hexsha
stringlengths 40
40
| size
int64 6
14.9M
| ext
stringclasses 1
value | lang
stringclasses 1
value | max_stars_repo_path
stringlengths 6
260
| max_stars_repo_name
stringlengths 6
119
| max_stars_repo_head_hexsha
stringlengths 40
41
| max_stars_repo_licenses
list | max_stars_count
int64 1
191k
⌀ | max_stars_repo_stars_event_min_datetime
stringlengths 24
24
⌀ | max_stars_repo_stars_event_max_datetime
stringlengths 24
24
⌀ | max_issues_repo_path
stringlengths 6
260
| max_issues_repo_name
stringlengths 6
119
| max_issues_repo_head_hexsha
stringlengths 40
41
| max_issues_repo_licenses
list | max_issues_count
int64 1
67k
⌀ | max_issues_repo_issues_event_min_datetime
stringlengths 24
24
⌀ | max_issues_repo_issues_event_max_datetime
stringlengths 24
24
⌀ | max_forks_repo_path
stringlengths 6
260
| max_forks_repo_name
stringlengths 6
119
| max_forks_repo_head_hexsha
stringlengths 40
41
| max_forks_repo_licenses
list | max_forks_count
int64 1
105k
⌀ | max_forks_repo_forks_event_min_datetime
stringlengths 24
24
⌀ | max_forks_repo_forks_event_max_datetime
stringlengths 24
24
⌀ | avg_line_length
float64 2
1.04M
| max_line_length
int64 2
11.2M
| alphanum_fraction
float64 0
1
| cells
list | cell_types
list | cell_type_groups
list |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
c5122cd9f6752a889bff43edc3ea8e61526f06aa
| 5,200 |
ipynb
|
Jupyter Notebook
|
doc/code/CodeForTutor/primer.ipynb
|
runawayhorse001/PythonTipsDS
|
e82a4be4774b56ff487644d2328fe7c6a782faeb
|
[
"MIT"
] | 26 |
2019-02-23T01:09:23.000Z
|
2021-11-25T21:50:27.000Z
|
doc/code/CodeForTutor/primer.ipynb
|
runawayhorse001/PythonTipsDS
|
e82a4be4774b56ff487644d2328fe7c6a782faeb
|
[
"MIT"
] | null | null | null |
doc/code/CodeForTutor/primer.ipynb
|
runawayhorse001/PythonTipsDS
|
e82a4be4774b56ff487644d2328fe7c6a782faeb
|
[
"MIT"
] | 8 |
2019-05-24T02:05:46.000Z
|
2021-11-25T20:52:01.000Z
| 17.04918 | 68 | 0.444038 |
[
[
[
"import random\nimport numpy as np",
"_____no_output_____"
]
],
[
[
"# Primer Functions",
"_____no_output_____"
],
[
"### *",
"_____no_output_____"
]
],
[
[
"my_list = [1,2,3]\nprint(my_list)",
"[1, 2, 3]\n"
],
[
"print(*my_list)",
"1 2 3\n"
]
],
[
[
"### Random ",
"_____no_output_____"
]
],
[
[
"import random\nrandom.random()",
"_____no_output_____"
],
[
"# (b - a) * random_sample() + a\nrandom.uniform(3,8)",
"_____no_output_____"
],
[
"np.random.random_sample()",
"_____no_output_____"
],
[
"np.random.random_sample(4)",
"_____no_output_____"
],
[
"np.random.random_sample([2,4])",
"_____no_output_____"
],
[
"# (b - a) * random_sample() + a\na = 3; b = 8\n(b-a)*np.random.random_sample([2,4])+a",
"_____no_output_____"
]
],
[
[
"### Round",
"_____no_output_____"
]
],
[
[
"np.round(np.random.random_sample([2,4]),2)",
"_____no_output_____"
]
],
[
[
"### range",
"_____no_output_____"
]
],
[
[
"print(range(5))\nprint(*range(5))\nprint(*range(3,8))",
"range(0, 5)\n0 1 2 3 4\n3 4 5 6 7\n"
],
[
"#range([start], stop[, step])\n\nfor i in range(3,8):\n print(i)",
"3\n4\n5\n6\n7\n"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
c51236fc1bee552e8c875f8294304bfe157137ac
| 551,823 |
ipynb
|
Jupyter Notebook
|
examples/webinars_conferences_etc/python_web_conf/NLU_crashcourse_py_web.ipynb
|
UPbook-innovations/nlu
|
2ae02ce7b6ca163f47271e98b71de109d38adefe
|
[
"Apache-2.0"
] | null | null | null |
examples/webinars_conferences_etc/python_web_conf/NLU_crashcourse_py_web.ipynb
|
UPbook-innovations/nlu
|
2ae02ce7b6ca163f47271e98b71de109d38adefe
|
[
"Apache-2.0"
] | 2 |
2021-09-28T05:55:05.000Z
|
2022-02-26T11:16:21.000Z
|
examples/webinars_conferences_etc/python_web_conf/NLU_crashcourse_py_web.ipynb
|
atdavidpark/nlu
|
619d07299e993323d83086c86506db71e2a139a9
|
[
"Apache-2.0"
] | 1 |
2021-09-13T10:06:20.000Z
|
2021-09-13T10:06:20.000Z
| 110.276379 | 73,646 | 0.754262 |
[
[
[
"\n\n[](https://colab.research.google.com/github/JohnSnowLabs/nlu/blob/master/examples/webinars_conferences_etc/python_web_conf/NLU_crashcourse_py_web.ipynb)\n\n\n<div>\n<img src=\"https://2021.pythonwebconf.com/images/pwcgenericlogo-opt2.jpg\" width=\"400\" height=\"250\" >\n</div>\n\n\n\n\n# NLU 20 Minutes Crashcourse - the fast Data Science route\nThis short notebook will teach you a lot of things!\n- Sentiment classification, binary, multi class and regressive\n- Extract Parts of Speech (POS)\n- Extract Named Entities (NER)\n- Extract Keywords (YAKE!)\n- Answer Open and Closed book questions with T5\n- Summarize text and more with Multi task T5\n- Translate text with Microsofts Marian Model\n- Train a Multi Lingual Classifier for 100+ languages from a dataset with just one language\n\n## More ressources \n- [Join our Slack](https://join.slack.com/t/spark-nlp/shared_invite/zt-lutct9gm-kuUazcyFKhuGY3_0AMkxqA)\n- [NLU Website](https://nlu.johnsnowlabs.com/)\n- [NLU Github](https://github.com/JohnSnowLabs/nlu)\n- [Many more NLU example tutorials](https://github.com/JohnSnowLabs/nlu/tree/master/examples)\n- [Overview of every powerful nlu 1-liner](https://nlu.johnsnowlabs.com/docs/en/examples)\n- [Checkout the Modelshub for an overview of all models](https://nlp.johnsnowlabs.com/models) \n- [Checkout the NLU Namespace where you can find every model as a tabel](https://nlu.johnsnowlabs.com/docs/en/namespace)\n- [Intro to NLU article](https://medium.com/spark-nlp/1-line-of-code-350-nlp-models-with-john-snow-labs-nlu-in-python-2f1c55bba619)\n- [Indepth and easy Sentence Similarity Tutorial, with StackOverflow Questions using BERTology embeddings](https://medium.com/spark-nlp/easy-sentence-similarity-with-bert-sentence-embeddings-using-john-snow-labs-nlu-ea078deb6ebf)\n- [1 line of Python code for BERT, ALBERT, ELMO, ELECTRA, XLNET, GLOVE, Part of Speech with NLU and t-SNE](https://medium.com/spark-nlp/1-line-of-code-for-bert-albert-elmo-electra-xlnet-glove-part-of-speech-with-nlu-and-t-sne-9ebcd5379cd)",
"_____no_output_____"
],
[
"# Install NLU\nYou need Java8, Pyspark and Spark-NLP installed, [see the installation guide for instructions](https://nlu.johnsnowlabs.com/docs/en/install). If you need help or run into troubles, [ping us on slack :)](https://join.slack.com/t/spark-nlp/shared_invite/zt-lutct9gm-kuUazcyFKhuGY3_0AMkxqA) ",
"_____no_output_____"
]
],
[
[
"import os\n! apt-get update -qq > /dev/null \n# Install java\n! apt-get install -y openjdk-8-jdk-headless -qq > /dev/null\nos.environ[\"JAVA_HOME\"] = \"/usr/lib/jvm/java-8-openjdk-amd64\"\nos.environ[\"PATH\"] = os.environ[\"JAVA_HOME\"] + \"/bin:\" + os.environ[\"PATH\"]\n! pip install nlu pyspark==2.4.7 > /dev/null \nimport nlu",
"_____no_output_____"
]
],
[
[
"# Simple NLU basics on Strings",
"_____no_output_____"
],
[
"## Context based spell Checking in 1 line\n\n",
"_____no_output_____"
]
],
[
[
"nlu.load('spell').predict('I also liek to live dangertus')",
"spellcheck_dl download started this may take some time.\nApproximate size to download 112.2 MB\n[OK!]\n"
]
],
[
[
"## Binary Sentiment classification in 1 Line\n\n",
"_____no_output_____"
]
],
[
[
"nlu.load('sentiment').predict('I love NLU and rainy days!')",
"analyze_sentiment download started this may take some time.\nApprox size to download 4.9 MB\n[OK!]\n"
]
],
[
[
"## Part of Speech (POS) in 1 line\n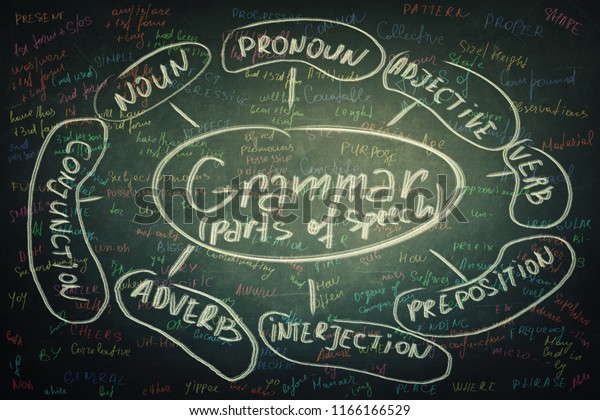\n\n|Tag |Description | Example|\n|------|------------|------|\n|CC| Coordinating conjunction | This batch of mushroom stew is savory **and** delicious |\n|CD| Cardinal number | Here are **five** coins |\n|DT| Determiner | **The** bunny went home |\n|EX| Existential there | **There** is a storm coming |\n|FW| Foreign word | I'm having a **déjà vu** |\n|IN| Preposition or subordinating conjunction | He is cleverer **than** I am |\n|JJ| Adjective | She wore a **beautiful** dress |\n|JJR| Adjective, comparative | My house is **bigger** than yours |\n|JJS| Adjective, superlative | I am the **shortest** person in my family |\n|LS| List item marker | A number of things need to be considered before starting a business **,** such as premises **,** finance **,** product demand **,** staffing and access to customers |\n|MD| Modal | You **must** stop when the traffic lights turn red |\n|NN| Noun, singular or mass | The **dog** likes to run |\n|NNS| Noun, plural | The **cars** are fast |\n|NNP| Proper noun, singular | I ordered the chair from **Amazon** |\n|NNPS| Proper noun, plural | We visted the **Kennedys** |\n|PDT| Predeterminer | **Both** the children had a toy |\n|POS| Possessive ending | I built the dog'**s** house |\n|PRP| Personal pronoun | **You** need to stop |\n|PRP$| Possessive pronoun | Remember not to judge a book by **its** cover |\n|RB| Adverb | The dog barks **loudly** |\n|RBR| Adverb, comparative | Could you sing more **quietly** please? |\n|RBS| Adverb, superlative | Everyone in the race ran fast, but John ran **the fastest** of all |\n|RP| Particle | He ate **up** all his dinner |\n|SYM| Symbol | What are you doing **?** |\n|TO| to | Please send it back **to** me |\n|UH| Interjection | **Wow!** You look gorgeous |\n|VB| Verb, base form | We **play** soccer |\n|VBD| Verb, past tense | I **worked** at a restaurant |\n|VBG| Verb, gerund or present participle | **Smoking** kills people |\n|VBN| Verb, past participle | She has **done** her homework |\n|VBP| Verb, non-3rd person singular present | You **flit** from place to place |\n|VBZ| Verb, 3rd person singular present | He never **calls** me |\n|WDT| Wh-determiner | The store honored the complaints, **which** were less than 25 days old |\n|WP| Wh-pronoun | **Who** can help me? |\n|WP\\$| Possessive wh-pronoun | **Whose** fault is it? |\n|WRB| Wh-adverb | **Where** are you going? |",
"_____no_output_____"
]
],
[
[
"nlu.load('pos').predict('POS assigns each token in a sentence a grammatical label')",
"pos_anc download started this may take some time.\nApproximate size to download 4.3 MB\n[OK!]\n"
]
],
[
[
"## Named Entity Recognition (NER) in 1 line\n\n\n\n|Type | \tDescription |\n|------|--------------|\n| PERSON | \tPeople, including fictional like **Harry Potter** |\n| NORP | \tNationalities or religious or political groups like the **Germans** |\n| FAC | \tBuildings, airports, highways, bridges, etc. like **New York Airport** |\n| ORG | \tCompanies, agencies, institutions, etc. like **Microsoft** |\n| GPE | \tCountries, cities, states. like **Germany** |\n| LOC | \tNon-GPE locations, mountain ranges, bodies of water. Like the **Sahara desert**|\n| PRODUCT | \tObjects, vehicles, foods, etc. (Not services.) like **playstation** |\n| EVENT | \tNamed hurricanes, battles, wars, sports events, etc. like **hurricane Katrina**|\n| WORK_OF_ART | \tTitles of books, songs, etc. Like **Mona Lisa** |\n| LAW | \tNamed documents made into laws. Like : **Declaration of Independence** |\n| LANGUAGE | \tAny named language. Like **Turkish**|\n| DATE | \tAbsolute or relative dates or periods. Like every second **friday**|\n| TIME | \tTimes smaller than a day. Like **every minute**|\n| PERCENT | \tPercentage, including ”%“. Like **55%** of workers enjoy their work |\n| MONEY | \tMonetary values, including unit. Like **50$** for those pants |\n| QUANTITY | \tMeasurements, as of weight or distance. Like this person weights **50kg** |\n| ORDINAL | \t“first”, “second”, etc. Like David placed **first** in the tournament |\n| CARDINAL | \tNumerals that do not fall under another type. Like **hundreds** of models are avaiable in NLU |\n",
"_____no_output_____"
]
],
[
[
"nlu.load('ner').predict(\"John Snow Labs congratulates the Amarican John Biden to winning the American election!\", output_level='chunk')",
"onto_recognize_entities_sm download started this may take some time.\nApprox size to download 159 MB\n[OK!]\n"
]
],
[
[
"# Let's apply NLU to a dataset!\n\n<div>\n<img src=\"http://ckl-it.de/wp-content/uploads/2021/02/crypto.jpeg \" width=\"400\" height=\"250\" >\n</div>\n",
"_____no_output_____"
]
],
[
[
"import pandas as pd \nimport nlu\n!wget http://ckl-it.de/wp-content/uploads/2020/12/small_btc.csv \ndf = pd.read_csv('/content/small_btc.csv').iloc[0:5000].title\ndf\n\n",
"--2021-03-24 09:32:01-- http://ckl-it.de/wp-content/uploads/2020/12/small_btc.csv\nResolving ckl-it.de (ckl-it.de)... 217.160.0.108, 2001:8d8:100f:f000::209\nConnecting to ckl-it.de (ckl-it.de)|217.160.0.108|:80... connected.\nHTTP request sent, awaiting response... 200 OK\nLength: 22244914 (21M) [text/csv]\nSaving to: ‘small_btc.csv’\n\nsmall_btc.csv 100%[===================>] 21.21M 6.62MB/s in 3.5s \n\n2021-03-24 09:32:04 (6.06 MB/s) - ‘small_btc.csv’ saved [22244914/22244914]\n\n"
]
],
[
[
"## NER on a Crypto News dataset\n### The **NER** model which you can load via `nlu.load('ner')` recognizes 18 different classes in your dataset.\nWe set output level to chunk, so that we get 1 row per NER class.\n\n\n#### Predicted entities:\n\n\nNER is avaiable in many languages, which you can [find in the John Snow Labs Modelshub](https://nlp.johnsnowlabs.com/models)",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
],
[
[
"ner_df = nlu.load('ner').predict(df, output_level = 'chunk')\nner_df ",
"onto_recognize_entities_sm download started this may take some time.\nApprox size to download 159 MB\n[OK!]\n"
]
],
[
[
"### Top 50 Named Entities",
"_____no_output_____"
]
],
[
[
"ner_df.entities.value_counts()[:100].plot.barh(figsize = (16,20))",
"_____no_output_____"
]
],
[
[
"### Top 50 Named Entities which are PERSONS",
"_____no_output_____"
]
],
[
[
"ner_df[ner_df.entities_class == 'PERSON'].entities.value_counts()[:50].plot.barh(figsize=(18,20), title ='Top 50 Occuring Persons in the dataset')",
"_____no_output_____"
]
],
[
[
"### Top 50 Named Entities which are Countries/Cities/States",
"_____no_output_____"
]
],
[
[
"ner_df[ner_df.entities_class == 'GPE'].entities.value_counts()[:50].plot.barh(figsize=(18,20),title ='Top 50 Countries/Cities/States Occuring in the dataset')",
"_____no_output_____"
]
],
[
[
"### Top 50 Named Entities which are PRODUCTS ",
"_____no_output_____"
]
],
[
[
"ner_df[ner_df.entities_class == 'PRODUCT'].entities.value_counts()[:50].plot.barh(figsize=(18,20),title ='Top 50 products occuring in the dataset')",
"_____no_output_____"
]
],
[
[
"### Top 50 Named Entities which are ORGANIZATIONS",
"_____no_output_____"
]
],
[
[
"ner_df[ner_df.entities_class == 'ORG'].entities.value_counts()[:50].plot.barh(figsize=(18,20),title ='Top 50 products occuring in the dataset')",
"_____no_output_____"
]
],
[
[
"## YAKE on a Crypto News dataset\n### The **YAKE!** model (Yet Another Keyword Extractor) is a **unsupervised** keyword extraction algorithm.\nYou can load it via which you can load via `nlu.load('yake')`. It has no weights and is very fast.\nIt has various parameters that can be configured to influence which keywords are beeing extracted, [here for an more indepth YAKE guide](https://github.com/JohnSnowLabs/nlu/blob/master/examples/webinars_conferences_etc/multi_lingual_webinar/1_NLU_base_features_on_dataset_with_YAKE_Lemma_Stemm_classifiers_NER_.ipynb)",
"_____no_output_____"
]
],
[
[
"yake_df = nlu.load('yake').predict(df)\nyake_df",
"_____no_output_____"
]
],
[
[
"### Top 50 extracted Keywords with YAKE!",
"_____no_output_____"
]
],
[
[
"yake_df.explode('keywords_classes').keywords_classes.value_counts()[0:50].plot.barh(figsize=(14,18))",
"_____no_output_____"
]
],
[
[
"## Binary Sentimental Analysis and Distribution on a dataset",
"_____no_output_____"
]
],
[
[
"sent_df = nlu.load('sentiment').predict(df)\nsent_df",
"analyze_sentiment download started this may take some time.\nApprox size to download 4.9 MB\n[OK!]\n"
],
[
"sent_df.sentiment.value_counts().plot.bar(title='Sentiment ')",
"_____no_output_____"
]
],
[
[
"## Emotional Analysis and Distribution of Headlines ",
"_____no_output_____"
]
],
[
[
"emo_df = nlu.load('emotion').predict(df)\nemo_df",
"classifierdl_use_emotion download started this may take some time.\nApproximate size to download 21.3 MB\n[OK!]\ntfhub_use download started this may take some time.\nApproximate size to download 923.7 MB\n[OK!]\n\n\n\n\n\n\n"
],
[
"emo_df.emotion.value_counts().plot.bar(title='Emotion Distribution')\n",
"_____no_output_____"
]
],
[
[
"**Make sure to restart your notebook again** before starting the next section",
"_____no_output_____"
]
],
[
[
"print(\"Please restart kernel if you are in google colab and run next cell after the restart to configure java 8 back\")\n1+'wait'\n",
"Please restart kernel if you are in google colab and run next cell after the restart to configure java 8 back\n"
],
[
"# This configures colab to use Java 8 again. \n# You need to run this in Google colab, because after restart it likes to set Java 11 as default, which will cause issues\n! echo 2 | update-alternatives --config java\nimport pandas as pd\nimport nlu ",
"There are 2 choices for the alternative java (providing /usr/bin/java).\n\n Selection Path Priority Status\n------------------------------------------------------------\n 0 /usr/lib/jvm/java-11-openjdk-amd64/bin/java 1111 auto mode\n 1 /usr/lib/jvm/java-11-openjdk-amd64/bin/java 1111 manual mode\n* 2 /usr/lib/jvm/java-8-openjdk-amd64/jre/bin/java 1081 manual mode\n\nPress <enter> to keep the current choice[*], or type selection number: "
]
],
[
[
"# Answer **Closed Book** and Open **Book Questions** with Google's T5!\n\n<!-- [T5]() -->\n\n\nYou can load the **question answering** model with `nlu.load('en.t5')`",
"_____no_output_____"
]
],
[
[
"# Load question answering T5 model\nt5_closed_question = nlu.load('en.t5')",
"google_t5_small_ssm_nq download started this may take some time.\nApproximate size to download 139 MB\n[OK!]\n"
]
],
[
[
"## Answer **Closed Book Questions** \nClosed book means that no additional context is given and the model must answer the question with the knowledge stored in it's weights",
"_____no_output_____"
]
],
[
[
"t5_closed_question.predict(\"Who is president of Nigeria?\")",
"_____no_output_____"
],
[
"t5_closed_question.predict(\"What is the most common language in India?\")",
"_____no_output_____"
],
[
"t5_closed_question.predict(\"What is the capital of Germany?\")",
"_____no_output_____"
]
],
[
[
"## Answer **Open Book Questions** \nThese are questions where we give the model some additional context, that is used to answer the question",
"_____no_output_____"
]
],
[
[
"t5_open_book = nlu.load('answer_question')",
"t5_base download started this may take some time.\nApproximate size to download 446 MB\n[OK!]\n"
],
[
"context = 'Peters last week was terrible! He had an accident and broke his leg while skiing!'\nquestion1 = 'Why was peters week so bad?' \nquestion2 = 'How did peter broke his leg?' \n\nt5_open_book.predict([question1+context, question2 + context]) ",
"_____no_output_____"
],
[
"# Ask T5 questions in the context of a News Article\nquestion1 = 'Who is Jack ma?'\nquestion2 = 'Who is founder of Alibaba Group?'\nquestion3 = 'When did Jack Ma re-appear?'\nquestion4 = 'How did Alibaba stocks react?'\nquestion5 = 'Whom did Jack Ma meet?'\nquestion6 = 'Who did Jack Ma hide from?'\n\n\n# from https://www.bbc.com/news/business-55728338 \nnews_article_context = \"\"\" context:\nAlibaba Group founder Jack Ma has made his first appearance since Chinese regulators cracked down on his business empire.\nHis absence had fuelled speculation over his whereabouts amid increasing official scrutiny of his businesses.\nThe billionaire met 100 rural teachers in China via a video meeting on Wednesday, according to local government media.\nAlibaba shares surged 5% on Hong Kong's stock exchange on the news.\n\"\"\"\n\nquestions = [\n question1+ news_article_context,\n question2+ news_article_context,\n question3+ news_article_context,\n question4+ news_article_context,\n question5+ news_article_context,\n question6+ news_article_context,]\n\n",
"_____no_output_____"
],
[
"t5_open_book.predict(questions)",
"_____no_output_____"
]
],
[
[
"# Multi Problem T5 model for Summarization and more\nThe main T5 model was trained for over 20 tasks from the SQUAD/GLUE/SUPERGLUE datasets. See [this notebook](https://github.com/JohnSnowLabs/nlu/blob/master/examples/webinars_conferences_etc/multi_lingual_webinar/7_T5_SQUAD_GLUE_SUPER_GLUE_TASKS.ipynb) for a demo of all tasks \n\n\n# Overview of every task available with T5\n[The T5 model](https://arxiv.org/pdf/1910.10683.pdf) is trained on various datasets for 17 different tasks which fall into 8 categories.\n\n\n\n1. Text summarization\n2. Question answering\n3. Translation\n4. Sentiment analysis\n5. Natural Language inference\n6. Coreference resolution\n7. Sentence Completion\n8. Word sense disambiguation\n\n### Every T5 Task with explanation:\n|Task Name | Explanation | \n|----------|--------------|\n|[1.CoLA](https://nyu-mll.github.io/CoLA/) | Classify if a sentence is gramaticaly correct|\n|[2.RTE](https://dl.acm.org/doi/10.1007/11736790_9) | Classify whether if a statement can be deducted from a sentence|\n|[3.MNLI](https://arxiv.org/abs/1704.05426) | Classify for a hypothesis and premise whether they contradict or contradict each other or neither of both (3 class).|\n|[4.MRPC](https://www.aclweb.org/anthology/I05-5002.pdf) | Classify whether a pair of sentences is a re-phrasing of each other (semantically equivalent)|\n|[5.QNLI](https://arxiv.org/pdf/1804.07461.pdf) | Classify whether the answer to a question can be deducted from an answer candidate.|\n|[6.QQP](https://www.quora.com/q/quoradata/First-Quora-Dataset-Release-Question-Pairs) | Classify whether a pair of questions is a re-phrasing of each other (semantically equivalent)|\n|[7.SST2](https://www.aclweb.org/anthology/D13-1170.pdf) | Classify the sentiment of a sentence as positive or negative|\n|[8.STSB](https://www.aclweb.org/anthology/S17-2001/) | Classify the sentiment of a sentence on a scale from 1 to 5 (21 Sentiment classes)|\n|[9.CB](https://ojs.ub.uni-konstanz.de/sub/index.php/sub/article/view/601) | Classify for a premise and a hypothesis whether they contradict each other or not (binary).|\n|[10.COPA](https://www.aaai.org/ocs/index.php/SSS/SSS11/paper/view/2418/0) | Classify for a question, premise, and 2 choices which choice the correct choice is (binary).|\n|[11.MultiRc](https://www.aclweb.org/anthology/N18-1023.pdf) | Classify for a question, a paragraph of text, and an answer candidate, if the answer is correct (binary),|\n|[12.WiC](https://arxiv.org/abs/1808.09121) | Classify for a pair of sentences and a disambigous word if the word has the same meaning in both sentences.|\n|[13.WSC/DPR](https://www.aaai.org/ocs/index.php/KR/KR12/paper/view/4492/0) | Predict for an ambiguous pronoun in a sentence what it is referring to. |\n|[14.Summarization](https://arxiv.org/abs/1506.03340) | Summarize text into a shorter representation.|\n|[15.SQuAD](https://arxiv.org/abs/1606.05250) | Answer a question for a given context.|\n|[16.WMT1.](https://arxiv.org/abs/1706.03762) | Translate English to German|\n|[17.WMT2.](https://arxiv.org/abs/1706.03762) | Translate English to French|\n|[18.WMT3.](https://arxiv.org/abs/1706.03762) | Translate English to Romanian|\n\n",
"_____no_output_____"
]
],
[
[
"# Load the Multi Task Model T5\nt5_multi = nlu.load('en.t5.base')",
"t5_base download started this may take some time.\nApproximate size to download 446 MB\n[OK!]\n"
],
[
"# https://www.reuters.com/article/instant-article/idCAKBN2AA2WF\ntext = \"\"\"(Reuters) - Mastercard Inc said on Wednesday it was planning to offer support for some cryptocurrencies on its network this year, joining a string of big-ticket firms that have pledged similar support.\n\nThe credit-card giant’s announcement comes days after Elon Musk’s Tesla Inc revealed it had purchased $1.5 billion of bitcoin and would soon accept it as a form of payment.\n\nAsset manager BlackRock Inc and payments companies Square and PayPal have also recently backed cryptocurrencies.\n\nMastercard already offers customers cards that allow people to transact using their cryptocurrencies, although without going through its network.\n\n\"Doing this work will create a lot more possibilities for shoppers and merchants, allowing them to transact in an entirely new form of payment. This change may open merchants up to new customers who are already flocking to digital assets,\" Mastercard said. (mstr.cd/3tLaPZM)\n\nMastercard specified that not all cryptocurrencies will be supported on its network, adding that many of the hundreds of digital assets in circulation still need to tighten their compliance measures.\n\nMany cryptocurrencies have struggled to win the trust of mainstream investors and the general public due to their speculative nature and potential for money laundering.\n\"\"\"\nt5_multi['t5'].setTask('summarize ') \nshort = t5_multi.predict(text)\nshort",
"_____no_output_____"
],
[
"print(f\"Original Length {len(short.document.iloc[0])} Summarized Length : {len(short.T5.iloc[0])} \\n summarized text :{short.T5.iloc[0]} \")\n",
"Original Length 1277 Summarized Length : 352 \n summarized text :mastercard said on Wednesday it was planning to offer support for some cryptocurrencies on its network this year . the credit-card giant’s announcement comes days after Elon Musk’s Tesla Inc revealed it had purchased $1.5 billion of bitcoin . asset manager blackrock and payments companies Square and PayPal have also recently backed cryptocurrencies . \n"
],
[
"short.T5.iloc[0]",
"_____no_output_____"
]
],
[
[
"**Make sure to restart your notebook again** before starting the next section",
"_____no_output_____"
]
],
[
[
"print(\"Please restart kernel if you are in google colab and run next cell after the restart to configure java 8 back\")\n1+'wait'\n",
"_____no_output_____"
],
[
"# This configures colab to use Java 8 again. \n# You need to run this in Google colab, because after restart it likes to set Java 11 as default, which will cause issues\n! echo 2 | update-alternatives --config java\n",
"_____no_output_____"
]
],
[
[
"# Translate between more than 200 Languages with [ Microsofts Marian Models](https://marian-nmt.github.io/publications/)\n\nMarian is an efficient, free Neural Machine Translation framework mainly being developed by the Microsoft Translator team (646+ pretrained models & pipelines in 192+ languages)\nYou need to specify the language your data is in as `start_language` and the language you want to translate to as `target_language`. \n The language references must be [ISO language codes](https://en.wikipedia.org/wiki/List_of_ISO_639-1_codes)\n\n`nlu.load('<start_language>.translate_to.<target_language>')` \n\n**Translate Turkish to English:** \n`nlu.load('tr.translate_to.en')`\n\n**Translate English to French:** \n`nlu.load('en.translate_to.fr')`\n\n\n**Translate French to Hebrew:** \n`nlu.load('fr.translate_to.he')`\n\n\n\n\n\n",
"_____no_output_____"
]
],
[
[
"import nlu\nimport pandas as pd\n!wget http://ckl-it.de/wp-content/uploads/2020/12/small_btc.csv \ndf = pd.read_csv('/content/small_btc.csv').iloc[0:20].title",
"_____no_output_____"
]
],
[
[
"## Translate to German",
"_____no_output_____"
]
],
[
[
"translate_pipe = nlu.load('en.translate_to.de')\ntranslate_pipe.predict(df)",
"translate_en_de download started this may take some time.\nApprox size to download 370.2 MB\n[OK!]\n"
]
],
[
[
"## Translate to Chinese",
"_____no_output_____"
]
],
[
[
"translate_pipe = nlu.load('en.translate_to.zh')\ntranslate_pipe.predict(df)",
"translate_en_zh download started this may take some time.\nApprox size to download 396.8 MB\n[OK!]\n"
]
],
[
[
"## Translate to Hindi",
"_____no_output_____"
]
],
[
[
"translate_pipe = nlu.load('en.translate_to.hi')\ntranslate_pipe.predict(df)",
"translate_en_hi download started this may take some time.\nApprox size to download 385.8 MB\n[OK!]\n"
]
],
[
[
"# Train a Multi Lingual Classifier for 100+ languages from a dataset with just one language\n\n[Leverage Language-agnostic BERT Sentence Embedding (LABSE) and acheive state of the art!](https://arxiv.org/abs/2007.01852) \n\nTraining a classifier with LABSE embeddings enables the knowledge to be transferred to 109 languages!\nWith the [SentimentDL model](https://nlp.johnsnowlabs.com/docs/en/annotators#sentimentdl-multi-class-sentiment-analysis-annotator) from Spark NLP you can achieve State Of the Art results on any binary class text classification problem.\n\n### Languages suppoted by LABSE\n\n\n",
"_____no_output_____"
]
],
[
[
"# Download French twitter Sentiment dataset https://www.kaggle.com/hbaflast/french-twitter-sentiment-analysis\n! wget http://ckl-it.de/wp-content/uploads/2021/02/french_tweets.csv\n\nimport pandas as pd\n\ntrain_path = '/content/french_tweets.csv'\n\ntrain_df = pd.read_csv(train_path)\n# the text data to use for classification should be in a column named 'text'\ncolumns=['text','y']\ntrain_df = train_df[columns]\ntrain_df = train_df.sample(frac=1).reset_index(drop=True)\ntrain_df",
"_____no_output_____"
]
],
[
[
"## Train Deep Learning Classifier using `nlu.load('train.sentiment')`\n\nAl you need is a Pandas Dataframe with a label column named `y` and the column with text data should be named `text`\n\nWe are training on a french dataset and can then predict classes correct **in 100+ langauges**",
"_____no_output_____"
]
],
[
[
"# Train longer!\ntrainable_pipe = nlu.load('xx.embed_sentence.labse train.sentiment')\ntrainable_pipe['sentiment_dl'].setMaxEpochs(60) \ntrainable_pipe['sentiment_dl'].setLr(0.005) \nfitted_pipe = trainable_pipe.fit(train_df.iloc[:2000])\n# predict with the trainable pipeline on dataset and get predictions\npreds = fitted_pipe.predict(train_df.iloc[:2000],output_level='document')\n\n#sentence detector that is part of the pipe generates sone NaNs. lets drop them first\npreds.dropna(inplace=True)\nprint(classification_report(preds['y'], preds['sentiment']))\n\npreds",
"labse download started this may take some time.\nApproximate size to download 1.7 GB\n[OK!]\n precision recall f1-score support\n\n negative 0.88 0.94 0.91 980\n positive 0.94 0.88 0.91 1020\n\n accuracy 0.91 2000\n macro avg 0.91 0.91 0.91 2000\nweighted avg 0.91 0.91 0.91 2000\n\n"
]
],
[
[
"### Test the fitted pipe on new example",
"_____no_output_____"
],
[
"#### The Model understands Englsih\n",
"_____no_output_____"
]
],
[
[
"fitted_pipe.predict(\"This was awful!\")",
"_____no_output_____"
],
[
"fitted_pipe.predict(\"This was great!\")",
"_____no_output_____"
]
],
[
[
"#### The Model understands German\n",
"_____no_output_____"
]
],
[
[
"# German for:' this movie was great!'\nfitted_pipe.predict(\"Der Film war echt klasse!\")",
"_____no_output_____"
],
[
"# German for: 'This movie was really boring'\nfitted_pipe.predict(\"Der Film war echt langweilig!\")",
"_____no_output_____"
]
],
[
[
"#### The Model understands Chinese\n",
"_____no_output_____"
]
],
[
[
"# Chinese for: \"This model was awful!\"\nfitted_pipe.predict(\"这部电影太糟糕了!\")",
"_____no_output_____"
],
[
"# Chine for : \"This move was great!\"\nfitted_pipe.predict(\"此举很棒!\")\n",
"_____no_output_____"
]
],
[
[
"#### Model understanda Afrikaans\n\n\n\n",
"_____no_output_____"
]
],
[
[
"# Afrikaans for 'This movie was amazing!'\nfitted_pipe.predict(\"Hierdie film was ongelooflik!\")\n",
"_____no_output_____"
],
[
"# Afrikaans for :'The movie made me fall asleep, it's awful!'\nfitted_pipe.predict('Die film het my aan die slaap laat raak, dit is verskriklik!')",
"_____no_output_____"
]
],
[
[
"#### The model understands Vietnamese\n",
"_____no_output_____"
]
],
[
[
"# Vietnamese for : 'The movie was painful to watch'\nfitted_pipe.predict('Phim đau điếng người xem')\n",
"_____no_output_____"
],
[
"\n# Vietnamese for : 'This was the best movie ever'\nfitted_pipe.predict('Đây là bộ phim hay nhất từ trước đến nay')",
"_____no_output_____"
]
],
[
[
"#### The model understands Japanese\n\n",
"_____no_output_____"
]
],
[
[
"\n# Japanese for : 'This is now my favorite movie!'\nfitted_pipe.predict('これが私のお気に入りの映画です!')",
"_____no_output_____"
],
[
"\n# Japanese for : 'I would rather kill myself than watch that movie again'\nfitted_pipe.predict('その映画をもう一度見るよりも自殺したい')",
"_____no_output_____"
]
],
[
[
"# There are many more models you can put to use in 1 line of code!\n## Checkout [the Modelshub](https://nlp.johnsnowlabs.com/models) and the [NLU Namespace](https://nlu.johnsnowlabs.com/docs/en/namespace) for more models\n\n\n### More ressources \n- [Join our Slack](https://join.slack.com/t/spark-nlp/shared_invite/zt-lutct9gm-kuUazcyFKhuGY3_0AMkxqA)\n- [NLU Website](https://nlu.johnsnowlabs.com/)\n- [NLU Github](https://github.com/JohnSnowLabs/nlu)\n- [Many more NLU example tutorials](https://github.com/JohnSnowLabs/nlu/tree/master/examples)\n- [Overview of every powerful nlu 1-liner](https://nlu.johnsnowlabs.com/docs/en/examples)\n- [Checkout the Modelshub for an overview of all models](https://nlp.johnsnowlabs.com/models) \n- [Checkout the NLU Namespace where you can find every model as a tabel](https://nlu.johnsnowlabs.com/docs/en/namespace)\n- [Intro to NLU article](https://medium.com/spark-nlp/1-line-of-code-350-nlp-models-with-john-snow-labs-nlu-in-python-2f1c55bba619)\n- [Indepth and easy Sentence Similarity Tutorial, with StackOverflow Questions using BERTology embeddings](https://medium.com/spark-nlp/easy-sentence-similarity-with-bert-sentence-embeddings-using-john-snow-labs-nlu-ea078deb6ebf)\n- [1 line of Python code for BERT, ALBERT, ELMO, ELECTRA, XLNET, GLOVE, Part of Speech with NLU and t-SNE](https://medium.com/spark-nlp/1-line-of-code-for-bert-albert-elmo-electra-xlnet-glove-part-of-speech-with-nlu-and-t-sne-9ebcd5379cd)",
"_____no_output_____"
]
],
[
[
"while 1 : 1 ",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
c512479ff28d562f3953384f7f2cf54a4f1fbeda
| 696,729 |
ipynb
|
Jupyter Notebook
|
flu-trained-models/florida/florida_flu_temp_nextWed.ipynb
|
tamjazad/ml-covid19
|
85130e22fd3ded2dc55ca1f773fb42a20318b75a
|
[
"MIT"
] | 1 |
2020-09-02T11:59:02.000Z
|
2020-09-02T11:59:02.000Z
|
flu-trained-models/florida/florida_flu_temp_nextWed.ipynb
|
tamjazad/ml-covid19
|
85130e22fd3ded2dc55ca1f773fb42a20318b75a
|
[
"MIT"
] | null | null | null |
flu-trained-models/florida/florida_flu_temp_nextWed.ipynb
|
tamjazad/ml-covid19
|
85130e22fd3ded2dc55ca1f773fb42a20318b75a
|
[
"MIT"
] | 1 |
2020-09-02T11:59:06.000Z
|
2020-09-02T11:59:06.000Z
| 402.500867 | 202,248 | 0.928896 |
[
[
[
"# Florida Single Weekly Predictions, trained on historical flu data and temperature\n\n> Once again, just like before in the USA flu model, I am going to index COVID weekly cases by Wednesdays",
"_____no_output_____"
]
],
[
[
"import tensorflow as tf\nphysical_devices = tf.config.list_physical_devices('GPU')\ntf.config.experimental.set_memory_growth(physical_devices[0], enable=True)\nimport matplotlib.pyplot as plt\nimport numpy as np\nimport pandas as pd\nimport sklearn\nfrom sklearn import preprocessing",
"_____no_output_____"
]
],
[
[
"### getting historical flu data",
"_____no_output_____"
]
],
[
[
"system = \"Windows\"\n\nif system == \"Windows\":\n flu_dir = \"..\\\\..\\\\..\\\\cdc-fludata\\\\us_national\\\\\"\nelse:\n flu_dir = \"../../../cdc-fludata/us_national/\"",
"_____no_output_____"
],
[
"flu_dictionary = {}\n\nfor year in range(1997, 2019):\n filepath = \"usflu_\"\n year_string = str(year) + \"-\" + str(year + 1)\n filepath = flu_dir + filepath + year_string + \".csv\"\n temp_df = pd.read_csv(filepath)\n flu_dictionary[year] = temp_df",
"_____no_output_____"
]
],
[
[
"### combining flu data into one chronological series of total cases",
"_____no_output_____"
]
],
[
[
"# getting total cases and putting them in a series by week\nflu_series_dict = {} \n\nfor year in flu_dictionary:\n temp_df = flu_dictionary[year]\n temp_df = temp_df.set_index(\"WEEK\")\n abridged_df = temp_df.iloc[:, 2:]\n \n try:\n abridged_df = abridged_df.drop(columns=\"PERCENT POSITIVE\")\n except:\n pass\n \n total_cases_series = abridged_df.sum(axis=1)\n flu_series_dict[year] = total_cases_series\n ",
"_____no_output_____"
],
[
"all_cases_series = pd.Series(dtype=\"int64\")\n\nfor year in flu_series_dict:\n temp_series = flu_series_dict[year]\n all_cases_series = all_cases_series.append(temp_series, ignore_index=True)",
"_____no_output_____"
],
[
"all_cases_series",
"_____no_output_____"
],
[
"all_cases_series.plot(grid=True, figsize=(60,20))",
"_____no_output_____"
]
],
[
[
"### Now, making a normalized series between 0, 1",
"_____no_output_____"
]
],
[
[
"norm_flu_series_dict = {}\n\nfor year in flu_series_dict:\n temp_series = flu_series_dict[year]\n temp_list = preprocessing.minmax_scale(temp_series)\n temp_series = pd.Series(temp_list)\n norm_flu_series_dict[year] = temp_series",
"_____no_output_____"
],
[
"all_cases_norm_series = pd.Series(dtype=\"int64\")\n\nfor year in norm_flu_series_dict:\n temp_series = norm_flu_series_dict[year]\n all_cases_norm_series = all_cases_norm_series.append(temp_series, ignore_index=True)",
"_____no_output_____"
],
[
"all_cases_norm_series.plot(grid=True, figsize=(60,5))\nall_cases_norm_series",
"_____no_output_____"
]
],
[
[
"## Getting COVID-19 Case Data",
"_____no_output_____"
]
],
[
[
"if system == \"Windows\":\n datapath = \"..\\\\..\\\\..\\\\COVID-19\\\\csse_covid_19_data\\\\csse_covid_19_time_series\\\\\"\nelse:\n datapath = \"../../../COVID-19/csse_covid_19_data/csse_covid_19_time_series/\"\n\n# Choose from \"US Cases\", \"US Deaths\", \"World Cases\", \"World Deaths\", \"World Recoveries\"\nkey = \"US Cases\" \n\nif key == \"US Cases\":\n datapath = datapath + \"time_series_covid19_confirmed_US.csv\"\nelif key == \"US Deaths\":\n datapath = datapath + \"time_series_covid19_deaths_US.csv\"\nelif key == \"World Cases\":\n datapath = datapath + \"time_series_covid19_confirmed_global.csv\"\nelif key == \"World Deaths\":\n datapath = datapath + \"time_series_covid19_deaths_global.csv\"\nelif key == \"World Recoveries\":\n datapath = datapath + \"time_series_covid19_recovered_global.csv\"",
"_____no_output_____"
],
[
"covid_df = pd.read_csv(datapath)",
"_____no_output_____"
],
[
"covid_df",
"_____no_output_____"
],
[
"florida_data = covid_df.loc[covid_df[\"Province_State\"] == \"Florida\"]",
"_____no_output_____"
],
[
"florida_cases = florida_data.iloc[:,11:]",
"_____no_output_____"
],
[
"florida_cases_total = florida_cases.sum(axis=0)",
"_____no_output_____"
],
[
"florida_cases_total.plot()",
"_____no_output_____"
]
],
[
[
"### convert daily data to weekly data",
"_____no_output_____"
]
],
[
[
"florida_weekly_cases = florida_cases_total.iloc[::7]",
"_____no_output_____"
],
[
"florida_weekly_cases",
"_____no_output_____"
],
[
"florida_weekly_cases.plot()",
"_____no_output_____"
]
],
[
[
"### Converting cumulative series to non-cumulative series",
"_____no_output_____"
]
],
[
[
"florida_wnew_cases = florida_weekly_cases.diff()\nflorida_wnew_cases[0] = 1.0\nflorida_wnew_cases",
"_____no_output_____"
],
[
"florida_wnew_cases.plot()",
"_____no_output_____"
]
],
[
[
"### normalizing weekly case data\n> This is going to be different for texas. This is because, the peak number of weekly new infections probably has not been reached yet. We need to divide everything by a guess for the peak number of predictions instead of min-max scaling.",
"_____no_output_____"
]
],
[
[
"# I'm guessing that the peak number of weekly cases will be about 60,000. Could definitely be wrong.\npeak_guess = 60000\n\nflorida_wnew_cases_norm = florida_wnew_cases / peak_guess\nflorida_wnew_cases_norm.plot()\nflorida_wnew_cases_norm",
"_____no_output_____"
]
],
[
[
"## getting temperature data\n> At the moment, this will be dummy data",
"_____no_output_____"
]
],
[
[
"flu_temp_data = np.full(len(all_cases_norm_series), 0.5)",
"_____no_output_____"
],
[
"training_data_df = pd.DataFrame({\n \"Temperature\" : flu_temp_data,\n \"Flu Cases\" : all_cases_norm_series\n})\ntraining_data_df",
"_____no_output_____"
],
[
"covid_temp_data = np.full(len(florida_wnew_cases_norm), 0.5)",
"_____no_output_____"
],
[
"testing_data_df = pd.DataFrame({\n \"Temperature\" : covid_temp_data,\n \"COVID Cases\" : florida_wnew_cases_norm\n})\ntesting_data_df",
"_____no_output_____"
],
[
"testing_data_df.shape",
"_____no_output_____"
],
[
"training_data_np = training_data_df.values\ntesting_data_np = testing_data_df.values",
"_____no_output_____"
]
],
[
[
"## Building Neural Net Model",
"_____no_output_____"
],
[
"### preparing model data",
"_____no_output_____"
]
],
[
[
"# this code is directly from https://www.tensorflow.org/tutorials/structured_data/time_series\n# much of below data formatting code is derived straight from same link\n\ndef multivariate_data(dataset, target, start_index, end_index, history_size,\n target_size, step, single_step=False):\n data = []\n labels = []\n\n start_index = start_index + history_size\n if end_index is None:\n end_index = len(dataset) - target_size\n\n for i in range(start_index, end_index):\n indices = range(i-history_size, i, step)\n data.append(dataset[indices])\n\n if single_step:\n labels.append(target[i+target_size])\n else:\n labels.append(target[i:i+target_size])\n\n return np.array(data), np.array(labels)",
"_____no_output_____"
],
[
"past_history = 22\nfuture_target = 0\nSTEP = 1\n\nx_train_single, y_train_single = multivariate_data(training_data_np, training_data_np[:, 1], 0,\n None, past_history,\n future_target, STEP,\n single_step=True)\nx_test_single, y_test_single = multivariate_data(testing_data_np, testing_data_np[:, 1],\n 0, None, past_history,\n future_target, STEP,\n single_step=True)",
"_____no_output_____"
],
[
"BATCH_SIZE = 300\nBUFFER_SIZE = 1000\n\ntrain_data_single = tf.data.Dataset.from_tensor_slices((x_train_single, y_train_single))\ntrain_data_single = train_data_single.cache().shuffle(BUFFER_SIZE).batch(BATCH_SIZE).repeat()\n\ntest_data_single = tf.data.Dataset.from_tensor_slices((x_test_single, y_test_single))\ntest_data_single = test_data_single.batch(1).repeat()",
"_____no_output_____"
]
],
[
[
"### designing actual model",
"_____no_output_____"
]
],
[
[
"# creating the neural network model\n\nlstm_prediction_model = tf.keras.Sequential([\n tf.keras.layers.LSTM(32, input_shape=x_train_single.shape[-2:]),\n tf.keras.layers.Dense(32),\n tf.keras.layers.Dense(1)\n])\n\nlstm_prediction_model.compile(optimizer=tf.keras.optimizers.RMSprop(), loss=\"mae\")",
"_____no_output_____"
],
[
"single_step_history = lstm_prediction_model.fit(train_data_single, epochs=10,\n steps_per_epoch=250,\n validation_data=test_data_single,\n validation_steps=50)",
"Train for 250 steps, validate for 50 steps\nEpoch 1/10\n250/250 [==============================] - 4s 17ms/step - loss: 0.0578 - val_loss: 0.1777\nEpoch 2/10\n250/250 [==============================] - 1s 4ms/step - loss: 0.0321 - val_loss: 0.1247\nEpoch 3/10\n250/250 [==============================] - 1s 4ms/step - loss: 0.0292 - val_loss: 0.1219\nEpoch 4/10\n250/250 [==============================] - 1s 4ms/step - loss: 0.0278 - val_loss: 0.1129\nEpoch 5/10\n250/250 [==============================] - 1s 4ms/step - loss: 0.0269 - val_loss: 0.1129\nEpoch 6/10\n250/250 [==============================] - 1s 4ms/step - loss: 0.0262 - val_loss: 0.1099\nEpoch 7/10\n250/250 [==============================] - 1s 4ms/step - loss: 0.0256 - val_loss: 0.1091\nEpoch 8/10\n250/250 [==============================] - 1s 4ms/step - loss: 0.0251 - val_loss: 0.1077\nEpoch 9/10\n250/250 [==============================] - 1s 4ms/step - loss: 0.0245 - val_loss: 0.1165\nEpoch 10/10\n250/250 [==============================] - 1s 4ms/step - loss: 0.0239 - val_loss: 0.1006\n"
],
[
"def create_time_steps(length):\n return list(range(-length, 0))\n\ndef show_plot(plot_data, delta, title):\n labels = ['History', 'True Future', 'Model Prediction']\n marker = ['.-', 'rx', 'go']\n time_steps = create_time_steps(plot_data[0].shape[0])\n if delta:\n future = delta\n else:\n future = 0\n\n plt.title(title)\n for i, x in enumerate(plot_data):\n if i:\n plt.plot(future, plot_data[i], marker[i], markersize=10,\n label=labels[i])\n else:\n plt.plot(time_steps, plot_data[i].flatten(), marker[i], label=labels[i])\n plt.legend()\n plt.xlim([time_steps[0], (future+5)*2])\n plt.xlabel('Week (defined by Wednesdays)')\n plt.ylabel('Normalized Cases')\n return plt",
"_____no_output_____"
],
[
"for x, y in train_data_single.take(10):\n #print(lstm_prediction_model.predict(x))\n plot = show_plot([x[0][:, 1].numpy(), y[0].numpy(),\n lstm_prediction_model.predict(x)[0]], 0,\n 'Training Data Prediction')\n plot.show()",
"_____no_output_____"
],
[
"for x, y in test_data_single.take(1):\n plot = show_plot([x[0][:, 1].numpy(), y[0].numpy(),\n lstm_prediction_model.predict(x)[0]], 0,\n 'Florida COVID Case Prediction, Single Week')\n plot.show()",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
c51252b9a6b53f76ff37e9b0153bd424ae9ea8ac
| 130,790 |
ipynb
|
Jupyter Notebook
|
Healthcare_Project/semi_proposal/Project_Figure_2.ipynb
|
vasudhathinks/Personal-Projects
|
7e59ddf4d3277efcb524f1e046528f7e9e8def18
|
[
"MIT"
] | null | null | null |
Healthcare_Project/semi_proposal/Project_Figure_2.ipynb
|
vasudhathinks/Personal-Projects
|
7e59ddf4d3277efcb524f1e046528f7e9e8def18
|
[
"MIT"
] | null | null | null |
Healthcare_Project/semi_proposal/Project_Figure_2.ipynb
|
vasudhathinks/Personal-Projects
|
7e59ddf4d3277efcb524f1e046528f7e9e8def18
|
[
"MIT"
] | null | null | null | 783.173653 | 125,204 | 0.956113 |
[
[
[
"### Background and Overview:\nThe [MIMIC-III](https://mimic.mit.edu/about/mimic/) (Medical Information Mart for Intensive Care) Clinical Database is comprised of deidentified health-related data associated with over 40,000 patients (available through request). Its 26 tables have a vast amount of information on the patients who stayed in critical care units of the Beth Israel Deaconess Medical Center between 2001 and 2012 ranging from patient demographics to lab reports to detailed clinical notes. ",
"_____no_output_____"
],
[
"#### Figure 2: Compares an Unstructured Data Field's Values for Alive and Deceased Patients\nThis analysis was done on 1,000 patients, with a 50/50 split between patients who were marked alive and those who were marked deceased. It explores the different reasons patients are admitted to the hospital. ",
"_____no_output_____"
]
],
[
[
"# import pandas as pd\n# import numpy as np\n# import matplotlib.pyplot as plt\n\nfrom IPython.display import Image",
"_____no_output_____"
],
[
"\"\"\"NOTE: This code block is commented out because I have not uploaded data files or source code; it is only here to show to the process.\"\"\"\n\n# # Set up path/s to file/s\n# path_to_data = '../data/'\n\n# # Read data\n# alive_admissions = pd.read_csv(path_to_data + 'alive_admissions.csv', header=None, \n# names=['patient_id', 'flag', 'type'])\n# deceased_admissions = pd.read_csv(path_to_data + 'deceased_admissions.csv', header=None,\n# names=['patient_id', 'flag', 'type'])\n\n# # Process data\n# alive_type_count = alive_admissions.groupby(['type']).size().reset_index()\n# deceased_type_count = deceased_admissions.groupby(['type']).size().reset_index()",
"_____no_output_____"
],
[
"\"\"\"NOTE: This code block is also commented out as above; it is only here to show to the process.\"\"\"\n\n# # Plot the data\n# n_bars = 3\n# index = np.arange(n_bars)\n# bar_width = 0.5\n\n# fig2 = plt.subplots()\n# plt.bar(index, alive_type_count[0], bar_width, alpha=0.5, label='alive')\n# plt.bar(index + bar_width / 2, deceased_type_count[0], bar_width, alpha=0.5, label='deceased')\n# plt.xticks(index + bar_width / 4, alive_type_count['type'])\n# plt.ylabel(\"Frequency\")\n# plt.xlabel(\"Admission Type\")\n# plt.legend(loc='upper right')\n# plt.title('Type of Admissions for Alive & Deceased Patients')\n\n# plt.tight_layout()\n# plt.show()",
"_____no_output_____"
],
[
"Image(filename='Figure_2.png')",
"_____no_output_____"
]
],
[
[
"**Figure 2** reviews the reasons behind why 1,000 patients were admitted to the hospital for each visit. The admission types are segmented by whether the patients were marked alive or deceased. Figure 2 shows that, overall, deceased patients tended to have more emergency-type visits, whereas alive patients tended to have more elective-based visits. This data field can be converted to another feature (e.g.: a normalized number of visits per patient, altogether or for each type of admission) to be added to the feature vectors for training a predictive model. ",
"_____no_output_____"
],
[
"**_Note:_** *as a starting point, I used 1,000 patients. I would like to expand this to a larger subset of the 46,000+ patients available through MIMIC-III. Additionally, I intend to review other unstructured data fields, such as diagnosis description and clinical notes.*",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
]
] |
c51266470d6094c9cc990ae321228a0ef6e97721
| 84,141 |
ipynb
|
Jupyter Notebook
|
04 - KNN & Scikit-learn/Part 1 - K Nearest Neighbor.ipynb
|
AdamArthurF/supervised_learning
|
4cb90c1503b57e685c9bb0721964c3fe93e274df
|
[
"MIT"
] | 1 |
2021-08-28T05:38:19.000Z
|
2021-08-28T05:38:19.000Z
|
04 - KNN & Scikit-learn/Part 1 - K Nearest Neighbor.ipynb
|
AdamArthurF/supervised_learning
|
4cb90c1503b57e685c9bb0721964c3fe93e274df
|
[
"MIT"
] | null | null | null |
04 - KNN & Scikit-learn/Part 1 - K Nearest Neighbor.ipynb
|
AdamArthurF/supervised_learning
|
4cb90c1503b57e685c9bb0721964c3fe93e274df
|
[
"MIT"
] | null | null | null | 858.581633 | 82,185 | 0.952698 |
[
[
[
"from luwiji.knn import illustration, demo",
"_____no_output_____"
],
[
"demo.knn()",
"_____no_output_____"
],
[
"illustration.knn_distance",
"_____no_output_____"
]
],
[
[
"### Other Distance Metric",
"_____no_output_____"
],
[
"https://scikit-learn.org/stable/modules/generated/sklearn.neighbors.DistanceMetric.html",
"_____no_output_____"
]
]
] |
[
"code",
"markdown"
] |
[
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
]
] |
c51266714a754c6f85570f619a3d59b12267998e
| 134,662 |
ipynb
|
Jupyter Notebook
|
docs/notebooks/Guide_for_Authors.ipynb
|
MaxCamillo/debuggingbook
|
9675706d2f089929aeb8211a508508b9d3e9348d
|
[
"MIT"
] | null | null | null |
docs/notebooks/Guide_for_Authors.ipynb
|
MaxCamillo/debuggingbook
|
9675706d2f089929aeb8211a508508b9d3e9348d
|
[
"MIT"
] | null | null | null |
docs/notebooks/Guide_for_Authors.ipynb
|
MaxCamillo/debuggingbook
|
9675706d2f089929aeb8211a508508b9d3e9348d
|
[
"MIT"
] | null | null | null | 49.727474 | 42,388 | 0.725691 |
[
[
[
"# Guide for Authors",
"_____no_output_____"
]
],
[
[
"print('Welcome to \"The Debugging Book\"!')",
"Welcome to \"The Debugging Book\"!\n"
]
],
[
[
"This notebook compiles the most important conventions for all chapters (notebooks) of \"The Debugging Book\".",
"_____no_output_____"
],
[
"## Organization of this Book",
"_____no_output_____"
],
[
"### Chapters as Notebooks\n\nEach chapter comes in its own _Jupyter notebook_. A single notebook (= a chapter) should cover the material (text and code, possibly slides) for a 90-minute lecture.\n\nA chapter notebook should be named `Topic.ipynb`, where `Topic` is the topic. `Topic` must be usable as a Python module and should characterize the main contribution. If the main contribution of your chapter is a class `FooDebugger`, for instance, then your topic (and notebook name) should be `FooDebugger`, such that users can state\n\n```python\nfrom FooDebugger import FooDebugger\n```\n\nSince class and module names should start with uppercase letters, all non-notebook files and folders start with lowercase letters. this may make it easier to differentiate them. The special notebook `index.ipynb` gets converted into the home pages `index.html` (on fuzzingbook.org) and `README.md` (on GitHub).\n\nNotebooks are stored in the `notebooks` folder.",
"_____no_output_____"
],
[
"### DebuggingBook and FuzzingBook\n\nThis project shares some infrastructure (and even chapters) with \"The Fuzzing Book\", established through _symbolic links_. Your file organization should be such that `debuggingbook` and `fuzzingbook` are checked out in the same folder; otherwise, sharing infrastructure will not work\n\n```\n<some folder>\n|- fuzzingbook\n|- debuggingbook (this project folder)\n```\n\nTo check whether the organization fits, check whether the `debuggingbook` `Makefile` properly points to `../fuzzingbook/Makefile` - that is, the `fuzzingbook` `Makefile`. If you can properly open the (shared) `Makefile` in both projects, things are set up properly.",
"_____no_output_____"
],
[
"### Output Formats\n\nThe notebooks by themselves can be used by instructors and students to toy around with. They can edit code (and text) as they like and even run them as a slide show.\n\nThe notebook can be _exported_ to multiple (non-interactive) formats:\n\n* HTML – for placing this material online.\n* PDF – for printing\n* Python – for coding\n* Slides – for presenting\n\nThe included Makefile can generate all of these automatically (and a few more).\n\nAt this point, we mostly focus on HTML and Python, as we want to get these out quickly; but you should also occasionally ensure that your notebooks can (still) be exported into PDF. Other formats (Word, Markdown) are experimental.",
"_____no_output_____"
],
[
"## Sites\n\nAll sources for the book end up on the [Github project page](https://github.com/uds-se/debuggingbook). This holds the sources (notebooks), utilities (Makefiles), as well as an issue tracker.\n\nThe derived material for the book ends up in the `docs/` folder, from where it is eventually pushed to the [debuggingbook website](http://www.debuggingbook.org/). This site allows to read the chapters online, can launch Jupyter notebooks using the binder service, and provides access to code and slide formats. Use `make publish` to create and update the site.",
"_____no_output_____"
],
[
"### The Book PDF\n\nThe book PDF is compiled automatically from the individual notebooks. Each notebook becomes a chapter; references are compiled in the final chapter. Use `make book` to create the book.",
"_____no_output_____"
],
[
"## Creating and Building",
"_____no_output_____"
],
[
"### Tools you will need\n\nTo work on the notebook files, you need the following:\n\n1. Jupyter notebook. The easiest way to install this is via the [Anaconda distribution](https://www.anaconda.com/download/).\n\n2. Once you have the Jupyter notebook installed, you can start editing and coding right away by starting `jupyter notebook` (or `jupyter lab`) in the topmost project folder.\n\n3. If (like me) you don't like the Jupyter Notebook interface, I recommend [Jupyter Lab](https://jupyterlab.readthedocs.io/en/stable/), the designated successor to Jupyter Notebook. Invoke it as `jupyter lab`. It comes with a much more modern interface, but misses autocompletion and a couple of extensions. I am running it [as a Desktop application](http://christopherroach.com/articles/jupyterlab-desktop-app/) which gets rid of all the browser toolbars.\nOn the Mac, there is also the [Pineapple app](https://nwhitehead.github.io/pineapple/), which integrates a nice editor with a local server. This is easy to use, but misses a few features; also, it hasn't seen updates since 2015.\n\n4. To create the entire book (with citations, references, and all), you also need the [ipybublish](https://github.com/chrisjsewell/ipypublish) package. This allows you to create the HTML files, merge multiple chapters into a single PDF or HTML file, create slides, and more. The Makefile provides the essential tools for creation.\n",
"_____no_output_____"
],
[
"### Version Control\n\nWe use git in a single strand of revisions. Feel free branch for features, but eventually merge back into the main \"master\" branch. Sync early; sync often. Only push if everything (\"make all\") builds and passes.\n\nThe Github repo thus will typically reflect work in progress. If you reach a stable milestone, you can push things on the fuzzingbook.org web site, using `make publish`.",
"_____no_output_____"
],
[
"#### nbdime\n\nThe [nbdime](https://github.com/jupyter/nbdime) package gives you tools such as `nbdiff` (and even better, `nbdiff-web`) to compare notebooks against each other; this ensures that cell _contents_ are compared rather than the binary format.\n\n`nbdime config-git --enable` integrates nbdime with git such that `git diff` runs the above tools; merging should also be notebook-specific.",
"_____no_output_____"
],
[
"#### nbstripout\n\nNotebooks in version control _should not contain output cells,_ as these tend to change a lot. (Hey, we're talking random output generation here!) To have output cells automatically stripped during commit, install the [nbstripout](https://github.com/kynan/nbstripout) package and use\n\n```\nnbstripout --install\n```\n\nto set it up as a git filter. The `notebooks/` folder comes with a `.gitattributes` file already set up for `nbstripout`, so you should be all set.\n\nNote that _published_ notebooks (in short, anything under the `docs/` tree _should_ have their output cells included, such that users can download and edit notebooks with pre-rendered output. This folder contains a `.gitattributes` file that should explicitly disable `nbstripout`, but it can't hurt to check.\n\nAs an example, the following cell \n\n1. _should_ have its output included in the [HTML version of this guide](https://www.debuggingbook.org/beta/html/Guide_for_Authors.html);\n2. _should not_ have its output included in [the git repo](https://github.com/uds-se/debuggingbook/blob/master/notebooks/Guide_for_Authors.ipynb) (`notebooks/`);\n3. _should_ have its output included in [downloadable and editable notebooks](https://github.com/uds-se/debuggingbook/blob/master/docs/beta/notebooks/Guide_for_Authors.ipynb) (`docs/notebooks/` and `docs/beta/notebooks/`).",
"_____no_output_____"
]
],
[
[
"import random",
"_____no_output_____"
],
[
"random.random()",
"_____no_output_____"
]
],
[
[
"### Inkscape and GraphViz\n\nCreating derived files uses [Inkscape](https://inkscape.org/en/) and [Graphviz](https://www.graphviz.org/) – through its [Python wrapper](https://pypi.org/project/graphviz/) – to process SVG images. These tools are not automatically installed, but are available on pip, _brew_ and _apt-get_ for all major distributions.",
"_____no_output_____"
],
[
"### LaTeX Fonts\n\nBy default, creating PDF uses XeLaTeX with a couple of special fonts, which you can find in the `fonts/` folder; install these fonts system-wide to make them accessible to XeLaTeX.\n\nYou can also run `make LATEX=pdflatex` to use `pdflatex` and standard LaTeX fonts instead.",
"_____no_output_____"
],
[
"### Creating Derived Formats (HTML, PDF, code, ...)\n\nThe [Makefile](../Makefile) provides rules for all targets. Type `make help` for instructions.\n\nThe Makefile should work with GNU make and a standard Jupyter Notebook installation. To create the multi-chapter book and BibTeX citation support, you need to install the [iPyPublish](https://github.com/chrisjsewell/ipypublish) package (which includes the `nbpublish` command).",
"_____no_output_____"
],
[
"### Creating a New Chapter\n\nTo create a new chapter for the book,\n\n1. Set up a new `.ipynb` notebook file as copy of [Template.ipynb](Template.ipynb).\n2. Include it in the `CHAPTERS` list in the `Makefile`.\n3. Add it to the git repository.",
"_____no_output_____"
],
[
"## Teaching a Topic\n\nEach chapter should be devoted to a central concept and a small set of lessons to be learned. I recommend the following structure:\n\n* Introduce the problem (\"We want to parse inputs\")\n* Illustrate it with some code examples (\"Here's some input I'd like to parse\")\n* Develop a first (possibly quick and dirty) solution (\"A PEG parser is short and often does the job\"_\n* Show that it works and how it works (\"Here's a neat derivation tree. Look how we can use this to mutate and combine expressions!\")\n* Develop a second, more elaborated solution, which should then become the main contribution. (\"Here's a general LR(1) parser that does not require a special grammar format. (You can skip it if you're not interested)\")\n* Offload non-essential extensions to later sections or to exercises. (\"Implement a universal parser, using the Dragon Book\")\n\nThe key idea is that readers should be able to grasp the essentials of the problem and the solution in the beginning of the chapter, and get further into details as they progress through it. Make it easy for readers to be drawn in, providing insights of value quickly. If they are interested to understand how things work, they will get deeper into the topic. If they just want to use the technique (because they may be more interested in later chapters), having them read only the first few examples should be fine for them, too.\n\nWhatever you introduce should be motivated first, and illustrated after. Motivate the code you'll be writing, and use plenty of examples to show what the code just introduced is doing. Remember that readers should have fun interacting with your code and your examples. Show and tell again and again and again.",
"_____no_output_____"
],
[
"### Special Sections",
"_____no_output_____"
],
[
"#### Quizzes",
"_____no_output_____"
],
[
"You can have _quizzes_ as part of the notebook. These are created using the `quiz()` function. Its arguments are\n\n* The question\n* A list of options\n* The correct answer(s) - either\n * the single number of the one single correct answer (starting with 1)\n * a list of numbers of correct answers (multiple choices)\n \nTo make the answer less obvious, you can specify it as a string containing an arithmetic expression evaluating to the desired number(s). The expression will remain in the code (and possibly be shown as hint in the quiz).",
"_____no_output_____"
]
],
[
[
"from bookutils import quiz",
"_____no_output_____"
],
[
"# A single-choice quiz\nquiz(\"The color of the sky is\",\n [\n \"blue\",\n \"red\",\n \"black\"\n ], '5 - 4')",
"_____no_output_____"
],
[
"# A multiple-choice quiz\nquiz(\"What is this book?\",\n [\n \"Novel\",\n \"Friendly\",\n \"Useful\"\n ], '[5 - 4, 1 + 1, 27 / 9]')",
"_____no_output_____"
]
],
[
[
"Cells that contain only the `quiz()` call will not be rendered (but the quiz will).",
"_____no_output_____"
],
[
"#### Synopsis",
"_____no_output_____"
],
[
"Each chapter should have a section named \"Synopsis\" at the very end:\n\n```markdown\n## Synopsis\n\nThis is the text of the synopsis.\n```",
"_____no_output_____"
],
[
"This section is evaluated at the very end of the notebook. It should summarize the most important functionality (classes, methods, etc.) together with examples. In the derived HTML and PDF files, it is rendered at the beginning, such that it can serve as a quick reference",
"_____no_output_____"
],
[
"#### Excursions",
"_____no_output_____"
],
[
"There may be longer stretches of text (and code!) that are too special, too boring, or too repetitve to read. You can mark such stretches as \"Excursions\" by enclosing them in MarkDown cells that state:\n\n```markdown\n#### Excursion: TITLE\n```\n\nand\n\n```markdown\n#### End of Excursion\n```",
"_____no_output_____"
],
[
"Stretches between these two markers get special treatment when rendering:\n\n* In the resulting HTML output, these blocks are set up such that they are shown on demand only.\n* In printed (PDF) versions, they will be replaced by a pointer to the online version.\n* In the resulting slides, they will be omitted right away.",
"_____no_output_____"
],
[
"Here is an example of an excursion:",
"_____no_output_____"
],
[
"#### Excursion: Fine points on Excursion Cells",
"_____no_output_____"
],
[
"Note that the `Excursion` and `End of Excursion` cells must be separate cells; they cannot be merged with others.",
"_____no_output_____"
],
[
"#### End of Excursion",
"_____no_output_____"
],
[
"### Ignored Code\n\nIf a code cell starts with\n```python\n# ignore\n```\nthen the code will not show up in rendered input. Its _output_ will, however. \nThis is useful for cells that create drawings, for instance - the focus should be on the result, not the code.\n\nThis also applies to cells that start with a call to `display()` or `quiz()`.",
"_____no_output_____"
],
[
"### Ignored Cells\n\nYou can have _any_ cell not show up at all (including its output) in any rendered input by adding the following meta-data to the cell:\n```json\n{\n \"ipub\": {\n \"ignore\": true\n}\n```\n",
"_____no_output_____"
],
[
"*This* text, for instance, does not show up in the rendered version.",
"_____no_output_____"
],
[
"## Coding",
"_____no_output_____"
],
[
"### Set up\n\nThe first code block in each notebook should be",
"_____no_output_____"
]
],
[
[
"import bookutils",
"_____no_output_____"
]
],
[
[
"This sets up stuff such that notebooks can import each other's code (see below). This import statement is removed in the exported Python code, as the .py files would import each other directly.",
"_____no_output_____"
],
[
"Importing `bookutils` also sets a fixed _seed_ for random number generation. This way, whenever you execute a notebook from scratch (restarting the kernel), you get the exact same results; these results will also end up in the derived HTML and PDF files. (If you run a notebook or a cell for the second time, you will get more random results.)",
"_____no_output_____"
],
[
"### Coding Style and Consistency\n\nHere's a few rules regarding coding style.",
"_____no_output_____"
],
[
"#### Use Python 3\n\nWe use Python 3 (specifically, Python 3.6) for all code. As of 2020, there is no need anymore to include compatibility hacks for Python 2.",
"_____no_output_____"
],
[
"#### Follow Python Coding Conventions\n\nWe use _standard Python coding conventions_ according to [PEP 8](https://www.python.org/dev/peps/pep-0008/).\n\nYour code must pass the `pycodestyle` style checks which you get by invoking `make style`. A very easy way to meet this goal is to invoke `make reformat`, which reformats all code accordingly. The `code prettify` notebook extension also allows you to automatically make your code (mostly) adhere to PEP 8.",
"_____no_output_____"
],
[
"#### One Cell per Definition\n\nUse one cell for each definition or example. During importing, this makes it easier to decide which cells to import (see below).",
"_____no_output_____"
],
[
"#### Identifiers\n\nIn the book, this is how we denote `variables`, `functions()` and `methods()`, `Classes`, `Notebooks`, `variables_and_constants`, `EXPORTED_CONSTANTS`, `files`, `folders/`, and `<grammar-elements>`.",
"_____no_output_____"
],
[
"#### Quotes\n\nIf you have the choice between quoting styles, prefer \n* double quotes (`\"strings\"`) around strings that are used for interpolation or that are natural language messages, and \n* single quotes (`'characters'`) for single characters and formal language symbols that a end user would not see.",
"_____no_output_____"
],
[
"#### Static Type Checking\n\nUse type annotations for all function definitions.",
"_____no_output_____"
],
[
"#### Documentation\n\nUse documentation strings for all public classes and methods.",
"_____no_output_____"
],
[
"#### Read More\n\nBeyond simple syntactical things, here's a [very nice guide](https://docs.python-guide.org/writing/style/) to get you started writing \"pythonic\" code.",
"_____no_output_____"
],
[
"### Importing Code from Notebooks\n\nTo import the code of individual notebooks, you can import directly from .ipynb notebook files.",
"_____no_output_____"
]
],
[
[
"from DeltaDebugger import DeltaDebugger",
"_____no_output_____"
],
[
"def fun(s: str) -> None:\n assert 'a' not in s\n\nwith DeltaDebugger() as dd:\n fun(\"abc\")\ndd",
"_____no_output_____"
]
],
[
[
"**Important**: When importing a notebook, the module loader will **only** load cells that start with\n\n* a function definition (`def`)\n* a class definition (`class`)\n* a variable definition if all uppercase (`ABC = 123`)\n* `import` and `from` statements\n\nAll other cells are _ignored_ to avoid recomputation of notebooks and clutter of `print()` output.",
"_____no_output_____"
],
[
"Exported Python code will import from the respective .py file instead. The exported Python code is set up such that only the above items will be imported.",
"_____no_output_____"
],
[
"If importing a module prints out something (or has other side effects), that is an error. Use `make check-imports` to check whether your modules import without output.",
"_____no_output_____"
],
[
"Import modules only as you need them, such that you can motivate them well in the text.",
"_____no_output_____"
],
[
"### Imports and Dependencies\n\nTry to depend on as few other notebooks as possible. This will not only ease construction and reconstruction of the code, but also reduce requirements for readers, giving then more flexibility in navigating through the book.\n\nWhen you import a notebook, this will show up as a dependency in the [Sitemap](00_Table_of_Contents.ipynb). If the imported module is not critical for understanding, and thus should not appear as a dependency in the sitemap, mark the import as \"minor dependency\" as follows:",
"_____no_output_____"
]
],
[
[
"from Intro_Debugging import remove_html_markup # minor dependency",
"_____no_output_____"
]
],
[
[
"### Design and Architecture\n\nStick to simple functions and data types. We want our readers to focus on functionality, not Python. You are encouraged to write in a \"pythonic\" style, making use of elegant Python features such as list comprehensions, sets, and more; however, if you do so, be sure to explain the code such that readers familiar with, say, C or Java can still understand things.",
"_____no_output_____"
],
[
"### Incomplete Examples\n\nWhen introducing examples for students to complete, use the ellipsis `...` to indicate where students should add code, as in here:",
"_____no_output_____"
]
],
[
[
"def student_example() -> None:\n x = some_computation() # type: ignore\n # Now, do something with x\n ...",
"_____no_output_____"
]
],
[
[
"The ellipsis is legal code in Python 3. (Actually, it is an `Ellipsis` object.)",
"_____no_output_____"
],
[
"### Introducing Classes\n\nDefining _classes_ can be a bit tricky, since all of a class must fit into a single cell. This defeats the incremental style preferred for notebooks. By defining a class _as a subclass of itself_, though, you can avoid this problem.",
"_____no_output_____"
],
[
"Here's an example. We introduce a class `Foo`:",
"_____no_output_____"
]
],
[
[
"class Foo:\n def __init__(self) -> None:\n pass\n\n def bar(self) -> None:\n pass",
"_____no_output_____"
]
],
[
[
"Now we could discuss what `__init__()` and `bar()` do, or give an example of how to use them:",
"_____no_output_____"
]
],
[
[
"f = Foo()\nf.bar()",
"_____no_output_____"
]
],
[
[
"We now can introduce a new `Foo` method by subclassing from `Foo` into a class which is _also_ called `Foo`:",
"_____no_output_____"
]
],
[
[
"class Foo(Foo):\n def baz(self) -> None:\n pass",
"_____no_output_____"
]
],
[
[
"This is the same as if we had subclassed `Foo` into `Foo_1` with `Foo` then becoming an alias for `Foo_1`. The original `Foo` class is overshadowed by the new one:",
"_____no_output_____"
]
],
[
[
"new_f = Foo()\nnew_f.baz()",
"_____no_output_____"
]
],
[
[
"Note, though, that _existing_ objects keep their original class:",
"_____no_output_____"
]
],
[
[
"from ExpectError import ExpectError",
"_____no_output_____"
],
[
"with ExpectError(AttributeError):\n f.baz() # type: ignore",
"Traceback (most recent call last):\n File \"<ipython-input-1-12b6020f98db>\", line 2, in <module>\n f.baz() # type: ignore\nAttributeError: 'Foo' object has no attribute 'baz' (expected)\n"
]
],
[
[
"## Helpers\n\nThere's a couple of notebooks with helpful functions, including [Timer](Timer.ipynb), [ExpectError and ExpectTimeout](ExpectError.ipynb). Also check out the [Tracer](Tracer.ipynb) class.",
"_____no_output_____"
],
[
"### Quality Assurance\n\nIn your code, make use of plenty of assertions that allow to catch errors quickly. These assertions also help your readers understand the code.",
"_____no_output_____"
],
[
"### Issue Tracker\n\nThe [Github project page](https://github.com/uds-se/debuggingbook) allows to enter and track issues.",
"_____no_output_____"
],
[
"## Writing Text\n\nText blocks use Markdown syntax. [Here is a handy guide](https://github.com/adam-p/markdown-here/wiki/Markdown-Cheatsheet).\n",
"_____no_output_____"
],
[
"### Sections\n\nAny chapter notebook must begin with `# TITLE`, and sections and subsections should then follow by `## SECTION` and `### SUBSECTION`.\n\nSections should start with their own block, to facilitate cross-referencing.\n",
"_____no_output_____"
],
[
"### Highlighting\n\nUse\n\n* _emphasis_ (`_emphasis_`) for highlighting,\n* *emphasis* (`*emphasis*`) for highlighting terms that will go into the index,\n* `backticks` for code and other verbatim elements.",
"_____no_output_____"
],
[
"### Hyphens and Dashes\n\nUse \"–\" for em-dashes, \"-\" for hyphens, and \"$-$\" for minus.",
"_____no_output_____"
],
[
"### Quotes\n\nUse standard typewriter quotes (`\"quoted string\"`) for quoted text. The PDF version will automatically convert these to \"smart\" (e.g. left and right) quotes.",
"_____no_output_____"
],
[
"### Lists and Enumerations\n\nYou can use bulleted lists:\n\n* Item A\n* Item B\n\nand enumerations:\n\n1. item 1\n1. item 2\n\nFor description lists, use a combination of bulleted lists and highlights:\n\n* **PDF** is great for reading offline\n* **HTML** is great for reading online\n",
"_____no_output_____"
],
[
"\n### Math\n\nLaTeX math formatting works, too.\n\n`$x = \\sum_{n = 1}^{\\infty}\\frac{1}{n}$` gets you\n$x = \\sum_{n = 1}^{\\infty}\\frac{1}{n}$.\n",
"_____no_output_____"
],
[
"### Inline Code\n\nPython code normally goes into its own cells, but you can also have it in the text:\n\n```python\ns = \"Python syntax highlighting\"\nprint(s)\n```",
"_____no_output_____"
],
[
"### Images\n\nTo insert images, use Markdown syntax `{width=100%}` inserts a picture from the `PICS` folder.",
"_____no_output_____"
],
[
"{width=100%}",
"_____no_output_____"
],
[
"All pictures go to `PICS/`, both in source as well as derived formats; both are stored in git, too. (Not all of us have all tools to recreate diagrams, etc.)",
"_____no_output_____"
],
[
"### Footnotes\n\nMarkdown supports footnotes, as in [^footnote]. These are rendered as footnotes in HTML and PDF, _but not within Jupyter_; hence, readers may find them confusing. So far, the book makes no use of footnotes, and uses parenthesized text instead.\n\n[^footnote]: Test, [Link](https://www.fuzzingbook.org).",
"_____no_output_____"
],
[
"### Floating Elements and References\n\n\\todo[inline]{I haven't gotten this to work yet -- AZ}\n\nTo produce floating elements in LaTeX and PDF, edit the metadata of the cell which contains it. (In the Jupyter Notebook Toolbar go to View -> Cell Toolbar -> Edit Metadata and a button will appear above each cell.) This allows you to control placement and create labels.",
"_____no_output_____"
],
[
"#### Floating Figures\n\nEdit metadata as follows:\n\n```json\n{\n\"ipub\": {\n \"figure\": {\n \"caption\": \"Figure caption.\",\n \"label\": \"fig:flabel\",\n \"placement\": \"H\",\n\t\"height\":0.4,\n \"widefigure\": false,\n }\n }\n}\n```\n\n- all tags are optional\n- height/width correspond to the fraction of the page height/width, only one should be used (aspect ratio will be maintained automatically)\n- `placement` is optional and constitutes using a placement arguments for the figure (e.g. \\begin{figure}[H]). See [Positioning_images_and_tables](https://www.sharelatex.com/learn/Positioning_images_and_tables).\n- `widefigure` is optional and constitutes expanding the figure to the page width (i.e. \\begin{figure*}) (placement arguments will then be ignored)",
"_____no_output_____"
],
[
"#### Floating Tables\n\nFor **tables** (e.g. those output by `pandas`), enter in cell metadata:\n\n```json\n{\n\"ipub\": {\n \"table\": {\n\t \"caption\": \"Table caption.\",\n\t \"label\": \"tbl:tlabel\",\n\t \"placement\": \"H\",\n \"alternate\": \"gray!20\"\n\t }\n }\n}\n```\n\n- `caption` and `label` are optional\n- `placement` is optional and constitutes using a placement arguments for the table (e.g. \\begin{table}[H]). See [Positioning_images_and_tables](https://www.sharelatex.com/learn/Positioning_images_and_tables).\n- `alternate` is optional and constitutes using alternating colors for the table rows (e.g. \\rowcolors{2}{gray!25}{white}). See (https://tex.stackexchange.com/a/5365/107738)[https://tex.stackexchange.com/a/5365/107738].\n- if tables exceed the text width, in latex, they will be shrunk to fit \n\n\n#### Floating Equations\n\nFor **equations** (e.g. those output by `sympy`), enter in cell metadata:\n\n```json\n{\n \"ipub\": {\n\t \"equation\": {\n \"environment\": \"equation\",\n\t \"label\": \"eqn:elabel\"\n\t }\n }\n}\n```\n\n- environment is optional and can be 'none' or any of those available in [amsmath](https://www.sharelatex.com/learn/Aligning_equations_with_amsmath); 'equation', 'align','multline','gather', or their \\* variants. Additionally, 'breqn' or 'breqn\\*' will select the experimental [breqn](https://ctan.org/pkg/breqn) environment to *smart* wrap long equations. \n- label is optional and will only be used if the equation is in an environment\n\n\n#### References\n\nTo reference a floating object, use `\\cref`, e.g. \\cref{eq:texdemo}",
"_____no_output_____"
],
[
"### Cross-Referencing\n\n#### Section References\n\n* To refer to sections in the same notebook, use the header name as anchor, e.g. \n`[Code](#Code)` gives you [Code](#Code). For multi-word titles, replace spaces by hyphens (`-`), as in [Using Notebooks as Modules](#Using-Notebooks-as-Modules).\n\n* To refer to cells (e.g. equations or figures), you can define a label as cell metadata. See [Floating Elements and References](#Floating-Elements-and-References) for details.\n\n* To refer to other notebooks, use a Markdown cross-reference to the notebook file, e.g. [the \"Debugger\" chapter](Debugger.ipynb). A special script will be run to take care of these links. Reference chapters by name, not by number.",
"_____no_output_____"
],
[
"### Citations\n\nTo cite papers, cite in LaTeX style. The text",
"_____no_output_____"
]
],
[
[
"print(r\"\\cite{Purdom1972}\")",
"\\cite{Purdom1972}\n"
]
],
[
[
"is expanded to \\cite{Purdom1972}, which in HTML and PDF should be a nice reference.\nThe keys refer to BibTeX entries in [fuzzingbook.bib](fuzzingbook.bib). \n* LaTeX/PDF output will have a \"References\" section appended.\n* HTML output will link to the URL field from the BibTeX entry. Be sure it points to the DOI.",
"_____no_output_____"
],
[
"### Todo's\n\n* To mark todo's, use `\\todo{Thing to be done}.` \\todo{Expand this}",
"_____no_output_____"
],
[
"### Tables\n\nTables with fixed contents can be produced using Markdown syntax:\n\n| Tables | Are | Cool |\n| ------ | ---:| ----:|\n| Zebra | 2 | 30 |\n| Gnu | 20 | 400 |\n",
"_____no_output_____"
],
[
"If you want to produce tables from Python data, the `PrettyTable` package (included in the book) allows to [produce tables with LaTeX-style formatting.](http://blog.juliusschulz.de/blog/ultimate-ipython-notebook)",
"_____no_output_____"
]
],
[
[
"from bookutils import PrettyTable as pt",
"_____no_output_____"
],
[
"import numpy as np",
"_____no_output_____"
],
[
"data = np.array([[1, 2, 30], [2, 3, 400]])\npt.PrettyTable(data, [r\"$\\frac{a}{b}$\", r\"$b$\",\n r\"$c$\"], print_latex_longtable=False)",
"_____no_output_____"
]
],
[
[
"### Plots and Data\n\nIt is possible to include plots in notebooks. Here is an example of plotting a function:",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\n\nimport matplotlib.pyplot as plt\n\nx = np.linspace(0, 3 * np.pi, 500)\nplt.plot(x, np.sin(x ** 2))\nplt.title('A simple chirp');",
"_____no_output_____"
]
],
[
[
"And here's an example of plotting data:",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\n\nimport matplotlib.pyplot as plt\ndata = [25, 36, 57]\nplt.plot(data)\nplt.title('Increase in data');",
"_____no_output_____"
]
],
[
[
"Plots are available in all derived versions (HTML, PDF, etc.) Plots with `plotly` are even nicer (and interactive, even in HTML), However, at this point, we cannot export them to PDF, so `matplotlib` it is.",
"_____no_output_____"
],
[
"## Slides\n\nYou can set up the notebooks such that they also can be presented as slides. In the browser, select View -> Cell Toolbar -> Slideshow. You can then select a slide type for each cell:\n\n* `New slide` starts a new slide with the cell (typically, every `## SECTION` in the chapter)\n* `Sub-slide` starts a new sub-slide which you navigate \"down\" to (anything in the section)\n* `Fragment` is a cell that gets revealed after a click (on the same slide)\n* `Skip` is skipped during the slide show (e.g. `import` statements; navigation guides)\n* `Notes` goes into presenter notes\n\nTo create slides, do `make slides`; to view them, change into the `slides/` folder and open the created HTML files. (The `reveal.js` package has to be in the same folder as the slide to be presented.)\n\nThe ability to use slide shows is a compelling argument for teachers and instructors in our audience.",
"_____no_output_____"
],
[
"(Hint: In a slide presentation, type `s` to see presenter notes.)",
"_____no_output_____"
],
[
"## Writing Tools\n\nWhen you're editing in the browser, you may find these extensions helpful:\n\n### Jupyter Notebook\n\n[Jupyter Notebook Extensions](https://github.com/ipython-contrib/jupyter_contrib_nbextensions) is a collection of productivity-enhancing tools (including spellcheckers).\n\nI found these extensions to be particularly useful:\n\n * Spell Checker (while you're editing)\n \n * Table of contents (for quick navigation)\n\n * Code prettify (to produce \"nice\" syntax)\n \n * Codefolding\n \n * Live Markdown Preview (while you're editing)\n\n### Jupyter Lab\n\nExtensions for _Jupyter Lab_ are much less varied and less supported, but things get better. I am running\n\n * [Spell Checker](https://github.com/ijmbarr/jupyterlab_spellchecker)\n \n * [Table of Contents](https://github.com/jupyterlab/jupyterlab-toc)\n\n * [JupyterLab-LSP](https://towardsdatascience.com/jupyterlab-2-0-edd4155ab897) providing code completion, signatures, style checkers, and more.\n",
"_____no_output_____"
],
[
"## Interaction\n\nIt is possible to include interactive elements in a notebook, as in the following example:",
"_____no_output_____"
],
[
"```python\ntry:\n from ipywidgets import interact, interactive, fixed, interact_manual\n\n x = interact(fuzzer, char_start=(32, 128), char_range=(0, 96))\nexcept ImportError:\n pass\n```",
"_____no_output_____"
],
[
"Note that such elements will be present in the notebook versions only, but not in the HTML and PDF versions, so use them sparingly (if at all). To avoid errors during production of derived files, protect against `ImportError` exceptions as in the above example.",
"_____no_output_____"
],
[
"## Read More\n\nHere is some documentation on the tools we use:\n\n1. [Markdown Cheatsheet](https://github.com/adam-p/markdown-here/wiki/Markdown-Cheatsheet) - general introduction to Markdown\n\n1. [iPyPublish](https://github.com/chrisjsewell/ipypublish) - rich set of tools to create documents with citations and references\n",
"_____no_output_____"
],
[
"\n\n## Alternative Tool Sets\n\nWe don't currently use these, but they are worth learning:\n\n1. [Making Publication-Ready Python Notebooks](http://blog.juliusschulz.de/blog/ultimate-ipython-notebook) - Another tool set on how to produce book chapters from notebooks\n\n1. [Writing academic papers in plain text with Markdown and Jupyter notebook](https://sylvaindeville.net/2015/07/17/writing-academic-papers-in-plain-text-with-markdown-and-jupyter-notebook/) - Alternate ways on how to generate citations\n\n1. [A Jupyter LaTeX template](https://gist.github.com/goerz/d5019bedacf5956bcf03ca8683dc5217#file-revtex-tplx) - How to define a LaTeX template\n\n1. [Boost Your Jupyter Notebook Productivity](https://towardsdatascience.com/jupyter-notebook-hints-1f26b08429ad) - a collection of hints for debugging and profiling Jupyter notebooks\n\n",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
c51272984b44b5a2c82fc7e5c19b2b0a64c0758d
| 38,093 |
ipynb
|
Jupyter Notebook
|
site/en/r2/guide/keras/custom_layers_and_models.ipynb
|
jackreeceejini/docs
|
ef0909c8037fccdf3ff88d6550bbe121f4e81738
|
[
"Apache-2.0"
] | 2 |
2021-07-05T19:07:31.000Z
|
2021-11-17T11:09:30.000Z
|
site/en/r2/guide/keras/custom_layers_and_models.ipynb
|
jackreeceejini/docs
|
ef0909c8037fccdf3ff88d6550bbe121f4e81738
|
[
"Apache-2.0"
] | null | null | null |
site/en/r2/guide/keras/custom_layers_and_models.ipynb
|
jackreeceejini/docs
|
ef0909c8037fccdf3ff88d6550bbe121f4e81738
|
[
"Apache-2.0"
] | null | null | null | 36.141366 | 298 | 0.507547 |
[
[
[
"##### Copyright 2019 The TensorFlow Authors.",
"_____no_output_____"
]
],
[
[
"#@title Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# https://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.",
"_____no_output_____"
]
],
[
[
"# Writing layers and models with TensorFlow Keras",
"_____no_output_____"
],
[
"<table class=\"tfo-notebook-buttons\" align=\"left\">\n <td>\n <a target=\"_blank\" href=\"https://www.tensorflow.org/alpha/guide/keras/custom_layers_and_models\"><img src=\"https://www.tensorflow.org/images/tf_logo_32px.png\" />View on TensorFlow.org</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://colab.research.google.com/github/tensorflow/docs/blob/master/site/en/r2/guide/keras/custom_layers_and_models.ipynb\"><img src=\"https://www.tensorflow.org/images/colab_logo_32px.png\" />Run in Google Colab</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://github.com/tensorflow/docs/blob/master/site/en/r2/guide/keras/custom_layers_and_models.ipynb\"><img src=\"https://www.tensorflow.org/images/GitHub-Mark-32px.png\" />View source on GitHub</a>\n </td>\n</table>",
"_____no_output_____"
],
[
"### Setup",
"_____no_output_____"
]
],
[
[
"from __future__ import absolute_import, division, print_function\n\n!pip install tensorflow-gpu==2.0.0-alpha0\nimport tensorflow as tf\n\ntf.keras.backend.clear_session() # For easy reset of notebook state.",
"_____no_output_____"
]
],
[
[
"## The Layer class\n\n",
"_____no_output_____"
],
[
"### Layers encapsulate a state (weights) and some computation\n\nThe main data structure you'll work with is the `Layer`.\nA layer encapsulates both a state (the layer's \"weights\")\nand a transformation from inputs to outputs (a \"call\", the layer's\nforward pass).\n\nHere's a densely-connected layer. It has a state: the variables `w` and `b`.\n",
"_____no_output_____"
]
],
[
[
"from tensorflow.keras import layers\n\n\nclass Linear(layers.Layer):\n\n def __init__(self, units=32, input_dim=32):\n super(Linear, self).__init__()\n w_init = tf.random_normal_initializer()\n self.w = tf.Variable(initial_value=w_init(shape=(input_dim, units),\n dtype='float32'),\n trainable=True)\n b_init = tf.zeros_initializer()\n self.b = tf.Variable(initial_value=b_init(shape=(units,),\n dtype='float32'),\n trainable=True)\n\n def call(self, inputs):\n return tf.matmul(inputs, self.w) + self.b\n\nx = tf.ones((2, 2))\nlinear_layer = Linear(4, 2)\ny = linear_layer(x)\nprint(y)",
"_____no_output_____"
]
],
[
[
"Note that the weights `w` and `b` are automatically tracked by the layer upon\nbeing set as layer attributes:",
"_____no_output_____"
]
],
[
[
"assert linear_layer.weights == [linear_layer.w, linear_layer.b]",
"_____no_output_____"
]
],
[
[
"Note you also have access to a quicker shortcut for adding weight to a layer: the `add_weight` method:\n\n",
"_____no_output_____"
]
],
[
[
"class Linear(layers.Layer):\n\n def __init__(self, units=32, input_dim=32):\n super(Linear, self).__init__()\n self.w = self.add_weight(shape=(input_dim, units),\n initializer='random_normal',\n trainable=True)\n self.b = self.add_weight(shape=(units,),\n initializer='zeros',\n trainable=True)\n\n def call(self, inputs):\n return tf.matmul(inputs, self.w) + self.b\n\nx = tf.ones((2, 2))\nlinear_layer = Linear(4, 2)\ny = linear_layer(x)\nprint(y)",
"_____no_output_____"
]
],
[
[
"#### Layers can have non-trainable weights\n\nBesides trainable weights, you can add non-trainable weights to a layer as well.\nSuch weights are meant not to be taken into account during backpropagation,\nwhen you are training the layer.\n\nHere's how to add and use a non-trainable weight:",
"_____no_output_____"
]
],
[
[
"class ComputeSum(layers.Layer):\n\n def __init__(self, input_dim):\n super(ComputeSum, self).__init__()\n self.total = tf.Variable(initial_value=tf.zeros((input_dim,)),\n trainable=False)\n\n def call(self, inputs):\n self.total.assign_add(tf.reduce_sum(inputs, axis=0))\n return self.total \n\nx = tf.ones((2, 2))\nmy_sum = ComputeSum(2)\ny = my_sum(x)\nprint(y.numpy())\ny = my_sum(x)\nprint(y.numpy())",
"_____no_output_____"
]
],
[
[
"It's part of `layer.weights`, but it gets categorized as a non-trainable weight:",
"_____no_output_____"
]
],
[
[
"print('weights:', len(my_sum.weights))\nprint('non-trainable weights:', len(my_sum.non_trainable_weights))\n\n# It's not included in the trainable weights:\nprint('trainable_weights:', my_sum.trainable_weights)",
"_____no_output_____"
]
],
[
[
"### Best practice: deferring weight creation until the shape of the inputs is known\n\nIn the logistic regression example above, our `Linear` layer took an `input_dim` argument\nthat was used to compute the shape of the weights `w` and `b` in `__init__`:",
"_____no_output_____"
]
],
[
[
"class Linear(layers.Layer):\n\n def __init__(self, units=32, input_dim=32):\n super(Linear, self).__init__()\n self.w = self.add_weight(shape=(input_dim, units),\n initializer='random_normal',\n trainable=True)\n self.b = self.add_weight(shape=(units,),\n initializer='random_normal',\n trainable=True)",
"_____no_output_____"
]
],
[
[
"In many cases, you may not know in advance the size of your inputs, and you would\nlike to lazily create weights when that value becomes known,\nsome time after instantiating the layer.\n\nIn the Keras API, we recommend creating layer weights in the `build(inputs_shape)` method of your layer.\nLike this:",
"_____no_output_____"
]
],
[
[
"class Linear(layers.Layer):\n\n def __init__(self, units=32):\n super(Linear, self).__init__()\n self.units = units\n\n def build(self, input_shape):\n self.w = self.add_weight(shape=(input_shape[-1], self.units),\n initializer='random_normal',\n trainable=True)\n self.b = self.add_weight(shape=(self.units,),\n initializer='random_normal',\n trainable=True)\n\n def call(self, inputs):\n return tf.matmul(inputs, self.w) + self.b",
"_____no_output_____"
]
],
[
[
"The `__call__` method of your layer will automatically run `build` the first time it is called.\nYou now have a layer that's lazy and easy to use:",
"_____no_output_____"
]
],
[
[
"linear_layer = Linear(32) # At instantiation, we don't know on what inputs this is going to get called\ny = linear_layer(x) # The layer's weights are created dynamically the first time the layer is called",
"_____no_output_____"
]
],
[
[
"\n### Layers are recursively composable\n\nIf you assign a Layer instance as attribute of another Layer,\nthe outer layer will start tracking the weights of the inner layer.\n\nWe recommend creating such sublayers in the `__init__` method (since the sublayers will typically have a `build` method, they will be built when the outer layer gets built).",
"_____no_output_____"
]
],
[
[
"# Let's assume we are reusing the Linear class\n# with a `build` method that we defined above.\n\nclass MLPBlock(layers.Layer):\n\n def __init__(self):\n super(MLPBlock, self).__init__()\n self.linear_1 = Linear(32)\n self.linear_2 = Linear(32)\n self.linear_3 = Linear(1)\n\n def call(self, inputs):\n x = self.linear_1(inputs)\n x = tf.nn.relu(x)\n x = self.linear_2(x)\n x = tf.nn.relu(x)\n return self.linear_3(x)\n \n\nmlp = MLPBlock()\ny = mlp(tf.ones(shape=(3, 64))) # The first call to the `mlp` will create the weights\nprint('weights:', len(mlp.weights))\nprint('trainable weights:', len(mlp.trainable_weights))",
"_____no_output_____"
]
],
[
[
"### Layers recursively collect losses created during the forward pass\n\nWhen writing the `call` method of a layer, you can create loss tensors that you will want to use later, when writing your training loop. This is doable by calling `self.add_loss(value)`:\n",
"_____no_output_____"
]
],
[
[
"# A layer that creates an activity regularization loss\nclass ActivityRegularizationLayer(layers.Layer):\n \n def __init__(self, rate=1e-2):\n super(ActivityRegularizationLayer, self).__init__()\n self.rate = rate\n \n def call(self, inputs):\n self.add_loss(self.rate * tf.reduce_sum(inputs))\n return inputs",
"_____no_output_____"
]
],
[
[
"These losses (including those created by any inner layer) can be retrieved via `layer.losses`.\nThis property is reset at the start of every `__call__` to the top-level layer, so that `layer.losses` always contains the loss values created during the last forward pass.",
"_____no_output_____"
]
],
[
[
"class OuterLayer(layers.Layer):\n\n def __init__(self):\n super(OuterLayer, self).__init__()\n self.activity_reg = ActivityRegularizationLayer(1e-2)\n\n def call(self, inputs):\n return self.activity_reg(inputs)\n\n\nlayer = OuterLayer()\nassert len(layer.losses) == 0 # No losses yet since the layer has never been called\n_ = layer(tf.zeros(1, 1))\nassert len(layer.losses) == 1 # We created one loss value\n\n# `layer.losses` gets reset at the start of each __call__\n_ = layer(tf.zeros(1, 1))\nassert len(layer.losses) == 1 # This is the loss created during the call above",
"_____no_output_____"
]
],
[
[
"In addition, the `loss` property also contains regularization losses created for the weights of any inner layer:",
"_____no_output_____"
]
],
[
[
"class OuterLayer(layers.Layer):\n\n def __init__(self):\n super(OuterLayer, self).__init__()\n self.dense = layers.Dense(32, kernel_regularizer=tf.keras.regularizers.l2(1e-3))\n \n def call(self, inputs):\n return self.dense(inputs)\n \n \nlayer = OuterLayer()\n_ = layer(tf.zeros((1, 1)))\n\n# This is `1e-3 * sum(layer.dense.kernel)`,\n# created by the `kernel_regularizer` above.\nprint(layer.losses)",
"_____no_output_____"
]
],
[
[
"These losses are meant to be taken into account when writing training loops, like this:\n\n\n```python\n# Instantiate an optimizer.\noptimizer = tf.keras.optimizers.SGD(learning_rate=1e-3)\nloss_fn = keras.losses.SparseCategoricalCrossentropy(from_logits=True)\n\n# Iterate over the batches of a dataset.\nfor x_batch_train, y_batch_train in train_dataset:\n with tf.GradientTape() as tape:\n logits = layer(x_batch_train) # Logits for this minibatch\n # Loss value for this minibatch\n loss_value = loss_fn(y_batch_train, logits))\n # Add extra losses created during this forward pass:\n loss_value += sum(model.losses)\n\n grads = tape.gradient(loss_value, model.trainable_variables)\n optimizer.apply_gradients(zip(grads, model.trainable_variables))\n```\n\nFor a detailed guide about writing training loops, see the second section of the [Guide to Training & Evaluation](./training_and_evaluation.ipynb).",
"_____no_output_____"
],
[
"### You can optionally enable serialization on your layers\n\nIf you need your custom layers to be serializable as part of a [Functional model](./functional.ipynb), you can optionally implement a `get_config` method:\n",
"_____no_output_____"
]
],
[
[
"class Linear(layers.Layer):\n\n def __init__(self, units=32):\n super(Linear, self).__init__()\n self.units = units\n\n def build(self, input_shape):\n self.w = self.add_weight(shape=(input_shape[-1], self.units),\n initializer='random_normal',\n trainable=True)\n self.b = self.add_weight(shape=(self.units,),\n initializer='random_normal',\n trainable=True)\n\n def call(self, inputs):\n return tf.matmul(inputs, self.w) + self.b\n\n def get_config(self):\n return {'units': self.units}\n \n\n# Now you can recreate the layer from its config:\nlayer = Linear(64)\nconfig = layer.get_config()\nprint(config)\nnew_layer = Linear.from_config(config)",
"_____no_output_____"
]
],
[
[
"Note that the `__init__` method of the base `Layer` class takes some keyword arguments, in particular a `name` and a `dtype`. It's good practice to pass these arguments to the parent class in `__init__` and to include them in the layer config:",
"_____no_output_____"
]
],
[
[
"class Linear(layers.Layer):\n\n def __init__(self, units=32, **kwargs):\n super(Linear, self).__init__(**kwargs)\n self.units = units\n\n def build(self, input_shape):\n self.w = self.add_weight(shape=(input_shape[-1], self.units),\n initializer='random_normal',\n trainable=True)\n self.b = self.add_weight(shape=(self.units,),\n initializer='random_normal',\n trainable=True)\n\n def call(self, inputs):\n return tf.matmul(inputs, self.w) + self.b\n\n def get_config(self):\n config = super(Linear, self).get_config()\n config.update({'units': self.units})\n return config\n \n\nlayer = Linear(64)\nconfig = layer.get_config()\nprint(config)\nnew_layer = Linear.from_config(config)",
"_____no_output_____"
]
],
[
[
"If you need more flexibility when deserializing the layer from its config, you can also override the `from_config` class method. This is the base implementation of `from_config`:\n\n```python\ndef from_config(cls, config):\n return cls(**config)\n```\n\nTo learn more about serialization and saving, see the complete [Guide to Saving and Serializing Models](./saving_and_serializing.ipynb).",
"_____no_output_____"
],
[
"### Privileged `training` argument in the `call` method\n\n\nSome layers, in particular the `BatchNormalization` layer and the `Dropout` layer, have different behaviors during training and inference. For such layers, it is standard practice to expose a `training` (boolean) argument in the `call` method.\n\nBy exposing this argument in `call`, you enable the built-in training and evaluation loops (e.g. `fit`) to correctly use the layer in training and inference.\n",
"_____no_output_____"
]
],
[
[
"class CustomDropout(layers.Layer):\n \n def __init__(self, rate, **kwargs):\n super(CustomDropout, self).__init__(**kwargs)\n self.rate = rate\n \n def call(self, inputs, training=None):\n return tf.cond(training,\n lambda: tf.nn.dropout(inputs, rate=self.rate),\n lambda: inputs)",
"_____no_output_____"
]
],
[
[
"## Building Models",
"_____no_output_____"
],
[
"\n\n### The Model class\n\nIn general, you will use the `Layer` class to define inner computation blocks,\nand will use the `Model` class to define the outer model -- the object you will train.\n\nFor instance, in a ResNet50 model, you would have several ResNet blocks subclassing `Layer`,\nand a single `Model` encompassing the entire ResNet50 network.\n\nThe `Model` class has the same API as `Layer`, with the following differences:\n- It exposes built-in training, evaluation, and prediction loops (`model.fit()`, `model.evaluate()`, `model.predict()`).\n- It exposes the list of its inner layers, via the `model.layers` property.\n- It exposes saving and serialization APIs.\n\nEffectively, the \"Layer\" class corresponds to what we refer to in the literature\nas a \"layer\" (as in \"convolution layer\" or \"recurrent layer\") or as a \"block\" (as in \"ResNet block\" or \"Inception block\").\n\nMeanwhile, the \"Model\" class corresponds to what is referred to in the literature\nas a \"model\" (as in \"deep learning model\") or as a \"network\" (as in \"deep neural network\").\n\nFor instance, we could take our mini-resnet example above, and use it to build a `Model` that we could\ntrain with `fit()`, and that we could save with `save_weights`:\n\n```python\nclass ResNet(tf.keras.Model):\n\n def __init__(self):\n super(ResNet, self).__init__()\n self.block_1 = ResNetBlock()\n self.block_2 = ResNetBlock()\n self.global_pool = layers.GlobalAveragePooling2D()\n self.classifier = Dense(num_classes)\n\n def call(self, inputs):\n x = self.block_1(inputs)\n x = self.block_2(x)\n x = self.global_pool(x)\n return self.classifier(x)\n\n\nresnet = ResNet()\ndataset = ...\nresnet.fit(dataset, epochs=10)\nresnet.save_weights(filepath)\n```\n",
"_____no_output_____"
],
[
"### Putting it all together: an end-to-end example\n\nHere's what you've learned so far:\n- A `Layer` encapsulate a state (created in `__init__` or `build`) and some computation (in `call`).\n- Layers can be recursively nested to create new, bigger computation blocks.\n- Layers can create and track losses (typically regularization losses).\n- The outer container, the thing you want to train, is a `Model`. A `Model` is just like a `Layer`, but with added training and serialization utilities.\n\nLet's put all of these things together into an end-to-end example: we're going to implement a Variational AutoEncoder (VAE). We'll train it on MNIST digits.\n\nOur VAE will be a subclass of `Model`, built as a nested composition of layers that subclass `Layer`. It will feature a regularization loss (KL divergence).",
"_____no_output_____"
]
],
[
[
"class Sampling(layers.Layer):\n \"\"\"Uses (z_mean, z_log_var) to sample z, the vector encoding a digit.\"\"\"\n\n def call(self, inputs):\n z_mean, z_log_var = inputs\n batch = tf.shape(z_mean)[0]\n dim = tf.shape(z_mean)[1]\n epsilon = tf.keras.backend.random_normal(shape=(batch, dim))\n return z_mean + tf.exp(0.5 * z_log_var) * epsilon\n\n\nclass Encoder(layers.Layer):\n \"\"\"Maps MNIST digits to a triplet (z_mean, z_log_var, z).\"\"\"\n \n def __init__(self,\n latent_dim=32,\n intermediate_dim=64,\n name='encoder',\n **kwargs):\n super(Encoder, self).__init__(name=name, **kwargs)\n self.dense_proj = layers.Dense(intermediate_dim, activation='relu')\n self.dense_mean = layers.Dense(latent_dim)\n self.dense_log_var = layers.Dense(latent_dim)\n self.sampling = Sampling()\n\n def call(self, inputs):\n x = self.dense_proj(inputs)\n z_mean = self.dense_mean(x)\n z_log_var = self.dense_log_var(x)\n z = self.sampling((z_mean, z_log_var))\n return z_mean, z_log_var, z\n\n \nclass Decoder(layers.Layer):\n \"\"\"Converts z, the encoded digit vector, back into a readable digit.\"\"\"\n \n def __init__(self,\n original_dim,\n intermediate_dim=64,\n name='decoder',\n **kwargs):\n super(Decoder, self).__init__(name=name, **kwargs)\n self.dense_proj = layers.Dense(intermediate_dim, activation='relu')\n self.dense_output = layers.Dense(original_dim, activation='sigmoid')\n \n def call(self, inputs):\n x = self.dense_proj(inputs)\n return self.dense_output(x)\n\n\nclass VariationalAutoEncoder(tf.keras.Model):\n \"\"\"Combines the encoder and decoder into an end-to-end model for training.\"\"\"\n \n def __init__(self,\n original_dim,\n intermediate_dim=64,\n latent_dim=32,\n name='autoencoder',\n **kwargs):\n super(VariationalAutoEncoder, self).__init__(name=name, **kwargs)\n self.original_dim = original_dim\n self.encoder = Encoder(latent_dim=latent_dim,\n intermediate_dim=intermediate_dim)\n self.decoder = Decoder(original_dim, intermediate_dim=intermediate_dim)\n \n def call(self, inputs):\n z_mean, z_log_var, z = self.encoder(inputs)\n reconstructed = self.decoder(z)\n # Add KL divergence regularization loss.\n kl_loss = - 0.5 * tf.reduce_mean(\n z_log_var - tf.square(z_mean) - tf.exp(z_log_var) + 1)\n self.add_loss(kl_loss)\n return reconstructed\n\n\noriginal_dim = 784\nvae = VariationalAutoEncoder(original_dim, 64, 32)\n\noptimizer = tf.keras.optimizers.Adam(learning_rate=1e-3)\nmse_loss_fn = tf.keras.losses.MeanSquaredError()\n\nloss_metric = tf.keras.metrics.Mean()\n\n(x_train, _), _ = tf.keras.datasets.mnist.load_data()\nx_train = x_train.reshape(60000, 784).astype('float32') / 255\n\ntrain_dataset = tf.data.Dataset.from_tensor_slices(x_train)\ntrain_dataset = train_dataset.shuffle(buffer_size=1024).batch(64)\n\n# Iterate over epochs.\nfor epoch in range(3):\n print('Start of epoch %d' % (epoch,))\n\n # Iterate over the batches of the dataset.\n for step, x_batch_train in enumerate(train_dataset):\n with tf.GradientTape() as tape:\n reconstructed = vae(x_batch_train)\n # Compute reconstruction loss\n loss = mse_loss_fn(x_batch_train, reconstructed)\n loss += sum(vae.losses) # Add KLD regularization loss\n \n grads = tape.gradient(loss, vae.trainable_variables)\n optimizer.apply_gradients(zip(grads, vae.trainable_variables))\n \n loss_metric(loss)\n \n if step % 100 == 0:\n print('step %s: mean loss = %s' % (step, loss_metric.result()))\n",
"_____no_output_____"
]
],
[
[
"Note that since the VAE is subclassing `Model`, it features built-in training loops. So you could also have trained it like this:",
"_____no_output_____"
]
],
[
[
"vae = VariationalAutoEncoder(784, 64, 32)\n\noptimizer = tf.keras.optimizers.Adam(learning_rate=1e-3)\n\nvae.compile(optimizer, loss=tf.keras.losses.MeanSquaredError())\nvae.fit(x_train, x_train, epochs=3, batch_size=64)",
"_____no_output_____"
]
],
[
[
"### Beyond object-oriented development: the Functional API\n\nWas this example too much object-oriented development for you? You can also build models using [the Functional API](./functional.ipynb). Importantly, choosing one style or another does not prevent you from leveraging components written in the other style: you can always mix-and-match.\n\nFor instance, the Functional API example below reuses the same `Sampling` layer we defined in the example above.",
"_____no_output_____"
]
],
[
[
"original_dim = 784\nintermediate_dim = 64\nlatent_dim = 32\n\n# Define encoder model.\noriginal_inputs = tf.keras.Input(shape=(original_dim,), name='encoder_input')\nx = layers.Dense(intermediate_dim, activation='relu')(original_inputs)\nz_mean = layers.Dense(latent_dim, name='z_mean')(x)\nz_log_var = layers.Dense(latent_dim, name='z_log_var')(x)\nz = Sampling()((z_mean, z_log_var))\nencoder = tf.keras.Model(inputs=original_inputs, outputs=z, name='encoder')\n\n# Define decoder model.\nlatent_inputs = tf.keras.Input(shape=(latent_dim,), name='z_sampling')\nx = layers.Dense(intermediate_dim, activation='relu')(latent_inputs)\noutputs = layers.Dense(original_dim, activation='sigmoid')(x)\ndecoder = tf.keras.Model(inputs=latent_inputs, outputs=outputs, name='decoder')\n\n# Define VAE model.\noutputs = decoder(z)\nvae = tf.keras.Model(inputs=original_inputs, outputs=outputs, name='vae')\n\n# Add KL divergence regularization loss.\nkl_loss = - 0.5 * tf.reduce_mean(\n z_log_var - tf.square(z_mean) - tf.exp(z_log_var) + 1)\nvae.add_loss(kl_loss)\n\n# Train.\noptimizer = tf.keras.optimizers.Adam(learning_rate=1e-3)\nvae.compile(optimizer, loss=tf.keras.losses.MeanSquaredError())\nvae.fit(x_train, x_train, epochs=3, batch_size=64)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
c512733caf5a4946169cdd046328777ef96865dc
| 100,746 |
ipynb
|
Jupyter Notebook
|
LECTURE_4.ipynb
|
Gennadiyev/SJTU-AI
|
75bea091c531d552fd63df379d1e567f39b6a3cc
|
[
"MIT"
] | 4 |
2021-07-02T00:41:44.000Z
|
2021-07-06T12:59:13.000Z
|
LECTURE_4.ipynb
|
Gennadiyev/SJTU-AI
|
75bea091c531d552fd63df379d1e567f39b6a3cc
|
[
"MIT"
] | 2 |
2021-07-05T19:14:40.000Z
|
2021-07-06T17:10:13.000Z
|
LECTURE_4.ipynb
|
Gennadiyev/SJTU-AI
|
75bea091c531d552fd63df379d1e567f39b6a3cc
|
[
"MIT"
] | 4 |
2021-07-02T08:05:45.000Z
|
2022-03-01T09:43:26.000Z
| 72.531317 | 24,564 | 0.696583 |
[
[
[
"# Using `matplotlib`",
"_____no_output_____"
]
],
[
[
"from IPython.display import set_matplotlib_formats\n%matplotlib inline\nset_matplotlib_formats('svg')\n\nimport numpy as np\nimport matplotlib.pyplot as plt\nplt.figure(num = 3, figsize = (8, 5))\n\nvecX = np.linspace(-10, 10, 100)\nvecY = vecX ** 2\nplt.plot(vecX, vecY)\nplt.show()",
"_____no_output_____"
],
[
"plt.figure(num = 3, figsize = (8, 5))\nplt.xlim((0, 5))\nplt.ylim(-5, 30) # Auto tuple\nplt.yticks([-10, 0, 15, 30])\nplt.plot(vecX, vecY)\nplt.xlabel('On Positive Axe')\nplt.show()",
"_____no_output_____"
],
[
"# Basic styling\nplt.figure(num = 3, figsize = (8, 5))\nax = plt.gca()\nax.spines['right'].set_color('none')\nax.spines['top'].set_color('none')\nax.spines['bottom'].set_color('none')\nax.spines['left'].set_color('none')\nplt.xlim((0, 5))\nplt.ylim(-5, 30) # Auto tuple\nplt.yticks([-10, 0, 15, 30])\nplt.plot(vecX, vecY)\nplt.xlabel('On Positive Axe')\nplt.show()",
"_____no_output_____"
],
[
"x = np.linspace(-1, 2, 50)\ny = x - 1\nplt.figure(num = 3, figsize = (8, 5))\nplt.plot(x, y, linewidth = 1.0, linestyle = \"dashed\")\nplt.xlim(-1, 2)\nplt.ylim(-2, 2)\nplt.xticks([-1, -0.5, 1], ['lost', 'far', 'pure'])\nax = plt.gca()\nax.spines['right'].set_color('none')\nax.spines['top'].set_color('none')\nax.spines['bottom'].set_position(('data', 0))\nax.spines['left'].set_position(('data', 0))\nplt.show()",
"_____no_output_____"
]
],
[
[
"> Note that Kun then ignores all that the teacher says and began to venture on her own. For more information see PROJECT_PLOT.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
c5127d176367fc5d363cd8679d72b36c63e395a6
| 228,392 |
ipynb
|
Jupyter Notebook
|
python_code/.ipynb_checkpoints/LODES_V5-checkpoint.ipynb
|
DistrictDataLabs/04-team3
|
14e399f639a047afcc796a05a383d3c805d68315
|
[
"Apache-2.0"
] | null | null | null |
python_code/.ipynb_checkpoints/LODES_V5-checkpoint.ipynb
|
DistrictDataLabs/04-team3
|
14e399f639a047afcc796a05a383d3c805d68315
|
[
"Apache-2.0"
] | null | null | null |
python_code/.ipynb_checkpoints/LODES_V5-checkpoint.ipynb
|
DistrictDataLabs/04-team3
|
14e399f639a047afcc796a05a383d3c805d68315
|
[
"Apache-2.0"
] | null | null | null | 40.989232 | 1,090 | 0.320348 |
[
[
[
"# LODES Data Analysis\n## Prepare Workbook",
"_____no_output_____"
]
],
[
[
"import numpy as np\nfrom pandas import Series, DataFrame\nimport pandas as pd\nimport urllib\nfrom urllib2 import urlopen\nfrom StringIO import StringIO\nimport gzip\n\nimport requests\nimport json\nimport os\nfrom copy import deepcopy\n\nfrom pandas.io.json import json_normalize",
"_____no_output_____"
],
[
"# Set create working folder and set as active directory\nos.chdir('C:\\Users\\dcapizzi\\Documents\\GitHub')\nif not os.path.exists('lodes'):\n os.makedirs('lodes')\n \nos.chdir('C:\\Users\\dcapizzi\\Documents\\GitHub\\lodes')",
"_____no_output_____"
]
],
[
[
"## Load initial LODES data",
"_____no_output_____"
]
],
[
[
"# Collect user input for the year and states to download for the analysis\nyear = raw_input('Enter a year: ')\ninput_list = raw_input(\"Enter states to include separated by commas (no spaces): \")\nstate_list = input_list.split(',')",
"Enter a year: 2014\nEnter states to include separated by commas (no spaces): MD\n"
],
[
"# Define final data frames to aggregate all state data\n\nlodes_columns = ['w_geocode', 'h_geocode', 'tot_jobs', 'age_29_bel_jobs',\n 'age_30_54_jobs', 'age_55_over_jobs', 'sal_1250_bel_jobs',\n 'sal_1250_3333_jobs', 'sal_3333_over_jobs', 'goods_prod_jobs',\n 'trade_transp_jobs', 'all_other_svc_jobs', 'createdate', 'state',\n 'w_block', 'h_block', 'w_2010_block', 'w_state', 'w_county_name',\n 'w_block_group_code', 'w_block_group_name', 'w_metro_name',\n 'w_zip_code', 'w_place_name', 'w_county_sub_name', 'w_createdate',\n 'h_2010_block', 'h_state', 'h_county_name', 'h_block_group_code',\n 'h_block_group_name', 'h_metro_name', 'h_zip_code', 'h_place_name',\n 'h_county_sub_name', 'h_createdate']\n\nlodes_data = pd.DataFrame([],columns=lodes_columns)",
"_____no_output_____"
],
[
"# Create dictionaries to house downloaded files\n\ndict_lodes = {}\ndict_xwalk = {}\n\n# Loop through all states selected by user, download the relevant files from the Census website, unzip, read, and load into dictionaries \n# Process takes some time, please be patient\n\nfor state in state_list:\n \n # Sets url for primary \"LODES\" data set - which provides data on the home Census block, work Census block, and commuters in between\n lodes_url = 'http://lehd.ces.census.gov/data/lodes/LODES7/' + state.lower() + '/od/' + state.lower() + '_od_main_JT00_' + year + '.csv.gz'\n \n # Sets url for \"cross-walk\" data with the city, state, ZIP, etc. for each Census block\n xwalk_url = 'http://lehd.ces.census.gov/data/lodes/LODES7/' + state.lower() + '/' + state.lower() + '_xwalk.csv.gz'\n \n # Names the files\n lodes_filename = 'lodes_' + state + \"_\" + year + '.csv.gz'\n xwalk_filename = 'xwalk_' + state + \"_\" + year + '.csv.gz'\n \n # Downloads the files\n urllib.urlretrieve(lodes_url, lodes_filename)\n urllib.urlretrieve(xwalk_url, xwalk_filename)\n \n print 'Data downloaded for '+state\n \n # Unzips the files\n unzip_lodes = gzip.open(lodes_filename, 'rb')\n unzip_xwalk = gzip.open(xwalk_filename, 'rb')\n \n # Reads the files to disk \n unzip_lodes = unzip_lodes.read()\n unzip_xwalk = unzip_xwalk.read()\n\n # Saves as objects in teh created dictionaries \n dict_lodes[state]=pd.read_csv(StringIO(unzip_lodes))\n dict_xwalk[state]=pd.read_csv(StringIO(unzip_xwalk))\n print 'Data tables created for '+state\n \n # Removes unnecessary fields and names the columns to consistent, human-readable names\n dict_lodes[state].columns = ['w_geocode','h_geocode','tot_jobs','age_29_bel_jobs',\n 'age_30_54_jobs','age_55_over_jobs','sal_1250_bel_jobs','sal_1250_3333_jobs','sal_3333_over_jobs',\n 'goods_prod_jobs','trade_transp_jobs','all_other_svc_jobs','createdate']\n\n dict_xwalk[state] = DataFrame(dict_xwalk[state],columns=['tabblk2010','stusps','ctyname', 'bgrp','bgrpname','cbsaname','zcta','stplcname','ctycsubname','createdate'])\n dict_xwalk[state].columns = ['2010_block', 'state', 'county_name', 'block_group_code', 'block_group_name','metro_name', 'zip_code','place_name', 'county_sub_name','createdate']\n \n print 'Column names defined for '+state\n \n # Creates 'block-group-level' field to join LODES to xwalk and centroid lat/longs (Census block group codes are the first 12 digits of Census block codes)\n left = lambda x: str(int(x))[:12]\n dict_lodes[state]['w_block'] = dict_lodes[state]['w_geocode'].apply(left)\n dict_lodes[state]['w_block'] = dict_lodes[state]['w_geocode'].apply(left)\n dict_lodes[state]['h_block'] = dict_lodes[state]['h_geocode'].apply(left)\n dict_xwalk[state]['block_group_code']= dict_xwalk[state]['block_group_code'].apply(left)\n \n dict_lodes[state]['state'] = state\n \n print 'New fields created for '+state\n \nprint 'Process complete!'",
"Data downloaded for MD\nData tables created for MD\nColumn names defined for MD\nNew fields created for MD\nProcess complete!\n"
],
[
"# Create blank dictionaries to join or merge cross-walk data with LODES data\n\ndict_xwalk_w = {}\ndict_xwalk_h = {}\n\n# Duplicay (copy) cross-walk data, with columns one for work, one for home\nfor state in dict_xwalk:\n dict_xwalk_w[state] = deepcopy(dict_xwalk[state]) \n dict_xwalk_h[state] = deepcopy(dict_xwalk[state]) \n dict_xwalk_w[state].rename(columns=lambda x: \"w_\"+x, inplace=\"True\")\n dict_xwalk_h[state].rename(columns=lambda x: \"h_\"+x, inplace=\"True\")",
"_____no_output_____"
],
[
"# For each state in dict_lodes, merge once on the \"work\" Census block (w_geocode) and once on the \"home\" Census block (h_geocode)\n# This data will provide an idea of the city/state/zip for both the work and home block code groups\n\nfor state in dict_lodes:\n dict_lodes[state] = pd.merge(dict_lodes[state], dict_xwalk_w[state], how='left', left_on='w_geocode', right_on='w_2010_block')\n dict_lodes[state] = pd.merge(dict_lodes[state], dict_xwalk_h[state], how='left', left_on='h_geocode', right_on='h_2010_block')\n lodes_data = lodes_data.append(dict_lodes[state])",
"_____no_output_____"
],
[
"lodes_data.columns",
"_____no_output_____"
]
],
[
[
"## Transform LODES data for analysis",
"_____no_output_____"
]
],
[
[
"# Create new field \"home to work\" with both home and work geocodes\nlodes_data['unique'] = lodes_data['h_geocode'].map('{0:f}'.format).astype(str).apply(lambda x: x[:15]) + ' to ' + lodes_data['w_geocode'].map('{0:f}'.format).astype(str).apply(lambda x: x[:15]) ",
"_____no_output_____"
],
[
"# Take new data set, and split into \"home\" and \"work\" tables to be flattened\n\nlodes_data_home = DataFrame(lodes_data, columns = ['unique','h_geocode', 'tot_jobs', 'age_29_bel_jobs',\n 'age_30_54_jobs', 'age_55_over_jobs', 'sal_1250_bel_jobs',\n 'sal_1250_3333_jobs', 'sal_3333_over_jobs', 'goods_prod_jobs',\n 'trade_transp_jobs', 'all_other_svc_jobs',\n 'h_block', 'h_state', 'h_county_name',\n 'h_block_group_code', 'h_block_group_name', 'h_metro_name',\n 'h_zip_code', 'h_place_name', 'h_county_sub_name'])\nlodes_data_home['type']='Home'\nlodes_data_home['path']=1\n\nlodes_data_work = DataFrame(lodes_data, columns = ['unique','w_geocode', 'tot_jobs', 'age_29_bel_jobs',\n 'age_30_54_jobs', 'age_55_over_jobs', 'sal_1250_bel_jobs',\n 'sal_1250_3333_jobs', 'sal_3333_over_jobs', 'goods_prod_jobs',\n 'trade_transp_jobs', 'all_other_svc_jobs',\n 'w_block', 'w_state', 'w_county_name',\n 'w_block_group_code', 'w_block_group_name', 'w_metro_name',\n 'w_zip_code', 'w_place_name', 'w_county_sub_name'])\n\nlodes_data_work['type']='Work'\nlodes_data_work['path']=2",
"_____no_output_____"
],
[
"# Rename columns to be the same for both new tables\nnew_columns = ['unique','geocode', 'tot_jobs', 'age_29_bel_jobs',\n 'age_30_54_jobs', 'age_55_over_jobs', 'sal_1250_bel_jobs',\n 'sal_1250_3333_jobs', 'sal_3333_over_jobs', 'goods_prod_jobs',\n 'trade_transp_jobs', 'all_other_svc_jobs',\n 'block', 'state', 'county_name',\n 'block_group_code', 'block_group_name', 'metro_name',\n 'zip_code', 'place_name', 'county_sub_name','type','path']\n\nlodes_data_home.columns = new_columns\nlodes_data_work.columns = new_columns",
"_____no_output_____"
],
[
"# Append both tables and sort by Path ID\nlodes_data_flat = lodes_data_home.append(lodes_data_work)\nlodes_data_flat = lodes_data_flat.sort(['unique','path']).reset_index(drop=True)\nlodes_data_flat[:3]",
"/Users/kruthika/anaconda/lib/python2.7/site-packages/ipykernel/__main__.py:3: FutureWarning: sort(columns=....) is deprecated, use sort_values(by=.....)\n app.launch_new_instance()\n"
]
],
[
[
"## Add additional data on latitude, longitude, and demographics into data set",
"_____no_output_____"
]
],
[
[
"# read in data with latitudes, longitudes, and other data sources\nlatlong = pd.read_csv('DDL_census_data.csv')\n\n# Rename columns\nlatlong.columns = ['state', 'county', 'tract', 'blockgrouppiece', 'full_geo_id', 'geoid',\n 'name', u'lsad', 'land_area', 'water_area', 'latitude', 'longitude', 'id',\n 'geoid2', 'geoid3', 'geo_display','median_income','moe_median_income',\n 'geoid4', 'geoid5', 'geo_display2', 'total','moe_total:',\n 'foodstamps','moe_foodstamps',\n 'foodstamps_disability','moe_foodstamps_disability','foodstamps_nodisability','moe_foodstamps_nodisability',\n 'nofoodstamps','moe_nofoodstamps',\n 'nofoodstamps_disability','moe_nofoodstamps_disability',\n 'nofoodstamps_nodisability','moe_nofoodstamps_nodisability']\n\n# Reformat columns\nlatlong['full_geo_id'] = latlong['full_geo_id'].apply(lambda x: x[9:])\n\n# Eliminate unnecessary columns\nlatlong = DataFrame(latlong, columns = ['full_geo_id', 'latitude', 'longitude',\n 'foodstamps','moe_foodstamps',\n 'foodstamps_disability','moe_foodstamps_disability','foodstamps_nodisability','moe_foodstamps_nodisability',\n 'nofoodstamps','moe_nofoodstamps',\n 'nofoodstamps_disability','moe_nofoodstamps_disability',\n 'nofoodstamps_nodisability','moe_nofoodstamps_nodisability'])",
"_____no_output_____"
],
[
"lodes_data_full = pd.merge(lodes_data_flat, latlong, how='left', left_on='block_group_code', right_on='full_geo_id') ",
"_____no_output_____"
]
],
[
[
"## Add additional data on transit for metro",
"_____no_output_____"
]
],
[
[
"lodes_data_full['category']='lodes'\nlodes_data_full",
"_____no_output_____"
],
[
"from sqlalchemy import create_engine\nsqlite_file = 'sqlite://///Users/Kruthika/Projects/DDL/04-team3/census.db'\nengine = create_engine(sqlite_file)\nfrom pandas.io import sql\nsql.execute('DROP TABLE IF EXISTS lodes_data',engine)\nlodes_data_full.to_sql('lodes_data', engine)",
"_____no_output_____"
],
[
"import requests\nimport json\nimport pandas as pd\nfrom pandas.io.json import json_normalize\nfrom urllib2 import urlopen",
"_____no_output_____"
],
[
"#Get station-level descriptive data from WMATA API, including latitude and longitude of stations and line codes\nr = requests.get('https://api.wmata.com/Rail.svc/json/jStations?api_key=fb7119a0d3464673825a26e94db74451')",
"_____no_output_____"
],
[
"data_list = []\nfor entrances in r.json()['Stations']:\n for e in entrances.keys():\n if e not in data_list:\n data_list.append(e)\nprint data_list",
"[u'Code', u'Name', u'StationTogether2', u'LineCode4', u'LineCode2', u'LineCode3', u'LineCode1', u'Lon', u'Address', u'Lat', u'StationTogether1']\n"
],
[
"metro_stations = json_normalize(r.json()['Stations'])\nmetro_stations.head(3)",
"_____no_output_____"
],
[
"metro_stations.to_csv('stations.csv')",
"_____no_output_____"
],
[
"#Get bus route descriptive data from WMATA API, including latitude and longitude of stations and route codes\nr1 = requests.get('https://api.wmata.com/Bus.svc/json/jStops?api_key=fb7119a0d3464673825a26e94db74451')",
"_____no_output_____"
],
[
"stops_list = []\nfor stops in r1.json()['Stops']:\n for s in stops.keys():\n if s not in stops_list:\n stops_list.append(s)\nprint stops_list",
"_____no_output_____"
],
[
"bus_stops = json_normalize(r1.json()['Stops'])\nbus_stops.head(3)",
"_____no_output_____"
],
[
"s = bus_stops.apply(lambda x: pd.Series(x['Routes']),axis=1).stack().reset_index(level=1, drop=True)",
"_____no_output_____"
],
[
"s.name = 'Routes'\nbus_routes = bus_stops.drop('Routes', axis=1).join(s)\nbus_routes['category'] = 'bus'\nbus_routes['type'] = 'bus'",
"_____no_output_____"
],
[
"bus_routes.columns = ['latitude','longitude','name','unique','detail','category','type']\nbus_routes[:6]",
"_____no_output_____"
],
[
"bus_routes.to_csv('busroutes.csv')",
"_____no_output_____"
],
[
"#Get path-level train data from WMATA API, including latitude and longitude of stations and line codes\nrblue = requests.get('https://api.wmata.com/Rail.svc/json/jPath?FromStationCode=J03&ToStationCode=G05&api_key=fb7119a0d3464673825a26e94db74451')\nrgreen = requests.get('https://api.wmata.com/Rail.svc/json/jPath?FromStationCode=F11&ToStationCode=E10&api_key=fb7119a0d3464673825a26e94db74451')\nrorange = requests.get('https://api.wmata.com/Rail.svc/json/jPath?FromStationCode=K08&ToStationCode=D13&api_key=fb7119a0d3464673825a26e94db74451')\nrred = requests.get('https://api.wmata.com/Rail.svc/json/jPath?FromStationCode=A15&ToStationCode=B11&api_key=fb7119a0d3464673825a26e94db74451')\nrsilver = requests.get('https://api.wmata.com/Rail.svc/json/jPath?FromStationCode=N06&ToStationCode=G05&api_key=fb7119a0d3464673825a26e94db74451')\nryellow = requests.get('https://api.wmata.com/Rail.svc/json/jPath?FromStationCode=C15&ToStationCode=E06&api_key=fb7119a0d3464673825a26e94db74451')",
"_____no_output_____"
],
[
"data_list = []\nfor paths in rblue.json()['Path']:\n for p in paths.keys():\n if p not in data_list:\n data_list.append(p)\nprint data_list\n\ndfblue = json_normalize(rblue.json()['Path'])\ndfgreen = json_normalize(rgreen.json()['Path'])\ndforange = json_normalize(rorange.json()['Path'])\ndfred = json_normalize(rred.json()['Path'])\ndfsilver = json_normalize(rsilver.json()['Path'])\ndfyellow = json_normalize(ryellow.json()['Path'])",
"[u'StationCode', u'SeqNum', u'LineCode', u'StationName', u'DistanceToPrev']\n"
],
[
"metro_lines = pd.concat([dfblue, dfgreen, dforange, dfred, dfsilver, dfyellow], ignore_index=True)",
"_____no_output_____"
],
[
"metro_lines.head(3)",
"_____no_output_____"
],
[
"metro_combined = pd.merge(metro_lines, metro_stations, how='left', left_on='StationCode', right_on='Code')\nmetro_combined.head(3)",
"_____no_output_____"
],
[
"metro_combined = DataFrame(metro_combined,columns=['LineCode','SeqNum', 'StationName','Address.City','Address.State','Address.Zip','Lat','Lon'])\nmetro_combined.columns = ['unique','path','name','metro_name','state','zip','latitude','longitude']\nmetro_combined['type']='train'\nmetro_combined['category']='train'\nmetro_combined.head(3)",
"_____no_output_____"
],
[
"metro_combined.to_csv('trainandroute.csv')",
"_____no_output_____"
]
],
[
[
"## Blend all data sets together",
"_____no_output_____"
]
],
[
[
"lodes_transit_data = pd.concat([lodes_data_full, bus_routes, metro_combined], ignore_index=True)\nlodes_transit_data[:3] ",
"_____no_output_____"
],
[
"lodes_transit_data [lodes_transit_data['category']=='train'][:5]",
"_____no_output_____"
],
[
"lodes_transit_data.to_csv('lodes_final_output.csv')",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
c5128f656b4f91d1728c9f8f30d55fc19fe750e2
| 140,780 |
ipynb
|
Jupyter Notebook
|
gradient-descent/GradientDescent.ipynb
|
one-for-all/deep-learning
|
0103f02c3183306e8a15967e2a98d2d7fd62f81c
|
[
"MIT"
] | null | null | null |
gradient-descent/GradientDescent.ipynb
|
one-for-all/deep-learning
|
0103f02c3183306e8a15967e2a98d2d7fd62f81c
|
[
"MIT"
] | null | null | null |
gradient-descent/GradientDescent.ipynb
|
one-for-all/deep-learning
|
0103f02c3183306e8a15967e2a98d2d7fd62f81c
|
[
"MIT"
] | null | null | null | 451.217949 | 104,124 | 0.936276 |
[
[
[
"# Implementing the Gradient Descent Algorithm\n\nIn this lab, we'll implement the basic functions of the Gradient Descent algorithm to find the boundary in a small dataset. First, we'll start with some functions that will help us plot and visualize the data.",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nimport numpy as np\nimport pandas as pd\n\n#Some helper functions for plotting and drawing lines\n\ndef plot_points(X, y):\n admitted = X[np.argwhere(y==1)]\n rejected = X[np.argwhere(y==0)]\n plt.scatter([s[0][0] for s in rejected], [s[0][1] for s in rejected], s = 25, color = 'blue', edgecolor = 'k')\n plt.scatter([s[0][0] for s in admitted], [s[0][1] for s in admitted], s = 25, color = 'red', edgecolor = 'k')\n\ndef display(m, b, color='g--'):\n plt.xlim(-0.05,1.05)\n plt.ylim(-0.05,1.05)\n x = np.arange(-10, 10, 0.1)\n plt.plot(x, m*x+b, color)",
"_____no_output_____"
]
],
[
[
"## Reading and plotting the data",
"_____no_output_____"
]
],
[
[
"data = pd.read_csv('data.csv', header=None)\nX = np.array(data[[0,1]])\ny = np.array(data[2])\nplot_points(X,y)\nplt.show()",
"_____no_output_____"
]
],
[
[
"## TODO: Implementing the basic functions\nHere is your turn to shine. Implement the following formulas, as explained in the text.\n- Sigmoid activation function\n\n$$\\sigma(x) = \\frac{1}{1+e^{-x}}$$\n\n- Output (prediction) formula\n\n$$\\hat{y} = \\sigma(w_1 x_1 + w_2 x_2 + b)$$\n\n- Error function\n\n$$Error(y, \\hat{y}) = - y \\log(\\hat{y}) - (1-y) \\log(1-\\hat{y})$$\n\n- The function that updates the weights\n\n$$ w_i \\longrightarrow w_i + \\alpha (y - \\hat{y}) x_i$$\n\n$$ b \\longrightarrow b + \\alpha (y - \\hat{y})$$",
"_____no_output_____"
]
],
[
[
"# Implement the following functions\n\n# Activation (sigmoid) function\ndef sigmoid(x):\n return 1/(1+np.exp(-x))\n\n# Output (prediction) formula\ndef output_formula(features, weights, bias):\n return sigmoid(np.matmul(features, weights)+bias)\n\n# Error (log-loss) formula\ndef error_formula(y, output):\n return -y*np.log(output)-(1-y)*np.log(1-output)\n\n# Gradient descent step\ndef update_weights(x, y, weights, bias, learnrate):\n output = output_formula(x, weights, bias)\n new_weights = weights + learnrate*(y-output)*x\n new_bias = bias + learnrate*(y-output)\n return new_weights, new_bias",
"_____no_output_____"
]
],
[
[
"## Training function\nThis function will help us iterate the gradient descent algorithm through all the data, for a number of epochs. It will also plot the data, and some of the boundary lines obtained as we run the algorithm.",
"_____no_output_____"
]
],
[
[
"np.random.seed(44)\n\nepochs = 100\nlearnrate = 0.01\n\ndef train(features, targets, epochs, learnrate, graph_lines=False):\n \n errors = []\n n_records, n_features = features.shape\n last_loss = None\n weights = np.random.normal(scale=1 / n_features**.5, size=n_features)\n bias = 0\n for e in range(epochs):\n del_w = np.zeros(weights.shape)\n for x, y in zip(features, targets):\n output = output_formula(x, weights, bias)\n error = error_formula(y, output)\n weights, bias = update_weights(x, y, weights, bias, learnrate)\n \n # Printing out the log-loss error on the training set\n out = output_formula(features, weights, bias)\n loss = np.mean(error_formula(targets, out))\n errors.append(loss)\n if e % (epochs / 10) == 0:\n print(\"\\n========== Epoch\", e,\"==========\")\n if last_loss and last_loss < loss:\n print(\"Train loss: \", loss, \" WARNING - Loss Increasing\")\n else:\n print(\"Train loss: \", loss)\n last_loss = loss\n predictions = out > 0.5\n accuracy = np.mean(predictions == targets)\n print(\"Accuracy: \", accuracy)\n if graph_lines and e % (epochs / 100) == 0:\n display(-weights[0]/weights[1], -bias/weights[1])\n \n\n # Plotting the solution boundary\n plt.title(\"Solution boundary\")\n display(-weights[0]/weights[1], -bias/weights[1], 'black')\n\n # Plotting the data\n plot_points(features, targets)\n plt.show()\n\n # Plotting the error\n plt.title(\"Error Plot\")\n plt.xlabel('Number of epochs')\n plt.ylabel('Error')\n plt.plot(errors)\n plt.show()",
"_____no_output_____"
]
],
[
[
"## Time to train the algorithm!\nWhen we run the function, we'll obtain the following:\n- 10 updates with the current training loss and accuracy\n- A plot of the data and some of the boundary lines obtained. The final one is in black. Notice how the lines get closer and closer to the best fit, as we go through more epochs.\n- A plot of the error function. Notice how it decreases as we go through more epochs.",
"_____no_output_____"
]
],
[
[
"train(X, y, epochs, learnrate, True)",
"\n========== Epoch 0 ==========\nTrain loss: 0.7135845195381634\nAccuracy: 0.4\n\n========== Epoch 10 ==========\nTrain loss: 0.6225835210454962\nAccuracy: 0.59\n\n========== Epoch 20 ==========\nTrain loss: 0.5548744083669508\nAccuracy: 0.74\n\n========== Epoch 30 ==========\nTrain loss: 0.501606141872473\nAccuracy: 0.84\n\n========== Epoch 40 ==========\nTrain loss: 0.4593334641861401\nAccuracy: 0.86\n\n========== Epoch 50 ==========\nTrain loss: 0.42525543433469976\nAccuracy: 0.93\n\n========== Epoch 60 ==========\nTrain loss: 0.3973461571671399\nAccuracy: 0.93\n\n========== Epoch 70 ==========\nTrain loss: 0.3741469765239074\nAccuracy: 0.93\n\n========== Epoch 80 ==========\nTrain loss: 0.35459973368161973\nAccuracy: 0.94\n\n========== Epoch 90 ==========\nTrain loss: 0.3379273658879921\nAccuracy: 0.94\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
c512bb1df92e20638268c4fdf58b34a0a74a1d33
| 9,487 |
ipynb
|
Jupyter Notebook
|
maldi_acquisition_queue.ipynb
|
andy-d-palmer/ims-direct-control
|
864d6de2b67f78dcd5e7abfef65a6c4adaa460b9
|
[
"MIT"
] | 4 |
2019-06-26T12:57:20.000Z
|
2021-10-03T22:17:05.000Z
|
maldi_acquisition_queue.ipynb
|
andy-d-palmer/ims-direct-control
|
864d6de2b67f78dcd5e7abfef65a6c4adaa460b9
|
[
"MIT"
] | null | null | null |
maldi_acquisition_queue.ipynb
|
andy-d-palmer/ims-direct-control
|
864d6de2b67f78dcd5e7abfef65a6c4adaa460b9
|
[
"MIT"
] | 1 |
2021-05-14T09:46:53.000Z
|
2021-05-14T09:46:53.000Z
| 23.482673 | 120 | 0.536734 |
[
[
[
"# MALDI acquisition of predefined areas",
"_____no_output_____"
],
[
"author: Alex Mattausch \nversion: 0.1.0",
"_____no_output_____"
]
],
[
[
"%load_ext autoreload\n%autoreload 2\n# \"%matplotlib widget\" is slightly better, but sometimes doesn't work\n# \"%matplotlib notebook\" or \"%matplotlib inline\" can be used as alternatives\n%matplotlib widget\nimport matplotlib.pyplot as plt\nimport remote_control.control as rc\nimport remote_control.utils as utils\n\n#from IPython.display import set_matplotlib_formats\n#set_matplotlib_formats('svg')\n\nfrom remote_control import acquisition\nfrom remote_control.control import configure_fly_at_fixed_z\nfrom itertools import product\n\nCONFIG_FN = 'remote_config.json'\n\n### IN CASE OF ERROR, make sure Jupyter is set to use the \"Python [conda env:maldi-control-notebooks]\" kernel",
"The autoreload extension is already loaded. To reload it, use:\n %reload_ext autoreload\n"
],
[
"qa = acquisition.QueueAquisition(\n config_fn = CONFIG_FN, \n datadir=\"./data\" # will save spatial position file here\n)",
"_____no_output_____"
],
[
"# For plates with recessed wells, configure this to move the slide away when moving between wells.\n# If the stage needs to move in the X/Y plane more than \"distance\", it will move the stage's Z axis\n# to the value of the \"z\" parameter. \n# configure_fly_at_fixed_z(distance=2000, z=3000) # Enable\nconfigure_fly_at_fixed_z(distance=None, z=None) # Disable",
"_____no_output_____"
]
],
[
[
"### 1. Define slide area",
"_____no_output_____"
]
],
[
[
"# Set up safety bounds (optional - comment this out if they're unwanted)\nqa.set_image_bounds(\n min_x=-15000,\n max_x=15000,\n min_y=-25000,\n max_y=25000,\n)",
"_____no_output_____"
]
],
[
[
"### 2. Add acquisition areas",
"_____no_output_____"
],
[
"Run this cell to clear areas and start over:",
"_____no_output_____"
]
],
[
[
"qa.clear_areas()",
"_____no_output_____"
],
[
"qa.add_area(\n name=\"well_1\", # <- Optional!\n line_start=(-10649, -18704, 3444),\n line_end=(-4149, -18704, 3444),\n perpendicular=(-9399, -24204, 3444),\n step_size_x=500,\n step_size_y=1000\n)",
"_____no_output_____"
],
[
"qa.add_area(\n name=\"well_2\",\n line_start=(-10729, -6580, 3444),\n line_end=(-8229, -6580, 3444),\n perpendicular=(-9479, -9080, 3444),\n step_size_x=25,\n step_size_y=25\n)",
"_____no_output_____"
],
[
"qa.add_area(\n name=\"well_4\",\n line_start=(-10729, 22000, 3444),\n line_end=(-8229, 22000, 3444),\n perpendicular=(-9479, 18000, 3444),\n step_size_x=250,\n step_size_y=250\n)",
"_____no_output_____"
],
[
"qa.plot_areas()",
"total areas: 3\n"
]
],
[
[
"**NOTE:** numbers in boxes indicate acquisition order!",
"_____no_output_____"
],
[
"### 3. Generate measurement positions from areas",
"_____no_output_____"
]
],
[
[
"qa.generate_targets()",
"_____no_output_____"
],
[
"plt.close('all')\nqa.plot_targets(annotate=True) ",
"_____no_output_____"
]
],
[
[
"### 4. Run acquistion\n\nOnce you are happy with plots above:\n- Launch Telnet in apsmaldi software\n- Press START on TUNE somputer\n- Run the following cell with dummy=True to test coordinates\n- Run the following cell with dummy=Fase, measure=True to perform acquisition",
"_____no_output_____"
]
],
[
[
"OUTPUT_DIR = 'D:\\\\imagingMS\\\\2021_08\\\\your name\\\\'\nIMZML_PREFIX = OUTPUT_DIR + '01052019_Mouse_DHB_pos_mz200-800_px50x50_LR'\n\nqa.acquire(\n filename=IMZML_PREFIX, # Prefix for output coordinates file used in ImzML conversion\n dummy=True, # False - send commands to MALDI, True - don't connect, just print commands\n measure=False, # False - move stage only, True - move stage & acquire data\n email_on_success='[email protected]', # Set to None to suppress\n email_on_failure='[email protected]', # Set to None to suppress\n)",
"_____no_output_____"
]
],
[
[
"### 5. Cleanup\nAfter imaging run the following cell to terminate Telnet",
"_____no_output_____"
]
],
[
[
"rc.close(quit=True)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
c512bfd8b6b81183ae7c24eb4846392f45241a64
| 1,172 |
ipynb
|
Jupyter Notebook
|
2021 Осенний семестр/Практическое задание 1/Бевз_Задание 1.ipynb
|
mosalov/Notebook_For_AI_Main
|
a693d29bf0bdcf824cb4f1eca86ff54b67ba7428
|
[
"MIT"
] | 6 |
2021-09-20T10:28:18.000Z
|
2022-03-14T18:39:17.000Z
|
2021 Осенний семестр/Практическое задание 1/Бевз_Задание 1.ipynb
|
mosalov/Notebook_For_AI_Main
|
a693d29bf0bdcf824cb4f1eca86ff54b67ba7428
|
[
"MIT"
] | 122 |
2020-09-07T11:57:57.000Z
|
2022-03-22T06:47:03.000Z
|
2021 Осенний семестр/Практическое задание 1/Бевз_Задание 1.ipynb
|
mosalov/Notebook_For_AI_Main
|
a693d29bf0bdcf824cb4f1eca86ff54b67ba7428
|
[
"MIT"
] | 97 |
2020-09-07T11:32:19.000Z
|
2022-03-31T10:27:38.000Z
| 18.903226 | 63 | 0.512799 |
[
[
[
"import numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\nimport sklearn as sk\n\nname = 'Aleksandr'\nsurname = 'Bevz'\n\nname_surname = name + ' ' + surname\n\nprint(name_surname)\nprint('Количество символов: ' + str(len(name_surname)))\n",
"Aleksandr Bevz\nКоличество символов: 14\n"
]
]
] |
[
"code"
] |
[
[
"code"
]
] |
c512c229c417821e8560175391903f413f109b3c
| 618,446 |
ipynb
|
Jupyter Notebook
|
gdp_growth.ipynb
|
K-O-N/World-s-Fastest-Growing-Economies-as-of-2018
|
4183b7d0df44535dca867592e06c1e4995a6bf6d
|
[
"Apache-2.0"
] | null | null | null |
gdp_growth.ipynb
|
K-O-N/World-s-Fastest-Growing-Economies-as-of-2018
|
4183b7d0df44535dca867592e06c1e4995a6bf6d
|
[
"Apache-2.0"
] | null | null | null |
gdp_growth.ipynb
|
K-O-N/World-s-Fastest-Growing-Economies-as-of-2018
|
4183b7d0df44535dca867592e06c1e4995a6bf6d
|
[
"Apache-2.0"
] | null | null | null | 339.246297 | 545,536 | 0.678455 |
[
[
[
"# World's Fastest Growing Economies as of 2018\n\nThis project seeks to find out countries with eceonomy growth. This data was gotten from <a>\"https://en.wikipedia.org/wiki/List_of_countries_by_real_GDP_growth_rate\"<a> by web scraping and loading the table from the website. \n## Let's dive in.",
"_____no_output_____"
]
],
[
[
"from bs4 import BeautifulSoup as bs\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nimport pandas as pd\nimport numpy as np\nimport requests\nimport csv\n\nurl = \"https://en.wikipedia.org/wiki/List_of_countries_by_real_GDP_growth_rate\"\nreq = requests.get(url)",
"_____no_output_____"
],
[
"req.status_code",
"_____no_output_____"
],
[
"req.text",
"_____no_output_____"
],
[
"soup = bs(req.text, 'html5lib')",
"_____no_output_____"
],
[
"#soup.title",
"_____no_output_____"
],
[
"#links = soup.find_all('a')",
"_____no_output_____"
],
[
"\"\"\"\nfor link in links:\n print('----------------------------------------------------------------')\n print(link.get('href'))\n print('----------------------------------------------------------------')\nprint(len(links)) \n\"\"\" ",
"_____no_output_____"
],
[
"df = pd.read_html(\"https://en.wikipedia.org/wiki/List_of_countries_by_real_GDP_growth_rate\")",
"_____no_output_____"
],
[
"gdp_growthrate = df[1]",
"_____no_output_____"
],
[
"gdp_growthrate.dtypes",
"_____no_output_____"
],
[
"gdp_growthrate.head(10)",
"_____no_output_____"
],
[
"gdp_growthrate.shape",
"_____no_output_____"
],
[
"growth_overtime = df[2]",
"_____no_output_____"
],
[
"growth_overtime.head()",
"_____no_output_____"
],
[
"growth_overtime.shape",
"_____no_output_____"
],
[
"growth_overtime",
"_____no_output_____"
]
],
[
[
"## Cleaning The Data",
"_____no_output_____"
]
],
[
[
"gdp_growthrate.isnull().sum()",
"_____no_output_____"
],
[
"#Identity Missing Value Index\n\nmissing_data = gdp_growthrate[gdp_growthrate.isnull().any(axis=1)].index.values.tolist()\n\n\n#To get the missing values in a dataframe\ngdp_growthrate.iloc[missing_data,:]",
"_____no_output_____"
],
[
"gdp_growthrate.drop([189], axis=0, inplace=True)\ngdp_growthrate.isnull().sum()",
"_____no_output_____"
],
[
"gdp_growthrate.dtypes",
"_____no_output_____"
],
[
"gdp_growthrate = gdp_growthrate.rename( columns={'Real GDP growthrate (%)[2]' : \"GDP_growthrate%\"})",
"_____no_output_____"
],
[
"change = [\"GDP_growthrate%\"]\nfor col in change:\n gdp_growthrate[col] = gdp_growthrate[col].str.split('|', expand=True)[0]\n gdp_growthrate[col] = gdp_growthrate[col].str.replace(' ', '')\n\n\ngdp_growthrate['GDP_growthrate%'] = gdp_growthrate['GDP_growthrate%'].astype(float)",
"_____no_output_____"
],
[
"gdp_growthrate.dtypes",
"_____no_output_____"
],
[
"growth_overtime.isnull().sum()",
"_____no_output_____"
],
[
"growth_overtime.dtypes",
"_____no_output_____"
],
[
"#Identity Missing Value Index\nmissing = growth_overtime[growth_overtime.isnull().any(axis=1)].index.values.tolist()\n\n\n#To get the missing values in a dataframe\ngrowth_overtime.iloc[missing,:]\n\n",
"_____no_output_____"
],
[
"growth_overtime.drop(missing, axis=0, inplace=True)\ngrowth_overtime.isnull().sum()",
"_____no_output_____"
],
[
"growth_overtime = growth_overtime.rename( columns={'2018[4]' : 2018})",
"_____no_output_____"
],
[
"growth_overtime.iloc[0, :]",
"_____no_output_____"
],
[
"growth_overtime['Country'] = growth_overtime['Country'].str.split('.', expand=True)[0]",
"_____no_output_____"
],
[
"growth_overtime.head(2)",
"_____no_output_____"
],
[
"growth_overtime['Country'] = growth_overtime['Country'].str.split(' ', expand=True)[0]",
"_____no_output_____"
],
[
"growth_overtime.head(2)",
"_____no_output_____"
]
],
[
[
"# Data Exploratory Analysis",
"_____no_output_____"
]
],
[
[
"#Country with least growth over the years\n\nleast_growth = growth_overtime.sort_values(by='Avg').head()\nleast_growth",
"_____no_output_____"
],
[
"#Countries with Higest growth Over the years \n\nhighest_growth = growth_overtime.sort_values(by='Avg', ascending=False).head()\nhighest_growth",
"_____no_output_____"
],
[
"Country = highest_growth['Country']\nGrowth = highest_growth['Avg']\n \n# Figure Size\nfig = plt.figure(figsize =(8, 6))\n \nplt.barh(Country, Growth)\n \n# Show Plot\nplt.show()",
"_____no_output_____"
],
[
"Country = least_growth['Country']\nGrowth = least_growth['Avg'].sort_values()\n \n# Figure Size\nfig = plt.figure(figsize =(8, 6))\n \nplt.barh(Country, Growth)\n \n# Show Plot\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Insights \n\nFrom my analysis and graphs above, it can be seen that as of 2018, the country's with highest growth are Ethopia, Ireland, Ivory, Djibouti and Turkmenistan with an average growth of 9.40, 8.60, 8.34, 7.66 and 7.62 respectively. While Yemen has the least average growth of -11.76 over the years. ",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
c512db2c6f1b64909c7653a2f252ef71caed97a5
| 688,112 |
ipynb
|
Jupyter Notebook
|
notebooks/basic_ml/06_Multilayer_Perceptrons.ipynb
|
Calinou/practicalAI
|
9d6b3533ddb2601e6c54c9c5202e08227b9ead65
|
[
"MIT"
] | 1 |
2020-02-17T13:22:26.000Z
|
2020-02-17T13:22:26.000Z
|
notebooks/basic_ml/06_Multilayer_Perceptrons.ipynb
|
wqxdoc/practicalAI
|
9d6b3533ddb2601e6c54c9c5202e08227b9ead65
|
[
"MIT"
] | 6 |
2020-09-25T22:22:55.000Z
|
2022-02-10T02:13:16.000Z
|
notebooks/basic_ml/06_Multilayer_Perceptrons.ipynb
|
wqxdoc/practicalAI
|
9d6b3533ddb2601e6c54c9c5202e08227b9ead65
|
[
"MIT"
] | 1 |
2020-04-07T22:06:35.000Z
|
2020-04-07T22:06:35.000Z
| 169.027757 | 92,554 | 0.81623 |
[
[
[
"<a href=\"https://practicalai.me\"><img src=\"https://raw.githubusercontent.com/practicalAI/images/master/images/rounded_logo.png\" width=\"100\" align=\"left\" hspace=\"20px\" vspace=\"20px\"></a>\n\n<img src=\"https://raw.githubusercontent.com/practicalAI/images/master/basic_ml/06_Multilayer_Perceptron/nn.png\" width=\"200\" vspace=\"10px\" align=\"right\">\n\n<div align=\"left\">\n<h1>Multilayer Perceptron (MLP)</h1>\n\nIn this lesson, we will explore multilayer perceptrons (MLPs) which are a basic type of neural network. We will implement them using Tensorflow with Keras.",
"_____no_output_____"
],
[
"<table align=\"center\">\n <td>\n<img src=\"https://raw.githubusercontent.com/practicalAI/images/master/images/rounded_logo.png\" width=\"25\"><a target=\"_blank\" href=\"https://practicalai.me\"> View on practicalAI</a>\n </td>\n <td>\n<img src=\"https://raw.githubusercontent.com/practicalAI/images/master/images/colab_logo.png\" width=\"25\"><a target=\"_blank\" href=\"https://colab.research.google.com/github/practicalAI/practicalAI/blob/master/notebooks/06_Multilayer_Perceptron.ipynb\"> Run in Google Colab</a>\n </td>\n <td>\n<img src=\"https://raw.githubusercontent.com/practicalAI/images/master/images/github_logo.png\" width=\"22\"><a target=\"_blank\" href=\"https://github.com/practicalAI/practicalAI/blob/master/notebooks/basic_ml/06_Multilayer_Perceptron.ipynb\"> View code on GitHub</a>\n </td>\n</table>",
"_____no_output_____"
],
[
"# Overview",
"_____no_output_____"
],
[
"* **Objective:** Predict the probability of class $y$ given the inputs $X$. Non-linearity is introduced to model the complex, non-linear data.\n* **Advantages:**\n * Can model non-linear patterns in the data really well.\n* **Disadvantages:**\n * Overfits easily.\n * Computationally intensive as network increases in size.\n * Not easily interpretable.\n* **Miscellaneous:** Future neural network architectures that we'll see use the MLP as a modular unit for feed forward operations (affine transformation (XW) followed by a non-linear operation).",
"_____no_output_____"
],
[
"Our goal is to learn a model 𝑦̂ that models 𝑦 given 𝑋 . You'll notice that neural networks are just extensions of the generalized linear methods we've seen so far but with non-linear activation functions since our data will be highly non-linear.\n\n<img src=\"https://raw.githubusercontent.com/practicalAI/images/master/basic_ml/06_Multilayer_Perceptron/nn.png\" width=\"550\">\n\n$z_1 = XW_1$\n\n$a_1 = f(z_1)$\n\n$z_2 = a_1W_2$\n\n$\\hat{y} = softmax(z_2)$ # classification\n\n* $X$ = inputs | $\\in \\mathbb{R}^{NXD}$ ($D$ is the number of features)\n* $W_1$ = 1st layer weights | $\\in \\mathbb{R}^{DXH}$ ($H$ is the number of hidden units in layer 1)\n* $z_1$ = outputs from first layer $\\in \\mathbb{R}^{NXH}$\n* $f$ = non-linear activation function\n*nn $a_1$ = activation applied first layer's outputs | $\\in \\mathbb{R}^{NXH}$\n* $W_2$ = 2nd layer weights | $\\in \\mathbb{R}^{HXC}$ ($C$ is the number of classes)\n* $z_2$ = outputs from second layer $\\in \\mathbb{R}^{NXH}$\n* $\\hat{y}$ = prediction | $\\in \\mathbb{R}^{NXC}$ ($N$ is the number of samples)",
"_____no_output_____"
],
[
"**Note**: We're going to leave out the bias terms $\\beta$ to avoid further crowding the backpropagation calculations.",
"_____no_output_____"
],
[
"### Training",
"_____no_output_____"
],
[
"1. Randomly initialize the model's weights $W$ (we'll cover more effective initalization strategies later in this lesson).\n2. Feed inputs $X$ into the model to do the forward pass and receive the probabilities.\n * $z_1 = XW_1$\n * $a_1 = f(z_1)$\n * $z_2 = a_1W_2$\n * $\\hat{y} = softmax(z_2)$\n3. Compare the predictions $\\hat{y}$ (ex. [0.3, 0.3, 0.4]]) with the actual target values $y$ (ex. class 2 would look like [0, 0, 1]) with the objective (cost) function to determine loss $J$. A common objective function for classification tasks is cross-entropy loss. \n * $J(\\theta) = - \\sum_i y_i ln (\\hat{y_i}) $\n * Since each input maps to exactly one class, our cross-entropy loss simplifies to: \n * $J(\\theta) = - \\sum_i ln(\\hat{y_i}) = - \\sum_i ln (\\frac{e^{X_iW_y}}{\\sum_j e^{X_iW}}) $\n4. Calculate the gradient of loss $J(\\theta)$ w.r.t to the model weights. \n * $\\frac{\\partial{J}}{\\partial{W_{2j}}} = \\frac{\\partial{J}}{\\partial{\\hat{y}}}\\frac{\\partial{\\hat{y}}}{\\partial{W_{2j}}} = - \\frac{1}{\\hat{y}}\\frac{\\partial{\\hat{y}}}{\\partial{W_{2j}}} = - \\frac{1}{\\frac{e^{W_{2y}a_1}}{\\sum_j e^{a_1W}}}\\frac{\\sum_j e^{a_1W}e^{a_1W_{2y}}0 - e^{a_1W_{2y}}e^{a_1W_{2j}}a_1}{(\\sum_j e^{a_1W})^2} = \\frac{a_1e^{a_1W_{2j}}}{\\sum_j e^{a_1W}} = a_1\\hat{y}$\n * $\\frac{\\partial{J}}{\\partial{W_{2y}}} = \\frac{\\partial{J}}{\\partial{\\hat{y}}}\\frac{\\partial{\\hat{y}}}{\\partial{W_{2y}}} = - \\frac{1}{\\hat{y}}\\frac{\\partial{\\hat{y}}}{\\partial{W_{2y}}} = - \\frac{1}{\\frac{e^{W_{2y}a_1}}{\\sum_j e^{a_1W}}}\\frac{\\sum_j e^{a_1W}e^{a_1W_{2y}}a_1 - e^{a_1W_{2y}}e^{a_1W_{2y}}a_1}{(\\sum_j e^{a_1W})^2} = \\frac{1}{\\hat{y}}(a_1\\hat{y} - a_1\\hat{y}^2) = a_1(\\hat{y}-1)$\n * $ \\frac{\\partial{J}}{\\partial{W_1}} = \\frac{\\partial{J}}{\\partial{\\hat{y}}} \\frac{\\partial{\\hat{y}}}{\\partial{a_2}} \\frac{\\partial{a_2}}{\\partial{z_2}} \\frac{\\partial{z_2}}{\\partial{W_1}} = W_2(\\partial{scores})(\\partial{ReLU})X $\n \n5. Update the weights $W$ using a small learning rate $\\alpha$. The updates will penalize the probabiltiy for the incorrect classes (j) and encourage a higher probability for the correct class (y).\n * $W_i = W_i - \\alpha\\frac{\\partial{J}}{\\partial{W_i}}$\n6. Repeat steps 2 - 4 until model performs well.",
"_____no_output_____"
],
[
"# Set up",
"_____no_output_____"
]
],
[
[
"# Use TensorFlow 2.x\n%tensorflow_version 2.x",
"TensorFlow 2.x selected.\n"
],
[
"import os\nimport numpy as np\nimport tensorflow as tf",
"_____no_output_____"
],
[
"# Arguments\nSEED = 1234\nSHUFFLE = True\nDATA_FILE = \"spiral.csv\"\nINPUT_DIM = 2\nNUM_CLASSES = 3\nNUM_SAMPLES_PER_CLASS = 500\nTRAIN_SIZE = 0.7\nVAL_SIZE = 0.15\nTEST_SIZE = 0.15\nNUM_EPOCHS = 10\nBATCH_SIZE = 32\nHIDDEN_DIM = 100\nLEARNING_RATE = 1e-2",
"_____no_output_____"
],
[
"# Set seed for reproducability\nnp.random.seed(SEED)\ntf.random.set_seed(SEED)",
"_____no_output_____"
]
],
[
[
"# Data",
"_____no_output_____"
],
[
"Download non-linear spiral data for a classification task.",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nimport pandas as pd\nimport urllib",
"_____no_output_____"
],
[
"# Upload data from GitHub to notebook's local drive\nurl = \"https://raw.githubusercontent.com/practicalAI/practicalAI/master/data/spiral.csv\"\nresponse = urllib.request.urlopen(url)\nhtml = response.read()\nwith open(DATA_FILE, 'wb') as fp:\n fp.write(html)",
"_____no_output_____"
],
[
"# Load data\ndf = pd.read_csv(DATA_FILE, header=0)\nX = df[['X1', 'X2']].values\ny = df['color'].values\ndf.head(5)",
"_____no_output_____"
],
[
"print (\"X: \", np.shape(X))\nprint (\"y: \", np.shape(y))",
"X: (1500, 2)\ny: (1500,)\n"
],
[
"# Visualize data\nplt.title(\"Generated non-linear data\")\ncolors = {'c1': 'red', 'c2': 'yellow', 'c3': 'blue'}\nplt.scatter(X[:, 0], X[:, 1], c=[colors[_y] for _y in y], edgecolors='k', s=25)\nplt.show()",
"_____no_output_____"
]
],
[
[
"# Split data",
"_____no_output_____"
]
],
[
[
"import collections\nimport json\nfrom sklearn.model_selection import train_test_split",
"_____no_output_____"
]
],
[
[
"### Components",
"_____no_output_____"
]
],
[
[
"def train_val_test_split(X, y, val_size, test_size, shuffle):\n \"\"\"Split data into train/val/test datasets.\n \"\"\"\n X_train, X_test, y_train, y_test = train_test_split(\n X, y, test_size=test_size, stratify=y, shuffle=shuffle)\n X_train, X_val, y_train, y_val = train_test_split(\n X_train, y_train, test_size=val_size, stratify=y_train, shuffle=shuffle)\n return X_train, X_val, X_test, y_train, y_val, y_test",
"_____no_output_____"
]
],
[
[
"### Operations",
"_____no_output_____"
]
],
[
[
"# Create data splits\nX_train, X_val, X_test, y_train, y_val, y_test = train_val_test_split(\n X=X, y=y, val_size=VAL_SIZE, test_size=TEST_SIZE, shuffle=SHUFFLE)\nclass_counts = dict(collections.Counter(y))\nprint (f\"X_train: {X_train.shape}, y_train: {y_train.shape}\")\nprint (f\"X_val: {X_val.shape}, y_val: {y_val.shape}\")\nprint (f\"X_test: {X_test.shape}, y_test: {y_test.shape}\")\nprint (f\"X_train[0]: {X_train[0]}\")\nprint (f\"y_train[0]: {y_train[0]}\")\nprint (f\"Classes: {class_counts}\")",
"X_train: (1083, 2), y_train: (1083,)\nX_val: (192, 2), y_val: (192,)\nX_test: (225, 2), y_test: (225,)\nX_train[0]: [ 0.23623443 -0.59618506]\ny_train[0]: c1\nClasses: {'c1': 500, 'c3': 500, 'c2': 500}\n"
]
],
[
[
"# Label encoder",
"_____no_output_____"
]
],
[
[
"import json\nfrom sklearn.preprocessing import LabelEncoder",
"_____no_output_____"
],
[
"# Output vectorizer\ny_tokenizer = LabelEncoder()",
"_____no_output_____"
],
[
"# Fit on train data\ny_tokenizer = y_tokenizer.fit(y_train)\nclasses = list(y_tokenizer.classes_)\nprint (f\"classes: {classes}\")",
"classes: ['c1', 'c2', 'c3']\n"
],
[
"# Convert labels to tokens\nprint (f\"y_train[0]: {y_train[0]}\")\ny_train = y_tokenizer.transform(y_train)\ny_val = y_tokenizer.transform(y_val)\ny_test = y_tokenizer.transform(y_test)\nprint (f\"y_train[0]: {y_train[0]}\")",
"y_train[0]: c1\ny_train[0]: 0\n"
],
[
"# Class weights\ncounts = collections.Counter(y_train)\nclass_weights = {_class: 1.0/count for _class, count in counts.items()}\nprint (f\"class counts: {counts},\\nclass weights: {class_weights}\")",
"class counts: Counter({0: 361, 2: 361, 1: 361}),\nclass weights: {0: 0.002770083102493075, 2: 0.002770083102493075, 1: 0.002770083102493075}\n"
]
],
[
[
"# Standardize data",
"_____no_output_____"
],
[
"We need to standardize our data (zero mean and unit variance) in order to optimize quickly. We're only going to standardize the inputs X because out outputs y are class values.",
"_____no_output_____"
]
],
[
[
"from sklearn.preprocessing import StandardScaler",
"_____no_output_____"
],
[
"# Standardize the data (mean=0, std=1) using training data\nX_scaler = StandardScaler().fit(X_train)",
"_____no_output_____"
],
[
"# Apply scaler on training and test data (don't standardize outputs for classification)\nstandardized_X_train = X_scaler.transform(X_train)\nstandardized_X_val = X_scaler.transform(X_val)\nstandardized_X_test = X_scaler.transform(X_test)",
"_____no_output_____"
],
[
"# Check\nprint (f\"standardized_X_train: mean: {np.mean(standardized_X_train, axis=0)[0]}, std: {np.std(standardized_X_train, axis=0)[0]}\")\nprint (f\"standardized_X_val: mean: {np.mean(standardized_X_val, axis=0)[0]}, std: {np.std(standardized_X_val, axis=0)[0]}\")\nprint (f\"standardized_X_test: mean: {np.mean(standardized_X_test, axis=0)[0]}, std: {np.std(standardized_X_test, axis=0)[0]}\")",
"standardized_X_train: mean: 2.0051673464144102e-16, std: 1.0000000000000002\nstandardized_X_val: mean: 0.10067009920283043, std: 1.020838918120433\nstandardized_X_test: mean: 0.1287588265908901, std: 0.9647767079653526\n"
]
],
[
[
"# Linear model",
"_____no_output_____"
],
[
"Before we get to our neural network, we're going to implement a generalized linear model (logistic regression) first to see why linear models won't suffice for our dataset. We will use Tensorflow with Keras to do this. ",
"_____no_output_____"
]
],
[
[
"import itertools\nimport matplotlib.pyplot as plt\nfrom sklearn.metrics import accuracy_score\nfrom sklearn.metrics import classification_report\nfrom sklearn.metrics import confusion_matrix\nfrom tensorflow.keras.layers import Dense\nfrom tensorflow.keras.layers import Input\nfrom tensorflow.keras.losses import SparseCategoricalCrossentropy\nfrom tensorflow.keras.models import Model\nfrom tensorflow.keras.optimizers import Adam",
"_____no_output_____"
]
],
[
[
"### Components",
"_____no_output_____"
]
],
[
[
"# Linear model\nclass LogisticClassifier(Model):\n def __init__(self, hidden_dim, num_classes):\n super(LogisticClassifier, self).__init__()\n self.fc1 = Dense(units=hidden_dim, activation='linear') # linear = no activation function\n self.fc2 = Dense(units=num_classes, activation='softmax')\n \n def call(self, x_in, training=False):\n \"\"\"Forward pass.\"\"\"\n z = self.fc1(x_in)\n y_pred = self.fc2(z)\n return y_pred\n \n def sample(self, input_shape):\n x_in = Input(shape=input_shape)\n return Model(inputs=x_in, outputs=self.call(x_in)).summary()",
"_____no_output_____"
],
[
"def plot_confusion_matrix(y_true, y_pred, classes, cmap=plt.cm.Blues):\n \"\"\"Plot a confusion matrix using ground truth and predictions.\"\"\"\n # Confusion matrix\n cm = confusion_matrix(y_true, y_pred)\n cm_norm = cm.astype('float') / cm.sum(axis=1)[:, np.newaxis]\n\n # Figure\n fig = plt.figure()\n ax = fig.add_subplot(111)\n cax = ax.matshow(cm, cmap=plt.cm.Blues)\n fig.colorbar(cax)\n\n # Axis\n plt.title(\"Confusion matrix\")\n plt.ylabel(\"True label\")\n plt.xlabel(\"Predicted label\")\n ax.set_xticklabels([''] + classes)\n ax.set_yticklabels([''] + classes)\n ax.xaxis.set_label_position('bottom') \n ax.xaxis.tick_bottom()\n\n # Values\n thresh = cm.max() / 2.\n for i, j in itertools.product(range(cm.shape[0]), range(cm.shape[1])):\n plt.text(j, i, f\"{cm[i, j]:d} ({cm_norm[i, j]*100:.1f}%)\",\n horizontalalignment=\"center\",\n color=\"white\" if cm[i, j] > thresh else \"black\")\n\n # Display\n plt.show()",
"_____no_output_____"
],
[
"def plot_multiclass_decision_boundary(model, X, y, savefig_fp=None):\n \"\"\"Plot the multiclass decision boundary for a model that accepts 2D inputs.\n\n Arguments:\n model {function} -- trained model with function model.predict(x_in).\n X {numpy.ndarray} -- 2D inputs with shape (N, 2).\n y {numpy.ndarray} -- 1D outputs with shape (N,).\n \"\"\"\n # Axis boundaries\n x_min, x_max = X[:, 0].min() - 0.1, X[:, 0].max() + 0.1\n y_min, y_max = X[:, 1].min() - 0.1, X[:, 1].max() + 0.1\n xx, yy = np.meshgrid(np.linspace(x_min, x_max, 101),\n np.linspace(y_min, y_max, 101))\n\n # Create predictions\n x_in = np.c_[xx.ravel(), yy.ravel()]\n y_pred = model.predict(x_in)\n y_pred = np.argmax(y_pred, axis=1).reshape(xx.shape)\n\n # Plot decision boundary\n plt.contourf(xx, yy, y_pred, cmap=plt.cm.Spectral, alpha=0.8)\n plt.scatter(X[:, 0], X[:, 1], c=y, s=40, cmap=plt.cm.RdYlBu)\n plt.xlim(xx.min(), xx.max())\n plt.ylim(yy.min(), yy.max())\n\n # Plot\n if savefig_fp:\n plt.savefig(savefig_fp, format='png')",
"_____no_output_____"
]
],
[
[
"### Operations",
"_____no_output_____"
]
],
[
[
"# Initialize the model\nmodel = LogisticClassifier(hidden_dim=HIDDEN_DIM,\n num_classes=NUM_CLASSES)\nmodel.sample(input_shape=(INPUT_DIM,))",
"Model: \"model\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\ninput_1 (InputLayer) [(None, 2)] 0 \n_________________________________________________________________\ndense (Dense) (None, 100) 300 \n_________________________________________________________________\ndense_1 (Dense) (None, 3) 303 \n=================================================================\nTotal params: 603\nTrainable params: 603\nNon-trainable params: 0\n_________________________________________________________________\n"
],
[
"# Compile\nmodel.compile(optimizer=Adam(lr=LEARNING_RATE),\n loss=SparseCategoricalCrossentropy(),\n metrics=['accuracy'])",
"_____no_output_____"
],
[
"# Training\nmodel.fit(x=standardized_X_train, \n y=y_train,\n validation_data=(standardized_X_val, y_val),\n epochs=NUM_EPOCHS,\n batch_size=BATCH_SIZE,\n class_weight=class_weights,\n shuffle=False,\n verbose=1)",
"Train on 1083 samples, validate on 192 samples\nEpoch 1/10\n1083/1083 [==============================] - 2s 2ms/sample - loss: 0.0022 - accuracy: 0.5199 - val_loss: 0.0021 - val_accuracy: 0.4948\nEpoch 2/10\n1083/1083 [==============================] - 0s 143us/sample - loss: 0.0021 - accuracy: 0.5254 - val_loss: 0.0021 - val_accuracy: 0.5052\nEpoch 3/10\n1083/1083 [==============================] - 0s 138us/sample - loss: 0.0021 - accuracy: 0.5263 - val_loss: 0.0021 - val_accuracy: 0.5104\nEpoch 4/10\n1083/1083 [==============================] - 0s 143us/sample - loss: 0.0021 - accuracy: 0.5254 - val_loss: 0.0021 - val_accuracy: 0.5104\nEpoch 5/10\n1083/1083 [==============================] - 0s 138us/sample - loss: 0.0021 - accuracy: 0.5254 - val_loss: 0.0021 - val_accuracy: 0.5000\nEpoch 6/10\n1083/1083 [==============================] - 0s 163us/sample - loss: 0.0021 - accuracy: 0.5254 - val_loss: 0.0021 - val_accuracy: 0.5000\nEpoch 7/10\n1083/1083 [==============================] - 0s 140us/sample - loss: 0.0021 - accuracy: 0.5235 - val_loss: 0.0021 - val_accuracy: 0.5000\nEpoch 8/10\n1083/1083 [==============================] - 0s 138us/sample - loss: 0.0021 - accuracy: 0.5226 - val_loss: 0.0021 - val_accuracy: 0.5000\nEpoch 9/10\n1083/1083 [==============================] - 0s 143us/sample - loss: 0.0021 - accuracy: 0.5235 - val_loss: 0.0021 - val_accuracy: 0.5052\nEpoch 10/10\n1083/1083 [==============================] - 0s 143us/sample - loss: 0.0021 - accuracy: 0.5245 - val_loss: 0.0021 - val_accuracy: 0.5052\n"
],
[
"# Predictions\npred_train = model.predict(standardized_X_train) \npred_test = model.predict(standardized_X_test)\nprint (f\"sample probability: {pred_test[0]}\")\npred_train = np.argmax(pred_train, axis=1)\npred_test = np.argmax(pred_test, axis=1)\nprint (f\"sample class: {pred_test[0]}\")",
"sample probability: [0.02820205 0.13940957 0.8323884 ]\nsample class: 2\n"
],
[
"# Accuracy\ntrain_acc = accuracy_score(y_train, pred_train)\ntest_acc = accuracy_score(y_test, pred_test)\nprint (f\"train acc: {train_acc:.2f}, test acc: {test_acc:.2f}\")",
"train acc: 0.53, test acc: 0.54\n"
],
[
"# Metrics\nplot_confusion_matrix(y_test, pred_test, classes=classes)\nprint (classification_report(y_test, pred_test))",
"_____no_output_____"
],
[
"# Visualize the decision boundary\nplt.figure(figsize=(12,5))\nplt.subplot(1, 2, 1)\nplt.title(\"Train\")\nplot_multiclass_decision_boundary(model=model, X=standardized_X_train, y=y_train)\nplt.subplot(1, 2, 2)\nplt.title(\"Test\")\nplot_multiclass_decision_boundary(model=model, X=standardized_X_test, y=y_test)\nplt.show()",
"_____no_output_____"
]
],
[
[
"# Activation functions",
"_____no_output_____"
],
[
"Using the generalized linear method (logistic regression) yielded poor results because of the non-linearity present in our data. We need to use an activation function that can allow our model to learn and map the non-linearity in our data. There are many different options so let's explore a few.",
"_____no_output_____"
]
],
[
[
"from tensorflow.keras.activations import relu\nfrom tensorflow.keras.activations import sigmoid\nfrom tensorflow.keras.activations import tanh",
"_____no_output_____"
],
[
"# Fig size\nplt.figure(figsize=(12,3))\n\n# Data\nx = np.arange(-5., 5., 0.1)\n\n# Sigmoid activation (constrain a value between 0 and 1.)\nplt.subplot(1, 3, 1)\nplt.title(\"Sigmoid activation\")\ny = sigmoid(x)\nplt.plot(x, y)\n\n# Tanh activation (constrain a value between -1 and 1.)\nplt.subplot(1, 3, 2)\ny = tanh(x)\nplt.title(\"Tanh activation\")\nplt.plot(x, y)\n\n# Relu (clip the negative values to 0)\nplt.subplot(1, 3, 3)\ny = relu(x)\nplt.title(\"ReLU activation\")\nplt.plot(x, y)\n\n# Show plots\nplt.show()",
"_____no_output_____"
]
],
[
[
"The ReLU activation function ($max(0,z)$) is by far the most widely used activation function for neural networks. But as you can see, each activation function has it's own contraints so there are circumstances where you'll want to use different ones. For example, if we need to constrain our outputs between 0 and 1, then the sigmoid activation is the best choice.",
"_____no_output_____"
],
[
"<img height=\"45\" src=\"http://bestanimations.com/HomeOffice/Lights/Bulbs/animated-light-bulb-gif-29.gif\" align=\"left\" vspace=\"20px\" hspace=\"10px\">\n\nIn some cases, using a ReLU activation function may not be sufficient. For instance, when the outputs from our neurons are mostly negative, the activation function will produce zeros. This effectively creates a \"dying ReLU\" and a recovery is unlikely. To mitigate this effect, we could lower the learning rate or use [alternative ReLU activations](https://medium.com/tinymind/a-practical-guide-to-relu-b83ca804f1f7), ex. leaky ReLU or parametric ReLU (PReLU), which have a small slope for negative neuron outputs. ",
"_____no_output_____"
],
[
"# From scratch\n\nNow let's create our multilayer perceptron (MLP) which is going to be exactly like the logistic regression model but with the activation function to map the non-linearity in our data. Before we use TensorFlow 2.0 + Keras we will implement our neural network from scratch using NumPy so we can:\n1. Absorb the fundamental concepts by implementing from scratch\n2. Appreciate the level of abstraction TensorFlow provides\n\n<div align=\"left\">\n<img src=\"https://raw.githubusercontent.com/practicalAI/images/master/images/lightbulb.gif\" width=\"45px\" align=\"left\" hspace=\"10px\">\n</div> \n\nIt's normal to find the math and code in this section slightly complex. You can still read each of the steps to build intuition for when we implement this using TensorFlow + Keras.\n",
"_____no_output_____"
]
],
[
[
"print (f\"X: {standardized_X_train.shape}\")\nprint (f\"y: {y_train.shape}\")",
"X: (1083, 2)\ny: (1083,)\n"
]
],
[
[
"Our goal is to learn a model 𝑦̂ that models 𝑦 given 𝑋 . You'll notice that neural networks are just extensions of the generalized linear methods we've seen so far but with non-linear activation functions since our data will be highly non-linear.\n\n$z_1 = XW_1$\n\n$a_1 = f(z_1)$\n\n$z_2 = a_1W_2$\n\n$\\hat{y} = softmax(z_2)$ # classification\n\n* $X$ = inputs | $\\in \\mathbb{R}^{NXD}$ ($D$ is the number of features)\n* $W_1$ = 1st layer weights | $\\in \\mathbb{R}^{DXH}$ ($H$ is the number of hidden units in layer 1)\n* $z_1$ = outputs from first layer $\\in \\mathbb{R}^{NXH}$\n* $f$ = non-linear activation function\n* $a_1$ = activation applied first layer's outputs | $\\in \\mathbb{R}^{NXH}$\n* $W_2$ = 2nd layer weights | $\\in \\mathbb{R}^{HXC}$ ($C$ is the number of classes)\n* $z_2$ = outputs from second layer $\\in \\mathbb{R}^{NXH}$\n* $\\hat{y}$ = prediction | $\\in \\mathbb{R}^{NXC}$ ($N$ is the number of samples)",
"_____no_output_____"
],
[
"1. Randomly initialize the model's weights $W$ (we'll cover more effective initalization strategies later in this lesson).",
"_____no_output_____"
]
],
[
[
"# Initialize first layer's weights\nW1 = 0.01 * np.random.randn(INPUT_DIM, HIDDEN_DIM)\nb1 = np.zeros((1, HIDDEN_DIM))\nprint (f\"W1: {W1.shape}\")\nprint (f\"b1: {b1.shape}\")",
"W1: (2, 100)\nb1: (1, 100)\n"
]
],
[
[
"2. Feed inputs $X$ into the model to do the forward pass and receive the probabilities.",
"_____no_output_____"
],
[
"First we pass the inputs into the first layer.\n * $z_1 = XW_1$",
"_____no_output_____"
]
],
[
[
"# z1 = [NX2] · [2X100] + [1X100] = [NX100]\nz1 = np.dot(standardized_X_train, W1) + b1\nprint (f\"z1: {z1.shape}\")",
"z1: (1083, 100)\n"
]
],
[
[
"Next we apply the non-linear activation function, ReLU ($max(0,z)$) in this case.\n * $a_1 = f(z_1)$",
"_____no_output_____"
]
],
[
[
"# Apply activation function\na1 = np.maximum(0, z1) # ReLU\nprint (f\"a_1: {a1.shape}\")",
"a_1: (1083, 100)\n"
]
],
[
[
"We pass the activations to the second layer to get our logits.\n * $z_2 = a_1W_2$",
"_____no_output_____"
]
],
[
[
"# Initialize second layer's weights\nW2 = 0.01 * np.random.randn(HIDDEN_DIM, NUM_CLASSES)\nb2 = np.zeros((1, NUM_CLASSES))\nprint (f\"W2: {W2.shape}\")\nprint (f\"b2: {b2.shape}\")",
"W2: (100, 3)\nb2: (1, 3)\n"
],
[
"# z2 = logits = [NX100] · [100X3] + [1X3] = [NX3]\nlogits = np.dot(a1, W2) + b2\nprint (f\"logits: {logits.shape}\")\nprint (f\"sample: {logits[0]}\")",
"logits: (1083, 3)\nsample: [ 0.0004134 0.0002782 -0.00118021]\n"
]
],
[
[
"We'll apply the softmax function to normalize the logits and btain class probabilities.\n * $\\hat{y} = softmax(z_2)$",
"_____no_output_____"
]
],
[
[
"# Normalization via softmax to obtain class probabilities\nexp_logits = np.exp(logits)\ny_hat = exp_logits / np.sum(exp_logits, axis=1, keepdims=True)\nprint (f\"y_hat: {y_hat.shape}\")\nprint (f\"sample: {y_hat[0]}\")",
"y_hat: (1083, 3)\nsample: [0.33352539 0.3334803 0.33299431]\n"
]
],
[
[
"3. Compare the predictions $\\hat{y}$ (ex. [0.3, 0.3, 0.4]]) with the actual target values $y$ (ex. class 2 would look like [0, 0, 1]) with the objective (cost) function to determine loss $J$. A common objective function for classification tasks is cross-entropy loss. \n * $J(\\theta) = - \\sum_i ln(\\hat{y_i}) = - \\sum_i ln (\\frac{e^{X_iW_y}}{\\sum_j e^{X_iW}}) $",
"_____no_output_____"
]
],
[
[
"# Loss\ncorrect_class_logprobs = -np.log(y_hat[range(len(y_hat)), y_train])\nloss = np.sum(correct_class_logprobs) / len(y_train)",
"_____no_output_____"
]
],
[
[
"4. Calculate the gradient of loss $J(\\theta)$ w.r.t to the model weights. \n \n The gradient of the loss w.r.t to W2 is the same as the gradients from logistic regression since $\\hat{y} = softmax(z_2)$.\n * $\\frac{\\partial{J}}{\\partial{W_{2j}}} = \\frac{\\partial{J}}{\\partial{\\hat{y}}}\\frac{\\partial{\\hat{y}}}{\\partial{W_{2j}}} = - \\frac{1}{\\hat{y}}\\frac{\\partial{\\hat{y}}}{\\partial{W_{2j}}} = - \\frac{1}{\\frac{e^{W_{2y}a_1}}{\\sum_j e^{a_1W}}}\\frac{\\sum_j e^{a_1W}e^{a_1W_{2y}}0 - e^{a_1W_{2y}}e^{a_1W_{2j}}a_1}{(\\sum_j e^{a_1W})^2} = \\frac{a_1e^{a_1W_{2j}}}{\\sum_j e^{a_1W}} = a_1\\hat{y}$\n * $\\frac{\\partial{J}}{\\partial{W_{2y}}} = \\frac{\\partial{J}}{\\partial{\\hat{y}}}\\frac{\\partial{\\hat{y}}}{\\partial{W_{2y}}} = - \\frac{1}{\\hat{y}}\\frac{\\partial{\\hat{y}}}{\\partial{W_{2y}}} = - \\frac{1}{\\frac{e^{W_{2y}a_1}}{\\sum_j e^{a_1W}}}\\frac{\\sum_j e^{a_1W}e^{a_1W_{2y}}a_1 - e^{a_1W_{2y}}e^{a_1W_{2y}}a_1}{(\\sum_j e^{a_1W})^2} = \\frac{1}{\\hat{y}}(a_1\\hat{y} - a_1\\hat{y}^2) = a_1(\\hat{y}-1)$\n\n The gradient of the loss w.r.t W1 is a bit trickier since we have to backpropagate through two sets of weights.\n * $ \\frac{\\partial{J}}{\\partial{W_1}} = \\frac{\\partial{J}}{\\partial{\\hat{y}}} \\frac{\\partial{\\hat{y}}}{\\partial{a_1}} \\frac{\\partial{a_1}}{\\partial{z_1}} \\frac{\\partial{z_1}}{\\partial{W_1}} = W_2(\\partial{scores})(\\partial{ReLU})X $",
"_____no_output_____"
]
],
[
[
"# dJ/dW2\ndscores = y_hat\ndscores[range(len(y_hat)), y_train] -= 1\ndscores /= len(y_train)\ndW2 = np.dot(a1.T, dscores)\ndb2 = np.sum(dscores, axis=0, keepdims=True)",
"_____no_output_____"
],
[
"# dJ/dW1\ndhidden = np.dot(dscores, W2.T)\ndhidden[a1 <= 0] = 0 # ReLu backprop\ndW1 = np.dot(standardized_X_train.T, dhidden)\ndb1 = np.sum(dhidden, axis=0, keepdims=True)",
"_____no_output_____"
]
],
[
[
"5. Update the weights $W$ using a small learning rate $\\alpha$. The updates will penalize the probabiltiy for the incorrect classes (j) and encourage a higher probability for the correct class (y).\n * $W_i = W_i - \\alpha\\frac{\\partial{J}}{\\partial{W_i}}$",
"_____no_output_____"
]
],
[
[
"# Update weights\nW1 += -LEARNING_RATE * dW1\nb1 += -LEARNING_RATE * db1\nW2 += -LEARNING_RATE * dW2\nb2 += -LEARNING_RATE * db2",
"_____no_output_____"
]
],
[
[
"6. Repeat steps 2 - 4 until model performs well.",
"_____no_output_____"
]
],
[
[
"# Initialize random weights\nW1 = 0.01 * np.random.randn(INPUT_DIM, HIDDEN_DIM)\nb1 = np.zeros((1, HIDDEN_DIM))\nW2 = 0.01 * np.random.randn(HIDDEN_DIM, NUM_CLASSES)\nb2 = np.zeros((1, NUM_CLASSES))\n\n# Training loop\nfor epoch_num in range(1000):\n\n # First layer forward pass [NX2] · [2X100] = [NX100]\n z1 = np.dot(standardized_X_train, W1) + b1\n\n # Apply activation function\n a1 = np.maximum(0, z1) # ReLU\n\n # z2 = logits = [NX100] · [100X3] = [NX3]\n logits = np.dot(a1, W2) + b2\n \n # Normalization via softmax to obtain class probabilities\n exp_logits = np.exp(logits)\n y_hat = exp_logits / np.sum(exp_logits, axis=1, keepdims=True)\n\n # Loss\n correct_class_logprobs = -np.log(y_hat[range(len(y_hat)), y_train])\n loss = np.sum(correct_class_logprobs) / len(y_train)\n\n # show progress\n if epoch_num%100 == 0:\n # Accuracy\n y_pred = np.argmax(logits, axis=1)\n accuracy = np.mean(np.equal(y_train, y_pred))\n print (f\"Epoch: {epoch_num}, loss: {loss:.3f}, accuracy: {accuracy:.3f}\")\n\n # dJ/dW2\n dscores = y_hat\n dscores[range(len(y_hat)), y_train] -= 1\n dscores /= len(y_train)\n dW2 = np.dot(a1.T, dscores)\n db2 = np.sum(dscores, axis=0, keepdims=True)\n\n # dJ/dW1\n dhidden = np.dot(dscores, W2.T)\n dhidden[a1 <= 0] = 0 # ReLu backprop\n dW1 = np.dot(standardized_X_train.T, dhidden)\n db1 = np.sum(dhidden, axis=0, keepdims=True)\n\n # Update weights\n W1 += -1e0 * dW1\n b1 += -1e0 * db1\n W2 += -1e0 * dW2\n b2 += -1e0 * db2",
"Epoch: 0, loss: 1.099, accuracy: 0.243\nEpoch: 100, loss: 0.551, accuracy: 0.680\nEpoch: 200, loss: 0.223, accuracy: 0.912\nEpoch: 300, loss: 0.131, accuracy: 0.951\nEpoch: 400, loss: 0.090, accuracy: 0.976\nEpoch: 500, loss: 0.069, accuracy: 0.985\nEpoch: 600, loss: 0.056, accuracy: 0.994\nEpoch: 700, loss: 0.048, accuracy: 0.996\nEpoch: 800, loss: 0.043, accuracy: 0.997\nEpoch: 900, loss: 0.039, accuracy: 0.997\n"
],
[
"class MLPFromScratch():\n def predict(self, x):\n z1 = np.dot(x, W1) + b1\n a1 = np.maximum(0, z1)\n logits = np.dot(a1, W2) + b2\n exp_logits = np.exp(logits)\n y_hat = exp_logits / np.sum(exp_logits, axis=1, keepdims=True)\n return y_hat",
"_____no_output_____"
],
[
"# Evaluation\nmodel = MLPFromScratch()\nlogits_train = model.predict(standardized_X_train)\npred_train = np.argmax(logits_train, axis=1)\nlogits_test = model.predict(standardized_X_test)\npred_test = np.argmax(logits_test, axis=1)",
"_____no_output_____"
],
[
"# Training and test accuracy\ntrain_acc = np.mean(np.equal(y_train, pred_train))\ntest_acc = np.mean(np.equal(y_test, pred_test))\nprint (f\"train acc: {train_acc:.2f}, test acc: {test_acc:.2f}\")",
"train acc: 1.00, test acc: 1.00\n"
],
[
"# Visualize the decision boundary\nplt.figure(figsize=(12,5))\nplt.subplot(1, 2, 1)\nplt.title(\"Train\")\nplot_multiclass_decision_boundary(model=model, X=standardized_X_train, y=y_train)\nplt.subplot(1, 2, 2)\nplt.title(\"Test\")\nplot_multiclass_decision_boundary(model=model, X=standardized_X_test, y=y_test)\nplt.show()",
"_____no_output_____"
]
],
[
[
"Credit for the plotting functions and the intuition behind all this is due to [CS231n](http://cs231n.github.io/neural-networks-case-study/), one of the best courses for machine learning. Now let's implement the MLP with TensorFlow + Keras.",
"_____no_output_____"
],
[
"# TensorFlow + Keras",
"_____no_output_____"
],
[
"### Components",
"_____no_output_____"
]
],
[
[
"# MLP\nclass MLP(Model):\n def __init__(self, hidden_dim, num_classes):\n super(MLP, self).__init__()\n self.fc1 = Dense(units=hidden_dim, activation='relu') # replaced linear with relu\n self.fc2 = Dense(units=num_classes, activation='softmax')\n \n def call(self, x_in, training=False):\n \"\"\"Forward pass.\"\"\"\n z = self.fc1(x_in)\n y_pred = self.fc2(z)\n return y_pred\n \n def sample(self, input_shape):\n x_in = Input(shape=input_shape)\n return Model(inputs=x_in, outputs=self.call(x_in)).summary()",
"_____no_output_____"
]
],
[
[
"### Operations",
"_____no_output_____"
]
],
[
[
"# Initialize the model\nmodel = MLP(hidden_dim=HIDDEN_DIM,\n num_classes=NUM_CLASSES)\nmodel.sample(input_shape=(INPUT_DIM,))",
"Model: \"model_1\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\ninput_2 (InputLayer) [(None, 2)] 0 \n_________________________________________________________________\ndense_2 (Dense) (None, 100) 300 \n_________________________________________________________________\ndense_3 (Dense) (None, 3) 303 \n=================================================================\nTotal params: 603\nTrainable params: 603\nNon-trainable params: 0\n_________________________________________________________________\n"
],
[
"# Compile\noptimizer = Adam(lr=LEARNING_RATE)\nmodel.compile(optimizer=optimizer,\n loss=SparseCategoricalCrossentropy(),\n metrics=['accuracy'])",
"_____no_output_____"
],
[
"# Training\nmodel.fit(x=standardized_X_train, \n y=y_train,\n validation_data=(standardized_X_val, y_val),\n epochs=NUM_EPOCHS,\n batch_size=BATCH_SIZE,\n class_weight=class_weights,\n shuffle=False,\n verbose=1)",
"Train on 1083 samples, validate on 192 samples\nEpoch 1/10\n1083/1083 [==============================] - 1s 769us/sample - loss: 0.0021 - accuracy: 0.5540 - val_loss: 0.0017 - val_accuracy: 0.5833\nEpoch 2/10\n1083/1083 [==============================] - 0s 147us/sample - loss: 0.0014 - accuracy: 0.7470 - val_loss: 0.0012 - val_accuracy: 0.8333\nEpoch 3/10\n1083/1083 [==============================] - 0s 137us/sample - loss: 8.5693e-04 - accuracy: 0.9049 - val_loss: 7.6481e-04 - val_accuracy: 0.8906\nEpoch 4/10\n1083/1083 [==============================] - 0s 137us/sample - loss: 5.4156e-04 - accuracy: 0.9501 - val_loss: 5.5680e-04 - val_accuracy: 0.9531\nEpoch 5/10\n1083/1083 [==============================] - 0s 140us/sample - loss: 3.8934e-04 - accuracy: 0.9649 - val_loss: 4.4130e-04 - val_accuracy: 0.9635\nEpoch 6/10\n1083/1083 [==============================] - 0s 145us/sample - loss: 3.0538e-04 - accuracy: 0.9741 - val_loss: 3.6685e-04 - val_accuracy: 0.9635\nEpoch 7/10\n1083/1083 [==============================] - 0s 138us/sample - loss: 2.5231e-04 - accuracy: 0.9778 - val_loss: 3.0834e-04 - val_accuracy: 0.9792\nEpoch 8/10\n1083/1083 [==============================] - 0s 145us/sample - loss: 2.1546e-04 - accuracy: 0.9815 - val_loss: 2.6535e-04 - val_accuracy: 0.9948\nEpoch 9/10\n1083/1083 [==============================] - 0s 138us/sample - loss: 1.8842e-04 - accuracy: 0.9834 - val_loss: 2.3150e-04 - val_accuracy: 0.9948\nEpoch 10/10\n1083/1083 [==============================] - 0s 144us/sample - loss: 1.6813e-04 - accuracy: 0.9843 - val_loss: 2.0421e-04 - val_accuracy: 0.9948\n"
],
[
"# Predictions\npred_train = model.predict(standardized_X_train) \npred_test = model.predict(standardized_X_test)\nprint (f\"sample probability: {pred_test[0]}\")\npred_train = np.argmax(pred_train, axis=1)\npred_test = np.argmax(pred_test, axis=1)\nprint (f\"sample class: {pred_test[0]}\")",
"sample probability: [1.2431189e-03 5.1021564e-04 9.9824667e-01]\nsample class: 2\n"
],
[
"# Accuracy\ntrain_acc = accuracy_score(y_train, pred_train)\ntest_acc = accuracy_score(y_test, pred_test)\nprint (f\"train acc: {train_acc:.2f}, test acc: {test_acc:.2f}\")",
"train acc: 0.99, test acc: 0.99\n"
],
[
"# Metrics\nplot_confusion_matrix(y_test, pred_test, classes=classes)\nprint (classification_report(y_test, pred_test))",
"_____no_output_____"
],
[
"# Visualize the decision boundary\nplt.figure(figsize=(12,5))\nplt.subplot(1, 2, 1)\nplt.title(\"Train\")\nplot_multiclass_decision_boundary(model=model, X=standardized_X_train, y=y_train)\nplt.subplot(1, 2, 2)\nplt.title(\"Test\")\nplot_multiclass_decision_boundary(model=model, X=standardized_X_test, y=y_test)\nplt.show()",
"_____no_output_____"
]
],
[
[
"# Inference",
"_____no_output_____"
]
],
[
[
"# Inputs for inference\nX_infer = pd.DataFrame([{'X1': 0.1, 'X2': 0.1}])\nX_infer.head()",
"_____no_output_____"
],
[
"# Standardize\nstandardized_X_infer = X_scaler.transform(X_infer)\nprint (standardized_X_infer)",
"[[0.30945845 0.30761858]]\n"
],
[
"# Predict\ny_infer = model.predict(standardized_X_infer)\n_class = np.argmax(y_infer)\nprint (f\"The probability that you have a class {classes[_class]} is {y_infer[0][_class]*100.0:.0f}%\")",
"The probability that you have a class c1 is 97%\n"
]
],
[
[
"# Initializing weights",
"_____no_output_____"
],
[
"So far we have been initializing weights with small random values and this isn't optimal for convergence during training. The objective is to have weights that are able to produce outputs that follow a similar distribution across all neurons. We can do this by enforcing weights to have unit variance prior the affine and non-linear operations.",
"_____no_output_____"
],
[
"<img height=\"45\" src=\"http://bestanimations.com/HomeOffice/Lights/Bulbs/animated-light-bulb-gif-29.gif\" align=\"left\" vspace=\"20px\" hspace=\"10px\">\n\nA popular method is to apply [xavier initialization](http://andyljones.tumblr.com/post/110998971763/an-explanation-of-xavier-initialization), which essentially initializes the weights to allow the signal from the data to reach deep into the network. You may be wondering why we don't do this for every forward pass and that's a great question. We'll look at more advanced strategies that help with optimization like batch/layer normalization, etc. in future lessons. Meanwhile you can check out other initializers [here](https://www.tensorflow.org/versions/r2.0/api_docs/python/tf/initializers).",
"_____no_output_____"
]
],
[
[
"from tensorflow.keras.initializers import glorot_normal",
"_____no_output_____"
],
[
"# MLP\nclass MLP(Model):\n def __init__(self, hidden_dim, num_classes):\n super(MLP, self).__init__()\n xavier_initializer = glorot_normal() # xavier glorot initiailization\n self.fc1 = Dense(units=hidden_dim,\n kernel_initializer=xavier_initializer,\n activation='relu')\n self.fc2 = Dense(units=num_classes, \n activation='softmax')\n \n def call(self, x_in, training=False):\n \"\"\"Forward pass.\"\"\"\n z = self.fc1(x_in)\n y_pred = self.fc2(z)\n return y_pred\n \n def sample(self, input_shape):\n x_in = Input(shape=input_shape)\n return Model(inputs=x_in, outputs=self.call(x_in)).summary()",
"_____no_output_____"
]
],
[
[
"# Dropout",
"_____no_output_____"
],
[
"A great technique to overcome overfitting is to increase the size of your data but this isn't always an option. Fortuntely, there are methods like regularization and dropout that can help create a more robust model. \n\nDropout is a technique (used only during training) that allows us to zero the outputs of neurons. We do this for `dropout_p`% of the total neurons in each layer and it changes every batch. Dropout prevents units from co-adapting too much to the data and acts as a sampling strategy since we drop a different set of neurons each time.\n\n<img src=\"https://raw.githubusercontent.com/practicalAI/images/master/basic_ml/06_Multilayer_Perceptron/dropout.png\" width=\"350\">\n\n* [Dropout: A Simple Way to Prevent Neural Networks from\nOverfitting](http://jmlr.org/papers/volume15/srivastava14a/srivastava14a.pdf)",
"_____no_output_____"
]
],
[
[
"from tensorflow.keras.layers import Dropout\nfrom tensorflow.keras.regularizers import l2",
"_____no_output_____"
]
],
[
[
"### Components",
"_____no_output_____"
]
],
[
[
"# MLP\nclass MLP(Model):\n def __init__(self, hidden_dim, lambda_l2, dropout_p, num_classes):\n super(MLP, self).__init__()\n self.fc1 = Dense(units=hidden_dim,\n kernel_regularizer=l2(lambda_l2), # adding L2 regularization\n activation='relu')\n self.dropout = Dropout(rate=dropout_p)\n self.fc2 = Dense(units=num_classes, \n activation='softmax')\n \n def call(self, x_in, training=False):\n \"\"\"Forward pass.\"\"\"\n z = self.fc1(x_in)\n if training:\n z = self.dropout(z, training=training) # adding dropout\n y_pred = self.fc2(z)\n return y_pred\n \n def sample(self, input_shape):\n x_in = Input(shape=input_shape)\n return Model(inputs=x_in, outputs=self.call(x_in)).summary()",
"_____no_output_____"
]
],
[
[
"### Operations",
"_____no_output_____"
]
],
[
[
"# Arguments\nDROPOUT_P = 0.1 # % of the neurons that are dropped each pass\nLAMBDA_L2 = 1e-4 # L2 regularization",
"_____no_output_____"
],
[
"# Initialize the model\nmodel = MLP(hidden_dim=HIDDEN_DIM,\n lambda_l2=LAMBDA_L2,\n dropout_p=DROPOUT_P,\n num_classes=NUM_CLASSES)\nmodel.sample(input_shape=(INPUT_DIM,))",
"Model: \"model_2\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\ninput_3 (InputLayer) [(None, 2)] 0 \n_________________________________________________________________\ndense_4 (Dense) (None, 100) 300 \n_________________________________________________________________\ndense_5 (Dense) (None, 3) 303 \n=================================================================\nTotal params: 603\nTrainable params: 603\nNon-trainable params: 0\n_________________________________________________________________\n"
]
],
[
[
"# Overfitting",
"_____no_output_____"
],
[
"Though neural networks are great at capturing non-linear relationships they are highly susceptible to overfitting to the training data and failing to generalize on test data. Just take a look at the example below where we generate completely random data and are able to fit a model with [$2*N*C + D$](https://arxiv.org/abs/1611.03530) hidden units. The training performance is good (~70%) but the overfitting leads to very poor test performance. We'll be covering strategies to tackle overfitting in future lessons.",
"_____no_output_____"
]
],
[
[
"# Arguments\nNUM_EPOCHS = 500\nNUM_SAMPLES_PER_CLASS = 50\nLEARNING_RATE = 1e-1\nHIDDEN_DIM = 2 * NUM_SAMPLES_PER_CLASS * NUM_CLASSES + INPUT_DIM # 2*N*C + D",
"_____no_output_____"
],
[
"# Generate random data\nX = np.random.rand(NUM_SAMPLES_PER_CLASS * NUM_CLASSES, INPUT_DIM)\ny = np.array([[i]*NUM_SAMPLES_PER_CLASS for i in range(NUM_CLASSES)]).reshape(-1)\nprint (\"X: \", format(np.shape(X)))\nprint (\"y: \", format(np.shape(y)))",
"X: (150, 2)\ny: (150,)\n"
],
[
"# Create data splits\nX_train, X_val, X_test, y_train, y_val, y_test = train_val_test_split(\n X, y, val_size=VAL_SIZE, test_size=TEST_SIZE, shuffle=SHUFFLE)\nprint (\"X_train:\", X_train.shape)\nprint (\"y_train:\", y_train.shape)\nprint (\"X_val:\", X_val.shape)\nprint (\"y_val:\", y_val.shape)\nprint (\"X_test:\", X_test.shape)\nprint (\"y_test:\", y_test.shape)",
"X_train: (107, 2)\ny_train: (107,)\nX_val: (20, 2)\ny_val: (20,)\nX_test: (23, 2)\ny_test: (23,)\n"
],
[
"# Standardize the inputs (mean=0, std=1) using training data\nX_scaler = StandardScaler().fit(X_train)\n# Apply scaler on training and test data (don't standardize outputs for classification)\nstandardized_X_train = X_scaler.transform(X_train)\nstandardized_X_val = X_scaler.transform(X_val)\nstandardized_X_test = X_scaler.transform(X_test)",
"_____no_output_____"
],
[
"# Initialize the model\nmodel = MLP(hidden_dim=HIDDEN_DIM,\n lambda_l2=0.0,\n dropout_p=0.0,\n num_classes=NUM_CLASSES)\nmodel.sample(input_shape=(INPUT_DIM,))",
"Model: \"model_3\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\ninput_4 (InputLayer) [(None, 2)] 0 \n_________________________________________________________________\ndense_6 (Dense) (None, 302) 906 \n_________________________________________________________________\ndense_7 (Dense) (None, 3) 909 \n=================================================================\nTotal params: 1,815\nTrainable params: 1,815\nNon-trainable params: 0\n_________________________________________________________________\n"
],
[
"# Compile\noptimizer = Adam(lr=LEARNING_RATE)\nmodel.compile(optimizer=optimizer,\n loss=SparseCategoricalCrossentropy(),\n metrics=['accuracy'])",
"_____no_output_____"
],
[
"# Training\nmodel.fit(x=standardized_X_train, \n y=y_train,\n validation_data=(standardized_X_val, y_val),\n epochs=NUM_EPOCHS,\n batch_size=BATCH_SIZE,\n class_weight=class_weights,\n shuffle=False,\n verbose=1)",
"Train on 107 samples, validate on 20 samples\nEpoch 1/500\n107/107 [==============================] - 1s 8ms/sample - loss: 0.0047 - accuracy: 0.2150 - val_loss: 0.0034 - val_accuracy: 0.3000\nEpoch 2/500\n107/107 [==============================] - 0s 362us/sample - loss: 0.0033 - accuracy: 0.3458 - val_loss: 0.0030 - val_accuracy: 0.3500\nEpoch 3/500\n107/107 [==============================] - 0s 341us/sample - loss: 0.0032 - accuracy: 0.4393 - val_loss: 0.0030 - val_accuracy: 0.4500\nEpoch 4/500\n107/107 [==============================] - 0s 312us/sample - loss: 0.0030 - accuracy: 0.4860 - val_loss: 0.0032 - val_accuracy: 0.3500\nEpoch 5/500\n107/107 [==============================] - 0s 315us/sample - loss: 0.0030 - accuracy: 0.4299 - val_loss: 0.0029 - val_accuracy: 0.4500\nEpoch 6/500\n107/107 [==============================] - 0s 549us/sample - loss: 0.0030 - accuracy: 0.4486 - val_loss: 0.0032 - val_accuracy: 0.3000\nEpoch 7/500\n107/107 [==============================] - 0s 323us/sample - loss: 0.0029 - accuracy: 0.4953 - val_loss: 0.0032 - val_accuracy: 0.3500\nEpoch 8/500\n107/107 [==============================] - 0s 377us/sample - loss: 0.0031 - accuracy: 0.5047 - val_loss: 0.0031 - val_accuracy: 0.4000\nEpoch 9/500\n107/107 [==============================] - 0s 347us/sample - loss: 0.0028 - accuracy: 0.5327 - val_loss: 0.0032 - val_accuracy: 0.4500\nEpoch 10/500\n107/107 [==============================] - 0s 327us/sample - loss: 0.0029 - accuracy: 0.4953 - val_loss: 0.0033 - val_accuracy: 0.4500\nEpoch 11/500\n107/107 [==============================] - 0s 345us/sample - loss: 0.0028 - accuracy: 0.5421 - val_loss: 0.0031 - val_accuracy: 0.4500\nEpoch 12/500\n107/107 [==============================] - 0s 350us/sample - loss: 0.0028 - accuracy: 0.5421 - val_loss: 0.0034 - val_accuracy: 0.4500\nEpoch 13/500\n107/107 [==============================] - 0s 340us/sample - loss: 0.0028 - accuracy: 0.5234 - val_loss: 0.0029 - val_accuracy: 0.5000\nEpoch 14/500\n107/107 [==============================] - 0s 372us/sample - loss: 0.0030 - accuracy: 0.4673 - val_loss: 0.0036 - val_accuracy: 0.3500\nEpoch 15/500\n107/107 [==============================] - 0s 371us/sample - loss: 0.0030 - accuracy: 0.4953 - val_loss: 0.0029 - val_accuracy: 0.5000\nEpoch 16/500\n107/107 [==============================] - 0s 347us/sample - loss: 0.0031 - accuracy: 0.4860 - val_loss: 0.0030 - val_accuracy: 0.4500\nEpoch 17/500\n107/107 [==============================] - 0s 338us/sample - loss: 0.0028 - accuracy: 0.4766 - val_loss: 0.0035 - val_accuracy: 0.4500\nEpoch 18/500\n107/107 [==============================] - 0s 348us/sample - loss: 0.0029 - accuracy: 0.4953 - val_loss: 0.0029 - val_accuracy: 0.4500\nEpoch 19/500\n107/107 [==============================] - 0s 351us/sample - loss: 0.0027 - accuracy: 0.5607 - val_loss: 0.0031 - val_accuracy: 0.6000\nEpoch 20/500\n107/107 [==============================] - 0s 331us/sample - loss: 0.0027 - accuracy: 0.5140 - val_loss: 0.0030 - val_accuracy: 0.5500\nEpoch 21/500\n107/107 [==============================] - 0s 312us/sample - loss: 0.0027 - accuracy: 0.5981 - val_loss: 0.0029 - val_accuracy: 0.6500\nEpoch 22/500\n107/107 [==============================] - 0s 314us/sample - loss: 0.0027 - accuracy: 0.5234 - val_loss: 0.0032 - val_accuracy: 0.4000\nEpoch 23/500\n107/107 [==============================] - 0s 311us/sample - loss: 0.0027 - accuracy: 0.5327 - val_loss: 0.0030 - val_accuracy: 0.5500\nEpoch 24/500\n107/107 [==============================] - 0s 294us/sample - loss: 0.0029 - accuracy: 0.5607 - val_loss: 0.0033 - val_accuracy: 0.3500\nEpoch 25/500\n107/107 [==============================] - 0s 322us/sample - loss: 0.0028 - accuracy: 0.4953 - val_loss: 0.0029 - val_accuracy: 0.6500\nEpoch 26/500\n107/107 [==============================] - 0s 400us/sample - loss: 0.0029 - accuracy: 0.5514 - val_loss: 0.0032 - val_accuracy: 0.4500\nEpoch 27/500\n107/107 [==============================] - 0s 313us/sample - loss: 0.0027 - accuracy: 0.5234 - val_loss: 0.0029 - val_accuracy: 0.6000\nEpoch 28/500\n107/107 [==============================] - 0s 308us/sample - loss: 0.0027 - accuracy: 0.5701 - val_loss: 0.0031 - val_accuracy: 0.6000\nEpoch 29/500\n107/107 [==============================] - 0s 273us/sample - loss: 0.0026 - accuracy: 0.5701 - val_loss: 0.0029 - val_accuracy: 0.6000\nEpoch 30/500\n107/107 [==============================] - 0s 337us/sample - loss: 0.0027 - accuracy: 0.5607 - val_loss: 0.0032 - val_accuracy: 0.4500\nEpoch 31/500\n107/107 [==============================] - 0s 337us/sample - loss: 0.0026 - accuracy: 0.5607 - val_loss: 0.0029 - val_accuracy: 0.6500\nEpoch 32/500\n107/107 [==============================] - 0s 304us/sample - loss: 0.0026 - accuracy: 0.5607 - val_loss: 0.0033 - val_accuracy: 0.4500\nEpoch 33/500\n107/107 [==============================] - 0s 313us/sample - loss: 0.0026 - accuracy: 0.5514 - val_loss: 0.0029 - val_accuracy: 0.6500\nEpoch 34/500\n107/107 [==============================] - 0s 312us/sample - loss: 0.0027 - accuracy: 0.5701 - val_loss: 0.0033 - val_accuracy: 0.4500\nEpoch 35/500\n107/107 [==============================] - 0s 312us/sample - loss: 0.0026 - accuracy: 0.5327 - val_loss: 0.0030 - val_accuracy: 0.7000\nEpoch 36/500\n107/107 [==============================] - 0s 306us/sample - loss: 0.0028 - accuracy: 0.5794 - val_loss: 0.0033 - val_accuracy: 0.5000\nEpoch 37/500\n107/107 [==============================] - 0s 306us/sample - loss: 0.0026 - accuracy: 0.5234 - val_loss: 0.0030 - val_accuracy: 0.6000\nEpoch 38/500\n107/107 [==============================] - 0s 280us/sample - loss: 0.0027 - accuracy: 0.5794 - val_loss: 0.0033 - val_accuracy: 0.6000\nEpoch 39/500\n107/107 [==============================] - 0s 290us/sample - loss: 0.0026 - accuracy: 0.5701 - val_loss: 0.0031 - val_accuracy: 0.5000\nEpoch 40/500\n107/107 [==============================] - 0s 290us/sample - loss: 0.0027 - accuracy: 0.5514 - val_loss: 0.0035 - val_accuracy: 0.5500\nEpoch 41/500\n107/107 [==============================] - 0s 292us/sample - loss: 0.0027 - accuracy: 0.5514 - val_loss: 0.0030 - val_accuracy: 0.5500\nEpoch 42/500\n107/107 [==============================] - 0s 327us/sample - loss: 0.0028 - accuracy: 0.5514 - val_loss: 0.0034 - val_accuracy: 0.5000\nEpoch 43/500\n107/107 [==============================] - 0s 310us/sample - loss: 0.0026 - accuracy: 0.5514 - val_loss: 0.0030 - val_accuracy: 0.6500\nEpoch 44/500\n107/107 [==============================] - 0s 313us/sample - loss: 0.0026 - accuracy: 0.5514 - val_loss: 0.0034 - val_accuracy: 0.5000\nEpoch 45/500\n107/107 [==============================] - 0s 311us/sample - loss: 0.0025 - accuracy: 0.5607 - val_loss: 0.0031 - val_accuracy: 0.6500\nEpoch 46/500\n107/107 [==============================] - 0s 296us/sample - loss: 0.0026 - accuracy: 0.5701 - val_loss: 0.0034 - val_accuracy: 0.6000\nEpoch 47/500\n107/107 [==============================] - 0s 312us/sample - loss: 0.0026 - accuracy: 0.5607 - val_loss: 0.0032 - val_accuracy: 0.5500\nEpoch 48/500\n107/107 [==============================] - 0s 299us/sample - loss: 0.0028 - accuracy: 0.5421 - val_loss: 0.0035 - val_accuracy: 0.5000\nEpoch 49/500\n107/107 [==============================] - 0s 326us/sample - loss: 0.0026 - accuracy: 0.5421 - val_loss: 0.0030 - val_accuracy: 0.6000\nEpoch 50/500\n107/107 [==============================] - 0s 304us/sample - loss: 0.0027 - accuracy: 0.5607 - val_loss: 0.0035 - val_accuracy: 0.5000\nEpoch 51/500\n107/107 [==============================] - 0s 281us/sample - loss: 0.0025 - accuracy: 0.5701 - val_loss: 0.0031 - val_accuracy: 0.6500\nEpoch 52/500\n107/107 [==============================] - 0s 294us/sample - loss: 0.0026 - accuracy: 0.5514 - val_loss: 0.0035 - val_accuracy: 0.4500\nEpoch 53/500\n107/107 [==============================] - 0s 305us/sample - loss: 0.0025 - accuracy: 0.5514 - val_loss: 0.0031 - val_accuracy: 0.6500\nEpoch 54/500\n107/107 [==============================] - 0s 360us/sample - loss: 0.0026 - accuracy: 0.5701 - val_loss: 0.0033 - val_accuracy: 0.6000\nEpoch 55/500\n107/107 [==============================] - 0s 378us/sample - loss: 0.0025 - accuracy: 0.5514 - val_loss: 0.0032 - val_accuracy: 0.6000\nEpoch 56/500\n107/107 [==============================] - 0s 335us/sample - loss: 0.0027 - accuracy: 0.5421 - val_loss: 0.0033 - val_accuracy: 0.5500\nEpoch 57/500\n107/107 [==============================] - 0s 330us/sample - loss: 0.0026 - accuracy: 0.5514 - val_loss: 0.0032 - val_accuracy: 0.6000\nEpoch 58/500\n107/107 [==============================] - 0s 326us/sample - loss: 0.0027 - accuracy: 0.5701 - val_loss: 0.0035 - val_accuracy: 0.5500\nEpoch 59/500\n107/107 [==============================] - 0s 359us/sample - loss: 0.0026 - accuracy: 0.5607 - val_loss: 0.0032 - val_accuracy: 0.6000\nEpoch 60/500\n107/107 [==============================] - 0s 372us/sample - loss: 0.0026 - accuracy: 0.5421 - val_loss: 0.0034 - val_accuracy: 0.6000\nEpoch 61/500\n107/107 [==============================] - 0s 378us/sample - loss: 0.0025 - accuracy: 0.5607 - val_loss: 0.0032 - val_accuracy: 0.7000\nEpoch 62/500\n107/107 [==============================] - 0s 326us/sample - loss: 0.0026 - accuracy: 0.5607 - val_loss: 0.0034 - val_accuracy: 0.5500\nEpoch 63/500\n107/107 [==============================] - 0s 387us/sample - loss: 0.0025 - accuracy: 0.5514 - val_loss: 0.0034 - val_accuracy: 0.5500\nEpoch 64/500\n107/107 [==============================] - 0s 334us/sample - loss: 0.0027 - accuracy: 0.5327 - val_loss: 0.0036 - val_accuracy: 0.6500\nEpoch 65/500\n107/107 [==============================] - 0s 333us/sample - loss: 0.0026 - accuracy: 0.5701 - val_loss: 0.0031 - val_accuracy: 0.6500\nEpoch 66/500\n107/107 [==============================] - 0s 333us/sample - loss: 0.0026 - accuracy: 0.5888 - val_loss: 0.0036 - val_accuracy: 0.6000\nEpoch 67/500\n107/107 [==============================] - 0s 372us/sample - loss: 0.0025 - accuracy: 0.5421 - val_loss: 0.0029 - val_accuracy: 0.7000\nEpoch 68/500\n107/107 [==============================] - 0s 324us/sample - loss: 0.0025 - accuracy: 0.5794 - val_loss: 0.0037 - val_accuracy: 0.4500\nEpoch 69/500\n107/107 [==============================] - 0s 341us/sample - loss: 0.0025 - accuracy: 0.5701 - val_loss: 0.0029 - val_accuracy: 0.7000\nEpoch 70/500\n107/107 [==============================] - 0s 316us/sample - loss: 0.0025 - accuracy: 0.6168 - val_loss: 0.0036 - val_accuracy: 0.4000\nEpoch 71/500\n107/107 [==============================] - 0s 301us/sample - loss: 0.0025 - accuracy: 0.5701 - val_loss: 0.0032 - val_accuracy: 0.7000\nEpoch 72/500\n107/107 [==============================] - 0s 329us/sample - loss: 0.0025 - accuracy: 0.6168 - val_loss: 0.0038 - val_accuracy: 0.4500\nEpoch 73/500\n107/107 [==============================] - 0s 312us/sample - loss: 0.0025 - accuracy: 0.5607 - val_loss: 0.0032 - val_accuracy: 0.6500\nEpoch 74/500\n107/107 [==============================] - 0s 310us/sample - loss: 0.0027 - accuracy: 0.5981 - val_loss: 0.0036 - val_accuracy: 0.5500\nEpoch 75/500\n107/107 [==============================] - 0s 303us/sample - loss: 0.0025 - accuracy: 0.5701 - val_loss: 0.0033 - val_accuracy: 0.6000\nEpoch 76/500\n107/107 [==============================] - 0s 319us/sample - loss: 0.0025 - accuracy: 0.5607 - val_loss: 0.0037 - val_accuracy: 0.6000\nEpoch 77/500\n107/107 [==============================] - 0s 306us/sample - loss: 0.0025 - accuracy: 0.5794 - val_loss: 0.0033 - val_accuracy: 0.6500\nEpoch 78/500\n107/107 [==============================] - 0s 311us/sample - loss: 0.0025 - accuracy: 0.5701 - val_loss: 0.0035 - val_accuracy: 0.6000\nEpoch 79/500\n107/107 [==============================] - 0s 323us/sample - loss: 0.0024 - accuracy: 0.5981 - val_loss: 0.0035 - val_accuracy: 0.6500\nEpoch 80/500\n107/107 [==============================] - 0s 290us/sample - loss: 0.0025 - accuracy: 0.5701 - val_loss: 0.0037 - val_accuracy: 0.5500\nEpoch 81/500\n107/107 [==============================] - 0s 293us/sample - loss: 0.0025 - accuracy: 0.5794 - val_loss: 0.0034 - val_accuracy: 0.6500\nEpoch 82/500\n107/107 [==============================] - 0s 435us/sample - loss: 0.0027 - accuracy: 0.5607 - val_loss: 0.0033 - val_accuracy: 0.6000\nEpoch 83/500\n107/107 [==============================] - 0s 352us/sample - loss: 0.0025 - accuracy: 0.5607 - val_loss: 0.0037 - val_accuracy: 0.5500\nEpoch 84/500\n107/107 [==============================] - 0s 311us/sample - loss: 0.0025 - accuracy: 0.5888 - val_loss: 0.0039 - val_accuracy: 0.7000\nEpoch 85/500\n107/107 [==============================] - 0s 389us/sample - loss: 0.0024 - accuracy: 0.5514 - val_loss: 0.0034 - val_accuracy: 0.6000\nEpoch 86/500\n107/107 [==============================] - 0s 383us/sample - loss: 0.0025 - accuracy: 0.5888 - val_loss: 0.0038 - val_accuracy: 0.6000\nEpoch 87/500\n107/107 [==============================] - 0s 342us/sample - loss: 0.0024 - accuracy: 0.5981 - val_loss: 0.0036 - val_accuracy: 0.7000\nEpoch 88/500\n107/107 [==============================] - 0s 349us/sample - loss: 0.0025 - accuracy: 0.5794 - val_loss: 0.0036 - val_accuracy: 0.5000\nEpoch 89/500\n107/107 [==============================] - 0s 326us/sample - loss: 0.0025 - accuracy: 0.5888 - val_loss: 0.0034 - val_accuracy: 0.7000\nEpoch 90/500\n107/107 [==============================] - 0s 332us/sample - loss: 0.0025 - accuracy: 0.6355 - val_loss: 0.0037 - val_accuracy: 0.5500\nEpoch 91/500\n107/107 [==============================] - 0s 325us/sample - loss: 0.0024 - accuracy: 0.5607 - val_loss: 0.0035 - val_accuracy: 0.5000\nEpoch 92/500\n107/107 [==============================] - 0s 333us/sample - loss: 0.0025 - accuracy: 0.5701 - val_loss: 0.0038 - val_accuracy: 0.6000\nEpoch 93/500\n107/107 [==============================] - 0s 340us/sample - loss: 0.0025 - accuracy: 0.5981 - val_loss: 0.0034 - val_accuracy: 0.7000\nEpoch 94/500\n107/107 [==============================] - 0s 340us/sample - loss: 0.0025 - accuracy: 0.5794 - val_loss: 0.0038 - val_accuracy: 0.6500\nEpoch 95/500\n107/107 [==============================] - 0s 324us/sample - loss: 0.0025 - accuracy: 0.5888 - val_loss: 0.0033 - val_accuracy: 0.7500\nEpoch 96/500\n107/107 [==============================] - 0s 356us/sample - loss: 0.0024 - accuracy: 0.6355 - val_loss: 0.0036 - val_accuracy: 0.5500\nEpoch 97/500\n107/107 [==============================] - 0s 329us/sample - loss: 0.0023 - accuracy: 0.5888 - val_loss: 0.0036 - val_accuracy: 0.7000\nEpoch 98/500\n107/107 [==============================] - 0s 324us/sample - loss: 0.0023 - accuracy: 0.6168 - val_loss: 0.0036 - val_accuracy: 0.7000\nEpoch 99/500\n107/107 [==============================] - 0s 327us/sample - loss: 0.0022 - accuracy: 0.5981 - val_loss: 0.0036 - val_accuracy: 0.7000\nEpoch 100/500\n107/107 [==============================] - 0s 331us/sample - loss: 0.0023 - accuracy: 0.5981 - val_loss: 0.0035 - val_accuracy: 0.7000\nEpoch 101/500\n107/107 [==============================] - 0s 325us/sample - loss: 0.0022 - accuracy: 0.6262 - val_loss: 0.0038 - val_accuracy: 0.7500\nEpoch 102/500\n107/107 [==============================] - 0s 322us/sample - loss: 0.0025 - accuracy: 0.6075 - val_loss: 0.0038 - val_accuracy: 0.4000\nEpoch 103/500\n107/107 [==============================] - 0s 328us/sample - loss: 0.0024 - accuracy: 0.6075 - val_loss: 0.0045 - val_accuracy: 0.6500\nEpoch 104/500\n107/107 [==============================] - 0s 310us/sample - loss: 0.0026 - accuracy: 0.5327 - val_loss: 0.0038 - val_accuracy: 0.5000\nEpoch 105/500\n107/107 [==============================] - 0s 286us/sample - loss: 0.0024 - accuracy: 0.5794 - val_loss: 0.0043 - val_accuracy: 0.6500\nEpoch 106/500\n107/107 [==============================] - 0s 276us/sample - loss: 0.0027 - accuracy: 0.5607 - val_loss: 0.0041 - val_accuracy: 0.7000\nEpoch 107/500\n107/107 [==============================] - 0s 277us/sample - loss: 0.0025 - accuracy: 0.5607 - val_loss: 0.0041 - val_accuracy: 0.4500\nEpoch 108/500\n107/107 [==============================] - 0s 289us/sample - loss: 0.0028 - accuracy: 0.6262 - val_loss: 0.0046 - val_accuracy: 0.6500\nEpoch 109/500\n107/107 [==============================] - 0s 391us/sample - loss: 0.0026 - accuracy: 0.5421 - val_loss: 0.0033 - val_accuracy: 0.5500\nEpoch 110/500\n107/107 [==============================] - 0s 343us/sample - loss: 0.0025 - accuracy: 0.5514 - val_loss: 0.0050 - val_accuracy: 0.6500\nEpoch 111/500\n107/107 [==============================] - 0s 298us/sample - loss: 0.0028 - accuracy: 0.5981 - val_loss: 0.0037 - val_accuracy: 0.6500\nEpoch 112/500\n107/107 [==============================] - 0s 306us/sample - loss: 0.0027 - accuracy: 0.5514 - val_loss: 0.0050 - val_accuracy: 0.4000\nEpoch 113/500\n107/107 [==============================] - 0s 304us/sample - loss: 0.0028 - accuracy: 0.5607 - val_loss: 0.0045 - val_accuracy: 0.6500\nEpoch 114/500\n107/107 [==============================] - 0s 303us/sample - loss: 0.0029 - accuracy: 0.5421 - val_loss: 0.0039 - val_accuracy: 0.6000\nEpoch 115/500\n107/107 [==============================] - 0s 323us/sample - loss: 0.0023 - accuracy: 0.6262 - val_loss: 0.0040 - val_accuracy: 0.5000\nEpoch 116/500\n107/107 [==============================] - 0s 324us/sample - loss: 0.0025 - accuracy: 0.5701 - val_loss: 0.0029 - val_accuracy: 0.6500\nEpoch 117/500\n107/107 [==============================] - 0s 299us/sample - loss: 0.0023 - accuracy: 0.6355 - val_loss: 0.0047 - val_accuracy: 0.6000\nEpoch 118/500\n107/107 [==============================] - 0s 319us/sample - loss: 0.0027 - accuracy: 0.5514 - val_loss: 0.0036 - val_accuracy: 0.5500\nEpoch 119/500\n107/107 [==============================] - 0s 275us/sample - loss: 0.0024 - accuracy: 0.6449 - val_loss: 0.0036 - val_accuracy: 0.6000\nEpoch 120/500\n107/107 [==============================] - 0s 305us/sample - loss: 0.0025 - accuracy: 0.6262 - val_loss: 0.0034 - val_accuracy: 0.5000\nEpoch 121/500\n107/107 [==============================] - 0s 285us/sample - loss: 0.0024 - accuracy: 0.5794 - val_loss: 0.0035 - val_accuracy: 0.7500\nEpoch 122/500\n107/107 [==============================] - 0s 297us/sample - loss: 0.0024 - accuracy: 0.5981 - val_loss: 0.0042 - val_accuracy: 0.6000\nEpoch 123/500\n107/107 [==============================] - 0s 270us/sample - loss: 0.0025 - accuracy: 0.6075 - val_loss: 0.0033 - val_accuracy: 0.7000\nEpoch 124/500\n107/107 [==============================] - 0s 303us/sample - loss: 0.0023 - accuracy: 0.6449 - val_loss: 0.0037 - val_accuracy: 0.5000\nEpoch 125/500\n107/107 [==============================] - 0s 338us/sample - loss: 0.0023 - accuracy: 0.6355 - val_loss: 0.0034 - val_accuracy: 0.7500\nEpoch 126/500\n107/107 [==============================] - 0s 290us/sample - loss: 0.0024 - accuracy: 0.5981 - val_loss: 0.0041 - val_accuracy: 0.6000\nEpoch 127/500\n107/107 [==============================] - 0s 317us/sample - loss: 0.0023 - accuracy: 0.6075 - val_loss: 0.0035 - val_accuracy: 0.7000\nEpoch 128/500\n107/107 [==============================] - 0s 321us/sample - loss: 0.0023 - accuracy: 0.6449 - val_loss: 0.0042 - val_accuracy: 0.5000\nEpoch 129/500\n107/107 [==============================] - 0s 310us/sample - loss: 0.0023 - accuracy: 0.5514 - val_loss: 0.0030 - val_accuracy: 0.7500\nEpoch 130/500\n107/107 [==============================] - 0s 293us/sample - loss: 0.0023 - accuracy: 0.6075 - val_loss: 0.0042 - val_accuracy: 0.6500\nEpoch 131/500\n107/107 [==============================] - 0s 338us/sample - loss: 0.0023 - accuracy: 0.5981 - val_loss: 0.0029 - val_accuracy: 0.7000\nEpoch 132/500\n107/107 [==============================] - 0s 316us/sample - loss: 0.0023 - accuracy: 0.6729 - val_loss: 0.0041 - val_accuracy: 0.6000\nEpoch 133/500\n107/107 [==============================] - 0s 305us/sample - loss: 0.0023 - accuracy: 0.6168 - val_loss: 0.0030 - val_accuracy: 0.7500\nEpoch 134/500\n107/107 [==============================] - 0s 281us/sample - loss: 0.0023 - accuracy: 0.5794 - val_loss: 0.0044 - val_accuracy: 0.7000\nEpoch 135/500\n107/107 [==============================] - 0s 303us/sample - loss: 0.0023 - accuracy: 0.6542 - val_loss: 0.0032 - val_accuracy: 0.5000\nEpoch 136/500\n107/107 [==============================] - 0s 299us/sample - loss: 0.0025 - accuracy: 0.5888 - val_loss: 0.0043 - val_accuracy: 0.6500\nEpoch 137/500\n107/107 [==============================] - 0s 332us/sample - loss: 0.0023 - accuracy: 0.6168 - val_loss: 0.0038 - val_accuracy: 0.6500\nEpoch 138/500\n107/107 [==============================] - 0s 354us/sample - loss: 0.0023 - accuracy: 0.5981 - val_loss: 0.0043 - val_accuracy: 0.7500\nEpoch 139/500\n107/107 [==============================] - 0s 343us/sample - loss: 0.0023 - accuracy: 0.6729 - val_loss: 0.0042 - val_accuracy: 0.6500\nEpoch 140/500\n107/107 [==============================] - 0s 324us/sample - loss: 0.0024 - accuracy: 0.5701 - val_loss: 0.0038 - val_accuracy: 0.6000\nEpoch 141/500\n107/107 [==============================] - 0s 294us/sample - loss: 0.0023 - accuracy: 0.6449 - val_loss: 0.0049 - val_accuracy: 0.6500\nEpoch 142/500\n107/107 [==============================] - 0s 335us/sample - loss: 0.0025 - accuracy: 0.5514 - val_loss: 0.0037 - val_accuracy: 0.5500\nEpoch 143/500\n107/107 [==============================] - 0s 345us/sample - loss: 0.0023 - accuracy: 0.6262 - val_loss: 0.0039 - val_accuracy: 0.7000\nEpoch 144/500\n107/107 [==============================] - 0s 271us/sample - loss: 0.0024 - accuracy: 0.6075 - val_loss: 0.0036 - val_accuracy: 0.6000\nEpoch 145/500\n107/107 [==============================] - 0s 279us/sample - loss: 0.0021 - accuracy: 0.6449 - val_loss: 0.0043 - val_accuracy: 0.6500\nEpoch 146/500\n107/107 [==============================] - 0s 320us/sample - loss: 0.0024 - accuracy: 0.5888 - val_loss: 0.0050 - val_accuracy: 0.5500\nEpoch 147/500\n107/107 [==============================] - 0s 363us/sample - loss: 0.0023 - accuracy: 0.6355 - val_loss: 0.0036 - val_accuracy: 0.7500\nEpoch 148/500\n107/107 [==============================] - 0s 299us/sample - loss: 0.0022 - accuracy: 0.6729 - val_loss: 0.0047 - val_accuracy: 0.5000\nEpoch 149/500\n107/107 [==============================] - 0s 301us/sample - loss: 0.0022 - accuracy: 0.5701 - val_loss: 0.0034 - val_accuracy: 0.7500\nEpoch 150/500\n107/107 [==============================] - 0s 339us/sample - loss: 0.0023 - accuracy: 0.6542 - val_loss: 0.0042 - val_accuracy: 0.6500\nEpoch 151/500\n107/107 [==============================] - 0s 309us/sample - loss: 0.0022 - accuracy: 0.6168 - val_loss: 0.0038 - val_accuracy: 0.7000\nEpoch 152/500\n107/107 [==============================] - 0s 312us/sample - loss: 0.0021 - accuracy: 0.6729 - val_loss: 0.0041 - val_accuracy: 0.6000\nEpoch 153/500\n107/107 [==============================] - 0s 342us/sample - loss: 0.0021 - accuracy: 0.6542 - val_loss: 0.0035 - val_accuracy: 0.7000\nEpoch 154/500\n107/107 [==============================] - 0s 324us/sample - loss: 0.0020 - accuracy: 0.7196 - val_loss: 0.0043 - val_accuracy: 0.6500\nEpoch 155/500\n107/107 [==============================] - 0s 309us/sample - loss: 0.0022 - accuracy: 0.6355 - val_loss: 0.0038 - val_accuracy: 0.7000\nEpoch 156/500\n107/107 [==============================] - 0s 283us/sample - loss: 0.0023 - accuracy: 0.7009 - val_loss: 0.0041 - val_accuracy: 0.5500\nEpoch 157/500\n107/107 [==============================] - 0s 306us/sample - loss: 0.0021 - accuracy: 0.6449 - val_loss: 0.0040 - val_accuracy: 0.7000\nEpoch 158/500\n107/107 [==============================] - 0s 298us/sample - loss: 0.0022 - accuracy: 0.6449 - val_loss: 0.0047 - val_accuracy: 0.6000\nEpoch 159/500\n107/107 [==============================] - 0s 292us/sample - loss: 0.0022 - accuracy: 0.6168 - val_loss: 0.0038 - val_accuracy: 0.6500\nEpoch 160/500\n107/107 [==============================] - 0s 318us/sample - loss: 0.0023 - accuracy: 0.6168 - val_loss: 0.0047 - val_accuracy: 0.6000\nEpoch 161/500\n107/107 [==============================] - 0s 284us/sample - loss: 0.0023 - accuracy: 0.6355 - val_loss: 0.0038 - val_accuracy: 0.7500\nEpoch 162/500\n107/107 [==============================] - 0s 312us/sample - loss: 0.0026 - accuracy: 0.6075 - val_loss: 0.0053 - val_accuracy: 0.5500\nEpoch 163/500\n107/107 [==============================] - 0s 289us/sample - loss: 0.0024 - accuracy: 0.6075 - val_loss: 0.0037 - val_accuracy: 0.7500\nEpoch 164/500\n107/107 [==============================] - 0s 292us/sample - loss: 0.0025 - accuracy: 0.6075 - val_loss: 0.0056 - val_accuracy: 0.5000\nEpoch 165/500\n107/107 [==============================] - 0s 295us/sample - loss: 0.0023 - accuracy: 0.5981 - val_loss: 0.0036 - val_accuracy: 0.6000\nEpoch 166/500\n107/107 [==============================] - 0s 304us/sample - loss: 0.0023 - accuracy: 0.6262 - val_loss: 0.0046 - val_accuracy: 0.6500\nEpoch 167/500\n107/107 [==============================] - 0s 334us/sample - loss: 0.0024 - accuracy: 0.6168 - val_loss: 0.0039 - val_accuracy: 0.5500\nEpoch 168/500\n107/107 [==============================] - 0s 365us/sample - loss: 0.0024 - accuracy: 0.6822 - val_loss: 0.0040 - val_accuracy: 0.7500\nEpoch 169/500\n107/107 [==============================] - 0s 298us/sample - loss: 0.0024 - accuracy: 0.6075 - val_loss: 0.0039 - val_accuracy: 0.6000\nEpoch 170/500\n107/107 [==============================] - 0s 299us/sample - loss: 0.0024 - accuracy: 0.6355 - val_loss: 0.0051 - val_accuracy: 0.5500\nEpoch 171/500\n107/107 [==============================] - 0s 301us/sample - loss: 0.0025 - accuracy: 0.6075 - val_loss: 0.0051 - val_accuracy: 0.6000\nEpoch 172/500\n107/107 [==============================] - 0s 299us/sample - loss: 0.0026 - accuracy: 0.6262 - val_loss: 0.0052 - val_accuracy: 0.6000\nEpoch 173/500\n107/107 [==============================] - 0s 354us/sample - loss: 0.0030 - accuracy: 0.6262 - val_loss: 0.0052 - val_accuracy: 0.5500\nEpoch 174/500\n107/107 [==============================] - 0s 329us/sample - loss: 0.0025 - accuracy: 0.6168 - val_loss: 0.0044 - val_accuracy: 0.6500\nEpoch 175/500\n107/107 [==============================] - 0s 382us/sample - loss: 0.0023 - accuracy: 0.6168 - val_loss: 0.0052 - val_accuracy: 0.7000\nEpoch 176/500\n107/107 [==============================] - 0s 351us/sample - loss: 0.0027 - accuracy: 0.6075 - val_loss: 0.0055 - val_accuracy: 0.7000\nEpoch 177/500\n107/107 [==============================] - 0s 359us/sample - loss: 0.0024 - accuracy: 0.6542 - val_loss: 0.0039 - val_accuracy: 0.5500\nEpoch 178/500\n107/107 [==============================] - 0s 395us/sample - loss: 0.0025 - accuracy: 0.6168 - val_loss: 0.0053 - val_accuracy: 0.4500\nEpoch 179/500\n107/107 [==============================] - 0s 336us/sample - loss: 0.0026 - accuracy: 0.5981 - val_loss: 0.0038 - val_accuracy: 0.6000\nEpoch 180/500\n107/107 [==============================] - 0s 343us/sample - loss: 0.0025 - accuracy: 0.5888 - val_loss: 0.0036 - val_accuracy: 0.7000\nEpoch 181/500\n107/107 [==============================] - 0s 354us/sample - loss: 0.0024 - accuracy: 0.6355 - val_loss: 0.0060 - val_accuracy: 0.4500\nEpoch 182/500\n107/107 [==============================] - 0s 314us/sample - loss: 0.0027 - accuracy: 0.5888 - val_loss: 0.0033 - val_accuracy: 0.7500\nEpoch 183/500\n107/107 [==============================] - 0s 319us/sample - loss: 0.0023 - accuracy: 0.6636 - val_loss: 0.0042 - val_accuracy: 0.5500\nEpoch 184/500\n107/107 [==============================] - 0s 293us/sample - loss: 0.0023 - accuracy: 0.6729 - val_loss: 0.0047 - val_accuracy: 0.6500\nEpoch 185/500\n107/107 [==============================] - 0s 289us/sample - loss: 0.0026 - accuracy: 0.6262 - val_loss: 0.0035 - val_accuracy: 0.5500\nEpoch 186/500\n107/107 [==============================] - 0s 287us/sample - loss: 0.0023 - accuracy: 0.6729 - val_loss: 0.0038 - val_accuracy: 0.7000\nEpoch 187/500\n107/107 [==============================] - 0s 318us/sample - loss: 0.0025 - accuracy: 0.6168 - val_loss: 0.0037 - val_accuracy: 0.6000\nEpoch 188/500\n107/107 [==============================] - 0s 372us/sample - loss: 0.0022 - accuracy: 0.6542 - val_loss: 0.0038 - val_accuracy: 0.7500\nEpoch 189/500\n107/107 [==============================] - 0s 354us/sample - loss: 0.0022 - accuracy: 0.6355 - val_loss: 0.0038 - val_accuracy: 0.6500\nEpoch 190/500\n107/107 [==============================] - 0s 332us/sample - loss: 0.0021 - accuracy: 0.6355 - val_loss: 0.0037 - val_accuracy: 0.7500\nEpoch 191/500\n107/107 [==============================] - 0s 321us/sample - loss: 0.0020 - accuracy: 0.6729 - val_loss: 0.0043 - val_accuracy: 0.6000\nEpoch 192/500\n107/107 [==============================] - 0s 338us/sample - loss: 0.0021 - accuracy: 0.6355 - val_loss: 0.0039 - val_accuracy: 0.7000\nEpoch 193/500\n107/107 [==============================] - 0s 316us/sample - loss: 0.0021 - accuracy: 0.7009 - val_loss: 0.0050 - val_accuracy: 0.6000\nEpoch 194/500\n107/107 [==============================] - 0s 357us/sample - loss: 0.0021 - accuracy: 0.6355 - val_loss: 0.0040 - val_accuracy: 0.7000\nEpoch 195/500\n107/107 [==============================] - 0s 381us/sample - loss: 0.0021 - accuracy: 0.7009 - val_loss: 0.0051 - val_accuracy: 0.4500\nEpoch 196/500\n107/107 [==============================] - 0s 346us/sample - loss: 0.0022 - accuracy: 0.6449 - val_loss: 0.0036 - val_accuracy: 0.7500\nEpoch 197/500\n107/107 [==============================] - 0s 326us/sample - loss: 0.0022 - accuracy: 0.6636 - val_loss: 0.0047 - val_accuracy: 0.4500\nEpoch 198/500\n107/107 [==============================] - 0s 332us/sample - loss: 0.0024 - accuracy: 0.6822 - val_loss: 0.0039 - val_accuracy: 0.6500\nEpoch 199/500\n107/107 [==============================] - 0s 305us/sample - loss: 0.0023 - accuracy: 0.6449 - val_loss: 0.0045 - val_accuracy: 0.5500\nEpoch 200/500\n107/107 [==============================] - 0s 274us/sample - loss: 0.0020 - accuracy: 0.6542 - val_loss: 0.0043 - val_accuracy: 0.7000\nEpoch 201/500\n107/107 [==============================] - 0s 291us/sample - loss: 0.0023 - accuracy: 0.5888 - val_loss: 0.0050 - val_accuracy: 0.5000\nEpoch 202/500\n107/107 [==============================] - 0s 314us/sample - loss: 0.0021 - accuracy: 0.6262 - val_loss: 0.0043 - val_accuracy: 0.6500\nEpoch 203/500\n107/107 [==============================] - 0s 387us/sample - loss: 0.0020 - accuracy: 0.6822 - val_loss: 0.0047 - val_accuracy: 0.6500\nEpoch 204/500\n107/107 [==============================] - 0s 341us/sample - loss: 0.0020 - accuracy: 0.6916 - val_loss: 0.0041 - val_accuracy: 0.7000\nEpoch 205/500\n107/107 [==============================] - 0s 287us/sample - loss: 0.0021 - accuracy: 0.6916 - val_loss: 0.0042 - val_accuracy: 0.5000\nEpoch 206/500\n107/107 [==============================] - 0s 276us/sample - loss: 0.0020 - accuracy: 0.6822 - val_loss: 0.0047 - val_accuracy: 0.6000\nEpoch 207/500\n107/107 [==============================] - 0s 296us/sample - loss: 0.0022 - accuracy: 0.6542 - val_loss: 0.0051 - val_accuracy: 0.5000\nEpoch 208/500\n107/107 [==============================] - 0s 312us/sample - loss: 0.0022 - accuracy: 0.6636 - val_loss: 0.0049 - val_accuracy: 0.6500\nEpoch 209/500\n107/107 [==============================] - 0s 330us/sample - loss: 0.0023 - accuracy: 0.6822 - val_loss: 0.0053 - val_accuracy: 0.4500\nEpoch 210/500\n107/107 [==============================] - 0s 285us/sample - loss: 0.0021 - accuracy: 0.6542 - val_loss: 0.0041 - val_accuracy: 0.7000\nEpoch 211/500\n107/107 [==============================] - 0s 314us/sample - loss: 0.0022 - accuracy: 0.6822 - val_loss: 0.0052 - val_accuracy: 0.5500\nEpoch 212/500\n107/107 [==============================] - 0s 301us/sample - loss: 0.0022 - accuracy: 0.6542 - val_loss: 0.0043 - val_accuracy: 0.6500\nEpoch 213/500\n107/107 [==============================] - 0s 294us/sample - loss: 0.0021 - accuracy: 0.6822 - val_loss: 0.0043 - val_accuracy: 0.5000\nEpoch 214/500\n107/107 [==============================] - 0s 292us/sample - loss: 0.0021 - accuracy: 0.6075 - val_loss: 0.0042 - val_accuracy: 0.7000\nEpoch 215/500\n107/107 [==============================] - 0s 326us/sample - loss: 0.0023 - accuracy: 0.5794 - val_loss: 0.0051 - val_accuracy: 0.6000\nEpoch 216/500\n107/107 [==============================] - 0s 356us/sample - loss: 0.0022 - accuracy: 0.6636 - val_loss: 0.0047 - val_accuracy: 0.6500\nEpoch 217/500\n107/107 [==============================] - 0s 333us/sample - loss: 0.0023 - accuracy: 0.6636 - val_loss: 0.0050 - val_accuracy: 0.5500\nEpoch 218/500\n107/107 [==============================] - 0s 333us/sample - loss: 0.0021 - accuracy: 0.6636 - val_loss: 0.0041 - val_accuracy: 0.6500\nEpoch 219/500\n107/107 [==============================] - 0s 312us/sample - loss: 0.0021 - accuracy: 0.6542 - val_loss: 0.0053 - val_accuracy: 0.6500\nEpoch 220/500\n107/107 [==============================] - 0s 287us/sample - loss: 0.0021 - accuracy: 0.6542 - val_loss: 0.0042 - val_accuracy: 0.6500\nEpoch 221/500\n107/107 [==============================] - 0s 282us/sample - loss: 0.0019 - accuracy: 0.7196 - val_loss: 0.0044 - val_accuracy: 0.5500\nEpoch 222/500\n107/107 [==============================] - 0s 323us/sample - loss: 0.0020 - accuracy: 0.7103 - val_loss: 0.0041 - val_accuracy: 0.6000\nEpoch 223/500\n107/107 [==============================] - 0s 412us/sample - loss: 0.0021 - accuracy: 0.6822 - val_loss: 0.0050 - val_accuracy: 0.5000\nEpoch 224/500\n107/107 [==============================] - 0s 326us/sample - loss: 0.0022 - accuracy: 0.6822 - val_loss: 0.0045 - val_accuracy: 0.6500\nEpoch 225/500\n107/107 [==============================] - 0s 315us/sample - loss: 0.0022 - accuracy: 0.7009 - val_loss: 0.0044 - val_accuracy: 0.6500\nEpoch 226/500\n107/107 [==============================] - 0s 315us/sample - loss: 0.0018 - accuracy: 0.6729 - val_loss: 0.0046 - val_accuracy: 0.7000\nEpoch 227/500\n107/107 [==============================] - 0s 354us/sample - loss: 0.0020 - accuracy: 0.5888 - val_loss: 0.0045 - val_accuracy: 0.6500\nEpoch 228/500\n107/107 [==============================] - 0s 320us/sample - loss: 0.0019 - accuracy: 0.6729 - val_loss: 0.0049 - val_accuracy: 0.6500\nEpoch 229/500\n107/107 [==============================] - 0s 302us/sample - loss: 0.0021 - accuracy: 0.6542 - val_loss: 0.0047 - val_accuracy: 0.6500\nEpoch 230/500\n107/107 [==============================] - 0s 277us/sample - loss: 0.0019 - accuracy: 0.6822 - val_loss: 0.0047 - val_accuracy: 0.6500\nEpoch 231/500\n107/107 [==============================] - 0s 315us/sample - loss: 0.0021 - accuracy: 0.6542 - val_loss: 0.0052 - val_accuracy: 0.5000\nEpoch 232/500\n107/107 [==============================] - 0s 309us/sample - loss: 0.0022 - accuracy: 0.6729 - val_loss: 0.0046 - val_accuracy: 0.6000\nEpoch 233/500\n107/107 [==============================] - 0s 277us/sample - loss: 0.0023 - accuracy: 0.6636 - val_loss: 0.0046 - val_accuracy: 0.5000\nEpoch 234/500\n107/107 [==============================] - 0s 270us/sample - loss: 0.0021 - accuracy: 0.6168 - val_loss: 0.0055 - val_accuracy: 0.6000\nEpoch 235/500\n107/107 [==============================] - 0s 316us/sample - loss: 0.0024 - accuracy: 0.6168 - val_loss: 0.0055 - val_accuracy: 0.6500\nEpoch 236/500\n107/107 [==============================] - 0s 301us/sample - loss: 0.0022 - accuracy: 0.6449 - val_loss: 0.0051 - val_accuracy: 0.6500\nEpoch 237/500\n107/107 [==============================] - 0s 314us/sample - loss: 0.0024 - accuracy: 0.6355 - val_loss: 0.0060 - val_accuracy: 0.5000\nEpoch 238/500\n107/107 [==============================] - 0s 285us/sample - loss: 0.0022 - accuracy: 0.6355 - val_loss: 0.0050 - val_accuracy: 0.6500\nEpoch 239/500\n107/107 [==============================] - 0s 283us/sample - loss: 0.0021 - accuracy: 0.6262 - val_loss: 0.0054 - val_accuracy: 0.7500\nEpoch 240/500\n107/107 [==============================] - 0s 291us/sample - loss: 0.0023 - accuracy: 0.6075 - val_loss: 0.0054 - val_accuracy: 0.6500\nEpoch 241/500\n107/107 [==============================] - 0s 289us/sample - loss: 0.0021 - accuracy: 0.6916 - val_loss: 0.0047 - val_accuracy: 0.5500\nEpoch 242/500\n107/107 [==============================] - 0s 308us/sample - loss: 0.0023 - accuracy: 0.6075 - val_loss: 0.0043 - val_accuracy: 0.6500\nEpoch 243/500\n107/107 [==============================] - 0s 316us/sample - loss: 0.0019 - accuracy: 0.7196 - val_loss: 0.0055 - val_accuracy: 0.7000\nEpoch 244/500\n107/107 [==============================] - 0s 276us/sample - loss: 0.0020 - accuracy: 0.6449 - val_loss: 0.0052 - val_accuracy: 0.5000\nEpoch 245/500\n107/107 [==============================] - 0s 293us/sample - loss: 0.0023 - accuracy: 0.6729 - val_loss: 0.0045 - val_accuracy: 0.7000\nEpoch 246/500\n107/107 [==============================] - 0s 276us/sample - loss: 0.0023 - accuracy: 0.6449 - val_loss: 0.0045 - val_accuracy: 0.5000\nEpoch 247/500\n107/107 [==============================] - 0s 305us/sample - loss: 0.0020 - accuracy: 0.6542 - val_loss: 0.0053 - val_accuracy: 0.7000\nEpoch 248/500\n107/107 [==============================] - 0s 323us/sample - loss: 0.0022 - accuracy: 0.6636 - val_loss: 0.0058 - val_accuracy: 0.6500\nEpoch 249/500\n107/107 [==============================] - 0s 304us/sample - loss: 0.0022 - accuracy: 0.6542 - val_loss: 0.0052 - val_accuracy: 0.6500\nEpoch 250/500\n107/107 [==============================] - 0s 320us/sample - loss: 0.0022 - accuracy: 0.6729 - val_loss: 0.0056 - val_accuracy: 0.5500\nEpoch 251/500\n107/107 [==============================] - 0s 321us/sample - loss: 0.0021 - accuracy: 0.6075 - val_loss: 0.0047 - val_accuracy: 0.6000\nEpoch 252/500\n107/107 [==============================] - 0s 449us/sample - loss: 0.0021 - accuracy: 0.6729 - val_loss: 0.0064 - val_accuracy: 0.6500\nEpoch 253/500\n107/107 [==============================] - 0s 345us/sample - loss: 0.0024 - accuracy: 0.6168 - val_loss: 0.0053 - val_accuracy: 0.7000\nEpoch 254/500\n107/107 [==============================] - 0s 400us/sample - loss: 0.0020 - accuracy: 0.7103 - val_loss: 0.0053 - val_accuracy: 0.5000\nEpoch 255/500\n107/107 [==============================] - 0s 339us/sample - loss: 0.0020 - accuracy: 0.6542 - val_loss: 0.0047 - val_accuracy: 0.6500\nEpoch 256/500\n107/107 [==============================] - 0s 338us/sample - loss: 0.0019 - accuracy: 0.7009 - val_loss: 0.0054 - val_accuracy: 0.6500\nEpoch 257/500\n107/107 [==============================] - 0s 315us/sample - loss: 0.0021 - accuracy: 0.6262 - val_loss: 0.0051 - val_accuracy: 0.7000\nEpoch 258/500\n107/107 [==============================] - 0s 371us/sample - loss: 0.0021 - accuracy: 0.6822 - val_loss: 0.0051 - val_accuracy: 0.5500\nEpoch 259/500\n107/107 [==============================] - 0s 391us/sample - loss: 0.0021 - accuracy: 0.6449 - val_loss: 0.0046 - val_accuracy: 0.6500\nEpoch 260/500\n107/107 [==============================] - 0s 341us/sample - loss: 0.0022 - accuracy: 0.6636 - val_loss: 0.0060 - val_accuracy: 0.5500\nEpoch 261/500\n107/107 [==============================] - 0s 347us/sample - loss: 0.0022 - accuracy: 0.6355 - val_loss: 0.0050 - val_accuracy: 0.6500\nEpoch 262/500\n107/107 [==============================] - 0s 351us/sample - loss: 0.0022 - accuracy: 0.6636 - val_loss: 0.0077 - val_accuracy: 0.4500\nEpoch 263/500\n107/107 [==============================] - 0s 350us/sample - loss: 0.0024 - accuracy: 0.6449 - val_loss: 0.0043 - val_accuracy: 0.6500\nEpoch 264/500\n107/107 [==============================] - 0s 367us/sample - loss: 0.0023 - accuracy: 0.6542 - val_loss: 0.0062 - val_accuracy: 0.5000\nEpoch 265/500\n107/107 [==============================] - 0s 372us/sample - loss: 0.0023 - accuracy: 0.6542 - val_loss: 0.0053 - val_accuracy: 0.6000\nEpoch 266/500\n107/107 [==============================] - 0s 383us/sample - loss: 0.0023 - accuracy: 0.6822 - val_loss: 0.0051 - val_accuracy: 0.5500\nEpoch 267/500\n107/107 [==============================] - 0s 351us/sample - loss: 0.0021 - accuracy: 0.6729 - val_loss: 0.0054 - val_accuracy: 0.6000\nEpoch 268/500\n107/107 [==============================] - 0s 338us/sample - loss: 0.0021 - accuracy: 0.6636 - val_loss: 0.0052 - val_accuracy: 0.7000\nEpoch 269/500\n107/107 [==============================] - 0s 364us/sample - loss: 0.0019 - accuracy: 0.6916 - val_loss: 0.0055 - val_accuracy: 0.6000\nEpoch 270/500\n107/107 [==============================] - 0s 458us/sample - loss: 0.0020 - accuracy: 0.6822 - val_loss: 0.0045 - val_accuracy: 0.6500\nEpoch 271/500\n107/107 [==============================] - 0s 359us/sample - loss: 0.0019 - accuracy: 0.7103 - val_loss: 0.0054 - val_accuracy: 0.6000\nEpoch 272/500\n107/107 [==============================] - 0s 354us/sample - loss: 0.0019 - accuracy: 0.7103 - val_loss: 0.0051 - val_accuracy: 0.5500\nEpoch 273/500\n107/107 [==============================] - 0s 365us/sample - loss: 0.0020 - accuracy: 0.7290 - val_loss: 0.0053 - val_accuracy: 0.6000\nEpoch 274/500\n107/107 [==============================] - 0s 354us/sample - loss: 0.0021 - accuracy: 0.7009 - val_loss: 0.0050 - val_accuracy: 0.5500\nEpoch 275/500\n107/107 [==============================] - 0s 360us/sample - loss: 0.0019 - accuracy: 0.7196 - val_loss: 0.0053 - val_accuracy: 0.6000\nEpoch 276/500\n107/107 [==============================] - 0s 460us/sample - loss: 0.0020 - accuracy: 0.7009 - val_loss: 0.0049 - val_accuracy: 0.6500\nEpoch 277/500\n107/107 [==============================] - 0s 394us/sample - loss: 0.0018 - accuracy: 0.7103 - val_loss: 0.0052 - val_accuracy: 0.6500\nEpoch 278/500\n107/107 [==============================] - 0s 397us/sample - loss: 0.0019 - accuracy: 0.7290 - val_loss: 0.0050 - val_accuracy: 0.6500\nEpoch 279/500\n107/107 [==============================] - 0s 350us/sample - loss: 0.0019 - accuracy: 0.7196 - val_loss: 0.0049 - val_accuracy: 0.7000\nEpoch 280/500\n107/107 [==============================] - 0s 331us/sample - loss: 0.0020 - accuracy: 0.7009 - val_loss: 0.0053 - val_accuracy: 0.4500\nEpoch 281/500\n107/107 [==============================] - 0s 360us/sample - loss: 0.0021 - accuracy: 0.6636 - val_loss: 0.0051 - val_accuracy: 0.6000\nEpoch 282/500\n107/107 [==============================] - 0s 358us/sample - loss: 0.0022 - accuracy: 0.6542 - val_loss: 0.0056 - val_accuracy: 0.5500\nEpoch 283/500\n107/107 [==============================] - 0s 421us/sample - loss: 0.0020 - accuracy: 0.6729 - val_loss: 0.0053 - val_accuracy: 0.6500\nEpoch 284/500\n107/107 [==============================] - 0s 353us/sample - loss: 0.0022 - accuracy: 0.6729 - val_loss: 0.0064 - val_accuracy: 0.6000\nEpoch 285/500\n107/107 [==============================] - 0s 366us/sample - loss: 0.0020 - accuracy: 0.6822 - val_loss: 0.0047 - val_accuracy: 0.6000\nEpoch 286/500\n107/107 [==============================] - 0s 375us/sample - loss: 0.0020 - accuracy: 0.7009 - val_loss: 0.0063 - val_accuracy: 0.7000\nEpoch 287/500\n107/107 [==============================] - 0s 355us/sample - loss: 0.0024 - accuracy: 0.6449 - val_loss: 0.0052 - val_accuracy: 0.6000\nEpoch 288/500\n107/107 [==============================] - 0s 349us/sample - loss: 0.0018 - accuracy: 0.6822 - val_loss: 0.0054 - val_accuracy: 0.5500\nEpoch 289/500\n107/107 [==============================] - 0s 357us/sample - loss: 0.0021 - accuracy: 0.7009 - val_loss: 0.0047 - val_accuracy: 0.6000\nEpoch 290/500\n107/107 [==============================] - 0s 342us/sample - loss: 0.0019 - accuracy: 0.6636 - val_loss: 0.0052 - val_accuracy: 0.6000\nEpoch 291/500\n107/107 [==============================] - 0s 354us/sample - loss: 0.0023 - accuracy: 0.6822 - val_loss: 0.0052 - val_accuracy: 0.6000\nEpoch 292/500\n107/107 [==============================] - 0s 366us/sample - loss: 0.0024 - accuracy: 0.6729 - val_loss: 0.0055 - val_accuracy: 0.5500\nEpoch 293/500\n107/107 [==============================] - 0s 385us/sample - loss: 0.0025 - accuracy: 0.6355 - val_loss: 0.0051 - val_accuracy: 0.5500\nEpoch 294/500\n107/107 [==============================] - 0s 347us/sample - loss: 0.0023 - accuracy: 0.6262 - val_loss: 0.0059 - val_accuracy: 0.4500\nEpoch 295/500\n107/107 [==============================] - 0s 336us/sample - loss: 0.0022 - accuracy: 0.6355 - val_loss: 0.0057 - val_accuracy: 0.5500\nEpoch 296/500\n107/107 [==============================] - 0s 416us/sample - loss: 0.0024 - accuracy: 0.6636 - val_loss: 0.0052 - val_accuracy: 0.6000\nEpoch 297/500\n107/107 [==============================] - 0s 361us/sample - loss: 0.0021 - accuracy: 0.7009 - val_loss: 0.0051 - val_accuracy: 0.5500\nEpoch 298/500\n107/107 [==============================] - 0s 397us/sample - loss: 0.0021 - accuracy: 0.6916 - val_loss: 0.0052 - val_accuracy: 0.4500\nEpoch 299/500\n107/107 [==============================] - 0s 371us/sample - loss: 0.0020 - accuracy: 0.6916 - val_loss: 0.0050 - val_accuracy: 0.7000\nEpoch 300/500\n107/107 [==============================] - 0s 420us/sample - loss: 0.0020 - accuracy: 0.6449 - val_loss: 0.0052 - val_accuracy: 0.6000\nEpoch 301/500\n107/107 [==============================] - 0s 479us/sample - loss: 0.0019 - accuracy: 0.7383 - val_loss: 0.0061 - val_accuracy: 0.5500\nEpoch 302/500\n107/107 [==============================] - 0s 361us/sample - loss: 0.0023 - accuracy: 0.6449 - val_loss: 0.0052 - val_accuracy: 0.5000\nEpoch 303/500\n107/107 [==============================] - 0s 343us/sample - loss: 0.0021 - accuracy: 0.7196 - val_loss: 0.0050 - val_accuracy: 0.6500\nEpoch 304/500\n107/107 [==============================] - 0s 338us/sample - loss: 0.0020 - accuracy: 0.7103 - val_loss: 0.0055 - val_accuracy: 0.6500\nEpoch 305/500\n107/107 [==============================] - 0s 389us/sample - loss: 0.0018 - accuracy: 0.7009 - val_loss: 0.0051 - val_accuracy: 0.6500\nEpoch 306/500\n107/107 [==============================] - 0s 369us/sample - loss: 0.0019 - accuracy: 0.6916 - val_loss: 0.0052 - val_accuracy: 0.6500\nEpoch 307/500\n107/107 [==============================] - 0s 357us/sample - loss: 0.0018 - accuracy: 0.7196 - val_loss: 0.0053 - val_accuracy: 0.6000\nEpoch 308/500\n107/107 [==============================] - 0s 422us/sample - loss: 0.0017 - accuracy: 0.7196 - val_loss: 0.0050 - val_accuracy: 0.6500\nEpoch 309/500\n107/107 [==============================] - 0s 408us/sample - loss: 0.0018 - accuracy: 0.7103 - val_loss: 0.0046 - val_accuracy: 0.6000\nEpoch 310/500\n107/107 [==============================] - 0s 345us/sample - loss: 0.0017 - accuracy: 0.7383 - val_loss: 0.0055 - val_accuracy: 0.6500\nEpoch 311/500\n107/107 [==============================] - 0s 330us/sample - loss: 0.0018 - accuracy: 0.7290 - val_loss: 0.0052 - val_accuracy: 0.6000\nEpoch 312/500\n107/107 [==============================] - 0s 387us/sample - loss: 0.0018 - accuracy: 0.7383 - val_loss: 0.0057 - val_accuracy: 0.5500\nEpoch 313/500\n107/107 [==============================] - 0s 368us/sample - loss: 0.0019 - accuracy: 0.7196 - val_loss: 0.0056 - val_accuracy: 0.6000\nEpoch 314/500\n107/107 [==============================] - 0s 387us/sample - loss: 0.0019 - accuracy: 0.7196 - val_loss: 0.0055 - val_accuracy: 0.6000\nEpoch 315/500\n107/107 [==============================] - 0s 307us/sample - loss: 0.0018 - accuracy: 0.7196 - val_loss: 0.0050 - val_accuracy: 0.6000\nEpoch 316/500\n107/107 [==============================] - 0s 368us/sample - loss: 0.0018 - accuracy: 0.7477 - val_loss: 0.0054 - val_accuracy: 0.6500\nEpoch 317/500\n107/107 [==============================] - 0s 395us/sample - loss: 0.0018 - accuracy: 0.7196 - val_loss: 0.0056 - val_accuracy: 0.6000\nEpoch 318/500\n107/107 [==============================] - 0s 374us/sample - loss: 0.0018 - accuracy: 0.7290 - val_loss: 0.0060 - val_accuracy: 0.6500\nEpoch 319/500\n107/107 [==============================] - 0s 345us/sample - loss: 0.0017 - accuracy: 0.7196 - val_loss: 0.0057 - val_accuracy: 0.6000\nEpoch 320/500\n107/107 [==============================] - 0s 339us/sample - loss: 0.0017 - accuracy: 0.7290 - val_loss: 0.0056 - val_accuracy: 0.6500\nEpoch 321/500\n107/107 [==============================] - 0s 331us/sample - loss: 0.0016 - accuracy: 0.7477 - val_loss: 0.0057 - val_accuracy: 0.6000\nEpoch 322/500\n107/107 [==============================] - 0s 371us/sample - loss: 0.0018 - accuracy: 0.7477 - val_loss: 0.0056 - val_accuracy: 0.6500\nEpoch 323/500\n107/107 [==============================] - 0s 402us/sample - loss: 0.0017 - accuracy: 0.7477 - val_loss: 0.0059 - val_accuracy: 0.6000\nEpoch 324/500\n107/107 [==============================] - 0s 362us/sample - loss: 0.0019 - accuracy: 0.7196 - val_loss: 0.0054 - val_accuracy: 0.5500\nEpoch 325/500\n107/107 [==============================] - 0s 393us/sample - loss: 0.0017 - accuracy: 0.7196 - val_loss: 0.0063 - val_accuracy: 0.6000\nEpoch 326/500\n107/107 [==============================] - 0s 475us/sample - loss: 0.0020 - accuracy: 0.6822 - val_loss: 0.0052 - val_accuracy: 0.6500\nEpoch 327/500\n107/107 [==============================] - 0s 377us/sample - loss: 0.0017 - accuracy: 0.7103 - val_loss: 0.0060 - val_accuracy: 0.6000\nEpoch 328/500\n107/107 [==============================] - 0s 353us/sample - loss: 0.0019 - accuracy: 0.7103 - val_loss: 0.0054 - val_accuracy: 0.6500\nEpoch 329/500\n107/107 [==============================] - 0s 377us/sample - loss: 0.0017 - accuracy: 0.7290 - val_loss: 0.0058 - val_accuracy: 0.6000\nEpoch 330/500\n107/107 [==============================] - 0s 404us/sample - loss: 0.0018 - accuracy: 0.7570 - val_loss: 0.0059 - val_accuracy: 0.6500\nEpoch 331/500\n107/107 [==============================] - 0s 364us/sample - loss: 0.0017 - accuracy: 0.7383 - val_loss: 0.0057 - val_accuracy: 0.6000\nEpoch 332/500\n107/107 [==============================] - 0s 362us/sample - loss: 0.0018 - accuracy: 0.7383 - val_loss: 0.0051 - val_accuracy: 0.6500\nEpoch 333/500\n107/107 [==============================] - 0s 355us/sample - loss: 0.0016 - accuracy: 0.7757 - val_loss: 0.0063 - val_accuracy: 0.6000\nEpoch 334/500\n107/107 [==============================] - 0s 356us/sample - loss: 0.0019 - accuracy: 0.7196 - val_loss: 0.0052 - val_accuracy: 0.6000\nEpoch 335/500\n107/107 [==============================] - 0s 356us/sample - loss: 0.0018 - accuracy: 0.7290 - val_loss: 0.0068 - val_accuracy: 0.6000\nEpoch 336/500\n107/107 [==============================] - 0s 361us/sample - loss: 0.0020 - accuracy: 0.6916 - val_loss: 0.0051 - val_accuracy: 0.6000\nEpoch 337/500\n107/107 [==============================] - 0s 397us/sample - loss: 0.0019 - accuracy: 0.7290 - val_loss: 0.0064 - val_accuracy: 0.6000\nEpoch 338/500\n107/107 [==============================] - 0s 349us/sample - loss: 0.0020 - accuracy: 0.6822 - val_loss: 0.0056 - val_accuracy: 0.6000\nEpoch 339/500\n107/107 [==============================] - 0s 362us/sample - loss: 0.0018 - accuracy: 0.7383 - val_loss: 0.0053 - val_accuracy: 0.6000\nEpoch 340/500\n107/107 [==============================] - 0s 361us/sample - loss: 0.0018 - accuracy: 0.7196 - val_loss: 0.0059 - val_accuracy: 0.6500\nEpoch 341/500\n107/107 [==============================] - 0s 338us/sample - loss: 0.0017 - accuracy: 0.7196 - val_loss: 0.0055 - val_accuracy: 0.6500\nEpoch 342/500\n107/107 [==============================] - 0s 334us/sample - loss: 0.0017 - accuracy: 0.7664 - val_loss: 0.0056 - val_accuracy: 0.6500\nEpoch 343/500\n107/107 [==============================] - 0s 354us/sample - loss: 0.0016 - accuracy: 0.7103 - val_loss: 0.0061 - val_accuracy: 0.6000\nEpoch 344/500\n107/107 [==============================] - 0s 386us/sample - loss: 0.0017 - accuracy: 0.7290 - val_loss: 0.0051 - val_accuracy: 0.6000\nEpoch 345/500\n107/107 [==============================] - 0s 380us/sample - loss: 0.0017 - accuracy: 0.7290 - val_loss: 0.0058 - val_accuracy: 0.6000\nEpoch 346/500\n107/107 [==============================] - 0s 362us/sample - loss: 0.0019 - accuracy: 0.6822 - val_loss: 0.0058 - val_accuracy: 0.5500\nEpoch 347/500\n107/107 [==============================] - 0s 363us/sample - loss: 0.0020 - accuracy: 0.6916 - val_loss: 0.0065 - val_accuracy: 0.6500\nEpoch 348/500\n107/107 [==============================] - 0s 356us/sample - loss: 0.0020 - accuracy: 0.6916 - val_loss: 0.0067 - val_accuracy: 0.5500\nEpoch 349/500\n107/107 [==============================] - 0s 459us/sample - loss: 0.0018 - accuracy: 0.6729 - val_loss: 0.0046 - val_accuracy: 0.7000\nEpoch 350/500\n107/107 [==============================] - 0s 473us/sample - loss: 0.0018 - accuracy: 0.6916 - val_loss: 0.0059 - val_accuracy: 0.6000\nEpoch 351/500\n107/107 [==============================] - 0s 380us/sample - loss: 0.0017 - accuracy: 0.6636 - val_loss: 0.0055 - val_accuracy: 0.6000\nEpoch 352/500\n107/107 [==============================] - 0s 375us/sample - loss: 0.0019 - accuracy: 0.7196 - val_loss: 0.0066 - val_accuracy: 0.6000\nEpoch 353/500\n107/107 [==============================] - 0s 368us/sample - loss: 0.0019 - accuracy: 0.6636 - val_loss: 0.0065 - val_accuracy: 0.5500\nEpoch 354/500\n107/107 [==============================] - 0s 352us/sample - loss: 0.0022 - accuracy: 0.7196 - val_loss: 0.0058 - val_accuracy: 0.5500\nEpoch 355/500\n107/107 [==============================] - 0s 379us/sample - loss: 0.0018 - accuracy: 0.6916 - val_loss: 0.0045 - val_accuracy: 0.6500\nEpoch 356/500\n107/107 [==============================] - 0s 363us/sample - loss: 0.0020 - accuracy: 0.6542 - val_loss: 0.0060 - val_accuracy: 0.6000\nEpoch 357/500\n107/107 [==============================] - 0s 454us/sample - loss: 0.0024 - accuracy: 0.7196 - val_loss: 0.0058 - val_accuracy: 0.5500\nEpoch 358/500\n107/107 [==============================] - 0s 355us/sample - loss: 0.0024 - accuracy: 0.6262 - val_loss: 0.0055 - val_accuracy: 0.6500\nEpoch 359/500\n107/107 [==============================] - 0s 371us/sample - loss: 0.0020 - accuracy: 0.6916 - val_loss: 0.0056 - val_accuracy: 0.7000\nEpoch 360/500\n107/107 [==============================] - 0s 410us/sample - loss: 0.0020 - accuracy: 0.7009 - val_loss: 0.0055 - val_accuracy: 0.6000\nEpoch 361/500\n107/107 [==============================] - 0s 359us/sample - loss: 0.0020 - accuracy: 0.7103 - val_loss: 0.0051 - val_accuracy: 0.6000\nEpoch 362/500\n107/107 [==============================] - 0s 354us/sample - loss: 0.0018 - accuracy: 0.6916 - val_loss: 0.0051 - val_accuracy: 0.6000\nEpoch 363/500\n107/107 [==============================] - 0s 353us/sample - loss: 0.0019 - accuracy: 0.7196 - val_loss: 0.0070 - val_accuracy: 0.5500\nEpoch 364/500\n107/107 [==============================] - 0s 343us/sample - loss: 0.0021 - accuracy: 0.6542 - val_loss: 0.0051 - val_accuracy: 0.6500\nEpoch 365/500\n107/107 [==============================] - 0s 390us/sample - loss: 0.0020 - accuracy: 0.7290 - val_loss: 0.0070 - val_accuracy: 0.5500\nEpoch 366/500\n107/107 [==============================] - 0s 383us/sample - loss: 0.0024 - accuracy: 0.6542 - val_loss: 0.0056 - val_accuracy: 0.6000\nEpoch 367/500\n107/107 [==============================] - 0s 353us/sample - loss: 0.0020 - accuracy: 0.6729 - val_loss: 0.0066 - val_accuracy: 0.6000\nEpoch 368/500\n107/107 [==============================] - 0s 348us/sample - loss: 0.0025 - accuracy: 0.6542 - val_loss: 0.0075 - val_accuracy: 0.5500\nEpoch 369/500\n107/107 [==============================] - 0s 391us/sample - loss: 0.0019 - accuracy: 0.7009 - val_loss: 0.0054 - val_accuracy: 0.7000\nEpoch 370/500\n107/107 [==============================] - 0s 380us/sample - loss: 0.0018 - accuracy: 0.7290 - val_loss: 0.0063 - val_accuracy: 0.6000\nEpoch 371/500\n107/107 [==============================] - 0s 340us/sample - loss: 0.0020 - accuracy: 0.6916 - val_loss: 0.0052 - val_accuracy: 0.5500\nEpoch 372/500\n107/107 [==============================] - 0s 403us/sample - loss: 0.0017 - accuracy: 0.7477 - val_loss: 0.0066 - val_accuracy: 0.6500\nEpoch 373/500\n107/107 [==============================] - 0s 350us/sample - loss: 0.0025 - accuracy: 0.6636 - val_loss: 0.0044 - val_accuracy: 0.7000\nEpoch 374/500\n107/107 [==============================] - 0s 337us/sample - loss: 0.0019 - accuracy: 0.7196 - val_loss: 0.0068 - val_accuracy: 0.5500\nEpoch 375/500\n107/107 [==============================] - 0s 502us/sample - loss: 0.0019 - accuracy: 0.6729 - val_loss: 0.0054 - val_accuracy: 0.6500\nEpoch 376/500\n107/107 [==============================] - 0s 413us/sample - loss: 0.0020 - accuracy: 0.7290 - val_loss: 0.0063 - val_accuracy: 0.5500\nEpoch 377/500\n107/107 [==============================] - 0s 379us/sample - loss: 0.0017 - accuracy: 0.7290 - val_loss: 0.0056 - val_accuracy: 0.6500\nEpoch 378/500\n107/107 [==============================] - 0s 348us/sample - loss: 0.0019 - accuracy: 0.7009 - val_loss: 0.0065 - val_accuracy: 0.5500\nEpoch 379/500\n107/107 [==============================] - 0s 355us/sample - loss: 0.0020 - accuracy: 0.6636 - val_loss: 0.0048 - val_accuracy: 0.6500\nEpoch 380/500\n107/107 [==============================] - 0s 348us/sample - loss: 0.0020 - accuracy: 0.6916 - val_loss: 0.0066 - val_accuracy: 0.6000\nEpoch 381/500\n107/107 [==============================] - 0s 371us/sample - loss: 0.0019 - accuracy: 0.6822 - val_loss: 0.0050 - val_accuracy: 0.6500\nEpoch 382/500\n107/107 [==============================] - 0s 357us/sample - loss: 0.0018 - accuracy: 0.7383 - val_loss: 0.0063 - val_accuracy: 0.5500\nEpoch 383/500\n107/107 [==============================] - 0s 385us/sample - loss: 0.0017 - accuracy: 0.7196 - val_loss: 0.0060 - val_accuracy: 0.6000\nEpoch 384/500\n107/107 [==============================] - 0s 370us/sample - loss: 0.0017 - accuracy: 0.7383 - val_loss: 0.0065 - val_accuracy: 0.6000\nEpoch 385/500\n107/107 [==============================] - 0s 383us/sample - loss: 0.0020 - accuracy: 0.6916 - val_loss: 0.0058 - val_accuracy: 0.7000\nEpoch 386/500\n107/107 [==============================] - 0s 375us/sample - loss: 0.0019 - accuracy: 0.6916 - val_loss: 0.0064 - val_accuracy: 0.6000\nEpoch 387/500\n107/107 [==============================] - 0s 363us/sample - loss: 0.0019 - accuracy: 0.7103 - val_loss: 0.0067 - val_accuracy: 0.6000\nEpoch 388/500\n107/107 [==============================] - 0s 352us/sample - loss: 0.0021 - accuracy: 0.7103 - val_loss: 0.0059 - val_accuracy: 0.6000\nEpoch 389/500\n107/107 [==============================] - 0s 365us/sample - loss: 0.0021 - accuracy: 0.7290 - val_loss: 0.0081 - val_accuracy: 0.5000\nEpoch 390/500\n107/107 [==============================] - 0s 339us/sample - loss: 0.0021 - accuracy: 0.6729 - val_loss: 0.0055 - val_accuracy: 0.5500\nEpoch 391/500\n107/107 [==============================] - 0s 373us/sample - loss: 0.0022 - accuracy: 0.6729 - val_loss: 0.0065 - val_accuracy: 0.6500\nEpoch 392/500\n107/107 [==============================] - 0s 374us/sample - loss: 0.0026 - accuracy: 0.6542 - val_loss: 0.0072 - val_accuracy: 0.5500\nEpoch 393/500\n107/107 [==============================] - 0s 373us/sample - loss: 0.0020 - accuracy: 0.6916 - val_loss: 0.0064 - val_accuracy: 0.5500\nEpoch 394/500\n107/107 [==============================] - 0s 376us/sample - loss: 0.0022 - accuracy: 0.6729 - val_loss: 0.0085 - val_accuracy: 0.5000\nEpoch 395/500\n107/107 [==============================] - 0s 386us/sample - loss: 0.0021 - accuracy: 0.6916 - val_loss: 0.0055 - val_accuracy: 0.6500\nEpoch 396/500\n107/107 [==============================] - 0s 383us/sample - loss: 0.0021 - accuracy: 0.7383 - val_loss: 0.0066 - val_accuracy: 0.6000\nEpoch 397/500\n107/107 [==============================] - 0s 410us/sample - loss: 0.0020 - accuracy: 0.7290 - val_loss: 0.0065 - val_accuracy: 0.6000\nEpoch 398/500\n107/107 [==============================] - 0s 357us/sample - loss: 0.0022 - accuracy: 0.7103 - val_loss: 0.0055 - val_accuracy: 0.5000\nEpoch 399/500\n107/107 [==============================] - 0s 495us/sample - loss: 0.0019 - accuracy: 0.7290 - val_loss: 0.0062 - val_accuracy: 0.6000\nEpoch 400/500\n107/107 [==============================] - 0s 382us/sample - loss: 0.0018 - accuracy: 0.6916 - val_loss: 0.0057 - val_accuracy: 0.5500\nEpoch 401/500\n107/107 [==============================] - 0s 369us/sample - loss: 0.0018 - accuracy: 0.7196 - val_loss: 0.0065 - val_accuracy: 0.6500\nEpoch 402/500\n107/107 [==============================] - 0s 379us/sample - loss: 0.0019 - accuracy: 0.6916 - val_loss: 0.0059 - val_accuracy: 0.5000\nEpoch 403/500\n107/107 [==============================] - 0s 361us/sample - loss: 0.0020 - accuracy: 0.7196 - val_loss: 0.0056 - val_accuracy: 0.6000\nEpoch 404/500\n107/107 [==============================] - 0s 360us/sample - loss: 0.0019 - accuracy: 0.7196 - val_loss: 0.0063 - val_accuracy: 0.5500\nEpoch 405/500\n107/107 [==============================] - 0s 381us/sample - loss: 0.0019 - accuracy: 0.6916 - val_loss: 0.0070 - val_accuracy: 0.5500\nEpoch 406/500\n107/107 [==============================] - 0s 407us/sample - loss: 0.0021 - accuracy: 0.6729 - val_loss: 0.0074 - val_accuracy: 0.5500\nEpoch 407/500\n107/107 [==============================] - 0s 346us/sample - loss: 0.0021 - accuracy: 0.6822 - val_loss: 0.0055 - val_accuracy: 0.6000\nEpoch 408/500\n107/107 [==============================] - 0s 338us/sample - loss: 0.0020 - accuracy: 0.7103 - val_loss: 0.0060 - val_accuracy: 0.5500\nEpoch 409/500\n107/107 [==============================] - 0s 347us/sample - loss: 0.0021 - accuracy: 0.7009 - val_loss: 0.0058 - val_accuracy: 0.5500\nEpoch 410/500\n107/107 [==============================] - 0s 365us/sample - loss: 0.0018 - accuracy: 0.7290 - val_loss: 0.0071 - val_accuracy: 0.5500\nEpoch 411/500\n107/107 [==============================] - 0s 327us/sample - loss: 0.0019 - accuracy: 0.7383 - val_loss: 0.0060 - val_accuracy: 0.6000\nEpoch 412/500\n107/107 [==============================] - 0s 398us/sample - loss: 0.0020 - accuracy: 0.7009 - val_loss: 0.0057 - val_accuracy: 0.6500\nEpoch 413/500\n107/107 [==============================] - 0s 369us/sample - loss: 0.0017 - accuracy: 0.7664 - val_loss: 0.0067 - val_accuracy: 0.6500\nEpoch 414/500\n107/107 [==============================] - 0s 342us/sample - loss: 0.0020 - accuracy: 0.7009 - val_loss: 0.0059 - val_accuracy: 0.6000\nEpoch 415/500\n107/107 [==============================] - 0s 375us/sample - loss: 0.0019 - accuracy: 0.7196 - val_loss: 0.0064 - val_accuracy: 0.6000\nEpoch 416/500\n107/107 [==============================] - 0s 402us/sample - loss: 0.0019 - accuracy: 0.7383 - val_loss: 0.0065 - val_accuracy: 0.6000\nEpoch 417/500\n107/107 [==============================] - 0s 401us/sample - loss: 0.0017 - accuracy: 0.7196 - val_loss: 0.0057 - val_accuracy: 0.6000\nEpoch 418/500\n107/107 [==============================] - 0s 368us/sample - loss: 0.0017 - accuracy: 0.7383 - val_loss: 0.0063 - val_accuracy: 0.6500\nEpoch 419/500\n107/107 [==============================] - 0s 378us/sample - loss: 0.0016 - accuracy: 0.7850 - val_loss: 0.0058 - val_accuracy: 0.6000\nEpoch 420/500\n107/107 [==============================] - 0s 378us/sample - loss: 0.0018 - accuracy: 0.7290 - val_loss: 0.0055 - val_accuracy: 0.6500\nEpoch 421/500\n107/107 [==============================] - 0s 410us/sample - loss: 0.0016 - accuracy: 0.7383 - val_loss: 0.0063 - val_accuracy: 0.6000\nEpoch 422/500\n107/107 [==============================] - 0s 352us/sample - loss: 0.0018 - accuracy: 0.7196 - val_loss: 0.0059 - val_accuracy: 0.6500\nEpoch 423/500\n107/107 [==============================] - 0s 446us/sample - loss: 0.0016 - accuracy: 0.7570 - val_loss: 0.0062 - val_accuracy: 0.6000\nEpoch 424/500\n107/107 [==============================] - 0s 358us/sample - loss: 0.0017 - accuracy: 0.7290 - val_loss: 0.0058 - val_accuracy: 0.6500\nEpoch 425/500\n107/107 [==============================] - 0s 339us/sample - loss: 0.0016 - accuracy: 0.7664 - val_loss: 0.0057 - val_accuracy: 0.6000\nEpoch 426/500\n107/107 [==============================] - 0s 346us/sample - loss: 0.0017 - accuracy: 0.7477 - val_loss: 0.0057 - val_accuracy: 0.6000\nEpoch 427/500\n107/107 [==============================] - 0s 360us/sample - loss: 0.0016 - accuracy: 0.7290 - val_loss: 0.0062 - val_accuracy: 0.6000\nEpoch 428/500\n107/107 [==============================] - 0s 341us/sample - loss: 0.0017 - accuracy: 0.7383 - val_loss: 0.0058 - val_accuracy: 0.6000\nEpoch 429/500\n107/107 [==============================] - 0s 382us/sample - loss: 0.0017 - accuracy: 0.7383 - val_loss: 0.0060 - val_accuracy: 0.6000\nEpoch 430/500\n107/107 [==============================] - 0s 400us/sample - loss: 0.0018 - accuracy: 0.7290 - val_loss: 0.0064 - val_accuracy: 0.6500\nEpoch 431/500\n107/107 [==============================] - 0s 375us/sample - loss: 0.0016 - accuracy: 0.7290 - val_loss: 0.0063 - val_accuracy: 0.6000\nEpoch 432/500\n107/107 [==============================] - 0s 353us/sample - loss: 0.0018 - accuracy: 0.7477 - val_loss: 0.0059 - val_accuracy: 0.6500\nEpoch 433/500\n107/107 [==============================] - 0s 369us/sample - loss: 0.0016 - accuracy: 0.7477 - val_loss: 0.0056 - val_accuracy: 0.6000\nEpoch 434/500\n107/107 [==============================] - 0s 350us/sample - loss: 0.0017 - accuracy: 0.7383 - val_loss: 0.0063 - val_accuracy: 0.6500\nEpoch 435/500\n107/107 [==============================] - 0s 345us/sample - loss: 0.0016 - accuracy: 0.7477 - val_loss: 0.0058 - val_accuracy: 0.6000\nEpoch 436/500\n107/107 [==============================] - 0s 343us/sample - loss: 0.0016 - accuracy: 0.7757 - val_loss: 0.0066 - val_accuracy: 0.6500\nEpoch 437/500\n107/107 [==============================] - 0s 360us/sample - loss: 0.0016 - accuracy: 0.7477 - val_loss: 0.0061 - val_accuracy: 0.6000\nEpoch 438/500\n107/107 [==============================] - 0s 356us/sample - loss: 0.0016 - accuracy: 0.7570 - val_loss: 0.0064 - val_accuracy: 0.6000\nEpoch 439/500\n107/107 [==============================] - 0s 375us/sample - loss: 0.0018 - accuracy: 0.7477 - val_loss: 0.0054 - val_accuracy: 0.6000\nEpoch 440/500\n107/107 [==============================] - 0s 359us/sample - loss: 0.0017 - accuracy: 0.7383 - val_loss: 0.0059 - val_accuracy: 0.6000\nEpoch 441/500\n107/107 [==============================] - 0s 373us/sample - loss: 0.0018 - accuracy: 0.7477 - val_loss: 0.0057 - val_accuracy: 0.6000\nEpoch 442/500\n107/107 [==============================] - 0s 386us/sample - loss: 0.0017 - accuracy: 0.7383 - val_loss: 0.0064 - val_accuracy: 0.6000\nEpoch 443/500\n107/107 [==============================] - 0s 350us/sample - loss: 0.0017 - accuracy: 0.7290 - val_loss: 0.0065 - val_accuracy: 0.6000\nEpoch 444/500\n107/107 [==============================] - 0s 361us/sample - loss: 0.0020 - accuracy: 0.7196 - val_loss: 0.0069 - val_accuracy: 0.6000\nEpoch 445/500\n107/107 [==============================] - 0s 361us/sample - loss: 0.0017 - accuracy: 0.7196 - val_loss: 0.0055 - val_accuracy: 0.6000\nEpoch 446/500\n107/107 [==============================] - 0s 314us/sample - loss: 0.0018 - accuracy: 0.7290 - val_loss: 0.0065 - val_accuracy: 0.6000\nEpoch 447/500\n107/107 [==============================] - 0s 327us/sample - loss: 0.0017 - accuracy: 0.7196 - val_loss: 0.0056 - val_accuracy: 0.6000\nEpoch 448/500\n107/107 [==============================] - 0s 346us/sample - loss: 0.0016 - accuracy: 0.7570 - val_loss: 0.0060 - val_accuracy: 0.6000\nEpoch 449/500\n107/107 [==============================] - 0s 438us/sample - loss: 0.0016 - accuracy: 0.7664 - val_loss: 0.0066 - val_accuracy: 0.5500\nEpoch 450/500\n107/107 [==============================] - 0s 345us/sample - loss: 0.0019 - accuracy: 0.6822 - val_loss: 0.0060 - val_accuracy: 0.6000\nEpoch 451/500\n107/107 [==============================] - 0s 336us/sample - loss: 0.0017 - accuracy: 0.7850 - val_loss: 0.0062 - val_accuracy: 0.6000\nEpoch 452/500\n107/107 [==============================] - 0s 313us/sample - loss: 0.0018 - accuracy: 0.7290 - val_loss: 0.0057 - val_accuracy: 0.6500\nEpoch 453/500\n107/107 [==============================] - 0s 342us/sample - loss: 0.0016 - accuracy: 0.7944 - val_loss: 0.0061 - val_accuracy: 0.5500\nEpoch 454/500\n107/107 [==============================] - 0s 333us/sample - loss: 0.0016 - accuracy: 0.7664 - val_loss: 0.0060 - val_accuracy: 0.6000\nEpoch 455/500\n107/107 [==============================] - 0s 350us/sample - loss: 0.0016 - accuracy: 0.7757 - val_loss: 0.0070 - val_accuracy: 0.6000\nEpoch 456/500\n107/107 [==============================] - 0s 362us/sample - loss: 0.0016 - accuracy: 0.7383 - val_loss: 0.0057 - val_accuracy: 0.7000\nEpoch 457/500\n107/107 [==============================] - 0s 319us/sample - loss: 0.0016 - accuracy: 0.7570 - val_loss: 0.0066 - val_accuracy: 0.6000\nEpoch 458/500\n107/107 [==============================] - 0s 325us/sample - loss: 0.0018 - accuracy: 0.7196 - val_loss: 0.0059 - val_accuracy: 0.6500\nEpoch 459/500\n107/107 [==============================] - 0s 317us/sample - loss: 0.0018 - accuracy: 0.7664 - val_loss: 0.0071 - val_accuracy: 0.5500\nEpoch 460/500\n107/107 [==============================] - 0s 308us/sample - loss: 0.0018 - accuracy: 0.7009 - val_loss: 0.0061 - val_accuracy: 0.6500\nEpoch 461/500\n107/107 [==============================] - 0s 311us/sample - loss: 0.0016 - accuracy: 0.7757 - val_loss: 0.0069 - val_accuracy: 0.6000\nEpoch 462/500\n107/107 [==============================] - 0s 303us/sample - loss: 0.0017 - accuracy: 0.7290 - val_loss: 0.0061 - val_accuracy: 0.7000\nEpoch 463/500\n107/107 [==============================] - 0s 329us/sample - loss: 0.0017 - accuracy: 0.7664 - val_loss: 0.0064 - val_accuracy: 0.6000\nEpoch 464/500\n107/107 [==============================] - 0s 299us/sample - loss: 0.0017 - accuracy: 0.7664 - val_loss: 0.0065 - val_accuracy: 0.6500\nEpoch 465/500\n107/107 [==============================] - 0s 288us/sample - loss: 0.0015 - accuracy: 0.8037 - val_loss: 0.0061 - val_accuracy: 0.6000\nEpoch 466/500\n107/107 [==============================] - 0s 307us/sample - loss: 0.0016 - accuracy: 0.7850 - val_loss: 0.0061 - val_accuracy: 0.6000\nEpoch 467/500\n107/107 [==============================] - 0s 300us/sample - loss: 0.0015 - accuracy: 0.7944 - val_loss: 0.0068 - val_accuracy: 0.6000\nEpoch 468/500\n107/107 [==============================] - 0s 275us/sample - loss: 0.0017 - accuracy: 0.7383 - val_loss: 0.0062 - val_accuracy: 0.6500\nEpoch 469/500\n107/107 [==============================] - 0s 309us/sample - loss: 0.0017 - accuracy: 0.7757 - val_loss: 0.0065 - val_accuracy: 0.6000\nEpoch 470/500\n107/107 [==============================] - 0s 319us/sample - loss: 0.0019 - accuracy: 0.7103 - val_loss: 0.0061 - val_accuracy: 0.6500\nEpoch 471/500\n107/107 [==============================] - 0s 278us/sample - loss: 0.0017 - accuracy: 0.7570 - val_loss: 0.0060 - val_accuracy: 0.6000\nEpoch 472/500\n107/107 [==============================] - 0s 284us/sample - loss: 0.0017 - accuracy: 0.7664 - val_loss: 0.0066 - val_accuracy: 0.6000\nEpoch 473/500\n107/107 [==============================] - 0s 302us/sample - loss: 0.0019 - accuracy: 0.7477 - val_loss: 0.0064 - val_accuracy: 0.5000\nEpoch 474/500\n107/107 [==============================] - 0s 297us/sample - loss: 0.0016 - accuracy: 0.7290 - val_loss: 0.0060 - val_accuracy: 0.6000\nEpoch 475/500\n107/107 [==============================] - 0s 296us/sample - loss: 0.0017 - accuracy: 0.7664 - val_loss: 0.0061 - val_accuracy: 0.6000\nEpoch 476/500\n107/107 [==============================] - 0s 330us/sample - loss: 0.0018 - accuracy: 0.7383 - val_loss: 0.0062 - val_accuracy: 0.7000\nEpoch 477/500\n107/107 [==============================] - 0s 350us/sample - loss: 0.0017 - accuracy: 0.7290 - val_loss: 0.0062 - val_accuracy: 0.5500\nEpoch 478/500\n107/107 [==============================] - 0s 311us/sample - loss: 0.0018 - accuracy: 0.7103 - val_loss: 0.0060 - val_accuracy: 0.6000\nEpoch 479/500\n107/107 [==============================] - 0s 300us/sample - loss: 0.0017 - accuracy: 0.7850 - val_loss: 0.0069 - val_accuracy: 0.6000\nEpoch 480/500\n107/107 [==============================] - 0s 271us/sample - loss: 0.0017 - accuracy: 0.7477 - val_loss: 0.0058 - val_accuracy: 0.6000\nEpoch 481/500\n107/107 [==============================] - 0s 282us/sample - loss: 0.0015 - accuracy: 0.7477 - val_loss: 0.0068 - val_accuracy: 0.5500\nEpoch 482/500\n107/107 [==============================] - 0s 282us/sample - loss: 0.0018 - accuracy: 0.7570 - val_loss: 0.0062 - val_accuracy: 0.6000\nEpoch 483/500\n107/107 [==============================] - 0s 398us/sample - loss: 0.0017 - accuracy: 0.7570 - val_loss: 0.0061 - val_accuracy: 0.6500\nEpoch 484/500\n107/107 [==============================] - 0s 345us/sample - loss: 0.0019 - accuracy: 0.7570 - val_loss: 0.0061 - val_accuracy: 0.6000\nEpoch 485/500\n107/107 [==============================] - 0s 321us/sample - loss: 0.0019 - accuracy: 0.7290 - val_loss: 0.0069 - val_accuracy: 0.6000\nEpoch 486/500\n107/107 [==============================] - 0s 299us/sample - loss: 0.0019 - accuracy: 0.7664 - val_loss: 0.0074 - val_accuracy: 0.6000\nEpoch 487/500\n107/107 [==============================] - 0s 302us/sample - loss: 0.0017 - accuracy: 0.7383 - val_loss: 0.0057 - val_accuracy: 0.6000\nEpoch 488/500\n107/107 [==============================] - 0s 293us/sample - loss: 0.0018 - accuracy: 0.7103 - val_loss: 0.0068 - val_accuracy: 0.6000\nEpoch 489/500\n107/107 [==============================] - 0s 297us/sample - loss: 0.0021 - accuracy: 0.6822 - val_loss: 0.0063 - val_accuracy: 0.5500\nEpoch 490/500\n107/107 [==============================] - 0s 287us/sample - loss: 0.0017 - accuracy: 0.7757 - val_loss: 0.0066 - val_accuracy: 0.6000\nEpoch 491/500\n107/107 [==============================] - 0s 300us/sample - loss: 0.0019 - accuracy: 0.7103 - val_loss: 0.0063 - val_accuracy: 0.5000\nEpoch 492/500\n107/107 [==============================] - 0s 318us/sample - loss: 0.0015 - accuracy: 0.7664 - val_loss: 0.0062 - val_accuracy: 0.6000\nEpoch 493/500\n107/107 [==============================] - 0s 315us/sample - loss: 0.0016 - accuracy: 0.7477 - val_loss: 0.0060 - val_accuracy: 0.5500\nEpoch 494/500\n107/107 [==============================] - 0s 309us/sample - loss: 0.0015 - accuracy: 0.7944 - val_loss: 0.0054 - val_accuracy: 0.6000\nEpoch 495/500\n107/107 [==============================] - 0s 330us/sample - loss: 0.0016 - accuracy: 0.7383 - val_loss: 0.0063 - val_accuracy: 0.6000\nEpoch 496/500\n107/107 [==============================] - 0s 299us/sample - loss: 0.0017 - accuracy: 0.7477 - val_loss: 0.0063 - val_accuracy: 0.6000\nEpoch 497/500\n107/107 [==============================] - 0s 308us/sample - loss: 0.0017 - accuracy: 0.7477 - val_loss: 0.0067 - val_accuracy: 0.6000\nEpoch 498/500\n107/107 [==============================] - 0s 291us/sample - loss: 0.0016 - accuracy: 0.7477 - val_loss: 0.0063 - val_accuracy: 0.6000\nEpoch 499/500\n107/107 [==============================] - 0s 294us/sample - loss: 0.0016 - accuracy: 0.7570 - val_loss: 0.0064 - val_accuracy: 0.5500\nEpoch 500/500\n107/107 [==============================] - 0s 366us/sample - loss: 0.0017 - accuracy: 0.7383 - val_loss: 0.0060 - val_accuracy: 0.6000\n"
],
[
"# Predictions\npred_train = model.predict(standardized_X_train) \npred_test = model.predict(standardized_X_test)\nprint (f\"sample probability: {pred_test[0]}\")\npred_train = np.argmax(pred_train, axis=1)\npred_test = np.argmax(pred_test, axis=1)\nprint (f\"sample class: {pred_test[0]}\")",
"sample probability: [0.00588787 0.7134166 0.28069556]\nsample class: 1\n"
],
[
"# Accuracy\ntrain_acc = accuracy_score(y_train, pred_train)\ntest_acc = accuracy_score(y_test, pred_test)\nprint (f\"train acc: {train_acc:.2f}, test acc: {test_acc:.2f}\")",
"train acc: 0.78, test acc: 0.26\n"
],
[
"# Classification report\nplot_confusion_matrix(y_true=y_test, y_pred=pred_test, classes=classes)\nprint (classification_report(y_test, pred_test))",
"_____no_output_____"
],
[
"# Visualize the decision boundary\nplt.figure(figsize=(12,5))\nplt.subplot(1, 2, 1)\nplt.title(\"Train\")\nplot_multiclass_decision_boundary(model=model, X=standardized_X_train, y=y_train)\nplt.subplot(1, 2, 2)\nplt.title(\"Test\")\nplot_multiclass_decision_boundary(model=model, X=standardized_X_test, y=y_test)\nplt.show()",
"_____no_output_____"
]
],
[
[
"It's important that we experiment, starting with simple models that underfit (high bias) and improve it towards a good fit. Starting with simple models (linear/logistic regression) let's us catch errors without the added complexity of more sophisticated models (neural networks). ",
"_____no_output_____"
],
[
"<img src=\"https://raw.githubusercontent.com/practicalAI/images/master/basic_ml/06_Multilayer_Perceptron/fit.png\" width=\"700\">",
"_____no_output_____"
],
[
"---\n<div align=\"center\">\n\nSubscribe to our <a href=\"https://practicalai.me/#newsletter\">newsletter</a> and follow us on social media to get the latest updates!\n\n<a class=\"ai-header-badge\" target=\"_blank\" href=\"https://github.com/practicalAI/practicalAI\">\n <img src=\"https://img.shields.io/github/stars/practicalAI/practicalAI.svg?style=social&label=Star\"></a> \n <a class=\"ai-header-badge\" target=\"_blank\" href=\"https://www.linkedin.com/company/practicalai-me\">\n <img src=\"https://img.shields.io/badge/style--5eba00.svg?label=LinkedIn&logo=linkedin&style=social\"></a> \n <a class=\"ai-header-badge\" target=\"_blank\" href=\"https://twitter.com/practicalAIme\">\n <img src=\"https://img.shields.io/twitter/follow/practicalAIme.svg?label=Follow&style=social\">\n </a>\n </div>\n\n</div>",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
c512eb2156d66c13ea01df2e6a99b5eef8f24218
| 1,243 |
ipynb
|
Jupyter Notebook
|
analysis/notebooks/Labs/polygon.ipynb
|
sigmantium/LiuAlgoTrader
|
640cc516910ac2eb025275994c7273a33adbd93f
|
[
"MIT"
] | null | null | null |
analysis/notebooks/Labs/polygon.ipynb
|
sigmantium/LiuAlgoTrader
|
640cc516910ac2eb025275994c7273a33adbd93f
|
[
"MIT"
] | null | null | null |
analysis/notebooks/Labs/polygon.ipynb
|
sigmantium/LiuAlgoTrader
|
640cc516910ac2eb025275994c7273a33adbd93f
|
[
"MIT"
] | null | null | null | 19.123077 | 74 | 0.555913 |
[
[
[
"from polygon import STOCKS_CLUSTER, RESTClient, WebSocketClient\nfrom liualgotrader.common import config",
"_____no_output_____"
],
[
"polygon_rest_client = RESTClient(config.polygon_api_key)",
"_____no_output_____"
]
],
[
[
"## snapshots",
"_____no_output_____"
]
],
[
[
"tickets = polygon_rest_client.stocks_equities_snapshot_all_tickers()",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code"
] |
[
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
c512f38e701aa58bcf370171e4452244a2eac3be
| 3,878 |
ipynb
|
Jupyter Notebook
|
EXERCISE - ATTRIBUTES AND METHODS.ipynb
|
Yanga93/JupyterWorkflow
|
24fb864fb8ac9456fad7f7b3d948f47cdf477aa2
|
[
"MIT"
] | null | null | null |
EXERCISE - ATTRIBUTES AND METHODS.ipynb
|
Yanga93/JupyterWorkflow
|
24fb864fb8ac9456fad7f7b3d948f47cdf477aa2
|
[
"MIT"
] | null | null | null |
EXERCISE - ATTRIBUTES AND METHODS.ipynb
|
Yanga93/JupyterWorkflow
|
24fb864fb8ac9456fad7f7b3d948f47cdf477aa2
|
[
"MIT"
] | null | null | null | 28.306569 | 108 | 0.506189 |
[
[
[
"# EXERCISE - ATTRIBUTES AND METHODS\n\n\nWrite an object oriented program to:\n\n1. create a precious stone.\n\n2. Not more than 5 precious stones can be held in possession at a\n given point of time. \n \n3. If there are more than 5 precious stones,\n delete the first stone.\n \n4. Store the new one.",
"_____no_output_____"
]
],
[
[
"class Stone:\n \n # instance attribute\n def __init__(self, name):\n self.name = name\n \n \nclass Container:\n def __init__(self, stone_list):\n self.stone_list = []\n \n #instance method\n def addStone(self):\n max_list = int(input('Maximum number of Stones to be created : '))\n i = 0\n \n while len(self.stone_list) < max_list:\n i += 1\n stone = input('Enter Precious Stone %d: '%i)\n if stone == '':\n print('Please type stone')\n self.stone_list.append(stone)\n print('List before delete the first Precious Stone', self.stone_list)\n return self.stone_list\n \n \n def delFirstStone(self):\n my_list = self.stone_list\n del my_list[0]\n print(\"List after deleted first Precious Stone\", my_list )\n return my_list\n \n def storeNewStone(self):\n my_list = self.stone_list\n stone = input('Store a new Precious Stone: ')\n my_list.append(stone)\n print('List after stored a new Precious Stone', my_list)\n return my_list\n \n \n# instantiate the Stone class\nstone = Stone(\"Gem\")\nstone.name\n\n# instantiate the Container class\nholder = Container(stone)\nholder.addStone()\nholder.delFirstStone()\nholder.storeNewStone()\n",
"Maximum number of Stones to be created : 5\nEnter Precious Stone 1: Diamond\nEnter Precious Stone 2: Emerald\nEnter Precious Stone 3: Garnet\nEnter Precious Stone 4: Paraiba\nEnter Precious Stone 5: Pearls\nList before delete the first Precious Stone ['Diamond', 'Emerald', 'Garnet', 'Paraiba', 'Pearls']\nList after deleted first Precious Stone ['Emerald', 'Garnet', 'Paraiba', 'Pearls']\nStore a new Precious Stone: Rubellite\nList after stored a new Precious Stone ['Emerald', 'Garnet', 'Paraiba', 'Pearls', 'Rubellite']\n"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
]
] |
c512f4fbd45479de3e1d6c42c3de19595b00f3fe
| 8,636 |
ipynb
|
Jupyter Notebook
|
finrl/datasets/datasets/setdata.ipynb
|
mh1208170/FinRL-Library
|
d1d454fbc1c2bce1d8bc6de8992bc9113ab28a5f
|
[
"MIT"
] | null | null | null |
finrl/datasets/datasets/setdata.ipynb
|
mh1208170/FinRL-Library
|
d1d454fbc1c2bce1d8bc6de8992bc9113ab28a5f
|
[
"MIT"
] | null | null | null |
finrl/datasets/datasets/setdata.ipynb
|
mh1208170/FinRL-Library
|
d1d454fbc1c2bce1d8bc6de8992bc9113ab28a5f
|
[
"MIT"
] | null | null | null | 29.37415 | 105 | 0.475683 |
[
[
[
"import pandas as pd\nimport swifter\nimport numpy as np\ndf = pd.read_csv(\"data/gc-1m.csv\", sep=\";\")#,nrows=1000)\ndf.columns=[\"Date\",\"Time\", \"Open\", \"High\", \"Low\", \"Close\", \"Volume\"]",
"_____no_output_____"
],
[
"#Wilder’s Smoothing function\ndef Wilder(data, periods):\n start = np.where(~np.isnan(data))[0][0] #Check if nans present in beginning\n Wilder = np.array([np.nan]*len(data))\n Wilder[start+periods-1] = data[start:(start+periods)].mean() #Simple Moving Average\n for i in range(start+periods,len(data)):\n Wilder[i] = (Wilder[i-1]*(periods-1) + data[i])/periods #Wilder Smoothing\n return(Wilder)",
"_____no_output_____"
],
[
"dt=(df['Date'] + ' ' + df['Time']).swifter.apply(pd.to_datetime)\ndf[\"Time\"]=dt\ndf.drop(columns=[\"Date\",\"Volume\"],inplace=True)\ndf.set_index([\"Time\"],inplace=True)",
"_____no_output_____"
],
[
"#Simple Moving Average (SMA)\ndf['SMA_5'] = df['Close'].transform(lambda x: x.rolling(window = 5).mean())\ndf['SMA_15'] = df['Close'].transform(lambda x: x.rolling(window = 15).mean())\ndf['SMA_ratio'] = df['SMA_15'] / df['SMA_5']",
"_____no_output_____"
],
[
"#Average True Range (ATR)\ndf['prev_close'] = df['Close'].shift(1)\ndf['TR'] = np.maximum((df['High'] - df['Low']),\n np.maximum(abs(df['High'] - df['prev_close']),\n abs(df['prev_close'] - df['Low'])))\n\nTR_data = df.copy()\ndf['ATR_5'] = Wilder(TR_data['TR'], 5)\ndf['ATR_15'] = Wilder(TR_data['TR'], 15)\ndf['ATR_Ratio'] = df['ATR_5'] / df['ATR_15']",
"_____no_output_____"
],
[
"#Average Directional Index (ADX)\ndf['prev_high'] = df['High'].shift(1)\ndf['prev_low'] = df['Low'].shift(1)\n\ndf['+DM'] = np.where(~np.isnan(df.prev_high),\n np.where((df['High'] > df['prev_high']) & \n (((df['High'] - df['prev_high']) > (df['prev_low'] - df['Low']))), \n df['High'] - df['prev_high'], \n 0),np.nan)\n\ndf['-DM'] = np.where(~np.isnan(df.prev_low),\n np.where((df['prev_low'] > df['Low']) & \n (((df['prev_low'] - df['Low']) > (df['High'] - df['prev_high']))), \n df['prev_low'] - df['Low'], \n 0),np.nan)\n\n\nADX_data = df.copy()\ndf['+DM_5'] = Wilder(ADX_data['+DM'], 5)\ndf['-DM_5'] = Wilder(ADX_data['-DM'], 5)\ndf['+DM_15'] = Wilder(ADX_data['+DM'], 15)\ndf['-DM_15'] = Wilder(ADX_data['-DM'], 15)\n\ndf['+DI_5'] = (df['+DM_5']/df['ATR_5'])*100\ndf['-DI_5'] = (df['-DM_5']/df['ATR_5'])*100\ndf['+DI_15'] = (df['+DM_15']/df['ATR_15'])*100\ndf['-DI_15'] = (df['-DM_15']/df['ATR_15'])*100\n\ndf['DX_5'] = (np.round(abs(df['+DI_5'] - df['-DI_5'])/(df['+DI_5'] + df['-DI_5']) * 100))\n\ndf['DX_15'] = (np.round(abs(df['+DI_15'] - df['-DI_15'])/(df['+DI_15'] + df['-DI_15']) * 100))\n\n\nADX_data = df.copy()\ndf['ADX_5'] = Wilder(ADX_data['DX_5'], 5)\ndf['ADX_15'] = Wilder(ADX_data['DX_15'], 15)",
"_____no_output_____"
],
[
"#Stochastic Oscillators\ndf['Lowest_5D'] = df['Low'].transform(lambda x: x.rolling(window = 5).min())\ndf['High_5D'] = df['High'].transform(lambda x: x.rolling(window = 5).max())\ndf['Lowest_15D'] = df['Low'].transform(lambda x: x.rolling(window = 15).min())\ndf['High_15D'] = df['High'].transform(lambda x: x.rolling(window = 15).max())\n\ndf['Stochastic_5'] = ((df['Close'] - df['Lowest_5D'])/(df['High_5D'] - df['Lowest_5D']))*100\ndf['Stochastic_15'] = ((df['Close'] - df['Lowest_15D'])/(df['High_15D'] - df['Lowest_15D']))*100\n\ndf['Stochastic_%D_5'] = df['Stochastic_5'].rolling(window = 5).mean()\ndf['Stochastic_%D_15'] = df['Stochastic_5'].rolling(window = 15).mean()\n\ndf['Stochastic_Ratio'] = df['Stochastic_%D_5']/df['Stochastic_%D_15']",
"_____no_output_____"
],
[
"#Relative Strength Index (RSI)\ndf['Diff'] = df['Close'].transform(lambda x: x.diff())\ndf['Up'] = df['Diff']\ndf.loc[(df['Up']<0), 'Up'] = 0\n\ndf['Down'] = df['Diff']\ndf.loc[(df['Down']>0), 'Down'] = 0 \ndf['Down'] = abs(df['Down'])\n\ndf['avg_5up'] = df['Up'].transform(lambda x: x.rolling(window=5).mean())\ndf['avg_5down'] = df['Down'].transform(lambda x: x.rolling(window=5).mean())\n\ndf['avg_15up'] = df['Up'].transform(lambda x: x.rolling(window=15).mean())\ndf['avg_15down'] = df['Down'].transform(lambda x: x.rolling(window=15).mean())\n\ndf['RS_5'] = df['avg_5up'] / df['avg_5down']\ndf['RS_15'] = df['avg_15up'] / df['avg_15down']\n\ndf['RSI_5'] = 100 - (100/(1+df['RS_5']))\ndf['RSI_15'] = 100 - (100/(1+df['RS_15']))\n\ndf['RSI_ratio'] = df['RSI_5']/df['RSI_15']",
"_____no_output_____"
],
[
"#Moving Average Convergence Divergence (MACD)\ndf['12Ewm'] = df['Close'].transform(lambda x: x.ewm(span=12, adjust=False).mean())\ndf['26Ewm'] = df['Close'].transform(lambda x: x.ewm(span=26, adjust=False).mean())\ndf['MACD'] = df['26Ewm'] - df['12Ewm']",
"_____no_output_____"
],
[
"#Bollinger Bands\ndf['15MA'] = df['Close'].transform(lambda x: x.rolling(window=15).mean())\ndf['SD'] = df['Close'].transform(lambda x: x.rolling(window=15).std())\ndf['upperband'] = df['15MA'] + 2*df['SD']\ndf['lowerband'] = df['15MA'] - 2*df['SD']",
"_____no_output_____"
],
[
"#Rate of Change\ndf['RC'] = df['Close'].transform(lambda x: x.pct_change(periods = 15))",
"_____no_output_____"
],
[
"df.interpolate()\ndf.dropna()\ndf.to_csv(\"data/gc-1m_all.csv\")",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
c512fc526c18272979937f7386abeef9ecb30a33
| 6,864 |
ipynb
|
Jupyter Notebook
|
dev/pytorch_dataset_loader/comparing_multi_thread.ipynb
|
nicolaseberle/flyTracker
|
387b64a7c92d7ecf503dfccde29d7db9efb07cfb
|
[
"MIT"
] | 2 |
2018-02-12T17:14:50.000Z
|
2020-05-26T16:58:19.000Z
|
dev/pytorch_dataset_loader/comparing_multi_thread.ipynb
|
nicolaseberle/flyTracker
|
387b64a7c92d7ecf503dfccde29d7db9efb07cfb
|
[
"MIT"
] | 12 |
2020-05-26T16:41:22.000Z
|
2021-03-15T09:09:12.000Z
|
dev/pytorch_dataset_loader/comparing_multi_thread.ipynb
|
nicolaseberle/flyTracker
|
387b64a7c92d7ecf503dfccde29d7db9efb07cfb
|
[
"MIT"
] | 1 |
2020-05-20T16:19:27.000Z
|
2020-05-20T16:19:27.000Z
| 27.902439 | 80 | 0.529138 |
[
[
[
"from flytracker.videoreader import VideoReader\nfrom flytracker.preprocessing import preprocessing\nimport cv2 as cv \nimport torch\nfrom flytracker.utils import FourArenasQRCodeMask\nfrom torch.utils.data import DataLoader\nimport numpy as np\n\n%load_ext autoreload\n%autoreload 2",
"_____no_output_____"
],
[
"torch.cuda.is_available()",
"_____no_output_____"
],
[
"class SingleThreadVideoDataset(torch.utils.data.IterableDataset):\n def __init__(self, path, mask):\n super().__init__()\n self.capture = cv.VideoCapture(path)\n self.mask = torch.tensor(mask, dtype=torch.bool)\n\n def __iter__(self):\n return self\n\n def __next__(self) -> torch.Tensor:\n succes, image = self.capture.read()\n if succes is False:\n raise StopIteration\n # because we use opencv\n image = np.moveaxis(image, -1, 0)\n processed_image = preprocessing(image, self.mask)\n return processed_image",
"_____no_output_____"
],
[
"class MultiThreadVideoDataset(torch.utils.data.IterableDataset):\n def __init__(self, path, mask):\n super().__init__()\n self.reader = VideoReader(path, max_queue=100)\n self.mask = torch.tensor(mask, dtype=torch.bool)\n\n def __iter__(self):\n return self\n\n def __next__(self) -> torch.Tensor:\n succes, image = self.reader.read()\n if succes is False:\n raise StopIteration\n # because we use opencv\n image = np.moveaxis(image, -1, 0)\n processed_image = preprocessing(image, self.mask)\n return processed_image",
"_____no_output_____"
],
[
"mask = FourArenasQRCodeMask().mask\npath = \"/home/gert-jan/Documents/flyTracker/data/movies/4arenas_QR.h264\"",
"_____no_output_____"
],
[
"%%time\ndataset = SingleThreadVideoDataset(path, mask)\nloader = DataLoader(dataset, batch_size=1, pin_memory=True)\nfor batch_idx, batch in enumerate(loader):\n batch = batch.cuda(non_blocking=True)\n if batch_idx % 100 == 0:\n print(f\"Loaded {batch_idx}, {batch.device}\")\n print(torch.mean(batch.type(torch.float32)))\n if batch_idx == 1000:\n break",
"Loaded 0, cuda:0\ntensor(235.7776, device='cuda:0')\nLoaded 100, cuda:0\ntensor(235.8470, device='cuda:0')\nLoaded 200, cuda:0\ntensor(235.7998, device='cuda:0')\nLoaded 300, cuda:0\ntensor(235.8143, device='cuda:0')\nLoaded 400, cuda:0\ntensor(235.8383, device='cuda:0')\nLoaded 500, cuda:0\ntensor(235.8459, device='cuda:0')\nLoaded 600, cuda:0\ntensor(235.7329, device='cuda:0')\nLoaded 700, cuda:0\ntensor(235.8555, device='cuda:0')\nLoaded 800, cuda:0\ntensor(235.8855, device='cuda:0')\nLoaded 900, cuda:0\ntensor(235.7736, device='cuda:0')\nLoaded 1000, cuda:0\ntensor(235.8390, device='cuda:0')\nCPU times: user 57.2 s, sys: 880 ms, total: 58.1 s\nWall time: 3.41 s\n"
],
[
"%%time\ndataset = MultiThreadVideoDataset(path, mask)\nloader = DataLoader(dataset, batch_size=1, pin_memory=True)\nfor batch_idx, batch in enumerate(loader):\n batch = batch.cuda(non_blocking=True)\n if batch_idx % 100 == 0:\n print(f\"Loaded {batch_idx}, {batch.device}\")\n print(torch.mean(batch.type(torch.float32)))\n if batch_idx == 1000:\n break\nloader.dataset.reader.stop()",
"Loaded 0, cuda:0\ntensor(235.7776, device='cuda:0')\nLoaded 100, cuda:0\ntensor(235.8470, device='cuda:0')\nLoaded 200, cuda:0\ntensor(235.7998, device='cuda:0')\nLoaded 300, cuda:0\ntensor(235.8143, device='cuda:0')\nLoaded 400, cuda:0\ntensor(235.8383, device='cuda:0')\nLoaded 500, cuda:0\ntensor(235.8459, device='cuda:0')\nLoaded 600, cuda:0\ntensor(235.7329, device='cuda:0')\nLoaded 700, cuda:0\ntensor(235.8555, device='cuda:0')\nLoaded 800, cuda:0\ntensor(235.8855, device='cuda:0')\nLoaded 900, cuda:0\ntensor(235.7736, device='cuda:0')\nLoaded 1000, cuda:0\ntensor(235.8390, device='cuda:0')\nCPU times: user 41 s, sys: 523 ms, total: 41.5 s\nWall time: 2.22 s\n"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
c51308a7bffa4e7a5e7c7af619ff90beb9a4ba14
| 846,080 |
ipynb
|
Jupyter Notebook
|
06/Copy_of_CS480_Assignment_6.ipynb
|
aayush9628/cs480student
|
8f5b295a2d34f0e1216026f7d3b2bcfb396246de
|
[
"MIT"
] | null | null | null |
06/Copy_of_CS480_Assignment_6.ipynb
|
aayush9628/cs480student
|
8f5b295a2d34f0e1216026f7d3b2bcfb396246de
|
[
"MIT"
] | null | null | null |
06/Copy_of_CS480_Assignment_6.ipynb
|
aayush9628/cs480student
|
8f5b295a2d34f0e1216026f7d3b2bcfb396246de
|
[
"MIT"
] | null | null | null | 744.133685 | 730,973 | 0.935836 |
[
[
[
"<a href=\"https://colab.research.google.com/github/aayush9628/cs480student/blob/main/06/Copy_of_CS480_Assignment_6.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"\n#Assignment 6",
"_____no_output_____"
]
],
[
[
"# In this assignment, we will train a U-Net classifer to detect mitochondria\n# in electron microscopy images!",
"_____no_output_____"
],
[
"### IMPORTANT ###\n#\n# Activate GPU support: Runtime -> Change Runtime Type\n# Hardware Accelerator: GPU\n#\n### IMPORTANT ###",
"_____no_output_____"
],
[
"# load numpy and matplotlib\n%pylab inline",
"Populating the interactive namespace from numpy and matplotlib\n"
],
[
"# load the unet tensorflow code\n!pip install git+https://github.com/jakeret/unet.git",
"Collecting git+https://github.com/jakeret/unet.git\n Cloning https://github.com/jakeret/unet.git to /tmp/pip-req-build-n_n78ang\n Running command git clone -q https://github.com/jakeret/unet.git /tmp/pip-req-build-n_n78ang\nBuilding wheels for collected packages: unet\n Building wheel for unet (setup.py) ... \u001b[?25l\u001b[?25hdone\n Created wheel for unet: filename=unet-0.0.post0.dev81+gf557a51-py2.py3-none-any.whl size=25688 sha256=164d08f0231f8bee2643a8e925620f93b3b79c5a6d8b1a811306724a968118c0\n Stored in directory: /tmp/pip-ephem-wheel-cache-vv7l33y1/wheels/22/00/93/ce57529ed355f160088cbb6ce086e55168770913b40cf624ba\nSuccessfully built unet\nInstalling collected packages: unet\nSuccessfully installed unet-0.0.post0.dev81+gf557a51\n"
],
[
"# .. and use it!\n# Note: There are a ton of U-Net implementations but this one is easier to use!\nimport unet",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
],
[
[
"**Task 1**: Study the unet circle example and learn about mitochondria! [15 Points]",
"_____no_output_____"
]
],
[
[
"# The unet package allows to train a U-Net classifier with little code.\n# As discussed in class, the U-Net can be used to label/segment/annotate images.",
"_____no_output_____"
],
[
"# TODO: Please study the Circles example \n# https://github.com/jakeret/unet/blob/master/notebooks/circles.ipynb",
"_____no_output_____"
],
[
"# TODO: ... and look how the (artificial) dataset is generated\n# https://github.com/jakeret/unet/blob/master/src/unet/datasets/circles.py",
"_____no_output_____"
],
[
"# 1) Question\n# TODO: Please describe what the U-Net is trained to do.\n#\n# TODO: YOUR ANSWER",
"_____no_output_____"
]
],
[
[
"U-Net is trained to predict the mask which will be used to segment the mitochondria in the input image.",
"_____no_output_____"
]
],
[
[
"# 2) Question\n# TODO: In circles.py, what does the following mean:\n# channels = 1\n# classes = 2\n#\n# TODO: YOUR ANSWER",
"_____no_output_____"
]
],
[
[
"In CNNs, channels are essentially concerned with capturing different features on the basis of which, distinction will be made and learned by the network between inputs. If the image is an RGB image, then the number of channels will be 3, one for each color. The variable \"channels\" has the value of 1 as grayscaled images are used. We have two classes one for foreground and one for background, so that's why the value of the \"classes\" variable is 2.",
"_____no_output_____"
]
],
[
[
"# 3) Question\n# TODO: What are mitochondria and what is their appearance in\n# electron microscopy data?\n# Hint: You can validate your answer in Task 4!\n#\n# TODO: YOUR ANSWER",
"_____no_output_____"
]
],
[
[
"Mitochondria is a cell organelle which is also known as the power house of the cell, is a bean shaped component of the cell. ",
"_____no_output_____"
],
[
"**Task 2**: Setup a datagenerator for mitochondria images! [45 Points]",
"_____no_output_____"
]
],
[
[
"# TODO: \n# Download https://cs480.org/data/mito.npz to your computer.\n# Then, please upload mito.npz using the file panel on the left.",
"_____no_output_____"
],
[
"# The code below is similar to the circles.py file from Task 1.\n# We follow Tensorflow conventions to design a tf.data.Dataset for training\n# the U-Net.\n#\n# TODO: Please add four different data augmentation methods in the code block\n# below. (image normalization to 0..1, horizontal data flip, vertical data flip,\n# rotation by 90 degrees)\n#\n# Hint: https://github.com/jakeret/unet/blob/master/src/unet/datasets/oxford_iiit_pet.py#L25",
"_____no_output_____"
],
[
"import tensorflow as tf\nimport tensorflow_datasets as tfds\nfrom tensorflow_datasets.core import DatasetInfo\nfrom typing import Tuple, List\nimport numpy as np\n\nIMAGE_SIZE = (256, 256)\nchannels = 1\nclasses = 2\n\ndef load_data(count:int, splits:Tuple[float]=(0.7, 0.2, 0.1), **kwargs) -> List[tf.data.Dataset]:\n return [tf.data.Dataset.from_tensor_slices(_build_samples(int(split * count), **kwargs))\n for split in splits]\n\n# loaded = np.load('mito.npz')\n# loadedimages = loaded['arr_0'][0].copy()\n# loadedmasks = loaded['arr_0'][1].copy()\n# print(loadedimages)\n# print(loadedmasks)\ndef _build_samples(sample_count:int, **kwargs) -> Tuple[np.array, np.array]:\n\n # here we load the mitochondria data\n loaded = np.load('mito.npz')\n loadedimages = loaded['arr_0'][0].copy()\n loadedmasks = loaded['arr_0'][1].copy()\n\n # now let's go to numpyland\n images = np.empty((sample_count, IMAGE_SIZE[0], IMAGE_SIZE[1], 1))\n labels = np.empty((sample_count, IMAGE_SIZE[0], IMAGE_SIZE[1], 2))\n for i in range(sample_count):\n image, mask = loadedimages[i], loadedmasks[i]\n\n image = image.reshape((IMAGE_SIZE[0], IMAGE_SIZE[1], 1)).astype(np.float)\n mask = mask.reshape((IMAGE_SIZE[0], IMAGE_SIZE[1], 1))\n\n #\n # TODO: Normalize the image to 0..1\n #\n # TODO: YOUR CODE\n image = tf.cast(image, tf.float32)/255.0\n #\n # TODO: Use Tensorflow to flip the image horizontally\n #\n if tf.random.uniform(()) > 0.5:\n #\n # TODO: YOUR CODE\n #\n image = tf.image.flip_left_right(image)\n mask = tf.image.flip_left_right(mask)\n #\n # TODO: Use Tensorflow to flip the image vertically\n #\n if tf.random.uniform(()) > 0.5:\n #\n # TODO: YOUR CODE\n #\n image = tf.image.flip_up_down(image)\n mask = tf.image.flip_up_down(mask)\n #\n # TODO: Use Tensorflow to rotate the image 90 degrees\n #\n if tf.random.uniform(()) > 0.5:\n #\n # TODO: YOUR CODE\n #\n image = tf.image.rot90(image)\n mask = tf.image.rot90(mask)\n\n # augmentation done, let's store the image\n images[i] = image\n\n # here we split the mask to background and foreground\n fg = np.zeros((IMAGE_SIZE[0], IMAGE_SIZE[1], 1), dtype=np.bool)\n fg[mask == 255] = 1\n bg = np.zeros((IMAGE_SIZE[0], IMAGE_SIZE[1], 1), dtype=np.bool)\n bg[mask == 0] = 1\n \n labels[i, :, :, 0] = bg[:,:,0]\n labels[i, :, :, 1] = fg[:,:,0]\n\n return images, labels",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
],
[
[
"**Task 3**: Let's train the U-Net! [25 Points]",
"_____no_output_____"
]
],
[
[
"#\n# We can now create our training, validation, and testing data by calling\n# our methods from Task 2.\n#\ntrain, val, test = load_data( 660, splits=(0.7, 0.2, 0.1) )",
"/usr/local/lib/python3.7/dist-packages/ipykernel_launcher.py:33: DeprecationWarning: `np.float` is a deprecated alias for the builtin `float`. To silence this warning, use `float` by itself. Doing this will not modify any behavior and is safe. If you specifically wanted the numpy scalar type, use `np.float64` here.\nDeprecated in NumPy 1.20; for more details and guidance: https://numpy.org/devdocs/release/1.20.0-notes.html#deprecations\n/usr/local/lib/python3.7/dist-packages/ipykernel_launcher.py:73: DeprecationWarning: `np.bool` is a deprecated alias for the builtin `bool`. To silence this warning, use `bool` by itself. Doing this will not modify any behavior and is safe. If you specifically wanted the numpy scalar type, use `np.bool_` here.\nDeprecated in NumPy 1.20; for more details and guidance: https://numpy.org/devdocs/release/1.20.0-notes.html#deprecations\n/usr/local/lib/python3.7/dist-packages/ipykernel_launcher.py:75: DeprecationWarning: `np.bool` is a deprecated alias for the builtin `bool`. To silence this warning, use `bool` by itself. Doing this will not modify any behavior and is safe. If you specifically wanted the numpy scalar type, use `np.bool_` here.\nDeprecated in NumPy 1.20; for more details and guidance: https://numpy.org/devdocs/release/1.20.0-notes.html#deprecations\n"
],
[
"# 1) Question\n# TODO: We have in total 660 images + 660 masks. Based on the code above,\n# how many images are used for training alone?\n#\n# TODO: YOUR ANSWER\n",
"_____no_output_____"
]
],
[
[
"There are around 462 images used for training.",
"_____no_output_____"
]
],
[
[
"#\n# Let's setup the U-Net!\n#\nLEARNING_RATE = 1e-3\n\nunet_model = unet.build_model(channels=channels,\n num_classes=classes,\n layer_depth=5,\n filters_root=64,\n padding=\"same\")\n\nunet.finalize_model(unet_model, learning_rate=LEARNING_RATE)",
"_____no_output_____"
],
[
"#\n# And, let's setup the trainer...\n#\ntrainer = unet.Trainer(checkpoint_callback=False,\n learning_rate_scheduler=unet.SchedulerType.WARMUP_LINEAR_DECAY,\n warmup_proportion=0.1,\n learning_rate=LEARNING_RATE)",
"_____no_output_____"
],
[
"#\n# ...and train the U-Net for 50 epochs with a batch_size of 10!\n#\n# TODO: Please complete the code below.\n# Hint: Don't forget to use training and validation data.\n# Hint 2: This will take roughly 30 minutes!\n#\ntrainer.fit(unet_model,\n train,\n val,\n epochs=50,\n batch_size=10\n )",
"Epoch 1/50\n47/47 [==============================] - 88s 2s/step - loss: 0.6020 - categorical_crossentropy: 0.6020 - categorical_accuracy: 0.7650 - mean_iou: 0.3192 - dice_coefficient: 0.5806 - auc: 0.7178 - learning_rate: 9.9911e-05 - val_loss: 0.2631 - val_categorical_crossentropy: 0.2631 - val_categorical_accuracy: 0.9434 - val_mean_iou: 0.4242 - val_dice_coefficient: 0.7955 - val_auc: 0.9847\nEpoch 2/50\n47/47 [==============================] - 66s 1s/step - loss: 0.3270 - categorical_crossentropy: 0.3270 - categorical_accuracy: 0.9115 - mean_iou: 0.4333 - dice_coefficient: 0.8091 - auc: 0.9307 - learning_rate: 2.9991e-04 - val_loss: 0.2913 - val_categorical_crossentropy: 0.2913 - val_categorical_accuracy: 0.9434 - val_mean_iou: 0.4109 - val_dice_coefficient: 0.7667 - val_auc: 0.9834\nEpoch 3/50\n47/47 [==============================] - 66s 1s/step - loss: 0.3153 - categorical_crossentropy: 0.3153 - categorical_accuracy: 0.9115 - mean_iou: 0.4421 - dice_coefficient: 0.8291 - auc: 0.9300 - learning_rate: 4.9991e-04 - val_loss: 0.2027 - val_categorical_crossentropy: 0.2027 - val_categorical_accuracy: 0.9434 - val_mean_iou: 0.4588 - val_dice_coefficient: 0.8638 - val_auc: 0.9832\nEpoch 4/50\n47/47 [==============================] - 66s 1s/step - loss: 0.3034 - categorical_crossentropy: 0.3034 - categorical_accuracy: 0.9115 - mean_iou: 0.4447 - dice_coefficient: 0.8312 - auc: 0.9322 - learning_rate: 6.9991e-04 - val_loss: 0.2251 - val_categorical_crossentropy: 0.2251 - val_categorical_accuracy: 0.9434 - val_mean_iou: 0.4452 - val_dice_coefficient: 0.8315 - val_auc: 0.9840\nEpoch 5/50\n47/47 [==============================] - 66s 1s/step - loss: 0.3091 - categorical_crossentropy: 0.3091 - categorical_accuracy: 0.9115 - mean_iou: 0.4434 - dice_coefficient: 0.8280 - auc: 0.9342 - learning_rate: 8.9991e-04 - val_loss: 0.2326 - val_categorical_crossentropy: 0.2326 - val_categorical_accuracy: 0.9434 - val_mean_iou: 0.4414 - val_dice_coefficient: 0.8220 - val_auc: 0.9839\nEpoch 6/50\n47/47 [==============================] - 66s 1s/step - loss: 0.3088 - categorical_crossentropy: 0.3088 - categorical_accuracy: 0.9115 - mean_iou: 0.4445 - dice_coefficient: 0.8302 - auc: 0.9349 - learning_rate: 9.8890e-04 - val_loss: 0.2480 - val_categorical_crossentropy: 0.2480 - val_categorical_accuracy: 0.9434 - val_mean_iou: 0.4337 - val_dice_coefficient: 0.8051 - val_auc: 0.9841\nEpoch 7/50\n47/47 [==============================] - 66s 1s/step - loss: 0.2974 - categorical_crossentropy: 0.2974 - categorical_accuracy: 0.9115 - mean_iou: 0.4487 - dice_coefficient: 0.8352 - auc: 0.9380 - learning_rate: 9.6668e-04 - val_loss: 0.2053 - val_categorical_crossentropy: 0.2053 - val_categorical_accuracy: 0.9434 - val_mean_iou: 0.4579 - val_dice_coefficient: 0.8496 - val_auc: 0.9846\nEpoch 8/50\n47/47 [==============================] - 66s 1s/step - loss: 0.2774 - categorical_crossentropy: 0.2774 - categorical_accuracy: 0.9115 - mean_iou: 0.4558 - dice_coefficient: 0.8415 - auc: 0.9465 - learning_rate: 9.4445e-04 - val_loss: 0.2041 - val_categorical_crossentropy: 0.2041 - val_categorical_accuracy: 0.9434 - val_mean_iou: 0.4603 - val_dice_coefficient: 0.8466 - val_auc: 0.9854\nEpoch 9/50\n47/47 [==============================] - 66s 1s/step - loss: 0.3281 - categorical_crossentropy: 0.3281 - categorical_accuracy: 0.9115 - mean_iou: 0.4484 - dice_coefficient: 0.8277 - auc: 0.9409 - learning_rate: 9.2223e-04 - val_loss: 0.1994 - val_categorical_crossentropy: 0.1994 - val_categorical_accuracy: 0.9434 - val_mean_iou: 0.4627 - val_dice_coefficient: 0.8531 - val_auc: 0.9851\nEpoch 10/50\n47/47 [==============================] - 65s 1s/step - loss: 0.2696 - categorical_crossentropy: 0.2696 - categorical_accuracy: 0.9115 - mean_iou: 0.4606 - dice_coefficient: 0.8445 - auc: 0.9503 - learning_rate: 9.0001e-04 - val_loss: 0.1806 - val_categorical_crossentropy: 0.1806 - val_categorical_accuracy: 0.9434 - val_mean_iou: 0.4771 - val_dice_coefficient: 0.8712 - val_auc: 0.9853\nEpoch 11/50\n47/47 [==============================] - 64s 1s/step - loss: 0.2592 - categorical_crossentropy: 0.2592 - categorical_accuracy: 0.9115 - mean_iou: 0.4677 - dice_coefficient: 0.8482 - auc: 0.9558 - learning_rate: 8.7779e-04 - val_loss: 0.1932 - val_categorical_crossentropy: 0.1932 - val_categorical_accuracy: 0.9434 - val_mean_iou: 0.4736 - val_dice_coefficient: 0.8527 - val_auc: 0.9856\nEpoch 12/50\n47/47 [==============================] - 64s 1s/step - loss: 0.2554 - categorical_crossentropy: 0.2554 - categorical_accuracy: 0.9115 - mean_iou: 0.4716 - dice_coefficient: 0.8500 - auc: 0.9580 - learning_rate: 8.5557e-04 - val_loss: 0.1915 - val_categorical_crossentropy: 0.1915 - val_categorical_accuracy: 0.9434 - val_mean_iou: 0.4772 - val_dice_coefficient: 0.8535 - val_auc: 0.9857\nEpoch 13/50\n47/47 [==============================] - 64s 1s/step - loss: 0.2505 - categorical_crossentropy: 0.2505 - categorical_accuracy: 0.9115 - mean_iou: 0.4755 - dice_coefficient: 0.8520 - auc: 0.9601 - learning_rate: 8.3334e-04 - val_loss: 0.1824 - val_categorical_crossentropy: 0.1824 - val_categorical_accuracy: 0.9434 - val_mean_iou: 0.4850 - val_dice_coefficient: 0.8621 - val_auc: 0.9859\nEpoch 14/50\n47/47 [==============================] - 64s 1s/step - loss: 0.2408 - categorical_crossentropy: 0.2408 - categorical_accuracy: 0.9115 - mean_iou: 0.4816 - dice_coefficient: 0.8554 - auc: 0.9639 - learning_rate: 8.1112e-04 - val_loss: 0.1885 - val_categorical_crossentropy: 0.1885 - val_categorical_accuracy: 0.9434 - val_mean_iou: 0.4847 - val_dice_coefficient: 0.8554 - val_auc: 0.9862\nEpoch 15/50\n47/47 [==============================] - 65s 1s/step - loss: 0.2401 - categorical_crossentropy: 0.2401 - categorical_accuracy: 0.9115 - mean_iou: 0.4837 - dice_coefficient: 0.8564 - auc: 0.9642 - learning_rate: 7.8890e-04 - val_loss: 0.1733 - val_categorical_crossentropy: 0.1733 - val_categorical_accuracy: 0.9434 - val_mean_iou: 0.4946 - val_dice_coefficient: 0.8705 - val_auc: 0.9863\nEpoch 16/50\n47/47 [==============================] - 65s 1s/step - loss: 0.2308 - categorical_crossentropy: 0.2308 - categorical_accuracy: 0.9115 - mean_iou: 0.4898 - dice_coefficient: 0.8591 - auc: 0.9672 - learning_rate: 7.6668e-04 - val_loss: 0.1753 - val_categorical_crossentropy: 0.1753 - val_categorical_accuracy: 0.9434 - val_mean_iou: 0.4958 - val_dice_coefficient: 0.8680 - val_auc: 0.9865\nEpoch 17/50\n47/47 [==============================] - 65s 1s/step - loss: 0.2277 - categorical_crossentropy: 0.2277 - categorical_accuracy: 0.9115 - mean_iou: 0.4929 - dice_coefficient: 0.8608 - auc: 0.9683 - learning_rate: 7.4445e-04 - val_loss: 0.1703 - val_categorical_crossentropy: 0.1703 - val_categorical_accuracy: 0.9434 - val_mean_iou: 0.5003 - val_dice_coefficient: 0.8730 - val_auc: 0.9866\nEpoch 18/50\n47/47 [==============================] - 65s 1s/step - loss: 0.2229 - categorical_crossentropy: 0.2229 - categorical_accuracy: 0.9115 - mean_iou: 0.4968 - dice_coefficient: 0.8628 - auc: 0.9697 - learning_rate: 7.2223e-04 - val_loss: 0.1688 - val_categorical_crossentropy: 0.1688 - val_categorical_accuracy: 0.9434 - val_mean_iou: 0.5024 - val_dice_coefficient: 0.8743 - val_auc: 0.9868\nEpoch 19/50\n47/47 [==============================] - 65s 1s/step - loss: 0.2196 - categorical_crossentropy: 0.2196 - categorical_accuracy: 0.9115 - mean_iou: 0.4996 - dice_coefficient: 0.8642 - auc: 0.9707 - learning_rate: 7.0001e-04 - val_loss: 0.1653 - val_categorical_crossentropy: 0.1653 - val_categorical_accuracy: 0.9434 - val_mean_iou: 0.5051 - val_dice_coefficient: 0.8779 - val_auc: 0.9870\nEpoch 20/50\n47/47 [==============================] - 65s 1s/step - loss: 0.2161 - categorical_crossentropy: 0.2161 - categorical_accuracy: 0.9115 - mean_iou: 0.5020 - dice_coefficient: 0.8661 - auc: 0.9717 - learning_rate: 6.7779e-04 - val_loss: 0.1623 - val_categorical_crossentropy: 0.1623 - val_categorical_accuracy: 0.9434 - val_mean_iou: 0.5081 - val_dice_coefficient: 0.8811 - val_auc: 0.9870\nEpoch 21/50\n47/47 [==============================] - 64s 1s/step - loss: 0.2132 - categorical_crossentropy: 0.2132 - categorical_accuracy: 0.9115 - mean_iou: 0.5052 - dice_coefficient: 0.8674 - auc: 0.9725 - learning_rate: 6.5557e-04 - val_loss: 0.1600 - val_categorical_crossentropy: 0.1600 - val_categorical_accuracy: 0.9434 - val_mean_iou: 0.5102 - val_dice_coefficient: 0.8835 - val_auc: 0.9872\nEpoch 22/50\n47/47 [==============================] - 65s 1s/step - loss: 0.2105 - categorical_crossentropy: 0.2105 - categorical_accuracy: 0.9115 - mean_iou: 0.5073 - dice_coefficient: 0.8689 - auc: 0.9733 - learning_rate: 6.3334e-04 - val_loss: 0.1580 - val_categorical_crossentropy: 0.1580 - val_categorical_accuracy: 0.9434 - val_mean_iou: 0.5119 - val_dice_coefficient: 0.8856 - val_auc: 0.9874\nEpoch 23/50\n47/47 [==============================] - 65s 1s/step - loss: 0.2083 - categorical_crossentropy: 0.2083 - categorical_accuracy: 0.9115 - mean_iou: 0.5089 - dice_coefficient: 0.8703 - auc: 0.9739 - learning_rate: 6.1112e-04 - val_loss: 0.1563 - val_categorical_crossentropy: 0.1563 - val_categorical_accuracy: 0.9434 - val_mean_iou: 0.5136 - val_dice_coefficient: 0.8875 - val_auc: 0.9875\nEpoch 24/50\n47/47 [==============================] - 65s 1s/step - loss: 0.2062 - categorical_crossentropy: 0.2062 - categorical_accuracy: 0.9115 - mean_iou: 0.5109 - dice_coefficient: 0.8714 - auc: 0.9744 - learning_rate: 5.8890e-04 - val_loss: 0.1543 - val_categorical_crossentropy: 0.1543 - val_categorical_accuracy: 0.9434 - val_mean_iou: 0.5156 - val_dice_coefficient: 0.8899 - val_auc: 0.9876\nEpoch 25/50\n47/47 [==============================] - 65s 1s/step - loss: 0.2042 - categorical_crossentropy: 0.2042 - categorical_accuracy: 0.9115 - mean_iou: 0.5127 - dice_coefficient: 0.8727 - auc: 0.9750 - learning_rate: 5.6668e-04 - val_loss: 0.1525 - val_categorical_crossentropy: 0.1525 - val_categorical_accuracy: 0.9434 - val_mean_iou: 0.5173 - val_dice_coefficient: 0.8918 - val_auc: 0.9877\nEpoch 26/50\n47/47 [==============================] - 70s 1s/step - loss: 0.2022 - categorical_crossentropy: 0.2022 - categorical_accuracy: 0.9115 - mean_iou: 0.5144 - dice_coefficient: 0.8740 - auc: 0.9755 - learning_rate: 5.4445e-04 - val_loss: 0.1511 - val_categorical_crossentropy: 0.1511 - val_categorical_accuracy: 0.9434 - val_mean_iou: 0.5188 - val_dice_coefficient: 0.8933 - val_auc: 0.9878\nEpoch 27/50\n47/47 [==============================] - 65s 1s/step - loss: 0.2005 - categorical_crossentropy: 0.2005 - categorical_accuracy: 0.9115 - mean_iou: 0.5162 - dice_coefficient: 0.8752 - auc: 0.9759 - learning_rate: 5.2223e-04 - val_loss: 0.1494 - val_categorical_crossentropy: 0.1494 - val_categorical_accuracy: 0.9434 - val_mean_iou: 0.5206 - val_dice_coefficient: 0.8952 - val_auc: 0.9880\nEpoch 28/50\n47/47 [==============================] - 65s 1s/step - loss: 0.1987 - categorical_crossentropy: 0.1987 - categorical_accuracy: 0.9115 - mean_iou: 0.5178 - dice_coefficient: 0.8762 - auc: 0.9763 - learning_rate: 5.0001e-04 - val_loss: 0.1478 - val_categorical_crossentropy: 0.1478 - val_categorical_accuracy: 0.9434 - val_mean_iou: 0.5224 - val_dice_coefficient: 0.8970 - val_auc: 0.9880\nEpoch 29/50\n47/47 [==============================] - 65s 1s/step - loss: 0.1969 - categorical_crossentropy: 0.1969 - categorical_accuracy: 0.9115 - mean_iou: 0.5196 - dice_coefficient: 0.8774 - auc: 0.9767 - learning_rate: 4.7779e-04 - val_loss: 0.1461 - val_categorical_crossentropy: 0.1461 - val_categorical_accuracy: 0.9434 - val_mean_iou: 0.5244 - val_dice_coefficient: 0.8990 - val_auc: 0.9881\nEpoch 30/50\n47/47 [==============================] - 65s 1s/step - loss: 0.1953 - categorical_crossentropy: 0.1953 - categorical_accuracy: 0.9115 - mean_iou: 0.5214 - dice_coefficient: 0.8785 - auc: 0.9771 - learning_rate: 4.5557e-04 - val_loss: 0.1449 - val_categorical_crossentropy: 0.1449 - val_categorical_accuracy: 0.9434 - val_mean_iou: 0.5260 - val_dice_coefficient: 0.9004 - val_auc: 0.9882\nEpoch 31/50\n47/47 [==============================] - 65s 1s/step - loss: 0.1940 - categorical_crossentropy: 0.1940 - categorical_accuracy: 0.9115 - mean_iou: 0.5230 - dice_coefficient: 0.8793 - auc: 0.9774 - learning_rate: 4.3334e-04 - val_loss: 0.1437 - val_categorical_crossentropy: 0.1437 - val_categorical_accuracy: 0.9434 - val_mean_iou: 0.5278 - val_dice_coefficient: 0.9017 - val_auc: 0.9883\nEpoch 32/50\n47/47 [==============================] - 64s 1s/step - loss: 0.1926 - categorical_crossentropy: 0.1926 - categorical_accuracy: 0.9115 - mean_iou: 0.5248 - dice_coefficient: 0.8802 - auc: 0.9776 - learning_rate: 4.1112e-04 - val_loss: 0.1428 - val_categorical_crossentropy: 0.1428 - val_categorical_accuracy: 0.9434 - val_mean_iou: 0.5295 - val_dice_coefficient: 0.9029 - val_auc: 0.9884\nEpoch 33/50\n47/47 [==============================] - 65s 1s/step - loss: 0.1915 - categorical_crossentropy: 0.1915 - categorical_accuracy: 0.9116 - mean_iou: 0.5266 - dice_coefficient: 0.8809 - auc: 0.9778 - learning_rate: 3.8890e-04 - val_loss: 0.1421 - val_categorical_crossentropy: 0.1421 - val_categorical_accuracy: 0.9434 - val_mean_iou: 0.5312 - val_dice_coefficient: 0.9037 - val_auc: 0.9884\nEpoch 34/50\n47/47 [==============================] - 65s 1s/step - loss: 0.1904 - categorical_crossentropy: 0.1904 - categorical_accuracy: 0.9118 - mean_iou: 0.5285 - dice_coefficient: 0.8816 - auc: 0.9780 - learning_rate: 3.6668e-04 - val_loss: 0.1415 - val_categorical_crossentropy: 0.1415 - val_categorical_accuracy: 0.9434 - val_mean_iou: 0.5332 - val_dice_coefficient: 0.9045 - val_auc: 0.9885\nEpoch 35/50\n47/47 [==============================] - 65s 1s/step - loss: 0.1893 - categorical_crossentropy: 0.1893 - categorical_accuracy: 0.9128 - mean_iou: 0.5306 - dice_coefficient: 0.8822 - auc: 0.9781 - learning_rate: 3.4445e-04 - val_loss: 0.1410 - val_categorical_crossentropy: 0.1410 - val_categorical_accuracy: 0.9436 - val_mean_iou: 0.5351 - val_dice_coefficient: 0.9051 - val_auc: 0.9885\nEpoch 36/50\n47/47 [==============================] - 64s 1s/step - loss: 0.1881 - categorical_crossentropy: 0.1881 - categorical_accuracy: 0.9151 - mean_iou: 0.5327 - dice_coefficient: 0.8829 - auc: 0.9784 - learning_rate: 3.2223e-04 - val_loss: 0.1405 - val_categorical_crossentropy: 0.1405 - val_categorical_accuracy: 0.9435 - val_mean_iou: 0.5372 - val_dice_coefficient: 0.9058 - val_auc: 0.9885\nEpoch 37/50\n47/47 [==============================] - 64s 1s/step - loss: 0.1871 - categorical_crossentropy: 0.1871 - categorical_accuracy: 0.9180 - mean_iou: 0.5351 - dice_coefficient: 0.8836 - auc: 0.9787 - learning_rate: 3.0001e-04 - val_loss: 0.1404 - val_categorical_crossentropy: 0.1404 - val_categorical_accuracy: 0.9423 - val_mean_iou: 0.5396 - val_dice_coefficient: 0.9061 - val_auc: 0.9885\nEpoch 38/50\n47/47 [==============================] - 65s 1s/step - loss: 0.1861 - categorical_crossentropy: 0.1861 - categorical_accuracy: 0.9198 - mean_iou: 0.5377 - dice_coefficient: 0.8844 - auc: 0.9790 - learning_rate: 2.7779e-04 - val_loss: 0.1403 - val_categorical_crossentropy: 0.1403 - val_categorical_accuracy: 0.9410 - val_mean_iou: 0.5419 - val_dice_coefficient: 0.9064 - val_auc: 0.9884\nEpoch 39/50\n47/47 [==============================] - 65s 1s/step - loss: 0.1852 - categorical_crossentropy: 0.1852 - categorical_accuracy: 0.9211 - mean_iou: 0.5400 - dice_coefficient: 0.8851 - auc: 0.9792 - learning_rate: 2.5557e-04 - val_loss: 0.1405 - val_categorical_crossentropy: 0.1405 - val_categorical_accuracy: 0.9402 - val_mean_iou: 0.5434 - val_dice_coefficient: 0.9062 - val_auc: 0.9883\nEpoch 40/50\n47/47 [==============================] - 64s 1s/step - loss: 0.1843 - categorical_crossentropy: 0.1843 - categorical_accuracy: 0.9219 - mean_iou: 0.5417 - dice_coefficient: 0.8857 - auc: 0.9794 - learning_rate: 2.3334e-04 - val_loss: 0.1407 - val_categorical_crossentropy: 0.1407 - val_categorical_accuracy: 0.9396 - val_mean_iou: 0.5446 - val_dice_coefficient: 0.9060 - val_auc: 0.9883\nEpoch 41/50\n47/47 [==============================] - 65s 1s/step - loss: 0.1837 - categorical_crossentropy: 0.1837 - categorical_accuracy: 0.9225 - mean_iou: 0.5432 - dice_coefficient: 0.8862 - auc: 0.9795 - learning_rate: 2.1112e-04 - val_loss: 0.1412 - val_categorical_crossentropy: 0.1412 - val_categorical_accuracy: 0.9390 - val_mean_iou: 0.5454 - val_dice_coefficient: 0.9053 - val_auc: 0.9883\nEpoch 42/50\n47/47 [==============================] - 64s 1s/step - loss: 0.1828 - categorical_crossentropy: 0.1828 - categorical_accuracy: 0.9230 - mean_iou: 0.5448 - dice_coefficient: 0.8869 - auc: 0.9797 - learning_rate: 1.8890e-04 - val_loss: 0.1419 - val_categorical_crossentropy: 0.1419 - val_categorical_accuracy: 0.9383 - val_mean_iou: 0.5458 - val_dice_coefficient: 0.9043 - val_auc: 0.9882\nEpoch 43/50\n47/47 [==============================] - 65s 1s/step - loss: 0.1822 - categorical_crossentropy: 0.1822 - categorical_accuracy: 0.9233 - mean_iou: 0.5459 - dice_coefficient: 0.8873 - auc: 0.9798 - learning_rate: 1.6668e-04 - val_loss: 0.1429 - val_categorical_crossentropy: 0.1429 - val_categorical_accuracy: 0.9376 - val_mean_iou: 0.5458 - val_dice_coefficient: 0.9030 - val_auc: 0.9881\nEpoch 44/50\n47/47 [==============================] - 65s 1s/step - loss: 0.1811 - categorical_crossentropy: 0.1811 - categorical_accuracy: 0.9237 - mean_iou: 0.5474 - dice_coefficient: 0.8881 - auc: 0.9800 - learning_rate: 1.4445e-04 - val_loss: 0.1439 - val_categorical_crossentropy: 0.1439 - val_categorical_accuracy: 0.9370 - val_mean_iou: 0.5458 - val_dice_coefficient: 0.9018 - val_auc: 0.9881\nEpoch 45/50\n47/47 [==============================] - 65s 1s/step - loss: 0.1800 - categorical_crossentropy: 0.1800 - categorical_accuracy: 0.9239 - mean_iou: 0.5489 - dice_coefficient: 0.8886 - auc: 0.9802 - learning_rate: 1.2223e-04 - val_loss: 0.1446 - val_categorical_crossentropy: 0.1446 - val_categorical_accuracy: 0.9365 - val_mean_iou: 0.5460 - val_dice_coefficient: 0.9011 - val_auc: 0.9880\nEpoch 46/50\n47/47 [==============================] - 65s 1s/step - loss: 0.1789 - categorical_crossentropy: 0.1789 - categorical_accuracy: 0.9241 - mean_iou: 0.5504 - dice_coefficient: 0.8892 - auc: 0.9805 - learning_rate: 1.0001e-04 - val_loss: 0.1446 - val_categorical_crossentropy: 0.1446 - val_categorical_accuracy: 0.9365 - val_mean_iou: 0.5464 - val_dice_coefficient: 0.9011 - val_auc: 0.9880\nEpoch 47/50\n47/47 [==============================] - 65s 1s/step - loss: 0.1778 - categorical_crossentropy: 0.1778 - categorical_accuracy: 0.9245 - mean_iou: 0.5517 - dice_coefficient: 0.8898 - auc: 0.9807 - learning_rate: 7.7788e-05 - val_loss: 0.1441 - val_categorical_crossentropy: 0.1441 - val_categorical_accuracy: 0.9368 - val_mean_iou: 0.5469 - val_dice_coefficient: 0.9018 - val_auc: 0.9880\nEpoch 48/50\n47/47 [==============================] - 65s 1s/step - loss: 0.1770 - categorical_crossentropy: 0.1770 - categorical_accuracy: 0.9248 - mean_iou: 0.5527 - dice_coefficient: 0.8901 - auc: 0.9809 - learning_rate: 5.5565e-05 - val_loss: 0.1433 - val_categorical_crossentropy: 0.1433 - val_categorical_accuracy: 0.9373 - val_mean_iou: 0.5475 - val_dice_coefficient: 0.9028 - val_auc: 0.9881\nEpoch 49/50\n47/47 [==============================] - 65s 1s/step - loss: 0.1760 - categorical_crossentropy: 0.1760 - categorical_accuracy: 0.9251 - mean_iou: 0.5537 - dice_coefficient: 0.8906 - auc: 0.9811 - learning_rate: 3.3343e-05 - val_loss: 0.1423 - val_categorical_crossentropy: 0.1423 - val_categorical_accuracy: 0.9380 - val_mean_iou: 0.5480 - val_dice_coefficient: 0.9040 - val_auc: 0.9882\nEpoch 50/50\n47/47 [==============================] - 65s 1s/step - loss: 0.1753 - categorical_crossentropy: 0.1753 - categorical_accuracy: 0.9255 - mean_iou: 0.5545 - dice_coefficient: 0.8909 - auc: 0.9812 - learning_rate: 1.1121e-05 - val_loss: 0.1413 - val_categorical_crossentropy: 0.1413 - val_categorical_accuracy: 0.9387 - val_mean_iou: 0.5484 - val_dice_coefficient: 0.9051 - val_auc: 0.9883\n"
],
[
"",
"_____no_output_____"
]
],
[
[
"**Task 4**: Let's predict mitochondria in the testing data! [15 Points]",
"_____no_output_____"
]
],
[
[
"# \n# After training, let's try the U-Net on our testing data.\n#\n# The code below displays the first 10 input images, the original masks by experts, \n# and the predicted masks from the U-Net.",
"_____no_output_____"
],
[
"rows = 10\nfig, axs = plt.subplots(rows, 3, figsize=(8, 30))\nfor ax, (image, label) in zip(axs, test.take(rows).batch(1)):\n \n prediction = unet_model.predict(image)\n # print(prediction.shape)\n ax[0].matshow(image[0, :, :, 0])\n ax[1].matshow(label[0, :, :, 1], cmap=\"gray\")\n ax[2].matshow(prediction[0].argmax(axis=-1), cmap=\"gray\")",
"_____no_output_____"
],
[
"# 1) Question\n# TODO: Why do we use the prediction[0].argmax(axis=-1) command\n# to display the prediction?\n# \n# TODO: YOUR ANSWER",
"_____no_output_____"
]
],
[
[
"As the shape of the prediction array is (1, 256, 256, 2), therefore picking the first value will result in all the predicted masks which have the dimensions of the input images to the network. The prediction results in probabilities and we want to pick the highest probability made by the model as that's what the model primarily implies. Hence the argmax in the code <code>prediction[0].argmax(axis=-1)</code> is used to pick the highest prediction.",
"_____no_output_____"
]
],
[
[
"# 2) Question\n# TODO: Is the quality of the segmentation good and how could we improve it?\n# Hint: Think along the lines of some traditional image processing rather than\n# increasing the training data size.\n#\n# TODO: YOUR ANSWER",
"_____no_output_____"
]
],
[
[
"There are few ways we can do this:\n<ol>\n<li>We can set a threshold and eliminate all the predicted masks which are less than that.</li>\n<li>We can incorporate shrink and expand algorithm to get rid of extraneous predicted masks.</li>\n</ol>",
"_____no_output_____"
]
],
[
[
"",
"_____no_output_____"
]
],
[
[
"**Bonus**: Compare against the state-of-the-art literature! [33 Points]",
"_____no_output_____"
]
],
[
[
"#\n# The following paper also uses a 2D U-Net to detect mitochondria:\n# \n# https://danielhaehn.com/papers/?casser2020fast\n#",
"_____no_output_____"
],
[
"# 1) Question\n#\n# TODO: How many learnable parameters does your trained unet_model have?\n# Hint: You can use TF's Model Summary function to answer this.\n#",
"_____no_output_____"
],
[
"# TODO: YOUR CODE\nunet_model.summary()",
"Model: \"unet\"\n__________________________________________________________________________________________________\n Layer (type) Output Shape Param # Connected to \n==================================================================================================\n inputs (InputLayer) [(None, None, None, 0 [] \n 1)] \n \n conv_block (ConvBlock) (None, None, None, 37568 ['inputs[0][0]'] \n 64) \n \n max_pooling2d (MaxPooling2D) (None, None, None, 0 ['conv_block[0][0]'] \n 64) \n \n conv_block_1 (ConvBlock) (None, None, None, 221440 ['max_pooling2d[0][0]'] \n 128) \n \n max_pooling2d_1 (MaxPooling2D) (None, None, None, 0 ['conv_block_1[0][0]'] \n 128) \n \n conv_block_2 (ConvBlock) (None, None, None, 885248 ['max_pooling2d_1[0][0]'] \n 256) \n \n max_pooling2d_2 (MaxPooling2D) (None, None, None, 0 ['conv_block_2[0][0]'] \n 256) \n \n conv_block_3 (ConvBlock) (None, None, None, 3539968 ['max_pooling2d_2[0][0]'] \n 512) \n \n max_pooling2d_3 (MaxPooling2D) (None, None, None, 0 ['conv_block_3[0][0]'] \n 512) \n \n conv_block_4 (ConvBlock) (None, None, None, 14157824 ['max_pooling2d_3[0][0]'] \n 1024) \n \n upconv_block (UpconvBlock) (None, None, None, 2097664 ['conv_block_4[0][0]'] \n 512) \n \n crop_concat_block (CropConcatB (None, None, None, 0 ['upconv_block[0][0]', \n lock) 1024) 'conv_block_3[0][0]'] \n \n conv_block_5 (ConvBlock) (None, None, None, 7078912 ['crop_concat_block[0][0]'] \n 512) \n \n upconv_block_1 (UpconvBlock) (None, None, None, 524544 ['conv_block_5[0][0]'] \n 256) \n \n crop_concat_block_1 (CropConca (None, None, None, 0 ['upconv_block_1[0][0]', \n tBlock) 512) 'conv_block_2[0][0]'] \n \n conv_block_6 (ConvBlock) (None, None, None, 1769984 ['crop_concat_block_1[0][0]'] \n 256) \n \n upconv_block_2 (UpconvBlock) (None, None, None, 131200 ['conv_block_6[0][0]'] \n 128) \n \n crop_concat_block_2 (CropConca (None, None, None, 0 ['upconv_block_2[0][0]', \n tBlock) 256) 'conv_block_1[0][0]'] \n \n conv_block_7 (ConvBlock) (None, None, None, 442624 ['crop_concat_block_2[0][0]'] \n 128) \n \n upconv_block_3 (UpconvBlock) (None, None, None, 32832 ['conv_block_7[0][0]'] \n 64) \n \n crop_concat_block_3 (CropConca (None, None, None, 0 ['upconv_block_3[0][0]', \n tBlock) 128) 'conv_block[0][0]'] \n \n conv_block_8 (ConvBlock) (None, None, None, 110720 ['crop_concat_block_3[0][0]'] \n 64) \n \n conv2d_18 (Conv2D) (None, None, None, 130 ['conv_block_8[0][0]'] \n 2) \n \n activation_22 (Activation) (None, None, None, 0 ['conv2d_18[0][0]'] \n 2) \n \n outputs (Activation) (None, None, None, 0 ['activation_22[0][0]'] \n 2) \n \n==================================================================================================\nTotal params: 31,030,658\nTrainable params: 31,030,658\nNon-trainable params: 0\n__________________________________________________________________________________________________\n"
]
],
[
[
"There are 31,030,658 training parameters.",
"_____no_output_____"
]
],
[
[
"",
"_____no_output_____"
],
[
"# 2) Question\n#\n# TODO: How many parameters do Casser et al. use?\n#",
"_____no_output_____"
],
[
"# TODO: YOUR ANSWER",
"_____no_output_____"
]
],
[
[
"The total number of trainable parameters are 1,178,480 + 780,053 = 1,958,533.",
"_____no_output_____"
]
],
[
[
"",
"_____no_output_____"
],
[
"# 3) Question\n#\n# TODO: How did Casser et al. reduce the parameters?\n#",
"_____no_output_____"
],
[
"# TODO: YOUR ANSWER",
"_____no_output_____"
]
],
[
[
"Casser et al. reduce the parameters by:\n\n\n1. Reducing number of convolutional filters throughout the network.\n2. Replacing the transpose convolutions with light weight bilinear upsampling layers in decoder.\n\n",
"_____no_output_____"
]
],
[
[
"",
"_____no_output_____"
],
[
"# 4) Question\n#\n# TODO: Why did Casser et al. reduce the parameters?\n#",
"_____no_output_____"
],
[
"# TODO: YOUR ANSWER",
"_____no_output_____"
]
],
[
[
"Upon investigation, it was discovered by Casser et al. that around 33% of the ReLU activations are dead, meaning the network is too complex to the problem of mitochondria mask prediction. Therefore, the authors, decided to design a new modified version of the U-Net which utilizes 99.7% of the network.",
"_____no_output_____"
]
],
[
[
"",
"_____no_output_____"
],
[
"#\n# You made it!!\n#\n# _ ___ /^^\\ /^\\ /^^\\_\n# _ _@)@) \\ ,,/ '` ~ `'~~ ', `\\.\n# _/o\\_ _ _ _/~`.`...'~\\ ./~~..,'`','',.,' ' ~:\n# / `,'.~,~.~ . , . , ~|, ,/ .,' , ,. .. ,,. `, ~\\_\n# ( ' _' _ '_` _ ' . , `\\_/ .' ..' ' ` ` `.. `, \\_\n# ~V~ V~ V~ V~ ~\\ ` ' . ' , ' .,.,''`.,.''`.,.``. ', \\_\n# _/\\ /\\ /\\ /\\_/, . ' , `_/~\\_ .' .,. ,, , _/~\\_ `. `. '., \\_\n# < ~ ~ '~`'~'`, ., . `_: ::: \\_ ' `_/ ::: \\_ `.,' . ', \\_\n# \\ ' `_ '`_ _ ',/ _::_::_ \\ _ _/ _::_::_ \\ `.,'.,`., \\-,-,-,_,_,\n# `'~~ `'~~ `'~~ `'~~ \\(_)(_)(_)/ `~~' \\(_)(_)(_)/ ~'`\\_.._,._,'_;_;_;_;_;\n#",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
c513154af03bcedbccfa0ddb858c10198cc33329
| 28,783 |
ipynb
|
Jupyter Notebook
|
x-archive-temp/m120-pandas/pandas-intro-demo.ipynb
|
UCBerkeley-SCET/DataX-Berkeley
|
f912d22c838b511d3ada4ecfa3548afd80437b74
|
[
"Apache-2.0"
] | 28 |
2020-06-15T23:53:36.000Z
|
2022-03-19T09:27:02.000Z
|
x-archive-temp/m120-pandas/pandas-intro-demo.ipynb
|
UCBerkeley-SCET/DataX-Berkeley
|
f912d22c838b511d3ada4ecfa3548afd80437b74
|
[
"Apache-2.0"
] | 4 |
2020-06-24T22:20:31.000Z
|
2022-02-28T01:37:36.000Z
|
x-archive-temp/m120-pandas/pandas-intro-demo.ipynb
|
UCBerkeley-SCET/DataX-Berkeley
|
f912d22c838b511d3ada4ecfa3548afd80437b74
|
[
"Apache-2.0"
] | 78 |
2020-06-19T09:41:01.000Z
|
2022-02-05T00:13:29.000Z
| 42.14202 | 15,492 | 0.76163 |
[
[
[
"\n\n---\n# Pandas Introduction \n\n**Author list:** Ikhlaq Sidhu & Alexander Fred Ojala\n\n**References / Sources:** \nIncludes examples from Wes McKinney and the 10 min intro to Pandas\n\n\n**License Agreement:** Feel free to do whatever you want with this code\n\n___",
"_____no_output_____"
],
[
"### Topics:\n1. Dataframe creation\n2. Reading data in DataFrames\n3. Data Manipulation",
"_____no_output_____"
],
[
"## Import package",
"_____no_output_____"
]
],
[
[
"# pandas\nimport pandas as pd",
"_____no_output_____"
],
[
"# Extra packages\nimport numpy as np\nimport matplotlib.pyplot as plt # for plotting\n\n# jupyter notebook magic to display plots in output\n%matplotlib inline\n\nplt.rcParams['figure.figsize'] = (10,6) # make the plots bigger",
"_____no_output_____"
]
],
[
[
"# Part 1: Creation of Pandas dataframes\n\n**Key Points:** Main data types in Pandas:\n* Series (similar to numpy arrays, but with index)\n* DataFrames (table or spreadsheet with Series in the columns)\n",
"_____no_output_____"
],
[
"\n\n### We use `pd.DataFrame()` and can insert almost any data type as an argument\n\n**Function:** `pd.DataFrame(data=None, index=None, columns=None, dtype=None, copy=False)`\n\nInput data can be a numpy ndarray (structured or homogeneous), dictionary, or DataFrame. \n",
"_____no_output_____"
],
[
"### 1.1 Create Dataframe using an array",
"_____no_output_____"
]
],
[
[
"# Try it with an array\nnp.random.seed(0) # set seed for reproducibility\n\na1 = np.random.randn(3)\na2 = np.random.randn(3)\na3 = np.random.randn(3)\n\nprint (a1)\nprint (a2)\nprint (a3)",
"_____no_output_____"
],
[
"# Create our first DataFrame w/ an np.array - it becomes a column\n",
"_____no_output_____"
],
[
"# Check type\n",
"_____no_output_____"
],
[
"# DataFrame from list of np.arrays\n\n# notice that there is no column label, only integer values,\n# and the index is set automatically",
"_____no_output_____"
],
[
"# We can set column and index names\n",
"_____no_output_____"
],
[
"# Add more columns to dataframe, like a dictionary, dimensions must match\n",
"_____no_output_____"
],
[
"# DataFrame from 2D np.array\nnp.random.seed(0)\narray_2d = np.array(np.random.randn(9)).reshape(3,3)\n",
"_____no_output_____"
],
[
"# Create df with labeled columns\n",
"_____no_output_____"
]
],
[
[
"### 1.2 Create Dataframe using an dictionary",
"_____no_output_____"
]
],
[
[
"# DataFrame from a Dictionary\ndict1 = {'a1': a1, 'a2':a2, 'a3':a3}\n",
"_____no_output_____"
],
[
"# Note that we now have columns without assignment\n\n",
"_____no_output_____"
],
[
"# We can add a list with strings and ints as a column \n",
"_____no_output_____"
]
],
[
[
"### Pandas Series object\nEvery column is a Series. Like an np.array, but we can combine data types and it has its own index",
"_____no_output_____"
]
],
[
[
"# Check type\n",
"_____no_output_____"
],
[
"# Dtype object\n",
"_____no_output_____"
],
[
"# Create a Series from a Python list, automatic index\n",
"_____no_output_____"
],
[
"# Specific index\n",
"_____no_output_____"
],
[
"# We can add the Series s to the DataFrame above as column Series\n# Remember to match indices\n",
"_____no_output_____"
],
[
"# We can also rename columns\n",
"_____no_output_____"
],
[
"# We can delete columns\n",
"_____no_output_____"
],
[
"# or drop columns, see axis = 1\n# does not change df1 if we don't set inplace=True\n",
"_____no_output_____"
],
[
"# Print df1",
"_____no_output_____"
],
[
"# Or drop rows\n",
"_____no_output_____"
]
],
[
[
"### 1.3 Indexing / Slicing a Pandas Datframe",
"_____no_output_____"
]
],
[
[
"# Example: view only one column\n",
"_____no_output_____"
],
[
"# Or view several column\n",
"_____no_output_____"
],
[
"# Slice of the DataFrame returned\n# this slices the first three rows first followed by first 2 rows of the sliced frame\n",
"_____no_output_____"
],
[
"# Lets print the five first 2 elements of column a1\n# This is a new Series (like a new table)\n",
"_____no_output_____"
],
[
"# Lets print the 2 column, and top 2 values- note the list of columns\n",
"_____no_output_____"
]
],
[
[
"### Instead of double indexing, we can use loc, iloc\n\n##### loc gets rows (or columns) with particular labels from the index.\n#### iloc gets rows (or columns) at particular positions in the index (so it only takes integers).",
"_____no_output_____"
],
[
"### .iloc()",
"_____no_output_____"
]
],
[
[
"# iloc\n",
"_____no_output_____"
],
[
"# Slice\n",
"_____no_output_____"
],
[
"# iloc will also accept 2 'lists' of position numbers\n",
"_____no_output_____"
],
[
"# Data only from row with index value '1'\n",
"_____no_output_____"
]
],
[
[
"### .loc()",
"_____no_output_____"
]
],
[
[
"# Usually we want to grab values by column names \n# Note: You have to know indices and columns\n",
"_____no_output_____"
],
[
"# Boolean indexing\n# Return full rows where a2>0\n",
"_____no_output_____"
],
[
"# Return column a3 values where a2 >0\n",
"_____no_output_____"
],
[
"# If you want the values in an np array\n",
"_____no_output_____"
]
],
[
[
"### More Basic Statistics",
"_____no_output_____"
]
],
[
[
"# Get basic statistics using .describe()",
"_____no_output_____"
],
[
"# Get specific statistics\n",
"_____no_output_____"
],
[
"# We can change the index sorting",
"_____no_output_____"
]
],
[
[
"#### For more functionalities check this notebook\nhttps://github.com/ikhlaqsidhu/data-x/blob/master/02b-tools-pandas_intro-mplib_afo/legacy/10-minutes-to-pandas-w-data-x.ipynb\n\n",
"_____no_output_____"
],
[
"# Part 2: Reading data in pandas Dataframe\n\n\n### Now, lets get some data in CSV format.\n\n#### Description:\nAggregate data on applicants to graduate school at Berkeley for the six largest departments in 1973 classified by admission and sex.\n\nhttps://vincentarelbundock.github.io/Rdatasets/doc/datasets/UCBAdmissions.html",
"_____no_output_____"
]
],
[
[
"# Read in the file\n",
"_____no_output_____"
],
[
"# Check statistics",
"_____no_output_____"
],
[
"# Columns\n",
"_____no_output_____"
],
[
"# Head\n",
"_____no_output_____"
],
[
"# Tail\n",
"_____no_output_____"
],
[
"# Groupby \n",
"_____no_output_____"
],
[
"# Describe\n",
"_____no_output_____"
],
[
"# Info\n",
"_____no_output_____"
],
[
"# Unique\n",
"_____no_output_____"
],
[
"# Total number of applicants to Dept A",
"_____no_output_____"
],
[
"# Groupby\n",
"_____no_output_____"
],
[
"# Plot using a bar graph\n",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
c5133777c21e988180ed60a84e9ad0322b5dfc9e
| 5,958 |
ipynb
|
Jupyter Notebook
|
Storytelling with Data/Cap.10 - Final thoughts.ipynb
|
luonishicastro/DataScienceTheory-OReillyExercises
|
c41cb4881e2d1a03b875e046bd63d8b9c5a525a0
|
[
"MIT"
] | null | null | null |
Storytelling with Data/Cap.10 - Final thoughts.ipynb
|
luonishicastro/DataScienceTheory-OReillyExercises
|
c41cb4881e2d1a03b875e046bd63d8b9c5a525a0
|
[
"MIT"
] | null | null | null |
Storytelling with Data/Cap.10 - Final thoughts.ipynb
|
luonishicastro/DataScienceTheory-OReillyExercises
|
c41cb4881e2d1a03b875e046bd63d8b9c5a525a0
|
[
"MIT"
] | null | null | null | 70.928571 | 527 | 0.725243 |
[
[
[
"<b>General Tips</b>",
"_____no_output_____"
],
[
"<i>\n<li>When the best course for visualizing certain data is unclear, start with a blank piece of paper.</li>\n<li>Sketch out potential views to see them side‐by‐side and determine what will work best for getting your message across to your audience.</li>\n<li>Create a version of the graph (let’s call it A), then make a copy of it (B) and make a single change. Then determine which looks better—A or B. Often, the practice of seeing slight variations next to each other makes it quickly clear which view is superior.</li>\n<li>At any point, if the best path is unclear, seek feedback. The fresh set of eyes that a friend or colleague can bring to the data visualization endeavor is invaluable.</li>\n<li>One of my biggest tips for success in storytelling with data is to allow adequate time for it. If we don’t consciously recognize that this takes time to do well and budget accordingly, our time can be entirely eaten up by the other parts of the analytical process.\n<li>Imitation really is the best form of flattery. If you see a data visualization or example of storytelling with data that you like, consider how you might adapt the approach for your own use. Pause to reflect on what makes it effective. Make a copy of it and create a visual library that you can add to over time and refer to for inspiration.</li>\n<li>There are a number of great blogs and resources on the topic of data visualization and communicating with data that contain many good examples. Here are a few of my current personal favorites:\n eagereyes.org / fivethirtyeight.com/datalab / flowingdata.com / thefunctionalart.com / theguardian.com/data / HelpMeViz.com / junkcharts.typepad.com / makeapowerfulpoint.com / perceptualedge.com / visualisingdata.com / vizwiz.blogspot.com / storytellingwithdata.com / wtfviz.net</li> \n<li>To the extent that it makes sense given the task at hand, don’t be afraid to let your own style develop and creativity come through when you communicate with data. Company brand can also play a role in developing a data visualization style; consider your company’s brand and whether there are opportunities to fold that into how you visualize and communicate with data. Just make sure that your approach and stylistic elements are making the information easier—not more difficult—for your audience to consume.</li>\n</i>",
"_____no_output_____"
],
[
"<b>Recap on this Book:</b>",
"_____no_output_____"
],
[
"<i>\n<ol>\n<li>Understand the context. Build a clear understanding of who you are communicating to, what you need them to know or do,\nhow you will communicate to them, and what data you have to back up your case. Employ concepts like the 3‐minute story, the\nBig Idea, and storyboarding to articulate your story and plan the desired content and flow.</li>\n<li>Choose an appropriate visual display. When highlighting a number or two, simple text is best. Line charts are usually best\nfor continuous data. Bar charts work great for categorical data and must have a zero baseline. Let the relationship you want to\nshow guide the type of chart you choose. Avoid pies, donuts, 3D, and secondary y‐axes due to difficulty of visual interpretation.</li>\n<li>Eliminate clutter. Identify elements that don’t add informative value and remove them from your visuals. Leverage the Gestalt principles to understand how people see and identify candidates for elimination. Use contrast strategically. Employ alignment of elements and maintain white space to help make the interpretation of your visuals a comfortable experience for your audience.</li>\n<li>Focus attention where you want it. Employ the power of preattentive attributes like color, size, and position to signal what’s important. Use these strategic attributes to draw attention to where you want your audience to look and guide your audience through your visual. Evaluate the effectiveness of preattentive attributes in your visual by applying the “where are your eyes drawn?” test.</li>\n<li>Think like a designer. Offer your audience visual affordances as cues for how to interact with your communication: highlight the important stuff, eliminate distractions, and create a visual hierarchy of information. Make your designs accessible by not overcomplicating and leveraging text to label and explain. Increase your audience’s tolerance of design issues by making your visuals aesthetically pleasing. Work to gain audience acceptance of your visual designs.</li>\n<li>Tell a story. Craft a story with clear beginning (plot), middle (twists), and end (call to action). Leverage conflict and tension to grab and maintain your audience’s attention. Consider the order and manner of your narrative. Utilize the power of repetition to help your stories stick. Employ tactics like vertical and horizontal logic, reverse storyboarding, and seeking a fresh perspective to ensure that your story comes across clearly in your communication.</li>\n</ol>\n</i>",
"_____no_output_____"
]
]
] |
[
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
c51346bd95e98ee414a941bbaf6b050c66dc01b6
| 222,513 |
ipynb
|
Jupyter Notebook
|
code/6_gpu_benchmarking.ipynb
|
Akshatha-Jagadish/DL_topics
|
98aa979dde2021a20e7b561b83230ac0a475cf5e
|
[
"MIT"
] | null | null | null |
code/6_gpu_benchmarking.ipynb
|
Akshatha-Jagadish/DL_topics
|
98aa979dde2021a20e7b561b83230ac0a475cf5e
|
[
"MIT"
] | null | null | null |
code/6_gpu_benchmarking.ipynb
|
Akshatha-Jagadish/DL_topics
|
98aa979dde2021a20e7b561b83230ac0a475cf5e
|
[
"MIT"
] | null | null | null | 182.987664 | 127,392 | 0.889197 |
[
[
[
"#used an environment with directml\n# with the help of https://www.youtube.com/watch?v=gjVFH7NHB9s\n#ref to choose the env in jupyter notebook: https://towardsdatascience.com/get-your-conda-environment-to-show-in-jupyter-notebooks-the-easy-way-17010b76e874\n\nimport tensorflow as tf\nfrom tensorflow import keras\nimport matplotlib.pyplot as plt\nimport numpy as np",
"_____no_output_____"
],
[
"tf.config.experimental.list_physical_devices()",
"_____no_output_____"
],
[
"tf.test.is_gpu_available()",
"_____no_output_____"
],
[
"from tensorflow.python.client import device_lib\n\ndevice_lib.list_local_devices()",
"_____no_output_____"
],
[
"(x_train, y_train), (x_test, y_test) = keras.datasets.cifar10.load_data()",
"_____no_output_____"
],
[
"x_train.shape",
"_____no_output_____"
],
[
"x_train[0].shape",
"_____no_output_____"
],
[
"y_train[0]",
"_____no_output_____"
],
[
"def plot_sample(index):\n plt.figure(figsize=(10,1))\n plt.imshow(x_train[index])",
"_____no_output_____"
],
[
"plot_sample(0)",
"_____no_output_____"
],
[
"plot_sample(3)",
"_____no_output_____"
],
[
"y_train[3]",
"_____no_output_____"
],
[
"classes = ['airplane','automobile','bird','cat','deer','dog','frog','horse','ship','truck']",
"_____no_output_____"
],
[
"classes[y_train[3][0]]",
"_____no_output_____"
],
[
"X_train_scaled = x_train/255\nX_test_scaled = x_test/255",
"_____no_output_____"
],
[
"# do one hot encoding\ny_train_categorical = keras.utils.to_categorical(\n y_train,num_classes=10,dtype='float32'\n)\ny_train_categorical[3]",
"_____no_output_____"
],
[
"y_test_categorical = keras.utils.to_categorical(\n y_test,num_classes=10,dtype='float32'\n)",
"_____no_output_____"
],
[
"#model building\nmodel = keras.Sequential([\n keras.layers.Flatten(input_shape=(32,32,3)),\n keras.layers.Dense(3000,activation='relu'),\n keras.layers.Dense(1000,activation='relu'),\n keras.layers.Dense(10,activation='sigmoid'),\n])\n\nmodel.compile(\n optimizer='SGD',\n loss='categorical_crossentropy', #if one hot encoded\n metrics=['accuracy']\n)\n\nmodel.fit(X_train_scaled,y_train_categorical,epochs=50)",
"Train on 50000 samples\nEpoch 1/50\n50000/50000 [==============================] - 110s 2ms/sample - loss: 1.8628 - acc: 0.3331\nEpoch 2/50\n50000/50000 [==============================] - 108s 2ms/sample - loss: 1.6579 - acc: 0.4125\nEpoch 3/50\n50000/50000 [==============================] - 108s 2ms/sample - loss: 1.5720 - acc: 0.4413s - loss: 1.5724 - acc:\nEpoch 4/50\n50000/50000 [==============================] - 109s 2ms/sample - loss: 1.5102 - acc: 0.4690\nEpoch 5/50\n50000/50000 [==============================] - 111s 2ms/sample - loss: 1.4602 - acc: 0.4840\nEpoch 6/50\n50000/50000 [==============================] - 111s 2ms/sample - loss: 1.4118 - acc: 0.5023s - loss: 1.4119 - ETA: 1s - loss: 1\nEpoch 7/50\n50000/50000 [==============================] - 121s 2ms/sample - loss: 1.3765 - acc: 0.5165\nEpoch 8/50\n50000/50000 [==============================] - 128s 3ms/sample - loss: 1.3399 - acc: 0.5271\nEpoch 9/50\n50000/50000 [==============================] - 122s 2ms/sample - loss: 1.3089 - acc: 0.5396\nEpoch 10/50\n50000/50000 [==============================] - 112s 2ms/sample - loss: 1.2781 - acc: 0.5515s - loss: 1.\nEpoch 11/50\n50000/50000 [==============================] - 112s 2ms/sample - loss: 1.2479 - acc: 0.5623\nEpoch 12/50\n50000/50000 [==============================] - 109s 2ms/sample - loss: 1.2173 - acc: 0.5710s - loss: 1.2163\nEpoch 13/50\n50000/50000 [==============================] - 110s 2ms/sample - loss: 1.1949 - acc: 0.5818\nEpoch 14/50\n50000/50000 [==============================] - ETA: 0s - loss: 1.1655 - acc: 0.594 - 110s 2ms/sample - loss: 1.1656 - acc: 0.5940\nEpoch 15/50\n50000/50000 [==============================] - 111s 2ms/sample - loss: 1.1409 - acc: 0.5995s - loss: 1.1404 - - ETA: 0s - loss: 1.1406 - acc: \nEpoch 16/50\n50000/50000 [==============================] - 111s 2ms/sample - loss: 1.1124 - acc: 0.6110\nEpoch 17/50\n50000/50000 [==============================] - 113s 2ms/sample - loss: 1.0890 - acc: 0.6202\nEpoch 18/50\n50000/50000 [==============================] - 119s 2ms/sample - loss: 1.0607 - acc: 0.6301\nEpoch 19/50\n50000/50000 [==============================] - 145s 3ms/sample - loss: 1.0375 - acc: 0.6384\nEpoch 20/50\n50000/50000 [==============================] - 112s 2ms/sample - loss: 1.0129 - acc: 0.6485s - loss: 1.0128 - acc: 0.6 - \nEpoch 21/50\n50000/50000 [==============================] - 128s 3ms/sample - loss: 0.9867 - acc: 0.6567\nEpoch 22/50\n50000/50000 [==============================] - 144s 3ms/sample - loss: 0.9626 - acc: 0.6657s - loss: 0.9624 - acc:\nEpoch 23/50\n50000/50000 [==============================] - 142s 3ms/sample - loss: 0.9421 - acc: 0.6739\nEpoch 24/50\n50000/50000 [==============================] - 143s 3ms/sample - loss: 0.9122 - acc: 0.6817\nEpoch 25/50\n50000/50000 [==============================] - 143s 3ms/sample - loss: 0.8919 - acc: 0.6905\nEpoch 26/50\n50000/50000 [==============================] - 143s 3ms/sample - loss: 0.8654 - acc: 0.7011s - lo\nEpoch 27/50\n50000/50000 [==============================] - 130s 3ms/sample - loss: 0.8390 - acc: 0.7111\nEpoch 28/50\n50000/50000 [==============================] - 127s 3ms/sample - loss: 0.8131 - acc: 0.7174\nEpoch 29/50\n50000/50000 [==============================] - 130s 3ms/sample - loss: 0.7924 - acc: 0.7258\nEpoch 30/50\n50000/50000 [==============================] - 123s 2ms/sample - loss: 0.7651 - acc: 0.7398\nEpoch 31/50\n50000/50000 [==============================] - 123s 2ms/sample - loss: 0.7455 - acc: 0.7446\nEpoch 32/50\n50000/50000 [==============================] - 122s 2ms/sample - loss: 0.7212 - acc: 0.7521\nEpoch 33/50\n50000/50000 [==============================] - 108s 2ms/sample - loss: 0.6926 - acc: 0.7631s - loss: 0.6916 - acc: 0.76 - - ETA: 1s - loss: 0. - ETA: 0s - loss: 0.6925 - acc\nEpoch 34/50\n50000/50000 [==============================] - 109s 2ms/sample - loss: 0.6713 - acc: 0.7728\nEpoch 35/50\n50000/50000 [==============================] - 108s 2ms/sample - loss: 0.6459 - acc: 0.7818\nEpoch 36/50\n50000/50000 [==============================] - 142s 3ms/sample - loss: 0.6181 - acc: 0.7910A: 3s - l - ETA: 1s - loss: 0\nEpoch 37/50\n50000/50000 [==============================] - 145s 3ms/sample - loss: 0.5971 - acc: 0.8004\nEpoch 38/50\n50000/50000 [==============================] - 146s 3ms/sample - loss: 0.5794 - acc: 0.8042 ETA:\nEpoch 39/50\n50000/50000 [==============================] - 145s 3ms/sample - loss: 0.5530 - acc: 0.8164\nEpoch 40/50\n50000/50000 [==============================] - 145s 3ms/sample - loss: 0.5326 - acc: 0.8230\nEpoch 41/50\n50000/50000 [==============================] - 145s 3ms/sample - loss: 0.5121 - acc: 0.8271\nEpoch 42/50\n50000/50000 [==============================] - 145s 3ms/sample - loss: 0.4905 - acc: 0.8382\nEpoch 43/50\n50000/50000 [==============================] - 146s 3ms/sample - loss: 0.4700 - acc: 0.8448s \nEpoch 44/50\n50000/50000 [==============================] - 145s 3ms/sample - loss: 0.4462 - acc: 0.8546s - loss: 0.4473 - acc: 0.854 - ETA: 3s - loss: 0.4470 - ac - ETA: 2s - lo\nEpoch 45/50\n50000/50000 [==============================] - 145s 3ms/sample - loss: 0.4285 - acc: 0.8607\nEpoch 46/50\n50000/50000 [==============================] - 145s 3ms/sample - loss: 0.4087 - acc: 0.8677\nEpoch 47/50\n50000/50000 [==============================] - 148s 3ms/sample - loss: 0.3851 - acc: 0.8770\nEpoch 48/50\n50000/50000 [==============================] - 145s 3ms/sample - loss: 0.3656 - acc: 0.8833\nEpoch 49/50\n50000/50000 [==============================] - 145s 3ms/sample - loss: 0.3468 - acc: 0.8915\nEpoch 50/50\n50000/50000 [==============================] - 132s 3ms/sample - loss: 0.3323 - acc: 0.8971\n"
],
[
"classes[y_test[1][0]]",
"_____no_output_____"
],
[
"classes[np.argmax(model.predict(np.array([X_test_scaled[1]])))]",
"_____no_output_____"
],
[
"def get_model():\n model = keras.Sequential([\n keras.layers.Flatten(input_shape=(32,32,3)),\n keras.layers.Dense(3000,activation='relu'),\n keras.layers.Dense(1000,activation='relu'),\n keras.layers.Dense(10,activation='sigmoid'),\n ])\n\n model.compile(\n optimizer='SGD',\n loss='categorical_crossentropy', #if one hot encoded\n metrics=['accuracy']\n )\n \n return model",
"_____no_output_____"
],
[
"%%timeit -n1 -r1\nwith tf.device('/DML:0'):\n cpu_model = get_model()\n cpu_model.fit(X_train_scaled, y_train_categorical, epochs=1)",
"Train on 50000 samples\n50000/50000 [==============================] - 103s 2ms/sample - loss: 2.3050 - acc: 0.09990s - loss: 2.3052 - - ETA: 9s - loss: 2.3052 - acc: 0.0 - ETA: 9s - \n1min 44s ± 0 ns per loop (mean ± std. dev. of 1 run, 1 loop each)\n"
],
[
"%%timeit -n1 -r1\nwith tf.device('/CPU:0'):\n cpu_model = get_model()\n cpu_model.fit(X_train_scaled, y_train_categorical, epochs=1)",
"Train on 50000 samples\n50000/50000 [==============================] - 104s 2ms/sample - loss: 1.8652 - acc: 0.3324\n1min 44s ± 0 ns per loop (mean ± std. dev. of 1 run, 1 loop each)\n"
],
[
"%%timeit -n1 -r1\nwith tf.device('/DML:1'):\n cpu_model = get_model()\n cpu_model.fit(X_train_scaled, y_train_categorical, epochs=1)",
"Train on 50000 samples\n50000/50000 [==============================] - 148s 3ms/sample - loss: 2.3053 - acc: 0.1000s - loss: 2. - ETA:\n2min 29s ± 0 ns per loop (mean ± std. dev. of 1 run, 1 loop each)\n"
]
],
[
[
"",
"_____no_output_____"
]
],
[
[
"fashion_mnist = keras.datasets.fashion_mnist\n\n(train_images, train_labels), (test_images, test_labels) = fashion_mnist.load_data()\nclass_names = ['T-shirt/top', 'Trouser', 'Pullover', 'Dress', 'Coat',\n 'Sandal', 'Shirt', 'Sneaker', 'Bag', 'Ankle boot']",
"Downloading data from https://storage.googleapis.com/tensorflow/tf-keras-datasets/train-labels-idx1-ubyte.gz\n32768/29515 [=================================] - 0s 4us/step\nDownloading data from https://storage.googleapis.com/tensorflow/tf-keras-datasets/train-images-idx3-ubyte.gz\n26427392/26421880 [==============================] - 5s 0us/step\nDownloading data from https://storage.googleapis.com/tensorflow/tf-keras-datasets/t10k-labels-idx1-ubyte.gz\n8192/5148 [===============================================] - 0s 0us/step\nDownloading data from https://storage.googleapis.com/tensorflow/tf-keras-datasets/t10k-images-idx3-ubyte.gz\n4423680/4422102 [==============================] - 1s 0us/step\n"
],
[
"train_images.shape",
"_____no_output_____"
],
[
"plt.imshow(train_images[0])",
"_____no_output_____"
],
[
"train_labels[0]",
"_____no_output_____"
],
[
"class_names[train_labels[0]]",
"_____no_output_____"
],
[
"plt.figure(figsize=(3,3))\nfor i in range(5):\n plt.imshow(train_images[i])\n plt.xlabel(class_names[train_labels[i]])\n plt.show()",
"_____no_output_____"
],
[
"train_images_scaled = train_images / 255.0\ntest_images_scaled = test_images / 255.0",
"_____no_output_____"
],
[
"def get_model(hidden_layers=1):\n layers = [\n keras.layers.Flatten(input_shape=(28,28)),\n keras.layers.Dense(hidden_layers,activation='relu'),\n keras.layers.Dense(10,activation='sigmoid')\n ]\n # Your code goes here-----------START\n # Create Flatten input layers\n # Create hidden layers that are equal to hidden_layers argument in this function\n # Create output \n # Your code goes here-----------END\n model = keras.Sequential(layers)\n \n model.compile(optimizer='adam',\n loss='sparse_categorical_crossentropy',\n metrics=['accuracy'])\n \n return model",
"_____no_output_____"
],
[
"model = get_model(1)\nmodel.fit(train_images_scaled, train_labels, epochs=5)",
"Train on 60000 samples\nEpoch 1/5\n60000/60000 [==============================] - 9s 150us/sample - loss: 2.0261 - acc: 0.1561\nEpoch 2/5\n60000/60000 [==============================] - 6s 108us/sample - loss: 1.8501 - acc: 0.2155\nEpoch 3/5\n60000/60000 [==============================] - 7s 110us/sample - loss: 1.8044 - acc: 0.2192\nEpoch 4/5\n60000/60000 [==============================] - 6s 108us/sample - loss: 1.7823 - acc: 0.2303\nEpoch 5/5\n60000/60000 [==============================] - 7s 112us/sample - loss: 1.7682 - acc: 0.2470\n"
],
[
"np.argmax(model.predict(test_images_scaled)[2])",
"_____no_output_____"
],
[
"test_labels[2]",
"_____no_output_____"
],
[
"tf.config.experimental.list_physical_devices() ",
"_____no_output_____"
]
],
[
[
"5 Epochs performance comparison for 1 hidden layer",
"_____no_output_____"
]
],
[
[
"%%timeit -n1 -r1\nwith tf.device('/CPU:0'):\n cpu_model = get_model(1)\n cpu_model.fit(train_images_scaled, train_labels, epochs=5)",
"Train on 60000 samples\nEpoch 1/5\n60000/60000 [==============================] - 2s 38us/sample - loss: 1.9082 - acc: 0.2306\nEpoch 2/5\n60000/60000 [==============================] - 2s 40us/sample - loss: 1.6805 - acc: 0.3092\nEpoch 3/5\n60000/60000 [==============================] - 2s 35us/sample - loss: 1.5632 - acc: 0.3745\nEpoch 4/5\n60000/60000 [==============================] - 2s 36us/sample - loss: 1.4933 - acc: 0.3985\nEpoch 5/5\n60000/60000 [==============================] - 2s 35us/sample - loss: 1.4493 - acc: 0.4060\n11.5 s ± 0 ns per loop (mean ± std. dev. of 1 run, 1 loop each)\n"
],
[
"%%timeit -n1 -r1\nwith tf.device('/DML:0'):\n cpu_model = get_model(1)\n cpu_model.fit(train_images_scaled, train_labels, epochs=5)",
"Train on 60000 samples\nEpoch 1/5\n60000/60000 [==============================] - 7s 117us/sample - loss: 1.9268 - acc: 0.2043\nEpoch 2/5\n60000/60000 [==============================] - 7s 114us/sample - loss: 1.7043 - acc: 0.2268\nEpoch 3/5\n60000/60000 [==============================] - 7s 116us/sample - loss: 1.6001 - acc: 0.2763\nEpoch 4/5\n60000/60000 [==============================] - 7s 112us/sample - loss: 1.5426 - acc: 0.2884\nEpoch 5/5\n60000/60000 [==============================] - 7s 113us/sample - loss: 1.5036 - acc: 0.2951\n34.8 s ± 0 ns per loop (mean ± std. dev. of 1 run, 1 loop each)\n"
],
[
"%%timeit -n1 -r1\nwith tf.device('/DML:1'):\n cpu_model = get_model(1)\n cpu_model.fit(train_images_scaled, train_labels, epochs=5)",
"Train on 60000 samples\nEpoch 1/5\n60000/60000 [==============================] - 6s 94us/sample - loss: 1.9546 - acc: 0.2045\nEpoch 2/5\n60000/60000 [==============================] - 5s 89us/sample - loss: 1.7216 - acc: 0.2928\nEpoch 3/5\n60000/60000 [==============================] - 6s 93us/sample - loss: 1.6140 - acc: 0.3615\nEpoch 4/5\n60000/60000 [==============================] - 5s 91us/sample - loss: 1.5558 - acc: 0.3714\nEpoch 5/5\n60000/60000 [==============================] - 5s 86us/sample - loss: 1.5205 - acc: 0.3772\n27.6 s ± 0 ns per loop (mean ± std. dev. of 1 run, 1 loop each)\n"
]
],
[
[
"5 Epocs performance comparison with 5 hidden layers",
"_____no_output_____"
]
],
[
[
"%%timeit -n1 -r1\nwith tf.device('/CPU:0'):\n cpu_model = get_model(5)\n cpu_model.fit(train_images_scaled, train_labels, epochs=5)",
"Train on 60000 samples\nEpoch 1/5\n60000/60000 [==============================] - 3s 42us/sample - loss: 1.0238 - acc: 0.6050\nEpoch 2/5\n60000/60000 [==============================] - 2s 41us/sample - loss: 0.6844 - acc: 0.7768\nEpoch 3/5\n60000/60000 [==============================] - 2s 41us/sample - loss: 0.6238 - acc: 0.7948\nEpoch 4/5\n60000/60000 [==============================] - 2s 41us/sample - loss: 0.5928 - acc: 0.8037\nEpoch 5/5\n60000/60000 [==============================] - 3s 47us/sample - loss: 0.5737 - acc: 0.8083\n13.2 s ± 0 ns per loop (mean ± std. dev. of 1 run, 1 loop each)\n"
],
[
"%%timeit -n1 -r1\nwith tf.device('/DML:0'):\n cpu_model = get_model(5)\n cpu_model.fit(train_images_scaled, train_labels, epochs=5)",
"Train on 60000 samples\nEpoch 1/5\n60000/60000 [==============================] - 10s 167us/sample - loss: 0.8139 - acc: 0.7135\nEpoch 2/5\n60000/60000 [==============================] - 8s 133us/sample - loss: 0.5806 - acc: 0.8010\nEpoch 3/5\n60000/60000 [==============================] - 7s 119us/sample - loss: 0.5464 - acc: 0.8145s - los\nEpoch 4/5\n60000/60000 [==============================] - 7s 118us/sample - loss: 0.5262 - acc: 0.8201\nEpoch 5/5\n60000/60000 [==============================] - 8s 129us/sample - loss: 0.5115 - acc: 0.8247\n40.5 s ± 0 ns per loop (mean ± std. dev. of 1 run, 1 loop each)\n"
],
[
"%%timeit -n1 -r1\nwith tf.device('/DML:1'):\n cpu_model = get_model(5)\n cpu_model.fit(train_images_scaled, train_labels, epochs=5)",
"Train on 60000 samples\nEpoch 1/5\n60000/60000 [==============================] - 6s 95us/sample - loss: 0.9885 - acc: 0.6005\nEpoch 2/5\n60000/60000 [==============================] - 6s 93us/sample - loss: 0.6275 - acc: 0.7890\nEpoch 3/5\n60000/60000 [==============================] - 6s 95us/sample - loss: 0.5740 - acc: 0.8067\nEpoch 4/5\n60000/60000 [==============================] - 5s 91us/sample - loss: 0.5478 - acc: 0.8147\nEpoch 5/5\n60000/60000 [==============================] - 5s 91us/sample - loss: 0.5264 - acc: 0.82050s - loss: 0.5266\n28.5 s ± 0 ns per loop (mean ± std. dev. of 1 run, 1 loop each)\n"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
c5134e50292439092e74d545fd1186f86ac24128
| 7,506 |
ipynb
|
Jupyter Notebook
|
Lectures notebooks/(Lectures notebooks) netology Machine learning/21. Syntactic analysis and keyword selection/HW1_banki_TM-and-classification.ipynb
|
Alex110117/data_analysis
|
3cac3aac63d617b9fbd862788c778c2858445622
|
[
"MIT"
] | 2 |
2020-07-22T07:33:17.000Z
|
2021-12-13T18:45:41.000Z
|
Lectures notebooks/(Lectures notebooks) netology Machine learning/21. Syntactic analysis and keyword selection/HW1_banki_TM-and-classification.ipynb
|
sibalex/data_analysis
|
3cac3aac63d617b9fbd862788c778c2858445622
|
[
"MIT"
] | null | null | null |
Lectures notebooks/(Lectures notebooks) netology Machine learning/21. Syntactic analysis and keyword selection/HW1_banki_TM-and-classification.ipynb
|
sibalex/data_analysis
|
3cac3aac63d617b9fbd862788c778c2858445622
|
[
"MIT"
] | null | null | null | 32.634783 | 396 | 0.598588 |
[
[
[
"import json\n\nimport bz2\nimport regex\nfrom tqdm import tqdm\nfrom scipy import sparse",
"_____no_output_____"
],
[
"import pandas as pd\nimport numpy as np\nimport nltk\nimport matplotlib.pyplot as plt\nimport seaborn as sns\n%matplotlib inline\n%pylab inline",
"Populating the interactive namespace from numpy and matplotlib\n"
],
[
"responses = []\nwith bz2.BZ2File('banki_responses.json.bz2', 'r') as thefile:\n for row in tqdm(thefile):\n resp = json.loads(row)\n if not resp['rating_not_checked'] and (len(resp['text'].split()) > 0):\n responses.append(resp)",
"201030it [02:13, 1503.24it/s]\n"
]
],
[
[
"# Домашнее задание по NLP # 1 [100 баллов]\n## Классификация по тональности \n\nВ этом домашнем задании вам предстоит классифицировать по тональности отзывы на банки с сайта banki.ru. [Ссылка на данные](https://drive.google.com/open?id=1CPKtX5HcgGWRpzbWZ2fMCyqgHGgk21l2).\n\nДанные содержат непосредственно тексты отзывов, некоторую дополнительную информацию, а также оценку по шкале от 1 до 5. \n\nТексты хранятся в json-ах в массиве responses.\n\nПосмотрим на пример отзыва:",
"_____no_output_____"
]
],
[
[
"responses[99]",
"_____no_output_____"
]
],
[
[
"## Часть 1. Анализ текстов [40/100]\n\n1. Посчитайте количество отзывов в разных городах и на разные банки\n2. Постройте гистограмы длин слов в символах и в словах (не обязательно)\n3. Найдите 10 самых частых:\n * слов\n * слов без стоп-слов\n * лемм \n* существительных\n4. Постройте кривую Ципфа\n5. Ответьте на следующие вопросы:\n * какое слово встречается чаще, \"сотрудник\" или \"клиент\"?\n * сколько раз встречается слова \"мошенничество\" и \"доверие\"?\n6. В поле \"rating_grade\" записана оценка отзыва по шкале от 1 до 5. Используйте меру $tf-idf$, для того, чтобы найти ключевые слова и биграмы для положительных отзывов (с оценкой 5) и отрицательных отзывов (с оценкой 1)",
"_____no_output_____"
],
[
"## Часть 2. Тематическое моделирование [20/100]\n\n1. Постройте несколько тематических моделей коллекции документов с разным числом тем. Приведите примеры понятных (интерпретируемых) тем.\n2. Найдите темы, в которых упомянуты конкретные банки (Сбербанк, ВТБ, другой банк). Можете ли вы их прокомментировать / объяснить?\n\nЭта часть задания может быть сделана с использованием gensim. ",
"_____no_output_____"
],
[
"## Часть 3. Классификация текстов [40/100]\n\nСформулируем для простоты задачу бинарной классификации: будем классифицировать на два класса, то есть, различать резко отрицательные отзывы (с оценкой 1) и положительные отзывы (с оценкой 5). \n\n1. Составьте обучающее и тестовое множество: выберите из всего набора данных N1 отзывов с оценкой 1 и N2 отзывов с оценкой 5 (значение N1 и N2 – на ваше усмотрение). Используйте ```sklearn.model_selection.train_test_split``` для разделения множества отобранных документов на обучающее и тестовое. \n2. Используйте любой известный вам алгоритм классификации текстов для решения задачи и получите baseline. Сравните разные варианты векторизации текста: использование только униграм, пар или троек слов или с использованием символьных $n$-грам. \n3. Сравните, как изменяется качество решения задачи при использовании скрытых тем в качестве признаков:\n * 1-ый вариант: $tf-idf$ преобразование (```sklearn.feature_extraction.text.TfidfTransformer```) и сингулярное разложение (оно же – латентый семантический анализ) (```sklearn.decomposition.TruncatedSVD```), \n * 2-ой вариант: тематические модели LDA (```sklearn.decomposition.LatentDirichletAllocation```). \n\n\nИспользуйте accuracy и F-measure для оценки качества классификации. \n\nНиже написан примерный Pipeline для классификации текстов. \n\nЭта часть задания может быть сделана с использованием sklearn. ",
"_____no_output_____"
]
],
[
[
"from sklearn.pipeline import Pipeline\nfrom sklearn.ensemble import RandomForestClassifier\n\n# !!! На каждом этапе Pipeline нужно указать свои параметры\n# 1-ый вариант: tf-idf + LSI\n# 2-ой вариант: LDA\n\n# clf = Pipeline([\n# ('vect', CountVectorizer(analyzer = 'char', ngram_range={4,6})),\n# ('clf', RandomForestClassifier()),\n# ])\n\n\n\nclf = Pipeline([ \n ('vect', CountVectorizer()), \n ('tfidf', TfidfTransformer()), \n ('tm', TruncatedSVD()), \n ('clf', RandomForestClassifier())\n])\n",
"_____no_output_____"
]
],
[
[
"## Бонус [20]\n\nИспользуйте для классификации эмбеддинги слов. Улучшилось ли качество?",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
c51350c309f0c07d6d00adac0d4f059b57214e25
| 312,103 |
ipynb
|
Jupyter Notebook
|
_notebooks/2021-11-12-deeplearning.ipynb
|
saahithirao/bios-823-blog
|
23644cb773a0bdea3810cc807fb15b40387ce625
|
[
"Apache-2.0"
] | null | null | null |
_notebooks/2021-11-12-deeplearning.ipynb
|
saahithirao/bios-823-blog
|
23644cb773a0bdea3810cc807fb15b40387ce625
|
[
"Apache-2.0"
] | null | null | null |
_notebooks/2021-11-12-deeplearning.ipynb
|
saahithirao/bios-823-blog
|
23644cb773a0bdea3810cc807fb15b40387ce625
|
[
"Apache-2.0"
] | null | null | null | 501.773312 | 268,277 | 0.935675 |
[
[
[
"# Dive into Deep Learning, Classifying images",
"_____no_output_____"
],
[
"The goal of this blog post is to explain the process of training a deep learning model to classify images (pixels) of insects: beetles, cockroaches, and dragonflies. The neural network (model) will be evaluated on how it classfied the images using Shapley Additive Explanations.",
"_____no_output_____"
],
[
"The first step is to import all of the necessary libraries. This neural network will be using the tensorflow package and specifically, the keras module.",
"_____no_output_____"
]
],
[
[
"import necessary libraries\nimport matplotlib.pyplot as plt\nimport numpy as np\nimport pandas as pd\n\n#import tensorflow as tf\n#from tensorflow import keras\n#import shap\n\n#from PIL import Image\n#import urllib\nimport io \n\nimport random\nimport os\n\nfrom keras.preprocessing.image import ImageDataGenerator,load_img\nfrom keras.utils import to_categorical\nfrom sklearn.model_selection import train_test_split\n\nfrom tensorflow.keras import Sequential\nfrom tensorflow.keras.layers import Conv2D, MaxPooling2D, Dropout, Flatten, Dense, Activation, BatchNormalization",
"Using TensorFlow backend.\n"
]
],
[
[
"The next step is to specify the image size because all the images need to be of the same size in the model and specify three color channels.",
"_____no_output_____"
]
],
[
[
"image specifications\nwidth=200\nheight=200\nsize=(width,height)\nchannels = 3",
"_____no_output_____"
]
],
[
[
"Now, we will create the training data by finding the file paths of each image. The file paths of the images will be added to a training file list and the corresponding image classifiation (type of insect) will be added to a categories list. These lists are appended to a training dataframe.",
"_____no_output_____"
]
],
[
[
"training data\ntrainfiles = []\ncategories = []\n\nfor path in os.listdir(\"insects/train/beetles\"):\n full_path = os.path.join(\"insects/train/beetles\", path)\n if os.path.isfile(full_path):\n trainfiles.append(full_path)\n categories.append(\"beetles\")\n \nfor path in os.listdir(\"insects/train/cockroach\"):\n full_path = os.path.join(\"insects/train/cockroach\", path)\n if os.path.isfile(full_path):\n trainfiles.append(full_path)\n categories.append(\"cockroach\")\n\nfor path in os.listdir(\"insects/train/dragonflies\"):\n full_path = os.path.join(\"insects/train/dragonflies\", path)\n if os.path.isfile(full_path):\n trainfiles.append(full_path)\n categories.append(\"dragonflies\")\n\ndf_train = pd.DataFrame({\n 'filename': trainfiles,\n 'category': categories\n}) ",
"_____no_output_____"
]
],
[
[
"Similarly, we will create the testing data. The file paths of the images will be added to a test file list and the corresponding image classifiation (type of insect) will be added to a categories list. These lists are appended to a test dataframe. ",
"_____no_output_____"
]
],
[
[
"test data\ntestfiles = []\ncategories = []\n\nfor path in os.listdir(\"insects/test/beetles\"):\n full_path = os.path.join(\"insects/test/beetles\", path)\n if os.path.isfile(full_path):\n testfiles.append(full_path)\n categories.append(\"beetles\")\n \nfor path in os.listdir(\"insects/test/cockroach\"):\n full_path = os.path.join(\"insects/test/cockroach\", path)\n if os.path.isfile(full_path):\n testfiles.append(full_path)\n categories.append(\"cockroach\")\n\nfor path in os.listdir(\"insects/test/dragonflies\"):\n full_path = os.path.join(\"insects/test/dragonflies\", path)\n if os.path.isfile(full_path):\n testfiles.append(full_path)\n categories.append(\"dragonflies\")\n \ndf_test = pd.DataFrame({\n 'filename': testfiles,\n 'category': categories\n}) ",
"_____no_output_____"
]
],
[
[
"This neural network will use a convolution 2D neural net architecture. We will add layers sequentially and each one has separate biases and weights. The output and shape of each layer is shown below. The loss function is 'categorical_crossentropy' and while training the model, this function will be minimized.",
"_____no_output_____"
]
],
[
[
"# neural net model\nmodel=Sequential()\nmodel.add(Conv2D(32,(3,3),activation='relu',input_shape=(width,height,channels)))\nmodel.add(BatchNormalization())\nmodel.add(MaxPooling2D(pool_size=(2,2)))\nmodel.add(Dropout(0.25))\nmodel.add(Conv2D(64,(3,3),activation='relu'))\nmodel.add(BatchNormalization())\nmodel.add(MaxPooling2D(pool_size=(2,2)))\nmodel.add(Dropout(0.25))\nmodel.add(Conv2D(128,(3,3),activation='relu'))\nmodel.add(BatchNormalization())\nmodel.add(MaxPooling2D(pool_size=(2,2)))\nmodel.add(Dropout(0.25))\nmodel.add(Flatten())\nmodel.add(Dense(512,activation='relu'))\nmodel.add(BatchNormalization())\nmodel.add(Dropout(0.2))\nmodel.add(Dense(3,activation='softmax'))\nmodel.compile(loss='categorical_crossentropy',\n optimizer='rmsprop',metrics=['accuracy'])",
"_____no_output_____"
],
[
"model.summary()",
"_____no_output_____"
]
],
[
[
"The training data will be split into training and validation sets to be used in the model.",
"_____no_output_____"
]
],
[
[
"train_df,validate_df = train_test_split(df_train,test_size=0.2,\n random_state=42)\n\ntotal_train=train_df.shape[0]\ntotal_validate=df_test.shape[0]\nbatch_size = 10\n\ntrain_df = train_df.reset_index(drop=True)\nvalidate_df = validate_df.reset_index(drop=True)",
"_____no_output_____"
]
],
[
[
"After some research, ImageDataGenerator seemed like the most optimal solution to create this image classification model as it can efficiently load images in batches. The output shows the number of images in each data set. ",
"_____no_output_____"
]
],
[
[
"train_datagen = ImageDataGenerator(rotation_range=15,\n rescale=1./255,\n shear_range=0.1,\n zoom_range=0.2,\n horizontal_flip=True,\n width_shift_range=0.1,\n height_shift_range=0.1)\n\ntrain_generator = train_datagen.flow_from_dataframe(train_df, x_col='filename',y_col='category',\n target_size=size,\n class_mode='categorical',\n batch_size=batch_size)\n\n\nvalidation_datagen = ImageDataGenerator(rescale=1./255)\n\nvalidation_generator = validation_datagen.flow_from_dataframe(\n validate_df, \n x_col='filename',\n y_col='category',\n target_size=size,\n class_mode='categorical',\n batch_size=batch_size)\n\ntest_datagen = ImageDataGenerator(rotation_range=15,\n rescale=1./255,\n shear_range=0.1,\n zoom_range=0.2,\n horizontal_flip=True,\n width_shift_range=0.1,\n height_shift_range=0.1)\n\ntest_generator = test_datagen.flow_from_dataframe(df_test,x_col='filename',y_col='category',\n target_size=size,\n class_mode='categorical',\n batch_size=batch_size)",
"Found 815 validated image filenames belonging to 3 classes.\nFound 204 validated image filenames belonging to 3 classes.\nFound 180 validated image filenames belonging to 3 classes.\n"
]
],
[
[
"Now, we will fit the model on the training data and validate it on the validation data. Various epochs and batch sizes were tried and due to the time it took to run the model, 3 and 10 were chosen respectively. ",
"_____no_output_____"
]
],
[
[
"hist = model.fit(\n train_generator, \n epochs=3,\n validation_data=validation_generator,\n batch_size=batch_size,\n verbose=1\n)",
"Epoch 1/3\n82/82 [==============================] - 52s 640ms/step - loss: 0.6923 - accuracy: 0.7485 - val_loss: 0.6357 - val_accuracy: 0.7500\nEpoch 2/3\n82/82 [==============================] - 52s 629ms/step - loss: 0.5808 - accuracy: 0.7779 - val_loss: 0.4552 - val_accuracy: 0.8235\nEpoch 3/3\n82/82 [==============================] - 51s 623ms/step - loss: 0.6468 - accuracy: 0.7521 - val_loss: 1.3222 - val_accuracy: 0.5392\n"
]
],
[
[
"As the plot illustrates, the model does not have a high accuracy. A lot of layers were added to the model including batch normalization and dropout layers to make the code run more efficiently. ",
"_____no_output_____"
]
],
[
[
"fig, axes = plt.subplots(1,2,figsize=(12, 4))\nfor ax, measure in zip(axes, ['loss', 'accuracy']):\n ax.plot(hist.history[measure], label=measure)\n ax.plot(hist.history['val_' + measure], label='val_' + measure)\n ax.legend()\nplt.show()",
"_____no_output_____"
],
[
"model.save(\"model1.h5\")",
"_____no_output_____"
]
],
[
[
"The test accuracy is about 63%. ",
"_____no_output_____"
]
],
[
[
"test_loss, test_acc = model.evaluate(test_generator)\ntest_acc",
"18/18 [==============================] - 2s 125ms/step - loss: 1.0333 - accuracy: 0.6500\n"
]
],
[
[
"The following code manipulates the test data frame to be used for predictions.",
"_____no_output_____"
]
],
[
[
"test_df = pd.DataFrame({\n 'filename': testfiles\n})\nnb_samples = test_df.shape[0]",
"_____no_output_____"
]
],
[
[
"The following code makes predictions from the model.",
"_____no_output_____"
]
],
[
[
"model.predict(test_generator, steps=np.ceil(nb_samples/batch_size))",
"_____no_output_____"
]
],
[
[
"Checking the prediction of one image by feeding an image of a beetle. The classification was a beetle. Yay!",
"_____no_output_____"
]
],
[
[
"results={\n 0:'beetles',\n 1:'cockroach',\n 2:'dragonflies'\n}\nfrom PIL import Image\nimport numpy as np\nim=Image.open(\"insects/test/beetles/5556745.jpg\")\nim=im.resize(size)\nim=np.expand_dims(im,axis=0)\nim=np.array(im)\nim=im/255\npred=model.predict_classes([im])[0]\nprint(pred,results[pred])",
"0 beetles\n"
]
],
[
[
"We will describe how well the model performed using shapley additive explanations.",
"_____no_output_____"
]
],
[
[
"import shap",
"_____no_output_____"
]
],
[
[
"I tried the following code and several other versions of what is below to convert the training and test sets into numpy arrays to be used in the gradient explainer and for the shapley values. After analyzing the output of the train and test generator (code above), I realized that the data was in batches and I would need to unbatch the data for this to work and plot the images correctly. So, I scratched this idea and skipped to what is shown below. ",
"_____no_output_____"
]
],
[
[
"# xtrain = []\n# ytrain = []\n\n# xtrain=np.concatenate([train_generator.next()[0] for i in range(train_generator.__len__())])\n# ytrain=np.concatenate([train_generator.next()[1] for i in range(train_generator.__len__())])\n\n# xtest = []\n# ytest = []\n\n# xtest=np.concatenate([test_generator.next()[0] for i in range(test_generator.__len__())])\n# ytest=np.concatenate([test_generator.next()[1] for i in range(test_generator.__len__())])\n\n# ytest = np.where(ytest == 0, \"beetles\", np.where(ytest == 1, \"cockroach\", \"dragonflies\"))\n\n# explainer = shap.GradientExplainer(model, xtrain)\n# shap_vals = explainer.shap_values(xtest[:3])",
"_____no_output_____"
]
],
[
[
"The following code produces the shapley values on the test data using a gradient from the model and background (training data). ",
"_____no_output_____"
]
],
[
[
"explainer = shap.GradientExplainer(model, train_generator[0][0])\nshap_vals, index = explainer.shap_values(test_generator[0][0], ranked_outputs = 3)",
"_____no_output_____"
]
],
[
[
"Now, we will create a numpy array to label the images. ",
"_____no_output_____"
]
],
[
[
"names = ['beetles', 'cockroach', 'dragonflies']\nindex_names = np.vectorize(lambda x: names[x])(index)",
"_____no_output_____"
]
],
[
[
"Finally, we will plot the images and see what parts of the images are most important in creating the image classification. ",
"_____no_output_____"
]
],
[
[
"# shap.image_plot(shap_vals, test_generator[0][0], labels = index_names, show = False)\n# plt.savefig('shap.jpg')",
"_____no_output_____"
],
[
"from IPython import display\ndisplay.Image(\"./shap.jpg\")",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
c5135158a1107ccd0f901e46637f02bbbf38e4f8
| 3,287 |
ipynb
|
Jupyter Notebook
|
Lesson 5b - Practice data structures and loops in Python.ipynb
|
cesalga/Accelerated_Intro_to_CompBio_Part_2
|
8e9f4ccba2d95c1f7205277dcef07754ba1a3161
|
[
"CC-BY-4.0"
] | null | null | null |
Lesson 5b - Practice data structures and loops in Python.ipynb
|
cesalga/Accelerated_Intro_to_CompBio_Part_2
|
8e9f4ccba2d95c1f7205277dcef07754ba1a3161
|
[
"CC-BY-4.0"
] | null | null | null |
Lesson 5b - Practice data structures and loops in Python.ipynb
|
cesalga/Accelerated_Intro_to_CompBio_Part_2
|
8e9f4ccba2d95c1f7205277dcef07754ba1a3161
|
[
"CC-BY-4.0"
] | null | null | null | 29.088496 | 218 | 0.488896 |
[
[
[
"# Practice data structures\n\nWe will create a data structure to hold our Germplasm data (I have updated it to be a little bit more complex... now a germplasm may hold TWO alleles - i.e. one germplasm has connections to more than one gene)\n\nRepresent these data in Python - create a **single variable** that contains all of this data (using lists and dictionaries).\n\nCreate loops over that data structure that can print the three tables to the screen.\n\n\n\n",
"_____no_output_____"
],
[
"\n\n<center>stock table</center>\n\ngermplasm_id | amount | date | location \n --- | --- | --- | --- \n 2 | 5 | 10/5/2013 | Room 2234 \n 1 | 9.8 | 12/1/2015 | Room 998 \n\n\n-----------------------------\n\n\n<center>germplasm table</center>\n\nid | taxonid | alleles | genes \n --- |--- | --- | ---\n 1 | 4150 | def-1 | DEF\n 2 | 3701 | ap3, ag | AP3, AG\n \n--------------------------------\n\n<center>gene table</center>\n\n gene | gene_name | embl\n --- | --- | --- \n DEF | Deficiens | https://www.ebi.ac.uk/ena/data/view/AB516402\n AP3 | Apetala3 | https://www.ebi.ac.uk/ena/data/view/AF056541\n AG | Agamous | https://www.ebi.ac.uk/ena/data/view/AL161549\n ",
"_____no_output_____"
]
],
[
[
"# put your code here... take it one step at a time!\n# do just one table\n",
"_____no_output_____"
]
],
[
[
" <pre> </pre>",
"_____no_output_____"
],
[
"<span style=\"visibility:hidden;\">\nDEF = {'id': 'DEF', 'name': 'Deficiens', 'embl': 'http://AB123434'}\nAP3 = {'id': 'AP3', 'name': 'Apetala3', 'embl': 'http://AB123434'}\nAG = {'id': 'AG', 'name': 'Agamous', 'embl': 'http://AB123434'}\ngp1 = {'id': 1, 'taxon': 4150, 'alleles': ['def1'], 'genes': [DEF]}\ngp2 = {'id': 2, 'taxon': 4150, 'alleles': ['ap3', 'ag'], 'genes': [AP3, AG]}\nstock1 = {'germplasm': gp2, 'amount': 5, 'date': '10/5/2013', 'location': 'Room 2234' }\nstock2 = {'germplasm': gp1, 'amount': 9.8, 'date': '12/1/2015', 'location': 'Room 998' }\ndatabase = [stock1, stock2]\nfor stock in database:\n print(stock['germplasm']['id'], \", \", stock['amount'],\",\",stock['date'],\",\",stock['location'])\n</span>",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
c51355489672a84c3e97a7a7d4b9b817989057fc
| 11,142 |
ipynb
|
Jupyter Notebook
|
7.24.ipynb
|
ygyw/yg
|
8d3efe5dedc01c2275fc03de494667808fc2c255
|
[
"Apache-2.0"
] | null | null | null |
7.24.ipynb
|
ygyw/yg
|
8d3efe5dedc01c2275fc03de494667808fc2c255
|
[
"Apache-2.0"
] | null | null | null |
7.24.ipynb
|
ygyw/yg
|
8d3efe5dedc01c2275fc03de494667808fc2c255
|
[
"Apache-2.0"
] | null | null | null | 18.235679 | 222 | 0.427841 |
[
[
[
"# 更多字符串和特殊方法\n- 前面我们已经学了类,在Python中还有一些特殊的方法起着非常重要的作用,这里会介绍一些特殊的方法和运算符的重载,以及使用特殊方法设计类",
"_____no_output_____"
],
[
"\n## str 类\n- 一个str对象是不可变的,也就是说,一旦创建了这个字符串,那么它的内容在认为不改变的情况下是不会变的\n- s1 = str()\n- s2 = str('welcome to Python')",
"_____no_output_____"
],
[
"## 创建两个对象,分别观察两者id\n- id为Python内存地址",
"_____no_output_____"
]
],
[
[
"a = id(100)\nb = id(12)",
"_____no_output_____"
],
[
"a is b",
"_____no_output_____"
]
],
[
[
"## 处理字符串的函数\n- len\n- max\n- min\n- 字符串一切是按照ASCII码值进行比较",
"_____no_output_____"
],
[
"## 下角标运算符 []\n- 一个字符串是一个字符序列,可以通过索引进行访问\n- 观察字符串是否是一个可迭代序列 \\__iter__",
"_____no_output_____"
],
[
"## 切片 [start: end]\n- start 默认值为0\n- end 默认值为-1",
"_____no_output_____"
],
[
"## 链接运算符 + 和复制运算符 *\n- \\+ 链接多个字符串,同时''.join()也是\n- \\* 复制多个字符串",
"_____no_output_____"
],
[
"## in 和 not in 运算符\n- in :判断某个字符是否在字符串内 \n- not in :判断某个字符是否不在字符串内\n- 返回的是布尔值",
"_____no_output_____"
]
],
[
[
"a3 = 'yg is Superman)'\n'yg' in a3",
"_____no_output_____"
],
[
"def ",
"_____no_output_____"
]
],
[
[
"## 比较字符串\n- ==, !=, >=, <=, >, <\n- 依照ASCII码值进行比较",
"_____no_output_____"
],
[
"## 测试字符串\n\n- 注意:\n> - isalnum() 中是不能包含空格,否则会返回False",
"_____no_output_____"
]
],
[
[
"a = '111'\ncount1 = 0\ncount2 = 0\ncount3 = 0\nfor i in a:\n if i.islower() is True:\n count1 +=1\n if i.isupper() is True:\n count2 +=1\n if i.isdigit() is True:\n count3 +=1\nelse:\n if count1 == 0:\n print('密码必须含有小写字母')\n if count2 == 0:\n print('密码必须含有大写')\n if count3 == 0:\n print('密码必须含有数字')\n if count1 !=0 and count2 !=0 and count3 !=0:\n print('密码设置成功')\nprint(count1,count2,count3)",
"密码必须含有小写字母\n密码必须含有大写\n0 0 3\n"
],
[
"n = 'a b'\nn.isalpha()",
"_____no_output_____"
]
],
[
[
"## 搜索子串\n",
"_____no_output_____"
],
[
"## 转换字符串\n",
"_____no_output_____"
],
[
"## 删除字符串\n",
"_____no_output_____"
]
],
[
[
"a = ' dsdss'\na.lstrip()",
"_____no_output_____"
],
[
"a = ' skjk kjk lk l;k; dkjsk '\na.replace(' ','')",
"_____no_output_____"
]
],
[
[
"## 格式化字符串\n",
"_____no_output_____"
],
[
"## EP:\n- 1\n\n- 2 \n 随机参数100个数字,将www.baidu.com/?page=进行拼接",
"_____no_output_____"
],
[
"## Python高级使用方法 -- 字符串\n- 我们经常使用的方法实际上就是调用Python的运算重载\n",
"_____no_output_____"
],
[
"# Homework\n- 1\n",
"_____no_output_____"
]
],
[
[
"ssn = input('输入安全号码:')\nyi = ssn[0:3]\ner = ssn[4:6]\nsan = ssn[7:11]\nif ssn.__len__() = 11:\n if yi.isdigit() and er.isdigit() and san.isdigit() is True:\n if ssn[3] == '-' and ssn[6] == '-' :\n print('Valid SSN')\n else:\n \n print('Invalid SSN')\n else:\n print('Invalid SSN')\nelse:\n print('Invalid SSN')",
"输入安全号码:123-12-1234\nValid SSN\n"
]
],
[
[
"- 2\n",
"_____no_output_____"
]
],
[
[
"a = 'dsds'\nb = 'sd'\na.find(b)",
"_____no_output_____"
]
],
[
[
"- 3\n",
"_____no_output_____"
]
],
[
[
"pwd = input('输入密码:')\ncount1 = 0\ncount3 = 0\nif pwd.__len__() >= 8:\n for i in pwd:\n if i.islower() or i.isupper() is True:\n count1 +=1\n if i.isdigit() is True:\n count3 +=1\n else:\n if count1 !=0 and count3 >=2:\n print('Valid password')\n else:\n print('invalid password')\nelse:\n print('invalid password')\n\n",
"输入密码:...adsadas\ninvalid password\n"
]
],
[
[
"- 4\n",
"_____no_output_____"
]
],
[
[
"\ndef countLetters(s):\n counts = 0\n for i in s:\n if i.islower() or i.isupper() is True:\n counts += 1\n print('字母个数为:'+ str(counts))",
"_____no_output_____"
],
[
"countLetters('123123a')",
"字母个数为:1\n"
]
],
[
[
"- 5\n",
"_____no_output_____"
]
],
[
[
"def getNumber(uppercaseLetter):\n number = uppercaseLetter\n ",
"_____no_output_____"
]
],
[
[
"- 6\n",
"_____no_output_____"
]
],
[
[
"def reverse(s):\n fanxiang = ''\n c = s.__len__()-1\n for i in range(s.__len__()):\n fanxiang = s[c]\n c -=1\n print(fanxiang,end='')",
"_____no_output_____"
],
[
"reverse('asdf')",
"fdsa"
]
],
[
[
"- 7\n",
"_____no_output_____"
],
[
"- 8\n",
"_____no_output_____"
]
],
[
[
"num = str(input('输入:'))\njiaoyan = 10 - (int(num[0]) + 3 * int(num[1]) + int(num[2]) + 3 * int(num[3]) + int(num[4]) + 3 * int(num[5]) + int(num[6]) + 3 * int(num[7]) + int(num[8]) + 3 * int(num[9]) + int(num[10]) + 3 * int(num[11])) % 10\nprint(num + str(jiaoyan))",
"输入:978013213080\n9780132130806\n"
]
],
[
[
"- 9\n",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
c51364da886f6a083bf6b4b2ae31f242d3949d24
| 6,236 |
ipynb
|
Jupyter Notebook
|
finance/basics/todos.ipynb
|
pangyuteng/aigonewrong
|
98a2c7a172be4664fc372d581cef5f23cf317b51
|
[
"MIT"
] | 8 |
2021-01-06T22:04:39.000Z
|
2022-02-22T19:38:14.000Z
|
finance/basics/todos.ipynb
|
pangyuteng/aigonewrong
|
98a2c7a172be4664fc372d581cef5f23cf317b51
|
[
"MIT"
] | null | null | null |
finance/basics/todos.ipynb
|
pangyuteng/aigonewrong
|
98a2c7a172be4664fc372d581cef5f23cf317b51
|
[
"MIT"
] | 5 |
2020-11-21T20:46:19.000Z
|
2021-08-08T08:47:19.000Z
| 38.257669 | 712 | 0.617223 |
[
[
[
"_= \"\"\"\nref https://www.reddit.com/r/algotrading/comments/e44pdd/list_of_stock_tickers_from_yahoo/\n\nhttps://old.nasdaq.com/screening/companies-by-name.aspx?letter=0&exchange=nasdaq&render=download\n\nAMEX\n\nhttps://old.nasdaq.com/screening/companies-by-name.aspx?letter=0&exchange=amex&render=download\n\nNYSE\n\nhttps://old.nasdaq.com/screening/companies-by-name.aspx?letter=0&exchange=nyse&render=download\n\"\"\"",
"_____no_output_____"
],
[
"# look for high vol high vol.\n# assume option would be too.\n# compute historical implied volatility?\n# suggest trading ideas.\n# depending on: vol and mean ret are relatively high or low points and their dp and ddp.",
"_____no_output_____"
],
[
"### Can you actually apply any of this to gain an edge when trading?\n\n+ Now I don't really buy technical analysis, but the immediate indicator that comes to mind is \"relative strength index\", obviously when you mention TA, we are implying trying to forcast a price trend, using historical data and pattern which has been proven to not be successful, albeit many successful traders swear by TAs. Thus here we will demonstrate how well/badly our 2 mean reverting time series can forecasting future price changes compared to buy-and-hold.\n\n+ Perhaps if you split the 2D distribution plot of rolling ret mean and hist volatility into 4 quandrants (show below) you can opt to deploy different option strategies accordingly.\n\n```\nlets contrain ourselves to the below per BAT's talk linked below!\nShort Strangle, Iron Condors, Credit Spread, Diagnal Spread, Ratio Spread, Broken Wing Butterfly\n```\n+ Tony Battisa, Tastytrade, How to Use Options Strategies & Key Mechanics https://www.youtube.com/watch?v=T6uA_XHunRc\n\n",
"_____no_output_____"
],
[
"#\n# ^ \n# high vol | high vol\n# low ret | high ret\n# -----------|---------->\n# low vol | low vol\n# low ret | high ret\n#\n# \n# high vol, low ret -> short put (or credit spread)\n# high vol, high ret -> short call (or credit spread) \n# high vol, mid ret -> short strangle (or iron condor)\n# mid vol, low ret -> Ratio Spread (sell 2 otm puts, buy 1 atm put)\n# mid vol, high ret -> Ratio Spread (sell 2 otm call, buy 1 atm call)\n# low vol, low ret -> Broken Wing Butter Fly \n# low vol, high ret -> Broken Wing Butter Fly \n# low vol, mid ret -> Diagnal to bet on vol increase.\n#\n# product idea. deploy below as a website, earn ad revenue.\n\n\n\n# since both signals are likely mean reverting\n# and assuming realized volatility tracks implied volatilityvol_change\n# \n# by sectioning the 2 changes to zones, we can accordingly decide what strategy to deploy\n\n# if vol increase, price increase - diagonal - short front month call, long back month call\n# if vol increase, price no-change - diagonal - short call strangle, long back month?\n# if vol increase, price decrease - diagonal - short front month put, long back month put\n\n# if vol decrease, price increase - short put\n# if vol decrease, price no-change - iron condor\n# if vol decrease, price decrease - short call\n\n# https://www.youtube.com/watch?v=T6uA_XHunRc, ratios spreads or broken wing butter fly\n# if vol no-change, price increase - short put ratio spread\n# if vol no-change, price no-change - iron condor ratio spreads?\n# if vol no-change, price decrease - short call ratio spread",
"_____no_output_____"
],
[
"# to simplify backtesting. we will just see if we can predict the trend\n# a win for each trade gets a +1 a loss for each trade gets a -1\n# for the same period, for buy and hold, +1 means price ret in that period is > 0.",
"_____no_output_____"
],
[
"np.random.rand(10,10)",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
c5136a2b7d41aef6b4e56275b4a9ff045eabf00d
| 13,840 |
ipynb
|
Jupyter Notebook
|
RandomForest.ipynb
|
Harish239/Unsupervised_Learning_Clustering
|
815fcd604e6db6e9876b6662899e3f8af27a8f46
|
[
"MIT"
] | null | null | null |
RandomForest.ipynb
|
Harish239/Unsupervised_Learning_Clustering
|
815fcd604e6db6e9876b6662899e3f8af27a8f46
|
[
"MIT"
] | null | null | null |
RandomForest.ipynb
|
Harish239/Unsupervised_Learning_Clustering
|
815fcd604e6db6e9876b6662899e3f8af27a8f46
|
[
"MIT"
] | null | null | null | 32.186047 | 100 | 0.456358 |
[
[
[
"import numpy as np\nimport pandas as pd\nimport random",
"_____no_output_____"
],
[
"def bootstrapdf(df):\n df = df.sample(frac=1, replace=True)\n return df",
"_____no_output_____"
],
[
"def check_for_leaf(df,counter, min_samples, max_depth):\n unique_classes = np.unique(df)\n if len(unique_classes) == 1 or len(df)<=min_samples or counter==max_depth:\n labelcol = df\n uniq_cls, cnt = np.unique(labelcol, return_counts=True)\n classification = unique_classes[cnt.argmax()]\n return classification\n else:\n return False ",
"_____no_output_____"
],
[
"def gini_imp_test(df, col_index):\n df.reset_index(inplace = True, drop = True)\n classes = df.iloc[:,-1]\n feature = df.iloc[:,col_index]\n if len(feature.unique()) == 2:\n gini_imp = 0\n for i in np.unique(feature):\n idx = np.where(feature == i)\n label = classes.loc[idx].values\n a, b = np.unique(label, return_counts = True)\n list1 = [(i/sum(b))**2 for i in b]\n prob = 1 - sum(list1)\n wt = len(idx[0]) / df.shape[0]\n gini_imp += wt * prob\n return gini_imp, i\n else:\n label = np.sort(feature.unique())[1:-1]\n best_gini_imp = float('inf')\n split_val = 0\n for i in label:\n idx1 = np.where(feature > i)\n idx2 = np.where(feature <= i)\n if len(idx1[0]) > 2 and len(idx2[0]) > 2:\n \n b1, b1cnt = np.unique(classes.loc[idx1].values, return_counts = True)\n b2, b2cnt = np.unique(classes.loc[idx2].values, return_counts = True)\n list1 = [(i/sum(b1cnt))**2 for i in b1cnt]\n list2 = [(i/sum(b2cnt))**2 for i in b2cnt]\n prob1 = 1 - sum(list1)\n prob2 = 1 - sum(list2)\n gini = ((sum(b1cnt)/df.shape[0])*prob1) + ((sum(b2cnt)/df.shape[0])*prob2) \n if gini < best_gini_imp:\n best_gini_imp = gini\n split_val = i\n else:\n continue \n return best_gini_imp, split_val",
"_____no_output_____"
],
[
"def best_node(df, col_list):\n best_gini_imp = float('inf')\n value = 0\n col = 0\n for i in col_list:\n gini, val = gini_imp_test(df, i) \n if gini < best_gini_imp:\n best_gini_imp = gini\n value = val\n col = i\n return col, value",
"_____no_output_____"
],
[
"def split_df(df, col_index, split_val):\n feature = df.iloc[:,col_index]\n if feature.dtypes == object:\n temp1 = df[df.iloc[:,col_index] == split_val]\n temp2 = df[df.iloc[:,col_index] != split_val]\n return temp1, temp2\n elif feature.dtypes != object:\n temp1 = df[df.iloc[:,col_index] <= split_val]\n temp2 = df[df.iloc[:,col_index] >= split_val]\n temp1.reset_index(inplace = True, drop = True)\n temp2.reset_index(inplace = True, drop = True)\n return temp1, temp2",
"_____no_output_____"
],
[
"def check_purity(data):\n \n label_column = data[:, -1]\n unique_classes = np.unique(label_column)\n\n if len(unique_classes) == 1:\n return True\n else:\n return False",
"_____no_output_____"
],
[
"def classify_data(data):\n \n label_column = data[:,-1]\n unique_classes, counts_unique_classes = np.unique(label_column, return_counts=True)\n index = counts_unique_classes.argmax()\n classification = unique_classes[index]\n return classification\n ",
"_____no_output_____"
],
[
"def metrics(ts_lb,answer):\n TN = 0\n TP = 0\n FN = 0\n FP = 0\n for i,j in zip(ts_lb,answer):\n if j==1 and i==1:\n TP += 1\n elif(j==1 and i==0):\n FN += 1\n elif(j==0 and i==1):\n FP += 1\n elif(j==0 and i==0):\n TN += 1\n Accuracy = (TP + TN)/(TP + FP + TN + FN)\n Precision = TP/(TP + FP)\n Recall = TP/(TP + FN)\n f1_score = (2*Precision*Recall)/(Precision + Recall)\n return Accuracy, Precision, Recall, f1_score",
"_____no_output_____"
],
[
"def decision_tree(df, columns, num_features, counter = 0, min_samples = 10, max_depth = 5):\n if (check_purity(df.values)) or (counter == max_depth) or (len(df) < min_samples):\n classification = classify_data(df.values)\n \n return classification\n \n else:\n counter += 1\n col_list = random.sample(columns, num_features)\n column, value = best_node(df, col_list)\n if df.iloc[:,column].dtype == object:\n columns.remove(column)\n branch1, branch2 = split_df(df, column, value)\n if len(branch1) == 0 or len(branch2) == 0:\n classification = classify_data(df.values)\n return classification\n \n query = \"{} <= {}\".format(column, value)\n branch = {query: []}\n\n left_branch = decision_tree(branch1, columns, num_features, counter)\n right_branch = decision_tree(branch2, columns, num_features, counter)\n\n if left_branch == right_branch:\n branch = left_branch\n else:\n branch[query].append(left_branch)\n branch[query].append(right_branch)\n return branch",
"_____no_output_____"
],
[
"def random_forest(df, num_trees, num_features):\n trees = []\n for i in range(num_trees):\n df = bootstrapdf(df)\n columns = list(df.iloc[:,:-1].columns)\n tree = decision_tree(df, columns, num_features)\n trees.append(tree)\n return trees",
"_____no_output_____"
],
[
"def predict(model, test_data):\n classes = []\n for tree in model:\n cls = []\n for i in range(len(test_data)):\n t = tree\n col,_,val = list(t.keys())[0].split()\n col = int(col)\n try:\n val = float(val)\n except:\n val = str(val)\n key = list(t.keys())[0]\n key_val = t[key]\n while True: \n if test_data.iloc[i,col] <= val:\n t = t[key][0]\n if type(t) != dict:\n cls.append(t)\n break\n else:\n col,_,val = list(t.keys())[0].split()\n col = int(col)\n try:\n val = float(val)\n except:\n val = str(val)\n key = list(t.keys())[0]\n key_val = t[key]\n else:\n t = t[key][1]\n if type(t) != dict:\n cls.append(t)\n break\n else:\n col,_,val = list(t.keys())[0].split()\n col = int(col)\n try:\n val = float(val)\n except:\n val = str(val)\n key = list(t.keys())[0]\n key_val = t[key]\n cls = [int(i) for i in cls]\n classes.append(cls)\n classes = np.array(classes)\n final_class = []\n for i in range(len(test_data)):\n unique_classes, counts_unique_classes = np.unique(classes[:,i], return_counts=True)\n index = counts_unique_classes.argmax()\n classification = unique_classes[index]\n final_class.append(classification)\n final_class\n test_data[\"Class\"] = final_class\n return test_data",
"_____no_output_____"
],
[
"def k_fold(df):\n num_trees = int(input(\"Enter number of trees: \"))\n num_features = int(input(\"Enter number of features for each split: \"))\n k = int(input(\"Enter k value: \"))\n metrics_list = []\n for i in range(k):\n splitdfs = np.array_split(df, k)\n test = splitdfs[i]\n del(splitdfs[i])\n train = pd.concat(splitdfs)\n test.reset_index(inplace = True, drop = True)\n train.reset_index(inplace = True, drop = True) \n actual = test.iloc[:,-1]\n test = test.iloc[:,:-1]\n model = random_forest(train, num_trees, num_features)\n results = predict(model, test)\n Accuracy, Precision, Recall, f1_score = metrics(actual, results[\"Class\"])\n metrics_list.append([Accuracy, Precision, Recall, f1_score])\n metrics_list = np.array(metrics_list)\n metrics_list = np.mean(metrics_list, axis = 0)\n print(\"Accuracy: \",metrics_list[0])\n print(\"Precision: \",metrics_list[1])\n print(\"Recall: \",metrics_list[2])\n print(\"f1_score: \",metrics_list[3])\n return metrics_list\n \n ",
"_____no_output_____"
],
[
"df1 = pd.read_csv(\"project3_dataset1.txt\", sep = '\\t', header=None)\nk_fold(df1)",
"Enter number of trees: 5\nEnter number of features for each split: 3\nEnter k value: 10\nAccuracy: 0.9348997493734335\nPrecision: 0.8793594721240956\nRecall: 0.9506969696969698\nf1_score: 0.9120824299912176\n"
],
[
"df2 = pd.read_csv(\"project3_dataset2.txt\", sep = '\\t', header=None)\nk_fold(df2)",
"Enter number of trees: 5\nEnter number of features for each split: 3\nEnter k value: 10\nAccuracy: 0.6406567992599445\nPrecision: 0.36184248500037974\nRecall: 0.4885966810966812\nf1_score: 0.39605076241748566\n"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
c5136a42d509aed62d3b2eca2f557e2f1057bbf4
| 104,881 |
ipynb
|
Jupyter Notebook
|
module1-decision-trees/LS_DS_411B_ipywidgets_Decision_Trees.ipynb
|
valogonor/DS-Unit-4-Sprint-1-Tree-Ensembles
|
ce80748bb17c8e3a4f061bc33b429a4b5764fe01
|
[
"MIT"
] | 71 |
2019-03-05T04:44:48.000Z
|
2022-03-24T09:47:48.000Z
|
13-Tree-Ensembles/01B-ipywidgets-Decision-Trees.ipynb
|
abdelrhman2023/Data-Science-Tutorial-By-Lambda-School
|
c145f5cc0559ee8ba7260b53e011c165e842fde0
|
[
"MIT"
] | 4 |
2019-03-18T14:36:24.000Z
|
2019-03-22T16:57:32.000Z
|
13-Tree-Ensembles/01B-ipywidgets-Decision-Trees.ipynb
|
abdelrhman2023/Data-Science-Tutorial-By-Lambda-School
|
c145f5cc0559ee8ba7260b53e011c165e842fde0
|
[
"MIT"
] | 56 |
2019-03-18T10:29:49.000Z
|
2021-02-14T12:20:33.000Z
| 357.955631 | 59,928 | 0.938416 |
[
[
[
"_Lambda School Data Science — Tree Ensembles_ \n\n# Decision Trees — with ipywidgets!",
"_____no_output_____"
],
[
"### Notebook requirements\n- [ipywidgets](https://ipywidgets.readthedocs.io/en/stable/examples/Using%20Interact.html): works in Jupyter but [doesn't work on Google Colab](https://github.com/googlecolab/colabtools/issues/60#issuecomment-462529981)\n- [mlxtend.plotting.plot_decision_regions](http://rasbt.github.io/mlxtend/user_guide/plotting/plot_decision_regions/): `pip install mlxtend`",
"_____no_output_____"
],
[
"## Regressing a wave",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nfrom sklearn.model_selection import train_test_split\n\n# Example from http://scikit-learn.org/stable/auto_examples/tree/plot_tree_regression.html\ndef make_data():\n import numpy as np\n rng = np.random.RandomState(1)\n X = np.sort(5 * rng.rand(80, 1), axis=0)\n y = np.sin(X).ravel()\n y[::5] += 2 * (0.5 - rng.rand(16))\n return X, y\n\nX, y = make_data()\n\nX_train, X_test, y_train, y_test = train_test_split(\n X, y, test_size=0.25, random_state=42)\n\nplt.scatter(X_train, y_train)\nplt.scatter(X_test, y_test);",
"_____no_output_____"
],
[
"from sklearn.tree import DecisionTreeRegressor\n\ndef regress_wave(max_depth):\n tree = DecisionTreeRegressor(max_depth=max_depth)\n tree.fit(X_train, y_train)\n print('Train R^2 score:', tree.score(X_train, y_train))\n print('Test R^2 score:', tree.score(X_test, y_test))\n plt.scatter(X_train, y_train)\n plt.scatter(X_test, y_test)\n plt.step(X, tree.predict(X))\n plt.show()",
"_____no_output_____"
],
[
"from ipywidgets import interact\ninteract(regress_wave, max_depth=(1,8,1));",
"_____no_output_____"
]
],
[
[
"## Classifying a curve",
"_____no_output_____"
]
],
[
[
"import numpy as np\ncurve_X = np.random.rand(1000, 2) \ncurve_y = np.square(curve_X[:,0]) + np.square(curve_X[:,1]) < 1.0\ncurve_y = curve_y.astype(int)",
"_____no_output_____"
],
[
"from sklearn.linear_model import LogisticRegression\nfrom mlxtend.plotting import plot_decision_regions\n\nlr = LogisticRegression(solver='lbfgs')\nlr.fit(curve_X, curve_y)\nplot_decision_regions(curve_X, curve_y, lr, legend=False)\nplt.axis((0,1,0,1));",
"_____no_output_____"
],
[
"from sklearn.tree import DecisionTreeClassifier\n\ndef classify_curve(max_depth):\n tree = DecisionTreeClassifier(max_depth=max_depth)\n tree.fit(curve_X, curve_y)\n plot_decision_regions(curve_X, curve_y, tree, legend=False)\n plt.axis((0,1,0,1))\n plt.show()",
"_____no_output_____"
],
[
"interact(classify_curve, max_depth=(1,8,1));",
"_____no_output_____"
]
],
[
[
"## Titanic survival, by age & fare",
"_____no_output_____"
]
],
[
[
"import seaborn as sns\nfrom sklearn.impute import SimpleImputer\n\ntitanic = sns.load_dataset('titanic')\nimputer = SimpleImputer()\ntitanic_X = imputer.fit_transform(titanic[['age', 'fare']])\ntitanic_y = titanic['survived'].values",
"_____no_output_____"
],
[
"from sklearn.linear_model import LogisticRegression\nfrom mlxtend.plotting import plot_decision_regions\n\nlr = LogisticRegression(solver='lbfgs')\nlr.fit(titanic_X, titanic_y)\nplot_decision_regions(titanic_X, titanic_y, lr, legend=False);\nplt.axis((0,75,0,175));",
"_____no_output_____"
],
[
"def classify_titanic(max_depth):\n tree = DecisionTreeClassifier(max_depth=max_depth)\n tree.fit(titanic_X, titanic_y)\n plot_decision_regions(titanic_X, titanic_y, tree, legend=False)\n plt.axis((0,75,0,175))\n plt.show()",
"_____no_output_____"
],
[
"interact(classify_titanic, max_depth=(1,8,1));",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
c51376d975c10c4af7170a32b885a2ac4946df54
| 94,932 |
ipynb
|
Jupyter Notebook
|
reports/Games.ipynb
|
ubco-mds-2020-labs/dashboard-project-games-dashboard
|
3dbf2c59e590e582a6c3677f57394c6beb0d27de
|
[
"MIT"
] | null | null | null |
reports/Games.ipynb
|
ubco-mds-2020-labs/dashboard-project-games-dashboard
|
3dbf2c59e590e582a6c3677f57394c6beb0d27de
|
[
"MIT"
] | 2 |
2021-03-08T13:55:12.000Z
|
2021-03-10T18:38:17.000Z
|
reports/Games.ipynb
|
ubco-mds-2020-labs/dashboard-project-games-dashboard
|
3dbf2c59e590e582a6c3677f57394c6beb0d27de
|
[
"MIT"
] | 2 |
2021-02-26T20:33:54.000Z
|
2021-03-07T19:47:41.000Z
| 46.741507 | 5,009 | 0.484663 |
[
[
[
"### Video Games Dataset: EDA\n#### 1. Describe Dataset\n- **Who:** The data was acquired from Kaggle and supplied by the user Gregory Smith (https://www.kaggle.com/gregorut/videogamesales). The data was scraped from www.vgchartz.com. \n- **What:** The dataset contains a list of video games with sales greater than 100,000 from 1980 to 2020. It contains information such as the platform the game was made available, year of release, genre, publisher, sales in NA, sales in JP, sales in EU, sales in the rest of the world and global sales (total). The data set also includes the rank of games in terms of overall sales. **NOTE: Sales are in millions**\n- **When:** The data set was last updated 4 years ago but contains games released from 1980 to seemingly 2020. \n- **Why:** The video game industry is a very competitive yet profitable industry. While big companies with large amounts of resources have an edge over smaller companies, we have recently seen many small companies finding huge success. Not only in game creation but in the case of streamers for example, playing a game before it becomes mainstream might give you an edge against bigger name streamers. With this data set, we are able to gain insight into general idea such as performance of companies, most popular titles and genres. We are also able to dive deeper and look at changing genre popularities over time, regional preference in game genres/platforms, upcoming developer etc. \n- **How:** The data set was scraped from the www.vgzchartz.com website using BeautifulSoup. The scraping script can be found here (https://github.com/GregorUT/vgchartzScrape)",
"_____no_output_____"
],
[
"#### 2. Load Dataset",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nimport altair as alt\nfrom altair_saver import save\nalt.renderers.enable('mimetype')\nalt.data_transformers.enable('data_server')",
"_____no_output_____"
],
[
"game = pd.read_csv(\"vgsales.csv\")",
"_____no_output_____"
]
],
[
[
"#### 3. Explore Dataset",
"_____no_output_____"
]
],
[
[
"game.info()\n\nprint(\"\\nPlatform:\\n\",game.Platform.unique(),\"\\nCount: \",game.Platform.nunique())\nprint(\"\\nYear\\n\",game.Year.unique(),\"\\nCount: \",game.Year.nunique())\nprint(\"\\nGenre\\n\",game.Genre.unique(),\"\\nCount: \",game.Genre.nunique())\nprint(\"\\nPublishers\\n\",game.Publisher.unique()[0:15],\"\\nCount: \",game.Publisher.nunique())\n\nprint(game.sort_values(\"NA_Sales\",ascending=False).head(5).iloc[:,0:6])\nprint(game.sort_values(\"EU_Sales\",ascending=False).head(5).iloc[:,0:6])\nprint(game.sort_values(\"JP_Sales\",ascending=False).head(5).iloc[:,0:6])\nprint(game.sort_values(\"Global_Sales\",ascending=False).head(5).iloc[:,0:6])\n\nprint(game.groupby(\"Year\").size())\nprint(game.groupby(\"Genre\").size())\nprint(game.groupby(\"Platform\").size())\n\nnulls = game[game.isna().any(axis=1)] #List of games with nulls in any field \ngame.sort_values(\"Name\").loc[game.Name.isin(game.Name[game.Name.duplicated()]),[\"Name\",\"Platform\"]].head(15) #Game titles that show up on multiple platforms ",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 16598 entries, 0 to 16597\nData columns (total 11 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 Rank 16598 non-null int64 \n 1 Name 16598 non-null object \n 2 Platform 16598 non-null object \n 3 Year 16327 non-null float64\n 4 Genre 16598 non-null object \n 5 Publisher 16540 non-null object \n 6 NA_Sales 16598 non-null float64\n 7 EU_Sales 16598 non-null float64\n 8 JP_Sales 16598 non-null float64\n 9 Other_Sales 16598 non-null float64\n 10 Global_Sales 16598 non-null float64\ndtypes: float64(6), int64(1), object(4)\nmemory usage: 1.4+ MB\n\nPlatform:\n ['Wii' 'NES' 'GB' 'DS' 'X360' 'PS3' 'PS2' 'SNES' 'GBA' '3DS' 'PS4' 'N64'\n 'PS' 'XB' 'PC' '2600' 'PSP' 'XOne' 'GC' 'WiiU' 'GEN' 'DC' 'PSV' 'SAT'\n 'SCD' 'WS' 'NG' 'TG16' '3DO' 'GG' 'PCFX'] \nCount: 31\n\nYear\n [2006. 1985. 2008. 2009. 1996. 1989. 1984. 2005. 1999. 2007. 2010. 2013.\n 2004. 1990. 1988. 2002. 2001. 2011. 1998. 2015. 2012. 2014. 1992. 1997.\n 1993. 1994. 1982. 2003. 1986. 2000. nan 1995. 2016. 1991. 1981. 1987.\n 1980. 1983. 2020. 2017.] \nCount: 39\n\nGenre\n ['Sports' 'Platform' 'Racing' 'Role-Playing' 'Puzzle' 'Misc' 'Shooter'\n 'Simulation' 'Action' 'Fighting' 'Adventure' 'Strategy'] \nCount: 12\n\nPublishers\n ['Nintendo' 'Microsoft Game Studios' 'Take-Two Interactive'\n 'Sony Computer Entertainment' 'Activision' 'Ubisoft' 'Bethesda Softworks'\n 'Electronic Arts' 'Sega' 'SquareSoft' 'Atari' '505 Games' 'Capcom'\n 'GT Interactive' 'Konami Digital Entertainment'] \nCount: 578\n Rank Name Platform Year Genre Publisher\n0 1 Wii Sports Wii 2006.0 Sports Nintendo\n1 2 Super Mario Bros. NES 1985.0 Platform Nintendo\n9 10 Duck Hunt NES 1984.0 Shooter Nintendo\n5 6 Tetris GB 1989.0 Puzzle Nintendo\n2 3 Mario Kart Wii Wii 2008.0 Racing Nintendo\n Rank Name Platform Year Genre \\\n0 1 Wii Sports Wii 2006.0 Sports \n2 3 Mario Kart Wii Wii 2008.0 Racing \n3 4 Wii Sports Resort Wii 2009.0 Sports \n10 11 Nintendogs DS 2005.0 Simulation \n16 17 Grand Theft Auto V PS3 2013.0 Action \n\n Publisher \n0 Nintendo \n2 Nintendo \n3 Nintendo \n10 Nintendo \n16 Take-Two Interactive \n Rank Name Platform Year Genre \\\n4 5 Pokemon Red/Pokemon Blue GB 1996.0 Role-Playing \n12 13 Pokemon Gold/Pokemon Silver GB 1999.0 Role-Playing \n1 2 Super Mario Bros. NES 1985.0 Platform \n6 7 New Super Mario Bros. DS 2006.0 Platform \n20 21 Pokemon Diamond/Pokemon Pearl DS 2006.0 Role-Playing \n\n Publisher \n4 Nintendo \n12 Nintendo \n1 Nintendo \n6 Nintendo \n20 Nintendo \n Rank Name Platform Year Genre Publisher\n0 1 Wii Sports Wii 2006.0 Sports Nintendo\n1 2 Super Mario Bros. NES 1985.0 Platform Nintendo\n2 3 Mario Kart Wii Wii 2008.0 Racing Nintendo\n3 4 Wii Sports Resort Wii 2009.0 Sports Nintendo\n4 5 Pokemon Red/Pokemon Blue GB 1996.0 Role-Playing Nintendo\nYear\n1980.0 9\n1981.0 46\n1982.0 36\n1983.0 17\n1984.0 14\n1985.0 14\n1986.0 21\n1987.0 16\n1988.0 15\n1989.0 17\n1990.0 16\n1991.0 41\n1992.0 43\n1993.0 60\n1994.0 121\n1995.0 219\n1996.0 263\n1997.0 289\n1998.0 379\n1999.0 338\n2000.0 349\n2001.0 482\n2002.0 829\n2003.0 775\n2004.0 763\n2005.0 941\n2006.0 1008\n2007.0 1202\n2008.0 1428\n2009.0 1431\n2010.0 1259\n2011.0 1139\n2012.0 657\n2013.0 546\n2014.0 582\n2015.0 614\n2016.0 344\n2017.0 3\n2020.0 1\ndtype: int64\nGenre\nAction 3316\nAdventure 1286\nFighting 848\nMisc 1739\nPlatform 886\nPuzzle 582\nRacing 1249\nRole-Playing 1488\nShooter 1310\nSimulation 867\nSports 2346\nStrategy 681\ndtype: int64\nPlatform\n2600 133\n3DO 3\n3DS 509\nDC 52\nDS 2163\nGB 98\nGBA 822\nGC 556\nGEN 27\nGG 1\nN64 319\nNES 98\nNG 12\nPC 960\nPCFX 1\nPS 1196\nPS2 2161\nPS3 1329\nPS4 336\nPSP 1213\nPSV 413\nSAT 173\nSCD 6\nSNES 239\nTG16 2\nWS 6\nWii 1325\nWiiU 143\nX360 1265\nXB 824\nXOne 213\ndtype: int64\n"
]
],
[
[
"#### 4. Initial thoughts?\n- We have null values in Year and Publisher (Total of 307 rows with NaN values)\n- Year is a float, we could probably turn it into an int to make it prettier\n- We have 31 unique Platforms\n- We have 39 unique years (one being NaN)\n- We have 12 unique genres\n- We have 578 unique publishers\n- Looking at the top games in each region EU and NA are more similar in taste than JP\n -JP loves their Pokemon\n -Nintendo is the dominating Publisher in all regions \n- Years with the most releases are 2006 to 2011 with a peak of 1431 releases in 2009.",
"_____no_output_____"
],
[
"#### 5. Wrangling\nThe main concern would be the NaN values. I feel the data is still valuable, we can remove/deal with those values on a need basis.",
"_____no_output_____"
]
],
[
[
"game_melt",
"_____no_output_____"
],
[
"game.Year = game.Year.astype(\"Int64\")\nprint(nulls.groupby(\"Year\").size())\nprint(nulls.groupby(\"Genre\").size())\nprint(nulls.groupby(\"Platform\").size())\n\n\n#For the analysis of sales - melting the NA,EU,JP,Other and Total columns\ngame_melt = game.melt(id_vars=[\"Rank\", \"Name\",\"Platform\",\"Year\",\"Genre\",\"Publisher\"], \n var_name=\"Region\", \n value_name=\"Sales\").reset_index(drop=True)",
"Year\n2004.0 19\n2005.0 5\n2007.0 1\n2010.0 2\n2011.0 3\n2012.0 2\n2014.0 2\n2016.0 2\ndtype: int64\nGenre\nAction 65\nAdventure 12\nFighting 12\nMisc 53\nPlatform 11\nPuzzle 12\nRacing 24\nRole-Playing 18\nShooter 28\nSimulation 19\nSports 42\nStrategy 11\ndtype: int64\nPlatform\n2600 17\n3DS 10\nDS 32\nGB 1\nGBA 36\nGC 14\nN64 3\nPC 22\nPS 7\nPS2 34\nPS3 25\nPSP 16\nPSV 3\nWii 35\nX360 31\nXB 21\ndtype: int64\n"
]
],
[
[
"#### 6. Research Questions/Visualization+Analysis",
"_____no_output_____"
]
],
[
[
"#1) Basic Exploratory visualisations of things we noted in the Initial Thoughts\n#Counts of number of games in each genre, platform and number of games released in each year\n#Genre and Platform counts are coloured by number of counts and sorted from largest to smallest\n#Year counts are coloured by year and sorted from largest to smallest \nsorted_genre_count = list(game.groupby(\"Genre\").size().sort_values(ascending=False).index)\nsorted_year_count = list(game.groupby(\"Year\").size().sort_values(ascending=False).index)\nsorted_platform_count = list(game.groupby(\"Platform\").size().sort_values(ascending=False).index)\n\ngenre_count = alt.Chart(game).mark_bar().encode(\n alt.X(\"Genre\",type=\"nominal\",sort=sorted_genre_count),\n alt.Y(\"count()\",title=\"Number of games\",type=\"quantitative\"),\n alt.Color(\"count()\",scale=alt.Scale(scheme='category20b'),legend=None),\n alt.Tooltip(\"count()\"))\n\nyear_count = alt.Chart(game).mark_bar().encode(\n alt.X(\"Year\",type=\"ordinal\",sort=sorted_year_count),\n alt.Y(\"count()\",title=\"Number of games\",type=\"quantitative\"),\n alt.Color(\"Year\",scale=alt.Scale(scheme='category20c')),\n alt.Tooltip(\"count()\"))\n\nplatform_count = alt.Chart(game).mark_bar().encode(\n alt.X(\"Platform\",type=\"nominal\",sort=sorted_platform_count),\n alt.Y(\"count()\",title=\"Number of games\",type=\"quantitative\"),\n alt.Color(\"count()\",scale=alt.Scale(scheme='category20b'),legend=None),\n alt.Tooltip(\"count()\"))\n\ncount_plots = ((genre_count.properties(width=333)|platform_count.properties(width=666)).resolve_scale(color='independent')\n &year_count.properties(width=1000)).resolve_scale(color='independent').configure_axis(\n labelFontSize=12,\n titleFontSize=13)\ncount_plots = count_plots.properties(title={\"text\": \"Number of games released by Genre, Platform and Year\", \n \"subtitle\": [\"Counts only include games that have sold more than 100,000 copies. Bars for the Genre and Platform plot are coloured by number of games.\",\"\"]}).configure_title(fontSize = 25,subtitleFontSize=15)\n\n#2) Lets look at sales across Genres for each Region\n#Genres are sorted by decreasing Global Sales (Action is most sales vs Strategy is least)\n#Notice we see Shooters - while having fewer games released, still sold a lot of copies meaning their titles seemed to do well and the same (to a larger extent) can be said about Platformers.\n#Looking at the means of each genre, we can see exacly as we noticed above with the mean number of sales in the Shooter/Platform genre now ahead of the rest. \n#It is also interesting to see the trend across genres. We see NA, EU and Other sale patters tend to be more similar while JP sale patterns are distinct from the other regions, with a large emphasis on RPG, Platformers. \nsales_data = game_melt.loc[game_melt.Region != \"Global_Sales\",:]\nsorted_genre_totalsales = list(game.groupby(\"Genre\").sum().sort_values(\"Global_Sales\",ascending=False).index)\n\ngenre_sales = alt.Chart(sales_data).mark_bar(opacity=0.5).encode(\n alt.X(\"Genre\",type=\"nominal\",sort=sorted_genre_totalsales),\n alt.Y(\"sum(Sales)\",title=\"Total Number of Sales (in millions)\",type=\"quantitative\",stack=None),\n alt.Color(\"Region\",scale=alt.Scale(scheme='set1'),type=\"nominal\"),\n alt.Tooltip(\"Region\"))\ngenre_sales = genre_sales+genre_sales.mark_circle()\ngenre_mean_sales = alt.Chart(sales_data).mark_bar(opacity=0.5).encode(\n alt.X(\"Genre\",type=\"nominal\",sort=sorted_genre_totalsales),\n alt.Y(\"mean(Sales)\",title=\"Average Number of Sales (in millions)\",type=\"quantitative\",stack=None),\n alt.Color(\"Region\",type=\"nominal\"),\n alt.Tooltip(\"Region\"))\n\ngenre_mean_sales = genre_mean_sales + genre_mean_sales.mark_circle()\ngenre_plots = (genre_sales | genre_mean_sales).properties(title={\"text\":\"Comparing sales in each region by genre.\",\"subtitle\":[\"Bars and points represent number of sales in millions and coloured by Region.\",\"\"]}).configure_axis(\n labelFontSize=12,\n titleFontSize=13).configure_title(fontSize = 25,subtitleFontSize=15) \n\n#3) Lets investigate what's going on in each region. We can do so by looking at individual title by region and color the points by genre. Scatter plot: Top 3 games in each genre for each region \n #This might help us explain and point out individual game titles that are dominating their genre.\nsorted_genre_NA = list(sales_data[sales_data.Region==\"NA_Sales\"].groupby(\"Genre\").sum().sort_values(\"Sales\",ascending=False).index) \nsorted_genre_EU = list(sales_data[sales_data.Region==\"EU_Sales\"].groupby(\"Genre\").sum().sort_values(\"Sales\",ascending=False).index) \nsorted_genre_JP = list(sales_data[sales_data.Region==\"JP_Sales\"].groupby(\"Genre\").sum().sort_values(\"Sales\",ascending=False).index) \nsorted_genre_OT = list(sales_data[sales_data.Region==\"Other_Sales\"].groupby(\"Genre\").sum().sort_values(\"Sales\",ascending=False).index) \n\ngenre_region = alt.Chart(sales_data).mark_bar(opacity=0.5).encode(\n alt.Y(\"Region\",type=\"nominal\",sort=sorted_genre_totalsales),\n alt.X(\"sum(Sales)\",title=\"Total Number of Sales (in millions)\",type=\"quantitative\",stack=None),\n alt.Color(\"Region\",scale=alt.Scale(scheme='set1'),type=\"nominal\",legend=None))\n\ntitle_NA =alt.Chart(sales_data[sales_data.Region==\"NA_Sales\"]).mark_circle(size=50).encode(\n alt.X(\"Genre\",sort=sorted_genre_NA,title=None),\n alt.Y(\"Sales:Q\",stack=None, title=\"Sales (in millions)\"),\n alt.Color(\"Genre\",scale=alt.Scale(scheme='category20')),\n alt.Tooltip(\"Name\"))\ntitle_NA = title_NA + alt.Chart(sales_data[sales_data.Region==\"NA_Sales\"].sort_values(\"Sales\",ascending=False).iloc[:5,]).mark_text(align = \"left\", dx=10).encode(\n alt.X(\"Genre\",sort=sorted_genre_NA),\n alt.Y(\"Sales:Q\"),\n text=\"Name\").properties(title=\"NA Sales\")\n\ntitle_JP =alt.Chart(sales_data[sales_data.Region==\"JP_Sales\"]).mark_circle(size=50).encode(\n alt.X(\"Genre\",sort=sorted_genre_JP,title=None),\n alt.Y(\"Sales:Q\",stack=None,title=None),\n alt.Color(\"Genre\",scale=alt.Scale(scheme='category20')),\n alt.Tooltip(\"Name\"))\ntitle_JP = title_JP + alt.Chart(sales_data[sales_data.Region==\"JP_Sales\"].sort_values(\"Sales\",ascending=False).iloc[:5,]).mark_text(align = \"left\", dx=10).encode(\n alt.X(\"Genre\",sort=sorted_genre_JP),\n alt.Y(\"Sales:Q\"),\n text=\"Name\").properties(title=\"JP Sales\")\n\ntitle_EU =alt.Chart(sales_data[sales_data.Region==\"EU_Sales\"]).mark_circle(size=50).encode(\n alt.X(\"Genre\",sort=sorted_genre_EU,title=None),\n alt.Y(\"Sales:Q\",stack=None,title=None),\n alt.Color(\"Genre\",scale=alt.Scale(scheme='category20')),\n alt.Tooltip(\"Name\"))\ntitle_EU = title_EU + alt.Chart(sales_data[sales_data.Region==\"EU_Sales\"].sort_values(\"Sales\",ascending=False).iloc[:10,]).mark_text(align = \"left\", dx=10).encode(\n alt.X(\"Genre\",sort=sorted_genre_EU),\n alt.Y(\"Sales:Q\"),\n text=\"Name\").properties(title=\"EU Sales\")\n\ntitle_OT =alt.Chart(sales_data[sales_data.Region==\"Other_Sales\"]).mark_circle(size=50).encode(\n alt.X(\"Genre\",sort=sorted_genre_OT,title=None),\n alt.Y(\"Sales:Q\",stack=None,title=None),\n alt.Color(\"Genre\",scale=alt.Scale(scheme='category20')),\n alt.Tooltip(\"Name\"))\n\ntitle_OT = title_OT + alt.Chart(sales_data[sales_data.Region==\"Other_Sales\"].sort_values(\"Sales\",ascending=False).iloc[:5,]).mark_text(align = \"left\", dx=10).encode(\n alt.X(\"Genre\",sort=sorted_genre_OT),\n alt.Y(\"Sales:Q\"),\n text=\"Name\").properties(title=\"Other Sales\")\n\nname_sales = (title_NA.properties(width=250) | title_EU.properties(width=250) | title_JP.properties(width=250) | title_OT.properties(width=250))\nname_sales = name_sales & genre_region.properties(width=1210).resolve_scale(color='independent')\nname_sales = name_sales.properties(title={\"text\":\"Distribution of games titles across each Region by Genre.\",\"subtitle\":[\"Top 5 games in each region are labelled. Overall Regional perfomance shown below.\",\"\"]}).configure_axis(\n labelFontSize=12,\n titleFontSize=13).configure_title(fontSize = 20,subtitleFontSize=15) \n\n#4) Growth of Game Genres, Publishers and Platforms over the Years\ngenre_data = pd.Series(game.groupby(['Year','Genre']).size().groupby('Year').size(), name='Genre')\npub_data = pd.Series(game.groupby(['Year','Publisher']).size().groupby('Year').size(), name='Publisher')\nplat_data = pd.Series(game.groupby(['Year','Platform']).size().groupby('Year').size(), name='Platform')\nrel_data = pd.concat([genre_data,pub_data,plat_data], axis=1).reset_index()\n\nplot4=alt.Chart(rel_data).transform_fold(['Genre','Publisher','Platform']\n).mark_bar(point=True).encode(\n x='Year:O',\n y=alt.Y('value:Q', axis=alt.Axis(title='Number of Genre/Publishers/Platforms')),\n color=alt.Color('key:N', legend=alt.Legend(title='Feature'))\n).properties(width=1000)\n\n#5) Maximum Annual Global Sales Numbers\nplot5=alt.Chart(game).mark_line(point=True).encode(\n x='Year:N',\n y=alt.Y('max(Global_Sales):Q', axis=alt.Axis(title='Maximum Annual Global Sales'))\n).properties(width=1000)",
"_____no_output_____"
]
],
[
[
"1) **Basic Exploratory visualizations of things we noted in the 'Initial Thoughts'**\n- Counts of number of games in each genre, platform and number of games released in each year\n- Genre and Platform counts are colored by number of counts and sorted from largest to smallest\n- Year counts are colored by year and sorted from largest to smallest \n- We see Action is quite ahead of other genres in terms of numbers of games released, followed by Sports\n- We see DS and PS2 have the highest number of games released in terms of Platform. \n- The years 2006-2011 have the highest number of game released. \n\nThere are a ton of different ways to analyze this data set, lets just look at Genres for now.",
"_____no_output_____"
]
],
[
[
"count_plots",
"_____no_output_____"
]
],
[
[
"2) **Sales across Genres for each Region**\n- Genres are sorted by decreasing Global Sales (Action is most sales vs Strategy is least)\n- Notice we see Shooters - while having fewer games released, still sold a lot of copies meaning their titles seemed to do well and the same (to a larger extent) can be said about Platformers.\n- Looking at the means of each genre, we can see exactly as we noticed above with the mean number of sales in the Shooter/Platform genre now ahead of the rest. \n- It is also interesting to see the trend across genres. We see NA, EU and Other sale patters tend to be more similar while JP sale patterns are distinct from the other regions, with a large emphasis on RPG, Platformers.\n\nLets investigate what's going on in each region. We can do so by looking at individual titles by region. This might help us explain and point out individual game titles that are dominating their genre.",
"_____no_output_____"
]
],
[
[
"genre_plots",
"_____no_output_____"
]
],
[
[
"3) **Distribution of game titles by Genre across each Region**\n- The top 5 titles in each region across genres are labeled. The genres are also sorted by most sales (left) to least sales (right). \n- Just by looking at the scales, it seems NA sells the most copies, followed by EU, JP and Other. This is also re-enforced by the bar plot below showing the total number of copies sold (in millions) by region. \n- Looking at genres across regions, we see NA, EU and Other are similar (as concluded from the plot before). While Action and Sports are still in the Top 3 genres for JP, we see RPG being first and Shooters being last.\n- We see NA, EU and Other having similar Top 5 games, all having Wii Sports and Mario Kart.\n- The JP region seems to love their RPG games which makes sense as they even have a subcategory named after them (JRPGs - Japanese RPGs)\n- We see the bulk of games in most regions (except Other) lie around or below the 5 million copies sold mark and relatively few titles make it above.",
"_____no_output_____"
]
],
[
[
"name_sales",
"_____no_output_____"
]
],
[
[
"4) **Growth of Game Genres, Publishers and Platforms over the Years**",
"_____no_output_____"
],
[
"- From our dataset we see that the year around 2010 were of highest games sales activity.\n- In 1980 we had only two game companies/publishers make five genre of games. \n- Lots of different publishers entered the gaming market with over 180 gaming companies around 2010 offering different gaming products.\n- Its interesting to see that the number of gaming consoles/platforms (like Xbox, PS5) have not increased. These consoles need bigger investment and only big gaming industry players held that market segment.",
"_____no_output_____"
]
],
[
[
"plot4",
"_____no_output_____"
]
],
[
[
"5) **Maximum Annual Global Sales Numbers**",
"_____no_output_____"
]
],
[
[
"plot5",
"_____no_output_____"
]
],
[
[
"#### 7. Future Studies\n1) **Revenue and marketing model of games** \nWhile this dataset can give us a general idea of popularity of games across years, genres, publishers and platforms, it does not necessarily tell us which genre is most profitable in terms of revenue. \nCurrently, one of the largest gaming market is `Mobile Gaming` which would not have a metric such as `copies sold` but rather `downloads`. We also know some of the biggest games such as League of Legends, and DOTA 2 run off a \"free to play\" model for PC so they would also not have a \"copies sold\" metric. On top of that, a game that is free, take League of Legends for example, will make money through micro transactions (for example they made $1.75 billion dollars in 2020). \nAs a more accurate analysis of how platforms, titles, developers and genres are performing in terms of revenue, popularity and activity, we might want to expand and improve the quality and type of data we are looking at. Certain things that we could use as better metrics: \n- Generic things such as: Game title, year of release, company, genre, platform\n- We could look at copies sold AND downloads \n- We could look at concurrent players (as to get a sense of the size of the active player base) *This would be very useful in comparing multi player games and even peak hours of gaming\n- The payment model of the game (single time purchase, subscription based, free-to-play)\n- Reported revenue (could be trickier to find)\n- We can also include things like development cost (could be trickier to find)\n- Reviews/General consensus \n\nThis could be very helpful from a companies perspective to narrow down on a target audience and possible find niche areas of the gaming community that show promise.\n\n2) **Viewership and Entertainment** \nWe could also expand our data set to analyze things in the streaming industry, where there is also big money to be had. Now a days, it seems like many people dream of being a streamer. Just to sit at home and play games all day while raking in large amounts of money, but it would be interesting to see certain stats such as, how much money do you need to break even (average living cost vs income from streaming broken down into views, subs, donations etc.) and to see how many people on platforms such as YouTube gaming, Twitch, Facebook gaming meet this threshold of 'success'. Things you would need would be: \n- List of streamers, platform they stream on, hours streamed, revenue\n \nYou could also look to see if certain genre of games or certain game titles themselves lead to better chances of success. For this you would need to further breakdown streamer information by game played, time streamed, views, donations and subscriptions received during that stream. \nObviously, I feel a lot of this data would be difficult to obtain and that the success of a streamer is heavily influenced by personality, but it would be interesting to see if there is anything that can be identified to give upcoming streamers a slight advantage against big names.",
"_____no_output_____"
]
],
[
[
"sales_data_platform",
"_____no_output_____"
],
[
"sales_data[(sales_data.Region==\"EU_Sales\") & (sales_data.Genre==\"Action\")].sort_values(\"Sales\",ascending=False)",
"_____no_output_____"
],
[
"max_year = 2020\nsales_data = game_melt.loc[game_melt.Region != \"Global_Sales\",:]\nsales_data_platform = sales_data.groupby([\"Platform\",\"Year\",\"Genre\",\"Region\"]).sum().reset_index()[[\"Platform\",\"Year\",\"Genre\",\"Region\",\"Sales\"]]\nsales_data_platform\nsales_data_platform[sales_data_platform.Year <= max_year].groupby([\"Platform\",\"Genre\",\"Region\"]).sum().reset_index()[[\"Platform\",\"Genre\",\"Region\",\"Sales\"]]\nlist(sales_data[(sales_data.Region==region_filter) & (sales_data.Year <= max_year)].groupby(\"Genre\").sum().sort_values(\"Sales\",ascending=False).index) ",
"_____no_output_____"
],
[
"\nsales_data_platform = sales_data.groupby([\"Platform\",\"Year\",\"Genre\",\"Region\"]).sum().reset_index()[[\"Platform\",\"Year\",\"Genre\",\"Region\",\"Sales\"]]\nmax_year = 1999\nregion_filter = \"NA_Sales\"\n\ndef platform_plot(region_filter,max_year=2020):\n sorted_genre = list(sales_data[(sales_data.Region==region_filter) & (sales_data.Year <= max_year)].groupby(\"Genre\").sum().sort_values(\"Sales\",ascending=False).index) \n filtered_set = sales_data_platform[sales_data_platform.Year <= max_year].groupby([\"Platform\",\"Genre\",\"Region\"]).sum().reset_index()[[\"Platform\",\"Genre\",\"Region\",\"Sales\"]]\n\n chart=alt.Chart(filtered_set[filtered_set.Region==region_filter]).mark_circle(size=50).encode(\n alt.X(\"Genre\",sort=sorted_genre,title=None),\n alt.Y(\"Sales:Q\",stack=None, title=\"Sales (in millions)\"),\n alt.Color(\"Genre\",scale=alt.Scale(scheme='category20')),\n alt.Tooltip(\"Platform\"))\n chart = chart + alt.Chart(filtered_set[filtered_set.Region==region_filter].sort_values(\"Sales\",ascending=False).iloc[:5,]).mark_text(align = \"left\", dx=10).encode(\n alt.X(\"Genre\",sort=sorted_genre),\n alt.Y(\"Sales:Q\"),\n text=\"Platform\")\n return chart\nx = platform_plot(region_filter,max_year)\nx\n# chart = chart + alt.Chart(sales_data_platform[sales_data_platform.Region==region_filter].sort_values(\"Sales\",ascending=False).iloc[:5,]).mark_text(align = \"left\", dx=10).encode(\n# alt.X(\"Genre\",sort=sorted_genre),\n# alt.Y(\"Sales:Q\"),\n# text=\"Name\")\n",
"_____no_output_____"
],
[
"sales_data_publisher = sales_data.groupby([\"Publisher\",\"Year\",\"Genre\",\"Region\"]).sum().reset_index()[[\"Publisher\",\"Year\",\"Genre\",\"Region\",\"Sales\"]]\nmax_year = 2020\nregion_filter = \"NA_Sales\"\n\ndef test(region_filter,max_year=2020):\n sorted_genre = list(sales_data[(sales_data.Region==region_filter) & (sales_data.Year <= max_year)].groupby(\"Genre\").sum().sort_values(\"Sales\",ascending=False).index) \n filtered_set = sales_data_publisher[sales_data_publisher.Year <= max_year].groupby([\"Publisher\",\"Genre\",\"Region\"]).sum().reset_index()[[\"Publisher\",\"Genre\",\"Region\",\"Sales\"]]\n\n chart=alt.Chart(filtered_set[filtered_set.Region==region_filter]).mark_circle(size=50).encode(\n alt.X(\"Genre\",sort=sorted_genre,title=None),\n alt.Y(\"Sales:Q\",stack=None, title=\"Sales (in millions)\"),\n alt.Color(\"Genre\",scale=alt.Scale(scheme='category20'),legend=None),\n alt.Tooltip(\"Publisher\"))\n chart = chart + alt.Chart(filtered_set[filtered_set.Region==region_filter].sort_values(\"Sales\",ascending=False).iloc[:5,]).mark_text(align = \"left\", dx=10).encode(\n alt.X(\"Genre\",sort=sorted_genre),\n alt.Y(\"Sales:Q\"),\n text=\"Publisher\")\n return chart\nx = test(region_filter,max_year)\nx",
"_____no_output_____"
],
[
"sales_data_publisher = sales_data.groupby([\"Publisher\",\"Year\",\"Genre\",\"Region\"]).sum().reset_index()[[\"Publisher\",\"Year\",\"Genre\",\"Region\",\"Sales\"]]\nsales_data_publisher",
"_____no_output_____"
],
[
"filtered_set = sales_data[sales_data.Year <= max_year].groupby([\"Name\",\"Genre\",\"Region\"]).sum().reset_index()[[\"Name\",\"Genre\",\"Region\",\"Sales\"]]\nfiltered_set",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
c51387d712bcbcd4e780906dc7e5f57ffb572324
| 77,922 |
ipynb
|
Jupyter Notebook
|
Lectures/02_NatureSignals.ipynb
|
bardet/GWU-MAE3120_2020
|
0b783b0550edd153231d19460c7575a2aa0a0675
|
[
"BSD-3-Clause"
] | 2 |
2019-12-12T02:24:13.000Z
|
2020-01-17T14:18:28.000Z
|
Lectures/02_NatureSignals.ipynb
|
bardet/GWU-MAE3120_2020
|
0b783b0550edd153231d19460c7575a2aa0a0675
|
[
"BSD-3-Clause"
] | null | null | null |
Lectures/02_NatureSignals.ipynb
|
bardet/GWU-MAE3120_2020
|
0b783b0550edd153231d19460c7575a2aa0a0675
|
[
"BSD-3-Clause"
] | 3 |
2019-12-18T15:56:16.000Z
|
2021-06-02T01:13:22.000Z
| 335.87069 | 26,832 | 0.923603 |
[
[
[
"# Nature of signals\n\nIn the context of this class, a signal is the data acquired by the measurement system. It contains much information that we need to be able to identify to extract knowledge about the system being tested and how to optimize the measurements. A signal caries also messages and information. We will use the content of this module for the other modules seen in the rest of the class.\n\n## Signal classification\n\nA signal can be characterized by its amplitude and frequency. __Amplitude__ is related to the strength of the signal and __frequency__ to the extent or duration of the signal. The time series of a signal is called a __waveform__. Multipe collection of the waveform is called an __ensemble__. \n\nSignals can be either __deterministic__ or __random__. \n\nDeterministic signals can be either __static__ (do not change in time) or __dynamic__. Dynamic signals can be decomposed into __periodic__ or __aperiodic__. A periodic signal repeats itself at regular interval. The smallest value over whih it repeats itself is the __fundamental period__, with an associated __fundamental frequency__. A __simple__ periodic signal has one period; it is a sine wave. A __complex__ has multiple periods and can be thought as the sum of several sinusoids (more on this in the next section). Aperiodic signals are typically __transient__ (such as step, ramp, or pulse responses). \n\nNondeterministic signals are an important class of signals that are often encountered in nature (think of turbulence, stock market, etc). They must be analyzed with satistical tools. They are classified as __nonstationary__ and __stationary__. This classification enables to select the proper statistical theory to analyze them. The properties of nondeterministic signals are computed with ensemble statistics of instantaneous properties. In particular, one computes the ensemble average, $\\mu(t_1)$, and ensemble autocorrelation function (more on the physical meaning of this function later), $R(t_1,t_1+\\tau)$.\n\n\\begin{align*}\n\\mu(t_1) & = \\frac{1}{N} \\sum_{i=0}^{N-1} x_i(t_1) \\\\\nR(t_1,t_1+\\tau) & = \\frac{1}{N} \\sum_{i=0}^{N-1} x_i(t_1)x_i(t_1+\\tau)\n\\end{align*}\n\nThe term ensemble means that we take N time series and perform statistics with the ensemble of the values at recorded time $t_1$.\n\nIf $\\mu(t_1) = \\mu$ and $R(t_1,t_1+\\tau) = R(\\tau)$, then the signal is considered (weakly) __stationary__ and nonstationary, otherwise. Stationarity introdcues a lot of simplification in the statistical analysis of the data (by using a lot of tools developed for time series analysis) and one should always start by checking for signal stationarity. Stationarity implies that signal ensemble-averaged statistical properties are independent of $t_1$. \n\nFor most stationary signals, the temporal and ensemble statistical properties are identical. The signal is then __ergodic__. Thus, from a _single_ time history of length $T_r$ one can calculate $\\mu$ and $R(\\tau)$ (which saves time in the acquisition and analysis):\n\n\\begin{align*}\n\\mu & = \\frac{1}{T_r} \\int_{0}^{T_r} x(t) dt \\\\\nR(\\tau) & = \\frac{1}{T_r} \\int_{0}^{T_r} x(t)x(t+\\tau) dt\n\\end{align*}\n\nThanks to statistical tools for ergodic processes, from a finite recording length of the signal, one can estimate population mean with confidence level.",
"_____no_output_____"
],
[
"## Signal variables\n\nMost signals can be decomposed as a sum of sines and cosines (more on this in the next module). Let's start with a simple periodic signal:\n\n\\begin{align*}\ny(t) = C \\sin (n \\omega t + \\phi) = C \\sin (n 2\\pi f t + \\phi)\n\\end{align*}\n\nWhen several sine and cosine waves are added, complext waveforms result. For example for second order dynamic system, the system response could take the form: \n\n\\begin{align*}\ny(t) = A \\cos (\\omega t) + B \\sin (\\omega t)\n\\end{align*}\n\nThis sum of a cosine and sine of same frequency can be rearranged as:\n\n\\begin{align*}\ny(t) = C \\cos (\\omega t - \\phi) = C \\cos (\\omega t - \\phi + \\pi/2) = C \\sin (\\omega t + \\phi')\n\\end{align*}\n\nwith:\n\\begin{align*}\nC & = \\sqrt{A^2 + B^2}\\\\\n\\phi & = \\tan^{-1} (B/A)\\\\\n\\phi' & = \\pi/2 - \\phi = \\tan^{-1} (A/B)\n\\end{align*}\n\nLet's look at some examples of simple and complex periodic signals.",
"_____no_output_____"
],
[
"First a simple function:\n\n\\begin{align*}\ny (t) = 2 \\sin (2\\pi t)\n\\end{align*}\n",
"_____no_output_____"
]
],
[
[
"import numpy\nfrom matplotlib import pyplot\n%matplotlib inline\n\nt=numpy.linspace(0.0,5.0,num=1000) # (s)\n\ny = 2 * numpy.sin(2*numpy.pi*t)\npyplot.plot(t, y, color='b', linestyle='-');",
"_____no_output_____"
]
],
[
[
"Now a complex function made of two frequencies (harmonics):\n\n\\begin{align*}\ny (t) = 2 \\sin (2\\pi t) + 1.2 \\sin (6 \\pi t)\n\\end{align*}\n\nThe signal has two frequencies: 1 and 3 Hz. 1 Hz is the lowest frequency and is the fundamental frequency with period 1 s. So the signal will repeat itself every second.",
"_____no_output_____"
]
],
[
[
"y = 2 * numpy.sin(2*numpy.pi*t) + 1.2 * numpy.sin(6*numpy.pi*t)\n\npyplot.plot(t, y, color='b', linestyle='-');",
"_____no_output_____"
]
],
[
[
"Let's now look at two sinusoidal with very close frequencies $\\Delta f$.\n\n\\begin{align*}\ny (t) = 2 \\sin (2\\pi t) + 1.2 \\sin ((2+0.2) \\pi t)\n\\end{align*}\n",
"_____no_output_____"
]
],
[
[
"t=numpy.linspace(0.0,20.0,num=1000) # (s)\ny = 2 * numpy.sin(2*numpy.pi*t) + 1.2 * numpy.sin((2+0.2)*numpy.pi*t)\npyplot.plot(t, y, color='b', linestyle='-');",
"_____no_output_____"
]
],
[
[
"Here the frequency difference is $\\Delta f = 0.2/2 = 0.1 Hz$. The resulting signal has a slow beat with __beat__ frequency $\\Delta f)$ or beat period $1/\\Delta f = 10$ s, i.e. the signal repepats itself every 10 s. Analytically (using trigonometric relations), one can show that the sum of two sine waves with close frequencies results in a signal modulated by $\\cos(\\Delta f/2)$.\n\n## Detection schemes\n\nThe mixing of two signals to produce a signal (wave) with a new frequency is called heterodyning and is commonly used in instrumentation to obtain very accurate measurements. __Heterodyne detection__ shifts the frequency content of a signal into a new range where it is easier to detected; in communucation it is called _frequency conversion_. Heterodyning is used in laser Doppler velocimetry, tuning of musical instruments, radio receivers, etc. \n\nIn contrast, __homodyne detection__ uses a single (homo) frequency and compares the signal with a standard oscillation that would be identical to the signal if it carried null information. and measures the amplitude and phase of a signal to gain information. It enables to extract information encoded as modulation of the phase and/or frequency of the signal. In optics, this results in interferometry. It is also the fundation behind lock-in amplifier to extract information for very weak or noisy signals.\n\nFinally in __magnitude detection__ one only records the amplitude of signals. This is the most common detection scheme used. ",
"_____no_output_____"
],
[
"## Statistical description of signals\n\n",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
c5138e3d2f2834bd63a19b32331ba5c1ecd7e136
| 13,487 |
ipynb
|
Jupyter Notebook
|
Deep Reinforcement Learning Nano Degree/navigation/Navigation.ipynb
|
farabi1038/Online-Course-Project
|
9471a6ed616a08cbacbf8db402cedbe465fe261f
|
[
"MIT"
] | 2 |
2020-08-07T07:59:53.000Z
|
2021-05-09T18:11:06.000Z
|
Deep Reinforcement Learning Nano Degree/navigation/Navigation.ipynb
|
farabi1038/Online-Course-Project
|
9471a6ed616a08cbacbf8db402cedbe465fe261f
|
[
"MIT"
] | null | null | null |
Deep Reinforcement Learning Nano Degree/navigation/Navigation.ipynb
|
farabi1038/Online-Course-Project
|
9471a6ed616a08cbacbf8db402cedbe465fe261f
|
[
"MIT"
] | null | null | null | 35.306283 | 258 | 0.554831 |
[
[
[
"# Navigation\n\n---\n\nYou are welcome to use this coding environment to train your agent for the project. Follow the instructions below to get started!\n\n### 1. Start the Environment\n\nRun the next code cell to install a few packages. This line will take a few minutes to run!",
"_____no_output_____"
]
],
[
[
"!pip -q install ./python",
"\u001b[31mtensorflow 1.7.1 has requirement numpy>=1.13.3, but you'll have numpy 1.12.1 which is incompatible.\u001b[0m\r\n\u001b[31mipython 6.5.0 has requirement prompt-toolkit<2.0.0,>=1.0.15, but you'll have prompt-toolkit 3.0.5 which is incompatible.\u001b[0m\r\n"
]
],
[
[
"The environment is already saved in the Workspace and can be accessed at the file path provided below. Please run the next code cell without making any changes.",
"_____no_output_____"
]
],
[
[
"from unityagents import UnityEnvironment\nimport numpy as np\n\n# please do not modify the line below\nenv = UnityEnvironment(file_name=\"/data/Banana_Linux_NoVis/Banana.x86_64\")\n\nimport gym\n!pip3 install box2d\nimport random\nimport torch\nimport numpy as np\nfrom collections import deque\nimport matplotlib.pyplot as plt\n%matplotlib inline\n\n!python -m pip install pyvirtualdisplay\nfrom pyvirtualdisplay import Display\ndisplay = Display(visible=0, size=(1400, 900))\ndisplay.start()\n\nis_ipython = 'inline' in plt.get_backend()\nif is_ipython:\n from IPython import display\n\nplt.ion()",
"INFO:unityagents:\n'Academy' started successfully!\nUnity Academy name: Academy\n Number of Brains: 1\n Number of External Brains : 1\n Lesson number : 0\n Reset Parameters :\n\t\t\nUnity brain name: BananaBrain\n Number of Visual Observations (per agent): 0\n Vector Observation space type: continuous\n Vector Observation space size (per agent): 37\n Number of stacked Vector Observation: 1\n Vector Action space type: discrete\n Vector Action space size (per agent): 4\n Vector Action descriptions: , , , \n"
]
],
[
[
"Environments contain **_brains_** which are responsible for deciding the actions of their associated agents. Here we check for the first brain available, and set it as the default brain we will be controlling from Python.",
"_____no_output_____"
]
],
[
[
"# get the default brain\nbrain_name = env.brain_names[0]\nbrain = env.brains[brain_name]",
"_____no_output_____"
]
],
[
[
"### 2. Examine the State and Action Spaces\n\nRun the code cell below to print some information about the environment.",
"_____no_output_____"
]
],
[
[
"# reset the environment\nenv_info = env.reset(train_mode=True)[brain_name]\n\n# number of agents in the environment\nprint('Number of agents:', len(env_info.agents))\n\n# number of actions\naction_size = brain.vector_action_space_size\nprint('Number of actions:', action_size)\n\n# examine the state space \nstate = env_info.vector_observations[0]\nprint('States look like:', state)\nstate_size = len(state)\nprint('States have length:', state_size)",
"Number of agents: 1\nNumber of actions: 4\nStates look like: [ 1. 0. 0. 0. 0.84408134 0. 0.\n 1. 0. 0.0748472 0. 1. 0. 0.\n 0.25755 1. 0. 0. 0. 0.74177343\n 0. 1. 0. 0. 0.25854847 0. 0.\n 1. 0. 0.09355672 0. 1. 0. 0.\n 0.31969345 0. 0. ]\nStates have length: 37\n"
]
],
[
[
"### 3. Take Random Actions in the Environment\n\nIn the next code cell, you will learn how to use the Python API to control the agent and receive feedback from the environment.\n\nNote that **in this coding environment, you will not be able to watch the agent while it is training**, and you should set `train_mode=True` to restart the environment.",
"_____no_output_____"
]
],
[
[
"env_info = env.reset(train_mode=True)[brain_name] # reset the environment\nstate = env_info.vector_observations[0] # get the current state\nscore = 0 # initialize the score\nwhile True:\n action = np.random.randint(action_size) # select an action\n env_info = env.step(action)[brain_name] # send the action to the environment\n next_state = env_info.vector_observations[0] # get the next state\n reward = env_info.rewards[0] # get the reward\n done = env_info.local_done[0] # see if episode has finished\n score += reward # update the score\n state = next_state # roll over the state to next time step\n if done: # exit loop if episode finished\n break\n \nprint(\"Score: {}\".format(score))",
"Score: 0.0\n"
]
],
[
[
"When finished, you can close the environment.",
"_____no_output_____"
],
[
"### 4. It's Your Turn!\n\nNow it's your turn to train your own agent to solve the environment! A few **important notes**:\n- When training the environment, set `train_mode=True`, so that the line for resetting the environment looks like the following:\n```python\nenv_info = env.reset(train_mode=True)[brain_name]\n```\n- To structure your work, you're welcome to work directly in this Jupyter notebook, or you might like to start over with a new file! You can see the list of files in the workspace by clicking on **_Jupyter_** in the top left corner of the notebook.\n- In this coding environment, you will not be able to watch the agent while it is training. However, **_after training the agent_**, you can download the saved model weights to watch the agent on your own machine! ",
"_____no_output_____"
],
[
"# Training the netowrk",
"_____no_output_____"
]
],
[
[
"from dqn_agent import Agent\n\nagent = Agent(state_size=37, action_size=4, seed=42)\nprint(type(state))",
"<class 'numpy.ndarray'>\n"
],
[
"def dqn(n_episodes=2000, max_t=1000, eps_start=1.0, eps_end=0.01, eps_decay=0.995):\n \"\"\"Deep Q-Learning.\n \n Params\n ======\n n_episodes (int): maximum number of training episodes\n max_t (int): maximum number of timesteps per episode\n eps_start (float): starting value of epsilon, for epsilon-greedy action selection\n eps_end (float): minimum value of epsilon\n eps_decay (float): multiplicative factor (per episode) for decreasing epsilon\n \"\"\"\n scores = [] # list containing scores from each episode\n scores_window = deque(maxlen=100) # last 100 scores\n eps = eps_start # initialize epsilon\n for i_episode in range(1, n_episodes+1):\n env_info = env.reset(train_mode=True)[brain_name]\n state = env_info.vector_observations[0]\n \n score = 0\n for t in range(max_t):\n action = agent.act(state, eps)\n env_info = env.step(action)[brain_name]\n next_state = env_info.vector_observations[0]\n reward = env_info.rewards[0]\n done = env_info.local_done[0] \n agent.step(state, action, reward, next_state, done)\n state = next_state\n score += reward\n if done:\n break \n scores_window.append(score) # save most recent score\n scores.append(score) # save most recent score\n eps = max(eps_end, eps_decay*eps) # decrease epsilon\n print('\\rEpisode {}\\tAverage Score: {:.2f}'.format(i_episode, np.mean(scores_window)), end=\"\")\n if i_episode % 100 == 0:\n print('\\rEpisode {}\\tAverage Score: {:.2f}'.format(i_episode, np.mean(scores_window)))\n if np.mean(scores_window)>=15.0:\n print('\\nEnvironment solved in {:d} episodes!\\tAverage Score: {:.2f}'.format(i_episode-100, np.mean(scores_window)))\n torch.save(agent.qnetwork_local.state_dict(), 'checkpoint.pth')\n break\n return scores\n\nscores = dqn()\n\n# plot the scores\nfig = plt.figure()\nax = fig.add_subplot(111)\nplt.plot(np.arange(len(scores)), scores)\nplt.ylabel('Score')\nplt.xlabel('Episode #')\nplt.show()",
"Episode 100\tAverage Score: 1.04\nEpisode 200\tAverage Score: 4.60\nEpisode 300\tAverage Score: 7.67\nEpisode 400\tAverage Score: 10.81\nEpisode 500\tAverage Score: 12.77\nEpisode 600\tAverage Score: 13.80\nEpisode 679\tAverage Score: 15.00\nEnvironment solved in 579 episodes!\tAverage Score: 15.00\n"
],
[
"env.close()",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
]
] |
c513a5eeefb4f5695f010f390145fe8e6de278ef
| 91,234 |
ipynb
|
Jupyter Notebook
|
MySQL/practice mysql.ipynb
|
TUCchkul/Database
|
8d98f7f8e85fd5abd3e805bb38a5520e26c6115d
|
[
"MIT"
] | null | null | null |
MySQL/practice mysql.ipynb
|
TUCchkul/Database
|
8d98f7f8e85fd5abd3e805bb38a5520e26c6115d
|
[
"MIT"
] | null | null | null |
MySQL/practice mysql.ipynb
|
TUCchkul/Database
|
8d98f7f8e85fd5abd3e805bb38a5520e26c6115d
|
[
"MIT"
] | null | null | null | 50.238987 | 1,695 | 0.531041 |
[
[
[
"!pip install mysql-connector-python",
"Requirement already satisfied: mysql-connector-python in c:\\programdata\\anaconda3\\lib\\site-packages (8.0.24)\nRequirement already satisfied: protobuf>=3.0.0 in c:\\programdata\\anaconda3\\lib\\site-packages (from mysql-connector-python) (3.15.8)\nRequirement already satisfied: six>=1.9 in c:\\programdata\\anaconda3\\lib\\site-packages (from protobuf>=3.0.0->mysql-connector-python) (1.15.0)\n"
],
[
"import mysql.connector as connection\n\ntry:\n mydb = connection.connect(host=\"localhost\",user=\"root\", passwd=\"mysql\",use_pure=True)\n # check if the connection is established\n\n query = \"SHOW DATABASES\"\n\n cursor = mydb.cursor() #create a cursor to execute queries\n cursor.execute(query)\n #print(cursor.fetchall())\n\nexcept Exception as e:\n mydb.close()\n print(str(e))",
"_____no_output_____"
],
[
"import mysql.connector as connection\n",
"_____no_output_____"
],
[
"conn = connection.connect(host=\"localhost\",user=\"root\", passwd=\"mysql\",use_pure=True)",
"_____no_output_____"
],
[
"cur = conn.cursor()",
"_____no_output_____"
],
[
"cur.execute(\"create database sudhanshu12345\")",
"_____no_output_____"
],
[
"res = cur.fetchall()",
"_____no_output_____"
],
[
"res",
"_____no_output_____"
],
[
"for i in res :\n print(i[0])",
"glassdata\nglassdata1\nglassdata11\nglassdata123\ninformation_schema\nmysql\nperformance_schema\nsakila\nstudent\nstudent1\nstudent12\nsudh\nsudh123\nsys\nworld\n"
],
[
"import mysql.connector as connection\n\ntry:\n mydb = connection.connect(host=\"localhost\", user=\"root\", passwd=\"mysql\",use_pure=True)\n # check if the connection is established\n print(mydb.is_connected())\n\n query = \"Create database Student;\"\n cursor = mydb.cursor() #create a cursor to execute queries\n cursor.execute(query)\n print(\"Database Created!!\")\n mydb.close()\nexcept Exception as e:\n mydb.close()\n print(str(e))",
"_____no_output_____"
],
[
"import mysql.connector as connection\n\ntry:\n mydb = connection.connect(host=\"localhost\", database = 'sudhanshu12345',user=\"root\", passwd=\"mysql\",use_pure=True)\n # check if the connection is established\n print(mydb.is_connected())\n\n query = \"CREATE TABLE StudentDetails (Studentid INT(10) AUTO_INCREMENT PRIMARY KEY,FirstName VARCHAR(60),\" \\\n \"LastName VARCHAR(60), RegistrationDate DATE,Class Varchar(20), Section Varchar(10))\"\n\n cursor = mydb.cursor() #create a cursor to execute queries\n cursor.execute(query)\n print(\"Table Created!!\")\n mydb.close()\nexcept Exception as e:\n mydb.close()\n print(str(e))",
"_____no_output_____"
],
[
" mydb = connection.connect(host=\"localhost\", database = 'sudhanshu12345',user=\"root\", passwd=\"mysql\",use_pure=True)",
"_____no_output_____"
],
[
"mydb.is_connected()",
"_____no_output_____"
],
[
"cur = mydb.cursor()\ncur.execute(\"create table test(x1 INT(5) , x2 VARCHAR(20) ,x3 DATE)\")\nmydb.close()",
"_____no_output_____"
],
[
"import mysql.connector as connection\n\ntry:\n mydb = connection.connect(host=\"localhost\", database = 'Student',user=\"root\", passwd=\"mysql\",use_pure=True)\n # check if the connection is established\n print(mydb.is_connected())\n query = \"INSERT INTO StudentDetails VALUES ('1132','Sachin','Kumar','1997-11-11','Eleventh','A')\"\n\n cursor = mydb.cursor() #create a cursor to execute queries\n cursor.execute(query)\n print(\"Values inserted into the table!!\")\n mydb.commit()\n mydb.close()\nexcept Exception as e:\n mydb.close()\n print(str(e))",
"_____no_output_____"
],
[
" mydb = connection.connect(host=\"localhost\", database = 'sudhanshu12345',user=\"root\", passwd=\"mysql\",use_pure=True)",
"_____no_output_____"
],
[
"cur = mydb.cursor()",
"_____no_output_____"
],
[
"cur.execute(\"insert into test values(4564,'sudh' , '2021-05-15')\") #yyyy-mm-dd",
"_____no_output_____"
],
[
"mydb.commit()",
"_____no_output_____"
],
[
"cur.execute(\"insert into test values(4545,'sudhdfgd' , '2021-05-15')\")\ncur.execute(\"insert into test values(4545,'sudhdfgd' , '2021-05-15')\")\ncur.execute(\"insert into test values(4545,'sudhdfgd' , '2021-05-15')\")\ncur.execute(\"insert into test values(4545,'sudhdfgd' , '2021-05-15')\")\ncur.execute(\"insert into test values(4545,'sudhdfgd' , '2021-05-15')\")\n\ncur.execute(\"insert into test values(4545,'sudhdfgd' , '2021-05-15')\")\ncur.execute(\"insert into test values(4545,'sudhdfgd' , '2021-05-15')\")\ncur.execute(\"insert into test values(4545,'sudhdfgd' , '2021-05-15')\")\ncur.execute(\"insert into test values(4545,'sudhdfgd' , '2021-05-15')\")\ncur.execute(\"insert into test values(4545,'sudhdfgd' , '2021-05-15')\")\ncur.execute(\"insert into test values(4545,'sudhdfgd' , '2021-05-15')\")\ncur.execute(\"insert into test values(4545,'sudhdfgd' , '2021-05-15')\")\ncur.execute(\"insert into test values(4545,'sudhdfgd' , '2021-05-15')\")\ncur.execute(\"insert into test values(4545,'sudhdfgd' , '2021-05-15')\")\ncur.execute(\"insert into test values(4545,'sudhdfgd' , '2021-05-15')\")\ncur.execute(\"insert into test values(4545,'sudhdfgd' , '2021-05-15')\")",
"_____no_output_____"
],
[
"import mysql.connector as connection\n\n\ntry:\n mydb = connection.connect(host=\"localhost\", database = 'GlassData',user=\"root\", passwd=\"mysql\",use_pure=True)\n #check if the connection is established\n print(mydb.is_connected())\n query = \"Select * from GlassData;\"\n cursor = mydb.cursor() #create a cursor to execute queries\n cursor.execute(query)\n for result in cursor.fetchall():\n print(result)\n mydb.close() #close the connection\n\n\nexcept Exception as e:\n #mydb.close()\n print(str(e))",
"_____no_output_____"
],
[
"cur1 = mydb.cursor()\ncur1.execute(\"select * from test\")",
"_____no_output_____"
],
[
"for i in cur1.fetchall():\n print(i)",
"(4564, 'sudh', datetime.date(2021, 5, 15))\n(4545, 'sudhdfgd', datetime.date(2021, 5, 15))\n(4545, 'sudhdfgd', datetime.date(2021, 5, 15))\n(4545, 'sudhdfgd', datetime.date(2021, 5, 15))\n(4545, 'sudhdfgd', datetime.date(2021, 5, 15))\n(4545, 'sudhdfgd', datetime.date(2021, 5, 15))\n(4545, 'sudhdfgd', datetime.date(2021, 5, 15))\n(4545, 'sudhdfgd', datetime.date(2021, 5, 15))\n(4545, 'sudhdfgd', datetime.date(2021, 5, 15))\n(4545, 'sudhdfgd', datetime.date(2021, 5, 15))\n(4545, 'sudhdfgd', datetime.date(2021, 5, 15))\n(4545, 'sudhdfgd', datetime.date(2021, 5, 15))\n(4545, 'sudhdfgd', datetime.date(2021, 5, 15))\n(4545, 'sudhdfgd', datetime.date(2021, 5, 15))\n(4545, 'sudhdfgd', datetime.date(2021, 5, 15))\n(4545, 'sudhdfgd', datetime.date(2021, 5, 15))\n(4545, 'sudhdfgd', datetime.date(2021, 5, 15))\n(4545, 'sudhdfgd', datetime.date(2021, 5, 15))\n(4545, 'sudhdfgd', datetime.date(2021, 5, 15))\n(4545, 'sudhdfgd', datetime.date(2021, 5, 15))\n(4545, 'sudhdfgd', datetime.date(2021, 5, 15))\n(4545, 'sudhdfgd', datetime.date(2021, 5, 15))\n(4545, 'sudhdfgd', datetime.date(2021, 5, 15))\n(4545, 'sudhdfgd', datetime.date(2021, 5, 15))\n(4545, 'sudhdfgd', datetime.date(2021, 5, 15))\n(4545, 'sudhdfgd', datetime.date(2021, 5, 15))\n(4545, 'sudhdfgd', datetime.date(2021, 5, 15))\n(4545, 'sudhdfgd', datetime.date(2021, 5, 15))\n(4545, 'sudhdfgd', datetime.date(2021, 5, 15))\n"
],
[
"cur = mydb.cursor()\ncur.execute(\"select x1, x2 from test\")",
"_____no_output_____"
],
[
"for i in cur.fetchall():\n print(i)",
"(4564, 'sudh')\n(4545, 'sudhdfgd')\n(4545, 'sudhdfgd')\n(4545, 'sudhdfgd')\n(4545, 'sudhdfgd')\n(4545, 'sudhdfgd')\n(4545, 'sudhdfgd')\n(4545, 'sudhdfgd')\n(4545, 'sudhdfgd')\n(4545, 'sudhdfgd')\n(4545, 'sudhdfgd')\n(4545, 'sudhdfgd')\n(4545, 'sudhdfgd')\n(4545, 'sudhdfgd')\n(4545, 'sudhdfgd')\n(4545, 'sudhdfgd')\n(4545, 'sudhdfgd')\n(4545, 'sudhdfgd')\n(4545, 'sudhdfgd')\n(4545, 'sudhdfgd')\n(4545, 'sudhdfgd')\n(4545, 'sudhdfgd')\n(4545, 'sudhdfgd')\n(4545, 'sudhdfgd')\n(4545, 'sudhdfgd')\n(4545, 'sudhdfgd')\n(4545, 'sudhdfgd')\n(4545, 'sudhdfgd')\n(4545, 'sudhdfgd')\n"
],
[
"import mysql.connector as connection\nimport pandas as pandas\n\ntry:\n\n mydb = connection.connect(host=\"localhost\", database='GlassData', user=\"root\", passwd=\"mysql\", use_pure=True)\n # check if the connection is established\n print(mydb.is_connected())\n query = \"Select * from GlassData;\"\n result_dataFrame = pandas.read_sql(query,mydb)\n print(result_dataFrame)\n\n mydb.close() # close the connection\n\nexcept Exception as e:\n #mydb.close()\n print(str(e))",
"_____no_output_____"
],
[
"import pandas as pd",
"_____no_output_____"
],
[
"mydb",
"_____no_output_____"
],
[
"a = pd.read_sql(\"select x3,x1 from test\",mydb)",
"_____no_output_____"
],
[
"a.to_csv(\"mydata.csv\")",
"_____no_output_____"
],
[
"mydb = connection.connect(host=\"localhost\", database='sudhanshu12345', user=\"root\", passwd=\"mysql\", use_pure=True)\nmydb",
"_____no_output_____"
],
[
"cur = mydb.cursor()\ncur.execute(\"create table mydata(n1 int(20) , n2 varchar(20) , n3 date)\")",
"_____no_output_____"
],
[
"data = pd.read_csv('mydata.csv')\ndata",
"_____no_output_____"
],
[
"data1 = pd.read_sql('select * from test' , mydb)",
"_____no_output_____"
],
[
"data1.to_sql('mydata1',mydb)",
"_____no_output_____"
],
[
"from sqlalchemy import create_engine\n>>> engine = create_engine('sqlite://', echo=False)\n\nCreate a table from scratch with 3 rows.\n\n>>> df = pd.DataFrame({'name' : ['User 1', 'User 2', 'User 3']})\n>>> df\n name\n0 User 1\n1 User 2\n2 User 3\n\n>>> df.to_sql('users', con=engine)\n>>> engine.execute(\"SELECT * FROM users\").fetchall()\n[(0, 'User 1'), (1, 'User 2'), (2, 'User 3')]\n\nAn `sqlalchemy.engine.Connection` can also be passed to to `con`:\n>>> with engine.begin() as connection:\n... df1 = pd.DataFrame({'name' : ['User 4', 'User 5']})\n... df1.to_sql('users', con=connection, if_exists='append')\n\nThis is allowed to support operations that require that the same\nDBAPI connection is used for the entire operation.\n\n>>> df2 = pd.DataFrame({'name' : ['User 6', 'User 7']})\n>>> df2.to_sql('users', con=engine, if_exists='append')\n>>> engine.execute(\"SELECT * FROM users\").fetchall()\n[(0, 'User 1'), (1, 'User 2'), (2, 'User 3'),\n (0, 'User 4'), (1, 'User 5'), (0, 'User 6'),\n (1, 'User 7')]\n\nOverwrite the table with just ``df2``.\n\n>>> df2.to_sql('users', con=engine, if_exists='replace',\n... index_label='id')\n>>> engine.execute(\"SELECT * FROM users\").fetchall()\n[(0, 'User 6'), (1, 'User 7')]\n\nSpecify the dtype (especially useful for integers with missing values).\nNotice that while pandas is forced to store the data as floating point,\nthe database supports nullable integers. When fetching the data with\nPython, we get back integer scalars.\n\n>>> df = pd.DataFrame({\"A\": [1, None, 2]})\n>>> df\n A\n0 1.0\n1 NaN\n2 2.0\n\n>>> from sqlalchemy.types import Integer\n>>> df.to_sql('integers', con=engine, index=False,\n... dtype={\"A\": Integer()})",
"_____no_output_____"
],
[
"import mysql.connector as connection\nimport pandas as pandas\nimport csv\n\ntry:\n mydb = connection.connect(host=\"localhost\", user=\"root\", passwd=\"mysql\",use_pure=True)\n #check if the connection is established\n print(mydb.is_connected())\n #create a new database\n query = \"Create database GlassData;\"\n cursor = mydb.cursor() #create a cursor to execute queries\n cursor.execute(query)\n print(\"Database Created!!\")\n mydb.close() #close the connection\n\n #Establish a new connection to the database created above\n mydb = connection.connect(host=\"localhost\", database = 'GlassData',user=\"root\", passwd=\"mysql\", use_pure=True)\n\n #create a new table to store glass data\n query = \"CREATE TABLE IF NOT EXISTS GlassData (Index_Number INT(10),RI float(10,5), Na float(10,5), Mg float(10,5),Al float(10,5),\" \\\n \" Si float(10,5), K float(10,5), Ca float(10,5), Ba float(10,5), Fe float(10,5), Class INT(5))\"\n cursor = mydb.cursor() # create a cursor to execute queries\n cursor.execute(query)\n print(\"Table Created!!\")\n\n #read from the file\n with open('glass.data', \"r\") as f:\n next(f)\n glass_data = csv.reader(f, delimiter=\"\\n\")\n for line in enumerate(glass_data):\n for list_ in (line[1]):\n cursor.execute('INSERT INTO GlassData values ({values})'.format(values=(list_)))\n print(\"Values inserted!!\")\n mydb.commit()\n cursor.close()\n mydb.close()\n\nexcept Exception as e:\n #mydb.close()\n print(str(e))",
"_____no_output_____"
],
[
"import mysql.connector as connection\nimport pandas as pandas\nimport csv\nmydb = connection.connect(host=\"localhost\", user=\"root\", passwd=\"mysql\",use_pure=True)\n",
"_____no_output_____"
],
[
"cur = mydb.cursor()\ncur.execute(\"CREATE TABLE sudhanshu12345.GlassData1 (Index_Number INT(10),RI float(10,5), Na float(10,5), Mg float(10,5),Al float(10,5),\" \\\n \" Si float(10,5), K float(10,5), Ca float(10,5), Ba float(10,5), Fe float(10,5), Class INT(5))\")",
"_____no_output_____"
],
[
"with open('glass.data',\"r\") as data :\n next(data)\n data_csv = csv.reader(data, delimiter= \"\\n\")\n print(data_csv)\n for i in enumerate(data_csv):\n print(i)\n for j in i[1] :\n cur.execute('insert into sudhanshu12345.GlassData1 values ({data})'.format(data=(j)))\n print(\"all the data inserted \")\nmydb.commit()",
"<_csv.reader object at 0x0000027292AB6CA0>\n(0, ['1,1.52101,13.64,4.49,1.10,71.78,0.06,8.75,0.00,0.00,1'])\n(1, ['2,1.51761,13.89,3.60,1.36,72.73,0.48,7.83,0.00,0.00,1'])\n(2, ['3,1.51618,13.53,3.55,1.54,72.99,0.39,7.78,0.00,0.00,1'])\n(3, ['4,1.51766,13.21,3.69,1.29,72.61,0.57,8.22,0.00,0.00,1'])\n(4, ['5,1.51742,13.27,3.62,1.24,73.08,0.55,8.07,0.00,0.00,1'])\n(5, ['6,1.51596,12.79,3.61,1.62,72.97,0.64,8.07,0.00,0.26,1'])\n(6, ['7,1.51743,13.30,3.60,1.14,73.09,0.58,8.17,0.00,0.00,1'])\n(7, ['8,1.51756,13.15,3.61,1.05,73.24,0.57,8.24,0.00,0.00,1'])\n(8, ['9,1.51918,14.04,3.58,1.37,72.08,0.56,8.30,0.00,0.00,1'])\n(9, ['10,1.51755,13.00,3.60,1.36,72.99,0.57,8.40,0.00,0.11,1'])\n(10, ['11,1.51571,12.72,3.46,1.56,73.20,0.67,8.09,0.00,0.24,1'])\n(11, ['12,1.51763,12.80,3.66,1.27,73.01,0.60,8.56,0.00,0.00,1'])\n(12, ['13,1.51589,12.88,3.43,1.40,73.28,0.69,8.05,0.00,0.24,1'])\n(13, ['14,1.51748,12.86,3.56,1.27,73.21,0.54,8.38,0.00,0.17,1'])\n(14, ['15,1.51763,12.61,3.59,1.31,73.29,0.58,8.50,0.00,0.00,1'])\n(15, ['16,1.51761,12.81,3.54,1.23,73.24,0.58,8.39,0.00,0.00,1'])\n(16, ['17,1.51784,12.68,3.67,1.16,73.11,0.61,8.70,0.00,0.00,1'])\n(17, ['18,1.52196,14.36,3.85,0.89,71.36,0.15,9.15,0.00,0.00,1'])\n(18, ['19,1.51911,13.90,3.73,1.18,72.12,0.06,8.89,0.00,0.00,1'])\n(19, ['20,1.51735,13.02,3.54,1.69,72.73,0.54,8.44,0.00,0.07,1'])\n(20, ['21,1.51750,12.82,3.55,1.49,72.75,0.54,8.52,0.00,0.19,1'])\n(21, ['22,1.51966,14.77,3.75,0.29,72.02,0.03,9.00,0.00,0.00,1'])\n(22, ['23,1.51736,12.78,3.62,1.29,72.79,0.59,8.70,0.00,0.00,1'])\n(23, ['24,1.51751,12.81,3.57,1.35,73.02,0.62,8.59,0.00,0.00,1'])\n(24, ['25,1.51720,13.38,3.50,1.15,72.85,0.50,8.43,0.00,0.00,1'])\n(25, ['26,1.51764,12.98,3.54,1.21,73.00,0.65,8.53,0.00,0.00,1'])\n(26, ['27,1.51793,13.21,3.48,1.41,72.64,0.59,8.43,0.00,0.00,1'])\n(27, ['28,1.51721,12.87,3.48,1.33,73.04,0.56,8.43,0.00,0.00,1'])\n(28, ['29,1.51768,12.56,3.52,1.43,73.15,0.57,8.54,0.00,0.00,1'])\n(29, ['30,1.51784,13.08,3.49,1.28,72.86,0.60,8.49,0.00,0.00,1'])\n(30, ['31,1.51768,12.65,3.56,1.30,73.08,0.61,8.69,0.00,0.14,1'])\n(31, ['32,1.51747,12.84,3.50,1.14,73.27,0.56,8.55,0.00,0.00,1'])\n(32, ['33,1.51775,12.85,3.48,1.23,72.97,0.61,8.56,0.09,0.22,1'])\n(33, ['34,1.51753,12.57,3.47,1.38,73.39,0.60,8.55,0.00,0.06,1'])\n(34, ['35,1.51783,12.69,3.54,1.34,72.95,0.57,8.75,0.00,0.00,1'])\n(35, ['36,1.51567,13.29,3.45,1.21,72.74,0.56,8.57,0.00,0.00,1'])\n(36, ['37,1.51909,13.89,3.53,1.32,71.81,0.51,8.78,0.11,0.00,1'])\n(37, ['38,1.51797,12.74,3.48,1.35,72.96,0.64,8.68,0.00,0.00,1'])\n(38, ['39,1.52213,14.21,3.82,0.47,71.77,0.11,9.57,0.00,0.00,1'])\n(39, ['40,1.52213,14.21,3.82,0.47,71.77,0.11,9.57,0.00,0.00,1'])\n(40, ['41,1.51793,12.79,3.50,1.12,73.03,0.64,8.77,0.00,0.00,1'])\n(41, ['42,1.51755,12.71,3.42,1.20,73.20,0.59,8.64,0.00,0.00,1'])\n(42, ['43,1.51779,13.21,3.39,1.33,72.76,0.59,8.59,0.00,0.00,1'])\n(43, ['44,1.52210,13.73,3.84,0.72,71.76,0.17,9.74,0.00,0.00,1'])\n(44, ['45,1.51786,12.73,3.43,1.19,72.95,0.62,8.76,0.00,0.30,1'])\n(45, ['46,1.51900,13.49,3.48,1.35,71.95,0.55,9.00,0.00,0.00,1'])\n(46, ['47,1.51869,13.19,3.37,1.18,72.72,0.57,8.83,0.00,0.16,1'])\n(47, ['48,1.52667,13.99,3.70,0.71,71.57,0.02,9.82,0.00,0.10,1'])\n(48, ['49,1.52223,13.21,3.77,0.79,71.99,0.13,10.02,0.00,0.00,1'])\n(49, ['50,1.51898,13.58,3.35,1.23,72.08,0.59,8.91,0.00,0.00,1'])\n(50, ['51,1.52320,13.72,3.72,0.51,71.75,0.09,10.06,0.00,0.16,1'])\n(51, ['52,1.51926,13.20,3.33,1.28,72.36,0.60,9.14,0.00,0.11,1'])\n(52, ['53,1.51808,13.43,2.87,1.19,72.84,0.55,9.03,0.00,0.00,1'])\n(53, ['54,1.51837,13.14,2.84,1.28,72.85,0.55,9.07,0.00,0.00,1'])\n(54, ['55,1.51778,13.21,2.81,1.29,72.98,0.51,9.02,0.00,0.09,1'])\n(55, ['56,1.51769,12.45,2.71,1.29,73.70,0.56,9.06,0.00,0.24,1'])\n(56, ['57,1.51215,12.99,3.47,1.12,72.98,0.62,8.35,0.00,0.31,1'])\n(57, ['58,1.51824,12.87,3.48,1.29,72.95,0.60,8.43,0.00,0.00,1'])\n(58, ['59,1.51754,13.48,3.74,1.17,72.99,0.59,8.03,0.00,0.00,1'])\n(59, ['60,1.51754,13.39,3.66,1.19,72.79,0.57,8.27,0.00,0.11,1'])\n(60, ['61,1.51905,13.60,3.62,1.11,72.64,0.14,8.76,0.00,0.00,1'])\n(61, ['62,1.51977,13.81,3.58,1.32,71.72,0.12,8.67,0.69,0.00,1'])\n(62, ['63,1.52172,13.51,3.86,0.88,71.79,0.23,9.54,0.00,0.11,1'])\n(63, ['64,1.52227,14.17,3.81,0.78,71.35,0.00,9.69,0.00,0.00,1'])\n(64, ['65,1.52172,13.48,3.74,0.90,72.01,0.18,9.61,0.00,0.07,1'])\n(65, ['66,1.52099,13.69,3.59,1.12,71.96,0.09,9.40,0.00,0.00,1'])\n(66, ['67,1.52152,13.05,3.65,0.87,72.22,0.19,9.85,0.00,0.17,1'])\n(67, ['68,1.52152,13.05,3.65,0.87,72.32,0.19,9.85,0.00,0.17,1'])\n(68, ['69,1.52152,13.12,3.58,0.90,72.20,0.23,9.82,0.00,0.16,1'])\n(69, ['70,1.52300,13.31,3.58,0.82,71.99,0.12,10.17,0.00,0.03,1'])\n(70, ['71,1.51574,14.86,3.67,1.74,71.87,0.16,7.36,0.00,0.12,2'])\n(71, ['72,1.51848,13.64,3.87,1.27,71.96,0.54,8.32,0.00,0.32,2'])\n(72, ['73,1.51593,13.09,3.59,1.52,73.10,0.67,7.83,0.00,0.00,2'])\n(73, ['74,1.51631,13.34,3.57,1.57,72.87,0.61,7.89,0.00,0.00,2'])\n(74, ['75,1.51596,13.02,3.56,1.54,73.11,0.72,7.90,0.00,0.00,2'])\n(75, ['76,1.51590,13.02,3.58,1.51,73.12,0.69,7.96,0.00,0.00,2'])\n(76, ['77,1.51645,13.44,3.61,1.54,72.39,0.66,8.03,0.00,0.00,2'])\n(77, ['78,1.51627,13.00,3.58,1.54,72.83,0.61,8.04,0.00,0.00,2'])\n(78, ['79,1.51613,13.92,3.52,1.25,72.88,0.37,7.94,0.00,0.14,2'])\n(79, ['80,1.51590,12.82,3.52,1.90,72.86,0.69,7.97,0.00,0.00,2'])\n(80, ['81,1.51592,12.86,3.52,2.12,72.66,0.69,7.97,0.00,0.00,2'])\n(81, ['82,1.51593,13.25,3.45,1.43,73.17,0.61,7.86,0.00,0.00,2'])\n(82, ['83,1.51646,13.41,3.55,1.25,72.81,0.68,8.10,0.00,0.00,2'])\n(83, ['84,1.51594,13.09,3.52,1.55,72.87,0.68,8.05,0.00,0.09,2'])\n(84, ['85,1.51409,14.25,3.09,2.08,72.28,1.10,7.08,0.00,0.00,2'])\n(85, ['86,1.51625,13.36,3.58,1.49,72.72,0.45,8.21,0.00,0.00,2'])\n(86, ['87,1.51569,13.24,3.49,1.47,73.25,0.38,8.03,0.00,0.00,2'])\n(87, ['88,1.51645,13.40,3.49,1.52,72.65,0.67,8.08,0.00,0.10,2'])\n(88, ['89,1.51618,13.01,3.50,1.48,72.89,0.60,8.12,0.00,0.00,2'])\n(89, ['90,1.51640,12.55,3.48,1.87,73.23,0.63,8.08,0.00,0.09,2'])\n(90, ['91,1.51841,12.93,3.74,1.11,72.28,0.64,8.96,0.00,0.22,2'])\n(91, ['92,1.51605,12.90,3.44,1.45,73.06,0.44,8.27,0.00,0.00,2'])\n(92, ['93,1.51588,13.12,3.41,1.58,73.26,0.07,8.39,0.00,0.19,2'])\n(93, ['94,1.51590,13.24,3.34,1.47,73.10,0.39,8.22,0.00,0.00,2'])\n(94, ['95,1.51629,12.71,3.33,1.49,73.28,0.67,8.24,0.00,0.00,2'])\n(95, ['96,1.51860,13.36,3.43,1.43,72.26,0.51,8.60,0.00,0.00,2'])\n(96, ['97,1.51841,13.02,3.62,1.06,72.34,0.64,9.13,0.00,0.15,2'])\n(97, ['98,1.51743,12.20,3.25,1.16,73.55,0.62,8.90,0.00,0.24,2'])\n(98, ['99,1.51689,12.67,2.88,1.71,73.21,0.73,8.54,0.00,0.00,2'])\n(99, ['100,1.51811,12.96,2.96,1.43,72.92,0.60,8.79,0.14,0.00,2'])\n(100, ['101,1.51655,12.75,2.85,1.44,73.27,0.57,8.79,0.11,0.22,2'])\n(101, ['102,1.51730,12.35,2.72,1.63,72.87,0.70,9.23,0.00,0.00,2'])\n(102, ['103,1.51820,12.62,2.76,0.83,73.81,0.35,9.42,0.00,0.20,2'])\n(103, ['104,1.52725,13.80,3.15,0.66,70.57,0.08,11.64,0.00,0.00,2'])\n(104, ['105,1.52410,13.83,2.90,1.17,71.15,0.08,10.79,0.00,0.00,2'])\n(105, ['106,1.52475,11.45,0.00,1.88,72.19,0.81,13.24,0.00,0.34,2'])\n(106, ['107,1.53125,10.73,0.00,2.10,69.81,0.58,13.30,3.15,0.28,2'])\n(107, ['108,1.53393,12.30,0.00,1.00,70.16,0.12,16.19,0.00,0.24,2'])\n(108, ['109,1.52222,14.43,0.00,1.00,72.67,0.10,11.52,0.00,0.08,2'])\n(109, ['110,1.51818,13.72,0.00,0.56,74.45,0.00,10.99,0.00,0.00,2'])\n(110, ['111,1.52664,11.23,0.00,0.77,73.21,0.00,14.68,0.00,0.00,2'])\n(111, ['112,1.52739,11.02,0.00,0.75,73.08,0.00,14.96,0.00,0.00,2'])\n(112, ['113,1.52777,12.64,0.00,0.67,72.02,0.06,14.40,0.00,0.00,2'])\n(113, ['114,1.51892,13.46,3.83,1.26,72.55,0.57,8.21,0.00,0.14,2'])\n(114, ['115,1.51847,13.10,3.97,1.19,72.44,0.60,8.43,0.00,0.00,2'])\n(115, ['116,1.51846,13.41,3.89,1.33,72.38,0.51,8.28,0.00,0.00,2'])\n(116, ['117,1.51829,13.24,3.90,1.41,72.33,0.55,8.31,0.00,0.10,2'])\n(117, ['118,1.51708,13.72,3.68,1.81,72.06,0.64,7.88,0.00,0.00,2'])\n(118, ['119,1.51673,13.30,3.64,1.53,72.53,0.65,8.03,0.00,0.29,2'])\n(119, ['120,1.51652,13.56,3.57,1.47,72.45,0.64,7.96,0.00,0.00,2'])\n(120, ['121,1.51844,13.25,3.76,1.32,72.40,0.58,8.42,0.00,0.00,2'])\n(121, ['122,1.51663,12.93,3.54,1.62,72.96,0.64,8.03,0.00,0.21,2'])\n(122, ['123,1.51687,13.23,3.54,1.48,72.84,0.56,8.10,0.00,0.00,2'])\n(123, ['124,1.51707,13.48,3.48,1.71,72.52,0.62,7.99,0.00,0.00,2'])\n(124, ['125,1.52177,13.20,3.68,1.15,72.75,0.54,8.52,0.00,0.00,2'])\n(125, ['126,1.51872,12.93,3.66,1.56,72.51,0.58,8.55,0.00,0.12,2'])\n(126, ['127,1.51667,12.94,3.61,1.26,72.75,0.56,8.60,0.00,0.00,2'])\n(127, ['128,1.52081,13.78,2.28,1.43,71.99,0.49,9.85,0.00,0.17,2'])\n(128, ['129,1.52068,13.55,2.09,1.67,72.18,0.53,9.57,0.27,0.17,2'])\n(129, ['130,1.52020,13.98,1.35,1.63,71.76,0.39,10.56,0.00,0.18,2'])\n(130, ['131,1.52177,13.75,1.01,1.36,72.19,0.33,11.14,0.00,0.00,2'])\n(131, ['132,1.52614,13.70,0.00,1.36,71.24,0.19,13.44,0.00,0.10,2'])\n(132, ['133,1.51813,13.43,3.98,1.18,72.49,0.58,8.15,0.00,0.00,2'])\n(133, ['134,1.51800,13.71,3.93,1.54,71.81,0.54,8.21,0.00,0.15,2'])\n(134, ['135,1.51811,13.33,3.85,1.25,72.78,0.52,8.12,0.00,0.00,2'])\n(135, ['136,1.51789,13.19,3.90,1.30,72.33,0.55,8.44,0.00,0.28,2'])\n(136, ['137,1.51806,13.00,3.80,1.08,73.07,0.56,8.38,0.00,0.12,2'])\n(137, ['138,1.51711,12.89,3.62,1.57,72.96,0.61,8.11,0.00,0.00,2'])\n(138, ['139,1.51674,12.79,3.52,1.54,73.36,0.66,7.90,0.00,0.00,2'])\n(139, ['140,1.51674,12.87,3.56,1.64,73.14,0.65,7.99,0.00,0.00,2'])\n(140, ['141,1.51690,13.33,3.54,1.61,72.54,0.68,8.11,0.00,0.00,2'])\n(141, ['142,1.51851,13.20,3.63,1.07,72.83,0.57,8.41,0.09,0.17,2'])\n(142, ['143,1.51662,12.85,3.51,1.44,73.01,0.68,8.23,0.06,0.25,2'])\n(143, ['144,1.51709,13.00,3.47,1.79,72.72,0.66,8.18,0.00,0.00,2'])\n(144, ['145,1.51660,12.99,3.18,1.23,72.97,0.58,8.81,0.00,0.24,2'])\n(145, ['146,1.51839,12.85,3.67,1.24,72.57,0.62,8.68,0.00,0.35,2'])\n(146, ['147,1.51769,13.65,3.66,1.11,72.77,0.11,8.60,0.00,0.00,3'])\n(147, ['148,1.51610,13.33,3.53,1.34,72.67,0.56,8.33,0.00,0.00,3'])\n(148, ['149,1.51670,13.24,3.57,1.38,72.70,0.56,8.44,0.00,0.10,3'])\n(149, ['150,1.51643,12.16,3.52,1.35,72.89,0.57,8.53,0.00,0.00,3'])\n(150, ['151,1.51665,13.14,3.45,1.76,72.48,0.60,8.38,0.00,0.17,3'])\n(151, ['152,1.52127,14.32,3.90,0.83,71.50,0.00,9.49,0.00,0.00,3'])\n(152, ['153,1.51779,13.64,3.65,0.65,73.00,0.06,8.93,0.00,0.00,3'])\n(153, ['154,1.51610,13.42,3.40,1.22,72.69,0.59,8.32,0.00,0.00,3'])\n(154, ['155,1.51694,12.86,3.58,1.31,72.61,0.61,8.79,0.00,0.00,3'])\n(155, ['156,1.51646,13.04,3.40,1.26,73.01,0.52,8.58,0.00,0.00,3'])\n(156, ['157,1.51655,13.41,3.39,1.28,72.64,0.52,8.65,0.00,0.00,3'])\n(157, ['158,1.52121,14.03,3.76,0.58,71.79,0.11,9.65,0.00,0.00,3'])\n(158, ['159,1.51776,13.53,3.41,1.52,72.04,0.58,8.79,0.00,0.00,3'])\n(159, ['160,1.51796,13.50,3.36,1.63,71.94,0.57,8.81,0.00,0.09,3'])\n(160, ['161,1.51832,13.33,3.34,1.54,72.14,0.56,8.99,0.00,0.00,3'])\n(161, ['162,1.51934,13.64,3.54,0.75,72.65,0.16,8.89,0.15,0.24,3'])\n(162, ['163,1.52211,14.19,3.78,0.91,71.36,0.23,9.14,0.00,0.37,3'])\n(163, ['164,1.51514,14.01,2.68,3.50,69.89,1.68,5.87,2.20,0.00,5'])\n(164, ['165,1.51915,12.73,1.85,1.86,72.69,0.60,10.09,0.00,0.00,5'])\n(165, ['166,1.52171,11.56,1.88,1.56,72.86,0.47,11.41,0.00,0.00,5'])\n(166, ['167,1.52151,11.03,1.71,1.56,73.44,0.58,11.62,0.00,0.00,5'])\n(167, ['168,1.51969,12.64,0.00,1.65,73.75,0.38,11.53,0.00,0.00,5'])\n(168, ['169,1.51666,12.86,0.00,1.83,73.88,0.97,10.17,0.00,0.00,5'])\n(169, ['170,1.51994,13.27,0.00,1.76,73.03,0.47,11.32,0.00,0.00,5'])\n(170, ['171,1.52369,13.44,0.00,1.58,72.22,0.32,12.24,0.00,0.00,5'])\n(171, ['172,1.51316,13.02,0.00,3.04,70.48,6.21,6.96,0.00,0.00,5'])\n(172, ['173,1.51321,13.00,0.00,3.02,70.70,6.21,6.93,0.00,0.00,5'])\n(173, ['174,1.52043,13.38,0.00,1.40,72.25,0.33,12.50,0.00,0.00,5'])\n(174, ['175,1.52058,12.85,1.61,2.17,72.18,0.76,9.70,0.24,0.51,5'])\n(175, ['176,1.52119,12.97,0.33,1.51,73.39,0.13,11.27,0.00,0.28,5'])\n(176, ['177,1.51905,14.00,2.39,1.56,72.37,0.00,9.57,0.00,0.00,6'])\n(177, ['178,1.51937,13.79,2.41,1.19,72.76,0.00,9.77,0.00,0.00,6'])\n(178, ['179,1.51829,14.46,2.24,1.62,72.38,0.00,9.26,0.00,0.00,6'])\n(179, ['180,1.51852,14.09,2.19,1.66,72.67,0.00,9.32,0.00,0.00,6'])\n(180, ['181,1.51299,14.40,1.74,1.54,74.55,0.00,7.59,0.00,0.00,6'])\n(181, ['182,1.51888,14.99,0.78,1.74,72.50,0.00,9.95,0.00,0.00,6'])\n(182, ['183,1.51916,14.15,0.00,2.09,72.74,0.00,10.88,0.00,0.00,6'])\n(183, ['184,1.51969,14.56,0.00,0.56,73.48,0.00,11.22,0.00,0.00,6'])\n(184, ['185,1.51115,17.38,0.00,0.34,75.41,0.00,6.65,0.00,0.00,6'])\n(185, ['186,1.51131,13.69,3.20,1.81,72.81,1.76,5.43,1.19,0.00,7'])\n(186, ['187,1.51838,14.32,3.26,2.22,71.25,1.46,5.79,1.63,0.00,7'])\n(187, ['188,1.52315,13.44,3.34,1.23,72.38,0.60,8.83,0.00,0.00,7'])\n(188, ['189,1.52247,14.86,2.20,2.06,70.26,0.76,9.76,0.00,0.00,7'])\n(189, ['190,1.52365,15.79,1.83,1.31,70.43,0.31,8.61,1.68,0.00,7'])\n(190, ['191,1.51613,13.88,1.78,1.79,73.10,0.00,8.67,0.76,0.00,7'])\n(191, ['192,1.51602,14.85,0.00,2.38,73.28,0.00,8.76,0.64,0.09,7'])\n(192, ['193,1.51623,14.20,0.00,2.79,73.46,0.04,9.04,0.40,0.09,7'])\n(193, ['194,1.51719,14.75,0.00,2.00,73.02,0.00,8.53,1.59,0.08,7'])\n(194, ['195,1.51683,14.56,0.00,1.98,73.29,0.00,8.52,1.57,0.07,7'])\n(195, ['196,1.51545,14.14,0.00,2.68,73.39,0.08,9.07,0.61,0.05,7'])\n(196, ['197,1.51556,13.87,0.00,2.54,73.23,0.14,9.41,0.81,0.01,7'])\n(197, ['198,1.51727,14.70,0.00,2.34,73.28,0.00,8.95,0.66,0.00,7'])\n(198, ['199,1.51531,14.38,0.00,2.66,73.10,0.04,9.08,0.64,0.00,7'])\n(199, ['200,1.51609,15.01,0.00,2.51,73.05,0.05,8.83,0.53,0.00,7'])\n(200, ['201,1.51508,15.15,0.00,2.25,73.50,0.00,8.34,0.63,0.00,7'])\n(201, ['202,1.51653,11.95,0.00,1.19,75.18,2.70,8.93,0.00,0.00,7'])\n(202, ['203,1.51514,14.85,0.00,2.42,73.72,0.00,8.39,0.56,0.00,7'])\n(203, ['204,1.51658,14.80,0.00,1.99,73.11,0.00,8.28,1.71,0.00,7'])\n(204, ['205,1.51617,14.95,0.00,2.27,73.30,0.00,8.71,0.67,0.00,7'])\n(205, ['206,1.51732,14.95,0.00,1.80,72.99,0.00,8.61,1.55,0.00,7'])\n(206, ['207,1.51645,14.94,0.00,1.87,73.11,0.00,8.67,1.38,0.00,7'])\n(207, ['208,1.51831,14.39,0.00,1.82,72.86,1.41,6.47,2.88,0.00,7'])\n(208, ['209,1.51640,14.37,0.00,2.74,72.85,0.00,9.45,0.54,0.00,7'])\n(209, ['210,1.51623,14.14,0.00,2.88,72.61,0.08,9.18,1.06,0.00,7'])\n(210, ['211,1.51685,14.92,0.00,1.99,73.06,0.00,8.40,1.59,0.00,7'])\n(211, ['212,1.52065,14.36,0.00,2.02,73.42,0.00,8.44,1.64,0.00,7'])\n(212, ['213,1.51651,14.38,0.00,1.94,73.61,0.00,8.48,1.57,0.00,7'])\n(213, ['214,1.51711,14.23,0.00,2.08,73.36,0.00,8.62,1.67,0.00,7'])\nall the data inserted \n"
],
[
"!ls",
"19.2.MySQL.ipynb\nimgs\nImp files\nmydata.csv\nMYSQL application.docx\nMYSQL application.pdf\nUntitled.ipynb\n"
],
[
"with open('glass.data',\"r\") as data :\n next(data)\n data_csv = csv.reader(data, delimiter= \"\\n\")\n print(data_csv)\n for i in enumerate(data_csv):\n print(i)\n for j in i[1] :\n print(type(j))\n cur.execute('insert into sudhanshu12345.GlassData1 values ({data})'.format(data=(j)))\n print(\"all the data inserted \")\nmydb.commit()",
"<_csv.reader object at 0x0000027293FE2460>\n(0, ['1,1.52101,13.64,4.49,1.10,71.78,0.06,8.75,0.00,0.00,1'])\n<class 'str'>\n(1, ['2,1.51761,13.89,3.60,1.36,72.73,0.48,7.83,0.00,0.00,1'])\n<class 'str'>\n(2, ['3,1.51618,13.53,3.55,1.54,72.99,0.39,7.78,0.00,0.00,1'])\n<class 'str'>\n(3, ['4,1.51766,13.21,3.69,1.29,72.61,0.57,8.22,0.00,0.00,1'])\n<class 'str'>\n(4, ['5,1.51742,13.27,3.62,1.24,73.08,0.55,8.07,0.00,0.00,1'])\n<class 'str'>\n(5, ['6,1.51596,12.79,3.61,1.62,72.97,0.64,8.07,0.00,0.26,1'])\n<class 'str'>\n(6, ['7,1.51743,13.30,3.60,1.14,73.09,0.58,8.17,0.00,0.00,1'])\n<class 'str'>\n(7, ['8,1.51756,13.15,3.61,1.05,73.24,0.57,8.24,0.00,0.00,1'])\n<class 'str'>\n(8, ['9,1.51918,14.04,3.58,1.37,72.08,0.56,8.30,0.00,0.00,1'])\n<class 'str'>\n(9, ['10,1.51755,13.00,3.60,1.36,72.99,0.57,8.40,0.00,0.11,1'])\n<class 'str'>\n(10, ['11,1.51571,12.72,3.46,1.56,73.20,0.67,8.09,0.00,0.24,1'])\n<class 'str'>\n(11, ['12,1.51763,12.80,3.66,1.27,73.01,0.60,8.56,0.00,0.00,1'])\n<class 'str'>\n(12, ['13,1.51589,12.88,3.43,1.40,73.28,0.69,8.05,0.00,0.24,1'])\n<class 'str'>\n(13, ['14,1.51748,12.86,3.56,1.27,73.21,0.54,8.38,0.00,0.17,1'])\n<class 'str'>\n(14, ['15,1.51763,12.61,3.59,1.31,73.29,0.58,8.50,0.00,0.00,1'])\n<class 'str'>\n(15, ['16,1.51761,12.81,3.54,1.23,73.24,0.58,8.39,0.00,0.00,1'])\n<class 'str'>\n(16, ['17,1.51784,12.68,3.67,1.16,73.11,0.61,8.70,0.00,0.00,1'])\n<class 'str'>\n(17, ['18,1.52196,14.36,3.85,0.89,71.36,0.15,9.15,0.00,0.00,1'])\n<class 'str'>\n(18, ['19,1.51911,13.90,3.73,1.18,72.12,0.06,8.89,0.00,0.00,1'])\n<class 'str'>\n(19, ['20,1.51735,13.02,3.54,1.69,72.73,0.54,8.44,0.00,0.07,1'])\n<class 'str'>\n(20, ['21,1.51750,12.82,3.55,1.49,72.75,0.54,8.52,0.00,0.19,1'])\n<class 'str'>\n(21, ['22,1.51966,14.77,3.75,0.29,72.02,0.03,9.00,0.00,0.00,1'])\n<class 'str'>\n(22, ['23,1.51736,12.78,3.62,1.29,72.79,0.59,8.70,0.00,0.00,1'])\n<class 'str'>\n(23, ['24,1.51751,12.81,3.57,1.35,73.02,0.62,8.59,0.00,0.00,1'])\n<class 'str'>\n(24, ['25,1.51720,13.38,3.50,1.15,72.85,0.50,8.43,0.00,0.00,1'])\n<class 'str'>\n(25, ['26,1.51764,12.98,3.54,1.21,73.00,0.65,8.53,0.00,0.00,1'])\n<class 'str'>\n(26, ['27,1.51793,13.21,3.48,1.41,72.64,0.59,8.43,0.00,0.00,1'])\n<class 'str'>\n(27, ['28,1.51721,12.87,3.48,1.33,73.04,0.56,8.43,0.00,0.00,1'])\n<class 'str'>\n(28, ['29,1.51768,12.56,3.52,1.43,73.15,0.57,8.54,0.00,0.00,1'])\n<class 'str'>\n(29, ['30,1.51784,13.08,3.49,1.28,72.86,0.60,8.49,0.00,0.00,1'])\n<class 'str'>\n(30, ['31,1.51768,12.65,3.56,1.30,73.08,0.61,8.69,0.00,0.14,1'])\n<class 'str'>\n(31, ['32,1.51747,12.84,3.50,1.14,73.27,0.56,8.55,0.00,0.00,1'])\n<class 'str'>\n(32, ['33,1.51775,12.85,3.48,1.23,72.97,0.61,8.56,0.09,0.22,1'])\n<class 'str'>\n(33, ['34,1.51753,12.57,3.47,1.38,73.39,0.60,8.55,0.00,0.06,1'])\n<class 'str'>\n(34, ['35,1.51783,12.69,3.54,1.34,72.95,0.57,8.75,0.00,0.00,1'])\n<class 'str'>\n(35, ['36,1.51567,13.29,3.45,1.21,72.74,0.56,8.57,0.00,0.00,1'])\n<class 'str'>\n(36, ['37,1.51909,13.89,3.53,1.32,71.81,0.51,8.78,0.11,0.00,1'])\n<class 'str'>\n(37, ['38,1.51797,12.74,3.48,1.35,72.96,0.64,8.68,0.00,0.00,1'])\n<class 'str'>\n(38, ['39,1.52213,14.21,3.82,0.47,71.77,0.11,9.57,0.00,0.00,1'])\n<class 'str'>\n(39, ['40,1.52213,14.21,3.82,0.47,71.77,0.11,9.57,0.00,0.00,1'])\n<class 'str'>\n(40, ['41,1.51793,12.79,3.50,1.12,73.03,0.64,8.77,0.00,0.00,1'])\n<class 'str'>\n(41, ['42,1.51755,12.71,3.42,1.20,73.20,0.59,8.64,0.00,0.00,1'])\n<class 'str'>\n(42, ['43,1.51779,13.21,3.39,1.33,72.76,0.59,8.59,0.00,0.00,1'])\n<class 'str'>\n(43, ['44,1.52210,13.73,3.84,0.72,71.76,0.17,9.74,0.00,0.00,1'])\n<class 'str'>\n(44, ['45,1.51786,12.73,3.43,1.19,72.95,0.62,8.76,0.00,0.30,1'])\n<class 'str'>\n(45, ['46,1.51900,13.49,3.48,1.35,71.95,0.55,9.00,0.00,0.00,1'])\n<class 'str'>\n(46, ['47,1.51869,13.19,3.37,1.18,72.72,0.57,8.83,0.00,0.16,1'])\n<class 'str'>\n(47, ['48,1.52667,13.99,3.70,0.71,71.57,0.02,9.82,0.00,0.10,1'])\n<class 'str'>\n(48, ['49,1.52223,13.21,3.77,0.79,71.99,0.13,10.02,0.00,0.00,1'])\n<class 'str'>\n(49, ['50,1.51898,13.58,3.35,1.23,72.08,0.59,8.91,0.00,0.00,1'])\n<class 'str'>\n(50, ['51,1.52320,13.72,3.72,0.51,71.75,0.09,10.06,0.00,0.16,1'])\n<class 'str'>\n(51, ['52,1.51926,13.20,3.33,1.28,72.36,0.60,9.14,0.00,0.11,1'])\n<class 'str'>\n(52, ['53,1.51808,13.43,2.87,1.19,72.84,0.55,9.03,0.00,0.00,1'])\n<class 'str'>\n(53, ['54,1.51837,13.14,2.84,1.28,72.85,0.55,9.07,0.00,0.00,1'])\n<class 'str'>\n(54, ['55,1.51778,13.21,2.81,1.29,72.98,0.51,9.02,0.00,0.09,1'])\n<class 'str'>\n(55, ['56,1.51769,12.45,2.71,1.29,73.70,0.56,9.06,0.00,0.24,1'])\n<class 'str'>\n(56, ['57,1.51215,12.99,3.47,1.12,72.98,0.62,8.35,0.00,0.31,1'])\n<class 'str'>\n(57, ['58,1.51824,12.87,3.48,1.29,72.95,0.60,8.43,0.00,0.00,1'])\n<class 'str'>\n(58, ['59,1.51754,13.48,3.74,1.17,72.99,0.59,8.03,0.00,0.00,1'])\n<class 'str'>\n(59, ['60,1.51754,13.39,3.66,1.19,72.79,0.57,8.27,0.00,0.11,1'])\n<class 'str'>\n(60, ['61,1.51905,13.60,3.62,1.11,72.64,0.14,8.76,0.00,0.00,1'])\n<class 'str'>\n(61, ['62,1.51977,13.81,3.58,1.32,71.72,0.12,8.67,0.69,0.00,1'])\n<class 'str'>\n(62, ['63,1.52172,13.51,3.86,0.88,71.79,0.23,9.54,0.00,0.11,1'])\n<class 'str'>\n(63, ['64,1.52227,14.17,3.81,0.78,71.35,0.00,9.69,0.00,0.00,1'])\n<class 'str'>\n(64, ['65,1.52172,13.48,3.74,0.90,72.01,0.18,9.61,0.00,0.07,1'])\n<class 'str'>\n(65, ['66,1.52099,13.69,3.59,1.12,71.96,0.09,9.40,0.00,0.00,1'])\n<class 'str'>\n(66, ['67,1.52152,13.05,3.65,0.87,72.22,0.19,9.85,0.00,0.17,1'])\n<class 'str'>\n(67, ['68,1.52152,13.05,3.65,0.87,72.32,0.19,9.85,0.00,0.17,1'])\n<class 'str'>\n(68, ['69,1.52152,13.12,3.58,0.90,72.20,0.23,9.82,0.00,0.16,1'])\n<class 'str'>\n(69, ['70,1.52300,13.31,3.58,0.82,71.99,0.12,10.17,0.00,0.03,1'])\n<class 'str'>\n(70, ['71,1.51574,14.86,3.67,1.74,71.87,0.16,7.36,0.00,0.12,2'])\n<class 'str'>\n(71, ['72,1.51848,13.64,3.87,1.27,71.96,0.54,8.32,0.00,0.32,2'])\n<class 'str'>\n(72, ['73,1.51593,13.09,3.59,1.52,73.10,0.67,7.83,0.00,0.00,2'])\n<class 'str'>\n(73, ['74,1.51631,13.34,3.57,1.57,72.87,0.61,7.89,0.00,0.00,2'])\n<class 'str'>\n(74, ['75,1.51596,13.02,3.56,1.54,73.11,0.72,7.90,0.00,0.00,2'])\n<class 'str'>\n(75, ['76,1.51590,13.02,3.58,1.51,73.12,0.69,7.96,0.00,0.00,2'])\n<class 'str'>\n(76, ['77,1.51645,13.44,3.61,1.54,72.39,0.66,8.03,0.00,0.00,2'])\n<class 'str'>\n(77, ['78,1.51627,13.00,3.58,1.54,72.83,0.61,8.04,0.00,0.00,2'])\n<class 'str'>\n(78, ['79,1.51613,13.92,3.52,1.25,72.88,0.37,7.94,0.00,0.14,2'])\n<class 'str'>\n(79, ['80,1.51590,12.82,3.52,1.90,72.86,0.69,7.97,0.00,0.00,2'])\n<class 'str'>\n(80, ['81,1.51592,12.86,3.52,2.12,72.66,0.69,7.97,0.00,0.00,2'])\n<class 'str'>\n(81, ['82,1.51593,13.25,3.45,1.43,73.17,0.61,7.86,0.00,0.00,2'])\n<class 'str'>\n(82, ['83,1.51646,13.41,3.55,1.25,72.81,0.68,8.10,0.00,0.00,2'])\n<class 'str'>\n(83, ['84,1.51594,13.09,3.52,1.55,72.87,0.68,8.05,0.00,0.09,2'])\n<class 'str'>\n(84, ['85,1.51409,14.25,3.09,2.08,72.28,1.10,7.08,0.00,0.00,2'])\n<class 'str'>\n(85, ['86,1.51625,13.36,3.58,1.49,72.72,0.45,8.21,0.00,0.00,2'])\n<class 'str'>\n(86, ['87,1.51569,13.24,3.49,1.47,73.25,0.38,8.03,0.00,0.00,2'])\n<class 'str'>\n(87, ['88,1.51645,13.40,3.49,1.52,72.65,0.67,8.08,0.00,0.10,2'])\n<class 'str'>\n(88, ['89,1.51618,13.01,3.50,1.48,72.89,0.60,8.12,0.00,0.00,2'])\n<class 'str'>\n(89, ['90,1.51640,12.55,3.48,1.87,73.23,0.63,8.08,0.00,0.09,2'])\n<class 'str'>\n(90, ['91,1.51841,12.93,3.74,1.11,72.28,0.64,8.96,0.00,0.22,2'])\n<class 'str'>\n(91, ['92,1.51605,12.90,3.44,1.45,73.06,0.44,8.27,0.00,0.00,2'])\n<class 'str'>\n(92, ['93,1.51588,13.12,3.41,1.58,73.26,0.07,8.39,0.00,0.19,2'])\n<class 'str'>\n(93, ['94,1.51590,13.24,3.34,1.47,73.10,0.39,8.22,0.00,0.00,2'])\n<class 'str'>\n(94, ['95,1.51629,12.71,3.33,1.49,73.28,0.67,8.24,0.00,0.00,2'])\n<class 'str'>\n(95, ['96,1.51860,13.36,3.43,1.43,72.26,0.51,8.60,0.00,0.00,2'])\n<class 'str'>\n(96, ['97,1.51841,13.02,3.62,1.06,72.34,0.64,9.13,0.00,0.15,2'])\n<class 'str'>\n(97, ['98,1.51743,12.20,3.25,1.16,73.55,0.62,8.90,0.00,0.24,2'])\n<class 'str'>\n(98, ['99,1.51689,12.67,2.88,1.71,73.21,0.73,8.54,0.00,0.00,2'])\n<class 'str'>\n(99, ['100,1.51811,12.96,2.96,1.43,72.92,0.60,8.79,0.14,0.00,2'])\n<class 'str'>\n(100, ['101,1.51655,12.75,2.85,1.44,73.27,0.57,8.79,0.11,0.22,2'])\n<class 'str'>\n(101, ['102,1.51730,12.35,2.72,1.63,72.87,0.70,9.23,0.00,0.00,2'])\n<class 'str'>\n(102, ['103,1.51820,12.62,2.76,0.83,73.81,0.35,9.42,0.00,0.20,2'])\n<class 'str'>\n(103, ['104,1.52725,13.80,3.15,0.66,70.57,0.08,11.64,0.00,0.00,2'])\n<class 'str'>\n(104, ['105,1.52410,13.83,2.90,1.17,71.15,0.08,10.79,0.00,0.00,2'])\n<class 'str'>\n(105, ['106,1.52475,11.45,0.00,1.88,72.19,0.81,13.24,0.00,0.34,2'])\n<class 'str'>\n(106, ['107,1.53125,10.73,0.00,2.10,69.81,0.58,13.30,3.15,0.28,2'])\n<class 'str'>\n(107, ['108,1.53393,12.30,0.00,1.00,70.16,0.12,16.19,0.00,0.24,2'])\n<class 'str'>\n(108, ['109,1.52222,14.43,0.00,1.00,72.67,0.10,11.52,0.00,0.08,2'])\n<class 'str'>\n(109, ['110,1.51818,13.72,0.00,0.56,74.45,0.00,10.99,0.00,0.00,2'])\n<class 'str'>\n(110, ['111,1.52664,11.23,0.00,0.77,73.21,0.00,14.68,0.00,0.00,2'])\n<class 'str'>\n(111, ['112,1.52739,11.02,0.00,0.75,73.08,0.00,14.96,0.00,0.00,2'])\n<class 'str'>\n(112, ['113,1.52777,12.64,0.00,0.67,72.02,0.06,14.40,0.00,0.00,2'])\n<class 'str'>\n(113, ['114,1.51892,13.46,3.83,1.26,72.55,0.57,8.21,0.00,0.14,2'])\n<class 'str'>\n(114, ['115,1.51847,13.10,3.97,1.19,72.44,0.60,8.43,0.00,0.00,2'])\n<class 'str'>\n(115, ['116,1.51846,13.41,3.89,1.33,72.38,0.51,8.28,0.00,0.00,2'])\n<class 'str'>\n(116, ['117,1.51829,13.24,3.90,1.41,72.33,0.55,8.31,0.00,0.10,2'])\n<class 'str'>\n(117, ['118,1.51708,13.72,3.68,1.81,72.06,0.64,7.88,0.00,0.00,2'])\n<class 'str'>\n(118, ['119,1.51673,13.30,3.64,1.53,72.53,0.65,8.03,0.00,0.29,2'])\n<class 'str'>\n(119, ['120,1.51652,13.56,3.57,1.47,72.45,0.64,7.96,0.00,0.00,2'])\n<class 'str'>\n(120, ['121,1.51844,13.25,3.76,1.32,72.40,0.58,8.42,0.00,0.00,2'])\n<class 'str'>\n(121, ['122,1.51663,12.93,3.54,1.62,72.96,0.64,8.03,0.00,0.21,2'])\n<class 'str'>\n(122, ['123,1.51687,13.23,3.54,1.48,72.84,0.56,8.10,0.00,0.00,2'])\n<class 'str'>\n(123, ['124,1.51707,13.48,3.48,1.71,72.52,0.62,7.99,0.00,0.00,2'])\n<class 'str'>\n(124, ['125,1.52177,13.20,3.68,1.15,72.75,0.54,8.52,0.00,0.00,2'])\n<class 'str'>\n(125, ['126,1.51872,12.93,3.66,1.56,72.51,0.58,8.55,0.00,0.12,2'])\n<class 'str'>\n(126, ['127,1.51667,12.94,3.61,1.26,72.75,0.56,8.60,0.00,0.00,2'])\n<class 'str'>\n(127, ['128,1.52081,13.78,2.28,1.43,71.99,0.49,9.85,0.00,0.17,2'])\n<class 'str'>\n(128, ['129,1.52068,13.55,2.09,1.67,72.18,0.53,9.57,0.27,0.17,2'])\n<class 'str'>\n(129, ['130,1.52020,13.98,1.35,1.63,71.76,0.39,10.56,0.00,0.18,2'])\n<class 'str'>\n(130, ['131,1.52177,13.75,1.01,1.36,72.19,0.33,11.14,0.00,0.00,2'])\n<class 'str'>\n(131, ['132,1.52614,13.70,0.00,1.36,71.24,0.19,13.44,0.00,0.10,2'])\n<class 'str'>\n(132, ['133,1.51813,13.43,3.98,1.18,72.49,0.58,8.15,0.00,0.00,2'])\n<class 'str'>\n(133, ['134,1.51800,13.71,3.93,1.54,71.81,0.54,8.21,0.00,0.15,2'])\n<class 'str'>\n(134, ['135,1.51811,13.33,3.85,1.25,72.78,0.52,8.12,0.00,0.00,2'])\n<class 'str'>\n(135, ['136,1.51789,13.19,3.90,1.30,72.33,0.55,8.44,0.00,0.28,2'])\n<class 'str'>\n(136, ['137,1.51806,13.00,3.80,1.08,73.07,0.56,8.38,0.00,0.12,2'])\n<class 'str'>\n(137, ['138,1.51711,12.89,3.62,1.57,72.96,0.61,8.11,0.00,0.00,2'])\n<class 'str'>\n(138, ['139,1.51674,12.79,3.52,1.54,73.36,0.66,7.90,0.00,0.00,2'])\n<class 'str'>\n(139, ['140,1.51674,12.87,3.56,1.64,73.14,0.65,7.99,0.00,0.00,2'])\n<class 'str'>\n(140, ['141,1.51690,13.33,3.54,1.61,72.54,0.68,8.11,0.00,0.00,2'])\n<class 'str'>\n(141, ['142,1.51851,13.20,3.63,1.07,72.83,0.57,8.41,0.09,0.17,2'])\n<class 'str'>\n(142, ['143,1.51662,12.85,3.51,1.44,73.01,0.68,8.23,0.06,0.25,2'])\n<class 'str'>\n(143, ['144,1.51709,13.00,3.47,1.79,72.72,0.66,8.18,0.00,0.00,2'])\n<class 'str'>\n(144, ['145,1.51660,12.99,3.18,1.23,72.97,0.58,8.81,0.00,0.24,2'])\n<class 'str'>\n(145, ['146,1.51839,12.85,3.67,1.24,72.57,0.62,8.68,0.00,0.35,2'])\n<class 'str'>\n(146, ['147,1.51769,13.65,3.66,1.11,72.77,0.11,8.60,0.00,0.00,3'])\n<class 'str'>\n(147, ['148,1.51610,13.33,3.53,1.34,72.67,0.56,8.33,0.00,0.00,3'])\n<class 'str'>\n(148, ['149,1.51670,13.24,3.57,1.38,72.70,0.56,8.44,0.00,0.10,3'])\n<class 'str'>\n(149, ['150,1.51643,12.16,3.52,1.35,72.89,0.57,8.53,0.00,0.00,3'])\n<class 'str'>\n(150, ['151,1.51665,13.14,3.45,1.76,72.48,0.60,8.38,0.00,0.17,3'])\n<class 'str'>\n(151, ['152,1.52127,14.32,3.90,0.83,71.50,0.00,9.49,0.00,0.00,3'])\n<class 'str'>\n(152, ['153,1.51779,13.64,3.65,0.65,73.00,0.06,8.93,0.00,0.00,3'])\n<class 'str'>\n(153, ['154,1.51610,13.42,3.40,1.22,72.69,0.59,8.32,0.00,0.00,3'])\n<class 'str'>\n(154, ['155,1.51694,12.86,3.58,1.31,72.61,0.61,8.79,0.00,0.00,3'])\n<class 'str'>\n(155, ['156,1.51646,13.04,3.40,1.26,73.01,0.52,8.58,0.00,0.00,3'])\n<class 'str'>\n(156, ['157,1.51655,13.41,3.39,1.28,72.64,0.52,8.65,0.00,0.00,3'])\n<class 'str'>\n(157, ['158,1.52121,14.03,3.76,0.58,71.79,0.11,9.65,0.00,0.00,3'])\n<class 'str'>\n(158, ['159,1.51776,13.53,3.41,1.52,72.04,0.58,8.79,0.00,0.00,3'])\n<class 'str'>\n(159, ['160,1.51796,13.50,3.36,1.63,71.94,0.57,8.81,0.00,0.09,3'])\n<class 'str'>\n(160, ['161,1.51832,13.33,3.34,1.54,72.14,0.56,8.99,0.00,0.00,3'])\n<class 'str'>\n(161, ['162,1.51934,13.64,3.54,0.75,72.65,0.16,8.89,0.15,0.24,3'])\n<class 'str'>\n(162, ['163,1.52211,14.19,3.78,0.91,71.36,0.23,9.14,0.00,0.37,3'])\n<class 'str'>\n(163, ['164,1.51514,14.01,2.68,3.50,69.89,1.68,5.87,2.20,0.00,5'])\n<class 'str'>\n(164, ['165,1.51915,12.73,1.85,1.86,72.69,0.60,10.09,0.00,0.00,5'])\n<class 'str'>\n(165, ['166,1.52171,11.56,1.88,1.56,72.86,0.47,11.41,0.00,0.00,5'])\n<class 'str'>\n(166, ['167,1.52151,11.03,1.71,1.56,73.44,0.58,11.62,0.00,0.00,5'])\n<class 'str'>\n(167, ['168,1.51969,12.64,0.00,1.65,73.75,0.38,11.53,0.00,0.00,5'])\n<class 'str'>\n(168, ['169,1.51666,12.86,0.00,1.83,73.88,0.97,10.17,0.00,0.00,5'])\n<class 'str'>\n(169, ['170,1.51994,13.27,0.00,1.76,73.03,0.47,11.32,0.00,0.00,5'])\n<class 'str'>\n(170, ['171,1.52369,13.44,0.00,1.58,72.22,0.32,12.24,0.00,0.00,5'])\n<class 'str'>\n(171, ['172,1.51316,13.02,0.00,3.04,70.48,6.21,6.96,0.00,0.00,5'])\n<class 'str'>\n(172, ['173,1.51321,13.00,0.00,3.02,70.70,6.21,6.93,0.00,0.00,5'])\n<class 'str'>\n(173, ['174,1.52043,13.38,0.00,1.40,72.25,0.33,12.50,0.00,0.00,5'])\n<class 'str'>\n(174, ['175,1.52058,12.85,1.61,2.17,72.18,0.76,9.70,0.24,0.51,5'])\n<class 'str'>\n(175, ['176,1.52119,12.97,0.33,1.51,73.39,0.13,11.27,0.00,0.28,5'])\n<class 'str'>\n(176, ['177,1.51905,14.00,2.39,1.56,72.37,0.00,9.57,0.00,0.00,6'])\n<class 'str'>\n(177, ['178,1.51937,13.79,2.41,1.19,72.76,0.00,9.77,0.00,0.00,6'])\n<class 'str'>\n(178, ['179,1.51829,14.46,2.24,1.62,72.38,0.00,9.26,0.00,0.00,6'])\n<class 'str'>\n(179, ['180,1.51852,14.09,2.19,1.66,72.67,0.00,9.32,0.00,0.00,6'])\n<class 'str'>\n(180, ['181,1.51299,14.40,1.74,1.54,74.55,0.00,7.59,0.00,0.00,6'])\n<class 'str'>\n(181, ['182,1.51888,14.99,0.78,1.74,72.50,0.00,9.95,0.00,0.00,6'])\n<class 'str'>\n(182, ['183,1.51916,14.15,0.00,2.09,72.74,0.00,10.88,0.00,0.00,6'])\n<class 'str'>\n(183, ['184,1.51969,14.56,0.00,0.56,73.48,0.00,11.22,0.00,0.00,6'])\n<class 'str'>\n(184, ['185,1.51115,17.38,0.00,0.34,75.41,0.00,6.65,0.00,0.00,6'])\n<class 'str'>\n(185, ['186,1.51131,13.69,3.20,1.81,72.81,1.76,5.43,1.19,0.00,7'])\n<class 'str'>\n(186, ['187,1.51838,14.32,3.26,2.22,71.25,1.46,5.79,1.63,0.00,7'])\n<class 'str'>\n(187, ['188,1.52315,13.44,3.34,1.23,72.38,0.60,8.83,0.00,0.00,7'])\n<class 'str'>\n(188, ['189,1.52247,14.86,2.20,2.06,70.26,0.76,9.76,0.00,0.00,7'])\n<class 'str'>\n(189, ['190,1.52365,15.79,1.83,1.31,70.43,0.31,8.61,1.68,0.00,7'])\n<class 'str'>\n(190, ['191,1.51613,13.88,1.78,1.79,73.10,0.00,8.67,0.76,0.00,7'])\n<class 'str'>\n(191, ['192,1.51602,14.85,0.00,2.38,73.28,0.00,8.76,0.64,0.09,7'])\n<class 'str'>\n(192, ['193,1.51623,14.20,0.00,2.79,73.46,0.04,9.04,0.40,0.09,7'])\n<class 'str'>\n(193, ['194,1.51719,14.75,0.00,2.00,73.02,0.00,8.53,1.59,0.08,7'])\n<class 'str'>\n(194, ['195,1.51683,14.56,0.00,1.98,73.29,0.00,8.52,1.57,0.07,7'])\n<class 'str'>\n(195, ['196,1.51545,14.14,0.00,2.68,73.39,0.08,9.07,0.61,0.05,7'])\n<class 'str'>\n(196, ['197,1.51556,13.87,0.00,2.54,73.23,0.14,9.41,0.81,0.01,7'])\n<class 'str'>\n(197, ['198,1.51727,14.70,0.00,2.34,73.28,0.00,8.95,0.66,0.00,7'])\n<class 'str'>\n(198, ['199,1.51531,14.38,0.00,2.66,73.10,0.04,9.08,0.64,0.00,7'])\n<class 'str'>\n(199, ['200,1.51609,15.01,0.00,2.51,73.05,0.05,8.83,0.53,0.00,7'])\n<class 'str'>\n(200, ['201,1.51508,15.15,0.00,2.25,73.50,0.00,8.34,0.63,0.00,7'])\n<class 'str'>\n(201, ['202,1.51653,11.95,0.00,1.19,75.18,2.70,8.93,0.00,0.00,7'])\n<class 'str'>\n(202, ['203,1.51514,14.85,0.00,2.42,73.72,0.00,8.39,0.56,0.00,7'])\n<class 'str'>\n(203, ['204,1.51658,14.80,0.00,1.99,73.11,0.00,8.28,1.71,0.00,7'])\n<class 'str'>\n(204, ['205,1.51617,14.95,0.00,2.27,73.30,0.00,8.71,0.67,0.00,7'])\n<class 'str'>\n(205, ['206,1.51732,14.95,0.00,1.80,72.99,0.00,8.61,1.55,0.00,7'])\n<class 'str'>\n(206, ['207,1.51645,14.94,0.00,1.87,73.11,0.00,8.67,1.38,0.00,7'])\n<class 'str'>\n(207, ['208,1.51831,14.39,0.00,1.82,72.86,1.41,6.47,2.88,0.00,7'])\n<class 'str'>\n(208, ['209,1.51640,14.37,0.00,2.74,72.85,0.00,9.45,0.54,0.00,7'])\n<class 'str'>\n(209, ['210,1.51623,14.14,0.00,2.88,72.61,0.08,9.18,1.06,0.00,7'])\n<class 'str'>\n(210, ['211,1.51685,14.92,0.00,1.99,73.06,0.00,8.40,1.59,0.00,7'])\n<class 'str'>\n(211, ['212,1.52065,14.36,0.00,2.02,73.42,0.00,8.44,1.64,0.00,7'])\n<class 'str'>\n(212, ['213,1.51651,14.38,0.00,1.94,73.61,0.00,8.48,1.57,0.00,7'])\n<class 'str'>\n(213, ['214,1.51711,14.23,0.00,2.08,73.36,0.00,8.62,1.67,0.00,7'])\n<class 'str'>\nall the data inserted \n"
],
[
"with open('glass.data',\"r\") as data :\n next(data)\n data_csv = csv.reader(data, delimiter= \"\\n\")\n print(data_csv)\n for j in data_csv :\n cur.execute(f'insert into sudhanshu12345.GlassData values (str(j[0]))')\n print(\"all the data inserted \")\nmydb.commit()",
"<_csv.reader object at 0x0000027293E88340>\n"
],
[
"https://archive.ics.uci.edu/ml/machine-learning-databases/00448/",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
c513b2371067823c230b7eb2c359edca7a87ff83
| 41,345 |
ipynb
|
Jupyter Notebook
|
Mini-Projects/IMDB Sentiment Analysis - XGBoost (Batch Transform).ipynb
|
AlbertoDeBlas/sagemaker_deployment
|
8e801206f86c499b6b23a2e1cd046788d4b25540
|
[
"MIT"
] | null | null | null |
Mini-Projects/IMDB Sentiment Analysis - XGBoost (Batch Transform).ipynb
|
AlbertoDeBlas/sagemaker_deployment
|
8e801206f86c499b6b23a2e1cd046788d4b25540
|
[
"MIT"
] | null | null | null |
Mini-Projects/IMDB Sentiment Analysis - XGBoost (Batch Transform).ipynb
|
AlbertoDeBlas/sagemaker_deployment
|
8e801206f86c499b6b23a2e1cd046788d4b25540
|
[
"MIT"
] | null | null | null | 46.507312 | 882 | 0.626001 |
[
[
[
"# Sentiment Analysis\n\n## Using XGBoost in SageMaker\n\n_Deep Learning Nanodegree Program | Deployment_\n\n---\n\nAs our first example of using Amazon's SageMaker service we will construct a random tree model to predict the sentiment of a movie review. You may have seen a version of this example in a pervious lesson although it would have been done using the sklearn package. Instead, we will be using the XGBoost package as it is provided to us by Amazon.\n\n## Instructions\n\nSome template code has already been provided for you, and you will need to implement additional functionality to successfully complete this notebook. You will not need to modify the included code beyond what is requested. Sections that begin with '**TODO**' in the header indicate that you need to complete or implement some portion within them. Instructions will be provided for each section and the specifics of the implementation are marked in the code block with a `# TODO: ...` comment. Please be sure to read the instructions carefully!\n\nIn addition to implementing code, there may be questions for you to answer which relate to the task and your implementation. Each section where you will answer a question is preceded by a '**Question:**' header. Carefully read each question and provide your answer below the '**Answer:**' header by editing the Markdown cell.\n\n> **Note**: Code and Markdown cells can be executed using the **Shift+Enter** keyboard shortcut. In addition, a cell can be edited by typically clicking it (double-click for Markdown cells) or by pressing **Enter** while it is highlighted.",
"_____no_output_____"
],
[
"## Step 1: Downloading the data\n\nThe dataset we are going to use is very popular among researchers in Natural Language Processing, usually referred to as the [IMDb dataset](http://ai.stanford.edu/~amaas/data/sentiment/). It consists of movie reviews from the website [imdb.com](http://www.imdb.com/), each labeled as either '**pos**itive', if the reviewer enjoyed the film, or '**neg**ative' otherwise.\n\n> Maas, Andrew L., et al. [Learning Word Vectors for Sentiment Analysis](http://ai.stanford.edu/~amaas/data/sentiment/). In _Proceedings of the 49th Annual Meeting of the Association for Computational Linguistics: Human Language Technologies_. Association for Computational Linguistics, 2011.\n\nWe begin by using some Jupyter Notebook magic to download and extract the dataset.",
"_____no_output_____"
]
],
[
[
"%mkdir ../data\n!wget -O ../data/aclImdb_v1.tar.gz http://ai.stanford.edu/~amaas/data/sentiment/aclImdb_v1.tar.gz\n!tar -zxf ../data/aclImdb_v1.tar.gz -C ../data",
"mkdir: cannot create directory ‘../data’: File exists\n--2020-04-07 15:21:14-- http://ai.stanford.edu/~amaas/data/sentiment/aclImdb_v1.tar.gz\nResolving ai.stanford.edu (ai.stanford.edu)... 171.64.68.10\nConnecting to ai.stanford.edu (ai.stanford.edu)|171.64.68.10|:80... connected.\nHTTP request sent, awaiting response... 200 OK\nLength: 84125825 (80M) [application/x-gzip]\nSaving to: ‘../data/aclImdb_v1.tar.gz’\n\n../data/aclImdb_v1. 100%[===================>] 80.23M 10.2MB/s in 11s \n\n2020-04-07 15:21:25 (7.32 MB/s) - ‘../data/aclImdb_v1.tar.gz’ saved [84125825/84125825]\n\n"
]
],
[
[
"## Step 2: Preparing the data\n\nThe data we have downloaded is split into various files, each of which contains a single review. It will be much easier going forward if we combine these individual files into two large files, one for training and one for testing.",
"_____no_output_____"
]
],
[
[
"import os\nimport glob\n\ndef read_imdb_data(data_dir='../data/aclImdb'):\n data = {}\n labels = {}\n \n for data_type in ['train', 'test']:\n data[data_type] = {}\n labels[data_type] = {}\n \n for sentiment in ['pos', 'neg']:\n data[data_type][sentiment] = []\n labels[data_type][sentiment] = []\n \n path = os.path.join(data_dir, data_type, sentiment, '*.txt')\n files = glob.glob(path)\n \n for f in files:\n with open(f) as review:\n data[data_type][sentiment].append(review.read())\n # Here we represent a positive review by '1' and a negative review by '0'\n labels[data_type][sentiment].append(1 if sentiment == 'pos' else 0)\n \n assert len(data[data_type][sentiment]) == len(labels[data_type][sentiment]), \\\n \"{}/{} data size does not match labels size\".format(data_type, sentiment)\n \n return data, labels",
"_____no_output_____"
],
[
"data, labels = read_imdb_data()\nprint(\"IMDB reviews: train = {} pos / {} neg, test = {} pos / {} neg\".format(\n len(data['train']['pos']), len(data['train']['neg']),\n len(data['test']['pos']), len(data['test']['neg'])))",
"IMDB reviews: train = 12500 pos / 12500 neg, test = 12500 pos / 12500 neg\n"
],
[
"from sklearn.utils import shuffle\n\ndef prepare_imdb_data(data, labels):\n \"\"\"Prepare training and test sets from IMDb movie reviews.\"\"\"\n \n #Combine positive and negative reviews and labels\n data_train = data['train']['pos'] + data['train']['neg']\n data_test = data['test']['pos'] + data['test']['neg']\n labels_train = labels['train']['pos'] + labels['train']['neg']\n labels_test = labels['test']['pos'] + labels['test']['neg']\n \n #Shuffle reviews and corresponding labels within training and test sets\n data_train, labels_train = shuffle(data_train, labels_train)\n data_test, labels_test = shuffle(data_test, labels_test)\n \n # Return a unified training data, test data, training labels, test labets\n return data_train, data_test, labels_train, labels_test",
"_____no_output_____"
],
[
"train_X, test_X, train_y, test_y = prepare_imdb_data(data, labels)\nprint(\"IMDb reviews (combined): train = {}, test = {}\".format(len(train_X), len(test_X)))",
"IMDb reviews (combined): train = 25000, test = 25000\n"
],
[
"train_X[100]",
"_____no_output_____"
]
],
[
[
"## Step 3: Processing the data\n\nNow that we have our training and testing datasets merged and ready to use, we need to start processing the raw data into something that will be useable by our machine learning algorithm. To begin with, we remove any html formatting that may appear in the reviews and perform some standard natural language processing in order to homogenize the data.",
"_____no_output_____"
]
],
[
[
"import nltk\nnltk.download(\"stopwords\")\nfrom nltk.corpus import stopwords\nfrom nltk.stem.porter import *\nstemmer = PorterStemmer()",
"[nltk_data] Downloading package stopwords to\n[nltk_data] /home/ec2-user/nltk_data...\n[nltk_data] Unzipping corpora/stopwords.zip.\n"
],
[
"import re\nfrom bs4 import BeautifulSoup\n\ndef review_to_words(review):\n text = BeautifulSoup(review, \"html.parser\").get_text() # Remove HTML tags\n text = re.sub(r\"[^a-zA-Z0-9]\", \" \", text.lower()) # Convert to lower case\n words = text.split() # Split string into words\n words = [w for w in words if w not in stopwords.words(\"english\")] # Remove stopwords\n words = [PorterStemmer().stem(w) for w in words] # stem\n \n return words",
"_____no_output_____"
],
[
"import pickle\n\ncache_dir = os.path.join(\"../cache\", \"sentiment_analysis\") # where to store cache files\nos.makedirs(cache_dir, exist_ok=True) # ensure cache directory exists\n\ndef preprocess_data(data_train, data_test, labels_train, labels_test,\n cache_dir=cache_dir, cache_file=\"preprocessed_data.pkl\"):\n \"\"\"Convert each review to words; read from cache if available.\"\"\"\n\n # If cache_file is not None, try to read from it first\n cache_data = None\n if cache_file is not None:\n try:\n with open(os.path.join(cache_dir, cache_file), \"rb\") as f:\n cache_data = pickle.load(f)\n print(\"Read preprocessed data from cache file:\", cache_file)\n except:\n pass # unable to read from cache, but that's okay\n \n # If cache is missing, then do the heavy lifting\n if cache_data is None:\n # Preprocess training and test data to obtain words for each review\n #words_train = list(map(review_to_words, data_train))\n #words_test = list(map(review_to_words, data_test))\n words_train = [review_to_words(review) for review in data_train]\n words_test = [review_to_words(review) for review in data_test]\n \n # Write to cache file for future runs\n if cache_file is not None:\n cache_data = dict(words_train=words_train, words_test=words_test,\n labels_train=labels_train, labels_test=labels_test)\n with open(os.path.join(cache_dir, cache_file), \"wb\") as f:\n pickle.dump(cache_data, f)\n print(\"Wrote preprocessed data to cache file:\", cache_file)\n else:\n # Unpack data loaded from cache file\n words_train, words_test, labels_train, labels_test = (cache_data['words_train'],\n cache_data['words_test'], cache_data['labels_train'], cache_data['labels_test'])\n \n return words_train, words_test, labels_train, labels_test",
"_____no_output_____"
],
[
"# Preprocess data\ntrain_X, test_X, train_y, test_y = preprocess_data(train_X, test_X, train_y, test_y)",
"Read preprocessed data from cache file: preprocessed_data.pkl\n"
]
],
[
[
"### Extract Bag-of-Words features\n\nFor the model we will be implementing, rather than using the reviews directly, we are going to transform each review into a Bag-of-Words feature representation. Keep in mind that 'in the wild' we will only have access to the training set so our transformer can only use the training set to construct a representation.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nfrom sklearn.feature_extraction.text import CountVectorizer\nfrom sklearn.externals import joblib\n# joblib is an enhanced version of pickle that is more efficient for storing NumPy arrays\n\ndef extract_BoW_features(words_train, words_test, vocabulary_size=5000,\n cache_dir=cache_dir, cache_file=\"bow_features.pkl\"):\n \"\"\"Extract Bag-of-Words for a given set of documents, already preprocessed into words.\"\"\"\n \n # If cache_file is not None, try to read from it first\n cache_data = None\n if cache_file is not None:\n try:\n with open(os.path.join(cache_dir, cache_file), \"rb\") as f:\n cache_data = joblib.load(f)\n print(\"Read features from cache file:\", cache_file)\n except:\n pass # unable to read from cache, but that's okay\n \n # If cache is missing, then do the heavy lifting\n if cache_data is None:\n # Fit a vectorizer to training documents and use it to transform them\n # NOTE: Training documents have already been preprocessed and tokenized into words;\n # pass in dummy functions to skip those steps, e.g. preprocessor=lambda x: x\n vectorizer = CountVectorizer(max_features=vocabulary_size,\n preprocessor=lambda x: x, tokenizer=lambda x: x) # already preprocessed\n features_train = vectorizer.fit_transform(words_train).toarray()\n\n # Apply the same vectorizer to transform the test documents (ignore unknown words)\n features_test = vectorizer.transform(words_test).toarray()\n \n # NOTE: Remember to convert the features using .toarray() for a compact representation\n \n # Write to cache file for future runs (store vocabulary as well)\n if cache_file is not None:\n vocabulary = vectorizer.vocabulary_\n cache_data = dict(features_train=features_train, features_test=features_test,\n vocabulary=vocabulary)\n with open(os.path.join(cache_dir, cache_file), \"wb\") as f:\n joblib.dump(cache_data, f)\n print(\"Wrote features to cache file:\", cache_file)\n else:\n # Unpack data loaded from cache file\n features_train, features_test, vocabulary = (cache_data['features_train'],\n cache_data['features_test'], cache_data['vocabulary'])\n \n # Return both the extracted features as well as the vocabulary\n return features_train, features_test, vocabulary",
"_____no_output_____"
],
[
"# Extract Bag of Words features for both training and test datasets\ntrain_X, test_X, vocabulary = extract_BoW_features(train_X, test_X)",
"Read features from cache file: bow_features.pkl\n"
]
],
[
[
"## Step 4: Classification using XGBoost\n\nNow that we have created the feature representation of our training (and testing) data, it is time to start setting up and using the XGBoost classifier provided by SageMaker.\n\n### (TODO) Writing the dataset\n\nThe XGBoost classifier that we will be using requires the dataset to be written to a file and stored using Amazon S3. To do this, we will start by splitting the training dataset into two parts, the data we will train the model with and a validation set. Then, we will write those datasets to a file and upload the files to S3. In addition, we will write the test set input to a file and upload the file to S3. This is so that we can use SageMakers Batch Transform functionality to test our model once we've fit it.",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nfrom sklearn.model_selection import train_test_split\n\n# TODO: Split the train_X and train_y arrays into the DataFrames val_X, train_X and val_y, train_y. Make sure that\n# val_X and val_y contain 10 000 entires while train_X and train_y contain the remaining 15 000 entries.\n\nX = train_X\ny = train_y\n\ntrain_X, val_X, train_y, val_y = train_test_split(X, y, test_size=10000, random_state=42)",
"_____no_output_____"
]
],
[
[
"The documentation for the XGBoost algorithm in SageMaker requires that the saved datasets should contain no headers or index and that for the training and validation data, the label should occur first for each sample.\n\nFor more information about this and other algorithms, the SageMaker developer documentation can be found on __[Amazon's website.](https://docs.aws.amazon.com/sagemaker/latest/dg/)__",
"_____no_output_____"
]
],
[
[
"# First we make sure that the local directory in which we'd like to store the training and validation csv files exists.\ndata_dir = '../data/xgboost'\nif not os.path.exists(data_dir):\n os.makedirs(data_dir)",
"_____no_output_____"
],
[
"# First, save the test data to test.csv in the data_dir directory. Note that we do not save the associated ground truth\n# labels, instead we will use them later to compare with our model output.\n\npd.DataFrame(test_X).to_csv(os.path.join(data_dir, 'test.csv'), header=False, index=False)\n\n# TODO: Save the training and validation data to train.csv and validation.csv in the data_dir directory.\n# Make sure that the files you create are in the correct format.\n\npd.concat([pd.DataFrame(train_y), pd.DataFrame(train_X)], axis=1).to_csv(os.path.join(data_dir, 'train.csv'), header=False, index=False)\n\npd.concat([pd.DataFrame(val_y), pd.DataFrame(val_X)], axis=1).to_csv(os.path.join(data_dir, 'validation.csv'), header=False, index=False)\n",
"_____no_output_____"
],
[
"# To save a bit of memory we can set text_X, train_X, val_X, train_y and val_y to None.\n\ntest_X = train_X = val_X = train_y = val_y = None",
"_____no_output_____"
]
],
[
[
"### (TODO) Uploading Training / Validation files to S3\n\nAmazon's S3 service allows us to store files that can be access by both the built-in training models such as the XGBoost model we will be using as well as custom models such as the one we will see a little later.\n\nFor this, and most other tasks we will be doing using SageMaker, there are two methods we could use. The first is to use the low level functionality of SageMaker which requires knowing each of the objects involved in the SageMaker environment. The second is to use the high level functionality in which certain choices have been made on the user's behalf. The low level approach benefits from allowing the user a great deal of flexibility while the high level approach makes development much quicker. For our purposes we will opt to use the high level approach although using the low-level approach is certainly an option.\n\nRecall the method `upload_data()` which is a member of object representing our current SageMaker session. What this method does is upload the data to the default bucket (which is created if it does not exist) into the path described by the key_prefix variable. To see this for yourself, once you have uploaded the data files, go to the S3 console and look to see where the files have been uploaded.\n\nFor additional resources, see the __[SageMaker API documentation](http://sagemaker.readthedocs.io/en/latest/)__ and in addition the __[SageMaker Developer Guide.](https://docs.aws.amazon.com/sagemaker/latest/dg/)__",
"_____no_output_____"
]
],
[
[
"import sagemaker\n\nsession = sagemaker.Session() # Store the current SageMaker session\n\n# S3 prefix (which folder will we use)\nprefix = 'sentiment-xgboost'\n\n# TODO: Upload the test.csv, train.csv and validation.csv files which are contained in data_dir to S3 using sess.upload_data().\ntest_location = session.upload_data(os.path.join(data_dir, 'test.csv'),key_prefix=prefix)\nval_location = session.upload_data(os.path.join(data_dir, 'validation.csv'),key_prefix=prefix)\ntrain_location = session.upload_data(os.path.join(data_dir, 'train.csv'),key_prefix=prefix)",
"_____no_output_____"
]
],
[
[
"### (TODO) Creating the XGBoost model\n\nNow that the data has been uploaded it is time to create the XGBoost model. To begin with, we need to do some setup. At this point it is worth discussing what a model is in SageMaker. It is easiest to think of a model of comprising three different objects in the SageMaker ecosystem, which interact with one another.\n\n- Model Artifacts\n- Training Code (Container)\n- Inference Code (Container)\n\nThe Model Artifacts are what you might think of as the actual model itself. For example, if you were building a neural network, the model artifacts would be the weights of the various layers. In our case, for an XGBoost model, the artifacts are the actual trees that are created during training.\n\nThe other two objects, the training code and the inference code are then used the manipulate the training artifacts. More precisely, the training code uses the training data that is provided and creates the model artifacts, while the inference code uses the model artifacts to make predictions on new data.\n\nThe way that SageMaker runs the training and inference code is by making use of Docker containers. For now, think of a container as being a way of packaging code up so that dependencies aren't an issue.",
"_____no_output_____"
]
],
[
[
"from sagemaker import get_execution_role\n\n# Our current execution role is require when creating the model as the training\n# and inference code will need to access the model artifacts.\nrole = get_execution_role()",
"_____no_output_____"
],
[
"# We need to retrieve the location of the container which is provided by Amazon for using XGBoost.\n# As a matter of convenience, the training and inference code both use the same container.\nfrom sagemaker.amazon.amazon_estimator import get_image_uri\n\ncontainer = get_image_uri(session.boto_region_name, 'xgboost')",
"WARNING:root:There is a more up to date SageMaker XGBoost image. To use the newer image, please set 'repo_version'='0.90-1'. For example:\n\tget_image_uri(region, 'xgboost', '0.90-1').\n"
],
[
"# TODO: Create a SageMaker estimator using the container location determined in the previous cell.\n# It is recommended that you use a single training instance of type ml.m4.xlarge. It is also\n# recommended that you use 's3://{}/{}/output'.format(session.default_bucket(), prefix) as the\n# output path.\n\ns3_output_location = 's3://{}/{}/output'.format(session.default_bucket(),prefix)\n\nxgb = sagemaker.estimator.Estimator(container,\n role, \n train_instance_count=1, \n train_instance_type='ml.m5.large',\n train_volume_size = 1,\n output_path=s3_output_location,\n sagemaker_session=sagemaker.Session())\n\n\n# TODO: Set the XGBoost hyperparameters in the xgb object. Don't forget that in this case we have a binary\n# label so we should be using the 'binary:logistic' objective.\n\nxgb.set_hyperparameters(max_depth=5,\n eta=0.2,\n gamma=4,\n min_child_weight=6,\n subsample=0.8,\n silent=0,\n objective='binary:logistic',\n early_stopping_rounds=10,\n num_round=500)\n",
"_____no_output_____"
]
],
[
[
"### Fit the XGBoost model\n\nNow that our model has been set up we simply need to attach the training and validation datasets and then ask SageMaker to set up the computation.",
"_____no_output_____"
]
],
[
[
"s3_input_train = sagemaker.s3_input(s3_data=train_location, content_type='csv')\ns3_input_validation = sagemaker.s3_input(s3_data=val_location, content_type='csv')",
"_____no_output_____"
],
[
"xgb.fit({'train': s3_input_train, 'validation': s3_input_validation})",
"2020-04-07 18:51:48 Starting - Starting the training job...\n2020-04-07 18:51:50 Starting - Launching requested ML instances......\n2020-04-07 18:52:49 Starting - Preparing the instances for training...\n2020-04-07 18:53:30 Downloading - Downloading input data...\n2020-04-07 18:54:09 Training - Downloading the training image..\u001b[34mArguments: train\u001b[0m\n\u001b[34m[2020-04-07:18:54:23:INFO] Running standalone xgboost training.\u001b[0m\n\u001b[34m[2020-04-07:18:54:23:INFO] File size need to be processed in the node: 1.87mb. Available memory size in the node: 207.81mb\u001b[0m\n\u001b[34m[2020-04-07:18:54:23:INFO] Determined delimiter of CSV input is ','\u001b[0m\n\u001b[34m[18:54:23] S3DistributionType set as FullyReplicated\u001b[0m\n\u001b[34m[18:54:23] 15000x36 matrix with 540000 entries loaded from /opt/ml/input/data/train?format=csv&label_column=0&delimiter=,\u001b[0m\n\u001b[34m[2020-04-07:18:54:23:INFO] Determined delimiter of CSV input is ','\u001b[0m\n\u001b[34m[18:54:23] S3DistributionType set as FullyReplicated\u001b[0m\n\u001b[34m[18:54:23] 10000x36 matrix with 360000 entries loaded from /opt/ml/input/data/validation?format=csv&label_column=0&delimiter=,\u001b[0m\n\u001b[34m[18:54:23] src/tree/updater_prune.cc:74: tree pruning end, 1 roots, 34 extra nodes, 8 pruned nodes, max_depth=5\u001b[0m\n\u001b[34m[0]#011train-error:0.4558#011validation-error:0.4752\u001b[0m\n\u001b[34mMultiple eval metrics have been passed: 'validation-error' will be used for early stopping.\n\u001b[0m\n\u001b[34mWill train until validation-error hasn't improved in 10 rounds.\u001b[0m\n\u001b[34m[18:54:23] src/tree/updater_prune.cc:74: tree pruning end, 1 roots, 32 extra nodes, 14 pruned nodes, max_depth=5\u001b[0m\n\u001b[34m[1]#011train-error:0.457067#011validation-error:0.4776\u001b[0m\n\u001b[34m[18:54:23] src/tree/updater_prune.cc:74: tree pruning end, 1 roots, 28 extra nodes, 12 pruned nodes, max_depth=5\u001b[0m\n\u001b[34m[2]#011train-error:0.453867#011validation-error:0.4749\u001b[0m\n\u001b[34m[18:54:23] src/tree/updater_prune.cc:74: tree pruning end, 1 roots, 22 extra nodes, 22 pruned nodes, max_depth=4\u001b[0m\n\u001b[34m[3]#011train-error:0.4512#011validation-error:0.4749\u001b[0m\n\u001b[34m[18:54:23] src/tree/updater_prune.cc:74: tree pruning end, 1 roots, 10 extra nodes, 20 pruned nodes, max_depth=4\u001b[0m\n\u001b[34m[4]#011train-error:0.4512#011validation-error:0.4748\u001b[0m\n\u001b[34m[18:54:23] src/tree/updater_prune.cc:74: tree pruning end, 1 roots, 38 extra nodes, 4 pruned nodes, max_depth=5\u001b[0m\n\u001b[34m[5]#011train-error:0.448#011validation-error:0.4762\u001b[0m\n\u001b[34m[18:54:23] src/tree/updater_prune.cc:74: tree pruning end, 1 roots, 28 extra nodes, 18 pruned nodes, max_depth=5\u001b[0m\n\u001b[34m[6]#011train-error:0.4452#011validation-error:0.4755\u001b[0m\n\u001b[34m[18:54:23] src/tree/updater_prune.cc:74: tree pruning end, 1 roots, 36 extra nodes, 14 pruned nodes, max_depth=5\u001b[0m\n\u001b[34m[7]#011train-error:0.442533#011validation-error:0.4793\u001b[0m\n\u001b[34m[18:54:23] src/tree/updater_prune.cc:74: tree pruning end, 1 roots, 18 extra nodes, 4 pruned nodes, max_depth=5\u001b[0m\n\u001b[34m[8]#011train-error:0.4414#011validation-error:0.4797\u001b[0m\n\u001b[34m[18:54:23] src/tree/updater_prune.cc:74: tree pruning end, 1 roots, 24 extra nodes, 12 pruned nodes, max_depth=5\u001b[0m\n\u001b[34m[9]#011train-error:0.441267#011validation-error:0.4788\u001b[0m\n\u001b[34m[18:54:23] src/tree/updater_prune.cc:74: tree pruning end, 1 roots, 22 extra nodes, 4 pruned nodes, max_depth=5\u001b[0m\n\u001b[34m[10]#011train-error:0.438467#011validation-error:0.4782\u001b[0m\n\u001b[34m[18:54:23] src/tree/updater_prune.cc:74: tree pruning end, 1 roots, 26 extra nodes, 8 pruned nodes, max_depth=5\u001b[0m\n\u001b[34m[11]#011train-error:0.438333#011validation-error:0.4791\u001b[0m\n\u001b[34m[18:54:23] src/tree/updater_prune.cc:74: tree pruning end, 1 roots, 12 extra nodes, 20 pruned nodes, max_depth=4\u001b[0m\n\u001b[34m[12]#011train-error:0.4382#011validation-error:0.4797\u001b[0m\n\u001b[34m[18:54:23] src/tree/updater_prune.cc:74: tree pruning end, 1 roots, 18 extra nodes, 14 pruned nodes, max_depth=5\u001b[0m\n\u001b[34m[13]#011train-error:0.436133#011validation-error:0.4789\u001b[0m\n\u001b[34m[18:54:23] src/tree/updater_prune.cc:74: tree pruning end, 1 roots, 26 extra nodes, 4 pruned nodes, max_depth=5\u001b[0m\n\u001b[34m[14]#011train-error:0.4362#011validation-error:0.4795\u001b[0m\n\u001b[34mStopping. Best iteration:\u001b[0m\n\u001b[34m[4]#011train-error:0.4512#011validation-error:0.4748\n\u001b[0m\n\n2020-04-07 18:54:35 Uploading - Uploading generated training model\n2020-04-07 18:54:35 Completed - Training job completed\nTraining seconds: 65\nBillable seconds: 65\n"
]
],
[
[
"### (TODO) Testing the model\n\nNow that we've fit our XGBoost model, it's time to see how well it performs. To do this we will use SageMakers Batch Transform functionality. Batch Transform is a convenient way to perform inference on a large dataset in a way that is not realtime. That is, we don't necessarily need to use our model's results immediately and instead we can peform inference on a large number of samples. An example of this in industry might be peforming an end of month report. This method of inference can also be useful to us as it means to can perform inference on our entire test set. \n\nTo perform a Batch Transformation we need to first create a transformer objects from our trained estimator object.",
"_____no_output_____"
]
],
[
[
"# TODO: Create a transformer object from the trained model. Using an instance count of 1 and an instance type of ml.m4.xlarge\n# should be more than enough.\nxgb_transformer = xgb.transformer(instance_count=1, instance_type='ml.m5.large')\n\n",
"_____no_output_____"
]
],
[
[
"Next we actually perform the transform job. When doing so we need to make sure to specify the type of data we are sending so that it is serialized correctly in the background. In our case we are providing our model with csv data so we specify `text/csv`. Also, if the test data that we have provided is too large to process all at once then we need to specify how the data file should be split up. Since each line is a single entry in our data set we tell SageMaker that it can split the input on each line.",
"_____no_output_____"
]
],
[
[
"# TODO: Start the transform job. Make sure to specify the content type and the split type of the test data.\nxgb_transformer.transform(test_location, content_type='text/csv', split_type='Line')",
"_____no_output_____"
]
],
[
[
"Currently the transform job is running but it is doing so in the background. Since we wish to wait until the transform job is done and we would like a bit of feedback we can run the `wait()` method.",
"_____no_output_____"
]
],
[
[
"xgb_transformer.wait()",
".....\u001b[34mArguments: serve\u001b[0m\n\u001b[34m[2020-04-07 19:01:02 +0000] [1] [INFO] Starting gunicorn 19.7.1\u001b[0m\n\u001b[34m[2020-04-07 19:01:02 +0000] [1] [INFO] Listening at: http://0.0.0.0:8080 (1)\u001b[0m\n\u001b[34m[2020-04-07 19:01:02 +0000] [1] [INFO] Using worker: gevent\u001b[0m\n\u001b[34m[2020-04-07 19:01:02 +0000] [37] [INFO] Booting worker with pid: 37\u001b[0m\n\u001b[34m[2020-04-07 19:01:02 +0000] [38] [INFO] Booting worker with pid: 38\u001b[0m\n\u001b[34m[2020-04-07:19:01:02:INFO] Model loaded successfully for worker : 37\u001b[0m\n\u001b[34m[2020-04-07:19:01:02:INFO] Model loaded successfully for worker : 38\u001b[0m\n\u001b[32m2020-04-07T19:01:27.686:[sagemaker logs]: MaxConcurrentTransforms=2, MaxPayloadInMB=6, BatchStrategy=MULTI_RECORD\u001b[0m\n\u001b[34m[2020-04-07:19:01:28:INFO] Sniff delimiter as ','\u001b[0m\n\u001b[34m[2020-04-07:19:01:28:INFO] Determined delimiter of CSV input is ','\u001b[0m\n\u001b[35m[2020-04-07:19:01:28:INFO] Sniff delimiter as ','\u001b[0m\n\u001b[35m[2020-04-07:19:01:28:INFO] Determined delimiter of CSV input is ','\u001b[0m\n\n"
]
],
[
[
"Now the transform job has executed and the result, the estimated sentiment of each review, has been saved on S3. Since we would rather work on this file locally we can perform a bit of notebook magic to copy the file to the `data_dir`.",
"_____no_output_____"
]
],
[
[
"!aws s3 cp --recursive $xgb_transformer.output_path $data_dir",
"Completed 256.0 KiB/365.3 KiB (3.7 MiB/s) with 1 file(s) remaining\rCompleted 365.3 KiB/365.3 KiB (5.1 MiB/s) with 1 file(s) remaining\rdownload: s3://sagemaker-eu-west-3-848439228145/xgboost-2020-04-07-18-58-12-761/test.csv.out to ../data/xgboost/test.csv.out\r\n"
]
],
[
[
"The last step is now to read in the output from our model, convert the output to something a little more usable, in this case we want the sentiment to be either `1` (positive) or `0` (negative), and then compare to the ground truth labels.",
"_____no_output_____"
]
],
[
[
"predictions = pd.read_csv(os.path.join(data_dir, 'test.csv.out'), header=None)\npredictions = [round(num) for num in predictions.squeeze().values]",
"_____no_output_____"
],
[
"from sklearn.metrics import accuracy_score\naccuracy_score(test_y, predictions)",
"_____no_output_____"
]
],
[
[
"## Optional: Clean up\n\nThe default notebook instance on SageMaker doesn't have a lot of excess disk space available. As you continue to complete and execute notebooks you will eventually fill up this disk space, leading to errors which can be difficult to diagnose. Once you are completely finished using a notebook it is a good idea to remove the files that you created along the way. Of course, you can do this from the terminal or from the notebook hub if you would like. The cell below contains some commands to clean up the created files from within the notebook.",
"_____no_output_____"
]
],
[
[
"# First we will remove all of the files contained in the data_dir directory\n!rm $data_dir/*\n\n# And then we delete the directory itself\n!rmdir $data_dir\n\n# Similarly we will remove the files in the cache_dir directory and the directory itself\n!rm $cache_dir/*\n!rmdir $cache_dir",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
c513b9fe64f84203bf14347aec28f2236cc1475d
| 6,437 |
ipynb
|
Jupyter Notebook
|
ejercicios/03/03_objects.ipynb
|
restom10/Hw2
|
4070d667f0346728b624eaf20c763953590091d1
|
[
"MIT"
] | 23 |
2015-05-23T13:49:41.000Z
|
2021-11-18T06:10:59.000Z
|
ejercicios/03/03_objects.ipynb
|
restom10/Hw2
|
4070d667f0346728b624eaf20c763953590091d1
|
[
"MIT"
] | 3 |
2015-11-23T09:11:23.000Z
|
2018-11-27T17:10:35.000Z
|
ejercicios/03/03_objects.ipynb
|
restom10/Hw2
|
4070d667f0346728b624eaf20c763953590091d1
|
[
"MIT"
] | 18 |
2016-01-21T16:08:24.000Z
|
2021-11-18T06:11:00.000Z
| 24.757692 | 192 | 0.537984 |
[
[
[
"# Objects\n\n*Python* is an object oriented language. As such it allows the definition of classes.\n\nFor instance lists are also classes, that's why there are methods associated with them (i.e. `append()`). Here we will see how to create classes and assign them attributes and methods.\n",
"_____no_output_____"
],
[
"## Definition and initialization\n\nA class gathers functions (called methods) and variables (called attributes).\nThe main of goal of having this kind of structure is that the methods can share a common\nset of inputs to operate and get the desired outcome by the programmer.\n\nIn *Python* classes are defined with the word `class` and are always initialized\nwith the method ``__init__``, which is a function that *always* must have as input argument the\nword `self`. The arguments that come after `self` are used to initialize the class attributes.\n\nIn the following example we create a class called ``Circle``.\n",
"_____no_output_____"
]
],
[
[
"class Circle:\n def __init__(self, radius):\n self.radius = radius #all attributes must be preceded by \"self.\"",
"_____no_output_____"
]
],
[
[
"To create an instance of this class we do it as follows",
"_____no_output_____"
]
],
[
[
"A = Circle(5.0)",
"_____no_output_____"
]
],
[
[
"We can check that the initialization worked out fine by printing its attributes",
"_____no_output_____"
]
],
[
[
"print(A.radius)",
"_____no_output_____"
]
],
[
[
"We now redefine the class to add new method called `area` that computes the area of the circle",
"_____no_output_____"
]
],
[
[
"class Circle:\n def __init__(self, radius):\n self.radius = radius #all attributes must be preceded by \"self.\"\n def area(self):\n import math\n return math.pi * self.radius * self.radius",
"_____no_output_____"
],
[
"A = Circle(1.0)",
"_____no_output_____"
],
[
"print(A.radius)\nprint(A.area())",
"_____no_output_____"
]
],
[
[
"### Exercise 3.1\n\nRedefine the class `Circle` to include a new method called `perimeter` that returns the value of the circle's perimeter.",
"_____no_output_____"
],
[
"We now want to define a method that returns a new Circle with twice the radius of the input Circle.",
"_____no_output_____"
]
],
[
[
"class Circle:\n def __init__(self, radius):\n self.radius = radius #all attributes must be preceded by \"self.\"\n def area(self):\n import math\n return math.pi * self.radius * self.radius\n def enlarge(self):\n return Circle(2.0*self.radius)",
"_____no_output_____"
],
[
"A = Circle(5.0) # Create a first circle\nB = A.enlarge() # Use the method to create a new Circle\nprint(B.radius) # Check that the radius is twice as the original one.",
"_____no_output_____"
]
],
[
[
"We now add a new method that takes as an input another element of the class `Circle`\nand returns the total area of the two circles",
"_____no_output_____"
]
],
[
[
"class Circle:\n def __init__(self, radius):\n self.radius = radius #all attributes must be preceded by \"self.\"\n def area(self):\n import math\n return math.pi * self.radius * self.radius\n def enlarge(self):\n return Circle(2.0*self.radius)\n def add_area(self, c):\n return self.area() + c.area()",
"_____no_output_____"
],
[
"A = Circle(1.0)\nB = Circle(2.0)\nprint(A.add_area(B))\nprint(B.add_area(A))",
"_____no_output_____"
]
],
[
[
"### Exercise 3.2\n\nDefine the class `Vector3D` to represent vectors in 3D.\nThe class must have\n\n* Three attributes: `x`, `y`, and `z`, to store the coordinates.\n\n* A method called `dot` that computes the dot product\n\n $$\\vec{v} \\cdot \\vec{w} = v_{x}w_{x} + v_{y}w_{y} + v_{z}w_{z}$$\n\n The method could then be used as follows\n \n```python\nv = Vector3D(2, 0, 1)\nw = Vector3D(1, -1, 3)\n```\n \n```python\nv.dot(w)\n5\n```",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
c513c51aa5104f1cc04bb01ede1c98c04e617fad
| 56,547 |
ipynb
|
Jupyter Notebook
|
EDA.ipynb
|
TUCchkul/EDA
|
7e5c8b5f8839d2914cae94d2bf69862ed0d99431
|
[
"MIT"
] | null | null | null |
EDA.ipynb
|
TUCchkul/EDA
|
7e5c8b5f8839d2914cae94d2bf69862ed0d99431
|
[
"MIT"
] | null | null | null |
EDA.ipynb
|
TUCchkul/EDA
|
7e5c8b5f8839d2914cae94d2bf69862ed0d99431
|
[
"MIT"
] | null | null | null | 53.095775 | 20,840 | 0.613171 |
[
[
[
"import numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\nimport seaborn as sns\n%matplotlib inline ",
"_____no_output_____"
],
[
"conda install -c anaconda xlrd",
"Collecting package metadata (current_repodata.json): ...working... done\nNote: you may need to restart the kernel to use updated packages.\nSolving environment: ...working... done\n\n## Package Plan ##\n\n environment location: D:\\DLCVANLP\\EDA\\env\n\n added / updated specs:\n - xlrd\n\n\nThe following packages will be downloaded:\n\n package | build\n ---------------------------|-----------------\n xlrd-1.2.0 | py37_0 190 KB anaconda\n ------------------------------------------------------------\n Total: 190 KB\n\n\nThe following NEW packages will be INSTALLED:\n\n xlrd anaconda/win-64::xlrd-1.2.0-py37_0\n\nThe following packages will be SUPERSEDED by a higher-priority channel:\n\n certifi pkgs/main::certifi-2021.10.8-py37haa9~ --> anaconda::certifi-2020.6.20-py37_0\n\n\n\nDownloading and Extracting Packages\n\nxlrd-1.2.0 | 190 KB | | 0% \nxlrd-1.2.0 | 190 KB | 8 | 8% \nxlrd-1.2.0 | 190 KB | ########## | 100% \nxlrd-1.2.0 | 190 KB | ########## | 100% \nPreparing transaction: ...working... done\nVerifying transaction: ...working... done\nExecuting transaction: ...working... done\n"
],
[
"df=pd.read_csv('zomato.csv', encoding='latin-1')\ndf.head()",
"_____no_output_____"
],
[
"df.columns",
"_____no_output_____"
],
[
"df.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 9551 entries, 0 to 9550\nData columns (total 21 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 Restaurant ID 9551 non-null int64 \n 1 Restaurant Name 9551 non-null object \n 2 Country Code 9551 non-null int64 \n 3 City 9551 non-null object \n 4 Address 9551 non-null object \n 5 Locality 9551 non-null object \n 6 Locality Verbose 9551 non-null object \n 7 Longitude 9551 non-null float64\n 8 Latitude 9551 non-null float64\n 9 Cuisines 9542 non-null object \n 10 Average Cost for two 9551 non-null int64 \n 11 Currency 9551 non-null object \n 12 Has Table booking 9551 non-null object \n 13 Has Online delivery 9551 non-null object \n 14 Is delivering now 9551 non-null object \n 15 Switch to order menu 9551 non-null object \n 16 Price range 9551 non-null int64 \n 17 Aggregate rating 9551 non-null float64\n 18 Rating color 9551 non-null object \n 19 Rating text 9551 non-null object \n 20 Votes 9551 non-null int64 \ndtypes: float64(3), int64(5), object(13)\nmemory usage: 1.5+ MB\n"
],
[
"df.describe()",
"_____no_output_____"
]
],
[
[
"## In data analysis What all things we do\n1. Missing values\n2. Explore about the neumericals variables\n3. Explore about categorical Variables\n4. Finding relationship between features",
"_____no_output_____"
]
],
[
[
"df.isnull().sum()",
"_____no_output_____"
],
[
"[features for features in df.columns if df[features].isnull().sum()>0]",
"_____no_output_____"
],
[
"sns.heatmap(df.isnull(),yticklabels=False,cbar=False, cmap='viridis')",
"_____no_output_____"
],
[
"df_country=pd.read_excel('Country-Code.xlsx')\ndf_country.head()",
"_____no_output_____"
],
[
"df.columns",
"_____no_output_____"
],
[
"final_df=pd.merge(df,df_country, on='Country Code', how='left')",
"_____no_output_____"
],
[
"final_df.head(2)",
"_____no_output_____"
],
[
"final_df.dtypes",
"_____no_output_____"
],
[
"final_df.Country.value_counts()",
"_____no_output_____"
],
[
"country_name=final_df.Country.value_counts().index",
"_____no_output_____"
],
[
"country_values=final_df.Country.value_counts().values",
"_____no_output_____"
],
[
"plt.pie(country_values[:3], labels=country_name[:3])",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
c513c924fc7e75fef50526cc687debdff422b560
| 57,808 |
ipynb
|
Jupyter Notebook
|
jupyter-samples/biomedical-integratedml-PyODBC-Copy1.ipynb
|
jrpereirajr/iris-sample-rest-angular
|
3a1b83f4f96289714519ae4f23d85fdee1f5603b
|
[
"MIT"
] | 1 |
2020-12-28T21:26:35.000Z
|
2020-12-28T21:26:35.000Z
|
jupyter-samples/biomedical-integratedml-PyODBC-Copy1.ipynb
|
jrpereirajr/iris-sample-rest-angular
|
3a1b83f4f96289714519ae4f23d85fdee1f5603b
|
[
"MIT"
] | 1 |
2020-12-25T06:05:59.000Z
|
2020-12-25T06:05:59.000Z
|
jupyter-samples/biomedical-integratedml-PyODBC-Copy1.ipynb
|
jrpereirajr/iris-sample-rest-angular
|
3a1b83f4f96289714519ae4f23d85fdee1f5603b
|
[
"MIT"
] | 1 |
2020-12-23T07:47:38.000Z
|
2020-12-23T07:47:38.000Z
| 51.339254 | 409 | 0.501194 |
[
[
[
"# IntegratedML applied to biomedical data, using PyODBC\nThis notebook demonstrates the following:\n- Connecting to InterSystems IRIS via PyODBC connection\n- Creating, Training and Executing (PREDICT() function) an IntegratedML machine learning model, applied to breast cancer tumor diagnoses\n- INSERTING machine learning predictions into a new SQL table\n- Executing a relatively complex SQL query containing IntegratedML PREDICT() and PROBABILITY() functions, and flexibly using the results to filter and sort the output",
"_____no_output_____"
],
[
"### ODBC and pyODBC Resources\nOften, connecting to a database is more than half the battle when developing SQL-heavy applications, especially if you are not familiar with the tools, or more importantly the particular database system. If this is the case, and you are just getting started using PyODBC and InterSystems IRIS, this notebook and these resources below may help you get up to speed!\n\nhttps://gettingstarted.intersystems.com/development-setup/odbc-connections/\n\nhttps://irisdocs.intersystems.com/irislatest/csp/docbook/DocBook.UI.Page.cls?KEY=BNETODBC_support#BNETODBC_support_pyodbc\n\nhttps://stackoverflow.com/questions/46405777/connect-docker-python-to-sql-server-with-pyodbc\n\nhttps://stackoverflow.com/questions/44527452/cant-open-lib-odbc-driver-13-for-sql-server-sym-linking-issue",
"_____no_output_____"
]
],
[
[
"# make the notebook full screen\nfrom IPython.core.display import display, HTML\ndisplay(HTML(\"<style>.container { width:100% !important; }</style>\"))",
"_____no_output_____"
]
],
[
[
"### 1. Install system packages for ODBC",
"_____no_output_____"
]
],
[
[
"!apt-get update\n!apt-get install gcc\n!apt-get install -y tdsodbc unixodbc-dev\n!apt install unixodbc-bin -y\n!apt-get clean ",
"Get:1 http://security.ubuntu.com/ubuntu bionic-security InRelease [88.7 kB] \nGet:2 http://archive.ubuntu.com/ubuntu bionic InRelease [242 kB]\nGet:3 http://archive.ubuntu.com/ubuntu bionic-updates InRelease [88.7 kB]\nGet:4 http://archive.ubuntu.com/ubuntu bionic-backports InRelease [74.6 kB]\nGet:5 http://security.ubuntu.com/ubuntu bionic-security/universe amd64 Packages [869 kB]\nGet:6 http://archive.ubuntu.com/ubuntu bionic/multiverse amd64 Packages [186 kB]\nGet:7 http://security.ubuntu.com/ubuntu bionic-security/main amd64 Packages [1003 kB]\nGet:8 http://archive.ubuntu.com/ubuntu bionic/main amd64 Packages [1344 kB] \nGet:9 http://security.ubuntu.com/ubuntu bionic-security/multiverse amd64 Packages [9282 B]\nGet:10 http://security.ubuntu.com/ubuntu bionic-security/restricted amd64 Packages [89.0 kB]\nGet:11 http://archive.ubuntu.com/ubuntu bionic/universe amd64 Packages [11.3 MB]\nGet:12 http://archive.ubuntu.com/ubuntu bionic/restricted amd64 Packages [13.5 kB]\nGet:13 http://archive.ubuntu.com/ubuntu bionic-updates/universe amd64 Packages [1406 kB]\nGet:14 http://archive.ubuntu.com/ubuntu bionic-updates/main amd64 Packages [1302 kB]\nGet:15 http://archive.ubuntu.com/ubuntu bionic-updates/multiverse amd64 Packages [13.6 kB]\nGet:16 http://archive.ubuntu.com/ubuntu bionic-updates/restricted amd64 Packages [102 kB]\nGet:17 http://archive.ubuntu.com/ubuntu bionic-backports/main amd64 Packages [8286 B]\nGet:18 http://archive.ubuntu.com/ubuntu bionic-backports/universe amd64 Packages [8158 B]\nFetched 18.2 MB in 1min 9s (264 kB/s) \nReading package lists... Done\nReading package lists... Done\nBuilding dependency tree \nReading state information... Done\ngcc is already the newest version (4:7.4.0-1ubuntu2.3).\ngcc set to manually installed.\n0 upgraded, 0 newly installed, 0 to remove and 25 not upgraded.\nReading package lists... Done\nBuilding dependency tree \nReading state information... Done\nThe following additional packages will be installed:\n autotools-dev freetds-common libltdl-dev libltdl7 libodbc1 libtool odbcinst\n odbcinst1debian2\nSuggested packages:\n libtool-doc libmyodbc odbc-postgresql unixodbc-bin autoconf automaken\n gfortran | fortran95-compiler gcj-jdk\nThe following NEW packages will be installed:\n autotools-dev freetds-common libltdl-dev libltdl7 libodbc1 libtool odbcinst\n odbcinst1debian2 tdsodbc unixodbc-dev\n0 upgraded, 10 newly installed, 0 to remove and 25 not upgraded.\nNeed to get 1094 kB of archives.\nAfter this operation, 5969 kB of additional disk space will be used.\nGet:1 http://archive.ubuntu.com/ubuntu bionic/main amd64 autotools-dev all 20180224.1 [39.6 kB]\nGet:2 http://archive.ubuntu.com/ubuntu bionic-updates/main amd64 freetds-common all 1.00.82-2ubuntu0.1 [24.7 kB]\nGet:3 http://archive.ubuntu.com/ubuntu bionic/main amd64 libltdl7 amd64 2.4.6-2 [38.8 kB]\nGet:4 http://archive.ubuntu.com/ubuntu bionic/main amd64 libltdl-dev amd64 2.4.6-2 [162 kB]\nGet:5 http://archive.ubuntu.com/ubuntu bionic/main amd64 libodbc1 amd64 2.3.4-1.1ubuntu3 [183 kB]\nGet:6 http://archive.ubuntu.com/ubuntu bionic/main amd64 libtool all 2.4.6-2 [194 kB]\nGet:7 http://archive.ubuntu.com/ubuntu bionic/main amd64 odbcinst1debian2 amd64 2.3.4-1.1ubuntu3 [40.4 kB]\nGet:8 http://archive.ubuntu.com/ubuntu bionic/main amd64 odbcinst amd64 2.3.4-1.1ubuntu3 [12.4 kB]\nGet:9 http://archive.ubuntu.com/ubuntu bionic-updates/main amd64 tdsodbc amd64 1.00.82-2ubuntu0.1 [183 kB]\nGet:10 http://archive.ubuntu.com/ubuntu bionic/main amd64 unixodbc-dev amd64 2.3.4-1.1ubuntu3 [217 kB]\nFetched 1094 kB in 3s (335 kB/s) \ndebconf: delaying package configuration, since apt-utils is not installed\nSelecting previously unselected package autotools-dev.\n(Reading database ... 17146 files and directories currently installed.)\nPreparing to unpack .../0-autotools-dev_20180224.1_all.deb ...\nUnpacking autotools-dev (20180224.1) ...\nSelecting previously unselected package freetds-common.\nPreparing to unpack .../1-freetds-common_1.00.82-2ubuntu0.1_all.deb ...\nUnpacking freetds-common (1.00.82-2ubuntu0.1) ...\nSelecting previously unselected package libltdl7:amd64.\nPreparing to unpack .../2-libltdl7_2.4.6-2_amd64.deb ...\nUnpacking libltdl7:amd64 (2.4.6-2) ...\nSelecting previously unselected package libltdl-dev:amd64.\nPreparing to unpack .../3-libltdl-dev_2.4.6-2_amd64.deb ...\nUnpacking libltdl-dev:amd64 (2.4.6-2) ...\nSelecting previously unselected package libodbc1:amd64.\nPreparing to unpack .../4-libodbc1_2.3.4-1.1ubuntu3_amd64.deb ...\nUnpacking libodbc1:amd64 (2.3.4-1.1ubuntu3) ...\nSelecting previously unselected package libtool.\nPreparing to unpack .../5-libtool_2.4.6-2_all.deb ...\nUnpacking libtool (2.4.6-2) ...\nSelecting previously unselected package odbcinst1debian2:amd64.\nPreparing to unpack .../6-odbcinst1debian2_2.3.4-1.1ubuntu3_amd64.deb ...\nUnpacking odbcinst1debian2:amd64 (2.3.4-1.1ubuntu3) ...\nSelecting previously unselected package odbcinst.\nPreparing to unpack .../7-odbcinst_2.3.4-1.1ubuntu3_amd64.deb ...\nUnpacking odbcinst (2.3.4-1.1ubuntu3) ...\nSelecting previously unselected package tdsodbc:amd64.\nPreparing to unpack .../8-tdsodbc_1.00.82-2ubuntu0.1_amd64.deb ...\nUnpacking tdsodbc:amd64 (1.00.82-2ubuntu0.1) ...\nSelecting previously unselected package unixodbc-dev:amd64.\nPreparing to unpack .../9-unixodbc-dev_2.3.4-1.1ubuntu3_amd64.deb ...\nUnpacking unixodbc-dev:amd64 (2.3.4-1.1ubuntu3) ...\nSetting up freetds-common (1.00.82-2ubuntu0.1) ...\nSetting up autotools-dev (20180224.1) ...\nSetting up libltdl7:amd64 (2.4.6-2) ...\nSetting up libtool (2.4.6-2) ...\nSetting up libodbc1:amd64 (2.3.4-1.1ubuntu3) ...\nSetting up libltdl-dev:amd64 (2.4.6-2) ...\nSetting up odbcinst1debian2:amd64 (2.3.4-1.1ubuntu3) ...\nSetting up unixodbc-dev:amd64 (2.3.4-1.1ubuntu3) ...\nSetting up odbcinst (2.3.4-1.1ubuntu3) ...\nSetting up tdsodbc:amd64 (1.00.82-2ubuntu0.1) ...\ndebconf: unable to initialize frontend: Dialog\ndebconf: (No usable dialog-like program is installed, so the dialog based frontend cannot be used. at /usr/share/perl5/Debconf/FrontEnd/Dialog.pm line 76.)\ndebconf: falling back to frontend: Readline\nProcessing triggers for libc-bin (2.27-3ubuntu1) ...\nReading package lists... Done\nBuilding dependency tree \nReading state information... Done\nThe following additional packages will be installed:\n fontconfig libaudio2 libice6 libjbig0 libmng2 libmysqlclient20\n libodbcinstq4-1 libqt4-dbus libqt4-declarative libqt4-network libqt4-script\n libqt4-sql libqt4-sql-mysql libqt4-xml libqt4-xmlpatterns libqtcore4\n libqtdbus4 libqtgui4 libsm6 libtiff5 libxt6 mysql-common qdbus qt-at-spi\n qtchooser qtcore4-l10n\nSuggested packages:\n nas libqt4-declarative-folderlistmodel libqt4-declarative-gestures\n libqt4-declarative-particles libqt4-declarative-shaders qt4-qmlviewer\n libqt4-dev libicu55 libthai0 qt4-qtconfig\nThe following NEW packages will be installed:\n fontconfig libaudio2 libice6 libjbig0 libmng2 libmysqlclient20\n libodbcinstq4-1 libqt4-dbus libqt4-declarative libqt4-network libqt4-script\n libqt4-sql libqt4-sql-mysql libqt4-xml libqt4-xmlpatterns libqtcore4\n libqtdbus4 libqtgui4 libsm6 libtiff5 libxt6 mysql-common qdbus qt-at-spi\n qtchooser qtcore4-l10n unixodbc-bin\n0 upgraded, 27 newly installed, 0 to remove and 25 not upgraded.\nNeed to get 12.0 MB of archives.\nAfter this operation, 47.6 MB of additional disk space will be used.\nGet:1 http://archive.ubuntu.com/ubuntu bionic/main amd64 fontconfig amd64 2.12.6-0ubuntu2 [169 kB]\nGet:2 http://archive.ubuntu.com/ubuntu bionic/universe amd64 libmng2 amd64 2.0.2-0ubuntu3 [169 kB]\nGet:3 http://archive.ubuntu.com/ubuntu bionic/main amd64 libice6 amd64 2:1.0.9-2 [40.2 kB]\nGet:4 http://archive.ubuntu.com/ubuntu bionic/main amd64 libsm6 amd64 2:1.2.2-1 [15.8 kB]\nGet:5 http://archive.ubuntu.com/ubuntu bionic/main amd64 libxt6 amd64 1:1.1.5-1 [160 kB]\nGet:6 http://archive.ubuntu.com/ubuntu bionic/main amd64 libaudio2 amd64 1.9.4-6 [50.3 kB]\nGet:7 http://archive.ubuntu.com/ubuntu bionic/main amd64 mysql-common all 5.8+1.0.4 [7308 B]\nGet:8 http://archive.ubuntu.com/ubuntu bionic-updates/main amd64 libmysqlclient20 amd64 5.7.30-0ubuntu0.18.04.1 [690 kB]\n"
]
],
[
[
"#### Use this command to troubleshoot a failed pyodbc installation:\n!pip install --upgrade --global-option=build_ext --global-option=\"-I/usr/local/include\" --global-option=\"-L/usr/local/lib\" pyodbc",
"_____no_output_____"
]
],
[
[
"!pip install pyodbc",
"Collecting pyodbc\n Downloading pyodbc-4.0.30.tar.gz (266 kB)\n\u001b[K |████████████████████████████████| 266 kB 781 kB/s eta 0:00:01\n\u001b[?25hBuilding wheels for collected packages: pyodbc\n Building wheel for pyodbc (setup.py) ... \u001b[?25ldone\n\u001b[?25h Created wheel for pyodbc: filename=pyodbc-4.0.30-cp36-cp36m-linux_x86_64.whl size=273428 sha256=9563b15d73734558f4d24d0724341ca82f0ca0486e3200ef68d2fee75a3f75ff\n Stored in directory: /root/.cache/pip/wheels/e3/3f/16/e11367542166d4f8a252c031ac3a4163d3b901b251ec71e905\nSuccessfully built pyodbc\nInstalling collected packages: pyodbc\nSuccessfully installed pyodbc-4.0.30\n"
],
[
"!rm /etc/odbcinst.ini\n!rm /etc/odbc.ini",
"_____no_output_____"
],
[
"!ln -s /tf/odbcinst.ini /etc/odbcinst.ini\n!ln -s /tf/odbc.ini /etc/odbc.ini",
"_____no_output_____"
],
[
"!cat /tf/odbcinst.ini",
"[InterSystems ODBC35]\r\r\nUsageCount=1\r\r\nDriver=/tf/libirisodbcu35.so\r\r\nSetup=/tf/libirisodbcu35.so\r\r\nSQLLevel=1\r\r\nFileUsage=0\r\r\nDriverODBCVer=02.10\r\r\nConnectFunctions=YYN\r\r\nAPILevel=1\r\r\nDEBUG=1\r\r\nCPTimeout=<not pooled>\r\r\n\r\r\n"
],
[
"!cat /tf/odbc.ini",
"[user]\r\r\nDriver=InterSystems ODBC35\r\r\nProtocol=TCP\r\r\nHost=irisimlsvr\r\r\nPort=51773\r\r\nNamespace=USER\r\r\nUID=SUPERUSER\r\r\nPassword=SYS\r\r\nDescription=Sample namespace\r\r\nQuery Timeout=0\r\r\nStatic Cursors=0\r\r\n\r\r\n"
],
[
"!odbcinst -j",
"unixODBC 2.3.4\r\nDRIVERS............: /etc/odbcinst.ini\r\nSYSTEM DATA SOURCES: /etc/odbc.ini\r\nFILE DATA SOURCES..: /etc/ODBCDataSources\r\nUSER DATA SOURCES..: /root/.odbc.ini\r\nSQLULEN Size.......: 8\r\nSQLLEN Size........: 8\r\nSQLSETPOSIROW Size.: 8\r\n"
]
],
[
[
"### 2. Verify you see \"InterSystems ODBC35\" in the drivers list",
"_____no_output_____"
]
],
[
[
"import pyodbc\nprint(pyodbc.drivers())",
"['InterSystems ODBC35']\n"
]
],
[
[
"### 3. Get an ODBC connection ",
"_____no_output_____"
]
],
[
[
"import pyodbc \nimport time\n\n\n#input(\"Hit any key to start\")\ndsn = 'IRIS QuickML demo via PyODBC'\nserver = 'irisimlsvr' #'192.168.99.101' \nport = '51773' #'9091'\ndatabase = 'USER' \nusername = 'SUPERUSER' \npassword = 'SYS' \ncnxn = pyodbc.connect('DRIVER={InterSystems ODBC35};SERVER='+server+';PORT='+port+';DATABASE='+database+';UID='+username+';PWD='+ password)\n\n### Ensure it read strings correctly.\ncnxn.setdecoding(pyodbc.SQL_CHAR, encoding='utf8')\ncnxn.setdecoding(pyodbc.SQL_WCHAR, encoding='utf8')\ncnxn.setencoding(encoding='utf8')",
"_____no_output_____"
]
],
[
[
"### 4. Get a cursor; start the timer",
"_____no_output_____"
]
],
[
[
"cursor = cnxn.cursor()\nstart= time.clock()",
"_____no_output_____"
]
],
[
[
"### 5. specify the training data, and give a model name",
"_____no_output_____"
]
],
[
[
"dataTable = 'SQLUser.BreastCancer'\ndataTablePredict = 'Result02'\ndataColumn = 'Diagnosis'\ndataColumnPredict = \"PredictedDiagnosis\"\nmodelName = \"bc\" #chose a name - must be unique in server end",
"_____no_output_____"
]
],
[
[
"### 6. Train and predict",
"_____no_output_____"
]
],
[
[
"cursor.execute(\"CREATE MODEL %s PREDICTING (%s) FROM %s\" % (modelName, dataColumn, dataTable))\ncursor.execute(\"TRAIN MODEL %s FROM %s\" % (modelName, dataTable))\ncursor.execute(\"Create Table %s (%s VARCHAR(100), %s VARCHAR(100))\" % (dataTablePredict, dataColumnPredict, dataColumn))\ncursor.execute(\"INSERT INTO %s SELECT TOP 20 PREDICT(%s) AS %s, %s FROM %s\" % (dataTablePredict, modelName, dataColumnPredict, dataColumn, dataTable)) \ncnxn.commit()",
"_____no_output_____"
]
],
[
[
"### 7. Show the predict result",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nfrom IPython.display import display\n\ndf1 = pd.read_sql(\"SELECT * from %s ORDER BY ID\" % dataTablePredict, cnxn)\ndisplay(df1)",
"_____no_output_____"
]
],
[
[
"### 8. Show a complicated query\nIntegratedML function PREDICT() and PROBABILITY() can appear virtually anywhere in a SQL query, for maximal flexibility!\nBelow we are SELECTing columns as well as the result of the PROBABILITY function, and then filtering on the result of the PREDICT function. To top it off, ORDER BY is using the output of PROBSBILITY for sorting.",
"_____no_output_____"
]
],
[
[
"df2 = pd.read_sql(\"SELECT ID, PROBABILITY(bc FOR 'M') AS Probability, Diagnosis FROM %s \\\n WHERE MeanArea BETWEEN 300 AND 600 AND MeanRadius > 5 AND PREDICT(%s) = 'M' \\\n ORDER BY Probability\" % (dataTable, modelName),cnxn) \ndisplay(df2)",
"_____no_output_____"
]
],
[
[
"### 9. Close and clean ",
"_____no_output_____"
]
],
[
[
"cnxn.close()\nend= time.clock()\nprint (\"Total elapsed time: \")\nprint (end-start)\n#input(\"Hit any key to end\")",
"Total elapsed time: \n0.6454539999999991\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
c513cbc5bdc0b42ab80651c0622cd43f4ec25473
| 293,635 |
ipynb
|
Jupyter Notebook
|
PandasPresentation.ipynb
|
Giuco/pandas-presentation
|
49471904771603b65e8641e7063b4f0858f2f782
|
[
"MIT"
] | 4 |
2020-04-23T13:07:01.000Z
|
2021-09-10T16:57:54.000Z
|
PandasPresentation.ipynb
|
Giuco/pandas-presentation
|
49471904771603b65e8641e7063b4f0858f2f782
|
[
"MIT"
] | null | null | null |
PandasPresentation.ipynb
|
Giuco/pandas-presentation
|
49471904771603b65e8641e7063b4f0858f2f782
|
[
"MIT"
] | 2 |
2020-04-23T13:57:39.000Z
|
2021-09-10T18:22:54.000Z
| 32.970469 | 1,222 | 0.326943 |
[
[
[
"# Advanced Data Wrangling with Pandas",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np",
"_____no_output_____"
]
],
[
[
"## Formas não usuais de se ler um dataset",
"_____no_output_____"
],
[
"Você não precisa que o arquivo com os seus dados esteja no seu disco local, o pandas está preparado para adquirir arquivos via http, s3, gs...",
"_____no_output_____"
]
],
[
[
"diamonds = pd.read_csv(\"https://raw.githubusercontent.com/mwaskom/seaborn-data/master/diamonds.csv\")\ndiamonds.head()",
"_____no_output_____"
]
],
[
[
"Você também pode crawlear uma tabela de uma página da internet de forma simples",
"_____no_output_____"
]
],
[
[
"clarity = pd.read_html(\"https://www.brilliantearth.com/diamond-clarity/\")\n\nclarity",
"_____no_output_____"
],
[
"clarity = clarity[0]\nclarity",
"_____no_output_____"
],
[
"clarity.columns = ['clarity', 'clarity_description']\nclarity",
"_____no_output_____"
]
],
[
[
"## Como explodir a coluna de um dataframe",
"_____no_output_____"
]
],
[
[
"clarity['clarity'] = clarity['clarity'].str.split()\nclarity",
"_____no_output_____"
],
[
"type(clarity.loc[0, 'clarity'])",
"_____no_output_____"
],
[
"clarity = clarity.explode(\"clarity\")\nclarity",
"_____no_output_____"
]
],
[
[
"## Como validar o merge\n\nEsse parametro serve para validar a relação entre as duas tabelas que você está juntando. Por exemplo, se a relação é 1 para 1, 1 para muitos, muitos para 1 ou muitos para muitos.",
"_____no_output_____"
]
],
[
[
"diamonds.merge(clarity, on='clarity', validate=\"m:1\")",
"_____no_output_____"
],
[
"clarity_with_problem = clarity.append(pd.Series({\"clarity\": \"SI2\", \"clarity_description\": \"slightly included\"}), ignore_index=True)\nclarity_with_problem",
"_____no_output_____"
],
[
"diamonds.merge(clarity_with_problem, on='clarity', validate=\"m:1\")",
"_____no_output_____"
],
[
"diamonds.merge(clarity_with_problem, on='clarity')",
"_____no_output_____"
]
],
[
[
"### Por que isso é importante?\n\nO que aconteceria seu tivesse keys duplicadas no meu depara. Ele duplicou as minhas linhas que tinham a key duplicada, o dataset foi de 53,940 linhas para 63,134 linhas",
"_____no_output_____"
],
[
"## Como usar o método `.assign`\n\nPara adicionar ou modificar colunas do dataframe. Você pode passar como argumento uma constante para a coluna ou um função que tenha como input um `pd.DataFrame` e output uma `pd.Series`.",
"_____no_output_____"
]
],
[
[
"diamonds.assign(foo=\"bar\", bar=\"foo\")",
"_____no_output_____"
],
[
"diamonds.assign(volume=lambda df: df['x'] * df['y'] * df['z'])",
"_____no_output_____"
],
[
"def calculate_volume(df):\n return df['x'] * df['y'] * df['z']\n\n\ndiamonds.assign(volume=calculate_volume)",
"_____no_output_____"
],
[
"diamonds['volume'] = diamonds['x'] * diamonds['y'] * diamonds['z']\ndiamonds",
"_____no_output_____"
]
],
[
[
"## Como usar o método `.query`\nPara filtrar. Tende a ser util quando você quer filtrar o dataframe baseado em algum estado intermediário",
"_____no_output_____"
]
],
[
[
"diamonds = pd.read_csv(\"https://raw.githubusercontent.com/mwaskom/seaborn-data/master/diamonds.csv\")\ndiamonds.head()",
"_____no_output_____"
],
[
"diamonds.describe()",
"_____no_output_____"
],
[
"diamonds[(diamonds['x'] == 0) | (diamonds['y'] == 0) | (diamonds['z'] == 0)]",
"_____no_output_____"
],
[
"diamonds.query(\"x == 0 | y == 0 | z == 0\")\n",
"_____no_output_____"
],
[
"x = diamonds \\\n .assign(volume=lambda df: df['x'] * df['y'] * df['z'])\n\nx = x[x['volume'] > 0]",
"_____no_output_____"
],
[
"diamonds = diamonds \\\n .assign(volume=lambda df: df['x'] * df['y'] * df['z']) \\\n .query(\"volume > 0\")\n\ndiamonds",
"_____no_output_____"
]
],
[
[
"Você também pode usar variáveis externas ao dataframe dentro da sua query, basta usar @ como marcador.",
"_____no_output_____"
]
],
[
[
"selected_cut = \"Premium\"\ndiamonds.query(\"cut == @selected_cut\")",
"_____no_output_____"
]
],
[
[
"Quase qualquer string que seria um código python válido, vai ser uma query valida",
"_____no_output_____"
]
],
[
[
"diamonds.query(\"clarity.str.startswith('SI')\")",
"_____no_output_____"
]
],
[
[
"Porém o parser do pandas tem algumas particularidades, como o `==` que também pode ser um `isin`",
"_____no_output_____"
]
],
[
[
"diamonds.query(\"color == ['E', 'J']\")",
"_____no_output_____"
],
[
"diamonds = diamonds.query(\"x != 0 & y != 0 & z != 0\")",
"_____no_output_____"
]
],
[
[
"Exemplo de que precisamos do estado intermediário para fazer um filtro. Você cria uma nova coluna e quer filtrar baseado nela sem precisar salvar esse resultado em uma variável intermerdiária",
"_____no_output_____"
],
[
"## Como usar o método `.loc` e `.iloc`\nUma das desvantagens do `.query` é que fica mais difícil fazer análise estática do código, os editores geralmente não suportam syntax highlighting. Um jeito de solucionar esse problemas é usando o `.loc` ou `.iloc`, que além de aceitarem mascaras, eles aceitam funções também.",
"_____no_output_____"
]
],
[
[
"diamonds.loc[[0, 1, 2], ['clarity', 'depth']]",
"_____no_output_____"
],
[
"diamonds.iloc[[0, 1, 2], [3, 4]]",
"_____no_output_____"
],
[
"diamonds.sort_values(\"depth\")",
"_____no_output_____"
],
[
"diamonds.sort_values(\"depth\").loc[[0, 1, 2]]",
"_____no_output_____"
],
[
"diamonds.sort_values(\"depth\").iloc[[0, 1, 2]]",
"_____no_output_____"
],
[
"diamonds.loc[diamonds[\"price\"] > 6000]",
"_____no_output_____"
],
[
"diamonds[\"price\"] > 6000",
"_____no_output_____"
],
[
"diamonds.loc[lambda x: x['price'] > 6000]",
"_____no_output_____"
],
[
"diamonds[diamonds['price'] > 10000]['price'] = 10000",
"/Users/giuliano.ferrari/Documents/Other/pandas-presentation/.env/lib/python3.7/site-packages/ipykernel_launcher.py:1: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\n \"\"\"Entry point for launching an IPython kernel.\n"
],
[
"diamonds.query(\"price > 10000\")",
"_____no_output_____"
],
[
"diamonds.loc[diamonds['price'] > 10000, 'price'] = 10000",
"_____no_output_____"
],
[
"diamonds.query(\"price > 10000\")",
"_____no_output_____"
]
],
[
[
"## O que o `.groupby(...) retorna`",
"_____no_output_____"
]
],
[
[
"diamonds = pd.read_csv(\"https://raw.githubusercontent.com/mwaskom/seaborn-data/master/diamonds.csv\") \\\n .assign(volume=lambda x: x['x'] * x['y'] * x['z']) \\\n .query(\"volume > 0\")\ndiamonds.head()",
"_____no_output_____"
],
[
"grouped_diamonds = diamonds.groupby(\"cut\")\ngrouped_diamonds",
"_____no_output_____"
],
[
"list(grouped_diamonds)",
"_____no_output_____"
]
],
[
[
"## Os N formatos de agregação do pandas",
"_____no_output_____"
],
[
"A função `.agg` é um *alias* da função `.aggregate`, então elas tem o mesmo resultado.\n\nO Pandas tem algumas funções padrão que permitem que você passe só o nome delas, ao invés do *callable*:\n* \"all\"\n* \"any\"\n* \"count\"\n* \"first\"\n* \"idxmax\"\n* \"idxmin\"\n* \"last\"\n* \"mad\"\n* \"max\"\n* \"mean\"\n* \"median\"\n* \"min\"\n* \"nunique\"\n* \"prod\"\n* \"sem\"\n* \"size\"\n* \"skew\"\n* \"std\"\n* \"sum\"\n* \"var\"",
"_____no_output_____"
],
[
"Você pode passar uma lista de callable e o pandas vai aplicar todas as funções para todas as colunas. Faz sentido se são muitas funções e poucas colunas. Um problema é que ele vai nomear as novas colunas com base na coluna anterior e na função, quando você usa uma lambda isso causa um problema.",
"_____no_output_____"
]
],
[
[
"diamonds.groupby('clarity').agg(['mean', 'sum', np.max, lambda x: x.min()])",
"_____no_output_____"
]
],
[
[
"Você também pode passar um dicionário de listas, assim você pode escolher qual função será aplicada em cada coluna, você ainda tem o problema de nome das novas colunas ao usar uma função anônima.",
"_____no_output_____"
]
],
[
[
"diamonds.groupby('clarity').agg({\"x\": 'mean', 'price': [np.max, 'max', max, lambda x: x.max()]})",
"_____no_output_____"
]
],
[
[
"A terceira opção é o NamedAgg foi lançada recentemente. Ela resolve o problema de nomes de colunas. Você passa como parâmetro uma tupla para cada agregação que você quer. O primeiro elemento é o nome da coluna e o segundo é a função.\n\n\\* *O Dask ainda não aceita esse tipo de agregação*",
"_____no_output_____"
]
],
[
[
"diamonds.groupby('clarity').agg(max_price=('price', 'max'), total_cost=('price', lambda x: x.sum()))",
"_____no_output_____"
]
],
[
[
"## `.groupby(...).apply(...)`",
"_____no_output_____"
],
[
"Um problema comum a todas essas abordagens é que você não consegue fazer uma agregação que depende de duas colunas. Para a maior parte dos casos existe uma forma razoável de resolver esse problema criando uma nova coluna e aplicando a agregação nela. Porém, se isso não foi possível, dá para usar o `.groupby(...).apply()`.",
"_____no_output_____"
]
],
[
[
"# Nesse caso ao invés da função de agregação receber a pd.Series relativa ao grupo,\n# ela vai receber o subset do grupo. Aqui vamos printar cada grupo do df de forma \n# separada\n\ndiamonds.groupby('cut').apply(lambda x: print(x.head().to_string() + \"\\n\"))",
" carat cut color clarity depth table price x y z volume\n8 0.22 Fair E VS2 65.1 61.0 337 3.87 3.78 2.49 36.425214\n91 0.86 Fair E SI2 55.1 69.0 2757 6.45 6.33 3.52 143.716320\n97 0.96 Fair F SI2 66.3 62.0 2759 6.27 5.95 4.07 151.837455\n123 0.70 Fair F VS2 64.5 57.0 2762 5.57 5.53 3.58 110.271518\n124 0.70 Fair F VS2 65.3 55.0 2762 5.63 5.58 3.66 114.980364\n\n carat cut color clarity depth table price x y z volume\n2 0.23 Good E VS1 56.9 65.0 327 4.05 4.07 2.31 38.076885\n4 0.31 Good J SI2 63.3 58.0 335 4.34 4.35 2.75 51.917250\n10 0.30 Good J SI1 64.0 55.0 339 4.25 4.28 2.73 49.658700\n17 0.30 Good J SI1 63.4 54.0 351 4.23 4.29 2.70 48.996090\n18 0.30 Good J SI1 63.8 56.0 351 4.23 4.26 2.71 48.833658\n\n carat cut color clarity depth table price x y z volume\n0 0.23 Ideal E SI2 61.5 55.0 326 3.95 3.98 2.43 38.202030\n11 0.23 Ideal J VS1 62.8 56.0 340 3.93 3.90 2.46 37.704420\n13 0.31 Ideal J SI2 62.2 54.0 344 4.35 4.37 2.71 51.515745\n16 0.30 Ideal I SI2 62.0 54.0 348 4.31 4.34 2.68 50.130472\n39 0.33 Ideal I SI2 61.8 55.0 403 4.49 4.51 2.78 56.294722\n\n carat cut color clarity depth table price x y z volume\n1 0.21 Premium E SI1 59.8 61.0 326 3.89 3.84 2.31 34.505856\n3 0.29 Premium I VS2 62.4 58.0 334 4.20 4.23 2.63 46.724580\n12 0.22 Premium F SI1 60.4 61.0 342 3.88 3.84 2.33 34.715136\n14 0.20 Premium E SI2 60.2 62.0 345 3.79 3.75 2.27 32.262375\n15 0.32 Premium E I1 60.9 58.0 345 4.38 4.42 2.68 51.883728\n\n carat cut color clarity depth table price x y z volume\n5 0.24 Very Good J VVS2 62.8 57.0 336 3.94 3.96 2.48 38.693952\n6 0.24 Very Good I VVS1 62.3 57.0 336 3.95 3.98 2.47 38.830870\n7 0.26 Very Good H SI1 61.9 55.0 337 4.07 4.11 2.53 42.321081\n9 0.23 Very Good H VS1 59.4 61.0 338 4.00 4.05 2.39 38.718000\n19 0.30 Very Good J SI1 62.7 59.0 351 4.21 4.27 2.66 47.818022\n\n"
]
],
[
[
"Esse formato de agregação introduz algumas complexidades, porque sua função pode retornar tanto um pd.DataFrame, pd.Series ou um escalar. O pandas vai tentar fazer um broadcasting do que você retorna para algo que ele acha que faz sentido. Exemplos:\n\nSe você retornar um escalar, o apply vai retornar uma `pd.Series` em que cada elemento corresponde a um grupo do .groupby",
"_____no_output_____"
]
],
[
[
"# Retornando um escalar\ndef returning_scalar(df: pd.DataFrame) -> float:\n return (df[\"x\"] * df[\"y\"] * df['z']).mean()\n\n\ndiamonds.groupby(\"cut\").apply(returning_scalar)",
"_____no_output_____"
]
],
[
[
"Se você retornar uma `pd.Series` nomeada, o apply vai retornar um `pd.DataFrame` em que cada linha corresponde a um grupo do `.groupby` e cada coluna corresponde a uma key do pd.Series que você retorna na sua função de agregação",
"_____no_output_____"
]
],
[
[
"def returning_named_series(df: pd.DataFrame) -> pd.Series:\n volume = (df[\"x\"] * df[\"y\"] * df['z'])\n price_to_volume = df['price'] / volume\n return pd.Series({\"mean_volume\": volume.mean(), \"mean_price_to_volume\": price_to_volume.mean()})\n\n\ndiamonds.groupby(\"cut\").apply(returning_named_series)",
"_____no_output_____"
]
],
[
[
"Se você retornar um `pd.DataFrame`, o apply vai retornar uma concatenação dos desses `pd.DataFrame`",
"_____no_output_____"
]
],
[
[
"def returning_dataframe(df: pd.DataFrame) -> pd.DataFrame:\n return df[df['volume'] >= df['volume'].median()]\n \n\ndiamonds.groupby(\"cut\").apply(returning_dataframe)",
"_____no_output_____"
]
],
[
[
"Se você retornar uma `pd.Series` não nomeada, o apply vai retornar uma `pd.Series` que é uma concatenação das `pd.Series` que você retorna da sua função",
"_____no_output_____"
]
],
[
[
"def returning_unnamed_series(df: pd.DataFrame) -> pd.Series:\n return df.loc[df['volume'] >= df['volume'].median(), 'volume']\n\n\ndiamonds.groupby(\"cut\").apply(returning_unnamed_series)",
"_____no_output_____"
]
],
[
[
"De forma resumida, o `.groupby(...).apply(...)` é extremamente flexível, ele consegue filtrar, agregar e tranformar. Mas é mais complicado de usar e é bem lento se comparado aos outros métodos de agregação. Só use se necessário.\n\n| Saída da Função | Saída do apply |\n|-----------------------|--------------------------------------------------------------------------------------------------------------------------------------------------------------------|\n| Escalar | Uma pd.Series em que cada elemento corresponde a um grupo do .groupby |\n| pd.Series nomeada | Um pd.DataFrame em que cada linha corresponde a um grupo do .groupby e cada coluna corresponde a uma key do pd.Series que você retorna na sua função de agregação |\n| pd.Series não nomeada | Uma `pd.Series` que é uma concatenação das `pd.Series` que você retorna da sua função |\n| pd.DataFrame | Uma concatenação dos desses `pd.DataFrame` |",
"_____no_output_____"
],
[
"## Como usar o método `.pipe`",
"_____no_output_____"
],
[
"O `.pipe` aplica uma função ao dataframe",
"_____no_output_____"
]
],
[
[
"def change_basis(df: pd.DataFrame, factor=10):\n df[['x', 'y', 'z']] = df[['x', 'y', 'z']] * factor\n return df\n\n\ndiamonds.pipe(change_basis)",
"_____no_output_____"
]
],
[
[
"Nós não atribuimos o resultado da nossa operação a nenhuma variável, então teoricamente se rodarmos de novo, o resultado vai ser o mesmo.",
"_____no_output_____"
]
],
[
[
"diamonds.pipe(change_basis)",
"_____no_output_____"
]
],
[
[
"Isso acontece porque a sua função está alterando o `pd.DataFrame` original ao invés de criar uma cópia, isso é um pouco contra intuitivo porque o Pandas por padrão faz as suas operações em copias da tabela. Para evitar isso podemos fazer uma cópia do dataframe manualmente",
"_____no_output_____"
]
],
[
[
"diamonds = pd.read_csv(\"https://raw.githubusercontent.com/mwaskom/seaborn-data/master/diamonds.csv\")",
"_____no_output_____"
],
[
"def change_basis(df: pd.DataFrame, factor=10):\n df = df.copy()\n df[['x', 'y', 'z']] = df[['x', 'y', 'z']] * factor\n return df\n\n\ndiamonds.pipe(change_basis, factor=10)",
"_____no_output_____"
],
[
"diamonds",
"_____no_output_____"
]
],
[
[
"## Como combinar o `.assign`, `.pipe`, `.query` e `.loc` para um Pandas mais idiomático\n\nOs métodos mais importantes para *Method Chaining* são\n* `.assign`\n* `.query`\n* `.loc`\n* `.pipe`",
"_____no_output_____"
]
],
[
[
"diamonds = pd.read_csv(\"https://raw.githubusercontent.com/mwaskom/seaborn-data/master/diamonds.csv\")\ndiamonds.head()",
"_____no_output_____"
],
[
"diamonds_cp = diamonds.copy()\ndiamonds_cp[['x', 'y', 'z']] = diamonds_cp[['x', 'y', 'z']] * 10\ndiamonds_cp['volume'] = diamonds_cp['x'] * diamonds_cp['y'] * diamonds_cp['z']\ndiamonds_cp = diamonds_cp[diamonds_cp['volume'] > 0]\ndiamonds_cp = pd.merge(diamonds_cp, clarity, on='clarity', how='left')\n\ndiamonds_cp",
"_____no_output_____"
],
[
"def change_basis(df: pd.DataFrame, factor=10):\n df = df.copy()\n df[['x', 'y', 'z']] = df[['x', 'y', 'z']] * factor\n return df\n\n\ndiamonds \\\n .copy() \\\n .pipe(change_basis, factor=10) \\\n .assign(volume=lambda df: df['x'] * df['y'] * df['z']) \\\n .query(\"volume > 0\") \\\n .merge(clarity, on='clarity', how='left')\n",
"_____no_output_____"
]
],
[
[
"Um problema que pode acontecer quando você usa o method chaining é você acabar com um bloco gigantesco que é impossível de debugar, uma boa prática é quebrar seus blocos por objetivos",
"_____no_output_____"
],
[
"## Como mandar um dataframe para a sua clipboard\nGeralmente isso não é uma boa pratica, mas as vezes é útil para enviar uma parte do dado por mensagem ou para colar em alguma planilha.",
"_____no_output_____"
]
],
[
[
"df = pd.DataFrame({'a':list('abc'), 'b':np.random.randn(3)})",
"_____no_output_____"
],
[
"df",
"_____no_output_____"
],
[
"df.to_clipboard()",
"_____no_output_____"
],
[
"df.to_csv(\"df.csv\")",
"_____no_output_____"
]
],
[
[
"Você também pode ler da sua *clipboard* com `pd.read_clipboard(...)`. O que é uma prática pior ainda, mas em alguns casos pode ser útil.",
"_____no_output_____"
],
[
"## Recursos\nhttps://pandas.pydata.org/docs/user_guide/cookbook.html\n\nhttps://tomaugspurger.github.io/modern-1-intro.html",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
]
] |
c513d271369659675fe886e440836f41905ac806
| 70,686 |
ipynb
|
Jupyter Notebook
|
07/pandas_cheatsheet.ipynb
|
M0nica/python-foundations-hw
|
fe5065d3af71511bdd0fcf437d1d9f15f9faf1ee
|
[
"MIT"
] | null | null | null |
07/pandas_cheatsheet.ipynb
|
M0nica/python-foundations-hw
|
fe5065d3af71511bdd0fcf437d1d9f15f9faf1ee
|
[
"MIT"
] | null | null | null |
07/pandas_cheatsheet.ipynb
|
M0nica/python-foundations-hw
|
fe5065d3af71511bdd0fcf437d1d9f15f9faf1ee
|
[
"MIT"
] | null | null | null | 62.333333 | 9,444 | 0.772515 |
[
[
[
"\n01: Building a pandas Cheat Sheet, Part 1\n\nUse the csv I've attached to answer the following questions\nImport pandas with the right name\n",
"_____no_output_____"
]
],
[
[
"# !workon dataanalysis\nimport pandas as pd",
"/Users/Monica/.virtualenvs/dataanalysis/lib/python3.5/site-packages/matplotlib/__init__.py:1035: UserWarning: Duplicate key in file \"/Users/Monica/.matplotlib/matplotlibrc\", line #2\n (fname, cnt))\n"
]
],
[
[
"Having matplotlib play nice with virtual environments\n\nThe matplotlib library has some issues when you’re using a Python 3 virtual environment. The error looks like this:\n\nRuntimeError: Python is not installed as a framework. The Mac OS X backend will not be able to function correctly if Python is not installed as a framework. See the Python documentation for more information on installing Python as a framework on Mac OS X. Please either reinstall Python as a framework, or try one of the other backends. If you are Working with Matplotlib in a virtual enviroment see ‘Working with Matplotlib in Virtual environments’ in the Matplotlib FAQ\nLuckily it’s an easy fix.\n\nmkdir -p ~/.matplotlib && echo 'backend: TkAgg' >> ~/.matplotlib/matplotlibrc (ADD THIS LINE TO TERMINAL) \n\nThis adds a line to the matplotlib startup script to set the backend to TkAgg, whatever that means.\n\n",
"_____no_output_____"
],
[
"Set all graphics from matplotlib to display inline",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\n#DISPLAY MOTPLOTLIB INLINE WITH THE NOTEBOOK AS OPPOSED TO POP UP WINDOW\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"Read the csv in (it should be UTF-8 already so you don't have to worry about encoding), save it with the proper boring name",
"_____no_output_____"
]
],
[
[
"df = pd.read_csv('07-hw-animals.csv')",
"_____no_output_____"
],
[
"df",
"_____no_output_____"
],
[
"# Display the names of the columns in the csv",
"_____no_output_____"
],
[
"df.columns",
"_____no_output_____"
]
],
[
[
"Display the first 3 animals.",
"_____no_output_____"
]
],
[
[
"df.head(3)",
"_____no_output_____"
],
[
"# Sort the animals to see the 3 longest animals.",
"_____no_output_____"
],
[
"df.sort_values('length', ascending = False).head(3)",
"_____no_output_____"
],
[
"# What are the counts of the different values of the \"animal\" column? a.k.a. how many cats and how many dogs.\n# Only select the dogs.",
"_____no_output_____"
],
[
"(df['animal'] == 'dog').value_counts()",
"_____no_output_____"
],
[
"# Display all of the animals that are greater than 40 cm.",
"_____no_output_____"
],
[
"df[df['length'] > 40]",
"_____no_output_____"
]
],
[
[
"'length' is the animal's length in cm. Create a new column called inches that is the length in inches.",
"_____no_output_____"
]
],
[
[
"length_in = df['length']* 0.3937\n\ndf['length (in.)'] = length_in",
"_____no_output_____"
]
],
[
[
"Save the cats to a separate variable called \"cats.\" Save the dogs to a separate variable called \"dogs.\"",
"_____no_output_____"
]
],
[
[
"dogs = df[df['animal'] == 'dog']\ncats = df[df['animal'] == 'cat']",
"_____no_output_____"
]
],
[
[
"Display all of the animals that are cats and above 12 inches long. First do it using the \"cats\" variable, then do it using your normal dataframe.",
"_____no_output_____"
]
],
[
[
"cats['length'] > 12",
"_____no_output_____"
],
[
"df[(df['length'] > 12) & (df['animal'] == 'cat')]",
"_____no_output_____"
]
],
[
[
"What's the mean length of a cat?",
"_____no_output_____"
]
],
[
[
"# cats.describe() displays all stats for length",
"_____no_output_____"
],
[
"cats['length'].mean()",
"_____no_output_____"
],
[
"#only shows mean length\ncats.mean()",
"_____no_output_____"
]
],
[
[
"What's the mean length of a dog?",
"_____no_output_____"
]
],
[
[
"dogs['length'].mean()",
"_____no_output_____"
],
[
"dogs['length'].describe()",
"_____no_output_____"
],
[
"dogs.mean()",
"_____no_output_____"
]
],
[
[
"Use groupby to accomplish both of the above tasks at once.",
"_____no_output_____"
]
],
[
[
"df.groupby('animal')['length (in.)'].mean()",
"_____no_output_____"
]
],
[
[
"Make a histogram of the length of dogs. I apologize that it is so boring.",
"_____no_output_____"
]
],
[
[
"dogs.plot(kind='hist', y = 'length (in.)') # all the same length \"/",
"_____no_output_____"
]
],
[
[
"Change your graphing style to be something else (anything else!)",
"_____no_output_____"
]
],
[
[
"df.plot(kind=\"bar\", x=\"name\", y=\"length\", color = \"red\", legend =False)",
"_____no_output_____"
],
[
"df.plot(kind=\"barh\", x=\"name\", y=\"length\", color = \"red\", legend =False)",
"_____no_output_____"
],
[
"dogs",
"_____no_output_____"
],
[
"dogs.plot(kind='bar')",
"_____no_output_____"
],
[
"# dogs.plot(kind='scatter', x='name', y='length (in.)')",
"_____no_output_____"
]
],
[
[
"Make a horizontal bar graph of the length of the animals, with their name as the label ",
"_____no_output_____"
]
],
[
[
"df.columns",
"_____no_output_____"
],
[
"dogs['name']",
"_____no_output_____"
],
[
"dogs.plot(kind='bar', x='name', y = 'length', legend=False)",
"_____no_output_____"
]
],
[
[
"Make a sorted horizontal bar graph of the cats, with the larger cats on top.",
"_____no_output_____"
]
],
[
[
"cats.sort_values('length').plot(kind='barh', x='name', y = 'length', legend = False)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
c513d607ad31aff0dea41175885f8fde0313d74f
| 17,548 |
ipynb
|
Jupyter Notebook
|
Programmeerelementen/Functies/0100_FunctiesExtraOefeningen.ipynb
|
dwengovzw/PythonNotebooks
|
633bea4b07efbd920349d6f1dc346522ce118b70
|
[
"CC0-1.0"
] | null | null | null |
Programmeerelementen/Functies/0100_FunctiesExtraOefeningen.ipynb
|
dwengovzw/PythonNotebooks
|
633bea4b07efbd920349d6f1dc346522ce118b70
|
[
"CC0-1.0"
] | 3 |
2021-09-30T11:38:24.000Z
|
2021-10-04T09:25:39.000Z
|
Programmeerelementen/Functies/0100_FunctiesExtraOefeningen.ipynb
|
dwengovzw/PythonNotebooks
|
633bea4b07efbd920349d6f1dc346522ce118b70
|
[
"CC0-1.0"
] | null | null | null | 33.616858 | 724 | 0.605311 |
[
[
[
"<img src=\"images/kiksmeisedwengougent.png\" alt=\"Banner\" width=\"1100\"/>",
"_____no_output_____"
],
[
"<div style='color: #690027;' markdown=\"1\">\n <h1>FUNCTIES EN STRUCTUREN</h1> \n</div>",
"_____no_output_____"
],
[
"<div class=\"alert alert-block alert-success\">\nPython kent heel wat ingebouwde functies, zoals <span style=\"background-color:whitesmoke; font-family:consolas; font-size:1em;\">print()</span>, <span style=\"background-color:whitesmoke; font-family:consolas; font-size:1em;\">input()</span>, <span style=\"background-color:whitesmoke; font-family:consolas; font-size:1em;\">type()</span>, <span style=\"background-color:whitesmoke; font-family:consolas; font-size:1em;\">int()</span> en <span style=\"background-color:whitesmoke; font-family:consolas; font-size:1em;\">len()</span>. <br><br>Je kan ook al zelf functies definiëren. Je kent ook al herhalingsstructuren en keuzestructuren. In deze notebook oefen je dat verder in. \n</div>",
"_____no_output_____"
],
[
"<div class=\"alert alert-block alert-info\">\nAls je bepaalde code meerdere keren wilt gebruiken, loont het de moeite om zelf een functie te definiëren.<br> Functies worden gedeclareerd met het sleutelwoord <b>def</b>. Via het sleutelwoord <b>return</b> geven ze een resultaat terug.<br>\nEen <b>docstring</b> verduidelijkt het doel van de functie. Een docstring staat tussen driedubbele aanhalingstekens, begint met een hoofletter en eindigt met een punt. \n</div>",
"_____no_output_____"
],
[
"<div style='color: #690027;' markdown=\"1\">\n <h2>1. Functie met keuzestructuur, invoer, uitvoer, docstring</h2> \n</div>",
"_____no_output_____"
],
[
"### Voorbeeld",
"_____no_output_____"
],
[
"Bij uitvoer van het volgende script wordt de gebruiker naar een willekeurig geheel getal gevraagd. <br>\nErna wordt 7 gedeeld door dat getal via de functie `zevendelen()`. Het quotiënt wordt getoond. Als de gebruiker het getal 0 invoert, is er echter geen quotiënt.<br>\nVoer het script enkele keren uit. Probeer verschillende getallen, vergeet 0 niet. ",
"_____no_output_____"
]
],
[
[
"def zevendelen(getal):\n \"\"\"Quotiënt van 7 met een getal.\"\"\"\n if getal != 0:\n resultaat = 7 / getal # als noemer niet 0 is, bereken quotiënt\n else:\n resultaat = \"Er is geen quotiënt want je kan niet delen door nul.\" # als noemer 0 is, dan is er geen quotiënt\n return resultaat\n\n# invoer\n# 7 is teller, in te voeren getal is noemer\nprint(\"Deel 7 door een geheel getal naar keuze.\") \nnoemer = int(input(\"Geef een geheel getal naar keuze waardoor 7 moet gedeeld worden: \")) # typecasting: string omzetten naar int\n\n# invoer verwerken\nquot = zevendelen(noemer) # quotiënt\n\n# uitvoer\nprint(quot)",
"_____no_output_____"
]
],
[
[
"<div class=\"alert alert-block alert-info\">\nBegrijp je wat er gebeurt?<br>\nEr wordt gevraagd naar een getal. Jij geeft dat getal in. Deze invoer wordt geïnterpreteerd als string, maar met typecasting omgezet naar een object dat type int heeft. De variabele <span style=\"background-color:whitesmoke; font-family:consolas; font-size:1em;\">noemer</span> refereert aan dat object met type int. <br>\nDe invoer wordt vervolgens verwerkt. De functie wordt opgeroepen: 7 wordt gedeeld door de waarde van <span style=\"background-color:whitesmoke; font-family:consolas; font-size:1em;\">noemer</span>, tenminste als die waarde niet nul is. Het quotiënt is een object dat type float heeft. De variabele <span style=\"background-color:whitesmoke; font-family:consolas; font-size:1em;\">resultaat</span> verwijst naar dat float-object. In het geval dat de invoer 0 is, verwijst de variabele <span style=\"background-color:whitesmoke; font-family:consolas; font-size:1em;\">resultaat</span> naar een object dat type <span style=\"background-color:whitesmoke; font-family:consolas; font-size:1em;\">string</span> heeft. <br>\nDe variabele <span style=\"background-color:whitesmoke; font-family:consolas; font-size:1em;\">quot</span> refereert aan hetzelfde object als de variabele <span style=\"background-color:whitesmoke; font-family:consolas; font-size:1em;\">resultaat</span>, die de functie teruggeeft. <br>\nDe waarde van dat object <span style=\"background-color:whitesmoke; font-family:consolas; font-size:1em;\">quot</span> wordt getoond. <br> <br>\n \nDe volgende zin vertelt wat het doel van de functie is: Quotiënt van 7 met een getal. Het is een <b>docstring</b>.\n \n</div>",
"_____no_output_____"
],
[
"<div class=\"alert alert-block alert-warning\"> \nMeer uitleg over <em>typecasting</em> vind je in de notebook 'Datastructuur'.\n</div>",
"_____no_output_____"
],
[
"### Oefening 1.1\n- Welk type heeft `noemer`?\n- Welk type heeft het object waarnaar `quot` verwijst?",
"_____no_output_____"
],
[
"Antwoord:",
"_____no_output_____"
],
[
"### Oefening 1.2\nSchrijf een script dat de vierkantswortel van een getal teruggeeft, indien dat mogelijk is. Anders verschijnt een boodschap met de reden waarom er geen is. ",
"_____no_output_____"
],
[
"<div style='color: #690027;' markdown=\"1\">\n <h2>2. Module Random - herhalingsstructuur</h2> \n</div>",
"_____no_output_____"
],
[
"*In de module NumPy* is er een *module Random* waarmee men toevalsgetallen kan genereren. <br>In het volgende script worden twee functies uit de module Random gebruikt: de functie `random()` om decimale getallen te genereren en de functie `randint()`, met twee parameters, om gehele getallen te genereren. Het is dus nodig om eerst de module NumPy te importeren.<br>\nDe getallen gegenereerd door `random()` liggen altijd in [0,1[ en de getallen gegenereerd door `randint()` liggen in het halfopen interval bepaald door de twee argumenten die je aan de functie meegeeft, de bovengrens niet inbegrepen. <br>\nOm deze functies op te roepen, laat je ze voorgaan door `np.random.`.",
"_____no_output_____"
],
[
"### Voorbeeld 2.1\nTest het volgende script eens uit:",
"_____no_output_____"
]
],
[
[
"import numpy as np\n\n# for-lus wordt 10 keer doorlopen\nfor teller in range(10):\n print(\"teller = \", teller)\n print(np.random.random()) # functie random() genereert decimaal getal in [0,1[\n print(np.random.randint(0, 4)) # functie randint(0,4) kiest willekeurig geheel getal uit [0,4[ \n print()",
"_____no_output_____"
]
],
[
[
"### Oefening 2.1: dobbelsteen",
"_____no_output_____"
],
[
"Jan en Piet willen samen een gezelschapsspel spelen. Diegene die met twee dobbelstenen het hoogste aantal ogen gooit, mag starten. Ze kunnen de dobbelstenen echter niet vinden. <br>Korneel schrijft een script dat het twee keer gooien met twee dobbelstenen nabootst en als boodschap meegeeft wie er mag starten.<br> Er wordt opnieuw geworpen, zolang Jan en Piet hetzelfde aantal ogen gooien. ",
"_____no_output_____"
],
[
"Vul het script aan en voer het uit. ",
"_____no_output_____"
]
],
[
[
"def worp():\n \"\"\"Aantal ogen na worp met twee dobbelstenen.\"\"\"\n dob1 = np.random.randint(..., ...)\n dob2 = np.random.randint(..., ...)\n aantal_ogen = dob1 + dob2\n return aantal_ogen\n\n# Jan werpt\nworp_jan = worp()\n\n# Piet werpt\n...\n\n# bij ex aequo opnieuw gooien\nwhile worp_jan == ....:\n worp_jan = worp()\n worp_piet = worp()\n \nprint(\"Jan werpt een\" + str(worp_jan) + \".\")\nprint(\"Piet werpt een\" + str(worp_piet) + \".\")\n\n# bepalen wat hoogste worp is en wie mag starten\ngrootste = max(worp_jan, worp_piet)\nif grootste == worp_jan:\n beginner = \"Jan\"\nelse:\n ...\n\n# output\nprint(..., \"mag starten.\")",
"_____no_output_____"
]
],
[
[
"### Voorbeeld 2.2\nMet de Random-functie `rand()` kan je ook een NumPy-lijst met een bepaalde lengte genereren. *De elementen behoren tot het halfopen interval [0,1[*.<br>\n\nMet de Random-functie `randint()` kan je er een genereren met gehele getallen.<br>\nTest dit uit via het volgende script.<br> Let goed op de parameters van deze functies en merk op dat er gewerkt wordt met een halfopen interval.",
"_____no_output_____"
]
],
[
[
"willekeurige_lijst = np.random.rand(8)\nwillekeurige_lijst_gehele_getallen = np.random.randint(10, 29, 4) \nprint(willekeurige_lijst)\nprint(willekeurige_lijst_gehele_getallen)",
"_____no_output_____"
]
],
[
[
"### Oefening 2.2\n- Genereer een NumPy-lijst met 15 elementen en waarvan de elementen gehele getallen zijn, gelegen in het interval [4,9]. <br>Laat de NumPy-lijst zien.",
"_____no_output_____"
],
[
"- Genereer een NumPy-lijst met 15 elementen en waarvan de elementen kommagetallen zijn, gelegen in het interval [0, 9[. <br>Laat de NumPy-lijst zien.",
"_____no_output_____"
],
[
"<div style='color: #690027;' markdown=\"1\">\n <h2>3. Repetitieve taken</h2> \n</div>",
"_____no_output_____"
],
[
"### Voorbeeld: DNA",
"_____no_output_____"
],
[
"Het erfelijke materiaal van een levend organisme is opgeslagen in het DNA. Een DNA-molecuul bestaat uit twee lange strengen van nucleotiden, die in de vorm van een dubbele helix met elkaar vervlochten zijn. Nucleotiden zijn een specifieke groep organische verbindingen. Een DNA-streng bevat vier verschillende nucleotiden met een van de nucleobasen adenine, thymine, guanine en cytosine als component. Deze nucleobasen worden afgekort tot respectievelijk de letters A, T, G en C. De DNA-sequentie is de volgorde van deze nucleotiden op een streng DNA. Er zijn zeer veel sequenties mogelijk. ",
"_____no_output_____"
],
[
"Met de functies `choice()` uit de module Random van de module NumPy en `join()` kun je de letters kiezen en samenbrengen. ",
"_____no_output_____"
],
[
"Met de volgende functie kan je een willekeurige DNA-sequentie met een gewenste lengte genereren:",
"_____no_output_____"
]
],
[
[
"def dna_sequentie(n):\n \"\"\"DNA-sequentie genereren met gewenste lengte.\"\"\"\n letters = [\"A\", \"C\", \"G\", \"T\"]\n keuze = [np.random.choice(letters) for i in range(n)] # kies n letters uit lijst letters en stop die in andere lijst\n # print(keuze)\n string = \"\".join(keuze) # elementen van keuze samenbrengen in een (samenhangende) string zonder keuze aan te passen\n return string",
"_____no_output_____"
]
],
[
[
"Genereer een DNA-sequentie van lengte 40.",
"_____no_output_____"
],
[
"Alternatief: je kan een module Random gebruiken (een andere, niet die van NumPy). Je kunt dan letters kiezen uit een string i.p.v. een lijst. \nMet de volgende functie kan je dan een willekeurige DNA-sequentie met een gewenste lengte genereren:",
"_____no_output_____"
]
],
[
[
"import random\n\ndef dna_sequentie_2(n):\n \"\"\"DNA-sequentie genereren met gewenste lengte.\"\"\"\n letters = \"ACGT\"\n keuze = [random.choice(letters) for i in range(n)] # kies n letters uit string letters en stop die in lijst\n # print(keuze)\n string = \"\".join(keuze) # elementen van keuze samenbrengen in een (samenhangende) string zonder keuze aan te passen\n return string",
"_____no_output_____"
]
],
[
[
"Genereer een DNA-sequentie van lengte 30 met deze tweede functie.",
"_____no_output_____"
],
[
"### Oefening 3.1",
"_____no_output_____"
],
[
"Stel een functie op om te tellen hoeveel keer een bepaalde nucleobase in een gegeven DNA-sequentie voorkomt.",
"_____no_output_____"
],
[
"Test de functie uit: laat tellen hoeveel keer de nucleobase \"T\" voorkomt in de DNA-sequentie \"ATGCGGACCTAT\".",
"_____no_output_____"
],
[
"### Oefening 3.2",
"_____no_output_____"
],
[
"Gebruik beide functies (een uit het voorbeeld en de functie die je zelf maakte) samen in een script om te tellen hoeveel keer elke nucleobase in een willekeurig gegeneerde DNA-sequentie voorkomt. ",
"_____no_output_____"
],
[
"<div class=\"alert alert-block alert-info\">De computer is razendsnel om repetitieve taken te doen, zoals een letter opsporen in een lange string. \n</div>",
"_____no_output_____"
],
[
"<div class=\"alert alert-block alert-warning\">Het neurale netwerk van het project 'KIKS' zal geen letter opsporen, maar wel een huidmondje. Het neurale netwerk doorloopt de volledige foto door er een vierkant venster over te laten glijden en na te gaan of er binnen het vierkantje een huidmondje te zien is.\n</div>",
"_____no_output_____"
],
[
"<img src=\"images/cclic.png\" alt=\"Banner\" align=\"left\" style=\"width:100px;\"/><br><br>\nNotebook KIKS, zie <a href=\"http://www.aiopschool.be\">AI Op School</a>, van F. wyffels & N. Gesquière is in licentie gegeven volgens een <a href=\"http://creativecommons.org/licenses/by-nc-sa/4.0/\">Creative Commons Naamsvermelding-NietCommercieel-GelijkDelen 4.0 Internationaal-licentie</a>. ",
"_____no_output_____"
],
[
"<div>\n <h2>Met steun van</h2> \n</div>",
"_____no_output_____"
],
[
"<img src=\"images/kikssteun.png\" alt=\"Banner\" width=\"800\"/>",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
c513dd6e08f3e6a49e2f9654160fe074bf190d41
| 2,200 |
ipynb
|
Jupyter Notebook
|
binder/index.ipynb
|
fbarresi/studious-funicular
|
ad58189449d70860a1200f2008aed834f491cd53
|
[
"MIT"
] | 1 |
2021-03-18T23:33:35.000Z
|
2021-03-18T23:33:35.000Z
|
binder/index.ipynb
|
fbarresi/studious-funicular
|
ad58189449d70860a1200f2008aed834f491cd53
|
[
"MIT"
] | 17 |
2020-01-28T22:33:27.000Z
|
2021-06-10T21:05:49.000Z
|
binder/index.ipynb
|
fbarresi/studious-funicular
|
ad58189449d70860a1200f2008aed834f491cd53
|
[
"MIT"
] | 1 |
2021-07-17T12:55:22.000Z
|
2021-07-17T12:55:22.000Z
| 29.333333 | 143 | 0.598636 |
[
[
[
"# Welcome to Jupyter!",
"_____no_output_____"
],
[
"This repo contains an introduction to [Jupyter](https://jupyter.org) and [IPython](https://ipython.org).\n\nOutline of some basics:\n\n* [Notebook Basics](../examples/Notebook/Notebook%20Basics.ipynb)\n* [IPython - beyond plain python](../examples/IPython%20Kernel/Beyond%20Plain%20Python.ipynb)\n* [Markdown Cells](../examples/Notebook/Working%20With%20Markdown%20Cells.ipynb)\n* [Rich Display System](../examples/IPython%20Kernel/Rich%20Output.ipynb)\n* [Custom Display logic](../examples/IPython%20Kernel/Custom%20Display%20Logic.ipynb)\n* [Running a Secure Public Notebook Server](../examples/Notebook/Running%20the%20Notebook%20Server.ipynb#Securing-the-notebook-server)\n* [How Jupyter works](../examples/Notebook/Multiple%20Languages%2C%20Frontends.ipynb) to run code in different languages.",
"_____no_output_____"
],
[
"You can also get this tutorial and run it on your laptop:\n\n git clone https://github.com/ipython/ipython-in-depth\n\nInstall IPython and Jupyter:\n\nwith [conda](https://www.anaconda.com/download):\n\n conda install ipython jupyter\n\nwith pip:\n\n # first, always upgrade pip!\n pip install --upgrade pip\n pip install --upgrade ipython jupyter\n\nStart the notebook in the tutorial directory:\n\n cd ipython-in-depth\n jupyter notebook",
"_____no_output_____"
]
]
] |
[
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown"
]
] |
c513f323fc3f62fa0295a04e7585d0c7454ce62e
| 7,430 |
ipynb
|
Jupyter Notebook
|
Kaggle_ML_Tutorials/IntermediateMachineLearning/DLtutorial.ipynb
|
ed-s-code/malwareClassificationChallenge
|
a1f6d2869880980473babe2f52cc33b3c01f048f
|
[
"MIT"
] | null | null | null |
Kaggle_ML_Tutorials/IntermediateMachineLearning/DLtutorial.ipynb
|
ed-s-code/malwareClassificationChallenge
|
a1f6d2869880980473babe2f52cc33b3c01f048f
|
[
"MIT"
] | null | null | null |
Kaggle_ML_Tutorials/IntermediateMachineLearning/DLtutorial.ipynb
|
ed-s-code/malwareClassificationChallenge
|
a1f6d2869880980473babe2f52cc33b3c01f048f
|
[
"MIT"
] | null | null | null | 7,430 | 7,430 | 0.601884 |
[
[
[
"# Code to load the data etc.\n\nimport pandas as pd\n\n# Read the data\n\ncredit_card_file_path = 'data/AER_credit_card_data.csv' # Set file path of the data.\ndata = pd.read_csv(credit_card_file_path, true_values = ['yes'], false_values = ['no']) # Read the data and store in a data frame.\n\n# Select target\ny = data.card\n\n# Select predictors\nX = data.drop(['card'], axis=1)\n\nprint(\"Number of rows in the dataset:\", X.shape[0])\nX.head()\n",
"Number of rows in the dataset: 1319\n"
],
[
"# Use cross-validation to ensure accurate measures of model quality (Small dataset)\n\nfrom sklearn.pipeline import make_pipeline\nfrom sklearn.ensemble import RandomForestClassifier\nfrom sklearn.model_selection import cross_val_score\n\n# Since there is no preprocessing, a piepleine is unnecessary, but it is good practice all the same!\nmy_pipeline = make_pipeline(RandomForestClassifier(n_estimators=100))\ncv_scores = cross_val_score(my_pipeline, X, y, \n cv=5,\n scoring='accuracy')\n\nprint(\"Cross-validation accuracy: %f\" % cv_scores.mean())\n\n# Example output: \"Cross-validation accuracy: 0.980292\"",
"Cross-validation accuracy: 0.980292\n"
],
[
"# Basic data comparisons, in order to try and detect data leakage\n\nexpenditures_cardholders = X.expenditure[y]\nexpenditures_noncardholders = X.expenditure[~y]\n\nprint('Fraction of those who did not receive a card and had no expenditures: %.2f' \\\n %((expenditures_noncardholders == 0).mean()))\nprint('Fraction of those who received a card and had no expenditures: %.2f' \\\n %(( expenditures_cardholders == 0).mean()))",
"Fraction of those who did not receive a card and had no expenditures: 1.00\nFraction of those who received a card and had no expenditures: 0.02\n"
],
[
"# As shown above, everyone who did not receive a card had no expenditures, while only 2% of those who received a card had no expenditures. It's not surprising that our model appeared to have a high accuracy. But this also seems to be a case of target leakage, where expenditures probably means expenditures on the card they applied for.\n\n# Since share is partially determined by expenditure, it should be excluded too. The variables active and majorcards are a little less clear, but from the description, they sound concerning. In most situations, it's better to be safe than sorry if you can't track down the people who created the data to find out more.\n\n# Above text is taken directly from the \"Data Leakage\" tutorial.",
"_____no_output_____"
],
[
"# Model without target leakage:\n\n# Drop leaky predictors from dataset (Established by analysing the data)\npotential_leaks = ['expenditure', 'share', 'active', 'majorcards']\nX2 = X.drop(potential_leaks, axis=1)\n\n# Evaluate the model with leaky predictors removed\ncv_scores = cross_val_score(my_pipeline, X2, y, \n cv=5,\n scoring='accuracy')\n\nprint(\"Cross-val accuracy: %f\" % cv_scores.mean())\n\n# Example output: Cross-val accuracy: 0.833201",
"Cross-val accuracy: 0.833201\n"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code"
]
] |
c513f9bb12e43bc1a2f8cf147404d5301cb29a28
| 576 |
ipynb
|
Jupyter Notebook
|
report/Imitation Learning.ipynb
|
madhawa-gunasekara/IL
|
4ca8cdc031863048d214bc148bc6e560df9984ee
|
[
"Apache-2.0"
] | 1 |
2021-05-22T06:48:05.000Z
|
2021-05-22T06:48:05.000Z
|
report/Imitation Learning.ipynb
|
madhawa-gunasekara/IL
|
4ca8cdc031863048d214bc148bc6e560df9984ee
|
[
"Apache-2.0"
] | null | null | null |
report/Imitation Learning.ipynb
|
madhawa-gunasekara/IL
|
4ca8cdc031863048d214bc148bc6e560df9984ee
|
[
"Apache-2.0"
] | null | null | null | 16.941176 | 34 | 0.524306 |
[] |
[] |
[] |
c514335326402dbf3bdc47897c56364d2429ad02
| 514,251 |
ipynb
|
Jupyter Notebook
|
Camera_Calibration/Camera_Calibration.ipynb
|
ozturkoguzhan/OpenCV-Projects
|
aae73c88ef998d60eb49b87f800a59e507d417e6
|
[
"MIT"
] | null | null | null |
Camera_Calibration/Camera_Calibration.ipynb
|
ozturkoguzhan/OpenCV-Projects
|
aae73c88ef998d60eb49b87f800a59e507d417e6
|
[
"MIT"
] | null | null | null |
Camera_Calibration/Camera_Calibration.ipynb
|
ozturkoguzhan/OpenCV-Projects
|
aae73c88ef998d60eb49b87f800a59e507d417e6
|
[
"MIT"
] | null | null | null | 1,856.501805 | 508,280 | 0.958619 |
[
[
[
"#I have implemented Camera Calibration using OpenCV\n#The source code is inspired from https://github.com/udacity/CarND-Camera-Calibration",
"_____no_output_____"
],
[
"#Import the libraries",
"_____no_output_____"
],
[
"import numpy as np\nimport cv2\nimport glob\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"# Prepare object points, like (0,0,0), (1,0,0), (2,0,0) ....,(6,5,0)",
"_____no_output_____"
],
[
"objp = np.zeros((6*8,3), np.float32)\nobjp[:,:2] = np.mgrid[0:8, 0:6].T.reshape(-1,2)",
"_____no_output_____"
],
[
"# Arrays to store object points and image points from all the images.",
"_____no_output_____"
],
[
"objpoints = [] # 3d points in real world space\nimgpoints = [] # 2d points in image plane.",
"_____no_output_____"
],
[
"# Make a list of calibration images",
"_____no_output_____"
],
[
"images = glob.glob('calibration_wide/GO*.jpg')",
"_____no_output_____"
],
[
"# Step through the list and search for chessboard corners",
"_____no_output_____"
],
[
"for idx, fname in enumerate(images):\n img = cv2.imread(fname)\n gray = cv2.cvtColor(img, cv2.COLOR_BGR2GRAY)\n\n # Find the chessboard corners\n ret, corners = cv2.findChessboardCorners(gray, (8,6), None)\n\n # If found, add object points, image points\n if ret == True:\n objpoints.append(objp)\n imgpoints.append(corners)\n\n # Draw and display the corners\n cv2.drawChessboardCorners(img, (8,6), corners, ret)\n #write_name = 'corners_found'+str(idx)+'.jpg'\n #cv2.imwrite(write_name, img)\n cv2.imshow('img', img)\n cv2.waitKey(100)\n\ncv2.destroyAllWindows()\nfor i in range (1,5):\n cv2.waitKey(1)",
"_____no_output_____"
],
[
"# Test undistortion on an image",
"_____no_output_____"
],
[
"import pickle\nimg = cv2.imread('calibration_wide/test_image.jpg')\nimg_size = (img.shape[1], img.shape[0])",
"_____no_output_____"
],
[
"# Do camera calibration given object points and image points",
"_____no_output_____"
],
[
"ret, mtx, dist, rvecs, tvecs = cv2.calibrateCamera(objpoints, imgpoints, img_size,None,None)",
"_____no_output_____"
],
[
"dst = cv2.undistort(img, mtx, dist, None, mtx)\ncv2.imwrite('calibration_wide/test_undist.jpg',dst)",
"_____no_output_____"
],
[
"# Save the camera calibration result for later use (we won't worry about rvecs / tvecs)",
"_____no_output_____"
],
[
"dist_pickle = {}\ndist_pickle[\"mtx\"] = mtx\ndist_pickle[\"dist\"] = dist\npickle.dump( dist_pickle, open( \"calibration_wide/wide_dist_pickle.p\", \"wb\" ) )",
"_____no_output_____"
],
[
"# Visualize undistortion",
"_____no_output_____"
],
[
"f, (ax1, ax2) = plt.subplots(1, 2, figsize=(20,10))\nax1.imshow(img)\nax1.set_title('Original Image', fontsize=30)\nax2.imshow(dst)\nax2.set_title('Undistorted Image', fontsize=30)",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
c51434f6c58efc0b2aa46086a23066c7b182a223
| 369,362 |
ipynb
|
Jupyter Notebook
|
3_Modelo_Medellin.ipynb
|
sefigueroacUNAL/predictiva-covid
|
6ae601908f89203d2ea7c921ed4e3397491c8144
|
[
"MIT"
] | null | null | null |
3_Modelo_Medellin.ipynb
|
sefigueroacUNAL/predictiva-covid
|
6ae601908f89203d2ea7c921ed4e3397491c8144
|
[
"MIT"
] | null | null | null |
3_Modelo_Medellin.ipynb
|
sefigueroacUNAL/predictiva-covid
|
6ae601908f89203d2ea7c921ed4e3397491c8144
|
[
"MIT"
] | null | null | null | 158.388508 | 48,804 | 0.863714 |
[
[
[
"# Modelo para la Ciudad de Medellín",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"##Sección de código para ejecutar el ejercicio en COLAB sin realizar ningún cambio adicional.\n#from google.colab import drive\n#drive.mount('/content/drive')\n#baseUrl = '/content/drive/Shared drives/Analitica Predictiva/covid-19-flr-analitica-predictiva'\n#os.chdir(baseUrl)",
"_____no_output_____"
]
],
[
[
"Librerias requeridas para realizar los modelos.",
"_____no_output_____"
]
],
[
[
"from sklearn.preprocessing import PolynomialFeatures, StandardScaler\nfrom sklearn.linear_model import Lasso\nfrom sklearn.pipeline import make_pipeline\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.metrics import mean_squared_error, mean_absolute_error",
"_____no_output_____"
]
],
[
[
"## Modelo Corto Plazo",
"_____no_output_____"
]
],
[
[
"ciudades = ['Bogotá D.C.','Medellín','Cali','Barranquilla', 'Cartagena de Indias']\nnames = ['Bogota','Medellin','Cali','Barranquilla','Cartagena']\nid_city = 1\nname = names[id_city]\nurlDataSet = 'Datos/data_{}.pickle'.format(name)",
"_____no_output_____"
]
],
[
[
"Se carga el dataset generado en la sección de preprocesamiento, se muestran los últimos 20 registros, para validar visualmente los ultimos días de información.",
"_____no_output_____"
]
],
[
[
"df_city = pd.read_pickle(urlDataSet)\ndf = df_city.copy()\ndf.tail(20)",
"_____no_output_____"
]
],
[
[
"### Activos",
"_____no_output_____"
],
[
"Para la predicción de casos activos se usó una Regressión Lasso con caracteristicas Polinomiales de grado 5; previamente se estandarizaron los datos, y se entrenaron con un conjunto del 70% de los datos; estos datos se toman aleatoriamente, pero al graficarlos se orden en secuencia.",
"_____no_output_____"
]
],
[
[
"totalDays = len(df['dias'].values)\nX = df['dias'].values[0:totalDays-5].reshape(-1,1) #Variable independiente.\ny = df['activos'].values[0:totalDays-5].reshape(-1,1) #Variable dependiente.\n#Datos de validación - nunca entran al modelo.\nX_v = df['dias'].values[totalDays-5:].reshape(-1,1)\ny_v = df['activos'].values[totalDays-5:].reshape(-1,1)\n#Se obtiene el 70% para entrenamiento y 30% para pruebas.\nX_train, X_test, y_train, y_test = train_test_split(X,y,test_size=0.3,random_state=123,shuffle=True)\n#Se crea un pipeline con el escalado, las caracteristicas polinomiales y la regresión lasso\npipe = make_pipeline(StandardScaler(),PolynomialFeatures(degree=5),Lasso(random_state=123))\n#Se realiza el entrenamiento.\npipe.fit(X_train,y_train)",
"_____no_output_____"
]
],
[
[
"Se realizan las predicciones sobre el dataset de pruebas, adicionalmente se establece el pico en el día 175, por lo cual para fines de prueba se realizan predicciones desde dicho día, hasta el día 200 de la pandemia, para probar la predicción futura de los modelos a corto plazo.",
"_____no_output_____"
]
],
[
[
"#Obtener predicciones para los datos de prueba.\ny_pred = pipe.predict(X_test)\n#Predicciones futuras.\nx_pred_future = np.arange(totalDays-10,totalDays,step=1)\ny_pred_future = pipe.predict(x_pred_future.reshape(-1,1))",
"_____no_output_____"
],
[
"def GetMax(*args):\n arr = np.array([[0]])\n for arg in args:\n s = [x for x in arg.shape]\n l = np.prod(s)\n a = np.array(arg.reshape(1,l))\n arr = np.concatenate((a,arr),axis=1)\n return arr.max()",
"_____no_output_____"
],
[
"def GetPeak(*args):\n x_arr = np.array([[0]])\n y_arr = np.array([[0]])\n for x,y in args:\n s = [x for x in x.shape]\n l = np.prod(s)\n xr = np.array(x.reshape(1,l))\n yr = np.array(y.reshape(1,l))\n x_arr = np.concatenate((x_arr,xr),axis=1)\n y_arr = np.concatenate((y_arr,yr),axis=1)\n print(x_arr)\n print(y_arr)\n print(y_arr.max())\n return x_arr[0][y_arr.argmax()]",
"_____no_output_____"
],
[
"Ymax = GetMax(y_train,y_test,y_pred,y_pred_future)\nXpeak = GetPeak((X_train,y_train))",
"[[ 0 20 157 28 13 104 24 133 93 117 116 85 60 7 30 163 147 29\n 35 0 169 45 173 40 59 136 46 5 16 155 65 74 61 180 38 27\n 54 121 154 167 132 87 9 6 146 149 107 134 181 143 89 21 11 67\n 3 86 122 91 177 48 168 44 51 1 81 18 12 162 56 156 50 25\n 161 101 43 129 103 14 115 184 70 15 22 58 75 64 69 178 131 76\n 34 94 118 97 102 92 124 99 105 135 172 159 182 39 84 2 55 49\n 68 175 164 78 179 153 111 32 73 47 183 113 96 57 123 106 83 17\n 98 66 126 109]]\n[[0.0000e+00 9.1000e+01 2.7017e+04 1.2900e+02 2.5000e+01 6.5100e+02\n 1.1200e+02 1.1611e+04 3.3600e+02 2.2550e+03 2.0330e+03 2.3800e+02\n 1.4600e+02 4.0000e+00 1.2100e+02 2.6202e+04 2.3756e+04 1.2300e+02\n 1.2200e+02 1.0000e+00 2.4527e+04 1.5500e+02 2.1653e+04 1.2900e+02\n 1.4700e+02 1.4537e+04 1.6100e+02 3.0000e+00 5.2000e+01 2.7068e+04\n 1.4700e+02 1.4800e+02 1.4400e+02 1.7481e+04 1.2400e+02 1.2700e+02\n 1.5500e+02 3.2480e+03 2.6937e+04 2.6096e+04 1.0689e+04 2.4200e+02\n 8.0000e+00 4.0000e+00 2.2900e+04 2.4964e+04 8.3300e+02 1.2441e+04\n 1.7016e+04 1.9636e+04 2.6700e+02 9.2000e+01 1.7000e+01 1.3700e+02\n 2.0000e+00 2.4300e+02 3.6870e+03 2.9600e+02 1.9547e+04 1.6400e+02\n 2.5241e+04 1.4600e+02 1.5900e+02 1.0000e+00 2.0900e+02 7.2000e+01\n 1.8000e+01 2.6250e+04 1.5600e+02 2.7008e+04 1.5800e+02 1.1500e+02\n 2.6991e+04 5.5600e+02 1.4700e+02 7.9070e+03 6.1900e+02 3.7000e+01\n 1.8420e+03 1.4785e+04 1.2800e+02 4.0000e+01 1.0200e+02 1.4800e+02\n 1.5500e+02 1.4700e+02 1.3300e+02 1.8702e+04 9.7140e+03 1.6400e+02\n 1.2300e+02 3.5000e+02 2.5030e+03 4.5100e+02 5.8800e+02 2.9400e+02\n 4.6550e+03 4.9100e+02 6.9200e+02 1.3433e+04 2.2343e+04 2.7087e+04\n 1.6306e+04 1.3000e+02 2.4000e+02 2.0000e+00 1.5400e+02 1.6400e+02\n 1.4100e+02 2.0848e+04 2.6423e+04 1.7500e+02 1.8025e+04 2.6550e+04\n 1.3020e+03 1.1800e+02 1.4500e+02 1.6500e+02 1.5495e+04 1.5560e+03\n 4.3900e+02 1.5100e+02 4.2370e+03 7.5500e+02 2.3200e+02 5.9000e+01\n 4.7500e+02 1.3900e+02 5.8380e+03 1.1120e+03]]\n27087.0\n"
]
],
[
[
"Se grafican los datos.",
"_____no_output_____"
]
],
[
[
"f = plt.figure(figsize=(18,8))\nplt.title('Predicción de casos activos')\nplt.grid()\nplt.plot(X_train[X_train.flatten().argsort(),0],y_train[X_train.flatten().argsort(),0],'ob',markersize=3,label='Train')\nplt.plot(X_test[X_test.flatten().argsort(),0],y_test[X_test.flatten().argsort(),0],'or',markersize=2,label='Test')\nplt.plot(X_v,y_v,'o',color='gray',markersize=2,label='Validation')\nplt.plot(X_test[X_test.flatten().argsort(),0],y_pred[X_test.flatten().argsort()],'--g',label='Predict')\nplt.plot(x_pred_future,y_pred_future,'--',color='orange',label='Future Predict')\nplt.plot([Xpeak,Xpeak],[-5000,Ymax],'--',color='lightseagreen', label='Peak') #Peak of cases.\nplt.ylim(-5000,Ymax)\nplt.legend(loc='upper left');\nplt.savefig('docs/images/pcp_{}_activos'.format(name))",
"_____no_output_____"
],
[
"\n",
"_____no_output_____"
],
[
"s = [y_pred_future.shape]\nl = np.prod(s)\n",
"_____no_output_____"
]
],
[
[
"Se calculan las métricas de la regresión, en este caso se calculan las metricas completas sobre el conjunto de prueba, adicionalmente se computan las metricas solo desde el pico en adelante como indicar de las predicciones futuras.",
"_____no_output_____"
]
],
[
[
"y_pred_val = pipe.predict(X_v)\nprint(' ---- Métricas ----')\nprint('Total RMSE:\\t\\t{:.0f}'.format(np.sqrt(mean_squared_error(y_test, y_pred))))\nprint('Total MAE:\\t\\t{:.0f}'.format(mean_absolute_error(y_test,y_pred)))\nprint('Future (5 days) RMSE:\\t{:.0f}'.format(np.sqrt(mean_squared_error(y_v, y_pred_val))))\nprint('Future (5 days) MAE:\\t{:.0f}'.format(mean_absolute_error(y_v,y_pred_val)))",
" ---- Métricas ----\nTotal RMSE:\t\t1507\nTotal MAE:\t\t1242\nFuture (5 days) RMSE:\t6409\nFuture (5 days) MAE:\t6146\n"
]
],
[
[
"### Recuperados\nPara la predicción de casos recuperados se usó una Regressión Lasso con caracteristicas Polinomiales de grado 4; previamente se estandarizaron los datos, y se entrenaron con un conjunto del 70% de los datos; estos datos se toman aleatoriamente, pero al graficarlos se orden en secuencia.",
"_____no_output_____"
]
],
[
[
"totalDays = len(df['dias'].values)\nX = df['dias'].values[0:totalDays-5].reshape(-1,1) #Variable independiente.\ny = df['acumulado_recuperados'].values[0:totalDays-5].reshape(-1,1) #Variable dependiente.\n#Datos de validación - nunca entran al modelo.\nX_v = df['dias'].values[totalDays-5:].reshape(-1,1)\ny_v = df['acumulado_recuperados'].values[totalDays-5:].reshape(-1,1)\n#Se obtiene el 70% para entrenamiento y 30% para pruebas.\nX_train, X_test, y_train, y_test = train_test_split(X,y,test_size=0.3,random_state=123,shuffle=True)\n#Se crea un pipeline con el escalado, las caracteristicas polinomiales y la regresión lasso\npipe = make_pipeline(StandardScaler(),PolynomialFeatures(degree=4),Lasso(random_state=123))\n#Se realiza el entrenamiento.\npipe.fit(X_train,y_train)",
"_____no_output_____"
]
],
[
[
"Se realizan las predicciones sobre el dataset de pruebas, adicionalmente se establece el pico en el día 175, por lo cual para fines de prueba se realizan predicciones desde dicho día, hasta el día 200 de la pandemia, para probar la predicción futura de los modelos a corto plazo.",
"_____no_output_____"
]
],
[
[
"#Obtener predicciones para los datos de prueba.\ny_pred = pipe.predict(X_test)\n#Predicciones futuras.\nx_pred_future = np.arange(totalDays-10,totalDays,step=1)\ny_pred_future = pipe.predict(x_pred_future.reshape(-1,1))",
"_____no_output_____"
],
[
"Ymax = GetMax(y_train,y_test,y_pred,y_pred_future)",
"_____no_output_____"
]
],
[
[
"Se grafican los datos.",
"_____no_output_____"
]
],
[
[
"f = plt.figure(figsize=(18,8))\nplt.title('Predicción de recuperados')\nplt.grid()\nplt.plot(X_train[X_train.flatten().argsort(),0],y_train[X_train.flatten().argsort(),0],'ob',markersize=3,label='Train')\nplt.plot(X_test[X_test.flatten().argsort(),0],y_test[X_test.flatten().argsort(),0],'or',markersize=2,label='Test')\nplt.plot(X_v,y_v,'o',color='gray',markersize=2,label='Validation')\nplt.plot(X_test[X_test.flatten().argsort(),0],y_pred[X_test.flatten().argsort()],'--g',label='Predict')\nplt.plot(x_pred_future,y_pred_future,'--',color='orange',label='Future Predict')\nplt.plot([Xpeak,Xpeak],[-5000,Ymax],'--',color='lightseagreen', label='Peak') #Peak of cases.\nplt.ylim(-5000,Ymax)\nplt.legend(loc='upper left');\nplt.savefig('docs/images/pcp_{}_recuperados'.format(name))",
"_____no_output_____"
]
],
[
[
"Se calculan las métricas de la regresión, en este caso se calculan las metricas completas sobre el conjunto de prueba, adicionalmente se computan las metricas solo desde el pico en adelante como indicar de las predicciones futuras.",
"_____no_output_____"
]
],
[
[
"y_pred_val = pipe.predict(X_v)\nprint(' ---- Métricas ----')\nprint('Total RMSE:\\t\\t{:.0f}'.format(np.sqrt(mean_squared_error(y_test, y_pred))))\nprint('Total MAE:\\t\\t{:.0f}'.format(mean_absolute_error(y_test,y_pred)))\nprint('Future (5 days) RMSE:\\t{:.0f}'.format(np.sqrt(mean_squared_error(y_v, y_pred_val))))\nprint('Future (5 days) MAE:\\t{:.0f}'.format(mean_absolute_error(y_v,y_pred_val)))\n",
" ---- Métricas ----\nTotal RMSE:\t\t932\nTotal MAE:\t\t763\nFuture (5 days) RMSE:\t3927\nFuture (5 days) MAE:\t3782\n"
]
],
[
[
"### Muertes\nPara la predicción de las muertes se usó una Regressión Lasso con caracteristicas Polinomiales de grado 6; previamente se estandarizaron los datos, y se entrenaron con un conjunto del 70% de los datos; estos datos se toman aleatoriamente, pero al graficarlos se orden en secuencia.",
"_____no_output_____"
]
],
[
[
"totalDays = len(df['dias'].values)\nX = df['dias'].values[0:totalDays-5].reshape(-1,1) #Variable independiente.\ny = df['acumulado_muertos'].values[0:totalDays-5].reshape(-1,1) #Variable dependiente.\n#Datos de validación - nunca entran al modelo.\nX_v = df['dias'].values[totalDays-5:].reshape(-1,1)\ny_v = df['acumulado_muertos'].values[totalDays-5:].reshape(-1,1)\n\nX_train, X_test, y_train, y_test = train_test_split(X,y,test_size=0.3,random_state=123,shuffle=True)\npipe = make_pipeline(StandardScaler(),PolynomialFeatures(degree=6),Lasso(random_state=123,max_iter=10000))\npipe.fit(X_train,y_train)",
"_____no_output_____"
],
[
"y_pred = pipe.predict(X_test)\nx_pred_future = np.arange(totalDays-10,totalDays,step=1)\ny_pred_future = pipe.predict(x_pred_future.reshape(-1,1))\nYmax = GetMax(y_train,y_test,y_pred,y_pred_future)",
"_____no_output_____"
],
[
"#Graph ordered data.\nf = plt.figure(figsize=(18,8))\nplt.title('Predicción de muertes')\nplt.grid()\nplt.plot(X_train[X_train.flatten().argsort(),0],y_train[X_train.flatten().argsort(),0],'ob',markersize=3,label='Train')\nplt.plot(X_test[X_test.flatten().argsort(),0],y_test[X_test.flatten().argsort(),0],'or',markersize=2,label='Test')\nplt.plot(X_v,y_v,'o',color='gray',markersize=2,label='Validation')\nplt.plot(X_test[X_test.flatten().argsort(),0],y_pred[X_test.flatten().argsort()],'--g',label='Predict')\nplt.plot(x_pred_future,y_pred_future,'--',color='orange',label='Future Predict')\nplt.plot([Xpeak,Xpeak],[-10,Ymax],'--',color='lightseagreen', label='Peak') #Peak of cases.\nplt.ylim(-10,Ymax)\nplt.legend(loc='upper left');\nplt.savefig('docs/images/pcp_{}_muertes'.format(name))",
"_____no_output_____"
],
[
"y_pred_val = pipe.predict(X_v)\nprint(' ---- Métricas ----')\nprint('Total RMSE:\\t\\t{:.0f}'.format(np.sqrt(mean_squared_error(y_test, y_pred))))\nprint('Total MAE:\\t\\t{:.0f}'.format(mean_absolute_error(y_test,y_pred)))\nprint('Future (5 days) RMSE:\\t{:.0f}'.format(np.sqrt(mean_squared_error(y_v, y_pred_val))))\nprint('Future (5 days) MAE:\\t{:.0f}'.format(mean_absolute_error(y_v,y_pred_val)))",
" ---- Métricas ----\nTotal RMSE:\t\t16\nTotal MAE:\t\t12\nFuture (5 days) RMSE:\t52\nFuture (5 days) MAE:\t50\n"
]
],
[
[
"### Infectados\nPara la predicción de los infectados se usó una Regressión Lasso con caracteristicas Polinomiales de grado 4; previamente se estandarizaron los datos, y se entrenaron con un conjunto del 70% de los datos; estos datos se toman aleatoriamente, pero al graficarlos se orden en secuencia.",
"_____no_output_____"
]
],
[
[
"totalDays = len(df['dias'].values)\nX = df['dias'].values[0:totalDays-5].reshape(-1,1) #Variable independiente.\ny = df['acumulado_infectados'].values[0:totalDays-5].reshape(-1,1) #Variable dependiente.\n#Datos de validación - nunca entran al modelo.\nX_v = df['dias'].values[totalDays-5:].reshape(-1,1)\ny_v = df['acumulado_infectados'].values[totalDays-5:].reshape(-1,1)\n\nX_train, X_test, y_train, y_test = train_test_split(X,y,test_size=0.3,random_state=123,shuffle=True)\npipe = make_pipeline(StandardScaler(),PolynomialFeatures(degree=5),Lasso(random_state=123))\npipe.fit(X_train,y_train)",
"_____no_output_____"
],
[
"y_pred = pipe.predict(X_test)\nx_pred_future = np.arange(totalDays-10,totalDays,step=1)\ny_pred_future = pipe.predict(x_pred_future.reshape(-1,1))\nYmax = GetMax(y_train,y_test,y_pred,y_pred_future)",
"_____no_output_____"
],
[
"#Graph ordered data.\nf = plt.figure(figsize=(18,8))\nplt.title('Predicción de infectados')\nplt.grid()\nplt.plot(X_train[X_train.flatten().argsort(),0],y_train[X_train.flatten().argsort(),0],'ob',markersize=3,label='Train')\nplt.plot(X_test[X_test.flatten().argsort(),0],y_test[X_test.flatten().argsort(),0],'or',markersize=2,label='Test')\nplt.plot(X_v,y_v,'o',color='gray',markersize=2,label='Validation')\nplt.plot(X_test[X_test.flatten().argsort(),0],y_pred[X_test.flatten().argsort()],'--g',label='Predict')\nplt.plot(x_pred_future,y_pred_future,'--',color='orange',label='Future Predict')\nplt.plot([Xpeak,Xpeak],[-10000,Ymax],'--',color='lightseagreen', label='Peak') #Peak of cases.\nplt.ylim(-10000,Ymax)\nplt.legend(loc='upper left');\nplt.savefig('docs/images/pcp_{}_infectados'.format(name))",
"_____no_output_____"
],
[
"y_pred_val = pipe.predict(X_v)\nprint(' ---- Métricas ----')\nprint('Total RMSE:\\t\\t{:.0f}'.format(np.sqrt(mean_squared_error(y_test, y_pred))))\nprint('Total MAE:\\t\\t{:.0f}'.format(mean_absolute_error(y_test,y_pred)))\nprint('Future (5 days) RMSE:\\t{:.0f}'.format(np.sqrt(mean_squared_error(y_v, y_pred_val))))\nprint('Future (5 days) MAE:\\t{:.0f}'.format(mean_absolute_error(y_v,y_pred_val)))",
" ---- Métricas ----\nTotal RMSE:\t\t994\nTotal MAE:\t\t783\nFuture (5 days) RMSE:\t515\nFuture (5 days) MAE:\t402\n"
]
],
[
[
"## Modelo Mediano Plazo\nPara los modelos de mediano plazo, se sigue la misma dinámica que en corto plazo, pero se trabaja con un conjunto de datos que se preprocesó para que tenga los datos acumulados semanalmente, esto permite obtener las predicciones de Infectados, recuperados y muertos por semana.",
"_____no_output_____"
]
],
[
[
"urlDataset_w = 'Datos/data_weekly_{}.pickle'.format(name)\ndf_w = pd.read_pickle(urlDataset_w)\ndf_w.tail(20)",
"_____no_output_____"
]
],
[
[
"### Activos",
"_____no_output_____"
]
],
[
[
"totalW = len(df_w.index.values)\nX = df_w.index.values[0:totalW-3].reshape(-1,1) #Variable independiente.\ny = df_w['activos'].values[0:totalW-3].reshape(-1,1) #Variable dependiente.\n#Datos de validación - nunca entran al modelo.\nX_v = df_w.index.values[totalW-3:].reshape(-1,1)\ny_v = df_w['activos'].values[totalW-3:].reshape(-1,1)\n\nX_train, X_test, y_train, y_test = train_test_split(X,y,test_size=0.3,random_state=123,shuffle=True)\npipe = make_pipeline(StandardScaler(),PolynomialFeatures(degree=5),Lasso(max_iter=10000,random_state=123))\npipe.fit(X_train,y_train)",
"_____no_output_____"
]
],
[
[
"El pico de la pandemia se grafica en la semana 22.",
"_____no_output_____"
]
],
[
[
"y_pred = pipe.predict(X_test)\nx_pred_future = np.arange(0,totalW,step=1)\ny_pred_future = pipe.predict(x_pred_future.reshape(-1,1))\nYmax = GetMax(y_train,y_test,y_pred,y_pred_future)\nXpeak = GetPeak((X_train,y_train))\nXpeak",
"[[ 0 4 3 24 12 16 9 14 20 0 1 10 19 17 6 23 2 13]]\n[[0.0000e+00 1.2300e+02 1.2700e+02 2.1304e+04 2.8100e+02 2.5030e+03\n 1.3300e+02 6.5100e+02 2.2900e+04 4.0000e+00 2.5000e+01 1.6400e+02\n 1.7100e+04 5.2290e+03 1.6400e+02 2.6096e+04 9.1000e+01 4.5100e+02]]\n26096.0\n"
],
[
"f = plt.figure(figsize=(18,8))\nplt.title('Predicción Semanal de casos activos')\nplt.grid()\nplt.plot(X_train[X_train.flatten().argsort(),0],y_train[X_train.flatten().argsort(),0],'ob',markersize=3,label='Train')\nplt.plot(X_test[X_test.flatten().argsort(),0],y_test[X_test.flatten().argsort(),0],'or',markersize=2,label='Test')\nplt.plot(X_v,y_v,'o',color='gray',markersize=2,label='Validation')\nplt.plot(x_pred_future[0:totalW-2],y_pred_future[0:totalW-2],'--g',label='Predict')\nplt.plot(x_pred_future[totalW-3:],y_pred_future[totalW-3:],'--',color='orange',label='Future Predict')\nplt.plot([Xpeak,Xpeak],[-5000,Ymax],'--',color='lightseagreen', label='Peak') #Peak of cases.\nplt.ylim(-5000,Ymax)\nplt.legend(loc='upper left');\nplt.savefig('docs/images/pmp_{}_activos'.format(name))",
"_____no_output_____"
],
[
"y_pred_val = pipe.predict(X_v)\nprint(' ---- Métricas ----')\nprint('Total RMSE:\\t\\t{:.0f}'.format(np.sqrt(mean_squared_error(y_test, y_pred))))\nprint('Total MAE:\\t\\t{:.0f}'.format(mean_absolute_error(y_test,y_pred)))\nprint('Future (3 weeks) RMSE:\\t{:.0f}'.format(np.sqrt(mean_squared_error(y_v, y_pred_val))))\nprint('Future (3 weeks) MAE:\\t{:.0f}'.format(mean_absolute_error(y_v,y_pred_val)))",
" ---- Métricas ----\nTotal RMSE:\t\t1181\nTotal MAE:\t\t1048\nFuture (3 weeks) RMSE:\t28750\nFuture (3 weeks) MAE:\t23424\n"
]
],
[
[
"### Recuperados",
"_____no_output_____"
]
],
[
[
"totalW = len(df_w.index.values)\nX = df_w.index.values[0:totalW-3].reshape(-1,1) #Variable independiente.\ny = df_w['acumulado_recuperados'].values[0:totalW-3].reshape(-1,1) #Variable dependiente.\n#Datos de validación - nunca entran al modelo.\nX_v = df_w.index.values[totalW-3:].reshape(-1,1)\ny_v = df_w['acumulado_recuperados'].values[totalW-3:].reshape(-1,1)\n\nX_train, X_test, y_train, y_test = train_test_split(X,y,test_size=0.3,random_state=123,shuffle=True)\npipe = make_pipeline(StandardScaler(),PolynomialFeatures(degree=5),Lasso(random_state=123,max_iter=10000))\npipe.fit(X_train,y_train)",
"_____no_output_____"
],
[
"y_pred = pipe.predict(X_test)\nx_pred_future = np.arange(0,totalW,step=1)\ny_pred_future = pipe.predict(x_pred_future.reshape(-1,1))\nYmax = GetMax(y_train,y_test,y_pred,y_pred_future)",
"_____no_output_____"
],
[
"#Graph ordered data.\nf = plt.figure(figsize=(18,8))\nplt.title('Predicción Semanal de recuperados')\nplt.grid()\nplt.plot(X_train[X_train.flatten().argsort(),0],y_train[X_train.flatten().argsort(),0],'ob',markersize=3,label='Train')\nplt.plot(X_test[X_test.flatten().argsort(),0],y_test[X_test.flatten().argsort(),0],'or',markersize=2,label='Test')\nplt.plot(X_v,y_v,'o',color='gray',markersize=2,label='Validation')\nplt.plot(x_pred_future[0:totalW-2],y_pred_future[0:totalW-2],'--g',label='Predict')\nplt.plot(x_pred_future[totalW-3:],y_pred_future[totalW-3:],'--',color='orange',label='Future Predict')\nplt.plot([Xpeak,Xpeak],[-5000,Ymax],'--',color='lightseagreen', label='Peak') #Peak of cases.\nplt.ylim(-5000,Ymax)\nplt.legend(loc='upper left');\nplt.savefig('docs/images/pmp_{}_recuperados'.format(name))",
"_____no_output_____"
],
[
"y_pred_val = pipe.predict(X_v)\nprint(' ---- Métricas ----')\nprint('Total RMSE:\\t\\t{:.0f}'.format(np.sqrt(mean_squared_error(y_test, y_pred))))\nprint('Total MAE:\\t\\t{:.0f}'.format(mean_absolute_error(y_test,y_pred)))\nprint('Future (3 weeks) RMSE:\\t{:.0f}'.format(np.sqrt(mean_squared_error(y_v, y_pred_val))))\nprint('Future (3 weeks) MAE:\\t{:.0f}'.format(mean_absolute_error(y_v,y_pred_val)))",
" ---- Métricas ----\nTotal RMSE:\t\t368\nTotal MAE:\t\t321\nFuture (3 weeks) RMSE:\t24555\nFuture (3 weeks) MAE:\t20548\n"
]
],
[
[
"### Muertos",
"_____no_output_____"
]
],
[
[
"totalW = len(df_w.index.values)\nX = df_w.index.values[0:totalW-3].reshape(-1,1) #Variable independiente.\ny = df_w['acumulado_muertos'].values[0:totalW-3].reshape(-1,1) #Variable dependiente.\n#Datos de validación - nunca entran al modelo.\nX_v = df_w.index.values[totalW-3:].reshape(-1,1)\ny_v = df_w['acumulado_muertos'].values[totalW-3:].reshape(-1,1)\n\nX_train, X_test, y_train, y_test = train_test_split(X,y,test_size=0.3,random_state=123,shuffle=True)\npipe = make_pipeline(StandardScaler(),PolynomialFeatures(degree=7),Lasso(random_state=123,max_iter=10000))\npipe.fit(X_train,y_train)",
"_____no_output_____"
],
[
"y_pred = pipe.predict(X_test)\nx_pred_future = np.arange(0,totalW,step=1)\ny_pred_future = pipe.predict(x_pred_future.reshape(-1,1))\nYmax = GetMax(y_train,y_test,y_pred,y_pred_future)",
"_____no_output_____"
],
[
"#Graph ordered data.\nf = plt.figure(figsize=(18,8))\nplt.title('Predicción Semanal de Muertes')\nplt.grid()\nplt.plot(X_train[X_train.flatten().argsort(),0],y_train[X_train.flatten().argsort(),0],'ob',markersize=3,label='Train')\nplt.plot(X_test[X_test.flatten().argsort(),0],y_test[X_test.flatten().argsort(),0],'or',markersize=2,label='Test')\nplt.plot(X_v,y_v,'o',color='gray',markersize=2,label='Validation')\nplt.plot(x_pred_future[0:totalW-2],y_pred_future[0:totalW-2],'--g',label='Predict')\nplt.plot(x_pred_future[totalW-3:],y_pred_future[totalW-3:],'--',color='orange',label='Future Predict')\nplt.plot([Xpeak,Xpeak],[-5000,Ymax],'--',color='lightseagreen', label='Peak') #Peak of cases.\nplt.ylim(-2000,Ymax)\nplt.legend(loc='upper left');\nplt.savefig('docs/images/pmp_{}_muertes'.format(name))",
"_____no_output_____"
],
[
"y_pred_val = pipe.predict(X_v)\nprint(' ---- Métricas ----')\nprint('Total RMSE:\\t\\t{:.0f}'.format(np.sqrt(mean_squared_error(y_test, y_pred))))\nprint('Total MAE:\\t\\t{:.0f}'.format(mean_absolute_error(y_test,y_pred)))\nprint('Future (3 weeks) RMSE:\\t{:.0f}'.format(np.sqrt(mean_squared_error(y_v, y_pred_val))))\nprint('Future (3 weeks) MAE:\\t{:.0f}'.format(mean_absolute_error(y_v,y_pred_val)))",
" ---- Métricas ----\nTotal RMSE:\t\t17\nTotal MAE:\t\t12\nFuture (3 weeks) RMSE:\t44\nFuture (3 weeks) MAE:\t44\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
c51446eed9ed560d48e32e625535c964637a7264
| 115,680 |
ipynb
|
Jupyter Notebook
|
my_autosleep_analysis.ipynb
|
edgarbc/my_autosleep_analysis
|
f27920e5d55fe21a68954aeb52d4bccdd0b68aaa
|
[
"MIT"
] | null | null | null |
my_autosleep_analysis.ipynb
|
edgarbc/my_autosleep_analysis
|
f27920e5d55fe21a68954aeb52d4bccdd0b68aaa
|
[
"MIT"
] | null | null | null |
my_autosleep_analysis.ipynb
|
edgarbc/my_autosleep_analysis
|
f27920e5d55fe21a68954aeb52d4bccdd0b68aaa
|
[
"MIT"
] | null | null | null | 194.09396 | 38,690 | 0.859258 |
[
[
[
"<a href=\"https://colab.research.google.com/github/edgarbc/my_autosleep_analysis/blob/main/my_autosleep_analysis.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport matplotlib.pyplot as plt\n",
"_____no_output_____"
],
[
"# get data from my google drive\nfrom google.colab import drive\ndrive.mount('/content/drive')\n!pwd",
"Mounted at /content/drive\n/content\n"
],
[
"!ls",
"drive sample_data\n"
],
[
"datadir = '/content/drive/My Drive/Colab Notebooks/autosleep_data/'\nfname = 'AutoSleep-20201118-to-20201215.csv'\nprint([datadir + fname])\ndf_data = pd.read_csv('/content/drive/My Drive/Colab Notebooks/autosleep_data/AutoSleep-20201118-to-20201215.csv') \n",
"['/content/drive/My Drive/Colab Notebooks/autosleep_data/AutoSleep-20201118-to-20201215.csv']\n"
],
[
"df_data.head()\n",
"_____no_output_____"
],
[
"print(df_data['fromDate'])\nfig, ax = plt.subplots()\nplt.plot_date(df_data['fromDate'],df_data['efficiency'])\nax.xaxis.set_tick_params(rotation=30, labelsize=10)\nplt.show()\n",
"0 Tuesday, Nov 17, 2020\n1 Wednesday, Nov 18, 2020\n2 Thursday, Nov 19, 2020\n3 Friday, Nov 20, 2020\n4 Saturday, Nov 21, 2020\n5 Sunday, Nov 22, 2020\n6 Monday, Nov 23, 2020\n7 Tuesday, Nov 24, 2020\n8 Wednesday, Nov 25, 2020\n9 Thursday, Nov 26, 2020\n10 Friday, Nov 27, 2020\n11 Saturday, Nov 28, 2020\n12 Monday, Nov 30, 2020\n13 Tuesday, Dec 1, 2020\n14 Wednesday, Dec 2, 2020\n15 Thursday, Dec 3, 2020\n16 Friday, Dec 4, 2020\n17 Saturday, Dec 5, 2020\n18 Sunday, Dec 6, 2020\n19 Monday, Dec 7, 2020\n20 Tuesday, Dec 8, 2020\n21 Thursday, Dec 10, 2020\n22 Friday, Dec 11, 2020\n23 Sunday, Dec 13, 2020\n24 Monday, Dec 14, 2020\nName: fromDate, dtype: object\n"
],
[
"# plot the amount of deep sleep across days\n\nnum_days = df_data['deep'].size\n\ndf_deep = pd.DataFrame()\nfor d in range(num_days):\n df_deep = df_deep.append({'mins':get_mins(df_data['deep'][d])}, ignore_index='True')\n \n# plot the deep sleep \nfig, ax = plt.subplots()\nplt.plot_date(df_data['fromDate'],df_deep['mins'])\nax.xaxis.set_tick_params(rotation=30, labelsize=10)\nplt.show()\n\n",
"_____no_output_____"
],
[
"def get_mins(time_str):\n h, m, s = time_str.split(':')\n return (int(h) * 3600 + int(m) * 60 + int(s))/60.0",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt\nfrom matplotlib.dates import (YEARLY, DateFormatter,\n rrulewrapper, RRuleLocator, drange)\nimport numpy as np\nimport datetime\n\n# Fixing random state for reproducibility\nnp.random.seed(19680801)\n\n\n# tick every 5th easter\nrule = rrulewrapper(YEARLY, byeaster=1, interval=5)\nloc = RRuleLocator(rule)\nformatter = DateFormatter('%m/%d/%y')\ndate1 = datetime.date(1952, 1, 1)\ndate2 = datetime.date(2004, 4, 12)\ndelta = datetime.timedelta(days=100)\n\ndates = drange(date1, date2, delta)\ns = np.random.rand(len(dates)) # make up some random y values\n\n\nfig, ax = plt.subplots()\nplt.plot_date(dates, s)\nax.xaxis.set_major_locator(loc)\nax.xaxis.set_major_formatter(formatter)\nax.xaxis.set_tick_params(rotation=30, labelsize=10)\n\nplt.show()",
"_____no_output_____"
],
[
"# Display data by week\n\n# take data starting by sunday\n\n# ",
"_____no_output_____"
],
[
"# Function to get data arranged by day\n\ndef get_weeks_of_mont(start_day, month, year):\n # \n\n\n# example\nimport calendar\nimport numpy as np\ncalendar.setfirstweekday(6)\n\ndef get_week_of_month(year, month, day):\n x = np.array(calendar.monthcalendar(year, month))\n week_of_month = np.where(x==day)[0][0] + 1\n return(week_of_month)\n\nget_week_of_month(2015,9,14)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
c5145052be6dc6eab4a17e642a8baa68923c2a08
| 16,016 |
ipynb
|
Jupyter Notebook
|
netpyne/tutorials/saving_loading_tut/saving_tut.ipynb
|
sanjayankur31/netpyne
|
d8b7e94cabeb27e23e30853ff17ae86518b35ac2
|
[
"MIT"
] | null | null | null |
netpyne/tutorials/saving_loading_tut/saving_tut.ipynb
|
sanjayankur31/netpyne
|
d8b7e94cabeb27e23e30853ff17ae86518b35ac2
|
[
"MIT"
] | null | null | null |
netpyne/tutorials/saving_loading_tut/saving_tut.ipynb
|
sanjayankur31/netpyne
|
d8b7e94cabeb27e23e30853ff17ae86518b35ac2
|
[
"MIT"
] | null | null | null | 33.366667 | 272 | 0.550637 |
[
[
[
"# Saving and Loading Tutorial",
"_____no_output_____"
],
[
"## Preparing a virtual environment\n\nFirst, you need to have `Python3` and `openmpi` installed and running on your machine.\n\nIn a new directory, here are the steps I took to create a virtual environment for this Jupyter notebook:\n\n\techo \"\" \n\techo \"Preparing a virtual environment for NetPyNE\" \n\techo \"=============================================================================\"\n\techo \"Using Python version:\"\n\tpython3 --version\n\techo \"Using Python from:\"\n\twhich python3\n\t\n\techo \"\"\n\techo \"Creating a virtual environment: python3 -m venv env\"\n\techo \"-----------------------------------------------------------------------------\"\n\tpython3 -m venv env\n\t\n\techo \"\"\n\techo \"Activating virtual environment: source env/bin/activate\"\n\techo \"-----------------------------------------------------------------------------\"\n\tsource env/bin/activate\n\t\n\techo \"\"\n\techo \"Updating pip: python3 -m pip install --upgrade pip\"\n\techo \"-----------------------------------------------------------------------------\"\n\tpython3 -m pip install --upgrade pip\n\n\techo \"\"\n\techo \"Installing wheel: python3 -m pip install --upgrade wheel\"\n\techo \"-----------------------------------------------------------------------------\"\n\tpython3 -m pip install --upgrade wheel\n\t\n\techo \"\"\n\techo \"Installing ipython: python3 -m pip install --upgrade ipython\"\n\techo \"-----------------------------------------------------------------------------\"\n\tpython3 -m pip install ipython\n\t\n\techo \"\"\n\techo \"Installing NEURON: python3 -m pip install --upgrade neuron\"\n\techo \"-----------------------------------------------------------------------------\"\n\tpython3 -m pip install --upgrade neuron \n\t\n\techo \"\"\n\techo \"Cloning NetPyNE: git clone https://github.com/Neurosim-lab/netpyne.git\"\n\techo \"-----------------------------------------------------------------------------\"\n\tgit clone https://github.com/Neurosim-lab/netpyne.git \n\t\n\techo \"\"\n\techo \"Installing NetPyNE: python3 -m pip install -e netpyne\"\n\techo \"-----------------------------------------------------------------------------\"\n\tpython3 -m pip install -e netpyne \n\t\n\techo \"\"\n\techo \"Installing ipykernel for Jupyter: python3 -m pip install --upgrade ipykernel\"\n\techo \"-----------------------------------------------------------------------------\"\n\tpython3 -m pip install --upgrade ipykernel \n\t\n\techo \"\"\n\techo \"Installing Jupyter: python3 -m pip install --upgrade jupyter\"\n\techo \"-----------------------------------------------------------------------------\"\n\tpython3 -m pip install --upgrade jupyter\n\t\n\techo \"\"\n\techo \"Creating a kernel for Jupyter: ipython kernel install --user --name=env\"\n\techo \"-----------------------------------------------------------------------------\"\n\tipython kernel install --user --name=env\n\n\techo \"\"\n\techo \"=============================================================================\"\n\techo \"Your virtual environment is ready for use.\"\n\techo \"\"\n\techo \"To deactivate, execute: deactivate\"\n\techo \"To reactivate, execute: source env/bin/activate\"\n\techo \"=============================================================================\"",
"_____no_output_____"
],
[
"## Copying this tutorial\n\nFor convenience, let's copy this tutorial's directory up to the directory we're working in and then change into that directory.\n\n pwd\n cp -r netpyne/netpyne/tutorials/saving_loading_tut .\n cd saving_loading_tut\n pwd",
"_____no_output_____"
],
[
"## Normal saving\n\nThen we'll run a simulation with normal saving, using `saving_netParams.py` (which is used by all simulations in this tutorial), `saving_normal_cfg.py`, and `saving_normal_init.py`.\n\nLet's take a look at `saving_normal_init.py`, to see the standard way to run and save a simulation:\n\n from netpyne import sim\n\n cfg, netParams = sim.readCmdLineArgs(\n simConfigDefault='saving_normal_cfg.py', \n netParamsDefault='saving_netParams.py')\n sim.initialize(simConfig=cfg, netParams=netParams)\n sim.net.createPops()\n sim.net.createCells()\n sim.net.connectCells()\n sim.net.addStims()\n sim.setupRecording()\n sim.runSim()\n sim.gatherData()\n sim.saveData()\n sim.analysis.plotData()\n \nWe could run this on a single core using `python3 saving_normal_init.py` (if we just want the output) or `ipython -i saving_normal_init.py` (if we wanted to interact with the simulation afterwards. But we will run this on multiple cores using the following command:",
"_____no_output_____"
]
],
[
[
"!mpiexec -n 4 nrniv -python -mpi saving_normal_init.py",
"_____no_output_____"
]
],
[
[
"This command does not currently exit to the system prompt, so you will have to restart your kernel. In the menu bar above, click on `Kernel`, then `Restart`, then `Restart`.\n\nThe `whos` in the next cell should return `Interactive namespace is empty.` after the Kernel has been cleared.",
"_____no_output_____"
]
],
[
[
"whos",
"_____no_output_____"
]
],
[
[
"The simulation should have produced a directory called `saving_normal_data` with three analysis plots and a data file named `saving_normal_data.pkl`. We are now going to load the simulation from this file and produce the same plots.",
"_____no_output_____"
]
],
[
[
"from netpyne import sim\nsim.loadAll('saving_normal_data/saving_normal_data.pkl')",
"_____no_output_____"
],
[
"sim.analysis.plotConn(saveFig='saving_normal_data/saving_normal_plot_conn_pop_strength_matrix_FROMFILE.png');\nsim.analysis.plotRaster(saveFig='saving_normal_data/saving_normal_raster_gid_FROMFILE.png');\nsim.analysis.plotTraces(saveFig='saving_normal_data/saving_normal_traces_FROMFILE.png');",
"_____no_output_____"
]
],
[
[
"Compare the plots, they should be identical. Congratulations! You have run a simulation, saved the data, then loaded it later to perform more analysis.\n\nNow restart your kernel and check the `whos`.",
"_____no_output_____"
]
],
[
[
"whos",
"_____no_output_____"
]
],
[
[
"## Distributed Saving\n\nIf you're running large sims, you may want to save the data from each node in a separate file, i.e. distributed saving.\n\nWe'll run a simulation using distributed saving and loading using `saving_netParams.py` (which is used by all simulations in this tutorial), `saving_dist_cfg.py`, and `saving_dist_init.py`.\n\nThe only changes to the cfg file are renaming the simulation:\n\n cfg.simLabel = 'saving_dist'\n\nOur init file for distributed saving looks like this:\n\nfrom netpyne import sim\n\n cfg, netParams = sim.readCmdLineArgs(\n simConfigDefault='saving_dist_cfg.py', \n netParamsDefault='saving_netParams.py')\n sim.initialize(simConfig=cfg, netParams=netParams)\n sim.net.createPops()\n sim.net.createCells()\n sim.net.connectCells()\n sim.net.addStims()\n sim.setupRecording()\n sim.runSim()\n #sim.gatherData()\n #sim.saveData()\n ##### new #####\n sim.saveDataInNodes()\n sim.gatherDataFromFiles()\n ##### end new #####\n sim.analysis.plotData()\n \nWe turned off `gatherData` and `saveData` and replaced those with `saveDataInNodes` and `gatherDataFromFiles`.\n\nLet's run the simulation now.",
"_____no_output_____"
]
],
[
[
"!mpiexec -n 4 nrniv -python -mpi saving_dist_init.py",
"_____no_output_____"
]
],
[
[
"That should have produced a directory `saving_dist_data` containing the same three analysis plots and a `node_data` directory containing a data file from each of the four nodes we used.\n\nNow restart your kernel so we can load the data from file analyze it again.\n\nThe `whos` in the next cell should return `Interactive namespace is empty.`",
"_____no_output_____"
]
],
[
[
"whos",
"_____no_output_____"
],
[
"from netpyne import sim\nsim.gatherDataFromFiles(simLabel='saving_dist')",
"_____no_output_____"
],
[
"sim.analysis.plotConn(saveFig='saving_dist_data/saving_dist_plot_conn_pop_strength_matrix_FROMFILE.png');\nsim.analysis.plotRaster(saveFig='saving_dist_data/saving_dist_raster_gid_FROMFILE.png');\nsim.analysis.plotTraces(saveFig='saving_dist_data/saving_dist_traces_FROMFILE.png');",
"_____no_output_____"
]
],
[
[
"Compare the plots, they should be identical except for the connectivity plot, which didn't retain the connectivity for the background inputs.\n\nNow restart your kernel and check the `whos`.",
"_____no_output_____"
]
],
[
[
"whos",
"_____no_output_____"
]
],
[
[
"## Interval Saving\n\nPerhaps you want to save data at intervals in case you have large, long simulations you're worried won't complete.\n\nWe'll run a simulation using interval saving and loading using `saving_netParams.py` (which is used by all simulations in this tutorial), `saving_int_cfg.py`, and `saving_int_init.py`.\n\nThe only changes to the cfg file are renaming the simulation:\n\n cfg.simLabel = 'saving_int'\n\nand turning back on the saving of the data into one file:\n\n cfg.savePickle = True\n\nOur init file for interval saving looks like this:\n\nfrom netpyne import sim\n\n from netpyne import sim\n\n cfg, netParams = sim.readCmdLineArgs(\n simConfigDefault='saving_int_cfg.py', \n netParamsDefault='saving_netParams.py')\n sim.initialize(simConfig=cfg, netParams=netParams)\n sim.net.createPops()\n sim.net.createCells()\n sim.net.connectCells()\n sim.net.addStims()\n sim.setupRecording()\n #sim.runSim()\n ##### new #####\n sim.runSimIntervalSaving(1000)\n ##### end new #####\n sim.gatherData()\n sim.saveData()\n sim.analysis.plotData()\n \nWe turned off `runSim` and replaced it with `runSimIntervalSaving(1000)`, which will save the simulation every 1000 ms.\n\nLet's run the simulation now. Remember you can run this without MPI using the command `python3 saving_int_init.py`.",
"_____no_output_____"
]
],
[
[
"!mpiexec -n 4 nrniv -python -mpi saving_int_init.py",
"_____no_output_____"
]
],
[
[
"That should have produced a directory `saving_int_data` containing the data file and the same three analysis plots (from the completed simulation) and an `interval_data` directory containing a data file for each 1000 ms of our 10,000 ms simulation.\n\nNow restart your kernel so we can load interval data from file.\n\nThe `whos` in the next cell should return `Interactive namespace is empty.`",
"_____no_output_____"
]
],
[
[
"whos",
"_____no_output_____"
]
],
[
[
"Now, let's assume our simulation timed out, and the last interval save we got was at 10000 ms. We can still analyze that partial data.",
"_____no_output_____"
]
],
[
[
"from netpyne import sim\nsim.loadAll('saving_int_data/interval_data/interval_10000.pkl', createNEURONObj=False)",
"_____no_output_____"
],
[
"sim.analysis.plotConn(saveFig='saving_int_data/saving_int_plot_conn_pop_strength_matrix_INTERVAL.png');\nsim.analysis.plotRaster(saveFig='saving_int_data/saving_int_raster_gid_INTERVAL.png');\nsim.analysis.plotTraces(saveFig='saving_int_data/saving_int_traces_INTERVAL.png');",
"_____no_output_____"
]
],
[
[
"The connectivity, traces and raster plots should be identical. You can see that we recovered partial data.\n\nCongratulations! You have successfully saved, loaded, and analyzed simulation data in a variety of ways.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
c5145133b4f9acf775ecfc216d961d6060eb04c5
| 74,856 |
ipynb
|
Jupyter Notebook
|
Code/.ipynb_checkpoints/FuzzyC-Means-checkpoint.ipynb
|
sammisetti2/Data-Mining-Cell-Services-in-India
|
15d6974960df0e1defa218b5c7b275b3182a20fe
|
[
"MIT"
] | null | null | null |
Code/.ipynb_checkpoints/FuzzyC-Means-checkpoint.ipynb
|
sammisetti2/Data-Mining-Cell-Services-in-India
|
15d6974960df0e1defa218b5c7b275b3182a20fe
|
[
"MIT"
] | null | null | null |
Code/.ipynb_checkpoints/FuzzyC-Means-checkpoint.ipynb
|
sammisetti2/Data-Mining-Cell-Services-in-India
|
15d6974960df0e1defa218b5c7b275b3182a20fe
|
[
"MIT"
] | null | null | null | 58.618637 | 17,286 | 0.54192 |
[
[
[
"import numpy\nimport random\nimport math\nimport numpy as np\nimport pandas as pd\nimport seaborn as sns\nimport matplotlib.pyplot as plt\n%matplotlib inline",
"_____no_output_____"
],
[
"address = \"\"\ndata = pd.read_csv(address); #To read csv file\ndf = pd.DataFrame(data)\ndf",
"_____no_output_____"
],
[
"#z-score normalization\ndf['signal_strength']=((df['signal_strength']-df['signal_strength'].min())/(df['signal_strength'].max()-df['signal_strength'].min()))*(100)\ndf['speed']=((df['speed']-df['speed'].min())/(df['speed'].max()-df['speed'].min()))*100\ndf.rename(columns={\"download/upload\": \"d_u\"}, inplace = True)\ndf",
"_____no_output_____"
],
[
"# Preparing data to be tested\ndf1 = df[(df['state'] == \"Delhi\") & (df['service_provider'] == \"VODAFONE\") & (df['Month'] == \"July\") & (df['Year'] == 2018) & (df['technology'] == \"4G\")]\n\n\ndf1 = df1[['speed','signal_strength']]\ndf1 = df1.values\ndf1",
"_____no_output_____"
],
[
"#1). Constructing the weight matrix\nres = [ [ 0 for i in range(2) ] for j in range(len(df1)) ]\n\nfor i in range(len(df1)):\n x = random.random()\n res[i][0] = x\n res[i][1] = 1-x\nprint(res)",
"[[0.7635504544755635, 0.23644954552443653], [0.9551649756600996, 0.04483502433990039], [0.7307093064275527, 0.2692906935724473], [0.7293246620251721, 0.2706753379748279], [0.11233591735239645, 0.8876640826476035], [0.6384291630252484, 0.36157083697475156], [0.5592782239416058, 0.44072177605839424], [0.08659473475137247, 0.9134052652486275], [0.4223644573202049, 0.5776355426797951], [0.6705809474581652, 0.3294190525418348], [0.37260467234560657, 0.6273953276543934], [0.6903484675263252, 0.30965153247367483], [0.9807841430751795, 0.01921585692482053], [0.012099833012576, 0.987900166987424], [0.3148443803052158, 0.6851556196947842], [0.10434730952586002, 0.89565269047414], [0.2797782545190668, 0.7202217454809332], [0.7046556452994718, 0.2953443547005282], [0.3900798444125908, 0.6099201555874092], [0.5813516953659587, 0.41864830463404135], [0.2809112224005613, 0.7190887775994387], [0.8404750720480213, 0.1595249279519787], [0.3857112878324912, 0.6142887121675088], [0.1110764650845456, 0.8889235349154544], [0.1607929213391649, 0.8392070786608351], [0.1218422667651734, 0.8781577332348266], [0.7631125038032863, 0.23688749619671368], [0.7384737165968251, 0.26152628340317485], [0.5068272903156729, 0.49317270968432714], [0.7348412576054245, 0.2651587423945755], [0.11582134319715154, 0.8841786568028485], [0.7155885348049887, 0.28441146519501126], [0.55189847754046, 0.44810152245954005], [0.04100792605463244, 0.9589920739453676], [0.11302777606556724, 0.8869722239344328], [0.6801069487535838, 0.3198930512464162], [0.531441506569552, 0.468558493430448], [0.07335659581268827, 0.9266434041873117], [0.3172140339155237, 0.6827859660844763], [0.6185070521104808, 0.38149294788951915], [0.6684845247843662, 0.33151547521563385], [0.4880034766346383, 0.5119965233653617], [0.10689298717480877, 0.8931070128251912], [0.48868628892992616, 0.5113137110700738], [0.5370661500979577, 0.46293384990204234], [0.05408094116255646, 0.9459190588374435], [0.9151496371906378, 0.0848503628093622], [0.8035722943703352, 0.19642770562966483], [0.4124975222840952, 0.5875024777159048], [0.42766738434421625, 0.5723326156557837], [0.7248433234266524, 0.27515667657334764], [0.7997027165183694, 0.20029728348163056], [0.03726528064502077, 0.9627347193549792], [0.9971064169100471, 0.0028935830899529114], [0.6403041267660751, 0.35969587323392493], [0.6685683366094306, 0.3314316633905694], [0.5534098933942265, 0.4465901066057735], [0.9247434716976625, 0.07525652830233753], [0.8609099284375101, 0.13909007156248987], [0.15792075157416385, 0.8420792484258361], [0.613658539625396, 0.38634146037460404], [0.17503821925219187, 0.8249617807478081], [0.36446588066989083, 0.6355341193301092], [0.07118641348529531, 0.9288135865147047], [0.36541346250110096, 0.634586537498899], [0.9150244790832633, 0.08497552091673666], [0.1419003604696919, 0.8580996395303081], [0.9653149022191645, 0.034685097780835505], [0.6773337937339856, 0.3226662062660144], [0.07084732093805235, 0.9291526790619477], [0.25766933444467766, 0.7423306655553223], [0.7426564457733331, 0.2573435542266669], [0.8474640042268295, 0.15253599577317045], [0.7463393499539107, 0.2536606500460893], [0.7990201987708814, 0.20097980122911863], [0.768430328948831, 0.231569671051169], [0.40636188753112734, 0.5936381124688727], [0.5473654475743456, 0.4526345524256544], [0.08618532008516921, 0.9138146799148308], [0.3778987125260602, 0.6221012874739398], [0.0026897412282965716, 0.9973102587717034], [0.2366980560870089, 0.7633019439129911], [0.06845998679257181, 0.9315400132074282], [0.32373649893007705, 0.676263501069923], [0.8587600888916491, 0.14123991110835088], [0.1974405254201259, 0.8025594745798741], [0.5214346347425569, 0.4785653652574431], [0.5060726303639167, 0.4939273696360833], [0.3837354540362645, 0.6162645459637355], [0.19809033686606248, 0.8019096631339375], [0.3827319339191648, 0.6172680660808352], [0.9072932893225495, 0.09270671067745051], [0.49238411777803626, 0.5076158822219637], [0.7671256443680462, 0.2328743556319538], [0.2150887177290418, 0.7849112822709582], [0.21128083091375516, 0.7887191690862448], [0.3772131981439787, 0.6227868018560213], [0.3418512317665754, 0.6581487682334246], [0.4384021732832004, 0.5615978267167996], [0.3832896062905926, 0.6167103937094074], [0.34313873257283556, 0.6568612674271644], [0.8373846584600153, 0.1626153415399847], [0.14546175280523754, 0.8545382471947625], [0.6676229010882657, 0.3323770989117343], [0.8778761119640752, 0.12212388803592478], [0.22008166085289382, 0.7799183391471062], [0.517191788093776, 0.482808211906224], [0.8497681875111127, 0.15023181248888728], [0.6657064514863239, 0.3342935485136761], [0.525007970746695, 0.47499202925330497], [0.5413773861999981, 0.4586226138000019], [0.35110251695676753, 0.6488974830432325], [0.5432095713063272, 0.4567904286936728], [0.9223478439082233, 0.07765215609177667], [0.8248070603203874, 0.17519293967961258], [0.06074374399651894, 0.9392562560034811], [0.0030624939164021203, 0.9969375060835979], [0.7258119873592362, 0.27418801264076376], [0.5611649003341495, 0.43883509966585055], [0.8686962691425533, 0.13130373085744673], [0.22638189094851047, 0.7736181090514895], [0.081834537756035, 0.918165462243965], [0.04480900761067963, 0.9551909923893204], [0.08554584518236497, 0.914454154817635], [0.4806642771361841, 0.5193357228638159], [0.5610567580210646, 0.4389432419789354], [0.03920037280437916, 0.9607996271956208], [0.6019041197042283, 0.3980958802957717], [0.9337028982419114, 0.06629710175808856], [0.5595680530019678, 0.44043194699803223], [0.4068438514956668, 0.5931561485043332], [0.23991303035834166, 0.7600869696416583], [0.7864038645505222, 0.21359613544947775], [0.32772952578194503, 0.672270474218055], [0.5113109628553845, 0.48868903714461553], [0.23661856875526055, 0.7633814312447395], [0.24604793628006139, 0.7539520637199386], [0.010662901909489486, 0.9893370980905105], [0.6062641958132415, 0.39373580418675846], [0.5682240407598295, 0.43177595924017054], [0.7987202418242727, 0.2012797581757273], [0.09754311155896833, 0.9024568884410317], [0.27649198516594753, 0.7235080148340525], [0.0199338288852855, 0.9800661711147145], [0.2597826895468629, 0.7402173104531371], [0.28821242314748574, 0.7117875768525143], [0.334756474517088, 0.665243525482912], [0.4764072318472843, 0.5235927681527157], [0.7448398240436009, 0.25516017595639906], [0.7322336488593902, 0.26776635114060976], [0.010570753102333241, 0.9894292468976668], [0.882931912865325, 0.11706808713467498], [0.6948684177182347, 0.3051315822817653], [0.47341612575906367, 0.5265838742409363], [0.018921877190943848, 0.9810781228090562], [0.7016718455665836, 0.2983281544334164], [0.5487329052352122, 0.45126709476478777], [0.0636075481367252, 0.9363924518632748], [0.9194036338782837, 0.08059636612171628], [0.47058263553408763, 0.5294173644659124], [0.5763496084967932, 0.4236503915032068], [0.030654692368164205, 0.9693453076318358], [0.004464090670473819, 0.9955359093295262], [0.7526631433496301, 0.24733685665036986], [0.011415827602383799, 0.9885841723976162], [0.2773539501093373, 0.7226460498906627], [0.13380315465050696, 0.866196845349493], [0.29130407264117375, 0.7086959273588262], [0.027708651714995036, 0.972291348285005], [0.8236470012214119, 0.17635299877858812], [0.8535411521336161, 0.14645884786638386], [0.7252792254757575, 0.2747207745242425], [0.08664649401383406, 0.9133535059861659], [0.7845088560800031, 0.2154911439199969], [0.38805166264654756, 0.6119483373534524], [0.4203760912134126, 0.5796239087865874], [0.48223404426760974, 0.5177659557323903], [0.8043978236844027, 0.1956021763155973], [0.04134278198891672, 0.9586572180110833], [0.3403486205079066, 0.6596513794920934], [0.12816253492666785, 0.8718374650733322], [0.667563407545745, 0.33243659245425505], [0.4723265872134438, 0.5276734127865562], [0.7007718501353634, 0.29922814986463664], [0.15957664491291346, 0.8404233550870865], [0.37888631173467735, 0.6211136882653226], [0.15603103669795704, 0.843968963302043], [0.06856832501353616, 0.9314316749864638], [0.29655304831089724, 0.7034469516891028], [0.9378024991485734, 0.06219750085142661], [0.11373915820587932, 0.8862608417941207], [0.6791091331626109, 0.3208908668373891], [0.08997508034442048, 0.9100249196555795], [0.6982782213406257, 0.30172177865937433], [0.7211714491184721, 0.2788285508815279], [0.009131745968703697, 0.9908682540312963], [0.08607832394523873, 0.9139216760547613], [0.8490966366549342, 0.15090336334506582], [0.7878671823686597, 0.2121328176313403], [0.3918402878867212, 0.6081597121132788], [0.8953251368543856, 0.10467486314561436], [0.35407452545253015, 0.6459254745474698], [0.7883109287877855, 0.21168907121221447], [0.26125027138675616, 0.7387497286132438], [0.30837155000645966, 0.6916284499935403], [0.376106995895216, 0.623893004104784], [0.7967895763311549, 0.20321042366884512], [0.16864853300159233, 0.8313514669984077], [0.97756706478623, 0.02243293521376999], [0.1441411018425094, 0.8558588981574906], [0.8934054083842194, 0.10659459161578055], [0.954923841538313, 0.045076158461687], [0.35265387536648585, 0.6473461246335142], [0.9704563920230691, 0.029543607976930897], [0.7219437801606471, 0.2780562198393529], [0.6502172665114737, 0.3497827334885263], [0.179953845124603, 0.820046154875397], [0.9240868721892, 0.07591312781079995], [0.9420077540717962, 0.057992245928203845], [0.8945897014648931, 0.10541029853510686], [0.33252566568772857, 0.6674743343122714], [0.9559400004005736, 0.044059999599426414], [0.00758768830314005, 0.99241231169686], [0.3868441277869824, 0.6131558722130176], [0.8643336291515501, 0.1356663708484499], [0.07754887675469668, 0.9224511232453033], [0.5066318664226204, 0.49336813357737963], [0.8702619421926703, 0.12973805780732972], [0.6125350565960318, 0.3874649434039682], [0.607560254384742, 0.39243974561525796], [0.1095583909044281, 0.8904416090955719], [0.28074555093743103, 0.719254449062569], [0.19998350099901419, 0.8000164990009858], [0.7464384469836105, 0.2535615530163895], [0.7145192218943565, 0.28548077810564354], [0.7150144863647722, 0.28498551363522784], [0.04349388432932799, 0.956506115670672], [0.0961276240454435, 0.9038723759545565], [0.11233249043751625, 0.8876675095624837], [0.6695996051601597, 0.3304003948398403], [0.33715621182967315, 0.6628437881703269], [0.847951390176185, 0.15204860982381496], [0.29134648018869214, 0.7086535198113079], [0.9156473787110861, 0.08435262128891385], [0.6329681785097812, 0.36703182149021885], [0.7420933809141818, 0.25790661908581825], [0.7431650658278973, 0.2568349341721027], [0.25922865995939104, 0.740771340040609], [0.8080737997916491, 0.19192620020835094], [0.5454796581306243, 0.45452034186937573], [0.5735390790999426, 0.4264609209000574], [0.2621378112550432, 0.7378621887449568], [0.08604687601938865, 0.9139531239806113], [0.15693121937721788, 0.8430687806227821], [0.07872760604541784, 0.9212723939545822], [0.17516183932706986, 0.8248381606729301], [0.5537861743746537, 0.4462138256253463], [0.7012134123632606, 0.2987865876367394], [0.3100320010609183, 0.6899679989390817], [0.0664862099176573, 0.9335137900823427], [0.15958109799401965, 0.8404189020059804], [0.7761593330602404, 0.22384066693975957], [0.7061635139574163, 0.2938364860425837], [0.34442422083744084, 0.6555757791625592], [0.28100443467140246, 0.7189955653285975], [0.35113948215695356, 0.6488605178430464], [0.7283379885323518, 0.27166201146764823], [0.4577397491921741, 0.5422602508078259], [0.9913264369276322, 0.008673563072367774], [0.8308950123072032, 0.1691049876927968], [0.9738067666142114, 0.026193233385788606], [0.11084213805530752, 0.8891578619446925], [0.02093200519010774, 0.9790679948098923], [0.16489784243277672, 0.8351021575672233], [0.47633830830984136, 0.5236616916901586], [0.6411001718003084, 0.3588998281996916], [0.06977717146598106, 0.9302228285340189], [0.8347618783092491, 0.16523812169075092], [0.6913213829588944, 0.30867861704110555], [0.5392946536131866, 0.4607053463868134], [0.0760652003633977, 0.9239347996366023], [0.7821535074706423, 0.21784649252935773], [0.053026075523165295, 0.9469739244768347], [0.5020623881663252, 0.4979376118336748], [0.9460401111169832, 0.05395988888301684], [0.6749637838074382, 0.32503621619256184], [0.8345181920545657, 0.16548180794543432], [0.019492778775749398, 0.9805072212242506], [0.07561820437454769, 0.9243817956254523], [0.8673158233049941, 0.13268417669500587], [0.33547153234066995, 0.66452846765933], [0.05455590785161579, 0.9454440921483842], [0.13672323105806483, 0.8632767689419352], [0.4220008413201283, 0.5779991586798717], [0.9729229299086957, 0.027077070091304334], [0.08154011044428111, 0.9184598895557189], [0.6204758479570552, 0.37952415204294476], [0.4180293809176747, 0.5819706190823253], [0.35505731298794185, 0.6449426870120581], [0.20203129992217372, 0.7979687000778263], [0.5789553144081504, 0.4210446855918496], [0.42280950403699613, 0.5771904959630039], [0.5362777057087412, 0.4637222942912588], [0.1978922887737503, 0.8021077112262497], [0.16970010200113705, 0.830299897998863], [0.8229939011943619, 0.1770060988056381], [0.9882280461856354, 0.011771953814364577], [0.9572677841137355, 0.042732215886264524], [0.4375770545933858, 0.5624229454066142], [0.11885899244422027, 0.8811410075557797], [0.2559724159971071, 0.7440275840028929], [0.12426924112554927, 0.8757307588744507], [0.8732728223459032, 0.1267271776540968], [0.09703633938234735, 0.9029636606176527], [0.4458151556991301, 0.5541848443008699], [0.2604897484855411, 0.7395102515144589], [0.6634025082493149, 0.3365974917506851], [0.02807026637387644, 0.9719297336261236], [0.408010011075422, 0.591989988924578], [0.2463758769392126, 0.7536241230607874], [0.2894266673393665, 0.7105733326606335], [0.2847474467786526, 0.7152525532213474], [0.3680943498146044, 0.6319056501853956], [0.8399552194310929, 0.16004478056890714], [0.007247546522780701, 0.9927524534772193], [0.6034633407799216, 0.3965366592200784], [0.6626388863366812, 0.3373611136633188], [0.7317755397605447, 0.26822446023945534], [0.09159870316628682, 0.9084012968337132], [0.18202341170795788, 0.8179765882920421], [0.217981441793214, 0.782018558206786], [0.4162265166966024, 0.5837734833033976], [0.29733095077502114, 0.7026690492249789], [0.39475691947961744, 0.6052430805203826], [0.628427362436635, 0.371572637563365], [0.9109161003473013, 0.0890838996526987], [0.4495767788883903, 0.5504232211116097], [0.5276761168788879, 0.47232388312111206], [0.794445453538318, 0.20555454646168203], [0.397607982384901, 0.602392017615099], [0.3151528647006506, 0.6848471352993494], [0.008835592689925287, 0.9911644073100747], [0.7181216252068351, 0.2818783747931649], [0.6458399359785946, 0.35416006402140543], [0.04219867866635185, 0.9578013213336481], [0.8059835339818991, 0.19401646601810085], [0.9623482612336226, 0.037651738766377396], [0.8958804521299983, 0.10411954787000166], [0.887292025176086, 0.11270797482391404], [0.8062837180994928, 0.19371628190050716], [0.5188823522189215, 0.48111764778107846], [0.03863519149475736, 0.9613648085052426], [0.09850525486639616, 0.9014947451336038], [0.9292067209662013, 0.07079327903379873], [0.4202180346835698, 0.5797819653164302], [0.9170214492419098, 0.08297855075809024], [0.990267348786435, 0.009732651213565036], [0.6812806406183938, 0.3187193593816062], [0.6042238388001243, 0.3957761611998757], [0.1707533047741171, 0.8292466952258829], [0.47330559195911115, 0.5266944080408889], [0.7713399445884918, 0.22866005541150825], [0.20867710344148271, 0.7913228965585173], [0.7145543704012978, 0.28544562959870223], [0.28178060975169394, 0.718219390248306], [0.22359468563282336, 0.7764053143671766], [0.5849617371067392, 0.41503826289326085], [0.7681954722265271, 0.23180452777347293], [0.8012727540514133, 0.1987272459485867], [0.44604993873932586, 0.5539500612606741], [0.8952361177684672, 0.10476388223153277], [0.2113291970170943, 0.7886708029829057], [0.6173993245871691, 0.38260067541283094], [0.9162791042083639, 0.08372089579163611], [0.9268748068711303, 0.07312519312886967], [0.300691482985435, 0.699308517014565], [0.8914529206549738, 0.10854707934502617], [0.4386641962850797, 0.5613358037149203], [0.0005251178081701058, 0.9994748821918299], [0.09220812470271311, 0.9077918752972869], [0.015580296841470997, 0.984419703158529], [0.7316040377586639, 0.2683959622413361], [0.19345623961467373, 0.8065437603853263], [0.8406969680323874, 0.15930303196761264], [0.9218919892521821, 0.07810801074781792], [0.7311750928632913, 0.2688249071367087], [0.13103948883878191, 0.8689605111612181], [0.633564103254742, 0.366435896745258], [0.7334490495281154, 0.2665509504718846], [0.9495618125229013, 0.05043818747709872], [0.530718257406079, 0.46928174259392097], [0.41722600487838524, 0.5827739951216148], [0.4913333930321008, 0.5086666069678992], [0.26938916421759773, 0.7306108357824023], [0.8746410042405967, 0.12535899575940335], [0.7567644258534273, 0.2432355741465727], [0.6885009173821264, 0.3114990826178736], [0.3998197681206562, 0.6001802318793438], [0.47598302576941864, 0.5240169742305814], [0.10843973424587372, 0.8915602657541263], [0.1466225664705394, 0.8533774335294606], [0.17455627187379774, 0.8254437281262023], [0.224418282927908, 0.775581717072092], [0.5375184013910634, 0.46248159860893656], [0.013356763403401417, 0.9866432365965986], [0.0469165405813019, 0.9530834594186981]]\n"
],
[
"#2). Calculating the respective centroids.\n#Centroid of cluster-1\nold_c1 = [0,0]\nold_c2 = [0,0]\nc1 = [0,0]\nc2 = [0,0]\nm = 2\nflag = 1\nval = 0\n\nwhile flag == 1:\n val += 1\n print(\"Presently, in iteration \",val)\n flag = 0\n c1 = [0,0]\n c2 = [0,0]\n weighted_sum_1 = 0\n weighted_sum_2 = 0\n \n #computing the centroids of each cluster\n for i in range(len(df1)):\n weighted_sum_1 += pow(res[i][0],m) # sum of powers of the membership function\n weighted_sum_2 += pow(res[i][1],m) # sum of powers of the membership function\n\n c1[0] += pow(res[i][0],m) * df1[i][0]\n c1[1] += pow(res[i][0],m) * df1[i][1]\n c2[0] += pow(res[i][1],m) * df1[i][0]\n c2[1] += pow(res[i][1],m) * df1[i][1]\n\n c1 = [x / weighted_sum_1 for x in c1]\n c2 = [x / weighted_sum_2 for x in c2]\n\n print(c1)\n print(c2)\n\n #4). Updating membership matrix\n for i in range(len(df1)):\n dist1 = math.sqrt((df1[i][0]-c1[0])**2 + (df1[i][1]-c1[1])**2) #distance from cluster-1\n dist2 = math.sqrt((df1[i][0]-c2[0])**2 + (df1[i][1]-c2[1])**2) #distance from cluster-2\n\n denominator = pow((1/dist1),(1/m-1)) + pow((1/dist2),(1/m-1))\n res[i][0] = pow((1/dist1),(1/m-1)) / denominator\n res[i][1] = pow((1/dist2),(1/m-1)) / denominator\n \n #checking if centroids changed or not\n if (c1 == old_c1) and (c2 == old_c2):\n continue\n else:\n flag = 1\n old_c1 = c1\n old_c2 = c2\n\nprint(\"Completed finding centroid\")",
"Presently, in iteration 1\n[3.7845634920980027, 46.88293933968423]\n[4.196266668664013, 48.81598852766707]\nPresently, in iteration 2\n[4.044818454790856, 48.349726421768914]\n[3.803914897867475, 47.016522716305495]\nPresently, in iteration 3\n[3.848182373336743, 47.21279412435468]\n[4.000208916600356, 48.139863089505276]\nPresently, in iteration 4\n[3.975065601365359, 47.997588588170586]\n[3.873884853218332, 47.351603436599575]\nPresently, in iteration 5\n[3.890070893145309, 47.4483265491207]\n[3.9591359981041667, 47.89922670875556]\nPresently, in iteration 6\n[3.9485210547330998, 47.83082693116146]\n[3.9007737012330175, 47.515935794995784]\nPresently, in iteration 7\n[3.9080731343432373, 47.56322530685141]\n[3.941273122267905, 47.783180677908376]\nPresently, in iteration 8\n[3.9362595406564203, 47.74994892901033]\n[3.9131147793342604, 47.596290871406204]\nPresently, in iteration 9\n[3.916617364985196, 47.61940561092451]\n[3.9327713788149534, 47.72675499479713]\nPresently, in iteration 10\n[3.9303384304655182, 47.710560708529975]\n[3.919057527874506, 47.63556169787477]\nPresently, in iteration 11\n[3.92075985136674, 47.64685277286946]\n[3.928639669940606, 47.69925109587356]\nPresently, in iteration 12\n[3.927453020547207, 47.69135168257781]\n[3.9219482497537514, 47.654743163097564]\nPresently, in iteration 13\n[3.9227781666477775, 47.660256759252825]\n[3.9266239597123263, 47.68583368808769]\nPresently, in iteration 14\n[3.9260446981496124, 47.68197896145688]\n[3.923357846629078, 47.66410934036956]\nPresently, in iteration 15\n[3.9237627830249058, 47.66680121223051]\n[3.9256399661268926, 47.67928604264911]\nPresently, in iteration 16\n[3.925357180164183, 47.67740471383418]\n[3.924045668779185, 47.6686820300809]\nPresently, in iteration 17\n[3.9242432978060173, 47.66999614031155]\n[3.9251595998559368, 47.6760903542058]\nPresently, in iteration 18\n[3.9250215534507302, 47.675172086606274]\n[3.924381367993953, 47.67091428617715]\nPresently, in iteration 19\n[3.924477829753991, 47.67155577326461]\n[3.9249251033001666, 47.67453054009795]\nPresently, in iteration 20\n[3.924857716023785, 47.67408232168826]\n[3.9245452226974, 47.672003962669514]\nPresently, in iteration 21\n[3.924592307019033, 47.67231709899297]\n[3.9248106344684226, 47.67376917120666]\nPresently, in iteration 22\n[3.9247777400529342, 47.6735503860953]\n[3.9246252027848185, 47.67253587719339]\nPresently, in iteration 23\n[3.9246481856889757, 47.672688730119546]\n[3.9247547578079125, 47.67339752979575]\nPresently, in iteration 24\n[3.9247387008988164, 47.673290735133996]\n[3.924664242919805, 47.67279552313459]\nPresently, in iteration 25\n[3.9246754614761987, 47.672870135619775]\n[3.924727482499468, 47.673216121845115]\nPresently, in iteration 26\n[3.9247196446042283, 47.67316399239685]\n[3.924683299448103, 47.672922264675655]\nPresently, in iteration 27\n[3.9246887755397677, 47.67295868535155]\n[3.924714168549984, 47.67312757152943]\nPresently, in iteration 28\n[3.9247103426304104, 47.67310212564271]\n[3.924692601477605, 47.67298413114474]\nPresently, in iteration 29\n[3.9246952745163024, 47.673001909185]\n[3.9247076696006324, 47.67308434755694]\nPresently, in iteration 30\n[3.924705802053107, 47.67307192667363]\n[3.924697142068192, 47.67301433004596]\nPresently, in iteration 31\n[3.9246984468568473, 47.6730230080424]\n[3.9247044972665632, 47.673063248666274]\nPresently, in iteration 32\n[3.924703585660461, 47.67305718566606]\n[3.9246993584639815, 47.67302907103726]\nPresently, in iteration 33\n[3.924699995369924, 47.673033307027126]\n[3.9247029487550447, 47.673052949673746]\nPresently, in iteration 34\n[3.924702503772829, 47.673049990143404]\n[3.9247004403523715, 47.673036266556046]\nPresently, in iteration 35\n[3.9247007512450507, 47.67303833426914]\n[3.9247021928802655, 47.67304792242975]\nPresently, in iteration 36\n[3.9247019756711743, 47.67304647779514]\n[3.9247009684542125, 47.67303977890352]\nPresently, in iteration 37\n[3.924701120210164, 47.673040788215836]\n[3.92470182391524, 47.67304546848259]\nPresently, in iteration 38\n[3.9247017178890027, 47.67304476331341]\n[3.9247012262364187, 47.67304149338498]\nPresently, in iteration 39\n[3.924701300313018, 47.673041986060404]\n[3.9247016438124103, 47.67304427063786]\nPresently, in iteration 40\n[3.9247015920578496, 47.67304392642386]\n[3.9247013520675904, 47.6730423302745]\nPresently, in iteration 41\n[3.924701388226581, 47.67304257076407]\n[3.924701555898855, 47.67304368593421]\nPresently, in iteration 42\n[3.924701530635912, 47.673043517913115]\n[3.9247014134895264, 47.6730427387852]\nPresently, in iteration 43\n[3.9247014311398045, 47.673042856175286]\n[3.92470151298563, 47.673043400523014]\nPresently, in iteration 44\n[3.9247015006540398, 47.67304331850698]\n[3.9247014434713985, 47.673042938191344]\nPresently, in iteration 45\n[3.924701452087025, 47.67304299549301]\n[3.9247014920384085, 47.67304326120534]\nPresently, in iteration 46\n[3.9247014860189937, 47.673043221170836]\n[3.9247014581064477, 47.67304303552748]\nPresently, in iteration 47\n[3.924701462311993, 47.67304306349813]\n[3.9247014818134462, 47.673043193200215]\nPresently, in iteration 48\n[3.924701478875187, 47.67304317365811]\n[3.9247014652502403, 47.673043083040135]\nPresently, in iteration 49\n[3.9247014673031013, 47.67304309669344]\n[3.9247014768223387, 47.67304316000484]\nPresently, in iteration 50\n[3.924701475388089, 47.673043150465766]\n[3.9247014687373545, 47.67304310623253]\nPresently, in iteration 51\n[3.92470146973941, 47.673043112897105]\n[3.924701474386028, 47.67304314380125]\nPresently, in iteration 52\n[3.924701473685927, 47.67304313914493]\n[3.924701470439512, 47.67304311755338]\nPresently, in iteration 53\n[3.92470147092865, 47.67304312080662]\n[3.924701473196788, 47.67304313589175]\nPresently, in iteration 54\n[3.9247014728550504, 47.673043133618854]\n[3.9247014712703847, 47.67304312307938]\nPresently, in iteration 55\n[3.9247014715091453, 47.67304312466743]\n[3.9247014726163005, 47.67304313203097]\nPresently, in iteration 56\n[3.9247014724494815, 47.67304313092148]\n[3.9247014716759594, 47.67304312577682]\nPresently, in iteration 57\n[3.9247014717925026, 47.673043126551995]\n[3.9247014723329343, 47.67304313014632]\nPresently, in iteration 58\n[3.9247014722515026, 47.67304312960476]\n[3.9247014718739393, 47.673043127093585]\nPresently, in iteration 59\n[3.924701471930827, 47.67304312747195]\n[3.9247014721946116, 47.67304312922635]\nPresently, in iteration 60\n[3.9247014721548683, 47.673043128962064]\n[3.9247014719705717, 47.673043127736214]\nPresently, in iteration 61\n[3.9247014719983344, 47.673043127920955]\n[3.924701472127097, 47.6730431287774]\nPresently, in iteration 62\n[3.924701472107705, 47.673043128648345]\n[3.9247014720177327, 47.67304312804991]\nPresently, in iteration 63\n[3.9247014720312845, 47.673043128140094]\n[3.9247014720941515, 47.67304312855822]\nPresently, in iteration 64\n[3.9247014720846867, 47.67304312849525]\n[3.9247014720407574, 47.67304312820306]\nPresently, in iteration 65\n[3.9247014720473694, 47.67304312824709]\n[3.9247014720780657, 47.67304312845119]\nPresently, in iteration 66\n[3.924701472073444, 47.673043128420495]\n[3.924701472051999, 47.67304312827785]\nPresently, in iteration 67\n[3.9247014720552262, 47.6730431282993]\n[3.924701472070213, 47.67304312839895]\nPresently, in iteration 68\n[3.924701472067951, 47.67304312838394]\n[3.9247014720574853, 47.673043128314326]\nPresently, in iteration 69\n[3.9247014720590596, 47.67304312832484]\n[3.9247014720663764, 47.67304312837346]\nPresently, in iteration 70\n[3.9247014720652724, 47.67304312836612]\n[3.9247014720601627, 47.67304312833216]\nPresently, in iteration 71\n[3.9247014720609354, 47.67304312833732]\n[3.9247014720645037, 47.67304312836101]\nPresently, in iteration 72\n[3.9247014720639672, 47.67304312835748]\n[3.9247014720614692, 47.67304312834085]\nPresently, in iteration 73\n[3.9247014720618467, 47.673043128343394]\n[3.9247014720635933, 47.67304312835494]\nPresently, in iteration 74\n[3.9247014720633264, 47.67304312835314]\n[3.924701472062112, 47.67304312834519]\nPresently, in iteration 75\n[3.9247014720622904, 47.67304312834626]\n[3.9247014720631492, 47.67304312835194]\nPresently, in iteration 76\n[3.9247014720630236, 47.67304312835112]\n[3.9247014720624174, 47.67304312834718]\nPresently, in iteration 77\n[3.9247014720625115, 47.6730431283478]\n[3.9247014720629285, 47.673043128350514]\nPresently, in iteration 78\n"
],
[
"print(res)",
"[[0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5], [0.5, 0.5]]\n"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
c514561b305834b0fc43e23596f5ed76d5387feb
| 41,177 |
ipynb
|
Jupyter Notebook
|
Chatbot/Chatbot.ipynb
|
Elyrie/Hacktoberfest-2021
|
1e197d851a367c2ac4b51874f0575fe25cee72eb
|
[
"MIT"
] | 14 |
2021-10-01T16:53:27.000Z
|
2021-10-17T13:15:44.000Z
|
Chatbot/Chatbot.ipynb
|
Elyrie/Hacktoberfest-2021
|
1e197d851a367c2ac4b51874f0575fe25cee72eb
|
[
"MIT"
] | 37 |
2021-10-01T17:14:52.000Z
|
2021-10-21T17:26:14.000Z
|
Chatbot/Chatbot.ipynb
|
Elyrie/Hacktoberfest-2021
|
1e197d851a367c2ac4b51874f0575fe25cee72eb
|
[
"MIT"
] | 38 |
2021-10-01T16:59:16.000Z
|
2021-10-30T16:05:31.000Z
| 44.133976 | 7,349 | 0.563518 |
[
[
[
"# Libraries needed for NLP\nimport nltk\nnltk.download('punkt')\nfrom nltk.stem.lancaster import LancasterStemmer\nstemmer = LancasterStemmer()\n\n# Libraries needed for Tensorflow processing\nimport tensorflow as tf\nimport numpy as np\nimport tflearn\nimport random\nimport json",
"[nltk_data] Downloading package punkt to /root/nltk_data...\n[nltk_data] Package punkt is already up-to-date!\n"
],
[
"from google.colab import files\nfiles.upload()",
"_____no_output_____"
],
[
"# import our chat-bot intents file\nwith open('intents.json') as json_data:\n intents = json.load(json_data)",
"_____no_output_____"
],
[
"intents",
"_____no_output_____"
],
[
"words = []\nclasses = []\ndocuments = []\nignore = ['?']\n# loop through each sentence in the intent's patterns\nfor intent in intents['intents']:\n for pattern in intent['patterns']:\n # tokenize each and every word in the sentence\n w = nltk.word_tokenize(pattern)\n # add word to the words list\n words.extend(w)\n # add word(s) to documents\n documents.append((w, intent['tag']))\n # add tags to our classes list\n if intent['tag'] not in classes:\n classes.append(intent['tag'])",
"_____no_output_____"
],
[
"# Perform stemming and lower each word as well as remove duplicates\nwords = [stemmer.stem(w.lower()) for w in words if w not in ignore]\nwords = sorted(list(set(words)))\n\n# remove duplicate classes\nclasses = sorted(list(set(classes)))\n\nprint (len(documents), \"documents\")\nprint (len(classes), \"classes\", classes)\nprint (len(words), \"unique stemmed words\", words)",
"31 documents\n9 classes ['deliveryoption', 'goodbye', 'greeting', 'hours', 'location', 'menu', 'payments', 'thanks', 'todaysmenu']\n57 unique stemmed words [\"'s\", 'acceiv', 'address', 'anyon', 'ar', 'bye', 'can', 'card', 'cash', 'cours', 'credit', 'day', 'del', 'delicy', 'delivery', 'dish', 'do', 'food', 'for', 'from', 'good', 'goodby', 'hello', 'help', 'hi', 'hom', 'hour', 'how', 'is', 'lat', 'loc', 'main', 'mastercard', 'me', 'menu', 'most', 'on', 'op', 'opt', 'provid', 'resta', 'see', 'serv', 'situ', 'spec', 'tak', 'tel', 'thank', 'that', 'the', 'ther', 'today', 'what', 'when', 'wher', 'yo', 'you']\n"
],
[
"# create training data\ntraining = []\noutput = []\n# create an empty array for output\noutput_empty = [0] * len(classes)\n\n# create training set, bag of words for each sentence\nfor doc in documents:\n # initialize bag of words\n bag = []\n # list of tokenized words for the pattern\n pattern_words = doc[0]\n # stemming each word\n pattern_words = [stemmer.stem(word.lower()) for word in pattern_words]\n # create bag of words array\n for w in words:\n bag.append(1) if w in pattern_words else bag.append(0)\n\n # output is '1' for current tag and '0' for rest of other tags\n output_row = list(output_empty)\n output_row[classes.index(doc[1])] = 1\n\n training.append([bag, output_row])\n\n# shuffling features and turning it into np.array\nrandom.shuffle(training)\ntraining = np.array(training)\n\n# creating training lists\ntrain_x = list(training[:,0])\ntrain_y = list(training[:,1])",
"_____no_output_____"
],
[
"# resetting underlying graph data\ntf.reset_default_graph()\n\n# Building neural network\nnet = tflearn.input_data(shape=[None, len(train_x[0])])\nnet = tflearn.fully_connected(net, 10)\nnet = tflearn.fully_connected(net, 10)\nnet = tflearn.fully_connected(net, len(train_y[0]), activation='softmax')\nnet = tflearn.regression(net)\n\n# Defining model and setting up tensorboard\nmodel = tflearn.DNN(net, tensorboard_dir='tflearn_logs')\n\n# Start training\nmodel.fit(train_x, train_y, n_epoch=1000, batch_size=8, show_metric=True)\nmodel.save('model.tflearn')",
"Training Step: 3999 | total loss: \u001b[1m\u001b[32m0.26208\u001b[0m\u001b[0m | time: 0.012s\n| Adam | epoch: 1000 | loss: 0.26208 - acc: 0.9708 -- iter: 24/31\nTraining Step: 4000 | total loss: \u001b[1m\u001b[32m0.24488\u001b[0m\u001b[0m | time: 0.016s\n| Adam | epoch: 1000 | loss: 0.24488 - acc: 0.9737 -- iter: 31/31\n--\n"
],
[
"import pickle\npickle.dump( {'words':words, 'classes':classes, 'train_x':train_x, 'train_y':train_y}, open( \"training_data\", \"wb\" ) )",
"_____no_output_____"
],
[
"# restoring all the data structures\ndata = pickle.load( open( \"training_data\", \"rb\" ) )\nwords = data['words']\nclasses = data['classes']\ntrain_x = data['train_x']\ntrain_y = data['train_y']",
"_____no_output_____"
],
[
"with open('intents.json') as json_data:\n intents = json.load(json_data)",
"_____no_output_____"
],
[
"# load the saved model\nmodel.load('./model.tflearn')",
"W0616 22:00:03.473513 140144447035264 deprecation.py:323] From /usr/local/lib/python3.6/dist-packages/tensorflow/python/training/saver.py:1276: checkpoint_exists (from tensorflow.python.training.checkpoint_management) is deprecated and will be removed in a future version.\nInstructions for updating:\nUse standard file APIs to check for files with this prefix.\n"
],
[
"def clean_up_sentence(sentence):\n # tokenizing the pattern\n sentence_words = nltk.word_tokenize(sentence)\n # stemming each word\n sentence_words = [stemmer.stem(word.lower()) for word in sentence_words]\n return sentence_words\n\n# returning bag of words array: 0 or 1 for each word in the bag that exists in the sentence\ndef bow(sentence, words, show_details=False):\n # tokenizing the pattern\n sentence_words = clean_up_sentence(sentence)\n # generating bag of words\n bag = [0]*len(words) \n for s in sentence_words:\n for i,w in enumerate(words):\n if w == s: \n bag[i] = 1\n if show_details:\n print (\"found in bag: %s\" % w)\n\n return(np.array(bag))",
"_____no_output_____"
],
[
"ERROR_THRESHOLD = 0.30\ndef classify(sentence):\n # generate probabilities from the model\n results = model.predict([bow(sentence, words)])[0]\n # filter out predictions below a threshold\n results = [[i,r] for i,r in enumerate(results) if r>ERROR_THRESHOLD]\n # sort by strength of probability\n results.sort(key=lambda x: x[1], reverse=True)\n return_list = []\n for r in results:\n return_list.append((classes[r[0]], r[1]))\n # return tuple of intent and probability\n return return_list\n\ndef response(sentence, userID='123', show_details=False):\n results = classify(sentence)\n # if we have a classification then find the matching intent tag\n if results:\n # loop as long as there are matches to process\n while results:\n for i in intents['intents']:\n # find a tag matching the first result\n if i['tag'] == results[0][0]:\n # a random response from the intent\n return print(random.choice(i['responses']))\n\n results.pop(0)",
"_____no_output_____"
],
[
"classify('What are you hours of operation?')",
"_____no_output_____"
],
[
"response('What are you hours of operation?')",
"Our hours are 9am-9pm every day\n"
],
[
"response('What is menu for today?')",
"Our speciality for today is Chicken Tikka\n"
],
[
"#Some of other context free responses.\nresponse('Do you accept Credit Card?')",
"We accept VISA, Mastercard and AMEX\n"
],
[
"response('Where can we locate you?')",
"We are situated at the intersection of London Alley and Bridge Avenue\n"
],
[
"response('That is helpful')",
"Happy to help!\n"
],
[
"response('Bye')",
"See you later, thanks for visiting\n"
],
[
"#Adding some context to the conversation i.e. Contexualization for altering question and intents etc.\n# create a data structure to hold user context\ncontext = {}\n\nERROR_THRESHOLD = 0.25\ndef classify(sentence):\n # generate probabilities from the model\n results = model.predict([bow(sentence, words)])[0]\n # filter out predictions below a threshold\n results = [[i,r] for i,r in enumerate(results) if r>ERROR_THRESHOLD]\n # sort by strength of probability\n results.sort(key=lambda x: x[1], reverse=True)\n return_list = []\n for r in results:\n return_list.append((classes[r[0]], r[1]))\n # return tuple of intent and probability\n return return_list\n\ndef response(sentence, userID='123', show_details=False):\n results = classify(sentence)\n # if we have a classification then find the matching intent tag\n if results:\n # loop as long as there are matches to process\n while results:\n for i in intents['intents']:\n # find a tag matching the first result\n if i['tag'] == results[0][0]:\n # set context for this intent if necessary\n if 'context_set' in i:\n if show_details: print ('context:', i['context_set'])\n context[userID] = i['context_set']\n\n # check if this intent is contextual and applies to this user's conversation\n if not 'context_filter' in i or \\\n (userID in context and 'context_filter' in i and i['context_filter'] == context[userID]):\n if show_details: print ('tag:', i['tag'])\n # a random response from the intent\n return print(random.choice(i['responses']))\n\n results.pop(0)",
"_____no_output_____"
],
[
"response('Can you please let me know the delivery options?')",
"We have home delivery options through UBER Eats and Zomato\n"
],
[
"response('What is menu for today?')",
"You can check various delicacies given in the food menu at www.mymenu.com\n"
],
[
"context",
"_____no_output_____"
],
[
"response(\"Hi there!\", show_details=True)",
"context: \ntag: greeting\nGood to see you again\n"
],
[
"response('What is menu for today?')",
"Today's special is Chicken Tikka\n"
],
[
"",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
c5145bb42b5724f22c888b777c45c1d31b11a6e8
| 42,520 |
ipynb
|
Jupyter Notebook
|
.ipynb_checkpoints/consolidated codes-checkpoint.ipynb
|
rvs36/Network_Science_Anlytics
|
d88cf6f3e1d280fb47ab03eec46a36aa19646874
|
[
"MIT"
] | null | null | null |
.ipynb_checkpoints/consolidated codes-checkpoint.ipynb
|
rvs36/Network_Science_Anlytics
|
d88cf6f3e1d280fb47ab03eec46a36aa19646874
|
[
"MIT"
] | null | null | null |
.ipynb_checkpoints/consolidated codes-checkpoint.ipynb
|
rvs36/Network_Science_Anlytics
|
d88cf6f3e1d280fb47ab03eec46a36aa19646874
|
[
"MIT"
] | 1 |
2020-03-30T16:40:27.000Z
|
2020-03-30T16:40:27.000Z
| 36.497854 | 1,031 | 0.590028 |
[
[
[
"## Predicting Missing links in a citation network",
"_____no_output_____"
]
],
[
[
"# global imports \nimport random \nimport numpy as np \nimport pandas as pd\nimport jgraph ## this was previously known as igraph\nimport csv \nimport matplotlib.pyplot as plt\n\n# machine learning imports\nfrom sklearn import svm \nfrom sklearn.feature_extraction.text import TfidfVectorizer\nfrom sklearn.metrics.pairwise import linear_kernel \nfrom sklearn import preprocessing \n\nimport spacy",
"_____no_output_____"
]
],
[
[
"### Import datasets",
"_____no_output_____"
]
],
[
[
"# function to read data from txt files\nnodes_info_df = pd.read_csv('./data/node_information.csv')\nrandom_preds_df = pd.read_csv('./data/random_predictions.csv') \ntest_set = pd.read_csv('./data/testing_set.txt', sep = ' ', header = None)\ntrain_set = pd.read_csv('./data/training_set.txt', sep = ' ', header = None)\ntest_set.columns = ['source_id', 'target_id']\ntrain_set.columns = ['source_id', 'target_id', 'label']\nnodes_info_df.columns = ['paper_id', 'publication_year', 'title', 'author', 'journal_name', 'abstract']",
"_____no_output_____"
]
],
[
[
"## Exploratory Analysis",
"_____no_output_____"
]
],
[
[
"print('Unique papers: ', len(set(nodes_info_df['paper_id'])))\nsym_diff = set(test_set['source_id'].append(test_set['target_id'])).symmetric_difference(set(nodes_info_df['paper_id']))\nprint('Unknown papers in test set (with nodes_info):', len(sym_diff))",
"_____no_output_____"
],
[
"# # get distribution of journal names \n# nodes_info_df['journal_name'] = nodes_info_df['journal_name'].fillna('unknown')\n# nodes_info_df.journal_name.value_counts()[:15]",
"_____no_output_____"
],
[
"# nodes_info_df.author",
"_____no_output_____"
]
],
[
[
"## Feature generation",
"_____no_output_____"
]
],
[
[
"#Load Spacy\nimport en_core_web_sm\nspacy_nlp = en_core_web_sm.load(disable=[\"tagger\", \"parser\",\"ner\",\"entity_linker\",\"textcat\",\"entity_ruler\",\"sentencizer\",\"merge_noun_chunks\",\"merge_entities\",\"merge_subtokens\"])",
"_____no_output_____"
]
],
[
[
"### Text features generation ",
"_____no_output_____"
]
],
[
[
"import re \nimport math\nfrom sklearn.feature_extraction.text import TfidfVectorizer\nfrom sklearn.metrics.pairwise import cosine_similarity\nfrom sklearn.decomposition import PCA\n\ndef isNaN(string):\n return string != string\n\ndef filter_bad(alphabet):\n bad = [',', None]\n\n if(alphabet in bad):\n return False\n else:\n return True\n \n## possible formats of authors:\n# several authors: separation via ','\n# sometimes mentions the university eg '(montpellier)'\n# sometimes mentions the first name \n# sometimes format is: firstname letter. lastname\ndef author_normalisation(authors):\n if isNaN(authors) == False:\n #print(authors)\n authors = authors.lower()\n final_authors = list()\n \n # remove universities and last space\n if '(' in authors:\n authors = re.sub(r'\\(+.*\\)', '', authors).strip() \n \n # remove extra spaces\n authors = authors.split()\n authors = ' '.join(filter(filter_bad, authors))\n \n # get all authors of one paper \n for author in authors.split(', '): \n author.strip() \n # get the names of an author\n names = author.split(' ')\n author_names = list() \n if len(names) == 2:\n # check if first element is 'letter.' format:\n if re.match('\\w\\.', names[0]):\n author_names.append(names[0])\n else:\n author_names.append(names[0][0] + '.')\n\n if len(names) == 3:\n if re.match('\\w\\.', names[0]):\n author_names.append(names[0])\n else:\n author_names.append(names[0][0] + '.')\n\n # skip the second middle name\n if re.match('\\w\\.', names[1]):\n pass\n #author_names.append(names[1])\n #else:\n # author_names.append(names[1][0] + '.')\n\n author_names.append(names[-1])\n if len(author_names) > 1:\n author_names = ' '.join(author_names)\n else:\n author_names = author_names[0]\n # append last name\n final_authors.append(author_names)\n\n\n number_of_authors = len(final_authors)\n if number_of_authors == 0:\n return np.NaN\n return final_authors\n \n return np.NaN\n\ndef common_authors(string1, string2):\n if isNaN(string1):\n return False\n if isNaN(string2):\n return False\n \n #a_set = set(string1.split(','))\n #b_set = set(string2.split(','))\n a_set = set(string1)\n b_set = set(string2)\n \n if (a_set & b_set): \n return True \n else: \n return False\n \ndef number_common_authors(string1, string2):\n pass\n\n\ndef remove_special_characters(string):\n string = re.sub(\"([^\\w]|[\\d_])+\", \" \", string)\n return string\n\ndef tokenize(string): \n # Code to tokenize\n spacy_tokens = spacy_nlp(string)\n # Code to remove punctuation tokens and create string tokens\n string_tokens = [token.lemma_ for token in spacy_tokens if not token.is_punct if not token.is_stop] \n return string_tokens \n\ndef recombining_tokens_into_a_string(list_of_tokens):\n return \" \".join(list_of_tokens)\n\n\ndef create_tf_idf(column,tf_idf):\n #if tf_idf doesn't exist\n if tf_idf==None:\n #create a TfidfVectorizer object\n tf_idf = TfidfVectorizer()\n #Vectorize the sample text\n X_tfidf_sample = tf_idf.fit_transform(column)\n #if tf_idf already exist use the same for the test\n else:\n X_tfidf_sample = tf_idf.transform(column)\n return X_tfidf_sample,tf_idf\n\ndef tf_idf_feature(column,dataset,tf_idf,author_or_not):\n #Remove special characters from the text\n dataset[column]=dataset[column].apply(lambda x: remove_special_characters(x))\n #if we deal with the column author\n if author_or_not==1:\n # Remove strings of size less than two\n column_cleaned= dataset[column].str.findall('\\w{2,}').str.join(' ')\n else:\n #Tokenize, extract lemmas and remove stop words\n tokenized=dataset[column].apply(lambda x: tokenize(x)) \n #Recombine tokens into a string\n column_cleaned=tokenized.apply(recombining_tokens_into_a_string)\n # Create the tf_idf matrix \n tf_idf_matrix,tf_idf=create_tf_idf(column_cleaned,tf_idf)\n return tf_idf_matrix,tf_idf\n\n# Compute the similarity between a column target and source\ndef compute_similarity(column,df_source,df_target,author_or_not):\n #Fill the Na's\n df_source[column].fillna(\"unknown\", inplace=True)\n df_target[column].fillna(\"unknown\", inplace=True)\n tf_idf=None\n #Create the tf_idf features\n tf_idf_title_source,tf_idf=tf_idf_feature(column,df_source,tf_idf,author_or_not)\n tf_idf_title_target,tf_idf=tf_idf_feature(column,df_target,tf_idf,author_or_not)\n #Calculate the similarities\n similarity=[]\n for i in range(tf_idf_title_source.shape[0]):\n cos_sim=cosine_similarity(tf_idf_title_source[i], tf_idf_title_target[i])\n similarity.append(cos_sim)\n #Convert the list as a DataFrame\n similarity_df=pd.DataFrame(np.vstack(similarity))\n return similarity_df\n\ndef reduce_matrix_width(source_df,target_df,n_components):\n # Apply a PCA to reduce the matrix width , we chose 15\n pca_train = PCA(n_components=n_components)\n #PCA on source feature\n pca_train.fit(source_df)\n matrix_source_reduced = pca_train.transform(source_df)\n print(sum(pca_train.explained_variance_ratio_)) # Percentage of initial matrix explained by reduced matrix\n #PCA on target feature\n pca_train.fit(target_df)\n matrix_target_reduced = pca_train.transform(target_df)\n print(sum(pca_train.explained_variance_ratio_)) # Percentage of initial matrix explained by reduced matrix\n return matrix_source_reduced,matrix_target_reduced\n\ndef journal_name_feature():\n #We first merge train and test to avoid a different number of features when one-hot-encoding\n #To keep trace of the train and test dataset\n train_source_info['train_test']=1\n train_target_info['train_test']=1\n test_source_info['train_test']=0\n test_target_info['train_test']=0\n # merging the two datasets together\n combined_source=pd.concat([train_source_info,test_source_info],ignore_index=True)\n combined_target=pd.concat([train_target_info,test_target_info],ignore_index=True)\n # One hot encoding\n journal_name_encoded_source=pd.get_dummies(combined_source['journal_name'])\n journal_name_encoded_target=pd.get_dummies(combined_target['journal_name'])\n #Apply PCA to reduce matrix with 15 components\n journal_name_encoded_source_reduced,journal_name_encoded_target_reduced =reduce_matrix_width(journal_name_encoded_source,journal_name_encoded_target,15)\n # Merge encoded dataset with the combine dataset\n combined_source=pd.concat([combined_source,pd.DataFrame(journal_name_encoded_source_reduced)],axis=1)\n combined_target=pd.concat([combined_target,pd.DataFrame(journal_name_encoded_target_reduced)],axis=1)\n #Separate train and test and keep only journal_name features\n train_source_journal=combined_source[combined_source[\"train_test\"]==1].drop(['abstract','author','journal_name','label','paper_id','publication_year','source_id','target_id','title','train_test'], axis=1)\n test_source_journal=combined_source[combined_source[\"train_test\"]==0].drop(['abstract','author','journal_name','label','paper_id','publication_year','source_id','target_id','title','train_test'], axis=1)\n train_target_journal=combined_target[combined_target[\"train_test\"]==1].drop(['abstract','author','journal_name','label','paper_id','publication_year','source_id','target_id','title','train_test'], axis=1)\n test_target_journal=combined_target[combined_target[\"train_test\"]==0].drop(['abstract','author','journal_name','label','paper_id','publication_year','source_id','target_id','title','train_test'], axis=1)\n #add prefix to columns names\n train_source_journal.columns=[str(col) + '_source' for col in train_source_journal.columns]\n test_source_journal.columns=[str(col) + '_source' for col in test_source_journal.columns]\n train_target_journal.columns=[str(col) + '_target' for col in train_target_journal.columns]\n test_target_journal.columns=[str(col) + '_target' for col in test_target_journal.columns]\n return train_source_journal,test_source_journal,train_target_journal,test_target_journal\n ",
"_____no_output_____"
],
[
"# reaye source and target info datasets\ntrain_source_info = train_set.merge(nodes_info_df, left_on='source_id', right_on='paper_id',how=\"left\")\ntrain_target_info = train_set.merge(nodes_info_df, left_on='target_id', right_on='paper_id',how=\"left\")\n\ntest_source_info = test_set.merge(nodes_info_df, left_on='source_id', right_on='paper_id',how=\"left\")\ntest_target_info = test_set.merge(nodes_info_df, left_on='target_id', right_on='paper_id',how=\"left\")\n",
"_____no_output_____"
],
[
"## apply the features to training set \ntrain_set['source_authors'] = train_source_info.author.apply(lambda x: author_normalisation(x))\ntrain_set['target_authors'] = train_target_info.author.apply(lambda x: author_normalisation(x))\n\ntrain_set['publication_year_diff'] = train_source_info.publication_year - train_target_info.publication_year\n\ntrain_set['source_journal'] = train_source_info.journal_name\ntrain_set['target_journal'] = train_target_info.journal_name\n\ntrain_set['same_journal'] = train_set.apply(lambda x: int(x.source_journal == x.target_journal), axis=1)\n\n## apply the features to test set\ntest_set['source_authors'] = test_source_info.author.apply(lambda x: author_normalisation(x))\ntest_set['target_authors'] = test_target_info.author.apply(lambda x: author_normalisation(x))\n\ntest_set['publication_year_diff'] = test_source_info.publication_year - test_target_info.publication_year\n\ntest_set['source_journal'] = test_source_info.journal_name\ntest_set['target_journal'] = test_target_info.journal_name\ntest_set['same_journal'] = test_set.apply(lambda x: int(x.source_journal == x.target_journal), axis=1)\n",
"_____no_output_____"
],
[
"#other features this might take some times to run\n## apply the features to training set\ntrain_set['similarity_title']=compute_similarity(\"title\",train_source_info,train_target_info,0)\ntrain_set['similarity_abstract']=compute_similarity(\"abstract\",train_source_info,train_target_info,0)\ntrain_set['similarity_author']=compute_similarity(\"author\",train_source_info,train_target_info,1)\n\n## apply features to test set\ntest_set['similarity_title']=compute_similarity(\"title\",test_source_info,test_target_info,0)\ntest_set['similarity_abstract']=compute_similarity(\"abstract\",test_source_info,test_target_info,0)\ntest_set['similarity_author']=compute_similarity(\"author\",test_source_info,test_target_info,1)",
"_____no_output_____"
],
[
"#journal_name feature\ntrain_source_journal,test_source_journal,train_target_journal,test_target_journal =journal_name_feature()",
"_____no_output_____"
],
[
"#Add journal_name to the train and test\ntrain_set=pd.concat([train_set,train_source_journal],axis=1,)\ntrain_set=pd.concat([train_set,train_target_journal],axis=1)\ntest_set=pd.concat([test_set,test_source_journal.reset_index().drop([\"index\"],axis=1)],axis=1)\ntest_set=pd.concat([test_set,test_target_journal.reset_index().drop([\"index\"],axis=1)],axis=1)",
"_____no_output_____"
]
],
[
[
"### Graph features generation ",
"_____no_output_____"
]
],
[
[
"import networkx as nx \n# get some elements and then assign the attributes -> this is shite so ignore it \ndef shortest_path_info(some_graph, source, target):\n if source not in some_graph.nodes():\n return -1 # not known \n if target not in some_graph.nodes():\n return -1 # not known \n if nx.has_path(some_graph, source, target):\n return nx.dijkstra_path_length(some_graph, source=source, target=target)\n \n return -2 # no path\n\ndef degree_centrality(some_graph):\n degree_dict = dict(some_graph.degree(some_graph.nodes()))\n return degree_dict\n\ndef get_in_out_degree(some_graph):\n in_degree_dict = dict(some_graph.in_degree(some_graph.nodes()))\n out_degree_dict = dict(some_graph.out_degree(some_graph.nodes()))\n return in_degree_dict, out_degree_dict\n \n\ndef common_neighs(some_graph, x, y):\n if x not in some_graph.nodes():\n return 0,[] # not known \n if y not in some_graph.nodes():\n return 0,[] # not known\n neighs = sorted(list(nx.common_neighbors(some_graph, x, y)))\n return len(neighs), neighs\n\ndef jac_index(g, x, y):\n if x not in g.nodes():\n return -1 # not known \n if y not in g.nodes():\n return -1 # not known\n preds = nx.jaccard_coefficient(g, [(x, y)])\n jacc = 0\n\n for u, v, p in preds:\n jacc = p\n return jacc\n\ndef pref_attachement(g, x, y):\n if x not in g.nodes():\n return -1 # not known \n if y not in g.nodes():\n return -1 # not known\n preds = nx.preferential_attachment(g, [(x, y)])\n pref = 0\n\n for u, v, p in preds:\n pref = p\n return pref\n\ndef aa_index(g, x, y):\n if x not in g.nodes():\n return -1 # not known \n if y not in g.nodes():\n return -1 # not known\n preds = nx.adamic_adar_index(g, [(x, y)])\n aa = 0\n\n for u, v, p in preds:\n aa = p\n return aa\n\n",
"_____no_output_____"
],
[
"# create the network \n# get network for when there is a connection in train set\n# edges = list(zip(train_set.loc[train_set.label == 1].source_id, train_set.loc[train_set.label == 1].target_id))\n# nodes = list(set(train_set.source_id + train_set.target_id))\n\n# train_G = nx.DiGraph()\n# train_G.add_nodes_from(nodes)\n# train_G.add_edges_from(edges)\n\ntrain_G = nx.from_pandas_edgelist(train_set, source='source_id', target='target_id', edge_attr=None,\n create_using=nx.DiGraph())\n\n# make sure you also have an undirected graph\ntrain_G_ud = train_G.to_undirected()\n\n# create some dictionaries to use later on\nclustering_coeff_dict = nx.clustering(train_G_ud)\navg_neigh_degree_dict = nx.average_neighbor_degree(train_G)\nout_degree_centrality = nx.out_degree_centrality(train_G)\nin_degree_centrality = nx.in_degree_centrality(train_G)\npage_rank = nx.pagerank_scipy(train_G)\nhub_score, authority_score = nx.hits(train_G)",
"_____no_output_____"
],
[
"# function to get features for graph of a single element\ndef get_features(directed_graph, ud_graph, source_id, target_id, label):\n # features for undirected graph\n jaccard_index = jac_index(ud_graph, source_id, target_id)\n preferencial_attachment = pref_attachement(ud_graph, source_id, target_id)\n number_common_neighbours, common_neighbours = common_neighs(ud_graph, source_id, target_id)\n adamic_adar_index = aa_index(ud_graph, source_id, target_id)\n #shortest_path = shortest_path_info(train_G, source_id, target_id)\n\n \n source_pr = page_rank[source_id]\n source_hub_score = hub_score[source_id]\n source_authority_score = authority_score[source_id]\n source_cluster_coeff = clustering_coeff_dict[source_id]\n source_out_centrality = out_degree_centrality[source_id]\n source_avg_neigh_degree = avg_neigh_degree_dict[source_id]\n \n target_pr = page_rank[target_id]\n target_hub_score = hub_score[target_id]\n target_authority_score = authority_score[target_id]\n target_cluster_coeff = clustering_coeff_dict[target_id]\n target_in_centrality = in_degree_centrality[target_id]\n target_avg_neigh_degree = avg_neigh_degree_dict[target_id]\n\n # no name feature but supposedly important \n feature_n = source_out_centrality * target_in_centrality\n \n return [source_id, target_id, label, jaccard_index, preferencial_attachment, \n number_common_neighbours, adamic_adar_index, source_pr, target_pr, \n source_hub_score, target_hub_score, source_authority_score, \n target_authority_score, source_cluster_coeff, target_cluster_coeff, \n source_out_centrality, target_in_centrality, source_avg_neigh_degree, \n target_avg_neigh_degree, feature_n]\n ",
"_____no_output_____"
]
],
[
[
"### IMPORTANT: add column names when adding new features to the dataset ",
"_____no_output_____"
]
],
[
[
"### add columns when you add Features\ncolumn_names = ['source_id', 'target_id', 'label', 'jaccard_index', 'preferential_attachement', \n 'number_common_neighbours', 'adamic_adar_index', 'source_pr',\n 'target_pr', 'source_hub_score', 'target_hub_score', 'source_authority_score',\n 'target_authority_score', 'source_cluster_coeff', 'target_cluster_coeff',\n 'source_out_centrality', 'target_in_centrality', 'source_avg_neigh_degree', \n 'target_avg_neigh_degree', 'feature_n']\nfinal_train_set = pd.DataFrame([[np.nan]*len(column_names)]* train_set.shape[0], columns=column_names)\nfinal_test_set = pd.DataFrame([[np.nan]*len(column_names)]* test_set.shape[0], columns=column_names)",
"_____no_output_____"
],
[
"# create the features for the train set\nfor idx, row in train_set.iterrows():\n features = get_features(train_G, train_G_ud, row.source_id, row.target_id, row.label)\n #update the features\n final_train_set.loc[idx] = features",
"_____no_output_____"
],
[
"#create the features for the test set\nfor idx, row in test_set.iterrows():\n features = get_features(train_G, train_G_ud, row.source_id, row.target_id, -1)\n #update the features\n final_test_set.loc[idx] = features",
"_____no_output_____"
],
[
"# merge graph and text features together \ntrain_set = train_set.merge(final_train_set, on=['source_id', 'target_id', 'label'], how='left') \ntest_set = test_set.merge(final_test_set, on=['source_id', 'target_id'], how='left')",
"_____no_output_____"
],
[
"from networkx import betweenness_centrality\nfrom networkx import edge_betweenness_centrality\nfrom networkx import load_centrality\nfrom networkx import eigenvector_centrality\n\ndef graph_features(directed_graph, dataframe_dataset):\n # betweenness\n between_centrality = betweenness_centrality(directed_graph) # shortest-path betweenness centrality for nodes\n # load centrality\n ld_centrality = load_centrality(directed_graph) # load centrality of a node is the fraction of all shortest paths that pass through that node\n #eigenvector centrality\n eig_centrality = eigenvector_centrality(directed_graph)\n \n # save features to training set \n dataframe_dataset['betweeness_centrality'] = pd.DataFrame.from_dict(dict(eig_centrality), orient='index')\n dataframe_dataset['load_centrality'] = pd.DataFrame.from_dict(dict(ld_centrality), orient='index')\n dataframe_dataset['eigen_centrality'] = pd.DataFrame.from_dict(dict(eig_centrality), orient='index')\n\n return dataframe_dataset",
"_____no_output_____"
],
[
"train_set = graph_features(train_G, train_set)\ntrain_set.betweeness_centrality.fillna(-1, inplace=True)\ntrain_set.load_centrality.fillna(-1, inplace=True)\ntrain_set.eigen_centrality.fillna(-1, inplace=True)\n\ntest_set = graph_features(train_G, test_set)\ntest_set.betweeness_centrality.fillna(-1, inplace=True)\ntest_set.load_centrality.fillna(-1, inplace=True)\ntest_set.eigen_centrality.fillna(-1, inplace=True)",
"_____no_output_____"
],
[
"# write out so that you do not have to run everything again\ntrain_set.to_csv('final_train.csv',index=False)\ntest_set.to_csv('final_test.csv', index=False)",
"_____no_output_____"
]
],
[
[
"### Can start from here as well when features were saved previously",
"_____no_output_____"
]
],
[
[
"test_set = pd.read_csv('final_test.csv')\ntrain_set = pd.read_csv('final_train.csv')",
"_____no_output_____"
]
],
[
[
"### Final clean (i.e replacing nans etc)",
"_____no_output_____"
]
],
[
[
"# fill nas in some way\ntrain_set.publication_year_diff.fillna(-24, inplace=True) # 24 is for unknown (?)\ntrain_set.fillna('unknown', inplace=True)\n\ntest_set.publication_year_diff.fillna(-24, inplace=True) # 24 is for unknown (?_)\ntest_set.fillna('unknown', inplace=True)",
"_____no_output_____"
],
[
"test_set.head()",
"_____no_output_____"
],
[
"train_set.head()",
"_____no_output_____"
],
[
"# check the types of each column (none should be object)\ntrain_set.dtypes",
"_____no_output_____"
],
[
"%matplotlib inline\n## Most interesting correlation is with label\nimport seaborn as sns\nimport matplotlib.pyplot as plt\n\nplt.figure(figsize=(14,12))\nsns.heatmap(train_set.corr(),\n vmax=0.5,\n square=True,\n annot=True)\n",
"_____no_output_____"
]
],
[
[
"## Learning Stuff",
"_____no_output_____"
]
],
[
[
"# separate features from labels:\nX = train_set.loc[:, (train_set.columns != 'label') & \n (train_set.columns != 'common_authors') & \n (train_set.columns != 'source_authors') & \n (train_set.columns != 'target_authors') & \n (train_set.columns != 'source_journal') & \n (train_set.columns != 'target_journal') \n ]\ny = train_set['label']\ny.astype(np.int)\n",
"_____no_output_____"
],
[
"# final feature correlation\nff = X.copy()\nff['label'] = y\nplt.figure(figsize=(14,12))\nsns.heatmap(X.corr(),\n vmax=0.5,\n square=True,\n annot=True)",
"_____no_output_____"
],
[
"## Train different models and compare the performance \nfrom sklearn.tree import DecisionTreeClassifier\nfrom sklearn.ensemble import AdaBoostClassifier, RandomForestClassifier, BaggingClassifier, GradientBoostingClassifier\nfrom sklearn.metrics import f1_score, confusion_matrix\nfrom sklearn.model_selection import cross_validate\n\nmodel = AdaBoostClassifier(n_estimators=75, learning_rate=1)\nscores = cross_validate(model, X, y, scoring='f1', \n cv=5) # n_jobs is the number of cpus to use -1 => all\nscores",
"_____no_output_____"
],
[
"# describe results from scores\nfrom scipy import stats \nstats.describe(scores['test_score'])",
"_____no_output_____"
],
[
"model = RandomForestClassifier()\nscores = cross_validate(model, X, y, scoring='f1', \n cv=5) # n_jobs is the number of cpus to use -1 => all\nscores",
"_____no_output_____"
],
[
"# describe results from scores\nfrom scipy import stats \nstats.describe(scores['test_score'])",
"_____no_output_____"
]
],
[
[
"### Recursive feature selection ",
"_____no_output_____"
]
],
[
[
"# ## ONLY RUN AT THE END FOR GRAPHS.. takes a v.long time to execute (been 3hours for now.. only execute on a virtual \n# # machine with GPUs (if possible))\n# from sklearn.feature_selection import RFECV\n\n# clf_rf_4 = model\n# rfecv = RFECV(estimator=clf_rf_4, step=1, cv=10,scoring='f1') #10-fold cross-validation\n# rfecv = rfecv.fit(X, y)\n\n# print('Optimal number of features :', rfecv.n_features_)\n# print('Best features :', X.columns[rfecv.support_])",
"_____no_output_____"
],
[
"# Plot number of features VS. cross-validation scores\nimport matplotlib.pyplot as plt\nplt.figure()\nplt.xlabel(\"Number of features selected\")\nplt.ylabel(\"Cross validation score of number of selected features\")\nplt.plot(range(1, len(rfecv.grid_scores_) + 1), rfecv.grid_scores_)\nplt.show()",
"_____no_output_____"
]
],
[
[
"## prior to authors:\nDescribeResult(nobs=10, minmax=(0.7092423428264374, 0.7505859928392963), mean=0.7330286516063008, variance=0.0002449243278408503, skewness=-0.16892931758355367, kurtosis=-1.5003847605685021)\n\nafter some basic graphs:\nDescribeResult(nobs=10, minmax=(0.9537111539570966, 0.9556853523477206), mean=0.9544708719147975, variance=4.3393884483164826e-07, skewness=0.7947367347642024, kurtosis=-0.6317507457312379)",
"_____no_output_____"
],
[
"### Comparing models",
"_____no_output_____"
],
[
"## 1. XG Boost\n\n1.1 XGboost base model",
"_____no_output_____"
]
],
[
[
"from xgboost.sklearn import XGBClassifier\n\n# making sure the test and the train files have same sequence of columns\n\ntest = test[X.columns]\n\n\n# defining the base model\nxgb_model_base = XGBClassifier(n_estimators = 100)\n\n# printing the cross validation scores for the classifier\nscores = cross_validate(xgb_model_base, X, y.values.ravel(), scoring='f1', \n cv=3,n_jobs = -1 ) # n_jobs is the number of cpus to use -1 => all\nscores\n\n\n# fitting on the training data\nxgb_model_base.fit(X, y.values.ravel())\n\n# predicting the outcome from the final \npredictions = xgb_model_base.predict(test)\n\n# write out\nout_df = test_set.copy()\ndata = {'id': list(out_df.index), 'category': predictions}\nfinal_df = pd.DataFrame(data)\n\n\n# 3: write file out\nfinal_df.to_csv('submission.csv',index=False, sep=',')",
"_____no_output_____"
]
],
[
[
"1.2 XgBosst with random search",
"_____no_output_____"
]
],
[
[
"# defining the search grid\n\nrandom_grid = {\n \"n_estimators\" : [int(x) for x in np.linspace(50, 600, num = 20)],\n \"learning_rate\" : [0.01, 0.02, 0.05, 0.10 ] ,\n \"max_depth\" : [ 6, 8, 10, 12, 15, 20],\n \"min_child_weight\" : [ 1, 3, 5, 7 ],\n \"gamma\" : [ 0.3, 0.4, 0.7, 0.9 ],\n \"colsample_bytree\" : [ 0.05, 0.1, 0.3, 0.4] }\n\n# Use the random grid to search for best hyperparameters\n\n# First create the base model to tune\nxgb_model = XGBClassifier()\n\n# Random search of parameters\nxgb_random = RandomizedSearchCV(estimator = xgb_model, param_distributions = random_grid,\nn_iter = 10, cv = 3, verbose=2, random_state=42 ,n_jobs = -1, scoring = 'f1_weighted')\n\noptimised_xgb_random = xgb_random.best_estimator_\n\n\n\n# printing the cross validation scores for the classifier\nscores = cross_validate(optimised_xgb_random, X, y.values.ravel(), scoring='f1', \n cv=3,n_jobs = -1 ) # n_jobs is the number of cpus to use -1 => all\nscores\n\n\n# fitting on the training data\nxgb_model_base.fit(X, y.values.ravel())\n\n# predicting the outcome from the final \noptimised_xgb_random.predict(test)\n\n# write out\nout_df = test_set.copy()\ndata = {'id': list(out_df.index), 'category': predictions}\nfinal_df = pd.DataFrame(data)\n\n\n# 3: write file out\nfinal_df.to_csv('submission.csv',index=False, sep=',')",
"_____no_output_____"
]
],
[
[
"## 2. Support Vector Machine",
"_____no_output_____"
]
],
[
[
"from sklearn.svm import LinearSVC\n\n# SVM has a zero tolerance towards null values, hence replacing them by 0\n\nXVM = X.fillna(value=0)\ntest_SVM = test.fillna(value=0)\n\nclf = LinearSVC( tol=1e-4)\n\n\n# printing the cross validation scores for the classifier\nscores = cross_validate(clf, XVM, y, scoring='f1', \n cv=10,n_jobs = -1 ) # n_jobs is the number of cpus to use -1 => all\nscores\n\n\n# fitting on the training data\nclf.fit(XVM, y)\n\n# predicting the outcome from the final \nprediction_clf = clf.predict(test_SVM)\n\n# write out\nout_df = test_set.copy()\ndata = {'id': list(out_df.index), 'category': predictions}\nfinal_df = pd.DataFrame(data)\n\n\n# 3: write file out\nfinal_df.to_csv('submission.csv',index=False, sep=',')",
"_____no_output_____"
]
],
[
[
"## 3. Random Forest",
"_____no_output_____"
]
],
[
[
"from sklearn.ensemble import RandomForestClassifier\n# 1: retrain the complete model -> don't forget to change this to optimal one @ end\nfinal_model = RandomForestClassifier()\nfinal_model.fit(X, y)",
"_____no_output_____"
],
[
"# 2: predict on the test set\nfinal_test_set = test_set.loc[:, (test_set.columns != 'source_authors') & (test_set.columns != 'common_authors') & (test_set.columns != 'target_authors')& (test_set.columns != 'label')& (test_set.columns != 'source_journal') & (test_set.columns != 'target_journal')]\npredictions = final_model.predict(final_test_set)\n\n# write out\nout_df = test_set.copy()\ndata = {'id': list(out_df.index), 'category': predictions}\nfinal_df = pd.DataFrame(data)\n\n\n# 3: write file out\nfinal_df.to_csv('submission.csv',index=False, sep=',')",
"_____no_output_____"
],
[
"# plot the feature importance\nfeat_importances = pd.Series(final_model.feature_importances_, index=X.columns)\nfeat_importances.nlargest(10).plot(kind='barh')\nplt.show()",
"_____no_output_____"
]
],
[
[
"## The end",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
]
] |
c51467c8ca5d63959514640a1d93ea31a4021749
| 9,967 |
ipynb
|
Jupyter Notebook
|
first.ipynb
|
DiptoChakrabarty/Data-Science-with-Ml
|
7fad91eec275935e26edc171c6a2169b79c44f42
|
[
"MIT"
] | 4 |
2019-03-11T05:30:32.000Z
|
2019-04-04T08:31:36.000Z
|
first.ipynb
|
DiptoChakrabarty/Data-Science-with-Ml
|
7fad91eec275935e26edc171c6a2169b79c44f42
|
[
"MIT"
] | null | null | null |
first.ipynb
|
DiptoChakrabarty/Data-Science-with-Ml
|
7fad91eec275935e26edc171c6a2169b79c44f42
|
[
"MIT"
] | null | null | null | 4,983.5 | 9,966 | 0.574696 |
[
[
[
"empty"
]
]
] |
[
"empty"
] |
[
[
"empty"
]
] |
c5147dfd33ad67a0efe432ebba25b9616cc314ab
| 181,572 |
ipynb
|
Jupyter Notebook
|
Model backlog/Train/123-jigsaw-fold1-xlm-roberta-ratio-1-2-optimizers.ipynb
|
dimitreOliveira/Jigsaw-Multilingual-Toxic-Comment-Classification
|
44422e6aeeff227e22dbb5c05101322e9d4aabbe
|
[
"MIT"
] | 4 |
2020-06-23T02:31:07.000Z
|
2020-07-04T11:50:08.000Z
|
Model backlog/Train/123-jigsaw-fold1-xlm-roberta-ratio-1-2-optimizers.ipynb
|
dimitreOliveira/Jigsaw-Multilingual-Toxic-Comment-Classification
|
44422e6aeeff227e22dbb5c05101322e9d4aabbe
|
[
"MIT"
] | null | null | null |
Model backlog/Train/123-jigsaw-fold1-xlm-roberta-ratio-1-2-optimizers.ipynb
|
dimitreOliveira/Jigsaw-Multilingual-Toxic-Comment-Classification
|
44422e6aeeff227e22dbb5c05101322e9d4aabbe
|
[
"MIT"
] | null | null | null | 127.418947 | 97,252 | 0.826273 |
[
[
[
"## Dependencies",
"_____no_output_____"
]
],
[
[
"import json, warnings, shutil, glob\nfrom jigsaw_utility_scripts import *\nfrom scripts_step_lr_schedulers import *\nfrom transformers import TFXLMRobertaModel, XLMRobertaConfig\nfrom tensorflow.keras.models import Model\nfrom tensorflow.keras import optimizers, metrics, losses, layers\n\nSEED = 0\nseed_everything(SEED)\nwarnings.filterwarnings(\"ignore\")\npd.set_option('max_colwidth', 120)\npd.set_option('display.float_format', lambda x: '%.4f' % x)",
"\u001b[34m\u001b[1mwandb\u001b[0m: \u001b[33mWARNING\u001b[0m W&B installed but not logged in. Run `wandb login` or set the WANDB_API_KEY env variable.\n"
]
],
[
[
"## TPU configuration",
"_____no_output_____"
]
],
[
[
"strategy, tpu = set_up_strategy()\nprint(\"REPLICAS: \", strategy.num_replicas_in_sync)\nAUTO = tf.data.experimental.AUTOTUNE",
"Running on TPU grpc://10.0.0.2:8470\nREPLICAS: 8\n"
]
],
[
[
"# Load data",
"_____no_output_____"
]
],
[
[
"database_base_path = '/kaggle/input/jigsaw-data-split-roberta-192-ratio-1-clean-polish/'\nk_fold = pd.read_csv(database_base_path + '5-fold.csv')\nvalid_df = pd.read_csv(\"/kaggle/input/jigsaw-multilingual-toxic-comment-classification/validation.csv\", \n usecols=['comment_text', 'toxic', 'lang'])\n\nprint('Train samples: %d' % len(k_fold))\ndisplay(k_fold.head())\nprint('Validation samples: %d' % len(valid_df))\ndisplay(valid_df.head())\n\nbase_data_path = 'fold_1/'\nfold_n = 1\n# Unzip files\n!tar -xf /kaggle/input/jigsaw-data-split-roberta-192-ratio-1-clean-polish/fold_1.tar.gz",
"Train samples: 267220\n"
]
],
[
[
"# Model parameters",
"_____no_output_____"
]
],
[
[
"base_path = '/kaggle/input/jigsaw-transformers/XLM-RoBERTa/'\n\nconfig = {\n \"MAX_LEN\": 192,\n \"BATCH_SIZE\": 128,\n \"EPOCHS\": 3,\n \"LEARNING_RATE\": 1e-5, \n \"ES_PATIENCE\": None,\n \"base_model_path\": base_path + 'tf-xlm-roberta-large-tf_model.h5',\n \"config_path\": base_path + 'xlm-roberta-large-config.json'\n}\n\nwith open('config.json', 'w') as json_file:\n json.dump(json.loads(json.dumps(config)), json_file)\n \nconfig",
"_____no_output_____"
]
],
[
[
"## Learning rate schedule",
"_____no_output_____"
]
],
[
[
"lr_min = 1e-7\nlr_start = 0\nlr_max = config['LEARNING_RATE']\nstep_size = (len(k_fold[k_fold[f'fold_{fold_n}'] == 'train']) * 2) // config['BATCH_SIZE']\ntotal_steps = config['EPOCHS'] * step_size\nhold_max_steps = 0\nwarmup_steps = total_steps * 0.1\ndecay = .9998\n\nrng = [i for i in range(0, total_steps, config['BATCH_SIZE'])]\ny = [exponential_schedule_with_warmup(tf.cast(x, tf.float32), warmup_steps=warmup_steps, \n hold_max_steps=hold_max_steps, lr_start=lr_start, \n lr_max=lr_max, lr_min=lr_min, decay=decay) for x in rng]\n\nsns.set(style=\"whitegrid\")\nfig, ax = plt.subplots(figsize=(20, 6))\nplt.plot(rng, y)\nprint(\"Learning rate schedule: {:.3g} to {:.3g} to {:.3g}\".format(y[0], max(y), y[-1]))",
"Learning rate schedule: 0 to 9.96e-06 to 1.66e-06\n"
]
],
[
[
"# Model",
"_____no_output_____"
]
],
[
[
"module_config = XLMRobertaConfig.from_pretrained(config['config_path'], output_hidden_states=False)\n\ndef model_fn(MAX_LEN):\n input_ids = layers.Input(shape=(MAX_LEN,), dtype=tf.int32, name='input_ids')\n attention_mask = layers.Input(shape=(MAX_LEN,), dtype=tf.int32, name='attention_mask')\n \n base_model = TFXLMRobertaModel.from_pretrained(config['base_model_path'], config=module_config)\n last_hidden_state, _ = base_model({'input_ids': input_ids, 'attention_mask': attention_mask})\n \n cls_token = last_hidden_state[:, 0, :]\n \n output = layers.Dense(1, activation='sigmoid', name='output')(cls_token)\n \n model = Model(inputs=[input_ids, attention_mask], outputs=output)\n \n return model",
"_____no_output_____"
]
],
[
[
"# Train",
"_____no_output_____"
]
],
[
[
"# Load data\nx_train = np.load(base_data_path + 'x_train.npy')\ny_train = np.load(base_data_path + 'y_train_int.npy').reshape(x_train.shape[1], 1).astype(np.float32)\nx_valid = np.load(base_data_path + 'x_valid.npy')\ny_valid = np.load(base_data_path + 'y_valid_int.npy').reshape(x_valid.shape[1], 1).astype(np.float32)\nx_valid_ml = np.load(database_base_path + 'x_valid.npy')\ny_valid_ml = np.load(database_base_path + 'y_valid.npy').reshape(x_valid_ml.shape[1], 1).astype(np.float32)\n\n#################### ADD TAIL ####################\nx_train_tail = np.load(base_data_path + 'x_train_tail.npy')\ny_train_tail = np.load(base_data_path + 'y_train_int_tail.npy').reshape(x_train_tail.shape[1], 1).astype(np.float32)\nx_train = np.hstack([x_train, x_train_tail])\ny_train = np.vstack([y_train, y_train_tail])\n\nstep_size = x_train.shape[1] // config['BATCH_SIZE']\nvalid_step_size = x_valid_ml.shape[1] // config['BATCH_SIZE']\nvalid_2_step_size = x_valid.shape[1] // config['BATCH_SIZE']\n\n# Build TF datasets\ntrain_dist_ds = strategy.experimental_distribute_dataset(get_training_dataset(x_train, y_train, config['BATCH_SIZE'], AUTO, seed=SEED))\nvalid_dist_ds = strategy.experimental_distribute_dataset(get_validation_dataset(x_valid_ml, y_valid_ml, config['BATCH_SIZE'], AUTO, repeated=True, seed=SEED))\nvalid_2_dist_ds = strategy.experimental_distribute_dataset(get_validation_dataset(x_valid, y_valid, config['BATCH_SIZE'], AUTO, repeated=True, seed=SEED))\ntrain_data_iter = iter(train_dist_ds)\nvalid_data_iter = iter(valid_dist_ds)\nvalid_2_data_iter = iter(valid_2_dist_ds)",
"_____no_output_____"
],
[
"# Step functions\[email protected]\ndef train_step(data_iter):\n def train_step_fn(x, y):\n with tf.GradientTape() as tape:\n probabilities = model(x, training=True)\n loss = loss_fn(y, probabilities)\n grads = tape.gradient(loss, model.trainable_variables)\n optimizer.apply_gradients(zip(grads, model.trainable_variables))\n train_auc.update_state(y, probabilities)\n train_loss.update_state(loss)\n for _ in tf.range(step_size):\n strategy.experimental_run_v2(train_step_fn, next(data_iter))\n\[email protected]\ndef valid_step(data_iter):\n def valid_step_fn(x, y):\n probabilities = model(x, training=False)\n loss = loss_fn(y, probabilities)\n valid_auc.update_state(y, probabilities)\n valid_loss.update_state(loss)\n for _ in tf.range(valid_step_size):\n strategy.experimental_run_v2(valid_step_fn, next(data_iter))\n\[email protected]\ndef valid_2_step(data_iter):\n def valid_step_fn(x, y):\n probabilities = model(x, training=False)\n loss = loss_fn(y, probabilities)\n valid_2_auc.update_state(y, probabilities)\n valid_2_loss.update_state(loss)\n for _ in tf.range(valid_2_step_size):\n strategy.experimental_run_v2(valid_step_fn, next(data_iter))",
"_____no_output_____"
],
[
"# Train model\nwith strategy.scope():\n model = model_fn(config['MAX_LEN'])\n \n lr = lambda: exponential_schedule_with_warmup(tf.cast(optimizer.iterations, tf.float32), \n warmup_steps=warmup_steps, hold_max_steps=hold_max_steps, \n lr_start=lr_start, lr_max=lr_max, lr_min=lr_min, decay=decay)\n \n optimizer = optimizers.Adam(learning_rate=lr)\n loss_fn = losses.binary_crossentropy\n train_auc = metrics.AUC()\n valid_auc = metrics.AUC()\n valid_2_auc = metrics.AUC()\n train_loss = metrics.Sum()\n valid_loss = metrics.Sum()\n valid_2_loss = metrics.Sum()\n\nmetrics_dict = {'loss': train_loss, 'auc': train_auc, \n 'val_loss': valid_loss, 'val_auc': valid_auc, \n 'val_2_loss': valid_2_loss, 'val_2_auc': valid_2_auc}\n\nhistory = custom_fit_2(model, metrics_dict, train_step, valid_step, valid_2_step, train_data_iter, \n valid_data_iter, valid_2_data_iter, step_size, valid_step_size, valid_2_step_size, \n config['BATCH_SIZE'], config['EPOCHS'], config['ES_PATIENCE'], save_last=False)\n# model.save_weights('model.h5')\n\n# Make predictions\n# x_train = np.load(base_data_path + 'x_train.npy')\n# x_valid = np.load(base_data_path + 'x_valid.npy')\nx_valid_ml_eval = np.load(database_base_path + 'x_valid.npy')\n\n# train_preds = model.predict(get_test_dataset(x_train, config['BATCH_SIZE'], AUTO))\n# valid_preds = model.predict(get_test_dataset(x_valid, config['BATCH_SIZE'], AUTO))\nvalid_ml_preds = model.predict(get_test_dataset(x_valid_ml_eval, config['BATCH_SIZE'], AUTO))\n\n# k_fold.loc[k_fold[f'fold_{fold_n}'] == 'train', f'pred_{fold_n}'] = np.round(train_preds)\n# k_fold.loc[k_fold[f'fold_{fold_n}'] == 'validation', f'pred_{fold_n}'] = np.round(valid_preds)\nvalid_df[f'pred_{fold_n}'] = valid_ml_preds",
"Train for 3340 steps, validate for 62 steps, validate_2 for 417 steps\n\nEPOCH 1/3\ntime: 1076.0s loss: 0.2593 auc: 0.9588 val_loss: 0.3138 val_auc: 0.9239 val_2_loss: 0.2067 val_2_auc: 0.9759\n\nEPOCH 2/3\ntime: 918.1s loss: 0.1674 auc: 0.9817 val_loss: 0.3362 val_auc: 0.9157 val_2_loss: 0.2212 val_2_auc: 0.9755\n\nEPOCH 3/3\ntime: 918.2s loss: 0.1349 auc: 0.9877 val_loss: 0.3602 val_auc: 0.9094 val_2_loss: 0.2385 val_2_auc: 0.9735\nTraining finished\n"
],
[
"# Fine-tune on validation set\n#################### ADD TAIL ####################\nx_valid_ml_tail = np.hstack([x_valid_ml, np.load(database_base_path + 'x_valid_tail.npy')])\ny_valid_ml_tail = np.vstack([y_valid_ml, y_valid_ml])\n\nvalid_step_size_tail = x_valid_ml_tail.shape[1] // config['BATCH_SIZE']\n\n# Build TF datasets\ntrain_ml_dist_ds = strategy.experimental_distribute_dataset(get_training_dataset(x_valid_ml_tail, y_valid_ml_tail, config['BATCH_SIZE'], AUTO, seed=SEED))\ntrain_ml_data_iter = iter(train_ml_dist_ds)",
"_____no_output_____"
],
[
"# Step functions\[email protected]\ndef train_ml_step(data_iter):\n def train_step_fn(x, y):\n with tf.GradientTape() as tape:\n probabilities = model(x, training=True)\n loss = loss_fn(y, probabilities)\n grads = tape.gradient(loss, model.trainable_variables)\n optimizer.apply_gradients(zip(grads, model.trainable_variables))\n train_auc.update_state(y, probabilities)\n train_loss.update_state(loss)\n for _ in tf.range(valid_step_size_tail):\n strategy.experimental_run_v2(train_step_fn, next(data_iter))",
"_____no_output_____"
],
[
"# Fine-tune on validation set\noptimizer = optimizers.Adam(learning_rate=config['LEARNING_RATE'])\n\nhistory_ml = custom_fit_2(model, metrics_dict, train_ml_step, valid_step, valid_2_step, train_ml_data_iter, \n valid_data_iter, valid_2_data_iter, valid_step_size_tail, valid_step_size, valid_2_step_size, \n config['BATCH_SIZE'], 2, config['ES_PATIENCE'], save_last=False)\n\n# Join history\nfor key in history_ml.keys():\n history[key] += history_ml[key]\n \nmodel.save_weights('model.h5')\n\n# Make predictions\nvalid_ml_preds = model.predict(get_test_dataset(x_valid_ml_eval, config['BATCH_SIZE'], AUTO))\nvalid_df[f'pred_ml_{fold_n}'] = valid_ml_preds\n\n### Delete data dir\nshutil.rmtree(base_data_path)",
"Train for 125 steps, validate for 62 steps, validate_2 for 417 steps\n\nEPOCH 1/2\ntime: 202.1s loss: 0.1897 auc: 0.9532 val_loss: 0.1017 val_auc: 0.9899 val_2_loss: 0.2474 val_2_auc: 0.9668\n\nEPOCH 2/2\ntime: 69.6s loss: 0.0937 auc: 0.9889 val_loss: 0.0268 val_auc: 0.9996 val_2_loss: 0.3438 val_2_auc: 0.9581\nTraining finished\n"
]
],
[
[
"## Model loss graph",
"_____no_output_____"
]
],
[
[
"plot_metrics_2(history)",
"_____no_output_____"
]
],
[
[
"# Model evaluation",
"_____no_output_____"
]
],
[
[
"# display(evaluate_model_single_fold(k_fold, fold_n, label_col='toxic_int').style.applymap(color_map))",
"_____no_output_____"
]
],
[
[
"# Confusion matrix",
"_____no_output_____"
]
],
[
[
"# train_set = k_fold[k_fold[f'fold_{fold_n}'] == 'train']\n# validation_set = k_fold[k_fold[f'fold_{fold_n}'] == 'validation'] \n# plot_confusion_matrix(train_set['toxic_int'], train_set[f'pred_{fold_n}'], \n# validation_set['toxic_int'], validation_set[f'pred_{fold_n}'])",
"_____no_output_____"
]
],
[
[
"# Model evaluation by language",
"_____no_output_____"
]
],
[
[
"display(evaluate_model_single_fold_lang(valid_df, fold_n).style.applymap(color_map))\n# ML fine-tunned preds\ndisplay(evaluate_model_single_fold_lang(valid_df, fold_n, pred_col='pred_ml').style.applymap(color_map))",
"_____no_output_____"
]
],
[
[
"# Visualize predictions",
"_____no_output_____"
]
],
[
[
"print('English validation set')\ndisplay(k_fold[['comment_text', 'toxic'] + [c for c in k_fold.columns if c.startswith('pred')]].head(10))\n\nprint('Multilingual validation set')\ndisplay(valid_df[['comment_text', 'toxic'] + [c for c in valid_df.columns if c.startswith('pred')]].head(10))",
"English validation set\n"
]
],
[
[
"# Test set predictions",
"_____no_output_____"
]
],
[
[
"x_test = np.load(database_base_path + 'x_test.npy')\ntest_preds = model.predict(get_test_dataset(x_test, config['BATCH_SIZE'], AUTO))",
"_____no_output_____"
],
[
"submission = pd.read_csv('/kaggle/input/jigsaw-multilingual-toxic-comment-classification/sample_submission.csv')\nsubmission['toxic'] = test_preds\nsubmission.to_csv('submission.csv', index=False)\n\ndisplay(submission.describe())\ndisplay(submission.head(10))",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
c5147f1d08596bedc0fb561efc9ed08427f4f0d3
| 287,169 |
ipynb
|
Jupyter Notebook
|
notebooks/Programacion/clf_cluster.ipynb
|
123972/PCA-nutricion
|
aff3c51a71c887c3fa367dbf9d599be5915c80cc
|
[
"MIT"
] | null | null | null |
notebooks/Programacion/clf_cluster.ipynb
|
123972/PCA-nutricion
|
aff3c51a71c887c3fa367dbf9d599be5915c80cc
|
[
"MIT"
] | 2 |
2021-05-11T16:00:55.000Z
|
2021-08-23T20:45:22.000Z
|
notebooks/Programacion/clf_cluster.ipynb
|
123972/PCA-nutricion
|
aff3c51a71c887c3fa367dbf9d599be5915c80cc
|
[
"MIT"
] | null | null | null | 186.958984 | 72,628 | 0.880757 |
[
[
[
"import pandas as pd\nimport numpy as np\nimport statsmodels.api as sm\nimport matplotlib.pyplot as plt\nimport pandas.util.testing as tm\nfrom sklearn.preprocessing import LabelEncoder\nfrom sklearn.tree import DecisionTreeClassifier\nfrom sklearn.model_selection import cross_val_score\nfrom sklearn.model_selection import train_test_split\nfrom sklearn import metrics\nfrom sklearn.metrics import accuracy_score, precision_score, recall_score\nfrom sklearn.preprocessing import StandardScaler\nfrom sklearn.metrics import classification_report\n\nimport seaborn as sns\nimport sys\n\nsys.path.append('./../../')\n\n# OPTIONAL: Load the \"autoreload\" extension so that code can change\n%load_ext autoreload\n\n#Own Library modules\nimport src.pca\n\n# OPTIONAL: always reload modules so that as you change code in src, it gets loaded\n%autoreload 2\n\nfrom src.pca import PCA_from_sklearn",
"C:\\Users\\Elizabeth\\Anaconda3\\lib\\site-packages\\statsmodels\\tools\\_testing.py:19: FutureWarning: pandas.util.testing is deprecated. Use the functions in the public API at pandas.testing instead.\n import pandas.util.testing as tm\n"
]
],
[
[
"#### 1. Árbol de decisión para clasificación",
"_____no_output_____"
],
[
"##### 1.1. Usando los 7 componentes principales que se obtuvieron con el criterio de eigenvalue ",
"_____no_output_____"
]
],
[
[
"#Se cargan los datos iniciales para obtener la target que es FoodGroup\ndf = pd.read_csv('../../data/nndb_flat.csv', encoding = \"L1\") ",
"_____no_output_____"
],
[
"df_1 = pd.DataFrame(df.FoodGroup)",
"_____no_output_____"
],
[
"df_1.FoodGroup.value_counts(dropna=False)",
"_____no_output_____"
],
[
"##Se codifica la variable FoodGroup usando LabelEncoder de sklearn\nlabelencoder = LabelEncoder()\ndf_1['FoodGroup'] = labelencoder.fit_transform(df_1.FoodGroup)",
"_____no_output_____"
],
[
"df_1.FoodGroup.value_counts(dropna=False)",
"_____no_output_____"
],
[
"##Se cargan los 7 componentes principales que se usaran en este análisis\ncomponentesprincipales_analisis_post = pd.read_csv('../../results/data_results/componentesprincipales_analisis_post.csv') ",
"_____no_output_____"
],
[
"classifier = DecisionTreeClassifier(random_state = 0)",
"_____no_output_____"
],
[
"classifier",
"_____no_output_____"
],
[
"###Se divide la muestra en entrenamiento y prueba, en 60% y 40%, respectivamente.\nX_train, X_test, y_train, y_test = train_test_split(componentesprincipales_analisis_post, df_1.FoodGroup, test_size=0.4, random_state=0)",
"_____no_output_____"
],
[
"###Desplegar el detalle de entrenamiento y prueba\nprint( \"Predictor - Training : \", X_train.shape, \"Predictor - Testing : \", X_test.shape )",
"Predictor - Training : (5170, 7) Predictor - Testing : (3448, 7)\n"
],
[
"###Se entrena el modelo\nclassifier = classifier.fit(X_train, y_train)",
"_____no_output_____"
],
[
"y_pred = pd.DataFrame(classifier.predict(X_test))",
"_____no_output_____"
],
[
"probs = classifier.predict_proba(X_test) ",
"_____no_output_____"
]
],
[
[
"- Métricas para evaluar el modelo",
"_____no_output_____"
]
],
[
[
"print(\"Accuracy:\",metrics.accuracy_score(y_test, y_pred))",
"Accuracy: 0.6244199535962877\n"
],
[
"print(\"Precision:\",metrics.precision_score(y_test, y_pred, average='macro'))",
"Precision: 0.5673195381729577\n"
],
[
"print(\"Recall:\",metrics.recall_score(y_test, y_pred, average='macro'))",
"Recall: 0.5557755290618943\n"
],
[
"print(metrics.classification_report(y_test, y_pred))",
" precision recall f1-score support\n\n 0 0.11 0.11 0.11 62\n 1 0.63 0.57 0.60 148\n 2 0.77 0.73 0.75 313\n 3 0.77 0.83 0.80 369\n 4 0.58 0.55 0.56 136\n 5 0.74 0.75 0.74 150\n 6 0.48 0.40 0.43 81\n 7 0.65 0.65 0.65 111\n 8 0.61 0.61 0.61 148\n 9 0.77 0.81 0.79 75\n 10 0.54 0.61 0.57 104\n 11 0.53 0.64 0.58 130\n 12 0.49 0.46 0.47 182\n 13 0.52 0.55 0.53 151\n 14 0.21 0.22 0.21 46\n 15 0.68 0.60 0.63 67\n 16 0.68 0.59 0.64 153\n 17 0.58 0.59 0.59 160\n 18 0.25 0.34 0.29 41\n 19 0.43 0.42 0.42 89\n 20 0.45 0.50 0.48 70\n 21 0.58 0.60 0.59 169\n 22 0.71 0.38 0.49 32\n 23 0.63 0.61 0.62 139\n 24 0.79 0.77 0.78 322\n\n accuracy 0.62 3448\n macro avg 0.57 0.56 0.56 3448\nweighted avg 0.63 0.62 0.62 3448\n\n"
],
[
"clf_matrix=metrics.confusion_matrix(y_test,y_pred)",
"_____no_output_____"
],
[
"f, ax = plt.subplots(figsize=(12,12))\nsns.heatmap(pd.DataFrame(clf_matrix), annot=True, fmt=\"g\", linewidths=.5, xticklabels=1, cmap=\"Greens\", yticklabels=False, cbar=True)",
"_____no_output_____"
]
],
[
[
"##### 1.2. Usando las variables originales ",
"_____no_output_____"
]
],
[
[
"##Se eligen las variables que se usaran en el modelo\nX = df.iloc[:, 7:45]",
"_____no_output_____"
],
[
"X.describe()",
"_____no_output_____"
],
[
"##Se escalan las variables\nX = StandardScaler().fit_transform(X)",
"_____no_output_____"
],
[
"df_1 = pd.DataFrame(df.FoodGroup)",
"_____no_output_____"
],
[
"df_1.FoodGroup.value_counts(dropna=False)",
"_____no_output_____"
],
[
"##Se codifica la variable FoodGroup usando LabelEncoder de sklearn\nlabelencoder = LabelEncoder()\ndf_1['FoodGroup'] = labelencoder.fit_transform(df_1.FoodGroup)",
"_____no_output_____"
],
[
"df_1.FoodGroup.value_counts(dropna=False)",
"_____no_output_____"
],
[
"classifier = DecisionTreeClassifier(random_state=0)",
"_____no_output_____"
],
[
"###Se divide la muestra en entrenamiento y prueba, en 60% y 40%, respectivamente.\nX_train, X_test, y_train, y_test = train_test_split(X, df_1.FoodGroup, test_size=0.4, random_state=0)",
"_____no_output_____"
],
[
"###Se entrena el modelo\nclassifier = classifier.fit(X_train, y_train)",
"_____no_output_____"
],
[
"y_pred = pd.DataFrame(classifier.predict(X_test))",
"_____no_output_____"
],
[
"probs = classifier.predict_proba(X_test) ",
"_____no_output_____"
]
],
[
[
"- Métricas para evaluar el modelo",
"_____no_output_____"
]
],
[
[
"print(\"Accuracy:\",metrics.accuracy_score(y_test, y_pred))",
"Accuracy: 0.7529002320185615\n"
],
[
"print(\"Precision:\",metrics.precision_score(y_test, y_pred, average='macro'))",
"Precision: 0.6991026307099126\n"
],
[
"print(\"Recall:\",metrics.recall_score(y_test, y_pred, average='macro'))",
"Recall: 0.6896707548628851\n"
],
[
"print(metrics.classification_report(y_test, y_pred))",
" precision recall f1-score support\n\n 0 0.33 0.37 0.35 62\n 1 0.77 0.73 0.75 148\n 2 0.80 0.82 0.81 313\n 3 0.91 0.92 0.91 369\n 4 0.61 0.66 0.64 136\n 5 0.86 0.79 0.82 150\n 6 0.68 0.72 0.70 81\n 7 0.76 0.68 0.71 111\n 8 0.68 0.71 0.70 148\n 9 0.84 0.83 0.83 75\n 10 0.80 0.77 0.78 104\n 11 0.58 0.72 0.64 130\n 12 0.77 0.83 0.80 182\n 13 0.75 0.77 0.76 151\n 14 0.36 0.43 0.39 46\n 15 0.75 0.73 0.74 67\n 16 0.89 0.78 0.83 153\n 17 0.83 0.85 0.84 160\n 18 0.68 0.56 0.61 41\n 19 0.65 0.65 0.65 89\n 20 0.57 0.56 0.57 70\n 21 0.69 0.71 0.70 169\n 22 0.40 0.25 0.31 32\n 23 0.64 0.57 0.60 139\n 24 0.87 0.84 0.85 322\n\n accuracy 0.75 3448\n macro avg 0.70 0.69 0.69 3448\nweighted avg 0.76 0.75 0.75 3448\n\n"
],
[
"clf_matrix=metrics.confusion_matrix(y_test,y_pred)",
"_____no_output_____"
],
[
"f, ax = plt.subplots(figsize=(12,12))\nsns.heatmap(pd.DataFrame(clf_matrix), annot=True, fmt=\"g\", linewidths=.5, xticklabels=1, cmap=\"Greens\", yticklabels=False, cbar=True)",
"_____no_output_____"
]
],
[
[
"### K - Means\n\n- Se utiliza este algoritmo para el análisis de conglomerados. \n- Se eligen los dos principales componentes",
"_____no_output_____"
]
],
[
[
"componentesprincipales_dos = componentesprincipales_analisis_post.loc[:, ['principal_component_1', 'principal_component_2']]",
"_____no_output_____"
],
[
"componentesprincipales_dos",
"_____no_output_____"
]
],
[
[
"Por lo que, para el primer componente principal se asocian más la vitamina Riboflavin o vitamina B2, Niacin o vitamina B3 y la vitamina B6. El porcentaje de varianza explicada de este componente es de 23.69%. Mientras que, para el segundo componente principal se asocian los carbohidratos, el azúcar y la vitamina B12. El porcentaje de varianza explicada de este segundo componente principal es de 11.38% (ver notebook [PCA_from_sklearn](https://github.com/123972/PCA-nutricion/blob/master/notebooks/Programacion/PCA_from_sklearn.ipynb)).",
"_____no_output_____"
]
],
[
[
"### Se cargan las funciones\nimport sklearn as sk\nfrom sklearn import preprocessing\nfrom sklearn.cluster import KMeans",
"_____no_output_____"
]
],
[
[
"- Gráfica que muestra los dos componentes principales.",
"_____no_output_____"
]
],
[
[
"plt.scatter(data=componentesprincipales_dos, x='principal_component_1', y='principal_component_2')\nplt.xlabel('Componente 1')\nplt.ylabel('Componente 2')",
"_____no_output_____"
]
],
[
[
"- Se obtiene el número de clusters óptimos a partir del punto de corte de la siguiente gráfica, el método que se uso fue el de Elbow. Sin embargo este proceso se tarda mucho y además nos arroja dos puntos de corte (2 y 5), por lo que se decidió paralelizar usando Dask.",
"_____no_output_____"
]
],
[
[
"wcss = []\nfor i in range(1, 11):\n kmeans = KMeans(n_clusters = i, init = 'k-means++', random_state = 42)\n kmeans.fit(componentesprincipales_dos)\n wcss.append(kmeans.inertia_)\n\n# Gráfica de la suma de las distancias al cuadrado\nplt.plot(range(1, 11), wcss)\nplt.xlabel('Número de clusters')\nplt.ylabel('Suma de las distancias al cuadrado')\nplt.show()",
"_____no_output_____"
]
],
[
[
"#### Cómputo en paralelo usando Dask",
"_____no_output_____"
]
],
[
[
"### Se cargan las librerías\nfrom dask.distributed import Client, progress\nimport dask_ml.cluster",
"_____no_output_____"
],
[
"client = Client()\nclient",
"C:\\Users\\Elizabeth\\Anaconda3\\lib\\site-packages\\distributed\\dashboard\\core.py:72: UserWarning: \nFailed to start diagnostics server on port 8787. [WinError 10048] Solo se permite un uso de cada dirección de socket (protocolo/dirección de red/puerto)\n warnings.warn(\"\\n\" + msg)\n"
]
],
[
[
"- Al dar click en el dashboard de arriba se muestra la página de status mediante bokeh, en el puerto 8787. A continuación se presenta el apartado de workers:",
"_____no_output_____"
]
],
[
[
"from IPython.display import Image, display\ndisplay(Image(filename='../../docs/img/dask_workers.png', embed=True))",
"_____no_output_____"
]
],
[
[
"- Se obtiene el número de cluster óptimos paralelizando mediante Dask.",
"_____no_output_____"
]
],
[
[
"wcss = []\nfor i in range(1, 11):\n kmeans = dask_ml.cluster.KMeans(n_clusters = i, init = 'k-means++', random_state = 42)\n kmeans.fit(componentesprincipales_dos)\n wcss.append(kmeans.inertia_)\n\n# Gráfica de la suma de las distancias al cuadrado\nplt.plot(range(1, 11), wcss)\nplt.xlabel('Número de clusters')\nplt.ylabel('Suma de las distancias al cuadrado')\nplt.show()",
"_____no_output_____"
]
],
[
[
"- En la gráfica se observa que el número de clusters óptimos es de 2, aunque se puede ver que también el 4 tiene un poco de inflexión, así que se usaremos K- Means para dos clusters y también se paraleliza. ",
"_____no_output_____"
]
],
[
[
"kmeans = dask_ml.cluster.KMeans(n_clusters = 2, init = 'k-means++', random_state = 42)\nk_means = kmeans.fit(componentesprincipales_dos)\ncenters = kmeans.cluster_centers_ ###centroides de los clusters\nlabels = kmeans.labels_",
"_____no_output_____"
],
[
"plt.scatter(data=componentesprincipales_dos, x='principal_component_1', y='principal_component_2', c=labels)\nplt.scatter(centers[:, 0], centers[:, 1], c='red', s=200, alpha=0.5)\nplt.xlabel('Componente 1 \\n (vitamina B2, vitamina B3, vitamina B6)')\nplt.ylabel('Componente 2 \\n (carbohidratos, azúcares, vitamina B12)')",
"_____no_output_____"
]
],
[
[
"##### Referencias:",
"_____no_output_____"
],
[
"- Palacios M. Erick, Notas de MNO 2020, [Cómputo en paralelo - Dask](https://github.com/ITAM-DS/analisis-numerico-computo-cientifico/blob/master/temas/II.computo_paralelo/2.2.Python_dask.ipynb)\n- Tipología_manejo_agrícola por [Irene Ramos](https://github.com/iramosp/tesis-paisajes/blob/master/Tipologia_manejo_agricola.ipynb)",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
]
] |
c5148d57a33f7740c684c3c3b2a7fb80e0ca6038
| 16,583 |
ipynb
|
Jupyter Notebook
|
notebooks/07.00 - Modeling - Select Cross Validation Rolling Window Size.ipynb
|
DMacGillivray/ontario-peak-power-forecasting
|
fb8d19bba3df07c065e5de46c088272b2387c58d
|
[
"MIT"
] | null | null | null |
notebooks/07.00 - Modeling - Select Cross Validation Rolling Window Size.ipynb
|
DMacGillivray/ontario-peak-power-forecasting
|
fb8d19bba3df07c065e5de46c088272b2387c58d
|
[
"MIT"
] | null | null | null |
notebooks/07.00 - Modeling - Select Cross Validation Rolling Window Size.ipynb
|
DMacGillivray/ontario-peak-power-forecasting
|
fb8d19bba3df07c065e5de46c088272b2387c58d
|
[
"MIT"
] | 1 |
2021-02-01T21:58:24.000Z
|
2021-02-01T21:58:24.000Z
| 34.620042 | 367 | 0.506302 |
[
[
[
"# 07.00 - Modeling - Prophet Model & Select Cross Validation Rolling Window Size",
"_____no_output_____"
],
[
" + We have data for each summer from 1994 to 2018\n + We initially decided that the minimum size of the hold out test data is 5 years from 2014 to 2018\n + We want to select a rolling window that extracts as much value as possible fom the data, but that leaves as much data as possible as hold-out data\n + Prophet seems to have good out of the box performance, and runs faster than statsmodels ARIMA\n + We beleive that there are some underlying structural changes that have changed cause and effect relationships between features and power demand between 1994 and 2018\n + The feature data is limited to weather. We do not have data for items such as air conditioner penetration, conserrvation growth (eg LEDs), population growth, housing stock types.\n + Therefore, I am going to make the assertion that next year's power demand pattern more closely resembles this year's pattern rather than last year's\n + We could introduce some sort of decay scheme where more recent data is weighted more heavily than older data. But this does not help us maximize the size of the held-out test data\n \n#### One approach could be:\n + We will use only the power data, and run a series of incrementally increasing cross validation windows across the data between 1994 and 2013\n + Based on the results we will select a window for the rolling time series cross validation to use in the rest of the modeling process. We will select the window by running prophet on an incremetally increasing sequence of rolling windows, and look for either a best size, or a size beyond which we get diminishing returns.\n + I realize that this is breaking some rules.If the window proves to be 3 years then to get 10 cross folds, my hold out data will be from 2008 to 2018. But, I will have already \"touched\" some of this data when I determined the size of the rolling window. \n\n#### Another approach could be:\n + Make a judgement as to a reasonable time period\n \n#### Making a judgement:\n + If I had to draw a chart of next year's demand by reviewing a chart of the last 100 years of data, I would draw a chart that looked exactly the same as last year + or - any obvious trend.\n + We are making a prediction for a single year ahead, using our cross validation scheme i.e the validation set comprises one year. If we only choose a single year of test data, then our model will miss out on trends, and will be working on a 50/50 train test split. Therefore, our training period should be greater than 1 year.\n + Two years of training data is not enough because a degree of randomness is introduced by the weather. ie. if we have a hot summer followed by a cold summer, this could be seen as a trend, but it is really randomness. Therefore, our training period should be greater than 2 years.\n + Twenty years seems too long because diverse undelying structural changes in the demand patterns mean that year 1 is not really the \"same\" as year 20\n + At this point, I have delayed making this decision long enough, and I am going to (semi-)arbitrarily select a training period of 5 years. This gives a train/ validation split of 83/17% which seems reasonable. My opinion is that this period is long enough to capture trends, and short enough to give a reasonably close representation of the validation data\n + I want to keep 10 cross folds in order to capture the uncertainty in the model\n + Therefore my data split will look like this:\n + Training Data - 1994 to 2009 with a 10 fold rolling tiome series cross validation\n + Test Data - 2010 to 2018 - 9 years",
"_____no_output_____"
],
[
"## Imports & setup",
"_____no_output_____"
]
],
[
[
"import pathlib\nimport warnings\nfrom datetime import datetime\nimport sys\nimport pickle\nimport joblib\nimport gc\n\nimport pandas as pd\nimport numpy as np\n\n# Plotting\nimport matplotlib as mpl\nimport matplotlib.pyplot as plt\nfrom pandas.plotting import register_matplotlib_converters\nregister_matplotlib_converters()\nfrom matplotlib.dates import DateFormatter\nimport matplotlib.dates as mdates\n\n\n# Imports\nsys.path.append(\"..\")\nfrom src.utils.utils import (AnnualTimeSeriesSplit,\n RollingAnnualTimeSeriesSplit,\n bound_precision,\n run_cross_val,\n run_data_split_cross_val,\n save_run_results)\nfrom src.features.features import CyclicalToCycle\nfrom src.models.models import SK_SARIMAX, SK_Prophet, SetTempAsPower, SK_Prophet_1\nfrom src.visualization.visualize import (plot_prediction,\n plot_joint_plot,\n residual_plots,\n print_residual_stats,\n resids_vs_preds_plot)\n#b # Packages\nfrom sklearn.pipeline import Pipeline\nfrom skoot.feature_selection import FeatureFilter\nfrom skoot.preprocessing import SelectiveRobustScaler\nfrom sklearn.metrics import mean_absolute_error\nfrom scipy.stats import norm\nfrom statsmodels.graphics.gofplots import qqplot\nfrom pandas.plotting import autocorrelation_plot\nfrom statsmodels.graphics.tsaplots import plot_acf\nimport statsmodels.api as sm\nfrom fbprophet import Prophet\n\n# Display\npd.set_option('display.max_columns', 500)\npd.set_option('display.width', 1000)\nfigsize=(15,7)\nwarnings.filterwarnings(action='ignore')\n%matplotlib inline\n\n# Data\nPROJECT_DIR = pathlib.Path.cwd().parent.resolve()\nCLEAN_DATA_DIR = PROJECT_DIR / 'data' / '05-clean'\nMODELS_DIR = PROJECT_DIR / 'data' / 'models'\nRESULTS_PATH = PROJECT_DIR / 'data' /'results' / 'results.csv'",
"_____no_output_____"
]
],
[
[
"## Load Daily Data & Inspect",
"_____no_output_____"
]
],
[
[
"df = pd.read_csv(CLEAN_DATA_DIR / 'clean-features.csv', parse_dates=True, index_col=0)",
"_____no_output_____"
],
[
"X = df.copy(deep=True)\nX = X.loc['1994': '2009']\ny = X.pop('daily_peak')\nX.head()",
"_____no_output_____"
],
[
"y.tail()",
"_____no_output_____"
]
],
[
[
"## Prophet Model \n\nRun using just the y data - the daily peak demand",
"_____no_output_____"
]
],
[
[
"n_splits=10\n\nprophet_model = SK_Prophet(pred_periods=96)\n \nratscv = RollingAnnualTimeSeriesSplit(n_splits=n_splits, goback_years=5)\n\nsteps = [('prophet', prophet_model)]\npipeline = Pipeline(steps)\nd = run_cross_val(X, y, ratscv, pipeline, scoring=['mae', 'bound_precision'])\nd",
"INFO:numexpr.utils:NumExpr defaulting to 4 threads.\n"
],
[
"# Take a look at the results on the validation data\nprint(np.mean(d['test']['mae']))\nprint(np.mean(d['test']['bound_precision']))",
"1841.1591618294005\n0.1\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
c51497c02b521251fb41b08078208753f9f4c672
| 30,758 |
ipynb
|
Jupyter Notebook
|
HW3.ipynb
|
whathelll/cs109-content
|
73dc67dadf0bc85884ee5e2794c090f3b1a3dd3e
|
[
"MIT"
] | null | null | null |
HW3.ipynb
|
whathelll/cs109-content
|
73dc67dadf0bc85884ee5e2794c090f3b1a3dd3e
|
[
"MIT"
] | null | null | null |
HW3.ipynb
|
whathelll/cs109-content
|
73dc67dadf0bc85884ee5e2794c090f3b1a3dd3e
|
[
"MIT"
] | null | null | null | 35.682135 | 764 | 0.59022 |
[
[
[
"empty"
]
]
] |
[
"empty"
] |
[
[
"empty"
]
] |
c514b873a11106703fd7f305ee640b75919a8c6a
| 253,879 |
ipynb
|
Jupyter Notebook
|
Lab-4/.ipynb_checkpoints/1_DT_Weather_Entropy-checkpoint.ipynb
|
yash-a-18/002_YashAmethiya
|
ab7e8bd8ebec553a0592b698dddc34c53b522967
|
[
"MIT"
] | null | null | null |
Lab-4/.ipynb_checkpoints/1_DT_Weather_Entropy-checkpoint.ipynb
|
yash-a-18/002_YashAmethiya
|
ab7e8bd8ebec553a0592b698dddc34c53b522967
|
[
"MIT"
] | null | null | null |
Lab-4/.ipynb_checkpoints/1_DT_Weather_Entropy-checkpoint.ipynb
|
yash-a-18/002_YashAmethiya
|
ab7e8bd8ebec553a0592b698dddc34c53b522967
|
[
"MIT"
] | null | null | null | 555.533917 | 217,248 | 0.942189 |
[
[
[
"**Aim: Implement Decsion Tree classifier**\n\n\n- Implement Decision Tree classifier using scikit learn library\n- Test the classifier for Weather dataset",
"_____no_output_____"
],
[
" Step 1: Import necessary libraries.",
"_____no_output_____"
]
],
[
[
"from sklearn import preprocessing\nfrom sklearn.tree import DecisionTreeClassifier",
"_____no_output_____"
]
],
[
[
" Step 2: Prepare dataset.",
"_____no_output_____"
]
],
[
[
"#Predictor variables\nOutlook = ['Rainy', 'Rainy', 'Overcast', 'Sunny', 'Sunny', 'Sunny', 'Overcast',\n 'Rainy', 'Rainy', 'Sunny', 'Rainy','Overcast', 'Overcast', 'Sunny']\nTemperature = ['Hot', 'Hot', 'Hot', 'Mild', 'Cool', 'Cool', 'Cool',\n 'Mild', 'Cool', 'Mild', 'Mild', 'Mild', 'Hot', 'Mild']\nHumidity = ['High', 'High', 'High', 'High', 'Normal', 'Normal', 'Normal',\n 'High', 'Normal', 'Normal', 'Normal', 'High', 'Normal', 'High']\nWind = ['False', 'True', 'False', 'False', 'False', 'True', 'True',\n 'False', 'False', 'False', 'True', 'True', 'False', 'True']\n\n#Class Label:\nPlay = ['No', 'No', 'Yes', 'Yes', 'Yes', 'No', 'Yes', 'No',\n'Yes', 'Yes', 'Yes', 'Yes', 'Yes', 'No']",
"_____no_output_____"
]
],
[
[
" Step 3: Digitize the data set using encoding",
"_____no_output_____"
]
],
[
[
"#creating labelEncoder\nle = preprocessing.LabelEncoder()\n\n# Converting string labels into numbers.\nOutlook_encoded = le.fit_transform(Outlook)\nOutlook_name_mapping = dict(zip(le.classes_, le.transform(le.classes_)))\nprint(\"Outllok mapping:\",Outlook_name_mapping)\n\nTemperature_encoded = le.fit_transform(Temperature)\nTemperature_name_mapping = dict(zip(le.classes_, le.transform(le.classes_)))\nprint(\"Temperature mapping:\",Temperature_name_mapping)\n\nHumidity_encoded = le.fit_transform(Humidity)\nHumidity_name_mapping = dict(zip(le.classes_, le.transform(le.classes_)))\nprint(\"Humidity mapping:\",Humidity_name_mapping)\n\nWind_encoded = le.fit_transform(Wind)\nWind_name_mapping = dict(zip(le.classes_, le.transform(le.classes_)))\nprint(\"Wind mapping:\",Wind_name_mapping)\n\nPlay_encoded = le.fit_transform(Play)\nPlay_name_mapping = dict(zip(le.classes_, le.transform(le.classes_)))\nprint(\"Play mapping:\",Play_name_mapping)\n\nprint(\"\\n\\n\")\nprint(\"Weather:\" ,Outlook_encoded)\nprint(\"Temerature:\" ,Temperature_encoded)\nprint(\"Humidity:\" ,Humidity_encoded)\nprint(\"Wind:\" ,Wind_encoded)\nprint(\"Play:\" ,Play_encoded)",
"Outllok mapping: {'Overcast': 0, 'Rainy': 1, 'Sunny': 2}\nTemperature mapping: {'Cool': 0, 'Hot': 1, 'Mild': 2}\nHumidity mapping: {'High': 0, 'Normal': 1}\nWind mapping: {'False': 0, 'True': 1}\nPlay mapping: {'No': 0, 'Yes': 1}\n\n\n\nWeather: [1 1 0 2 2 2 0 1 1 2 1 0 0 2]\nTemerature: [1 1 1 2 0 0 0 2 0 2 2 2 1 2]\nHumidity: [0 0 0 0 1 1 1 0 1 1 1 0 1 0]\nWind: [0 1 0 0 0 1 1 0 0 0 1 1 0 1]\nPlay: [0 0 1 1 1 0 1 0 1 1 1 1 1 0]\n"
]
],
[
[
" Step 4: Merge different features to prepare dataset",
"_____no_output_____"
]
],
[
[
"features = tuple(zip(Outlook_encoded, Temperature_encoded, Humidity_encoded, Wind_encoded))\nfeatures",
"_____no_output_____"
]
],
[
[
" Step 5: Train ’Create and Train DecisionTreeClassifier’",
"_____no_output_____"
]
],
[
[
"#Create a Decision Tree Classifier (using Entropy)\nDT = DecisionTreeClassifier(criterion = \"entropy\")\n\n# Train the model using the training sets\nfinal_model = DT.fit(features, Play_encoded) #(features, Class_label)",
"_____no_output_____"
]
],
[
[
" Step 6: Predict Output for new data",
"_____no_output_____"
]
],
[
[
"#Predict Output\nprediction = DT.predict([[0, 1, 1, 0],[2, 2, 1, 1], [2, 2, 0, 1]]) # last is from training dataset and expected o/p was 0 and it is\nprint(\"Predicted Values for Playing: \", prediction) #No:0 ; Yes:1",
"Predicted Values for Playing: [1 1 0]\n"
]
],
[
[
" Step 7: Display Decsion Tree Created\n \n - This step requires graphviz and tkinter packages installed ",
"_____no_output_____"
]
],
[
[
"from sklearn.tree import export_graphviz\nexport_graphviz(DT,out_file='tree_entropy.dot',\n feature_names=['outlook','temperature','humidity','wind'],\n class_names=['play_no','play_yes'], \n filled=True)\n\n# Convert to png\nfrom subprocess import call\ncall(['dot', '-Tpng', 'tree_entropy.dot', '-o', 'tree_entropy.png', '-Gdpi=600'], shell=True)\n\n# Display in python\nimport matplotlib.pyplot as plt\nplt.figure(figsize = (14, 18))\nplt.imshow(plt.imread('tree_entropy.png'))\nplt.axis('off');\nplt.show();",
"_____no_output_____"
],
[
"from sklearn.tree import plot_tree\nprint(plot_tree(DT, class_names = [\"Yes\", \"No\"]))",
"[Text(111.60000000000001, 195.696, 'X[0] <= 0.5\\nentropy = 0.94\\nsamples = 14\\nvalue = [5, 9]\\nclass = No'), Text(74.4, 152.208, 'entropy = 0.0\\nsamples = 4\\nvalue = [0, 4]\\nclass = No'), Text(148.8, 152.208, 'X[2] <= 0.5\\nentropy = 1.0\\nsamples = 10\\nvalue = [5, 5]\\nclass = Yes'), Text(74.4, 108.72, 'X[0] <= 1.5\\nentropy = 0.722\\nsamples = 5\\nvalue = [4, 1]\\nclass = Yes'), Text(37.2, 65.232, 'entropy = 0.0\\nsamples = 3\\nvalue = [3, 0]\\nclass = Yes'), Text(111.60000000000001, 65.232, 'X[3] <= 0.5\\nentropy = 1.0\\nsamples = 2\\nvalue = [1, 1]\\nclass = Yes'), Text(74.4, 21.744, 'entropy = 0.0\\nsamples = 1\\nvalue = [0, 1]\\nclass = No'), Text(148.8, 21.744, 'entropy = 0.0\\nsamples = 1\\nvalue = [1, 0]\\nclass = Yes'), Text(223.20000000000002, 108.72, 'X[3] <= 0.5\\nentropy = 0.722\\nsamples = 5\\nvalue = [1, 4]\\nclass = No'), Text(186.0, 65.232, 'entropy = 0.0\\nsamples = 3\\nvalue = [0, 3]\\nclass = No'), Text(260.40000000000003, 65.232, 'X[1] <= 1.0\\nentropy = 1.0\\nsamples = 2\\nvalue = [1, 1]\\nclass = Yes'), Text(223.20000000000002, 21.744, 'entropy = 0.0\\nsamples = 1\\nvalue = [1, 0]\\nclass = Yes'), Text(297.6, 21.744, 'entropy = 0.0\\nsamples = 1\\nvalue = [0, 1]\\nclass = No')]\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
c514cc08db6d542eb7669e340f069abbc0f0dd6a
| 53,244 |
ipynb
|
Jupyter Notebook
|
scraping/kbo.ipynb
|
WhiteHyun/Lab
|
d162b5a4674fc8b0514dba8bfdd17af01eef8eca
|
[
"MIT"
] | null | null | null |
scraping/kbo.ipynb
|
WhiteHyun/Lab
|
d162b5a4674fc8b0514dba8bfdd17af01eef8eca
|
[
"MIT"
] | null | null | null |
scraping/kbo.ipynb
|
WhiteHyun/Lab
|
d162b5a4674fc8b0514dba8bfdd17af01eef8eca
|
[
"MIT"
] | null | null | null | 32.665031 | 101 | 0.302438 |
[
[
[
"https://fenderist.tistory.com/168\n타자 \nAVG(Batting Average) : 타율\nG (Game) : 참여경기수(경기)\nPA(Plate Appearances) : 타석수( 타자가 타석에 선 횟수 ), 한게임 평균 3~4타석 슴\nAB(At Bat) : 타수 ( 타격을 완료한 횟수, 볼넷, 희생번트, 타격 방해등은 포함 X)\nR(Runs) : 득점 ( 홈플레이트를 밟아 팀에 점수가 올랐을때 기록됨 )\nH(Hits) : 안타 \n2B(double) : 2루타\n3B(Triple) : 3루타\nHR(Home Run) : 홈런\nTB(Total Bases) : 총 루타, 계산법(H+2B+(2*3B)+(3*HR) or 1B+(2*2B)+(3*3B)+(4*HR))\nTBI(Runs Batted In) : 타점\nSAC(Sacrifice Bunt) : 희생번트\nSF(sacrifice Flying): 희생플라이\nBB(Base on Balls) : 볼넷(4구)\nIBB(Intentional Base on Balls) : 고의 4구\nHBP(Bit By Pitch) : 사구(몸에 맞는 볼)\nSO(Strike Out) : 삼진\nGDP(Groinded Into Double Play) : 병살타 (주자와 타자 모두 아웃되는 타격, 2아웃 이상)\nSLG(Slugging Percentage) : 장타율\nOBP(On-Base Percentage) : 출루율\nOPS( OBP + SLG ) : OPS\nMH(Multi Hits) : 멀티히트\nRISP(Batting Average with Runners in Scoring Position) : 득점권 타율, 2루나 3루에 주자 진출해있을때 타자가 안타칠확률\nPH-BA(Pinch Batting Average) : 대타 타율\n\n투수\nERA(Earned Run Average) : 9이닝당 평균 자책점\nG : 경기수\nW(Wins) : 승리\nL(Losses) : 패배\nSV(Save) : 세이브 \nHLD(Hold) : 홀드\nWPCT(Win Percentage) : 승률\nIP(Innings Pitched) : 던진 이닝\nH(Hits) : 피안타\nHR(Home Run) : 피홈런\nBB(Base on Balls) : 볼넷\nHBP(Hit By Pitch) : 사구(몸에 맞는 공)\nSO(Strike Out) : 삼진\nR(Runs): 실점\nER(Earned Runs) :자책점(야수 실책이 아닌, 투수가 내준 점수)\nWHIP(Walks plus Hits divided by Innings Pitched) : 이닝당 출루허용률\nCG(Completed Games) : 완투승 ( 9이닝까지 투수가 전부 다 던져서 이긴 게임수)\nSHO(Shutouts) : 완봉승( 9이닝까지 투수가 무실점으로 던져서 이긴 게임수)\nQS(Quality Start) : 퀄리티스타트 수 , 6이닝이상 3실점 이하로 던졌을때 퀄리티 스타트 라고한다.\nBSV(Blown Saves) : 블론 세이브, 세이브/홀드 기회에서 동점 또는 역전 허용시 블론 세이브라고함.\nTBF(Total Batters Faced) : 상대 타자수\nNP(Number of Pitchs) : 투구수\nAVG(Batting Average) : 피안타율\n2B(Double) : 2루타\n3B(Triple) : 3루타\nSAC(Sacrifice Bunt): 희생번트\nSF(sacrifice Flying) : 희생플라이\nIBB(Intentional Base on Balls) : 고의 사구\nWP(Wild Pitches ) : 폭투 , 투구 에러 \nBK(Balks) : 보크, 주자가 있는 상황에서 투수의 반칙 행위\n",
"_____no_output_____"
]
],
[
[
"from bs4 import BeautifulSoup\nfrom selenium import webdriver\nimport requests\nimport time\nimport csv",
"_____no_output_____"
],
[
"driver = webdriver.Chrome(\"./chromedriver\")\ndriver.get(\"https://www.koreabaseball.com/Record/Player/HitterBasic/Basic1.aspx\")\n# driver.get(\"https://www.koreabaseball.com/Record/Player/Defense/Basic.aspx\")\n# driver.get(\"https://www.koreabaseball.com/Record/Player/Runner/Basic.aspx\")\n",
"_____no_output_____"
],
[
"driver = webdriver.Chrome(\"./chromedriver\")\ndriver.get(\"https://www.koreabaseball.com/Record/Player/HitterBasic/Basic1.aspx\")\n\nwith open(\"kbo.csv\", \"w\", encoding=\"utf-8\") as f:\n writer = csv.writer(f)\n\n soup = BeautifulSoup(driver.page_source, \"lxml\")\n hitter_table = soup.find(\"table\", attrs={\"class\": \"tData01 tt\"})\n hitter_thead = hitter_table.find(\"thead\")\n data_rows = hitter_thead.find_all(\"tr\")\n for row in data_rows:\n columns = row.find_all(\"th\")\n data = [column.text for column in columns]\n writer.writerow(data)\n\n for i in range(1, 3):\n time.sleep(2)\n driver.find_element_by_link_text(f\"{i}\").click()\n\n time.sleep(0.5)\n\n soup = BeautifulSoup(driver.page_source, \"lxml\")\n\n hitter_table = soup.find(\"table\", attrs={\"class\": \"tData01 tt\"})\n hitter_tbody = hitter_table.find(\"tbody\")\n data_rows = hitter_tbody.find_all(\"tr\")\n \n\n for row in data_rows:\n columns = row.find_all(\"td\")\n data = [column.text for column in columns]\n writer.writerow(data)\n\ndriver.quit()\n",
"_____no_output_____"
],
[
"driver = webdriver.Chrome(\"./chromedriver\")\ndriver.get(\"https://www.koreabaseball.com/Record/Player/HitterBasic/Basic2.aspx\")\n\nwith open(\"kbo2.csv\", \"w\") as f:\n writer = csv.writer(f)\n\n # --------- thead 과정\n soup = BeautifulSoup(driver.page_source, \"lxml\")\n hitter_table = soup.find(\"table\", attrs={\"class\": \"tData01 tt\"})\n hitter_thead = hitter_table.find(\"thead\")\n data_rows = hitter_thead.find_all(\"tr\")\n\n for row in data_rows:\n columns = row.find_all(\"th\")\n data = [column.text for column in columns]\n writer.writerow(data)\n # ---------\n\n # --------- tbody\n for i in range(1, 3):\n time.sleep(2)\n driver.find_element_by_link_text(f\"{i}\").click()\n\n time.sleep(0.5)\n\n soup = BeautifulSoup(driver.page_source, \"lxml\")\n\n hitter_table = soup.find(\"table\", attrs={\"class\": \"tData01 tt\"})\n hitter_tbody = hitter_table.find(\"tbody\")\n data_rows = hitter_tbody.find_all(\"tr\")\n\n for row in data_rows:\n columns = row.find_all(\"td\")\n data = [column.text for column in columns]\n writer.writerow(data)\n # ---------\ndriver.quit()\n",
"_____no_output_____"
],
[
"import pandas as pd",
"_____no_output_____"
],
[
"df_1 = pd.read_csv(\"kbo.csv\")\ndf_2 = pd.read_csv(\"kbo2.csv\")",
"_____no_output_____"
],
[
"df_3 = pd.concat([df_1, df_2], axis=1, join=\"inner\")",
"_____no_output_____"
],
[
"df_3.to_csv(\"kbo3.csv\")",
"_____no_output_____"
],
[
"renew_df = df_3.loc[:, ~df_3.T.duplicated()]\nrenew_df.to_csv(\"kbo4.csv\")",
"_____no_output_____"
],
[
"renew_df",
"_____no_output_____"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
c514ccefd1904c41798470f1245ec540bdc2bcb1
| 381,285 |
ipynb
|
Jupyter Notebook
|
LinearAlgebra.ipynb
|
SevdanurGENC/Machine-Learning-Lecture-Notes
|
77aed2ec5db8e87a4d79863dcb0685851cce2e6e
|
[
"MIT"
] | null | null | null |
LinearAlgebra.ipynb
|
SevdanurGENC/Machine-Learning-Lecture-Notes
|
77aed2ec5db8e87a4d79863dcb0685851cce2e6e
|
[
"MIT"
] | null | null | null |
LinearAlgebra.ipynb
|
SevdanurGENC/Machine-Learning-Lecture-Notes
|
77aed2ec5db8e87a4d79863dcb0685851cce2e6e
|
[
"MIT"
] | null | null | null | 586.592308 | 367,968 | 0.950835 |
[
[
[
"# Linear Algebra Review",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"- **Scalar:** Any single numerical value.\n- **Vector:** An array of numbers(data) is a vector. \n- **Matrix:** A matrix is a 2-D array of shape (m×n) with m rows and n columns.\n- **Tensor:** Generally, an n-dimensional array where n>2 is called a Tensor. But a matrix or a vector is also a valid tensor.",
"_____no_output_____"
]
],
[
[
"import numpy as np",
"_____no_output_____"
]
],
[
[
"### Creating Vector",
"_____no_output_____"
]
],
[
[
"arr_1 = np.array([1,2,3,4,5])\narr_1",
"_____no_output_____"
],
[
"print(f\"Type: {type(arr_1)}\")\nprint(f\"Shape: {arr_1.shape}\")\nprint(f\"Dimension: {arr_1.ndim}\")",
"Type: <class 'numpy.ndarray'>\nShape: (5,)\nDimension: 1\n"
]
],
[
[
"### Creating Matrice",
"_____no_output_____"
]
],
[
[
"arr_2 = np.array([[1,2,3,4],[5,6,7,8]]) \narr_2",
"_____no_output_____"
],
[
"print(f\"Type: {type(arr_2)}\")\nprint(f\"Shape: {arr_2.shape}\")\nprint(f\"Dimension: {arr_2.ndim}\")",
"Type: <class 'numpy.ndarray'>\nShape: (2, 4)\nDimension: 2\n"
]
],
[
[
"# Addition and Scalar Multiplication ",
"_____no_output_____"
],
[
"## Addition",
"_____no_output_____"
],
[
" Two matrices may be added or subtracted only if they have the same dimension; that is, they must have the same number of rows and columns. Addition or subtraction is accomplished by adding or subtracting corresponding elements. ",
"_____no_output_____"
]
],
[
[
"matrice_1 = np.array([[1, 2, 3, 4], \n [5, 6, 7, 8], \n [9, 8, 6, 5]])\n\nmatrice_2 = np.array([[-1, 4, 3, 5],\n [1, 4, 7, 9],\n [-6, 5, 11, -4]])\n\nprint(f\"Matrice_1: \\n{matrice_1}\")\nprint(f\"\\nMatrice_2: \\n{matrice_2}\")",
"Matrice_1: \n[[1 2 3 4]\n [5 6 7 8]\n [9 8 6 5]]\n\nMatrice_2: \n[[-1 4 3 5]\n [ 1 4 7 9]\n [-6 5 11 -4]]\n"
]
],
[
[
"### Adding two matrices",
"_____no_output_____"
]
],
[
[
"matrice_1 + matrice_2",
"_____no_output_____"
]
],
[
[
"---",
"_____no_output_____"
],
[
"## Multiplication",
"_____no_output_____"
],
[
"### Scalar Multiplication",
"_____no_output_____"
],
[
" The term scalar multiplication refers to the product of a real number and a matrix. In scalar multiplication, each entry in the matrix is multiplied by the given scalar.",
"_____no_output_____"
]
],
[
[
"matrice_1 * matrice_2",
"_____no_output_____"
]
],
[
[
"### Matrix-Vector Multiplication ",
"_____no_output_____"
],
[
" Multiplication between a matrix \"M\" and a vector \"v\", we need to view the vector as a column matrix. We define the matrix-vector product only for the case when the number of columns in M equals the number of rows in v. So, if M is an m×n matrix (i.e., with n columns), then the product Mv is defined for n×1 column vectors x. If we let Mv=r, then r is an m×1 column vector. ",
"_____no_output_____"
]
],
[
[
"M = np.array([[ 6, 1 ,3], \n [ -1, 1 ,1], \n [ 1, 3 ,2]])\n\nv = np.array([1, 2, 3])",
"_____no_output_____"
]
],
[
[
"#### Option 1:",
"_____no_output_____"
]
],
[
[
"M.dot(v)",
"_____no_output_____"
]
],
[
[
"#### Option 2:",
"_____no_output_____"
]
],
[
[
"np.dot(M,v)",
"_____no_output_____"
]
],
[
[
"### Matrix-Matrix Multiplication ",
"_____no_output_____"
],
[
"Matrix-Matrix multiplication, the number of columns in the first matrix must be equal to the number of rows in the second matrix. The resulting matrix, known as the matrix product, has the number of rows of the first and the number of columns of the second matrix.",
"_____no_output_____"
]
],
[
[
"C = np.array([[1, 2, 3, 4],\n [5, 6, 7, 8],\n [9, 8, 6, 5]])\n\nD = np.array([[-1, 4, 3, 5],\n [1, 4, 7, 9],\n [-6, 5, 11, -4]]).reshape(4,3) ",
"_____no_output_____"
],
[
"C.dot(D)",
"_____no_output_____"
],
[
"np.dot(C,D)",
"_____no_output_____"
]
],
[
[
"## Matrix Multiplication Properties",
"_____no_output_____"
],
[
"1. The commutative property of multiplication $AB \\neq BA$\n\n2. Associative property of multiplication $(AB)C = A(BC)$\n\n3. Distributive properties $A(B+C) = AB+AC$\n\n4. Multiplicative identity property $ IA =A\\, \\& \\, AI=A$\n\n5. Multiplicative property of zero $ I0 =0 \\, \\& \\, A0=0$\n\n6. Dimension property",
"_____no_output_____"
],
[
"# Inverse and Transpose",
"_____no_output_____"
],
[
"## Inverse",
"_____no_output_____"
],
[
"In linear algebra, an n-by-n square matrix A is called invertible (also nonsingular or nondegenerate), if there exists an n-by-n square matrix B such that\n\n $ AB=BA=I $ where In denotes the n-by-n identity matrix and the multiplication used is ordinary matrix multiplication. If this is the case, then the matrix B is uniquely determined by A, and is called the (multiplicative) inverse of A, denoted by A−1.",
"_____no_output_____"
]
],
[
[
"x = np.array([[4, 9],\n [25, 36]])\n\ny = np.array([[8, 5],\n [1, 2]])",
"_____no_output_____"
],
[
"x_inv = np.linalg.inv(x)",
"_____no_output_____"
],
[
"x.dot(x_inv)",
"_____no_output_____"
]
],
[
[
"## Transpose",
"_____no_output_____"
],
[
"In linear algebra, the transpose of a matrix is an operator which flips a matrix over its diagonal; that is, it switches the row and column indices of the matrix $A$ by producing another matrix, often denoted by $A^T$(among other notations).",
"_____no_output_____"
]
],
[
[
"x",
"_____no_output_____"
],
[
"x_trans = x.T\nx_trans",
"_____no_output_____"
],
[
"A = np.random.randint(1, 10, size=(5, 3))\n\nprint(f\"Matrice: \\n{A}\")\nprint(f\"\\nShape: {A.shape}\")",
"Matrice: \n[[8 3 3]\n [9 2 8]\n [6 8 1]\n [3 9 5]\n [7 5 2]]\n\nShape: (5, 3)\n"
],
[
"A_t = A.T\n\nprint(f\"Matrice: \\n{A_t}\")\nprint(f\"\\nShape: {A_t.shape}\")",
"Matrice: \n[[1 2 6 4 7]\n [7 5 7 3 5]\n [6 5 4 2 1]]\n\nShape: (3, 5)\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
c514ce0b912e32e4dab110a314567f90bb34cee4
| 50,668 |
ipynb
|
Jupyter Notebook
|
data_codes/dataset EDA.ipynb
|
dudtjakdl/OpenNMT-Korean-To-English
|
32fcdb860906f40f84375ec17a23ae32cb90baa0
|
[
"Apache-2.0"
] | 11 |
2020-01-27T02:17:07.000Z
|
2021-06-29T08:58:08.000Z
|
data_codes/dataset EDA.ipynb
|
dudtjakdl/OpenNMT-Korean-To-English
|
32fcdb860906f40f84375ec17a23ae32cb90baa0
|
[
"Apache-2.0"
] | null | null | null |
data_codes/dataset EDA.ipynb
|
dudtjakdl/OpenNMT-Korean-To-English
|
32fcdb860906f40f84375ec17a23ae32cb90baa0
|
[
"Apache-2.0"
] | 4 |
2020-02-10T05:32:22.000Z
|
2022-02-04T13:14:11.000Z
| 33.801201 | 1,315 | 0.429976 |
[
[
[
"## 1. 데이터 불러오기",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nimport random",
"_____no_output_____"
],
[
"data1 = pd.read_csv('C:/Users/Soyoung Cho/Desktop/NMT Project/dataset/train.csv')\ndata2 = pd.read_csv('C:/Users/Soyoung Cho/Desktop/NMT Project/dataset/test.csv')\ndata3 = pd.read_csv('C:/Users/Soyoung Cho/Desktop/NMT Project/dataset/tatoeba_data.csv')",
"_____no_output_____"
],
[
"print(\"data1: \", len(data1))\nprint(\"data2: \", len(data2))\nprint(\"data3: \", len(data3))",
"data1: 1282167\ndata2: 320541\ndata3: 3318\n"
],
[
"data = pd.concat([data1, data2, data3])",
"_____no_output_____"
],
[
"data = data.reset_index(drop=True) # 0~20만, 0~20만 이런 인덱스까지 concat 되었던 것을 초기화, 다시 인덱스 주었다",
"_____no_output_____"
],
[
"data",
"_____no_output_____"
],
[
"Kor_list = list(data['Korean']) #모든 한글 문장이 담긴 리스트\nEng_list = list(data['English']) #모든 영어 문장이 담긴 리스트\n\nprint(Kor_list[:5])\nprint(\"\\n\")\nprint(Eng_list[:5])",
"['경기도가 평화시대를 맞아 통일문제를 보다 쉽고 재미있게 다가갈 수 있도록 통일전문 팟캐스트 <남북상열지사>와 콜라보레이션을 진행한다.', '우선 현재 강원 동해안을 제외한 중부 지방과 호남 서해안에 약하게 눈이 내리거나 눈발이 조금 날리고 있습니다.', '아직 구체적인 의제가 확정된 것은 아니지만 지난 19일 한미 국방부가 발표한 UFG 연합훈련중단에 따른 세부협의를 진행할 것으로 관측된다.', '이시기의 남자아이에게는 레고 시티나 레고 닌자고 시리즈를 추천합니다.', '손 대표로선 이날 지도부 회의에서 최고위원들을 중심으로 “사퇴하라”는 요구가 나온 가운데 정면 돌파를 택한 셈이다.']\n\n\n['Gyeonggi-do Province will collaborate with the unification special podcast <Love Song Between North and South Korea> in order to approach the issue of unification in easier and more fun ways in the time of peace.', 'First of all, it is currently snowing slightly or snow flutters lightly son the central region and the west coast of Honam except the east coast of Gangwon.', 'The specific agenda has not been decided yet, but it is expected to proceed with detailed consultations following the suspension of the UFG coalition training announced by the US Department of Defense on the 19th.', 'Lego City and the series Lego Ninjago are recommended to the boys of this age.', 'The representive Sohn apparently chose to make a head-on breakthrough during the leadership meeting amid the calls for his resignation led by the supreme council members.']\n"
],
[
"result = list(zip(Kor_list,Eng_list))\n\nrandom.shuffle(result)\nresult \n\nKor_list, Eng_list = zip(*result)",
"_____no_output_____"
],
[
"dict_ = {\"Korean\": [], \"English\" : []}\ndict_[\"Korean\"] = Kor_list\ndict_[\"English\"] = Eng_list",
"_____no_output_____"
],
[
"data = pd.DataFrame(dict_)\ndata",
"_____no_output_____"
]
],
[
[
"## 2. 데이터 중복 검사 및 제거",
"_____no_output_____"
]
],
[
[
"data.describe()",
"_____no_output_____"
],
[
"data.duplicated().sum()",
"_____no_output_____"
],
[
"data = data.drop_duplicates()",
"_____no_output_____"
],
[
"data.duplicated().sum()",
"_____no_output_____"
],
[
"data = data.reset_index(drop=True) # 0~20만, 0~20만 이런 인덱스까지 concat 되었던 것을 초기화, 다시 인덱스 주었다",
"_____no_output_____"
],
[
"data",
"_____no_output_____"
],
[
"#data.to_csv(\"datalist.csv\", encoding = 'utf-8-sig', index = False, mode = \"w\")",
"_____no_output_____"
]
],
[
[
"## 3. 문장별 단어 개수 파악 & 문제 파악",
"_____no_output_____"
]
],
[
[
"kor_word_cnt = []\neng_word_cnt = []\nfor i in range(len(data)):\n kor_word_cnt.append(len(data['Korean'][i].split(\" \")))\n eng_word_cnt.append(len(data['English'][i].split(\" \")))",
"_____no_output_____"
],
[
"data[\"Korean_word_count\"] = kor_word_cnt\ndata[\"English_word_count\"] = eng_word_cnt",
"_____no_output_____"
]
],
[
[
"### (1) 단어 개수 별 정렬해 데이터 확인 & 문제 수정",
"_____no_output_____"
]
],
[
[
"kor_sorted = data.sort_values(by=['Korean_word_count'], axis=0, ascending=False)",
"_____no_output_____"
],
[
"kor_sorted = kor_sorted.reset_index(drop=True)\nkor_sorted.head()",
"_____no_output_____"
],
[
"kor_sorted[0:10]",
"_____no_output_____"
],
[
"kor_sorted[-10:]",
"_____no_output_____"
]
],
[
[
"#### 문제 발견 및 수정, 데이터 재저장",
"_____no_output_____"
]
],
[
[
"kor_sorted[\"Korean\"][1603523]",
"_____no_output_____"
],
[
"kor_sorted[\"Korean\"][1603524]",
"_____no_output_____"
],
[
"kor_sorted[\"Korean\"][1603515]",
"_____no_output_____"
]
],
[
[
"중간의 \\xa0 때문에 한 문장이 한 단어로 인식 되었었다. 전체 dataset의 \\xa0 를 \" \" 로 대체해준다. \n다시 word를 카운트 해준다.",
"_____no_output_____"
]
],
[
[
"data.replace(\"\\xa0\", \" \", regex=True, inplace=True)",
"_____no_output_____"
],
[
"#data",
"_____no_output_____"
]
],
[
[
"---",
"_____no_output_____"
],
[
"---",
"_____no_output_____"
],
[
"### (2) 한글 문장 단어 개수 파악 ",
"_____no_output_____"
]
],
[
[
"kor_sorted = data.sort_values(by=['Korean_word_count'], axis=0, ascending=False)",
"_____no_output_____"
],
[
"kor_sorted = kor_sorted.reset_index(drop=True)\nkor_sorted.head()",
"_____no_output_____"
],
[
"kor_sorted[-110:-90]",
"_____no_output_____"
],
[
"kor_sorted[\"Korean\"][1603427]",
"_____no_output_____"
],
[
"kor_sorted[-130:-110]",
"_____no_output_____"
]
],
[
[
"위와 같이 띄어쓰기가 아예 이루어지지 않은 문장은 어떻게 해야할지 고민 필요.",
"_____no_output_____"
]
],
[
[
"kor_sorted[0:20]",
"_____no_output_____"
]
],
[
[
"### (3) 영어 문장 단어 개수 파악 ",
"_____no_output_____"
]
],
[
[
"eng_sorted = data.sort_values(by=['English_word_count'], axis=0, ascending=False)",
"_____no_output_____"
],
[
"eng_sorted = eng_sorted.reset_index(drop=True)",
"_____no_output_____"
],
[
"eng_sorted[:20]",
"_____no_output_____"
],
[
"eng_sorted['English'][0]",
"_____no_output_____"
],
[
"eng_sorted['English'][1]",
"_____no_output_____"
],
[
"eng_sorted['English'][2]",
"_____no_output_____"
],
[
"eng_sorted['English'][3]",
"_____no_output_____"
],
[
"len(eng_sorted['English'][3].split(\" \"))",
"_____no_output_____"
],
[
"len('\"We will play the role of a hub for inter-Korean exchanges of performing arts and culture,\" said Kim Cheol-ho, 66, the new director of the National Theater of Korea, at an inaugural press conference held at a restaurant in Jongro-gu, Seoul on the 8th and he said, \"We can invite North Korean national art troupe for the festival that will be held in 2020 for the 70th anniversary of our foundation.\" '.split(\" \"))",
"_____no_output_____"
],
[
"eng_sorted[-30:]",
"_____no_output_____"
]
],
[
[
"한글 데이터는 짧은 문장의 경우 띄어쓰기가 잘 안 이루어져 있음.\n영어 데이터는 긴 문장의 경우 띄어쓰기가 지나치게 많이 들어가 있음. \n짧은 문장의 경우 검수 안된 문장들 . 혹은 x 등 많음.",
"_____no_output_____"
],
[
"## 4. 박스플롯 그려보기",
"_____no_output_____"
]
],
[
[
"print(\"한글 문장 중 가장 적은 단어 개수 가진 문장은 \", min(kor_word_cnt))\nprint(\"한글 문장 중 가장 많은 단어 개수 가진 문장은 \", max(kor_word_cnt))\nprint(\"영어 문장 중 가장 적은 단어 개수 가진 문장은 \", min(eng_word_cnt))\nprint(\"영어 문장 중 가장 많은 단어 개수 가진 문장은 \", max(eng_word_cnt))",
"_____no_output_____"
],
[
"fig, ax = plt.subplots(figsize = (12,8))\nsns.boxplot(kor_word_cnt)\nplt.show()",
"_____no_output_____"
],
[
"fig, ax = plt.subplots(figsize = (12,8))\nsns.boxplot(eng_word_cnt)\nplt.show()",
"_____no_output_____"
]
],
[
[
"## 5. 데이터 저장하기",
"_____no_output_____"
]
],
[
[
"del data['Korean_word_count']\ndel data['English_word_count']\n\n#data.to_csv(\"C:/Users/Soyoung Cho/Desktop/NMT Project/dataset/datalist_modified.csv\", encoding = 'utf-8-sig', index = False, mode = \"w\")",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
c514d30d8d04c33b489016ae971db79a01f911ee
| 273,171 |
ipynb
|
Jupyter Notebook
|
investigate-a-dataset-template.ipynb
|
anjalinagel12/movieRecommender
|
985a6915a26ec87e26d9f4c46de063d3ef66dccb
|
[
"MIT"
] | null | null | null |
investigate-a-dataset-template.ipynb
|
anjalinagel12/movieRecommender
|
985a6915a26ec87e26d9f4c46de063d3ef66dccb
|
[
"MIT"
] | null | null | null |
investigate-a-dataset-template.ipynb
|
anjalinagel12/movieRecommender
|
985a6915a26ec87e26d9f4c46de063d3ef66dccb
|
[
"MIT"
] | null | null | null | 60.826319 | 19,156 | 0.567604 |
[
[
[
"> **Tip**: Welcome to the Investigate a Dataset project! You will find tips in quoted sections like this to help organize your approach to your investigation. Before submitting your project, it will be a good idea to go back through your report and remove these sections to make the presentation of your work as tidy as possible. First things first, you might want to double-click this Markdown cell and change the title so that it reflects your dataset and investigation.\n\n# Project: Investigate a Dataset (Replace this with something more specific!)\n\n## Table of Contents\n<ul>\n<li><a href=\"#intro\">Introduction</a></li>\n<li><a href=\"#wrangling\">Data Wrangling</a></li>\n<li><a href=\"#eda\">Exploratory Data Analysis</a></li>\n<li><a href=\"#conclusions\">Conclusions</a></li>\n</ul>",
"_____no_output_____"
],
[
"<a id='intro'></a>\n## Introduction\n\n> **Tip**: In this section of the report, provide a brief introduction to the dataset you've selected for analysis. At the end of this section, describe the questions that you plan on exploring over the course of the report. Try to build your report around the analysis of at least one dependent variable and three independent variables.\n>\n> If you haven't yet selected and downloaded your data, make sure you do that first before coming back here. If you're not sure what questions to ask right now, then make sure you familiarize yourself with the variables and the dataset context for ideas of what to explore.",
"_____no_output_____"
]
],
[
[
"#The dataset which is selected is tmdb-movies.csv i.e. movies dataset which contains data on movies and ratings.\n#Revenue,Runtime and Popularity is tend to be explored.Over a period span reveneue v/s runtime, runtime v/s popularity and popularity v/s revenue is to be explored.\n#Questions which will be answered are: \n# 1.Over the decades, what are the popular runtimes?\n# 2.Spanning the time periods, is revenue proportional to popularity?\n# 3.Does runtime affect popularity?\n#only visualization and basic correlations are attempted in this project.And any investigation and exploratory are tentative at its best.",
"_____no_output_____"
],
[
"# Use this cell to set up import statements for all of the packages that you\n# plan to use.\n\n# Remember to include a 'magic word' so that your visualizations are plotted\n# inline with the notebook. See this page for more:\n# http://ipython.readthedocs.io/en/stable/interactive/magics.html\n\nimport pandas as pd\nimport numpy as np\nimport csv\nimport datetime as datetime\nimport matplotlib.pyplot as plt\n% matplotlib inline",
"_____no_output_____"
]
],
[
[
"<a id='wrangling'></a>\n## Data Wrangling\n\n> **Tip**: In this section of the report, you will load in the data, check for cleanliness, and then trim and clean your dataset for analysis. Make sure that you document your steps carefully and justify your cleaning decisions.\n\n### General Properties",
"_____no_output_____"
]
],
[
[
"# Load your data and print out a few lines. Perform operations to inspect data\n# types and look for instances of missing or possibly errant data.\n\ndf=pd.read_csv('tmdb-movies.csv')\n",
"_____no_output_____"
],
[
"df.info()\n",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 10866 entries, 0 to 10865\nData columns (total 21 columns):\nid 10866 non-null int64\nimdb_id 10856 non-null object\npopularity 10866 non-null float64\nbudget 10866 non-null int64\nrevenue 10866 non-null int64\noriginal_title 10866 non-null object\ncast 10790 non-null object\nhomepage 2936 non-null object\ndirector 10822 non-null object\ntagline 8042 non-null object\nkeywords 9373 non-null object\noverview 10862 non-null object\nruntime 10866 non-null int64\ngenres 10843 non-null object\nproduction_companies 9836 non-null object\nrelease_date 10866 non-null object\nvote_count 10866 non-null int64\nvote_average 10866 non-null float64\nrelease_year 10866 non-null int64\nbudget_adj 10866 non-null float64\nrevenue_adj 10866 non-null float64\ndtypes: float64(4), int64(6), object(11)\nmemory usage: 1.7+ MB\n"
],
[
"df.head()\n",
"_____no_output_____"
],
[
"#df.drop_duplicates() Return DataFrame with duplicate rows removed, optionally only considering certain columns\n\nsum(df.duplicated())",
"_____no_output_____"
],
[
"df.drop_duplicates(inplace=True)\ndf.info()\n",
"<class 'pandas.core.frame.DataFrame'>\nInt64Index: 10865 entries, 0 to 10865\nData columns (total 21 columns):\nid 10865 non-null int64\nimdb_id 10855 non-null object\npopularity 10865 non-null float64\nbudget 10865 non-null int64\nrevenue 10865 non-null int64\noriginal_title 10865 non-null object\ncast 10789 non-null object\nhomepage 2936 non-null object\ndirector 10821 non-null object\ntagline 8041 non-null object\nkeywords 9372 non-null object\noverview 10861 non-null object\nruntime 10865 non-null int64\ngenres 10842 non-null object\nproduction_companies 9835 non-null object\nrelease_date 10865 non-null object\nvote_count 10865 non-null int64\nvote_average 10865 non-null float64\nrelease_year 10865 non-null int64\nbudget_adj 10865 non-null float64\nrevenue_adj 10865 non-null float64\ndtypes: float64(4), int64(6), object(11)\nmemory usage: 1.8+ MB\n"
],
[
"#isnull Return a boolean same-sized object indicating if the values are NA.\n#sum of those values are taken\n\ndf.isnull().sum()",
"_____no_output_____"
]
],
[
[
"> **Tip**: You should _not_ perform too many operations in each cell. Create cells freely to explore your data. One option that you can take with this project is to do a lot of explorations in an initial notebook. These don't have to be organized, but make sure you use enough comments to understand the purpose of each code cell. Then, after you're done with your analysis, create a duplicate notebook where you will trim the excess and organize your steps so that you have a flowing, cohesive report.\n\n> **Tip**: Make sure that you keep your reader informed on the steps that you are taking in your investigation. Follow every code cell, or every set of related code cells, with a markdown cell to describe to the reader what was found in the preceding cell(s). Try to make it so that the reader can then understand what they will be seeing in the following cell(s).\n\n### Data Cleaning (Replace this with more specific notes!)",
"_____no_output_____"
]
],
[
[
"# After discussing the structure of the data and any problems that need to be\n# cleaned, perform those cleaning steps in the second part of this section.\n\n\n\ndf.info()",
"<class 'pandas.core.frame.DataFrame'>\nInt64Index: 10865 entries, 0 to 10865\nData columns (total 21 columns):\nid 10865 non-null int64\nimdb_id 10855 non-null object\npopularity 10865 non-null float64\nbudget 10865 non-null int64\nrevenue 10865 non-null int64\noriginal_title 10865 non-null object\ncast 10789 non-null object\nhomepage 2936 non-null object\ndirector 10821 non-null object\ntagline 8041 non-null object\nkeywords 9372 non-null object\noverview 10861 non-null object\nruntime 10865 non-null int64\ngenres 10842 non-null object\nproduction_companies 9835 non-null object\nrelease_date 10865 non-null object\nvote_count 10865 non-null int64\nvote_average 10865 non-null float64\nrelease_year 10865 non-null int64\nbudget_adj 10865 non-null float64\nrevenue_adj 10865 non-null float64\ndtypes: float64(4), int64(6), object(11)\nmemory usage: 1.8+ MB\n"
],
[
"#earlier, we have removed the single duplicate record.\n# Here, we are removing rows with null values in imdb_id column\ndf.dropna(subset=['imdb_id'], inplace=True) \ndf.info()",
"<class 'pandas.core.frame.DataFrame'>\nInt64Index: 10855 entries, 0 to 10865\nData columns (total 21 columns):\nid 10855 non-null int64\nimdb_id 10855 non-null object\npopularity 10855 non-null float64\nbudget 10855 non-null int64\nrevenue 10855 non-null int64\noriginal_title 10855 non-null object\ncast 10779 non-null object\nhomepage 2934 non-null object\ndirector 10815 non-null object\ntagline 8038 non-null object\nkeywords 9368 non-null object\noverview 10852 non-null object\nruntime 10855 non-null int64\ngenres 10834 non-null object\nproduction_companies 9830 non-null object\nrelease_date 10855 non-null object\nvote_count 10855 non-null int64\nvote_average 10855 non-null float64\nrelease_year 10855 non-null int64\nbudget_adj 10855 non-null float64\nrevenue_adj 10855 non-null float64\ndtypes: float64(4), int64(6), object(11)\nmemory usage: 1.8+ MB\n"
],
[
"df.head()\n",
"_____no_output_____"
],
[
"# write dataframe 'df' to a csv file 'data_imdb.csv' to use it for the next session\ndf.to_csv('data_imdb.csv', index=False)",
"_____no_output_____"
]
],
[
[
"<a id='eda'></a>\n## Exploratory Data Analysis\n\n> **Tip**: Now that you've trimmed and cleaned your data, you're ready to move on to exploration. Compute statistics and create visualizations with the goal of addressing the research questions that you posed in the Introduction section. It is recommended that you be systematic with your approach. Look at one variable at a time, and then follow it up by looking at relationships between variables.\n\n### Research Question 1 (Replace this header name!)",
"_____no_output_____"
]
],
[
[
"# Use this, and more code cells, to explore your data. Don't forget to add\n# Markdown cells to document your observations and findings.\n",
"_____no_output_____"
],
[
"# Importing data from newly cleaned dataset\ndf_imdb = pd.read_csv('data_imdb.csv')",
"_____no_output_____"
],
[
"df_imdb.info()\n",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 10855 entries, 0 to 10854\nData columns (total 21 columns):\nid 10855 non-null int64\nimdb_id 10855 non-null object\npopularity 10855 non-null float64\nbudget 10855 non-null int64\nrevenue 10855 non-null int64\noriginal_title 10855 non-null object\ncast 10779 non-null object\nhomepage 2934 non-null object\ndirector 10815 non-null object\ntagline 8038 non-null object\nkeywords 9368 non-null object\noverview 10852 non-null object\nruntime 10855 non-null int64\ngenres 10834 non-null object\nproduction_companies 9830 non-null object\nrelease_date 10855 non-null object\nvote_count 10855 non-null int64\nvote_average 10855 non-null float64\nrelease_year 10855 non-null int64\nbudget_adj 10855 non-null float64\nrevenue_adj 10855 non-null float64\ndtypes: float64(4), int64(6), object(11)\nmemory usage: 1.7+ MB\n"
],
[
"df_imdb\n",
"_____no_output_____"
],
[
"df_new = df_imdb.groupby('release_year').mean()\n",
"_____no_output_____"
],
[
"df_new\n",
"_____no_output_____"
],
[
"df_new.plot(kind='bar')\n",
"_____no_output_____"
],
[
"df_new.plot(kind='hist')\n",
"_____no_output_____"
],
[
"df_new.describe()\n",
"_____no_output_____"
],
[
"df_new.hist()\n",
"_____no_output_____"
]
],
[
[
"### Research Question 2 (Replace this header name!)",
"_____no_output_____"
]
],
[
[
"# Continue to explore the data to address your additional research\n# questions. Add more headers as needed if you have more questions to\n# investigate.\n",
"_____no_output_____"
],
[
"#The given below are observation Popularity,Revenue and Runtime\ndf_new['popularity'].hist()\nplt.xlabel('Popularity')\nplt.title('Popularity Over the Years');",
"_____no_output_____"
],
[
"df_new['revenue'].hist()\nplt.xlabel('Revenue')\nplt.title('Revenue Over the Years');",
"_____no_output_____"
],
[
"df_new['runtime'].hist()\nplt.xlabel('Runtime')\nplt.title('Runtime Over the Years');\n",
"_____no_output_____"
],
[
"df_new['popularity'].describe()\n",
"_____no_output_____"
],
[
"df_new['revenue'].describe()\n",
"_____no_output_____"
],
[
"df_new['runtime'].describe()\n",
"_____no_output_____"
],
[
"# Continue to explore the data to address your additional research\n# questions. Add more headers as needed if you have more questions to\n# investigate.\n\n#We can see that distribution is left skewed.\n#Most movie revenues fall in the 3.257984e+07 to 4.293171e+07 ranges. \n\n",
"_____no_output_____"
]
],
[
[
"<a id='conclusions'></a>\n## Conclusions\n\n> **Tip**: Finally, summarize your findings and the results that have been performed. Make sure that you are clear with regards to the limitations of your exploration. If you haven't done any statistical tests, do not imply any statistical conclusions. And make sure you avoid implying causation from correlation!\n\n> **Tip**: Once you are satisfied with your work, you should save a copy of the report in HTML or PDF form via the **File** > **Download as** submenu. Before exporting your report, check over it to make sure that the flow of the report is complete. You should probably remove all of the \"Tip\" quotes like this one so that the presentation is as tidy as possible. Congratulations!",
"_____no_output_____"
]
],
[
[
"from subprocess import call\ncall(['python', '-m', 'nbconvert', 'Investigate_a_Dataset.ipynb'])",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
c514d38f3b8033376cc3a2293df7ab9c5b0825c4
| 89,376 |
ipynb
|
Jupyter Notebook
|
jupyter_notebook/ModelHyperparameterTesting.ipynb
|
Keith-Tachibana/Machine_Learning_Income_Predictor
|
55e2151b170012b10274d8d77ffe35b5201ded44
|
[
"MIT"
] | null | null | null |
jupyter_notebook/ModelHyperparameterTesting.ipynb
|
Keith-Tachibana/Machine_Learning_Income_Predictor
|
55e2151b170012b10274d8d77ffe35b5201ded44
|
[
"MIT"
] | null | null | null |
jupyter_notebook/ModelHyperparameterTesting.ipynb
|
Keith-Tachibana/Machine_Learning_Income_Predictor
|
55e2151b170012b10274d8d77ffe35b5201ded44
|
[
"MIT"
] | null | null | null | 214.330935 | 20,936 | 0.897422 |
[
[
[
"import data_loader as dl\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nimport pandas as pd\nnp.random.seed(0)\n\ntrain_df, valid_df = dl.load_train_data(\"adult.data\")\ntest_df = dl.load_test_data(\"adult.test\")\ncolumn_names = ['age', 'workclass', 'fnlwgt', 'education', 'education.num', 'martial.status',\n 'occupation', 'relationship', 'race', 'sex', 'capital.gain', 'capital.loss',\n 'hours.per.week', 'native.country', 'income']\ntrain_df.columns = column_names\ntrain_df.replace(' ?', np.nan, inplace=True)\nfrom sklearn.impute import SimpleImputer\nimp_mode = SimpleImputer(missing_values=np.nan, strategy='most_frequent')\nimp_mode.fit(train_df)\nimp_train_df = imp_mode.transform(train_df)\ntrain_df = pd.DataFrame(imp_train_df, columns = column_names)\n\nfor col in ['age', 'fnlwgt', 'education.num', 'capital.gain', 'capital.loss', 'hours.per.week']:\n train_df[col] = train_df[col].astype('int64')\nfor col in ['workclass', 'education', 'martial.status', 'occupation', 'relationship', 'race', \n 'sex', 'native.country', 'income']:\n train_df[col] = train_df[col].astype('category')\ntrain_df['capital.gain.loss'] = train_df['capital.gain'] - train_df['capital.loss']\nnum_features = ['age', 'fnlwgt', 'education.num', 'capital.gain', 'capital.loss', 'hours.per.week', 'capital.gain.loss']\ncat_features = ['workclass', 'education', 'martial.status', 'occupation', 'relationship', 'race', 'sex', 'native.country', 'income']\ncat_features.remove(\"education\")\ncat_features.remove(\"relationship\")\ncat_features.remove(\"income\")\n\nfor cat in cat_features:\n train_df = pd.concat([train_df,pd.get_dummies(train_df[cat], prefix='is')],axis=1)\n train_df.drop([cat],axis=1, inplace=True)\ntrain_df['income.prediction'] = train_df.income.cat.codes\ntrain_df.drop(labels=[\"fnlwgt\", \"education\",\"relationship\", \"income\"], axis = 1, inplace = True)\n\nvalid_df.columns = column_names\nvalid_df.replace(' ?', np.nan, inplace=True)\nimp_train_df = imp_mode.transform(valid_df)\nvalid_df = pd.DataFrame(imp_train_df, columns = column_names)\n\nfor col in ['age', 'fnlwgt', 'education.num', 'capital.gain', 'capital.loss', 'hours.per.week']:\n valid_df[col] = valid_df[col].astype('int64')\nfor col in ['workclass', 'education', 'martial.status', 'occupation', 'relationship', 'race', \n 'sex', 'native.country', 'income']:\n valid_df[col] = valid_df[col].astype('category')\nvalid_df['capital.gain.loss'] = valid_df['capital.gain'] - valid_df['capital.loss']\nnum_features = ['age', 'fnlwgt', 'education.num', 'capital.gain', 'capital.loss', 'hours.per.week', 'capital.gain.loss']\ncat_features = ['workclass', 'education', 'martial.status', 'occupation', 'relationship', 'race', 'sex', 'native.country', 'income']\ncat_features.remove(\"education\")\ncat_features.remove(\"relationship\")\ncat_features.remove(\"income\")\n\nfor cat in cat_features:\n valid_df = pd.concat([valid_df,pd.get_dummies(valid_df[cat], prefix='is')],axis=1)\n valid_df.drop([cat],axis=1, inplace=True)\nvalid_df['income.prediction'] = valid_df.income.cat.codes\nvalid_df.drop(labels=[\"fnlwgt\", \"education\",\"relationship\", \"income\"], axis = 1, inplace = True)\nmissing_cols = set( train_df.columns ) - set( valid_df.columns )\nfor c in missing_cols:\n valid_df[c] = 0\nvalid_df = valid_df[train_df.columns]\n\ntest_df.columns = column_names\ntest_df.replace(' ?', np.nan, inplace=True)\nimp_train_df = imp_mode.transform(test_df)\ntest_df = pd.DataFrame(imp_train_df, columns = column_names)\n\nfor col in ['age', 'fnlwgt', 'education.num', 'capital.gain', 'capital.loss', 'hours.per.week']:\n test_df[col] = test_df[col].astype('int64')\nfor col in ['workclass', 'education', 'martial.status', 'occupation', 'relationship', 'race', \n 'sex', 'native.country', 'income']:\n test_df[col] = test_df[col].astype('category')\ntest_df['capital.gain.loss'] = test_df['capital.gain'] - test_df['capital.loss']\nnum_features = ['age', 'fnlwgt', 'education.num', 'capital.gain', 'capital.loss', 'hours.per.week', 'capital.gain.loss']\ncat_features = ['workclass', 'education', 'martial.status', 'occupation', 'relationship', 'race', 'sex', 'native.country', 'income']\ncat_features.remove(\"education\")\ncat_features.remove(\"relationship\")\ncat_features.remove(\"income\")\n\nfor cat in cat_features:\n test_df = pd.concat([test_df,pd.get_dummies(test_df[cat], prefix='is')],axis=1)\n test_df.drop([cat],axis=1, inplace=True)\ntest_df['income.prediction'] = test_df.income.cat.codes\ntest_df.drop(labels=[\"fnlwgt\", \"education\",\"relationship\", \"income\"], axis = 1, inplace = True)\nmissing_cols = set( train_df.columns ) - set( test_df.columns )\nfor c in missing_cols:\n test_df[c] = 0\ntest_df = test_df[train_df.columns]",
"_____no_output_____"
],
[
"from sklearn.linear_model import LogisticRegression\nfrom sklearn.neighbors import KNeighborsClassifier\nfrom sklearn.tree import DecisionTreeClassifier\nfrom sklearn.ensemble import RandomForestClassifier\nfrom sklearn.ensemble import GradientBoostingClassifier\nfrom sklearn.ensemble import AdaBoostClassifier\nfrom sklearn.metrics import confusion_matrix\nfrom sklearn.metrics import accuracy_score\nfrom sklearn.metrics import roc_auc_score\nfrom collections import namedtuple\n\nModel = namedtuple('Model', 'name model')\n\ntrain = train_df.values\nX, Y= train[:,:-1], train[:,-1]\nvalid = valid_df.values\nXval, Yval = valid[:,:-1], valid[:,-1]\ntest = test_df.values\nXtst, Ytst = test[:,:-1], test[:,-1]\n\nmodels = []\nmodels.append(Model('LR', LogisticRegression(solver='liblinear')))\nmodels.append(Model('KNN', KNeighborsClassifier()))\nmodels.append(Model('DT', DecisionTreeClassifier()))\nmodels.append(Model('RF', RandomForestClassifier()))\nmodels.append(Model('GB', GradientBoostingClassifier()))\n\nfor m in models:\n m.model.fit(X, Y)\n print(m.name)\n Yhat = m.model.predict(Xval)\n auc = roc_auc_score(Yval, Yhat)\n print(\"AUC: {}\".format(auc))\n",
"LR\nAUC: 0.7562438021911996\nKNN\nAUC: 0.7629646861150472\nDT\nAUC: 0.7479104910126788\n"
],
[
"kList = [1, 3, 5, 10, 25, 50, 100]\ntr_auc = np.zeros(len(kList))\nva_auc = np.zeros(len(kList))\n\n\nfor i, kValue in enumerate(kList):\n model = KNeighborsClassifier(n_neighbors=kValue)\n model.fit(X,Y)\n Yhat = model.predict(X)\n tr_auc[i] = roc_auc_score(Y, Yhat)\n Yhat = model.predict(Xval)\n va_auc[i] = roc_auc_score(Yval, Yhat)\n \nplt.plot(kList, tr_auc, 'r', label='Training AUC', marker='o')\nplt.plot(kList, va_auc, 'g', label='Validation AUC', marker='s')\nplt.title(\"Training and Validation AUC vs K\")\nplt.xlabel(\"K\")\nplt.ylabel(\"AUC\")\nplt.xticks(kList)\nplt.legend(framealpha=0.75)\nplt.show()\nprint(\"Train AUC: {}\".format(tr_auc))\nprint(\"Validation AUC: {}\".format(va_auc))",
"_____no_output_____"
],
[
"maxDepth = [1, 3, 5, 10, 25, 30, 40, 50, 100]\ntr_auc = np.zeros(len(maxDepth))\nva_auc = np.zeros(len(maxDepth))\n\nfor i, d in enumerate(maxDepth):\n model = DecisionTreeClassifier(max_depth=d)\n model.fit(X,Y)\n Yhat = model.predict(X)\n tr_auc[i] = roc_auc_score(Y, Yhat)\n Yhat = model.predict(Xval)\n va_auc[i] = roc_auc_score(Yval, Yhat)\n \nplt.plot(maxDepth, tr_auc, 'r', label='Training AUC', marker='o')\nplt.plot(maxDepth, va_auc, 'g', label='Validation AUC', marker='s')\nplt.title(\"Training and Validation AUC vs Max Depth\")\nplt.xlabel(\"Max Depth\")\nplt.ylabel(\"AUC\")\nplt.xticks(kList)\nplt.legend(framealpha=0.75)\nplt.show()\nprint(\"Train AUC: {}\".format(tr_auc))\nprint(\"Validation AUC: {}\".format(va_auc))",
"_____no_output_____"
],
[
"trees = [1, 3, 5, 10, 25, 30, 40, 50, 100]\ntr_auc = np.zeros(len(trees))\nva_auc = np.zeros(len(trees))\n\nfor i, t in enumerate(trees):\n model = RandomForestClassifier(n_estimators=t)\n model.fit(X,Y)\n Yhat = model.predict(X)\n tr_auc[i] = roc_auc_score(Y, Yhat)\n Yhat = model.predict(Xval)\n va_auc[i] = roc_auc_score(Yval, Yhat)\n \nplt.plot(trees, tr_auc, 'r', label='Training AUC', marker='o')\nplt.plot(trees, va_auc, 'g', label='Validation AUC', marker='s')\nplt.title(\"Training and Validation AUC vs Number of Trees\")\nplt.xlabel(\"Number of Trees\")\nplt.ylabel(\"AUC\")\nplt.xticks(trees)\nplt.legend(framealpha=0.75)\nplt.show()\nprint(\"Train AUC: {}\".format(tr_auc))\nprint(\"Validation AUC: {}\".format(va_auc))",
"_____no_output_____"
],
[
"nEstimators = [1, 3, 5, 10, 25, 30, 40, 50, 100, 250, 500, 1000]\ntr_auc = np.zeros(len(nEstimators))\nva_auc = np.zeros(len(nEstimators))\n\nfor i, n in enumerate(nEstimators):\n model = GradientBoostingClassifier(n_estimators=n)\n model.fit(X,Y)\n Yhat = model.predict(X)\n tr_auc[i] = roc_auc_score(Y, Yhat)\n Yhat = model.predict(Xval)\n va_auc[i] = roc_auc_score(Yval, Yhat)\n \nplt.plot(nEstimators, tr_auc, 'r', label='Training AUC', marker='o')\nplt.plot(nEstimators, va_auc, 'g', label='Validation AUC', marker='s')\nplt.title(\"Training and Validation AUC vs Number of Boosting Stages\")\nplt.xlabel(\"Number of Boosting Stages\")\nplt.ylabel(\"AUC\")\nplt.xticks(nEstimators)\nplt.legend(framealpha=0.75)\nplt.show()\nprint(\"Train AUC: {}\".format(tr_auc))\nprint(\"Validation AUC: {}\".format(va_auc))",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
c514dff4c529a8a70fd8a3a04948fd0303662010
| 369,161 |
ipynb
|
Jupyter Notebook
|
Google Play store Apps - Exploratory Data Analysis.ipynb
|
subhasushi/Google_Playstore_Apps
|
62668e1580ff2b9cea5e3789f238b0d8a8aeae3c
|
[
"CC-BY-3.0"
] | 1 |
2020-11-22T04:13:56.000Z
|
2020-11-22T04:13:56.000Z
|
Google Play store Apps - Exploratory Data Analysis.ipynb
|
subhasushi/Google_Playstore_Apps
|
62668e1580ff2b9cea5e3789f238b0d8a8aeae3c
|
[
"CC-BY-3.0"
] | null | null | null |
Google Play store Apps - Exploratory Data Analysis.ipynb
|
subhasushi/Google_Playstore_Apps
|
62668e1580ff2b9cea5e3789f238b0d8a8aeae3c
|
[
"CC-BY-3.0"
] | 2 |
2019-11-15T17:35:52.000Z
|
2020-04-10T00:53:47.000Z
| 745.779798 | 173,600 | 0.946162 |
[
[
[
"#import all the dependencies\nimport os\nimport csv\nimport pandas as pd\nimport matplotlib.pyplot as plt\nimport seaborn as sns",
"_____no_output_____"
],
[
"#read the csv files to view the data\ngoogle_apps = pd.read_csv(\"googleplaystore.csv\")\ngoogle_apps.shape",
"_____no_output_____"
]
],
[
[
"# Data Cleaning",
"_____no_output_____"
]
],
[
[
"#Check for number of apps in total\nno_apps = google_apps[\"App\"].nunique()\nprint(f\"Total number of unique apps: {no_apps}\")\n\n#dropping all the duplicate apps from the dataframe\ngoogle_apps.drop_duplicates(subset = \"App\", inplace = True)\ngoogle_apps\n\n#size of the apps are not consistent so convert all to same measure and replace any other values with nan and \"\"\ngoogle_apps[\"Size\"] = google_apps[\"Size\"].apply(lambda x: str(x).replace(\",\", \"\") if \",\" in str(x) else x)\ngoogle_apps[\"Size\"] = google_apps[\"Size\"].apply(lambda x: str(x).replace('M', '') if 'M' in str(x) else x)\ngoogle_apps[\"Size\"] = google_apps[\"Size\"].apply(lambda x: str(x).replace(\"Varies with device\", \"NAN\") if \"Varies with device\" in str(x) else x)\ngoogle_apps[\"Size\"] = google_apps[\"Size\"].apply(lambda x: float(str(x).replace('k', '')) / 1000 if 'k' in str(x) else x)\n\n#convert all the sizes to float\n# google_apps = google_apps.drop([10472])\ngoogle_apps[\"Size\"] = google_apps[\"Size\"].apply(lambda x:float(x))\n\n#Install column has '+' sign so removing that will help easy computation\ngoogle_apps[\"Installs\"] = google_apps[\"Installs\"].apply(lambda x:x.replace(\"+\",\"\")if \"+\" in str(x) else x)\ngoogle_apps[\"Installs\"] = google_apps[\"Installs\"].apply(lambda x: x.replace(\",\",\"\") if \",\" in str(x) else x)\ngoogle_apps[\"Installs\"] = google_apps[\"Installs\"].apply(lambda x:float(x))\n\n#Make the price column consistent by removing the '$' symbol and replacing \"Free\" with 0\ngoogle_apps[\"Price\"] = google_apps[\"Price\"].apply(lambda x: x.replace(\"Free\",0) if \"Free\" in str(x) else x)\ngoogle_apps[\"Price\"] = google_apps[\"Price\"].apply(lambda x:x.replace(\"$\",\"\") if \"$\" in str(x)else x)\ngoogle_apps[\"Price\"] = google_apps[\"Price\"].apply(lambda x: float(x))\n \ngoogle_apps[\"Price\"].dtype \n",
"Total number of unique apps: 9659\n"
]
],
[
[
"# Exploratory Analysis",
"_____no_output_____"
]
],
[
[
"#Basic pie chart to view distribution of apps across various categories\nfig, ax = plt.subplots(figsize=(10, 10), subplot_kw=dict(aspect=\"equal\"))\nnumber_of_apps = google_apps[\"Category\"].value_counts()\nlabels = number_of_apps.index\nsizes = number_of_apps.values\nax.pie(sizes,labeldistance=2,autopct='%1.1f%%')\nax.legend(labels=labels,loc=\"right\",bbox_to_anchor=(0.9, 0, 0.5, 1))\nax.axis(\"equal\")\nplt.show()",
"_____no_output_____"
],
[
"#looking at the number of installs in the top 5 categories and their geners\nno_of_apps_category = google_apps[\"Category\"].value_counts()\nno_of_apps_category[0:5]\nnumber_of_installs = google_apps[\"Installs\"].groupby(google_apps[\"Category\"]).sum()\nprint(f\"Number of installs in family: {number_of_installs.loc['FAMILY']}\")\nprint(f\"Number of installs in Game: {number_of_installs.loc['GAME']}\")\nprint(f\"Number of installs in Tools: {number_of_installs.loc['TOOLS']}\")\n\n#Plotting a simple bar graph to represent the number of installs in each category\nplt.figure(figsize=(10,8))\nsns.barplot(x=\"Category\", y=\"Installs\", data=google_apps,\n label=\"Total Installs\", color=\"b\")\nplt.xticks(rotation=90)\nplt.show()\nprint(\"Top 3 categories in terms of number of installations are: Communication,Video Players and Entertainment\")\n",
"Number of installs in family: 4427941505.0\nNumber of installs in Game: 13878924415.0\nNumber of installs in Tools: 8001771915.0\n"
],
[
"#Let's look at why family even though has lot of apps does not have the highest number of installs. Price could be one of the factors\npaid_apps = google_apps[google_apps[\"Price\"] != 0.0]\n\npaid_family_apps = paid_apps[paid_apps[\"Category\"]==\"FAMILY\"]\npaid_family_apps.count()\n\npaid_communications_apps = paid_apps[paid_apps[\"Category\"]==\"COMMUNICATION\"]\npaid_communications_apps.count()\n\n#Let's visualize this in the form of a simple bar graph\nplt.figure(figsize=(10,8))\nsns.barplot(x=\"Category\", y=\"Price\", data=paid_apps,\n label=\"Total Paid Apps in Each Category\")\nplt.xticks(rotation=90)\nplt.show()\n",
"D:\\Su_Data_Downloads\\Python\\lib\\site-packages\\scipy\\stats\\stats.py:1713: FutureWarning: Using a non-tuple sequence for multidimensional indexing is deprecated; use `arr[tuple(seq)]` instead of `arr[seq]`. In the future this will be interpreted as an array index, `arr[np.array(seq)]`, which will result either in an error or a different result.\n return np.add.reduce(sorted[indexer] * weights, axis=axis) / sumval\n"
],
[
"#Ratings of the apps over various categories\navg_rating = google_apps[\"Rating\"].mean()\nprint(avg_rating)\nplt.figure(figsize=(10,8))\nsns.boxplot('Category','Rating',data=google_apps)\nplt.title(\"Distribution of Categorywise Ratings\")\nplt.ylabel(\"Rating\")\nplt.xlabel(\"Category\")\nplt.xticks(rotation=90)\n# plt.savefig('data_images/plot3a_income.png',bbox_inches='tight')\nplt.show();",
"4.173243045387998\n"
],
[
"#Paid Vs free and the number of installs\ninstalls_greater_1000 = google_apps[google_apps[\"Installs\"]>1000]\ninstalls_greater_1000 = installs_greater_1000.sort_values(['Price'])",
"_____no_output_____"
],
[
"plt.figure(figsize=(20,20))\nsns.catplot(x=\"Installs\", y=\"Price\",data=installs_greater_1000);\nplt.xticks(rotation=90)\n# plt.ytick.direction('out')\nplt.show()",
"_____no_output_____"
],
[
"#take a deeper look at the apps priced more than $100\nexpensive_apps = google_apps[google_apps[\"Price\"]>100]\nexpensive_apps[\"Installs\"].groupby(expensive_apps[\"App\"]).sum()",
"_____no_output_____"
],
[
"#number of installs Vs price Vs Category\nsns.relplot(x=\"Installs\", y=\"Price\", hue=\"Category\", size=\"Rating\",\n sizes=(200, 400), alpha=1,\n height=5, data=expensive_apps)\nplt.show()",
"_____no_output_____"
]
],
[
[
"# Conclusions\n",
"_____no_output_____"
]
],
[
[
"print(f\"The Top three category of Apps based on the number of Apps are\")\nprint(f\" - Family\")\nprint(f\" - Game\")\nprint(f\" - Tool\")\n\nprint(f\"The bottom three category of Apps based on the number of Apps are\")\nprint(f\" - Parenting\")\nprint(f\" - Comics\")\nprint(f\" - Beauty\")\n\nprint(f\"This is not the case when we look at the number of intalls. Based on number of installs, Communication,Video players and entertainment are the top 3 categories\")\nprint(f\"To find out why, I looked at the price of paid apps in each category and clearly, communication was priced less than the family apps. This could be one of the reasons\")\n\nprint(f\"-----------------------------------------------------------------------------------------------------------------------------\")\nprint(f\"The Average rating of the apps across all the categories is 4.17\")\nprint(f\"-----------------------------------------------------------------------------------------------------------------------------\")\nprint(f\"Users tend to download more free apps compared to paid apps. This being said, there are people who are willing to pay more than $100 for an app\")\nprint(f\"-----------------------------------------------------------------------------------------------------------------------------\")\n\nprint(f\"Based on the data, Users tend to buy apps which are priced $1 - $30 compared to other expensive apps\")\nprint(f\"-----------------------------------------------------------------------------------------------------------------------------\")\nprint(f\"There are 20 apps which cost above $100. Finance, Lifestyle and family being the top 3 categories.\")\nprint(f\"-----------------------------------------------------------------------------------------------------------------------------\")\n\n\n\nprint(f\"Among the most expensive apps, 'I am Rich' is the most popular app with the most number of installs\")\n",
"The Top three category of Apps based on the number of Apps are\n - Family\n - Game\n - Tool\nThe bottom three category of Apps based on the number of Apps are\n - Parenting\n - Comics\n - Beauty\nThis is not the case when we look at the number of intalls. Based on number of installs, Communication,Video players and entertainment are the top 3 categories\nTo find out why, I looked at the price of paid apps in each category and clearly, communication was priced less than the family apps. This could be one of the reasons\n-----------------------------------------------------------------------------------------------------------------------------\nThe Average rating of the apps across all the categories is 4.17\n-----------------------------------------------------------------------------------------------------------------------------\nUsers tend to download more free apps compared to paid apps. This being said, there are people who are willing to pay more than $100 for an app\n-----------------------------------------------------------------------------------------------------------------------------\nBased on the data, Users tend to buy apps which are priced $1 - $30 compared to other expensive apps\n-----------------------------------------------------------------------------------------------------------------------------\nThere are 20 apps which cost above $100. Finance, Lifestyle and family being the top 3 categories.\n-----------------------------------------------------------------------------------------------------------------------------\nAmong the most expensive apps, 'I am Rich' is the most popular app with the most number of installs\n"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
c514e11104e6aafd9f78db9ebe8e675850ec7dce
| 19,622 |
ipynb
|
Jupyter Notebook
|
Assignment.ipynb
|
Abhishek5456/Assignments_Session_1_To_Session_5
|
86add7c5d5bca7fde7426b472aecd093e346b68d
|
[
"Unlicense"
] | null | null | null |
Assignment.ipynb
|
Abhishek5456/Assignments_Session_1_To_Session_5
|
86add7c5d5bca7fde7426b472aecd093e346b68d
|
[
"Unlicense"
] | null | null | null |
Assignment.ipynb
|
Abhishek5456/Assignments_Session_1_To_Session_5
|
86add7c5d5bca7fde7426b472aecd093e346b68d
|
[
"Unlicense"
] | null | null | null | 23.929268 | 838 | 0.482163 |
[
[
[
"<center><h2>Assignment</h2></center>",
"_____no_output_____"
],
[
"<h3>2.1. Problem Statement: PYTHON 1</h3>",
"_____no_output_____"
],
[
"<b>1. Install Jupyter notebook and run the first program and share the screenshot of the output.</b>",
"_____no_output_____"
]
],
[
[
"str = \"Hello Python. This is my First Program\";\nprint(str);",
"Hello Python. This is my First Program\n"
]
],
[
[
"<b>2. Write a program which will find all such numbers which are divisible by 7 but are not a\nmultiple of 5, between 2000 and 3200 (both included). The numbers obtained should be printed\nin a comma-separated sequence on a single line.</b>",
"_____no_output_____"
]
],
[
[
"lst = [];\nfor i in range(2000,3200):\n if(i%7 == 0 and i%5 != 0):\n lst.append(i);\nprint(lst);",
"[2002, 2009, 2016, 2023, 2037, 2044, 2051, 2058, 2072, 2079, 2086, 2093, 2107, 2114, 2121, 2128, 2142, 2149, 2156, 2163, 2177, 2184, 2191, 2198, 2212, 2219, 2226, 2233, 2247, 2254, 2261, 2268, 2282, 2289, 2296, 2303, 2317, 2324, 2331, 2338, 2352, 2359, 2366, 2373, 2387, 2394, 2401, 2408, 2422, 2429, 2436, 2443, 2457, 2464, 2471, 2478, 2492, 2499, 2506, 2513, 2527, 2534, 2541, 2548, 2562, 2569, 2576, 2583, 2597, 2604, 2611, 2618, 2632, 2639, 2646, 2653, 2667, 2674, 2681, 2688, 2702, 2709, 2716, 2723, 2737, 2744, 2751, 2758, 2772, 2779, 2786, 2793, 2807, 2814, 2821, 2828, 2842, 2849, 2856, 2863, 2877, 2884, 2891, 2898, 2912, 2919, 2926, 2933, 2947, 2954, 2961, 2968, 2982, 2989, 2996, 3003, 3017, 3024, 3031, 3038, 3052, 3059, 3066, 3073, 3087, 3094, 3101, 3108, 3122, 3129, 3136, 3143, 3157, 3164, 3171, 3178, 3192, 3199]\n"
]
],
[
[
"<b>3. Write a Python program to accept the user's first and last name and then getting them\nprinted in the the reverse order with a space between first name and last name.</b>",
"_____no_output_____"
]
],
[
[
"firstName = input(\"Enter your first Name: \");\nlastName = input(\"Enter your last Name: \");\n\nprint(firstName[::-1] +' '+ lastName[::-1]);",
"Enter your first Name: Abhishek\nEnter your last Name: Trivedi\nkehsihbA idevirT\n"
]
],
[
[
"<b>4. Write a Python program to find the volume of a sphere with diameter 12 cm.</b>",
"_____no_output_____"
]
],
[
[
"pie = 3.14\ndiameter = 12\nvolume = (4/3)*pie*(12/2)**3\nprint(\"volume: {0:.2f}\".format(volume))",
"volume: 904.32\n"
]
],
[
[
"<h3>2.2. Problem Statement: PYTHON 2</h3>",
"_____no_output_____"
],
[
"<b>1. Write a program which accepts a sequence of comma-separated numbers from console and\ngenerate a list.</b>",
"_____no_output_____"
]
],
[
[
"lst = input(\"Enter sequence: \")\nprint(lst.split(','))",
"Enter sequence: 4,5,6,10\n['4', '5', '6', '10']\n"
]
],
[
[
"<b>2 Create the below pattern using nested for loop in Python.\n \n *\n * *\n * * *\n * * * *\n * * * * *\n * * * *\n * * *\n * *\n *\n</b>",
"_____no_output_____"
]
],
[
[
"for i in range(1, 10):\n if i < 6:\n print('* '*i);\n else :\n print('* '*(10-i));",
"* \n* * \n* * * \n* * * * \n* * * * * \n* * * * \n* * * \n* * \n* \n"
]
],
[
[
"<b>3. Write a Python program to reverse a word after accepting the input from the user.<br/>\nSample Input - AcadGild <br/>\nSample Output - dliGdacA\n</b>",
"_____no_output_____"
]
],
[
[
"input_str = input(\"Write Something:\")\nprint(input_str[::-1])",
"Write Something:AcadGild\ndliGdacA\n"
]
],
[
[
"<b>4. Write a Python Program to print the given string in the format specified in the<br/>\nSample Input - WE, THE PEOPLE OF INDIA, having solemnly resolved to constitute India into a SOVEREIGN,\nSOCIALIST, SECULAR, DEMOCRATIC REPUBLIC and to secure to all its citizens<br/>\n<br/>\nSample Output - <br/>\n WE, THE PEOPLE OF INDIA,<br/>\n having solemnly resolved to constitute India into a SOVEREIGN,<br/>\n SOCIALIST, SECULAR, DEMOCRATIC<br/>\n REPUBLIC and to secure to all its citizens\n</b>",
"_____no_output_____"
]
],
[
[
"print(\"WE, THE PEOPLE OF INDIA,\");\nprint(\"\\thaving solemnly resolved to constitute India into a SOVEREIGN,\")\nprint(\"\\t\\tSOCIALIST, SECULAR, DEMOCRATIC\")\nprint(\"\\t\\t\\tREPUBLIC and to secure to all its citizens\")",
"WE, THE PEOPLE OF INDIA,\n\thaving solemnly resolved to constitute India into a SOVEREIGN,\n\t\tSOCIALIST, SECULAR, DEMOCRATIC\n\t\t\tREPUBLIC and to secure to all its citizens\n"
]
],
[
[
"<h3>2.3. Problem Statement: PYTHON 3</h3>",
"_____no_output_____"
],
[
"<b>1.1. Write a Python Program to implement your own myreduce() function which works exactly\nlike Python's built-in function reduce()</b>",
"_____no_output_____"
]
],
[
[
"def myreduce(function, data):\n result = data[0]\n for item in data[1:]:\n result = function(result,item)\n return result\n\n#driver test\ndef sum(x,y):\n return x+y;\nprint(myreduce(sum, [1,2,3,4,5]))",
"15\n"
]
],
[
[
"<b>1.2. Write a Python program to implement your own myfilter() function which works exactly\nlike Python's built-in function filter()</b>",
"_____no_output_____"
]
],
[
[
"def myfilter(function, data):\n result = []\n for item in data:\n if function(item):\n result.append(item)\n return result\n\n#driver test\ndef isGreaterthan5(x):\n if x>5:\n return True\n else:\n return False\nprint(myfilter(isGreaterthan5, [2,6,1,8,9]))",
"[6, 8, 9]\n"
]
],
[
[
"<b>2. Write List comprehensions to produce the following Lists.</b>",
"_____no_output_____"
],
[
"<b>['A', 'C', 'A', 'D', 'G', 'I', ’L’, ‘ D’]</b>",
"_____no_output_____"
]
],
[
[
"lst = [letter for letter in \"ACADGILD\"]\nprint(lst)",
"['A', 'C', 'A', 'D', 'G', 'I', 'L', 'D']\n"
]
],
[
[
"<b>['x', 'xx', 'xxx', 'xxxx', 'y', 'yy', 'yyy', 'yyyy', 'z', 'zz', 'zzz', 'zzzz']</b>",
"_____no_output_____"
]
],
[
[
"Char_lst = [i*j for i in ['x', 'y', 'z'] for j in range(1,5)]\nprint(Char_lst)",
"['x', 'xx', 'xxx', 'xxxx', 'y', 'yy', 'yyy', 'yyyy', 'z', 'zz', 'zzz', 'zzzz']\n"
]
],
[
[
"<b>['x', 'y', 'z', 'xx', 'yy', 'zz', 'xxx', 'yyy', 'zzz', 'xxxx', 'yyyy', 'zzzz']</b>",
"_____no_output_____"
]
],
[
[
"Char_lst = [i*j for j in range(1,5) for i in ['x', 'y', 'z']]\nprint(Char_lst)",
"['x', 'y', 'z', 'xx', 'yy', 'zz', 'xxx', 'yyy', 'zzz', 'xxxx', 'yyyy', 'zzzz']\n"
]
],
[
[
"<b>[[2], [3], [4], [3], [4], [5], [4], [5], [6]]</b>",
"_____no_output_____"
]
],
[
[
"lst = [[x+y] for x in range(2,5) for y in range(3)]\nprint(lst)",
"[[2], [3], [4], [3], [4], [5], [4], [5], [6]]\n"
]
],
[
[
"<b>[[2, 3, 4, 5], [3, 4, 5, 6], [4,5, 6, 7], [5, 6, 7, 8]]</b>",
"_____no_output_____"
]
],
[
[
"list_of_lst = [list(map(lambda x:x+i, [1,2,3,4])) for i in range(1,5)]\nprint(list_of_lst)",
"[[2, 3, 4, 5], [3, 4, 5, 6], [4, 5, 6, 7], [5, 6, 7, 8]]\n"
]
],
[
[
"<b>[(1, 1), (2, 1), (3, 1), (1, 2), (2, 2), (3, 2), (1, 3), (2, 3), (3, 3)]</b>",
"_____no_output_____"
]
],
[
[
"Pairs = [(y,x) for x in [1,2,3] for y in [1,2,3]]\nprint(Pairs)",
"[(1, 1), (2, 1), (3, 1), (1, 2), (2, 2), (3, 2), (1, 3), (2, 3), (3, 3)]\n"
]
],
[
[
"<b>3. Implement a function longestWord() that takes a list of words and returns the longest one.</b>",
"_____no_output_____"
]
],
[
[
"def longestWord(word_list):\n word_len = []\n for item in word_list :\n word_len.append((len(item), item))\n word_len.sort()\n return word_len[-1][1]\n\nprint(longestWord([\"Final\", \"Finally\", \"Finalize\"]))",
"Finalize\n"
]
],
[
[
"<h3>2.4. Problem Statement: PYTHON 4</h3>",
"_____no_output_____"
],
[
"<b>\n1.1 Write a Python Program(with class concepts) to find the area of the triangle using the below\nformula.<br/>\narea = (s*(s-a)*(s-b)*(s-c)) ** 0.5<br/>\nFunction to take the length of the sides of triangle from user should be defined in the parent<br/>\nclass and function to calculate the area should be defined in subclass.\n</b>",
"_____no_output_____"
]
],
[
[
"class Triangle:\n ''' Class to initialize Triangle object '''\n def __init__(self, a,b,c):\n self.a = a\n self.b = b\n self.c = c\n\nclass Area(Triangle):\n def __init__(self, *args):\n super(Area, self).__init__(*args)\n self.parimeter = (self.a+self.b+self.c)/2\n \n def getArea(self):\n return (self.parimeter*(self.parimeter-self.a)*(self.parimeter-self.b)*(self.parimeter-self.c))**0.5",
"_____no_output_____"
],
[
"C = Area(2,4,5)\nprint(\"Area of Triangle: {0:.2f}\".format(C.getArea()))",
"Area of Triangle: 3.80\n"
]
],
[
[
"<b>1.2 Write a function filter_long_words() that takes a list of words and an integer n and returns\nthe list of words that are longer than n.</b>",
"_____no_output_____"
]
],
[
[
"def filter_long_words(words_list, n):\n result = []\n for item in words_list :\n if len(item) > n:\n result.append(item)\n \n return result\n\n#driver test\nwords = [\"Apple\", \"Orange\", \"PineApple\", \"Guava\"]\nlength =5\nprint(\"Words in {} greater than length {} are {}\".format(words, length, filter_long_words(words,length)))",
"Words in ['Apple', 'Orange', 'PineApple', 'Guava'] greater than length 5 are ['Orange', 'PineApple']\n"
]
],
[
[
"<b>2.1 Write a Python program using function concept that maps list of words into a list of integers\nrepresenting the lengths of the corresponding words.</b>",
"_____no_output_____"
]
],
[
[
"def maplength(words_list):\n result = []\n for item in words_list:\n result.append(len(item))\n return result\n\n#driver test\nwords = [\"Apple\", \"Orange\", \"PineApple\", \"Guava\"]\nprint(\"length of words in {} are {}\".format(words, maplength(words)))",
"length of words in ['Apple', 'Orange', 'PineApple', 'Guava'] are [5, 6, 9, 5]\n"
]
],
[
[
"<b>2.2 Write a Python function which takes a character (i.e. a string of length 1) and returns True if\nit is a vowel, False otherwise.<b/>",
"_____no_output_____"
]
],
[
[
"def checkVowel(char):\n if len(char) > 1:\n return False\n elif char.lower() in ['a', 'e', 'i', 'o', 'u']:\n return True\n else:\n return False\n \n#driver test\nchar = 'a'\nprint(\"Is {} vowel?: {}\".format(char, checkVowel(char)))\nchar = 'b'\nprint(\"Is {} vowel?: {}\".format(char, checkVowel(char)))",
"Is a vowel?: True\nIs b vowel?: False\n"
]
],
[
[
"<h3>2.5. Problem Statement: PYTHON 5</h3>",
"_____no_output_____"
],
[
"<b>1. Write a function to compute 5/0 and use try/except to catch the exceptions.</b>",
"_____no_output_____"
]
],
[
[
"def Compute():\n try:\n print(5/0)\n except ZeroDivisionError:\n print(\"You cannot divide by 0\")\n else:\n print(\"Division done successfully\")\n \nCompute(); ",
"You cannot divide by 0\n"
]
],
[
[
"<b>2. Implement a Python program to generate all sentences where subject is in [\"Americans\",\n\"Indians\"] and verb is in [\"Play\", \"watch\"] and the object is in [\"Baseball\",\"cricket\"].</b>",
"_____no_output_____"
]
],
[
[
"subjects = [\"Americans\", \"Indians\"]\nverbs = [\"play\", \"watch\"]\nobjects = [\"Baseball\", \"Cricket\"]\n\nfor sub in subjects:\n for verb in verbs:\n for obj in objects:\n print(sub+\" \"+verb+\" \"+obj)",
"Americans play Baseball\nAmericans play Cricket\nAmericans watch Baseball\nAmericans watch Cricket\nIndians play Baseball\nIndians play Cricket\nIndians watch Baseball\nIndians watch Cricket\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
c514e5e3aa7c9a189d72f397c30070b5db134b7b
| 2,703 |
ipynb
|
Jupyter Notebook
|
module02_intermediate_python/02_00answer_maze_comprehension.ipynb
|
mmlchang/rse-course
|
86baedd4b949109efad12534ec15a9a4698e259e
|
[
"CC-BY-3.0"
] | null | null | null |
module02_intermediate_python/02_00answer_maze_comprehension.ipynb
|
mmlchang/rse-course
|
86baedd4b949109efad12534ec15a9a4698e259e
|
[
"CC-BY-3.0"
] | null | null | null |
module02_intermediate_python/02_00answer_maze_comprehension.ipynb
|
mmlchang/rse-course
|
86baedd4b949109efad12534ec15a9a4698e259e
|
[
"CC-BY-3.0"
] | null | null | null | 22.338843 | 174 | 0.471328 |
[
[
[
"# Solution",
"_____no_output_____"
],
[
"With this maze structure:",
"_____no_output_____"
]
],
[
[
"house = {\n \"living\": {\n \"exits\": {\"north\": \"kitchen\", \"outside\": \"garden\", \"upstairs\": \"bedroom\"},\n \"people\": [\"James\"],\n \"capacity\": 2,\n },\n \"kitchen\": {\"exits\": {\"south\": \"living\"}, \"people\": [], \"capacity\": 1},\n \"garden\": {\"exits\": {\"inside\": \"living\"}, \"people\": [\"Sue\"], \"capacity\": 3},\n \"bedroom\": {\n \"exits\": {\"downstairs\": \"living\", \"jump\": \"garden\"},\n \"people\": [],\n \"capacity\": 1,\n },\n}",
"_____no_output_____"
]
],
[
[
"We can get a simpler dictionary with just capacities like this:",
"_____no_output_____"
]
],
[
[
"{name: room[\"capacity\"] for name, room in house.items()}",
"_____no_output_____"
]
],
[
[
"To get the current number of occupants, we can use a similar dictionary comprehension. Remember that we can *filter* (only keep certain rooms) by adding an `if` clause:",
"_____no_output_____"
]
],
[
[
"{name: len(room[\"people\"]) for name, room in house.items() if len(room[\"people\"]) > 0}",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
c514e5e73aba10498535ca41e8b483a3e5f85c23
| 24,612 |
ipynb
|
Jupyter Notebook
|
PowerPlant/kafka.ipynb
|
biljiang/pyprojects
|
10095c6b8f2f32831e8a36e122d1799f135dc5df
|
[
"MIT"
] | null | null | null |
PowerPlant/kafka.ipynb
|
biljiang/pyprojects
|
10095c6b8f2f32831e8a36e122d1799f135dc5df
|
[
"MIT"
] | null | null | null |
PowerPlant/kafka.ipynb
|
biljiang/pyprojects
|
10095c6b8f2f32831e8a36e122d1799f135dc5df
|
[
"MIT"
] | null | null | null | 61.53 | 1,630 | 0.623842 |
[
[
[
"from pykafka import KafkaClient\nclient = KafkaClient(hosts=\"192.168.0.93:9092,192.168.0.94:9092,192.168.0.95:9092\")",
"_____no_output_____"
],
[
"client = KafkaClient(hosts=\"127.0.0.1:9092,127.0.0.1:9093,127.0.0.1:9094\")",
"_____no_output_____"
],
[
"client.topics\ntopic = client.topics['Tran_detail_analyzed']",
"_____no_output_____"
],
[
"client.topics\ntopic = client.topics[\"my-replicated-topic\"]",
"_____no_output_____"
],
[
"consumer = topic.get_simple_consumer()\nfor message in consumer:\n if message is not None:\n print (message.offset, message.value)",
"0 b'my test message1'\n1 b'my test message2'\n2 b'what a fail over mechanic'\n3 b'It is very nice of you to find kafa\\x08k'\n4 b'this is kafka test with pythn'\n5 b'this is test for cumsume function'\n6 b'is it effective'\n7 b'what is the bolck mechanics'\n8 b'this is very interesting'\n"
],
[
"from pykafka.common import OffsetType\nconsumer = topic.get_simple_consumer(\n consumer_group=\"console-consumer-74603\",\n #auto_offset_reset=OffsetType.EARLIEST,\n #reset_offset_on_start=True\n )",
"_____no_output_____"
],
[
"from datetime import datetime\nprint(\"Start time:\",datetime.strftime(datetime.now(),\"%Y-%m-%d %H:%M:%S\"))\nfor i in range(1000000):\n consume=consumer.consume() \nprint(\"Start time:\",datetime.strftime(datetime.now(),\"%Y-%m-%d %H:%M:%S\"))",
"Start time: 2018-11-15 12:21:40\nStart time: 2018-11-15 12:23:29\n"
],
[
"import time\ntime.strftime(\"%Y-%m-%d %H:%M:%S\",time.localtime())",
"_____no_output_____"
],
[
"from datetime import datetime\ndatetime.now()\ndatetime.strftime(datetime.now(),\"%Y-%m-%d %H:%M:%S\")",
"_____no_output_____"
],
[
"\nfor message in consumer:\n if message is None:\n\n break\n #print (message.offset, message.value)\n",
"Start time: 2018-11-15 11:55:14\n"
],
[
"consumer.fetch_offsets()",
"_____no_output_____"
],
[
"topic",
"_____no_output_____"
],
[
"consume.value",
"_____no_output_____"
],
[
"message.timestamp",
"_____no_output_____"
],
[
"consumer.partition_cycle?",
"_____no_output_____"
],
[
"datetime.fromtimestamp(1542340357024/1000)",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
c514e7e04745b00982f64e940cdc37185b627028
| 170,801 |
ipynb
|
Jupyter Notebook
|
ml-h/h2-tfnn.ipynb
|
vishwakarmarhl/mlearn
|
01c86d02517a9c03eaf3330cd6d237de17464256
|
[
"MIT"
] | 1 |
2018-04-17T08:11:51.000Z
|
2018-04-17T08:11:51.000Z
|
ml-h/h2-tfnn.ipynb
|
vishwakarmarhl/mlearn
|
01c86d02517a9c03eaf3330cd6d237de17464256
|
[
"MIT"
] | null | null | null |
ml-h/h2-tfnn.ipynb
|
vishwakarmarhl/mlearn
|
01c86d02517a9c03eaf3330cd6d237de17464256
|
[
"MIT"
] | null | null | null | 161.133019 | 31,036 | 0.860241 |
[
[
[
"###### ECE 283: Homework 2\n###### Topics: Classification using neural networks\n###### Due: Monday April 30\n\n- Neural networks; Tensorflow\n- 2D synthetic gaussian mixture data for binary classification",
"_____no_output_____"
],
[
"### Report\n\n----------------------------------------\n\n##### 1. Tensorflow based neural network\n\n- 2D Gaussian mixture is synthesized based on the provided mean, covariances for class 0 and 1.\n- Training, validation and test sample counts are 70, 20, and 10 respectively\n\n##### (a) One hidden layer: Implementation code below In[7] : oneHiddenNeuralNetwork()\n\nBelow are the parameters that are used to run training for this network.\nThe validation data is used to compute loss/accuracy in order to tune the hyper parameters.\n```\n Hyper Parameters\n learning_rate = 0.001\n num_steps = 1000\n batch_size = 1000\n display_step = 100\n reg_const_lambda = 0.01\n\n Network Parameters\n n_hidden_1 = 9 # 1st layer number of neurons\n num_input = 2 # data input (shape: 2 * 70)\n num_classes = 1 # total classes (0 or 1 based on the value)\n```\n###### Execution:\n\n1. Without input preprocessing: Single Layer Network\n\n > Log\n > - Trn Step 1, Minibatch Loss= 2.3662, Accuracy= 49.500\n > - Val Step 1, Minibatch Loss= 2.4016, Accuracy= 48.800\n > - Trn Step 100, Minibatch Loss= 1.8325, Accuracy= 58.437\n > - Val Step 100, Minibatch Loss= 1.8935, Accuracy= 57.050\n > - Trn Step 1000, Minibatch Loss= 0.6166, Accuracy= 79.854\n > - Val Step 1000, Minibatch Loss= 0.6331, Accuracy= 79.000\n\n > - Test Accuracy: 80.800\n > - Diff Error: 192/1000\n\n2. With input preprocessing: Single Layer Network \n\n > Log\n > - Trn Step 1, Minibatch Loss= 1.3303, Accuracy= 30.100\n > - Val Step 1, Minibatch Loss= 1.6977, Accuracy= 33.150\n > - Trn Step 100, Minibatch Loss= 1.0398, Accuracy= 36.600\n > - Val Step 100, Minibatch Loss= 1.2065, Accuracy= 37.400\n > - Trn Step 1000, Minibatch Loss= 0.5143, Accuracy= 80.700\n > - Val Step 1000, Minibatch Loss= 0.5572, Accuracy= 76.700\n\n > - Test Accuracy: 77.100\n > - Diff Error: 229/1000\n\n\n\n###### Observations: Q 1,2,3,4\n\n1. The number of neurons here are 10 which provided more accuracy over single neuron. Upon changing the number of neurons from 1 to 10 we see a jump of accuracy from 50% to 75%. However growing neurons beyond 10 does not provide much benefit/accuracy change on the validation data. Which says that training further may be overfitting to the training dataset.\n\n2. Training samples are 70% and validation samples are 20%. When we run for 1000 steps/epoch with batch size 1000 on a learning rate of 0.001. We see that training loss converges towards 0.5572, while training accuracy converges from 30% to 80%. The validation values appear to be peaks at 77%. Training was stopped when we saw consistent convergence and similar accuracy on the validation and the test dataset.\n \n > - Upon changing the learning rate to a higher value like 1 we see that convergence is an issue. This was observed since the data kept alternating between two values consistently, irrespective of the iterations ran. When learning rate is of the order 10**(-3) then we see the convergence in the data.\n > - The L2 regularization constant will penalize the square value of the weights and it is set to 0.01 here. When we changed the value to say 10 it will allow for a higher order coefficient to affect and may cause over fitting. However, it does not seem to affect the results here and it may be due to the fact that the higher order coefficient do not affect this data. \n > - Final Test Accuracy: 77.1%\n\n3. Input pre-processing and Weight Initialization\n\n > Normalization/input-preprocessing is achieved by subracting the mean and scaling with standard deviation. \n > - The function getNextTrainBatch() was without normalization and gave the results in sections 1 and 2 above. Upon using normalized batch training data using getNextNormalizedTrainBatch() function we have the following observations,\n > - The convergence was relatively faster than before (1 Step)\n > - The batch loss reduced to ~0.5572 while the accuracy on test was around 77%\n\n > Weight initialization has a major impact since these multipliers lead to vanishing or exploding gradients issue. \n > - In the current scenario we have used random_normal distribution for initialization. In the currnt scenario since the convergence is fast and data is separable we do not see any difference by using uniform initialization. However there are datasets that demonstrate the empirical benefit of using a uniform distribution for initializing weights.\n\n4. Comparing the performance of neural network from HW1 (MAP, Kernelized Logistic Regression and Logistic Regression with feature engg)\n\n > - We observed a probability of error around 23% here.\n > - Misclassification rate in MAP was around 16% for class0 and 47% for class1\n > - Misclassification in Kernelized Logistic regression was slightly better than the MAP\n > - Misclassification for Logistic regression by feature engineering was around 56% for class0 and 10% for class1\nHowever if we see the overall misclassification error rate we get a great accuracy of about 77%-81% using the neural network technique here. We can be sure that upon learning more data this technique will provide a better accuracy.\n\n\n##### (b) Two hidden layer: Implementation code below In[11] : twoHiddenNeuralNetwork()\n\nBelow are the parameters that are used to run training for this network\n```\n Hyper Parameters\n learning_rate = 0.001\n num_steps = 1000\n batch_size = 1000\n display_step = 100\n reg_const_lambda = 0.01\n\n Network Parameters\n n_hidden_1 = 4 # 1st layer number of neurons\n n_hidden_2 = 4 # 2nd layer number of neurons\n num_input = 2 # data input (shape: 2 * 70)\n num_classes = 1 # total classes (0 or 1 based on the value)\n\n```\n###### Execution:\n\n1. Without input preprocessing: Two Layer Network\n\n > Log\n > - Trn Step 1, Minibatch Loss= 1.8265, Accuracy= 67.295\n > - Val Step 1, Minibatch Loss= 1.9003, Accuracy= 66.800\n > - Trn Step 100, Minibatch Loss= 1.2101, Accuracy= 80.126\n > - Val Step 100, Minibatch Loss= 1.2648, Accuracy= 80.550\n > - Trn Step 1000, Minibatch Loss= 1.0394, Accuracy= 83.812\n > - Val Step 1000, Minibatch Loss= 1.0760, Accuracy= 83.750\n\n > - Test Accuracy: 83.600\n > - Diff Error: 164/1000\n\n2. With input preprocessing: Two Layer Network\n\n > Log\n > - Trn Step 1, Minibatch Loss= 2.0676, Accuracy= 30.800\n > - Val Step 1, Minibatch Loss= 2.1635, Accuracy= 27.600\n > - Trn Step 100, Minibatch Loss= 0.8971, Accuracy= 51.700\n > - Val Step 100, Minibatch Loss= 1.0530, Accuracy= 51.000\n > - Trn Step 1000, Minibatch Loss= 0.6649, Accuracy= 80.600\n > - Val Step 1000, Minibatch Loss= 0.6496, Accuracy= 83.100\n\n > - Test Accuracy: 81.900\n > - Diff Error: 181/1000\n\n\n\n###### Observations: Additional observations for two layer network only\n\n1. In this case we see a better result when compared to the single network but that may not always the case. Upon increasing the number of layers to two we add more capacity. By doing this we allow for complex fitting of the weights which leads to good results on the training data and I.I.D. test data. \n2. On increasing the learning rate we see that the convergence is quick around the loss value of 0.6496\n\n > - Final Test Accuracy: 81.900\n > - All the same observations as described for a single layer network as above. However here we see that due to higher capacity a better linear accuracy is observed at every neuron count.\n\n3. The number of neurons here are 4 each and provides a peak accuracy at that value. An overfitting may be occurring beyond that.\n\n > - The test accuracy is 82% but we also see that the training and validation accuracy are 81% and 83% respectively. This is data specific and can be improved by increasing the size of the training data. 10000 test samples is not a great sample to compute a general accuracy for the entire dataset.\n\n\n\n\n\n",
"_____no_output_____"
],
[
"# Code Section",
"_____no_output_____"
]
],
[
[
"# -*- coding: utf-8 -*-\n\nimport tensorflow as tf\nimport numpy as np\nfrom math import *\nimport matplotlib.pyplot as plt\nfrom matplotlib import cm\nfrom mpl_toolkits.mplot3d import Axes3D\n\nfrom scipy.stats import norm\nfrom IPython.display import Image, display, Math, Latex\n\n\n# Params\nn_inpoints = 10000",
"_____no_output_____"
],
[
"def generateClass0():\n theta0 = 0\n lmb01 = 2\n lmb02 = 1\n m0 = (0, 0)\n # computing u * u.T and later multiplying with lambda\n cov01 = [[(cos(theta0))**2, cos(theta0)*sin(theta0)],\n [(sin(theta0))*cos(theta0), (sin(theta0))**2]]\n cov02 = [[(sin(theta0))**2, -(cos(theta0)*sin(theta0))],\n [-(cos(theta0)*sin(theta0)), (cos(theta0))**2]]\n cov0 = lmb01*np.matrix(cov01) + lmb02*np.matrix(cov02)\n cov0_det = np.linalg.det(cov0)\n x0, y0 = np.random.multivariate_normal(m0, cov0, int(n_inpoints/2)).T\n return x0,y0\n\nx0, y0 = generateClass0()\nplt.scatter(x0, y0, color = 'r',marker='x', label = 'Cl 0')\nplt.legend()\nplt.title('Distribution of Class 0')\nplt.show()",
"_____no_output_____"
],
[
"def generateClass1():\n # Mixture A\n theta1a = -3*pi/4\n lmb1a1 = 2\n lmb1a2 = 1/4\n m1a = (-2, 1)\n cov1a = [[(cos(theta1a))**2, cos(theta1a)*sin(theta1a)],\n [(sin(theta1a))*cos(theta1a), (sin(theta1a))**2]]\n cov2a = [[(sin(theta1a))**2, -(cos(theta1a)*sin(theta1a))],\n [-(cos(theta1a)*sin(theta1a)), (cos(theta1a))**2]]\n cov1a = lmb1a1*np.matrix(cov1a) + lmb1a2*np.matrix(cov2a)\n cov1a_det = np.linalg.det(cov1a)\n x1a, y1a = np.random.multivariate_normal(m1a, cov1a, int(n_inpoints/2)).T\n #print('Shape: ',x1a.shape,', ',y1a.shape,', ',cov1a)\n\n # Mixture B\n theta1b = pi/4\n lmb1b1 = 3\n lmb1b2 = 1\n m1b = (3, 2)\n cov1b = [[(cos(theta1b))**2, cos(theta1b)*sin(theta1b)],\n [(sin(theta1b))*cos(theta1b), (sin(theta1b))**2]]\n cov2b = [[(sin(theta1b))**2, -(cos(theta1b)*sin(theta1b))],\n [-(cos(theta1b)*sin(theta1b)), (cos(theta1b))**2]]\n cov1b = lmb1b1*np.matrix(cov1b) + lmb1b2*np.matrix(cov2b)\n cov1b_det = np.linalg.det(cov1b)\n x1b, y1b = np.random.multivariate_normal(m1b, cov1b, int(n_inpoints/2)).T\n #print('Shape: ',x1b.shape,', ',y1b.shape,', ',cov1b)\n\n # Class 1 (A * 0.33 +B * 0.66)\n y1 = np.array(y1a)* (1 / 3)+np.array(y1b)* (2 / 3)\n x1 = np.array(x1a)* (1 / 3)+np.array(x1b)* (2 / 3)\n return x1,y1\n\nx1, y1 = generateClass1()\nplt.scatter(x1, y1, color = 'b',marker='^', label = 'Cl 1')\nplt.title('Distribution of Class 1')\nplt.legend()\nplt.show()",
"_____no_output_____"
],
[
"x = np.concatenate((x0, x1))\ny = np.concatenate((y0, y1))\nprint('Shape; X:',x.shape,', Y:',y.shape)\nplt.scatter(x0, y0, color = 'r',marker='x', label = 'Cl 0')\nplt.scatter(x1, y1, color = 'b',marker='^', label = 'Cl 1')\nplt.legend()\nplt.show()\n\nc0 = np.vstack((x0, y0)).T\nc1 = np.vstack((x1, y1)).T\n# ----------------------------------------\n# Set up the [xi, yi] training data vector\n# ----------------------------------------\nX = np.concatenate((c0,c1), axis = 0)\nY = np.array([0]*int(n_inpoints/2) + [1]*int(n_inpoints/2)).reshape(n_inpoints,1)",
"Shape; X: (10000,) , Y: (10000,)\n"
]
],
[
[
"### Training, test and validation sets (70:20:10)\n\n##### Without Normalization",
"_____no_output_____"
]
],
[
[
"# Divide the data into Train Valid, Test\ntot_count = n_inpoints\ntrn_count = int(0.7 * tot_count)\nval_count = int(0.2 * tot_count)\ntst_count = int(0.1 * tot_count)\n\n# Shuffle X & Y values\nsfl_idx = np.arange(0,tot_count)\nnp.random.shuffle(sfl_idx)\n\nXc0 = X[:,0]\nXc1 = X[:,1]\nXc0 = Xc0.reshape(tot_count,1)\nXc1 = Xc1.reshape(tot_count,1)\nprint(Xc1.shape)\n\ntrain_X0 = Xc0[sfl_idx[np.arange(0,trn_count)]]\ntrain_X1 = Xc1[sfl_idx[np.arange(0,trn_count)]]\ntrain_Y = Y[sfl_idx[np.arange(0,trn_count)]]\nn_samples = train_X1.shape[0]\n\nvalid_X0 = Xc0[sfl_idx[np.arange(trn_count,trn_count+val_count)]]\nvalid_X1 = Xc1[sfl_idx[np.arange(trn_count,trn_count+val_count)]]\nvalid_X = np.vstack((valid_X0.T, valid_X1.T))\nvalid_Y = Y[sfl_idx[np.arange(trn_count,trn_count+val_count)]]\n\ntests_X0 = Xc0[sfl_idx[np.arange(trn_count+val_count, tot_count)]]\ntests_X1 = Xc1[sfl_idx[np.arange(trn_count+val_count, tot_count)]]\ntests_X = np.vstack((tests_X0.T, tests_X1.T))\ntests_Y = Y[sfl_idx[np.arange(trn_count+val_count, tot_count)]]\n\nbatchIndex = 0\ndef getNextTrainBatch(size):\n global batchIndex\n if((batchIndex + size) >= trn_count):\n size = trn_count-1\n batchIndex = 0 # recycle the batches from start\n \n #trn_sfl_idx = np.arange(0,trn_count)\n #np.random.shuffle(trn_sfl_idx)\n trn_X0_r1 = train_X0[np.arange(batchIndex, batchIndex + size)]\n trn_X1_r1 = train_X1[np.arange(batchIndex, batchIndex + size)]\n trn_Y_r1 = train_Y[np.arange(batchIndex, batchIndex + size)]\n #print(trn_X0_r1.shape)\n trn_X = np.vstack((trn_X0_r1.T, trn_X1_r1.T))\n #print((trn_X.T).shape)\n batchIndex = batchIndex + size\n return trn_X.T, trn_Y_r1\n\nprint('Train: ',train_X0.shape, train_Y.shape)\nprint('Valid: ',valid_X.shape, valid_Y.shape)\nprint('Tests: ',tests_X.shape, tests_Y.shape)",
"(10000, 1)\nTrain: (7000, 1) (7000, 1)\nValid: (2, 2000) (2000, 1)\nTests: (2, 1000) (1000, 1)\n"
]
],
[
[
"##### With Normalization",
"_____no_output_____"
]
],
[
[
"# ------------------- \n# Normalize the data\n# ------------------- \n\n# Mean\ntrain_X0_mean = np.mean(train_X0)\ntrain_X1_mean = np.mean(train_X1)\n\n# Standard deviation\ntrain_X0_stddev = np.std(train_X0)\ntrain_X1_stddev = np.std(train_X1)\n\n# Normalization by scaling using standard deviation\ntrain_X0_nrm = (train_X0 - train_X0_mean)/train_X0_stddev\ntrain_X1_nrm = (train_X1 - train_X1_mean)/train_X1_stddev\n\nprint(train_X0_nrm.shape)\nprint(train_X1_nrm.shape)\n\ntrain_X_nrm = np.vstack((train_X0_nrm.T, train_X1_nrm.T))\n\ndef getNextNormalizedTrainBatch(size):\n global batchIndex\n batchIndex = 0\n if((batchIndex + size) >= trn_count):\n size = trn_count-1\n batchIndex = 0 # recycle the batches from start\n # Shuffle the dataset each time\n trn_sfl_idx = np.arange(batchIndex, batchIndex + size)\n np.random.shuffle(trn_sfl_idx)\n trn_X0_r1 = train_X0_nrm[trn_sfl_idx[np.arange(batchIndex, batchIndex + size)]]\n trn_X1_r1 = train_X1_nrm[trn_sfl_idx[np.arange(batchIndex, batchIndex + size)]]\n trn_Y_r1 = train_Y[trn_sfl_idx[np.arange(batchIndex, batchIndex + size)]]\n #print(trn_X0_r1.shape)\n trn_X = np.vstack((trn_X0_r1.T, trn_X1_r1.T))\n #print((trn_X.T).shape)\n batchIndex = batchIndex + size\n return trn_X.T, trn_Y_r1\n\nprint('Train: ',train_X_nrm.shape, train_Y.shape)\n#print('Valid: ',valid_X.shape, valid_Y.T)\n#print('Tests: ',tests_X.shape, tests_Y.T)",
"(7000, 1)\n(7000, 1)\nTrain: (2, 7000) (7000, 1)\n"
],
[
"def linearRegression():\n # Parameters\n learning_rate = 0.01\n training_epochs = 500\n display_step = 50\n rng = np.random\n\n # tf Graph Input\n Xtf = tf.placeholder(tf.float32, [None, 1])\n Ytf = tf.placeholder(tf.float32, [None, 1])\n\n # Set model weights\n # figure tf.rand\n # tf.keras.initializer\n Wtf = tf.Variable(np.zeros([1,1]), dtype=tf.float32, name=\"weight\")\n btf = tf.Variable(np.zeros([1,1]), dtype=tf.float32, name=\"bias\")\n\n # Construct a linear model\n predtf = tf.add(tf.matmul(Xtf, Wtf), btf)\n\n # Mean squared error\n costtf = tf.reduce_sum(tf.pow(predtf-Ytf, 2))/(2*n_samples)\n # Gradient descent\n optimizer = tf.train.GradientDescentOptimizer(learning_rate).minimize(costtf)\n\n # Initialize the variables (i.e. assign their default value)\n init = tf.global_variables_initializer()\n\n # Start training\n with tf.Session() as sess:\n sess.run(init)\n\n # Fit all training data\n for epoch in range(training_epochs):\n sess.run(optimizer, feed_dict={Xtf: train_X1, Ytf: train_Y})\n #Display logs per epoch step\n if (epoch+1) % display_step == 0:\n c = sess.run(costtf, feed_dict={Xtf: train_X1, Ytf:train_Y})\n print(\"Epoch:\", '%04d' % (epoch+1), \"cost=\", \"{:.9f}\".format(c), \\\n \"W=\", sess.run(Wtf), \"b=\", sess.run(btf))\n print(\"Optimization Finished!\")\n training_cost = sess.run(costtf, feed_dict={Xtf: train_X1, Ytf: train_Y})\n print(\"Training cost=\", training_cost, \"W=\", sess.run(Wtf), \"b=\", sess.run(btf), '\\n')\n\n #Graphic display plt.plot(train_X, train_Y, 'ro', label='Original data')\n plt.scatter(x0, y0, color = 'r',marker='x', label = 'Cl 0')\n plt.scatter(x1, y1, color = 'b',marker='^', label = 'Cl 1')\n plt.plot(train_X1, sess.run(Wtf) * train_X1 + sess.run(btf), label='Fitted line')\n plt.legend()\n plt.show()\n sess.close()\n\n# Run Linear Regression\nlinearRegression()",
"Epoch: 0050 cost= 0.086520113 W= [[0.22172657]] b= [[0.14955907]]\nEpoch: 0100 cost= 0.075541042 W= [[0.26486838]] b= [[0.20467149]]\nEpoch: 0150 cost= 0.073922656 W= [[0.2683425]] b= [[0.23227467]]\nEpoch: 0200 cost= 0.073287055 W= [[0.26424062]] b= [[0.2495297]]\nEpoch: 0250 cost= 0.072956562 W= [[0.25965545]] b= [[0.2615027]]\nEpoch: 0300 cost= 0.072778456 W= [[0.25591362]] b= [[0.27013934]]\nEpoch: 0350 cost= 0.072682224 W= [[0.25307462]] b= [[0.2764516]]\nEpoch: 0400 cost= 0.072630167 W= [[0.2509661]] b= [[0.28108466]]\nEpoch: 0450 cost= 0.072602026 W= [[0.24941045]] b= [[0.28448996]]\nEpoch: 0500 cost= 0.072586790 W= [[0.2482652]] b= [[0.286994]]\nOptimization Finished!\nTraining cost= 0.07258679 W= [[0.2482652]] b= [[0.286994]] \n\n"
]
],
[
[
"### Neural Network implementation\n- 1.(a) One hidden layer",
"_____no_output_____"
]
],
[
[
"def oneHiddenNeuralNetwork():\n # Parameters\n learning_rate = 0.001\n num_steps = 1000\n batch_size = 1000\n display_step = 100\n reg_const_lambda = 0.01\n\n # Network Parameters\n n_hidden_1 = 9 # 1st layer number of neurons\n num_input = 2 # data input (shape: 2 * 70)\n num_classes = 1 # total classes (0 or 1 based on the value)\n\n # tf Graph input\n X = tf.placeholder(\"float\", [None, num_input])\n Y = tf.placeholder(\"float\", [None, num_classes])\n\n # Store layers weight & bias (initializing using random nromal)\n weights = {\n 'h1': tf.Variable(tf.random_normal([num_input, n_hidden_1])),\n 'out': tf.Variable(tf.random_normal([n_hidden_1, num_classes]))\n }\n biases = {\n 'b1': tf.Variable(tf.random_normal([n_hidden_1])),\n 'out': tf.Variable(tf.random_normal([num_classes]))\n }\n\n # Create model\n def one_neural_net(x):\n # Hidden fully connected layer, a1 \n layer_1 = tf.add(tf.matmul(x, weights['h1']), biases['b1'])\n #layer_1 = tf.nn.relu(layer_1)\n # Output fully connected layer with a neuron for each class \n out_layer = tf.matmul(layer_1, weights['out']) + biases['out']\n return out_layer\n\n # Construct model\n logits = one_neural_net(X)\n output = tf.sigmoid(logits) # Convert output to a probability\n\n # Define loss and optimizer\n cel_loss_op = tf.reduce_mean(tf.nn.sigmoid_cross_entropy_with_logits(logits=logits, labels=Y))\n reg_loss = tf.nn.l2_loss(weights['h1']) + tf.nn.l2_loss(weights['out']) # L2 regularization\n loss_op = tf.reduce_mean(cel_loss_op + reg_const_lambda*reg_loss) \n \n optimizer = tf.train.GradientDescentOptimizer(learning_rate=learning_rate)\n train_op = optimizer.minimize(loss_op)\n\n # Evaluate model (with test logits, for dropout to be disabled)\n # keep in mind boolean to float32 tensor output\n #correct_pred = tf.equal(tf.argmax(logits, 1), tf.argmax(Y, 1))\n correct_pred = tf.cast(tf.greater(output, 0.5), tf.float32)\n accuracy = tf.reduce_mean(tf.cast(tf.equal(correct_pred, Y), tf.float32))\n\n # Initialize the variables (i.e. assign their default value)\n init = tf.global_variables_initializer()\n\n # Start training\n batchIndex = 0\n with tf.Session() as sess:\n # Run the initializer\n sess.run(init)\n for step in range(1, num_steps+1):\n batch_x, batch_y = getNextNormalizedTrainBatch(batch_size)\n # Run optimization op (backprop)\n # print(batch_x)\n # print(batch_y)\n sess.run(train_op, feed_dict={X: batch_x, Y: batch_y})\n if step % display_step == 0 or step == 1:\n # Training batch loss and accuracy\n loss, acc, pred = sess.run([loss_op, accuracy, correct_pred], feed_dict={X: batch_x,\n Y: batch_y})\n print(\"Trn Step \" + str(step) + \", Minibatch Loss= \" + \\\n \"{:.4f}\".format(loss) + \", Accuracy= \" + \\\n \"{:.3f}\".format(100*acc)) \n #print(\"actuals:\", batch_y.T)\n #print(\"predict:\", pred.T)\n print(\"differr:\", (pred.T != batch_y.T).sum())\n # Validation accuracy\n loss_v, acc_v, pred_v = sess.run([loss_op, accuracy, correct_pred], feed_dict={X: valid_X.T,\n Y: valid_Y})\n print(\"Val Step \" + str(step) + \", Minibatch Loss= \" + \\\n \"{:.4f}\".format(loss_v) + \", Accuracy= \" + \\\n \"{:.3f}\".format(100*acc_v))\n #print(\"actuals:\", valid_Y.T)\n #print(\"predict:\", pred_v.T)\n print(\"differr:\", (pred_v.T != valid_Y.T).sum())\n\n print(\"Optimization Finished!\")\n # Calculate accuracy for test data\n acc_t, pred_t = sess.run([accuracy,correct_pred], feed_dict={X: tests_X.T, Y: tests_Y})\n print(\"Test Accuracy:\", \"{:.3f}\".format(100*acc_t))\n print(\"actuals:\", tests_Y.shape)\n print(\"predict:\", pred_t.shape)\n print(\"differr:\", (pred_t.T != tests_Y.T).sum())\n sess.close()\n\n# Execute\noneHiddenNeuralNetwork() ",
"Trn Step 1, Minibatch Loss= 1.3303, Accuracy= 30.100\ndifferr: 699\nVal Step 1, Minibatch Loss= 1.6977, Accuracy= 33.150\ndifferr: 1337\nTrn Step 100, Minibatch Loss= 1.0398, Accuracy= 36.600\ndifferr: 634\nVal Step 100, Minibatch Loss= 1.2065, Accuracy= 37.400\ndifferr: 1252\nTrn Step 200, Minibatch Loss= 0.8524, Accuracy= 47.100\ndifferr: 529\nVal Step 200, Minibatch Loss= 0.8916, Accuracy= 48.650\ndifferr: 1027\nTrn Step 300, Minibatch Loss= 0.7363, Accuracy= 59.500\ndifferr: 405\nVal Step 300, Minibatch Loss= 0.7172, Accuracy= 64.550\ndifferr: 709\nTrn Step 400, Minibatch Loss= 0.6629, Accuracy= 65.900\ndifferr: 341\nVal Step 400, Minibatch Loss= 0.6273, Accuracy= 73.700\ndifferr: 526\nTrn Step 500, Minibatch Loss= 0.6144, Accuracy= 71.800\ndifferr: 282\nVal Step 500, Minibatch Loss= 0.5822, Accuracy= 76.900\ndifferr: 462\nTrn Step 600, Minibatch Loss= 0.5808, Accuracy= 75.400\ndifferr: 246\nVal Step 600, Minibatch Loss= 0.5606, Accuracy= 77.900\ndifferr: 442\nTrn Step 700, Minibatch Loss= 0.5567, Accuracy= 76.900\ndifferr: 231\nVal Step 700, Minibatch Loss= 0.5517, Accuracy= 78.200\ndifferr: 436\nTrn Step 800, Minibatch Loss= 0.5387, Accuracy= 78.800\ndifferr: 212\nVal Step 800, Minibatch Loss= 0.5500, Accuracy= 77.450\ndifferr: 451\nTrn Step 900, Minibatch Loss= 0.5250, Accuracy= 80.000\ndifferr: 200\nVal Step 900, Minibatch Loss= 0.5525, Accuracy= 77.250\ndifferr: 455\nTrn Step 1000, Minibatch Loss= 0.5143, Accuracy= 80.700\ndifferr: 193\nVal Step 1000, Minibatch Loss= 0.5572, Accuracy= 76.700\ndifferr: 466\nOptimization Finished!\nTest Accuracy: 77.100\nactuals: (1000, 1)\npredict: (1000, 1)\ndifferr: 229\n"
]
],
[
[
"\n- 1.(b) Two hidden layer",
"_____no_output_____"
]
],
[
[
"def twoHiddenNeuralNetwork():\n # Parameters\n learning_rate = 0.01\n num_steps = 1000\n batch_size = 1000\n display_step = 100\n reg_const_lambda = 0.01\n\n # Network Parameters\n n_hidden_1 = 4 # 1st layer number of neurons\n n_hidden_2 = 4 # 2nd layer number of neurons\n num_input = 2 # data input (shape: 2 * 70)\n num_classes = 1 # total classes (0 or 1 based on the value)\n\n # tf Graph input\n X = tf.placeholder(\"float\", [None, num_input])\n Y = tf.placeholder(\"float\", [None, num_classes])\n\n # Store layers weight & bias (initializing using random nromal)\n weights = {\n 'h1': tf.Variable(tf.random_normal([num_input, n_hidden_1])),\n 'h2': tf.Variable(tf.random_normal([n_hidden_1, n_hidden_2])),\n 'out': tf.Variable(tf.random_normal([n_hidden_2, num_classes]))\n }\n biases = {\n 'b1': tf.Variable(tf.random_normal([n_hidden_1])),\n 'b2': tf.Variable(tf.random_normal([n_hidden_2])),\n 'out': tf.Variable(tf.random_normal([num_classes]))\n }\n\n # Create model\n def two_neural_net(x):\n # Hidden fully connected layer, a1 \n layer_1 = tf.add(tf.matmul(x, weights['h1']), biases['b1'])\n layer_1 = tf.nn.relu(layer_1)\n layer_2 = tf.add(tf.matmul(layer_1, weights['h2']), biases['b2'])\n layer_2 = tf.nn.relu(layer_2)\n # Output fully connected layer with a neuron for each class \n out_layer = tf.matmul(layer_2, weights['out']) + biases['out']\n return out_layer\n\n # Construct model\n logits = two_neural_net(X)\n output = tf.sigmoid(logits) # Convert output to a probability\n\n # Define loss and optimizer\n cel_loss_op = tf.reduce_mean(tf.nn.sigmoid_cross_entropy_with_logits(logits=logits, labels=Y))\n reg_loss = tf.nn.l2_loss(weights['h1']) + tf.nn.l2_loss(weights['h2']) + tf.nn.l2_loss(weights['out']) # L2 regularization\n loss_op = tf.reduce_mean(cel_loss_op + reg_const_lambda*reg_loss) \n \n optimizer = tf.train.GradientDescentOptimizer(learning_rate=learning_rate)\n train_op = optimizer.minimize(loss_op)\n \n # Evaluate model (with test logits, for dropout to be disabled)\n # keep in mind boolean to float32 tensor output\n #correct_pred = tf.equal(tf.argmax(logits, 1), tf.argmax(Y, 1))\n correct_pred = tf.cast(tf.greater(output, 0.5), tf.float32)\n accuracy = tf.reduce_mean(tf.cast(tf.equal(correct_pred, Y), tf.float32))\n\n # Initialize the variables (i.e. assign their default value)\n init = tf.global_variables_initializer()\n\n # Start training\n batchIndex = 0\n with tf.Session() as sess:\n # Run the initializer\n sess.run(init)\n for step in range(1, num_steps+1):\n batch_x, batch_y = getNextNormalizedTrainBatch(batch_size)\n # Run optimization op (backprop)\n # print(batch_x)\n # print(batch_y)\n sess.run(train_op, feed_dict={X: batch_x, Y: batch_y})\n if step % display_step == 0 or step == 1:\n # Training batch loss and accuracy\n loss, acc, pred = sess.run([loss_op, accuracy, correct_pred], feed_dict={X: batch_x,\n Y: batch_y})\n print(\"Trn Step \" + str(step) + \", Minibatch Loss= \" + \\\n \"{:.4f}\".format(loss) + \", Accuracy= \" + \\\n \"{:.3f}\".format(100*acc)) \n #print(\"actuals:\", batch_y.T)\n #print(\"predict:\", pred.T)\n print(\"differr:\", (pred.T != batch_y.T).sum())\n # Validation accuracy\n loss_v, acc_v, pred_v = sess.run([loss_op, accuracy, correct_pred], feed_dict={X: valid_X.T,\n Y: valid_Y})\n print(\"Val Step \" + str(step) + \", Minibatch Loss= \" + \\\n \"{:.4f}\".format(loss_v) + \", Accuracy= \" + \\\n \"{:.3f}\".format(100*acc_v))\n #print(\"actuals:\", valid_Y.T)\n #print(\"predict:\", pred_v.T)\n print(\"differr:\", (pred_v.T != valid_Y.T).sum())\n\n print(\"Optimization Finished!\")\n # Calculate accuracy for test data\n acc_t, pred_t = sess.run([accuracy,correct_pred], feed_dict={X: tests_X.T, Y: tests_Y})\n print(\"Test Accuracy:\", \"{:.3f}\".format(100*acc_t))\n print(\"actuals:\", tests_Y.shape)\n print(\"predict:\", pred_t.shape)\n print(\"differr:\", (pred_t.T != tests_Y.T).sum())\n sess.close()\n\n# Execute\ntwoHiddenNeuralNetwork() ",
"Trn Step 1, Minibatch Loss= 2.0676, Accuracy= 30.800\ndifferr: 692\nVal Step 1, Minibatch Loss= 2.1635, Accuracy= 27.600\ndifferr: 1448\nTrn Step 100, Minibatch Loss= 0.8971, Accuracy= 51.700\ndifferr: 483\nVal Step 100, Minibatch Loss= 1.0530, Accuracy= 51.000\ndifferr: 980\nTrn Step 200, Minibatch Loss= 0.8193, Accuracy= 63.700\ndifferr: 363\nVal Step 200, Minibatch Loss= 0.8234, Accuracy= 67.850\ndifferr: 643\nTrn Step 300, Minibatch Loss= 0.7975, Accuracy= 67.100\ndifferr: 329\nVal Step 300, Minibatch Loss= 0.7820, Accuracy= 73.050\ndifferr: 539\nTrn Step 400, Minibatch Loss= 0.7781, Accuracy= 69.600\ndifferr: 304\nVal Step 400, Minibatch Loss= 0.7564, Accuracy= 76.400\ndifferr: 472\nTrn Step 500, Minibatch Loss= 0.7584, Accuracy= 71.600\ndifferr: 284\nVal Step 500, Minibatch Loss= 0.7352, Accuracy= 78.150\ndifferr: 437\nTrn Step 600, Minibatch Loss= 0.7395, Accuracy= 73.900\ndifferr: 261\nVal Step 600, Minibatch Loss= 0.7148, Accuracy= 79.250\ndifferr: 415\nTrn Step 700, Minibatch Loss= 0.7202, Accuracy= 76.200\ndifferr: 238\nVal Step 700, Minibatch Loss= 0.6950, Accuracy= 81.150\ndifferr: 377\nTrn Step 800, Minibatch Loss= 0.7005, Accuracy= 77.900\ndifferr: 221\nVal Step 800, Minibatch Loss= 0.6776, Accuracy= 82.000\ndifferr: 360\nTrn Step 900, Minibatch Loss= 0.6820, Accuracy= 78.800\ndifferr: 212\nVal Step 900, Minibatch Loss= 0.6624, Accuracy= 82.850\ndifferr: 343\nTrn Step 1000, Minibatch Loss= 0.6649, Accuracy= 80.600\ndifferr: 194\nVal Step 1000, Minibatch Loss= 0.6496, Accuracy= 83.100\ndifferr: 338\nOptimization Finished!\nTest Accuracy: 81.900\nactuals: (1000, 1)\npredict: (1000, 1)\ndifferr: 181\n"
]
],
[
[
"### Results",
"_____no_output_____"
]
],
[
[
"num_neurons = np.arange(0, 15)\naccuracy_1_net = [50,66,57,72,75,72,74,69,77,75,74,70,70,74,75]\naccuracy_2_net = [74,67,78,82,73,78,79,75,78,79,80,80,80,78,80]\nplt.plot(num_neurons, accuracy_2_net, c = 'red' , label = 'Two Layer Network')\nplt.plot(num_neurons, accuracy_1_net, c = 'blue' , label = 'One Layer Network')\nplt.legend()\nplt.title(\"Number of Neurons vs Accuracy\")\nplt.show()",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
c514f0a947e6e930244a61b1f5c8120842f78034
| 35,025 |
ipynb
|
Jupyter Notebook
|
notebooks/Visualizations_Iteration_2.ipynb
|
Spotifier/Data-science
|
a054107cb21d743a9878ce5d14c6b007aa9a022f
|
[
"MIT"
] | 4 |
2019-09-28T02:39:06.000Z
|
2022-03-29T11:07:15.000Z
|
notebooks/Visualizations_Iteration_2.ipynb
|
Spotifier/Data-science
|
a054107cb21d743a9878ce5d14c6b007aa9a022f
|
[
"MIT"
] | 4 |
2020-03-24T18:21:18.000Z
|
2021-08-23T20:44:48.000Z
|
notebooks/Visualizations_Iteration_2.ipynb
|
Build-Week-Spotify-Song-Suggester/Data-science
|
4ba8e52f361995ebc408d42bc43321b49ab80143
|
[
"MIT"
] | 4 |
2019-09-21T20:00:15.000Z
|
2019-09-27T05:59:48.000Z
| 97.835196 | 4,864 | 0.840428 |
[
[
[
"import sys\nimport pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\nfrom sklearn.preprocessing import StandardScaler\nfrom sklearn.neighbors import KDTree\nfrom sklearn.decomposition import PCA \n\n#### Visulization imports\nimport pandas_profiling\nimport plotly.express as px\nimport seaborn as sns\nimport plotly.graph_objs as go\nfrom plotly.offline import download_plotlyjs, init_notebook_mode, plot, iplot",
"_____no_output_____"
],
[
"df_april_19 = pd.read_csv('../data/SpotifyAudioFeaturesApril2019.csv')\ndf_nov_18 = pd.read_csv('../data/SpotifyAudioFeaturesNov2018.csv')",
"_____no_output_____"
],
[
"df = pd.concat([df_april_19, df_nov_18], ignore_index=True)\nprint(df.shape)\nassert df.shape[0] == (df_april_19.shape[0] + df_nov_18.shape[0])",
"_____no_output_____"
],
[
"df = df.drop_duplicates(subset = 'track_id', keep='first')\nprint(df.shape)",
"_____no_output_____"
],
[
"# number_of_songs = 200\n # remove categoricals\ndf_numerics = df.drop(columns=['track_id', 'track_name', 'artist_name'])\n \n # Scale Data To Cluster More Accurately, and fit clustering model\ndf_scaled = StandardScaler().fit_transform(df_numerics)\ndf_modeled = KDTree(df_scaled)\n \n # Querying the model for the 15 Nearest Neighbors\ndist, ind = df_modeled.query(df_scaled, k=(number_of_songs+1))\n \n # Putting the Results into a Dataframe\ndist_df = pd.DataFrame(dist)\n \n # Calculating the Distances\nscores = (1 - ((dist - dist.min()) / (dist.max() - dist.min()))) * 100\n \n # Creating A New Dataframe for the Distances\ncolumns = ['Searched_Song']\nfor i in range(number_of_songs):\n columns.append(f'Nearest_Song{i}')\n\ndist_score = pd.DataFrame(scores.tolist(), columns = columns)\n \n # An Array of all indices of the nearest neighbors\nind[:(number_of_songs+1)]\n \n # Making an array of the Track IDs\nsong_ids = np.array(df.track_id)\n \n # A function that creates list of the each song with its nearest neighbors\ndef find_similars(song_ids, ind):\n similars = []\n for row in ind:\n ids = [song_ids[i] for i in row]\n similars.append(ids)\n\n return similars \n \n # using the above function\nnearest_neighbors = find_similars(song_ids, ind)\n \n # putting the results into a dataframe\nnearest_neighbors_df = pd.DataFrame(nearest_neighbors, columns=columns)\n \n",
"_____no_output_____"
]
],
[
[
"## 3D Representation of a Random Sample From Dataset, Visualized Spacially",
"_____no_output_____"
]
],
[
[
"fig = px.scatter_3d(df.sample(n=5000, random_state=69), x='acousticness', y='liveness', z='tempo', color='loudness', size='popularity',\n opacity=.7, hover_name='track_name', color_discrete_sequence=px.colors.sequential.Plasma[-2::-1],\n template=\"plotly_dark\")\nfig.show()",
"_____no_output_____"
]
],
[
[
"# A variety of Song Selections along with 200 Song recommendations\n## Notice how they generally follow the same trajectory along the path across the features\nThis helping to Visually convey how Songs are recommended based on songs nearest to in terms of quantifable Audio Features such as accoustiness, danceability, energy etc.",
"_____no_output_____"
]
],
[
[
"id_numbers = ''' 16UKw34UY9w40Vc7TOkPpA\t7LYb6OuJcmMsBXnBHacrZE\t0Lpsmg0pmlm1h1SJyWPGN2\t6T8CFjCR5G83Ew3EILL60q\t5ba3vTyegTVbMoLDniANWy\t6VK3ZdppJW3Q6I1plyADxX\t47nZUjQa9NZb7Nheg8gSj0\t5P42OvFcCn5hZm8lzXqNJZ\t77RsQL1RDECVnB3LL7zhTF\t2vqZnmBn0REOMmNp5pMTJz\t1dLHaoG70esepC2eC0ykV4\t4SUQbrebZgvSX8i3aYHMB6\t4D0Xgaln0O8K8LK2gjwpr8\t5ipjhrirlnBV7BMY7QV3H5\t2lvkak4Ik64c4vlAQyek12\t0t4JgAUj8ZCbWOwSU9h4nt\t1RjYRvWpZeh9vMjjKzpH3w\t0YELRuijk4XsKWvyoWY7jI\t3Xn791JUhuITZdLsIuKuQQ\t1Y2wWhbLCHW0WfTczmuA2X\t65CE7YGQzGY4p1MqnfWYZt\t6a6zG2o8geJvBVJkDkFCHQ\t4Vcqv8zsfoNpxr7dWEJi48\t2sfcE3uPqDObs5COsvk7QJ\t2gz8HI5hZew7abJ9gcLY7J\t2UFpXorq5JOIctCwcmDyZ5\t7pNNFcYN2N1T0lOKMHL8u9\t7deuaj4pjJqxWVky0jcFrd\t2eCdpRpnYLp4fj0iMNra3p\t5WyXaXmMpo1fJds5pzmS4c\t2HLNwAHYH7Ejs2rZLLyrmj\t0wXjzthQdMd7SZu2kNwsVC\t3EnzqTwdFWe68x0OTxR9T5\t50rPhDfxSL2kmEovmXqTNf\t3VY3JjW7T0f49JqdFlvqIV\t458Cn793jgrNc6miDUSAiK\t40XOJ16Zc7pqgqYq9o7wjS\t0QuuDvOB9fZ49pZ2cIdEdw\t1f5aQjgYy4mKjA7EgJJvLY\t1QJjIWHLf05mUQPq3N2hxZ\t0wrhAauh8QSw2DFDi6ZHFV\t2K55wT0q49n54mZmA3hqS8\t6glST22VPJZRTKvxecHSp6\t0lvEyZrkTDg0vK9luhcjZg\t5YaV62mxj62GSlXvwzgG3J\t6yC44aQAf9AALUyJPimZ11\t1frCKo4D3lktaPHfkyEuHo\t3hXsGl1WdOuKye1aHo6pF7\t40NAjxDw25daUXVt1b0A0D\t0bkPHOwWOIG6ffwJISGNUr\t6w3401sQAMkeKdQ3z3RPXt\t56UwCbkvU1p3vHTnlbv3kS\t04MkdoV7vxprPhtYA0Cx5y\t7AesCHBrKOy4Npkxt907mG\t5B7w6neMDX6BYPJdb6ikRE\t4AowP9TvejSnEpxxJigpyn\t4M9onsaj8IxHJEFVezMRoA\t2DRNLTuiZr3MdFNfEHzWfz\t4Wo5LyWddbPCogBIBrkhlt\t0UJmSMFB05CyY3dTps6g2c\t7nZR4x2aHeIyzAtrMi4Wua\t6UZVW9DjfRKrcIVco5uwc1\t2O1FwU85kgG0SJGJhszkB0\t4OK4tHSUnCXpBfIusCOdAo\t0MfWpTp3GrJ51bNxLanyy1\t5DVsV3ZetLbmDUak9z0d1E\t3ki056t9qL4g9GHWkPFJYe\t4WCNiW7DJFE6h94q5NPZmZ\t3N0Q5ce0Q3v6MmcNwaGG2p\t7rQFDOKqUEaXE6X6Of4HTw\t0wi0Hn8puUPmYdZ0JvpG2H\t5wMD46niyehV3y5HfeQpNf\t1nTn4pZhcgfRPobs43xrvL\t0NxPZvt6UYWLgTbvjCJd2n\t7fdHvtur1uLx5crFzAfWJ2\t5AZt6HoqpUdHyhia36Khtc\t1exbNAnvvYLYsEFESsCjDO\t27ZfYwqic7RnwuitxJZiE9\t2iPvO3ctXFGlkzOsx6iWyn\t2w8g5LJzKqez8mENuk2pbL\t3aBmFnfx9QfLB3knrKr1Mo\t4UUA76EBTJzcICr2nNyhnV\t4aV1txuotqBFGLB2jwiogo\t7ASmnEp32JgxgH76TAaWwo\t344WuUSk6SRQd9849fkAct\t7aXH7YjPAixvHIPxCKxwIo\t1CakWoqY0bPK9Ov8UocFTR\t2B9VQlYlq6CUH0VXdQqB4y\t3gCPlZpymjidx564rWcPHX\t691J2jGivJasVLkWU11dpU\t0ulEzQTIdtZGvYH3mkK84G\t2XpxTgvloEbIIVfEt4XUKt\t4dqcedp9451K9DvxYugrTt\t2Y6IAs1aCdb4rzFfGjONUo\t7LDtRLCz9D5DOR31jQZ65m\t0oliuZWC43aafuxqNlGuxy\t0Ks2NJH2PCxyWAFPlI4p9B\t7oLqoswT2hfCG90crbiToe\t11wZ39zESerUTPXKWhx7QE\t4HWfA0iD0gXuL6gVreNYTL\t5EFw2MVleUknhnPzfrCrTq\t2drp4ajf2V2xUvV79EmzMw\t6KL8uR3Y3JjFpzzLQFBzQa\t0SYo2aRh2MYfBoJAFOYtNs\t6Iq5a3BvMSx6X7auul0yDE\t6TZUjNnW4qHI9wPrO54L5o\t4v3s1AdtPSBxFK93PNMFSg\t7FM6VwHNF3EWQTyiloogTV\t3FNbf1Qt2ycepS4fasCuOm\t2qK9xZkbBrTRiDw2dnJul8\t5ozbdCZw5MZmJryCOyDYO1\t0M82DdRxHFedS7fg7Gk2qB\t6k1Epe9JbePsbdq0EZCc4i\t63TMt7zR9YLpNBpzRYLG5I\t6tbdFaJWas52BT8DZH76Xj\t4V7gH33fKlEhX4d1uk2xYB\t6jY7PeOZ4P6ww8XuzCyGfO\t3m4nvQbC1n3dm6SbYIDbDR\t6J5ArwJqeLHFKNfHcDP6OG\t4RlzULwFEYBjTJNhc7frWm\t1kZ0mav2lhlhXf4fWjw5Nc\t0gJBsp5q8Ro6zXxKzT4DiQ\t0CWuF6SrEXmfM0EDIERBS1\t0ogRPfqHhhZuaeeVt02L0Z\t4AEJ6dqjb3uo7K9R2xKGJ0\t0b4akisi6edx4RkU3VO1XW\t2xLzmImDWvk0jw92tTsnHk\t2PFvERcsENO2mSXV2abmMW\t57miVDdQOiOx7ZNaEjGaFC\t0LdkVfBGmZUOKf8sway0tM\t5GtQkJTQ01zxZ9xRuIBRyY\t1LX7SGrc4FIE6LnzV498Ow\t2l3OlYqGIiJrPByZNx8Ll6\t1yCb0FSeO48efDRg80Turo\t3r5OR32RDkcp3eIQ2ylF5o\t3grKLoUX87NaEkvouW0vmz\t7ts8ZBKNCtJvd0ijGxTgCw\t6LSlTgBUF1T8rBrTKtzbWB\t0VCTFk3PtHHTbCdiI2SNf6\t5flKCotkqTK0SRHyu9ywOE\t7FNVvZKIFb5VIwyY4tCMXt\t1mc6PrRRhSipTHKSLRuv5B\t1s7X6ZKOMhP2luohWVXNNP\t5WPjMN7nxk2HqcPfewseyz\t2rX3PbfV6OrObng2YL9Osd\t6ahWJqh8GQag4OWmyRbcnE\t3ZYN2cfyCFn4NuWxEW9tuh\t3DchJOgF4JUzQJyoAVePa7\t1fhnlsDdCLs1Oi5X3oVCTD\t3T0UOBcMTeytq7RmFDZMbu\t14gtLymOStY8niLakJlbf8\t677SnHIc0M92Nb6XUnaSCT\t1t2hs48AduLr9wik6nF0pw\t3QavdjzqIxMUPeSXgoA4Di\t4LK5o7buDJB9A3aL86y5dR\t1JAGP2PPls6WXahoN9IM14\t0uteQpEpt2XpZ99ZT7m0eA\t0zm5v1li5HwBcFJZzXz2Iq\t7epZd4ZUwXGq5CTOwW9EO7\t1R8ihhEOnbscF8kheDNC0H\t5gYUBAE3o6k5yBv2Ni7KwQ\t4EuW6g3eq56jUDqdNbUryM\t727FY7suhFAVmwP3tsg6uG\t2j9tX4ubo2WISo9GIJLySx\t3QUtbFgjjnAHTtLup31xVa\t6viaOSezCxDApUQlIc8mhA\t3J0ZbecfqYszqlQJKYswVV\t10aAr61dsWKA9RRdAmk2CM\t7gE8QvR9Pxl7G2ey8XFtwa\t6RF6zRVTz1FUYzBhop3jen\t2stJA4LcpvwPHIRa1Gxp2P\t0yrFVbIvtPU6bb4YMD2Vcr\t68Hwxn8KEb3cXjv3w3eHtV\t6aTdoiCwo5eYrl6ik4jRYH\t3FWU0Aq3QHHkslDWD5sXvJ\t3ckyP4jOXNBskOGeM1E4WY\t137Lgw0gey9uw6hDKI6Los\t4FrbvIGxud4J9DeWC5OYrd\t0d29ZVNUaxWOtUFzElL3B9\t7AvTgaX6gs7L0f1O0qSlDf\t3C3pZzGJJR8wwuh6npPvHv\t3YcmUK7BiWMBJoRWC5p0vi\t3gBPhTsYDm9xtuOt4iFjMW\t6QotxMJ0VE8eh1rvm2alsC\t1fh5YKCSpo4OvC6usURns4\t11bs6ROtD5D1VfDcCje9Sy\t2DLcXvfFrQRm9D1GzMbgMg\t1HqOKMf8bNLaEPvd8NXx3c\t3tN1favTAEXAadxfygjNmG\t7F8ip8rt5cfD18wUTgE7us\t08pFqsZZZYeFbiTGPQj1J8\t512JyhHrndIxZ81JmYZLmP\t5Df1IuQ5AqKIrK1Rplsr9p\t52MsPDozAb8oy9IjsndB6v\t4tYja8TMtjBAejK7pzP2y4\t3s9BUjzYDIesX8PXqcWno3\t4jAbuuhObXbHrJP5ShVOZ8\t7ezSDJfiOAmSt5nYe00VaQ\t1p6BhKjxF03jOd00W6io6O\t56b6kZuturLKiFl9v29tEp\t3YGG0dmOCgA60bQts3J0C2 '''.split('\t')\n\nto_be_parallel_coordinated = df.query('track_id == @id_numbers')\nlen(to_be_parallel_coordinated)\n\npx.parallel_coordinates(to_be_parallel_coordinated, template=\"plotly_dark\")",
"_____no_output_____"
],
[
"id_numbers = ''' 3Rx1zM3nDFQzAOYg9Hd0D4\t67AHtK2jFq8dlOLSSSbQ7T\t2ystp6xiBASPJkFR16gJON\t5VNGj3qgKC1n28B9etIoJv\t6OarwT6HBT8jW6MsnVwn58\t61VbbeUd8bXxzyZYrg4djH\t21rvKibsH3WmojUZh5H3Gm\t11wxWExHmqBNKIo6zK9NEn\t5ZGXAHp0YPYFUMbyMqDQH9\t4BMPGmOzi4H3S31B2Ckx0u\t1VcVGJ4sqRv2Iruxc8CfYf\t1xOoqWTv2wLhUeLtXZTm9q\t4SV8h3RlcuQc9jE9MUQfFF\t5c1Hz72Bc8VMbghi4MJQus\t0iZOviuGDLFc8vSrB4RI2T\t7JRV17HtiiXksxDdTdpYTy\t7apGuGr4Zf6t9JkATkolAI\t0Mw9dLno600aQgA0Gf9Usr\t6jUXJaXtxOhBLeWbpR2kN5\t1nASmYf1d9HiiIgEOPhYQR\t5LAe0lSl7rMle11o6Et9WI\t5LZu2syDoQNaA0NptU1YIs\t0lz57CGwAyuYdMk7BO72XI\t3MDnGMGGC00nbpkLP1r6cN\t4QZpmKzjC5t1OxEKCvL7Ft\t15sVDXzpwJLfHM99VeP7mR\t3Yeb5nDeWTvXfJ4TdlTtIP\t56Tuc3GqQrByXDZu82TfN2\t2jyrDZbZoScSdiTxVRlzb3\t5RHZg80sV4QFq3alySZABa\t3IYkFudbmV1sgbz4riV73P\t0xtEwGTNW1fjQVJM6PZ3U2\t5zllzp3gvXWq2cmyBZReij\t43hjTh4WF2cICX1JhwfE9x\t7BCPy7FIt6MIZwIYjgwHUc\t3HRLlKWdmzXfAmbcrOkevH\t5zTE3LjI0vXoNs5sXe1wBd\t5ijr9nCHXMTb9qYvn3taSg\t0R9HIKNmfmn44AYsSux8Qs\t4AtiPcMHA5VPbNlO4EdB4T\t0Ica23299eon0SQ5GMcJYc\t2xkcKjB8CYW1wXiZ4haZfu\t1kcNoS77udN6sSUWq9mI60\t2kWUZwAhXDQmEvxv6zAxsx\t6a5vpD5O3gMZH7G8xwOv5X\t2mg15L7RUwpaymfobUFHOM\t6HMKAeNDeWkPaHVEwvf6OJ\t6zZeZcCSnugaVt5mCiCCP0\t58xiGZhGtgJGCBDlXwCTbe\t5O4MkYjbKpC3UH7oE7YRQa\t6NBheB7uq3KuwjrriafhSy\t6Tdyv7xZrcnHmO9iQoysKS\t6GJh9XXO7e9D16Eyw0RIuz\t3ayOojGZYT6wNtFC0NDQTm\t79wTeGSVlONiNfZTdyGUNq\t43w1mfDBN6MHueSkUjN7D8\t4HqgpQdgUT12xACerT4yS6\t3XRfdbb65XE1bfrAwlRu28\t3Cv56grsf8F5iWn4BHtZx8\t3YG5WGhUOj8Qzj4q9hF4TE\t2MpCXZtBR02QWKP6xwRqy8\t1WmKw3lMhA5YU869ilylyn\t0vOSZ7hAUxocDU7qPh0VCo\t3rnjAdt1duHuVV5AjavYk2\t3uUzHjzRxKewzg1bE4TJwq\t7M3e3QMHiGgWaGqwaRS0oH\t6JtZVLdOzT6GeTgPzSoGAA\t5u7UqEwOyaEIoA1TLLFpz9\t0TWdTb7si8hunDhLmynRsr\t0fzEYa7EiGDTU9wz976bAX\t1HybrAhpKs9bm4ol6UR8bZ\t4dp22919ccLK9SpvAEfTbA\t4dhR3lLe5XLiR1TDNuGJ25\t2Ovrl3OYjw4Ys4UJJQZaVT\t0KU1n705y9CXC2F6fBOWej\t4sPQHt3Tk3zz2TxBv6iSwu\t1IdFop8kheQ8DF0rFhHiqa\t4Ex2Fk2vc5JOsYptDUBtJA\t1slZlNfFpMAfNiqtf9uYto\t5ykg5P1kKcYCVqF5cHXjYu\t6IGRNK7vC8fuhncF7YXXg9\t1gZRSXSFGgZ2FfTClxI2A9\t46BanJsjr1hqamrvLBYkng\t5IwncSTQf2nC5aTktUNJFQ\t58iaGunPax6nehU5K3AlCO\t5vEwDx78onSExtBl8Q44Qf\t65fd6IOZZjFYkuApCdbGxR\t0G69NybuKLFtOulxwW348d\t1z0b8KGrWldcZLakynC9Hc\t2iaJ69ql68l3uCFtP6Rz0w\t525g3ZvALoI6eTwOnE0dvh\t54Amn3maW5gDB20vIkOzMK\t3ZSj7F0vNEUmr0pJX3ROcD\t0DbubpYjXBCGCrbcVl6YCY\t6gdYVynIAdcSMWIaK3x7lW\t23NI7LEZNcNgqMQ4MtNZPf\t3sVNfmjOawrMVBxZ5HR992\t4CCFVqakDhrAqEBbIeebgw\t4VRoNouo8soGhl3GaFLmdr\t5Mtb2rpcBkZEbNqLx06qfp\t2m2Si8RtoOGPfbIjDx9Ug7\t64SrUvSXvi2DCqwnScNQ87\t7boSAJxzyyCJbP3LcDzssT\t0SgncrTJSvH5xrvkllBZWj\t23ptyiin2PKgaHZW6F0mMa\t6gpomTTKog3SU0if4XT8E3\t71jN5pqWqS1Gq2UXg8IabB\t0yItuTAWCQ4JRvo9a081uD\t0TSzNyWeCGVz9VdwFLWc2k\t4gq34v5gzCtdaL4o8drPBx\t3IR6Za6YHTAeikVF8w1DvK\t2pkluglrMGfygP1yVADsX6\t6sQyFRXaDU3MmLORr6EdNv\t4QtS332yh4ex5KFgcMA40E\t5t6GgWRjcigpk0pXpcwzSO\t1bHaP4ZOPgtpoZ3CN6bIML\t2zT9xdBcvSo1CO8RZ8Tcqj\t0GgFwGjaAdqVga8j3ZKCtl\t7m5LVVSaWzik4h332VqvbN\t1P3RGzIqmcHKvH68e5nkBW\t6uIYA3RVNgr1btPAtr1XXy\t79pqKla5Q9IiAQfK4jalAO\t3KDZxrjgFLKWs7ds2rvVcW\t3yiT9hyDinSAvubb3XZ8S5\t4byppJf1BVIEYj0FV48uN7\t1PihJ1fLjU2wkTatRudSyE\t1rVYJMGey3MZapQwCx6xXn\t3X1MK1cg0in1bV5s8BvI4O\t6xDEZCZm0Ehbzgj1HAqLIe\t5fDXSKPlZQlaq1jC3izCkd\t3JOdpt3Msi1e20Nxmor4o5\t7gLSX6HlNso7WkoWPCGNGr\t0PswjCzT2lZY8EDjVRPrPc\t3XXbyMFA9F4adfcnEjMKHM\t5jM3bDFV7UuyhHA5264QAs\t1KRiMLHjthCAhWqDunAJOV\t79ojwy5zomoWoQNuaOWbKh\t7qbUjczokcnGFIwx68aBqV\t5IKtH5C078QBjDSniwdTXj\t2LfM9NwbQkBFV8XKAwhuTo\t7A2lPmhXhtlZlsRMz0ShNs\t3nSvqC1W3IEhdubx1538g6\t5pFoVXWo5sCBfC5pLZu1Gg\t1XCccHjyDRUdOVrEOpLzoH\t6LeiYw9DsrS6fTGG329tK4\t7md22n0LputBo41lYOG7tA\t6YPafAdayjyjcoPoKIxn6y\t5Tpbw8WbGEwI2pzjxXrGvm\t6ummA8cVxCDnjT9382Ui8G\t3m9yfMVIpEYvNLQZl2f8YF\t37S7watyULcdUTc7z8Opha\t2uOPEftUSMDJK4UpsUjGPO\t2Xv0TmNKxLIV0cVRwM2HFz\t246dN8gCiMv5nHi5wR2Anr\t6i05cmZT3PHtSriKFWxTPn\t06M77pQeFWvFiVn1Be6XsI\t6WW4VgC1CHJjrWxYOtvayZ\t06qD1C1Tcd0mYdRBBmYuTx\t02ZFCSXPFgFPEahuN88kOQ\t06QqCHpEStp7fwJYK4qoB1\t3XuQifZguMGzjZJ7zHw7O8\t7bXHynjjhieyUVyq8PfjHg\t5WGOhaEiVJzjeUbjgPK2ww\t4FXamUtTru5LlMNoCjlBRH\t5oi0T9CsacaGLVECLBKWq5\t5ulm5IhULY27ehqTSrQeLB\t4L0RXCGs4SP8CkrBbZxsfS\t5jYACoLz1e0r07W9G7oqOi\t5PbIFyF34gCASgnG7yi0AG\t0iZU8XzmveXaRiWBpE1ZTI\t4pvwyXkwtXdrKIXpOc0keI\t4wILZuKMKmJZIQxW30u960\t3DrjcLyxLSG3aOh3MvXnUF\t6Zm6DJFgghFMnMw7xBIwyn\t02MMgyaLCvnIBw4skXmZ9V\t1kVyvQzqxOZz4BgAWOY8ps\t6U3j5OkhwwHlVeVgZlyl7n\t6wdOphejlm1hNfFhXmzT0l\t5rNFuymSOcCW8nTfd3vYJn\t7kfZsjQgEApwNuceCzJIp8\t4AhUSi91kDdC4G51qwvDlD\t5Oi4T8e7vZK1xfJgBEWDdd\t5Q5POfYGAdWGSSYLtkVQ4T\t1KgOw1rCe9YWTFbFJYuYjD\t2Z40xmLbAGbv1vQno1YMvJ\t4PgpYEtlH6VfWmds9jVDoT\t0ERjKxvwU91tthphZGgLFn\t45b5fAvIFHBWmEcBGytul1\t5biNqsTCkccqUfmzRFVIPO\t1fdwOBuqrsjf95i8rAMUCC\t0Sm76b6hQobYvHebmCa49H\t73A5MOZ2MJyKw5sigQe64R\t56rBa1McCcF8Q6cyPOAWji\t76B1zH5bbarUGH4CYLfvbS\t1bUQorCYDuyQhIyDYWzNyz\t0eOAeqbD5sxU77qdHSYLOY\t26VXbBYVzPXvl0wAAEppnr\t5DK7vMKUkq3ejNQK1SP2I0\t1E3e15pztQETb3hysHnuDy\t6yl56wrtGJVrnhFJjQvIVS\t1xWDs7mhV3YbENkbEkmvH8 '''.split('\t')\n\nto_be_parallel_coordinated = df.query('track_id == @id_numbers')\nlen(to_be_parallel_coordinated)\n\npx.parallel_coordinates(to_be_parallel_coordinated, template=\"plotly_dark\")",
"_____no_output_____"
],
[
"id_numbers = ''' 6bwTuNxmVEOQw0dXdmgLjC\t4rTVdzMKkbRtcJtbHCtTKm\t09m4moKIXDyQNZDkoDqjNk\t74VJWMSZHMcvkHQhyFmsXk\t6CE0gR4USBQnxKj9vWiotk\t3REJFRU6OZmqWk5neOLPXd\t1jEH3K14qOijd64Sa052fn\t5Z5YYYAFiSsfwOm3EMmWJY\t58bs4VQUlgyZcMKJVjpZ6o\t78EsU5Njik3K2b1Os6zwLV\t0BdUgqNA6b63BXGDu4PeKN\t4PdEXwNLZrPK0BxuJwr0nJ\t4kKREED4rj50B72mZFuIip\t14houuG4FrK5ZHlzVccj3I\t5gH7dn57qXFVoeY2IKULtY\t2bJs4cwj40fPxm3m94ORe7\t0KE6mugI11bbF8kBYC41R3\t2PWUpPMK2GeLxLm6boZjto\t60bhcR1KCbE3KXx0zDv0XY\t1zl1cnISd42IeaGjcnQNAD\t07jABQKHpIpXKCOcqWtDpV\t1kdgim6R7kqUAOOakjyWGq\t5NiqIB4BwRpoU1V6U195OU\t1oNvNkTsX2YtpPpYQHL9Zv\t038Cff0ZD16m5byH6ohfVM\t0dgHfb4WaQAzBdS7n4SPmN\t2Us0EFBMreM3VlE8AS9srv\t6K3E77Wxm5oH9kEI7Qb6rv\t2IAvDrAdvPDiz7Z9ABphO5\t2m0pE0vX5h4NahhFsPMwnr\t2jaKU9jN3X2auwOGjukuE3\t5MtAIjUBeWqQ4ZUsb66vEZ\t4CvRCtSjUTYksvMiHsT0CV\t537UFrFPasLdnwe4Rk0ROO\t2UBg1GC3tMTnw0VzwmLelz\t4dVWz5zq7XXigjOfrAfI19\t3Ek6sWpamhmmtk032Uhg2V\t7oYH3VjR13Kmtj7o7xLEZr\t5wZxmzrLNDTcw2JNyaKHS1\t7EsGSHSaobePkf3Lsqre6s\t1pe3AGBuipdklcKbJKDP9u\t4IDNf4oDocAj6dufznifao\t0rjX0ul1dfUmtNDAUXIPup\t46Pk9K4Ta26lFiUs5thsU0\t2OP7W1lsZkSWGBPdnO3mgk\t3jrcoA3eEMZGKzF11VzxO2\t1XbzwdyDW4YohbntjCdso4\t78XVcxI67oXSzfV6YAODtr\t3BWTnYtojgn68TZSkGeaZw\t6pVGYwDiMSfrEAMdIVSoLt\t0S3f2G3nuCWHmmSbck4i9C\t58yF5Yqokn4NxABBmpK8Yi\t0cEL1Cg68zorMS2hFq0JJI\t536PcP6LHChvhsH64QVBhq\t4gRH3vcS741pSZW66LQK4P\t6ULiCxVUaWBG0Gw2UAg8Dz\t5QkHEhAJcVrsTKSZFJDzwX\t5bQygUkLEUYEWSk6rA59QU\t4XdhTfbWbD11U3fTW4EHcj\t1rS24VudoY628mdFumzVcI\t32iYiowgoEfTsWQkcwTRlX\t7HcbJJxIaZbbPIRb1CyZ3m\t27do8NxmUa0D1O9Mfi7qJN\t4MpCSQSpk2yLnfrOSHsZxq\t0PkKfT55z3nNSVhII0tZdN\t20QnKWlncgqaX5NYOybhgy\t5gFjlxAUKTqM1GUlFNKw0S\t0CkMQnSzNWzx30BaLnllr9\t30ZIabSNa8EbZT49b6HdFO\t0hrdCoV5LPC0ni1ahSbAID\t3FfWjwjwjVDZWlddoQ7jP9\t1RDif5mDdaGro37AxOVYoJ\t5rfLztZGbpbF2qC2sU0LZq\t6bcIIzSu0niVuplUk7t7LB\t4khYVmGHZz4JWpFlOMXanb\t3xXqlPnnVXRsxfz7UGVi71\t5a26fblCJE2O4kEJSJxU5h\t3up1JsYa4JNZBakiWP41s0\t3WOFMQnYvfcGFxA13J1e55\t6On8OnESrMsfScviCLu0ac\t2vVVMFMLolbasmvpkyEF8K\t2GgiRBztrAUC3SHmBxAgdB\t0aCwjJMzkOdxUZfAjKtmuY\t5k3DQ5XZGBc5a0Rwbwc8hW\t3DOm109bpm8LVlGrPj8601\t6uSQ61RK297rMcatNDbUqW\t4kcM8vye44jgsRMus1UjER\t3umDgMGgONpKVH6KzpCcho\t6CqEVY16aBgIMzKmHOBLAy\t3x2Xk59n3Ey2703JJX8ss7\t0ajlXtd6JWlrEGt1Cb2gRH\t5YE0jwzEgR55ngUvtAzEG3\t31Z3tkTDOaYAIJt37DG7lW\t0v5tTD8cCbNsuSPdZq4ppU\t62tQ11UnK9za7j0dyqT7Hs\t5h53e771faNluczmIdNTqd\t2lhWPS4vdx7F0kkwfLmAwG\t7oLLKRFfOyE6FnIbbpXsyR\t16Hf2J1HuPbNPWFvNZzYPs\t6i1fuTteHcDcO64tGAnGeh\t0URolWwoi4SSkoNHXDrTpO\t6KiZqNhZtkdB219BIJkxNJ\t1XKMWyhXlzu54mHfQuLUlf\t064OyTlK7wUeK3D0OcCNcp\t53APvcivoxGrAmK2b0Givf\t2qKCyrQ61bmJqoV0cCl6eW\t2mpINSrBUHvmP5oYSZ1ZFV\t5K7gKm344eKOkDPHQPKAzd\t0utSnGPZthEAuKH2kUfTcj\t1FC2CEy48qcygiudnhS11x\t2uGcDgpKyKBIIOfGwTd6bu\t3CgPWIPgiLM0fuYQSPV3Vb\t3cQCiT1PvddSKI8pRk4ygK\t7rPm8nyaZMDzrt7HDFC1IA\t6FS6mOlzpyIWMz9o7pZoWo\t5bOGB5m6V5yWR0tGhbBhX6\t6HnJLuczohJYWkDGgYmm0u\t1BZe0OJ0eEjJloBAvg6aJJ\t5avuMjb46hBDucxFvxn0zo\t2Z0q1138jfn6aSMB7O8o4w\t1sVtiUcsOJTWYjucbPoVnN\t1QSdwCcfv00YVFjlMFzlo9\t4IRGT4KQBDfevJfYgUuZvP\t3zM11n3Po3s6eBH9QAqcNr\t5w6y38iH5HdSNk0EtjAdW9\t5BZNTeEo1t1HXVucObfYSp\t66bWbHHVd9Zi5xNAKQjTmS\t4NlYgUpDS3K7m7mw4lsTM0\t1NBksoTuYxMACF2v9OVDMB\t4jomQr6ARl89f4ZguNlIQm\t3lQ1IPdzulBHfTrqLYH4vX\t7gsd2pg4vXfmAnMuXRxTEE\t56Sz3MTf0cGyjYwTJOZVRY\t7aw7h5j6BK5KvzSPNpKNRj\t3woUcMUIeew0PfIlEAGUcH\t3j1jNAZIgr4vhBfI6sgfxC\t7zhc7NI9JHyPmcOaDcHCVn\t6lGe38gKVRfF6cKeXmhidF\t0XUZDGgOioOehdcstP1hU6\t4aILeLn5yHT6AsB1W7bEHG\t6DdGyHy8hlqylxfaDRpVcK\t2Kt3W0rl0PjPCOjAsf9mjX\t0sAuFhtMq2SKZ3jZeU59Yn\t6ldSXWJYVt1Qig7mDm3fXv\t2YlIQsylMAOcqI7aLas6zj\t4G96MmIt9XmoVPn9XzgtSy\t4gPw3HZ18KN0UOniw4UEm3\t5n0mpjpvR5iWWkiQL4kgRX\t2pX3YMabAIjH2yQxb56n9l\t4p3zss13iYj3TcxUgjmrKM\t3QuoES16r0kfiewaKeYYnJ\t6Cz0v9MHjAdviUGTtzO3Dq\t0DdCjDmCzioT6W6nIhMOgA\t4ZNj2L44lvkGZ58SaSql7O\t04ENoZKEACEkrcc7v9EjnY\t3xYgJpdnAuKPBSA0LHtg4I\t4Xds70hJW0HNo0K7OKJbl7\t1AIYotQAJnVXpyfAznXK8y\t1Ez2SpFr05CspgDgHSja91\t0si5v3WiNFDgQUcbkgRp3o\t0HRQMiz9Ua969JXOPVLlcB\t51XnpBsO8S8utaHscyhOnP\t5myMjEVTHoBQrvatNM0kyy\t58b7PzFbREarz0Os8GRBZK\t4sX6evSOdSL04HR40EcEN1\t4fubn0dRFW1WMa7yiYIZSs\t1OKVJpL9RPeLjFGJUzeXv6\t33gjPr3rzp1dylPMPgvLYV\t2qeEyuDUaucAe63BoqJqoS\t5v44Md1bcJYN0rL5kpWfd7\t6PSyaM5jEbwLXm1RsKZyWE\t0hLPDVYwODPeJfkHSol5aI\t4OPPSKaowfmIiUEVNyh0l2\t682gIKe9M4YJeDbw0Uqimn\t5aGZpag8gyQf8bYu1RhYZe\t42o454bTsMf9g1A0cwGxke\t40vqauqc0VQpvTGYYH8ad1\t6oxVrlxeTwhmOroYJkrAad\t3AVBA0GTpnMFh1Rv6Xqymu\t1VZmjJ3WV1nc3ojykNVxFa\t4Nclo8xnQeuX54AGKOybbM\t7Dba82QckMfi9xvgeePc72\t6PFiq41950kSI58ILz7uGO\t2jJUHXFaFdvtxCOVW7q8bd\t2lEmjaR8rQqsQqe6CLXtdz\t3lPO5WuqFNY12UGkZzZ4Xf\t1o1tRS1Vzt9RZDJSDJUzSC\t5D7erlQmTndO42J9VuvBW0\t1kjxPdNwFKldrMVxlO7lio\t3l7DVkePu6bBxBXTl8cIDc\t6pTMJuynSqNQXuGar4Skno\t7oGEP1UfFPnJOFeE38Erjr\t6tIXXMXvOi3XNHdRTwYFOl\t5lYAexg45DfNm7LfJNYMva\t2wgL4gIm8InPw4IPaOBp8h\t1CzXfJbCKcHb33F28SyGv2\t4nHMoGnvsDsCMHmwfSVWop\t2R3ifU5sK0FygVOZpk1yJW\t7yeO78qI0fxnz6gjTZEp7i\t68SS7wcjzSTXcifbplZztH\t6fbTH5few6yjRaQuD0tqfA '''.split('\t')\n\nto_be_parallel_coordinated = df.query('track_id == @id_numbers')\nlen(to_be_parallel_coordinated)\n\npx.parallel_coordinates(to_be_parallel_coordinated, template=\"plotly_dark\")",
"_____no_output_____"
],
[
"id_numbers = ''' 16VsMwJDmhKf8rzvIHB1lJ\t4DdgOzDiK3VocRlOpgyZvI\t5smmdqbHwTdVJI1VlnBizP\t6lyFgQE2nJwT34DYJO0gr9\t6C7oT5ZSNyy7ljnkgwRH6E\t4YSO3y5EkzXDiBW2JSsXyk\t2PktIwDOLDNRntJHjghIZj\t2OKbnAB4LIw93b8IXJr34m\t6drCDhqlK6cZ7LKDi3SB18\t0ZsWvJXGaHqKUHrvBjZxSy\t4hnq2TnTGgiLG1qFAFQtQG\t40OCjuNPJQUTjSnTqFc9u5\t2J3vblLOe0NKOJvHXxmvuu\t2NGl2ljBxtvl5duT5U0Rgc\t07iwjTrXQsfQRJ65rEConJ\t4Mjn1iv3fhTtDt1ZRnUvn7\t77MM047j6loQsPsUFntTiC\t1oTmjppGp1ITPZCKsYNqs9\t1DJUNsDTNuMWGrxfJmNGnm\t5ZTiNyy1YtvyBEwDWoVOsa\t20iBwNgEMH8b63MZ7wmN2F\t6HgNAjt5zvGy3YQfib9hbC\t4zG58gSipyazhsiVdS84lM\t4NDw0ExQPFKQNkkFKvPh32\t5ghFFUCCEspRulW23d3Awc\t6FCl5VIhI3c6StmRgieLKu\t1IeEYWlLBatGhtSTVRdOgJ\t5MzQStKKOo666peyPoltxh\t6D2KvMGxjFMk47D6CbCEaT\t0DVnlsmBltpcWafM3TScIu\t6jwmlu44QMMDesyUIFLQS9\t4lUz3IxMsXYpsrbV6SVQAM\t01y9jiO8FHCzv9iLmYpw4F\t5XIkSMJ9sODfZoHUJYoi1g\t7atUBpdQv34PNmYix84wzR\t6vhOg0jBNyCzQo7nlotVeH\t0m0ndzeNd7bTNWpgeGoQcP\t1NBBs5Ym76El2gojyE4EvP\t0R5S8PHmsl3TzHdMUx1oiM\t1b35m5XbZpyNAx9atEDaDH\t3aCIbAoc0CTE46enUrDmuu\t2Y88xiM3oe4DFYX0jLLSON\t7DcVWzeud5tqtNTZKQWvhz\t6DdG99q2hNKrSHZ7hL6pBt\t7ESz0yGdmhiWp85j5z09Ub\t3xmwsqwkhI9gbvmapDO9S0\t2N9LsBQMtLyMZL0LeydiLW\t1sGGodtsPFq1JC2w3vXZLv\t150NZIcOF5CtN93dp72A6g\t1COgmyz8tnpvBoZvqqZqCL\t314QsKiXd2SgDXPYNsKu0N\t57p3QcWwIjVwvAcQpu4hkr\t5IYNm9xiOZkLjGJYH0kqsR\t6z2Rtx1CjQGaEEC1xzqtIT\t247ye33xXOEhnjN2rCdj8I\t32ccjDeiYYtombISVtse9U\t5eEZLIu17HRBwt0Beldd0j\t30DnQCN64v8xBpGZpLgb6l\t0PrPfp5FbP87rTk39MUKcc\t14EblrVdzyjpAWaedKO7x8\t1l5CriNdYpEL3NoJxKA9uA\t45ZTQl9GbmdM418qgLZvQZ\t3dgf8JT9Ya3QAfWaJTNuI6\t6ga6wioJAkB7MtOwremcSe\t3HUsmE6j4afm7zWM3bprkW\t7Jcf74UJvImsHrGOqSS0tG\t7he1eOKQBxz1JK66afUzzD\t2jtaAeW1k3qgbpQxT8Y4lm\t3C9ZhZSSd2ki6Ko4Zj4sOo\t3KuP7KttXAKmsjCLx9gKeM\t6I5FyefGR36b9OF8rFkxVK\t6YNIvsHK5fdy0ROHDuFpm4\t0M7ZzCZ75sAUBq6Rkwpu09\t5soDoRuEEmx9BriBtoWbr4\t0zjLqMGvY7j7TuBkh2MIVd\t4YfWZTRKOt0Lp1x1TkgsJz\t3xhxhvEYDY0Txl8jUqbH0p\t05FSDW170E4Brk3Et2Tsn9\t64sixBk8xj9Eaz1VmdbenU\t2KcO2wBpD9kfEUq7K5L8NU\t5lpIW3pxLBGZ47LhXmHuH7\t3aayFmSl21VgL3vybq2EAe\t1nhZ34zdByR7TKRNLi6jXH\t1WU3fG5GlEsQSsxj4SlGn2\t6mAMDridbMDlW2ovdyPDUy\t4yKqq31wiiTYlzsTspc9bF\t5BgjDdJGaa7iB3kQfj6QMh\t0AYTA3nevKu9S6LpeJwG7B\t2q1mQzjkmrUINRWiyvctSi\t2OIGt6nkvpYyTCsgqgosut\t4nHpPnnYddn9KhXWKcVcPS\t1aeKIPo431ykCa62MFpVxO\t6J0LsDeQEMbXNCJCsPEnPx\t4U4UKccQf96YM2pVVehbDd\t0iInUMrkWaGGUkPwIY1Ntk\t5kM4TGc7A3VyX1AmnIznGx\t5ByZw9BY1See6eYgqUiB1x\t1odwlrTdOkOVUoJhlE25Dx\t4zsYOCkDiS14hdCc7gJX1Q\t3XnpqyDY1Jo53Tgod58Mxf\t5w3peXuUoDQIRWJbtK4kYi\t1LWhjl461aekeNdmQk2JuJ\t18zmtkXBaSHd7G3xobWIEJ\t45vdRv1YwLbpbVeJ8BO2pR\t1K6WHHqLXlqyGxX2lUMQr3\t7gIS4JjropHYqNq3UzjHNB\t2wklaFrsGnIfvLggxQhwQB\t68WhMF4gKml7wKQcpILei6\t2NVoGLBsrbQrH9c8bRDQu7\t5gxxz91fYTlkR2cqmDkPWP\t0tewjlNbotxqF2obibsg36\t55hoUnXPjk2xma2eYSbltW\t2iGTayx2t62y1J0XOInyfX\t6ScbJrUjGIWS76VXsK8UEp\t6M1W8DojBHXnjenYcn7H7M\t4VyvzQoIfG49xiNuYVYBiv\t1dMabx7tqxUpeDYQAu8c7S\t2bQN2bSNXxpGTnVKpKXl2R\t1FCueyFK8jtU0zmxQZyVtJ\t0sMph7dbpLD4DlzEEfJlpX\t5rW3anmLNKDA81nVJvW50H\t0w71NjrPNzBsa6yO0of2CZ\t76hmKWewz3vGnKLbY2nPRh\t3BIyzKK2U5O4Ij19G9z51J\t5OLQw1i9uk8Je39V0SJ2GR\t6FAPlqbXTuXOPM1UmJj1X3\t1kAJBuEhXnXHNA64DDO0Bq\t2H5cbxbGjC00Zqe8IqKHm7\t6wd1MrcFIjgblPkTvm0veJ\t2BfTod61ST4H3K9jxPg9mp\t4Uq8jQxsADt7piVcuwYgVJ\t3z8VNabIASkrBxq94cP3TL\t4c86vSmmzcIO4x21LuD7XM\t6gqoJC9MUub1AbISMFCuWr\t7s4SSLsUwBjEJzNVODbV8z\t1zXA806qSJVWnHpGWQ3UUC\t57E1gf3WclWxUuLcwYYyU4\t33azw14HJcaClFGZ5kW6Nn\t1izLAQzCTkTCTpu3l9TFzB\t754UYs1LuDtaEKKfaDkx7Y\t6sNMSl0MAqzvlGEt4Y072v\t4aAZVfU1M4cm7XqTnzhCnr\t28Val6Yko2x2iJQ9YlG789\t4RwLQseJrBm0Pjl6vQcY5D\t4TZvXowrJenK3OCEbmJzUT\t1I3iCPuCId7Vkg5rlqYDrp\t7hWa53fOj9Fh0X790Bl32B\t1JMkYhhLa7KPDd8i3sPGOL\t355ezvqbe2QtgMf70xXBE6\t0KlGGlCwuBw9cPcjq7xjgf\t5kwDBRZrCvDtN27XtT2wzA\t7oMJTXLhm8TAkk6K3j8u1E\t0ELWm49HJEJqIvqzTdZK3n\t6VziOL8abdt5gchEEBCMRg\t0XUHYxHOOctkSXReILAaJV\t3wMVhcD7YbfOFqhgYiN9hp\t30VCkYXm8pkZ1rOg5yC4LL\t1NE1ljBeJzmk6wZZ4uUdRT\t6FWhcFQApH24r8AgaOLrFw\t5z4mf1xZt0z0u89ntbWN5z\t05Tz6QuSWq66WaqpHGK6iw\t6xq7BAoiGiXC27rW6RH3ww\t47AJA4geNelnpulvvfZjdn\t0BOhco72YhbPpJIqDEZNmA\t1ciJCLzKzezhHbBtii28UD\t63IkPNf3Z4xHLASIyhxS1R\t0BNWj55u3tfVB3hozoC5lY\t55FD4r3EgXRMKP79hDbt5y\t3SatXFFuUyX2IlV9JbaWp2\t0L4u2qg18ieitQkA2HBXgq\t5OmUVlZP8zQ5zGCX9wsD3p\t38ueylzenb5JK5JHDGnWuO\t7FLUgR5esAR2m8kl6CSQ32\t7KOOHzDAxzl87i8VYk1iO2\t47jAQrNH7CLIcYu1lqE7pZ\t7ve96Lk22N2ZGVqVq8EJOf\t6F6MrtUbHqf7AASOXDMlMp\t78E3QFSTlLijRUrukdbXK8\t5wMlr2ncg0SoPOKEs0Pc85\t0rfSwqjq0k20rVZLzATVwP\t0PYPlbP5Vdz5ivIfC0jAmf\t4UWkS1obHdt123rtx5v9cx\t5RpMFAJcf116DGFBcK5Ny8\t6i4o7jn033PDiNab3Yc3jY\t6FCWOKBTjzHsHpa0cF0br6\t2b3Xo30P9KFEqBvsTRQTM6\t1b903k5gadxEFXhbGHAoWD\t5tA3oQh58iYSdJWhSw0yJV\t4f01YssEopYUrYIO6YZmjZ\t3960gvUO5yuDJtI6VtPqYS\t7fc3kOECAsJoCbsV2p64rt\t3CboU4vdisSItbjfbx6SqO\t745VS3h8id3zcLh7Gd6gGa\t5JQlQR9REVJmP34AqI7Tpc\t5K4LPGFKqKO7YSbUdSQAZH\t18vjAkuAMaSxfAf2EAcjP5\t7is6wEBQ4zPEcjust2rB7u\t1PxJV79Px9gFHPLvFO9ZOS\t7cgt4TZJH3HDdmHQhfVmzx\t3bl6n1sBma0Lp7etqjx5j6\t76rLK2XhT6waumcLkLNTID '''.split('\t')\n\nto_be_parallel_coordinated = df.query('track_id == @id_numbers')\nlen(to_be_parallel_coordinated)\n\npx.parallel_coordinates(to_be_parallel_coordinated, template=\"plotly_dark\")",
"_____no_output_____"
],
[
"id_numbers = ''' 6eZ4ivJPxbK7I6QToXVPTU\t6V37apVtCiUpEKcAUyUjoA\t5SxlhL1idBgsfYBfR1KEcR\t0C0XJ2JYr9jEGAt89JyZqJ\t1XsqZ0mMrIRMAktdnEuFF8\t5SUMNsXNVtR4ujz84sWEWe\t1xfTdLDg10CJfhcR4Yis0z\t5zHgA4J4CrOaUvQ9UD219j\t1XO9zgpDMkwhmAijuYBCxb\t1U6vwXAvc7VvbhqNyedGEG\t2T9ZyRnW6omzsVDLo4I72l\t0UBDke5y1kqTgTkgmyHiwj\t23tftAc7uJnxEfy5AGS9lr\t0n2gtAOGT6Pxu5cEeaugym\t0nqRtO4jdv4K6AJ7hYmDW6\t2wsVeO1Hqx6IqM48UXGWSO\t7mmqxoKWTFZB8tHXfQpmk4\t336ihMIODpi6nlL1ytSEm6\t4w2lb0V0qHGwj1GR2f52c5\t7cKSdtwLEayFd8MuLdZR85\t44q1XQgawoP50HHMiMMWCq\t4iPaNKCg8kY3rwUK3CnUw3\t5EvsUz8wsUh0dP7HaixMh8\t6A1prRyHlB113go9En4cX7\t7iylYXaOUTO3BixPecSjhP\t52pvmjSRaV7k0TCqJK5sKn\t5ATIMj2gOKsj06UvoTkFxe\t6Isu6pTUwBa3ftiyOpKf7s\t6lajHnTKM9Fiv10kzUpD90\t37VDfyF70jTo1HqGQOsrRR\t3RYMOo7YF9gCkVZomhOPrK\t1ZIQ5girZEdA70xIkevkrt\t76C7vN5uEcuF1BXvUJMvjk\t3v8Zu57HCIauve733J6PjR\t0KfjaQSlDL0r7dLaXNDMv5\t7sRTfvTV5EUhDY4e4LjlVS\t5wI6LhywYSgmHNMVERAJpe\t4K0hPQgmWzx4jGM2Q4tNQN\t0WmyLH7XemypvsAHuIOCp7\t2YbZbmqqxrCysQDc4AkIIX\t1UegIYDIgDicEBuHhWY026\t3gdHLVZqeU2mHNggC6Tzwr\t1uYAog8LWWeVnqNWItZaHc\t4LpsUDYp9D7VvzU0iRTCq3\t2akKNicOhUSp1QHQEQDTbC\t4zHo8J0WbUDDiHTAURs6kO\t32Q6wqR85WhBeoqZwMRwnV\t5iofFSJRoRDyiKD4kWTpf9\t7owI1qTHoXGBVznJod7yuh\t6rbiT8DV9h50NBjPxkDygF\t5twkCu1ET6objhnLfQtgJQ\t7gGLo0dwMbJhRy0JVJP00p\t2ZWv2tklegv3gwKeLD35o9\t7sLsIr2vhjYeR6rniJj5dj\t5IOozjD7gJOOhTV1lDXrXl\t2cC2PIXKFjnY8sbuS8spzw\t4PHM9PG5J6IQ8fumsJuSYJ\t0WcGdMWl75v33B27KafycK\t6K4pZ32MorbsHeqtAwaWHW\t0h0jNccol3eyMQ2mIcNcBp\t2MfFjRh4gv4lU0vtYH0GaZ\t3uEFKAtU1hdfcgFC60yt84\t0slfqpTh3q10bNfAYb73RS\t7dg0pRcn7R5VVekBryq583\t082bDyzPxizG0gIqArJoQ7\t73OC95krAM3n1u2LcKraBX\t3qpm5w0qS99qUN0q8MzvlL\t1NywSw2TUrdnpnNtGu8KL8\t1zSqLFmuL6mDCVbZNj7hTR\t7kPsDSN7eFLbzNF0xEchjc\t2qw3xeuKWfsV8GynO2peHr\t6tEeqhvdmOVU2iQqnLk2zg\t5K7VRObcsBDfKnyVbVhwTx\t78WeKIDpoVu6r0TziQwl3y\t4ZYir67KzcmiNKTmFVqNf8\t22BJjJeknJ7ff8vGGzPB98\t0b81xIMQLSdUpeGv1oStXH\t4u00iLhEPkbLlclQDYuIHV\t1p8QusGejMBctlhsZ3jtSF\t2FzI0rp4FsSvx7N1GFs4HB\t1XKqzLGxhIcpEXv8SoA8tu\t6T3yaivZB0v5AODCyaR67G\t4WOPKEtVmSAZvWXtyApl3h\t3xvtJJiFdTR6d5N8PaFb8f\t4ZAjZHxvrzKZMXdHmg0DFz\t3ekvh2GPv2ebjPHYKhuIXG\t0bv1k0dLjgp9f9rj5dBScM\t1MQio3srmAmDC0c32Xh56A\t0BZ7rkI4prRAbfkO3jo2OB\t5Vu5DPFMNAJc0eoq7i8skM\t1zE9o1WK0Vpocnf1H5nssQ\t3zdIn3IbbJAddtf9Qo6i0D\t3huj9hX9ECvhipWIGNObFl\t1rFMpIUb6Hs66ypS32MOOb\t1Qmb5p0mK08hxMjWJvCfBw\t3C6fiBrM14YAynsEeRZXWv\t4t8WpwzDLTYwMulJBavljv\t7vqMKsg985FFLyK5DN9uq1\t5yqoXxgDIQ9fPOcSAQUjUq\t2D0FmjFP7dxrin4XanSnbo\t4Yuux4zVxXI0KVHil24U9L\t5MzGtEojUtMsLueJ55hRn3\t2RDFWx08YULhklhS0DyVtj\t4yEdofTvNsL7PnBJNDN1Sf\t4n9SsVwbc7Y4tn5UfPTNn4\t29ldunhjkUfuB5k1gXlqFS\t6VFAILGN7uOz24elIyt4vB\t2361cLjSnEpolPC3Mb0yv1\t0T19N334CPKgpMpxh36KiE\t3RjuP7n7x8DaOVN62TXFke\t3V5LrENP5AgplQwvGeTIIU\t4SNbrw7KNj3rupRnXzV31d\t5XdtGPF22knBwy1fAzjSCK\t3GE6KLTgmCxsNzhp0nI3Zf\t75iGW6GTfBU7j6ldQNAvu4\t1FvxqWCDg1xYdg0eXOr9FU\t3NmVag0g3N0B4nDT0ypVk4\t07jMNENLpJ60ej30L1BFPD\t4KVybsvg26UiPJEVynN3qE\t4k304lkj8Ga9Kp0p82cii2\t1HVwhAQMU71rg7GVlQVxNz\t6nYTfmQEE9ZYYFzdLRWP8Z\t5QdTBAXXaFZDhsBqPT0GBI\t3QElxQCbZjCqAG8yLRwLsm\t5yvF3kvaX2ufVt3VvWbGP2\t52uwpMhSoReK5wQ3Yxr2eC\t1awdo11NQFC6THLXQAaDjV\t6n6Wrf6HRSgTXwyWugKDwf\t5MXF8IhBY1z63VZVRvFZUK\t6NjMv3rcXwyQg4Dtr3WpoE\t0JsAUsmagEqYQo8FZUkpBE\t36Kumm8Qj49ABflKCvltIH\t078Sr3upDQIPRIAc2IpSxy\t2wJdo21bsx5HfTnwPJ3p92\t0WWk0UiErQiR8EAnSjll1o\t1Fs2986kJPeJR94vCqRGha\t5eImJYwPyrdhUqZ4gTO6Qs\t6bXr647nkFkrphCoA3L2KK\t1counClRuzpBxsb8gkTCmO\t7yCtrkXdQEVJQyk7pFxGyq\t4sGN5db8sJsecYNWoxLPky\t4EbVxLV394SADIDf5zFTHY\t0tZvlW8YxwnPS7Ui7pzF9q\t69LAIJUcPbsw6G8F1vCv1y\t4wzeevLrnqs87z6FrcFNKu\t2fKvOnZPwh4gz24MjM5hWp\t3Hbl4FnRkj8TK88Jg37Omt\t2mSHfW689yTYIZCu0k1Frb\t00MLppbVubwv4Rbf46CCfg\t1MvhXhNkwRJDH94ZloFU4c\t7oM8U222NuBLUun8aFjhKu\t2veD2T9UElKuePBt6FW4nO\t4Bulfi18OkBRXehhVg1SzI\t6M9bTZutc2QtXWl2p5TQ1I\t4fM8cupzQbc6qNeDK9FXu3\t7xktbw9wyJyJbwS3y4LZFg\t63PP8XGwgRI7gIruMO7IG3\t3C0Kxh2lnOTmlSCD1rB15W\t0YFoUawskWM6iKHSyQgeNZ\t1HEzYfexDpgfwyceOWvNz8\t2zKB5hjGfqoYZUi7B3LAK0\t3mEnnPSXvKoVouByyUqhUX\t0dC2glrlKpld5xY5BAX9lK\t0XXvMZGbrz60taMwPbVGgK\t2y2xE0gB5lVIGbdAnHNUIz\t6Ech2zanuCQ2ihfXDOLtID\t6rEcPr1jbReCGcT7LD2cB1\t0gn77iNwUHN2pScHbqttN8\t5NH0w0LSvcjiMjWnTwhm2u\t19HDqVwakevUkynlB1Ztut\t0g5kny7FqZlnS1bGMPQFWR\t02PBxJsA9YIhdbiXMNN9Cd\t0tpRok1p8ooccX7DQqy1BZ\t1P5uhYSYMDxXpcYgpMYnkg\t3UTt7dSBf9MG6833z9gNUV\t0Si0HsULu8gFAtYm0BwqXI\t4sO0deplZf1WJnXwrEVNUt\t1fTuKuiLtYmckVKwtoT812\t0hMOYGKQK3m2ipKTZKUbrI\t6nsyzCRGHluwU3QIDSQr6d\t5y3HyzqdypXCRFz2V8OpOF\t0mPvAhvAA0IyrcbUh9KEQv\t3n5N1ECcHzZDvAzHLpJULT\t5Wo8dHK8N9pMyDdXI4WWsZ\t7KvGuebu3RAtH0FSY8RG6l\t6XEfmMikJLYbYZ3ZL4l7yK\t5ijg8Z5M9WNI2VLXDaxrAz\t0FGiZTL9LSSzdO05Vtgg9U\t1tYLrptJ56VWore4o9Mj50\t4EI3t79hsPIQJLdHitvB2A\t0uwIsRVkvzZTzxqCQHlgiz\t4dM9Vju1O76L2V79EebLsj\t20XscF3HtxEGo8ghFhOgCx\t0QPSeBG4P39z9KOihZARLf\t7wbsdw0VnVe421V68sNwDk\t75nO71NiNoIaGVIqYTqSvN\t6Jk8VFFPoUyr7zCXIGcUQS\t1UdTsJcI4MwzKIxCP5HHXG\t53oWCQ8bcFSFzcQd0Xggl8\t4iFYF17QReVxN6bQoKE4NM\t4uAg8KXLiGu0kIvICmdUR0 '''.split('\t')\n\nto_be_parallel_coordinated = df.query('track_id == @id_numbers')\nlen(to_be_parallel_coordinated)\n\npx.parallel_coordinates(to_be_parallel_coordinated, template=\"plotly_dark\")",
"_____no_output_____"
],
[
"fig = px.line_polar(df.sample(n=1000, random_state=42), theta = 'tempo',\n color_discrete_sequence=px.colors.sequential.Plasma[-2::-1],\n template=\"plotly_dark\")\nfig.show()",
"_____no_output_____"
],
[
"# Make a PCA like the one I did on the Iris, but make it 2d and 3d because that's cool\n",
"_____no_output_____"
],
[
"pd.set_option('display.max_columns', None)\nnearest_neighbors_df.iloc[[69000]]",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
c514fc24984a8bc881627ed0e32957f3c8180473
| 114,415 |
ipynb
|
Jupyter Notebook
|
HW/Home_Credit_default_risk_HW.ipynb
|
Shaheer-Khan/AISem3
|
911f5fedc581dc9e4bbb1e2e75e8919a1af9fa3e
|
[
"MIT"
] | null | null | null |
HW/Home_Credit_default_risk_HW.ipynb
|
Shaheer-Khan/AISem3
|
911f5fedc581dc9e4bbb1e2e75e8919a1af9fa3e
|
[
"MIT"
] | null | null | null |
HW/Home_Credit_default_risk_HW.ipynb
|
Shaheer-Khan/AISem3
|
911f5fedc581dc9e4bbb1e2e75e8919a1af9fa3e
|
[
"MIT"
] | null | null | null | 36.104449 | 6,937 | 0.403146 |
[
[
[
"<a href=\"https://colab.research.google.com/github/Shaheer-Khan/AISem3/blob/master/HW/Home_Credit_default_risk_HW.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"!pip install xgboost",
"Requirement already satisfied: xgboost in /usr/local/lib/python3.6/dist-packages (0.90)\nRequirement already satisfied: numpy in /usr/local/lib/python3.6/dist-packages (from xgboost) (1.18.5)\nRequirement already satisfied: scipy in /usr/local/lib/python3.6/dist-packages (from xgboost) (1.4.1)\n"
],
[
"#data.set_index(\"SK_ID_CURR\",\n# drop=True,\n# inplace=True)\n\n#test.set_index(\"SK_ID_CURR\",\n# drop=True,\n# inplace=True)\n\n#full_set = pd.concat([data, test])",
"_____no_output_____"
],
[
"!pip install kaggle",
"_____no_output_____"
],
[
"from google.colab import files\nfiles.upload()",
"_____no_output_____"
],
[
"!mkdir -p ~/.kaggle\n!cp kaggle.json ~/.kaggle/\n\n!chmod 600 ~/.kaggle/kaggle.json",
"_____no_output_____"
],
[
"!pip uninstall -y kaggle\n!pip install —upgrade pip\n!pip install kaggle==1.5.6\n!kaggle -v",
"Uninstalling kaggle-1.5.6:\n Successfully uninstalled kaggle-1.5.6\n\u001b[31mERROR: Invalid requirement: '—upgrade'\u001b[0m\nCollecting kaggle==1.5.6\n\u001b[?25l Downloading https://files.pythonhosted.org/packages/62/ab/bb20f9b9e24f9a6250f95a432f8d9a7d745f8d24039d7a5a6eaadb7783ba/kaggle-1.5.6.tar.gz (58kB)\n\u001b[K |████████████████████████████████| 61kB 2.2MB/s \n\u001b[?25hRequirement already satisfied: urllib3<1.25,>=1.21.1 in /usr/local/lib/python3.6/dist-packages (from kaggle==1.5.6) (1.24.3)\nRequirement already satisfied: six>=1.10 in /usr/local/lib/python3.6/dist-packages (from kaggle==1.5.6) (1.12.0)\nRequirement already satisfied: certifi in /usr/local/lib/python3.6/dist-packages (from kaggle==1.5.6) (2020.6.20)\nRequirement already satisfied: python-dateutil in /usr/local/lib/python3.6/dist-packages (from kaggle==1.5.6) (2.8.1)\nRequirement already satisfied: requests in /usr/local/lib/python3.6/dist-packages (from kaggle==1.5.6) (2.23.0)\nRequirement already satisfied: tqdm in /usr/local/lib/python3.6/dist-packages (from kaggle==1.5.6) (4.41.1)\nRequirement already satisfied: python-slugify in /usr/local/lib/python3.6/dist-packages (from kaggle==1.5.6) (4.0.0)\nRequirement already satisfied: chardet<4,>=3.0.2 in /usr/local/lib/python3.6/dist-packages (from requests->kaggle==1.5.6) (3.0.4)\nRequirement already satisfied: idna<3,>=2.5 in /usr/local/lib/python3.6/dist-packages (from requests->kaggle==1.5.6) (2.9)\nRequirement already satisfied: text-unidecode>=1.3 in /usr/local/lib/python3.6/dist-packages (from python-slugify->kaggle==1.5.6) (1.3)\nBuilding wheels for collected packages: kaggle\n Building wheel for kaggle (setup.py) ... \u001b[?25l\u001b[?25hdone\n Created wheel for kaggle: filename=kaggle-1.5.6-cp36-none-any.whl size=72859 sha256=22da01d62f836f9d67c688c4361c04f8a68a16c0736ce4fed7cffa96e6b4a7b7\n Stored in directory: /root/.cache/pip/wheels/57/4e/e8/bb28d035162fb8f17f8ca5d42c3230e284c6aa565b42b72674\nSuccessfully built kaggle\nInstalling collected packages: kaggle\nSuccessfully installed kaggle-1.5.6\nKaggle API 1.5.6\n"
],
[
"!kaggle competitions download -c home-credit-default-risk",
"Downloading home-credit-default-risk.zip to /content\n100% 688M/688M [00:26<00:00, 42.9MB/s]\n100% 688M/688M [00:26<00:00, 26.8MB/s]\n"
],
[
"from zipfile import ZipFile\n\nfile_name = \"home-credit-default-risk.zip\"\n\nwith ZipFile(file_name,\"r\") as zip:\n zip.extractall()\n print('done')",
"done\n"
],
[
"import numpy as np \nimport pandas as pd",
"_____no_output_____"
],
[
"train = pd.read_csv('application_train.csv')\nbureau = pd.read_csv('bureau.csv')\nbureau_balance = pd.read_csv('bureau_balance.csv')\ncredit_card_balance = pd.read_csv('credit_card_balance.csv')\ninstallments_payments = pd.read_csv('installments_payments.csv')\nprevious_application = pd.read_csv('previous_application.csv')\nPOS_CASH_balance = pd.read_csv('POS_CASH_balance.csv')\n\ntest = pd.read_csv('application_test.csv')\nsample_sub = pd.read_csv('sample_submission.csv')",
"_____no_output_____"
],
[
"print('Train data', train.shape)\nprint('bureau data', bureau.shape)\nprint('bureau_balance data', bureau_balance.shape)\nprint('previous_application data', previous_application.shape)\nprint('POS_CASH_balance data', POS_CASH_balance.shape)\nprint('installments_payments data', installments_payments.shape)\nprint('credit_card_balance data', credit_card_balance.shape)\nprint('Test data shape: ', test.shape)",
"Train data (307511, 122)\nbureau data (1716428, 17)\nbureau_balance data (27299925, 3)\nprevious_application data (1670214, 37)\nPOS_CASH_balance data (10001358, 8)\ninstallments_payments data (13605401, 8)\ncredit_card_balance data (3840312, 23)\nTest data shape: (48744, 121)\n"
],
[
"train['TARGET'].value_counts()",
"_____no_output_____"
],
[
"train.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 307511 entries, 0 to 307510\nColumns: 122 entries, SK_ID_CURR to AMT_REQ_CREDIT_BUREAU_YEAR\ndtypes: float64(65), int64(41), object(16)\nmemory usage: 286.2+ MB\n"
],
[
"train.select_dtypes('float64').apply(pd.Series.nunique, axis = 0)",
"_____no_output_____"
],
[
"train.select_dtypes('int64').apply(pd.Series.nunique, axis = 0)",
"_____no_output_____"
],
[
"train.select_dtypes('object').apply(pd.Series.nunique, axis = 0)",
"_____no_output_____"
]
],
[
[
"no null values in vraiables with dtypes int64",
"_____no_output_____"
]
],
[
[
"train.select_dtypes('object').isnull().sum()",
"_____no_output_____"
],
[
"((train.select_dtypes('object').isnull().sum())/train.select_dtypes('object').shape[0])*100",
"_____no_output_____"
],
[
"train['NAME_CONTRACT_TYPE'].unique()",
"_____no_output_____"
],
[
"train['ORGANIZATION_TYPE'].unique()",
"_____no_output_____"
]
],
[
[
"As the secondary tables have many to one relationship with the main table (application train/test) hierarchy is: <br>\n1. application train/test | key = SK_ID_CURR to | (bureau and previous_application) <br>\n2. bureau | key = SK_ID_BUREAU to | bureau_balance <br>\n3. previous_application | key = SK_ID_PREV to | (POS_CASH_balance, installments_payments and credit_card_balance) <br> <br>\nMerge the tables",
"_____no_output_____"
],
[
"2. bureau_merge",
"_____no_output_____"
]
],
[
[
"bureau.dtypes",
"_____no_output_____"
],
[
"bureau.isnull().sum()",
"_____no_output_____"
],
[
"((bureau.select_dtypes('float64').isnull().sum())/bureau.select_dtypes('float64').shape[0])*100",
"_____no_output_____"
],
[
"bureau.fillna(bureau.select_dtypes('float64').mean(), inplace=True)",
"_____no_output_____"
],
[
"bureau.isnull().sum()",
"_____no_output_____"
],
[
"bureau_balance.isnull().sum()",
"_____no_output_____"
],
[
"bureau_merge = pd.merge(left=bureau,\n right=bureau_balance,\n how=\"left\",\n left_on=\"SK_ID_BUREAU\",\n right_index=True)",
"_____no_output_____"
],
[
"print(bureau.shape)\nprint(bureau_balance.shape)\nprint(bureau_merge.shape)",
"(1716428, 17)\n(27299925, 3)\n(1716428, 20)\n"
]
],
[
[
"3. previous_application_merge",
"_____no_output_____"
]
],
[
[
"credit_card_balance.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 3840312 entries, 0 to 3840311\nData columns (total 23 columns):\n # Column Dtype \n--- ------ ----- \n 0 SK_ID_PREV int64 \n 1 SK_ID_CURR int64 \n 2 MONTHS_BALANCE int64 \n 3 AMT_BALANCE float64\n 4 AMT_CREDIT_LIMIT_ACTUAL int64 \n 5 AMT_DRAWINGS_ATM_CURRENT float64\n 6 AMT_DRAWINGS_CURRENT float64\n 7 AMT_DRAWINGS_OTHER_CURRENT float64\n 8 AMT_DRAWINGS_POS_CURRENT float64\n 9 AMT_INST_MIN_REGULARITY float64\n 10 AMT_PAYMENT_CURRENT float64\n 11 AMT_PAYMENT_TOTAL_CURRENT float64\n 12 AMT_RECEIVABLE_PRINCIPAL float64\n 13 AMT_RECIVABLE float64\n 14 AMT_TOTAL_RECEIVABLE float64\n 15 CNT_DRAWINGS_ATM_CURRENT float64\n 16 CNT_DRAWINGS_CURRENT int64 \n 17 CNT_DRAWINGS_OTHER_CURRENT float64\n 18 CNT_DRAWINGS_POS_CURRENT float64\n 19 CNT_INSTALMENT_MATURE_CUM float64\n 20 NAME_CONTRACT_STATUS object \n 21 SK_DPD int64 \n 22 SK_DPD_DEF int64 \ndtypes: float64(15), int64(7), object(1)\nmemory usage: 673.9+ MB\n"
],
[
"credit_card_balance.isnull().sum()",
"_____no_output_____"
],
[
"credit_card_balance.fillna(credit_card_balance.select_dtypes('float64').mean(), inplace=True)",
"_____no_output_____"
],
[
"credit_card_balance.isnull().sum()",
"_____no_output_____"
],
[
"POS_CASH_balance.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 10001358 entries, 0 to 10001357\nData columns (total 8 columns):\n # Column Dtype \n--- ------ ----- \n 0 SK_ID_PREV int64 \n 1 SK_ID_CURR int64 \n 2 MONTHS_BALANCE int64 \n 3 CNT_INSTALMENT float64\n 4 CNT_INSTALMENT_FUTURE float64\n 5 NAME_CONTRACT_STATUS object \n 6 SK_DPD int64 \n 7 SK_DPD_DEF int64 \ndtypes: float64(2), int64(5), object(1)\nmemory usage: 610.4+ MB\n"
],
[
"POS_CASH_balance.isnull().sum()",
"_____no_output_____"
],
[
"POS_CASH_balance.fillna(POS_CASH_balance.select_dtypes('float64').mean(), inplace=True)",
"_____no_output_____"
],
[
"POS_CASH_balance.isnull().sum()",
"_____no_output_____"
],
[
"installments_payments.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 13605401 entries, 0 to 13605400\nData columns (total 8 columns):\n # Column Dtype \n--- ------ ----- \n 0 SK_ID_PREV int64 \n 1 SK_ID_CURR int64 \n 2 NUM_INSTALMENT_VERSION float64\n 3 NUM_INSTALMENT_NUMBER int64 \n 4 DAYS_INSTALMENT float64\n 5 DAYS_ENTRY_PAYMENT float64\n 6 AMT_INSTALMENT float64\n 7 AMT_PAYMENT float64\ndtypes: float64(5), int64(3)\nmemory usage: 830.4 MB\n"
],
[
"installments_payments.isnull().sum()",
"_____no_output_____"
],
[
"installments_payments.fillna(installments_payments.select_dtypes('float64').mean(), inplace=True)",
"_____no_output_____"
],
[
"installments_payments.isnull().sum()",
"_____no_output_____"
],
[
"installments_payments_POS_merge = pd.merge(left=POS_CASH_balance,\n right=installments_payments,\n how=\"inner\",\n left_on=\"SK_ID_PREV\",\n right_index=True)",
"_____no_output_____"
],
[
"print(installments_payments.shape)\nprint(POS_CASH_balance.shape)\nprint(installments_payments_POS_merge.shape)",
"(13605401, 8)\n(10001358, 8)\n(10001358, 17)\n"
],
[
"installments_payments_POS_merge.dtypes",
"_____no_output_____"
],
[
"installments_POS_credit_merge = pd.merge(left=installments_payments_POS_merge,\n right=credit_card_balance,\n how=\"left\",\n left_on=\"SK_ID_PREV\",\n right_index=True)",
"_____no_output_____"
],
[
"print(credit_card_balance.shape)\nprint(POS_CASH_balance.shape)\nprint(installments_payments.shape)\nprint(installments_POS_credit_merge.shape)",
"(3840312, 23)\n(10001358, 8)\n(13605401, 8)\n(10001358, 40)\n"
],
[
"previous_application_merge = pd.merge(left=previous_application,\n right=installments_POS_credit_merge,\n how=\"left\",\n left_on=\"SK_ID_PREV\",\n right_index=True)",
"_____no_output_____"
],
[
"print(previous_application.shape)\nprint(credit_card_balance.shape)\nprint(POS_CASH_balance.shape)\nprint(installments_payments.shape)\nprint(previous_application_merge.shape)",
"(1670214, 37)\n(3840312, 23)\n(10001358, 8)\n(13605401, 8)\n(1670214, 77)\n"
]
],
[
[
"1. application train/test (bureau and previous_application) <br>\ni.e. bureau_merge and previous_application_merge",
"_____no_output_____"
]
],
[
[
"bureau_pre_application_merge = pd.merge(left=bureau_merge,\n right=previous_application_merge,\n how=\"inner\",\n left_on=\"SK_ID_CURR\",\n right_index=True)",
"_____no_output_____"
],
[
"print(bureau_merge.shape)\nprint(previous_application_merge.shape)\nprint(bureau_pre_application_merge.shape)",
"(1716428, 20)\n(1670214, 77)\n(1716428, 97)\n"
],
[
"application_merge = pd.merge(left=train,\n right=bureau_pre_application_merge,\n how=\"inner\",\n left_on=\"SK_ID_CURR\",\n right_index=True)",
"_____no_output_____"
],
[
"print(train.shape)\nprint(bureau_pre_application_merge.shape)\nprint(application_merge.shape)",
"(307511, 122)\n(1716428, 97)\n(307511, 220)\n"
],
[
"application_merge.head(10)",
"_____no_output_____"
]
],
[
[
"i was able to merge the file but because of the size of the file processing any thing on the final dataframe the colab server crashes so i would be modelling over the train set instead.",
"_____no_output_____"
]
],
[
[
"train.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 307511 entries, 0 to 307510\nColumns: 122 entries, SK_ID_CURR to AMT_REQ_CREDIT_BUREAU_YEAR\ndtypes: float64(65), int64(41), object(16)\nmemory usage: 286.2+ MB\n"
],
[
"from sklearn.preprocessing import LabelEncoder\nle=LabelEncoder()\n\nfor i in train:\n if train[i].dtype=='object':\n if len(list(train[i].unique()))<=2:\n le.fit(train[i])\n train[i]=le.transform(train[i])\n test[i]=le.transform(test[i])",
"_____no_output_____"
],
[
"train=pd.get_dummies(train)\ntest=pd.get_dummies(test)",
"_____no_output_____"
],
[
"x=train.drop(columns=['TARGET'])\ny=train['TARGET']",
"_____no_output_____"
],
[
"from sklearn.model_selection import train_test_split\nx_train,x_test,y_train,y_test = train_test_split(x,y,test_size =0.2,random_state =10)",
"_____no_output_____"
],
[
"from sklearn.preprocessing import MinMaxScaler\nfrom sklearn.impute import SimpleImputer\n\nimputer=SimpleImputer(strategy = 'median')\n\nx_train=imputer.fit_transform(x_train)\nx_test=imputer.transform(x_test)\n\nscaler=MinMaxScaler(feature_range = (0, 1))\nx_train_scale=scaler.fit_transform(x_train)\nx_test_scale=scaler.transform(x_test)",
"_____no_output_____"
],
[
"from sklearn.linear_model import LogisticRegression\nmodel = LogisticRegression(solver='liblinear', random_state=0)\nmodel.fit(x_train,y_train)\n",
"_____no_output_____"
],
[
"pred = model.predict(x_train)",
"_____no_output_____"
],
[
"model.score(x_train, y_train)",
"_____no_output_____"
],
[
"from sklearn.metrics import classification_report\nprint(classification_report(y_train, pred))",
" precision recall f1-score support\n\n 0 0.92 1.00 0.96 226148\n 1 0.00 0.00 0.00 19860\n\n accuracy 0.92 246008\n macro avg 0.46 0.50 0.48 246008\nweighted avg 0.85 0.92 0.88 246008\n\n"
],
[
"from sklearn.ensemble import RandomForestClassifier\nrandomforest=RandomForestClassifier(max_depth=4, random_state=10)\nrandomforest.fit(x_train_scale,y_train)\n\nfrom sklearn import metrics\ny_pred=randomforest.predict(x_test)\nprint(metrics.accuracy_score(y_test,y_pred))",
"0.9192722306228964\n"
],
[
"import xgboost as xgb\nfrom sklearn.metrics import mean_squared_error\n\ndmatrix = xgb.DMatrix(data=x,label=y)",
"_____no_output_____"
],
[
"xg_reg = xgb.XGBRegressor(objective ='reg:linear', colsample_bytree = 0.3, learning_rate = 0.1,max_depth = 5, alpha = 10, n_estimators = 10)",
"_____no_output_____"
],
[
"xg_reg.fit(x_train,y_train)\n\npred = xg_reg.predict(x_test)",
"[18:02:07] WARNING: /workspace/src/objective/regression_obj.cu:152: reg:linear is now deprecated in favor of reg:squarederror.\n"
],
[
"import numpy as np\nr_mean_square_error = np.sqrt(mean_squared_error(y_test, pred))\nprint(\"RMSE: %f\" % (r_mean_square_error))",
"RMSE: 0.305382\n"
],
[
"from sklearn.metrics import roc_auc_score\nscore = roc_auc_score(y_test,pred)\nprint(score)",
"0.7293659012571534\n"
],
[
"",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
c51517602ef0e37f75d875561150aaefb93de069
| 13,912 |
ipynb
|
Jupyter Notebook
|
MeasuringPROVProvenanceWebofData.ipynb
|
pgroth/prov-wod-analysis
|
1c5e0ceb6c143d41061c3c019e0f28da0632c08d
|
[
"Apache-2.0"
] | 2 |
2016-06-30T21:15:31.000Z
|
2017-06-16T01:50:32.000Z
|
MeasuringPROVProvenanceWebofData.ipynb
|
pgroth/prov-wod-analysis
|
1c5e0ceb6c143d41061c3c019e0f28da0632c08d
|
[
"Apache-2.0"
] | null | null | null |
MeasuringPROVProvenanceWebofData.ipynb
|
pgroth/prov-wod-analysis
|
1c5e0ceb6c143d41061c3c019e0f28da0632c08d
|
[
"Apache-2.0"
] | null | null | null | 38.969188 | 868 | 0.591288 |
[
[
[
"# Measuring PROV Provenance on the Web of Data\n\n* Authors: \n * [Paul Groth](http://pgroth.com), [Elsevier Labs](http://labs.elsevier.com)\n * [Wouter Beek](http://www.wouterbeek.com), Vrije Universiteit Amsterdam\n* Date: May 11, 2016\n\nOne of the motivations behind the original charter for the [W3C Provenance Incubator group](https://www.w3.org/2005/Incubator/prov/charter) was the need for provenance information for Semantic Web and Linked Data applications. Thus, a question to ask, three years after the introduction of the [W3C PROV family of documents](https://www.w3.org/TR/prov-overview/), is what is the adoption of PROV by the Semantic Web community.\n\nA proxy for this adoption is measuring how often PROV is used within Linked Data. In this work, we begin to do such a measurement. Our analytics are based on the [LOD Laundromat](http://lodlaundromat.org/) (Beek et al. 2014). The LOD Laudromat crawls and cleans over 650 thousand linked data documents representing over 38 billion triples. LOD Laudromat has been used in the past to do large scale analysis of linked data (Rietveld et al. 2015). \n\nHere, we focus on core statistics based around what [PROV-DM](http://www.w3.org/TR/prov-dm/) refers to as core structures. We only look at directly asserted information about resources in the dataset (i.e. no inference was performed before calculating these statistics).\n\n",
"_____no_output_____"
]
],
[
[
"from IPython.display import HTML\ndisplay(HTML('''<script>\ncode_show=true; \nfunction code_toggle() {\n if (code_show){\n $('div.input').hide();\n } else {\n $('div.input').show();\n }\n code_show = !code_show\n} \n$( document ).ready(code_toggle);\n</script>\nWe note that the code for our analysis is embeded within this document but is by default hidden for easier reading.\nTo toggle on/off the raw code, click <a href=\"javascript:code_toggle()\">here</a>.'''))",
"_____no_output_____"
]
],
[
[
"Additionally, all code is available [online](https://github.com/pgroth/prov-wod-analysis)",
"_____no_output_____"
]
],
[
[
"import requests\nnsr = requests.get(\"http://index.lodlaundromat.org/ns2d/\", params={\"uri\":\"http://www.w3.org/ns/prov#\"})\ntotal_prov_docs = nsr.json()[\"totalResults\"]",
"_____no_output_____"
],
[
"nsr = requests.get(\"http://index.lodlaundromat.org/ns2d/\", params={\"uri\":\"http://www.w3.org/ns/prov#\",\"limit\":total_prov_docs} )",
"_____no_output_____"
],
[
"import io\nfrom rdflib.namespace import RDFS, RDF\nfrom rdflib.namespace import Namespace\nfrom rdflib import Graph\nfrom rdflib import URIRef\nPROV = Namespace('http://www.w3.org/ns/prov#')",
"_____no_output_____"
],
[
"entitySubclasses = []\nactivitySubclasses = []\nagentSubclasses = []\ntotalNumberOfEntities = 0\ntotalNumberOfActivities = 0\ntotalNumberOfAgents = 0\nnumWasDerivedFrom = 0\nnumUsed = 0\nnumWGB = 0\nnumWAW = 0\nnumWasAttributedTo = 0\n\nfor doc in nsr.json()[\"results\"]:\n #print(doc)\n headers = {'Accept': 'text/turtle'}\n x = requests.get(\"http://ldf.lodlaundromat.org/\" + doc, headers=headers)\n txt_res = x.text\n tmpGraph = Graph()\n tmpGraph.parse(io.StringIO(txt_res), format=\"turtle\")\n #print(doc + \" \" + str(len(tmpGraph)))\n for entityClass in tmpGraph.subjects(RDFS.subClassOf, PROV.Entity):\n #print(entityClass)\n entitySubclasses.append(entityClass)\n for entity in tmpGraph.subjects(RDF.type, PROV.Entity):\n totalNumberOfEntities = totalNumberOfEntities + 1\n \n for activityClass in tmpGraph.subjects(RDFS.subClassOf, PROV.Activity):\n #print(activityClass)\n activitySubclasses.append(activityClass)\n \n for activity in tmpGraph.subjects(RDF.type, PROV.Activity):\n totalNumberOfActivities = totalNumberOfActivities + 1\n \n for agentClass in tmpGraph.subjects(RDFS.subClassOf, PROV.Agent):\n #print(agentClass)\n agentSubclasses.append(agentClass)\n \n for agent in tmpGraph.subjects(RDF.type, PROV.Agent):\n totalNumberOfAgents = totalNumberOfAgents + 1\n \n ##look at relations\n \n for s,p,o in tmpGraph.triples( (None, PROV.wasDerivedFrom, None )):\n numWasDerivedFrom = numWasDerivedFrom + 1\n\n for s,p,o in tmpGraph.triples( (None, PROV.used, None )):\n numUsed = numUsed + 1\n \n for s,p,o in tmpGraph.triples( (None, PROV.wasGeneratedBy, None )):\n numWGB = numWGB + 1\n \n for s,p,o in tmpGraph.triples( (None, PROV.wasAssociatedWith, None )):\n numWAW = numWAW + 1\n \n for s,p,o in tmpGraph.triples( (None, PROV.wasAttributedTo, None) ): \n numWasAttributedTo = numWasAttributedTo + 1\n \n \n \n \n ",
"_____no_output_____"
],
[
"from IPython.display import display, Markdown\n\noutput = \"### Statistics \\n\"\noutput += \"We first look at how many times both the namespace is declared and how many resources are of a given core type.\\n\"\noutput += \"* The PROV namespace occurs in \" + str(total_prov_docs) + \" documents.\\n\" \noutput += \"* Number of Entites: \" + str(totalNumberOfEntities) + \"\\n\"\noutput += \"* Number of Activities: \" + str(totalNumberOfActivities) + \"\\n\"\noutput += \"* Number of Agents: \" + str(totalNumberOfAgents) + \"\\n\\n\"\n\noutput += \"We also looked at the number of PROV edges that were used with the various documents.\\n\"\noutput += \"* Number of wasDerivedFrom edges: \" + str(numWasDerivedFrom) + \"\\n\"\noutput += \"* Number of used edges: \" + str(numUsed) + \"\\n\"\noutput += \"* Number of wasGeneratedBy edges: \" + str(numWGB) + \"\\n\"\noutput += \"* Number of wasAssociatedWith edges: \" + str(numWAW) + \"\\n\"\noutput += \"* Number of wasAttributedTo edges: \" + str(numWasAttributedTo) + \"\\n\\n\"\n\ndisplay(Markdown(output))",
"_____no_output_____"
]
],
[
[
"We also note that PROV has been extended by 8 other ontologies as calculated by manual inspection of the extensions of the various core classes as listed in the appendix.\n\n### Conclusion\nThis initial analysis shows some uptake within the Semantic Web community. However, while PROV is widely referenced within the community's literature, it appears, that direct usage of the standard could be improved (at least within the dataset represented by the LOD Laudromat). It should be noted that our analysis is preliminary and there is a much room for further work. In particular, we aim to look at the indirect usage of PROV through usage by ontologies that extend it (e.g. The Provenance Vocabulary) or that map to it such as Dublin Core or [PAV](http://pav-ontology.github.io/pav/). Understanding such indirect usage will help us better understand the true state of provenance interoperability within Linked Data. Likewise, it would be interesting to perform network analysis to understand the role that PROV plays within the Linked Data network. \n\n\n### References\n\n* Beek, W. & Rietveld, L & Bazoobandi, H.R. & Wielemaker, J. & Schlobach, S.: LOD Laundromat: A Uniform Way of Publishing Other People's Dirty Data. Proceedings of the International Semantic Web Conference (2014).\n* Rietveld, L. & Beek, W. & Schlobach, S.: LOD Lab: Experiments at LOD Scale. Proceedings of the International Semantic Web Conference (2015).\n\n",
"_____no_output_____"
],
[
"### Appendix: Classes that subclass a PROV core class",
"_____no_output_____"
]
],
[
[
"print(\"Subclasses of Entity\")\nfor i in entitySubclasses:\n print(i)\nprint(\"Subclasses of Activity\")\nfor i in activitySubclasses:\n print(i)\nprint(\"Subclasses of Agent\")\nfor i in agentSubclasses:\n print(i)",
"Subclasses of Entity\nhttp://www.gsi.dit.upm.es/ontologies/marl/ns#Opinion\nhttp://purl.org/net/p-plan#Entity\nhttp://www.w3.org/ns/prov#Plan\nhttp://www.w3.org/ns/prov#Bundle\nhttp://www.w3.org/ns/prov#Collection\nhttp://www.opmw.org/ontology/WorkflowExecutionArtifact\nhttp://purl.org/twc/vocab/vsr#Color\nhttp://www.co-ode.org/ontologies/ont.owl#Graphic\nhttp://purl.org/twc/vocab/vsr#Root\nhttp://purl.org/twc/vocab/vsr#Color\nhttp://www.co-ode.org/ontologies/ont.owl#Graphic\nhttp://purl.org/twc/vocab/vsr#Root\nhttp://purl.org/net/provenance/ns#DataItem\nhttp://purl.org/net/provenance/ns#File\nhttp://purl.org/net/provenance/ns#Immutable\nhttp://purl.org/net/provenance/ns#File\nhttp://purl.org/net/provenance/ns#Immutable\nhttp://purl.org/net/provenance/ns#DataItem\nSubclasses of Activity\nhttp://www.gsi.dit.upm.es/ontologies/marl/ns#SentimentAnalysis\nhttp://spitfire-project.eu/ontology/ns/Activity\nhttp://www.w3.org/ns/org#ChangeEvent\nhttp://purl.org/net/p-plan#Activity\nhttp://www.opmw.org/ontology/WorkflowExecutionProcess\nhttp://www.w3.org/ns/org#ChangeEvent\nhttp://purl.org/net/provenance/ns#DataCreation\nhttp://purl.org/net/provenance/ns#DataAccess\nhttp://purl.org/net/provenance/ns#DataCreation\nhttp://purl.org/net/provenance/ns#DataAccess\nSubclasses of Agent\nhttp://spitfire-project.eu/ontology/ns/Agent\nhttp://purl.org/net/provenance/types#DataCreator\nhttp://purl.org/net/provenance/ns#HumanAgent\nhttp://purl.org/net/provenance/ns#HumanAgent\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
]
] |
c515188d73db6e99fa9c17c14adf8fe30553cadf
| 14,038 |
ipynb
|
Jupyter Notebook
|
Practice/GraphCNN/mlp_kora.ipynb
|
pradm007/Neural-Networks-Practice
|
d0887c39e0292fd94651b13abd18ba47be5277be
|
[
"Apache-2.0"
] | null | null | null |
Practice/GraphCNN/mlp_kora.ipynb
|
pradm007/Neural-Networks-Practice
|
d0887c39e0292fd94651b13abd18ba47be5277be
|
[
"Apache-2.0"
] | null | null | null |
Practice/GraphCNN/mlp_kora.ipynb
|
pradm007/Neural-Networks-Practice
|
d0887c39e0292fd94651b13abd18ba47be5277be
|
[
"Apache-2.0"
] | null | null | null | 36.557292 | 193 | 0.530489 |
[
[
[
"try:\n # %tensorflow_version only exists in Colab.\n %tensorflow_version 2.x\nexcept Exception:\n !pip install -q tensorflow-gpu>=2.0.0",
"_____no_output_____"
],
[
"!pip install --quiet neural-structured-learning",
"_____no_output_____"
],
[
"from __future__ import absolute_import, division, print_function, unicode_literals\n\nimport neural_structured_learning as nsl\n\nimport tensorflow as tf\n\n# Resets notebook state\ntf.keras.backend.clear_session()\n\nprint(\"Version: \", tf.__version__)\nprint(\"Eager mode: \", tf.executing_eagerly())\nprint(\"GPU is\", \"available\" if tf.test.is_gpu_available() else \"NOT AVAILABLE\")",
"Version: 2.0.0\nEager mode: True\nGPU is NOT AVAILABLE\n"
],
[
"!wget --quiet -P /tmp https://linqs-data.soe.ucsc.edu/public/lbc/cora.tgz\n!tar -C /tmp -xvzf /tmp/cora.tgz",
"cora/\ncora/README\ncora/cora.content\ncora/cora.cites\n"
],
[
"!wget https://raw.githubusercontent.com/tensorflow/neural-structured-learning/master/neural_structured_learning/examples/preprocess/cora/preprocess_cora_dataset.py\n\n!python preprocess_cora_dataset.py \\\n--input_cora_content=/tmp/cora/cora.content \\\n--input_cora_graph=/tmp/cora/cora.cites \\\n--max_nbrs=5 \\\n--output_train_data=/tmp/cora/train_merged_examples.tfr \\\n--output_test_data=/tmp/cora/test_examples.tfr",
"--2019-10-24 05:06:03-- https://raw.githubusercontent.com/tensorflow/neural-structured-learning/master/neural_structured_learning/examples/preprocess/cora/preprocess_cora_dataset.py\nResolving raw.githubusercontent.com (raw.githubusercontent.com)...151.101.0.133, 151.101.64.133, 151.101.128.133, ...\nConnecting to raw.githubusercontent.com (raw.githubusercontent.com)|151.101.0.133|:443... connected.\nHTTP request sent, awaiting response... 200 OK\nLength: 11419 (11K) [text/plain]\nSaving to: ‘preprocess_cora_dataset.py.1’\n\npreprocess_cora_dat 100%[===================>] 11.15K --.-KB/s in 0s \n\n2019-10-24 05:06:03 (67.9 MB/s) - ‘preprocess_cora_dataset.py.1’ saved [11419/11419]\n\npreprocess_cora_dataset.py:130: DeprecationWarning: 'U' mode is deprecated\n with open(in_file, 'rU') as cora_content:\nReading graph file: /tmp/cora/cora.cites...\nDone reading 5429 edges from: /tmp/cora/cora.cites (0.02 seconds).\nMaking all edges bi-directional...\nDone (0.01 seconds). Total graph nodes: 2708\nJoining seed and neighbor tf.train.Examples with graph edges...\nDone creating and writing 2155 merged tf.train.Examples (1.90 seconds).\nOut-degree histogram: [(1, 386), (2, 468), (3, 452), (4, 309), (5, 540)]\nOutput training data written to TFRecord file: /tmp/cora/train_merged_examples.tfr.\nOutput test data written to TFRecord file: /tmp/cora/test_examples.tfr.\nTotal running time: 0.07 minutes.\n"
],
[
"### Experiment dataset\nTRAIN_DATA_PATH = '/tmp/cora/train_merged_examples.tfr'\nTEST_DATA_PATH = '/tmp/cora/test_examples.tfr'\n\n### Constants used to identify neighbor features in the input.\nNBR_FEATURE_PREFIX = 'NL_nbr_'\nNBR_WEIGHT_SUFFIX = '_weight'",
"_____no_output_____"
],
[
"class HParams(object):\n \"\"\"Hyperparameters used for training.\"\"\"\n def __init__(self):\n ### dataset parameters\n self.num_classes = 7\n self.max_seq_length = 1433\n ### neural graph learning parameters\n self.distance_type = nsl.configs.DistanceType.L2\n self.graph_regularization_multiplier = 0.1\n self.num_neighbors = 1\n ### model architecture\n self.num_fc_units = [50, 50]\n ### training parameters\n self.train_epochs = 100\n self.batch_size = 128\n self.dropout_rate = 0.5\n ### eval parameters\n self.eval_steps = None # All instances in the test set are evaluated.\n\nHPARAMS = HParams()",
"_____no_output_____"
],
[
"def parse_example(example_proto):\n \"\"\"Extracts relevant fields from the `example_proto`.\n\n Args:\n example_proto: An instance of `tf.train.Example`.\n\n Returns:\n A pair whose first value is a dictionary containing relevant features\n and whose second value contains the ground truth labels.\n \"\"\"\n # The 'words' feature is a multi-hot, bag-of-words representation of the\n # original raw text. A default value is required for examples that don't\n # have the feature.\n feature_spec = {\n 'words':\n tf.io.FixedLenFeature([HPARAMS.max_seq_length],\n tf.int64,\n default_value=tf.constant(\n 0,\n dtype=tf.int64,\n shape=[HPARAMS.max_seq_length])),\n 'label':\n tf.io.FixedLenFeature((), tf.int64, default_value=-1),\n }\n # We also extract corresponding neighbor features in a similar manner to\n # the features above.\n for i in range(HPARAMS.num_neighbors):\n nbr_feature_key = '{}{}_{}'.format(NBR_FEATURE_PREFIX, i, 'words')\n nbr_weight_key = '{}{}{}'.format(NBR_FEATURE_PREFIX, i, NBR_WEIGHT_SUFFIX)\n feature_spec[nbr_feature_key] = tf.io.FixedLenFeature(\n [HPARAMS.max_seq_length],\n tf.int64,\n default_value=tf.constant(\n 0, dtype=tf.int64, shape=[HPARAMS.max_seq_length]))\n\n # We assign a default value of 0.0 for the neighbor weight so that\n # graph regularization is done on samples based on their exact number\n # of neighbors. In other words, non-existent neighbors are discounted.\n feature_spec[nbr_weight_key] = tf.io.FixedLenFeature(\n [1], tf.float32, default_value=tf.constant([0.0]))\n\n features = tf.io.parse_single_example(example_proto, feature_spec)\n\n labels = features.pop('label')\n return features, labels\n\n\ndef make_dataset(file_path, training=False):\n \"\"\"Creates a `tf.data.TFRecordDataset`.\n\n Args:\n file_path: Name of the file in the `.tfrecord` format containing\n `tf.train.Example` objects.\n training: Boolean indicating if we are in training mode.\n\n Returns:\n An instance of `tf.data.TFRecordDataset` containing the `tf.train.Example`\n objects.\n \"\"\"\n dataset = tf.data.TFRecordDataset([file_path])\n if training:\n dataset = dataset.shuffle(10000)\n dataset = dataset.map(parse_example)\n dataset = dataset.batch(HPARAMS.batch_size)\n return dataset\n\n\ntrain_dataset = make_dataset(TRAIN_DATA_PATH, training=True)\ntest_dataset = make_dataset(TEST_DATA_PATH)",
"_____no_output_____"
],
[
"for feature_batch, label_batch in train_dataset.take(1):\n print('Feature list:', list(feature_batch.keys()))\n print('Batch of inputs:', feature_batch['words'])\n nbr_feature_key = '{}{}_{}'.format(NBR_FEATURE_PREFIX, 0, 'words')\n nbr_weight_key = '{}{}{}'.format(NBR_FEATURE_PREFIX, 0, NBR_WEIGHT_SUFFIX)\n print('Batch of neighbor inputs:', feature_batch[nbr_feature_key])\n print('Batch of neighbor weights:',\n tf.reshape(feature_batch[nbr_weight_key], [-1]))\n print('Batch of labels:', label_batch)",
"Feature list: ['NL_nbr_0_weight', 'NL_nbr_0_words', 'words']\nBatch of inputs: tf.Tensor(\n[[0 0 0 ... 0 0 0]\n [0 0 0 ... 0 0 0]\n [1 0 0 ... 0 0 0]\n ...\n [0 0 0 ... 0 0 0]\n [0 0 0 ... 0 0 0]\n [0 0 0 ... 0 0 0]], shape=(128, 1433), dtype=int64)\nBatch of neighbor inputs: tf.Tensor(\n[[0 0 0 ... 0 0 0]\n [0 0 0 ... 0 0 0]\n [0 0 0 ... 0 0 0]\n ...\n [0 0 0 ... 0 0 0]\n [0 0 0 ... 0 0 0]\n [0 0 0 ... 0 0 0]], shape=(128, 1433), dtype=int64)\nBatch of neighbor weights: tf.Tensor(\n[1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1.\n 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1.\n 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1.\n 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1.\n 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1.\n 1. 1. 1. 1. 1. 1. 1. 1.], shape=(128,), dtype=float32)\nBatch of labels: tf.Tensor(\n[3 5 0 4 3 2 1 2 6 2 2 2 5 4 2 1 5 6 3 2 1 0 5 4 3 5 5 0 3 1 1 2 6 5 2 2 2\n 2 2 1 6 1 6 2 0 1 1 3 6 1 5 4 5 3 4 1 1 1 3 6 2 3 2 4 3 2 0 1 2 2 1 0 2 6\n 1 1 0 3 1 2 2 6 6 3 6 2 6 2 4 2 6 2 0 6 1 1 4 2 5 4 2 1 4 2 3 5 1 0 4 2 0\n 3 2 0 4 2 2 2 2 4 0 1 3 2 1 2 3 1], shape=(128,), dtype=int64)\n"
],
[
"for feature_batch, label_batch in test_dataset.take(1):\n print('Feature list:', list(feature_batch.keys()))\n print('Batch of inputs:', feature_batch['words'])\n nbr_feature_key = '{}{}_{}'.format(NBR_FEATURE_PREFIX, 0, 'words')\n nbr_weight_key = '{}{}{}'.format(NBR_FEATURE_PREFIX, 0, NBR_WEIGHT_SUFFIX)\n print('Batch of neighbor inputs:', feature_batch[nbr_feature_key])\n print('Batch of neighbor weights:',\n tf.reshape(feature_batch[nbr_weight_key], [-1]))\n print('Batch of labels:', label_batch)",
"Feature list: ['NL_nbr_0_weight', 'NL_nbr_0_words', 'words']\nBatch of inputs: tf.Tensor(\n[[0 0 0 ... 0 0 0]\n [0 0 0 ... 0 0 0]\n [0 0 0 ... 0 0 0]\n ...\n [0 0 0 ... 0 0 0]\n [0 0 0 ... 0 0 0]\n [0 0 0 ... 0 0 0]], shape=(128, 1433), dtype=int64)\nBatch of neighbor inputs: tf.Tensor(\n[[0 0 0 ... 0 0 0]\n [0 0 0 ... 0 0 0]\n [0 0 0 ... 0 0 0]\n ...\n [0 0 0 ... 0 0 0]\n [0 0 0 ... 0 0 0]\n [0 0 0 ... 0 0 0]], shape=(128, 1433), dtype=int64)\nBatch of neighbor weights: tf.Tensor(\n[0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.\n 0. 0. 0. 0. 0. 0. 0. 0.], shape=(128,), dtype=float32)\nBatch of labels: tf.Tensor(\n[5 2 2 2 1 2 6 3 2 3 6 1 3 6 4 4 2 3 3 0 2 0 5 2 1 0 6 3 6 4 2 2 3 0 4 2 2\n 2 2 3 2 2 2 0 2 2 2 2 4 2 3 4 0 2 6 2 1 4 2 0 0 1 4 2 6 0 5 2 2 3 2 5 2 5\n 2 3 2 2 2 2 2 6 6 3 2 4 2 6 3 2 2 6 2 4 2 2 1 3 4 6 0 0 2 4 2 1 3 6 6 2 6\n 6 6 1 4 6 4 3 6 6 0 0 2 6 2 4 0 0], shape=(128,), dtype=int64)\n"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
c5151a642f7c85779c7daeeceed027425831bbcc
| 576,027 |
ipynb
|
Jupyter Notebook
|
Analysis.ipynb
|
Mtale/Coursera_Capstone
|
8883eff7c323b9655b135ca26697e8c874dbd89c
|
[
"MIT"
] | null | null | null |
Analysis.ipynb
|
Mtale/Coursera_Capstone
|
8883eff7c323b9655b135ca26697e8c874dbd89c
|
[
"MIT"
] | null | null | null |
Analysis.ipynb
|
Mtale/Coursera_Capstone
|
8883eff7c323b9655b135ca26697e8c874dbd89c
|
[
"MIT"
] | null | null | null | 382.488048 | 74,828 | 0.923799 |
[
[
[
"\n# Import libraries and packages",
"_____no_output_____"
]
],
[
[
"import folium\nimport numpy as np\nimport pandas as pd\nimport seaborn as sns\n\nfrom geopy.geocoders import Nominatim\nfrom matplotlib import pyplot as plt\nfrom scipy import stats\nfrom sklearn.linear_model import LinearRegression\nfrom sklearn.pipeline import Pipeline\nfrom sklearn.preprocessing import MinMaxScaler\n\n# Custom modules\n# We will use top_20_cities dictionary from here\nfrom openstreetmap import openstreetmap as osm ",
"_____no_output_____"
]
],
[
[
"# Set parameters",
"_____no_output_____"
]
],
[
[
"color = 'goldenrod'\nsns.set_style(\"whitegrid\")",
"_____no_output_____"
]
],
[
[
"# Data preparation",
"_____no_output_____"
],
[
"## Data import",
"_____no_output_____"
]
],
[
[
"# Read the data and sort it by total population\ndata = pd.read_csv('tidy_data.csv', sep=';')\ndata = data.sort_values(by='Population on the 1st of January, total', ascending=False).reset_index(drop=True)\n\n# Rename the total population column\ndata.rename(columns={'Population on the 1st of January, total':'total_population'}, inplace=True)\n\n# Replace unicode characters due to rendering issue in Folium\ndata = data.replace(to_replace={'ü':'u','ö':'o'}, regex=True)\n\n# print(data.shape)\n# data.tail()",
"_____no_output_____"
],
[
"# Confirm the venue_id is unique\nlen(data['venue_id'].unique()) == data.shape[0]",
"_____no_output_____"
]
],
[
[
"## Add counts to venue data",
"_____no_output_____"
]
],
[
[
"# Add ratings count per city to tidy data\ndata['ratings_count'] = data.rating.notnull().groupby(data['city']).transform('sum').astype(int)\n\n# Add likes_count per city to tidy data\ndata['likes_count'] = data.likes_cnt.groupby(data['city']).transform('sum').astype(int)",
"_____no_output_____"
]
],
[
[
"## Create DataFrame to carry counts per city",
"_____no_output_____"
]
],
[
[
"# Count ratings to distinct dataframe\ndata_counts = pd.DataFrame(data.rating.notnull().groupby(data['city'], sort=False).sum().astype(int).reset_index())\ndata_counts = data_counts.merge(data[['city', 'total_population']], on='city') \\\n .drop_duplicates() \\\n .reset_index(drop=True)\ndata_counts.columns = ['city', 'ratings_count', 'total_population']\n# print(data_counts.shape)\n# data_counts",
"_____no_output_____"
],
[
"# Count likes to distinct dataframe\nlikes_counts = pd.DataFrame(data.likes_cnt.groupby(data['city'], sort=False).sum().astype(int).reset_index())\nlikes_counts.columns = ['city','likes_count']\ndata_counts = data_counts.merge(likes_counts, on='city')\n# data_counts",
"_____no_output_____"
],
[
"# Count number of biergartens per city\nno_of_biergartens_city = pd.DataFrame(data.groupby('city', sort=False).count().venue_id).reset_index()\nno_of_biergartens_city.columns = ['city', 'biergarten_count']\n\n# Join to count data\ndata_counts = data_counts.merge(no_of_biergartens_city, on='city')\n# data_counts",
"_____no_output_____"
],
[
"# Count no of biergartens per 100,000 people\ndata_counts['biergarten_count_100k'] = data_counts['biergarten_count']/data_counts['total_population']*100000\n# data_counts",
"_____no_output_____"
],
[
"# Add rank variables to dataset\ndata_counts['biergarten_rank'] = data_counts['biergarten_count'].rank()\ndata_counts['biergarten_100k_rank'] = data_counts['biergarten_count_100k'].rank()\ndata_counts",
"_____no_output_____"
]
],
[
[
"# Where can you find most biergartens?",
"_____no_output_____"
]
],
[
[
"g = sns.PairGrid(data_counts, y_vars=[\"city\"], x_vars=[\"biergarten_count\", \"biergarten_count_100k\"]\n ,height=6\n ,corner=False\n ,despine=True)\ng.map(sns.barplot, color=color, order=data_counts['city'])\ng.axes[0,0].grid(True)\ng.axes[0,1].grid(True)\ng.axes[0,0].set_ylabel('')\ng.axes[0,0].set_xlabel('No of biergartens', fontdict={'fontsize':16})\ng.axes[0,1].set_xlabel('No of biergartens per 100,000 people', fontdict={'fontsize':16})\n",
"_____no_output_____"
],
[
"# Plot ranks\nplt.figure(figsize=(10,10)) \nax = sns.scatterplot(data=data_counts\n , x='biergarten_rank'\n , y='biergarten_100k_rank'\n , size='total_population'\n , sizes=(90,1080) # Population/10,000*3\n , legend=False\n , color=color)\n\nfor line in range(0, data_counts.shape[0]):\n ax.text(x=data_counts.biergarten_rank[line]-0.4\n , y=data_counts.biergarten_100k_rank[line]\n , s=data_counts.city[line]\n , horizontalalignment='right'\n , verticalalignment='baseline'\n , size='small'\n , color='black')\n \nax.set_ylabel('Rank of number of biergartens per 100,000 people', fontdict={'fontsize':16})\nax.set_xlabel('Rank of number of biergartens', fontdict={'fontsize':16})\nax.set_xticks(range(0,22,2))\nax.set_yticks(range(0,22,2))",
"_____no_output_____"
]
],
[
[
"# Are biergartens equally popular in different regions?",
"_____no_output_____"
]
],
[
[
"# Get coordinates for Germany to center the map\ngeolocator = Nominatim(user_agent=\"germany_explorer\")\naddress = 'Germany'\nlocation = geolocator.geocode(address)\ngermany_latitude = location.latitude\ngermany_longitude = location.longitude\nprint('The geograpical coordinate of Germany are {}, {}.'.format(germany_latitude, germany_longitude))",
"The geograpical coordinate of Germany are 51.0834196, 10.4234469.\n"
],
[
"# Create empty dataframe to store coordinates to\ngermany_city_coordinates = pd.DataFrame()\n\n# Get coordinates for cities to be plotted\ngeolocator = Nominatim(user_agent=\"germany_explorer\")\nfor city in osm.top20_cities.keys():\n address = city + ', Germany'\n location = geolocator.geocode(address)\n d = {\n 'city': city,\n 'latitude': location.latitude,\n 'longitude': location.longitude,\n }\n germany_city_coordinates = germany_city_coordinates.append(d, ignore_index=True)\n \n# Replace unicode characters due to rendering issue in Folium and to match rest of the data\ngermany_city_coordinates = germany_city_coordinates.replace(to_replace={'ü':'u','ö':'o'}, regex=True)\n# germany_city_coordinates",
"_____no_output_____"
],
[
"# Join coordinates to counts data\ndata_counts = data_counts.merge(germany_city_coordinates, on='city')\n# data_counts",
"_____no_output_____"
],
[
"# Join coordinates to venue data\ndata = data.merge(germany_city_coordinates, on='city')",
"_____no_output_____"
],
[
"# Inititate map of Germany\nmap_germany = folium.Map(location=[germany_latitude, germany_longitude], zoom_start=6)\n\n# Loop through data_counts\nfor city, lat, lng, pop, cnt, cnt_100k, rank, rank_100k in zip(data_counts['city']\n , data_counts['latitude']\n , data_counts['longitude']\n , data_counts['total_population']\n , data_counts['biergarten_count']\n , data_counts['biergarten_count_100k']\n , data_counts['biergarten_rank']\n , data_counts['biergarten_100k_rank']):\n \n # Generate html to include data in popup\n label = (\n \"{city}<br>\"\n \"Population: {pop}<br>\"\n \"No of biergartens: {cnt}<br>\"\n \"No of biergartens per 100,000 people: {cnt_100k}<br>\"\n ).format(city=city.upper(),\n pop=str(int(pop)),\n cnt=str(int(cnt)),\n cnt_100k=str(round(cnt_100k, 1)),\n )\n \n # Set marker color based on the biergarten_count_100k\n if cnt_100k > 5:\n colour = 'darkpurple'\n elif cnt_100k > 4:\n colour = 'red'\n elif cnt_100k > 3:\n colour = 'orange'\n elif cnt_100k > 2:\n colour = 'pink'\n else:\n colour = 'lightgray'\n \n # Add marker\n map_germany.add_child(folium.Marker(\n location=[lat, lng],\n popup=label,\n icon=folium.Icon(\n color=colour,\n prefix='fa',\n icon='circle')))\n\n# Create a legent to map\nlegend_html = \"\"\"\n <div style=\"position: fixed; bottom: 50px; left: 50px; width: 150px; height: 200px; \\\n border:2px solid grey; z-index:9999; font-size:14px;\" >\n No of biergartens <br>\n per 100,000 people <br>\n 5 + <i class=\"fa fa-map-marker fa-2x\"\n style=\"color:darkpurple\"></i><br>\n 4-5 <i class=\"fa fa-map-marker fa-2x\"\n style=\"color:red\"></i><br>\n 3-4 <i class=\"fa fa-map-marker fa-2x\"\n style=\"color:orange\"></i><br>\n 2-3 <i class=\"fa fa-map-marker fa-2x\"\n style=\"color:pink\"></i><br>\n 0-2 <i class=\"fa fa-map-marker fa-2x\"\n style=\"color:lightgray\"></i></div>\n \"\"\"\nmap_germany.get_root().html.add_child(folium.Element(legend_html))\n \n# Show the map\nmap_germany",
"_____no_output_____"
]
],
[
[
"# Do biergarten reviews hint where to go to?",
"_____no_output_____"
]
],
[
[
"# Plot likes\nplt.figure(figsize=(6,8)) \nax=sns.barplot(y='city', x='likes_count', data=data_counts, color=color)\nax.set_ylabel('')\nax.set_xlabel('Count of likes in Foursquare', fontdict={'fontsize':16})",
"_____no_output_____"
],
[
"# Plot ratings\nplt.figure(figsize=(6,10)) \ng = sns.boxplot(data=data, y='city', x='rating'\n , order=data_counts['city']\n , hue=None\n , color='goldenrod'\n , saturation=1.0\n , fliersize=0.0\n )\ng.axes.set_ylabel('')\ng.axes.set_xlabel('Foursquare rating', fontdict={'fontsize':16})\n\n# Calculate number of obs per group & median to position labels\nmedians = data.groupby(['city'], sort=False)['rating'].median().values\nnobs = data_counts['ratings_count']\nnobs = [str(x) for x in nobs.tolist()]\nnobs = [\"n: \" + i for i in nobs]\n\n# Add it to the plot\npos = range(len(nobs))\nfor tick, label in zip(pos, g.get_yticklabels()):\n g.text(x=4.72\n , y=pos[tick]\n , s=nobs[tick]\n , horizontalalignment='left'\n , verticalalignment='center'\n , size='small'\n , color='black'\n , weight='normal')\n\n",
"_____no_output_____"
]
],
[
[
"# Does population structure explain density of biergartens?",
"_____no_output_____"
]
],
[
[
"# Create modeling dataset\nX_cols = [\n 'latitude', \n 'longitude',\n 'Proportion of population aged 0-4 years',\n 'Proportion of population aged 5-9 years',\n 'Proportion of population aged 10-14 years',\n 'Proportion of population aged 15-19 years',\n 'Proportion of population aged 20-24 years',\n 'Proportion of population aged 25-34 years',\n 'Proportion of population aged 35-44 years',\n 'Proportion of population aged 45-54 years',\n 'Proportion of total population aged 55-64',\n 'Proportion of population aged 65-74 years',\n 'Proportion of population aged 75 years and over',\n 'Women per 100 men',\n# 'Young-age dependency ratio (population aged 0-19 to population 20-64 years)',\n 'Nationals as a proportion of population']\ncity_df = pd.DataFrame(data['city'])\nX = data[X_cols].drop_duplicates().reset_index(drop=True)\n# X.rename(columns={'Proportion of total population aged 55-64':'Proportion of population aged 55-64 years'}, inplace=True)\n\n# Create target variable\ny = data_counts['biergarten_count_100k']",
"_____no_output_____"
],
[
"# Create correlation matrix\ncorr_matrix = X.corr().abs()\n\n# Select upper triangle of correlation matrix\nupper = corr_matrix.where(np.triu(np.ones(corr_matrix.shape), k=1).astype(np.bool))\n\n# Find index of feature columns with correlation greater than 0.8\nto_drop = [column for column in upper.columns if any(upper[column] > 0.8)]\n\n# Drop features \nX.drop(X[to_drop], axis=1, inplace=True)",
"_____no_output_____"
],
[
"# Pipeline for linear regression\nlin_reg_pipe = Pipeline([('minmax', MinMaxScaler(feature_range=(-1,1)))\n ,('lin_reg', LinearRegression(fit_intercept=True))])\n\n# Train the regression model\nlin_reg_pipe.fit(X,y)",
"_____no_output_____"
],
[
"# Plot regression coeffiecient\nplt.figure(figsize=(6,8)) \nbarplot_data = pd.concat([pd.Series(X.columns.to_list()), pd.Series(lin_reg_pipe['lin_reg'].coef_)], axis=1)\nbarplot_data.columns = ['variable', 'coef']\nax=sns.barplot(y='variable', x='coef', data=barplot_data, color=color)\nax.axes.set_ylabel('')\nax.axes.set_xlabel('Regression coefficient', fontdict={'fontsize':16})\nplt.show()\n\n# Print regression measures\nprint('Intercept: {}'.format(lin_reg_pipe['lin_reg'].intercept_))\nprint('R^2: {}'.format(lin_reg_pipe.score(X,y)))\n\n# Plot predictions and actuals\nslope, intercept, r_value, p_value, std_err = stats.linregress(lin_reg_pipe.predict(X),y)\nplt.figure(figsize=(10,10)) \ng = sns.regplot(lin_reg_pipe.predict(X), y, color=color\n , scatter_kws={'s':data_counts['total_population']/5000})\ng.axes.set_xlabel('Predicted number of biergartens per 100,000 people', fontdict={'fontsize':16})\ng.axes.set_ylabel('Actual number of biergartens per 100,000 people', fontdict={'fontsize':16})\ng.text(1.7, 5, r'$R^2:{0:.2f}$'.format(r_value**2), fontdict={'fontsize':14})\ng.set_xticks(np.arange(0.5,7,1))\ng.set_yticks(np.arange(0.5,7,1))\n\nfor line in range(0, data_counts.shape[0]):\n g.text(x=lin_reg_pipe.predict(X)[line]-0.1\n , y=y[line]\n , s=data_counts.city[line]\n , horizontalalignment='right'\n , verticalalignment='baseline'\n , size='small'\n , color='black')\n \nplt.show()",
"_____no_output_____"
]
],
[
[
"# Does local living standard explain biergarten density in region?",
"_____no_output_____"
]
],
[
[
"# Create modeling dataset\nX_cols = [\n 'latitude',\n 'longitude',\n 'Activity rate',\n 'Employment (jobs) in agriculture, fishery (NACE Rev. 2, A)',\n 'Employment (jobs) in arts, entertainment and recreation; other service activities; activities of household and extra-territorial organizations and bodies (NACE Rev. 2, R to U)',\n 'Employment (jobs) in construction (NACE Rev. 2, F)',\n 'Employment (jobs) in financial and insurance activities (NACE Rev. 2, K)',\n 'Employment (jobs) in information and communication (NACE Rev. 2, J)',\n 'Employment (jobs) in mining, manufacturing, energy (NACE Rev. 2, B-E)',\n 'Employment (jobs) in professional, scientific and technical activities; administrative and support service activities (NACE Rev. 2, M and N)',\n 'Employment (jobs) in public administration, defence, education, human health and social work activities (NACE Rev. 2, O to Q)',\n 'Employment (jobs) in real estate activities (NACE Rev. 2, L)',\n 'Employment (jobs) in trade, transport, hotels, restaurants (NACE Rev. 2, G to I)',\n 'Proportion of employment in industries (NACE Rev.1.1 C-E)',\n 'Unemployment rate, female', \n 'Unemployment rate, male']\ncity_df = pd.DataFrame(data['city'])\nX = data[X_cols].drop_duplicates().reset_index(drop=True)\nX.rename(columns={'Employment (jobs) in agriculture, fishery (NACE Rev. 2, A)':'Jobs in agriculture, fishery',\n 'Employment (jobs) in arts, entertainment and recreation; other service activities; activities of household and extra-territorial organizations and bodies (NACE Rev. 2, R to U)': 'Jobs in arts, entertainment and recreation; other service',\n 'Employment (jobs) in construction (NACE Rev. 2, F)':'Jobs in construction',\n 'Employment (jobs) in financial and insurance activities (NACE Rev. 2, K)':'Jobs in financial and insurance activities',\n 'Employment (jobs) in information and communication (NACE Rev. 2, J)':'Jobs in information and communication',\n 'Employment (jobs) in mining, manufacturing, energy (NACE Rev. 2, B-E)':'Jobs in mining, manufacturing, energy',\n 'Employment (jobs) in professional, scientific and technical activities; administrative and support service activities (NACE Rev. 2, M and N)':'Jobs in professional, scientific and technical; administrative and support service',\n 'Employment (jobs) in public administration, defence, education, human health and social work activities (NACE Rev. 2, O to Q)':'Jobs in public administration, defence, education, human health and social work',\n 'Employment (jobs) in real estate activities (NACE Rev. 2, L)':'Jobs in real estate',\n 'Employment (jobs) in trade, transport, hotels, restaurants (NACE Rev. 2, G to I)':'Jobs in trade, transport, hotels, restaurants',\n 'Proportion of employment in industries (NACE Rev.1.1 C-E)':'Proportion of employment in industries'}\n , inplace=True)\n\n# Create target variable\ny = data_counts['biergarten_count_100k']",
"_____no_output_____"
],
[
"# Create correlation matrix\ncorr_matrix = X.corr().abs()\n\n# Select upper triangle of correlation matrix\nupper = corr_matrix.where(np.triu(np.ones(corr_matrix.shape), k=1).astype(np.bool))\n\n# Find index of feature columns with correlation greater than 0.8\nto_drop = [column for column in upper.columns if any(upper[column] > 0.8)]\n\n# Drop features \nX.drop(X[to_drop], axis=1, inplace=True)",
"_____no_output_____"
],
[
"# Pipeline for linear regression\nlin_reg_pipe = Pipeline([('minmax', MinMaxScaler(feature_range=(-1,1)))\n ,('lin_reg', LinearRegression(fit_intercept=True))])\n\n# Train the regression model\nlin_reg_pipe.fit(X,y)",
"_____no_output_____"
],
[
"# Plot regression coeffiecient\nplt.figure(figsize=(6,8)) \nbarplot_data = pd.concat([pd.Series(X.columns.to_list()), pd.Series(lin_reg_pipe['lin_reg'].coef_)], axis=1)\nbarplot_data.columns = ['variable', 'coef']\nax=sns.barplot(y='variable', x='coef', data=barplot_data, color=color)\nax.axes.set_ylabel('')\nax.axes.set_xlabel('Regression coefficient', fontdict={'fontsize':16})\nplt.show()\n\n# Print regression measures\nprint('Intercept: {}'.format(lin_reg_pipe['lin_reg'].intercept_))\nprint('R^2: {}'.format(lin_reg_pipe.score(X,y)))\n\n# Plot predictions and actuals\nslope, intercept, r_value, p_value, std_err = stats.linregress(lin_reg_pipe.predict(X),y)\nplt.figure(figsize=(10,10)) \ng = sns.regplot(lin_reg_pipe.predict(X), y, color=color\n , scatter_kws={'s':data_counts['total_population']/5000})\ng.axes.set_xlabel('Predicted number of biergartens per 100,000 people', fontdict={'fontsize':16})\ng.axes.set_ylabel('Actual number of biergartens per 100,000 people', fontdict={'fontsize':16})\ng.text(1.7, 5, r'$R^2:{0:.2f}$'.format(r_value**2), fontdict={'fontsize':14})\ng.set_xticks(np.arange(0.5,7,1))\ng.set_yticks(np.arange(0.5,7,1))\n\nfor line in range(0, data_counts.shape[0]):\n g.text(x=lin_reg_pipe.predict(X)[line]-0.1\n , y=y[line]\n , s=data_counts.city[line]\n , horizontalalignment='right'\n , verticalalignment='baseline'\n , size='small'\n , color='black')\n \nplt.show()",
"_____no_output_____"
],
[
"# Create modeling dataset\nX_cols = [\n 'latitude',\n 'longitude',\n 'Proportion of population aged 0-4 years',\n 'Proportion of population aged 5-9 years',\n 'Proportion of population aged 10-14 years',\n 'Proportion of population aged 15-19 years',\n 'Proportion of population aged 20-24 years',\n 'Proportion of population aged 25-34 years',\n 'Proportion of population aged 35-44 years',\n 'Proportion of population aged 45-54 years',\n 'Proportion of total population aged 55-64',\n 'Proportion of population aged 65-74 years',\n 'Proportion of population aged 75 years and over',\n 'Women per 100 men',\n 'Young-age dependency ratio (population aged 0-19 to population 20-64 years)',\n 'Nationals as a proportion of population',\n 'Activity rate',\n 'Employment (jobs) in agriculture, fishery (NACE Rev. 2, A)',\n 'Employment (jobs) in arts, entertainment and recreation; other service activities; activities of household and extra-territorial organizations and bodies (NACE Rev. 2, R to U)',\n 'Employment (jobs) in construction (NACE Rev. 2, F)',\n 'Employment (jobs) in financial and insurance activities (NACE Rev. 2, K)',\n 'Employment (jobs) in information and communication (NACE Rev. 2, J)',\n 'Employment (jobs) in mining, manufacturing, energy (NACE Rev. 2, B-E)',\n 'Employment (jobs) in professional, scientific and technical activities; administrative and support service activities (NACE Rev. 2, M and N)',\n 'Employment (jobs) in public administration, defence, education, human health and social work activities (NACE Rev. 2, O to Q)',\n 'Employment (jobs) in real estate activities (NACE Rev. 2, L)',\n 'Employment (jobs) in trade, transport, hotels, restaurants (NACE Rev. 2, G to I)',\n 'Proportion of employment in industries (NACE Rev.1.1 C-E)',\n 'Unemployment rate, female', \n 'Unemployment rate, male']\ncity_df = pd.DataFrame(data['city'])\nX = data[X_cols].drop_duplicates().reset_index(drop=True)\nX.rename(columns={'Proportion of total population aged 55-64':'Proportion of population aged 55-64 years','Employment (jobs) in agriculture, fishery (NACE Rev. 2, A)':'Jobs in agriculture, fishery',\n 'Employment (jobs) in arts, entertainment and recreation; other service activities; activities of household and extra-territorial organizations and bodies (NACE Rev. 2, R to U)': 'Jobs in arts, entertainment and recreation; other service',\n 'Employment (jobs) in construction (NACE Rev. 2, F)':'Jobs in construction',\n 'Employment (jobs) in financial and insurance activities (NACE Rev. 2, K)':'Jobs in financial and insurance activities',\n 'Employment (jobs) in information and communication (NACE Rev. 2, J)':'Jobs in information and communication',\n 'Employment (jobs) in mining, manufacturing, energy (NACE Rev. 2, B-E)':'Jobs in mining, manufacturing, energy',\n 'Employment (jobs) in professional, scientific and technical activities; administrative and support service activities (NACE Rev. 2, M and N)':'Jobs in professional, scientific and technical; administrative and support service',\n 'Employment (jobs) in public administration, defence, education, human health and social work activities (NACE Rev. 2, O to Q)':'Jobs in public administration, defence, education, human health and social work',\n 'Employment (jobs) in real estate activities (NACE Rev. 2, L)':'Jobs in real estate',\n 'Employment (jobs) in trade, transport, hotels, restaurants (NACE Rev. 2, G to I)':'Jobs in trade, transport, hotels, restaurants',\n 'Proportion of employment in industries (NACE Rev.1.1 C-E)':'Proportion of employment in industries'}, inplace=True)\n\n# Create target variable\ny = data_counts['biergarten_count_100k']",
"_____no_output_____"
],
[
"# Create correlation matrix\ncorr_matrix = X.corr().abs()\n\n# Select upper triangle of correlation matrix\nupper = corr_matrix.where(np.triu(np.ones(corr_matrix.shape), k=1).astype(np.bool))\n\n# Find index of feature columns with correlation greater than 0.7\nto_drop = [column for column in upper.columns if any(upper[column] > 0.7)]\n\n# Drop features \nX.drop(X[to_drop], axis=1, inplace=True)",
"_____no_output_____"
],
[
"# Pipeline for linear regression\nlin_reg_pipe = Pipeline([\n ('minmax', MinMaxScaler(feature_range=(-1,1))),\n ('lin_reg', LinearRegression(fit_intercept=True))])\n\n# Train the regression model\nlin_reg_pipe.fit(X,y)",
"_____no_output_____"
],
[
"# Plot regression coeffiecient\nplt.figure(figsize=(6,8)) \nbarplot_data = pd.concat([pd.Series(X.columns.to_list()), pd.Series(lin_reg_pipe['lin_reg'].coef_)], axis=1)\nbarplot_data.columns = ['variable', 'coef']\nax=sns.barplot(y='variable', x='coef', data=barplot_data, color=color)\nax.axes.set_ylabel('')\nax.axes.set_xlabel('Regression coefficient', fontdict={'fontsize':16})\nplt.show()\n\n# Print regression measures\nprint('Intercept: {}'.format(lin_reg_pipe['lin_reg'].intercept_))\nprint('R^2: {}'.format(lin_reg_pipe.score(X,y)))\n\n# Plot predictions and actuals\nslope, intercept, r_value, p_value, std_err = stats.linregress(lin_reg_pipe.predict(X),y)\nplt.figure(figsize=(10,10)) \ng = sns.regplot(lin_reg_pipe.predict(X), y, color=color\n , scatter_kws={'s':data_counts['total_population']/5000})\ng.axes.set_xlabel('Predicted number of biergartens per 100,000 people', fontdict={'fontsize':16})\ng.axes.set_ylabel('Actual number of biergartens per 100,000 people', fontdict={'fontsize':16})\ng.text(1.7, 5, r'$R^2:{0:.2f}$'.format(r_value**2), fontdict={'fontsize':14})\ng.set_xticks(np.arange(0.5,7,1))\ng.set_yticks(np.arange(0.5,7,1))\n\nfor line in range(0, data_counts.shape[0]):\n g.text(x=lin_reg_pipe.predict(X)[line]-0.1\n , y=y[line]\n , s=data_counts.city[line]\n , horizontalalignment='right'\n , verticalalignment='baseline'\n , size='small'\n , color='black')\n \nplt.show()",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
c51521c5204ecc57b706d6cd0d284b9884c8adaa
| 50,259 |
ipynb
|
Jupyter Notebook
|
Jupyter Notebooks/Bagging and Boosting - Machine Learning Ensemble Techniques.ipynb
|
akshaydnicator/ensemble-techniques
|
8686f925ccfe4cca5f91e5585904585d7164abb2
|
[
"Apache-2.0"
] | 1 |
2020-08-15T08:54:17.000Z
|
2020-08-15T08:54:17.000Z
|
Jupyter Notebooks/Bagging and Boosting - Machine Learning Ensemble Techniques.ipynb
|
akshaydnicator/ensemble-techniques-bagging-boosting-stacking
|
8686f925ccfe4cca5f91e5585904585d7164abb2
|
[
"Apache-2.0"
] | null | null | null |
Jupyter Notebooks/Bagging and Boosting - Machine Learning Ensemble Techniques.ipynb
|
akshaydnicator/ensemble-techniques-bagging-boosting-stacking
|
8686f925ccfe4cca5f91e5585904585d7164abb2
|
[
"Apache-2.0"
] | null | null | null | 32.509056 | 322 | 0.519947 |
[
[
[
"# Import required modules\n\nimport pandas as pd\nimport numpy as np\nfrom sklearn.model_selection import train_test_split, GridSearchCV\nfrom sklearn.ensemble import AdaBoostClassifier, GradientBoostingClassifier, BaggingClassifier, RandomForestClassifier, VotingClassifier\nfrom sklearn.tree import DecisionTreeClassifier\nfrom sklearn.svm import LinearSVC\nimport xgboost as xgb\nimport lightgbm as lgb\nfrom sklearn import metrics\nfrom catboost import CatBoostClassifier\nfrom sklearn.neighbors import KNeighborsClassifier\nfrom sklearn.linear_model import LogisticRegression",
"_____no_output_____"
],
[
"# Import original train set and Principal Components (PCs) obtained from PCA done in other notebook\ndf = pd.read_csv('train.csv')\npca_train = pd.read_csv('pca_train.csv')\npca_train.head()",
"_____no_output_____"
],
[
"# Convert the Categorical Y/N target variable 'Loan_Status' for binary 1/0 classification\ndf['Loan_Status'] = df['Loan_Status'].map(lambda x: 1 if x == 'Y' else 0)",
"_____no_output_____"
],
[
"# Set X and y for ML model training do train-test split using sklearn module\n\nX = pca_train.values\ny = df['Loan_Status']\n\nX_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.15, random_state=1)",
"_____no_output_____"
],
[
"y_test.shape",
"_____no_output_____"
],
[
"X_train.shape",
"_____no_output_____"
],
[
"# Initiate a new Adaptive Classifier, an ensemble boosting algorithm\nada = AdaBoostClassifier()\n\n# Create a dictionary of all values we want to test for selected model parameters of the respective algorithm\nparams_ada = {'n_estimators': np.arange(1, 10)}\n\n# Use GridSearchCV to test all values for selected model parameters\nada_gs = GridSearchCV(ada, params_ada, cv=10, verbose=1, n_jobs=-1, pre_dispatch='128*n_jobs')\n\n# Fit model to training data\nada_gs.fit(X_train, y_train)",
"Fitting 10 folds for each of 9 candidates, totalling 90 fits\n"
],
[
"# Save the best model\nada_best = ada_gs.best_estimator_\n\n# Check the value of the best selected model parameter(s)\nprint(ada_gs.best_params_)",
"{'n_estimators': 1}\n"
],
[
"# Print the accuracy score on the test data using best model\nprint('ada: {}'.format(ada_best.score(X_test, y_test)))",
"ada: 0.8064516129032258\n"
],
[
"# Initiate a new Gradient Boosting Classifier, an ensemble boosting algorithm\ngbc = GradientBoostingClassifier(learning_rate=0.005,warm_start=True)\n\n# Create a dictionary of all values we want to test for selected model parameters of the respective algorithm\nparams_gbc = {'n_estimators': np.arange(1, 200)}\n\n# Use GridSearchCV to test all values for selected model parameters\ngbc_gs = GridSearchCV(gbc, params_gbc, cv=10, verbose=1, n_jobs=-1, pre_dispatch='128*n_jobs')\n\n# Fit model to training data\ngbc_gs.fit(X_train, y_train)",
"Fitting 10 folds for each of 199 candidates, totalling 1990 fits\n"
],
[
"# Save the best model\ngbc_best = gbc_gs.best_estimator_\n\n# Check the value of the best selected model parameter(s)\nprint(gbc_gs.best_params_)",
"{'n_estimators': 176}\n"
],
[
"# Print the accuracy score on the test data using best model\nprint('gbc: {}'.format(gbc_best.score(X_test, y_test)))",
"gbc: 0.8064516129032258\n"
],
[
"# Initiate a new Bagging Classifier, an ensemble bagging algorithm \nbcdt = BaggingClassifier(DecisionTreeClassifier(random_state=1))\n\n# Create a dictionary of all values we want to test for selected model parameters of the respective algorithm\nparams_bcdt = {'n_estimators': np.arange(1, 100)}\n\n# Use GridSearchCV to test all values for selected model parameters\nbcdt_gs = GridSearchCV(bcdt, params_bcdt, cv=10, verbose=1, n_jobs=-1, pre_dispatch='128*n_jobs')\n\n# Fit model to training data\nbcdt_gs.fit(X_train, y_train)",
"Fitting 10 folds for each of 99 candidates, totalling 990 fits\n"
],
[
"# Save the best model\nbcdt_best = bcdt_gs.best_estimator_\n\n# Check the value of the best selected model parameter(s)\nprint(bcdt_gs.best_params_)",
"{'n_estimators': 15}\n"
],
[
"# Print the accuracy score on the test data using best model\nprint('bcdt: {}'.format(bcdt_best.score(X_test, y_test)))",
"bcdt: 0.7741935483870968\n"
],
[
"# Initiate a new Decision Tree Classifier and follow the similar process as mentioned in comments above\ndt = DecisionTreeClassifier(random_state=1)\nparams_dt = {}\ndt_gs = GridSearchCV(dt, params_dt, cv=10, verbose=1, n_jobs=-1, pre_dispatch='128*n_jobs')\ndt_gs.fit(X_train, y_train)",
"Fitting 10 folds for each of 1 candidates, totalling 10 fits\n"
],
[
"# Save the best model and check best model parameters\ndt_best = dt_gs.best_estimator_\nprint(dt_gs.best_params_)",
"{}\n"
],
[
"# Print the accuracy score on the test data using best model\nprint('dt: {}'.format(dt_best.score(X_test, y_test)))",
"dt: 0.7311827956989247\n"
],
[
"# Initiate a new Support Vector Classifier and follow the similar process as mentioned in comments above\nsvc = LinearSVC(random_state=1)\nparams_svc = {}\nsvc_gs = GridSearchCV(svc, params_svc, cv=10,verbose=1,n_jobs=-1,pre_dispatch='128*n_jobs')\nsvc_gs.fit(X_train, y_train)",
"Fitting 10 folds for each of 1 candidates, totalling 10 fits\n"
],
[
"# Save the best model and check best model parameters\nsvc_best = svc_gs.best_estimator_\nprint(svc_gs.best_params_)",
"{}\n"
],
[
"# Print the accuracy score on the test data using best model\nprint('svc: {}'.format(svc_best.score(X_test, y_test)))",
"svc: 0.7956989247311828\n"
],
[
"# Initiate a new XG Boost Classifier, an ensemble boosting algorithm and follow the similar process as mentioned in comments above\nxg = xgb.XGBClassifier(random_state=1,learning_rate=0.005)\nparams_xg = {'max_depth': np.arange(2,5), 'n_estimators': np.arange(1, 100)}\nxg_gs = GridSearchCV(xg, params_xg, cv=10, verbose=1, n_jobs=-1, pre_dispatch='128*n_jobs')\nxg_gs.fit(X_train, y_train)",
"Fitting 10 folds for each of 297 candidates, totalling 2970 fits\n"
],
[
"# Save the best model and check best model parameters\nxg_best = xg_gs.best_estimator_\nprint(xg_gs.best_params_)\n\n# Print the accuracy score on the test data using best model\nprint('xg: {}'.format(xg_best.score(X_test, y_test)))",
"{'max_depth': 2, 'n_estimators': 2}\nxg: 0.8064516129032258\n"
],
[
"# Initiate a new Light Gradient Boosted Machine, an ensemble boosting algorithm\n\n# Set the train data and initiate ML training\ntrain_data = lgb.Dataset(X_train,label=y_train)\nparams = {'learning_rate':0.01}\nlgbm = lgb.train(params, train_data, 100) \n\ny_pred = lgbm.predict(X_test)\n\nfor i in range(0,y_test.shape[0]):\n if y_pred[i]>=0.5:\n y_pred[i]=1\n else:\n y_pred[i]=0",
"_____no_output_____"
],
[
"# Print the overall accuracy\nprint(metrics.accuracy_score(y_test,y_pred))",
"0.8064516129032258\n"
],
[
"# Initiate a new Cat Boost Classifier, an ensemble boosting algorithm and fit on train data\ncbc = CatBoostClassifier(random_state=1, iterations=100)\ncbc.fit(X_train, y_train)",
"Learning rate set to 0.06442\n0:\tlearn: 0.6730757\ttotal: 57.5ms\tremaining: 5.69s\n1:\tlearn: 0.6557088\ttotal: 61.4ms\tremaining: 3.01s\n2:\tlearn: 0.6397677\ttotal: 65.3ms\tremaining: 2.11s\n3:\tlearn: 0.6248063\ttotal: 69.1ms\tremaining: 1.66s\n4:\tlearn: 0.6109242\ttotal: 73ms\tremaining: 1.39s\n5:\tlearn: 0.5993601\ttotal: 76.9ms\tremaining: 1.21s\n6:\tlearn: 0.5879616\ttotal: 80.8ms\tremaining: 1.07s\n7:\tlearn: 0.5767795\ttotal: 84.6ms\tremaining: 973ms\n8:\tlearn: 0.5673600\ttotal: 88.4ms\tremaining: 894ms\n9:\tlearn: 0.5582118\ttotal: 92.3ms\tremaining: 831ms\n10:\tlearn: 0.5509164\ttotal: 96ms\tremaining: 777ms\n11:\tlearn: 0.5433785\ttotal: 99.6ms\tremaining: 731ms\n12:\tlearn: 0.5365546\ttotal: 103ms\tremaining: 692ms\n13:\tlearn: 0.5301993\ttotal: 107ms\tremaining: 658ms\n14:\tlearn: 0.5234929\ttotal: 111ms\tremaining: 629ms\n15:\tlearn: 0.5179924\ttotal: 115ms\tremaining: 602ms\n16:\tlearn: 0.5125370\ttotal: 118ms\tremaining: 578ms\n17:\tlearn: 0.5078143\ttotal: 122ms\tremaining: 557ms\n18:\tlearn: 0.5035376\ttotal: 126ms\tremaining: 537ms\n19:\tlearn: 0.4988116\ttotal: 130ms\tremaining: 519ms\n20:\tlearn: 0.4950427\ttotal: 134ms\tremaining: 505ms\n21:\tlearn: 0.4916040\ttotal: 138ms\tremaining: 490ms\n22:\tlearn: 0.4883079\ttotal: 142ms\tremaining: 475ms\n23:\tlearn: 0.4847853\ttotal: 146ms\tremaining: 461ms\n24:\tlearn: 0.4809036\ttotal: 149ms\tremaining: 448ms\n25:\tlearn: 0.4781385\ttotal: 153ms\tremaining: 435ms\n26:\tlearn: 0.4755684\ttotal: 157ms\tremaining: 423ms\n27:\tlearn: 0.4727789\ttotal: 161ms\tremaining: 413ms\n28:\tlearn: 0.4697539\ttotal: 165ms\tremaining: 403ms\n29:\tlearn: 0.4671146\ttotal: 169ms\tremaining: 393ms\n30:\tlearn: 0.4652867\ttotal: 172ms\tremaining: 384ms\n31:\tlearn: 0.4634438\ttotal: 176ms\tremaining: 374ms\n32:\tlearn: 0.4618740\ttotal: 180ms\tremaining: 365ms\n33:\tlearn: 0.4599415\ttotal: 183ms\tremaining: 356ms\n34:\tlearn: 0.4580530\ttotal: 187ms\tremaining: 347ms\n35:\tlearn: 0.4563005\ttotal: 190ms\tremaining: 338ms\n36:\tlearn: 0.4549877\ttotal: 194ms\tremaining: 330ms\n37:\tlearn: 0.4532695\ttotal: 198ms\tremaining: 323ms\n38:\tlearn: 0.4517009\ttotal: 201ms\tremaining: 315ms\n39:\tlearn: 0.4504325\ttotal: 205ms\tremaining: 307ms\n40:\tlearn: 0.4487633\ttotal: 208ms\tremaining: 300ms\n41:\tlearn: 0.4474635\ttotal: 212ms\tremaining: 293ms\n42:\tlearn: 0.4461672\ttotal: 216ms\tremaining: 286ms\n43:\tlearn: 0.4445100\ttotal: 219ms\tremaining: 279ms\n44:\tlearn: 0.4436834\ttotal: 223ms\tremaining: 273ms\n45:\tlearn: 0.4426211\ttotal: 227ms\tremaining: 266ms\n46:\tlearn: 0.4415297\ttotal: 230ms\tremaining: 260ms\n47:\tlearn: 0.4400346\ttotal: 234ms\tremaining: 253ms\n48:\tlearn: 0.4389770\ttotal: 237ms\tremaining: 247ms\n49:\tlearn: 0.4382394\ttotal: 241ms\tremaining: 241ms\n50:\tlearn: 0.4368076\ttotal: 245ms\tremaining: 235ms\n51:\tlearn: 0.4358796\ttotal: 248ms\tremaining: 229ms\n52:\tlearn: 0.4350246\ttotal: 252ms\tremaining: 223ms\n53:\tlearn: 0.4339755\ttotal: 255ms\tremaining: 217ms\n54:\tlearn: 0.4330911\ttotal: 259ms\tremaining: 212ms\n55:\tlearn: 0.4324006\ttotal: 262ms\tremaining: 206ms\n56:\tlearn: 0.4314883\ttotal: 266ms\tremaining: 201ms\n57:\tlearn: 0.4308050\ttotal: 270ms\tremaining: 196ms\n58:\tlearn: 0.4298615\ttotal: 274ms\tremaining: 191ms\n59:\tlearn: 0.4291668\ttotal: 278ms\tremaining: 185ms\n60:\tlearn: 0.4282103\ttotal: 282ms\tremaining: 180ms\n61:\tlearn: 0.4275142\ttotal: 286ms\tremaining: 175ms\n62:\tlearn: 0.4268833\ttotal: 290ms\tremaining: 170ms\n63:\tlearn: 0.4262309\ttotal: 294ms\tremaining: 165ms\n64:\tlearn: 0.4251077\ttotal: 298ms\tremaining: 160ms\n65:\tlearn: 0.4242704\ttotal: 302ms\tremaining: 155ms\n66:\tlearn: 0.4238925\ttotal: 305ms\tremaining: 150ms\n67:\tlearn: 0.4230932\ttotal: 309ms\tremaining: 146ms\n68:\tlearn: 0.4222016\ttotal: 313ms\tremaining: 141ms\n69:\tlearn: 0.4217784\ttotal: 317ms\tremaining: 136ms\n70:\tlearn: 0.4209833\ttotal: 320ms\tremaining: 131ms\n71:\tlearn: 0.4201607\ttotal: 325ms\tremaining: 126ms\n72:\tlearn: 0.4196217\ttotal: 329ms\tremaining: 122ms\n73:\tlearn: 0.4189834\ttotal: 332ms\tremaining: 117ms\n74:\tlearn: 0.4180649\ttotal: 336ms\tremaining: 112ms\n75:\tlearn: 0.4174125\ttotal: 340ms\tremaining: 107ms\n76:\tlearn: 0.4165338\ttotal: 344ms\tremaining: 103ms\n77:\tlearn: 0.4161339\ttotal: 348ms\tremaining: 98.2ms\n78:\tlearn: 0.4157503\ttotal: 352ms\tremaining: 93.5ms\n79:\tlearn: 0.4150722\ttotal: 356ms\tremaining: 89ms\n80:\tlearn: 0.4148293\ttotal: 360ms\tremaining: 84.5ms\n81:\tlearn: 0.4139804\ttotal: 365ms\tremaining: 80.1ms\n82:\tlearn: 0.4134270\ttotal: 369ms\tremaining: 75.6ms\n83:\tlearn: 0.4127417\ttotal: 373ms\tremaining: 71.1ms\n84:\tlearn: 0.4121085\ttotal: 377ms\tremaining: 66.5ms\n85:\tlearn: 0.4114559\ttotal: 381ms\tremaining: 62ms\n86:\tlearn: 0.4107664\ttotal: 385ms\tremaining: 57.5ms\n87:\tlearn: 0.4103529\ttotal: 388ms\tremaining: 53ms\n88:\tlearn: 0.4098923\ttotal: 392ms\tremaining: 48.5ms\n89:\tlearn: 0.4091367\ttotal: 398ms\tremaining: 44.2ms\n90:\tlearn: 0.4083762\ttotal: 402ms\tremaining: 39.8ms\n91:\tlearn: 0.4078581\ttotal: 406ms\tremaining: 35.3ms\n92:\tlearn: 0.4073294\ttotal: 410ms\tremaining: 30.8ms\n93:\tlearn: 0.4065949\ttotal: 414ms\tremaining: 26.4ms\n94:\tlearn: 0.4060081\ttotal: 417ms\tremaining: 22ms\n95:\tlearn: 0.4055657\ttotal: 421ms\tremaining: 17.5ms\n96:\tlearn: 0.4051619\ttotal: 425ms\tremaining: 13.1ms\n97:\tlearn: 0.4049740\ttotal: 428ms\tremaining: 8.74ms\n98:\tlearn: 0.4043446\ttotal: 432ms\tremaining: 4.36ms\n99:\tlearn: 0.4035927\ttotal: 436ms\tremaining: 0us\n"
],
[
"# Print the overall accuracy\nprint('cbc: {}'.format(cbc.score(X_test, y_test)))",
"cbc: 0.8064516129032258\n"
],
[
"# Initiate a new KNeighbors Classifier and follow the similar process as mentioned in previous comments\nknn = KNeighborsClassifier()\nparams_knn = {'n_neighbors': np.arange(1, 25)}\nknn_gs = GridSearchCV(knn, params_knn, cv=10, verbose=1, n_jobs=-1, pre_dispatch='128*n_jobs')\nknn_gs.fit(X_train, y_train)",
"Fitting 10 folds for each of 24 candidates, totalling 240 fits\n"
],
[
"# Save the best model and check best model parameters\nknn_best = knn_gs.best_estimator_\nprint(knn_gs.best_params_)\n\n# Print the overall accuracy\nprint('knn: {}'.format(knn_best.score(X_test, y_test)))",
"{'n_neighbors': 15}\nknn: 0.7956989247311828\n"
],
[
"# Initiate a new Random Forest Classifier, an ensemble bagging algorithm and follow the similar process as mentioned in previous comments\nrf = RandomForestClassifier()\nparams_rf = {'n_estimators': [100, 150, 200, 250, 300, 350, 400, 450, 500]}\nrf_gs = GridSearchCV(rf, params_rf, cv=10, verbose=1, n_jobs=-1, pre_dispatch='128*n_jobs')\nrf_gs.fit(X_train, y_train)",
"Fitting 10 folds for each of 9 candidates, totalling 90 fits\n"
],
[
"# Save the best model and check best model parameters\nrf_best = rf_gs.best_estimator_\nprint(rf_gs.best_params_)\n\n# Print the overall accuracy\nprint('rf: {}'.format(rf_best.score(X_test, y_test)))",
"{'n_estimators': 100}\nrf: 0.7526881720430108\n"
],
[
"# Create a new Logistic Regression model and fit on train data\nlog_reg = LogisticRegression(solver='lbfgs')\nlog_reg.fit(X_train, y_train)",
"_____no_output_____"
],
[
"# Print the overall accuracy\nprint('log_reg: {}'.format(log_reg.score(X_test, y_test)))",
"log_reg: 0.7956989247311828\n"
],
[
"# Print the overall accuracy score for all the 11 best classification models trained earlier\nprint('Overall Accuracy of best selected models on X_test dataset\\n')\nprint('knn: {}'.format(knn_best.score(X_test, y_test)))\nprint('rf: {}'.format(rf_best.score(X_test, y_test)))\nprint('log_reg: {}'.format(log_reg.score(X_test, y_test)))\nprint('ada: {}'.format(ada_best.score(X_test, y_test)))\nprint('gbc: {}'.format(gbc_best.score(X_test, y_test)))\nprint('bcdt: {}'.format(bcdt_best.score(X_test, y_test)))\nprint('dt: {}'.format(dt_best.score(X_test, y_test)))\nprint('svc: {}'.format(svc_best.score(X_test, y_test)))\nprint('xg: {}'.format(xg_best.score(X_test, y_test)))\nprint('lgbm: {}'.format(metrics.accuracy_score(y_test,y_pred)))\nprint('cbc: {}'.format(cbc.score(X_test, y_test)))",
"Overall Accuracy of best selected models on X_test dataset\n\nknn: 0.7956989247311828\nrf: 0.7526881720430108\nlog_reg: 0.7956989247311828\nada: 0.8064516129032258\ngbc: 0.8064516129032258\nbcdt: 0.7741935483870968\ndt: 0.7311827956989247\nsvc: 0.7956989247311828\nxg: 0.8064516129032258\nlgbm: 0.8064516129032258\ncbc: 0.8064516129032258\n"
],
[
"# Create a dictionary of our models\nestimators=[('knn', knn_best), ('rf', rf_best), ('log_reg', log_reg), ('ada', ada_best), ('gbc', gbc_best), ('bcdt', bcdt_best), ('dt', dt_best), ('xg', xg_best), ('cbc', cbc)]\n\n# Create a voting classifier, input the dictionary of our models as estimators for the ensemble\nensemble = VotingClassifier(estimators, voting='soft', n_jobs=-1, flatten_transform=True, weights=[1/9,1/9,1/9,1/9,1/9,1/9,1/9,1/9,1/9])",
"_____no_output_____"
],
[
"# Fit the Final Ensemble Model on train data\nensemble.fit(X_train, y_train)\n\n# Test our final model on the test data and print our final accuracy score for the Ensemble made using Bagging and Boosting techniques\nensemble.score(X_test, y_test)",
"_____no_output_____"
],
[
"# Import the PCs of test data for final predictions\ndft = pd.read_csv('pca_test.csv')\ndft.head()",
"_____no_output_____"
],
[
"# Assign the PCs dft to test_X\ntest_X = dft.values\nprint(len(test_X))",
"367\n"
],
[
"# Make final predictions on the test data\ntest_predictions = ensemble.predict(test_X)\ntest_predictions",
"_____no_output_____"
],
[
"# Import original test file for Loan_IDs and assign the test_predictions to a new column 'Loan_Status'\ndft2 = pd.read_csv('test.csv')\ndft2['Loan_Status'] = test_predictions",
"_____no_output_____"
],
[
"# Drop unnecessary columns\ndft2 = dft2.drop(['Gender','Married','Dependents','Education','Self_Employed','ApplicantIncome','CoapplicantIncome','LoanAmount','Loan_Amount_Term','Credit_History','Property_Area'],axis=1)\ndft2.head()",
"_____no_output_____"
],
[
"# Convert binary 1/0 targets back to Categorical Y/N alphabets\ndft2['Loan_Status'] = dft2['Loan_Status'].map(lambda x: 'Y' if x == 1 else 'N')\ndft2.head()",
"_____no_output_____"
],
[
"# Save the predictions from the final Ensemble on local disk\ndft2.to_csv('Ensemble.csv', index=False)",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
c515250fcdbdc08b03871b7d352ac440b3514b17
| 5,958 |
ipynb
|
Jupyter Notebook
|
Sequence models & Attention mechanism.ipynb
|
reata/DeepLearning
|
7920096b3853f1e8a3a16632e3e042ad28618d22
|
[
"MIT"
] | null | null | null |
Sequence models & Attention mechanism.ipynb
|
reata/DeepLearning
|
7920096b3853f1e8a3a16632e3e042ad28618d22
|
[
"MIT"
] | null | null | null |
Sequence models & Attention mechanism.ipynb
|
reata/DeepLearning
|
7920096b3853f1e8a3a16632e3e042ad28618d22
|
[
"MIT"
] | 1 |
2018-10-08T15:37:07.000Z
|
2018-10-08T15:37:07.000Z
| 28.103774 | 213 | 0.596509 |
[
[
[
"# 序列模型和注意机制 Sequence Models & Attention Mechanism",
"_____no_output_____"
],
[
"## 1. 常见的序列到序列架构 Various sequence to sequence architectures",
"_____no_output_____"
],
[
"### 1.1 基本模型 Basic Models\n\n\n\n",
"_____no_output_____"
],
[
"### 1.2 挑选概率最高的句子 Picking the most likely sentence\n\n序列到序列的机器翻译模型,和语言模型,有一定相同的地方,它们都需要挑选概率最高的句子。但二者之间也存在一些显著的不同。\n\n序列到序列的机器翻译模型,可以看做条件概率下的语言模型:\n\n\n有了这个条件概率的语言模型,它可以生成不同的翻译,并且判定各个翻译的概率。但对于机器翻译问题,我们不需要随机生成的句子,而是需要找到概率最大的句子:\n\n\n使用贪心算法,每一步找到最大概率的下一个词,可能导致无法找到全局最大值;而穷举搜索是不现实的,计算量太大:\n",
"_____no_output_____"
],
[
"### 1.3 定向搜索 Beam Search\n\n定向搜索不一定保证能找到全局最优解,但通常能达到不错的效果。\n\n第一步:预先设定好**定向宽度 beam width**的参数B,计算encoder部分的值 $x$,根据 $x$ 就可以计算 $P(y^{<1>}|x)$ 的值。取条件概率最大的B个词,并保存其对应的条件概率。\n\n\n\n第二步:将上一步选出的B个词,分别作为 $y^{<1>}$,将词典中所有的词作为 $y^{<2>}$,计算 $P(y^{<1>},y^{<2>}|x) = P(y^{<1>}|x)P(y^{<2>}|x, y^{<1>})$。这样假设词典中有10000个词,就可以获得 10000×B 组 $y^{<1>},y^{<1>}$ 的组合。再一次从中挑选出概率最大的B个 $y^{<1>},y^{<1>}$ 组合。\n\n\n\n第三步:类似上一步。继续重复上面的步骤,知道句子结束。注意当 $B=1$ 时,定向搜索就是贪心搜索。\n\n",
"_____no_output_____"
],
[
"### 1.4 改进定向搜索 Refinements to Beam Search\n\n长度归一化:多个概率值相乘,可能导致堆栈下溢,丢失精度。转为求对数的最大值。然后求平均,从而对长度进行归一,否则之前的优化目标会对长句子有过多的惩罚。\n\n\n\n",
"_____no_output_____"
],
[
"### 1.5 定向搜索的误差分析 Error analysis in beam search\n\n误差分析,定向搜索出来的结果不好,应该归因为定向搜索,还是归因为RNN模型。\n\n\n\n\n\n从训练集中找出所有翻译错误,和人工翻译进行比较。判定定向搜索和RNN模型各自对造成最终误差的占比。如果是RNN,可以再进一步分析,是增加正则化,获取更多数据,还是更换神经网络的架构。\n\n\n",
"_____no_output_____"
],
[
"### 1.6 BLEU评分\n\n[BLEU: a Method for Automatic Evaluation of Machine Translation](http://www.aclweb.org/anthology/P02-1040)\n\n正确的翻译可能有很多种,要在多个正确的翻译之间取舍,就需要一个评分机制。\n\n\n\n",
"_____no_output_____"
],
[
"### 1.7 注意力模型的直观理解 Attention model intuition\n\n目前介绍的机器翻译模型,是编码解码架构,编码和解码分别训练两个不同的RNN,整个句子通过编码RNN输出一个预测值,然后根据这个预测值,解码RNN将其翻译出来。而实际上人类在进行翻译的时候,尤其对于长句子,不会整个看完之后再开始翻译。因为这种传统架构,随着句子变长,其Bleu评分会缓缓下降。\n\n\n\n[NEURAL MACHINE TRANSLATION BY JOINTLY LEARNING TO ALIGN AND TRANSLATE](https://arxiv.org/pdf/1409.0473.pdf)\n\n",
"_____no_output_____"
],
[
"### 1.8 注意力模型 Attention model\n\n\n\n\n\n\n",
"_____no_output_____"
],
[
"## 2. 语音数据 Audio Data",
"_____no_output_____"
],
[
"### 2.1 语音识别 Speech Recognition\n\n\n\n\n\n",
"_____no_output_____"
],
[
"### 2.2 触发字检测 Trigger Word Detection\n\n\n\n",
"_____no_output_____"
]
]
] |
[
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
c51533c9a878b3aba80e2417359cb5620ea7e38a
| 31,992 |
ipynb
|
Jupyter Notebook
|
DSE/RecommendationSystems/Movies.ipynb
|
f14-bertolotti/labs
|
f0068f71cac82a0042d1286f7f22a8b53a676462
|
[
"MIT"
] | 1 |
2022-03-16T12:56:58.000Z
|
2022-03-16T12:56:58.000Z
|
DSE/RecommendationSystems/Movies.ipynb
|
f14-bertolotti/labs
|
f0068f71cac82a0042d1286f7f22a8b53a676462
|
[
"MIT"
] | null | null | null |
DSE/RecommendationSystems/Movies.ipynb
|
f14-bertolotti/labs
|
f0068f71cac82a0042d1286f7f22a8b53a676462
|
[
"MIT"
] | null | null | null | 37.950178 | 517 | 0.545511 |
[
[
[
"# Predicting movie ratings\n\nOne of the most common uses of big data is to predict what users want. This allows Google to show you relevant ads, Amazon to recommend relevant products, and Netflix to recommend movies that you might like. This lab will demonstrate how we can use Apache Spark to recommend movies to a user. We will start with some basic techniques, and then use the mllib library's Alternating Least Squares method to make more sophisticated predictions.\n",
"_____no_output_____"
],
[
"## 1. Data Setup\n\nBefore starting with the recommendation systems, we need to download the dataset and we need to do a little bit of pre-processing. \n\n### 1.1 Download\nLet's begin with downloading the dataset. If you have already a copy of the dataset you can skip this part. For this lab, we will use [movielens 25M stable benchmark rating dataset](https://files.grouplens.org/datasets/movielens/ml-25m.zip). \n\n\n",
"_____no_output_____"
]
],
[
[
"# let's start by downloading the dataset.\nimport wget\nwget.download(url = \"https://files.grouplens.org/datasets/movielens/ml-25m.zip\", out = \"dataset.zip\")",
"100% [......................................................................] 261978986 / 261978986"
],
[
"# let's unzip the dataset\nimport zipfile\nwith zipfile.ZipFile(\"dataset.zip\", \"r\") as zfile:\n zfile.extractall()",
"_____no_output_____"
]
],
[
[
"### 1.2 Dataset Format\n\nThe following table highlights some data from `ratings.csv` (with comma-separated elements):\n\n| UserID | MovieID | Rating | Timestamp |\n|--------|---------|--------|------------|\n|...|...|...|...|\n|3022|152836|5.0|1461788770|\n|3023|169|5.0|1302559971|\n|3023|262|5.0|1302559918|\n|...|...|...|...|\n\nThe following table highlights some data from `movies.csv` (with comma-separated elements):\n\n| MovieID | Title | Genres | \n|---------|---------|--------|\n|...|...|...|\n| 209133 |The Riot and the Dance (2018) | (no genres listed) |\n| 209135 |Jane B. by Agnès V. (1988) | Documentary\\|Fantasy |\n|...|...|...|\n\nThe `Genres` field has the format\n\n`Genres1|Genres2|Genres3|...` or `(no generes listed)`\n\nThe format of these files is uniform and simple, so we can easily parse them using python:\n- For each line in the rating dataset, we create a tuple of (UserID, MovieID, Rating). We drop\nthe timestamp because we do not need it for this exercise.\n- For each line in the movies dataset, we create a tuple of (MovieID, Title). We drop the Genres\nbecause we do not need them for this exercise.\n\n### 1.3 Preprocessing\n\nWe can begin to preprocess our data. This step includes: \n1) We should drop the timestamp, we do not need it.\n2) We should drop the genres, we do not need them.\n3) We should parse data according to their intended type. For example, the elements of rating should be floats.\n4) Each line should encode data in an easily processable format, like a tuple. \n5) We should filter the first line of both datasets (the header).",
"_____no_output_____"
]
],
[
[
"# let's intialize the spark session\n\nimport pyspark\nfrom pyspark.sql import SparkSession\n\nspark = SparkSession.builder \\\n .appName(\"Python Spark SQL basic example\") \\\n .getOrCreate()\nspark",
"WARNING: An illegal reflective access operation has occurred\nWARNING: Illegal reflective access by org.apache.spark.unsafe.Platform (file:/home/f14/Devel/labs/DSE/RecommendationSystems/.venv-RS/lib/python3.10/site-packages/pyspark/jars/spark-unsafe_2.12-3.2.1.jar) to constructor java.nio.DirectByteBuffer(long,int)\nWARNING: Please consider reporting this to the maintainers of org.apache.spark.unsafe.Platform\nWARNING: Use --illegal-access=warn to enable warnings of further illegal reflective access operations\nWARNING: All illegal access operations will be denied in a future release\nUsing Spark's default log4j profile: org/apache/spark/log4j-defaults.properties\nSetting default log level to \"WARN\".\nTo adjust logging level use sc.setLogLevel(newLevel). For SparkR, use setLogLevel(newLevel).\n22/02/22 09:40:41 WARN NativeCodeLoader: Unable to load native-hadoop library for your platform... using builtin-java classes where applicable\n"
]
],
[
[
"#### 1.3.1 Load The Data\n\nWe can start by loading the dataset formatted as raw text. ",
"_____no_output_____"
]
],
[
[
"from pprint import pprint\nratings_rdd = spark.sparkContext.textFile(name = \"ml-25m/ratings.csv\", minPartitions = 2)\nmovies_rdd = spark.sparkContext.textFile(name = \"ml-25m/movies.csv\" , minPartitions = 2)\n\n# let's have a peek a our dataset\nprint(\"ratings --->\")\npprint(ratings_rdd.take(5))\n\nprint(\"\\nmovies --->\")\npprint(movies_rdd.take(5))",
"ratings --->\n"
]
],
[
[
"#### 1.3.2 SubSampling\nSince we have limited resources in terms of computation, sometimes, it is useful to work with only a fraction of the whole dataset.",
"_____no_output_____"
]
],
[
[
"ratings_rdd = ratings_rdd.sample(withReplacement=False, fraction=1/25, seed=14).cache()\nmovies_rdd = movies_rdd .sample(withReplacement=False, fraction=1, seed=14).cache()\n\nprint(f\"ratings_rdd: {ratings_rdd.count()}, movies_rdd {movies_rdd.count()}\")",
" \r"
]
],
[
[
"#### 1.3.2 Parsing\nHere, we do the real preprocessing: dropping columns, parsing elements, and filtering the heading.",
"_____no_output_____"
]
],
[
[
"def string2rating(line):\n \"\"\" Parse a line in the ratings dataset.\n Args:\n line (str): a line in the ratings dataset in the form of UserID,MovieID,Rating,Timestamp\n Returns:\n tuple[int,int,float]: (UserID, MovieID, Rating)\n \"\"\"\n userID, movieID, rating, *others = line.split(\",\")\n try: return int(userID), int(movieID), float(rating),\n except ValueError: return None\n\ndef string2movie(line):\n \"\"\" Parse a line in the movies dataset.\n Args:\n line (str): a line in the movies dataset in the form of MovieID,Title,Genres. \n Genres in the form of Genre1|Genre2|...\n Returns:\n tuple[int,str,list[str]]: (MovieID, Title, Genres)\n \"\"\"\n movieID, title, *others = line.split(\",\")\n try: return int(movieID), title\n except ValueError: return None\n\nratings_rdd = ratings_rdd.map(string2rating).filter(lambda x:x!=None).cache()\nmovies_rdd = movies_rdd .map(string2movie ).filter(lambda x:x!=None).cache()",
"_____no_output_____"
],
[
"print(f\"There are {ratings_rdd.count()} ratings and {movies_rdd.count()} movies in the datasets\")\nprint(f\"Ratings: ---> \\n{ratings_rdd.take(3)}\")\nprint(f\"Movies: ---> \\n{movies_rdd.take(3)}\")",
" \r"
]
],
[
[
"## 2. Basic Raccomandations\n\n\n### 2.1 Highest Average Rating.\n\nOne way to recommend movies is to always recommend the movies with the highest average rating. In this section, we will use Spark to find the name, number of ratings, and the average rating of the 20 movies with the highest average rating and more than 500 reviews. We want to filter our movies with high ratings but fewer than or equal to 500 reviews because movies with few reviews may not have broad appeal to everyone.",
"_____no_output_____"
]
],
[
[
"def averageRating(ratings):\n \"\"\" Computes the average rating.\n Args:\n tuple[int, list[float]]: a MovieID with its list of ratings\n Returns:\n tuple[int, float]: returns the the MovieID with its average rating.\n \"\"\" \n return (ratings[0], sum(ratings[1]) / len(ratings[1]))\n\nrdd = ratings_rdd.map(lambda x:(x[0], x[2])).groupByKey() # group by MovieID\nrdd = rdd.filter(lambda x:len(x[1])>500) # filter movies with less than 500 reviews \nrdd = rdd.map(averageRating) # computes the average Rating\nrdd = rdd.sortBy(lambda x:x[1], ascending=False)\n\nrdd.take(5)",
" \r"
]
],
[
[
"Ok, now we have the best (according to the average) popular (according to the number of reviews) movies. However, we can only see their MovieID. Let's convert the IDs into titles.",
"_____no_output_____"
]
],
[
[
"rdd.join(movies_rdd)\\\n .map(lambda x:(x[1][1],x[1][0]))\\\n .sortBy(lambda x:x[1], ascending=False)\\\n .take(20)",
" \r"
]
],
[
[
"### 2.2 Collaborative Filtering\n\nWe are going to use a technique called collaborative filtering. Collaborative filtering is a method of making automatic predictions (filtering) about the interests of a user by collecting preferences or taste information from many users (collaborating). The underlying assumption of the collaborative filtering approach is that if a person A has the same opinion as a person B on an issue, A is more likely to have B's opinion on a different issue x than to have the opinion on x of a person chosen randomly.\n\nAt first, people rate different items (like videos, images, games). After that, the system is making predictions about a user's rating for an item, which the user has not rated yet. These predictions are built upon the existing ratings of other users, who have similar ratings with the active user.\n\n#### 2.2.1 Creating a Training Set\n\nBefore we jump into using machine learning, we need to break up the `ratings_rdd` dataset into three pieces:\n\n* a training set (RDD), which we will use to train models,\n* a validation set (RDD), which we will use to choose the best model,\n* a test set (RDD), which we will use for estimating the predictive power of the recommender system.\n\nTo randomly split the dataset into multiple groups, we can use the pyspark [randomSplit] transformation, which takes a list of splits with a seed and returns multiple RDDs.\n\n[randomSplit]:https://spark.apache.org/docs/3.1.1/api/python/reference/api/pyspark.RDD.randomSplit.html?highlight=randomsplit#pyspark.RDD.randomSplit",
"_____no_output_____"
]
],
[
[
"training_rdd, validation_rdd, test_rdd = ratings_rdd.randomSplit([6, 2, 2], seed=14)\n\nprint(f\"Training: {training_rdd.count()}, validation: {validation_rdd.count()}, test: {test_rdd .count()}\")\n\nprint(\"training samples: \", training_rdd .take(3))\nprint(\"validation samples: \", validation_rdd.take(3))\nprint(\"test samples: \", test_rdd .take(3))",
" \r"
]
],
[
[
"#### 2.2.2 Alternating Least Square Errors\n\nFor movie recommendations, we start with a matrix whose entries are movie ratings by users. Each column represents a user and each row represents a particular movie.\n\nSince not all users have rated all movies, we do not know all of the entries in this matrix, which is precisely why we need collaborative filtering. For each user, we have ratings for only a subset of the movies. With collaborative filtering, the idea is to approximate the rating matrix by factorizing it as the product of two matrices: one that describes properties of each user, and one that describes properties of each movie.\n\nWe want to select these two matrices such that the error for the users/movie pairs where we know the correct ratings is minimized. The *Alternating Least Squares* algorithm does this by first randomly filling the user matrix with values and then optimizing the value of the movies such that the error is minimized. Then, it holds the movies matrix constant and optimizes the value of the user's matrix. This alternation between which matrix to optimize is the reason for the \"alternating\" in the name.\n",
"_____no_output_____"
]
],
[
[
"from pyspark.mllib.recommendation import ALS\n\n# thanks to modern libraries training an ALS model is as easy as\nmodel = ALS.train(training_rdd, rank = 4, seed = 14, iterations = 5, lambda_ = 0.1)\n\n# let's have a peek to few predictions\nmodel.predictAll(validation_rdd.map(lambda x:(x[0],x[1]))).take(5)",
"22/02/22 09:41:26 WARN InstanceBuilder$NativeBLAS: Failed to load implementation from:dev.ludovic.netlib.blas.JNIBLAS\n22/02/22 09:41:26 WARN InstanceBuilder$NativeBLAS: Failed to load implementation from:dev.ludovic.netlib.blas.ForeignLinkerBLAS\n22/02/22 09:41:27 WARN InstanceBuilder$NativeLAPACK: Failed to load implementation from:dev.ludovic.netlib.lapack.JNILAPACK\n \r"
]
],
[
[
"#### 2.2.3 Root Mean Square Error (RMSE)\n\nNext, we need to evaluate our model: is it good or is it bad?\n\nTo score the model, we will use RMSE (often called also Root Mean Square Deviation (RMSD)). You can think of RMSE as a distance function that measures the distance between the predictions and the ground truths. It is computed as follows: \n\n$$ RMSE(f, \\mathcal{D}) = \\sqrt{\\frac{\\sum_{(x,y) \\in \\mathcal{D}} (f(x) - y)^2}{|\\mathcal{D}|}}$$\n\nWhere:\n* $\\mathcal{D}$ is our dataset it contains samples alongside their predictions. Formally, $\\mathcal{D} \\subseteq \\mathcal{X} \\times \\mathcal{Y}$. Where:\n * $\\mathcal{X}$ is the set of all input samples.\n * $\\mathcal{Y}$ is the set of all possible predictions. \n* $f : \\mathcal{X} \\rightarrow \\mathcal{Y}$ is the model we wish to evaluate. Given an input $x$ (from $\\mathcal{X}$, the set of possible inputs) it returns a value $f(x)$ (from $\\mathcal{Y}$, the set of possible outputs). \n* $x$ represents an input. \n* $f(x)$ represents the prediction of $x$.\n* $y$ represents the ground truth.\n\nAs you can imagine $f(x)$ and $y$ can be different, i.e our model is wrong. With $RMSE(f, \\mathcal{D})$, we want to measure the degree to which our model, $f$, is wrong on the dataset $\\mathcal{D}$. The higher is $RMSE(f, \\mathcal{D})$ the higher is the degree to which $f$ is wrong. The smaller is $RMSE(f, \\mathcal{D})$ the more accurate $f$ is. \n\nTo better understand the RMSE consider the following facts:\n* When $f(x)$ is close to $y$ our model is accurate. In the same case $(f(x) - y)^2$ is small.\n* When $f(x)$ is far from $y$ our model is inaccurate. In the same case $(f(x) - y)^2$ is high.\n* If our model is accurate, it will be often accurate in $\\mathcal{D}$. Therefore, it will make often small errors which will amount to a small RMSE. \n* If our model is inaccurate, it will be often inaccurate in $\\mathcal{D}$. Therefore, it will make often big errors which will amount to a large RMSE.\n\nLet's make a function to compute the RMSE so that we can use it multiple times easily.",
"_____no_output_____"
]
],
[
[
"\ndef RMSE(predictions_rdd, truths_rdd):\n \"\"\" Compute the root mean squared error between predicted and actual\n Args:\n predictions_rdd: predicted ratings for each movie and each user where each entry is in the form (UserID, MovieID, Rating).\n truths_rdd: actual ratings where each entry is in the form (UserID, MovieID, Rating).\n Returns:\n RSME (float): computed RSME value\n \"\"\"\n # Transform predictions and truths into the tuples of the form ((UserID, MovieID), Rating)\n predictions = predictions_rdd.map(lambda i: ((i[0], i[1]), i[2]))\n truths = truths_rdd .map(lambda i: ((i[0], i[1]), i[2]))\n\n # Compute the squared error for each matching entry (i.e., the same (User ID, Movie ID) in each\n # RDD) in the reformatted RDDs using RDD transformtions - do not use collect()\n squared_errors = predictions.join(truths)\\\n .map(lambda i: (i[1][0] - i[1][1])**2)\n \n\n total_squared_error = squared_errors.sum()\n total_ratings = squared_errors.count()\n mean_squared_error = total_squared_error / total_ratings\n root_mean_squared_error = mean_squared_error ** (1/2)\n \n return root_mean_squared_error\n",
"_____no_output_____"
],
[
"# let's evaluate the trained models\n\nRMSE(predictions_rdd = model.predictAll(validation_rdd.map(lambda x:(x[0],x[1]))),\n truths_rdd = validation_rdd)\n",
" \r"
]
],
[
[
"#### 2.2.4 HyperParameters Tuning\n\nCan we do better? \n\nWhen training the ALS model there were few parameters to set. However, we do not know which is the best configuration. On these occasions, we want to try a few combinations to obtain even better results. In this section, we will search a few parameters. We will perform a so-called **grid search**. We will proceed as follows:\n\n1) We decide the parameters to tune.\n2) We train with all possible configurations. \n3) We evaluate a trained model with all possible configurations on the validation set.\n4) We evaluate the best model on the test set.",
"_____no_output_____"
]
],
[
[
"\nHyperParameters = {\n \"rank\" : [4, 8, 12],\n \"seed\" : [14],\n \"iterations\" : [5, 10], \n \"lambda\" : [0.05, 0.1, 0.25]\n}\n\nbest_model = None\nbest_error = float(\"inf\")\nbest_conf = dict()\n\n# how many training are we doing ?\nfor rank in HyperParameters[\"rank\"]: #\n for seed in HyperParameters[\"seed\"]: # I consider these nested for-loops an anti-pattern.\n for iterations in HyperParameters[\"iterations\"]: # However, We can leave as it is for sake of simplicity. \n for lambda_ in HyperParameters[\"lambda\"]: # \n \n model = ALS.train(training_rdd, rank = rank, seed = seed, iterations = iterations, lambda_ = lambda_)\n validation_error = RMSE(predictions_rdd = model.predictAll(validation_rdd.map(lambda x:(x[0],x[1]))),\n truths_rdd = validation_rdd)\n \n if validation_error < best_error: \n best_model, best_error = model, validation_error\n best_conf = {\"rank\":rank, \"seed\":seed, \"iterations\":iterations, \"lambda\":lambda_}\n print(f\"current best validation error {best_error} with configuration {best_conf}\")\n\ntest_error = RMSE(predictions_rdd = model.predictAll(test_rdd.map(lambda x:(x[0],x[1]))), truths_rdd = test_rdd)\nprint(f\"test error {test_error} with configuration {best_conf}\")",
" \r"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
c515375050e21eeb2fd8ae853ec0415a4f043fe1
| 46,842 |
ipynb
|
Jupyter Notebook
|
gen_train_data.ipynb
|
fromjupiter/ReverbNN
|
19c078e0d37c4f6bf85e749a31c40f36f3562145
|
[
"MIT"
] | null | null | null |
gen_train_data.ipynb
|
fromjupiter/ReverbNN
|
19c078e0d37c4f6bf85e749a31c40f36f3562145
|
[
"MIT"
] | null | null | null |
gen_train_data.ipynb
|
fromjupiter/ReverbNN
|
19c078e0d37c4f6bf85e749a31c40f36f3562145
|
[
"MIT"
] | null | null | null | 264.644068 | 17,000 | 0.88284 |
[
[
[
"import librosa\nimport librosa.display\nimport matplotlib.pyplot as plt\nfrom scipy import signal\nimport soundfile\nimport numpy as np\nimport os\n\nSR = 44100\n\n# Update the path to your IR here\nir_verb, sr = librosa.load('./dataset/gill-heads-mine/b-format/mine_site1_1way_bformat.wav',sr=SR)\nir_verb = librosa.resample(ir_verb, sr, 44100)\nir_verb = np.multiply(ir_verb, 1.0/np.max(ir_verb))\n\n\nfor root, dirs, files in os.walk(\"./dataset/guitarset\"):\n for name in files:\n audio, sr = librosa.load(os.path.join(root,name),sr=SR)\n filtered = signal.convolve(audio, ir_verb, method='fft')\n soundfile.write(os.path.join('./dataset/mine_guitarset/', name), filtered, SR)\n print(\"Generated file for {}\".format(os.path.join(root, name)))",
"Generated file for ./dataset/guitarset/00_Rock3-117-Bb_solo_mix.wav\nGenerated file for ./dataset/guitarset/00_Rock1-90-C#_solo_mix.wav\nGenerated file for ./dataset/guitarset/00_Jazz2-187-F#_solo_mix.wav\nGenerated file for ./dataset/guitarset/00_Funk1-97-C_solo_mix.wav\nGenerated file for ./dataset/guitarset/00_Rock3-148-C_solo_mix.wav\nGenerated file for ./dataset/guitarset/00_Jazz3-150-C_solo_mix.wav\nGenerated file for ./dataset/guitarset/00_Funk2-119-G_solo_mix.wav\nGenerated file for ./dataset/guitarset/00_BN1-129-Eb_solo_mix.wav\nGenerated file for ./dataset/guitarset/00_Funk2-108-Eb_solo_mix.wav\nGenerated file for ./dataset/guitarset/00_Funk3-98-A_solo_mix.wav\nGenerated file for ./dataset/guitarset/00_BN1-147-Gb_solo_mix.wav\nGenerated file for ./dataset/guitarset/00_Rock2-142-D_solo_mix.wav\nGenerated file for ./dataset/guitarset/00_Rock1-130-A_solo_mix.wav\nGenerated file for ./dataset/guitarset/00_BN3-119-G_solo_mix.wav\nGenerated file for ./dataset/guitarset/00_Funk1-114-Ab_solo_mix.wav\nGenerated file for ./dataset/guitarset/00_Rock2-85-F_solo_mix.wav\nGenerated file for ./dataset/guitarset/00_Funk3-112-C#_solo_mix.wav\nGenerated file for ./dataset/guitarset/00_BN2-166-Ab_solo_mix.wav\nGenerated file for ./dataset/guitarset/00_BN3-154-E_solo_mix.wav\nGenerated file for ./dataset/guitarset/00_Jazz1-200-B_solo_mix.wav\nGenerated file for ./dataset/guitarset/00_Jazz3-137-Eb_solo_mix.wav\nGenerated file for ./dataset/guitarset/00_Jazz1-130-D_solo_mix.wav\n"
],
[
"verb = y#[:96000]\naudio, sr = librosa.load('./data/guitarset/00_BN1-129-Eb_solo_mix.wav',sr=None)\nprint(sr)\nfiltered = signal.convolve(audio, ir_filter, method='fft')\n\nsoundfile.write('./filtered_guitar.wav', filtered, SR)",
"44100\n"
],
[
"librosa.display.waveplot(filtered, sr=sr)\nplt.show()",
"_____no_output_____"
],
[
"\nlibrosa.display.waveplot(audio, sr=sr)\nplt.show()",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code"
]
] |
c51543e0e4bfabcaa081699416db90c362a0386f
| 912,566 |
ipynb
|
Jupyter Notebook
|
demo/tables.ipynb
|
jo-mueller/napari-skimage-regionprops
|
648ce9c9e7ea1670cd5adb842d486982278ff89b
|
[
"BSD-3-Clause"
] | null | null | null |
demo/tables.ipynb
|
jo-mueller/napari-skimage-regionprops
|
648ce9c9e7ea1670cd5adb842d486982278ff89b
|
[
"BSD-3-Clause"
] | null | null | null |
demo/tables.ipynb
|
jo-mueller/napari-skimage-regionprops
|
648ce9c9e7ea1670cd5adb842d486982278ff89b
|
[
"BSD-3-Clause"
] | null | null | null | 1,554.626917 | 60,411 | 0.956633 |
[
[
[
"# Table widgets in the napari viewer\nBefore we talk about tables and widgets in napari, let's create a viewer, a simple test image and a labels layer:",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport napari\nimport pandas\nfrom napari_skimage_regionprops import regionprops_table, add_table, get_table",
"_____no_output_____"
],
[
"viewer = napari.Viewer()",
"c:\\structure\\code\\napari-tools-menu\\napari_tools_menu\\__init__.py:168: FutureWarning: Public access to Window.qt_viewer is deprecated and will be removed in\nv0.5.0. It is considered an \"implementation detail\" of the napari\napplication, not part of the napari viewer model. If your use case\nrequires access to qt_viewer, please open an issue to discuss.\n self.tools_menu = ToolsMenu(self, self.qt_viewer.viewer)\n"
],
[
"viewer.add_image(np.asarray([[1,2],[2,2]]))\nviewer.add_labels(np.asarray([[1,2],[3,3]]))",
"_____no_output_____"
]
],
[
[
"Now, let's perform a measurement of `size` and `intensity` of the labeled objects in the given image. A table with results will be automatically added to the viewer",
"_____no_output_____"
]
],
[
[
"regionprops_table(\n viewer.layers[0].data,\n viewer.layers[1].data,\n viewer,\n size=True,\n intensity=True\n)",
"_____no_output_____"
],
[
"napari.utils.nbscreenshot(viewer)",
"_____no_output_____"
]
],
[
[
"We can also get the widget representing the table:",
"_____no_output_____"
]
],
[
[
"# The table is associated with a given labels layer:\nlabels = viewer.layers[1]\n\ntable = get_table(labels, viewer)\ntable",
"_____no_output_____"
]
],
[
[
"You can also read the content from the table as a dictionary. It is recommended to convert it into a pandas `DataFrame`:",
"_____no_output_____"
]
],
[
[
"content = pandas.DataFrame(table.get_content())\ncontent",
"_____no_output_____"
]
],
[
[
"The content of this table can be changed programmatically. This also changes the `properties` of the associated layer.",
"_____no_output_____"
]
],
[
[
"new_values = {'A': [1, 2, 3],\n 'B': [4, 5, 6]\n }\ntable.set_content(new_values)\nnapari.utils.nbscreenshot(viewer)",
"_____no_output_____"
]
],
[
[
"You can also append data to an existing table through the `append_content()` function: Suppose you have another measurement for the labels in your image, i.e. the \"double area\":",
"_____no_output_____"
]
],
[
[
"table.set_content(content.to_dict('list'))\ndouble_area = {'label': content['label'].to_numpy(),\n 'Double area': content['area'].to_numpy() * 2.0}",
"_____no_output_____"
]
],
[
[
"You can now append this as a new column to the existing table:",
"_____no_output_____"
]
],
[
[
"table.append_content(double_area)\nnapari.utils.nbscreenshot(viewer)",
"_____no_output_____"
]
],
[
[
"*Note*: If the added data has columns in common withh the exisiting table (for instance, the labels columns), the tables will be merged on the commonly available columns. If no common columns exist, the data will simply be added to the table and the non-intersecting row/columns will be filled with NaN:",
"_____no_output_____"
]
],
[
[
"tripple_area = {'Tripple area': content['area'].to_numpy() * 3.0}\ntable.append_content(tripple_area)\nnapari.utils.nbscreenshot(viewer)",
"_____no_output_____"
]
],
[
[
"Note: Changing the label's `properties` does not invoke changes of the table...",
"_____no_output_____"
]
],
[
[
"new_values = {'C': [6, 7, 8],\n 'D': [9, 10, 11]\n }\n\nlabels.properties = new_values\nnapari.utils.nbscreenshot(viewer)",
"_____no_output_____"
]
],
[
[
"But you can refresh the content:",
"_____no_output_____"
]
],
[
[
"table.update_content()\nnapari.utils.nbscreenshot(viewer)",
"_____no_output_____"
]
],
[
[
"You can remove the table from the viewer like this:",
"_____no_output_____"
]
],
[
[
"viewer.window.remove_dock_widget(table)\nnapari.utils.nbscreenshot(viewer)",
"_____no_output_____"
]
],
[
[
"Afterwards, the `get_table` method will return None:",
"_____no_output_____"
]
],
[
[
"get_table(labels, viewer)",
"_____no_output_____"
]
],
[
[
"To add the table again, just call `add_table` again. Note that the content of the properties of the labels have not been changed.",
"_____no_output_____"
]
],
[
[
"add_table(labels, viewer)\nnapari.utils.nbscreenshot(viewer)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
c515452d4c2a5feb0e61f6e37ae453c973d780de
| 74,214 |
ipynb
|
Jupyter Notebook
|
stract/stract/.ipynb_checkpoints/stract_nb-checkpoint.ipynb
|
crossminer/maracas
|
662a44542065a896195e0ae719624550ae4dd39d
|
[
"MIT"
] | 6 |
2020-06-13T19:46:29.000Z
|
2021-12-13T13:17:13.000Z
|
stract/stract/.ipynb_checkpoints/stract_nb-checkpoint.ipynb
|
crossminer/maracas
|
662a44542065a896195e0ae719624550ae4dd39d
|
[
"MIT"
] | 29 |
2019-08-19T13:50:19.000Z
|
2021-01-26T16:01:44.000Z
|
stract/stract/.ipynb_checkpoints/stract_nb-checkpoint.ipynb
|
crossminer/maracas
|
662a44542065a896195e0ae719624550ae4dd39d
|
[
"MIT"
] | null | null | null | 163.107692 | 17,096 | 0.874256 |
[
[
[
"# Getting started",
"_____no_output_____"
]
],
[
[
"pip install numpy pandas matplotlib seaborn",
"Requirement already satisfied: numpy in /usr/local/lib/python3.7/site-packages (1.16.4)\nRequirement already satisfied: pandas in /usr/local/lib/python3.7/site-packages (0.25.0)\nRequirement already satisfied: matplotlib in /usr/local/lib/python3.7/site-packages (3.1.1)\nRequirement already satisfied: seaborn in /usr/local/lib/python3.7/site-packages (0.9.0)\nRequirement already satisfied: pytz>=2017.2 in /usr/local/lib/python3.7/site-packages (from pandas) (2019.1)\nRequirement already satisfied: python-dateutil>=2.6.1 in /Users/ochoa/Library/Python/3.7/lib/python/site-packages (from pandas) (2.8.0)\nRequirement already satisfied: cycler>=0.10 in /usr/local/lib/python3.7/site-packages (from matplotlib) (0.10.0)\nRequirement already satisfied: pyparsing!=2.0.4,!=2.1.2,!=2.1.6,>=2.0.1 in /usr/local/lib/python3.7/site-packages (from matplotlib) (2.4.1.1)\nRequirement already satisfied: kiwisolver>=1.0.1 in /usr/local/lib/python3.7/site-packages (from matplotlib) (1.1.0)\nRequirement already satisfied: scipy>=0.14.0 in /usr/local/lib/python3.7/site-packages (from seaborn) (1.3.0)\nRequirement already satisfied: six>=1.5 in /Users/ochoa/Library/Python/3.7/lib/python/site-packages (from python-dateutil>=2.6.1->pandas) (1.12.0)\nRequirement already satisfied: setuptools in /usr/local/lib/python3.7/site-packages (from kiwisolver>=1.0.1->matplotlib) (40.8.0)\nNote: you may need to restart the kernel to use updated packages.\n"
],
[
"import numpy as np\nimport pandas as pd\nimport seaborn as sb\nimport matplotlib.pyplot as mplot",
"_____no_output_____"
],
[
"# Load Quaatlas matrix and create dataframe\nquaatlas = pd.read_csv('./../data/quaatlas/matrix.csv')",
"_____no_output_____"
],
[
"# Let's check first rows of the dataframe\nquaatlas.head()",
"_____no_output_____"
]
],
[
[
"# Visualizations",
"_____no_output_____"
]
],
[
[
"# Set size of the figure\nmplot.figure(figsize=(9,5), dpi=100)\n\n# Set title and labels\nmplot.title('Declarations per Project')\nmplot.xlabel('Number of declarations')\nmplot.ylabel('Number of projects')\n\n# Create histogram\nmplot.hist(quaatlas['declarations'], rwidth=0.9, bins=77)\nmplot.show()",
"_____no_output_____"
],
[
"sb.distplot(quaatlas['declarations'], bins=60, kde=False, rug=True);",
"_____no_output_____"
],
[
"sb.distplot(quaatlas['types'], bins=60, kde=False, rug=True);",
"_____no_output_____"
],
[
"sb.distplot(quaatlas['methods'], bins=60, kde=False, rug=True);",
"_____no_output_____"
],
[
"sb.distplot(quaatlas['fields'], bins=60, kde=False, rug=True);",
"_____no_output_____"
],
[
"sb.relplot(x=\"declarations\", y=\"classes\", data=quaatlas)",
"_____no_output_____"
],
[
"fig, axs = mplot.subplots(ncols=2)\nsb.distplot(quaatlas['declarations'], bins=60, kde=False, rug=True, ax=axs[0][0]);\nsb.distplot(quaatlas['types'], bins=60, kde=False, rug=True, ax=axs[1][0]);\nsb.distplot(quaatlas['methods'], bins=60, kde=False, rug=True, ax=axs[0][1]);\nsb.distplot(quaatlas['fields'], bins=60, kde=False, rug=True, ax=axs[1][1]);",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
c5154d62cf38a1936e94cca7e0dbbf8e169347a0
| 297,041 |
ipynb
|
Jupyter Notebook
|
test_fftanalysis.ipynb
|
gmweir/PYFFT
|
e2404fb99871bb81f9fac9829965e9a8ed3cc60e
|
[
"MIT"
] | 1 |
2022-03-19T13:30:03.000Z
|
2022-03-19T13:30:03.000Z
|
test_fftanalysis.ipynb
|
gmweir/PYFFT
|
e2404fb99871bb81f9fac9829965e9a8ed3cc60e
|
[
"MIT"
] | null | null | null |
test_fftanalysis.ipynb
|
gmweir/PYFFT
|
e2404fb99871bb81f9fac9829965e9a8ed3cc60e
|
[
"MIT"
] | null | null | null | 986.847176 | 34,132 | 0.957141 |
[
[
[
"#%load_ext autoreload\n#%autoreload 2",
"_____no_output_____"
],
[
"import numpy as _np\nimport matplotlib.pyplot as _plt\nimport FFT as _fft",
"G:\\Workshop\\pyanalysis\\pybaseutils\\plt_utils.py:15: MatplotlibDeprecationWarning: \nThe mpl_toolkits.axes_grid module was deprecated in Matplotlib 2.1 and will be removed two minor releases later. Use mpl_toolkits.axes_grid1 and mpl_toolkits.axisartist, which provide the same functionality instead.\n from mpl_toolkits.axes_grid.inset_locator import inset_axes\n"
],
[
"_fft.fft.test_fftpwelch(plotit=False, verbose=False)",
"_____no_output_____"
],
[
"_fft.fft.test_fftanal(plotit=False)",
"Using a periodic Hamming window function\n(SLDR~f**-1, PSLL=-42.7dB, ROV=50.0%, AF=1.000, PF=0.761, OC=0.234)\nUsing a periodic Hamming window function\n(SLDR~f**-1, PSLL=-42.7dB, ROV=50.0%, AF=1.000, PF=0.761, OC=0.234)\nusing home-brew functions for spectra/coherence calculations\n"
],
[
"_fft.spectrogram.test_stft()\n_fft.spectrogram.test_specgram()",
"Please give at least a time-vector [s] and a signal vector [a.u.]\nUsing a periodic Rectangular window function\n(SLDR~f**-1, PSLL=-13.3dB, ROV=0.0%, AF=0, PF=1, OC=0)\nnWindows = 39\n"
],
[
"_fft.hilbert.test_hilbert(plotit=True, verbose=False)",
"_____no_output_____"
],
[
"_fft.laplace.test_laplace()",
"_____no_output_____"
],
[
"_fft.fft.test_fft_deriv(modified=True)",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
c5154ecf277441450f1973a6aac65b1ce6c4ddf9
| 802,845 |
ipynb
|
Jupyter Notebook
|
inteligencia_artificial/000-neural-networks-regression.ipynb
|
edwinb-ai/intelicompu
|
7ed4c51c789d0b71aac5f507d800cc57ba752fe4
|
[
"Apache-2.0"
] | 1 |
2020-01-10T03:22:50.000Z
|
2020-01-10T03:22:50.000Z
|
inteligencia_artificial/000-neural-networks-regression.ipynb
|
edwinb-ai/intelicompu
|
7ed4c51c789d0b71aac5f507d800cc57ba752fe4
|
[
"Apache-2.0"
] | null | null | null |
inteligencia_artificial/000-neural-networks-regression.ipynb
|
edwinb-ai/intelicompu
|
7ed4c51c789d0b71aac5f507d800cc57ba752fe4
|
[
"Apache-2.0"
] | null | null | null | 1,635.1222 | 786,028 | 0.959063 |
[
[
[
"# Regresión con Redes Neuronales\n\nEmpleando diferentes *funciones de pérdida* y *funciones de activación* las **redes neuronales** pueden resolver\nefectivamente problemas de **regresión.**\nEn esta libreta se estudia el ejemplo de [California Housing](http://www.spatial-statistics.com/pace_manuscripts/spletters_ms_dir/statistics_prob_lets/html/ms_sp_lets1.html)\ndonde el propósito es predecir el valor medio de una casa según 8 atributos.",
"_____no_output_____"
],
[
"## Descripción general del conjunto de datos\n\nEl conjunto de datos `California Housing` está hecho de 9 variables numéricas, donde 8 son las *características* y 1 es la variable objetivo.\nEste conjunto de datos fue creado en 1990 basándose en el censo poblacional realizado por el gobierno de EUA. La estructura del conjunto de datos\nes simple: cada línea en el archivo de datos cuenta por un **bloque** poblacional que consta de entre 600 y 3000 personas. Por cada *bloque*\nse tienen 8 características de cada casa y su costo medio.\n\nEmpleando *redes neuronales* se pretende predecir el costo de las casas por bloque.",
"_____no_output_____"
],
[
"## Atributos del conjunto de datos\n\nEste conjunto de datos cuenta con 8 *atributos*, descritos a continuación, con la etiqueta como viene en el conjunto de datos de `scikit-learn`:\n\n- **MedInc**, *Ingresos promedio por bloque*\n- **HouseAge**, *Antigüedad promedio por casa en el bloque*\n- **AveRooms**, *Número promedio de cuartos por casa en el bloque*\n- **AveBedrms**, *Número promedio de recámaras por casa en el bloque*\n- **Population**, *Población total del bloque*\n- **AveOccup**, *Ocupancia promedio por casa en el bloque*\n- **Latitude**, *Latitud del bloque*\n- **Longitude**, *Longitud del bloque*\n\nY la *variable respuesta* es:\n\n- **MedValue**, *Costo promedio por casa en el distrito*\n",
"_____no_output_____"
]
],
[
[
"import tensorflow as tf\nfrom sklearn import datasets, metrics, model_selection, preprocessing\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nimport pandas as pd",
"_____no_output_____"
],
[
"# Importar el conjunto de datos California Housing\ncali_data = datasets.fetch_california_housing()",
"_____no_output_____"
]
],
[
[
"## Visualización de datos\n",
"_____no_output_____"
]
],
[
[
"# Realizar una visualización general de la relación entre atributos del conjunto de datos\nsns.pairplot(pd.DataFrame(cali_data.data, columns=cali_data.feature_names))\nplt.show()",
"_____no_output_____"
]
],
[
[
"Con estas figuras se pueden observar algunas características interesantes:\n\n- *Primero*, todas las variables importan en el modelo. Esto significa que el modelo de regresión viene pesado por todas las\n características y se requiere que el modelo sea *robusto* ante esta situación.\n- *Segundo*, hay algunas características que tienen relación *lineal* entre ellas, como lo es **AveRooms** y **AveBedrms**.\n Esto puede ayudar a discriminar ciertas características que no tienen mucho peso sobre el modelo y solamente utilizar\n aquellas que influyen mucho más. A esta parte del *procesamiento de datos* se le conoce como **selección de características**\n y es una rama específica de la *inteligencia computacional.*\n- *Tercero*, la línea diagonal muestra la relación *distribución* de cada una de las características. Esto es algo importante\n de estudiar dado que algunas características muestran *distribuciones* conocidas y este hecho se puede utilizar para\n emplear técnicas estadísticas más avanzadas en el **análisis de regresión.**\n\nSin embargo, en toda esta libreta se dejarán las 8 características para que sean pesadas en el modelo final.\n",
"_____no_output_____"
]
],
[
[
"# Separar todos los datos y estandarizarlos\nX = cali_data.data\ny = cali_data.target\n# Crear el transformador para estandarización\nstd = preprocessing.StandardScaler()\nX = std.fit_transform(X)\nX = np.array(X).astype(np.float32)\ny = std.fit_transform(y.reshape(-1, 1))\ny = np.array(y).astype(np.float32)",
"_____no_output_____"
]
],
[
[
"Dado que los datos vienen en diferentes unidades y escalas, siempre se debe estandarizar los datos de alguna forma. En particular\nen esta libreta se emplea la normalización de los datos, haciendo que tengan *media* $\\mu = 0$ y *desviación estándar* $\\sigma = 1$.\n",
"_____no_output_____"
]
],
[
[
"# Separar en conjunto de entrenamiento y prueba\nx_train, x_test, y_train, y_test = model_selection.train_test_split(\n X, y, test_size=0.2, random_state=49\n)",
"_____no_output_____"
],
[
"# Definir parámetros generales de la Red Neuronal\npasos_entrenamiento = 1000\ntam_lote = 30\nratio_aprendizaje = 0.01",
"_____no_output_____"
]
],
[
[
"## Estructura o *topología* de la red neuronal\n\nPara esta regresión se pretende utilizar una *red neuronal* de **dos capas ocultas**, con *funciones de activación* **ReLU**,\nla **primera** capa oculta cuenta con 25 neuronas mientras que la **segunda** cuenta con 50.\nLa **capa de salida** *no* tiene función de activación, por lo que el modelo lineal queda de la siguiente forma\n\n$$ \\hat{y}(x) = \\sum_{i=1}^{8} \\alpha_i \\cdot x_i + \\beta_i$$\n\ndonde $\\alpha_i$ son los *pesos* de la *capa de salida*, mientras que $\\beta_i$ son los *sesgos*.\n",
"_____no_output_____"
]
],
[
[
"# Parámetros para la estructura general de la red\n# Número de neuronas por capa\nn_capa_oculta_1 = 25\nn_capa_oculta_2 = 50\nn_entrada = X.shape[1]\nn_salida = 1",
"_____no_output_____"
],
[
"# Definir las entradas de la red neuronal\nx_entrada = tf.placeholder(tf.float32, shape=[None, n_entrada])\ny_entrada = tf.placeholder(tf.float32, shape=[None, n_salida])",
"_____no_output_____"
],
[
"# Diccionario de pesos\npesos = {\n \"o1\": tf.Variable(tf.random_normal([n_entrada, n_capa_oculta_1])),\n \"o2\": tf.Variable(tf.random_normal([n_capa_oculta_1, n_capa_oculta_2])),\n \"salida\": tf.Variable(tf.random_normal([n_capa_oculta_2, n_salida])),\n}\n# Diccionario de sesgos\nsesgos = {\n \"b1\": tf.Variable(tf.random_normal([n_capa_oculta_1])),\n \"b2\": tf.Variable(tf.random_normal([n_capa_oculta_2])),\n \"salida\": tf.Variable(tf.random_normal([n_salida])),\n}",
"WARNING:tensorflow:From /home/edwin/anaconda3/lib/python3.7/site-packages/tensorflow/python/framework/op_def_library.py:263: colocate_with (from tensorflow.python.framework.ops) is deprecated and will be removed in a future version.\nInstructions for updating:\nColocations handled automatically by placer.\n"
],
[
"def propagacion_adelante(x):\n # Capa oculta 1\n # Esto es la mismo que Ax + b, un modelo lineal\n capa_1 = tf.add(tf.matmul(x, pesos[\"o1\"]), sesgos[\"b1\"])\n # ReLU como función de activación\n capa_1 = tf.nn.relu(capa_1)\n # Capa oculta 1\n # Esto es la mismo que Ax + b, un modelo lineal\n capa_2 = tf.add(tf.matmul(capa_1, pesos[\"o2\"]), sesgos[\"b2\"])\n # ReLU como función de activación\n capa_2 = tf.nn.relu(capa_2)\n\n # Capa de salida\n # Nuevamente, un modelo lineal\n capa_salida = tf.add(tf.matmul(capa_2, pesos[\"salida\"]), sesgos[\"salida\"])\n\n return capa_salida",
"_____no_output_____"
],
[
"# Implementar el modelo y sus capas\ny_prediccion = propagacion_adelante(x_entrada)",
"_____no_output_____"
]
],
[
[
"## Función de pérdida\n\nPara la función de pérdida se emplea la [función de Huber](https://en.wikipedia.org/wiki/Huber_loss) definida como\n\n\\begin{equation}\n L_{\\delta} \\left( y, f(x) \\right) = \n \\begin{cases}\n \\frac{1}{2} \\left( y - f(x) \\right)^2 & \\text{para} \\vert y - f(x) \\vert \\leq \\delta, \\\\\n \\delta \\vert y - f(x) \\vert - \\frac{1}{2} \\delta^2 & \\text{en cualquier otro caso.}\n \\end{cases}\n\\end{equation}\n\nEsta función es [robusta](https://en.wikipedia.org/wiki/Robust_regression) lo cual está hecha para erradicar el peso de posibles\nvalores atípicos y puede encontrar la verdadera relación entre las características sin tener que recurrir a metodologías paramétricas\ny no paramétricas.",
"_____no_output_____"
],
[
"## Nota\n\nEs importante mencionar que el valor de $\\delta$ en la función de Huber es un **hiperparámetro** que debe de ser ajustado mediante *validación cruzada*\npero no es realiza en esta libreta por limitaciones de equipo y rendimiento en la ejecución de esta libreta.\n",
"_____no_output_____"
]
],
[
[
"# Definir la función de costo\nf_costo = tf.reduce_mean(tf.losses.huber_loss(y_entrada, y_prediccion, delta=2.0))\n# f_costo = tf.reduce_mean(tf.square(y_entrada - y_prediccion))\noptimizador = tf.train.AdamOptimizer(learning_rate=ratio_aprendizaje).minimize(f_costo)",
"WARNING:tensorflow:From /home/edwin/anaconda3/lib/python3.7/site-packages/tensorflow/python/ops/losses/losses_impl.py:448: to_float (from tensorflow.python.ops.math_ops) is deprecated and will be removed in a future version.\nInstructions for updating:\nUse tf.cast instead.\n"
],
[
"# Primero, inicializar las variables\ninit = tf.global_variables_initializer()",
"_____no_output_____"
],
[
"# Función para evaluar la precisión de clasificación\ndef precision(prediccion, real):\n return tf.sqrt(tf.losses.mean_squared_error(real, prediccion))",
"_____no_output_____"
]
],
[
[
"## Precisión del modelo\n\nPara evaluar la precisión del modelo se emplea la función [RMSE](https://en.wikipedia.org/wiki/Root-mean-square_deviation) (Root Mean Squared Error)\ndefinida por la siguiente función:\n\n$$ RMSE = \\sqrt{\\frac{\\sum_{i=1}^{N} \\left( \\hat{y}_i - y_i \\right)^2}{N}} $$",
"_____no_output_____"
],
[
"Para crear un mejor estimado, se empleará validación cruzada de 5 pliegues.\n",
"_____no_output_____"
]
],
[
[
"# Crear el plegador para el conjunto de datos\nkf = model_selection.KFold(n_splits=5)\nkf_val_score_train = []\nkf_val_score_test = []",
"_____no_output_____"
],
[
"# Crear un grafo de computación\nwith tf.Session() as sess:\n # Inicializar las variables\n sess.run(init)\n\n for tr_idx, ts_idx in kf.split(x_train):\n # Comenzar los pasos de entrenamiento\n # solamente con el conjunto de datos de entrenamiento\n for p in range(pasos_entrenamiento):\n # Minimizar la función de costo\n minimizacion = sess.run(\n optimizador,\n feed_dict={x_entrada: x_train[tr_idx], y_entrada: y_train[tr_idx]},\n )\n # Cada tamaño de lote, calcular la precisión del modelo\n if p % tam_lote == 0:\n prec_entrenamiento = sess.run(\n precision(y_prediccion, y_entrada),\n feed_dict={x_entrada: x_train[tr_idx], y_entrada: y_train[tr_idx]},\n )\n kf_val_score_train.append(prec_entrenamiento)\n prec_prueba = sess.run(\n precision(y_prediccion, y_entrada),\n feed_dict={x_entrada: x_train[ts_idx], y_entrada: y_train[ts_idx]},\n )\n kf_val_score_test.append(prec_prueba)\n # Prediccion final, una vez entrenado el modelo\n pred_final = sess.run(\n precision(y_prediccion, y_entrada),\n feed_dict={x_entrada: x_test, y_entrada: y_test},\n )\n pred_report = sess.run(y_prediccion, feed_dict={x_entrada: x_test})\n print(\"Precisión final: {0}\".format(pred_final))",
"Precisión final: 0.5226657390594482\n"
],
[
"print(\"Precisión RMSE para entrenamiento: {0}\".format(np.mean(kf_val_score_train)))\nprint(\"Precisión RMSE para entrenamiento: {0}\".format(np.mean(kf_val_score_test)))",
"Precisión RMSE para entrenamiento: 0.9006941914558411\nPrecisión RMSE para entrenamiento: 0.9006157517433167\n"
]
],
[
[
"Aquí se muestra el valor de *RMSE* final para cada parte, entrenamiento y prueba. Se puede observar que hay muy poco sobreajuste, \ny si se quisiera corregir se puede realizar aumentando el número de neuronas, de capas, cambiando las funciones de activación,\nentre muchas otras cosas.\n\n",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
]
] |
c51561b8715743f36d1e6ed6ef4370e002ed027c
| 68,485 |
ipynb
|
Jupyter Notebook
|
testing/.ipynb_checkpoints/Untitled1-checkpoint.ipynb
|
wsbuck/DogeNet
|
4ad0917cddd0baddb452021f2a7dc61cf895b6ce
|
[
"MIT"
] | null | null | null |
testing/.ipynb_checkpoints/Untitled1-checkpoint.ipynb
|
wsbuck/DogeNet
|
4ad0917cddd0baddb452021f2a7dc61cf895b6ce
|
[
"MIT"
] | null | null | null |
testing/.ipynb_checkpoints/Untitled1-checkpoint.ipynb
|
wsbuck/DogeNet
|
4ad0917cddd0baddb452021f2a7dc61cf895b6ce
|
[
"MIT"
] | null | null | null | 165.024096 | 43,369 | 0.909367 |
[
[
[
"import tensorflow as tf\nfrom tensorflow import keras\n\nfrom tensorflow.keras.preprocessing import image\n\nimport numpy as np\nimport os\n\nfrom IPython.display import Image",
"_____no_output_____"
],
[
"from tensorflow.keras.applications.mobilenet import preprocess_input, decode_predictions",
"_____no_output_____"
],
[
"mobile = keras.applications.mobilenet.MobileNet()",
"WARNING:tensorflow:From /media/HDD/DogeNetTFModel/lib/python3.6/site-packages/tensorflow/python/ops/resource_variable_ops.py:435: colocate_with (from tensorflow.python.framework.ops) is deprecated and will be removed in a future version.\nInstructions for updating:\nColocations handled automatically by placer.\nWARNING:tensorflow:From /media/HDD/DogeNetTFModel/lib/python3.6/site-packages/tensorflow/python/keras/layers/core.py:143: calling dropout (from tensorflow.python.ops.nn_ops) with keep_prob is deprecated and will be removed in a future version.\nInstructions for updating:\nPlease use `rate` instead of `keep_prob`. Rate should be set to `rate = 1 - keep_prob`.\n"
],
[
"def prepare_image(file):\n img_path = ''\n img = image.load_img(img_path + file, target_size=(224, 224))\n img_array = image.img_to_array(img)\n img_array_expanded_dims = np.expand_dims(img_array, axis=0)\n return keras.applications.mobilenet.preprocess_input(img_array_expanded_dims)",
"_____no_output_____"
],
[
"data_path = \"../data/Images/n02097474-Tibetan_terrier/\"",
"_____no_output_____"
],
[
"os.listdir(data_path)[:10]",
"_____no_output_____"
],
[
"Image(filename=os.path.join(data_path, \"n02097474_1023.jpg\"))",
"_____no_output_____"
],
[
"preprocessed_image = prepare_image(os.path.join(data_path, \"n02097474_1023.jpg\"))\npredictions = mobile.predict(preprocessed_image)\nresults = decode_predictions(predictions)\nresults",
"_____no_output_____"
],
[
"data_path = \"../data/Images/\"",
"_____no_output_____"
],
[
"Image(filename=os.path.join(data_path, \"n02113978-Mexican_hairless/n02113978_3375.jpg\"))",
"_____no_output_____"
],
[
"preprocessed_image = prepare_image(os.path.join(data_path, \"n02113978-Mexican_hairless/n02113978_3375.jpg\"))\npredictions = mobile.predict(preprocessed_image)\nresults = decode_predictions(predictions)\nresults",
"_____no_output_____"
]
],
[
[
"## Retrain MobileNet",
"_____no_output_____"
]
],
[
[
"from tensorflow.keras.layers import Dense, GlobalAveragePooling2D\nfrom tensorflow.keras.applications.mobilenet import MobileNet",
"_____no_output_____"
],
[
"from tensorflow.keras.models import Model",
"_____no_output_____"
],
[
"# imports the mobilenet model and discards the last 1000 neuron layer.\nbase_model = MobileNet(weights='imagenet', include_top=False)\n\nx = base_model.output\nx = GlobalAveragePooling2D()(x)\nx = Dense(1024, activation='relu')(x)\nx = Dense(1024, activation='relu')(x)\nx = Dense(512, activation='relu')(x)\npreds = Dense(2, activation='softmax')(x)",
"_____no_output_____"
],
[
"model = Model(inputs=base_model.input, outputs=preds)",
"_____no_output_____"
],
[
"for i, layer in enumerate(model.layers):\n print(i,layer.name)",
"0 input_3\n1 conv1_pad\n2 conv1\n3 conv1_bn\n4 conv1_relu\n5 conv_dw_1\n6 conv_dw_1_bn\n7 conv_dw_1_relu\n8 conv_pw_1\n9 conv_pw_1_bn\n10 conv_pw_1_relu\n11 conv_pad_2\n12 conv_dw_2\n13 conv_dw_2_bn\n14 conv_dw_2_relu\n15 conv_pw_2\n16 conv_pw_2_bn\n17 conv_pw_2_relu\n18 conv_dw_3\n19 conv_dw_3_bn\n20 conv_dw_3_relu\n21 conv_pw_3\n22 conv_pw_3_bn\n23 conv_pw_3_relu\n24 conv_pad_4\n25 conv_dw_4\n26 conv_dw_4_bn\n27 conv_dw_4_relu\n28 conv_pw_4\n29 conv_pw_4_bn\n30 conv_pw_4_relu\n31 conv_dw_5\n32 conv_dw_5_bn\n33 conv_dw_5_relu\n34 conv_pw_5\n35 conv_pw_5_bn\n36 conv_pw_5_relu\n37 conv_pad_6\n38 conv_dw_6\n39 conv_dw_6_bn\n40 conv_dw_6_relu\n41 conv_pw_6\n42 conv_pw_6_bn\n43 conv_pw_6_relu\n44 conv_dw_7\n45 conv_dw_7_bn\n46 conv_dw_7_relu\n47 conv_pw_7\n48 conv_pw_7_bn\n49 conv_pw_7_relu\n50 conv_dw_8\n51 conv_dw_8_bn\n52 conv_dw_8_relu\n53 conv_pw_8\n54 conv_pw_8_bn\n55 conv_pw_8_relu\n56 conv_dw_9\n57 conv_dw_9_bn\n58 conv_dw_9_relu\n59 conv_pw_9\n60 conv_pw_9_bn\n61 conv_pw_9_relu\n62 conv_dw_10\n63 conv_dw_10_bn\n64 conv_dw_10_relu\n65 conv_pw_10\n66 conv_pw_10_bn\n67 conv_pw_10_relu\n68 conv_dw_11\n69 conv_dw_11_bn\n70 conv_dw_11_relu\n71 conv_pw_11\n72 conv_pw_11_bn\n73 conv_pw_11_relu\n74 conv_pad_12\n75 conv_dw_12\n76 conv_dw_12_bn\n77 conv_dw_12_relu\n78 conv_pw_12\n79 conv_pw_12_bn\n80 conv_pw_12_relu\n81 conv_dw_13\n82 conv_dw_13_bn\n83 conv_dw_13_relu\n84 conv_pw_13\n85 conv_pw_13_bn\n86 conv_pw_13_relu\n87 global_average_pooling2d_2\n88 dense_3\n89 dense_4\n90 dense_5\n91 dense_6\n"
],
[
"for layer in model.layers:\n layer.trainable = False\n\nfor layer in model.layers[:20]:\n layer.trainable = False\n\nfor layer in model.layers[20:]:\n layer.trainable = True",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
c5156e9c00c443693a8507866cc2acc9811569e2
| 26,953 |
ipynb
|
Jupyter Notebook
|
jupyter/notebooks/example_train.ipynb
|
refit-ml/refit-local
|
9181ae91a92ece7243b9351467cd8e538f003a15
|
[
"Apache-2.0"
] | 1 |
2020-12-11T01:59:26.000Z
|
2020-12-11T01:59:26.000Z
|
jupyter/notebooks/example_train.ipynb
|
refit-ml/refit-local
|
9181ae91a92ece7243b9351467cd8e538f003a15
|
[
"Apache-2.0"
] | null | null | null |
jupyter/notebooks/example_train.ipynb
|
refit-ml/refit-local
|
9181ae91a92ece7243b9351467cd8e538f003a15
|
[
"Apache-2.0"
] | null | null | null | 35.371391 | 227 | 0.46774 |
[
[
[
"!refit_init",
"Requirement already satisfied: pycryptodome==3.9.8 in /opt/conda/lib/python3.8/site-packages (from -r refit/requirements.txt (line 1)) (3.9.8)\nRequirement already satisfied: numpy==1.18.5 in /opt/conda/lib/python3.8/site-packages (from -r refit/requirements.txt (line 2)) (1.18.5)\nRequirement already satisfied: pandas==1.1.0 in /opt/conda/lib/python3.8/site-packages (from -r refit/requirements.txt (line 3)) (1.1.0)\nRequirement already satisfied: seaborn==0.10.1 in /opt/conda/lib/python3.8/site-packages (from -r refit/requirements.txt (line 4)) (0.10.1)\nRequirement already satisfied: matplotlib==3.3.0 in /opt/conda/lib/python3.8/site-packages (from -r refit/requirements.txt (line 5)) (3.3.0)\nRequirement already satisfied: scipy==1.5.2 in /opt/conda/lib/python3.8/site-packages (from -r refit/requirements.txt (line 6)) (1.5.2)\nRequirement already satisfied: statsmodels==0.11.1 in /opt/conda/lib/python3.8/site-packages (from -r refit/requirements.txt (line 7)) (0.11.1)\nRequirement already satisfied: sklearn==0.0 in /opt/conda/lib/python3.8/site-packages (from -r refit/requirements.txt (line 8)) (0.0)\nRequirement already satisfied: scikit-learn==0.23.2 in /opt/conda/lib/python3.8/site-packages (from -r refit/requirements.txt (line 9)) (0.23.2)\nRequirement already satisfied: cassandra-driver==3.24.0 in /opt/conda/lib/python3.8/site-packages (from -r refit/requirements.txt (line 10)) (3.24.0)\nRequirement already satisfied: keras2onnx==1.6.0 in /opt/conda/lib/python3.8/site-packages (from -r refit/requirements.txt (line 11)) (1.6.0)\nRequirement already satisfied: Keras==2.2.5 in /opt/conda/lib/python3.8/site-packages (from -r refit/requirements.txt (line 12)) (2.2.5)\nRequirement already satisfied: protobuf==3.8.0 in /opt/conda/lib/python3.8/site-packages (from -r refit/requirements.txt (line 13)) (3.8.0)\nRequirement already satisfied: onnxmltools==1.6.0 in /opt/conda/lib/python3.8/site-packages (from -r refit/requirements.txt (line 14)) (1.6.0)\nRequirement already satisfied: pyyaml==5.3.1 in /opt/conda/lib/python3.8/site-packages (from -r refit/requirements.txt (line 15)) (5.3.1)\nRequirement already satisfied: minio==6.0.0 in /opt/conda/lib/python3.8/site-packages (from -r refit/requirements.txt (line 16)) (6.0.0)\nRequirement already satisfied: python-dateutil>=2.7.3 in /opt/conda/lib/python3.8/site-packages (from pandas==1.1.0->-r refit/requirements.txt (line 3)) (2.8.1)\nRequirement already satisfied: pytz>=2017.2 in /opt/conda/lib/python3.8/site-packages (from pandas==1.1.0->-r refit/requirements.txt (line 3)) (2020.1)\nRequirement already satisfied: pyparsing!=2.0.4,!=2.1.2,!=2.1.6,>=2.0.3 in /opt/conda/lib/python3.8/site-packages (from matplotlib==3.3.0->-r refit/requirements.txt (line 5)) (2.4.7)\nRequirement already satisfied: cycler>=0.10 in /opt/conda/lib/python3.8/site-packages (from matplotlib==3.3.0->-r refit/requirements.txt (line 5)) (0.10.0)\nRequirement already satisfied: kiwisolver>=1.0.1 in /opt/conda/lib/python3.8/site-packages (from matplotlib==3.3.0->-r refit/requirements.txt (line 5)) (1.2.0)\nRequirement already satisfied: pillow>=6.2.0 in /opt/conda/lib/python3.8/site-packages (from matplotlib==3.3.0->-r refit/requirements.txt (line 5)) (7.2.0)\nRequirement already satisfied: patsy>=0.5 in /opt/conda/lib/python3.8/site-packages (from statsmodels==0.11.1->-r refit/requirements.txt (line 7)) (0.5.1)\nRequirement already satisfied: threadpoolctl>=2.0.0 in /opt/conda/lib/python3.8/site-packages (from scikit-learn==0.23.2->-r refit/requirements.txt (line 9)) (2.1.0)\nRequirement already satisfied: joblib>=0.11 in /opt/conda/lib/python3.8/site-packages (from scikit-learn==0.23.2->-r refit/requirements.txt (line 9)) (0.16.0)\nRequirement already satisfied: geomet<0.3,>=0.1 in /opt/conda/lib/python3.8/site-packages (from cassandra-driver==3.24.0->-r refit/requirements.txt (line 10)) (0.2.1.post1)\nRequirement already satisfied: six>=1.9 in /opt/conda/lib/python3.8/site-packages (from cassandra-driver==3.24.0->-r refit/requirements.txt (line 10)) (1.15.0)\nRequirement already satisfied: onnxconverter-common>=1.6.0 in /opt/conda/lib/python3.8/site-packages (from keras2onnx==1.6.0->-r refit/requirements.txt (line 11)) (1.7.0)\nRequirement already satisfied: requests in /opt/conda/lib/python3.8/site-packages (from keras2onnx==1.6.0->-r refit/requirements.txt (line 11)) (2.24.0)\nRequirement already satisfied: fire in /opt/conda/lib/python3.8/site-packages (from keras2onnx==1.6.0->-r refit/requirements.txt (line 11)) (0.3.1)\nRequirement already satisfied: onnx in /opt/conda/lib/python3.8/site-packages (from keras2onnx==1.6.0->-r refit/requirements.txt (line 11)) (1.7.0)\nRequirement already satisfied: keras-applications>=1.0.8 in /opt/conda/lib/python3.8/site-packages (from Keras==2.2.5->-r refit/requirements.txt (line 12)) (1.0.8)\nRequirement already satisfied: h5py in /opt/conda/lib/python3.8/site-packages (from Keras==2.2.5->-r refit/requirements.txt (line 12)) (2.10.0)\nRequirement already satisfied: keras-preprocessing>=1.1.0 in /opt/conda/lib/python3.8/site-packages (from Keras==2.2.5->-r refit/requirements.txt (line 12)) (1.1.2)\nRequirement already satisfied: setuptools in /opt/conda/lib/python3.8/site-packages (from protobuf==3.8.0->-r refit/requirements.txt (line 13)) (49.6.0.post20200917)\nRequirement already satisfied: skl2onnx in /opt/conda/lib/python3.8/site-packages (from onnxmltools==1.6.0->-r refit/requirements.txt (line 14)) (1.7.0)\nRequirement already satisfied: configparser in /opt/conda/lib/python3.8/site-packages (from minio==6.0.0->-r refit/requirements.txt (line 16)) (5.0.0)\nRequirement already satisfied: certifi in /opt/conda/lib/python3.8/site-packages (from minio==6.0.0->-r refit/requirements.txt (line 16)) (2020.6.20)\nRequirement already satisfied: urllib3 in /opt/conda/lib/python3.8/site-packages (from minio==6.0.0->-r refit/requirements.txt (line 16)) (1.25.10)\nRequirement already satisfied: click in /opt/conda/lib/python3.8/site-packages (from geomet<0.3,>=0.1->cassandra-driver==3.24.0->-r refit/requirements.txt (line 10)) (7.1.2)\nRequirement already satisfied: idna<3,>=2.5 in /opt/conda/lib/python3.8/site-packages (from requests->keras2onnx==1.6.0->-r refit/requirements.txt (line 11)) (2.10)\nRequirement already satisfied: chardet<4,>=3.0.2 in /opt/conda/lib/python3.8/site-packages (from requests->keras2onnx==1.6.0->-r refit/requirements.txt (line 11)) (3.0.4)\nRequirement already satisfied: termcolor in /opt/conda/lib/python3.8/site-packages (from fire->keras2onnx==1.6.0->-r refit/requirements.txt (line 11)) (1.1.0)\nRequirement already satisfied: typing-extensions>=3.6.2.1 in /opt/conda/lib/python3.8/site-packages (from onnx->keras2onnx==1.6.0->-r refit/requirements.txt (line 11)) (3.7.4.3)\nRefit Installed, You can now run the following to import REFIT\nfrom refit.api.refit import Refit\n"
],
[
"#Example Notebook\nfrom sklearn.datasets import load_iris\nfrom datetime import datetime\nfrom refit.api.refit import Refit\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.ensemble import RandomForestClassifier\nfrom sklearn import metrics\nfrom refit.enums.ModelFormat import ModelFormat\nfrom skl2onnx.common.data_types import FloatTensorType, BooleanTensorType\n",
"_____no_output_____"
],
[
"project_guid = \"b6ee5bab-08dd-49b0-98b6-45cd0a28b12f\"\nrefit = Refit(project_guid)\n\nstart = datetime(2020, 6, 27)\nend = datetime(2020, 6, 29)\nsensors = list(\n map(lambda x: str(x),\n range(5160, 5170))) #['5163']\n\nsensors",
"/home/docker_worker/notebooks/refit/util/Schema.py:23: YAMLLoadWarning: calling yaml.load() without Loader=... is deprecated, as the default Loader is unsafe. Please read https://msg.pyyaml.org/load for full details.\n dict = yaml.load(schema_yaml)\n"
],
[
"# Getting Sensor Data with the training window flag incorporated\ndf = refit.sensor_data_with_flag(start, end, sensors=sensors)\ndf",
"_____no_output_____"
],
[
"skip_columns = ['project_guid', 'sensor_id', 'partition_key', 'timestamp', 'operable']\nfeature_columns = list(filter( lambda x: x not in skip_columns, df.columns.array))\nfeature_columns",
"_____no_output_____"
],
[
"df['temperature'] = df['temperature'].astype(float)\ndf['pressure'] = df['pressure'].astype(float)\ndf['wind'] = df['wind'].astype(float)\n\n# In the real world we would take the operable flag that REFIT gave us, \n# but since this is synthetic data, we are going to create a fake trend\ndf['operable'] = (df.temperature < 75.0)\ndf['operable'] = df['operable'].astype(int)\ndf",
"_____no_output_____"
],
[
"x = df[feature_columns]\ny = df['operable']",
"_____no_output_____"
],
[
"X_train, X_test, y_train, y_test = train_test_split(x, y, test_size=0.3) # 70% training and 30% test",
"_____no_output_____"
],
[
"clf=RandomForestClassifier(n_estimators=100)\nclf.fit(X_train,y_train)",
"_____no_output_____"
],
[
"y_pred=clf.predict(X_test)\ny_pred",
"_____no_output_____"
],
[
"print(\"Accuracy:\",metrics.accuracy_score(y_test, y_pred))",
"Accuracy: 1.0\n"
],
[
"initial_types = [('input', FloatTensorType([None,len(feature_columns)]))]",
"_____no_output_____"
],
[
"refit.save(clf, ModelFormat.SK_LEARN, initial_types=initial_types)",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
c51583fd2c0ccb52bbf09a805eaceb1fc108339a
| 352,619 |
ipynb
|
Jupyter Notebook
|
first ML project.ipynb
|
Aniket29-shiv/Real-Estate-Price-Prediction-App
|
c020254ff93eb066b27bad2b3effb50456268154
|
[
"MIT"
] | null | null | null |
first ML project.ipynb
|
Aniket29-shiv/Real-Estate-Price-Prediction-App
|
c020254ff93eb066b27bad2b3effb50456268154
|
[
"MIT"
] | null | null | null |
first ML project.ipynb
|
Aniket29-shiv/Real-Estate-Price-Prediction-App
|
c020254ff93eb066b27bad2b3effb50456268154
|
[
"MIT"
] | null | null | null | 174.13284 | 154,092 | 0.853808 |
[
[
[
"## First ML project",
"_____no_output_____"
]
],
[
[
"import pandas as pd",
"_____no_output_____"
],
[
"housing = pd.read_csv(\"data.csv\")",
"_____no_output_____"
],
[
"housing.head()",
"_____no_output_____"
],
[
"housing.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 506 entries, 0 to 505\nData columns (total 14 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 CRIM 506 non-null float64\n 1 ZN 506 non-null float64\n 2 INDUS 506 non-null float64\n 3 CHAS 506 non-null int64 \n 4 NOX 506 non-null float64\n 5 RM 501 non-null float64\n 6 AGE 506 non-null float64\n 7 DIS 506 non-null float64\n 8 RAD 506 non-null int64 \n 9 TAX 506 non-null int64 \n 10 PTRATIO 506 non-null float64\n 11 B 506 non-null float64\n 12 LSTAT 506 non-null float64\n 13 MEDV 506 non-null float64\ndtypes: float64(11), int64(3)\nmemory usage: 55.4 KB\n"
],
[
"housing['AGE'].value_counts()\n",
"_____no_output_____"
],
[
"housing.describe()",
"_____no_output_____"
],
[
"%matplotlib inline",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"housing.hist(bins=50, figsize=(20,15))",
"_____no_output_____"
],
[
"import numpy as np\ndef split_train_test(data , test_ratio):\n np.random.seed(42) # this fixes the shuffled value of data\n shuffled = np.random.permutation(len(data))\n print(shuffled)\n test_set_size = int(len(data)*test_ratio)\n test_indices = shuffled[:test_set_size]\n train_indices = shuffled[test_set_size:]\n return data.iloc[train_indices],data.iloc[test_indices]",
"_____no_output_____"
],
[
"train_set,test_set = split_train_test(housing , 0.2)",
"[173 274 491 72 452 76 316 140 471 500 218 9 414 78 323 473 124 388\n 195 448 271 278 30 501 421 474 79 454 210 497 172 320 375 362 467 153\n 2 336 208 73 496 307 204 68 90 390 33 70 470 0 11 281 22 101\n 268 485 442 290 84 245 63 55 229 18 351 209 395 82 39 456 46 481\n 444 355 77 398 104 203 381 489 69 408 255 392 312 234 460 324 93 137\n 176 417 131 346 365 132 371 412 436 411 86 75 477 15 332 423 19 325\n 335 56 437 409 334 181 227 434 180 25 493 238 244 250 418 117 42 322\n 347 182 155 280 126 329 31 113 148 432 338 57 194 24 17 298 66 211\n 404 94 154 441 23 225 433 447 5 116 45 16 468 360 3 405 185 60\n 110 321 265 29 262 478 26 7 492 108 37 157 472 118 114 175 192 272\n 144 373 383 356 277 220 450 141 369 67 361 168 499 394 400 193 249 109\n 420 145 92 152 222 304 83 248 165 163 199 231 74 311 455 253 119 284\n 302 483 357 403 228 261 237 386 476 36 196 139 368 247 287 378 59 111\n 89 266 6 364 503 341 158 150 177 397 184 318 10 384 103 81 38 317\n 167 475 299 296 198 377 146 396 147 428 289 123 490 96 143 239 275 97\n 353 122 183 202 246 484 301 354 410 399 286 125 305 223 422 219 129 424\n 291 331 380 480 358 297 294 370 438 112 179 310 342 333 487 457 233 314\n 164 136 197 258 232 115 120 352 224 406 340 127 285 415 107 374 449 133\n 367 44 495 65 283 85 242 186 425 159 12 35 28 170 142 402 349 221\n 95 51 240 376 382 178 41 440 391 206 282 254 416 4 256 453 100 226\n 431 213 426 171 98 292 215 61 47 32 267 327 200 451 27 393 230 260\n 288 162 429 138 62 135 128 482 8 326 469 64 300 14 156 40 379 465\n 407 216 279 439 504 337 236 207 212 295 462 251 494 464 303 350 269 201\n 161 43 217 401 190 309 259 105 53 389 1 446 488 49 419 80 205 34\n 430 263 427 366 91 339 479 52 345 264 241 13 315 88 387 273 166 328\n 498 134 306 486 319 243 54 363 50 461 174 445 189 502 463 187 169 58\n 48 344 235 252 21 313 459 160 276 443 191 385 293 413 343 257 308 149\n 130 151 359 99 372 87 458 330 214 466 121 505 20 188 71 106 270 348\n 435 102]\n"
],
[
"print(f\"Rows in train set:{len(train_set)}\\nRows in test set:{len(test_set)}\")",
"Rows in train set:405\nRows in test set:101\n"
],
[
"from sklearn.model_selection import train_test_split\ntrain_set,test_set = train_test_split(housing , test_size=0.2,random_state=42)\nprint(f\"Rows in train set:{len(train_set)}\\nRows in test set:{len(test_set)}\")",
"Rows in train set:404\nRows in test set:102\n"
],
[
"from sklearn.model_selection import StratifiedShuffleSplit\nsplit = StratifiedShuffleSplit(n_splits=1, test_size=0.2, random_state=42)\nfor train_index, test_index in split.split(housing, housing['CHAS']):\n strat_train_set = housing.loc[train_index]\n strat_test_set = housing.loc[test_index]",
"_____no_output_____"
],
[
"strat_test_set['AGE'].value_counts()",
"_____no_output_____"
],
[
"strat_train_set['AGE'].value_counts()",
"_____no_output_____"
],
[
"housing = strat_train_set.copy()",
"_____no_output_____"
]
],
[
[
"## Finding Correlations",
"_____no_output_____"
]
],
[
[
"corr_matrix = housing.corr()\ncorr_matrix['MEDV'].sort_values(ascending=False)",
"_____no_output_____"
],
[
"from pandas.plotting import scatter_matrix\nattributes = [\"MEDV\",\"RM\",\"ZN\",\"LSTAT\"]\nscatter_matrix(housing[attributes],figsize=[12,8])",
"_____no_output_____"
],
[
"housing.plot(kind=\"scatter\",x=\"RM\",y=\"MEDV\",alpha=0.9)",
"_____no_output_____"
]
],
[
[
"## Try out attributes",
"_____no_output_____"
]
],
[
[
"housing[\"TAXRM\"]= housing[\"TAX\"]/housing[\"RM\"]",
"_____no_output_____"
],
[
"housing.head()",
"_____no_output_____"
],
[
"corr_matrix = housing.corr()\ncorr_matrix['MEDV'].sort_values(ascending=False)\nhousing.plot(kind=\"scatter\",x=\"TAXRM\",y=\"MEDV\",alpha=0.9)",
"_____no_output_____"
],
[
"housing = strat_train_set.drop(\"MEDV\", axis=1)\nhousing_labels = strat_train_set[\"MEDV\"].copy()",
"_____no_output_____"
]
],
[
[
"## Missing Attributes",
"_____no_output_____"
]
],
[
[
"# To take care of missing attributes, you have three options:\n# 1. Get rid of the missing data points\n# 2. Get rid of the whole attribute\n# 3. Set the value to some value(0, mean or median)",
"_____no_output_____"
],
[
"a = housing.dropna(subset=[\"RM\"]) #option 1 Here the missing attributes are dropped\na.shape",
"_____no_output_____"
],
[
"housing.drop(\"RM\", axis=1).shape # option 2 here the attribure column of RM is dropped",
"_____no_output_____"
],
[
"median = housing[\"RM\"].median() #Compute median for option 3",
"_____no_output_____"
],
[
"housing[\"RM\"].fillna(median)# option 3",
"_____no_output_____"
],
[
"housing.describe() # before we started imputing this was RM with 501 value",
"_____no_output_____"
],
[
"from sklearn.impute import SimpleImputer\nimputer = SimpleImputer(strategy = \"median\")\nimputer.fit(housing)\n",
"_____no_output_____"
],
[
"imputer.statistics_",
"_____no_output_____"
],
[
"X = imputer.transform(housing)",
"_____no_output_____"
],
[
"housing_tr = pd.DataFrame(X, columns=housing.columns)",
"_____no_output_____"
],
[
"housing_tr.describe() # Now after imputing we hv got the value of RM as 506",
"_____no_output_____"
]
],
[
[
"## Creating a pipeline",
"_____no_output_____"
]
],
[
[
"from sklearn.pipeline import Pipeline\nfrom sklearn.preprocessing import StandardScaler\nmy_pipeline = Pipeline([\n ('imputer',SimpleImputer(strategy=\"median\")),\n ('std_scaler', StandardScaler()),\n])\n\n",
"_____no_output_____"
],
[
"housing_num_tr = my_pipeline.fit_transform(housing)\n",
"_____no_output_____"
],
[
"housing_num_tr.shape\n",
"_____no_output_____"
]
],
[
[
"## Selecting the model for dragon real Estate",
"_____no_output_____"
]
],
[
[
"from sklearn.linear_model import LinearRegression\nfrom sklearn.tree import DecisionTreeRegressor\nfrom sklearn.ensemble import RandomForestRegressor\n\nmodel=RandomForestRegressor()\nmodel.fit(housing_num_tr,housing_labels)",
"_____no_output_____"
],
[
"some_data = housing.iloc[:5]",
"_____no_output_____"
],
[
"some_labels = housing_labels.iloc[:5]",
"_____no_output_____"
],
[
"prepared_data = my_pipeline.transform(some_data)",
"_____no_output_____"
],
[
"model.predict(prepared_data)",
"_____no_output_____"
],
[
"list(some_labels)",
"_____no_output_____"
]
],
[
[
"## Evaluating the model",
"_____no_output_____"
]
],
[
[
"from sklearn.metrics import mean_squared_error\nhousing_predictions = model.predict(housing_num_tr)\nmse = mean_squared_error(housing_labels, housing_predictions)\nrmse = np.sqrt(mse)",
"_____no_output_____"
],
[
"rmse",
"_____no_output_____"
]
],
[
[
"## Using better evaluation techniques - CrossValidation",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import cross_val_score\nscores = cross_val_score(model, housing_num_tr, housing_labels, scoring=\"neg_mean_squared_error\", cv=10)\nrmse_scores = np.sqrt(-scores)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
c5158728fec23320ee8507ef2e63eadb889ac8a7
| 1,746 |
ipynb
|
Jupyter Notebook
|
analysis/notebooks/portfolio_current.ipynb
|
ElliotVilhelm/LiuAlgoTrader
|
3bedfc2d2dac886a4b51c798eaf1a70487b22e48
|
[
"MIT"
] | null | null | null |
analysis/notebooks/portfolio_current.ipynb
|
ElliotVilhelm/LiuAlgoTrader
|
3bedfc2d2dac886a4b51c798eaf1a70487b22e48
|
[
"MIT"
] | null | null | null |
analysis/notebooks/portfolio_current.ipynb
|
ElliotVilhelm/LiuAlgoTrader
|
3bedfc2d2dac886a4b51c798eaf1a70487b22e48
|
[
"MIT"
] | 1 |
2021-08-31T04:03:15.000Z
|
2021-08-31T04:03:15.000Z
| 18.978261 | 61 | 0.542955 |
[
[
[
"## Take Action: Select portfolio for analysis",
"_____no_output_____"
]
],
[
[
"portfolio_id = \"f5d0ab5b-56af-4b99-8891-ba5258d25374\"",
"_____no_output_____"
]
],
[
[
"### Imports",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nimport empyrical as ep\nimport math\nimport pandas as pd\nimport numpy as np\nimport quantstats as qs\nfrom IPython.display import HTML, display, Markdown\nfrom liualgotrader.analytics import analysis\nimport matplotlib.ticker as mtick\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"### Load data details",
"_____no_output_____"
]
],
[
[
"portfolio = analysis.get_portfolio_equity(portfolio_id)",
"_____no_output_____"
],
[
"portfolio",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
c5158d2cfcb6b4683fcf378a155c6a556c884919
| 60,272 |
ipynb
|
Jupyter Notebook
|
docs/cruncher_demo.ipynb
|
hdoupe/Tax-Cruncher
|
e263bcf8643d747d85855bfc2af2faba519a9ace
|
[
"MIT"
] | 10 |
2019-07-16T20:33:15.000Z
|
2021-07-26T19:33:02.000Z
|
docs/cruncher_demo.ipynb
|
hdoupe/Tax-Cruncher
|
e263bcf8643d747d85855bfc2af2faba519a9ace
|
[
"MIT"
] | 31 |
2019-07-16T18:52:30.000Z
|
2022-02-01T20:59:41.000Z
|
docs/cruncher_demo.ipynb
|
hdoupe/Tax-Cruncher
|
e263bcf8643d747d85855bfc2af2faba519a9ace
|
[
"MIT"
] | 5 |
2019-05-03T14:39:33.000Z
|
2019-07-11T12:58:21.000Z
| 38.935401 | 455 | 0.328876 |
[
[
[
"# Batch analysis\n\nThe `Batch:` class allows users to analyze the tax liabilities of a batch of filing units under current law and under a tax policy reform. \n\n## Inputs\n\nThe class is initialized with a file path to CSV input -- each column contains data on a different tax information variable (e.g. year, marital status, wages, etc.) and each row represents a different filing unit. A detailed discussion of the inputs is [here](https://github.com/PSLmodels/Tax-Cruncher/blob/master/docs/INPUT_INSTRUCTIONS.md).",
"_____no_output_____"
]
],
[
[
"from taxcrunch.multi_cruncher import Batch\nimport os",
"_____no_output_____"
],
[
"# path to input csv file\nCURRENT_PATH = os.path.abspath(os.path.dirname('file'))\npath = os.path.join(CURRENT_PATH, \"example_input.csv\")\n\n# create Batch object\nb = Batch(path)",
"_____no_output_____"
]
],
[
[
"## Analyzing tax data under current law\n\nThe `create_table()` method returns a Pandas DataFrame in which each row is a filing unit and each column is an aspect of its tax burden under current law.",
"_____no_output_____"
]
],
[
[
"b.create_table()",
"_____no_output_____"
]
],
[
[
"## Analyzing tax data under a policy reform\n\nTo analyze a policy reform, specify the `reform_file` argument in the `create_table()` method. You may enter the file path to a json reform file, a Python dictionary, or the URL to a reform in the [Tax-Calculator repository](https://github.com/PSLmodels/Tax-Calculator/tree/master/taxcalc/reforms). Make sure that your reform adheres to Tax-Calculator [guidelines](https://github.com/PSLmodels/Tax-Calculator/blob/master/taxcalc/reforms/REFORMS.md).",
"_____no_output_____"
]
],
[
[
"reform_path = os.path.join(CURRENT_PATH, \"../taxcrunch/tests/test_reform.json\")\nb.create_table(reform_file=reform_path)",
"_____no_output_____"
],
[
"# you can also create a difference table\nb.create_diff_table(reform_file=reform_path)",
"_____no_output_____"
]
],
[
[
"## Behavioral Responses\n\nFinally, the `Batch` class gives users the option to incorporate partial-equilibrium behavioral responses to a tax policy reform. The three optional arguments a dynamic analysis are:\n\n- `be_sub`: substitution elasticity of taxable income\n- `be_inc`: income elasticity of taxable income\n- `be_cg`: semi-elasticity of long-term capital gains\n\nThe implementation of these elasticities is carried out by the [Behavioral-Responses](https://github.com/PSLmodels/Behavioral-Responses) project. Further discussion on the definition of the elasticities can be found [here](https://github.com/PSLmodels/Behavioral-Responses/blob/master/behresp/behavior.py)",
"_____no_output_____"
]
],
[
[
"b.create_table(reform_file=reform_path, be_sub=0.25)",
"_____no_output_____"
],
[
"b.create_diff_table(reform_file=reform_path, be_sub=0.25)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
c5158db9045ad62f4f6505e5347baa81ccdcad76
| 94,197 |
ipynb
|
Jupyter Notebook
|
Ass 7&8.ipynb
|
Ann-ah/ADS-Assignment-7-8
|
ee9ba704d770ab5352acbc549499b7f72ce3016f
|
[
"MIT"
] | null | null | null |
Ass 7&8.ipynb
|
Ann-ah/ADS-Assignment-7-8
|
ee9ba704d770ab5352acbc549499b7f72ce3016f
|
[
"MIT"
] | null | null | null |
Ass 7&8.ipynb
|
Ann-ah/ADS-Assignment-7-8
|
ee9ba704d770ab5352acbc549499b7f72ce3016f
|
[
"MIT"
] | null | null | null | 57.227825 | 8,460 | 0.729291 |
[
[
[
"conda install pandas",
"Collecting package metadata (current_repodata.json): ...working... done\nSolving environment: ...working... done\n\n# All requested packages already installed.\n\n\nNote: you may need to restart the kernel to use updated packages.\n"
],
[
"conda install numpy",
"Collecting package metadata (current_repodata.json): ...working... done\nSolving environment: ...working... done\n\n# All requested packages already installed.\n\n\nNote: you may need to restart the kernel to use updated packages.\n"
],
[
"conda install matplotlib",
"Collecting package metadata (current_repodata.json): ...working... done\nNote: you may need to restart the kernel to use updated packages.\n\nSolving environment: ...working... done\n\n# All requested packages already installed.\n\n"
],
[
"pip install plotly",
"Requirement already satisfied: plotly in c:\\users\\admin\\anaconda3\\lib\\site-packages (5.1.0)\nRequirement already satisfied: tenacity>=6.2.0 in c:\\users\\admin\\anaconda3\\lib\\site-packages (from plotly) (8.0.1)\nRequirement already satisfied: six in c:\\users\\admin\\anaconda3\\lib\\site-packages (from plotly) (1.15.0)\nNote: you may need to restart the kernel to use updated packages.\n"
],
[
"import pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nimport plotly.express as px\nfrom scipy import stats\nimport warnings\n\nwarnings.filterwarnings(\"ignore\")\n",
"_____no_output_____"
],
[
"df = pd.read_csv(\"insurance.csv\")\ndf",
"_____no_output_____"
],
[
"df.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 1338 entries, 0 to 1337\nData columns (total 7 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 age 1338 non-null int64 \n 1 sex 1338 non-null object \n 2 bmi 1338 non-null float64\n 3 children 1338 non-null int64 \n 4 smoker 1338 non-null object \n 5 region 1338 non-null object \n 6 charges 1338 non-null float64\ndtypes: float64(2), int64(2), object(3)\nmemory usage: 57.6+ KB\n"
],
[
"df.shape",
"_____no_output_____"
],
[
"df.columns",
"_____no_output_____"
],
[
"df.describe()",
"_____no_output_____"
]
],
[
[
"1. Import the data set, ‘insurance’. The column ‘charges’ should be considered as your target label.",
"_____no_output_____"
]
],
[
[
"X_df = df.drop(\"charges\", axis=1)\nX_df.shape",
"_____no_output_____"
],
[
"y_df = df[\"charges\"]\ny_df.shape",
"_____no_output_____"
]
],
[
[
"2. Explore the data using at least 3 data exploratory tools of your choosing in pandas and interpret your observation in a markdown cell of what form of predictive analysis that can be conducted on the data.",
"_____no_output_____"
]
],
[
[
"#Total charges of insuarance\ndf[\"charges\"].sum()",
"_____no_output_____"
],
[
"#best region in terms of insuarance sales\nbest_region = df.groupby([\"region\"]).sum().sort_values(by = \"charges\")\nbest_region",
"_____no_output_____"
]
],
[
[
"3. Visualize the age distribution for the column ‘age’ and comment on the results in a markdown cell as well. (Ensure your visualization is of an appropriate size for effective analysis)",
"_____no_output_____"
]
],
[
[
"plt.hist(df[\"age\"], bins = 50, histtype = \"bar\", rwidth = 0.5)\nplt.title (\"Visualisation of age\")\nplt.show()",
"_____no_output_____"
]
],
[
[
"Majority of the people recorded, were below the age of 20. Above 20years, there was a balance in the recorded ages.",
"_____no_output_____"
],
[
"4. Isolate all the continuous and discrete columns into their respective lists named ‘numerical_continuous’ and ‘numerical_discrete’ respectively.",
"_____no_output_____"
]
],
[
[
"df.nunique()",
"_____no_output_____"
],
[
"numerical_continuous = []\nfor column in df.columns:\n if df[column].dtypes != \"object\":\n if df[column].nunique() >= 10:\n numerical_continuous.append(column)\nnumerical_continuous.remove(\"charges\")\nnumerical_continuous",
"_____no_output_____"
],
[
"numerical_discreet = []\nfor column in df.columns:\n if df[column].dtypes != \"object\":\n if df[column].nunique() < 10:\n numerical_discreet.append(column)\nnumerical_discreet",
"_____no_output_____"
]
],
[
[
"5. Visually identify if there is presence of any outliers in the numerical_continuous columns and resolve them using a zscore test and a threshold of your choosing.",
"_____no_output_____"
]
],
[
[
"sns.boxplot(data = df[numerical_continuous], orient = \"v\", palette = \"Oranges\")",
"_____no_output_____"
],
[
"threshold = 0.375\nzscore = np.abs(stats.zscore(df[[\"bmi\"]]))\ndf[(zscore > threshold).all(axis = 1)][numerical_continuous].plot(kind = \"box\", figsize = (10,5))",
"_____no_output_____"
]
],
[
[
"6. Validate that your analysis above was successful by visualizing the value distribution in the resulting columns using an appropriate visualization method.",
"_____no_output_____"
]
],
[
[
"df = df[(zscore > threshold).all(axis = 1)]\ndf",
"_____no_output_____"
],
[
"plt.hist(df[numerical_continuous], bins = 15, rwidth = 0.5)\nplt.show()",
"_____no_output_____"
]
],
[
[
"7. Isolate all the categorical column names into a list named ‘categorical’",
"_____no_output_____"
]
],
[
[
"categorical = []\nfor column in df.columns:\n if df[column].dtypes == \"object\":\n categorical.append(column)\ncategorical",
"_____no_output_____"
]
],
[
[
"8. Visually identify the outliers in the discrete and categorical features and resolve them using the combined rare levels method.",
"_____no_output_____"
]
],
[
[
"sns.boxplot(data = df[numerical_discreet], orient = \"v\", palette = \"Oranges\")",
"_____no_output_____"
],
[
"for column in numerical_discreet + categorical:\n (df[column].value_counts()/df.shape[0]).plot(kind = \"bar\")\n plt.title(column)\n plt.show()\n",
"_____no_output_____"
],
[
"df[\"children\"] = df[\"children\"].replace([3,4,5], \"Rare\")\ndf[\"children\"]",
"_____no_output_____"
]
],
[
[
"9. Encode the discrete and categorical features with one of the measures of central tendency of your choosing.",
"_____no_output_____"
]
],
[
[
"#mode\n#median\n#mean\n\nencoded_features = {}\nfor column in numerical_discreet + categorical:\n encoded_features[column] = df.groupby([column])[\"charges\"].median().to_dict()\n df[column] = df[column].map(encoded_features[column])\n \n \n",
"_____no_output_____"
]
],
[
[
"10. Separate your features from the target appropriately. Narrow down the number of features to 5 using the most appropriate and accurate method. Which feature had to be dropped and what inference would you give as the main contributor of dropping the given feature.",
"_____no_output_____"
]
],
[
[
"X = df.drop(\"charges\", axis =1)\ny = df[\"charges\"]",
"_____no_output_____"
],
[
"from sklearn.linear_model import LinearRegression\nfrom sklearn.feature_selection import RFE\nfrom sklearn.model_selection import train_test_split\nmodel = LinearRegression()\nrfe = RFE(model, 5)\nX_rfe = rfe.fit_transform(X, y) \nmodel.fit(X_rfe, y)\nprint(pd.Series(rfe.support_, index = X.columns))",
"age True\nsex True\nbmi True\nchildren False\nsmoker True\nregion True\ndtype: bool\n"
]
],
[
[
"8) 1.Convert the target labels to their respective log values and give 2 reasons why this step may be useful as we train the machine learning model. (Explain in a markdown cell.)",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import train_test_split, cross_val_score\nfrom sklearn.preprocessing import StandardScaler\nfrom sklearn.metrics import mean_squared_error, mean_absolute_error, r2_score\nfrom sklearn.linear_model import ElasticNet\nfrom sklearn.tree import DecisionTreeRegressor\nfrom sklearn.ensemble import BaggingRegressor, AdaBoostRegressor",
"_____no_output_____"
],
[
"y_log = np.log(y)\ny_log",
"_____no_output_____"
]
],
[
[
"Handles any outliers in the target coumn.",
"_____no_output_____"
],
[
"8) 2.Slice the selected feature columns and the labels into the training and testing set. Also ensure your features are normalized.",
"_____no_output_____"
]
],
[
[
"X_train, X_test, y_train, y_test = train_test_split(X_rfe, y_log, test_size = 0.2, random_state = 0)",
"_____no_output_____"
]
],
[
[
"8) 3.Use at least 4 different regression based machine learning methods and use the training and testing cross accuracy and divergence to identify the best model",
"_____no_output_____"
]
],
[
[
"scaler = StandardScaler()\n\nX_train = scaler.fit_transform(X_train)\nX_test = scaler.transform(X_test)",
"_____no_output_____"
],
[
"regular_reg = ElasticNet()\ndt_reg = DecisionTreeRegressor(random_state = 0)\nbag_reg = BaggingRegressor(random_state = 0) \nboost_reg = AdaBoostRegressor(random_state = 0)\n\nmodels = {'ElasticNet': regular_reg, 'DecisionTreeRegressor': dt_reg, 'BaggingRegressor': bag_reg, \n 'AdaBoostRegressor': boost_reg}",
"_____no_output_____"
],
[
"def cross_valid(models, X, y, process = 'Training'):\n print(f'Process: {process}')\n for model_name, model in models.items():\n scores = cross_val_score(model, X, y, cv = 5)\n print(f'Model: {model_name}')\n print(f'Cross validation mean score: {round(np.mean(scores), 4)}')\n print(f'Cross validation deviation: {round(np.std(scores), 4)}')\n print('\\n')",
"_____no_output_____"
],
[
"cross_valid(models, X_train, y_train, process = 'Training')",
"Process: Training\nModel: ElasticNet\nCross validation mean score: 0.1033\nCross validation deviation: 0.0263\n\n\nModel: DecisionTreeRegressor\nCross validation mean score: 0.603\nCross validation deviation: 0.039\n\n\nModel: BaggingRegressor\nCross validation mean score: 0.7559\nCross validation deviation: 0.0252\n\n\nModel: AdaBoostRegressor\nCross validation mean score: 0.7403\nCross validation deviation: 0.0312\n\n\n"
],
[
"cross_valid(models, X_test, y_test, process = 'Testing')",
"Process: Testing\nModel: ElasticNet\nCross validation mean score: 0.0684\nCross validation deviation: 0.015\n\n\nModel: DecisionTreeRegressor\nCross validation mean score: 0.5241\nCross validation deviation: 0.2098\n\n\nModel: BaggingRegressor\nCross validation mean score: 0.6946\nCross validation deviation: 0.1292\n\n\nModel: AdaBoostRegressor\nCross validation mean score: 0.6887\nCross validation deviation: 0.1018\n\n\n"
]
],
[
[
"8) 4. After identifying the best model, train it with the training data again. Using at least 3 model evaluation metrics in regression, evaluate the models training and testing score. Also ensure as you test the models, the predicted and actual targets have been converted back to the original values using antilog. (Hint: Antilog function is equal to Exponential)",
"_____no_output_____"
]
],
[
[
"bag_reg.fit(X_train, y_train)",
"_____no_output_____"
],
[
"def model_evaluation(model, X, y):\n y_predict = np.exp(model.predict(X))\n y = np.exp(y)\n print(f'Mean Squared Error: {mean_squared_error(y, y_predict)}')\n print(f'Mean Absolute Error: {mean_absolute_error(y, y_predict)}')\n print(f'R2 Score: {r2_score(y, y_predict)}')",
"_____no_output_____"
],
[
"model_evaluation(bag_reg, X_train, y_train)",
"Mean Squared Error: 6516459.587248233\nMean Absolute Error: 1136.6169348240364\nR2 Score: 0.9563185771898398\n"
],
[
"model_evaluation(bag_reg, X_test, y_test)",
"Mean Squared Error: 18879664.249590866\nMean Absolute Error: 2302.334042744642\nR2 Score: 0.867393735541934\n"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
c51591b6ebeb71b96a536f536baf56f9aea78849
| 223,877 |
ipynb
|
Jupyter Notebook
|
Figure1_performance_analysis.ipynb
|
p-koo/residualbind
|
0af8d81c49d535a891474929016a871a27665625
|
[
"MIT"
] | 8 |
2020-08-29T15:53:18.000Z
|
2022-03-15T00:16:31.000Z
|
Figure1_performance_analysis.ipynb
|
p-koo/residualbind
|
0af8d81c49d535a891474929016a871a27665625
|
[
"MIT"
] | 1 |
2021-05-18T07:58:16.000Z
|
2021-05-18T07:58:16.000Z
|
Figure1_performance_analysis.ipynb
|
p-koo/residualbind
|
0af8d81c49d535a891474929016a871a27665625
|
[
"MIT"
] | 3 |
2020-08-21T00:07:58.000Z
|
2021-06-01T18:40:31.000Z
| 329.715758 | 76,724 | 0.928179 |
[
[
[
"import os\nimport numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\n%matplotlib inline",
"_____no_output_____"
],
[
"data_path = '../results/results.csv'\ndf = pd.read_csv(data_path, delimiter='\\t')\nray = df['Ray_et_al'].to_numpy()\nmatrixreduce = df['MatrixREDUCE'].to_numpy()\nrnacontext = df['RNAcontext'].to_numpy()\ndeepbind = df['DeepBind'].to_numpy()\ndlprb = df['DLPRB'].to_numpy()\nrck = df['RCK'].to_numpy()\ncdeepbind = df['cDeepbind'].to_numpy()\nthermonet = df['ThermoNet'].to_numpy()\nresidualbind = df['ResidualBind'].to_numpy()",
"_____no_output_____"
]
],
[
[
"# Plot box-violin plot",
"_____no_output_____"
]
],
[
[
"names = ['Ray et al.', 'MatrixREDUCE', 'RNAcontext', 'DeepBind', 'DLPRB', 'RCK', 'cDeepbind', 'ThermoNet', 'ResidualBind']\ndata = [ray, matrixreduce, rnacontext, deepbind, rck, dlprb, cdeepbind, thermonet, residualbind]\n\nfig = plt.figure(figsize=(12,5))\nvplot = plt.violinplot(data, \n showextrema=False);\n\ndata = [ray, matrixreduce, rnacontext, deepbind, rck, dlprb, cdeepbind, thermonet, residualbind]\n\n\nimport matplotlib.cm as cm\ncmap = cm.ScalarMappable(cmap='tab10')\ntest_mean = [0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, 0.9]\n\n\n \nfor patch, color in zip(vplot['bodies'], cmap.to_rgba(test_mean)):\n patch.set_facecolor(color)\n patch.set_edgecolor('black')\n \n\nmedianprops = dict(color=\"red\",linewidth=2)\n \nbplot = plt.boxplot(data, \n notch=True, patch_artist=True, \n widths=0.2,\n medianprops=medianprops);\n\nfor patch, color in zip(bplot['boxes'], cmap.to_rgba(test_mean)):\n patch.set_facecolor(color)\n patch.set_edgecolor('black')\n #patch.set(color=colors[i])\nplt.xticks(range(1,len(names)+1), names, rotation=40, fontsize=14, ha='right');\nax = plt.gca();\nplt.setp(ax.get_yticklabels(),fontsize=14)\nplt.ylabel('Pearson correlation', fontsize=14);\n\n\nplot_path = '../results/rnacompete_2013/'\noutfile = os.path.join(plot_path, 'Performance_comparison.pdf')\nfig.savefig(outfile, format='pdf', dpi=200, bbox_inches='tight')",
"_____no_output_____"
]
],
[
[
"# plot comparison between ResidualBind and ThermoNet",
"_____no_output_____"
]
],
[
[
"\nfig = plt.figure(figsize=(3,3))\nax = plt.subplot(111)\nplt.hist(residualbind-thermonet, bins=20);\n\nplt.setp(ax.get_yticklabels(),fontsize=14)\nplt.ylabel('Counts', fontsize=14);\nplt.setp(ax.get_xticklabels(),fontsize=14)\nplt.xlabel('$\\Delta$ Pearson r', fontsize=14);\n\n\nplot_path = '../results/rnacompete_2013/'\noutfile = os.path.join(plot_path, 'Performance_comparison_hist.pdf')\nfig.savefig(outfile, format='pdf', dpi=200, bbox_inches='tight')\n",
"_____no_output_____"
],
[
"from scipy import stats\nstats.wilcoxon(residualbind, thermonet)",
"_____no_output_____"
]
],
[
[
"# Compare performance based on binding score normalization and different input features",
"_____no_output_____"
]
],
[
[
"data_path = '../results/rnacompete_2013/clip_norm_seq_performance.tsv'\ndf = pd.read_csv(data_path, delimiter='\\t')\nclip_norm_seq = df['Pearson score'].to_numpy()\n\ndata_path = '../results/rnacompete_2013/clip_norm_pu_performance.tsv'\ndf = pd.read_csv(data_path, delimiter='\\t')\nclip_norm_pu = df['Pearson score'].to_numpy()\n\ndata_path = '../results/rnacompete_2013/clip_norm_struct_performance.tsv'\ndf = pd.read_csv(data_path, delimiter='\\t')\nclip_norm_struct = df['Pearson score'].to_numpy()\n\ndata_path = '../results/rnacompete_2013/log_norm_seq_performance.tsv'\ndf = pd.read_csv(data_path, delimiter='\\t')\nlog_norm_seq = df['Pearson score'].to_numpy()\n\ndata_path = '../results/rnacompete_2013/log_norm_pu_performance.tsv'\ndf = pd.read_csv(data_path, delimiter='\\t')\nlog_norm_pu = df['Pearson score'].to_numpy()\n\ndata_path = '../results/rnacompete_2013/log_norm_struct_performance.tsv'\ndf = pd.read_csv(data_path, delimiter='\\t')\nlog_norm_struct = df['Pearson score'].to_numpy()",
"_____no_output_____"
],
[
"names = ['Clip-norm', 'Log-norm']\ndata = [clip_norm_seq, log_norm_seq]\n\nfig = plt.figure(figsize=(3,3))\nvplot = plt.violinplot(data, \n showextrema=False);\n\n\nimport matplotlib.cm as cm\ncmap = cm.ScalarMappable(cmap='viridis')\ntest_mean = [0.1, 0.5, 0.9]\n\n \nfor patch, color in zip(vplot['bodies'], cmap.to_rgba(test_mean)):\n patch.set_facecolor(color)\n patch.set_edgecolor('black')\n \n\nmedianprops = dict(color=\"red\",linewidth=2)\n \nbplot = plt.boxplot(data, \n notch=True, patch_artist=True, \n widths=0.2,\n medianprops=medianprops);\n\nfor patch, color in zip(bplot['boxes'], cmap.to_rgba(test_mean)):\n patch.set_facecolor(color)\n patch.set_edgecolor('black')\n #patch.set(color=colors[i])\nplt.xticks(range(1,len(names)+1), names, rotation=40, fontsize=14, ha='right');\nax = plt.gca();\nplt.setp(ax.get_yticklabels(),fontsize=14)\nplt.ylabel('Pearson correlation', fontsize=14);\n\n\nplot_path = '../results/rnacompete_2013/'\noutfile = os.path.join(plot_path, 'Performance_comparison_clip_vs_log.pdf')\nfig.savefig(outfile, format='pdf', dpi=200, bbox_inches='tight')",
"_____no_output_____"
],
[
"names = ['Sequence', 'Sequence + PU', 'Sequence + PHIME']\ndata = [clip_norm_seq, clip_norm_pu, clip_norm_struct]\n\nfig = plt.figure(figsize=(5,5))\nvplot = plt.violinplot(data, \n showextrema=False);\n\n\nimport matplotlib.cm as cm\ncmap = cm.ScalarMappable(cmap='viridis')\ntest_mean = [0.1, 0.5, 0.9]\n\n \nfor patch, color in zip(vplot['bodies'], cmap.to_rgba(test_mean)):\n patch.set_facecolor(color)\n patch.set_edgecolor('black')\n \n\nmedianprops = dict(color=\"red\",linewidth=2)\n \nbplot = plt.boxplot(data, \n notch=True, patch_artist=True, \n widths=0.2,\n medianprops=medianprops);\n\nfor patch, color in zip(bplot['boxes'], cmap.to_rgba(test_mean)):\n patch.set_facecolor(color)\n patch.set_edgecolor('black')\n #patch.set(color=colors[i])\nplt.xticks(range(1,len(names)+1), names, rotation=40, fontsize=14, ha='right');\nax = plt.gca();\nplt.setp(ax.get_yticklabels(),fontsize=14)\nplt.ylabel('Pearson correlation', fontsize=14);\n\n\nplot_path = '../results/rnacompete_2013/'\noutfile = os.path.join(plot_path, 'Performance_comparison_clip_structure.pdf')\nfig.savefig(outfile, format='pdf', dpi=200, bbox_inches='tight')",
"_____no_output_____"
],
[
"names = ['Sequence', 'Sequence + PU', 'Sequence + PHIME']\ndata = [log_norm_seq, log_norm_pu, log_norm_struct]\n\nfig = plt.figure(figsize=(5,3))\nvplot = plt.violinplot(data, \n showextrema=False);\n\n\nimport matplotlib.cm as cm\ncmap = cm.ScalarMappable(cmap='viridis')\ntest_mean = [0.1, 0.5, 0.9]\n\n \nfor patch, color in zip(vplot['bodies'], cmap.to_rgba(test_mean)):\n patch.set_facecolor(color)\n patch.set_edgecolor('black')\n \n\nmedianprops = dict(color=\"red\",linewidth=2)\n \nbplot = plt.boxplot(data, \n notch=True, patch_artist=True, \n widths=0.2,\n medianprops=medianprops);\n\nfor patch, color in zip(bplot['boxes'], cmap.to_rgba(test_mean)):\n patch.set_facecolor(color)\n patch.set_edgecolor('black')\n #patch.set(color=colors[i])\nplt.xticks(range(1,len(names)+1), names, rotation=40, fontsize=14, ha='right');\nax = plt.gca();\nplt.setp(ax.get_yticklabels(),fontsize=14)\nplt.ylabel('Pearson correlation', fontsize=14);\n\n\nplot_path = '../results/rnacompete_2013/'\noutfile = os.path.join(plot_path, 'Performance_comparison_log_structure.pdf')\nfig.savefig(outfile, format='pdf', dpi=200, bbox_inches='tight')",
"_____no_output_____"
],
[
"data = [clip_norm_seq, clip_norm_pu, clip_norm_struct, log_norm_seq, log_norm_pu, log_norm_struct]\nname = ['clip_norm_seq', 'clip_norm_pu', 'clip_norm_struct', 'log_norm_seq', 'log_norm_pu', 'log_norm_struct']\n\nfor n,x in zip(name, data):\n print(n, np.mean(x), np.std(x))\n",
"clip_norm_seq 0.688619262295082 0.16975689898179339\nclip_norm_pu 0.690047131147541 0.17045535408919055\nclip_norm_struct 0.6886290983606558 0.17097206323661115\nlog_norm_seq 0.6862725609756097 0.17848520377502483\nlog_norm_pu 0.6837483606557376 0.18308362249306023\nlog_norm_struct 0.6827598360655739 0.1832164039652027\n"
]
],
[
[
"# compare PHIME vs seq only",
"_____no_output_____"
]
],
[
[
"fig = plt.figure(figsize=(3,3))\nax = plt.subplot(111)\nplt.hist(clip_norm_seq-clip_norm_struct, bins=15)\n\nplt.setp(ax.get_yticklabels(),fontsize=14)\nplt.ylabel('Counts', fontsize=14);\nplt.setp(ax.get_xticklabels(),fontsize=14)\nplt.xlabel('$\\Delta$ Pearson r', fontsize=14);\n\n\nplot_path = '../results/rnacompete_2013/'\noutfile = os.path.join(plot_path, 'Performance_comparison_hist_seq_vs_struct.pdf')\nfig.savefig(outfile, format='pdf', dpi=200, bbox_inches='tight')",
"_____no_output_____"
]
],
[
[
"# 2009 RNAcompete analysis",
"_____no_output_____"
]
],
[
[
"data_path = '../results/rnacompete_2009/log_norm_seq_performance.tsv'\ndf = pd.read_csv(data_path, delimiter='\\t')\nlog_norm_seq = df['Pearson score'].to_numpy()\n\ndata_path = '../results/rnacompete_2009/log_norm_pu_performance.tsv'\ndf = pd.read_csv(data_path, delimiter='\\t')\nlog_norm_pu = df['Pearson score'].to_numpy()\n",
"_____no_output_____"
],
[
"names = ['Sequence', 'Sequence + PU']\ndata = [log_norm_seq, log_norm_pu]\n\nfig = plt.figure(figsize=(5,5))\nvplot = plt.violinplot(data, \n showextrema=False);\n\n\nimport matplotlib.cm as cm\ncmap = cm.ScalarMappable(cmap='viridis')\ntest_mean = [0.1, 0.5, 0.9]\n\n \nfor patch, color in zip(vplot['bodies'], cmap.to_rgba(test_mean)):\n patch.set_facecolor(color)\n patch.set_edgecolor('black')\n \n\nmedianprops = dict(color=\"red\",linewidth=2)\n \nbplot = plt.boxplot(data, \n notch=True, patch_artist=True, \n widths=0.2,\n medianprops=medianprops);\n\nfor patch, color in zip(bplot['boxes'], cmap.to_rgba(test_mean)):\n patch.set_facecolor(color)\n patch.set_edgecolor('black')\n #patch.set(color=colors[i])\nplt.xticks(range(1,len(names)+1), names, rotation=40, fontsize=14, ha='right');\nax = plt.gca();\nplt.setp(ax.get_yticklabels(),fontsize=14)\nplt.ylabel('Pearson correlation', fontsize=14);\n\n\nplot_path = '../results/rnacompete_2013/'\noutfile = os.path.join(plot_path, 'Performance_comparison_log_structure_2009.pdf')\nfig.savefig(outfile, format='pdf', dpi=200, bbox_inches='tight')",
"_____no_output_____"
]
],
[
[
"# Compare log vs clip as a scatter plot",
"_____no_output_____"
]
],
[
[
"\ndata_path = '../data/RNAcompete_2013/rnacompete2013.h5'\nresults_path = helper.make_directory('../results', 'rnacompete_2013')\n\nexperiment = 'RNCMPT00169'\nrbp_index = helper.find_experiment_index(data_path, experiment)\n",
"_____no_output_____"
],
[
"normalization = 'clip_norm' # 'log_norm' or 'clip_norm'\nss_type = 'seq' # 'seq', 'pu', or 'struct'\nsave_path = helper.make_directory(results_path, normalization+'_'+ss_type)\n\n# load rbp dataset\ntrain, valid, test = helper.load_rnacompete_data(data_path, \n ss_type=ss_type, \n normalization=normalization, \n rbp_index=rbp_index)\n\n# load residualbind model\ninput_shape = list(train['inputs'].shape)[1:]\nweights_path = os.path.join(save_path, experiment + '_weights.hdf5') \nmodel = ResidualBind(input_shape, weights_path)\n\n# load pretrained weights\nmodel.load_weights()\n\n# get predictions for test sequences\npredictions_clip = model.predict(test['inputs'])\ny = test['targets']\n\n\nfig = plt.figure(figsize=(3,3))\nplt.scatter(predictions_clip, y, alpha=0.5, rasterized=True)\nplt.plot([-2,9],[-2,9],'--k')\nplt.xlabel('Predicted binding scores', fontsize=14)\nplt.ylabel('Experimental binding scores', fontsize=14)\nplt.xticks([-2, 0, 2, 4, 6, 8], fontsize=14)\nplt.yticks([-2, 0, 2, 4, 6, 8], fontsize=14)\noutfile = os.path.join(results_path, experiment+'_scatter_clip.pdf')\nfig.savefig(outfile, format='pdf', dpi=600, bbox_inches='tight')\n",
"_____no_output_____"
],
[
"normalization = 'log_norm' # 'log_norm' or 'clip_norm'\nss_type = 'seq' # 'seq', 'pu', or 'struct'\nsave_path = helper.make_directory(results_path, normalization+'_'+ss_type)\n\n# load rbp dataset\ntrain, valid, test = helper.load_rnacompete_data(data_path, \n ss_type=ss_type, \n normalization=normalization, \n rbp_index=rbp_index)\n\n# load residualbind model\ninput_shape = list(train['inputs'].shape)[1:]\nweights_path = os.path.join(save_path, experiment + '_weights.hdf5') \nmodel = ResidualBind(input_shape, weights_path)\n\n# load pretrained weights\nmodel.load_weights()\n\n# get predictions for test sequences\npredictions_log = model.predict(test['inputs'])\n\ny2 = test['targets']\n\n\nfig = plt.figure(figsize=(3,3))\nplt.scatter(predictions_log, y2, alpha=0.5, rasterized=True)\nplt.plot([-2,9],[-2,9],'--k')\nplt.xlabel('Predicted binding scores', fontsize=14)\nplt.ylabel('Experimental binding scores', fontsize=14)\nplt.xticks([-2, 0, 2, 4, 6, 8,], fontsize=14)\nplt.yticks([-2, 0, 2, 4, 6, 8], fontsize=14)\noutfile = os.path.join(results_path, experiment+'_scatter_log.pdf')\nfig.savefig(outfile, format='pdf', dpi=600, bbox_inches='tight')\n\n",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
c515947dc457db39062eb16a4af8a3b718d46a3c
| 142,808 |
ipynb
|
Jupyter Notebook
|
project/reports/fisheries_monitoring/4.Fish_classificator.ipynb
|
vinceHardy/learning
|
941e5979d471567411e7593c36617ef4a8e47f70
|
[
"MIT"
] | 1 |
2019-11-05T06:17:40.000Z
|
2019-11-05T06:17:40.000Z
|
project/reports/fisheries_monitoring/4.Fish_classificator.ipynb
|
vinceHardy/learning
|
941e5979d471567411e7593c36617ef4a8e47f70
|
[
"MIT"
] | null | null | null |
project/reports/fisheries_monitoring/4.Fish_classificator.ipynb
|
vinceHardy/learning
|
941e5979d471567411e7593c36617ef4a8e47f70
|
[
"MIT"
] | null | null | null | 408.022857 | 126,254 | 0.511876 |
[
[
[
"## Fish classification\n\nIn this notebook the fish classification is done. We are going to classify in four classes: Tuna fish (TUNA), LAG, DOL and SHARK. The detector will save the cropped image of a fish. Here we will take this image and we will use a CNN to classify it.\n\nIn the original Kaggle competition there are six classes of fish: ALB, BET, YFT, DOL, LAG and SHARK. We started trying to classify them all, but three of them are vey similar: ALB, BET and YFT. In fact, they are all different tuna species, while the other fishes come from different families. Therefore, the classification of those species was difficult and the results were not too good. We will make a small comparison of both on the presentation, but here we will only upload the clsifier with four classes.\n",
"_____no_output_____"
]
],
[
[
"from PIL import Image\nimport tensorflow as tf\nimport numpy as np\nimport scipy\nimport os\nimport cv2\nfrom sklearn.preprocessing import LabelEncoder\nfrom sklearn.metrics import log_loss\nfrom keras.utils import np_utils\nfrom keras.models import Sequential\nfrom keras.layers.convolutional import Convolution2D\nfrom keras.layers.convolutional import MaxPooling2D\nfrom keras.layers.core import Activation\nfrom keras.layers.core import Flatten\nfrom keras.layers.core import Dense\nfrom keras.layers.core import Dropout\nfrom keras import backend as K\nimport matplotlib.pyplot as plt",
"Using TensorFlow backend.\n"
],
[
"#Define some values and constants\nfish_classes = ['TUNA','DOL','SHARK','LAG']\nfish_classes_test = fish_classes\nnumber_classes = len(fish_classes)\nmain_path_train = '../train_cut_oversample'\nmain_path_test = '../test'\nchannels = 3\nROWS_RESIZE = 100\nCOLS_RESIZE = 100",
"_____no_output_____"
]
],
[
[
"Now we read the data from the file where the fish detection part has stored the images.\n\nWe also preprocess slightly the images to convert them to the same size (100x100). The aspect ratio of the images is important, so instead of just resizing the image, we have created the function resize(im). This function takes an image and resizes its longest side to 100, keeping the aspect ratio. In other words, the short side of the image will be smaller than 100 poixels. This image is pasted onto the middle of a white layer that is 100x100. So, our image will have white pixels on two of its sides. This is not optimum, but it is still better than changing the aspect ratio. We have also tried with other colors, but the best results were achieved with white.",
"_____no_output_____"
]
],
[
[
"# Get data and preproccess it\n\ndef resize(image):\n rows = image.shape[0]\n cols = image.shape[1]\n dominant = max(rows,cols)\n ratio = ROWS_RESIZE/float(dominant)\n im_res = scipy.misc.imresize(image,ratio)\n rows = im_res.shape[0]\n cols = im_res.shape[1]\n im_res = Image.fromarray(im_res)\n layer = Image.new('RGB',[ROWS_RESIZE,COLS_RESIZE],(255,255,255))\n if rows > cols:\n layer.paste(im_res,(COLS_RESIZE/2-cols/2,0))\n if cols > rows:\n layer.paste(im_res,(0,ROWS_RESIZE/2-rows/2))\n if rows == cols:\n layer.paste(im_res,(0,0)) \n return np.array(layer)\n\n\nX_train = []\ny_labels = []\nfor classes in fish_classes:\n path_class = os.path.join(main_path_train,classes)\n y_class = np.tile(classes,len(os.listdir(path_class)))\n y_labels.extend(y_class)\n for image in os.listdir(path_class):\n path = os.path.join(path_class,image)\n im = scipy.misc.imread(path)\n im = resize(im)\n X_train.append(np.array(im))\n \nX_train = np.array(X_train)\n\n# Convert labels into one hot vectors\ny_labels = LabelEncoder().fit_transform(y_labels)\ny_train = np_utils.to_categorical(y_labels)\n\n\nX_test = []\ny_test = []\nfor classes in fish_classes_test:\n path_class = os.path.join(main_path_test,classes)\n y_class = np.tile(classes,len(os.listdir(path_class)))\n y_test.extend(y_class)\n for image in os.listdir(path_class):\n path = os.path.join(path_class,image)\n im = scipy.misc.imread(path)\n im = resize(im)\n X_test.append(np.array(im))\n \nX_test = np.array(X_test)\n\n# Convert labels into one hot vectors\ny_test = LabelEncoder().fit_transform(y_test)\ny_test = np_utils.to_categorical(y_test)\n\n\n\nX_train = np.reshape(X_train,(X_train.shape[0],ROWS_RESIZE,COLS_RESIZE,channels))\nX_test = np.reshape(X_test,(X_test.shape[0],ROWS_RESIZE,COLS_RESIZE,channels))\nprint('X_train shape: ',X_train.shape)\nprint('y_train shape: ',y_train.shape)\nprint('X_test shape: ',X_test.shape)\nprint('y_test shape: ',y_test.shape)\n\n\n\n",
"('X_train shape: ', (23581, 100, 100, 3))\n('y_train shape: ', (23581, 4))\n('X_test shape: ', (400, 100, 100, 3))\n('y_test shape: ', (400, 4))\n"
]
],
[
[
"The data is now organized in the following way:\n\n-The training has been done with 23581 images of size 100x100x3 (rgb).\n\n-There are 4 possible classes: LAG, SHARK, DOL and TUNA.\n\n-The test has been done with 400 images of the same size, 100 per class.\n\n\nWe are now ready to build and train the classifier. Th CNN has 7 convolutional layers, 4 pooling layers and three fully connected layers at the end. Dropout has been used in the fully connected layers to avoid overfitting. The loss function used is multi class logloss because is the one used by Kaggle in the competition. The optimizeer is gradient descent.",
"_____no_output_____"
]
],
[
[
"def center_normalize(x):\n return (x-K.mean(x))/K.std(x)\n# Convolutional net\n\nmodel = Sequential()\n\nmodel.add(Activation(activation=center_normalize,input_shape=(ROWS_RESIZE,COLS_RESIZE,channels)))\n\nmodel.add(Convolution2D(6,20,20,border_mode='same',activation='relu',dim_ordering='tf'))\nmodel.add(MaxPooling2D(pool_size=(2,2),dim_ordering='tf'))\n\nmodel.add(Convolution2D(12,10,10,border_mode='same',activation='relu',dim_ordering='tf'))\nmodel.add(Convolution2D(12,10,10,border_mode='same',activation='relu',dim_ordering='tf'))\nmodel.add(MaxPooling2D(pool_size=(2,2),dim_ordering='tf'))\n\nmodel.add(Convolution2D(24,5,5,border_mode='same',activation='relu',dim_ordering='tf'))\nmodel.add(Convolution2D(24,5,5,border_mode='same',activation='relu',dim_ordering='tf'))\nmodel.add(MaxPooling2D(pool_size=(2,2),dim_ordering='tf'))\n\nmodel.add(Convolution2D(24,5,5,border_mode='same',activation='relu',dim_ordering='tf'))\nmodel.add(Convolution2D(24,5,5,border_mode='same',activation='relu',dim_ordering='tf'))\nmodel.add(MaxPooling2D(pool_size=(2,2),dim_ordering='tf'))\n\nmodel.add(Flatten())\nmodel.add(Dense(4092,activation='relu'))\nmodel.add(Dropout(0.5))\nmodel.add(Dense(1024,activation='relu'))\nmodel.add(Dropout(0.5))\n\nmodel.add(Dense(number_classes))\nmodel.add(Activation('softmax'))\n\nprint(model.summary())\n\nmodel.compile(optimizer='sgd',loss='categorical_crossentropy',metrics=['accuracy'])\nmodel.fit(X_train,y_train,nb_epoch=1,verbose=1)\n",
"____________________________________________________________________________________________________\nLayer (type) Output Shape Param # Connected to \n====================================================================================================\nactivation_1 (Activation) (None, 100, 100, 3) 0 activation_input_1[0][0] \n____________________________________________________________________________________________________\nconvolution2d_1 (Convolution2D) (None, 100, 100, 6) 7206 activation_1[0][0] \n____________________________________________________________________________________________________\nmaxpooling2d_1 (MaxPooling2D) (None, 50, 50, 6) 0 convolution2d_1[0][0] \n____________________________________________________________________________________________________\nconvolution2d_2 (Convolution2D) (None, 50, 50, 12) 7212 maxpooling2d_1[0][0] \n____________________________________________________________________________________________________\nconvolution2d_3 (Convolution2D) (None, 50, 50, 12) 14412 convolution2d_2[0][0] \n____________________________________________________________________________________________________\nmaxpooling2d_2 (MaxPooling2D) (None, 25, 25, 12) 0 convolution2d_3[0][0] \n____________________________________________________________________________________________________\nconvolution2d_4 (Convolution2D) (None, 25, 25, 24) 7224 maxpooling2d_2[0][0] \n____________________________________________________________________________________________________\nconvolution2d_5 (Convolution2D) (None, 25, 25, 24) 14424 convolution2d_4[0][0] \n____________________________________________________________________________________________________\nmaxpooling2d_3 (MaxPooling2D) (None, 12, 12, 24) 0 convolution2d_5[0][0] \n____________________________________________________________________________________________________\nconvolution2d_6 (Convolution2D) (None, 12, 12, 24) 14424 maxpooling2d_3[0][0] \n____________________________________________________________________________________________________\nconvolution2d_7 (Convolution2D) (None, 12, 12, 24) 14424 convolution2d_6[0][0] \n____________________________________________________________________________________________________\nmaxpooling2d_4 (MaxPooling2D) (None, 6, 6, 24) 0 convolution2d_7[0][0] \n____________________________________________________________________________________________________\nflatten_1 (Flatten) (None, 864) 0 maxpooling2d_4[0][0] \n____________________________________________________________________________________________________\ndense_1 (Dense) (None, 4092) 3539580 flatten_1[0][0] \n____________________________________________________________________________________________________\ndropout_1 (Dropout) (None, 4092) 0 dense_1[0][0] \n____________________________________________________________________________________________________\ndense_2 (Dense) (None, 1024) 4191232 dropout_1[0][0] \n____________________________________________________________________________________________________\ndropout_2 (Dropout) (None, 1024) 0 dense_2[0][0] \n____________________________________________________________________________________________________\ndense_3 (Dense) (None, 4) 4100 dropout_2[0][0] \n____________________________________________________________________________________________________\nactivation_2 (Activation) (None, 4) 0 dense_3[0][0] \n====================================================================================================\nTotal params: 7,814,238\nTrainable params: 7,814,238\nNon-trainable params: 0\n____________________________________________________________________________________________________\nNone\nEpoch 1/1\n23581/23581 [==============================] - 3622s - loss: 1.0135 - acc: 0.5487 \b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\n"
]
],
[
[
"Since there are a lot of images the training takes around one hour. Once it is done we can pass the test set to the classifier and measure its accuracy.",
"_____no_output_____"
]
],
[
[
"(loss,accuracy) = model.evaluate(X_test,y_test,verbose=1)\nprint('accuracy',accuracy)\n",
"400/400 [==============================] - 22s \b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\n('accuracy', 0.69750000000000001)\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
c515969ca1efbea7979cd0e37e1876a43a2f3022
| 565,567 |
ipynb
|
Jupyter Notebook
|
notebooks/raw_notebooks/mfreilich_C_O2_heat_storage.ipynb
|
pedmondlerner/cmip6hack-ocean-bgc
|
217d30c2d50fc9895e4f8edd3d0bfb37a9649103
|
[
"MIT"
] | 2 |
2019-10-17T15:30:10.000Z
|
2020-02-12T15:50:19.000Z
|
notebooks/raw_notebooks/mfreilich_C_O2_heat_storage.ipynb
|
pedmondlerner/cmip6hack-ocean-bgc
|
217d30c2d50fc9895e4f8edd3d0bfb37a9649103
|
[
"MIT"
] | 13 |
2019-10-16T17:14:05.000Z
|
2020-03-04T14:42:03.000Z
|
notebooks/raw_notebooks/mfreilich_C_O2_heat_storage.ipynb
|
pedmondlerner/cmip6hack-ocean-bgc
|
217d30c2d50fc9895e4f8edd3d0bfb37a9649103
|
[
"MIT"
] | 13 |
2019-10-16T17:13:01.000Z
|
2019-10-18T15:39:53.000Z
| 725.085897 | 309,592 | 0.949304 |
[
[
[
"# Uptake of carbon, heat, and oxygen\n\nPlotting a global map of carbon, heat, and oxygen uptake",
"_____no_output_____"
]
],
[
[
"from dask.distributed import Client\n\nclient = Client(\"tcp://10.32.15.112:32829\")\nclient",
"_____no_output_____"
],
[
"%matplotlib inline\n\nimport xarray as xr\nimport intake\nimport numpy as np\nfrom cmip6_preprocessing.preprocessing import read_data\nfrom cmip6_preprocessing.parse_static_metrics import parse_static_thkcello\nfrom cmip6_preprocessing.preprocessing import rename_cmip6\nimport warnings\nimport matplotlib.pyplot as plt\n\n# util.py is in the local directory\n# it contains code that is common across project notebooks\n# or routines that are too extensive and might otherwise clutter\n# the notebook design\nimport util ",
"_____no_output_____"
],
[
"def _compute_slope(y):\n \"\"\"\n Private function to compute slopes at each grid cell using\n polyfit. \n \"\"\"\n x = np.arange(len(y))\n return np.polyfit(x, y, 1)[0] # return only the slope\n\ndef compute_slope(da):\n \"\"\"\n Computes linear slope (m) at each grid cell.\n \n Args:\n da: xarray DataArray to compute slopes for\n \n Returns:\n xarray DataArray with slopes computed at each grid cell.\n \"\"\"\n # apply_ufunc can apply a raw numpy function to a grid.\n # \n # vectorize is only needed for functions that aren't already\n # vectorized. You don't need it for polyfit in theory, but it's\n # good to use when using things like np.cov.\n #\n # dask='parallelized' parallelizes this across dask chunks. It requires\n # an output_dtypes of the numpy array datatype coming out.\n #\n # input_core_dims should pass the dimension that is being *reduced* by this operation,\n # if one is being reduced.\n slopes = xr.apply_ufunc(_compute_slope,\n da,\n vectorize=True,\n dask='parallelized', \n input_core_dims=[['time']],\n output_dtypes=[float],\n )\n return slopes",
"_____no_output_____"
],
[
"if util.is_ncar_host():\n col = intake.open_esm_datastore(\"../catalogs/glade-cmip6.json\")\nelse:\n col = intake.open_esm_datastore(\"../catalogs/pangeo-cmip6_update_2019_10_18.json\")",
"_____no_output_____"
],
[
"cat = col.search(experiment_id=['historical'], table_id='Omon', variable_id=['dissic'], grid_label='gr')\n\nimport pprint \nuni_dict = col.unique(['source_id', 'experiment_id', 'table_id'])\n#pprint.pprint(uni_dict, compact=True)",
"_____no_output_____"
],
[
"models = set(uni_dict['source_id']['values']) # all the models\n\nfor experiment_id in ['historical']:\n query = dict(experiment_id=experiment_id, table_id=['Omon','Ofx'], \n variable_id=['dissic'], grid_label=['gn','gr']) \n cat = col.search(**query)\n models = models.intersection({model for model in cat.df.source_id.unique().tolist()})\n\n# for oxygen, ensure the CESM2 models are not included (oxygen was erroneously submitted to the archive)\n# UKESM has an issue with the attributes\nmodels = models - {'UKESM1-0-LL','GISS-E2-1-G-CC','GISS-E2-1-G','MCM-UA-1-0'}\n\nmodels = list(models)\nmodels",
"_____no_output_____"
],
[
"# read all data with thickness and DIC for DIC storage\nwith warnings.catch_warnings(): # these lines just make sure that the warnings dont clutter your notebook\n warnings.simplefilter(\"ignore\")\n data_dict_thk = read_data(col,\n experiment_id=['historical'],\n grid_label='gn',\n variable_id=['thkcello','dissic'],\n table_id = ['Omon'],\n source_id = models,\n #member_id = 'r1i1p1f1', # so that this runs faster for testing\n required_variable_id = ['thkcello','dissic']\n )\n#data_dict_thk['IPSL-CM6A-LR'] = data_dict_thk['IPSL-CM6A-LR'].rename({'olevel':'lev'})",
"--> The keys in the returned dictionary of datasets are constructed as follows:\n\t'activity_id.institution_id.source_id.experiment_id.table_id.grid_label'\n\n--> There will be 6 group(s)\n--> The keys in the returned dictionary of datasets are constructed as follows:\n\t'activity_id.institution_id.source_id.experiment_id.table_id.grid_label'\n\n--> There will be 5 group(s)\nCNRM-ESM2-1\nNON-REFERENCE MODE. This should just be used for a bunch of variables on the same grid\nIPSL-CM6A-LR\nNON-REFERENCE MODE. This should just be used for a bunch of variables on the same grid\n"
],
[
"# read all data with volume and oxygen\nwith warnings.catch_warnings(): # these lines just make sure that the warnings dont clutter your notebook\n warnings.simplefilter(\"ignore\")\n data_dict_dic = read_data(col,\n experiment_id=['historical'],\n grid_label='gn',\n variable_id=['dissic'],\n table_id = ['Omon'],\n source_id = models,\n #member_id = 'r1i1p1f1', # so that this runs faster for testing\n required_variable_id = ['dissic']\n )\ndata_dict_dic['IPSL-CM6A-LR'] = data_dict_dic['IPSL-CM6A-LR'].rename({'olevel_bounds':'lev_bounds'})",
"--> The keys in the returned dictionary of datasets are constructed as follows:\n\t'activity_id.institution_id.source_id.experiment_id.table_id.grid_label'\n\n--> There will be 5 group(s)\nCanESM5\nNON-REFERENCE MODE. This should just be used for a bunch of variables on the same grid\nCNRM-ESM2-1\nNON-REFERENCE MODE. This should just be used for a bunch of variables on the same grid\nIPSL-CM6A-LR\nNON-REFERENCE MODE. This should just be used for a bunch of variables on the same grid\nMIROC-ES2L\nNON-REFERENCE MODE. This should just be used for a bunch of variables on the same grid\nCESM2\nNON-REFERENCE MODE. This should just be used for a bunch of variables on the same grid\n"
],
[
"#data_dict_dic['IPSL-CM6A-LR'] = data_dict_dic['IPSL-CM6A-LR'].rename({'olevel_bounds':'lev_bounds'})\ndata_dict_dic['MIROC-ES2L'] = data_dict_dic['MIROC-ES2L'].rename({'zlev_bnds':'lev_bounds'})",
"_____no_output_____"
],
[
"data_dict_dic_thk = {k: parse_static_thkcello(ds) for k, ds in data_dict_dic.items()}",
"_____no_output_____"
]
],
[
[
"### Loading data\n\n`intake-esm` enables loading data directly into an [xarray.Dataset](http://xarray.pydata.org/en/stable/api.html#dataset).\n\nNote that data on the cloud are in \n[zarr](https://zarr.readthedocs.io/en/stable/) format and data on \n[glade](https://www2.cisl.ucar.edu/resources/storage-and-file-systems/glade-file-spaces) are stored as \n[netCDF](https://www.unidata.ucar.edu/software/netcdf/) files. This is opaque to the user.\n\n`intake-esm` has rules for aggegating datasets; these rules are defined in the collection-specification file.",
"_____no_output_____"
]
],
[
[
"#cat = col.search(experiment_id=['historical'], table_id='Omon', \n# variable_id=['dissic'], grid_label='gn', source_id=models)",
"_____no_output_____"
],
[
"#dset_dict_dic_gn = cat.to_dataset_dict(zarr_kwargs={'consolidated': True, 'decode_times': False}, \n# cdf_kwargs={'chunks': {'time' : 20}, 'decode_times': False})",
"--> The keys in the returned dictionary of datasets are constructed as follows:\n\t'activity_id.institution_id.source_id.experiment_id.table_id.grid_label'\n\n--> There will be 5 group(s)\n"
]
],
[
[
"### Plotting DIC storage",
"_____no_output_____"
]
],
[
[
"data_dict_dic.keys()",
"_____no_output_____"
],
[
"fig, ax = plt.subplots(ncols=3, nrows=2,figsize=[15, 10])\nA = 0\nfor model_key in data_dict_dic.keys():\n dsC = data_dict_dic[model_key]\n ds = dsC['dissic'].isel(lev = 0).chunk({'time': -1, 'x': 110, 'y': 110, 'member_id': 10})\n #dz = dsC['thkcello'].isel(member_id=0)\n #DICstore_slope = (ds.isel(time=-np.arange(10*12)).mean('time')*dz-ds.isel(time=np.arange(10*12)).mean('time')*dz).sum('lev')\n slope = compute_slope(ds)\n slope = slope.compute()\n slope = slope.mean('member_id')*12 # in mol/m^3/year\n A1 = int(np.floor(A/3))\n A2 = np.mod(A,3)\n slope.plot(ax = ax[A1][A2],vmax = 0.001)\n ax[A1][A2].title.set_text(model_key)\n A += 1\nfig.tight_layout()\nfig.savefig('rate_of_change_DIC_surface_historical.png')",
"/srv/conda/envs/notebook/lib/python3.7/site-packages/xarray/core/nanops.py:140: RuntimeWarning: Mean of empty slice\n return np.nanmean(a, axis=axis, dtype=dtype)\n/srv/conda/envs/notebook/lib/python3.7/site-packages/xarray/core/nanops.py:140: RuntimeWarning: Mean of empty slice\n return np.nanmean(a, axis=axis, dtype=dtype)\n/srv/conda/envs/notebook/lib/python3.7/site-packages/xarray/core/nanops.py:140: RuntimeWarning: Mean of empty slice\n return np.nanmean(a, axis=axis, dtype=dtype)\n/srv/conda/envs/notebook/lib/python3.7/site-packages/xarray/core/nanops.py:140: RuntimeWarning: Mean of empty slice\n return np.nanmean(a, axis=axis, dtype=dtype)\n"
],
[
"fig, ax = plt.subplots(ncols=3, nrows=2,figsize=[15, 10])\nA = 0\nfor model_key in data_dict_thk.keys():\n dsC = data_dict_thk[model_key]\n ds = dsC['dissic']\n dz = dsC['thkcello']\n DICstore = (ds*dz).sum('lev').chunk({'time': -1, 'x': 110, 'y': 110, 'member_id': 10})\n slope = compute_slope(DICstore)\n slope = slope.compute()\n slope = slope.mean('member_id')*12 # in mol/m^3/year\n A1 = int(np.floor(A/3))\n A2 = np.mod(A,3)\n slope.plot(ax = ax[A1][A2],vmax = 0.8)\n ax[A1][A2].title.set_text(model_key)\n A += 1\nfig.tight_layout()\nfig.savefig('rate_of_change_DIC_content_historical.png')",
"_____no_output_____"
]
],
[
[
"# Load heat content",
"_____no_output_____"
]
],
[
[
"cat = col.search(experiment_id=['historical'], table_id='Omon', variable_id=['thetao','thkcello'], grid_label='gn')\n\nimport pprint \nuni_dict = col.unique(['source_id', 'experiment_id', 'table_id'])\n#pprint.pprint(uni_dict, compact=True)",
"_____no_output_____"
],
[
"models = set(uni_dict['source_id']['values']) # all the models\n\nfor experiment_id in ['historical']:\n query = dict(experiment_id=experiment_id, table_id=['Omon','Ofx'], \n variable_id=['thetao','thkcello'], grid_label='gn') \n cat = col.search(**query)\n models = models.intersection({model for model in cat.df.source_id.unique().tolist()})\n\n# for oxygen, ensure the CESM2 models are not included (oxygen was erroneously submitted to the archive)\n# UKESM has an issue with the attributes\nmodels = models - {'HadGEM3-GC31-LL','UKESM1-0-LL'}\n#{'UKESM1-0-LL','GISS-E2-1-G-CC','GISS-E2-1-G','MCM-UA-1-0'}\n\nmodels = list(models)\nmodels",
"_____no_output_____"
],
[
"# read all data with thickness and DIC for DIC storage\nwith warnings.catch_warnings(): # these lines just make sure that the warnings dont clutter your notebook\n warnings.simplefilter(\"ignore\")\n data_dict_heat_thk = read_data(col,\n experiment_id=['historical'],\n grid_label='gn',\n variable_id=['thkcello','thetao'],\n table_id = ['Omon'],\n source_id = models,\n #member_id = 'r1i1p1f1', # so that this runs faster for testing\n required_variable_id = ['thkcello','thetao']\n )\n#data_dict_heat_thk['IPSL-CM6A-LR'] = data_dict_heat_thk['IPSL-CM6A-LR'].rename({'olevel':'lev'})",
"--> The keys in the returned dictionary of datasets are constructed as follows:\n\t'activity_id.institution_id.source_id.experiment_id.table_id.grid_label'\n\n--> There will be 5 group(s)\n--> The keys in the returned dictionary of datasets are constructed as follows:\n\t'activity_id.institution_id.source_id.experiment_id.table_id.grid_label'\n\n--> There will be 21 group(s)\nBCC-CSM2-MR\nNON-REFERENCE MODE. This should just be used for a bunch of variables on the same grid\nBCC-ESM1\nNON-REFERENCE MODE. This should just be used for a bunch of variables on the same grid\nCNRM-CM6-1\nNON-REFERENCE MODE. This should just be used for a bunch of variables on the same grid\nCNRM-ESM2-1\nNON-REFERENCE MODE. This should just be used for a bunch of variables on the same grid\nIPSL-CM6A-LR\nNON-REFERENCE MODE. This should just be used for a bunch of variables on the same grid\n"
]
],
[
[
"# Plot heat content",
"_____no_output_____"
]
],
[
[
"data_dict_heat_thk.keys()",
"_____no_output_____"
],
[
"fig, ax = plt.subplots(ncols=3, nrows=2,figsize=[15, 10])\nA = 0\nfor model_key in data_dict_heat_thk.keys():\n dsC = data_dict_heat_thk[model_key]\n ds = (dsC['thetao']+273.15)*4.15*1e6/1025 # heat content (assume constant density and heat capacity)\n dz = dsC['thkcello'].isel(member_id=0)\n DICstore = (ds*dz).sum('lev').chunk({'time': -1, 'x': 110, 'y': 110, 'member_id': 10})\n slope = compute_slope(DICstore)\n slope = slope.compute()\n slope = slope.mean('member_id')*12 # in mol/m^3/year\n A1 = int(np.floor(A/3))\n A2 = np.mod(A,3)\n slope.plot(ax = ax[A1][A2],vmax = 80000)\n ax[A1][A2].title.set_text(model_key)\n A += 1\nfig.tight_layout()\nfig.savefig('rate_of_change_heat_content_historical.png')",
"_____no_output_____"
]
],
[
[
"# Load oxygen content",
"_____no_output_____"
]
],
[
[
"cat = col.search(experiment_id=['piControl'], table_id='Omon', variable_id=['o2','thkcello'], grid_label='gn')\n\nimport pprint \nuni_dict = col.unique(['source_id', 'experiment_id', 'table_id'])\n#pprint.pprint(uni_dict, compact=True)",
"_____no_output_____"
],
[
"models = set(uni_dict['source_id']['values']) # all the models\n\nfor experiment_id in ['historical']:\n query = dict(experiment_id=experiment_id, table_id=['Omon','Ofx'], \n variable_id=['o2','thkcello'], grid_label='gn') \n cat = col.search(**query)\n models = models.intersection({model for model in cat.df.source_id.unique().tolist()})\n\n# for oxygen, ensure the CESM2 models are not included (oxygen was erroneously submitted to the archive)\n# UKESM has an issue with the attributes\nmodels = models - {'UKESM1-0-LL'}\n#{'UKESM1-0-LL','GISS-E2-1-G-CC','GISS-E2-1-G','MCM-UA-1-0'}\n\nmodels = list(models)\nmodels",
"_____no_output_____"
],
[
"# read all data with thickness and o2 for o2 content\nwith warnings.catch_warnings(): # these lines just make sure that the warnings dont clutter your notebook\n warnings.simplefilter(\"ignore\")\n data_dict_o2_thk = read_data(col,\n experiment_id=['historical'],\n grid_label='gn',\n variable_id=['thkcello','o2'],\n table_id = ['Omon'],\n source_id = models,\n #member_id = 'r1i1p1f1', # so that this runs faster for testing\n required_variable_id = ['thkcello','o2']\n )\n#data_dict_o2_thk['IPSL-CM6A-LR'] = data_dict_o2_thk['IPSL-CM6A-LR'].rename({'olevel':'lev'})",
"_____no_output_____"
]
],
[
[
"# Plot O2 content",
"_____no_output_____"
]
],
[
[
"data_dict_o2_thk.keys()",
"_____no_output_____"
],
[
"fig, ax = plt.subplots(ncols=2, nrows=1,figsize=[10, 5])\nA = 0\nfor model_key in data_dict_o2_thk.keys():\n dsC = data_dict_o2_thk[model_key]\n ds = dsC['o2']\n dz = dsC['thkcello'].isel(member_id=0)\n DICstore = (ds*dz).sum('lev').chunk({'time': -1, 'x': 110, 'y': 110, 'member_id': 10})\n slope = compute_slope(DICstore)\n slope = slope.compute()\n slope = slope.mean('member_id')*12 # in mol/m^3/year\n slope.plot(ax = ax[A],vmax = 0.8)\n ax[A].title.set_text(model_key)\n A += 1\nfig.tight_layout()\nfig.savefig('rate_of_change_o2_content_historical.png')",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
c515b52018be6f9a7d96909a7699dbad2adefc82
| 430,926 |
ipynb
|
Jupyter Notebook
|
Winston_Lee_DS_Unit_1_Sprint_Challenge_4.ipynb
|
wel51x/DS-Unit-1-Sprint-4-Statistical-Tests-and-Experiments
|
105c5c0caba5d2deec3d2d89037b16baef7012c4
|
[
"MIT"
] | null | null | null |
Winston_Lee_DS_Unit_1_Sprint_Challenge_4.ipynb
|
wel51x/DS-Unit-1-Sprint-4-Statistical-Tests-and-Experiments
|
105c5c0caba5d2deec3d2d89037b16baef7012c4
|
[
"MIT"
] | null | null | null |
Winston_Lee_DS_Unit_1_Sprint_Challenge_4.ipynb
|
wel51x/DS-Unit-1-Sprint-4-Statistical-Tests-and-Experiments
|
105c5c0caba5d2deec3d2d89037b16baef7012c4
|
[
"MIT"
] | null | null | null | 253.934001 | 264,844 | 0.864278 |
[
[
[
"<a href=\"https://colab.research.google.com/github/wel51x/DS-Unit-1-Sprint-4-Statistical-Tests-and-Experiments/blob/master/Winston_Lee_DS_Unit_1_Sprint_Challenge_4.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# Data Science Unit 1 Sprint Challenge 4\n\n## Exploring Data, Testing Hypotheses\n\nIn this sprint challenge you will look at a dataset of people being approved or rejected for credit.\n\nhttps://archive.ics.uci.edu/ml/datasets/Credit+Approval\n\nData Set Information: This file concerns credit card applications. All attribute names and values have been changed to meaningless symbols to protect confidentiality of the data. This dataset is interesting because there is a good mix of attributes -- continuous, nominal with small numbers of values, and nominal with larger numbers of values. There are also a few missing values.\n\nAttribute Information:\n- A1: b, a.\n- A2: continuous.\n- A3: continuous.\n- A4: u, y, l, t.\n- A5: g, p, gg.\n- A6: c, d, cc, i, j, k, m, r, q, w, x, e, aa, ff.\n- A7: v, h, bb, j, n, z, dd, ff, o.\n- A8: continuous.\n- A9: t, f.\n- A10: t, f.\n- A11: continuous.\n- A12: t, f.\n- A13: g, p, s.\n- A14: continuous.\n- A15: continuous.\n- A16: +,- (class attribute)\n\nYes, most of that doesn't mean anything. A16 (the class attribute) is the most interesting, as it separates the 307 approved cases from the 383 rejected cases. The remaining variables have been obfuscated for privacy - a challenge you may have to deal with in your data science career.\n\nSprint challenges are evaluated based on satisfactory completion of each part. It is suggested you work through it in order, getting each aspect reasonably working, before trying to deeply explore, iterate, or refine any given step. Once you get to the end, if you want to go back and improve things, go for it!",
"_____no_output_____"
],
[
"## Part 1 - Load and validate the data\n\n- Load the data as a `pandas` data frame.\n- Validate that it has the appropriate number of observations (you can check the raw file, and also read the dataset description from UCI).\n- UCI says there should be missing data - check, and if necessary change the data so pandas recognizes it as na\n- Make sure that the loaded features are of the types described above (continuous values should be treated as float), and correct as necessary\n\nThis is review, but skills that you'll use at the start of any data exploration. Further, you may have to do some investigation to figure out which file to load from - that is part of the puzzle.",
"_____no_output_____"
]
],
[
[
"# TODO\n# imports & defaults\nimport pandas as pd\nimport numpy as np\nimport random\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nimport scipy.stats as stats\nfrom scipy.stats import chisquare\n\npd.set_option('display.width', 162)",
"_____no_output_____"
],
[
"# Load data, changing ? to na\nheaders = [\"A1\",'A2','A3','A4','A5','A6','A7','A8','A9','A10','A11','A12','A13','A14','A15','A16']\ndf = pd.read_csv(\"https://archive.ics.uci.edu/ml/machine-learning-databases/credit-screening/crx.data\",\n na_values='?',\n names = headers)\ndf.describe(include='all')",
"_____no_output_____"
],
[
"df.isnull().sum()",
"_____no_output_____"
],
[
"# Replace nulls randomly from other values in column\ndf = df.apply(lambda x: np.where(x.isnull(), x.dropna().sample(len(x), replace=True), x))",
"_____no_output_____"
],
[
"df.dtypes",
"_____no_output_____"
],
[
"# change A11, A15 to float\ndf['A11'] = df['A11'].astype(float)\ndf['A15'] = df['A15'].astype(float)",
"_____no_output_____"
],
[
"# change A16: '-' => 0, '+' => 1\ndf['A16'] = df['A16'].replace(\"+\", 1)\ndf['A16'] = df['A16'].replace('-', 0)\ndf.describe(include='all')",
"_____no_output_____"
]
],
[
[
"## Part 2 - Exploring data, Testing hypotheses\n\nThe only thing we really know about this data is that A16 is the class label. Besides that, we have 6 continuous (float) features and 9 categorical features.\n\nExplore the data: you can use whatever approach (tables, utility functions, visualizations) to get an impression of the distributions and relationships of the variables. In general, your goal is to understand how the features are different when grouped by the two class labels (`+` and `-`).\n\nFor the 6 continuous features, how are they different when split between the two class labels? Choose two features to run t-tests (again split by class label) - specifically, select one feature that is *extremely* different between the classes, and another feature that is notably less different (though perhaps still \"statistically significantly\" different). You may have to explore more than two features to do this.\n\nFor the categorical features, explore by creating \"cross tabs\" (aka [contingency tables](https://en.wikipedia.org/wiki/Contingency_table)) between them and the class label, and apply the Chi-squared test to them. [pandas.crosstab](http://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.crosstab.html) can create contingency tables, and [scipy.stats.chi2_contingency](https://docs.scipy.org/doc/scipy/reference/generated/scipy.stats.chi2_contingency.html) can calculate the Chi-squared statistic for them.\n\nThere are 9 categorical features - as with the t-test, try to find one where the Chi-squared test returns an extreme result (rejecting the null that the data are independent), and one where it is less extreme.\n\n**NOTE** - \"less extreme\" just means smaller test statistic/larger p-value. Even the least extreme differences may be strongly statistically significant.\n\nYour *main* goal is the hypothesis tests, so don't spend too much time on the exploration/visualization piece. That is just a means to an end - use simple visualizations, such as boxplots or a scatter matrix (both built in to pandas), to get a feel for the overall distribution of the variables.\n\nThis is challenging, so manage your time and aim for a baseline of at least running two t-tests and two Chi-squared tests before polishing. And don't forget to answer the questions in part 3, even if your results in this part aren't what you want them to be.",
"_____no_output_____"
]
],
[
[
"# TODO\n",
"_____no_output_____"
],
[
"# Create lists for continuous & categorical vars\n#cont = list(df.select_dtypes(include=['float64']))\ncontinuous_col_list = ['A16', 'A2', 'A3', 'A8', 'A11', 'A14', 'A15']\ncategorical_col_list = [obj for obj in list(df) if obj not in continuous_col_list]\ncategorical_col_list = ['A16'] + categorical_col_list\n#print(continuous_col_list, categorical_col_list)",
"['A16', 'A2', 'A3', 'A8', 'A11', 'A14', 'A15'] ['A16', 'A1', 'A4', 'A5', 'A6', 'A7', 'A9', 'A10', 'A12', 'A13']\n"
],
[
"# Now create cont df\ndf_continuous = df[continuous_col_list]\ndf_continuous.describe(include='all')",
"_____no_output_____"
],
[
"# and categ df\ndf_categorical = df[categorical_col_list]\ndf_categorical.describe(include='all')",
"_____no_output_____"
],
[
"# Subset continuous for reject/accept\ndf_continuous_rej = df_continuous[df_continuous['A16'] == 0].drop('A16', axis = 1)\ndf_continuous_acc = df_continuous[df_continuous['A16'] == 1].drop('A16', axis = 1)\nprint(\"Rejected\")\nprint(df_continuous_rej.describe(include='all'))\nprint(\"Accepted\")\nprint(df_continuous_acc.describe(include='all'))",
"Rejected\n A2 A3 A8 A11 A14 A15\ncount 383.000000 383.000000 383.000000 383.000000 383.000000 383.000000\nmean 29.846997 3.839948 1.257924 0.631854 201.159269 198.605744\nstd 11.184626 4.337662 2.120481 1.900049 181.698900 671.608839\nmin 15.170000 0.000000 0.000000 0.000000 0.000000 0.000000\n25% 22.000000 0.835000 0.125000 0.000000 100.000000 0.000000\n50% 27.250000 2.210000 0.415000 0.000000 170.000000 1.000000\n75% 34.830000 5.000000 1.500000 0.000000 278.000000 67.000000\nmax 80.250000 26.335000 13.875000 20.000000 2000.000000 5552.000000\nAccepted\n A2 A3 A8 A11 A14 A15\ncount 307.000000 307.000000 307.000000 307.000000 307.000000 307.000000\nmean 33.694104 5.904951 3.427899 4.605863 165.925081 2038.859935\nstd 12.775016 5.471485 4.120792 6.320242 161.075970 7659.763941\nmin 13.750000 0.000000 0.000000 0.000000 0.000000 0.000000\n25% 23.210000 1.500000 0.750000 0.000000 0.000000 0.000000\n50% 30.500000 4.460000 2.000000 3.000000 120.000000 221.000000\n75% 41.330000 9.520000 5.000000 7.000000 280.000000 1209.000000\nmax 76.750000 28.000000 28.500000 67.000000 840.000000 100000.000000\n"
],
[
"# Same for categ\ndf_categorical_rej = df_categorical[df_categorical['A16'] == 0].drop('A16', axis = 1)\ndf_categorical_acc = df_categorical[df_categorical['A16'] == 1].drop('A16', axis = 1)\nprint(\"Rejected\")\nprint(df_categorical_rej.describe(include='all'))\nprint(\"Accepted\")\nprint(df_categorical_acc.describe(include='all'))",
"Rejected\n A1 A4 A5 A6 A7 A9 A10 A12 A13\ncount 383 383 383 383 383 383 383 383 383\nunique 2 2 2 14 9 2 2 2 3\ntop b u g c v f f f g\nfreq 267 264 265 75 234 306 297 213 338\nAccepted\n A1 A4 A5 A6 A7 A9 A10 A12 A13\ncount 307 307 307 307 307 307 307 307 307\nunique 2 3 3 14 9 2 2 2 3\ntop b u g c v t t f g\nfreq 207 260 258 62 173 284 209 161 287\n"
],
[
"g = sns.PairGrid(data=df, hue='A16')\ng.map(plt.scatter)",
"_____no_output_____"
],
[
"# I'm wrong...these don't produce much of interest\nfor i in categorical_col_list:\n df_categorical[i].value_counts().plot(kind='hist')\n plt.title(i)\n plt.show()",
"_____no_output_____"
],
[
"# Continuous tests\nfor col in continuous_col_list[1:]:\n t_stat, p_val = stats.ttest_ind(df_continuous_acc[col],\n df_continuous_rej[col],\n equal_var = False)\n print(col, \"has t-statistic =\", t_stat, \"and pvalue =\", p_val, \"when comparing accepted vs rejected\")\n",
"A2 has t-statistic = 4.152740636486119 and pvalue = 3.751707252379698e-05 when comparing accepted vs rejected\nA3 has t-statistic = 5.392530906223675 and pvalue = 1.0158807568737146e-07 when comparing accepted vs rejected\nA8 has t-statistic = 8.38006008674286 and pvalue = 7.425348666782441e-16 when comparing accepted vs rejected\nA11 has t-statistic = 10.6384190682749 and pvalue = 4.310254123415665e-23 when comparing accepted vs rejected\nA14 has t-statistic = -2.6966924711429936 and pvalue = 0.007176339456028075 when comparing accepted vs rejected\nA15 has t-statistic = 4.196600236397611 and pvalue = 3.5433798702024966e-05 when comparing accepted vs rejected\n"
],
[
"# Categorical tests\ndf_categorical.sample(11)\nfor col in categorical_col_list[1:]:\n xtab = pd.crosstab(df_categorical[\"A16\"], df_categorical[col])\n ar = np.array(xtab).T\n chi_stat, p_val = chisquare(ar, axis=None)\n print(col, \"has chi statistic\", chi_stat, \"and p_value\", p_val)\n",
"A1 has chi statistic 107.64637681159421 and p_value 3.522132844308837e-23\nA4 has chi statistic 644.6608695652174 and p_value 4.513423235115485e-137\nA5 has chi statistic 639.7913043478261 and p_value 5.0932454456708715e-136\nA6 has chi statistic 385.60579710144935 and p_value 4.231522219000745e-65\nA7 has chi statistic 1899.7565217391302 and p_value 0.0\nA9 has chi statistic 357.82608695652175 and p_value 3.01326407188219e-77\nA10 has chi statistic 173.1304347826087 and p_value 2.6842170765821316e-37\nA12 has chi statistic 14.382608695652173 and p_value 0.0024280203352694887\nA13 has chi statistic 1037.2695652173913 and p_value 5.124924990548949e-222\n"
]
],
[
[
"## Part 3 - Analysis and Interpretation\n\nNow that you've looked at the data, answer the following questions:\n\n- Interpret and explain the two t-tests you ran - what do they tell you about the relationships between the continuous features you selected and the class labels?\n- Interpret and explain the two Chi-squared tests you ran - what do they tell you about the relationships between the categorical features you selected and the class labels?\n- What was the most challenging part of this sprint challenge?\n\nAnswer with text, but feel free to intersperse example code/results or refer to it from earlier.",
"_____no_output_____"
],
[
"**T-tests**\n\nI ran stats.ttest_ind() on all the continuous variables, comparing accepteds (col A16 = '+') vs rejecteds('-'). The best I can say is that :\n\nA14 , with t-statistic = -2.696 and pvalue = 0.007 have a slight relationship, but with the pvalue < .01, still not enough to reject the hypothesis that they are independent\n\nA11, with t-statistic = 10.638 and pvalue = pvalue = 4.310e-23 is the most dependent\n\nA8, with t-statistic = 8.380 and pvalue = pvalue = 7.425e-16 also makes a lot of difference are accepted or rejected\n\nEssentially all six continuous variables to a lesser or greater degree make a difference whether one was accepted or rejected. Inspecting the data seems to confirm a relationship between wherther one was accepted or rejected for columns A8, A11 and A15. These have rejected means of 1.257, 0.631 and 198.605, and accepted means of 3.427, 4.605 and 2038.859, respectively\n\n**Chi-squared tests**\n\nI ran scipy.stats.chisquare() on all the categorical variables\n\nA12 had the lowest statistic (14.382) and p_value 0.002\n\nA13 had the highest statistic (1037.269) and p_value 5.124e-222\n\nA7 has chi statistic 1899.7565217391302, but p_value = 0.0, which is very weird\n\nLooking at the data, it appears that for columns A9 and, to a lesser degree, A10 (both binary t/f items) make a difference as whether one is accepted or rejected. For A9, 306 of 383 rejects had a value of 'f', whereas 284 of 307 accepts had a value of 't'. For A10 the comparative figures are 'f': 297/383 for rejects and 't': 209/307 for accepts.\n\n**What was the most challenging part of this sprint challenge?**\n\nRealizing that data without context is a pain. For example, I suspect A1 is sex, A2 is age, A14 is level of debt and A15 level of assets or income. Also, I didn't feel I got much intelligence - no comments from the peanut gallery, please - from the ChiSq tests.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
]
] |
c515ba772cfe551dba7c4f1cb21dadd0ee62554d
| 774,142 |
ipynb
|
Jupyter Notebook
|
Week3_Titanic_Model.ipynb
|
naseerfaheem/Titanic
|
68d8451cc65a52833eca1dddb66882f6abcb5dd7
|
[
"MIT"
] | null | null | null |
Week3_Titanic_Model.ipynb
|
naseerfaheem/Titanic
|
68d8451cc65a52833eca1dddb66882f6abcb5dd7
|
[
"MIT"
] | null | null | null |
Week3_Titanic_Model.ipynb
|
naseerfaheem/Titanic
|
68d8451cc65a52833eca1dddb66882f6abcb5dd7
|
[
"MIT"
] | null | null | null | 224.129126 | 79,788 | 0.892392 |
[
[
[
"## Getting the Data from Kaggle Using the Kaggle API",
"_____no_output_____"
]
],
[
[
"#!kaggle competitions download -c titanic",
"_____no_output_____"
],
[
"# Unzip the folder\n#!unzip 'titanic.zip' -d data/titanic/",
"_____no_output_____"
]
],
[
[
"# Setup",
"_____no_output_____"
]
],
[
[
"# Load the train file to pandas\nimport pandas as pd\nimport numpy as np\nimport missingno as msno \nfrom collections import Counter\nimport re\nimport seaborn as sns\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.model_selection import cross_val_score\nfrom sklearn.model_selection import GridSearchCV\nfrom sklearn.ensemble import RandomForestClassifier\nfrom sklearn.metrics import accuracy_score\nfrom sklearn.svm import SVC\n\nfrom subprocess import check_output\nsns.set(style='white', context='notebook', palette='deep')\nimport matplotlib.pyplot as plt\n%matplotlib inline\n\nsns.set()",
"_____no_output_____"
]
],
[
[
"# Load Data",
"_____no_output_____"
]
],
[
[
"data_dict = pd.read_csv(\"data/titanic/data_dictionary.csv\")\ndata_dict",
"_____no_output_____"
],
[
"titanic_train = pd.read_csv(\"data/titanic/train.csv\")\ntitanic_test = pd.read_csv(\"data/titanic/test.csv\")\n\n# Getting the passengerID for test dataset so that we can split the \n# dataframe later by it. \ntitanic_test_ID = titanic_test['PassengerId']",
"_____no_output_____"
]
],
[
[
"## Descriptive Statistics of the Dataset",
"_____no_output_____"
]
],
[
[
"# Checking the distribution of each feature:\ntitanic_train.hist(figsize=(15,10));",
"_____no_output_____"
],
[
"titanic_train[\"Age\"].hist(figsize=(15,10));",
"_____no_output_____"
],
[
"# Scatter Matrix to see the correlation between some of the features\nfrom pandas.plotting import scatter_matrix\nattributes = [ \"Pclass\", \"Age\", \"Fare\"]\nscatter_matrix(titanic_train[attributes], figsize=(15,10));",
"_____no_output_____"
],
[
"titanic_train.plot(kind=\"scatter\", x=\"Age\", y=\"Fare\", alpha=0.9, figsize=(15,10));",
"'c' argument looks like a single numeric RGB or RGBA sequence, which should be avoided as value-mapping will have precedence in case its length matches with 'x' & 'y'. Please use a 2-D array with a single row if you really want to specify the same RGB or RGBA value for all points.\n"
],
[
"titanic_train.describe()",
"_____no_output_____"
],
[
"# Looking for missing values\ntitanic_train.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 891 entries, 0 to 890\nData columns (total 12 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 PassengerId 891 non-null int64 \n 1 Survived 891 non-null int64 \n 2 Pclass 891 non-null int64 \n 3 Name 891 non-null object \n 4 Sex 891 non-null object \n 5 Age 714 non-null float64\n 6 SibSp 891 non-null int64 \n 7 Parch 891 non-null int64 \n 8 Ticket 891 non-null object \n 9 Fare 891 non-null float64\n 10 Cabin 204 non-null object \n 11 Embarked 889 non-null object \ndtypes: float64(2), int64(5), object(5)\nmemory usage: 83.7+ KB\n"
]
],
[
[
"## Detecting Outliers\n\nIn this section, I am going to define a function that helps detect outliers in teh dataset (anything that falls out of 1.5* the IQR range). ",
"_____no_output_____"
]
],
[
[
"\ndef detect_outliers(df,n,features):\n \"\"\"\n Takes a dataframe df of features and returns a list of the indices\n corresponding to the observations containing more than n outliers according\n to the Tukey method.\n \"\"\"\n outlier_indices = []\n \n # iterate over features(columns)\n for col in features:\n # 1st quartile (25%)\n Q1 = np.percentile(df[col], 25)\n # 3rd quartile (75%)\n Q3 = np.percentile(df[col],75)\n # Interquartile range (IQR)\n IQR = Q3 - Q1\n \n # outlier step\n outlier_step = 1.5 * IQR\n \n # Determine a list of indices of outliers for feature col\n outlier_list_col = df[(df[col] < Q1 - outlier_step) | (df[col] > Q3 + outlier_step )].index\n \n # append the found outlier indices for col to the list of outlier indices \n outlier_indices.extend(outlier_list_col)\n \n # select observations containing more than 2 outliers\n outlier_indices = Counter(outlier_indices) \n multiple_outliers = list( k for k, v in outlier_indices.items() if v > n )\n \n return multiple_outliers \n\n# detect outliers from Age, SibSp , Parch and Fare\nOutliers_to_drop = detect_outliers(titanic_train,2,[\"Age\",\"SibSp\",\"Parch\",\"Fare\"])",
"_____no_output_____"
],
[
"titanic_train.loc[Outliers_to_drop]",
"_____no_output_____"
],
[
"titanic_train = titanic_train.drop(Outliers_to_drop, axis = 0).reset_index(drop=True)",
"_____no_output_____"
]
],
[
[
"### Joining Train and Test datasets\n\nHere, I am going to join both dataset for feature engineering and will later split them back using the titanic_test_ID. ",
"_____no_output_____"
]
],
[
[
"# Getting the length of the train dataset\nlen_titanic_train = len(titanic_train)\n# We are stacking two datasets, so it's important to remember the order\ndf = pd.concat(objs=[titanic_train, titanic_test], axis=0).reset_index(drop=True)",
"_____no_output_____"
]
],
[
[
"### Missing Values",
"_____no_output_____"
],
[
"It looks like the Age, Embarked and Cabin columns having missing values. I assume that Age and Embarked columns could be more relevant than the cabin, so I am going to impute the Age column with the mean of the column grouped by PClass. ",
"_____no_output_____"
]
],
[
[
"# Inspecting some of the missing Age rows\ndf[df['Age'].isnull()]",
"_____no_output_____"
],
[
" # Visualize missing values as a matrix \nmsno.matrix(df) ",
"_____no_output_____"
],
[
"msno.bar(df) ",
"_____no_output_____"
]
],
[
[
"Let's see if there a correlation among the missing values in the data using the heatmap function of the missinno library.",
"_____no_output_____"
]
],
[
[
"msno.heatmap(df);",
"_____no_output_____"
]
],
[
[
"From the heatmap above, it looks there is not a significant correlation between the missing values. ",
"_____no_output_____"
],
[
"### Imputing Missing Values",
"_____no_output_____"
],
[
"##### Impute Age",
"_____no_output_____"
],
[
"For the Age column, we can impute the missing values by the mean value of each group by Sex and Pclass. ",
"_____no_output_____"
]
],
[
[
"# Filling missing value of Age \n\n## Fill Age with the median age of similar rows according to Pclass, Parch and SibSp\n# Index of NaN age rows\nindex_NaN_age = list(df[\"Age\"][df[\"Age\"].isnull()].index)\n\nfor i in index_NaN_age :\n age_med = df[\"Age\"].median()\n age_pred = df[\"Age\"][((df['SibSp'] == df.iloc[i][\"SibSp\"]) & (df['Parch'] == df.iloc[i][\"Parch\"]) & (df['Pclass'] == df.iloc[i][\"Pclass\"]))].median()\n if not np.isnan(age_pred) :\n df['Age'].iloc[i] = age_pred\n else :\n df['Age'].iloc[i] = age_med",
"_____no_output_____"
]
],
[
[
"##### Impute Fare",
"_____no_output_____"
]
],
[
[
"# Let's fill the null values with the median value\ndf['Fare'] = df['Fare'].fillna(df['Fare'].median())",
"_____no_output_____"
]
],
[
[
"##### Impute Cabin",
"_____no_output_____"
]
],
[
[
"df['Cabin_mapped'] = df['Cabin'].astype(str).str[0]\n# this transforms the letters into numbers\ncabin_dict = {k:i for i, k in enumerate(df.Cabin_mapped.unique())} \ndf.loc[:, 'Cabin_mapped'] = df.loc[:, 'Cabin_mapped'].map(cabin_dict)",
"_____no_output_____"
],
[
"# Let's inspect cabins and see how they are labeled\ndf['Cabin'].unique()",
"_____no_output_____"
],
[
"df['Cabin'].isnull().sum()",
"_____no_output_____"
],
[
"# We can try to replace the Cabin with X for missing\n# Replace the Cabin number by the type of cabin 'X' if not\ndf[\"Cabin\"] = pd.Series([i[0] if not pd.isnull(i) else 'X' for i in df['Cabin'] ])",
"_____no_output_____"
]
],
[
[
"##### Impute Embarked",
"_____no_output_____"
]
],
[
[
"# Let's inspect the embarked column and see which rows have missing records\ndf[df['Embarked'].isnull()]",
"_____no_output_____"
],
[
"# Embarked\n# We can impute this feature with the mode which is S\ndf['Embarked'] = df['Embarked'].fillna(df['Embarked'].mode()[0])",
"_____no_output_____"
],
[
"# Checking to see if the above function worked: \ndf.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 1309 entries, 0 to 1308\nData columns (total 13 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 PassengerId 1309 non-null int64 \n 1 Survived 891 non-null float64\n 2 Pclass 1309 non-null int64 \n 3 Name 1309 non-null object \n 4 Sex 1309 non-null object \n 5 Age 1309 non-null float64\n 6 SibSp 1309 non-null int64 \n 7 Parch 1309 non-null int64 \n 8 Ticket 1309 non-null object \n 9 Fare 1309 non-null float64\n 10 Cabin 1309 non-null object \n 11 Embarked 1309 non-null object \n 12 Cabin_mapped 1309 non-null int64 \ndtypes: float64(3), int64(5), object(5)\nmemory usage: 133.1+ KB\n"
],
[
"\ndef draw_heatmap(df, y_variable, no_features):\n \"\"\" This Function takes three arguments; \n 1. The dataframe that we want to draw the heatmap for\n 2. The variable that we want to see the correlation of with other features for example the y-variable.\n 3. The top_n. For example for top 10 variables, type 10.\"\"\"\n # Calculate the correlation matrix\n cor = df.corr()\n # Get the columns for n largetst features\n columns = cor.nlargest(no_features, y_variable)[y_variable].index\n cm = np.corrcoef(df[columns].values.T)\n sns.set(font_scale=1)\n fig = plt.figure(num=None, figsize=(10, 10), dpi=80, facecolor='w', edgecolor='k')\n # Define the color pallet\n cmap = sns.cm.vlag_r\n heat_map = sns.heatmap(cm, cbar=True, annot=True, square=True, fmt=\".2f\", annot_kws={'size':12}, \n yticklabels=columns.values, xticklabels=columns.values,\n linewidths=.2,vmax=1, center=0, cmap= cmap)\n return plt.show();",
"_____no_output_____"
],
[
"draw_heatmap(titanic_train, 'Survived', 10)",
"_____no_output_____"
]
],
[
[
"We could see that that Fare and age has higher negative correlations. This means that there might hidden patterns within each feature and some feature engineering, that we could see different heatmap",
"_____no_output_____"
],
[
"# Feature Analysis\n\nFor Feature Analysis, I am going to define three helper functions that are going to help in drawing plots. I am planning ot Seaborn's Factoplots and Pandas' Barcharts. ",
"_____no_output_____"
]
],
[
[
"def plot_factorplot(df, x, y='Survived', hue=None):\n import warnings\n warnings.simplefilter(action='ignore', category=Warning)\n plt.figure(figsize=(12,10))\n g = sns.factorplot(x=x,y=y,data=df,kind=\"bar\", size = 6 , hue=hue, palette = \"muted\")\n g.despine(left=True)\n g = g.set_ylabels(\"Survival Probability\")\n g = g.set_xlabels(\"{}\".format(x))",
"_____no_output_____"
],
[
"def plot_barchart(df, feature):\n \"\"\" This functions takes the feature that we want to plot against survivors\"\"\"\n survived = df[df['Survived']==1][feature].value_counts()\n not_survived = df[df['Survived']==0][feature].value_counts()\n df = pd.DataFrame([survived,not_survived])\n df.index=['Survived','Not Survived']\n df.plot(kind='bar',stacked=False,title=\"Stacked Chart for \"+feature, figsize=(12,10))",
"_____no_output_____"
],
[
"def plot_distribution(df, col, **options):\n from scipy.stats import norm\n\n \"\"\"\n This function helps draw a distribution plot for the desired colum.\n Input args: \n 1. df : Dataframe that we want to pick the column from. \n 2. col : Column of the dataframe that we want to display.\n 3. options:\n a. kde : optional, boolian - Whether to plot a gaussian kernel density estimate.\n b. fit : An object with `fit` method, returning a tuple that can be passed to a `pdf` method a \n positional arguments following a grid of values to evaluate the pdf on.\n \"\"\"\n plt.figure(figsize=(12,10))\n plt.ylabel(\"Frequency\")\n plt.title(\"{} Distribution\".format(col))\n if options.get(\"kde\")==True:\n sns.distplot(df[col], kde=True, color=\"#2b7bba\");\n if options.get(\"fit\")== \"norm\":\n (mu, sigma) = norm.fit(df[col])\n sns.distplot(df[col], fit=norm, color=\"#2b7bba\");\n plt.legend([\"Normal dist. ($\\mu=$ {:.2f} and $\\sigma=$ {:.2f} )\".format(mu, sigma)],\n loc='best');\n \n if (options.get(\"fit\")== \"norm\") & (options.get(\"kde\")==True):\n sns.distplot(df[col], fit=norm, kde=True, color=\"#2b7bba\");\n else:\n sns.distplot(df[col], color=\"#2b7bba\");",
"_____no_output_____"
]
],
[
[
"##### Sex",
"_____no_output_____"
]
],
[
[
"survivors_data = titanic_train[titanic_train.Survived==True]\nnon_survivors_data = titanic_train[titanic_train.Survived==False]",
"_____no_output_____"
],
[
"Gender = pd.crosstab(titanic_train['Survived'],titanic_train['Sex'])\nGender",
"_____no_output_____"
],
[
"plot_barchart(titanic_train,\"Sex\")",
"_____no_output_____"
]
],
[
[
"We could see that females had a higher chance of survival than the males. It looks like Sex might be an important factor in determining the chance of survival. We can create some features for that. \n",
"_____no_output_____"
],
[
"##### Pclass",
"_____no_output_____"
]
],
[
[
"Pclass = pd.crosstab(titanic_train['Survived'],titanic_train['Pclass'])\nPclass",
"_____no_output_____"
],
[
"plot_barchart(titanic_train, \"Pclass\")",
"_____no_output_____"
]
],
[
[
"We can see in the above chart that passengers with with tickets in class 3 had a less survival chance.",
"_____no_output_____"
]
],
[
[
"# Explore Pclass vs Survived by Sex\nplot_factorplot(titanic_train, \"Pclass\", hue='Sex')",
"_____no_output_____"
]
],
[
[
"We could see that Pclass and sex both have role in determining survival. We could see that within females, the ones with the ticket class 1 and 2 had a higher survival chance. ",
"_____no_output_____"
],
[
"##### Fare\n\nLet's see the distribution of the fare",
"_____no_output_____"
]
],
[
[
"#Explore Fare distribution \nplot_distribution(df, \"Fare\", kde=True)",
"_____no_output_____"
]
],
[
[
"We can see that the fare is skewed positively. We can fix this by transforming the fare feature with a logarithmic transformation function",
"_____no_output_____"
],
[
"#### Transform Fare",
"_____no_output_____"
]
],
[
[
"df['Fare'] = np.log1p(df['Fare'])",
"_____no_output_____"
],
[
"# Let's display the distribution after log transformation\nplot_distribution(df, \"Fare\", kde=True, fit=\"norm\")",
"_____no_output_____"
]
],
[
[
"##### Age",
"_____no_output_____"
]
],
[
[
"# Explore Age distibution \nfig = plt.figure(figsize=(12,10))\ng = sns.kdeplot(titanic_train[\"Age\"][(titanic_train[\"Survived\"] == 0) & (titanic_train[\"Age\"].notnull())], color=\"Red\", shade = True)\ng = sns.kdeplot(titanic_train[\"Age\"][(titanic_train[\"Survived\"] == 1) & (titanic_train[\"Age\"].notnull())], ax =g, color=\"Green\", shade= True)\ng.set_xlabel(\"Age\")\ng.set_ylabel(\"Frequency\")\ng = g.legend([\"Did Not Survived\",\"Survived\"])",
"_____no_output_____"
]
],
[
[
"After plotting the survival by age, we can see that there high survial for teens and also on the right tail we can see that people above 70 have survived higher. ",
"_____no_output_____"
],
[
"##### SibSP",
"_____no_output_____"
]
],
[
[
"plot_barchart(titanic_train, \"SibSp\")",
"_____no_output_____"
]
],
[
[
"It looks like the passengers having more siblings/spouses had a higher chance of not surviving. On the other hand, single passengers were more likely to survive. ",
"_____no_output_____"
],
[
"##### Parch",
"_____no_output_____"
]
],
[
[
"plot_barchart(titanic_train, \"Parch\")",
"_____no_output_____"
],
[
"# Explore Parch feature vs Survived\nplot_factorplot(titanic_train, 'Parch')",
"_____no_output_____"
]
],
[
[
"Small families have more chance to survive, more than single (Parch 0), medium (Parch 3,4) and large families (Parch 5,6 ).",
"_____no_output_____"
],
[
"##### Embarked",
"_____no_output_____"
]
],
[
[
"plot_factorplot(titanic_train, 'Embarked')",
"_____no_output_____"
]
],
[
[
"We can see that passengers embarking the ship from Southhampton (S) had the lowest survival rate, however, passengers embarking from Cherbourg(C) had the highest chance of survival. \n\nLet's look a little deeper and see if the passengers from C had more Class 1 tickets.",
"_____no_output_____"
]
],
[
[
"plot_factorplot(titanic_train,'Embarked', hue='Pclass')",
"_____no_output_____"
]
],
[
[
"We can see that passengers from C had more 1st class tickets compared those those from S.",
"_____no_output_____"
],
[
"## Feature Engineering",
"_____no_output_____"
],
[
"#### Pclass\nWe can convert Sex to categorical and then to dummy. ",
"_____no_output_____"
]
],
[
[
"# Create categorical values for Pclass\ndf[\"Pclass\"] = df[\"Pclass\"].astype(\"category\")\ndf = pd.get_dummies(df, columns = [\"Pclass\"],prefix=\"Pc\")",
"_____no_output_____"
]
],
[
[
"#### Sex\nWe can convert Sex to categorical",
"_____no_output_____"
]
],
[
[
"df['Sex'] = df['Sex'].map({'male': 0, 'female':1})",
"_____no_output_____"
]
],
[
[
"#### Family Size\n\nWe can try to calculate a feature called family size where we are adding Parch, SibSp and 1 for the passenger him/herself",
"_____no_output_____"
]
],
[
[
"df['Fam_size'] = 1 + df['Parch'] + df['SibSp']",
"_____no_output_____"
],
[
"plot_factorplot(df, 'Fam_size')",
"_____no_output_____"
]
],
[
[
"We can see that that familyi size have some effect on the survival. ",
"_____no_output_____"
]
],
[
[
"# Create new feature of family size\ndf['Single'] = df['Fam_size'].map(lambda s: 1 if s == 1 else 0)\ndf['SmallF'] = df['Fam_size'].map(lambda s: 1 if s == 2 else 0)\ndf['MedF'] = df['Fam_size'].map(lambda s: 1 if 3 <= s <= 4 else 0)\ndf['LargeF'] = df['Fam_size'].map(lambda s: 1 if s >= 5 else 0)",
"_____no_output_____"
]
],
[
[
"#### Title\n\nWe can see that some of the passenger names have titles in fron them. This may add predicive power to the survival rate. Let's extract their titles and convert it into a dummy variable.",
"_____no_output_____"
]
],
[
[
"df['Name'].head()",
"_____no_output_____"
],
[
"def get_title(name):\n title_search = re.search(' ([A-Za-z]+)\\.', name)\n if title_search:\n return title_search.group(1)\n return \"\"",
"_____no_output_____"
],
[
"df['Title']=df['Name'].apply(get_title)\ntitle_lev=list(df['Title'].value_counts().reset_index()['index'])",
"_____no_output_____"
],
[
"df['Title']=pd.Categorical(df['Title'], categories=title_lev)",
"_____no_output_____"
],
[
"g = sns.countplot(x=\"Title\",data=df)\ng = plt.setp(g.get_xticklabels(), rotation=45)",
"_____no_output_____"
],
[
"df = pd.get_dummies(df, columns=['Title'], drop_first=True, prefix=\"Title\")",
"_____no_output_____"
],
[
"df.columns",
"_____no_output_____"
],
[
"# Drop the name column\ndf = df.drop(['Name'], axis=1)",
"_____no_output_____"
]
],
[
[
"We can that passengers with Miss-Mrs had a higher chance of survival. ",
"_____no_output_____"
]
],
[
[
"df.columns",
"_____no_output_____"
]
],
[
[
"#### Ticket\n\nWe can try to extract some information from the ticket feature by extracting it's prefix. We can use X for those that don't have a prefix. ",
"_____no_output_____"
]
],
[
[
"df['Ticket']",
"_____no_output_____"
],
[
"## Treat Ticket by extracting the ticket prefix. When there is no prefix it returns X. \n\nTicket = []\nfor i in list(df.Ticket):\n if not i.isdigit() :\n Ticket.append(i.replace(\".\",\"\").replace(\"/\",\"\").strip().split(' ')[0]) #Take prefix\n else:\n Ticket.append(\"X\")\n \ndf[\"Ticket\"] = Ticket\ndf[\"Ticket\"].head()",
"_____no_output_____"
],
[
"df = pd.get_dummies(df, columns = [\"Ticket\"], prefix=\"T\")\n",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
]
],
[
[
"#### Embarked\n\nLet's convert this categorical to numerical using Pandas' get_dummies function",
"_____no_output_____"
]
],
[
[
"df = pd.get_dummies(df, columns=['Embarked'], prefix=\"Embarked\")",
"_____no_output_____"
]
],
[
[
"#### Cabin\n\nLet's convert this categorical to numerical using Pandas' get_dummies function",
"_____no_output_____"
]
],
[
[
"df['HasCabin'] = df['Cabin'].apply(lambda x: 0 if x==0 else 1)\ndf = pd.get_dummies(df, columns=['Cabin'], prefix=\"Cabin\")",
"_____no_output_____"
],
[
"df = pd.get_dummies(df, columns=['HasCabin'], prefix=\"CabinBol\")",
"_____no_output_____"
],
[
"df.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 1299 entries, 0 to 1298\nData columns (total 82 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 PassengerId 1299 non-null int64 \n 1 Survived 881 non-null float64\n 2 Sex 1299 non-null int64 \n 3 Age 1299 non-null float64\n 4 SibSp 1299 non-null int64 \n 5 Parch 1299 non-null int64 \n 6 Fare 1299 non-null float64\n 7 Pc_1 1299 non-null uint8 \n 8 Pc_2 1299 non-null uint8 \n 9 Pc_3 1299 non-null uint8 \n 10 Fam_size 1299 non-null int64 \n 11 Single 1299 non-null int64 \n 12 SmallF 1299 non-null int64 \n 13 MedF 1299 non-null int64 \n 14 LargeF 1299 non-null int64 \n 15 Title_Miss 1299 non-null uint8 \n 16 Title_Mrs 1299 non-null uint8 \n 17 Title_Master 1299 non-null uint8 \n 18 Title_Rev 1299 non-null uint8 \n 19 Title_Dr 1299 non-null uint8 \n 20 Title_Col 1299 non-null uint8 \n 21 Title_Mlle 1299 non-null uint8 \n 22 Title_Major 1299 non-null uint8 \n 23 Title_Ms 1299 non-null uint8 \n 24 Title_Don 1299 non-null uint8 \n 25 Title_Lady 1299 non-null uint8 \n 26 Title_Mme 1299 non-null uint8 \n 27 Title_Countess 1299 non-null uint8 \n 28 Title_Dona 1299 non-null uint8 \n 29 Title_Jonkheer 1299 non-null uint8 \n 30 Title_Sir 1299 non-null uint8 \n 31 Title_Capt 1299 non-null uint8 \n 32 T_A 1299 non-null uint8 \n 33 T_A4 1299 non-null uint8 \n 34 T_A5 1299 non-null uint8 \n 35 T_AQ3 1299 non-null uint8 \n 36 T_AQ4 1299 non-null uint8 \n 37 T_AS 1299 non-null uint8 \n 38 T_C 1299 non-null uint8 \n 39 T_CA 1299 non-null uint8 \n 40 T_CASOTON 1299 non-null uint8 \n 41 T_FC 1299 non-null uint8 \n 42 T_FCC 1299 non-null uint8 \n 43 T_Fa 1299 non-null uint8 \n 44 T_LINE 1299 non-null uint8 \n 45 T_LP 1299 non-null uint8 \n 46 T_PC 1299 non-null uint8 \n 47 T_PP 1299 non-null uint8 \n 48 T_PPP 1299 non-null uint8 \n 49 T_SC 1299 non-null uint8 \n 50 T_SCA3 1299 non-null uint8 \n 51 T_SCA4 1299 non-null uint8 \n 52 T_SCAH 1299 non-null uint8 \n 53 T_SCOW 1299 non-null uint8 \n 54 T_SCPARIS 1299 non-null uint8 \n 55 T_SCParis 1299 non-null uint8 \n 56 T_SOC 1299 non-null uint8 \n 57 T_SOP 1299 non-null uint8 \n 58 T_SOPP 1299 non-null uint8 \n 59 T_SOTONO2 1299 non-null uint8 \n 60 T_SOTONOQ 1299 non-null uint8 \n 61 T_SP 1299 non-null uint8 \n 62 T_STONO 1299 non-null uint8 \n 63 T_STONO2 1299 non-null uint8 \n 64 T_STONOQ 1299 non-null uint8 \n 65 T_SWPP 1299 non-null uint8 \n 66 T_WC 1299 non-null uint8 \n 67 T_WEP 1299 non-null uint8 \n 68 T_X 1299 non-null uint8 \n 69 Embarked_C 1299 non-null uint8 \n 70 Embarked_Q 1299 non-null uint8 \n 71 Embarked_S 1299 non-null uint8 \n 72 Cabin_A 1299 non-null uint8 \n 73 Cabin_B 1299 non-null uint8 \n 74 Cabin_C 1299 non-null uint8 \n 75 Cabin_D 1299 non-null uint8 \n 76 Cabin_E 1299 non-null uint8 \n 77 Cabin_F 1299 non-null uint8 \n 78 Cabin_G 1299 non-null uint8 \n 79 Cabin_T 1299 non-null uint8 \n 80 Cabin_X 1299 non-null uint8 \n 81 CabinBol_1 1299 non-null uint8 \ndtypes: float64(3), int64(9), uint8(70)\nmemory usage: 210.7 KB\n"
],
[
"df.columns",
"_____no_output_____"
],
[
"df = df.drop(labels = [\"PassengerId\", \"Parch\", \"Fam_size\"],axis = 1)",
"_____no_output_____"
],
[
"df.columns",
"_____no_output_____"
],
[
"cols = ['Pclass', 'SibSp', 'Parch', 'Fare', 'Sex','Cabin_mapped', 'Embarked', 'Survived', 'Age']\ndf = df[cols]",
"_____no_output_____"
],
[
"df = pd.get_dummies(df, columns=['Sex', 'Cabin_mapped', 'Embarked'],drop_first=True)",
"_____no_output_____"
]
],
[
[
"# Modeling",
"_____no_output_____"
]
],
[
[
"# Let's split the train and test data sets\ntrain = df[:len_titanic_train]\ntest = df[len_titanic_train:]\n\n# Drop the empty Survived column from the test dataset. \ntest.drop(labels=['Survived'], axis=1, inplace=True)",
"_____no_output_____"
],
[
"## Separate train features and label \n\ntrain[\"Survived\"] = train[\"Survived\"].astype(int)\n\ny = train[\"Survived\"]\n\nX = train.drop(labels = [\"Survived\"],axis = 1)",
"_____no_output_____"
]
],
[
[
"#### Split Test Train Data\n\nHere, I am going to split the data into training and validation sets using Scikit-Learn. ",
"_____no_output_____"
]
],
[
[
"X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3, random_state=42)",
"_____no_output_____"
]
],
[
[
"#### Model",
"_____no_output_____"
],
[
"For the first run, I am going to try Random Forest Classifier using GridSearch. ",
"_____no_output_____"
]
],
[
[
"# Istentiate the model\nrfc=RandomForestClassifier(random_state=42, n_jobs=4)",
"_____no_output_____"
],
[
"# Parameter for our classifier\nparam_grid = { \n 'n_estimators': [100,150, 200, 500, 600],\n 'max_features': ['auto', 'sqrt', 'log2'],\n 'max_depth' : [2, 4,5,6,7,8, 9, 10, 11, 12, 13, 14, 18],\n 'criterion' :['gini', 'entropy']\n}",
"_____no_output_____"
],
[
"# Defining our Gridsearch cross validation\nCV_rfc = GridSearchCV(estimator=rfc, param_grid=param_grid, cv= 5)",
"_____no_output_____"
],
[
"# Fitting the GridSearch to training and testing. \nCV_rfc.fit(X_train, y_train)",
"_____no_output_____"
],
[
"# Looking the best parameters. \nCV_rfc.best_params_",
"_____no_output_____"
],
[
"# Now, we can use the parameters above to define our model. \nrfc1=RandomForestClassifier(random_state=42, max_features='auto', n_estimators= 500, \n max_depth=12, criterion='entropy', n_jobs=6)",
"_____no_output_____"
],
[
"rfc1.fit(X_train, y_train)",
"_____no_output_____"
],
[
"test_predictions = rfc1.predict(test)",
"_____no_output_____"
],
[
"submission = pd.DataFrame()\nsubmission['PassengerId'] = titanic_test['PassengerId']\nsubmission['Survived'] = test_predictions",
"_____no_output_____"
],
[
"submission.to_csv(\"data/titanic/submission.csv\", index=False)\n",
"_____no_output_____"
]
],
[
[
"#### XGBoost",
"_____no_output_____"
]
],
[
[
"import warnings\nwarnings.filterwarnings('ignore')\nfrom datetime import datetime\nfrom sklearn.model_selection import RandomizedSearchCV, GridSearchCV\nfrom sklearn.metrics import roc_auc_score\nfrom sklearn.model_selection import StratifiedKFold\nfrom xgboost import XGBClassifier",
"_____no_output_____"
],
[
"# A parameter grid for XGBoost\nparams = {\n 'min_child_weight': [1, 5, 10],\n 'gamma': [0.5, 1, 1.5, 2, 5],\n 'subsample': [0.6, 0.8, 1.0],\n 'colsample_bytree': [0.6, 0.8, 1.0],\n 'max_depth': [3, 4, 5]\n }",
"_____no_output_____"
],
[
"xgb = XGBClassifier(learning_rate=0.02, n_estimators=600, objective='binary:logistic',\n silent=True, nthread=1)",
"_____no_output_____"
],
[
"folds = 3\nparam_comb = 5\n\nskf = StratifiedKFold(n_splits=folds, shuffle = True, random_state = 42)\n\nrandom_search = RandomizedSearchCV(xgb, param_distributions=params, n_iter=param_comb, scoring='accuracy', \n n_jobs=4, cv=skf.split(X,y), verbose=3, random_state=1001 )\n\n# Here we go\nrandom_search.fit(X, y)\n#roc_auc",
"Fitting 3 folds for each of 5 candidates, totalling 15 fits\n"
],
[
"submission = pd.DataFrame()\nsubmission['PassengerId'] = titanic_test['PassengerId']\nsubmission['Survived'] = test_predictions\nsubmission.to_csv(\"data/titanic/submission.csv\", index=False)",
"_____no_output_____"
]
],
[
[
"### Ongoing work! \n\nI am still trying to improve my Kaggle Score. I will continue using the following models. ",
"_____no_output_____"
]
],
[
[
"import xgboost as xgb\nfrom sklearn.model_selection import RandomizedSearchCV\n\n# Create the parameter grid: gbm_param_grid \ngbm_param_grid = {\n 'n_estimators': range(8, 20),\n 'max_depth': range(6, 10),\n 'learning_rate': [.4, .45, .5, .55, .6],\n 'colsample_bytree': [.6, .7, .8, .9, 1]\n}\n\n# Instantiate the regressor: gbm\ngbm = XGBClassifier(n_estimators=10)\n\n# Perform random search: grid_mse\nxgb_random = RandomizedSearchCV(param_distributions=gbm_param_grid, \n estimator = gbm, scoring = \"accuracy\", \n verbose = 1, n_iter = 50, cv = 4)\n\n\n# Fit randomized_mse to the data\nxgb_random.fit(X, y)\n\n# Print the best parameters and lowest RMSE\nprint(\"Best parameters found: \", xgb_random.best_params_)\nprint(\"Best accuracy found: \", xgb_random.best_score_)",
"Fitting 4 folds for each of 50 candidates, totalling 200 fits\n"
],
[
"xgb_pred = xgb_random.predict(test)",
"_____no_output_____"
],
[
"submission = pd.DataFrame()\nsubmission['PassengerId'] = titanic_test['PassengerId']\nsubmission['Survived'] = xgb_pred\nsubmission.to_csv(\"data/titanic/submission.csv\", index=False)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
c515bad20825d145d2d16f3f82cddfcf842d46b3
| 14,911 |
ipynb
|
Jupyter Notebook
|
scripts/notebooks/halo/plot_halos.ipynb
|
Hoseung/pyRamAn
|
f9386fa5a9f045f98590039988d3cd50bc488dc2
|
[
"MIT"
] | 1 |
2021-11-25T16:11:56.000Z
|
2021-11-25T16:11:56.000Z
|
scripts/notebooks/halo/plot_halos.ipynb
|
Hoseung/pyRamAn
|
f9386fa5a9f045f98590039988d3cd50bc488dc2
|
[
"MIT"
] | 6 |
2020-02-17T13:44:43.000Z
|
2020-06-25T15:35:05.000Z
|
scripts/notebooks/halo/plot_halos.ipynb
|
Hoseung/pyRamAn
|
f9386fa5a9f045f98590039988d3cd50bc488dc2
|
[
"MIT"
] | 1 |
2021-11-25T16:11:56.000Z
|
2021-11-25T16:11:56.000Z
| 44.510448 | 3,533 | 0.572195 |
[
[
[
"### Halo check\nPlot halos to see if halofinders work well",
"_____no_output_____"
]
],
[
[
"#import os\n#base = os.path.abspath('/home/hoseung/Work/data/05427/')\n\n#base = base + '/'\n\n# basic parameters\n# Directory, file names, snapshots, scale, npix\nbase = '/home/hoseung/Work/data/05427/'\ncluster_name = base.split('/')[-2]\nfrefine= 'refine_params.txt'\nfnml = input(\"type namelist file name (enter = cosmo_200.nml):\")\nif fnml ==\"\":\n fnml = 'cosmo_200.nml'\nnout_ini=int(input(\"Starting nout?\"))\nnout_fi=int(input(\"ending nout?\"))\nnouts = range(nout_ini,nout_fi+1)\n\nscale = input(\"Scale?: \")\nif scale==\"\":\n scale = 0.3\nscale = float(scale)\n\nnpix = input(\"npix (enter = 400)\")\nif npix == \"\":\n npix = 400\nnpix = int(npix)\n\n# data loading parameters\nptype=[\"star pos mass\"]\nrefine_params = True\ndmo=False\ndraw=True\ndraw_halos=True\ndraw_part = True\ndraw_hydro = False\nif draw_hydro:\n lmax=input(\"maximum level\")\n if lmax==\"\":\n lmax=19\n lmax = int(lmax)",
"\ntype namelist file name (enter = cosmo_200.nml):\nStarting nout?186\nending nout?187\nScale?: 1.0\nnpix (enter = 400)800\n"
],
[
"import load\nimport utils.sampling as smp\nimport utils.match as mtc\nimport draw\nimport pickle\n\n\nfor nout in nouts:\n snout = str(nout).zfill(3)\n\n if refine_params:\n # instead of calculating zoomin region, just load it from the refine_params.txt file.\n # region = s.part.search_zoomin(scale=0.5, load=True)\n\n rr = load.info.RefineParam()\n rr.loadRegion(base + frefine)\n \n nn = load.info.Nml(fname=base + fnml)\n \n aexp = nn.aout[nout-1]\n i_aexp = mtc.closest(aexp, rr.aexp)\n \n x_refine = rr.x_refine[i_aexp]\n y_refine = rr.y_refine[i_aexp]\n z_refine = rr.z_refine[i_aexp]\n r_refine = rr.r_refine[i_aexp] * 0.5\n \n region = smp.set_region(xc = x_refine, yc = y_refine, zc = z_refine,\n radius = r_refine * scale)\n else: \n region = smp.set_region(xc=0.5, yc=0.5, zc=0.5, radius=0.1) \n \n s = load.sim.Sim(nout, base, dmo=dmo, ranges=region[\"ranges\"], setup=True)\n imgs = draw.img_obj.MapSet(info=s.info, region=region)\n imgp = draw.img_obj.MapImg(info=s.info, proj='z', npix=npix, ptype=ptype)\n imgp.set_region(region)\n\n #%%\n if draw_part:\n s.add_part(ptype)\n s.part.load()\n part = getattr(s.part, s.part.pt[0])\n \n x = part['x']\n y = part['y']\n z = part['y']\n m = part['m'] * s.info.msun # part must be normalized already!\n \n #imgp.set_data(draw.pp.den2d(x, y, z, m, npix, s.info, cic=True, norm_integer=True))\n imgp.set_data(draw.pp.den2d(x, y, z, m, npix, region, cic=True, norm_integer=True))\n imgs.ptden2d = imgp\n# imgp.show_data()\n\n #%%\n if draw_hydro:\n s.add_hydro()\n s.hydro.amr2cell(lmax=lmax)\n field = draw.pp.pp_cell(s.hydro.cell, npix, s.info, verbose=True)\n ptype = 'gas_den'\n imgh = draw.img_obj.MapImg(info=s.info, proj='z', npix=npix, ptype=ptype)\n imgh.set_data(field)\n imgh.set_region(region)\n # imgh.show_data()\n imgs.hydro = imgh\n \n #%%\n fdump = base + snout + 'map.pickle'\n with open(fdump, 'wb') as f:\n pickle.dump(imgs, f)\n if draw:\n if draw_part:\n imgs.ptden2d.plot_2d_den(save= base + cluster_name + snout +'star.png', dpi=400, show=False)\n if draw_hydro: \n imgs.hydro.plot_2d_den(save= base + cluster_name +snout + 'hydro.png',vmax=15,vmin=10, show=False,\n dpi=400)",
"Ranges = [[0.486934514119, 0.526934514119], [0.292903416014, 0.33290341601400003], [0.283048251159, 0.32304825115900004]]\n\nNo AMR instance,\nLoading one...\nAn AMR instance is created\n\nUpdating info.cpus\nAn AMR instance is created\n\nUpdating info.cpus\n Simulation set up.\nUpdating data\nTypes of particles you want to load are: ['star pos mass']\nNo AMR instance,\nLoading one...\nAn AMR instance is created\n\nUpdating info.cpus\nNo info._set_cpus attribute??\nA particle instance is created\n\nUse part.load() to load particle\nLoading particle... \n ranges: [[0.486934514119, 0.526934514119], [0.292903416014, 0.33290341601400003], [0.283048251159, 0.32304825115900004]]\nnpart_arr: [529265, 150869, 20003, 22575, 43984, 37461, 36154, 48006, 57590, 9513, 17105, 27692, 22877, 11515, 26766, 522364, 19630, 24029, 24154, 22484, 11173, 9271, 824362, 56354, 475406, 254975, 29430, 23149, 5520, 8118, 7391, 9926, 15295, 15177, 28856, 30638, 27114, 33751, 7464, 204523, 309474, 19159, 43451, 8224, 35226, 21942, 11894, 17735, 20877, 21723, 15979, 70024, 5921, 9949, 10020, 16786, 11333, 21722, 14847, 19518, 17132, 21256, 9394, 27007, 26537, 21641, 27645, 24015, 17073, 7606, 34677, 8870, 30942, 20329, 30404, 7629, 8131, 30725, 20468, 10113, 19699, 12102, 19664, 38313, 38244, 69874, 109368, 24489, 11307, 20466, 52467, 19660, 13548, 33961, 13332, 45313, 47917, 18428, 23815, 11196, 7319, 41786, 43866, 32716, 26223, 16488, 26614, 20561, 38232, 38293, 26603, 28724, 16690, 52383, 33255, 45442, 32104, 11932, 17118, 17921, 84654, 18490, 36028, 44233, 33678, 9302, 23399, 10536, 32452, 26232, 38902, 27886, 32312, 58857, 21297, 34302, 26282, 23167, 47750, 76078, 59090, 70214, 20422, 41138, 88852, 82494, 31957, 23277, 73332, 43356, 33305, 8980, 53595, 50352, 47958, 73418, 80016, 38782, 13865, 28989, 49620, 60289, 94562, 92784, 66015, 52305, 56445, 48663, 41591, 85647, 52880, 86496, 894, 4107, 44887, 60310, 22963, 33196, 17234, 37436, 44829, 93569, 128584, 4726, 4381, 14496, 25216, 32565, 91911, 28984, 17981, 27571, 69113, 35791, 2762, 46956, 64625, 64696, 3215, 4447, 39170, 28169, 22718, 6339, 43872, 40548, 41209, 31514, 7657, 34874, 1874, 42945, 32000, 43026, 17290, 41553, 28773, 48842, 31395, 32984, 15848, 43440, 53971, 68887, 39082, 53564, 34854, 38711, 29219, 43339, 50797, 30328, 19171, 23300, 24489, 31471, 22893, 12415, 223497, 15420, 15723, 6762, 46117, 35463, 43322, 584027, 31820, 7328, 9258, 10080, 8547, 19347, 14112, 5431, 16543, 10737, 17104, 15103, 22363, 3922, 20825, 9516, 10737, 21872, 18589, 1100, 34399, 48714, 44292, 37897, 20593, 10549, 43452, 42166, 23834, 17400, 24698, 38343, 38517, 25552, 16764, 28588, 23986, 21962, 25219, 18429, 18746, 15464, 18246, 53176, 11873, 34195, 13824, 34217, 36625, 26595, 29859, 23787, 14560, 29480, 18774, 10195, 59160, 26622, 36086, 54378, 40566, 47629, 39171, 30037, 46922, 48545, 65365, 58790, 69937, 25395, 68465, 35501, 66447, 31385, 50651, 25449, 10812, 20569, 35861, 26726, 15062, 30619, 53629, 37574, 27291, 8216, 17065, 61571, 20034, 62319, 60138, 57448, 30817, 23710, 13328, 49182, 53300, 74505, 81985, 52126, 29145, 24000, 39309, 57308, 30143, 41471, 58924, 68170, 85980, 45847, 58099, 62767, 14864, 13621, 46571, 66094, 84409, 57987, 91566, 45780, 55043, 53591, 29607, 13157, 326, 3101, 96341, 47163, 34352, 31671, 47208, 16953, 11941, 14103, 25071, 52109, 43121, 45686, 5310, 40943, 26962, 28618, 50700, 20850, 6761, 46515, 18665, 36488, 45376, 47810, 49104, 9329, 51838, 31268, 17730, 45549, 34136, 57291, 21198, 24641, 37536, 37781, 27909, 7314, 44860, 49944, 50581, 35038, 63620, 8057, 45088, 16808, 11595, 16316, 67988, 26044, 41473, 28600, 18396, 37386, 13051, 24468, 29761, 45706, 22299, 15762, 47005, 17895, 13167, 11750, 14427, 12007, 21804, 27693, 48735, 19316, 10829, 11816, 18551, 5868, 15858, 25487, 20857, 7633, 11421, 5150, 577, 10568, 78677, 19543, 19636, 46467, 14042, 12867, 41928, 56219, 51409, 39481, 21861, 21116, 14889, 19717, 26649, 20875, 9941, 20052, 3511, 27773, 23280, 17796, 39632, 35024, 11286, 14843, 19374, 25490, 12914, 45499, 40331, 58693, 20762, 1991, 15542, 29187, 40573, 48939, 10857, 27722, 22813, 6546, 40336, 54689, 31695, 32252, 374216, 43405, 17217, 64585, 333731, 228090, 1142615]\nTotal DM particle 3986787\nTotal star particle 10414404\nTotal sink particle 1594404\nResampling 10294369 values to a 800 by 800 by 1 grid\nUpdating field vals\nUpdating field vals\nUpdating field vals\nUpdating field vals\nminmax field after crop and converting into physical unit 54131815.8091 2.34816405446e+19\nUpdating data\n"
],
[
"import matplotlib.pyplot as plt\nfig = plt.figure()\nax1 = fig.add_subplot(111)\nsnout = str(nout).zfill(3)\n\nfin = base + snout + 'map.pickle'\nwith open(fin, 'rb') as f:\n img = pickle.load(f)\n\nptimg = img.ptden2d\nfout = base + snout + \"dmmap_\" + ptimg.proj + \".png\"\nimg.ptden2d.plot_2d_den(save=False, show=False, vmin=1e13, vmax=1e20, dpi=400, axes=ax1)\n\nimport tree\nimport numpy as np\n\n#s = load.sim.Sim(nout, base_dir)\ninfo = load.info.Info(nout=nout, base=base, load=True)\nhall = tree.halomodule.Halo(nout=nout, base=base, halofinder=\"HM\", info=info, load=True)\n\ni_center = np.where(hall.data['np'] == max(hall.data['np']))\nh = tree.halomodule.Halo()\nh.derive_from(hall, [i_center]) \n\n#region = smp.set_region(xc=h.data.x, yc=h.data.y, zc=h.data.z, radius = h.data.rvir * 2) \n\n#%%\nfrom draw import pp\n\nind = np.where(hall.data.mvir > 5e10)\nh_sub = tree.halomodule.Halo()\nh_sub.derive_from(hall, ind) \n#x = hall.data.x#[ind]\n#y = hall.data.y#[ind]\n#r = hall.data.rvir#[ind]\n#pp.circle_scatter(ax1, x*npix, y*npix, r*30, facecolor='none', edgecolor='b', label='555')\n\n#ax1.set_xlim(right=npix).\n#ax1.set_ylim(top=npix)\npp.pp_halo(h_sub, npix, region=img.ptden2d.region, axes=ax1, rscale=3, name=True)\n\nplt.show()",
"Are these parameters right?\nAre these parameters right?\n"
]
],
[
[
"##### Load halofinder result\n\n##### get position and virial radius \n\n##### load particles data (star or DM) and draw density map\n##### plot halos on top of particle density map",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
]
] |
c515bb7f695d566f173a0bdd5748fc7a60389f24
| 134 |
ipynb
|
Jupyter Notebook
|
stevensmiley1989_MoNuSAC_Entire_Code_5_15_2021.ipynb
|
stevensmiley1989/MoNuSAC_Grand_Challenge_S4xUNet
|
56c69bd09d6e9a81d4f67905baf5bda19240412c
|
[
"MIT"
] | 1 |
2021-07-15T08:13:24.000Z
|
2021-07-15T08:13:24.000Z
|
stevensmiley1989_MoNuSAC_Entire_Code_5_15_2021.ipynb
|
stevensmiley1989/MoNuSAC_Grand_Challenge_S4xUNet
|
56c69bd09d6e9a81d4f67905baf5bda19240412c
|
[
"MIT"
] | 1 |
2021-05-20T16:05:42.000Z
|
2021-05-20T16:05:42.000Z
|
stevensmiley1989_MoNuSAC_Entire_Code_5_15_2021.ipynb
|
stevensmiley1989/MoNuSAC_Grand_Challenge_S4xUNet
|
56c69bd09d6e9a81d4f67905baf5bda19240412c
|
[
"MIT"
] | 1 |
2022-03-23T15:17:21.000Z
|
2022-03-23T15:17:21.000Z
| 33.5 | 75 | 0.88806 |
[
[
[
"empty"
]
]
] |
[
"empty"
] |
[
[
"empty"
]
] |
c515d65c8bcaf039302928925fede77f490a286d
| 132,404 |
ipynb
|
Jupyter Notebook
|
jupyter/chap12.ipynb
|
SadPuffin/ModSimPy
|
6cac03b114a0bf0893aa160048ba6dae460f5f86
|
[
"MIT"
] | null | null | null |
jupyter/chap12.ipynb
|
SadPuffin/ModSimPy
|
6cac03b114a0bf0893aa160048ba6dae460f5f86
|
[
"MIT"
] | null | null | null |
jupyter/chap12.ipynb
|
SadPuffin/ModSimPy
|
6cac03b114a0bf0893aa160048ba6dae460f5f86
|
[
"MIT"
] | null | null | null | 127.55684 | 23,128 | 0.880434 |
[
[
[
"# Chapter 12",
"_____no_output_____"
],
[
"*Modeling and Simulation in Python*\n\nCopyright 2021 Allen Downey\n\nLicense: [Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International](https://creativecommons.org/licenses/by-nc-sa/4.0/)",
"_____no_output_____"
]
],
[
[
"# check if the libraries we need are installed\n\ntry:\n import pint\nexcept ImportError:\n !pip install pint\n \ntry:\n import modsim\nexcept ImportError:\n !pip install modsimpy",
"_____no_output_____"
]
],
[
[
"### Code\n\nHere's the code from the previous notebook that we'll need.",
"_____no_output_____"
]
],
[
[
"from modsim import State, System\n\ndef make_system(beta, gamma):\n \"\"\"Make a system object for the SIR model.\n \n beta: contact rate in days\n gamma: recovery rate in days\n \n returns: System object\n \"\"\"\n init = State(S=89, I=1, R=0)\n init /= sum(init)\n\n t0 = 0\n t_end = 7 * 14\n\n return System(init=init, t0=t0, t_end=t_end,\n beta=beta, gamma=gamma)",
"_____no_output_____"
],
[
"def update_func(state, t, system):\n \"\"\"Update the SIR model.\n \n state: State with variables S, I, R\n t: time step\n system: System with beta and gamma\n \n returns: State object\n \"\"\"\n s, i, r = state\n\n infected = system.beta * i * s \n recovered = system.gamma * i\n \n s -= infected\n i += infected - recovered\n r += recovered\n \n return State(S=s, I=i, R=r)",
"_____no_output_____"
],
[
"from numpy import arange\nfrom modsim import TimeFrame\n\ndef run_simulation(system, update_func):\n \"\"\"Runs a simulation of the system.\n \n system: System object\n update_func: function that updates state\n \n returns: TimeFrame\n \"\"\"\n frame = TimeFrame(columns=system.init.index)\n frame.loc[system.t0] = system.init\n \n for t in arange(system.t0, system.t_end):\n frame.loc[t+1] = update_func(frame.loc[t], t, system)\n \n return frame",
"_____no_output_____"
]
],
[
[
"In the previous chapter I presented the SIR model of infectious disease and used it to model the Freshman Plague at Olin. In this chapter we'll consider metrics intended to quantify the effects of the disease and interventions intended to reduce those effects.",
"_____no_output_____"
],
[
"## Immunization\n\nModels like this are useful for testing \"what if?\" scenarios. As an\nexample, we'll consider the effect of immunization.\n\nSuppose there is a vaccine that causes a student to become immune to the Freshman Plague without being infected. How might you modify the model to capture this effect?\n\nOne option is to treat immunization as a shortcut from susceptible to\nrecovered without going through infectious. We can implement this\nfeature like this:",
"_____no_output_____"
]
],
[
[
"def add_immunization(system, fraction):\n system.init.S -= fraction\n system.init.R += fraction",
"_____no_output_____"
]
],
[
[
"`add_immunization` moves the given fraction of the population from `S`\nto `R`. ",
"_____no_output_____"
]
],
[
[
"tc = 3 # time between contacts in days \ntr = 4 # recovery time in days\n\nbeta = 1 / tc # contact rate in per day\ngamma = 1 / tr # recovery rate in per day\n\nsystem = make_system(beta, gamma)\nresults = run_simulation(system, update_func)",
"_____no_output_____"
]
],
[
[
"If we assume that 10% of students are vaccinated at the\nbeginning of the semester, and the vaccine is 100% effective, we can\nsimulate the effect like this:",
"_____no_output_____"
]
],
[
[
"system2 = make_system(beta, gamma)\nadd_immunization(system2, 0.1)\nresults2 = run_simulation(system2, update_func)",
"_____no_output_____"
]
],
[
[
"The following figure shows `S` as a function of time, with and\nwithout immunization.",
"_____no_output_____"
]
],
[
[
"results.S.plot(label='No immunization')\nresults2.S.plot(label='10% immunization')\n\ndecorate(xlabel='Time (days)',\n ylabel='Fraction of population')",
"_____no_output_____"
]
],
[
[
"## Metrics\n\nWhen we plot a time series, we get a view of everything that happened\nwhen the model ran, but often we want to boil it down to a few numbers\nthat summarize the outcome. These summary statistics are called\n**metrics**, as we saw in Section xxx.\n\nIn the SIR model, we might want to know the time until the peak of the\noutbreak, the number of people who are sick at the peak, the number of\nstudents who will still be sick at the end of the semester, or the total number of students who get sick at any point.\n\nAs an example, I will focus on the last one --- the total number of sick students --- and we will consider interventions intended to minimize it.\n\nWhen a person gets infected, they move from `S` to `I`, so we can get\nthe total number of infections by computing the difference in `S` at the beginning and the end:",
"_____no_output_____"
]
],
[
[
"def calc_total_infected(results, system):\n s_0 = results.S[system.t0]\n s_end = results.S[system.t_end]\n return s_0 - s_end",
"_____no_output_____"
],
[
"calc_total_infected(results, system)",
"_____no_output_____"
],
[
"calc_total_infected(results2, system2)",
"_____no_output_____"
]
],
[
[
"Without immunization, almost 47% of the population gets infected at some point. With 10% immunization, only 31% get infected. That's pretty good.",
"_____no_output_____"
],
[
"## Sweeping Immunization\n\nNow let's see what happens if we administer more vaccines. This\nfollowing function sweeps a range of immunization rates:",
"_____no_output_____"
]
],
[
[
"def sweep_immunity(immunize_array):\n sweep = SweepSeries()\n\n for fraction in immunize_array:\n sir = make_system(beta, gamma)\n add_immunization(sir, fraction)\n results = run_simulation(sir, update_func)\n sweep[fraction] = calc_total_infected(results, sir)\n\n return sweep",
"_____no_output_____"
]
],
[
[
"The parameter of `sweep_immunity` is an array of immunization rates. The\nresult is a `SweepSeries` object that maps from each immunization rate\nto the resulting fraction of students ever infected.\n\nThe following figure shows a plot of the `SweepSeries`. Notice that\nthe x-axis is the immunization rate, not time.",
"_____no_output_____"
]
],
[
[
"immunize_array = linspace(0, 1, 21)\ninfected_sweep = sweep_immunity(immunize_array)",
"_____no_output_____"
],
[
"infected_sweep.plot()\n\ndecorate(xlabel='Fraction immunized',\n ylabel='Total fraction infected',\n title='Fraction infected vs. immunization rate')",
"_____no_output_____"
]
],
[
[
"As the immunization rate increases, the number of infections drops\nsteeply. If 40% of the students are immunized, fewer than 4% get sick.\nThat's because immunization has two effects: it protects the people who get immunized (of course) but it also protects the rest of the\npopulation.\n\nReducing the number of \"susceptibles\" and increasing the number of\n\"resistants\" makes it harder for the disease to spread, because some\nfraction of contacts are wasted on people who cannot be infected. This\nphenomenon is called **herd immunity**, and it is an important element\nof public health (see <http://modsimpy.com/herd>).",
"_____no_output_____"
],
[
"The steepness of the curve is a blessing and a curse. It's a blessing\nbecause it means we don't have to immunize everyone, and vaccines can\nprotect the \"herd\" even if they are not 100% effective.\n\nBut it's a curse because a small decrease in immunization can cause a\nbig increase in infections. In this example, if we drop from 80%\nimmunization to 60%, that might not be too bad. But if we drop from 40% to 20%, that would trigger a major outbreak, affecting more than 15% of the population. For a serious disease like measles, just to name one, that would be a public health catastrophe.\n\nOne use of models like this is to demonstrate phenomena like herd\nimmunity and to predict the effect of interventions like vaccination.\nAnother use is to evaluate alternatives and guide decision making. We'll see an example in the next section.",
"_____no_output_____"
],
[
"## Hand washing\n\nSuppose you are the Dean of Student Life, and you have a budget of just \\$1200 to combat the Freshman Plague. You have two options for spending this money:\n\n1. You can pay for vaccinations, at a rate of \\$100 per dose.\n\n2. You can spend money on a campaign to remind students to wash hands\n frequently.\n\nWe have already seen how we can model the effect of vaccination. Now\nlet's think about the hand-washing campaign. We'll have to answer two\nquestions:\n\n1. How should we incorporate the effect of hand washing in the model?\n\n2. How should we quantify the effect of the money we spend on a\n hand-washing campaign?\n\nFor the sake of simplicity, let's assume that we have data from a\nsimilar campaign at another school showing that a well-funded campaign\ncan change student behavior enough to reduce the infection rate by 20%.\n\nIn terms of the model, hand washing has the effect of reducing `beta`.\nThat's not the only way we could incorporate the effect, but it seems\nreasonable and it's easy to implement.",
"_____no_output_____"
],
[
"Now we have to model the relationship between the money we spend and the\neffectiveness of the campaign. Again, let's suppose we have data from\nanother school that suggests:\n\n- If we spend \\$500 on posters, materials, and staff time, we can\n change student behavior in a way that decreases the effective value of `beta` by 10%.\n\n- If we spend \\$1000, the total decrease in `beta` is almost 20%.\n\n- Above \\$1000, additional spending has little additional benefit.",
"_____no_output_____"
],
[
"### Logistic function",
"_____no_output_____"
],
[
"To model the effect of a hand-washing campaign, I'll use a [generalized logistic function](https://en.wikipedia.org/wiki/Generalised_logistic_function) (GLF), which is a convenient function for modeling curves that have a generally sigmoid shape. The parameters of the GLF correspond to various features of the curve in a way that makes it easy to find a function that has the shape you want, based on data or background information about the scenario.",
"_____no_output_____"
]
],
[
[
"from numpy import exp\n\ndef logistic(x, A=0, B=1, C=1, M=0, K=1, Q=1, nu=1):\n \"\"\"Computes the generalize logistic function.\n \n A: controls the lower bound\n B: controls the steepness of the transition \n C: not all that useful, AFAIK\n M: controls the location of the transition\n K: controls the upper bound\n Q: shift the transition left or right\n nu: affects the symmetry of the transition\n \n returns: float or array\n \"\"\"\n exponent = -B * (x - M)\n denom = C + Q * exp(exponent)\n return A + (K-A) / denom ** (1/nu)",
"_____no_output_____"
]
],
[
[
"The following array represents the range of possible spending.",
"_____no_output_____"
]
],
[
[
"spending = linspace(0, 1200, 21)",
"_____no_output_____"
]
],
[
[
"`compute_factor` computes the reduction in `beta` for a given level of campaign spending.\n\n`M` is chosen so the transition happens around \\$500.\n\n`K` is the maximum reduction in `beta`, 20%.\n\n`B` is chosen by trial and error to yield a curve that seems feasible.",
"_____no_output_____"
]
],
[
[
"def compute_factor(spending):\n \"\"\"Reduction factor as a function of spending.\n \n spending: dollars from 0 to 1200\n \n returns: fractional reduction in beta\n \"\"\"\n return logistic(spending, M=500, K=0.2, B=0.01)",
"_____no_output_____"
]
],
[
[
"Here's what it looks like.",
"_____no_output_____"
]
],
[
[
"percent_reduction = compute_factor(spending) * 100\n\nplot(spending, percent_reduction)\n\ndecorate(xlabel='Hand-washing campaign spending (USD)',\n ylabel='Percent reduction in infection rate',\n title='Effect of hand washing on infection rate')",
"_____no_output_____"
]
],
[
[
"The result is the following function, which\ntakes spending as a parameter and returns `factor`, which is the factor\nby which `beta` is reduced:",
"_____no_output_____"
]
],
[
[
"def compute_factor(spending):\n return logistic(spending, M=500, K=0.2, B=0.01)",
"_____no_output_____"
]
],
[
[
"I use `compute_factor` to write `add_hand_washing`, which takes a\n`System` object and a budget, and modifies `system.beta` to model the\neffect of hand washing:",
"_____no_output_____"
]
],
[
[
"def add_hand_washing(system, spending):\n factor = compute_factor(spending)\n system.beta *= (1 - factor)",
"_____no_output_____"
]
],
[
[
"Now we can sweep a range of values for `spending` and use the simulation\nto compute the effect:",
"_____no_output_____"
]
],
[
[
"def sweep_hand_washing(spending_array):\n sweep = SweepSeries()\n \n for spending in spending_array:\n system = make_system(beta, gamma)\n add_hand_washing(system, spending)\n results = run_simulation(system, update_func)\n sweep[spending] = calc_total_infected(results, system)\n \n return sweep",
"_____no_output_____"
]
],
[
[
"Here's how we run it:",
"_____no_output_____"
]
],
[
[
"from numpy import linspace\n\nspending_array = linspace(0, 1200, 20)\ninfected_sweep2 = sweep_hand_washing(spending_array)",
"_____no_output_____"
]
],
[
[
"The following figure shows the result. ",
"_____no_output_____"
]
],
[
[
"infected_sweep2.plot()\n\ndecorate(xlabel='Hand-washing campaign spending (USD)',\n ylabel='Total fraction infected',\n title='Effect of hand washing on total infections')",
"_____no_output_____"
]
],
[
[
"Below \\$200, the campaign has little effect. \n\nAt \\$800 it has a substantial effect, reducing total infections from more than 45% to about 20%. \n\nAbove \\$800, the additional benefit is small.",
"_____no_output_____"
],
[
"## Optimization\n\nLet's put it all together. With a fixed budget of \\$1200, we have to\ndecide how many doses of vaccine to buy and how much to spend on the\nhand-washing campaign.\n\nHere are the parameters:",
"_____no_output_____"
]
],
[
[
"num_students = 90\nbudget = 1200\nprice_per_dose = 100\nmax_doses = int(budget / price_per_dose)",
"_____no_output_____"
]
],
[
[
"The fraction `budget/price_per_dose` might not be an integer. `int` is a\nbuilt-in function that converts numbers to integers, rounding down.\n\nWe'll sweep the range of possible doses:",
"_____no_output_____"
]
],
[
[
"dose_array = arange(max_doses+1)",
"_____no_output_____"
]
],
[
[
"In this example we call `linrange` with only one argument; it returns a\nNumPy array with the integers from 0 to `max_doses`. With the argument\n`endpoint=True`, the result includes both endpoints.\n\nThen we run the simulation for each element of `dose_array`:",
"_____no_output_____"
]
],
[
[
"def sweep_doses(dose_array):\n sweep = SweepSeries()\n \n for doses in dose_array:\n fraction = doses / num_students\n spending = budget - doses * price_per_dose\n \n system = make_system(beta, gamma)\n add_immunization(system, fraction)\n add_hand_washing(system, spending)\n \n results = run_simulation(system, update_func)\n sweep[doses] = calc_total_infected(results, system)\n\n return sweep",
"_____no_output_____"
]
],
[
[
"For each number of doses, we compute the fraction of students we can\nimmunize, `fraction` and the remaining budget we can spend on the\ncampaign, `spending`. Then we run the simulation with those quantities\nand store the number of infections.\n\nThe following figure shows the result.",
"_____no_output_____"
]
],
[
[
"infected_sweep3 = sweep_doses(dose_array)",
"_____no_output_____"
],
[
"infected_sweep3.plot()\n\ndecorate(xlabel='Doses of vaccine',\n ylabel='Total fraction infected',\n title='Total infections vs. doses')",
"_____no_output_____"
]
],
[
[
"If we buy no doses of vaccine and spend the entire budget on the campaign, the fraction infected is around 19%. At 4 doses, we have \\$800 left for the campaign, and this is the optimal point that minimizes the number of students who get sick.\n\nAs we increase the number of doses, we have to cut campaign spending,\nwhich turns out to make things worse. But interestingly, when we get\nabove 10 doses, the effect of herd immunity starts to kick in, and the\nnumber of sick students goes down again.",
"_____no_output_____"
],
[
"## Summary",
"_____no_output_____"
],
[
"### Exercises\n\n**Exercise:** Suppose the price of the vaccine drops to $50 per dose. How does that affect the optimal allocation of the spending?",
"_____no_output_____"
],
[
"**Exercise:** Suppose we have the option to quarantine infected students. For example, a student who feels ill might be moved to an infirmary, or a private dorm room, until they are no longer infectious.\n\nHow might you incorporate the effect of quarantine in the SIR model?",
"_____no_output_____"
]
],
[
[
"# Solution\n\n\"\"\"There is no unique best answer to this question,\nbut one simple option is to model quarantine as an\neffective reduction in gamma, on the assumption that\nquarantine reduces the number of infectious contacts\nper infected student.\n\nAnother option would be to add a fourth compartment\nto the model to track the fraction of the population\nin quarantine at each point in time. This approach\nwould be more complex, and it is not obvious that it\nis substantially better.\n\nThe following function could be used, like \nadd_immunization and add_hand_washing, to adjust the\nparameters in order to model various interventions.\n\nIn this example, `high` is the highest duration of\nthe infection period, with no quarantine. `low` is\nthe lowest duration, on the assumption that it takes\nsome time to identify infectious students.\n\n`fraction` is the fraction of infected students who \nare quarantined as soon as they are identified.\n\"\"\"\n\ndef add_quarantine(system, fraction):\n \"\"\"Model the effect of quarantine by adjusting gamma.\n \n system: System object\n fraction: fraction of students quarantined\n \"\"\"\n # `low` represents the number of days a student \n # is infectious if quarantined.\n # `high` is the number of days they are infectious\n # if not quarantined\n low = 1\n high = 4\n tr = high - fraction * (high-low)\n system.gamma = 1 / tr",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
]
] |
c515d9ce58e37e534ca83e9208353471ae1b6554
| 92,897 |
ipynb
|
Jupyter Notebook
|
joining_data_with_pandas/2_merging_tables_with_different_join_types.ipynb
|
vhsenna/datacamp-courses
|
dad9982bf7e90061efcbecc3cce97b7a5d14dd80
|
[
"MIT"
] | null | null | null |
joining_data_with_pandas/2_merging_tables_with_different_join_types.ipynb
|
vhsenna/datacamp-courses
|
dad9982bf7e90061efcbecc3cce97b7a5d14dd80
|
[
"MIT"
] | 1 |
2022-02-19T17:18:22.000Z
|
2022-02-19T21:51:45.000Z
|
joining_data_with_pandas/2_merging_tables_with_different_join_types.ipynb
|
vhsenna/datacamp-courses
|
dad9982bf7e90061efcbecc3cce97b7a5d14dd80
|
[
"MIT"
] | null | null | null | 131.768794 | 33,240 | 0.656717 |
[
[
[
"import pandas as pd\nimport matplotlib.pyplot as plt",
"_____no_output_____"
]
],
[
[
"## Counting missing rows with left join\nThe Movie Database is supported by volunteers going out into the world, collecting data, and entering it into the database. This includes financial data, such as movie budget and revenue. If you wanted to know which movies are still missing data, you could use a left join to identify them. Practice using a left join by merging the `movies` table and the `financials` table.\n\nThe `movies` and `financials` tables have been loaded for you.\n\nInstructions\n\n- What column is likely the best column to merge the two tables on?\n- Merge the `movies` table, as the left table, with the `financials` table using a left join, and save the result to `movies_financials`.\n- Count the number of rows in `movies_financials` with a null value in the `budget` column.",
"_____no_output_____"
]
],
[
[
"# Import the DataFrames\nmovies = pd.read_pickle('movies.pkl')\nfinancials = pd.read_pickle('financials.pkl')\n\ndisplay(movies.head())\ndisplay(movies.shape)\ndisplay(financials.head())\ndisplay(financials.shape)",
"_____no_output_____"
],
[
"# R:\non='id'",
"_____no_output_____"
],
[
"# Merge movies and financials with a left join\nmovies_financials = movies.merge(financials, on='id', how='left')",
"_____no_output_____"
],
[
"# Count the number of rows in the budget column that are missing\nnumber_of_missing_fin = movies_financials['budget'].isnull().sum()\n\n# Print the number of movies missing financials\nnumber_of_missing_fin",
"_____no_output_____"
]
],
[
[
"## Enriching a dataset\nSetting `how='left'` with the `.merge()` method is a useful technique for enriching or enhancing a dataset with additional information from a different table. In this exercise, you will start off with a sample of movie data from the movie series Toy Story. Your goal is to enrich this data by adding the marketing tag line for each movie. You will compare the results of a left join versus an inner join.\n\nThe `toy_story` DataFrame contains the Toy Story movies. The `toy_story` and `taglines` DataFrames have been loaded for you.\n\nInstructions\n- Merge `toy_story` and `taglines` on the `id` column with a **left join**, and save the result as `toystory_tag`.\n- With `toy_story` as the left table, merge to it `taglines` on the `id` column with an **inner join**, and save as `toystory_tag`.",
"_____no_output_____"
]
],
[
[
"# Import the DataFrames\ntoy_story = pd.read_csv('toy_story.csv')\ntaglines = pd.read_pickle('taglines.pkl')",
"_____no_output_____"
],
[
"# Merge the toy_story and taglines tables with a left join\ntoystory_tag = toy_story.merge(taglines, on='id', how='left')\n\n# Print the rows and shape of toystory_tag\nprint(toystory_tag)\nprint(toystory_tag.shape)",
" id title popularity release_date tagline\n0 10193 Toy Story 3 59.995418 2010-06-16 No toy gets left behind.\n1 863 Toy Story 2 73.575118 1999-10-30 The toys are back!\n2 862 Toy Story 73.640445 1995-10-30 NaN\n(3, 5)\n"
],
[
"# Merge the toy_story and taglines tables with a inner join\ntoystory_tag = toy_story.merge(taglines, on='id')\n\n# Print the rows and shape of toystory_tag\nprint(toystory_tag)\nprint(toystory_tag.shape)",
" id title popularity release_date tagline\n0 10193 Toy Story 3 59.995418 2010-06-16 No toy gets left behind.\n1 863 Toy Story 2 73.575118 1999-10-30 The toys are back!\n(2, 5)\n"
]
],
[
[
"## Right join to find unique movies\nMost of the recent big-budget science fiction movies can also be classified as action movies. You are given a table of science fiction movies called `scifi_movies` and another table of action movies called `action_movies`. Your goal is to find which movies are considered only science fiction movies. Once you have this table, you can merge the `movies` table in to see the movie names. Since this exercise is related to science fiction movies, use a right join as your superhero power to solve this problem.\n\nThe `movies`, `scifi_movies`, and `action_movie`s tables have been loaded for you.\n\nInstructions\n\n- Merge `action_movies` and `scifi_movies` tables with a **right join** on `movie_id`. Save the result as `action_scifi`.\n- Update the merge to add suffixes, where `'_act'` and `'_sci'` are suffixes for the left and right tables, respectively.\n- From `action_scifi`, subset only the rows where the `genre_act` column is null.\n- Merge `movies` and `scifi_only` using the `id` column in the left table and the `movie_id` column in the right table with an inner join.",
"_____no_output_____"
]
],
[
[
"# Import the DataFrames\nmovies = pd.read_pickle('movies.pkl')\nscifi_movies = pd.read_pickle('scifi_movies')\naction_movies = pd.read_pickle('action_movies')",
"_____no_output_____"
],
[
"# Merge action_movies to scifi_movies with right join\naction_scifi = action_movies.merge(scifi_movies, on='movie_id', how='right')",
"_____no_output_____"
],
[
"# Merge action_movies to scifi_movies with right join\naction_scifi = action_movies.merge(scifi_movies, on='movie_id', how='right',\n suffixes=['_act', '_sci'])\n\n# Print the first few rows of action_scifi to see the structure\nprint(action_scifi.head())",
"_____no_output_____"
],
[
"# From action_scifi, select only the rows where the genre_act column is null\nscifi_only = action_scifi[action_scifi['genre_act'].isnull()]",
"_____no_output_____"
],
[
"# Merge the movies and scifi_only tables with an inner join\nmovies_and_scifi_only = movies.merge(scifi_only, how='inner',\n left_on='id', right_on='movie_id')\n\n# Print the first few rows and shape of movies_and_scifi_only\nprint(movies_and_scifi_only.head())\nprint(movies_and_scifi_only.shape)",
"_____no_output_____"
]
],
[
[
"## Popular genres with right join\nWhat are the genres of the most popular movies? To answer this question, you need to merge data from the `movies` and `movie_to_genres` tables. In a table called `pop_movies`, the top 10 most popular movies in the movies table have been selected. To ensure that you are analyzing all of the popular movies, merge it with the `movie_to_genres` table using a right join. To complete your analysis, count the number of different genres. Also, the two tables can be merged by the movie ID. However, in `pop_movies` that column is called `id`, and in `movies_to_genres` it's called `movie_id`.\n\nThe `pop_movies` and `movie_to_genres` tables have been loaded for you.\n\nInstructions\n\n- Merge `movie_to_genres` and `pop_movies` using a right join. Save the results as `genres_movies`.\n- Group `genres_movies` by `genre` and count the number of `id` values.",
"_____no_output_____"
]
],
[
[
"# Import the DataFrames\nmovies = pd.read_pickle('movies.pkl')\npop_movies = pd.read_csv('pop_movies.csv')\nmovie_to_genres = pd.read_pickle('movie_to_genres.pkl')",
"_____no_output_____"
],
[
"# Use right join to merge the movie_to_genres and pop_movies tables\ngenres_movies = movie_to_genres.merge(pop_movies, how='right', \n left_on='movie_id', right_on='id')\n\n# Count the number of genres\ngenre_count = genres_movies.groupby('genre').agg({'id':'count'})\n\n# Plot a bar chart of the genre_count\ngenre_count.plot(kind='bar')\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Using outer join to select actors\nOne cool aspect of using an outer join is that, because it returns all rows from both merged tables and null where they do not match, you can use it to find rows that do not have a match in the other table. To try for yourself, you have been given two tables with a list of actors from two popular movies: Iron Man 1 and Iron Man 2. Most of the actors played in both movies. Use an outer join to find actors who **did not** act in both movies.\n\nThe Iron Man 1 table is called `iron_1_actors`, and Iron Man 2 table is called `iron_2_actors`. Both tables have been loaded for you and a few rows printed so you can see the structure.\n\n\nInstructions\n\n- Save to `iron_1_and_2` the merge of `iron_1_actors` (left) with `iron_2_actors` tables with an outer join on the `id` column, and set suffixes to `('_1','_2')`.\n- Create an index that returns `True` if `name_1` or `name_2` are null, and `False` otherwise.",
"_____no_output_____"
]
],
[
[
"# Import the DataFrames\niron_1_actors = pd.read_csv('iron_1_actors.csv', index_col=0)\niron_2_actors = pd.read_csv('iron_2_actors.csv', index_col=0)",
"_____no_output_____"
],
[
"# Merge iron_1_actors to iron_2_actors on id with outer join using suffixes\niron_1_and_2 = iron_1_actors.merge(iron_2_actors, how='outer', on='id',\n suffixes=['_1', '_2'])\n\n# Create an index that returns true if name_1 or name_2 are null\nm = ((iron_1_and_2['name_1'].isnull()) | (iron_1_and_2['name_2'].isnull()))\n\n# Print the first few rows of iron_1_and_2\niron_1_and_2[m].head()",
"_____no_output_____"
]
],
[
[
"## Self join\nMerging a table to itself can be useful when you want to compare values in a column to other values in the same column. In this exercise, you will practice this by creating a table that for each movie will list the movie director and a member of the crew on one row. You have been given a table called `crews`, which has columns `id`, `job`, and `name`. First, merge the table to itself using the movie ID. This merge will give you a larger table where for each movie, every job is matched against each other. Then select only those rows with a director in the left table, and avoid having a row where the director's job is listed in both the left and right tables. This filtering will remove job combinations that aren't with the director.\n\nThe `crews` table has been loaded for you.\n\nInstructions\n\n- To a variable called `crews_self_merged`, merge the `crews` table to itself on the `id` column using an inner join, setting the suffixes to `'_dir'` and `'_crew'` for the left and right tables respectively.\n- Create a Boolean index, named `boolean_filter`, that selects rows from the left table with the job of `'Director'` and avoids rows with the job of `'Director'` in the right table.\n- Use the `.head()` method to print the first few rows of `direct_crews`.",
"_____no_output_____"
]
],
[
[
"# Import the DataFrame\ncrews = pd.read_pickle('crews.pkl')\ncrews.head()",
"_____no_output_____"
],
[
"# Merge the crews table to itself\ncrews_self_merged = crews.merge(crews, on='id', how='inner',\n suffixes=('_dir','_crew'))\n\ncrews_self_merged.head()",
"_____no_output_____"
],
[
"# Create a Boolean index to select the appropriate rows\nboolean_filter = ((crews_self_merged['job_dir'] == 'Director') & \n (crews_self_merged['job_crew'] != 'Director'))\n\ndirect_crews = crews_self_merged[boolean_filter]",
"_____no_output_____"
],
[
"# Print the first few rows of direct_crews\ndirect_crews.head()",
"_____no_output_____"
]
],
[
[
"## Index merge for movie ratings\nTo practice merging on indexes, you will merge `movies` and a table called `ratings` that holds info about movie ratings. Make sure your merge returns **all** of the rows from the `movies` table and not all the rows of `ratings` table need to be included in the result.\n\nThe `movies` and `ratings` tables have been loaded for you.\n\nInstructions\n\n- Merge `movies` and `ratings` on the index and save to a variable called `movies_ratings`, ensuring that all of the rows from the `movies` table are returned.",
"_____no_output_____"
]
],
[
[
"# Import the DataFrames\nmovies = pd.read_pickle('movies.pkl')\nratings = pd.read_pickle('ratings.pkl')",
"_____no_output_____"
],
[
"# Merge to the movies table the ratings table on the index\nmovies_ratings = movies.merge(ratings, how='left', on='id')\n\n# Print the first few rows of movies_ratings\nmovies_ratings.head()",
"_____no_output_____"
]
],
[
[
"## Do sequels earn more?\nIt is time to put together many of the aspects that you have learned in this chapter. In this exercise, you'll find out which movie sequels earned the most compared to the original movie. To answer this question, you will merge a modified version of the `sequels` and `financials` tables where their index is the movie ID. You will need to choose a merge type that will return all of the rows from the `sequels` table and not all the rows of `financials` table need to be included in the result. From there, you will join the resulting table to itself so that you can compare the revenue values of the original movie to the sequel. Next, you will calculate the difference between the two revenues and sort the resulting dataset.\n\nThe `sequels` and `financials` tables have been provided.\n\nInstructions\n\n- With the `sequels` table on the left, merge to it the `financials` table on index named `id`, ensuring that all the rows from the `sequels` are returned and some rows from the other table may not be returned, Save the results to `sequels_fin`.\n- Merge the `sequels_fin` table to itself with an inner join, where the left and right tables merge on `sequel` and `id` respectively with suffixes equal to `('_org','_seq')`, saving to `orig_seq`.\n- Select the `title_org`, `title_seq`, and `diff` columns of `orig_seq` and save this as `titles_diff`.\n- Sort by `titles_diff` by `diff` in descending order and print the first few rows.",
"_____no_output_____"
]
],
[
[
"# Import the DataFrames\nsequels = pd.read_pickle('sequels.pkl')\nfinancials = pd.read_pickle('financials.pkl')",
"_____no_output_____"
],
[
"# Merge sequels and financials on index id\nsequels_fin = sequels.merge(financials, on='id', how='left')",
"_____no_output_____"
],
[
"# Self merge with suffixes as inner join with left on sequel and right on id\norig_seq = sequels_fin.merge(sequels_fin, how='inner', left_on='sequel', right_on='id',\n right_index=True, suffixes=('_org', '_seq'))\n\n# Add calculation to subtract revenue_org from revenue_seq \norig_seq['diff'] = orig_seq['revenue_seq'] - orig_seq['revenue_org']",
"_____no_output_____"
],
[
"# Select the title_org, title_seq, and diff \ntitles_diff = orig_seq[['title_org','title_seq','diff']]",
"_____no_output_____"
],
[
"# Print the first rows of the sorted titles_diff\ntitles_diff.sort_values('diff', ascending=False).head()",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
c515ee96a95414e03ee7e2632c7550ae4e1d2eb0
| 74,631 |
ipynb
|
Jupyter Notebook
|
19_training_and_deploying_at_scale.ipynb
|
astridgutierrez/handson-ml2
|
568d9887be5417bc332e8d78ea31a5b38a426984
|
[
"Apache-2.0"
] | null | null | null |
19_training_and_deploying_at_scale.ipynb
|
astridgutierrez/handson-ml2
|
568d9887be5417bc332e8d78ea31a5b38a426984
|
[
"Apache-2.0"
] | null | null | null |
19_training_and_deploying_at_scale.ipynb
|
astridgutierrez/handson-ml2
|
568d9887be5417bc332e8d78ea31a5b38a426984
|
[
"Apache-2.0"
] | null | null | null | 35.932114 | 1,523 | 0.528735 |
[
[
[
"**Chapter 19 – Training and Deploying TensorFlow Models at Scale**",
"_____no_output_____"
],
[
"_This notebook contains all the sample code in chapter 19._",
"_____no_output_____"
],
[
"<table align=\"left\">\n <td>\n <a href=\"https://colab.research.google.com/github/ageron/handson-ml2/blob/master/19_training_and_deploying_at_scale.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://kaggle.com/kernels/welcome?src=https://github.com/ageron/handson-ml2/blob/master/19_training_and_deploying_at_scale.ipynb\"><img src=\"https://kaggle.com/static/images/open-in-kaggle.svg\" /></a>\n </td>\n</table>",
"_____no_output_____"
],
[
"# Setup\nFirst, let's import a few common modules, ensure MatplotLib plots figures inline and prepare a function to save the figures. We also check that Python 3.5 or later is installed (although Python 2.x may work, it is deprecated so we strongly recommend you use Python 3 instead), as well as Scikit-Learn ≥0.20 and TensorFlow ≥2.0.\n",
"_____no_output_____"
]
],
[
[
"# Python ≥3.5 is required\nimport sys\nassert sys.version_info >= (3, 5)\n\n# Is this notebook running on Colab or Kaggle?\nIS_COLAB = \"google.colab\" in sys.modules\nIS_KAGGLE = \"kaggle_secrets\" in sys.modules\n\nif IS_COLAB or IS_KAGGLE:\n !echo \"deb http://storage.googleapis.com/tensorflow-serving-apt stable tensorflow-model-server tensorflow-model-server-universal\" > /etc/apt/sources.list.d/tensorflow-serving.list\n !curl https://storage.googleapis.com/tensorflow-serving-apt/tensorflow-serving.release.pub.gpg | apt-key add -\n !apt update && apt-get install -y tensorflow-model-server\n %pip install -q -U tensorflow-serving-api\n\n# Scikit-Learn ≥0.20 is required\nimport sklearn\nassert sklearn.__version__ >= \"0.20\"\n\n# TensorFlow ≥2.0 is required\nimport tensorflow as tf\nfrom tensorflow import keras\nassert tf.__version__ >= \"2.0\"\n\nif not tf.config.list_physical_devices('GPU'):\n print(\"No GPU was detected. CNNs can be very slow without a GPU.\")\n if IS_COLAB:\n print(\"Go to Runtime > Change runtime and select a GPU hardware accelerator.\")\n if IS_KAGGLE:\n print(\"Go to Settings > Accelerator and select GPU.\")\n\n# Common imports\nimport numpy as np\nimport os\n\n# to make this notebook's output stable across runs\nnp.random.seed(42)\ntf.random.set_seed(42)\n\n# To plot pretty figures\n%matplotlib inline\nimport matplotlib as mpl\nimport matplotlib.pyplot as plt\nmpl.rc('axes', labelsize=14)\nmpl.rc('xtick', labelsize=12)\nmpl.rc('ytick', labelsize=12)\n\n# Where to save the figures\nPROJECT_ROOT_DIR = \".\"\nCHAPTER_ID = \"deploy\"\nIMAGES_PATH = os.path.join(PROJECT_ROOT_DIR, \"images\", CHAPTER_ID)\nos.makedirs(IMAGES_PATH, exist_ok=True)\n\ndef save_fig(fig_id, tight_layout=True, fig_extension=\"png\", resolution=300):\n path = os.path.join(IMAGES_PATH, fig_id + \".\" + fig_extension)\n print(\"Saving figure\", fig_id)\n if tight_layout:\n plt.tight_layout()\n plt.savefig(path, format=fig_extension, dpi=resolution)",
"_____no_output_____"
]
],
[
[
"# Deploying TensorFlow models to TensorFlow Serving (TFS)\nWe will use the REST API or the gRPC API.",
"_____no_output_____"
],
[
"## Save/Load a `SavedModel`",
"_____no_output_____"
]
],
[
[
"(X_train_full, y_train_full), (X_test, y_test) = keras.datasets.mnist.load_data()\nX_train_full = X_train_full[..., np.newaxis].astype(np.float32) / 255.\nX_test = X_test[..., np.newaxis].astype(np.float32) / 255.\nX_valid, X_train = X_train_full[:5000], X_train_full[5000:]\ny_valid, y_train = y_train_full[:5000], y_train_full[5000:]\nX_new = X_test[:3]",
"_____no_output_____"
],
[
"np.random.seed(42)\ntf.random.set_seed(42)\n\nmodel = keras.models.Sequential([\n keras.layers.Flatten(input_shape=[28, 28, 1]),\n keras.layers.Dense(100, activation=\"relu\"),\n keras.layers.Dense(10, activation=\"softmax\")\n])\nmodel.compile(loss=\"sparse_categorical_crossentropy\",\n optimizer=keras.optimizers.SGD(learning_rate=1e-2),\n metrics=[\"accuracy\"])\nmodel.fit(X_train, y_train, epochs=10, validation_data=(X_valid, y_valid))",
"Epoch 1/10\n1719/1719 [==============================] - 2s 1ms/step - loss: 1.1140 - accuracy: 0.7066 - val_loss: 0.3715 - val_accuracy: 0.9024\nEpoch 2/10\n1719/1719 [==============================] - 1s 713us/step - loss: 0.3695 - accuracy: 0.8981 - val_loss: 0.2990 - val_accuracy: 0.9144\nEpoch 3/10\n1719/1719 [==============================] - 1s 718us/step - loss: 0.3154 - accuracy: 0.9100 - val_loss: 0.2651 - val_accuracy: 0.9272\nEpoch 4/10\n1719/1719 [==============================] - 1s 706us/step - loss: 0.2765 - accuracy: 0.9223 - val_loss: 0.2436 - val_accuracy: 0.9334\nEpoch 5/10\n1719/1719 [==============================] - 1s 711us/step - loss: 0.2556 - accuracy: 0.9276 - val_loss: 0.2257 - val_accuracy: 0.9364\nEpoch 6/10\n1719/1719 [==============================] - 1s 715us/step - loss: 0.2367 - accuracy: 0.9321 - val_loss: 0.2121 - val_accuracy: 0.9396\nEpoch 7/10\n1719/1719 [==============================] - 1s 729us/step - loss: 0.2198 - accuracy: 0.9390 - val_loss: 0.1970 - val_accuracy: 0.9454\nEpoch 8/10\n1719/1719 [==============================] - 1s 716us/step - loss: 0.2057 - accuracy: 0.9425 - val_loss: 0.1880 - val_accuracy: 0.9476\nEpoch 9/10\n1719/1719 [==============================] - 1s 704us/step - loss: 0.1940 - accuracy: 0.9459 - val_loss: 0.1777 - val_accuracy: 0.9524\nEpoch 10/10\n1719/1719 [==============================] - 1s 711us/step - loss: 0.1798 - accuracy: 0.9482 - val_loss: 0.1684 - val_accuracy: 0.9546\n"
],
[
"np.round(model.predict(X_new), 2)",
"_____no_output_____"
],
[
"model_version = \"0001\"\nmodel_name = \"my_mnist_model\"\nmodel_path = os.path.join(model_name, model_version)\nmodel_path",
"_____no_output_____"
],
[
"!rm -rf {model_name}",
"_____no_output_____"
],
[
"tf.saved_model.save(model, model_path)",
"INFO:tensorflow:Assets written to: my_mnist_model/0001/assets\n"
],
[
"for root, dirs, files in os.walk(model_name):\n indent = ' ' * root.count(os.sep)\n print('{}{}/'.format(indent, os.path.basename(root)))\n for filename in files:\n print('{}{}'.format(indent + ' ', filename))",
"my_mnist_model/\n 0001/\n saved_model.pb\n variables/\n variables.data-00000-of-00001\n variables.index\n assets/\n"
],
[
"!saved_model_cli show --dir {model_path}",
"The given SavedModel contains the following tag-sets:\r\n'serve'\r\n"
],
[
"!saved_model_cli show --dir {model_path} --tag_set serve",
"The given SavedModel MetaGraphDef contains SignatureDefs with the following keys:\r\nSignatureDef key: \"__saved_model_init_op\"\r\nSignatureDef key: \"serving_default\"\r\n"
],
[
"!saved_model_cli show --dir {model_path} --tag_set serve \\\n --signature_def serving_default",
"The given SavedModel SignatureDef contains the following input(s):\r\n inputs['flatten_input'] tensor_info:\r\n dtype: DT_FLOAT\r\n shape: (-1, 28, 28, 1)\r\n name: serving_default_flatten_input:0\r\nThe given SavedModel SignatureDef contains the following output(s):\r\n outputs['dense_1'] tensor_info:\r\n dtype: DT_FLOAT\r\n shape: (-1, 10)\r\n name: StatefulPartitionedCall:0\r\nMethod name is: tensorflow/serving/predict\r\n"
],
[
"!saved_model_cli show --dir {model_path} --all",
"\nMetaGraphDef with tag-set: 'serve' contains the following SignatureDefs:\n\nsignature_def['__saved_model_init_op']:\n The given SavedModel SignatureDef contains the following input(s):\n The given SavedModel SignatureDef contains the following output(s):\n outputs['__saved_model_init_op'] tensor_info:\n dtype: DT_INVALID\n shape: unknown_rank\n name: NoOp\n Method name is: \n\nsignature_def['serving_default']:\n The given SavedModel SignatureDef contains the following input(s):\n inputs['flatten_input'] tensor_info:\n dtype: DT_FLOAT\n shape: (-1, 28, 28, 1)\n name: serving_default_flatten_input:0\n The given SavedModel SignatureDef contains the following output(s):\n outputs['dense_1'] tensor_info:\n dtype: DT_FLOAT\n shape: (-1, 10)\n name: StatefulPartitionedCall:0\n Method name is: tensorflow/serving/predict\n\nDefined Functions:\n Function Name: '__call__'\n Option #1\n Callable with:\n Argument #1\n flatten_input: TensorSpec(shape=(None, 28, 28, 1), dtype=tf.float32, name='flatten_input')\n Argument #2\n DType: bool\n Value: True\n Argument #3\n<<45 more lines>>\n DType: bool\n Value: False\n Argument #3\n DType: NoneType\n Value: None\n Option #2\n Callable with:\n Argument #1\n inputs: TensorSpec(shape=(None, 28, 28, 1), dtype=tf.float32, name='inputs')\n Argument #2\n DType: bool\n Value: True\n Argument #3\n DType: NoneType\n Value: None\n Option #3\n Callable with:\n Argument #1\n flatten_input: TensorSpec(shape=(None, 28, 28, 1), dtype=tf.float32, name='flatten_input')\n Argument #2\n DType: bool\n Value: True\n Argument #3\n DType: NoneType\n Value: None\n Option #4\n Callable with:\n Argument #1\n flatten_input: TensorSpec(shape=(None, 28, 28, 1), dtype=tf.float32, name='flatten_input')\n Argument #2\n DType: bool\n Value: False\n Argument #3\n DType: NoneType\n Value: None\n"
]
],
[
[
"Let's write the new instances to a `npy` file so we can pass them easily to our model:",
"_____no_output_____"
]
],
[
[
"np.save(\"my_mnist_tests.npy\", X_new)",
"_____no_output_____"
],
[
"input_name = model.input_names[0]\ninput_name",
"_____no_output_____"
]
],
[
[
"And now let's use `saved_model_cli` to make predictions for the instances we just saved:",
"_____no_output_____"
]
],
[
[
"!saved_model_cli run --dir {model_path} --tag_set serve \\\n --signature_def serving_default \\\n --inputs {input_name}=my_mnist_tests.npy",
"2021-02-18 22:15:30.294109: I tensorflow/compiler/jit/xla_cpu_device.cc:41] Not creating XLA devices, tf_xla_enable_xla_devices not set\n2021-02-18 22:15:30.294306: I tensorflow/core/platform/cpu_feature_guard.cc:142] This TensorFlow binary is optimized with oneAPI Deep Neural Network Library (oneDNN) to use the following CPU instructions in performance-critical operations: AVX2 FMA\nTo enable them in other operations, rebuild TensorFlow with the appropriate compiler flags.\nWARNING:tensorflow:From /Users/ageron/miniconda3/envs/tf2/lib/python3.7/site-packages/tensorflow/python/tools/saved_model_cli.py:445: load (from tensorflow.python.saved_model.loader_impl) is deprecated and will be removed in a future version.\nInstructions for updating:\nThis function will only be available through the v1 compatibility library as tf.compat.v1.saved_model.loader.load or tf.compat.v1.saved_model.load. There will be a new function for importing SavedModels in Tensorflow 2.0.\nINFO:tensorflow:Restoring parameters from my_mnist_model/0001/variables/variables\n2021-02-18 22:15:30.323498: I tensorflow/compiler/mlir/mlir_graph_optimization_pass.cc:196] None of the MLIR optimization passes are enabled (registered 0 passes)\nResult for output key dense_1:\n[[1.1347984e-04 1.5187356e-07 9.7032893e-04 2.7640699e-03 3.7826971e-06\n 7.6876910e-05 3.9140293e-08 9.9559116e-01 5.3502394e-05 4.2665208e-04]\n [8.2443521e-04 3.5493889e-05 9.8826385e-01 7.0466995e-03 1.2957400e-07\n 2.3389691e-04 2.5639210e-03 9.5886099e-10 1.0314899e-03 8.7952529e-08]\n [4.4693781e-05 9.7028232e-01 9.0526715e-03 2.2641101e-03 4.8766597e-04\n 2.8800720e-03 2.2714981e-03 8.3753867e-03 4.0439744e-03 2.9759688e-04]]\n"
],
[
"np.round([[1.1347984e-04, 1.5187356e-07, 9.7032893e-04, 2.7640699e-03, 3.7826971e-06,\n 7.6876910e-05, 3.9140293e-08, 9.9559116e-01, 5.3502394e-05, 4.2665208e-04],\n [8.2443521e-04, 3.5493889e-05, 9.8826385e-01, 7.0466995e-03, 1.2957400e-07,\n 2.3389691e-04, 2.5639210e-03, 9.5886099e-10, 1.0314899e-03, 8.7952529e-08],\n [4.4693781e-05, 9.7028232e-01, 9.0526715e-03, 2.2641101e-03, 4.8766597e-04,\n 2.8800720e-03, 2.2714981e-03, 8.3753867e-03, 4.0439744e-03, 2.9759688e-04]], 2)",
"_____no_output_____"
]
],
[
[
"## TensorFlow Serving",
"_____no_output_____"
],
[
"Install [Docker](https://docs.docker.com/install/) if you don't have it already. Then run:\n\n```bash\ndocker pull tensorflow/serving\n\nexport ML_PATH=$HOME/ml # or wherever this project is\ndocker run -it --rm -p 8500:8500 -p 8501:8501 \\\n -v \"$ML_PATH/my_mnist_model:/models/my_mnist_model\" \\\n -e MODEL_NAME=my_mnist_model \\\n tensorflow/serving\n```\nOnce you are finished using it, press Ctrl-C to shut down the server.",
"_____no_output_____"
],
[
"Alternatively, if `tensorflow_model_server` is installed (e.g., if you are running this notebook in Colab), then the following 3 cells will start the server:",
"_____no_output_____"
]
],
[
[
"os.environ[\"MODEL_DIR\"] = os.path.split(os.path.abspath(model_path))[0]",
"_____no_output_____"
],
[
"%%bash --bg\nnohup tensorflow_model_server \\\n --rest_api_port=8501 \\\n --model_name=my_mnist_model \\\n --model_base_path=\"${MODEL_DIR}\" >server.log 2>&1",
"_____no_output_____"
],
[
"!tail server.log",
"2021-02-16 22:33:09.323538: I external/org_tensorflow/tensorflow/cc/saved_model/reader.cc:93] Reading SavedModel debug info (if present) from: /models/my_mnist_model/0001\n2021-02-16 22:33:09.323642: I external/org_tensorflow/tensorflow/core/platform/cpu_feature_guard.cc:142] This TensorFlow binary is optimized with oneAPI Deep Neural Network Library (oneDNN) to use the following CPU instructions in performance-critical operations: AVX2 FMA\nTo enable them in other operations, rebuild TensorFlow with the appropriate compiler flags.\n2021-02-16 22:33:09.360572: I external/org_tensorflow/tensorflow/cc/saved_model/loader.cc:206] Restoring SavedModel bundle.\n2021-02-16 22:33:09.361764: I external/org_tensorflow/tensorflow/core/platform/profile_utils/cpu_utils.cc:112] CPU Frequency: 2200000000 Hz\n2021-02-16 22:33:09.387713: I external/org_tensorflow/tensorflow/cc/saved_model/loader.cc:190] Running initialization op on SavedModel bundle at path: /models/my_mnist_model/0001\n2021-02-16 22:33:09.392739: I external/org_tensorflow/tensorflow/cc/saved_model/loader.cc:277] SavedModel load for tags { serve }; Status: success: OK. Took 71106 microseconds.\n2021-02-16 22:33:09.393390: I tensorflow_serving/servables/tensorflow/saved_model_warmup_util.cc:59] No warmup data file found at /models/my_mnist_model/0001/assets.extra/tf_serving_warmup_requests\n2021-02-16 22:33:09.393847: I tensorflow_serving/core/loader_harness.cc:87] Successfully loaded servable version {name: my_mnist_model version: 1}\n2021-02-16 22:33:09.398470: I tensorflow_serving/model_servers/server.cc:371] Running gRPC ModelServer at 0.0.0.0:8500 ...\n[warn] getaddrinfo: address family for nodename not supported\n2021-02-16 22:33:09.405622: I tensorflow_serving/model_servers/server.cc:391] Exporting HTTP/REST API at:localhost:8501 ...\n[evhttp_server.cc : 238] NET_LOG: Entering the event loop ...\n"
],
[
"import json\n\ninput_data_json = json.dumps({\n \"signature_name\": \"serving_default\",\n \"instances\": X_new.tolist(),\n})",
"_____no_output_____"
],
[
"repr(input_data_json)[:1500] + \"...\"",
"_____no_output_____"
]
],
[
[
"Now let's use TensorFlow Serving's REST API to make predictions:",
"_____no_output_____"
]
],
[
[
"import requests\n\nSERVER_URL = 'http://localhost:8501/v1/models/my_mnist_model:predict'\nresponse = requests.post(SERVER_URL, data=input_data_json)\nresponse.raise_for_status() # raise an exception in case of error\nresponse = response.json()",
"_____no_output_____"
],
[
"response.keys()",
"_____no_output_____"
],
[
"y_proba = np.array(response[\"predictions\"])\ny_proba.round(2)",
"_____no_output_____"
]
],
[
[
"### Using the gRPC API",
"_____no_output_____"
]
],
[
[
"from tensorflow_serving.apis.predict_pb2 import PredictRequest\n\nrequest = PredictRequest()\nrequest.model_spec.name = model_name\nrequest.model_spec.signature_name = \"serving_default\"\ninput_name = model.input_names[0]\nrequest.inputs[input_name].CopyFrom(tf.make_tensor_proto(X_new))",
"_____no_output_____"
],
[
"import grpc\nfrom tensorflow_serving.apis import prediction_service_pb2_grpc\n\nchannel = grpc.insecure_channel('localhost:8500')\npredict_service = prediction_service_pb2_grpc.PredictionServiceStub(channel)\nresponse = predict_service.Predict(request, timeout=10.0)",
"_____no_output_____"
],
[
"response",
"_____no_output_____"
]
],
[
[
"Convert the response to a tensor:",
"_____no_output_____"
]
],
[
[
"output_name = model.output_names[0]\noutputs_proto = response.outputs[output_name]\ny_proba = tf.make_ndarray(outputs_proto)\ny_proba.round(2)",
"_____no_output_____"
]
],
[
[
"Or to a NumPy array if your client does not include the TensorFlow library:",
"_____no_output_____"
]
],
[
[
"output_name = model.output_names[0]\noutputs_proto = response.outputs[output_name]\nshape = [dim.size for dim in outputs_proto.tensor_shape.dim]\ny_proba = np.array(outputs_proto.float_val).reshape(shape)\ny_proba.round(2)",
"_____no_output_____"
]
],
[
[
"## Deploying a new model version",
"_____no_output_____"
]
],
[
[
"np.random.seed(42)\ntf.random.set_seed(42)\n\nmodel = keras.models.Sequential([\n keras.layers.Flatten(input_shape=[28, 28, 1]),\n keras.layers.Dense(50, activation=\"relu\"),\n keras.layers.Dense(50, activation=\"relu\"),\n keras.layers.Dense(10, activation=\"softmax\")\n])\nmodel.compile(loss=\"sparse_categorical_crossentropy\",\n optimizer=keras.optimizers.SGD(learning_rate=1e-2),\n metrics=[\"accuracy\"])\nhistory = model.fit(X_train, y_train, epochs=10, validation_data=(X_valid, y_valid))",
"Epoch 1/10\n1719/1719 [==============================] - 1s 748us/step - loss: 1.1567 - accuracy: 0.6691 - val_loss: 0.3418 - val_accuracy: 0.9042\nEpoch 2/10\n1719/1719 [==============================] - 1s 697us/step - loss: 0.3376 - accuracy: 0.9032 - val_loss: 0.2674 - val_accuracy: 0.9242\nEpoch 3/10\n1719/1719 [==============================] - 1s 676us/step - loss: 0.2779 - accuracy: 0.9187 - val_loss: 0.2227 - val_accuracy: 0.9368\nEpoch 4/10\n1719/1719 [==============================] - 1s 669us/step - loss: 0.2362 - accuracy: 0.9318 - val_loss: 0.2032 - val_accuracy: 0.9432\nEpoch 5/10\n1719/1719 [==============================] - 1s 670us/step - loss: 0.2109 - accuracy: 0.9389 - val_loss: 0.1833 - val_accuracy: 0.9482\nEpoch 6/10\n1719/1719 [==============================] - 1s 675us/step - loss: 0.1951 - accuracy: 0.9430 - val_loss: 0.1740 - val_accuracy: 0.9498\nEpoch 7/10\n1719/1719 [==============================] - 1s 667us/step - loss: 0.1799 - accuracy: 0.9474 - val_loss: 0.1605 - val_accuracy: 0.9540\nEpoch 8/10\n1719/1719 [==============================] - 1s 673us/step - loss: 0.1654 - accuracy: 0.9519 - val_loss: 0.1543 - val_accuracy: 0.9558\nEpoch 9/10\n1719/1719 [==============================] - 1s 671us/step - loss: 0.1570 - accuracy: 0.9554 - val_loss: 0.1460 - val_accuracy: 0.9572\nEpoch 10/10\n1719/1719 [==============================] - 1s 672us/step - loss: 0.1420 - accuracy: 0.9583 - val_loss: 0.1359 - val_accuracy: 0.9616\n"
],
[
"model_version = \"0002\"\nmodel_name = \"my_mnist_model\"\nmodel_path = os.path.join(model_name, model_version)\nmodel_path",
"_____no_output_____"
],
[
"tf.saved_model.save(model, model_path)",
"INFO:tensorflow:Assets written to: my_mnist_model/0002/assets\n"
],
[
"for root, dirs, files in os.walk(model_name):\n indent = ' ' * root.count(os.sep)\n print('{}{}/'.format(indent, os.path.basename(root)))\n for filename in files:\n print('{}{}'.format(indent + ' ', filename))",
"my_mnist_model/\n 0001/\n saved_model.pb\n variables/\n variables.data-00000-of-00001\n variables.index\n assets/\n 0002/\n saved_model.pb\n variables/\n variables.data-00000-of-00001\n variables.index\n assets/\n"
]
],
[
[
"**Warning**: You may need to wait a minute before the new model is loaded by TensorFlow Serving.",
"_____no_output_____"
]
],
[
[
"import requests\n\nSERVER_URL = 'http://localhost:8501/v1/models/my_mnist_model:predict'\n \nresponse = requests.post(SERVER_URL, data=input_data_json)\nresponse.raise_for_status()\nresponse = response.json()",
"_____no_output_____"
],
[
"response.keys()",
"_____no_output_____"
],
[
"y_proba = np.array(response[\"predictions\"])\ny_proba.round(2)",
"_____no_output_____"
]
],
[
[
"# Deploy the model to Google Cloud AI Platform",
"_____no_output_____"
],
[
"Follow the instructions in the book to deploy the model to Google Cloud AI Platform, download the service account's private key and save it to the `my_service_account_private_key.json` in the project directory. Also, update the `project_id`:",
"_____no_output_____"
]
],
[
[
"project_id = \"onyx-smoke-242003\"",
"_____no_output_____"
],
[
"import googleapiclient.discovery\n\nos.environ[\"GOOGLE_APPLICATION_CREDENTIALS\"] = \"my_service_account_private_key.json\"\nmodel_id = \"my_mnist_model\"\nmodel_path = \"projects/{}/models/{}\".format(project_id, model_id)\nmodel_path += \"/versions/v0001/\" # if you want to run a specific version\nml_resource = googleapiclient.discovery.build(\"ml\", \"v1\").projects()",
"_____no_output_____"
],
[
"def predict(X):\n input_data_json = {\"signature_name\": \"serving_default\",\n \"instances\": X.tolist()}\n request = ml_resource.predict(name=model_path, body=input_data_json)\n response = request.execute()\n if \"error\" in response:\n raise RuntimeError(response[\"error\"])\n return np.array([pred[output_name] for pred in response[\"predictions\"]])",
"_____no_output_____"
],
[
"Y_probas = predict(X_new)\nnp.round(Y_probas, 2)",
"_____no_output_____"
]
],
[
[
"# Using GPUs",
"_____no_output_____"
],
[
"**Note**: `tf.test.is_gpu_available()` is deprecated. Instead, please use `tf.config.list_physical_devices('GPU')`.",
"_____no_output_____"
]
],
[
[
"#tf.test.is_gpu_available() # deprecated\ntf.config.list_physical_devices('GPU')",
"_____no_output_____"
],
[
"tf.test.gpu_device_name()",
"_____no_output_____"
],
[
"tf.test.is_built_with_cuda()",
"_____no_output_____"
],
[
"from tensorflow.python.client.device_lib import list_local_devices\n\ndevices = list_local_devices()\ndevices",
"_____no_output_____"
]
],
[
[
"# Distributed Training",
"_____no_output_____"
]
],
[
[
"keras.backend.clear_session()\ntf.random.set_seed(42)\nnp.random.seed(42)",
"_____no_output_____"
],
[
"def create_model():\n return keras.models.Sequential([\n keras.layers.Conv2D(filters=64, kernel_size=7, activation=\"relu\",\n padding=\"same\", input_shape=[28, 28, 1]),\n keras.layers.MaxPooling2D(pool_size=2),\n keras.layers.Conv2D(filters=128, kernel_size=3, activation=\"relu\",\n padding=\"same\"), \n keras.layers.Conv2D(filters=128, kernel_size=3, activation=\"relu\",\n padding=\"same\"),\n keras.layers.MaxPooling2D(pool_size=2),\n keras.layers.Flatten(),\n keras.layers.Dense(units=64, activation='relu'),\n keras.layers.Dropout(0.5),\n keras.layers.Dense(units=10, activation='softmax'),\n ])",
"_____no_output_____"
],
[
"batch_size = 100\nmodel = create_model()\nmodel.compile(loss=\"sparse_categorical_crossentropy\",\n optimizer=keras.optimizers.SGD(learning_rate=1e-2),\n metrics=[\"accuracy\"])\nmodel.fit(X_train, y_train, epochs=10,\n validation_data=(X_valid, y_valid), batch_size=batch_size)",
"Epoch 1/10\n550/550 [==============================] - 11s 18ms/step - loss: 1.8163 - accuracy: 0.3979 - val_loss: 0.3446 - val_accuracy: 0.9010\nEpoch 2/10\n550/550 [==============================] - 9s 17ms/step - loss: 0.4949 - accuracy: 0.8482 - val_loss: 0.1962 - val_accuracy: 0.9458\nEpoch 3/10\n550/550 [==============================] - 10s 17ms/step - loss: 0.3345 - accuracy: 0.9012 - val_loss: 0.1343 - val_accuracy: 0.9622\nEpoch 4/10\n550/550 [==============================] - 10s 17ms/step - loss: 0.2537 - accuracy: 0.9267 - val_loss: 0.1049 - val_accuracy: 0.9718\nEpoch 5/10\n550/550 [==============================] - 10s 17ms/step - loss: 0.2099 - accuracy: 0.9394 - val_loss: 0.0875 - val_accuracy: 0.9752\nEpoch 6/10\n550/550 [==============================] - 10s 17ms/step - loss: 0.1901 - accuracy: 0.9439 - val_loss: 0.0797 - val_accuracy: 0.9772\nEpoch 7/10\n550/550 [==============================] - 10s 18ms/step - loss: 0.1672 - accuracy: 0.9506 - val_loss: 0.0745 - val_accuracy: 0.9780\nEpoch 8/10\n550/550 [==============================] - 10s 18ms/step - loss: 0.1537 - accuracy: 0.9554 - val_loss: 0.0700 - val_accuracy: 0.9804\nEpoch 9/10\n550/550 [==============================] - 10s 18ms/step - loss: 0.1384 - accuracy: 0.9592 - val_loss: 0.0641 - val_accuracy: 0.9818\nEpoch 10/10\n550/550 [==============================] - 10s 18ms/step - loss: 0.1358 - accuracy: 0.9602 - val_loss: 0.0611 - val_accuracy: 0.9818\n"
],
[
"keras.backend.clear_session()\ntf.random.set_seed(42)\nnp.random.seed(42)\n\ndistribution = tf.distribute.MirroredStrategy()\n\n# Change the default all-reduce algorithm:\n#distribution = tf.distribute.MirroredStrategy(\n# cross_device_ops=tf.distribute.HierarchicalCopyAllReduce())\n\n# Specify the list of GPUs to use:\n#distribution = tf.distribute.MirroredStrategy(devices=[\"/gpu:0\", \"/gpu:1\"])\n\n# Use the central storage strategy instead:\n#distribution = tf.distribute.experimental.CentralStorageStrategy()\n\n#if IS_COLAB and \"COLAB_TPU_ADDR\" in os.environ:\n# tpu_address = \"grpc://\" + os.environ[\"COLAB_TPU_ADDR\"]\n#else:\n# tpu_address = \"\"\n#resolver = tf.distribute.cluster_resolver.TPUClusterResolver(tpu_address)\n#tf.config.experimental_connect_to_cluster(resolver)\n#tf.tpu.experimental.initialize_tpu_system(resolver)\n#distribution = tf.distribute.experimental.TPUStrategy(resolver)\n\nwith distribution.scope():\n model = create_model()\n model.compile(loss=\"sparse_categorical_crossentropy\",\n optimizer=keras.optimizers.SGD(learning_rate=1e-2),\n metrics=[\"accuracy\"])",
"INFO:tensorflow:Using MirroredStrategy with devices ('/job:localhost/replica:0/task:0/device:GPU:0', '/job:localhost/replica:0/task:0/device:GPU:1')\nINFO:tensorflow:Reduce to /job:localhost/replica:0/task:0/device:CPU:0 then broadcast to ('/job:localhost/replica:0/task:0/device:CPU:0',).\nINFO:tensorflow:Reduce to /job:localhost/replica:0/task:0/device:CPU:0 then broadcast to ('/job:localhost/replica:0/task:0/device:CPU:0',).\nINFO:tensorflow:Reduce to /job:localhost/replica:0/task:0/device:CPU:0 then broadcast to ('/job:localhost/replica:0/task:0/device:CPU:0',).\nINFO:tensorflow:Reduce to /job:localhost/replica:0/task:0/device:CPU:0 then broadcast to ('/job:localhost/replica:0/task:0/device:CPU:0',).\n"
],
[
"batch_size = 100 # must be divisible by the number of workers\nmodel.fit(X_train, y_train, epochs=10,\n validation_data=(X_valid, y_valid), batch_size=batch_size)",
"Epoch 1/10\nINFO:tensorflow:batch_all_reduce: 10 all-reduces with algorithm = nccl, num_packs = 1\nINFO:tensorflow:Reduce to /job:localhost/replica:0/task:0/device:CPU:0 then broadcast to ('/job:localhost/replica:0/task:0/device:CPU:0',).\nINFO:tensorflow:Reduce to /job:localhost/replica:0/task:0/device:CPU:0 then broadcast to ('/job:localhost/replica:0/task:0/device:CPU:0',).\nINFO:tensorflow:Reduce to /job:localhost/replica:0/task:0/device:CPU:0 then broadcast to ('/job:localhost/replica:0/task:0/device:CPU:0',).\nINFO:tensorflow:Reduce to /job:localhost/replica:0/task:0/device:CPU:0 then broadcast to ('/job:localhost/replica:0/task:0/device:CPU:0',).\nINFO:tensorflow:batch_all_reduce: 10 all-reduces with algorithm = nccl, num_packs = 1\nINFO:tensorflow:Reduce to /job:localhost/replica:0/task:0/device:CPU:0 then broadcast to ('/job:localhost/replica:0/task:0/device:CPU:0',).\nINFO:tensorflow:Reduce to /job:localhost/replica:0/task:0/device:CPU:0 then broadcast to ('/job:localhost/replica:0/task:0/device:CPU:0',).\n550/550 [==============================] - 14s 16ms/step - loss: 1.8193 - accuracy: 0.3957 - val_loss: 0.3366 - val_accuracy: 0.9102\nEpoch 2/10\n550/550 [==============================] - 7s 13ms/step - loss: 0.4886 - accuracy: 0.8497 - val_loss: 0.1865 - val_accuracy: 0.9478\nEpoch 3/10\n550/550 [==============================] - 7s 13ms/step - loss: 0.3305 - accuracy: 0.9008 - val_loss: 0.1344 - val_accuracy: 0.9616\nEpoch 4/10\n550/550 [==============================] - 7s 13ms/step - loss: 0.2472 - accuracy: 0.9282 - val_loss: 0.1115 - val_accuracy: 0.9696\nEpoch 5/10\n550/550 [==============================] - 7s 13ms/step - loss: 0.2020 - accuracy: 0.9425 - val_loss: 0.0873 - val_accuracy: 0.9748\nEpoch 6/10\n550/550 [==============================] - 7s 13ms/step - loss: 0.1865 - accuracy: 0.9458 - val_loss: 0.0783 - val_accuracy: 0.9764\nEpoch 7/10\n550/550 [==============================] - 8s 14ms/step - loss: 0.1633 - accuracy: 0.9512 - val_loss: 0.0771 - val_accuracy: 0.9776\nEpoch 8/10\n550/550 [==============================] - 8s 14ms/step - loss: 0.1422 - accuracy: 0.9570 - val_loss: 0.0705 - val_accuracy: 0.9786\nEpoch 9/10\n550/550 [==============================] - 7s 13ms/step - loss: 0.1408 - accuracy: 0.9603 - val_loss: 0.0627 - val_accuracy: 0.9830\nEpoch 10/10\n550/550 [==============================] - 7s 13ms/step - loss: 0.1293 - accuracy: 0.9618 - val_loss: 0.0605 - val_accuracy: 0.9836\n"
],
[
"model.predict(X_new)",
"_____no_output_____"
]
],
[
[
"Custom training loop:",
"_____no_output_____"
]
],
[
[
"keras.backend.clear_session()\ntf.random.set_seed(42)\nnp.random.seed(42)\n\nK = keras.backend\n\ndistribution = tf.distribute.MirroredStrategy()\n\nwith distribution.scope():\n model = create_model()\n optimizer = keras.optimizers.SGD()\n\nwith distribution.scope():\n dataset = tf.data.Dataset.from_tensor_slices((X_train, y_train)).repeat().batch(batch_size)\n input_iterator = distribution.make_dataset_iterator(dataset)\n \[email protected]\ndef train_step():\n def step_fn(inputs):\n X, y = inputs\n with tf.GradientTape() as tape:\n Y_proba = model(X)\n loss = K.sum(keras.losses.sparse_categorical_crossentropy(y, Y_proba)) / batch_size\n\n grads = tape.gradient(loss, model.trainable_variables)\n optimizer.apply_gradients(zip(grads, model.trainable_variables))\n return loss\n\n per_replica_losses = distribution.experimental_run(step_fn, input_iterator)\n mean_loss = distribution.reduce(tf.distribute.ReduceOp.SUM,\n per_replica_losses, axis=None)\n return mean_loss\n\nn_epochs = 10\nwith distribution.scope():\n input_iterator.initialize()\n for epoch in range(n_epochs):\n print(\"Epoch {}/{}\".format(epoch + 1, n_epochs))\n for iteration in range(len(X_train) // batch_size):\n print(\"\\rLoss: {:.3f}\".format(train_step().numpy()), end=\"\")\n print()",
"INFO:tensorflow:Using MirroredStrategy with devices ('/job:localhost/replica:0/task:0/device:GPU:0', '/job:localhost/replica:0/task:0/device:GPU:1')\nWARNING:tensorflow:From <ipython-input-9-acb7c62c8738>:36: DistributedIteratorV1.initialize (from tensorflow.python.distribute.input_lib) is deprecated and will be removed in a future version.\nInstructions for updating:\nUse the iterator's `initializer` property instead.\nEpoch 1/10\nINFO:tensorflow:batch_all_reduce: 10 all-reduces with algorithm = nccl, num_packs = 1\nINFO:tensorflow:batch_all_reduce: 10 all-reduces with algorithm = nccl, num_packs = 1\nLoss: 0.380\nEpoch 2/10\nLoss: 0.302\nEpoch 3/10\nLoss: 0.285\nEpoch 4/10\nLoss: 0.294\nEpoch 5/10\nLoss: 0.304\nEpoch 6/10\nLoss: 0.310\nEpoch 7/10\nLoss: 0.310\nEpoch 8/10\nLoss: 0.306\nEpoch 9/10\nLoss: 0.303\nEpoch 10/10\nLoss: 0.298\n"
]
],
[
[
"## Training across multiple servers",
"_____no_output_____"
],
[
"A TensorFlow cluster is a group of TensorFlow processes running in parallel, usually on different machines, and talking to each other to complete some work, for example training or executing a neural network. Each TF process in the cluster is called a \"task\" (or a \"TF server\"). It has an IP address, a port, and a type (also called its role or its job). The type can be `\"worker\"`, `\"chief\"`, `\"ps\"` (parameter server) or `\"evaluator\"`:\n* Each **worker** performs computations, usually on a machine with one or more GPUs.\n* The **chief** performs computations as well, but it also handles extra work such as writing TensorBoard logs or saving checkpoints. There is a single chief in a cluster, typically the first worker (i.e., worker #0).\n* A **parameter server** (ps) only keeps track of variable values, it is usually on a CPU-only machine.\n* The **evaluator** obviously takes care of evaluation. There is usually a single evaluator in a cluster.\n\nThe set of tasks that share the same type is often called a \"job\". For example, the \"worker\" job is the set of all workers.\n\nTo start a TensorFlow cluster, you must first define it. This means specifying all the tasks (IP address, TCP port, and type). For example, the following cluster specification defines a cluster with 3 tasks (2 workers and 1 parameter server). It's a dictionary with one key per job, and the values are lists of task addresses:",
"_____no_output_____"
]
],
[
[
"cluster_spec = {\n \"worker\": [\n \"machine-a.example.com:2222\", # /job:worker/task:0\n \"machine-b.example.com:2222\" # /job:worker/task:1\n ],\n \"ps\": [\"machine-c.example.com:2222\"] # /job:ps/task:0\n}",
"_____no_output_____"
]
],
[
[
"Every task in the cluster may communicate with every other task in the server, so make sure to configure your firewall to authorize all communications between these machines on these ports (it's usually simpler if you use the same port on every machine).\n\nWhen a task is started, it needs to be told which one it is: its type and index (the task index is also called the task id). A common way to specify everything at once (both the cluster spec and the current task's type and id) is to set the `TF_CONFIG` environment variable before starting the program. It must be a JSON-encoded dictionary containing a cluster specification (under the `\"cluster\"` key), and the type and index of the task to start (under the `\"task\"` key). For example, the following `TF_CONFIG` environment variable defines the same cluster as above, with 2 workers and 1 parameter server, and specifies that the task to start is worker #1:",
"_____no_output_____"
]
],
[
[
"import os\nimport json\n\nos.environ[\"TF_CONFIG\"] = json.dumps({\n \"cluster\": cluster_spec,\n \"task\": {\"type\": \"worker\", \"index\": 1}\n})\nos.environ[\"TF_CONFIG\"]",
"_____no_output_____"
]
],
[
[
"Some platforms (e.g., Google Cloud ML Engine) automatically set this environment variable for you.",
"_____no_output_____"
],
[
"TensorFlow's `TFConfigClusterResolver` class reads the cluster configuration from this environment variable:",
"_____no_output_____"
]
],
[
[
"import tensorflow as tf\n\nresolver = tf.distribute.cluster_resolver.TFConfigClusterResolver()\nresolver.cluster_spec()",
"_____no_output_____"
],
[
"resolver.task_type",
"_____no_output_____"
],
[
"resolver.task_id",
"_____no_output_____"
]
],
[
[
"Now let's run a simpler cluster with just two worker tasks, both running on the local machine. We will use the `MultiWorkerMirroredStrategy` to train a model across these two tasks.\n\nThe first step is to write the training code. As this code will be used to run both workers, each in its own process, we write this code to a separate Python file, `my_mnist_multiworker_task.py`. The code is relatively straightforward, but there are a couple important things to note:\n* We create the `MultiWorkerMirroredStrategy` before doing anything else with TensorFlow.\n* Only one of the workers will take care of logging to TensorBoard and saving checkpoints. As mentioned earlier, this worker is called the *chief*, and by convention it is usually worker #0.",
"_____no_output_____"
]
],
[
[
"%%writefile my_mnist_multiworker_task.py\n\nimport os\nimport numpy as np\nimport tensorflow as tf\nfrom tensorflow import keras\nimport time\n\n# At the beginning of the program\ndistribution = tf.distribute.MultiWorkerMirroredStrategy()\n\nresolver = tf.distribute.cluster_resolver.TFConfigClusterResolver()\nprint(\"Starting task {}{}\".format(resolver.task_type, resolver.task_id))\n\n# Only worker #0 will write checkpoints and log to TensorBoard\nif resolver.task_id == 0:\n root_logdir = os.path.join(os.curdir, \"my_mnist_multiworker_logs\")\n run_id = time.strftime(\"run_%Y_%m_%d-%H_%M_%S\")\n run_dir = os.path.join(root_logdir, run_id)\n callbacks = [\n keras.callbacks.TensorBoard(run_dir),\n keras.callbacks.ModelCheckpoint(\"my_mnist_multiworker_model.h5\",\n save_best_only=True),\n ]\nelse:\n callbacks = []\n\n# Load and prepare the MNIST dataset\n(X_train_full, y_train_full), (X_test, y_test) = keras.datasets.mnist.load_data()\nX_train_full = X_train_full[..., np.newaxis] / 255.\nX_valid, X_train = X_train_full[:5000], X_train_full[5000:]\ny_valid, y_train = y_train_full[:5000], y_train_full[5000:]\n\nwith distribution.scope():\n model = keras.models.Sequential([\n keras.layers.Conv2D(filters=64, kernel_size=7, activation=\"relu\",\n padding=\"same\", input_shape=[28, 28, 1]),\n keras.layers.MaxPooling2D(pool_size=2),\n keras.layers.Conv2D(filters=128, kernel_size=3, activation=\"relu\",\n padding=\"same\"), \n keras.layers.Conv2D(filters=128, kernel_size=3, activation=\"relu\",\n padding=\"same\"),\n keras.layers.MaxPooling2D(pool_size=2),\n keras.layers.Flatten(),\n keras.layers.Dense(units=64, activation='relu'),\n keras.layers.Dropout(0.5),\n keras.layers.Dense(units=10, activation='softmax'),\n ])\n model.compile(loss=\"sparse_categorical_crossentropy\",\n optimizer=keras.optimizers.SGD(learning_rate=1e-2),\n metrics=[\"accuracy\"])\n\nmodel.fit(X_train, y_train, validation_data=(X_valid, y_valid),\n epochs=10, callbacks=callbacks)",
"Overwriting my_mnist_multiworker_task.py\n"
]
],
[
[
"In a real world application, there would typically be a single worker per machine, but in this example we're running both workers on the same machine, so they will both try to use all the available GPU RAM (if this machine has a GPU), and this will likely lead to an Out-Of-Memory (OOM) error. To avoid this, we could use the `CUDA_VISIBLE_DEVICES` environment variable to assign a different GPU to each worker. Alternatively, we can simply disable GPU support, like this:",
"_____no_output_____"
]
],
[
[
"os.environ[\"CUDA_VISIBLE_DEVICES\"] = \"-1\"",
"_____no_output_____"
]
],
[
[
"We are now ready to start both workers, each in its own process, using Python's `subprocess` module. Before we start each process, we need to set the `TF_CONFIG` environment variable appropriately, changing only the task index:",
"_____no_output_____"
]
],
[
[
"import subprocess\n\ncluster_spec = {\"worker\": [\"127.0.0.1:9901\", \"127.0.0.1:9902\"]}\n\nfor index, worker_address in enumerate(cluster_spec[\"worker\"]):\n os.environ[\"TF_CONFIG\"] = json.dumps({\n \"cluster\": cluster_spec,\n \"task\": {\"type\": \"worker\", \"index\": index}\n })\n subprocess.Popen(\"python my_mnist_multiworker_task.py\", shell=True)",
"_____no_output_____"
]
],
[
[
"That's it! Our TensorFlow cluster is now running, but we can't see it in this notebook because it's running in separate processes (but if you are running this notebook in Jupyter, you can see the worker logs in Jupyter's server logs).\n\nSince the chief (worker #0) is writing to TensorBoard, we use TensorBoard to view the training progress. Run the following cell, then click on the settings button (i.e., the gear icon) in the TensorBoard interface and check the \"Reload data\" box to make TensorBoard automatically refresh every 30s. Once the first epoch of training is finished (which may take a few minutes), and once TensorBoard refreshes, the SCALARS tab will appear. Click on this tab to view the progress of the model's training and validation accuracy.",
"_____no_output_____"
]
],
[
[
"%load_ext tensorboard\n%tensorboard --logdir=./my_mnist_multiworker_logs --port=6006",
"The tensorboard extension is already loaded. To reload it, use:\n %reload_ext tensorboard\n"
]
],
[
[
"That's it! Once training is over, the best checkpoint of the model will be available in the `my_mnist_multiworker_model.h5` file. You can load it using `keras.models.load_model()` and use it for predictions, as usual:",
"_____no_output_____"
]
],
[
[
"from tensorflow import keras\n\nmodel = keras.models.load_model(\"my_mnist_multiworker_model.h5\")\nY_pred = model.predict(X_new)\nnp.argmax(Y_pred, axis=-1)",
"_____no_output_____"
]
],
[
[
"And that's all for today! Hope you found this useful. 😊",
"_____no_output_____"
],
[
"# Exercise Solutions",
"_____no_output_____"
],
[
"## 1. to 8.\n\nSee Appendix A.",
"_____no_output_____"
],
[
"## 9.\n_Exercise: Train a model (any model you like) and deploy it to TF Serving or Google Cloud AI Platform. Write the client code to query it using the REST API or the gRPC API. Update the model and deploy the new version. Your client code will now query the new version. Roll back to the first version._",
"_____no_output_____"
],
[
"Please follow the steps in the <a href=\"#Deploying-TensorFlow-models-to-TensorFlow-Serving-(TFS)\">Deploying TensorFlow models to TensorFlow Serving</a> section above.",
"_____no_output_____"
],
[
"# 10.\n_Exercise: Train any model across multiple GPUs on the same machine using the `MirroredStrategy` (if you do not have access to GPUs, you can use Colaboratory with a GPU Runtime and create two virtual GPUs). Train the model again using the `CentralStorageStrategy `and compare the training time._",
"_____no_output_____"
],
[
"Please follow the steps in the [Distributed Training](#Distributed-Training) section above.",
"_____no_output_____"
],
[
"# 11.\n_Exercise: Train a small model on Google Cloud AI Platform, using black box hyperparameter tuning._",
"_____no_output_____"
],
[
"Please follow the instructions on pages 716-717 of the book. You can also read [this documentation page](https://cloud.google.com/ai-platform/training/docs/hyperparameter-tuning-overview) and go through the example in this nice [blog post](https://towardsdatascience.com/how-to-do-bayesian-hyper-parameter-tuning-on-a-blackbox-model-882009552c6d) by Lak Lakshmanan.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
c515eee70e05a61ed64366712ba726efa8b7d321
| 28,091 |
ipynb
|
Jupyter Notebook
|
test_convert_h2o_py1.0.ipynb
|
edenau/Water-Vapour-Unit-Conversion
|
9a95d51c189dd1c08e84ca335381ac2daeebba12
|
[
"MIT"
] | null | null | null |
test_convert_h2o_py1.0.ipynb
|
edenau/Water-Vapour-Unit-Conversion
|
9a95d51c189dd1c08e84ca335381ac2daeebba12
|
[
"MIT"
] | null | null | null |
test_convert_h2o_py1.0.ipynb
|
edenau/Water-Vapour-Unit-Conversion
|
9a95d51c189dd1c08e84ca335381ac2daeebba12
|
[
"MIT"
] | null | null | null | 44.098901 | 1,541 | 0.5825 |
[
[
[
"Test accuracy of convert_h2o.py by comparing its result with convert_h2o.m \n\ncode_standard = stage-6",
"_____no_output_____"
],
[
"# py1.0 - STABLE - 13 June 2019 - tested by Eden Au",
"_____no_output_____"
],
[
"# 1. Setup",
"_____no_output_____"
],
[
"## 1.1 Initialization",
"_____no_output_____"
]
],
[
[
"import warnings\nwarnings.filterwarnings('ignore')\n\nimport os\nimport numpy as np\nimport xarray as xr\nprint('Packages imported.')",
"Packages imported.\n"
]
],
[
[
"## 1.2 Data Import",
"_____no_output_____"
]
],
[
[
"from utils_import_export import get_path_from_main, get_DS\n\n# Get clean ARM data\npath = get_path_from_main(suffix='data/clean/stage-6/arm_prec_sum_1_clean.cdf')\nDS = get_DS(path)\nDS",
"_____no_output_____"
],
[
"# Pick the first instance\nt = DS.sel(time = DS.time[0]).T_p.values\np = DS.sel(time = DS.time[0]).p.values\nrh = DS.sel(time = DS.time[0]).rh_p.values",
"_____no_output_____"
],
[
"rh",
"_____no_output_____"
]
],
[
[
"# 2. Testing",
"_____no_output_____"
]
],
[
[
"from convert_h2o import convert_h2o",
"_____no_output_____"
]
],
[
[
"## 2.1 Errors",
"_____no_output_____"
]
],
[
[
"# Wrong input flag okay\nconvert_h2o(p,t,rh,'wrong_input','N')",
"_____no_output_____"
],
[
"# Wrong output flag okay\nconvert_h2o(p,t,rh,'N','wrong_output')",
"_____no_output_____"
],
[
"# Inconsistent input dim okay\nconvert_h2o(np.concatenate((p,p)),t,rh,'N','N')",
"_____no_output_____"
]
],
[
[
"## 2.2 'H' - Relative Humidity & 'N' - Number Density",
"_____no_output_____"
],
[
"Checks were conducted by comparing the outputs between MATLAB and Python scripts ",
"_____no_output_____"
],
[
"We only have relative humidity ('H') data in hand, so let's convert it to number density ('N') first",
"_____no_output_____"
]
],
[
[
"# H2N okay\ndennum = convert_h2o(p,t,rh,'H','N')\ndennum",
"_____no_output_____"
]
],
[
[
"Test number density ('N') to itself",
"_____no_output_____"
]
],
[
[
"# N2N okay\nconvert_h2o(p,t,dennum,'N','N')",
"_____no_output_____"
],
[
"# N2H okay\nnp.abs(convert_h2o(p,t,dennum,'N','H') - rh).sum()",
"_____no_output_____"
]
],
[
[
"## 2.3 'C' - Columnar Content (one-way)",
"_____no_output_____"
],
[
"Test number density ('N') to columnar content ('C'). This is one-way street as columnar content is a scalar and cannot convert to a vertical profile.",
"_____no_output_____"
]
],
[
[
"# N2C okay\nconvert_h2o(p,t,dennum,'N','C')",
"_____no_output_____"
]
],
[
[
"## 2.4 Other Variables",
"_____no_output_____"
],
[
"Convert ('N') to some unit, then convert it back to ('N'), and cross-check it with the MATLAB version",
"_____no_output_____"
],
[
"### 2.4.1 'M' Mass Mixing Ratio",
"_____no_output_____"
]
],
[
[
"# N2M okay\nmmr = convert_h2o(p,t,dennum,'N','M')\nmmr",
"_____no_output_____"
],
[
"# M2N okay\nconvert_h2o(p,t,mmr,'M','N')",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
]
] |
c515fccd6fc87e989fc4c286f9abe4082cea6e6f
| 9,977 |
ipynb
|
Jupyter Notebook
|
samples/src/main/databricks/ADB-pre-process-json-2-ADX.ipynb
|
hau-mal/azure-kusto-spark
|
11c3a362eea5f93f6649f6b6c777fc2ba2b820e9
|
[
"Apache-2.0"
] | null | null | null |
samples/src/main/databricks/ADB-pre-process-json-2-ADX.ipynb
|
hau-mal/azure-kusto-spark
|
11c3a362eea5f93f6649f6b6c777fc2ba2b820e9
|
[
"Apache-2.0"
] | null | null | null |
samples/src/main/databricks/ADB-pre-process-json-2-ADX.ipynb
|
hau-mal/azure-kusto-spark
|
11c3a362eea5f93f6649f6b6c777fc2ba2b820e9
|
[
"Apache-2.0"
] | null | null | null | 4,988.5 | 9,976 | 0.733186 |
[
[
[
"# Data pre-processing for Azure Data Explorer\n\n<img src=\"https://github.com/Azure/azure-kusto-spark/raw/master/kusto_spark.png\" style=\"border: 1px solid #aaa; border-radius: 10px 10px 10px 10px; box-shadow: 5px 5px 5px #aaa\"/>\n\nWe often see customer scenarios where historical data has to be migrated to Azure Data Explorer (ADX). Although ADX has very powerful data-transformation capabilities via [update policies](https://docs.microsoft.com/azure/data-explorer/kusto/management/updatepolicy), sometimes more or less complex data engineering tasks must be done upfront. This happens if the original data structure is too complex or just single data elements being too big, hitting data explorer limits of dynamic columns of 1 MB or maximum ingest file-size of 1 GB for uncompressed data (see also [Comparing ingestion methods and tools](https://docs.microsoft.com/azure/data-explorer/ingest-data-overview#comparing-ingestion-methods-and-tools)) .\n\nLet' s think about an Industrial Internet-of-Things (IIoT) use-case where you get data from several production lines. In the production line several devices read humidity, pressure, etc. The following example shows a scenario where a one-to-many relationship is implemented within an array. With this you might get very large columns (with millions of device readings per production line) that might exceed the limit of 1 MB in Azure Data Explorer for dynamic columns.\nIn this case you need to do some pre-processing.\n\n\nData has already been uploaded to Azure storage. You will start reading the json-data into a data frame:",
"_____no_output_____"
]
],
[
[
"inputpath = \"wasbs://[email protected]/*.json\"\n\n# optional, for the output to Azure Storage:\n#outputpath = \"<your-storage-path>\"\n\ndf = spark.read.format(\"json\").load(inputpath)",
"_____no_output_____"
]
],
[
[
"The notebook has a parameter IngestDate, this will be used setting the extentsCreationtime. You can call this notebook from Azure Data Factory for all days you want to load to Azure Data Explorer.\nAlternatively you can make use of a partitioning policy.",
"_____no_output_____"
]
],
[
[
"dbutils.widgets.text(\"wIngestDate\", \"2021-08-06T00:00:00.000Z\", \"Ingestion Date\")\nIngestDate = dbutils.widgets.get(\"wIngestDate\")",
"_____no_output_____"
],
[
"display (df)",
"_____no_output_____"
]
],
[
[
"We see that the dataframe has some complex datatypes. The only thing that we want to change here is getting rid of the array, so having the resulting dataset a row for every entry in the measurement array. \n\n*How can we achieve this?*\n\npyspark-sql has some very powerful functions for transformations of complex datatypes. We will make use of the [explode-function](https://spark.apache.org/docs/latest/api/python/reference/api/pyspark.sql.functions.explode.html). In this case explode (\"measurement\") will give us a resulting dataframe with single rows per array-element. Finally we only have to drop the original measurement-column (it is the original structure):",
"_____no_output_____"
]
],
[
[
"from pyspark.sql.functions import *\n\ndf_explode = df.select(\"*\", explode(\"measurement\").alias(\"device\")).drop(\"measurement\")",
"_____no_output_____"
]
],
[
[
"With this we alreadyhave done the necessary data transformation with one line of code. Let' s do some final \"prettyfying\". \nAs we are already preprocessing the data and want to get rid of the complex data types we select the struct elements to get a simplified table:",
"_____no_output_____"
]
],
[
[
"df_all_in_column = df_explode.select (\"header.*\", \"device.header.*\", \"device.*\", \"ProdLineData.*\").drop(\"header\")",
"_____no_output_____"
],
[
"display (df_all_in_column)",
"_____no_output_____"
]
],
[
[
"We are setting the extentsCreationTime to the notebook-parameter *IngestDate*. For other ingestion properties see [here](https://github.com/Azure/azure-kusto-spark/blob/master/samples/src/main/python/pyKusto.py).",
"_____no_output_____"
]
],
[
[
"extentsCreationTime = sc._jvm.org.joda.time.DateTime.parse(IngestDate)\nsp = sc._jvm.com.microsoft.kusto.spark.datasink.SparkIngestionProperties(\n False, None, None, None, None, extentsCreationTime, None, None)",
"_____no_output_____"
]
],
[
[
"Finally, we write the resulting dataframe back to to Azure Data Explorer. Prerequisite doing this is: \n* the target table created in the target database (.create table measurement (ProductionLineId : string, deviceId:string, enqueuedTime:datetime, humidity:real, humidity_unit:string, temperature:real, temperature_unit:string, pressure:real, pressure_unit:string, reading : dynamic))\n* having created a service principal for the ADX access\n* the service principal (AAD-application) accessing ADX has sufficient permissions (add the ingestor and viewer role)\n* Install the latest Kusto library from maven see also the [Azure Data Explorer Connector for Apache Spark documentation](https://github.com/Azure/azure-kusto-spark#usage)",
"_____no_output_____"
]
],
[
[
"df_all_in_column.write. \\\n format(\"com.microsoft.kusto.spark.datasource\"). \\\n option(\"kustoCluster\", \"https://<yourcluster>\"). \\\n option(\"kustoDatabase\", \"your-database\"). \\\n option(\"kustoTable\", \"<your-table>\"). \\\n option(\"sparkIngestionPropertiesJson\", sp.toString()). \\\n option(\"kustoAadAppId\", \"<app-id>\"). \\\n option(\"kustoAadAppSecret\",dbutils.secrets.get(scope=\"<scope-name>\",key=\"<service-credential-key-name>\"). \\\n option(\"kustoAadAuthorityID\", \"<tenant-id>\"). \\\n mode(\"Append\"). \\\n save()",
"_____no_output_____"
]
],
[
[
"You might also consider writing the data to Azure Storage (this might be also make sense for mor complex tranformation pipelines as an intermediate staging step):",
"_____no_output_____"
]
],
[
[
"# df_all_in_column.write.mode('overwrite').json(outputpath) ",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
c5160482003fca0d5eb63e2481630c2468d47d71
| 23,604 |
ipynb
|
Jupyter Notebook
|
gans/wgan.ipynb
|
elbum/gan
|
5ed4409a48f95e55257236ba847c8b6f2d7d9e40
|
[
"Apache-2.0"
] | 35 |
2019-03-18T08:38:44.000Z
|
2021-07-28T08:49:37.000Z
|
gans/wgan.ipynb
|
elbum/gan
|
5ed4409a48f95e55257236ba847c8b6f2d7d9e40
|
[
"Apache-2.0"
] | 1 |
2020-02-06T02:06:25.000Z
|
2020-02-06T02:06:25.000Z
|
gans/wgan.ipynb
|
elbum/gan
|
5ed4409a48f95e55257236ba847c8b6f2d7d9e40
|
[
"Apache-2.0"
] | 7 |
2019-04-09T04:53:00.000Z
|
2021-01-13T11:42:19.000Z
| 29.61606 | 201 | 0.567573 |
[
[
[
"# WGAN with MNIST (or Fashion MNIST)\n\n* `Wasserstein GAN`, [arXiv:1701.07875](https://arxiv.org/abs/1701.07875)\n * Martin Arjovsky, Soumith Chintala, and L ́eon Bottou\n \n* This code is available to tensorflow version 2.0\n* Implemented by [`tf.keras.layers`](https://www.tensorflow.org/versions/r2.0/api_docs/python/tf/keras/layers) [`tf.losses`](https://www.tensorflow.org/versions/r2.0/api_docs/python/tf/losses)\n* Use `transposed_conv2d` and `conv2d` for Generator and Discriminator, respectively.\n * I do not use `dense` layer for model architecture consistency. (So my architecture is different from original dcgan structure)\n* based on DCGAN model",
"_____no_output_____"
],
[
"## Import modules",
"_____no_output_____"
]
],
[
[
"from __future__ import absolute_import\nfrom __future__ import division\nfrom __future__ import print_function\nfrom __future__ import unicode_literals\n\nimport os\nimport sys\nimport time\nimport glob\n\nimport numpy as np\nimport matplotlib.pyplot as plt\n%matplotlib inline\n\nimport PIL\nimport imageio\nfrom IPython import display\n\nimport tensorflow as tf\nfrom tensorflow.keras import layers\n\nsys.path.append(os.path.dirname(os.path.abspath('.')))\nfrom utils.image_utils import *\nfrom utils.ops import *\n\nos.environ[\"CUDA_VISIBLE_DEVICES\"]=\"0\"",
"_____no_output_____"
]
],
[
[
"## Setting hyperparameters",
"_____no_output_____"
]
],
[
[
"# Training Flags (hyperparameter configuration)\nmodel_name = 'wgan'\ntrain_dir = os.path.join('train', model_name, 'exp1')\ndataset_name = 'mnist'\nassert dataset_name in ['mnist', 'fashion_mnist']\n\nmax_epochs = 100\nsave_model_epochs = 10\nprint_steps = 200\nsave_images_epochs = 1\nbatch_size = 64\nlearning_rate_D = 5e-5\nlearning_rate_G = 5e-5\nk = 5 # the number of step of learning D before learning G (Not used in this code)\nnum_examples_to_generate = 25\nnoise_dim = 100\nclip_value = 0.01 # cliping value for D weights in order to implement `1-Lipshitz function`",
"_____no_output_____"
]
],
[
[
"## Load the MNIST dataset",
"_____no_output_____"
]
],
[
[
"# Load training and eval data from tf.keras\nif dataset_name == 'mnist':\n (train_images, train_labels), _ = \\\n tf.keras.datasets.mnist.load_data()\nelse:\n (train_images, train_labels), _ = \\\n tf.keras.datasets.fashion_mnist.load_data()\n\ntrain_images = train_images.reshape(-1, MNIST_SIZE, MNIST_SIZE, 1).astype('float32')\n#train_images = train_images / 255. # Normalize the images to [0, 1]\ntrain_images = (train_images - 127.5) / 127.5 # Normalize the images to [-1, 1]",
"_____no_output_____"
]
],
[
[
"## Set up dataset with `tf.data`\n\n### create input pipeline with `tf.data.Dataset`",
"_____no_output_____"
]
],
[
[
"#tf.random.set_seed(219)\n# for train\nN = len(train_images)\ntrain_dataset = tf.data.Dataset.from_tensor_slices(train_images)\ntrain_dataset = train_dataset.shuffle(buffer_size=N)\ntrain_dataset = train_dataset.batch(batch_size=batch_size, drop_remainder=True)\nprint(train_dataset)",
"_____no_output_____"
]
],
[
[
"## Create the generator and discriminator models",
"_____no_output_____"
]
],
[
[
"class Generator(tf.keras.Model):\n \"\"\"Build a generator that maps latent space to real space.\n G(z): z -> x\n \"\"\"\n def __init__(self):\n super(Generator, self).__init__()\n self.conv1 = ConvTranspose(256, 3, padding='valid')\n self.conv2 = ConvTranspose(128, 3, padding='valid')\n self.conv3 = ConvTranspose(64, 4)\n self.conv4 = ConvTranspose(1, 4, apply_batchnorm=False, activation='tanh')\n\n def call(self, inputs, training=True):\n \"\"\"Run the model.\"\"\"\n # inputs: [1, 1, 100]\n conv1 = self.conv1(inputs, training=training) # conv1: [3, 3, 256]\n conv2 = self.conv2(conv1, training=training) # conv2: [7, 7, 128]\n conv3 = self.conv3(conv2, training=training) # conv3: [14, 14, 64]\n generated_images = self.conv4(conv3, training=training) # generated_images: [28, 28, 1]\n \n return generated_images",
"_____no_output_____"
],
[
"class Discriminator(tf.keras.Model):\n \"\"\"Build a discriminator that discriminate real image x whether real or fake.\n D(x): x -> [0, 1]\n \"\"\"\n def __init__(self):\n super(Discriminator, self).__init__()\n self.conv1 = Conv(64, 4, 2, apply_batchnorm=False, activation='leaky_relu')\n self.conv2 = Conv(128, 4, 2, activation='leaky_relu')\n self.conv3 = Conv(256, 3, 2, padding='valid', activation='leaky_relu')\n self.conv4 = Conv(1, 3, 1, padding='valid', apply_batchnorm=False, activation='none')\n\n def call(self, inputs, training=True):\n \"\"\"Run the model.\"\"\"\n # inputs: [28, 28, 1]\n conv1 = self.conv1(inputs) # conv1: [14, 14, 64]\n conv2 = self.conv2(conv1) # conv2: [7, 7, 128]\n conv3 = self.conv3(conv2) # conv3: [3, 3, 256]\n conv4 = self.conv4(conv3) # conv4: [1, 1, 1]\n discriminator_logits = tf.squeeze(conv4, axis=[1, 2]) # discriminator_logits: [1,]\n \n return discriminator_logits",
"_____no_output_____"
],
[
"generator = Generator()\ndiscriminator = Discriminator()",
"_____no_output_____"
]
],
[
[
"### Plot generated image via generator network",
"_____no_output_____"
]
],
[
[
"noise = tf.random.normal([1, 1, 1, noise_dim])\ngenerated_image = generator(noise, training=False)\n\nplt.imshow(generated_image[0, :, :, 0], cmap='gray')",
"_____no_output_____"
]
],
[
[
"### Test discriminator network\n\n* **CAUTION**: the outputs of discriminator is **logits** (unnormalized probability) NOT probabilites",
"_____no_output_____"
]
],
[
[
"decision = discriminator(generated_image)\nprint(decision)",
"_____no_output_____"
]
],
[
[
"## Define the loss functions and the optimizer",
"_____no_output_____"
]
],
[
[
"# use logits for consistency with previous code I made\n# `tf.losses` and `tf.keras.losses` are the same API (alias)\nbce = tf.losses.BinaryCrossentropy(from_logits=True)\nmse = tf.losses.MeanSquaredError()",
"_____no_output_____"
],
[
"def WGANLoss(logits, is_real=True):\n \"\"\"Computes Wasserstain GAN loss\n\n Args:\n logits (`2-rank Tensor`): logits\n is_real (`bool`): boolean, Treu means `-` sign, False means `+` sign.\n\n Returns:\n loss (`0-rank Tensor`): the WGAN loss value.\n \"\"\"\n loss = tf.reduce_mean(logits)\n if is_real:\n loss = -loss\n\n return loss",
"_____no_output_____"
],
[
"def GANLoss(logits, is_real=True, use_lsgan=True):\n \"\"\"Computes standard GAN or LSGAN loss between `logits` and `labels`.\n\n Args:\n logits (`2-rank Tensor`): logits.\n is_real (`bool`): True means `1` labeling, False means `0` labeling.\n use_lsgan (`bool`): True means LSGAN loss, False means standard GAN loss.\n\n Returns:\n loss (`0-rank Tensor`): the standard GAN or LSGAN loss value. (binary_cross_entropy or mean_squared_error)\n \"\"\"\n if is_real:\n labels = tf.ones_like(logits)\n else:\n labels = tf.zeros_like(logits)\n \n if use_lsgan:\n loss = mse(labels, tf.nn.sigmoid(logits))\n else:\n loss = bce(labels, logits)\n \n return loss",
"_____no_output_____"
],
[
"def discriminator_loss(real_logits, fake_logits):\n # losses of real with label \"1\"\n real_loss = WGANLoss(logits=real_logits, is_real=True)\n # losses of fake with label \"0\"\n fake_loss = WGANLoss(logits=fake_logits, is_real=False)\n \n return real_loss + fake_loss",
"_____no_output_____"
],
[
"def generator_loss(fake_logits):\n # losses of Generator with label \"1\" that used to fool the Discriminator\n return WGANLoss(logits=fake_logits, is_real=True)",
"_____no_output_____"
],
[
"discriminator_optimizer = tf.keras.optimizers.RMSprop(learning_rate_D)\ngenerator_optimizer = tf.keras.optimizers.RMSprop(learning_rate_G)",
"_____no_output_____"
]
],
[
[
"## Checkpoints (Object-based saving)",
"_____no_output_____"
]
],
[
[
"checkpoint_dir = train_dir\nif not tf.io.gfile.exists(checkpoint_dir):\n tf.io.gfile.makedirs(checkpoint_dir)\ncheckpoint_prefix = os.path.join(checkpoint_dir, \"ckpt\")\ncheckpoint = tf.train.Checkpoint(generator_optimizer=generator_optimizer,\n discriminator_optimizer=discriminator_optimizer,\n generator=generator,\n discriminator=discriminator)",
"_____no_output_____"
]
],
[
[
"## Training",
"_____no_output_____"
]
],
[
[
"# keeping the random vector constant for generation (prediction) so\n# it will be easier to see the improvement of the gan.\n# To visualize progress in the animated GIF\nconst_random_vector_for_saving = tf.random.uniform([num_examples_to_generate, 1, 1, noise_dim],\n minval=-1.0, maxval=1.0)",
"_____no_output_____"
]
],
[
[
"### Define training one step function",
"_____no_output_____"
]
],
[
[
"# Notice the use of `tf.function`\n# This annotation causes the function to be \"compiled\".\[email protected]\ndef discriminator_train_step(images):\n # generating noise from a uniform distribution\n noise = tf.random.uniform([batch_size, 1, 1, noise_dim], minval=-1.0, maxval=1.0)\n\n with tf.GradientTape() as disc_tape:\n generated_images = generator(noise, training=True)\n\n real_logits = discriminator(images, training=True)\n fake_logits = discriminator(generated_images, training=True)\n\n gen_loss = generator_loss(fake_logits)\n disc_loss = discriminator_loss(real_logits, fake_logits) \n\n gradients_of_discriminator = disc_tape.gradient(disc_loss, discriminator.trainable_variables)\n discriminator_optimizer.apply_gradients(zip(gradients_of_discriminator, discriminator.trainable_variables))\n \n # clip the weights for discriminator to implement 1-Lipshitz function\n for var in discriminator.trainable_variables:\n var.assign(tf.clip_by_value(var, -clip_value, clip_value))\n \n return gen_loss, disc_loss",
"_____no_output_____"
],
[
"# Notice the use of `tf.function`\n# This annotation causes the function to be \"compiled\".\[email protected]\ndef generator_train_step():\n # generating noise from a uniform distribution\n noise = tf.random.uniform([batch_size, 1, 1, noise_dim], minval=-1.0, maxval=1.0)\n\n with tf.GradientTape() as gen_tape:\n generated_images = generator(noise, training=True)\n\n fake_logits = discriminator(generated_images, training=True)\n gen_loss = generator_loss(fake_logits)\n\n gradients_of_generator = gen_tape.gradient(gen_loss, generator.trainable_variables)\n generator_optimizer.apply_gradients(zip(gradients_of_generator, generator.trainable_variables))",
"_____no_output_____"
]
],
[
[
"### Train full steps",
"_____no_output_____"
]
],
[
[
"print('Start Training.')\nnum_batches_per_epoch = int(N / batch_size)\nglobal_step = tf.Variable(0, trainable=False)\nnum_learning_critic = 0\n\nfor epoch in range(max_epochs):\n\n for step, images in enumerate(train_dataset):\n start_time = time.time()\n \n if num_learning_critic < k:\n gen_loss, disc_loss = discriminator_train_step(images)\n num_learning_critic += 1\n global_step.assign_add(1)\n else:\n generator_train_step()\n num_learning_critic = 0\n \n if global_step.numpy() % print_steps == 0:\n epochs = epoch + step / float(num_batches_per_epoch)\n duration = time.time() - start_time\n examples_per_sec = batch_size / float(duration)\n display.clear_output(wait=True)\n print(\"Epochs: {:.2f} global_step: {} Wasserstein distance: {:.3g} loss_G: {:.3g} ({:.2f} examples/sec; {:.3f} sec/batch)\".format(\n epochs, global_step.numpy(), -disc_loss, gen_loss, examples_per_sec, duration))\n random_vector_for_sampling = tf.random.uniform([num_examples_to_generate, 1, 1, noise_dim],\n minval=-1.0, maxval=1.0)\n sample_images = generator(random_vector_for_sampling, training=False)\n print_or_save_sample_images(sample_images.numpy(), num_examples_to_generate)\n\n if (epoch + 1) % save_images_epochs == 0:\n display.clear_output(wait=True)\n print(\"This images are saved at {} epoch\".format(epoch+1))\n sample_images = generator(const_random_vector_for_saving, training=False)\n print_or_save_sample_images(sample_images.numpy(), num_examples_to_generate,\n is_square=True, is_save=True, epoch=epoch+1,\n checkpoint_dir=checkpoint_dir)\n\n # saving (checkpoint) the model every save_epochs\n if (epoch + 1) % save_model_epochs == 0:\n checkpoint.save(file_prefix=checkpoint_prefix)\n \nprint('Training Done.')",
"_____no_output_____"
],
[
"# generating after the final epoch\ndisplay.clear_output(wait=True)\nsample_images = generator(const_random_vector_for_saving, training=False)\nprint_or_save_sample_images(sample_images.numpy(), num_examples_to_generate,\n is_square=True, is_save=True, epoch=epoch+1,\n checkpoint_dir=checkpoint_dir)",
"_____no_output_____"
]
],
[
[
"## Restore the latest checkpoint",
"_____no_output_____"
]
],
[
[
"# restoring the latest checkpoint in checkpoint_dir\ncheckpoint.restore(tf.train.latest_checkpoint(checkpoint_dir))",
"_____no_output_____"
]
],
[
[
"## Display an image using the epoch number",
"_____no_output_____"
]
],
[
[
"display_image(max_epochs, checkpoint_dir=checkpoint_dir)",
"_____no_output_____"
]
],
[
[
"## Generate a GIF of all the saved images.",
"_____no_output_____"
]
],
[
[
"filename = model_name + '_' + dataset_name + '.gif'\ngenerate_gif(filename, checkpoint_dir)",
"_____no_output_____"
],
[
"display.Image(filename=filename + '.png')",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
c5160f41dec8250dcb6f59a2b028a79de75af4c7
| 4,597 |
ipynb
|
Jupyter Notebook
|
BiometricSystem.ipynb
|
VirajShah30/Fingerprint-verification-using-SIFT
|
081853f1eb879e70b92732b026bb449ae939415d
|
[
"MIT"
] | null | null | null |
BiometricSystem.ipynb
|
VirajShah30/Fingerprint-verification-using-SIFT
|
081853f1eb879e70b92732b026bb449ae939415d
|
[
"MIT"
] | null | null | null |
BiometricSystem.ipynb
|
VirajShah30/Fingerprint-verification-using-SIFT
|
081853f1eb879e70b92732b026bb449ae939415d
|
[
"MIT"
] | null | null | null | 32.373239 | 163 | 0.510333 |
[
[
[
"import cv2\nimport numpy as np\nimport os\nimport gradio as gr",
"_____no_output_____"
],
[
"def match(action, name, fingerprint_scan):\n result = fingerprint_scan\n x = \"No matches found.\"\n\n if(action == \"Enrollment\"):\n dir = \"./database/\"\n filename = dir + name + '.bmp'\n cv2.imwrite(filename, fingerprint_scan)\n x = \"Succesfully Registered as \" + name\n sift = cv2.xfeatures2d.SIFT_create()\n keypoints, descriptors = sift.detectAndCompute(fingerprint_scan, None)\n cv2.drawKeypoints(fingerprint_scan, keypoints, fingerprint_scan)\n result = fingerprint_scan\n return result,x\n\n else:\n for file in os.listdir(\"./database/\"):\n fingerprint_database_image = cv2.imread(\"./database/\"+file)\n sift = cv2.xfeatures2d.SIFT_create()\n keypoints_1, descriptors_1 = sift.detectAndCompute(fingerprint_scan, None)\n keypoints_2, descriptors_2 = sift.detectAndCompute(fingerprint_database_image, None)\n matches = cv2.FlannBasedMatcher(dict(algorithm=1, trees=10), dict()).knnMatch(descriptors_1, descriptors_2, k=2)\n match_points = []\n for p, q in matches:\n if p.distance < 0.1*q.distance:\n match_points.append(p)\n keypoints = 0\n if len(keypoints_1) <= len(keypoints_2):\n keypoints = len(keypoints_1) \n else:\n keypoints = len(keypoints_2)\n \n if (len(match_points) / keypoints)>0.95:\n result = cv2.drawMatches(fingerprint_scan, keypoints_1, fingerprint_database_image, keypoints_2, match_points, None) \n a = len(match_points) / keypoints * 100\n x = \"%.2f\" % a + \"% match with \" + str(file).split(\".\")[0]\n result = cv2.resize(result, None, fx=2.5, fy=2.5)\n break\n return result,x",
"_____no_output_____"
],
[
"iface = gr.Interface(fn=match,\n title=\"Fingerprint Recognition System\", \n inputs=[gr.inputs.Radio([\"Enrollment\", \"Identification\"],label=\"Action\"),gr.inputs.Textbox(label=\"Name\"), gr.inputs.Image()], \n outputs=[gr.outputs.Image(label=\"Flann-Based Matching Result on SIFT Features\"), gr.outputs.Textbox(label=\"Match Percentage\")], \n allow_screenshot=False, \n allow_flagging=False)\niface.launch(inbrowser=True)",
"Running locally at: http://127.0.0.1:7860/\nTo create a public link, set `share=True` in `launch()`.\n"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code"
]
] |
c5161eb78efa48d886da12c4d472a2f4d49b609a
| 172,553 |
ipynb
|
Jupyter Notebook
|
notebooks/Profile code.ipynb
|
gschivley/ElectricityLCI
|
1c1c1b69705d3ffab1e1e844aaf7379e4f51198e
|
[
"CC0-1.0"
] | 1 |
2019-04-15T18:11:16.000Z
|
2019-04-15T18:11:16.000Z
|
notebooks/Profile code.ipynb
|
gschivley/ElectricityLCI
|
1c1c1b69705d3ffab1e1e844aaf7379e4f51198e
|
[
"CC0-1.0"
] | 3 |
2019-05-07T19:04:22.000Z
|
2019-09-30T21:29:59.000Z
|
notebooks/Profile code.ipynb
|
gschivley/ElectricityLCI
|
1c1c1b69705d3ffab1e1e844aaf7379e4f51198e
|
[
"CC0-1.0"
] | null | null | null | 80.934803 | 1,773 | 0.496543 |
[
[
[
"# OPTIONAL: Load the \"autoreload\" extension so that code can change\n%load_ext autoreload\n\n# OPTIONAL: always reload modules so that as you change code in src, it gets loaded\n%autoreload 2\n\n%load_ext line_profiler",
"_____no_output_____"
],
[
"import random\nfrom stewicombo import combineInventoriesforFacilitiesinOneInventory\nfrom stewicombo.overlaphandler import aggregate_and_remove_overlap, reliablity_weighted_sum, join_with_underscore, get_first_item, get_by_preference\nfrom electricitylci.globals import inventories_of_interest\nimport pandas as pd",
"_____no_output_____"
],
[
"df = pd.DataFrame({'a': ['a', 'a', 'b', 'c', 'd', 'e'], \n 'b': range(6)})\nINVENTORY_PREFERENCE_BY_COMPARTMENT = {\"air\":[\"eGRID\",\"GHGRP\",\"NEI\",\"TRI\"],\n \"water\":[\"DMR\", \"TRI\"],\n \"soil\":[\"TRI\"],\n \"waste\":[\"RCRAInfo\",\"TRI\"]}",
"_____no_output_____"
],
[
"%%timeit\nrandom.choice(list(INVENTORY_PREFERENCE_BY_COMPARTMENT))",
"1.08 µs ± 20.2 ns per loop (mean ± std. dev. of 7 runs, 1000000 loops each)\n"
],
[
"df.head()",
"_____no_output_____"
],
[
"def get_by_preference1(group):\n prefs = random.choice(list(INVENTORY_PREFERENCE_BY_COMPARTMENT))\n \n for pref in prefs:\n for index, row in group.iterrows():\n a = row['a']\n \ndef get_by_preference2(group):\n if len(group) > 1:\n prefs = random.choice(list(INVENTORY_PREFERENCE_BY_COMPARTMENT))\n \n for pref in prefs:\n for index, row in group.iterrows():\n a = index\n else:\n a = group['a']",
"_____no_output_____"
],
[
"%%timeit\ndf.groupby('a').apply(get_by_preference1)",
"5.05 ms ± 33.7 µs per loop (mean ± std. dev. of 7 runs, 100 loops each)\n"
],
[
"%%timeit\ndf.groupby('a').apply(get_by_preference2)",
"2.77 ms ± 53.1 µs per loop (mean ± std. dev. of 7 runs, 100 loops each)\n"
],
[
"inventories_of_interest",
"_____no_output_____"
],
[
"inventories_of_interest.pop('RCRAInfo')",
"_____no_output_____"
],
[
"%pdb",
"Automatic pdb calling has been turned ON\n"
],
[
"from stewicombo.globals import COL_FUNC_DEFAULT\nCOL_FUNC_DEFAULT",
"_____no_output_____"
],
[
"%lprun -f get_by_preference -f reliablity_weighted_sum -f aggregate_and_remove_overlap combineInventoriesforFacilitiesinOneInventory(\"eGRID\",inventories_of_interest,filter_for_LCI=True)",
"Aggregating inventories...\nAdding any rows with NaN FRS_ID or SRS_ID\nOverlap removed.\n"
],
[
"%lprun -f get_by_preference -f get_first_item -f reliablity_weighted_sum -f aggregate_and_remove_overlap combineInventoriesforFacilitiesinOneInventory(\"eGRID\",inventories_of_interest,filter_for_LCI=True)",
"Aggregating inventories...\nAdding any rows with NaN FRS_ID or SRS_ID\nOverlap removed.\n"
],
[
"%prun combineInventoriesforFacilitiesinOneInventory(\"eGRID\",inventories_of_interest,filter_for_LCI=True)",
"Aggregating inventories...\nAdding any rows with NaN FRS_ID or SRS_ID\nOverlap removed.\n"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.