitely there! And the uncertainties on the corrections have an effect also.",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
]
] |
c50fdcfda2806a8eb8e50cea851315445ce79c12
| 20,702 |
ipynb
|
Jupyter Notebook
|
Jupyter/stock/stock1.ipynb
|
mr4jay/IA
|
14cedadbe3d17eb86d328bc00f123839a6891d40
|
[
"MIT"
] | null | null | null |
Jupyter/stock/stock1.ipynb
|
mr4jay/IA
|
14cedadbe3d17eb86d328bc00f123839a6891d40
|
[
"MIT"
] | null | null | null |
Jupyter/stock/stock1.ipynb
|
mr4jay/IA
|
14cedadbe3d17eb86d328bc00f123839a6891d40
|
[
"MIT"
] | 1 |
2021-06-06T07:51:02.000Z
|
2021-06-06T07:51:02.000Z
| 184.839286 | 17,876 | 0.903149 |
[
[
[
"import csv\nimport numpy as np\nfrom sklearn.svm import SVR\nimport matplotlib.pyplot as plt\n\ndates=[]\nprices=[]\n\ndef get_data(filename):\n with open(filename,'r') as csvfile:\n csvFileReader=csv.reader(csvfile)\n next(csvFileReader)\n for row in csvFileReader:\n dates.append(int(row[0].split('-')[0]))\n prices.append(float(row[1]))\n return\n\ndef predict_prices(dates, prices, x):\n dates=np.reshape(dates,(len(dates),1))\n \n svr_lin=SVR(kernel='linear', C=1e3)\n svr_poly=SVR(kernel='poly', C=1e3, degree=2)\n svr_rbf=SVR(kernel='rbf', C=1e3, gamma=0.1)\n \n svr_lin.fit(dates,prices)\n svr_poly.fit(dates,prices)\n svr_rbf.fit(dates,prices)\n \n plt.scatter(dates,prices, color='black', label='Data')\n plt.plot(dates, svr_rbf.predict(dates), color='red', label='RBF model') \n plt.plot(dates, svr_lin.predict(dates), color='green', label='Linear model') \n plt.plot(dates, svr_poly.predict(dates), color='blue', label='Polynomial model')\n plt.xlabel('Date')\n plt.ylabel('Price')\n plt.title('Support Vector Regression')\n plt.legend()\n plt.show()\n \n return svr_rbf.predict(x)[0], svr_lin.predict(x)[0], svr_poly.predict(x)[0]\n\nget_data('aapl.csv')\n\npredicted_price = predict_prices(dates,prices,29)\n\nprint(predicted_price)\n ",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code"
]
] |
c50fe9ba54de815b73474e3d3262be8283558b06
| 979,223 |
ipynb
|
Jupyter Notebook
|
codes/Hesthaven_1D.ipynb
|
CorbinFoucart/FEMexperiment
|
9bad34d9ed7cbdd740e3a4b67f433779dd53b264
|
[
"MIT"
] | 2 |
2018-05-26T22:09:32.000Z
|
2018-06-25T21:46:32.000Z
|
codes/Hesthaven_1D.ipynb
|
CorbinFoucart/FEMexperiment
|
9bad34d9ed7cbdd740e3a4b67f433779dd53b264
|
[
"MIT"
] | 16 |
2018-05-17T21:38:44.000Z
|
2022-03-11T23:21:25.000Z
|
codes/Hesthaven_1D.ipynb
|
CorbinFoucart/FEMexperiment
|
9bad34d9ed7cbdd740e3a4b67f433779dd53b264
|
[
"MIT"
] | null | null | null | 41.322657 | 567 | 0.496046 |
[
[
[
"## setup and notebook configuration",
"_____no_output_____"
]
],
[
[
"# scientific python stack\nimport numpy as np\nimport scipy as sp\nimport sympy as sym\nimport orthopy, quadpy",
"_____no_output_____"
],
[
"# matplotlib, plotting setup\nimport matplotlib as mpl\nimport matplotlib.pyplot as plt\nimport matplotlib.tri as mtri # delaunay triangulation\nfrom mpl_toolkits.mplot3d import Axes3D # surface plotting\nimport seaborn as sns # nice plotting defaults\nimport cmocean as cmo # ocean colormaps\nsym.init_printing(use_latex='mathjax')\nsns.set()\n%matplotlib inline\n%config InlineBackend.figure_format = 'svg'",
"/Users/corbin/virtual_envs/FEMexperiment_env/lib/python3.6/site-packages/cmocean/tools.py:76: MatplotlibDeprecationWarning: The is_string_like function was deprecated in version 2.1.\n if not mpl.cbook.is_string_like(rgbin[0]):\n"
],
[
"%load_ext autoreload\n%autoreload 2",
"_____no_output_____"
],
[
"# local imports\nimport src.fem_base.master.mk_basis_nodal as mbn\nimport src.fem_base.master.mk_master as mkm\nimport src.fem_maps.fem_map as fem_map",
"_____no_output_____"
]
],
[
[
"# creation of 1D nodal bases",
"_____no_output_____"
],
[
"We define the master 1D element as $I\\in[-1, 1]$",
"_____no_output_____"
]
],
[
[
"b = mbn.Basis_nodal(order=1, dim=1, element=0)\nb.plot_elm()",
"_____no_output_____"
],
[
"fig, ax = plt.subplots(1, 5, figsize = (10, 1))\nxx = np.linspace(-1, 1, 100)\npts = [[x, 0, 0] for x in xx]\n\nfor order in range(5):\n b = mbn.Basis_nodal(order=order, dim=1, element=0)\n yy = b.eval_at_pts(pts=pts)\n for basis_fn in range(b.nb):\n ax[order].plot(xx, yy[:, basis_fn])\n ax[order].set_title(r'$p = {}$'.format(order))",
"_____no_output_____"
]
],
[
[
"## construction of vandermonde matrices",
"_____no_output_____"
]
],
[
[
"x = np.linspace(-1, 1, 100)\nn_polys = 4\nvals = orthopy.line_segment.tree_jacobi(x, n=n_polys, alpha=0, beta=0, standardization='normal')\nfor i in range(n_polys):\n plt.plot(x, vals[i], label='P_{}'.format(i))\nplt.legend()\n\nplt.title('normalized Legendre polynomials')\nplt.show()",
"_____no_output_____"
]
],
[
[
"These polynomials agree with the explicitly listed polynomials in Hesthaven, so we know that they are orthonormalized correctly.",
"_____no_output_____"
]
],
[
[
"def Jacobi_Poly(r, alpha, beta, N):\n \"\"\" wraps orthopy to return Jacobi polynomial \"\"\"\n return orthopy.line_segment.tree_jacobi(r, n=N-1, alpha=alpha, beta=beta, standardization='normal')\n\ndef P_tilde(r, N):\n P = np.zeros((len(r), N))\n polyvals = Jacobi_Poly(r, alpha=0, beta=0, N=N)\n for j in range(N):\n P[:, j] = polyvals[j]\n return P.T\n \ndef Vandermonde1D(N, x):\n \"\"\" initialize 1D vandermonde Matrix Vij = phi_j(x_i)\"\"\"\n V1D = np.zeros((len(x), N))\n JacobiP = Jacobi_Poly(x, alpha=0, beta=0, N=N)\n for j, polyvals in enumerate(JacobiP):\n V1D[:, j] = polyvals\n return V1D",
"_____no_output_____"
],
[
"def LegendreGaussLobatto(N):\n GL = quadpy.line_segment.GaussLobatto(N, a=0., b=0.)\n return GL.points, GL.weights\n\ndef GaussLegendre(N):\n GL = quadpy.line_segment.GaussLegendre(N)\n return GL.points, GL.weights",
"_____no_output_____"
]
],
[
[
"An important conceptual point is that the Vandermonde matrix here is NOT the shape function matrix, it's the Vandermonde matrix of the Orthonormal polynomial basis. We will see this later as we have to create the shape function matrices.",
"_____no_output_____"
],
[
"## properties / conditioning of vandermonde matrices",
"_____no_output_____"
]
],
[
[
"equi_det, LGL_det = [], []\nfor N in range(2, 35):\n nb = N + 1\n equi_pts = np.linspace(-1, 1, nb)\n V = Vandermonde1D(nb, equi_pts)\n equi_det.append(np.linalg.det(V))\n \n LGL_pts, _ = LegendreGaussLobatto(nb)\n V = Vandermonde1D(nb, LGL_pts)\n LGL_det.append(np.linalg.det(V))",
"_____no_output_____"
],
[
"plt.semilogy(list(range(2, 35)), equi_det, label='equidistant')\nplt.semilogy(list(range(2, 35)), LGL_det, label='LGL nodes')\nplt.legend()\nplt.show()",
"_____no_output_____"
]
],
[
[
"This result agrees with Hesthaven.",
"_____no_output_____"
]
],
[
[
"# construct generic lagrange interpolant\nfrom scipy.interpolate import lagrange\n\ndef lagrange_polys(pts):\n lagrange_polys = []\n for i, pt in enumerate(pts):\n data = np.zeros_like(pts)\n data[i] = 1\n lagrange_polys.append(lagrange(pts, data))\n return lagrange_polys\n\ndef lagrange_basis_at_pts(lagrange_polys, eval_pts):\n \"\"\" evaluates lagrange polynomials at eval_pts\"\"\"\n result = np.zeros((len(lagrange_polys) ,len(eval_pts)))\n for i, poly in enumerate(lagrange_polys):\n result[i, :] = lagrange_polys[i](eval_pts)\n return result",
"_____no_output_____"
]
],
[
[
"plot lagrange polys over equally spaced vs LGL points",
"_____no_output_____"
]
],
[
[
"N = 5\nlp = np.linspace(-1, 1, N)\nlpolys = lagrange_polys(lp)\n\nvN = 100\nview_pts = np.linspace(-1, 1, vN)\nli = lagrange_basis_at_pts(lpolys, view_pts)\nplt.plot(view_pts, li.T)\nplt.title('lagrange polynomials over equally spaced points')\nplt.show()",
"_____no_output_____"
],
[
"N = 5\nlp, _ = LegendreGaussLobatto(N)\nlpolys = lagrange_polys(lp)\n\nvN = 100\nview_pts = np.linspace(-1, 1, vN)\nli = lagrange_basis_at_pts(lpolys, view_pts)\nplt.plot(view_pts, li.T)\nplt.title('lagrange polynomials over LGL points')\nplt.show()",
"_____no_output_____"
]
],
[
[
"So beautiful! By moving the lagrange data points to the nodal points, our basis functions don't exceed 1, unlike in the above plot, where we are already seeing a slight Runge phenomenon.",
"_____no_output_____"
],
[
"## vandermonde relations",
"_____no_output_____"
],
[
"### relationship between vandermonde $V$, basis polynomials $\\tilde{\\mathbf{P}}$, and lagrange basis functions (shape functions) $\\ell$",
"_____no_output_____"
],
[
"Hesthaven makes the claim that $V^T \\mathbf{\\ell}(r) = \\tilde{\\mathbf{P}}(r)$ in (3.3).\n\nIn Hesthaven's notation, $N$ denotes the polynomial order, $N_p$ denotes the number of nodal points (we would call $nb$), and let's call the number of \"view points\" `xx`, which are arbitrary.\n\nThen the shapes of the Hesthaven structures are:\n- $\\mathbf{\\ell}$, $\\tilde{\\mathbf{P}}$, $V$ are all (`nb`, `xx`)\n- $V^T \\ell$ is (`xx`, `nb`) x (`nb`, `xx`) $\\rightarrow$ (`xx`, `xx`) where rows contain the values of polynomials $\\tilde{\\mathbf{P}}$\n\nThis works for either equidistant points or the LGL points.",
"_____no_output_____"
]
],
[
[
"N = 5\nlp, _ = LegendreGaussLobatto(N) \n#lp = np.linspace(-1, 1, N)\nview_pts = np.linspace(-1, 1, 50)\n\nl_polys = lagrange_polys(pts=lp)\nℓ = lagrange_basis_at_pts(l_polys, eval_pts=view_pts)\nV = Vandermonde1D(N=len(view_pts), x=lp)\nP = np.dot(V.T, ℓ)\n\n# plot the result\nplt.plot(view_pts, ℓ.T, '--')\nplt.plot(view_pts, P[0:3,:].T)\nplt.show()",
"_____no_output_____"
]
],
[
[
"We see that indeed we recover the Legendre polynomials.",
"_____no_output_____"
],
[
"More directly, we can invert the relation to find that\n\n$$\\ell = (V^T)^{-1} \\tilde{\\mathbf{P}}$$\n\nwhich allows us to create our nodal shape functions.",
"_____no_output_____"
]
],
[
[
"nb = 4\nnodal_pts, _ = LegendreGaussLobatto(nb)\nview_pts = np.linspace(-1, 1, 50)\n\n# create the Vandermonde, P matrices\nV = Vandermonde1D(N=nb, x=nodal_pts)\nVti = np.linalg.inv(V.T)\nP = P_tilde(r=view_pts, N=nb)\nprint('shape of Vandermonde: {}'.format(V.shape))\nprint('shape of P: {}'.format(P.shape))\n\nyy = np.dot(Vti, P)\nplt.plot(view_pts, yy.T)\nplt.title('nodal shape functions generated from orthogonal basis polynomials')\nplt.show()",
"shape of Vandermonde: (4, 4)\nshape of P: (4, 50)\n"
]
],
[
[
"### relationship between vandermonde $V$ and mass matrix",
"_____no_output_____"
],
[
"We can build on the relationship developed in the section above to form the mass matrix for a nodal basis. We note that \n\n$M_{ij} = \\int_{-1}^{1}\\ell_i(r)\\, \\ell_j(r) \\,dr = (\\ell_i, \\ell_j)_I$, and if we expand out $\\ell = (V^T)^{-1}\\tilde{\\mathbf{P}}$, it turns out (page 51)\n\n$$M = (V V^T)^{-1}$$\n\nbecause of the orthogonal nature of our choice of basis function; the implication is that we can compute the integrals over the master element without the explicit need for quadrature points or weights. Note first that $\\phi_i(\\xi) = \\sum_{n=1}^{nb} (V^T)_{in}^{-1} \\tilde{P}_{n-1}(\\xi)$. Then\n\n\\begin{align}\nM_{ij} &= \\int^{1}_{-1} \\phi_i(\\xi)\\,\\phi_j(\\xi)\\,d\\xi \n = \\int^{1}_{-1}\\left[\\sum_{k=1}^{nb} (V^T)_{ik}^{-1} \\tilde{P}_{k-1}(\\xi)\n \\sum_{m=1}^{nb} (V^T)_{jm}^{-1} \\tilde{P}_{m-1}(\\xi) \\right]\\, d\\xi \\\\\n &= \\sum_{k=1}^{nb} \\sum_{m=1}^{nb} (V^T)_{ik}^{-1} \n (V^T)_{jm}^{-1} \\int^{1}_{-1}\\tilde{P}_{k-1}(\\xi) \\tilde{P}_{m-1}(\\xi)\n =\\sum_{k=1}^{nb} \\sum_{m=1}^{nb} (V^T)_{ik}^{-1}\n (V^T)_{jm}^{-1} \\delta_{km} \\\\\n &=\\sum_{k=1}^{nb} (V^T)_{im}^{-1} \n (V^T)_{jm}^{-1} = \\sum_{k=1}^{nb} (V^T)_{mi}^{-1} (V)_{mj}^{-1} \\\\\n &= (V^{T})^{-1} V^{-1} = (VV^T)^{-1}\n\\end{align}\n\nWhere note we've used the cute trick that $\\int_{-1}^1 \\tilde{P}_m \\tilde{P}_n = \\delta_{mn}$, since we chose an __orthonormal__ modal basis. Orthogonal wouldn't have done it, but an orthonormal modal basis has this property.\n\nWe can check this relation against the more traditional way of constructing the mass matrix with quadrature. `master.shap` has dimensions of (`n_quad`, `nb`)",
"_____no_output_____"
]
],
[
[
"order = 3\nm1d = mkm.Master_nodal(order=order, dim=1, element=0)\nxq, wq = m1d.cube_pts, m1d.cube_wghts\nshap = m1d.shap\nshapw = np.dot(np.diag(wq), m1d.shap)\nM_quadrature = np.dot(shap.T, shapw)\n\n\nNp = order + 1\nnodal_points, _ = LegendreGaussLobatto(Np)\nV = Vandermonde1D(N=Np, x=nodal_points)\nM_vand = np.linalg.inv(np.dot(V, V.T))\n\n# this will throw an error if not correct\nassert(np.allclose(M_quadrature, M_vand))",
"_____no_output_____"
]
],
[
[
"## efficient computation of derivatives of the basis functions",
"_____no_output_____"
],
[
"### derivatives of Legendre polynomials",
"_____no_output_____"
],
[
"In order to compute the derivatives of the shape functions (which are expressed via the vandermonde matrix $V$), we must take the derivatives with respect to the orthogonal basis polynomials. There is an identity (Hesthaven, p. 52)\n\n$$ \\frac{d \\tilde{P}_n}{d r} = \\sqrt{n(n+1)}\\,\\tilde{P}^{(1,1)}_{n-1}$$\n\nThis is in contrast to directly differentiating either the coefficients of $\\tilde{P}$ or more directly the nodal shape functions $\\ell$ if the explicit polynomial form is known (like in `scipy`, but this becomes trickier in multiple dimensions). As it turns out, the first approach is a very efficient way to compute these operators.",
"_____no_output_____"
]
],
[
[
"def Jacobi_Poly_Derivative(r, alpha, beta, N):\n \"\"\" take a derivative of Jacobi Poly, more general than above\n copy the format of orthopy (list of arrays)\n \"\"\"\n dp = [np.zeros_like(r)]\n Jacobi_P = Jacobi_Poly(r, alpha + 1, beta + 1, N)\n for n in range(1, N+1):\n gamma = np.sqrt(n * (n + alpha + beta + 1))\n dp.append(gamma * Jacobi_P[n-1])\n return dp\n\n#def dP_tilde(r, N):\n# P = np.zeros((len(r), N))\n# polyvals = Jacobi_Poly_Derivative(r, alpha=0, beta=0, N=N)\n# for j in range(N):\n# P[:, j] = polyvals[j]\n# return P",
"_____no_output_____"
]
],
[
[
"We can examine some of the derivatives of the Legendre polynomials.",
"_____no_output_____"
]
],
[
[
"# some unit testing \n# first jacobi poly is const, so der should be 0\nxx = np.linspace(-1, 1, 50)\njpd = Jacobi_Poly_Derivative(xx, alpha=0, beta=0, N=3)\nfor i, polyder in enumerate(jpd):\n plt.plot(xx, polyder, label=r'$P_{}^\\prime(x)$'.format(i))\nplt.legend()\nplt.show()",
"_____no_output_____"
]
],
[
[
"These look good. The derivative of the first Legendre polynomial is analytically 0, $P_1^\\prime = \\sqrt{3/2}$, $P_2^\\prime$ should be linear, $P_3^\\prime$ should be quadratic.",
"_____no_output_____"
],
[
"### discrete derivative operators",
"_____no_output_____"
],
[
"We can declare the derivative Vandermonde matrix, and invert it in the same manner to obtain the derivatives of the nodal shape functions.\n\nThis works because \n$$V^T \\ell = P \\Rightarrow V^T \\frac{d}{dx} \\ell = \\frac{d}{dx}P$$\n\nHence $$V_r\\equiv V^T D_r^T, \\qquad {V_r}_{(ij)} = \\frac{d \\tilde{P}_j(r_i)}{d x} $$\n\nand finally $D_r = V_r V^{-1}$ (see Hesthaven, p. 53), as well as $S = M D_r $, where $S_{ij} = \\left(\\phi_i, \\frac{d\\phi_j}{dx}\\right)_I$, and where $M$ is the mass matrix.",
"_____no_output_____"
]
],
[
[
"def GradVandermonde1D(N, x):\n Vr = np.zeros((len(x), N))\n dJacobi_P = Jacobi_Poly_Derivative(x, alpha=0, beta=0, N=N-1)\n for j, polyder in enumerate(dJacobi_P):\n Vr[:,j] = polyder\n return Vr",
"_____no_output_____"
],
[
"p = 3\nnb = p+1\nnodal_pts, _ = LegendreGaussLobatto(nb)\n#nodal_pts = np.linspace(-1, 1, nb)\nview_pts = np.linspace(-1, 1, 50)\n\n# grad vandermonde\nV = Vandermonde1D(N=nb, x=nodal_pts)\nVr = GradVandermonde1D(N=nb, x=view_pts)\nVi = np.linalg.inv(V)\nDr = np.dot(Vr, Vi)\n\nprint('shape Vr: {}'.format(Vr.shape))\nprint('shape V inv: {}'.format(Vi.shape))\nprint('shape Dr: {}'.format(Dr.shape))\n\n# shape functions\nV = Vandermonde1D(N=nb, x=nodal_pts)\nVti = np.linalg.inv(V.T)\nP = P_tilde(r=view_pts, N=nb)\nshap = np.dot(Vti, P)\n\n# shape functions at view points\nfig, ax = plt.subplots(1, 2, figsize=(8, 4))\nax[0].plot(view_pts, shap.T, '--')\nax[0].set_title(r'nodal shape functions $\\phi$')\nax[1].plot(view_pts, Dr)\nax[1].set_title(r'derivatives of nodal shape functions $\\frac{d \\phi}{dx}$')\nplt.show()",
"shape Vr: (50, 4)\nshape V inv: (4, 4)\nshape Dr: (50, 4)\n"
]
],
[
[
"As a remark, we can once again show the effect of using Legendre Gauss Lobatto points vs equally spaced nodal points.",
"_____no_output_____"
]
],
[
[
"N = 8\nnb = N+1\nnodal_pts_LGL, _ = LegendreGaussLobatto(nb)\nnodal_pts_equi = np.linspace(-1, 1, nb)\nview_pts = np.linspace(-1, 1, 100)\n\n# shape functions at view points\nfig, ax = plt.subplots(figsize=(8, 8))\nax.set_yticks([])\nax.set_xticks([])\nlabels = ['LGL nodes', 'uniform nodes']\nfor idx, nodal_pts in enumerate([nodal_pts_LGL, nodal_pts_equi]):\n # grad vandermonde\n V = Vandermonde1D(N=nb, x=nodal_pts)\n Vr = GradVandermonde1D(N=nb, x=view_pts)\n Vi = np.linalg.inv(V)\n Dr = np.dot(Vr, Vi)\n\n # shape functions\n V = Vandermonde1D(N=nb, x=nodal_pts)\n Vti = np.linalg.inv(V.T)\n P = P_tilde(r=view_pts, N=nb)\n shap = np.dot(Vti, P)\n \n # plot\n ax = fig.add_subplot(2, 2, idx*2+1)\n ax.plot(view_pts, shap.T)\n ax.set_yticks([0, 1])\n ax.set_title(r' $\\phi$, {}'.format(labels[idx]))\n ax = fig.add_subplot(2, 2, idx*2+2)\n ax.plot(view_pts, Dr)\n ax.set_title(r'$\\f{}$, {}'.format('rac{d \\phi}{dx}',labels[idx]))\nplt.subplots_adjust(wspace=0.2, hspace=0.2)\nfig.suptitle(r'$\\phi$ and $\\frac{d\\phi}{d x}$, LGL vs uniformly-spaced nodes')\nplt.show()",
"_____no_output_____"
]
],
[
[
"### remarks on discrete derivative operators",
"_____no_output_____"
],
[
"Suppose we compute the derivative matrix $D_r$ at the nodal points for some order $p$. There are some interesting properties to understand about these derivative matrices.",
"_____no_output_____"
],
[
"#### annihilation of constant vectors",
"_____no_output_____"
],
[
"Note that if we represent a function nodally, i.e., $u = c_i \\phi_i(x)$, then\n\n$$ \\frac{du}{dx} = \\frac{d}{dx}(c_i \\phi_i(x)) = c_i \\frac{d\\phi_i}{dx} $$\n\nTherefore, if we want to discretely take a derivative of a function (we'll represent the function on the master element for now, but trivially, we could map it to some other region), it suffices to multiply the derivative operator with the nodal vector $D_r u$. It should be clear, then, that $D_r$ will annihilate any constant vector.",
"_____no_output_____"
]
],
[
[
"p = 2\nnb = p+1\nnodal_pts, _ = LegendreGaussLobatto(nb)\nu = np.ones_like(nodal_pts)\n\n# grad vandermonde\nV = Vandermonde1D(N=nb, x=nodal_pts)\nVr = GradVandermonde1D(N=nb, x=nodal_pts)\nVi = np.linalg.inv(V)\nDr = np.dot(Vr, Vi)\n\nduh = np.dot(Dr, u)\nprint(np.max(np.abs(duh)))",
"1.1102230246251565e-16\n"
]
],
[
[
"#### exponential convergence in $p$ for smooth functions\n\nIf the function of which we are attempting to take the discrete derivative is smooth (in the sense of infinitely differentiable), then we will see exponential convergence to the analytical solution w/r/t polynomial order of the nodal basis.",
"_____no_output_____"
]
],
[
[
"ps = [1, 2, 3, 4, 8, 12, 16, 18, 24, 32]\nerrs = []\nfig, ax = plt.subplots(1, 3, figsize=(8, 3))\nfor p in ps:\n nb = p+1\n nodal_pts, _ = LegendreGaussLobatto(nb)\n view_pts = np.linspace(-1, 1, 100)\n\n # grad vandermonde\n V = Vandermonde1D(N=nb, x=nodal_pts)\n Vr = GradVandermonde1D(N=nb, x=nodal_pts)\n Vi = np.linalg.inv(V)\n Dr = np.dot(Vr, Vi)\n\n # nodal shap\n V = Vandermonde1D(N=nb, x=nodal_pts)\n Vti = np.linalg.inv(V.T)\n P = P_tilde(r=view_pts, N=nb)\n view_shap = np.dot(Vti, P)\n \n u = np.sin(nodal_pts-np.pi/4.)\n du = np.cos(view_pts-np.pi/4)\n duh = np.dot(Dr, u)\n\n view_duh = np.dot(duh, view_shap)\n err = np.max(np.abs(view_duh - du))\n errs.append(err)\n \n # plot first few\n if p < 4:\n ax[p-1].plot(view_pts, np.sin(view_pts), label=r'$u$')\n ax[p-1].plot(view_pts, du, label=r'$u^\\prime$')\n ax[p-1].plot(view_pts, view_duh, '--', label=r'$du_h$') \n ax[p-1].set_title(r'$p={}$'.format(p))\n ax[p-1].legend()\nplt.show()",
"_____no_output_____"
],
[
"fig, ax = plt.subplots()\nax.semilogy(ps, errs)\nax.set_xticks(ps)\nax.set_ylabel(r'$||du - du_h||_{L_\\infty}$')\nax.set_xlabel('polynomial order p')\nax.set_title('exponential convergence of discrete derivative')\nplt.show()",
"_____no_output_____"
]
],
[
[
"# creation of a 1D master element",
"_____no_output_____"
],
[
"Define a simple nodal basis object for 1D problems -- no need to be fancy, pretty much the only thing we need this for is to get nodal shape functions and their derivatives efficiently. The underlying orthonormal Legendre polynomial basis is hidden to the user.",
"_____no_output_____"
]
],
[
[
"class NodalBasis1D(object):\n \"\"\" minimalist nodal basis object: \n efficiently computes shape functions and their derivatives \n \"\"\"\n def __init__(self, p, node_spacing='GAUSS_LOBATTO'):\n self.nb = p + 1\n if node_spacing == 'GAUSS_LOBATTO':\n self.nodal_pts, _ = LegendreGaussLobatto(self.nb)\n elif node_spacing == 'EQUIDISTANT':\n self.nodal_pts = np.linspace(-1, 1, self.nb)\n else: raise ValueError('node_spacing {} not recognized'.format(node_spacing))\n \n def shape_functions_at_pts(self, pts):\n \"\"\" computes shape functions evaluated at pts on [-1, 1]\n @retval shap (len(pts), nb) phi_j(pts[i])\n \"\"\"\n V = Vandermonde1D(N=self.nb, x=self.nodal_pts)\n VTinv = np.linalg.inv(V.T)\n P = P_tilde(r=pts, N=self.nb)\n shap = np.dot(VTinv, P)\n return shap.T\n \n def shape_function_derivatives_at_pts(self, pts):\n \"\"\" computes shape function derivatives w/r/t x on [-1, 1]\n @retval shap_der, (Dr in Hesthaven), (len(pts), nb) d/dx phi_j(pts[i])\n \"\"\"\n V = Vandermonde1D(N=self.nb, x=self.nodal_pts)\n Vx = GradVandermonde1D(N=self.nb, x=pts)\n Vinv = np.linalg.inv(V)\n shap_der = np.dot(Vx, Vinv)\n return shap_der",
"_____no_output_____"
]
],
[
[
"Define a 1D master element, which is built on top of the 1D basis. \n- Precompute shape functions at the nodal points and Gauss Legendre quadrature points, both are useful for different types of schemes. We use Gauss Legendre points instead of Gauss Lobatto points because they can integrate degree $2n-1$ polynomials exactly instead of $2n - 3$, where $n$ is the number of integration points. We would like to integrate $(\\phi_i, \\phi_j)_{\\hat{K}}$, which is order 2$p$, so to integrate the mass matrix exactly, we need $2p + 1$ points, and common practice is $2p+2$. Since quadrature in 1D is cheap, we opt for the latter.\n- Precompute mass matrix $M_{ij} = (\\phi_i, \\phi_j)$ and stiffness matrices $S_{ij} = \\left(\\phi_i, \\frac{d\\phi_j}{dx}\\right)$, $K_{ij} = \\left(\\frac{d\\phi_i}{dx}, \\frac{d\\phi_j}{dx}\\right)$. Additionally, store $M^{-1}$, as it is commonly used. Although Hesthaven's method for mass and stiffness matrices are elegant, they rely on the underlying choice of an orthanormal modal basis. Since this class could be overloaded to work with other choices of basis, better to simply compute these matrices with quadrature.",
"_____no_output_____"
]
],
[
[
"class Master1D(object):\n \"\"\" minimalist 1D master object \"\"\"\n def __init__(self, p, nquad_pts=None, *args, **kwargs):\n self.p, self.nb = p, p+1\n self.basis = NodalBasis1D(p=p, **kwargs)\n self.nodal_pts = self.basis.nodal_pts\n self.nq = 2*self.p + 2 if nquad_pts is None else nquad_pts\n self.quad_pts, self.wghts = GaussLegendre(self.nq)\n \n # shape functions at nodal and quadrature points\n self.shap_nodal, self.dshap_nodal = self.mk_shap_and_dshap_at_pts(self.nodal_pts)\n self.shap_quad, self.dshap_quad = self.mk_shap_and_dshap_at_pts(self.quad_pts)\n \n # mass, stiffness matrices\n self.M, self.S, self.K = self.mk_M(), self.mk_S(), self.mk_K()\n self.Minv = np.linalg.inv(self.M)\n \n # lifting permuatation matrix L (0s, 1s)\n self.L = self.mk_L()\n \n def mk_shap_and_dshap_at_pts(self, pts):\n shap = self.basis.shape_functions_at_pts(pts)\n dshap = self.basis.shape_function_derivatives_at_pts(pts)\n return shap, dshap\n \n def mk_M(self):\n shapw = np.dot(np.diag(self.wghts), self.shap_quad)\n M = np.dot(self.shap_quad.T, shapw)\n return M\n \n def mk_S(self):\n dshapw = np.dot(np.diag(self.wghts), self.dshap_quad)\n S = np.dot(self.shap_quad.T, dshapw)\n return S\n \n def mk_K(self):\n dshapw = np.dot(np.diag(self.wghts), self.dshap_quad)\n K = np.dot(self.dshap_quad.T, dshapw)\n return K\n \n def mk_L(self):\n L = np.zeros((self.nb, 2))\n L[0, 0] = 1\n L[-1, 1] = 1\n return L \n \n @property\n def shap_der(self): \n \"\"\" return the shape derivatives for apps expecting 2, 3D\"\"\"\n return [self.dshap_quad]",
"_____no_output_____"
]
],
[
[
"# creation of 1D mesh and DOF handler",
"_____no_output_____"
],
[
"## 1D mappings",
"_____no_output_____"
],
[
"For 1D problems, the mapping from the master element to physical space elements is somewhat trivial, since there's no reason for the transformation to be anything except affine. Note though, that when the 1D elements are embedded in 2D, then the transformations may be non-affine, in which case we must handle isoparametric mappings and the like. We defer this until later. For an affine mapping, we have the simple mapping\n\n$$x(\\xi) = x_L^k + \\frac{1 + \\xi}{2}(x_R^k - x_L^k)$$\n\nWith which we can move the nodal master points to their physical space coordinates.",
"_____no_output_____"
]
],
[
[
"# build T and P arrays\nP = np.linspace(2, 4, 5)",
"_____no_output_____"
],
[
"class Mesh1D(object):\n def __init__(self, P):\n \"\"\" @param P vertex points, sorted by x position \"\"\"\n self.verts = P\n self.nElm, self.nEdges = len(self.verts) - 1, len(self.verts)\n self.connectivity = self.build_T()\n connected_one_side = np.bincount(self.connectivity.ravel()) == 1\n self.boundary_verts = np.where(connected_one_side)[0]\n \n def build_T(self):\n \"\"\" element connectivity array from 1D vertex list \"\"\"\n T = np.zeros((self.nElm, 2), dtype=int)\n T[:,0] = np.arange(self.nElm)\n T[:,1] = np.arange(self.nElm) + 1\n return T",
"_____no_output_____"
],
[
"class dofh_1D(object): pass\n\nclass DG_dofh_1D(dofh_1D):\n def __init__(self, mesh, master):\n self.mesh, self.master = mesh, master\n self.n_dof = self.master.nb * self.mesh.nElm\n self.dgnodes = self.mk_dgnodes()\n self.lg = self.mk_lg()\n self.lg_PM = self.mk_minus_plus_lg()\n self.nb, self.nElm = self.master.nb, self.mesh.nElm\n self.ed2elm = self.mk_ed2elm()\n \n def mk_dgnodes(self):\n \"\"\" map master nodal pts to element vertices def'd in self.mesh \"\"\"\n dgn = np.zeros((self.master.nb, self.mesh.nElm))\n master_nodal_pts = np.squeeze(self.master.nodal_pts)\n for elm, elm_verts in enumerate(self.mesh.connectivity):\n elm_vert_pts = self.mesh.verts[elm_verts]\n elm_width = elm_vert_pts[1] - elm_vert_pts[0]\n mapped_pts = elm_vert_pts[0] + (1+master_nodal_pts)/2.*(elm_width)\n dgn[:, elm] = mapped_pts \n return dgn\n \n def mk_lg(self):\n \"\"\" number all dof sequentially by dgnodes \"\"\"\n node_numbers = np.arange(np.size(self.dgnodes))\n lg = node_numbers.reshape(self.dgnodes.shape, order='F')\n return lg\n \n def mk_minus_plus_lg(self):\n \"\"\" (-) denotes element interior, (+) denotes exterior\"\"\"\n lg_PM = dict()\n lg_PM['-'] = self.lg[[0, -1], :].ravel(order='F')\n lgP = self.lg[[0, -1],:]\n lgP[0, 1: ] -= 1 # shift nodes to left of first\n lgP[1, :-1] += 1 # shift nodes to right of last\n lg_PM['+'] = lgP.ravel(order='F')\n return lg_PM \n \n def mk_ed2elm(self):\n \"\"\" internal map holding the indicies to reshape vector of values on faces to \n element edge space (2, nElm), duplicating the values on either side of interior faces\n \"\"\"\n f2elm = np.zeros((2, self.nElm))\n faces = np.arange(self.mesh.nEdges)\n # numpy magic is doing the following:\n #\n # [[0, 1, 2, 3]\n # [0, 1, 2, 3]] - ravel('F') -> [0, 0, 1, 1, 2, 2, 3, 3]\n # \n # close, but ends duplicated. => trim the ends and reshape to f2elm shape\n # \n # [[0, 1, 2]\n # [1, 2, 3]]\n #\n f2elm = np.vstack((faces, faces)).ravel(\n order='F')[1:-1].reshape(f2elm.shape, order='F')\n return f2elm\n \n def edge2elm_ed(self, arr):\n \"\"\" internal method to move edge values (defined on the interfaces)\n to values on the \"element edge space\", the edge dof interior to each element\n @param arr array formatted on edge space (nFaces,)\n @retval elmEdArr array formatted on \"element edge space\" (2, nElm)\n \"\"\"\n return arr[self.ed2elm]",
"_____no_output_____"
]
],
[
[
"# computation of fluxes",
"_____no_output_____"
],
[
"The 'back end' of an explicit DG computation is the unrolled vector of all the problem unknowns. The front end that we'd like to interact with is the dgnodes data structure",
"_____no_output_____"
]
],
[
[
"def plot_solution(ax, u, dofh):\n \"\"\" u formatted like dgnodes \"\"\"\n for elm in range(dofh.nElm):\n nodal_pts = dofh.dgnodes[:, elm]\n nodal_values = u[:, elm]\n ax.plot(nodal_pts, nodal_values)\n return ax",
"_____no_output_____"
],
[
"# Low storage Runge-Kutta coefficients LSERK\nrk4a = np.array([\n 0.0,\n -567301805773.0/1357537059087.0,\n -2404267990393.0/2016746695238.0,\n -3550918686646.0/2091501179385.0,\n -1275806237668.0/842570457699.0])\n\nrk4b = np.array([ \n 1432997174477.0/9575080441755.0,\n 5161836677717.0/13612068292357.0,\n 1720146321549.0/2090206949498.0,\n 3134564353537.0/4481467310338.0,\n 2277821191437.0/14882151754819.0])\n\nrk4c = np.array([\n 0.0,\n 1432997174477.0/9575080441755.0,\n 2526269341429.0/6820363962896.0,\n 2006345519317.0/3224310063776.0,\n 2802321613138.0/2924317926251.0])",
"_____no_output_____"
],
[
"# constants\nπ = np.pi\n\n# geometry set up\nP = np.linspace(0, 2*π, 10)\nmesh1d = Mesh1D(P)\nmaster = Master1D(p=2)\ndofh = DG_dofh_1D(mesh1d, master)\nmapdgn = np.zeros((dofh.dgnodes.shape[0], 1, dofh.dgnodes.shape[1]))\nmapdgn[:,0,:] = dofh.dgnodes\n_map = fem_map.Affine_Mapping(master=[master], dgnodes=[mapdgn])",
"_____no_output_____"
]
],
[
[
"We choose numerical fluxes of the form\n\n$$\\widehat{au} =\\left\\{\\!\\!\\left\\{au\\right\\}\\!\\!\\right\\} + (1-\\alpha)\\frac{|a|}{2} \\left[\\!\\!\\left[u\\right]\\!\\!\\right]$$\n\nWhere $\\alpha = 0$ represents an upwinded flux and $\\alpha=1$ represents a central flux. These are shown in Hesthaven to be stable for the equation we are interested in solving.",
"_____no_output_____"
]
],
[
[
"def compute_interior_flux(u, norm, dofh, α):\n \"\"\" computes the numerical flux at all of the element interfaces\n @param u the current solution u, unrolled to a vector\n NOTE: boundary interfaces will be filled with garbage, and must be corrected\n \"\"\"\n pm = dofh.lg_PM\n \n # equivalent to the flux\n # \\hat{au} = {{au}} + (1-α) * |a|/2 * [[u]]\n # at element interfaces. First and last interface will have garbage.\n flux = a/2*(u[pm['-']] + u[pm['+']]) + (1-α)*np.abs(norm*a)/2.*(u[pm['+']] - u[pm['-']])\n return flux",
"_____no_output_____"
]
],
[
[
"# semi-discrete scheme",
"_____no_output_____"
],
[
"Considering the \"weak\" DG-FEM form, we have the semi-discrete element local equation\n\n\\begin{align}\n \\int_K \\frac{\\partial u_h}{\\partial t} v \\, dK\n +\\int_{K} (au_h) \\frac{\\partial v}{\\partial x} \\, dK = \n -\\int_{\\partial K} \\hat{n}\\cdot \\widehat{au} v \\, d\\partial K\n\\end{align}\n\nChoosing a representation $u=u_i\\phi_i$ piecewise polynomial over each element, and the same test space, we have, for a given choice of numerical flux $\\widehat{au}$, and noting that in 1D, the normal vectors are simply (-1, +1):\n\n\\begin{align}\n \\int_K \\frac{\\partial}{\\partial t} (u_i(t) \\phi_i(x)) \\phi_j(x) \\, dx\n +\\int_{K} a(u_i(t)\\phi_i(x)) \\frac{\\partial \\phi_j(x)}{\\partial x} \\, dx = \n -(\\widehat{au}(x_R) - \\widehat{au}(x_L))\n\\end{align}\n\ntransforming the integrals to the reference element:\n\n\\begin{align}\n \\int_{\\hat{K}} \\frac{\\partial}{\\partial t} (u_i(t) \\phi_i(\\xi)) \\phi_j(\\xi) \\,|det(J)|\\, d\\xi\n +\\int_{\\hat{K}} a(u_i(t)\\phi_i(\\xi)) \\frac{\\partial \\phi_j(\\xi)}{\\partial \\xi} \\, |det(J)|\\, d\\xi = \n -(\\widehat{au}(x_R) - \\widehat{au}(x_L))\n\\end{align}\n\nThis completes the description of the semi-discrete scheme, and we have a choice as to how to compute these integrals. The important part is that since the coefficients $u_i$ vary in time but are constants with respect to space, we can write \n\n\\begin{align}\n &\\frac{\\partial u_i(t)}{\\partial t} \\int_{\\hat{K}} \\phi_i(\\xi) \\phi_j(\\xi) \\,|det(J)|\\, d\\xi\n +au_i\\int_{\\hat{K}} \\phi_i(\\xi) \\left(\\frac{d\\xi}{dx}\\right)\\frac{\\partial \\phi_j(\\xi)}{\\partial \\xi} \\, |det(J)|\\, d\\xi = \n -(\\widehat{au}(x_R) - \\widehat{au}(x_L)) \\\\\n &\\Rightarrow M_K \\vec{\\frac{du_h}{dt}} + a S_K \\vec{u_h} \n = - L\\, (\\widehat{au}(x_R) - \\widehat{au}(x_L))\n\\end{align}\n\nWhere we have computed $M_K$ and $S_K$, the mass and stiffness matrices for element $K$. Although we would normally do this with a quadrature rule, we can take advantage of the fact that in 1D (and indeed under any affine mapping from reference to physical element), $J^{-1}$ and $|\\det(J)|$ will be constant over the entire element (also note that in 1D, $J^{-1}$ is a 1x1 matrix)<sup>1</sup>. In that case, we can treat both as constants, precompute $M_{\\hat{K}}, S_{\\hat{K}}$, and multiply the entire element-local equation by $M^{-1}$, giving\n\n\\begin{align}\n\\vec{\\frac{du_h}{dt}} &= - a \\frac{\\det(J)_K}{\\det(J)_K}\\, J^{-1}_K M_{\\hat{K}}^{-1}S^T_{\\hat{K}} \\vec{u_h} \n - \\frac{1}{\\det(J)_K} M^{-1}_K L\\, (\\widehat{au}(x_R) - \\widehat{au}(x_L)) \\\\\n &= - a \\, J^{-1}_K M_{\\hat{K}}^{-1}S^T_{\\hat{K}} \\vec{u_h} \n - \\frac{1}{\\det(J)_K} M^{-1}_K L\\, (\\widehat{au}(x_R) - \\widehat{au}(x_L))\n\\end{align}\n\nWhich is a good form for a black box integrator, since we have a \"naked\" $\\frac{du_h}{dt}$, and because the scheme is explicit.\n\nnote<sup>1</sup>: $J, J^{-1}$ are 1x1 matrices, and $\\det{J}$ is simply $J_{11}$; $J^{-1} = 1/J_{11}$. It's important for the clarity of explicit schemes to understand where these cancellations occur.",
"_____no_output_____"
]
],
[
[
"def advect_rhs_1D(u, t_local, a, dofh, _map, master, flux_fn, gD, norm):\n \n return u",
"_____no_output_____"
],
[
"# final time\nT = 10\n\n# compute time step size, irrelevant for backward euler\nCFL = 0.75\nΔx = dofh.dgnodes[1,0] - dofh.dgnodes[0,0]\nΔt = CFL/(2*π)*Δx\nΔt = Δt / 2\n\n# number of timesteps needed\nsteps = int(np.ceil(T/Δt))\n\n# initial condition, advection speed\nsolution = np.zeros((steps, *dofh.dgnodes.shape))\na = 2 * np.pi\nsolution[0,::] = np.sin(dofh.dgnodes)\n\nLSERK_stages = [0, 1, 2, 3, 4]\nt = 0\n\ngD = lambda t: -np.sin(a*t)\n\n# normal vectors, all positive\nnorm = np.ones((2, dofh.nElm))\nnorm[0,:] *= -1\n\n# function pointer to something that can compute fluxes\nflux_fn = compute_interior_flux\n\n# time loop\nRK_resid = np.zeros_like(dofh.dgnodes)\nfor tstep in range(3):\n u = solution[tstep, ::]\n for s in LSERK_stages:\n t_local = t + rk4c[s]*Δt\n rhsu = advect_rhs_1D(u, t_local, a, dofh, _map, master, flux_fn, gD, norm)\n RK_resid = rk4a[s]*RK_resid + Δt*rhsu\n u += rk4b[s]*RK_resid\n t += Δt",
"_____no_output_____"
],
[
"u0 = solution[0,:,:]\nfix, ax = plt.subplots()\nax = plot_solution(ax, u0, dofh)",
"_____no_output_____"
],
[
"pm = dofh.lg_PM\nu = u0.ravel()\n\n# normal vectors on interfaces, all positive\nnorm_faces = np.ones(pm['-'].shape[0])\n\nα = 0\n\n# compute interior fluxes\nflux = compute_interior_flux(u, norm_faces, dofh, α)\n\n# compute boundary fluxes\nflux[0] = gD(t_local)\nflux[-1] = flux[0]\n\ndofh.edge2elm_ed(flux)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
c51001443abb637989c43452deb028ee14168057
| 311,342 |
ipynb
|
Jupyter Notebook
|
2. Define the Network Architecture.ipynb
|
mexxik/cv-project-1
|
09a382f41c190970945cbf496d940e5d0fa133b9
|
[
"MIT"
] | null | null | null |
2. Define the Network Architecture.ipynb
|
mexxik/cv-project-1
|
09a382f41c190970945cbf496d940e5d0fa133b9
|
[
"MIT"
] | null | null | null |
2. Define the Network Architecture.ipynb
|
mexxik/cv-project-1
|
09a382f41c190970945cbf496d940e5d0fa133b9
|
[
"MIT"
] | 1 |
2019-05-10T10:26:29.000Z
|
2019-05-10T10:26:29.000Z
| 253.329536 | 37,616 | 0.910626 |
[
[
[
"## Define the Convolutional Neural Network\n\nAfter you've looked at the data you're working with and, in this case, know the shapes of the images and of the keypoints, you are ready to define a convolutional neural network that can *learn* from this data.\n\nIn this notebook and in `models.py`, you will:\n1. Define a CNN with images as input and keypoints as output\n2. Construct the transformed FaceKeypointsDataset, just as before\n3. Train the CNN on the training data, tracking loss\n4. See how the trained model performs on test data\n5. If necessary, modify the CNN structure and model hyperparameters, so that it performs *well* **\\***\n\n**\\*** What does *well* mean?\n\n\"Well\" means that the model's loss decreases during training **and**, when applied to test image data, the model produces keypoints that closely match the true keypoints of each face. And you'll see examples of this later in the notebook.\n\n---\n",
"_____no_output_____"
],
[
"## CNN Architecture\n\nRecall that CNN's are defined by a few types of layers:\n* Convolutional layers\n* Maxpooling layers\n* Fully-connected layers\n\nYou are required to use the above layers and encouraged to add multiple convolutional layers and things like dropout layers that may prevent overfitting. You are also encouraged to look at literature on keypoint detection, such as [this paper](https://arxiv.org/pdf/1710.00977.pdf), to help you determine the structure of your network.\n\n\n### TODO: Define your model in the provided file `models.py` file\n\nThis file is mostly empty but contains the expected name and some TODO's for creating your model.\n\n---",
"_____no_output_____"
],
[
"## PyTorch Neural Nets\n\nTo define a neural network in PyTorch, you define the layers of a model in the function `__init__` and define the feedforward behavior of a network that employs those initialized layers in the function `forward`, which takes in an input image tensor, `x`. The structure of this Net class is shown below and left for you to fill in.\n\nNote: During training, PyTorch will be able to perform backpropagation by keeping track of the network's feedforward behavior and using autograd to calculate the update to the weights in the network.\n\n#### Define the Layers in ` __init__`\nAs a reminder, a conv/pool layer may be defined like this (in `__init__`):\n```\n# 1 input image channel (for grayscale images), 32 output channels/feature maps, 3x3 square convolution kernel\nself.conv1 = nn.Conv2d(1, 32, 3)\n\n# maxpool that uses a square window of kernel_size=2, stride=2\nself.pool = nn.MaxPool2d(2, 2) \n```\n\n#### Refer to Layers in `forward`\nThen referred to in the `forward` function like this, in which the conv1 layer has a ReLu activation applied to it before maxpooling is applied:\n```\nx = self.pool(F.relu(self.conv1(x)))\n```\n\nBest practice is to place any layers whose weights will change during the training process in `__init__` and refer to them in the `forward` function; any layers or functions that always behave in the same way, such as a pre-defined activation function, should appear *only* in the `forward` function.",
"_____no_output_____"
],
[
"#### Why models.py\n\nYou are tasked with defining the network in the `models.py` file so that any models you define can be saved and loaded by name in different notebooks in this project directory. For example, by defining a CNN class called `Net` in `models.py`, you can then create that same architecture in this and other notebooks by simply importing the class and instantiating a model:\n```\n from models import Net\n net = Net()\n```",
"_____no_output_____"
]
],
[
[
"# import the usual resources\nimport matplotlib.pyplot as plt\nimport numpy as np\n\n# watch for any changes in model.py, if it changes, re-load it automatically\n%load_ext autoreload\n%autoreload 2",
"_____no_output_____"
],
[
"## TODO: Define the Net in models.py\n\nimport torch\nimport torch.nn as nn\nimport torch.nn.functional as F\nimport torch\n\ndevice = \"cpu\"\nFloatTensor = torch.FloatTensor\n\n#if torch.cuda.is_available():\n# device = \"cuda:0\"\n# FloatTensor = torch.cuda.FloatTensor\n\n## TODO: Once you've define the network, you can instantiate it\n# one example conv layer has been provided for you\nfrom models import NaimishNet\n\nnet = NaimishNet().to(device)\nprint(net)",
"NaimishNet(\n (conv_1): Conv2d(1, 32, kernel_size=(4, 4), stride=(1, 1))\n (conv_2): Conv2d(32, 64, kernel_size=(3, 3), stride=(1, 1))\n (conv_3): Conv2d(64, 128, kernel_size=(2, 2), stride=(1, 1))\n (conv_4): Conv2d(128, 256, kernel_size=(1, 1), stride=(1, 1))\n (pool): MaxPool2d(kernel_size=2, stride=2, padding=0, dilation=1, ceil_mode=False)\n (dropout_1): Dropout(p=0.1)\n (dropout_2): Dropout(p=0.2)\n (dropout_3): Dropout(p=0.3)\n (dropout_4): Dropout(p=0.4)\n (dropout_5): Dropout(p=0.5)\n (dropout_6): Dropout(p=0.6)\n (fc_1): Linear(in_features=43264, out_features=1000, bias=True)\n (fc_2): Linear(in_features=1000, out_features=500, bias=True)\n (fc_3): Linear(in_features=500, out_features=136, bias=True)\n)\n"
]
],
[
[
"## Transform the dataset \n\nTo prepare for training, create a transformed dataset of images and keypoints.\n\n### TODO: Define a data transform\n\nIn PyTorch, a convolutional neural network expects a torch image of a consistent size as input. For efficient training, and so your model's loss does not blow up during training, it is also suggested that you normalize the input images and keypoints. The necessary transforms have been defined in `data_load.py` and you **do not** need to modify these; take a look at this file (you'll see the same transforms that were defined and applied in Notebook 1).\n\nTo define the data transform below, use a [composition](http://pytorch.org/tutorials/beginner/data_loading_tutorial.html#compose-transforms) of:\n1. Rescaling and/or cropping the data, such that you are left with a square image (the suggested size is 224x224px)\n2. Normalizing the images and keypoints; turning each RGB image into a grayscale image with a color range of [0, 1] and transforming the given keypoints into a range of [-1, 1]\n3. Turning these images and keypoints into Tensors\n\nThese transformations have been defined in `data_load.py`, but it's up to you to call them and create a `data_transform` below. **This transform will be applied to the training data and, later, the test data**. It will change how you go about displaying these images and keypoints, but these steps are essential for efficient training.\n\nAs a note, should you want to perform data augmentation (which is optional in this project), and randomly rotate or shift these images, a square image size will be useful; rotating a 224x224 image by 90 degrees will result in the same shape of output.",
"_____no_output_____"
]
],
[
[
"from torch.utils.data import Dataset, DataLoader\nfrom torchvision import transforms, utils\n\n# the dataset we created in Notebook 1 is copied in the helper file `data_load.py`\nfrom data_load import FacialKeypointsDataset\n# the transforms we defined in Notebook 1 are in the helper file `data_load.py`\nfrom data_load import Rescale, RandomCrop, Normalize, ToTensor\n\n\n## TODO: define the data_transform using transforms.Compose([all tx's, . , .])\n# order matters! i.e. rescaling should come before a smaller crop\ndata_transform = transforms.Compose([Rescale((250, 250)),\n RandomCrop((224, 224)),\n Normalize(), \n ToTensor()])\n\n# testing that you've defined a transform\nassert(data_transform is not None), 'Define a data_transform'",
"_____no_output_____"
],
[
"# create the transformed dataset\ntransformed_dataset = FacialKeypointsDataset(csv_file='data/training_frames_keypoints.csv',\n root_dir='data/training/',\n transform=data_transform)\n\n\nprint('Number of images: ', len(transformed_dataset))\n\n# iterate through the transformed dataset and print some stats about the first few samples\nfor i in range(4):\n sample = transformed_dataset[i]\n print(i, sample['image'].size(), sample['keypoints'].size())",
"Number of images: 3462\n0 torch.Size([1, 224, 224]) torch.Size([68, 2])\n1 torch.Size([1, 224, 224]) torch.Size([68, 2])\n2 torch.Size([1, 224, 224]) torch.Size([68, 2])\n3 torch.Size([1, 224, 224]) torch.Size([68, 2])\n"
]
],
[
[
"## Batching and loading data\n\nNext, having defined the transformed dataset, we can use PyTorch's DataLoader class to load the training data in batches of whatever size as well as to shuffle the data for training the model. You can read more about the parameters of the DataLoader, in [this documentation](http://pytorch.org/docs/master/data.html).\n\n#### Batch size\nDecide on a good batch size for training your model. Try both small and large batch sizes and note how the loss decreases as the model trains.\n\n**Note for Windows users**: Please change the `num_workers` to 0 or you may face some issues with your DataLoader failing.",
"_____no_output_____"
]
],
[
[
"# load training data in batches\nbatch_size = 8\n\ntrain_loader = DataLoader(transformed_dataset, \n batch_size=batch_size,\n shuffle=True, \n num_workers=0)\n",
"_____no_output_____"
]
],
[
[
"## Before training\n\nTake a look at how this model performs before it trains. You should see that the keypoints it predicts start off in one spot and don't match the keypoints on a face at all! It's interesting to visualize this behavior so that you can compare it to the model after training and see how the model has improved.\n\n#### Load in the test dataset\n\nThe test dataset is one that this model has *not* seen before, meaning it has not trained with these images. We'll load in this test data and before and after training, see how your model performs on this set!\n\nTo visualize this test data, we have to go through some un-transformation steps to turn our images into python images from tensors and to turn our keypoints back into a recognizable range. ",
"_____no_output_____"
]
],
[
[
"# load in the test data, using the dataset class\n# AND apply the data_transform you defined above\n\n# create the test dataset\ntest_data_transform = transforms.Compose([Rescale((224, 224)), \n Normalize(), \n ToTensor()])\n\ntest_dataset = FacialKeypointsDataset(csv_file='data/test_frames_keypoints.csv',\n root_dir='data/test/',\n transform=test_data_transform)\n\n",
"_____no_output_____"
],
[
"# load test data in batches\nbatch_size = 10\n\ntest_loader = DataLoader(test_dataset, \n batch_size=batch_size,\n shuffle=True, \n num_workers=4)",
"_____no_output_____"
]
],
[
[
"## Apply the model on a test sample\n\nTo test the model on a test sample of data, you have to follow these steps:\n1. Extract the image and ground truth keypoints from a sample\n2. Make sure the image is a FloatTensor, which the model expects.\n3. Forward pass the image through the net to get the predicted, output keypoints.\n\nThis function test how the network performs on the first batch of test data. It returns the images, the transformed images, the predicted keypoints (produced by the model), and the ground truth keypoints.",
"_____no_output_____"
]
],
[
[
"# test the model on a batch of test images\n\ndef net_sample_output():\n \n # iterate through the test dataset\n for i, sample in enumerate(test_loader):\n \n # get sample data: images and ground truth keypoints\n images = sample['image']\n key_pts = sample['keypoints']\n\n # convert images to FloatTensors\n images = images.type(FloatTensor)\n\n # forward pass to get net output\n output_pts = net(images)\n \n # reshape to batch_size x 68 x 2 pts\n output_pts = output_pts.view(output_pts.size()[0], 68, -1)\n \n # break after first image is tested\n if i == 0:\n return images, output_pts, key_pts\n ",
"_____no_output_____"
]
],
[
[
"#### Debugging tips\n\nIf you get a size or dimension error here, make sure that your network outputs the expected number of keypoints! Or if you get a Tensor type error, look into changing the above code that casts the data into float types: `images = images.type(torch.FloatTensor)`.",
"_____no_output_____"
]
],
[
[
"# call the above function\n# returns: test images, test predicted keypoints, test ground truth keypoints\ntest_images, test_outputs, gt_pts = net_sample_output()\n\n# print out the dimensions of the data to see if they make sense\nprint(test_images.data.size())\nprint(test_outputs.data.size())\nprint(gt_pts.size())",
"torch.Size([10, 1, 224, 224])\ntorch.Size([10, 68, 2])\ntorch.Size([10, 68, 2])\n"
]
],
[
[
"## Visualize the predicted keypoints\n\nOnce we've had the model produce some predicted output keypoints, we can visualize these points in a way that's similar to how we've displayed this data before, only this time, we have to \"un-transform\" the image/keypoint data to display it.\n\nNote that I've defined a *new* function, `show_all_keypoints` that displays a grayscale image, its predicted keypoints and its ground truth keypoints (if provided).",
"_____no_output_____"
]
],
[
[
"def show_all_keypoints(image, predicted_key_pts, gt_pts=None):\n \"\"\"Show image with predicted keypoints\"\"\"\n # image is grayscale\n plt.imshow(image, cmap='gray')\n plt.scatter(predicted_key_pts[:, 0], predicted_key_pts[:, 1], s=20, marker='.', c='m')\n # plot ground truth points as green pts\n if gt_pts is not None:\n plt.scatter(gt_pts[:, 0], gt_pts[:, 1], s=20, marker='.', c='g')\n",
"_____no_output_____"
]
],
[
[
"#### Un-transformation\n\nNext, you'll see a helper function. `visualize_output` that takes in a batch of images, predicted keypoints, and ground truth keypoints and displays a set of those images and their true/predicted keypoints.\n\nThis function's main role is to take batches of image and keypoint data (the input and output of your CNN), and transform them into numpy images and un-normalized keypoints (x, y) for normal display. The un-transformation process turns keypoints and images into numpy arrays from Tensors *and* it undoes the keypoint normalization done in the Normalize() transform; it's assumed that you applied these transformations when you loaded your test data.",
"_____no_output_____"
]
],
[
[
"# visualize the output\n# by default this shows a batch of 10 images\ndef visualize_output(test_images, test_outputs, gt_pts=None, batch_size=10):\n\n for i in range(batch_size):\n plt.figure(figsize=(20,10))\n ax = plt.subplot(1, batch_size, i+1)\n\n # un-transform the image data\n image = test_images[i].data # get the image from it's wrapper\n image = image.numpy() # convert to numpy array from a Tensor\n image = np.transpose(image, (1, 2, 0)) # transpose to go from torch to numpy image\n\n # un-transform the predicted key_pts data\n predicted_key_pts = test_outputs[i].data\n predicted_key_pts = predicted_key_pts.numpy()\n # undo normalization of keypoints \n predicted_key_pts = predicted_key_pts*50.0+100\n \n # plot ground truth points for comparison, if they exist\n ground_truth_pts = None\n if gt_pts is not None:\n ground_truth_pts = gt_pts[i] \n ground_truth_pts = ground_truth_pts*50.0+100\n \n # call show_all_keypoints\n show_all_keypoints(np.squeeze(image), predicted_key_pts, ground_truth_pts)\n \n plt.axis('off')\n\n plt.show()\n \n# call it\nvisualize_output(test_images, test_outputs, gt_pts)",
"_____no_output_____"
]
],
[
[
"## Training\n\n#### Loss function\nTraining a network to predict keypoints is different than training a network to predict a class; instead of outputting a distribution of classes and using cross entropy loss, you may want to choose a loss function that is suited for regression, which directly compares a predicted value and target value. Read about the various kinds of loss functions (like MSE or L1/SmoothL1 loss) in [this documentation](http://pytorch.org/docs/master/_modules/torch/nn/modules/loss.html).\n\n### TODO: Define the loss and optimization\n\nNext, you'll define how the model will train by deciding on the loss function and optimizer.\n\n---",
"_____no_output_____"
]
],
[
[
"## TODO: Define the loss and optimization\nimport torch.optim as optim\n\ncriterion = nn.SmoothL1Loss() #nn.MSELoss()\n\noptimizer = optim.Adam(net.parameters(), lr=0.001, betas=(0.9, 0.999), eps=1e-08)\n",
"_____no_output_____"
]
],
[
[
"## Training and Initial Observation\n\nNow, you'll train on your batched training data from `train_loader` for a number of epochs. \n\nTo quickly observe how your model is training and decide on whether or not you should modify it's structure or hyperparameters, you're encouraged to start off with just one or two epochs at first. As you train, note how your the model's loss behaves over time: does it decrease quickly at first and then slow down? Does it take a while to decrease in the first place? What happens if you change the batch size of your training data or modify your loss function? etc. \n\nUse these initial observations to make changes to your model and decide on the best architecture before you train for many epochs and create a final model.",
"_____no_output_____"
]
],
[
[
"import sys\nimport math\n\n\ndef process_data(data_loader, train=False):\n if train:\n net.train()\n else:\n net.eval()\n\n losses = []\n average_loss = 0.0\n #running_loss = 0.0\n\n batch_count = len(data_loader)\n\n for batch_i, data in enumerate(data_loader):\n # get the input images and their corresponding labels\n images = data['image']\n key_pts = data['keypoints']\n\n # flatten pts\n key_pts = key_pts.view(key_pts.size(0), -1)\n\n # convert variables to floats for regression loss\n key_pts = key_pts.type(FloatTensor)\n images = images.type(FloatTensor)\n\n if train:\n # forward pass to get outputs\n output_pts = net(images)\n else:\n with torch.no_grad():\n output_pts = net(images)\n\n # calculate the loss between predicted and target keypoints\n loss = criterion(output_pts, key_pts)\n\n if train:\n # zero the parameter (weight) gradients\n optimizer.zero_grad()\n # backward pass to calculate the weight gradients\n loss.backward()\n # update the weights\n optimizer.step()\n\n # print loss statistics\n # to convert loss into a scalar and add it to the running_loss, use .item()\n running_loss = loss.item()\n losses.append(running_loss)\n\n # average_training_loss = total_training_loss / (batch_i + 1)\n #if batch_i % 10 == 9: # print every 10 batches\n action = \"training\" if train else \"validating\"\n print('\\r{} batch: {}/{}, current loss: {:.3f}'.format(action, batch_i + 1, batch_count, running_loss), end=\"\")\n sys.stdout.flush()\n #running_loss = 0.0\n\n average_loss = np.mean(losses)\n\n return average_loss\n\n\ndef train_net(n_epochs, patience):\n last_train_loss = math.inf\n last_valid_loss = math.inf\n best_valid_loss = last_valid_loss\n epochs_without_improvement = 0\n\n for epoch in range(n_epochs):\n print(\"\\n--------------------------------------------------------------------------\")\n print(\"Epoch: {}, last train loss: {:.3f}, last valid loss: {:.3f}, best valid loss: {:.3f}\".format(\n epoch + 1, last_train_loss, last_valid_loss, best_valid_loss\n ))\n last_train_loss = process_data(train_loader, train=True)\n test_loss = process_data(test_loader, train=False)\n\n last_valid_loss = test_loss\n if last_valid_loss < best_valid_loss:\n best_valid_loss = last_valid_loss\n torch.save(net.state_dict(), \"saved_models/keypoints_model.pth\")\n else:\n epochs_without_improvement += 1\n\n if epochs_without_improvement > patience:\n print(\"\\nstopping early\")\n break",
"_____no_output_____"
],
[
"# train your network\nn_epochs = 50\npatience = 3\n\ntrain_net(n_epochs, patience)",
"\n--------------------------------------------------------------------------\nEpoch: 1, last train loss: inf, last valid loss: inf, best valid loss: inf\nvalidating batch: 97/97, current loss: 0.0682.2970\n--------------------------------------------------------------------------\nEpoch: 2, last train loss: 1327781.065, last valid loss: 0.069, best valid loss: 0.069\nvalidating batch: 97/97, current loss: 0.060.984\n--------------------------------------------------------------------------\nEpoch: 3, last train loss: 51.834, last valid loss: 0.035, best valid loss: 0.035\nvalidating batch: 97/97, current loss: 0.046.4088\n--------------------------------------------------------------------------\nEpoch: 4, last train loss: 454.936, last valid loss: 0.028, best valid loss: 0.028\nvalidating batch: 97/97, current loss: 0.0309.023\n--------------------------------------------------------------------------\nEpoch: 5, last train loss: 239.124, last valid loss: 0.027, best valid loss: 0.027\nvalidating batch: 97/97, current loss: 0.015\n--------------------------------------------------------------------------\nEpoch: 6, last train loss: 0.049, last valid loss: 0.027, best valid loss: 0.027\nvalidating batch: 97/97, current loss: 0.025\n--------------------------------------------------------------------------\nEpoch: 7, last train loss: 0.049, last valid loss: 0.028, best valid loss: 0.027\nvalidating batch: 97/97, current loss: 0.115\n--------------------------------------------------------------------------\nEpoch: 8, last train loss: 0.048, last valid loss: 0.028, best valid loss: 0.027\nvalidating batch: 97/97, current loss: 0.029\nstopping early\n"
]
],
[
[
"## Test data\n\nSee how your model performs on previously unseen, test data. We've already loaded and transformed this data, similar to the training data. Next, run your trained model on these images to see what kind of keypoints are produced. You should be able to see if your model is fitting each new face it sees, if the points are distributed randomly, or if the points have actually overfitted the training data and do not generalize.",
"_____no_output_____"
],
[
"## Test data\n\nSee how your model performs on previously unseen, test data. We've already loaded and transformed this data, similar to the training data. Next, run your trained model on these images to see what kind of keypoints are produced. You should be able to see if your model is fitting each new face it sees, if the points are distributed randomly, or if the points have actually overfitted the training data and do not generalize.",
"_____no_output_____"
],
[
"## Test data\n\nSee how your model performs on previously unseen, test data. We've already loaded and transformed this data, similar to the training data. Next, run your trained model on these images to see what kind of keypoints are produced. You should be able to see if your model is fitting each new face it sees, if the points are distributed randomly, or if the points have actually overfitted the training data and do not generalize.",
"_____no_output_____"
],
[
"## Test data\n\nSee how your model performs on previously unseen, test data. We've already loaded and transformed this data, similar to the training data. Next, run your trained model on these images to see what kind of keypoints are produced. You should be able to see if your model is fitting each new face it sees, if the points are distributed randomly, or if the points have actually overfitted the training data and do not generalize.",
"_____no_output_____"
],
[
"## Test data\n\nSee how your model performs on previously unseen, test data. We've already loaded and transformed this data, similar to the training data. Next, run your trained model on these images to see what kind of keypoints are produced. You should be able to see if your model is fitting each new face it sees, if the points are distributed randomly, or if the points have actually overfitted the training data and do not generalize.",
"_____no_output_____"
]
],
[
[
"net.load_state_dict(torch.load(\"saved_models/keypoints_model.pth\", map_location=\"cpu\"))\n\n# get a sample of test data again\ntest_images, test_outputs, gt_pts = net_sample_output()\n\nprint(test_images.data.size())\nprint(test_outputs.data.size())\nprint(gt_pts.size())",
"torch.Size([10, 1, 224, 224])\ntorch.Size([10, 68, 2])\ntorch.Size([10, 68, 2])\n"
],
[
"## TODO: visualize your test output\n# you can use the same function as before, by un-commenting the line below:\n\nvisualize_output(test_images, test_outputs, gt_pts)\n",
"_____no_output_____"
]
],
[
[
"Once you've found a good model (or two), save your model so you can load it and use it later!",
"_____no_output_____"
]
],
[
[
"## TODO: change the name to something uniqe for each new model\nmodel_dir = 'saved_models/'\nmodel_name = 'keypoints_model_1.pt'\n\n# after training, save your model parameters in the dir 'saved_models'\ntorch.save(net.state_dict(), model_dir+model_name)",
"_____no_output_____"
]
],
[
[
"After you've trained a well-performing model, answer the following questions so that we have some insight into your training and architecture selection process. Answering all questions is required to pass this project.",
"_____no_output_____"
],
[
"### Question 1: What optimization and loss functions did you choose and why?\n",
"_____no_output_____"
],
[
"**Answer**: I tried 2 loss functions: Mean Square Error and Smooth L1. As for the optimizer, the recommended by the original Paper Adam was selected.\n\nFew expreriements were conducted with different hyper-paramers sets. Check report on that below. ",
"_____no_output_____"
],
[
"### Question 2: What kind of network architecture did you start with and how did it change as you tried different architectures? Did you decide to add more convolutional layers or any layers to avoid overfitting the data?",
"_____no_output_____"
],
[
"**Answer**: Naimish archtecture was selected as in Facial Key Points Detection using Deep Convolutional Neural Network paper.",
"_____no_output_____"
],
[
"### Question 3: How did you decide on the number of epochs and batch_size to train your model?",
"_____no_output_____"
],
[
"\n**Answer**: There were attempts to train the model on both CPU and GPU. In a GPU case the batch size was selected based on the GPU memory available. \n\nAs you can see above, a regularization technique Early Stopping was implemented with patience 3 which monitored test set loss. As soon training stopped improving the training loss for more than 3 epochs, the processed is stopped. Besides, the function saves the model with the lowest training loss.\n\n(check results.txt for more detailed info about training)\n\n### CPU with MSE loss and 8 batch size\nThis attempt reached test loss 0.698 on the 21st epoch and stopped on the 23rd.\nWe consider this try as a baseline\n\n### CPU with MSE loss and 128 batch size\nIncreasing batch size significantly. Test loss reached 23.369 on 11th epoch and stopped shotly. We consider this test failed.\n\n### GPU with MSE loss and 8 batch size\nTrying to repoduce CPU's result on GPU to see if it is any better. It reached 0.691 value on a test set on the 19th epoch and stopped. We can say that the performance is the same as on CPU.\n\n### GPU with Smooth L1 loss and 8 batch size\nIt reached test loss value 0.027 on 5th epoch! This is the fastest and the most effecient set of parameters.",
"_____no_output_____"
],
[
"## Feature Visualization\n\nSometimes, neural networks are thought of as a black box, given some input, they learn to produce some output. CNN's are actually learning to recognize a variety of spatial patterns and you can visualize what each convolutional layer has been trained to recognize by looking at the weights that make up each convolutional kernel and applying those one at a time to a sample image. This technique is called feature visualization and it's useful for understanding the inner workings of a CNN.",
"_____no_output_____"
],
[
"In the cell below, you can see how to extract a single filter (by index) from your first convolutional layer. The filter should appear as a grayscale grid.",
"_____no_output_____"
]
],
[
[
"# Get the weights in the first conv layer, \"conv1\"\n# if necessary, change this to reflect the name of your first conv layer\nweights1 = net.conv_1.weight.data\n\nw = weights1.numpy()\n\nfilter_index = 0\n\nprint(w[filter_index][0])\nprint(w[filter_index][0].shape)\n\n# display the filter weights\nplt.imshow(w[filter_index][0], cmap='gray')\n",
"[[ 0.4506664 0.82551289 0.13585635 0.96562827]\n [-0.00785176 0.36505774 0.94039446 0.9160589 ]\n [ 0.15764125 0.63777268 0.11388673 0.69258046]\n [ 0.32289311 0.82186466 0.55420667 0.24580522]]\n(4, 4)\n"
]
],
[
[
"## Feature maps\n\nEach CNN has at least one convolutional layer that is composed of stacked filters (also known as convolutional kernels). As a CNN trains, it learns what weights to include in it's convolutional kernels and when these kernels are applied to some input image, they produce a set of **feature maps**. So, feature maps are just sets of filtered images; they are the images produced by applying a convolutional kernel to an input image. These maps show us the features that the different layers of the neural network learn to extract. For example, you might imagine a convolutional kernel that detects the vertical edges of a face or another one that detects the corners of eyes. You can see what kind of features each of these kernels detects by applying them to an image. One such example is shown below; from the way it brings out the lines in an the image, you might characterize this as an edge detection filter.\n\n<img src='images/feature_map_ex.png' width=50% height=50%/>\n\n\nNext, choose a test image and filter it with one of the convolutional kernels in your trained CNN; look at the filtered output to get an idea what that particular kernel detects.\n\n### TODO: Filter an image to see the effect of a convolutional kernel\n---",
"_____no_output_____"
]
],
[
[
"import cv2\n\n\nweights1 = net.conv_1.weight.data\n\nw = weights1.numpy()\n\nfilter_index = 0\n\nprint(w[filter_index][0])\nprint(w[filter_index][0].shape)\n\nidx_img = 0\nimg = np.squeeze(test_images[idx_img].data.numpy())\n\nplt.imshow(w[filter_index][0], cmap='gray')\n\nfiltered_img = cv2.filter2D(img, -1, w[filter_index][0])\nplt.imshow(filtered_img, cmap=\"gray\")",
"[[ 0.4506664 0.82551289 0.13585635 0.96562827]\n [-0.00785176 0.36505774 0.94039446 0.9160589 ]\n [ 0.15764125 0.63777268 0.11388673 0.69258046]\n [ 0.32289311 0.82186466 0.55420667 0.24580522]]\n(4, 4)\n"
]
],
[
[
"### Question 4: Choose one filter from your trained CNN and apply it to a test image; what purpose do you think it plays? What kind of feature do you think it detects?\n",
"_____no_output_____"
],
[
"**Answer**: I seems like this filter blurs an image a bit.",
"_____no_output_____"
],
[
"---\n## Moving on!\n\nNow that you've defined and trained your model (and saved the best model), you are ready to move on to the last notebook, which combines a face detector with your saved model to create a facial keypoint detection system that can predict the keypoints on *any* face in an image!",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
c51003433d502a05bf831021d7cdc826a34e61c1
| 148,727 |
ipynb
|
Jupyter Notebook
|
LegalBERT-th/Legalbert_finetuning.ipynb
|
WiratchawaKannika/bert
|
174cb4714436be1e8e2f11cfa485d0233ee79905
|
[
"Apache-2.0"
] | null | null | null |
LegalBERT-th/Legalbert_finetuning.ipynb
|
WiratchawaKannika/bert
|
174cb4714436be1e8e2f11cfa485d0233ee79905
|
[
"Apache-2.0"
] | null | null | null |
LegalBERT-th/Legalbert_finetuning.ipynb
|
WiratchawaKannika/bert
|
174cb4714436be1e8e2f11cfa485d0233ee79905
|
[
"Apache-2.0"
] | null | null | null | 98.690776 | 23,920 | 0.721133 |
[
[
[
"# **Legal BERT-th (FineTuning)**",
"_____no_output_____"
]
],
[
[
"%cd bert_finetuning",
"/home/jupyter/bert_finetuning\n"
],
[
"pwd",
"_____no_output_____"
]
],
[
[
"> Install and import libraries",
"_____no_output_____"
]
],
[
[
"!pip install tensorflow-gpu==1.15",
"Collecting tensorflow-gpu==1.15\n Using cached tensorflow_gpu-1.15.0-cp37-cp37m-manylinux2010_x86_64.whl (411.5 MB)\nRequirement already satisfied: wrapt>=1.11.1 in /opt/conda/lib/python3.7/site-packages (from tensorflow-gpu==1.15) (1.12.1)\nRequirement already satisfied: tensorflow-estimator==1.15.1 in /opt/conda/lib/python3.7/site-packages (from tensorflow-gpu==1.15) (1.15.1)\nRequirement already satisfied: keras-preprocessing>=1.0.5 in /opt/conda/lib/python3.7/site-packages (from tensorflow-gpu==1.15) (1.1.2)\nRequirement already satisfied: gast==0.2.2 in /opt/conda/lib/python3.7/site-packages (from tensorflow-gpu==1.15) (0.2.2)\nRequirement already satisfied: keras-applications>=1.0.8 in /opt/conda/lib/python3.7/site-packages (from tensorflow-gpu==1.15) (1.0.8)\nRequirement already satisfied: numpy<2.0,>=1.16.0 in /opt/conda/lib/python3.7/site-packages (from tensorflow-gpu==1.15) (1.19.4)\nRequirement already satisfied: tensorboard<1.16.0,>=1.15.0 in /opt/conda/lib/python3.7/site-packages (from tensorflow-gpu==1.15) (1.15.0)\nRequirement already satisfied: grpcio>=1.8.6 in /opt/conda/lib/python3.7/site-packages (from tensorflow-gpu==1.15) (1.34.0)\nRequirement already satisfied: six>=1.10.0 in /opt/conda/lib/python3.7/site-packages (from tensorflow-gpu==1.15) (1.15.0)\nRequirement already satisfied: absl-py>=0.7.0 in /opt/conda/lib/python3.7/site-packages (from tensorflow-gpu==1.15) (0.11.0)\nRequirement already satisfied: protobuf>=3.6.1 in /opt/conda/lib/python3.7/site-packages (from tensorflow-gpu==1.15) (3.14.0)\nRequirement already satisfied: wheel>=0.26 in /opt/conda/lib/python3.7/site-packages (from tensorflow-gpu==1.15) (0.36.2)\nRequirement already satisfied: termcolor>=1.1.0 in /opt/conda/lib/python3.7/site-packages (from tensorflow-gpu==1.15) (1.1.0)\nRequirement already satisfied: opt-einsum>=2.3.2 in /opt/conda/lib/python3.7/site-packages (from tensorflow-gpu==1.15) (3.3.0)\nRequirement already satisfied: google-pasta>=0.1.6 in /opt/conda/lib/python3.7/site-packages (from tensorflow-gpu==1.15) (0.2.0)\nRequirement already satisfied: astor>=0.6.0 in /opt/conda/lib/python3.7/site-packages (from tensorflow-gpu==1.15) (0.8.1)\nRequirement already satisfied: h5py in /opt/conda/lib/python3.7/site-packages (from keras-applications>=1.0.8->tensorflow-gpu==1.15) (3.1.0)\nRequirement already satisfied: setuptools>=41.0.0 in /opt/conda/lib/python3.7/site-packages (from tensorboard<1.16.0,>=1.15.0->tensorflow-gpu==1.15) (51.1.0)\nRequirement already satisfied: werkzeug>=0.11.15 in /opt/conda/lib/python3.7/site-packages (from tensorboard<1.16.0,>=1.15.0->tensorflow-gpu==1.15) (1.0.1)\nRequirement already satisfied: markdown>=2.6.8 in /opt/conda/lib/python3.7/site-packages (from tensorboard<1.16.0,>=1.15.0->tensorflow-gpu==1.15) (3.3.3)\nRequirement already satisfied: importlib-metadata in /opt/conda/lib/python3.7/site-packages (from markdown>=2.6.8->tensorboard<1.16.0,>=1.15.0->tensorflow-gpu==1.15) (3.3.0)\nRequirement already satisfied: cached-property in /opt/conda/lib/python3.7/site-packages (from h5py->keras-applications>=1.0.8->tensorflow-gpu==1.15) (1.5.2)\nRequirement already satisfied: typing-extensions>=3.6.4 in /opt/conda/lib/python3.7/site-packages (from importlib-metadata->markdown>=2.6.8->tensorboard<1.16.0,>=1.15.0->tensorflow-gpu==1.15) (3.7.4.3)\nRequirement already satisfied: zipp>=0.5 in /opt/conda/lib/python3.7/site-packages (from importlib-metadata->markdown>=2.6.8->tensorboard<1.16.0,>=1.15.0->tensorflow-gpu==1.15) (3.4.0)\nInstalling collected packages: tensorflow-gpu\n\u001b[31mERROR: Could not install packages due to an EnvironmentError: [Errno 13] Permission denied: '/opt/conda/lib/python3.7/site-packages/tensorflow/__init__.py'\nConsider using the `--user` option or check the permissions.\n\u001b[0m\n"
],
[
"import tensorflow\nprint(tensorflow.__version__)",
"1.15.4\n"
],
[
"# Install sentencepiece >> used for tokenizing Thai senetences\r\n!pip install sentencepiece",
"Requirement already satisfied: sentencepiece in /opt/conda/lib/python3.7/site-packages (0.1.95)\n"
],
[
"# Install gdown for downloading files from google drive\r\n!pip install gdown",
"Requirement already satisfied: gdown in /opt/conda/lib/python3.7/site-packages (3.12.2)\nRequirement already satisfied: filelock in /opt/conda/lib/python3.7/site-packages (from gdown) (3.0.12)\nRequirement already satisfied: six in /opt/conda/lib/python3.7/site-packages (from gdown) (1.15.0)\nRequirement already satisfied: tqdm in /opt/conda/lib/python3.7/site-packages (from gdown) (4.55.0)\nRequirement already satisfied: requests[socks] in /opt/conda/lib/python3.7/site-packages (from gdown) (2.25.1)\nRequirement already satisfied: chardet<5,>=3.0.2 in /opt/conda/lib/python3.7/site-packages (from requests[socks]->gdown) (3.0.4)\nRequirement already satisfied: certifi>=2017.4.17 in /opt/conda/lib/python3.7/site-packages (from requests[socks]->gdown) (2020.12.5)\nRequirement already satisfied: urllib3<1.27,>=1.21.1 in /opt/conda/lib/python3.7/site-packages (from requests[socks]->gdown) (1.26.2)\nRequirement already satisfied: idna<3,>=2.5 in /opt/conda/lib/python3.7/site-packages (from requests[socks]->gdown) (2.10)\nRequirement already satisfied: PySocks!=1.5.7,>=1.5.6 in /opt/conda/lib/python3.7/site-packages (from requests[socks]->gdown) (1.7.1)\n"
],
[
"import os\r\nimport numpy as np\r\nimport pandas as pd\r\nfrom sklearn.model_selection import train_test_split\r\nfrom sklearn.metrics import confusion_matrix\r\nimport matplotlib.pyplot as plt",
"_____no_output_____"
]
],
[
[
"# **Download required files**\r\n> Thai Pretrained BERT model - bert_base_th (ThAIKeras)",
"_____no_output_____"
]
],
[
[
"!gdown --id 1J3uuXZr_Se_XIFHj7zlTJ-C9wzI9W_ot # use only id in the link https://drive.google.com/uc?id=1J3uuXZr_Se_XIFHj7zlTJ-C9wzI9W_ot",
"Downloading...\nFrom: https://drive.google.com/uc?id=1J3uuXZr_Se_XIFHj7zlTJ-C9wzI9W_ot\nTo: /home/jupyter/bert_finetuning/bert_base_th.zip\n1.17GB [00:05, 230MB/s] \n"
],
[
"!unzip bert_base_th.zip",
"Archive: bert_base_th.zip\n creating: bert_base_th/\n inflating: bert_base_th/model.ckpt.index \n inflating: bert_base_th/model.ckpt.meta \n inflating: bert_base_th/bert_config.json \n inflating: bert_base_th/model.ckpt.data-00000-of-00001 \n"
]
],
[
[
"> th_wiki_bpe",
"_____no_output_____"
]
],
[
[
"!gdown --id 1F7pCgt3vPlarI9RxKtOZUrC_67KMNQ1W",
"Downloading...\nFrom: https://drive.google.com/uc?id=1F7pCgt3vPlarI9RxKtOZUrC_67KMNQ1W\nTo: /home/jupyter/bert_finetuning/th_wiki_bpe.zip\n100%|████████████████████████████████████████| 560k/560k [00:00<00:00, 86.3MB/s]\n"
],
[
"os.mkdir('th_wiki_bpe')",
"_____no_output_____"
],
[
"!unzip th_wiki_bpe.zip -d th_wiki_bpe",
"Archive: th_wiki_bpe.zip\n inflating: th_wiki_bpe/th.wiki.bpe.op25000.model \n inflating: th_wiki_bpe/th.wiki.bpe.op25000.vocab \n"
]
],
[
[
"> BERT classifier finetuner modified for Thai\r\n\r\nhttps://github.com/ThAIKeras/bert",
"_____no_output_____"
]
],
[
[
"!gdown https://github.com/ThAIKeras/bert.git",
"Downloading...\nFrom: https://github.com/ThAIKeras/bert.git\nTo: /home/jupyter/bert_finetuning/bert.git\n139kB [00:00, 17.1MB/s]\n"
],
[
"!git clone https://github.com/ThAIKeras/bert.git",
"Cloning into 'bert'...\nremote: Enumerating objects: 275, done.\u001b[K\nremote: Total 275 (delta 0), reused 0 (delta 0), pack-reused 275\u001b[K\nReceiving objects: 100% (275/275), 201.44 KiB | 6.50 MiB/s, done.\nResolving deltas: 100% (151/151), done.\n"
],
[
"!https://github.com/ThAIKeras/bert.git",
"/bin/bash: https://github.com/ThAIKeras/bert.git: No such file or directory\n"
]
],
[
[
"Now you should have these folders in the directory\r\n\r\n|-- bert\r\n\r\n|-- bert_base_th\r\n\r\n|-- th_wiki_bpe\r\n\r\n|-- truevoice-intent",
"_____no_output_____"
],
[
"# Finetune the model\n\n> SAVE MODEL AS .pb using\n> ## --do_eval=true",
"_____no_output_____"
]
],
[
[
"os.mkdir('model_finetuning')",
"_____no_output_____"
],
[
"# Declare path to parse when finetuning\nos.environ['BPE_DIR'] = 'th_wiki_bpe' #'/content/th_wiki_bpe'\nos.environ['DATA_DIR'] = 'law_dataset' #'/content/law_data'\nos.environ['OUTPUT_DIR'] = 'model_finetuning' #'/content/model'\nos.environ['BERT_BASE_DIR'] = 'Legalbert_th' #'/content/bert_base_th'",
"_____no_output_____"
],
[
"# Run finetuning(Classes)\n!python bert/law_classifier.py \\--task_name=legaldoc \\--do_train=true \\--do_eval=true \\--do_export=true \\--data_dir=$DATA_DIR \\--vocab_file=$BPE_DIR/th.wiki.bpe.op25000.vocab \\--bert_config_file=$BERT_BASE_DIR/bert_config.json \\--init_checkpoint=$BERT_BASE_DIR/model.ckpt-20 \\--max_seq_length=128 \\--train_batch_size=32 \\--learning_rate=5e-5 \\--num_train_epochs=3.0 \\--output_dir=$OUTPUT_DIR \\--spm_file=$BPE_DIR/th.wiki.bpe.op25000.model",
"python: can't open file 'bert/law_classifier.py': [Errno 2] No such file or directory\n"
]
],
[
[
"# BERT to Predict on Test Data",
"_____no_output_____"
]
],
[
[
"os.mkdir('output_predict2')",
"_____no_output_____"
],
[
"os.environ['BPE_DIR'] = 'th_wiki_bpe' #'/content/th_wiki_bpe'\nos.environ['DATA_DIR'] = 'law_dataset' #'/content/law_data'\nos.environ['OUTPUT_DIR'] = 'output_predict2' #'/content/model'\nos.environ['BERT_FINE_DIR'] = 'model_finetuning2' #'/content/bert_base_th'",
"_____no_output_____"
],
[
"# Run predict(Classes)\n!python bert/law_classifier.py \\--task_name=legaldoc \\--do_predict=true \\--data_dir=$DATA_DIR \\--vocab_file=$BPE_DIR/th.wiki.bpe.op25000.vocab \\--bert_config_file=$BERT_BASE_DIR/bert_config.json \\--init_checkpoint=$BERT_FINE_DIR/model.ckpt-456 \\--max_seq_length=128 \\--output_dir=$OUTPUT_DIR \\--spm_file=$BPE_DIR/th.wiki.bpe.op25000.model ",
"2021-01-23 13:57:27.671096: I tensorflow/stream_executor/platform/default/dso_loader.cc:48] Successfully opened dynamic library libcudart.so.11.0\nWARNING:tensorflow:From /home/jupyter/bert_finetuning/bert/optimization.py:84: The name tf.train.Optimizer is deprecated. Please use tf.compat.v1.train.Optimizer instead.\n\nWARNING:tensorflow:From bert/law_classifier.py:1069: The name tf.app.run is deprecated. Please use tf.compat.v1.app.run instead.\n\nWARNING:tensorflow:From bert/law_classifier.py:874: The name tf.logging.set_verbosity is deprecated. Please use tf.compat.v1.logging.set_verbosity instead.\n\nW0123 13:57:29.128298 140674925967168 module_wrapper.py:139] From bert/law_classifier.py:874: The name tf.logging.set_verbosity is deprecated. Please use tf.compat.v1.logging.set_verbosity instead.\n\nWARNING:tensorflow:From bert/law_classifier.py:874: The name tf.logging.INFO is deprecated. Please use tf.compat.v1.logging.INFO instead.\n\nW0123 13:57:29.128467 140674925967168 module_wrapper.py:139] From bert/law_classifier.py:874: The name tf.logging.INFO is deprecated. Please use tf.compat.v1.logging.INFO instead.\n\nWARNING:tensorflow:From /home/jupyter/bert_finetuning/bert/modeling.py:92: The name tf.gfile.GFile is deprecated. Please use tf.io.gfile.GFile instead.\n\nW0123 13:57:29.128630 140674925967168 module_wrapper.py:139] From /home/jupyter/bert_finetuning/bert/modeling.py:92: The name tf.gfile.GFile is deprecated. Please use tf.io.gfile.GFile instead.\n\nWARNING:tensorflow:From bert/law_classifier.py:897: The name tf.gfile.MakeDirs is deprecated. Please use tf.io.gfile.makedirs instead.\n\nW0123 13:57:29.129191 140674925967168 module_wrapper.py:139] From bert/law_classifier.py:897: The name tf.gfile.MakeDirs is deprecated. Please use tf.io.gfile.makedirs instead.\n\nWARNING:tensorflow:\nThe TensorFlow contrib module will not be included in TensorFlow 2.0.\nFor more information, please see:\n * https://github.com/tensorflow/community/blob/master/rfcs/20180907-contrib-sunset.md\n * https://github.com/tensorflow/addons\n * https://github.com/tensorflow/io (for I/O related ops)\nIf you depend on functionality not listed there, please file an issue.\n\nW0123 13:57:29.251199 140674925967168 lazy_loader.py:50] \nThe TensorFlow contrib module will not be included in TensorFlow 2.0.\nFor more information, please see:\n * https://github.com/tensorflow/community/blob/master/rfcs/20180907-contrib-sunset.md\n * https://github.com/tensorflow/addons\n * https://github.com/tensorflow/io (for I/O related ops)\nIf you depend on functionality not listed there, please file an issue.\n\nWARNING:tensorflow:Estimator's model_fn (<function model_fn_builder.<locals>.model_fn at 0x7ff0d5d8c0e0>) includes params argument, but params are not passed to Estimator.\nW0123 13:57:29.767994 140674925967168 estimator.py:1994] Estimator's model_fn (<function model_fn_builder.<locals>.model_fn at 0x7ff0d5d8c0e0>) includes params argument, but params are not passed to Estimator.\nINFO:tensorflow:Using config: {'_model_dir': 'output_predict2', '_tf_random_seed': None, '_save_summary_steps': 100, '_save_checkpoints_steps': 1000, '_save_checkpoints_secs': None, '_session_config': allow_soft_placement: true\ngraph_options {\n rewrite_options {\n meta_optimizer_iterations: ONE\n }\n}\n, '_keep_checkpoint_max': 5, '_keep_checkpoint_every_n_hours': 10000, '_log_step_count_steps': None, '_train_distribute': None, '_device_fn': None, '_protocol': None, '_eval_distribute': None, '_experimental_distribute': None, '_experimental_max_worker_delay_secs': None, '_session_creation_timeout_secs': 7200, '_service': None, '_cluster_spec': <tensorflow.python.training.server_lib.ClusterSpec object at 0x7ff0d5d38750>, '_task_type': 'worker', '_task_id': 0, '_global_id_in_cluster': 0, '_master': '', '_evaluation_master': '', '_is_chief': True, '_num_ps_replicas': 0, '_num_worker_replicas': 1, '_tpu_config': TPUConfig(iterations_per_loop=1000, num_shards=8, num_cores_per_replica=None, per_host_input_for_training=3, tpu_job_name=None, initial_infeed_sleep_secs=None, input_partition_dims=None, eval_training_input_configuration=2, experimental_host_call_every_n_steps=1), '_cluster': None}\nI0123 13:57:29.768618 140674925967168 estimator.py:212] Using config: {'_model_dir': 'output_predict2', '_tf_random_seed': None, '_save_summary_steps': 100, '_save_checkpoints_steps': 1000, '_save_checkpoints_secs': None, '_session_config': allow_soft_placement: true\ngraph_options {\n rewrite_options {\n meta_optimizer_iterations: ONE\n }\n}\n, '_keep_checkpoint_max': 5, '_keep_checkpoint_every_n_hours': 10000, '_log_step_count_steps': None, '_train_distribute': None, '_device_fn': None, '_protocol': None, '_eval_distribute': None, '_experimental_distribute': None, '_experimental_max_worker_delay_secs': None, '_session_creation_timeout_secs': 7200, '_service': None, '_cluster_spec': <tensorflow.python.training.server_lib.ClusterSpec object at 0x7ff0d5d38750>, '_task_type': 'worker', '_task_id': 0, '_global_id_in_cluster': 0, '_master': '', '_evaluation_master': '', '_is_chief': True, '_num_ps_replicas': 0, '_num_worker_replicas': 1, '_tpu_config': TPUConfig(iterations_per_loop=1000, num_shards=8, num_cores_per_replica=None, per_host_input_for_training=3, tpu_job_name=None, initial_infeed_sleep_secs=None, input_partition_dims=None, eval_training_input_configuration=2, experimental_host_call_every_n_steps=1), '_cluster': None}\nINFO:tensorflow:_TPUContext: eval_on_tpu True\nI0123 13:57:29.768902 140674925967168 tpu_context.py:220] _TPUContext: eval_on_tpu True\nWARNING:tensorflow:eval_on_tpu ignored because use_tpu is False.\nW0123 13:57:29.769019 140674925967168 tpu_context.py:222] eval_on_tpu ignored because use_tpu is False.\nWARNING:tensorflow:From bert/law_classifier.py:190: The name tf.gfile.Open is deprecated. Please use tf.io.gfile.GFile instead.\n\nW0123 13:57:29.769159 140674925967168 module_wrapper.py:139] From bert/law_classifier.py:190: The name tf.gfile.Open is deprecated. Please use tf.io.gfile.GFile instead.\n\nWARNING:tensorflow:From bert/law_classifier.py:551: The name tf.python_io.TFRecordWriter is deprecated. Please use tf.io.TFRecordWriter instead.\n\nW0123 13:57:29.821521 140674925967168 module_wrapper.py:139] From bert/law_classifier.py:551: The name tf.python_io.TFRecordWriter is deprecated. Please use tf.io.TFRecordWriter instead.\n\nWARNING:tensorflow:From bert/law_classifier.py:555: The name tf.logging.info is deprecated. Please use tf.compat.v1.logging.info instead.\n\nW0123 13:57:29.821841 140674925967168 module_wrapper.py:139] From bert/law_classifier.py:555: The name tf.logging.info is deprecated. Please use tf.compat.v1.logging.info instead.\n\nINFO:tensorflow:Writing example 0 of 2109\nI0123 13:57:29.821932 140674925967168 law_classifier.py:555] Writing example 0 of 2109\nINFO:tensorflow:*** Example ***\nI0123 13:57:29.853045 140674925967168 law_classifier.py:530] *** Example ***\nINFO:tensorflow:guid: test-1\nI0123 13:57:29.853245 140674925967168 law_classifier.py:531] guid: test-1\nINFO:tensorflow:tokens: [CLS] ▁เรื่อง มี อยู ่ว่า พ่อ ฉัน เป็น ขี้ เมา ประจํา หมู่บ้าน ตามหลัก ก็คือ คน ไม่ดี ซัก เท่า ไหร ่ หร อก เวลา เมา ชอบ หา เรื่อง อยาก จะมี เรื่อง ก กับ ชาวบ้าน เดิน ด ่า ไป เรื่อย ตาม ทาง แต่ก็ แค่ ปาก พูด ไม่เคย ทํา อะไร ใคร จิง เวลา พ่อ ฉัน เดิน กลับ บ้าน ก็ จะต้อง เดิน ผ่าน บ้าน คนอื่น ก่อน มัน เป็นทาง ประจํา ของ พ่อ แต่ บ้าน นี้ เค้า จะเปิด บ้าน เช่า เป็น หลัง ๆ ไว้ ให้ กรรม กร อยู่ เพราะ บ้าน ฉัน เป็นแหล่ง อุตสาหกรรม พ่อ เดินทาง นี้ มาก่อน เรา เกิด อีก ประมาณปี และ เมื่อ วันอาทิตย์ที่ ผ่านมา พ่อ ฉัน ก็ เม ามา ตาก ปกติ พ่อ เดิน ผ่าน บ้าน นี้ เหมือนเดิม และ ปาก ก็ ไม่ดี เหมือน ทุกวัน มี บ้าน เช่า อยู่ หล ัว หนึ่ง พ่อ ฉัน [SEP]\nI0123 13:57:29.853435 140674925967168 law_classifier.py:533] tokens: [CLS] ▁เรื่อง มี อยู ่ว่า พ่อ ฉัน เป็น ขี้ เมา ประจํา หมู่บ้าน ตามหลัก ก็คือ คน ไม่ดี ซัก เท่า ไหร ่ หร อก เวลา เมา ชอบ หา เรื่อง อยาก จะมี เรื่อง ก กับ ชาวบ้าน เดิน ด ่า ไป เรื่อย ตาม ทาง แต่ก็ แค่ ปาก พูด ไม่เคย ทํา อะไร ใคร จิง เวลา พ่อ ฉัน เดิน กลับ บ้าน ก็ จะต้อง เดิน ผ่าน บ้าน คนอื่น ก่อน มัน เป็นทาง ประจํา ของ พ่อ แต่ บ้าน นี้ เค้า จะเปิด บ้าน เช่า เป็น หลัง ๆ ไว้ ให้ กรรม กร อยู่ เพราะ บ้าน ฉัน เป็นแหล่ง อุตสาหกรรม พ่อ เดินทาง นี้ มาก่อน เรา เกิด อีก ประมาณปี และ เมื่อ วันอาทิตย์ที่ ผ่านมา พ่อ ฉัน ก็ เม ามา ตาก ปกติ พ่อ เดิน ผ่าน บ้าน นี้ เหมือนเดิม และ ปาก ก็ ไม่ดี เหมือน ทุกวัน มี บ้าน เช่า อยู่ หล ัว หนึ่ง พ่อ ฉัน [SEP]\nINFO:tensorflow:input_ids: 1 1722 35 2499 3761 2565 5414 38 6274 7495 1154 3096 18602 5693 325 15105 11654 2632 24436 24727 85 74 879 7495 2185 832 420 5935 1402 420 24725 104 8922 1302 24736 73 157 5092 395 107 13730 4511 1345 1292 9505 212 6883 4629 7793 879 2565 5414 1302 1486 738 606 4318 1302 746 738 9286 961 845 19848 1154 34 2565 340 738 138 9155 20641 738 9932 38 421 24802 1135 166 586 186 125 2895 738 5414 10125 2526 2565 2182 138 5214 3267 214 568 19057 30 188 13660 6859 2565 5414 606 46 1215 4187 1671 2565 1302 746 738 138 23535 30 1345 606 15105 1681 2832 35 738 9932 125 72 118 229 2565 5414 2\nI0123 13:57:29.853657 140674925967168 law_classifier.py:534] input_ids: 1 1722 35 2499 3761 2565 5414 38 6274 7495 1154 3096 18602 5693 325 15105 11654 2632 24436 24727 85 74 879 7495 2185 832 420 5935 1402 420 24725 104 8922 1302 24736 73 157 5092 395 107 13730 4511 1345 1292 9505 212 6883 4629 7793 879 2565 5414 1302 1486 738 606 4318 1302 746 738 9286 961 845 19848 1154 34 2565 340 738 138 9155 20641 738 9932 38 421 24802 1135 166 586 186 125 2895 738 5414 10125 2526 2565 2182 138 5214 3267 214 568 19057 30 188 13660 6859 2565 5414 606 46 1215 4187 1671 2565 1302 746 738 138 23535 30 1345 606 15105 1681 2832 35 738 9932 125 72 118 229 2565 5414 2\nINFO:tensorflow:input_mask: 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1\nI0123 13:57:29.853788 140674925967168 law_classifier.py:535] input_mask: 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1\nINFO:tensorflow:segment_ids: 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0\nI0123 13:57:29.853874 140674925967168 law_classifier.py:536] segment_ids: 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0\nINFO:tensorflow:label: family (id = 1)\nI0123 13:57:29.853921 140674925967168 law_classifier.py:537] label: family (id = 1)\nINFO:tensorflow:*** Example ***\nI0123 13:57:29.855584 140674925967168 law_classifier.py:530] *** Example ***\nINFO:tensorflow:guid: test-2\nI0123 13:57:29.855707 140674925967168 law_classifier.py:531] guid: test-2\nINFO:tensorflow:tokens: [CLS] ▁อายุ ความ คดี อาญา ที่เกี่ยวกับ เช ็ค จะเริ่ม นับ จาก เมื่อ ไหร ่ และ หมด อายุ เมื่อ ไหร ่ เริ่ม นับ จาก วันที่ ออก หมาย จับ หรือ ว่าน ับ จาก วันที่ รู้ว่า เช ็ค เด้ ง [SEP]\nI0123 13:57:29.855775 140674925967168 law_classifier.py:533] tokens: [CLS] ▁อายุ ความ คดี อาญา ที่เกี่ยวกับ เช ็ค จะเริ่ม นับ จาก เมื่อ ไหร ่ และ หมด อายุ เมื่อ ไหร ่ เริ่ม นับ จาก วันที่ ออก หมาย จับ หรือ ว่าน ับ จาก วันที่ รู้ว่า เช ็ค เด้ ง [SEP]\nINFO:tensorflow:input_ids: 1 6217 165 2724 18123 9750 211 2423 17888 2297 80 188 24436 24727 30 653 1397 188 24436 24727 855 2297 80 129 183 256 2036 135 6754 32 80 129 19381 211 2423 15523 24726 2 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0\nI0123 13:57:29.855863 140674925967168 law_classifier.py:534] input_ids: 1 6217 165 2724 18123 9750 211 2423 17888 2297 80 188 24436 24727 30 653 1397 188 24436 24727 855 2297 80 129 183 256 2036 135 6754 32 80 129 19381 211 2423 15523 24726 2 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0\nINFO:tensorflow:input_mask: 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0\nI0123 13:57:29.855949 140674925967168 law_classifier.py:535] input_mask: 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0\nINFO:tensorflow:segment_ids: 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0\nI0123 13:57:29.856072 140674925967168 law_classifier.py:536] segment_ids: 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0\nINFO:tensorflow:label: family (id = 1)\nI0123 13:57:29.856195 140674925967168 law_classifier.py:537] label: family (id = 1)\nINFO:tensorflow:*** Example ***\nI0123 13:57:29.867826 140674925967168 law_classifier.py:530] *** Example ***\nINFO:tensorflow:guid: test-3\nI0123 13:57:29.867919 140674925967168 law_classifier.py:531] guid: test-3\nINFO:tensorflow:tokens: [CLS] ▁คือ ดิ ฉัน เป็น พนักงาน บริษัท แห่งหนึ่ง ซึ่ง เช่า พื้นที่ ของทาง ห้าง ในการ ขาย สินค้า เรื่อง มี อยู ่ว่า ตอน เย็น วันที่ ตอน เลิก งาน มี พนักงาน อีก ร้าน ใน ห้าง เดียวกัน ซึ่ง รู้สึก จะเป็น วันหยุด ของเขา เข้ามา ทําร้าย ร่างกาย ดิ ฉัน ใน ลาน จอด รถ พนักงาน ซึ่ง ดิ ฉัน ก็ ไม่ได้ โต้ ตอบ แต่อย่างใด จึง นํา เรื่องนี้ เข้า แจ้ง ความ ที่ โรง พัก แล้ว ทําการ ตรวจ ร่างกาย ประกอบ พอ มา อีก วัน ทาง ห้าง ทราบ ข่าว ก็ เรียก เข้าไป สอบสวน เขา ถาม ว่ามี เรื่อง อะ ไ รก ัน มาก่อน หรือ ปล ่าว ดิ ฉัน ก็ บอกว่า เคยมี ตอนต้น เดือน เพราะ เขา ไม่พอใจ ที่เรา โทร หา แฟน เขา ซึ่ง เรื่อง นั้น มัน ก็ ล่วง เลย มา พอสมควร แล้วแต่ ดิ ฉัน คิดว่า เรื่อง ทําร้าย ร่างกาย มัน คนละ [SEP]\nI0123 13:57:29.867994 140674925967168 law_classifier.py:533] tokens: [CLS] ▁คือ ดิ ฉัน เป็น พนักงาน บริษัท แห่งหนึ่ง ซึ่ง เช่า พื้นที่ ของทาง ห้าง ในการ ขาย สินค้า เรื่อง มี อยู ่ว่า ตอน เย็น วันที่ ตอน เลิก งาน มี พนักงาน อีก ร้าน ใน ห้าง เดียวกัน ซึ่ง รู้สึก จะเป็น วันหยุด ของเขา เข้ามา ทําร้าย ร่างกาย ดิ ฉัน ใน ลาน จอด รถ พนักงาน ซึ่ง ดิ ฉัน ก็ ไม่ได้ โต้ ตอบ แต่อย่างใด จึง นํา เรื่องนี้ เข้า แจ้ง ความ ที่ โรง พัก แล้ว ทําการ ตรวจ ร่างกาย ประกอบ พอ มา อีก วัน ทาง ห้าง ทราบ ข่าว ก็ เรียก เข้าไป สอบสวน เขา ถาม ว่ามี เรื่อง อะ ไ รก ัน มาก่อน หรือ ปล ่าว ดิ ฉัน ก็ บอกว่า เคยมี ตอนต้น เดือน เพราะ เขา ไม่พอใจ ที่เรา โทร หา แฟน เขา ซึ่ง เรื่อง นั้น มัน ก็ ล่วง เลย มา พอสมควร แล้วแต่ ดิ ฉัน คิดว่า เรื่อง ทําร้าย ร่างกาย มัน คนละ [SEP]\nINFO:tensorflow:input_ids: 1 353 283 5414 38 8213 1088 5133 681 9932 763 12303 10618 546 1191 3038 420 35 2499 3761 679 3474 129 679 3411 301 35 8213 568 3208 17 10618 1355 681 8306 1550 14115 1668 2399 16133 3005 283 5414 17 4695 7623 620 8213 681 283 5414 606 1680 4950 3471 14158 1075 533 7781 521 5232 165 13 608 3024 559 1981 3071 3005 467 2675 78 568 70 107 10618 4159 2156 606 900 2829 17244 477 4845 4165 420 610 24760 146 15 5214 135 181 671 283 5414 606 23315 16641 6826 849 2895 477 20269 16939 548 832 4382 477 681 420 489 845 606 11892 3179 78 13268 19850 283 5414 14666 420 16133 3005 845 10884 2\nI0123 13:57:29.868093 140674925967168 law_classifier.py:534] input_ids: 1 353 283 5414 38 8213 1088 5133 681 9932 763 12303 10618 546 1191 3038 420 35 2499 3761 679 3474 129 679 3411 301 35 8213 568 3208 17 10618 1355 681 8306 1550 14115 1668 2399 16133 3005 283 5414 17 4695 7623 620 8213 681 283 5414 606 1680 4950 3471 14158 1075 533 7781 521 5232 165 13 608 3024 559 1981 3071 3005 467 2675 78 568 70 107 10618 4159 2156 606 900 2829 17244 477 4845 4165 420 610 24760 146 15 5214 135 181 671 283 5414 606 23315 16641 6826 849 2895 477 20269 16939 548 832 4382 477 681 420 489 845 606 11892 3179 78 13268 19850 283 5414 14666 420 16133 3005 845 10884 2\nINFO:tensorflow:input_mask: 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1\nI0123 13:57:29.868165 140674925967168 law_classifier.py:535] input_mask: 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1\nINFO:tensorflow:segment_ids: 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0\nI0123 13:57:29.868245 140674925967168 law_classifier.py:536] segment_ids: 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0\nINFO:tensorflow:label: family (id = 1)\nI0123 13:57:29.868290 140674925967168 law_classifier.py:537] label: family (id = 1)\nINFO:tensorflow:*** Example ***\nI0123 13:57:29.868684 140674925967168 law_classifier.py:530] *** Example ***\nINFO:tensorflow:guid: test-4\nI0123 13:57:29.868764 140674925967168 law_classifier.py:531] guid: test-4\nINFO:tensorflow:tokens: [CLS] ▁การ รับ โอน มรดก ชื่อ พ่อ [SEP]\nI0123 13:57:29.868849 140674925967168 law_classifier.py:533] tokens: [CLS] ▁การ รับ โอน มรดก ชื่อ พ่อ [SEP]\nINFO:tensorflow:input_ids: 1 470 205 3665 2065 136 2565 2 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0\nI0123 13:57:29.868916 140674925967168 law_classifier.py:534] input_ids: 1 470 205 3665 2065 136 2565 2 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0\nINFO:tensorflow:input_mask: 1 1 1 1 1 1 1 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0\nI0123 13:57:29.868983 140674925967168 law_classifier.py:535] input_mask: 1 1 1 1 1 1 1 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0\nINFO:tensorflow:segment_ids: 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0\nI0123 13:57:29.869046 140674925967168 law_classifier.py:536] segment_ids: 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0\nINFO:tensorflow:label: family (id = 1)\nI0123 13:57:29.869091 140674925967168 law_classifier.py:537] label: family (id = 1)\nINFO:tensorflow:*** Example ***\nI0123 13:57:29.881726 140674925967168 law_classifier.py:530] *** Example ***\nINFO:tensorflow:guid: test-5\nI0123 13:57:29.881819 140674925967168 law_classifier.py:531] guid: test-5\nINFO:tensorflow:tokens: [CLS] ▁ ข้อส อบ ถาม หน ่อย นะ คะ พอดี โดน ข้อ ห าล ัก ทรัพย์ กับ ห้างสรรพสินค้า แห่งหนึ่ง โดย ได้ทําการ ลัก กระเป ๋ ามา ใบ เป็นจํานวน เงิน บาท แต่ ถูก ทาง ห้าง จับ ได้ แล้วนํา ตัวไป สอบสวน จาก นั้น ให้ เขียน ประวัติ ของตัวเอง ถ่าย สํา เนา บัตร ประจําตัว ประชาชน แล้ว ก็ ถ่ายรูป ทุกอย่าง เป็น หลักฐาน พร้อม ทั้ง ให้ เขียน สํานวน ใน คดี ลัก ทรัพย์ ซึ่งได้ ยอมรับ ทุก ข้อก ล ่าว หาว่า เป็น ความจริง สาร ภาพ ทุกอย่าง จาก นั้น ทาง ห้าง ก็ได้ ให้ ญาติ มารับ ตัว พร้อมกับ จ่าย ค่า เสียหาย สินค้า ที่ได้ ขโมย ไป อย ากร ู้ ว่า ทาง ห้าง จะ ดําเนิน คดี กับ เรา อย่างไร ต่อไป คะ แล้ว ประวัติ ของเรา ที่เขียน ไป นั้นจะ ทํา อย่างไร ต่อ ถ้า เก็บ ไว้ จะมี อายุ ความ กี่ ปี จะ กระทบ [SEP]\nI0123 13:57:29.881896 140674925967168 law_classifier.py:533] tokens: [CLS] ▁ ข้อส อบ ถาม หน ่อย นะ คะ พอดี โดน ข้อ ห าล ัก ทรัพย์ กับ ห้างสรรพสินค้า แห่งหนึ่ง โดย ได้ทําการ ลัก กระเป ๋ ามา ใบ เป็นจํานวน เงิน บาท แต่ ถูก ทาง ห้าง จับ ได้ แล้วนํา ตัวไป สอบสวน จาก นั้น ให้ เขียน ประวัติ ของตัวเอง ถ่าย สํา เนา บัตร ประจําตัว ประชาชน แล้ว ก็ ถ่ายรูป ทุกอย่าง เป็น หลักฐาน พร้อม ทั้ง ให้ เขียน สํานวน ใน คดี ลัก ทรัพย์ ซึ่งได้ ยอมรับ ทุก ข้อก ล ่าว หาว่า เป็น ความจริง สาร ภาพ ทุกอย่าง จาก นั้น ทาง ห้าง ก็ได้ ให้ ญาติ มารับ ตัว พร้อมกับ จ่าย ค่า เสียหาย สินค้า ที่ได้ ขโมย ไป อย ากร ู้ ว่า ทาง ห้าง จะ ดําเนิน คดี กับ เรา อย่างไร ต่อไป คะ แล้ว ประวัติ ของเรา ที่เขียน ไป นั้นจะ ทํา อย่างไร ต่อ ถ้า เก็บ ไว้ จะมี อายุ ความ กี่ ปี จะ กระทบ [SEP]\nINFO:tensorflow:input_ids: 1 24719 12522 171 4845 57 718 1512 1468 12947 6410 1577 24745 122 39 2709 104 20017 5133 77 11963 3722 14249 24821 1215 941 14999 1289 1204 340 410 107 10618 2036 61 15416 9162 17244 80 489 166 793 1434 7358 1864 315 7213 4937 11367 1565 559 606 23375 11830 38 4636 2336 243 166 793 14283 17 2724 3722 2709 19421 3814 986 9693 24732 671 16639 38 9441 581 167 11830 80 489 107 10618 2807 166 6080 8978 231 4347 4836 1407 6285 3038 2264 12659 157 66 597 111 121 107 10618 177 1779 2724 104 3267 12190 1552 1468 559 1434 11920 5424 157 10821 212 12190 275 5123 2466 1135 1402 1397 165 6803 89 177 6463 2\nI0123 13:57:29.881996 140674925967168 law_classifier.py:534] input_ids: 1 24719 12522 171 4845 57 718 1512 1468 12947 6410 1577 24745 122 39 2709 104 20017 5133 77 11963 3722 14249 24821 1215 941 14999 1289 1204 340 410 107 10618 2036 61 15416 9162 17244 80 489 166 793 1434 7358 1864 315 7213 4937 11367 1565 559 606 23375 11830 38 4636 2336 243 166 793 14283 17 2724 3722 2709 19421 3814 986 9693 24732 671 16639 38 9441 581 167 11830 80 489 107 10618 2807 166 6080 8978 231 4347 4836 1407 6285 3038 2264 12659 157 66 597 111 121 107 10618 177 1779 2724 104 3267 12190 1552 1468 559 1434 11920 5424 157 10821 212 12190 275 5123 2466 1135 1402 1397 165 6803 89 177 6463 2\nINFO:tensorflow:input_mask: 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1\nI0123 13:57:29.882070 140674925967168 law_classifier.py:535] input_mask: 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1\nINFO:tensorflow:segment_ids: 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0\nI0123 13:57:29.882134 140674925967168 law_classifier.py:536] segment_ids: 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0\nINFO:tensorflow:label: family (id = 1)\nI0123 13:57:29.882179 140674925967168 law_classifier.py:537] label: family (id = 1)\nINFO:tensorflow:***** Running prediction*****\nI0123 13:57:46.162699 140674925967168 law_classifier.py:1023] ***** Running prediction*****\nINFO:tensorflow: Num examples = 2109\nI0123 13:57:46.162866 140674925967168 law_classifier.py:1024] Num examples = 2109\nINFO:tensorflow: Batch size = 8\nI0123 13:57:46.163048 140674925967168 law_classifier.py:1025] Batch size = 8\nWARNING:tensorflow:From bert/law_classifier.py:581: The name tf.FixedLenFeature is deprecated. Please use tf.io.FixedLenFeature instead.\n\nW0123 13:57:46.163182 140674925967168 module_wrapper.py:139] From bert/law_classifier.py:581: The name tf.FixedLenFeature is deprecated. Please use tf.io.FixedLenFeature instead.\n\nINFO:tensorflow:***** Predict results *****\nI0123 13:57:46.163298 140674925967168 law_classifier.py:1045] ***** Predict results *****\nINFO:tensorflow:Could not find trained model in model_dir: output_predict2, running initialization to predict.\nI0123 13:57:46.163626 140674925967168 estimator.py:615] Could not find trained model in model_dir: output_predict2, running initialization to predict.\nWARNING:tensorflow:From /opt/conda/lib/python3.7/site-packages/tensorflow_core/python/ops/resource_variable_ops.py:1630: calling BaseResourceVariable.__init__ (from tensorflow.python.ops.resource_variable_ops) with constraint is deprecated and will be removed in a future version.\nInstructions for updating:\nIf using Keras pass *_constraint arguments to layers.\nW0123 13:57:46.170173 140674925967168 deprecation.py:506] From /opt/conda/lib/python3.7/site-packages/tensorflow_core/python/ops/resource_variable_ops.py:1630: calling BaseResourceVariable.__init__ (from tensorflow.python.ops.resource_variable_ops) with constraint is deprecated and will be removed in a future version.\nInstructions for updating:\nIf using Keras pass *_constraint arguments to layers.\nWARNING:tensorflow:From bert/law_classifier.py:616: map_and_batch (from tensorflow.contrib.data.python.ops.batching) is deprecated and will be removed in a future version.\nInstructions for updating:\nUse `tf.data.experimental.map_and_batch(...)`.\nW0123 13:57:46.185888 140674925967168 deprecation.py:323] From bert/law_classifier.py:616: map_and_batch (from tensorflow.contrib.data.python.ops.batching) is deprecated and will be removed in a future version.\nInstructions for updating:\nUse `tf.data.experimental.map_and_batch(...)`.\nWARNING:tensorflow:From /opt/conda/lib/python3.7/site-packages/tensorflow_core/contrib/data/python/ops/batching.py:276: map_and_batch (from tensorflow.python.data.experimental.ops.batching) is deprecated and will be removed in a future version.\nInstructions for updating:\nUse `tf.data.Dataset.map(map_func, num_parallel_calls)` followed by `tf.data.Dataset.batch(batch_size, drop_remainder)`. Static tf.data optimizations will take care of using the fused implementation.\nW0123 13:57:46.186037 140674925967168 deprecation.py:323] From /opt/conda/lib/python3.7/site-packages/tensorflow_core/contrib/data/python/ops/batching.py:276: map_and_batch (from tensorflow.python.data.experimental.ops.batching) is deprecated and will be removed in a future version.\nInstructions for updating:\nUse `tf.data.Dataset.map(map_func, num_parallel_calls)` followed by `tf.data.Dataset.batch(batch_size, drop_remainder)`. Static tf.data optimizations will take care of using the fused implementation.\nWARNING:tensorflow:From /opt/conda/lib/python3.7/site-packages/tensorflow_core/python/autograph/converters/directives.py:119: The name tf.parse_single_example is deprecated. Please use tf.io.parse_single_example instead.\n\nW0123 13:57:46.241840 140674925967168 module_wrapper.py:139] From /opt/conda/lib/python3.7/site-packages/tensorflow_core/python/autograph/converters/directives.py:119: The name tf.parse_single_example is deprecated. Please use tf.io.parse_single_example instead.\n\nWARNING:tensorflow:From bert/law_classifier.py:596: to_int32 (from tensorflow.python.ops.math_ops) is deprecated and will be removed in a future version.\nInstructions for updating:\nUse `tf.cast` instead.\nW0123 13:57:46.325631 140674925967168 deprecation.py:323] From bert/law_classifier.py:596: to_int32 (from tensorflow.python.ops.math_ops) is deprecated and will be removed in a future version.\nInstructions for updating:\nUse `tf.cast` instead.\nINFO:tensorflow:Calling model_fn.\nI0123 13:57:46.336562 140674925967168 estimator.py:1148] Calling model_fn.\nINFO:tensorflow:Running infer on CPU\nI0123 13:57:46.336713 140674925967168 tpu_estimator.py:3124] Running infer on CPU\nINFO:tensorflow:*** Features ***\nI0123 13:57:46.336985 140674925967168 law_classifier.py:693] *** Features ***\nINFO:tensorflow: name = input_ids, shape = (?, 128)\nI0123 13:57:46.337093 140674925967168 law_classifier.py:695] name = input_ids, shape = (?, 128)\nINFO:tensorflow: name = input_mask, shape = (?, 128)\nI0123 13:57:46.337166 140674925967168 law_classifier.py:695] name = input_mask, shape = (?, 128)\nINFO:tensorflow: name = label_ids, shape = (?,)\nI0123 13:57:46.337230 140674925967168 law_classifier.py:695] name = label_ids, shape = (?,)\nINFO:tensorflow: name = segment_ids, shape = (?, 128)\nI0123 13:57:46.337292 140674925967168 law_classifier.py:695] name = segment_ids, shape = (?, 128)\nWARNING:tensorflow:From /home/jupyter/bert_finetuning/bert/modeling.py:172: The name tf.variable_scope is deprecated. Please use tf.compat.v1.variable_scope instead.\n\nW0123 13:57:46.339661 140674925967168 module_wrapper.py:139] From /home/jupyter/bert_finetuning/bert/modeling.py:172: The name tf.variable_scope is deprecated. Please use tf.compat.v1.variable_scope instead.\n\nWARNING:tensorflow:From /home/jupyter/bert_finetuning/bert/modeling.py:411: The name tf.get_variable is deprecated. Please use tf.compat.v1.get_variable instead.\n\nW0123 13:57:46.340750 140674925967168 module_wrapper.py:139] From /home/jupyter/bert_finetuning/bert/modeling.py:411: The name tf.get_variable is deprecated. Please use tf.compat.v1.get_variable instead.\n\nWARNING:tensorflow:From /home/jupyter/bert_finetuning/bert/modeling.py:680: dense (from tensorflow.python.layers.core) is deprecated and will be removed in a future version.\nInstructions for updating:\nUse keras.layers.Dense instead.\nW0123 13:57:46.402619 140674925967168 deprecation.py:323] From /home/jupyter/bert_finetuning/bert/modeling.py:680: dense (from tensorflow.python.layers.core) is deprecated and will be removed in a future version.\nInstructions for updating:\nUse keras.layers.Dense instead.\nWARNING:tensorflow:From /opt/conda/lib/python3.7/site-packages/tensorflow_core/python/layers/core.py:187: Layer.apply (from tensorflow.python.keras.engine.base_layer) is deprecated and will be removed in a future version.\nInstructions for updating:\nPlease use `layer.__call__` method instead.\nW0123 13:57:46.403153 140674925967168 deprecation.py:323] From /opt/conda/lib/python3.7/site-packages/tensorflow_core/python/layers/core.py:187: Layer.apply (from tensorflow.python.keras.engine.base_layer) is deprecated and will be removed in a future version.\nInstructions for updating:\nPlease use `layer.__call__` method instead.\nWARNING:tensorflow:From /home/jupyter/bert_finetuning/bert/modeling.py:277: The name tf.erf is deprecated. Please use tf.math.erf instead.\n\nW0123 13:57:46.481572 140674925967168 module_wrapper.py:139] From /home/jupyter/bert_finetuning/bert/modeling.py:277: The name tf.erf is deprecated. Please use tf.math.erf instead.\n\nWARNING:tensorflow:From bert/law_classifier.py:708: The name tf.trainable_variables is deprecated. Please use tf.compat.v1.trainable_variables instead.\n\nW0123 13:57:47.741535 140674925967168 module_wrapper.py:139] From bert/law_classifier.py:708: The name tf.trainable_variables is deprecated. Please use tf.compat.v1.trainable_variables instead.\n\nWARNING:tensorflow:From bert/law_classifier.py:722: The name tf.train.init_from_checkpoint is deprecated. Please use tf.compat.v1.train.init_from_checkpoint instead.\n\nW0123 13:57:47.745567 140674925967168 module_wrapper.py:139] From bert/law_classifier.py:722: The name tf.train.init_from_checkpoint is deprecated. Please use tf.compat.v1.train.init_from_checkpoint instead.\n\nINFO:tensorflow:**** Trainable Variables ****\nI0123 13:57:48.301690 140674925967168 law_classifier.py:724] **** Trainable Variables ****\nINFO:tensorflow: name = bert/embeddings/word_embeddings:0, shape = (25004, 768), *INIT_FROM_CKPT*\nI0123 13:57:48.301911 140674925967168 law_classifier.py:730] name = bert/embeddings/word_embeddings:0, shape = (25004, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/embeddings/token_type_embeddings:0, shape = (2, 768), *INIT_FROM_CKPT*\nI0123 13:57:48.302073 140674925967168 law_classifier.py:730] name = bert/embeddings/token_type_embeddings:0, shape = (2, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/embeddings/position_embeddings:0, shape = (512, 768), *INIT_FROM_CKPT*\nI0123 13:57:48.302183 140674925967168 law_classifier.py:730] name = bert/embeddings/position_embeddings:0, shape = (512, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/embeddings/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.302312 140674925967168 law_classifier.py:730] name = bert/embeddings/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/embeddings/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.302407 140674925967168 law_classifier.py:730] name = bert/embeddings/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_0/attention/self/query/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nI0123 13:57:48.302494 140674925967168 law_classifier.py:730] name = bert/encoder/layer_0/attention/self/query/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_0/attention/self/query/bias:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.302589 140674925967168 law_classifier.py:730] name = bert/encoder/layer_0/attention/self/query/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_0/attention/self/key/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nI0123 13:57:48.302674 140674925967168 law_classifier.py:730] name = bert/encoder/layer_0/attention/self/key/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_0/attention/self/key/bias:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.302769 140674925967168 law_classifier.py:730] name = bert/encoder/layer_0/attention/self/key/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_0/attention/self/value/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nI0123 13:57:48.302857 140674925967168 law_classifier.py:730] name = bert/encoder/layer_0/attention/self/value/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_0/attention/self/value/bias:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.302946 140674925967168 law_classifier.py:730] name = bert/encoder/layer_0/attention/self/value/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_0/attention/output/dense/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nI0123 13:57:48.303030 140674925967168 law_classifier.py:730] name = bert/encoder/layer_0/attention/output/dense/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_0/attention/output/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.303120 140674925967168 law_classifier.py:730] name = bert/encoder/layer_0/attention/output/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_0/attention/output/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.303215 140674925967168 law_classifier.py:730] name = bert/encoder/layer_0/attention/output/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_0/attention/output/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.303298 140674925967168 law_classifier.py:730] name = bert/encoder/layer_0/attention/output/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_0/intermediate/dense/kernel:0, shape = (768, 3072), *INIT_FROM_CKPT*\nI0123 13:57:48.303376 140674925967168 law_classifier.py:730] name = bert/encoder/layer_0/intermediate/dense/kernel:0, shape = (768, 3072), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_0/intermediate/dense/bias:0, shape = (3072,), *INIT_FROM_CKPT*\nI0123 13:57:48.303459 140674925967168 law_classifier.py:730] name = bert/encoder/layer_0/intermediate/dense/bias:0, shape = (3072,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_0/output/dense/kernel:0, shape = (3072, 768), *INIT_FROM_CKPT*\nI0123 13:57:48.303538 140674925967168 law_classifier.py:730] name = bert/encoder/layer_0/output/dense/kernel:0, shape = (3072, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_0/output/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.303621 140674925967168 law_classifier.py:730] name = bert/encoder/layer_0/output/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_0/output/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.303699 140674925967168 law_classifier.py:730] name = bert/encoder/layer_0/output/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_0/output/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.303777 140674925967168 law_classifier.py:730] name = bert/encoder/layer_0/output/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_1/attention/self/query/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nI0123 13:57:48.303863 140674925967168 law_classifier.py:730] name = bert/encoder/layer_1/attention/self/query/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_1/attention/self/query/bias:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.303955 140674925967168 law_classifier.py:730] name = bert/encoder/layer_1/attention/self/query/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_1/attention/self/key/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nI0123 13:57:48.304034 140674925967168 law_classifier.py:730] name = bert/encoder/layer_1/attention/self/key/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_1/attention/self/key/bias:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.304116 140674925967168 law_classifier.py:730] name = bert/encoder/layer_1/attention/self/key/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_1/attention/self/value/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nI0123 13:57:48.304195 140674925967168 law_classifier.py:730] name = bert/encoder/layer_1/attention/self/value/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_1/attention/self/value/bias:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.304282 140674925967168 law_classifier.py:730] name = bert/encoder/layer_1/attention/self/value/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_1/attention/output/dense/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nI0123 13:57:48.304367 140674925967168 law_classifier.py:730] name = bert/encoder/layer_1/attention/output/dense/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_1/attention/output/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.304486 140674925967168 law_classifier.py:730] name = bert/encoder/layer_1/attention/output/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_1/attention/output/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.304581 140674925967168 law_classifier.py:730] name = bert/encoder/layer_1/attention/output/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_1/attention/output/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.304673 140674925967168 law_classifier.py:730] name = bert/encoder/layer_1/attention/output/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_1/intermediate/dense/kernel:0, shape = (768, 3072), *INIT_FROM_CKPT*\nI0123 13:57:48.304766 140674925967168 law_classifier.py:730] name = bert/encoder/layer_1/intermediate/dense/kernel:0, shape = (768, 3072), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_1/intermediate/dense/bias:0, shape = (3072,), *INIT_FROM_CKPT*\nI0123 13:57:48.304867 140674925967168 law_classifier.py:730] name = bert/encoder/layer_1/intermediate/dense/bias:0, shape = (3072,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_1/output/dense/kernel:0, shape = (3072, 768), *INIT_FROM_CKPT*\nI0123 13:57:48.304960 140674925967168 law_classifier.py:730] name = bert/encoder/layer_1/output/dense/kernel:0, shape = (3072, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_1/output/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.305073 140674925967168 law_classifier.py:730] name = bert/encoder/layer_1/output/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_1/output/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.305145 140674925967168 law_classifier.py:730] name = bert/encoder/layer_1/output/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_1/output/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.305210 140674925967168 law_classifier.py:730] name = bert/encoder/layer_1/output/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_2/attention/self/query/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nI0123 13:57:48.305279 140674925967168 law_classifier.py:730] name = bert/encoder/layer_2/attention/self/query/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_2/attention/self/query/bias:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.305346 140674925967168 law_classifier.py:730] name = bert/encoder/layer_2/attention/self/query/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_2/attention/self/key/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nI0123 13:57:48.305410 140674925967168 law_classifier.py:730] name = bert/encoder/layer_2/attention/self/key/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_2/attention/self/key/bias:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.305476 140674925967168 law_classifier.py:730] name = bert/encoder/layer_2/attention/self/key/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_2/attention/self/value/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nI0123 13:57:48.305551 140674925967168 law_classifier.py:730] name = bert/encoder/layer_2/attention/self/value/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_2/attention/self/value/bias:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.305629 140674925967168 law_classifier.py:730] name = bert/encoder/layer_2/attention/self/value/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_2/attention/output/dense/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nI0123 13:57:48.305698 140674925967168 law_classifier.py:730] name = bert/encoder/layer_2/attention/output/dense/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_2/attention/output/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.305755 140674925967168 law_classifier.py:730] name = bert/encoder/layer_2/attention/output/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_2/attention/output/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.305809 140674925967168 law_classifier.py:730] name = bert/encoder/layer_2/attention/output/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_2/attention/output/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.305862 140674925967168 law_classifier.py:730] name = bert/encoder/layer_2/attention/output/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_2/intermediate/dense/kernel:0, shape = (768, 3072), *INIT_FROM_CKPT*\nI0123 13:57:48.305915 140674925967168 law_classifier.py:730] name = bert/encoder/layer_2/intermediate/dense/kernel:0, shape = (768, 3072), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_2/intermediate/dense/bias:0, shape = (3072,), *INIT_FROM_CKPT*\nI0123 13:57:48.305972 140674925967168 law_classifier.py:730] name = bert/encoder/layer_2/intermediate/dense/bias:0, shape = (3072,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_2/output/dense/kernel:0, shape = (3072, 768), *INIT_FROM_CKPT*\nI0123 13:57:48.306025 140674925967168 law_classifier.py:730] name = bert/encoder/layer_2/output/dense/kernel:0, shape = (3072, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_2/output/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.306082 140674925967168 law_classifier.py:730] name = bert/encoder/layer_2/output/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_2/output/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.306155 140674925967168 law_classifier.py:730] name = bert/encoder/layer_2/output/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_2/output/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.306212 140674925967168 law_classifier.py:730] name = bert/encoder/layer_2/output/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_3/attention/self/query/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nI0123 13:57:48.306289 140674925967168 law_classifier.py:730] name = bert/encoder/layer_3/attention/self/query/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_3/attention/self/query/bias:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.306351 140674925967168 law_classifier.py:730] name = bert/encoder/layer_3/attention/self/query/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_3/attention/self/key/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nI0123 13:57:48.306408 140674925967168 law_classifier.py:730] name = bert/encoder/layer_3/attention/self/key/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_3/attention/self/key/bias:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.306468 140674925967168 law_classifier.py:730] name = bert/encoder/layer_3/attention/self/key/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_3/attention/self/value/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nI0123 13:57:48.306525 140674925967168 law_classifier.py:730] name = bert/encoder/layer_3/attention/self/value/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_3/attention/self/value/bias:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.306585 140674925967168 law_classifier.py:730] name = bert/encoder/layer_3/attention/self/value/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_3/attention/output/dense/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nI0123 13:57:48.306641 140674925967168 law_classifier.py:730] name = bert/encoder/layer_3/attention/output/dense/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_3/attention/output/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.306701 140674925967168 law_classifier.py:730] name = bert/encoder/layer_3/attention/output/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_3/attention/output/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.306758 140674925967168 law_classifier.py:730] name = bert/encoder/layer_3/attention/output/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_3/attention/output/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.306814 140674925967168 law_classifier.py:730] name = bert/encoder/layer_3/attention/output/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_3/intermediate/dense/kernel:0, shape = (768, 3072), *INIT_FROM_CKPT*\nI0123 13:57:48.306871 140674925967168 law_classifier.py:730] name = bert/encoder/layer_3/intermediate/dense/kernel:0, shape = (768, 3072), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_3/intermediate/dense/bias:0, shape = (3072,), *INIT_FROM_CKPT*\nI0123 13:57:48.306931 140674925967168 law_classifier.py:730] name = bert/encoder/layer_3/intermediate/dense/bias:0, shape = (3072,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_3/output/dense/kernel:0, shape = (3072, 768), *INIT_FROM_CKPT*\nI0123 13:57:48.306988 140674925967168 law_classifier.py:730] name = bert/encoder/layer_3/output/dense/kernel:0, shape = (3072, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_3/output/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.307062 140674925967168 law_classifier.py:730] name = bert/encoder/layer_3/output/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_3/output/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.307117 140674925967168 law_classifier.py:730] name = bert/encoder/layer_3/output/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_3/output/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.307170 140674925967168 law_classifier.py:730] name = bert/encoder/layer_3/output/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_4/attention/self/query/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nI0123 13:57:48.307223 140674925967168 law_classifier.py:730] name = bert/encoder/layer_4/attention/self/query/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_4/attention/self/query/bias:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.307284 140674925967168 law_classifier.py:730] name = bert/encoder/layer_4/attention/self/query/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_4/attention/self/key/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nI0123 13:57:48.307338 140674925967168 law_classifier.py:730] name = bert/encoder/layer_4/attention/self/key/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_4/attention/self/key/bias:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.307394 140674925967168 law_classifier.py:730] name = bert/encoder/layer_4/attention/self/key/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_4/attention/self/value/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nI0123 13:57:48.307446 140674925967168 law_classifier.py:730] name = bert/encoder/layer_4/attention/self/value/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_4/attention/self/value/bias:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.307502 140674925967168 law_classifier.py:730] name = bert/encoder/layer_4/attention/self/value/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_4/attention/output/dense/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nI0123 13:57:48.307556 140674925967168 law_classifier.py:730] name = bert/encoder/layer_4/attention/output/dense/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_4/attention/output/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.307612 140674925967168 law_classifier.py:730] name = bert/encoder/layer_4/attention/output/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_4/attention/output/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.307666 140674925967168 law_classifier.py:730] name = bert/encoder/layer_4/attention/output/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_4/attention/output/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.307719 140674925967168 law_classifier.py:730] name = bert/encoder/layer_4/attention/output/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_4/intermediate/dense/kernel:0, shape = (768, 3072), *INIT_FROM_CKPT*\nI0123 13:57:48.307772 140674925967168 law_classifier.py:730] name = bert/encoder/layer_4/intermediate/dense/kernel:0, shape = (768, 3072), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_4/intermediate/dense/bias:0, shape = (3072,), *INIT_FROM_CKPT*\nI0123 13:57:48.307828 140674925967168 law_classifier.py:730] name = bert/encoder/layer_4/intermediate/dense/bias:0, shape = (3072,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_4/output/dense/kernel:0, shape = (3072, 768), *INIT_FROM_CKPT*\nI0123 13:57:48.307882 140674925967168 law_classifier.py:730] name = bert/encoder/layer_4/output/dense/kernel:0, shape = (3072, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_4/output/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.307970 140674925967168 law_classifier.py:730] name = bert/encoder/layer_4/output/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_4/output/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.308024 140674925967168 law_classifier.py:730] name = bert/encoder/layer_4/output/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_4/output/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.308077 140674925967168 law_classifier.py:730] name = bert/encoder/layer_4/output/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_5/attention/self/query/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nI0123 13:57:48.308130 140674925967168 law_classifier.py:730] name = bert/encoder/layer_5/attention/self/query/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_5/attention/self/query/bias:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.308187 140674925967168 law_classifier.py:730] name = bert/encoder/layer_5/attention/self/query/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_5/attention/self/key/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nI0123 13:57:48.308244 140674925967168 law_classifier.py:730] name = bert/encoder/layer_5/attention/self/key/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_5/attention/self/key/bias:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.308301 140674925967168 law_classifier.py:730] name = bert/encoder/layer_5/attention/self/key/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_5/attention/self/value/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nI0123 13:57:48.308354 140674925967168 law_classifier.py:730] name = bert/encoder/layer_5/attention/self/value/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_5/attention/self/value/bias:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.308410 140674925967168 law_classifier.py:730] name = bert/encoder/layer_5/attention/self/value/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_5/attention/output/dense/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nI0123 13:57:48.308484 140674925967168 law_classifier.py:730] name = bert/encoder/layer_5/attention/output/dense/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_5/attention/output/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.308544 140674925967168 law_classifier.py:730] name = bert/encoder/layer_5/attention/output/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_5/attention/output/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.308601 140674925967168 law_classifier.py:730] name = bert/encoder/layer_5/attention/output/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_5/attention/output/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.308657 140674925967168 law_classifier.py:730] name = bert/encoder/layer_5/attention/output/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_5/intermediate/dense/kernel:0, shape = (768, 3072), *INIT_FROM_CKPT*\nI0123 13:57:48.308713 140674925967168 law_classifier.py:730] name = bert/encoder/layer_5/intermediate/dense/kernel:0, shape = (768, 3072), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_5/intermediate/dense/bias:0, shape = (3072,), *INIT_FROM_CKPT*\nI0123 13:57:48.308772 140674925967168 law_classifier.py:730] name = bert/encoder/layer_5/intermediate/dense/bias:0, shape = (3072,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_5/output/dense/kernel:0, shape = (3072, 768), *INIT_FROM_CKPT*\nI0123 13:57:48.308828 140674925967168 law_classifier.py:730] name = bert/encoder/layer_5/output/dense/kernel:0, shape = (3072, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_5/output/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.308889 140674925967168 law_classifier.py:730] name = bert/encoder/layer_5/output/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_5/output/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.308945 140674925967168 law_classifier.py:730] name = bert/encoder/layer_5/output/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_5/output/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.309001 140674925967168 law_classifier.py:730] name = bert/encoder/layer_5/output/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_6/attention/self/query/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nI0123 13:57:48.309069 140674925967168 law_classifier.py:730] name = bert/encoder/layer_6/attention/self/query/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_6/attention/self/query/bias:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.309125 140674925967168 law_classifier.py:730] name = bert/encoder/layer_6/attention/self/query/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_6/attention/self/key/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nI0123 13:57:48.309178 140674925967168 law_classifier.py:730] name = bert/encoder/layer_6/attention/self/key/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_6/attention/self/key/bias:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.309233 140674925967168 law_classifier.py:730] name = bert/encoder/layer_6/attention/self/key/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_6/attention/self/value/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nI0123 13:57:48.309292 140674925967168 law_classifier.py:730] name = bert/encoder/layer_6/attention/self/value/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_6/attention/self/value/bias:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.309348 140674925967168 law_classifier.py:730] name = bert/encoder/layer_6/attention/self/value/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_6/attention/output/dense/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nI0123 13:57:48.309401 140674925967168 law_classifier.py:730] name = bert/encoder/layer_6/attention/output/dense/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_6/attention/output/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.309457 140674925967168 law_classifier.py:730] name = bert/encoder/layer_6/attention/output/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_6/attention/output/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.309511 140674925967168 law_classifier.py:730] name = bert/encoder/layer_6/attention/output/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_6/attention/output/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.309564 140674925967168 law_classifier.py:730] name = bert/encoder/layer_6/attention/output/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_6/intermediate/dense/kernel:0, shape = (768, 3072), *INIT_FROM_CKPT*\nI0123 13:57:48.309616 140674925967168 law_classifier.py:730] name = bert/encoder/layer_6/intermediate/dense/kernel:0, shape = (768, 3072), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_6/intermediate/dense/bias:0, shape = (3072,), *INIT_FROM_CKPT*\nI0123 13:57:48.309673 140674925967168 law_classifier.py:730] name = bert/encoder/layer_6/intermediate/dense/bias:0, shape = (3072,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_6/output/dense/kernel:0, shape = (3072, 768), *INIT_FROM_CKPT*\nI0123 13:57:48.309726 140674925967168 law_classifier.py:730] name = bert/encoder/layer_6/output/dense/kernel:0, shape = (3072, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_6/output/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.309783 140674925967168 law_classifier.py:730] name = bert/encoder/layer_6/output/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_6/output/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.309837 140674925967168 law_classifier.py:730] name = bert/encoder/layer_6/output/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_6/output/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.309889 140674925967168 law_classifier.py:730] name = bert/encoder/layer_6/output/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_7/attention/self/query/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nI0123 13:57:48.309942 140674925967168 law_classifier.py:730] name = bert/encoder/layer_7/attention/self/query/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_7/attention/self/query/bias:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.309998 140674925967168 law_classifier.py:730] name = bert/encoder/layer_7/attention/self/query/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_7/attention/self/key/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nI0123 13:57:48.310050 140674925967168 law_classifier.py:730] name = bert/encoder/layer_7/attention/self/key/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_7/attention/self/key/bias:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.310105 140674925967168 law_classifier.py:730] name = bert/encoder/layer_7/attention/self/key/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_7/attention/self/value/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nI0123 13:57:48.310158 140674925967168 law_classifier.py:730] name = bert/encoder/layer_7/attention/self/value/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_7/attention/self/value/bias:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.310214 140674925967168 law_classifier.py:730] name = bert/encoder/layer_7/attention/self/value/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_7/attention/output/dense/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nI0123 13:57:48.310285 140674925967168 law_classifier.py:730] name = bert/encoder/layer_7/attention/output/dense/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_7/attention/output/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.310343 140674925967168 law_classifier.py:730] name = bert/encoder/layer_7/attention/output/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_7/attention/output/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.310397 140674925967168 law_classifier.py:730] name = bert/encoder/layer_7/attention/output/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_7/attention/output/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.310451 140674925967168 law_classifier.py:730] name = bert/encoder/layer_7/attention/output/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_7/intermediate/dense/kernel:0, shape = (768, 3072), *INIT_FROM_CKPT*\nI0123 13:57:48.310503 140674925967168 law_classifier.py:730] name = bert/encoder/layer_7/intermediate/dense/kernel:0, shape = (768, 3072), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_7/intermediate/dense/bias:0, shape = (3072,), *INIT_FROM_CKPT*\nI0123 13:57:48.310559 140674925967168 law_classifier.py:730] name = bert/encoder/layer_7/intermediate/dense/bias:0, shape = (3072,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_7/output/dense/kernel:0, shape = (3072, 768), *INIT_FROM_CKPT*\nI0123 13:57:48.310613 140674925967168 law_classifier.py:730] name = bert/encoder/layer_7/output/dense/kernel:0, shape = (3072, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_7/output/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.310669 140674925967168 law_classifier.py:730] name = bert/encoder/layer_7/output/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_7/output/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.310723 140674925967168 law_classifier.py:730] name = bert/encoder/layer_7/output/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_7/output/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.310793 140674925967168 law_classifier.py:730] name = bert/encoder/layer_7/output/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_8/attention/self/query/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nI0123 13:57:48.310850 140674925967168 law_classifier.py:730] name = bert/encoder/layer_8/attention/self/query/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_8/attention/self/query/bias:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.310909 140674925967168 law_classifier.py:730] name = bert/encoder/layer_8/attention/self/query/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_8/attention/self/key/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nI0123 13:57:48.310966 140674925967168 law_classifier.py:730] name = bert/encoder/layer_8/attention/self/key/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_8/attention/self/key/bias:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.311025 140674925967168 law_classifier.py:730] name = bert/encoder/layer_8/attention/self/key/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_8/attention/self/value/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nI0123 13:57:48.311082 140674925967168 law_classifier.py:730] name = bert/encoder/layer_8/attention/self/value/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_8/attention/self/value/bias:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.311141 140674925967168 law_classifier.py:730] name = bert/encoder/layer_8/attention/self/value/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_8/attention/output/dense/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nI0123 13:57:48.311198 140674925967168 law_classifier.py:730] name = bert/encoder/layer_8/attention/output/dense/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_8/attention/output/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.311272 140674925967168 law_classifier.py:730] name = bert/encoder/layer_8/attention/output/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_8/attention/output/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.311344 140674925967168 law_classifier.py:730] name = bert/encoder/layer_8/attention/output/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_8/attention/output/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.311400 140674925967168 law_classifier.py:730] name = bert/encoder/layer_8/attention/output/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_8/intermediate/dense/kernel:0, shape = (768, 3072), *INIT_FROM_CKPT*\nI0123 13:57:48.311456 140674925967168 law_classifier.py:730] name = bert/encoder/layer_8/intermediate/dense/kernel:0, shape = (768, 3072), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_8/intermediate/dense/bias:0, shape = (3072,), *INIT_FROM_CKPT*\nI0123 13:57:48.311527 140674925967168 law_classifier.py:730] name = bert/encoder/layer_8/intermediate/dense/bias:0, shape = (3072,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_8/output/dense/kernel:0, shape = (3072, 768), *INIT_FROM_CKPT*\nI0123 13:57:48.311581 140674925967168 law_classifier.py:730] name = bert/encoder/layer_8/output/dense/kernel:0, shape = (3072, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_8/output/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.311638 140674925967168 law_classifier.py:730] name = bert/encoder/layer_8/output/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_8/output/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.311691 140674925967168 law_classifier.py:730] name = bert/encoder/layer_8/output/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_8/output/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.311744 140674925967168 law_classifier.py:730] name = bert/encoder/layer_8/output/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_9/attention/self/query/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nI0123 13:57:48.311797 140674925967168 law_classifier.py:730] name = bert/encoder/layer_9/attention/self/query/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_9/attention/self/query/bias:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.311853 140674925967168 law_classifier.py:730] name = bert/encoder/layer_9/attention/self/query/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_9/attention/self/key/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nI0123 13:57:48.311906 140674925967168 law_classifier.py:730] name = bert/encoder/layer_9/attention/self/key/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_9/attention/self/key/bias:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.311961 140674925967168 law_classifier.py:730] name = bert/encoder/layer_9/attention/self/key/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_9/attention/self/value/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nI0123 13:57:48.312014 140674925967168 law_classifier.py:730] name = bert/encoder/layer_9/attention/self/value/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_9/attention/self/value/bias:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.312069 140674925967168 law_classifier.py:730] name = bert/encoder/layer_9/attention/self/value/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_9/attention/output/dense/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nI0123 13:57:48.312122 140674925967168 law_classifier.py:730] name = bert/encoder/layer_9/attention/output/dense/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_9/attention/output/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.312177 140674925967168 law_classifier.py:730] name = bert/encoder/layer_9/attention/output/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_9/attention/output/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.312230 140674925967168 law_classifier.py:730] name = bert/encoder/layer_9/attention/output/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_9/attention/output/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.312288 140674925967168 law_classifier.py:730] name = bert/encoder/layer_9/attention/output/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_9/intermediate/dense/kernel:0, shape = (768, 3072), *INIT_FROM_CKPT*\nI0123 13:57:48.312341 140674925967168 law_classifier.py:730] name = bert/encoder/layer_9/intermediate/dense/kernel:0, shape = (768, 3072), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_9/intermediate/dense/bias:0, shape = (3072,), *INIT_FROM_CKPT*\nI0123 13:57:48.312397 140674925967168 law_classifier.py:730] name = bert/encoder/layer_9/intermediate/dense/bias:0, shape = (3072,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_9/output/dense/kernel:0, shape = (3072, 768), *INIT_FROM_CKPT*\nI0123 13:57:48.312451 140674925967168 law_classifier.py:730] name = bert/encoder/layer_9/output/dense/kernel:0, shape = (3072, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_9/output/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.312506 140674925967168 law_classifier.py:730] name = bert/encoder/layer_9/output/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_9/output/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.312560 140674925967168 law_classifier.py:730] name = bert/encoder/layer_9/output/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_9/output/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.312613 140674925967168 law_classifier.py:730] name = bert/encoder/layer_9/output/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_10/attention/self/query/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nI0123 13:57:48.312666 140674925967168 law_classifier.py:730] name = bert/encoder/layer_10/attention/self/query/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_10/attention/self/query/bias:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.312723 140674925967168 law_classifier.py:730] name = bert/encoder/layer_10/attention/self/query/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_10/attention/self/key/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nI0123 13:57:48.312776 140674925967168 law_classifier.py:730] name = bert/encoder/layer_10/attention/self/key/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_10/attention/self/key/bias:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.312832 140674925967168 law_classifier.py:730] name = bert/encoder/layer_10/attention/self/key/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_10/attention/self/value/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nI0123 13:57:48.312886 140674925967168 law_classifier.py:730] name = bert/encoder/layer_10/attention/self/value/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_10/attention/self/value/bias:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.312943 140674925967168 law_classifier.py:730] name = bert/encoder/layer_10/attention/self/value/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_10/attention/output/dense/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nI0123 13:57:48.312996 140674925967168 law_classifier.py:730] name = bert/encoder/layer_10/attention/output/dense/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_10/attention/output/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.313071 140674925967168 law_classifier.py:730] name = bert/encoder/layer_10/attention/output/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_10/attention/output/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.313155 140674925967168 law_classifier.py:730] name = bert/encoder/layer_10/attention/output/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_10/attention/output/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.313211 140674925967168 law_classifier.py:730] name = bert/encoder/layer_10/attention/output/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_10/intermediate/dense/kernel:0, shape = (768, 3072), *INIT_FROM_CKPT*\nI0123 13:57:48.313272 140674925967168 law_classifier.py:730] name = bert/encoder/layer_10/intermediate/dense/kernel:0, shape = (768, 3072), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_10/intermediate/dense/bias:0, shape = (3072,), *INIT_FROM_CKPT*\nI0123 13:57:48.313332 140674925967168 law_classifier.py:730] name = bert/encoder/layer_10/intermediate/dense/bias:0, shape = (3072,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_10/output/dense/kernel:0, shape = (3072, 768), *INIT_FROM_CKPT*\nI0123 13:57:48.313389 140674925967168 law_classifier.py:730] name = bert/encoder/layer_10/output/dense/kernel:0, shape = (3072, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_10/output/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.313450 140674925967168 law_classifier.py:730] name = bert/encoder/layer_10/output/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_10/output/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.313506 140674925967168 law_classifier.py:730] name = bert/encoder/layer_10/output/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_10/output/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.313561 140674925967168 law_classifier.py:730] name = bert/encoder/layer_10/output/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_11/attention/self/query/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nI0123 13:57:48.313617 140674925967168 law_classifier.py:730] name = bert/encoder/layer_11/attention/self/query/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_11/attention/self/query/bias:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.313677 140674925967168 law_classifier.py:730] name = bert/encoder/layer_11/attention/self/query/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_11/attention/self/key/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nI0123 13:57:48.313734 140674925967168 law_classifier.py:730] name = bert/encoder/layer_11/attention/self/key/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_11/attention/self/key/bias:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.313805 140674925967168 law_classifier.py:730] name = bert/encoder/layer_11/attention/self/key/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_11/attention/self/value/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nI0123 13:57:48.313858 140674925967168 law_classifier.py:730] name = bert/encoder/layer_11/attention/self/value/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_11/attention/self/value/bias:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.313914 140674925967168 law_classifier.py:730] name = bert/encoder/layer_11/attention/self/value/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_11/attention/output/dense/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nI0123 13:57:48.313987 140674925967168 law_classifier.py:730] name = bert/encoder/layer_11/attention/output/dense/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_11/attention/output/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.314047 140674925967168 law_classifier.py:730] name = bert/encoder/layer_11/attention/output/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_11/attention/output/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.314104 140674925967168 law_classifier.py:730] name = bert/encoder/layer_11/attention/output/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_11/attention/output/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.314171 140674925967168 law_classifier.py:730] name = bert/encoder/layer_11/attention/output/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_11/intermediate/dense/kernel:0, shape = (768, 3072), *INIT_FROM_CKPT*\nI0123 13:57:48.314236 140674925967168 law_classifier.py:730] name = bert/encoder/layer_11/intermediate/dense/kernel:0, shape = (768, 3072), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_11/intermediate/dense/bias:0, shape = (3072,), *INIT_FROM_CKPT*\nI0123 13:57:48.314300 140674925967168 law_classifier.py:730] name = bert/encoder/layer_11/intermediate/dense/bias:0, shape = (3072,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_11/output/dense/kernel:0, shape = (3072, 768), *INIT_FROM_CKPT*\nI0123 13:57:48.314354 140674925967168 law_classifier.py:730] name = bert/encoder/layer_11/output/dense/kernel:0, shape = (3072, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_11/output/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.314461 140674925967168 law_classifier.py:730] name = bert/encoder/layer_11/output/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_11/output/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.314524 140674925967168 law_classifier.py:730] name = bert/encoder/layer_11/output/LayerNorm/beta:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/encoder/layer_11/output/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.314586 140674925967168 law_classifier.py:730] name = bert/encoder/layer_11/output/LayerNorm/gamma:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/pooler/dense/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nI0123 13:57:48.314649 140674925967168 law_classifier.py:730] name = bert/pooler/dense/kernel:0, shape = (768, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = bert/pooler/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nI0123 13:57:48.314715 140674925967168 law_classifier.py:730] name = bert/pooler/dense/bias:0, shape = (768,), *INIT_FROM_CKPT*\nINFO:tensorflow: name = output_weights:0, shape = (5, 768), *INIT_FROM_CKPT*\nI0123 13:57:48.314777 140674925967168 law_classifier.py:730] name = output_weights:0, shape = (5, 768), *INIT_FROM_CKPT*\nINFO:tensorflow: name = output_bias:0, shape = (5,), *INIT_FROM_CKPT*\nI0123 13:57:48.314842 140674925967168 law_classifier.py:730] name = output_bias:0, shape = (5,), *INIT_FROM_CKPT*\nINFO:tensorflow:Done calling model_fn.\nI0123 13:57:48.315157 140674925967168 estimator.py:1150] Done calling model_fn.\nWARNING:tensorflow:From /opt/conda/lib/python3.7/site-packages/tensorflow_core/python/ops/array_ops.py:1475: where (from tensorflow.python.ops.array_ops) is deprecated and will be removed in a future version.\nInstructions for updating:\nUse tf.where in 2.0, which has the same broadcast rule as np.where\nW0123 13:57:48.431097 140674925967168 deprecation.py:323] From /opt/conda/lib/python3.7/site-packages/tensorflow_core/python/ops/array_ops.py:1475: where (from tensorflow.python.ops.array_ops) is deprecated and will be removed in a future version.\nInstructions for updating:\nUse tf.where in 2.0, which has the same broadcast rule as np.where\nINFO:tensorflow:Graph was finalized.\nI0123 13:57:48.682827 140674925967168 monitored_session.py:240] Graph was finalized.\n2021-01-23 13:57:48.689899: I tensorflow/core/platform/profile_utils/cpu_utils.cc:94] CPU Frequency: 2200000000 Hz\n2021-01-23 13:57:48.690996: I tensorflow/compiler/xla/service/service.cc:168] XLA service 0x558acf13a390 initialized for platform Host (this does not guarantee that XLA will be used). Devices:\n2021-01-23 13:57:48.691025: I tensorflow/compiler/xla/service/service.cc:176] StreamExecutor device (0): Host, Default Version\n2021-01-23 13:57:48.693355: I tensorflow/stream_executor/platform/default/dso_loader.cc:48] Successfully opened dynamic library libcuda.so.1\n2021-01-23 13:57:49.416640: I tensorflow/stream_executor/cuda/cuda_gpu_executor.cc:983] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero\n2021-01-23 13:57:49.417386: I tensorflow/compiler/xla/service/service.cc:168] XLA service 0x558acb181620 initialized for platform CUDA (this does not guarantee that XLA will be used). Devices:\n2021-01-23 13:57:49.417421: I tensorflow/compiler/xla/service/service.cc:176] StreamExecutor device (0): Tesla V100-SXM2-16GB, Compute Capability 7.0\n2021-01-23 13:57:49.417681: I tensorflow/stream_executor/cuda/cuda_gpu_executor.cc:983] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero\n2021-01-23 13:57:49.418281: I tensorflow/core/common_runtime/gpu/gpu_device.cc:1639] Found device 0 with properties: \nname: Tesla V100-SXM2-16GB major: 7 minor: 0 memoryClockRate(GHz): 1.53\npciBusID: 0000:00:04.0\n2021-01-23 13:57:49.418319: I tensorflow/stream_executor/platform/default/dso_loader.cc:48] Successfully opened dynamic library libcudart.so.11.0\n2021-01-23 13:57:49.421084: I tensorflow/stream_executor/platform/default/dso_loader.cc:48] Successfully opened dynamic library libcublas.so.11\n2021-01-23 13:57:49.422316: I tensorflow/stream_executor/platform/default/dso_loader.cc:48] Successfully opened dynamic library libcufft.so.10\n2021-01-23 13:57:49.422616: I tensorflow/stream_executor/platform/default/dso_loader.cc:48] Successfully opened dynamic library libcurand.so.10\n2021-01-23 13:57:49.425567: I tensorflow/stream_executor/platform/default/dso_loader.cc:48] Successfully opened dynamic library libcusolver.so.10\n2021-01-23 13:57:49.426309: I tensorflow/stream_executor/platform/default/dso_loader.cc:48] Successfully opened dynamic library libcusparse.so.11\n2021-01-23 13:57:49.426476: I tensorflow/stream_executor/platform/default/dso_loader.cc:48] Successfully opened dynamic library libcudnn.so.8\n2021-01-23 13:57:49.426584: I tensorflow/stream_executor/cuda/cuda_gpu_executor.cc:983] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero\n2021-01-23 13:57:49.427260: I tensorflow/stream_executor/cuda/cuda_gpu_executor.cc:983] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero\n2021-01-23 13:57:49.427805: I tensorflow/core/common_runtime/gpu/gpu_device.cc:1767] Adding visible gpu devices: 0\n2021-01-23 13:57:49.427838: I tensorflow/stream_executor/platform/default/dso_loader.cc:48] Successfully opened dynamic library libcudart.so.11.0\n2021-01-23 13:57:49.774299: I tensorflow/core/common_runtime/gpu/gpu_device.cc:1180] Device interconnect StreamExecutor with strength 1 edge matrix:\n2021-01-23 13:57:49.774352: I tensorflow/core/common_runtime/gpu/gpu_device.cc:1186] 0 \n2021-01-23 13:57:49.774361: I tensorflow/core/common_runtime/gpu/gpu_device.cc:1199] 0: N \n2021-01-23 13:57:49.774705: I tensorflow/stream_executor/cuda/cuda_gpu_executor.cc:983] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero\n2021-01-23 13:57:49.775448: I tensorflow/stream_executor/cuda/cuda_gpu_executor.cc:983] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero\n2021-01-23 13:57:49.776091: I tensorflow/core/common_runtime/gpu/gpu_device.cc:1325] Created TensorFlow device (/job:localhost/replica:0/task:0/device:GPU:0 with 14963 MB memory) -> physical GPU (device: 0, name: Tesla V100-SXM2-16GB, pci bus id: 0000:00:04.0, compute capability: 7.0)\nINFO:tensorflow:Running local_init_op.\nI0123 13:57:50.966366 140674925967168 session_manager.py:500] Running local_init_op.\nINFO:tensorflow:Done running local_init_op.\nI0123 13:57:51.024328 140674925967168 session_manager.py:502] Done running local_init_op.\n2021-01-23 13:57:51.772977: I tensorflow/stream_executor/platform/default/dso_loader.cc:48] Successfully opened dynamic library libcublas.so.11\nINFO:tensorflow:prediction_loop marked as finished\nI0123 13:57:58.185830 140674925967168 error_handling.py:101] prediction_loop marked as finished\nINFO:tensorflow:prediction_loop marked as finished\nI0123 13:57:58.186083 140674925967168 error_handling.py:101] prediction_loop marked as finished\n"
]
],
[
[
"# Accuracy and Result",
"_____no_output_____"
],
[
"**could test its accuracy by setting do_eval=true when finetuning. Here, I added a multi-class confusion matrix to give more information about the prediction in order to tune the model or come up with improvement strategi**",
"_____no_output_____"
]
],
[
[
"def plot_confusion_matrix(y_true, y_pred, classes, destination=None, normalize=False, title=None, cmap=plt.cm.Blues):\n \"\"\"\n This function prints and plots the confusion matrix.\n Normalization can be applied by setting `normalize=True`.\n \"\"\"\n if not title:\n if normalize:\n title = 'Normalized confusion matrix'\n else:\n title = 'Confusion matrix, without normalization'\n\n # Compute confusion matrix\n #cm = np.array([[ 11, 2 , 40 , 2], [ 0 , 0 , 0 , 0], [ 37 , 8 ,114 , 4], [ 11 , 0 ,33 , 1]])\n cm = confusion_matrix(y_true, y_pred)\n # Only use the labels that appear in the data\n #classes = classes[unique_labels(y_true, y_pred)]\n if normalize:\n cm = cm.astype('float') / cm.sum(axis=1)[:, np.newaxis]\n print(\"Normalized confusion matrix\")\n else:\n print('Confusion matrix, without normalization')\n\n print(cm)\n\n fig, ax = plt.subplots()\n im = ax.imshow(cm, interpolation='nearest', cmap=cmap)\n ax.figure.colorbar(im, ax=ax)\n # We want to show all ticks...\n ax.set(xticks=np.arange(cm.shape[1]),\n yticks=np.arange(cm.shape[0]),\n # ... and label them with the respective list entries\n xticklabels=classes, yticklabels=classes,\n title=title,\n ylabel='True label',\n xlabel='Predicted label')\n\n # Rotate the tick labels and set their alignment.\n plt.setp(ax.get_xticklabels(), rotation=30, ha=\"right\",\n rotation_mode=\"anchor\")\n\n # Loop over data dimensions and create text annotations.\n fmt = '.2f' if normalize else 'd'\n thresh = cm.max() / 2.\n for i in range(cm.shape[0]):\n for j in range(cm.shape[1]):\n ax.text(j, i, format(cm[i, j], fmt),\n ha=\"center\", va=\"center\",\n color=\"white\" if cm[i, j] > thresh else \"black\")\n fig.tight_layout()\n #plt.show()\n try:\n plt.savefig(destination)\n except:\n pass\n \n return ax",
"_____no_output_____"
],
[
"def compare_answer(row):\n '''\n Compares 2 columns of actual and predicted class. \n \n Returns:\n -- 1 if they are the same\n -- 0 otherwise.\n '''\n \n if row['prediction'] == row['actual']:\n return 1\n else:\n return 0",
"_____no_output_____"
],
[
"def benchmark(actual_path, pred_path, confusion_matrix_path):\n '''\n Calculates model accuracy and confusion matrix\n\n Arguments:\n actual_path -- path of test file which contains actual labels.\n pred_path -- path of prediction file which contains predicted labels.\n\n Returns:\n accuracy -- accuracy of the prediction\n confusion matrix -- save as image file\n '''\n\n # Read files to dataframes\n actual = pd.read_csv(actual_path, sep='\\t')\n #actual = pd.read_csv(actual_path, error_bad_lines=False)\n pred = pd.read_csv(pred_path, sep='\\t')\n\n # Create result dataframe\n result = pred.copy()\n result['prediction'] = result.idxmax(axis=1)\n result['actual'] = actual['labels']\n result['correct'] = result.apply(lambda row: compare_answer(row), axis=1)\n\n # Calculate accuracy\n accuracy = sum(result['correct'])/len(result)\n\n # Confusion Matrix\n confusion_matrix = plot_confusion_matrix(result['actual'], result['prediction'], classes=['Violation', 'family', 'labor', 'contract', 'criminal'], destination=confusion_matrix_path, normalize=False, title='Confusion Matrix')\n\n return accuracy, result",
"_____no_output_____"
],
[
"# Benchmark accuracy\nactual_path = 'law_dataset/test_labels.tsv'\npred_path = 'output_predict2/test_results.tsv'\nconfusion_matrix_path = 'output_predict2/confusion_matrix.png'",
"_____no_output_____"
],
[
"accuracy, result = benchmark(actual_path, pred_path, confusion_matrix_path)",
"Confusion matrix, without normalization\n[[ 74 17 13 3 3]\n [ 6 733 17 22 16]\n [ 9 12 266 1 9]\n [ 0 22 1 582 0]\n [ 0 8 2 3 290]]\n"
],
[
"print(accuracy)",
"0.9222380275011854\n"
],
[
"print(result)",
" Violation family labor contract criminal prediction actual \\\n0 0.957460 0.026618 0.005343 0.001329 0.009250 Violation Violation \n1 0.000926 0.000286 0.000432 0.010299 0.988057 criminal contract \n2 0.031536 0.000375 0.964034 0.001094 0.002960 labor labor \n3 0.000159 0.998271 0.000599 0.000487 0.000483 family family \n4 0.000818 0.000231 0.000272 0.000455 0.998223 criminal criminal \n... ... ... ... ... ... ... ... \n2104 0.000286 0.000148 0.000166 0.999152 0.000248 contract contract \n2105 0.000134 0.999341 0.000187 0.000157 0.000181 family family \n2106 0.000105 0.999364 0.000158 0.000234 0.000139 family family \n2107 0.000116 0.999404 0.000183 0.000159 0.000139 family family \n2108 0.000808 0.000230 0.000380 0.000377 0.998205 criminal criminal \n\n correct \n0 1 \n1 0 \n2 1 \n3 1 \n4 1 \n... ... \n2104 1 \n2105 1 \n2106 1 \n2107 1 \n2108 1 \n\n[2109 rows x 8 columns]\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
c51017eb044084cd41f0378f62b120692a265c6d
| 231,777 |
ipynb
|
Jupyter Notebook
|
jupyter/2019-05-06-tidycensus.ipynb
|
mabalam/mabalam.github.io
|
8b7279b58e3eb442561bbb21f250dba3137522e5
|
[
"MIT"
] | null | null | null |
jupyter/2019-05-06-tidycensus.ipynb
|
mabalam/mabalam.github.io
|
8b7279b58e3eb442561bbb21f250dba3137522e5
|
[
"MIT"
] | null | null | null |
jupyter/2019-05-06-tidycensus.ipynb
|
mabalam/mabalam.github.io
|
8b7279b58e3eb442561bbb21f250dba3137522e5
|
[
"MIT"
] | null | null | null | 836.740072 | 100,056 | 0.942078 |
[
[
[
"## Spatial data visualization with `tidycensus`",
"_____no_output_____"
],
[
"**Location! Location! Location!** \n\nThe location people live in tells us a lot about the space itself as well as the people who live in there. This demo is about spatial data visualization with `tidycensus` R package with two variables of interest -- population and race distribution. First we will get the big picture at the Virginia state scale, then will zoom in on northern Virginia in Washington DC metro area.\n\n\n### Tools\nWe do not have to start from scratch. As always the case, someone has already done the hard work for us so we can stand on their shoulders. \n\n[Kyle Walker](https://walkerke.github.io/) in this case has developed an `rstat` package called [`tidycensus`](https://github.com/walkerke/tidycensus). This package allows for easy access, analysis and visualization of Census Beureau data on hundreds of variables. \n\nNot that in Python you can not do spatial analysis/visualization of census data, but certainly not as easily as in R because of some excellent rstats packages available and tailored for this purpose. \n\nTo be able to use `tidycensus` you'll need your own Census API Key. If you do not have one, [get one](https://www.census.gov/developers/). \n\nThe only other library you'll need is `tidyverse`; and this is it! If you like interactive visialization of maps (i.e. zoom in zoom out etc.) you will need additional libraries and codes. \n\n---\nOkay, so here we go ... ",
"_____no_output_____"
]
],
[
[
"# import libraries\nlibrary(tidycensus)\nlibrary(tidyverse)\noptions(tigris_use_cache = TRUE)\n\n# get your Census Bureau API key\ncensus_api_key(\"xxxx\")",
"To install your API key for use in future sessions, run this function with `install = TRUE`.\n"
],
[
"# geting VA population data of counties\nvapop <- get_acs(state = \"VA\", geography = \"county\", \n variables = \"B19013_001\", geometry = TRUE)\n# plot VA population\nvapop %>%\n ggplot(aes(fill = estimate)) + \n geom_sf(color = NA) + \n coord_sf(crs = \"+init=epsg:4326\") + # original 26911 \n scale_fill_viridis_c(option = \"viridis\", direction=-1) # original: scale_fill_viridis_c(option = \"magma\") ",
"Getting data from the 2012-2016 5-year ACS\n"
],
[
"# specify races\nraces <- c(White = \"P005003\", \n Black = \"P005004\", \n Asian = \"P005006\", \n Hispanic = \"P004003\")\n\n# get decennial data on races\nvarace <- get_decennial(geography = \"county\", variables = races, \n state = \"VA\", geometry = TRUE,\n summary_var = \"P001001\") \n\n# plot race variables as a percent of total population\nvarace %>%\n mutate(percent = 100 * (value / summary_value)) %>% # create a calculated column of percent value\n ggplot(aes(fill = percent)) +\n facet_wrap(~variable) +\n geom_sf(color = NA) +\n coord_sf(crs = \"+init=epsg:4326\") + \n scale_fill_viridis_c(option = \"magma\", direction=-1)\n",
"Getting data from the 2010 decennial Census\n"
]
],
[
[
"## Zooming in on Northern Virginia",
"_____no_output_____"
]
],
[
[
"# specify counties that constitute Northern Virginia\nNOVA = c(\"Fairfax County\", \"Fairfax City\", \"Manassas Park City\", \"Arlington County\", \"Loudoun County\", \"Alexandria City\", \"Falls Church City\", \"Prince William County\", \"Manassas City\")\n\n# get population data of NOVA cunties\nnovapop <- get_acs(state = \"VA\", county = NOVA, geography = \"tract\", \n variables = \"B19013_001\", geometry = TRUE)\n\n# plot NOVA population\n\nnovapop %>%\n ggplot(aes(fill = estimate)) + \n geom_sf(color = NA) + \n coord_sf(crs = \"+init=epsg:4326\") + \n scale_fill_viridis_c(option = \"viridis\", direction=-1) # original: scale_fill_viridis_c(option = \"magma\") ",
"Getting data from the 2012-2016 5-year ACS\n"
],
[
"# get decennial data on races\nnovarace <- get_decennial(geography = \"tract\", variables = races, \n state = \"VA\", county = NOVA, geometry = TRUE,\n summary_var = \"P001001\") \n\n# plot race variables as a percent of total population\nnovarace %>%\n mutate(percent = 100 * (value / summary_value)) %>% # create a calculated column of percent value\n ggplot(aes(fill = percent)) +\n facet_wrap(~variable) +\n geom_sf(color = NA) +\n coord_sf(crs = \"+init=epsg:4326\") + \n scale_fill_viridis_c(option = \"magma\", direction=-1)",
"Getting data from the 2010 decennial Census\nGetting data from the 2010 decennial Census\nGetting data from the 2010 decennial Census\nGetting data from the 2010 decennial Census\nGetting data from the 2010 decennial Census\nGetting data from the 2010 decennial Census\nGetting data from the 2010 decennial Census\nGetting data from the 2010 decennial Census\nGetting data from the 2010 decennial Census\nGetting data from the 2010 decennial Census\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
c5101b5e537e5e3697f7f2480e1dd2b11fa46d5d
| 5,901 |
ipynb
|
Jupyter Notebook
|
Plotly/Plotly_Create_Linechart.ipynb
|
srini047/awesome-notebooks
|
2a5b771b37b62090de5311d61dce8495fae7b59f
|
[
"BSD-3-Clause"
] | null | null | null |
Plotly/Plotly_Create_Linechart.ipynb
|
srini047/awesome-notebooks
|
2a5b771b37b62090de5311d61dce8495fae7b59f
|
[
"BSD-3-Clause"
] | null | null | null |
Plotly/Plotly_Create_Linechart.ipynb
|
srini047/awesome-notebooks
|
2a5b771b37b62090de5311d61dce8495fae7b59f
|
[
"BSD-3-Clause"
] | null | null | null | 20.925532 | 284 | 0.508388 |
[
[
[
"<img width=\"10%\" alt=\"Naas\" src=\"https://landen.imgix.net/jtci2pxwjczr/assets/5ice39g4.png?w=160\"/>\n",
"_____no_output_____"
],
[
"# Plotly - Create Linechart\n<a href=\"https://app.naas.ai/user-redirect/naas/downloader?url=https://raw.githubusercontent.com/jupyter-naas/awesome-notebooks/master/Plotly/Plotly_Create_Linechart.ipynb\" target=\"_parent\"><img src=\"https://naasai-public.s3.eu-west-3.amazonaws.com/open_in_naas.svg\"/></a>",
"_____no_output_____"
],
[
"**Tags:** #plotly #chart #linechart #trend #dataviz #yahoofinance #naas_drivers #snippet",
"_____no_output_____"
],
[
"**Author:** [Jeremy Ravenel](https://www.linkedin.com/in/ACoAAAJHE7sB5OxuKHuzguZ9L6lfDHqw--cdnJg/)",
"_____no_output_____"
],
[
"Learn more on the Plotly doc : https://plotly.com/python/line-charts/",
"_____no_output_____"
],
[
"## Input",
"_____no_output_____"
],
[
"### Import libraries",
"_____no_output_____"
]
],
[
[
"import naas\nfrom naas_drivers import yahoofinance, plotly",
"_____no_output_____"
]
],
[
[
"### Variables",
"_____no_output_____"
]
],
[
[
"title = \"Linechart\"\n\n# Output paths\noutput_image = f\"{title}.png\"\noutput_html = f\"{title}.html\"",
"_____no_output_____"
]
],
[
[
"### Get data",
"_____no_output_____"
]
],
[
[
"date_from = -360 # Date can be number or date or today\ndate_to = \"today\"\ndf = yahoofinance.get(\"TSLA\",\n date_from=date_from,\n date_to=date_to)\ndf",
"_____no_output_____"
]
],
[
[
"## Model",
"_____no_output_____"
]
],
[
[
"fig = plotly.linechart(df,\n x=\"Date\",\n y=[\"Open\", \"Close\"],\n title=title,\n yaxis_title=\"Price in $\")",
"_____no_output_____"
]
],
[
[
"## Output",
"_____no_output_____"
],
[
"### Export in PNG and HTML",
"_____no_output_____"
]
],
[
[
"fig.write_image(output_image, width=1200)\nfig.write_html(output_html)",
"_____no_output_____"
]
],
[
[
"### Generate shareable assets",
"_____no_output_____"
]
],
[
[
"link_image = naas.asset.add(output_image)\nlink_html = naas.asset.add(output_html, {\"inline\":True})\n\n#-> Uncomment the line below to remove your assets\n# naas.asset.delete(output_image)\n# naas.asset.delete(output_html)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
c51027c280ce4abf7226ea5bc4178bc62162b37e
| 8,945 |
ipynb
|
Jupyter Notebook
|
report/chapter-3/noise/1-create.ipynb
|
ccberg/LA
|
df3929c9ab4b7cbfa38749363c5ccced010f3002
|
[
"MIT"
] | 1 |
2021-12-28T03:27:42.000Z
|
2021-12-28T03:27:42.000Z
|
report/chapter-3/noise/1-create.ipynb
|
ccberg/LA
|
df3929c9ab4b7cbfa38749363c5ccced010f3002
|
[
"MIT"
] | null | null | null |
report/chapter-3/noise/1-create.ipynb
|
ccberg/LA
|
df3929c9ab4b7cbfa38749363c5ccced010f3002
|
[
"MIT"
] | null | null | null | 31.607774 | 129 | 0.566909 |
[
[
[
"import os\n\nimport numpy as np\nfrom tqdm import tqdm\n\nfrom src.data.loaders.ascad import ASCADData\nfrom src.dlla.berg import make_mlp\nfrom src.dlla.hw import prepare_traces_dl, dlla_known_p\nfrom src.pollution.gaussian_noise import gaussian_noise\nfrom src.tools.cache import cache_np\nfrom src.trace_set.database import Database\nfrom src.trace_set.pollution import Pollution, PollutionType\nfrom src.trace_set.set_hw import TraceSetHW\nfrom src.trace_set.window import get_windows, extract_traces",
"_____no_output_____"
],
[
"# Source [EDIT]\nDB = Database.ascad_none\n\n# Limit number of traces [EDIT]\nLIMIT_PROF = None\nLIMIT_ATT = 1000\n\n# Select targets and noise parameters\n\nRAW_TRACES, WINDOW_JITTER_PARAMS, GAUSS_PARAMS, LIMIT_RAW = [None] * 4\n\nif DB is Database.ascad_none or DB is Database.ascad:\n TARGET_ROUND = 0\n TARGET_BYTE = 0\n\n WINDOW_JITTER_PARAMS = np.arange(0, 205, 5)\n GAUSS_PARAMS = np.arange(0, 205, 5)\n\n RAW_TRACES = ASCADData.raw()['traces']\n LIMIT_RAW = -1\n\nif DB is Database.ascad:\n TARGET_BYTE = 2\n\n WINDOW_JITTER_PARAMS = np.arange(0, 2.05, .05)\n GAUSS_PARAMS = np.arange(0, 5.1, .1)\n\nif DB is Database.aisy:\n TARGET_ROUND = 4\n TARGET_BYTE = 0\n\n WINDOW_JITTER_PARAMS = np.arange(0, 460, 10)\n GAUSS_PARAMS = np.arange(0, 4100, 100)\n\n RAW_TRACES = cache_np(\"aisy_traces\")\n\n# Select targets\nTRACE_SET = TraceSetHW(DB)\nSAMPLE_TRACE = TRACE_SET.profile()[0][0]\nWINDOW, WINDOW_CXT = get_windows(RAW_TRACES, SAMPLE_TRACE)\n\n# Isolate context trace for window jitter.\n# Gets cached, as this procedure takes some time (depending on disk read speed)\nX_CXT = cache_np(f\"{DB.name}_x_cxt\", extract_traces, RAW_TRACES, WINDOW_CXT)[:LIMIT_RAW]",
"_____no_output_____"
],
[
"PROFILING_MASK = np.ones(len(X_CXT), dtype=bool)\nPROFILING_MASK[2::3] = 0",
"_____no_output_____"
],
[
"X_PROF, Y_PROF = TRACE_SET.profile_states()\nX_ATT, Y_ATT = TRACE_SET.attack_states()\n\nX_PROF_CXT = X_CXT[PROFILING_MASK]\nX_ATT_CXT = X_CXT[~PROFILING_MASK]",
"_____no_output_____"
],
[
"def verify(db: Database, pollution: Pollution):\n \"\"\"\n Assess leakage from database by\n \"\"\"\n trace_set = TraceSetHW(db, pollution, (LIMIT_PROF, LIMIT_ATT))\n x9, y9, x9_att, y9_att = prepare_traces_dl(*trace_set.profile(), *trace_set.attack())\n mdl9 = make_mlp(x9, y9, progress=False)\n dlla9_p = dlla_known_p(mdl9, x9_att, y9_att)\n\n print(f\"Pollution {pollution.type} ({pollution.parameter}): p-value ({dlla9_p}).\")",
"_____no_output_____"
],
[
"def desync(traces: np.ndarray, window: (int, int), sigma: float):\n start, end = window\n num_traces = len(traces)\n num_sample_points = end - start\n\n permutations = np.round(np.random.normal(scale=sigma, size=num_traces)).astype(int)\n\n if np.max(np.abs(permutations)) >= num_sample_points:\n raise Exception(f\"Window jitter parameter ({sigma}) too high. PoI is not always within the resulting traces.\")\n\n permutations += start\n\n res = np.ones((num_traces, num_sample_points), dtype=traces.dtype)\n\n for ix in tqdm(range(num_traces), f\"Trace desynchronization, sigma={sigma}\"):\n permutation = permutations[ix]\n res[ix] = traces[ix, permutation:permutation + num_sample_points]\n\n return res\n\ndef apply_desync(db, x_prof_cxt, y_prof, x_att_cxt, y_att, window: (int, int), params: list):\n for param in params:\n\n pollution = Pollution(PollutionType.desync, param)\n out = TraceSetHW(db, pollution, (LIMIT_PROF, LIMIT_ATT))\n\n if not os.path.exists(out.path):\n xn = desync(x_prof_cxt, window, param)\n xn_att = desync(x_att_cxt, window, param)\n\n out.create(xn, y_prof, xn_att, y_att)\n\n verify(db, pollution)",
"_____no_output_____"
],
[
"apply_desync(DB, X_PROF_CXT, Y_PROF, X_ATT_CXT, Y_ATT, WINDOW, WINDOW_JITTER_PARAMS)",
"_____no_output_____"
],
[
"def apply_gauss(db, params: list):\n for param in params:\n pollution = Pollution(PollutionType.gauss, param)\n default = TraceSetHW(db)\n out = TraceSetHW(db, pollution, (LIMIT_PROF, LIMIT_ATT))\n\n x_prof, y_prof = default.profile_states()\n x_att, y_att = default.attack_states()\n\n if not os.path.exists(out.path):\n xn = gaussian_noise(x_prof, param)\n xn_att = gaussian_noise(x_att, param)\n\n out.create(xn, y_prof, xn_att, y_att)\n\n verify(db, pollution)\n\napply_gauss(DB, GAUSS_PARAMS)",
"Pollution PollutionType.gauss (5): p-value (4.676300196748799e-201).\nPollution PollutionType.gauss (15): p-value (4.718244810262959e-102).\nPollution PollutionType.gauss (25): p-value (1.861315463054157e-36).\nPollution PollutionType.gauss (35): p-value (1.736725741787256e-23).\nPollution PollutionType.gauss (45): p-value (8.150609351398289e-16).\nPollution PollutionType.gauss (55): p-value (3.829841782334295e-09).\nPollution PollutionType.gauss (65): p-value (3.5357833007055315e-06).\nPollution PollutionType.gauss (75): p-value (0.0005559614850769871).\nPollution PollutionType.gauss (85): p-value (0.3535435021488311).\nPollution PollutionType.gauss (95): p-value (0.006780517900180397).\nPollution PollutionType.gauss (105): p-value (0.001164332651326126).\nPollution PollutionType.gauss (115): p-value (0.011099771169440794).\nPollution PollutionType.gauss (125): p-value (0.05676775957039404).\nPollution PollutionType.gauss (135): p-value (0.3465120016471147).\nPollution PollutionType.gauss (145): p-value (0.2208807382520106).\nPollution PollutionType.gauss (155): p-value (0.337703129827742).\nPollution PollutionType.gauss (165): p-value (0.03813751032199153).\nPollution PollutionType.gauss (175): p-value (0.11361421794157522).\nPollution PollutionType.gauss (185): p-value (0.9969132888530265).\nPollution PollutionType.gauss (195): p-value (0.9234597479259983).\n"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
c5103e33baa6485f1034d00fb3e72071b1084a22
| 12,856 |
ipynb
|
Jupyter Notebook
|
A8-Triangulation/MV_A8.ipynb
|
Haoban/Machine-Vision
|
077ad258f5fdce5495bd80478c9191fd6ed4bd9e
|
[
"BSD-4-Clause-UC"
] | null | null | null |
A8-Triangulation/MV_A8.ipynb
|
Haoban/Machine-Vision
|
077ad258f5fdce5495bd80478c9191fd6ed4bd9e
|
[
"BSD-4-Clause-UC"
] | null | null | null |
A8-Triangulation/MV_A8.ipynb
|
Haoban/Machine-Vision
|
077ad258f5fdce5495bd80478c9191fd6ed4bd9e
|
[
"BSD-4-Clause-UC"
] | null | null | null | 34.191489 | 694 | 0.504356 |
[
[
[
"# Machine Vision<br>Assignment 8 - Triangulation\n\n## Personal details\n\n* **Name(s):** ``\n* **Student ID(s):** ``\n\n## 1. Introduction\n\nIn this assignment we will use a pair of stereo images to triangulate points in 3D. Let us first display the test images and 2D point correspondences. We also load 3D points mainly for testing purposes. The data is from __http://www.robots.ox.ac.uk/~vgg/data/data-mview.html__.",
"_____no_output_____"
]
],
[
[
"import cv2\nimport numpy as np\nimport matplotlib.pyplot as plt\nfrom mpl_toolkits.mplot3d import Axes3D\n%matplotlib inline\n\n# Load image pair and convert to RGB\nleft = cv2.imread('left.jpg')\nright = cv2.imread('right.jpg')\nleft = cv2.cvtColor(left,cv2.COLOR_BGR2RGB)\nright = cv2.cvtColor(right,cv2.COLOR_BGR2RGB)\n\n# Load 2D points (2xN matrices)\npts1 = np.load('pts1.npy')\npts2 = np.load('pts2.npy')\n\n# Load 3D points (3xN matrix)\npts3D = np.load('pts3D.npy')\n\n# Display 2D point correspondences\nplt.figure(figsize=(14,8))\nplt.subplot(121)\nplt.imshow(left)\nplt.plot(pts1[0,:],pts1[1,:], '.r')\nplt.title('Left image')\nplt.axis('off')\nplt.subplot(122)\nplt.imshow(right)\nplt.plot(pts2[0,:],pts2[1,:], '.r')\nplt.title('Right image')\nplt.axis('off')",
"_____no_output_____"
]
],
[
[
"The camera projection matrices $\\mathbf{C}$ and $\\mathbf{C}'$ are also provided for both views.",
"_____no_output_____"
]
],
[
[
"# Load 3x4 camera projection matrices\nC1 = np.load('C1.npy')\nC2 = np.load('C2.npy')",
"_____no_output_____"
]
],
[
[
"## 2. From 3D points to 2D points\n\nWith the camera matrix $\\mathbf{C} = \\mathbf{K}[\\mathbf{R} | \\mathbf{t}]$ we can project 3D points to the 2D points on the image plane:\n\n$$\ns \\begin{pmatrix}u \\\\ v \\\\ 1 \\end{pmatrix} = \\mathbf{C} \\begin{pmatrix} X \\\\ Y \\\\ Z \\\\ 1 \\end{pmatrix} \\qquad \\qquad (1)\n$$\n\nNote that we are using homogeneous coordinates.\n\n**2.1. Project 3D points** $\\quad$ <font color=red>(0.50 points)</font>\n\nComplete the function `projectPts()` by following the instructions below. The function should project 3D points `pts3D` to 2D image points. The result should look like the previous figure.\n",
"_____no_output_____"
]
],
[
[
"# INPUT \n# pts3D : 3D points (X,Y,Z) (3xN matrix)\n# C : Camera projection matrix (3x4 matrix)\n#\n# OUTPUT \n# pts2D : 2D points (x,y) (2xN matrix)\n#\ndef projectPts(pts3D,C):\n \n N = pts3D.shape[1] # Number of points\n \n # ---------- YOUR CODE STARTS HERE -----------\n \n # This line can be removed\n pts2D = np.zeros((2,N), dtype=np.float_)\n \n # 1. Convert 3D points from Euclidean to \n # homogeneous coordinates (4xN matrix)\n newarray = np.append(pts3D,np.ones((1,N), dtype=np.float_))\n hom3D = newarray.reshape(pts3D.shape[0]+1,N)\n \n # 2. Project points using Equation 1\n projectpoints = np.matmul(C,hom3D)\n for i in range(N):\n projectpoints[0,i] = np.divide(projectpoints[0,i],projectpoints[2,i])\n projectpoints[1,i] = np.divide(projectpoints[1,i],projectpoints[2,i])\n projectpoints[2,i] = 1\n \n # 3. Covert 2D points from homogeneous to \n # Euclidean coordinates (2xN matrix)\n hom2D = np.append(projectpoints[0,:],projectpoints[1,:])\n pts2D = hom2D.reshape(2,N)\n \n # ----------- YOUR CODE ENDS HERE ------------\n\n return pts2D\n\n\n# Project 3D points and visualize the result\npoints1 = projectPts(pts3D,C1)\npoints2 = projectPts(pts3D,C2)\n\nplt.figure(figsize=(14,8))\nplt.subplot(121)\nplt.imshow(left)\nplt.plot(points1[0,:],points1[1,:], '.r')\nplt.title('Left image')\nplt.axis('off')\nplt.subplot(122)\nplt.imshow(right)\nplt.plot(points2[0,:],points2[1,:], '.r')\nplt.title('Right image')\nplt.axis('off')",
"_____no_output_____"
]
],
[
[
"## 3. From 2D points to 3D points\n\nThe lecture notes describe a linear method to triangulate a point observed in two cameras (slide 196).\nGiven a point $\\mathbf{X} = (X,Y,Z)^{\\top}$ the projection equations are:\n\n$$\n\\begin{pmatrix}\ns u \\\\ \ns v \\\\\ns \\end{pmatrix} = \n\\begin{pmatrix}\nc_{11} & c_{12} & c_{13} & c_{14} \\\\ \nc_{21} & c_{22} & c_{23} & c_{24} \\\\\nc_{31} & c_{32} & c_{33} & c_{34} \\end{pmatrix}\n\\begin{pmatrix}\nX \\\\ \nY \\\\\nZ \\\\\n1 \\end{pmatrix}\n$$\n\n$$\n\\begin{pmatrix}\nt u' \\\\ \nt v' \\\\\nt \\end{pmatrix} = \n\\begin{pmatrix}\nc_{11}' & c_{12}' & c_{13}' & c_{14}' \\\\ \nc_{21}' & c_{22}' & c_{23}' & c_{24}' \\\\\nc_{31}' & c_{32}' & c_{33}' & c_{34}' \\end{pmatrix}\n\\begin{pmatrix}\nX \\\\ \nY \\\\\nZ \\\\\n1 \\end{pmatrix}\n$$\n\nHere we have used the same notation as in the exercises. Eliminating $s$ and $t$ we obtain the system of equations:\n\n$$\n(c_{31} u - c_{11}) X + (c_{32} u - c_{12}) Y + (c_{33} u - c_{13}) Z = c_{14} - c_{34} u \\\\\n(c_{31} v - c_{21}) X + (c_{32} v - c_{22}) Y + (c_{33} v - c_{23}) Z = c_{24} - c_{34} v \\\\\n(c_{31}' u' - c_{11}') X + (c_{32}' u' - c_{12}') Y + (c_{33}' u' - c_{13}') Z = c_{14}' - c_{34}' u' \\\\\n(c_{31}' v' - c_{21}') X + (c_{32}' v' - c_{22}') Y + (c_{33}' v' - c_{23}') Z = c_{24}' - c_{34}' v'\n$$\n\nwhich can be expressed in a linear system of the form: $\\mathbf{Ax} = \\mathbf{b}$ and solved using the least squares method. *See the Exercise 8 (Q2)*. <br>Notice also that equations are different from the ones presented in the lecture slides.",
"_____no_output_____"
],
[
"**3.1. Triangulate** $\\quad$ <font color=red>(1.50 points)</font>\n\nComplete the function `triangulatePts()`. Estimate 3D points given 2D points `pts1` and `pts2` and projection matrices `C1` and `C2`. For each point, form a linear system $\\mathbf{Ax} = \\mathbf{b}$, where $\\mathbf{A}$ is a $4 \\times 3$ matrix and $\\mathbf{b}$ is a $4 \\times 1$ vector. The least-squares solution can be obtained by $\\hat{\\mathbf{x}} = (\\mathbf{A}^{\\top} \\mathbf{A})^{-1} \\mathbf{A}^{\\top} \\mathbf{b}$, where $\\hat{\\mathbf{x}}$ contains the coordinates of the 3D point $(X,Y,Z)$. Once you have completed the function, execute the following code cell. The implementation is correct if the estimated 3D points `points3D` overlap with given 3D points `pts3D`.",
"_____no_output_____"
]
],
[
[
"# INPUT \n# pts1 : 2D points from the first image (2xN matrix)\n# pts2 : 2D points from the second image (2xN matrix)\n# C1 : Camera matrix for the first image (3x4 matrix)\n# C2 : Camera matrix for the second image (3x4 matrix)\n#\n# OUTPUT \n# pts3D : Triangulated 3D points (X,Y,Z) (3xN matrix)\n#\nfrom numpy.linalg import inv\ndef triangulatePts(pts1,pts2,C1,C2):\n \n N = pts1.shape[1] # Number of points\n pts3D = np.zeros((3,N),dtype=np.float_)\n \n # ---------- YOUR CODE STARTS HERE -----------\n \n # 1. For each point i, form A and b\n \n A = np.zeros((4,3),dtype=np.float_)\n \n b = np.zeros((4,1),dtype=np.float_)\n \n \n for i in range(N):\n \n A[0,0]=C1[2][0]*pts1[0,i]-C1[0][0]\n A[0,1]=C1[2][1]*pts1[0,i]-C1[0][1]\n A[0,2]=C1[2][2]*pts1[0,i]-C1[0][2]\n \n \n A[1,0]=C1[2][0] * pts1[1,i]-C1[1][0]\n A[1,1]=C1[2][1] * pts1[1,i]-C1[1][1]\n A[1,2]=C1[2][2] * pts1[1,i]-C1[1][2]\n \n A[2,0]=C2[2][0]*pts2[0,i]-C2[0][0]\n A[2,1]=C2[2][1]*pts2[0,i]-C2[0][1]\n A[2,2]=C2[2][2]*pts2[0,i]-C2[0][2]\n \n A[3,0]=C2[2][0] * pts2[1,i]-C2[1][0]\n A[3,1]=C2[2][1] * pts2[1,i]-C2[1][1]\n A[3,2]=C2[2][2] * pts2[1,i]-C2[1][2]\n \n b[0,0]=C1[0][3]-C1[2][3]*pts1[0,i]\n b[1,0]=C1[1][3]-C1[2][3]*pts1[1,i]\n b[2,0]=C2[0][3]-C2[2][3]*pts2[0,i]\n b[3,0]=C2[1][3]-C2[2][3]*pts2[1,i]\n \n # 2. Find solution to linear system Ax=b\n\n points = np.zeros((3,1),dtype=np.float_) \n points = np.matmul( np.matmul(inv(np.matmul(np.transpose(A) , A )), np.transpose(A)), b)\n \n # 3. Save triangulated point to pts3D[i,:]\n pts3D[0,i]=points[0]\n pts3D[1,i]=points[1]\n pts3D[2,i]=points[2] \n \n\n # ----------- YOUR CODE ENDS HERE ------------\n\n return pts3D\n\n\n# Triangulate points and compare to given 3D points\npoints3D = triangulatePts(pts1,pts2,C1,C2)\n\nfig = plt.figure(figsize=(12,12))\nax = fig.add_subplot(111, projection='3d')\nax.scatter(-pts3D[1,:],pts3D[2,:],-pts3D[0,:], \n color='blue', label='Given 3D points')\nax.scatter(-points3D[1,:],points3D[2,:],-points3D[0,:], \n color='red', label='Estimated 3D points')\nax.set_xlabel('Y')\nax.set_ylabel('Z')\nax.set_zlabel('X')\nax.set_title('Comparison of 3D points')\nax.legend()\n",
"_____no_output_____"
]
],
[
[
"# Aftermath\nFinally, fill your answers to the following questions:",
"_____no_output_____"
],
[
"**How much time did you need to complete this exercise?**\n\n`2 hours.`",
"_____no_output_____"
],
[
"**Did you experience any problems with the exercise? Was there enough help available? Should this notebook be more (or less) detailed?**\n\n`None.`",
"_____no_output_____"
],
[
"# References\n`None!`",
"_____no_output_____"
],
[
"# Submission\n\n1. Click on the menu `Kernel -> Restart & Clear Output` to clear all outputs and variables, etc.\n2. Compress the resulting Jupyter notebook (`MV_A8.ipynb` file) into **`MV_A8_[student number(s)].zip`** (e.g. `MV_A8_1234567.zip` if solo work or `MV_A8_1234567-7654321.zip` if pair work).\n3. Send an email to [email protected] with the subject line `MV_A8_[student number(s)]`. Remember to attach the .zip file.\n\n**Deadline of the assignment 10.3.2019**",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
c5103f64050ad21c1ae819810fbf7d038774b29d
| 129 |
ipynb
|
Jupyter Notebook
|
01-Lesson-Plans/05-APIs/3/Activities/01-Ins_Getting_into_Probability_Distributions/Unsolved/stock_price_normal_distribution.ipynb
|
tatianegercina/FinTech
|
b40687aa362d78674e223eb15ecf14bc59f90b62
|
[
"ADSL"
] | 1 |
2021-04-13T07:14:34.000Z
|
2021-04-13T07:14:34.000Z
|
01-Lesson-Plans/05-APIs/3/Activities/01-Ins_Getting_into_Probability_Distributions/Unsolved/stock_price_normal_distribution.ipynb
|
tatianegercina/FinTech
|
b40687aa362d78674e223eb15ecf14bc59f90b62
|
[
"ADSL"
] | 2 |
2021-06-02T03:14:19.000Z
|
2022-02-11T23:21:24.000Z
|
01-Lesson-Plans/05-APIs/3/Activities/01-Ins_Getting_into_Probability_Distributions/Unsolved/stock_price_normal_distribution.ipynb
|
tatianegercina/FinTech
|
b40687aa362d78674e223eb15ecf14bc59f90b62
|
[
"ADSL"
] | 1 |
2021-05-07T13:26:50.000Z
|
2021-05-07T13:26:50.000Z
| 32.25 | 75 | 0.883721 |
[
[
[
"empty"
]
]
] |
[
"empty"
] |
[
[
"empty"
]
] |
c51059268816b1800377a94645953e24a5eba4c2
| 19,448 |
ipynb
|
Jupyter Notebook
|
notebooks/Ch07_Analyzing_Movie_Reviews_Sentiment/Sentiment Analysis - Unsupervised Lexical.ipynb
|
guptasanjeev/practical-machine-learning-with-python
|
bc42a88552128bc19c6852e1f4be1d718710a291
|
[
"Apache-2.0"
] | 1,989 |
2017-11-29T02:34:45.000Z
|
2022-03-30T18:18:51.000Z
|
notebooks/Ch07_Analyzing_Movie_Reviews_Sentiment/Sentiment Analysis - Unsupervised Lexical.ipynb
|
guptasanjeev/practical-machine-learning-with-python
|
bc42a88552128bc19c6852e1f4be1d718710a291
|
[
"Apache-2.0"
] | 29 |
2018-01-21T04:17:23.000Z
|
2022-03-17T10:59:50.000Z
|
notebooks/Ch07_Analyzing_Movie_Reviews_Sentiment/Sentiment Analysis - Unsupervised Lexical.ipynb
|
guptasanjeev/practical-machine-learning-with-python
|
bc42a88552128bc19c6852e1f4be1d718710a291
|
[
"Apache-2.0"
] | 1,617 |
2017-12-22T16:13:58.000Z
|
2022-03-31T15:58:08.000Z
| 33.415808 | 215 | 0.488585 |
[
[
[
"# Import necessary depencencies",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nimport text_normalizer as tn\nimport model_evaluation_utils as meu\n\nnp.set_printoptions(precision=2, linewidth=80)",
"_____no_output_____"
]
],
[
[
"# Load and normalize data",
"_____no_output_____"
]
],
[
[
"dataset = pd.read_csv(r'movie_reviews.csv')\n\nreviews = np.array(dataset['review'])\nsentiments = np.array(dataset['sentiment'])\n\n# extract data for model evaluation\ntest_reviews = reviews[35000:]\ntest_sentiments = sentiments[35000:]\nsample_review_ids = [7626, 3533, 13010]\n\n# normalize dataset\nnorm_test_reviews = tn.normalize_corpus(test_reviews)",
"_____no_output_____"
]
],
[
[
"# Sentiment Analysis with AFINN",
"_____no_output_____"
]
],
[
[
"from afinn import Afinn\n\nafn = Afinn(emoticons=True) ",
"_____no_output_____"
]
],
[
[
"## Predict sentiment for sample reviews",
"_____no_output_____"
]
],
[
[
"for review, sentiment in zip(test_reviews[sample_review_ids], test_sentiments[sample_review_ids]):\n print('REVIEW:', review)\n print('Actual Sentiment:', sentiment)\n print('Predicted Sentiment polarity:', afn.score(review))\n print('-'*60)",
"REVIEW: no comment - stupid movie, acting average or worse... screenplay - no sense at all... SKIP IT!\nActual Sentiment: negative\nPredicted Sentiment polarity: -7.0\n------------------------------------------------------------\nREVIEW: I don't care if some people voted this movie to be bad. If you want the Truth this is a Very Good Movie! It has every thing a movie should have. You really should Get this one.\nActual Sentiment: positive\nPredicted Sentiment polarity: 3.0\n------------------------------------------------------------\nREVIEW: Worst horror film ever but funniest film ever rolled in one you have got to see this film it is so cheap it is unbeliaveble but you have to see it really!!!! P.s watch the carrot\nActual Sentiment: positive\nPredicted Sentiment polarity: -3.0\n------------------------------------------------------------\n"
]
],
[
[
"## Predict sentiment for test dataset",
"_____no_output_____"
]
],
[
[
"sentiment_polarity = [afn.score(review) for review in test_reviews]\npredicted_sentiments = ['positive' if score >= 1.0 else 'negative' for score in sentiment_polarity]",
"_____no_output_____"
]
],
[
[
"## Evaluate model performance",
"_____no_output_____"
]
],
[
[
"meu.display_model_performance_metrics(true_labels=test_sentiments, predicted_labels=predicted_sentiments, \n classes=['positive', 'negative'])",
"Model Performance metrics:\n------------------------------\nAccuracy: 0.71\nPrecision: 0.73\nRecall: 0.71\nF1 Score: 0.71\n\nModel Classification report:\n------------------------------\n precision recall f1-score support\n\n positive 0.67 0.85 0.75 7510\n negative 0.79 0.57 0.67 7490\n\navg / total 0.73 0.71 0.71 15000\n\n\nPrediction Confusion Matrix:\n------------------------------\n Predicted: \n positive negative\nActual: positive 6376 1134\n negative 3189 4301\n"
]
],
[
[
"# Sentiment Analysis with SentiWordNet",
"_____no_output_____"
]
],
[
[
"from nltk.corpus import sentiwordnet as swn\n\nawesome = list(swn.senti_synsets('awesome', 'a'))[0]\nprint('Positive Polarity Score:', awesome.pos_score())\nprint('Negative Polarity Score:', awesome.neg_score())\nprint('Objective Score:', awesome.obj_score())",
"Positive Polarity Score: 0.875\nNegative Polarity Score: 0.125\nObjective Score: 0.0\n"
]
],
[
[
"## Build model",
"_____no_output_____"
]
],
[
[
"def analyze_sentiment_sentiwordnet_lexicon(review,\n verbose=False):\n\n # tokenize and POS tag text tokens\n tagged_text = [(token.text, token.tag_) for token in tn.nlp(review)]\n pos_score = neg_score = token_count = obj_score = 0\n # get wordnet synsets based on POS tags\n # get sentiment scores if synsets are found\n for word, tag in tagged_text:\n ss_set = None\n if 'NN' in tag and list(swn.senti_synsets(word, 'n')):\n ss_set = list(swn.senti_synsets(word, 'n'))[0]\n elif 'VB' in tag and list(swn.senti_synsets(word, 'v')):\n ss_set = list(swn.senti_synsets(word, 'v'))[0]\n elif 'JJ' in tag and list(swn.senti_synsets(word, 'a')):\n ss_set = list(swn.senti_synsets(word, 'a'))[0]\n elif 'RB' in tag and list(swn.senti_synsets(word, 'r')):\n ss_set = list(swn.senti_synsets(word, 'r'))[0]\n # if senti-synset is found \n if ss_set:\n # add scores for all found synsets\n pos_score += ss_set.pos_score()\n neg_score += ss_set.neg_score()\n obj_score += ss_set.obj_score()\n token_count += 1\n \n # aggregate final scores\n final_score = pos_score - neg_score\n norm_final_score = round(float(final_score) / token_count, 2)\n final_sentiment = 'positive' if norm_final_score >= 0 else 'negative'\n if verbose:\n norm_obj_score = round(float(obj_score) / token_count, 2)\n norm_pos_score = round(float(pos_score) / token_count, 2)\n norm_neg_score = round(float(neg_score) / token_count, 2)\n # to display results in a nice table\n sentiment_frame = pd.DataFrame([[final_sentiment, norm_obj_score, norm_pos_score, \n norm_neg_score, norm_final_score]],\n columns=pd.MultiIndex(levels=[['SENTIMENT STATS:'], \n ['Predicted Sentiment', 'Objectivity',\n 'Positive', 'Negative', 'Overall']], \n labels=[[0,0,0,0,0],[0,1,2,3,4]]))\n print(sentiment_frame)\n \n return final_sentiment",
"_____no_output_____"
]
],
[
[
"## Predict sentiment for sample reviews",
"_____no_output_____"
]
],
[
[
"for review, sentiment in zip(test_reviews[sample_review_ids], test_sentiments[sample_review_ids]):\n print('REVIEW:', review)\n print('Actual Sentiment:', sentiment)\n pred = analyze_sentiment_sentiwordnet_lexicon(review, verbose=True) \n print('-'*60)",
"REVIEW: no comment - stupid movie, acting average or worse... screenplay - no sense at all... SKIP IT!\nActual Sentiment: negative\n SENTIMENT STATS: \n Predicted Sentiment Objectivity Positive Negative Overall\n0 negative 0.76 0.09 0.15 -0.06\n------------------------------------------------------------\nREVIEW: I don't care if some people voted this movie to be bad. If you want the Truth this is a Very Good Movie! It has every thing a movie should have. You really should Get this one.\nActual Sentiment: positive\n SENTIMENT STATS: \n Predicted Sentiment Objectivity Positive Negative Overall\n0 positive 0.74 0.2 0.06 0.14\n------------------------------------------------------------\nREVIEW: Worst horror film ever but funniest film ever rolled in one you have got to see this film it is so cheap it is unbeliaveble but you have to see it really!!!! P.s watch the carrot\nActual Sentiment: positive\n SENTIMENT STATS: \n Predicted Sentiment Objectivity Positive Negative Overall\n0 positive 0.8 0.14 0.07 0.07\n------------------------------------------------------------\n"
]
],
[
[
"## Predict sentiment for test dataset",
"_____no_output_____"
]
],
[
[
"predicted_sentiments = [analyze_sentiment_sentiwordnet_lexicon(review, verbose=False) for review in norm_test_reviews]",
"_____no_output_____"
]
],
[
[
"## Evaluate model performance",
"_____no_output_____"
]
],
[
[
"meu.display_model_performance_metrics(true_labels=test_sentiments, predicted_labels=predicted_sentiments, \n classes=['positive', 'negative'])",
"Model Performance metrics:\n------------------------------\nAccuracy: 0.69\nPrecision: 0.69\nRecall: 0.69\nF1 Score: 0.68\n\nModel Classification report:\n------------------------------\n precision recall f1-score support\n\n positive 0.66 0.76 0.71 7510\n negative 0.72 0.61 0.66 7490\n\navg / total 0.69 0.69 0.68 15000\n\n\nPrediction Confusion Matrix:\n------------------------------\n Predicted: \n positive negative\nActual: positive 5742 1768\n negative 2932 4558\n"
]
],
[
[
"# Sentiment Analysis with VADER",
"_____no_output_____"
]
],
[
[
"from nltk.sentiment.vader import SentimentIntensityAnalyzer",
"C:\\Program Files\\Anaconda3\\lib\\site-packages\\nltk\\twitter\\__init__.py:20: UserWarning: The twython library has not been installed. Some functionality from the twitter package will not be available.\n warnings.warn(\"The twython library has not been installed. \"\n"
]
],
[
[
"## Build model",
"_____no_output_____"
]
],
[
[
"def analyze_sentiment_vader_lexicon(review, \n threshold=0.1,\n verbose=False):\n # pre-process text\n review = tn.strip_html_tags(review)\n review = tn.remove_accented_chars(review)\n review = tn.expand_contractions(review)\n \n # analyze the sentiment for review\n analyzer = SentimentIntensityAnalyzer()\n scores = analyzer.polarity_scores(review)\n # get aggregate scores and final sentiment\n agg_score = scores['compound']\n final_sentiment = 'positive' if agg_score >= threshold\\\n else 'negative'\n if verbose:\n # display detailed sentiment statistics\n positive = str(round(scores['pos'], 2)*100)+'%'\n final = round(agg_score, 2)\n negative = str(round(scores['neg'], 2)*100)+'%'\n neutral = str(round(scores['neu'], 2)*100)+'%'\n sentiment_frame = pd.DataFrame([[final_sentiment, final, positive,\n negative, neutral]],\n columns=pd.MultiIndex(levels=[['SENTIMENT STATS:'], \n ['Predicted Sentiment', 'Polarity Score',\n 'Positive', 'Negative', 'Neutral']], \n labels=[[0,0,0,0,0],[0,1,2,3,4]]))\n print(sentiment_frame)\n \n return final_sentiment",
"_____no_output_____"
]
],
[
[
"## Predict sentiment for sample reviews",
"_____no_output_____"
]
],
[
[
"for review, sentiment in zip(test_reviews[sample_review_ids], test_sentiments[sample_review_ids]):\n print('REVIEW:', review)\n print('Actual Sentiment:', sentiment)\n pred = analyze_sentiment_vader_lexicon(review, threshold=0.4, verbose=True) \n print('-'*60)",
"REVIEW: no comment - stupid movie, acting average or worse... screenplay - no sense at all... SKIP IT!\nActual Sentiment: negative\n SENTIMENT STATS: \n Predicted Sentiment Polarity Score Positive Negative Neutral\n0 negative -0.8 0.0% 40.0% 60.0%\n------------------------------------------------------------\nREVIEW: I don't care if some people voted this movie to be bad. If you want the Truth this is a Very Good Movie! It has every thing a movie should have. You really should Get this one.\nActual Sentiment: positive\n SENTIMENT STATS: \n Predicted Sentiment Polarity Score Positive Negative Neutral\n0 negative -0.16 16.0% 14.000000000000002% 69.0%\n------------------------------------------------------------\nREVIEW: Worst horror film ever but funniest film ever rolled in one you have got to see this film it is so cheap it is unbeliaveble but you have to see it really!!!! P.s watch the carrot\nActual Sentiment: positive\n SENTIMENT STATS: \n Predicted Sentiment Polarity Score Positive Negative Neutral\n0 positive 0.49 11.0% 11.0% 77.0%\n------------------------------------------------------------\n"
]
],
[
[
"## Predict sentiment for test dataset",
"_____no_output_____"
]
],
[
[
"predicted_sentiments = [analyze_sentiment_vader_lexicon(review, threshold=0.4, verbose=False) for review in test_reviews]",
"_____no_output_____"
]
],
[
[
"## Evaluate model performance",
"_____no_output_____"
]
],
[
[
"meu.display_model_performance_metrics(true_labels=test_sentiments, predicted_labels=predicted_sentiments, \n classes=['positive', 'negative'])",
"Model Performance metrics:\n------------------------------\nAccuracy: 0.71\nPrecision: 0.72\nRecall: 0.71\nF1 Score: 0.71\n\nModel Classification report:\n------------------------------\n precision recall f1-score support\n\n positive 0.67 0.83 0.74 7510\n negative 0.78 0.59 0.67 7490\n\navg / total 0.72 0.71 0.71 15000\n\n\nPrediction Confusion Matrix:\n------------------------------\n Predicted: \n positive negative\nActual: positive 6235 1275\n negative 3068 4422\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
c5105e195b0a113c2c7bfa24e676154ed3d86327
| 76,965 |
ipynb
|
Jupyter Notebook
|
bonston_housing_project/Gradient Descent .ipynb
|
taareek/machine_learning
|
e9e7cf3636a3adf8572e69346c08e65cfcdb1100
|
[
"MIT"
] | null | null | null |
bonston_housing_project/Gradient Descent .ipynb
|
taareek/machine_learning
|
e9e7cf3636a3adf8572e69346c08e65cfcdb1100
|
[
"MIT"
] | null | null | null |
bonston_housing_project/Gradient Descent .ipynb
|
taareek/machine_learning
|
e9e7cf3636a3adf8572e69346c08e65cfcdb1100
|
[
"MIT"
] | null | null | null | 106.599723 | 20,368 | 0.823426 |
[
[
[
"import numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\n%matplotlib inline \nimport seaborn as sns\nsns.set_style(\"whitegrid\")",
"_____no_output_____"
],
[
"# Applying gradient descent algorithm \n\ntheta= 3\nalpha = 0.1\ndata = []\n\nfor i in range(0,10):\n res = alpha * 2 * theta # update rule \n print(\"{0:.4f} {1:.4f}\".format(theta, res))\n data.append([theta, theta**2])\n theta = theta - res",
"3.0000 0.6000\n2.4000 0.4800\n1.9200 0.3840\n1.5360 0.3072\n1.2288 0.2458\n0.9830 0.1966\n0.7864 0.1573\n0.6291 0.1258\n0.5033 0.1007\n0.4027 0.0805\n"
],
[
"tmp = pd.DataFrame(data)\ntmp",
"_____no_output_____"
],
[
"# plotting \nplt.figure(figsize=(8,6))\nplt.plot(np.linspace(-2, 4, 100), np.linspace(-2, 4, 100) **2);\nplt.scatter(tmp.iloc[:,0], tmp.iloc[:,1], marker= 'X');\nplt.xlabel('theta');\nplt.ylabel('J(theta)');",
"_____no_output_____"
]
],
[
[
"#### Second Example\n",
"_____no_output_____"
],
[
"$$J(\\theta) = \\theta^4 + \\theta^2$$\n$$\\frac{d}{d\\theta}.J(\\theta) = 4\\theta^3 + 2\\theta$$\n$$\\theta:= \\theta-\\alpha . (4\\theta^3 + 2\\theta)$$",
"_____no_output_____"
]
],
[
[
"theta = 3\nalpha = 0.01\ndata = []\n\nfor i in range(0,10):\n res = alpha * (4* theta **3 + 2*theta) # update function \n print(\"{0:.4f} {1:.4f}\".format(theta, res))\n data.append([theta, theta**4 + theta**2])\n theta = theta - res",
"3.0000 1.1400\n1.8600 0.2946\n1.5654 0.1847\n1.3807 0.1329\n1.2478 0.1027\n1.1451 0.0830\n1.0621 0.0692\n0.9930 0.0590\n0.9339 0.0513\n0.8827 0.0452\n"
],
[
"tmp = pd.DataFrame(data)\ntmp",
"_____no_output_____"
],
[
"# plotting \nplt.figure(figsize=(8,6))\nx_grid = np.linspace(-2, 4, 100)\nplt.plot(x_grid, x_grid**4 + x_grid**2);\nplt.scatter(tmp.iloc[:,0], tmp.iloc[:,1], marker='X');\nplt.xlabel('theta')\nplt.ylabel('J(theta)')",
"_____no_output_____"
]
],
[
[
"# Applying Gradient Descent Algorithm to Boston House Data",
"_____no_output_____"
]
],
[
[
"from sklearn.datasets import load_boston\n# loading boston house data\nboston_data = load_boston()\n# making a dataframae for boston house data\ndf = pd.DataFrame(boston_data.data, columns= boston_data.feature_names)\n# visualize dataset\ndf.head()",
"_____no_output_____"
],
[
"# taking one feature \nX = df[['LSTAT']].values\n# targets\ny = boston_data.target",
"_____no_output_____"
],
[
"from sklearn.preprocessing import StandardScaler\nsc_x = StandardScaler()\nsc_y = StandardScaler()\nX_std = sc_x.fit_transform(X)\ny_std = sc_y.fit_transform(y.reshape(-1, 1)).flatten()",
"_____no_output_____"
],
[
"print(\"For X: \")\nfor i in range(5):\n print(X[i])\nprint('=================================================')\nprint(\"For X_std: \")\nfor i in range(5):\n print(X_std[i])\nprint('=================================================')\nprint(\"For y: \")\nfor i in range(5):\n print(y[i])\nprint('=================================================')\nprint(\"For y_std: \")\nfor i in range(5):\n print(y_std[i])\nprint('=================================================')\nprint(\"Shape of X_std: \", X_std.shape)\nprint(\"Shape of y_std: \", y_std.shape)",
"For X: \n[4.98]\n[9.14]\n[4.03]\n[2.94]\n[5.33]\n=================================================\nFor X_std: \n[-1.0755623]\n[-0.49243937]\n[-1.2087274]\n[-1.36151682]\n[-1.02650148]\n=================================================\nFor y: \n24.0\n21.6\n34.7\n33.4\n36.2\n=================================================\nFor y_std: \n0.1596856587150597\n-0.1015242873296184\n1.3242466681642504\n1.1827579473900494\n1.4875028844421743\n=================================================\nShape of X_std: (506, 1)\nShape of y_std: (506,)\n"
],
[
"alpha = 0.0001\nw_ = np.zeros(1+ X_std.shape[1])\ncost_ = []\nn_ = 100\n\nfor i in range(n_):\n y_pred = np.dot(X_std, w_[1:]) + w_[0]\n errors = (y_std - y_pred)\n \n # updatig weights \n w_[1:] += alpha * X_std.T.dot(errors)\n w_[0] += alpha * errors.sum()\n \n cost = (errors**2).sum() / 2.0\n cost_.append(cost)\n ",
"_____no_output_____"
],
[
"# plot \nplt.figure(figsize=(10,8))\nplt.plot(range(1, n_ + 1), cost_);\nplt.ylabel('SSE')\nplt.xlabel('Epoch')",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
c510628cfdf1fd8286ab7bd197cab45989af2227
| 662,321 |
ipynb
|
Jupyter Notebook
|
Notebooks/RadarCOVID-Report/Daily/RadarCOVID-Report-2020-11-22.ipynb
|
pvieito/Radar-STATS
|
9ff991a4db776259bc749a823ee6f0b0c0d38108
|
[
"Apache-2.0"
] | 9 |
2020-10-14T16:58:32.000Z
|
2021-10-05T12:01:56.000Z
|
Notebooks/RadarCOVID-Report/Daily/RadarCOVID-Report-2020-11-22.ipynb
|
pvieito/Radar-STATS
|
9ff991a4db776259bc749a823ee6f0b0c0d38108
|
[
"Apache-2.0"
] | 3 |
2020-10-08T04:48:35.000Z
|
2020-10-10T20:46:58.000Z
|
Notebooks/RadarCOVID-Report/Daily/RadarCOVID-Report-2020-11-22.ipynb
|
Radar-STATS/Radar-STATS
|
61d8b3529f6bbf4576d799e340feec5b183338a3
|
[
"Apache-2.0"
] | 3 |
2020-09-27T07:39:26.000Z
|
2020-10-02T07:48:56.000Z
| 91.835968 | 191,964 | 0.759304 |
[
[
[
"# RadarCOVID-Report",
"_____no_output_____"
],
[
"## Data Extraction",
"_____no_output_____"
]
],
[
[
"import datetime\nimport json\nimport logging\nimport os\nimport shutil\nimport tempfile\nimport textwrap\nimport uuid\n\nimport matplotlib.pyplot as plt\nimport matplotlib.ticker\nimport numpy as np\nimport pandas as pd\nimport retry\nimport seaborn as sns\n\n%matplotlib inline",
"_____no_output_____"
],
[
"current_working_directory = os.environ.get(\"PWD\")\nif current_working_directory:\n os.chdir(current_working_directory)\n\nsns.set()\nmatplotlib.rcParams[\"figure.figsize\"] = (15, 6)\n\nextraction_datetime = datetime.datetime.utcnow()\nextraction_date = extraction_datetime.strftime(\"%Y-%m-%d\")\nextraction_previous_datetime = extraction_datetime - datetime.timedelta(days=1)\nextraction_previous_date = extraction_previous_datetime.strftime(\"%Y-%m-%d\")\nextraction_date_with_hour = datetime.datetime.utcnow().strftime(\"%Y-%m-%d@%H\")\ncurrent_hour = datetime.datetime.utcnow().hour\nare_today_results_partial = current_hour != 23",
"_____no_output_____"
]
],
[
[
"### Constants",
"_____no_output_____"
]
],
[
[
"from Modules.ExposureNotification import exposure_notification_io\n\nspain_region_country_code = \"ES\"\ngermany_region_country_code = \"DE\"\n\ndefault_backend_identifier = spain_region_country_code\n\nbackend_generation_days = 7 * 2\ndaily_summary_days = 7 * 4 * 3\ndaily_plot_days = 7 * 4\ntek_dumps_load_limit = daily_summary_days + 1",
"_____no_output_____"
]
],
[
[
"### Parameters",
"_____no_output_____"
]
],
[
[
"environment_backend_identifier = os.environ.get(\"RADARCOVID_REPORT__BACKEND_IDENTIFIER\")\nif environment_backend_identifier:\n report_backend_identifier = environment_backend_identifier\nelse:\n report_backend_identifier = default_backend_identifier\nreport_backend_identifier",
"_____no_output_____"
],
[
"environment_enable_multi_backend_download = \\\n os.environ.get(\"RADARCOVID_REPORT__ENABLE_MULTI_BACKEND_DOWNLOAD\")\nif environment_enable_multi_backend_download:\n report_backend_identifiers = None\nelse:\n report_backend_identifiers = [report_backend_identifier]\n\nreport_backend_identifiers",
"_____no_output_____"
],
[
"environment_invalid_shared_diagnoses_dates = \\\n os.environ.get(\"RADARCOVID_REPORT__INVALID_SHARED_DIAGNOSES_DATES\")\nif environment_invalid_shared_diagnoses_dates:\n invalid_shared_diagnoses_dates = environment_invalid_shared_diagnoses_dates.split(\",\")\nelse:\n invalid_shared_diagnoses_dates = []\n\ninvalid_shared_diagnoses_dates",
"_____no_output_____"
]
],
[
[
"### COVID-19 Cases",
"_____no_output_____"
]
],
[
[
"report_backend_client = \\\n exposure_notification_io.get_backend_client_with_identifier(\n backend_identifier=report_backend_identifier)",
"_____no_output_____"
],
[
"@retry.retry(tries=10, delay=10, backoff=1.1, jitter=(0, 10))\ndef download_cases_dataframe_from_ecdc():\n return pd.read_csv(\n \"https://opendata.ecdc.europa.eu/covid19/casedistribution/csv/data.csv\")\n\nconfirmed_df_ = download_cases_dataframe_from_ecdc()",
"_____no_output_____"
],
[
"confirmed_df = confirmed_df_.copy()\nconfirmed_df = confirmed_df[[\"dateRep\", \"cases\", \"geoId\"]]\nconfirmed_df.rename(\n columns={\n \"dateRep\":\"sample_date\",\n \"cases\": \"new_cases\",\n \"geoId\": \"country_code\",\n },\n inplace=True)\nconfirmed_df[\"sample_date\"] = pd.to_datetime(confirmed_df.sample_date, dayfirst=True)\nconfirmed_df[\"sample_date\"] = confirmed_df.sample_date.dt.strftime(\"%Y-%m-%d\")\nconfirmed_df.sort_values(\"sample_date\", inplace=True)\nconfirmed_df.tail()",
"_____no_output_____"
],
[
"def sort_source_regions_for_display(source_regions: list) -> list:\n if report_backend_identifier in source_regions:\n source_regions = [report_backend_identifier] + \\\n list(sorted(set(source_regions).difference([report_backend_identifier])))\n else:\n source_regions = list(sorted(source_regions))\n return source_regions",
"_____no_output_____"
],
[
"report_source_regions = report_backend_client.source_regions_for_date(\n date=extraction_datetime.date())\nreport_source_regions = sort_source_regions_for_display(\n source_regions=report_source_regions)\nreport_source_regions",
"_____no_output_____"
],
[
"confirmed_days = pd.date_range(\n start=confirmed_df.iloc[0].sample_date,\n end=extraction_datetime)\nconfirmed_days_df = pd.DataFrame(data=confirmed_days, columns=[\"sample_date\"])\nconfirmed_days_df[\"sample_date_string\"] = \\\n confirmed_days_df.sample_date.dt.strftime(\"%Y-%m-%d\")\nconfirmed_days_df.tail()",
"_____no_output_____"
],
[
"source_regions_at_date_df = confirmed_days_df.copy()\nsource_regions_at_date_df[\"source_regions_at_date\"] = \\\n source_regions_at_date_df.sample_date.apply(\n lambda x: report_backend_client.source_regions_for_date(date=x))\nsource_regions_at_date_df.sort_values(\"sample_date\", inplace=True)\nsource_regions_at_date_df[\"_source_regions_group\"] = source_regions_at_date_df. \\\n source_regions_at_date.apply(lambda x: \",\".join(sort_source_regions_for_display(x)))\nsource_regions_at_date_df.tail()",
"_____no_output_____"
],
[
"source_regions_for_summary_df = \\\n source_regions_at_date_df[[\"sample_date\", \"_source_regions_group\"]].copy()\nsource_regions_for_summary_df.rename(columns={\"_source_regions_group\": \"source_regions\"}, inplace=True)\nsource_regions_for_summary_df.tail()",
"_____no_output_____"
],
[
"confirmed_output_columns = [\"sample_date\", \"new_cases\", \"covid_cases\"]\nconfirmed_output_df = pd.DataFrame(columns=confirmed_output_columns)\n\nfor source_regions_group, source_regions_group_series in \\\n source_regions_at_date_df.groupby(\"_source_regions_group\"):\n source_regions_set = set(source_regions_group.split(\",\"))\n confirmed_source_regions_set_df = \\\n confirmed_df[confirmed_df.country_code.isin(source_regions_set)].copy()\n confirmed_source_regions_group_df = \\\n confirmed_source_regions_set_df.groupby(\"sample_date\").new_cases.sum() \\\n .reset_index().sort_values(\"sample_date\")\n confirmed_source_regions_group_df[\"covid_cases\"] = \\\n confirmed_source_regions_group_df.new_cases.rolling(7, min_periods=0).mean().round()\n confirmed_source_regions_group_df = \\\n confirmed_source_regions_group_df[confirmed_output_columns]\n confirmed_source_regions_group_df.fillna(method=\"ffill\", inplace=True)\n confirmed_source_regions_group_df = \\\n confirmed_source_regions_group_df[\n confirmed_source_regions_group_df.sample_date.isin(\n source_regions_group_series.sample_date_string)]\n confirmed_output_df = confirmed_output_df.append(confirmed_source_regions_group_df)\n\nconfirmed_df = confirmed_output_df.copy()\nconfirmed_df.tail()",
"_____no_output_____"
],
[
"confirmed_df.rename(columns={\"sample_date\": \"sample_date_string\"}, inplace=True)\nconfirmed_df = confirmed_days_df[[\"sample_date_string\"]].merge(confirmed_df, how=\"left\")\nconfirmed_df.sort_values(\"sample_date_string\", inplace=True)\nconfirmed_df.fillna(method=\"ffill\", inplace=True)\nconfirmed_df.tail()",
"_____no_output_____"
],
[
"confirmed_df[[\"new_cases\", \"covid_cases\"]].plot()",
"_____no_output_____"
]
],
[
[
"### Extract API TEKs",
"_____no_output_____"
]
],
[
[
"raw_zip_path_prefix = \"Data/TEKs/Raw/\"\nfail_on_error_backend_identifiers = [report_backend_identifier]\nmulti_backend_exposure_keys_df = \\\n exposure_notification_io.download_exposure_keys_from_backends(\n backend_identifiers=report_backend_identifiers,\n generation_days=backend_generation_days,\n fail_on_error_backend_identifiers=fail_on_error_backend_identifiers,\n save_raw_zip_path_prefix=raw_zip_path_prefix)\nmulti_backend_exposure_keys_df[\"region\"] = multi_backend_exposure_keys_df[\"backend_identifier\"]\nmulti_backend_exposure_keys_df.rename(\n columns={\n \"generation_datetime\": \"sample_datetime\",\n \"generation_date_string\": \"sample_date_string\",\n },\n inplace=True)\nmulti_backend_exposure_keys_df.head()",
"WARNING:root:NoKeysFoundException(\"No exposure keys found on endpoint 'https://radarcovidpre.covid19.gob.es/dp3t/v1/gaen/exposed/1606003200000' (parameters: {'generation_date': '2020-11-22', 'endpoint_identifier_components': ['2020-11-22'], 'backend_identifier': 'ES@PRE', 'server_endpoint_url': 'https://radarcovidpre.covid19.gob.es/dp3t'}).\")\n"
],
[
"early_teks_df = multi_backend_exposure_keys_df[\n multi_backend_exposure_keys_df.rolling_period < 144].copy()\nearly_teks_df[\"rolling_period_in_hours\"] = early_teks_df.rolling_period / 6\nearly_teks_df[early_teks_df.sample_date_string != extraction_date] \\\n .rolling_period_in_hours.hist(bins=list(range(24)))",
"_____no_output_____"
],
[
"early_teks_df[early_teks_df.sample_date_string == extraction_date] \\\n .rolling_period_in_hours.hist(bins=list(range(24)))",
"_____no_output_____"
],
[
"multi_backend_exposure_keys_df = multi_backend_exposure_keys_df[[\n \"sample_date_string\", \"region\", \"key_data\"]]\nmulti_backend_exposure_keys_df.head()",
"_____no_output_____"
],
[
"active_regions = \\\n multi_backend_exposure_keys_df.groupby(\"region\").key_data.nunique().sort_values().index.unique().tolist()\nactive_regions",
"_____no_output_____"
],
[
"multi_backend_summary_df = multi_backend_exposure_keys_df.groupby(\n [\"sample_date_string\", \"region\"]).key_data.nunique().reset_index() \\\n .pivot(index=\"sample_date_string\", columns=\"region\") \\\n .sort_index(ascending=False)\nmulti_backend_summary_df.rename(\n columns={\"key_data\": \"shared_teks_by_generation_date\"},\n inplace=True)\nmulti_backend_summary_df.rename_axis(\"sample_date\", inplace=True)\nmulti_backend_summary_df = multi_backend_summary_df.fillna(0).astype(int)\nmulti_backend_summary_df = multi_backend_summary_df.head(backend_generation_days)\nmulti_backend_summary_df.head()",
"_____no_output_____"
],
[
"def compute_keys_cross_sharing(x):\n teks_x = x.key_data_x.item()\n common_teks = set(teks_x).intersection(x.key_data_y.item())\n common_teks_fraction = len(common_teks) / len(teks_x)\n return pd.Series(dict(\n common_teks=common_teks,\n common_teks_fraction=common_teks_fraction,\n ))\n\nmulti_backend_exposure_keys_by_region_df = \\\n multi_backend_exposure_keys_df.groupby(\"region\").key_data.unique().reset_index()\nmulti_backend_exposure_keys_by_region_df[\"_merge\"] = True\nmulti_backend_exposure_keys_by_region_combination_df = \\\n multi_backend_exposure_keys_by_region_df.merge(\n multi_backend_exposure_keys_by_region_df, on=\"_merge\")\nmulti_backend_exposure_keys_by_region_combination_df.drop(\n columns=[\"_merge\"], inplace=True)\nif multi_backend_exposure_keys_by_region_combination_df.region_x.nunique() > 1:\n multi_backend_exposure_keys_by_region_combination_df = \\\n multi_backend_exposure_keys_by_region_combination_df[\n multi_backend_exposure_keys_by_region_combination_df.region_x !=\n multi_backend_exposure_keys_by_region_combination_df.region_y]\nmulti_backend_exposure_keys_cross_sharing_df = \\\n multi_backend_exposure_keys_by_region_combination_df \\\n .groupby([\"region_x\", \"region_y\"]) \\\n .apply(compute_keys_cross_sharing) \\\n .reset_index()\nmulti_backend_cross_sharing_summary_df = \\\n multi_backend_exposure_keys_cross_sharing_df.pivot_table(\n values=[\"common_teks_fraction\"],\n columns=\"region_x\",\n index=\"region_y\",\n aggfunc=lambda x: x.item())\nmulti_backend_cross_sharing_summary_df",
"<ipython-input-24-4e21708c19d8>:2: FutureWarning: `item` has been deprecated and will be removed in a future version\n teks_x = x.key_data_x.item()\n<ipython-input-24-4e21708c19d8>:3: FutureWarning: `item` has been deprecated and will be removed in a future version\n common_teks = set(teks_x).intersection(x.key_data_y.item())\n"
],
[
"multi_backend_without_active_region_exposure_keys_df = \\\n multi_backend_exposure_keys_df[multi_backend_exposure_keys_df.region != report_backend_identifier]\nmulti_backend_without_active_region = \\\n multi_backend_without_active_region_exposure_keys_df.groupby(\"region\").key_data.nunique().sort_values().index.unique().tolist()\nmulti_backend_without_active_region",
"_____no_output_____"
],
[
"exposure_keys_summary_df = multi_backend_exposure_keys_df[\n multi_backend_exposure_keys_df.region == report_backend_identifier]\nexposure_keys_summary_df.drop(columns=[\"region\"], inplace=True)\nexposure_keys_summary_df = \\\n exposure_keys_summary_df.groupby([\"sample_date_string\"]).key_data.nunique().to_frame()\nexposure_keys_summary_df = \\\n exposure_keys_summary_df.reset_index().set_index(\"sample_date_string\")\nexposure_keys_summary_df.sort_index(ascending=False, inplace=True)\nexposure_keys_summary_df.rename(columns={\"key_data\": \"shared_teks_by_generation_date\"}, inplace=True)\nexposure_keys_summary_df.head()",
"/opt/hostedtoolcache/Python/3.8.6/x64/lib/python3.8/site-packages/pandas/core/frame.py:4110: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\n return super().drop(\n"
]
],
[
[
"### Dump API TEKs",
"_____no_output_____"
]
],
[
[
"tek_list_df = multi_backend_exposure_keys_df[\n [\"sample_date_string\", \"region\", \"key_data\"]].copy()\ntek_list_df[\"key_data\"] = tek_list_df[\"key_data\"].apply(str)\ntek_list_df.rename(columns={\n \"sample_date_string\": \"sample_date\",\n \"key_data\": \"tek_list\"}, inplace=True)\ntek_list_df = tek_list_df.groupby(\n [\"sample_date\", \"region\"]).tek_list.unique().reset_index()\ntek_list_df[\"extraction_date\"] = extraction_date\ntek_list_df[\"extraction_date_with_hour\"] = extraction_date_with_hour\n\ntek_list_path_prefix = \"Data/TEKs/\"\ntek_list_current_path = tek_list_path_prefix + f\"/Current/RadarCOVID-TEKs.json\"\ntek_list_daily_path = tek_list_path_prefix + f\"Daily/RadarCOVID-TEKs-{extraction_date}.json\"\ntek_list_hourly_path = tek_list_path_prefix + f\"Hourly/RadarCOVID-TEKs-{extraction_date_with_hour}.json\"\n\nfor path in [tek_list_current_path, tek_list_daily_path, tek_list_hourly_path]:\n os.makedirs(os.path.dirname(path), exist_ok=True)\n\ntek_list_df.drop(columns=[\"extraction_date\", \"extraction_date_with_hour\"]).to_json(\n tek_list_current_path,\n lines=True, orient=\"records\")\ntek_list_df.drop(columns=[\"extraction_date_with_hour\"]).to_json(\n tek_list_daily_path,\n lines=True, orient=\"records\")\ntek_list_df.to_json(\n tek_list_hourly_path,\n lines=True, orient=\"records\")\ntek_list_df.head()",
"_____no_output_____"
]
],
[
[
"### Load TEK Dumps",
"_____no_output_____"
]
],
[
[
"import glob\n\ndef load_extracted_teks(mode, region=None, limit=None) -> pd.DataFrame:\n extracted_teks_df = pd.DataFrame(columns=[\"region\"])\n file_paths = list(reversed(sorted(glob.glob(tek_list_path_prefix + mode + \"/RadarCOVID-TEKs-*.json\"))))\n if limit:\n file_paths = file_paths[:limit]\n for file_path in file_paths:\n logging.info(f\"Loading TEKs from '{file_path}'...\")\n iteration_extracted_teks_df = pd.read_json(file_path, lines=True)\n extracted_teks_df = extracted_teks_df.append(\n iteration_extracted_teks_df, sort=False)\n extracted_teks_df[\"region\"] = \\\n extracted_teks_df.region.fillna(spain_region_country_code).copy()\n if region:\n extracted_teks_df = \\\n extracted_teks_df[extracted_teks_df.region == region]\n return extracted_teks_df",
"_____no_output_____"
],
[
"daily_extracted_teks_df = load_extracted_teks(\n mode=\"Daily\",\n region=report_backend_identifier,\n limit=tek_dumps_load_limit)\ndaily_extracted_teks_df.head()",
"_____no_output_____"
],
[
"exposure_keys_summary_df_ = daily_extracted_teks_df \\\n .sort_values(\"extraction_date\", ascending=False) \\\n .groupby(\"sample_date\").tek_list.first() \\\n .to_frame()\nexposure_keys_summary_df_.index.name = \"sample_date_string\"\nexposure_keys_summary_df_[\"tek_list\"] = \\\n exposure_keys_summary_df_.tek_list.apply(len)\nexposure_keys_summary_df_ = exposure_keys_summary_df_ \\\n .rename(columns={\"tek_list\": \"shared_teks_by_generation_date\"}) \\\n .sort_index(ascending=False)\nexposure_keys_summary_df = exposure_keys_summary_df_\nexposure_keys_summary_df.head()",
"_____no_output_____"
]
],
[
[
"### Daily New TEKs",
"_____no_output_____"
]
],
[
[
"tek_list_df = daily_extracted_teks_df.groupby(\"extraction_date\").tek_list.apply(\n lambda x: set(sum(x, []))).reset_index()\ntek_list_df = tek_list_df.set_index(\"extraction_date\").sort_index(ascending=True)\ntek_list_df.head()",
"_____no_output_____"
],
[
"def compute_teks_by_generation_and_upload_date(date):\n day_new_teks_set_df = tek_list_df.copy().diff()\n try:\n day_new_teks_set = day_new_teks_set_df[\n day_new_teks_set_df.index == date].tek_list.item()\n except ValueError:\n day_new_teks_set = None\n if pd.isna(day_new_teks_set):\n day_new_teks_set = set()\n day_new_teks_df = daily_extracted_teks_df[\n daily_extracted_teks_df.extraction_date == date].copy()\n day_new_teks_df[\"shared_teks\"] = \\\n day_new_teks_df.tek_list.apply(lambda x: set(x).intersection(day_new_teks_set))\n day_new_teks_df[\"shared_teks\"] = \\\n day_new_teks_df.shared_teks.apply(len)\n day_new_teks_df[\"upload_date\"] = date\n day_new_teks_df.rename(columns={\"sample_date\": \"generation_date\"}, inplace=True)\n day_new_teks_df = day_new_teks_df[\n [\"upload_date\", \"generation_date\", \"shared_teks\"]]\n day_new_teks_df[\"generation_to_upload_days\"] = \\\n (pd.to_datetime(day_new_teks_df.upload_date) -\n pd.to_datetime(day_new_teks_df.generation_date)).dt.days\n day_new_teks_df = day_new_teks_df[day_new_teks_df.shared_teks > 0]\n return day_new_teks_df\n\nshared_teks_generation_to_upload_df = pd.DataFrame()\nfor upload_date in daily_extracted_teks_df.extraction_date.unique():\n shared_teks_generation_to_upload_df = \\\n shared_teks_generation_to_upload_df.append(\n compute_teks_by_generation_and_upload_date(date=upload_date))\nshared_teks_generation_to_upload_df \\\n .sort_values([\"upload_date\", \"generation_date\"], ascending=False, inplace=True)\nshared_teks_generation_to_upload_df.tail()",
"<ipython-input-32-827222b35590>:4: FutureWarning: `item` has been deprecated and will be removed in a future version\n day_new_teks_set = day_new_teks_set_df[\n"
],
[
"today_new_teks_df = \\\n shared_teks_generation_to_upload_df[\n shared_teks_generation_to_upload_df.upload_date == extraction_date].copy()\ntoday_new_teks_df.tail()",
"_____no_output_____"
],
[
"if not today_new_teks_df.empty:\n today_new_teks_df.set_index(\"generation_to_upload_days\") \\\n .sort_index().shared_teks.plot.bar()",
"_____no_output_____"
],
[
"generation_to_upload_period_pivot_df = \\\n shared_teks_generation_to_upload_df[\n [\"upload_date\", \"generation_to_upload_days\", \"shared_teks\"]] \\\n .pivot(index=\"upload_date\", columns=\"generation_to_upload_days\") \\\n .sort_index(ascending=False).fillna(0).astype(int) \\\n .droplevel(level=0, axis=1)\ngeneration_to_upload_period_pivot_df.head()",
"_____no_output_____"
],
[
"new_tek_df = tek_list_df.diff().tek_list.apply(\n lambda x: len(x) if not pd.isna(x) else None).to_frame().reset_index()\nnew_tek_df.rename(columns={\n \"tek_list\": \"shared_teks_by_upload_date\",\n \"extraction_date\": \"sample_date_string\",}, inplace=True)\nnew_tek_df.tail()",
"_____no_output_____"
],
[
"shared_teks_uploaded_on_generation_date_df = shared_teks_generation_to_upload_df[\n shared_teks_generation_to_upload_df.generation_to_upload_days == 0] \\\n [[\"upload_date\", \"shared_teks\"]].rename(\n columns={\n \"upload_date\": \"sample_date_string\",\n \"shared_teks\": \"shared_teks_uploaded_on_generation_date\",\n })\nshared_teks_uploaded_on_generation_date_df.head()",
"_____no_output_____"
],
[
"estimated_shared_diagnoses_df = shared_teks_generation_to_upload_df \\\n .groupby([\"upload_date\"]).shared_teks.max().reset_index() \\\n .sort_values([\"upload_date\"], ascending=False) \\\n .rename(columns={\n \"upload_date\": \"sample_date_string\",\n \"shared_teks\": \"shared_diagnoses\",\n })\ninvalid_shared_diagnoses_dates_mask = \\\n estimated_shared_diagnoses_df.sample_date_string.isin(invalid_shared_diagnoses_dates)\nestimated_shared_diagnoses_df[invalid_shared_diagnoses_dates_mask] = 0\nestimated_shared_diagnoses_df.head()",
"_____no_output_____"
]
],
[
[
"### Hourly New TEKs",
"_____no_output_____"
]
],
[
[
"hourly_extracted_teks_df = load_extracted_teks(\n mode=\"Hourly\", region=report_backend_identifier, limit=25)\nhourly_extracted_teks_df.head()",
"_____no_output_____"
],
[
"hourly_new_tek_count_df = hourly_extracted_teks_df \\\n .groupby(\"extraction_date_with_hour\").tek_list. \\\n apply(lambda x: set(sum(x, []))).reset_index().copy()\nhourly_new_tek_count_df = hourly_new_tek_count_df.set_index(\"extraction_date_with_hour\") \\\n .sort_index(ascending=True)\n\nhourly_new_tek_count_df[\"new_tek_list\"] = hourly_new_tek_count_df.tek_list.diff()\nhourly_new_tek_count_df[\"new_tek_count\"] = hourly_new_tek_count_df.new_tek_list.apply(\n lambda x: len(x) if not pd.isna(x) else 0)\nhourly_new_tek_count_df.rename(columns={\n \"new_tek_count\": \"shared_teks_by_upload_date\"}, inplace=True)\nhourly_new_tek_count_df = hourly_new_tek_count_df.reset_index()[[\n \"extraction_date_with_hour\", \"shared_teks_by_upload_date\"]]\nhourly_new_tek_count_df.head()",
"_____no_output_____"
],
[
"hourly_summary_df = hourly_new_tek_count_df.copy()\nhourly_summary_df.set_index(\"extraction_date_with_hour\", inplace=True)\nhourly_summary_df = hourly_summary_df.fillna(0).astype(int).reset_index()\nhourly_summary_df[\"datetime_utc\"] = pd.to_datetime(\n hourly_summary_df.extraction_date_with_hour, format=\"%Y-%m-%d@%H\")\nhourly_summary_df.set_index(\"datetime_utc\", inplace=True)\nhourly_summary_df = hourly_summary_df.tail(-1)\nhourly_summary_df.head()",
"_____no_output_____"
]
],
[
[
"### Data Merge",
"_____no_output_____"
]
],
[
[
"result_summary_df = exposure_keys_summary_df.merge(\n new_tek_df, on=[\"sample_date_string\"], how=\"outer\")\nresult_summary_df.head()",
"_____no_output_____"
],
[
"result_summary_df = result_summary_df.merge(\n shared_teks_uploaded_on_generation_date_df, on=[\"sample_date_string\"], how=\"outer\")\nresult_summary_df.head()",
"_____no_output_____"
],
[
"result_summary_df = result_summary_df.merge(\n estimated_shared_diagnoses_df, on=[\"sample_date_string\"], how=\"outer\")\nresult_summary_df.head()",
"_____no_output_____"
],
[
"result_summary_df = confirmed_df.tail(daily_summary_days).merge(\n result_summary_df, on=[\"sample_date_string\"], how=\"left\")\nresult_summary_df.head()",
"_____no_output_____"
],
[
"result_summary_df[\"sample_date\"] = pd.to_datetime(result_summary_df.sample_date_string)\nresult_summary_df = result_summary_df.merge(source_regions_for_summary_df, how=\"left\")\nresult_summary_df.set_index([\"sample_date\", \"source_regions\"], inplace=True)\nresult_summary_df.drop(columns=[\"sample_date_string\"], inplace=True)\nresult_summary_df.sort_index(ascending=False, inplace=True)\nresult_summary_df.head()",
"_____no_output_____"
],
[
"with pd.option_context(\"mode.use_inf_as_na\", True):\n result_summary_df = result_summary_df.fillna(0).astype(int)\n result_summary_df[\"teks_per_shared_diagnosis\"] = \\\n (result_summary_df.shared_teks_by_upload_date / result_summary_df.shared_diagnoses).fillna(0)\n result_summary_df[\"shared_diagnoses_per_covid_case\"] = \\\n (result_summary_df.shared_diagnoses / result_summary_df.covid_cases).fillna(0)\n\nresult_summary_df.head(daily_plot_days)",
"_____no_output_____"
],
[
"weekly_result_summary_df = result_summary_df \\\n .sort_index(ascending=True).fillna(0).rolling(7).agg({\n \"covid_cases\": \"sum\",\n \"shared_teks_by_generation_date\": \"sum\",\n \"shared_teks_by_upload_date\": \"sum\",\n \"shared_diagnoses\": \"sum\"\n}).sort_index(ascending=False)\n\nwith pd.option_context(\"mode.use_inf_as_na\", True):\n weekly_result_summary_df = weekly_result_summary_df.fillna(0).astype(int)\n weekly_result_summary_df[\"teks_per_shared_diagnosis\"] = \\\n (weekly_result_summary_df.shared_teks_by_upload_date / weekly_result_summary_df.shared_diagnoses).fillna(0)\n weekly_result_summary_df[\"shared_diagnoses_per_covid_case\"] = \\\n (weekly_result_summary_df.shared_diagnoses / weekly_result_summary_df.covid_cases).fillna(0)\n\nweekly_result_summary_df.head()",
"_____no_output_____"
],
[
"last_7_days_summary = weekly_result_summary_df.to_dict(orient=\"records\")[1]\nlast_7_days_summary",
"_____no_output_____"
]
],
[
[
"## Report Results",
"_____no_output_____"
]
],
[
[
"display_column_name_mapping = {\n \"sample_date\": \"Sample\\u00A0Date\\u00A0(UTC)\",\n \"source_regions\": \"Source Countries\",\n \"datetime_utc\": \"Timestamp (UTC)\",\n \"upload_date\": \"Upload Date (UTC)\",\n \"generation_to_upload_days\": \"Generation to Upload Period in Days\",\n \"region\": \"Backend\",\n \"region_x\": \"Backend\\u00A0(A)\",\n \"region_y\": \"Backend\\u00A0(B)\",\n \"common_teks\": \"Common TEKs Shared Between Backends\",\n \"common_teks_fraction\": \"Fraction of TEKs in Backend (A) Available in Backend (B)\",\n \"covid_cases\": \"COVID-19 Cases in Source Countries (7-day Rolling Average)\",\n \"shared_teks_by_generation_date\": \"Shared TEKs by Generation Date\",\n \"shared_teks_by_upload_date\": \"Shared TEKs by Upload Date\",\n \"shared_diagnoses\": \"Shared Diagnoses (Estimation)\",\n \"teks_per_shared_diagnosis\": \"TEKs Uploaded per Shared Diagnosis\",\n \"shared_diagnoses_per_covid_case\": \"Usage Ratio (Fraction of Cases in Source Countries Which Shared Diagnosis)\",\n \"shared_teks_uploaded_on_generation_date\": \"Shared TEKs Uploaded on Generation Date\",\n}",
"_____no_output_____"
],
[
"summary_columns = [\n \"covid_cases\",\n \"shared_teks_by_generation_date\",\n \"shared_teks_by_upload_date\",\n \"shared_teks_uploaded_on_generation_date\",\n \"shared_diagnoses\",\n \"teks_per_shared_diagnosis\",\n \"shared_diagnoses_per_covid_case\",\n]",
"_____no_output_____"
]
],
[
[
"### Daily Summary Table",
"_____no_output_____"
]
],
[
[
"result_summary_df_ = result_summary_df.copy()\nresult_summary_df = result_summary_df[summary_columns]\nresult_summary_with_display_names_df = result_summary_df \\\n .rename_axis(index=display_column_name_mapping) \\\n .rename(columns=display_column_name_mapping)\nresult_summary_with_display_names_df",
"_____no_output_____"
]
],
[
[
"### Daily Summary Plots",
"_____no_output_____"
]
],
[
[
"result_plot_summary_df = result_summary_df.head(daily_plot_days)[summary_columns] \\\n .droplevel(level=[\"source_regions\"]) \\\n .rename_axis(index=display_column_name_mapping) \\\n .rename(columns=display_column_name_mapping)\nsummary_ax_list = result_plot_summary_df.sort_index(ascending=True).plot.bar(\n title=f\"Daily Summary\",\n rot=45, subplots=True, figsize=(15, 22), legend=False)\nax_ = summary_ax_list[-1]\nax_.get_figure().tight_layout()\nax_.get_figure().subplots_adjust(top=0.95)\nax_.yaxis.set_major_formatter(matplotlib.ticker.PercentFormatter(1.0))\n_ = ax_.set_xticklabels(sorted(result_plot_summary_df.index.strftime(\"%Y-%m-%d\").tolist()))",
"_____no_output_____"
]
],
[
[
"### Daily Generation to Upload Period Table",
"_____no_output_____"
]
],
[
[
"display_generation_to_upload_period_pivot_df = \\\n generation_to_upload_period_pivot_df \\\n .head(backend_generation_days)\ndisplay_generation_to_upload_period_pivot_df \\\n .head(backend_generation_days) \\\n .rename_axis(columns=display_column_name_mapping) \\\n .rename_axis(index=display_column_name_mapping)",
"_____no_output_____"
],
[
"fig, generation_to_upload_period_pivot_table_ax = plt.subplots(\n figsize=(12, 1 + 0.6 * len(display_generation_to_upload_period_pivot_df)))\ngeneration_to_upload_period_pivot_table_ax.set_title(\n \"Shared TEKs Generation to Upload Period Table\")\nsns.heatmap(\n data=display_generation_to_upload_period_pivot_df\n .rename_axis(columns=display_column_name_mapping)\n .rename_axis(index=display_column_name_mapping),\n fmt=\".0f\",\n annot=True,\n ax=generation_to_upload_period_pivot_table_ax)\ngeneration_to_upload_period_pivot_table_ax.get_figure().tight_layout()",
"_____no_output_____"
]
],
[
[
"### Hourly Summary Plots ",
"_____no_output_____"
]
],
[
[
"hourly_summary_ax_list = hourly_summary_df \\\n .rename_axis(index=display_column_name_mapping) \\\n .rename(columns=display_column_name_mapping) \\\n .plot.bar(\n title=f\"Last 24h Summary\",\n rot=45, subplots=True, legend=False)\nax_ = hourly_summary_ax_list[-1]\nax_.get_figure().tight_layout()\nax_.get_figure().subplots_adjust(top=0.9)\n_ = ax_.set_xticklabels(sorted(hourly_summary_df.index.strftime(\"%Y-%m-%d@%H\").tolist()))",
"_____no_output_____"
]
],
[
[
"### Publish Results",
"_____no_output_____"
]
],
[
[
"def get_temporary_image_path() -> str:\n return os.path.join(tempfile.gettempdir(), str(uuid.uuid4()) + \".png\")\n\ndef save_temporary_plot_image(ax):\n if isinstance(ax, np.ndarray):\n ax = ax[0]\n media_path = get_temporary_image_path()\n ax.get_figure().savefig(media_path)\n return media_path\n\ndef save_temporary_dataframe_image(df):\n import dataframe_image as dfi\n media_path = get_temporary_image_path()\n dfi.export(df, media_path)\n return media_path",
"_____no_output_____"
],
[
"github_repository = os.environ.get(\"GITHUB_REPOSITORY\")\nif github_repository is None:\n github_repository = \"pvieito/Radar-STATS\"\n\ngithub_project_base_url = \"https://github.com/\" + github_repository\n\ndisplay_formatters = {\n display_column_name_mapping[\"teks_per_shared_diagnosis\"]: lambda x: f\"{x:.2f}\",\n display_column_name_mapping[\"shared_diagnoses_per_covid_case\"]: lambda x: f\"{x:.2%}\",\n}\ndaily_summary_table_html = result_summary_with_display_names_df \\\n .head(daily_plot_days) \\\n .rename_axis(index=display_column_name_mapping) \\\n .rename(columns=display_column_name_mapping) \\\n .to_html(formatters=display_formatters)\nmulti_backend_summary_table_html = multi_backend_summary_df \\\n .head(daily_plot_days) \\\n .rename_axis(columns=display_column_name_mapping) \\\n .rename(columns=display_column_name_mapping) \\\n .rename_axis(index=display_column_name_mapping) \\\n .to_html(formatters=display_formatters)\n\ndef format_multi_backend_cross_sharing_fraction(x):\n if pd.isna(x):\n return \"-\"\n elif round(x * 100, 1) == 0:\n return \"\"\n else:\n return f\"{x:.1%}\"\n\nmulti_backend_cross_sharing_summary_table_html = multi_backend_cross_sharing_summary_df \\\n .rename_axis(columns=display_column_name_mapping) \\\n .rename(columns=display_column_name_mapping) \\\n .rename_axis(index=display_column_name_mapping) \\\n .to_html(\n classes=\"table-center\",\n formatters=display_formatters,\n float_format=format_multi_backend_cross_sharing_fraction)\nmulti_backend_cross_sharing_summary_table_html = \\\n multi_backend_cross_sharing_summary_table_html \\\n .replace(\"<tr>\",\"<tr style=\\\"text-align: center;\\\">\")\n\nextraction_date_result_summary_df = \\\n result_summary_df[result_summary_df.index.get_level_values(\"sample_date\") == extraction_date]\nextraction_date_result_hourly_summary_df = \\\n hourly_summary_df[hourly_summary_df.extraction_date_with_hour == extraction_date_with_hour]\n\ncovid_cases = \\\n extraction_date_result_summary_df.covid_cases.item()\nshared_teks_by_generation_date = \\\n extraction_date_result_summary_df.shared_teks_by_generation_date.item()\nshared_teks_by_upload_date = \\\n extraction_date_result_summary_df.shared_teks_by_upload_date.item()\nshared_diagnoses = \\\n extraction_date_result_summary_df.shared_diagnoses.item()\nteks_per_shared_diagnosis = \\\n extraction_date_result_summary_df.teks_per_shared_diagnosis.item()\nshared_diagnoses_per_covid_case = \\\n extraction_date_result_summary_df.shared_diagnoses_per_covid_case.item()\n\nshared_teks_by_upload_date_last_hour = \\\n extraction_date_result_hourly_summary_df.shared_teks_by_upload_date.sum().astype(int)\n\ndisplay_source_regions = \", \".join(report_source_regions)\nif len(report_source_regions) == 1:\n display_brief_source_regions = report_source_regions[0]\nelse:\n display_brief_source_regions = f\"{len(report_source_regions)} 🇪🇺\"",
"<ipython-input-58-bd16b454de40>:49: FutureWarning: `item` has been deprecated and will be removed in a future version\n extraction_date_result_summary_df.covid_cases.item()\n<ipython-input-58-bd16b454de40>:51: FutureWarning: `item` has been deprecated and will be removed in a future version\n extraction_date_result_summary_df.shared_teks_by_generation_date.item()\n<ipython-input-58-bd16b454de40>:53: FutureWarning: `item` has been deprecated and will be removed in a future version\n extraction_date_result_summary_df.shared_teks_by_upload_date.item()\n<ipython-input-58-bd16b454de40>:55: FutureWarning: `item` has been deprecated and will be removed in a future version\n extraction_date_result_summary_df.shared_diagnoses.item()\n<ipython-input-58-bd16b454de40>:57: FutureWarning: `item` has been deprecated and will be removed in a future version\n extraction_date_result_summary_df.teks_per_shared_diagnosis.item()\n<ipython-input-58-bd16b454de40>:59: FutureWarning: `item` has been deprecated and will be removed in a future version\n extraction_date_result_summary_df.shared_diagnoses_per_covid_case.item()\n"
],
[
"summary_plots_image_path = save_temporary_plot_image(\n ax=summary_ax_list)\nsummary_table_image_path = save_temporary_dataframe_image(\n df=result_summary_with_display_names_df)\nhourly_summary_plots_image_path = save_temporary_plot_image(\n ax=hourly_summary_ax_list)\nmulti_backend_summary_table_image_path = save_temporary_dataframe_image(\n df=multi_backend_summary_df)\ngeneration_to_upload_period_pivot_table_image_path = save_temporary_plot_image(\n ax=generation_to_upload_period_pivot_table_ax)",
"_____no_output_____"
]
],
[
[
"### Save Results",
"_____no_output_____"
]
],
[
[
"report_resources_path_prefix = \"Data/Resources/Current/RadarCOVID-Report-\"\nresult_summary_df.to_csv(\n report_resources_path_prefix + \"Summary-Table.csv\")\nresult_summary_df.to_html(\n report_resources_path_prefix + \"Summary-Table.html\")\nhourly_summary_df.to_csv(\n report_resources_path_prefix + \"Hourly-Summary-Table.csv\")\nmulti_backend_summary_df.to_csv(\n report_resources_path_prefix + \"Multi-Backend-Summary-Table.csv\")\nmulti_backend_cross_sharing_summary_df.to_csv(\n report_resources_path_prefix + \"Multi-Backend-Cross-Sharing-Summary-Table.csv\")\ngeneration_to_upload_period_pivot_df.to_csv(\n report_resources_path_prefix + \"Generation-Upload-Period-Table.csv\")\n_ = shutil.copyfile(\n summary_plots_image_path,\n report_resources_path_prefix + \"Summary-Plots.png\")\n_ = shutil.copyfile(\n summary_table_image_path,\n report_resources_path_prefix + \"Summary-Table.png\")\n_ = shutil.copyfile(\n hourly_summary_plots_image_path,\n report_resources_path_prefix + \"Hourly-Summary-Plots.png\")\n_ = shutil.copyfile(\n multi_backend_summary_table_image_path,\n report_resources_path_prefix + \"Multi-Backend-Summary-Table.png\")\n_ = shutil.copyfile(\n generation_to_upload_period_pivot_table_image_path,\n report_resources_path_prefix + \"Generation-Upload-Period-Table.png\")",
"_____no_output_____"
]
],
[
[
"### Publish Results as JSON",
"_____no_output_____"
]
],
[
[
"def generate_summary_api_results(df: pd.DataFrame) -> list:\n api_df = df.reset_index().copy()\n api_df[\"sample_date_string\"] = \\\n api_df[\"sample_date\"].dt.strftime(\"%Y-%m-%d\")\n api_df[\"source_regions\"] = \\\n api_df[\"source_regions\"].apply(lambda x: x.split(\",\"))\n return api_df.to_dict(orient=\"records\")\n\nsummary_api_results = \\\n generate_summary_api_results(df=result_summary_df)\ntoday_summary_api_results = \\\n generate_summary_api_results(df=extraction_date_result_summary_df)[0]\n\nsummary_results = dict(\n backend_identifier=report_backend_identifier,\n source_regions=report_source_regions,\n extraction_datetime=extraction_datetime,\n extraction_date=extraction_date,\n extraction_date_with_hour=extraction_date_with_hour,\n last_hour=dict(\n shared_teks_by_upload_date=shared_teks_by_upload_date_last_hour,\n shared_diagnoses=0,\n ),\n today=today_summary_api_results,\n last_7_days=last_7_days_summary,\n daily_results=summary_api_results)\n\nsummary_results = \\\n json.loads(pd.Series([summary_results]).to_json(orient=\"records\"))[0]\n\nwith open(report_resources_path_prefix + \"Summary-Results.json\", \"w\") as f:\n json.dump(summary_results, f, indent=4)",
"_____no_output_____"
]
],
[
[
"### Publish on README",
"_____no_output_____"
]
],
[
[
"with open(\"Data/Templates/README.md\", \"r\") as f:\n readme_contents = f.read()\n\nreadme_contents = readme_contents.format(\n extraction_date_with_hour=extraction_date_with_hour,\n github_project_base_url=github_project_base_url,\n daily_summary_table_html=daily_summary_table_html,\n multi_backend_summary_table_html=multi_backend_summary_table_html,\n multi_backend_cross_sharing_summary_table_html=multi_backend_cross_sharing_summary_table_html,\n display_source_regions=display_source_regions)\n\nwith open(\"README.md\", \"w\") as f:\n f.write(readme_contents)",
"_____no_output_____"
]
],
[
[
"### Publish on Twitter",
"_____no_output_____"
]
],
[
[
"enable_share_to_twitter = os.environ.get(\"RADARCOVID_REPORT__ENABLE_PUBLISH_ON_TWITTER\")\ngithub_event_name = os.environ.get(\"GITHUB_EVENT_NAME\")\n\nif enable_share_to_twitter and github_event_name == \"schedule\" and \\\n (shared_teks_by_upload_date_last_hour or not are_today_results_partial):\n import tweepy\n\n twitter_api_auth_keys = os.environ[\"RADARCOVID_REPORT__TWITTER_API_AUTH_KEYS\"]\n twitter_api_auth_keys = twitter_api_auth_keys.split(\":\")\n auth = tweepy.OAuthHandler(twitter_api_auth_keys[0], twitter_api_auth_keys[1])\n auth.set_access_token(twitter_api_auth_keys[2], twitter_api_auth_keys[3])\n\n api = tweepy.API(auth)\n\n summary_plots_media = api.media_upload(summary_plots_image_path)\n summary_table_media = api.media_upload(summary_table_image_path)\n generation_to_upload_period_pivot_table_image_media = api.media_upload(generation_to_upload_period_pivot_table_image_path)\n media_ids = [\n summary_plots_media.media_id,\n summary_table_media.media_id,\n generation_to_upload_period_pivot_table_image_media.media_id,\n ]\n\n if are_today_results_partial:\n today_addendum = \" (Partial)\"\n else:\n today_addendum = \"\"\n\n status = textwrap.dedent(f\"\"\"\n #RadarCOVID – {extraction_date_with_hour}\n\n Source Countries: {display_brief_source_regions}\n\n Today{today_addendum}:\n - Uploaded TEKs: {shared_teks_by_upload_date:.0f} ({shared_teks_by_upload_date_last_hour:+d} last hour)\n - Shared Diagnoses: ≤{shared_diagnoses:.0f}\n - Usage Ratio: ≤{shared_diagnoses_per_covid_case:.2%}\n\n Last 7 Days:\n - Shared Diagnoses: ≤{last_7_days_summary[\"shared_diagnoses\"]:.0f}\n - Usage Ratio: ≤{last_7_days_summary[\"shared_diagnoses_per_covid_case\"]:.2%}\n\n Info: {github_project_base_url}#documentation\n \"\"\")\n status = status.encode(encoding=\"utf-8\")\n api.update_status(status=status, media_ids=media_ids)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
c5106597a72d8024ab178138be18997ef3d5f4f5
| 4,936 |
ipynb
|
Jupyter Notebook
|
EnvironmentSensorModules/Air Quality vs Weather Correlation Module.ipynb
|
kevinlmao/dosenet-analysis
|
d19008b63d2bf5cffda6bdf49ddcc70f7b84be0e
|
[
"MIT"
] | 2 |
2018-06-20T00:21:31.000Z
|
2020-06-22T23:50:52.000Z
|
EnvironmentSensorModules/Air Quality vs Weather Correlation Module.ipynb
|
kevinlmao/dosenet-analysis
|
d19008b63d2bf5cffda6bdf49ddcc70f7b84be0e
|
[
"MIT"
] | 3 |
2016-08-02T17:59:12.000Z
|
2016-09-23T18:12:31.000Z
|
EnvironmentSensorModules/Air Quality vs Weather Correlation Module.ipynb
|
kevinlmao/dosenet-analysis
|
d19008b63d2bf5cffda6bdf49ddcc70f7b84be0e
|
[
"MIT"
] | 16 |
2016-05-24T16:23:03.000Z
|
2020-08-14T20:50:21.000Z
| 31.044025 | 168 | 0.478323 |
[
[
[
"# Plotting the Correlation between Air Quality and Weather",
"_____no_output_____"
]
],
[
[
"# If done right, this program should\n# Shoutout to my bois at StackOverflow - you da real MVPs\n\n# Shoutout to my bois over at StackOverflow - couldn't've done it without you\n\nimport pandas as pd\nimport numpy as np\nfrom bokeh.plotting import figure\nfrom bokeh.io import show\nfrom bokeh.models import HoverTool, Label\nimport scipy.stats\n\n\nweatherfile = input(\"Which weather file would you like to use? \")\ndf = pd.read_csv(weatherfile)\n\ntemp = df.as_matrix(columns=df.columns[3:4])\ntemp = temp.ravel()\n\nhumidity = df.as_matrix(columns=df.columns[4:5])\nhumidity = humidity.ravel()\n\npressure = df.as_matrix(columns=df.columns[5:])\npressure = pressure.ravel()\n\nunix_timeweather = df.as_matrix(columns=df.columns[2:3])\n\ni = 0\n\nw_used = eval(raw_input(\"Which data set do you want? temp, humidity, or pressure? \"))\n\n######################################################################################\naqfile = input(\"Which air quality file would you like to use? \")\ndf2 = pd.read_csv(aqfile)\n\nPM25 = df2.as_matrix(columns=df2.columns[4:5])\nPM1 = df2.as_matrix(columns=df2.columns[3:4])\nPM10 = df2.as_matrix(columns=df2.columns[5:])\n\nunix_timeaq = df2.as_matrix(columns=df2.columns[2:3])\n\naq_used = eval(raw_input(\"Which data set do you want? PM1, PM25, or PM10? \"))\n\n\n######################################################################################\n\ndef find_nearest(array, value):\n\n array = np.asarray(array)\n idx = (np.abs(array - value)).argmin()\n if np.abs(array[idx]-value) <= 30:\n # print str(value) + \"Vs\" + str(array[idx])\n return idx\n else:\n return None\n\n#######################################################################################\n\ndef make_usable(array1, array):\n i = len(array1) - 1\n while i > 0:\n if np.isnan(array[i]) or np.isnan(array1[i]):\n del array[i]\n del array1[i]\n i = i - 1\n\n#######################################################################################\n\nweatherarr = []\naqarr = []\n\ni = 0\n\nwhile i < len(aq_used):\n\n aqarr.append(float(aq_used[i]))\n\n nearest_time = find_nearest(unix_timeweather, unix_timeaq[i])\n\n if nearest_time is None:\n weatherarr.append(np.nan)\n else:\n weatherarr.append(float(w_used[nearest_time]))\n\n i = i+1\n\n\n# Plot the arrays #####################################################################\n\nmake_usable(weatherarr,aqarr)\n\n\n\nhoverp = HoverTool(tooltips=[(\"(x,y)\", \"($x, $y)\")])\n\n\np = figure(tools = [hoverp])\ncorrelation = Label(x=50, y=50, x_units='screen', y_units='screen', text=\"Pearson r and p: \"+ str(scipy.stats.pearsonr(weatherarr, aqarr)),render_mode='css',\n border_line_color='black', border_line_alpha=1.0,\n background_fill_color='white', background_fill_alpha=1.0)\n\np.add_layout(correlation)\np.circle(x = weatherarr, y = aqarr, color = \"firebrick\")\n\nshow(p)",
"Which weather file would you like to use? 'chs_os_weather.csv'\nWhich data set do you want? temp, humidity, or pressure? temp\nWhich air quality file would you like to use? 'chs_os_aq.csv'\nWhich data set do you want? PM1, PM25, or PM10? PM10\n"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
]
] |
c510693ee9cef75ca32df1e7c9add35df33a454c
| 27,391 |
ipynb
|
Jupyter Notebook
|
examples/optimisation-snes.ipynb
|
iamleeg/pints
|
bd1c11472ff3ec0990f3d55f0b2f20d92397926d
|
[
"BSD-3-Clause"
] | null | null | null |
examples/optimisation-snes.ipynb
|
iamleeg/pints
|
bd1c11472ff3ec0990f3d55f0b2f20d92397926d
|
[
"BSD-3-Clause"
] | null | null | null |
examples/optimisation-snes.ipynb
|
iamleeg/pints
|
bd1c11472ff3ec0990f3d55f0b2f20d92397926d
|
[
"BSD-3-Clause"
] | null | null | null | 182.606667 | 22,844 | 0.884853 |
[
[
[
"# Optimisation: SNES\n\nThis example shows you how to run a global optimisation with [SNES](http://pints.readthedocs.io/en/latest/optimisers/snes.html).\n\nFor a more elaborate example of an optimisation, see: [basic optimisation example](./optimisation-first-example.ipynb).",
"_____no_output_____"
]
],
[
[
"from __future__ import print_function\nimport pints\nimport pints.toy as toy\nimport numpy as np\nimport matplotlib.pyplot as pl\n\n# Load a forward model\nmodel = toy.LogisticModel()\n\n# Create some toy data\nreal_parameters = [0.015, 500]\ntimes = np.linspace(0, 1000, 1000)\nvalues = model.simulate(real_parameters, times)\n\n# Add noise\nvalues += np.random.normal(0, 10, values.shape)\n\n# Create an object with links to the model and time series\nproblem = pints.SingleOutputProblem(model, times, values)\n\n# Select a score function\nscore = pints.SumOfSquaresError(problem)\n\n# Select some boundaries\nboundaries = pints.RectangularBoundaries([0, 400], [0.03, 600])\n\n# Perform an optimization with boundaries and hints\nx0 = 0.01, 450\nsigma0 = [0.01, 100]\nfound_parameters, found_value = pints.optimise(\n score,\n x0,\n sigma0,\n boundaries,\n method=pints.SNES,\n)\n\n# Show score of true solution\nprint('Score at true solution: ')\nprint(score(real_parameters))\n\n# Compare parameters with original\nprint('Found solution: True parameters:' )\nfor k, x in enumerate(found_parameters):\n print(pints.strfloat(x) + ' ' + pints.strfloat(real_parameters[k]))\n\n# Show quality of fit\npl.figure()\npl.xlabel('Time')\npl.ylabel('Value')\npl.plot(times, values, label='Nosiy data')\npl.plot(times, problem.evaluate(found_parameters), label='Fit')\npl.legend()\npl.show()",
"Minimising error measure\nusing Seperable Natural Evolution Strategy (SNES)\nRunning in sequential mode.\nIter. Eval. Best Time m:s\n0 6 1032189 0:03.3\n1 12 1032189 0:03.3\n2 18 1032189 0:03.3\n3 24 415890.9 0:03.3\n20 126 99951.86 0:03.3\n40 246 97790.04 0:03.3\n60 366 97712.96 0:03.4\n80 486 97712.92 0:03.4\n100 606 97712.92 0:03.4\n120 726 97712.92 0:03.4\n140 846 97712.92 0:03.4\n160 966 97712.92 0:03.4\n180 1086 97712.92 0:03.5\n200 1206 97712.92 0:03.5\n220 1326 97712.92 0:03.5\n240 1446 97712.92 0:03.5\n260 1566 97712.92 0:03.5\n280 1686 97712.92 0:03.5\n300 1806 97712.92 0:03.6\n320 1926 97712.92 0:03.6\n340 2046 97712.92 0:03.6\n360 2166 97712.92 0:03.6\n380 2286 97712.92 0:03.6\n381 2286 97712.92 0:03.6\nHalting: No significant change for 200 iterations.\nScore at true solution: \n97762.3046752\nFound solution: True parameters:\n 1.49887429771769151e-02 1.49999999999999994e-02\n 5.00225855587396723e+02 5.00000000000000000e+02\n"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
]
] |
c5109b21e0f31262a4f95cb258dd752e0746b12d
| 10,717 |
ipynb
|
Jupyter Notebook
|
Preliminaries/Mathematics For ML - ICL/1. Linear Algebra/SpecialMatrices.ipynb
|
MarcosSalib/Cocktail_MOOC
|
46279c2ec642554537c639702ed8e540ea49afdf
|
[
"MIT"
] | null | null | null |
Preliminaries/Mathematics For ML - ICL/1. Linear Algebra/SpecialMatrices.ipynb
|
MarcosSalib/Cocktail_MOOC
|
46279c2ec642554537c639702ed8e540ea49afdf
|
[
"MIT"
] | null | null | null |
Preliminaries/Mathematics For ML - ICL/1. Linear Algebra/SpecialMatrices.ipynb
|
MarcosSalib/Cocktail_MOOC
|
46279c2ec642554537c639702ed8e540ea49afdf
|
[
"MIT"
] | null | null | null | 34.022222 | 285 | 0.488476 |
[
[
[
"# Identifying special matrices\n## Instructions\nIn this assignment, you shall write a function that will test if a 4×4 matrix is singular, i.e. to determine if an inverse exists, before calculating it.\n\nYou shall use the method of converting a matrix to echelon form, and testing if this fails by leaving zeros that can’t be removed on the leading diagonal.\n\nDon't worry if you've not coded before, a framework for the function has already been written.\nLook through the code, and you'll be instructed where to make changes.\nWe'll do the first two rows, and you can use this as a guide to do the last two.\n\n### Matrices in Python\nIn the *numpy* package in Python, matrices are indexed using zero for the top-most column and left-most row.\nI.e., the matrix structure looks like this:\n```python\nA[0, 0] A[0, 1] A[0, 2] A[0, 3]\nA[1, 0] A[1, 1] A[1, 2] A[1, 3]\nA[2, 0] A[2, 1] A[2, 2] A[2, 3]\nA[3, 0] A[3, 1] A[3, 2] A[3, 3]\n```\nYou can access the value of each element individually using,\n```python\nA[n, m]\n```\nwhich will give the n'th row and m'th column (starting with zero).\nYou can also access a whole row at a time using,\n```python\nA[n]\n```\nWhich you will see will be useful when calculating linear combinations of rows.\n\nA final note - Python is sensitive to indentation.\nAll the code you should complete will be at the same level of indentation as the instruction comment.\n\n### How to submit\nEdit the code in the cell below to complete the assignment.\nOnce you are finished and happy with it, press the *Submit Assignment* button at the top of this notebook.\n\nPlease don't change any of the function names, as these will be checked by the grading script.\n\nIf you have further questions about submissions or programming assignments, here is a [list](https://www.coursera.org/learn/linear-algebra-machine-learning/discussions/weeks/1/threads/jB4klkn5EeibtBIQyzFmQg) of Q&A. You can also raise an issue on the discussion forum. Good luck!",
"_____no_output_____"
]
],
[
[
"# GRADED FUNCTION\nimport numpy as np\n\n# Our function will go through the matrix replacing each row in order turning it into echelon form.\n# If at any point it fails because it can't put a 1 in the leading diagonal,\n# we will return the value True, otherwise, we will return False.\n# There is no need to edit this function.\ndef isSingular(A) :\n B = np.array(A, dtype=np.float_) # Make B as a copy of A, since we're going to alter it's values.\n try:\n fixRowZero(B)\n fixRowOne(B)\n fixRowTwo(B)\n fixRowThree(B)\n except MatrixIsSingular:\n return True\n return False\n\n# This next line defines our error flag. For when things go wrong if the matrix is singular.\n# There is no need to edit this line.\nclass MatrixIsSingular(Exception): pass\n\n# For Row Zero, all we require is the first element is equal to 1.\n# We'll divide the row by the value of A[0, 0].\n# This will get us in trouble though if A[0, 0] equals 0, so first we'll test for that,\n# and if this is true, we'll add one of the lower rows to the first one before the division.\n# We'll repeat the test going down each lower row until we can do the division.\n# There is no need to edit this function.\ndef fixRowZero(A) :\n if A[0,0] == 0 :\n A[0] = A[0] + A[1]\n if A[0,0] == 0 :\n A[0] = A[0] + A[2]\n if A[0,0] == 0 :\n A[0] = A[0] + A[3]\n if A[0,0] == 0 :\n raise MatrixIsSingular()\n # otherwise, \n A[0] = A[0] / A[0,0]\n return A\n\n# First we'll set the sub-diagonal elements to zero, i.e. A[1,0].\n# Next we want the diagonal element to be equal to one.\n# We'll divide the row by the value of A[1, 1].\n# Again, we need to test if this is zero.\n# If so, we'll add a lower row and repeat setting the sub-diagonal elements to zero.\n# There is no need to edit this function.\ndef fixRowOne(A) :\n A[1] = A[1] - A[1,0] * A[0]\n if A[1,1] == 0 :\n A[1] = A[1] + A[2]\n A[1] = A[1] - A[1,0] * A[0]\n if A[1,1] == 0 :\n A[1] = A[1] + A[3]\n A[1] = A[1] - A[1,0] * A[0]\n if A[1,1] == 0 :\n raise MatrixIsSingular()\n A[1] = A[1] / A[1,1]\n return A\n\n# This is the first function that you should complete.\n# Follow the instructions inside the function at each comment.\ndef fixRowTwo(A) :\n # Insert code below to set the sub-diagonal elements of row two to zero (there are two of them).\n A[2] = A[2] - A[2,0] * A[0]\n A[2] = A[2] - A[2,1] * A[1]\n # Next we'll test that the diagonal element is not zero.\n if A[2,2] == 0 :\n # Insert code below that adds a lower row to row 2.\n A[2] = A[2] + A[3]\n # Now repeat your code which sets the sub-diagonal elements to zero.\n A[2] = A[2] - A[2,0] * A[0]\n A[2] = A[2] - A[2,1] * A[1]\n \n if A[2,2] == 0 :\n raise MatrixIsSingular()\n # Finally set the diagonal element to one by dividing the whole row by that element.\n A[2] = A[2] / A[2,2]\n return A\n\n# You should also complete this function\n# Follow the instructions inside the function at each comment.\ndef fixRowThree(A) :\n # Insert code below to set the sub-diagonal elements of row three to zero.\n A[3] = A[3] - A[3,0] * A[0]\n A[3] = A[3] - A[3,1] * A[1]\n A[3] = A[3] - A[3,2] * A[2]\n \n # Complete the if statement to test if the diagonal element is zero.\n if A[3,3] == 0:\n raise MatrixIsSingular()\n # Transform the row to set the diagonal element to one.\n A[3] = A[3] / A[3,3]\n return A",
"_____no_output_____"
]
],
[
[
"## Test your code before submission\nTo test the code you've written above, run the cell (select the cell above, then press the play button [ ▶| ] or press shift-enter).\nYou can then use the code below to test out your function.\nYou don't need to submit this cell; you can edit and run it as much as you like.\n\nTry out your code on tricky test cases!",
"_____no_output_____"
]
],
[
[
"A = np.array([\n [2, 0, 0, 0],\n [0, 3, 0, 0],\n [0, 0, 4, 4],\n [0, 0, 5, 5]\n ], dtype=np.float_)\nisSingular(A)",
"_____no_output_____"
],
[
"A = np.array([\n [0, 7, -5, 3],\n [2, 8, 0, 4],\n [3, 12, 0, 5],\n [1, 3, 1, 3]\n ], dtype=np.float_)\nfixRowZero(A)",
"_____no_output_____"
],
[
"fixRowOne(A)",
"_____no_output_____"
],
[
"fixRowTwo(A)",
"_____no_output_____"
],
[
"fixRowThree(A)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
c5109f3a7de0bc3e5bf7b521900bb8c0c8a64e12
| 337,293 |
ipynb
|
Jupyter Notebook
|
exploration.ipynb
|
supremethapa/Data-Project-7-Communicate-Data-Findings
|
4512de3e7fdd26d71bb1414fc8acd6a7448f4f16
|
[
"MIT"
] | 3 |
2020-06-19T10:31:54.000Z
|
2021-07-15T21:16:55.000Z
|
exploration.ipynb
|
supremethapa/Data-Project-7-Communicate-Data-Findings
|
4512de3e7fdd26d71bb1414fc8acd6a7448f4f16
|
[
"MIT"
] | null | null | null |
exploration.ipynb
|
supremethapa/Data-Project-7-Communicate-Data-Findings
|
4512de3e7fdd26d71bb1414fc8acd6a7448f4f16
|
[
"MIT"
] | 3 |
2020-09-09T00:44:20.000Z
|
2021-12-31T09:52:16.000Z
| 147.418269 | 79,836 | 0.82707 |
[
[
[
"# Prosper Loan Data Exploration\n## By Abhishek Tiwari",
"_____no_output_____"
],
[
"# Preliminary Wrangling\nThis data set contains information on peer to peer loans facilitated by credit company Prosper",
"_____no_output_____"
]
],
[
[
"# import all packages\nimport pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport seaborn as sns\n\n%matplotlib inline",
"_____no_output_____"
],
[
"df = pd.read_csv('prosperLoanData.csv')\ndf.head()",
"_____no_output_____"
],
[
"df.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 113937 entries, 0 to 113936\nData columns (total 81 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 ListingKey 113937 non-null object \n 1 ListingNumber 113937 non-null int64 \n 2 ListingCreationDate 113937 non-null object \n 3 CreditGrade 28953 non-null object \n 4 Term 113937 non-null int64 \n 5 LoanStatus 113937 non-null object \n 6 ClosedDate 55089 non-null object \n 7 BorrowerAPR 113912 non-null float64\n 8 BorrowerRate 113937 non-null float64\n 9 LenderYield 113937 non-null float64\n 10 EstimatedEffectiveYield 84853 non-null float64\n 11 EstimatedLoss 84853 non-null float64\n 12 EstimatedReturn 84853 non-null float64\n 13 ProsperRating (numeric) 84853 non-null float64\n 14 ProsperRating (Alpha) 84853 non-null object \n 15 ProsperScore 84853 non-null float64\n 16 ListingCategory (numeric) 113937 non-null int64 \n 17 BorrowerState 108422 non-null object \n 18 Occupation 110349 non-null object \n 19 EmploymentStatus 111682 non-null object \n 20 EmploymentStatusDuration 106312 non-null float64\n 21 IsBorrowerHomeowner 113937 non-null bool \n 22 CurrentlyInGroup 113937 non-null bool \n 23 GroupKey 13341 non-null object \n 24 DateCreditPulled 113937 non-null object \n 25 CreditScoreRangeLower 113346 non-null float64\n 26 CreditScoreRangeUpper 113346 non-null float64\n 27 FirstRecordedCreditLine 113240 non-null object \n 28 CurrentCreditLines 106333 non-null float64\n 29 OpenCreditLines 106333 non-null float64\n 30 TotalCreditLinespast7years 113240 non-null float64\n 31 OpenRevolvingAccounts 113937 non-null int64 \n 32 OpenRevolvingMonthlyPayment 113937 non-null float64\n 33 InquiriesLast6Months 113240 non-null float64\n 34 TotalInquiries 112778 non-null float64\n 35 CurrentDelinquencies 113240 non-null float64\n 36 AmountDelinquent 106315 non-null float64\n 37 DelinquenciesLast7Years 112947 non-null float64\n 38 PublicRecordsLast10Years 113240 non-null float64\n 39 PublicRecordsLast12Months 106333 non-null float64\n 40 RevolvingCreditBalance 106333 non-null float64\n 41 BankcardUtilization 106333 non-null float64\n 42 AvailableBankcardCredit 106393 non-null float64\n 43 TotalTrades 106393 non-null float64\n 44 TradesNeverDelinquent (percentage) 106393 non-null float64\n 45 TradesOpenedLast6Months 106393 non-null float64\n 46 DebtToIncomeRatio 105383 non-null float64\n 47 IncomeRange 113937 non-null object \n 48 IncomeVerifiable 113937 non-null bool \n 49 StatedMonthlyIncome 113937 non-null float64\n 50 LoanKey 113937 non-null object \n 51 TotalProsperLoans 22085 non-null float64\n 52 TotalProsperPaymentsBilled 22085 non-null float64\n 53 OnTimeProsperPayments 22085 non-null float64\n 54 ProsperPaymentsLessThanOneMonthLate 22085 non-null float64\n 55 ProsperPaymentsOneMonthPlusLate 22085 non-null float64\n 56 ProsperPrincipalBorrowed 22085 non-null float64\n 57 ProsperPrincipalOutstanding 22085 non-null float64\n 58 ScorexChangeAtTimeOfListing 18928 non-null float64\n 59 LoanCurrentDaysDelinquent 113937 non-null int64 \n 60 LoanFirstDefaultedCycleNumber 16952 non-null float64\n 61 LoanMonthsSinceOrigination 113937 non-null int64 \n 62 LoanNumber 113937 non-null int64 \n 63 LoanOriginalAmount 113937 non-null int64 \n 64 LoanOriginationDate 113937 non-null object \n 65 LoanOriginationQuarter 113937 non-null object \n 66 MemberKey 113937 non-null object \n 67 MonthlyLoanPayment 113937 non-null float64\n 68 LP_CustomerPayments 113937 non-null float64\n 69 LP_CustomerPrincipalPayments 113937 non-null float64\n 70 LP_InterestandFees 113937 non-null float64\n 71 LP_ServiceFees 113937 non-null float64\n 72 LP_CollectionFees 113937 non-null float64\n 73 LP_GrossPrincipalLoss 113937 non-null float64\n 74 LP_NetPrincipalLoss 113937 non-null float64\n 75 LP_NonPrincipalRecoverypayments 113937 non-null float64\n 76 PercentFunded 113937 non-null float64\n 77 Recommendations 113937 non-null int64 \n 78 InvestmentFromFriendsCount 113937 non-null int64 \n 79 InvestmentFromFriendsAmount 113937 non-null float64\n 80 Investors 113937 non-null int64 \ndtypes: bool(3), float64(50), int64(11), object(17)\nmemory usage: 60.7+ MB\n"
],
[
"df.describe()",
"_____no_output_____"
],
[
"df.sample(10)",
"_____no_output_____"
]
],
[
[
"Note that this data set contains 81 columns. For the purpose of this analysis I’ve took the following columns (variables):",
"_____no_output_____"
]
],
[
[
"target_columns = [\n 'Term', 'LoanStatus', 'BorrowerRate', 'ProsperRating (Alpha)', 'ListingCategory (numeric)', 'EmploymentStatus',\n 'DelinquenciesLast7Years', 'StatedMonthlyIncome', 'TotalProsperLoans', 'LoanOriginalAmount',\n 'LoanOriginationDate', 'Recommendations', 'Investors'\n]",
"_____no_output_____"
],
[
"target_df = df[target_columns]",
"_____no_output_____"
],
[
"target_df.sample(10)",
"_____no_output_____"
]
],
[
[
"Since Prosper use their own proprietary Prosper Rating only since 2009, we have a lot of missing values in ProsperRating column. Let's drop these missing values:",
"_____no_output_____"
]
],
[
[
"target_df.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 113937 entries, 0 to 113936\nData columns (total 13 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 Term 113937 non-null int64 \n 1 LoanStatus 113937 non-null object \n 2 BorrowerRate 113937 non-null float64\n 3 ProsperRating (Alpha) 84853 non-null object \n 4 ListingCategory (numeric) 113937 non-null int64 \n 5 EmploymentStatus 111682 non-null object \n 6 DelinquenciesLast7Years 112947 non-null float64\n 7 StatedMonthlyIncome 113937 non-null float64\n 8 TotalProsperLoans 22085 non-null float64\n 9 LoanOriginalAmount 113937 non-null int64 \n 10 LoanOriginationDate 113937 non-null object \n 11 Recommendations 113937 non-null int64 \n 12 Investors 113937 non-null int64 \ndtypes: float64(4), int64(5), object(4)\nmemory usage: 9.6+ MB\n"
],
[
"target_df.describe()",
"_____no_output_____"
]
],
[
[
"Since Prosper use their own proprietary Prosper Rating only since 2009, we have a lot of missing values in ProsperRating column. Let's drop these missing values:",
"_____no_output_____"
]
],
[
[
"target_df = target_df.dropna(subset=['ProsperRating (Alpha)']).reset_index()",
"_____no_output_____"
]
],
[
[
"Convert LoanOriginationDate to datetime datatype:",
"_____no_output_____"
]
],
[
[
"target_df['LoanOriginationDate'] = pd.to_datetime(target_df['LoanOriginationDate'])",
"_____no_output_____"
],
[
"target_df['TotalProsperLoans'] = target_df['TotalProsperLoans'].fillna(0)",
"_____no_output_____"
],
[
"target_df.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 84853 entries, 0 to 84852\nData columns (total 14 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 index 84853 non-null int64 \n 1 Term 84853 non-null int64 \n 2 LoanStatus 84853 non-null object \n 3 BorrowerRate 84853 non-null float64 \n 4 ProsperRating (Alpha) 84853 non-null object \n 5 ListingCategory (numeric) 84853 non-null int64 \n 6 EmploymentStatus 84853 non-null object \n 7 DelinquenciesLast7Years 84853 non-null float64 \n 8 StatedMonthlyIncome 84853 non-null float64 \n 9 TotalProsperLoans 84853 non-null float64 \n 10 LoanOriginalAmount 84853 non-null int64 \n 11 LoanOriginationDate 84853 non-null datetime64[ns]\n 12 Recommendations 84853 non-null int64 \n 13 Investors 84853 non-null int64 \ndtypes: datetime64[ns](1), float64(4), int64(6), object(3)\nmemory usage: 8.1+ MB\n"
]
],
[
[
"### What is/are the main feature(s) of interest in your dataset?\n> Trying to figure out what features can be used to predict default on credit. Also i would like to check what are major factors connected with prosper credit rating.\n\n### What features in the dataset do you think will help support your investigation into your feature(s) of interest?\n> I think that the borrowers Prosper rating will have the highest impact on chances of default. Also I expect that loan amount will play a major role and maybe the category of credit. Prosper rating will depend on stated income and employment status.",
"_____no_output_____"
],
[
"## Univariate Exploration",
"_____no_output_____"
],
[
"### Loan status",
"_____no_output_____"
]
],
[
[
"# setting color\nbase_color = sns.color_palette()[0]\nplt.xticks(rotation=90)\nsns.countplot(data = target_df, x = 'LoanStatus', color = base_color);",
"_____no_output_____"
]
],
[
[
"Observation 1: \n* Most of the loans in the data set are actually current loans. \n* Past due loans are split in several groups based on the length of payment delay.\n* Other big part is completed loans, defaulted loans compromise a minority, however chargedoff loans also comporomise a substanial amount.\n",
"_____no_output_____"
],
[
"### Employment Status",
"_____no_output_____"
]
],
[
[
"sns.countplot(data = target_df, x = 'EmploymentStatus', color = base_color);\nplt.xticks(rotation = 90);",
"_____no_output_____"
]
],
[
[
"Observation 2: \n* The majority of borrowers are employed and all other categories as small part of borrowers. \n* In small Group full time has highest, after that self empolyed are there and so on.",
"_____no_output_____"
],
[
"### Stated Monthly Income",
"_____no_output_____"
]
],
[
[
"plt.hist(data=target_df, x='StatedMonthlyIncome', bins=1000);",
"_____no_output_____"
]
],
[
[
"(**Note**: Distribution of stated monthly income is highly skewed to the right. so, we have to check how many outliers are there)",
"_____no_output_____"
]
],
[
[
"income_std = target_df['StatedMonthlyIncome'].std()\nincome_mean = target_df['StatedMonthlyIncome'].mean()\nboundary = income_mean + income_std * 3\nlen(target_df[target_df['StatedMonthlyIncome'] >= boundary])",
"_____no_output_____"
]
],
[
[
"**After Zooming the Graph We Get This**",
"_____no_output_____"
]
],
[
[
"plt.hist(data=target_df, x='StatedMonthlyIncome', bins=1000);\nplt.xlim(0, boundary);",
"_____no_output_____"
]
],
[
[
"Observation 3:\n* With a boundary of mean and 3 times standard deviations distribution of monthly income still has noticeable right skew but now we can see that mode is about 5000. ",
"_____no_output_____"
],
[
"### Discuss the distribution(s) of your variable(s) of interest. Were there any unusual points? Did you need to perform any transformations?\n> Distribution of monthly stated income is very awkward: with a lot of outliers and very large range but still it was right skew. The majority of borrowers are employed and all other categories as small part of borrowers and most of the loans in the data set are actually current loans.\n\n### Of the features you investigated, were there any unusual distributions? Did you perform any operations on the data to tidy, adjust, or change the form of the data? If so, why did you do this?\n> The majority of loans are actually current loans. Since our main goal is to define driving factors of outcome of loan we are not interested in any current loans.\n",
"_____no_output_____"
],
[
"## Bivariate Exploration",
"_____no_output_____"
]
],
[
[
"#I'm just adjusting the form of data\ncondition = (target_df['LoanStatus'] == 'Completed') | (target_df['LoanStatus'] == 'Defaulted') |\\\n (target_df['LoanStatus'] == 'Chargedoff')\ntarget_df = target_df[condition]\n\ndef change_to_defaulted(row):\n if row['LoanStatus'] == 'Chargedoff':\n return 'Defaulted'\n else:\n return row['LoanStatus']\n \ntarget_df['LoanStatus'] = target_df.apply(change_to_defaulted, axis=1)\ntarget_df['LoanStatus'].value_counts()",
"_____no_output_____"
]
],
[
[
"**After transforming dataset we have 19664 completed loans and 6341 defaulted.**",
"_____no_output_____"
]
],
[
[
"categories = {1: 'Debt Consolidation', 2: 'Home Improvement', 3: 'Business', 6: 'Auto', 7: 'Other'}\ndef reduce_categorie(row):\n loan_category = row['ListingCategory (numeric)']\n if loan_category in categories:\n return categories[loan_category]\n else:\n return categories[7]\n \ntarget_df['ListingCategory (numeric)'] = target_df.apply(reduce_categorie, axis=1)\ntarget_df['ListingCategory (numeric)'].value_counts()",
"_____no_output_____"
]
],
[
[
"Variable Listing Category is set up as numeric and most of the values have very `low frequency`, for the easier visualization so we have change it to `categorical and reduce the number of categories`. ",
"_____no_output_____"
],
[
"### Status and Prosper Rating:",
"_____no_output_____"
]
],
[
[
"sns.countplot(data = target_df, x = 'LoanStatus', hue = 'ProsperRating (Alpha)', palette = 'Blues')",
"_____no_output_____"
]
],
[
[
"Observation 1:\n* The `most frequent` rating among defaulted loans is actually `D`.\n* And the `most frequent` rating among Completed is also` D `and second highest is A and so on.",
"_____no_output_____"
],
[
"### Credit Start with Listing Category:",
"_____no_output_____"
]
],
[
[
"sns.countplot(data = target_df, x = 'LoanStatus', hue = 'ListingCategory (numeric)', palette = 'Blues');",
"_____no_output_____"
]
],
[
[
"Observation 2:\n* In both of the Graphs the `debt Consolidation` have `most frequency among all of them`.",
"_____no_output_____"
],
[
"## Loan Status and Loan Amount",
"_____no_output_____"
]
],
[
[
"sns.boxplot(data = target_df, x = 'LoanStatus', y = 'LoanOriginalAmount', color = base_color);",
"_____no_output_____"
]
],
[
[
"Observation 3:\n* As from Above Graph we can state that `defaulted credits` tend to be `smaller` than `completed credits` onces. ",
"_____no_output_____"
],
[
"## Prosper Rating and Employment Status",
"_____no_output_____"
]
],
[
[
"plt.figure(figsize = [12, 10])\nsns.countplot(data = target_df, x = 'ProsperRating (Alpha)', hue = 'EmploymentStatus', palette = 'Blues');",
"_____no_output_____"
]
],
[
[
"Observation 4:\n* Lower ratings seem to have greater proportions of individuals with employment status Not Employed, Self-employed, Retired and Part-Time. ",
"_____no_output_____"
],
[
"## Talk about some of the relationships you observed in this part of the investigation. How did the feature(s) of interest vary with other features in the dataset?\n> In Loan status vs Loan amount defaulted credits tend to be smaller than completed credits onces. Employment status of individuals with lower ratings tends to be 'Not employed', 'Self-employed', 'Retired' or 'Part-time'.\n\n## Did you observe any interesting relationships between the other features (not the main feature(s) of interest)?\n> Prosper rating D is the most frequent rating among defaulted credits.",
"_____no_output_____"
],
[
"## Multivariate Exploration",
"_____no_output_____"
],
[
"## Rating, Loan Amount and Loan Status",
"_____no_output_____"
]
],
[
[
"plt.figure(figsize = [12, 8])\nsns.boxplot(data=target_df, x='ProsperRating (Alpha)', y='LoanOriginalAmount', hue='LoanStatus');",
"_____no_output_____"
]
],
[
[
"Observation 1:\n* Except for the lowest ratings defaulted credits tend to be larger than completed.\n* Most of the defaulted credits comes from individuals with low Prosper rating.",
"_____no_output_____"
],
[
"## Relationships between Credit category, Credit rating and outcome of Credit.",
"_____no_output_____"
]
],
[
[
"sns.catplot(x = 'ProsperRating (Alpha)', hue = 'LoanStatus', col = 'ListingCategory (numeric)',\n data = target_df, kind = 'count', palette = 'Blues', col_wrap = 3);",
"_____no_output_____"
]
],
[
[
"Observation 2:\n* There are 5 graphs in the second one has much up and downs in it other than all of them.\n* There is no substantial difference for default rates in different categories broken up by ratings.",
"_____no_output_____"
],
[
"## Amount, Listing Category Loan and Loan Status Interact",
"_____no_output_____"
]
],
[
[
"plt.figure(figsize = [12, 8])\nsns.violinplot(data=target_df, x='ListingCategory (numeric)', y='LoanOriginalAmount', hue='LoanStatus');",
"_____no_output_____"
]
],
[
[
"Observation 3:\n* Except for Auto, Business and Home Improvemrnt dont have nearly equal mean amoong all of them.\n* Business category tend to have larger amount.",
"_____no_output_____"
],
[
"## Talk about some of the relationships you observed in this part of the investigation. Were there features that strengthened each other in terms of looking at your feature(s) of interest?\n> Our initial assumptions were strengthened. Most of the defaulted credits comes from individuals with low Prosper rating and Business category tend to have larger amount.\n\n## Were there any interesting or surprising interactions between features?\n> Interesting find was that defaulted credits for individuals with high Prosper ratings tend to be larger than completed credits.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
c510b9c0def92637616456daf52160dcbb1517b7
| 16,524 |
ipynb
|
Jupyter Notebook
|
semlog_mongo/example/Add lines and texts.ipynb
|
robcog-iai/semlog_web
|
dde8478f8fc2a273c94ce8f55faf8b0b80a8213c
|
[
"BSD-3-Clause"
] | null | null | null |
semlog_mongo/example/Add lines and texts.ipynb
|
robcog-iai/semlog_web
|
dde8478f8fc2a273c94ce8f55faf8b0b80a8213c
|
[
"BSD-3-Clause"
] | null | null | null |
semlog_mongo/example/Add lines and texts.ipynb
|
robcog-iai/semlog_web
|
dde8478f8fc2a273c94ce8f55faf8b0b80a8213c
|
[
"BSD-3-Clause"
] | null | null | null | 35.459227 | 119 | 0.453522 |
[
[
[
"import pandas as pd\nimport cv2\nimport random\nimport os\nfrom matplotlib import pyplot as plt",
"_____no_output_____"
],
[
"color1=(255,0,0)\nroot_folder=\"T\"\nroot_name=\"t\"\ndf=pd.read_csv(\"new.csv\")",
"_____no_output_____"
],
[
"df=df[df.type==\"Color\"]\ndf.head(15)\n",
"_____no_output_____"
],
[
"grouped_df=df.groupby(['file_id','class'])",
"_____no_output_____"
],
[
"coordinate_names=['x_max','x_min','y_max','y_min']\nfor name, group in grouped_df:\n img_name,class_name=name\n bb_list=group[coordinate_names].values.astype(int)\n draw_label_on_image(\"T\",\"t\",img_name,class_name,(0,0,255),bb_list)\n\ndef draw_all_labels(df,root_folder_path,root_folder_name):\n grouped_df=df.groupby(['file_id','class'])\n coordinate_names=['x_max','x_min','y_max','y_min']\n for name, group in grouped_df:\n img_name,class_name=name\n bb_list=group[coordinate_names].values.astype(int)\n draw_label_on_image(root_folder_path,root_folder_name,img_name,class_name,(0,0,255),bb_list)",
"_____no_output_____"
],
[
"def draw_label_on_image(root_folder_path,root_folder_name,img_name,class_name,bb_color,bb_list):\n img_path=os.path.join(root_folder_path,root_folder_name,\"Color\",img_name+\".png\")\n img=cv2.imread(img_path)\n for each_bb in bb_list:\n cv2.rectangle(img,(each_bb[0],each_bb[2]),(each_bb[1],each_bb[3]),bb_color,3)\n cv2.putText(img,class_name,(each_bb[0],each_bb[2]),cv2.FONT_HERSHEY_SIMPLEX,1,(255,0,0),2,cv2.LINE_AA)\n cv2.imwrite(img_path,img)",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
c510bbbbbbfec75a94796e4e7ff95c64f46ee393
| 281,811 |
ipynb
|
Jupyter Notebook
|
Fig03a-d - length & frequency of sleep stages .ipynb
|
margoliashlab/budgie-sleep-paper-2020
|
be4ee5f992c3b2628fd63b8b234ca4701c137f24
|
[
"MIT"
] | null | null | null |
Fig03a-d - length & frequency of sleep stages .ipynb
|
margoliashlab/budgie-sleep-paper-2020
|
be4ee5f992c3b2628fd63b8b234ca4701c137f24
|
[
"MIT"
] | null | null | null |
Fig03a-d - length & frequency of sleep stages .ipynb
|
margoliashlab/budgie-sleep-paper-2020
|
be4ee5f992c3b2628fd63b8b234ca4701c137f24
|
[
"MIT"
] | null | null | null | 144.964506 | 64,604 | 0.875988 |
[
[
[
"# Setup",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\n\nimport numpy as np\nimport scipy.signal as sig\nimport scipy.stats as stat\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nimport os\nimport h5py\nimport datetime\nimport pandas as pd\n\nfrom pandas import DataFrame,Series,read_table",
"_____no_output_____"
]
],
[
[
"General info",
"_____no_output_____"
]
],
[
[
"savePlots = True # whether or not to save plots\nsaveData = True # whether or not to save csv files\n\nsaveAsPath = './Fig 03/'\nif not os.path.exists(saveAsPath):\n os.mkdir(saveAsPath)\nsaveAsName = 'Fig3'",
"_____no_output_____"
],
[
"#path = '/Users/svcanavan/Dropbox/Coding in progress/00_BudgieSleep/Data_copies/'\n\nbirdPaths = ['../data_copies/01_PreprocessedData/01_BudgieFemale_green1/00_Baseline_night/',\n '../data_copies/01_PreprocessedData/02_BudgieMale_yellow1/00_Baseline_night/',\n '../data_copies/01_PreprocessedData/03_BudgieFemale_white1/00_Baseline_night/',\n '../data_copies/01_PreprocessedData/04_BudgieMale_yellow2/00_Baseline_night/',\n '../data_copies/01_PreprocessedData/05_BudgieFemale_green2/00_Baseline_night/']\n\narfFilePaths = ['EEG 2 scored/',\n 'EEG 3 scored/',\n 'EEG 3 scored/',\n 'EEG 4 scored/',\n 'EEG 4 scored/']\n\n### load BEST EEG channels - as determined during manual scoring ####\nchannelsToLoadEEG_best = [['6 LEEGm-LEEGp', '5 LEEGf-LEEGp'], #, '9 REEGp-LEEGp'], # extra channel to represent R hemisphere\n ['5 LEEGf-LEEGm', '4 LEEGf-Fgr'], #, '9 REEGf-REEGm'], # extra channel to represent R hemisphere\n ['9REEGm-REEGp', '4LEEGf-LEEGp'], \n ['6LEEGm-LEEGf', '9REEGf-REEGp'], \n ['7REEGf-REEGp', '4LEEGf-LEEGp']]\n\n\n### load ALL of EEG channels ####\nchannelsToLoadEEG = [['4 LEEGf-Fgr', '5 LEEGf-LEEGp', '6 LEEGm-LEEGp', '7 LEEGp-Fgr', '8 REEGp-Fgr','9 REEGp-LEEGp'],\n ['4 LEEGf-Fgr','5 LEEGf-LEEGm', '6 LEEGm-LEEGp', '7 REEGf-Fgr', '8 REEGm-Fgr', '9 REEGf-REEGm'],\n ['4LEEGf-LEEGp', '5LEEGf-LEEGm', '6LEEGm-LEEGp', '7REEGf-REEGp', '8REEGf-REEGm', '9REEGm-REEGp'],\n ['4LEEGf-LEEGp', '5LEEGm-LEEGp', '6LEEGm-LEEGf', '7REEGf-Fgr', '8REEGf-REEGm','9REEGf-REEGp',],\n ['4LEEGf-LEEGp', '5LEEGf-LEEGm', '6LEEGm-LEEGp', '7REEGf-REEGp', '8REEGf-REEGm', '9REEGm-REEGp']]\n\n\nchannelsToLoadEOG = [['1 LEOG-Fgr', '2 REOG-Fgr'],\n ['2 LEOG-Fgr', '3 REOG-Fgr'],\n ['2LEOG-Fgr', '3REOG-Fgr'],\n ['2LEOG-Fgr', '3REOG-Fgr'],\n ['2LEOG-Fgr', '3REOG-Fgr']]\n\nbirds_LL = [1,2,3]\nnBirds_LL = len(birds_LL)\n\nbirdPaths_LL = ['../data_copies/01_PreprocessedData/02_BudgieMale_yellow1/01_Constant_light/',\n '../data_copies/01_PreprocessedData/03_BudgieFemale_white1/01_Constant_light/',\n '../data_copies/01_PreprocessedData/04_BudgieMale_yellow2/01_Constant_light/',]\n\narfFilePaths_LL = ['EEG 2 preprocessed/',\n 'EEG 2 preprocessed/',\n 'EEG 2 preprocessed/']\n\nlightsOffSec = np.array([7947, 9675, 9861 + 8*3600, 9873, 13467]) # lights off times in seconds from beginning of file\nlightsOnSec = np.array([46449, 48168, 48375+ 8*3600, 48381, 52005]) # Bird 3 gets 8 hours added b/c file starts at 8:00 instead of 16:00\n\nepochLength = 3\nsr = 200\nscalingFactor = (2**15)*0.195 # scaling/conversion factor from amplitude to uV (when recording arf from jrecord)\n\nstages = ['w','d','u','i','s','r'] # wake, drowsy, unihem sleep, intermediate sleep, SWS, REM\nstagesSleep = ['u','i','s','r']\n\nstagesVideo = ['m','q','d','s','u'] # moving wake, quiet wake, drowsy, sleep, unclear\n\n## Path to scores formatted as CSVs\nformatted_scores_path = '../formatted_scores/'\n\n## Path to detect SW ands EM events: use folder w/ EMs and EM artifacts detected during non-sleep\nevents_path = '../data_copies/SWs_EMs_and_EMartifacts/'",
"_____no_output_____"
],
[
"colors = sns.color_palette(np.array([[234,103,99],\n[218,142,60],\n[174,174,62],\n[97,188,101],\n[140,133,232],\n[225,113,190]])\n/255)\n\nsns.palplot(colors)\n\n# colorpalette from iWantHue",
"_____no_output_____"
],
[
"colors_birds = [np.repeat(.4, 3),\n np.repeat(.5, 3),\n np.repeat(.6, 3),\n np.repeat(.7, 3),\n np.repeat(.8, 3)]\n\nsns.palplot(colors_birds)\n",
"_____no_output_____"
]
],
[
[
"Plot-specific info",
"_____no_output_____"
]
],
[
[
"sns.set_context(\"notebook\", font_scale=1.5)\nsns.set_style(\"white\")\n\n# Markers for legends of EEG scoring colors\nlegendMarkersEEG = []\nfor stage in range(len(stages)):\n legendMarkersEEG.append(plt.Line2D([0],[0], color=colors[stage], marker='o', linestyle='', alpha=0.7)) ",
"_____no_output_____"
]
],
[
[
"Calculate general variables",
"_____no_output_____"
]
],
[
[
"lightsOffEp = lightsOffSec / epochLength\nlightsOnEp = lightsOnSec / epochLength\n\nnBirds = len(birdPaths)\n\nepochLengthPts = epochLength*sr\n\nnStages = len(stagesSleep)",
"_____no_output_____"
]
],
[
[
"## LEGEND: bird colors",
"_____no_output_____"
]
],
[
[
"# Markers for bird colors\nlegendMarkers_birds = []\nfor b in range(nBirds):\n legendMarkers_birds.append(plt.Line2D([0],[0], marker='o', color=colors_birds[b], linestyle='', alpha=0.7)) \nplt.legend(legendMarkers_birds, ['Bird 1', 'Bird 2', 'Bird 3', 'Bird 4', 'Bird 5'])\nsns.despine(bottom=True, left=True)\nplt.yticks([])\nplt.xticks([])\n\n\nif savePlots:\n plt.savefig(saveAsPath + saveAsName + \"a-d_bird_color_legend.pdf\")",
"_____no_output_____"
]
],
[
[
"## Load formatted scores",
"_____no_output_____"
]
],
[
[
"AllScores = {}\nfor b in range(nBirds):\n bird_name = 'Bird ' + str(b+1)\n file = formatted_scores_path + 'All_scores_' + bird_name + '.csv'\n data = pd.read_csv(file, index_col=0)\n AllScores[bird_name] = data",
"_____no_output_____"
]
],
[
[
"## Calculate lights off in Zeitgeber time (s and hrs)\nLights on is 0 ",
"_____no_output_____"
]
],
[
[
"lightsOffDatetime = np.array([], dtype='datetime64')\nlightsOnDatetime = np.array([], dtype='datetime64')\n\nfor b_num in range(nBirds):\n b_name = 'Bird ' + str(b_num+1)\n Scores = AllScores[b_name]\n startDatetime = np.datetime64(Scores.index.values[0])\n\n # Calc lights off & on using datetime formats\n lightsOffTimedelta = lightsOffSec[b_num].astype('timedelta64[s]')\n lightsOffDatetime = np.append(lightsOffDatetime, startDatetime + lightsOffTimedelta)\n lightsOnTimedelta = lightsOnSec[b_num].astype('timedelta64[s]')\n lightsOnDatetime = np.append(lightsOnDatetime, startDatetime + lightsOnTimedelta)",
"_____no_output_____"
],
[
"lightsOffZeit_s = lightsOffSec - lightsOnSec\nlightsOffZeit_hr = lightsOffZeit_s / 3600",
"_____no_output_____"
]
],
[
[
"# With all sleep stages separately",
"_____no_output_____"
],
[
"## Merge continuous epochs of the same stage, same length & start time",
"_____no_output_____"
]
],
[
[
"ScoresMerged = {}\n\nfor key in AllScores.keys():\n\n scores = AllScores[key]\n \n # add a \"stop\" column\n scores['Stop (s)'] = scores['Time (s)'] + epochLength\n \n original_end_s = scores['Stop (s)'].iloc[-1]\n \n # add a dummy row wherever there is a gap between epochs\n gaps = np.where(scores['Stop (s)'] != scores['Time (s)'].shift(-1))[0]\n gaps = gaps[0:-1] # don't include the last row of the file as a gap\n gap_datetimes = scores.iloc[gaps].index.astype('datetime64') + np.timedelta64(3, 's')\n\n lines = pd.DataFrame({'Time (s)': scores['Stop (s)'].iloc[gaps].values,\n 'Stop (s)': scores['Time (s)'].iloc[gaps+1].values,\n 'Label': 'gap'}, index=gap_datetimes.astype('str'))\n scores = scores.append(lines, ignore_index=False)\n scores = scores.sort_index()\n \n # add a datetime column\n scores['datetime'] = scores.index\n \n # add a column to keep track of consecutive epochs of the same stage\n scores['episode #'] = (scores['Label'] != scores['Label'].shift(1)).astype(int).cumsum()\n # don't worry about the gaps where the video recording restarted\n # if the behavior is the same before and after the gap, count it as a continuous episode\n # if you want to split episodes where the gap occurs, add:\n # (scores['Stop (s)'] == scores['Time (s)'].shift(-1))\n \n # combine all epochs of the same episode # and use the values from the first epoch of that episode\n merged_scores = scores.groupby(scores['episode #'], sort=False).aggregate('first')\n \n # calculate length of each episode\n lengths = merged_scores['Time (s)'].shift(-1) - merged_scores['Time (s)']\n lengths.iloc[-1] = original_end_s - merged_scores['Time (s)'].iloc[-1]\n \n merged_scores['Length (s)'] = lengths\n \n # set index back to datetime\n merged_scores.index = merged_scores['datetime']\n \n gap_rows =merged_scores[merged_scores['Label']=='gap'].index\n merged_scores = merged_scores.drop(gap_rows)\n \n ScoresMerged[key] = merged_scores",
"/Users/svcanavan/anaconda3/lib/python3.7/site-packages/pandas/core/indexes/base.py:346: FutureWarning: Passing in 'datetime64' dtype with no precision is deprecated\nand will raise in a future version. Please pass in\n'datetime64[ns]' instead.\n data, copy=copy, name=name, dtype=dtype, **kwargs\n/Users/svcanavan/anaconda3/lib/python3.7/site-packages/pandas/core/frame.py:7123: FutureWarning: Sorting because non-concatenation axis is not aligned. A future version\nof pandas will change to not sort by default.\n\nTo accept the future behavior, pass 'sort=False'.\n\nTo retain the current behavior and silence the warning, pass 'sort=True'.\n\n sort=sort,\n"
],
[
"# Check lengths\nprint(len(AllScores['Bird 5']) * epochLength/3600) # original\n\nprint(ScoresMerged['Bird 5']['Length (s)'].sum()/3600) # merged\n",
"23.905833333333334\n23.905833333333334\n"
]
],
[
[
"### Save to csv",
"_____no_output_____"
]
],
[
[
"if saveData:\n for b in range(nBirds):\n\n b_name = 'Bird ' + str(b+1)\n\n scores = ScoresMerged[b_name]\n scores.to_csv(saveAsPath + saveAsName + '_scores_merged_' + b_name + '.csv')",
"_____no_output_____"
]
],
[
[
"## Frequency of episodes of each stage per bin",
"_____no_output_____"
]
],
[
[
"binSize_min = 60\n\nbinSize_s = np.timedelta64(int(binSize_min*60), 's')\n\nstageProportions_whole_night_all = {}\n\nfor b in range(nBirds):\n nBins = int(np.ceil(np.min(lightsOnSec - lightsOffSec)/(60*binSize_min)))\n\n stageProportions = DataFrame([], columns=range(len(stages)))\n\n b_name = 'Bird ' + str(b+1)\n Scores = ScoresMerged[b_name]\n\n for bn in range(nBins):\n\n start_time = str(lightsOffDatetime[b] + bn*binSize_s).replace('T', ' ')\n end_time = str(lightsOffDatetime[b] + (bn+1)*binSize_s).replace('T', ' ')\n\n bn_scores = Scores[str(start_time):str(end_time)]\n bn_stage_frequencies = bn_scores['Label (#)'].value_counts(sort=False)\n \n stageProportions = stageProportions.append(bn_stage_frequencies, ignore_index=True)\n \n # Replace NaNs with 0\n stageProportions = stageProportions.fillna(0)\n\n # Add to dictionary\n stageProportions_whole_night_all[b] = stageProportions\n ",
"_____no_output_____"
]
],
[
[
"### Save to csv",
"_____no_output_____"
]
],
[
[
"if saveData:\n for b in range(nBirds):\n b_name = 'Bird ' + str(b+1)\n\n stageProportions = stageProportions_whole_night_all[b]\n stageProportions.to_csv(saveAsPath + saveAsName + 'c_stage_frequencies_' + b_name + '.csv')",
"_____no_output_____"
]
],
[
[
"### FIGURE 3C: Plot",
"_____no_output_____"
]
],
[
[
"figsize = (8,6)\naxis_label_fontsize = 24\n\n# Line formatting\nlinewidth = 5\nlinealpha = .7\nmarker = 'o'\nmarkersize = 10\nerr_capsize = 3\nerr_capthick = 3\nelinewidth = 3\n\n# Bar formatting\nbar_linewidth = 4",
"_____no_output_____"
],
[
"plt.figure(figsize=figsize)\n\nbird_means_by_stage = pd.DataFrame([])\nAll_per_bin_means = pd.DataFrame([])\n\nfor st in range(len(stages[3:6])):\n\n st_freqs = np.zeros((nBins, nBirds))\n bird_means = np.zeros(nBirds)\n for b in range(nBirds):\n\n stageProportions = stageProportions_whole_night_all[b]\n\n st_freqs[:,b] = stageProportions[st+3]\n \n bird_means[b] = np.mean(stageProportions[st+3])\n\n nighttime_mean = np.mean(st_freqs)\n per_bin_mean = np.mean(st_freqs, axis=1)\n per_bin_sd = np.std(st_freqs, axis=1)\n per_bin_sem = per_bin_sd / np.sqrt(nBirds)\n \n # save to dataframe\n All_per_bin_means[st+3] = per_bin_mean\n\n plt.errorbar(range(nBins), per_bin_mean, yerr=per_bin_sem,\n color=colors[3:6][st], linewidth=linewidth, alpha=linealpha,\n marker=marker, markersize=markersize,\n capsize=err_capsize, capthick=err_capthick, elinewidth=elinewidth);\n \n # Dots marking nighttime mean of each bird\n plt.scatter(np.ones(nBirds)*(nBins+2+(st*2)), bird_means, 50, color=colors[3:6][st]);\n \n # Bar graph of mean across all birds\n plt.bar(nBins+2+(st*2), np.mean(bird_means), width=2, color='none',edgecolor=colors[3:6][st], linewidth=bar_linewidth);\n \n print(stages[3:6][st] + ' : mean ' + str(np.mean(bird_means)) + ', SD ' + str(np.std(bird_means)))\n bird_means_by_stage[stages[3:6][st]] = bird_means\n \n# Dots color coded by bird\nfor b in range(nBirds):\n plt.scatter(nBins+(np.arange(0,3)*2)+2, \n bird_means_by_stage.loc[b], 50, color=colors_birds[b], alpha=.5)\n \n \nplt.ylim(0,225)\nplt.xlim(-.5,19)\n\n# x tick labels: label each bar of the bar graph separately\nc = (0,0,0)\nplt.xticks([0,2,4,6,8,10,13,15,17], [0,2,4,6,8,10, 'IS', 'SWS','REM']);\nax = plt.gca()\n[t.set_color(i) for (i,t) in zip([c,c,c,c,c,c,colors[3],colors[4],colors[5]],ax.xaxis.get_ticklabels())]\n\nplt.ylabel('Number of episodes / hour', fontsize=axis_label_fontsize)\nplt.xlabel('Hour of night Total', fontsize=axis_label_fontsize)\n \nsns.despine()\n \nif savePlots:\n plt.savefig(saveAsPath + saveAsName + \"c_frequencies.pdf\")",
"i : mean 164.6727272727273, SD 25.31027949142175\ns : mean 102.9090909090909, SD 26.014172869469537\nr : mean 79.98181818181817, SD 10.878502570750175\n"
]
],
[
[
"### FIGURE 3C: STATISTICS",
"_____no_output_____"
]
],
[
[
"# One-way ANOVA: mean frequencies\nstat.f_oneway(bird_means_by_stage['i'],bird_means_by_stage['s'], bird_means_by_stage['r'])",
"_____no_output_____"
],
[
"# Mean frequencies: IS vs REM\nstat.ttest_rel(bird_means_by_stage['i'], bird_means_by_stage['r'])",
"_____no_output_____"
],
[
"# Mean frequencies: SWS vs REM\nstat.ttest_rel(bird_means_by_stage['s'], bird_means_by_stage['r'])",
"_____no_output_____"
],
[
"# Mean frequencies: IS vs SWS\nstat.ttest_rel(bird_means_by_stage['i'], bird_means_by_stage['s'])",
"_____no_output_____"
],
[
"# IS: regression with hour of night\ntest = All_per_bin_means[3]\n\nslope, intercept, r_value, p_value, std_err = stat.linregress(test.index.values, test.values)\nprint('slope =', slope, ', r2 =', r_value**2, ', p =', p_value)",
"slope = -0.0018181818181822317 , r2 = 4.207263250987887e-08 , p = 0.9995224466780807\n"
],
[
"# IS: regression with hour of night\ntest = All_per_bin_means[3][1:] # FIRST HOUR OMITTED\n\nslope, intercept, r_value, p_value, std_err = stat.linregress(test.index.values, test.values)\nprint('slope =', slope, ', r2 =', r_value**2, ', p =', p_value)",
"slope = -4.900606060606061 , r2 = 0.732465049967923 , p = 0.0015818264642422173\n"
],
[
"# SWS: regression with hour of night\ntest = All_per_bin_means[4]\n\nslope, intercept, r_value, p_value, std_err = stat.linregress(test.index.values, test.values)\nprint('slope =', slope, ', r2 =', r_value**2, ', p =', p_value)",
"slope = -3.6327272727272715 , r2 = 0.1726966520589901 , p = 0.20369015541009844\n"
],
[
"# SWS: regression with hour of night\ntest = All_per_bin_means[4][1:] # FIRST HOUR OMITTED\n\nslope, intercept, r_value, p_value, std_err = stat.linregress(test.index.values, test.values)\nprint('slope =', slope, ', r2 =', r_value**2, ', p =', p_value)",
"slope = -7.664242424242423 , r2 = 0.7528924210890774 , p = 0.0011392692535196122\n"
],
[
"# REM: regression with hour of night\ntest = All_per_bin_means[5]\n\nslope, intercept, r_value, p_value, std_err = stat.linregress(test.index.values, test.values)\nprint('slope =', slope, ', r2 =', r_value**2, ', p =', p_value)",
"slope = 3.698181818181817 , r2 = 0.2252302847909687 , p = 0.14022155928550747\n"
],
[
"# REM: regression with hour of night\ntest = All_per_bin_means[5][1:] # FIRST HOUR OMITTED\n\nslope, intercept, r_value, p_value, std_err = stat.linregress(test.index.values, test.values)\nprint('slope =', slope, ', r2 =', r_value**2, ', p =', p_value)",
"slope = 0.838787878787878 , r2 = 0.022897306014779267 , p = 0.6764663179233157\n"
]
],
[
[
"## Duration of episodes of each stage per bin",
"_____no_output_____"
]
],
[
[
"binSize_min = 60\n\nbinSize_s = np.timedelta64(int(binSize_min*60), 's')\n\nstageProportions_whole_night_all = {}\n\nfor b in range(nBirds):\n nBins = int(np.ceil(np.min(lightsOnSec - lightsOffSec)/(60*binSize_min)))\n\n stageProportions = DataFrame([], columns=range(len(stages)))\n\n b_name = 'Bird ' + str(b+1)\n Scores = ScoresMerged[b_name]\n\n for bn in range(nBins):\n\n start_time = str(lightsOffDatetime[b] + bn*binSize_s).replace('T', ' ')\n end_time = str(lightsOffDatetime[b] + (bn+1)*binSize_s).replace('T', ' ')\n\n bn_scores = Scores[start_time:end_time]\n \n bn_stage_lengths = np.array([])\n for st in range(len(stages)):\n bn_st_episodes = bn_scores[bn_scores['Label (#)'] == st]\n if len(bn_st_episodes) > 0:\n bn_avg_length = bn_st_episodes['Length (s)'].mean(0)\n else:\n bn_avg_length = np.nan\n bn_stage_lengths = np.append(bn_stage_lengths, bn_avg_length)\n \n stageProportions.loc[bn] = bn_stage_lengths\n\n # Add to dictionary\n stageProportions_whole_night_all[b] = stageProportions\n ",
"_____no_output_____"
]
],
[
[
"### FIGURE 3A: Plot",
"_____no_output_____"
]
],
[
[
"plt.figure(figsize=figsize)\n\nbird_means_by_stage = pd.DataFrame([]) # init\nAll_per_bin_means = pd.DataFrame([])\n\nfor st in range(len(stages[3:6])):\n\n st_lengths = np.zeros((nBins, nBirds))\n bird_means = np.zeros(nBirds)\n for b in range(nBirds):\n\n stageProportions = stageProportions_whole_night_all[b]\n\n st_lengths[:,b] = stageProportions[st+3]\n \n bird_means[b] = np.mean(stageProportions[st+3])\n\n nighttime_mean = np.mean(st_lengths)\n per_bin_mean = np.mean(st_lengths, axis=1)\n per_bin_sd = np.std(st_lengths, axis=1)\n per_bin_sem = per_bin_sd / np.sqrt(nBirds)\n \n # save to dataframe\n All_per_bin_means[st+3] = per_bin_mean\n\n plt.errorbar(range(nBins), per_bin_mean, yerr=per_bin_sem, color=colors[3:6][st],\n linewidth=linewidth, alpha=linealpha,\n marker=marker, markersize=markersize,\n capsize=err_capsize, capthick=err_capthick, elinewidth=elinewidth);\n \n # Dots marking nighttime mean of each bird\n plt.scatter(np.ones(nBirds)*(nBins+(st*2)+2), bird_means, 50, color=colors[3:6][st]);\n \n # Bar graph of mean across all birds\n plt.bar(nBins+(st*2)+2, np.mean(bird_means), width=2, color='none',edgecolor=colors[3:6][st], linewidth=bar_linewidth);\n \n print(stages[3:6][st] + ' : mean ' + str(np.mean(bird_means)) + ', SD ' + str(np.std(bird_means)))\n bird_means_by_stage[stages[3:6][st]] = bird_means\n \n# Dots color coded by bird\nfor b in range(nBirds):\n plt.scatter(nBins+(np.arange(0,3)*2)+2, \n bird_means_by_stage.loc[b], 50, color=colors_birds[b], alpha=.5)\n\nplt.ylim(0,25)\nplt.xlim(-.5,19)\n\n# x tick labels: label each bar of the bar graph separately\nc = (0,0,0)\nplt.xticks([0,2,4,6,8,10,13,15,17], [0,2,4,6,8,10,'IS', 'SWS','REM']);\nax = plt.gca()\n[t.set_color(i) for (i,t) in zip([c,c,c,c,c,c,colors[3],colors[4],colors[5]],ax.xaxis.get_ticklabels())]\n\nplt.ylabel('Mean duration of episodes (s)', fontsize=axis_label_fontsize)\nplt.xlabel('Hour of night Total', fontsize=axis_label_fontsize)\n\nsns.despine()\n\nif savePlots:\n plt.savefig(saveAsPath + saveAsName + \"a_durations.pdf\")",
"i : mean 9.246904631604412, SD 1.250965921913051\ns : mean 5.134153837328954, SD 0.789065355060224\nr : mean 11.26150728060531, SD 5.060989008212911\n"
]
],
[
[
"### FIGURE 3A STATISTICS",
"_____no_output_____"
]
],
[
[
"# One-way ANOVA: mean durations\nstat.f_oneway(bird_means_by_stage['i'],bird_means_by_stage['s'], bird_means_by_stage['r'])",
"_____no_output_____"
],
[
"# Mean durations: IS vs REM\nstat.ttest_rel(bird_means_by_stage['i'], bird_means_by_stage['r'])",
"_____no_output_____"
],
[
"# Mean durations: SWS vs REM\nstat.ttest_rel(bird_means_by_stage['s'], bird_means_by_stage['r'])",
"_____no_output_____"
],
[
"# Mean durations: SWS vs IS\nstat.ttest_rel(bird_means_by_stage['s'], bird_means_by_stage['i'])",
"_____no_output_____"
],
[
"# IS: regression with hour of night\ntest = All_per_bin_means[3]\n\nslope, intercept, r_value, p_value, std_err = stat.linregress(test.index.values, test.values)\nprint('slope =', slope, ', r2 =', r_value**2, ', p =', p_value)",
"slope = 0.3606111319295407 , r2 = 0.6813777712756139 , p = 0.0017529352370463235\n"
],
[
"# SWS: regression with hour of night\ntest = All_per_bin_means[4]\n\nslope, intercept, r_value, p_value, std_err = stat.linregress(test.index.values, test.values)\nprint('slope =', slope, ', r2 =', r_value**2, ', p =', p_value)",
"slope = -0.24056236891664864 , r2 = 0.7080237477666089 , p = 0.0011658502864583798\n"
],
[
"# REM: regression with hour of night\ntest = All_per_bin_means[5]\n\nslope, intercept, r_value, p_value, std_err = stat.linregress(test.index.values, test.values)\nprint('slope =', slope, ', r2 =', r_value**2, ', p =', p_value)",
"slope = 0.8017961761027387 , r2 = 0.7649437977969713 , p = 0.00042632071896564987\n"
]
],
[
[
"## Save as csv",
"_____no_output_____"
]
],
[
[
"if saveData:\n for b in range(nBirds):\n b_name = 'Bird ' + str(b+1)\n\n stageProportions = stageProportions_whole_night_all[b]\n stageProportions.to_csv(saveAsPath + saveAsName + 'a_stage_durations_' + b_name + '.csv')",
"_____no_output_____"
]
],
[
[
"# With NREM stages lumped",
"_____no_output_____"
]
],
[
[
"# THIS NOTEBOOK ONLY:\nstages = ['w','d','u', 'n', 'n','r'] # wake, drowsy, unihem sleep, NREM, REM\nstagesSleep = ['u','n', 'n','r']",
"_____no_output_____"
],
[
"# THIS NOTEBOOK ONLY - LUMP NREM:\nNREM_color = np.median(np.array([[97,188,101],\n[140,133,232]]),axis=0)\n\ncolors = sns.color_palette(np.array([[234,103,99],\n[218,142,60],\n[174,174,62],\nNREM_color,\nNREM_color, \n[225,113,190]])\n/255)\n\nsns.palplot(colors)\n\n# colorpalette from iWantHue",
"_____no_output_____"
]
],
[
[
"## Reload formatted scores and replace IS and SWS with just \"NREM\"",
"_____no_output_____"
]
],
[
[
"AllScores = {}\nfor b in range(nBirds):\n bird_name = 'Bird ' + str(b+1)\n file = formatted_scores_path + 'All_scores_' + bird_name + '.csv'\n data = pd.read_csv(file, index_col=0)\n \n labels = data['Label'].replace(to_replace=np.nan, value='u').values\n label_nums = data['Label (#)'].values\n indsNREM = [x for x in range(int(len(labels))) if ('i' in labels[x])|('s' in labels[x])]\n for ind in indsNREM:\n labels[ind] = 'n' \n label_nums[ind] = 4\n data['Label'] = labels\n data['Label (#)'] = label_nums\n \n AllScores[bird_name] = data",
"_____no_output_____"
]
],
[
[
"## Calculate lights off in Zeitgeber time (s and hrs)\nLights on is 0 ",
"_____no_output_____"
]
],
[
[
"lightsOffDatetime = np.array([], dtype='datetime64')\nlightsOnDatetime = np.array([], dtype='datetime64')\n\nfor b_num in range(nBirds):\n b_name = 'Bird ' + str(b_num+1)\n Scores = AllScores[b_name]\n startDatetime = np.datetime64(Scores.index.values[0])\n\n # Calc lights off & on using datetime formats\n lightsOffTimedelta = lightsOffSec[b_num].astype('timedelta64[s]')\n lightsOffDatetime = np.append(lightsOffDatetime, startDatetime + lightsOffTimedelta)\n lightsOnTimedelta = lightsOnSec[b_num].astype('timedelta64[s]')\n lightsOnDatetime = np.append(lightsOnDatetime, startDatetime + lightsOnTimedelta)",
"_____no_output_____"
],
[
"lightsOffZeit_s = lightsOffSec - lightsOnSec\nlightsOffZeit_hr = lightsOffZeit_s / 3600",
"_____no_output_____"
]
],
[
[
"## Merge continuous epochs of the same stage, same length & start time",
"_____no_output_____"
]
],
[
[
"ScoresMerged = {}\n\nfor key in AllScores.keys():\n\n scores = AllScores[key]\n \n # add a \"stop\" column\n scores['Stop (s)'] = scores['Time (s)'] + epochLength\n \n original_end_s = scores['Stop (s)'].iloc[-1]\n \n # add a dummy row wherever there is a gap between epochs\n gaps = np.where(scores['Stop (s)'] != scores['Time (s)'].shift(-1))[0]\n gaps = gaps[0:-1] # don't include the last row of the file as a gap\n gap_datetimes = scores.iloc[gaps].index.astype('datetime64') + np.timedelta64(3, 's')\n\n lines = pd.DataFrame({'Time (s)': scores['Stop (s)'].iloc[gaps].values,\n 'Stop (s)': scores['Time (s)'].iloc[gaps+1].values,\n 'Label': 'gap'}, index=gap_datetimes.astype('str'))\n scores = scores.append(lines, ignore_index=False)\n scores = scores.sort_index()\n \n # add a datetime column\n scores['datetime'] = scores.index\n \n # add a column to keep track of consecutive epochs of the same stage\n scores['episode #'] = (scores['Label'] != scores['Label'].shift(1)).astype(int).cumsum()\n # don't worry about the gaps where the video recording restarted\n # if the behavior is the same before and after the gap, count it as a continuous episode\n # if you want to split episodes where the gap occurs, add:\n # (scores['Stop (s)'] == scores['Time (s)'].shift(-1))\n \n # combine all epochs of the same episode # and use the values from the first epoch of that episode\n merged_scores = scores.groupby(scores['episode #'], sort=False).aggregate('first')\n \n # calculate length of each episode\n lengths = merged_scores['Time (s)'].shift(-1) - merged_scores['Time (s)']\n lengths.iloc[-1] = original_end_s - merged_scores['Time (s)'].iloc[-1]\n \n merged_scores['Length (s)'] = lengths\n \n # set index back to datetime\n merged_scores.index = merged_scores['datetime']\n \n gap_rows =merged_scores[merged_scores['Label']=='gap'].index\n merged_scores = merged_scores.drop(gap_rows)\n \n ScoresMerged[key] = merged_scores",
"/Users/svcanavan/anaconda3/lib/python3.7/site-packages/pandas/core/indexes/base.py:346: FutureWarning: Passing in 'datetime64' dtype with no precision is deprecated\nand will raise in a future version. Please pass in\n'datetime64[ns]' instead.\n data, copy=copy, name=name, dtype=dtype, **kwargs\n/Users/svcanavan/anaconda3/lib/python3.7/site-packages/pandas/core/frame.py:7123: FutureWarning: Sorting because non-concatenation axis is not aligned. A future version\nof pandas will change to not sort by default.\n\nTo accept the future behavior, pass 'sort=False'.\n\nTo retain the current behavior and silence the warning, pass 'sort=True'.\n\n sort=sort,\n"
],
[
"# Check lengths\nprint(len(AllScores['Bird 5']) * epochLength/3600) # original\n\nprint(ScoresMerged['Bird 5']['Length (s)'].sum()/3600) # merged\n",
"23.905833333333334\n23.905833333333334\n"
]
],
[
[
"## Frequency of episodes of each stage per bin",
"_____no_output_____"
]
],
[
[
"binSize_min = 60\n\nbinSize_s = np.timedelta64(int(binSize_min*60), 's')\n\nstageProportions_whole_night_all = {}\n\n\nfor b in range(nBirds):\n nBins = int(np.ceil(np.min(lightsOnSec - lightsOffSec)/(60*binSize_min)))\n\n stageProportions = DataFrame([], columns=range(len(stages)))\n\n b_name = 'Bird ' + str(b+1)\n Scores = ScoresMerged[b_name]\n\n for bn in range(nBins):\n\n start_time = str(lightsOffDatetime[b] + bn*binSize_s).replace('T', ' ')\n end_time = str(lightsOffDatetime[b] + (bn+1)*binSize_s).replace('T', ' ')\n\n bn_scores = Scores[str(start_time):str(end_time)]\n bn_stage_frequencies = bn_scores['Label (#)'].value_counts(sort=False)\n \n stageProportions = stageProportions.append(bn_stage_frequencies, ignore_index=True)\n \n # Replace NaNs with 0\n stageProportions = stageProportions.fillna(0)\n\n # Add to dictionary\n stageProportions_whole_night_all[b] = stageProportions\n ",
"_____no_output_____"
]
],
[
[
"### save to csv",
"_____no_output_____"
]
],
[
[
"if saveData:\n for b in range(nBirds):\n b_name = 'Bird ' + str(b+1)\n\n stageProportions = stageProportions_whole_night_all[b]\n stageProportions.to_csv(saveAsPath + saveAsName + 'd_NREM_lumped_stage_frequencies_' + b_name + '.csv')",
"_____no_output_____"
]
],
[
[
"### FIGURE 3D Plot",
"_____no_output_____"
]
],
[
[
"figsize = (8,6)\naxis_label_fontsize = 24\n\n# Line formatting\nlinewidth = 5\nlinealpha = .7\nmarker = 'o'\nmarkersize = 10\nerr_capsize = 3\nerr_capthick = 3\nelinewidth = 3\n\n# Bar formatting\nbar_linewidth = 4",
"_____no_output_____"
],
[
"plt.figure(figsize=figsize)\n\nbird_means_by_stage = pd.DataFrame([])\nAll_per_bin_means = pd.DataFrame([])\n\nfor st in range(len(stages[4:6])):\n\n st_freqs = np.zeros((nBins, nBirds))\n bird_means = np.zeros(nBirds)\n for b in range(nBirds):\n\n stageProportions = stageProportions_whole_night_all[b]\n\n st_freqs[:,b] = stageProportions[st+4]\n \n bird_means[b] = np.mean(stageProportions[st+4])\n\n nighttime_mean = np.mean(st_freqs)\n per_bin_mean = np.mean(st_freqs, axis=1)\n per_bin_sd = np.std(st_freqs, axis=1)\n per_bin_sem = per_bin_sd / np.sqrt(nBirds)\n \n # save to dataframe\n All_per_bin_means[st+4] = per_bin_mean\n\n plt.errorbar(range(nBins), per_bin_mean, yerr=per_bin_sem,\n color=colors[4:6][st], linewidth=linewidth, alpha=linealpha,\n marker=marker, markersize=markersize,\n capsize=err_capsize, capthick=err_capthick, elinewidth=elinewidth);\n \n # Dots marking nighttime mean of each bird\n plt.scatter(np.ones(nBirds)*(nBins+2+(st*2)), bird_means, 50, color=colors[4:6][st]);\n \n # Bar graph of mean across all birds\n plt.bar(nBins+2+(st*2), np.mean(bird_means), width=2, color='none',edgecolor=colors[4:6][st], linewidth=bar_linewidth);\n \n print(stages[4:6][st] + ' : mean ' + str(np.mean(bird_means)) + ', SD ' + str(np.std(bird_means)))\n bird_means_by_stage[stages[4:6][st]] = bird_means\n\n \n# Dots color coded by bird\nfor b in range(nBirds):\n plt.scatter(nBins+(np.arange(0,2)*2)+2, \n bird_means_by_stage.loc[b], 50, color=colors_birds[b], alpha=.5)\n \nplt.ylim(0,150)\nplt.xlim(-.5,19)\n\n# x tick labels: label each bar of the bar graph separately\nc = (0,0,0)\nplt.xticks([0,2,4,6,8,10,13,15], [0,2,4,6,8,10,'nonREM','REM']);\nax = plt.gca()\n[t.set_color(i) for (i,t) in zip([c,c,c,c,c,c,colors[4],colors[5]],ax.xaxis.get_ticklabels())]\n\nplt.ylabel('Number of episodes / hour', fontsize=axis_label_fontsize)\nplt.xlabel('Hour of night Total', fontsize=axis_label_fontsize)\n\nsns.despine()\n\nif savePlots:\n plt.savefig(saveAsPath + saveAsName + \"d_NREM_lumped_frequencies.pdf\")",
"n : mean 90.14545454545456, SD 7.945808188510155\nr : mean 79.98181818181817, SD 10.878502570750175\n"
]
],
[
[
"### FIGURE 3D: STATISTICS",
"_____no_output_____"
]
],
[
[
"# Mean frequencies: NREM vs REM\nstat.ttest_rel(bird_means_by_stage['n'], bird_means_by_stage['r'])",
"_____no_output_____"
],
[
"# NREM: regression with hour of night\ntest = All_per_bin_means[4]\n\nslope, intercept, r_value, p_value, std_err = stat.linregress(test.index.values, test.values)\nprint('slope =', slope, ', r2 =', r_value**2, ', p =', p_value)",
"slope = 2.3854545454545453 , r2 = 0.21350373835948772 , p = 0.15247522575946523\n"
]
],
[
[
"## Duration of episodes of each stage per bin",
"_____no_output_____"
]
],
[
[
"binSize_min = 60\n\nbinSize_s = np.timedelta64(int(binSize_min*60), 's')\n\nstageProportions_whole_night_all = {}\n\nfor b in range(nBirds):\n nBins = int(np.ceil(np.min(lightsOnSec - lightsOffSec)/(60*binSize_min)))\n\n stageProportions = DataFrame([], columns=range(len(stages)))\n\n b_name = 'Bird ' + str(b+1)\n Scores = ScoresMerged[b_name]\n\n for bn in range(nBins):\n\n start_time = str(lightsOffDatetime[b] + bn*binSize_s).replace('T', ' ')\n end_time = str(lightsOffDatetime[b] + (bn+1)*binSize_s).replace('T', ' ')\n\n bn_scores = Scores[start_time:end_time]\n \n bn_stage_lengths = np.array([])\n for st in range(len(stages)):\n bn_st_episodes = bn_scores[bn_scores['Label (#)'] == st]\n if len(bn_st_episodes) > 0:\n bn_avg_length = bn_st_episodes['Length (s)'].mean(0)\n else:\n bn_avg_length = np.nan\n bn_stage_lengths = np.append(bn_stage_lengths, bn_avg_length)\n \n stageProportions.loc[bn] = bn_stage_lengths\n\n # Add to dictionary\n stageProportions_whole_night_all[b] = stageProportions\n ",
"_____no_output_____"
]
],
[
[
"### FIGURE 3B: Plot",
"_____no_output_____"
]
],
[
[
"plt.figure(figsize=figsize)\n\nbird_means_by_stage = pd.DataFrame([])\nAll_per_bin_means = pd.DataFrame([])\n\nfor st in range(len(stages[4:6])):\n\n st_lengths = np.zeros((nBins, nBirds))\n bird_means = np.zeros(nBirds)\n for b in range(nBirds):\n\n stageProportions = stageProportions_whole_night_all[b]\n\n st_lengths[:,b] = stageProportions[st+4]\n \n bird_means[b] = np.mean(stageProportions[st+4])\n\n nighttime_mean = np.mean(st_lengths)\n per_bin_mean = np.mean(st_lengths, axis=1)\n per_bin_sd = np.std(st_lengths, axis=1)\n per_bin_sem = per_bin_sd / np.sqrt(nBirds)\n\n # save to dataframe\n All_per_bin_means[st+4] = per_bin_mean\n \n plt.errorbar(range(nBins), per_bin_mean, yerr=per_bin_sem, color=colors[4:6][st],\n linewidth=linewidth, alpha=linealpha,\n marker=marker, markersize=markersize,\n capsize=err_capsize, capthick=err_capthick, elinewidth=elinewidth);\n \n # Dots marking nighttime mean of each bird\n plt.scatter(np.ones(nBirds)*(nBins+(st*2)+2), bird_means, 50, color=colors[4:6][st]);\n \n # Bar graph of mean across all birds\n plt.bar(nBins+(st*2)+2, np.mean(bird_means), width=2, color='none',edgecolor=colors[4:6][st], linewidth=bar_linewidth);\n \n print(stages[4:6][st] + ' : mean ' + str(np.mean(bird_means)) + ', SD ' + str(np.std(bird_means)))\n bird_means_by_stage[stages[4:6][st]] = bird_means\n \n# Dots color coded by bird\nfor b in range(nBirds):\n plt.scatter(nBins+(np.arange(0,2)*2)+2, \n bird_means_by_stage.loc[b], 50, color=colors_birds[b], alpha=.5)\n \n \nplt.ylim(0,60)\nplt.xlim(-.5,19)\n\n# x tick labels: label each bar of the bar graph separately\nc = (0,0,0)\nplt.xticks([0,2,4,6,8,10,13,15], [0,2,4,6,8,10,'nonREM','REM']);\nax = plt.gca()\n[t.set_color(i) for (i,t) in zip([c,c,c,c,c,c,colors[4],colors[5]],ax.xaxis.get_ticklabels())]\n\nplt.ylabel('Mean duration of episodes (s)', fontsize=axis_label_fontsize)\nplt.xlabel('Hour of night Total', fontsize=axis_label_fontsize)\n\nsns.despine()\n\nif savePlots:\n plt.savefig(saveAsPath + saveAsName + \"b_NREM_lumped_durations.pdf\")\n ",
"n : mean 25.059823683308498, SD 6.556666048684886\nr : mean 11.26150728060531, SD 5.060989008212911\n"
]
],
[
[
"### FIGURE 3B STATISTICS",
"_____no_output_____"
]
],
[
[
"# Mean durations: NREM vs REM\nstat.ttest_rel(bird_means_by_stage['n'], bird_means_by_stage['r'])",
"_____no_output_____"
],
[
"# NREM: regression with hour of night\ntest = All_per_bin_means[4]\n\nslope, intercept, r_value, p_value, std_err = stat.linregress(test.index.values, test.values)\nprint('slope =', slope, ', r2 =', r_value**2, ', p =', p_value)",
"slope = -1.0748672504921193 , r2 = 0.3375036895087704 , p = 0.060896633283737865\n"
],
[
"# NREM: regression with hour of night\ntest = All_per_bin_means[4][1:] # ONLY SIGNIFICANT IF FIRST HOUR OMITTED\n\nslope, intercept, r_value, p_value, std_err = stat.linregress(test.index.values, test.values)\nprint('slope =', slope, ', r2 =', r_value**2, ', p =', p_value)",
"slope = -1.3562427985389598 , r2 = 0.4045730640042001 , p = 0.04805153416194467\n"
]
],
[
[
"## Save as csv",
"_____no_output_____"
]
],
[
[
"if saveData:\n for b in range(nBirds):\n b_name = 'Bird ' + str(b+1)\n\n stageProportions = stageProportions_whole_night_all[b]\n stageProportions.to_csv(saveAsPath + saveAsName + 'b_NREM_lumped_stage_durations_' + b_name + '.csv')",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
c510e4bf7285cc553697b24896ab644d09f73575
| 8,462 |
ipynb
|
Jupyter Notebook
|
Redfish/Current/RedfishAnsible/1-RetrieveOneViewToken.ipynb
|
donzef/JupyterNotebooks
|
76c69e6e0e21120f3b88e7c991be6e946bc156d5
|
[
"Apache-2.0"
] | 1 |
2021-05-04T19:31:17.000Z
|
2021-05-04T19:31:17.000Z
|
Redfish/Current/RedfishAnsible/1-RetrieveOneViewToken.ipynb
|
donzef/JupyterNotebooks
|
76c69e6e0e21120f3b88e7c991be6e946bc156d5
|
[
"Apache-2.0"
] | null | null | null |
Redfish/Current/RedfishAnsible/1-RetrieveOneViewToken.ipynb
|
donzef/JupyterNotebooks
|
76c69e6e0e21120f3b88e7c991be6e946bc156d5
|
[
"Apache-2.0"
] | null | null | null | 37.114035 | 736 | 0.627866 |
[
[
[
"empty"
]
]
] |
[
"empty"
] |
[
[
"empty"
]
] |
c510eb3ef37a77d419e7215f84b6240463f885d2
| 225,136 |
ipynb
|
Jupyter Notebook
|
n_day_python/the_spotify_api/notebooks/3 - Use Access Token.ipynb
|
macknilan/python-examples-and-exercises
|
be3d60b8c6a4c6a570a7acff4a0f2b358e794e75
|
[
"MIT"
] | null | null | null |
n_day_python/the_spotify_api/notebooks/3 - Use Access Token.ipynb
|
macknilan/python-examples-and-exercises
|
be3d60b8c6a4c6a570a7acff4a0f2b358e794e75
|
[
"MIT"
] | null | null | null |
n_day_python/the_spotify_api/notebooks/3 - Use Access Token.ipynb
|
macknilan/python-examples-and-exercises
|
be3d60b8c6a4c6a570a7acff4a0f2b358e794e75
|
[
"MIT"
] | null | null | null | 27.617272 | 141 | 0.235289 |
[
[
[
"client_id = ''\nclient_secret = ''",
"_____no_output_____"
],
[
"import base64\nimport requests\nimport datetime\nfrom urllib.parse import urlencode",
"_____no_output_____"
],
[
"class SpotifyAPI(object):\n access_token = None\n access_token_expires = datetime.datetime.now()\n access_token_did_expire = True\n client_id = None\n client_secret = None\n token_url = \"https://accounts.spotify.com/api/token\"\n \n def __init__(self, client_id, client_secret, *args, **kwargs):\n super().__init__(*args, **kwargs)\n self.client_id = client_id\n self.client_secret = client_secret\n \n def get_client_credentials(self):\n \"\"\"\n RETURNS A BASE 64 ENCODE STRING\n \"\"\"\n client_id = self.client_id\n client_secret = self.client_secret\n if client_id == None or client_secret == None:\n raise Exception(\"TOU MUST SET client_id AND client_secret\")\n client_creds= f\"{client_id}:{client_secret}\"\n client_creds_b64 = base64.b64encode(client_creds.encode())\n return client_creds_b64.decode()\n \n def get_token_headers(self):\n client_creds_b64 = self.get_client_credentials()\n return {\n \"Authorization\": f\"Basic {client_creds_b64}\"\n }\n \n def get_token_data(self):\n return {\n \"grant_type\": \"client_credentials\"\n }\n \n def perform_auth(self):\n token_url = self.token_url\n token_data = self.get_token_data()\n token_headers = self.get_token_headers()\n r = requests.post(token_url, data=token_data, headers=token_headers)\n # print(r.json())\n if r.status_code not in range(200, 299):\n return False\n data = r.json()\n now = datetime.datetime.now()\n access_token = data['access_token']\n expires_in = data['expires_in']\n expires = now + datetime.timedelta(seconds=expires_in)\n self.access_token = access_token\n self.access_token_expires = expires\n self.access_token_did_expire = expires < now\n # print(f\"access_token -> {access_token}\")\n # print(f\"now -> {now}\")\n # print(f\"expires_in -> {expires_in}\")\n # print(f\"expires -> {expires}\")\n return True",
"_____no_output_____"
],
[
"spotify = SpotifyAPI(client_id, client_secret)",
"_____no_output_____"
],
[
"spotify.perform_auth()",
"_____no_output_____"
],
[
"# spotify.search",
"_____no_output_____"
],
[
"headers = {\n \"Authorization\": f\"Bearer {spotify.access_token}\"\n}\nendpoint_search = \"https://api.spotify.com/v1/search\"\ndata = urlencode({\"q\": \"Time\", \"type\":\"track\"})\nprint(data)\nlookup_url = f\"{endpoint_search}?{data}\"\nprint(lookup_url)\nr = requests.get(lookup_url, headers=headers)\nprint(r.status_code)",
"q=Time&type=track\nhttps://api.spotify.com/v1/search?q=Time&type=track\n200\n"
],
[
"r.json()",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
c510efe2673d7444e7785c6e72086f6c4b2ad72d
| 26,677 |
ipynb
|
Jupyter Notebook
|
Exercise04/Exercise04.ipynb
|
Develop-Packt/Plotting-Geospatial-Data
|
c8909ed208f59ae3f4cf58fb9e60ff07fad25497
|
[
"MIT"
] | null | null | null |
Exercise04/Exercise04.ipynb
|
Develop-Packt/Plotting-Geospatial-Data
|
c8909ed208f59ae3f4cf58fb9e60ff07fad25497
|
[
"MIT"
] | null | null | null |
Exercise04/Exercise04.ipynb
|
Develop-Packt/Plotting-Geospatial-Data
|
c8909ed208f59ae3f4cf58fb9e60ff07fad25497
|
[
"MIT"
] | 1 |
2021-02-25T16:50:33.000Z
|
2021-02-25T16:50:33.000Z
| 33.13913 | 175 | 0.443716 |
[
[
[
"## Exercise 04: Plotting the Movement of an Aircraft with a Custom Layer",
"_____no_output_____"
],
[
"In this exercise, we will take a look at how to create custom layers that allow you to not only display geo-spatial data but also animate your datapoints over time. \nWe'll get a deeper understanding of how geoplotlib works and how layers are created and drawn.\n\nOur dataset does not only contain spatial but also temporal information which enables us to plot flights over time on our map. \nThere is an example on how to do this with taxis in the examples folder of geoplotlib. \nhttps://github.com/andrea-cuttone/geoplotlib/blob/master/examples/taxi.py\n\n**Note:** \nThe dataset can be found here: \nhttps://datamillnorth.org/dataset/flight-tracking",
"_____no_output_____"
],
[
"#### Loading the dataset",
"_____no_output_____"
],
[
"This time our dataset contains flight data recorded from different machines. \nEach entry is assigned to a unique plane through a `hex_ident`. \nEach location is related to a specific timestamp that consists of a `date` and a `time`.",
"_____no_output_____"
]
],
[
[
"# importing the necessary dependencies\nimport pandas as pd",
"_____no_output_____"
],
[
"# loading the dataset from the csv file\ndataset = pd.read_csv('../../Datasets/flight_tracking.csv')",
"_____no_output_____"
],
[
"# displaying the first 5 rows of the dataset\ndataset.head()",
"_____no_output_____"
]
],
[
[
"Rename the latitude and longitude columns to lat and lon by using the rename method provided by pandas.",
"_____no_output_____"
]
],
[
[
"# renaming columns latitude to lat and longitude to lon\ndataset = dataset.rename(index=str, columns={\"latitude\": \"lat\", \"longitude\": \"lon\"})",
"_____no_output_____"
]
],
[
[
"**Note:** \nRemember that geoplotlib needs columns that are named `lat` and `lon`. You will encounter an error if that is not the case.",
"_____no_output_____"
]
],
[
[
"# displaying the first 5 rows of the dataset\ndataset.head()",
"_____no_output_____"
]
],
[
[
"---",
"_____no_output_____"
],
[
"#### Adding an unix timestamp",
"_____no_output_____"
],
[
"The easiest way to work with and handle time is to use a unix timestamp. \nIn previous activities, we've already seen how to create a new column in our dataset by applying a function to it. \nWe are using the datatime library to parse the date and time columns of our dataset and use it to create a unix timestamp.\n\nCombine date and time into a timestamp, using the, already provided, to_epoch method.",
"_____no_output_____"
]
],
[
[
"# method to convert date and time to an unix timestamp\nfrom datetime import datetime\n\ndef to_epoch(date, time):\n try:\n timestamp = round(datetime.strptime('{} {}'.format(date, time), '%Y/%m/%d %H:%M:%S.%f').timestamp())\n return timestamp\n except ValueError:\n return round(datetime.strptime('2017/09/11 17:02:06.418', '%Y/%m/%d %H:%M:%S.%f').timestamp())",
"_____no_output_____"
]
],
[
[
"Use to_epoch and the apply method provided by the pandas DataFrame to create a new column called timestamp that holds the unix timestamp.",
"_____no_output_____"
]
],
[
[
"# creating a new column called timestamp with the to_epoch method applied\ndataset['timestamp'] = dataset.apply(lambda x: to_epoch(x['date'], x['time']), axis=1)",
"_____no_output_____"
],
[
"# displaying the first 5 rows of the dataset\ndataset.head()",
"_____no_output_____"
]
],
[
[
"**Note:** \nWe round up the miliseconds in our `to_epoch` method since epoch is the number of seconds (not miliseconds) that have passes since January 1st 1970. \nOf course we loose some precision here, but we want to focus on creating our own custom layer instead of wasting a lot of time with our dataset.",
"_____no_output_____"
],
[
"---",
"_____no_output_____"
],
[
"#### Writing our custom layer",
"_____no_output_____"
],
[
"After preparing our dataset, we can now start writing our custom layer. \nAs mentioned at the beginning of this activity, it will be based on the taxi example of geoplotlib. \n\nWe want to have a layer `TrackLayer` that takes an argument, dataset, which contains `lat` and `lon` data in combination with a `timestamp`. \nGiven this data, we want to plot each point for each timestamp on the map, creating a tail behind the newest position of the plane.\nThe geoplotlib colorbrewer is used to give each plane a color based on their unique `hex_ident`. \nThe view (bounding box) of our visualization will be set to the city Leeds and a text information with the current timestamp is displayed in the upper right corner.",
"_____no_output_____"
]
],
[
[
"# custom layer creation\nimport geoplotlib\nfrom geoplotlib.layers import BaseLayer\nfrom geoplotlib.core import BatchPainter\nfrom geoplotlib.colors import colorbrewer\nfrom geoplotlib.utils import epoch_to_str, BoundingBox\n\nclass TrackLayer(BaseLayer):\n\n def __init__(self, dataset, bbox=BoundingBox.WORLD):\n self.data = dataset\n self.cmap = colorbrewer(self.data['hex_ident'], alpha=200)\n self.time = self.data['timestamp'].min()\n self.painter = BatchPainter()\n self.view = bbox\n\n\n def draw(self, proj, mouse_x, mouse_y, ui_manager):\n self.painter = BatchPainter()\n df = self.data.where((self.data['timestamp'] > self.time) & (self.data['timestamp'] <= self.time + 180))\n\n for element in set(df['hex_ident']):\n grp = df.where(df['hex_ident'] == element)\n self.painter.set_color(self.cmap[element])\n x, y = proj.lonlat_to_screen(grp['lon'], grp['lat'])\n self.painter.points(x, y, 15, rounded=True)\n\n self.time += 1\n\n if self.time > self.data['timestamp'].max():\n self.time = self.data['timestamp'].min()\n\n self.painter.batch_draw()\n ui_manager.info('Current timestamp: {}'.format(epoch_to_str(self.time)))\n \n # bounding box that gets used when layer is created\n def bbox(self):\n return self.view",
"_____no_output_____"
]
],
[
[
"---",
"_____no_output_____"
],
[
"#### Visualization with of the custom layer",
"_____no_output_____"
],
[
"After creating the custom layer, using it is as simple as using any other layer in geoplotlib. \nWe can use the `add_layer` method and pass in our custom layer class with the parameters needed.",
"_____no_output_____"
],
[
"Our data is focused on the UK and specifically Leeds. \nSo we want to adjust our bounding box to exactly this area.",
"_____no_output_____"
]
],
[
[
"# bounding box for our view on leeds\nfrom geoplotlib.utils import BoundingBox\n\nleeds_bbox = BoundingBox(north=53.8074, west=-3, south=53.7074 , east=0)",
"_____no_output_____"
]
],
[
[
"Use Geoplotlib to convert any pandas DataFrame into a DataAccessObject.",
"_____no_output_____"
]
],
[
[
"# displaying our custom layer using add_layer\nfrom geoplotlib.utils import DataAccessObject\n\ndata = DataAccessObject(dataset)\n\ngeoplotlib.add_layer(TrackLayer(data, bbox=leeds_bbox))\ngeoplotlib.show()",
"_____no_output_____"
]
],
[
[
"**Note:** \nIn order to avoid any errors associated with the library, we have to convert our pandas dataframe to a geoplotlib DataAccessObject. \nThe creator of geoplotlib provides a handy interface for this conversion.",
"_____no_output_____"
],
[
"When looking at the upper right hand corner, we can clearly see the temporal aspect of this visualization. \nThe first observation we make is that our data is really sparse, we sometimes only have a single data point for a plane, seldomly a whole path is drawn. \n\nEven though it is so sparse, we can already get a feeling about where the planes are flying most.\n\n**Note:** \nIf you're interested in what else can be achieved with this custom layer approach, there are more examples in the geoplotlib repository. \n- https://github.com/andrea-cuttone/geoplotlib/blob/master/examples/follow_camera.py\n- https://github.com/andrea-cuttone/geoplotlib/blob/master/examples/quadtree.py\n- https://github.com/andrea-cuttone/geoplotlib/blob/master/examples/kmeans.py",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
c51103321555f9150d937bd51c80fc3c018a5103
| 20,254 |
ipynb
|
Jupyter Notebook
|
figures/compile-LSA-zscores-perMOA.ipynb
|
broadinstitute/cell-painting-vae
|
7c1762b9e8d4d1318130924f0cd7d8a275a3a539
|
[
"BSD-3-Clause"
] | 3 |
2021-09-19T15:19:07.000Z
|
2022-03-22T09:46:47.000Z
|
figures/compile-LSA-zscores-perMOA.ipynb
|
broadinstitute/cell-painting-vae
|
7c1762b9e8d4d1318130924f0cd7d8a275a3a539
|
[
"BSD-3-Clause"
] | 4 |
2020-07-23T20:14:54.000Z
|
2022-01-13T16:41:13.000Z
|
figures/compile-LSA-zscores-perMOA.ipynb
|
broadinstitute/cell-painting-vae
|
7c1762b9e8d4d1318130924f0cd7d8a275a3a539
|
[
"BSD-3-Clause"
] | 2 |
2020-07-17T22:55:08.000Z
|
2020-07-23T18:56:21.000Z
| 32.82658 | 112 | 0.434334 |
[
[
[
"## Compile per MOA p value for shuffled comparison",
"_____no_output_____"
]
],
[
[
"import pathlib\nimport numpy as np\nimport pandas as pd\nimport scipy.stats",
"_____no_output_____"
],
[
"# Load L2 distances per MOA\ncp_l2_file = pathlib.Path(\"..\", \"cell-painting\", \"3.application\", \"L2_distances_with_moas.csv\")\ncp_l2_df = pd.read_csv(cp_l2_file).assign(shuffled=\"real\")\n\ncp_l2_df.loc[cp_l2_df.Model.str.contains(\"Shuffled\"), \"shuffled\"] = \"shuffled\"\ncp_l2_df = cp_l2_df.assign(\n architecture=[x[-1] for x in cp_l2_df.Model.str.split(\" \")],\n assay=\"CellPainting\",\n metric=\"L2 distance\"\n).rename(columns={\"L2 Distance\": \"metric_value\"})\n\nprint(cp_l2_df.shape)\ncp_l2_df.head()",
"(4620, 7)\n"
],
[
"# Load Pearson correlations per MOA\ncp_file = pathlib.Path(\"..\", \"cell-painting\", \"3.application\", \"pearson_with_moas.csv\")\ncp_pearson_df = pd.read_csv(cp_file).assign(shuffled=\"real\")\n\ncp_pearson_df.loc[cp_pearson_df.Model.str.contains(\"Shuffled\"), \"shuffled\"] = \"shuffled\"\ncp_pearson_df = cp_pearson_df.assign(\n architecture=[x[-1] for x in cp_pearson_df.Model.str.split(\" \")],\n assay=\"CellPainting\",\n metric=\"Pearson correlation\"\n).rename(columns={\"Pearson\": \"metric_value\"})\n\nprint(cp_pearson_df.shape)\ncp_pearson_df.head()",
"(4620, 7)\n"
],
[
"# Combine data\ncp_df = pd.concat([cp_l2_df, cp_pearson_df]).reset_index(drop=True)\n\nprint(cp_df.shape)\ncp_df.head()",
"(9240, 7)\n"
],
[
"all_moas = cp_df.MOA.unique().tolist()\nprint(len(all_moas))\nall_metrics = cp_df.metric.unique().tolist()\nall_architectures = cp_df.architecture.unique().tolist()\nall_architectures",
"84\n"
],
[
"results_df = []\nfor metric in all_metrics:\n for moa in all_moas:\n for arch in all_architectures:\n # subset data to include moa per architecture\n sub_cp_df = (\n cp_df\n .query(f\"metric == '{metric}'\")\n .query(f\"architecture == '{arch}'\")\n .query(f\"MOA == '{moa}'\")\n .reset_index(drop=True)\n )\n\n real_ = sub_cp_df.query(\"shuffled == 'real'\").loc[:, \"metric_value\"].tolist()\n shuff_ = sub_cp_df.query(\"shuffled != 'real'\").loc[:, \"metric_value\"].tolist()\n\n # Calculate zscore consistently with other experiments\n zscore_result = scipy.stats.zscore(shuff_ + real_)[-1]\n results_df.append([moa, arch, zscore_result, metric])\n\n# Compile results\nresults_df = pd.DataFrame(results_df, columns=[\"MOA\", \"model\", \"zscore\", \"metric\"])\n\nprint(results_df.shape)\nresults_df.head()",
"(840, 4)\n"
],
[
"# Output data\noutput_file = pathlib.Path(\"data\", \"MOA_LSA_metrics.tsv\")\nresults_df.to_csv(output_file, sep=\"\\t\", index=False)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
c5111324787b97f73328dfccf961cf516396cb85
| 3,698 |
ipynb
|
Jupyter Notebook
|
python/to_black_and_white.ipynb
|
Giorat/pocketbotanist
|
1d918bcead6cbe2822f9c04c42a62f315aabceb9
|
[
"MIT"
] | 3 |
2018-06-08T05:44:16.000Z
|
2019-02-27T19:28:04.000Z
|
python/to_black_and_white.ipynb
|
riccardogiorato/pocketbotanist
|
1d918bcead6cbe2822f9c04c42a62f315aabceb9
|
[
"MIT"
] | 8 |
2018-06-08T06:43:45.000Z
|
2019-05-08T06:37:28.000Z
|
python/to_black_and_white.ipynb
|
riccardogiorato/pocketbotanist
|
1d918bcead6cbe2822f9c04c42a62f315aabceb9
|
[
"MIT"
] | null | null | null | 39.763441 | 208 | 0.604922 |
[
[
[
"!pip install scikit-image",
"Requirement already satisfied: scikit-image in c:\\users\\giora\\appdata\\local\\programs\\python\\python36\\lib\\site-packages (0.14.0)\nRequirement already satisfied: six>=1.10.0 in c:\\users\\giora\\appdata\\local\\programs\\python\\python36\\lib\\site-packages (from scikit-image) (1.11.0)\nRequirement already satisfied: dask[array]>=0.9.0 in c:\\users\\giora\\appdata\\local\\programs\\python\\python36\\lib\\site-packages (from scikit-image) (0.17.5)\nRequirement already satisfied: PyWavelets>=0.4.0 in c:\\users\\giora\\appdata\\local\\programs\\python\\python36\\lib\\site-packages (from scikit-image) (0.5.2)\nRequirement already satisfied: pillow>=4.3.0 in c:\\users\\giora\\appdata\\local\\programs\\python\\python36\\lib\\site-packages (from scikit-image) (5.1.0)\nRequirement already satisfied: networkx>=1.8 in c:\\users\\giora\\appdata\\local\\programs\\python\\python36\\lib\\site-packages (from scikit-image) (2.1)\nRequirement already satisfied: cloudpickle>=0.2.1 in c:\\users\\giora\\appdata\\local\\programs\\python\\python36\\lib\\site-packages (from scikit-image) (0.5.3)\nRequirement already satisfied: numpy>=1.11.0; extra == \"array\" in c:\\users\\giora\\appdata\\local\\programs\\python\\python36\\lib\\site-packages (from dask[array]>=0.9.0->scikit-image) (1.14.1)\nRequirement already satisfied: toolz>=0.7.3; extra == \"array\" in c:\\users\\giora\\appdata\\local\\programs\\python\\python36\\lib\\site-packages (from dask[array]>=0.9.0->scikit-image) (0.9.0)\nRequirement already satisfied: decorator>=4.1.0 in c:\\users\\giora\\appdata\\local\\programs\\python\\python36\\lib\\site-packages (from networkx>=1.8->scikit-image) (4.3.0)\n"
],
[
"from skimage import io, color\nimport os\nimport imghdr\n\nsource = r'flowers\\waterlily' # Source Folder\ndestination = r'bw\\waterlily' # Destination Folder\n\nimage_files = [os.path.join(root, filename) \n for root, dirs, files in os.walk(source) \n for filename in files \n if imghdr.what(os.path.join(root, filename))]\n\nfor fn in image_files:\n rgb = io.imread(fn)\n grey = color.rgb2gray(rgb)\n head, tail = os.path.split(fn)\n io.imsave(os.path.join(destination, tail), grey)",
"c:\\users\\giora\\appdata\\local\\programs\\python\\python36\\lib\\site-packages\\skimage\\util\\dtype.py:130: UserWarning: Possible precision loss when converting from float64 to uint8\n .format(dtypeobj_in, dtypeobj_out))\n"
]
]
] |
[
"code"
] |
[
[
"code",
"code"
]
] |
c5112e5575d4112259fd762644b14182423060ff
| 127,885 |
ipynb
|
Jupyter Notebook
|
fraud-detection/notebooks/yet_another_model.ipynb
|
thinkmorestupidless/pipelines-model-serving
|
c6e87794396e71ee8df44138888692a6a34fe604
|
[
"Apache-2.0"
] | null | null | null |
fraud-detection/notebooks/yet_another_model.ipynb
|
thinkmorestupidless/pipelines-model-serving
|
c6e87794396e71ee8df44138888692a6a34fe604
|
[
"Apache-2.0"
] | null | null | null |
fraud-detection/notebooks/yet_another_model.ipynb
|
thinkmorestupidless/pipelines-model-serving
|
c6e87794396e71ee8df44138888692a6a34fe604
|
[
"Apache-2.0"
] | null | null | null | 70.421256 | 22,608 | 0.715541 |
[
[
[
"# This Python 3 environment comes with many helpful analytics libraries installed\n# It is defined by the kaggle/python docker image: https://github.com/kaggle/docker-python\n# For example, here's several helpful packages to load in \n\nimport numpy as np # linear algebra\nimport pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)\n\n# Input data files are available in the \"../input/\" directory.\n# For example, running this (by clicking run or pressing Shift+Enter) will list the files in the input directory\n\nfrom subprocess import check_output\nprint(check_output([\"ls\", \"./data\"]).decode(\"utf8\"))\n\n# Any results you write to the current directory are saved as output.\n\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nfrom sklearn.preprocessing import MinMaxScaler\nfrom sklearn.metrics import accuracy_score,precision_score,recall_score,f1_score\\\n ,fbeta_score,classification_report,confusion_matrix,precision_recall_curve,roc_auc_score\\\n ,roc_curve",
"creditcard.csv\n\n"
],
[
"df_full = pd.read_csv('./data/creditcard.csv')\ndf_full.head()",
"_____no_output_____"
],
[
"df_full.Class.value_counts()",
"_____no_output_____"
],
[
"df_full.sort_values(by='Class', ascending=False, inplace=True) #easier for stratified sampling\ndf_full.drop('Time', axis=1, inplace = True)",
"_____no_output_____"
],
[
"df_full.head()",
"_____no_output_____"
],
[
"df_sample = df_full.iloc[:3000,:]\ndf_sample.Class.value_counts()",
"_____no_output_____"
],
[
"feature = np.array(df_sample.values[:,0:29])\nlabel = np.array(df_sample.values[:,-1])",
"_____no_output_____"
],
[
"from sklearn.utils import shuffle\n\nshuffle_df = shuffle(df_sample, random_state=42)\n\ndf_train = shuffle_df[0:2400]\ndf_test = shuffle_df[2400:]",
"_____no_output_____"
],
[
"train_feature = np.array(df_train.values[:,0:29])\ntrain_label = np.array(df_train.values[:,-1])\ntest_feature = np.array(df_test.values[:,0:29])\ntest_label = np.array(df_test.values[:,-1])",
"_____no_output_____"
],
[
"train_feature.shape",
"_____no_output_____"
],
[
"train_label.shape",
"_____no_output_____"
],
[
"from sklearn.preprocessing import MinMaxScaler\n\nscaler = MinMaxScaler()\n\nscaler.fit(train_feature)\ntrain_feature_trans = scaler.transform(train_feature)\ntest_feature_trans = scaler.transform(test_feature)",
"_____no_output_____"
],
[
"from keras.models import Sequential\nfrom keras.layers import Dense\nfrom keras.layers import Dropout\n\nimport matplotlib.pyplot as plt \ndef show_train_history(train_history,train,validation):\n plt.plot(train_history.history[train])\n plt.plot(train_history.history[validation])\n plt.title('Train History')\n plt.ylabel(train)\n plt.xlabel('Epoch')\n plt.legend(['train', 'validation'], loc='best')\n plt.show()\n\nmodel = Sequential() #一層一層到底,按順序\n\n#輸入層(隱藏層1)\nmodel.add(Dense(units=200, \n input_dim=29, \n kernel_initializer='uniform', \n activation='relu'))\nmodel.add(Dropout(0.5))\n\n#隱藏層2,不用寫input_dim,因為就是前一層的units\nmodel.add(Dense(units=200, \n kernel_initializer='uniform', \n activation='relu'))\nmodel.add(Dropout(0.5))\n\n#輸出層\nmodel.add(Dense(units=1, #輸出一個數字 \n kernel_initializer='uniform',\n activation='sigmoid'))\n\nprint(model.summary()) #可以清楚看到model還有參數數量\n\nmodel.compile(loss='binary_crossentropy', #二元用binary\n optimizer='adam', metrics=['accuracy'])\n\ntrain_history = model.fit(x=train_feature_trans, y=train_label, #上面多分割一步在keras是內建的\n validation_split=0.8, epochs=200, \n batch_size=500, verbose=2) #verbose=2表示顯示訓練過程\n\n######################### 訓練過程視覺化\nshow_train_history(train_history,'acc','val_acc')\nshow_train_history(train_history,'loss','val_loss')\n\n\n######################### 實際測驗得分\nscores = model.evaluate(test_feature_trans, test_label)\nprint('\\n')\nprint('accuracy=',scores[1])\n\n######################### 紀錄模型預測情形(答案卷)\nprediction = model.predict_classes(test_feature_trans)\n\n#儲存訓練結果\nmodel.save_weights(\"Keras_CreditCardFraud_MLP.h5\")\nprint('model saved to disk')",
"Using TensorFlow backend.\nWARNING: Logging before flag parsing goes to stderr.\nW1007 11:34:22.086395 4668208576 deprecation_wrapper.py:119] From /Users/trevorburton-mccreadie/anaconda3/envs/fraud/lib/python3.7/site-packages/keras/backend/tensorflow_backend.py:74: The name tf.get_default_graph is deprecated. Please use tf.compat.v1.get_default_graph instead.\n\nW1007 11:34:22.120425 4668208576 deprecation_wrapper.py:119] From /Users/trevorburton-mccreadie/anaconda3/envs/fraud/lib/python3.7/site-packages/keras/backend/tensorflow_backend.py:517: The name tf.placeholder is deprecated. Please use tf.compat.v1.placeholder instead.\n\nW1007 11:34:22.123545 4668208576 deprecation_wrapper.py:119] From /Users/trevorburton-mccreadie/anaconda3/envs/fraud/lib/python3.7/site-packages/keras/backend/tensorflow_backend.py:4138: The name tf.random_uniform is deprecated. Please use tf.random.uniform instead.\n\nW1007 11:34:22.145820 4668208576 deprecation_wrapper.py:119] From /Users/trevorburton-mccreadie/anaconda3/envs/fraud/lib/python3.7/site-packages/keras/backend/tensorflow_backend.py:133: The name tf.placeholder_with_default is deprecated. Please use tf.compat.v1.placeholder_with_default instead.\n\nW1007 11:34:22.157491 4668208576 deprecation.py:506] From /Users/trevorburton-mccreadie/anaconda3/envs/fraud/lib/python3.7/site-packages/keras/backend/tensorflow_backend.py:3445: calling dropout (from tensorflow.python.ops.nn_ops) with keep_prob is deprecated and will be removed in a future version.\nInstructions for updating:\nPlease use `rate` instead of `keep_prob`. Rate should be set to `rate = 1 - keep_prob`.\nW1007 11:34:22.232460 4668208576 deprecation_wrapper.py:119] From /Users/trevorburton-mccreadie/anaconda3/envs/fraud/lib/python3.7/site-packages/keras/optimizers.py:790: The name tf.train.Optimizer is deprecated. Please use tf.compat.v1.train.Optimizer instead.\n\nW1007 11:34:22.263739 4668208576 deprecation_wrapper.py:119] From /Users/trevorburton-mccreadie/anaconda3/envs/fraud/lib/python3.7/site-packages/keras/backend/tensorflow_backend.py:3376: The name tf.log is deprecated. Please use tf.math.log instead.\n\nW1007 11:34:22.356359 4668208576 deprecation.py:323] From /Users/trevorburton-mccreadie/anaconda3/envs/fraud/lib/python3.7/site-packages/tensorflow/python/ops/nn_impl.py:180: add_dispatch_support.<locals>.wrapper (from tensorflow.python.ops.array_ops) is deprecated and will be removed in a future version.\nInstructions for updating:\nUse tf.where in 2.0, which has the same broadcast rule as np.where\n"
],
[
"df_ans = pd.DataFrame({'Real Class' :test_label})\ndf_ans['Prediction'] = prediction",
"_____no_output_____"
],
[
"df_ans[ df_ans['Real Class'] != df_ans['Prediction'] ]",
"_____no_output_____"
],
[
"df_ans['Prediction'].value_counts()",
"_____no_output_____"
],
[
"df_ans['Real Class'].value_counts()",
"_____no_output_____"
],
[
"import seaborn as sns\n%matplotlib inline\n\ncols = ['Real_Class_1','Real_Class_0'] #Gold standard\nrows = ['Prediction_1','Prediction_0'] #diagnostic tool (our prediction)\n\nB1P1 = len(df_ans[(df_ans['Prediction'] == df_ans['Real Class']) & (df_ans['Real Class'] == 1)])\nB1P0 = len(df_ans[(df_ans['Prediction'] != df_ans['Real Class']) & (df_ans['Real Class'] == 1)])\nB0P1 = len(df_ans[(df_ans['Prediction'] != df_ans['Real Class']) & (df_ans['Real Class'] == 0)])\nB0P0 = len(df_ans[(df_ans['Prediction'] == df_ans['Real Class']) & (df_ans['Real Class'] == 0)])\n\nconf = np.array([[B1P1,B0P1],[B1P0,B0P0]])\ndf_cm = pd.DataFrame(conf, columns = [i for i in cols], index = [i for i in rows])\n\nf, ax= plt.subplots(figsize = (5, 5))\nsns.heatmap(df_cm, annot=True, ax=ax, fmt='d') \nax.xaxis.set_ticks_position('top') #Making x label be on top is common in textbooks.\n\nprint('total test case number: ', np.sum(conf))",
"total test case number: 600\n"
],
[
"def model_efficacy(conf):\n total_num = np.sum(conf)\n sen = conf[0][0]/(conf[0][0]+conf[1][0])\n spe = conf[1][1]/(conf[1][0]+conf[1][1])\n false_positive_rate = conf[0][1]/(conf[0][1]+conf[1][1])\n false_negative_rate = conf[1][0]/(conf[0][0]+conf[1][0])\n \n print('total_num: ',total_num)\n print('G1P1: ',conf[0][0]) #G = gold standard; P = prediction\n print('G0P1: ',conf[0][1])\n print('G1P0: ',conf[1][0])\n print('G0P0: ',conf[1][1])\n print('##########################')\n print('sensitivity: ',sen)\n print('specificity: ',spe)\n print('false_positive_rate: ',false_positive_rate)\n print('false_negative_rate: ',false_negative_rate)\n \n return total_num, sen, spe, false_positive_rate, false_negative_rate\n\nmodel_efficacy(conf)",
"total_num: 600\nG1P1: 74\nG0P1: 5\nG1P0: 7\nG0P0: 514\n##########################\nsensitivity: 0.9135802469135802\nspecificity: 0.9865642994241842\nfalse_positive_rate: 0.009633911368015413\nfalse_negative_rate: 0.08641975308641975\n"
],
[
"df_sample2 = df_full.iloc[:,:] #由於都是label=0,就不shuffle了\n\nfeature2 = np.array(df_sample2.values[:,0:29])\nlabel2 = np.array(df_sample2.values[:,-1])\n\nfeature2_trans = scaler.transform(feature2) #using the same scaler as above\n\n######################### 實際測驗得分\nscores = model.evaluate(feature2_trans, label2)\nprint('\\n')\nprint('accuracy=',scores[1])\n\n######################### 紀錄模型預測情形(答案卷)\nprediction2 = model.predict_classes(feature2_trans)",
"284807/284807 [==============================] - 11s 39us/step\n\n\naccuracy= 0.9864504734785311\n"
],
[
"prediction2_list = prediction2.reshape(-1).astype(int)\nlabel2_list = label2.astype(int)\n\nprint(classification_report(label2_list, prediction2_list))\nprint(confusion_matrix(label2_list, prediction2_list))",
" precision recall f1-score support\n\n 0 1.00 0.99 0.99 284315\n 1 0.10 0.88 0.18 492\n\n accuracy 0.99 284807\n macro avg 0.55 0.93 0.59 284807\nweighted avg 1.00 0.99 0.99 284807\n\n[[280514 3801]\n [ 58 434]]\n"
],
[
"conf = confusion_matrix(label2_list, prediction2_list)\nf, ax= plt.subplots(figsize = (5, 5))\nsns.heatmap(conf, annot=True, ax=ax, fmt='d') \nax.xaxis.set_ticks_position('top') #Making x label be on top is common in textbooks.",
"_____no_output_____"
],
[
"def model_efficacy(conf):\n total_num = np.sum(conf)\n sen = conf[0][0]/(conf[0][0]+conf[1][0])\n spe = conf[1][1]/(conf[1][0]+conf[1][1])\n false_positive_rate = conf[0][1]/(conf[0][1]+conf[1][1])\n false_negative_rate = conf[1][0]/(conf[0][0]+conf[1][0])\n \n print('total_num: ',total_num)\n print('G1P1: ',conf[0][0]) #G = gold standard; P = prediction\n print('G0P1: ',conf[0][1])\n print('G1P0: ',conf[1][0])\n print('G0P0: ',conf[1][1])\n print('##########################')\n print('sensitivity: ',sen)\n print('specificity: ',spe)\n print('false_positive_rate: ',false_positive_rate)\n print('false_negative_rate: ',false_negative_rate)\n \n return total_num, sen, spe, false_positive_rate, false_negative_rate\n\nmodel_efficacy(conf)",
"total_num: 284807\nG1P1: 280514\nG0P1: 3801\nG1P0: 58\nG0P0: 434\n##########################\nsensitivity: 0.9997932794434227\nspecificity: 0.8821138211382114\nfalse_positive_rate: 0.8975206611570248\nfalse_negative_rate: 0.00020672055657727784\n"
],
[
"input_nodes = [node.op.name for node in model.inputs]\noutput_nodes = [node.op.name for node in model.outputs]\n\nprint('input nodes => ' + str(input_nodes))\nprint('output nodes => ' + str(output_nodes))",
"input nodes => ['dense_1_input']\noutput nodes => ['dense_3/Sigmoid']\n"
],
[
"import keras\nimport tensorflow as tf\nfrom tensorflow.python.tools import freeze_graph\nfrom tensorflow.python.tools import optimize_for_inference_lib\n\nMODEL_NAME = 'yetAnotherModel'\nMODEL_PATH = './output/' + MODEL_NAME + '/'\nOPTIMISED_MODEL_PATH = '../pipelines/src/main/resources/models/'\nOPTIMISED_MODEL_NAME = 'optimised_' + MODEL_NAME + '.pb'\nOPTIMISED_MODEL_TXT_NAME = 'optimised_' + MODEL_NAME + '.pbtxt'\n\ncheckpoint_path = MODEL_PATH + MODEL_NAME + '.ckpt'\nmodel_frozen_path = MODEL_PATH + 'frozen_' + MODEL_NAME + '.pb'\n\nsess = keras.backend.get_session()",
"_____no_output_____"
],
[
"# Save produced model\nsaver = tf.train.Saver()\n\nsave_path = saver.save(sess, checkpoint_path)\nprint (\"Saved model at \", save_path)\n\ngraph_path = tf.train.write_graph(sess.graph_def, MODEL_PATH, MODEL_NAME + \".pb\", as_text=True)\nprint (\"Saved graph at :\", graph_path)",
"Saved model at ./output/yetAnotherModel/yetAnotherModel.ckpt\nSaved graph at : ./output/yetAnotherModel/yetAnotherModel.pb\n"
],
[
"# Now freeze the graph (put variables into graph)\n\ninput_saver_def_path = \"\"\ninput_binary = False\noutput_node_names = ', '.join(output_nodes)\nrestore_op_name = \"save/restore_all\"\nfilename_tensor_name = \"save/Const:0\"\nclear_devices = True\n\n\nfreeze_graph.freeze_graph(graph_path, input_saver_def_path,\n input_binary, save_path, output_node_names,\n restore_op_name, filename_tensor_name,\n model_frozen_path, clear_devices, \"\")\n\nprint (\"Model is frozen\")",
"W1007 11:41:05.555653 4668208576 deprecation.py:323] From /Users/trevorburton-mccreadie/anaconda3/envs/fraud/lib/python3.7/site-packages/tensorflow/python/tools/freeze_graph.py:127: checkpoint_exists (from tensorflow.python.training.checkpoint_management) is deprecated and will be removed in a future version.\nInstructions for updating:\nUse standard file APIs to check for files with this prefix.\nW1007 11:41:06.142923 4668208576 deprecation.py:323] From /Users/trevorburton-mccreadie/anaconda3/envs/fraud/lib/python3.7/site-packages/tensorflow/python/tools/freeze_graph.py:233: convert_variables_to_constants (from tensorflow.python.framework.graph_util_impl) is deprecated and will be removed in a future version.\nInstructions for updating:\nUse `tf.compat.v1.graph_util.convert_variables_to_constants`\nW1007 11:41:06.144479 4668208576 deprecation.py:323] From /Users/trevorburton-mccreadie/anaconda3/envs/fraud/lib/python3.7/site-packages/tensorflow/python/framework/graph_util_impl.py:270: extract_sub_graph (from tensorflow.python.framework.graph_util_impl) is deprecated and will be removed in a future version.\nInstructions for updating:\nUse `tf.compat.v1.graph_util.extract_sub_graph`\n"
],
[
"# optimizing graph\n\ninput_graph_def = tf.GraphDef()\nwith tf.gfile.Open(model_frozen_path, \"rb\") as f:\n data = f.read()\n input_graph_def.ParseFromString(data)\n\n\noutput_graph_def = optimize_for_inference_lib.optimize_for_inference(\n input_graph_def,\n input_nodes, # an array of the input node(s)\n output_nodes, # an array of output nodes\n tf.float32.as_datatype_enum)",
"W1007 11:41:27.923722 4668208576 deprecation.py:323] From /Users/trevorburton-mccreadie/anaconda3/envs/fraud/lib/python3.7/site-packages/tensorflow/python/tools/optimize_for_inference_lib.py:113: remove_training_nodes (from tensorflow.python.framework.graph_util_impl) is deprecated and will be removed in a future version.\nInstructions for updating:\nUse `tf.compat.v1.graph_util.remove_training_nodes`\n"
],
[
"tf.train.write_graph(output_graph_def, OPTIMISED_MODEL_PATH, OPTIMISED_MODEL_NAME, as_text=False)",
"_____no_output_____"
],
[
"tf.train.write_graph(output_graph_def, MODEL_PATH, OPTIMISED_MODEL_TXT_NAME, as_text=True)",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
c5113193d4a01d9ded0482a164d06234867856d3
| 17,712 |
ipynb
|
Jupyter Notebook
|
Chapter 1 - Machine Learning Toolkit/Exercise 9 - Lambda Functions.ipynb
|
doc-E-brown/Applied-Supervised-Learning-with-Python
|
f125cecde1af4f77017302c3393acf9c2415ce9a
|
[
"MIT"
] | 2 |
2021-06-08T18:00:07.000Z
|
2021-10-08T06:31:38.000Z
|
Chapter 1 - Machine Learning Toolkit/Exercise 9 - Lambda Functions.ipynb
|
TrainingByPackt/Applied-Supervised-Learning-with-Python
|
f125cecde1af4f77017302c3393acf9c2415ce9a
|
[
"MIT"
] | null | null | null |
Chapter 1 - Machine Learning Toolkit/Exercise 9 - Lambda Functions.ipynb
|
TrainingByPackt/Applied-Supervised-Learning-with-Python
|
f125cecde1af4f77017302c3393acf9c2415ce9a
|
[
"MIT"
] | 16 |
2019-06-04T22:22:17.000Z
|
2022-01-02T06:43:44.000Z
| 32.739372 | 159 | 0.33593 |
[
[
[
"## Exercise 9: Lambda Functions",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\n\ndf = pd.read_csv('titanic.csv')\nembarked_grouped = df.groupby('Embarked')",
"_____no_output_____"
]
],
[
[
"Lambda or inline functions are really useful when applying **agg** as they provide a quick and convenient method of applying the function across the data",
"_____no_output_____"
]
],
[
[
"embarked_grouped.agg(lambda x: x.values[0])",
"_____no_output_____"
]
],
[
[
"We can also apply multiple functions at once:",
"_____no_output_____"
]
],
[
[
"embarked_grouped.agg([lambda x: x.values[0], np.mean, np.std])",
"_____no_output_____"
]
],
[
[
"We can also use a dictionary to specific different functions to apply to different columns:",
"_____no_output_____"
]
],
[
[
"embarked_grouped.agg({\n 'Fare': np.sum,\n 'Age': lambda x: x.values[0]\n})",
"_____no_output_____"
]
],
[
[
"We can also group by more than one column by providing a list of columns",
"_____no_output_____"
]
],
[
[
"age_embarked_grouped = df.groupby(['Sex', 'Embarked'])\nage_embarked_grouped.groups\n",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
c51131eff52badbc39c8999a1b3b86a4ccd963c0
| 139,488 |
ipynb
|
Jupyter Notebook
|
siamese_contrastive_1D.ipynb
|
epigenetic-ml/siamese-sprint
|
ed64f0ea9d5f76327feaff0b6be007b735dc402b
|
[
"Apache-2.0"
] | 1 |
2022-03-03T13:51:39.000Z
|
2022-03-03T13:51:39.000Z
|
siamese_contrastive_1D.ipynb
|
epigenetic-ml/siamese-sprint
|
ed64f0ea9d5f76327feaff0b6be007b735dc402b
|
[
"Apache-2.0"
] | null | null | null |
siamese_contrastive_1D.ipynb
|
epigenetic-ml/siamese-sprint
|
ed64f0ea9d5f76327feaff0b6be007b735dc402b
|
[
"Apache-2.0"
] | null | null | null | 134.51109 | 23,072 | 0.857744 |
[
[
[
"# Image similarity estimation using a Siamese Network with a contrastive loss\n\n**Author:** Mehdi<br>\n**Date created:** 2021/05/06<br>\n**Last modified:** 2021/05/06<br>\n**ORIGINAL SOURCE:** https://github.com/keras-team/keras-io/blob/master/examples/vision/ipynb/siamese_contrastive.ipynb<br>\n**Description:** Similarity learning using a siamese network trained with a contrastive loss.",
"_____no_output_____"
],
[
"### NOTE:\n**We adapted the code for 1D data.**",
"_____no_output_____"
],
[
"## Introduction\n\n[Siamese Networks](https://en.wikipedia.org/wiki/Siamese_neural_network)\nare neural networks which share weights between two or more sister networks,\neach producing embedding vectors of its respective inputs.\n\nIn supervised similarity learning, the networks are then trained to maximize the\ncontrast (distance) between embeddings of inputs of different classes, while minimizing the distance between\nembeddings of similar classes, resulting in embedding spaces that reflect\nthe class segmentation of the training inputs.",
"_____no_output_____"
],
[
"## Setup",
"_____no_output_____"
]
],
[
[
"import random\nimport numpy as np\nimport tensorflow as tf\nfrom tensorflow import keras\nfrom tensorflow.keras import layers\nimport matplotlib.pyplot as plt",
"_____no_output_____"
]
],
[
[
"## Load the MNIST dataset",
"_____no_output_____"
]
],
[
[
"(x_train_val, y_train_val), (x_test_2D, y_test) = keras.datasets.mnist.load_data()",
"_____no_output_____"
],
[
"# Change the data type to a floating point format\nx_train_val = x_train_val.astype(\"float32\")\nx_test_2D = x_test_2D.astype(\"float32\")",
"_____no_output_____"
]
],
[
[
"## Define training and validation sets",
"_____no_output_____"
]
],
[
[
"# Keep 50% of train_val in validation set\nx_train_2D, x_val_2D = x_train_val[:30000], x_train_val[30000:]\ny_train, y_val = y_train_val[:30000], y_train_val[30000:]\ndel x_train_val, y_train_val\n",
"_____no_output_____"
]
],
[
[
"## Convert 2D to 1D ",
"_____no_output_____"
]
],
[
[
"print(x_train_2D.shape, x_val_2D.shape, x_test_2D.shape)",
"(30000, 28, 28) (30000, 28, 28) (10000, 28, 28)\n"
],
[
"# convert 2D image to 1D image\nsize = 28*28\nx_train = x_train_2D.reshape(x_train_2D.shape[0], size)\nx_val = x_val_2D.reshape(x_val_2D.shape[0], size)\nx_test = x_test_2D.reshape(x_test_2D.shape[0], size)",
"_____no_output_____"
],
[
"print(x_train.shape, x_val.shape, x_test.shape)",
"(30000, 784) (30000, 784) (10000, 784)\n"
]
],
[
[
"## Create pairs of images\n\nWe will train the model to differentiate between digits of different classes. For\nexample, digit `0` needs to be differentiated from the rest of the\ndigits (`1` through `9`), digit `1` - from `0` and `2` through `9`, and so on.\nTo carry this out, we will select N random images from class A (for example,\nfor digit `0`) and pair them with N random images from another class B\n(for example, for digit `1`). Then, we can repeat this process for all classes\nof digits (until digit `9`). Once we have paired digit `0` with other digits,\nwe can repeat this process for the remaining classes for the rest of the digits\n(from `1` until `9`).",
"_____no_output_____"
]
],
[
[
"\ndef make_pairs(x, y):\n \"\"\"Creates a tuple containing image pairs with corresponding label.\n\n Arguments:\n x: List containing images, each index in this list corresponds to one image.\n y: List containing labels, each label with datatype of `int`.\n\n Returns:\n Tuple containing two numpy arrays as (pairs_of_samples, labels),\n where pairs_of_samples' shape is (2len(x), 2,n_features_dims) and\n labels are a binary array of shape (2len(x)).\n \"\"\"\n\n num_classes = max(y) + 1\n digit_indices = [np.where(y == i)[0] for i in range(num_classes)]\n\n pairs = []\n labels = []\n\n for idx1 in range(len(x)):\n # add a matching example\n x1 = x[idx1]\n label1 = y[idx1]\n idx2 = random.choice(digit_indices[label1])\n x2 = x[idx2]\n\n pairs += [[x1, x2]]\n labels += [1]\n\n # add a non-matching example\n label2 = random.randint(0, num_classes - 1)\n while label2 == label1:\n label2 = random.randint(0, num_classes - 1)\n\n idx2 = random.choice(digit_indices[label2])\n x2 = x[idx2]\n\n pairs += [[x1, x2]]\n labels += [0]\n\n return np.array(pairs), np.array(labels).astype(\"float32\")\n\n\n# make train pairs\npairs_train, labels_train = make_pairs(x_train, y_train)\n\n# make validation pairs\npairs_val, labels_val = make_pairs(x_val, y_val)\n\n# make test pairs\npairs_test, labels_test = make_pairs(x_test, y_test)",
"_____no_output_____"
],
[
"print(pairs_train.shape, pairs_val.shape, pairs_test.shape)",
"(60000, 2, 784) (60000, 2, 784) (20000, 2, 784)\n"
]
],
[
[
"We get:\n\n- We have 60,000 pairs\n- Each pair contains 2 images\n- Each image has shape `(784)`",
"_____no_output_____"
],
[
"Split the training pairs",
"_____no_output_____"
]
],
[
[
"x_train_1 = pairs_train[:, 0] \nx_train_2 = pairs_train[:, 1]",
"_____no_output_____"
]
],
[
[
"Split the validation pairs",
"_____no_output_____"
]
],
[
[
"x_val_1 = pairs_val[:, 0] \nx_val_2 = pairs_val[:, 1]",
"_____no_output_____"
]
],
[
[
"Split the test pairs",
"_____no_output_____"
]
],
[
[
"x_test_1 = pairs_test[:, 0] # x_test_1.shape = (20000, 784)\nx_test_2 = pairs_test[:, 1]\n",
"_____no_output_____"
]
],
[
[
"## Visualize pairs and their labels",
"_____no_output_____"
]
],
[
[
"\ndef visualize(pairs, labels, to_show=6, num_col=3, predictions=None, test=False):\n \"\"\"Creates a plot of pairs and labels, and prediction if it's test dataset.\n\n Arguments:\n pairs: Numpy Array, of pairs to visualize, having shape\n (Number of pairs, 2, 28, 28).\n to_show: Int, number of examples to visualize (default is 6)\n `to_show` must be an integral multiple of `num_col`.\n Otherwise it will be trimmed if it is greater than num_col,\n and incremented if if it is less then num_col.\n num_col: Int, number of images in one row - (default is 3)\n For test and train respectively, it should not exceed 3 and 7.\n predictions: Numpy Array of predictions with shape (to_show, 1) -\n (default is None)\n Must be passed when test=True.\n test: Boolean telling whether the dataset being visualized is\n train dataset or test dataset - (default False).\n\n Returns:\n None.\n \"\"\"\n\n # Define num_row\n # If to_show % num_col != 0\n # trim to_show,\n # to trim to_show limit num_row to the point where\n # to_show % num_col == 0\n #\n # If to_show//num_col == 0\n # then it means num_col is greater then to_show\n # increment to_show\n # to increment to_show set num_row to 1\n num_row = to_show // num_col if to_show // num_col != 0 else 1\n\n # `to_show` must be an integral multiple of `num_col`\n # we found num_row and we have num_col\n # to increment or decrement to_show\n # to make it integral multiple of `num_col`\n # simply set it equal to num_row * num_col\n to_show = num_row * num_col\n\n # Plot the images\n fig, axes = plt.subplots(num_row, num_col, figsize=(5, 5))\n for i in range(to_show):\n\n # If the number of rows is 1, the axes array is one-dimensional\n if num_row == 1:\n ax = axes[i % num_col]\n else:\n ax = axes[i // num_col, i % num_col]\n\n ax.imshow(tf.concat([pairs[i][0].reshape(28,28), pairs[i][1].reshape(28,28)], axis=1), cmap=\"gray\")\n ax.set_axis_off()\n if test:\n ax.set_title(\"True: {} | Pred: {:.5f}\".format(labels[i], predictions[i][0][0])) # TODO: check shape of predictions\n else:\n ax.set_title(\"Label: {}\".format(labels[i]))\n if test:\n plt.tight_layout(rect=(0, 0, 1.9, 1.9), w_pad=0.0)\n else:\n plt.tight_layout(rect=(0, 0, 1.5, 1.5))\n plt.show()\n",
"_____no_output_____"
]
],
[
[
"Inspect training pairs",
"_____no_output_____"
]
],
[
[
"visualize(pairs_train[:-1], labels_train[:-1], to_show=4, num_col=4)",
"2022-02-02 17:50:02.736125: I tensorflow/core/platform/cpu_feature_guard.cc:151] This TensorFlow binary is optimized with oneAPI Deep Neural Network Library (oneDNN) to use the following CPU instructions in performance-critical operations: AVX2 FMA\nTo enable them in other operations, rebuild TensorFlow with the appropriate compiler flags.\n"
]
],
[
[
"Inspect validation pairs",
"_____no_output_____"
]
],
[
[
"visualize(pairs_val[:-1], labels_val[:-1], to_show=4, num_col=4)",
"_____no_output_____"
]
],
[
[
"Inspect test pairs",
"_____no_output_____"
]
],
[
[
"visualize(pairs_test[:-1], labels_test[:-1], to_show=4, num_col=4)",
"_____no_output_____"
]
],
[
[
"## Define the model\n\nThere are be two input layers, each leading to its own network, which\nproduces embeddings. A `Lambda` layer then merges them using an\n[Euclidean distance](https://en.wikipedia.org/wiki/Euclidean_distance) and the\nmerged output is fed to the final network.",
"_____no_output_____"
]
],
[
[
"# Provided two tensors t1 and t2\n# Euclidean distance = sqrt(sum(square(t1-t2)))\ndef euclidean_distance(vects):\n \"\"\"Find the Euclidean distance between two vectors.\n\n Arguments:\n vects: List containing two tensors of same length.\n\n Returns:\n Tensor containing euclidean distance\n (as floating point value) between vectors.\n \"\"\"\n\n x, y = vects\n sum_square = tf.math.reduce_sum(tf.math.square(x - y), axis=1, keepdims=True)\n return tf.math.sqrt(tf.math.maximum(sum_square, tf.keras.backend.epsilon()))\n",
"_____no_output_____"
],
[
"def make_model(input_shape):\n \n input = layers.Input(input_shape)\n x = tf.keras.layers.BatchNormalization()(input)\n \n x = layers.Conv1D(4, 5, activation=\"tanh\")(x)\n x = layers.AveragePooling1D(pool_size=2)(x)\n \n x = layers.Conv1D(16, 5, activation=\"tanh\")(x)\n x = layers.AveragePooling1D(pool_size=2)(x)\n\n x = layers.Conv1D(32, 5, activation=\"tanh\")(x)\n x = layers.AveragePooling1D(pool_size=2)(x)\n \n x = tf.keras.layers.BatchNormalization()(x)\n x = layers.Dense(10, activation=\"tanh\")(x)\n embedding_network = keras.Model(input, x)\n\n\n input_1 = layers.Input(input_shape)\n input_2 = layers.Input(input_shape)\n\n # As mentioned above, Siamese Network share weights between\n # tower networks (sister networks). To allow this, we will use\n # same embedding network for both tower networks.\n tower_1 = embedding_network(input_1)\n tower_2 = embedding_network(input_2)\n\n merge_layer = layers.Lambda(euclidean_distance)([tower_1, tower_2])\n normal_layer = tf.keras.layers.BatchNormalization()(merge_layer)\n output_layer = layers.Dense(1, activation=\"sigmoid\")(normal_layer)\n siamese = keras.Model(inputs=[input_1, input_2], outputs=output_layer)\n \n return siamese, embedding_network",
"_____no_output_____"
],
[
"siamese, embedding_network = make_model((784,1))",
"_____no_output_____"
],
[
"embedding_network.summary()",
"Model: \"model\"\n_________________________________________________________________\n Layer (type) Output Shape Param # \n=================================================================\n input_1 (InputLayer) [(None, 784, 1)] 0 \n \n batch_normalization (BatchN (None, 784, 1) 4 \n ormalization) \n \n conv1d (Conv1D) (None, 780, 4) 24 \n \n average_pooling1d (AverageP (None, 390, 4) 0 \n ooling1D) \n \n conv1d_1 (Conv1D) (None, 386, 16) 336 \n \n average_pooling1d_1 (Averag (None, 193, 16) 0 \n ePooling1D) \n \n conv1d_2 (Conv1D) (None, 189, 32) 2592 \n \n average_pooling1d_2 (Averag (None, 94, 32) 0 \n ePooling1D) \n \n batch_normalization_1 (Batc (None, 94, 32) 128 \n hNormalization) \n \n dense (Dense) (None, 94, 10) 330 \n \n=================================================================\nTotal params: 3,414\nTrainable params: 3,348\nNon-trainable params: 66\n_________________________________________________________________\n"
],
[
"siamese.summary()",
"Model: \"model_1\"\n__________________________________________________________________________________________________\n Layer (type) Output Shape Param # Connected to \n==================================================================================================\n input_2 (InputLayer) [(None, 784, 1)] 0 [] \n \n input_3 (InputLayer) [(None, 784, 1)] 0 [] \n \n model (Functional) (None, 94, 10) 3414 ['input_2[0][0]', \n 'input_3[0][0]'] \n \n lambda (Lambda) (None, 1, 10) 0 ['model[0][0]', \n 'model[1][0]'] \n \n batch_normalization_2 (BatchNo (None, 1, 10) 40 ['lambda[0][0]'] \n rmalization) \n \n dense_1 (Dense) (None, 1, 1) 11 ['batch_normalization_2[0][0]'] \n \n==================================================================================================\nTotal params: 3,465\nTrainable params: 3,379\nNon-trainable params: 86\n__________________________________________________________________________________________________\n"
]
],
[
[
"## Define the constrastive Loss",
"_____no_output_____"
]
],
[
[
"\ndef loss(margin=1):\n \"\"\"Provides 'constrastive_loss' an enclosing scope with variable 'margin'.\n\n Arguments:\n margin: Integer, defines the baseline for distance for which pairs\n should be classified as dissimilar. - (default is 1).\n\n Returns:\n 'constrastive_loss' function with data ('margin') attached.\n \"\"\"\n\n # Contrastive loss = mean( (1-true_value) * square(prediction) +\n # true_value * square( max(margin-prediction, 0) ))\n def contrastive_loss(y_true, y_pred):\n \"\"\"Calculates the constrastive loss.\n\n Arguments:\n y_true: List of labels, each label is of type float32.\n y_pred: List of predictions of same length as of y_true,\n each label is of type float32.\n\n Returns:\n A tensor containing constrastive loss as floating point value.\n \"\"\"\n\n square_pred = tf.math.square(y_pred)\n margin_square = tf.math.square(tf.math.maximum(margin - (y_pred), 0))\n return tf.math.reduce_mean(\n (1 - y_true) * square_pred + (y_true) * margin_square\n )\n\n return contrastive_loss\n",
"_____no_output_____"
]
],
[
[
"## Hyperparameters",
"_____no_output_____"
]
],
[
[
"epochs = 10\nbatch_size = 16\nmargin = 1 # Margin for constrastive loss.",
"_____no_output_____"
]
],
[
[
"## Compile the model with the contrastive loss",
"_____no_output_____"
]
],
[
[
"siamese.compile(loss=loss(margin=margin), optimizer=\"RMSprop\", metrics=[\"accuracy\"])\n",
"_____no_output_____"
]
],
[
[
"## Train the model",
"_____no_output_____"
]
],
[
[
"history = siamese.fit(\n [x_train_1, x_train_2],\n labels_train,\n validation_data=([x_val_1, x_val_2], labels_val),\n batch_size=batch_size,\n epochs=epochs,\n)",
"Epoch 1/10\n3750/3750 [==============================] - 48s 12ms/step - loss: 0.2509 - accuracy: 0.5030 - val_loss: 0.2501 - val_accuracy: 0.5000\nEpoch 2/10\n3750/3750 [==============================] - 44s 12ms/step - loss: 0.2501 - accuracy: 0.4993 - val_loss: 0.2500 - val_accuracy: 0.5000\nEpoch 3/10\n3750/3750 [==============================] - 44s 12ms/step - loss: 0.2500 - accuracy: 0.4978 - val_loss: 0.2501 - val_accuracy: 0.5000\nEpoch 4/10\n3750/3750 [==============================] - 44s 12ms/step - loss: 0.2500 - accuracy: 0.4995 - val_loss: 0.2501 - val_accuracy: 0.5000\nEpoch 5/10\n3750/3750 [==============================] - 44s 12ms/step - loss: 0.2500 - accuracy: 0.5018 - val_loss: 0.2501 - val_accuracy: 0.5000\nEpoch 6/10\n3750/3750 [==============================] - 44s 12ms/step - loss: 0.2500 - accuracy: 0.5004 - val_loss: 0.2500 - val_accuracy: 0.5000\nEpoch 7/10\n3750/3750 [==============================] - 44s 12ms/step - loss: 0.2500 - accuracy: 0.4980 - val_loss: 0.2500 - val_accuracy: 0.5000\nEpoch 8/10\n3750/3750 [==============================] - 44s 12ms/step - loss: 0.2500 - accuracy: 0.4949 - val_loss: 0.2500 - val_accuracy: 0.5000\nEpoch 9/10\n3750/3750 [==============================] - 45s 12ms/step - loss: 0.2500 - accuracy: 0.4997 - val_loss: 0.2500 - val_accuracy: 0.5000\nEpoch 10/10\n3750/3750 [==============================] - 45s 12ms/step - loss: 0.2500 - accuracy: 0.4981 - val_loss: 0.2500 - val_accuracy: 0.5000\n"
]
],
[
[
"## Visualize results",
"_____no_output_____"
]
],
[
[
"\ndef plt_metric(history, metric, title, has_valid=True):\n \"\"\"Plots the given 'metric' from 'history'.\n\n Arguments:\n history: history attribute of History object returned from Model.fit.\n metric: Metric to plot, a string value present as key in 'history'.\n title: A string to be used as title of plot.\n has_valid: Boolean, true if valid data was passed to Model.fit else false.\n\n Returns:\n None.\n \"\"\"\n plt.plot(history[metric])\n if has_valid:\n plt.plot(history[\"val_\" + metric])\n plt.legend([\"train\", \"validation\"], loc=\"upper left\")\n plt.title(title)\n plt.ylabel(metric)\n plt.xlabel(\"epoch\")\n plt.show()\n\n\n# Plot the accuracy\nplt_metric(history=history.history, metric=\"accuracy\", title=\"Model accuracy\")\n\n# Plot the constrastive loss\nplt_metric(history=history.history, metric=\"loss\", title=\"Constrastive Loss\")",
"_____no_output_____"
]
],
[
[
"## Evaluate the model",
"_____no_output_____"
]
],
[
[
"results = siamese.evaluate([x_test_1, x_test_2], labels_test)\nprint(\"test loss, test acc:\", results)",
"625/625 [==============================] - 3s 4ms/step - loss: 0.2500 - accuracy: 0.5000\ntest loss, test acc: [0.25001052021980286, 0.5]\n"
]
],
[
[
"## Visualize the predictions",
"_____no_output_____"
]
],
[
[
"predictions = siamese.predict([x_test_1, x_test_2])\nvisualize(pairs_test, labels_test, to_show=3, predictions=predictions, test=True)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
c5113fe80b4fc710ec3c2cf4638d5e17514c2592
| 78,331 |
ipynb
|
Jupyter Notebook
|
test/output/data_summary_notebook_properties_2016.ipynb
|
rvuillez/pydqc
|
006fb578601fd9dae45253f4da3b08ec731678cc
|
[
"MIT"
] | 276 |
2017-12-29T11:32:18.000Z
|
2022-03-24T06:25:36.000Z
|
test/output/data_summary_notebook_properties_2016.ipynb
|
rvuillez/pydqc
|
006fb578601fd9dae45253f4da3b08ec731678cc
|
[
"MIT"
] | 17 |
2018-01-02T15:20:12.000Z
|
2020-08-08T23:33:22.000Z
|
test/output/data_summary_notebook_properties_2016.ipynb
|
rvuillez/pydqc
|
006fb578601fd9dae45253f4da3b08ec731678cc
|
[
"MIT"
] | 71 |
2018-01-02T14:16:59.000Z
|
2022-03-15T17:50:55.000Z
| 22.957503 | 129 | 0.526931 |
[
[
[
"empty"
]
]
] |
[
"empty"
] |
[
[
"empty"
]
] |
c51143dde1e56dfafff5b0c86ba90c74ba744ba2
| 54,158 |
ipynb
|
Jupyter Notebook
|
site/en/r2/guide/ragged_tensors.ipynb
|
ecrows/docs
|
d169cb190d4907044f750d669303bd9af6b7650b
|
[
"Apache-2.0"
] | 1 |
2019-09-14T03:09:24.000Z
|
2019-09-14T03:09:24.000Z
|
site/en/r2/guide/ragged_tensors.ipynb
|
ecrows/docs
|
d169cb190d4907044f750d669303bd9af6b7650b
|
[
"Apache-2.0"
] | null | null | null |
site/en/r2/guide/ragged_tensors.ipynb
|
ecrows/docs
|
d169cb190d4907044f750d669303bd9af6b7650b
|
[
"Apache-2.0"
] | 23 |
2019-06-13T16:03:21.000Z
|
2019-09-13T20:37:37.000Z
| 32.507803 | 306 | 0.491654 |
[
[
[
"##### Copyright 2018 The TensorFlow Authors.",
"_____no_output_____"
]
],
[
[
"#@title Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# https://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.",
"_____no_output_____"
]
],
[
[
"# Ragged Tensors\n\n<table class=\"tfo-notebook-buttons\" align=\"left\">\n <td>\n <a target=\"_blank\" href=\"https://www.tensorflow.org/beta/guide/ragged_tensors\"><img src=\"https://www.tensorflow.org/images/tf_logo_32px.png\" />View on TensorFlow.org</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://colab.research.google.com/github/tensorflow/docs/blob/master/site/en/r2/guide/ragged_tensors.ipynb\"><img src=\"https://www.tensorflow.org/images/colab_logo_32px.png\" />Run in Google Colab</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://github.com/tensorflow/docs/blob/master/site/en/r2/guide/ragged_tensors.ipynb\"><img src=\"https://www.tensorflow.org/images/GitHub-Mark-32px.png\" />View source on GitHub</a>\n </td>\n <td>\n <a href=\"https://storage.googleapis.com/tensorflow_docs/site/en/r2/guide/ragged_tensors.ipynb\"><img src=\"https://www.tensorflow.org/images/download_logo_32px.png\" />Download notebook</a>\n </td>\n</table>",
"_____no_output_____"
],
[
"## Setup",
"_____no_output_____"
]
],
[
[
"from __future__ import absolute_import, division, print_function, unicode_literals\n\nimport math\n!pip install tensorflow==2.0.0-beta0\nimport tensorflow as tf",
"_____no_output_____"
]
],
[
[
"## Overview\n\nYour data comes in many shapes; your tensors should too.\n*Ragged tensors* are the TensorFlow equivalent of nested variable-length\nlists. They make it easy to store and process data with non-uniform shapes,\nincluding:\n\n* Variable-length features, such as the set of actors in a movie.\n* Batches of variable-length sequential inputs, such as sentences or video\n clips.\n* Hierarchical inputs, such as text documents that are subdivided into\n sections, paragraphs, sentences, and words.\n* Individual fields in structured inputs, such as protocol buffers.\n\n### What you can do with a ragged tensor\n\nRagged tensors are supported by more than a hundred TensorFlow operations,\nincluding math operations (such as `tf.add` and `tf.reduce_mean`), array operations\n(such as `tf.concat` and `tf.tile`), string manipulation ops (such as\n`tf.substr`), and many others:\n",
"_____no_output_____"
]
],
[
[
"digits = tf.ragged.constant([[3, 1, 4, 1], [], [5, 9, 2], [6], []])\nwords = tf.ragged.constant([[\"So\", \"long\"], [\"thanks\", \"for\", \"all\", \"the\", \"fish\"]])\nprint(tf.add(digits, 3))\nprint(tf.reduce_mean(digits, axis=1))\nprint(tf.concat([digits, [[5, 3]]], axis=0))\nprint(tf.tile(digits, [1, 2]))\nprint(tf.strings.substr(words, 0, 2))",
"_____no_output_____"
]
],
[
[
"There are also a number of methods and operations that are\nspecific to ragged tensors, including factory methods, conversion methods,\nand value-mapping operations.\nFor a list of supported ops, see the `tf.ragged` package\ndocumentation.\n\nAs with normal tensors, you can use Python-style indexing to access specific\nslices of a ragged tensor. For more information, see the section on\n**Indexing** below.",
"_____no_output_____"
]
],
[
[
"print(digits[0]) # First row",
"_____no_output_____"
],
[
"print(digits[:, :2]) # First two values in each row.",
"_____no_output_____"
],
[
"print(digits[:, -2:]) # Last two values in each row.",
"_____no_output_____"
]
],
[
[
"And just like normal tensors, you can use Python arithmetic and comparison\noperators to perform elementwise operations. For more information, see the section on\n**Overloaded Operators** below.",
"_____no_output_____"
]
],
[
[
"print(digits + 3)",
"_____no_output_____"
],
[
"print(digits + tf.ragged.constant([[1, 2, 3, 4], [], [5, 6, 7], [8], []]))",
"_____no_output_____"
]
],
[
[
"If you need to perform an elementwise transformation to the values of a `RaggedTensor`, you can use `tf.ragged.map_flat_values`, which takes a function plus one or more arguments, and applies the function to transform the `RaggedTensor`'s values.",
"_____no_output_____"
]
],
[
[
"times_two_plus_one = lambda x: x * 2 + 1\nprint(tf.ragged.map_flat_values(times_two_plus_one, digits))",
"_____no_output_____"
]
],
[
[
"### Constructing a ragged tensor\n\nThe simplest way to construct a ragged tensor is using\n`tf.ragged.constant`, which builds the\n`RaggedTensor` corresponding to a given nested Python `list`:",
"_____no_output_____"
]
],
[
[
"sentences = tf.ragged.constant([\n [\"Let's\", \"build\", \"some\", \"ragged\", \"tensors\", \"!\"],\n [\"We\", \"can\", \"use\", \"tf.ragged.constant\", \".\"]])\nprint(sentences)",
"_____no_output_____"
],
[
"paragraphs = tf.ragged.constant([\n [['I', 'have', 'a', 'cat'], ['His', 'name', 'is', 'Mat']],\n [['Do', 'you', 'want', 'to', 'come', 'visit'], [\"I'm\", 'free', 'tomorrow']],\n])\nprint(paragraphs)",
"_____no_output_____"
]
],
[
[
"Ragged tensors can also be constructed by pairing flat *values* tensors with\n*row-partitioning* tensors indicating how those values should be divided into\nrows, using factory classmethods such as `tf.RaggedTensor.from_value_rowids`,\n`tf.RaggedTensor.from_row_lengths`, and\n`tf.RaggedTensor.from_row_splits`.\n\n#### `tf.RaggedTensor.from_value_rowids`\nIf you know which row each value belongs in, then you can build a `RaggedTensor` using a `value_rowids` row-partitioning tensor:\n\n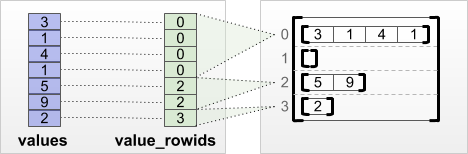",
"_____no_output_____"
]
],
[
[
"print(tf.RaggedTensor.from_value_rowids(\n values=[3, 1, 4, 1, 5, 9, 2, 6],\n value_rowids=[0, 0, 0, 0, 2, 2, 2, 3]))",
"_____no_output_____"
]
],
[
[
"#### `tf.RaggedTensor.from_row_lengths`\n\nIf you know how long each row is, then you can use a `row_lengths` row-partitioning tensor:\n\n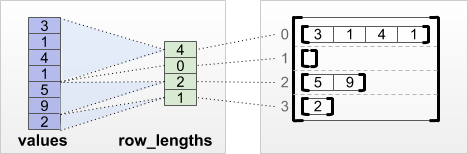",
"_____no_output_____"
]
],
[
[
"print(tf.RaggedTensor.from_row_lengths(\n values=[3, 1, 4, 1, 5, 9, 2, 6],\n row_lengths=[4, 0, 3, 1]))",
"_____no_output_____"
]
],
[
[
"#### `tf.RaggedTensor.from_row_splits`\n\nIf you know the index where each row starts and ends, then you can use a `row_splits` row-partitioning tensor:\n\n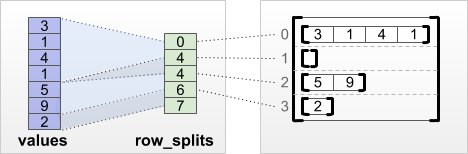",
"_____no_output_____"
]
],
[
[
"print(tf.RaggedTensor.from_row_splits(\n values=[3, 1, 4, 1, 5, 9, 2, 6],\n row_splits=[0, 4, 4, 7, 8]))",
"_____no_output_____"
]
],
[
[
"See the `tf.RaggedTensor` class documentation for a full list of factory methods.",
"_____no_output_____"
],
[
"### What you can store in a ragged tensor\n\nAs with normal `Tensor`s, the values in a `RaggedTensor` must all have the same\ntype; and the values must all be at the same nesting depth (the *rank* of the\ntensor):",
"_____no_output_____"
]
],
[
[
"print(tf.ragged.constant([[\"Hi\"], [\"How\", \"are\", \"you\"]])) # ok: type=string, rank=2",
"_____no_output_____"
],
[
"print(tf.ragged.constant([[[1, 2], [3]], [[4, 5]]])) # ok: type=int32, rank=3",
"_____no_output_____"
],
[
"try:\n tf.ragged.constant([[\"one\", \"two\"], [3, 4]]) # bad: multiple types\nexcept ValueError as exception:\n print(exception)",
"_____no_output_____"
],
[
"try:\n tf.ragged.constant([\"A\", [\"B\", \"C\"]]) # bad: multiple nesting depths\nexcept ValueError as exception:\n print(exception)",
"_____no_output_____"
]
],
[
[
"### Example use case\n\nThe following example demonstrates how `RaggedTensor`s can be used to construct\nand combine unigram and bigram embeddings for a batch of variable-length\nqueries, using special markers for the beginning and end of each sentence.\nFor more details on the ops used in this example, see the `tf.ragged` package documentation.",
"_____no_output_____"
]
],
[
[
"queries = tf.ragged.constant([['Who', 'is', 'Dan', 'Smith'],\n ['Pause'],\n ['Will', 'it', 'rain', 'later', 'today']])\n\n# Create an embedding table.\nnum_buckets = 1024\nembedding_size = 4\nembedding_table = tf.Variable(\n tf.random.truncated_normal([num_buckets, embedding_size],\n stddev=1.0 / math.sqrt(embedding_size)))\n\n# Look up the embedding for each word.\nword_buckets = tf.strings.to_hash_bucket_fast(queries, num_buckets)\nword_embeddings = tf.ragged.map_flat_values(\n tf.nn.embedding_lookup, embedding_table, word_buckets) # ①\n\n# Add markers to the beginning and end of each sentence.\nmarker = tf.fill([queries.nrows(), 1], '#')\npadded = tf.concat([marker, queries, marker], axis=1) # ②\n\n# Build word bigrams & look up embeddings.\nbigrams = tf.strings.join([padded[:, :-1],\n padded[:, 1:]],\n separator='+') # ③\n\nbigram_buckets = tf.strings.to_hash_bucket_fast(bigrams, num_buckets)\nbigram_embeddings = tf.ragged.map_flat_values(\n tf.nn.embedding_lookup, embedding_table, bigram_buckets) # ④\n\n# Find the average embedding for each sentence\nall_embeddings = tf.concat([word_embeddings, bigram_embeddings], axis=1) # ⑤\navg_embedding = tf.reduce_mean(all_embeddings, axis=1) # ⑥\nprint(avg_embedding)",
"_____no_output_____"
]
],
[
[
"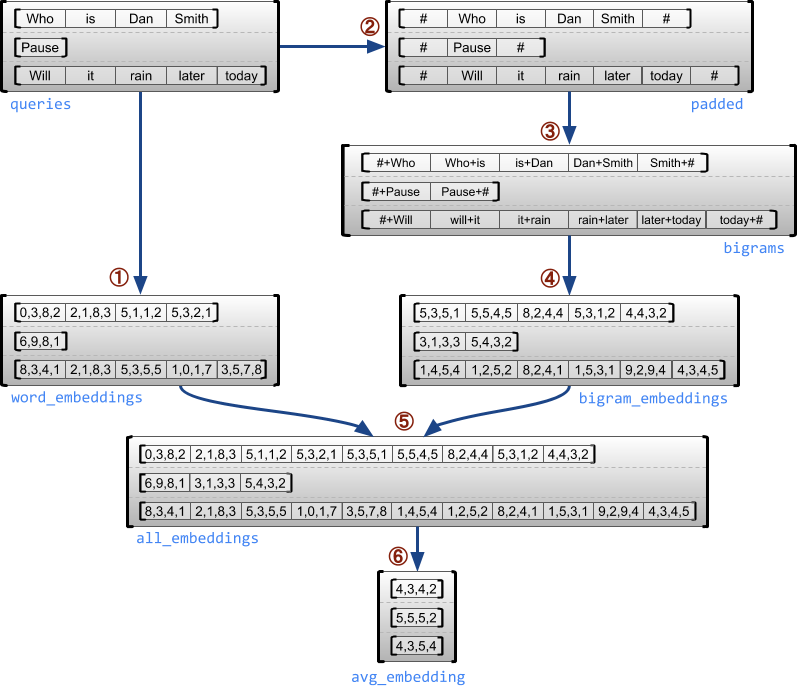",
"_____no_output_____"
],
[
"## Ragged tensors: definitions\n\n### Ragged and uniform dimensions\n\nA *ragged tensor* is a tensor with one or more *ragged dimensions*,\nwhich are dimensions whose slices may have different lengths. For example, the\ninner (column) dimension of `rt=[[3, 1, 4, 1], [], [5, 9, 2], [6], []]` is\nragged, since the column slices (`rt[0, :]`, ..., `rt[4, :]`) have different\nlengths. Dimensions whose slices all have the same length are called *uniform\ndimensions*.\n\nThe outermost dimension of a ragged tensor is always uniform, since it consists\nof a single slice (and so there is no possibility for differing slice lengths).\nIn addition to the uniform outermost dimension, ragged tensors may also have\nuniform inner dimensions. For example, we might store the word embeddings for\neach word in a batch of sentences using a ragged tensor with shape\n`[num_sentences, (num_words), embedding_size]`, where the parentheses around\n`(num_words)` indicate that the dimension is ragged.\n\n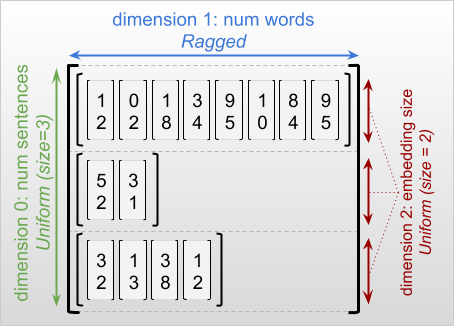\n\nRagged tensors may have multiple ragged dimensions. For example, we could store\na batch of structured text documents using a tensor with shape `[num_documents,\n(num_paragraphs), (num_sentences), (num_words)]` (where again parentheses are\nused to indicate ragged dimensions).\n\n#### Ragged tensor shape restrictions\n\nThe shape of a ragged tensor is currently restricted to have the following form:\n\n* A single uniform dimension\n* Followed by one or more ragged dimensions\n* Followed by zero or more uniform dimensions.\n\nNote: These restrictions are a consequence of the current implementation, and we\nmay relax them in the future.\n\n### Rank and ragged rank\n\nThe total number of dimensions in a ragged tensor is called its ***rank***, and\nthe number of ragged dimensions in a ragged tensor is called its ***ragged\nrank***. In graph execution mode (i.e., non-eager mode), a tensor's ragged rank\nis fixed at creation time: it can't depend\non runtime values, and can't vary dynamically for different session runs.\nA ***potentially ragged tensor*** is a value that might be\neither a `tf.Tensor` or a `tf.RaggedTensor`. The\nragged rank of a `tf.Tensor` is defined to be zero.\n\n### RaggedTensor shapes\n\nWhen describing the shape of a RaggedTensor, ragged dimensions are indicated by\nenclosing them in parentheses. For example, as we saw above, the shape of a 3-D\nRaggedTensor that stores word embeddings for each word in a batch of sentences\ncan be written as `[num_sentences, (num_words), embedding_size]`.\nThe `RaggedTensor.shape` attribute returns a `tf.TensorShape` for a\nragged tensor, where ragged dimensions have size `None`:\n",
"_____no_output_____"
]
],
[
[
"tf.ragged.constant([[\"Hi\"], [\"How\", \"are\", \"you\"]]).shape",
"_____no_output_____"
]
],
[
[
"The method `tf.RaggedTensor.bounding_shape` can be used to find a tight\nbounding shape for a given `RaggedTensor`:",
"_____no_output_____"
]
],
[
[
"print(tf.ragged.constant([[\"Hi\"], [\"How\", \"are\", \"you\"]]).bounding_shape())",
"_____no_output_____"
]
],
[
[
"## Ragged vs sparse tensors\n\nA ragged tensor should *not* be thought of as a type of sparse tensor, but\nrather as a dense tensor with an irregular shape.\n\nAs an illustrative example, consider how array operations such as `concat`,\n`stack`, and `tile` are defined for ragged vs. sparse tensors. Concatenating\nragged tensors joins each row to form a single row with the combined length:\n\n\n",
"_____no_output_____"
]
],
[
[
"ragged_x = tf.ragged.constant([[\"John\"], [\"a\", \"big\", \"dog\"], [\"my\", \"cat\"]])\nragged_y = tf.ragged.constant([[\"fell\", \"asleep\"], [\"barked\"], [\"is\", \"fuzzy\"]])\nprint(tf.concat([ragged_x, ragged_y], axis=1))",
"_____no_output_____"
]
],
[
[
"\nBut concatenating sparse tensors is equivalent to concatenating the corresponding dense tensors,\nas illustrated by the following example (where Ø indicates missing values):\n\n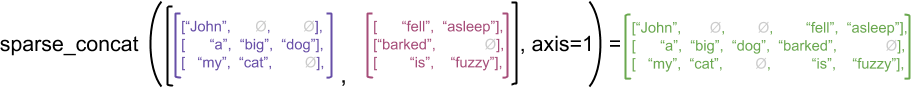\n",
"_____no_output_____"
]
],
[
[
"sparse_x = ragged_x.to_sparse()\nsparse_y = ragged_y.to_sparse()\nsparse_result = tf.sparse.concat(sp_inputs=[sparse_x, sparse_y], axis=1)\nprint(tf.sparse.to_dense(sparse_result, ''))",
"_____no_output_____"
]
],
[
[
"For another example of why this distinction is important, consider the\ndefinition of “the mean value of each row” for an op such as `tf.reduce_mean`.\nFor a ragged tensor, the mean value for a row is the sum of the\nrow’s values divided by the row’s width.\nBut for a sparse tensor, the mean value for a row is the sum of the\nrow’s values divided by the sparse tensor’s overall width (which is\ngreater than or equal to the width of the longest row).\n",
"_____no_output_____"
],
[
"## Overloaded operators\n\nThe `RaggedTensor` class overloads the standard Python arithmetic and comparison\noperators, making it easy to perform basic elementwise math:",
"_____no_output_____"
]
],
[
[
"x = tf.ragged.constant([[1, 2], [3], [4, 5, 6]])\ny = tf.ragged.constant([[1, 1], [2], [3, 3, 3]])\nprint(x + y)",
"_____no_output_____"
]
],
[
[
"Since the overloaded operators perform elementwise computations, the inputs to\nall binary operations must have the same shape, or be broadcastable to the same\nshape. In the simplest broadcasting case, a single scalar is combined\nelementwise with each value in a ragged tensor:",
"_____no_output_____"
]
],
[
[
"x = tf.ragged.constant([[1, 2], [3], [4, 5, 6]])\nprint(x + 3)",
"_____no_output_____"
]
],
[
[
"For a discussion of more advanced cases, see the section on\n**Broadcasting**.\n\nRagged tensors overload the same set of operators as normal `Tensor`s: the unary\noperators `-`, `~`, and `abs()`; and the binary operators `+`, `-`, `*`, `/`,\n`//`, `%`, `**`, `&`, `|`, `^`, `<`, `<=`, `>`, and `>=`. Note that, as with\nstandard `Tensor`s, binary `==` is not overloaded; you can use\n`tf.equal()` to check elementwise equality.",
"_____no_output_____"
],
[
"## Indexing\n\nRagged tensors support Python-style indexing, including multidimensional\nindexing and slicing. The following examples demonstrate ragged tensor indexing\nwith a 2-D and a 3-D ragged tensor.\n\n### Indexing a 2-D ragged tensor with 1 ragged dimension",
"_____no_output_____"
]
],
[
[
"queries = tf.ragged.constant(\n [['Who', 'is', 'George', 'Washington'],\n ['What', 'is', 'the', 'weather', 'tomorrow'],\n ['Goodnight']])\nprint(queries[1])",
"_____no_output_____"
],
[
"print(queries[1, 2]) # A single word",
"_____no_output_____"
],
[
"print(queries[1:]) # Everything but the first row",
"_____no_output_____"
],
[
"print(queries[:, :3]) # The first 3 words of each query",
"_____no_output_____"
],
[
"print(queries[:, -2:]) # The last 2 words of each query",
"_____no_output_____"
]
],
[
[
"### Indexing a 3-D ragged tensor with 2 ragged dimensions",
"_____no_output_____"
]
],
[
[
"rt = tf.ragged.constant([[[1, 2, 3], [4]],\n [[5], [], [6]],\n [[7]],\n [[8, 9], [10]]])",
"_____no_output_____"
],
[
"print(rt[1]) # Second row (2-D RaggedTensor)",
"_____no_output_____"
],
[
"print(rt[3, 0]) # First element of fourth row (1-D Tensor)",
"_____no_output_____"
],
[
"print(rt[:, 1:3]) # Items 1-3 of each row (3-D RaggedTensor)",
"_____no_output_____"
],
[
"print(rt[:, -1:]) # Last item of each row (3-D RaggedTensor)",
"_____no_output_____"
]
],
[
[
"`RaggedTensor`s supports multidimensional indexing and slicing, with one\nrestriction: indexing into a ragged dimension is not allowed. This case is\nproblematic because the indicated value may exist in some rows but not others.\nIn such cases, it's not obvious whether we should (1) raise an `IndexError`; (2)\nuse a default value; or (3) skip that value and return a tensor with fewer rows\nthan we started with. Following the\n[guiding principles of Python](https://www.python.org/dev/peps/pep-0020/)\n(\"In the face\nof ambiguity, refuse the temptation to guess\" ), we currently disallow this\noperation.",
"_____no_output_____"
],
[
"## Tensor Type Conversion\n\nThe `RaggedTensor` class defines methods that can be used to convert\nbetween `RaggedTensor`s and `tf.Tensor`s or `tf.SparseTensors`:",
"_____no_output_____"
]
],
[
[
"ragged_sentences = tf.ragged.constant([\n ['Hi'], ['Welcome', 'to', 'the', 'fair'], ['Have', 'fun']])\nprint(ragged_sentences.to_tensor(default_value=''))",
"_____no_output_____"
],
[
"print(ragged_sentences.to_sparse())",
"_____no_output_____"
],
[
"x = [[1, 3, -1, -1], [2, -1, -1, -1], [4, 5, 8, 9]]\nprint(tf.RaggedTensor.from_tensor(x, padding=-1))",
"_____no_output_____"
],
[
"st = tf.SparseTensor(indices=[[0, 0], [2, 0], [2, 1]],\n values=['a', 'b', 'c'],\n dense_shape=[3, 3])\nprint(tf.RaggedTensor.from_sparse(st))",
"_____no_output_____"
]
],
[
[
"## Evaluating ragged tensors\n\n### Eager execution\n\nIn eager execution mode, ragged tensors are evaluated immediately. To access the\nvalues they contain, you can:\n\n* Use the\n `tf.RaggedTensor.to_list()`\n method, which converts the ragged tensor to a Python `list`.",
"_____no_output_____"
]
],
[
[
"rt = tf.ragged.constant([[1, 2], [3, 4, 5], [6], [], [7]])\nprint(rt.to_list())",
"_____no_output_____"
]
],
[
[
"* Use Python indexing. If the tensor piece you select contains no ragged\n dimensions, then it will be returned as an `EagerTensor`. You can then use\n the `numpy()` method to access the value directly.",
"_____no_output_____"
]
],
[
[
"print(rt[1].numpy())",
"_____no_output_____"
]
],
[
[
"* Decompose the ragged tensor into its components, using the\n `tf.RaggedTensor.values`\n and\n `tf.RaggedTensor.row_splits`\n properties, or row-paritioning methods such as `tf.RaggedTensor.row_lengths()`\n and `tf.RaggedTensor.value_rowids()`.",
"_____no_output_____"
]
],
[
[
"print(rt.values)",
"_____no_output_____"
],
[
"print(rt.row_splits)",
"_____no_output_____"
]
],
[
[
"### Broadcasting\n\nBroadcasting is the process of making tensors with different shapes have\ncompatible shapes for elementwise operations. For more background on\nbroadcasting, see:\n\n* [Numpy: Broadcasting](https://docs.scipy.org/doc/numpy/user/basics.broadcasting.html)\n* `tf.broadcast_dynamic_shape`\n* `tf.broadcast_to`\n\nThe basic steps for broadcasting two inputs `x` and `y` to have compatible\nshapes are:\n\n1. If `x` and `y` do not have the same number of dimensions, then add outer\n dimensions (with size 1) until they do.\n\n2. For each dimension where `x` and `y` have different sizes:\n\n * If `x` or `y` have size `1` in dimension `d`, then repeat its values\n across dimension `d` to match the other input's size.\n\n * Otherwise, raise an exception (`x` and `y` are not broadcast\n compatible).",
"_____no_output_____"
],
[
"Where the size of a tensor in a uniform dimension is a single number (the size\nof slices across that dimension); and the size of a tensor in a ragged dimension\nis a list of slice lengths (for all slices across that dimension).\n\n#### Broadcasting examples",
"_____no_output_____"
]
],
[
[
"# x (2D ragged): 2 x (num_rows)\n# y (scalar)\n# result (2D ragged): 2 x (num_rows)\nx = tf.ragged.constant([[1, 2], [3]])\ny = 3\nprint(x + y)",
"_____no_output_____"
],
[
"# x (2d ragged): 3 x (num_rows)\n# y (2d tensor): 3 x 1\n# Result (2d ragged): 3 x (num_rows)\nx = tf.ragged.constant(\n [[10, 87, 12],\n [19, 53],\n [12, 32]])\ny = [[1000], [2000], [3000]]\nprint(x + y)",
"_____no_output_____"
],
[
"# x (3d ragged): 2 x (r1) x 2\n# y (2d ragged): 1 x 1\n# Result (3d ragged): 2 x (r1) x 2\nx = tf.ragged.constant(\n [[[1, 2], [3, 4], [5, 6]],\n [[7, 8]]],\n ragged_rank=1)\ny = tf.constant([[10]])\nprint(x + y)",
"_____no_output_____"
],
[
"# x (3d ragged): 2 x (r1) x (r2) x 1\n# y (1d tensor): 3\n# Result (3d ragged): 2 x (r1) x (r2) x 3\nx = tf.ragged.constant(\n [\n [\n [[1], [2]],\n [],\n [[3]],\n [[4]],\n ],\n [\n [[5], [6]],\n [[7]]\n ]\n ],\n ragged_rank=2)\ny = tf.constant([10, 20, 30])\nprint(x + y)",
"_____no_output_____"
]
],
[
[
"Here are some examples of shapes that do not broadcast:",
"_____no_output_____"
]
],
[
[
"# x (2d ragged): 3 x (r1)\n# y (2d tensor): 3 x 4 # trailing dimensions do not match\nx = tf.ragged.constant([[1, 2], [3, 4, 5, 6], [7]])\ny = tf.constant([[1, 2, 3, 4], [5, 6, 7, 8], [9, 10, 11, 12]])\ntry:\n x + y\nexcept tf.errors.InvalidArgumentError as exception:\n print(exception)",
"_____no_output_____"
],
[
"# x (2d ragged): 3 x (r1)\n# y (2d ragged): 3 x (r2) # ragged dimensions do not match.\nx = tf.ragged.constant([[1, 2, 3], [4], [5, 6]])\ny = tf.ragged.constant([[10, 20], [30, 40], [50]])\ntry:\n x + y\nexcept tf.errors.InvalidArgumentError as exception:\n print(exception)",
"_____no_output_____"
],
[
"# x (3d ragged): 3 x (r1) x 2\n# y (3d ragged): 3 x (r1) x 3 # trailing dimensions do not match\nx = tf.ragged.constant([[[1, 2], [3, 4], [5, 6]],\n [[7, 8], [9, 10]]])\ny = tf.ragged.constant([[[1, 2, 0], [3, 4, 0], [5, 6, 0]],\n [[7, 8, 0], [9, 10, 0]]])\ntry:\n x + y\nexcept tf.errors.InvalidArgumentError as exception:\n print(exception)",
"_____no_output_____"
]
],
[
[
"## RaggedTensor encoding\n\nRagged tensors are encoded using the `RaggedTensor` class. Internally, each\n`RaggedTensor` consists of:\n\n* A `values` tensor, which concatenates the variable-length rows into a\n flattened list.\n* A `row_splits` vector, which indicates how those flattened values are\n divided into rows. In particular, the values for row `rt[i]` are stored in\n the slice `rt.values[rt.row_splits[i]:rt.row_splits[i+1]]`.\n\n\n\n",
"_____no_output_____"
]
],
[
[
"rt = tf.RaggedTensor.from_row_splits(\n values=[3, 1, 4, 1, 5, 9, 2],\n row_splits=[0, 4, 4, 6, 7])\nprint(rt)",
"_____no_output_____"
]
],
[
[
"### Multiple ragged dimensions\n\nA ragged tensor with multiple ragged dimensions is encoded by using a nested\n`RaggedTensor` for the `values` tensor. Each nested `RaggedTensor` adds a single\nragged dimension.\n\n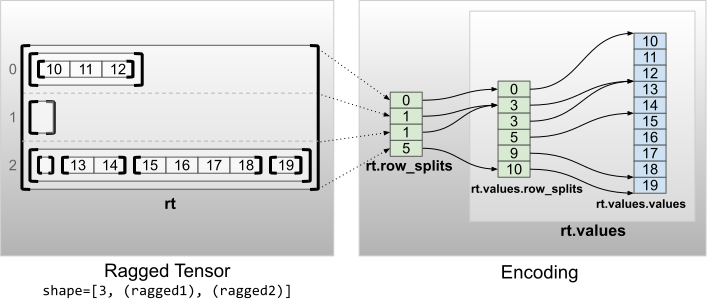",
"_____no_output_____"
]
],
[
[
"rt = tf.RaggedTensor.from_row_splits(\n values=tf.RaggedTensor.from_row_splits(\n values=[10, 11, 12, 13, 14, 15, 16, 17, 18, 19],\n row_splits=[0, 3, 3, 5, 9, 10]),\n row_splits=[0, 1, 1, 5])\nprint(rt)\nprint(\"Shape: {}\".format(rt.shape))\nprint(\"Number of ragged dimensions: {}\".format(rt.ragged_rank))",
"_____no_output_____"
]
],
[
[
"The factory function `tf.RaggedTensor.from_nested_row_splits` may be used to construct a\nRaggedTensor with multiple ragged dimensions directly, by providing a list of\n`row_splits` tensors:",
"_____no_output_____"
]
],
[
[
"rt = tf.RaggedTensor.from_nested_row_splits(\n flat_values=[10, 11, 12, 13, 14, 15, 16, 17, 18, 19],\n nested_row_splits=([0, 1, 1, 5], [0, 3, 3, 5, 9, 10]))\nprint(rt)",
"_____no_output_____"
]
],
[
[
"### Uniform Inner Dimensions\n\nRagged tensors with uniform inner dimensions are encoded by using a\nmultidimensional `tf.Tensor` for `values`.\n\n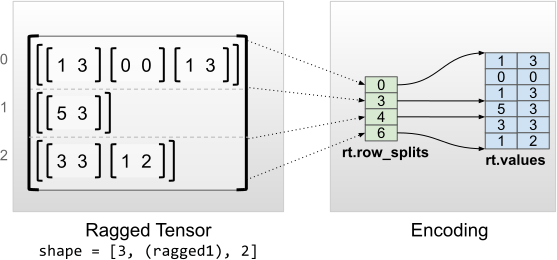",
"_____no_output_____"
]
],
[
[
"rt = tf.RaggedTensor.from_row_splits(\n values=[[1, 3], [0, 0], [1, 3], [5, 3], [3, 3], [1, 2]],\n row_splits=[0, 3, 4, 6])\nprint(rt)\nprint(\"Shape: {}\".format(rt.shape))\nprint(\"Number of ragged dimensions: {}\".format(rt.ragged_rank))",
"_____no_output_____"
]
],
[
[
"### Alternative row-partitioning schemes\n\nThe `RaggedTensor` class uses `row_splits` as the primary mechanism to store\ninformation about how the values are partitioned into rows. However,\n`RaggedTensor` also provides support for four alternative row-partitioning\nschemes, which can be more convenient to use depending on how your data is\nformatted. Internally, `RaggedTensor` uses these additional schemes to improve\nefficiency in some contexts.\n\n<dl>\n <dt>Row lengths</dt>\n <dd>`row_lengths` is a vector with shape `[nrows]`, which specifies the\n length of each row.</dd>\n\n <dt>Row starts</dt>\n <dd>`row_starts` is a vector with shape `[nrows]`, which specifies the start\n offset of each row. Equivalent to `row_splits[:-1]`.</dd>\n\n <dt>Row limits</dt>\n <dd>`row_limits` is a vector with shape `[nrows]`, which specifies the stop\n offset of each row. Equivalent to `row_splits[1:]`.</dd>\n\n <dt>Row indices and number of rows</dt>\n <dd>`value_rowids` is a vector with shape `[nvals]`, corresponding\n one-to-one with values, which specifies each value's row index. In\n particular, the row `rt[row]` consists of the values `rt.values[j]` where\n `value_rowids[j]==row`. \\\n `nrows` is an integer that specifies the number of rows in the\n `RaggedTensor`. In particular, `nrows` is used to indicate trailing empty\n rows.</dd>\n</dl>\n\nFor example, the following ragged tensors are equivalent:",
"_____no_output_____"
]
],
[
[
"values = [3, 1, 4, 1, 5, 9, 2, 6]\nprint(tf.RaggedTensor.from_row_splits(values, row_splits=[0, 4, 4, 7, 8, 8]))\nprint(tf.RaggedTensor.from_row_lengths(values, row_lengths=[4, 0, 3, 1, 0]))\nprint(tf.RaggedTensor.from_row_starts(values, row_starts=[0, 4, 4, 7, 8]))\nprint(tf.RaggedTensor.from_row_limits(values, row_limits=[4, 4, 7, 8, 8]))\nprint(tf.RaggedTensor.from_value_rowids(\n values, value_rowids=[0, 0, 0, 0, 2, 2, 2, 3], nrows=5))",
"_____no_output_____"
]
],
[
[
"The RaggedTensor class defines methods which can be used to construct\neach of these row-partitioning tensors.",
"_____no_output_____"
]
],
[
[
"rt = tf.ragged.constant([[3, 1, 4, 1], [], [5, 9, 2], [6], []])\nprint(\" values: {}\".format(rt.values))\nprint(\" row_splits: {}\".format(rt.row_splits))\nprint(\" row_lengths: {}\".format(rt.row_lengths()))\nprint(\" row_starts: {}\".format(rt.row_starts()))\nprint(\" row_limits: {}\".format(rt.row_limits()))\nprint(\"value_rowids: {}\".format(rt.value_rowids()))",
"_____no_output_____"
]
],
[
[
"(Note that `tf.RaggedTensor.values` and `tf.RaggedTensors.row_splits` are properties, while the remaining row-partitioning accessors are all methods. This reflects the fact that the `row_splits` are the primary underlying representation, and the other row-partitioning tensors must be computed.)",
"_____no_output_____"
],
[
"Some of the advantages and disadvantages of the different row-partitioning\nschemes are:\n\n+ **Efficient indexing**:\n The `row_splits`, `row_starts`, and `row_limits` schemes all enable\n constant-time indexing into ragged tensors. The `value_rowids` and\n `row_lengths` schemes do not.\n\n+ **Small encoding size**:\n The `value_rowids` scheme is more efficient when storing ragged tensors that\n have a large number of empty rows, since the size of the tensor depends only\n on the total number of values. On the other hand, the other four encodings\n are more efficient when storing ragged tensors with longer rows, since they\n require only one scalar value for each row.\n\n+ **Efficient concatenation**:\n The `row_lengths` scheme is more efficient when concatenating ragged\n tensors, since row lengths do not change when two tensors are concatenated\n together (but row splits and row indices do).\n\n+ **Compatibility**:\n The `value_rowids` scheme matches the\n [segmentation](../api_guides/python/math_ops.md#Segmentation)\n format used by operations such as `tf.segment_sum`. The `row_limits` scheme\n matches the format used by ops such as `tf.sequence_mask`.",
"_____no_output_____"
]
],
[
[
"",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
]
] |
c5116cb03af28a686d988679dd2ed790532bee16
| 27,622 |
ipynb
|
Jupyter Notebook
|
guardian_article_words.ipynb
|
Nasrin63/guardian_article_words
|
8e83de0f3be916426b74736dc13bc8af50a4664b
|
[
"MIT"
] | null | null | null |
guardian_article_words.ipynb
|
Nasrin63/guardian_article_words
|
8e83de0f3be916426b74736dc13bc8af50a4664b
|
[
"MIT"
] | null | null | null |
guardian_article_words.ipynb
|
Nasrin63/guardian_article_words
|
8e83de0f3be916426b74736dc13bc8af50a4664b
|
[
"MIT"
] | null | null | null | 33.400242 | 829 | 0.414561 |
[
[
[
"#make a list of words in an article\nfrom urllib import request\nfrom bs4 import BeautifulSoup\nfrom collections import Counter \nimport spacy\nurl = \"https://www.theguardian.com/commentisfree/2019/jun/24/boris-johnson-prime-minister-tory-party-britain\"",
"_____no_output_____"
],
[
"html = request.urlopen(url).read().decode('utf8')",
"_____no_output_____"
],
[
"soup = BeautifulSoup(html, 'html.parser')\n# soup.get_text()",
"_____no_output_____"
],
[
"articleBody = soup.find(\"div\", attrs=dict(itemprop=\"articleBody\"))",
"_____no_output_____"
],
[
"text = articleBody.getText()",
"_____no_output_____"
],
[
"print(text)",
"\nSix years ago, the Cambridge historian Christopher Clark published a study of the outbreak of the first world war, titled The Sleepwalkers. Though Clark is a fine scholar, I was unconvinced by his title, which suggested that the great powers stumbled mindlessly to disaster. On the contrary, the maddest aspect of 1914 was that each belligerent government convinced itself that it was acting rationally.\nIt would be fanciful to liken the ascent of Boris Johnson to the outbreak of global war, but similar forces are in play. There is room for debate about whether he is a scoundrel or mere rogue, but not much about his moral bankruptcy, rooted in a contempt for truth. Nonetheless, even before the Conservative national membership cheers him in as our prime minister – denied the option of Nigel Farage, whom some polls suggest they would prefer – Tory MPs have thronged to do just that.\n\n\n\n\n \n\nHe would not recognise the truth, whether about his private or political life, if confronted by it in an identity parade\n\n\nI have known Johnson since the 1980s, when I edited the Daily Telegraph and he was our flamboyant Brussels correspondent. I have argued for a decade that, while he is a brilliant entertainer who made a popular maître d’ for London as its mayor, he is unfit for national office, because it seems he cares for no interest save his own fame and gratification.\nTory MPs have launched this country upon an experiment in celebrity government, matching that taking place in Ukraine and the US, and it is unlikely to be derailed by the latest headlines. The Washington Post columnist George Will observes that Donald Trump does what his political base wants “by breaking all the china”. We can’t predict what a Johnson government will do, because its prospective leader has not got around to thinking about this. But his premiership will almost certainly reveal a contempt for rules, precedent, order and stability.\nA few admirers assert that, in office, Johnson will reveal an accession of wisdom and responsibility that have hitherto eluded him, not least as foreign secretary. This seems unlikely, as the weekend’s stories emphasised. Dignity still matters in public office, and Johnson will never have it. Yet his graver vice is cowardice, reflected in a willingness to tell any audience, whatever he thinks most likely to please, heedless of the inevitability of its contradiction an hour later.\nLike many showy personalities, he is of weak character. I recently suggested to a radio audience that he supposes himself to be Winston Churchill, while in reality being closer to Alan Partridge. Churchill, for all his wit, was a profoundly serious human being. Far from perceiving anything glorious about standing alone in 1940, he knew that all difficult issues must be addressed with allies and partners.\nChurchill’s self-obsession was tempered by a huge compassion for humanity, or at least white humanity, which Johnson confines to himself. He has long been considered a bully, prone to making cheap threats. My old friend Christopher Bland, when chairman of the BBC, once described to me how he received an angry phone call from Johnson, denouncing the corporation’s “gross intrusion upon my personal life” for its coverage of one of his love affairs.\n“We know plenty about your personal life that you would not like to read in the Spectator,” the then editor of the magazine told the BBC’s chairman, while demanding he order the broadcaster to lay off his own dalliances.\nBland told me he replied: “Boris, think about what you have just said. There is a word for it, and it is not a pretty one.”\nHe said Johnson blustered into retreat, but in my own files I have handwritten notes from our possible next prime minister, threatening dire consequences in print if I continued to criticise him.\nJohnson would not recognise truth, whether about his private or political life, if confronted by it in an identity parade. In a commonplace book the other day, I came across an observation made in 1750 by a contemporary savant, Bishop Berkeley: “It is impossible that a man who is false to his friends and neighbours should be true to the public.” Almost the only people who think Johnson a nice guy are those who do not know him.\nThere is, of course, a symmetry between himself and Jeremy Corbyn. Corbyn is far more honest, but harbours his own extravagant delusions. He may yet prove to be the only possible Labour leader whom Johnson can defeat in a general election. If the opposition was led by anybody else, the Tories would be deservedly doomed, because we would all vote for it. As it is, the Johnson premiership could survive for three or four years, shambling from one embarrassment and debacle to another, of which Brexit may prove the least.\n\n\n\n\nBoris Johnson row: top Tory party donor joins calls for explanation\n\n\n\n\n\n\n \n\n\nRead more\n\n\n\n\n\n\nFor many of us, his elevation will signal Britain’s abandonment of any claim to be a serious country. It can be claimed that few people realised what a poor prime minister Theresa May would prove until they saw her in Downing Street. With Boris, however, what you see now is almost assuredly what we shall get from him as ruler of Britain.\nWe can scarcely strip the emperor’s clothes from a man who has built a career, or at least a lurid love life, out of strutting without them. The weekend stories of his domestic affairs are only an aperitif for his future as Britain’s leader. I have a hunch that Johnson will come to regret securing the prize for which he has struggled so long, because the experience of the premiership will lay bare his absolute unfitness for it.\nIf the Johnson family had stuck to showbusiness like the Osmonds, Marx Brothers or von Trapp family, the world would be a better place. Yet the Tories, in their terror, have elevated a cavorting charlatan to the steps of Downing Street, and they should expect to pay a full forfeit when voters get the message. If the price of Johnson proves to be Corbyn, blame will rest with the Conservative party, which is about to foist a tasteless joke upon the British people – who will not find it funny for long.\n• Max Hastings is a former editor of the Daily Telegraph and the London Evening Standard\n\n\nTopics\n\n\n\n\nConservative leadership\n\n\n\n\nOpinion\n\n\n\n\n\n\n\n\nBoris Johnson\n\n\n\n\nBrexit\n\n\n\n\nConservatives\n\n\n\n\nNewspapers\n\n\n\n\nDaily Telegraph\n\n\n\n\ncomment\n\n\n\n\n\n\n\n\nShare on Facebook\n\n\n\n\n\n\n\n\n\nShare on Twitter\n\n\n\n\n\n\n\n\n\nShare via Email\n\n\n\n\n\n\n\n\n\nShare on LinkedIn\n\n\n\n\n\n\n\n\n\nShare on Pinterest\n\n\n\n\n\n\n\n\n\nShare on WhatsApp\n\n\n\n\n\n\n\n\n\nShare on Messenger\n\n\n\n\n\n\n\n\n\n\n\n\n\nReuse this content\n\n\n\n\n\n\n"
],
[
"len(text)",
"_____no_output_____"
],
[
"nlp = spacy.load('en_core_web_sm')",
"_____no_output_____"
],
[
"doc = nlp(text)",
"_____no_output_____"
],
[
"tokens = [t.text for t in doc if t.is_alpha and not t.is_stop]",
"_____no_output_____"
],
[
"counts = Counter(tokens)",
"_____no_output_____"
],
[
"counts",
"_____no_output_____"
],
[
"result = filter(lambda token: len(token) == 5, tokens)\n ",
"_____no_output_____"
],
[
"result_list = list(result) ",
"_____no_output_____"
],
[
"print(result_list)",
"['years', 'Clark', 'study', 'world', 'Clark', 'title', 'great', 'liken', 'Boris', 'rogue', 'moral', 'truth', 'prime', 'Nigel', 'polls', 'truth', 'known', 'Daily', 'mayor', 'unfit', 'cares', 'place', 'Trump', 'wants', 'china', 'rules', 'order', 'later', 'showy', 'radio', 'human', 'white', 'bully', 'prone', 'cheap', 'Bland', 'angry', 'phone', 'gross', 'order', 'Bland', 'Boris', 'think', 'files', 'notes', 'prime', 'print', 'truth', 'false', 'think', 'prove', 'years', 'prove', 'Boris', 'party', 'donor', 'joins', 'calls', 'claim', 'prime', 'prove', 'Boris', 'shall', 'ruler', 'strip', 'built', 'lurid', 'hunch', 'prize', 'stuck', 'Trapp', 'world', 'place', 'steps', 'price', 'blame', 'party', 'foist', 'funny', 'Daily', 'Boris', 'Daily', 'Share', 'Share', 'Share', 'Email', 'Share', 'Share', 'Share', 'Share', 'Reuse']\n"
],
[
"most_common = counts.most_common(20)",
"_____no_output_____"
],
[
"most_common",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
c5117f1466ef0a4a9c20557f69adf3db98f18877
| 11,710 |
ipynb
|
Jupyter Notebook
|
site/en-snapshot/hub/tutorials/text_to_video_retrieval_with_s3d_milnce.ipynb
|
ilyaspiridonov/docs-l10n
|
a061a44e40d25028d0a4458094e48ab717d3565c
|
[
"Apache-2.0"
] | 1 |
2020-07-22T05:03:55.000Z
|
2020-07-22T05:03:55.000Z
|
site/en-snapshot/hub/tutorials/text_to_video_retrieval_with_s3d_milnce.ipynb
|
ilyaspiridonov/docs-l10n
|
a061a44e40d25028d0a4458094e48ab717d3565c
|
[
"Apache-2.0"
] | null | null | null |
site/en-snapshot/hub/tutorials/text_to_video_retrieval_with_s3d_milnce.ipynb
|
ilyaspiridonov/docs-l10n
|
a061a44e40d25028d0a4458094e48ab717d3565c
|
[
"Apache-2.0"
] | 1 |
2020-08-03T21:04:29.000Z
|
2020-08-03T21:04:29.000Z
| 39.96587 | 298 | 0.540393 |
[
[
[
"# Copyright 2019 The TensorFlow Hub Authors. All Rights Reserved.\n#\n# Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# http://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.\n# ==============================================================================",
"_____no_output_____"
]
],
[
[
"# Text-to-Video retrieval with S3D MIL-NCE",
"_____no_output_____"
],
[
"<table class=\"tfo-notebook-buttons\" align=\"left\">\n <td>\n <a target=\"_blank\" href=\"https://www.tensorflow.org/hub/tutorials/text_to_video_retrieval_with_s3d_milnce\"><img src=\"https://www.tensorflow.org/images/tf_logo_32px.png\" />View on TensorFlow.org</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://colab.research.google.com/github/tensorflow/hub/blob/master/examples/colab/text_to_video_retrieval_with_s3d_milnce.ipynb\"><img src=\"https://www.tensorflow.org/images/colab_logo_32px.png\" />Run in Google Colab</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://github.com/tensorflow/hub/blob/master/examples/colab/text_to_video_retrieval_with_s3d_milnce.ipynb\"><img src=\"https://www.tensorflow.org/images/GitHub-Mark-32px.png\" />View source on GitHub</a>\n </td>\n <td>\n <a href=\"https://storage.googleapis.com/tensorflow_docs/hub/examples/colab/text_to_video_retrieval_with_s3d_milnce.ipynb\"><img src=\"https://www.tensorflow.org/images/download_logo_32px.png\" />Download notebook</a>\n </td>\n</table>",
"_____no_output_____"
]
],
[
[
"!pip install -q opencv-python\n\nimport os\n\nimport tensorflow.compat.v2 as tf\nimport tensorflow_hub as hub\n\nimport numpy as np\nimport cv2\nfrom IPython import display\nimport math",
"_____no_output_____"
]
],
[
[
"## Import TF-Hub model\n\nThis tutorial demonstrates how to use the [S3D MIL-NCE model](https://tfhub.dev/deepmind/mil-nce/s3d/1) from TensorFlow Hub to do **text-to-video retrieval** to find the most similar videos for a given text query.\n\nThe model has 2 signatures, one for generating *video embeddings* and one for generating *text embeddings*. We will use these embedding to find the nearest neighbors in the embedding space.",
"_____no_output_____"
]
],
[
[
"# Load the model once from TF-Hub.\nhub_handle = 'https://tfhub.dev/deepmind/mil-nce/s3d/1'\nhub_model = hub.load(hub_handle)\n\ndef generate_embeddings(model, input_frames, input_words):\n \"\"\"Generate embeddings from the model from video frames and input words.\"\"\"\n # Input_frames must be normalized in [0, 1] and of the shape Batch x T x H x W x 3\n vision_output = model.signatures['video'](tf.constant(tf.cast(input_frames, dtype=tf.float32)))\n text_output = model.signatures['text'](tf.constant(input_words))\n return vision_output['video_embedding'], text_output['text_embedding']",
"_____no_output_____"
],
[
"# @title Define video loading and visualization functions { display-mode: \"form\" }\n\n# Utilities to open video files using CV2\ndef crop_center_square(frame):\n y, x = frame.shape[0:2]\n min_dim = min(y, x)\n start_x = (x // 2) - (min_dim // 2)\n start_y = (y // 2) - (min_dim // 2)\n return frame[start_y:start_y+min_dim,start_x:start_x+min_dim]\n\n\ndef load_video(video_url, max_frames=32, resize=(224, 224)):\n path = tf.keras.utils.get_file(os.path.basename(video_url)[-128:], video_url)\n cap = cv2.VideoCapture(path)\n frames = []\n try:\n while True:\n ret, frame = cap.read()\n if not ret:\n break\n frame = crop_center_square(frame)\n frame = cv2.resize(frame, resize)\n frame = frame[:, :, [2, 1, 0]]\n frames.append(frame)\n\n if len(frames) == max_frames:\n break\n finally:\n cap.release()\n frames = np.array(frames)\n if len(frames) < max_frames:\n n_repeat = int(math.ceil(max_frames / float(len(frames))))\n frames = frames.repeat(n_repeat, axis=0)\n frames = frames[:max_frames]\n return frames / 255.0\n\ndef display_video(urls):\n html = '<table>'\n html += '<tr><th>Video 1</th><th>Video 2</th><th>Video 3</th></tr><tr>'\n for url in urls:\n html += '<td>'\n html += '<img src=\"{}\" height=\"224\">'.format(url)\n html += '</td>'\n html += '</tr></table>'\n return display.HTML(html)\n\ndef display_query_and_results_video(query, urls, scores):\n \"\"\"Display a text query and the top result videos and scores.\"\"\"\n sorted_ix = np.argsort(-scores)\n html = ''\n html += '<h2>Input query: <i>{}</i> </h2><div>'.format(query)\n html += 'Results: <div>'\n html += '<table>'\n html += '<tr><th>Rank #1, Score:{:.2f}</th>'.format(scores[sorted_ix[0]])\n html += '<th>Rank #2, Score:{:.2f}</th>'.format(scores[sorted_ix[1]])\n html += '<th>Rank #3, Score:{:.2f}</th></tr><tr>'.format(scores[sorted_ix[2]])\n for i, idx in enumerate(sorted_ix):\n url = urls[sorted_ix[i]];\n html += '<td>'\n html += '<img src=\"{}\" height=\"224\">'.format(url)\n html += '</td>'\n html += '</tr></table>'\n return html\n",
"_____no_output_____"
],
[
"# @title Load example videos and define text queries { display-mode: \"form\" }\n\nvideo_1_url = 'https://upload.wikimedia.org/wikipedia/commons/b/b0/YosriAirTerjun.gif' # @param {type:\"string\"}\nvideo_2_url = 'https://upload.wikimedia.org/wikipedia/commons/e/e6/Guitar_solo_gif.gif' # @param {type:\"string\"}\nvideo_3_url = 'https://upload.wikimedia.org/wikipedia/commons/3/30/2009-08-16-autodrift-by-RalfR-gif-by-wau.gif' # @param {type:\"string\"}\n\nvideo_1 = load_video(video_1_url)\nvideo_2 = load_video(video_2_url)\nvideo_3 = load_video(video_3_url)\nall_videos = [video_1, video_2, video_3]\n\nquery_1_video = 'waterfall' # @param {type:\"string\"}\nquery_2_video = 'playing guitar' # @param {type:\"string\"}\nquery_3_video = 'car drifting' # @param {type:\"string\"}\nall_queries_video = [query_1_video, query_2_video, query_3_video]\nall_videos_urls = [video_1_url, video_2_url, video_3_url]\ndisplay_video(all_videos_urls)",
"_____no_output_____"
]
],
[
[
"## Demonstrate text to video retrieval\n",
"_____no_output_____"
]
],
[
[
"# Prepare video inputs.\nvideos_np = np.stack(all_videos, axis=0)\n\n# Prepare text input.\nwords_np = np.array(all_queries_video)\n\n# Generate the video and text embeddings.\nvideo_embd, text_embd = generate_embeddings(hub_model, videos_np, words_np)\n\n# Scores between video and text is computed by dot products.\nall_scores = np.dot(text_embd, tf.transpose(video_embd))",
"_____no_output_____"
],
[
"# Display results.\nhtml = ''\nfor i, words in enumerate(words_np):\n html += display_query_and_results_video(words, all_videos_urls, all_scores[i, :])\n html += '<br>'\ndisplay.HTML(html)",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
c5119095b8d52e96056afc28efe698b28381499e
| 119,994 |
ipynb
|
Jupyter Notebook
|
sample/PyQuil_adder_Vedral.ipynb
|
gines-carrascal/jupyter-pyquil
|
e06af7cfb976a3f5a3518c809d914ce9ef46aa72
|
[
"Apache-2.0"
] | null | null | null |
sample/PyQuil_adder_Vedral.ipynb
|
gines-carrascal/jupyter-pyquil
|
e06af7cfb976a3f5a3518c809d914ce9ef46aa72
|
[
"Apache-2.0"
] | null | null | null |
sample/PyQuil_adder_Vedral.ipynb
|
gines-carrascal/jupyter-pyquil
|
e06af7cfb976a3f5a3518c809d914ce9ef46aa72
|
[
"Apache-2.0"
] | null | null | null | 36.97812 | 12,213 | 0.546061 |
[
[
[
"# PyQuil example: Ripple adder\nBased on Vedral, Barenco, and Ekert (1996).\n\n* Gines Carrascal, Complutense University of Madrid\n\nhttps://github.com/gines-carrascal/jupyter-pyquil",
"_____no_output_____"
]
],
[
[
"from pyquil import get_qc, Program\nfrom pyquil.gates import X, CNOT, CCNOT, MEASURE\nfrom pyquil.latex import to_latex, display\nfrom pyquil.quil import address_qubits\nfrom pyquil.quilatom import QubitPlaceholder",
"_____no_output_____"
],
[
"sumando_1 = input(\"Primer sumando en binario (4 bits)\")\nsumando_2 = input(\"Segundo sumando en binario(4 bits)\")",
"Primer sumando en binario (4 bits)0001\nSegundo sumando en binario(4 bits)0001\n"
],
[
"n = 4\n\np = Program()\n\n\na = QubitPlaceholder.register(n)\nb = QubitPlaceholder.register(n+1)\nc = QubitPlaceholder.register(n)\n\nro = p.declare('ro', 'BIT', n+1)\n\n\nfor i in range(n):\n if sumando_1[i] == \"1\":\n p += X(a[n - (i+1)])\nfor i in range(n):\n if sumando_2[i] == \"1\":\n p += X(b[n - (i+1)])\n \nfor i in range(n-1):\n p += CCNOT(a[i], b[i], c[i+1])\n p += CNOT(a[i], b[i])\n p += CCNOT(a[i], b[i], c[i+1])\n\np += CCNOT(a[n-1], b[n-1], b[n])\np += CNOT(a[n-1], b[n-1])\np += CCNOT(a[n-1], b[n-1], b[n]) \n\np += CNOT(c[n-1], b[n-1])\n\nfor i in range(n-1):\n p += CCNOT(c[(n-2)-i], b[(n-2)-i], c[(n-1)-i])\n p += CNOT(a[(n-2)-i], b[(n-2)-i])\n p += CCNOT(a[(n-2)-i], b[(n-2)-i], c[(n-1)-i])\n \n p += CNOT(c[(n-2)-i], b[(n-2)-i])\n p += CNOT(a[(n-2)-i], b[(n-2)-i])\n\n\nfor i in range(n+1):\n p += MEASURE(b[i], ro[i])\n\np.wrap_in_numshots_loop(20)\n\nprint(address_qubits(p)) ",
"DECLARE ro BIT[5]\nX 0\nX 1\nCCNOT 0 1 2\nCNOT 0 1\nCCNOT 0 1 2\nCCNOT 3 4 5\nCNOT 3 4\nCCNOT 3 4 5\nCCNOT 6 7 8\nCNOT 6 7\nCCNOT 6 7 8\nCCNOT 9 10 11\nCNOT 9 10\nCCNOT 9 10 11\nCNOT 8 10\nCCNOT 5 7 8\nCNOT 6 7\nCCNOT 6 7 8\nCNOT 5 7\nCNOT 6 7\nCCNOT 2 4 5\nCNOT 3 4\nCCNOT 3 4 5\nCNOT 2 4\nCNOT 3 4\nCCNOT 12 1 2\nCNOT 0 1\nCCNOT 0 1 2\nCNOT 12 1\nCNOT 0 1\nMEASURE 1 ro[0]\nMEASURE 4 ro[1]\nMEASURE 7 ro[2]\nMEASURE 10 ro[3]\nMEASURE 11 ro[4]\n\n"
],
[
"from pyquil import list_quantum_computers\nprint(list_quantum_computers())",
"['Aspen-7-28Q-A', 'Aspen-7-14Q-C', 'Aspen-7-13Q-C', 'Aspen-4-13Q-E', 'Aspen-7-12Q-C', 'Aspen-7-11Q-C', 'Aspen-7-10Q-C', 'Aspen-4-9Q-E', 'Aspen-7-9Q-C', 'Aspen-7-8Q-C', 'Aspen-4-8Q-F', 'Aspen-4-7Q-F', 'Aspen-7-7Q-C', 'Aspen-7-6Q-C', 'Aspen-4-6Q-F', 'Aspen-7-5Q-C', 'Aspen-4-5Q-E', 'Aspen-7-4Q-D', 'Aspen-4-4Q-D', 'Aspen-4-4Q-F', 'Aspen-7-3Q-C', 'Aspen-4-3Q-F', 'Aspen-4-3Q-G', 'Aspen-7-2Q-C', 'Aspen-4-2Q-C', 'Aspen-4-2Q-H', '9q-square-qvm', '9q-square-noisy-qvm']\n"
],
[
"#qvm = get_qc(str(2*n+1)+\"q-qvm\")\n\nqvm = get_qc(\"Aspen-7-28Q-A\", as_qvm=True)",
"_____no_output_____"
],
[
"cp = qvm.compile(address_qubits(p),protoquil=True)",
"_____no_output_____"
],
[
"print(cp.program)",
"DECLARE ro BIT[5]\nRX(pi) 32\nCZ 32 31\nRX(pi) 16\nCZ 16 17\nRX(pi/2) 12\nRZ(-3.0761091086471977) 12\nRX(-pi/2) 12\nRZ(-2.35406633988804) 12\nRX(pi/2) 12\nRZ(0.9317843330464246) 12\nRX(-pi/2) 12\nRX(pi/2) 13\nRZ(-2.3009408748502995) 13\nRX(-pi/2) 13\nRZ(-0.5719412675491293) 13\nRX(pi/2) 13\nRZ(-2.9833635973204427) 13\nRX(-pi/2) 13\nCZ 13 12\nRZ(-pi/2) 12\nRX(pi/2) 12\nRZ(-pi/2) 13\nRX(-pi/2) 13\nCZ 13 12\nRX(-pi/2) 12\nRX(pi/2) 13\nCZ 13 12\nRX(pi) 14\nRZ(pi) 31\nRZ(-pi/2) 32\nRX(-pi/2) 32\nRZ(-pi/4) 32\nRX(pi/2) 32\nCZ 32 33\nRZ(-pi/2) 16\nRX(-pi/2) 16\nRZ(-pi/4) 16\nRX(pi/2) 16\nCZ 16 15\nRZ(pi) 17\nRZ(pi/2) 12\nRX(pi/2) 12\nRZ(-1.7290253830642477) 12\nRX(-pi/2) 12\nRZ(-2.569651386040664) 12\nRX(pi/2) 12\nRZ(0.8406517787394918) 12\nRX(-pi/2) 12\nRX(pi/2) 13\nRZ(2.5525617757224426) 13\nRX(-pi/2) 13\nRZ(-1.0167476369539092) 13\nRX(pi/2) 13\nRZ(-2.30345703570484) 13\nRX(pi/2) 13\nCZ 14 13\nRX(-pi/2) 32\nRZ(pi/4) 32\nRX(pi/2) 32\nCZ 32 31\nRZ(pi) 33\nRZ(pi) 15\nRX(-pi/2) 16\nRZ(pi/4) 16\nRX(pi/2) 16\nCZ 16 17\nRX(-pi/2) 13\nRZ(-pi/4) 13\nRX(pi/2) 13\nCZ 13 12\nRZ(pi) 14\nRZ(pi) 12\nRX(-pi/2) 13\nRZ(pi/4) 13\nRX(pi/2) 13\nCZ 13 14\nRZ(3*pi/4) 31\nRX(pi/2) 31\nRZ(pi/2) 31\nRX(pi/2) 32\nRZ(pi/4) 32\nRX(-pi/2) 32\nRZ(2.6977093785143182) 32\nRX(-pi/2) 32\nRZ(0.19115829914713522) 33\nRX(pi/2) 33\nCZ 33 32\nRZ(pi) 32\nRX(pi/2) 32\nRZ(pi) 33\nRX(pi/2) 33\nCZ 33 32\nRZ(0.793598945054252) 15\nRX(-pi/2) 15\nRX(pi/2) 16\nRZ(-3*pi/4) 16\nRX(-pi/2) 16\nRZ(-2.3643952718491485) 16\nRX(pi/2) 16\nCZ 16 15\nRZ(pi) 15\nRX(pi/2) 15\nRZ(pi) 16\nRX(pi/2) 16\nCZ 16 15\nRZ(3*pi/4) 17\nRX(pi/2) 17\nRZ(pi/2) 17\nRZ(-3.141353431656798) 12\nRX(-pi/2) 12\nRX(pi/2) 13\nCZ 13 12\nRZ(pi) 12\nRX(pi/2) 12\nRZ(pi/4) 13\nRX(pi/2) 13\nCZ 13 12\nRZ(3*pi/4) 14\nRX(pi/2) 14\nRZ(pi/2) 14\nRZ(0.9274165021124094) 31\nRX(pi) 31\nRX(pi/2) 32\nCZ 31 32\nRX(-pi/2) 33\nRZ(2.6977093785143174) 33\nRX(-pi/2) 33\nRZ(pi/4) 33\nRX(-pi/2) 33\nRZ(-1.2365784406849656) 33\nRX(-pi/2) 33\nRZ(-1.4190325161734523) 33\nRX(pi/2) 33\nRZ(2.7838915531496973) 33\nRX(-pi/2) 33\nRX(pi/2) 15\nRZ(0.7771973817406423) 15\nRX(-pi/2) 15\nRZ(-3*pi/4) 15\nRX(-pi/2) 15\nRZ(-0.9809307985653112) 15\nRX(-pi/2) 15\nRZ(-0.6137822307922566) 15\nRX(pi/2) 15\nRZ(0.9315720589381091) 15\nRX(-pi/2) 15\nRX(pi/2) 16\nCZ 17 16\nRZ(-pi/4) 12\nRX(pi/2) 12\nRZ(-0.16263847489248415) 12\nRX(-pi/2) 12\nRZ(-2.4826734982987237) 12\nRX(pi/2) 12\nRZ(-2.2567642980594442) 12\nRX(-pi/2) 12\nRX(pi/2) 13\nCZ 14 13\nRZ(pi) 31\nRX(pi/2) 31\nRZ(0.8960110174983944) 31\nRX(-pi/2) 31\nRZ(-2.008869589694714) 31\nRX(pi/2) 31\nRZ(2.0583079868598912) 31\nRX(-pi/2) 31\nRZ(3.0381738350343443) 32\nRX(pi/2) 32\nRZ(-1.9361798097685459) 32\nRX(-pi/2) 32\nRZ(-1.551289366420918) 32\nRX(pi/2) 32\nRZ(-0.8192704177455041) 32\nRX(-pi/2) 32\nCZ 33 32\nRZ(-pi/2) 32\nRX(pi/2) 32\nRZ(-pi/2) 33\nRX(-pi/2) 33\nCZ 33 32\nRX(-pi/2) 32\nRX(pi/2) 33\nCZ 33 32\nRZ(-2.707904748544218) 16\nRX(pi/2) 16\nRZ(1.031118454503258) 16\nRX(-pi/2) 16\nRZ(-1.0500054320435226) 16\nRX(pi/2) 16\nRZ(-0.25760091708066346) 16\nRX(-pi/2) 16\nCZ 16 15\nRZ(-pi/2) 15\nRX(pi/2) 15\nRZ(-pi/2) 16\nRX(-pi/2) 16\nCZ 16 15\nRX(-pi/2) 15\nRX(pi/2) 16\nCZ 16 15\nRZ(-pi/2) 17\nRX(-pi/2) 17\nRZ(pi/4) 17\nRX(pi/2) 32\nRZ(0.26021415583235336) 32\nRX(-pi/2) 32\nRZ(-1.3106061954738266) 32\nRX(pi/2) 32\nRZ(-1.9370846838728044) 32\nRX(pi/2) 32\nCZ 31 32\nRZ(pi/2) 33\nRX(pi/2) 33\nRZ(1.9284974272349926) 33\nRX(-pi/2) 33\nRZ(-1.7225601374163413) 33\nRX(pi/2) 33\nRZ(1.9050142129048275) 33\nRX(-pi/2) 33\nRZ(pi/2) 15\nRX(pi/2) 15\nRZ(-0.8152042950748697) 15\nRX(-pi/2) 15\nRZ(-0.8757898209368592) 15\nRX(pi/2) 15\nRZ(1.8209560715414543) 15\nRX(-pi/2) 15\nRX(pi/2) 16\nRZ(2.743689562438258) 16\nRX(-pi/2) 16\nRZ(-1.0902716175127205) 16\nRX(pi/2) 16\nRZ(0.1480695852195293) 16\nRX(pi/2) 16\nRZ(-pi/2) 17\nRX(pi) 17\nCZ 17 16\nRX(pi/2) 13\nRZ(-0.0755404315407737) 13\nRX(-pi/2) 13\nRZ(-1.522741021871032) 13\nRX(pi/2) 13\nRZ(2.547404627696741) 13\nRX(-pi/2) 13\nCZ 13 12\nRZ(-pi/2) 12\nRX(pi/2) 12\nRZ(-pi/2) 13\nRX(-pi/2) 13\nCZ 13 12\nRX(-pi/2) 12\nRX(pi/2) 13\nCZ 13 12\nRZ(-pi/2) 14\nRX(-pi/2) 14\nRZ(pi/4) 14\nRZ(pi) 31\nRX(-pi/2) 32\nRZ(-3*pi/4) 32\nRX(-pi/2) 32\nCZ 32 33\nRZ(-pi/2) 16\nRX(pi/2) 16\nRZ(pi/4) 16\nRX(pi/2) 16\nCZ 16 15\nRZ(-pi/2) 17\nRX(pi) 17\nRX(pi/2) 12\nRZ(0.7164576833098726) 12\nRX(-pi/2) 12\nRZ(-2.0472898241413495) 12\nRX(pi/2) 12\nRZ(1.187747569064694) 12\nRX(-pi/2) 12\nRX(pi/2) 13\nRZ(-0.4560236761258309) 13\nRX(-pi/2) 13\nRZ(-2.0643661658727472) 13\nRX(pi/2) 13\nRZ(-2.210907576608408) 13\nRX(pi/2) 13\nRZ(-pi/2) 14\nRX(pi) 14\nCZ 14 13\nRX(-pi/2) 32\nRZ(pi/4) 32\nRX(pi/2) 32\nCZ 32 31\nRZ(pi) 33\nRZ(pi) 15\nRX(pi/2) 16\nRZ(-3*pi/4) 16\nRX(pi/2) 16\nCZ 16 17\nRZ(-pi/2) 13\nRX(pi/2) 13\nRZ(pi/4) 13\nRX(pi/2) 13\nCZ 12 13\nRZ(-pi/2) 14\nRX(pi) 14\nRZ(pi) 12\nRX(pi/2) 13\nRZ(-3*pi/4) 13\nRX(pi/2) 13\nCZ 13 14\nRZ(3*pi/4) 31\nRX(pi/2) 31\nRZ(pi/2) 31\nRX(pi/2) 32\nRZ(pi/4) 32\nRX(-pi/2) 32\nRZ(2.352974177644855) 32\nRX(-pi/2) 32\nRZ(-0.7821778508499602) 33\nRX(pi/2) 33\nCZ 33 32\nRZ(pi) 32\nRX(pi/2) 32\nRZ(pi) 33\nRX(pi/2) 33\nCZ 33 32\nRX(-pi/2) 33\nRZ(2.352974177644855) 33\nRX(-pi/2) 33\nRZ(-3*pi/4) 33\nRX(-pi/2) 33\nRZ(-1.0621773177865779) 33\nRX(-pi/2) 33\nRZ(-2.0840157525609144) 33\nRX(pi/2) 33\nRZ(-0.9895995496572645) 33\nRX(-pi/2) 33\nRZ(pi) 34\nRX(pi/2) 34\nRZ(-2.632973644581474) 34\nRX(-pi/2) 34\nRZ(-2.084015752560915) 34\nRX(pi/2) 34\nRZ(-0.9895995496572638) 34\nRX(-pi/2) 34\nRZ(0.2214938587060693) 15\nRX(-pi/2) 15\nRX(pi/2) 16\nRZ(-3*pi/4) 16\nRX(-pi/2) 16\nRZ(2.903201199539261) 16\nRX(-pi/2) 16\nCZ 16 15\nRZ(pi) 15\nRX(pi/2) 15\nRZ(pi) 16\nRX(pi/2) 16\nCZ 16 15\nRZ(3*pi/4) 17\nRX(pi/2) 17\nRZ(pi/2) 17\nRZ(-1.1800936637845592) 12\nRX(-pi/2) 12\nRX(pi/2) 13\nRZ(pi/4) 13\nRX(-pi/2) 13\nRZ(-1.1782368562744168) 13\nRX(pi/2) 13\nCZ 13 12\nRZ(pi) 12\nRX(pi/2) 12\nRZ(pi) 13\nRX(pi/2) 13\nCZ 13 12\nRZ(3*pi/4) 14\nRX(pi/2) 14\nRZ(pi/2) 14\nRZ(pi) 10\nRX(pi/2) 10\nRZ(2.0585522864068317) 10\nRX(-pi/2) 10\nRZ(-0.999694420870997) 10\nRX(pi/2) 10\nRZ(2.339424819156606) 10\nRX(-pi/2) 10\nRX(pi/2) 16\nCZ 17 16\nRZ(-pi/2) 17\nRX(-pi/2) 17\nRZ(-pi/4) 17\nRX(pi/2) 17\nCZ 17 16\nCZ 34 33\nRZ(-pi/2) 33\nRX(pi/2) 33\nRZ(-pi/2) 34\nRX(-pi/2) 34\nCZ 34 33\nRX(-pi/2) 33\nRX(pi/2) 34\nCZ 34 33\nRX(pi) 36\nCZ 36 35\nRZ(-pi/2) 36\nRX(-pi/2) 36\nRZ(-pi/4) 36\nRX(pi/2) 36\nCZ 36 21\nRX(-pi/2) 36\nRZ(pi/4) 36\nRX(pi/2) 36\nCZ 36 35\nRZ(1.7391117323457639) 21\nRX(-pi/2) 21\nRX(pi/2) 36\nRZ(-3*pi/4) 36\nRX(-pi/2) 36\nRZ(-1.6491561319419867) 36\nRX(pi/2) 36\nCZ 21 36\nRZ(pi) 21\nRX(pi/2) 21\nRZ(pi) 36\nRX(pi/2) 36\nCZ 21 36\nRZ(-pi/4) 35\nRX(pi/2) 35\nRX(pi/2) 36\nRZ(1.4024809212440283) 36\nCZ 35 36\nRX(pi/2) 21\nRZ(-1.4133203837870587) 21\nRX(-pi/2) 21\nRZ(-2.342160871123152) 21\nRX(pi/2) 21\nRZ(0.8427233520432797) 21\nRX(-pi/2) 21\nRZ(1.1466579245372337) 21\nRX(-pi/2) 21\nRZ(pi/4) 36\nRX(pi/2) 36\nRZ(1.3699349620081573) 36\nRX(-pi/2) 36\nRZ(-0.6253621093545758) 36\nRX(pi/2) 36\nRZ(2.676700651579777) 36\nRX(-pi/2) 36\nCZ 36 21\nRZ(-pi/2) 21\nRX(pi/2) 21\nRZ(-pi/2) 36\nRX(-pi/2) 36\nCZ 36 21\nRX(-pi/2) 21\nRX(pi/2) 36\nCZ 36 21\nRX(pi/2) 35\nRZ(0.09764351479329392) 35\nRX(pi) 35\nRX(pi/2) 36\nRZ(0.38097020954719635) 36\nRX(-pi/2) 36\nRZ(-0.851284815245248) 36\nRX(pi/2) 36\nRZ(-0.729580265549445) 36\nRX(pi/2) 36\nCZ 35 36\nRX(pi/2) 21\nRZ(-0.5731386193658733) 21\nRX(-pi/2) 21\nRZ(-1.305240021327016) 21\nRX(pi/2) 21\nRZ(2.5545160568125773) 21\nRX(pi/2) 21\nRZ(-1.6684398415881896) 36\nRX(-pi/2) 36\nRZ(3*pi/4) 36\nRX(pi/2) 36\nCZ 21 36\nRX(-pi/2) 36\nRZ(-pi/4) 36\nRX(-pi/2) 36\nCZ 36 35\nRZ(0.7188903724715816) 21\nRX(-pi/2) 21\nRX(pi/2) 36\nRZ(-3*pi/4) 36\nRX(-pi/2) 36\nRZ(0.851905954323315) 36\nRX(pi/2) 36\nCZ 36 21\nRZ(pi) 21\nRX(pi/2) 21\nRZ(pi) 36\nRX(pi/2) 36\nCZ 36 21\nRZ(pi/2) 17\nRX(-pi/2) 17\nRZ(-2.421270596155203) 17\nRX(-pi/2) 17\nRZ(-1.720398034666342) 17\nRX(pi/2) 17\nRZ(1.376424033150382) 17\nRX(-pi/2) 17\nCZ 17 10\nRZ(-pi/2) 10\nRX(pi/2) 10\nRZ(-pi/2) 17\nRX(-pi/2) 17\nCZ 17 10\nRX(-pi/2) 10\nRX(pi/2) 17\nCZ 17 10\nRZ(pi) 11\nRX(pi/2) 11\nRZ(1.5402270321593616) 11\nRX(-pi/2) 11\nRZ(-0.5567213682107837) 11\nRX(pi/2) 11\nRZ(2.628174481531998) 11\nRX(-pi/2) 11\nRX(pi/2) 13\nRZ(3.0538347782469053) 13\nRX(pi) 13\nRZ(-pi/2) 14\nRX(-pi/2) 14\nRZ(2.0532497025248944) 14\nRX(-pi/2) 14\nCZ 14 13\nRX(pi/2) 14\nRZ(pi/4) 14\nRX(-pi/2) 14\nCZ 14 13\nRX(-pi/2) 15\nRZ(2.903201199539262) 15\nRX(-pi/2) 15\nRZ(3*pi/4) 15\nRX(pi/2) 15\nRZ(1.6950603377091138) 15\nRX(-pi/2) 15\nRZ(-1.555861634755871) 15\nRX(pi/2) 15\nRZ(-1.3318739764707335) 15\nRX(-pi/2) 15\nRX(pi/2) 33\nRZ(1.5761185037364298) 33\nRX(-pi/2) 33\nRZ(-1.1969932805389485) 33\nRX(pi/2) 33\nRZ(2.3684005417539247) 33\nRX(-pi/2) 33\nRZ(0.09764351479329392) 35\nRX(pi/2) 35\nRX(-pi/2) 36\nCZ 35 36\nRX(-pi/2) 35\nRZ(-pi/4) 35\nRX(pi/2) 35\nCZ 35 36\nRZ(1.0883429510649005) 13\nRX(-pi/2) 13\nRZ(-1.6330180431047234) 13\nRX(-pi/2) 13\nRZ(-0.5662652067537542) 13\nRX(pi/2) 13\nRZ(-2.718472747125277) 13\nRX(-pi/2) 13\nRZ(-pi/2) 14\nRX(pi/2) 14\nRZ(1.300394787839315) 14\nRX(-pi/2) 14\nRZ(2.0532497025248926) 14\nRX(pi/2) 14\nCZ 15 14\nRX(pi/2) 14\nRZ(-pi/2) 15\nRX(-pi/2) 15\nCZ 15 14\nRX(-pi/2) 14\nRX(pi/2) 15\nCZ 15 14\nRZ(-pi/2) 31\nRX(pi/2) 31\nRZ(2.9933658042136138) 31\nRX(pi/2) 31\nRX(pi/2) 32\nRZ(1.7211068301910277) 32\nCZ 31 32\nRX(pi/2) 31\nRZ(pi/4) 31\nRX(-pi/2) 31\nCZ 31 32\nRX(pi/2) 34\nRZ(0.4476079641754716) 34\nRX(-pi/2) 34\nRZ(-1.5362327088720684) 34\nRX(pi/2) 34\nRZ(-2.8579468965980466) 34\nRX(pi/2) 34\nRZ(2.70732621147865) 35\nRX(pi) 35\nCZ 35 34\nRX(pi/2) 12\nRZ(1.9633557973153763) 12\nRX(-pi/2) 12\nRZ(-3*pi/4) 12\nRX(-pi/2) 12\nRZ(-1.8254351871869416) 12\nRX(-pi/2) 12\nRZ(-1.604372667016842) 12\nRX(pi/2) 12\nRZ(1.8316229748255886) 12\nRX(-pi/2) 12\nRX(pi/2) 13\nRZ(2.4117172184489064) 13\nRX(-pi/2) 13\nRZ(-0.6464734056448967) 13\nRX(pi/2) 13\nRZ(-1.2389015759001385) 13\nRX(-pi/2) 13\nRZ(-1.6432860034521584) 13\nRX(pi/2) 13\nRZ(-1.3385899450628604) 13\nRX(-pi/2) 13\nRX(pi/2) 14\nRZ(-1.5212942257629865) 14\nRX(-pi/2) 14\nRZ(-2.061278996618876) 14\nRX(pi/2) 14\nRZ(1.9647073228985443) 14\nRX(-pi/2) 14\nRZ(-1.6432860034521584) 14\nRX(pi/2) 14\nRZ(-1.3385899450628607) 14\nRX(-pi/2) 14\nCZ 13 14\nRZ(-pi/2) 13\nRX(-pi/2) 13\nRZ(-pi/2) 14\nRX(pi/2) 14\nCZ 13 14\nRX(pi/2) 13\nRX(-pi/2) 14\nCZ 13 14\nRX(pi/2) 31\nRZ(-1.7190231761710768) 31\nRZ(pi/2) 34\nRX(pi/2) 34\nRZ(0.44683284287478925) 34\nRX(-pi/2) 34\nRZ(-2.70732621147865) 34\nRX(pi/2) 34\nCZ 34 33\nRZ(-pi/2) 33\nRX(pi/2) 33\nRX(-pi/2) 34\nCZ 34 33\nRX(-pi/2) 33\nRX(pi/2) 34\nCZ 34 33\nRX(pi/2) 12\nRZ(0.92168047365704) 12\nRX(-pi/2) 12\nRZ(-2.3202083356456664) 12\nRX(pi/2) 12\nRZ(-1.7647277127823262) 12\nRX(-pi/2) 12\nRZ(-3.0572743578665857) 12\nRX(pi/2) 12\nRZ(-1.1266841196202588) 12\nRX(-pi/2) 12\nRX(pi/2) 13\nRZ(1.6146884247291156) 13\nRX(-pi/2) 13\nRZ(-1.4571955443331277) 13\nRX(pi/2) 13\nRZ(1.4002834550860184) 13\nRX(-pi/2) 13\nRZ(-3.0572743578665853) 13\nRX(pi/2) 13\nRZ(-1.1266841196202586) 13\nRX(-pi/2) 13\nCZ 12 13\nRZ(-pi/2) 12\nRX(-pi/2) 12\nRZ(-pi/2) 13\nRX(pi/2) 13\nCZ 12 13\nRX(pi/2) 12\nRX(-pi/2) 13\nCZ 12 13\nRX(pi/2) 25\nRZ(0.025876173771659558) 25\nRX(-pi/2) 25\nRZ(-2.283526438428502) 25\nRX(pi/2) 25\nRZ(2.494730549438146) 25\nRX(-pi/2) 25\nRZ(pi/2) 13\nRX(pi/2) 13\nRZ(2.697480446415148) 13\nRX(-pi/2) 13\nRZ(-0.08431829572320709) 13\nRX(pi/2) 13\nRZ(1.9353611367181234) 13\nRX(-pi/2) 13\nRZ(-0.795290020436582) 13\nRX(pi/2) 13\nRZ(1.9282721047999978) 13\nRX(pi/2) 13\nRZ(-1.287744838582695) 13\nRX(pi/2) 13\nRZ(-1.5826163357309502) 13\nRX(-pi/2) 13\nRZ(pi/2) 14\nRX(pi/2) 14\nRZ(2.9093862718577577) 14\nRX(-pi/2) 14\nRZ(-1.4983066501376348) 14\nRX(pi/2) 14\nRZ(1.7277548032990437) 14\nRX(-pi/2) 14\nRZ(-1.1965816552765813) 14\nRX(pi/2) 14\nRZ(0.7518412465151476) 14\nRX(pi/2) 14\nRZ(-1.2877448385826948) 14\nRX(pi/2) 14\nRZ(-1.5826163357309502) 14\nRX(-pi/2) 14\nRX(pi/2) 24\nRZ(-1.3820627380461623) 24\nRX(-pi/2) 24\nRZ(-0.9818656448436526) 24\nRX(pi/2) 24\nRZ(-2.9528590648410598) 24\nRX(-pi/2) 24\nRX(pi/2) 12\nRZ(-2.724864317420739) 12\nRX(-pi/2) 12\nRZ(-2.2970480201395675) 12\nRX(pi/2) 12\nRZ(2.7600944984198246) 12\nRX(-pi/2) 12\nRZ(-2.2261794440597478) 12\nRX(pi/2) 12\nRZ(-2.6003232170370616) 12\nRX(-pi/2) 12\nRX(pi/2) 25\nRZ(-0.5846034652565888) 25\nRX(-pi/2) 25\nRZ(-1.381093141754414) 25\nRX(pi/2) 25\nRZ(-2.752820941189089) 25\nRX(-pi/2) 25\nCZ 25 12\nRZ(-pi/2) 12\nRX(pi/2) 12\nRZ(-pi/2) 25\nRX(-pi/2) 25\nCZ 25 12\nRX(-pi/2) 12\nRX(pi/2) 25\nCZ 25 12\nRX(pi/2) 10\nRZ(-1.0459953081708746) 10\nRX(-pi/2) 10\nRZ(-1.2630463825689127) 10\nRX(pi/2) 10\nRZ(-1.283337261912755) 10\nRX(-pi/2) 10\nRZ(-0.8354070151452242) 10\nRX(pi/2) 10\nRZ(-0.009220221078568347) 10\nRX(-pi/2) 10\nRX(pi/2) 11\nRZ(0.9923198694530337) 11\nRX(-pi/2) 11\nRZ(-0.9249849226746982) 11\nRX(pi/2) 11\nRZ(-1.2275662250810857) 11\nRX(-pi/2) 11\nCZ 11 10\nRZ(-pi/2) 10\nRX(pi/2) 10\nRZ(-pi/2) 11\nRX(-pi/2) 11\nCZ 11 10\nRX(-pi/2) 10\nRX(pi/2) 11\nCZ 11 10\nRZ(pi/2) 12\nRX(pi/2) 12\nRZ(-0.8792195194716462) 12\nRX(-pi/2) 12\nRZ(-0.8654491676796116) 12\nRX(pi/2) 12\nRZ(-0.4791400058038784) 12\nRX(-pi/2) 12\nRZ(-0.6319164958615521) 12\nRX(pi/2) 12\nRZ(0.5394926968732916) 12\nRX(pi/2) 12\nRZ(-0.7468555982934713) 12\nRX(pi/2) 12\nRZ(-0.5537505296172055) 12\nRX(-pi/2) 12\nCZ 14 13\nRZ(-pi/2) 13\nRX(pi/2) 13\nRZ(-pi/2) 14\nRX(-pi/2) 14\nCZ 14 13\nRX(-pi/2) 13\nRX(pi/2) 14\nCZ 14 13\nRZ(pi/2) 15\nRX(pi/2) 15\nRZ(-0.23892235032416306) 15\nRX(-pi/2) 15\nRZ(-1.585731018833922) 15\nRX(pi/2) 15\nRZ(1.5197950947812302) 15\nRX(pi/2) 15\nRZ(-0.7778764458884829) 15\nRX(pi/2) 15\nRZ(-2.9663274106619095) 15\nRX(-pi/2) 15\nRX(pi/2) 23\nRZ(-0.35080737237156195) 23\nRX(-pi/2) 23\nRZ(-2.5004084664607418) 23\nRX(pi/2) 23\nRZ(-2.259457567519501) 23\nRX(-pi/2) 23\nRX(pi/2) 24\nRZ(0.26425383172522887) 24\nRX(-pi/2) 24\nRZ(-1.5759258556911668) 24\nRX(pi/2) 24\nRZ(2.8094715759326583) 24\nRX(-pi/2) 24\nRX(pi/2) 25\nRZ(1.6037442276493585) 25\nRX(-pi/2) 25\nRZ(-1.5251992227345033) 25\nRX(pi/2) 25\nRZ(1.3216309085583897) 25\nRX(-pi/2) 25\nRZ(-1.6802875394981998) 25\nRX(pi/2) 25\nRZ(1.8707099859282785) 25\nRX(-pi/2) 25\nCZ 24 25\nRZ(-pi/2) 24\nRX(-pi/2) 24\nRZ(-pi/2) 25\nRX(pi/2) 25\nCZ 24 25\nRX(pi/2) 24\nRX(-pi/2) 25\nCZ 24 25\nRZ(pi) 22\nRX(pi/2) 22\nRZ(-1.1344199050892758) 22\nRX(-pi/2) 22\nRZ(-2.3034913738557) 22\nRX(pi/2) 22\nRZ(1.3619106284254077) 22\nRX(-pi/2) 22\nRX(pi/2) 23\nRZ(1.6876135609209981) 23\nRX(-pi/2) 23\nRZ(-1.0027417258031517) 23\nRX(pi/2) 23\nRZ(-1.260960540366467) 23\nRX(-pi/2) 23\nRZ(-2.83352214166156) 23\nRX(pi/2) 23\nRZ(0.12589221900795167) 23\nRX(-pi/2) 23\nRX(pi/2) 24\nRZ(2.0957937574983476) 24\nRX(-pi/2) 24\nRZ(-2.105739201148076) 24\nRX(pi/2) 24\nRZ(-0.15799107368789844) 24\nRX(-pi/2) 24\nRZ(-1.584694074280446) 24\nRX(pi/2) 24\nRZ(0.2855114858454803) 24\nRX(-pi/2) 24\nCZ 23 24\nRZ(-pi/2) 23\nRX(-pi/2) 23\nRZ(-pi/2) 24\nRX(pi/2) 24\nCZ 23 24\nRX(pi/2) 23\nRX(-pi/2) 24\nCZ 23 24\nRZ(0.5639043046913788) 16\nRX(pi/2) 16\nRZ(1.7122872309354404) 16\nRX(-pi/2) 16\nRZ(-0.3551340183241856) 16\nRX(pi/2) 16\nRZ(-3.0001017494492483) 16\nRX(-pi/2) 16\nRX(pi/2) 15\nRZ(-2.878179693435657) 15\nRX(-pi/2) 15\nRZ(-1.070545432691663) 15\nRX(pi/2) 15\nRZ(1.6319191530422277) 15\nRX(-pi/2) 15\nRZ(-1.2463384751001279) 15\nRX(pi/2) 15\nRZ(1.1140483037934459) 15\nRX(-pi/2) 15\nRX(pi/2) 16\nRZ(0.29459672077920973) 16\nRX(-pi/2) 16\nRZ(-0.9122761366772631) 16\nRX(pi/2) 16\nRZ(2.540169576152308) 16\nRX(-pi/2) 16\nRZ(-2.123048125996565) 16\nRX(pi/2) 16\nRZ(3.0660200482474886) 16\nRX(-pi/2) 16\nCZ 16 15\nRZ(-pi/2) 15\nRX(pi/2) 15\nRZ(-pi/2) 16\nRX(-pi/2) 16\nCZ 16 15\nRX(-pi/2) 15\nRX(pi/2) 16\nCZ 16 15\nRX(pi/2) 17\nRZ(0.7751111341382971) 17\nRX(-pi/2) 17\nRZ(-2.9825941050063856) 17\nRX(pi/2) 17\nRZ(2.4070304077566753) 17\nRX(-pi/2) 17\nRX(pi/2) 22\nRZ(-1.977059374084002) 22\nRX(-pi/2) 22\nRZ(-2.1188282330966417) 22\nRX(pi/2) 22\nRZ(0.3366197086729812) 22\nRX(-pi/2) 22\nRZ(-1.5509921304164147) 22\nRX(pi/2) 22\nRZ(-3.0068768600141977) 22\nRX(-pi/2) 22\nRX(pi/2) 23\nRZ(-0.47400794994760276) 23\nRX(-pi/2) 23\nRZ(-1.2793719642933878) 23\nRX(pi/2) 23\nRZ(-0.7500380870153244) 23\nRX(-pi/2) 23\nRZ(-1.089575128027193) 23\nRX(pi/2) 23\nRZ(1.6032297126640547) 23\nRX(-pi/2) 23\nCZ 22 23\nRZ(-pi/2) 22\nRX(-pi/2) 22\nRZ(-pi/2) 23\nRX(pi/2) 23\nCZ 22 23\nRX(pi/2) 22\nRX(-pi/2) 23\nCZ 22 23\nRZ(1.1365298846837533) 35\nRX(pi/2) 35\nRZ(1.6415858762748954) 35\nRX(-pi/2) 35\nRZ(-1.317343401556909) 35\nRX(pi/2) 35\nRZ(-1.3597504555907551) 35\nRX(-pi/2) 35\nRZ(pi/2) 34\nRX(pi/2) 34\nRZ(2.8637925886036344) 34\nRX(-pi/2) 34\nRZ(-1.3879707580392253) 34\nRX(pi/2) 34\nRZ(-1.94781093288591) 34\nRX(pi/2) 34\nRZ(-1.3592570829851311) 34\nRX(pi/2) 34\nRZ(-1.3586767993884306) 34\nRX(-pi/2) 34\nRX(pi/2) 22\nRZ(0.5451471674758638) 22\nRX(-pi/2) 22\nRZ(-0.9484754894300755) 22\nRX(pi/2) 22\nRZ(1.3783553780890188) 22\nRX(-pi/2) 22\nRZ(-1.5046725788565871) 22\nRX(pi/2) 22\nRZ(-1.2854115185790813) 22\nRX(-pi/2) 22\nRX(pi/2) 35\nRZ(-3.1135280121590614) 35\nRX(-pi/2) 35\nRZ(-2.4077312639910016) 35\nRX(pi/2) 35\nRZ(-0.9555951730897747) 35\nRX(-pi/2) 35\nRZ(-1.5046725788565871) 35\nRX(pi/2) 35\nRZ(-1.285411518579081) 35\nRX(-pi/2) 35\nCZ 35 22\nRZ(-pi/2) 22\nRX(pi/2) 22\nRZ(-pi/2) 35\nRX(-pi/2) 35\nCZ 35 22\nRX(-pi/2) 22\nRX(pi/2) 35\nCZ 35 22\nRZ(pi/2) 10\nRX(pi/2) 10\nRZ(-1.561576105716328) 10\nRX(-pi/2) 10\nRZ(-0.8354070151452242) 10\nRX(pi/2) 10\nRZ(-0.5651384288691665) 10\nRX(-pi/2) 10\nRZ(-2.1245625101593606) 10\nRX(pi/2) 10\nRZ(-2.9853110872666315) 10\nRX(pi/2) 10\nRZ(-1.2062010741893727) 10\nRX(pi/2) 10\nRZ(-1.3854835901982347) 10\nRX(-pi/2) 10\nRX(pi/2) 16\nRZ(-0.82276754480832) 16\nRX(-pi/2) 16\nRZ(-2.108833625283388) 16\nRX(pi/2) 16\nRZ(2.446490455438198) 16\nRX(-pi/2) 16\nRZ(-0.8238479117383929) 16\nRX(pi/2) 16\nRZ(-1.2478661341200414) 16\nRX(-pi/2) 16\nRX(pi/2) 17\nRZ(-0.6897262009276717) 17\nRX(-pi/2) 17\nRZ(-1.1102474536404516) 17\nRX(pi/2) 17\nRZ(-2.613739575836709) 17\nRX(-pi/2) 17\nCZ 17 16\nRZ(-pi/2) 16\nRX(pi/2) 16\nRZ(-pi/2) 17\nRX(-pi/2) 17\nCZ 17 16\nRX(-pi/2) 16\nRX(pi/2) 17\nCZ 17 16\nRZ(-pi/2) 32\nRX(-pi/2) 32\nRZ(1.8132486803847225) 32\nRX(-pi/2) 32\nRZ(2.9933658042136124) 32\nRX(pi) 32\nRZ(pi/2) 33\nRX(pi/2) 33\nRZ(2.8637925886036344) 33\nRX(-pi/2) 33\nRZ(-1.7536218955505676) 33\nRX(pi/2) 33\nRZ(-2.4489449427962056) 33\nRX(pi/2) 33\nRZ(-0.3233479707845904) 33\nRX(pi/2) 33\nRZ(-1.772783460017437) 33\nRX(-pi/2) 33\nCZ 33 32\nRZ(-pi/2) 32\nRX(pi/2) 32\nRZ(-pi/2) 33\nRX(-pi/2) 33\nCZ 33 32\nRX(-pi/2) 32\nRX(pi/2) 33\nCZ 33 32\nRX(pi/2) 34\nRZ(0.22271332758155726) 34\nRX(-pi/2) 34\nRZ(-1.784874340589417) 34\nRX(pi/2) 34\nRZ(-1.8374539059188946) 34\nRX(-pi/2) 34\nRZ(-2.1111957032090425) 34\nRX(pi/2) 34\nRZ(-2.827919909849622) 34\nRX(-pi/2) 34\nRX(pi/2) 35\nRZ(-1.5671787289216244) 35\nRX(-pi/2) 35\nRZ(-1.864279700737517) 35\nRX(pi/2) 35\nRZ(2.767169309728218) 35\nRX(-pi/2) 35\nRZ(-2.2095442539250434) 35\nRX(pi/2) 35\nRZ(2.1849594223088302) 35\nRX(-pi/2) 35\nCZ 34 35\nRZ(-pi/2) 34\nRX(-pi/2) 34\nRZ(-pi/2) 35\nRX(pi/2) 35\nCZ 34 35\nRX(pi/2) 34\nRX(-pi/2) 35\nCZ 34 35\nRX(pi/2) 32\nRZ(-1.7190231761710761) 32\nRX(-pi/2) 32\nRZ(-1.8132486803847214) 32\nRX(pi/2) 32\nRZ(-1.8972243458130091) 32\nRX(pi/2) 32\nCZ 31 32\nRX(pi/2) 33\nRZ(-0.6040184031551078) 33\nRX(-pi/2) 33\nRZ(-1.2749625054538452) 33\nRX(pi/2) 33\nRZ(-0.6916768203289241) 33\nRX(-pi/2) 33\nRX(pi/2) 34\nRZ(1.400097135766543) 34\nRX(-pi/2) 34\nRZ(-1.7396919628267542) 34\nRX(pi/2) 34\nRZ(1.8969446240343615) 34\nRX(-pi/2) 34\nCZ 34 33\nRZ(-pi/2) 33\nRX(pi/2) 33\nRZ(-pi/2) 34\nRX(-pi/2) 34\nCZ 34 33\nRX(-pi/2) 33\nRX(pi/2) 34\nCZ 34 33\nRX(-pi/2) 32\nRZ(-pi/4) 32\nRX(pi/2) 32\nRZ(pi/2) 33\nRX(pi/2) 33\nRZ(-2.971156252998421) 33\nRX(-pi/2) 33\nRZ(-0.9495883321700593) 33\nRX(pi/2) 33\nRZ(2.374316973588387) 33\nRX(-pi/2) 33\nCZ 32 33\nRX(-pi/2) 32\nRZ(pi/4) 32\nRX(pi/2) 32\nCZ 32 31\nRX(pi/2) 32\nRZ(-3*pi/4) 32\nRX(-pi/2) 32\nRZ(2.4340910083852147) 32\nRX(-pi/2) 32\nRZ(2.596347967216115) 33\nRX(pi/2) 33\nCZ 33 32\nRZ(pi) 32\nRX(pi/2) 32\nRZ(pi) 33\nRX(pi/2) 33\nCZ 33 32\nRZ(pi/2) 34\nRX(pi/2) 34\nRZ(-0.5848040016933371) 34\nRX(-pi/2) 34\nRZ(-1.910083751249015) 34\nRX(pi/2) 34\nRZ(-0.32371931194287384) 34\nRX(pi/2) 34\nRZ(-0.6363813990751102) 34\nRX(pi/2) 34\nRZ(2.0840911329446508) 34\nRX(-pi/2) 34\nRZ(-pi/4) 31\nRX(pi/2) 31\nRX(pi/2) 32\nRZ(2.596347967216113) 32\nRX(pi) 32\nCZ 31 32\nRX(-pi/2) 31\nRZ(-pi/4) 31\nRX(pi/2) 31\nCZ 31 32\nRX(-pi/2) 33\nRZ(-0.2717533934759903) 33\nRX(-pi/2) 33\nRZ(-2.3072209018279355) 33\nRX(pi/2) 33\nRZ(-1.9040970430449198) 33\nRX(pi/2) 33\nRZ(2.08409113294465) 33\nRX(-pi/2) 33\nCZ 34 33\nRZ(-pi/2) 33\nRX(pi/2) 33\nRZ(-pi/2) 34\nRX(-pi/2) 34\nCZ 34 33\nRX(-pi/2) 33\nRX(pi/2) 34\nCZ 34 33\nRZ(-3*pi/4) 32\nRX(pi/2) 32\nRZ(-1.9758509332949856) 32\nRX(-pi/2) 32\nRZ(-1.9821354992939246) 32\nRX(pi/2) 32\nRZ(2.19670328622247) 32\nRX(-pi/2) 32\nRZ(pi/2) 33\nRX(pi/2) 33\nRZ(-0.513294806149753) 33\nRX(-pi/2) 33\nRZ(-2.5052112545146827) 33\nRX(pi/2) 33\nRZ(-1.4837917155114457) 33\nRX(pi/2) 33\nRZ(-1.9821354992939249) 33\nRX(pi/2) 33\nRZ(2.196703286222471) 33\nRX(-pi/2) 33\nCZ 33 32\nRZ(-pi/2) 32\nRX(pi/2) 32\nRZ(-pi/2) 33\nRX(-pi/2) 33\nCZ 33 32\nRX(-pi/2) 32\nRX(pi/2) 33\nCZ 33 32\nRZ(pi/2) 34\nRX(pi/2) 34\nRZ(2.6282978474400402) 34\nRX(-pi/2) 34\nRZ(-2.5052112545146836) 34\nRX(pi/2) 34\nRZ(0.3345347352532291) 34\nRX(-pi/2) 34\nRZ(-1.7826307014421676) 34\nRX(pi/2) 34\nRZ(1.605008038526728) 34\nRX(-pi/2) 34\nRX(pi/2) 32\nRZ(-2.058963958159833) 32\nRX(-pi/2) 32\nRZ(-2.4264463901032696) 32\nRX(pi/2) 32\nRZ(0.1022959231693985) 32\nRX(pi/2) 32\nCZ 31 32\nRX(pi/2) 33\nRZ(-0.6197098188362428) 33\nRX(-pi/2) 33\nRZ(-2.372206567025024) 33\nRX(pi/2) 33\nRZ(2.8835850502425906) 33\nRX(-pi/2) 33\nRX(pi/2) 34\nRZ(-2.694749962670786) 34\nRX(-pi/2) 34\nRZ(-2.2365266513783313) 34\nRX(pi/2) 34\nRZ(-3.0198889015276995) 34\nRX(-pi/2) 34\nRZ(-0.8871843246860479) 34\nRX(pi/2) 34\nRZ(-1.811688538850085) 34\nRX(-pi/2) 34\nCZ 33 34\nRZ(-pi/2) 33\nRX(-pi/2) 33\nRZ(-pi/2) 34\nRX(pi/2) 34\nCZ 33 34\nRX(pi/2) 33\nRX(-pi/2) 34\nCZ 33 34\nRZ(pi/2) 32\nRX(pi/2) 32\nRZ(2.8960604075888505) 32\nRX(-pi/2) 32\nRZ(1.9144158626879468) 32\nRX(pi/2) 32\nRZ(pi/2) 33\nRX(pi/2) 33\nRZ(0.24089221205518727) 33\nRX(-pi/2) 33\nRZ(-2.254408328903745) 33\nRX(pi/2) 33\nRZ(1.4350054876557705) 33\nRX(-pi/2) 33\nRZ(-2.961912714792631) 33\nRX(pi/2) 33\nRZ(2.0094361503835048) 33\nRX(-pi/2) 33\nCZ 33 32\nRX(pi/2) 32\nRZ(-pi/2) 33\nRX(-pi/2) 33\nCZ 33 32\nRX(-pi/2) 32\nRX(pi/2) 33\nCZ 33 32\nRZ(pi) 31\nRX(pi/2) 31\nRX(pi/2) 32\nRZ(-1.7988475430423838) 32\nRX(-pi/2) 32\nRZ(-0.23323261130110745) 32\nRX(pi/2) 32\nRZ(0.13998555069250607) 32\nRX(pi/2) 32\nCZ 31 32\nRZ(-pi/2) 32\nRX(-pi/2) 32\nRZ(-3*pi/4) 32\nRX(pi/2) 32\nRX(pi/2) 33\nRZ(0.031611654385909765) 33\nRX(-pi/2) 33\nRZ(-1.3507425167680207) 33\nRX(pi/2) 33\nRZ(0.23672291923785949) 33\nRX(pi/2) 33\nCZ 33 32\nRX(pi/2) 32\nRZ(3*pi/4) 32\nRX(-pi/2) 32\nCZ 32 31\nRX(pi/2) 32\nRZ(pi/4) 32\nRX(-pi/2) 32\nRZ(-1.3893980560875967) 32\nRX(pi/2) 32\nRZ(0.8242320959844265) 33\nRX(pi/2) 33\nCZ 33 32\nRZ(pi) 32\nRX(pi/2) 32\nRZ(pi) 33\nRX(pi/2) 33\nCZ 33 32\nRX(pi/2) 34\nRZ(0.42483498522905566) 34\nRX(-pi/2) 34\nRZ(-0.7976718863972023) 34\nRX(pi/2) 34\nRZ(2.6863376886070913) 34\nRX(-pi/2) 34\nRZ(-0.5105616545654783) 31\nRX(-pi/2) 31\nRX(pi/2) 32\nRZ(-3.116827892786848) 32\nRX(pi) 32\nCZ 31 32\nRX(pi/2) 31\nRZ(pi/4) 31\nRX(-pi/2) 31\nCZ 31 32\nRX(pi/2) 33\nRZ(1.0463904424174573) 33\nRX(-pi/2) 33\nRZ(-0.9202351994945497) 33\nRX(pi/2) 33\nRZ(0.4858606087865507) 33\nRX(pi/2) 33\nRZ(-1.3400985392794258) 33\nRX(-pi/2) 33\nCZ 34 33\nRZ(-pi/2) 33\nRX(pi/2) 33\nRZ(-pi/2) 34\nRX(-pi/2) 34\nCZ 34 33\nRX(-pi/2) 33\nRX(pi/2) 34\nCZ 34 33\nRZ(pi/2) 32\nRX(-pi/2) 32\nRZ(-0.6130492611300983) 32\nRX(-pi/2) 32\nRZ(-0.9517152255972785) 32\nRX(pi/2) 32\nRZ(-1.6613900279561802) 32\nRX(-pi/2) 32\nRX(pi/2) 33\nRZ(0.662539499466291) 33\nRX(-pi/2) 33\nRZ(-0.4248862827976883) 33\nRX(pi/2) 33\nRZ(2.563413881704646) 33\nRX(-pi/2) 33\nCZ 33 32\nRZ(-pi/2) 32\nRX(pi/2) 32\nRZ(-pi/2) 33\nRX(-pi/2) 33\nCZ 33 32\nRX(-pi/2) 32\nRX(pi/2) 33\nCZ 33 32\nRX(pi/2) 31\nRZ(-1.8456328356268483) 31\nRX(-pi/2) 31\nRZ(-0.4094924588591954) 31\nRX(pi/2) 32\nRZ(0.2874557199054342) 32\nRX(-pi/2) 32\nRZ(-1.4955942875047104) 32\nRX(pi/2) 32\nRZ(0.9782745553517493) 32\nRX(pi/2) 32\nCZ 32 31\nRZ(pi/2) 32\nRX(pi/2) 32\nRZ(0.7039694428085634) 32\nRX(-pi/2) 32\nRZ(1.1613038679357004) 32\nRX(pi) 32\nRZ(pi/2) 33\nRX(pi/2) 33\nRZ(-3.041139900074036) 33\nRX(-pi/2) 33\nRZ(-0.9632453955025345) 33\nRX(pi/2) 33\nRZ(-2.5153783022672815) 33\nRX(pi/2) 33\nRZ(-0.8722527799549666) 33\nRX(pi/2) 33\nRZ(-0.6565624764336044) 33\nRX(-pi/2) 33\nCZ 33 32\nRZ(-pi/2) 32\nRX(pi/2) 32\nRZ(-pi/2) 33\nRX(-pi/2) 33\nCZ 33 32\nRX(-pi/2) 32\nRX(pi/2) 33\nCZ 33 32\nRX(pi/2) 34\nRZ(-1.0526246016400678) 34\nRX(-pi/2) 34\nRZ(-0.8264261242723349) 34\nRX(pi/2) 34\nRZ(2.321216747459439) 34\nRX(-pi/2) 34\nRZ(pi/2) 22\nRX(pi/2) 22\nRZ(2.856207845373978) 22\nRX(-pi/2) 22\nRZ(-1.6369200747332058) 22\nRX(pi/2) 22\nRZ(-2.871578856339742) 22\nRX(-pi/2) 22\nRZ(-1.6034529103438913) 22\nRX(pi/2) 22\nRZ(1.8331385119974963) 22\nRX(pi/2) 22\nRZ(-2.0250287820538646) 22\nRX(pi/2) 22\nRZ(-0.13357158197067776) 22\nRX(-pi/2) 22\nRX(pi/2) 33\nRZ(1.9554108039554994) 33\nRX(-pi/2) 33\nRZ(-2.1809785924504927) 33\nRX(pi/2) 33\nRZ(1.82530344276992) 33\nRX(-pi/2) 33\nRZ(-2.4822788909774784) 33\nRX(pi/2) 33\nRZ(-1.2239757540038785) 33\nRX(-pi/2) 33\nRX(pi/2) 34\nRZ(2.403756306567167) 34\nRX(-pi/2) 34\nRZ(-1.1844315818854247) 34\nRX(pi/2) 34\nRZ(-1.157727844293817) 34\nRX(-pi/2) 34\nRZ(-1.5610711211749335) 34\nRX(pi/2) 34\nRZ(-1.3386984615395077) 34\nRX(-pi/2) 34\nCZ 34 33\nRZ(-pi/2) 33\nRX(pi/2) 33\nRZ(-pi/2) 34\nRX(-pi/2) 34\nCZ 34 33\nRX(-pi/2) 33\nRX(pi/2) 34\nCZ 34 33\nRX(pi/2) 34\nRZ(-0.7658421894064993) 34\nRX(-pi/2) 34\nRZ(-1.6218499572521543) 34\nRX(pi/2) 34\nRZ(-1.576272859556072) 34\nRX(-pi/2) 34\nRZ(-2.0994006411029815) 34\nRX(pi/2) 34\nRZ(1.721164996253035) 34\nRX(-pi/2) 34\nRX(pi/2) 35\nRZ(-0.461074370587815) 35\nRX(-pi/2) 35\nRZ(-0.5568421080735692) 35\nRX(pi/2) 35\nRZ(-2.2093037829714404) 35\nRX(-pi/2) 35\nRZ(-1.7472914477865673) 35\nRX(pi/2) 35\nRZ(-2.4541385373025735) 35\nRX(-pi/2) 35\nCZ 35 34\nRZ(-pi/2) 34\nRX(pi/2) 34\nRZ(-pi/2) 35\nRX(-pi/2) 35\nCZ 35 34\nRX(-pi/2) 34\nRX(pi/2) 35\nCZ 35 34\nRX(pi/2) 23\nRZ(0.7550511722641996) 23\nRX(-pi/2) 23\nRZ(-1.5482112128494268) 23\nRX(pi/2) 23\nRZ(2.443336238231783) 23\nRX(-pi/2) 23\nRX(pi/2) 22\nRZ(0.35107570099437874) 22\nRX(-pi/2) 22\nRZ(-2.351942587632102) 22\nRX(pi/2) 22\nRZ(0.5426169618621635) 22\nRX(-pi/2) 22\nRZ(-1.0344459579307617) 22\nRX(pi/2) 22\nRZ(1.871870526067365) 22\nRX(-pi/2) 22\nRX(pi/2) 35\nRZ(1.818722868887314) 35\nRX(-pi/2) 35\nRZ(-1.810771251843158) 35\nRX(pi/2) 35\nRZ(-1.9557657398433146) 35\nRX(-pi/2) 35\nRZ(-2.893449984296033) 35\nRX(pi/2) 35\nRZ(1.3695264614363234) 35\nRX(-pi/2) 35\nCZ 22 35\nRZ(-pi/2) 22\nRX(-pi/2) 22\nRZ(-pi/2) 35\nRX(pi/2) 35\nCZ 22 35\nRX(pi/2) 22\nRX(-pi/2) 35\nCZ 22 35\nRX(pi/2) 24\nRZ(0.4842633611830364) 24\nRX(-pi/2) 24\nRZ(-0.4551148102090614) 24\nRX(pi/2) 24\nRZ(1.9755565382438354) 24\nRX(-pi/2) 24\nRX(pi/2) 22\nRZ(-2.265329317336565) 22\nRX(-pi/2) 22\nRZ(-0.9766242902060458) 22\nRX(pi/2) 22\nRZ(-0.40579644166064055) 22\nRX(-pi/2) 22\nRZ(-1.9982870141786468) 22\nRX(pi/2) 22\nRZ(1.771397963276348) 22\nRX(-pi/2) 22\nRX(pi/2) 23\nRZ(0.4995242433873427) 23\nRX(-pi/2) 23\nRZ(-2.203448077898284) 23\nRX(pi/2) 23\nRZ(2.827589270564385) 23\nRX(-pi/2) 23\nCZ 23 22\nRZ(-pi/2) 22\nRX(pi/2) 22\nRZ(-pi/2) 23\nRX(-pi/2) 23\nCZ 23 22\nRX(-pi/2) 22\nRX(pi/2) 23\nCZ 23 22\nRZ(pi/2) 25\nRX(pi/2) 25\nRZ(2.8416789944564114) 25\nRX(-pi/2) 25\nRZ(-1.6802875394982) 25\nRX(pi/2) 25\nRZ(-2.8573066315407294) 25\nRX(-pi/2) 25\nRZ(-1.0016332012145894) 25\nRX(pi/2) 25\nRZ(2.3416109563619787) 25\nRX(pi/2) 25\nRZ(-1.968544191919183) 25\nRX(pi/2) 25\nRZ(-1.2702582281583492) 25\nRX(-pi/2) 25\nRX(pi/2) 11\nRZ(0.5082503773561751) 11\nRX(-pi/2) 11\nRZ(-2.3348625926370983) 11\nRX(pi/2) 11\nRZ(-2.2430139321043523) 11\nRX(-pi/2) 11\nCZ 12 11\nRZ(-pi/2) 11\nRX(pi/2) 11\nRZ(-pi/2) 12\nRX(-pi/2) 12\nCZ 12 11\nRX(-pi/2) 11\nRX(pi/2) 12\nCZ 12 11\nRX(pi/2) 23\nRZ(1.4877089420127783) 23\nRX(-pi/2) 23\nRZ(-1.698422376215833) 23\nRX(pi/2) 23\nRZ(-1.203624492523817) 23\nRX(-pi/2) 23\nRZ(-0.7975524455744859) 23\nRX(pi/2) 23\nRZ(0.2623183676359151) 23\nRX(-pi/2) 23\nRX(pi/2) 24\nRZ(-1.7335604894280447) 24\nRX(-pi/2) 24\nRZ(-0.8150108898915912) 24\nRX(pi/2) 24\nRZ(2.7974445701799793) 24\nRX(-pi/2) 24\nRZ(-1.955742713046001) 24\nRX(pi/2) 24\nRZ(1.6713696043584108) 24\nRX(-pi/2) 24\nCZ 24 23\nRZ(-pi/2) 23\nRX(pi/2) 23\nRZ(-pi/2) 24\nRX(-pi/2) 24\nCZ 24 23\nRX(-pi/2) 23\nRX(pi/2) 24\nCZ 24 23\nRX(pi/2) 24\nRZ(0.9685136099445191) 24\nRX(-pi/2) 24\nRZ(-2.649460436733263) 24\nRX(pi/2) 24\nRZ(-2.716466743619449) 24\nRX(-pi/2) 24\nCZ 25 24\nRZ(-pi/2) 24\nRX(pi/2) 24\nRZ(-pi/2) 25\nRX(-pi/2) 25\nCZ 25 24\nRX(-pi/2) 24\nRX(pi/2) 25\nCZ 25 24\nRX(pi/2) 13\nRZ(0.8097674520809302) 13\nRX(-pi/2) 13\nRZ(-2.0925620120009194) 13\nRX(pi/2) 13\nRZ(2.2648059137500036) 13\nRX(-pi/2) 13\nRX(pi/2) 12\nRZ(-2.39152295206011) 12\nRX(-pi/2) 12\nRZ(-2.3024722347149096) 12\nRX(pi/2) 12\nRZ(0.5407821619144744) 12\nRX(-pi/2) 12\nRX(pi/2) 25\nRZ(1.0186777305086236) 25\nRX(-pi/2) 25\nRZ(-1.1114965113842776) 25\nRX(pi/2) 25\nRZ(-0.2882900887211721) 25\nRX(pi/2) 25\nCZ 25 12\nRZ(pi) 12\nRX(pi/2) 12\nRZ(pi) 25\nRX(pi/2) 25\nCZ 25 12\nRX(pi/2) 14\nRZ(-2.097175371303689) 14\nRX(-pi/2) 14\nRZ(-0.8122218065516321) 14\nRX(pi/2) 14\nRZ(2.8550376861939784) 14\nRX(-pi/2) 14\nRZ(-0.9866315317093275) 14\nRX(pi/2) 14\nRZ(-0.306371908100858) 14\nRX(-pi/2) 14\nRX(pi/2) 12\nRZ(0.8693384597466656) 12\nRX(-pi/2) 12\nRZ(-0.8641678658928522) 12\nRX(pi/2) 12\nRZ(1.9433441362734571) 12\nRX(-pi/2) 12\nCZ 13 12\nRZ(-pi/2) 12\nRX(pi/2) 12\nRZ(-pi/2) 13\nRX(-pi/2) 13\nCZ 13 12\nRX(-pi/2) 12\nRX(pi/2) 13\nCZ 13 12\nRX(pi/2) 13\nRZ(-1.5227683493820354) 13\nRX(-pi/2) 13\nRZ(-2.5702669566745415) 13\nRX(pi/2) 13\nRZ(1.7669264150142343) 13\nRX(pi/2) 13\nRZ(-1.6361823752120586) 14\nRX(pi) 14\nCZ 14 13\nRX(pi/2) 12\nRZ(1.7190831823189199) 12\nRX(-pi/2) 12\nRZ(-1.8841954034214825) 12\nRX(pi/2) 12\nRZ(0.28309859087576505) 12\nRX(-pi/2) 12\nRZ(-1.4413828176848102) 12\nRX(pi/2) 12\nRZ(2.0826316277009655) 12\nRX(-pi/2) 12\nRX(pi/2) 13\nRZ(-1.3667447948842588) 13\nRX(-pi/2) 13\nRZ(-1.7778591106944233) 13\nRX(pi/2) 13\nRZ(-0.1453754800620084) 13\nRX(-pi/2) 13\nRZ(-1.7002098359049833) 13\nRX(pi/2) 13\nRZ(-1.0589610258888278) 13\nRX(-pi/2) 13\nCZ 13 12\nRZ(-pi/2) 12\nRX(pi/2) 12\nRZ(-pi/2) 13\nRX(-pi/2) 13\nCZ 13 12\nRX(-pi/2) 12\nRX(pi/2) 13\nCZ 13 12\nRX(-pi/2) 25\nRZ(-2.103648393287246) 25\nRX(pi/2) 12\nRZ(-0.4345158513503502) 12\nRX(-pi/2) 12\nRZ(-1.9544018900374875) 12\nRX(pi/2) 12\nRZ(2.4642710164484765) 12\nRX(pi/2) 12\nRZ(-pi/2) 25\nRX(pi) 25\nCZ 12 25\nRX(pi/2) 13\nRZ(-0.21264430861676634) 13\nRX(-pi/2) 13\nRZ(-0.24442473138327092) 13\nRX(pi/2) 13\nRZ(-2.929582380659726) 13\nRX(-pi/2) 13\nRZ(-2.1874849109866674) 13\nRX(pi/2) 13\nRZ(2.081739707654205) 13\nRX(-pi/2) 13\nRX(pi/2) 14\nRZ(-1.8526195229505678) 14\nRX(-pi/2) 14\nRZ(-1.9315511346770804) 14\nRX(pi/2) 14\nRZ(0.6688997020438777) 14\nRX(-pi/2) 14\nRZ(-1.9638114101937196) 14\nRX(pi/2) 14\nRZ(-2.6057189986656457) 14\nRX(-pi/2) 14\nCZ 14 13\nRZ(-pi/2) 13\nRX(pi/2) 13\nRZ(-pi/2) 14\nRX(-pi/2) 14\nCZ 14 13\nRX(-pi/2) 13\nRX(pi/2) 14\nCZ 14 13\nRX(pi/2) 12\nRZ(-pi/4) 12\nRX(-pi/2) 12\nRZ(0.3491191261777571) 12\nRX(-pi/2) 12\nRX(pi/2) 13\nRZ(0.8230240976060454) 13\nRX(-pi/2) 13\nRZ(-0.6856921350527333) 13\nRX(pi/2) 13\nRZ(2.2723097839424042) 13\nRX(pi/2) 13\nCZ 13 12\nRZ(pi) 12\nRX(pi/2) 12\nRZ(pi) 13\nRX(pi/2) 13\nCZ 13 12\nRZ(-pi/4) 25\nRX(-pi/2) 25\nRZ(pi/2) 25\nRX(pi/2) 12\nRZ(2.490703264764803) 12\nRX(-pi/2) 12\nRZ(-pi/2) 25\nRX(-pi/2) 25\nRZ(1.8093097114347323) 25\nRX(pi) 25\nCZ 12 25\nRZ(pi) 12\nRX(-pi/2) 12\nRZ(-pi/2) 25\nRX(pi/2) 25\nCZ 12 25\nRZ(pi/4) 12\nRX(pi/2) 12\nRX(-pi/2) 25\nCZ 12 25\nRX(-pi/2) 13\nRZ(-2.792473527412035) 13\nRX(-pi/2) 13\nRZ(3*pi/4) 13\nRX(pi/2) 13\nRZ(-2.2078078721496057) 13\nRX(-pi/2) 13\nRZ(-1.4603380589331292) 13\nRX(pi/2) 13\nRZ(-1.5969047865979578) 13\nRX(-pi/2) 13\nRX(-pi/2) 12\nRZ(1.9464126769739878) 12\nRX(-pi/2) 12\nRZ(-0.938286537397745) 12\nRX(pi/2) 12\nRZ(3.0341013669520356) 12\nRX(-pi/2) 12\nCZ 13 12\nRZ(-pi/2) 12\nRX(pi/2) 12\nRZ(-pi/2) 13\nRX(-pi/2) 13\nCZ 13 12\nRX(-pi/2) 12\nRX(pi/2) 13\nCZ 13 12\nRX(pi/2) 14\nRZ(-1.633426416730553) 14\nRX(-pi/2) 14\nRZ(-1.7597025382772835) 14\nRX(pi/2) 14\nRZ(-1.9543079588773333) 14\nRX(-pi/2) 14\nRZ(-1.4437911391574627) 14\nRX(pi/2) 14\nRZ(1.211370736512162) 14\nRX(-pi/2) 14\nRX(pi/2) 15\nRZ(-0.4469187011597193) 15\nRX(-pi/2) 15\nRZ(-1.2662905179216106) 15\nRX(pi/2) 15\nRZ(0.7721216937434701) 15\nRX(-pi/2) 15\nRZ(-2.3275580439210843) 15\nRX(pi/2) 15\nRZ(2.8962288670731136) 15\nRX(-pi/2) 15\nCZ 15 14\nRZ(-pi/2) 14\nRX(pi/2) 14\nRZ(-pi/2) 15\nRX(-pi/2) 15\nCZ 15 14\nRX(-pi/2) 14\nRX(pi/2) 15\nCZ 15 14\nRX(pi/2) 13\nRZ(0.9050959348245454) 13\nRX(-pi/2) 13\nRZ(-1.2534010281989196) 13\nRX(pi/2) 13\nRZ(0.07697366228673452) 13\nRX(pi/2) 13\nRX(pi/2) 14\nRZ(-1.746092549711173) 14\nRX(-pi/2) 14\nRZ(-2.9396968559154795) 14\nRX(pi/2) 14\nRZ(-1.4095843573254627) 14\nRX(pi/2) 14\nCZ 14 13\nRX(pi/2) 12\nRZ(-0.9355242223072686) 12\nRX(-pi/2) 12\nRZ(-1.4841581351549407) 12\nRX(pi/2) 12\nRZ(0.5523749021099489) 12\nRX(-pi/2) 12\nRZ(-2.736446137112887) 13\nRX(-pi/2) 13\nRZ(2.332817144787261) 13\nRX(pi/2) 13\nCZ 12 13\nRZ(pi) 12\nRX(pi/2) 12\nRZ(pi) 13\nRX(pi/2) 13\nCZ 12 13\nRX(-pi/2) 13\nRZ(0.47897721534170934) 13\nRX(-pi/2) 13\nRZ(3*pi/4) 13\nRX(pi/2) 13\nCZ 13 14\nRX(pi/2) 12\nRX(-pi/2) 13\nRZ(pi/4) 13\nRX(pi/2) 13\nCZ 13 12\nRX(pi/2) 13\nRZ(-3*pi/4) 13\nRX(-pi/2) 13\nRZ(-2.5025253953470656) 13\nRZ(-2.3865896914869342) 14\nRX(pi) 14\nCZ 14 13\nRX(pi/2) 12\nRZ(0.5642658520110123) 12\nRX(-pi/2) 12\nRZ(-1.0069331407327016) 12\nRX(pi/2) 12\nRZ(3.024836277777915) 12\nRX(-pi/2) 12\nRX(pi/2) 13\nRZ(-1.3082206173965414) 13\nRX(pi/2) 13\nCZ 13 12\nRZ(-pi/2) 12\nRX(pi/2) 12\nRZ(2.7309458053746605) 13\nRX(-pi/2) 13\nCZ 13 12\nRX(-pi/2) 12\nRX(pi/2) 13\nCZ 13 12\nRX(pi/2) 13\nRZ(-2.5726755223628466) 13\nRX(-pi/2) 13\nRZ(-1.259328636059486) 13\nRX(pi/2) 13\nRZ(1.1595626385928068) 13\nRX(-pi/2) 13\nRZ(1.307042917440481) 14\nRX(pi/2) 14\nCZ 14 13\nRX(pi/2) 14\nRZ(pi/4) 14\nRX(-pi/2) 14\nCZ 14 13\nRZ(-0.6355452284111163) 25\nRZ(pi/2) 11\nRX(pi/2) 11\nRZ(2.1245468564121017) 11\nRX(-pi/2) 11\nRZ(-2.3947370552963214) 11\nRX(pi/2) 11\nRZ(3.1045613179688183) 11\nRX(pi/2) 11\nRZ(-2.3832930169497493) 11\nRX(pi/2) 11\nRZ(1.1094893601905866) 11\nRX(-pi/2) 11\nRX(pi/2) 12\nRZ(0.32921073990638905) 12\nRX(-pi/2) 12\nRZ(-1.7290655087767561) 12\nRX(pi/2) 12\nRZ(-1.2993047155538484) 12\nRX(-pi/2) 12\nRZ(pi) 13\nRX(-pi/2) 13\nRZ(-2.313773007650722) 13\nRX(-pi/2) 13\nRZ(-2.063577819397147) 13\nRX(pi/2) 13\nRZ(1.1608628418986264) 13\nRX(-pi/2) 13\nCZ 13 12\nRZ(-pi/2) 12\nRX(pi/2) 12\nRZ(-pi/2) 13\nRX(-pi/2) 13\nCZ 13 12\nRX(-pi/2) 12\nRX(pi/2) 13\nCZ 13 12\nRX(pi/2) 17\nRZ(0.4413312808013779) 17\nRX(-pi/2) 17\nRZ(-1.5718010850131154) 17\nRX(pi/2) 17\nRZ(-3.12401725506822) 17\nRX(-pi/2) 17\nCZ 17 10\nRZ(-pi/2) 10\nRX(pi/2) 10\nRZ(-pi/2) 17\nRX(-pi/2) 17\nCZ 17 10\nRX(-pi/2) 10\nRX(pi/2) 17\nCZ 17 10\nRX(pi/2) 12\nRZ(1.8141550608122614) 12\nRX(-pi/2) 12\nRZ(-0.7568891281602017) 12\nRX(pi/2) 12\nRZ(-0.7948714430478994) 12\nRX(-pi/2) 12\nRZ(0.8627195871688373) 25\nRX(pi/2) 25\nCZ 25 12\nRZ(pi) 12\nRX(pi/2) 12\nRZ(pi) 25\nRX(pi/2) 25\nCZ 25 12\nRX(pi/2) 12\nRZ(-0.13535530275854218) 12\nRX(-pi/2) 12\nRZ(-1.6271562696753878) 12\nRX(pi/2) 12\nRZ(-2.082575004807808) 12\nRX(-pi/2) 12\nCZ 12 11\nRZ(-pi/2) 11\nRX(pi/2) 11\nRZ(-pi/2) 12\nRX(-pi/2) 12\nCZ 12 11\nRX(-pi/2) 11\nRX(pi/2) 12\nCZ 12 11\nRX(pi/2) 10\nRZ(2.884322182524435) 10\nRX(-pi/2) 10\nRZ(-1.8875840406944406) 10\nRX(pi/2) 10\nRZ(-0.2601273808691591) 10\nRX(pi/2) 10\nRX(pi/2) 11\nRZ(2.608022996304862) 11\nRX(-pi/2) 11\nRZ(-1.796834858572197) 11\nRX(pi/2) 11\nRZ(2.037153642063654) 11\nRX(pi/2) 11\nCZ 10 11\nRZ(-pi/2) 11\nRX(-pi/2) 11\nRZ(-pi/4) 11\nRX(pi/2) 11\nRX(pi/2) 12\nRZ(-2.7425836298853103) 12\nRX(-pi/2) 12\nRZ(-2.2342035637345217) 12\nRX(pi/2) 12\nRZ(2.3717924634972114) 12\nRX(pi/2) 12\nCZ 12 11\nRX(pi/2) 11\nRZ(-pi/4) 11\nRX(-pi/2) 11\nCZ 11 10\nRX(-pi/2) 25\nRZ(1.8072609586929698) 25\nRX(pi/2) 25\nRZ(1.3434152713716006) 25\nRX(-pi/2) 25\nRZ(-0.8974428919033961) 25\nRX(pi/2) 25\nRZ(1.966402122657653) 25\nRX(-pi/2) 25\nRZ(-3*pi/4) 10\nRX(pi/2) 10\nRZ(pi/2) 10\nRX(pi/2) 11\nRZ(pi/4) 11\nRX(-pi/2) 11\nRZ(-2.724188351024705) 11\nRX(pi/2) 11\nRZ(-0.31227091009399666) 12\nRX(-pi/2) 12\nCZ 11 12\nRZ(pi) 11\nRX(pi/2) 11\nRZ(pi) 12\nRX(pi/2) 12\nCZ 11 12\nRX(pi/2) 11\nRZ(-2.8293217434957967) 11\nCZ 10 11\nRZ(-pi/2) 10\nRX(-pi/2) 10\nRZ(-pi/4) 10\nRX(pi/2) 10\nCZ 10 11\nRZ(pi/2) 17\nRX(pi/2) 17\nRZ(-0.18531273659666261) 17\nRX(-pi/2) 17\nRZ(-1.9353915794004208) 17\nRX(pi/2) 17\nRZ(2.3069829574695637) 17\nRX(pi/2) 17\nRZ(-0.5410793375618874) 17\nRX(pi/2) 17\nRZ(-2.197497809698847) 17\nRX(-pi/2) 17\nRX(pi/2) 12\nRZ(2.3818854769735385) 12\nRX(-pi/2) 12\nRZ(-0.9696736415355677) 12\nRX(pi/2) 12\nRZ(0.9776510328345869) 12\nRX(-pi/2) 12\nCZ 25 12\nRZ(-pi/2) 12\nRX(pi/2) 12\nRZ(-pi/2) 25\nRX(-pi/2) 25\nCZ 25 12\nRX(-pi/2) 12\nRX(pi/2) 25\nCZ 25 12\nRX(pi/2) 14\nRZ(0.9322916022581644) 14\nRX(pi/2) 15\nRZ(-1.441296019798911) 15\nRX(-pi/2) 15\nRZ(-1.5169645845635826) 15\nRX(pi/2) 15\nRZ(0.391746196136614) 15\nRX(-pi/2) 15\nRZ(-1.7936982852461991) 15\nRX(pi/2) 15\nRZ(-1.2255516491706204) 15\nRX(-pi/2) 15\nRX(pi/2) 16\nRZ(2.9697150407535076) 16\nRX(-pi/2) 16\nRZ(-1.0625771287259098) 16\nRX(pi/2) 16\nRZ(0.5672270856045549) 16\nRX(-pi/2) 16\nRZ(-1.7390977582838365) 16\nRX(pi/2) 16\nRZ(-1.242498061584269) 16\nRX(-pi/2) 16\nCZ 15 16\nRZ(-pi/2) 15\nRX(-pi/2) 15\nRZ(-pi/2) 16\nRX(pi/2) 16\nCZ 15 16\nRX(pi/2) 15\nRX(-pi/2) 16\nCZ 15 16\nRX(pi/2) 10\nRZ(-0.3066292510918109) 10\nRX(-pi/2) 10\nRZ(-2.0239062963509653) 10\nRX(pi/2) 10\nRZ(-2.8236613044608614) 10\nRX(-pi/2) 10\nCZ 17 10\nRZ(-pi/2) 10\nRX(pi/2) 10\nRZ(-pi/2) 17\nRX(-pi/2) 17\nCZ 17 10\nRX(-pi/2) 10\nRX(pi/2) 17\nCZ 17 10\nRZ(pi/2) 10\nRX(pi/2) 10\nRZ(-2.514891170685843) 10\nRX(-pi/2) 10\nRZ(-2.6005133160279055) 10\nRX(pi/2) 10\nRZ(-1.712717757047584) 10\nRX(pi/2) 10\nRZ(-1.7321333365137435) 10\nRX(pi/2) 10\nRZ(-0.8911238510814623) 10\nRX(-pi/2) 10\nRX(pi/2) 16\nRZ(-1.8287170114196354) 16\nRX(-pi/2) 16\nRZ(-2.0833842627918515) 16\nRX(pi/2) 16\nRZ(2.407507349837072) 16\nRX(pi/2) 16\nRX(pi/2) 17\nRZ(-1.5682576649151605) 17\nRX(-pi/2) 17\nRZ(-2.0258084806092636) 17\nRX(pi/2) 17\nRZ(-1.2948456020812886) 17\nRX(pi/2) 17\nCZ 16 17\nRZ(pi/2) 15\nRX(pi/2) 15\nRZ(-0.32829826521062805) 15\nRX(-pi/2) 15\nRZ(-1.4024948953059566) 15\nRX(pi/2) 15\nRZ(0.40867816325980416) 15\nRX(-pi/2) 15\nRZ(-2.5625258135946596) 15\nRX(pi/2) 15\nRZ(1.8936776380731997) 15\nRX(pi/2) 15\nRZ(-2.494585328886366) 15\nRX(pi/2) 15\nRZ(-2.070765359125314) 15\nRX(-pi/2) 15\nRZ(1.6640365554422294) 16\nRX(pi/2) 16\nRZ(-2.9116157733605728) 16\nRX(-pi/2) 16\nRZ(-0.9085236389082955) 16\nRX(pi/2) 16\nRZ(-0.8401873081494577) 16\nRX(-pi/2) 16\nRZ(pi/2) 17\nRX(pi/2) 17\nRZ(-0.8533482663851909) 17\nRX(-pi/2) 17\nRZ(-1.6640365554422292) 17\nRX(-pi/2) 17\nCZ 17 10\nRZ(-pi/2) 10\nRX(pi/2) 10\nRX(-pi/2) 17\nCZ 17 10\nRX(-pi/2) 10\nRX(pi/2) 17\nCZ 17 10\nRX(pi/2) 11\nRZ(-1.2089067217440184) 11\nRX(-pi/2) 11\nRZ(-1.65542222491992) 11\nRX(pi/2) 11\nRZ(2.143096931069474) 11\nRX(-pi/2) 11\nRZ(pi/2) 12\nRX(pi/2) 12\nRZ(-0.39560579586275596) 12\nRX(-pi/2) 12\nRZ(-2.2441497616863972) 12\nRX(pi/2) 12\nRZ(1.5781159256543265) 12\nRX(pi/2) 12\nRZ(-0.6337811733919919) 12\nRX(pi/2) 12\nRZ(1.1710503440747402) 12\nRX(-pi/2) 12\nRZ(pi/2) 13\nRX(pi/2) 13\nRZ(0.8504237674418025) 13\nRX(-pi/2) 13\nRZ(-0.5722619176994139) 13\nRX(pi/2) 13\nRZ(2.979441435338898) 13\nRX(pi/2) 13\nRZ(-2.2643708065993824) 13\nRX(pi/2) 13\nRZ(2.233378464703351) 13\nRX(-pi/2) 13\nCZ 13 12\nRZ(-pi/2) 12\nRX(pi/2) 12\nRZ(-pi/2) 13\nRX(-pi/2) 13\nCZ 13 12\nRX(-pi/2) 12\nRX(pi/2) 13\nCZ 13 12\nCZ 16 15\nRZ(-pi/2) 15\nRX(pi/2) 15\nRZ(-pi/2) 16\nRX(-pi/2) 16\nCZ 16 15\nRX(-pi/2) 15\nRX(pi/2) 16\nCZ 16 15\nRX(pi/2) 10\nRZ(-2.95709973388311) 10\nRX(-pi/2) 10\nRZ(-0.8704158755663063) 10\nRX(pi/2) 10\nRZ(-0.3421440654604135) 10\nRX(-pi/2) 10\nRZ(-1.5584556497791477) 10\nRX(pi/2) 10\nRZ(-0.9576027501683757) 10\nRX(-pi/2) 10\nRX(pi/2) 11\nRZ(0.7368383549406338) 11\nRX(-pi/2) 11\nRZ(-2.831750654405329) 11\nRX(pi/2) 11\nRZ(2.961249066740188) 11\nRX(-pi/2) 11\nCZ 11 10\nRZ(-pi/2) 10\nRX(pi/2) 10\nRZ(-pi/2) 11\nRX(-pi/2) 11\nCZ 11 10\nRX(-pi/2) 10\nRX(pi/2) 11\nCZ 11 10\nRX(pi/2) 12\nRZ(0.303825493424186) 12\nRX(-pi/2) 12\nRZ(-0.7504285357250252) 12\nRX(pi/2) 12\nRZ(2.1215842065949957) 12\nRX(-pi/2) 12\nRX(pi/2) 10\nRZ(0.6203003149390425) 10\nRX(-pi/2) 10\nRZ(-1.4250621049977954) 10\nRX(pi/2) 10\nRZ(1.915143826856644) 10\nRX(-pi/2) 10\nRX(pi/2) 11\nRZ(1.1735609949677743) 11\nRX(-pi/2) 11\nRZ(-1.2525220850007466) 11\nRX(pi/2) 11\nRZ(2.750356408540993) 11\nRX(-pi/2) 11\nRX(pi/2) 12\nRZ(-0.8656293009580467) 12\nRX(-pi/2) 12\nRZ(-2.0118068317695394) 12\nRX(pi/2) 12\nRZ(0.9853193082756118) 12\nRX(-pi/2) 12\nCZ 11 12\nRZ(-pi/2) 11\nRX(-pi/2) 11\nRZ(-pi/2) 12\nRX(pi/2) 12\nCZ 11 12\nRX(pi/2) 11\nRX(-pi/2) 12\nCZ 11 12\nRZ(pi/2) 12\nRX(pi/2) 12\nRZ(-1.4006941647092157) 12\nRX(-pi/2) 12\nRZ(-1.785807814037351) 12\nRX(pi/2) 12\nRZ(1.8690128816736102) 12\nRX(-pi/2) 12\nRZ(-1.4950341158730833) 12\nRX(pi/2) 12\nRZ(1.9296932553387043) 12\nRX(-pi/2) 12\nRX(pi/2) 25\nRZ(-2.691571113657532) 25\nRX(-pi/2) 25\nRZ(-0.7650377806710601) 25\nRX(pi/2) 25\nRZ(2.778884198798357) 25\nRX(-pi/2) 25\nRZ(-1.3323645658160557) 12\nRX(-pi/2) 12\nRZ(1.1121360076009923) 25\nRX(-pi/2) 25\nCZ 25 12\nRZ(pi) 12\nRX(pi/2) 12\nRZ(pi) 25\nRX(pi/2) 25\nCZ 25 12\nRZ(pi/2) 13\nRX(pi/2) 13\nRZ(-2.7418466708696365) 13\nRX(-pi/2) 13\nRZ(-2.507811480197801) 13\nRX(pi/2) 13\nRZ(-0.2347006542827259) 13\nRX(-pi/2) 13\nCZ 13 14\nRX(-pi/2) 12\nRZ(-0.12182152891615727) 12\nRX(-pi/2) 12\nRZ(-1.8015754720573127) 12\nRX(pi/2) 12\nRZ(-1.6582504202897281) 12\nRX(-pi/2) 12\nRX(-pi/2) 13\nRZ(-1.1932941208690242) 13\nRX(-pi/2) 13\nRZ(-1.0610721662629612) 13\nRX(pi/2) 13\nRZ(-1.9292944310363485) 13\nRX(-pi/2) 13\nCZ 13 12\nRZ(-pi/2) 12\nRX(pi/2) 12\nRZ(-pi/2) 13\nRX(-pi/2) 13\nCZ 13 12\nRX(-pi/2) 12\nRX(pi/2) 13\nCZ 13 12\nRX(pi/2) 14\nRZ(-0.9279439770093677) 14\nRX(-pi/2) 14\nRZ(-0.9989020248904217) 14\nRX(pi/2) 14\nRZ(2.2964047084426973) 14\nRX(-pi/2) 14\nRZ(pi/2) 15\nRX(pi/2) 15\nRZ(2.4109836349443547) 15\nRX(-pi/2) 15\nRZ(-2.2330690146814978) 15\nRX(pi/2) 15\nRZ(-2.1255565449678335) 15\nRX(pi/2) 15\nRZ(-1.8845618725173814) 15\nRX(pi/2) 15\nRZ(1.370924885868175) 15\nRX(-pi/2) 15\nCZ 15 14\nRZ(-pi/2) 14\nRX(pi/2) 14\nRZ(-pi/2) 15\nRX(-pi/2) 15\nCZ 15 14\nRX(-pi/2) 14\nRX(pi/2) 15\nCZ 15 14\nRX(pi/2) 13\nRZ(2.415007132392796) 13\nRX(-pi/2) 13\nRZ(-2.2876822125586136) 13\nRX(pi/2) 13\nRZ(0.8256466059260408) 13\nRX(pi/2) 13\nRX(pi/2) 14\nRZ(-0.8663741937803923) 14\nRX(-pi/2) 14\nRZ(-0.533263889977049) 14\nRX(pi/2) 14\nRZ(0.14557467422572223) 14\nRX(pi/2) 14\nCZ 13 14\nRX(pi/2) 12\nRZ(-3.1242137877855933) 12\nRX(-pi/2) 12\nRZ(-1.6009141001432352) 12\nRX(pi/2) 12\nRZ(2.147333804401222) 12\nRX(-pi/2) 12\nRZ(-0.9294442497866636) 12\nRX(pi/2) 12\nRZ(2.939674860141322) 12\nRX(-pi/2) 12\nRX(pi/2) 13\nRZ(-1.8955981295229216) 13\nRX(-pi/2) 13\nRZ(-2.3786074150359906) 13\nRX(pi/2) 13\nRZ(1.8293940560285864) 13\nRX(-pi/2) 13\nRZ(-2.0868845286249864) 13\nRX(pi/2) 13\nRZ(0.7969370781966514) 13\nRX(-pi/2) 13\nCZ 13 12\nRZ(-pi/2) 12\nRX(pi/2) 12\nRZ(-pi/2) 13\nRX(-pi/2) 13\nCZ 13 12\nRX(-pi/2) 12\nRX(pi/2) 13\nCZ 13 12\nRX(pi/2) 25\nRZ(1.3323645658160541) 25\nRX(pi/2) 12\nRZ(0.046077823109343004) 12\nRX(-pi/2) 12\nRZ(-1.3618288165867545) 12\nRX(pi/2) 12\nRZ(-0.9901342856364916) 12\nRX(pi/2) 12\nCZ 12 25\nRZ(pi/2) 13\nRX(pi/2) 13\nRZ(1.7727141202433678) 13\nRX(-pi/2) 13\nRZ(-2.2121484038031296) 13\nRX(pi/2) 13\nRZ(-1.7027396750997799) 13\nRX(-pi/2) 13\nRZ(-2.219292405251225) 13\nRX(pi/2) 13\nRZ(1.5276436957413608) 13\nRX(-pi/2) 13\nRZ(2.2498349842619736) 14\nRX(pi/2) 14\nRZ(-2.4591715349765155) 14\nRX(-pi/2) 14\nRZ(-1.9714263150167577) 14\nRX(pi/2) 14\nRZ(-1.2796750536165185) 14\nRX(-pi/2) 14\nCZ 14 13\nRZ(-pi/2) 13\nRX(pi/2) 13\nRZ(-pi/2) 14\nRX(-pi/2) 14\nCZ 14 13\nRX(-pi/2) 13\nRX(pi/2) 14\nCZ 14 13\nRX(pi/2) 10\nRZ(-2.305841661245038) 10\nRX(-pi/2) 10\nRZ(-1.2595374408570195) 10\nRX(pi/2) 10\nRZ(2.8024706004322013) 10\nRX(-pi/2) 10\nRX(pi/2) 11\nRZ(1.514868576742307) 11\nRX(-pi/2) 11\nRZ(-0.45554811032787196) 11\nRX(pi/2) 11\nRZ(-2.013571503412182) 11\nRX(-pi/2) 11\nRZ(-1.5121872212279117) 11\nRX(pi/2) 11\nRZ(1.8157024979397907) 11\nRX(-pi/2) 11\nCZ 11 10\nRZ(-pi/2) 10\nRX(pi/2) 10\nRZ(-pi/2) 11\nRX(-pi/2) 11\nCZ 11 10\nRX(-pi/2) 10\nRX(pi/2) 11\nCZ 11 10\nRX(pi/2) 12\nRZ(-0.8371877672784243) 12\nRX(-pi/2) 12\nRZ(-1.259688423927694) 12\nRX(pi/2) 12\nRZ(1.930599298416177) 12\nRX(-pi/2) 12\nRX(pi/2) 13\nRZ(-2.1904856118997764) 13\nRX(-pi/2) 13\nRZ(-1.5579299123759487) 13\nRX(pi/2) 13\nRZ(-0.5228407385323754) 13\nRX(pi/2) 13\nCZ 13 12\nRZ(pi) 12\nRX(pi/2) 12\nRZ(pi) 13\nRX(pi/2) 13\nCZ 13 12\nRX(pi/2) 12\nRZ(1.1420947522924827) 25\nRX(-pi/2) 25\nCZ 25 12\nRX(pi/2) 25\nRZ(pi/4) 25\nRX(-pi/2) 25\nCZ 25 12\nRZ(pi/2) 11\nRX(pi/2) 11\nRZ(-0.6803317912920921) 11\nRX(-pi/2) 11\nRZ(-2.8314495072977324) 11\nRX(pi/2) 11\nRZ(2.1094740454093572) 11\nRX(-pi/2) 11\nRZ(-1.6372314406918238) 11\nRX(pi/2) 11\nRZ(2.01920522208883) 11\nRX(-pi/2) 11\nRZ(-0.3138722909134002) 12\nRX(-pi/2) 12\nRZ(1.8676919273662065) 12\nRX(-pi/2) 12\nRZ(0.3566965888950355) 12\nRX(-pi/2) 12\nCZ 12 11\nRZ(-pi/2) 11\nRX(pi/2) 11\nRZ(-pi/2) 12\nRX(-pi/2) 12\nCZ 12 11\nRX(-pi/2) 11\nRX(pi/2) 12\nCZ 12 11\nRX(pi/2) 12\nRZ(-0.34719823152722634) 12\nRX(-pi/2) 12\nRZ(-1.481782937835571) 12\nRX(pi/2) 12\nRZ(0.34185037548076913) 12\nRX(pi/2) 12\nRX(pi/2) 25\nRZ(-1.9274929156899334) 25\nRX(-pi/2) 25\nRZ(-3.039705944867549) 25\nCZ 25 12\nRZ(pi/2) 11\nRX(pi/2) 11\nRZ(-3.010535364892442) 11\nRX(-pi/2) 11\nRZ(-0.5783530037994004) 11\nRX(pi/2) 11\nRZ(-0.5099212145867467) 11\nRX(pi/2) 11\nRZ(-2.5507929130113594) 11\nRX(pi/2) 11\nRZ(-2.012101536874198) 11\nRX(-pi/2) 11\nRZ(pi/2) 12\nRX(pi/2) 12\nRZ(2.3901570083874732) 12\nRX(-pi/2) 12\nRZ(3.0397059448675487) 12\nRX(pi) 12\nCZ 12 11\nRZ(-pi/2) 11\nRX(pi/2) 11\nRZ(-pi/2) 12\nRX(-pi/2) 12\nCZ 12 11\nRX(-pi/2) 11\nRX(pi/2) 12\nCZ 12 11\nRX(pi/2) 12\nRZ(2.5909847589902815) 12\nRX(-pi/2) 12\nRZ(-0.7010445956599257) 12\nRX(pi/2) 12\nRZ(-2.5013042534105936) 12\nRX(pi/2) 12\nCZ 12 25\nRZ(-2.732100194730598) 31\nRX(pi) 31\nRZ(pi/2) 32\nRX(pi/2) 32\nRZ(2.2273588032285017) 32\nRX(-pi/2) 32\nRZ(-2.2693398736348267) 32\nRX(pi/2) 32\nRZ(3.122752742605404) 32\nRX(-pi/2) 32\nCZ 31 32\nRZ(pi/2) 10\nRX(pi/2) 10\nRZ(-0.24490617114489371) 10\nRX(-pi/2) 10\nRZ(-1.6294054323618814) 10\nRX(pi/2) 10\nRZ(-2.559224622081924) 10\nRX(-pi/2) 10\nRZ(-1.3972721112972175) 10\nRX(pi/2) 10\nRZ(0.04417051323203314) 10\nRX(-pi/2) 10\nRX(pi/2) 11\nRZ(0.5506715189497419) 11\nRX(-pi/2) 11\nRZ(-2.0868362702386776) 11\nRX(pi/2) 11\nRZ(0.9974830438019158) 11\nRX(-pi/2) 11\nRZ(2.7431975221986096) 12\nRX(pi) 12\nRX(-pi/2) 13\nRZ(0.0324375080326589) 13\nRX(-pi/2) 13\nRZ(3*pi/4) 13\nRX(-pi/2) 13\nRZ(pi/2) 13\nRX(pi/2) 14\nRZ(-2.5459966822321562) 14\nRX(-pi/2) 14\nRZ(-0.4905582374157596) 14\nRX(pi/2) 14\nRZ(-3.114279646550637) 14\nRX(-pi/2) 14\nRZ(pi/2) 15\nRX(pi/2) 15\nRZ(2.4159842719419915) 15\nRX(-pi/2) 15\nRZ(-2.1426906286993717) 15\nRX(pi/2) 15\nRZ(-0.927943977009368) 15\nRX(-pi/2) 15\nRZ(pi/2) 16\nRX(pi/2) 16\nRZ(0.499969032330418) 16\nRX(-pi/2) 16\nRZ(-0.6470073247034271) 16\nRX(pi/2) 16\nRZ(-0.031206993338134037) 16\nRX(-pi/2) 16\nRZ(pi/2) 17\nRX(pi/2) 17\nRZ(-0.6796724757134349) 17\nRX(-pi/2) 17\nRZ(-1.4094593170760492) 17\nRX(pi/2) 17\nRZ(-0.4130312710484465) 17\nRX(-pi/2) 17\nRX(pi/2) 21\nRZ(-2.2896866992664755) 21\nRX(-pi/2) 21\nRZ(-3*pi/4) 21\nRX(pi/2) 21\nRZ(pi/2) 21\nRZ(pi/2) 22\nRX(pi/2) 22\nRZ(-1.5828292003940743) 22\nRX(-pi/2) 22\nRZ(-2.3846898633538034) 22\nRX(pi/2) 22\nRZ(0.3941123423692211) 22\nRX(-pi/2) 22\nRZ(-2.4415270115610297) 22\nRX(pi/2) 22\nRZ(-2.1274062884536873) 22\nRX(-pi/2) 22\nRZ(pi/2) 23\nRX(pi/2) 23\nRZ(-0.10057327756351464) 23\nRX(-pi/2) 23\nRZ(-1.1858499405437926) 23\nRX(pi/2) 23\nRZ(-1.7220907407505557) 23\nRX(-pi/2) 23\nRZ(-0.5209127938088676) 23\nRX(pi/2) 23\nRZ(-1.28360650199139) 23\nRX(-pi/2) 23\nRZ(pi/2) 24\nRX(pi/2) 24\nRZ(2.841054554953246) 24\nRX(-pi/2) 24\nRZ(-1.1730484616706094) 24\nRX(pi/2) 24\nRZ(-2.105915898022002) 24\nRX(-pi/2) 24\nRZ(3.0397059448675483) 25\nRX(pi/2) 25\nRZ(-pi/2) 25\nRX(-pi/2) 31\nRZ(pi/2) 31\nRZ(pi) 32\nRZ(pi/2) 33\nRX(pi/2) 33\nRZ(2.9094947883344044) 33\nRX(-pi/2) 33\nRZ(-1.5805215324148598) 33\nRX(pi/2) 33\nRZ(-1.8405640452862602) 33\nRX(-pi/2) 33\nRZ(-1.977381822134548) 33\nRX(pi/2) 33\nRZ(1.8414959455792692) 33\nRX(-pi/2) 33\nRZ(pi/2) 34\nRX(pi/2) 34\nRZ(-2.2582504430821166) 34\nRX(-pi/2) 34\nRZ(-1.3943012058032247) 34\nRX(pi/2) 34\nRZ(0.4751096729050288) 34\nRX(-pi/2) 34\nRZ(-1.9391850154928505) 34\nRX(pi/2) 34\nRZ(0.7897077387076045) 34\nRX(-pi/2) 34\nRZ(pi/2) 35\nRX(pi/2) 35\nRZ(-0.30107419927246726) 35\nRX(-pi/2) 35\nRZ(-2.107146695659032) 35\nRX(pi/2) 35\nRZ(0.3369189669423358) 35\nRX(-pi/2) 35\nRZ(-1.8903357166978296) 35\nRX(pi/2) 35\nRZ(-2.63341112863067) 35\nRX(-pi/2) 35\nRZ(1.5042885358690317) 36\nMEASURE 21 ro[4]\nMEASURE 35 ro[3]\nMEASURE 31 ro[2]\nMEASURE 14 ro[1]\nMEASURE 25 ro[0]\n\n"
],
[
"print(cp.attributes)",
"{'native_quil_metadata': NativeQuilMetadata(final_rewiring=[12, 25, 13, 15, 14, 10, 32, 31, 33, 36,...], gate_depth=923, gate_volume=2875, multiqubit_gate_depth=181, program_duration=57453.17000000016, program_fidelity=0.23057478508104318, qpu_runtime_estimation=4886.46826171875, topological_swaps=81), 'num_shots': 20}\n"
],
[
"print(len(cp.attributes['native_quil_metadata'].final_rewiring))",
"38\n"
],
[
"qvm = get_qc(\"Aspen-7-14Q-C\", as_qvm=True)\ncp = qvm.compile(address_qubits(p),protoquil=True)\nprint(cp.attributes)",
"{'native_quil_metadata': NativeQuilMetadata(final_rewiring=[12, 13, 15, 17, 1, 10, 7, 0, 16, 6,...], gate_depth=858, gate_volume=2632, multiqubit_gate_depth=165, program_duration=52353.01000000009, program_fidelity=0.203549302452065, qpu_runtime_estimation=4489.91845703125, topological_swaps=75), 'num_shots': 20}\n"
],
[
"qvm.run(cp)",
"_____no_output_____"
],
[
"display(cp, width=300)",
"_____no_output_____"
],
[
"from pyquil.gates import CCNOT\np = Program()\np += CCNOT(0,1,2)\ndisplay(p, width=300)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
c5119d0e22d4123fabf8ec0211619e525964a557
| 11,955 |
ipynb
|
Jupyter Notebook
|
code/plot_SST_pretty.ipynb
|
jtomfarrar/OpenCTD_testing
|
c2530f241d3f6861d5380ee6a8fd8c329090232a
|
[
"MIT"
] | null | null | null |
code/plot_SST_pretty.ipynb
|
jtomfarrar/OpenCTD_testing
|
c2530f241d3f6861d5380ee6a8fd8c329090232a
|
[
"MIT"
] | null | null | null |
code/plot_SST_pretty.ipynb
|
jtomfarrar/OpenCTD_testing
|
c2530f241d3f6861d5380ee6a8fd8c329090232a
|
[
"MIT"
] | 1 |
2021-08-30T19:15:14.000Z
|
2021-08-30T19:15:14.000Z
| 38.317308 | 978 | 0.59105 |
[
[
[
"## Plots of SST for S-MODE region\n",
"_____no_output_____"
]
],
[
[
"cd C:\\users\\jtomf\\Documents\\Python\\S-MODE_analysis\\code",
"[WinError 3] The system cannot find the path specified: 'C:\\\\users\\\\jtomf\\\\Documents\\\\Python\\\\S-MODE_analysis\\\\code'\nC:\\Users\\Home\\Documents\\GitHub\\OpenCTD_testing\\code\n"
],
[
"import xarray as xr\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport matplotlib\nimport cftime\n\nimport cartopy.crs as ccrs # import projections\nimport cartopy\nimport gsw\n# For great circle distance between two points, use gsw.geostrophy.distance(lon, lat, p=0, axis=-1)\n\nimport functions # requires functions.py from this repository",
"_____no_output_____"
],
[
"# %matplotlib inline\n%matplotlib qt5\nplt.rcParams['figure.figsize'] = (6,5)\nplt.rcParams['figure.dpi'] = 300\nplt.rcParams['savefig.dpi'] = 700\nplt.close('all')\n\n__figdir__ = '../plots/' + 'SMODE_'\nsavefig_args = {'bbox_inches':'tight', 'pad_inches':0.2}\nplotfiletype='png'",
"_____no_output_____"
],
[
"savefig = True\nzoom = True\nif zoom:\n xmin, xmax = (-126,-122)\n ymin, ymax = (36,39)\n levels = np.linspace(13.5,15.5,21)+.5\n zoom_str='zoom'\nelse:\n xmin, xmax = (-126,-121)\n ymin, ymax = (35, 39)\n levels = np.linspace(12.5,16,21)+0\n zoom_str='wide'\n ",
"_____no_output_____"
],
[
"def plot_ops_area(ax,**kwargs):\n \"\"\" Add polygon to show S-MODE pilot operations area\n Inputs\n - matplotlib.pyplot.plot kwargs\n\n return\n - exit code (True if OK)\n\n \"\"\"\n\n # Add S-MODE pilot operations area\n ''' \n\n New corners of pentagon:\n 38° 05.500’ N, 125° 22.067’ W\n 37° 43.000’ N, 124° 00.067’ W\n 37° 45.000’ N, 123° 26.000‘ W\n 36° 58.000’ N, 122° 57.000’ W\n 36° 20.000’ N, 124° 19.067’ W \n '''\n \n coord = [[-(125+22.067/60),38+5.5/60], [-(124+0.067/60),37+43/60], [-(123+26/60),37+45/60], [-(122+57/60),36+58/60], [-(124+19.067/60),36+20/60]]\n coord.append(coord[0]) #repeat the first point to create a 'closed loop'\n\n xs, ys = zip(*coord) #create lists of x and y values\n\n if ax is None:\n ax = plt.gca() \n # ax.plot(xs,ys,transform=ccrs.PlateCarree()) \n ax.plot(xs,ys,**kwargs) \n \n SF_lon=-(122+25/60)\n SF_lat= 37+47/60\n \n # mark a known place to help us geo-locate ourselves\n ax.plot(SF_lon, SF_lat, 'o', markersize=3, zorder=10, **kwargs)\n ax.text(SF_lon-5/60, SF_lat+5/60, 'San Francisco', fontsize=8, zorder=10, **kwargs)\n # ax.text(np.mean(xs)-.6, np.mean(ys)-.3, 'S-MODE ops area', fontsize=8, **kwargs)\n print(kwargs)\n \n return(xs,ys,ax)\n",
"_____no_output_____"
],
[
"#url = 'http://smode.whoi.edu:8080/thredds/dodsC/satellite/AVHRR_METOPB/AVHRR_METOPB_20210930T060000Z.nc'\n#url = 'http://smode.whoi.edu:8080/thredds/dodsC/satellite/MODIS_Terra/MODIS_Terra_20210930T065001Z.nc'3 \n# url = 'http://smode.whoi.edu:8080/thredds/dodsC/satellite/VIIRS_NRT/VIIRS_NRT_20210929T213000Z.nc' #This one is good\n\n#url = 'http://smode.whoi.edu:8080/thredds/dodsC/satellite/VIIRS_NRT/VIIRS_NRT_20211016T095000Z.nc' # good one\n# url = 'http://smode.whoi.edu:8080/thredds/fileServer/satellite/AVHRR_METOPA/AVHRR_METOPA_20211019T031000Z.nc#bytes' #Really great!\n# url = 'http://smode.whoi.edu:8080/thredds/fileServer/satellite/VIIRS_NRT/VIIRS_NRT_20211019T103001Z.nc#mode=bytes' #Not awesome, but coincident with B200 flight on 10/19\n#url = 'http://smode.whoi.edu:8080/thredds/dodsC/satellite/MODIS_Terra/MODIS_Terra_20211011T233459Z.nc'\n# url = 'http://smode.whoi.edu:8080/thredds/dodsC/satellite/VIIRS_NRT/VIIRS_NRT_20211022T111000Z.nc'\n#url = 'http://smode.whoi.edu:8080/thredds/dodsC/satellite/AVHRR_METOPA/AVHRR_METOPA_20211022T161000Z.nc' # good one!\n\n#Best sequence\n#url = 'http://smode.whoi.edu:8080/thredds/dodsC/satellite/AVHRR_METOPC/AVHRR_METOPC_20211019T052000Z.nc' # prettiest of all!!\n#url = 'http://smode.whoi.edu:8080/thredds/dodsC/satellite/VIIRS_NRT/VIIRS_NRT_20211025T214001Z.nc'\n\n# url = 'http://smode.whoi.edu:8080/thredds/dodsC/satellite/AVHRR_METOPC/AVHRR_METOPC_20211025T050000Z.nc' #not good, but looking for front...\n# url = 'http://smode.whoi.edu:8080/thredds/dodsC/satellite/VIIRS_NRT/VIIRS_NRT_20211025T102000Z.nc'\n# url = 'http://smode.whoi.edu:8080/thredds/dodsC/satellite/VIIRS_NRT/VIIRS_NRT_20211025T214001Z.nc'\n\n#url = 'http://smode.whoi.edu:8080/thredds/dodsC/satellite/AVHRR_METOPA/AVHRR_METOPA_20211028T033000Z.nc'\nurl = 'http://smode.whoi.edu:8080/thredds/dodsC/satellite/VIIRS_NRT/VIIRS_NRT_20211104T103001Z.nc'\nds = xr.open_dataset(url)\n",
"_____no_output_____"
],
[
"ds",
"_____no_output_____"
],
[
"fig = plt.figure()\nax = plt.axes(projection = ccrs.PlateCarree(central_longitude=200)) # Orthographic\nextent = [xmin, xmax, ymin, ymax]\nax.set_extent(extent, crs=ccrs.PlateCarree())\n'''daystr=ds.time.dt.day.astype(str).values[0]\nmonstr=ds.time.dt.month.astype(str).values[0]\nyrstr=ds.time.dt.year.astype(str).values[0]\nday_str = monstr+'-'+daystr+'-'+yrstr\n'''\nday_str=ds.time.dt.strftime(\"%a, %b %d %H:%M\").values[0]\nday_str2=ds.time.dt.strftime(\"%m-%d-%Y\").values[0]\n\nax.set_title('SST, ' + day_str, size = 10.)\n\n#plt.set_cmap(cmap=plt.get_cmap('nipy_spectral'))\nplt.set_cmap(cmap=plt.get_cmap('turbo'))\ngl = ax.gridlines(draw_labels=True, dms=True, x_inline=False, y_inline=False, alpha=0.5, linestyle='--')\ngl.top_labels = False\ngl.ylabels_right = False\n#gl.xlocator = matplotlib.ticker.MaxNLocator(10)\n#gl.xlocator = matplotlib.ticker.AutoLocator\n# gl.xlocator = matplotlib.ticker.FixedLocator(np.arange(0, 360 ,30))\n\nax.coastlines()\nax.add_feature(cartopy.feature.LAND, zorder=3, facecolor=[.6,.6,.6], edgecolor='black')\n# cs = ax.contourf(ds.lon,ds.lat,np.squeeze(ds.sea_surface_temperature)-273.15, levels, extend='both', transform=ccrs.PlateCarree())\ncs = ax.pcolormesh(ds.lon,ds.lat,np.squeeze(ds.sea_surface_temperature)-273.15, vmin=levels[0], vmax=levels[-1], transform=ccrs.PlateCarree())\n# cb = plt.colorbar(cs,ax=ax,shrink=.8,pad=.05)\ncb = plt.colorbar(cs,fraction = 0.022,extend='both')\ncb.set_label('SST [$\\circ$C]',fontsize = 10)\nplot_ops_area(ax,transform=ccrs.PlateCarree(),color='w')\n\n# Add a 10 km scale bar\nkm_per_deg_lat=gsw.geostrophy.distance((125,125), (37,38))/1000\ndeg_lat_equal_10km=10/km_per_deg_lat\nx0 = -125\ny0 = 37.75\nax.plot(x0+np.asarray([0, 0]),y0+np.asarray([0.,deg_lat_equal_10km]),transform=ccrs.PlateCarree(),color='k')\nax.text(x0+2/60, y0-.5/60, '10 km', fontsize=6,transform=ccrs.PlateCarree())\n\n",
"_____no_output_____"
],
[
"# saildrones.plot('longitude','latitude','scatter',markersize=3,transform=ccrs.PlateCarree())\n#ax.plot(saildrones['longitude'],saildrones['latitude'],'ko',markersize=3,transform=ccrs.PlateCarree())\nsaildrones=functions.get_current_position('saildrone')#,'ko', markersize=3,transform=ccrs.PlateCarree())\nax.plot(saildrones['longitude'],saildrones['latitude'],'ko',markersize=1,transform=ccrs.PlateCarree())\nwg=functions.get_current_position('waveglider')#,'ko', markersize=3,transform=ccrs.PlateCarree())\nhWG = ax.plot(wg['longitude'],wg['latitude'],'mo',markersize=1,transform=ccrs.PlateCarree())\n",
"_____no_output_____"
],
[
"if savefig:\n plt.savefig(__figdir__+'SST_' + day_str2 + '_' + zoom_str + 'pretty2' +'.' +plotfiletype,**savefig_args)",
"_____no_output_____"
],
[
"pwd",
"_____no_output_____"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
c511a027541424cb119e97e8f80e8a6e07a755d5
| 130 |
ipynb
|
Jupyter Notebook
|
01-Lesson-Plans/12-NLP/2/Activities/04-Stu_Crisis_Voice/Solved/voice_crisis.ipynb
|
tatianegercina/FinTech
|
b40687aa362d78674e223eb15ecf14bc59f90b62
|
[
"ADSL"
] | 1 |
2021-04-13T07:14:34.000Z
|
2021-04-13T07:14:34.000Z
|
01-Lesson-Plans/12-NLP/2/Activities/04-Stu_Crisis_Voice/Solved/voice_crisis.ipynb
|
tatianegercina/FinTech
|
b40687aa362d78674e223eb15ecf14bc59f90b62
|
[
"ADSL"
] | 2 |
2021-06-02T03:14:19.000Z
|
2022-02-11T23:21:24.000Z
|
01-Lesson-Plans/12-NLP/2/Activities/04-Stu_Crisis_Voice/Solved/voice_crisis.ipynb
|
tatianegercina/FinTech
|
b40687aa362d78674e223eb15ecf14bc59f90b62
|
[
"ADSL"
] | 1 |
2021-05-07T13:26:50.000Z
|
2021-05-07T13:26:50.000Z
| 32.5 | 75 | 0.884615 |
[
[
[
"empty"
]
]
] |
[
"empty"
] |
[
[
"empty"
]
] |
c511b2e4ee6039305234690ec174163f616cff99
| 11,853 |
ipynb
|
Jupyter Notebook
|
social-tags/notebooks/movielens-imdb/tf-lda-knn.ipynb
|
queirozfcom/auto-tagger
|
d9c0339648562ceca2d7cd10a02aaf56d353ae7b
|
[
"MIT"
] | null | null | null |
social-tags/notebooks/movielens-imdb/tf-lda-knn.ipynb
|
queirozfcom/auto-tagger
|
d9c0339648562ceca2d7cd10a02aaf56d353ae7b
|
[
"MIT"
] | 1 |
2016-02-19T03:08:47.000Z
|
2016-02-19T03:08:47.000Z
|
social-tags/notebooks/movielens-imdb/tf-lda-knn.ipynb
|
queirozfcom/auto-tagger
|
d9c0339648562ceca2d7cd10a02aaf56d353ae7b
|
[
"MIT"
] | null | null | null | 28.909756 | 125 | 0.58964 |
[
[
[
"import pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport gc\nimport os\nimport re\nimport pickle\nimport sklearn\nimport sys\nimport string\n\nfrom sklearn.decomposition import LatentDirichletAllocation\nfrom sklearn.metrics import f1_score, precision_score, recall_score\nfrom sklearn.metrics.pairwise import cosine_similarity,cosine_distances\nfrom sklearn.model_selection import cross_val_score, GridSearchCV,ParameterGrid, train_test_split\nfrom sklearn.multiclass import OneVsRestClassifier\nfrom sklearn.preprocessing import MultiLabelBinarizer\nfrom sklearn.feature_extraction.text import CountVectorizer\nfrom sklearn.feature_extraction.text import TfidfTransformer,TfidfVectorizer\nfrom sklearn.pipeline import Pipeline\nfrom sklearn.neighbors import KNeighborsClassifier,NearestNeighbors\nfrom sklearn.svm import LinearSVC\n\nfrom tqdm import *\n\n%matplotlib inline\n%load_ext autoreload\n%autoreload 1",
"_____no_output_____"
],
[
"src_dir = os.path.join(os.getcwd(), os.pardir, '../src')\nsys.path.append(src_dir)",
"_____no_output_____"
],
[
"%aimport data.movielens_20m_imdb\n%aimport helpers.labels,helpers.neighbours",
"_____no_output_____"
],
[
"from data.movielens_20m_imdb import load_or_get_from_cache\nfrom helpers.labels import truncate_labels\nfrom helpers.neighbours import get_predicted_labels_from_neighbours",
"_____no_output_____"
],
[
"INTERIM_DATA_ROOT = os.path.abspath(\"../../data/interim/movielens-ml20m-imdb/\")\nML_ROOT = \"/media/felipe/SAMSUNG/movielens/ml-20m/\"\nIMDB_ROOT = \"/media/felipe/SAMSUNG/imdb/\"\n\nPATH_TO_MOVIES = ML_ROOT + \"/movies.csv\"\nPATH_TO_TAG_ASSIGNMENTS = ML_ROOT + \"/tags.csv\"\nPATH_TO_MOVIE_PLOTS = IMDB_ROOT+\"/plot.list\"\n\n# CONFIGS\nMAX_NB_WORDS = 4000\nNB_NEIGHBOURS = 3\nDISTANCE_METRIC='cosine'\nWEIGHTS='distance'\nPREPROC=None\nSTOP_WORDS='english'\nNB_COMPONENTS = 30",
"_____no_output_____"
],
[
"docs_df = load_or_get_from_cache(PATH_TO_MOVIES,PATH_TO_TAG_ASSIGNMENTS,PATH_TO_MOVIE_PLOTS,INTERIM_DATA_ROOT)",
"_____no_output_____"
],
[
"data = docs_df['plot'].values\nlabelsets = docs_df[\"unique_tags\"].map(lambda tagstring: tagstring.split(\",\")).values",
"_____no_output_____"
],
[
"mlb = MultiLabelBinarizer()\nmlb.fit(labelsets)",
"_____no_output_____"
],
[
"# I can't put this into a pipeline because NearestNeighbors is not a normal classifier, I think\n# I need to customize the pipeline object to be able to call the methods for that class.\nvect = CountVectorizer(max_features=MAX_NB_WORDS, preprocessor=PREPROC, stop_words=STOP_WORDS)\n\n# arsg taken from http://scikit-learn.org/stable/auto_examples/applications/plot_topics_extraction_with_nmf_lda.html\nlda = LatentDirichletAllocation(n_components=NB_COMPONENTS, max_iter=5,\n learning_method='online',\n learning_offset=50.)\n\nnbrs = NearestNeighbors(n_neighbors=NB_NEIGHBOURS, metric=DISTANCE_METRIC)",
"_____no_output_____"
],
[
"X_train, X_test, y_train, y_test = train_test_split(data,labelsets,test_size=0.25)",
"_____no_output_____"
],
[
"y_train = mlb.transform(y_train)\ny_test = mlb.transform(y_test)",
"_____no_output_____"
],
[
"# train\nX_train = vect.fit_transform(X_train)\nX_train = lda.fit_transform(X_train)\nnbrs.fit(X_train)",
"_____no_output_____"
],
[
"# test\nX_test = vect.transform(X_test)\nX_test = lda.transform(X_test)",
"_____no_output_____"
],
[
"X_train.shape,X_test.shape",
"_____no_output_____"
],
[
"y_train.shape,y_test.shape",
"_____no_output_____"
],
[
"y_preds = []\ny_trues = []\n\ndistances_matrix, indices_matrix = nbrs.kneighbors(X_test)\n\nneighbour_labels_tensor = y_train[indices_matrix] \n\ndistances_matrix.shape, indices_matrix.shape, neighbour_labels_tensor.shape",
"_____no_output_____"
],
[
"for i in tqdm(range(distances_matrix.shape[0])):\n \n distances = distances_matrix[i].ravel() \n \n neighbour_labels = neighbour_labels_tensor[i]\n \n y_pred = get_predicted_labels_from_neighbours(neighbour_labels, distances)\n \n y_true = y_test[i]\n \n y_preds.append(y_pred)\n y_trues.append(y_true)\n \ny_preds = np.array(y_preds)\ny_trues = np.array(y_trues)",
"100%|██████████| 1578/1578 [01:52<00:00, 14.30it/s]\n"
],
[
"f1_score(y_trues,y_preds,average='micro')",
"_____no_output_____"
],
[
"def print_top_words(model, feature_names, n_top_words):\n for topic_idx, topic in enumerate(model.components_):\n message = \"Topic #%d: \" % topic_idx\n message += \" \".join([feature_names[i]\n for i in topic.argsort()[:-n_top_words - 1:-1]])\n print(message)\n print()\n",
"_____no_output_____"
],
[
"tf_feature_names = vect.get_feature_names()\nprint_top_words(lda, tf_feature_names, 10)",
"Topic #0: max miss albert girls maya amanda network electric ginger mall\nTopic #1: david jake elizabeth john holly casino vince zoo diane jsackste\nTopic #2: war american world army men president qv government british com\nTopic #3: school high park cindy football party seth prom angel friends\nTopic #4: maggie fiona fairy nathan swamp pig phillips shrek jonah twin\nTopic #5: sam larry annie jason terry loh carol edu doc radio\nTopic #6: mike ted marcus russian hank pat barbara jennifer spy sully\nTopic #7: new life man com time friend money help way wants\nTopic #8: roy jeremy vegas molly perkins sean las lola caesar ac\nTopic #9: town johnny henry students local sister small sheriff helen teacher\nTopic #10: ben linda phil junior raymond bernie elsa liz jewel dylan\nTopic #11: family life father mother love year old new relationship son\nTopic #12: film tom movie documentary rock music tommy band ray world\nTopic #13: george jane animals circus jungle ned animal braddock gene brooks\nTopic #14: arthur lucy au sir cs oz chapman murray muzzle uq\nTopic #15: rocky la karen lisa kit di vienna mickey ma il\nTopic #16: andrew project rebecca lillian thought recording tape experiences destroying centuries\nTopic #17: kenneth chisholm moon kchishol murphy rogers detroit norman jan beverly\nTopic #18: love john nick meets woman life mary married hotel falls\nTopic #19: gang food emily carl roger leo japanese japan woody jacob\nTopic #20: mexico workers restaurant mexican border union rosa chef cooking rat\nTopic #21: police murder harry killer frank detective case wife crime paul\nTopic #22: vampire church stephan ed priest harold vampires edu cc wwu\nTopic #23: billy kate christmas eve kelly fred sidney santa joey angela\nTopic #24: joe king richard baby story god princess prince son india\nTopic #25: tony dance club dancer dancing brooklyn ballet valentine contest jasmine\nTopic #26: earth dr world evil human ship bond planet com crew\nTopic #27: susan al jesse laura foster ranch pearl willy danny chocolate\nTopic #28: jack peter charlie ed tracy uncle mindspring flint sutton esutton\nTopic #29: dave percy glory gwen link crash annie train shadow brings\n\n"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
c511b9dcd66acc563ef501fe2f79f2ae3b3b1c42
| 3,790 |
ipynb
|
Jupyter Notebook
|
code/main_cls_htmlResult.ipynb
|
sjchoi86/choicenet
|
0b30f8aba0abde800ebd38b0b6987f268c0778cc
|
[
"MIT"
] | 129 |
2018-05-23T06:52:07.000Z
|
2021-09-19T17:13:21.000Z
|
code/main_cls_htmlResult.ipynb
|
sjchoi86/choicenet
|
0b30f8aba0abde800ebd38b0b6987f268c0778cc
|
[
"MIT"
] | null | null | null |
code/main_cls_htmlResult.ipynb
|
sjchoi86/choicenet
|
0b30f8aba0abde800ebd38b0b6987f268c0778cc
|
[
"MIT"
] | 9 |
2018-06-20T14:20:46.000Z
|
2019-06-01T08:29:26.000Z
| 26.690141 | 84 | 0.490237 |
[
[
[
"# Make a webpage for summarizing classification results",
"_____no_output_____"
]
],
[
[
"import os,scipy.misc,nbloader#,cv2\nimport numpy as np\nimport matplotlib.pyplot as plt\nfrom glob import glob\nfrom main_cls_config import errTypeList,outlierRatioList\n%matplotlib inline \n%config InlineBackend.figure_format = 'retina'\nprint (\"Packages loaded.\")",
"Packages loaded.\n"
]
],
[
[
"### Make a webpage",
"_____no_output_____"
]
],
[
[
"html_name = 'cls_results'\npath2save = '../html'\nif not os.path.exists(path2save): os.makedirs(path2save) \nindex_path = os.path.join(('../html/%s.html'%(html_name)))\nindex = open(index_path, \"w\")\n\nindex.write(\"<html>\")\nfor _mode in errTypeList: # For all types \n \n if _mode == 'rs':\n errType = 'Random Shuffle'\n elif _mode == 'rp':\n errType = 'Permutation';\n elif _mode == 'b':\n errType = 'Label Bias'; \n else:\n print (\"Unknown [%s]\"%(_mode))\n\n index.write(\"<body><table>\")\n index.write(\"<caption> Error type: [%s] </caption>\" % (errType))\n index.write(\"<tr>\")\n index.write(\"<th> name </th>\")\n index.write(\"<th> Result </th>\")\n index.write(\"</tr>\")\n\n for outlierRatio in outlierRatioList: # For all outlier lists\n if _mode == 'rp':\n outlierRatio /= 2\n elif _mode == 'b':\n outlierRatio /= 2\n \n index.write(\"<tr>\")\n # Name\n index.write(\"<td>Outlier Rate: %.1f%%</td>\" % (outlierRatio*100))\n # Data\n imgpath = '../fig/fig_mnistRes_%s_%d.png'%(_mode,outlierRatio*100)\n print (imgpath)\n index.write(\"<td><img src='%s'></td>\" % (imgpath))\n index.write(\"</tr>\")\n # Finish table\n index.write(\"<br>\")\n index.write(\"</body></table>\")\nindex.write(\"</html>\")\nindex.close()\n\nprint (\"%s saved.\"%(index_path))",
"../fig/fig_mnistRes_rp_25.png\n../fig/fig_mnistRes_rp_45.png\n../fig/fig_mnistRes_rp_47.png\n../fig/fig_mnistRes_rs_50.png\n../fig/fig_mnistRes_rs_90.png\n../fig/fig_mnistRes_rs_95.png\n../fig/fig_mnistRes_b_25.png\n../fig/fig_mnistRes_b_45.png\n../fig/fig_mnistRes_b_47.png\n../html/cls_results.html saved.\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
c511cb1354ae8af8d9854073d15fb4f1413c646f
| 8,388 |
ipynb
|
Jupyter Notebook
|
assignment11.ipynb
|
PGE323M-Fall2018/assignment11
|
c6c5aad28fb09c1c81cd57fefa2544720fcb2a10
|
[
"Apache-2.0"
] | null | null | null |
assignment11.ipynb
|
PGE323M-Fall2018/assignment11
|
c6c5aad28fb09c1c81cd57fefa2544720fcb2a10
|
[
"Apache-2.0"
] | null | null | null |
assignment11.ipynb
|
PGE323M-Fall2018/assignment11
|
c6c5aad28fb09c1c81cd57fefa2544720fcb2a10
|
[
"Apache-2.0"
] | 1 |
2020-07-29T15:20:44.000Z
|
2020-07-29T15:20:44.000Z
| 40.133971 | 1,157 | 0.541488 |
[
[
[
"# Assignment 11\n\nConsider the reservoir shown below with the given properties that has been discretized into 4 equal grid blocks.\n\n\n\nBelow is a skeleton of a Python class that can be used to solve for the pressures in the reservoir. The class is actually written generally enough that it can account for an arbitrary number of grid blocks, but we will only test cases with 4. The class takes a Python dictonary of input parameters as an initialization argument. An example of a complete set of input parameters is shown in the `setup()` function of the tests below.\n\nSeveral simple useful functions are already implemented, your task is to implement the functions `compute_transmisibility()`, `compute_accumulation()`, `fill_matrices()` and `solve_one_step()`. `fill_matrices()` should correctly populate the $\\mathbf{T}$, $\\mathbf{B}$ matrices as well as the vector $\\vec{Q}$. These should also correctly account for the application of boundary conditions. Only the boundary conditions shown in the figure will be tested, but in preparation for future assignments, you may wish to add the logic to the code such that arbitrary pressure/no flow boundary conditions can be applied to either side of the one-dimensional reservoir. You may need to use the `'conversion factor'` for the transmissibilities. `solve_one_step()` should solve a single time step for either the explicit or implicit methods depending on which is specified in the input parameters. The $\\vec{p}{}^{n+1}$ values should be stored in the class attribute `self.p`. If this is implemented correctly, you will be able to then use the `solve()` function to solve the problem up to the `'number of time steps'` value in the input parameters.\n\nThis time, in preparation for solving much larger systems of equations in the future, use the `scipy.sparse` module to create sparse matrix data structures for $\\mathbf{T}$ and $\\mathbf{B}$. The sparsity of the matrix $\\mathbf{T}$ is tested, so please assign this matrix to a class attribute named exactly `T`. Use `scipy.sparse.linalg.spsolve()` for the linear solution of the `'implicit'` method implementation.\n\nOnce you have the tests passing, you might like to experiment with viewing several plots with different time steps, explicit vs. implicit, number of grid blocks, etc. To assist in giving you a feel for how they change the character of the approximate solution. I have implemented a simple plot function that might help for this.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport yaml\nimport scipy.sparse\nimport scipy.sparse.linalg\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"class OneDimReservoir():\n \n def __init__(self, inputs):\n '''\n Class for solving one-dimensional reservoir problems with\n finite differences.\n '''\n \n #stores input dictionary as class attribute, either read from a yaml file\n #or directly from a Python dictonary\n if isinstance(inputs, str):\n with open(inputs) as f:\n self.inputs = yaml.load(f, yaml.FullLoader)\n else:\n self.inputs = inputs\n \n #computes delta_x\n self.Nx = self.inputs['numerical']['number of grids']['x']\n self.N = self.Nx\n self.delta_x = self.inputs['reservoir']['length'] / float(self.Nx)\n \n #gets delta_t from inputs\n self.delta_t = self.inputs['numerical']['time step']\n \n #applies the initial reservoir pressues to self.p\n self.apply_initial_conditions()\n \n #calls fill matrix method (must be completely implemented to work)\n self.fill_matrices()\n \n #create an empty list for storing data if plots are requested\n if 'plots' in self.inputs:\n self.p_plot = []\n \n return\n \n def compute_transmissibility(self):\n '''\n Computes the transmissibility.\n '''\n # Complete implementation here\n \n \n return \n \n \n def compute_accumulation(self):\n '''\n Computes the accumulation.\n '''\n # Complete implementation here\n \n return \n \n def fill_matrices(self):\n '''\n Fills the matrices T, B, and \\vec{Q} and applies boundary\n conditions.\n '''\n # Complete implementation here\n \n return\n \n \n def apply_initial_conditions(self):\n '''\n Applies initial pressures to self.p\n '''\n \n N = self.N\n \n self.p = np.ones(N) * self.inputs['initial conditions']['pressure']\n \n return\n \n \n def solve_one_step(self):\n '''\n Solve one time step using either the implicit or explicit method\n '''\n # Complete implementation here\n \n return\n \n \n def solve(self):\n '''\n Solves until \"number of time steps\"\n '''\n \n for i in range(self.inputs['numerical']['number of time steps']):\n self.solve_one_step()\n \n if i % self.inputs['plots']['frequency'] == 0:\n self.p_plot += [self.get_solution()]\n \n return\n \n def plot(self):\n '''\n Crude plotting function. Plots pressure as a function of grid block #\n '''\n \n if self.p_plot is not None:\n for i in range(len(self.p_plot)):\n plt.plot(self.p_plot[i])\n \n return\n \n def get_solution(self):\n '''\n Returns solution vector\n '''\n return self.p",
"_____no_output_____"
]
],
[
[
"# Example code execution\n\nIf you'd like to run your code in the notebook, perhaps creating a crude plot of the output, you can uncomment the following lines of code in the cell below. You can also inspect the contents of `inputs.yml` and change the parameters to see how the solution is affected.",
"_____no_output_____"
]
],
[
[
"#import matplotlib.pyplot as plt\n#implicit = OneDimReservoir('inputs.yml')\n#implicit.solve()\n#implicit.plot()",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
c511d5b9d42f75d480d7d334691861731ed0dee8
| 505,261 |
ipynb
|
Jupyter Notebook
|
SDD/debug/model_camera_colibration.ipynb
|
thomascong121/SocialDistance
|
c8e6302bc854fcaac0c8503f2b97632cdd948cc9
|
[
"MIT"
] | 2 |
2021-04-12T14:16:33.000Z
|
2021-05-14T13:33:22.000Z
|
SDD/debug/model_camera_colibration.ipynb
|
thomascong121/SocialDistance
|
c8e6302bc854fcaac0c8503f2b97632cdd948cc9
|
[
"MIT"
] | 1 |
2021-05-03T18:54:59.000Z
|
2021-11-24T01:23:48.000Z
|
SDD/debug/model_camera_colibration.ipynb
|
thomascong121/SocialDistance
|
c8e6302bc854fcaac0c8503f2b97632cdd948cc9
|
[
"MIT"
] | 1 |
2021-04-09T09:10:08.000Z
|
2021-04-09T09:10:08.000Z
| 589.569428 | 191,946 | 0.934687 |
[
[
[
"<a href=\"https://colab.research.google.com/github/thomascong121/SocialDistance/blob/master/model_camera_colibration.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"from google.colab import drive\ndrive.mount('/content/drive')",
"Go to this URL in a browser: https://accounts.google.com/o/oauth2/auth?client_id=947318989803-6bn6qk8qdgf4n4g3pfee6491hc0brc4i.apps.googleusercontent.com&redirect_uri=urn%3aietf%3awg%3aoauth%3a2.0%3aoob&response_type=code&scope=email%20https%3a%2f%2fwww.googleapis.com%2fauth%2fdocs.test%20https%3a%2f%2fwww.googleapis.com%2fauth%2fdrive%20https%3a%2f%2fwww.googleapis.com%2fauth%2fdrive.photos.readonly%20https%3a%2f%2fwww.googleapis.com%2fauth%2fpeopleapi.readonly\n\nEnter your authorization code:\n··········\nMounted at /content/drive\n"
],
[
"%%capture\n!pip install gluoncv\n!pip install mxnet-cu101",
"_____no_output_____"
],
[
"import gluoncv\nfrom gluoncv import model_zoo, data, utils\nfrom matplotlib import pyplot as plt\nimport numpy as np\nfrom collections import defaultdict\n\nfrom mxnet import nd\nimport mxnet as mx\nfrom skimage import io\n\nimport cv2\nimport os\nfrom copy import deepcopy\nfrom tqdm import tqdm",
"_____no_output_____"
],
[
"!ls '/content/drive/My Drive/social distance/0.png'",
"'/content/drive/My Drive/social distance/0.png'\n"
],
[
"!nvidia-smi",
"Wed Apr 29 01:15:38 2020 \n+-----------------------------------------------------------------------------+\n| NVIDIA-SMI 440.64.00 Driver Version: 418.67 CUDA Version: 10.1 |\n|-------------------------------+----------------------+----------------------+\n| GPU Name Persistence-M| Bus-Id Disp.A | Volatile Uncorr. ECC |\n| Fan Temp Perf Pwr:Usage/Cap| Memory-Usage | GPU-Util Compute M. |\n|===============================+======================+======================|\n| 0 Tesla P4 Off | 00000000:00:04.0 Off | 0 |\n| N/A 39C P8 7W / 75W | 0MiB / 7611MiB | 0% Default |\n+-------------------------------+----------------------+----------------------+\n \n+-----------------------------------------------------------------------------+\n| Processes: GPU Memory |\n| GPU PID Type Process name Usage |\n|=============================================================================|\n| No running processes found |\n+-----------------------------------------------------------------------------+\n"
],
[
"!nvcc --version",
"nvcc: NVIDIA (R) Cuda compiler driver\nCopyright (c) 2005-2019 NVIDIA Corporation\nBuilt on Sun_Jul_28_19:07:16_PDT_2019\nCuda compilation tools, release 10.1, V10.1.243\n"
],
[
"img_path = '/content/drive/My Drive/social distance/0.png'\nimg = io.imread(img_path)",
"_____no_output_____"
],
[
"video_path = '/content/drive/My Drive/social distance/TownCentreXVID.avi'",
"_____no_output_____"
],
[
"io.imshow(img)\nio.show()",
"_____no_output_____"
],
[
"class Bird_eye_view_Transformer:\n def __init__(self, keypoints, keypoints_birds_eye_view, actual_length, actual_width):\n '''\n keypoints input order \n 0 1\n\n 3 2\n '''\n self.keypoint = np.float32(keypoints)\n self.keypoints_birds_eye_view = np.float32(keypoints_birds_eye_view)\n self.M = cv2.getPerspectiveTransform(self.keypoint, self.keypoints_birds_eye_view)\n self.length_ratio = actual_width/(keypoints_birds_eye_view[3][1] - keypoints_birds_eye_view[0][1])\n self.width_ratio = actual_length/(keypoints_birds_eye_view[1][0] - keypoints_birds_eye_view[0][0])\n print(self.length_ratio, self.width_ratio)\n\n def imshow(self, img):\n dst_img = cv2.warpPerspective(img, self.M, (img.shape[1], img.shape[0]))\n plt.imshow(dst_img)\n plt.show()\n\n def __call__(self, points):\n h = points.shape[0]\n points = np.concatenate([points, np.ones((h, 1))], axis = 1)\n temp = self.M.dot(points.T)\n return (temp[:2]/temp[2]).T\n \n def distance(self, p0, p1):\n return ((p0[0] - p1[0])*self.width_ratio)**2 \\\n + ((p0[1] - p1[1])*self.length_ratio)**2 \n\n",
"_____no_output_____"
],
[
"keypoints = [(1175, 189), (1574, 235), (976, 831), (364, 694)]\nkeypoints_birds_eye_view = [(700, 400), (1200, 400), (1200, 900), (700, 900)]\nactual_length = 10\nactual_width = 5",
"_____no_output_____"
],
[
"transformer = Bird_eye_view_Transformer(keypoints, keypoints_birds_eye_view, actual_length, actual_width)",
"0.01 0.02\n"
],
[
"transformer.imshow(img)",
"_____no_output_____"
],
[
"'''\nstep0 install gluoncv\npip install --upgrade mxnet gluoncv\n'''\nclass Model_Zoo:\n def __init__(self,selected_model, transformer, device):\n self.device = device\n self.transformer = transformer\n self.net = model_zoo.get_model(selected_model, pretrained=True, ctx = self.device)\n\n def __call__(self,image,display=False):\n '''get bbox for input image'''\n image = nd.array(image)\n x, orig_img = data.transforms.presets.yolo.transform_test(image)\n self.shape = orig_img.shape[:2]\n self.benchmark = max(orig_img.shape[:2])\n x = x.copyto(self.device)\n box_ids, scores, bboxes = self.net(x)\n bboxes = bboxes * (image.shape[0]/orig_img.shape[0])\n person_index = []\n\n #check person class\n for i in range(box_ids.shape[1]):\n if box_ids[0][i][0] == 14 and scores[0][i][0] > 0.7:\n person_index.append(i)\n #select bbox of person\n #p1:bbox id of person\n #p2:confidence score\n #p3:bbox location\n # print('======{0} bbox of persons are detected===='.format(len(person_index)))\n p1,p2,p3 = box_ids[0][[person_index],:],scores[0][[person_index],:],bboxes[0][[person_index],:]\n #calaulate bbox coordinate\n bbox_center = self.bbox_center(p3)\n #img with bbox \n\n img_with_bbox = utils.viz.cv_plot_bbox(image.astype('uint8'), p3[0], p2[0], p1[0], colors={14: (0,255,0)},class_names = self.net.classes, linewidth=1)\n result_img = self.bbox_distance(bbox_center,img_with_bbox)\n if display:\n plt.imshow(result_img)\n plt.show()\n return result_img, p1, p2, p3, bbox_center\n\n def show(self, img, p1, p2, p3, bbox_center, resize = None):\n if resize is not None:\n img = mx.image.imresize(nd.array(img).astype('uint8'), self.shape[1], self.shape[0])\n else:\n img = nd.array(img).astype('uint8')\n img_with_bbox = utils.viz.cv_plot_bbox(img, p3[0], p2[0], p1[0], colors={14: (0,255,0)},class_names = self.net.classes, linewidth=1)\n return self.bbox_distance(bbox_center,img_with_bbox)\n\n def bbox_center(self,bbox_location):\n '''calculate center coordinate for each bbox'''\n rst = None\n for loc in range(bbox_location[0].shape[0]):\n (xmin, ymin, xmax, ymax) = bbox_location[0][loc].copyto(mx.cpu())\n center_x = (xmin+xmax)/2\n center_y = ymax\n if rst is not None:\n rst = nd.concatenate([rst, nd.stack(center_x, center_y, axis = 1)])\n else:\n rst = nd.stack(center_x, center_y, axis = 1)\n\n return rst.asnumpy()\n\n def bbox_distance(self,bbox_coord,img, max_detect = 4, safe=2):\n '''\n calculate distance between each bbox, \n if distance < safe, draw a red line\n '''\n #draw the center\n safe = safe**2\n max_detect = max_detect**2\n for coor in range(len(bbox_coord)):\n cv2.circle(img,(int(bbox_coord[coor][0]),int(bbox_coord[coor][1])),5,(0, 0, 255),-1)\n\n bird_eye_view = self.transformer(deepcopy(bbox_coord))\n # print(bird_eye_view)\n # self.transformer.imshow(img)\n\n for i in range(0, len(bbox_coord)):\n for j in range(i+1, len(bbox_coord)):\n dist = self.transformer.distance(bird_eye_view[i], bird_eye_view [j])\n # print(bird_eye_view[i], bird_eye_view [j],dist)\n if dist < safe:\n cv2.line(img,(bbox_coord[i][0],bbox_coord[i][1]),(bbox_coord[j][0],bbox_coord[j][1]),(255, 0, 0), 2)\n elif dist < max_detect:\n cv2.line(img,(bbox_coord[i][0],bbox_coord[i][1]),(bbox_coord[j][0],bbox_coord[j][1]),(0, 255, 0), 2)\n return img\n",
"_____no_output_____"
],
[
"pretrained_models = 'yolo3_darknet53_voc'\ndetect_model = Model_Zoo(pretrained_models, transformer, mx.gpu())",
"Downloading /root/.mxnet/models/yolo3_darknet53_voc-f5ece5ce.zip from https://apache-mxnet.s3-accelerate.dualstack.amazonaws.com/gluon/models/yolo3_darknet53_voc-f5ece5ce.zip...\n"
],
[
"%%time\nrst = detect_model(img,display=True)",
"_____no_output_____"
],
[
"rst",
"_____no_output_____"
],
[
"class Detector:\n def __init__(self, model, save_path = './detections', batch_size = 60, interval = None):\n self.detector = model\n self.save_path = save_path\n self.interval = interval\n self.batch_size = batch_size\n\n def __call__(self, filename):\n v_cap = cv2.VideoCapture(filename)\n v_len = int(v_cap.get(cv2.CAP_PROP_FRAME_COUNT))\n frame_size = (v_cap.get(cv2.CAP_PROP_FRAME_WIDTH), v_cap.get(cv2.CAP_PROP_FRAME_HEIGHT))\n \n fourcc = cv2.VideoWriter_fourcc(*'MP4V')\n fps = v_cap.get(cv2.CAP_PROP_FPS)\n if not os.path.exists(self.save_path):\n os.mkdir(self.save_path)\n print(f'{self.save_path}/{filename.split(\"/\")[-1]}')\n out = cv2.VideoWriter(f'{self.save_path}/{filename.split(\"/\")[-1]}', fourcc, fps,\\\n (int(frame_size[0]), int(frame_size[1]))) \n \n if self.interval is None:\n sample = np.arange(0, v_len)\n else:\n sample = np.arange(0, v_len, self.interval)\n frame = p1 = p2 = p3 = bbox_center =None\n for i in tqdm(range(v_len)):\n success = v_cap.grab()\n \n success, frame = v_cap.retrieve()\n if not success:\n continue\n frame = cv2.cvtColor(frame, cv2.COLOR_BGR2RGB)\n\n if i in sample:\n frame, p1, p2, p3, bbox_center = self.detector(frame)\n else:\n frame = self.detector.show(frame, p1, p2, p3, bbox_center)\n # plt.imshow(frame)\n # plt.show()\n frame = cv2.cvtColor(frame, cv2.COLOR_RGB2BGR)\n out.write(frame)\n\n v_cap.release()\n return out\n\n \n",
"_____no_output_____"
],
[
"detector = Detector(detect_model, interval = 10)",
"_____no_output_____"
],
[
"%%time\ndetector(video_path)",
"\r 0%| | 0/7500 [00:00<?, ?it/s]"
],
[
"!ls ./detections",
"TownCentreXVID.avi\n"
],
[
"",
"_____no_output_____"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
c511d91d12fa5c90e3c58eb053107f5898a42a48
| 154,840 |
ipynb
|
Jupyter Notebook
|
CNN/Exercise3_transferlearning.ipynb
|
scifiswapnil/DeepLearningExperiments
|
1f6c23a7f2b1898352dd3c218effc6c3ae3fa60c
|
[
"BSD-3-Clause"
] | null | null | null |
CNN/Exercise3_transferlearning.ipynb
|
scifiswapnil/DeepLearningExperiments
|
1f6c23a7f2b1898352dd3c218effc6c3ae3fa60c
|
[
"BSD-3-Clause"
] | null | null | null |
CNN/Exercise3_transferlearning.ipynb
|
scifiswapnil/DeepLearningExperiments
|
1f6c23a7f2b1898352dd3c218effc6c3ae3fa60c
|
[
"BSD-3-Clause"
] | null | null | null | 93.502415 | 265 | 0.602564 |
[
[
[
"<a href=\"https://colab.research.google.com/github/scifiswapnil/DeepLearningExperiments/blob/master/CNN/Exercise3_transferlearning.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"#@title Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# https://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.",
"_____no_output_____"
],
[
"# Import all the necessary files!\nimport os\nimport tensorflow as tf\nfrom tensorflow.keras import layers\nfrom tensorflow.keras import Model",
"_____no_output_____"
],
[
"# Download the inception v3 weights\n!wget --no-check-certificate \\\n https://storage.googleapis.com/mledu-datasets/inception_v3_weights_tf_dim_ordering_tf_kernels_notop.h5 \\\n -O /tmp/inception_v3_weights_tf_dim_ordering_tf_kernels_notop.h5\n\n# Import the inception model \nfrom tensorflow.keras.applications.inception_v3 import InceptionV3\n\n# Create an instance of the inception model from the local pre-trained weights\nlocal_weights_file = '/tmp/inception_v3_weights_tf_dim_ordering_tf_kernels_notop.h5'\n\npre_trained_model = InceptionV3(input_shape = (150, 150, 3), \n include_top = False, \n weights = None)\n\npre_trained_model.load_weights(local_weights_file)\n\n# Make all the layers in the pre-trained model non-trainable\nfor layer in pre_trained_model.layers:\n layer.trainable = False\n \n# Print the model summary\npre_trained_model.summary()\n\n# Expected Output is extremely large, but should end with:\n\n#batch_normalization_v1_281 (Bat (None, 3, 3, 192) 576 conv2d_281[0][0] \n#__________________________________________________________________________________________________\n#activation_273 (Activation) (None, 3, 3, 320) 0 batch_normalization_v1_273[0][0] \n#__________________________________________________________________________________________________\n#mixed9_1 (Concatenate) (None, 3, 3, 768) 0 activation_275[0][0] \n# activation_276[0][0] \n#__________________________________________________________________________________________________\n#concatenate_5 (Concatenate) (None, 3, 3, 768) 0 activation_279[0][0] \n# activation_280[0][0] \n#__________________________________________________________________________________________________\n#activation_281 (Activation) (None, 3, 3, 192) 0 batch_normalization_v1_281[0][0] \n#__________________________________________________________________________________________________\n#mixed10 (Concatenate) (None, 3, 3, 2048) 0 activation_273[0][0] \n# mixed9_1[0][0] \n# concatenate_5[0][0] \n# activation_281[0][0] \n#==================================================================================================\n#Total params: 21,802,784\n#Trainable params: 0\n#Non-trainable params: 21,802,784",
"--2020-05-19 17:27:39-- https://storage.googleapis.com/mledu-datasets/inception_v3_weights_tf_dim_ordering_tf_kernels_notop.h5\nResolving storage.googleapis.com (storage.googleapis.com)... 64.233.189.128, 2404:6800:4008:c07::80\nConnecting to storage.googleapis.com (storage.googleapis.com)|64.233.189.128|:443... connected.\nHTTP request sent, awaiting response... 200 OK\nLength: 87910968 (84M) [application/x-hdf]\nSaving to: ‘/tmp/inception_v3_weights_tf_dim_ordering_tf_kernels_notop.h5’\n\n/tmp/inception_v3_w 100%[===================>] 83.84M 46.0MB/s in 1.8s \n\n2020-05-19 17:27:41 (46.0 MB/s) - ‘/tmp/inception_v3_weights_tf_dim_ordering_tf_kernels_notop.h5’ saved [87910968/87910968]\n\nModel: \"inception_v3\"\n__________________________________________________________________________________________________\nLayer (type) Output Shape Param # Connected to \n==================================================================================================\ninput_1 (InputLayer) [(None, 150, 150, 3) 0 \n__________________________________________________________________________________________________\nconv2d (Conv2D) (None, 74, 74, 32) 864 input_1[0][0] \n__________________________________________________________________________________________________\nbatch_normalization (BatchNorma (None, 74, 74, 32) 96 conv2d[0][0] \n__________________________________________________________________________________________________\nactivation (Activation) (None, 74, 74, 32) 0 batch_normalization[0][0] \n__________________________________________________________________________________________________\nconv2d_1 (Conv2D) (None, 72, 72, 32) 9216 activation[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_1 (BatchNor (None, 72, 72, 32) 96 conv2d_1[0][0] \n__________________________________________________________________________________________________\nactivation_1 (Activation) (None, 72, 72, 32) 0 batch_normalization_1[0][0] \n__________________________________________________________________________________________________\nconv2d_2 (Conv2D) (None, 72, 72, 64) 18432 activation_1[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_2 (BatchNor (None, 72, 72, 64) 192 conv2d_2[0][0] \n__________________________________________________________________________________________________\nactivation_2 (Activation) (None, 72, 72, 64) 0 batch_normalization_2[0][0] \n__________________________________________________________________________________________________\nmax_pooling2d (MaxPooling2D) (None, 35, 35, 64) 0 activation_2[0][0] \n__________________________________________________________________________________________________\nconv2d_3 (Conv2D) (None, 35, 35, 80) 5120 max_pooling2d[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_3 (BatchNor (None, 35, 35, 80) 240 conv2d_3[0][0] \n__________________________________________________________________________________________________\nactivation_3 (Activation) (None, 35, 35, 80) 0 batch_normalization_3[0][0] \n__________________________________________________________________________________________________\nconv2d_4 (Conv2D) (None, 33, 33, 192) 138240 activation_3[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_4 (BatchNor (None, 33, 33, 192) 576 conv2d_4[0][0] \n__________________________________________________________________________________________________\nactivation_4 (Activation) (None, 33, 33, 192) 0 batch_normalization_4[0][0] \n__________________________________________________________________________________________________\nmax_pooling2d_1 (MaxPooling2D) (None, 16, 16, 192) 0 activation_4[0][0] \n__________________________________________________________________________________________________\nconv2d_8 (Conv2D) (None, 16, 16, 64) 12288 max_pooling2d_1[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_8 (BatchNor (None, 16, 16, 64) 192 conv2d_8[0][0] \n__________________________________________________________________________________________________\nactivation_8 (Activation) (None, 16, 16, 64) 0 batch_normalization_8[0][0] \n__________________________________________________________________________________________________\nconv2d_6 (Conv2D) (None, 16, 16, 48) 9216 max_pooling2d_1[0][0] \n__________________________________________________________________________________________________\nconv2d_9 (Conv2D) (None, 16, 16, 96) 55296 activation_8[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_6 (BatchNor (None, 16, 16, 48) 144 conv2d_6[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_9 (BatchNor (None, 16, 16, 96) 288 conv2d_9[0][0] \n__________________________________________________________________________________________________\nactivation_6 (Activation) (None, 16, 16, 48) 0 batch_normalization_6[0][0] \n__________________________________________________________________________________________________\nactivation_9 (Activation) (None, 16, 16, 96) 0 batch_normalization_9[0][0] \n__________________________________________________________________________________________________\naverage_pooling2d (AveragePooli (None, 16, 16, 192) 0 max_pooling2d_1[0][0] \n__________________________________________________________________________________________________\nconv2d_5 (Conv2D) (None, 16, 16, 64) 12288 max_pooling2d_1[0][0] \n__________________________________________________________________________________________________\nconv2d_7 (Conv2D) (None, 16, 16, 64) 76800 activation_6[0][0] \n__________________________________________________________________________________________________\nconv2d_10 (Conv2D) (None, 16, 16, 96) 82944 activation_9[0][0] \n__________________________________________________________________________________________________\nconv2d_11 (Conv2D) (None, 16, 16, 32) 6144 average_pooling2d[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_5 (BatchNor (None, 16, 16, 64) 192 conv2d_5[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_7 (BatchNor (None, 16, 16, 64) 192 conv2d_7[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_10 (BatchNo (None, 16, 16, 96) 288 conv2d_10[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_11 (BatchNo (None, 16, 16, 32) 96 conv2d_11[0][0] \n__________________________________________________________________________________________________\nactivation_5 (Activation) (None, 16, 16, 64) 0 batch_normalization_5[0][0] \n__________________________________________________________________________________________________\nactivation_7 (Activation) (None, 16, 16, 64) 0 batch_normalization_7[0][0] \n__________________________________________________________________________________________________\nactivation_10 (Activation) (None, 16, 16, 96) 0 batch_normalization_10[0][0] \n__________________________________________________________________________________________________\nactivation_11 (Activation) (None, 16, 16, 32) 0 batch_normalization_11[0][0] \n__________________________________________________________________________________________________\nmixed0 (Concatenate) (None, 16, 16, 256) 0 activation_5[0][0] \n activation_7[0][0] \n activation_10[0][0] \n activation_11[0][0] \n__________________________________________________________________________________________________\nconv2d_15 (Conv2D) (None, 16, 16, 64) 16384 mixed0[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_15 (BatchNo (None, 16, 16, 64) 192 conv2d_15[0][0] \n__________________________________________________________________________________________________\nactivation_15 (Activation) (None, 16, 16, 64) 0 batch_normalization_15[0][0] \n__________________________________________________________________________________________________\nconv2d_13 (Conv2D) (None, 16, 16, 48) 12288 mixed0[0][0] \n__________________________________________________________________________________________________\nconv2d_16 (Conv2D) (None, 16, 16, 96) 55296 activation_15[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_13 (BatchNo (None, 16, 16, 48) 144 conv2d_13[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_16 (BatchNo (None, 16, 16, 96) 288 conv2d_16[0][0] \n__________________________________________________________________________________________________\nactivation_13 (Activation) (None, 16, 16, 48) 0 batch_normalization_13[0][0] \n__________________________________________________________________________________________________\nactivation_16 (Activation) (None, 16, 16, 96) 0 batch_normalization_16[0][0] \n__________________________________________________________________________________________________\naverage_pooling2d_1 (AveragePoo (None, 16, 16, 256) 0 mixed0[0][0] \n__________________________________________________________________________________________________\nconv2d_12 (Conv2D) (None, 16, 16, 64) 16384 mixed0[0][0] \n__________________________________________________________________________________________________\nconv2d_14 (Conv2D) (None, 16, 16, 64) 76800 activation_13[0][0] \n__________________________________________________________________________________________________\nconv2d_17 (Conv2D) (None, 16, 16, 96) 82944 activation_16[0][0] \n__________________________________________________________________________________________________\nconv2d_18 (Conv2D) (None, 16, 16, 64) 16384 average_pooling2d_1[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_12 (BatchNo (None, 16, 16, 64) 192 conv2d_12[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_14 (BatchNo (None, 16, 16, 64) 192 conv2d_14[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_17 (BatchNo (None, 16, 16, 96) 288 conv2d_17[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_18 (BatchNo (None, 16, 16, 64) 192 conv2d_18[0][0] \n__________________________________________________________________________________________________\nactivation_12 (Activation) (None, 16, 16, 64) 0 batch_normalization_12[0][0] \n__________________________________________________________________________________________________\nactivation_14 (Activation) (None, 16, 16, 64) 0 batch_normalization_14[0][0] \n__________________________________________________________________________________________________\nactivation_17 (Activation) (None, 16, 16, 96) 0 batch_normalization_17[0][0] \n__________________________________________________________________________________________________\nactivation_18 (Activation) (None, 16, 16, 64) 0 batch_normalization_18[0][0] \n__________________________________________________________________________________________________\nmixed1 (Concatenate) (None, 16, 16, 288) 0 activation_12[0][0] \n activation_14[0][0] \n activation_17[0][0] \n activation_18[0][0] \n__________________________________________________________________________________________________\nconv2d_22 (Conv2D) (None, 16, 16, 64) 18432 mixed1[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_22 (BatchNo (None, 16, 16, 64) 192 conv2d_22[0][0] \n__________________________________________________________________________________________________\nactivation_22 (Activation) (None, 16, 16, 64) 0 batch_normalization_22[0][0] \n__________________________________________________________________________________________________\nconv2d_20 (Conv2D) (None, 16, 16, 48) 13824 mixed1[0][0] \n__________________________________________________________________________________________________\nconv2d_23 (Conv2D) (None, 16, 16, 96) 55296 activation_22[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_20 (BatchNo (None, 16, 16, 48) 144 conv2d_20[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_23 (BatchNo (None, 16, 16, 96) 288 conv2d_23[0][0] \n__________________________________________________________________________________________________\nactivation_20 (Activation) (None, 16, 16, 48) 0 batch_normalization_20[0][0] \n__________________________________________________________________________________________________\nactivation_23 (Activation) (None, 16, 16, 96) 0 batch_normalization_23[0][0] \n__________________________________________________________________________________________________\naverage_pooling2d_2 (AveragePoo (None, 16, 16, 288) 0 mixed1[0][0] \n__________________________________________________________________________________________________\nconv2d_19 (Conv2D) (None, 16, 16, 64) 18432 mixed1[0][0] \n__________________________________________________________________________________________________\nconv2d_21 (Conv2D) (None, 16, 16, 64) 76800 activation_20[0][0] \n__________________________________________________________________________________________________\nconv2d_24 (Conv2D) (None, 16, 16, 96) 82944 activation_23[0][0] \n__________________________________________________________________________________________________\nconv2d_25 (Conv2D) (None, 16, 16, 64) 18432 average_pooling2d_2[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_19 (BatchNo (None, 16, 16, 64) 192 conv2d_19[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_21 (BatchNo (None, 16, 16, 64) 192 conv2d_21[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_24 (BatchNo (None, 16, 16, 96) 288 conv2d_24[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_25 (BatchNo (None, 16, 16, 64) 192 conv2d_25[0][0] \n__________________________________________________________________________________________________\nactivation_19 (Activation) (None, 16, 16, 64) 0 batch_normalization_19[0][0] \n__________________________________________________________________________________________________\nactivation_21 (Activation) (None, 16, 16, 64) 0 batch_normalization_21[0][0] \n__________________________________________________________________________________________________\nactivation_24 (Activation) (None, 16, 16, 96) 0 batch_normalization_24[0][0] \n__________________________________________________________________________________________________\nactivation_25 (Activation) (None, 16, 16, 64) 0 batch_normalization_25[0][0] \n__________________________________________________________________________________________________\nmixed2 (Concatenate) (None, 16, 16, 288) 0 activation_19[0][0] \n activation_21[0][0] \n activation_24[0][0] \n activation_25[0][0] \n__________________________________________________________________________________________________\nconv2d_27 (Conv2D) (None, 16, 16, 64) 18432 mixed2[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_27 (BatchNo (None, 16, 16, 64) 192 conv2d_27[0][0] \n__________________________________________________________________________________________________\nactivation_27 (Activation) (None, 16, 16, 64) 0 batch_normalization_27[0][0] \n__________________________________________________________________________________________________\nconv2d_28 (Conv2D) (None, 16, 16, 96) 55296 activation_27[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_28 (BatchNo (None, 16, 16, 96) 288 conv2d_28[0][0] \n__________________________________________________________________________________________________\nactivation_28 (Activation) (None, 16, 16, 96) 0 batch_normalization_28[0][0] \n__________________________________________________________________________________________________\nconv2d_26 (Conv2D) (None, 7, 7, 384) 995328 mixed2[0][0] \n__________________________________________________________________________________________________\nconv2d_29 (Conv2D) (None, 7, 7, 96) 82944 activation_28[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_26 (BatchNo (None, 7, 7, 384) 1152 conv2d_26[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_29 (BatchNo (None, 7, 7, 96) 288 conv2d_29[0][0] \n__________________________________________________________________________________________________\nactivation_26 (Activation) (None, 7, 7, 384) 0 batch_normalization_26[0][0] \n__________________________________________________________________________________________________\nactivation_29 (Activation) (None, 7, 7, 96) 0 batch_normalization_29[0][0] \n__________________________________________________________________________________________________\nmax_pooling2d_2 (MaxPooling2D) (None, 7, 7, 288) 0 mixed2[0][0] \n__________________________________________________________________________________________________\nmixed3 (Concatenate) (None, 7, 7, 768) 0 activation_26[0][0] \n activation_29[0][0] \n max_pooling2d_2[0][0] \n__________________________________________________________________________________________________\nconv2d_34 (Conv2D) (None, 7, 7, 128) 98304 mixed3[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_34 (BatchNo (None, 7, 7, 128) 384 conv2d_34[0][0] \n__________________________________________________________________________________________________\nactivation_34 (Activation) (None, 7, 7, 128) 0 batch_normalization_34[0][0] \n__________________________________________________________________________________________________\nconv2d_35 (Conv2D) (None, 7, 7, 128) 114688 activation_34[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_35 (BatchNo (None, 7, 7, 128) 384 conv2d_35[0][0] \n__________________________________________________________________________________________________\nactivation_35 (Activation) (None, 7, 7, 128) 0 batch_normalization_35[0][0] \n__________________________________________________________________________________________________\nconv2d_31 (Conv2D) (None, 7, 7, 128) 98304 mixed3[0][0] \n__________________________________________________________________________________________________\nconv2d_36 (Conv2D) (None, 7, 7, 128) 114688 activation_35[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_31 (BatchNo (None, 7, 7, 128) 384 conv2d_31[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_36 (BatchNo (None, 7, 7, 128) 384 conv2d_36[0][0] \n__________________________________________________________________________________________________\nactivation_31 (Activation) (None, 7, 7, 128) 0 batch_normalization_31[0][0] \n__________________________________________________________________________________________________\nactivation_36 (Activation) (None, 7, 7, 128) 0 batch_normalization_36[0][0] \n__________________________________________________________________________________________________\nconv2d_32 (Conv2D) (None, 7, 7, 128) 114688 activation_31[0][0] \n__________________________________________________________________________________________________\nconv2d_37 (Conv2D) (None, 7, 7, 128) 114688 activation_36[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_32 (BatchNo (None, 7, 7, 128) 384 conv2d_32[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_37 (BatchNo (None, 7, 7, 128) 384 conv2d_37[0][0] \n__________________________________________________________________________________________________\nactivation_32 (Activation) (None, 7, 7, 128) 0 batch_normalization_32[0][0] \n__________________________________________________________________________________________________\nactivation_37 (Activation) (None, 7, 7, 128) 0 batch_normalization_37[0][0] \n__________________________________________________________________________________________________\naverage_pooling2d_3 (AveragePoo (None, 7, 7, 768) 0 mixed3[0][0] \n__________________________________________________________________________________________________\nconv2d_30 (Conv2D) (None, 7, 7, 192) 147456 mixed3[0][0] \n__________________________________________________________________________________________________\nconv2d_33 (Conv2D) (None, 7, 7, 192) 172032 activation_32[0][0] \n__________________________________________________________________________________________________\nconv2d_38 (Conv2D) (None, 7, 7, 192) 172032 activation_37[0][0] \n__________________________________________________________________________________________________\nconv2d_39 (Conv2D) (None, 7, 7, 192) 147456 average_pooling2d_3[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_30 (BatchNo (None, 7, 7, 192) 576 conv2d_30[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_33 (BatchNo (None, 7, 7, 192) 576 conv2d_33[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_38 (BatchNo (None, 7, 7, 192) 576 conv2d_38[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_39 (BatchNo (None, 7, 7, 192) 576 conv2d_39[0][0] \n__________________________________________________________________________________________________\nactivation_30 (Activation) (None, 7, 7, 192) 0 batch_normalization_30[0][0] \n__________________________________________________________________________________________________\nactivation_33 (Activation) (None, 7, 7, 192) 0 batch_normalization_33[0][0] \n__________________________________________________________________________________________________\nactivation_38 (Activation) (None, 7, 7, 192) 0 batch_normalization_38[0][0] \n__________________________________________________________________________________________________\nactivation_39 (Activation) (None, 7, 7, 192) 0 batch_normalization_39[0][0] \n__________________________________________________________________________________________________\nmixed4 (Concatenate) (None, 7, 7, 768) 0 activation_30[0][0] \n activation_33[0][0] \n activation_38[0][0] \n activation_39[0][0] \n__________________________________________________________________________________________________\nconv2d_44 (Conv2D) (None, 7, 7, 160) 122880 mixed4[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_44 (BatchNo (None, 7, 7, 160) 480 conv2d_44[0][0] \n__________________________________________________________________________________________________\nactivation_44 (Activation) (None, 7, 7, 160) 0 batch_normalization_44[0][0] \n__________________________________________________________________________________________________\nconv2d_45 (Conv2D) (None, 7, 7, 160) 179200 activation_44[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_45 (BatchNo (None, 7, 7, 160) 480 conv2d_45[0][0] \n__________________________________________________________________________________________________\nactivation_45 (Activation) (None, 7, 7, 160) 0 batch_normalization_45[0][0] \n__________________________________________________________________________________________________\nconv2d_41 (Conv2D) (None, 7, 7, 160) 122880 mixed4[0][0] \n__________________________________________________________________________________________________\nconv2d_46 (Conv2D) (None, 7, 7, 160) 179200 activation_45[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_41 (BatchNo (None, 7, 7, 160) 480 conv2d_41[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_46 (BatchNo (None, 7, 7, 160) 480 conv2d_46[0][0] \n__________________________________________________________________________________________________\nactivation_41 (Activation) (None, 7, 7, 160) 0 batch_normalization_41[0][0] \n__________________________________________________________________________________________________\nactivation_46 (Activation) (None, 7, 7, 160) 0 batch_normalization_46[0][0] \n__________________________________________________________________________________________________\nconv2d_42 (Conv2D) (None, 7, 7, 160) 179200 activation_41[0][0] \n__________________________________________________________________________________________________\nconv2d_47 (Conv2D) (None, 7, 7, 160) 179200 activation_46[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_42 (BatchNo (None, 7, 7, 160) 480 conv2d_42[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_47 (BatchNo (None, 7, 7, 160) 480 conv2d_47[0][0] \n__________________________________________________________________________________________________\nactivation_42 (Activation) (None, 7, 7, 160) 0 batch_normalization_42[0][0] \n__________________________________________________________________________________________________\nactivation_47 (Activation) (None, 7, 7, 160) 0 batch_normalization_47[0][0] \n__________________________________________________________________________________________________\naverage_pooling2d_4 (AveragePoo (None, 7, 7, 768) 0 mixed4[0][0] \n__________________________________________________________________________________________________\nconv2d_40 (Conv2D) (None, 7, 7, 192) 147456 mixed4[0][0] \n__________________________________________________________________________________________________\nconv2d_43 (Conv2D) (None, 7, 7, 192) 215040 activation_42[0][0] \n__________________________________________________________________________________________________\nconv2d_48 (Conv2D) (None, 7, 7, 192) 215040 activation_47[0][0] \n__________________________________________________________________________________________________\nconv2d_49 (Conv2D) (None, 7, 7, 192) 147456 average_pooling2d_4[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_40 (BatchNo (None, 7, 7, 192) 576 conv2d_40[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_43 (BatchNo (None, 7, 7, 192) 576 conv2d_43[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_48 (BatchNo (None, 7, 7, 192) 576 conv2d_48[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_49 (BatchNo (None, 7, 7, 192) 576 conv2d_49[0][0] \n__________________________________________________________________________________________________\nactivation_40 (Activation) (None, 7, 7, 192) 0 batch_normalization_40[0][0] \n__________________________________________________________________________________________________\nactivation_43 (Activation) (None, 7, 7, 192) 0 batch_normalization_43[0][0] \n__________________________________________________________________________________________________\nactivation_48 (Activation) (None, 7, 7, 192) 0 batch_normalization_48[0][0] \n__________________________________________________________________________________________________\nactivation_49 (Activation) (None, 7, 7, 192) 0 batch_normalization_49[0][0] \n__________________________________________________________________________________________________\nmixed5 (Concatenate) (None, 7, 7, 768) 0 activation_40[0][0] \n activation_43[0][0] \n activation_48[0][0] \n activation_49[0][0] \n__________________________________________________________________________________________________\nconv2d_54 (Conv2D) (None, 7, 7, 160) 122880 mixed5[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_54 (BatchNo (None, 7, 7, 160) 480 conv2d_54[0][0] \n__________________________________________________________________________________________________\nactivation_54 (Activation) (None, 7, 7, 160) 0 batch_normalization_54[0][0] \n__________________________________________________________________________________________________\nconv2d_55 (Conv2D) (None, 7, 7, 160) 179200 activation_54[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_55 (BatchNo (None, 7, 7, 160) 480 conv2d_55[0][0] \n__________________________________________________________________________________________________\nactivation_55 (Activation) (None, 7, 7, 160) 0 batch_normalization_55[0][0] \n__________________________________________________________________________________________________\nconv2d_51 (Conv2D) (None, 7, 7, 160) 122880 mixed5[0][0] \n__________________________________________________________________________________________________\nconv2d_56 (Conv2D) (None, 7, 7, 160) 179200 activation_55[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_51 (BatchNo (None, 7, 7, 160) 480 conv2d_51[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_56 (BatchNo (None, 7, 7, 160) 480 conv2d_56[0][0] \n__________________________________________________________________________________________________\nactivation_51 (Activation) (None, 7, 7, 160) 0 batch_normalization_51[0][0] \n__________________________________________________________________________________________________\nactivation_56 (Activation) (None, 7, 7, 160) 0 batch_normalization_56[0][0] \n__________________________________________________________________________________________________\nconv2d_52 (Conv2D) (None, 7, 7, 160) 179200 activation_51[0][0] \n__________________________________________________________________________________________________\nconv2d_57 (Conv2D) (None, 7, 7, 160) 179200 activation_56[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_52 (BatchNo (None, 7, 7, 160) 480 conv2d_52[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_57 (BatchNo (None, 7, 7, 160) 480 conv2d_57[0][0] \n__________________________________________________________________________________________________\nactivation_52 (Activation) (None, 7, 7, 160) 0 batch_normalization_52[0][0] \n__________________________________________________________________________________________________\nactivation_57 (Activation) (None, 7, 7, 160) 0 batch_normalization_57[0][0] \n__________________________________________________________________________________________________\naverage_pooling2d_5 (AveragePoo (None, 7, 7, 768) 0 mixed5[0][0] \n__________________________________________________________________________________________________\nconv2d_50 (Conv2D) (None, 7, 7, 192) 147456 mixed5[0][0] \n__________________________________________________________________________________________________\nconv2d_53 (Conv2D) (None, 7, 7, 192) 215040 activation_52[0][0] \n__________________________________________________________________________________________________\nconv2d_58 (Conv2D) (None, 7, 7, 192) 215040 activation_57[0][0] \n__________________________________________________________________________________________________\nconv2d_59 (Conv2D) (None, 7, 7, 192) 147456 average_pooling2d_5[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_50 (BatchNo (None, 7, 7, 192) 576 conv2d_50[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_53 (BatchNo (None, 7, 7, 192) 576 conv2d_53[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_58 (BatchNo (None, 7, 7, 192) 576 conv2d_58[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_59 (BatchNo (None, 7, 7, 192) 576 conv2d_59[0][0] \n__________________________________________________________________________________________________\nactivation_50 (Activation) (None, 7, 7, 192) 0 batch_normalization_50[0][0] \n__________________________________________________________________________________________________\nactivation_53 (Activation) (None, 7, 7, 192) 0 batch_normalization_53[0][0] \n__________________________________________________________________________________________________\nactivation_58 (Activation) (None, 7, 7, 192) 0 batch_normalization_58[0][0] \n__________________________________________________________________________________________________\nactivation_59 (Activation) (None, 7, 7, 192) 0 batch_normalization_59[0][0] \n__________________________________________________________________________________________________\nmixed6 (Concatenate) (None, 7, 7, 768) 0 activation_50[0][0] \n activation_53[0][0] \n activation_58[0][0] \n activation_59[0][0] \n__________________________________________________________________________________________________\nconv2d_64 (Conv2D) (None, 7, 7, 192) 147456 mixed6[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_64 (BatchNo (None, 7, 7, 192) 576 conv2d_64[0][0] \n__________________________________________________________________________________________________\nactivation_64 (Activation) (None, 7, 7, 192) 0 batch_normalization_64[0][0] \n__________________________________________________________________________________________________\nconv2d_65 (Conv2D) (None, 7, 7, 192) 258048 activation_64[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_65 (BatchNo (None, 7, 7, 192) 576 conv2d_65[0][0] \n__________________________________________________________________________________________________\nactivation_65 (Activation) (None, 7, 7, 192) 0 batch_normalization_65[0][0] \n__________________________________________________________________________________________________\nconv2d_61 (Conv2D) (None, 7, 7, 192) 147456 mixed6[0][0] \n__________________________________________________________________________________________________\nconv2d_66 (Conv2D) (None, 7, 7, 192) 258048 activation_65[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_61 (BatchNo (None, 7, 7, 192) 576 conv2d_61[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_66 (BatchNo (None, 7, 7, 192) 576 conv2d_66[0][0] \n__________________________________________________________________________________________________\nactivation_61 (Activation) (None, 7, 7, 192) 0 batch_normalization_61[0][0] \n__________________________________________________________________________________________________\nactivation_66 (Activation) (None, 7, 7, 192) 0 batch_normalization_66[0][0] \n__________________________________________________________________________________________________\nconv2d_62 (Conv2D) (None, 7, 7, 192) 258048 activation_61[0][0] \n__________________________________________________________________________________________________\nconv2d_67 (Conv2D) (None, 7, 7, 192) 258048 activation_66[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_62 (BatchNo (None, 7, 7, 192) 576 conv2d_62[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_67 (BatchNo (None, 7, 7, 192) 576 conv2d_67[0][0] \n__________________________________________________________________________________________________\nactivation_62 (Activation) (None, 7, 7, 192) 0 batch_normalization_62[0][0] \n__________________________________________________________________________________________________\nactivation_67 (Activation) (None, 7, 7, 192) 0 batch_normalization_67[0][0] \n__________________________________________________________________________________________________\naverage_pooling2d_6 (AveragePoo (None, 7, 7, 768) 0 mixed6[0][0] \n__________________________________________________________________________________________________\nconv2d_60 (Conv2D) (None, 7, 7, 192) 147456 mixed6[0][0] \n__________________________________________________________________________________________________\nconv2d_63 (Conv2D) (None, 7, 7, 192) 258048 activation_62[0][0] \n__________________________________________________________________________________________________\nconv2d_68 (Conv2D) (None, 7, 7, 192) 258048 activation_67[0][0] \n__________________________________________________________________________________________________\nconv2d_69 (Conv2D) (None, 7, 7, 192) 147456 average_pooling2d_6[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_60 (BatchNo (None, 7, 7, 192) 576 conv2d_60[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_63 (BatchNo (None, 7, 7, 192) 576 conv2d_63[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_68 (BatchNo (None, 7, 7, 192) 576 conv2d_68[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_69 (BatchNo (None, 7, 7, 192) 576 conv2d_69[0][0] \n__________________________________________________________________________________________________\nactivation_60 (Activation) (None, 7, 7, 192) 0 batch_normalization_60[0][0] \n__________________________________________________________________________________________________\nactivation_63 (Activation) (None, 7, 7, 192) 0 batch_normalization_63[0][0] \n__________________________________________________________________________________________________\nactivation_68 (Activation) (None, 7, 7, 192) 0 batch_normalization_68[0][0] \n__________________________________________________________________________________________________\nactivation_69 (Activation) (None, 7, 7, 192) 0 batch_normalization_69[0][0] \n__________________________________________________________________________________________________\nmixed7 (Concatenate) (None, 7, 7, 768) 0 activation_60[0][0] \n activation_63[0][0] \n activation_68[0][0] \n activation_69[0][0] \n__________________________________________________________________________________________________\nconv2d_72 (Conv2D) (None, 7, 7, 192) 147456 mixed7[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_72 (BatchNo (None, 7, 7, 192) 576 conv2d_72[0][0] \n__________________________________________________________________________________________________\nactivation_72 (Activation) (None, 7, 7, 192) 0 batch_normalization_72[0][0] \n__________________________________________________________________________________________________\nconv2d_73 (Conv2D) (None, 7, 7, 192) 258048 activation_72[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_73 (BatchNo (None, 7, 7, 192) 576 conv2d_73[0][0] \n__________________________________________________________________________________________________\nactivation_73 (Activation) (None, 7, 7, 192) 0 batch_normalization_73[0][0] \n__________________________________________________________________________________________________\nconv2d_70 (Conv2D) (None, 7, 7, 192) 147456 mixed7[0][0] \n__________________________________________________________________________________________________\nconv2d_74 (Conv2D) (None, 7, 7, 192) 258048 activation_73[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_70 (BatchNo (None, 7, 7, 192) 576 conv2d_70[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_74 (BatchNo (None, 7, 7, 192) 576 conv2d_74[0][0] \n__________________________________________________________________________________________________\nactivation_70 (Activation) (None, 7, 7, 192) 0 batch_normalization_70[0][0] \n__________________________________________________________________________________________________\nactivation_74 (Activation) (None, 7, 7, 192) 0 batch_normalization_74[0][0] \n__________________________________________________________________________________________________\nconv2d_71 (Conv2D) (None, 3, 3, 320) 552960 activation_70[0][0] \n__________________________________________________________________________________________________\nconv2d_75 (Conv2D) (None, 3, 3, 192) 331776 activation_74[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_71 (BatchNo (None, 3, 3, 320) 960 conv2d_71[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_75 (BatchNo (None, 3, 3, 192) 576 conv2d_75[0][0] \n__________________________________________________________________________________________________\nactivation_71 (Activation) (None, 3, 3, 320) 0 batch_normalization_71[0][0] \n__________________________________________________________________________________________________\nactivation_75 (Activation) (None, 3, 3, 192) 0 batch_normalization_75[0][0] \n__________________________________________________________________________________________________\nmax_pooling2d_3 (MaxPooling2D) (None, 3, 3, 768) 0 mixed7[0][0] \n__________________________________________________________________________________________________\nmixed8 (Concatenate) (None, 3, 3, 1280) 0 activation_71[0][0] \n activation_75[0][0] \n max_pooling2d_3[0][0] \n__________________________________________________________________________________________________\nconv2d_80 (Conv2D) (None, 3, 3, 448) 573440 mixed8[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_80 (BatchNo (None, 3, 3, 448) 1344 conv2d_80[0][0] \n__________________________________________________________________________________________________\nactivation_80 (Activation) (None, 3, 3, 448) 0 batch_normalization_80[0][0] \n__________________________________________________________________________________________________\nconv2d_77 (Conv2D) (None, 3, 3, 384) 491520 mixed8[0][0] \n__________________________________________________________________________________________________\nconv2d_81 (Conv2D) (None, 3, 3, 384) 1548288 activation_80[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_77 (BatchNo (None, 3, 3, 384) 1152 conv2d_77[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_81 (BatchNo (None, 3, 3, 384) 1152 conv2d_81[0][0] \n__________________________________________________________________________________________________\nactivation_77 (Activation) (None, 3, 3, 384) 0 batch_normalization_77[0][0] \n__________________________________________________________________________________________________\nactivation_81 (Activation) (None, 3, 3, 384) 0 batch_normalization_81[0][0] \n__________________________________________________________________________________________________\nconv2d_78 (Conv2D) (None, 3, 3, 384) 442368 activation_77[0][0] \n__________________________________________________________________________________________________\nconv2d_79 (Conv2D) (None, 3, 3, 384) 442368 activation_77[0][0] \n__________________________________________________________________________________________________\nconv2d_82 (Conv2D) (None, 3, 3, 384) 442368 activation_81[0][0] \n__________________________________________________________________________________________________\nconv2d_83 (Conv2D) (None, 3, 3, 384) 442368 activation_81[0][0] \n__________________________________________________________________________________________________\naverage_pooling2d_7 (AveragePoo (None, 3, 3, 1280) 0 mixed8[0][0] \n__________________________________________________________________________________________________\nconv2d_76 (Conv2D) (None, 3, 3, 320) 409600 mixed8[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_78 (BatchNo (None, 3, 3, 384) 1152 conv2d_78[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_79 (BatchNo (None, 3, 3, 384) 1152 conv2d_79[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_82 (BatchNo (None, 3, 3, 384) 1152 conv2d_82[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_83 (BatchNo (None, 3, 3, 384) 1152 conv2d_83[0][0] \n__________________________________________________________________________________________________\nconv2d_84 (Conv2D) (None, 3, 3, 192) 245760 average_pooling2d_7[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_76 (BatchNo (None, 3, 3, 320) 960 conv2d_76[0][0] \n__________________________________________________________________________________________________\nactivation_78 (Activation) (None, 3, 3, 384) 0 batch_normalization_78[0][0] \n__________________________________________________________________________________________________\nactivation_79 (Activation) (None, 3, 3, 384) 0 batch_normalization_79[0][0] \n__________________________________________________________________________________________________\nactivation_82 (Activation) (None, 3, 3, 384) 0 batch_normalization_82[0][0] \n__________________________________________________________________________________________________\nactivation_83 (Activation) (None, 3, 3, 384) 0 batch_normalization_83[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_84 (BatchNo (None, 3, 3, 192) 576 conv2d_84[0][0] \n__________________________________________________________________________________________________\nactivation_76 (Activation) (None, 3, 3, 320) 0 batch_normalization_76[0][0] \n__________________________________________________________________________________________________\nmixed9_0 (Concatenate) (None, 3, 3, 768) 0 activation_78[0][0] \n activation_79[0][0] \n__________________________________________________________________________________________________\nconcatenate (Concatenate) (None, 3, 3, 768) 0 activation_82[0][0] \n activation_83[0][0] \n__________________________________________________________________________________________________\nactivation_84 (Activation) (None, 3, 3, 192) 0 batch_normalization_84[0][0] \n__________________________________________________________________________________________________\nmixed9 (Concatenate) (None, 3, 3, 2048) 0 activation_76[0][0] \n mixed9_0[0][0] \n concatenate[0][0] \n activation_84[0][0] \n__________________________________________________________________________________________________\nconv2d_89 (Conv2D) (None, 3, 3, 448) 917504 mixed9[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_89 (BatchNo (None, 3, 3, 448) 1344 conv2d_89[0][0] \n__________________________________________________________________________________________________\nactivation_89 (Activation) (None, 3, 3, 448) 0 batch_normalization_89[0][0] \n__________________________________________________________________________________________________\nconv2d_86 (Conv2D) (None, 3, 3, 384) 786432 mixed9[0][0] \n__________________________________________________________________________________________________\nconv2d_90 (Conv2D) (None, 3, 3, 384) 1548288 activation_89[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_86 (BatchNo (None, 3, 3, 384) 1152 conv2d_86[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_90 (BatchNo (None, 3, 3, 384) 1152 conv2d_90[0][0] \n__________________________________________________________________________________________________\nactivation_86 (Activation) (None, 3, 3, 384) 0 batch_normalization_86[0][0] \n__________________________________________________________________________________________________\nactivation_90 (Activation) (None, 3, 3, 384) 0 batch_normalization_90[0][0] \n__________________________________________________________________________________________________\nconv2d_87 (Conv2D) (None, 3, 3, 384) 442368 activation_86[0][0] \n__________________________________________________________________________________________________\nconv2d_88 (Conv2D) (None, 3, 3, 384) 442368 activation_86[0][0] \n__________________________________________________________________________________________________\nconv2d_91 (Conv2D) (None, 3, 3, 384) 442368 activation_90[0][0] \n__________________________________________________________________________________________________\nconv2d_92 (Conv2D) (None, 3, 3, 384) 442368 activation_90[0][0] \n__________________________________________________________________________________________________\naverage_pooling2d_8 (AveragePoo (None, 3, 3, 2048) 0 mixed9[0][0] \n__________________________________________________________________________________________________\nconv2d_85 (Conv2D) (None, 3, 3, 320) 655360 mixed9[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_87 (BatchNo (None, 3, 3, 384) 1152 conv2d_87[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_88 (BatchNo (None, 3, 3, 384) 1152 conv2d_88[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_91 (BatchNo (None, 3, 3, 384) 1152 conv2d_91[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_92 (BatchNo (None, 3, 3, 384) 1152 conv2d_92[0][0] \n__________________________________________________________________________________________________\nconv2d_93 (Conv2D) (None, 3, 3, 192) 393216 average_pooling2d_8[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_85 (BatchNo (None, 3, 3, 320) 960 conv2d_85[0][0] \n__________________________________________________________________________________________________\nactivation_87 (Activation) (None, 3, 3, 384) 0 batch_normalization_87[0][0] \n__________________________________________________________________________________________________\nactivation_88 (Activation) (None, 3, 3, 384) 0 batch_normalization_88[0][0] \n__________________________________________________________________________________________________\nactivation_91 (Activation) (None, 3, 3, 384) 0 batch_normalization_91[0][0] \n__________________________________________________________________________________________________\nactivation_92 (Activation) (None, 3, 3, 384) 0 batch_normalization_92[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_93 (BatchNo (None, 3, 3, 192) 576 conv2d_93[0][0] \n__________________________________________________________________________________________________\nactivation_85 (Activation) (None, 3, 3, 320) 0 batch_normalization_85[0][0] \n__________________________________________________________________________________________________\nmixed9_1 (Concatenate) (None, 3, 3, 768) 0 activation_87[0][0] \n activation_88[0][0] \n__________________________________________________________________________________________________\nconcatenate_1 (Concatenate) (None, 3, 3, 768) 0 activation_91[0][0] \n activation_92[0][0] \n__________________________________________________________________________________________________\nactivation_93 (Activation) (None, 3, 3, 192) 0 batch_normalization_93[0][0] \n__________________________________________________________________________________________________\nmixed10 (Concatenate) (None, 3, 3, 2048) 0 activation_85[0][0] \n mixed9_1[0][0] \n concatenate_1[0][0] \n activation_93[0][0] \n==================================================================================================\nTotal params: 21,802,784\nTrainable params: 0\nNon-trainable params: 21,802,784\n__________________________________________________________________________________________________\n"
],
[
"last_layer = pre_trained_model.get_layer('mixed7')\nprint('last layer output shape: ', last_layer.output_shape)\nlast_output = last_layer.output\n\n# Expected Output:\n# ('last layer output shape: ', (None, 7, 7, 768))",
"last layer output shape: (None, 7, 7, 768)\n"
],
[
"# Define a Callback class that stops training once accuracy reaches 99.9%\nclass myCallback(tf.keras.callbacks.Callback):\n def on_epoch_end(self, epoch, logs={}):\n if(logs.get('acc')>0.999):\n print(\"\\nReached 99.9% accuracy so cancelling training!\")\n self.model.stop_training = True",
"_____no_output_____"
],
[
"from tensorflow.keras.optimizers import RMSprop\n\n# Flatten the output layer to 1 dimension\nx = layers.Flatten()(last_output)\n# Add a fully connected layer with 1,024 hidden units and ReLU activation\nx = layers.Dense(1024, activation='relu')(x)\n# Add a dropout rate of 0.2\nx = layers.Dropout(0.2)(x) \n# Add a final sigmoid layer for classification\nx = layers.Dense (1, activation='sigmoid')(x) \n\nmodel = Model( pre_trained_model.input, x) \n\nmodel.compile(optimizer = RMSprop(lr=0.0001), \n loss = 'binary_crossentropy', \n metrics = ['acc'])\n\nmodel.summary()\n\n# Expected output will be large. Last few lines should be:\n\n# mixed7 (Concatenate) (None, 7, 7, 768) 0 activation_248[0][0] \n# activation_251[0][0] \n# activation_256[0][0] \n# activation_257[0][0] \n# __________________________________________________________________________________________________\n# flatten_4 (Flatten) (None, 37632) 0 mixed7[0][0] \n# __________________________________________________________________________________________________\n# dense_8 (Dense) (None, 1024) 38536192 flatten_4[0][0] \n# __________________________________________________________________________________________________\n# dropout_4 (Dropout) (None, 1024) 0 dense_8[0][0] \n# __________________________________________________________________________________________________\n# dense_9 (Dense) (None, 1) 1025 dropout_4[0][0] \n# ==================================================================================================\n# Total params: 47,512,481\n# Trainable params: 38,537,217\n# Non-trainable params: 8,975,264",
"Model: \"model\"\n__________________________________________________________________________________________________\nLayer (type) Output Shape Param # Connected to \n==================================================================================================\ninput_1 (InputLayer) [(None, 150, 150, 3) 0 \n__________________________________________________________________________________________________\nconv2d (Conv2D) (None, 74, 74, 32) 864 input_1[0][0] \n__________________________________________________________________________________________________\nbatch_normalization (BatchNorma (None, 74, 74, 32) 96 conv2d[0][0] \n__________________________________________________________________________________________________\nactivation (Activation) (None, 74, 74, 32) 0 batch_normalization[0][0] \n__________________________________________________________________________________________________\nconv2d_1 (Conv2D) (None, 72, 72, 32) 9216 activation[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_1 (BatchNor (None, 72, 72, 32) 96 conv2d_1[0][0] \n__________________________________________________________________________________________________\nactivation_1 (Activation) (None, 72, 72, 32) 0 batch_normalization_1[0][0] \n__________________________________________________________________________________________________\nconv2d_2 (Conv2D) (None, 72, 72, 64) 18432 activation_1[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_2 (BatchNor (None, 72, 72, 64) 192 conv2d_2[0][0] \n__________________________________________________________________________________________________\nactivation_2 (Activation) (None, 72, 72, 64) 0 batch_normalization_2[0][0] \n__________________________________________________________________________________________________\nmax_pooling2d (MaxPooling2D) (None, 35, 35, 64) 0 activation_2[0][0] \n__________________________________________________________________________________________________\nconv2d_3 (Conv2D) (None, 35, 35, 80) 5120 max_pooling2d[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_3 (BatchNor (None, 35, 35, 80) 240 conv2d_3[0][0] \n__________________________________________________________________________________________________\nactivation_3 (Activation) (None, 35, 35, 80) 0 batch_normalization_3[0][0] \n__________________________________________________________________________________________________\nconv2d_4 (Conv2D) (None, 33, 33, 192) 138240 activation_3[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_4 (BatchNor (None, 33, 33, 192) 576 conv2d_4[0][0] \n__________________________________________________________________________________________________\nactivation_4 (Activation) (None, 33, 33, 192) 0 batch_normalization_4[0][0] \n__________________________________________________________________________________________________\nmax_pooling2d_1 (MaxPooling2D) (None, 16, 16, 192) 0 activation_4[0][0] \n__________________________________________________________________________________________________\nconv2d_8 (Conv2D) (None, 16, 16, 64) 12288 max_pooling2d_1[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_8 (BatchNor (None, 16, 16, 64) 192 conv2d_8[0][0] \n__________________________________________________________________________________________________\nactivation_8 (Activation) (None, 16, 16, 64) 0 batch_normalization_8[0][0] \n__________________________________________________________________________________________________\nconv2d_6 (Conv2D) (None, 16, 16, 48) 9216 max_pooling2d_1[0][0] \n__________________________________________________________________________________________________\nconv2d_9 (Conv2D) (None, 16, 16, 96) 55296 activation_8[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_6 (BatchNor (None, 16, 16, 48) 144 conv2d_6[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_9 (BatchNor (None, 16, 16, 96) 288 conv2d_9[0][0] \n__________________________________________________________________________________________________\nactivation_6 (Activation) (None, 16, 16, 48) 0 batch_normalization_6[0][0] \n__________________________________________________________________________________________________\nactivation_9 (Activation) (None, 16, 16, 96) 0 batch_normalization_9[0][0] \n__________________________________________________________________________________________________\naverage_pooling2d (AveragePooli (None, 16, 16, 192) 0 max_pooling2d_1[0][0] \n__________________________________________________________________________________________________\nconv2d_5 (Conv2D) (None, 16, 16, 64) 12288 max_pooling2d_1[0][0] \n__________________________________________________________________________________________________\nconv2d_7 (Conv2D) (None, 16, 16, 64) 76800 activation_6[0][0] \n__________________________________________________________________________________________________\nconv2d_10 (Conv2D) (None, 16, 16, 96) 82944 activation_9[0][0] \n__________________________________________________________________________________________________\nconv2d_11 (Conv2D) (None, 16, 16, 32) 6144 average_pooling2d[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_5 (BatchNor (None, 16, 16, 64) 192 conv2d_5[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_7 (BatchNor (None, 16, 16, 64) 192 conv2d_7[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_10 (BatchNo (None, 16, 16, 96) 288 conv2d_10[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_11 (BatchNo (None, 16, 16, 32) 96 conv2d_11[0][0] \n__________________________________________________________________________________________________\nactivation_5 (Activation) (None, 16, 16, 64) 0 batch_normalization_5[0][0] \n__________________________________________________________________________________________________\nactivation_7 (Activation) (None, 16, 16, 64) 0 batch_normalization_7[0][0] \n__________________________________________________________________________________________________\nactivation_10 (Activation) (None, 16, 16, 96) 0 batch_normalization_10[0][0] \n__________________________________________________________________________________________________\nactivation_11 (Activation) (None, 16, 16, 32) 0 batch_normalization_11[0][0] \n__________________________________________________________________________________________________\nmixed0 (Concatenate) (None, 16, 16, 256) 0 activation_5[0][0] \n activation_7[0][0] \n activation_10[0][0] \n activation_11[0][0] \n__________________________________________________________________________________________________\nconv2d_15 (Conv2D) (None, 16, 16, 64) 16384 mixed0[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_15 (BatchNo (None, 16, 16, 64) 192 conv2d_15[0][0] \n__________________________________________________________________________________________________\nactivation_15 (Activation) (None, 16, 16, 64) 0 batch_normalization_15[0][0] \n__________________________________________________________________________________________________\nconv2d_13 (Conv2D) (None, 16, 16, 48) 12288 mixed0[0][0] \n__________________________________________________________________________________________________\nconv2d_16 (Conv2D) (None, 16, 16, 96) 55296 activation_15[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_13 (BatchNo (None, 16, 16, 48) 144 conv2d_13[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_16 (BatchNo (None, 16, 16, 96) 288 conv2d_16[0][0] \n__________________________________________________________________________________________________\nactivation_13 (Activation) (None, 16, 16, 48) 0 batch_normalization_13[0][0] \n__________________________________________________________________________________________________\nactivation_16 (Activation) (None, 16, 16, 96) 0 batch_normalization_16[0][0] \n__________________________________________________________________________________________________\naverage_pooling2d_1 (AveragePoo (None, 16, 16, 256) 0 mixed0[0][0] \n__________________________________________________________________________________________________\nconv2d_12 (Conv2D) (None, 16, 16, 64) 16384 mixed0[0][0] \n__________________________________________________________________________________________________\nconv2d_14 (Conv2D) (None, 16, 16, 64) 76800 activation_13[0][0] \n__________________________________________________________________________________________________\nconv2d_17 (Conv2D) (None, 16, 16, 96) 82944 activation_16[0][0] \n__________________________________________________________________________________________________\nconv2d_18 (Conv2D) (None, 16, 16, 64) 16384 average_pooling2d_1[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_12 (BatchNo (None, 16, 16, 64) 192 conv2d_12[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_14 (BatchNo (None, 16, 16, 64) 192 conv2d_14[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_17 (BatchNo (None, 16, 16, 96) 288 conv2d_17[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_18 (BatchNo (None, 16, 16, 64) 192 conv2d_18[0][0] \n__________________________________________________________________________________________________\nactivation_12 (Activation) (None, 16, 16, 64) 0 batch_normalization_12[0][0] \n__________________________________________________________________________________________________\nactivation_14 (Activation) (None, 16, 16, 64) 0 batch_normalization_14[0][0] \n__________________________________________________________________________________________________\nactivation_17 (Activation) (None, 16, 16, 96) 0 batch_normalization_17[0][0] \n__________________________________________________________________________________________________\nactivation_18 (Activation) (None, 16, 16, 64) 0 batch_normalization_18[0][0] \n__________________________________________________________________________________________________\nmixed1 (Concatenate) (None, 16, 16, 288) 0 activation_12[0][0] \n activation_14[0][0] \n activation_17[0][0] \n activation_18[0][0] \n__________________________________________________________________________________________________\nconv2d_22 (Conv2D) (None, 16, 16, 64) 18432 mixed1[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_22 (BatchNo (None, 16, 16, 64) 192 conv2d_22[0][0] \n__________________________________________________________________________________________________\nactivation_22 (Activation) (None, 16, 16, 64) 0 batch_normalization_22[0][0] \n__________________________________________________________________________________________________\nconv2d_20 (Conv2D) (None, 16, 16, 48) 13824 mixed1[0][0] \n__________________________________________________________________________________________________\nconv2d_23 (Conv2D) (None, 16, 16, 96) 55296 activation_22[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_20 (BatchNo (None, 16, 16, 48) 144 conv2d_20[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_23 (BatchNo (None, 16, 16, 96) 288 conv2d_23[0][0] \n__________________________________________________________________________________________________\nactivation_20 (Activation) (None, 16, 16, 48) 0 batch_normalization_20[0][0] \n__________________________________________________________________________________________________\nactivation_23 (Activation) (None, 16, 16, 96) 0 batch_normalization_23[0][0] \n__________________________________________________________________________________________________\naverage_pooling2d_2 (AveragePoo (None, 16, 16, 288) 0 mixed1[0][0] \n__________________________________________________________________________________________________\nconv2d_19 (Conv2D) (None, 16, 16, 64) 18432 mixed1[0][0] \n__________________________________________________________________________________________________\nconv2d_21 (Conv2D) (None, 16, 16, 64) 76800 activation_20[0][0] \n__________________________________________________________________________________________________\nconv2d_24 (Conv2D) (None, 16, 16, 96) 82944 activation_23[0][0] \n__________________________________________________________________________________________________\nconv2d_25 (Conv2D) (None, 16, 16, 64) 18432 average_pooling2d_2[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_19 (BatchNo (None, 16, 16, 64) 192 conv2d_19[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_21 (BatchNo (None, 16, 16, 64) 192 conv2d_21[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_24 (BatchNo (None, 16, 16, 96) 288 conv2d_24[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_25 (BatchNo (None, 16, 16, 64) 192 conv2d_25[0][0] \n__________________________________________________________________________________________________\nactivation_19 (Activation) (None, 16, 16, 64) 0 batch_normalization_19[0][0] \n__________________________________________________________________________________________________\nactivation_21 (Activation) (None, 16, 16, 64) 0 batch_normalization_21[0][0] \n__________________________________________________________________________________________________\nactivation_24 (Activation) (None, 16, 16, 96) 0 batch_normalization_24[0][0] \n__________________________________________________________________________________________________\nactivation_25 (Activation) (None, 16, 16, 64) 0 batch_normalization_25[0][0] \n__________________________________________________________________________________________________\nmixed2 (Concatenate) (None, 16, 16, 288) 0 activation_19[0][0] \n activation_21[0][0] \n activation_24[0][0] \n activation_25[0][0] \n__________________________________________________________________________________________________\nconv2d_27 (Conv2D) (None, 16, 16, 64) 18432 mixed2[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_27 (BatchNo (None, 16, 16, 64) 192 conv2d_27[0][0] \n__________________________________________________________________________________________________\nactivation_27 (Activation) (None, 16, 16, 64) 0 batch_normalization_27[0][0] \n__________________________________________________________________________________________________\nconv2d_28 (Conv2D) (None, 16, 16, 96) 55296 activation_27[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_28 (BatchNo (None, 16, 16, 96) 288 conv2d_28[0][0] \n__________________________________________________________________________________________________\nactivation_28 (Activation) (None, 16, 16, 96) 0 batch_normalization_28[0][0] \n__________________________________________________________________________________________________\nconv2d_26 (Conv2D) (None, 7, 7, 384) 995328 mixed2[0][0] \n__________________________________________________________________________________________________\nconv2d_29 (Conv2D) (None, 7, 7, 96) 82944 activation_28[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_26 (BatchNo (None, 7, 7, 384) 1152 conv2d_26[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_29 (BatchNo (None, 7, 7, 96) 288 conv2d_29[0][0] \n__________________________________________________________________________________________________\nactivation_26 (Activation) (None, 7, 7, 384) 0 batch_normalization_26[0][0] \n__________________________________________________________________________________________________\nactivation_29 (Activation) (None, 7, 7, 96) 0 batch_normalization_29[0][0] \n__________________________________________________________________________________________________\nmax_pooling2d_2 (MaxPooling2D) (None, 7, 7, 288) 0 mixed2[0][0] \n__________________________________________________________________________________________________\nmixed3 (Concatenate) (None, 7, 7, 768) 0 activation_26[0][0] \n activation_29[0][0] \n max_pooling2d_2[0][0] \n__________________________________________________________________________________________________\nconv2d_34 (Conv2D) (None, 7, 7, 128) 98304 mixed3[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_34 (BatchNo (None, 7, 7, 128) 384 conv2d_34[0][0] \n__________________________________________________________________________________________________\nactivation_34 (Activation) (None, 7, 7, 128) 0 batch_normalization_34[0][0] \n__________________________________________________________________________________________________\nconv2d_35 (Conv2D) (None, 7, 7, 128) 114688 activation_34[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_35 (BatchNo (None, 7, 7, 128) 384 conv2d_35[0][0] \n__________________________________________________________________________________________________\nactivation_35 (Activation) (None, 7, 7, 128) 0 batch_normalization_35[0][0] \n__________________________________________________________________________________________________\nconv2d_31 (Conv2D) (None, 7, 7, 128) 98304 mixed3[0][0] \n__________________________________________________________________________________________________\nconv2d_36 (Conv2D) (None, 7, 7, 128) 114688 activation_35[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_31 (BatchNo (None, 7, 7, 128) 384 conv2d_31[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_36 (BatchNo (None, 7, 7, 128) 384 conv2d_36[0][0] \n__________________________________________________________________________________________________\nactivation_31 (Activation) (None, 7, 7, 128) 0 batch_normalization_31[0][0] \n__________________________________________________________________________________________________\nactivation_36 (Activation) (None, 7, 7, 128) 0 batch_normalization_36[0][0] \n__________________________________________________________________________________________________\nconv2d_32 (Conv2D) (None, 7, 7, 128) 114688 activation_31[0][0] \n__________________________________________________________________________________________________\nconv2d_37 (Conv2D) (None, 7, 7, 128) 114688 activation_36[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_32 (BatchNo (None, 7, 7, 128) 384 conv2d_32[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_37 (BatchNo (None, 7, 7, 128) 384 conv2d_37[0][0] \n__________________________________________________________________________________________________\nactivation_32 (Activation) (None, 7, 7, 128) 0 batch_normalization_32[0][0] \n__________________________________________________________________________________________________\nactivation_37 (Activation) (None, 7, 7, 128) 0 batch_normalization_37[0][0] \n__________________________________________________________________________________________________\naverage_pooling2d_3 (AveragePoo (None, 7, 7, 768) 0 mixed3[0][0] \n__________________________________________________________________________________________________\nconv2d_30 (Conv2D) (None, 7, 7, 192) 147456 mixed3[0][0] \n__________________________________________________________________________________________________\nconv2d_33 (Conv2D) (None, 7, 7, 192) 172032 activation_32[0][0] \n__________________________________________________________________________________________________\nconv2d_38 (Conv2D) (None, 7, 7, 192) 172032 activation_37[0][0] \n__________________________________________________________________________________________________\nconv2d_39 (Conv2D) (None, 7, 7, 192) 147456 average_pooling2d_3[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_30 (BatchNo (None, 7, 7, 192) 576 conv2d_30[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_33 (BatchNo (None, 7, 7, 192) 576 conv2d_33[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_38 (BatchNo (None, 7, 7, 192) 576 conv2d_38[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_39 (BatchNo (None, 7, 7, 192) 576 conv2d_39[0][0] \n__________________________________________________________________________________________________\nactivation_30 (Activation) (None, 7, 7, 192) 0 batch_normalization_30[0][0] \n__________________________________________________________________________________________________\nactivation_33 (Activation) (None, 7, 7, 192) 0 batch_normalization_33[0][0] \n__________________________________________________________________________________________________\nactivation_38 (Activation) (None, 7, 7, 192) 0 batch_normalization_38[0][0] \n__________________________________________________________________________________________________\nactivation_39 (Activation) (None, 7, 7, 192) 0 batch_normalization_39[0][0] \n__________________________________________________________________________________________________\nmixed4 (Concatenate) (None, 7, 7, 768) 0 activation_30[0][0] \n activation_33[0][0] \n activation_38[0][0] \n activation_39[0][0] \n__________________________________________________________________________________________________\nconv2d_44 (Conv2D) (None, 7, 7, 160) 122880 mixed4[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_44 (BatchNo (None, 7, 7, 160) 480 conv2d_44[0][0] \n__________________________________________________________________________________________________\nactivation_44 (Activation) (None, 7, 7, 160) 0 batch_normalization_44[0][0] \n__________________________________________________________________________________________________\nconv2d_45 (Conv2D) (None, 7, 7, 160) 179200 activation_44[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_45 (BatchNo (None, 7, 7, 160) 480 conv2d_45[0][0] \n__________________________________________________________________________________________________\nactivation_45 (Activation) (None, 7, 7, 160) 0 batch_normalization_45[0][0] \n__________________________________________________________________________________________________\nconv2d_41 (Conv2D) (None, 7, 7, 160) 122880 mixed4[0][0] \n__________________________________________________________________________________________________\nconv2d_46 (Conv2D) (None, 7, 7, 160) 179200 activation_45[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_41 (BatchNo (None, 7, 7, 160) 480 conv2d_41[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_46 (BatchNo (None, 7, 7, 160) 480 conv2d_46[0][0] \n__________________________________________________________________________________________________\nactivation_41 (Activation) (None, 7, 7, 160) 0 batch_normalization_41[0][0] \n__________________________________________________________________________________________________\nactivation_46 (Activation) (None, 7, 7, 160) 0 batch_normalization_46[0][0] \n__________________________________________________________________________________________________\nconv2d_42 (Conv2D) (None, 7, 7, 160) 179200 activation_41[0][0] \n__________________________________________________________________________________________________\nconv2d_47 (Conv2D) (None, 7, 7, 160) 179200 activation_46[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_42 (BatchNo (None, 7, 7, 160) 480 conv2d_42[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_47 (BatchNo (None, 7, 7, 160) 480 conv2d_47[0][0] \n__________________________________________________________________________________________________\nactivation_42 (Activation) (None, 7, 7, 160) 0 batch_normalization_42[0][0] \n__________________________________________________________________________________________________\nactivation_47 (Activation) (None, 7, 7, 160) 0 batch_normalization_47[0][0] \n__________________________________________________________________________________________________\naverage_pooling2d_4 (AveragePoo (None, 7, 7, 768) 0 mixed4[0][0] \n__________________________________________________________________________________________________\nconv2d_40 (Conv2D) (None, 7, 7, 192) 147456 mixed4[0][0] \n__________________________________________________________________________________________________\nconv2d_43 (Conv2D) (None, 7, 7, 192) 215040 activation_42[0][0] \n__________________________________________________________________________________________________\nconv2d_48 (Conv2D) (None, 7, 7, 192) 215040 activation_47[0][0] \n__________________________________________________________________________________________________\nconv2d_49 (Conv2D) (None, 7, 7, 192) 147456 average_pooling2d_4[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_40 (BatchNo (None, 7, 7, 192) 576 conv2d_40[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_43 (BatchNo (None, 7, 7, 192) 576 conv2d_43[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_48 (BatchNo (None, 7, 7, 192) 576 conv2d_48[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_49 (BatchNo (None, 7, 7, 192) 576 conv2d_49[0][0] \n__________________________________________________________________________________________________\nactivation_40 (Activation) (None, 7, 7, 192) 0 batch_normalization_40[0][0] \n__________________________________________________________________________________________________\nactivation_43 (Activation) (None, 7, 7, 192) 0 batch_normalization_43[0][0] \n__________________________________________________________________________________________________\nactivation_48 (Activation) (None, 7, 7, 192) 0 batch_normalization_48[0][0] \n__________________________________________________________________________________________________\nactivation_49 (Activation) (None, 7, 7, 192) 0 batch_normalization_49[0][0] \n__________________________________________________________________________________________________\nmixed5 (Concatenate) (None, 7, 7, 768) 0 activation_40[0][0] \n activation_43[0][0] \n activation_48[0][0] \n activation_49[0][0] \n__________________________________________________________________________________________________\nconv2d_54 (Conv2D) (None, 7, 7, 160) 122880 mixed5[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_54 (BatchNo (None, 7, 7, 160) 480 conv2d_54[0][0] \n__________________________________________________________________________________________________\nactivation_54 (Activation) (None, 7, 7, 160) 0 batch_normalization_54[0][0] \n__________________________________________________________________________________________________\nconv2d_55 (Conv2D) (None, 7, 7, 160) 179200 activation_54[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_55 (BatchNo (None, 7, 7, 160) 480 conv2d_55[0][0] \n__________________________________________________________________________________________________\nactivation_55 (Activation) (None, 7, 7, 160) 0 batch_normalization_55[0][0] \n__________________________________________________________________________________________________\nconv2d_51 (Conv2D) (None, 7, 7, 160) 122880 mixed5[0][0] \n__________________________________________________________________________________________________\nconv2d_56 (Conv2D) (None, 7, 7, 160) 179200 activation_55[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_51 (BatchNo (None, 7, 7, 160) 480 conv2d_51[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_56 (BatchNo (None, 7, 7, 160) 480 conv2d_56[0][0] \n__________________________________________________________________________________________________\nactivation_51 (Activation) (None, 7, 7, 160) 0 batch_normalization_51[0][0] \n__________________________________________________________________________________________________\nactivation_56 (Activation) (None, 7, 7, 160) 0 batch_normalization_56[0][0] \n__________________________________________________________________________________________________\nconv2d_52 (Conv2D) (None, 7, 7, 160) 179200 activation_51[0][0] \n__________________________________________________________________________________________________\nconv2d_57 (Conv2D) (None, 7, 7, 160) 179200 activation_56[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_52 (BatchNo (None, 7, 7, 160) 480 conv2d_52[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_57 (BatchNo (None, 7, 7, 160) 480 conv2d_57[0][0] \n__________________________________________________________________________________________________\nactivation_52 (Activation) (None, 7, 7, 160) 0 batch_normalization_52[0][0] \n__________________________________________________________________________________________________\nactivation_57 (Activation) (None, 7, 7, 160) 0 batch_normalization_57[0][0] \n__________________________________________________________________________________________________\naverage_pooling2d_5 (AveragePoo (None, 7, 7, 768) 0 mixed5[0][0] \n__________________________________________________________________________________________________\nconv2d_50 (Conv2D) (None, 7, 7, 192) 147456 mixed5[0][0] \n__________________________________________________________________________________________________\nconv2d_53 (Conv2D) (None, 7, 7, 192) 215040 activation_52[0][0] \n__________________________________________________________________________________________________\nconv2d_58 (Conv2D) (None, 7, 7, 192) 215040 activation_57[0][0] \n__________________________________________________________________________________________________\nconv2d_59 (Conv2D) (None, 7, 7, 192) 147456 average_pooling2d_5[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_50 (BatchNo (None, 7, 7, 192) 576 conv2d_50[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_53 (BatchNo (None, 7, 7, 192) 576 conv2d_53[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_58 (BatchNo (None, 7, 7, 192) 576 conv2d_58[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_59 (BatchNo (None, 7, 7, 192) 576 conv2d_59[0][0] \n__________________________________________________________________________________________________\nactivation_50 (Activation) (None, 7, 7, 192) 0 batch_normalization_50[0][0] \n__________________________________________________________________________________________________\nactivation_53 (Activation) (None, 7, 7, 192) 0 batch_normalization_53[0][0] \n__________________________________________________________________________________________________\nactivation_58 (Activation) (None, 7, 7, 192) 0 batch_normalization_58[0][0] \n__________________________________________________________________________________________________\nactivation_59 (Activation) (None, 7, 7, 192) 0 batch_normalization_59[0][0] \n__________________________________________________________________________________________________\nmixed6 (Concatenate) (None, 7, 7, 768) 0 activation_50[0][0] \n activation_53[0][0] \n activation_58[0][0] \n activation_59[0][0] \n__________________________________________________________________________________________________\nconv2d_64 (Conv2D) (None, 7, 7, 192) 147456 mixed6[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_64 (BatchNo (None, 7, 7, 192) 576 conv2d_64[0][0] \n__________________________________________________________________________________________________\nactivation_64 (Activation) (None, 7, 7, 192) 0 batch_normalization_64[0][0] \n__________________________________________________________________________________________________\nconv2d_65 (Conv2D) (None, 7, 7, 192) 258048 activation_64[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_65 (BatchNo (None, 7, 7, 192) 576 conv2d_65[0][0] \n__________________________________________________________________________________________________\nactivation_65 (Activation) (None, 7, 7, 192) 0 batch_normalization_65[0][0] \n__________________________________________________________________________________________________\nconv2d_61 (Conv2D) (None, 7, 7, 192) 147456 mixed6[0][0] \n__________________________________________________________________________________________________\nconv2d_66 (Conv2D) (None, 7, 7, 192) 258048 activation_65[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_61 (BatchNo (None, 7, 7, 192) 576 conv2d_61[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_66 (BatchNo (None, 7, 7, 192) 576 conv2d_66[0][0] \n__________________________________________________________________________________________________\nactivation_61 (Activation) (None, 7, 7, 192) 0 batch_normalization_61[0][0] \n__________________________________________________________________________________________________\nactivation_66 (Activation) (None, 7, 7, 192) 0 batch_normalization_66[0][0] \n__________________________________________________________________________________________________\nconv2d_62 (Conv2D) (None, 7, 7, 192) 258048 activation_61[0][0] \n__________________________________________________________________________________________________\nconv2d_67 (Conv2D) (None, 7, 7, 192) 258048 activation_66[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_62 (BatchNo (None, 7, 7, 192) 576 conv2d_62[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_67 (BatchNo (None, 7, 7, 192) 576 conv2d_67[0][0] \n__________________________________________________________________________________________________\nactivation_62 (Activation) (None, 7, 7, 192) 0 batch_normalization_62[0][0] \n__________________________________________________________________________________________________\nactivation_67 (Activation) (None, 7, 7, 192) 0 batch_normalization_67[0][0] \n__________________________________________________________________________________________________\naverage_pooling2d_6 (AveragePoo (None, 7, 7, 768) 0 mixed6[0][0] \n__________________________________________________________________________________________________\nconv2d_60 (Conv2D) (None, 7, 7, 192) 147456 mixed6[0][0] \n__________________________________________________________________________________________________\nconv2d_63 (Conv2D) (None, 7, 7, 192) 258048 activation_62[0][0] \n__________________________________________________________________________________________________\nconv2d_68 (Conv2D) (None, 7, 7, 192) 258048 activation_67[0][0] \n__________________________________________________________________________________________________\nconv2d_69 (Conv2D) (None, 7, 7, 192) 147456 average_pooling2d_6[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_60 (BatchNo (None, 7, 7, 192) 576 conv2d_60[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_63 (BatchNo (None, 7, 7, 192) 576 conv2d_63[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_68 (BatchNo (None, 7, 7, 192) 576 conv2d_68[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_69 (BatchNo (None, 7, 7, 192) 576 conv2d_69[0][0] \n__________________________________________________________________________________________________\nactivation_60 (Activation) (None, 7, 7, 192) 0 batch_normalization_60[0][0] \n__________________________________________________________________________________________________\nactivation_63 (Activation) (None, 7, 7, 192) 0 batch_normalization_63[0][0] \n__________________________________________________________________________________________________\nactivation_68 (Activation) (None, 7, 7, 192) 0 batch_normalization_68[0][0] \n__________________________________________________________________________________________________\nactivation_69 (Activation) (None, 7, 7, 192) 0 batch_normalization_69[0][0] \n__________________________________________________________________________________________________\nmixed7 (Concatenate) (None, 7, 7, 768) 0 activation_60[0][0] \n activation_63[0][0] \n activation_68[0][0] \n activation_69[0][0] \n__________________________________________________________________________________________________\nflatten (Flatten) (None, 37632) 0 mixed7[0][0] \n__________________________________________________________________________________________________\ndense (Dense) (None, 1024) 38536192 flatten[0][0] \n__________________________________________________________________________________________________\ndropout (Dropout) (None, 1024) 0 dense[0][0] \n__________________________________________________________________________________________________\ndense_1 (Dense) (None, 1) 1025 dropout[0][0] \n==================================================================================================\nTotal params: 47,512,481\nTrainable params: 38,537,217\nNon-trainable params: 8,975,264\n__________________________________________________________________________________________________\n"
],
[
"# Get the Horse or Human dataset\n!wget --no-check-certificate https://storage.googleapis.com/laurencemoroney-blog.appspot.com/horse-or-human.zip -O /tmp/horse-or-human.zip\n\n# Get the Horse or Human Validation dataset\n!wget --no-check-certificate https://storage.googleapis.com/laurencemoroney-blog.appspot.com/validation-horse-or-human.zip -O /tmp/validation-horse-or-human.zip \n \nfrom tensorflow.keras.preprocessing.image import ImageDataGenerator\n\nimport os\nimport zipfile\n\nlocal_zip = '//tmp/horse-or-human.zip'\nzip_ref = zipfile.ZipFile(local_zip, 'r')\nzip_ref.extractall('/tmp/training')\nzip_ref.close()\n\nlocal_zip = '//tmp/validation-horse-or-human.zip'\nzip_ref = zipfile.ZipFile(local_zip, 'r')\nzip_ref.extractall('/tmp/validation')\nzip_ref.close()",
"--2020-05-19 17:28:46-- https://storage.googleapis.com/laurencemoroney-blog.appspot.com/horse-or-human.zip\nResolving storage.googleapis.com (storage.googleapis.com)... 64.233.189.128, 2404:6800:4008:c07::80\nConnecting to storage.googleapis.com (storage.googleapis.com)|64.233.189.128|:443... connected.\nHTTP request sent, awaiting response... 200 OK\nLength: 149574867 (143M) [application/zip]\nSaving to: ‘/tmp/horse-or-human.zip’\n\n/tmp/horse-or-human 100%[===================>] 142.65M 64.1MB/s in 2.2s \n\n2020-05-19 17:28:49 (64.1 MB/s) - ‘/tmp/horse-or-human.zip’ saved [149574867/149574867]\n\n--2020-05-19 17:28:51-- https://storage.googleapis.com/laurencemoroney-blog.appspot.com/validation-horse-or-human.zip\nResolving storage.googleapis.com (storage.googleapis.com)... 64.233.189.128, 2404:6800:4008:c05::80\nConnecting to storage.googleapis.com (storage.googleapis.com)|64.233.189.128|:443... connected.\nHTTP request sent, awaiting response... 200 OK\nLength: 11480187 (11M) [application/zip]\nSaving to: ‘/tmp/validation-horse-or-human.zip’\n\n/tmp/validation-hor 100%[===================>] 10.95M 26.1MB/s in 0.4s \n\n2020-05-19 17:28:52 (26.1 MB/s) - ‘/tmp/validation-horse-or-human.zip’ saved [11480187/11480187]\n\n"
],
[
"train_dir = '/tmp/training'\nvalidation_dir = '/tmp/validation'\n\ntrain_horses_dir = os.path.join(train_dir, 'horses') # Directory with our training horse pictures\ntrain_humans_dir = os.path.join(train_dir, 'humans') # Directory with our training humans pictures\nvalidation_horses_dir = os.path.join(validation_dir, 'horses') # Directory with our validation horse pictures\nvalidation_humans_dir = os.path.join(validation_dir, 'humans')# Directory with our validation humanas pictures\n\ntrain_horses_fnames = os.listdir(train_horses_dir)\ntrain_humans_fnames = os.listdir(train_humans_dir)\nvalidation_horses_fnames = os.listdir(validation_horses_dir)\nvalidation_humans_fnames = os.listdir(validation_humans_dir)\n\nprint(len(train_horses_fnames))\nprint(len(train_humans_fnames))\nprint(len(validation_horses_fnames))\nprint(len(validation_humans_fnames))\n\n# Expected Output:\n# 500\n# 527\n# 128\n# 128",
"500\n527\n128\n128\n"
],
[
"# Define our example directories and files\ntrain_dir = '/tmp/training'\nvalidation_dir = '/tmp/validation'\n\n# Add our data-augmentation parameters to ImageDataGenerator\ntrain_datagen = ImageDataGenerator(rescale = 1./255.,\n rotation_range = 40,\n width_shift_range = 0.2,\n height_shift_range = 0.2,\n shear_range = 0.2,\n zoom_range = 0.2,\n horizontal_flip = True)\n\n# Note that the validation data should not be augmented!\ntest_datagen = ImageDataGenerator( rescale = 1.0/255. )\n\n# Flow training images in batches of 20 using train_datagen generator\ntrain_generator = train_datagen.flow_from_directory(train_dir,\n batch_size = 100,\n class_mode = 'binary', \n target_size = (150, 150)) \n\n# Flow validation images in batches of 20 using test_datagen generator\nvalidation_generator = test_datagen.flow_from_directory( validation_dir,\n batch_size = 100,\n class_mode = 'binary', \n target_size = (150, 150))\n\n# Expected Output:\n# Found 1027 images belonging to 2 classes.\n# Found 256 images belonging to 2 classes.",
"Found 1027 images belonging to 2 classes.\nFound 256 images belonging to 2 classes.\n"
],
[
"# Run this and see how many epochs it should take before the callback\n# fires, and stops training at 99.9% accuracy\n# (It should take less than 100 epochs)\ncallbacks = myCallback()\nhistory = model.fit_generator(\n train_generator,\n validation_data = validation_generator,\n steps_per_epoch = 10,\n epochs = 10,\n validation_steps = 10,\n verbose = 2,\n callbacks=[callbacks])",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt\nacc = history.history['acc']\nval_acc = history.history['val_acc']\nloss = history.history['loss']\nval_loss = history.history['val_loss']\n\nepochs = range(len(acc))\n\nplt.plot(epochs, acc, 'r', label='Training accuracy')\nplt.plot(epochs, val_acc, 'b', label='Validation accuracy')\nplt.title('Training and validation accuracy')\nplt.legend(loc=0)\nplt.figure()\n\n\nplt.show()",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
c511df1135b29ff5427f552e0cb04b50060bc993
| 11,821 |
ipynb
|
Jupyter Notebook
|
Hands-on lab/notebooks/02 Deploy Summarizer Web Service.ipynb
|
Rodrigossz/MCW-Cognitive-services-and-deep-learning
|
6705499d0b91d23cd451a3152bc4bd91b07b79f0
|
[
"MIT"
] | 1 |
2019-12-03T04:32:36.000Z
|
2019-12-03T04:32:36.000Z
|
Hands-on lab/notebooks/02 Deploy Summarizer Web Service.ipynb
|
pwaila/MCW-Cognitive-services-and-deep-learning
|
315df8de84b4d25def9ff313789dffccfb193768
|
[
"MIT"
] | null | null | null |
Hands-on lab/notebooks/02 Deploy Summarizer Web Service.ipynb
|
pwaila/MCW-Cognitive-services-and-deep-learning
|
315df8de84b4d25def9ff313789dffccfb193768
|
[
"MIT"
] | null | null | null | 61.248705 | 1,201 | 0.649691 |
[
[
[
"# Azure Machine Learning Setup\nTo begin, you will need to provide the following information about your Azure Subscription.\n\n**If you are using your own Azure subscription, please provide names for subscription_id, resource_group, workspace_name and workspace_region to use.** Note that the workspace needs to be of type [Machine Learning Workspace](https://docs.microsoft.com/en-us/azure/machine-learning/service/setup-create-workspace).\n\n**If an enviorment is provided to you be sure to replace XXXXX in the values below with your unique identifier.**\n\nIn the following cell, be sure to set the values for `subscription_id`, `resource_group`, `workspace_name` and `workspace_region` as directed by the comments (*these values can be acquired from the Azure Portal*).\n\nTo get these values, do the following:\n1. Navigate to the Azure Portal and login with the credentials provided.\n2. From the left hand menu, under Favorites, select `Resource Groups`.\n3. In the list, select the resource group with the name similar to `XXXXX`.\n4. From the Overview tab, capture the desired values.\n\nExecute the following cell by selecting the `>|Run` button in the command bar above.",
"_____no_output_____"
]
],
[
[
"#Provide the Subscription ID of your existing Azure subscription\nsubscription_id = \"\" #\"<your-azure-subscription-id>\"\n\n#Provide a name for the Resource Group that will contain Azure ML related services \nresource_group = \"mcw-ai-lab-XXXXX\" #\"<your-subscription-group-name>\"\n\n# Provide the name and region for the Azure Machine Learning Workspace that will be created\nworkspace_name = \"mcw-ai-lab-ws-XXXXX\"\nworkspace_region = \"eastus\" # eastus2, eastus, westcentralus, southeastasia, australiaeast, westeurope",
"_____no_output_____"
]
],
[
[
"## Create and connect to an Azure Machine Learning Workspace\n\nThe Azure Machine Learning Python SDK is required for leveraging the experimentation, model management and model deployment capabilities of Azure Machine Learning services. Run the following cell to create a new Azure Machine Learning **Workspace** and save the configuration to disk. The configuration file named `config.json` is saved in a folder named `.azureml`. \n\n**Important Note**: You will be prompted to login in the text that is output below the cell. Be sure to navigate to the URL displayed and enter the code that is provided. Once you have entered the code, return to this notebook and wait for the output to read `Workspace configuration succeeded`.",
"_____no_output_____"
]
],
[
[
"import azureml.core\nprint('azureml.core.VERSION: ', azureml.core.VERSION)\n\n# import the Workspace class and check the azureml SDK version\nfrom azureml.core import Workspace\n\nws = Workspace.create(\n name = workspace_name,\n subscription_id = subscription_id,\n resource_group = resource_group, \n location = workspace_region, \n exist_ok = True)\n\nws.write_config()\nprint('Workspace configuration succeeded')",
"_____no_output_____"
]
],
[
[
"Take a look at the contents of the generated configuration file by running the following cell:",
"_____no_output_____"
]
],
[
[
"!cat .azureml/config.json",
"_____no_output_____"
]
],
[
[
"# Deploy model to Azure Container Instance (ACI)\n\nIn this section, you will deploy a web service that uses Gensim as shown in `01 Summarize` to summarize text. The web service will be hosted in Azure Container Service.",
"_____no_output_____"
],
[
"## Create the scoring web service\n\nWhen deploying models for scoring with Azure Machine Learning services, you need to define the code for a simple web service that will load your model and use it for scoring. By convention this service has two methods init which loads the model and run which scores data using the loaded model.\n\nThis scoring service code will later be deployed inside of a specially prepared Docker container.",
"_____no_output_____"
]
],
[
[
"%%writefile summarizer_service.py\n\nimport re\nimport nltk\nimport unicodedata\nfrom gensim.summarization import summarize, keywords\n\ndef clean_and_parse_document(document):\n if isinstance(document, str):\n document = document\n elif isinstance(document, unicode):\n return unicodedata.normalize('NFKD', document).encode('ascii', 'ignore')\n else:\n raise ValueError(\"Document is not string or unicode.\")\n document = document.strip()\n sentences = nltk.sent_tokenize(document)\n sentences = [sentence.strip() for sentence in sentences]\n return sentences\n\ndef summarize_text(text, summary_ratio=None, word_count=30):\n sentences = clean_and_parse_document(text)\n cleaned_text = ' '.join(sentences)\n summary = summarize(cleaned_text, split=True, ratio=summary_ratio, word_count=word_count)\n return summary \n\ndef init(): \n nltk.download('all')\n return\n\ndef run(input_str):\n try:\n return summarize_text(input_str)\n except Exception as e:\n return (str(e))",
"_____no_output_____"
]
],
[
[
"## Create a Conda dependencies environment file\n\nYour web service can have dependencies installed by using a Conda environment file. Items listed in this file will be conda or pip installed within the Docker container that is created and thus be available to your scoring web service logic.",
"_____no_output_____"
]
],
[
[
"from azureml.core.conda_dependencies import CondaDependencies \n\nmyacienv = CondaDependencies.create(pip_packages=['gensim','nltk'])\n\nwith open(\"mydeployenv.yml\",\"w\") as f:\n f.write(myacienv.serialize_to_string())",
"_____no_output_____"
]
],
[
[
"## Deployment\n\nIn the following cells you will use the Azure Machine Learning SDK to package the model and scoring script in a container, and deploy that container to an Azure Container Instance.\n\nRun the following cells.",
"_____no_output_____"
]
],
[
[
"from azureml.core.webservice import AciWebservice, Webservice\n\naci_config = AciWebservice.deploy_configuration(\n cpu_cores = 1, \n memory_gb = 1, \n tags = {'name':'Summarization'}, \n description = 'Summarizes text.')",
"_____no_output_____"
]
],
[
[
"Next, build up a container image configuration that names the scoring service script, the runtime, and provides the conda file.",
"_____no_output_____"
]
],
[
[
"service_name = \"summarizer\"\nruntime = \"python\"\ndriver_file = \"summarizer_service.py\"\nconda_file = \"mydeployenv.yml\"\n\nfrom azureml.core.image import ContainerImage\n\nimage_config = ContainerImage.image_configuration(execution_script = driver_file,\n runtime = runtime,\n conda_file = conda_file)",
"_____no_output_____"
]
],
[
[
"Now you are ready to begin your deployment to the Azure Container Instance.\n\nRun the following cell. This may take between 5-15 minutes to complete.\n\nYou will see output similar to the following when your web service is ready: `SucceededACI service creation operation finished, operation \"Succeeded\"`",
"_____no_output_____"
]
],
[
[
"webservice = Webservice.deploy(\n workspace=ws, \n name=service_name, \n model_paths=[],\n deployment_config=aci_config,\n image_config=image_config, \n )\n\nwebservice.wait_for_deployment(show_output=True)",
"_____no_output_____"
]
],
[
[
"## Test the deployed service\n\nNow you are ready to test scoring using the deployed web service. The following cell invokes the web service.\n\nRun the following cells to test scoring using a single input row against the deployed web service.",
"_____no_output_____"
]
],
[
[
"example_document = \"\"\"\nI was driving down El Camino and stopped at a red light.\nIt was about 3pm in the afternoon. \nThe sun was bright and shining just behind the stoplight.\nThis made it hard to see the lights.\nThere was a car on my left in the left turn lane.\nA few moments later another car, a black sedan pulled up behind me. \nWhen the left turn light changed green, the black sedan hit me thinking \nthat the light had changed for us, but I had not moved because the light \nwas still red.\nAfter hitting my car, the black sedan backed up and then sped past me.\nI did manage to catch its license plate. \nThe license plate of the black sedan was ABC123. \n\"\"\"",
"_____no_output_____"
],
[
"result = webservice.run(input_data = example_document)\nprint(result)",
"_____no_output_____"
]
],
[
[
"## Capture the scoring URI\n\nIn order to call the service from a REST client, you need to acquire the scoring URI. Run the following cell to retrieve the scoring URI and take note of this value, you will need it in the last notebook.",
"_____no_output_____"
]
],
[
[
"webservice.scoring_uri",
"_____no_output_____"
]
],
[
[
"The default settings used in deploying this service result in a service that does not require authentication, so the scoring URI is the only value you need to call this service.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
c511e0be343a0b49bf5ff320fafa206fe03e0cb0
| 16,446 |
ipynb
|
Jupyter Notebook
|
Assignment2_DelosReyes.ipynb
|
jereyel/LinearAlgebra
|
1280b98f316f433df3ce9ad3754690e93658eac8
|
[
"Apache-2.0"
] | null | null | null |
Assignment2_DelosReyes.ipynb
|
jereyel/LinearAlgebra
|
1280b98f316f433df3ce9ad3754690e93658eac8
|
[
"Apache-2.0"
] | null | null | null |
Assignment2_DelosReyes.ipynb
|
jereyel/LinearAlgebra
|
1280b98f316f433df3ce9ad3754690e93658eac8
|
[
"Apache-2.0"
] | null | null | null | 20.455224 | 240 | 0.388909 |
[
[
[
"<a href=\"https://colab.research.google.com/github/jereyel/LinearAlgebra/blob/main/Assignment2_DelosReyes.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# Welcome to Python Fundamentals\nIn this module, we are going to establish our skills in Python Programming. In this notebook we are going to cover:\n\n\n\n\n* Variables and Data Types\n* Operations\n* Input and Output Operations\n* Iterables\n* Functions\n\n",
"_____no_output_____"
],
[
"## Variables and Data Types",
"_____no_output_____"
]
],
[
[
"x = 1\na, b = 3, -2",
"_____no_output_____"
],
[
"type (x)",
"_____no_output_____"
],
[
"y = 3.0\ntype(y)",
"_____no_output_____"
],
[
"x = float(x)\ntype(x)",
"_____no_output_____"
],
[
"s, t, u = \"1\", '3', 'three'\ntype(s)",
"_____no_output_____"
]
],
[
[
"## Operations",
"_____no_output_____"
],
[
"### Arithmetic\n",
"_____no_output_____"
]
],
[
[
"w, x, y, z = 4.0, -3.0, 1, -32",
"_____no_output_____"
],
[
"### Addition\nS = w + x",
"_____no_output_____"
],
[
"### Subtractions\nD = y - z",
"_____no_output_____"
],
[
"### Multiplication\nP = w*z",
"_____no_output_____"
],
[
"### Division\nQ = y/x",
"_____no_output_____"
],
[
"### Floor Division\nQf = w//z\nQf",
"_____no_output_____"
],
[
"### Exponentiation\nE = w**w\nE",
"_____no_output_____"
],
[
"### Modulo\nmod = z%x\nmod",
"_____no_output_____"
]
],
[
[
"### Assignment\n",
"_____no_output_____"
]
],
[
[
"A, B, C, D, E = 0, 100, 2, 1, 2",
"_____no_output_____"
],
[
"A += w",
"_____no_output_____"
],
[
"B -= x",
"_____no_output_____"
],
[
"C *= w",
"_____no_output_____"
],
[
"D /= x\n",
"_____no_output_____"
],
[
"E **= y\nE",
"_____no_output_____"
]
],
[
[
"### Comparators",
"_____no_output_____"
]
],
[
[
"size_1, size_2, size_3 = 1, 2.0, \"1\"\ntrue_size = 1.0",
"_____no_output_____"
],
[
"## Equality\nsize_1 == true_size",
"_____no_output_____"
],
[
"## Non-Equality\nsize_2 != true_size",
"_____no_output_____"
],
[
"## Inequality\ns1 = size_1 > size_2\ns2 = size_1 < size_2/2\ns3 = true_size <= size_1\ns4 = size_2 <= true_size",
"_____no_output_____"
]
],
[
[
"### Logical",
"_____no_output_____"
]
],
[
[
"size_1 == true_size\nsize_1",
"_____no_output_____"
],
[
"size_1 is true_size",
"_____no_output_____"
],
[
"size_1 is not true_size",
"_____no_output_____"
],
[
"P, Q = True, False\nconj = P and Q",
"_____no_output_____"
],
[
"disj = P or Q\ndisj",
"_____no_output_____"
],
[
"nand = not (P and Q)\nnand",
"_____no_output_____"
],
[
"xor = (not P and Q) or (P and not Q)\nxor",
"_____no_output_____"
]
],
[
[
"## Input and Output",
"_____no_output_____"
]
],
[
[
"print(\"Helllo World!\")",
"_____no_output_____"
],
[
"cnt = 14000",
"_____no_output_____"
],
[
"string = \"Hello World!\"\nprint(string, \", Current COVID count is\", cnt)\ncnt += 10000",
"_____no_output_____"
],
[
"print(f\"{string}, current count is: {cnt}\")",
"_____no_output_____"
],
[
"sem_grade = 85.25\nname = \"jerbox\"\nprint(\"Hello {}, your semestral grade is: {}\".format(name, sem_grade))",
"_____no_output_____"
],
[
"pg, mg, fg = 0.3, 0.3, 0.4\nprint(\"The weights of your semestral grade are:\\\n\\n\\t {:.2%} for Prelims\\\n\\n\\t {:.2%} for Midterms, and\\\n\\n\\t {:.2%} for Finals.\".format(pg, mg, fg))\n",
"_____no_output_____"
],
[
"e = input(\"Enter a number: \")",
"_____no_output_____"
],
[
"name = input(\"Enter your name: \");\npg = input(\"Enter prelim grade: \")\nmg = input(\"Enter midterm grade: \")\nfg = input(\"Enter final grade: \")\nsem_grade = None\nprint(\"Hello {}, your semestral grade is: {}\".format(name, sem_grade))",
"_____no_output_____"
]
],
[
[
"### Looping Statements",
"_____no_output_____"
],
[
"## While",
"_____no_output_____"
]
],
[
[
"i, j = 0, 10\nwhile(i<=j):\n print(f\"{i}\\t|\\t{j}\")\n i += 1",
"_____no_output_____"
]
],
[
[
"## For",
"_____no_output_____"
]
],
[
[
"i = 0\nfor i in range(11):\n print(i)",
"0\n1\n2\n3\n4\n5\n6\n7\n8\n9\n10\n"
],
[
"playlist = [\"Bahay Kubo\", \"Magandang Kanta\", \"Kubo\"]\nprint('Now playing:\\n')\nfor song in playlist:\n print(song)",
"Now playing:\n\nBahay Kubo\nMagandang Kanta\nKubo\n"
]
],
[
[
"##Flow Control",
"_____no_output_____"
],
[
"###Condition Statements",
"_____no_output_____"
]
],
[
[
"num_1, num_2 = 14, 12\nif(num_1 == num_2):\n print(\"HAHA\")\nelif(num_1>num_2):\n print(\"HOHO\")\nelse:\n print(\"HUHU\")",
"_____no_output_____"
]
],
[
[
"##Functions",
"_____no_output_____"
]
],
[
[
"# void Deleteuser (int userid){\n# delete(userid);\n# }\n\ndef delete_user (userid):\n print(\"Successfully deleted user: {}\".format(userid))",
"_____no_output_____"
],
[
"addend1, addend2 = 5, 6",
"_____no_output_____"
],
[
"def add(addend1, addend2):\n sum = addend1 + addend2\n return sum",
"_____no_output_____"
],
[
"add(5, 4)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
c511eece7759ef6ff99c8a0151c2216d1855b6ef
| 66,048 |
ipynb
|
Jupyter Notebook
|
[Part 3]Train_license_character_recognition.ipynb
|
kylenannig/Plate_detect_and_recognize
|
015fd7c7f5f1d132d079e548d28355b3cc9eea18
|
[
"MIT"
] | null | null | null |
[Part 3]Train_license_character_recognition.ipynb
|
kylenannig/Plate_detect_and_recognize
|
015fd7c7f5f1d132d079e548d28355b3cc9eea18
|
[
"MIT"
] | 2 |
2020-10-23T19:27:41.000Z
|
2020-10-23T19:32:33.000Z
|
[Part 3]Train_license_character_recognition.ipynb
|
kylenannig/Plate_detect_and_recognize
|
015fd7c7f5f1d132d079e548d28355b3cc9eea18
|
[
"MIT"
] | null | null | null | 114.666667 | 45,052 | 0.836952 |
[
[
[
"#### This notebook is used to train a character recongition from input image using MobileNets ",
"_____no_output_____"
]
],
[
[
"# ignore warning \nimport os\nos.environ['TF_CPP_MIN_LOG_LEVEL'] = '3'\n\nfrom keras.preprocessing.image import ImageDataGenerator\nfrom keras.applications import MobileNetV2\nfrom keras.layers import AveragePooling2D\nfrom keras.layers import Dropout\nfrom keras.layers import Flatten\nfrom keras.layers import Dense\nfrom keras.layers import Input\nfrom keras.models import Model\nfrom keras.optimizers import Adam\nfrom keras.preprocessing.image import img_to_array\nfrom keras.preprocessing.image import load_img\nfrom keras.utils import to_categorical\nfrom keras.callbacks import ModelCheckpoint, EarlyStopping\nfrom keras.models import model_from_json\nfrom sklearn.preprocessing import LabelEncoder\nfrom sklearn.model_selection import train_test_split\nimport matplotlib.pyplot as plt\nimport matplotlib.gridspec as gridspec\nimport glob\nimport numpy as np",
"_____no_output_____"
]
],
[
[
"### Visualize dataset",
"_____no_output_____"
]
],
[
[
"dataset_paths = glob.glob(\"dataset_characters/**/*.jpg\")\n\ncols=4\nrows=3\nfig = plt.figure(figsize=(10,8))\nplt.rcParams.update({\"font.size\":14})\ngrid = gridspec.GridSpec(ncols=cols,nrows=rows,figure=fig)\n\n# create a random list of images will be displayed\nnp.random.seed(45)\nrand = np.random.randint(0,len(dataset_paths),size=(cols*rows))\n\n# Plot image\nfor i in range(cols*rows):\n fig.add_subplot(grid[i])\n image = load_img(dataset_paths[rand[i]])\n label = dataset_paths[rand[i]].split(os.path.sep)[-2]\n plt.title('\"{:s}\"'.format(label))\n plt.axis(False)\n plt.imshow(image)\n\nplt.savefig(\"Visualize_dataset.jpg\",dpi=300) \n",
"_____no_output_____"
]
],
[
[
"## Data pre-processing",
"_____no_output_____"
]
],
[
[
"# Arange input data and corresponding labels\nX=[]\nlabels=[]\n\nfor image_path in dataset_paths:\n label = image_path.split(os.path.sep)[-2]\n image=load_img(image_path,target_size=(80,80))\n image=img_to_array(image)\n\n X.append(image)\n labels.append(label)\n\nX = np.array(X,dtype=\"float16\")\nlabels = np.array(labels)\n\nprint(\"[INFO] Find {:d} images with {:d} classes\".format(len(X),len(set(labels))))\n\n\n# perform one-hot encoding on the labels\nlb = LabelEncoder()\nlb.fit(labels)\nlabels = lb.transform(labels)\ny = to_categorical(labels)\n\n# save label file so we can use in another script\nnp.save('license_character_classes.npy', lb.classes_)",
"_____no_output_____"
],
[
"# split 10% of data as validation set\n(trainX, testX, trainY, testY) = train_test_split(X, y, test_size=0.10, stratify=y, random_state=42)",
"_____no_output_____"
],
[
"# data augumentation\nimage_gen = ImageDataGenerator(rotation_range=10,\n width_shift_range=0.1,\n height_shift_range=0.1,\n shear_range=0.1,\n zoom_range=0.1,\n fill_mode=\"nearest\"\n )",
"_____no_output_____"
]
],
[
[
"## Initialize MobileNets architecture with pre-trained weight",
"_____no_output_____"
]
],
[
[
"# Create our model with pre-trained MobileNetV2 architecture from imagenet\ndef create_model(lr=1e-4,decay=1e-4/25, training=False,output_shape=y.shape[1]):\n baseModel = MobileNetV2(weights=\"imagenet\", \n include_top=False,\n input_tensor=Input(shape=(80, 80, 3)))\n\n headModel = baseModel.output\n headModel = AveragePooling2D(pool_size=(3, 3))(headModel)\n headModel = Flatten(name=\"flatten\")(headModel)\n headModel = Dense(128, activation=\"relu\")(headModel)\n headModel = Dropout(0.5)(headModel)\n headModel = Dense(output_shape, activation=\"softmax\")(headModel)\n \n model = Model(inputs=baseModel.input, outputs=headModel)\n \n if training:\n # define trainable lalyer\n for layer in baseModel.layers:\n layer.trainable = True\n # compile model\n optimizer = Adam(lr=lr, decay = decay)\n model.compile(loss=\"categorical_crossentropy\", optimizer=optimizer,metrics=[\"accuracy\"]) \n \n return model",
"_____no_output_____"
],
[
"# initilaize initial hyperparameter\nINIT_LR = 1e-4\nEPOCHS = 30\n\nmodel = create_model(lr=INIT_LR, decay=INIT_LR/EPOCHS,training=True)",
"/usr/local/lib/python3.6/dist-packages/keras_applications/mobilenet_v2.py:294: UserWarning: `input_shape` is undefined or non-square, or `rows` is not in [96, 128, 160, 192, 224]. Weights for input shape (224, 224) will be loaded as the default.\n warnings.warn('`input_shape` is undefined or non-square, '\n"
]
],
[
[
"## Train model",
"_____no_output_____"
]
],
[
[
"BATCH_SIZE = 64\n\nmy_checkpointer = [\n EarlyStopping(monitor='val_loss', patience=5, verbose=0),\n ModelCheckpoint(filepath=\"License_character_recognition.h5\", verbose=1, save_weights_only=True)\n ]\n\nresult = model.fit(image_gen.flow(trainX, trainY, batch_size=BATCH_SIZE), \n steps_per_epoch=len(trainX) // BATCH_SIZE, \n validation_data=(testX, testY), \n validation_steps=len(testX) // BATCH_SIZE, \n epochs=EPOCHS, callbacks=my_checkpointer)",
"Epoch 1/30\n486/486 [==============================] - 147s 303ms/step - loss: 1.5441 - accuracy: 0.5989 - val_loss: 1.5866 - val_accuracy: 0.6090\n\nEpoch 00001: saving model to License_character_recognition.h5\nEpoch 2/30\n486/486 [==============================] - 124s 256ms/step - loss: 0.4201 - accuracy: 0.8930 - val_loss: 0.4698 - val_accuracy: 0.8788\n\nEpoch 00002: saving model to License_character_recognition.h5\nEpoch 3/30\n486/486 [==============================] - 125s 258ms/step - loss: 0.2769 - accuracy: 0.9281 - val_loss: 0.2617 - val_accuracy: 0.9352\n\nEpoch 00003: saving model to License_character_recognition.h5\nEpoch 4/30\n486/486 [==============================] - 125s 257ms/step - loss: 0.2103 - accuracy: 0.9454 - val_loss: 0.1744 - val_accuracy: 0.9563\n\nEpoch 00004: saving model to License_character_recognition.h5\nEpoch 5/30\n486/486 [==============================] - 126s 259ms/step - loss: 0.1642 - accuracy: 0.9554 - val_loss: 0.1491 - val_accuracy: 0.9592\n\nEpoch 00005: saving model to License_character_recognition.h5\nEpoch 6/30\n486/486 [==============================] - 127s 261ms/step - loss: 0.1429 - accuracy: 0.9610 - val_loss: 0.1245 - val_accuracy: 0.9656\n\nEpoch 00006: saving model to License_character_recognition.h5\nEpoch 7/30\n486/486 [==============================] - 125s 257ms/step - loss: 0.1311 - accuracy: 0.9654 - val_loss: 0.1258 - val_accuracy: 0.9641\n\nEpoch 00007: saving model to License_character_recognition.h5\nEpoch 8/30\n486/486 [==============================] - 125s 256ms/step - loss: 0.1125 - accuracy: 0.9693 - val_loss: 0.1202 - val_accuracy: 0.9688\n\nEpoch 00008: saving model to License_character_recognition.h5\nEpoch 9/30\n486/486 [==============================] - 124s 256ms/step - loss: 0.1024 - accuracy: 0.9730 - val_loss: 0.1038 - val_accuracy: 0.9714\n\nEpoch 00009: saving model to License_character_recognition.h5\nEpoch 10/30\n486/486 [==============================] - 126s 260ms/step - loss: 0.0924 - accuracy: 0.9751 - val_loss: 0.0972 - val_accuracy: 0.9746\n\nEpoch 00010: saving model to License_character_recognition.h5\nEpoch 11/30\n486/486 [==============================] - 126s 259ms/step - loss: 0.0825 - accuracy: 0.9772 - val_loss: 0.0869 - val_accuracy: 0.9751\n\nEpoch 00011: saving model to License_character_recognition.h5\nEpoch 12/30\n486/486 [==============================] - 125s 257ms/step - loss: 0.0821 - accuracy: 0.9782 - val_loss: 0.0872 - val_accuracy: 0.9786\n\nEpoch 00012: saving model to License_character_recognition.h5\nEpoch 13/30\n486/486 [==============================] - 125s 257ms/step - loss: 0.0736 - accuracy: 0.9803 - val_loss: 0.0601 - val_accuracy: 0.9844\n\nEpoch 00013: saving model to License_character_recognition.h5\nEpoch 14/30\n486/486 [==============================] - 125s 256ms/step - loss: 0.0761 - accuracy: 0.9800 - val_loss: 0.0747 - val_accuracy: 0.9800\n\nEpoch 00014: saving model to License_character_recognition.h5\nEpoch 15/30\n486/486 [==============================] - 126s 260ms/step - loss: 0.0631 - accuracy: 0.9829 - val_loss: 0.0598 - val_accuracy: 0.9853\n\nEpoch 00015: saving model to License_character_recognition.h5\nEpoch 16/30\n486/486 [==============================] - 126s 260ms/step - loss: 0.0563 - accuracy: 0.9847 - val_loss: 0.0873 - val_accuracy: 0.9803\n\nEpoch 00016: saving model to License_character_recognition.h5\nEpoch 17/30\n486/486 [==============================] - 125s 258ms/step - loss: 0.0565 - accuracy: 0.9848 - val_loss: 0.0726 - val_accuracy: 0.9832\n\nEpoch 00017: saving model to License_character_recognition.h5\nEpoch 18/30\n486/486 [==============================] - 126s 259ms/step - loss: 0.0534 - accuracy: 0.9851 - val_loss: 0.0762 - val_accuracy: 0.9803\n\nEpoch 00018: saving model to License_character_recognition.h5\nEpoch 19/30\n486/486 [==============================] - 126s 259ms/step - loss: 0.0516 - accuracy: 0.9861 - val_loss: 0.0979 - val_accuracy: 0.9792\n\nEpoch 00019: saving model to License_character_recognition.h5\nEpoch 20/30\n486/486 [==============================] - 127s 261ms/step - loss: 0.0462 - accuracy: 0.9873 - val_loss: 0.0749 - val_accuracy: 0.9847\n\nEpoch 00020: saving model to License_character_recognition.h5\n"
]
],
[
[
"## Visualize training result",
"_____no_output_____"
]
],
[
[
"\nfig = plt.figure(figsize=(14,5))\ngrid=gridspec.GridSpec(ncols=2,nrows=1,figure=fig)\nfig.add_subplot(grid[0])\nplt.plot(result.history['accuracy'], label='training accuracy')\nplt.plot(result.history['val_accuracy'], label='val accuracy')\nplt.title('Accuracy')\nplt.xlabel('epochs')\nplt.ylabel('accuracy')\nplt.legend()\n\nfig.add_subplot(grid[1])\nplt.plot(result.history['loss'], label='training loss')\nplt.plot(result.history['val_loss'], label='val loss')\nplt.title('Loss')\nplt.xlabel('epochs')\nplt.ylabel('loss')\nplt.legend()\n\n#plt.savefig(\"Training_result.jpg\",dpi=300)",
"_____no_output_____"
],
[
"# save model architectur as json file\nmodel_json = model.to_json()\nwith open(\"MobileNets_character_recognition.json\", \"w\") as json_file:\n json_file.write(model_json)",
"_____no_output_____"
],
[
"print(trainX)",
"_____no_output_____"
],
[
"print(testX)",
"_____no_output_____"
]
],
[
[
"## The End!",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
c511f2740446af5d733956bd87b30459fc08cb6d
| 6,259 |
ipynb
|
Jupyter Notebook
|
notebooks/book1/17/gpKernelPlot.ipynb
|
patel-zeel/pyprobml
|
027ef3c13a2a63d958e05fdedb68fd7b8f0e0261
|
[
"MIT"
] | null | null | null |
notebooks/book1/17/gpKernelPlot.ipynb
|
patel-zeel/pyprobml
|
027ef3c13a2a63d958e05fdedb68fd7b8f0e0261
|
[
"MIT"
] | 1 |
2022-03-27T04:59:50.000Z
|
2022-03-27T04:59:50.000Z
|
notebooks/book1/17/gpKernelPlot.ipynb
|
patel-zeel/pyprobml
|
027ef3c13a2a63d958e05fdedb68fd7b8f0e0261
|
[
"MIT"
] | 2 |
2022-03-26T11:52:36.000Z
|
2022-03-27T05:17:48.000Z
| 40.380645 | 121 | 0.511424 |
[
[
[
"empty"
]
]
] |
[
"empty"
] |
[
[
"empty"
]
] |
c512150a9280d5dbcaad20a029ad6ee29a374fe6
| 36,089 |
ipynb
|
Jupyter Notebook
|
Big-Data-Clusters/CU8/Public/content/sample/sam001a-load-sample-data-into-bdc.ipynb
|
meenal-gupta141/tigertoolbox
|
5c432392f7cab091121a8879ea886b39c54f519b
|
[
"MIT"
] | 541 |
2019-05-07T11:41:25.000Z
|
2022-03-29T17:33:19.000Z
|
Big-Data-Clusters/CU8/Public/content/sample/sam001a-load-sample-data-into-bdc.ipynb
|
sqlworldwide/tigertoolbox
|
2abcb62a09daf0116ab1ab9c9dd9317319b23297
|
[
"MIT"
] | 89 |
2019-05-09T14:23:52.000Z
|
2022-01-13T20:21:04.000Z
|
Big-Data-Clusters/CU8/Public/content/sample/sam001a-load-sample-data-into-bdc.ipynb
|
sqlworldwide/tigertoolbox
|
2abcb62a09daf0116ab1ab9c9dd9317319b23297
|
[
"MIT"
] | 338 |
2019-05-08T05:45:16.000Z
|
2022-03-28T15:35:03.000Z
| 60.349498 | 2,915 | 0.447616 |
[
[
[
"SAM001a - Query Storage Pool from SQL Server Master Pool (1 of 3) - Load sample data\n====================================================================================\n\nDescription\n-----------\n\nIn this 3 part tutorial, load data into the Storage Pool (HDFS) using\n`azdata`, convert it into Parquet (using Spark) and the in the 3rd part,\nquery the data using the Master Pool (SQL Server)\n\n### Common functions\n\nDefine helper functions used in this notebook.",
"_____no_output_____"
]
],
[
[
"# Define `run` function for transient fault handling, suggestions on error, and scrolling updates on Windows\nimport sys\nimport os\nimport re\nimport json\nimport platform\nimport shlex\nimport shutil\nimport datetime\n\nfrom subprocess import Popen, PIPE\nfrom IPython.display import Markdown\n\nretry_hints = {} # Output in stderr known to be transient, therefore automatically retry\nerror_hints = {} # Output in stderr where a known SOP/TSG exists which will be HINTed for further help\ninstall_hint = {} # The SOP to help install the executable if it cannot be found\n\nfirst_run = True\nrules = None\ndebug_logging = False\n\ndef run(cmd, return_output=False, no_output=False, retry_count=0, base64_decode=False, return_as_json=False):\n \"\"\"Run shell command, stream stdout, print stderr and optionally return output\n\n NOTES:\n\n 1. Commands that need this kind of ' quoting on Windows e.g.:\n\n kubectl get nodes -o jsonpath={.items[?(@.metadata.annotations.pv-candidate=='data-pool')].metadata.name}\n\n Need to actually pass in as '\"':\n\n kubectl get nodes -o jsonpath={.items[?(@.metadata.annotations.pv-candidate=='\"'data-pool'\"')].metadata.name}\n\n The ' quote approach, although correct when pasting into Windows cmd, will hang at the line:\n \n `iter(p.stdout.readline, b'')`\n\n The shlex.split call does the right thing for each platform, just use the '\"' pattern for a '\n \"\"\"\n MAX_RETRIES = 5\n output = \"\"\n retry = False\n\n global first_run\n global rules\n\n if first_run:\n first_run = False\n rules = load_rules()\n\n # When running `azdata sql query` on Windows, replace any \\n in \"\"\" strings, with \" \", otherwise we see:\n #\n # ('HY090', '[HY090] [Microsoft][ODBC Driver Manager] Invalid string or buffer length (0) (SQLExecDirectW)')\n #\n if platform.system() == \"Windows\" and cmd.startswith(\"azdata sql query\"):\n cmd = cmd.replace(\"\\n\", \" \")\n\n # shlex.split is required on bash and for Windows paths with spaces\n #\n cmd_actual = shlex.split(cmd)\n\n # Store this (i.e. kubectl, python etc.) to support binary context aware error_hints and retries\n #\n user_provided_exe_name = cmd_actual[0].lower()\n\n # When running python, use the python in the ADS sandbox ({sys.executable})\n #\n if cmd.startswith(\"python \"):\n cmd_actual[0] = cmd_actual[0].replace(\"python\", sys.executable)\n\n # On Mac, when ADS is not launched from terminal, LC_ALL may not be set, which causes pip installs to fail\n # with:\n #\n # UnicodeDecodeError: 'ascii' codec can't decode byte 0xc5 in position 4969: ordinal not in range(128)\n #\n # Setting it to a default value of \"en_US.UTF-8\" enables pip install to complete\n #\n if platform.system() == \"Darwin\" and \"LC_ALL\" not in os.environ:\n os.environ[\"LC_ALL\"] = \"en_US.UTF-8\"\n\n # When running `kubectl`, if AZDATA_OPENSHIFT is set, use `oc`\n #\n if cmd.startswith(\"kubectl \") and \"AZDATA_OPENSHIFT\" in os.environ:\n cmd_actual[0] = cmd_actual[0].replace(\"kubectl\", \"oc\")\n\n # To aid supportability, determine which binary file will actually be executed on the machine\n #\n which_binary = None\n\n # Special case for CURL on Windows. The version of CURL in Windows System32 does not work to\n # get JWT tokens, it returns \"(56) Failure when receiving data from the peer\". If another instance\n # of CURL exists on the machine use that one. (Unfortunately the curl.exe in System32 is almost\n # always the first curl.exe in the path, and it can't be uninstalled from System32, so here we\n # look for the 2nd installation of CURL in the path)\n if platform.system() == \"Windows\" and cmd.startswith(\"curl \"):\n path = os.getenv('PATH')\n for p in path.split(os.path.pathsep):\n p = os.path.join(p, \"curl.exe\")\n if os.path.exists(p) and os.access(p, os.X_OK):\n if p.lower().find(\"system32\") == -1:\n cmd_actual[0] = p\n which_binary = p\n break\n\n # Find the path based location (shutil.which) of the executable that will be run (and display it to aid supportability), this\n # seems to be required for .msi installs of azdata.cmd/az.cmd. (otherwise Popen returns FileNotFound) \n #\n # NOTE: Bash needs cmd to be the list of the space separated values hence shlex.split.\n #\n if which_binary == None:\n which_binary = shutil.which(cmd_actual[0])\n\n # Display an install HINT, so the user can click on a SOP to install the missing binary\n #\n if which_binary == None:\n print(f\"The path used to search for '{cmd_actual[0]}' was:\")\n print(sys.path)\n\n if user_provided_exe_name in install_hint and install_hint[user_provided_exe_name] is not None:\n display(Markdown(f'HINT: Use [{install_hint[user_provided_exe_name][0]}]({install_hint[user_provided_exe_name][1]}) to resolve this issue.'))\n\n raise FileNotFoundError(f\"Executable '{cmd_actual[0]}' not found in path (where/which)\")\n else: \n cmd_actual[0] = which_binary\n\n start_time = datetime.datetime.now().replace(microsecond=0)\n\n print(f\"START: {cmd} @ {start_time} ({datetime.datetime.utcnow().replace(microsecond=0)} UTC)\")\n print(f\" using: {which_binary} ({platform.system()} {platform.release()} on {platform.machine()})\")\n print(f\" cwd: {os.getcwd()}\")\n\n # Command-line tools such as CURL and AZDATA HDFS commands output\n # scrolling progress bars, which causes Jupyter to hang forever, to\n # workaround this, use no_output=True\n #\n\n\n # Work around a infinite hang when a notebook generates a non-zero return code, break out, and do not wait\n #\n wait = True \n\n try:\n if no_output:\n p = Popen(cmd_actual)\n else:\n p = Popen(cmd_actual, stdout=PIPE, stderr=PIPE, bufsize=1)\n with p.stdout:\n for line in iter(p.stdout.readline, b''):\n line = line.decode()\n if return_output:\n output = output + line\n else:\n if cmd.startswith(\"azdata notebook run\"): # Hyperlink the .ipynb file\n regex = re.compile(' \"(.*)\"\\: \"(.*)\"') \n match = regex.match(line)\n if match:\n if match.group(1).find(\"HTML\") != -1:\n display(Markdown(f' - \"{match.group(1)}\": \"{match.group(2)}\"'))\n else:\n display(Markdown(f' - \"{match.group(1)}\": \"[{match.group(2)}]({match.group(2)})\"'))\n\n wait = False\n break # otherwise infinite hang, have not worked out why yet.\n else:\n print(line, end='')\n if rules is not None:\n apply_expert_rules(line)\n\n if wait:\n p.wait()\n except FileNotFoundError as e:\n if install_hint is not None:\n display(Markdown(f'HINT: Use {install_hint} to resolve this issue.'))\n\n raise FileNotFoundError(f\"Executable '{cmd_actual[0]}' not found in path (where/which)\") from e\n\n exit_code_workaround = 0 # WORKAROUND: azdata hangs on exception from notebook on p.wait()\n\n if not no_output:\n for line in iter(p.stderr.readline, b''):\n try:\n line_decoded = line.decode()\n except UnicodeDecodeError:\n # NOTE: Sometimes we get characters back that cannot be decoded(), e.g.\n #\n # \\xa0\n #\n # For example see this in the response from `az group create`:\n #\n # ERROR: Get Token request returned http error: 400 and server \n # response: {\"error\":\"invalid_grant\",# \"error_description\":\"AADSTS700082: \n # The refresh token has expired due to inactivity.\\xa0The token was \n # issued on 2018-10-25T23:35:11.9832872Z\n #\n # which generates the exception:\n #\n # UnicodeDecodeError: 'utf-8' codec can't decode byte 0xa0 in position 179: invalid start byte\n #\n print(\"WARNING: Unable to decode stderr line, printing raw bytes:\")\n print(line)\n line_decoded = \"\"\n pass\n else:\n\n # azdata emits a single empty line to stderr when doing an hdfs cp, don't\n # print this empty \"ERR:\" as it confuses.\n #\n if line_decoded == \"\":\n continue\n \n print(f\"STDERR: {line_decoded}\", end='')\n\n if line_decoded.startswith(\"An exception has occurred\") or line_decoded.startswith(\"ERROR: An error occurred while executing the following cell\"):\n exit_code_workaround = 1\n\n # inject HINTs to next TSG/SOP based on output in stderr\n #\n if user_provided_exe_name in error_hints:\n for error_hint in error_hints[user_provided_exe_name]:\n if line_decoded.find(error_hint[0]) != -1:\n display(Markdown(f'HINT: Use [{error_hint[1]}]({error_hint[2]}) to resolve this issue.'))\n\n # apply expert rules (to run follow-on notebooks), based on output\n #\n if rules is not None:\n apply_expert_rules(line_decoded)\n\n # Verify if a transient error, if so automatically retry (recursive)\n #\n if user_provided_exe_name in retry_hints:\n for retry_hint in retry_hints[user_provided_exe_name]:\n if line_decoded.find(retry_hint) != -1:\n if retry_count < MAX_RETRIES:\n print(f\"RETRY: {retry_count} (due to: {retry_hint})\")\n retry_count = retry_count + 1\n output = run(cmd, return_output=return_output, retry_count=retry_count)\n\n if return_output:\n if base64_decode:\n import base64\n\n return base64.b64decode(output).decode('utf-8')\n else:\n return output\n\n elapsed = datetime.datetime.now().replace(microsecond=0) - start_time\n\n # WORKAROUND: We avoid infinite hang above in the `azdata notebook run` failure case, by inferring success (from stdout output), so\n # don't wait here, if success known above\n #\n if wait: \n if p.returncode != 0:\n raise SystemExit(f'Shell command:\\n\\n\\t{cmd} ({elapsed}s elapsed)\\n\\nreturned non-zero exit code: {str(p.returncode)}.\\n')\n else:\n if exit_code_workaround !=0 :\n raise SystemExit(f'Shell command:\\n\\n\\t{cmd} ({elapsed}s elapsed)\\n\\nreturned non-zero exit code: {str(exit_code_workaround)}.\\n')\n\n print(f'\\nSUCCESS: {elapsed}s elapsed.\\n')\n\n if return_output:\n if base64_decode:\n import base64\n\n return base64.b64decode(output).decode('utf-8')\n else:\n return output\n\ndef load_json(filename):\n \"\"\"Load a json file from disk and return the contents\"\"\"\n\n with open(filename, encoding=\"utf8\") as json_file:\n return json.load(json_file)\n\ndef load_rules():\n \"\"\"Load any 'expert rules' from the metadata of this notebook (.ipynb) that should be applied to the stderr of the running executable\"\"\"\n\n # Load this notebook as json to get access to the expert rules in the notebook metadata.\n #\n try:\n j = load_json(\"sam001a-load-sample-data-into-bdc.ipynb\")\n except:\n pass # If the user has renamed the book, we can't load ourself. NOTE: Is there a way in Jupyter, to know your own filename?\n else:\n if \"metadata\" in j and \\\n \"azdata\" in j[\"metadata\"] and \\\n \"expert\" in j[\"metadata\"][\"azdata\"] and \\\n \"expanded_rules\" in j[\"metadata\"][\"azdata\"][\"expert\"]:\n\n rules = j[\"metadata\"][\"azdata\"][\"expert\"][\"expanded_rules\"]\n\n rules.sort() # Sort rules, so they run in priority order (the [0] element). Lowest value first.\n\n # print (f\"EXPERT: There are {len(rules)} rules to evaluate.\")\n\n return rules\n\ndef apply_expert_rules(line):\n \"\"\"Determine if the stderr line passed in, matches the regular expressions for any of the 'expert rules', if so\n inject a 'HINT' to the follow-on SOP/TSG to run\"\"\"\n\n global rules\n\n for rule in rules:\n notebook = rule[1]\n cell_type = rule[2]\n output_type = rule[3] # i.e. stream or error\n output_type_name = rule[4] # i.e. ename or name \n output_type_value = rule[5] # i.e. SystemExit or stdout\n details_name = rule[6] # i.e. evalue or text \n expression = rule[7].replace(\"\\\\*\", \"*\") # Something escaped *, and put a \\ in front of it!\n\n if debug_logging:\n print(f\"EXPERT: If rule '{expression}' satisfied', run '{notebook}'.\")\n\n if re.match(expression, line, re.DOTALL):\n\n if debug_logging:\n print(\"EXPERT: MATCH: name = value: '{0}' = '{1}' matched expression '{2}', therefore HINT '{4}'\".format(output_type_name, output_type_value, expression, notebook))\n\n match_found = True\n\n display(Markdown(f'HINT: Use [{notebook}]({notebook}) to resolve this issue.'))\n\n\n\n\nprint('Common functions defined successfully.')\n\n# Hints for binary (transient fault) retry, (known) error and install guide\n#\nretry_hints = {'kubectl': ['A connection attempt failed because the connected party did not properly respond after a period of time, or established connection failed because connected host has failed to respond'], 'azdata': ['Endpoint sql-server-master does not exist', 'Endpoint livy does not exist', 'Failed to get state for cluster', 'Endpoint webhdfs does not exist', 'Adaptive Server is unavailable or does not exist', 'Error: Address already in use', 'Login timeout expired (0) (SQLDriverConnect)', 'SSPI Provider: No Kerberos credentials available', 'ERROR: No credentials were supplied, or the credentials were unavailable or inaccessible.']}\nerror_hints = {'kubectl': [['no such host', 'TSG010 - Get configuration contexts', '../monitor-k8s/tsg010-get-kubernetes-contexts.ipynb'], ['No connection could be made because the target machine actively refused it', 'TSG056 - Kubectl fails with No connection could be made because the target machine actively refused it', '../repair/tsg056-kubectl-no-connection-could-be-made.ipynb']], 'azdata': [['The token is expired', 'SOP028 - azdata login', '../common/sop028-azdata-login.ipynb'], ['Reason: Unauthorized', 'SOP028 - azdata login', '../common/sop028-azdata-login.ipynb'], ['Max retries exceeded with url: /api/v1/bdc/endpoints', 'SOP028 - azdata login', '../common/sop028-azdata-login.ipynb'], ['Look at the controller logs for more details', 'TSG027 - Observe cluster deployment', '../diagnose/tsg027-observe-bdc-create.ipynb'], ['provided port is already allocated', 'TSG062 - Get tail of all previous container logs for pods in BDC namespace', '../log-files/tsg062-tail-bdc-previous-container-logs.ipynb'], ['Create cluster failed since the existing namespace', 'SOP061 - Delete a big data cluster', '../install/sop061-delete-bdc.ipynb'], ['Failed to complete kube config setup', 'TSG067 - Failed to complete kube config setup', '../repair/tsg067-failed-to-complete-kube-config-setup.ipynb'], ['Error processing command: \"ApiError', 'TSG110 - Azdata returns ApiError', '../repair/tsg110-azdata-returns-apierror.ipynb'], ['Error processing command: \"ControllerError', 'TSG036 - Controller logs', '../log-analyzers/tsg036-get-controller-logs.ipynb'], ['ERROR: 500', 'TSG046 - Knox gateway logs', '../log-analyzers/tsg046-get-knox-logs.ipynb'], ['Data source name not found and no default driver specified', 'SOP069 - Install ODBC for SQL Server', '../install/sop069-install-odbc-driver-for-sql-server.ipynb'], [\"Can't open lib 'ODBC Driver 17 for SQL Server\", 'SOP069 - Install ODBC for SQL Server', '../install/sop069-install-odbc-driver-for-sql-server.ipynb'], ['Control plane upgrade failed. Failed to upgrade controller.', 'TSG108 - View the controller upgrade config map', '../diagnose/tsg108-controller-failed-to-upgrade.ipynb'], [\"[Errno 2] No such file or directory: '..\\\\\\\\\", 'TSG053 - ADS Provided Books must be saved before use', '../repair/tsg053-save-book-first.ipynb'], [\"NameError: name 'azdata_login_secret_name' is not defined\", 'SOP013 - Create secret for azdata login (inside cluster)', '../common/sop013-create-secret-for-azdata-login.ipynb'], ['ERROR: No credentials were supplied, or the credentials were unavailable or inaccessible.', \"TSG124 - 'No credentials were supplied' error from azdata login\", '../repair/tsg124-no-credentials-were-supplied.ipynb'], ['Please accept the license terms to use this product through', \"TSG126 - azdata fails with 'accept the license terms to use this product'\", '../repair/tsg126-accept-license-terms.ipynb']]}\ninstall_hint = {'kubectl': ['SOP036 - Install kubectl command line interface', '../install/sop036-install-kubectl.ipynb'], 'azdata': ['SOP063 - Install azdata CLI (using package manager)', '../install/sop063-packman-install-azdata.ipynb']}",
"_____no_output_____"
]
],
[
[
"### Get the Kubernetes namespace for the big data cluster\n\nGet the namespace of the Big Data Cluster use the kubectl command line\ninterface .\n\n**NOTE:**\n\nIf there is more than one Big Data Cluster in the target Kubernetes\ncluster, then either:\n\n- set \\[0\\] to the correct value for the big data cluster.\n- set the environment variable AZDATA\\_NAMESPACE, before starting\n Azure Data Studio.",
"_____no_output_____"
]
],
[
[
"# Place Kubernetes namespace name for BDC into 'namespace' variable\n\nif \"AZDATA_NAMESPACE\" in os.environ:\n namespace = os.environ[\"AZDATA_NAMESPACE\"]\nelse:\n try:\n namespace = run(f'kubectl get namespace --selector=MSSQL_CLUSTER -o jsonpath={{.items[0].metadata.name}}', return_output=True)\n except:\n from IPython.display import Markdown\n print(f\"ERROR: Unable to find a Kubernetes namespace with label 'MSSQL_CLUSTER'. SQL Server Big Data Cluster Kubernetes namespaces contain the label 'MSSQL_CLUSTER'.\")\n display(Markdown(f'HINT: Use [TSG081 - Get namespaces (Kubernetes)](../monitor-k8s/tsg081-get-kubernetes-namespaces.ipynb) to resolve this issue.'))\n display(Markdown(f'HINT: Use [TSG010 - Get configuration contexts](../monitor-k8s/tsg010-get-kubernetes-contexts.ipynb) to resolve this issue.'))\n display(Markdown(f'HINT: Use [SOP011 - Set kubernetes configuration context](../common/sop011-set-kubernetes-context.ipynb) to resolve this issue.'))\n raise\n\nprint(f'The SQL Server Big Data Cluster Kubernetes namespace is: {namespace}')",
"_____no_output_____"
]
],
[
[
"### Create a temporary directory to stage files",
"_____no_output_____"
]
],
[
[
"# Create a temporary directory to hold configuration files\n\nimport tempfile\n\ntemp_dir = tempfile.mkdtemp()\n\nprint(f\"Temporary directory created: {temp_dir}\")",
"_____no_output_____"
]
],
[
[
"### Helper function to save configuration files to disk",
"_____no_output_____"
]
],
[
[
"# Define helper function 'save_file' to save configuration files to the temporary directory created above\nimport os\nimport io\n\ndef save_file(filename, contents):\n with io.open(os.path.join(temp_dir, filename), \"w\", encoding='utf8', newline='\\n') as text_file:\n text_file.write(contents)\n\n print(\"File saved: \" + os.path.join(temp_dir, filename))\n\nprint(\"Function `save_file` defined successfully.\")",
"_____no_output_____"
]
],
[
[
"### Get the controller username and password\n\nGet the controller username and password from the Kubernetes Secret\nStore and place in the required AZDATA\\_USERNAME and AZDATA\\_PASSWORD\nenvironment variables.",
"_____no_output_____"
]
],
[
[
"# Place controller secret in AZDATA_USERNAME/AZDATA_PASSWORD environment variables\n\nimport os, base64\n\nos.environ[\"AZDATA_USERNAME\"] = run(f'kubectl get secret/controller-login-secret -n {namespace} -o jsonpath={{.data.username}}', return_output=True, base64_decode=True)\nos.environ[\"AZDATA_PASSWORD\"] = run(f'kubectl get secret/controller-login-secret -n {namespace} -o jsonpath={{.data.password}}', return_output=True, base64_decode=True)\n\nprint(f\"Controller username '{os.environ['AZDATA_USERNAME']}' and password stored in environment variables\")",
"_____no_output_____"
]
],
[
[
"### Steps\n\nUpload this data into HDFS.",
"_____no_output_____"
]
],
[
[
"import os\n\nitems = [\n [1, \"Eldon Base for stackable storage shelf platinum\", \"Muhammed MacIntyre\", 3, -213.25, 38.94, 35, \"Nunavut\", \"Storage & Organization\", 0.8],\n [2, \"1.7 Cubic Foot Compact \"\"Cube\"\" Office Refrigerators\", \"Barry French\", 293, 457.81, 208.16, 68.02, \"Nunavut\", \"Appliances\", 0.58], \n [3, \"Cardinal Slant-D Ring Binder Heavy Gauge Vinyl\", \"Barry French\", 293,46.71, 8.69, 2.99, \"Nunavut\", \"Binders and Binder Accessories\", 0.39],\n [4, \"R380\", \"Clay Rozendal\", 483, 1198.97, 195.99, 3.99, \"Nunavut\", \"Telephones and Communication\", 0.58],\n [5, \"Holmes HEPA Air Purifier\", \"Carlos Soltero\", 515, 30.94, 21.78, 5.94, \"Nunavut\", \"Appliances\", 0.5],\n [6, \"G.E. Longer-Life Indoor Recessed Floodlight Bulbs\", \"Carlos Soltero\", 515, 4.43, 6.64, 4.95, \"Nunavut\", \"Office Furnishings\", 0.37],\n [7, \"Angle-D Binders with Locking Rings Label Holders\", \"Carl Jackson\", 613, -54.04, 7.3, 7.72, \"Nunavut\", \"Binders and Binder Accessories\", 0.38],\n [8, \"SAFCO Mobile Desk Side File Wire Frame\", \"Carl Jackson\", 613, 127.7, 42.76, 6.22, \"Nunavut\", \"Storage & Organization\", ],\n [9, \"SAFCO Commercial Wire Shelving Black\", \"Monica Federle\", 643, -695.26, 138.14, 35, \"Nunavut\", \"Storage & Organization\", ],\n [10, \"Xerox 198\", \"Dorothy Badders\", 678, -226.36, 4.98, 8.33, \"Nunavut\", \"Paper\", 0.38]\n]\n\nsrc = os.path.join(temp_dir, \"items.csv\")\ndest = \"/tmp/clickstream_data/datasampleCS.csv\"\n\ns = \"\"\nfor item in items:\n s = s + str(item)[1:-1] + \"\\n\"\n\nsave_file(src, s)\n\nrun(f'azdata bdc hdfs rm --path {dest}')\n\nsrc = src.replace(\"\\\\\", \"\\\\\\\\\")\n\nrun(f'azdata bdc hdfs rm --path hdfs:{dest}')\nrun(f'azdata bdc hdfs cp --from-path {src} --to-path hdfs:{dest}')\n\nprint (f\"CSV uploaded to HDFS: {dest}\")",
"_____no_output_____"
]
],
[
[
"### Clean up temporary directory for staging configuration files",
"_____no_output_____"
]
],
[
[
"# Delete the temporary directory used to hold configuration files\n\nimport shutil\n\nshutil.rmtree(temp_dir)\n\nprint(f'Temporary directory deleted: {temp_dir}')",
"_____no_output_____"
],
[
"print('Notebook execution complete.')",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
c5121e1b1ba9795c9b02f18bcee0019252852e1a
| 7,123 |
ipynb
|
Jupyter Notebook
|
Image/composite_bands.ipynb
|
mllzl/earthengine-py-notebooks
|
cade6a81dd4dbbfb1b9b37aaf6955de42226cfc5
|
[
"MIT"
] | 1 |
2020-03-26T04:21:15.000Z
|
2020-03-26T04:21:15.000Z
|
Image/composite_bands.ipynb
|
mllzl/earthengine-py-notebooks
|
cade6a81dd4dbbfb1b9b37aaf6955de42226cfc5
|
[
"MIT"
] | null | null | null |
Image/composite_bands.ipynb
|
mllzl/earthengine-py-notebooks
|
cade6a81dd4dbbfb1b9b37aaf6955de42226cfc5
|
[
"MIT"
] | null | null | null | 46.555556 | 1,031 | 0.596378 |
[
[
[
"<table class=\"ee-notebook-buttons\" align=\"left\">\n <td><a target=\"_blank\" href=\"https://github.com/giswqs/earthengine-py-notebooks/tree/master/Image/composite_bands.ipynb\"><img width=32px src=\"https://www.tensorflow.org/images/GitHub-Mark-32px.png\" /> View source on GitHub</a></td>\n <td><a target=\"_blank\" href=\"https://nbviewer.jupyter.org/github/giswqs/earthengine-py-notebooks/blob/master/Image/composite_bands.ipynb\"><img width=26px src=\"https://upload.wikimedia.org/wikipedia/commons/thumb/3/38/Jupyter_logo.svg/883px-Jupyter_logo.svg.png\" />Notebook Viewer</a></td>\n <td><a target=\"_blank\" href=\"https://mybinder.org/v2/gh/giswqs/earthengine-py-notebooks/master?filepath=Image/composite_bands.ipynb\"><img width=58px src=\"https://mybinder.org/static/images/logo_social.png\" />Run in binder</a></td>\n <td><a target=\"_blank\" href=\"https://colab.research.google.com/github/giswqs/earthengine-py-notebooks/blob/master/Image/composite_bands.ipynb\"><img src=\"https://www.tensorflow.org/images/colab_logo_32px.png\" /> Run in Google Colab</a></td>\n</table>",
"_____no_output_____"
],
[
"## Install Earth Engine API and geemap\nInstall the [Earth Engine Python API](https://developers.google.com/earth-engine/python_install) and [geemap](https://github.com/giswqs/geemap). The **geemap** Python package is built upon the [ipyleaflet](https://github.com/jupyter-widgets/ipyleaflet) and [folium](https://github.com/python-visualization/folium) packages and implements several methods for interacting with Earth Engine data layers, such as `Map.addLayer()`, `Map.setCenter()`, and `Map.centerObject()`.\nThe following script checks if the geemap package has been installed. If not, it will install geemap, which automatically installs its [dependencies](https://github.com/giswqs/geemap#dependencies), including earthengine-api, folium, and ipyleaflet.\n\n**Important note**: A key difference between folium and ipyleaflet is that ipyleaflet is built upon ipywidgets and allows bidirectional communication between the front-end and the backend enabling the use of the map to capture user input, while folium is meant for displaying static data only ([source](https://blog.jupyter.org/interactive-gis-in-jupyter-with-ipyleaflet-52f9657fa7a)). Note that [Google Colab](https://colab.research.google.com/) currently does not support ipyleaflet ([source](https://github.com/googlecolab/colabtools/issues/60#issuecomment-596225619)). Therefore, if you are using geemap with Google Colab, you should use [`import geemap.eefolium`](https://github.com/giswqs/geemap/blob/master/geemap/eefolium.py). If you are using geemap with [binder](https://mybinder.org/) or a local Jupyter notebook server, you can use [`import geemap`](https://github.com/giswqs/geemap/blob/master/geemap/geemap.py), which provides more functionalities for capturing user input (e.g., mouse-clicking and moving).",
"_____no_output_____"
]
],
[
[
"# Installs geemap package\nimport subprocess\n\ntry:\n import geemap\nexcept ImportError:\n print('geemap package not installed. Installing ...')\n subprocess.check_call([\"python\", '-m', 'pip', 'install', 'geemap'])\n\n# Checks whether this notebook is running on Google Colab\ntry:\n import google.colab\n import geemap.eefolium as emap\nexcept:\n import geemap as emap\n\n# Authenticates and initializes Earth Engine\nimport ee\n\ntry:\n ee.Initialize()\nexcept Exception as e:\n ee.Authenticate()\n ee.Initialize() ",
"_____no_output_____"
]
],
[
[
"## Create an interactive map \nThe default basemap is `Google Satellite`. [Additional basemaps](https://github.com/giswqs/geemap/blob/master/geemap/geemap.py#L13) can be added using the `Map.add_basemap()` function. ",
"_____no_output_____"
]
],
[
[
"Map = emap.Map(center=[40,-100], zoom=4)\nMap.add_basemap('ROADMAP') # Add Google Map\nMap",
"_____no_output_____"
]
],
[
[
"## Add Earth Engine Python script ",
"_____no_output_____"
]
],
[
[
"# Add Earth Engine dataset\n# There are many fine places to look here is one. Comment\n# this out if you want to twiddle knobs while panning around.\nMap.setCenter(-61.61625, -11.64273, 14)\n\n# Grab a sample L7 image and pull out the RGB and pan bands\n# in the range (0, 1). (The range of the pan band values was\n# chosen to roughly match the other bands.)\nimage1 = ee.Image('LANDSAT/LE7/LE72300681999227EDC00')\n\nrgb = image1.select('B3', 'B2', 'B1').unitScale(0, 255)\ngray = image1.select('B8').unitScale(0, 155)\n\n# Convert to HSV, swap in the pan band, and convert back to RGB.\nhuesat = rgb.rgbToHsv().select('hue', 'saturation')\nupres = ee.Image.cat(huesat, gray).hsvToRgb()\n\n# Display before and after layers using the same vis parameters.\nvisparams = {'min': [.15, .15, .25], 'max': [1, .9, .9], 'gamma': 1.6}\nMap.addLayer(rgb, visparams, 'Orignal')\nMap.addLayer(upres, visparams, 'Pansharpened')\n",
"_____no_output_____"
]
],
[
[
"## Display Earth Engine data layers ",
"_____no_output_____"
]
],
[
[
"Map.addLayerControl() # This line is not needed for ipyleaflet-based Map.\nMap",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
c5121fc22a09b97ceccd8565b90c3d434693f23b
| 3,902 |
ipynb
|
Jupyter Notebook
|
python-note/python_notebooks-master/types/real.ipynb
|
ii6uu99/ipynb
|
d924a6926838ca5e563620cd324368a07d2c2521
|
[
"MIT"
] | 1 |
2021-01-27T09:01:33.000Z
|
2021-01-27T09:01:33.000Z
|
python-note/python_notebooks-master/types/real.ipynb
|
ii6uu99/ipynb
|
d924a6926838ca5e563620cd324368a07d2c2521
|
[
"MIT"
] | null | null | null |
python-note/python_notebooks-master/types/real.ipynb
|
ii6uu99/ipynb
|
d924a6926838ca5e563620cd324368a07d2c2521
|
[
"MIT"
] | 1 |
2021-01-26T13:22:21.000Z
|
2021-01-26T13:22:21.000Z
| 20.322917 | 275 | 0.536392 |
[
[
[
"# real",
"_____no_output_____"
],
[
"A **real** is a double-precision floating-point number, not single precision.",
"_____no_output_____"
]
],
[
[
"from numpy import pi\n\nradius = 5.6\narea = pi * (radius * radius)\narea",
"_____no_output_____"
]
],
[
[
"The maximum amount of _reals_ can be calculated: `2 ** 64`",
"_____no_output_____"
]
],
[
[
"pow(2, 64) # '64' indicates exponent",
"_____no_output_____"
]
],
[
[
"The `sys.float_info` function in the `sys` module returns data which indicates how floating point numbers will behave.",
"_____no_output_____"
]
],
[
[
"import sys\n\nsys.float_info",
"_____no_output_____"
]
],
[
[
"Since there is not enough memory available to represent every single floating point decimal value efficiently, nearest representable numbers are approximated. Which leads to approximation issues. For example, the following equation—in reality—should evaluate to `True`.",
"_____no_output_____"
]
],
[
[
"(7 * 0.1) - 0.7 == 0.0",
"_____no_output_____"
]
],
[
[
"Notice the following miscalculation.",
"_____no_output_____"
]
],
[
[
"7 * 0.1",
"_____no_output_____"
]
],
[
[
"To avoid approximation issues, financial calculations should involve [decimal](https://docs.python.org/2/library/decimal.html#module-decimal) type instead of [float](https://docs.python.org/2/library/functions.html#float) type.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
c51223f8c569a3473a2f2ecdd5fb6d63980f9f9f
| 68,749 |
ipynb
|
Jupyter Notebook
|
Maximum_likelihood.ipynb
|
bmswgnp/Python-for-Signal-Processing
|
fa91188f764ef7cb0d8b9f1ccbe26ab23691e4ec
|
[
"CC-BY-3.0"
] | 3 |
2017-05-19T16:28:42.000Z
|
2017-12-21T02:15:06.000Z
|
Maximum_likelihood.ipynb
|
yokoxue/Python-for-Signal-Processing
|
fa91188f764ef7cb0d8b9f1ccbe26ab23691e4ec
|
[
"CC-BY-3.0"
] | null | null | null |
Maximum_likelihood.ipynb
|
yokoxue/Python-for-Signal-Processing
|
fa91188f764ef7cb0d8b9f1ccbe26ab23691e4ec
|
[
"CC-BY-3.0"
] | 1 |
2022-03-22T07:39:21.000Z
|
2022-03-22T07:39:21.000Z
| 133.752918 | 24,392 | 0.827925 |
[
[
[
"empty"
]
]
] |
[
"empty"
] |
[
[
"empty"
]
] |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.